
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Immunol. , 08 December 2022
Sec. B Cell Biology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.1072702
This article is part of the Research Topic Mechanisms and Strategies of Unconventional Antibody Diversification for Greater Immune Adaptability View all 5 articles
The diversity of three hypervariable loops in antibody heavy chain and light chain, termed the complementarity-determining regions (CDRs), defines antibody’s binding affinity and specificity owing to the direct contact between the CDRs and antigens. These CDR regions typically contain tyrosine (Tyr) residues that are known to engage in both nonpolar and pi stacking interaction with antigens through their complementary aromatic ring side chains. Nearly two decades ago, sulfotyrosine residue (sTyr), a negatively charged Tyr formed by Golgi-localized membrane-bound tyrosylprotein sulfotransferases during protein trafficking, were also found in the CDR regions and shown to play an important role in modulating antibody-antigen interaction. This breakthrough finding demonstrated that antibody repertoire could be further diversified through post-translational modifications, in addition to the conventional genetic recombination. This review article summarizes the current advances in the understanding of the Tyr-sulfation modification mechanism and its application in potentiating protein-protein interaction for antibody engineering and production. Challenges and opportunities are also discussed.
Antibody therapeutics represent the largest group of biologic drugs with over 100 approved products on the market, providing a significant treatment impact on cancer, immune-mediated disorders, infectious diseases, and cardiovascular/hemostasis disorders (1, 2). Antibodies have favorable attributes such as metabolic stability, specificity, and potency, which can be further enhanced by protein engineering (3–5). A new wave of multispecific innovation (6–10) as well as recent speedy development of antibody products targeting SARS-CoV-2 (2, 11, 12) have demonstrated antibody therapeutics’ important roles in addressing complex disease pathobiology and global health challenges.
Human immunoglobulin G (IgG) is the dominant antibody drug format for recombinant therapeutic antibodies, among the isotypes of IgA, IgD, IgE and IgM (13, 14). IgGs are heterodimers with two identical heavy chains (HC) and light chains (LC) containing constant domains and variable domains (VH or VL). An intact antibody molecule consists of two fragment antigen binding domains (Fabs) and the fragment crystallizable (Fc) that carries out effector function through binding to different Fc receptor proteins on effector cells or by activating immune mediators like complements (13–16). In the Fab regions, the domains of VL and VH are paired to form the antigen binding site, in which three regions of sequence variability termed the complementarity-determining regions (CDRs) are the hypervariable loops in contact with antigens (13, 14). The paratope at these six CDRs from HC and LC determines antibody’s binding affinity and specificity to the epitope on the antigens.
The diversity of antibody CDRs is mainly generated through the genetic processes of V(variable), D (diversity), and J (joining) recombination and somatic hypermutation, occurred during B cell development in the bone marrow and upon encounters of antigen in the periphery (17–20). The introduction of combinatorial and junctional diversity in the V regions, as well as point mutations in the rearranged V regions after contact of naïve or memory B cells with antigens, create the diversity of antibody repertoires (19–21). Further investigations have revealed that the immune systems can employ post-translational modifications (PTMs) to increase antibody diversification. The first such concrete example is tyrosine (Tyr) sulfation in which a negatively charged sulfo group is added to the phenol group of Tyr through an O4-sulfate ester (22–24) (Figure 1A). Choe and colleagues first reported that anti-HIV-1 gp120 antibodies’ CDRH3s were Tyr-sulfated that modulated specifically the antibody-antigen interaction (29). This finding demonstrated that Tyr sulfation could be part of the natural humoral immune response to viral infection (30). Subsequently, several other types of unconventional mechanisms which utilize various PTMs to regulate humoral immune responses and antibody diversification have been discovered [reviewed by (31)]. Over the past twenty years, a significant understanding upon the molecular mechanisms of Tyr-sulfation modification and the biological function of the resulting sulfo-Tyr (sTyr) has been shown in protein-protein interaction for peptide hormones, cell surface receptors, virus entry, chemokine signaling, and blood clotting enzymes [see recent reviews, (23–26, 32, 33)]. New examples and mechanistic insights for the role of sTyr in modulating antibody diversification have been reported (28, 34–47). This review focuses on the comprehensive summary of these recent progresses and their potential impacts on applying sTyr in antibody engineering and production. Challenges and perspective for therapeutics development are discussed.
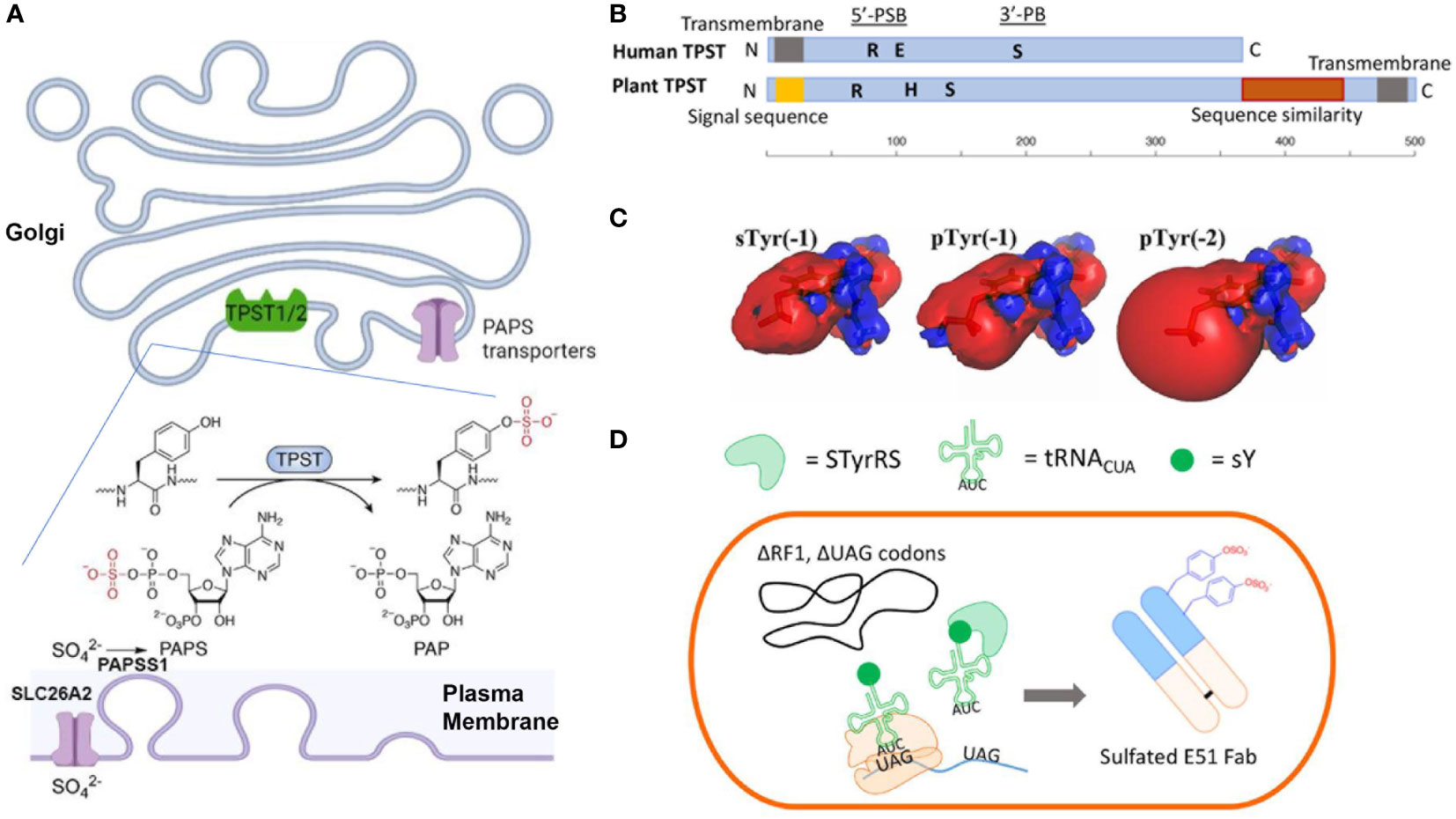
Figure 1 (A) Mechanisms of Tyr-sulfation in mammalian cells. (B) Primary structures of human TPST and plant TPST (25, 26) (created with BioRender.com). Conserved residue Lys (R) in 5’-PSB, Ser (S) in 3’-PB, and catalytic base residue His (H) and Glu (E) are depicted. (C) Comparison of electrostatic potentials for sTyr, pTyr (-1) and pTyr (-2) (27). (D) Production of sTyr-containing Fab with extended genetic codon technologies (28).
The tyrosylprotein sulfotransferases (TPSTs) pathway (48, 49) is the key synthesis pathway for Tyr-sulfation in antibody. Biological utilization of inorganic sulfate for sTyr, and other sulfated biomolecules (sulfated carbohydrates, or neurotransmitters), requires the metabolic activation into adenosine-5’-phosphosulfate (APS) by ATP sulfurylase (ATPS) and further into 3’-phosphoadenosine-5’-phosphosulfate (PAPS) by APS kinase (22, 23, 25, 26, 32, 33, 50). In animal cells, the APS and ATPS are fused to form a single bifunctional PAPS synthase (PAPSS). The inorganic sulfate ion for the PAPS synthesis is transported from outside into the cytosol through plasma membrane transporter SLC26A2 (Figure 1A) (51, 52). Then the PAPS is translocated into the Golgi lumen through the PAPS transporters SLC35B2 and SLC35B3 (53, 54). PAPS is the main activated sulfate group donor for sulfotransferases that catalyze biological sulfation. TPSTs is a major member of sulfotransferases, which also include cytosolic sulfotransferases for sulfation of hormones, neurotransmitters, as well as drugs and xenobiotics (32, 55–57) and membrane-bound carbohydrate sulfotransferase for the sulfation of glycolipid, glycoproteins, and proteoglycans (56, 58). The Golgi-localized membrane-bound TPSTs mediates Tyr-sulfation in antibodies, many other secreted proteins and integral membrane proteins (48, 49, 59, 60).
Tyr sulfation was first reported nearly seventy years ago (61), yet its physiological significance and link to diseases haven’t begun to be appreciated until the cloning of TPST genes more than two decades ago (48, 49). TPSTs are widely expressed in most tissues, and conserved from worm to mammals, and plants but not in yeast and most of the prokaryotes (22, 23, 25, 26). Human TPST1 and 2 exhibit different expression patterns, e.g., TPST1 is more in testis and TPST2 is more in the blood, trachea, thyroid gland, and several other organs (25). These two isoform enzymes also have different substrate specificities (62–64) and distinct mouse knockout phenotypes (65, 66), indicating a difference in specific biological functions (65–67). Animal TPSTs are type II membrane glycoproteins with a length of around 370 amino acids (25, 26). Like those of the cytoplasmic sulfotransferases and saccharide sulfotransferases, the enzymatic domain of TPSTs contains two short sequence motifs, termed 5’-phosphosulfate binding motif (PSB) and 3’-phosphate binding (PB) motif, and a catalytic base residue (His or Glu). They also contain spacing sequences with varying lengths among the functional domains (25, 26, 68) (Figure 1B). These enzymes are optimally active at slightly acidic pHs, consistent with their Golgi localization.
There is no unambiguous consensus sequence motif defined for prediction of Tyr sulfation sites in proteins. The common predictors include Tyr residues flanked by acidic residues, large accessible surface areas, and flexible or disordered secondary structure regions fitting well into the cleft of TPSTs. There are several in silico algorithm tools for predicting sulfation, such as the Sulfinator algorithm (69), the SulfoSite (70), The PredSulSite (71), and the Sulfotyrosine site (72). The current publicly available active online tool is Sulfinator (http://web.expasy.org/sulfinator) that has been frequently utilized. Antibodies are underrepresented in these machine learning algorithms, and the data of accuracy in predicting Tyr-sulfation for antibodies remain limited. It is well known that sTyr residues are neighboring with acidic amino acid (73). Presence of acidic residues, glutamic and aspartic acids, within +5 to -5 positions of the Tyr increase the activity of TPSTs (74, 75). These residues make multiple electrotactic interactions proximal to the active site of TPSTs (76–78). A basic residue in the amino-terminal (-1) position of the Tyr abolishes sulfation 76), as the negative charge of glutamic acid at this position is recognized by the backbone amide nitrogen of TSPTs (78). The presence of Gly or Asn residues surrounding sTyr tends to increase sulfation (62). The role of structural flexibility in sulfation regions could imply that the solvent-exposed and flexible CDRs is more likely Tyr-sulfated than the framework regions in antibodies (79).
Through determining protein-protein interaction, Tyr-sulfation has been involved in many biological processes, including blood coagulation, leukocyte rolling, complement cascade, hormonal regulation, viral infection, chemokine signaling, collagen binding, and Wnt signaling (22, 23, 25, 26, 80). These examples in nature illustrate the intricate mechanisms through which sTyr is utilized in protein-protein interactions, providing a guiding principle for the application of sTyr residue [For details see a recent review by (26)].
As a bulky amino acid that can facilitate hydrophobic interaction, Tyr residue is frequently found in CDR regions of antibodies and sulfation can presumably occur to some of these Tyr residues in the CDR loops. Hence discovering sTyr in antibody repertoire shouldn’t be a surprise in principle, yet the finding didn’t happen until 20 years ago. This is probably due to the fact that sTyr is heat labile and can be rapidly hydrolyzed under strong acidic condition. These features made sTyr residue difficult to be identified reliably with analytical tools such as standard Edman sequencing and mass spectrometry (23, 26). The conclusive identification of sTyr residues in antibody CDRs by Choe and colleagues was through traditional radioactive labeling using 35S-sulfate (29). A set of patient-derived human monoclonal antibodies against HIV-1 envelope glycoprotein gp120 was metabolically labeled with 35S-sulfate in transformed human cell lines during recombinant expression. Only the HCs were labeled and subsequently Tyr residues in CDRH3 were found sulfated. These sTyr residues contribute directly to antibody engagement with gp120, as in the case of CCR5 with overlapping regions in gp120. Despite structural differences between the Fab and CCR5 N-terminus, one sTyr residue in either the Fab or CCR5 is recognized in a similar manner by gp120 (35), indicating that the antibody mimics mechanistically the CCR5 for interacting with gp120. Interestingly, these sulfated antibodies were obtained from three different individuals. sTyr residues in two of these sulfated antibodies were originated from a common VH gene V1-69, whereas those in other three sulfated antibodies were from the longest of six HC joining gene JH6. This landmark finding demonstrated for the first time that Tyr-sulfation can contribute to the potency and diversity of human antibody repertoire.
There are additional antibodies with sTyr being reported (Table 1). Consistent with the knowledge that CDRH3 is much more diverse because of the VDJ gene rearrangement versus V gene only for CDRH1/2, CDRL1/2, and V gene and J gene recombination for CDRL3 (21), there are more sTyr found in the CDRH3. There are a number of examples of clustered sTyr found in CDRH3 (Table 1). Some of them were not functional whereas some contributed to the potency and diversity of the antibody. Huang et al. (34) hypothesized that the CDRH3 could be the only CDR long enough and diverse enough to be a substrate for Tyr sulfation. This is further supported by the fact that Tyr is often a bias outcome of the VDJ rearrangement as many IGHD genes have nucleotide sequence TAC (codon for Tyr) at their 3’ end (21), which could further skew toward a potential sTyr site. Interestingly, the non-functional Tyr-sulfation is ordered whereas those functional are disordered and therefore might be flexible enough for stereochemical orientation. This is consistent with the fact that there are quite a few Fab alone structures but no high-resolution structure of functional sTyr-containing Fabs complexed with the target antigens (Figure 2A). Regarding the activity, both antibody PG9 and PG16 contained sTyr residue in a unique hammerhead CDRH3 subdomain (Table 1), which showed significantly more potency on neutralization than its unsulfated form (36). The specific electrostatic interactions were made between cationic residues of gp120 and sTyr at antibody PG9, as the Tyr sulfate provides a closer match than the standard acidic Asp and Glu side chains (37).
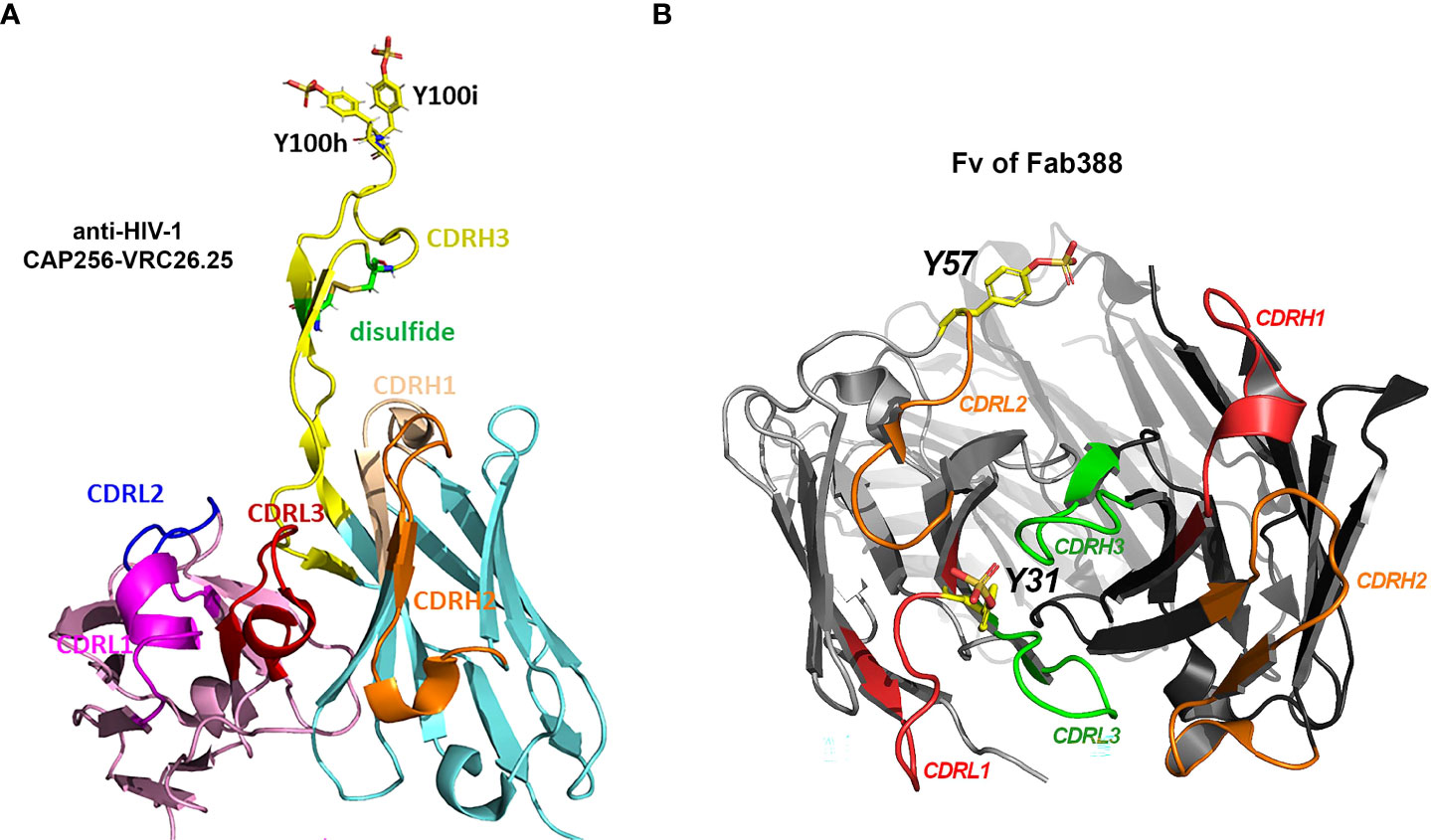
Figure 2 Three types of Tyr sulfation modification at antibody CDRs. (A) Structural display of sTyr residues at CDRH3 of anti-HIV-1 antibody CAP256-VRC26.25 (Gorman, Chuang et al. 2020). PDB 5DT1 (47, 81). (B) sTyr residues at CDRL1 (Y31) (40) and CDRL2 (Y57) (41) are displayed on antibody Fab structure (Fv of Fab388). Sulfated tyrosine residues were modeled on the corresponding Fab structure PDB 5I1A (14, 82) using Pymol molecular graphics v2.5.4.
CAP256-VRC26.25 (Figure 2A and Table 1), one of the most potent antibodies targeting V1V2 apex of gp120, contains two sTyr in CDRH3 (43), and uses an extended CDRH3 loop to insert sTyr into the hole at the V1V2 apex. One recent study (47) further indicates that an increased level of Tyr sulfation on this anti-HIV-1 antibody corresponded to more robust antigen binding to the V1V2 domain of gp120 protein. Molecular binding efficacy dropped significantly with a loss of only one sulfo group. Without the sulfation, nearly no binding was detectable. Full sulfation occupancy in the antibody CDRH3 is important for effective high antigen binding and should be regarded as a critical quality attribute for this antibody (47).
Several recent reports provide other examples of Tyr sulfated antibody at the varied CDRs. Different from the modification sites in CDRH3 for anti-HIV-1 antibodies, the Tyr sulfation sites located in the short CDRL1 region (40) or CDRL2 (41) have been recently discovered (Figure 2B and Table 1). One human antibody produced by stable CHO cells was 40% Tyr-sulfated at its LC CDR1 region (40). The modified Tyr residue (Y31) is surrounded with Glu or Asp residue at -1, +1, +3, +5, supporting its being a substrate for TPSTs. This is the first example in which Tyr-sulfation is on an antibody’s CDRL1 region. During the immunoglobulin rearrangement, CDRL1 contains only V gene fragments. In human germline V-segment loci, there are 51 VH and 70 Vκ/λ (83). Therefore it is much less diverse than CDRL3 (with additional J fragment recombination) or CDRH3 (with additional D or J recombination). The biological function of this sTyr is not disclosed in the paper (40), but the fact that this modification was intentionally preserved during stable CHO production suggested its possibly important role in antigen binding (45).
Tyshchuk and colleagues reported that a human IgG1 expressed in stable CHO cells was 21% Tyr-sulfated at its LC CDR-2 region (Figure 2B and Table 1) (41). The sTyr residue (Y57) is nearby with Asp residue at -1 and -3 position whereas a similar antibody with unsulfated Tyr has a Thr residue at -3 position, supporting the important role of acidic residue on amino-terminal site of Tyr for sulfation. Even though this sTyr is not essential for antigen binding (41), one would expect to see more functional examples of sTyr in antibody CDRs being uncovered, as the functional role of sTyr catches a wider attention.
sTyr extends the vocabulary of protein synthesis beyond the standard 20 amino acids (26, 34). It has an important implication in antibody engineering for protein-protein interaction, especially under the fact that Tyr-sulfation is not a rare event, estimated to be ~7% of mammalian proteins and ~1% of all Tyr residues in the eukaryotic proteome (22, 25, 74, 84). sTyr heightens protein-protein interactions via two mechanisms. The first one is that Tyr’s aromatic ring engages nonpolar and pi stacking interactions with various binding residues. The second one is that the sulfate group can make electrostatic interactions with positive-charged Arg or Lys residues on the surface of the binding partner. For instance, replacing the sTyr12-interacting-Lys27 of chemokine CXCL12 with Ala or Glu resulted in 3.9- and 181.7-fold reduction in CXCR4 receptor activation respectively (85).
One advantage of sTyr is that its sulfate ester is anionic at physiological pH, providing an electrostatic component to specific interaction without acting as a base or nucleophile. Tyr sulfate also provides a longer electrostatic arm than the standard acidic Asp and Glu side chains. Sulfate esters are often found in clusters which is an efficient way of generating unique structures to increase interaction affinity. Another advantage of sTyr is that the sulfate ester is stable in physiological conditions. Other than the sulfatases for steroids or carbohydrates (86), no sulfoprotein sulfatase is yet reported for mammalian cells and tissues. sTyr is therefore considered as a long-lasting PTM.
With a nearly identical mass, sTyr and phospho-Tyr (pTyr) has been compared both biologically and biophysically (Figure 1C). Both the sulfate group and the phosphate group are PTMs with the addition of an oxoanion functional group (27, 87). Both groups are fully ionized at neutral pH and can increase side-chain polarity. Comparing to pTyr, sTyr makes weaker hydrogen bonds, attributed to the reduced electrostatic potentials (-1 for sTyr vs -2 for pTyr) and smaller dipole moment (27). sTyr sulfate can create distinct ionic contact (88, 89) and provide a unique interaction specificity, even though for some protein-protein-interactions pTyr can partially replace sTyr (90–92). There are cases that pTyr cannot substitute sTyr (93). For CCR7 signaling with CCL21, pTyr can only replace sTyr at position 8 of CCR7 but at postion17 sTyr provides a drastic effect over pTyr (92).
From a protein engineering point of view, sTyr interactions with Arg are weaker significantly than that of pTyr, and even not the same interactions made by the Glu residue. However, sTyr has a greater flexibility than pTyr, attributing to a weaker hydrogen bonding interaction. It provides a unique interaction specificity determinant, as in the example that phosphate can’t replace sulfate for interacting with Lys145 of CCR5 (34). Many secreted proteins contain basic patches on their protein surfaces. Charge-charge interaction between sulfate group and Lys/Arg residues on these proteins might provide a way of affinity up-regulation for Tyr-sulfated antibodies. This mechanism might have been utilized by human antibody repertoire for targeting this kind of antigen.
As a known player for protein-protein interaction, sTyr usually mediates the interactions through relatively short protein segments, e.g., amino-termini of cell surface proteins and receptors. This feature fits ideally with the length of the CDR loops in antibodies, even though not all sTyr are important for protein-interaction, with some being detrimental (28). Interestingly, even a peptide derived from the Tyr-sulfated CDRH3 of E51 antibody is sufficient to engage with virus spike protein and neutralize virus isolates (94, 95). Recently Siguna Mueller wrote an article on rarely-recognized antibody diversification against SARS-CoV and SAR-CoV-2 with a potential role of sulfated antibodies (96), as the basic residue stretches on the surface of the viral spike protein (97) can be targeted by the negatively-charged Tyr-sulfate. More novel examples in protein engineering with sTyr are expected to appear in the literatures.
Several sTyr-containing antibodies such as PG9, PG16, and CAP256-VCR26 have entered clinical trials or preclinical stages as broadly neutralizing antibodies for the treatment and prevention of HIV infection (98–100). These sTyr-containing antibodies have been successfully produced by stable CHO cells (45, 47) or in plants (38, 44).
A major challenge for Tyr sulfation is the difficulty of expressing target proteins in a homogenously sulfated state. Chemical synthetic approaches have been developed for peptide synthesis, but not for large proteins (101). The genetic code expansion technology (Figure 1D) utilizes an engineered tyrosyl-tRNA synthetase/tRNA pair that co-translationally incorporates sTyr into the UAG codons in bacterial (102) and mammalian cells (103, 104). These new methods provide a promising way to incorporate sTyr into proteins site-specifically for evaluating the roles of individual sulfations.
Sulfated antibodies have been routinely produced by mammalian cells, even though different sulfation levels were observed (37, 45). This is likely due to different gene expression in sulfation pathway like those of TPSTs, which can be rescued by TPST overexpression. Ectopic expression of either TPST1 or TPST2 can result in a high percentage of Tyr-sulfated antibodies, yet producing a full Tyr-sulfation occupancy on modified sites remains to be a challenging task (47).
Plant-based expression system has been employed for cost-effective antibody production (38, 44, 46). Plants encode a single TPST with a carboxyl-terminal transmembrane domain which is different from the amino-terminal segment for human TPSTs (Figure 1B) (26, 105). Plant TPST has putative 5’-PSB ad 3’-PB motifs even though the similarity with human counterparts is low. It shares additional carboxyl-terminal sequence similarity with heparan sulfate 6-O-sulfotransferase (105). Initial sulfo-antibody expression in plant did not produce detectable sTyr, indicating that mammalian-type of sulfation does not naturally occur in plant (38, 39). When human TPST1 was engineered in plants, a plant Golgi-targeting sequence was needed for targeting the enzyme to late Golgi compartment (38). A drastic improvement for Tyr sulfation was subsequently observed. Several sTyr-containing antibodies have been produced successfully by this engineered plant-based system for anti-viral therapy (44, 44, 46).
PTMs like Tyr sulfation in mammalian cells provide de novo synthesized proteins with additional level of structural diversity beyond their primary sequences. These PTMs impose new biological functions and activity modulations as well as further physiological consequences. For Tyr sulfation modification, it results in versatile interaction motifs that are critical for numerous high-affinity physiological interactions. It is fascinating that sTyr also has a role in regulating humoral immune responses and contributing to the extent of the antibody diversification as first uncovered nearly two decades ago. This PTM has provided a new strategy for therapeutic antibody engineering and diversification.
However, there are still significant challenges being presented to Tyr sulfation. Specificity determinants for Tyr sulfation remain to be well defined. Better understanding how TPSTs engage with their substrates is essential for evaluating the location and function of sTyr at CDRs. Moreover, feasible and reliable analytical methods for detecting sTyr are needed for understanding many under-documented protein substrates in protein database. Elucidating the biological mechanisms for these newly identified sulfoproteins are challenging, whereas accumulating these physiological knowledges is important for designing new sTyr-containing antibodies and therapeutic proteins. For instance, three sTyr residues (Y46, Y48 & Y51) of P-selectin glycoprotein ligand-1 (PSGL-1) make ionic interactions with protonated His residues (H154, H153, & H100) of the PSGL-1 ligand VISTA in acidic tumor microenvironments, which is disrupted by the imidazole sidechain deprotonation at the physiological pH7.4 (106). This finding reveals a novel role of sulfation in a pH-selective binding interaction for future protein engineering design.
Developing new technologies in producing site-specific sulfo-antibodies is helpful in determining the structure-function relationship. While the genetic codon expanding technology is promising, its bioprocessing bottlenecks need to be overcome for a large-scale production. In addition, producing sTyr-containing secreted full-length intact antibody proteins with this methodology remains to be demonstrated. Further understanding of sTyr as a critical drug product attribute is also warranted for sulfo-biotherapeutics. sTyr is known to work together with N- or O-glycans (24), which are also present at CDRs for enhancing antigen binding (107, 108). With more knowledge being gathered for the roles of these PTMs at CDRs, one expects molecular insights and mechanisms of these unconventional strategies for antibody diversification will be further revealed.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
XZ and AD conceptualized, wrote, and edited the manuscript. All authors contributed to the article and approved the submitted version.
The authors would like to thank James Apgar, Melvin Zhang, Julie Lee, Tao He, Laura Lin, and Will Somers for discussion.
The authors declare competing financial interests. Both authors are Pfizer employees.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Hodgson J. Refreshing the biologic pipeline 2020. Nat Biotechnol (2021) 39(2):135–43. doi: 10.1038/s41587-021-00814-w
2. Kaplon H, Chenoweth A, Crescioli S, Reichert JM. Antibodies to watch in 2022. MAbs (2022) 14(1):2014296. doi: 10.1080/19420862.2021.2014296
3. Chiu ML, Gilliland GL. Engineering antibody therapeutics. Curr Opin Struct Biol (2016) 38:163–73. doi: 10.1016/j.sbi.2016.07.012
4. Goulet DR, Atkins WM. Considerations for the design of antibody-based therapeutics. J Pharm Sci (2020) 109(1):74–103. doi: 10.1016/j.xphs.2019.05.031
5. Carter PJ, Rajpal A. Designing antibodies as therapeutics. Cell (2022) 185(15):2789–805. doi: 10.1016/j.cell.2022.05.029
6. Brinkmann U, Kontermann RE. The making of bispecific antibodies. MAbs (2017) 9(2):182–212. doi: 10.1080/19420862.2016.1268307
7. Labrijn AF, Janmaat ML, Reichert JM, Parren P. Bispecific antibodies a mechanistic review of the pipeline. Nat Rev Drug Discov (2019) 18(8):585–608. doi: 10.1038/s41573-019-0028-1
8. Deshaies RJ. Multispecific drugs herald a new era of biopharmaceutical innovation. Nature (2020) 580(7803):329–38. doi: 10.1038/s41586-020-2168-1
9. Goebeler ME, Bargou RC. T Cell-engaging therapies - BiTEs and beyond. Nat Rev Clin Oncol (2020) 17(7):418–34. doi: 10.1038/s41571-020-0347-5
10. Zhong X, D’Antona AM. Recent advances in the molecular design and applications of multispecific biotherapeutics. Antibodies (Basel) (2021) 10(2):1–26. doi: 10.3390/antib10020013
11. Corti D, Purcell LA, Snell G, Veesler D. Tackling COVID-19 with neutralizing monoclonal antibodies. Cell (2021) 184(12):3086–108. doi: 10.1016/j.cell.2021.05.005
12. Almagro JC, Mellado-Sanchez G, Pedraza-Escalona M, Perez-Tapia SM. Evolution of anti-SARS-CoV-2 therapeutic antibodies. Int J Mol Sci (2022) 23(17):1–18. doi: 10.3390/ijms23179763
13. Schroeder HW Jr., Cavacini L. Structure and function of immunoglobulins. J Allergy Clin Immunol (2010) 125(2 Suppl 2):S41–52. doi: 10.1016/j.jaci.2009.09.046
14. Chiu ML, Goulet DR, Teplyakov A, Gilliland GL. Antibody structure and function the basis for engineering therapeutics. Antibodies (Basel) (2019) 8(4):1–80. doi: 10.3390/antib8040055
15. Nimmerjahn F, Ravetch JV. Fcgamma receptors as regulators of immune responses. Nat Rev Immunol (2008) 8(1):34–47. doi: 10.1038/nri2206
16. Bruhns P, Jonsson F. Mouse and human FcR effector functions. Immunol Rev (2015) 268(1):25–51. doi: 10.1111/imr.12350
17. Tonegawa S. Somatic generation of antibody diversity. Nature (1983) 302(5909):575–81. doi: 10.1038/302575a0
18. Alt FW, Yancopoulos GD, Blackwell TK, Wood C, Thomas E, Boss M, et al. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J (1984) 3(6):1209–19. doi: 10.1002/j.1460-2075.1984.tb01955.x
19. de Villartay JP, Fischer A, Durandy A. The mechanisms of immune diversification and their disorders. Nat Rev Immunol (2003) 3(12):962–72. doi: 10.1038/nri1247
20. Westra ER, Sunderhauf D, Landsberger M, Buckling A. Mechanisms and consequences of diversity-generating immune strategies. Nat Rev Immunol (2017) 17(11):719–28. doi: 10.1038/nri.2017.78
21. Jackson KJ, Kidd MJ, Wang Y, Collins AM. The shape of the lymphocyte receptor repertoire lessons from the b cell receptor. Front Immunol (2013) 4:263. doi: 10.3389/fimmu.2013.00263
22. Moore KL. The biology and enzymology of protein tyrosine O-sulfation. J Biol Chem (2003) 278(27):24243–6. doi: 10.1074/jbc.R300008200
23. Sasaki N. Current status and future prospects for research on tyrosine sulfation. Curr Pharm Biotechnol (2012) 13(14):2632–41. doi: 10.2174/138920101314151120122922
24. Mehta AY, Heimburg-Molinaro J, Cummings RD, Goth CK. Emerging patterns of tyrosine sulfation and O-glycosylation cross-talk and co-localization. Curr Opin Struct Biol (2020) 62:102–11. doi: 10.1016/j.sbi.2019.12.002
25. Stone MJ, Chuang S, Hou X, Shoham M, Zhu JZ. Tyrosine sulfation an increasingly recognised post-translational modification of secreted proteins. N Biotechnol (2009) 25(5):299–317. doi: 10.1016/j.nbt.2009.03.011
26. Stewart V, Ronald PC. Sulfotyrosine residues interaction specificity determinants for extracellular protein-protein interactions. J Biol Chem (2022) 298(8):102232. doi: 10.1016/j.jbc.2022.102232
27. Rapp C, Klerman H, Levine E, McClendon CL. Hydrogen bond strengths in phosphorylated and sulfated amino acid residues. PloS One (2013) 8(3):e57804. doi: 10.1371/journal.pone.0057804
28. Li X, Hitomi J, Liu CC. Characterization of a sulfated anti-HIV antibody using an expanded genetic code. Biochemistry (2018) 57(20):2903–7. doi: 10.1021/acs.biochem.8b00374
29. Choe H, Li W, Wright PL, Vasilieva N, Venturi M, Huang CC, et al. Tyrosine sulfation of human antibodies contributes to recognition of the CCR5 binding region of HIV-1 gp120. Cell (2003) 114(2):161–70. doi: 10.1016/S0092-8674(03)00508-7
30. Lin G, Hoxie JA. CCR5 mimicry by sulfated human anti-HIV-1 antibodies. Cell (2003) 114(2):147–8. doi: 10.1016/S0092-8674(03)00564-6
31. Kanyavuz A, Marey-Jarossay A, Lacroix-Desmazes S, Dimitrov JD. Breaking the law unconventional strategies for antibody diversification. Nat Rev Immunol (2019) 19(6):355–68. doi: 10.1038/s41577-019-0126-7
32. Yang YS, Wang CC, Chen BH, Hou YH, Hung KS, Mao YC. Tyrosine sulfation as a protein post-translational modification. Molecules (2015) 20(2):2138–64. doi: 10.3390/molecules20022138
33. Gunal S, Hardman R, Kopriva S, Mueller JW. Sulfation pathways from red to green. J Biol Chem (2019) 294(33):12293–312. doi: 10.1074/jbc.REV119.007422
34. Huang CC, Venturi M, Majeed S, Moore MJ, Phogat S, Zhang MY, et al. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc Natl Acad Sci U.S.A. (2004) 101(9):2706–11. doi: 10.1073/pnas.0308527100
35. Huang CC, Lam SN, Acharya P, Tang M, Xiang SH, Hussan SS, et al. Structures of the CCR5 n terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science (2007) 317(5846):1930–4. doi: 10.1126/science.1145373
36. Pejchal R, Walker LM, Stanfield RL, Phogat SK, Koff WC, Poignard P, et al. Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proc Natl Acad Sci USA (2010) 107(25):11483–8. doi: 10.1073/pnas.1004600107
37. McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature (2011) 480(7377):336–43. doi: 10.1038/nature10696
38. Loos A, Gach JS, Hackl T, Maresch D, Henkel T, Porodko A, et al. Glycan modulation and sulfoengineering of anti-HIV-1 monoclonal antibody PG9 in plants. Proc Natl Acad Sci USA (2015) 112(41):12675–80. doi: 10.1073/pnas.1509090112
39. Rosenberg Y, Sack M, Montefiori D, Labranche C, Lewis M, Urban L, et al. Pharmacokinetics and immunogenicity of broadly neutralizing HIV monoclonal antibodies in macaques. PloS One (2015) 10(3):e0120451. doi: 10.1371/journal.pone.0120451
40. Zhao J, Saunders J, Schussler SD, Rios S, Insaidoo FK, Fridman AL, et al. Characterization of a novel modification of a CHO-produced mAb evidence for the presence of tyrosine sulfation. MAbs (2017) 9(6):985–95. doi: 10.1080/19420862.2017.1332552
41. Tyshchuk O, Gstottner C, Funk D, Nicolardi S, Frost S, Klostermann S, et al. Characterization and prediction of positional 4-hydroxyproline and sulfotyrosine, two post-translational modifications that can occur at substantial levels in CHO cells-expressed biotherapeutics. MAbs (2019) 11(7):1219–32. doi: 10.1080/19420862.2019.1635865
42. Yang Z, Wang H, Liu AZ, Gristick HB, Bjorkman PJ. Asymmetric opening of HIV-1 env bound to CD4 and a coreceptor-mimicking antibody. Nat Struct Mol Biol (2019) 26(12):1167–75. doi: 10.1038/s41594-019-0344-5
43. Gorman J, Chuang GY, Lai YT, Shen CH, Boyington JC, Druz A, et al. Structure of super-potent antibody CAP256-VRC26.25 in complex with HIV-1 envelope reveals a combined mode of trimer-apex recognition. Cell Rep (2020) 31(1):107488. doi: 10.1016/S0021-9258(19)50666-4
44. Singh AA, Pooe O, Kwezi L, Lotter-Stark T, Stoychev SH, Alexandra K, et al. Plant-based production of highly potent anti-HIV antibodies with engineered posttranslational modifications. Sci Rep (2020) 10(1):6201. doi: 10.1038/s41598-020-63052-1
45. Liu R, Zhang Y, Kumar A, Huhn S, Hullinger L, Du Z. Modulating tyrosine sulfation of recombinant antibodies in CHO cell culture by host selection and sodium chlorate supplementation. Biotechnol J (2021) 16(9):e2100142. doi: 10.1002/biot.202100142
46. Singh AA, Pillay P, Kwezi L, Tsekoa TL. A plant-biotechnology approach for producing highly potent anti-HIV antibodies for antiretroviral therapy consideration. J Genet Eng Biotechnol (2021) 19(1):180. doi: 10.1186/s43141-021-00279-z
47. Cai CX, Doria-Rose NA, Schneck NA, Ivleva VB, Tippett B, Shadrick WR, et al. Tyrosine O-sulfation proteoforms affect HIV-1 monoclonal antibody potency. Sci Rep (2022) 12(1):8433. doi: 10.1038/s41598-022-12423-x
48. Beisswanger R, Corbeil D, Vannier C, Thiele C, Dohrmann U, Kellner R, et al. Existence of distinct tyrosylprotein sulfotransferase genes molecular characterization of tyrosylprotein sulfotransferase-2. Proc Natl Acad Sci U.S.A. (1998) 95(19):11134–9. doi: 10.1073/pnas.95.19.11134
49. Ouyang YB, Moore KL. Molecular cloning and expression of human and mouse tyrosylprotein sulfotransferase-2 and a tyrosylprotein sulfotransferase homologue in caenorhabditis elegans. J Biol Chem (1998) 273(38):24770–4. doi: 10.1074/jbc.273.38.24770
50. Mueller JW, Shafqat N. Adenosine-5’-phosphosulfate–a multifaceted modulator of bifunctional 3’-phospho-adenosine-5’-phosphosulfate synthases and related enzymes. FEBS J (2013) 280(13):3050–7. doi: 10.1111/febs.12252
51. Hastbacka J, de la Chapelle A, Mahtani MM, Clines G, Reeve-Daly MP, Daly M, et al. The diastrophic dysplasia gene encodes a novel sulfate transporter positional cloning by fine-structure linkage disequilibrium mapping. Cell (1994) 78(6):1073–87. doi: 10.1016/0092-8674(94)90281-X
52. Ohana E, Shcheynikov N, Park M, Muallem S. Solute carrier family 26 member a2 (Slc26a2) protein functions as an electroneutral SOFormula/OH-/Cl- exchanger regulated by extracellular cl. J Biol Chem (2012) 287(7):5122–32. doi: 10.1074/jbc.M111.297192
53. Kamiyama S, Suda T, Ueda R, Suzuki M, Okubo R, Kikuchi N, et al. Molecular cloning and identification of 3’-phosphoadenosine 5’-phosphosulfate transporter. J Biol Chem (2003) 278(28):25958–63. doi: 10.1074/jbc.M302439200
54. Kamiyama S, Sasaki N, Goda E, Ui-Tei K, Saigo K, Narimatsu H, et al. Molecular cloning and characterization of a novel 3’-phosphoadenosine 5’-phosphosulfate transporter, PAPST2. J Biol Chem (2006) 281(16):10945–53. doi: 10.1074/jbc.M508991200
55. Blanchard RL, Freimuth RR, Buck J, Weinshilboum RM, Coughtrie MW. A proposed nomenclature system for the cytosolic sulfotransferase (SULT) superfamily. Pharmacogenetics (2004) 14(3):199–211. doi: 10.1097/00008571-200403000-00009
56. Chapman E, Best MD, Hanson SR, Wong CH. Sulfotransferases structure, mechanism, biological activity, inhibition, and synthetic utility. Angew Chem Int Ed Engl (2004) 43(27):3526–48. doi: 10.1002/anie.200300631
57. Rath VL, Verdugo D, Hemmerich S. Sulfotransferase structural biology and inhibitor discovery. Drug Discov Today (2004) 9(23):1003–11. doi: 10.1016/S1359-6446(04)03273-8
58. Bowman KG, Bertozzi CR. Carbohydrate sulfotransferases mediators of extracellular communication. Chem Biol (1999) 6(1):R9–R22. doi: 10.1016/S1074-5521(99)80014-3
59. Baeuerle PA, Huttner WB. Tyrosine sulfation is a trans-golgi-specific protein modification. J Cell Biol (1987) 105(6 Pt 1):2655–64. doi: 10.1083/jcb.105.6.2655
60. Ouyang Y, Lane WS, Moore KL. Tyrosylprotein sulfotransferase purification and molecular cloning of an enzyme that catalyzes tyrosine O-sulfation, a common posttranslational modification of eukaryotic proteins. Proc Natl Acad Sci USA (1998) 95(6):2896–901. doi: 10.1073/pnas.95.6.2896
61. Bettelheim FR. Tyrosine O-sulfate in a peptide from fibrinogen. J Am Chem Soc (1954) 76:2838–9. doi: 10.1021/ja01639a073
62. Niehrs C, Kraft M, Lee RW, Huttner WB. Analysis of the substrate specificity of tyrosylprotein sulfotransferase using synthetic peptides. J Biol Chem (1990) 265(15):8525–32. doi: 10.1016/S0021-9258(19)38920-3
63. Seibert C, Cadene M, Sanfiz A, Chait BT, Sakmar TP. Tyrosine sulfation of CCR5 n-terminal peptide by tyrosylprotein sulfotransferases 1 and 2 follows a discrete pattern and temporal sequence. Proc Natl Acad Sci U.S.A. (2002) 99(17):11031–6. doi: 10.1073/pnas.172380899
64. Yu Y, Hoffhines AJ, Moore KL, Leary JA. Determination of the sites of tyrosine O-sulfation in peptides and proteins. Nat Methods (2007) 4(7):583–8. doi: 10.1038/nmeth1056
65. Ouyang YB, Crawley JT, Aston CE, Moore KL. Reduced body weight and increased postimplantation fetal death in tyrosylprotein sulfotransferase-1-deficient mice. J Biol Chem (2002) 277(26):23781–7. doi: 10.1074/jbc.M202420200
66. Borghei A, Ouyang YB, Westmuckett AD, Marcello MR, Landel CP, Evans JP, et al. Targeted disruption of tyrosylprotein sulfotransferase-2, an enzyme that catalyzes post-translational protein tyrosine O-sulfation, causes male infertility. J Biol Chem (2006) 281(14):9423–31. doi: 10.1074/jbc.M513768200
67. Westmuckett AD, Hoffhines AJ, Borghei A, Moore KL. Early postnatal pulmonary failure and primary hypothyroidism in mice with combined TPST-1 and TPST-2 deficiency. Gen Comp Endocrinol (2008) 156(1):145–53. doi: 10.1016/j.ygcen.2007.12.006
68. Kakuta Y, Pedersen LG, Pedersen LC, Negishi M. Conserved structural motifs in the sulfotransferase family. Trends Biochem Sci (1998) 23(4):129–30. doi: 10.1016/S0968-0004(98)01182-7
69. Monigatti F, Gasteiger E, Bairoch A, Jung E. The sulfinator predicting tyrosine sulfation sites in protein sequences. Bioinformatics (2002) 18(5):769–70. doi: 10.1093/bioinformatics/18.5.769
70. Chang WC, Lee TY, Shien DM, Hsu JB, Horng JT, Hsu PC, et al. Incorporating support vector machine for identifying protein tyrosine sulfation sites. J Comput Chem (2009) 30(15):2526–37. doi: 10.1002/jcc.21258
71. Huang SY, Shi SP, Qiu JD, Sun XY, Suo SB, Liang RP. PredSulSite prediction of protein tyrosine sulfation sites with multiple features and analysis. Anal Biochem (2012) 428(1):16–23. doi: 10.1016/j.ab.2012.06.003
72. Yang ZR. Predicting sulfotyrosine sites using the random forest algorithm with significantly improved prediction accuracy. BMC Bioinf (2009) 10:361. doi: 10.1186/1471-2105-10-361
73. Hortin G, Folz R, Gordon JI, Strauss AW. Characterization of sites of tyrosine sulfation in proteins and criteria for predicting their occurrence. Biochem Biophys Res Commun (1986) 141(1):326–33. doi: 10.1016/S0006-291X(86)80372-2
74. Huttner WB. Tyrosine sulfation and the secretory pathway. Annu Rev Physiol (1988) 50:363–76. doi: 10.1146/annurev.ph.50.030188.002051
75. Rosenquist GL, Nicholas HB Jr. Analysis of sequence requirements for protein tyrosine sulfation. Protein Sci (1993) 2(2):215–22. doi: 10.1002/pro.5560020210
76. Lin WH, Larsen K, Hortin GL, Roth JA. Recognition of substrates by tyrosylprotein sulfotransferase. determination of affinity by acidic amino acids near the target sites. J Biol Chem (1992) 267(5):2876–9. doi: 10.1016/S0021-9258(19)50666-4
77. Bundgaard JR, Vuust J, Rehfeld JF. New consensus features for tyrosine O-sulfation determined by mutational analysis. J Biol Chem (1997) 272(35):21700–5. doi: 10.1074/jbc.272.35.21700
78. Teramoto T, Fujikawa Y, Kawaguchi Y, Kurogi K, Soejima M, Adachi R, et al. Crystal structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction. Nat Commun (2013) 4:1572. doi: 10.1038/ncomms2593
79. Vatsa S. In silico prediction of post-translational modifications in therapeutic antibodies. MAbs (2022) 14(1):2023938. doi: 10.1080/19420862.2021.2023938
80. Zhong X, Desilva T, Lin L, Bodine P, Bhat RA, Presman E, et al. Regulation of secreted frizzled-related protein-1 by heparin. J Biol Chem (2007) 282(28):20523–33. doi: 10.1074/jbc.M609096200
81. Doria-Rose NA, Bhiman JN, Roark RS, Schramm CA, Gorman J, Chuang GY, et al. New member of the V1V2-directed CAP256-VRC26 lineage that shows increased breadth and exceptional potency. J Virol (2016) 90(1):76–91. doi: 10.1128/JVI.01791-15
82. Teplyakov A, Obmolova G, Malia TJ, Luo J, Muzammil S, Sweet R, et al. Structural diversity in a human antibody germline library. MAbs (2016) 8(6):1045–63. doi: 10.1080/19420862.2016.1190060
83. Glanville J, Zhai W, Berka J, Telman D, Huerta G, Mehta GR, et al. Precise determination of the diversity of a combinatorial antibody library gives insight into the human immunoglobulin repertoire. Proc Natl Acad Sci U.S.A. (2009) 106(48):20216–21. doi: 10.1073/pnas.0909775106
84. Seibert C, Sakmar TP. Toward a framework for sulfoproteomics synthesis and characterization of sulfotyrosine-containing peptides. Biopolymers (2008) 90(3):459–77. doi: 10.1002/bip.20821
85. Veldkamp CT, Seibert C, Peterson FC, de la Cruz NB, Haugner JC, Basnet H, et al. Structural basis of CXCR4 sulfotyrosine recognition by the chemokine SDF-1/CXCL12. Sci Signal (2008) 1(37):ra4. doi: 10.1126/scisignal.1160755
86. Hanson SR, Best MD, Wong CH. Sulfatases structure, mechanism, biological activity, inhibition, and synthetic utility. Angew Chem Int Ed Engl (2004) 43(43):5736–63. doi: 10.1002/anie.200300632
87. Lima MA, Rudd TR, Fernig DG, Yates EA. Phosphorylation and sulfation share a common biosynthetic pathway, but extend biochemical and evolutionary diversity of biological macromolecules in distinct ways. J R Soc Interface (2022) 19(193):20220391. doi: 10.1098/rsif.2022.0391
88. Kanyo ZF, Christianson DW. Biological recognition of phosphate and sulfate. J Biol Chem (1991) 266(7):4264–8. doi: 10.1016/S0021-9258(20)64316-2
89. Quiocho FA. Atomic basis of the exquisite specificity of phosphate and sulfate transport receptors. Kidney Int (1996) 49(4):943–6. doi: 10.1038/ki.1996.132
90. Hofsteenge J, Stone SR, Donella-Deana A, Pinna LA. The effect of substituting phosphotyrosine for sulphotyrosine on the activity of hirudin. Eur J Biochem (1990) 188(1):55–9. doi: 10.1111/j.1432-1033.1990.tb15370.x
91. Lechtenberg BC, Freund SM, Huntington JA. GpIbalpha interacts exclusively with exosite II of thrombin. J Mol Biol (2014) 426(4):881–93. doi: 10.1016/j.jmb.2013.11.027
92. Phillips AJ, Taleski D, Koplinski CA, Getschman AE, Moussouras NA, Richard AM, et al. CCR7 sulfotyrosine enhances CCL21 binding. Int J Mol Sci (2017) 18(9):1–13. doi: 10.3390/ijms18091857
93. Cormier EG, Persuh M, Thompson DA, Lin SW, Sakmar TP, Olson WC, et al. Specific interaction of CCR5 amino-terminal domain peptides containing sulfotyrosines with HIV-1 envelope glycoprotein gp120. Proc Natl Acad Sci U.S.A. (2000) 97(11):5762–7. doi: 10.1073/pnas.97.11.5762
94. Dorfman T, Moore MJ, Guth AC, Choe H, Farzan M. A tyrosine-sulfated peptide derived from the heavy-chain CDR3 region of an HIV-1-neutralizing antibody binds gp120 and inhibits HIV-1 infection. J Biol Chem (2006) 281(39):28529–35. doi: 10.1074/jbc.M602732200
95. Gardner MR, Kattenhorn LM, Kondur HR, von Schaewen M, Dorfman T, Chiang JJ, et al. AAV-expressed eCD4-ig provides durable protection from multiple SHIV challenges. Nature (2015) 519(7541):87–91. doi: 10.1038/nature14264
96. Mueller S. Rarely recognized antibody diversification in covid-19 evolution to counteract advanced SARS-CoV-2 evasion strategies, and implications for prophylactic treatment. Front Physiol (2021) 12:624675. doi: 10.3389/fphys.2021.624675
97. Sorensen B, Susrud A, Dalgleish AG. Biovacc-19 a candidate vaccine for covid-19 (SARS-CoV-2) developed from analysis of its general method of action for infectivity. QRB Discovery (2020) 1:e6. doi: 10.1017/qrd.2020.8
98. Caskey M. Broadly neutralizing antibodies for the treatment and prevention of HIV infection. Curr Opin HIV AIDS (2020) 15(1):49–55. doi: 10.1097/COH.0000000000000600
99. Mahomed S, Garrett N, Karim QA, Zuma NY, Capparelli E, Baxter C, et al. Assessing the safety and pharmacokinetics of the anti-HIV monoclonal antibody CAP256V2LS alone and in combination with VRC07-523LS and PGT121 in south African women study protocol for the first-in-human CAPRISA 012B phase I clinical trial. BMJ Open (2020) 10(11):e042247. doi: 10.1136/bmjopen-2020-042247
100. Spencer DA, Shapiro MB, Haigwood NL, Hessell AJ. Advancing HIV broadly neutralizing antibodies from discovery to the clinic. Front Public Health (2021) 9:690017. doi: 10.3389/fpubh.2021.690017
101. Stone SR, Hofsteenge J. Kinetics of the inhibition of thrombin by hirudin. Biochemistry (1986) 25(16):4622–8. doi: 10.1021/bi00364a025
102. Liu CC, Schultz PG. Recombinant expression of selectively sulfated proteins in escherichia coli. Nat Biotechnol (2006) 24(11):1436–40. doi: 10.1038/nbt1254
103. He X, Chen Y, Beltran DG, Kelly M, Ma B, Lawrie J, et al. Functional genetic encoding of sulfotyrosine in mammalian cells. Nat Commun (2020) 11(1):4820. doi: 10.1038/s41467-020-18629-9
104. Italia JS, Peeler JC, Hillenbrand CM, Latour C, Weerapana E, Chatterjee A. Genetically encoded protein sulfation in mammalian cells. Nat Chem Biol (2020) 16(4):379–82. doi: 10.1038/s41589-020-0493-1
105. Komori R, Amano Y, Ogawa-Ohnishi M, Matsubayashi Y. Identification of tyrosylprotein sulfotransferase in arabidopsis. Proc Natl Acad Sci USA (2009) 106(35):15067–72. doi: 10.1073/pnas.0902801106
106. Johnston RJ, Su LJ, Pinckney J, Critton D, Boyer E, Krishnakumar A, et al. VISTA is an acidic pH-selective ligand for PSGL-1. Nature (2019) 574(7779):565–70. doi: 10.1038/s41586-019-1674-5
107. Song R, Oren DA, Franco D, Seaman MS, Ho DD. Strategic addition of an n-linked glycan to a monoclonal antibody improves its HIV-1-neutralizing activity. Nat Biotechnol (2013) 31(11):1047–52. doi: 10.1038/nbt.2677
Keywords: antibody repertoire diversification, tyrosine sulfation, complementarity determination region (CDR), antigen-binding affinity, biotherapeutic engineering and production
Citation: Zhong X and D’Antona AM (2022) A potential antibody repertoire diversification mechanism through tyrosine sulfation for biotherapeutics engineering and production. Front. Immunol. 13:1072702. doi: 10.3389/fimmu.2022.1072702
Received: 17 October 2022; Accepted: 23 November 2022;
Published: 08 December 2022.
Edited by:
Prabakaran Ponraj, Sanofi, United StatesReviewed by:
Ramya Yarlagadda, EXUMA Biotech, United StatesCopyright © 2022 Zhong and D’Antona. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaotian Zhong, eGlhb3RpYW4uemhvbmdAcGZpemVyLmNvbQ==; Aaron M. D’Antona, YWFyb24uZGFudG9uYUBwZml6ZXIuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.