
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol., 11 October 2022
Sec. Molecular Innate Immunity
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.1027403
This article is part of the Research TopicImmunomodulatory Role of Metalloproteases in Chronic Inflammatory DiseasesView all 13 articles
 Yongfang Wang
Yongfang Wang Huiying Wang*
Huiying Wang*Histone deacetylase 1 (HDAC1) is a unique member of the classes I HDACs and helps to regulate acute and chronic adaptation to environmental stimuli such as allergen, stress. Allergic diseases are complex diseases resulting from the effect of multiple genetic and interacting foreign substances. Epigenetics play an important role in both pathological and immunomodulatory conditions of allergic diseases. To be consistent with this role, recent evidence strongly suggests that histone deacetylase 1 (HDAC1) plays a critical role in allergic response. HDAC1 expression is stimulated by allergen and attributes to increase T helper 2 (Th2) cytokine levels, decrease Th1/Th17 cells and anti-inflammatory cytokine Interleukin-10 (IL-10), and TWIK-related potassium channel-1 (Trek-1) expression. This review focuses on the contribution of HDAC1 and the regulatory role in characterizing allergic endotypes with common molecular pathways and understanding allergic multimorbidity relationships, as well as addressing their potential as therapeutic targets for these conditions.
Epigenetics includes the heritable alterations in gene expression without any changes in a deoxyribonucleic acid (DNA) sequence, which is crucial in the pathophysiology of many diseases (1, 2). Multiple enzymes have been extensively studied that induce epigenetic changes, such as DNA methylation and histone acetylation of DNA regions. Histone deacetylases (HDACs) are the enzymes that catalyze lysine deacetylation of both histone and non-histone proteins. HDACs increase the positive charge on histones after removing acetyl groups from lysine residues, thus increasing the affinity of positively charged histones for negatively charged DNA (3). HDACs lead to the condensation of the chromatin and then reduces the accessibility of transcriptase, and finally leads to an overall suppression of gene transcription (4). HDAC family has four subclasses including I, II, III and IV. Classes I, II, and IV HDACs utilize a zinc-dependent mechanism and belong to the Zn2+ superfamily, while class III HDACs require nicotinamide adenine dinucleotide(NAD)+ for catalytic activity.
Histone deacetylase 1 (HDAC1) is a unique member of the classes I HDACs that has been shown to be involved in gene transcription, transcriptional regulation, cell cycle progression and developmental events by controlling both enzyme activity and epigenetics of key proteins (5). HDAC1 is the most abundant member of the class I HDACs in pulmonary endothelial cells (6), regulating the enzymatic activity and epigenetics of key proteins to adapt to external stimuli. It can efficiently decrotonylate this relatively less abundant histone modification (7). Moreover, HDAC1 is the key regulators of T cell subset differentiation and T cell-mediated immune diseases (8) that helps to regulate acute and chronic adaptation to environmental stimuli such as allergen, stress (9). Allergic diseases represent a collection of disorders such as allergic rhinitis, asthma, that mostly characterized by a type 2 immune response involving Th2 cells, eosinophils and mast cells, and M2 macrophages. T cell specific loss of HDAC1 leads to an increase in Th2 type allergic airway inflammation, such as enhanced secretion of Th2 type cytokines, eosinophil recruitment to the lung (10).For example, HDAC1 is highly expressed and the most abundant member of the class I HDACs in allergic rhinitis and severe asthma (3, 11, 12). Studies show that HDAC1 is localized within most airway cells and infiltrating inflammatory cells of asthmatic lung tissues (13). HDAC1 is significantly upregulated in the murine AR model while H3 acetylation is decreased at lysine 9 (H3AcK9) (14). The HDAC1 inhibitor sodium butyrate exhibits a preventive effect by decreasing HDAC1 expression and increasing H3 acetylation at lysine 9. Herein, we made a thorough review of recent studies and summarized the emerging functions of HDAC1 by regulating histone modifications and gene transcription in allergic disease.
Generalized allergic diseases include allergic rhinitis, asthma, Immunoglobulin E(IgE)-mediated food allergy, eosinophilic esophagitis, drug allergy, atopic dermatitis, and urticaria/angioedema. These different allergic diseases share several overlapping inflammatory pathways concerning with the hypersensitivity of the individual to foreign substances (15–18). Allergic diseases are a type 2 immune disorder classically characterized by high levels of IgE-mediated inflammation and Th1/Th2 cells imbalance (19–21). The Th 2 immune response involves Th2 cells, type 2 innate lymphoid cells, mast cells, eosinophils, and M2 macrophages (22). Th2 cytokines, particularly IL-4, are essential in the pathophysiology of allergic rhinitis and asthma (23, 24). In type I immediate allergic responses, naïve T cells is activated by dendritic cells to differentiate, proliferate and clonally expand into Th2 cells (23, 25). Enhanced Th2 cytokines induce IgE synthesis in B cells in an indirect manner (26, 27). In turn, IgE can also enhance Th2-cell response after sensitization (28). However, the aberrant immune responses in atopic disorders are not fully understood yet.
Epigenetics plays a major pathogenetic role in the development and management of allergic diseases by superimposing its effects above the DNA molecule through interaction with susceptibility genes, environmental factors, and immunologic influences (29). Epigenetics holds the key to unravel the complex associations between phenotypes and endotypes of allergic disease by identifying effective therapies and diagnosis (30). Epigenetic modifications of genes are contributing to asthma induced by allergens, such as DNA methylation changes in DCs, can be passed to future generations (31, 32). Histone modifications and DNA methylation represent the classical epigenetic mechanisms. Histone modifications participate in airway remodeling by regulation of T cells and macrophages. Inhibitors of histone-modifying enzymes may potentially be used as anti-allergic drugs (33).
HDAC1 displays compensatory or specific roles in different cell types or in response to different stimuli and signaling pathways of atopic disorders. The expression level of HDAC1 in the nasal epithelia is elevated in allergic rhinitis (34), and HDAC1 inhibitors reduce the symptoms of allergic rhinitis (3, 12, 35). Immunohistochemical results also demonstrate the high HDAC1 expression in nasal epithelium of patients with sinusitis and nasal polyps (36). The differentially expressed genes (DEGs) analysis of 1,662 nasal−epithelium tissue samples and 572 DEGs from peripheral blood samples shows that HDAC1 is hub genes and serves an important role in the process of asthma (37). HDAC1 expression is enhanced in patients with severe asthma compared with healthy volunteers (11). Moreover, expression of HDAC1 is upregulated by the stimulation of dermatophagoides pteronyssinus allergen (Der p 1) in peripheral blood mononuclear cells of patients with severe and non-severe asthma (38). Animal models of allergic asthma exhibits significantly higher expression of HDAC1 compared to control. Selective targeting of HDAC1 may improve therapeutic effects of asthma (39). One single nucleotide polymorphism (SNP) in HDAC1 (rs1741981) is closely associated to asthma severity in a recessive model and increases the sensitivity to systemic corticosteroids treatment in asthmatic patients (40, 41). Besides, in epidermal keratinocytes, HDAC1 expression and activity are upregulated by the aryl hydrocarbon receptor nuclear translocator (ARNT or HIF1β) (42).
A number of studies have shown that exposure to allergens would increase HDAC1 expression, leading to significantly advanced Th2 cytokine levels, reduced Th1/Th17 cells and anti-inflammatory cytokine IL-10, and Trek-1 expression (Figure 1). In the mouse model of allergic rhinitis, epigenetic regulation of HDAC1 produce an imbalance in Th1/Th2 by decreasing the secretion of interferon(IFN)-γ, increasing the secretion of IL-4 and IL-6 (14). Moreover, the transcriptional activity of forkhead box P3(Foxp3) is restrained that decreases T regulatory cells (43). As the number of Th1 cells decreases, the number of Th2 cells correspondingly increases, and subsequently the secretion of IL-4 increases to promote the activation of IgE released by B cells (44). Additionally, murine models of asthma confirm the upregulation of HDAC1 could increase airway inflammation, Th2 cytokine level, IgE and goblet cell metaplasia dramatically (45). Indeed, treatment with HDAC1 inhibitor trichostatin A(TSA) significantly attenuate airway hyper-responsiveness, mucus occlusions in lung tissue and the numbers of eosinophils and lymphocytes in bronchoalveolar lavage fluid. The infiltration of CD4+ and the expression of IL-4, IL-5, and IgE in BALF are also restrained by TSA (13). Particularly, Th2 cytokine interleukin 4 (IL-4) plays a key role in the pathogenesis of allergic disorders (46). HDAC1 can be recruited to the IL-4 gene locus in CD4(+) T cells, thereby promoting the immunoactivity of CD4 positive T cells to increase Th2 cytokine levels (47–49). The IL-4-induced rat nasal epithelial barrier dysfunction is blocked by HDAC1 inhibitor (Trichostatin A), or sodium butyrate (NaB), or administration of Clostridium Butyricum (Table 1) (14, 62). A non-secreted IL-4 variant (IL-4δ13) expression in human γδ T-cells is also stimulated by another HDAC inhibitor valproic acid (VPA) (Table 1) (58). The Induction of IL-4δ13 increases cytoplasmic IL-4Rα and decreases mature IL-4 (59). Along with the role of HDAC1 in altering the Th2 cytokine profile, it is reported that HDAC1 is recruited to change the euchromatin into tightly-packed heterochromatin to repress its expression in Th17 cells through production of cytokine IL17 (63). HDAC1 inhibitor sodium butyrate increases IL−17、interleukin 2 (IL−2) and interferon γ and decreases the expression of IL−4 and IL−5 (50). HDAC1 regulates the retinoic acid-related orphan receptor-mediated transcriptional activation of IL-17 (64).
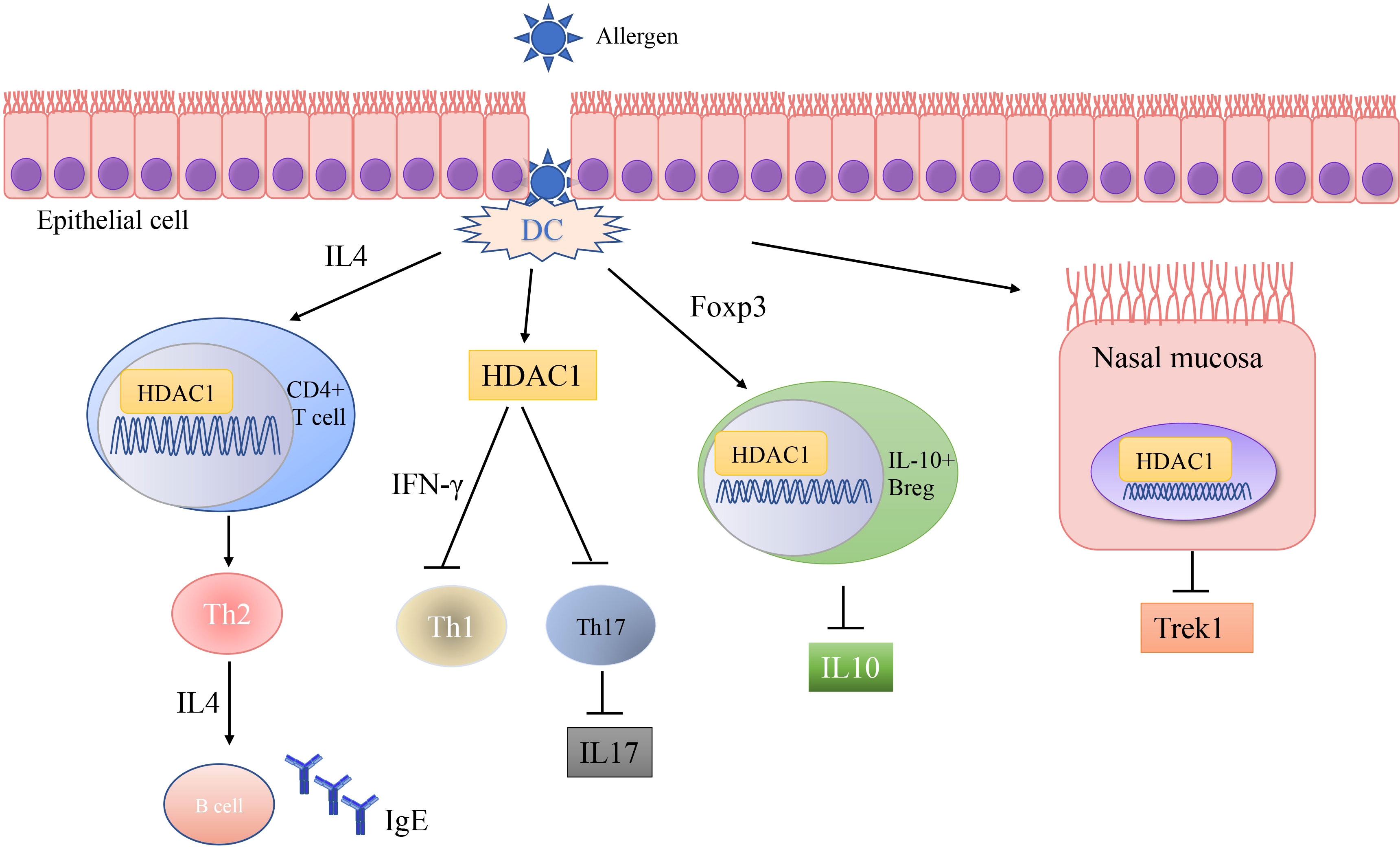
Figure 1 Schematic representations of HDAC1 related mechanism in allergic diseases. Allergic disease patients have an epithelial barrier suffering from allergen stimulation. Exposure to allergens activate dendritic cell and increase HDAC1 expression, leading to significantly increase Th2 cytokine levels, decrease Th1/Th17 cells and anti-inflammatory cytokine IL-10, and Trek-1 expression.
Apart from the studies showing the Th1/Th2 imbalance and inhibition of IL17, histone deacetylation is an important mechanism that regulates the expression of anti-inflammatory cytokine IL-10 (65). HDAC1 represses IL-10 transcription activity by reducing chromatin accessibility and recruiting histone H3 acetylation at IL-10 regulatory regions (66). Sodium butyrate restrains the activation of HDAC1 in the antigen specific B cells to induce the expression of IL-10 and decrease the production of IgE in allergic rhinitis model (51). Another HDAC inhibitor entinostat stimulates the formation of IL-10 positive Breg cells to suppress contact hypersensitivity in vivo (54). Indeed, the administration with Clostridium butyricum (C. butyricum) enforces the effect of specific immunotherapy on intestinal allergic inflammation by increasing the phosphorylation of HDAC1, the expression of IL-10 and the IgE-producing plasma cells (67).
There are some studies documenting the role of Trek-1 in the maintenance of epithelial cell barrier function (62, 68). The allergic responses induce an insufficiency of Trek1 expression (69). Enhanced IL-4 markedly suppresses the expression of Trek1 via upregulating the expression of the HDAC1 in the nasal mucosa of allergic rhinitis (62). The treatment with antigen-specific immunotherapy and administration of probiotic C. butyricum reduce the serum levels of Th2 cytokines by increasing Trek-1 expression levels and decreasing HDAC1 in the nasal mucosa of allergic rhinitis patients (23). Allergic responses markedly suppress the expression of Trek1 in the intestinal epithelia via increasing the expression of HDAC1 (70).
Different stimuli includes temperature, particles containing hazardous chemicals, and small chemical molecules that exhibits an impact on the expression of HDAC1. Particulate matter (PM) 2.5 exposure and cold stress (PMCS) exposures promote inflammation and redox levels in asthmatic mice through increasing the percentage of Th2 T cells and decreasing Th1 T cells, thereby decreasing HDAC1 expression and hyperacetylation of H3K9 and H3K14 in IL-4 gene promoter of CD4+T cells (71). Mechanically, HDAC1 helps maintain DNA-binding sites (response elements) for redox-sensitive transcription factors by co-repressor complexes (72). Besides, exposure to diesel exhaust particulate matter (DEP) causes degradation of histone deacetylase 1 (HDAC1), thus recruiting histone acetyltransferase (HAT) p300 to the promoter of the Cyclooxygenase-2 (COX-2) gene in vitro human bronchial epithelial cell line (BEAS-2B) (73). In addition, chronic exposure to alcohol decreases HDAC1 expression (74). Trichostatin A alleviates tissue damage that is caused by cigarette smoke exposure (75, 76).
On the other hand, HDAC1 is modulated by upstream transcription factors and signaling pathway in allergic diseases. Previous studies have shown that the transcription factor c-Myc-interacting zinc finger protein-1 (Miz1) was upregulated in allergic asthma, which in turn prevented the pro-Th1 skewing through the recruitment of histone deacetylase 1 (HDAC1) and transcriptional repression of IL-12 (77). HDAC1 expression is also increased by the advanced glycation end products via the phosphatidylinositol 3-kinase(PI3K)/AKT pathway through promoting the airway inflammation (45).
Moreover, gut microbiome is associated with allergic diseases (78–81). Sodium butyrate treatments lead to increase the richness in the stomach and colon and modify colonic microbial composition in pigs by decreasing HDAC1 (82, 83). The intestinal epithelial cells specific HDAC1 support intestinal homeostasis by controlling specific biological processes including oxidation-reduction, survival and translation processes, differentiation and lipid-related metabolic pathways via Janus kinase(JAK)/signal transducer and activator of transcription (STAT) pathway and steroid receptor pathway (84–86).
A large body of evidence shows that HDAC1 is a potential clinical target for treatment of allergic diseases. At present, numerous questions remain regarding to the precise functions of HDAC1 in allergic inflammation. The HDAC inhibitors such as trichostatin A (TSA) have a bidentate cheator, which binds to catalytic Zn2+ (87). The broad-spectrum HDAC1 inhibitor trichostatin A has a hydroxamic acid based structure that affects the expression of thousand genes in the human genome. There is still no clinical application of these HDAC1 inhibitors. Thus, there is an ongoing discussion whether selective HDAC inhibitors have advantage for clinical use. These small-molecule compounds targeting HDAC1 have no serious toxicities.
There are many HDAC inhibitors in ongoing clinical trials (Table 1). The study on the tolerance of trichostatin A in patients with recurrent or refractory hematological malignancies is still in progress. Genetic and pharmacological studies have confirmed that HDAC1 is the key enzyme to reverse tumor immune escape. Entinostat selectively promotes the immune editing of new tumor antigens, leading effectively reshaping the tumor immune microenvironment (55). The randomized phase III trial of endocrine therapy confirms target inhibition in entinostat-treated breast cancer patients (56). Valproic acid and entinostat exhibit synergy in preclinical models when combined with rituximab in Non-Hodgkin’s lymphoma (57). On the other hand, Valproic acid is the first-line drug for tonic clonic seizures (60). Besides, Valproic acid induces apoptosis of activated T cells to maintain immune homeostasis, which may be a safe and effective treatment for autoimmune diseases, such as multiple sclerosis (61). Entinostat and valproic acid can potentially be repurposed for treating asthma (88). However, there is no clinical trials to determine the role of entinostat and valproic acid in asthma. These findings highlight the need for further exploration of HDAC inhibitors in allergic diseases.
Sodium butyrate therapy during shigellosis leads to early reduction of inflammation and enhanced antimicrobial peptides (LL-37) expression in the rectal epithelia (52). The double‐blind randomized controlled trial shows that sodium-butyrate supplementation in 49 inflammatory bowel diseases patients increases the growth of bacteria able to produce short‐chain fatty acids (SCFA) with potentially anti-inflammatory action (53). These results support the potential effect of sodium butyrate in modulating gut microbiota, which anyway requires further confirmatory data including more patients. In considering future potential clinical application in allergic diseases, more studies are still needed to develop new HDAC1 specific selective inhibitors. HDAC1 specific selective inhibitors may provide a new starting point for the treatment of allergic diseases.
Allergic diseases comprise some of the most common chronic disorders in both childhood and adulthood. Allergic conditions are influenced by epigenetic elements which ultimately affect multiple molecular pathways (89, 90). Accumulating evidences have established in HDAC1 as a critical regulator of immune response in terms of imbalance in Th1/Th2, change in anti-inflammatory cytokine IL-10/IL-17 and Trek-1 expression. Over the past decades, histone deacetylase inhibitors are being evaluated in clinical trials for their safety and efficacy (91, 92). HDAC1 has become an attractive target to treat a wide range of diseases. However, these HDAC inhibitors do not display high selectivity and may restrain related HDACs. The potential side effects due to inhibition of systemic immune response are an urgent problem to be solved. Besides, additional work is required to examine the expression and activity of HDAC1 in allergic diseases. The development of selective HDAC1 inhibitors may lead to new therapeutic agents for allergic diseases, particularly in situations where current therapies are suboptimal.
All authors contributed to the search and collation of literature. YW contributed to the manuscript preparation and the revision of the manuscript. All authors read and approved the final manuscript.
This work is supported by the National Natural Science Foundation of China (NSFC) (Grant 81902331).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Nagy C, Vaillancourt K, Turecki G. A role for activity-dependent epigenetics in the development and treatment of major depressive disorder. Genes Brain Behav (2018) 17:e12446. doi: 10.1111/gbb.12446
2. Lockett GA, Patil VK, Soto-Ramírez N, Ziyab AH, Holloway JW, Karmaus W. Epigenomics and allergic disease. Epigenomics (2013) 5:685–99. doi: 10.2217/epi.13.68
3. Yang J, Zhong W, Xue K, Wang Z. Epigenetic changes: An emerging potential pharmacological target in allergic rhinitis. Int Immunopharmacol (2019) 71:76–83. doi: 10.1016/j.intimp.2019.03.004
4. Hutt DM, Roth DM, Marchal C, Bouchecareilh M. Using histone deacetylase inhibitors to analyze the relevance of HDACs for translation. Methods Mol Biol (2017) 1510:77–91. doi: 10.1007/978-1-4939-6527-4_6
5. Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene (2005) 363:15–23. doi: 10.1016/j.gene.2005.09.010
6. Dunaway LS, Pollock JS. HDAC1: An environmental sensor regulating endothelial function. Cardiovasc Res (2022) 118:1885–903. doi: 10.1093/cvr/cvab198
7. Fellows R, Denizot J, Stellato C, Cuomo A, Jain P, Stoyanova E, et al. Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nat Commun (2018) 9:105. doi: 10.1038/s41467-017-02651-5
8. Göschl L, Preglej T, Boucheron N, Saferding V, Müller L, Platzer A, et al. Histone deacetylase 1 (HDAC1): A key player of T cell-mediated arthritis. J Autoimmun (2020) 108:102379. doi: 10.1016/j.jaut.2019.102379
9. Ma P, Schultz RM. HDAC1 and HDAC2 in mouse oocytes and preimplantation embryos: Specificity versus compensation. Cell Death Differ (2016) 23:1119–27. doi: 10.1038/cdd.2016.31
10. Grausenburger R, Bilic I, Boucheron N, Zupkovitz G, El-Housseiny L, Tschismarov R, et al. Conditional deletion of histone deacetylase 1 in T cells leads to enhanced airway inflammation and increased Th2 cytokine production. J Immunol (2010) 185:3489–97. doi: 10.4049/jimmunol.0903610
11. Butler CA, McQuaid S, Taggart CC, Weldon S, Carter R, Skibinski G, et al. Glucocorticoid receptor β and histone deacetylase 1 and 2 expression in the airways of severe asthma. Thorax (2012) 67:392–8. doi: 10.1136/thoraxjnl-2011-200760
12. Wang Y, Lv L, Zang H, Gao Z, Zhang F, Wang X, et al. Regulation of Trek1 expression in nasal mucosa with allergic rhinitis by specific immunotherapy. Cell Biochem Funct (2015) 33:23–8. doi: 10.1002/cbf.3075
13. Choi JH, Oh SW, Kang MS, Kwon HJ, Oh GT, Kim DY. Trichostatin a attenuates airway inflammation in mouse asthma model. Clin Exp Allergy (2005) 35:89–96. doi: 10.1111/j.1365-2222.2004.02006.x
14. Wang J, Wen L, Wang Y, Chen F. Therapeutic effect of histone deacetylase inhibitor, sodium butyrate, on allergic rhinitis in vivo. DNA Cell Biol (2016) 35:203–8. doi: 10.1089/dna.2015.3037
15. Saunders SP, Moran T, Floudas A, Wurlod F, Kaszlikowska A, Salimi M, et al. Spontaneous atopic dermatitis is mediated by innate immunity, with the secondary lung inflammation of the atopic march requiring adaptive immunity. J Allergy Clin Immunol (2016) 137:482–91. doi: 10.1016/j.jaci.2015.06.045
16. Krempski JW, Dant C, Nadeau KC. The origins of allergy from a systems approach. Ann Allergy Asthma Immunol (2020) 125:507–16. doi: 10.1016/j.anai.2020.07.013
17. Peroni DG, Nuzzi G, Trambusti I, Di Cicco ME, Comberiati P. Microbiome composition and its impact on the development of allergic diseases. Front Immunol (2020) 11:700. doi: 10.3389/fimmu.2020.00700
18. Luscher E. Allergic disorders of the upper respiratory and esophageal tracts and of the hearing organ with some general remarks on the so-called allergic diseases in humans. Schweiz Med Wochenschr (1953) 83:619–22.
19. Zhu TH, Zhu TR, Tran KA, Sivamani RK, Shi VY. Epithelial barrier dysfunctions in atopic dermatitis: a skin-gut-lung model linking microbiome alteration and immune dysregulation. Br J Dermatol (2018) 179:570–81. doi: 10.1111/bjd.16734
20. Yao Y, Chen CL, Yu D, Liu Z. Roles of follicular helper and regulatory T cells in allergic diseases and allergen immunotherapy. Allergy (2021) 76:456–70. doi: 10.1111/all.14639
21. Robinson DS, Larché M, Durham SR. Tregs and allergic disease. J Clin Invest (2004) 114:1389–97. doi: 10.1172/JCI200423595
22. Ogulur I, Pat Y, Ardicli O, Barletta E, Cevhertas L, Fernandez-Santamaria R, et al. Advances and highlights in biomarkers of allergic diseases. Allergy (2021) 76:3659–86. doi: 10.1111/all.15089
23. Shamji MH, Valenta R, Jardetzky T, Verhasselt V, Durham SR, Würtzen PA, et al. The role of allergen-specific IgE, IgG and IgA in allergic disease. Allergy (2021) 76:3627–41. doi: 10.1111/all.14908
24. Averbeck M, Gebhardt C, Emmrich F, Treudler R, Simon JC. Immunologic principles of allergic disease. J Dtsch Dermatol Ges (2007) 5:1015–28. doi: 10.1111/j.1610-0387.2007.06538.x
25. Akdis M, Akdis CA. Mechanisms of allergen-specific immunotherapy. J Allergy Clin Immunol (2007) 119:780–91. doi: 10.1016/j.jaci.2007.01.022
26. Wypych TP, Marzi R, Wu GF, Lanzavecchia A, Sallusto F. Role of b cells in T(H) cell responses in a mouse model of asthma. J Allergy Clin Immunol (2018) 141:1395–410. doi: 10.1016/j.jaci.2017.09.001
27. Takhar P, Corrigan CJ, Smurthwaite L, O’Connor BJ, Durham SR, Lee TH, et al. Class switch recombination to IgE in the bronchial mucosa of atopic and nonatopic patients with asthma. J Allergy Clin Immunol (2007) 119:213–8. doi: 10.1016/j.jaci.2006.09.045
28. van der Heijden FL, Joost van Neerven RJ, van Katwijk M, Bos JD, Kapsenberg ML. Serum-IgE-facilitated allergen presentation in atopic disease. J Immunol (1993) 150:3643–50.
29. Bellanti JA. Genetics/epigenetics/allergy: The gun is loaded … but what pulls the trigger? Allergy Asthma Proc (2019) 40:76–83. doi: 10.2500/aap.2019.40.4205
30. Bellanti JA, Settipane RA. Genetics, epigenetics, and allergic disease: A gun loaded by genetics and a trigger pulled by epigenetics. Allergy Asthma Proc (2019) 40:73–5. doi: 10.2500/aap.2019.40.4206
31. Gregory DJ, Kobzik L, Yang Z, McGuire CC, Fedulov AV. Transgenerational transmission of asthma risk after exposure to environmental particles during pregnancy. Am J Physiol Lung Cell Mol Physiol (2017) 313:L395–l405. doi: 10.1152/ajplung.00035.2017
32. Long A, Bunning B, Sampath V, DeKruyff RH, Nadeau KC. Epigenetics and the environment in airway disease: Asthma and allergic rhinitis. Adv Exp Med Biol (2020) 1253:153–81. doi: 10.1007/978-981-15-3449-2_6
33. Alaskhar Alhamwe B, Khalaila R, Wolf J, von Bülow V, Harb H, Alhamdan F, et al. Histone modifications and their role in epigenetics of atopy and allergic diseases. Allergy Asthma Clin Immunol (2018) 14:39. doi: 10.1186/s13223-018-0259-4
34. Siti Sarah CO, Md Shukri N, Mohd Ashari NS, Wong KK. Zonula occludens and nasal epithelial barrier integrity in allergic rhinitis. PeerJ (2020) 8:e9834. doi: 10.7717/peerj.9834
35. Lanza M, Casili G, Filippone A, Campolo M, Paterniti I, Cuzzocrea S, et al. Evaluating the protective properties of a xyloglucan-based nasal spray in a mouse model of allergic rhinitis. Int J Mol Sci (2021) 22:10472. doi: 10.3390/ijms221910472
36. Kaneko Y, Kohno T, Kakuki T, Takano KI, Ogasawara N, Miyata R, et al. The role of transcriptional factor p63 in regulation of epithelial barrier and ciliogenesis of human nasal epithelial cells. Sci Rep (2017) 7:10935. doi: 10.1038/s41598-017-11481-w
37. Wang C, Li H, Cao L, Wang G. Identification of differentially expressed genes associated with asthma in children based on the bioanalysis of the regulatory network. Mol Med Rep (2018) 18:2153–63. doi: 10.3892/mmr.2018.9205
38. Pniewska-Dawidczyk E, Kupryś-Lipińska I, Turek G, Kacprzak D, Wieczfinska J, Kleniewska P, et al. Expression of cPLA(2)γ mRNA and protein differs the response of PBMC from severe and non-severe asthmatics to bacterial lipopolysaccharide and house dust mite allergen. Int J Immunopathol Pharmacol (2021) 35:2058738421990952. doi: 10.1177/2058738421990952
39. Su XM, Ren Y, Li ML, Zhao X, Kong LF, Kang J. Performance evaluation of histone deacetylases in lungs of mice exposed to ovalbumin aerosols. J Physiol Pharmacol 69 (2018). doi: 10.26402/jpp.2018.2.12
40. Kim MH, Kim SH, Kim YK, Hong SJ, Min KU, Cho SH, et al. A polymorphism in the histone deacetylase 1 gene is associated with the response to corticosteroids in asthmatics. Korean J Intern Med (2013) 28:708–14. doi: 10.3904/kjim.2013.28.6.708
41. Ito K, Caramori G, Lim S, Oates T, Chung KF, Barnes PJ, et al. Expression and activity of histone deacetylases in human asthmatic airways. Am J Respir Crit Care Med (2002) 166:392–6. doi: 10.1164/rccm.2110060
42. Robertson ED, Weir L, Romanowska M, Leigh IM, Panteleyev AA. ARNT controls the expression of epidermal differentiation genes through HDAC- and EGFR-dependent pathways. J Cell Sci (2012) 125:3320–32. doi: 10.1242/jcs.095125
43. Swamy RS, Reshamwala N, Hunter T, Vissamsetti S, Santos CB, Baroody FM, et al. Epigenetic modifications and improved regulatory T-cell function in subjects undergoing dual sublingual immunotherapy. J Allergy Clin Immunol (2012) 130:215–24.e7. doi: 10.1016/j.jaci.2012.04.021
44. Yanagihara Y, Ikizawa K, Kajiwara K, Koshio T, Basaki Y, Akiyama K. Functional significance of IL-4 receptor on b cells in IL-4-induced human IgE production. J Allergy Clin Immunol (1995) 96:1145–51. doi: 10.1016/S0091-6749(95)70199-0
45. Peng X, Huang M, Zhao W, Lan Z, Wang X, Yuan Y, et al. RAGE mediates airway inflammation via the HDAC1 pathway in a toluene diisocyanate-induced murine asthma model. BMC Pulm Med (2022) 22:61. doi: 10.1186/s12890-022-01832-3
46. Akdis CA, Arkwright PD, Brüggen MC, Busse W, Gadina M, Guttman-Yassky E, et al. Type 2 immunity in the skin and lungs. Allergy (2020) 75:1582–605. doi: 10.1111/all.14318
47. Chen J, Peng L, Zhao Z, Yang Q, Yin F, Liu M, et al. HDAC1 potentiates CD4 + T cell activation by inhibiting miR-124 and promoting IRF1 in systemic lupus erythematosus. Cell Immunol (2021) 362:104284. doi: 10.1016/j.cellimm.2021.104284
48. Hamminger P, Marchetti L, Preglej T, Platzer R, Zhu C, Kamnev A, et al. Histone deacetylase 1 controls CD4(+) T cell trafficking in autoinflammatory diseases. J Autoimmun (2021) 119:102610. doi: 10.1016/j.jaut.2021.102610
49. Ellmeier W. Molecular control of CD4(+) T cell lineage plasticity and integrity. Int Immunopharmacol (2015) 28:813–7. doi: 10.1016/j.intimp.2015.03.050
50. Wang J, Cui M, Sun F, Zhou K, Fan B, Qiu JH, et al. HDAC inhibitor sodium butyrate prevents allergic rhinitis and alters lncRNA and mRNA expression profiles in the nasal mucosa of mice. Int J Mol Med (2020) 45:1150–62. doi: 10.3892/ijmm.2020.4489
51. Leus NG, van den Bosch T, Wouden PEvd, Krist K, Ourailidou ME, Eleftheriadis N, et al. HDAC1-3 inhibitor MS-275 enhances IL10 expression in RAW264.7 macrophages and reduces cigarette smoke-induced airway inflammation in mice. Sci Rep (2017) 7:45047. doi: 10.1038/srep45047
52. Raqib R, Sarker P, Mily A, Alam NH, Arifuzzaman AS, Rekha RS, et al. Efficacy of sodium butyrate adjunct therapy in shigellosis: a randomized, double-blind, placebo-controlled clinical trial. BMC Infect Dis (2012) 12:111. doi: 10.1186/1471-2334-12-111
53. Facchin S, Vitulo N, Calgaro M, Buda A, Romualdi C, Pohl D, et al. Microbiota changes induced by microencapsulated sodium butyrate in patients with inflammatory bowel disease. Neurogastroenterol Motil (2020) 32:e13914. doi: 10.1111/nmo.13914
54. Min KY, Lee MB, Hong SH, Lee D, Jo MG, Lee JE, et al. Entinostat, a histone deacetylase inhibitor, increases the population of IL-10(+) regulatory b cells to suppress contact hypersensitivity. BMB Rep (2021) 54:534–9. doi: 10.5483/BMBRep.2021.54.10.092
55. Truong AS, Zhou M, Krishnan B, Utsumi T, Manocha U, Stewart KG, et al. Entinostat induces antitumor immune responses through immune editing of tumor neoantigens. J Clin Invest (2021) 131. doi: 10.1172/JCI138560
56. Connolly RM, Zhao F, Miller KD, Lee MJ, Piekarz RL, Smith KL, et al. E2112: Randomized phase III trial of endocrine therapy plus entinostat or placebo in hormone receptor-positive advanced breast cancer. a trial of the ECOG-ACRIN cancer research group. J Clin Oncol (2021) 39:3171–81. doi: 10.1200/JCO.21.00944
57. Apuri S, Sokol L. An overview of investigational histone deacetylase inhibitors (HDACis) for the treatment of non-hodgkin’s lymphoma. Expert Opin Investig Drugs (2016) 25:687–96. doi: 10.1517/13543784.2016.1164140
58. Royce SG, Dang W, Ververis K, De Sampayo N, El-Osta A, Tang ML, et al. Protective effects of valproic acid against airway hyperresponsiveness and airway remodeling in a mouse model of allergic airways disease. Epigenetics (2011) 6:1463–70. doi: 10.4161/epi.6.12.18396
59. Bhat J, Sosna J, Fritsch J, Quabius ES, Schütze S, Zeissig S, et al. Expression of non-secreted IL-4 is associated with HDAC inhibitor-induced cell death, histone acetylation and c-jun regulation in human gamma/delta T-cells. Oncotarget (2016) 7:64743–56. doi: 10.18632/oncotarget.11462
60. Glauser TA, Cnaan A, Shinnar S, Hirtz DG, Dlugos D, Masur D, et al. Ethosuximide, valproic acid, and lamotrigine in childhood absence epilepsy. N Engl J Med (2010) 362:790–9. doi: 10.1056/NEJMoa0902014
61. Lv J, Du C, Wei W, Wu Z, Zhao G, Li Z, et al. The antiepileptic drug valproic acid restores T cell homeostasis and ameliorates pathogenesis of experimental autoimmune encephalomyelitis. J Biol Chem (2012) 287:28656–65. doi: 10.1074/jbc.M112.356584
62. Jiang J, Liu JQ, Li J, Li M, Chen HB, Yan H, et al. Trek1 contributes to maintaining nasal epithelial barrier integrity. Sci Rep (2015) 5:9191. doi: 10.1038/srep09191
63. Roy D, Bose S, Pati S, Guin A, Banerjee K, Saha S, et al. GFI1/HDAC1-axis differentially regulates immunosuppressive CD73 in human tumor-associated FOXP3(+) Th17 and inflammation-linked Th17 cells. Eur J Immunol (2021) 51:1206–17. doi: 10.1002/eji.202048892
64. Wu Q, Nie J, Gao Y, Xu P, Sun Q, Yang J, et al. Reciprocal regulation of RORγt acetylation and function by p300 and HDAC1. Sci Rep (2015) 5:16355. doi: 10.1038/srep16355
65. Munro SK, Balakrishnan B, Lissaman AC, Gujral P, Ponnampalam AP. Cytokines and pregnancy: Potential regulation by histone deacetylases. Mol Reprod Dev (2021) 88:321–37. doi: 10.1002/mrd.23430
66. Lee CG, Kwon HK, Sahoo A, Hwang W, So JS, Hwang JS, et al. Interaction of ets-1 with HDAC1 represses IL-10 expression in Th1 cells. J Immunol (2012) 188:2244–53. doi: 10.4049/jimmunol.1101614
67. Shi Y, Xu LZ, Peng K, Wu W, Wu R, Liu ZQ, et al. Specific immunotherapy in combination with clostridium butyricum inhibits allergic inflammation in the mouse intestine. Sci Rep (2015) 5:17651. doi: 10.1038/srep17651
68. Brune K, Frank J, Schwingshackl A, Finigan J, Sidhaye VK. Pulmonary epithelial barrier function: some new players and mechanisms. Am J Physiol Lung Cell Mol Physiol (2015) 308:L731–45. doi: 10.1152/ajplung.00309.2014
69. Kim HK, Kim JH, Kim HJ, Kim TH, Lee SH. Role of TWIK-related potassium channel-1 in chronic rhinosinusitis. J Allergy Clin Immunol (2018) 141:1124–1127.e6. doi: 10.1016/j.jaci.2017.10.012
70. Huang H, Liu JQ, Yu Y, Mo LH, Ge RT, Zhang HP, et al. Regulation of TWIK-related potassium channel-1 (Trek1) restitutes intestinal epithelial barrier function. Cell Mol Immunol (2016) 13:110–8. doi: 10.1038/cmi.2014.137
71. Zhou J, Geng F, Xu J, Peng L, Ye X, Yang D, et al. PM(2.5) exposure and cold stress exacerbates asthma in mice by increasing histone acetylation in IL-4 gene promoter in CD4(+) T cells. Toxicol Lett (2019) 316:147–53. doi: 10.1016/j.toxlet.2019.09.011
72. Doyle K, Fitzpatrick FA. Redox signaling, alkylation (carbonylation) of conserved cysteines inactivates class I histone deacetylases 1, 2, and 3 and antagonizes their transcriptional repressor function. J Biol Chem (2010) 285:17417–24. doi: 10.1074/jbc.M109.089250
73. Cao D, Bromberg PA, Samet JM. COX-2 expression induced by diesel particles involves chromatin modification and degradation of HDAC1. Am J Respir Cell Mol Biol (2007) 37:232–9. doi: 10.1165/rcmb.2006-0449OC
74. Guo F, Zhang YF, Liu K, Huang X, Li RX, Wang SY, et al. Chronic exposure to alcohol inhibits new myelin generation in adult mouse brain. Front Cell Neurosci (2021) 15:732602. doi: 10.3389/fncel.2021.732602
75. Li F, Ding J, Cong Y, Liu B, Miao J, Wu D, et al. Trichostatin a alleviated ovarian tissue damage caused by cigarette smoke exposure. Reprod Toxicol (2020) 93:89–98. doi: 10.1016/j.reprotox.2020.01.006
76. Ding J, Liu B, Han P, Cong Y, Wu D, Miao J, et al. Trichostatin a inhibits uterine histomorphology alterations induced by cigarette smoke exposure in mice. Life Sci (2019) 228:112–20. doi: 10.1016/j.lfs.2019.04.069
77. Do-Umehara HC, Chen C, Zhang Q, Schleimer RP, Budinger GRS, Liu J. Suppression of allergic asthma by loss of function of Miz1-mediated Th1 skewing. Am J Respir Cell Mol Biol (2022)67 (3). doi: 10.1165/rcmb.2022-0135OC
78. Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science (2012) 336:489–93. doi: 10.1126/science.1219328
79. Trivedi R, Barve K. Gut microbiome a promising target for management of respiratory diseases. Biochem J (2020) 477:2679–96. doi: 10.1042/BCJ20200426
80. de Vrese M, Schrezenmeir J. Probiotics, prebiotics, and synbiotics. Adv Biochem Eng Biotechnol (2008) 111:1–66. doi: 10.1007/10_2008_097
81. Zhang X, Zhang J, Chu Z, Shi L, Geng S, Guo K. Gut microbiome alterations and functional prediction in chronic spontaneous urticaria patients. J Microbiol Biotechnol (2021) 31:747–55. doi: 10.4014/jmb.2012.12022
82. Chen X, Xu J, Su Y, Zhu W. Effects of intravenous infusion with sodium butyrate on colonic microbiota, intestinal development- and mucosal immune-related gene expression in normal growing pigs. Front Microbiol (2018) 9:1652. doi: 10.3389/fmicb.2018.01652
83. Xu J, Chen X, Yu S, Su Y, Zhu W. Effects of early intervention with sodium butyrate on gut microbiota and the expression of inflammatory cytokines in neonatal piglets. PloS One (2016) 11:e0162461. doi: 10.1371/journal.pone.0162461
84. Gonneaud A, Turgeon N, Boisvert FM, Boudreau F, Asselin C. JAK-STAT pathway inhibition partially restores intestinal homeostasis in Hdac1- and Hdac2-intestinal epithelial cell-deficient mice. Cells (2021) 10:224. doi: 10.3390/cells10020224
85. Turgeon N, Gagné JM, Blais M, Gendron FP, Boudreau F, Asselin C. The acetylome regulators Hdac1 and Hdac2 differently modulate intestinal epithelial cell dependent homeostatic responses in experimental colitis. Am J Physiol Gastrointest Liver Physiol (2014) 306:G594–605. doi: 10.1152/ajpgi.00393.2013
86. Gonneaud A, Turgeon N, Jones C, Couture C, Lévesque D, Boisvert FM, et al. HDAC1 and HDAC2 independently regulate common and specific intrinsic responses in murine enteroids. Sci Rep (2019) 9:5363. doi: 10.1038/s41598-019-41842-6
87. Pan H, Cao J, Xu W. Selective histone deacetylase inhibitors. Anticancer Agents Med Chem (2012) 12:247–70. doi: 10.2174/187152012800228814
88. Ghosh D, Ding L, Bernstein JA, Mersha TB. The utility of resolving asthma molecular signatures using tissue-specific transcriptome data. G3 (Bethesda) (2020) 10:4049–62. doi: 10.1534/g3.120.401718
89. Paller AS, Spergel JM, Mina-Osorio P, Irvine AD. The atopic march and atopic multimorbidity: Many trajectories, many pathways. J Allergy Clin Immunol (2019) 143:46–55. doi: 10.1016/j.jaci.2018.11.006
90. Kemter AM, Nagler CR. Influences on allergic mechanisms through gut, lung, and skin microbiome exposures. J Clin Invest (2019) 129:1483–92. doi: 10.1172/JCI124610
91. Ribrag V, Kim WS, Bouabdallah R, Lim ST, Coiffier B, Illes A, et al. Safety and efficacy of abexinostat, a pan-histone deacetylase inhibitor, in non-Hodgkin lymphoma and chronic lymphocytic leukemia: results of a phase II study. Haematologica (2017) 102:903–9. doi: 10.3324/haematol.2016.154377
Keywords: epigenetic modifications, allergic diseases, HDAC1, Th2 cytokines, IL-10, Trek-1
Citation: Wang Y and Wang H (2022) The emerging role of histone deacetylase 1 in allergic diseases. Front. Immunol. 13:1027403. doi: 10.3389/fimmu.2022.1027403
Received: 25 August 2022; Accepted: 27 September 2022;
Published: 11 October 2022.
Edited by:
Guan-Jun Yang, Ningbo University, ChinaReviewed by:
Jessie Song, University of Macau, ChinaCopyright © 2022 Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huiying Wang, bWFyeXdhbmdAemp1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.