
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 07 November 2022
Sec. Cancer Immunity and Immunotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.1028013
CD160 is a glycosylphosphatidylinositol (GPI)-anchored cell surface glycoprotein expressed on cytotoxic natural killer (NK) cells and T-cell subsets. It plays a crucial role in the activation of NK-cell cytotoxicity and cytokine production. It also modulates the immune system and is involved in some pathologies, such as cancer. CD160 is abnormally expressed in B-cell chronic lymphocytic leukemia (CLL) but not expressed in normal B lymphocytes. Its expression in CLL enhances tumor cell proliferation and resistance to apoptosis. CD160 is also a potential prognostic marker for the detection of minimal residual disease (MRD) in CLL, which is important for the clinical management of CLL, the prevention of disease relapse, and the achievement of complete remission. In this review, we present an overview of CD160 and its involvement in the pathophysiology of CLL. We also discuss its use as a prognostic marker for the assessment of MRD in CLL.
CD160 is a glycophosphatidylinositol (GPI)-anchored surface membrane protein that was initially identified as BY55 by Maïza et al. on cytotoxic natural killer (NK) cells, where it activates NK-cell cytotoxicity and cytokine production (1–3). CD160 is also expressed by some subsets of T cells and activated endothelial cells, in which it regulates cell activation and apoptosis, respectively (4–7). Thus, CD160 is involved in antitumor immunity (3) and protection during chronic infections (8, 9). In general, CD160 has been reported to be involved in the development of some pathologies, including autoimmune diseases (10), inflammatory diseases (11), atherosclerosis (12), retinal vascular diseases (7), chronic viral infections (8, 13–15), and cancer (16–18).
CD160 is abnormally overexpressed in B-chronic lymphocytic leukemia (CLL), which is the most frequently diagnosed leukemia in developed countries (19, 20). The disease is characterized by the clonal expansion and accumulation of small mature-like CD5+ CD23+ B cells in the blood, bone marrow, and secondary lymphoid tissues (21). Different factors have been implicated in CLL progression. First, the genetic and epigenetic profiles of CLL B cells are altered (22, 23). Second, CLL B cells interact with neighboring cells in their microenvironment, such as nurse-like cells (24–28), endothelial cells (29–31), mesenchymal stromal cells (32), and T cells (33–35). These cells play a crucial role in the maintenance of CLL B-cell proliferation and CLL progression, the enhancement of prosurvival signaling, and the induction of resistance to drug-induced apoptosis (36). The microenvironment is also characterized by exhausted cytotoxic T cells and NK cells, allowing CLL B cells to escape the immune system (33, 37–39). Third, B-cell receptor (BCR) signaling plays a key pathological role by activating the signaling pathways implicated in CLL cell survival, metabolism, proliferation, and resistance to apoptosis (40–42). Furthermore, CLL is characterized by the expression of several markers specific to certain immune cells, such as ζ-associated protein kinase 70 (ZAP-70) (43) and lymphocyte-specific tyrosine kinase (Lck) (44), which favor CLL cell survival, and CD5, which maintains CLL cell anergy (45). CD160 is also a crucial marker of CLL and a key activator of CLL cell survival and resistance to apoptosis (46).
Nonetheless, CLL remains an incurable disease. It can be treated by chemoimmunotherapy (CIT) using a combination of fludarabine, cyclophosphamide, and rituximab (FCR) or chlorambucil and obinutuzumab (CLBO) (47) or by targeted therapy using small-molecule inhibitors targeting various mediators involved in BCR signaling, such as BTK inhibitors (BTKis) (ibrutinib and acalabrutinib) (48), PI3K inhibitors (PI3Kis) (idelalisib and duvelisib) (49), and Bcl-2 (B-cell lymphoma 2) antagonists (venetoclax) (50). These molecules can be prescribed as frontline or second-line treatments depending on the existence/type of IGHV and p53 mutations (47). They have improved the prognosis of patients for whom CIT is not suited (47). However, CLL patients can develop resistance to these treatments due to secondary mutations in the proteins targeted by the drugs and the development of alternative bypass pathways (51). In addition, resistance can be caused by the activation of survival and antiapoptotic signaling pathways triggered through crosstalk between CLL components and surrounding cells in the microenvironment (51). Though these treatments can improve survival rates, some of them have adverse effects (52, 53). Therefore, for clinical management of this disease, it is important to establish a good follow-up strategy to analyze the development or regression of CLL in patients after treatment. Hence, the assessment of minimal residual disease remains crucial for the achievement of complete remission, relapse prevention, and prolonged survival.
The restricted expression of CD160 in CLL and lack of CD160 expression in normal B cells make it a good potential diagnostic marker but also a prognostic marker for the detection of minimal residual disease in CLL (54). In this review, we discuss the characteristics of CD160, its expression in the CLL tumor microenvironment, and its roles in the pathophysiological processes of CLL. We also report the use of CD160 as a potential prognostic marker for the assessment of minimal residual disease in CLL.
CD160 is a receptor that is capable of delivering both stimulatory and inhibitory signals depending on the type of cell in which it is expressed (4, 55). Its gene is located on the human 1q21.1 chromosome. It contains 6 exons, with exons 1 and 2 being untranslated (56) (Figure 1). A core promoter sequence containing a single transcription start site (TSS) is located upstream of exon 1 (56). This sequence is homologous to three highly conserved transcription factor-binding sites for FREAC-4, SOX17, and acute myelogenous leukemia-1 (AML-1) (also called RUNX1 or CBFα). The AML-1 binding site is the only one that has been implicated in the regulation of CD160 expression (56). However, data explaining the functional properties of this binding site and the mechanism by which CD160 expression is controlled in immune cells are needed.
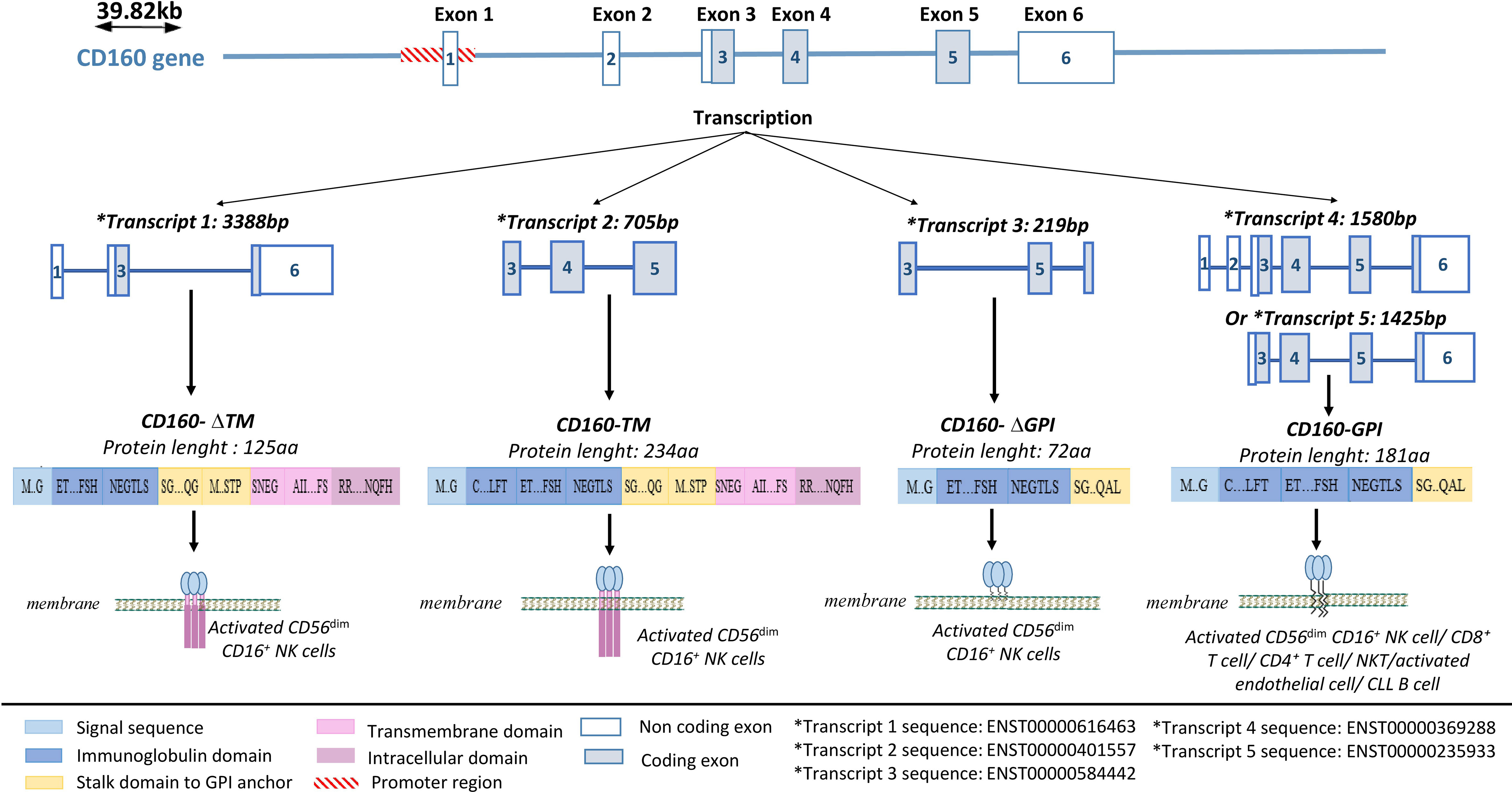
Figure 1 CD160 gene structure and isoforms. CD160 has four isoforms: a transmembrane isoform (CD160-TM), a glycophosphatidylinositol-anchored isoform (CD160-GPI), and two truncated isoforms (CD160-ΔIg-GPI and CD160-ΔIg-TM) that are generated through alternative splicing. Adapted from (57) and analyzed on Ensembl.
Four CD160 isoforms are generated by alternative splicing, and they differ based on the presence or absence of a GPI anchor, an Ig domain, and transmembrane and cytoplasmic domains (57); a transmembrane form (CD160-TM), two truncated forms (CD160-ΔIg-GPI and CD160-ΔIg-TM), a GPI-anchored membrane form (CD160-GPI), and a soluble form (sCD160) (57, 58) (Figure 1).
CD160-GPI has a single Ig-like domain that is weakly homologous to the KIR2DL4 receptor and is expressed in peripheral blood (PB) CD56dim CD16+ NK-cell subsets (59, 60), cytotoxic CD8+ T cells (59), activated endothelial cells (7, 61), a small fraction of CD4+ T cells (4) and γδ-T cells (1). CD160-TM is selectively expressed by activated CD56dim CD16+ NK cells (57). Its expression is activation-dependent and amplifies NK-cell cytotoxicity (57). CD4+ and CD8+ T cells can also express CD160-TM, but only at the transcriptional level and not at the protein level (5). Soluble CD160 can be secreted by human PB-NK cells (58), murine splenic CD8+ T cells (62), and human and murine mast cells (63). In some pathological conditions, sCD160 can also be expressed by tumor cells, such as melanoma cells (18) (Table 1).
Both human and murine CD160 exhibit a broad but weak specificity for classical and nonclassical MHC-I molecules (73). For instance, human CD160 preferentially binds to HLA-C (3) and soluble HLA-G (61). CD160 can also bind to HLA-A2 tetramers, HLA-E, and HLA-B7 (74). In addition, CD160 binds to herpesvirus entry mediator (HVEM), which is in the tumor necrosis factor (TNF) superfamily (75). The CD160-TM isoform binds less robustly to HVEM (5).
CD160 plays a major role in the induction of CD56dim CD16+ NK-cell cytotoxicity and degranulation (60) (Table 1). However, it is not expressed on CD56bright CD16⁻ NK cells, which exert immunoregulatory functions through the expression of high levels of cytokines and chemokines (60). Upon engagement with MHC-I molecules, HVEM or agonistic monoclonal anti-CD160 antibodies, including BY55 and CL1-R2, on PB-CD56dim CD16+ NK cells, CD160-GPI activates cytotoxic functions and enhances the production of cytokines, including interferon-γ (IFN-γ), TNF-α, interleukin-6 (IL-6), IL-8 and MIP-1b, which induce target cell death (3, 60, 64). The CD160 cytotoxic effect does not depend on cross-linking with other activating NK-cell receptors, such as activating killer Ig-like receptors (KIRs) or CD94/NK2C, and its effect is similar to that mediated by CD16 (60). In addition, the expression of the CD160-TM isoform by activated NK cells can exacerbate toxicity (57). CD160 mediates cytotoxicity through the recruitment of spleen-associated tyrosine kinase (Syk) (55), which activates the PI3K/Akt/mTORC1 signaling pathway, promoting translation through the upregulation of 4EBP1 and S6 ribosomal kinase expression (55, 76). Furthermore, the PI3K/Akt pathway can also activate NK-cell migration through the phosphorylation of MEK1/2-ERK, which has been implicated in actin reorganization and cell polarization (55, 77). CD160-GPI does not contain immunoreceptor tyrosine-based motifs (ITAMs), and it is not yet clear how it mediates signal transduction (60). Le Bouteiller et al. suggested that CD160 may associate with adaptor proteins containing ITAMs in lipid rafts to initiate its downstream signaling pathway (60). However, Rabot et al. demonstrated that CD160 was unable to associate with DAP10 or DAP12 signaling adaptors (55). In addition, it has been reported that CD160-TM activates NK-cell-activating signaling pathways through its phosphorylated Y225 residue located on its intracellular motif, which can interact with intracellular signaling proteins (57). However, further studies are required to determine how CD160 signaling is mediated. Furthermore, the function of the two truncated isoforms, CD160-ΔIg-GPI and CD160-ΔIg-TM, has not yet been described. Both isoforms have only been detected at the transcriptional level. Therefore, we still do not know whether CD160-ΔIg-GPI and CD160-ΔIg-TM are expressed on the cell surface (57).
Moreover, CD160 is also implicated in the regulation of NK-cell metabolism. It was reported that CD160 expression is positively correlated with the expression of the glucose transporter (GLUT1) and glucose uptake in NK cells, which is indispensable for NK-cell functioning and IFN-γ secretion (14, 78). CD160 expression in NK cells upregulates glucose metabolism through the activation of the AKT/mTOR/s6k signaling pathway (14). In addition, mTORC1 signaling was previously demonstrated to be required for the upregulation of GLUT1 expression and the expression of glycolysis rate-limiting enzymes such hexokinase 2 and lactate dehydrogenase A, which enhance glucose uptake and glycolysis and increase NK-cell functioning (79). Furthermore, CD160 plays an important role in activating the regulatory function of liver-resident CD56bright NK cells against allogenic CD8+ T cells (80). Its binding with HLA molecules expressed on activated allogenic T cells activates cytotoxic hepatic NK-cell functions and consequently the killing of activated allogenic T cells. Hence, it has a role in immune tolerance in liver transplantation (80).
Furthermore, NK cells play a crucial role in the antitumor immune response due to their capacity to spontaneously detect and lyse transformed or stressed cells (81). Their function is mediated by different activating receptors, such as NKG2D, CD160, and NKp30, and inhibitory receptors, such as killer cell Ig-like receptors (KIRs) and the CD94/NKG2A heterodimer (82, 83). Synergetic stimulation from a combination of receptors elicits the activation of NK-cell cytotoxicity against tumor cells (84). However, an imbalance of these receptors (that is, upregulation of inhibitory receptors and downregulation of activating receptors) impairs NK-cell function and permits tumor evasion (85, 86). In line with these findings, it was reported that downregulation of CD160 in intrahepatic NK cells induces NK-cell impairment in hepatocellular carcinoma (87). This reduced expression is associated with disease aggressiveness, tumor metastasis, and poorer outcomes (87). In addition, intratumoural CD160+ NK cells were found to be more exhausted than peritumoural CD160+ NK cells in hepatocellular carcinoma and to produce less IFNγ (87). This exhausted phenotype is due to the expression of TGF-β by tumor cells and inhibits NK-cell functions (87). Similarly, the elevated levels of TGF-β1 in human immunodeficiency virus (HIV)-infected individuals decrease the expression of CD160 in NK cells and enhance HIV disease progression (14). This downregulation may be mediated through the upregulation of specific miRNAs that inhibit CD160 transcription by TGF-β1 (14).
CD160-GPI is expressed in activated endothelial cells in retinal vessels (7). Its expression is elevated in retinal blood vessels of patients with vascular retinal diseases compared with normal individuals (7). CD160-GPI is also increased in endothelial cells of newly formed blood vessels in human colon carcinoma and mouse B16 melanoma but is absent in the vessels of healthy tissues (17). The interaction of CD160 with soluble HLA-G1 induces endothelial cell apoptosis and inhibits fibroblast growth factor 2 (FGF2)- and vascular endothelial growth factor (VEGF)-induced neoangiogenesis in vivo (61). Therefore, CD160-GPI has been utilized as a therapeutic target in ocular diseases through the use of anti-CD160-GPI mAbs. These mAbs trigger caspase-dependent apoptosis in endothelial cells and prevent pathological neovascularization (7, 17, 61, 66). Treatment with CL1-R2, an anti-CD160 IgG1 mAb, in a rabbit corneal neovascularization (CNV) model led to significant regression of neovessels (66). Additionally, the use of ELB01101, a humanized anti-CD160 IgG4 mAb, in a monkey model of choroidal neovascularization (ChNV) decreased the number of clinically relevant lesions by 50% and has shown good safety and tolerability (66). Furthermore, the use of CL1-R2 in combination with cyclophosphamide chemotherapy in B16 melanoma-bearing mice decreased size of the tumor vasculature (17). Hence, anti-CD160 mAbs can be used as a potential antiangiogenic treatment in ocular diseases and cancer (17, 61, 66).
Assessments of CD160 function in T cells have produced contradictory results, as CD160 has been reported to both activate and inhibit T cell functions. In previous studies, we demonstrated that CD160 has a coactivating function with the CD3 receptor in a minor subset of CD4+ CD160+ T cells isolated from inflammatory skin lesions in dermatitis and psoriasis samples (67). This minor subset corresponds to effector memory cytotoxic T lymphocytes characterized by the expression of CD3, CD4, CD160, CD8, CD244, and perforin but lacking CD28 expression (68). In contrast, other studies have demonstrated that CD160 functions as a negative regulator of CD4+ T cells and NKT cells (4, 5, 69, 72). CD160 inhibits CD4+ T-cell activation and decreases the production of cytokines through its engagement with HVEM (4, 69–71). CD160 receptor acts as a bidirectional switch of T-cell activation since it can produce both positive and negative signals depending on its binding to costimulatory ligands (lymphotoxin-α/LIGHT) or coinhibitory ligands (BTLA/CD160) (4). The inactivation of CD4+ T cells is induced by the cooperation of BTLA and CD160 (70). These two proteins bind with the same affinity to cysteine-rich domain 1 (CRD1) of HVEM, with CD160 having a stronger inhibitory signaling function and slower dissociation constant than BTLA (70). CD160-GPI does not contain an ITIM domain. Hence, it does not mediate T-cell inhibition through the phosphorylation of the tyrosine phosphatases SHP-1 or SHP-2 in the same way as BTLA upon its engagement with HVEM (88). However, it was suggested that CD160 induces CD4+ T-cell inhibition by modulating binding complexes that translocate to lipid rafts and by reducing the tyrosine phosphorylation of different proteins, such as CD3ζ (4, 70). However, the mechanism underlying CD160-mediated negative signaling is still poorly understood (72). In addition, contradictory reports have indicated that CD160 can activate (9, 89) or inhibit CD8+ T-cell functions (65, 90, 91). Nikolova and his colleagues have demonstrated that CD160 acts as a coreceptor in TCR signal transduction in human circulating CD8+ T cells (6). It plays a protective role during chronic viral infections by enhancing CD8+ T-cell functions (8). It synergizes with TCR signaling to enhance the activation of the PI3K-AKT and MEK-ERK signaling pathways, and it promotes T-cell proliferation and degranulation capacities during HIV infections (8). Similarly, Tan and his colleagues demonstrated that CD160 activates CD8+ T-cell effector functions against Listeria monocytogenes and allows the secretion of granzyme B, IFN-γ, and TNF-α (9). In contrast, others demonstrated that CD160 interaction with HVEM inhibits CD160+ CD8+ T cells in patients infected with human T-lymphotropic virus type 1 (HTLV-1), and blockade of this interaction by anti-CD160 mAbs or anti-HVEM mAbs reactivates T-cell function and enhances cytokine production (15). Additionally, CD160+ CD8+ T cells have lower cytotoxicity in pancreatic cancer patients than CD160ˉ CD8+ T cells (65). Additionally, the levels of CD160+ CD8+ T cells were found to be inversely associated with patient survival. This could be related to the expression of other inhibitory receptors, such as PD-1 and TIM-3, that intrinsically impair the function of T cells in CD160+ CD8+ T cells (65). Moreover, a recent study showed that CD160 is upregulated in Th1-like cells through IL-23 activity and is implicated in the induction of intestinal inflammation during colitis (11). In addition, in vivo studies have demonstrated that recipient animals that receive CD160-deficient Th1-like cells have strong protection against colitis and less inflammation (11). Furthermore, loss of CD160 contributes to the maintenance of a naïve-like phenotype and prevents the differentiation of T cells into an effector phenotype (11). In summary, CD160 mediates cell activation during inflammation and presents an important therapeutic target.
Soluble CD160 plays an important role in the regulation of the immune system. It can be released by activated CD56dim CD16+ NK cells and mast cells through proteolytic cleavage of membrane-bound CD160-GPI (58, 63). Soluble CD160 regulates adaptive immunity functions, modulates helper T-cell functions and inhibits CD8+ T-cell cytotoxicity by preventing the interaction of CD160 with HVEM or MHC-I (58, 63) (Table 1). Soluble CD160 also prevents the interaction of the beta subunit of the CD8+ T-cell coreceptor with the alpha 3 domain of MHC class I, which inhibits cytotoxic T-cell activity (67). Under pathological conditions, sCD160 is secreted by cancer cells to allow tumor immune escape (18). It was recently reported that only melanoma cells and not normal melanocytes constitutively secrete sCD160 (18). Secreted sCD160 binds to HLA molecules and HVEM on target cells and inhibits the activation of NK-cell cytotoxicity toward their target cells (18).
In addition to its important role in the regulation of different immune cells, CD160 has been implicated in the pathophysiological processes of different diseases, including autoimmune diseases (10), atherosclerosis (12), retinal vascular diseases (7), and chronic viral infections (13). It has also been implicated in the physiopathology of various cancer types, such as melanoma and B-chronic lymphocytic leukemia (CLL) (18, 46).
CLL is a clonal lymphoproliferative disorder characterized by the proliferation of small, mature-appearing CD5+ CD23+ B lymphocytes that accumulate in the blood, bone marrow, and secondary lymphoid tissues (21). It is classified into two main subsets depending on the mutational status of the immunoglobulin heavy chain variable region (IGHV) genes of the B-cell receptor (BCR), which influences the prognosis of the disease and predicts overall survival (92, 93). IGHV-mutated CLL (M-CLL) represents the indolent form of the disease and is associated with a favorable prognosis (94). IGHV-unmutated CLL (U-CLL) is an aggressive form of the disease and is associated with a worse prognosis (95).
BCR-mediated signaling is a crucial factor in CLL development and CLL cell survival and proliferation (96). Under physiological conditions, BCR targeting activates different signaling pathways that mediate cell activation, proliferation, survival, and migration through the recruitment of kinases, such as lyn, fyn, and syk, to BCR (96) (Figure 2). In CLL, malignant B cells express constitutively activated kinases such as Syk and Btk, which induce continuous activation of the cell and upregulation of downstream signaling pathways in the absence of BCR ligands (96–98). In addition to these kinases, CLL cells abnormally express different T-cell markers that are not expressed by normal B cells, such as Lck, Src kinase (a homolog of Lyn) (99), and ZAP70, a protein tyrosine kinase (PTK) associated with the TCR ζ chain (100) (Figure 2).
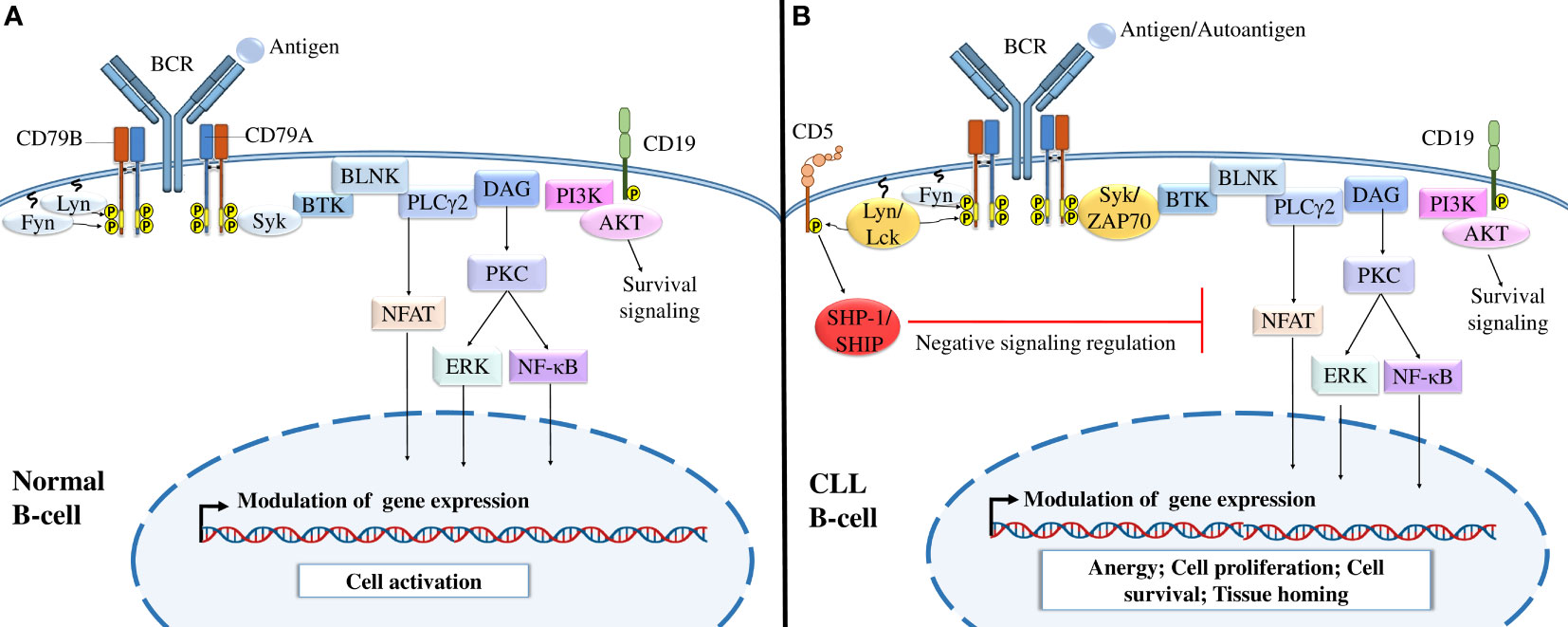
Figure 2 B-cell receptor signaling in normal B cells and CLL cells. (A) BCR signaling in normal B cells. The interaction of normal B lymphocyte BCRs with antigens induces the tyrosine phosphorylation of ITAMs by Src-family kinases such as Syk, Fyn, and Lyn, activating different downstream signaling pathways, including the PI3K/Akt, NFAT, NF-κB, and MAPK signaling pathways, which modulate gene expression and activate B cells. (B) BCR signaling in CLL B cells. After the interaction of CLL BCRs with antigens or autoantigens, Src-family kinases, such as Lck and ZAP-70 kinases, enhance the activation of different downstream signaling pathways necessary for B-cell survival. Malignant B cells also express CD5, which, upon activation by Lyn kinase recruits SHP-1 or SHIP to dephosphorylate signaling molecules and maintain B-cell anergy. Adapted from (36, 43).
In addition to the prosurvival activity of BCR, CLL cells express other surface markers to survive and proliferate. Phenotypically, CLL cells express different markers of the B lymphocyte lineage, such as CD19, CD23, and CD20 (101, 102). CLL cells are also characterized by low levels of surface IgM expression, which may be a result of a defect in glycosylation and folding of the μIg and CD79a chains and their retention in the endoplasmic reticulum (103). CLL cells also express CD5, a T-cell inhibitory marker (10). CD5 is associated with immunoreceptor tyrosine inhibitory motifs (ITIMs) that, once phosphorylated (104), recruit the tyrosine phosphatases SHP-1 and SHIP, leading to downregulation of BCR-mediated signaling events and continued cell anergy (105).
CD160 is another surface marker that is aberrantly expressed by CLL cells at all disease stages (19). This marker is normally specific for NK cells and some T-cell subsets (64, 65). It plays a dual role in CLL by triggering both prosurvival and anti-apoptotic signals, which favor cytokine production and cell survival and decrease spontaneous cell death (46) (Figure 3). CD160 expression activates prosurvival signaling through the upregulation of PI3K/Akt signaling pathway and increases the secretion of cytokines, mainly the proinflammatory cytokine IL-6 (46); this interleukin activates signal transducer and activator of transcription 3 (STAT3) and NF-κB, which regulate the expression of several genes implicated in CLL cell proliferation and survival (106). CD160 also decreases apoptosis through the downregulation of proapoptotic caspase (caspase-3, -9, and -8) expression and the upregulation of the expression of antiapoptotic proteins, including Bcl-2, Bcl-xL, and Mcl-1, blocking both mitochondria-dependent and mitochondria-independent apoptotic pathways (46, 107). Thus, CD160 prevents apoptosis by blocking cytochrome c release from the outer mitochondrial membrane and inhibits mitochondrial membrane potential decrease and caspase activation (46). However, to understand the complete mechanism underlying CD160-mediated anti-apoptotic effects, further investigation is needed. Additionally, a recent study demonstrated that CD160 hypomethylation in blood cells was correlated with breast cancer in a Chinese population (16). Therefore, further studies should be conducted to demonstrate the mechanisms regulating CD160 expression in both CLL B cells and the neighboring cells that reside in the microenvironment (e.g., T cells and NK cells) and whether this expression is mediated by genetic or epigenetic alterations.
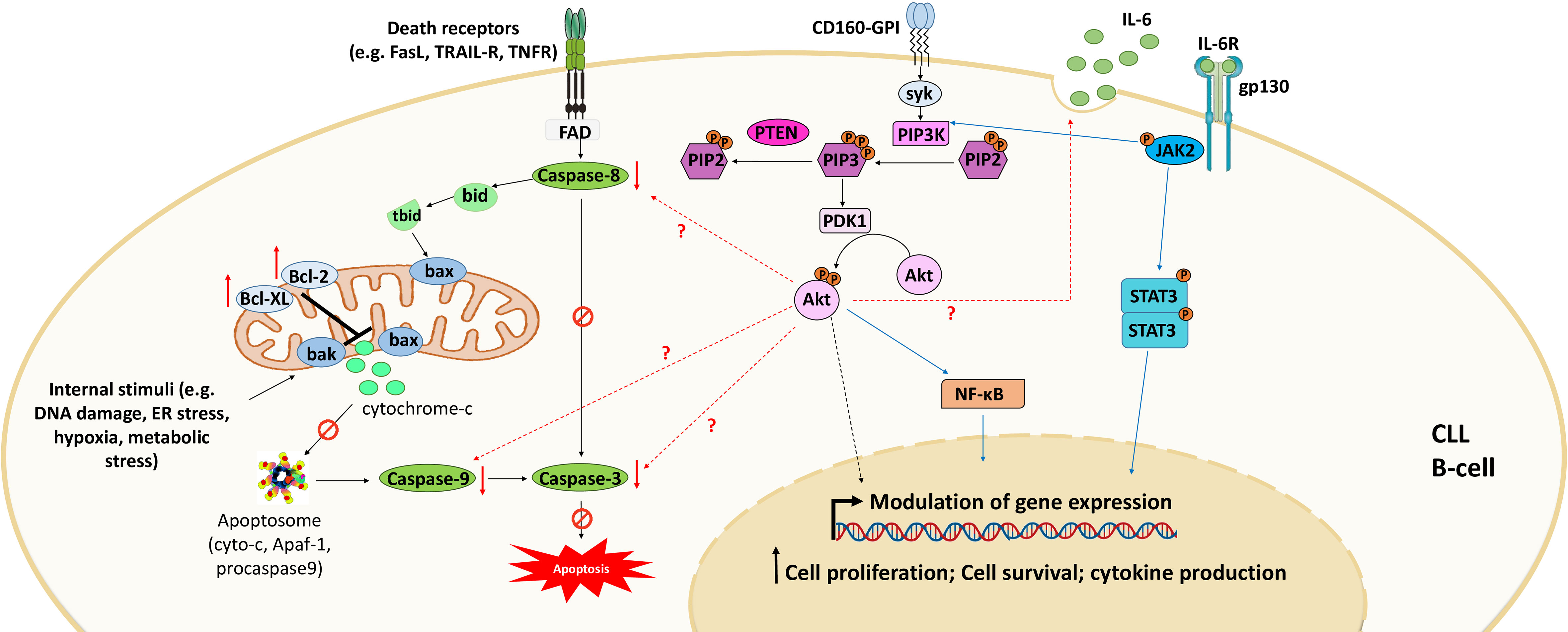
Figure 3 CD160 signaling in CLL. Engagement of CD160-GPI via its receptors (MHC-I or HVEM) or monoclonal antibodies leads to activation of the PI3K/Akt signaling pathway to modulate gene expression. This activation also downregulates the expression of pro-apoptotic proteins and upregulates the expression of anti-apoptotic proteins. Consequently, CD160 dysregulates both intrinsic apoptotic pathways (mediated by internal stimuli) and extrinsic apoptotic pathways (triggered by death receptors such as FasR, TRAIL-R, and TNFR). CD160 also activates the expression of cytokines such as IL-6 that activate STAT3 and NF-κB via the JAK2/PI3K/Akt axis. Adapted from (46).
CLL cell survival and disease progression are highly influenced by the surrounding microenvironment. It has been reported that tumor B cells are more sensitive to apoptosis when cultured in vitro (108). However, they resist apoptosis in vivo, which confirms the importance of the microenvironment in CLL cell survival (108). Malignant B cells constantly interact with different cell types in the microenvironment, including T cells (109), mesenchymal stromal cells (32), lymphoma-associated macrophages (110), endothelial cells (29–31), follicular dendritic cells (111), and nurse-like cells (24–28), which negatively or positively influence each other. Furthermore, these neighboring cells in the tumor microenvironment are crucial during all CLL development stages. They can express various chemokines, integrins, cytokines, soluble ligands, adhesion molecules, and survival factors that deliver growth and survival signals to CLL cells, enhance their clonal expansion, and induce drug resistance (36). CLL cells deliver chemokine gradients into lymph nodes, where they form proliferation centers and interact closely with tumor microenvironment components (112). This interaction favors strong activation of BCR-mediated signaling and enhances CLL cell proliferation (112). In addition, CLL cells can also create a favorable microenvironment by regulating the functions of neighboring cells either through the expression of soluble molecules or through direct contact with these cells, inducing their proliferation and survival (110) (Figure 4).
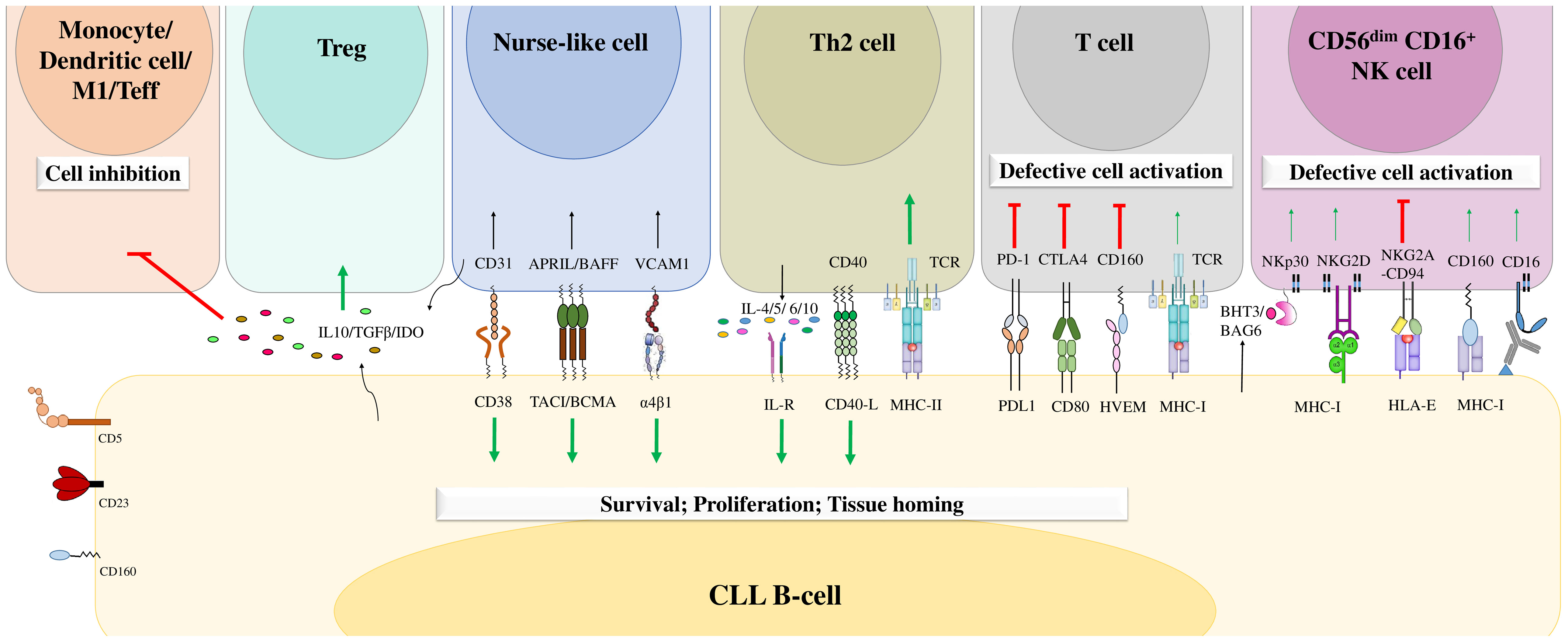
Figure 4 The B-cell chronic lymphocytic leukemia microenvironment. CLL cells engage in crosstalk with different cells in the microenvironment through different molecular interactions or through the release of soluble factors. This crosstalk favors the delivery of activating signals to promote CLL B-cell survival, proliferation, and tissue homing and decreases the numbers of cytotoxic immune cells, leading to tumor cell escape from immune surveillance. Adapted from (36, 113).
Moreover, the antitumor immune response that is mainly mediated by T cells and NK cells is impaired in the CLL microenvironment (114–116). CLL cells overexpress different inhibitory ligands, such as programmed cell death 1 ligand (PD-L1) and HVEM, which bind to PD1 and BTLA, respectively, expressed on cytotoxic T cells and NK cells. These ligands alter T-cell activation, actin polymerization, and immune synapse formation and induce immunosuppression (114–116). This exhaustion phenotype has been described more frequently in T cells located in secondary lymphoid organs than in those circulating in peripheral blood (33). In addition, extracellular vesicles secreted by CLL cells induce phenotypic, functional and transcriptional dysfunction in chimeric antigen receptor (CAR) T cells, leading to their exhaustion (117). Moreover, the exhausted CD8+ T cells in patients with CLL express high levels of inhibitory molecules such as CD160, PD-1 and TIGIT (91). This increased expression of inhibitory receptors decreases T-cell proliferation and cytotoxic functions (91). This impairment is also illustrated by the low production of TNF-α and IFN-γ by CD160+ CD8+ T cells, which are critical cytokines for antitumoral immunity (91). Moreover, the expression of CD160 in CD8+ T cells in patients with CLL was found to be correlated with the oversecretion of IL-16 by CLL cells. Hence, Bozorgmehr et al. suggested that IL-16 secretion by CLL cells may have a role in the upregulation of CD160 expression in T cells and the induction of their exhaustion (91). However, further studies should be conducted to confirm these results and analyze the impact of IL-16 on the expression of CD160 in other cell types, such as NK cells.
NK cells remain defective in CLL patients and show impaired production of soluble cytotoxic mediators and reduced degranulation and lysis capacities (113, 118). Under physiological conditions, NK cells express different inhibitory and activating molecules that regulate their own activity and functions (119). However, in CLL, NK-cell exhaustion could result from inhibitory signaling mediated by the engagement of both NKG2A and ILT2 by HLA-E and HLA-G, respectively (120, 121). In addition, other inhibitory receptors are overexpressed in NK cells, including the killer cell Ig-like receptors (KIRs) KIR2DL1 and KIR3DL1 (37), which strengthen the inhibitory signals delivered to these cells. In contrast, NK cells exhibit decreased expression of activating receptors such as CD16, NKG2D, and NKp30, which could contribute to the hyporeactivity of NK cells in CLL (120). However, the expression level of CD160 in NK cells in patients with CLL has not yet been determined. In addition, the expression of CD160 in both NK cells and CLL B cells suggests potential future therapeutic avenues. Future studies should be conducted to evaluate the targeting of CD160 by monoclonal antibodies in CLL, the potential effect of such agents on the reactivation of NK cells, and their induction of antibody-dependent cellular cytotoxicity (ADCC).
Minimal residual disease (MRD) is the remaining cancer cells that can be detected after treatment (122). The amount of remaining cells can indicate the efficacy of the treatment and the probability of disease relapse (122). Moreover, the use of targeted therapy as a monotherapy induces partial remission and resistance (123). Therefore, the assessment of MRD is essential for the establishment of an effective therapeutic protocol for CLL and the prevention of relapse, especially after a first-line therapeutic regimen.
MRD assessment in CLL can be performed through the use of three sensitive techniques: real-time polymerase chain reaction (PCR), next-generation sequencing and flow cytometry (124, 125). The European Research Initiative for CLL (ERIC) has developed a gold standard flow cytometry assay that combines 8 antibodies, namely, antibodies against CD5, CD3, CD19, CD20, CD22, CD43, CD79b, and CD81 (126). Novel assays have been developed by combining new prognostic markers such as CD160 into a simple single flow cytometry analysis. The restricted expression of CD160 in CLL and its lack of expression in normal B cells make it a good prognostic marker for the detection of MRD in CLL (54). A single tube flow cytometry assay detecting CD160 (CD160FCA) was developed for the assessment of CLL MRD (54). This assay incorporates six markers, namely, CD160, CD2, CD5, CD19, CD23, and CD45. It allows the quantification of MRD to a level of 10ˉ4 (1 malignant cell per 10 000 normal PB cells) in both peripheral blood and bone marrow (54). It also has a high concordance with the gold standard assay (p<0.01). In addition, MRD assessed by CD160FCA after first-line or second-line treatment was found to be correlated with event-free survival (EFS) (54). After first-line treatment, patients with CR and MRD negativity had a prolonged EFS (63 months n=32) compared with patients in CR with MRD positivity (16 months, n=11). Similarly, after second-line treatment, patients with CR and MRD negativity had prolonged EFS (48 months) compared with patients with CR and MRD positivity (24 months) (54).
Another assay combining CD160 and receptor tyrosine kinase-like orphan receptor 1 (ROR1) in a single tube flow cytometry experiment (CD160-ROR1FCA) was developed for the detection of MRD. ROR1 is a tumor-specific antigen of malignant B cells. It is constitutively phosphorylated in CLL and is associated with disease progression (127, 128). CD160-ROR1FCA also includes monoclonal antibodies against CD2, CD5, CD19, CD23, and CD45. The assay showed high sensitivity and had a limit of detection of 0.001%. It also had good correlation (R = 0.98, p < 0.01) with the ERIC gold-standard assay (129). Both assays detect MRD to a level of 10-5 (1 malignant cell per 100 000 normal PB cells) (129).
Emerging evidence has demonstrated that CD160 participates in CLL pathogenesis by activating PI3K/Akt-independent pathways and inhibiting apoptosis, consequently activating tumor proliferation and inducing resistance to apoptosis. However, further characterization of CD160 signaling pathways in CLL is needed to fully understand its role in CLL pathogenesis. In addition, abnormal expression of CD160 in only CLL cells and its lack of expression in normal B cells make it a potential prognostic marker for the assessment of MRD in CLL. Approaches have been recently developed to assess MRD in CLL through the detection of CD160 and other markers using flow cytometry. These approaches have demonstrated high sensitivity and accuracy, proving the usefulness of CD160 as a prognostic marker. However, the therapeutic use of CD160 in CLL has not been assessed; thus, further related studies should be conducted. Nevertheless, the role of CD160 in CLL pathogenesis and the impairment of antitumor immune cells in the CLL microenvironment makes it a promising target in CLL, and relevant therapeutic approaches may be developed. One potential target that should be assessed is NK cells. These cells are known for their capacity to spontaneously detect and kill tumor cells. Nevertheless, the CLL microenvironment hinders their cytotoxic and antitumor immune functions. Hence, reactivation of exhausted NK cells in the CLL microenvironment is an important therapeutic strategy. One therapeutic approach that could be tested is the use of engineered anti-CD160-GPI mAbs that can engage both CD160-GPI receptors expressed on NK cells and CLL-B cells and FcγRs expressed on NK cells. The engagement of CD160-GPI receptors on NK cells might mediate the reactivation of NK-cell cytotoxicity and the production of cytokines, including IFN-γ and TNF-α. IFN-γ is known for its antitumor activity. It can induce tumor cell cycle arrest and activate the polarization of macrophages into the tumoricidal M1 macrophage subtype (130). It also activates the NADPH-dependent phagocyte oxidase system and upregulates nitric oxide production, contributing to tumor cell death. Moreover, IFN-γ upregulates lysosomal enzymes such as cathepsin B, which enhances cell surface MHC expression on antigen-presenting cells (APCs) and favors the recruitment and activation of effector cells, including CD8 cytotoxic T cells and NK cells (130). IFN-γ also inhibits the differentiation of protumoral immune cells, namely, Treg cells, Th2 cells and Th17 cells (130). Furthermore, IFN-γ can also have direct antitumor activity by activating proapoptotic proteins such as caspase-1, 3, and 8 and upregulating FAS production and the expression of TNF-related apoptosis-inducing ligand (TRAIL) (130). Additionally, the use of anti-CD160 mAbs may also trigger ADCC through the engagement of FcγRs. Moreover, the FcγRs present on phagocytic cells may also recognize anti-CD160 mAbs and activate ADCP against tumor cells. However, this approach may have some limitations. In particular, the use of anti-CD160-GPI mAbs may enhance the proliferation and resistance to apoptosis of CLL B cells rather than activating exhausted NK cells. In addition, the dual function of CD160 in T cells makes it difficult to predict its impact on the activation or inhibition of immune cells in the CLL microenvironment. Hence, proof of concept studies are needed to investigate the effect of anti-CD160 mAbs in the CLL microenvironment.
LO: conceptualization, manuscript writing, figure artwork. AA: review, editing. AB: conceptualization, review & editing, supervision, project administration; SB: conceptualization, writing - review & editing, supervision, project administration. All authors contributed to the article and approved the submitted version.
This work was financially supported by the Mohammed VI Polytechnic University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.

1. Maïza H, Leca G, Mansur IG, Schiavon V, Boumsell L, Bensussan A. A novel 80-kD cell surface structure identifies human circulating lymphocytes with natural killer activity. J Exp Med (1993) 178:1121–6. doi: 10.1084/jem.178.3.1121
2. Le Bouteiller P, Barakonyi A, Giustiniani J, Lenfant F, Marie-Cardine A, Aguerre-Girr M, et al. Engagement of CD160 receptor by HLA-c is a triggering mechanism used by circulating natural killer (NK) cells to mediate cytotoxicity. Proc Natl Acad Sci (2002) 99:16963–8. doi: 10.1073/pnas.012681099
3. Barakonyi A, Rabot M, Marie-Cardine A, Aguerre-Girr M, Polgar B, Schiavon V, et al. Cutting edge: Engagement of CD160 by its HLA-c physiological ligand triggers a unique cytokine profile secretion in the cytotoxic peripheral blood NK cell subset. J Immunol (2004) 173:5349–54. doi: 10.4049/jimmunol.173.9.5349
4. Cai G, Anumanthan A, Brown JA, Greenfield EA, Zhu B, Freeman GJ. CD160 inhibits activation of human CD4+ T cells through interaction with herpesvirus entry mediator. Nat Immunol (2008) 9:176–85. doi: 10.1038/ni1554
5. El-Far M, Pellerin C, Pilote L, Fortin J-F, Lessard IAD, Peretz Y, et al. CD160 isoforms and regulation of CD4 and CD8 T-cell responses. J Transl Med (2014) 12:217. doi: 10.1186/s12967-014-0217-y
6. Nikolova M, Marie-Cardine A, Boumsell L, Bensussan A. BY55/CD160 acts as a co-receptor in TCR signal transduction of a human circulating cytotoxic effector T lymphocyte subset lacking CD28 expression. Int Immunol (2002) 14:445–51. doi: 10.1093/intimm/14.5.445
7. Henry A, Boulagnon-Rombi C, Menguy T, Giustiniani J, Garbar C, Mascaux C, et al. CD160 expression in retinal vessels is associated with retinal neovascular diseases. Investig Opthalmol Vis Sci (2018) 59:2679. doi: 10.1167/iovs.18-24021
8. Zhang L, Zhang A, Xu J, Qiu C, Zhu L, Qiu C, et al. CD160 plays a protective role during chronic infection by enhancing both functionalities and proliferative capacity of CD8+ T cells. Front Immunol (2020) 11:2188. doi: 10.3389/fimmu.2020.02188
9. Tan CL, Peluso MJ, Drijvers JM, Mera CM, Grande SM, Brown KE, et al. CD160 stimulates CD8 + T cell responses and is required for optimal protective immunity to listeria monocytogenes. ImmunoHorizons (2018) 2:238–50. doi: 10.4049/immunohorizons.1800039
10. He W, Wang B, Li Q, Yao Q, Jia X, Song R, et al. Aberrant expressions of Co-stimulatory and Co-inhibitory molecules in autoimmune diseases. Front Immunol (2019) 10:261. doi: 10.3389/fimmu.2019.00261
11. Pawlak M, DeTomaso D, Schnell A, Meyer zu Horste G, Lee Y, Nyman J, et al. Induction of a colitogenic phenotype in Th1-like cells depends on interleukin-23 receptor signaling. Immunity (2022) 55:1663–1679.e6. doi: 10.1016/j.immuni.2022.08.007
12. Zuo J, Shan Z, Zhou L, Yu J, Liu X, Gao Y. Increased CD160 expression on circulating natural killer cells in atherogenesis. J Transl Med (2015) 13:188. doi: 10.1186/s12967-015-0564-3
13. Peretz Y, He Z, Shi Y, Yassine-Diab B, Goulet J-P, Bordi R, et al. CD160 and PD-1 Co-expression on HIV-specific CD8 T cells defines a subset with advanced dysfunction. PloS Pathog (2012) 8:e1002840. doi: 10.1371/journal.ppat.1002840
14. Sun Z, Li Y, Zhang Z, Fu Y, Han X, Hu Q, et al. CD160 promotes NK cell functions by upregulating glucose metabolism and negatively correlates with HIV disease progression. Front Immunol (2022) 13:854432. doi: 10.3389/fimmu.2022.854432
15. Chibueze CE, Yoshimitsu M, Arima N. CD160 expression defines a uniquely exhausted subset of T lymphocytes in HTLV-1 infection. Biochem Biophys Res Commun (2014) 453:379–84. doi: 10.1016/j.bbrc.2014.09.084
16. Liu C, Zhou X, Jin J, Zhu Q, Li L, Yin Q, et al. The association between breast cancer and blood-based methylation of CD160, ISYNA1 and RAD51B in the Chinese population. Front Genet (2022) 13:927519. doi: 10.3389/fgene.2022.927519
17. Chabot S, Jabrane-Ferrat N, Bigot K, Tabiasco J, Provost A, Golzio M, et al. A novel antiangiogenic and vascular normalization therapy targeted against human CD160 receptor. J Exp Med (2011) 208:973–86. doi: 10.1084/jem.20100810
18. Gauci M-L, Giustiniani J, Lepelletier C, Garbar C, Thonnart N, Dumaz N, et al. The soluble form of CD160 acts as a tumor mediator of immune escape in melanoma. Cancer Immunol Immunother (2022) 71:2731–42. doi: 10.1007/s00262-022-03199-0
19. Farren TW, Giustiniani J, Liu FT, Tsitsikas DA, Macey MG, Cavenagh JD, et al. Differential and tumor-specific expression of CD160 in b-cell malignancies. Blood (2011) 118:2174–83. doi: 10.1182/blood-2011-02-334326
20. Hallek M, Shanafelt TD, Eichhorst B. Chronic lymphocytic leukaemia. Lancet (2018) 391:1524–37. doi: 10.1016/S0140-6736(18)30422-7
21. Kipps TJ, Stevenson FK, Wu CJ, Croce CM, Packham G, Wierda WG, et al. Chronic lymphocytic leukaemia. Nat Rev Dis Prim (2017) 3:16096. doi: 10.1038/nrdp.2016.96
22. Lazarian G, Guièze R, Wu CJ. Clinical implications of novel genomic discoveries in chronic lymphocytic leukemia. J Clin Oncol (2017) 35:984–93. doi: 10.1200/JCO.2016.71.0822
23. Beekman R, Chapaprieta V, Russiñol N, Vilarrasa-Blasi R, Verdaguer-Dot N, Martens JHA, et al. The reference epigenome and regulatory chromatin landscape of chronic lymphocytic leukemia. Nat Med (2018) 24:868–80. doi: 10.1038/s41591-018-0028-4
24. Burger JA, Tsukada N, Burger M, Zvaifler NJ, Dell’aquila M, Kipps TJ. Blood-derived nurse-like cells protect chronic lymphocytic leukemia b cells from spontaneous apoptosis through stromal cell-derived factor-1. Blood (2000) 96:2655–63. doi: 10.1182/blood.V96.8.2655
25. Nishio M, Endo T, Tsukada N, Ohata J, Kitada S, Reed JC, et al. Nurselike cells express BAFF and APRIL, which can promote survival of chronic lymphocytic leukemia cells via a paracrine pathway distinct from that of SDF-1α. Blood (2005) 106:1012–20. doi: 10.1182/blood-2004-03-0889
26. Chen Y, Chen L, Yu J, Zhang L, Rassenti LZ KT. Nurse-like cells express high-levels of Wnt5a, which induces ROR1-dependent signaling that promotes migration and survival of neoplastic cells in chronic lymphocytic leukemia. Blood (2017) 130:3007. doi: 10.1182/blood.V130.Suppl_1.3007.3007
27. Talbot H, Saada S, Barthout E, Gallet P-F, Gachard N, Abraham J, et al. BDNF belongs to the nurse-like cell secretome and supports survival of b chronic lymphocytic leukemia cells. Sci Rep (2020) 10:12572. doi: 10.1038/s41598-020-69307-1
28. Vaca AM, Ioannou N, Sivina M, Vlachonikola E, Clise-Dwyer K, Kim E, et al. Activation and expansion of T-follicular helper cells in chronic lymphocytic leukemia nurselike cell co-cultures. Leukemia (2022) 36:1324–35. doi: 10.1038/s41375-022-01519-y
29. Buggins AGS, Pepper C, Patten PEM, Hewamana S, Gohil S, Moorhead J, et al. Interaction with vascular endothelium enhances survival in primary chronic lymphocytic leukemia cells via NF-κB activation and de novo gene transcription. Cancer Res (2010) 70:7523–33. doi: 10.1158/0008-5472.CAN-10-1634
30. Cols M, Barra CM, He B, Puga I, Xu W, Chiu A, et al. Stromal endothelial cells establish a bidirectional crosstalk with chronic lymphocytic leukemia cells through the TNF-related factors BAFF, APRIL, and CD40L. J Immunol (2012) 188:6071–83. doi: 10.4049/jimmunol.1102066
31. Badoux X, Bueso-Ramos C, Harris D, Li P, Liu Z, Burger J, et al. Cross-talk between chronic lymphocytic leukemia cells and bone marrow endothelial cells: role of signal transducer and activator of transcription 3. Hum Pathol (2011) 42:1989–2000. doi: 10.1016/j.humpath.2011.02.027
32. Ding W, Knox TR, Tschumper RC, Wu W, Schwager SM, Boysen JC, et al. Platelet-derived growth factor (PDGF)–PDGF receptor interaction activates bone marrow–derived mesenchymal stromal cells derived from chronic lymphocytic leukemia: implications for an angiogenic switch. Blood (2010) 116:2984–93. doi: 10.1182/blood-2010-02-269894
33. Hanna BS, Roessner PM, Yazdanparast H, Colomer D, Campo E, Kugler S, et al. Control of chronic lymphocytic leukemia development by clonally-expanded CD8+ T-cells that undergo functional exhaustion in secondary lymphoid tissues. Leukemia (2019) 33:625–37. doi: 10.1038/s41375-018-0250-6
34. Catakovic K, Gassner FJ, Ratswohl C, Zaborsky N, Rebhandl S, Schubert M, et al. TIGIT expressing CD4+T cells represent a tumor-supportive T cell subset in chronic lymphocytic leukemia. Oncoimmunology (2018) 7:e1371399. doi: 10.1080/2162402X.2017.1371399
35. Lewinsky H, Barak AF, Huber V, Kramer MP, Radomir L, Sever L, et al. CD84 regulates PD-1/PD-L1 expression and function in chronic lymphocytic leukemia. J Clin Invest (2018) 128:5465–78. doi: 10.1172/JCI96610
36. ten Hacken E, Burger JA. Microenvironment interactions and b-cell receptor signaling in chronic lymphocytic leukemia: Implications for disease pathogenesis and treatment. Biochim Biophys Acta - Mol Cell Res (2016) 1863:401–13. doi: 10.1016/j.bbamcr.2015.07.009
37. MacFarlane AW, Jillab M, Smith MR, Alpaugh RK, Cole ME, Litwin S, et al. NK cell dysfunction in chronic lymphocytic leukemia is associated with loss of the mature cells expressing inhibitory killer cell ig-like receptors. Oncoimmunology (2017) 6:e1330235. doi: 10.1080/2162402X.2017.1330235
38. Roessner PM, Seiffert M. T-Cells in chronic lymphocytic leukemia: Guardians or drivers of disease? Leukemia (2020) 34:2012–24. doi: 10.1038/s41375-020-0873-2
39. Llaó Cid L, Hanna BS, Iskar M, Roessner PM, Öztürk S, Lichter P, et al. CD8 + T-cells of CLL-bearing mice acquire a transcriptional program of T-cell activation and exhaustion. Leuk Lymphoma (2020) 61:351–6. doi: 10.1080/10428194.2019.1660972
40. Muzio M, Apollonio B, Scielzo C, Frenquelli M, Vandoni I, Boussiotis V, et al. Constitutive activation of distinct BCR-signaling pathways in a subset of CLL patients: a molecular signature of anergy. Blood (2008) 112:188–95. doi: 10.1182/blood
41. Vangapandu HV, Havranek O, Ayres ML, Kaipparettu BA, Balakrishnan K, Wierda WG, et al. B-cell receptor signaling regulates metabolism in chronic lymphocytic leukemia. Mol Cancer Res (2017) 15:1692–703. doi: 10.1158/1541-7786.MCR-17-0026
42. Schleiss C, Ilias W, Tahar O, Güler Y, Miguet L, Mayeur-Rousse C, et al. BCR-associated factors driving chronic lymphocytic leukemia cells proliferation ex vivo. Sci Rep (2019) 9:1–12. doi: 10.1038/s41598-018-36853-8
43. Chen J, Sathiaseelan V, Moore A, Tan S, Chilamakuri CSR, Roamio Franklin VN, et al. ZAP-70 constitutively regulates gene expression and protein synthesis in chronic lymphocytic leukemia. Blood (2021) 137:3629–40. doi: 10.1182/blood.2020009960
44. Talab F, Allen JC, Thompson V, Lin K, Slupsky JR. LCK is an important mediator of b-cell receptor signaling in chronic lymphocytic leukemia cells. Mol Cancer Res (2013) 11:541–54. doi: 10.1158/1541-7786.MCR-12-0415-T
45. Garaud S, Taher TE, Debant M, Burgos M, Melayah S, Berthou C, et al. CD5 expression promotes IL-10 production through activation of the MAPK/Erk pathway and upregulation of TRPC1 channels in b lymphocytes. Cell Mol Immunol (2018) 15:158–70. doi: 10.1038/cmi.2016.42
46. Liu F-T, Giustiniani J, Farren T, Jia L, Bensussan A, Gribben JG, et al. CD160 signaling mediates PI3K-dependent survival and growth signals in chronic lymphocytic leukemia. Blood (2010) 115:3079–88. doi: 10.1182/blood-2009-08
47. Eichhorst B, Robak T, Montserrat E, Ghia P, Niemann CU, Kater AP, et al. Chronic lymphocytic leukaemia: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol (2021) 32:23–33. doi: 10.1016/j.annonc.2020.09.019
48. Lin VS, Lew TE, Handunnetti SM, Blombery P, Nguyen T, Westerman DA, et al. BTK inhibitor therapy is effective in patients with CLL resistant to venetoclax. Blood (2020) 135:2266–70. doi: 10.1182/blood.2020004782
49. Brown JR, Byrd JC, Coutre SE, Benson DM, Flinn IW, Wagner-Johnston ND, et al. Idelalisib, an inhibitor of phosphatidylinositol 3-kinase p110δ, for relapsed/refractory chronic lymphocytic leukemia. Blood (2014) 123:3390–7. doi: 10.1182/blood-2013-11-535047
50. Roberts AW, Davids MS, Pagel JM, Kahl BS, Puvvada SD, Gerecitano JF, et al. Targeting BCL2 with venetoclax in relapsed chronic lymphocytic leukemia. N Engl J Med (2016) 374:311–22. doi: 10.1056/NEJMoa1513257
51. Skånland SS, Mato AR. Overcoming resistance to targeted therapies in chronic lymphocytic leukemia. Blood Adv (2021) 5:334–43. doi: 10.1182/bloodadvances.2020003423
52. Kajüter H, Wellmann I, Khil L, Jöckel K-H, Zhang C, Fink A-M, et al. Survival of patients with chronic lymphocytic leukemia before and after the introduction of chemoimmunotherapy in Germany. Blood Cancer J (2021) 11:174. doi: 10.1038/s41408-021-00556-7
53. Kaur V, Swami A. Ibrutinib in CLL: a focus on adverse events, resistance, and novel approaches beyond ibrutinib. Ann Hematol (2017) 96:1175–84. doi: 10.1007/s00277-017-2973-2
54. Farren TW, Giustiniani J, Fanous M, Liu F, Macey MG, Wright F, et al. Minimal residual disease detection with tumor-specific CD160 correlates with event-free survival in chronic lymphocytic leukemia. Blood Cancer J (2015) 5:e273. doi: 10.1038/bcj.2014.92
55. Rabot M, El Costa H, Polgar B, Marie-Cardine A, Aguerre-Girr M, Barakonyi A, et al. CD160-activating NK cell effector functions depend on the phosphatidylinositol 3-kinase recruitment. Int Immunol (2007) 19:401–9. doi: 10.1093/intimm/dxm005
56. Schmitt C, Ghazi B, Bellier F, Bensussan A. Identification and analysis of the human CD160 promoter: implication of a potential AML-1 binding site in promoter activation. Genes Immun (2009) 10:616–23. doi: 10.1038/gene.2009.52
57. Giustiniani J, Bensussan A, Marie-Cardine A. Identification and characterization of a transmembrane isoform of CD160 (CD160-TM), a unique activating receptor selectively expressed upon human NK cell activation. J Immunol (2009) 182:63–71. doi: 10.4049/jimmunol.182.1.63
58. Giustiniani J, Marie-Cardine A, Bensussan A. A soluble form of the MHC class I-specific CD160 receptor is released from human activated NK lymphocytes and inhibits cell-mediated cytotoxicity. J Immunol (2007) 178:1293–300. doi: 10.4049/jimmunol.178.3.1293
59. Anumanthan A, Bensussan A, Boumsell L, Christ AD, Blumberg RS, Voss SD, et al. Cloning of BY55, a novel ig superfamily member expressed on NK cells, CTL, and intestinal intraepithelial lymphocytes. J Immunol (1998) 161:2780–90.
60. Le Bouteiller P, Tabiasco J, Polgar B, Kozma N, Giustiniani J, Siewiera J, et al. CD160: A unique activating NK cell receptor. Immunol Lett (2011) 138:93–6. doi: 10.1016/j.imlet.2011.02.003
61. Fons P, Chabot S, Cartwright JE, Lenfant F, L’Faqihi F, Giustiniani J, et al. Soluble HLA-G1 inhibits angiogenesis through an apoptotic pathway and by direct binding to CD160 receptor expressed by endothelial cells. Blood (2006) 108:2608–15. doi: 10.1182/blood-2005-12-019919
62. Tsujimura K, Obata Y, Matsudaira Y, Nishida K, Akatsuka Y, Ito Y, et al. Characterization of murine CD160+ CD8+ T lymphocytes. Immunol Lett (2006) 106:48–56. doi: 10.1016/j.imlet.2006.04.006
63. Ortonne N, Ram-Wolff C, Giustiniani J, Marie-Cardine A, Bagot M, Mecheri S, et al. Human and mouse mast cells express and secrete the GPI-anchored isoform of CD160. J Invest Dermatol (2011) 131:916–24. doi: 10.1038/jid.2010.412
64. Tu TC, Brown NK, Kim T-J, Wroblewska J, Yang X, Guo X, et al. CD160 is essential for NK-mediated IFN-γ production. J Exp Med (2015) 212:415–29. doi: 10.1084/jem.20131601
65. Liu S, Zhang W, Liu K, Wang Y. CD160 expression on CD8+ T cells is associated with active effector responses but limited activation potential in pancreatic cancer. Cancer Immunol Immunother (2020) 69:789–97. doi: 10.1007/s00262-020-02500-3
66. Menguy T, Briaux A, Jeunesse E, Giustiniani J, Calcei A, Guyon T, et al. Anti-CD160, alone or in combination with bevacizumab, is a potent inhibitor of ocular neovascularization in rabbit and monkey models. Investig Opthalmol Vis Sci (2018) 59:2687. doi: 10.1167/iovs.18-24024
67. Abecassis S, Giustiniani J, Meyer N, Schiavon V, Ortonne N, Campillo JA, et al. Identification of a novel CD160+CD4+ T-lymphocyte subset in the skin: A possible role for CD160 in skin inflammation. J Invest Dermatol (2007) 127:1161–6. doi: 10.1038/sj.jid.5700680
68. Sako N, Schiavon V, Bounfour T, Dessirier V, Ortonne N, Olive D, et al. Membrane expression of NK receptors CD160 and CD158k contributes to delineate a unique CD4 + T-lymphocyte subset in normal and mycosis fungoides skin. Cytom Part A (2014) 85:869–82. doi: 10.1002/cyto.a.22512
69. Kaye J. CD160 and BTLA: LIGHTs out for CD4+ T cells. Nat Immunol (2008) 9:122–4. doi: 10.1038/ni0208-122
70. Kojima R, Kajikawa M, Shiroishi M, Kuroki K, Maenaka K. Molecular basis for herpesvirus entry mediator recognition by the human immune inhibitory receptor CD160 and its relationship to the cosignaling molecules BTLA and LIGHT. J Mol Biol (2011) 413:762–72. doi: 10.1016/j.jmb.2011.09.018
71. Rodriguez-Barbosa JI, Schneider P, Weigert A, Lee K-M, Kim T-J, Perez-Simon J-A, et al. HVEM, a cosignaling molecular switch, and its interactions with BTLA, CD160 and LIGHT. Cell Mol Immunol (2019) 16:679–82. doi: 10.1038/s41423-019-0241-1
72. Kim T-J, Park G, Kim J, Lim SA, Kim J, Im K, et al. CD160 serves as a negative regulator of NKT cells in acute hepatic injury. Nat Commun (2019) 10:3258. doi: 10.1038/s41467-019-10320-y
73. Maeda M, Carpenito C, Russell RC, Dasanjh J, Veinotte LL, Ohta H, et al. Murine CD160, ig-like receptor on NK cells and NKT cells, recognizes classical and nonclassical MHC class I and regulates NK cell activation. J Immunol (2005) 175:4426–32. doi: 10.4049/jimmunol.175.7.4426
74. Agrawal S, Marquet J, Freeman GJ, Tawab A, Bouteiller P, Roth P, et al. Cutting edge: MHC class I triggering by a novel cell surface ligand costimulates proliferation of activated human T cells. J Immunol (1999) 162:1223–6.
75. Liu W, Garrett SC, Fedorov EV, Ramagopal UA, Garforth SJ, Bonanno JB, et al. Structural basis of CD160:HVEM recognition. Structure (2019) 27:1286–1295.e4. doi: 10.1016/j.str.2019.05.010
76. Ali AK, Nandagopal N, Lee S-H. IL-15–PI3K–AKT–mTOR: A critical pathway in the life journey of natural killer cells. Front Immunol (2015) 6:355. doi: 10.3389/fimmu.2015.00355
77. Jiang K, Zhong B, Gilvary DL, Corliss BC, Hong-Geller E, Wei S, et al. Pivotal role of phosphoinositide-3 kinase in regulation of cytotoxicity in natural killer cells. Nat Immunol (2000) 1:419–25. doi: 10.1038/80859
78. Keating SE, Zaiatz-Bittencourt V, Loftus RM, Keane C, Brennan K, Finlay DK, et al. Metabolic reprogramming supports IFN-γ production by CD56 bright NK cells. J Immunol (2016) 196:2552–60. doi: 10.4049/jimmunol.1501783
79. Donnelly RP, Loftus RM, Keating SE, Liou KT, Biron CA, Gardiner CM, et al. mTORC1-dependent metabolic reprogramming is a prerequisite for NK cell effector function. J Immunol (2014) 193:4477–84. doi: 10.4049/jimmunol.1401558
80. Jameson G, Harmon C, Santiago RM, Houlihan DD, Gallagher TK, Lynch L, et al. Human hepatic CD56bright NK cells display a tissue-resident transcriptional profile and enhanced ability to kill allogenic CD8+ T cells. Front Immunol (2022) 13:921212. doi: 10.3389/fimmu.2022.921212
81. Huntington ND, Cursons J, Rautela J. The cancer–natural killer cell immunity cycle. Nat Rev Cancer (2020) 20:437–54. doi: 10.1038/s41568-020-0272-z
82. Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol (2008) 9:503–10. doi: 10.1038/ni1582
83. Vivier E. What is natural in natural killer cells? Immunol Lett (2006) 107:1–7. doi: 10.1016/j.imlet.2006.07.004
84. Long EO, Sik Kim H, Liu D, Peterson ME, Rajagopalan S. Controlling natural killer cell responses: Integration of signals for activation and inhibition. Annu Rev Immunol (2013) 31:227–58. doi: 10.1146/annurev-immunol-020711-075005
85. Sandoval-Borrego D, Moreno-Lafont MC, Vazquez-Sanchez EA, Gutierrez-Hoya A, López-Santiago R, Montiel-Cervantes LA, et al. Overexpression of CD158 and NKG2A inhibitory receptors and underexpression of NKG2D and NKp46 activating receptors on NK cells in acute myeloid leukemia. Arch Med Res (2016) 47:55–64. doi: 10.1016/j.arcmed.2016.02.001
86. Parry HM, Stevens T, Oldreive C, Zadran B, McSkeane T, Rudzki Z, et al. NK cell function is markedly impaired in patients with chronic lymphocytic leukaemia but is preserved in patients with small lymphocytic lymphoma. Oncotarget (2016) 7:68513–26. doi: 10.18632/ONCOTARGET.12097
87. Sun H, Xu J, Huang Q, Huang M, Li K, Qu K, et al. Reduced CD160 expression contributes to impaired NK-cell function and poor clinical outcomes in patients with HCC. Cancer Res (2018) 78:6581–93. doi: 10.1158/0008-5472.CAN-18-1049
88. Watanabe N, Gavrieli M, Sedy JR, Yang J, Fallarino F, Loftin SK, et al. BTLA is a lymphocyte inhibitory receptor with similarities to CTLA-4 and PD-1. Nat Immunol (2003) 4:670–9. doi: 10.1038/ni944
89. Rey J, Giustiniani J, Mallet F, Schiavon V, Boumsell L, Bensussan A, et al. The co-expression of 2B4 (CD244) and CD160 delineates a subpopulation of human CD8+ T cells with a potent CD160-mediated cytolytic effector function. Eur J Immunol (2006) 36:2359–66. doi: 10.1002/eji.200635935
90. Paulos CM, June CH. Putting the brakes on BTLA in T cell-mediated cancer immunotherapy. J Clin Invest (2010) 120:76–80. doi: 10.1172/JCI41811
91. Bozorgmehr N, Okoye I, Oyegbami O, Xu L, Fontaine A, Cox-Kennett N, et al. Expanded antigen-experienced CD160+CD8+effector T cells exhibit impaired effector functions in chronic lymphocytic leukemia. J Immunother Cancer (2021) 9:e002189. doi: 10.1136/jitc-2020-002189
92. Delgado J, Doubek M, Baumann T, Kotaskova J, Molica S, Mozas P, et al. Chronic lymphocytic leukemia: A prognostic model comprising only two biomarkers ( IGHV mutational status and FISH cytogenetics) separates patients with different outcome and simplifies the CLL-IPI. Am J Hematol (2017) 92:375–80. doi: 10.1002/ajh.24660
93. Crombie J, Davids MS. IGHV mutational status testing in chronic lymphocytic leukemia. Am J Hematol (2017) 92:1393–7. doi: 10.1002/ajh.24808
94. Tobin G, Thunberg U, Johnson A, Thoürn I, Soüderberg O, Hultdin M, et al. Somatically mutated ig VH3-21 genes characterize a new subset of chronic lymphocytic leukemia. Blood (2002) 99:2262–4. doi: 10.1182/blood.V99.6.2262
95. Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated ig V h genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood (1999) 94:1848–54. doi: 10.1182/blood.V94.6.1848
96. Burger JA, Chiorazzi N. B cell receptor signaling in chronic lymphocytic leukemia. Trends Immunol (2013) 34:592–601. doi: 10.1016/j.it.2013.07.002
97. Baudot AD, Jeandel PY, Mouska X, Maurer U, Tartare-Deckert S, Raynaud SD, et al. The tyrosine kinase syk regulates the survival of chronic lymphocytic leukemia b cells through PKC and proteasome-dependent regulation of mcl-1 expression. Oncogene (2009) 28:3261–73. doi: 10.1038/onc.2009.179
98. Woyach JA, Furman RR, Liu T-M, Ozer HG, Zapatka M, Ruppert AS, et al. Resistance mechanisms for the bruton’s tyrosine kinase inhibitor ibrutinib. N Engl J Med (2014) 370:2286–94. doi: 10.1056/NEJMoa1400029
99. Majolini MB, D’Elios MM, Galieni P, Boncristiano M, Lauria F, Del Prete G, et al. Expression of the T-Cell–specific tyrosine kinase lck in normal b-1 cells and in chronic lymphocytic leukemia b cells. Blood (1998) 91:3390–6. doi: 10.1182/blood.V91.9.3390
100. Wiestner A, Rosenwald A, Barry TS, Wright G, Davis RE, Henrickson SE, et al. ZAP-70 expression identifies a chronic lymphocytic leukemia subtype with unmutated immunoglobulin genes, inferior clinical outcome, and distinct gene expression profile. Blood (2003) 101:4944–51. doi: 10.1182/blood
101. Ginaldi L, De Martinis M, Matutes E, Farahat N, Morilla R, Catovsky D. Levels of expression of CD19 and CD20 in chronic b cell leukaemias. J Clin Pathol (1998) 51:364–9. doi: 10.1136/jcp.51.5.364
102. Fournier S, Delespesse G, Rubio M, Biron G, Sarfati M. CD23 antigen regulation and signaling in chronic lymphocytic leukemia. J Clin Invest (1992) 89:1312–21. doi: 10.1172/JCI115717
103. Vuillier F, Dumas G, Magnac C, Prevost M-C, Iné Lalanne A, Oppezzo P, et al. Lower levels of surface b-cell-receptor expression in chronic lymphocytic leukemia are associated with glycosylation and folding defects of the and CD79a chains. Blood (2005) 105:2933–40. doi: 10.1182/blood
104. Tibaldi E, Brunati AM, Zonta F, Frezzato F, Gattazzo C, Zambello R, et al. Lyn-Mediated SHP-1 recruitment to CD5 contributes to resistance to apoptosis of b-cell chronic lymphocytic leukemia cells. Leukemia (2011) 25:1768–81. doi: 10.1038/leu.2011.152
105. Hippen KL, Tze LE, Behrens TW. CD5 maintains tolerance in anergic b cells. J Exp Med (2000) 191:883–9. doi: 10.1084/jem.191.5.883
106. Wang H-Q, Jia L, Li Y-T, Farren T, Agrawal SG, Liu F-T. Increased autocrine interleukin-6 production is significantly associated with worse clinical outcome in patients with chronic lymphocytic leukemia. J Cell Physiol (2019) 234:13994–4006. doi: 10.1002/jcp.28086
107. Zhou H, Li XM, Meinkoth J, Pittman RN. Akt regulates cell survival and apoptosis at a postmitochondrial level. J Cell Biol (2000) 151:483–94. doi: 10.1083/jcb.151.3.483
108. Caligaris-Cappio F. Role of the microenvironment in chronic lymphocytic leukaemia. Br J Haematol (2003) 123:380–8. doi: 10.1046/j.1365-2141.2003.04679.x
109. Vlachonikola E, Stamatopoulos K, Chatzidimitriou A. T Cells in chronic lymphocytic leukemia: A two-edged sword. Front Immunol (2021) 11:612244. doi: 10.3389/fimmu.2020.612244
110. Burger JA, Gribben JG. The microenvironment in chronic lymphocytic leukemia (CLL) and other b cell malignancies: Insight into disease biology and new targeted therapies. Semin Cancer Biol (2014) 24:71–81. doi: 10.1016/j.semcancer.2013.08.011
111. Heinig K, Gätjen M, Grau M, Stache V, Anagnostopoulos I, Gerlach K, et al. Access to follicular dendritic cells is a pivotal step in murine chronic lymphocytic leukemia b-cell activation and proliferation. Cancer Discovery (2014) 4:1448–65. doi: 10.1158/2159-8290.CD-14-0096
112. Herishanu Y, Pérez-Galán P, Liu D, Biancotto A, Pittaluga S, Vire B, et al. The lymph node microenvironment promotes b-cell receptor signaling, NF-κB activation, and tumor proliferation in chronic lymphocytic leukemia. Blood (2011) 117:563–74. doi: 10.1182/blood-2010-05-284984
113. Sportoletti P, De Falco F, Del Papa B, Baldoni S, Guarente V, Marra A, et al. Nk cells in chronic lymphocytic leukemia and their therapeutic implications. Int J Mol Sci (2021):226665. doi: 10.3390/ijms22136665
114. Ramsay AG, Clear AJ, Fatah R, Gribben JG. Multiple inhibitory ligands induce impaired T-cell immunologic synapse function in chronic lymphocytic leukemia that can be blocked with lenalidomide: Establishing a reversible immune evasion mechanism in human cancer. Blood (2012) 120:1412–21. doi: 10.1182/blood-2012-02-411678
115. Brusa D, Serra S, Coscia M, Rossi D, D’Arena G, Laurenti L, et al. The PD-1/PD-L1 axis contributes to T-cell dysfunction in chronic lymphocytic leukemia. Haematologica (2013) 98:953–63. doi: 10.3324/haematol.2012.077537
116. Karabon L, Partyka A, Ciszak L, Pawlak-Adamska E, Tomkiewicz A, Bojarska-Junak A, et al. Abnormal expression of BTLA and CTLA-4 immune checkpoint molecules in chronic lymphocytic leukemia patients. J Immunol Res (2020) 2020:1–12. doi: 10.1155/2020/6545921
117. Cox MJ, Lucien F, Sakemura R, Boysen JC, Kim Y, Horvei P, et al. Leukemic extracellular vesicles induce chimeric antigen receptor T cell dysfunction in chronic lymphocytic leukemia. Mol Ther (2021) 29:1529–40. doi: 10.1016/j.ymthe.2020.12.033
118. Katrinakis G, Kyriakou D, Papadaki H, Kalokyri I, Markidou F, Eliopoulos GD. Defective natural killer cell activity in b-cell chronic lymphocytic leukaemia is associated with impaired release of natural killer cytotoxic factor(s) but not of tumour necrosis factor-α. Acta Haematol (1996) 96:16–23. doi: 10.1159/000203709
119. Vivier E, Nunès JA, Veíly F. Natural killer cell signaling pathways. Sci (80- ) (2004) 306:1517–9. doi: 10.1126/science.1103478
120. Veuillen C, Aurran-Schleinitz T, Castellano R, Rey J, Mallet F, Orlanducci F, et al. Primary b-CLL resistance to NK cell cytotoxicity can be overcome in vitro and in vivo by priming NK cells and monoclonal antibody therapy. J Clin Immunol (2012) 32:632–46. doi: 10.1007/s10875-011-9624-5
121. Maki G, Hayes GM, Naji A, Tyler T, Carosella ED, Rouas-Freiss N, et al. NK resistance of tumor cells from multiple myeloma and chronic lymphocytic leukemia patients: implication of HLA-G. Leukemia (2008) 22:998–1006. doi: 10.1038/leu.2008.15
122. Tomuleasa C, Selicean C, Cismas S, Jurj A, Marian M, Dima D, et al. Minimal residual disease in chronic lymphocytic leukemia: A consensus paper that presents the clinical impact of the presently available laboratory approaches. Crit Rev Clin Lab Sci (2018) 55:329–45. doi: 10.1080/10408363.2018.1463508
123. Burger JA, O’Brien S. Evolution of CLL treatment — from chemoimmunotherapy to targeted and individualized therapy. Nat Rev Clin Oncol (2018) 15:510–27. doi: 10.1038/s41571-018-0037-8
124. Rawstron AC, Villamor N, Ritgen M, Böttcher S, Ghia P, Zehnder JL, et al. International standardized approach for flow cytometric residual disease monitoring in chronic lymphocytic leukaemia. Leukemia (2007) 21:956–64. doi: 10.1038/sj.leu.2404584
125. Thompson PA, Srivastava J, Peterson C, Strati P, Jorgensen JL, Hether T, et al. Minimal residual disease undetectable by next-generation sequencing predicts improved outcome in CLL after chemoimmunotherapy. Blood (2019) 134:1951–9. doi: 10.1182/blood.2019001077
126. Dowling AK, Liptrot SD, O’Brien D, Vandenberghe E. Optimization and validation of an 8-color single-tube assay for the sensitive detection of minimal residual disease in b-cell chronic lymphocytic leukemia detected via flow cytometry. Lab Med (2016) 47:103–11. doi: 10.1093/labmed/lmw006
127. Hojjat-Farsangi M, Khan AS, Daneshmanesh AH, Moshfegh A, Sandin Å, Mansouri L, et al. The tyrosine kinase receptor ROR1 is constitutively phosphorylated in chronic lymphocytic leukemia (CLL) cells. PloS One (2013) 8:e78339. doi: 10.1371/journal.pone.0078339
128. Cui B, Ghia EM, Chen L, Rassenti LZ, DeBoever C, Widhopf GF, et al. High-level ROR1 associates with accelerated disease progression in chronic lymphocytic leukemia. Blood (2016) 128:2931–40. doi: 10.1182/blood-2016-04-712562
129. Farren TW, Sadanand KS, Agrawal SG. Highly sensitive and accurate assessment of minimal residual disease in chronic lymphocytic leukemia using the novel CD160-ROR1 assay. Front Oncol (2020) 10:597730. doi: 10.3389/fonc.2020.597730
Keywords: chronic lymphocytic leukemia, CD160, NK cells, minimal residual disease, prognosis
Citation: Oumeslakht L, Aziz A-i, Bensussan A and Ben Mkaddem S (2022) CD160 receptor in CLL: Current state and future avenues. Front. Immunol. 13:1028013. doi: 10.3389/fimmu.2022.1028013
Received: 25 August 2022; Accepted: 19 October 2022;
Published: 07 November 2022.
Edited by:
Laura Patrussi, University of Siena, ItalyReviewed by:
Tiziana Vaisitti, University of Turin, ItalyCopyright © 2022 Oumeslakht, Aziz, Bensussan and Ben Mkaddem. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sanae Ben Mkaddem, c2FuYWUuYmVubWthZGRlbUB1bTZwLm1h; Armand Bensussan, YXJtYW5kLmJlbnN1c3NhbkBpbnNlcm0uZnI=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.