- 1Manchester Fungal Infection Group, School of Biological Sciences, Faculty of Biology, Medicine and Health, The University of Manchester, Manchester, United Kingdom
- 2Department of Medicine and Surgery, University of Perugia, Perugia, Italy
The first perspective: The host genetics
The outcome of fungal disease is determined by complex interactions between fungal pathogens, human hosts and their environment including the host microbiome (1–4). Morbidity and mortality in fungal disease remain very high despite recent advances in the diagnostic and treatment of these conditions (5–7). There are only three classes of antifungal drugs available to treat these disease and, antifungal resistance linked to the use of agricultural use of triazole fungicides is on the rise (8). The development of new antifungal drugs to treat human fungal disease is challenging as both, host and pathogen are eukaryotes and, there are different potential druggable targets exposed at different points of fungal morphogenesis.
So far, the identification of high-risk patients for fungal disease has relied on the use of clinical scores that combine the use of clinical and host factors to predict the risk of subsequent disease (9–11). However, the prevalence of opportunistic fungal diseases within at-risk population, ranges from 0.1 – 20% (12). In the last decades, individual genetic variation has been recognised as a major contribution of functional immune responses against fungal pathogens. Several monogenic defects and polymorphisms in genes regulating antifungal immunity or pathogen sensing have been associated with susceptibility to aspergillosis, cryptococcosis and candidiasis (13) (Figure 1).
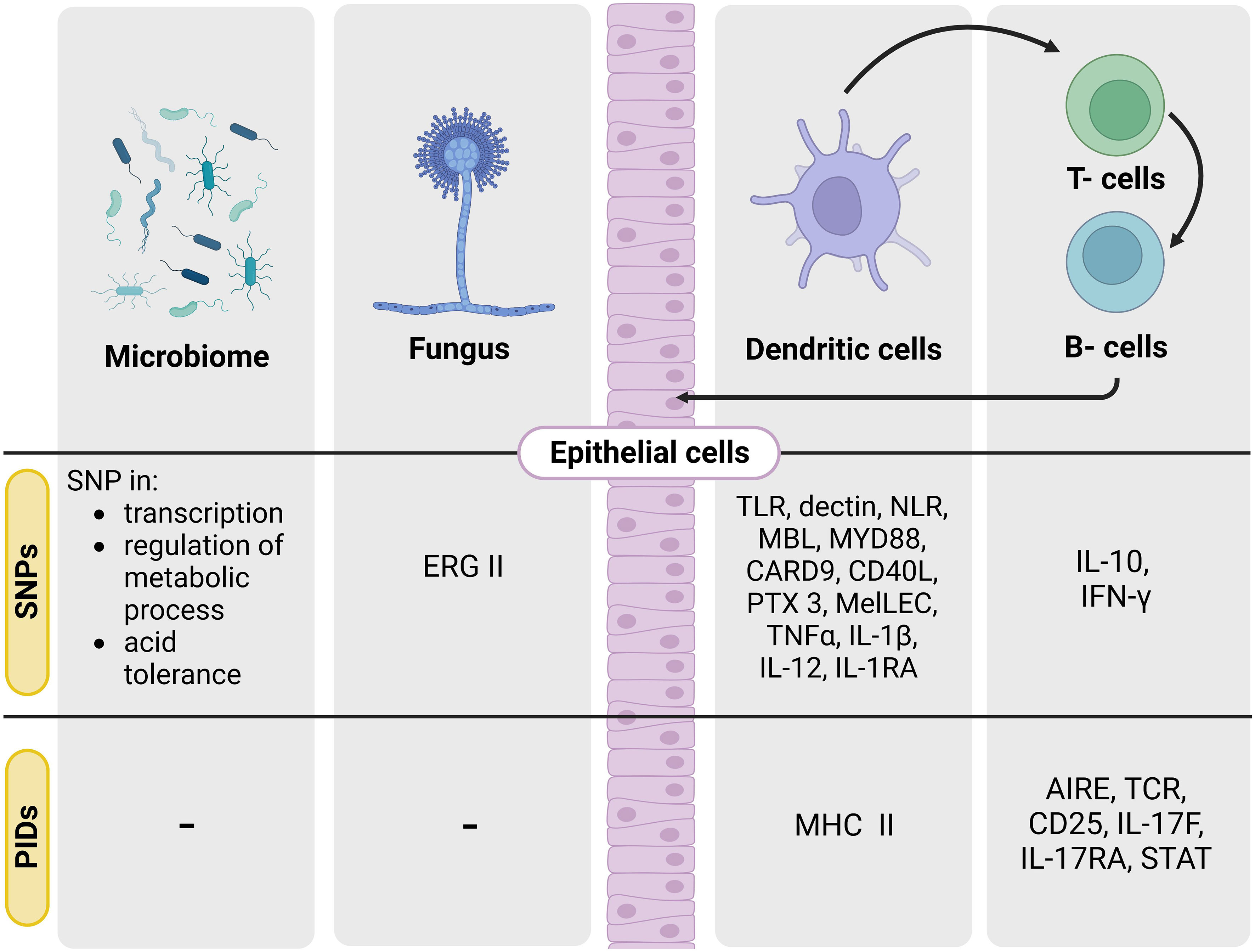
Figure 1 SNPs and PIDs known in the ‘genetic triangle’ leading to opportunistic human fungal infections. Scheme of the main target genes involved in SNPs and PIDs leading to the main described human fungal diseases. Single nucleotide polymorphisms (SNPs), primary immunodeficiencies (PIDs).
Sensing of human fungal pathogens by the host immune system requires the interplay between pathogen-associated molecular patterns (PAMPs), mostly located in the cell wall of fungal pathogens, and pattern recognition receptors (PRRs) (14–17). The interaction between PRRs and PAMPs, leads to the regulation of uptake of fungal pathogens by immune cells. In addition to membrane receptors, soluble PRRs such as pentraxins or mannose binding lectins (MBLs) are also critical for pathogen sensing and efficient phagocytosis (18, 19).
To date, polymorphisms in PTX3 have been reported in different clinical settings as a risk factor for invasive pulmonary aspergillosis in haematopoietic stem cell transplant recipients (20), solid organ transplants (21) and chronic obstructive pulmonary disease (22). Using ex vitro an in vivo models of disease it has been reported that PTX3 deficiency increases susceptibility to A. fumigatus infection due to impaired neutrophil function (20, 23).
Polymorphisms in PRRs and other immune pathways have been reported in different patient cohorts (24–28). Nevertheless, none of these polymorphisms (except in the case of primary immunodeficiencies) allow to predict risk of fungal disease with high specificity suggesting the genetic basis of these diseases may be polygenic.
Despite genetic replication studies are in general scarce, the link between genetic polymorphisms in PTX3 (rs1840680) and rs7309123 (CLEC7a) and aspergillosis risk have been successfully replicated (29). White et al. (30) recently explored whether screening for genetic variants in genes previously linked with susceptibility to invasive aspergillosis alongside clinical factors and mycological evidence could be used to improve aspergillosis risk stratification in patients undergoing allogeneic stem cell transplantation. In their model, they reported that mutations in Dectin-1, DC-SIGN, allogeneic stem cell transplantation, current respiratory viral infection and Aspergillus-specific positive PCR were all high-risk factors for the development of invasive disease.
An increasing number of case studies and family studies have reported fungal disease in children with primary immunodeficiencies. For example, invasive aspergillosis has been linked with inborn errors in patients with chronic granulomatous disease, severe congenital neutropenia or leukocyte adhesion deficiency type I (31, 32). Moreover, other less common congenital immunodeficiencies (e.g., CARD9 immunity, IL-12/interferon (IFN)-γ axis or IL-17 immunity) have been described to increase susceptibility risk to invasive candidiasis, dermatophytosis, chronic mucocutaneous candidiasis or endemic mycoses (Figure 1) (33–36).
Genome-wide association studies (GWAS) have allowed us to identify a number of novel genetic loci affecting susceptibility to fungal infections (37, 38). A GWAS study of patients with common infections revealed a significant association between DSG1 variants and susceptibility to vulvovaginal candidiasis. DSG1 encodes for a desmoglein, a critical protein involved in maintaining the integrity of the epithelial compartment (37, 39).
GWAS in patients with candidemia revealed a strong association between the genetic variant rs8028958 in the PLA2GB4 gene and susceptibility to disease (40). PLA2GB4 encodes a cytosolic phospholipase A2 involved in lipid metabolism, affecting cytokine production in the presence of Candida in the bloodstream (40).
Combining the use of GWAS, bulk RNA-seq and scRNA-seq from human PBMCs upon Candida stimulation, a recent study suggested a critical role of LY86 in susceptibility to candidemia. LY86 encodes for Lymphocyte Antigen 86, mainly expressed in monocytes. LY86 silencing impairs monocyte migration, increasing susceptibility to candidemia (38).
The role of genetic variation in genes encoding for host cytokine responses has been extensively studied including data in the Human Functional Genomics Project (41). In this study, 17 new genome-wide significant loci that influence cytokine production were identified (41). In vitro studies of human PBMCs challenged with fungi demonstrated a high inter-individual variation in cytokine release (IL-6, TNF-α, IL-1β) (39). Thus, suggesting many genome-wide quantitative trait locus (QTLs) might contribute to susceptibility to infectious. Interestingly, this study shows that the QTLs are not affecting adaptive cytokines as IL-17 (41).
How far are we from implementing host genetic screening in the diagnostic pipelines for fungal disease? Studies aiming to characterise the genetic basis of fungal disease have been based on association studies with either disease and common polymorphisms in genes known to be important for efficient antifungal responses such as those involved in antigen presentation, pathogen sensing, or regulation of immune pathways. Even though these associations are not surprisingly significant, they are present in the general population. In addition, rigorous clinical definitions for some diseases such as allergic and chronic forms of aspergillosis or more recently viral-associated fungal disease have not been available until recently thus, hampering the usefulness of genetic risk to predict susceptibility to fungal disease. To overcome this issue, whole genome exome or genome sequencing studies might be useful (42–44). However, a joint effort from the scientific community should be made to optimise and simplify bioinformatic pipelines. Finally, implementation of host genetic screening in the diagnostic pipelines for fungal disease would require validation in large and well-characterised cohort of patients with different genetic backgrounds and the development of point of care testing approaches that would allow the transference of these technologies to those regions where the prevalence of fungal disease is particularly high.
The second perspective: The pathogen genetics
Most of what we know about the pathogenicity mechanisms used by fungal species to cause disease has arisen from in vivo or in vitro infection models in which a particular fungal species, clinical strain or deletion mutant is assessed for virulence. However, results are very much dependent on the animal strain used, the model of disease (e.g., immunosuppression vs no immunosuppression), the dose or route of infection, or the cell population assessed thus, results are not always translated into human disease. In addition, to understand the opportunistic nature of most fungal human pathogens, it is important to consider that genetic drivers of virulence have probably been developed so fungal pathogens can survive in their natural environments (45). In fact, in a recent publication using population genomics, it was observed that human infections caused by drug resistant A. fumigatus have their origin in the environment (46).
With the increasing number of sequenced fungal genomes it has been observed that pathogenicity emerged in different lineages in the fungal kingdom (43). However, there is a huge variation in fungal drivers of human disease among pathogens but also strains from the same pathogen. For example, virulence of A. fumigatus strains is significantly different depending on the infection model of disease used (47). Nevertheless, it seems that there is a link between the capacity of a fungal pathogen to adapt to extreme environments and their capacity to cause disease.
The human mould pathogen A. fumigatus can cause invasive, chronic or allergic diseases in immunosuppressed patients or those with a chronic respiratory condition (48). In fact, Snelders et al. (49),using whole genome sequencing of fungal isolates from patients with cystic fibrosis and chronic pulmonary aspergillosis demonstrated than parasexual recombination is critical for A. fumigatus adaptation and might also be a driver for the development of azole resistance beyond the occurrence of point mutations in the CYP51 (Erg11) (Figure 1) (50). Similarly, Ballard et al. (51) reported that long-term Aspergillus infection in patients with chronic granulomatous disease is driven by host microevolution (51). Moreover, recent analyses of fungal pangenomes has shown that A. fumigatus environmental isolates do not differ in their gene content (52). However, it has been shown an increased number of accessory genes in clinical isolates compared to environmental that might help to better understand human disease (53).
Fungal species of the same genera can also cause disease to different populations as for Cryptococcus neoformans and Cryptococcus gattii. However, within each of these species there is a significant genomic and phenotypic heterogeneity (53, ) that can be linked with disease outcomes. Similarly, there is a significant genomic and phenotypic variability within C. albicans and some loss of function mutations might help to better understand genetic drivers of disease (54).
Recently, we have discovered a new model where the metabolic route of tryptophan degradation, as well as the total amount available of tryptophan, differently affect fungal virulence. Fungi express the tryptophan degrading enzyme Indoleamine 2,3-dioxygenases that degrade l-tryptophan to kynurenines. Aspergillus fumigatus possesses three ido genes that are expressed under hypoxia or tryptophan abundance. Loss of ido genes increases fungal pathogenicity due to the activation of the tryptophan-degrading enzyme AroH (55).
The third wheel: The host microbiome genetics
Until recently, the contribution of the environment to the development of fungal diseases has been mainly linked to ecological factors such geographic distribution, climate or the existence of a possible zoonotic reservoir (45, 56). However, there is an increasing number of studies suggesting that the host microbiome, is crucial in driving resistance against fungal disease (57). In particular, host xenobiotic receptors (XRs) activated by metabolism may affect susceptibility to fungal infection (58, 59). Indeed, several factors such as tissue microenvironment, diet, nutrient availability or antibiotic exposure are known to affect the microbiome evolution and microbial SNPs (60, 61). Probiotics may also acquire SNPs when several stressors act in particular microbiome niches (Figure 1) (62).
Fungal-bacteria interactions in clinically relevant contexts such as oral, gut and respiratory dysbiosis have been increasingly studied and, both synergistic and antagonistic interactions have been reported. Several studies have shown that Candida albicans germination and virulence can be directly or indirectly regulated by bacteria such as Lactobacillus spp. For example, lactobacilli release quorum sensing molecules or antifungal molecules (e.g., hydrogen peroxide or organic acids) to prevent fungal growth (63). Similarly, we recently found that Lactobacillus reuteri reduces C. albicans gut colonization via metabolic activation of specific bacterial gene cluster and the release of indole-derivatives (59). A similar phenotype has been shown in vulvovaginal candidiasis (64). In oral mucositis, Candida spp adheres to Streptococcus biofilms by increasing the expression of Als1 or Als5 genes (65). In addition, C. albicans and S. mutants interact in biofilm formation in which C. albicans-induced expression of S. mutans glucosyltransferase B, facilitating pathogen-pathogen binding (66, 67). L. crispatus SNUV220 and L. fermentum SNUV175 supernatant downregulates the expression of the hypha-related genes ALS3, ECE1, SAP5 and HWP1 in C. albicans (68). In an independent study, it was shown that L. plantarum SD5870, L. helveticus CBS N116411 and S. salivarius DSM 14685 also inhibit Candida yeast-hypha transition (69).
The combinatorial impact of host genetic variation and pathogen genetics in the outcome of fungal disease has been overlooked. Only recently, these two approaches have been integrated by using the dual RNA sequencing in infectious diseases (70). This approach, that has previously been used for plant-host interaction studies, consists in performing parallel transcriptomic analysis of pathogens and their eukaryotic host cells (71). Thus, multi-organism RNA-seq, may be applied to the human population bearing opportunistic fungal infections, eventually co-infected with other pathogens. Moreover, human genetic variants may be analysed alongside fungal genetic variability by using integrated GWAS approaches as reported for meningitis (72) and, it could be potentially expanded to define microbiome genetic variants. This systems biology approach will enable us to define the role of genetic variation in the host, microbiome and the pathogen with a view to improving our understanding of the complexity of the human ecosystem during infection (Figure 2).
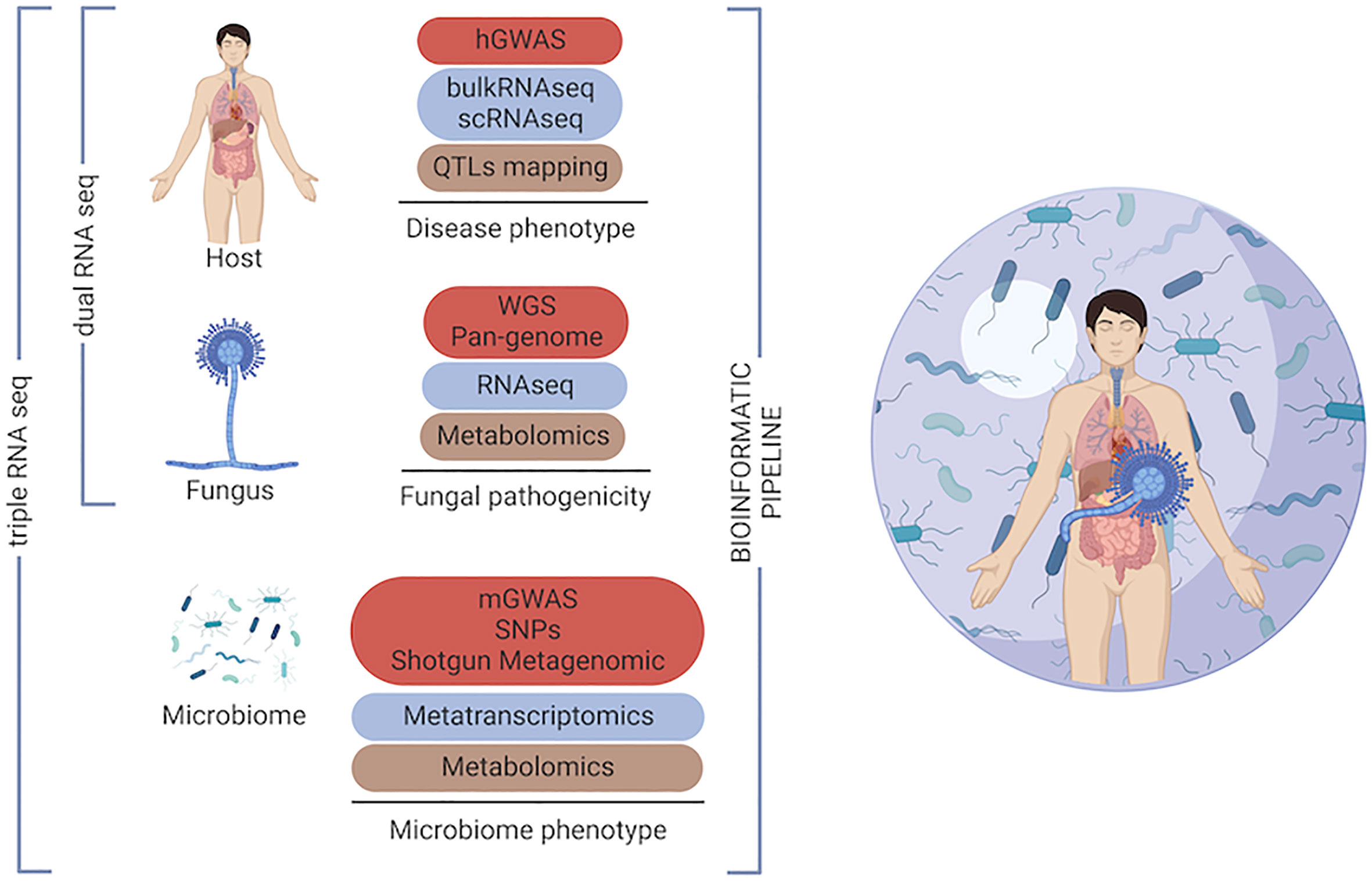
Figure 2 Omics toolkit for investigating the ‘genetic triangle’ in the human host during fungal infection. Multi-omics approaches, which enable intermediate phenotypes into the host, the fungus and the host microbiome to be measured by different -omics technologies. Genome-wide association studies (GWAS), quantitative trait locus (QTLs), single nucleotide polymorphism (SNP).
Author contributions
MM, CF critically read, analyzed, and discussed the literature and conceived the outline of the manuscript. SG and TZ wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
SG was co-funded by the National Institute for Health Research (NIHR) Manchester Biomedical Research Centre (https://www.manchesterbrc.nihr.ac.uk/), the Fungal Infection Trust and the Dowager Countess Eleanor Peel Trust. TZ was supported by the Fondazione per la Ricerca sulla Fibrosi Cistica Onlus (FFC#15/2022). Figures created with Biorender.
Acknowledgments
We thank Dott. Leonardo Brizzi for digital art and editorial assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Ingrosso G, Saldi S, Marani S, Wong AYW, Bertelli M, Aristei C, et al. Breakdown of symbiosis in radiation-induced oral mucositis. J Fungi (Basel) (2021) 7(4):290. doi: 10.3390/jof7040290
2. Hong S, Sun Y, Sun D, Wang C. Microbiome assembly on drosophila body surfaces benefits the flies to combat fungal infections. iScience (2022) 25:104408. doi: 10.1016/j.isci.2022.104408
3. Roudbary M, Kumar S, Kumar A, Cernakova L, Nikoomanesh F, Rodrigues CF. Overview on the prevalence of fungal infections, immune response, and microbiome role in COVID-19 patients. J Fungi (Basel) (2021) 7(9):720. doi: 10.3390/jof7090720
4. Kwok AJ, Mentzer A, Knight JC. Host genetics and infectious disease: new tools, insights and translational opportunities. Nat Rev Genet (2021) 22:137–53. doi: 10.1038/s41576-020-00297-6
5. Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. Hidden killers: human fungal infections. Sci Transl Med (2012) 4:165rv113. doi: 10.1126/scitranslmed.3004404
6. Jenks JD, Cornely OA, Chen SC, Thompson GR. 3rd, hoenigl m: Breakthrough invasive fungal infections: Who is at risk? Mycoses (2020) 63:1021–32. doi: 10.1111/myc.13148
7. Fisher MC, Alastruey-Izquierdo A, Berman J, Bicanic T, Bignell EM, Bowyer P, et al. Tackling the emerging threat of antifungal resistance to human health. Nat Rev Microbiol (2022) 20(9):557–71. doi: 10.1038/s41579-022-00720-1
8. Langfeldt A, Gold JAW, Chiller T. Emerging fungal infections: from the fields to the clinic, resistant aspergillus fumigatus and dermatophyte species: a one health perspective on an urgent public health problem. Curr Clin Microbiol Rep (2022) 9:46–51. doi: 10.1007/s40588-022-00181-3
9. Donnelly JP, Chen SC, Kauffman CA, Steinbach WJ, Baddley JW, Verweij PE, et al. Revision and update of the consensus definitions of invasive fungal disease from the European organization for research and treatment of cancer and the mycoses study group education and research consortium. Clin Infect Dis (2020) 71:1367–76. doi: 10.1093/cid/ciz1008
10. Leon C, Ruiz-Santana S, Saavedra P, Almirante B, Nolla-Salas J, Alvarez-Lerma F, et al. A bedside scoring system (“Candida score”) for early antifungal treatment in nonneutropenic critically ill patients with candida colonization. Crit Care Med (2006) 34:730–7. doi: 10.1097/01.CCM.0000202208.37364.7D
11. Sun Y, Hu J, Huang H, Chen J, Li J, Ma J, et al. Clinical risk score for predicting invasive fungal disease after allogeneic hematopoietic stem cell transplantation: Analysis of the China assessment of antifungal therapy in hematological diseases (CAESAR) study. Transpl Infect Dis (2021) 23:e13611. doi: 10.1111/tid.13611
12. Bongomin F, Gago S, Oladele RO, Denning DW. Global and multi-national prevalence of fungal diseases-estimate precision. J Fungi (Basel) (2017) 3(4):57. doi: 10.3390/jof3040057
13. Romani L, Puccetti P. Protective tolerance to fungi: the role of IL-10 and tryptophan catabolism. Trends Microbiol (2006) 14:183–9. doi: 10.1016/j.tim.2006.02.003
14. Bojang E, Ghuman H, Kumwenda P, Hall RA. Immune sensing of candida albicans. J Fungi (Basel) (2021) 7(2):119. doi: 10.3390/jof7020119
15. Campuzano A, Wormley FL. Innate immunity against cryptococcus, from recognition to elimination. J Fungi (Basel) (2018) 4(1):33. doi: 10.3390/jof4010033
16. van de Veerdonk FL, Gresnigt MS, Romani L, Netea MG, Latge JP. Aspergillus fumigatus morphology and dynamic host interactions. Nat Rev Microbiol (2017) 15:661–74. doi: 10.1038/nrmicro.2017.90
17. Patin EC, Thompson A, Orr SJ. Pattern recognition receptors in fungal immunity. Semin Cell Dev Biol (2019) 89:24–33. doi: 10.1016/j.semcdb.2018.03.003
18. Parente R, Possetti V, Erreni M, D’Autilia F, Bottazzi B, Garlanda C, et al. Complementary roles of short and long pentraxins in the complement-mediated immune response to aspergillus fumigatus infections. Front Immunol (2021) 12:785883. doi: 10.3389/fimmu.2021.785883
19. Riwes MM, Leather H, Neal D, Bennett C, Sugrue M, Cline C, et al. Association of mannose-binding lectin levels and invasive fungal disease in hematologic malignancy patients receiving myelosuppressive chemotherapy or allogeneic hematopoietic stem cell transplantation. Bone Marrow Transplant (2016) 51:1228–32. doi: 10.1038/bmt.2016.92
20. Cunha C, Aversa F, Lacerda JF, Busca A, Kurzai O, Grube M, et al. Genetic PTX3 deficiency and aspergillosis in stem-cell transplantation. N Engl J Med (2014) 370:421–32. doi: 10.1056/NEJMoa1211161
21. Wojtowicz A, Lecompte TD, Bibert S, Manuel O, Rueger S, Berger C, et al. PTX3 polymorphisms and invasive mold infections after solid organ transplant. Clin Infect Dis (2015) 61:619–22. doi: 10.1093/cid/civ386
22. He Q, Li H, Rui Y, Liu L, He B, Shi Y, et al. Pentraxin 3 gene polymorphisms and pulmonary aspergillosis in chronic obstructive pulmonary disease patients. Clin Infect Dis (2018) 66:261–7. doi: 10.1093/cid/cix749
23. Garlanda C, Hirsch E, Bozza S, Salustri A, De Acetis M, Nota R, et al. Non-redundant role of the long pentraxin PTX3 in anti-fungal innate immune response. Nature (2002) 420:182–6. doi: 10.1038/nature01195
24. Cunha C, Carvalho A. Genetic defects in fungal recognition and susceptibility to invasive pulmonary aspergillosis. Med Mycol (2019) 57:S211–8. doi: 10.1093/mmy/myy057
25. Kang Y, Yu Y, Lu L. The role of pentraxin 3 in aspergillosis: Reality and prospects. Mycobiology (2020) 48:1–8. doi: 10.1080/12298093.2020.1722576
26. Maskarinec SA, Johnson MD, Perfect JR. Genetic susceptibility to fungal infections: What is in the genes? Curr Clin Microbiol Rep (2016) 3:81–91. doi: 10.1007/s40588-016-0037-3
27. Onyishi CU, May RC. Human immune polymorphisms associated with the risk of cryptococcal disease. Immunology (2022) 165:143–57. doi: 10.1111/imm.13425
28. Smeekens SP, van de Veerdonk FL, Kullberg BJ, Netea MG. Genetic susceptibility to candida infections. EMBO Mol Med (2013) 5:805–13. doi: 10.1002/emmm.201201678
29. Fisher CE, Hohl TM, Fan W, Storer BE, Levine DM, Zhao LP, et al. Validation of single nucleotide polymorphisms in invasive aspergillosis following hematopoietic cell transplantation. Blood (2017) 129:2693–701. doi: 10.1182/blood-2016-10-743294
30. White PL, Parr C, Barnes RA. Predicting invasive aspergillosis in hematology patients by combining clinical and genetic risk factors with early diagnostic biomarkers. J Clin Microbiol (2018) 56(1):e01122–17. doi: 10.1128/JCM.01122-17
31. Lanternier F, Cypowyj S, Picard C, Bustamante J, Lortholary O, Casanova JL, et al. Primary immunodeficiencies underlying fungal infections. Curr Opin Pediatr (2013) 25:736–47. doi: 10.1097/MOP.0000000000000031
32. Lionakis MS. Primary immunodeficiencies and invasive fungal infection: when to suspect and how to diagnose and manage. Curr Opin Infect Dis (2019) 32:531–7. doi: 10.1097/QCO.0000000000000593
33. Egri N, Esteve-Sole A, Deya-Martinez A, Ortiz de Landazuri I, Vlagea A, Garcia AP, et al. Primary immunodeficiency and chronic mucocutaneous candidiasis: pathophysiological, diagnostic, and therapeutic approaches. Allergol Immunopathol (Madr) (2021) 49:118–27. doi: 10.15586/aei.v49i1.20
34. Corvilain E, Casanova JL, Puel A. Inherited CARD9 deficiency: Invasive disease caused by ascomycete fungi in previously healthy children and adults. J Clin Immunol (2018) 38:656–93. doi: 10.1007/s10875-018-0539-2
35. Johnson ME, Rojas-Moreno C, Salzer W, Regunath H. Disseminated histoplasmosis in a patient with common variable immunodeficiency: A coincidence or the result of T cell defects? IDCases (2017) 10:105–7. doi: 10.1016/j.idcr.2017.10.004
36. Rouzaud C, Hay R, Chosidow O, Dupin N, Puel A, Lortholary O, et al. Severe dermatophytosis and acquired or innate immunodeficiency: A review. J Fungi (Basel) (2015) 2(1):4. doi: 10.3390/jof2010004
37. Tian C, Hromatka BS, Kiefer AK, Eriksson N, Noble SM, Tung JY, et al. Genome-wide association and HLA region fine-mapping studies identify susceptibility loci for multiple common infections. Nat Commun (2017) 8:599. doi: 10.1038/s41467-017-00257-5
38. de Vries DH, Matzaraki V, Bakker OB, Brugge H, Westra HJ, Netea MG, et al. Integrating GWAS with bulk and single-cell RNA-sequencing reveals a role for LY86 in the anti-candida host response. PloS Pathog (2020) 16:e1008408. doi: 10.1371/journal.ppat.1008408
39. Fan W, Kan H, Liu HY, Wang TL, He YN, Zhang M, et al. Association between human genetic variants and the vaginal bacteriome of pregnant women. mSystems (2021) 6:e0015821. doi: 10.1128/mSystems.00158-21
40. Jaeger M, Matzaraki V, Aguirre-Gamboa R, Gresnigt MS, Chu X, Johnson MD, et al. A genome-wide functional genomics approach identifies susceptibility pathways to fungal bloodstream infection in humans. J Infect Dis (2019) 220:862–72. doi: 10.1093/infdis/jiz206
41. Li Y, Oosting M, Smeekens SP, Jaeger M, Aguirre-Gamboa R, Le KTT, et al. A functional genomics approach to understand variation in cytokine production in humans. Cell (2016) 167:1099–1110 e1014. doi: 10.1016/j.cell.2016.10.017
42. Kannambath S, Jarvis JN, Wake RM, Longley N, Loyse A, Matzaraki V, et al. Genome-wide association study identifies novel colony stimulating factor 1 locus conferring susceptibility to cryptococcosis in human immunodeficiency virus-infected south africans. Open Forum Infect Dis (2020) 7:ofaa489. doi: 10.1093/ofid/ofaa489
43. Gago S, Overton NLD, Ben-Ghazzi N, Novak-Frazer L, Read ND, Denning DW, et al. Lung colonization by aspergillus fumigatus is controlled by ZNF77. Nat Commun (2018) 9:3835. doi: 10.1038/s41467-018-06148-7
44. Overton NLD, Brakhage AA, Thywissen A, Denning DW, Bowyer P. Mutations in EEA1 are associated with allergic bronchopulmonary aspergillosis and affect phagocytosis of aspergillus fumigatus by human macrophages. PloS One (2018) 13:e0185706. doi: 10.1371/journal.pone.0185706
45. Rokas A. Evolution of the human pathogenic lifestyle in fungi. Nat Microbiol (2022) 7:607–19. doi: 10.1038/s41564-022-01112-0
46. Rhodes J, Abdolrasouli A, Dunne K, Sewell TR, Zhang Y, Ballard E, et al. Population genomics confirms acquisition of drug-resistant aspergillus fumigatus infection by humans from the environment. Nat Microbiol (2022) 7:663–74. doi: 10.1038/s41564-022-01091-2
47. Keizer EM, Valdes ID, Forn-Cuni G, Klijn E, Meijer AH, Hillman F, et al. Variation of virulence of five aspergillus fumigatus isolates in four different infection models. PloS One (2021) 16:e0252948. doi: 10.1371/journal.pone.0252948
48. Kosmidis C, Denning DW. The clinical spectrum of pulmonary aspergillosis. Thorax (2015) 70:270–7. doi: 10.1136/thoraxjnl-2014-206291
49. Engel T, Verweij PE, van den Heuvel J, Wangmo D, Zhang J, Debets AJM, et al. Parasexual recombination enables aspergillus fumigatus to persist in cystic fibrosis. ERJ Open Res (2020) 6(4):00020–2020. doi: 10.1183/23120541.00020-2020
50. Zhang J, Li L, Lv Q, Yan L, Wang Y, Jiang Y. The fungal CYP51s: Their functions, structures, related drug resistance, and inhibitors. Front Microbiol (2019) 10:691. doi: 10.3389/fmicb.2019.00691
51. Ballard E, Melchers WJG, Zoll J, Brown AJP, Verweij PE, Warris A. In-host microevolution of aspergillus fumigatus: A phenotypic and genotypic analysis. Fungal Genet Biol (2018) 113:1–13. doi: 10.1016/j.fgb.2018.02.003
52. Horta MAC, Steenwyk JL, Mead ME, Dos Santos LHB, Zhao S, Gibbons JG, et al. Examination of genome-wide ortholog variation in clinical and environmental isolates of the fungal pathogen aspergillus fumigatus. mBio (2022) 13(4):e0151922. doi: 10.1101/2022.03.23.485522
53. Barber AE, Sae-Ong T, Kang K, Seelbinder B, Li J, Walther G, et al. Aspergillus fumigatus pan-genome analysis identifies genetic variants associated with human infection. Nat Microbiol (2021) 6:1526–36. doi: 10.1038/s41564-021-00993-x
54. Hirakawa MP, Martinez DA, Sakthikumar S, Anderson MZ, Berlin A, Gujja S, et al. Genetic and phenotypic intra-species variation in candida albicans. Genome Res (2015) 25:413–25. doi: 10.1101/gr.174623.114
55. Zelante T, Choera T, Beauvais A, Fallarino F, Paolicelli G, Pieraccini G, et al. Aspergillus fumigatus tryptophan metabolic route differently affects host immunity. Cell Rep (2021) 34:108673. doi: 10.1016/j.celrep.2020.108673
56. Rodrigues AM, de Hoog GS, de Camargo ZP. Sporothrix species causing outbreaks in animals and humans driven by animal-animal transmission. PloS Pathog (2016) 12:e1005638. doi: 10.1371/journal.ppat.1005638
57. Huang D, Li H, Lin Y, Lin J, Li C, Kuang Y, et al. Antibiotic-induced depletion of clostridium species increases the risk of secondary fungal infections in preterm infants. Front Cell Infect Microbiol (2022) 12:981823. doi: 10.3389/fcimb.2022.981823
58. Lee WJ, Hase K. Gut microbiota-generated metabolites in animal health and disease. Nat Chem Biol (2014) 10:416–24. doi: 10.1038/nchembio.1535
59. Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity (2013) 39:372–85. doi: 10.1016/j.immuni.2013.08.003
60. Zhu X, Qin J, Tan C, Ning K. The seasonal changes of the gut microbiome of the population living in traditional lifestyles are represented by characteristic species-level and functional-level SNP enrichment patterns. BMC Genomics (2021) 22:83. doi: 10.1186/s12864-021-07372-0
61. Hua X, Song L, Yu G, Vogtmann E, Goedert JJ, Abnet CC, et al. MicrobiomeGWAS: A tool for identifying host genetic variants associated with microbiome composition. Genes (Basel) (2022) 13(7):1224. doi: 10.3390/genes13071224
62. Huang S, Jiang S, Huo D, Allaband C, Estaki M, Cantu V, et al. Candidate probiotic lactiplantibacillus plantarum HNU082 rapidly and convergently evolves within human, mice, and zebrafish gut but differentially influences the resident microbiome. Microbiome (2021) 9:151. doi: 10.1186/s40168-021-01102-0
63. MacAlpine J, Daniel-Ivad M, Liu Z, Yano J, Revie NM, Todd RT, et al. A small molecule produced by lactobacillus species blocks candida albicans filamentation by inhibiting a DYRK1-family kinase. Nat Commun (2021) 12:6151. doi: 10.1038/s41467-021-26390-w
64. Coudeyras S, Jugie G, Vermerie M, Forestier C. Adhesion of human probiotic lactobacillus rhamnosus to cervical and vaginal cells and interaction with vaginosis-associated pathogens. Infect Dis Obstet Gynecol (2008) 2008:549640. doi: 10.1155/2008/549640
65. Klotz SA, Gaur NK, De Armond R, Sheppard D, Khardori N, Edwards JE Jr., et al. Candida albicans als proteins mediate aggregation with bacteria and yeasts. Med Mycol (2007) 45:363–70. doi: 10.1080/13693780701299333
66. Hwang G, Liu Y, Kim D, Li Y, Krysan DJ, Koo H. Candida albicans mannans mediate streptococcus mutans exoenzyme GtfB binding to modulate cross-kingdom biofilm development in vivo. PloS Pathog (2017) 13:e1006407. doi: 10.1371/journal.ppat.1006407
67. Strus M, Kucharska A, Kukla G, Brzychczy-Wloch M, Maresz K, Heczko PB. The in vitro activity of vaginal lactobacillus with probiotic properties against candida. Infect Dis Obstet Gynecol (2005) 13:69–75. doi: 10.1080/10647440400028136
68. Jang SJ, Lee K, Kwon B, You HJ, Ko G. Vaginal lactobacilli inhibit growth and hyphae formation of candida albicans. Sci Rep (2019) 9:8121. doi: 10.1038/s41598-019-44579-4
69. James KM, MacDonald KW, Chanyi RM, Cadieux PA, Burton JP. Inhibition of candida albicans biofilm formation and modulation of gene expression by probiotic cells and supernatant. J Med Microbiol (2016) 65:328–36. doi: 10.1099/jmm.0.000226
70. Guan Y, Chen M, Ma Y, Du Z, Yuan N, Li Y, et al. Whole-genome and time-course dual RNA-seq analyses reveal chronic pathogenicity-related gene dynamics in the ginseng rusty root rot pathogen ilyonectria robusta. Sci Rep (2020) 10:1586. doi: 10.1038/s41598-020-58342-7
71. Seelbinder B, Wallstabe J, Marischen L, Weiss E, Wurster S, Page L, et al. Triple RNA-seq reveals synergy in a human virus-fungus Co-infection model. Cell Rep (2020) 33:108389. doi: 10.1016/j.celrep.2020.108389
Keywords: Fungal infections, Aspergillus fumigatus, Candida albicans, microbiome, immunodeficiency
Citation: Gago S, Mandarano M, Floridi C and Zelante T (2023) Host, pathogenic fungi and the microbiome: A genetic triangle in infection. Front. Immunol. 13:1078014. doi: 10.3389/fimmu.2022.1078014
Received: 23 October 2022; Accepted: 28 December 2022;
Published: 17 January 2023.
Edited by:
Agostinho Carvalho, University of Minho, PortugalReviewed by:
Anutthaman Parthasarathy, University of Bradford, United KingdomCopyright © 2023 Gago, Mandarano, Floridi and Zelante. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Teresa Zelante, dGVyZXNhLnplbGFudGVAdW5pcGcuaXQ=
 Sara Gago
Sara Gago Martina Mandarano
Martina Mandarano Claudia Floridi2
Claudia Floridi2 Teresa Zelante
Teresa Zelante