
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 05 January 2023
Sec. Molecular Innate Immunity
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.1091543
Despite noteworthy progress made in the management and treatment of HIV/AIDS-related disease, including the introduction of the now almost ubiquitous HAART, there remains much to understand with respect to HIV infection. Although some roles that miRNAs play in some diseases have become more obvious of late, the roles of miRNAs in the context of HIV pathogenesis have not, as yet, been elucidated, and require further investigations. miRNAs can either be beneficial or harmful to the host, depending upon the genes they target. Some miRNAs target the 3′ UTR of viral mRNAs to accomplish restriction of viral infection. However, upon HIV-1 infection, there are several dysregulated host miRNAs which target their respective host factors to either facilitate or abrogate viral infection. In this review, we discuss the miRNAs which play roles in various aspects of viral pathogenesis. We describe in detail the various mechanisms thereby miRNAs either directly or indirectly regulate HIV-1 infection. Moreover, the predictive roles of miRNAs in various aspects of the HIV viral life cycle are also discussed. Contemporary antiretroviral therapeutic drugs have received much attention recently, due to their success in the treatment of HIV/AIDS; therefore, miRNA involvement in various aspects of antiretroviral therapeutics are also elaborated upon herein. The therapeutic potential of miRNAs are discussed, and we also propose herein that the therapeutic potential of one specific miRNA, miR-34a, warrants further exploration, as this miRNA is known to target three host proteins to promote HIV-1 pathogenesis. Finally, future perspectives and some controversy around the expression of miRNAs by HIV-1 are also discussed.
MicroRNAs (miRNAs) refer to non-coding RNAs (ncRNAs) which are approximately 22 nucleotides in length, and usually bind to the 3′ untranslated region (3′ UTR) of their target messenger RNA (mRNAs), to either promote mRNA degradation or to repress translation (1). miRNA biogenesis and maturation is a stepwise process that starts in the nucleus and ends in cytoplasm. Most miRNA genes are transcribed by RNA polymerase II (Pol II) into long primary transcripts of miRNA called pri-miRNAs harboring a hairpin structure (2–4). The pri-miRNA are processed within the nucleus by Drosha, an RNAse III enzyme, into stem-loop intermediates called precursor-miRNA (pre-miRNAs) (2). These pre-miRNAs are then transported from the nucleus to the cytoplasm where they are further processed by the Dicer enzyme. Dicer converts them into their mature forms, called miRNAs, which are ultimately responsible for the phenomenon of RNA silencing (2, 5).
miRNAs are involved in the regulation of several human diseases including cancer, liver diseases, neurodegenerative diseases, and autoimmune diseases, and therefore affects many developmental pathways and cellular processes (1, 6–9). Furthermore, miRNAs are known to play regulatory roles in the pathogenesis of different viral diseases, including in human immunodeficiency virus (HIV)-related disease (10).
HIV is a retrovirus with a single-stranded RNA genome. After entering the host cell, single-stranded RNA is reverse transcribed into DNA, which then integrates into host DNA (11, 12). HIV DNA integration is the crucial step of the viral life cycle which renders the infection challenging to cure, as the infected cell remains infected for the life of the cell (12). Host enzymes and miRNAs are actively involved in all processes of the HIV life cycle, from attachment to the host cell until the production of viable HIV virions (13–16).
Past evidence has demonstrated that miRNAs are involved not only in HIV pathogenesis before treatment, but also in the immune recovery process during antiretroviral therapy (ART), and may serve as biomarkers and even as predictive markers for immune responses after initiation of ART (17–19). This review aims to summarize major advancements made in the field of miRNAs and HIV. This work presents details of the functions of cell-encoded and viral-encoded miRNAs, and their regulation during the pathophysiological processes of HIV-1. Furthermore, the controversy surrounding HIV’s ability to express miRNA and the therapeutic potential of miRNAs for HIV-1-related disease are discussed. Finally, a chapter introducing future perspectives in this specific area of scientific endeavor concludes our review.
Host miRNAs play important roles in regulating HIV-1 infection, and the biological crosstalk between HIV-1 and host miRNAs has been studied in the past (10, 20). miRNAs regulate HIV-1 infection either directly, by interfering with the HIV genome, or indirectly, by regulating host factors which regulate HIV-1 pathogenesis (14, 21, 22).
The latent presence of HIV-1 virions in resting primary CD4+ T-cells is the major hurdle for the eradication of the HIV in patients on highly active antiretroviral therapy (HAART). The potential role that miRNAs may play in revealing the potential mechanism/s of this latency in resting CD4+ T-cells has been discussed previously (23). The 3′ UTR region of HIV-1 mRNAs contains binding sites for miR-28, miR-125b, miR-150, miR-223, and miR-382 (Figure 1) (Table 1). Microarray analysis indicates that these miRNAs are highly abundant in resting CD4+ T-cells, compared to activated CD4+ T-cells. However, when resting CD4+ T-cells are activated, this results in the downregulation of all the above-mentioned miRNAs. Therefore, the higher expression level of these miRNAs in resting CD4+ T-cells may be seen to be responsible for maintaining HIV-1 latency (23). These miRNAs have been found to be enriched in monocytes as opposed to macrophages, because upon differentiation, macrophages suppress the expression of miRNAs, to promote HIV-1 infection (26). In a publicly available re-analysis of the above data set, only miR-223 was confirmed to be down-regulated in macrophages, while the other miRNAs were either highly expressed or remained unchanged in macrophages, relative to monocytes (27). The differences in results from the above studies indicate that more sophisticated experimental approaches are required in the future to confidently make valid conclusions regarding these processes. Furthermore, another study also identified miR-196b and miR-1290, which targets the 3′ UTR of HIV-1 to regulate HIV-1 latency (Figure 1) (16). These studies suggest that a large variety of miRNAs may be responsible for regulating latency in HIV-1-infected cells.
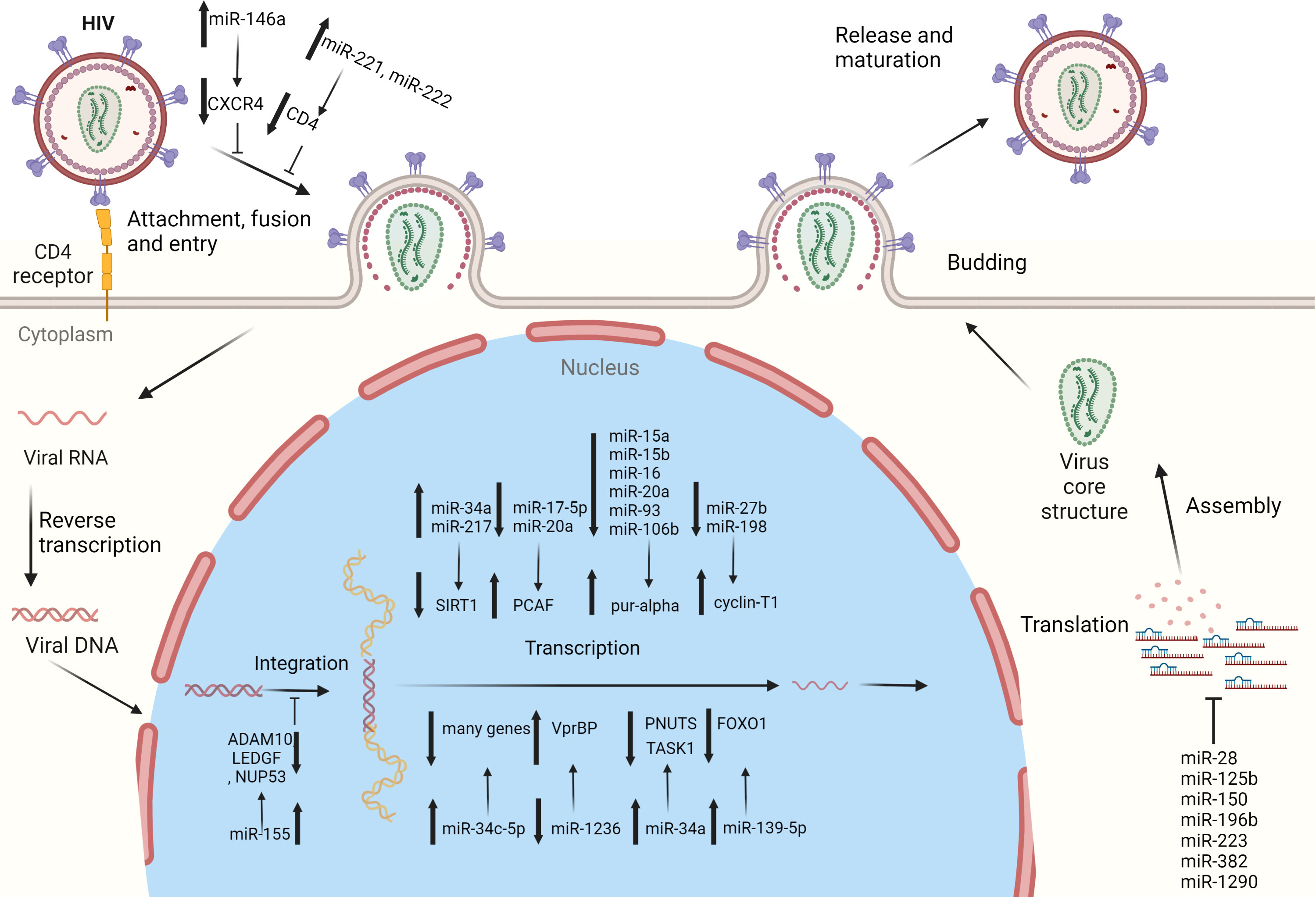
Figure 1 Cellular miRNAs regulating HIV-1 replication: several miRNAs have been identified which regulate HIV-1 replication and pathogenesis. These miRNAs target the specific host genes to regulate specific steps of the HIV-1 life cycle, i.e., cell entry, integration, replication, and latency.
Nef is expressed early during the HIV-1 life cycle, and is indispensable for the progression of HIV-1 infection and for the release of infectious virions (28, 29). Bioinformatics tools have identified miR-29a and miR-29b to have binding sites in the Nef transcript (Table 1) and inhibition of miR-29a increases the replication of HIV-1 (24), indicating that miR-29a plays a role in inhibiting HIV-1 replication.
Some miRNAs are well known to regulate host proteins that contribute to the HIV-1 life cycle, e.g., those miRNAs indirectly targeting HIV-1 infection by regulating host proteins.
CXCR4 is a co-receptor of the CD4 molecule, which aids HIV-1 entry into myeloid cells and T-lymphocytes. MiR-146a directly targets CXCR4 while Promyelocytic leukemia zinc finger (PLZF) is responsible for down-regulating the expression of miR-146a (Figure 1) and therefore, silencing PLZF will lead to up-regulation of miR-146a. Consequently, levels of CXCR4 are decreased, which prevents HIV-1 infection of the leukemic monocyte cell line and CD4+ T- lymphocytes (Table 2) (36). Thus, a novel therapeutic strategy could potentially be devised by utilizing the preceding mechanism to prevent HIV-1 entry into cells via exploitation of the PLZF/miR-146a axis.
CD4 receptors that are required for viral entry are expressed on macrophages (41). However, CD4 expression by macrophages is 20-fold less, compared to CD4+ T-cells. RNA-seq data showed that two miRNAs, miR-221 and miR-222, are up-regulated in bystander macrophages and were found to be negative regulators of CD4 and CD4-mediated HIV-1 entry (Figure 1) (Table 2). TNF-α, which is an inhibitor of CD4, enhances the expression of both miRNAs. Inhibition of miR-221/222 enhances CD4 expression, and allows HIV-1 entry into TNF-α-treated macrophages. Therefore, both miR-221 and miR-222 restricts HIV-1 entry, and act as effectors of the antiviral host response activated during infection of macrophages (14).
HIV-1 relies on host dependency factors (HDFs) to facilitate infection. miR-155 targets HIV-1 HDFs before its genome is integrated into the host genome (15). miR-155 expression levels are increased when primary human macrophages are induced by TL3 or TLR4, and this subsequently results in decreased HIV-1 infectivity. miR-155 targets three HDFs i.e., ADAM metallopeptidase domain 10 (ADAM10), transcriptional co-activator lens epithelium-derived growth factor (LEDGF)/p75, and nucleoporin 153 (NUP153) in primary macrophages at their 3′ UTR (Table 2). Since all these three HDFs are involved in the pre-integration process of viral genome with host DNA and miR-155 targets HDFs, increased expression of miR-155 will result in anti-HIV-1 effects (Figure 1) (15).
Purine-rich element binding protein α (Pur-α) is an RNA and DNA binding protein which binds to Tat and TAR elements to augment HIV-1 transcription. Levels of Pur-α are lower in monocytes compared to monocyte-derived dendritic cells (DCs) because of high expression levels of miR-15a, miR-15b, miR-16, miR-20a, miR-93, and miR-106b in monocytes, which target the 3’ UTR of Pur-α (Table 2). Over-expression of these miRNAs significantly suppresses HIV-1 infection of monocyte-derived DCs by down-regulating Pur-α, while inhibition of these miRNAs increases Pur-α levels and removes the restriction of HIV-1 infection in monocytes (Figure 1) (33). Therefore, host miRNAs regulate differentiation dependent susceptibility of host cells towards HIV-1 infection.
HIV-1 Tat is required for the initiation of HIV genome transcription, and is one of the first HIV proteins that are expressed after infection starts. Tat regulatory protein requires two domains for its activity, i.e., the activation domain (AD) and the arginine rich motif (ARM). The AD is regulated by the acetylation of p300/CBP-associated factor (PCAF) at Lys-28 (42), which is an important co-factor of Tat in HIV-1 replication and is targeted by the polycistronic miRNA cluster-miR-17/92. When the miR-17/92 cluster is knocked down, HIV-1 production is increased in the Jurkat cell line (Table 2). More specifically, it is miR-17-5p and miR-20a of the miR-17/92 cluster that down-regulates PCAF protein and HIV-1 production (Figure 1). Transfection of PBMCs with miR-11-5p and miR-20a significantly reduced both PCAF expression and HIV-1 production, and transfection of anti-miR-17-5p and anti-miR-20a increased both PCAF expression and HIV-1 production (32, 42).
The positive transcription elongation factor B (P-TEFb) is known to play a role in viral transcription by interacting with HIV-1’s Tat protein and TAR element, and is composed of two subunits, viz., cyclin dependent kinase 9 (CDK9) and cyclin T1 (cycT1). P-TEFb expression is decreased by the degradation of cycT1 or CDK9, and as a result, HIV-1 replication is inhibited in HeLa cells (43). miR-27b has been identified to target cycT1 in resting CD4+ T-cells (Table 2). The activated CD4+ T-cells result in down-regulation of miR-27b and a subsequent up-regulation of cycT1 (Figure 1). Over expression of this miRNA results in reduced HIV-1 reporter virus viral gene expression levels and replication of the NL4.3 strain by down-regulation of cycT1 (31). Subsequently, in another study, miR-198 was identified to target cycT1 and suppress its protein levels (Table 2). Additionally, over expression of miR-198 decreases the replication of HIV-1 in monocytes by repressing cycT1 expression (Figure 1) (30).
miR-34a and miR-217 target sirtuin 1 (SIRT1) to enhance HIV-1 infection (34, 35). The Tat protein is activated when it is acetylated; however, SIRT1 mediates the regulation of Tat transactivation and HIV-1 transcription by deacetylating Tat. Tat expression leads to up-regulation of miR-34a and miR-217, where they target the 3′ UTR of SIRT1 mRNA to inhibit its expression (Table 2) (34, 35). Thus, inhibition of SIRT1 due to miR-34a and miR-217 will ease the restriction in terms of deacetylation on Tat, and in turn will enhance HIV-1 infection (Figure 1).
miRNA microarray studies suggest that miR-34a is up-regulated in response to HIV-1 infection in T-cells. Furthermore, miR-34a modulates HIV-1 replication in T-cells by regulating a host protein, i.e., phosphatase 1 nuclear targeting subunit (PNUTS) (Table 2). miR-34a therefore can be seen to target PNUTS to promote viral replication in cells (Figure 1) (37).
The innate host immune response delays the spread of HIV. The innate inhibitory mechanism includes several host proteins which play roles in HIV-1 infection inhibition (40). miRNA expression profiling has been performed in HIV-1-infected T-cell lines, which show that let‐7c, miR‐34a, and miR‐124a are up-regulated. The let-7c targets and down-regulates p21 (CDKN1), while miR-34a and miR-124a target and down-regulate TASK1 (KCNK3), which is among several proteins that restrict viral spread (Table 2). HIV-1 infection up-regulates the expression of these miRNAs, which decreases the expression levels of p21 and TASK1, and as a result, virion release and copy numbers are increased. By inhibiting these miRNAs, the levels of p21 and TASK1 are increased, resulting in decreased viral replication and spread. HIV-1 becomes more infectious by inducing miRNAs that target and down-regulate host innate immune system proteins (40).
Specific T-cell receptors (TCR) on CD4+ T-cells prime different gene expression profiles responsible for either cell activation, proliferation, and differentiation into effectors and regulatory cells, which are under the control of miRNAs (44, 45). Next generation sequencing has shown miR-34c-5p to be up-regulated in naïve CD4+ T-cells in response to TCR stimulation, which subsequently targets several genes involved in TCR signaling and cell activation (Figure 1) (Table 2). Moreover, both HIV-1 and HIV-2 have the capacity to reduce the induction of this miRNA, which was also found to be responsible for activation of cells and HIV-1 replication. These findings suggest that the down-regulation of miR-34c-5p during HIV infection delineates its anti-viral host response (20).
Extracellular vesicles (EVs) contain extra-chromosomal DNA, RNA, and various proteins (46). miR-139-5p was found to be significantly up-regulated in plasma (p) EVs from HIV-1 patients on ART, HIV-1 patients who were ART-naïve, and even in healthy controls (HCs) (22). FOXO1 inhibition promotes HIV-1 infection in resting T-cells (47). FOXO1 was observed to be down-regulated in plasma extracellular vesicles (pEVs) in treated J-Lat 10.6 cells obtained from HIV ART-naïve patients and from HIV patients on ART, but not in HCs. This indicates that pEV-associated activation of virus, especially by ART-naïve pEVs and HIV patients on ART in J-Lat 10.6 cells, occurs with the help of the miR-139-5p/FOXO1 axis (Figure 1) (Table 2) (22).
Chemokine (C-C motif) ligand 5 (CCL5) plays an important role in HIV-1 pathogenesis (21). HIV-1-infected macrophages express higher levels of miR-146a compared to uninfected cells. CCL5 is the direct target of miR-146a and is down-regulated when mir-146a is over expressed, and vice versa. Furthermore, CCL5-induced monocyte migration is reduced by miR-146a, and thus this miRNA contributes to HIV-1 pathogenesis (Table 2) (21).
Generally, astrocytes are able to restrict HIV infection, and this is regulated by sterile alpha motif and histidine/aspartic acid domain-containing protein 1 (SAMHD1). miR-155/miR-181a directly targets SAMHD1 to decrease SAMHD1 expression levels and hence reactivates viral replication in astrocytes, while inhibiting these miRNAs results in increased SAMHD1 levels, and hence restricts viral replication (Table 2) (38).
HIV-1 replicates in monocyte-derived DCs and macrophages; however, this replication is restricted in monocytes (39, 48). HIV-1 viral protein binding protein (VprBP) expression is necessary to promote HIV-1 infection. DCs express elevated levels of VprBP to facilitate viral replication. VprBP levels are undetectable in monocytes; however, VprBP mRNA levels in monocytes are even higher than levels in DCs. This suggests post-transcriptional inhibition of VprBP in monocytes. Furthermore, the expression of miR-1236 is higher in monocytes compared to DCs, and this miRNA directly targets the 3’UTR of VprBP to inhibit its expression, and restrict HIV-1 infection in monocytes (Table 2). Interestingly, inhibiting miR-1236 increases the translation of VprBP and HIV-1 infection in monocytes (Figure 1). Similarly, the overexpression of miR-1236 decreases the translation of VprBP and HIV-1 infection in DCs. Thus, VprBP, a host factor targeted by miR-1236, regulates monocyte/DC susceptibility to HIV-1 infection (39).
Various stimuli can alter miRNA expression within cells to alter HIV-1 infectivity. Drugs of abuse are co-related to the HIV-1 pandemic, and affect various aspects of HIV disease. Morphine has been found to enhance HIV replication by downregulating anti-HIV miRNAs, i.e., miR-28, mir-125b, miR-150, and miR-382 in monocytes, compared to untreated cells. Heroin addicts have lower levels of expression of these miRNAs in PBMCs, compared to healthy controls. Further, monocytes treated with morphine has been shown to impair type I interferon (IFN)-induced anti-HIV miRNA expression. These findings suggest that the use of opioids compromises intracellular innate anti-HIV mechanisms in monocytes, and therefore enhances cellular susceptibility to HIV infection. (49). Similarly, cocaine serves as a cofactor to enable HIV-1 infection to progress to acquired immunodeficiency syndrome (AIDS). Cocaine contributes to HIV-1 replication in primary CD4+ T-cells isolated from human PBMCs. miR-125b has been shown to be significantly down-regulated in cocaine-treated CD4+ T-cells. The down-regulation of this miRNA enhances HIV-1 replication, while overexpression of miR-125b results in decreased viral replication (50).
Other stimuli of anti-HIV-1 miRNAs include Toll-like receptor (TLR) ligands and cytokines. TLR3, type I interferon (IFN-I), IFN-α, and IFN-β induction have been found to be responsible for the up-regulation of miR-28, mir-125b, miR-150, and miR-382 in primary macrophages to exert their anti-HIV-1 effects (51, 52).
Apart from the direct or indirect roles of miRNAs in HIV-1 pathogenesis, some studies describe different miRNAs being involved in the pathogenesis of different diseases, for example, in cancers, hepatitis C virus (HCV) infection, dengue virus (DENV) infection, influenza, and vesicular stomatitis virus (VSV) infection (53–56). Moreover, they target genes which have known functions in HIV-1 infection. Therefore, these miRNAs could reliably be predicted to play active roles in HIV-1 pathogenesis as well.
HIV-1 entry into the host cell is the first step in causing successful infection. A host factor, chemokine receptor 5 (CCR5), plays an indispensable role in HIV internalization (57). Interestingly, a deletion of 32 base pairs (bp) in the CCR5 gene has been observed in some individuals, and these individuals produce lower levels of CCR5 on the surface of their CD4+ T-cells, which positively correlates with resistance to HIV internalization. Therefore, CCR5 expression is pivotal in regulating HIV replication. In another study, miR-107 was found to directly target the CCR5 gene by binding to its 3′ UTR (Table 3) (65). It is, therefore, reasonable to assume that miR-107 expression is responsible for the regulation of HIV internalization via modulation of CCR5 levels.
HIV-1 hijacks host miRNAs to enhance infection. miR-132 is the first miRNA known to enhance HIV-1 replication (66). This miRNA is highly up-regulated in activated CD4+ T-cells. Upon over expression of miR-132, methyl-CpG binding protein 2 (MeCP2), the known target of miR-132, is down-regulated in Jurkat cells (Table 3) (66). The role of MeCP2 in the regulation of HIV-1 replication has been demonstrated previously, where its inhibition enhances HIV-1 replication (67). Therefore, the possible link is that miR-132 up-regulation suppresses MeCP2 levels, and thereby increases HIV-1 replication.
A component of the IFN signaling pathway, tumor necrosis factor receptor-associated factor 6 (TRAF6) expression is decreased in primary human macrophages after HIV-1 infection. Inhibition of expression of TRAF6 in macrophages increases HIV-1 replication, indicating that TRAF6 regulates HIV-1 production (68). In another two independent studies, miR-144 was found to decrease the replication of influenza virus, encephalomyocarditis virus (EMCV), and VSV, while miR-146a was found to decrease the replication of DENV (Table 3) (53, 55). Both these miRNAs directly target TRAF6, and therefore alter the antiviral landscape in cells and contribute to viral replication (Table 3). The assumptive deduction from the findings of these studies is that miRNA-144 and miR-146a may target TRAF6 and induce the replication of HIV-1 as well.
The members of the family of E3 ubiquitin ligases, the tripartite motif (TRIM) proteins i.e. TRIM11, TRIM22, and TRIM28, have all been shown to have anti-HIV activity (61–63, 79). miR-24-3p suppresses the expression of TRIM11 in colon cancer, and therefore inhibits apoptosis to promote cellular proliferation (Table 3) (56). In HIV infection, TRIM22, through ubiquitylation, inhibits the processing of viral particles and budding. TRIM22 also has anti-HCV activity, which is regulated by miR-215 in con1b and Huh7.5.1 cells (Table 3) (54). Similarly, miR-491 targets TRIM28 in the pathogenesis of glioblastoma multiforme (GBM) (Table 3) (64). Therefore, determining the fundamental roles of miR-24-3p, miR-215, and miR- 491 with respect to HIV-related disease is likely to be a rewarding area of investigation for future study.
Intracellular adhesion molecule 1 (ICAM1), a host factor, augments viral infectivity by directly inserting into mature HIV virions (58, 59). It has also been shown that miR-296-3p inhibits the expression of ICAM1 by directly binding to it in the malignant M12 cell line and prostate cancer cells (Table 3) (60). It would, therefore, be an interesting exercise to experimentally attempt to determine whether miR-296-3p reduces HIV-1 infectivity.
The predictions made from a few other examples include the over expression of interferon regulatory factor 7 (IRF7) resulting in increased HIV-1 replication in macrophages while inhibiting IRF7, which results in reduced viral replication (68). In another study, miR-541 was shown to promote abnormal proliferation and invasion of vascular smooth muscle cells (VSMCs). miR-541 targets IRF7, and down-regulation of IRF7 increases VSMC proliferation (Table 3) (69). In one study, Viperin (RSAD2) was observed to inhibit HIV-1 production (Table 3) (71), while in another study it was found that miR-200a, miR-200b, and miR-429 promote podocyte differentiation by directly targeting RSAD2, and represses its expression (Table 3) (70). Interferon stimulated gene 15 (ISG15) inhibits HIV-1 replication (Table 3) (72), while miR-138 inhibits the expression of ISG15 in oral squamous cell carcinoma (OSCC) cells (Table 3) (73). TLR3 reduces HIV-1 infection and miR-26a ameliorates Pristane-induced arthritis in rats by targeting TLR3 (Table 3) (15, 74). SET domain, bifurcated 1 (SETDB1) restricts HIV-1 replication, while another study showed that miR-381-3p suppresses SETDB1 to inhibit breast cancer progression (Table 3) (75, 76). Rab27a was shown to promote HIV-1 assembly by regulating plasma membrane levels of phosphatidylinositol 4,5-bisphosphate, while another study showed miR-134-3p to inhibit proliferation and cell cycle progression in human OCSCs by inhibiting the expression of Rab27a (Table 3) (77, 78). Therefore, the miRNAs identified in these studies targeting mRNAs that have effects on HIV infection, seem quite likely to play roles in HIV pathogenesis as well, and warrant investigative exploration in the future.
Approximately 10-40% of HIV-infected patients fail to achieve meaningful or sustained improvement in immune function after ART. Therefore, investigation of potential roles that miRNAs may play in immune recovery during or after antiretroviral therapy (ART) in HIV-1 patients and immunological non-responders (INRs) has already been initiated (17, 19, 80). miRNA sponges are RNA transcripts which include long non coding RNAs (lncRNAs) and circular RNAs (circRNAs), that associate with and sequester miRNAs, thus not allowing them to interact with their target mRNAs (81). circRNAs contain several miRNA binding sites, and inhibit miRNA function by acting as miRNA sponges. In an attempt to determine the existence of biological crosstalk in competing endogenous RNA (ceRNA) networks, RNA sequencing (RNA-seq) and miRNA sequencing (miR-seq) have been performed on PBMCs from HAART-naïve patients, early HIV-infected (EHI) patients, and 3 healthy controls (HCs) (80). miR-seq analysis showed that 30 miRNAs are differentially expressed (DE) between the two groups, and among them 12 miRNAs were up-regulated and 18 miRNAs were down-regulated. Moreover, in the same study, ceRNA networks were constructed which showed that 21 DE miRNAs share miRNA response elements (MREs) with 516 DE circRNAs and 903 DE mRNAs. Furthermore, the RNA-seq data indicates that the mRNAs, CCNK, CDKN1A, and IL-15 are DE, and interestingly, these three mRNAs have already been found to play roles in HIV-1 replication. Among the DE miRNAs, miR-27b-3p, miR-542-3p, miR-101-3p, let-7c-5p, and miR-548ah-3p have been predicted to have MRE binding sites on the preceding mRNAs. Similarly, 67 DE circRNAs were predicted to have shared MREs with CCNK, CDKN1A, and IL-15 mRNAs. Among these 67 circRNAs, 18 were predicted to share MREs of miR-27b-3p with CCNK, 40 were predicted to share MREs of let-7c-5p, miR-101-3p, and miR-542-3p with CDKN1A, and 9 were predicted to share MREs of miR-548ah-3p targeting IL-15. Therefore, HIV replication seems to be regulated by miRNA sponging activity in PBMCs in HAART-naïve patients.
In another study, the association between miRNAs and immunological nonresponse was assessed by investigating the miRNA profiles in immunologically reconstituted HIV-1 patients and immunological HIV-1 non-responders (17). miRNAs were analyzed using next-generation sequencing, and differences in expression profiles at baseline and after 24 months of maintaining virological suppression (VS) were measured between immunological responders (IR) and INRs. It was observed that, compared to IRs, a substantial proportion of miRNAs were downregulated in INRs. The miRNA, let-7d-5p, was downregulated in 9 INRs (total INRs=11), but only in 2 IRs (total IRs=13). There is therefore the potential for the miRNA let-7d-5p to be used as a potential biomarker for INRs (17).
One study attempted to investigate miRNAs as predictive markers that may be useful in identifying patients with poor immune response earlier (19). Plasma samples were taken before ART initiation for miRNA isolation and sequencing. It was observed that miR-580, miR-627, miR-138-5p, miR-16-5p, and miR-323-3p are upregulated in INRs, in contrast to observations in IRs. The expression levels of miRNAs negatively correlate with both CD4+ T-cell counts and increased proportions of CD4+ T cells after one year of ART, and could potentially be used to differentiate IRs from INRs. Therefore, miRNAs could possibly be used to accurately predict immune response after ART (19).
Several viruses encode viral miRNAs (vmiRNAs) which play roles in viral replication, host immune evasion, and survival within a host (82, 83). However, whether RNA viruses actually encode miRNAs at all is debatable (84, 85). One of the reasons for this controversy is because RNA-induced silencing complex- (RISC)-mediated degradation of the non-coding region responsible for a functional miRNA will result in the degradation of the entire viral RNA, and therefore there is an absence of a non-coding region (86). Another reason could be that RNA viruses replicate within the cytoplasm of cells, and are therefore not processed by the Drosha enzyme and transactivation response element RNA binding protein (TRBP). Nevertheless, HIV-1, being a retrovirus, can be reverse transcribed into double stranded DNA and imported into the nucleus where it integrates into the host genome. Therefore, HIV-1 has access to the RNA interference (RNAi) machinery of both the nucleus and the cytoplasm, similar to host miRNAs.
Bioinformatics tools have suggested that HIV-1 encodes five pre-miRNA candidates (87). Moreover, Nef, the viral protein, is also supposed to encode a vmiRNA (Nef-U3-miR-N367) that degrades Nef and decreases viral replication (88, 89). Another vmiRNA, hiv1-miR-H1, encoded in the 3′-end of the HIV-1 genome, has been reported to target cellular miR-149 that can target Vpr of HIV-1 (90). Similarly, the protein coding regions of Gag-Pol and Env proteins of HIV-1 can encode several miRNA-like sequences (91). In HIV-infected T-lymphocytes, viral RNA accumulation has been analyzed via a sensitive SOLID™ 3Plus System, and several small non-coding RNAs (sncRNAs) derived from the genome of HIV-1 have been detected. These sncRNAs were either vmiRNAs or viral short interference RNAs (vsiRNAs) (92). In another study, a sequence-targeted enrichment strategy was used to identify sncRNAs derived from HIV-1 infected primary CD4+ T-lymphocytes and macrophages and hundreds of sncRNAs from 16 to 89 nucleotides in length were detected (93). Moreover, the transactivation RNA (TAR) element, which recruits Tat in infected cells, has been determined to interact with Dicer, and is processed by Dicer, and generates vmiRNAs that regulate chromatin remodeling of viral LTR (87, 94). Two other vmiRNAs, vmiR88 and vmiR99, are encoded by HIV-1 LTR through a non-canonical miRNA pathway, and are able to trigger tumor necrosis factor alpha (TNF-α) release by macrophages (95).
Evidence endorsing the fact that HIV-1 does not encode short interference RNA (siRNAs) or miRNAs in infected T-cells, primary peripheral blood mononuclear cells (PBMCs), and macrophages is provided in other studies (96, 97). Advanced and improved detection methods are therefore indispensable to identify HIV-1 encoded miRNAs.
Different phases of the HIV viral life cycle have been targeted in developing the multiple antiretroviral drugs utilized in modern ART. ART has been successful in controlling viral replication in the majority of HIV-infected patients (98). Extensive progress has been made with ART; however, infection by HIV-1 remains a global menace, and has the real and unavoidable risk of being incurable in most patients. Besides, all ART drugs have adverse effects and the potential for drug toxicity, and the emergence of HIV drug resistance poses a substantial threat to the overall success of current ART regimens (99–101). These concerns have compelled investigators to attempt to identify novel therapeutics in the quest to cure HIV-1.
RNA-based therapeutics have garnered much attention in recent times because of its reported success in treating several types of diseases. Potential miRNA drugs, with reported success in treating many types of infectious and non-infectious diseases, have been reported in the literature (102–106). Recent studies have shown that dysregulation in miRNA profiles affect HIV replication (17, 62, 71, 72, 107, 108). Despite being aware of the diverse roles of the many miRNAs intricately woven into the fabric of the HIV life cycle, the practical application of miRNAs as interventional therapeutics and diagnostics remains a largely unexplored and untapped area of HIV/AIDS research. The use of miRNAs as a treatment strategy is an emerging field of research endeavor, and very few miRNA-associated drugs have thus far been approved for clinical applications by the United States Food and Drug Administration (FDA) (109). For future study, it would be of considerable interest to explore the therapeutic potential of miR-34a, as this particular miRNA targets three host proteins (PNUTS, SIRT1, and TASK1) to promote HIV-1 infection (Figure 1).
To sum up, numerous miRNAs target HIV-1 either directly or indirectly and play significant roles in the pathogenesis of HIV infection and progression. These miRNAs are involved in different processes of HIV-1 infection and effect different impacts on the pathogenesis of HIV-1 infection. For example, miR-146a, miR-221 and miR-222 down regulate host factors to prevent HIV-1 entry into host cells; miR-155 down regulates host proteins to inhibit HIV-1 genome from integrating into host genome; miR-34a, miR-139-5p, miR-155, mir-181a and miR-217 suppress different host proteins to increase HIV-1 replication; and miR-17/92 cluster, miR-27b and miR-198 down regulate host proteins to decrease HIV-1 replication; miR-34a targets three host proteins to promote HIV-1 pathogenesis, whereas miR-24-3p, miR-107, miR-132, miR-144, miR-146a, miR-215, miR-396-3p and miR-491 may play predictive roles in HIV-1 pathogenesis by targeting genes that are favorable to HIV-1 infection. Furthermore, some miRNAs, such as let-7d-5p, are associated with immune reconstitution in HIV-infected patients and could be used as a potential biomarker for INRs. However, much remains unclear and warrants further study regarding the involvement of miRNAs in HIV-1 pathogenesis. Firstly, although it is already known that cell-type-specific expression of miRNAs and HDFs contribute to HIV-1 susceptibility, it is not known whether cell differentiation and cell activation status are involved in the process. Secondly, even though it is known that numerous miRNAs play roles in HIV-1 life cycle and pathogenesis, the potency and expression levels of miRNAs remain largely unknown. Thirdly, it is known that some miRNAs contribute to the establishment of HIV-1 latency, however, the degree of participation of other factors such as transcriptional inefficiencies have not been determined. Fourthly, researchers found that miR-27b repressed HIV-1 replication by down regulating cyclin T1, however, the role of miR-27a, which differed from miR-27b only by one nucleotide at the 3′ end, was undermined (31). And fifthly, in the studies describing the association of miRNAs and immune reconstitution in HIV-infected patients (17, 80), the sample sizes were small and specific cells types were not used for analysis, indicating that the study conclusions warrant further investigation.
In the future, the potential application of miRNAs may be of great interest to investigators, and miRNA mimics and inhibitors which specifically target their respective mRNAs could be used to inhibit HIV-1 replication, infectivity and pathogenicity, or to boost immune reconstitution of people living with HIV. In addition, differentially expressed miRNAs during HIV-1 infection could also be used to guide researchers to explore host proteins that may be manipulated for therapeutic benefits. However, some issues need to be fully addressed in order to achieve these goals. Firstly, one miRNA may target several host genes. For example, miR-34a can target three different host genes, i.e. SIRT1, PNUTS and TASK1, in its regulation of HIV-1 infection (35, 37, 40), and therefore, it is reasonable to speculate that miR-34a mimics may result in off-target effects when used in vivo, which may pose a challenging obstacle for its potential future clinical application. Secondly, a potential Achilles’ heel of a potential miRNA-based HIV therapy is related to the lack of a targeted delivery system. miRNAs, by nature, are inherently structurally unstable, and therefore, it is unimaginable for them to be able to strike their targets with a high degree of accuracy without an efficient, targeted delivery system that can substantially enhance their half-life and can accurately guide them to their targets. And finally, another compelling research area in the future that warrants future investigation is that miRNAs may be used as biomarkers or predictive markers of immune response after ART. We believe that as research deepens in the future, the therapeutic application of miRNAs will eventually come true.
FR and YC conceptualize the study. FR wrote the manuscript. SZ revised the manuscript and figure. FS and YC revised the manuscript and approved the final draft. All authors contributed to the article and approved the submitted version.
This work was supported by the Chongqing Talent Cultivation Program (cstc2021ycjh-bgzxm0275) and the Chongqing Key Public Health Disciplines Improvement Project.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Rashid F, Awan HM, Shah A, Chen L, Shan G. Induction of miR-3648 upon ER stress and its regulatory role in cell proliferation. Int J Mol Sci (2017) 18. doi: 10.3390/ijms18071375
2. Menegazzi M, Gotte G. Role of the ribonuclease ONCONASE in miRNA biogenesis and tRNA processing: Focus on cancer and viral infections. Int J Mol Sci (2022) 23:6556. doi: 10.3390/ijms23126556
3. Michlewski G, Cáceres JF. Post-transcriptional control of miRNA biogenesis. RNA (2019) 25:1–16. doi: 10.1261/rna.068692.118
4. Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res (2004) 14:1902–10. doi: 10.1101/gr.2722704
5. Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III drosha initiates microRNA processing. Nature (2003) 425:415–9. doi: 10.1038/nature01957
7. Khodakarimi S, Zarebkohan A, Kahroba H, Omrani M, Sepasi T, Mohaddes G, et al. The role of miRNAs in the regulation of autophagy in autoimmune diseases. Life Sci (2021) 287:119726. doi: 10.1016/j.lfs.2021.119726
8. Kinoshita C, Kubota N, Aoyama K. Interplay of RNA-binding proteins and microRNAs in neurodegenerative diseases. Int J Mol Sci (2021) 22:5292. doi: 10.3390/ijms22105292
9. Wang X, He Y, Mackowiak B, Gao B. MicroRNAs as regulators, biomarkers and therapeutic targets in liver diseases. Gut (2021) 70:784–95. doi: 10.1136/gutjnl-2020-322526
10. Bruscella P, Bottini S, Baudesson C, Pawlotsky J-M, Feray C, Trabucchi M. Viruses and miRNAs: More friends than foes. Front Microbiol (2017) 8:824. doi: 10.3389/fmicb.2017.00824
11. McLaren PJ, Fellay J. HIV-1 and human genetic variation. Nat Rev Genet (2021) 22:645–57. doi: 10.1038/s41576-021-00378-0
12. Roberts JD, Bebenek K, Kunkel TA. The accuracy of reverse transcriptase from HIV-1. science (80-. ). Science (1988) 242:1171–3. doi: 10.1126/science.2460925
13. Deeks SG, Overbaugh J, Phillips A, Buchbinder S. HIV Infection. Nat Rev Dis Prim (2015) 1:15035. doi: 10.1038/nrdp.2015.35
14. Lodge R, Ferreira Barbosa JA, Lombard-Vadnais F, Gilmore JC, Deshiere A, Gosselin A, et al. Host MicroRNAs-221 and -222 inhibit HIV-1 entry in macrophages by targeting the CD4 viral receptor. Cell Rep (2017) 21:141–53. doi: 10.1016/j.celrep.2017.09.030
15. Swaminathan G, Rossi F, Sierra L-J, Gupta A, Navas-Martín S, Martín-García J. A role for microRNA-155 modulation in the anti-HIV-1 effects of toll-like receptor 3 stimulation in macrophages. PloS Pathog (2012) 8:e1002937. doi: 10.1371/journal.ppat.1002937
16. Wang P, Qu X, Zhou X, Shen Y, Ji H, Fu Z, et al. Two cellular microRNAs, miR-196b and miR-1290, contribute to HIV-1 latency. Virology (2015) 486:228–38. doi: 10.1016/j.virol.2015.09.016
17. Fu Y, Liu Y, Liu Z, Liu L, Yuan L, An X, et al. The impact of microRNA regulation on immune recovery in HIV-1-Infected patients treated during acute infection: A pilot study. BioMed Res Int (2020) 2020:1–12. doi: 10.1155/2020/5782927
18. Leo CG, Mincarone P, Tumolo MR, Panico A, Guido M, Zizza A, et al. MiRNA expression profiling in HIV pathogenesis, disease progression and response to treatment: a systematic review. Epigenomics (2021) 13:1653–71. doi: 10.2217/epi-2021-0237
19. Lv J-N, Li J-Q, Cui Y-B, Ren Y-Y, Fu Y-J, Jiang Y-J, et al. Plasma MicroRNA signature panel predicts the immune response after antiretroviral therapy in HIV-infected patients. Front Immunol (2021) 12:753044. doi: 10.3389/fimmu.2021.753044
20. Amaral AJ, Andrade J, Foxall RB, Matoso P, Matos AM, Soares RS, et al. miRNA profiling of human naive CD4 T cells links miR-34c-5p to cell activation and HIV replication. EMBO J (2017) 36:346–60. doi: 10.15252/embj.201694335
21. Huang Q, Chen L, Luo M, Lv H, Luo D, Li T, et al. HIV-1-Induced miR-146a attenuates monocyte migration by targeting CCL5 in human primary macrophages. AIDS Res Hum Retroviruses (2018) 34:580–9. doi: 10.1089/AID.2017.0217
22. Okoye I, Xu L, Oyegbami O, Shahbaz S, Pink D, Gao P, et al. Plasma extracellular vesicles enhance HIV-1 infection of activated CD4+ T cells and promote the activation of latently infected J-Lat10.6 cells via miR-139-5p transfer. Front Immunol (2021) 12:697604. doi: 10.3389/fimmu.2021.697604
23. Huang J, Wang F, Argyris E, Chen K, Liang Z, Tian H, et al. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat Med (2007) 13:1241–7. doi: 10.1038/nm1639
24. Ahluwalia JK, Khan SZ, Soni K, Rawat P, Gupta A, Hariharan M, et al. Human cellular microRNA hsa-miR-29a interferes with viral nef protein expression and HIV-1 replication. Retrovirology (2008) 5:117. doi: 10.1186/1742-4690-5-117
25. Nathans R, Chu C-Y, Serquina AK, Lu C-C, Cao H, Rana TM. Cellular microRNA and p bodies modulate host-HIV-1 interactions. Mol Cell (2009) 34:696–709. doi: 10.1016/j.molcel.2009.06.003
26. Wang X, Ye L, Hou W, Zhou Y, Wang Y-J, Metzger DS, et al. Cellular microRNA expression correlates with susceptibility of monocytes/macrophages to HIV-1 infection. Blood (2009) 113:671–4. doi: 10.1182/blood-2008-09-175000
27. Sisk JM, Clements JE, Witwer KW. miRNA profiles of monocyte-lineage cells are consistent with complicated roles in HIV-1 restriction. Viruses (2012) 4:1844–64. doi: 10.3390/v4101844
28. Gorry PR, McPhee DA, Verity E, Dyer WB, Wesselingh SL, Learmont J, et al. Pathogenicity and immunogenicity of attenuated, nef-deleted HIV-1 strains in vivo. Retrovirology (2007) 4:66. doi: 10.1186/1742-4690-4-66
29. Ross TM, Oran AE, Cullen BR. Inhibition of HIV-1 progeny virion release by cell-surface CD4 is relieved by expression of the viral nef protein. Curr Biol (1999) 9:613–21. doi: 10.1016/s0960-9822(99)80283-8
30. Sung T-L, Rice AP. miR-198 inhibits HIV-1 gene expression and replication in monocytes and its mechanism of action appears to involve repression of cyclin T1. PloS Pathog (2009) 5:e1000263. doi: 10.1371/journal.ppat.1000263
31. Chiang K, Sung T-L, Rice AP. Regulation of cyclin T1 and HIV-1 replication by microRNAs in resting CD4+ T lymphocytes. J Virol (2012) 86:3244–52. doi: 10.1128/JVI.05065-11
32. Triboulet R, Mari B, Lin Y-L, Chable-Bessia C, Bennasser Y, Lebrigand K, et al. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science (2007) 315:1579–82. doi: 10.1126/science.1136319
33. Shen C, Jia Y, Tian R, Ding M, Zhang C, Wang J. Translation of pur-α is targeted by cellular miRNAs to modulate the differentiation-dependent susceptibility of monocytes to HIV-1 infection. FASEB J (2012) 26:4755–64. doi: 10.1096/fj.12-209023
34. Zhang H-S, Wu T-C, Sang W-W, Ruan Z. MiR-217 is involved in tat-induced HIV-1 long terminal repeat (LTR) transactivation by down-regulation of SIRT1. biochim. biophys. Acta (2012) 1823:1017–23. doi: 10.1016/j.bbamcr.2012.02.014
35. Zhang H-S, Chen X-Y, Wu T-C, Sang W-W, Ruan Z. MiR-34a is involved in tat-induced HIV-1 long terminal repeat (LTR) transactivation through the SIRT1/NFκB pathway. FEBS Lett (2012) 586:4203–7. doi: 10.1016/j.febslet.2012.10.023
36. Quaranta MT, Olivetta E, Sanchez M, Spinello I, Paolillo R, Arenaccio C, et al. miR-146a controls CXCR4 expression in a pathway that involves PLZF and can be used to inhibit HIV-1 infection of CD4(+) T lymphocytes. Virology (2015) 478:27–38. doi: 10.1016/j.virol.2015.01.016
37. Kapoor R, Arora S, Ponia SS, Kumar B, Maddika S, Banerjea AC. The miRNA miR - 34a enhances HIV-1 replication by targeting PNUTS/PPP1R10, which negatively regulates HIV-1 transcriptional complex formation. Biochem J (2015) 470:293–302. doi: 10.1042/BJ20150700
38. Pilakka-Kanthikeel S, Raymond A, Atluri VSR, Sagar V, Saxena SK, Diaz P, et al. Sterile alpha motif and histidine/aspartic acid domain-containing protein 1 (SAMHD1)-facilitated HIV restriction in astrocytes is regulated by miRNA-181a. J Neuroinflamm (2015) 12:66. doi: 10.1186/s12974-015-0285-9
39. Ma L, Shen C-J, Cohen É.A., Xiong S-D, Wang J-H. miRNA-1236 inhibits HIV-1 infection of monocytes by repressing translation of cellular factor VprBP. PloS One (2014) 9:e99535. doi: 10.1371/journal.pone.0099535
40. Farberov L, Herzig E, Modai S, Isakov O, Hizi A, Shomron N. MicroRNA regulation of p21 and TASK1 cellular restriction-factors enhances HIV-1 infection. J Cell Sci (2015) 128(8):1607–16. doi: 10.1242/jcs.167817
41. Pagani I, Demela P, Ghezzi S, Vicenzi E, Pizzato M, Poli G. Host restriction factors modulating HIV latency and replication in macrophages. Int J Mol Sci (2022) 23:3021. doi: 10.3390/ijms23063021
42. D’Orso I, Frankel AD. Tat acetylation modulates assembly of a viral-host RNA-protein transcription complex. Proc Natl Acad Sci U S A (2009) 106:3101–6. doi: 10.1073/pnas.0900012106
43. Chiu Y-L, Cao H, Jacque J-M, Stevenson M, Rana TM. Inhibition of human immunodeficiency virus type 1 replication by RNA interference directed against human transcription elongation factor p-TEFb (CDK9/CyclinT1). J Virol (2004) 78:2517–29. doi: 10.1128/jvi.78.5.2517-2529.2004
44. Pagani M, Rossetti G, Panzeri I, de Candia P, Bonnal RJP, Rossi RL, et al. Role of microRNAs and long-non-coding RNAs in CD4(+) T-cell differentiation. Immunol Rev (2013) 253:82–96. doi: 10.1111/imr.12055
45. Yamane H, Paul WE. Early signaling events that underlie fate decisions of naive CD4(+) T cells toward distinct T-helper cell subsets. Immunol Rev (2013) 252:12–23. doi: 10.1111/imr.12032
46. Robbins PD, Morelli AE. Regulation of immune responses by extracellular vesicles. Nat Rev Immunol (2014) 14:195–208. doi: 10.1038/nri3622
47. Roux A, Leroy H, De Muylder B, Bracq L, Oussous S, Dusanter-Fourt I, et al. FOXO1 transcription factor plays a key role in T cell-HIV-1 interaction. PloS Pathog (2019) 15:e1007669. doi: 10.1371/journal.ppat.1007669
48. Ouyang J, Xiong Y, Shang H, Liang G. LAPTM5 restricts HIV-1 infection in dendritic cells and is counteracted by vpr. Microbiol Spectr (2022) 10. doi: 10.1128/spectrum.01382-21
49. Wang X, Ye L, Zhou Y, Liu M-Q, Zhou D-J, Ho W-Z. Inhibition of anti-HIV microRNA expression: a mechanism for opioid-mediated enhancement of HIV infection of monocytes. Am J Pathol (2011) 178:41–7. doi: 10.1016/j.ajpath.2010.11.042
50. Mantri CK, Pandhare Dash J, Mantri JV, Dash CCV. Cocaine enhances HIV-1 replication in CD4+ T cells by down-regulating MiR-125b. PloS One (2012) 7:e51387. doi: 10.1371/journal.pone.0051387
51. Cobos Jiménez V, Booiman T, de Taeye SW, van Dort KA, Rits MAN, Hamann J, et al. Differential expression of HIV-1 interfering factors in monocyte-derived macrophages stimulated with polarizing cytokines or interferons. Sci Rep (2012) 2:763. doi: 10.1038/srep00763
52. Zhou Y, Wang X, Liu M, Hu Q, Song L, Ye L, et al. A critical function of toll-like receptor-3 in the induction of anti-human immunodeficiency virus activities in macrophages. Immunology (2010) 131:40–9. doi: 10.1111/j.1365-2567.2010.03270.x
53. Rosenberger CM, Podyminogin RL, Diercks AH, Treuting PM, Peschon JJ, Rodriguez D, et al. miR-144 attenuates the host response to influenza virus by targeting the TRAF6-IRF7 signaling axis. PloS Pathog (2017) 13:e1006305. doi: 10.1371/journal.ppat.1006305
54. Tian H, He Z. miR-215 enhances HCV replication by targeting TRIM22 and inactivating NF-κB signaling. Yonsei Med J (2018) 59:511. doi: 10.3349/ymj.2018.59.4.511
55. Wu S, He L, Li Y, Wang T, Feng L, Jiang L, et al. miR-146a facilitates replication of dengue virus by dampening interferon induction by targeting TRAF6. J Infect (2013) 67:329–41. doi: 10.1016/j.jinf.2013.05.003
56. Yin Y, Zhong J, Li S-W, Li J-Z, Zhou M, Chen Y, et al. TRIM11, a direct target of miR-24-3p, promotes cell proliferation and inhibits apoptosis in colon cancer. Oncotarget (2016) 7:86755–65. doi: 10.18632/oncotarget.13550
57. Lederman MM, Penn-Nicholson A, Cho M, Mosier D. Biology of CCR5 and its role in HIV infection and treatment. JAMA (2006) 296:815. doi: 10.1001/jama.296.7.815
58. Bounou S, Leclerc JE, Tremblay MJ. Presence of host ICAM-1 in laboratory and clinical strains of human immunodeficiency virus type 1 increases virus infectivity and CD4(+)-t-cell depletion in human lymphoid tissue, a major site of replication in vivo. J Virol (2002) 76:1004–14. doi: 10.1128/jvi.76.3.1004-1014.2002
59. Fortin JF, Cantin R, Lamontagne G, Tremblay M. Host-derived ICAM-1 glycoproteins incorporated on human immunodeficiency virus type 1 are biologically active and enhance viral infectivity. J Virol (1997) 71:3588–96. doi: 10.1128/JVI.71.5.3588-3596.1997
60. Liu X, Chen Q, Yan J, Wang Y, Zhu C, Chen C, et al. MiRNA-296-3p-ICAM-1 axis promotes metastasis of prostate cancer by possible enhancing survival of natural killer cell-resistant circulating tumour cells. Cell Death Dis (2013) 4:e928–8. doi: 10.1038/cddis.2013.458
61. Yuan T, Yao W, Tokunaga K, Yang R, Sun B. An HIV-1 capsid binding protein TRIM11 accelerates viral uncoating. Retrovirology (2016) 13:72. doi: 10.1186/s12977-016-0306-5
62. Barr SD, Smiley JR, Bushman FD. The interferon response inhibits HIV particle production by induction of TRIM22. PloS Pathog (2008) 4:e1000007. doi: 10.1371/journal.ppat.1000007
63. Allouch A, Di Primio C, Alpi E, Lusic M, Arosio D, Giacca M, et al. The TRIM family protein KAP1 inhibits HIV-1 integration. Cell Host Microbe (2011) 9:484–95. doi: 10.1016/j.chom.2011.05.004
64. Qi Z, Cai S, Cai J, Chen L, Yao Y, Chen L, et al. miR-491 regulates glioma cells proliferation by targeting TRIM28 in vitro. BMC Neurol (2016) 16:248. doi: 10.1186/s12883-016-0769-y
65. CHE L-F, SHAO S-F, WANG L-X. Downregulation of CCR5 inhibits the proliferation and invasion of cervical cancer cells and is regulated by microRNA-107. Exp Ther Med (2016) 11:503–9. doi: 10.3892/etm.2015.2911
66. Chiang K, Liu H, Rice AP. miR-132 enhances HIV-1 replication. Virology (2013) 438:1–4. doi: 10.1016/j.virol.2012.12.016
67. Leoh LS, van Heertum B, De Rijck J, Filippova M, Rios-Colon L, Basu A, et al. The stress oncoprotein LEDGF/p75 interacts with the methyl CpG binding protein MeCP2 and influences its transcriptional activity. Mol Cancer Res (2012) 10:378–91. doi: 10.1158/1541-7786.MCR-11-0314
68. Sirois M, Robitaille L, Allary R, Shah M, Woelk CH, Estaquier J, et al. TRAF6 and IRF7 control HIV replication in macrophages. PloS One (2011) 6:e28125. doi: 10.1371/journal.pone.0028125
69. Yang F, Xu Z, Duan S, Luo M. MicroRNA-541 promotes the proliferation of vascular smooth muscle cells by targeting IRF7. Am J Transl Res (2016) 8:506–15.
70. Li Z, Yin H, Hao S, Wang L, Gao J, Tan X, et al. miR-200 family promotes podocyte differentiation through repression of RSAD2. Sci Rep (2016) 6:27105. doi: 10.1038/srep27105
71. Raposo RAS, Abdel-Mohsen M, Bilska M, Montefiori DC, Nixon DF, Pillai SK. Effects of cellular activation on anti-HIV-1 restriction factor expression profile in primary cells. J Virol (2013) 87:11924–9. doi: 10.1128/JVI.02128-13
72. Pincetic A, Kuang Z, Seo EJ, Leis J. The interferon-induced gene ISG15 blocks retrovirus release from cells late in the budding process. J Virol (2010) 84:4725–36. doi: 10.1128/JVI.02478-09
73. Zhang Q, He Y, Nie M, Cai W. Roles of miR-138 and ISG15 in oral squamous cell carcinoma. Exp Ther Med (2017) 14:2329–34. doi: 10.3892/etm.2017.4720
74. Jiang C, Zhu W, Xu J, Wang B, Hou W, Zhang R, et al. MicroRNA-26a negatively regulates toll-like receptor 3 expression of rat macrophages and ameliorates pristane induced arthritis in rats. Arthritis Res Ther (2014) 16:R9. doi: 10.1186/ar4435
75. Liu L, Oliveira NMM, Cheney KM, Pade C, Dreja H, Bergin A-MH, et al. A whole genome screen for HIV restriction factors. Retrovirology (2011) 8:94. doi: 10.1186/1742-4690-8-94
76. Wu M, Fan B, Guo Q, Li Y, Chen R, Lv N, et al. Knockdown of SETDB1 inhibits breast cancer progression by miR-381-3p-related regulation. Biol Res (2018) 51:39. doi: 10.1186/s40659-018-0189-0
77. Chang C, Liu T, Huang Y, Qin W, Yang H, Chen J. MicroRNA-134-3p is a novel potential inhibitor of human ovarian cancer stem cells by targeting RAB27A. Gene (2017) 605:99–107. doi: 10.1016/j.gene.2016.12.030
78. Gerber PP, Cabrini M, Jancic C, Paoletti L, Banchio C, von Bilderling C, et al. Rab27a controls HIV-1 assembly by regulating plasma membrane levels of phosphatidylinositol 4,5-bisphosphate. J Cell Biol (2015) 209:435–52. doi: 10.1083/jcb.201409082
79. van Gent M, Sparrer KMJ, Gack MU. TRIM proteins and their roles in antiviral host defenses. Annu Rev Virol (2018) 5:385–405. doi: 10.1146/annurev-virology-092917-043323
80. Zhang Y, Zhang H, An M, Zhao B, Ding H, Zhang Z, et al. Crosstalk in competing endogenous RNA networks reveals new circular RNAs involved in the pathogenesis of early HIV infection. J. Transl Med (2018) 16:332. doi: 10.1186/s12967-018-1706-1
81. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature (2013) 495:384–8. doi: 10.1038/nature11993
82. Kincaid RP, Sullivan CS. Virus-encoded microRNAs: an overview and a look to the future. PloS Pathog (2012) 8:e1003018. doi: 10.1371/journal.ppat.1003018
83. Nair V, Zavolan M. Virus-encoded microRNAs: novel regulators of gene expression. trends microbiol. Trends Microbiol (2006) 14:169–75. doi: 10.1016/j.tim.2006.02.007
84. Nanbo A, Furuyama W, Lin Z. RNA Virus-encoded miRNAs: Current insights and future challenges. Front Microbiol (2021) 12:679210. doi: 10.3389/fmicb.2021.679210
85. Swaminathan G, Navas-Martín S, Martín-García J. MicroRNAs and HIV-1 infection: Antiviral activities and beyond. J Mol Biol (2014) 426:1178–97. doi: 10.1016/j.jmb.2013.12.017
86. Cullen BR. Five questions about viruses and microRNAs. PloS Pathog (2010) 6:e1000787. doi: 10.1371/journal.ppat.1000787
87. Bennasser Y, Le S-Y, Yeung ML, Jeang K-T. HIV-1 encoded candidate micro-RNAs and their cellular targets. Retrovirology (2004) 1:43. doi: 10.1186/1742-4690-1-43
88. Omoto S, Fujii YR. Regulation of human immunodeficiency virus 1 transcription by nef microRNA. J Gen Virol (2005) 86:751–5. doi: 10.1099/vir.0.80449-0
89. Omoto S, Ito M, Tsutsumi Y, Ichikawa Y, Okuyama H, Brisibe EA, et al. HIV-1 nef suppression by virally encoded microRNA. Retrovirology (2004) 144. doi: 10.1186/1742-4690-1-44
90. Kaul D, Ahlawat A, Gupta SD. HIV-1 genome-encoded hiv1-mir-H1 impairs cellular responses to infection. Mol Cell Biochem (2009) 323:143–8. doi: 10.1007/s11010-008-9973-4
91. Holland B, Wong J, Li M, Rasheed S. Identification of human microRNA-like sequences embedded within the protein-encoding genes of the human immunodeficiency virus. PloS One (2013) 8:e58586. doi: 10.1371/journal.pone.0058586
92. Schopman NCT, Willemsen M, Liu YP, Bradley T, van Kampen A, Baas F, et al. Deep sequencing of virus-infected cells reveals HIV-encoded small RNAs. Nucleic Acids Res (2012) 40:414–27. doi: 10.1093/nar/gkr719
93. Althaus CF, Vongrad V, Niederöst B, Joos B, Di Giallonardo F, Rieder P, et al. Tailored enrichment strategy detects low abundant small noncoding RNAs in HIV-1 infected cells. Retrovirology (2012) 9:27. doi: 10.1186/1742-4690-9-27
94. Klase Z, Kale P, Winograd R, Gupta MV, Heydarian M, Berro R, et al. HIV-1 TAR element is processed by dicer to yield a viral micro-RNA involved in chromatin remodeling of the viral LTR. BMC Mol Biol (2007) 8:63. doi: 10.1186/1471-2199-8-63
95. Bernard MA, Zhao H, Yue SC, Anandaiah A, Koziel H, Tachado SD. Novel HIV-1 miRNAs stimulate TNFα release in human macrophages via TLR8 signaling pathway. PloS One (2014) 9:e106006. doi: 10.1371/journal.pone.0106006
96. Lin J, Cullen BR. Analysis of the interaction of primate retroviruses with the human RNA interference machinery. J Virol (2007) 81:12218–26. doi: 10.1128/JVI.01390-07
97. Whisnant AW, Bogerd HP, Flores O, Ho P, Powers JG, Sharova N, et al. In-depth analysis of the interaction of HIV-1 with cellular microRNA biogenesis and effector mechanisms. MBio (2013) 4:e000193. doi: 10.1128/mBio.00193-13
98. Board NL, Moskovljevic M, Wu F, Siliciano RF, Siliciano JD. Engaging innate immunity in HIV-1 cure strategies. Nat Rev Immunol (2022) 22:499–512. doi: 10.1038/s41577-021-00649-1
99. Bertagnolio S, De Luca A, Vitoria M, Essajee S, Penazzato M, Hong SY, et al. Determinants of HIV drug resistance and public health implications in low- and middle-income countries. Antivir Ther (2012) 17:941–53. doi: 10.3851/IMP2320
100. Le Douce V, Janossy A, Hallay H, Ali S, Riclet R, Rohr O, et al. Achieving a cure for HIV infection: do we have reasons to be optimistic? J Antimicrob Chemother (2012) 67:1063–74. doi: 10.1093/jac/dkr599
101. Stadeli KM, Richman DD. Rates of emergence of HIV drug resistance in resource-limited settings: a systematic review. Antivir Ther (2013) 18:115–23. doi: 10.3851/IMP2437
102. Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E, et al. Modulation of K-ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell (2010) 18:282–93. doi: 10.1016/j.ccr.2010.08.013
103. Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis c virus RNA abundance by a liver-specific MicroRNA. Science (2005) 309:1577–81. doi: 10.1126/science.1113329
104. Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis c virus infection. Science (2010) 327:198–201. doi: 10.1126/science.1178178
105. Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, Thannickal VJ, et al. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med (2010) 207:1589–97. doi: 10.1084/jem.20100035
106. Steele CW, Oien KA, McKay CJ, Jamieson NB. Clinical potential of microRNAs in pancreatic ductal adenocarcinoma. Pancreas (2011) 40:1165–71. doi: 10.1097/MPA.0b013e3182218ffb
107. Doyle T, Goujon C, Malim MH. HIV-1 and interferons: who’s interfering with whom? Nat Rev Microbiol (2015) 13:403–13. doi: 10.1038/nrmicro3449
108. Witwer KW, Watson AK, Blankson JN, Clements JE. Relationships of PBMC microRNA expression, plasma viral load, and CD4+ T-cell count in HIV-1-infected elite suppressors and viremic patients. Retrovirology (2012) 9:5. doi: 10.1186/1742-4690-9-5
Keywords: HIV-1, miRNA, mRNAs, HAART, therapeutic potential
Citation: Rashid F, Zaongo SD, Song F and Chen Y (2023) The diverse roles of miRNAs in HIV pathogenesis: Current understanding and future perspectives. Front. Immunol. 13:1091543. doi: 10.3389/fimmu.2022.1091543
Received: 07 November 2022; Accepted: 14 December 2022;
Published: 05 January 2023.
Edited by:
Velpandi Ayyavoo, University of Pittsburgh, United StatesReviewed by:
Ihtisham Bukhari, Fifth Affiliated Hospital of Zhengzhou University, ChinaCopyright © 2023 Rashid, Zaongo, Song and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaokai Chen, eWFva2FpY2hlbkBob3RtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.