
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol., 20 December 2022
Sec. NK and Innate Lymphoid Cell Biology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.1040624
This article is part of the Research TopicRising Stars in NK and Innate Lymphoid Cell Biology 2021View all 3 articles
The family of innate lymphoid cells (ILCs), consisting of Group 1 ILCs (natural killer cells and ILC1), ILC2, and ILC3, are critical effectors of innate immunity, inflammation, and homeostasis post-natally, but also exert essential functions before birth. Recent studies during critical developmental periods in the embryo have hinted at complex waves of tissue colonization, and highlighted the breadth of multipotent and committed ILC progenitors from both classic fetal hematopoietic organs such as the liver, as well as tissue sites such as the lung, thymus, and intestine. Assessment of the mechanisms driving cell fate and function of the ILC family in the embryo will be vital to the understanding ILC biology throughout fetal life and beyond.
Predominantly known as tissue-resident sentinels, innate lymphoid cells (ILCs) are innate lymphocytes that, in large part, share transcriptional identities and effector function profiles with their adaptive immune system counterparts, T cells. The ILC family consists of three primary lineages: Group 1 ILCs, ILC2, and Group 3 ILCs (as reviewed in Vivier et al., 2018) (1). Group 1 ILCs comprise both T-bet+ ILC1 and Eomes+ natural killer (NK) cells, which are the counterparts of TH1 and Tcytotoxic cells, respectively, mount an anti-viral and anti-tumor type 1 response and secrete IFNγ. ILC2 express the lineage-identifying transcription factor GATA3, reflecting TH2 cells, and are part of a type 2 allergic response through secretion of IL-4, IL-5, and IL-13. Finally, Group 3 ILCs are identified by the hallmark transcription factor (TF) RORγt, and are classified into two subsets: lymphoid tissue inducer (LTi) cells, which are of particular importance during embryonic development, as discussed later in-depth, and ILC3, which are a counterpart to TH17 cells and have an anti-bacterial type 3 response through production of IL-17 and IL-22. Whereas adaptive lymphocytes are found primarily in the circulation and lymph tissues, ILCs are mainly tissue-resident and particularly enriched at mucosal surfaces, such as the lung and intestine. There, they play a significant role in regulating tissue homeostasis in the adult, together with their role as regulators of immunity. However, the developmental history of ILCs begins early on in life, even before adaptive lymphocytes develop.
As all lymphocytes, ILCs originate through the process of lymphopoiesis, whereby lymphoid cells arise from hematopoietic progenitor cells (HPCs) in a series of sequentially restrictive steps as they ultimately commit to a single lineage (reviewed in Doulatov et al., 2012; Golub and Cumano, 2013) (2, 3). The first wave of progenitors in the mouse originates within the yolk-sac (YS) from embryonic gestation day 7 (E7) and from the aorta-gonad-mesonephros (AGM) from E10.5. Here, hemogenic endothelial cells transform into cells with hematopoietic potential which travel through the vascular system to colonize the fetal liver, the primary site of hematopoiesis (4–7). The equivalent in humans occurs at around weeks 3-4 of gestation (8). From HPCs rise the common lymphoid progenitor (CLP), which has lost myeloid potential but has the ability to generate ILC subsets, as well as T cells and B cells (9, 10). In the adult, the transition from HPC to CLP in the mouse is dependent on IL-7 (11, 12), a cytokine that plays a vital role in lymphoid development and maintenance, particularly that of ILC2 and ILC3 as shown in Il7-/- mice (13–18). IL-7R-expressing progenitors can be identified in the YS as early as E9.5 (19). Downstream of the CLP, ID2-expressing as well as PLZF+/- precursors have been identified in fetal liver and newly generated mouse strains have highlighted their potential to generate all ILC subsets (17, 20–25). Of note, we will use the simplified term innate lymphoid cell progenitor (ILCP) to describe ILC-restricted precursor populations including subsets that were originally named EILP, αLP or ChILP in the primary publications. Various investigations have highlighted their complex transcriptional network regulating ILC ontogeny during embryogenesis, including lineage-specific transcripts such as Tbx21, Eomes, Rorc, and Gata3 (26–30). Interestingly, these lineage-identifying transcription factors in the fetal ILCP pool are often co-expressed before commitment to a single lineage (21, 31). Although human fetal ILC-poiesis studies naturally lag behind that of the mouse, the era of single-cell analyses has also allowed for fetal landscape interrogation in humans. Human fetal liver samples ranging from 4 to 17 weeks post-conception (PCW) have been assessed by various groups at single-cell resolution (32–34). In these studies, human fetal liver ILCP have been described as a heterogeneous population expressing KIT, IL7R, and KLRB1 (encoding CD161, a human ILC marker). This follows the description of a circulating ILCP in both adult and fetal tissues in 2017 (35). Liu and colleagues further detected low expression of CD34, marking their potential progenitor properties more directly (32). In humans, PLZF is expressed by fetal and circulating ILCP, as well as differentiated ILCs, making it ineffective as a marker of progenitors (32, 35, 36). Interestingly, these circulating ILCP have a substantial type 3 signature reminiscent of group 3 ILCs, including RORC transcripts as well as KIT, LTB, LTA, and CXCR5, as shown in the online data repositories (32, 33). It remains an open question whether there is a transient upregulation of RORC starting from the ILCP during human ILC maturation or whether the RORC-expressing ILCP described in these studies rather represent progenitors with LTi potential. Specifically, LTB, LTA, and CXCR5, which encode functional molecules during lymph node (LN) development, have been described in connection with an LTiP signature (21, 31). Future studies are needed to reveal these cell populations’ precise progenitor-progeny relationship and functionality.
The overhanging dogma regarding circulating ILC progenitors was that they home to the bone marrow (BM) from the fetal liver shortly before birth, where they remain into adulthood as the primary site of hematopoiesis. However, multiple studies over the past years have suggested that ILCs arise from fetal ILCP in distinct waves of differentiation throughout embryogenesis, as summarized in Figure 1. ILC layered ontogeny generates specialized tissue subsets from local ILC progenitors as shown in the lung, thymus, intestine, or liver (31, 37–42). This suggests that ILC progenitors may seed tissues during embryogenesis and that hematopoiesis in the embryo is not restricted to the fetal liver or BM. In 2015, Bando et al. showed a fetal intestinal precursor expressing Arginase 1 (encoded by Arg1) that has the capacity to give rise to ILC1, ILC2, and ILC3, suggesting a model where fetal progenitors may seed peripheral organs and differentiate in situ during embryogenesis (43). This fetal progenitor seeds the intestine at day E13.5, prior to Peyer’s patches organogenesis. By single-cell RNAseq analysis, we have recently identified progenitors with CLP-like signatures and an IL-7Rα+ α4β7+ PLZFhi ILCP in the fetal intestine, likely at least partially overlapping with the Arg1+ ILCP described by Bando et al. ( 43). The transcriptional profile of this fetal ILCP correlated with that reported in the BM and fetal liver and included ILCP-associated genes Zbtb16, Tcf7, Tox, Id2, and Arg1, as well as ILC-lineage transcripts such as Rora, Ahr, Il7r, Il2rb, and CXCR6 (31). Similarly, CLP, as well as ILCP populations, could be found in the embryonic periphery, enriched in LN Anlagen (39). In the embryonic thymus, studies have revealed a first wave of early thymic progenitors (ETPs) from E13 and E15.5 capable of giving rise to ILCs as well as T cells (19, 40). In concordance, progenitors with T cell and ILC potential have been described using single-cell in vitro cultures of sorted early T cell progenitors from E13 thymi on OP9 stromal cells (19). In vivo adoptive transfer of E13 ETPs into irradiated neonatal Rag2-/-Il2rg-/- mice produced all ILC lineages as assessed five weeks later in the intestine of host mice, showing their potential to generate innate lymphocyte lineages. Together, these studies show that tissue-specific progenitor seeding gives rise to differentiated ILCs throughout embryogenesis, where they remain in their tissue niche or have the potential to seed other tissues. As such, tissue progenitors have been described in lung tissue of adult mice by Zeis et al. and Ghaedi et al., that also investigated postnatal day 4 and 12 (44, 45). These tissue progenitors were found to express IL-18R, α4β7 and PD1 and presented gene expression of Tcf7, Zbtb16, Rora and Gata3, thereby displaying core similarities with BM-derived ILCPs. Importantly, a similar population was also identified in human blood and lung (45).
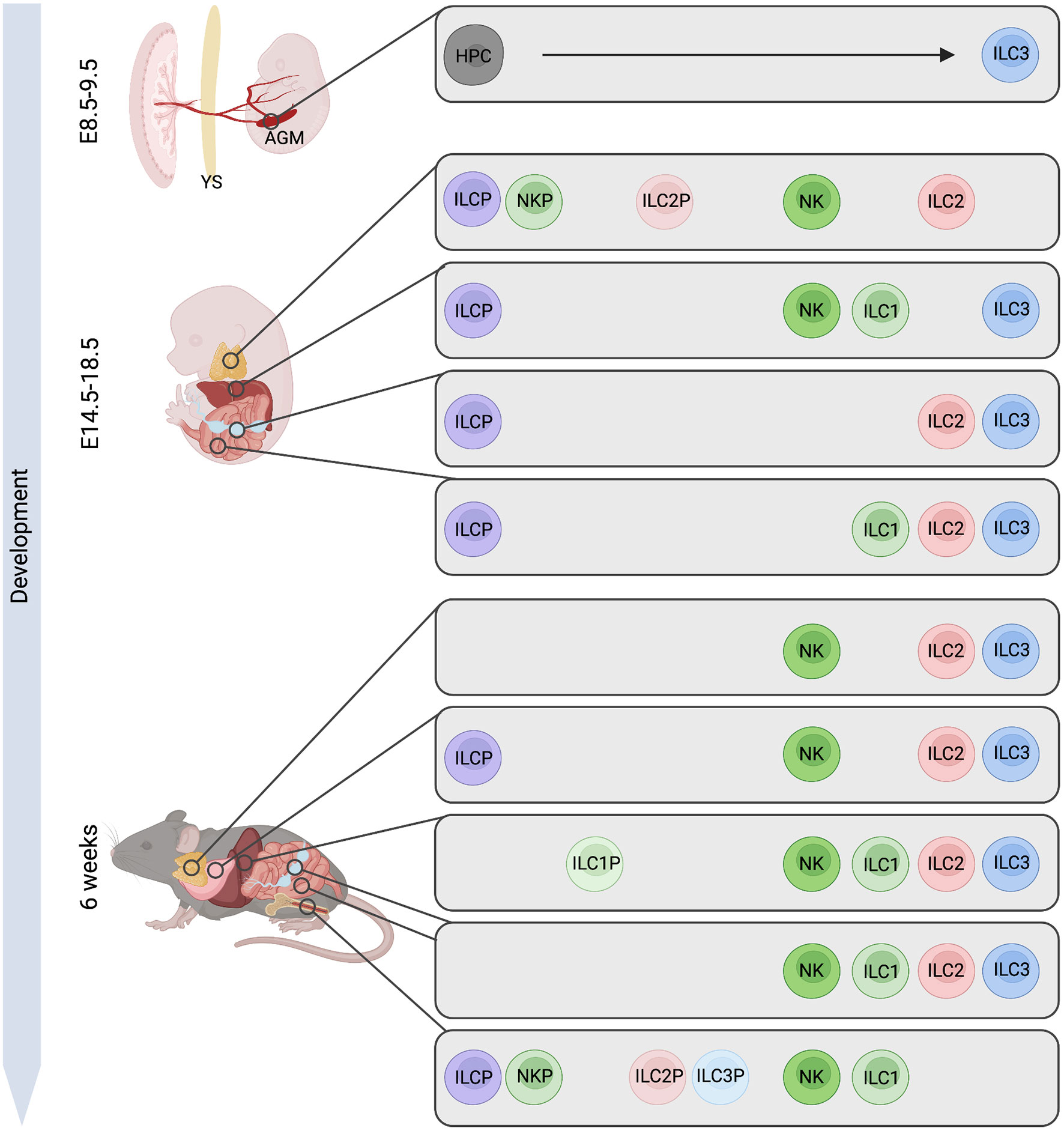
Figure 1 Murine ILC-poiesis from embryo to adult. Early hematopoietic progenitor cells (HPCs) arise from hemogenic endothelial cells within the aorta gonad mesonephros (AGM) region giving rise to ILC3. Later in ontogeny, innate lymphoid cell progenitors (ILCP) are detected in fetal liver but also in lymph node (LN) Anlagen or small intestine, while this population in the adult has only been described in the bone marrow. Further, precursors to specific ILC lineages (NKP, ILC1P, ILC2P, ILC3P) are present in the embryonic thymus, in fetal liver and LN while these cells are mostly confined to the adult bone marrow with exceptions of ILC1P and ILC2P also being found in liver and lung tissue. Figure created with BioRender.com.
Interestingly, the significant presence of human fetal lymphoid progenitors (CD34+ CD127+) in the fetal intestine suggests it is a site of hematopoiesis in the human as well (32). Liu and colleagues analyzed human fetal tissues from 8, 10, and 12 weeks PCW, specifically the liver, thymus, spleen, intestine, skin, and lung, and followed the heterogeneous CD34+ progenitor pool at single-cell resolution (32). To which extent these cells might resemble the CD34+ ILC- and ILC3-committed progenitors previously characterized in human tonsil and intestine need to be ascertained (46, 47). In contrast, the CD34- ILCP described by Lim et al. in adult tissues lack genes associated with general hematopoiesis such as CD34, and instead express genes shared with fetal ILCP such as GATA3, CD7 and IL7R, which are also expressed by differentiated ILCs (32, 35). Comprehensive analyses of the differential potential of these precursors, consisting of functional in vitro assays, in vivo transfers, and in-depth transcriptomic and chromatin accessibility comparisons are required to untangle the differentiation stages of human fetal ILCs.
The main site of NK cell development in adult mice and humans is the BM. However, in the murine fetal liver, the emergence of CD122+ progenitors at E13.5, which upregulate DX5 and NK1.1 at E15.5, has been reported (48, 49). By E15/16.5, these cells are suggested to be functionally competent as assessed by cytotoxicity chromium release assay against YAC-1 target cells, or NK1.1-activation and subsequent degranulation. In OP9 co-cultures of E14.5 progenitors, these cells efficiently generated not only NK cells but also T cells in vitro, suggesting that these progenitors remain bipotential (48). In addition, it has recently been demonstrated that the fetal liver harbors a Lin- Sca-1+ Mac-1+ (LSM) population which largely expresses the CLP-associated markers CD34 and Flt3 and that in the adult liver includes CD122+ CD49a+ cells (50). Although T-bet or Eomes expression were not assessed, these cells are suggested to give rise to ILC1, a process that depends on an IFNγ-feedback loop from NKp46+ cells (50). Along these lines, two recent studies utilizing inducible fate mapping models and in vivo transfers have also identified an embryonic wave of ILC1 generation (41, 42). Chen et al. found that ILC1 are present at E16.5 in fetal liver with highly cytotoxic Ly49E+ subsets residing into adulthood in contrast to Ly49E- ILC1 that are gradually replenished by hematopoietic progenitors after birth (41), in concordance with earlier reports by Filtjens et al. ( 51). Accordingly, T-bet+ Eomes- ILC1 were detected in the E16 fetal liver predominating over NK cells with promiscuous DX5 expression, which is subsequently lost after birth (52). During neonatal infection with murine cytomegalovirus (CMV), embryonic-derived ILC1 were potent IFNγ-producers and Ly49E+ subsets displayed higher cytotoxic molecules perforin and granzyme B reducing viral loads in adoptive transfer settings into Tbx21-/- mice (41). Besides the fetal liver, the embryonic thymus has also been identified to harbor Group 1 lineage precursors defined as Id2+ Bcl11b- as early as E13.5. These cells expressed Eomes and perforin after in vitro culture in IL-2/IL-5/IL-18 (40). Group 1 ILCs displaying transcripts of Klrb1c, Klrk1, and Tbx21 have been described in parallel through a detailed kinetic of thymic cells from E12.5 to postnatal day one at single-cell resolution, suggesting that ILC1-like cells are particularly present at early time points (E12.5-E15.5) (53). Moreover, as ILC1 also reside in the adult intestine, they can be readily detected at E18.5 in the fetal intestine, possibly originating in situ from the intestinal PLZF+ ILCP (31). In humans, CD3ε- CD56+ cells can be detected as early as 6 PCW, gradually increasing in the human fetal liver and in fetal spleens of 15-24 PCW (54). Between 18-36 PCW, NK cells are frequently represented not only in the liver but also in the lung and spleen, and to lower frequencies in fetal BM and mLN, and display organ-specific variations in killer-cell immunoglobulin-like receptor (KIR) expression (55). NK cell-restricted progenitors defined as CD34+ CD38+ cells expressing CD45RA, CD10, and CD7 but lacking CD127 were found in human fetal liver from gestational week 6 onwards, and also in fetal BM from gestational week 12 (56). Further extending our knowledge on human tissue hematopoiesis, early progenitors defined as CD34+ CD38dim with NK lineage potential have also been detected in the fetal human thymus (54, 57). Interestingly, CD127+ ILC1, as analyzed by the lack of ILC2- and ILC3-specific surface markers (CRTH2- CD117-), are preferentially found in the embryonic thymus but also in the spleen, skin, lung, intestine, and liver with tissue-preferred distributions of subsets (32). Of note, since Eomes transcripts were ubiquitously expressed in Group 1 ILCs, distinct NK/ILC1 identities could not be clearly dissected. Within the embryonic intestine, frequencies of ILC1 in humans were very low compared to adult gut (58–60). However, due to the unclear differentiation of NK and ILC1 in particular tissues, during inflammation, or in general in humans, further investigations focusing on their identities are necessary to tease out fetal developmental pathways. Regarding type 1 immunity, the placental-fetal interface represents a protective barrier. Still, infection with a variety of pathogens inducing type 1 responses, including viruses such as CMV, bacteria like Listeria monocytogenes, or the intracellular parasite Toxoplasma gondii may cause vertical transmission (61). In the case of CMV infection, NK cells isolated from cord blood of newborn children with congenital CMV infection showed increased expression of cytotoxic molecules granzyme B, perforin, and granulysin, suggestive of in utero activation (62). Notably, NKG2C, LILRB1, and CD57 – hallmarks of CMV infection in adults – were lowly expressed in all infants, including non-infected controls, possibly due to a lack of proinflammatory cytokines IL-12 and IL-15, as reported to be reduced in neonatal DC populations (63). For granzyme C-expressing ILC1, a perforin-dependent lethal immunopathology affecting the salivary gland and liver of neonates has been suggested under enhanced STAT5 signaling (64).
The tissue-resident adult pool of ILC2 has been shown to be composed of cells originating from several phases during ontogeny (37). Id2 and Arg1 reporter mice, in combination with complementary fate-mapping methodologies, revealed that tissue ILC2 are generated in three primary waves. In this study, Thy1+ IL-7R+ ST2+ cells were detected as early as day E17.5 in fetal lung, skin, and small intestine, with the majority being replaced by eight weeks of age with tissue-specific signatures of de novo replacements. Early postnatal differentiation waves generated the majority of ILC2 in lung and adipose tissue establishing tissue-specific pools of ILC2. Interestingly, the BM did not display any fetal-derived ILC2 suggesting that this niche is only populated post-natally. However, whether peripheral ILC2 in adults derive from distinct fetal or adult hematopoietic stem cell progenitors remains unknown. Interestingly, tissue signals implicated in ILC2 differentiation and maturation have been described in the fetal and adult mesentery where PDGFRα+ gp38+ mesenchymal cells support terminal ILC2 differentiation from the ILCP but not CLP stage (65). Through single-cell sequencing of E15.5 and E19.5 CD44+ CD25+/- cells, ILC2-precursors were detected in the developing embryonic thymus, where RORα expressed by a common progenitor has been identified as a critical checkpoint in controlling T cell versus ILC2 commitment within the sorted double-negative stage 1 and 2 compartments. ATAC-seq and RORα-ChIP-seq analyses revealed the binding of RORα to ILC2-associated genes such as Il5, Il13, and Arg1, as well as to regulatory elements of the loci regulating Id2 and Nfil3, restricting T cell commitment while promoting ILC2 development. In two experimental settings that used either adoptive transfer or ectopic E15.5 thymi transplantation under the kidney capsule, thymus-derived ILC2 are able to migrate to the intestine of host Rag2-/-Il2rg-/- mice exposing their capability to seed extrathymic tissues (40). Together, these data expose a common thymic precursor that has the potential to generate T cells as well as ILC2 that seed the periphery. Accordingly, the human embryonic thymus harbors a substantial fraction of ILC2, while lower frequencies can be found in the skin, lung, liver, and intestine (32). Pooling CRTH2+ ILC2 from each organ at various time points further exposed a CCR9+ Bcl11b+ subset, a T cell gene-enriched population (paralleling the murine studies of Ferreira et al., 2021) (40), as well as precursor cells which, however, lack functional validation. In contrast to adult tissues, ILC2 are readily detected in human fetal mLN and intestine as well as in lung (66, 67). The functions of fetal ILC2 have not been suitably addressed; nevertheless, an interesting study by Saluzzo et al. reports a postnatal influx of ILC2 into lung tissue due to first breath-induced tissue damage and its associated IL-33 epithelial response, resulting in alveolar macrophage polarization into a regulatory M2 phenotype and increased resistance to Streptococcus pneumaoniae (68). This perinatal IL-33/ILC2 axis has also been reported to confer a risk for developing asthma later in life if neonatal allergen exposure in the developing lung, such as house dust mite exposure, leads to hyperactivation enhancing type 2 immunity involving not only ILC2, but also dendritic cells as well as Th2 cells (69). Lastly, early-life respiratory viral infections such as infection with respiratory syncytial virus (RSV) and rhinovirus (RV) may also cause ILC2 activation and long-term lung alterations and lead to immunopathology (reviewed in Fonseca et al., 2021) (70).
ILC3 are amongst the earliest lymphocytes to develop during embryogenesis, and include LTi cells which play a fundamental role in the development of lymphoid tissues prenatally. In addition to RORγt, murine LTi cells express CD4 on their surface, as well as functional molecules such as α4β7, CXCR5, RANKL, or CCR6 enabling them to interact with stromal cells to orchestrate LN development (71–73). CD127+ CD4+ RORγt+ LTi cells are detected as early as E12.5 in murine cervical LN anlagen and can also be found in other peripheral LN anlagen, fetal liver, spleen, and intestine (39, 74–77). Challenging the fetal liver as the origin of ILC3 lineages within the embryo, the group of van de Pavert observed that hematopoietic progenitors arise in the hemogenic endothelial sites by E7.5-9 by using Cxcr4 and Cdh5 fate-mapping models (39). Interestingly, they furthermore note that in contrast to peripheral LN sites, progenitors and LTi cells within the fetal liver had no direct connection in ontogeny, suggesting that LTi maturation occurs in situ. In line with these findings, we observed the presence of an LTi lineage-skewed progenitor population together with mature progeny in the embryonic intestinal tissue (31). Over the last years, several factors influencing the differentiation program of ILC3 have been described, and their spatial availability might control specific maturational steps at different locations. In the fetal liver, Notch signaling has been proposed to regulate the expression of Id2 within progenitors while subsequent expression of RORγt is inhibited by Notch, blocking final hepatic maturation (75). Along these lines, Golub and colleagues suggest that BM-derived CLP from adult mice depend on Notch signaling in the periphery to mature into RORγt+ cells but are dispensable for the development of fetal RORγt+ ILCs (78). Additional data exposed that, contrary to the fetal liver, developing progenitors in the periphery respond to maternal-derived retinoids to upregulate RORγt, and retinoic acid signaling even controls the size of the secondary lymphoid organs (79). Of note, not only RORγt expression in ILCs is regulated by retinoic acid signaling, but also the expression of the chemokine CXCL13 by stromal lymphoid tissue organizer cells to attract CXCR5+ ILC3. Altogether this data demonstrates the importance of retinoic acid availability during embryonic life for the organogenesis of lymphoid tissues (80). Along the lines of maternally transferred signals, Agüero and Ganal-Vonarburg et al. describe that, although the embryo develops in a sterile environment during pregnancy, maternal microbial-derived metabolites involving Ahr ligands can be transferred to the embryonic offspring thereby shaping at least the neonatal ILC3 pool (81). Notably, while Ahr regulates apoptosis and IL-22 production in ILC3 isolated from adult mice, there is no main effect on fetal intestinal ILC3 (82). Moreover, IL-7 is known to play a key role in LN organogenesis by not only regulating the pool size of IL-7R+ LTi cells, but also their Lymphotoxin (LT) α1β2 production (83–87). Besides the aforementioned tissues, comparably to ILC1-like and ILC2 subsets, ILC3 could also be detected by analysis of E16 thymic organ cultures (88). However, whether the cells analyzed originated from mature ILC3 lineages or from immature progenitors within the embryonic thymus has not been addressed in this study. Similar observations to those in mice have also been found in human fetal mLN, liver, and intestine by Teichmann and colleagues, who elegantly describe ILCP and LTi compartments with profound corresponding transcriptional profiles and in addition, using single-molecule fluorescence in situ hybridization, place CXCR5 and RORC-expressing LTi cells adjacent to CXCL13-expressing lymphoid tissue organizer cells in proximal gut mucosa (89). Spatially, human CRTH2- CD117+ ILC3 among Lin- CD34- CD45+ cells can be detected in liver, intestine, spleen, skin, lung, and thymus at 12 PCW displaying LTi-related as well as non-LTi gene expression patterns in concordance with flow cytometric analysis by Marquardt et al (32, 90). Of note, NKp44 expression was only observed from week 12 onward in the intestine and liver (90–92). Importantly, as CD4 is not expressed by human LTi cells (93), Nrp-1 has been suggested to identify human but also mouse LTi cells instead (94, 95). As such, Nrp-1-expressing CD117+ ILC3 can be detected in human fetal gut, mLN and spleen (94). Tissue seeding of ILCs is initiated already during fetal development, which on the one hand facilitates balanced homeostatic conditions early in ontogeny, and on the other confers the first line of defense before the adaptive immune system is generated. In murine neonates, it has been suggested that ILC3 promote IL-22-mediated resistance to pneumonia induced by intranasal challenge with Streptococcus pneumoniae, an effect which is dependent on postnatal colonization by commensal bacteria (96). Similarly, in the neonate, Oherle et al. show that lung ILC progenitors can differentiate towards ILC3 in situ, and importantly, that this process is dependent on tissue signals (38). Using lineage-specific IGF1 or IGF1 receptor ablation, they showed that IGF1 producing alveolar fibroblasts generate a special niche in the neonatal lung to influence differentiation of PLZF+ ILCP thereby regulating ILC3 differentiation including IL-22-mediated immunity in the lung (38).
Characteristic markers of tissue-derived ILC progenitors are listed in Table 1.
Several aspects of ILC-poiesis in the fetus remain to be clarified. Revealing the embryonic developmental timing of various ILC populations (Figure 1) in particular will require advanced lineage tracing techniques and sensitive mouse models. The exact waves of ILC ontogeny in various organs for example, as well as their contribution to shaping organ architecture and innate immunity post-birth, remains unclear. Whether diverse embryonic waves exclusively produce heterogeneous ILC subsets that remain in the adult potentially locating to specific sites, as shown for innate-like lymphocytes such as B-1a B cells in the peritoneal and pleural cavities, or certain γδ T cell subsets in gut and skin, remains to be addressed in future studies. In this context, it will be an important area of further investigation to reveal the impact of embryonic lymphocyte waves not only on the establishment of immune function and disease susceptibility but also its impact on the development of the physiological functioning of various organs. As in vitro cell differentiation and cultivation technologies are rapidly evolving, so does the ability to address the main questions of ILC ontogeny also in humans. The use of comprehensive platforms for in vitro ILC generation from CD34+ progenitors from various tissues (104), which allow for an accessible resource of ILCs for interrogation and genome editing, as well as the complex microphysiological systems becoming available (105), would allow for the dynamic investigation of human lymphoid developmental pathways and their regulators within tissue niches. As disruption of ILCs and their development can have enduring consequences on the immunity and health of the host, it is critical to understand local tissue ILC progenitor differentiation and maintenance, and the tissue signals which regulate these processes.
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) grants SPP 1937 (RO3565/4-2) and DFG-SFB TRR241 B02, as well as the Leibniz-Science Campus Chronic Inflammation and Leibniz-Kooperative Exzellenz K259/2019.
We thank Chiara Romagnani, as well as Timo Rückert and Nils Christian Müller, for critical reading of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Vivier E, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate lymphoid cells: 10 years on. Cell (2018) 174:1054–66. doi: 10.1016/j.cell.2018.07.017
2. Doulatov S, Notta F, Laurenti E, Dick JEE. Hematopoiesis: A human perspective. Cell Stem Cell (2012) 10:120–36. doi: 10.1016/j.stem.2012.01.006
3. Golub R, Cumano A. Embryonic hematopoiesis. Blood Cells Mol Dis (2013) 51:226–31. doi: 10.1016/j.bcmd.2013.08.004
4. Zhu Q, Gao P, Tober J, Bennett L, Chen C, Uzun Y, et al. Developmental trajectory of prehematopoietic stem cell formation from endothelium. Blood (2020) 136:845–56. doi: 10.1182/blood.2020004801
5. Tober J, Koniski A, McGrath KE, Vemishetti R, Emerson R, De Mesy-Bentley KKL, et al. The megakaryocyte lineage originates from hemangioblast precursors and is an integral component both of primitive and of definitive hematopoiesis. Blood (2007) 109:1433–41. doi: 10.1182/BLOOD-2006-06-031898
6. Palis J, Robertson S, Kennedy M, Wall C, Keller G. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development (1999) 126:5073–84. doi: 10.1242/DEV.126.22.5073
7. Lancrin C, Sroczynska Stephenson P, Allen C, Kouskoff T, Lacaud V, G, et al. The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature (2009) 457:892–5. doi: 10.1038/nature07679
9. Mebius RE, Miyamoto T, Christensen J, Domen J, Cupedo T, Weissman IL, et al. The fetal liver counterpart of adult common lymphoid progenitors gives rise to all lymphoid lineages, CD45 + CD4 + CD3 – cells, as well as macrophages. J Immunol (2001) 166:6593–601. doi: 10.4049/jimmunol.166.11.6593
10. Palis J, Yoder MC. Yolk-sac hematopoiesis: The first blood cells of mouse and man. Exp Hematol (2001) 29:927–36. doi: 10.1016/S0301-472X(01)00669-5
11. Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell (1997) 91:661–72. doi: 10.1016/S0092-8674(00)80453-5
12. Peschon JJ, Morrissey PJ, Grabstein KH, Ramsdell FJ, Maraskovsky E, Gliniak BC, et al. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J Exp Med (1994) 180:1955–60. doi: 10.1084/JEM.180.5.1955
13. Satoh-Takayama N, Lesjean-Pottier S, Vieira P, Sawa S, Eberl G, Vosshenrich CAJ, et al. IL-7 and IL-15 independently program the differentiation of intestinal CD3–NKp46+ cell subsets from Id2-dependent precursors. J Exp Med (2010) 207:273–80. doi: 10.1084/JEM.20092029
14. Puel A, Ziegler SF, Buckley RH, Leonard WJ. Defective IL7R expression in T-B+NK+ severe combined immunodeficiency. Nat Genet (1998) 20:394–7. doi: 10.1038/3877
15. Vonarbourg C, Mortha A, Bui VL, Hernandez PP, Kiss EA, Hoyler T, et al. Regulated expression of nuclear receptor RORγt confers distinct functional fates to NK cell receptor-expressing RORγt+ innate lymphocytes. Immunity (2010) 33:736–51. doi: 10.1016/j.immuni.2010.10.017
16. Mazzucchelli R, Durum SK. Interleukin-7 receptor expression: Intelligent design. Nat Rev Immunol (2007) 7:144–54. doi: 10.1038/nri2023
17. Klose CSN, Flach M, Mohle L, Rogell L, Hoyler T, Ebert K, et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell (2014) 157:340–56. doi: 10.1016/j.cell.2014.03.030
18. Moro K, Yamada T, Tanabe M, Takeuchi T, Ikawa T, Kawamoto H, et al. Innate production of T(H)2 cytokines by adipose tissue-associated c-Kit(+)Sca-1(+) lymphoid cells. Nature (2010) 463:540–4. doi: 10.1038/nature08636
19. Elsaid R, Meunier S, Burlen-Defranoux O, Soares-da-Silva F, Perchet T, Iturri L, et al. A wave of bipotent T/ILC-restricted progenitors shapes the embryonic thymus microenvironment in a time-dependent manner. Blood (2021) 137. doi: 10.1182/BLOOD.2020006779
20. Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nat (2014) 508:397–401. doi: 10.1038/nature13047
21. Ishizuka IE, Chea S, Gudjonson H, Constantinides MG, Dinner AR, Bendelac A, et al. Single-cell analysis defines the divergence between the innate lymphoid cell lineage and lymphoid tissue–inducer cell lineage. Nat Immunol (2016) 17:269–76. doi: 10.1038/ni.3344
22. Chea S, Possot C, Perchet T, Petit M, Cumano A, Golub R, et al. CXCR6 expression is important for retention and circulation of ILC precursors. Mediators Inflammation (2015) 2015:368427. doi: 10.1155/2015/368427
23. Walker JA, Clark PA, Crisp A, Barlow JL, Szeto A, Ferreira ACF, et al. Polychromic reporter mice reveal unappreciated innate lymphoid cell progenitor heterogeneity and elusive ILC3 progenitors in bone marrow. Immunity (2019) 51:104–118.e7. doi: 10.1016/j.immuni.2019.05.002
24. Zook EC, Li ZY, Xu Y, de Pooter RF, Verykokakis M, Beaulieu A, et al. Transcription factor ID2 prevents e proteins from enforcing a naïve T lymphocyte gene program during NK cell development. Sci Immunol (2018) 3. doi: 10.1126/SCIIMMUNOL.AAO2139
25. Xu W, Cherrier DE, Chea S, Vosshenrich C, Serafini N, Petit M, et al. An Id2RFP-reporter mouse redefines innate lymphoid cell precursor potentials. Immunity (2019) 50:1054–1068.e3. doi: 10.1016/j.immuni.2019.02.022
26. Serafini N, Klein Wolterink RGJ, Satoh-Takayama N, Xu W, Vosshenrich CAJ, Hendriks RW, Di Santo JP, et al, et al. Gata3 drives development of RORγt+ group 3 innate lymphoid cells. J Exp Med (2014) 211:199–208. doi: 10.1084/jem.20131038
27. Yagi R, Zhong C, Northrup DLL, Yu F, Bouladoux N, Spencer S, Hu G, Barron L, et al, et al. The transcription factor GATA3 is critical for the development of all IL-7Rα-Expressing innate lymphoid cells. Immunity (2014) 40:378–88. doi: 10.1016/j.immuni.2014.01.012
28. Townsend MJ, Weinmann AS, Matsuda JL, Salomon R, Farnham PJ, Biron CA, et al. T-Bet regulates the terminal maturation and homeostasis of NK and Vα14i NKT cells. Immunity (2004) 20:477–94. doi: 10.1016/S1074-7613(04)00076-7
29. Sun Z, Unutmaz D, Zou YR, Sunshine MJ, Pierani Brenner-Morton S, Mebius RE, et al. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science (2000) 288:2369–73. doi: 10.1126/science.288.5475.2369
30. Eberl G, Marmon S, Sunshine MJ, Rennert PD, Choi Y, Littmann DR, et al. An essential function for the nuclear receptor RORγt in the generation of fetal lymphoid tissue inducer cells. Nat Immunol (2004) 5:64–73. doi: 10.1038/ni1022
31. Stehle C, Rückert T, Fiancette R, Gajdasik DW, Willis C, Ulbricht C, et al. T-Bet and RORα control lymph node formation by regulating embryonic innate lymphoid cell differentiation. Nat Immunol (2021) 22:1231–44. doi: 10.1038/s41590-021-01029-6
32. Liu C, Gong Y, Zhang H, Yang H, Zeng Y, Bian Z, et al. Delineating spatiotemporal and hierarchical development of human fetal innate lymphoid cells. Cell Res (2021) 31:1106–22. doi: 10.1038/s41422-021-00529-2
33. Popescu D-M, Botting RA, Stephenson E, Green K, Webb S, Jardine L, et al. Decoding human fetal liver haematopoiesis. In: Nature(2019) 574:365–71. doi: 10.1038/s41586-019-1652-y
34. Suo C, Dann E, Goh I, Jardine L, Kleshchevnikov V, Park J-E, et al. Mapping the developing human immune system across organs. Science (2022) 376:eabo0510. doi: 10.1126/science.abo0510
35. Lim AI, Li Y, Lopez-Lastra S, Stadhouders R, Paul F, Casrouge A, et al. Systemic human ILC precursors provide a substrate for tissue ILC differentiation. Cell (2017) 168:1086–1100.e10. doi: 10.1016/J.CELL.2017.02.021
36. Björklund Å.K, Forkel M, Picelli S, Konya V, Theorell J, Friberg D, et al. The heterogeneity of human CD127(+) innate lymphoid cells revealed by single-cell RNA sequencing. Nat Immunol (2016) 17:451–60. doi: 10.1038/ni.3368
37. Schneider C, Lee J, Koga S, Ricardo-Gonzalez RR, Nussbaum JC, Smith LK, et al. Tissue-resident group 2 innate lymphoid cells differentiate by layered ontogeny and In situ perinatal priming. Immunity (2019) 50:1425–1438.e5. doi: 10.1016/J.IMMUNI.2019.04.019
38. Oherle K, Acker E, Bonfield M, Wang T, Gray J, Lang I, et al. Insulin-like growth factor 1 supports a pulmonary niche that promotes type 3 innate lymphoid cell development in newborn lungs. Immunity (2020) 52:275–294.e9. doi: 10.1016/J.IMMUNI.2020.01.005
39. Simic M, Manosalva I, Spinelli L, Gentek R, Shayan RR, Siret C, et al. Distinct waves from the hemogenic endothelium give rise to layered lymphoid tissue inducer cell ontogeny. Cell Rep (2020) 32:108004. doi: 10.1016/j.celrep.2020.108004
40. Ferreira ACF, Szeto ACH, Heycock MWD, Clark PA, Walker JA, Crisp A, et al. RORα is a critical checkpoint for T cell and ILC2 commitment in the embryonic thymus. Nat Immunol (2021) 22:166–78. doi: 10.1038/s41590-020-00833-w
41. Chen Y, Wang X, Hao X, Li B, Tao W, Zhu S, et al. Ly49E separates liver ILC1s into embryo-derived and postnatal subsets with different functions. J Exp Med (2022) 219. doi: 10.1084/jem.20211805
42. Sparano C, Solís-Sayago D, Vijaykumar A, Rickenbach C, Vermeer M, Ingelfinger F, et al. Embryonic and neonatal waves generate distinct populations of hepatic ILC1s. Sci Immunol (2022) 7:eabo6641. doi: 10.1126/sciimmunol.abo6641
43. Bando JK, Liang H-EE, Locksley RM. Identification and distribution of developing innate lymphoid cells in the fetal mouse intestine. Nat Immunol (2015) 16:153–60. doi: 10.1038/ni.3057
44. Ghaedi M, Shen ZY, Orangi M, Martinez-Gonzalez I, Wei L, Lu X, et al. Single-cell analysis of RORα tracer mouse lung reveals ILC progenitors and effector ILC2 subsets. J Exp Med (2020) 217. doi: 10.1084/jem.20182293
45. Zeis P, Lian M, Fan X, Herman JS, Hernandez DC, Gentek R, et al. In situ maturation and tissue adaptation of type 2 innate lymphoid cell progenitors. Immunity (2020) 53:775–792.e9. doi: 10.1016/j.immuni.2020.09.002
46. Montaldo E, Teixeira-Alves LG, Glatzer T, Durek P, Stervbo U, Hamann W, et al. Human RORγt+CD34+ cells are lineage-specified progenitors of group 3 RORγt+ innate lymphoid cells. Immunity (2014) 41:988–1000. doi: 10.1016/j.immuni.2014.11.010
47. Scoville SD, Mundy-Bosse BL, Zhang MH, Chen LL, Zhang X, Keller KA, et al. A progenitor cell expressing transcription factor RORgt generates all human innate lymphoid cell subsets. Immunity (2016) 44:1140–50. doi: 10.1016/j.immuni.2016.04.007
48. Tang Y, Peitzsch C, Charoudeh HN, Cheng M, Chaves P, Jacobsen SEW, et al. Emergence of NK-cell progenitors and functionally competent NK-cell lineage subsets in the early mouse embryo. Blood (2012) 120:63–75. doi: 10.1182/BLOOD-2011-02-337980
49. Carlyle JR, Michie AM, Cho SK, Zúñiga-Pflücker JC. Natural killer cell development and function precede αβ T cell differentiation in mouse fetal thymic ontogeny. J Immunol (1998) 160:744–53. doi:
50. Bai L, Vienne M, Tang L, Kerdiles Y, Etiennot M, Escalière B, et al. Liver type 1 innate lymphoid cells develop locally via an interferon-γ-dependent loop. Science (2021) 371. doi: 10.1126/science.aba4177
51. Filtjens J, Taveirne S, Van Acker A, Van Ammel E, Vanhees M, Kerre T, et al. Abundant stage-dependent Ly49E expression by liver NK cells is not essential for their differentiation and function. J Leukoc Biol (2013) 93:699–711. doi: 10.1189/jlb.0812378
52. Constantinides MG, Gudjonson H, McDonald BD, Ishizuka IE, Verhoef PA, Dinner AR, et al. PLZF expression maps the early stages of ILC1 lineage development. Proc Natl Acad Sci USA (2015) 112:5123–8. doi: 10.1073/PNAS.1423244112/SUPPL_FILE/PNAS.201423244SI.PDF
53. Kernfeld EM, Genga RMJ, Neherin K, Magaletta ME, Xu P, Maehr R, et al. A single-cell transcriptomic atlas of thymus organogenesis resolves cell types and developmental maturation. Immunity (2018) 48:1258–1270.e6. doi: 10.1016/J.IMMUNI.2018.04.015
54. Sánchez MJ, Muench MO, Roncarolo MG, Lanier LL, Phillips JH. Identification of a common t/natural killer cell progenitor in human fetal thymus. J Exp Med (1994) 180:569–76. doi: 10.1084/JEM.180.2.569
55. Ivarsson MA, Loh L, Marquardt N, Kekäläinen E, Berglin L, Björkström NK, et al. Differentiation and functional regulation of human fetal NK cells. J Clin Invest (2013) 123:3889–901. doi: 10.1172/JCI68989
56. Renoux VM, Zriwil A, Peitzsch C, Michaëlsson J, Friberg D, Soneji S, et al. Identification of a human natural killer cell lineage-restricted progenitor in fetal and adult tissues. Immunity (2015) 43:394–407. doi: 10.1016/J.IMMUNI.2015.07.011
57. Res P, Martínez-Cáceres E, Jaleco AC, Staal F, Noteboom E, Weijer K, Spits H, et al, et al. CD34+CD38dim cells in the human thymus can differentiate into T, natural killer, and dendritic cells but are distinct from pluripotent stem cells. Blood (1996) 87:5196–206. doi: 10.1182/BLOOD.V87.12.5196.BLOODJOURNAL87125196
58. Bernink JH, Krabbendam L, Germar K, de Jong E, Gronke K, Kofoed-Nielsen M, et al. Interleukin-12 and -23 control plasticity of CD127+ group 1 and group 3 innate lymphoid cells in the intestinal lamina propria. Immunity (2015) 43:146–60. doi: 10.1016/J.IMMUNI.2015.06.019
59. Bernink JH, Peters CP, Munneke M, Te Velde AA, Meijer SL, Weijer K, et al. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat Immunol (2013) 14:221–9. doi: 10.1038/ni.2534
60. Collins A, Rothman N, Liu K, Reiner SL. Eomesodermin and T-bet mark developmentally distinct human natural killer cells. JCI Insight (2017) 2:90063. doi: 10.1172/JCI.INSIGHT.90063
61. Arora N, Sadovsky Y, Dermody TS, Coyne CB. Microbial vertical transmission during human pregnancy. Cell Host Microbe (2017) 21:561–7. doi: 10.1016/J.CHOM.2017.04.007
62. Vaaben A, Levan J, Nguyen CBT, Callaway PC, Prahl M, Warrier L, et al. In utero activation of NK cells in congenital CMV infection. J Infect Dis (2022) 2066:566–75. doi: 10.1093/INFDIS/JIAC307
63. Kollmann TR, Crabtree J, Rein-Weston A, Blimkie D, Thommai F, Wang XY, et al. Neonatal innate TLR-mediated responses are distinct from those of adults. J Immunol (2009) 183:7150–60. doi: 10.4049/JIMMUNOL.0901481
64. Nixon BG, Chou C, Krishna C, Dadi S, Michel AO, Cornish AE, et al. Cytotoxic granzyme c-expressing ILC1s contribute to antitumor immunity and neonatal autoimmunity. Sci Immunol (2022) 7:1–17. doi: 10.1126/sciimmunol.abi8642
65. Koga S, Hozumi K, Hirano K-I, Yazawa M, Terooatea T, Minoda A, et al. Peripheral PDGFRα+gp38+ mesenchymal cells support the differentiation of fetal liver-derived ILC2. J Exp Med jem.20172310 (2018) 215:1609–26. doi: 10.1084/jem.20172310
66. Bal SM, Bernink JH, Nagasawa M, Groot J, Shikhagaie MM, Golebski K, et al. IL-1β, IL-4 and IL-12 control the fate of group 2 innate lymphoid cells in human airway inflammation in the lungs. Nat Immunol (2016) 17:636–45. doi: 10.1038/ni.3444
67. Mjösberg JM, Trifari S, Crellin NK, Peters CP, Van Drunen CM, Piet B, et al. Human IL-25-and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat Immunol (2011) 12:1055–62. doi: 10.1038/ni.2104
68. Saluzzo S, Gorki AD, Rana BMJ, Martins R, Scanlon S, Starkl P, et al. First-Breath-Induced type 2 pathways shape the lung immune environment. Cell Rep (2017) 18:1893. doi: 10.1016/J.CELREP.2017.01.071
69. de Kleer IM, Kool M, de Bruijn MJW, Willart M, van Moorleghem J, Schuijs MJ, et al. Perinatal activation of the interleukin-33 pathway promotes type 2 immunity in the developing lung. Immunity (2016) 45:1285–98. doi: 10.1016/J.IMMUNI.2016.10.031
70. Fonseca W, Lukacs NW, Elesela S, Malinczak CA. Role of ILC2 in viral-induced lung pathogenesis. Front Immunol (2021) 12. doi: 10.3389/FIMMU.2021.675169
71. Mebius RE, Rennert P, Weissman IL. Developing lymph nodes collect CD4+CD3– LTβ+ cells that can differentiate to APC, NK cells, and follicular cells but not T or b cells. Immunity (1997) 7:493–504. doi: 10.1016/S1074-7613(00)80371-4
72. Ansel KM, Ngo VN, Hyman PL, Luther SA, Förster R, Sedgwlck JD, et al. A chemokine-driven positive feedback loop organizes lymphoid follicles. Nature (2000) 406:309–14. doi: 10.1038/35018581
73. Kim D, Mebius RE, MacMicking JD, Jung S, Cupedo T, Castellanos Y, et al. Regulation of peripheral lymph node genesis by the tumor necrosis factor family member TRANCE. J Exp Med (2000) 192:1467–78. doi: 10.1084/JEM.192.10.1467
74. Sawa S, Cherrier M, Lochner M, Satoh-Takayama N, Fehling HJ, Langa F, et al. Lineage relationship analysis of RORγt+ innate lymphoid cells. Science (2010) 330:665–9. doi: 10.1126/science.1194597
75. Cherrier M, Sawa S, Eberl G. Notch, Id2, and RORγt sequentially orchestrate the fetal development of lymphoid tissue inducer cells. J Exp Med (2012) 209:729–40. doi: 10.1084/jem.20111594
76. Yoshida H, Honda K, Shinkura R, Adachi S, Nishikawa S, Maki K, et al. IL-7 receptor α+ CD3– cells in the embryonic intestine induces the organizing center of peyer’s patches. Int Immunol (1999) 11:643–55. doi: 10.1093/INTIMM/11.5.643
77. Eberl G, Marmon S, Sunshine M-J, Rennert PD, Choi Y, Littman DR, et al. An essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nat Immunol (2004) 5:64–73. doi: 10.1038/ni1022
78. Possot C, Schmutz S, Chea S, Boucontet L, Louise A, Cumano A, et al. Notch signaling is necessary for adult, but not fetal, development of RORγt+ innate lymphoid cells. Nat Immunol (2011) 12:949–58. doi: 10.1038/ni.2105
79. Van De Pavert SA, Ferreira M, Domingues RG, Ribeiro H, Molenaar R, Moreira-Santos L, et al. Maternal retinoids control type 3 innate lymphoid cells and set the offspring immunity. Nature (2014) 508:123–7. doi: 10.1038/NATURE13158
80. Van De Pavert SA, Olivier BJ, Goverse G, Vondenhoff MF, Greuter M, Beke P, et al. Chemokine CXCL13 is essential for lymph node initiation and is induced by retinoic acid and neuronal stimulation. Nat Immunol (2009) 10:1193–9. doi: 10.1038/NI.1789
81. De Agüero MG, Ganal-Vonarburg SC, Fuhrer T, Rupp S, Uchimura Y, Li H, et al. The maternal microbiota drives early postnatal innate immune development. Science (2016) 351:1296–302. doi: 10.1126/SCIENCE.AAD2571/SUPPL_FILE/DEAGUERO.SM.PDF
82. Qiu J, Heller JJ, Guo X, Chen ZME, Fish K, Fu YX, et al. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity (2012) 36:92–104. doi: 10.1016/j.immuni.2011.11.011
83. Meier D, Bornmann C, Chappaz S, Schmutz S, Otten LA, Ceredig R, et al. Ectopic lymphoid-organ development occurs through interleukin 7-mediated enhanced survival of lymphoid-tissue-inducer cells. Immunity (2007) 26:643–54. doi: 10.1016/J.IMMUNI.2007.04.009
84. Luther SA, Ansel KM, Cyster JG. Overlapping roles of CXCL13, interleukin 7 receptor alpha, and CCR7 ligands in lymph node development. J Exp Med (2003) 197:1191–8. doi: 10.1084/JEM.20021294
85. Cao X, Shores EW, Hu-Li J, Anver MR, Kelsail BL, Russell SM, et al. Defective lymphoid development in mice lacking expression of the common cytokine receptor gamma chain. Immunity (1995) 2:223–38. doi: 10.1016/1074-7613(95)90047-0
86. Adachi S, Yoshida H, Honda K, Maki K, Saijo K, Ikuta K, et al. Essential role of IL-7 receptor alpha in the formation of peyer’s patch anlage. Int Immunol (1998) 10:1–6. doi: 10.1093/INTIMM/10.1.1
87. Chappaz S, Finke D. The IL-7 signaling pathway regulates lymph node development independent of peripheral lymphocytes. J Immunol (2010) 184:3562–9. doi: 10.4049/jimmunol.0901647
88. Jones R, Cosway EJ, Willis C, White AJ, Jenkinson WE, Fehling HJ, et al. Dynamic changes in intrathymic ILC populations during murine neonatal development. Eur J Immunol (2018) 48:1481–91. doi: 10.1002/EJI.201847511
89. Elmentaite R, Kumasaka N, Roberts K, Fleming A, Dann E, King HW, et al. Cells of the human intestinal tract mapped across space and time. Nature (2021) 597:250–5. doi: 10.1038/s41586-021-03852-1
90. Marquardt N, Ivarsson MA, Sundström E, Åkesson E, Martini E, Eidsmo L, et al. Fetal CD103 + IL-17–producing group 3 innate lymphoid cells represent the dominant lymphocyte subset in human amniotic fluid. J Immunol (2016) 197:3069–75. doi: 10.4049/JIMMUNOL.1502204/-/DCSUPPLEMENTAL
91. Hoorweg K, Peters CP, Cornelissen F, Aparicio-Domingo P, Papazian N, Kazemier G, et al. Functional differences between human NKp44- and NKp44+ RORC+ innate lymphoid cells. Front Immunol (2012) 3:72. doi: 10.3389/FIMMU.2012.00072/BIBTEX
92. Forkel M, Berglin L, Kekäläinen E, Carlsson A, Svedin E, Michaëlsson J, et al. Composition and functionality of the intrahepatic innate lymphoid cell-compartment in human nonfibrotic and fibrotic livers. Eur J Immunol (2017) 47:1280–94. doi: 10.1002/EJI.201646890
93. Cupedo T, Crellin NK, Papazian N, Rombouts EJ, Weijer K, Grogan JL, et al. Human fetal lymphoid tissue-inducer cells are interleukin 17-producing precursors to RORC+ CD127+ natural killer-like cells. Nat Immunol (2009) 10:66–74. doi: 10.1038/ni.1668
94. Shikhagaie MM, Björklund ÅK, Mjösberg J, Erjefält JS, Cornelissen AS, Ros XR, et al. Neuropilin-1 is expressed on lymphoid tissue residing LTi-like group 3 innate lymphoid cells and associated with ectopic lymphoid aggregates. Cell Rep (2017) 18:1761–73. doi: 10.1016/j.celrep.2017.01.063
95. Robinette ML, Fuchs A, Cortez VS, Lee JS, Wang Y, Durum SK, et al. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat Immunol (2015) 16:306–17. doi: 10.1038/NI.3094
96. Gray J, Oehrle K, Worthen G, Alenghat T, Whitsett J, Deshmukh H, et al. Intestinal commensal bacteria mediate lung mucosal immunity and promote resistance of newborn mice to infection. Sci Transl Med (2017) 9. doi: 10.1126/scitranslmed.aaf9412
97. Chea S, Schmutz S, Berthault C, Perchet T, Petit M, Burlen-Defranoux O, et al. Single-cell gene expression analyses reveal heterogeneous responsiveness of fetal innate lymphoid progenitors to notch signaling. Cell Rep (2016). doi: 10.1016/j.celrep.2016.01.015
98. Yang Q, Li F, Harly C, Xing S, Ye L, Xia X, et al. TCF-1 upregulation identifies early innate lymphoid progenitors in the bone marrow. Nat Immunol (2015) 16:1044. doi: 10.1038/NI.3248
99. Harly C, Kenney D, Ren G, Lai B, Raabe T, Yang Q, et al. The transcription factor TCF-1 enforces commitment to the innate lymphoid cell lineage. Nat Immunol (2019) 20:1150–60. doi: 10.1038/s41590-019-0445-7
100. Seillet C, Mielke LA, Amann-Zalcenstein DB, Su S, Gao J, Almeida FF, et al. Deciphering the innate lymphoid cell transcriptional program. Cell Rep (2016) 17:436–47. doi: 10.1016/j.celrep.2016.09.025
101. Fathman JW, Bhattacharya D, Inlay MA, Seita J, Karsunky H, Weissman IL, et al. Identification of the earliest natural killer cell–committed progenitor in murine bone marrow. Blood (2011) 118:5439–47. doi: 10.1182/BLOOD-2011-04-348912
102. Carotta S, Pang SHM, Nutt SL, Belz GT. Identification of the earliest NK-cell precursor in the mouse BM. Blood (2011) 117:5449–52. doi: 10.1182/BLOOD-2010-11-318956
103. Hoyler T, Klose CSN, Souabni A, Turqueti-Neves A, Pfeifer D, Rawlins EL, et al. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity (2012) 37:634–48. doi: 10.1016/j.immuni.2012.06.020
104. Hernández DC, Juelke K, Müller NC, Durek P, Ugursu B, Mashreghi MF, et al. An in vitro platform supports generation of human innate lymphoid cells from CD34 + hematopoietic progenitors that recapitulate ex vivo identity. Immunity (2021) 54:2417–2432.e5. doi: 10.1016/J.IMMUNI.2021.07.019
Keywords: innate lymphoid cells (ILCs), progenitors cells, embryonic development, in situ differentiation, prenatal immunity
Citation: Hernández-Torres DC and Stehle C (2022) Embryonic ILC-poiesis across tissues. Front. Immunol. 13:1040624. doi: 10.3389/fimmu.2022.1040624
Received: 09 September 2022; Accepted: 02 December 2022;
Published: 20 December 2022.
Edited by:
Ana Stojanovic, Heidelberg University, GermanyReviewed by:
Christin Friedrich, University of Würzburg, GermanyCopyright © 2022 Hernández-Torres and Stehle. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniela Carolina Hernández-Torres, aGVybmFuZGV6QGRyZnouZGU=; Christina Stehle, Y2hyaXN0aW5hLnN0ZWhsZUBkcmZ6LmRl
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.