- 1Division of Pharmaceutical Sciences, School of Pharmacy, University of Wisconsin–Madison, Madison, WI, United States
- 2Pharmacology and Toxicology Program, School of Pharmacy, University of Wisconsin–Madison, Madison, WI, United States
Interleukin-1β (IL-1β) is a potent proinflammatory cytokine that plays a vital role in the innate immune system. To observe the innate immune response in vivo, several transgenic zebrafish lines have been developed to model IL-1β-induced inflammation and to visualize immune cell migration and proliferation in real time. However, our understanding of the IL-1β response in zebrafish is limited due to an incomplete genome annotation and a lack of functional data for the cytokine receptors involved in the inflammatory process. Here, we use a combination of database mining, genetic analyses, and functional assays to identify zebrafish Interleukin-1 receptor, type 1 (Il1r1). We identified putative zebrafish il1r1 candidate genes that encode proteins with predicted structures similar to human IL1R1. To examine functionality of these candidates, we designed highly effective morpholinos to disrupt gene expression in a zebrafish model of embryonic Il-1β-induced systemic inflammation. In this double transgenic model, ubb:Gal4-EcR, uas:il1βmat, the zebrafish ubiquitin b (ubb) promoter drives expression of the modified Gal4 transcription factor fused to the ecdysone receptor (EcR), which in turn drives the tightly-regulated expression and secretion of mature Il-1β only in the presence of the ecdysone analog tebufenozide (Teb). Application of Teb to ubb:Gal4-EcR, uas:il1βmat embryos causes premature death, fin degradation, substantial neutrophil expansion, and generation of reactive oxygen species (ROS). To rescue these deleterious phenotypes, we injected ubb:Gal4-EcR, uas:il1βmat embryos with putative il1r1 morpholinos and found that knockdown of only one candidate gene prevented the adverse effects caused by Il-1β. Mosaic knockout of il1r1 using the CRISPR/Cas9 system phenocopied these results. Taken together, our study identifies the functional zebrafish Il1r1 utilizing a genetic model of Il-1β-induced inflammation and provides valuable new insights to study inflammatory conditions specifically driven by Il-1β or related to Il1r1 function in zebrafish.
Introduction
Zebrafish (Danio rerio) have emerged as a valuable vertebrate model to study inflammation due to several important features including rapid development of the innate immune system, optical transparency for in vivo imaging, and the accessibility of genetic tools and experimental methods (1–3). The relative ease of transgenesis in zebrafish (4) has given rise to several genetic models of inflammation driven by the expression of the potent proinflammatory cytokine Interleukin-1β (Il-1β). These models include heat-shock-inducible Il-1β (5), cell-specific expression of Il-1β (6), and a doxycycline-inducible model (7). A variety of non-genetic zebrafish inflammation models have also been characterized, including infection models using bacteria (8–10), viruses (11), fungi (12, 13), and lipopolysaccharide (LPS) (14) as well as several wound-induced models (15–20). Similar to the genetic models, wound-induced models stimulate Il-1β expression and promote Il-1β dependent migration of neutrophils to the site of injury (5, 21). Additionally, morpholino knockdown of zebrafish Il-1β decreases the recruitment of neutrophils demonstrating that Il-1β plays a specific role in regulating neutrophil migration during injury-induced inflammation (5). More recently, we generated a genetically inducible model of systemic inflammation using the Gal4-EcR/UAS system (22). In this model, the mature form of Il-1β is secreted in response to the ecdysone analog tebufenozide resulting in dose-dependent neutrophil expansion, reactive oxygen species (ROS) generation, morbidity, and mortality (23).
During an innate immune response to foreign antigens, IL-1β is produced as an inactive precursor that is processed by caspase-1 and the inflammasome to produce mature IL-1β for secretion (24). Once secreted, IL-1β (as well as IL-1α) binds specifically to the Interleukin-1 receptor, type 1 (IL1R1) causing a conformational change that allows for binding of the co-receptor Interleukin-1 receptor accessory protein (IL1RAP; also known as IL1R3) that forms a trimeric complex that promotes a strong proinflammatory signal (25). To demonstrate functionality, previous studies showed that targeted disruption of the murine genes encoding either IL1R1 (26, 27) or IL1RAP (28) block IL-1 signaling, indicating that IL1R1 and IL1RAP are both required to elicit an immune response. While IL1R1 and IL1RAP share some structural similarities, both receptors possess unique primary sequences and distinct genomic locations in both human and mouse. As IL1R1 binds specifically to IL-1β and serves as a primary therapeutic target for treating inflammation in a broad spectrum of diseases, our current study focuses on the identification of the zebrafish Interleukin-1 receptor, type 1 (Il1r1).
Given the prominent role of Il-1β in the inflammatory response, it is particularly surprising that the zebrafish Il1r1 has not been identified. Indeed, the discovery and validation of functional orthologs in zebrafish can be challenging for a variety of reasons. While comparison of the human and zebrafish reference genomes reveals that approximately 70% of human genes have at least one obvious zebrafish ortholog, the sequence divergence of many genes is so great that they cannot be recognized or confirmed as orthologs without experimental validation (29). Furthermore, members of the Teleostei infraclass, including zebrafish, underwent a teleost-specific whole-genome duplication event (30). This further complicates the identification of human - zebrafish orthologs due to the possibility of 1) functionally redundant paralogs, 2) mutually exclusive, overlapping, or redundant expression patterns of the paralogs, or 3) the presence of pseudogenes.
In this study, we perform an in silico analysis and identify putative zebrafish Il1r1 orthologs with low sequence identities but with predicted protein structures highly similar to human IL1R1. To determine if these proteins function as the receptor for Il-1β, we utilize our validated in vivo zebrafish model of Il-1β-induced systemic inflammation (23). Here, we knockdown the expression of putative receptors and examine the phenotypic rescue of inflammation by performing multiple functional assays. Using this experimental approach, we identify the functional ortholog for zebrafish Il1r1 and demonstrate that it is absolutely required for the Il-1β-driven inflammatory response and the associated disease phenotypes. Given the increasing interest in zebrafish models of inflammation, our findings provide valuable new tools to dissect signaling pathways, cellular responses, and disease models that are specifically driven by or related to Il1r1 function.
Materials and methods
Zebrafish husbandry
Zebrafish lines were maintained using guidelines established in The Zebrafish Book (31). The AB wild-type (WT) strain was acquired from the Zebrafish International Resource Center. Transgenic lines Tg(ubb:IVS2GVEcR, cmcl2:EGFP), herein abbreviated as ubb:Gal4-EcR, and Tg(uas:GSP-Il1βmat, cmlc2:mCherry), herein abbreviated as uas:Il1βmat were previously generated in our lab (23). The Tg(mpx:mCherry)uwm7 transgenic line, herein abbreviated as mpx:mCherry, was a gift from Dr. Anna Huttenlocher (UW-Madison). Embryos were maintained at 28.5°C in egg water (0.03% Instant Ocean reconstituted in reverse osmosis water). For imaging, 0.003% phenylthiourea (PTU) was used to inhibit melanin production. All experiments were performed in accordance with the University of Wisconsin-Madison Institutional Animal Care and Use Committee.
Database mining for putative zebrafish interleukin-1 receptors
To identify putative zebrafish Interleukin-1 receptor(s), type 1 (Il1r1), we performed database searches for zebrafish homologs. The human IL1R1 protein sequence (accession number P14778) was used to search the zebrafish genome at Ensembl (assembly GRCz11) using the TBLASTN search tool. Sequence identities and phylogenetic analysis of protein sequences was performed using the Clustal V Method in MegAlign (DNASTAR). Predicted protein structures for human IL1R1 (https://alphafold.ebi.ac.uk/entry/P14778), zebrafish CABZ01054965 (https://alphafold.ebi.ac.uk/entry/E7FGC6), and zebrafish ZMP:0000000936 (https://alphafold.ebi.ac.uk/entry/E7F5V6) were identified using AlphaFold (32, 33).
Morpholino antisense oligonucleotide design and microinjection
Morpholino antisense oligonucleotides (MO) were designed against zebrafish cabz01054965 and zmp:0000000936 using the manufacturers recommendations (GeneTools). The cabz01054965 splice donor morpholino sequence 5’-TGTGCATCAGGGTTTACCTTTCGC-3’ was designed against the splice donor site (underlined) located between exon 4 and intron 5. The zmp:0000000936 splice donor morpholino sequence 5’-GTGATATGAAAGGCTCACCCTGCAC-3’ was designed against the splice donor site (underlined) located between exon 5 and intron 6. The zmp:0000000936 start site morpholino sequence 5’-ACCAATCGACCCATATCTACAGCCG-3’ was designed to span across the translation start site (underlined). For control morpholino injections, the standard GeneTools control oligo was used 5’-CCTCTTACCTCAGTTACAATTTATA-3’. Uninjected embryos were used as controls for some experiments as indicated.
Morpholinos were resuspended in dH2O at a stock concentration of 2 mM. For microinjection, morpholinos were diluted to 0.1-0.5 mM in dH2O containing phenol red (0.05%) as an injection tracer. Microinjection needles were fabricated from 1.2 mm thin wall glass capillaries (WPI; TW120F-4) using a Sutter Instrument Flaming/Brown Micropipette Puller (Model P-97). Approximately 2 nl were microinjected into the yolk of single-celled embryos from pair-wise crosses of the relevant genotypes. Damaged embryos identified with gross morphological defects as a result of microinjection were removed from the study prior to analyses. All morpholinos were found to be effective at a concentration of 0.25 mM (~2 ng) without any obvious off-target effects.
Generation of Il-1β-induced embryonic systemic inflammation
The transgenic lines ubb:Gal4-EcR and uas:Il1βmat were used to generate Il-1β-induced embryonic systemic inflammation in the presence of the ecdysone analog, tebufenozide (Teb), as previously described (23). Teb was prepared as a 1 mM stock solution in 100% DMSO. Adults carrying the transgenes were bred, embryos were injected with morpholino at the one-cell stage or left uninjected, and then treated at 1 or 2 days postfertilization (dpf) with 1 µM Teb or 0.1% DMSO (untreated controls). Embryos were selected for both transgenesis markers cmlc2:EGFP (green heart) and cmlc2:mCherry (red heart) using a Nikon SMZ18 epifluorescence stereomicroscope prior to further analyses. For neutrophil experiments, the ubb:Gal4-EcR and uas:Il1βmat lines were bred to mpx:mCherry. All experimental embryos were heterozygous for all transgenes.
Mosiac rescue of Il-1β-induced embryonic systemic inflammation with CRISPR/Cas9
To phenocopy the zmp splice-donor and start-site morpholino rescue of Il-1β-induced embryonic systemic inflammation, we used a CRISPR/Cas9 strategy utilizing the crRNA:tracrRNA duplex format with recombinant S. Pyogenes Cas9 nuclease (Cas9) from Integrated DNA Technologies (IDT). Using the Alt-R Custom Cas9 crRNA Design Tool (IDT), we designed two CRISPR RNAs (crRNAs): cr1.zmp:0000000936.ex8 5’-/AltR1/ucgacugcuggacaccagacguuuuagagcuaugcu/AltR2/-3’and cr2.zmp:0000000936.ex9 5’-/AltR1/uuaagguggagcuggucuuaguuuuagagcuaugcu/AltR2/-3’ against exon 8 and exon 9, respectively. For CRISPR-Cas9 ribonucleoprotein (RNP) preparation and microinjection, we followed the IDT demonstrated protocol “Zebrafish embryo microinjection” modified from Dr. Jeffrey Essner (Iowa State University). The crRNAs and transactivating crRNA (tracrRNA) were resuspended to 100 µM in nuclease-free TE buffer, pH 8.0, the individual crRNAs were combined with tracrRNA at 1:1 molar ratio in nuclease-free Duplex Buffer (IDT) to create 3 µM gRNA complexes (cr1 and cr2). The solutions were heated to 95°C for 5 min, then cooled to room temperature. Recombinant Cas9, glycerol-free (IDT) was diluted to 0.5 µg/µl in PBS, pH 7.4. The RNP complex was assembled by combining 3 µl of gRNA complexes with 3 µl of diluted Cas9, incubated at 37°C for 10 min, then cooled to room temperature. Approximately 2 nl of the RNP complexes with either cr1, cr2, or combined cr1/cr2 (1:1) was microinjected into ubb:Gal4-EcR, uas:Il1βmat single-cell embryos and monitored for morbidity and mortality. PCR was performed on individual embryos using zmp:0000000936-specific primers: forward primer 5’-tatgtgttcctcttgcagCG-3’ and reverse primer 5’-tgtttatacgagcacCTGTGG-3’ located in intron 7 and intron 9, respectively. Percentages of survival (alive), morbidity (sick), and mortality (dead) were plotted for each group using Excel (Microsoft). To test for differences between groups, a chi-square test of independence was performed, followed by a post hoc test using adjusted residuals and a p-value Bonferroni adjustment.
RNA extraction, cDNA synthesis, and RT-PCR
Embryos from 0-3 dpf (20-30 per group in duplicate) were anesthetized in 0.02% Tricaine, transferred into RNase/DNase-free 1.5 ml microcentrifuge tubes with fitted pestle (Kontes), homogenized in TRIzol, and total RNA was extracted according to the manufacturer’s protocol (Invitrogen). cDNA was synthesized by reverse transcription using the SuperScript IV First-Strand Synthesis System using Oligo(dT) primers according to the manufacturer’s protocol (Invitrogen). Reverse transcription polymerase chain reaction (RT-PCR) amplified a 578 bp fragment of cabz01054965 using forward primer 5’-ACGCACCTGACACATCGTAA-3’ and reverse primer 5’-GTTTGACTTGGCTTCGGGTA-3’, a 577 bp fragment of zmp:0000000936 using forward primer 5’-GCGAGATGACCTCAGAAACC-3’ and reverse primer 5’-TCCTCCGACACATGAGACAC-3’, and a 932 bp fragment of actin, beta 1 (actb1) as an RT-PCR control using forward primer 5’-CCCTCCATTGTTGGACGAC-3’ and reverse primer 5’-CCGATCCAGACGGAGTATTTG -3’. All primers were designed using Primer3 (34).
Whole-mount in situ hybridization
WISH was performed using protocols adapted from Vauti et al. and Thisse and Thisse (35, 36). This new protocol afforded greater probe penetration into the central nervous system (CNS) and trunk of 1-3 dpf animals used for this study. Briefly, WT embryos from 1-3 dpf were fixed in 4% paraformaldehyde/PBS at 4°C overnight, dehydrated in 100% methanol, then stored at −20°C. For probe synthesis, cabz01054965 (forward primer 5’-AATTAACCCTCACTAAAGGGGCGAGATGACCTCAGAAACC-3’; reverse primer 5’-TAATACGACTCACTATAGGGACCTCCTCCTCCTCTTCCAG-3’) and zmp:0000000936 (forward primer 5’-AATTAACCCTCACTAAAGGGGCGAGATGACCTCAGAAACC-3’; reverse primer 5’-TAATACGACTCACTATAGGGACCTCCTCCTCCTCTTCCAG-3’) were PCR amplified from WT zebrafish cDNA to produce products of 1,227 and 1,293 bp, respectively. Both forward primers contained a 5’-T3 RNA polymerase binding site (italics) and both reverse primers contained a 5’-T7 RNA polymerase binding site (italics). PCR products were then purified using the QIAquick PCR purification kit (Qiagen). Approximately 1 µg of purified PCR product was used to synthesize sense and antisense digoxigenin (DIG)-labeled RNA probes with T3 and T7 RNA polymerase, respectively, using a DIG RNA Labeling Kit (Roche). Prepared embryos (n=8 per group) were transitioned to ethanol, treated with ethanol/xylol (1:1 vol/vol), rehydrated in H2O with 0.1% Tween, permeabilized in 80% Acetone, and bleached in H2O2, according to Vauti et al., 2020 (36). Embryos were prehybridized at 70°C for 2 hr, then hybridized with DIG-labeled RNA probes at 70°C overnight. After washing and blocking, the embryos were incubated with anti-DIG-AP Fab fragment (Roche) at 4°C overnight then stained with BM Purple, AP precipitating substrate (Roche), containing NBT and BCIP, until the desired signal intensity appeared (35). Images were captured using a Nikon SMZ18 stereomicroscope equipped with a Nikon DS-Fi2 color camera and Nikon NIS-Elements software.
Survival analysis
To examine Il-1β-induced mortality, double transgenic ubb:Gal4-EcR, uas:Il1βmat embryos were induced with 1 µM Teb at 1 dpf and survival was monitored until 6 dpf. All survival assays were performed in 100x15 mm petri dishes (Falcon). In our previous survival studies, Teb (concentration range: 10 nM-10 µM) was added at 2 dpf, which caused mortality beginning at 5 dpf (23). Therefore, in the current study, we added Teb at 1 dpf to expedite mortality as morpholino effectiveness may be reduced past 3 dpf (37). Survival was tallied daily as dead embryos were identified and removed from the petri dishes. Teb was replaced daily with freshly prepared solution. Kaplan-Meier curves were made using Excel (Microsoft) and log rank tests were used to evaluate statistical significance.
Gross morphology
Transgenic ubb:Gal4-EcR and uas:Il1βmat embryos (10-20 per group) were induced with 1 µM Teb at 2 dpf and maintained in fresh Teb until 4 dpf. Three embryos per group (except for cabz +Teb; n=2), were randomly selected and imaged. Lateral images were acquired by confocal microscopy using the TD channel at 4 dpf. Regions of interest were quantified using FIJI ROI selection and measurement tools. Mean cross sectional area + standard deviation was plotted using Excel (Microsoft).
Neutrophil quantification
The neutrophil reporter line mpx:mCherry was bred to ubb:Gal4-EcR, uas:Il1βmat double transgenics to produce triple heterozygous transgenic embryos. Systemic inflammation was induced at 2 dpf with 1 µM Teb (10-20 per group). Three embryos per group (except for cabz +Teb; n=2), were randomly selected and neutrophils were imaged by confocal microscopy at 4 dpf. Neutrophils were quantified by manually counting the number of mCherry-positive cells throughout the entire volume rendered image using FIJI Cell Counter Plugin (38). Mean + standard deviation was plotted using Excel (Microsoft).
Reactive oxygen species analysis
Transgenic ubb:Gal4-EcR, uas:Il1βmat embryos (10-20 per group) were induced with 1 µM Teb at 2 dpf and maintained in Teb until 3 dpf. CM-H2DCFDA (Invitrogen), a fluorescent cell-permeant indicator for ROS, was freshly prepared as a 10 mM stock in DMSO. Embryos were incubated with 2.5 μM CM-H2DCFDA (Invitrogen) for 30 min in the dark, then washed in egg water (3 x 10 minutes each wash) prior to imaging. Three embryos per group (except for cabz +Teb; n=2), were randomly selected and imaged by confocal microscopy at 3 dpf. Quantification of fluorescent signal was completed on 2D projections of 3D confocal z-stacks, created using the Nikon NIS-Elements Maximum Intensity Projection algorithm. Regions of interest were identified using FIJI ROI selection tools, then the fluorescence was quantified using the FIJI ‘mean grey value’ measurement tool. Values were normalized to uninjected no Teb controls and normalized mean + standard deviation was plotted using Excel (Microsoft).
Confocal laser scanning microscopy
Zebrafish from 1 to 4 dpf were anesthetized in 0.02% Tricaine and immobilized in 1.2% low melting point agarose (Invitrogen) in glass bottom culture dishes (MatTek). Confocal microscopy was performed using a Nikon Eclipse Ti microscope equipped with a Nikon A1R. For images of whole embryos and larvae, large images (4 x 1 mm) were captured and stitched together with a 15% overlap. All images are 2D projections of 3D confocal z-stacks using the Nikon NIS-Elements Maximum Intensity Projection algorithm or are single frame lateral TD images. All image manipulation for brightness or contrast (via Nikon NIS-Elements software) was applied to all pixels, equally, and does not affect interpretation of data.
Statistical analysis
Statistical differences of mean values among multiple groups were determined using one-way analysis of variance (ANOVA) followed by Tukey’s HSD post-hoc test. The criterion for statistical significance was set at P < 0.05. Values represent means ± standard deviation. Error bars show +1 standard deviation.
Results
Identification of the putative zebrafish interleukin-1 receptor, type 1
Using the human IL1R1 protein sequence (accession number: P14778), we performed a TBLASTN against the zebrafish genome at Ensembl. We identified three genes, cabz01054965, (accession number: ENSDARG00000090844), zmp:0000000936 (accession number: ENSDARG00000088672), and cu855885, (accession number: ENSDARG00000101527) on zebrafish chromosome (chr.) 9 with sequence similarities to human IL1R1. In addition, these three genes showed partial synteny to the IL-1 Receptor Cluster on human chr. 2q (39) and mouse chr. 1 (Figure 1A). The cu855885 gene has previously been identified as the zebrafish ortholog for human Interleukin-1 receptor-like 1 (IL1RL1) (40), herein referred to as zebrafish il1rl1. Protein sequence alignment using the Clustal V Method in MegAlign (DNASTAR) indicated percent identities to the human IL1R1 protein sequence of 19.6%, 21.4%, and 19.1% for the proteins encoded by the zebrafish genes cabz01054965, zmp:0000000936, and il1rl1, respectively.
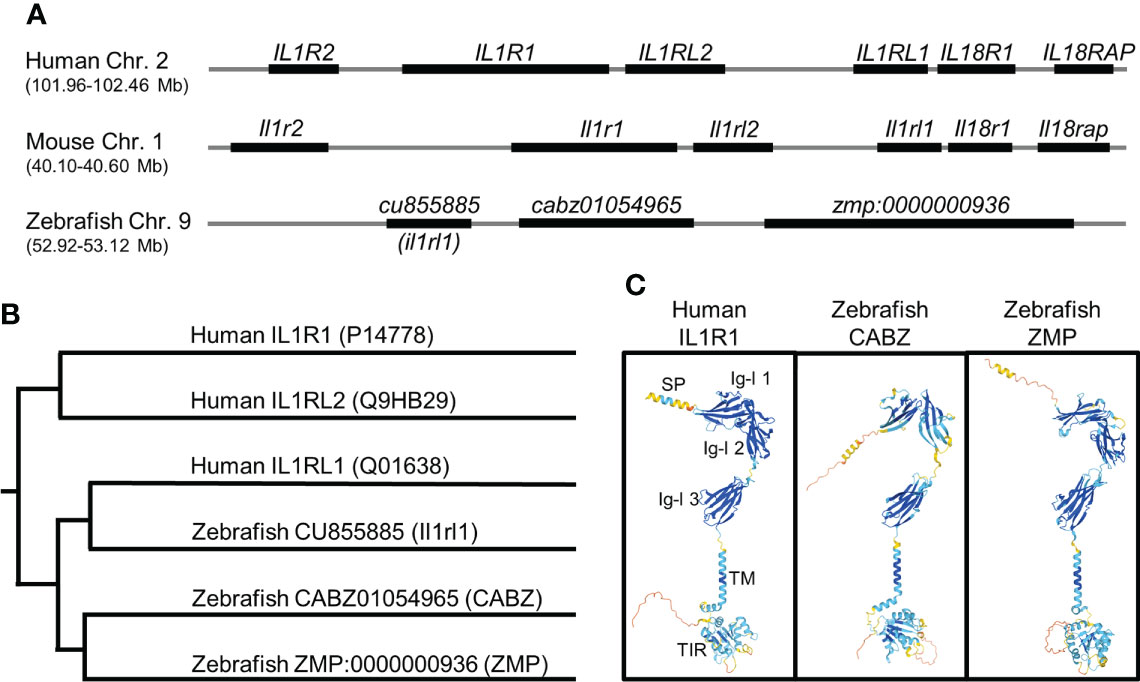
Figure 1 In silico identification of the putative zebrafish interleukin-1 receptor, type 1 (Il1r1). (A) Alignment of human, mouse, and zebrafish genomic regions showing conserved synteny. Shown here are the human and mouse IL-1 receptor clusters and the putative zebrafish IL-1 receptor cluster (not to scale). (B) Phylogenetic alignment of human IL1R protein sequences with putative zebrafish Il1r protein sequences. (C) Predicted protein structures of human IL1R1, zebrafish CABZ (CABZ01054965), and zebrafish ZMP (ZMP:0000000936) showing conserved domains: N-terminal signal peptide (SP), 3 extracellular immunoglobulin-like (Ig-l) domains (Ig-l 1, Ig-l 2, and Ig-l 3), a single transmembrane domain (TM), and an intracellular Toll/Interleukin-1 receptor (TIR).
Phylogenetic analysis of human IL1R1 (P14778), human IL1RL1 (Q01638), human IL1RL2 (Q9HB29), zebrafish CABZ01054965, zebrafish ZMP:0000000936, and zebrafish Il1rl1 was also performed using the Clustal V Method in MegAlign (Figure 1B). As predicted, zebrafish Il1rl1 was found to be most similar to human IL1RL1, confirming the previously established annotation (40). However, our analysis was unable to definitively distinguish whether CABZ01054965 or ZMP:0000000936 were functional zebrafish orthologs or paralogs for either human IL1R1 or IL1RL2.
We next examined the structural similarities between human IL1R1, zebrafish CABZ01054965 (abbreviated herein as CABZ), and ZMP:0000000936 (abbreviated herein as ZMP) using the AlphaFold Protein Structure Database (32, 33). As shown in Figure 1C, human IL1R1 consists of an N-terminal signal peptide (SP), 3 extracellular immunoglobulin-like (Ig-l) domains, a single transmembrane domain (TM), and an intracellular Toll/Interleukin-1 receptor (TIR) domain (41–43). Despite the low sequence similarities between the human and zebrafish proteins, AlphaFold predicted conserved protein structures and domains for human IL1R1, zebrafish CABZ, and zebrafish ZMP (Figure 1C). However, these structures did not distinguish functionality between the two zebrafish proteins.
We also examined the zebrafish genome for IL1RAP and the three additional IL1R genes found within the human and mouse IL-1 receptor clusters (i.e. IL1R2, IL18R1, and IL18RAP). As IL-1 signaling requires the association of IL1RAP with the IL-1β/IL1R1 complex, we mined the zebrafish genome for the gene encoding the zebrafish ortholog for Il1rap. BLAST analysis using human IL1RAP localized the putative zebrafish il1rap gene (cabz01068246) to chr. 15 near the zebrafish fgf12b gene (data not shown). This genomic region in zebrafish shares conserved synteny with both the human and mouse genomes, which show colocalization of the IL1RAP and FGF12 genes. In addition, BLAST analysis using human IL1R2 localized the putative zebrafish il1r2 (cabz01078737) near the proximal telomere of chr. 9 adjacent to zebrafish map4K4 (data not shown). This genomic region shows conserved synteny between il1r2 and map4K4 in human, mouse, and zebrafish, albeit at a distant genomic region in the zebrafish genome, with respect to the zebrafish IL-1 receptor cluster. No obvious zebrafish orthologs for human or mouse IL18R1 or IL18RAP were identified using BLAST analysis. However, the previously annotated zebrafish gene products Il1rapl1a, Il1rapl1b, and Il1rapl2 (44) share partial sequence alignment to human and mouse IL18R1 and IL18RAP, indicating the potential for conserved function.
Developmental expression of putative zebrafish interleukin-1 receptors
To examine the developmental expression of the zebrafish cabz and zmp transcripts, we performed reverse-transcription polymerase chain reaction (RT-PCR) and whole-mount in situ hybridization (WISH). No antibodies against the zebrafish proteins are currently available for protein expression analysis, and no expression data for either zebrafish transcript is currently available. For RT-PCR, we extracted total RNA from embryos at 0, 1, 2, and 3 days postfertilization (dpf), synthesized cDNA, and PCR amplified using sequence-specific DNA primers (see Materials and Methods). As shown in Figure 2A, the cabz transcript was not detected maternally, whereas the zmp transcript demonstrated maternally derived expression at 0 dpf. Both the cabz and zmp transcripts showed robust expression from 1-3 dpf, indicating that both transcripts are developmentally expressed (Figure 2A).
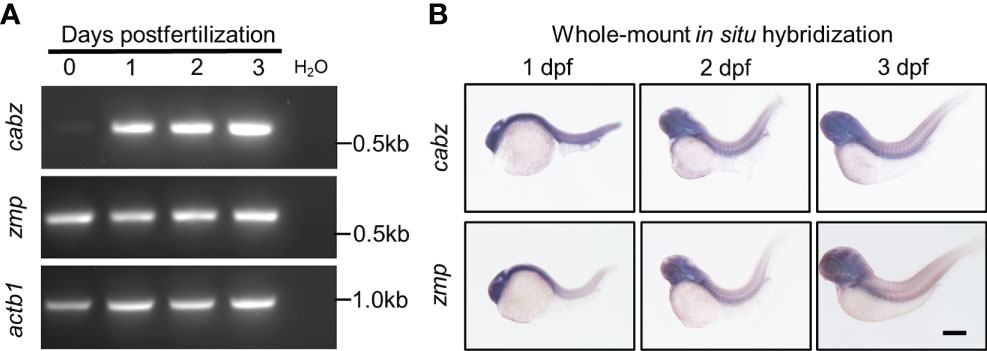
Figure 2 Developmental expression of putative zebrafish Il1r1. (A) RT-PCR from WT embryos at 0-3 dpf showed expression of cabz beginning at 1 dpf, maternal expression of zmp at 0 dpf, and actin beta 1 (actb1) transcript as an experimental control. Molecular weight markers are shown on the right. (B) Whole-mount in situ hybridization using antisense DIG-labeled riboprobes against cabz or zmp transcripts showed unrestricted expression of both transcripts at 1, 2, and 3 dpf. Scale bar in (B) is 200 µm.
To examine the spatiotemporal expression of the zebrafish cabz and zmp transcripts, we performed WISH. Using wild-type (WT) embryos at 1, 2, and 3 dpf, we analyzed transcript expression using antisense and sense digoxigenin (DIG)-labeled RNA probes. As expected, sense probes did not detect any expression (data not shown). We found that the cabz and zmp antisense probes exhibited unrestricted expression of both transcripts at all three developmental stages examined (Figure 2B). The unrestricted expression of these zebrafish il1r genes is consistent with low tissue specificity of the human IL1R1 transcript as demonstrated by RNAseq, microarray, and SAGE analysis (see https://www.proteinatlas.org/ENSG00000115594-IL1R1/tissue and https://www.genecards.org/cgi-bin/carddisp.pl?gene=IL1R1#expression).
Morpholino knockdown of the putative zebrafish interleukin-1 receptors
Based upon sequence homology and transcript expression results, we were unable to conclusively identify the functional homolog(s) for zebrafish Il1r1. Therefore, we designed morpholino oligonucleotides (MO) against splice donor sites to functionally knockdown the expression of both CABZ and ZMP by disrupting pre-mRNA splicing (45). Morpholinos were injected into WT embryos at the single-cell stage. The resulting morphants were raised to 2 dpf, total RNA was extracted, then morphant transcripts were analyzed by RT-PCR using DNA primers designed to span the splice donor sites of cabz and zmp, or the actin beta 1 (actb1) transcript as a control. We found that injection of the cabz splice donor morpholino resulted in the loss of the WT transcript (578 bp) and the formation of a single alternatively spliced transcript of approximately 400 bp (Figure 3A; top panel). In addition, we found that injection of the zmp splice donor morpholino resulted in the loss of the WT transcript (577 bp) and the formation of three aberrantly spliced transcripts of approximately 2,100, 570, and 400 bp (Figure 3A; middle panel). As expected, the standard control morpholino (Con.) supplied by GeneTools showed no effect on the zmp, cabz, or actb1 transcripts. To further examine the splicing effects of both morpholinos, we gel purified and sequenced the aberrantly spliced transcripts. For the cabz morphants, we found that the ~400 bp band was due to exclusion of exon 4 (-ex4), which was predicted to result in a non-functional product (Figures 3A, B). For the zmp morphants, we found that the ~2,100 bp band was due to the inclusion of intron 5 (+in5), the ~570 bp band was due to a cryptic splicing site (css) in exon 5 twelve bases upstream of the authentic splice-donor site, and the ~400 bp band was due to exclusion of exon 5 (-ex5) (Figures 3A, C). All three alternatively spliced zmp morphant transcripts were predicted to result in non-functional products. Importantly, the ~570 bp band from the zmp splice-site morphants is slightly smaller than the WT band and no WT transcript was detected in the zmp splice-site morphants using DNA sequence analysis. In fact, no WT transcripts were detected in either of the cabz or zmp morphants at the developmental stages examined. Thus, both the cabz and zmp morpholinos provide valuable genetic tools that disrupt the expression of these transcripts.
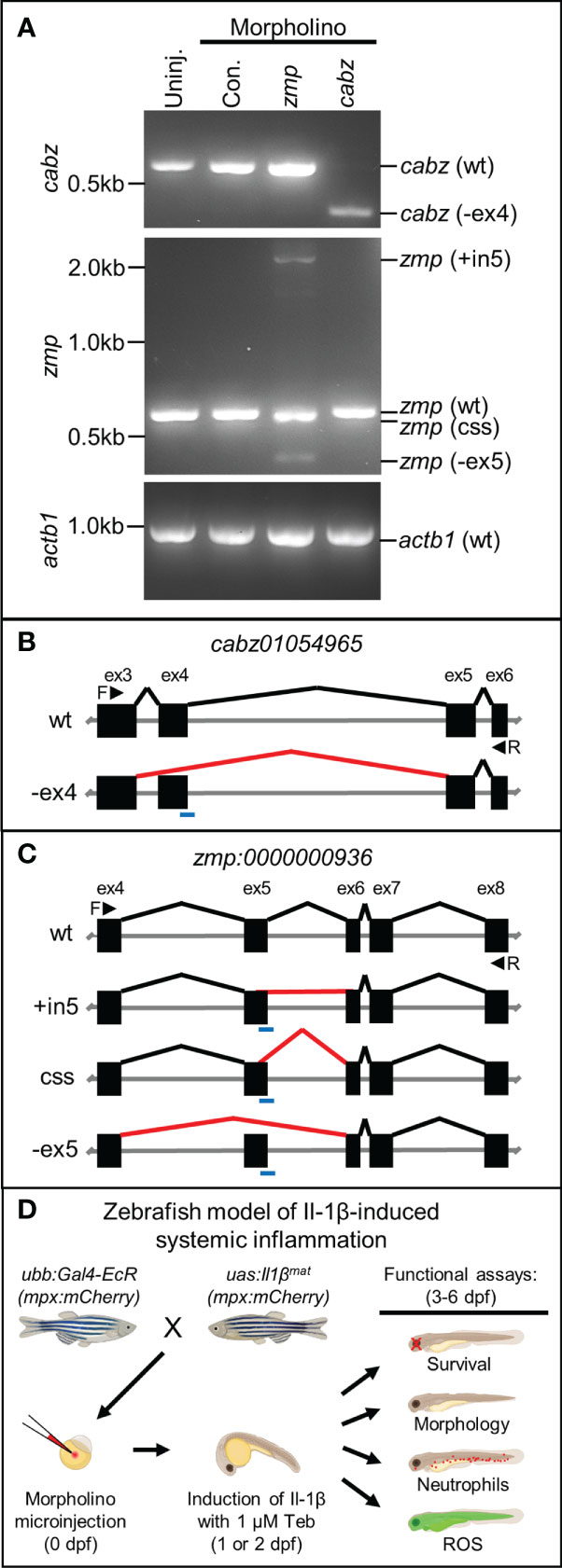
Figure 3 Morpholino knockdown of putative zebrafish Il1r1. (A) RT-PCR of cabz, zmp, and actb1 transcripts from uninjected embryos, control morpholino injected embryos, and cabz and zmp morphants. (B) Schematic of the cabz01054965 (cabz) gene showing exons 3-6, forward (F) and reverse (R) primers used for analysis (black arrowheads), the location of the splice donor site morpholino (blue line), normal splicing of wt (black lines), and aberrant splicing with the cabz morpholino (red line) that excludes exon 4 (-ex4). (C) Schematic of the zmp:0000000936 (zmp) gene showing exons 4-8, forward (F) and reverse (R) primers used for analysis (black arrowheads), the location of the splice donor site morpholino (blue line), normal splicing of wt (black lines), and aberrant splicing with the zmp morpholino (red lines) that includes intron 5 (+in5), introduce a cryptic splice sites (css), or excludes exon 5 (-ex5). (D) Diagram of the experimental paradigm used to functionally identify zebrafish Il1r1. A zebrafish model of Il-1β-induced systemic inflammation was generated by breeding transgenic lines ubb:Gal4-EcR and uas:Ilβmat with or without the neutrophil reporter line mpx:mCherry. Single-celled embryos were injected with morpholino, inflammation was induced with 1 µM Tebufenozide (Teb) at 1 or 2 dpf, and functional analyses were performed from 3-6 dpf to assess morpholino rescue of Il-1β-induced inflammation.
To test the functional consequences of the cabz and zmp morpholinos, we utilized our transgenic zebrafish model of Il-1β-induced systemic inflammation (23). We previously generated two transgenic zebrafish lines Tg(ubb:IVS2GVEcR, cmcl2:EGFP), herein abbreviated as ubb:Gal4-EcR, and Tg(uas:GSP-Il1βmat, cmlc2:mCherry), herein abbreviated as uas:Il1βmat. This model uses the Gal4-EcR/UAS system (22), where mature Il-1β (Il1βmat) is ubiquitously secreted only in the presence of the ecdysone analog, Tebufenozide (Teb). In order to identify the functional Il1r1 in zebrafish, we used our model to examine 1) survival, 2) morphology, 3) neutrophils (mpx:mCherry), and 4) reactive oxygen species (ROS) in the putative Il1r1 morphants (Figure 3D).
Survival analysis of Il-1β-induced mortality
Our previous studies established Il-1β-induced mortality in double transgenic ubb:Gal4-EcR, uas:Il1βmat larvae in the presence of Teb in a dose-dependent manner (23). To accelerate mortality in our current study, we modified the original experimental paradigm by generating inflammation at 1 dpf, which coincides with the onset of innate immunity in zebrafish (10, 46, 47). Here, double transgenic ubb:Gal4-EcR, uas:Il1βmat embryos were 1) injected with control, cabz, or zmp morpholinos at the single-cell stage or left uninjected, 2) treated with 1 µM Teb or no Teb (0.1% DMSO) at 1 dpf, and then 3) observed daily for survival up to 6 dpf. As shown in Figure 4A, all untreated ubb:Gal4-EcR, uas:Il1βmat controls (ubb/uas Uninj No Teb) and all wild type controls (WT Uninj No Teb and WT Uninj + 1 µM Teb) survived to 6 dpf and beyond. Moreover, none of the morpholino injections or Teb treatments significantly reduced survival in WT embryos at the concentrations used (Supplemental Figure 1). In contrast, when ubb:Gal4-EcR, uas:Il1βmat embryos were treated with 1 µM Teb, the majority of uninjected controls (ubb/uas Uninj + 1 µM Teb; n=32) and control morphants (ubb/uas Con MO + 1 µM Teb; n=36) died by 3 dpf (Figure 4A). As control MO showed no effect on morbidity and mortality, the remainder of controlled experiments were conducted with uninjected embryos only. These results demonstrate that Teb-treated ubb:Gal4-EcR, uas:Il1βmat embryos are extremely susceptible to mortality and that this experimental paradigm provides a high level of reproducibility.
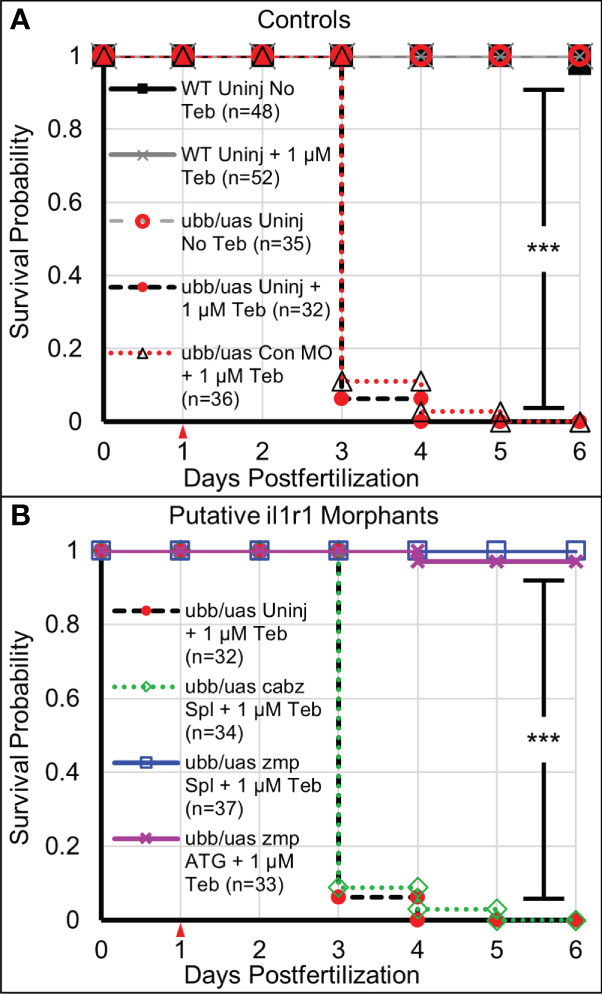
Figure 4 Survival analysis of Il-1β-induced mortality. (A) Kaplan-Meier representations of the survival of WT larvae left untreated (WT Uninj No Teb), or Teb treated (WT Uninj + 1 µM Teb), ubb:Gal4-EcR, uas:Ilβmat larvae with no Teb treatment (ubb/uas Uninj No Teb), with Teb (ubb/uas Uninj + 1 µM Teb), or injected with control morpholino with Teb (ubb/uas Con MO + 1 µM Teb). (B) Kaplan-Meier representations of the survival of ubb:Gal4-EcR, uas:Ilβmat larvae left uninjected with teb treatment (ubb/uas Uninj + 1 µM Teb) injected with the cabz splice donor morpholino with Teb (ubb/uas cabz Spl + 1 µM Teb), zmp splice donor morpholino with Teb (ubb/uas zmp Spl + 1 µ M Teb), or zmp start site morpholino with Teb (ubb/uas zmp ATG + 1 µM Teb). Teb was added to the embryos at 1 dpf (red arrowhead). Note that only the zmp morpholinos rescued Il-1β-induced death. ***p<0.001.
Concurrently, we monitored survival of the putative il1r1 morphants. We reasoned that if Il-1β-induced mortality is mediated by the activation of zebrafish Il1r1, then morpholino knockdown of the appropriate receptor(s) would reduce death and increase survival. As shown in Figure 4B, we found that the cabz splice-site morphants treated with 1 µM Teb (ubb/uas cabz Spl + 1 µM Teb; n=34) died at a rate comparable to the uninjected and control MO injected Teb treated embryos, indicating that Il-1β-induced death is not mediated through zebrafish CABZ. In contrast, the zmp splice-site morphants treated with 1 µM Teb (ubb/uas zmp Spl + 1 µM Teb; n=37) survived equally to No Teb embryos (Figures 4A, B). To confirm the effects of the zmp splice-site morpholino, we designed and injected a zmp start-site morpholino (zmp ATG) to block translation of the zmp transcript (48). As with the zmp splice morphants, the zmp start morphants (ubb/uas zmp ATG + 1 µM Teb; n=33) survived at a frequency comparable to No Teb embryos (Figures 4A, B).
To further verify and phenocopy the zmp morpholino rescue of Il-1β-induced morbidity and mortality, we microinjected CRISPR-Cas9 ribonucleoprotein (RNP) complexes targeting the zmp gene into ubb:Gal4-EcR, uas:Il1βmat embryos to generate mosaic knockouts. Two RNPs, cr1 and cr2, targeting exon 8 and exon 9, respectively, were injected independently or in combination at 1:1 molar ratio (Supplemental Figure 2A) then treated with 1 µM Teb (+ Teb) or untreated (No Teb). As shown in Supplemental Figure 2B, uninjected control embryos (Con.) treated with Teb at 1 dpf showed ~80% dead, ~15% sick (fin degradation and general morbidity), and ~5% alive by 4 dpf. In contrast, the RNP-injected embryos showed a dramatic increase in survival with ~85% of cr1/cr2, ~94% of cr1, and ~50% of cr2 alive at 4 dpf. Injection of RNPs had no effect on survival in the absence of Teb treatment. As cr1/cr2 injection could result in a deletion mutation, we analyzed crispants by PCR. We found that the controls, cr1, and cr2 produced a predicted 508 bp band, whereas the cr1/cr2 combination resulted in the 508 bp band as well as a smaller ~160 bp band in three of the four samples shown (Supplemental Figure 2C). As the protospacer adjacent motif (PAM) site of cr1 and cr2 are ~350 bp apart, this smaller band likely represents a genomic deletion between cr1 and cr2. We also conclude that the cr1 and cr2 crispants must cause in a small inactivating insertion or deletion (indel) in il1r1 as both rescued Il-1β-induced mortality.
Our data demonstrate that knockdown of zebrafish ZMP function by disrupting pre-mRNA splicing, blocking translation, or mosaic CRISPR/Cas9 knockout rescues Il-1β-dependent mortality. Since both zmp splice-site and zmp start-site morpholinos equally rescued death and worked as effectively as mosaic knockout with CRISPR/Cas9, we used the splice-site morpholinos for the remainder of experiments described below.
Rescue of Il-1β-induced gross morphological defects
Double transgenic ubb:Gal4-EcR, uas:Il1βmat embryos were 1) uninjected, injected with cabz morpholino (cabz), or injected with zmp morpholino (zmp), 2) untreated (No Teb) or treated with 1 µM Teb at 2 dpf, and then 3) imaged for gross morphology at 4 dpf. We previously demonstrated that Il-1β-induced systemic inflammation results in fin degradation and general morbidity prior to death (23). As shown in Figure 5A, untreated larvae (No Teb) maintained normal morphology, whereas Teb-treated larvae (+Teb) showed extensive fin degradation and an overall unhealthy phenotype (Figure 5A; top panels). Untreated cabz morphants also displayed normal fin morphology, whereas Teb-treated embryos showed extensive fin degradation similar to the uninjected controls (Figure 5A; middle panels). In contrast, zmp morphants displayed normal fin morphology with and without Teb treatment, indicating rescue of the inflammation phenotype (Figure 5A; bottom panels). Furthermore, quantification of fin degradation revealed that the zmp morpholino, but not the cabz morpholino, prevented Il-1β-dependent morbidity in our model. We found that the total fin area was significantly decreased with Teb treatment in the uninjected and the cabz morphants, whereas the zmp morphants maintained normal fin area similar to untreated larvae (Figure 5B). We also measured the effects on total body area between the different groups. We found that Teb-induced larvae had a smaller cross-sectional area compared to untreated controls (Figure 5C). However, all cross-sectional area differences were a consequence of fin degradation only (Figure 5D). No other gross morphological differences were observed between the groups. In addition, Teb-treatment (even up to 10 µM Teb) showed no effect on morphology in WT embryos without the ubb:Gal4-EcR, uas:Il1βmat transgenes (Supplemental Figure 3).
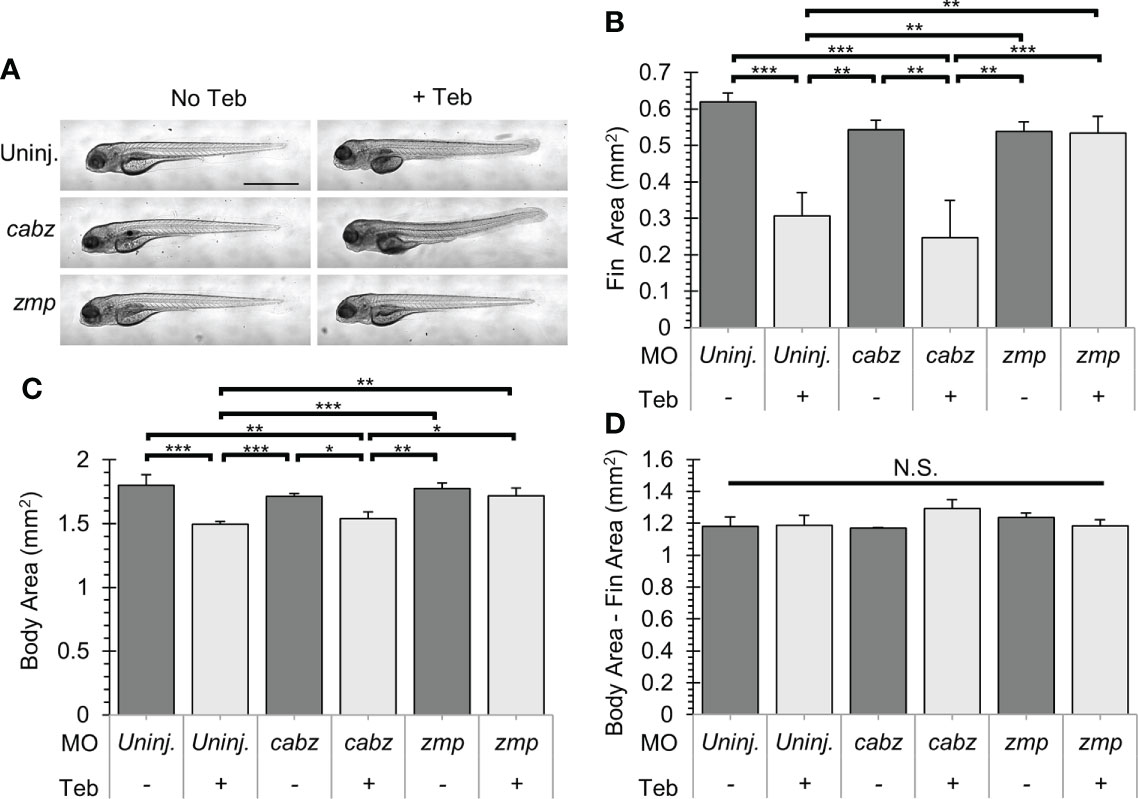
Figure 5 Gross morphological analysis of Il-1β-induced morbidity. (A) Representative images of ubb:Gal4-EcR, uas:Ilβmat larvae showing gross morphology. Shown here are uninjected larvae, cabz morphants, and zmp morphants without Teb (No Teb) and with Teb treatment (+Teb). Note that zmp morphants +Teb showed normal morphology with no gross defects. (B–D) Mean Cross-Sectional Fin Area (B), Body Area (C), and the product of Body Area – Fin Area (D) was plotted in mm2 for each group. Error Bars are +1 standard deviation. Asterisks indicate significant differences *p < 0.05 **p < 0.01 ***p < 0.001; N.S., not significant; by one-way ANOVA followed by Tukey’s HSD Test. Scale bar in (A) is 1 mm.
Inhibition of Il-1β-induced neutrophil expansion
Neutrophils are innate immune cells responsive to proinflammatory cytokines such at Il-1β. In zebrafish, neutrophils become functional during early development (10, 46), after which they can expand and migrate in response to inflammatory events such as tissue damage or microbial infection (18, 49). Here, we investigated whether our putative il1r1 morpholinos were able to block Il-1β-dependent neutrophil activity. Triple transgenic ubb:Gal4-EcR, uas:Il1βmat, mpx:mCherry embryos were 1) uninjected, injected with cabz morpholino (cabz), or injected with zmp morpholino (zmp), 2) untreated (No Teb) or treated with 1 µM Teb at 2 dpf, and then 3) imaged and quantified at 4 dpf. As shown in Figure 6A, untreated larvae (No Teb), showed a stereotypical distribution of neutrophils primarily located in the caudal hematopoietic tissue (CHT) with few cells dispersed throughout the larvae (left panels). When treated with Teb (+Teb), uninjected larvae and cabz morphants showed an extensive expansion of neutrophils within the CHT and throughout the larvae, whereas zmp morphants were phenotypically similar to untreated (No Teb) larvae. Teb showed no effects on neutrophil numbers or distribution in mpx:mCherry embryos (Supplemental Figure 4). To quantify these results, we counted total neutrophils in the whole larvae (Figure 6B). In uninjected larvae and cabz morphants, Teb treatment caused a significant increase in the total number of neutrophils. In contrast, the Teb-treated zmp morphants showed no significant neutrophil expansion, indicating complete phenotypic rescue by the zmp morpholino.
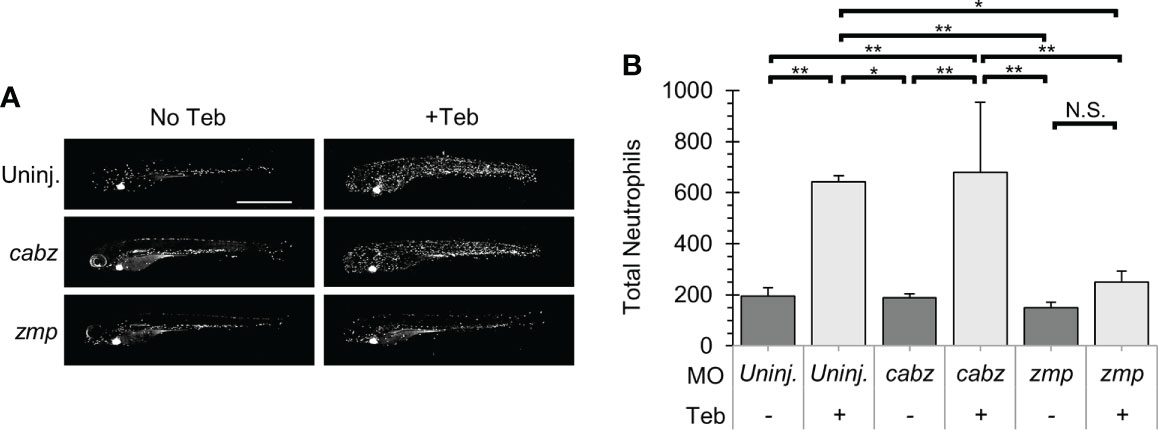
Figure 6 Il-1β-induced neutrophil recruitment. (A) Representative confocal images of ubb:Gal4-EcR, uas:Ilβmat, mpx:mCherry larvae. Shown here are uninjected larvae, cabz morphants, and zmp morphants without Teb (No Teb) and with Teb treatment (+Teb). (B) Neutrophils were counted using FIJI Cell Counter and Total Neutrophil counts were plotted for each group. Error Bars are +1 standard deviation. Asterisks indicate significant differences *p < 0.05 **p < 0.01; N.S., not significant; by one-way ANOVA followed by Tukey’s HSD Test. Scale bar in (A) is 1 mm.
Prevention of Il-1β-induced reactive oxygen species
ROS are central to the progression of many inflammatory diseases and are produced by cells, such as neutrophils, that are involved in the host-defense response to various cytokines (50). For this study, we examined whether our putative il1r1 morpholinos were capable of blocking Il-1β-dependent ROS production. Double transgenic ubb:Gal4-EcR, uas:Il1βmat embryos were 1) uninjected, injected with cabz morpholino (cabz), or injected with zmp morpholino (zmp), 2) untreated (No Teb) or treated with 1 µM Teb at 2 dpf, 3) exposed to CM-H2DCFDA, a fluorescent cell-permeant indicator for ROS, at 4 dpf, and 4) imaged and quantified for relative fluorescence. Here, we show an overlay of brightfield and fluorescent images (Figure 7A; left panels) and the area of fluorescence quantified (Figure 7A; right panels, white boundaries). As untreated larvae showed fluorescence in the yolk, gut, and heart from autofluorescence, CM-H2DCFDA cleavage, and cmlc2:EGFP, respectively, we excluded these regions from quantification. To quantify the signal, we measured the fluorescence within the white boundaries using FIJI Mean Gray Value Measurement as described in Methods. As shown in Figure 7B, addition of Teb caused a significant increase in the fluorescent signal in the uninjected controls, demonstrating Il-1β-dependent ROS production. Similarly, the cabz morphants showed a dramatic increase in ROS, indicating that the cabz morpholino did not block this process. In contrast, the zmp morphants showed almost no fluorescent signal, equivalent to the uninjected larvae without Teb. These data indicate that the zmp morpholino blocks Il-1β-dependent ROS production.
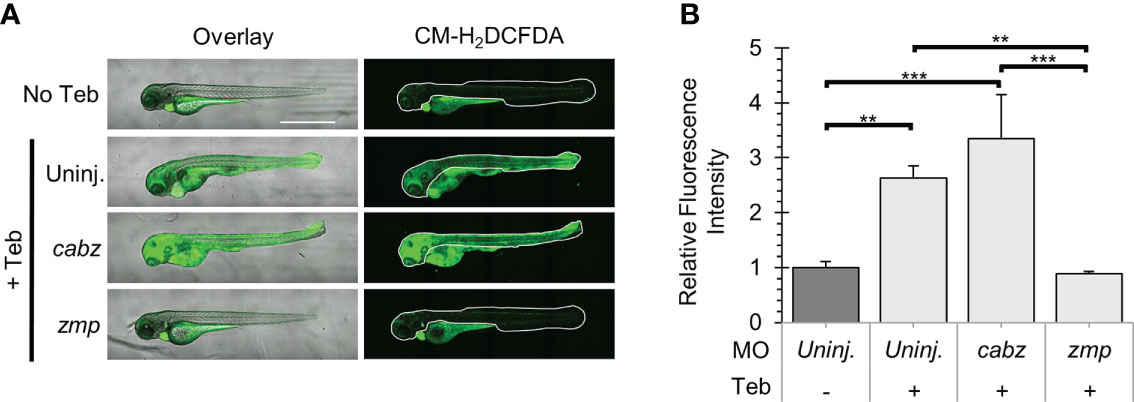
Figure 7 Analysis of Reactive Oxygen Species in Il-1β-induced inflammation. (A) Representative confocal images of ubb:Gal4-EcR, uas:Ilβmat larvae treated with CM-H2DCFDA. Shown here are the overlay of brightfield and CM-H2DCFDA fluorescence (left panels) and CM-H2DCFDA fluorescence only (right panels). Fluorescence signal was quantified from the region of interest outlined with a white border in the CM-H2DCFDA images. (B) Relative Fluorescence Intensity (RFI) was quantified using FIJI ‘mean grey value’ measurement. All mean RFIs were normalized to uninjected No Teb mean values. Error Bars are +1 standard deviation. Asterisks indicate significant differences **p < 0.01 ***p < 0.001 by one-way ANOVA followed by Tukey’s HSD Test. Scale bar in (A) is 1 mm.
Given that 1) the zmp gene shares conserved synteny between the human, mouse, and zebrafish genomes, 2) zmp morpholinos rescued mortality, gross morphology, neutrophil counts, and prevented ROS in our model of Il-1β-induced embryonic systemic inflammation, and 3) mosaic CRISPR/Cas9 knockout phenocopies the morpholino rescue, we have genetically and functionally demonstrated that the zmp:0000000936 gene encodes zebrafish Il1r1.
Discussion
In this study, we use a combination of database mining, genetic analyses, and functional assays to identify zebrafish Il1r1. Although our in silico analysis revealed several putative genes that encode for proteins with partial alignment to human IL1R1, we discovered two putative candidate genes with conserved synteny to the human and mouse genomes. The proteins encoded by these genes show relatively low sequence identities to human IL1R1 but have highly similar predicted protein structures. To examine functionality of these candidates, we designed highly effective morpholinos that disrupted normal splicing or transcription with no obvious off-target effects. We found that knockdown of only one candidate rescued neutrophil expansion, ROS generation, morbidity, and mortality in our zebrafish model of embryonic Il-1β-induced systemic inflammation. We also designed effective gRNAs for mosaic knockout using the CRISPR/Cas9 system that phenocopied the knockdown effects of the morpholinos. We therefore conclude that the zebrafish genome contains only one functional ortholog of Il1r1. Our study highlights the importance of functional data to accurately identify and annotate zebrafish orthologs of inflammation genes and provides essential insights to the inflammatory response driven by Il-1β.
As a potential caveat of our results, Il1rap is also required for IL-1 signaling and our study has not conclusively eliminated the possibility that zmp:0000000936 could encode Il1rap. For example, knockdown of either zebrafish Il1r1 or Il1rap could both result in the rescue of Il-1β-induced inflammation as previously demonstrated by knockouts of either mouse Il1r1 or Il1rap (26–28). However, based upon conserved synteny and protein alignments, we predict that zebrafish Il1rap is most likely encoded by the cabz01068246 gene (see Results), although we do not present functional data for this observation. In addition, by process of elimination, we predict that the cabz01054965 gene, which is adjacent to zmp:0000000936, likely encodes the zebrafish Il1rl2 ortholog as shown by conserved synteny. Based upon our collective data, we are confident that we have identified zebrafish Il1r1, but have not conclusively demonstrated receptor functionality without a formal biochemical analysis of receptor/ligand binding.
Prior to our study, zebrafish Il1r1 was not accurately annotated in the zebrafish genome. Previous studies used the zebrafish genomic sequence to identify putative genes encoding Toll-like receptors (TLR) and interleukin receptors that contain a Toll/IL-1 receptor (TIR) domain. For example, Meijer et al. identified several predicted TLR proteins, one putative Il1r, and one putative Il18r (51). This study confirmed the expression of the predicted transcripts by RT-PCR and investigated their expression in Mycobacterium-infected zebrafish. Ultimately, the authors concluded that other il1r and il18r homologues may exist as the expression levels of the putative il1r and il18r transcripts were unaffected by Mycobacterium infection. On review of this work, we determined that the transcript-specific primers used to amplify il1r and il18r by RT-PCR are actually against cu855885 (annotated as il1rl1) located on chr. 9 and il18r (annotated as il1rap12) located on chr. 14, respectively. More recently, Frame et al. described the identification of the zebrafish gene encoding Interleukin-1 receptor-like 1 (Il1rl1), a predicted homolog of zebrafish Il1r1. This study found upregulation of zebrafish il1rl1 in Flk1+ cMyb+ hematopoietic stem and progenitor cells (HSPCs) in response to Il-1β stimulation by macrophages. The authors also found reduced runx1/cmyb expression and reduced numbers of CD41+ HSPCs following morpholino knockdown of il1rl1 and suggest that Il-1β promotes Il1rl1+ HSPC production via inflammasome activity (40). Based upon the results we present in our study; it would be interesting to examine the expression and knockdown of zebrafish il1r1 in this model of HSPC production.
To identify the functional zebrafish Il1r1, we implemented our validated transgenic model of embryonic Il-1β-induced systemic inflammation and used morpholino oligonucleotides to knockdown the expression of putative Il1r1 orthologs. In our model, the mature form of Il-1β is secreted only in the presence of the ecdysone analog tebufenozide (Teb), resulting in adverse inflammation-related phenotypes. Other inducible transgenic models, such as Tet-On/Tet-Off and Cre-ER recombinase often have unintended consequences due to antibiotic- and estrogen receptor-mediated effects that may complicate data interpretation (52). In contrast, we used the Teb-inducible UAS/Gal4-EcR system as there is no endogenous ecdysone receptor in vertebrates, no known off-target effects, and markedly low toxicity (22). Further, our inflammation model is driven by a single proinflammatory cytokine, Il-1β. Unlike wound-induced or infection-based models that cause inflammation via a myriad of damage-associated molecular patterns (DAMPs) and pathogen-associated molecular patterns (PAMPs), the use of a single molecular effector allowed for our targeted, morpholino-based approach. To demonstrate morpholino effectiveness in our model, we showed 1) disrupted pre-mRNA splicing as demonstrated by RT-PCR and DNA sequencing, 2) phenocopy of the splice-site morpholino using an independent il1r1 start-site morpholino, 3) no obvious off-target phenotypes, and 4) complete rescue of the deleterious phenotypes caused by induced expression of Il-1β. Thus, our experimental paradigm, with appropriate caveats considered, follows the basic guidelines for morpholino use in zebrafish as described by Stainier et al. (53).
While newer technologies enable targeted gene disruption in zebrafish, including the CRISPR/Cas9 system, transcription activator-like effector nucleases (TALENs), and zinc finger nucleases (ZFNs) (54), these strategies require significant time and resources compared to validated, highly effective morpholinos. For example, the successful design and generation of CRISPR/Cas9 knockouts in combination with breeding schemes to produce double or triple transgenic zebrafish in a mutant background could take years to successfully accomplish. Furthermore, these analyses generally target one gene at a time, so if two paralogs were to have overlapping function, both genes would need to be targeted and bred into the appropriate background to observe the desired phenotypes. These analyses could be further complicated by compensatory networks that buffer against deleterious mutations (55). Given these limitations, we were surprised to find that mosaic CRISPR/Cas9 knockout of zebrafish Il1r1 effectively phenocopied the knockdown effects of the morpholinos. Thus, our study highlights the effective use of morpholinos and mosaic CRISPR/Cas9 to identify gene function in a relevant embryonic disease model simply by rescuing the deleterious phenotypes.
In conclusion, our study provides important new tools and insights for investigating the role of Il-1β and Il1r1 in a broad spectrum of diseases, such as autoinflammatory diseases, metabolic syndromes, acute and chronic inflammation, and malignancies (25, 56). As an example, neonatal-onset multisystem inflammatory disease (NOMID), a rare congenital inflammatory disorder, is caused by autosomal dominant mutations in the NLRP3 gene, also known as Cryopyrin or CIAS1 (57). Clinical manifestations in NOMID, similar to our zebrafish inflammation model, are caused by increased release of Il-1β. Current therapies for NOMID and related disorders primarily involve biologics against IL1R1 signaling such as Anakinra (a recombinant IL1R antagonist), Canakinumab (an antibody targeting Il-1β), and Rilonacept (a soluble decoy receptor). Given that many inflammatory conditions are driven by Il-1β and that there are no known small molecule inhibitors of IL1R1 (25, 56), identification of Il1r1 in our zebrafish model provides a novel platform to identify new drugs for the treatment of Il-1β-related diseases.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was reviewed and approved by University of Wisconsin-Madison, Institutional Animal Care and Use Committee.
Author contributions
DS conducted the experiments, collected, assembled, analyzed the data, and wrote the manuscript. AF provided experimental support, helped in data analysis, and edited the manuscript. KP provided experimental support and helped in data analysis. MT conceived the study, designed experiments, interpreted data, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
Funding was provided by NIH RO1NS116043. AF was supported by the UW-Madison Biotechnology Training Program, NIH T32GM008349 and T32GM135066.
Acknowledgments
We thank Dr. Anna Huttenlocher (University of Wisconsin-Madison) for providing the Tg(mpx:mCherry)uwm7 transgenic line. We also wish to thank Randall Kopielski and Jessica Fairbanks for assistance with zebrafish husbandry and animal facility care and maintenance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.1039161/full#supplementary-material
References
1. Campos-Sanchez JC, Esteban MA. Review of inflammation in fish and value of the zebrafish model. J Fish Dis (2021) 44(2):123–39. doi: 10.1111/jfd.13310
2. Xie Y, Meijer AH, Schaaf MJM. Modeling inflammation in zebrafish for the development of anti-inflammatory drugs. Front Cell Dev Biol (2020) 8:620984. doi: 10.3389/fcell.2020.620984
3. Zanandrea R, Bonan CD, Campos MM. Zebrafish as a model for inflammation and drug discovery. Drug Discovery Today (2020) 25(12):2201–11. doi: 10.1016/j.drudis.2020.09.036
4. Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, et al. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn (2007) 236(11):3088–99. doi: 10.1002/dvdy.21343
5. Yan B, Han P, Pan L, Lu W, Xiong J, Zhang M, et al. IL-1beta and reactive oxygen species differentially regulate neutrophil directional migration and basal random motility in a zebrafish injury-induced inflammation model. J Immunol (2014) 192(12):5998–6008. doi: 10.4049/jimmunol.1301645
6. Delgadillo-Silva LF, Tsakmaki A, Akhtar N, Franklin ZJ, Konantz J, Bewick GA, et al. Modelling pancreatic beta-cell inflammation in zebrafish identifies the natural product wedelolactone for human islet protection. Dis Model Mech (2019) 12(1):1–12. doi: 10.1242/dmm.036004
7. Ibrahim S, Harris-Kawano A, Haider I, Mirmira RG, Sims EK, Anderson RM. A novel cre-enabled tetracycline-inducible transgenic system for tissue-specific cytokine expression in the zebrafish: CETI-PIC3. Dis Model Mech (2020) 13(6):1–12. doi: 10.1242/dmm.042556
8. Deng Q, Harvie EA, Huttenlocher A. Distinct signalling mechanisms mediate neutrophil attraction to bacterial infection and tissue injury. Cell Microbiol (2012) 14(4):517–28. doi: 10.1111/j.1462-5822.2011.01738.x
9. Hall CJ, Flores MV, Oehlers SH, Sanderson LE, Lam EY, Crosier KE, et al. Infection-responsive expansion of the hematopoietic stem and progenitor cell compartment in zebrafish is dependent upon inducible nitric oxide. Cell Stem Cell (2012) 10(2):198–209. doi: 10.1016/j.stem.2012.01.007
10. Le Guyader D, Redd MJ, Colucci-Guyon E, Murayama E, Kissa K, Briolat V, et al. Origins and unconventional behavior of neutrophils in developing zebrafish. Blood (2008) 111(1):132–41. doi: 10.1182/blood-2007-06-095398
11. Goody M, Jurczyszak D, Kim C, Henry C. Influenza a virus infection damages zebrafish skeletal muscle and exacerbates disease in zebrafish modeling duchenne muscular dystrophy. PloS Curr (2017) 9. doi: 10.1371/currents.md.8a7e35c50fa2b48156799d3c39788175
12. Davis JM, Huang M, Botts MR, Hull CM, Huttenlocher A. A zebrafish model of cryptococcal infection reveals roles for macrophages, endothelial cells, and neutrophils in the establishment and control of sustained fungemia. Infect Immun (2016) 84(10):3047–62. doi: 10.1128/IAI.00506-16
13. Voelz K, Gratacap RL, Wheeler RT. A zebrafish larval model reveals early tissue-specific innate immune responses to mucor circinelloides. Dis Model Mech (2015) 8(11):1375–88. doi: 10.1242/dmm.019992
14. Philip AM, Wang Y, Mauro A, El-Rass S, Marshall JC, Lee WL, et al. Development of a zebrafish sepsis model for high-throughput drug discovery. Mol Med (2017) 23:134–48. doi: 10.2119/molmed.2016.00188
15. Cvejic A, Hall C, Bak-Maier M, Flores MV, Crosier P, Redd MJ, et al. Analysis of WASp function during the wound inflammatory response–live-imaging studies in zebrafish larvae. J Cell Sci (2008) 121(Pt 19):3196–206. doi: 10.1242/jcs.032235
16. Hasegawa T, Hall CJ, Crosier PS, Abe G, Kawakami K, Kudo A, et al. Transient inflammatory response mediated by interleukin-1beta is required for proper regeneration in zebrafish fin fold. Elife (2017) 6:1–22 doi: 10.7554/eLife.22716
17. Mathias JR, Perrin BJ, Liu TX, Kanki J, Look AT, Huttenlocher A. Resolution of inflammation by retrograde chemotaxis of neutrophils in transgenic zebrafish. J Leukoc Biol (2006) 80(6):1281–8. doi: 10.1189/jlb.0506346
18. Renshaw SA, Loynes CA, Trushell DM, Elworthy S, Ingham PW, Whyte MK. A transgenic zebrafish model of neutrophilic inflammation. Blood (2006) 108(13):3976–8. doi: 10.1182/blood-2006-05-024075
19. Sipka T, Peroceschi R, Hassan-Abdi R, Gross M, Ellett F, Begon-Pescia C, et al. Damage-induced calcium signaling and reactive oxygen species mediate macrophage activation in zebrafish. Front Immunol (2021) 12:636585. doi: 10.3389/fimmu.2021.636585
20. Yoo SK, Huttenlocher A. Spatiotemporal photolabeling of neutrophil trafficking during inflammation in live zebrafish. J Leukoc Biol (2011) 89(5):661–7. doi: 10.1189/jlb.1010567
21. Ogryzko NV, Hoggett EE, Solaymani-Kohal S, Tazzyman S, Chico TJ, Renshaw SA, et al. Zebrafish tissue injury causes upregulation of interleukin-1 and caspase-dependent amplification of the inflammatory response. Dis Model Mech (2014) 7(2):259–64. doi: 10.1242/dmm.013029
22. Esengil H, Chang V, Mich JK, Chen JK. Small-molecule regulation of zebrafish gene expression. Nat Chem Biol (2007) 3(3):154–5. doi: 10.1038/nchembio858
23. Lanham KA, Nedden ML, Wise VE, Taylor MR. Genetically inducible and reversible zebrafish model of systemic inflammation. Biol Open (2022) 11(3):1–12. doi: 10.1242/bio.058559
24. Lopez-Castejon G, Brough D. Understanding the mechanism of IL-1beta secretion. Cytokine Growth Factor Rev (2011) 22(4):189–95. doi: 10.1016/j.cytogfr.2011.10.001
25. Kaneko N, Kurata M, Yamamoto T, Morikawa S, Masumoto J. The role of interleukin-1 in general pathology. Inflamm Regen (2019) 39:12. doi: 10.1186/s41232-019-0101-5
26. Glaccum MB, Stocking KL, Charrier K, Smith JL, Willis CR, Maliszewski C, et al. Phenotypic and functional characterization of mice that lack the type I receptor for IL-1. J Immunol (1997) 159(7):3364–71.
27. Labow M, Shuster D, Zetterstrom M, Nunes P, Terry R, Cullinan EB, et al. Absence of IL-1 signaling and reduced inflammatory response in IL-1 type I receptor-deficient mice. J Immunol (1997) 159(5):2452–61.
28. Cullinan EB, Kwee L, Nunes P, Shuster DJ, Ju G, McIntyre KW, et al. IL-1 receptor accessory protein is an essential component of the IL-1 receptor. J Immunol (1998) 161(10):5614–20.
29. Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature (2013) 496(7446):498–503. doi: 10.1038/nature12111
30. Meyer A, Schartl M. Gene and genome duplications in vertebrates: the one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr Opin Cell Biol (1999) 11(6):699–704. doi: 10.1016/S0955-0674(99)00039-3
31. Westerfield M. The zebrafish book: University of Oregon press. (Eugene, Oregon: University of Oregon Press) (1995).
32. Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature (2021) 596(7873):583–9. doi: 10.1038/s41586-021-03819-2
33. Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, et al. AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res (2022) 50(D1):D439–D44. doi: 10.1093/nar/gkab1061
34. Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3–new capabilities and interfaces. Nucleic Acids Res (2012) 40(15):e115. doi: 10.1093/nar/gks596
35. Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc (2008) 3(1):59–69. doi: 10.1038/nprot.2007.514
36. Vauti F, Stegemann LA, Vogele V, Koster RW. All-age whole mount in situ hybridization to reveal larval and juvenile expression patterns in zebrafish. PloS One (2020) 15(8):e0237167. doi: 10.1371/journal.pone.0237167
37. Bill BR, Petzold AM, Clark KJ, Schimmenti LA, Ekker SC. A primer for morpholino use in zebrafish. Zebrafish (2009) 6(1):69–77. doi: 10.1089/zeb.2008.0555
38. Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods (2012) 9(7):676–82. doi: 10.1038/nmeth.2019
39. Dale M, Nicklin MJ. Interleukin-1 receptor cluster: gene organization of IL1R2, IL1R1, IL1RL2 (IL-1Rrp2), IL1RL1 (T1/ST2), and IL18R1 (IL-1Rrp) on human chromosome 2q. Genomics (1999) 57(1):177–9. doi: 10.1006/geno.1999.5767
40. Frame JM, Kubaczka C, Long TL, Esain V, Soto RA, Hachimi M, et al. Metabolic regulation of inflammasome activity controls embryonic hematopoietic stem and progenitor cell production. Dev Cell (2020) 55(2):133–49 e6. doi: 10.1016/j.devcel.2020.07.015
41. Fields JK, Gunther S, Sundberg EJ. Structural basis of IL-1 family cytokine signaling. Front Immunol (2019) 10:1412. doi: 10.3389/fimmu.2019.01412
42. O’Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in toll-like receptor signalling. Nat Rev Immunol (2007) 7(5):353–64. doi: 10.1038/nri2079
43. Riva F, Bonavita E, Barbati E, Muzio M, Mantovani A, Garlanda C. TIR8/SIGIRR is an interleukin-1 Receptor/Toll like receptor family member with regulatory functions in inflammation and immunity. Front Immunol (2012) 3:322. doi: 10.3389/fimmu.2012.00322
44. Yoshida T, Mishina M. Zebrafish orthologue of mental retardation protein IL1RAPL1 regulates presynaptic differentiation. Mol Cell Neurosci (2008) 39(2):218–28. doi: 10.1016/j.mcn.2008.06.013
45. Draper BW, Morcos PA, Kimmel CB. Inhibition of zebrafish fgf8 pre-mRNA splicing with morpholino oligos: a quantifiable method for gene knockdown. Genesis (2001) 30(3):154–6. doi: 10.1002/gene.1053
46. Bader A, Gao J, Riviere T, Schmid B, Walzog B, Maier-Begandt D. Molecular insights into neutrophil biology from the zebrafish perspective: Lessons from CD18 deficiency. Front Immunol (2021) 12:677994. doi: 10.3389/fimmu.2021.677994
47. Herbomel P, Thisse B, Thisse C. Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development (1999) 126(17):3735–45. doi: 10.1242/dev.126.17.3735
48. Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet (2000) 26(2):216–20. doi: 10.1038/79951
49. Harvie EA, Huttenlocher A. Neutrophils in host defense: new insights from zebrafish. J Leukoc Biol (2015) 98(4):523–37. doi: 10.1189/jlb.4MR1114-524R
50. Mittal M, Siddiqui MR, Tran K, Reddy SP, Malik AB. Reactive oxygen species in inflammation and tissue injury. Antioxid Redox Signal (2014) 20(7):1126–67. doi: 10.1089/ars.2012.5149
51. Meijer AH, Gabby Krens SF, Medina Rodriguez IA, He S, Bitter W, Ewa Snaar-Jagalska B, et al. Expression analysis of the toll-like receptor and TIR domain adaptor families of zebrafish. Mol Immunol (2004) 40(11):773–83. doi: 10.1016/j.molimm.2003.10.003
52. Wust RCI, Houtkooper RH, Auwerx J. Confounding factors from inducible systems for spatiotemporal gene expression regulation. J Cell Biol (2020) 219(7):1–4 doi: 10.1083/jcb.202003031
53. Stainier DYR, Raz E, Lawson ND, Ekker SC, Burdine RD, Eisen JS, et al. Guidelines for morpholino use in zebrafish. PloS Genet (2017) 13(10):e1007000. doi: 10.1371/journal.pgen.1007000
54. Lawson ND. Reverse genetics in zebrafish: Mutants, morphants, and moving forward. Trends Cell Biol (2016) 26(2):77–9. doi: 10.1016/j.tcb.2015.11.005
55. Rossi A, Kontarakis Z, Gerri C, Nolte H, Holper S, Kruger M, et al. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature (2015) 524(7564):230–3. doi: 10.1038/nature14580
56. Dinarello CA, Simon A, van der Meer JW. Treating inflammation by blocking interleukin-1 in a broad spectrum of diseases. Nat Rev Drug Discov (2012) 11(8):633–52. doi: 10.1038/nrd3800
Keywords: zebrafish, interleukin - 1 β, interleukin 1 receptor type 1, inflammation, neutrophils, ROS - reactive oxygen species, UAS/Gal4
Citation: Sebo DJ, Fetsko AR, Phipps KK and Taylor MR (2022) Functional identification of the zebrafish Interleukin-1 receptor in an embryonic model of Il-1β-induced systemic inflammation. Front. Immunol. 13:1039161. doi: 10.3389/fimmu.2022.1039161
Received: 07 September 2022; Accepted: 14 October 2022;
Published: 26 October 2022.
Edited by:
Jun-ichi Hikima, University of Miyazaki, JapanReviewed by:
Diana Boraschi, Shenzhen Institute of Advanced Technology (SIAT) (CAS), ChinaSylwia Dominika Tyrkalska, University of Murcia, Spain
Copyright © 2022 Sebo, Fetsko, Phipps and Taylor. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael R. Taylor, bWljaGFlbC50YXlsb3JAd2lzYy5lZHU=
 Dylan J. Sebo
Dylan J. Sebo Audrey R. Fetsko
Audrey R. Fetsko Kallie K. Phipps
Kallie K. Phipps Michael R. Taylor
Michael R. Taylor