- Department of Anesthesiology, Shanghai Pulmonary Hospital, School of Medicine, Tongji University, Shanghai, China
Background: Autophagy refers to the process in which cells wrap their damaged organelles or unwanted proteins into a double-membrane structure and direct them to lysosomes for degradation. Autophagy can regulate many lung diseases such as pulmonary hypertension, acute lung injury, and lung cancer. However, few bibliometric studies on autophagy are available. The aim of the present study was to clarify the role of autophagy in lung diseases by bibliometric analysis.
Methods: Publications were retrieved from the 2012–2021 Science Citation Index Expanded of Web of Science Core Collection on 20 September 2022. Bibliometrix package in R software was used for data retrieval. VOSviewer and CiteSpace were used to visualize the research focus and trend regarding the effect of autophagy on lung disease.
Results: A total of 4,522 original articles and reviews on autophagy in lung diseases published between 2012 and 2021 were identified. China had the largest number of published papers and citations, whereas the United States (US) ranked first in the H-index and G-index. Moreover, cooperation network analysis showed close cooperation between the US, China, and some European countries, and the top 10 affiliates were all from these countries and regions. Bibliometric analysis showed that “autophagy” and “apoptosis” were the keywords with the highest frequency. During the past decade, most studies were concerned with basic research on pathways related to the regulatory role of autophagy in the inhibition and attenuation of lung diseases.
Conclusion: The study of autophagy in lung diseases is still in the development stage. The information published in these articles has helped researchers understand further the hot spots and development trends in the field more and learn about the collaboration network information regarding authors, countries, and institutions, as well as the paper citation correlation. More studies have been performed to gain deeper insights into the pathogenesis of autophagy by focusing on the links and effects between various diseases. More recently, research in this field has paid increasing attention to the function of autophagy in COVID-19–related lung diseases.
Introduction
Autophagy (“self-eating” in Greek) refers to the process in which cells wrap their damaged organelles or unwanted proteins into a double-membrane structure and direct them to lysosomes for degradation (1, 2). Autophagy exhibits a strong conservative property in all eukaryotes and is essential for maintaining cellular homeostasis. Hence, nearly all cell types present a basal autophagy level. Normally, autophagy takes charge of regulating the physiological functions, degrading cellular components, and forming building blocks and cell response to stress (RTS) (2). Autophagy can be easily induced by environmental insults, and the autophagic cycle rate (flux) can commonly change in RTS (3). Upon most occasions, induction of autophagy in RTS is a cytoprotective mechanism. Despite this, undue self-degradation results in cell death (4–6). The lung is a complex organ mainly functioning in gas exchange and is composed by many cell types including endothelial, epithelial, inflammatory, and mesenchymal cell types (7). Autophagy is an inducible RTS in lung cells. Agents triggering autophagy present a special association with lung cell biology, including hypoxia, particle and cigarette smoke exposure, pro-inflammatory states, and conditions that promote endoplasmic reticulum (ER) stress or oxidative stress. An increasing number of lung-related diseases are emerging, such as chronic obstructive pulmonary disease (COPD), idiopathic pulmonary fibrosis, pulmonary hypertension, acute lung injury, and lung cancer, which form a crux in the field of malignancy (8), causing high mortality (9, 10). Although some progress has been made in research of the effect of autophagy on lung diseases, many issues need to be further explored.
Bibliometric analysis pays attention to the literature systems and literature characteristics and broadly serves for understanding the knowledge structure as well as exploring the developmental trends by virtue of the qualitative and quantitative scientific literature analysis (11). Bibliometric analysis quantitatively measures the study domain outline distribution as well as the relationship and clustering. It describes and predicts the future development of a certain research field on the one hand and compares the contributions made by different authors, institutions, countries, and journals on the other hand. As bibliometric analysis can help develop guidelines, understand research hot spots, and evaluate the research trends (12), it has been applied to the research of digestive system diseases (13), cancer (14), rheumatic system diseases (15), and nervous system diseases (16). However, there are basically no bibliometric studies investigating the autophagy in lung diseases. Hence, the aim of this study is to make a systematical analysis of the research on the autophagy in lung diseases and to evaluate the related research state and trend.
Materials and methods
Data sources and search strategies
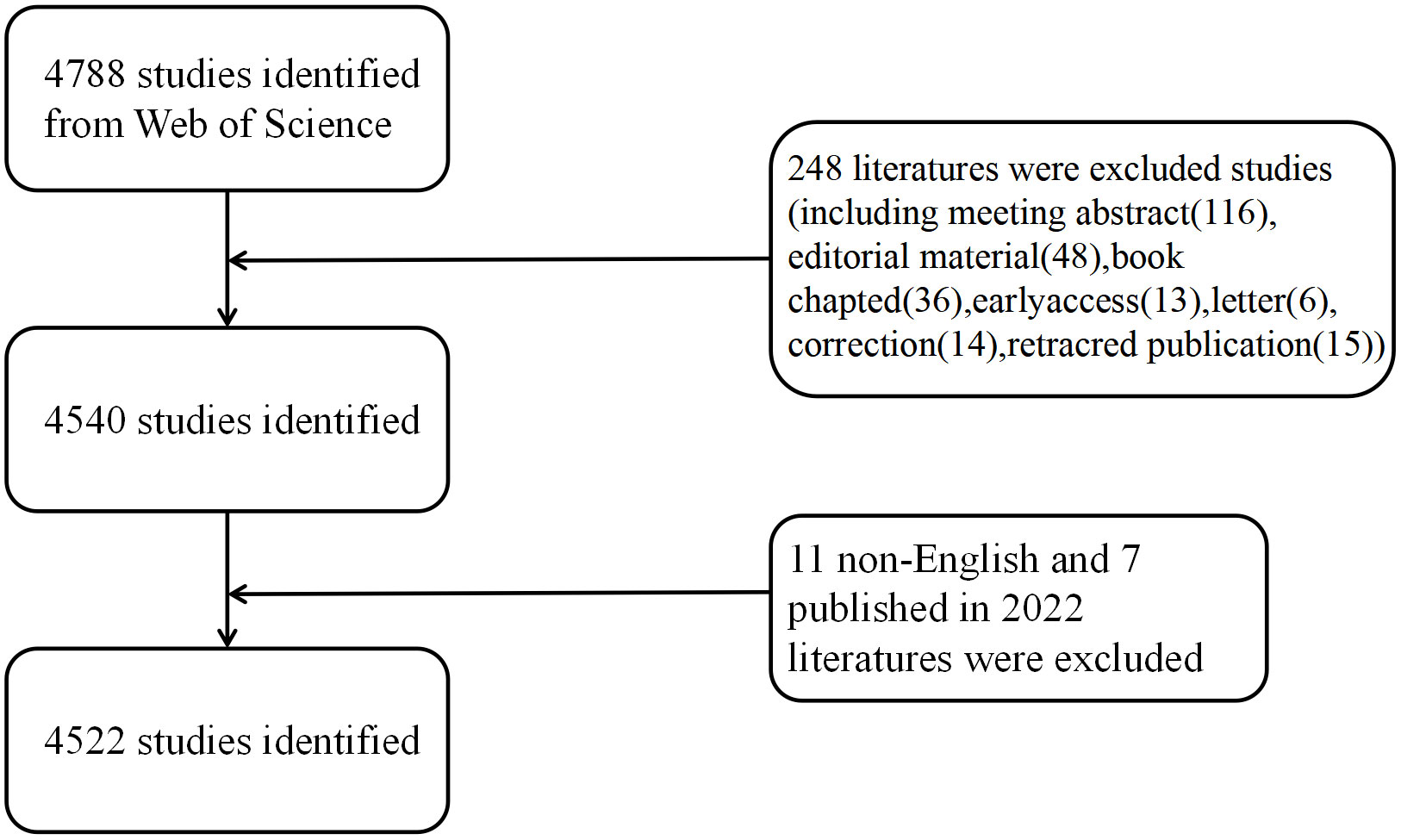
We performed a systematic search for the Science Citation Index Expanded (SCI-Expanded) of Web of Science Core Collection (WoSCC) in 2012–2021 during 1 January 2012 and 31 December 2021, and we downloaded the data on 20 September 2022 for avoiding deviations. The search terms included: TS = (autophagy) AND TS= (“pulmonary” OR “lung”). Two reviewers (FH Lin and JX Yuan) took charge of independently identifying this data search and discussing underlying differences, with the final agreement of 0.90 (17). After excluding the online publication time of 2022 and articles written in non-English and by limiting the publication type to reviews and original articles, we finally obtained 4,522 original articles and reviews that met the criteria for inclusion in the analysis (Figure 1).
Data collection and cleaning
First, the original data were extracted from the SCI-Expanded database of WoSCC. The recorded information included the number of papers and citations, H-index, publication year, countries/regions, affiliations, authors, journals, references, and keywords. Afterward, duplicate authors and misspelled elements were moved artificially. Although inaccurate analysis may not be avoided completely due to multiple versions of cited references, the same abbreviated names of different authors, and different forms of cited journals, we pointed out the first extracted original data from the SCI-Expanded database. The recorded information was paper and citation number, H-index, G-index publication year, countries/regions, affiliations, authors, journal, references, and keywords. Then, we removed misspelled elements and duplicate authors. Cited references have different versions, different authors may have the same abbreviated name, and cited journals have different forms, which may lead to inaccurate analysis. Despite this, most raw data were accurate. We removed the misspelled elements and duplicate authors and adopted a thesaurus file for merging duplicates into one word, deleting useless words and correcting the misspelled elements. Later, we imported the cleansed data to VOSviewer (version 1.6.18.0), CiteSpace (version 6.1. R3), and the “bibliometrix package 4.0.1” of R software (version 4.2.1) for the subsequent bibliometric analysis.
Bibliometric analysis
The number of papers and citations is the bibliometric indicator commonly used to represent the bibliographic material. In general, the research level can be evaluated from the perspective of productivity and impact, which, in the paper, were respectively measured via the number of publications (NP) and the number of citations without self-citations (NC). Sometimes, H-index is used to evaluate the individual academic achievements or the publication output (18). G-index refers to the highest number of papers that receive H-index or more citations (19). In addition, the article value can also be indicated by the impact factor (IF) from the latest version of Journal Citation Reports (20, 21). We visualized the results for the number of papers per year, country, organization, author, citation, and other relevant aspects. Prior to data analysis, the R4.2.1-based Bibliometrix package was used to store, count, and clean up data. As important indicators of the research, H-index, G-index, and IF were all included in the analysis. Subsequently, the following software was used for bibliometric analysis.
R (version 4.2.1) stands for the language and environment specific to the statistical computing and graphics. It presents a strong extensibility, capable of automating the analyses and creating new functions. The cleansed data received bibliometric analysis under the assistance of bibliometrix package in R (22). To further explain the annual document quantity alternation, we employed the fitting polynomial model for predicting the annual NP. Variable f (x) refers to the annual study number, and x stands for the publication year. Moreover, VOSviewer software assisted in constructing bibliometric maps for obtaining more extensive information regarding the results considering co-citation (CC) and co-occurrence (CO) (23, 24). Co-citation refers to the situation that two items are simultaneously cited by the third one. Keyword co-occurrence takes charge of measuring the keywords appearing in the same documents most frequently (25). Analysis on the keywords serves for demonstrating the autophagy-related research in lung disease.
VOSviewer, CiteSpace, and R (version 4.2.1) were used for the statistical computing and graphics. VOSviewer uses data from WoSCC for establishing the bibliometric maps (23), capable of assisting in comprehensively viewing the bibliometric maps in detail considering the collaborative data. CiteSpace focuses on analyzing the underlying knowledge in the scientific literature and visualizing the collected data (26).
Results
An overview of publications on autophagy in lung disease
The number of retrieved publications was 4,522, including 3,772 original research articles and 750 reviews, with total NC of 114,589, a mean NC/article ratio of 27.91, and a mean H-index of 130.
The annual trend of paper publication quantity
Figure 2A displays the annual NP associated with autophagy in lung diseases. The annual paper number indicates a fast increase from 119 in 2012 to 785 in 2021, and the NP reached the peak in 2021. From 2012, annual NP did not change significantly in the United States (US) and Korea but indicated a fast elevation in China, and there has been a surge since 2015. The annual NP exhibited an obvious relation to the publication year, with the correlation coefficient R2 of 0.9872 (Figure 2A). The rapidly increasing NP demonstrates the increasing number of scholars who placed emphasis on this field. There were more than 4,500 publications in 2021, demonstrating the increasing number of scholars who conducted relevant research and the flourish of lung disease-related autophagy research theories (Figure 2B).
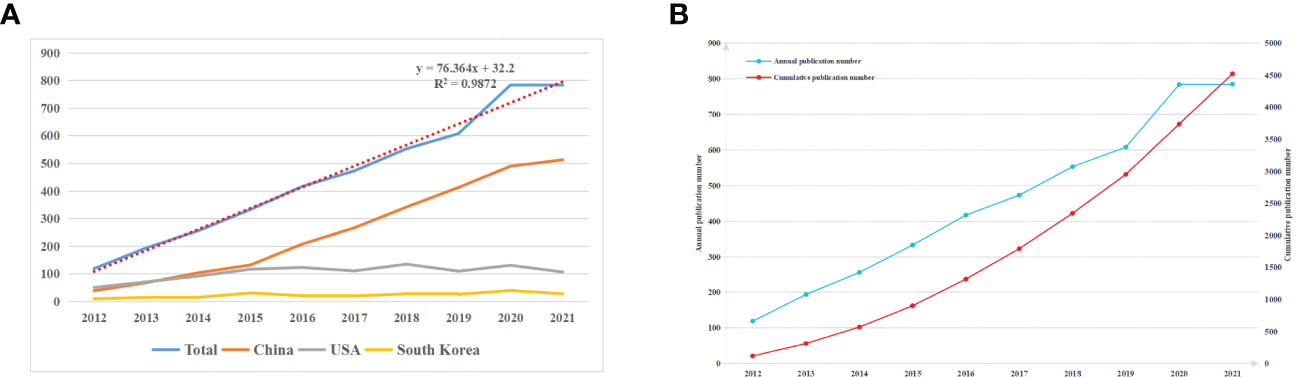
Figure 2 (A) The total NP and top three countries by number of published papers during 2012 and 2021. (B) The annual NP and accumulation during 2012 and 2021.
Contributions of countries/regions to global publications
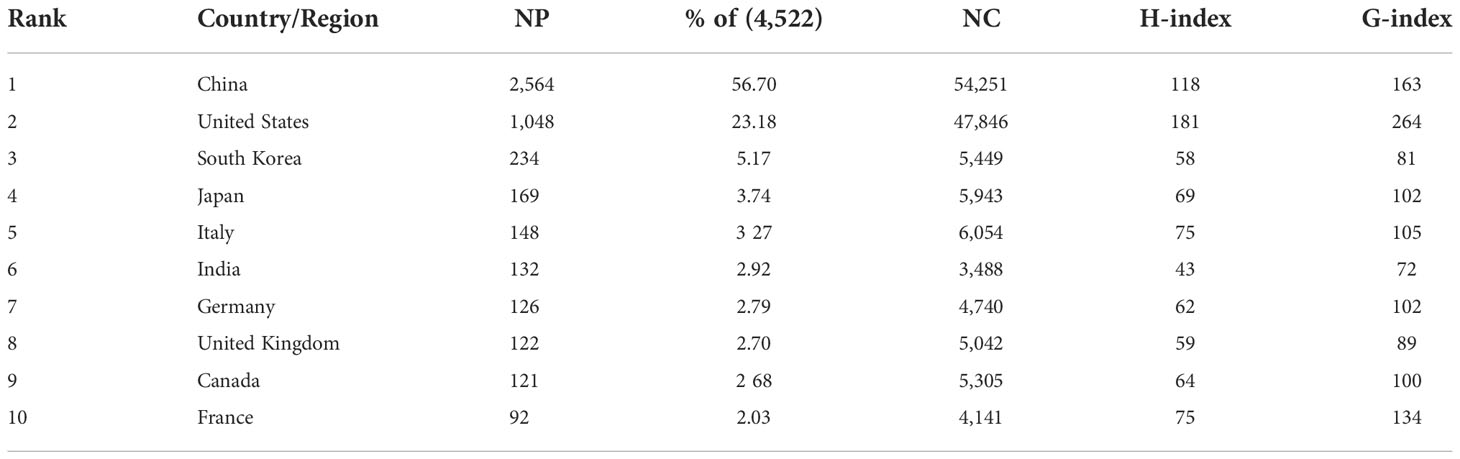
The 10 high-output countries/regions with the highest output were ranked according to NP of all authors. China published the most papers (2,564, 56.7%), followed by the US (1,048, 23.18%) and South Korea (234, 5.17%) (Figures 3A, B; Table 1). NC was 23,791 for China, accounting for 38.2% of the total number of citations, followed by the US (47,846) and South Korea (5,449). However, the US enjoyed the highest H-index (181) and G-index (264), about three times that of South Korea (58 of 81).
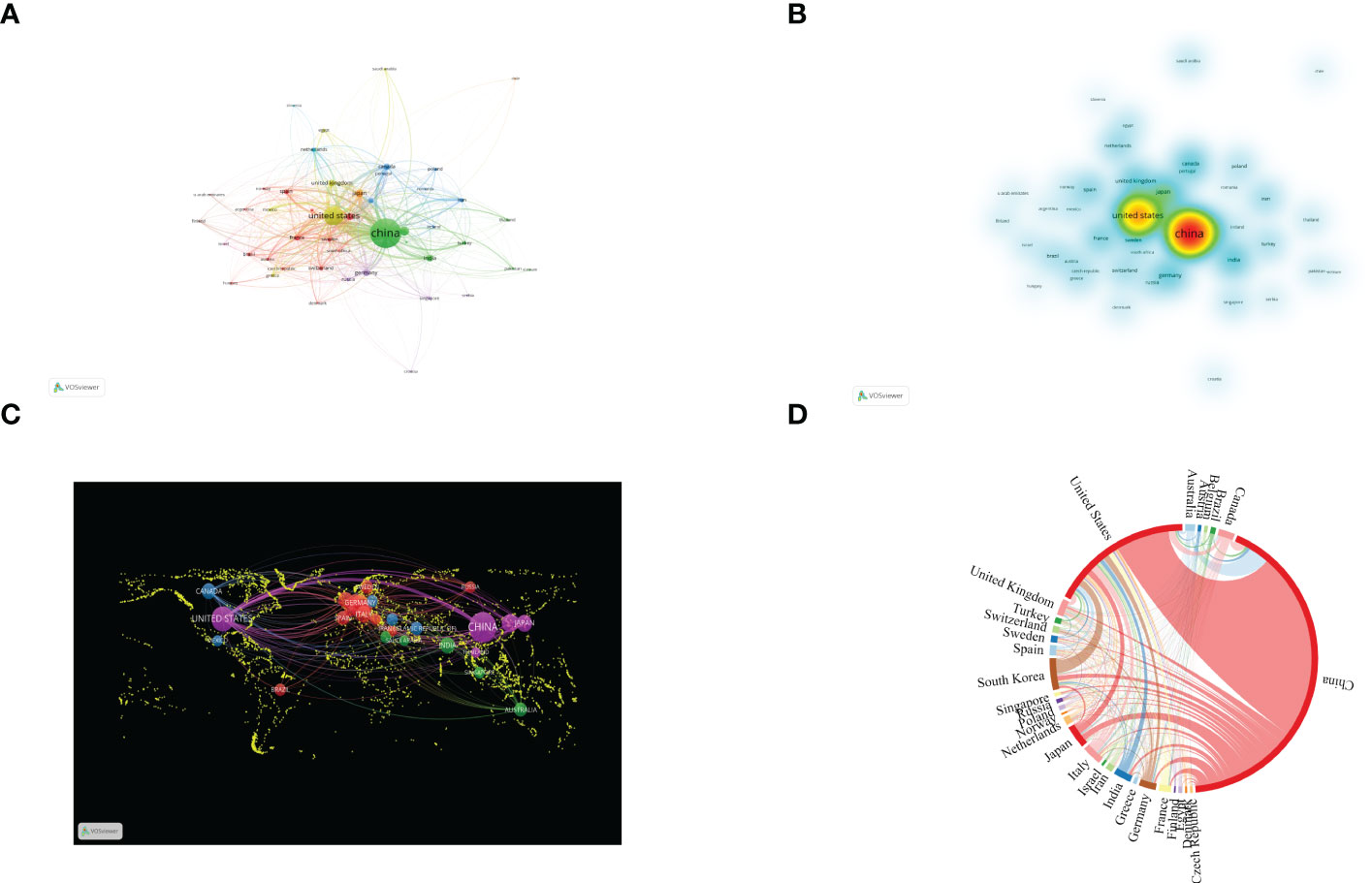
Figure 3 Contributions of various countries to the research of autophagy in lung diseases. (A) Country collaboration map of autophagy research in lung disease. Circles denote countries, and lines denote their collaborations. (B) Countries’ density map. (C) World map of countries’ cooperation density. (D) A circle diagram evaluating the international collaboration between clusters.
Figures 3C, D display the publication distribution in various countries and regions. Related cooperation was mainly concentrated upon China and the US, and cooperation with other countries was relatively weak. As shown in Figure 3C, cooperation between countries and regions significantly promoted the development of scientific research. The lines in the figure indicate cooperation: The wider the lines, the stronger the cooperation. Nevertheless, lines cannot be found in most countries, meaning that these countries had no stable communication and cooperation.
Analysis of authors
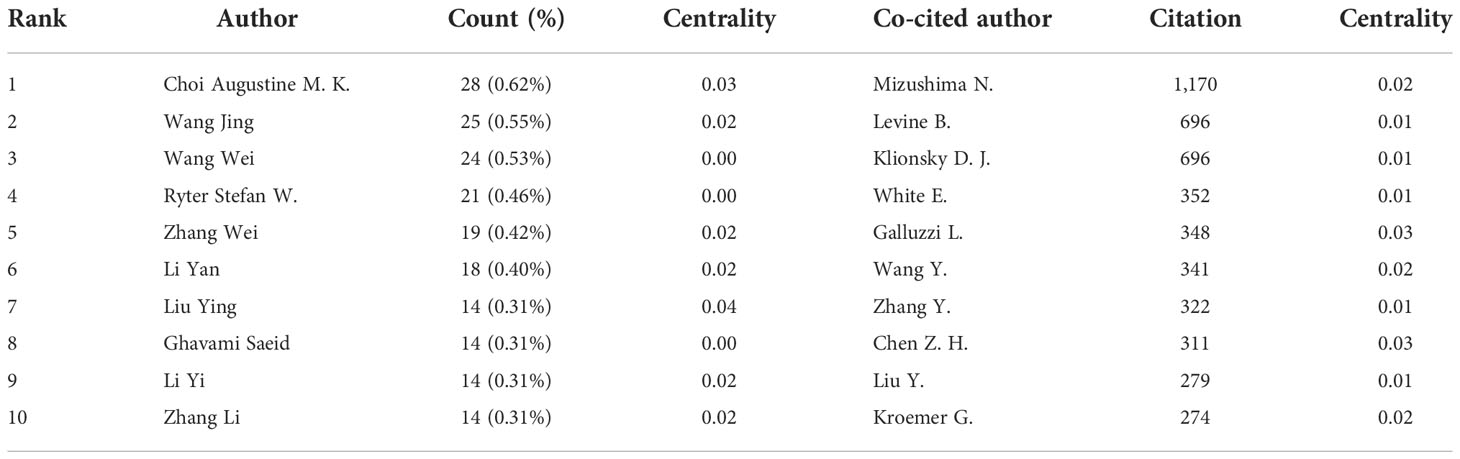
According to the author visualization chart by VOSviewer, 78 authors performed autophagy-related studies. In terms of the NP, Choi Augustine M. K. (28 publications) ranked first, followed by Ryter Stefan W. (28 publications), Wang Jing (25articles), and Wang Wei (24 articles) (Figure 4A; Table 2). Obviously, there was a relatively low author centrality (≤0.04), demonstrating the necessity to enhance the autophagy impact in lung disease. Co-cited authors refer to those (at least two) who are cited at the same time (Figure 4B). Only six had a frequency of citation of over 400 times amid the 103,587 co-cited authors. The lines between nodes stand for authors’ collaboration, and the collaboration graph means that authors’ collaboration indeed existed in the field.
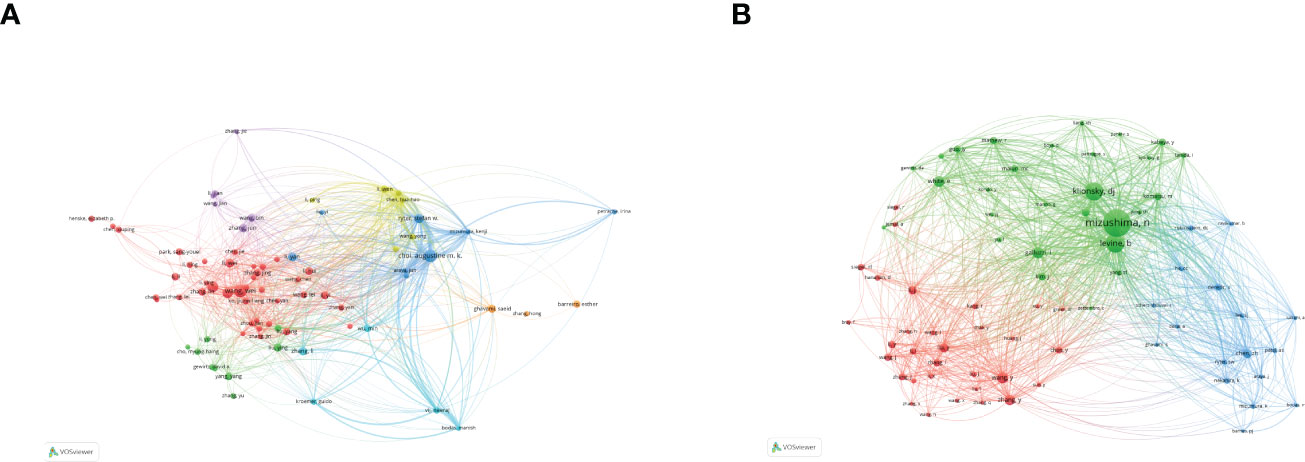
Figure 4 Authors related to the research on autophagy in lung disease. (A) Author co-occurrence. (B) VOSviewer visualization of co-cited authors. Circle size means the number of authors’ published articles; the connecting lines stand for authors’ mutual communication and interaction.
Analysis of affiliations
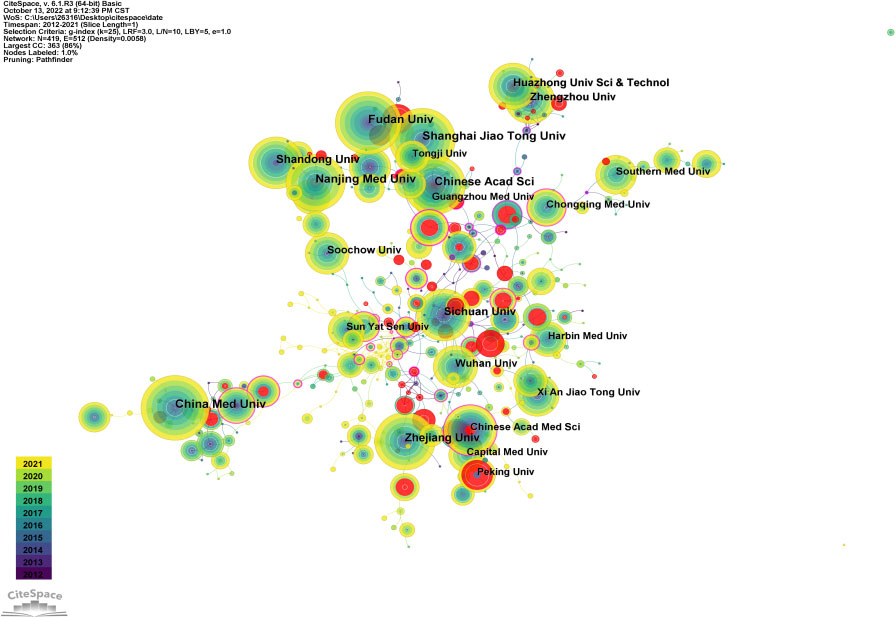
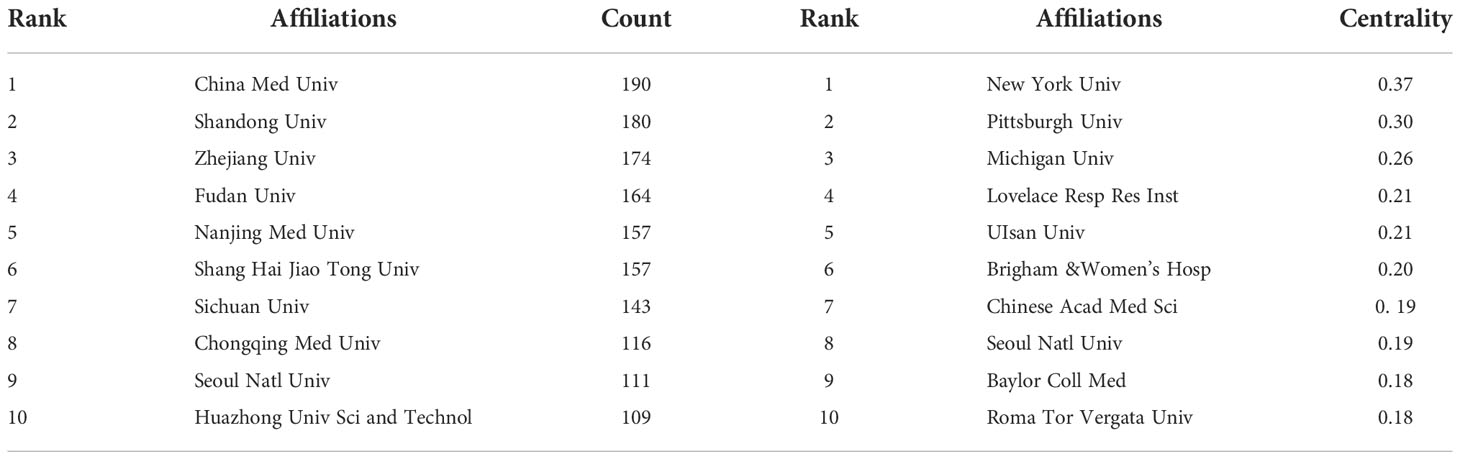
Figure 5 is an institutional co-occurrence map built with CiteSpace, listing the top 10 institutions considering the output and centrality in autophagy-related lung disease studies. Nodes stand for institutions: The larger the node, the more output the institution made. Links between nodes mean institutional collaboration, and the color and thickness denote the collaboration duration and intensity, respectively. China Medical University (n = 190) was in the leading position considering the output, followed by Shandong University (n =180) and Zhejiang University (n = 174). The top three institutions considering the centrality were New York University (0.37), Pittsburgh University (0.30), and Michigan University (0.26) (Table 3). As the data show, institutions were closely collaborated with each other.
Analysis of journals
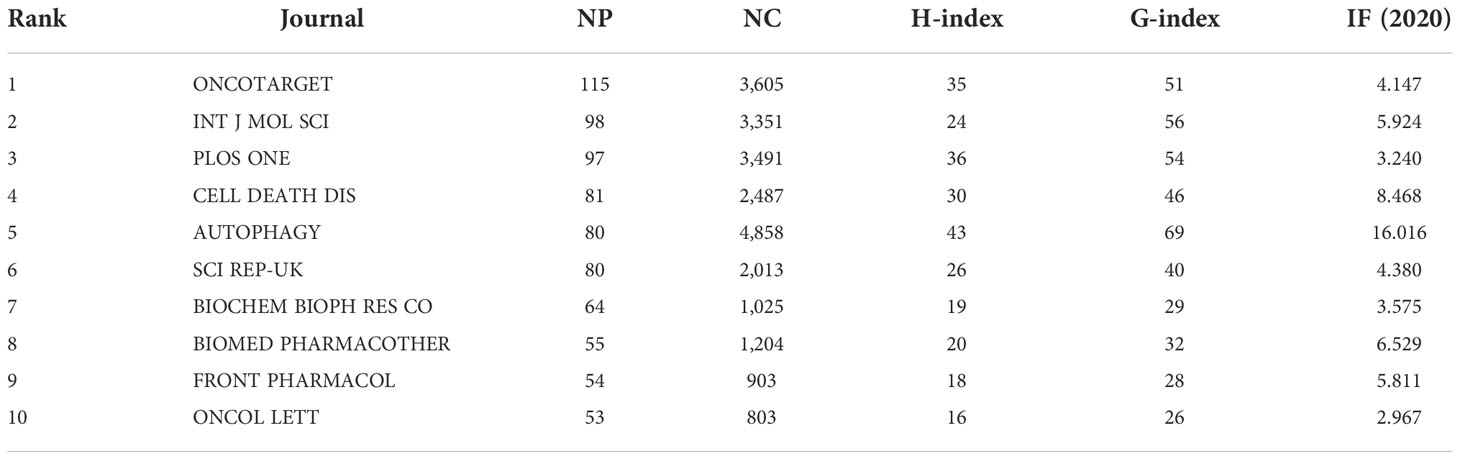
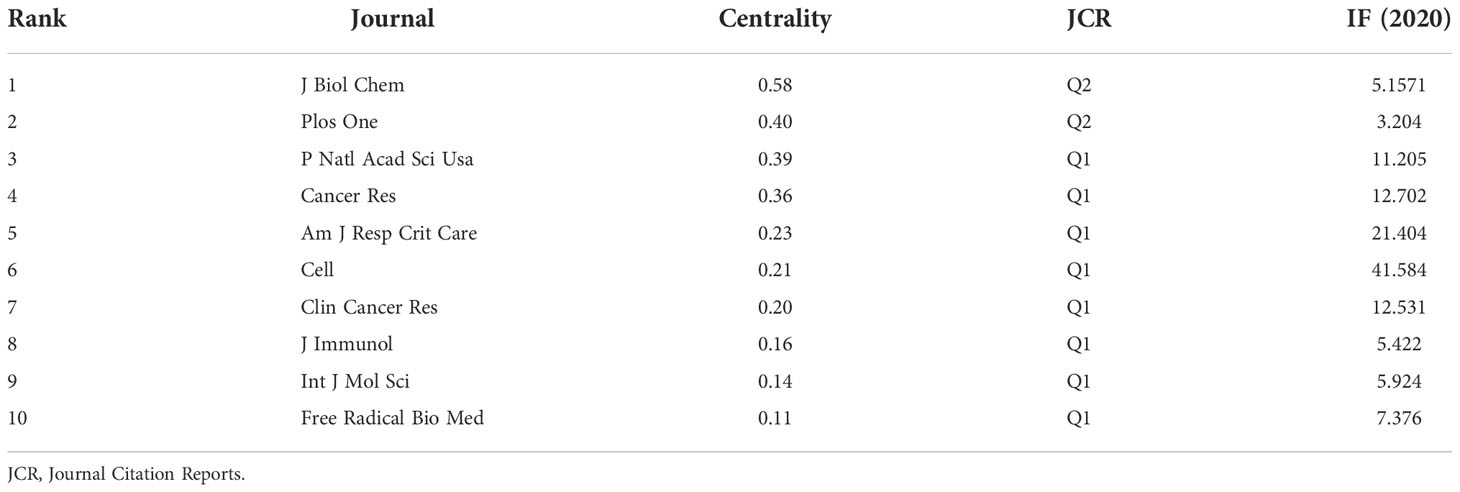
Table 4 gives the top 10 journals considering the research number of lung disease–related autophagy and the impact indicators, namely, H-index, G-index, and IF eigenfactor score. These journals had a larger possibility in accepting articles regarding autophagy in lung disease, given their largest NP of relevant topics. Scholars in the autophagy area are suggested to place emphasis on these journals. ONCOTARGET (115 publications, IF = 4.147) had the highest NP, followed by INT J MOL SCI (98 publications, IF = 5.924), PLOS ONE (97 publications, IF = 3.248), CELL DEATH DIS (81 publications, IF = 8.468), and AUTOPHAGY (80 publications, IF = 16.016). Notably, the AUTOPHAGY had higher citations, H-index, and G-index, demonstrating that the quality of many studies was not high. In Figure 6A, the analysis of journals revealed that PLOS ONE (2,677 total citations) presented the highest citation frequency, followed by AUTOPHAGY (2,626 total citations) and J BIOL CHEM (2,440 citations). J BIOL CHEM (0.58) enjoyed the highest centrality, followed by PLOS ONE (0.40) and P NATL ACAD SCI USA (0.39) (Table 5), all of which had a high centrality, demonstrating their stronger impact in this field. In the domain dimension, superposition graph analysis was carried out for the fields involved in the literature (Figure 6B). Among them, the top fields of paper production were oncology, pharmacology and pharmacy, chemistry medicinal, multidisciplinary sciences, and chemistry multidisciplinary. Therefore, in the study of autophagy in lung diseases, oncology and pharmacochemistry are currently the focused areas.
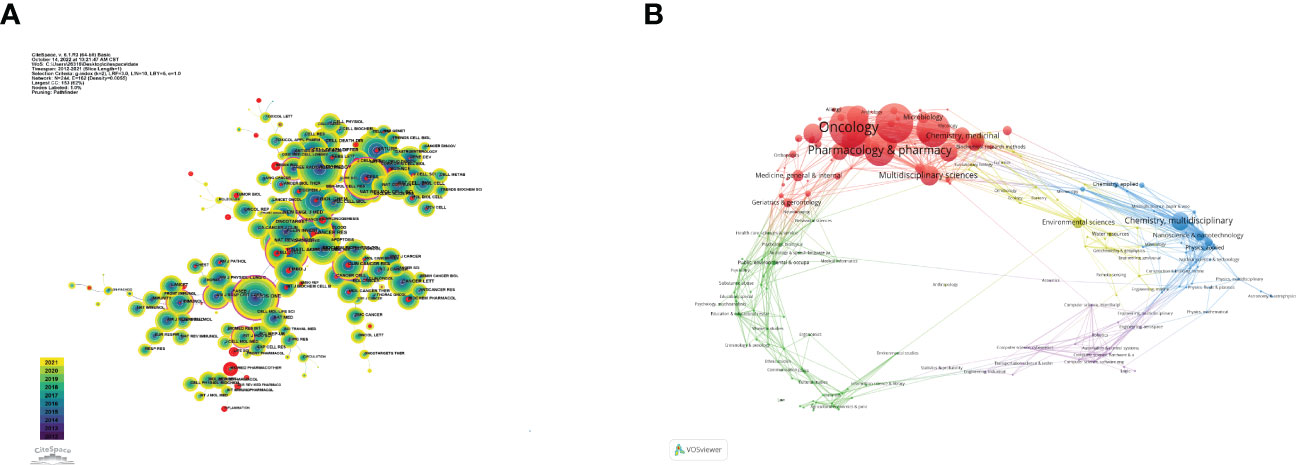
Figure 6 Visualization of co-cited journals on lung disease-related autophagy. (A) Circles denote co-cited journals; connecting lines stand for journal interaction. (B) Superposition analysis of research fields.
CiteSpace assisted in constructing the subject distribution regarding academic journals by virtue of the biplot overlay function (Figure 7). According to the journal biplot overlay, there were mainly two citation paths. Journals publishing articles were basically in medical sciences covering the pharmaceutical, clinical, molecular, biological, and immunological fields, and most cited articles were published in journals in molecular, biological, immunologic, genetic, dermatological, health, nursing, rehabilitation, dental, surgical, sports, neurological, pharmaceutical, and ophthalmological fields.
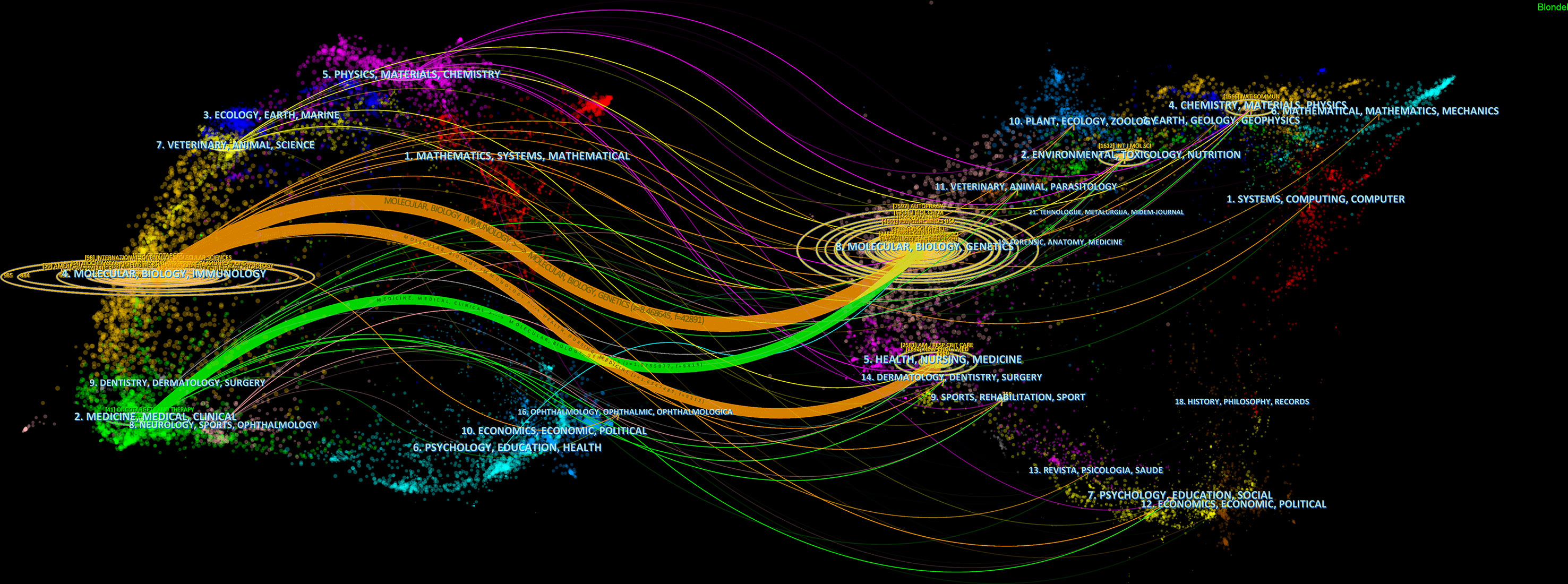
Figure 7 A biplot overlay of journals on autophagy research in lung disease (left and right sides refer to the citing journal areas and the cited journal areas, respectively).
Co-cited reference clusters analysis
A co-citation network refers to a reference network simultaneously co-cited by at least one paper. Conceptual clusters were created when a set of manuscripts was cited together repeatedly (27). As shown in Figures 8A, B, the most cited article was written by Basant A. Abdulrahman who pointed out that rapamycin, an autophagy stimulator, enhanced the clearance of C. coronans by inducing autophagy and significantly reduced the infection of C. coronans in vitro.
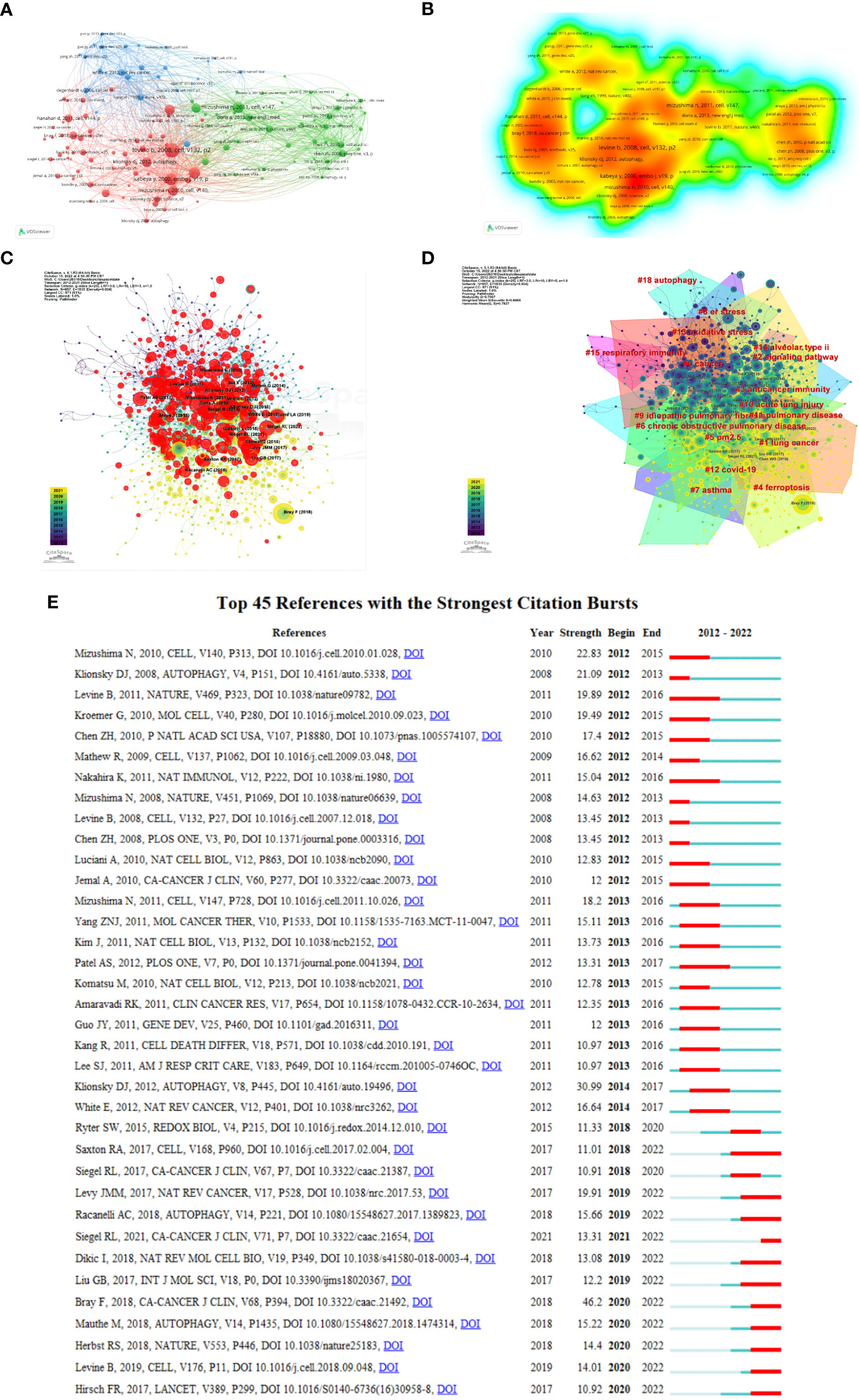
Figure 8 Visualization of co-cited references on autophagy research in lung disease. (A) Reference co-citation network. Circles are co-cited literature. (B) Co-cited reference density visualization. (C) Network visualization diagram of cited references. (D) The clustered network map of lung disease autophagy-related co-cited references. (E) The 45 references having the highest citation burst.
Rapamycin was found capable of lowering the bacterial burden in the mouse lung in vitro and markedly eliminating lung inflammation signs (28). Figure 8C demonstrates a network visualization map regarding cited references, with nodes denoting the cited reference, where the connecting lines stand for the co-citation relationship. The different colors varying from purple to yellow mean different years in the range of 2012 to 2021. Node size denotes the number of co-citation number. Figure 8D displays the 16 relevant clusters divided by CiteSpace: cancer, lung cancer, signaling pathway, anticancer immunity, ferroptosis, particulate matter (PM2.5), COPD, asthma, ER stress, idiopathic pulmonary, acute lung injury, pulmonary disease, COVID-19, oxidative stress, alveolar type II, respiratory immunity, and autophagy. Citation burst reference refers to the sudden citation increase of specific articles within a certain period, which serves for finding the latest high-profile research topics in relative fields (29). We obtained 674 references with the highest citation burst and selected the top 25 (Figure 8E). The blue line denotes the timeline, and the red sections denote the burst interval, respectively, showing the beginning and end of the year, and the burst duration.
Analysis of keywords
Keywords demonstrate a paper’s principal ideas and theme concepts, briefly describing the certain research hot spots. VOSviewer and CiteSpace were used for drawing charts. We identified words that appeared over 13 times in the process of analysis as keywords and finally obtained 155 keywords (Figure 9A; Table 6). As shown in Figure 9C, the closer the keyword was closer to yellow, the more frequently it appeared in the year. According to Figure 9A, clusters 1 and 5 were mainly composed of autophagy, apoptosis, and death, which paid attention to relevant mechanisms in the programmed cell death. Cluster 2 stands for “oxidative stress”, “mitochondrial dysfunction”, and “lung disease”. Oxidative stress, the oxidation-antioxidant imbalance in the body, usually causes mitochondrial dysfunction and lung disease. Cluster 3 emphasizes the function of autophagy in lung diseases caused by COVID-19. Clusters 4 and 5 mainly reflect the cellular mechanism and molecular pathway of lung diseases caused by autophagy. In overlay visualization, the color of keywords depends on the average publication year (APY). As shown in Figure 9B, the VOSviewer colors all keywords on the basis of APY. The most recent keyword was “sars-cov-2” (cluster 3, APY: 2020.53), followed by “COVID-19” (cluster 3, APY: 2020.45), both closely related to COVID-19. The color division confirms that the most recent keywords in the research field were “pyroptosis“, “immunity”, “COVID-19”, and “lung cancer”, which are proven to be the recent research hot spots.
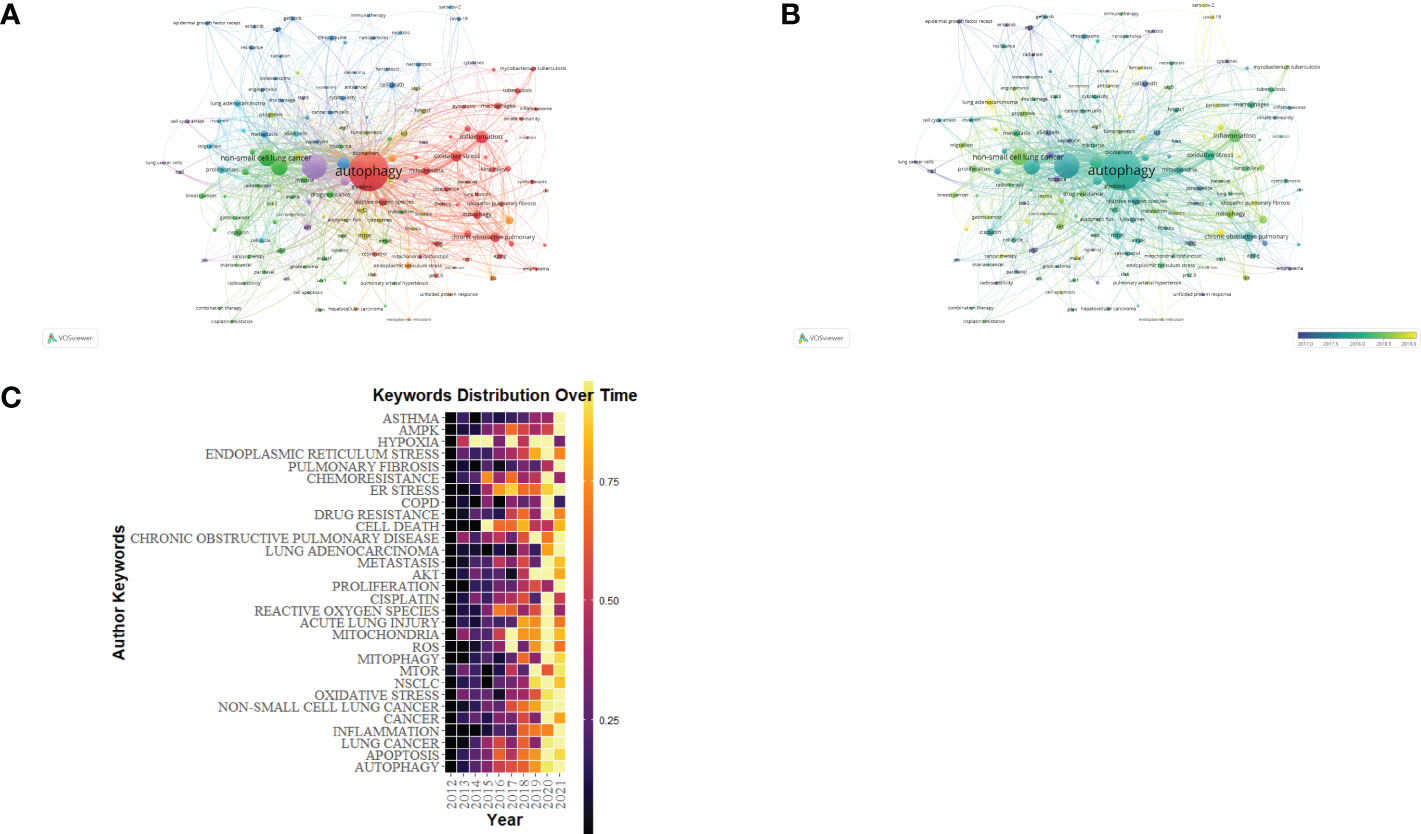
Figure 9 The keyword mapping of autophagy research in lung disease. (A) The 155 keywords appearing over 13 times fell into five clusters based on colors: Cluster 1, 2, 3, 4, and 5 are, respectively, red, green, blue, yellow, and purple. The node size denotes the occurrence frequency. (B) Keywords visualization based on the APY. Keywords in cluster 4 occurred later than those in cluster 3. (C) Yearly occurrences of the top keywords.
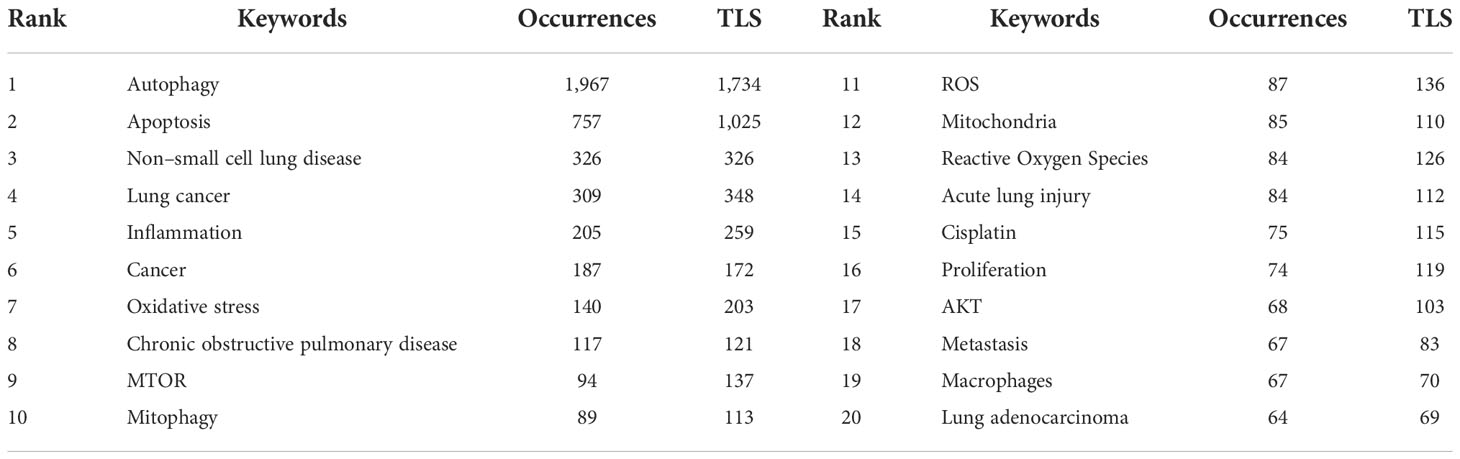
Table 6 Top 20 keywords of publications regarding autophagy research in lung disease based on occurrences.
Analysis of research for autophagy caused by COVID-19
Following the COVID-19 epidemic outbreak in December 2019, research on lung disease increased markedly in 2020 (Figure 10A). Our study screened out and analyzed 76 papers on lung diseases and autophagy related to COVID-19 from the retrieved literature in an attempt to reveal the research focuses and trends in the association between severe lung diseases and autophagy resulting from COVID-19 (Figure 10B). The terms inflammation, immunology, cytokine storm, oxidative stress, and macrophages all had the highest frequency as revealed in the keyword visualization analysis, in addition to COVID-19, SARS-COV-2, and lung-related diseases, explaining how autophagy exerted its function in severe lung disease caused by the pathogenesis of COVID-19. In addition, the APY-based keyword color classification by VOSviewer confirmed “inflammation” and “oxidative stress” as the top two recent keywords, meeting previous findings on autophagy in lung diseases. In addition, Figure 10 shows the involvement of autophagy in the pathogenesis of COVID-19. We also found that the mechanism of anti-inflammatory treatment and immunomodulation therapy for COVID-19–related lung disease was through inhibiting autophagy activation (Figure 10).
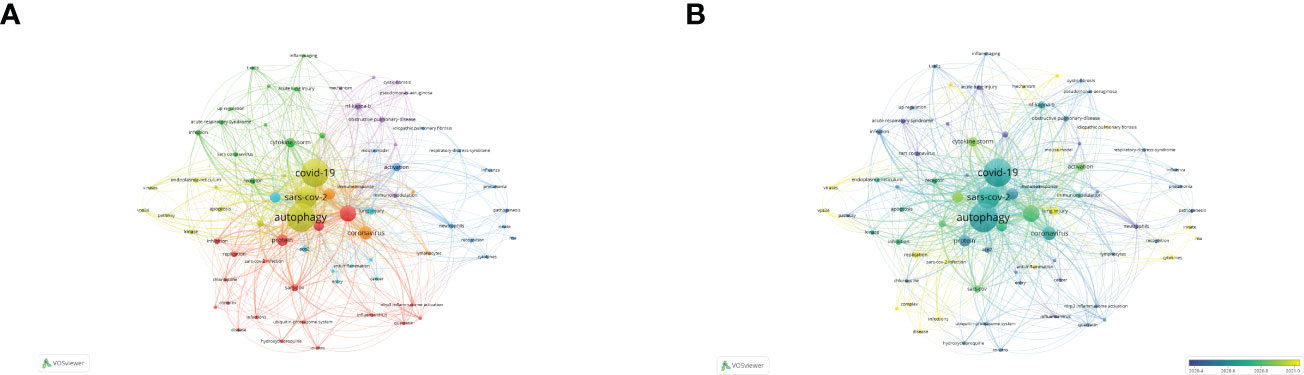
Figure 10 Visualization analysis of autophagy research in lung disease resulted from COVID-19. (A) Keyword network mapping. A total of 76 keywords appearing for over two times fell into six clusters based on colors. (B) APY-based keyword visualization. Keywords in yellow occurred later than those in blue (color version can be found online).
Discussion
In this study, we used VOSviewer, CiteSpace, and R to investigate the research dynamics and hot spots of autophagy in lung diseases in the SCI-Expanded. A total of 4,522 publications were obtained from WoSCC. It was found that there was a substantial increase in the number of annual publications and citations during 2012 and 2021, particularly after 2015. China, which published the most papers, has made greater contributions to the study of autophagy in lung diseases. Moreover, nine of the top 10 productive institutions were from China, including China Medical University, Shandong University, and Zhejiang University, indicating that autophagy has drawn increasing attention from Chinese research institutions in the study of lung diseases recently. However, the H-index and G-index of the US were relatively high compared with that of China. The US ranked first among all countries in both H-index and G-index, demonstrating that the quality of documents from the US is high. Despite China’s rapid development and dominant position in the field, it is necessary to improve the collaboration and academic influence in regions. The international distribution of current research in this field is imbalanced.
Choi Augustine M. K. is the most published and cited author in this field. He has long studied the relationship between acute and chronic lung diseases and molecular, cellular, and genetic triggers. He proposed that lung disease may be associated with a pathological accumulation of mutated α1-antitrypsin and that autophagy may be a clearance mechanism for this disease (3). A proper method for monitoring autophagy has been established by Klionsky et al., who suggested that autophagy defects could be induced in favor of lung tumor formation by modulating the tumor microenvironment (28). Noboru Mizushima is also a very influential writer whose works are widely cited. In 1998, Noboru Mizushima reported the homologous gene of Atg12 in human and clarified autophagy (cell self-digestion) as a cellular pathway capable of assisting in degrading proteins and organelles, which is deeply related to human diseases and physiology (30). Yoshinori OhSumi, winner of the 2016 Nobel Prize in Physiology or Medicine, made excellent achievements regarding the function had by autophagy (31). The results of his work have already assisted us in better understanding the function and the effect of autophagy on disease and health (32). The above authors are well respected as they more comprehensively elucidate the mechanism regarding autophagy and contribute to the prevention of lung diseases.
Usually, there is a close relation between the impact of a journal and the impact of its articles (33). Amid the top 10 journals that had the most published papers, AUTOPHAGY exhibited the highest IF (16.016), which was first published in 2005 and is now a leading journal in the field. ONCOTARGET was the most prolific journal, with an IF of 4.147. The journal is sponsored by the US and focuses on articles related to cancer, tumors, autophagy, and cellular immunity. In addition, most active journals are in the professional category. Several multidisciplinary journals, such as CELL DEATH DIS and BIOMED PHARMACOTHER, have also published high-quality studies investigating autophagy in lung disease, which is an emerging and developing research topic and involves molecular biology, cell biology, biochemistry, and medicine. Journals of various disciplines have published advances in such research topic, demonstrating the wide attention paid to lung disease pathogenesis and treatment. In addition, multidisciplinary journals have a wider readership (34), more benefiting interdisciplinary cooperation.
As the research continuously progresses, researchers have revealed the signal pathway that affects different mechanisms. On the basis of keyword mapping, oxidative stress critically impacts the pathogenesis of autophagy, regulating the progression of related lung diseases. The activation of autophagy in response to oxidative stress is mainly for avoiding cell apoptosis (35, 36), In contrast, autophagy inhibition increase oxidative stress damage and even cell death. When ROS is elevated, the HIF-1α transcription factor, p53, FOXO3, and Nrf2 will be activated, which then triggers BNIP3 and NIX, TIGAR, LC3, and p62 (37) to transcribe. The oxidized ATG4 can facilitate LC3 lipidation to initiate autophagy and autophagosome formation. According to a recent report, angiotensin II (Ang II) vasoconstriction can induce autophagy, revealing the ability of Ang II to form autophagic vesicles that contain LC3 and to elevate the ATG12–ATG5, ATG7, and ATG4, thereby simulating the elongation of phagophores (38). As for human glomerular mesangial cells, oxidative stress triggered by Ang II can use mTORC1 signaling for enhancing autophagy, leading to early-stage senescence (39). ROS was found capable of regulating autophagy through pathways dependent of mTOR in the cytoplasm (40, 41). ROS inhibits the PI3K-Akt-mTOR pathway or activates AMPK for suppressing the mTOR signaling pathway, thereby resulting in autophagy activation (42). The increase in ROS oxidizing phosphatase and tensin homolog helps inhibit Akt-mTOR activity (43). Similarly, excess H2O2 is capable of triggering autophagy dependent of AMPK, coupled with weakened mTORC1 activity (44). Thioredoxin-interacting protein (TXNIP) takes charge of regulating the autophagy of rats with diabetic nephropathy via the mTOR signaling pathway (45). In lung diseases, hypoxia caused by the weakened lung functions primarily stimulates the autophagy induction (45). As for the primary human lung vascular endothelial and smooth muscle-cells, hypoxia could induce BECN1 expression, activate LC3, promote autophagosome formation, and stimulate the autophagic flux, thereby protecting mice from pulmonary vascular disease (46). Therefore, this provides a clear path for us to further explore the autophagy-lung disease relationship.
When we talk about the impact of autophagy on various lung diseases, COVID-19 is an unavoidable topic associated with autophagy-mediated lung disease (Figure 9A). The SARS-CoV-2 virus causing COVID-19 exhibits a strong infectivity and can trigger cytokine-storm, inducing acute respiratory distress syndrome (ARDS)–like lung injury that can develop into pneumonia and severe lung damage, causing high risk of mortality, especially in susceptible populations (47–49). According to recent studies, SARS-CoV-2 is capable of restricting autophagy, which, as anticipated, is an underlying mechanism for serious COVID-19 lung disease as it impairs the viral clearance ability and causes immune dysfunction (50, 51). Recently, researchers have revealed the stronger affinity exhibited by NSP6 protein of SARS-CoV-2 in binding with the ER (52). Because of the genetic alternation, virus is capable of restricting autophagy through damaged autophagosome processing, where viral particles can be less degraded by lysosome (52). In addition, relying on PLP2 overexpression in SARS-CoV and MERS-CoV cell lines, the virus is capable of preventing autophagolysosomal formation and suppressing autophagy flux, which may serve for inhibiting autophagy in SARS-CoV-2 (53). These studies reveal the potential ways for SARS-CoV-2 to inhibit autophagy infection or avoid the pathogenic clearance of host cells, meanwhile restricting the sufficient immune response, which is similar to other types of coronaviruses, and need to be further investigated. Eliminating viruses by autophagy (also called virophagy) can well serve for various viral infections (54, 55). Despite the various ways through viruses get access to cells, autophagy augmentation can promote virus clearance, thus reducing the viral load from a strategic perspective (56, 57). To prove this from a conceptual perspective, some recent studies have investigated how the three different drugs (spermidine, MK02206, and niclosamide) induced by autophagy restrict the propagation of SARS-CoV-2 (58). Autophagy induction together with relevant overall immunity upregulation assists in combating the exacerbations, which may prove to be a strategy to enhance immunity and prevent COVID-19 infection (59). Autophagy induction can enhance immunity, restrict viral load, support SARS-CoV-2 clearance, and simultaneously assist in treating COVID-19 infection and preventing negative results, although further investigation is required to confirm these effects.
The pathogenesis regarding severe COVID-19 mediated by SARS-CoV-2 is pointed out to be associated with the activation of many pro-inflammatory cytokines that constitute the cytokine storm, resulting in a hyper-inflammatory state. In addition to destroying the lung and COVID-19–related ARDS, such inflammatory response is also capable of damaging the cardiovascular, nervous, and gastrointestinal systems and bringing into direct and long-term outcomes (60). To support these findings, studies have revealed how autophagy affects the inflammatory response in organ systems including the lung (61, 62). Accordingly, autophagy induction is capable of attenuating lung inflammation in the event of being exposed to pathogens. Hence, except for infection, using autophagy induction for restricting the inflammatory response has huge potentials for treating COVID-19 and decreasing relevant morbidity and mortality.
In this study, we employed the bibliometric approach for visualizing the research on autophagy-mediated lung diseases, thereby gaining a better understanding about the hot spots and trends in this field. Nevertheless, it suffers many limitations (1). The study only included English articles and reviews from the SCI-Expanded. (2) Currently, the limitation of software-based scientific measurement added the difficulty in combining at least two databases for the analysis. Hence, the study only adopted the WoSCC database for the screening and might have missed the other related studies in the literature. Therefore, more databases should be employed for a more rigorous analysis in the future. (3) Open-source journals may have an impact on both citation volume and publication volume. (4) The combination of CiteSpace and VOSviewer for literature analysis may miss information during analysis. Specifically, VOSviewer keyword analysis clustering might not have been able to extract key information, because color was only used to divide the range.
Conclusions
According to bibliometric analysis, the research on autophagy in the field of lung diseases has broad prospects and rapid development. We identified the major contributors, regions, and publications in the research area. China published the largest NP and the US made more huge breakthroughs. The latest studies have been published and new progression regarding the field has been made on AUTOPHAGY and ONCOTARGET. Recently, oxidative stress has been revealed to have significant impact on the pathogenesis of autophagy-mediated lung diseases. In addition, the mechanism of autophagy induction may prove crucial as a treatment strategy for COVID-19 over classical antivirals and provide a tool to end the pandemic. Attenuating the progression of COVID-19–associated lung diseases by inhibiting autophagy may become a new research hotspot in this field.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Author contributions
FL, YC and WM did the bibliometrics analysis and wrote the manuscript. HZ, ZX, SH, XS, ML, JW and WZ participated in the experimental design and manuscript writing. SW and XL designed this study and organized the manuscript writing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81871601), the Program of Shanghai Academic Research Leader (No. 21XD1402800), the Shanghai Natural Science Foundation (22ZR1452200), the Development Fund for the Department of Anesthesiology, Shanghai Pulmonary Hospital.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science (2000) 290(5497):1717–21. doi: 10.1126/science.290.5497.1717
2. Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell (2011) 147(4):728–41. doi: 10.1016/j.cell.2011.10.026
3. Klionsky DJ, Petroni G, Amaravadi RK, Baehrecke EH, Ballabio A, Boya P, et al. Autophagy in major human diseases. EMBO J (2021) 40(19):e108863. doi: 10.15252/embj.2021108863
4. Verzella D, Pescatore A, Capece D, Vecchiotti D, Ursini MV, Franzoso G, et al. Life, death, and autophagy in cancer: NF-κB turns up everywhere. Cell Death Dis (2020) 11(3):210. doi: 10.1038/s41419-020-2399-y
5. Galluzzi L, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cecconi F, et al. Molecular definitions of autophagy and related processes. EMBO J (2017) 36(13):1811–36. doi: 10.15252/embj.201796697
6. Mizushima N. A brief history of autophagy from cell biology to physiology and disease. Nat Cell Biol (2018) 20(5):521–7. doi: 10.1038/s41556-018-0092-5
7. Petersson J, Glenny RW. Gas exchange and ventilation-perfusion relationships in the lung. Eur Respir J (2014) 44(4):1023–41. doi: 10.1183/09031936.00037014
8. Nasim F, Sabath BF, Eapen GA. Lung cancer. Med Clin North Am (2019) 103(3):463–73. doi: 10.1016/j.mcna.2018.12.006
9. Ranieri VM, Rubenfeld GD, Thompson BT, Ferguson ND, Caldwell E, Fan E, et al. Acute respiratory distress syndrome: the Berlin definition. JAMA (2012) 307(23):2526–33. doi: 10.1001/jama.2012.5669
10. Beitler JR, Thompson BT, Baron RM, Bastarache JA, Denlinger LC, Esserman L, et al. Advancing precision medicine for acute respiratory distress syndrome. Lancet Respir Med (2022) 10(1):107–20. doi: 10.1016/S2213-2600(21)00157-0
11. Ma C, Su H, Li H. Global research trends on prostate diseases and erectile dysfunction: A bibliometric and visualized study. Front Oncol (2020) 10:627891. doi: 10.3389/fonc.2020.627891
12. Guler AT, Waaijer CJ, Palmblad M. Scientific workflows for bibliometrics. Scientometrics (2016) 107:385–98. doi: 10.1007/s11192-016-1885-6
13. Huang X, Fan X, Ying J, Chen S. Emerging trends and research foci in gastrointestinal microbiome. J Transl Med (2019) 17(1):67. doi: 10.1186/s12967-019-1810-x
14. Zhang T, Yin X, Yang X, Man J, He Q, Wu Q, et al. Research trends on the relationship between microbiota and gastric cancer: A bibliometric analysis from 2000 to 2019. J Cancer (2020) 11(16):4823–31. doi: 10.7150/jca.44126
15. Liang M, Meng Y, Zhou S, Tao Z, Tao L. Research hotspots and trends analysis of ankylosing spondylitis: a bibliometric and scientometric analysis from 2009 to 2018. Ann Transl Med (2020) 8(21):1445. doi: 10.21037/atm-20-1259
16. Martynov I, Klima-Frysch J, Schoenberger J. A scientometric analysis of neuroblastoma research. BMC Cancer (2020) 20(1):486. doi: 10.1186/s12885-020-06974-3
17. Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics (1977) 33(1):159–74. doi: 10.2307/2529310
18. Hirsch JE. An index to quantify an individual's scientific research output. Proc Natl Acad Sci U S A. (2005) 102(46):16569–72. doi: 10.1073/pnas.0507655102
19. Abbas AM. Bounds and inequalities relating h-index, g-index, e-index and generalized impact factor: an improvement over existing models. PloS One (2012) 7(4):e33699. doi: 10.1371/journal.pone.0033699
20. Jones T, Huggett S, Kamalski J. Finding a way through the scientific literature: indexes and measures. World Neurosurg (2011) 76(1-2):36–8. doi: 10.1016/j.wneu.2011.01.015
21. Roldan-Valadez E, Salazar-Ruiz SY, Ibarra-Contreras R, Rios C. Current concepts on bibliometrics: a brief review about impact factor, eigenfactor score, CiteScore, SCImago journal rank, source-normalised impact per paper, h-index, and alternative metrics. Ir J Med Sci (2019) 188(3):939–51. doi: 10.1007/s11845-018-1936-5
22. Campra M, Riva P, Oricchio G, Brescia V. Bibliometrix analysis of medical tourism. Health Serv Manage Res (2022) 35(3):172–88. doi: 10.1177/09514848211011738
23. van Eck NJ, Waltman L. Software survey: VOSviewer, a computer program for bibliometric mapping. Scientometrics (2010) 84(2):523–38. doi: 10.1007/s11192-009-0146-3
24. Yao L, Hui L, Yang Z, Chen X, Xiao A. Freshwater microplastics pollution: Detecting and visualizing emerging trends based on citespace II. Chemosphere (2020) 245:125627. doi: 10.1016/j.chemosphere.2019.125627
25. Zou X, Vu HL, Huang H. Fifty years of accident analysis & prevention: A bibliometric and scientometric overview. Accid Anal Prev (2020) 144:105568. doi: 10.1016/j.aap.2020.105568
26. Chen C. Searching for intellectual turning points: progressive knowledge domain visualization. Proc Natl Acad Sci U S A. (2004) 101(Suppl 1):5303–10. doi: 10.1073/pnas.0307513100
27. Shi X, Wang S, Wu Y, Li Q, Zhang T, Min K, et al. A bibliometric analysis of the innate immune DNA sensing cGAS-STING pathway from 2013 to 2021. Front Immunol (2022) 13:916383. doi: 10.3389/fimmu.2022.916383
28. Abdulrahman BA, Khweek AA, Akhter A, Caution K, Kotrange S, Abdelaziz DH, et al. Autophagy stimulation by rapamycin suppresses lung inflammation and infection by burkholderia cenocepacia in a model of cystic fibrosis. Autophagy (2011) 7(11):1359–70. doi: 10.4161/auto.7.11.17660
29. Miao L, Zhang J, Zhang Z, Wang S, Tang F, Teng M, et al. A bibliometric and knowledge-map analysis of CAR-T cells from 2009 to 2021. Front Immunol (2022) 13:840956. doi: 10.3389/fimmu.2022.840956
30. Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature (2008) 451(7182):1069–75. doi: 10.1038/nature06639
31. Martinet W, De Meyer GR, Andries L, Herman AG, Kockx MM. In situ detection of starvation-induced autophagy. J Histochem Cytochem (2006) 54(1):85–96. doi: 10.1369/jhc.5A6743.2005
32. Ohsumi Y. Historical landmarks of autophagy research. Cell Res (2014) 24(1):9–23. doi: 10.1038/cr.2013.169
33. Callaham M, Wears RL, Weber E. Journal prestige, publication bias, and other characteristics associated with citation of published studies in peer-reviewed journals. JAMA (2002) 287(21):2847–50. doi: 10.1001/jama.287.21.2847
34. Bolli R. Ten years at the helm of circulation research. Circ Res (2019) 124(12):1707–17. doi: 10.1161/CIRCRESAHA.119.315287
35. Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell (2008) 132(1):27–42. doi: 10.1016/j.cell.2007.12.018
36. Choi AM, Alam J. Heme oxygenase-1: function, regulation, and implication of a novel stress-inducible protein in oxidant-induced lung injury. Am J Respir Cell Mol Biol (1996) 15(1):9–19. doi: 10.1165/ajrcmb.15.1.8679227
37. Lee J, Giordano S, Zhang J. Autophagy, mitochondria and oxidative stress: cross-talk and redox signalling. Biochem J (2012) 441(2):523–40.
38. Mondaca-Ruff D, Riquelme JA, Quiroga C, Norambuena-Soto I, Sanhueza-Olivares F, Villar-Fincheira P, et al. Angiotensin II-regulated autophagy is required for vascular smooth muscle cell hypertrophy. Front Pharmacol (2018) 9:1553. doi: 10.3389/fphar.2018.01553
39. Yang S, Sun D, Wang L, Wang X, Shi M, Jiang X, et al. The role of STAT3/mTOR-regulated autophagy in angiotensin II-induced senescence of human glomerular mesangial cells. Cell Signal (2019) 53:327–38. doi: 10.1016/j.cellsig.2018.10.021
40. Zhang L, Wang H, Xu J, Zhu J, Ding K. Inhibition of cathepsin s induces autophagy and apoptosis in human glioblastoma cell lines through ROS-mediated PI3K/AKT/mTOR/p70S6K and JNK signaling pathways. Toxicol Lett (2014) 228(3):248–59. doi: 10.1016/j.toxlet.2014.05.015
41. Rahman I, Biswas SK, Kode A. Oxidant and antioxidant balance in the airways and airway diseases. Eur J Pharmacol (2006) 533(1-3):222–39. doi: 10.1016/j.ejphar.2005.12.087
42. Zhang J, Kim J, Alexander A, Cai S, Tripathi DN, Dere R, et al. A tuberous sclerosis complex signalling node at the peroxisome regulates mTORC1 and autophagy in response to ROS. Nat Cell Biol (2013) 15(10):1186–96. doi: 10.1038/ncb2822
43. Salmeen A, Barford D. Functions and mechanisms of redox regulation of cysteine-based phosphatases. Antioxid Redox Signal (2005) 7(5-6):560–77. doi: 10.1089/ars.2005.7.560
44. Koga H, Cuervo AM. Chaperone-mediated autophagy dysfunction in the pathogenesis of neurodegeneration. Neurobiol Dis (2011) 43(1):29–37. doi: 10.1016/j.nbd.2010.07.006
45. Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouysségur J, et al. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol (2009) 29(10):2570–81. doi: 10.1128/MCB.00166-09
46. Lee SJ, Smith A, Guo L, Alastalo TP, Li M, Sawada H, et al. Autophagic protein LC3B confers resistance against hypoxia-induced pulmonary hypertension. Am J Respir Crit Care Med (2011) 183(5):649–58. doi: 10.1164/rccm.201005-0746OC
47. Li X, Ma X. Acute respiratory failure in COVID-19: is it "typical" ARDS. Crit Care (2020) 24(1):198. doi: 10.1186/s13054-020-02911-9
48. Guo T, Fan Y, Chen M, Wu X, Zhang L, He T, et al. Cardiovascular implications of fatal outcomes of patients with coronavirus disease 2019 (COVID-19). JAMA Cardiol (2020) 5(7):811–8. doi: 10.1001/jamacardio.2020.1017
49. Chaimayo C, Kaewnaphan B, Tanlieng N, Athipanyasilp N, Sirijatuphat R, Chayakulkeeree M, et al. Rapid SARS-CoV-2 antigen detection assay in comparison with real-time RT-PCR assay for laboratory diagnosis of COVID-19 in Thailand. Virol J (2020) 17(1):177. doi: 10.1186/s12985-020-01452-5
50. Mijaljica D, Klionsky DJ. Autophagy/virophagy: a "disposal strategy" to combat COVID-19. Autophagy (2020) 16(12):2271–2. doi: 10.1080/15548627.2020.1782022
51. Bonam SR, Muller S, Bayry J, Klionsky DJ. Autophagy as an emerging target for COVID-19: lessons from an old friend, chloroquine. Autophagy (2020) 16(12):2260–6. doi: 10.1080/15548627.2020.1779467
52. Benvenuto D, Angeletti S, Giovanetti M, Bianchi M, Pascarella S, Cauda R, et al. Evolutionary analysis of SARS-CoV-2: how mutation of non-structural protein 6 (NSP6) could affect viral autophagy. J Infect (2020) 81(1):e24–7. doi: 10.1016/j.jinf.2020.03.058
53. Chen X, Wang K, Xing Y, Tu J, Yang X, Zhao Q, et al. Coronavirus membrane-associated papain-like proteases induce autophagy through interacting with Beclin1 to negatively regulate antiviral innate immunity. Protein Cell (2014) 5(12):912–27. doi: 10.1007/s13238-014-0104-6
54. Abdoli A, Alirezaei M, Mehrbod P, Forouzanfar F. Autophagy: The multi-purpose bridge in viral infections and host cells. Rev Med Virol (2018) 28(4):e1973. doi: 10.1002/rmv.1973
55. Mehrbod P, Ande SR, Alizadeh J, Rahimizadeh S, Shariati A, Malek H, et al. The roles of apoptosis, autophagy and unfolded protein response in arbovirus, influenza virus, and HIV infections. Virulence (2019) 10(1):376–413. doi: 10.1080/21505594.2019.1605803
56. Carmona-Gutierrez D, Bauer MA, Zimmermann A, Kainz K, Hofer SJ, Kroemer G, et al. Digesting the crisis: autophagy and coronaviruses. Microb Cell (2020) 7(5):119–28. doi: 10.15698/mic2020.05.715
57. Yang N, Shen HM. Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID-19. Int J Biol Sci (2020) 16(10):1724–31. doi: 10.7150/ijbs.45498
58. Woodby B, Arnold MM, Valacchi G. SARS-CoV-2 infection, COVID-19 pathogenesis, and exposure to air pollution: What is the connection. Ann N Y Acad Sci (2021) 1486(1):15–38. doi: 10.1111/nyas.14512
59. Hannan MA, Rahman MA, Rahman MS, Sohag AAM, Dash R, Hossain KS, et al. Intermittent fasting, a possible priming tool for host defense against SARS-CoV-2 infection: Crosstalk among calorie restriction, autophagy and immune response. Immunol Lett (2020) 226:38–45. doi: 10.1016/j.imlet.2020.07.001
60. Jamwal S, Gautam A, Elsworth J, Kumar M, Chawla R, Kumar P. An updated insight into the molecular pathogenesis, secondary complications and potential therapeutics of COVID-19 pandemic. Life Sci (2020) 257:118105. doi: 10.1016/j.lfs.2020.118105
61. Painter JD, Galle-Treger L, Akbari O. Role of autophagy in lung inflammation. Front Immunol (2020) 11:1337. doi: 10.3389/fimmu.2020.01337
Keywords: autophagy, lung, bibliometrics, bibliometrix, VOSviewer, citespace
Citation: Lin F, Chen Y, Mo W, Zhou H, Xiao Z, Hu S, Shi X, Liu M, Wei J, Zhu W, Wang S and Lv X (2022) A bibliometric analysis of autophagy in lung diseases from 2012 to 2021. Front. Immunol. 13:1092575. doi: 10.3389/fimmu.2022.1092575
Received: 08 November 2022; Accepted: 28 November 2022;
Published: 16 December 2022.
Edited by:
Kai Wang, Southwest Medical University, ChinaReviewed by:
Milad Shirvaliloo, Tabriz University of Medical Sciences, IranMd. Abdul Alim Al-Bari, Rajshahi University, Bangladesh
Copyright © 2022 Lin, Chen, Mo, Zhou, Xiao, Hu, Shi, Liu, Wei, Zhu, Wang and Lv. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin Lv, eGlubHZnQDEyNi5jb20=; Sheng Wang, d3MyMDE1bWFAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Feihong Lin
Feihong Lin Yong Chen
Yong Chen Wei Mo
Wei Mo Huanping Zhou
Huanping Zhou Zhuoran Xiao
Zhuoran Xiao Song Hu
Song Hu Xuan Shi
Xuan Shi Meiyun Liu
Meiyun Liu Juan Wei
Juan Wei Wanli Zhu
Wanli Zhu Sheng Wang
Sheng Wang Xin Lv
Xin Lv