- 1Department of Experimental Oncology, Translational Immunology Unit, Fondazione IRCCS Istituto Nazionale dei Tumori, Milan, Italy
- 2Department of Pathology, Fondazione IRCCS Istituto Nazionale dei Tumori, Milan, Italy
- 3Melanoma and Sarcoma Surgery Unit, Fondazione IRCCS Istituto Nazionale dei Tumori, Milan, Italy
- 4Department of Medicine and Surgery, University of Milano-Bicocca, Monza, Italy
Immunotherapy with immune checkpoint inhibitors can induce durable clinical responses in different human malignancies but the number of responding patients remains globally modest. The limited therapeutic efficacy of ICI depends on multiple factors, among which the immune suppressive features of the tumor microenvironment play a key role. For this reason, experimental models that enable dissection of the immune-hostile tumor milieu components are required to unravel how to overcome resistance and obtain full-fledged anti-tumor immunity. Recent evidence supports the usefulness of 3D ex vivo systems in retaining features of tumor microenvironment to elucidate molecular and immunologic mechanisms of response and resistance to immune checkpoint blockade. In this perspective article we discuss the recent advances in patient-derived 3D tumor models and their potential in support of treatment decision making in clinical setting. We will also share our experience with dynamic bioreactor tumor explant culture of samples from melanoma and sarcoma patients as a reliable and promising platform to unravel immune responses to immune checkpoint inhibitors.
Introduction
Rapid scientific and technological advance is revolutionizing treatment options and research tools in the era of personalized cancer therapy. The advent of immune checkpoint inhibitors (ICI) targeting the immune system has found wide application across cancer histotypes and represents the principal therapeutic intervention for advanced cancer patients (1). However, intrinsic and on-treatment resistance development remains a challenge of this therapeutic approach and major effort is dedicated to elucidate the involved mechanisms (2). In depth analysis of tumor biopsies by multi omics approaches has provided insight into the complex scenario prevailing at tumor site, but these approaches have a major limitation as they give only a snapshot of the actual conditions and ignore tumor evolution during treatment (3, 4). Co-clinical studies based on systems allowing the in-toto culture of tumor biopsies and the maintenance of the genomic and morphological characteristics of the original tumor may contribute filling this gap. In particular, ex vivo 3D culture platforms, which maintain most of the features of tumor immune microenvironment (TIME) (5) and capture drug-mediated dynamic modulations, can elucidate molecular and immune-related mechanisms underlying clinical response or resistance to therapy (6). This approach could help gathering a comprehensive mechanistic overview and define predictive biomarkers of clinical outcome in patients treated with ICI (7). Here, we provide an overview of the relevance and potential implications of the tumor explant culture system in studying ICI responses. In addition, we show the advantages of a 3D platform of dynamic culture of tumor explants in a Bioreactor for profiling molecular and immunologic mechanisms induced by ICI in human metastatic melanoma (MM) and soft tissue sarcoma (STS) samples.
Patient-derived models for cancer drug testing
The contribution of newly developed targeted drugs and immunotherapies to improve patient survival has been limited by the lack of appropriate experimental models maintaining original tumor microenvironment (TME) architecture and composition. So far, patient-derived xenografts (PDX) have become the preferred tool for drug testing, to identify novel tumor markers and therapeutic targets and to translate findings aimed at optimizing treatment of cancer patients. PDX models generated from freshly resected tumor specimens or from tumor-derived organoids transplanted into immunodeficient mice preserve the tumor intrinsic heterogeneity and can recapitulate the interactions of cancer cells with the surrounding live environment, but not with the human immune system (8). Humanized mice models grafted with human immune cells have been generated to bypass this problem (9). Nonetheless, animal models are time consuming and have high costs, and thus may not represent the rational choice for real-time precision cancer therapy. Moreover, the advent of immune based-therapies requires models able to mimic native TIME. In this scenario 3D culture systems represent a valid alternative.
3D models are classified into spheroids, organoids and patient-derived tumor explants (PDE), based on the structural complexity. Spheroids are aggregates of cells obtained from cancer cell lines or tumor biopsies, self-assembling in an environment that prevents attachment to a flat surface. They are of low complexity in mirroring tumor organization, but they may retain their endogenous extracellular matrix and many metabolic similarities to the original tissue (10). Organoids are mini-organs reconstituted and embedded in an extracellular matrix reproducing many structural and functional aspects of the parental organ. In addition to the use of pluripotent stem cells, tissue-derived tumors from patients can also be established as organoids, namely patient-derived organoids (PDO) (11). PDO typically recapitulate features and genetic characteristics of the parental tumor, as observed in PDO from colorectal and gastroesophageal cancer patients (12). Of note, organoids and PDO can be expanded and cryopreserved, a fundamental feature especially for rare cancer types (13). However, these models are characterized by the inability of preserving the native features of TIME, and even the exogenous addition of selected immune cell populations as a co-culture is insufficient to reproduce the required complexity to evaluate the response to immunotherapy.
A further level in TME complexity in 3D models is represented by PDE, that consist in the ex vivo culture of freshly-resected human tumor fragments. PDE recapitulate tissue architecture, TME and preserve the human immune system components, thus allowing the evaluation of drug responses in a 3D context. PDE preserve tumor-specific genetic alterations, transcriptomic profiles and histopathology of individual patients, allowing personalized drug screening and the identification of drug resistance mechanisms (14). LeBlanc and coworkers reported that glioblastoma-derived PDE largely retain genetic and transcriptomic heterogeneity of parent tumors, enabling the dissection of glioblastoma heterogeneity evolution during disease progression and treatment response (15). In a recent study, the inhibition of NOTCH signaling pathway in PDO of BRAFV600E/K601Q MM enhanced the sensitivity to the MEK inhibitor cobimetinib, thereby supporting the contribution of PDO models to identify therapy resistance mechanisms (16).
PDE, endowed with the native TIME, represent ideal ex vivo models for immunotherapy studies. In fact, deciphering the dynamic interactions between tumor and immune cells provides insights into the mechanisms regulating sensitivity or resistance to immune-based therapies including ICI. A further PDE model consisting of patient-derived tumor tissues embedded in collagen and termed organotypic tumor spheroids (PDOTS) maintains immune cell composition when cultured in microfluidic chips, and anticipates clinical benefit of PD-1 blockade based on cyto/chemokine release (17, 18). The addition of anti-PD-1 antibody to air-liquid interface (ALI) culture of PDO obtained from different cancer types, including MM, activates the PD-1-expressing CD3+ T cell compartment and induces cytotoxicity. The bystander increase in distance between CD8+ effector T cells and regulatory T cells (Treg) contributes avoiding Treg-mediated suppression of resident CD8+ T cells (19). Similarly, ICI efficacy in restoring anti-tumor activity of γδ T cell-based immunotherapy has been demonstrated in melanoma PDO (20, 21). In this setting, the secretion of cyto/chemokines, along with T cell activation levels measured in a PDE model of patient-derived tumor fragments (PDTF) from different tumor types embedded into an artificial extracellular matrix, could predict clinical response to PD-1 blockade (22). Similar observations were recorded following PD-1 inhibition in explant cultures of head and neck cancer, gastric and gastroesophageal adenocarcinoma (23, 24). Importantly, the PDTF platform can be also applied to identify suitable neoadjuvant treatment strategies in patients with early-stage cancer, as recently described for anti-CTLA-4 plus anti-PD-1 combined with IL-2 in PDE of checkpoint inhibitor-resistant melanoma patients (25).
3D culture models for personalized melanoma and sarcoma therapies
MM is generally characterized by a strong immune infiltrate. Its characteristically very high tumor mutational burden (TMB) and consequent tumor neoantigen expression determines its high responsiveness to ICI. However, the majority of patients demonstrate resistance either by lack of an initial response or by resistance acquisition during treatment (26). Even if tumor PD-L1 expression and TMB have been found to correlate with clinical responses to ICI in melanoma, they cannot accurately predict patient outcome due to the complex mechanisms driving immune responses (27). Multi omics analysis of tumor biopsies have identified other biomarkers predicting response of MM patients to ICI (3, 28). However, tumor biopsy does not represent a suitable system to intercept early ICI-derived immune responses, as selected modifications of resident immune cell phenotype and function possibly occur in the tumor evolving during therapy. This limitation could be overcome by TIME preserving melanoma ex vivo 3D platforms. So far, melanoma 3D culture models such as spheroids and organoids, skin reconstructs and melanoma-on-chip models that incorporate TME elements represent helpful tools to screen therapeutic agents (29). Nonetheless, limited experience exists in utilizing melanoma organoids for drug screening and preclinical studies, despite organoids can be readily generated from melanoma biopsies and can be supplemented with autologous immune cells for evaluation of immunotherapeutic responses (30). On the other hand, melanoma spheroids can be useful for lead-compound testing, as shown by the evaluation of the spheroid morphometric parameters in the screening of novel compounds like photodynamic therapy (31). To overcome MAPK-mediated radioresistance, 3D BRAF- and NRAS-mutant melanoma cell spheroids have been exploited to evaluate the efficacy of combined targeted radionuclide therapy and MEK inhibitor (32). Despite the lack of pre-existing immune infiltration, human organotypic skin melanoma cultures can be also used for the investigation of early events that regulate tumor-immunological mechanisms. In fact, this system consists in a reconstructed TME that closely resembles tumor growth, allowing the study of host-malignant cell interactions within a multicellular tissue architecture (33). Finally, melanoma PDO cultures in microfluidic 3D devices or embedded in collagen extracellular matrix represent a valid model to evaluate the effect of ICI and to predict patient clinical response (17, 22, 25).
STS are rare tumors of mesenchymal origin and encompass more than 80 histologies. In the last years, clinical trials testing ICI in advanced STS showed limited clinical activity in an unselected population of patients, with anecdotal durable responses to PD-1 blockade observed in undifferentiated pleomorphic STS and dedifferentiated liposarcoma. These STS histologies are characterized by a complex altered karyotype and are thus supposedly more immunogenic, more infiltrated by immune cells and therefore more responsive to ICI compared to translocated STS (34, 35). However, STS dichotomization in simple (skSTS) and complex karyotypes (ckSTS) does not reflect a sharp demarcation of immunogenicity. Recent studies highlighted unexpected associations between genetic and/or epigenetic features of tumor and immune composition of the TME, revealing a great heterogeneity also within the same STS type (36). Tertiary lymphoid structures (TLS), present in 20% of STS, and high tumor immune infiltration have been recently described as potential biomarkers of response to anti-PD-1 therapy (37). However, ICI resistance can be observed also in TLS-positive STS, and TME analysis revealed an enrichment of Treg in resistant TLS-positive STS compared to the sensitive counterpart (38). This indicates that a fine and systematic TME dissection is required to identify STS determinants of ICI response and resistance. Growing evidence promotes the suitability of 3D cultures biology and therapeutic efficacy studies in both STS and bone sarcoma. In Ewing-Sarcoma, spheroids enabled the evaluation of chemotherapeutic and targeted drugs and the identification of antigens for immunotherapy (39). Voissiere et al. showed in 2D and 3D chondrosarcoma models that doxorubicin resistant spheroids were sensitive to TH-302, a pro-drug activated in hypoxia, and reported that the sensitivity increased with spheroid dimensions (40). Similarly, STS organoids obtained from surgical specimens generated knowledge about STS biology, while synovial sarcoma PDO studies revealed epigenetic dysregulations (41). Of note, PDO from epithelioid sarcomas could be also established by ALI culture method (42). To evaluate efficacy of Wee1 inhibitor MK-1775, alone and in combination with gemcitabine, PDE models of undifferentiated high-grade sarcoma, malignant peripheral nerve sheath tumors, pleomorphic spindle cell sarcoma and osteosarcoma were successfully applied (43). Recently, PDO derived from STS subtypes were generated by mixing tumor cell suspensions with extracellular matrix hydrogel and with autologous immune cells to perform chemotherapy and immunotherapy screening, thus highlighting the potential application of PDO also in STS (44).
PDE as an optimal 3D platform to investigate ICI treatment efficacy in melanoma and sarcoma
PDE implementation in clinical practice is still hampered by several limitations. They include the relative short-term viability of the cultures, as evidenced for endometrial PDE that remained viable only for one day, and the lack of a functional vascular system, impacting oxygen and nutrient diffusion and waste removal (14). To circumvent these limitations, an innovative 3D dynamic culture system, based on the use of the microgravity technology provided by the Rotary Cell Culture System (RCCS) Bioreactor was developed, enabling long-term culture of tissue explants.
This device was first dedicated to 3D tissue engineering of cartilage and human collagen bone matrix formation (45, 46), demonstrating how the RCCS-based fluid dynamic culture conditions influence tissue growth and regeneration. Such conditions favor 3D assembly of single cells into aggregates, as shown in the case of astrocyte-like or neuronal-like models reproducing neuronal features (47, 48). RCCS Bioreactor culture was further validated for its ability to preserve the morphological and functional features in multiple myeloma tissue components. These studies showed that seven days treatment with Bortezomib of a peritoneal myeloma PDE retrieved from a responsive patient decreased tumor viability and release of the pro-angiogenic factors VEGF and angiopoietin-2. Conversely, no effect was observed in a myeloma PDE retrieved from a non-responding patient, suggesting that RCCS Bioreactor-based cultures may reflect the drug response observed in vivo (49). Moreover, the simulated microgravity in RCCS Bioreactor-based cultures could modify the ultrastructure of human breast cancer cells and was accompanied by increased apoptosis and inhibition of their migration ability (50). In addition, simulated microgravity influenced metabolic pathways, such as lipid metabolism and the Krebs cycle in human breast cancer cells, osteoblasts, oligodendrocytes and gastric cancer cells (51–54). Of note, metabolomic analysis of supernatants of Erdheim-Chester disease (ECD) biopsies cultured for 4 days in RCCS Bioreactor revealed that the immunometabolic changes of trained immunity occurring in ECD lesions in vivo, such as increased glycolysis and accumulation of fumarate and malate, can be dampened by MAPK pathway inhibition (55). A recent study in gastric cancer showed that PDE culture in RCCS bioreactor recapitulates the action of chemotherapy in patients by affecting cell survival and by weakening the action of drug resistance-associated genes (56).
3D dynamic cultures of PDE in the RCCS Bioreactor can be reproducibly applied to study the effects of exposure to drugs, resistance mechanisms and molecular signaling occurring during the interactions between tumor cells and immune infiltrate (57). The absence of an ALI interface and the hydrodynamic forces acting inside the RCCS Bioreactor culture vessels permit efficient gas inter-exchange, thereby optimizing oxygen and nutrient supply, catabolite removal and drug tissue distribution. While 3D cultures are typically short-term cultures and thus amenable only to short-term drug treatments, one fundamental asset of this approach is the reliable long-term maintenance of the tissue, which permits up to fourteen days culture of differentiated tissues like tumors (46–49). This allows to investigate the modulating activities of drugs acting on cancer and/or TME and TIME.
We set up a RCCS Bioreactor PDE platform at our unit to investigate mechanisms associated with response/resistance to ICI, with a particular focus on MM and STS. Samples obtained at surgical tumor resection were processed by a 3 mm biopsy puncher within 24 hours upon storage at 4°C in tissue storage solution to obtain PDE. PDE were cultured in RCCS Bioreactor upon drug treatment and then analyzed for drug response by gene expression profiling, tissue immunohistochemistry, cyto-chemokine release in culture supernatants (Figure 1A). PDE can be frozen in cell cryopreservation medium and banked in liquid nitrogen for prospective studies (Supplementary materials and methods). Several PDE can be obtained from one clinical sample, allowing comparative testing of different drugs and analysis of modifications at different time points. A particular feature of RCCS Bioreactor 3D cultures is the uniform tissue distribution of drugs, which is guaranteed by the low shear force in the rotating vessels (Figure 1B).
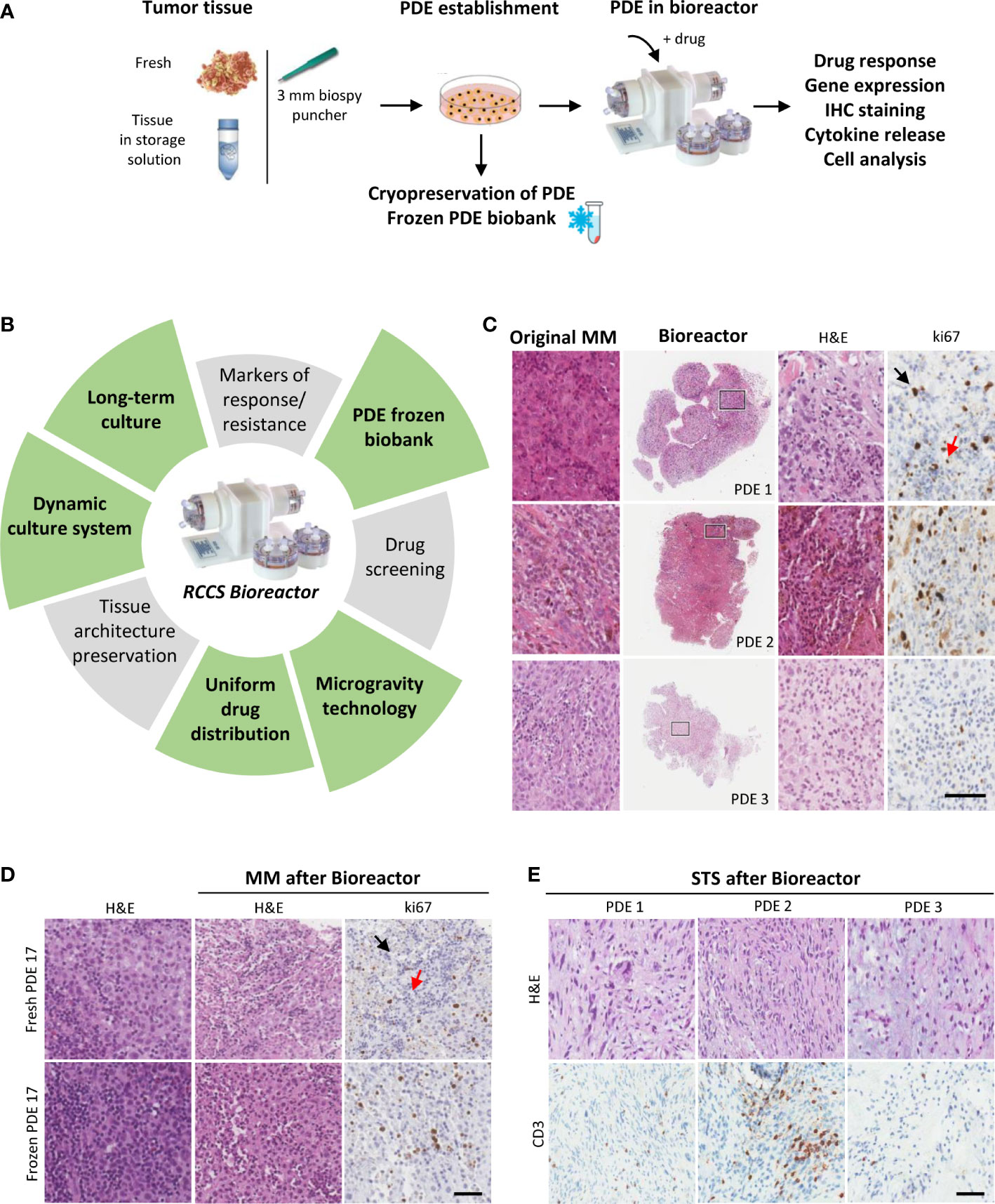
Figure 1 Dynamic 3D culture of PDE in RCCS Bioreactor preserves TIME architecture of MM and STS. (A) Strategy to establish tumor PDE to evaluate drug effects after culture in Bioreactor. (B) Main features common of PDE cultures (grey sectors) and those specific of PDE cultured in RCCS Bioreactor (green sectors). (C) Representative histological images of MM PDE after 3 days culture in Bioreactor compared to the original tumors. Right panels show higher magnification of the marked areas. (D) Representative images of ki67 stained fresh or frozen MM PDE before and after culture in Bioreactor. (E) Representative images of CD3 staining of frozen STS PDE after culture in Bioreactor. PDE 1: ckSTS, a myxofibrosarcoma of the extremities; PDE2: ckSTS, a retroperitoneal dedifferentiated liposarcoma; PDE 3: skSTS, a myxoid liposarcoma of the extremities. Ki67 staining of proliferating tumor cells and lymphocytes are indicated by black and red arrows, respectively. H&E, hematoxylin and eosin. Scale bar 80 μM (C), and 200 μM (D, E).
Our experiments show that melanoma PDE cultured for three days in RCCS Bioreactor vessels maintain the characteristics of the original tumors, including TME/TIME architecture and cellularity, viability of tumor cells and infiltrating lymphocytes, as indicated by ki67 staining of both cell types (Figure 1C). Moreover, this culture system prevented the efflux of infiltrating immune cells evidenced by the absence of CD45+ cells in culture supernatants (data not shown). A clear advantage of the RCCS Bioreactor PDE model system is the preservation of the original TME/TIME viability in banked tumor fragments frozen in vitality-preserving freezing buffer (Figure 1D). We could observe the preservation of tumor tissue histo-architecture and TME also in STS samples, as demonstrated in two representative cases of ckSTS, a myxofibrosarcoma of the extremities and a retroperitoneal dedifferentiated liposarcoma, and in one case of skSTS, a myxoid liposarcoma of the extremities (Figure 1E). These features of RCCS Bioreactor allow the creation of a biobank for simultaneous testing and retrospective evaluation. Exposure to BRAF/MEK inhibitors of PDE deriving from treatment naïve melanoma patients revealed determined a reduced number of ki67 proliferating melanoma cells (57). This setting also enabled us to evaluate T cell function after exposure to anti-PD-1 antibody of PDE obtained at baseline from patients clinically responding to adjuvant nivolumab, as demonstrated by the significant upregulation of a gene signature associated to immune response activation (IFNG, GZMB, PRF1, CD8, TIM3, PDCD1) (Figure 2A). PDE tissue immunostaining confirmed the increase of Granzyme B positive cells compared to untreated control samples (2-5 fold increase), together with a reduction of ki67 positive proliferating melanoma cells (Figure 2B). The analysis of PDE culture supernatants revealed also a boost in the release of Granzyme B and Fractalkine/CX3CL1 and the decrease of IL-8, confirming the immune modulation after PD-1 blocking (Figure 2C). Similar results were obtained in STS samples, where treatment with nivolumab increased the gene expression of the T cell activation markers IFNγ, Granzyme B and TNFα (Figure 2D). As in MM, also in STS, banking of tumor samples for 3D testing should be included in prospective studies to allow investigations of personalized therapies.
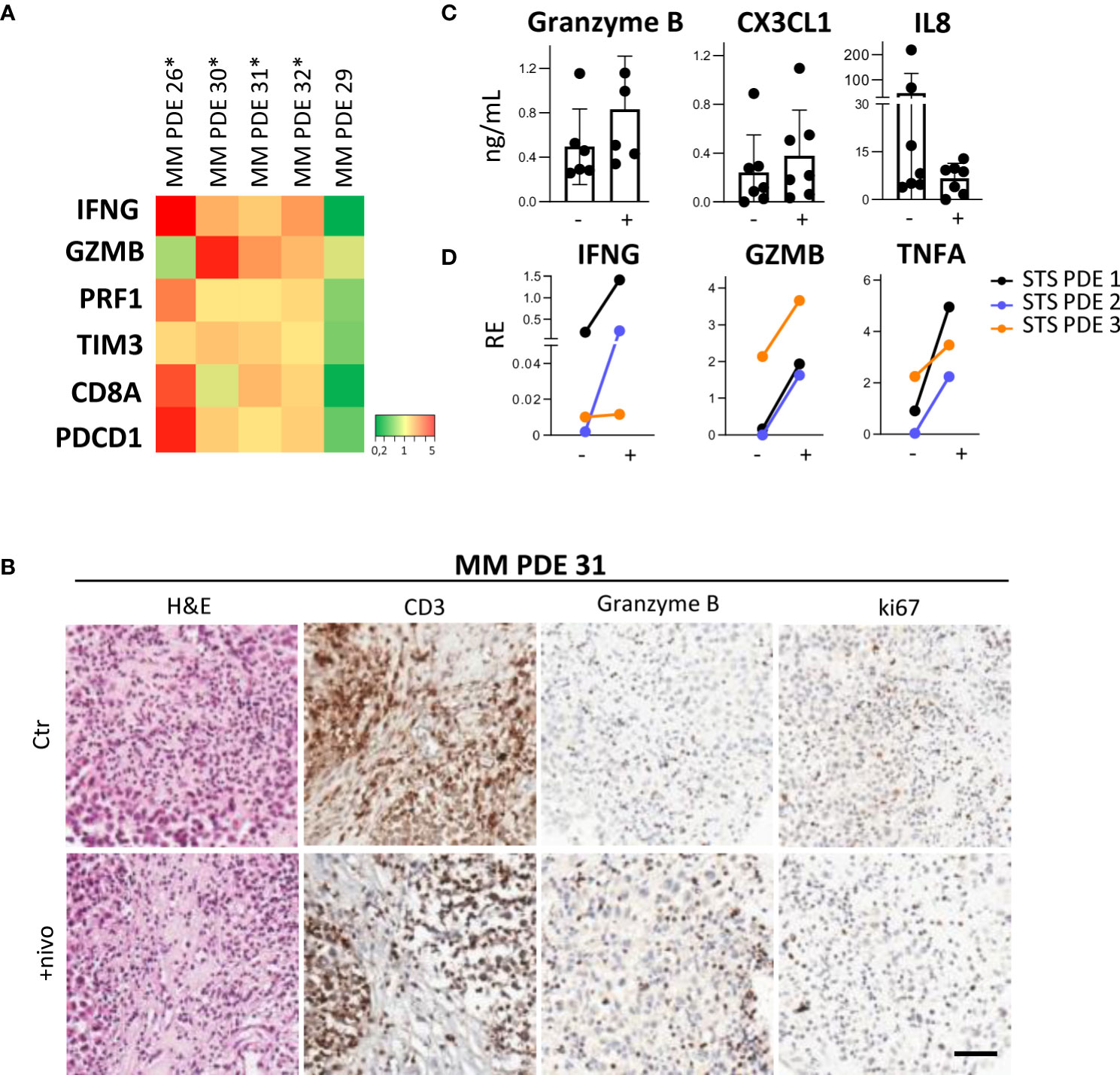
Figure 2 Ex-vivo treatment with nivolumab of 3D PDE in RCCS Bioreactor. (A) Heatmap showing gene expression levels in MM PDE deriving from lymph node metastases of treatment-naïve melanoma patients. Ratio of PDE treated with nivolumab and untreated control, p=0.013 by MANOVA. *: patients responding to adjuvant immunotherapy (absence of relapse at one year). (B) IHC staining of CD3, Granzyme B and ki67 in a representative case of MM PDE. Ctr: Control; +nivo: +nivolumab. (C) Granzyme B, CX3CL1 and IL8 in culture supernatants from MM PDE. (D) Expression of IFNG, Granzyme B (GZMB) and TNFA transcripts in PDE from STS after treatment compared to untreated control by qRT-PCR. p < 0.05 by paired Student’s t test. RE, Relative Expression. -, untreated control; +, nivolumab.
Discussion and conclusions
In comparison to PDX models, PDE, by maintaining the phenotype and TME of the individual tumor, may provide a realistic environment to assess the functional ex vivo response to drugs. Several studies highlighted the usefulness of PDE-based drug screening and their predictive value or association with clinical responses, confirming the usefulness of this system to prospectively assess individual patient response (7, 13, 58).
Our findings indicate that 3D RCCS Bioreactor-based culture of PDE can be exploited as a preclinical approach to investigate melanoma and sarcoma clinical response to current targeted and immune therapies (57). Of note, in a subset of melanoma explants from nivolumab responders, we measured increased IFNγ and Granzyme B levels, suggesting the activation of infiltrating T cell effector functions. We observed analogous effects in PDE from STS, representative of both ckSTS and skSTS. Our data suggest that ex vivo ICI treatment of PDE simulates an active antitumor immune response, thereby providing a proof-of-concept strategy to identify patients who might benefit from such kind of treatment. Our results are in line with the numerous other investigators operating in the 3D cancer model field (15, 17, 20, 22, 24, 25) and we encourage the establishment in the clinics of personalized ex vivo platforms to test immunotherapy efficacy, define prediction markers and develop combination strategies. We underline that preclinical or co-clinical testing of immunotherapeutic agents must rely on 3D models, such as PDO (59), PDOTS (17) or PDTF (22), representing the most accurate patient-tailored TME replicas. By leveraging cryopreserved PDE, this system permits comparing retrospective samples from immunotherapy responders vs non-responders to unravel the local immunomodulatory effects and identify predictive markers. Based on our experience, cultures of frozen PDE retain the same morphology characteristics as cultures derived from fresh PDE regardless of freezing and storage time, supporting the creation of multicentric PDE biobanks. In a desirably close future, routine 3D personalized testing may improve accuracy and efficiency of personalized therapeutic approaches ultimately impacting clinical outcome of cancer patients.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The study was approved by the Independent Ethic Committee and by the review board of Fondazione IRCCS Istituto Nazionale dei Tumori (protocol INT92/17 and INT85/10) and written informed consent was obtained from patients.
Author contributions
MR and EV set up the system, supervised the experiments, wrote and revised the manuscript. MC, BEL, and GG selected and analyzed specimens. EV performed experiments. EV, VV, and MR analyzed the results. VH and LR wrote and revised the manuscript. All authors critically revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by Italian Ministry of Health, RCR-2021 to Alleanza Contro il Cancro Melanoma Working Group, Italian Ministry of Health (RF-2019-12370923), AIRC-IG 25078 to VH, AIRC IG 20752 to LR, CARIPLO Foundation to VV (2015-0911).
Acknowledgments
We sincerely thank Barbara Vergani, Simona Frigerio, Francesca Rini and Agata Cova for their excellent technical support and Sandro Pasquali, Paola Collini and Laura Bergamaschi for helpful suggestions and discussion.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.1068091/full#supplementary-material
References
1. Ugurel S, Röhmel J, Ascierto PA, Becker JC, Flaherty KT, Grob JJ, et al. Survival of patients with advanced metastatic melanoma: The impact of MAP kinase pathway inhibition and immune checkpoint inhibition - update 2019. Eur J Cancer (2020) 130:126–38. doi: 10.1016/j.ejca.2020.02.021
2. Wang S, Xie K, Liu T. Cancer immunotherapies: From efficacy to resistance mechanisms - not only checkpoint matters. Front Immunol (2021) 12:690112. doi: 10.3389/fimmu.2021.690112
3. Huang AC, Zappasodi R. A decade of checkpoint blockade immunotherapy in melanoma: understanding the molecular basis for immune sensitivity and resistance. Nat Immunol (2022) 23:660–70. doi: 10.1038/s41590-022-01141-1
4. Newell F, Pires da Silva I, Johansson PA, Menzies AM, Wilmott JS, Addala V, et al. Multiomic profiling of checkpoint inhibitor-treated melanoma: Identifying predictors of response and resistance, and markers of biological discordance. Cancer Cell (2022) 40(1):88–102.e7. doi: 10.1016/j.ccell.2021.11.012
5. Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med (2018) 24(5):541–50. doi: 10.1038/s41591-018-0014-x
6. Wensink GE, Elias SG, Mullenders J, Koopman M, Boj SF, Kranenburg OW, et al. Patient-derived organoids as a predictive biomarker for treatment response in cancer patients. NPJ Precis Oncol (2021) 5(1):30. doi: 10.1038/s41698-021-00168-1
7. Sun CP, Lan HR, Fang XL, Yang XY, Jin KT. Organoid models for precision cancer immunotherapy. Front Immunol (2022) 13:770465. doi: 10.3389/fimmu.2022.770465
8. Abdolahi S, Ghazvinian Z, Muhammadnejad S, Saleh M, Asadzadeh Aghdaei H, Baghaei K. Patient-derived xenograft (PDX) models, applications and challenges in cancer research. J Transl Med (2022) 20(1):206. doi: 10.1186/s12967-022-03405-8
9. De La Rochere P, Guil-Luna S, Decaudin D, Azar G, Sidhu SS, Piaggio E. Humanized mice for the study of immuno-oncology. Trends Immunol (2018) 39(9):748–63. doi: 10.1016/j.it.2018.07.001
10. Nath S, Devi GR. Three-dimensional culture systems in cancer research: Focus on tumor spheroid model. Pharmacol Ther (2016) 163:94–108. doi: 10.1016/j.pharmthera.2016.03.013
11. Drost J, Clevers H. Organoids in cancer research. Nat Rev Cancer (2018) 18(7):407–18. doi: 10.1038/s41568-018-0007-6
12. Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernández-Mateos J, Khan K, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science (2018) 359(6378):920–6. doi: 10.1126/science.aao2774
13. Wang J, Chen C, Wang L, Xie M, Ge X, Wu S, et al. Patient-derived tumor organoids: New progress and opportunities to facilitate precision cancer immunotherapy. Front Oncol (2022) 12:872531. doi: 10.3389/fonc.2022.872531
14. Powley IR, Patel M, Miles G, Pringle H, Howells L, Thomas A, et al. Patient-derived explants (PDEs) as a powerful preclinical platform for anti-cancer drug and biomarker discovery. Br J Cancer (2020) 122:735–44. doi: 10.1038/s41416-019-0672-6
15. LeBlanc VG, Trinh DL, Aslanpour S, Hughes M, Livingstone D, Jin D, et al. Single-cell landscapes of primary glioblastomas and matched explants and cell lines show variable retention of inter- and intratumor heterogeneity. Cancer Cell (2022) 40(4):379–92.e9. doi: 10.1016/j.ccell.2022.02.016
16. Porcelli L, Di Fonte R, Pierri CL, Fucci L, Saponaro C, Armenio A, et al. BRAFV600E;K601Q metastatic melanoma patient-derived organoids and docking analysis to predict the response to targeted therapy. Pharmacol Res (2022) 182:106323. doi: 10.1016/j.phrs.2022.106323
17. Jenkins RW, Aref AR, Lizotte PH, Ivanova E, Stinson S, ZhouJenkins CW, et al. Ex vivo profiling of PD-1 blockade using organotypic tumor spheroids. Cancer Discovery (2018) 8:196–215. doi: 10.1158/2159-8290.CD-17-0833
18. Cui X, Ma C, Vasudevaraja V, Serrano J, Tong J, Peng Y, et al. Dissecting the immunosuppressive tumor microenvironments in glioblastoma-on-a-Chip for optimized PD-1 immunotherapy. Elife (2020) 9:e52253. doi: 10.7554/eLife.52253
19. Neal JT, Li X, Zhu J, Giangarra V, Grzeskowiak CL, Ju J, et al. Organoid modeling of the tumor immune microenvironment. Cell (2018) 175(7):1972–88.e16. doi: 10.1016/j.cell.2018.11.021
20. Ou L, Wang H, Huang H, Zhou Z, Lin Q, Guo Y, et al. Preclinical platforms to study therapeutic efficacy of human γδ T cells. Clin Transl Med (2022) 12(6):e814. doi: 10.1002/ctm2.814
21. Huber V, Vallacchi V, Daveri E, Vergani E. 3D models for melanoma γδ T cell-based immunotherapy. Clin Transl Med (2022) 12(6):e926. doi: 10.1002/ctm2.926
22. Voabil P, de Bruijn M, Roelofsen LM, Hendriks SH, Brokamp S, van den Braber M, et al. An ex vivo tumor fragment platform to dissect response to PD-1 blockade in cancer. Nat Med (2021) 27:1250–61. doi: 10.1038/s41591-021-01398-3
23. Sharon S, Duhen T, Bambina S, Baird J, Leidner R, Bell B, et al. Explant modeling of the immune environment of head and neck cancer. Front Oncol (2021) 11:611365. doi: 10.3389/fonc.2021.611365
24. Hußtegge M, Hoang NA, Rebstock J, Monecke A, Gockel I, Weimann A, et al. PD-1 inhibition in patient derived tissue cultures of human gastric and gastroesophageal adenocarcinoma. Oncoimmunology (2021) 10(1):1960729. doi: 10.1080/2162402X.2021.1960729
25. Kaptein P, Jacoberger-Foissac C, Dimitriadis P, Voabil P, de Bruijn M, Brokamp S, et al. Addition of interleukin-2 overcomes resistance to neoadjuvant CTLA4 and PD1 blockade in ex vivo patient tumors. Sci Transl Med (2022) 14(642):eabj9779. doi: 10.1126/scitranslmed.abj9779
26. Abbott CW, Boyle SM, Pyke RM, McDaniel LD, Levy E, Navarro FCP, et al. Prediction of immunotherapy response in melanoma through combined modeling of neoantigen burden and immune-related resistance mechanisms. Clin Cancer Res (2021) 27(15):4265–76. doi: 10.1158/1078-0432.CCR-20-4314
27. Yarchoan M, Albacker LA, Hopkins AC, Montesion M, Murugesan K, Vithayathil TT, et al. PD-L1 expression and tumor mutational burden are independent biomarkers in most cancers. JCI Insight (2019) 4(6):e126908. doi: 10.1172/jci.insight.126908
28. Patterson A, Auslander N. Mutated processes predict immune checkpoint inhibitor therapy benefit in metastatic melanoma. Nat Commun (2022) 13:5151. doi: 10.1038/s41467-022-32838-4
29. Marconi A, Quadri M, Saltari A, Pincelli C. Progress in melanoma modelling in vitro. Exp Dermatol (2018) 27(5)::578–586. doi: 10.1111/exd.13670
30. Baker K. Organoids provide an important window on inflammation in cancer. Cancers (Basel) (2018) 10(5):151. doi: 10.3390/cancers10050151
31. Aguilar Cosme JR, Gagui DC, Bryant HE, Claeyssens F. Morphological response in cancer spheroids for screening photodynamic therapy parameters. Front Mol Biosci (2021) 8:784962. doi: 10.3389/fmolb.2021.784962
32. Akil H, Quintana M, Raymond JH, Billoux T, Benboubker V, Besse S, et al. Efficacy of targeted radionuclide therapy using [131I]ICF01012 in 3D pigmented BRAF- and NRAS-mutant melanoma models and In vivo NRAS-mutant melanoma. Cancers (Basel) (2021) 13(6):1421. doi: 10.3390/cancers13061421
33. Di Blasio S, van Wigcheren GF, Becker A, van Duffelen A, Gorris M, Verrijp K, et al. The tumour microenvironment shapes dendritic cell plasticity in a human organotypic melanoma culture. Nat Commun (2020) 11(1):2749. doi: 10.1038/s41467-020-16583-0
34. Birdi HK, Jirovec A, Cortés-Kaplan S, Werier J, Nessim C, Diallo JS, et al. Immunotherapy for sarcomas: new frontiers and unveiled opportunities. J Immunother Cancer (2021) 9(2):e001580. doi: 10.1136/jitc-2020-001580
35. Roulleaux Dugage M, Nassif EF, Italiano A, Bahleda R. Improving immunotherapy efficacy in soft-tissue sarcomas: A biomarker driven and histotype tailored review. Front Immunol (2021) 12:775761. doi: 10.3389/fimmu.2021.775761
36. Tazzari M, Bergamaschi L, De Vita A, Collini P, Barisella M, Bertolotti A, et al. Molecular determinants of soft tissue sarcoma immunity: Targets for immune intervention. Int J Mol Sci (2021) 22(14):7518. doi: 10.3390/ijms22147518
37. Petitprez F, de Reyniès A, Keung EZ, Chen TW, Sun CM, Calderaro J, et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature (2020) 577(7791):556–60. doi: 10.1038/s41586-019-1906-8
38. Italiano A, Bessede A, Pulido M, Bompas E, Piperno-Neumann S, Chevreau C, et al. Pembrolizumab in soft-tissue sarcomas with tertiary lymphoid structures: a phase 2 PEMBROSARC trial cohort. Nat Med (2022) 28(6):1199–206. doi: 10.1038/s41591-022-01821-3
39. Lamhamedi-Cherradi SE, Santoro M, Ramammoorthy V, Menegaz BA, Bartholomeusz G, Iles LR, et al. 3D tissue-engineered model of ewing's sarcoma. Adv Drug Delivery Rev (2014) 79-80:155–71. doi: 10.1016/j.addr.2014.07.012
40. Voissiere A, Jouberton E, Maubert E, Degoul F, Peyrode C, Chezal JM, et al. Development and characterization of a human three-dimensional chondrosarcoma culture for in vitro drug testing. PloS One (2017) 12(7):e0181340. doi: 10.1371/journal.pone.0181340
41. Boulay G, Cironi L, Garcia SP, Rengarajan S, Xing YH, Lee L, et al. The chromatin landscape of primary synovial sarcoma organoids is linked to specific epigenetic mechanisms and dependencies. Life Sci Alliance (2020) 4(2):e202000808. doi: 10.26508/lsa.202000808
42. Wakamatsu T, Ogawa H, Yoshida K, Matsuoka Y, Shizuma K, Imura Y, et al. Establishment of organoids from human epithelioid sarcoma with the air-liquid interface organoid cultures. Front Oncol (2022) 12:893592. doi: 10.3389/fonc.2022.893592
43. Kreahling JM, Foroutan P, Reed D, Martinez G, Razabdouski T, Bui MM, et al. Wee1 inhibition by MK-1775 leads to tumor inhibition and enhances efficacy of gemcitabine in human sarcomas. PloS One (2013) 8(3):e57523. doi: 10.1371/journal.pone.0057523
44. Forsythe SD, Sivakumar H, Erali RA, Wajih N, Li W, Shen P, et al. Patient-specific sarcoma organoids for personalized translational research: Unification of the operating room with rare cancer research and clinical implications. Ann Surg Oncol (2022) 29(12):7354–67. doi: 10.1245/s10434-022-12086-y
45. Cinbiz MN, Tığli RS, Beşkardeş IG, Gümüşderelioğlu M, Colak U. Computational fluid dynamics modeling of momentum transport in rotating wall perfused bioreactor for cartilage tissue engineering. J Biotechnol (2010) 150(3):389–95. doi: 10.1016/j.jbiotec.2010.09.950
46. Mazzoleni G, Boukhechbab F, Steimberga N, Boniottia J, Boulerc JM and Rochetd N. Impact of dynamic culture in the RCCS bioreactor on a three-dimensional model of bone matrix formation. Proc Engineering (2011) 10:3662–7. doi: 10.1016/j.proeng.2011.04.603
47. Morabito C, Steimberg N, Mazzoleni G, Guarnieri S, Fanò-Illic G, Mariggiò MA. RCCS bioreactor-based modelled microgravity induces significant changes on in vitro 3D neuroglial cell cultures. BioMed Res Int (2015) 2015:754283. doi: 10.1155/2015/754283
48. Cui Y, Yin Y, Zou Y, Zhao Y, Han J, Xu B, et al. The rotary cell culture system increases NTRK3 expression and promotes neuronal differentiation and migratory ability of neural stem cells cultured on collagen sponge. Stem Cell Res Ther (2021) 12(1):298. doi: 10.1186/s13287-021-02381-y
49. Ferrarini M, Steimberg N, Ponzoni M, Belloni D, Berenzi A, Girlanda S, et al. Ex-vivo dynamic 3-d culture of human tissues in the RCCS™ bioreactor allows the study of multiple myeloma biology and response to therapy. PLoS One (2013) 8(8):e71613. doi: 10.1371/journal.pone.0071613
50. Jiang N, Chen Z, Li B, Guo S, Li A, Zhang T, et al. Effects of rotary cell culture system-simulated microgravity on the ultrastructure and biological behavior of human MDA-MB-231 breast cancer cells. Precis Radiat Oncol (2019) 3:87–93. doi: 10.1002/pro6.1074
51. Chen L, Yang X, Cui X, Jiang MM, Gui Y, Zhang YN, et al. Adrenomedullin is a key protein mediating rotary cell culture system that induces the effects of simulated microgravity on human breast cancer cells. Microgravity Sci Technol (2015) 27:417–26. doi: 10.1007/s12217-015-9434-0
52. Michaletti A, Gioia M, Tarantino U, Zolla L. Effects of microgravity on osteoblast mitochondria: A proteomic and metabolomics profile. Sci Rep (2017) 7:15376. doi: 10.1038/s41598-017-15612-1
53. Espinosa-Jeffrey A, Nguyen K, Kumar S, Toshimasa O, Hirose R, Reue K, et al. Simulated microgravity enhances oligodendrocyte mitochondrial function and lipid metabolism. J Neurosci Res (2016) 94:1434–50. doi: 10.1002/jnr.23958
54. Chen ZY, Jiang N, Guo S, Li BB, Yang JQ, Chai SB, et al. Effect of simulated microgravity on metabolism of HGC-27 gastric cancer cells. Oncol Lett (2020) 19(5):3439–50. doi: 10.3892/ol.2020.11451
55. Molteni R, Biavasco R, Stefanoni D, Nemkov T, Domínguez-Andrés J, Arts RJ, et al. Oncogene-induced maladaptive activation of trained immunity in the pathogenesis and treatment of erdheim-Chester disease. Blood (2021) 138(17):1554–69. doi: 10.1182/blood.2020009594
56. Rembiałkowska N, Baczyńska D, Dubińska-Magiera M, Choromańska A, Bieżuńska-Kusiak K, Gajewska-Naryniecka A, et al. RCCS bioreactor-based modeled microgravity affects gastric cancer cells and improves the chemotherapeutic effect. Membranes (Basel) (2022) 12(5):448. doi: 10.3390/membranes12050448
57. Vergani E, Dugo M, Cossa M, Frigerio S, Di Guardo L, Gallino G, et al. miR-146a-5p impairs melanoma resistance to kinase inhibitors by targeting COX2 and regulating NFkB-mediated inflammatory mediators. Cell Commun Signal (2020) 18:156. doi: 10.1186/s12964-020-00601-1
58. Guzzeloni V, Veschini L, Pedica F, Ferrero E, Ferrarini M. 3D models as a tool to assess the anti-tumor efficacy of therapeutic antibodies: Advantages and limitations. Antibodies (Basel) (2022) 11(3):46. doi: 10.3390/antib11030046
Keywords: patient-derived 3D tumor explants, bioreactor, immunotherapy, melanoma, sarcoma
Citation: Rodolfo M, Huber V, Cossa M, Gallino G, Leone BE, Vallacchi V, Rivoltini L and Vergani E (2022) 3D tumor explant as a novel platform to investigate therapeutic pathways and predictive biomarkers in cancer patients. Front. Immunol. 13:1068091. doi: 10.3389/fimmu.2022.1068091
Received: 12 October 2022; Accepted: 30 November 2022;
Published: 14 December 2022.
Edited by:
Marcel Deckert, U1065 Centre Meíditerraneíen de Meídecine Moleículaire (INSERM), FranceReviewed by:
Fernanda Silva, Universidade NOVA de Lisboa, PortugalCopyright © 2022 Rodolfo, Huber, Cossa, Gallino, Leone, Vallacchi, Rivoltini and Vergani. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Monica Rodolfo, monica.rodolfo@istitutotumori.mi.it
 Monica Rodolfo
Monica Rodolfo