
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 13 December 2022
Sec. Viral Immunology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.1031200
This article is part of the Research TopicHost-Virus Interaction at the Omics and Ecology LevelsView all 7 articles
N6‐methyladenosine (m6A) is the most abundant RNA chemical modification in eukaryotes and is also found in the RNAs of many viruses. In recent years, m6A RNA modification has been reported to have a role not only in the replication of numerous viruses but also in the innate immune escape process. In this review, we describe the viruses that contain m6A in their genomes or messenger RNAs (mRNAs), and summarize the effects of m6A on the replication of different viruses. We also discuss how m6A modification helps viral RNAs escape recognition by exogenous RNA sensors, such as retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), during viral invasion. Overall, the goal of our review is to summarize how m6A regulates viral replication and facilitates innate immune escape. Furthermore, we elaborate on the potential of m6A as a novel antiviral target.
N6-methyladenosine (m6A) was the first internal RNA modification identified in mRNAs of mammalian cells in 1974 (1). However, our understanding of m6A is currently limited. In 1994, 20 years after the discovery of m6A, methyltransferase was identified as a protein complex, and methyltransferase-like 3 (METTL3) was identified as an S−adenosyl methionine (SAM)-binding protein with methyltransferase capacity (2, 3). M6A is the most abundant and well-characterized RNA modification (4, 5). Additionally, m6A is a reversible chemical modification that affects nearly all aspects of RNA biology, including RNA folding structure, mRNA maturation, nuclear export, translation, and mRNA decay (6–20).
M6A was once termed as the fifth base in mRNA. It was first identified by chromatography (1). Methylated RNA immunoprecipitation sequencing (MeRIP-seq), also named m6A-seq, is the most widely used sequencing method for RNA m6A profiling, but it cannot precisely identify which adenosines are modified (21). M6A individual-nucleotide resolution crosslinking and immunoprecipitation sequencing (miCLIP-m6A-seq) and photo-crosslinking-assisted m6A sequencing (PA-m6A-seq) can detect and characterize m6A in RNA with pinpoint accuracy (22). In addition to mRNA of mammalian cells, m6A has been identified in a wide range of viral RNAs, including DNA and RNA viruses (as shown in Table 1). The replication of many viruses can be modulated by m6A, and in-depth studies have revealed that m6A exhibits contrary functions in the replication process of different viruses. Additionally, m6A affects the recognition of viral RNAs by RLRs (38–40).
Innate immune responses function as the primary antiviral strategy when host cells are invaded by viruses. RLRs are key sensors among pattern recognition receptors (PRRs). RLRs can recognize exogenous viral RNAs and stimulate the production of type I interferons (IFNs), which can result in the upregulation of antiviral proteins, such as RNA-dependent protein kinase (PKR), 2′,5′-oligoadenylate synthetase (OAS), 2’,5’-oligoadenylate-dependent ribonuclease L (RNase L), and Mx proteins (41, 42). RLRs include three components: RIG-I, melanoma differentiation-associated protein 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2) (43, 44). RIG-I recognizes double-stranded RNAs (dsRNAs) (<300 bp) containing either a 5’-triphosphate or 5’-diphosphate (45–47); MDA5, which shares a similar structure with RIG-I, senses long dsRNAs (>1,000 bp) (48, 49); and LGP2, which lacks the caspase recruitment domain (CARD), is a regulator of RIG-I and MDA5, and exhibits different regulatory functions (50–52). RIG-I and MDA5, which are sensors of exogenous viral RNAs, can sense RNAs generated by both DNA and RNA viruses.
Interestingly, m6A modifications in the RNAs of different viruses exhibit many differences during the replication process. Moreover, m6A modifications in viral RNAs play a significant role in RLR recognition after viral infection. Here, we review the function of m6A in viral replication and the innate immune sensing of RLRs.
Eukaryotic cell mRNA has many internal chemical modifications, including m6A, 5-methylcytosine (m5C), N1-methyladenosine (m1A), and pseudouridine (Ψ) (1, 53–56); among these, m6A is the most abundant modification (Figure 1A). In addition, m6A is a reversible chemical modification (Figure 1B). The RNA transferases, including METTL3, methyltransferase-like 14 (METTL14), WT1-associated protein (WTAP), KIAA1429 (also known as vir-like m6A methyltransferase-associated protein [VIRMA]), zinc finger CCCH domain-containing protein 1 (ZC3H13), RNA-binding motif protein 15 (RBM15), and methyltransferase-like 16 (METTL16) are termed as ‘writers’. METTL3, METTL14, and WTAP, which are the most well-known ‘writers’, can form a protein complex. This protein complex can recognize the consensus DRA*CH ([A/G/U], [A/G], A*, C, [A/C/U]) motifs and add a methyl to the specific N6 position of adenosine (22, 57). As previously mentioned, AlkB homolog 5 (ALKBH5) and fat mass and obesity-associated protein (FTO), which function as demethylases, are known as ‘erasers’, and can remove the methyl of m6A. ‘Readers’, including YTH N6-methyladenosine RNA-binding protein 1, 2, and 3 (YTHDF1, YTHDF2, and YTHDF3), YTH Domain Containing 1 and 2 (YTHDC1 and YTHDC2), eukaryotic initiation factor 3 (eIF3), insulin-like growth factor 2 mRNA-binding protein 1, 2, and 3 (IGF2BP1, IGF2BP2, and IGF2BP3), fragile X mental retardation protein (FMRP), and heterogeneous nuclear ribonucleoproteins A2/B1 (hnRNPA2/B1), recognize the m6A modifications in RNAs and regulate several biological processes of RNAs, such as translation, decay, and translocation. Owing to the lack of research techniques, there was limited knowledge about the function of m6A until the Chinese-American scientist Chuan He proposed the concept of RNA epigenetics in the early 2010s (58). Since then, significant progress has been made in the study of m6A modification. In addition to eukaryotic mRNAs, m6A is also found in many viral mRNAs, viral genomes, and intermediate RNAs produced during the viral replication process. In 1976, only 2 years after m6A was identified in eukaryotic mRNAs, influenza virus mRNA was found to contain internal m6A modifications (59). Although a few articles on m6A modifications of viral RNAs have been published to date, it seems that the study of viral RNA epigenetics is poised for a major expansion and has the potential to change our understanding of how viruses regulate their life cycle.
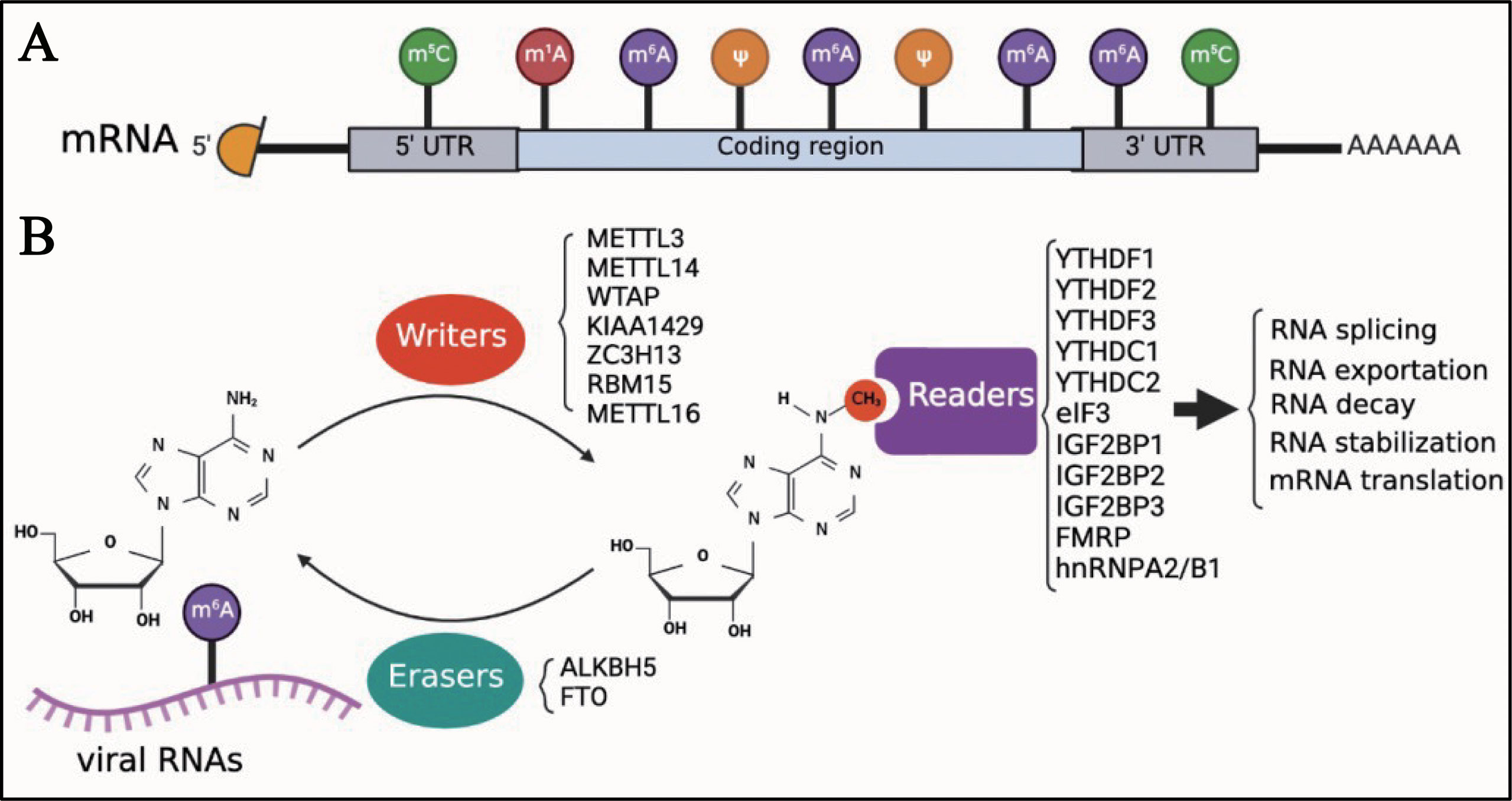
Figure 1 Introduction of m6A modifications. (A) M6A is one of the first identified and most abundant internal modifications in the mRNA of eukaryotic cells. (B) M6A is a reversible chemical modification in RNAs; ‘writers’ add a methyl to the N6 position of adenosine; ‘erasers’ remove the methyl of m6A; after adding m6A in RNAs, ‘readers’ recognize the modified RNAs and regulate the biological process of RNAs, including translation, decay, and translocation.
DNA viruses (with DNA genomes) can also produce RNAs (which contain m6A) during replication (Figure 2). Further studies have shown that m6A modifications have different regulatory functions in the life cycle of different viruses (Table 1). M6A in viral RNAs promotes the replication of herpes simplex virus 1 (HSV‐1), simian vacuolating virus 40 (SV40), adenovirus (AdV), and human papillomavirus 16 (HPV-16) (30–32, 34). However, m6A functions as a suppressive regulator of Bombyx mori nucleopolyhedrovirus (BmNPV) replication (33). Furthermore, m6A in the RNA of Kaposi’s sarcoma‐associated herpesvirus (KSHV) adversely affects replication in different cells. Interestingly, m6A functions as a positive regulator of KSHV in iSLK.219, iSLK.BAC16, and B cells, but as a negative regulator in KiSLK and TREx BCBL1-Rta cells (23–26). Different researchers have different opinions regarding the function of m6A. M6A functions adversely during the replication process of Epstein–Barr virus (EBV) and hepatitis B virus (HBV) (27, 28, 35–37). Although there is no direct sequencing evidence to prove the presence of m6A in the RNA of human cytomegalovirus (HCMV), METTL3 and METTL14 small interfering RNAs (siRNAs) inhibit HCMV reproduction, indicating that m6A may function as a positive regulator of the life cycle of HCMV (60, 61).
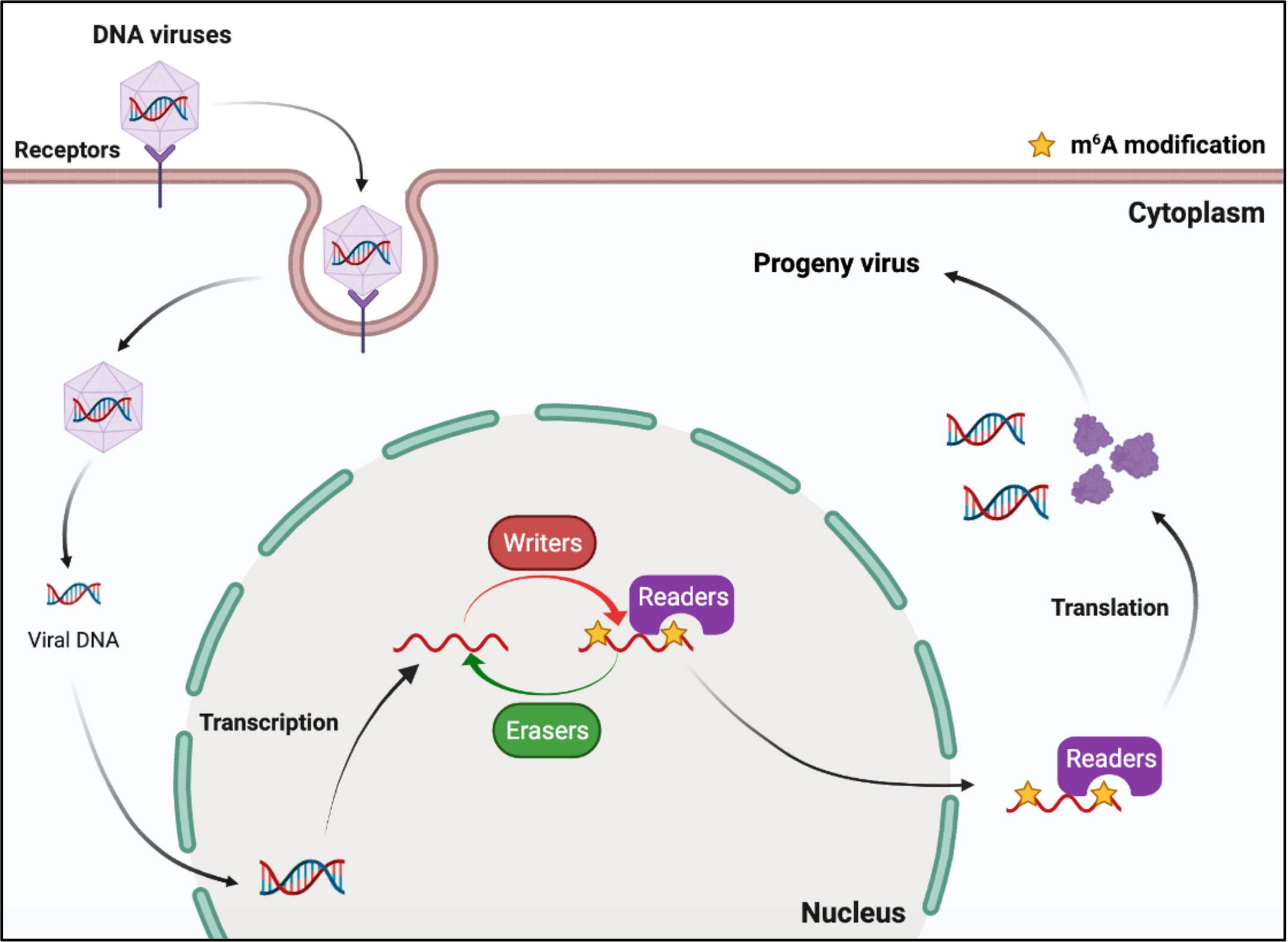
Figure 2 M6A modifications of viral RNAs during the replication process of DNA viruses. DNA viruses produce mRNAs during replication. The viral mRNAs can be m6A modified, and these modifications can modulate the replication of DNA viruses.
RNA viruses have RNA genomes, most of which replicate in the cytoplasm. As shown in Figure 3, ‘writers’ and ‘erasers’ are located in the nucleus under a steady state; however, they may also be detected in the cytoplasm after infection, suggesting that these proteins can shuttle between the nucleus and the cytoplasm (62–66). The positive-sense RNA genome of cytoplasmic RNA viruses, such as Flaviviridae, Coronavirus, and Picornaviridae, functions as mRNA, and can produce negative-sense complementary RNA (cRNA) by serving as a replication template during the replication process. The genomes of the Pneumoviridate and Rhabdoviridae families consist of negative-sense RNA, and both cRNA and mRNA are produced during transcription. Positive-sense cRNA functions as a template for viral genome replication. M6A modifications exist in the viral genome RNA, cRNA, and mRNA of many RNA viruses, and m6A plays different roles in different viruses (Table 2). Replication of Flaviviridae, including Zika virus (ZIKV), dengue virus (DENV), and hepatitis C virus (HCV), is deeply modulated by m6A, and m6A in the RNA of these viruses is a suppressive regulator of viral replication (62, 63, 67). With the spread of the SARS-CoV-2 infection since 2019, scientists have been paying great attention to the study of coronaviruses. Liu’s work indicated that both positive and negative RNAs of SARS-CoV-2 contain m6A modifications, and m6A negatively regulates SARS-CoV-2 infection, as overexpression of METTL3 can inhibit its replication (66, 68–70). Porcine epidemic diarrhea virus (PEDV), a member of the Coronaviridate, also contains m6A in its genomic RNA, and m6A suppresses its replication (71). Enterovirus 71 (EV71) also possesses a positive-sense RNA genome; however, m6A promotes the replication of EV71, which contrasts with its function in the replication of coronavirus. Human respiratory syncytial virus (HRSV, a member of the Pneumoviridae family), human metapneumovirus (HMPV, a member of the Paramyxoviridae family), and vesicular stomatitis virus (VSV, a member of the Rhabdoviridae family) all have negative-sense RNA genomes and share a similar life cycle in the cytoplasm. M6A is also found in the genomes of these viruses and plays a positive role in the replication process, as it can promote viral protein expression and help viral RNAs escape RIG-I recognition (72–74).
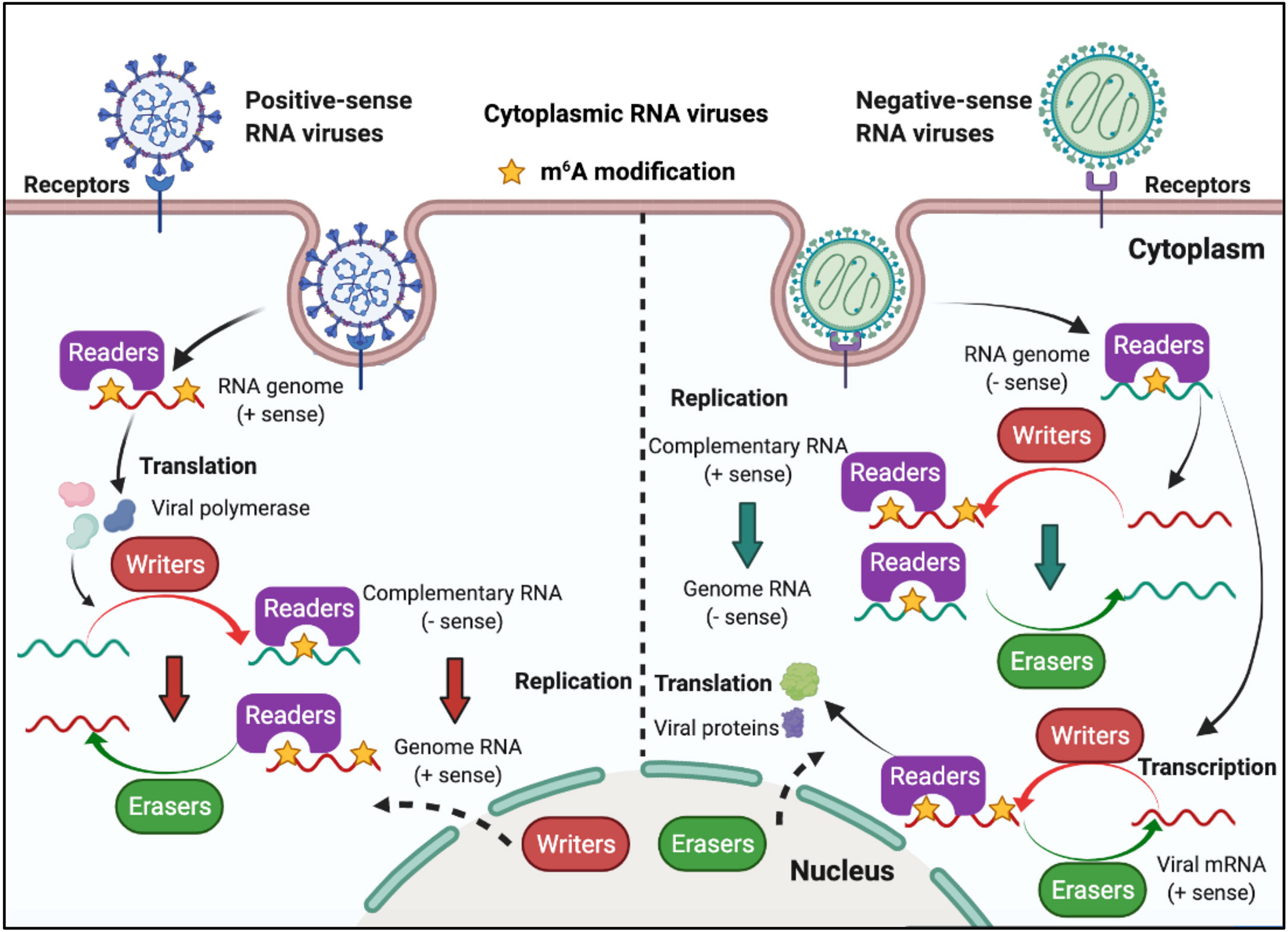
Figure 3 M6A modifications of viral RNAs during the replication process of cytoplasmic RNA viruses. Some of the cytoplasmic RNA viruses possess a positive RNA genome, whereas some possess a negative RNA genome. Cytoplasmic RNA viruses can produce mRNA (the positive-sense RNA genome of cytoplasmic RNA viruses functions as mRNA) and cRNA. M6A is present in the viral genome RNA, cRNA, and mRNA, and can modulate viral protein expression and viral replication. ‘Writers’ and ‘erasers’ can translocate from the nucleus to the cytoplasm after viral infection.
Although most RNA viruses replicate in the cytoplasm, for some of them, replication occurs in the nucleus. Influenza virus (belonging to the Orthomyxoviridae family) possesses a segmented negative-sense single-stranded RNA genome, and is a strict intranuclear replication RNA virus. When influenza virus encounters host cells, hemagglutinin (HA) proteins bind to α-(2, 3)-linked or α-(2, 6)-linked sialic acid on the cell surface membrane, and endocytosis of viral particles is triggered. Once influenza virus penetrates the cells, the viral genome can be released from uncoated virion particles and translocated to the nucleus for genome replication; cRNA and mRNA are synthesized in the nucleus during transcription; cRNA acts as a template for the virion RNA (vRNA) replication process, which also occurs in the nucleus; mRNA of the influenza virus is exported to the cytoplasm and serves as a template for the synthesis of viral proteins. Finally, progeny virus assembly and budding is completed in the plasma membrane (Figure 4).
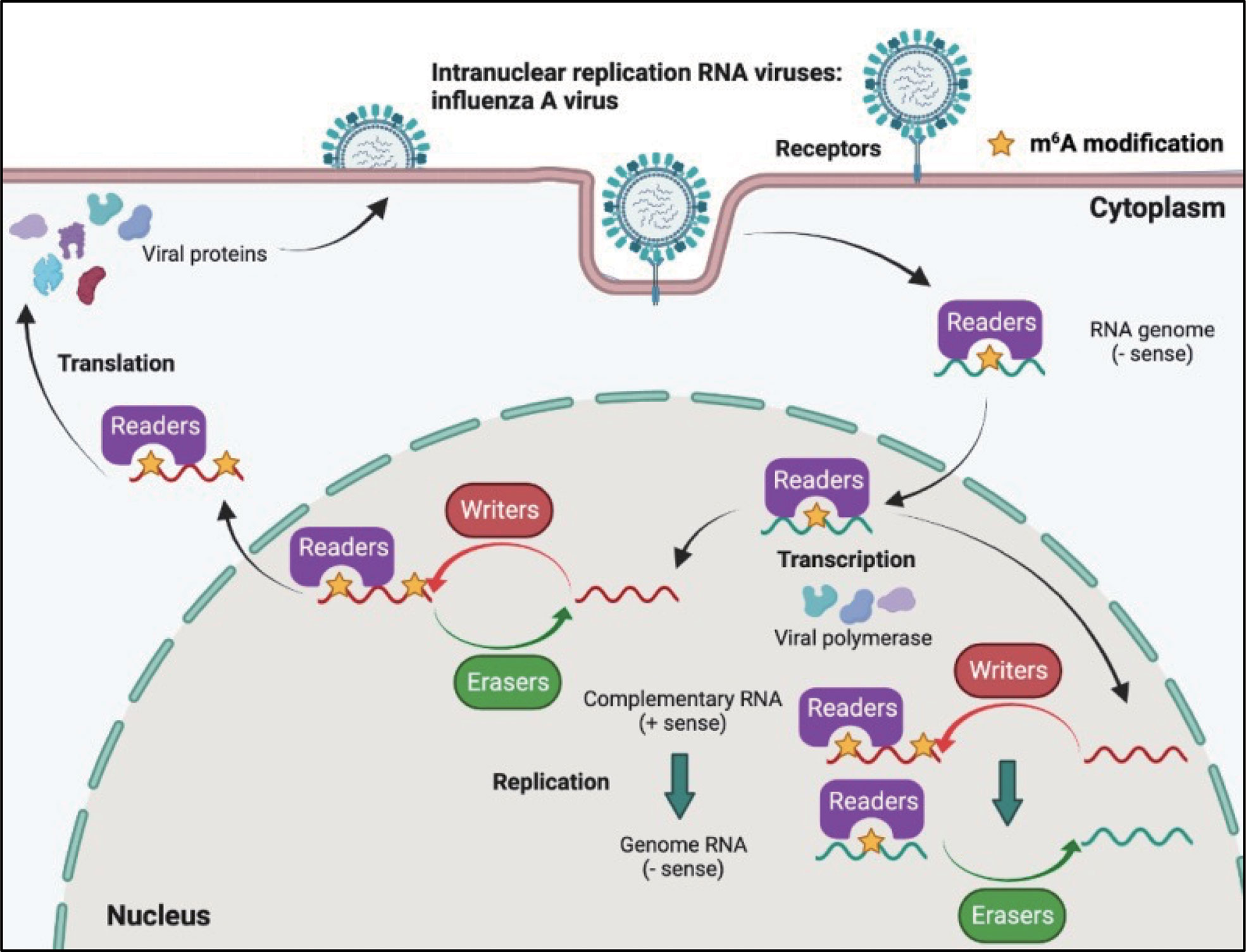
Figure 4 M6A modifications of viral RNAs during the replication process of intranuclear RNA viruses. After binding to the receptors, the genome of intranuclear virus enters the nucleus, where replication and transcription occur. M6A is present in the viral genome RNA, cRNA, and mRNA. Both RNA translocation and mRNA translation are modulated by m6A modification.
The influenza virus was the first confirmed to contain m6A, and the replication of influenza virus is modulated by m6A (Table 2). At first, Krug discovered that m6A was present in the mRNA of influenza virus (59); more than 40 years later, Courtney revealed that vRNA and cRNA also contained m6A modifications (75). Further studies have indicated that m6A promotes influenza replication, and that METTL3 and YTHDF2 play an important role in the replication process (75).
Retroviruses are a family of RNA viruses that have a reverse transcriptase capable of making a complementary DNA copy of the viral genomic RNA, which is then integrated into the host cell’s DNA. M6A can also be found in the viral genomic RNA and mRNA of retroviruses (Figure 5). Human immunodeficiency virus type 1 (HIV-1), Rous sarcoma virus (RSV), and feline leukemia virus (FeLV) all belong to groups of retroviruses that contain m6A modifications in their RNAs (76, 77, 82–84). It was discovered that m6A plays different roles in the replication of retroviruses (Table 2). Many research groups have revealed that post-transcriptional m6A modification of HIV-1 mRNAs enhances viral gene expression, whereas Lu et al. discovered that YTHD proteins could bind to the genomic RNA of HIV-1 and inhibit viral reverse transcription after viral entry (76–80). A recent study has revealed that m6A functions as a suppressive regulator of the life cycle of endogenous retroviruses (ERVs). Host cells can recognize m6A modifications in the mRNAs of intracisternal A-particles (IAPs) and related ERVK elements, and the m6A-modified RNAs can restrain their ability to trigger inflammatory responses, such as those observed in human neurodegenerative diseases (81, 85).
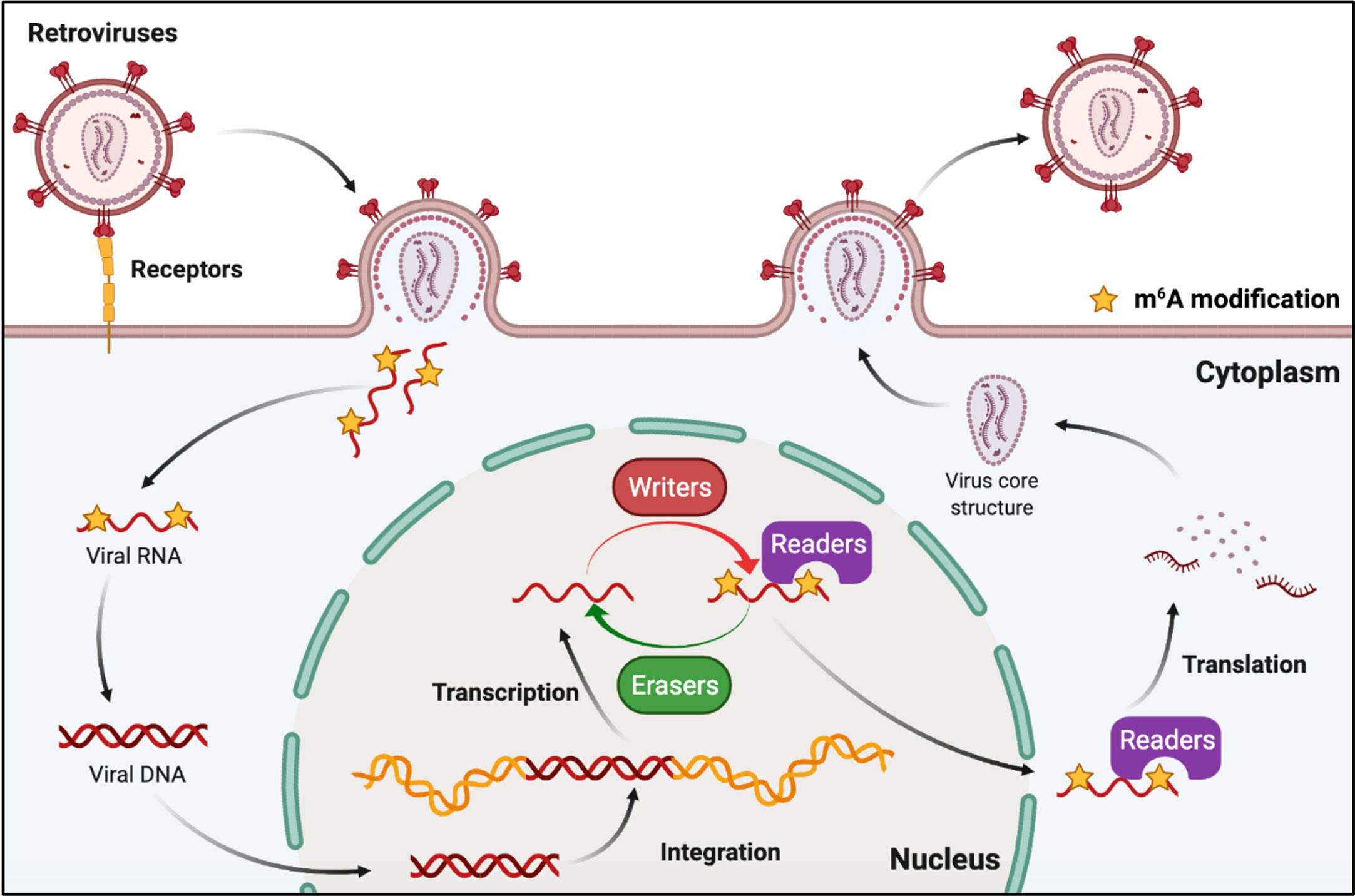
Figure 5 M6A modifications of viral RNAs during the replication process of retroviruses. Retroviruses possess an RNA genome and have reverse transcription activity. M6A is present in both genomic RNA and mRNA of retroviruses, and plays roles in reverse transcription, RNA transcription, and mRNA translation.
Toll-like receptors (TLRs) and RLRs are the main receptors of PRRs that can sense viral RNAs. Studies of PRR sensing of m6A-modified viral RNA have been mainly focused on RLRs. RIG-I and MDA5 are the main sensors of RLRs, and their primary function is to recognize exogenous RNA and stimulate the expression of type I IFNs when host cells are invaded by viruses (86). The RNAs produced in the replication process of both DNA and RNA viruses can be recognized by RLRs, and some studies have indicated that m6A modification helps exogenous viral RNA escape recognition by RLRs (Figure 6). Although HCMV is a DNA virus, it can trigger RLR sensing activity, and m6A might play a key role in this process (60, 87). However, the interaction between m6A and RLR stimulation has not yet been thoroughly clarified, and there is no direct evidence to indicate that the mRNA of HCMV contains m6A (60). This study suggests that m6A might play a role in the recognition of HCMV viral mRNA. HBV is another well-known DNA virus; but its life cycle produces an RNA intermediate termed ‘pregenomic’ RNA (pgRNA). Furthermore, pgRNA is modified by m6A, which reduces the sensing activity of RIG-I (38, 88). RLRs mainly recognize viral RNA from RNA viruses during viral infection, and m6A in the RNA genomes of HIV-1, HCV, SARS-CoV-2, HMPV, and VSV can help viral RNA escape RIG-I recognition and inhibit the expression of type I IFNs (38, 39, 69, 73, 74). RNAs containing chemically modified nucleotides fail to trigger RLRs, and m6A is a functional modification (40, 89, 90). However, the mechanism of how m6A-modified RNA escapes RLR sensing remains unclear. Qiu suggested that m6A modification impairs the conformation of duplex structures in viral RNAs and interferes with sensing by intracellular receptor RLRs; finally, m6A attenuates innate immune response and facilitates immune invasion (74).
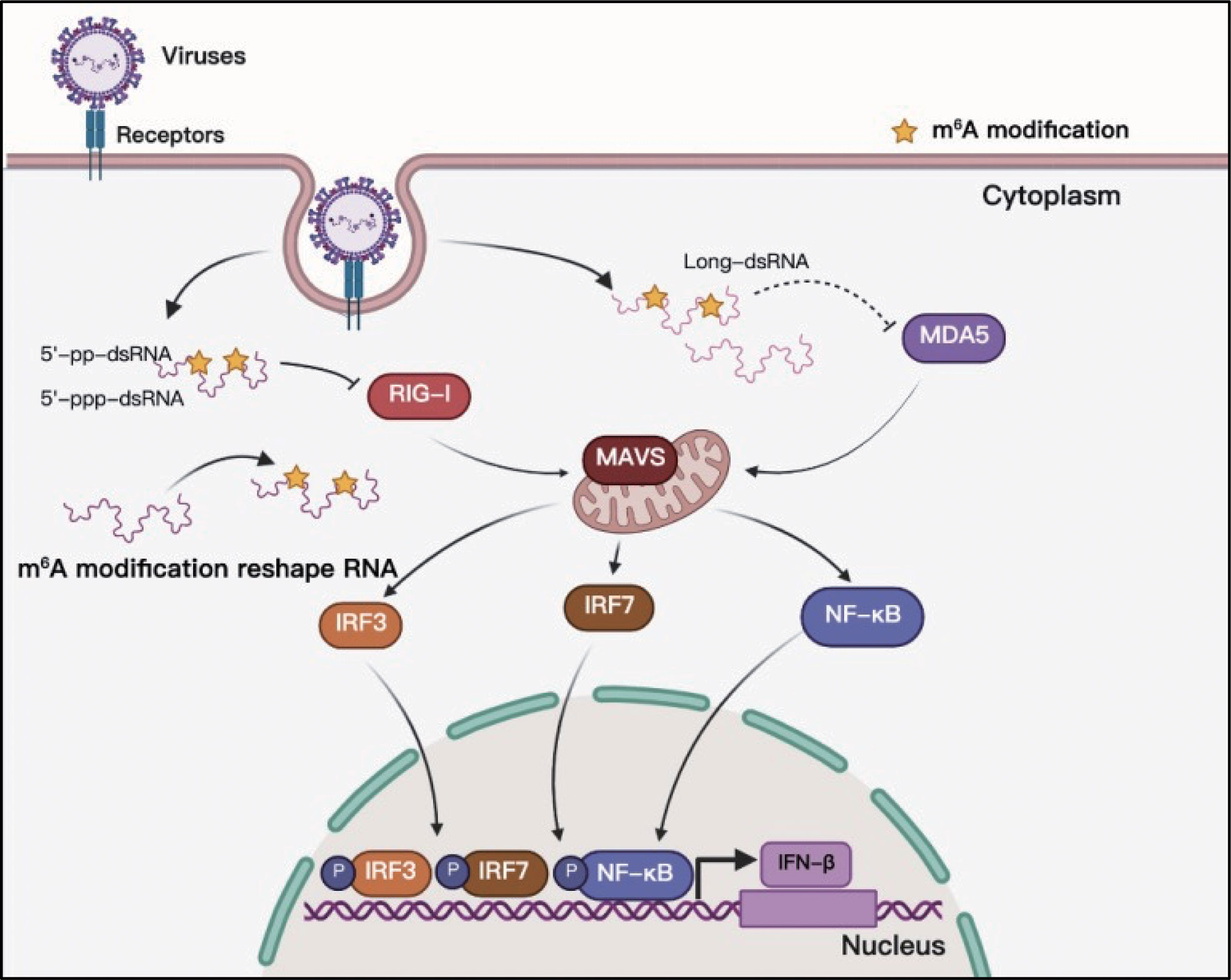
Figure 6 M6A modifications of viral RNAs and their function in RLR sensing. Viral RNAs can form complicated secondary structures, and RLRs can recognize the double-stranded component. Adding m6A to viral RNAs can reshape RNA structure and enable viral RNAs to escape RLR recognition.
We have concluded from previous studies that the addition of m6A to viral RNAs has both promotive and suppressive functions in the viral life cycle and plays an important role in immune escape from RLRs. M6A promotes the replication of DNA viruses, including HSV-1, SV40, AdV, and HPV-16 (30–32, 34), and it has also been shown to positively regulate infection by many RNA viruses, such as EV71, HRSV, HMPV, VSV, and IAV (64, 65, 72–75). By contrast, m6A suppresses the replication of DNA viruses, such as BmNPV, and RNA viruses, including ERVs, Flaviviridae, and Coronaviridae (62, 63, 66, 67, 71, 81). Importantly, m6A can function as both a proviral and antiviral regulator in the life cycle of some viruses, such as KSHV, EBV, HBV, and HIV-1 (23, 25–28, 35–37, 76–80). The reason why the effect of m6A varies between different viruses is uncertain. We think that this is because RNAs of different viruses interact with different ‘readers’.
To date, studies have indicated that m6A in viral RNAs could reduce the sensing activity of RLRs and help viruses escape innate immune recognition during viral invasion (38–40, 69, 73, 74). However, other studies have revealed that viral invasion can cause changes in the expression of ‘writers’, ‘readers’, or ‘erasers’, resulting in expression changes in immunoregulatory proteins and eventually influencing IFN production. M6A targeting of IFN-β can enhance the destabilization of IFN-β mRNA and restrict the duration of the antiviral response (61). Degradation of WTAP induced by viral infection reduces the m6A levels of interferon-regulatory factor 3 (IRF3) and interferon α/β receptor subunit 1 (IFNAR1) mRNAs, resulting in the suppression of IRF3 translation and destabilization of IFNAR1 mRNA (91). Kastan’s work revealed that the RNA-binding protein YTHDF3 promotes the production of interferon-stimulated genes (ISGs); however, Zhang’s work indicated that YTHDF3 functions as a negative regulator of antiviral immunity by promoting the translation of FOXO3 mRNA (92, 93). METTL3, METTL14, and YTHDF1 promote the expression of interferon-induced transmembrane 1 (IFITM1), a well-known ISG (94). A recent study by You’s group reported that m6A can stabilize IRF3 mRNA, and Zhu’s group demonstrated that m6A can increase the stability of interferon-regulatory factor 7 (IRF7) mRNA (95, 96). As a result, the expression of type I IFNs is enhanced. Therefore, m6A has multiple functions in the viral replication process and modulates the antiviral response of type I IFNs.
As m6A is present in the life cycle of many viruses, drugs targeting this pathway may have the potential to act as antiviral drugs. For example, 3-deazaadenosine (DAA), an m6A modification inhibitor, inhibits the replication of various viruses in vitro or in vivo, including HRSV, parainfluenza virus type 3 (PIV3), Ebola virus, HIV, and IAV (75, 76, 97, 98). The SARS-CoV-2 pandemic is still ongoing, and studies have provided a proof of concept suggesting that targeting of the cellular components of the m6A RNA modification pathway could lead to novel therapeutic opportunities to control this viral pathogen.
In general, the study of viral m6A epitranscriptomics, which started in the early 1970s, has rapidly evolved in the past 5 years, and indicates that m6A modification is an important component in viral infections and innate immunity recognition. Importantly, there is a need for a clear mechanistic understanding of m6A modifications in viral RNAs to determine their function in viral replication, and to explore their potential as antiviral targets.
HL, ML, and WQ conceptualized the review. HL and YG wrote the manuscript. HL, WQ, and ML revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (32102662), the China Postdoctoral Science Foundation Funded Project (2021M691082), and the Young Scholars of Yangtze River Scholar Professor Program (2019, WQ).
All the pictures were created using BioRender (https://biorender.com/), and we express our special appreciation to the creators of this application.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from novikoff hepatoma cells. Proc Natl Acad Sci USA (1974) 71(10):3971–5. doi: 10.1073/pnas.71.10.3971
2. Bokar JA, Rath-Shambaugh ME, Ludwiczak R, Narayan P, Rottman F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. internal mRNA methylation requires a multisubunit complex. J Biol Chem (1994) 269(26):17697–704. doi: 10.1016/s0021-9258(17)32497-3
3. Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA (1997) 3(11):1233–47.
4. Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat Rev Genet (2014) 15(5):293–306. doi: 10.1038/nrg3724
5. Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol (2014) 15(5):313–26. doi: 10.1038/nrm3785
6. Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol Life Sci (2001) 58(2):266–77. doi: 10.1007/PL00000854
7. David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-myc deregulate pyruvate kinase mRNA splicing in cancer. Nature (2010) 463(7279):364–8. doi: 10.1038/nature08697
8. Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol (2010) 17(7):909–15. doi: 10.1038/nsmb.1838
9. Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, et al. RNA-Methylation-dependent RNA processing controls the speed of the circadian clock. Cell (2013) 155(4):793–806. doi: 10.1016/j.cell.2013.10.026
10. Schwartz S, Agarwala SD, Mumbach MR, Jovanovic M, Mertins P, Shishkin A, et al. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell (2013) 155(6):1409–21. doi: 10.1016/j.cell.2013.10.047
11. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell (2013) 49(1):18–29. doi: 10.1016/j.molcel.2012.10.015
12. Wan Y, Qu K, Zhang QC, Flynn RA, Manor O, Ouyang Z, et al. Landscape and variation of RNA secondary structure across the human transcriptome. Nature (2014) 505(7485):706–9. doi: 10.1038/nature12946
13. Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature (2014) 505(7481):117–20. doi: 10.1038/nature12730
14. Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science (2015) 347(6225):1002–6. doi: 10.1126/science.1261417
15. Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A, et al. A majority of m6A residues are in the last exons, allowing the potential for 3' UTR regulation. Genes Dev (2015) 29(19):2037–53. doi: 10.1101/gad.269415.115
16. Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, et al. 5' UTR m(6)A promotes cap-independent translation. Cell (2015) 163(4):999–1010. doi: 10.1016/j.cell.2015.10.012
17. Roost C, Lynch SR, Batista PJ, Qu K, Chang HY, Kool ET. Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. J Am Chem Soc (2015) 137(5):2107–15. doi: 10.1021/ja513080v
18. Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung JW, et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature (2015) 519(7544):486–90. doi: 10.1038/nature14263
19. Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun (2016) 7:12626. doi: 10.1038/ncomms12626
20. Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell (2016) 62(3):335–45. doi: 10.1016/j.molcel.2016.03.021
21. McIntyre ABR, Gokhale NS, Cerchietti L, Jaffrey SR, Horner SM, Mason CE. Limits in the detection of m(6)A changes using MeRIP/m(6)A-seq. Sci Rep (2020) 10(1):6590. doi: 10.1038/s41598-020-63355-3
22. Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods (2015) 12(8):767–72. doi: 10.1038/nmeth.3453
23. Ye F, Chen ER, Nilsen TW. Kaposi's sarcoma-associated herpesvirus utilizes and manipulates RNA N(6)-adenosine methylation to promote lytic replication. J Virol (2017) 91(16):e00466–17. doi: 10.1128/JVI.00466-17
24. Hesser CR, Karijolich J, Dominissini D, He C, Glaunsinger BA. N6-methyladenosine modification and the YTHDF2 reader protein play cell type specific roles in lytic viral gene expression during kaposi's sarcoma-associated herpesvirus infection. PLoS Pathog (2018) 14(4):e1006995. doi: 10.1371/journal.ppat.1006995
25. Tan B, Liu H, Zhang S, da Silva SR, Zhang L, Meng J, et al. Viral and cellular N(6)-methyladenosine and N(6),2'-o-dimethyladenosine epitranscriptomes in the KSHV life cycle. Nat Microbiol (2018) 3(1):108–20. doi: 10.1038/s41564-017-0056-8
26. Baquero-Perez B, Antanaviciute A, Yonchev ID, Carr IM, Wilson SA, Whitehouse A. The Tudor SND1 protein is an m(6)A RNA reader essential for replication of kaposi's sarcoma-associated herpesvirus. Elife (2019) 8:e47261. doi: 10.7554/eLife.47261
27. Lang F, Singh RK, Pei Y, Zhang S, Sun K, Robertson ES. EBV epitranscriptome reprogramming by METTL14 is critical for viral-associated tumorigenesis. PLoS Pathog (2019) 15(6):e1007796. doi: 10.1371/journal.ppat.1007796
28. Zheng X, Wang J, Zhang X, Fu Y, Peng Q, Lu J, et al. RNA m(6) a methylation regulates virus-host interaction and EBNA2 expression during Epstein-Barr virus infection. Immun Inflamm Dis (2021) 9(2):351–62. doi: 10.1002/iid3.396
29. Xia TL, Li X, Wang X, Zhu YJ, Zhang H, Cheng W, et al. N(6)-methyladenosine-binding protein YTHDF1 suppresses EBV replication and promotes EBV RNA decay. EMBO Rep (2021) 22(4):e50128. doi: 10.15252/embr.202050128
30. Feng Z, Zhou F, Tan M, Wang T, He ML. Targeting m6A modification inhibits herpes virus 1 infection. Genes Dis (2021) 9(4):1114–1128. doi: 10.1016/j.gendis.2021.02.004
31. Tsai K, Courtney DG, Cullen BR. Addition of m6A to SV40 late mRNAs enhances viral structural gene expression and replication. PLoS Pathog (2018) 14(2):e1006919. doi: 10.1371/journal.ppat.1006919
32. Price AM, Hayer KE, McIntyre ABR, Gokhale NS, Abebe JS, Della Fera AN, et al. Direct RNA sequencing reveals m(6)A modifications on adenovirus RNA are necessary for efficient splicing. Nat Commun (2020) 11(1):6016. doi: 10.1038/s41467-020-19787-6
33. Zhang X, Zhang Y, Dai K, Liang Z, Zhu M, Pan J, et al. N (6)-methyladenosine level in silkworm Midgut/Ovary cell line is associated with bombyx mori nucleopolyhedrovirus infection. Front Microbiol (2019) 10:2988. doi: 10.3389/fmicb.2019.02988
34. Cui X, Nilsson K, Kajitani N, Schwartz S. Overexpression of m6A-factors METTL3, ALKBH5, and YTHDC1 alters HPV16 mRNA splicing. Virus Genes. (2022) 58(2):98–112. doi: 10.1007/s11262-022-01889-6
35. Imam H, Khan M, Gokhale NS, McIntyre ABR, Kim GW, Jang JY, et al. N6-methyladenosine modification of hepatitis b virus RNA differentially regulates the viral life cycle. Proc Natl Acad Sci USA (2018) 115(35):8829–34. doi: 10.1073/pnas.1808319115
36. Kim GW, Imam H, Siddiqui A. The RNA binding proteins YTHDC1 and FMRP regulate the nuclear export of N(6)-Methyladenosine-Modified hepatitis b virus transcripts and affect the viral life cycle. J Virol (2021) 95(13):e0009721. doi: 10.1128/JVI.00097-21
37. Imam H, Kim GW, Mir SA, Khan M, Siddiqui A. Interferon-stimulated gene 20 (ISG20) selectively degrades N6-methyladenosine modified hepatitis b virus transcripts. PLoS Pathog (2020) 16(2):e1008338. doi: 10.1371/journal.ppat.1008338
38. Kim GW, Imam H, Khan M, Siddiqui A. N (6)-methyladenosine modification of hepatitis b and c viral RNAs attenuates host innate immunity via RIG-I signaling. J Biol Chem (2020) 295(37):13123–33. doi: 10.1074/jbc.RA120.014260
39. Chen S, Kumar S, Espada CE, Tirumuru N, Cahill MP, Hu L, et al. N6-methyladenosine modification of HIV-1 RNA suppresses type-I interferon induction in differentiated monocytic cells and primary macrophages. PLoS Pathog (2021) 17(3):e1009421. doi: 10.1371/journal.ppat.1009421
40. Lu M, Xue M, Wang HT, Kairis EL, Ahmad S, Wei J, et al. Nonsegmented negative-sense RNA viruses utilize n (6)-methyladenosine (m(6)A) as a common strategy to evade host innate immunity. J Virol (2021) 95(9):e01939–20. doi: 10.1128/JVI.01939-20
41. Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev (2001) 14(4):778–809. doi: 10.1128/CMR.14.4.778-809.2001
42. Hartmann G. Nucleic acid immunity. Adv Immunol (2017) 133:121–69. doi: 10.1016/bs.ai.2016.11.001
43. Yoneyama M, Fujita T. Structural mechanism of RNA recognition by the RIG-i-like receptors. Immunity (2008) 29(2):178–81. doi: 10.1016/j.immuni.2008.07.009
44. Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev (2009) 227(1):75–86. doi: 10.1111/j.1600-065X.2008.00737.x
45. Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, et al. RIG-i-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science (2006) 314(5801):997–1001. doi: 10.1126/science.1132998
46. Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, et al. Cytosolic viral sensor RIG-I is a 5'-triphosphate-dependent translocase on. Science (2009) 323(5917):1070–4. doi: 10.1126/science.1168352
47. Goubau D, Schlee M, Deddouche S, Pruijssers AJ, Zillinger T, Goldeck M, et al. Antiviral immunity via RIG-i-mediated recognition of RNA bearing 5'-diphosphates. Nature (2014) 514(7522):372–5. doi: 10.1038/nature13590
48. Berke IC, Modis Y. MDA5 cooperatively forms dimers and ATP-sensitive filaments upon binding double-stranded RNA. EMBO J (2012) 31(7):1714–26. doi: 10.1038/emboj.2012.19
49. Peisley A, Jo MH, Lin C, Wu B, Orme-Johnson M, Walz T, et al. Kinetic mechanism for viral dsRNA length discrimination by MDA5 filaments. Proc Natl Acad Sci USA (2012) 109(49):E3340–49. doi: 10.1073/pnas.1208618109
50. Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, et al. LGP2 is a positive regulator of RIG-i- and MDA5-mediated antiviral responses. Proc Natl Acad Sci USA (2010) 107(4):1512–7. doi: 10.1073/pnas.0912986107
51. Bruns AM, Leser GP, Lamb RA, Horvath CM. The innate immune sensor LGP2 activates antiviral signaling by regulating MDA5-RNA interaction and filament assembly. Mol Cell (2014) 55(5):771–81. doi: 10.1016/j.molcel.2014.07.003
52. Duic I, Tadakuma H, Harada Y, Yamaue R, Deguchi K, Suzuki Y, et al. Viral RNA recognition by LGP2 and MDA5, and activation of signaling through step-by-step conformational changes. Nucleic Acids Res (2020) 48(20):11664–74. doi: 10.1093/nar/gkaa935
53. Sommer S, Salditt-Georgieff M, Bachenheimer S, Darnell JE, Furuichi Y, Morgan M, et al. The methylation of adenovirus-specific nuclear and cytoplasmic RNA. Nucleic Acids Res (1976) 3(3):749–65. doi: 10.1093/nar/3.3.749
54. Anderson J, Phan L, Cuesta R, Carlson BA, Pak M, Asano K, et al. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev (1998) 12(23):3650–62. doi: 10.1101/gad.12.23.3650
55. Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature (2014) 515(7525):143–6. doi: 10.1038/nature13802
56. Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, Leon-Ricardo BX, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell (2014) 159(1):148–62. doi: 10.1016/j.cell.2014.08.028
57. Patil DP, Pickering BF, Jaffrey SR. Reading m(6)A in the transcriptome: m(6)A-binding proteins. Trends Cell Biol (2018) 28(2):113–27. doi: 10.1016/j.tcb.2017.10.001
58. He C. Grand challenge commentary: RNA epigenetics? Nat Chem Biol (2010) 6(12):863–5. doi: 10.1038/nchembio.482
59. Krug RM, Morgan MA, Shatkin AJ. Influenza viral mRNA contains internal N6-methyladenosine and 5'-terminal 7-methylguanosine in cap structures. J Virol (1976) 20(1):45–53. doi: 10.1128/JVI.20.1.45-53.1976
60. Rubio RM, Depledge DP, Bianco C, Thompson L, Mohr I. RNA m(6) a modification enzymes shape innate responses to DNA by regulating interferon beta. Genes Dev (2018) 32(23-24):1472–84. doi: 10.1101/gad.319475.118
61. Winkler R, Gillis E, Lasman L, Safra M, Geula S, Soyris C, et al. m(6)A modification controls the innate immune response to infection by targeting type I interferons. Nat Immunol (2019) 20(2):173–82. doi: 10.1038/s41590-018-0275-z
62. Gokhale NS, McIntyre ABR, McFadden MJ, Roder AE, Kennedy EM, Gandara JA, et al. N6-methyladenosine in flaviviridae viral RNA genomes regulates infection. Cell Host Microbe (2016) 20(5):654–65. doi: 10.1016/j.chom.2016.09.015
63. Lichinchi G, Zhao BS, Wu Y, Lu Z, Qin Y, He C, et al. Dynamics of human and viral RNA methylation during zika virus infection. Cell Host Microbe (2016) 20(5):666–73. doi: 10.1016/j.chom.2016.10.002
64. Hao H, Hao S, Chen H, Chen Z, Zhang Y, Wang J, et al. N6-methyladenosine modification and METTL3 modulate enterovirus 71 replication. Nucleic Acids Res (2019) 47(1):362–74. doi: 10.1093/nar/gky1007
65. Yao M, Dong Y, Wang Y, Liu H, Ma H, Zhang H, et al. N(6)-methyladenosine modifications enhance enterovirus 71 ORF translation through METTL3 cytoplasmic distribution. Biochem Biophys Res Commun (2020) 527(1):297–304. doi: 10.1016/j.bbrc.2020.04.088
66. Liu J, Xu YP, Li K, Ye Q, Zhou HY, Sun H, et al. The m(6)A methylome of SARS-CoV-2 in host cells. Cell Res (2021) 31(4):404–14. doi: 10.1038/s41422-020-00465-7
67. Gokhale NS, McIntyre ABR, Mattocks MD, Holley CL, Lazear HM, Mason CE, et al. Altered m(6)A modification of specific cellular transcripts affects flaviviridae infection. Mol Cell (2020) 77(3):542–55.e548. doi: 10.1016/j.molcel.2019.11.007
68. Burgess HM, Depledge DP, Thompson L, Srinivas KP, Grande RC, Vink EI, et al. Targeting the m(6)A RNA modification pathway blocks SARS-CoV-2 and HCoV-OC43 replication. Genes Dev (2021) 35(13-14):1005–19. doi: 10.1101/gad.348320.121
69. Li N, Hui H, Bray B, Gonzalez GM, Zeller M, Anderson KG, et al. METTL3 regulates viral m6A RNA modification and host cell innate immune responses during SARS-CoV-2 infection. Cell Rep (2021) 35(6):109091. doi: 10.1016/j.celrep.2021.109091
70. Campos JHC, Maricato JT, Braconi CT, Antoneli F, Janini LMR, Briones MRS. Direct RNA sequencing reveals SARS-CoV-2 m6A sites and possible differential DRACH motif methylation among variants. Viruses (2021) 13(11):2108. doi: 10.3390/v13112108
71. Chen J, Jin L, Wang Z, Wang L, Chen Q, Cui Y, et al. N6-methyladenosine regulates PEDV replication and host gene expression. Virology (2020) 548:59–72. doi: 10.1016/j.virol.2020.06.008
72. Xue M, Zhao BS, Zhang Z, Lu M, Harder O, Chen P, et al. Viral N(6)-methyladenosine upregulates replication and pathogenesis of human respiratory syncytial virus. Nat Commun (2019) 10(1):4595. doi: 10.1038/s41467-019-12504-y
73. Lu M, Zhang Z, Xue M, Zhao BS, Harder O, Li A, et al. N(6)-methyladenosine modification enables viral RNA To escape recognition by RNA sensor RIG-I. Nat Microbiol (2020) 5(4):584–98. doi: 10.1038/s41564-019-0653-9
74. Qiu W, Zhang Q, Zhang R, Lu Y, Wang X, Tian H, et al. N(6)-methyladenosine RNA modification suppresses antiviral innate sensing pathways via reshaping. Nat Commun (2021) 12(1):1582. doi: 10.1038/s41467-021-21904-y
75. Courtney DG, Kennedy EM, Dumm RE, Bogerd HP, Tsai K, Heaton NS, et al. Epitranscriptomic enhancement of influenza a virus gene expression and replication. Cell Host Microbe (2017) 22(3):377–86.e375. doi: 10.1016/j.chom.2017.08.004
76. Kennedy EM, Bogerd HP, Kornepati AV, Kang D, Ghoshal D, Marshall JB, et al. Posttranscriptional m(6)A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microbe (2016) 19(5):675–85. doi: 10.1016/j.chom.2016.04.002
77. Lichinchi G, Gao S, Saletore Y, Gonzalez GM, Bansal V, Wang Y, et al. Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat Microbiol (2016) 1:16011. doi: 10.1038/nmicrobiol.2016.11
78. Tirumuru N, Zhao BS, Lu W, Lu Z, He C, Wu L. N(6)-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 gag protein expression. Elife (2016) 5:e15528. doi: 10.7554/eLife.15528
79. Jurczyszak D, Zhang W, Terry SN, Kehrer T, Bermudez Gonzalez MC, McGregor E, et al. HIV Protease cleaves the antiviral m6A reader protein YTHDF3 in the viral particle. PLoS Pathog (2020) 16(2):e1008305. doi: 10.1371/journal.ppat.1008305
80. Lu W, Tirumuru N, St Gelais C, Koneru PC, Liu C, Kvaratskhelia M, et al. N(6)-methyladenosine-binding proteins suppress HIV-1 infectivity and viral production. J Biol Chem (2018) 293(34):12992–3005. doi: 10.1074/jbc.RA118.004215
81. Chelmicki T, Roger E, Teissandier A, Dura M, Bonneville L, Rucli S, et al. m(6)A RNA methylation regulates the fate of endogenous retroviruses. Nature (2021) 591(7849):312–6. doi: 10.1038/s41586-020-03135-1
82. Thomason AR, Brian DA, Velicer LF, Rottman FM. Methylation of high-molecular-weight subunit RNA of feline leukemia virus. J Virol (1976) 20(1):123–32. doi: 10.1128/JVI.20.1.123-132.1976
83. Dimock K, Stoltzfus CM. Sequence specificity of internal methylation in B77 avian sarcoma virus RNA subunits. Biochemistry (1977) 16(3):471–8. doi: 10.1021/bi00622a021
84. Kane SE, Beemon K. Precise localization of m6A in rous sarcoma virus RNA reveals clustering of methylation sites: implications for RNA processing. Mol Cell Biol (1985) 5(9):2298–306. doi: 10.1128/mcb.5.9.2298-2306.1985
85. Johnson WE. Origins and evolutionary consequences of ancient endogenous retroviruses. Nat Rev Microbiol (2019) 17(6):355–70. doi: 10.1038/s41579-019-0189-2
86. Di Giorgio E, Xodo LE. Endogenous retroviruses (ERVs) : Does (RIG-I-Like receptors)-MAVS pathway directly control senescence and aging as a consequence of ERV de-repression? Front Immunol (2022) 13:917998. doi: 10.3389/fimmu.2022.917998
87. Scott I. Degradation of RIG-I following cytomegalovirus infection is independent of apoptosis. Microbes Infect (2009) 11(12):973–9. doi: 10.1016/j.micinf.2009.07.001
88. Kostyusheva A, Brezgin S, Glebe D, Kostyushev D, Chulanov V. Host-cell interactions in HBV infection and pathogenesis: the emerging role of m6A modification. Emerg Microbes Infect (2021) 10(1):2264–75. doi: 10.1080/22221751.2021.2006580
89. Durbin AF, Wang C, Marcotrigiano J, Gehrke L. RNAs containing modified nucleotides fail to trigger RIG-I conformational changes for innate immune signaling. mBio (2016) 7(5):e00833–16. doi: 10.1128/mBio.00833-16
90. Tong J, Zhang W, Chen Y, Yuan Q, Qin NN, Qu G. The emerging role of RNA modifications in the regulation of antiviral innate immunity. Front Microbiol (2022) 13:845625. doi: 10.3389/fmicb.2022.845625
91. Ge Y, Ling T, Wang Y, Jia X, Xie X, Chen R, et al. Degradation of WTAP blocks antiviral responses by reducing the m(6) a levels of IRF3 and IFNAR1 mRNA. EMBO Rep (2021) 22(11):e52101. doi: 10.15252/embr.202052101
92. Zhang Y, Wang X, Zhang X, Wang J, Ma Y, Zhang L, et al. RNA-Binding protein YTHDF3 suppresses interferon-dependent antiviral responses by promoting FOXO3 translation. Proc Natl Acad Sci USA (2019) 116(3):976–81. doi: 10.1073/pnas.1812536116
93. Kastan JP, Tremblay MW, Brown MC, Trimarco JD, Dobrikova EY, Dobrikov MI, et al. Enterovirus 2A(pro) cleavage of the YTHDF m(6)A readers implicates YTHDF3 as a mediator of type I interferon-driven JAK/STAT signaling. mBio (2021) 12(2):e00116–21. doi: 10.1128/mBio.00116-21
94. McFadden MJ, McIntyre ABR, Mourelatos H, Abell NS, Gokhale NS, Ipas H, et al. Post-transcriptional regulation of antiviral gene expression by N6-methyladenosine. Cell Rep (2021) 34(9):108798. doi: 10.1016/j.celrep.2021.108798
95. Chen J, Wei X, Wang X, Liu T, Zhao Y, Chen L, et al. TBK1-METTL3 axis facilitates antiviral immunity. Cell Rep (2022) 38(7):110373. doi: 10.1016/j.celrep.2022.110373
96. Wang A, Tao W, Tong J, Gao J, Wang J, Hou G, et al. m6A modifications regulate intestinal immunity and rotavirus infection. Elife (2022) 11:e73628. doi: 10.7554/eLife.73628
97. Wyde PR, Ambrose MW, Meyer HL, Zolinski CL, Gilbert BE. Evaluation of the toxicity and antiviral activity of carbocyclic 3-deazaadenosine against respiratory syncytial and parainfluenza type 3 viruses in tissue culture and in cotton rats. Antiviral Res (1990) 14(4-5):215–25. doi: 10.1016/0166-3542(90)90003-p
Keywords: m6A, viruses, replication, RIG-I-like receptors, innate immune escape
Citation: Li H, Guo Y, Qi W and Liao M (2022) N6-methyladenosine modification of viral RNA and its role during the recognition process of RIG-I-like receptors. Front. Immunol. 13:1031200. doi: 10.3389/fimmu.2022.1031200
Received: 29 August 2022; Accepted: 17 November 2022;
Published: 13 December 2022.
Edited by:
Rúbia Marília De Medeiros, Federal University of Rio Grande do Sul, BrazilReviewed by:
Elias A. Said, Sultan Qaboos University, OmanCopyright © 2022 Li, Guo, Qi and Liao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenbao Qi, cWl3ZW5iYW9Ac2NhdS5lZHUuY24=; Ming Liao, bWxpYW9Ac2NhdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.