
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol. , 28 October 2022
Sec. Cancer Immunity and Immunotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.1050982
This article is part of the Research Topic Community Series in Manipulating the Immunological Tumor Microenvironment - Volume II View all 20 articles
 Huimin Liu1,2†
Huimin Liu1,2† Dongxu Wang3†
Dongxu Wang3† Zhijing Yang1,2
Zhijing Yang1,2 Shuangji Li1,2
Shuangji Li1,2 Han Wu1,2
Han Wu1,2 Jingcheng Xiang1,2
Jingcheng Xiang1,2 Shaoning Kan1,2
Shaoning Kan1,2 Ming Hao1,2
Ming Hao1,2 Weiwei Liu1,2*
Weiwei Liu1,2*Head and neck tumours are common malignancies that are associated with high mortality. The low rate of early diagnosis and the high rates of local recurrence and distant metastasis are the main reasons for treatment failure. Recent studies have established that the tumour microenvironment (TME) can affect the proliferation and metastasis of head and neck tumours via several mechanisms, including altered expressions of certain genes and cytokines. Increasing evidence has shown that epigenetic modifications, such as DNA methylation, histone modification, RNA modification, and non-coding RNAs, can regulate the head and neck TME and thereby influence tumour development. Epigenetic modifications can regulate the expression of different genes and subsequently alter the TME to affect the progression of head and neck tumours. In addition, the cell components in the TME are regulated by epigenetic modifications, which, in turn, affect the behaviour of head and neck tumour cells. In this review, we have discussed the functions of epigenetic modifications in the head and neck TME. We have further examined the roles of such modifications in the malignancy and metastasis of head and neck tumours.
Globally, head and neck tumours are the most common malignant tumours (1). More than 430,000 people die from the disease annually (1), and 90% of the cases involve head and neck squamous cell carcinoma (HNSCC) (2). It is considered the dominant phenotype and is associated with cervical lymph node metastasis and the progression of malignancy (3). Surgical resection is the mainstay of HNSCC treatment; however, it is often unsatisfactory owing to the uncertainty of tumour boundaries and the potential for aggressive lymphatic metastasis (4). Many therapies are available for head and neck tumour microenvironment (TME), including immunotherapy and anti-angiogenic therapy (5, 6). Immunological monotherapy and combination therapy have been shown to prolong the survival of patients (7), and anti-angiogenic therapy is an attractive option for overcoming hypoxia and radiation in head and neck tumours (6). Both immunotherapy and anti-angiogenic therapy are closely linked to alterations in the TME (8, 9). Therefore, it is vital to explore the impact of TME on head and neck tumours.
The TME includes both cellular and non-cellular components, such as stromal cells, immune cells, and chemokines (10). The processes of proliferation, apoptosis resistance, invasion, migration, and immune evasion of cancer cells are considered to be related to TME (11-15). The acidic and hypoxic TME creates a unique growth environment for cancer cells, which enables them to resist the immune response (16, 17). The TME can influence tumour progression via multiple mechanisms, such as Notch and STAT3 signalling pathways (18, 19). Furthermore, m6A modifications are involved in the regulation of TME. For example, the m6A demethylase ALKBH5 can regulate the tumour immune microenvironment (TIME) of HNSCC via the RIG-I/IFNα signalling pathway (20), which suggests that epigenetic modifications are involved in the regulation of the TME in head and neck tumours. Therefore, the characteristics of the TME should be explored, and the mechanisms regulating epigenetic modifications of the TME in HNSCC must be clarified.
Epigenetic alterations lead to abnormal gene expression in the cells in the TME and are linked to the development of cancer (21). Recent studies have shown that TME is regulated by epigenetic modifications (22). In this review, we have summarised the interactions between epigenetic modifications and the TME as well as their effects on the fate of HNSCC. These insights may help in identifying new potential targets for the effective treatment of head and neck tumours.
The microenvironment of head and neck cancer is a complex system that is composed of non-tumour cells, an extracellular matrix, and a vascular system (23). The TME is characterized by hypoxia, high angiogenic factor content, and immunosuppression and is involved in tumour growth, metastasis, and invasion (6, 24, 25). In addition to conventional approaches, such as surgical treatment, radiotherapy, and chemotherapy, anti-angiogenic and immunosuppressive therapies are novel directions for treating these tumours. Therefore, we focussed on the role of the TME in angiogenesis and immune responses (Figure 1).
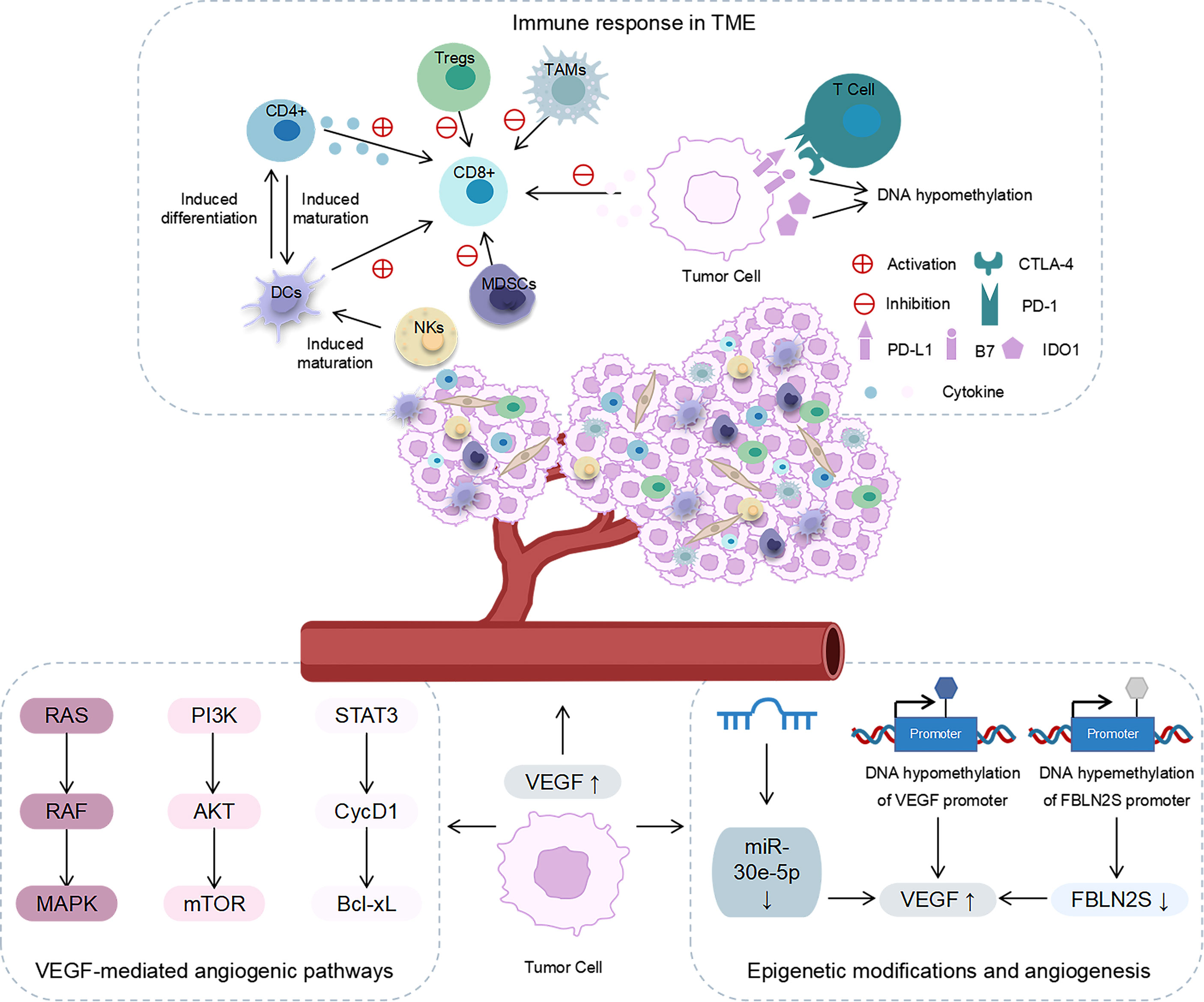
Figure 1 The role of angiogenesis and immune response in the TME. Immune response in the TME: NKs and CD4+ induced DC maturation; DCs promoted the activation of CD8+T cells and induced CD4+T cells to differentiate into antigen-specific effector T cells; CD4+ secreted cytokines to activate CD8+T cells; MDSCs, Tregs and TAMs inhibited the tumour-killing effect of CD8+T cells; cancer cells released cytokines to inhibit T-cell proliferation and the effector function; PD-1/PD-L1 interaction inhibited the function of effector T cells; CTLA-4 binds to B7 ligands to inhibit T cell activation; IDO1 inhibited the function of T cells; the expression levels of CTLA-4 and IDO1 were regulated by DNA methylation. Angiogenesis in the TME: VEGF affected angiogenesis through RAS/RAF/MAPK, PI3K/AKT/mTOR, and STAT3/cycD1/Bcl-xL signalling pathways. Epigenetic modification affected the expression of VEGF and the angiogenesis in the TME.
The rapid proliferation of cells in malignant tumours requires a supply of oxygen and nutrients via the bloodstream (26). Angiogenesis is a key factor that affects the progression of head and neck cancers (27, 28). Microvessel density (MVD) is an indirect marker of tumour angiogenesis and is associated with poor prognosis in HNSCC (29). Disease-free survival and overall survival are significantly reduced in patients with high CD105+ MVD (29). A study has reported that increased MVD in head and neck tumours promotes the proliferation of cancer cells and leads to poor tissue differentiation and lymph node metastasis (30). Therefore, the mechanism of angiogenesis in head and neck cancer needs to be explored for developing targeted angiogenic therapy.
The downstream signalling pathway of angiogenesis is mainly mediated by vascular endothelial growth factor (VEGF) (31). It affects angiogenesis chiefly via three signalling pathways, namely, RAS/RAF/MAPK, PI3K/AKT/mTOR, and STAT3/cycD1/Bcl-xL (6). In head and neck tumours, VEGF-C has been observed to promote cell growth and migration (32). Recent studies have revealed that epigenetic modifications can directly or indirectly regulate the expression of VEGF and, thus, play a role in the angiogenesis in head and neck tumours. For instance, sevoflurane can reduce the angiogenic ability of tongue squamous cell carcinoma by enhancing DNA methylation in the VEGF promoter region (33). In nasopharyngeal carcinoma, high DNA methylation of the FBLN2S promoter inhibits the expression of FBLN2S, while overexpression of FBLN2S downregulates the expression of angiogenesis-related factors such as VEGF165 and VEGF189 and inhibits angiogenesis (34). Moreover, non-coding RNAs (ncRNAs) play a vital role in angiogenesis. A study has suggested that upregulating the expression of miR-30e-5p can inhibit mRNA expression of VEGF and inhibit angiogenesis in HNSCC in vivo, thereby reducing tumour invasion and metastasis (35). Based on these findings, it is clear that epigenetic modifications are instrumental in the angiogenesis in head and neck tumours and may provide a new target for anti-angiogenic treatment.
The role of the immune system in tumour development is widely accepted, and therapeutic modalities targeting tumour immunity have been frequently reported (36). To facilitate the development of immunotherapy for head and neck cancer, the components involved in immunotherapy, such as immune cells and immune checkpoints, deserve to be investigated.
The TME of solid tumours contains a variety of immune cells, such as CD8+ T cells, CD4+ T cells, natural killer cells (NKs), and dendritic cells (DCs), which act as anti-tumour agents. The TME also contains myeloid suppressor cells (MDSCs), regulatory T cells (Tregs), and tumour-associated macrophages (TAMs), which act as immunosuppressive agents (37, 38). These immune cells, along with cytokines, constitute TIME. DCs promote the activation of CD8+ T cells and induce CD4+ T cells to differentiate into antigen-specific effector T cells (39). CD4+ T cells and NKs induce the maturation of DCs and contribute to the activation of CD8+ T cells (40, 41). Furthermore, CD4+ T cells exert their tumour-killing effect by secreting cytokines that activate CD8+ T cells (42). The TME of solid tumours presents a highly immunosuppressed state. MDSCs, Tregs, and TAMs inhibit the tumour killing effect of CD8+ T cells and effectively achieve immunosuppression (43-45). Moreover, cancer-associated fibroblasts (CAFs), a major cellular component of the TME matrix, are involved in the immune processes of head and neck tumours. For example, in nasopharyngeal carcinoma, there is a significant correlation between the densities of CAFs and M2 TAMs (46). Furthermore, tumour cells can release immunosuppressive mediators to exert their immunosuppressive effects. Head and neck tumour cells can avoid detection by T cells and NKs by secreting various cytokines, including TGF-β, IL-6, and IL-10, which can inhibit T-cell proliferation and effector function (47–49). Another study has reported that exosomes from peripheral blood of head and neck patients containing cyclooxygenase 2, TGF-β, programmed death 1 (PD-1), and cytotoxic T lymphocyte antigen 4 (CTLA-4). These exosomes promote CD8+ T-cell apoptosis, inhibit CD4+ T-cell proliferation, upregulate Tregs and impair the function of NKs (50).
Some immune checkpoints play a role in the immune escape of head and neck tumours, including PD-1/PD-L1, CTLA-4, and indoleamine 2,3-dioxygenase 1 (IDO1). In HNSCC, the expressions of PD1 and PD-L1 are upregulated, which represents one of the key immune checkpoints in HNSCC (51–53). PD-1 is mainly expressed on the surface of T cells and interacts with PD-L1 and PD-L2 ligands (54). The interaction of PD-1 and PD-L1 can inhibit the function of effector T cells and promote immunological tolerance (55). Furthermore, CTLA-4 can bind to B7 ligands on cancer cells, which results in the inhibition of T-cell activation and the promotion of HNSCC immune escape (56). IDO1 is a new immunosuppressive locus whose increased expression can inhibit the function of anti-tumour T cells (57). Recent studies have stated that epigenetic modifications are also involved in the immunomodulatory process in head and neck cancers. Investigations have suggested that CTLA-4 and IDO1 expression levels in head and neck tumours are epigenetically regulated via DNA methylation (57, 58). This finding implies that epigenetic modifications are involved in the TME of head and neck tumours. Therefore, the role of epigenetics in the TME of head and neck cancers must be clarified.
Multiple studies have suggested that DNA methylation is involved in genetic alterations in tumour cells and in activities in the TME (Figure S1) (59, 60). A recent study showed that inhibiting the expression of DNA methyltransferase 1 can reduce MDSCs and increase tumour-infiltrating T cells to prevent tumour growth in the TME of oral squamous cell carcinoma (OSCC) (61). Furthermore, altered DNA methylation of certain genes can affect immune cells and, therefore, the TME. In HNSCC, squalene cyclooxygenase is demethylated and overexpressed, which inhibits the activation of CD8+ T cells and leads to immunity evasion (62). Another study observed that the TME of patients with ornithine aminotransferase hypomethylation had a higher degree of immune cell infiltration and ornithine aminotransferase hypermethylation exhibited higher radiosensitivity (63).
One study indicated that immune checkpoints in the TME of head and neck tumours were regulated by DNA methylation (58). According to a recent study on oral cancer, the expressions of the immune checkpoint CTLA-4 and its function-related molecules CD28, ICOS, CD80, and CD86 were regulated by DNA methylation (58). Expression of CTLA4 is negatively correlated with DNA methylation, whereas mRNA levels of CD28, ICOS, CD80, and CD86 are positively correlated (58). This evidence suggests that DNA methylation directly or indirectly influences the regulation of the TME in head and neck tumours.
Histones comprise the core histones H2A, H2B, H3, and H4 and the connective histone H1 (64). Acetylation of histone H4-Lys16 and trimethylation deletion of H4-Lys20 are common markers of human cancer (65). Many studies have shown that histone deacetylase inhibitors (HDACis) can reshape the TME and enhance the ability of the immune system to kill tumour cells (66–69). A recent study opined that head and neck tumours sustain low levels of histone acetylation, which may be the reason for the accumulation and maintenance of cancer stem cells (70). The above evidence implies that histone acetylation has an essential role in the TME of head and neck tumours.
m1A modification is closely related to the TIME in several cancers (71, 72). m1A modification has been documented to affect the TME in HNSCC and thereby influence its prognosis. A recent study has shown that the m1A gene mutation may be associated with the TME of OSCC and that it could potentially predict its prognosis (73). Researchers have analysed the methylation pattern of m1A, which revealed that the expressions of all m1A regulatory factors were significantly upregulated in 502 patients with OSCC compared with the normal control group. This upregulation was closely linked to poor prognosis in the patients (73). Moreover, m1A modification was shown to be negatively correlated with immune checkpoints, angiogenesis, and CD8+ T cells in the TME (73). A study has reported that this modification can affect the TME in head and neck tumours by influencing the long non-coding RNAs (lncRNAs). Furthermore, m1A-related lncRNA has been observed to be closely related to the prognosis, TME, and tumour mutation burden in HNSCC (74). In summary, m1A modification exerts immunomodulatory effects in the TME of HNSCC.
m5C modification is a key player in several biological and pathological processes, such as cell proliferation and differentiation, tumorigenesis, malignant tumour progression, and tumour immunity (75, 76). Moreover, m5C regulatory factors can regulate the TIME (77). A recent study has stated that the activities of most immune cells in the TME are significantly reduced in patients with a high expression of m5C regulators (78). This observation implies a correlation between the TME and m5C in patients with OSCC (78). Furthermore, m5C can influence the TME via lncRNAs. Another study has suggested the presence of a close relationship between m5C-related lncRNA and the HNSCC TME (74). In HNSCC, m5C regulators inhibit immune cell activity, which means that m5C can regulate the TME and influence its fate.
m6A modification is the most common mRNA modification in eukaryotes (79). This modification is involved in the regulation of RNA stability, localisation, output, splicing, and translation and is closely linked to several cellular activities (80). Abnormal m6A modification is associated with a TME phenotype of non-inflammation and immunorejection (81). For instance, low expression of the m6A writer METTL3 promotes the production of IL-8. Tumour-associated neutrophils are recruited to the TME for immunosuppression, which promotes the progression of papillary thyroid carcinoma (82). Moreover, m6A eraser acts in the TME, with a recent study reporting that the expression of ALKBH5 is upregulated in HNSCC (20). A subsequent study has shown that ALKBH5 downregulates the expression of RIG-I and reduces the secretion of interferon-α in the TME via the IKK-α/Tbk1/IRF3 pathway. Ultimately, the infiltration of immune killer cells is inhibited, and immune escape is promoted (20). Furthermore, m6A readers YTHDF1 and IGF2BP2 have been found to be significantly correlated with different immunological states in HNSCC. These readers may regulate the TME by blocking the expression of specific genes related to antigen recognition, signal transduction, and effector T-cell proliferation and activation (83). YTHDC2 is associated with the degree of immune infiltration of B cells, CD8+ T cells, CD4+ T cells, neutrophils, and DCs in HNSCC (84). High expression of YTHDC2 is accompanied by high immune infiltration (84). Moreover, m6A is associated with immune checkpoints in the TME. A report has suggested that the upregulation of PD-L1 expression is associated with m6A methylation (85). Also, m6A methylation and the PI3K/AKT/mTOR signalling pathway may be involved in the regulation of the HNSCC immune microenvironment (85). These reports suggest that m6A modifications are primarily associated with immune regulation of the TME in head and neck tumours.
ncRNAs, including circular RNAs (circRNAs), microRNAs (miRNAs), and lncRNAs, interact with the components of the TME, thereby affecting tumorigenesis and progression (Table S1) (86–88). These ncRNAs, which are enriched in exosomes of CAFs, are transmitted to the cancer cells to regulate their biological characteristics. The decreased expressions of miR-34a-5p and miR-3188 in the CAF exosomes augment the metastatic potential of head and neck tumours (89, 90). miR-382-5p transported by CAF exosomes can promote migration and invasion in OSCC (91). In addition, miR-196a transported by CAF exosomes can augment cisplatin resistance in head and neck tumours by targeting CDKN1B and ING5 (92). Moreover, miRNAs from cancer cells can influence the TME. miR-192/215 transported by exosomes in head and neck tumour cells could promote remodelling of the hypoxic TME (93). Another study has reported that deletion of the p53 gene results in the reduced expression of miR-34a, which promotes adrenergic trans-differentiation of tumour-associated sensory nerves and head and neck tumour progression (94). Furthermore, the downregulation of miR-34a promotes immune escape in head and neck tumours via upregulation of MET expression (95). Low levels of miR-9 can promote tumour growth by upregulating MDK expression and regulating the PDK/AKT signalling pathway to enhance angiogenesis in the TME of nasopharyngeal carcinoma (96). Therefore, miRNA plays a key role in the TME of head and neck tumours.
The role of lncRNAs in the microenvironment of head and neck cancer is yet to be elucidated. Like miRNAs, lncRNAs from the TME can influence the fate of the tumour. The lncRNA H19 is upregulated in CAF and participates in the glycolytic pathway of CAF via the miR-675-5p/PFKFB3 axis, which promotes the progression of oral cancer (97). The lncRNA FLJ22447 is significantly upregulated in CAFs and promotes the transformation of CAF into OSCC by upregulating IL-33 (87). Furthermore, lncRNA transported by tumour cell exosomes can act on components in the TME to regulate tumour progression. Oral leukoplasia (OL) is a precancerous state of OSCC (98). A study has shown that the lncRNA IFITM4P can induce PD-L1 expression, thereby activating the immunosuppressive process and immune escape of OL cells in the cytoplasm (98). The expression of the lncRNA DCST1-AS1 in OSCC is significantly increased, and the polarization of M2 macrophages is promoted by regulation of the NF-κB pathway to enhance tumour progression (99). These results suggest that ncRNA can regulate the TME of head and neck tumours and might serve as a potential therapeutic target.
Head and neck tumours are associated with a poor prognosis because of their high degree of malignancy and the high rate of lymph node metastasis (3). At present, the major treatment for these tumours is surgical resection (4). However, owing to their location, surgical resection of head and neck tumours often results in physiological disorders that affect swallowing, mastication, or pronunciation. Moreover, it can exert a negative influence on the facial appearance and mental health of patients (100, 101). Therefore, therapies that are more efficient and less damaging should be immediately developed. Several studies have shown that the TME can regulate tumorigenesis, growth, and metastasis of head and neck tumours (102–105). As mentioned above, angiogenesis promotes the invasion and metastasis of head and neck tumour, and increased immunosuppression promotes the immune escape of tumour cells (8, 32). Therefore, TME is extremely important for the pathological progression of head and neck tumours. Anti-angiogenic drugs and immunotherapies that target the TME of head and neck tumours are promising alternative therapies (103, 106). However, these treatment modalities require further research to refine them. Therefore, elucidating the role of the TME in head and neck tumours is the need of the hour.
Epigenetic mechanisms are involved in a variety of pathological processes and play an essential role in tumour progression (21). These alterations can influence the TME of head and neck tumours and regulate cancer progression (Figure 2). In this regard, DNA methylation can affect tumour progression by regulating immune infiltration and immune checkpoints in the TME of head and neck tumours (58, 61, 62). Furthermore, histone acetylation can weaken the immune-killing capability of the TME in head and neck tumours and promote their growth (70). RNA modification predominantly regulates the level of angiogenesis, immune activity, and immune infiltration of immune cells in the TME of head and neck tumours and participates in tumour progression (20, 73, 78, 82–85). ncRNA secreted by certain cells in the TME of head and neck tumours can influence the behaviour of cancer cells, including invasion, metastasis, and drug resistance (89, 97). Furthermore, ncRNA in tumour cells can participate in the immune regulation of the TME in head and neck tumours and promote their progression (95, 96, 99). Succinctly, epigenetic modification holds promising potential in the regulation of the TME in head and neck tumours and is expected to provide targets for their treatment. Key proteins in epigenetic modifications can affect angiogenesis, immune responses in TME and can further affect tumour growth (Table S2) (61, 73, 78, 82). Numerous drugs targeting different epigenetic modifications have been used to treat head and neck tumours. The major DNA methylation drugs include zebularine, 5-azacytidine, 5-aza-2’-deoxycytidine, aloe-emodin, and procaine (107–109). Trichostatin A, suberoylanilide hydroxamic acid, M344 (an analogue of hydroxamic acid), and cyclic tetrapeptide are potent HDAC inhibitors that have been reported to promote radiosensitivity in head and neck tumours (110). Additionally, the HDAC inhibitor LBH589 promotes p21 expression and induces cell death (111). Therefore, the development of drugs targeting epigenetic modifications in the TME may offer new prospects for the treatment of head and neck tumours.
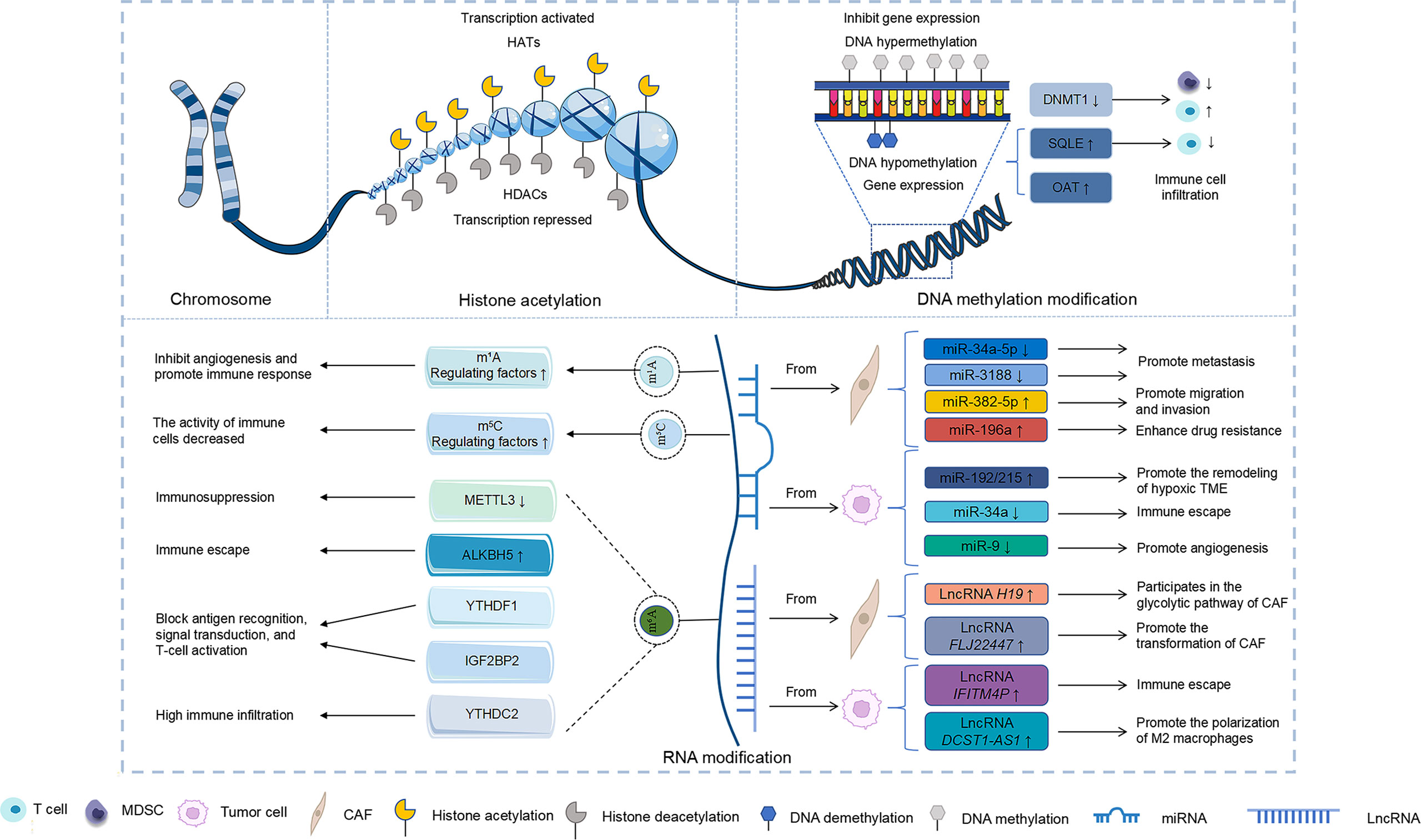
Figure 2 Effect of epigenetic modifications on the TME of head and neck tumours. DNA methylation, RNA modification, and ncRNA modification resulted in different gene expression changes that affected the TME of head and neck tumours.
The data summarized herein establishes that the TME can affect the malignant development of head and neck tumours, including their growth, metastasis, and drug resistance. Epigenetic modifications are involved in these processes. It is essential to gain more knowledge about the molecular mechanisms involved in epigenetic modifications of TME in head and neck tumours. Therefore, the development of new drugs effectively targeting epigenetic modifications can be envisaged in the near future. In general, clarifying the role of epigenetic modifications in the TME can provide a novel therapeutic target for head and neck tumours.
HL and DW wrote the manuscript. HL, WL, ZY, SL, HW, JX, SK, and MH collected the references and prepared figures. All authors reviewed the manuscript. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
This work was supported by the Fundamental Research Funds for the Jilin Province Department of Finance (grant no. jcsz2021893-13), the Changchun Scientific and Technological Development Program (grant no. 21ZY26) and the Jilin Province Scientific and Technological Development Program (grant no. 20200801077GH, 20210204013YY, 20200504005YY, 20220505033ZP).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.1050982/full#supplementary-material
Supplementary Figure 1 | The role of DNA methylation in TME of head and neck tumours. (A) DNA hypomethylation of VEGF and DNA hypermethylation of FBLN2S promote angiogenesis. (B) Inhibition of DNMT1 expression decreased the percentage of MDSCs and increased CD8+T cell infiltration. (C) Hypomethylation at CpG S_Shore site of SQLE promotes immune escape. (D) OAT hypomethylation promotes immune cell infiltration, and hypermethylation tumour is more sensitive to radiotherapy.
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2018) 68:394–424. doi: 10.3322/caac.21492
2. Curado MP, Hashibe M. Recent Changes in the Epidemiology ofHead and Neck Cancer. Curr Opin Oncol (2009) 21(3):194–200. doi:10.1097/CCO.0b013e32832a68ca
3. Cramer JD, Burtness B, Le QT, Ferris RL. The changing therapeutic landscape of head and neck cancer. Nat Rev Clin Oncol (2019) 16:669–83. doi: 10.1038/s41571-019-0227-z
4. Chinn SB, Myers JN. Oral cavity carcinoma: Current management, controversies, and future directions. J Clin Oncol (2015) 33:3269–76. doi: 10.1200/JCO.2015.61.2929
5. Ferris RL, Blumenschein GB Jr, Fayette J, Guigay J, Colevas AD, Licitra L, et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N Engl J Med (2016) 375:1856–67. doi: 10.1056/NEJMoa1602252
6. Hsu HW, Wall NR, Hsueh CT, Kim S, Ferris RL, Chen CS, et al. Combination antiangiogenic therapy and radiation in head and neck cancers. Oral Oncol (2014) 50:19–26. doi: 10.1016/j.oraloncology.2013.10.003
7. Mei Z, Huang J, Qiao B, Lam AK. Immune checkpoint pathways in immunotherapy for head and neck squamous cell carcinoma. Int J Oral Sci (2020) 12:16. doi: 10.1038/s41368-020-0084-8
8. Ferris RL. Immunology and immunotherapy of head and neck cancer. J Clin Oncol (2015) 33:3293–304. doi: 10.1200/JCO.2015.61.1509
9. Zhu C, Gu LQ, Liu ZL, Li J, Yao MF, Fang CG, et al. Correlation between vascular endothelial growth factor pathway and immune microenvironment in head and neck squamous cell carcinoma. BMC Cancer (2021) 21:836. doi: 10.1186/s12885-021-08547-4
10. Kim J, Bae JS. Tumor-associated macrophages and neutrophils in tumor microenvironment. Mediators Inflammation (2016) 2016:6058147. doi: 10.1155/2016/6058147
11. Xiao Y, Yu D. Tumor Microenvironment as a Therapeutic Target in Cancer. Pharmacol Ther (2021) 221:107753. doi: 10.1016/j.pharmthera.2020.107753.
12. Biffi G, Tuveson DA. Diversity and Biology of Cancer-Associated Fibroblasts. Physiol Rev (2021) 101(1):147–76. doi: 10.1152/physrev.00048.2019
13. Yaacoub K, Pedeux R, Tarte K, Guillaudeux T. Role of the Tumor Microenvironment in Regulating Apoptosis andCancer Progression. Cancer Lett (2016) 378(2):150–9. doi: 10.1016/j.canlet.2016.05.012
14. Jing X, Yang F, Shao C, Wei K, Xie M, Shen H, et al. Role of Hypoxia in Cancer Therapy by Regulating the TumorMicroenvironment. Mol Cancer (2019) 18(1):157. doi: 10.1186/s12943-019-1089-9
15. Lei X, Lei Y, Li JK, Du WX, Li RG, Yang J, et al. Immune cells within the tumor microenvironment: biological functions and roles in cancer immunotherapy. Cancer Lett (2020) 470:126–33. doi: 10.1016/j.canlet.2019.11.009
16. Chang CH, Qiu J, O'Sullivan D, Buck MD, Noguchi T, Curtis JD, et al. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell (2015) 162:1229–41. doi: 10.1016/j.cell.2015.08.016
17. Buck MD, Sowell RT, Kaech SM, Pearce EL. Metabolic instruction of immunity. Cell (2017) 169:570–86. doi: 10.1016/j.cell.2017.04.004
18. Meurette O, Mehlen P. Notch signaling in the tumor microenvironment. Cancer Cell (2018) 34:536–48. doi: 10.1016/j.ccell.2018.07.009
19. Zou S, Tong QY, Liu BW, Huang W, Tian Y, Fu XH, et al. Targeting STAT3 in cancer immunotherapy. Mol Cancer (2020) 19:145. doi: 10.1186/s12943-020-01258-7
20. Jin S, Li M, Chang H, Wang R, Zhang Z, Zhang J, et al. The m6A demethylase ALKBH5 promotes tumor progression by inhibiting RIG-I expression and interferon alpha production through the IKKepsilon/TBK1/IRF3 pathway in head and neck squamous cell carcinoma. Mol Cancer (2022) 21:97. doi: 10.1186/s12943-022-01572-2
21. Hu M, Cai JYL, Bachman KE, Brûle FVD, Velculescu V, Polyak K, et al. Distinct epigenetic changes in the stromal cells of breast cancers. Nat Genet (2005) 37:899–905. doi: 10.1038/ng1596
22. Gu Y, Wu X, Zhang J, Fang Y, Pan Y, Shu Y, et al. The evolving landscape of N(6)-methyladenosine modification in the tumor microenvironment. Mol Ther (2021) 29:1703–15. doi: 10.1016/j.ymthe.2021.04.009
23. Chen SMY, Krinsky AL, Woolaver RA, Wang X, Chen Z, Wang JH, et al. Tumor immune microenvironment in head and neck cancers. Mol Carcinog (2020) 59:766–74. doi: 10.1002/mc.23162
24. Meng W, Hao Y, He C, Li L, Zhu G. Exosome-Orchestrated Hypoxic TumorMicroenvironment. Mol Cancer (2019) 18(1):57. doi: 10.1186/s12943-019-0982-6
25. Gavrielatou N, Doumas S, Economopoulou P, Foukas PG, Psyrri A. Biomarkers for immunotherapy response in head and neck cancer. Cancer Treat Rev (2020) 84:101977. doi: 10.1016/j.ctrv.2020.101977
26. Carmeliet P, Jain RK. Molecular mechanisms and clinical applications of angiogenesis. Nature (2011) 473:298–307. doi: 10.1038/nature10144
27. Lugano R, Ramachandran M, Dimberg A. Tumor angiogenesis: causes, consequences, challenges and opportunities. Cell Mol Life Sci (2020) 77:1745–70. doi: 10.1007/s00018-019-03351-7
28. Carla C, Daris F, Cecilia B, Francesca B, Francesca C, Paolo F, et al. Angiogenesis in head and neck cancer: a review of the literature. J Oncol (2012) 2012:358472. doi: 10.1155/2012/358472
29. Martone T, Rosso P, Albera R, Migliaretti G, Fraire F, Pignataro L, et al. Prognostic relevance of CD105+ microvessel density in HNSCC patient outcome. Oral Oncol (2005) 41:147–55. doi: 10.1016/j.oraloncology.2004.08.001
30. Gallo O, Masini E, Morbidelli L, Franchi A, Fini-Storchi I, Vergari WA, Ziche M, et al, et al. Role of nitric oxide in angiogenesis and tumor progression in head and neck cancer. J Natl Cancer Inst (1998) 90:587–96. doi: 10.1093/jnci/90.8.587
31. Ferrara N, Gerber HP, LeCouter J. The biology of VEGF and its receptors. Nat Med (2003) 9:669–76. doi: 10.1038/nm0603-669
32. Benke EM, Ji Y, Patel V, Wang H, Miyazaki H, Yeudall WA, et al. VEGF-c contributes to head and neck squamous cell carcinoma growth and motility. Oral Oncol (2010) 46:e19–24. doi: 10.1016/j.oraloncology.2010.02.006
33. Lu Y, Wang J, Yan J, Yang Y, Sun Y, Huang Y, et al. Sevoflurane attenuate hypoxia-induced VEGF level in tongue squamous cell carcinoma cell by upregulating the DNA methylation states of the promoter region. BioMed Pharmacother (2015) 71:139–45. doi: 10.1016/j.biopha.2015.02.032
34. Law EW, Cheung AKL, Kashuba VI, Pavlova TV, Zabarovsky ER, Lung HL, et al. Anti-angiogenic and tumor-suppressive roles of candidate tumor-suppressor gene, fibulin-2, in nasopharyngeal carcinoma. Oncogene (2012) 31:728–38. doi: 10.1038/onc.2011.272
35. Zhang S, Li G, Liu C, Lu S, Jing Q, Chen X, Zheng H, et al. miR-30e-5p represses angiogenesis and metastasis by directly targeting AEG-1 in squamous cell carcinoma of the head and neck. Cancer Sci (2020) 111:356–68. doi: 10.1111/cas.14259
36. Wolchok J. Putting the immunologic brakes on cancer. Cell (2018) 175:1452–4. doi: 10.1016/j.cell.2018.11.006
37. Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression. Cancer Res (2019) 79:4557–66. doi: 10.1158/0008-5472.CAN-18-3962
38. Farhood B, Najafi M, Mortezaee K. CD8(+) cytotoxic T lymphocytes in cancer immunotherapy: A review. J Cell Physiol (2019) 234:8509–21. doi: 10.1002/jcp.27782
39. Palucka K, Banchereau J. Cancer immunotherapy via dendritic cells. Nat Rev Cancer (2012) 12(4):265–77. doi: 10.1038/nrc3258
40. Gottschalk C, Mettke E, Kurts C. The role of invariant natural killer T cells in dendritic cell licensing, cross-priming, and memory CD8(+) T cell generation. Front Immunol (2015) 6:379. doi: 10.3389/fimmu.2015.00379
41. Srivastava RM, Lee SC, Filho PAA, Lord CA, Jie HB, Davidson HC, et al. Cetuximab-activated natural killer and dendritic cells collaborate to trigger tumor antigen-specific T-cell immunity in head and neck cancer patients. Clin Cancer Res (2013) 19:1858–72. doi: 10.1158/1078-0432.CCR-12-2426
42. Kershaw MH, Westwood JA, Darcy PK. Gene-engineered T cells for cancer therapy. Nat Rev Cancer (2013) 13:525–41. doi: 10.1038/nrc3565
43. Joseph R, Soundararajan R, Vasaikar S, Yang F, Allton KL, Tian L, et al. Cd8(+) T Cells InhibitMetastasis and Cxcl4 Regulates Its Function. Br J Cancer (2021) 125(2):176–89. doi: 10.1038/s41416-021-01338-5
44. Noyes D, Bag A, Oseni S, Semidey-Hurtado J, Cen L, Sarnaik AA, et al. Tumor-Associated TregsObstruct Antitumor Immunity by Promoting T Cell Dysfunction and Restricting Clonal Diversity inTumor-Infiltrating Cd8+ T Cells. J Immunother Cancer (2022) 10(5). doi: 10.1136/jitc-2022-004605
45. Qiu SQ, Waaijer SJH, Zwager MC, de Vries EGE, van der Vegt B, Schroder CP. Tumor-AssociatedMacrophages in Breast Cancer: Innocent Bystander or Important Player? Cancer Treat Rev (2018)70:178–89. doi: 10.1016/j.ctrv.2018.08.010
46. Yu Y, Ke L, Lv X, Ling YH, Lu J, Liang H, et al. The prognostic significance of carcinoma-associated fibroblasts and tumor-associated macrophages in nasopharyngeal carcinoma. Cancer Manag Res (2018) 10:1935–46. doi: 10.2147/CMAR.S167071
47. Lu SL, Reh D, Li AG, Woods J, Corless CL, Kulesz-Martin M, et al. Overexpression of transforming growth factor beta1 in head and neck epithelia results in inflammation, angiogenesis, and epithelial hyperproliferation. Cancer Res (2004) 64:4405–10. doi: 10.1158/0008-5472.CAN-04-1032
48. Davis RJ, Van Waes C, Allen CT. Overcoming barriers to effective immunotherapy: MDSCs, TAMs, and tregs as mediators of the immunosuppressive microenvironment in head and neck cancer. Oral Oncol (2016) 58:59–70. doi: 10.1016/j.oraloncology.2016.05.002
49. Lathers DM, Young MR Increased Aberrance of Cytokine Expression inPlasma of Patients with More Advanced Squamous Cell Carcinoma of the Headand Neck. Cytokine (2004) 25(5):220–8. doi: 10.1016/j.cyto.2003.11.005
50. Ludwig S, Floros T, Theodoraki MN, Hong CS, Jackson EK, Lang S, et al. Suppression ofLymphocyte Functions by Plasma Exosomes Correlates with Disease Activity in Patients with Head and Neck Cancer. Clin Cancer Res (2017) 23(16):4843–54. doi: 10.1158/1078-0432.CCR-16-2819.
51. Qiao XW, Jiang J, Pang X, Huang MC, Tang YJ, Liang XH, et al. The evolving landscape of PD-1/PD-L1 pathway in head and neck cancer. Front Immunol (2020) 11:1721. doi: 10.3389/fimmu.2020.01721
52. Allen CT, Clavijo PE, Van Waes C, Chen Z. Anti-tumor immunity in head and neck cancer: Understanding the evidence, how tumors escape and immunotherapeutic approaches. Cancers (Basel) (2015) 7:2397–414. doi: 10.3390/cancers7040900
53. Zandberg DP, Strome SE. The role of the PD-L1:PD-1 pathway in squamous cell carcinoma of the head and neck. Oral Oncol (2014) 50:627–32. doi: 10.1016/j.oraloncology.2014.04.003
54. Ansell SM. Where do programmed death-1 inhibitors fit in the management of malignant lymphoma? J Oncol Pract (2016) 12(2):101–6. doi: 10.1200/JOP.2015.009191
55. Sharpe AH, Wherry EJ, Ahmed R, Freeman GJ. The function of programmed cell death 1 and its ligands in regulating autoimmunity and infection. Nat Immunol (2007) 8:239–45. doi: 10.1038/ni1443
56. Kaidar-Person O, Gil Z, Billan S. Precision medicine in head and neck cancer. Drug Resist Update (2018) 40:13–6. doi: 10.1016/j.drup.2018.09.001
57. Sailer V, Sailer U, Bawden EG, Zarbl R, Wiek C, Vogt TJ, et al. DNA Methylation of indoleamine 2,3-dioxygenase 1 (IDO1) in head and neck squamous cell carcinomas correlates with IDO1 expression, HPV status, patients' survival, immune cell infiltrates, mutational load, and interferon gamma signature. EBioMedicine (2019) 48:341–52. doi: 10.1016/j.ebiom.2019.09.038
58. de Vos L, Grunwald I, Bawden EG, Dietrich J, Scheckenbach K, Wiek C, et al. The landscape of CD28, CD80, CD86, CTLA4, and ICOS DNA methylation in head and neck squamous cell carcinomas. Epigenetics (2020) 15:1195–212. doi: 10.1080/15592294.2020.1754675
59. Li D, Zhao W, Zhang X, Lv H, Li C, Sun L, et al. NEFM DNA methylation correlates with immune infiltration and survival in breast cancer. Clin Epigenet (2021) 13:112. doi: 10.1186/s13148-021-01096-4
60. Mitra S, Lauss M, Cabrita R, Choi J, Zhang T, Isaksson K, et al. Analysis of DNA methylation patterns in the tumor immune microenvironment of metastatic melanoma. Mol Oncol (2020) 14:933–50. doi: 10.1002/1878-0261.12663
61. Yang SC, Wang WY, Zhou JJ, Wu L, Zhang MJ, Yang QC, et al. Inhibition of DNMT1 potentiates antitumor immunity in oral squamous cell carcinoma. Int Immunopharmacol (2022) 111:109113. doi: 10.1016/j.intimp.2022.109113
62. Liu Y, Fang L, Liu W. High SQLE expression and gene amplification correlates with poor prognosis in head and neck squamous cell carcinoma. Cancer Manag Res (2021) 13:4709–23. doi: 10.2147/CMAR.S305719
63. Sun Y, Yang J, Cai H, Liu J, Liu Y, Luo J, et al. Differential OAT methylation correlates with cell infiltration in tumor microenvironment and overall survival postradiotherapy in oral squamous cell carcinoma patient. J Oral Pathol Med (2022) 51:611–9. doi: 10.1111/jop.13328
64. Audia JE, Campbell RM. Histone modifications and cancer. Cold Spring Harb Perspect Biol (2016) 8:a019521. doi: 10.1101/cshperspect.a019521
65. Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet (2005) 37:391–400. doi: 10.1038/ng1531
66. Ritter C, Fan K, Paschen A, Reker Hardrup S, Ferrone S, Nghiem P, et al. Epigenetic priming restores the HLA class-I antigen processing machinery expression in merkel cell carcinoma. Sci Rep (2017) 7:2290. doi: 10.1038/s41598-017-02608-0
67. Souri Z, Jochemsen AG, Versluis M, Wierenga APA, Nemati F, van der Velden PAc, et al. HDAC inhibition increases HLA class I expression in uveal melanoma. Cancers (Basel) (2020) 12:3690. doi: 10.3390/cancers12123690
68. Jiang Y, Ortega-Molina A, Geng H, Ying HY, Hatzi K, Parsa S, et al. CREBBP inactivation promotes the development of HDAC3-dependent lymphomas. Cancer Discovery (2017) 7:38–53. doi: 10.1158/2159-8290.CD-16-0975
69. Buglio D, Khaskhely NM, Voo KS, Martinez-Valdez H, Liu YJ, Younes A, et al. HDAC11 plays an essential role in regulating OX40 ligand expression in Hodgkin lymphoma. Blood (2011) 117:2910–7. doi: 10.1182/blood-2010-08-303701
70. Giudice FS, Pinto DS Jr., Nor JE, Squarize CH, Castilho RM. Inhibition of histone deacetylase impacts cancer stem cells and induces epithelial-mesenchyme transition of head and neck cancer. PLoS One (2013) 8:e58672. doi: 10.1371/journal.pone.0058672
71. Zhao M, Shen S. and Xue, C. A novel m1A-score model correlated with the immune microenvironment predicts prognosis in hepatocellular carcinoma. Front Immunol (2022) 13:805967. doi: 10.3389/fimmu.2022.805967
72. Gao Y, Wang H, Li H, Ye X, Xia Y, Yuan S, et al. Integrated analyses of m(1)A regulator-mediated modification patterns in tumor microenvironment-infiltrating immune cells in colon cancer. Oncoimmunology (2021) 10:1936758. doi: 10.1080/2162402X.2021.1936758
73. Gao L, Chen R, Sugimoto M, Mizuta M, Kishimoto Y, Omori K., et al. The impact of m1A methylation modification patterns on tumor immune microenvironment and prognosis in oral squamous cell carcinoma. Int J Mol Sci (2021) 22(19):10302. doi: 10.3390/ijms221910302
74. Wang E, Li Y, Ming R, Wei J, Du P, Zhou P, et al. The prognostic value and immune landscapes of a m(6)A/m(5)C/m(1)A-related LncRNAs signature in head and neck squamous cell carcinoma. Front Cell Dev Biol (2021) 9:718974. doi: 10.3389/fcell.2021.718974
75. Song P, Tayier S, Cai Z, Jia G. RNA Methylation in mammalian development and cancer. Cell Biol Toxicol (2021) 37:811–31. doi: 10.1007/s10565-021-09627-8
76. Zhang M, Song J, Yuan W, Zhang W, Sun Z. Roles of RNA methylation on tumor immunity and clinical implications. Front Immunol (2021) 12:641507. doi: 10.3389/fimmu.2021.641507
77. Pan J, Huang Z, Xu Y. m5C RNA methylation regulators predict prognosis and regulate the immune microenvironment in lung squamous cell carcinoma. Front Oncol (2021) 11:657466. doi: 10.3389/fonc.2021.657466
78. Gao L, Chen R, Sugimoto M, Mizuta M, Zhou L, Kishimoto Y, et al. The RNA methylation modification 5-methylcytosine impacts immunity characteristics, prognosis and progression of oral squamous cell carcinoma by bioinformatics analysis. Front Bioeng Biotechnol (2021) 9:760724. doi: 10.3389/fbioe.2021.760724
79. Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol (2014) 10:93–5. doi: 10.1038/nchembio.1432
80. Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell (2015) 161:1388–99. doi: 10.1016/j.cell.2015.05.014
81. Qin S, Mao Y, Wang H, Duan Y, Zhao L. The interplay between m6A modification and non-coding RNA in cancer stemness modulation: mechanisms, signaling pathways, and clinical implications. Int J Biol Sci (2021) 17:2718–36. doi: 10.7150/ijbs.60641
82. He J, Zhou M, Yin J, Wan J, Chu J, Jia J, et al. METTL3 restrains papillary thyroid cancer progression via m(6)A/c-Rel/IL-8-mediated neutrophil infiltration. Mol Ther (2021) 29:1821–37. doi: 10.1016/j.ymthe.2021.01.019
83. Li S, Wu Q, Liu J, Zhong Y. Identification of two m6A readers YTHDF1 and IGF2BP2 as immune biomarkers in head and neck squamous cell carcinoma. Front Genet (2022) 13:903634. doi: 10.3389/fgene.2022.903634
84. Li Y, Zheng JN, Wang EH, Gong CJ, Lan KF, Ding X. The m6A reader protein YTHDC2 is a potential biomarker and associated with immune infiltration in head and neck squamous cell carcinoma. PeerJ (2020) 8:e10385. doi: 10.7717/peerj.10385
85. Yi L, Wu G, Guo L, Zou X, Huang P. Comprehensive analysis of the PD-L1 and immune infiltrates of m(6)A RNA methylation regulators in head and neck squamous cell carcinoma. Mol Ther Nucleic Acids (2020) 21:299–314. doi: 10.1016/j.omtn.2020.06.001
86. Shiah SG, Chou ST, Chang JY. MicroRNAs: Their role in metabolism, tumor microenvironment, and therapeutic implications in head and neck squamous cell carcinoma. Cancers (Basel) (2021) 13(22):5604. doi: 10.3390/cancers13225604
87. Ding L, Ren J, Zhang D, Li Y, Huang X, Hu Q, et al. A novel stromal lncRNA signature reprograms fibroblasts to promote the growth of oral squamous cell carcinoma via LncRNA-CAF/interleukin-33. Carcinogenesis (2018) 39:397–406. doi: 10.1093/carcin/bgy006
88. Jia L, Wang Y, Wang CY. Circfat1 promotes cancer stemness and immune evasion by promoting stat3 activation. Adv Sci (Weinh) (2021) 8(13):2003376. doi: 10.1002/advs.202003376.
89. Li YY, Tao YW, Gao S, Li P, Zheng JM, Zhang SE, et al. Cancer-associated fibroblasts contribute to oral cancer cells proliferation and metastasis via exosome-mediated paracrine miR-34a-5p. EBioMedicine (2018) 36:209–20. doi: 10.1016/j.ebiom.2018.09.006
90. Wang X, Qin X, Yan M, Shi J, Xu Q, Li Z, et al. Loss of exosomal miR-3188 in cancer-associated fibroblasts contributes to HNC progression. J Exp Clin Cancer Res (2019) 38:151. doi: 10.1186/s13046-019-1144-9
91. Sun LP, Xu K, Cui J, Yuan DY, Zou B, Li J, et al. Cancerassociated fibroblastderived exosomal miR3825p promotes the migration and invasion of oral squamous cell carcinoma. Oncol Rep (2019) 42:1319–28. doi: 10.3892/or.2019.7255
92. Qin X, Guo H, Wang X, Zhu X, Yan M, Wang X, et al. Exosomal miR-196a derived from cancer-associated fibroblasts confers cisplatin resistance in head and neck cancer through targeting CDKN1B and ING5. Genome Biol (2019) 20:12. doi: 10.1186/s13059-018-1604-0
93. Zhu G, Cao B, Liang X, Li L, Hao Y, Meng W, et al. Small extracellular vesicles containing miR-192/215 mediate hypoxia-induced cancer-associated fibroblast development in head and neck squamous cell carcinoma. Cancer Lett (2021) 506:11–22. doi: 10.1016/j.canlet.2021.01.006
94. Amit M, Takahashi H, Dragomir MP, Lindemann A, Gleber-Netto FO, Pickering CR, et al. Loss of p53 drives neuron reprogramming in head and neck cancer. Nature (2020) 578:449–54. doi: 10.1038/s41586-020-1996-3
95. Wu X, Cheng YL, Matthen M, Yoon A, Schwartz GK, Bala S, et al. Down-regulation of the tumor suppressor miR-34a contributes to head and neck cancer by up-regulating the MET oncogene and modulating tumor immune evasion. J Exp Clin Cancer Res (2021) 40:70. doi: 10.1186/s13046-021-01865-2
96. Lu J, Liu QH, Wang F, Tan JJ, Deng YQ, Peng XH, et al. Exosomal miR-9 inhibits angiogenesis by targeting MDK and regulating PDK/AKT pathway in nasopharyngeal carcinoma. J Exp Clin Cancer Res (2018) 37:147. doi: 10.1186/s13046-018-0814-3
97. Yang J, Shi X, Yang M, Luo J, Gao Q, Wang X, et al. Glycolysis reprogramming in cancer-associated fibroblasts promotes the growth of oral cancer through the lncRNA H19/miR-675-5p/PFKFB3 signaling pathway. Int J Oral Sci (2021) 13:12. doi: 10.1038/s41368-021-00115-7
98. Shi L, Yang Y, Li M, Li C, Zhou Z, Tang G, et al. LncRNA IFITM4P promotes immune escape by up-regulating PD-L1. via dual Mech Oral carcinogenesis. Mol Ther (2022) 30:1564–77. doi: 10.1016/j.ymthe.2022.01.003
99. Ai Y, Liu S, Luo H, Wu S, Wei H, Tang Z, et al. lncRNA DCST1-AS1 facilitates oral squamous cell carcinoma by promoting M2 macrophage polarization through activating NF-kappaB signaling. J Immunol Res (2021) 2021:5524231. doi: 10.1155/2021/5524231
100. Murphy BA, Ridner S, Wells N, Dietrich M. Quality of life research in head and neck cancer: a review of the current state of the science. Crit Rev Oncol Hematol (2007) 62:251–67. doi: 10.1016/j.critrevonc.2006.07.005
101. Pandey M, Devi N, Ramdas K, Krishnan R, Kumar V. Higher distress relates to poor quality of life in patients with head and neck cancer. Int J Oral Maxillofac Surg (2009) 38:955–9. doi: 10.1016/j.ijom.2009.04.004
102. Canning M, Guo G, Yu M, Myint C, Groves MW, Byrd JK, et al. Heterogeneity of the head and neck squamous cell carcinoma immune landscape and its impact on immunotherapy. Front Cell Dev Biol (2019) 7:52. doi: 10.3389/fcell.2019.00052
103. Horton JD, Knochelmann HM, Day TA, Paulos CM, Neskey DM. Immune evasion by head and neck cancer: Foundations for combination therapy. Trends Cancer (2019) 5:208–32. doi: 10.1016/j.trecan.2019.02.007
104. Oliva M, Spreafico A, Taberna M, Alemany L, Coburn B, Mesia R, et al. Immune biomarkers of response to immune-checkpoint inhibitors in head and neck squamous cell carcinoma. Ann Oncol (2019) 30:57–67. doi: 10.1093/annonc/mdy507
105. Santuray RT, Johnson DE, Grandis JR. New therapies in head and neck cancer. Trends Cancer (2018) 4:385–96. doi: 10.1016/j.trecan.2018.03.006
106. Hua Y, Dong R, Jin T, Jin Q, Chen X. Anti-PD-1 monoclonal antibody combined with anti-VEGF agent is safe and effective in patients with Recurrent/Metastatic head and neck squamous cancer as second-line or beyond treatment. Front Oncol (2022) 12:781348. doi: 10.3389/fonc.2022.781348
107. Flausino CS, Daniel FI, Modolo F. DNA Methylation in oral squamous cell carcinoma: from its role in carcinogenesis to potential inhibitor drugs. Crit Rev Oncol Hematol (2021) 164:103399. doi: 10.1016/j.critrevonc.2021.103399
108. Notarstefano V, Belloni A, Sabbatini S, Pro C, Orilisi G, Monterubbianesi R, et al. Cytotoxic effects of 5-azacytidine on primary tumour cells and cancer stem cells from oral squamous cell carcinoma: An In vitro FTIRM analysis. Cells (2021) 10(8):2127. doi: 10.3390/cells10082127
109. Bhol CS, Mishra SR, Patil S, Sahu K, Kirtana R, Manna S, et al. Pax9 reactivation by inhibiting DNA methyltransferase triggers antitumor effect in oral squamous cell carcinoma. Biochim Biophys Acta Mol Basis Dis (2022) 1868(9):166428. doi: 10.1016/j.bbadis.2022.166428.
110. Zhang Y, Jung M, Dritschilo A, Jung M. Enhancement of radiation sensitivity of human squamous carcinoma cells by histone deacetylase inhibitors. Radiat Res (2004) 161:667–74. doi: 10.1667/rr3192
Keywords: DNA methylation, histone modification, RNA modifications, ncRNAs, head and neck tumours, tumour microenvironment
Citation: Liu H, Wang D, Yang Z, Li S, Wu H, Xiang J, Kan S, Hao M and Liu W (2022) Regulation of epigenetic modifications in the head and neck tumour microenvironment. Front. Immunol. 13:1050982. doi: 10.3389/fimmu.2022.1050982
Received: 22 September 2022; Accepted: 14 October 2022;
Published: 28 October 2022.
Edited by:
Peng Qu, National Institutes of Health (NIH), United StatesReviewed by:
Yunyun Chen, University of Texas MD Anderson Cancer Center, United StatesCopyright © 2022 Liu, Wang, Yang, Li, Wu, Xiang, Kan, Hao and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weiwei Liu, bGl1d2Vpd0BqbHUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.