- 1School of Medicine, Xiamen University, Xiamen, China
- 2Central Laboratory, Clinical Medical College & Affiliated Hospital of Chengdu University, Chengdu University, Chengdu, China
- 3Department of Critical Care Medicine, The Affiliated Hospital of Putian University, Putian, China
- 4School of Medicine, Sun Yat-Sen University, Shenzhen, China
- 5Department of Radiation Oncology, Shanghai Chest Hospital, Shanghai Jiao Tong University, Shanghai, China
Background: The F-box and WD repeat domain containing (FBXW) family of SCF E3 complexes has 10 members that are responsible for ubiquitination and degradation of substrate proteins involved in cell cycle regulation and tumorigenesis. Among them, FBXW1 (also called b-TrCP1/BTRC) and FBXW7 are the central proteins in this category. However, there is still a lack of elaborate exploration of the contribution of FBXW family members, especially FBXW1 and FBXW7, in various tumor types.
Methods: In this present study, we preliminarily analyzed the genetic structure characteristics of the FBXW family, and systematically investigated their expression patterns and clinical correlations based on the TCGA pan-cancer data. Survival analysis of FBXWs was also conducted through the Kaplan-Meier method. In addition, we assessed their immune infiltration level through immune-related algorithms like Timer and xCell.
Results: There were obvious genetic heterogeneity and different clinical traits in FBXW family members. Moreover, we found that FBXW family genes may be useful in predicting prognosis and therapeutic efficacy using survival analysis. In addition, the immune infiltration of FBXW family was also clearly illustrated in this study. The results showed these genes were closely involved in immune components such as immune score, immune subtypes, tumor-infiltrating lymphocytes and immune checkpoints. Notedly, FBXW1 as an oncogene and FBXW7 as a tumor suppressor gene also show opposite relationships on immune cells.
Conclusion: Our results provided valuable strategies to guide the therapeutic orientation concerning the role of FBXW family genes in cancer.
Introduction
Cancer is a crucial cause of death and a great barrier to improving life expectancy and quality (1, 2). Based on the predictions of Global Cancer Statistics 2020, there are almost 19.3 million new cancer illness and 10.0 million carcinoma-related deaths (3). In addition, the incidence and mortality of cancer are growing dramatically. Ubiquitin Proteasome System (UPS), which can promote protein binding to ubiquitin (Ub) and transfer to proteasome for proteolysis, has been involved in the progression and development of cancers as the UPS controls the degradation of numerous tumor suppressors and oncogenes (4, 5).UPS with highly conserved protein degradation mechanisms plays critical roles in various physiological processes, including signal transduction, cell cycle, DNA replication, transcriptional regulation, as well as cell death and immune response (6). Dysregulation of this system has been implicated in several human malignant transformation (7). The process of degradation is carried out in three strides by multiple components which includes Ub activating E1 enzyme, Ub conjugating E2 enzyme, and Ub E3 ligase (4, 8).
Multitude of E3 ligases have been broadly involved in the cancer-related processes, due to the characteristic of their specific substrate recognition (9). A representative example of E3 enzyme is a conserved ubiquitin ligase complex known as SCF E3 ligases which is composed of three invariable elements- SKP1, Cullin, RBX1 and a variable component -FBP (10). The 69 FBPs family members can be classified as three subclasses as follows: FBXW, FBXL, and FBXO. The FBXWs containing WD40 repeats play a pivotal role in targeting proteins associated with cell cycle regulation and tumorigenesis (11). The FBXW family has ten members: FBXW1(also known as β-TrCP1), FBXW2, FBXW4, FBXW5, FBXW7, FBXW8, FBXW9, FBXW10, FBXW11(also known as β-TrCP2), and FBXW12, in which β-TrCP and FBXW7 are central proteins (4, 12). FBXW1 and FBXW11 are two β-TrCP homologs with similar biological effects which mainly function as adaptors to recognize specific substrates such as β-catenin, CDC25A, IκB and DEPTOR (12, 13). β-TrCPs have been demonstrated to role as oncogene or tumor suppressor depending on the diversity of substrates. Susanne et al. discovered that overexpression of FBXW1 was vital mediator of constitutive NF-κB activation leading to chemoresistance in pancreatic cancer cells (14). In particular, FBXW1 targets Snail and EZH2 ubiquitination which are involved in epithelial-to-mesenchymal transition in cancers (15). FBXW7 roles as a tumor suppressor on account of promoting the degradation of various oncogenes including Cyclin E, c-Jun, Notch, mTOR, c-Myc, etc. (16). Zhan et al. studied that FBXW7 as a tumor suppressor negatively regulates ENO1-induced cell proliferation and migration in colorectal cancer cells (17). In addition, the high frequency mutation of FBXW7 was found in multiple cancers. For example, the rate of mutation is approximately 35% in cholangiocarcinomas and 30% in T-cell acute lymphocytic leukemia (18, 19). Additionally, FBXW1 and FBXW7 co-coordinate the apoptotic process of cells by ubiquitinating MCL1 (20). However, the full picture of FBXW family in pan-cancer, especially these central proteins, has yet to be explained.
In this study, we primarily investigated the structural of FBXW members and comprehensively analyzed the expression levels and prognostic value of FBXWs in pan-cancer via the Cancer Genome Atlas (TCGA) database. Furthermore, the relationship between FBXWs expression and tumor microenvironment (TME) were evaluated by TIMER database. Besides, we also explored the correlation between FBXWs expression levels and immune subtype, drug sensitivity in multiple cancers. FBXW1/7 as central proteins, their relationship with immune infiltration was also conducted in this research. Together, our study revealed the detailed expression patterns and overall picture of the immune infiltration of FBXW gene family in pan-cancer, which provide further insights for FBXWs as potential therapy targets in Pan-cancer.
Materials and methods
Analysis of the main characteristics of FBXW family members in Homo sapien
The phylogenetic tree of the FBXWs was constructed in this study using MEGA 7.0.26 with the neighbour-joining (NJ) method and 1000 bootstrap replications (21). Then, it was visualized by the online tool TBtools (22). GFF3 files (General Feature Format Version 3) were downloaded in the ensemble database for the structural analysis of FBXWs. The characteristic motifs of FBXWs were conducted by MEME (http://meme-suite.org/tools/meme) software. Furthermore, TBtools was also used for analyzing the gene structure of all FBXWs and SWISS-MODEL Interactive Workshop was employed for the prediction of the tertiary structure of FBXW proteins (23, 24).
TCGA pan-cancer atlas data profile
We used the UCSC Xena (https://xenabrowser.net/) to analyse the TCGA pan-cancer data of FBXW family members, including gene expression, clinicopathological data, molecular subtype, survival data and stemness score (25). The perl software was used to integrate the FBXW family expression and the Wilcox test was also taken to assess the difference between normal and tumor tissues. “*”, “**”, “***”, indicate P-value <0.05, <0.01, <0.001, respectively. “Ggpubr” and “pheatmap” packages were used for illustrating the expression pattern of FBXWs with a form of box plot and heatmap. Correlation analysis among FBXW family genes was performed by R-package “corrplot”. Furthermore, “ggplot2” package was use for the examination the relationship between the clinical stages and grades and FBXW family, and the GSCA database was performed to analysis the correlation between FBXW family expression and the subtypes of various tumors.
Survival and cox analysis of expression of FBXW family
The Kaplan-Meier method and the log-rank test were employed to evaluate the survival situation of FBXW members in various tumors. “survival” and “survminer” R package were used for evaluation the prognosis role of these genes. Besides, we also conducted a COX analysis to establish the association between FBXWs expression and prognosis of pan-cancer. Finally, the forest plot was drawn using “survival” and “forestplot” packages.
Genetic alteration analysis
cBioPortal platform integrates data from comprehensive tumor genome studies, including TCGA and ICGC large tumor projects, with data from more than 28,000 samples (26). In this study, we used the cBioPortal database to explore the mutation frequency and form of FBXW family members.
Tumor microenvironment analysis
“Estimate” and “limma” R packages were applied to investigate the immune score, estimate score, and stromal score of different tumor cases based on TCGA expression data. Spearman test was applied to demonstrate the relationship between FBXW family genes and these scores. Furthermore, an association analysis between FBXWs expression and RNA stemness score (RNAss), DNA stemness score (DNAss) was established using the Spearman’s method and “limma” package.
Immune infiltration cells and immune checkpoints correlation analysis
Immuneeconv, which is able to integrate the two latest algorithms, TIMER and xCell for reliable immune score evaluation, was used in this study for evaluating the immune infiltration level of FBXWs in Pan-cancer. A spearman correlation analysis of the immune score or immune infiltration cells and FBXW family genes expression in various tumors was also conducted in this research. Furthermore, the associations of FBXW family genes with representative immune checkpoints selected were also generated using spearman correlation test. ∗P< 0.05, ∗∗P< 0.01, and ∗∗∗P< 0.001.
Immune subtype and drug sensitivity correlation analysis of FBXW family genes
The “limma” “ggplot2” and “reshape2” R package were employed for the conduction of the immune subtype analysis of FBXW family genes. Additionally, CellMiner™ (https://discover.nci.nih.gov/cellminer/home.do) database was applied for the drug sensitivity analysis of FBXWs, and “impute”, “limma”, “ggplot2”, and “ggpubr” R packages were used for the data process and visualization.
Cell culture
All cells used in the study were purchased from the American Type Tissue Collection (ATCC) and cultured according to the manufacturer’s instructions (27). Cell mediums were commercially obtained from Solarbio Science & Technology Co., Ltd (Beijing, China).
qRT-PCR analysis
Total RNA extraction and cDNA reverse transcription were elaborated in previous studies (28). The above steps were taken by the SPARKscript IISYBR Green qRT-PCR Kit (Shandong Sparkjade Biotechnology Co., Ltd.) instructions. Syber Green (Yeasen Biotechnology Co., Ltd.) was used for the qRT-PCR process with a 10ul reaction system.
FBXW1 forward, 5′- TCTCGAAGGCCGCTTACT -3′,
FBXW1 reverse, 5′- ATACCTGGATGCCAAATCA-3′;
FBXW7 forward, 5′- ACAACGCACAGTGGAAGTA -3′,
FBXW7 reverse, 5′-AGTGGGACATACAGGTGGA-3′
The 2−ΔΔCt method was applied to calculate the relative expression levels of targeted genes. Dunnett’s t test was employed for the comparison between the experimental group and the normal group in this study. Each experiment was conducted in triplicate.
Results
Gene structure and motif composition of FBXWs in Homo sapiens
The F-box and WD repeat domain containing (FBXW) family, a key component of SCF-type (Skp1-Cullin1-F-box) ubiquitin ligase, specifically recognize the substrates and perform ubiquitination degradation, thereby affecting cell growth, differentiation, apoptosis and tumorigenesis (29, 30). In this study, we firstly investigated characteristics of 10 members of the FBXW family in Homo sapiens using uniport and GeneCards database, and the results showed that most of them located in cell cytoplasm and cytosol. Besides, the acquired Go term by the “pathway” module of GeneCards tool illustrated FBXW family members are involved in different biological response processes. For example, FBXW1 and FBXW4 were related to Protein polyubiquitination and Wnt signaling pathway, whereas FBXW7 was mainly involved in Vasculogenesis, DNA repair, Sister chromatid cohesion and Protein ubiquitination (Table 1). Besides, we found that these FBXWs shared same F-box and WD repeat domain with close interactions, and the enrichment analysis results demonstrated that they were primarily involved in ubiquitination-mediated degradation pathway (sFigure 1).
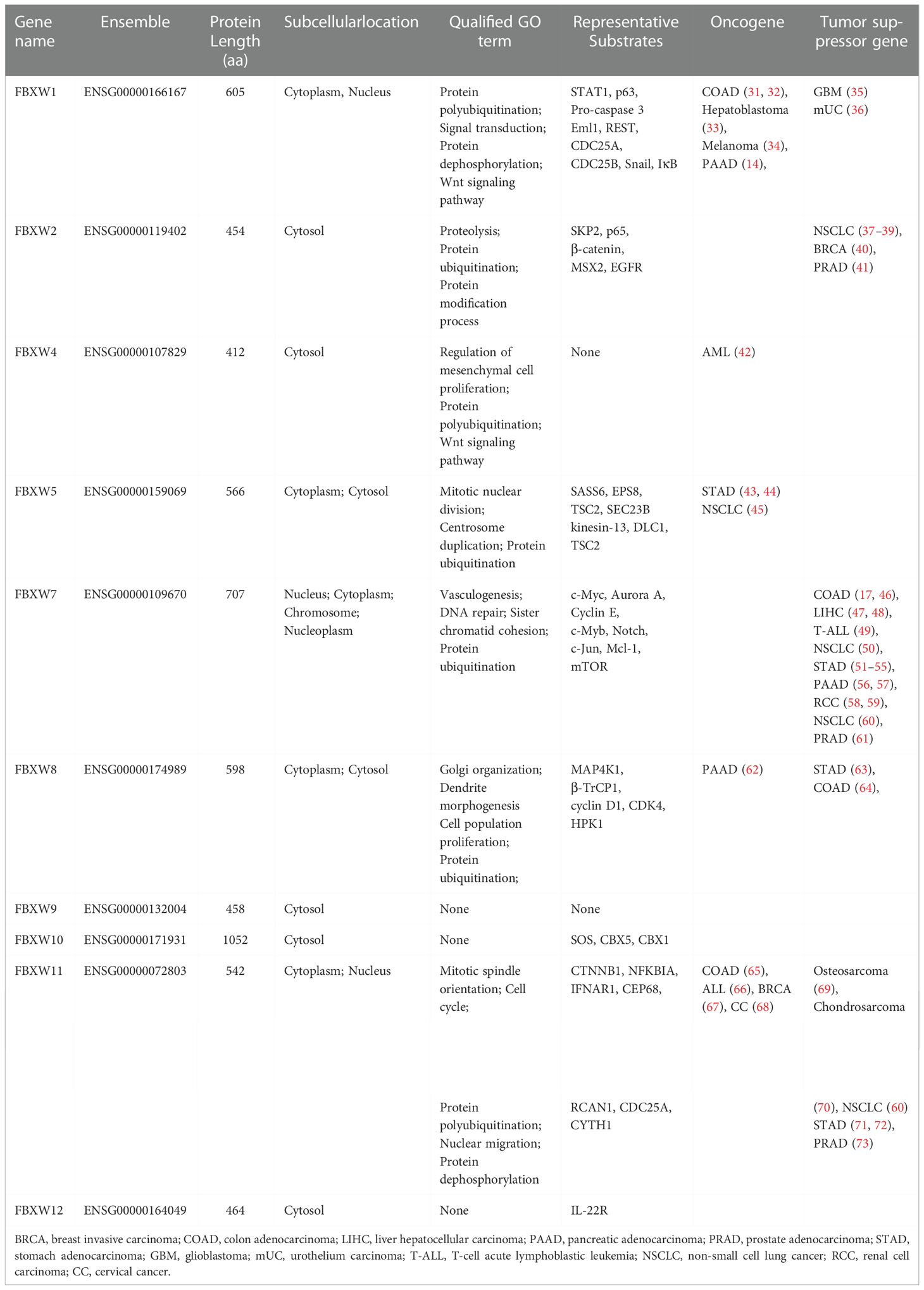
Table 1 Characteristics of 10 members of the FBXW family in Homo sapiens using uniport and GeneCards database.
To further understand the phylogenetic relationship of human FBXW family, 10 amino acid sequences of FBXWs were used to construct phylogenetic tree in the research, and the gene family was divided into three groups. Group a was the largest members of the FBXW family, however, group b has only one member FBXW10 (Figure 1A). To explore the function of FBXWs, the conserved motifs of these 10 proteins were illustrated. The results showed 20 conserved motifs were detected in the FBXW family members (Figure 1B). Notedly, FBXW1 and FBXW7 had the same Motifs 2, 3, 5 and 6, suggesting that they may have similar functions in some biological processes. Furthermore, motifs 14 was discovered only in group a, and group b had the largest number of motifs. The reason of functional differences in the FBXWs of Homo sapiens was probably the clade-specific distribution of conserved motifs. Furthermore, the exon-intron patterns and tertiary structures of the 10 FBXWs were also explored in this study, suggesting the FBXW family members are structurally distinct (Figure 1C; sFigure 2).
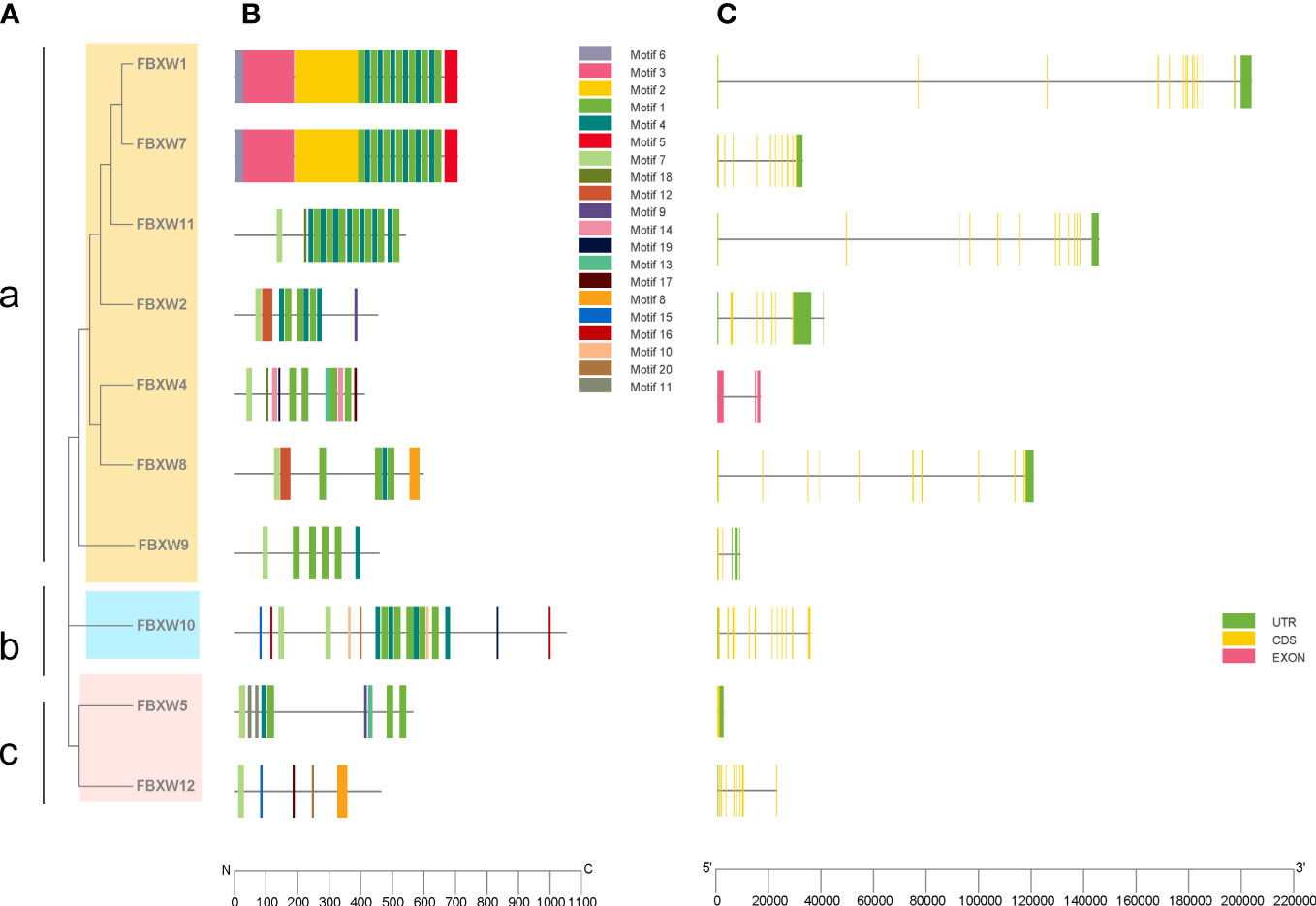
Figure 1 Phylogenetic relationships, conserved protein motifs and gene structures of the FBXW family members. (A) The phylogenetic tree was constructed based on the full-length sequences of Homo sapiens FBXW proteins. (B) Motifs distribution of the FBXW proteins. (C) Exon-intron structures of the FBXW genes. Green boxes indicate untranslated region; yellow boxes indicate coding sequences; pink boxes indicate exons and black lines indicate introns. a, b and c represent the three groups, respectively.
The expression levels of FBXW family in pan-cancer
We estimated the FBXWs differential expressions in Pan-cancer with RNA sequencing data of TCGA database. Our results indicated that FBXW1 was more expressed in colon adenocarcinoma (COAD), cholangiocarcinoma (CHOL), lung adenocarcinoma (LUAD), liver hepatocellular carcinoma (LIHC), Prostate adenocarcinoma (PRAD), and stomach adenocarcinoma (STAD). In contrast, a lower expression was found in kidney renal papillary cell carcinoma (KIRP), kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), glioblastoma multiforme (GBM), thyroid carcinoma (THCA) and uterine corpus endometrial carcinoma (UCEC) (Figure 2A). FBXW2 was more expressed in various cancers including CHOL, COAD, neck squamous cell carcinoma (HNSC), KICH, LIHC, PRAD, and STAD (Figure 2B).Meanwhile, lower expression was found in KIRC, LUAD, Lung squamous cell carcinoma (LUSC), THCA, and UCEC. The higher expression of FBXW4 was discovered in CHOL, KIRP, LIHC, PRAD, and UCEC. However, the lower level was in breast invasive carcinoma (BRCA), COAD, GBM, LUAD, LUSC, HNSC and STAD (Figure 2C). Additionally, FBXW5 was more expressed in BRCA, CHOL, LIHC, LUAD, LUSC, KICH, KIRC, PRAD, THCA and UCEC. The lower expression was in COAD, HNSC and STAD (Figure 2D). FBXW7 was higher expressed in CHOL, KIRC, KIRP, LIHC, LUAD, and THCA. Nevertheless, FBXW7 was lower expressed in BRCA, GBM, HNSC, PRAD, and UCEC (Figure 2E). We also analyzed that the higher expression level of FBXW8 was in several cancers, containing Bladder Urothelial Carcinoma (BLCA), BRCA, GBM, CHOL, COAD, HNSC, KIRP, LIHC, LUSC, STAD, PRAD and READ. However, the lower expression of FBXW8 was found was in LUSC, PRAD, READ, and STAD (Figure 2F). Besides, a higher FBXW9 expression was analyzed in BLCA, BRCA, CHOL, COAD, ESCA, GBM, LIHC, LUAD, LUSC, KIRP, PRAD, STAD, THCA, READ and UCEC. A lower FBXW9 expression was only in KICH (Figure 2G). We further explored that the higher expression of FBXW10 in several cancers including LUAD, LUSC, LIHC, PRAD and STAD compared with normal tissues. Meanwhile, the FBXW10 expression reduced in KICH, KIRC, and THCA (Figure 2H). In addition, the lower FBXW11 expression level was discovered in BLCA, BRCA, GBM, LUSC, HNSC, KIRC, KIRP and UCEC. The higher expression level of FBXW11 was found in CHOL and LIHC (Figure 2I). We also discovered FBXW12 was more expressed in BLCA, BRCA, LIHC, LUSC, CHOL, COAD, STAD, PRAD and UCEC. In contrast, the lower FBXW12 was found in GBM, KICH, KIRC, KIRP, and THCA (Figure 2J). Since FBXW1/7 are the central genes of this family and have same motifs, we also analyzed transcriptional expression of these two genes in four common cancer cell lines (breast, lung, colorectal and renal cancer). The results showed FBXW1 expression in COAD was higher than matched normal cell and the expression of FBXW7 in COAD was not different from that in normal group. However, the transcription levels of the other three cancer cells were not completely consistent with the TCGA database results. A small number of cell lines selected may be one of the reasons (sFigure 3).
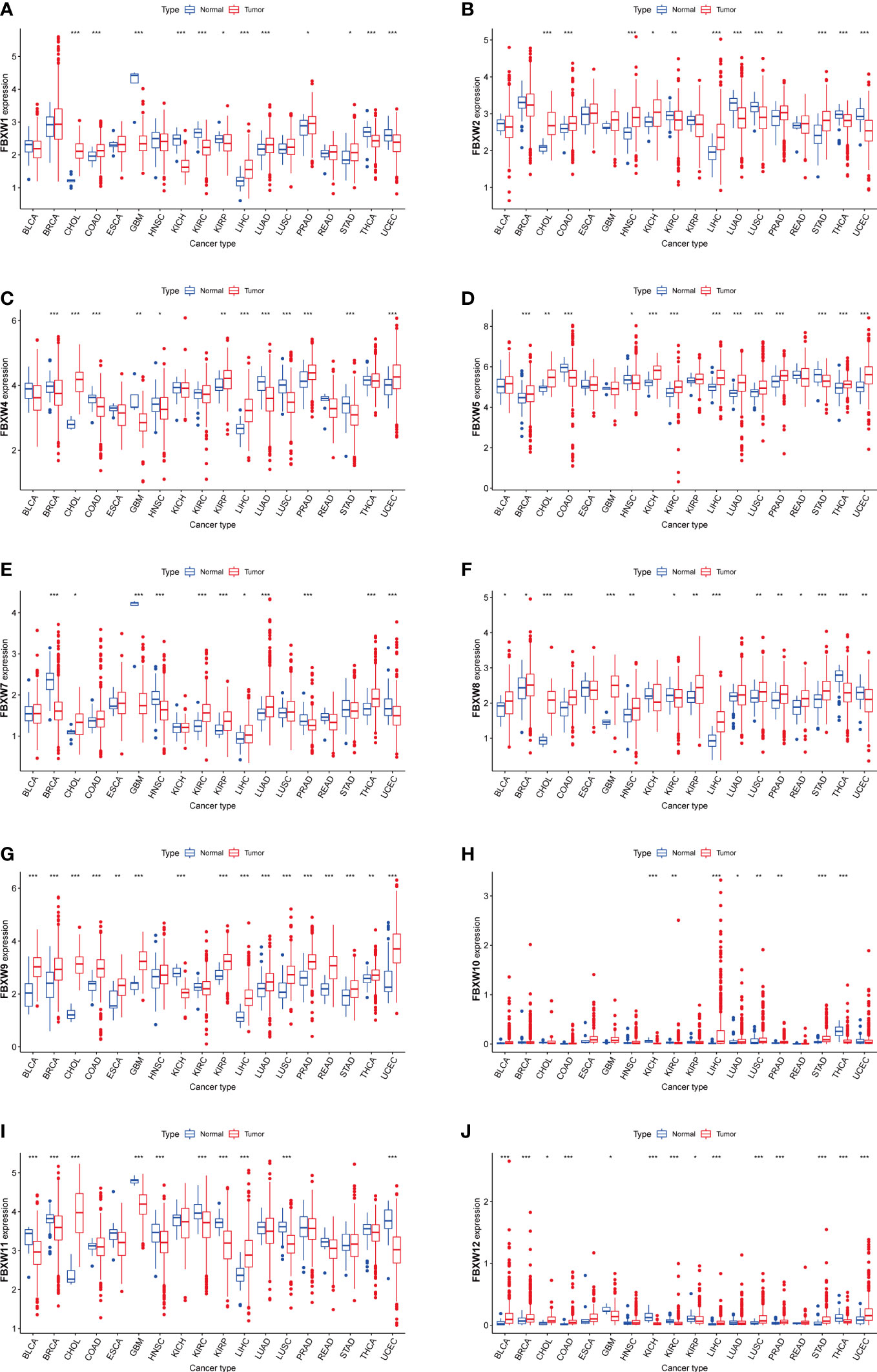
Figure 2 Boxplot of FBXW family (A–J) differential expression between cancer and adjacent normal tissues. The blue boxplots indicate the normal tissues. The red boxplots indicate the cancer tissues. ∗P< 0.05, ∗∗P< 0.01, and ∗∗∗P< 0.001.
We further analyzed FBXW gene family expression and correlation in various cancer types by TCGA database. The results were shown that FBXW1/4/9/11 were highest expression in CHOL (Figure 3A). we further investigated that FBXW5 was highly expressed, FBXW10 and FBXW12 were lowly expressed, and other members were moderately expressed in pan-cancer (Figure 3B). In terms of all these 18 types of cancer, two genes with most significant positive correlation were FBXW1 and FBXW11, whereas the correlation of FBXW5 and FBXW7 was completely opposite (Figure 3C). For the above researched cancers (BRCA, COAD, LUAD and KIRC), FBXWs as a whole showed a positive relationship, including FBXW1 and FBXW7 (sFigure 4) (These findings confirmed the synergistic relationships among FBXW members and revealed that the SCF complex may have a more competitive and complicated mechanism than we thought.
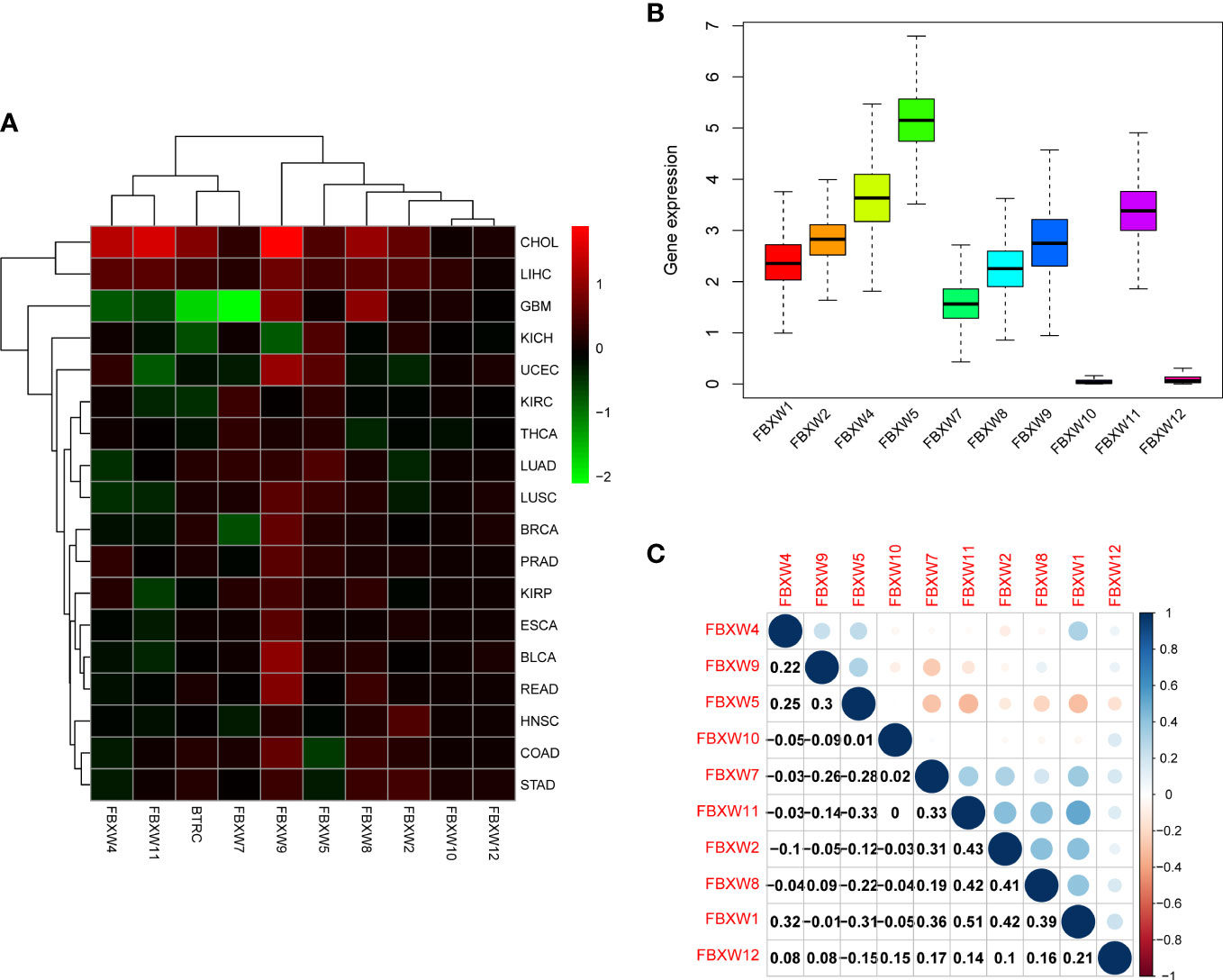
Figure 3 FBXW family gene expression levels and correlation in different cancer types from TCGA. (A) Heatmap showing the difference of FBXW family gene expression in 18 cancer types from TCGA database. The red and green indicate the high or low expression, respectively. (B) Boxplot illustrating the distribution of FBXW family gene expression in various cancers. (C) The correlation between FBXW family genes. The blue dot indicated the positive correlation. The red dot indicated the negative correlation.
The association between clinical characteristics, tumor subtypes and FBXWs expression
In the examination on the tumor stage relevance, we discovered that some FBXW genes expression significantly increased in tumor early stage, such as FBXW1/12 in KIRC, FBXW4 in pancreatic adenocarcinoma (PAAD), and FBXW11 in LUSC. Whereas, other genes expression predominantly increased in tumor advanced stage, including FBXW2 in ACC, FBXW2/9 in ILHC, FBXW5 in LUAD, FBXW7 in STAD, and FBXW10 in KIRP/Esophageal Squamous Cell Carcinoma (ESCC) (Figure 4).Moreover, we further explored the relationship between FBXWs expression levels and tumor grade. The result was indicated that FBXW4/7/11 expressions were significantly increased in KIRC early grade. In contrast, the lower expressions of FBXW1/8/9 were found in LIHC early grade, And the lower FBXW2/5/11 expressions were analyzed in the early grade of HNSC (sFigure 5). In addition, we analyzed the FBXWs expression levels in different subtypes of nine cancers. As shown in Figure 5A, the protein expression levels of most FBXWs were obviously different in BRCA and KIRC. Furthermore, we found that the expression of FBXW1/7 were significantly different in four subtypes of BRCA containing Basal, Her2, LumA, LumB compared with normal-like subtype. However, there were no notable difference of FBXW2/4/5/10/11/12 expression levels between LumA/LumB and Normal-like subtype (Figures 5B–K).
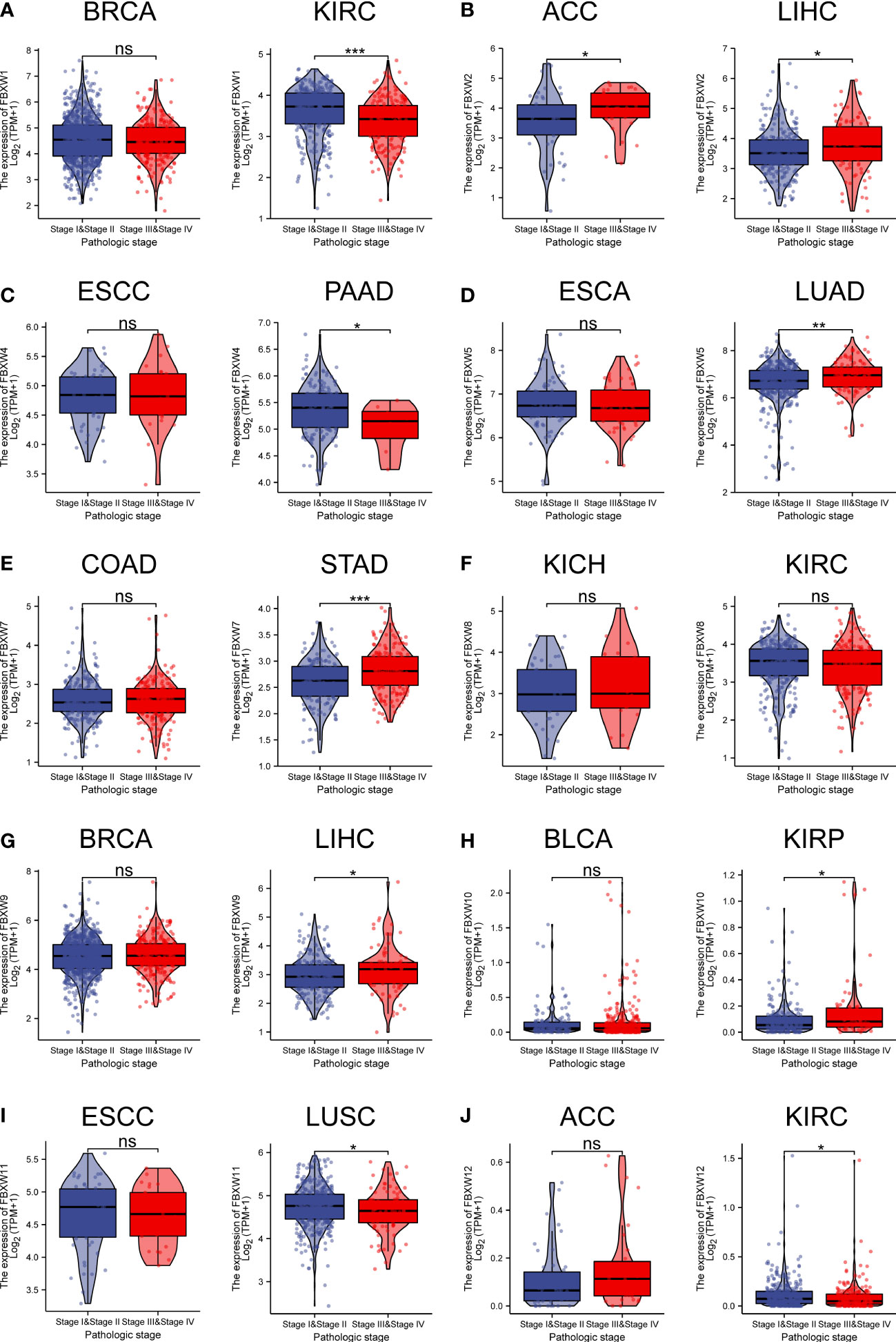
Figure 4 Association of FBXW family (A–J) gene expression with the clinical stages for different cancer types. ∗P< 0.05, ∗∗P< 0.01, and ∗∗∗P< 0.001, ns, No statistical significant.
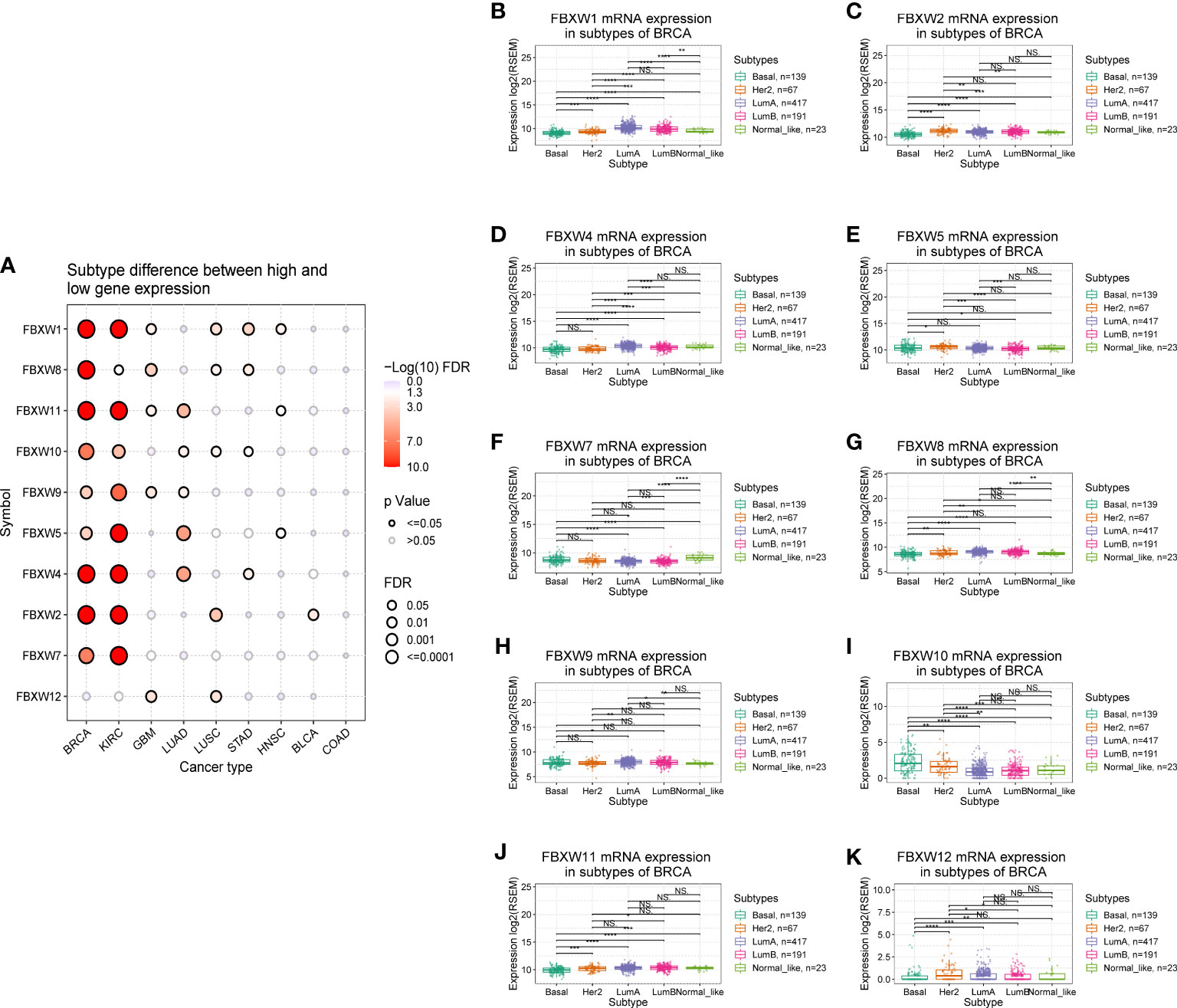
Figure 5 Correlation analysis between FBXW family gene expression and the subtypes of various tumors. (A) Subtype differences between high and low gene expression of FBXWs. (B–K) the relationship between FBXWs expression and the subtype of BRCA. ∗P< 0.05, ∗∗P< 0.01, ∗∗∗P< 0.001 and ****P<0.0001.
Prognostic value of FBXWs in pan-cancer
Aiming to investigate the relationship between FBXWs expression level and prognosis value, significant survival analysis with Kaplan-Meier survival curves was performed in the research (Figure 6; sFigure 6). FBXW1 was a beneficial factor in LGG, KIRC, MESO, and UVM. Besides, FBXW5 was a protective factor in KICH and UCEC. Nevertheless, FBXW5 was a high-risk factor in LUAD and ACC. The results also shown that among patients with PAAD, STAD, HNSC and SKCM, high FBXW7 expression was related with longer survival times. FBXW9 played a protective role in three different cancers, which contained KIRP, DLBC and HNSC. While, FBXW9 had a detrimental role in other three cancers including LIHC, ACC, and LGG. FBXW12 was a protective prognostic gene in LAML, READ, and UVM. FBXW12 was a high-risk gene in THYM. In addition, we found that FBXW1/2/11 played a protective role in KIRC. In contrast, FBXW10 was a dangerous factor in KIRC. On the one hand, FBXW1/11 were low-risk factors in LGG patients. On the other hand, high expressions of FBXW2/9 were associated with worse in LGG.
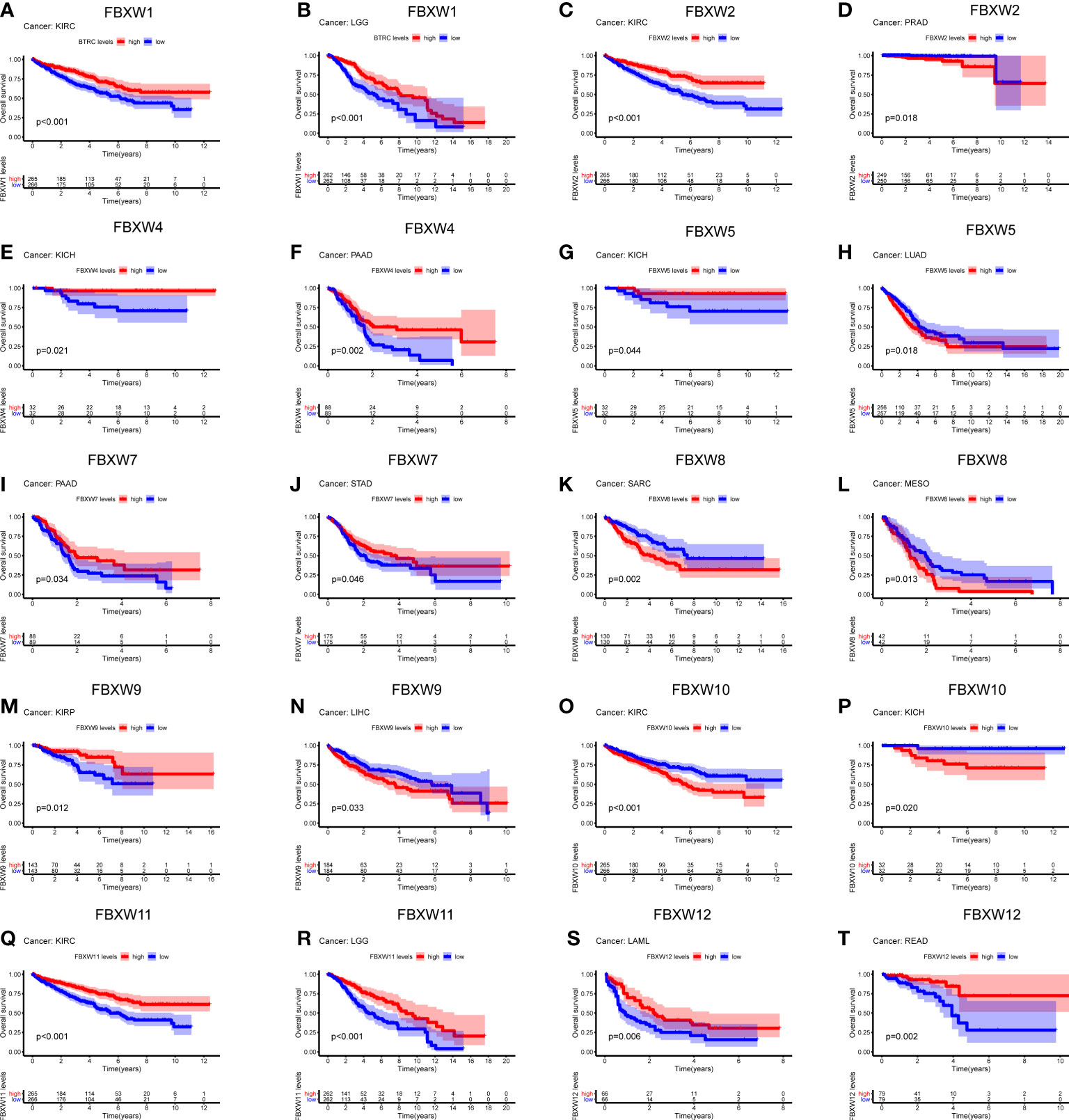
Figure 6 Survival analysis of FBXW family genes (A–T) across multiple cancer types. The red line in the photos indicates high expression and the blue line in the photos indicates low expression. P value less than 0.05 is considered as difference.
Then, we explored the prognosis risk of FBXWs via COX analysis. As shown in Figure 7, FBXW1 was a protective gene in LGG. FBXW2 had protective roles in KIRC. However, FBXW2 was high-risk factor in ACC, LGG, and PRAD. FBXW4 was a low-risk factor in KICH and LGG. For another, FBXW4 was a detrimental factor in LAML. FBXW5 acted as a detrimental factor in ACC, PCPG, and THYM. FBXW7 played a detrimental role in PRAD. FBXW8 functioned as a high-risk gene in KICH. FBXW9 had a protective effect in UVM. In contrast, FBXW9 acted as a high-risk factor in KICH and LGG. Among patients with KIRP, THCA, and THYM, high FBXW10 expression was associated with worse prognosis. While in patients with KIRC, LGG, and MESO, those with high FBXW11 expression had better prognosis. Our results also indicated that FBXW12 played a protective role in LAML and UVM. Contrarily, FBXW12 was a high-risk prognosis factor in DLBC, KICH, and PRAD (Figure 7). These results indicated that FBXWs in different cancers may play a different role in prognosis value.
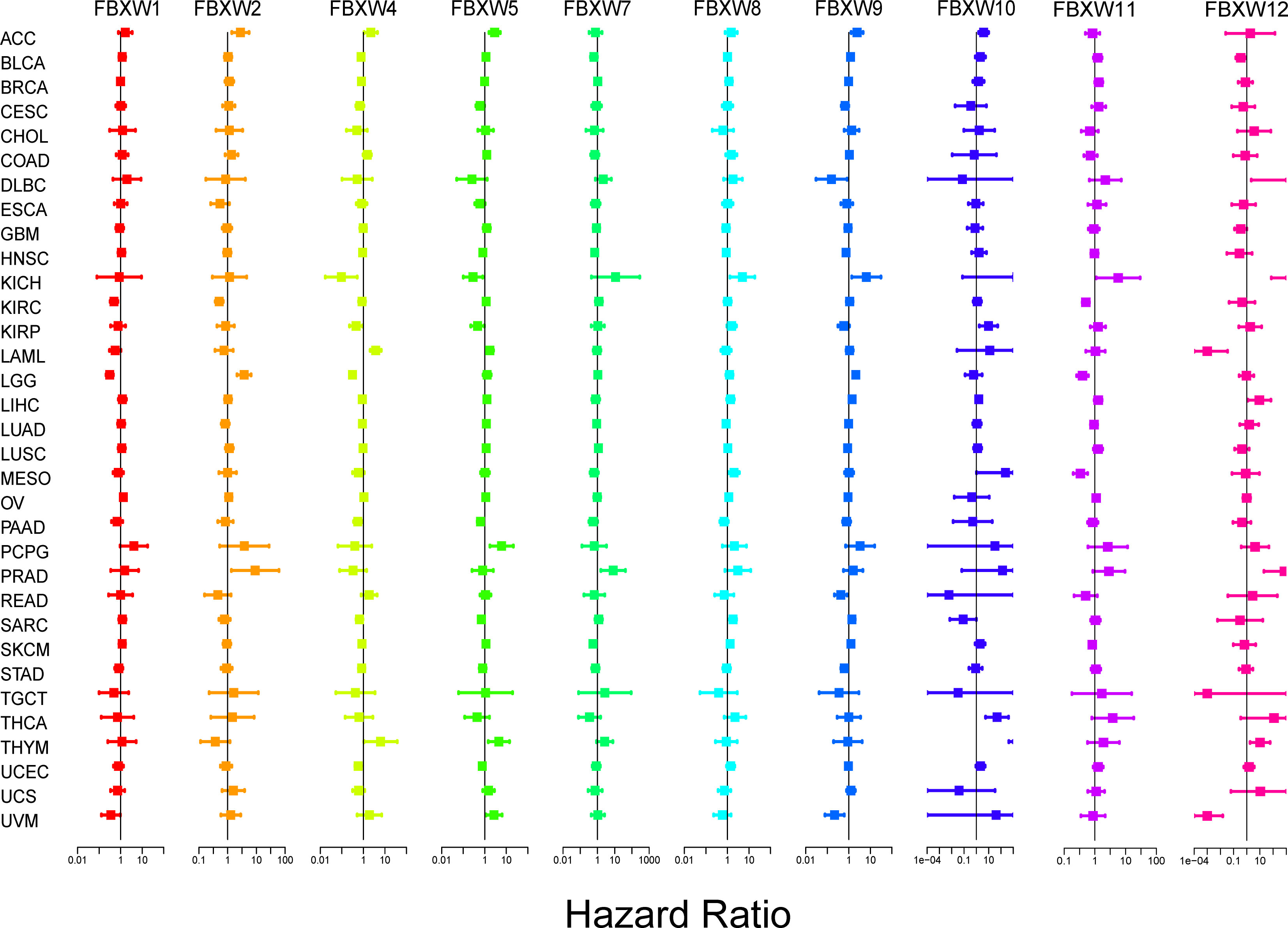
Figure 7 Correlation analysis of FBXW family gene expression with survival by the COX method in different types of cancers. Different colored lines indicate the risk value of different genes in tumors, hazard ratio <1 represents low risk and hazard ratio >1 represents high risk.
Genetic alterations of FBXW gene family in pan-cancer
We further analyzed the variation frequency and types of the FBXW gene family in 2922 cancer patients via cBioPortal database. The results indicated that gene change rates of FBXW7/9/11 were approximately 7%, which was highest in the FBXW family. Nevertheless, FBXW1/4/12 had lowest variation rates, which was only 3% (Figure 8A). Besides, we found that the main genetic alteration types containing amplification, mRNA high expression, mutation and deep deletion. The FBXW2/5/8/9/10/11 possessed the highest amplification rates. Meanwhile, the change type of FBXW1/4/7/12 were mainly mRNA high expression (Figure 8B). In conclusion, FBXW family members were likely to mutate in various cancer types.
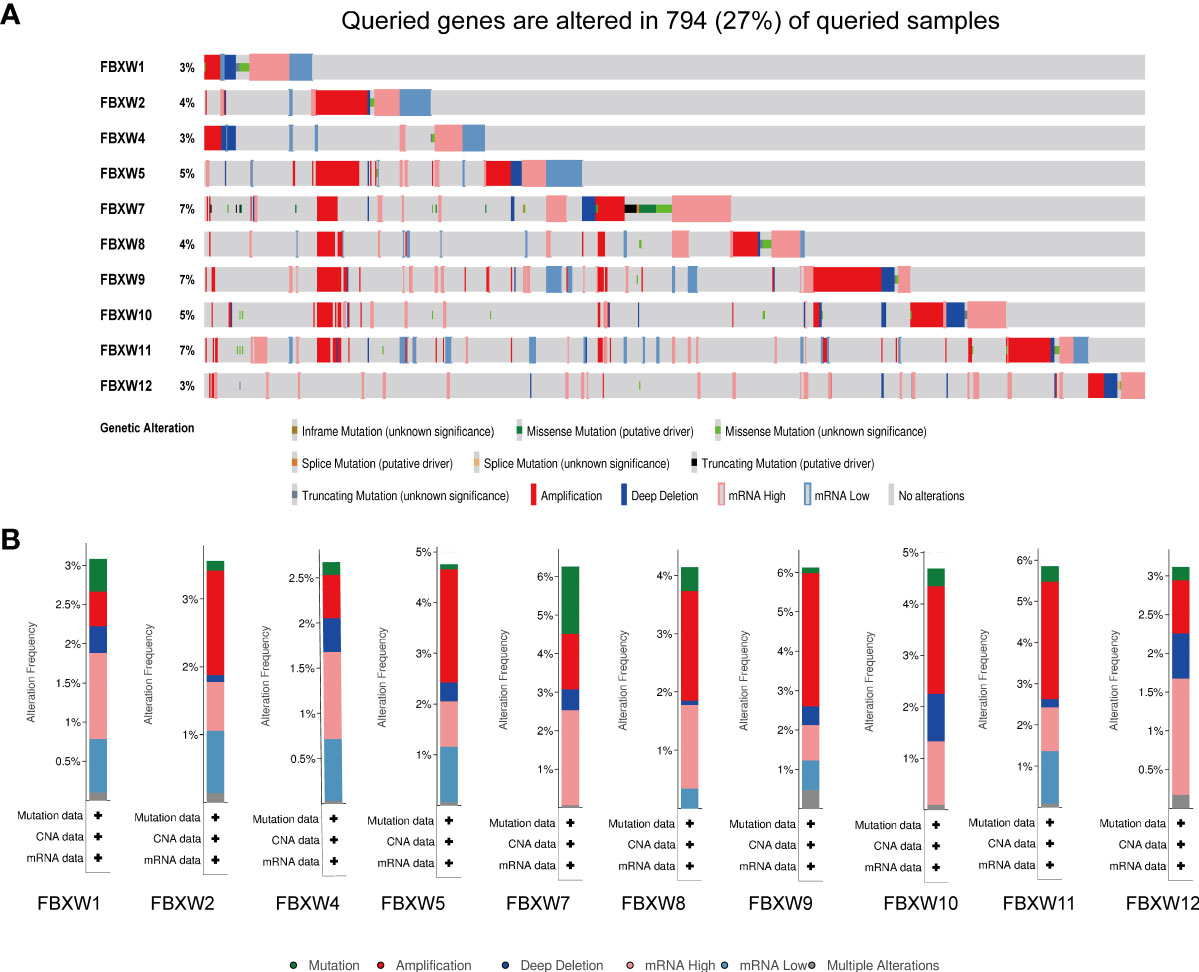
Figure 8 Genetic alterations and correlation analysis of FBXW family members in Pan-cancer. (A) summary of alteration rates for FBXW family from TCGA database using cBioPortal. (B) Genetic alteration frequency data of FBXW family members in pan-cancer, respectively.
Relationship between FBXWs expression and tumor microenvironment in pan-cancer
Cancer stem cell played a key role in tumor proliferation, migration, metastasis, epithelial-to-mesenchymal recurrence, and therapy resistance (74). Therefore, we explored the association between FBXWs expression and stemness score. As we can see from Figure 9, the expression of FBXW family had a positive or negative relationship with DNAss and RNAss in pan-cancer. For example, FBXW1/8/10 expression were negatively related with DNAss in OV. At the same time, we found that FBXW2 was positively associated with DNAss in OV. Besides, FBXW1 had a significantly positive relation with DNAss in TCGA and THYM (Figure 9A). During the analysis of RNAss, we observed that FBXW1/8/10/11 were negatively associated with RNAss in THYM. FBXW7 was negatively related with RNAss in several cancers such as CHOL, KIRC, and KIRP (Figure 9B). Furthermore, the tumor microenvironment related score results indicated most of FBXWs expression was significantly negatively associated with stromal score, immune score, and estimate score and positively related to tumor purity (Figures 9C–F). Specifically, we could find that FBXW5 appeared to have a consistent positive relationship across all cancers in these scores; on the other hand, FBXW12 had a consistent negative relationship. Moreover, we also discovered there was an obvious relation between immune score and FBXWs in ACC, GBM and LAML. These results demonstrated that FBXW members vary in their ability to regulate the immune microenvironment in different cancers.
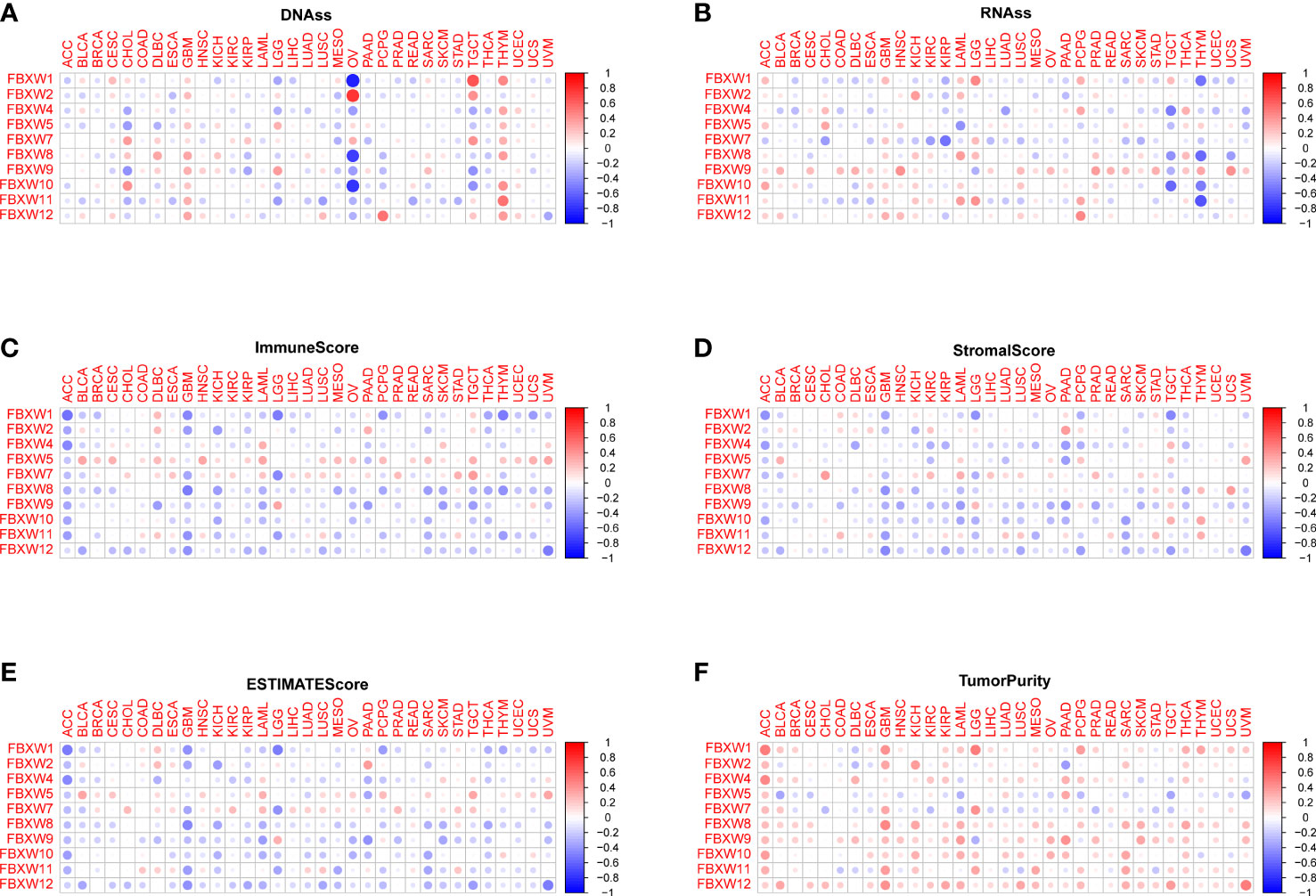
Figure 9 Association between FBXW family gene expression and tumor micro-environment factors and stemness score in pan-cancer. The FBXW family gene associated immune score (A), stromal score (B), estimate score (C), DNAss (D), RNAss (E) and tumor purity (F) are illustrated.
Correlation between immune infiltration and FBXWs expression
Then, we analyzed the relation between FBXWs and immune subtype. As was shown in Figure 10, The expression of all FBXW gene family members were significantly associated with immune subtype C1(wound healing), C2 (IFN-gamma dominant), C3
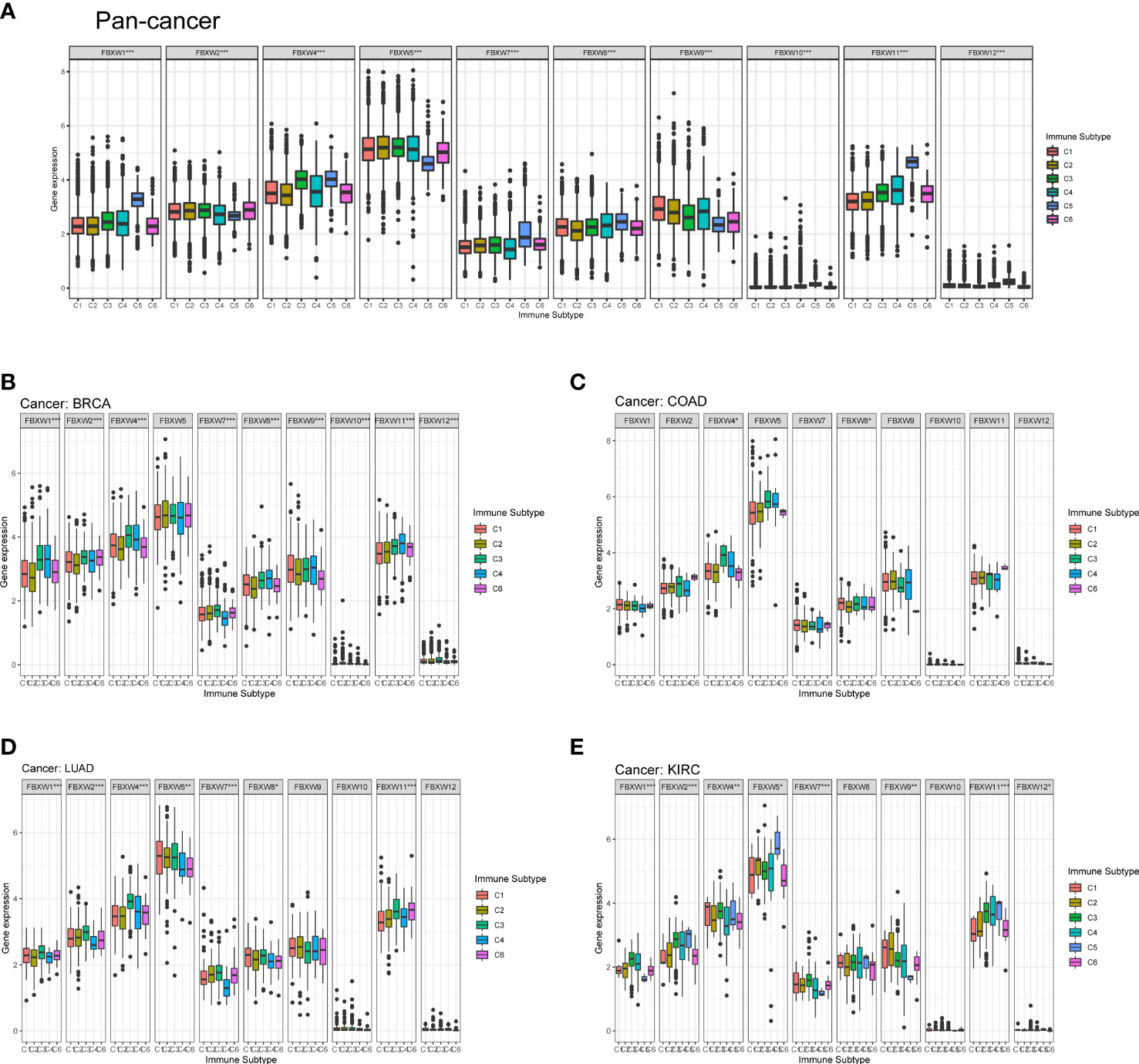
Figure 10 FBXW family gene expression level of different immune subtype in pan-cancer, and specific four cancer types. (A) In pan-cancer. (B) In BRCA. (C) In COAD. (D) In LUAD. (E) In KIRC. X axis represents immune subtype, Y axis represents gene expression. * P<0.05; ** P<0.01; *** P<0.001. C1: wound healing, C2: IFN-gamma dominant, C3: inflammatory, C4: lymphocyte depleted, C5: immunological quiet, and C6: TGF-beta dominant.
(inflammatory), C4 (lymphocyte depleted), C5 (immunological quiet), and C6 (TGF-beta dominant) in pan-cancer (Figure 10A). Then, we selected four cancer types to analyze including BRCA, COAD, LUAD, and KIRC. FBXW1 was higher expressed in C3 and C4, the expression of FBXW7 up-regulated in C2 and C3 (Figure 10B). FBXW4 and FBXW8 were significantly related to the immune subtype in COAD (Figure 10C). Furthermore, we found that FBXW1/2/4/5/7/8/11 were significantly different in LUAD, and these genes overexpressed in C3 (Figure 10D). The Figure 10E indicated that the expressions of FBXW1/2/4/5/7/9/11/12 were associated with immune subtype in KIRC. FBXW1/2/4/7 were higher expressed in C3. Meanwhile, the expression of FBXW5 was higher in C5 (Figure 10E). However, only FBXW5 and FBXW9 on the relation of immune subtypes have statistical significance in ACC, and FBXW11/12 was the same in GBM (sFigure 7). These results once again demonstrated the specificity of FBXW members in various tumors.
Previous study had demonstrated that FBXW1/7 were involved in the immune process in different ways (75–77). To further verify their immune role on the influence of tumor development and outcome, the association between immune cell infiltration levels and FBXWs expression was also evaluated. As indicated in Figure 11, most of FBXWs expression had significantly positive or negative relationship in these cancers, especially in BRCA and KIRC. FBXW1 expression was significantly negative association with immune-active cells such as NK, CD8-T, cytotoxic, and Th1, etc. However, FBXW7 as tumor suppressor factor had a positive correlation with immune-active cells and negative association with immune-suppressive cells such as Exhausted T cells, Th17, and macrophage (Figures 11A–D). Based on the above TME analysis results, we also investigated the correlation between FBXWs and infiltration score in ACC, GBM and LAML and found that it was not obvious (sFigure 8) In order to validate the results, we further investigate the relationship between FBXW1/7 expression and immune cells in pan-cancer. FBXW1 as an oncogene and FBXW7 as a tumor suppressive gene also show opposite relationships on immune infiltration cells (s.Figure 9). In addition, the TCGA database was used to conduct co-expression analysis, which can reveal the association between FBXW1/7 expression and immune checkpoints in pan-cancer. As shown in Figure 12A, FBXW1 was negatively correlated with these representative immune markers in ACC, BLCA, BRCA, PCPG, UCEC and UCS. However, FBXW7 was positively associated with these immune checkpoints in most cancers (Figure 12B). The opposite relationship between FBXW1 and FBXW7 in immune infiltration may be the reason for the different prognosis of cancer patients.
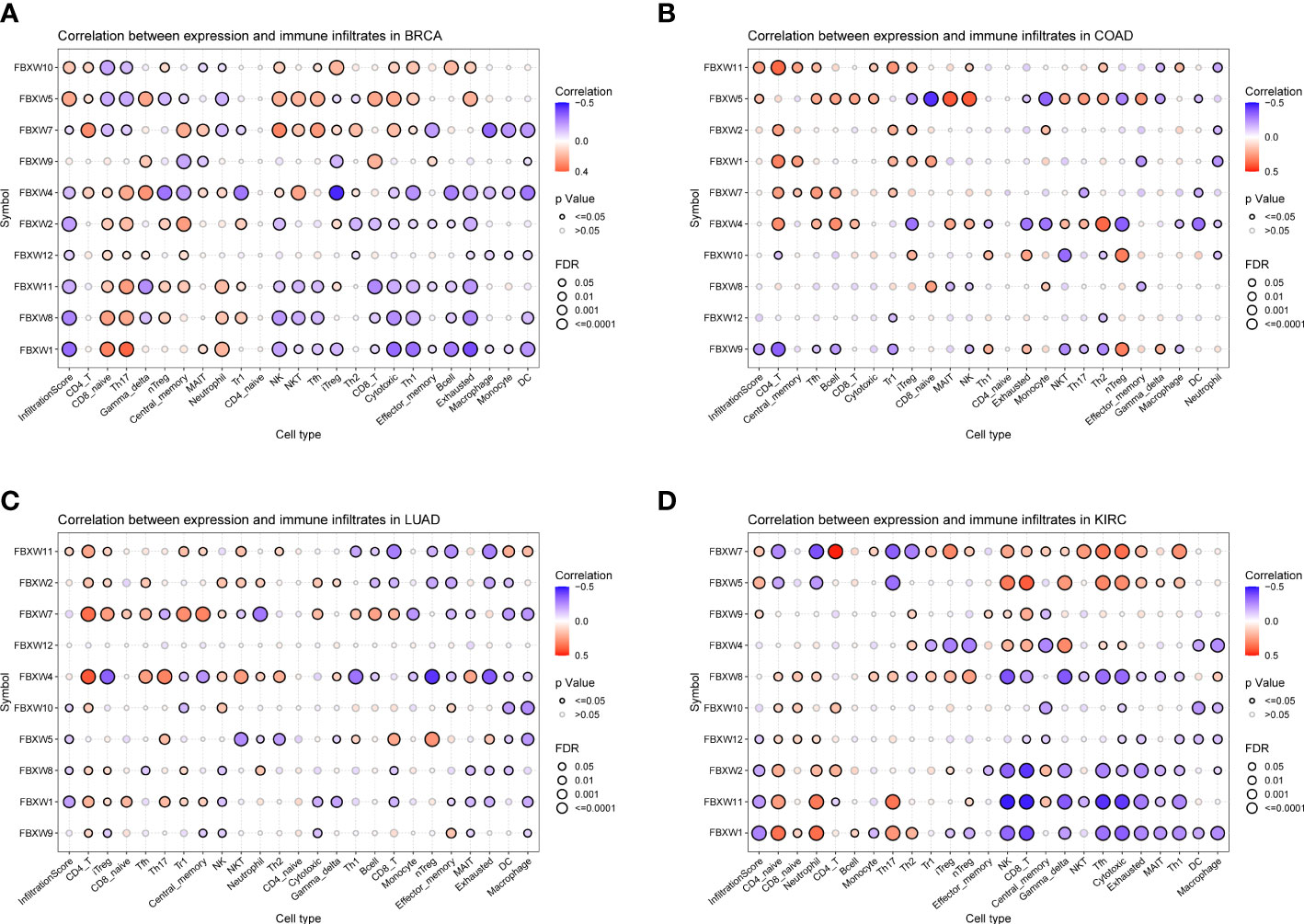
Figure 11 Association analysis of FBXW family gene expression with the immune-infiltration cells in four types of cancer. (A) in BRCA. (B) in COAD. (C) in LUAD. (D) in KIRC.
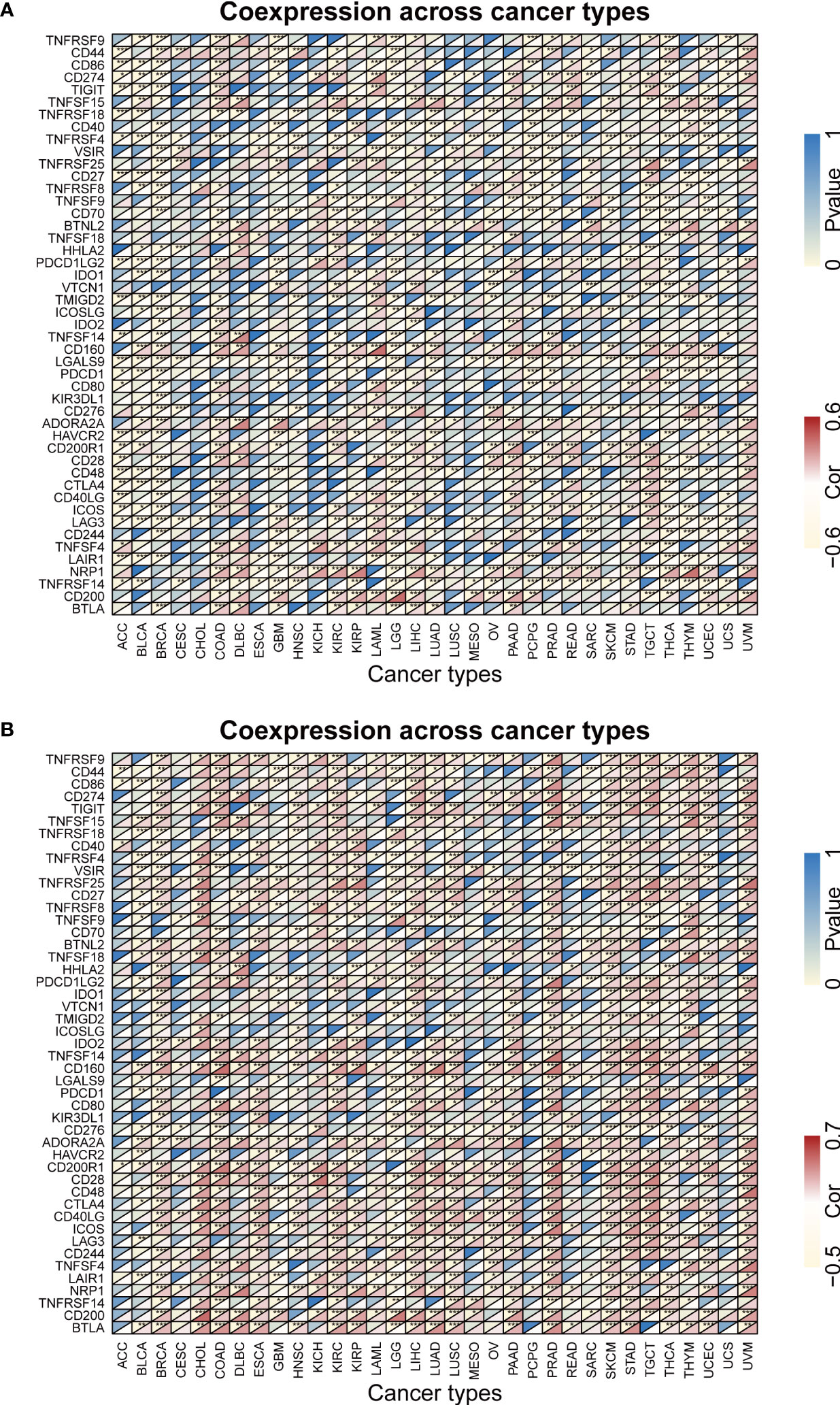
Figure 12 The relationship between FBXW1/FBXW7 and immune checkpoints. (A) Heatmap illustrating the relationship between FBXW1 and known immune checkpoints. (B) Heatmap illustrating the relationship between FBXW7 and known immune checkpoints. ∗P< 0.05, ∗∗P< 0.01, and ∗∗∗P< 0.001.
Drug sensitivity analysis of FBXW gene family
Drug resistance has always been an insurmountable problem in clinic treatment, and also a prominent factor in the development of adverse outcomes for many patients. Previous study reported that FBXWs has been validated to involve in drug resistance development through proteolytic regulation of substrates (20). For example, abnormal expression of FBXW7 could increase doxorubicin resistance in COAD, and in tamoxifen-resistant breast cells, FBXW1 expression was significantly upregulated.
Aiming to further explore the role of FBXWs in chemotherapy or targeted therapy, the integrated analysis between drug sensitivity and FBXW gene expression data was also performed in this study. The result from CellMiner™ was shown that FBXW7 expression had a positive relationship with drug sensitivity of Nelarabine and Chelerythrine. In addition, FBXW11 expression had a significantly negative relation with the drug sensitivity of Imatinib, LDK-378, and Palbociclib. FBXW9 expression was negatively associated with Dasatinib’s drug sensitivity and positively related to drug sensitivity of tfdu (sFigure 10). These results may bring some implications for clinical medication based on FBXWs drug sensitivity.
Discussion
In consideration of the vital roles of FBXW gene family in progression of numerous cancers, it is of great importance to study the expression patterns and prognostic effect of FBXW gene family in multiple cancers, which could facilitate tumor early diagnosis (78). In our study, we firstly performed gene structure and motif analysis of the FBXWs, and found that they have obvious individual differences which may lead to functional differences in FBXW genes. Then, we also summarized previous reports on the function of the FBXW family in multiple cancers, and found that FBXW1/5 act as oncogenes in many cancers, however, FBXW2/7 exert the antitumor properties (Table 1). Specifically, from the perspective of the transcription level, we found that FBXW1 was significantly up-regulated in six different cancers, which was consistent with most previous studies (79). Whereas the abnormal expression of FBXW2 was related to twelve different cancers, seven of which were upregulated and five were down-regulated. FBXW5/9/12 were overexpressed in most cancers, however, FBXW11 expression in pan-cancer was opposite. Notedly, FBXW7, as a tumor suppressor, was highly expressed in six cancers (CHOL, KIRC, KIRP, LIHC, LUAD and THCA) which may be one of the reasons affecting the outcome of these cancers. Besides, we analyzed the FBXWs expression in different tumor stages and grades. The results indicated the expression of most genes was significantly different in the development of various cancers, which inspired us to consider the roles of FBXW family genes as diagnosis and prognosis markers in pan-cancer.
We also summarized all meaningful survival-related results and found that FBXW members accounted for the most significant proportion in KIRC among the above mentioned four common cancers (BRCA, COAD, LUAD and KIRC) (Supplementary Table 1).Additionally, we also demonstrated that FBXW1 was a protective factor in LGG, which was consistent in different database. The overexpression of FBXW2 was related to worse OS in ACC, LGG, and PRAD. However, Zhou et al.’s study indicated that FBXW2 could be a tumor suppressor of PRAD by promoting EGFR ubiquitination and degradation (41), which seems to be contrary to our results. The reason for this phenomenon may be that FBXW2 itself is prone to mutation which could change the overall prognosis tendency (37), or it may be that FBXW2 needs to be combined with other ligand proteins to form SCF complexes, which determine carcinogenicity depending on the kind of ubiquitinated substrates, thereby playing itself function and maintaining itself activity (80–82). Therefore, artificial obvious changes in the expression level of FBXW2 in the tumor microenvironment may lead to different prognosis compared with the database analysis. Additionally, High expression of FBXW7 was significantly associated with longer survival in PAAD, STAD, HNSC, SKCM. Previous studies indicated that low expression of FBXW7 was a major cause of carcinogenesis and poor prognosis in Gastric cancer (51, 83), which was consistent with our findings. Up-regulation of FBXW11 expression was related to better prognosis in KIRC, LGG, and MESO. In short, these results suggested that FBXWs could be great prognosis-related markers in pan-cancer.
An increasing number of studies have showed that TME could exert great influence on tumor proliferation, metastasis, micro angiogenesis, and even immune escape (84–86). However, to date, the role of FBXWs in TME has not been elucidated. Our results indicated FBXW7 expression had a positive relationship with the levels of stromal score, immune score and estimate score in multiple tumors, suggesting FBXW7 is expected to efficient immunomodulatory factor. Furthermore, the correlation between immune subtypes and FBXWs expression was also examined in this study and found that most of FBXWs have a close relation with BRCA, LUAD and KIRC, however, in other cancers such as ACC and GBM, the correlations was weakened obviously. Similarly, the relation between FBXWs and infiltration score of immune cells in BRCA and KIRC was the most significant, suggesting FBXW family may be potential orientation in the immune-therapy of the mentioned four cancers. Then, we selected two most representative members FBXW1/7 for further immune infiltration analysis, and the reverse results on immune checkpoints and cells showed that they may induce diverse immune response and thereby cause markedly different endpoint of tumor progression.
In conclusion, our research unveiled complicated and comprehensive roles of FBXW family members expression in cancer progression and clinical outcome that warrant further investigation, suggesting FBXWs could be promising prognostic biomarkers in special tumors. More importantly, we identified the relationship between immune infiltration and individual members of the FBXW family in different cancers, thus providing a few direction for immunotherapy. This study, however, has some limitations. Our results derives mainly from the computational analysis of genomic data, and detailed functional mechanisms of FBXWs by vivo and vitro experiments are needed to build in the future. In addition, our study does have a limitation on the connection of FBXW family with immunotherapy. The role of FBXWs in immunotherapy should be further validated in clinical trials and cell experiments.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
ZY and ZYS designed the overall programming of this study and conducted comprehensive guidance. TH and XO drafted and revised the manuscript. TH and JL performed the data analysis. BS and ZDS participated in the data collection. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by grants from the National Natural Science Foundation of China (No. 81803044).
Acknowledgments
We thank the support of the grants from the National Natural Science Foundation of China (No. 81803044) and School of Medicine, Xiamen University, Xiamen, China.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.1084339/full#supplementary-material
References
1. Bray F, Laversanne M, Weiderpass E, Soerjomataram I. The ever-increasing importance of cancer as a leading cause of premature death worldwide. Cancer (2021) 127(16):3029–30. doi: 10.1002/cncr.33587
2. Cheek DM, Naxerova K. Mapping the long road to cancer. Cell (2022) 185(6):939–40. doi: 10.1016/j.cell.2022.02.020
3. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
4. Uddin S, Bhat AA, Krishnankutty R, Mir F, Kulinski M, Mohammad RM. Involvement of f-box proteins in progression and development of human malignancies. Semin Cancer Biol (2016) 36:18–32. doi: 10.1016/j.semcancer.2015.09.008
5. Zhang X, Linder S, Bazzaro M. Drug development targeting the ubiquitin-proteasome system (Ups) for the treatment of human cancers. Cancers (Basel) (2020) 12(4):902. doi: 10.3390/cancers12040902
6. Bonacci T, Emanuele MJ. Dissenting degradation: Deubiquitinases in cell cycle and cancer. Semin Cancer Biol (2020) 67(Pt 2):145–58. doi: 10.1016/j.semcancer.2020.03.008
7. Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem (1998) 67(1):425–79. doi: 10.1146/annurev.biochem.67.1.425
8. Park J, Cho J, Song EJ. Ubiquitin-proteasome system (Ups) as a target for anticancer treatment. Arch Pharm Res (2020) 43(11):1144–61. doi: 10.1007/s12272-020-01281-8
9. Burger AM, Seth AK. The ubiquitin-mediated protein degradation pathway in cancer: Therapeutic implications. Eur J Cancer (2004) 40(15):2217–29. doi: 10.1016/j.ejca.2004.07.006
10. Deshaies RJ. Scf and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol (1999) 15(1):435–67. doi: 10.1146/annurev.cellbio.15.1.435
11. Smith TF, Gaitatzes C, Saxena K, Neer EJ. The wd repeat: A common architecture for diverse functions. Trends Biochem Sci (1999) 24(5):181–5. doi: 10.1016/s0968-0004(99)01384-5
12. Wang Z, Liu P, Inuzuka H, Wei W. Roles of f-box proteins in cancer. Nat Rev Cancer (2014) 14(4):233–47. doi: 10.1038/nrc3700
13. Lau AW, Fukushima H, Wei W. The Fbw7 and betatrcp E3 ubiquitin ligases and their roles in tumorigenesis. Front Biosci (Landmark Ed) (2012) 17(6):2197–212. doi: 10.2741/4045
14. Müerköster S, Arlt A, Sipos B, Witt M, Grossmann M, Klöppel G, et al. Increased expression of the E3-ubiquitin ligase receptor subunit Betatrcp1 relates to constitutive nuclear factor-kappab activation and chemoresistance in pancreatic carcinoma cells. Cancer Res (2005) 65(4):1316–24. doi: 10.1158/0008-5472.Can-04-1626
15. Díaz VM, de Herreros AG. F-box proteins: Keeping the epithelial-to-Mesenchymal transition (Emt) in check. Semin Cancer Biol (2016) 36:71–9. doi: 10.1016/j.semcancer.2015.10.003
16. Wang Z, Inuzuka H, Zhong J, Wan L, Fukushima H, Sarkar FH, et al. Tumor suppressor functions of Fbw7 in cancer development and progression. FEBS Lett (2012) 586(10):1409–18. doi: 10.1016/j.febslet.2012.03.017
17. Zhan P, Wang Y, Zhao S, Liu C, Wang Y, Wen M, et al. Fbxw7 negatively regulates Eno1 expression and function in colorectal cancer. Lab Invest (2015) 95(9):995–1004. doi: 10.1038/labinvest.2015.71
18. O’Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, et al. Fbw7 mutations in leukemic cells mediate notch pathway activation and resistance to gamma-secretase inhibitors. J Exp Med (2007) 204(8):1813–24. doi: 10.1084/jem.20070876
19. Chang CC, Lin HH, Lin JK, Lin CC, Lan YT, Wang HS, et al. Fbxw7 mutation analysis and its correlation with clinicopathological features and prognosis in colorectal cancer patients. Int J Biol Markers (2015) 30(1):e88–95. doi: 10.5301/jbm.5000125
20. Tekcham DS, Chen D, Liu Y, Ling T, Zhang Y, Chen H, et al. F-box proteins and cancer: An update from functional and regulatory mechanism to therapeutic clinical prospects. Theranostics (2020) 10(9):4150–67. doi: 10.7150/thno.42735
21. Sun Y, Liu C, Liu Z, Zhao T, Jiang J, Li J, et al. Genome-wide identification, characterization and expression analysis of the jaz gene family in resistance to Gray leaf spots in tomato. Int J Mol Sci (2021) 22(18):9974. doi: 10.3390/ijms22189974
22. Han Y, Luthe D. Identification and evolution analysis of the jaz gene family in maize. BMC Genomics (2021) 22(1):256. doi: 10.1186/s12864-021-07522-4
23. Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, et al. Swiss-Model: Homology modelling of protein structures and complexes. Nucleic Acids Res (2018) 46(W1):W296–w303. doi: 10.1093/nar/gky427
24. Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, et al. Tbtools: An integrative toolkit developed for interactive analyses of big biological data. Mol Plant (2020) 13(8):1194–202. doi: 10.1016/j.molp.2020.06.009
25. Goldman MJ, Craft B, Hastie M, Repečka K, McDade F, Kamath A, et al. Visualizing and interpreting cancer genomics data Via the xena platform. Nat Biotechnol (2020) 38(6):675–8. doi: 10.1038/s41587-020-0546-8
26. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cbioportal. Sci Signal (2013) 6(269):pl1. doi: 10.1126/scisignal.2004088
27. Qin G, Wang X, Ye S, Li Y, Chen M, Wang S, et al. Npm1 upregulates the transcription of pd-L1 and suppresses T cell activity in triple-negative breast cancer. Nat Commun (2020) 11(1):1669. doi: 10.1038/s41467-020-15364-z
28. Dou XW, Liang YK, Lin HY, Wei XL, Zhang YQ, Bai JW, et al. Notch3 maintains luminal phenotype and suppresses tumorigenesis and metastasis of breast cancer Via trans-activating estrogen receptor-A. Theranostics (2017) 7(16):4041–56. doi: 10.7150/thno.19989
29. Kipreos ET, Pagano M. The f-box protein family. Genome Biol (2000) 1(5):Reviews3002. doi: 10.1186/gb-2000-1-5-reviews3002
30. Cardozo T, Pagano M. The scf ubiquitin ligase: Insights into a molecular machine. Nat Rev Mol Cell Biol (2004) 5(9):739–51. doi: 10.1038/nrm1471
31. Ougolkov A, Zhang B, Yamashita K, Bilim V, Mai M, Fuchs SY, et al. Associations among beta-trcp, an E3 ubiquitin ligase receptor, beta-catenin, and nf-kappab in colorectal cancer. J Natl Cancer Inst (2004) 96(15):1161–70. doi: 10.1093/jnci/djh219
32. Spiegelman VS, Slaga TJ, Pagano M, Minamoto T, Ronai Z, Fuchs SY. Wnt/Beta-catenin signaling induces the expression and activity of betatrcp ubiquitin ligase receptor. Mol Cell (2000) 5(5):877–82. doi: 10.1016/s1097-2765(00)80327-5
33. Koch A, Waha A, Hartmann W, Hrychyk A, Schüller U, Waha A, et al. Elevated expression of wnt antagonists is a common event in hepatoblastomas. Clin Cancer Res (2005) 11(12):4295–304. doi: 10.1158/1078-0432.Ccr-04-1162
34. Liu J, Suresh Kumar KG, Yu D, Molton SA, McMahon M, Herlyn M, et al. Oncogenic braf regulates beta-trcp expression and nf-kappab activity in human melanoma cells. Oncogene (2007) 26(13):1954–8. doi: 10.1038/sj.onc.1209994
35. Wei P, Jiang J, Xiao M, Zeng M, Liu X, Zhao B, et al. The transcript Enst00000444125 of lncrna Linc01503 promotes cancer stem cell properties of glioblastoma cells Via reducing Fbxw1 mediated Gli2 degradation. Exp Cell Res (2022) 412(1):113009. doi: 10.1016/j.yexcr.2022.113009
36. Belaïdouni N, Peuchmaur M, Perret C, Florentin A, Benarous R, Besnard-Guérin C. Overexpression of human beta Trcp1 deleted of its f box induces tumorigenesis in transgenic mice. Oncogene (2005) 24(13):2271–6. doi: 10.1038/sj.onc.1208418
37. Xu J, Zhou W, Yang F, Chen G, Li H, Zhao Y, et al. The B-Trcp-Fbxw2-Skp2 axis regulates lung cancer cell growth with Fbxw2 acting as a tumour suppressor. Nat Commun (2017) 8:14002. doi: 10.1038/ncomms14002
38. Yang F, Sun Y. Fbxw2 suppresses proliferation and invasion of lung cancer cells by targeting Skp2 and B-catenin. Mol Cell Oncol (2019) 6(4):1607458. doi: 10.1080/23723556.2019.1607458
39. Yang F, Xu J, Li H, Tan M, Xiong X, Sun Y. Fbxw2 suppresses migration and invasion of lung cancer cells Via promoting B-catenin ubiquitylation and degradation. Nat Commun (2019) 10(1):1382. doi: 10.1038/s41467-019-09289-5
40. Ren C, Han X, Lu C, Yang T, Qiao P, Sun Y, et al. Ubiquitination of nf-Kb P65 by Fbxw2 suppresses breast cancer stemness, tumorigenesis, and paclitaxel resistance. Cell Death differ (2022) 29(2):381–92. doi: 10.1038/s41418-021-00862-4
41. Zhou T, Chen T, Lai B, Zhang W, Luo X, Xia D, et al. Fbxw2 inhibits prostate cancer proliferation and metastasis Via promoting egfr ubiquitylation and degradation. Cell Mol Life Sci (2022) 79(5):268. doi: 10.1007/s00018-022-04320-3
42. Han Q, Zhang Q, Song H, Bamme Y, Song C, Ge Z. Fbxw4 is highly expressed and associated with poor survival in acute myeloid leukemia. Front Oncol (2020) 10:149. doi: 10.3389/fonc.2020.00149
43. Yeo MS, Subhash VV, Suda K, Balcıoğlu HE, Zhou S, Thuya WL, et al. Fbxw5 promotes tumorigenesis and metastasis in gastric cancer via activation of the fak-src signaling pathway. Cancers (Basel) (2019) 11(6):836. doi: 10.3390/cancers11060836
44. Yao Y, Liu Z, Huang S, Huang C, Cao Y, Li L, et al. The E3 ubiquitin ligase, Fbxw5, promotes the migration and invasion of gastric cancer through the dysregulation of the hippo pathway. Cell Death Discovery (2022) 8(1):79. doi: 10.1038/s41420-022-00868-y
45. Kim TY, Jackson S, Xiong Y, Whitsett TG, Lobello JR, Weiss GJ, et al. Crl4a-Fbxw5-Mediated degradation of Dlc1 rho gtpase-activating protein tumor suppressor promotes non-small cell lung cancer cell growth. Proc Natl Acad Sci United States America (2013) 110(42):16868–73. doi: 10.1073/pnas.1306358110
46. Li L, Sarver AL, Khatri R, Hajeri PB, Kamenev I, French AJ, et al. Sequential expression of mir-182 and mir-503 cooperatively targets Fbxw7, contributing to the malignant transformation of colon adenoma to adenocarcinoma. J Pathol (2014) 234(4):488–501. doi: 10.1002/path.4407
47. Wang X, Zhang J, Zhou L, Sun W, Zheng ZG, Lu P, et al. Fbxw7 regulates hepatocellular carcinoma migration and invasion Via Notch1 signaling pathway. Int J Oncol (2015) 47(1):231–43. doi: 10.3892/ijo.2015.2981
48. Zhou L, Wang DS, Li QJ, Sun W, Zhang Y, Dou KF. The down-regulation of Notch1 inhibits the invasion and migration of hepatocellular carcinoma cells by inactivating the cyclooxygenase-2/Snail/E-Cadherin pathway in vitro. Dig Dis Sci (2013) 58(4):1016–25. doi: 10.1007/s10620-012-2434-7
49. Matsuoka S, Oike Y, Onoyama I, Iwama A, Arai F, Takubo K, et al. Fbxw7 acts as a critical fail-safe against premature loss of hematopoietic stem cells and development of T-all. Genes Dev (2008) 22(8):986–91. doi: 10.1101/gad.1621808
50. Yokobori T, Yokoyama Y, Mogi A, Endoh H, Altan B, Kosaka T, et al. Fbxw7 mediates chemotherapeutic sensitivity and prognosis in nsclcs. Mol Cancer Res (2014) 12(1):32–7. doi: 10.1158/1541-7786.Mcr-13-0341
51. Yokobori T, Mimori K, Iwatsuki M, Ishii H, Onoyama I, Fukagawa T, et al. P53-altered Fbxw7 expression determines poor prognosis in gastric cancer cases. Cancer Res (2009) 69(9):3788–94. doi: 10.1158/0008-5472.Can-08-2846
52. Milne AN, Leguit R, Corver WE, Morsink FH, Polak M, de Leng WW, et al. Loss of Cdc4/Fbxw7 in gastric carcinoma. Cell Oncol (2010) 32(5-6):347–59. doi: 10.3233/clo-2010-523
53. Li J, Guo Y, Liang X, Sun M, Wang G, De W, et al. Microrna-223 functions as an oncogene in human gastric cancer by targeting Fbxw7/Hcdc4. J Cancer Res Clin Oncol (2012) 138(5):763–74. doi: 10.1007/s00432-012-1154-x
54. Eto K, Iwatsuki M, Watanabe M, Ishimoto T, Ida S, Imamura Y, et al. The sensitivity of gastric cancer to trastuzumab is regulated by the mir-223/Fbxw7 pathway. Int J Cancer (2015) 136(7):1537–45. doi: 10.1002/ijc.29168
55. Gong J, Cui Z, Li L, Ma Q, Wang Q, Gao Y, et al. Microrna-25 promotes gastric cancer proliferation, invasion, and migration by directly targeting f-box and wd-40 domain protein 7, Fbxw7. Tumour Biol (2015) 36(10):7831–40. doi: 10.1007/s13277-015-3510-3
56. Calhoun ES, Jones JB, Ashfaq R, Adsay V, Baker SJ, Valentine V, et al. Braf and Fbxw7 (Cdc4, Fbw7, ago, Sel10) mutations in distinct subsets of pancreatic cancer: Potential therapeutic targets. Am J Pathol (2003) 163(4):1255–60. doi: 10.1016/s0002-9440(10)63485-2
57. Gao J, Azmi AS, Aboukameel A, Kauffman M, Shacham S, Abou-Samra AB, et al. Nuclear retention of Fbw7 by specific inhibitors of nuclear export leads to Notch1 degradation in pancreatic cancer. Oncotarget (2014) 5(11):3444–54. doi: 10.18632/oncotarget.1813
58. Kuiper RP, Vreede L, Venkatachalam R, Ricketts C, Kamping E, Verwiel E, et al. The tumor suppressor gene Fbxw7 is disrupted by a constitutional T(3;4)(Q21;Q31) in a patient with renal cell cancer. Cancer Genet cytogenet (2009) 195(2):105–11. doi: 10.1016/j.cancergencyto.2009.07.001
59. Fu Y, Lin Y, Yang Z, Yang G, Li G, Liu Y, et al. Fbxw7 overexpression suppresses renal cancer cell proliferation and induces apoptosis. Med Oncol (Northwood London England) (2015) 32(8):215. doi: 10.1007/s12032-015-0656-1
60. Chang H, Liu YH, Wang LL, Wang J, Zhao ZH, Qu JF, et al. Mir-182 promotes cell proliferation by suppressing Fbxw7 and Fbxw11 in non-small cell lung cancer. Am J Trans Res (2018) 10(4):1131–42.
61. Koh MS, Ittmann M, Kadmon D, Thompson TC, Leach FS. Cdc4 gene expression as potential biomarker for targeted therapy in prostate cancer. Cancer Biol Ther (2006) 5(1):78–83. doi: 10.4161/cbt.5.1.2290
62. Wang H, Chen Y, Lin P, Li L, Zhou G, Liu G, et al. The Cul7/F-box and wd repeat domain containing 8 (Cul7/Fbxw8) ubiquitin ligase promotes degradation of hematopoietic progenitor kinase 1. J Biol Chem (2014) 289(7):4009–17. doi: 10.1074/jbc.M113.520106
63. Wu P, Wang F, Wang Y, Men H, Zhu X, He G, et al. Significance of Fbx8 in progression of gastric cancer. Exp Mol Pathol (2015) 98(3):360–6. doi: 10.1016/j.yexmp.2015.03.015
64. Zheng Z, Hong D, Zhang X, Chang Y, Sun N, Lin Z, et al. Uc.77- downregulation promotes colorectal cancer cell proliferation by inhibiting Fbxw8-mediated Cdk4 protein degradation. Front Oncol (2021) 11:673223. doi: 10.3389/fonc.2021.673223
65. Yao J, Yang J, Yang Z, Wang XP, Yang T, Ji B, et al. Fbxw11 contributes to stem-Cell-Like features and liver metastasis through regulating Hic1-mediated Sirt1 transcription in colorectal cancer. Cell Death Dis (2021) 12(10):930. doi: 10.1038/s41419-021-04185-7
66. Wang L, Feng W, Yang X, Yang F, Wang R, Ren Q, et al. Fbxw11 promotes the proliferation of lymphocytic leukemia cells through the concomitant activation of nf-Kb and B-Catenin/Tcf signaling pathways. Cell Death Dis (2018) 9(4):427. doi: 10.1038/s41419-018-0440-1
67. Fuchs SY, Spiegelman VS, Kumar KG. The many faces of beta-trcp E3 ubiquitin ligases: Reflections in the magic mirror of cancer. Oncogene (2004) 23(11):2028–36. doi: 10.1038/sj.onc.1207389
68. Zhang Q, Zheng J, Liu L. The long noncoding RNA Pcgem1 promotes cell proliferation, migration and invasion Via targeting the mir-182/Fbxw11 axis in cervical cancer. Cancer Cell Int (2019) 19:304. doi: 10.1186/s12935-019-1030-8
69. Zhang Q, Yin X, Zhang Y. Microrna-221 promotes cell proliferation and inhibits apoptosis in osteosarcoma cells by directly targeting Fbxw11 and regulating wnt signaling. Arch Med Res (2021) 52(2):191–9. doi: 10.1016/j.arcmed.2020.10.017
70. Chen C, Zhou H, Zhang X, Liu Z, Ma X. Association of Fbxw11 levels with tumor development and prognosis in chondrosarcoma. Cancer biomark: Sect A Dis Markers (2022) 1–9. doi: 10.3233/cbm-210426
71. Kim CJ, Song JH, Cho YG, Kim YS, Kim SY, Nam SW, et al. Somatic mutations of the beta-trcp gene in gastric cancer. Apmis (2007) 115(2):127–33. doi: 10.1111/j.1600-0463.2007.apm_562.x
72. Saitoh T, Katoh M. Expression profiles of Betatrcp1 and Betatrcp2, and mutation analysis of Betatrcp2 in gastric cancer. Int J Oncol (2001) 18(5):959–64.
73. Wang S, Ji J, Song J, Li X, Han S, Lian W, et al. Microrna-182 promotes pancreatic cancer cell proliferation and migration by targeting B-Trcp2. Acta Biochim Biophys Sin (Shanghai) (2016) 48(12):1085–93. doi: 10.1093/abbs/gmw105
74. Maiuthed A, Chantarawong W, Chanvorachote P. Lung cancer stem cells and cancer stem cell-targeting natural compounds. Anticancer Res (2018) 38(7):3797–809. doi: 10.21873/anticanres.12663
75. Li X, Wen D, Li X, Yao C, Chong W, Chen H. Identification of an immune signature predicting prognosis risk and lymphocyte infiltration in colon cancer. Front Immunol (2020) 11:1678. doi: 10.3389/fimmu.2020.01678
76. Shen JZ, Qiu Z, Wu Q, Zhang G, Harris R, Sun D, et al. A Fbxo7/Eya2-Scf(Fbxw7) axis promotes axl-mediated maintenance of mesenchymal and immune evasion phenotypes of cancer cells. Mol Cell (2022) 82(6):1123–39.e8. doi: 10.1016/j.molcel.2022.01.022
77. Li CW, Lim SO, Xia W, Lee HH, Chan LC, Kuo CW, et al. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat Commun (2016) 7:12632. doi: 10.1038/ncomms12632
78. Lan R, Jin B, Liu YZ, Zhang K, Niu T, You Z. Genome and transcriptome profiling of fbxw family in human prostate cancer. Am J Clin Exp Urol (2020) 8(4):116–28.
79. Frescas D, Pagano M. Deregulated proteolysis by the f-box proteins Skp2 and B-trcp: Tipping the scales of cancer. Nat Rev Cancer (2008) 8(6):438–49. doi: 10.1038/nrc2396
80. Yang CS, Yu C, Chuang HC, Chang CW, Chang GD, Yao TP, et al. Fbw2 targets gcma to the ubiquitin-proteasome degradation system. J Biol Chem (2005) 280(11):10083–90. doi: 10.1074/jbc.M413986200
81. Chiang MH, Chen LF, Chen H. Ubiquitin-conjugating enzyme Ube2d2 is responsible for Fbxw2 (F-box and wd repeat domain containing 2)-mediated human Gcm1 (Glial cell missing homolog 1) ubiquitination and degradation. Biol Reprod (2008) 79(5):914–20. doi: 10.1095/biolreprod.108.071407
82. Zhang F, Zhao Y, Sun Y. Usp2 is an Skp2 deubiquitylase that stabilizes both Skp2 and its substrates. J Biol Chem (2021) 297(4):101109. doi: 10.1016/j.jbc.2021.101109
83. Akhoondi S, Sun D, von der Lehr N, Apostolidou S, Klotz K, Maljukova A, et al. Fbxw7/Hcdc4 is a general tumor suppressor in human cancer. Cancer Res (2007) 67(19):9006–12. doi: 10.1158/0008-5472.Can-07-1320
84. Altorki NK, Markowitz GJ, Gao D, Port JL, Saxena A, Stiles B, et al. The lung microenvironment: An important regulator of tumour growth and metastasis. Nat Rev Cancer (2019) 19(1):9–31. doi: 10.1038/s41568-018-0081-9
85. Fane M, Weeraratna AT. How the ageing microenvironment influences tumour progression. Nat Rev Cancer (2020) 20(2):89–106. doi: 10.1038/s41568-019-0222-9
Keywords: FBXW family, pan-cancer, prognosis, immune infiltration, therapeutic target
Citation: Huang T, OuYang X, Li J, Shi B, Shan Z, Shi Z and Yang Z (2022) Pan-cancer analysis of FBXW family with potential implications in prognosis and immune infiltration. Front. Immunol. 13:1084339. doi: 10.3389/fimmu.2022.1084339
Received: 30 October 2022; Accepted: 06 December 2022;
Published: 14 December 2022.
Edited by:
Jinghua Pan, Jinan University, ChinaReviewed by:
Mi Deng, Peking University, ChinaSuresh Veeramani, The University of Iowa, United States
Copyright © 2022 Huang, OuYang, Li, Shi, Shan, Shi and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiyuan Shi, cXkyMDEwc2hpemhpeXVhbkAxMjYuY29t; Zhangru Yang, YXJpZXM4NzAzQDE2My5jb20=
†These authors have contributed equally to this work
‡Present address: XIaoxiao OuYang, Department of pathology, Sichuan Academy of Medical Sciences & Sichuan Provincial People's Hospital, Chengdu, China
 Tingting Huang
Tingting Huang XIaoxiao OuYang
XIaoxiao OuYang Jiwei Li
Jiwei Li Bingbing Shi3
Bingbing Shi3 Zhiyuan Shi
Zhiyuan Shi Zhangru Yang
Zhangru Yang