- Shandong Provincial Key Laboratory of Animal Resistance Biology, College of Life Sciences, Shandong Normal University, Jinan, China
The common carp (Cyprinus carpio L.) is an important farmed species worldwide. Mucosal-associated lymphoid tissues play an essential role in the fight against pathogen infection. Spring viremia of carp virus (SVCV) poses a serious threat to the common carp aquaculture industry. Understanding the molecular mechanisms driving mucosal immune responses to SVCV infection is critical. In this study, the mucosal tissues (gills, foregut and hindgut) were collected from normal and infected fishes for transcriptome analysis. A total of 932,378,600 clean reads were obtained, of which approximately 80% were successfully mapped to the common carp genome. 577, 1,054 and 1,014 differential expressed genes (DEGs) were identified in the gills, foregut and hindgut, respectively. A quantitative polymerase chain reaction assay indicated that the DEGs expression in the foregut following SVCV infection was consistent with the transcriptome results. Among them, two key genes of the retinoic acid-inducible gene I (RIG-I)-like receptor family, melanoma-differentiation-associated gene 5 (MDA5) and laboratory of genetics and physiology 2 (LGP2) (i.e., CcMDA5 and CcLGP2), underwent further analysis. Overexpression of CcMDA5 or CcLGP2 increased phosphorylation of TANK-binding kinase 1 and interferon regulatory factor 3 and the expression of interferon-1 (ifn-1), myxovirus resistance (mx), viperin and interferon-stimulated gene 15 (isg15), and inhibited SVCV replication in epithelioma papulosum cyprini cells. Furthermore, CcLGP2 significantly upregulated the CcMDA5-induced ifn-1 mRNA expression and the activation of the ifn-1 promoter. Finally, confocal microscopy and coimmunoprecipitation experiments revealed that CcLGP2 colocalizes and interacts with CcMDA5 via the C-terminal regulatory domain. This study provides essential gene resources for understanding the fish immune response to SVCV infection and sheds light on the potential role of fish LGP2 in the MDA5 regulation.
Introduction
Spring viremia of carp virus (SVCV) belongs to the Vesiculovirus genus of the Rhabdoviridae family (1). It is a linear, negative-sense, single-stranded RNA virus that is believed to be the cause of spring viremia of carp (2). Affected fish suffer from acute hemorrhagic illness and have a high mortality rate (3). Once infected, it is difficult to eradicate the virus from affected ponds (4). Currently, no specific medicine is approved for the treatment of SVCV-induced diseases, nevertheless, safe and effective vaccines are being developed. Hence, it is critical to explore the intricate mechanisms of host-virus interaction.
The innate immune cells utilize pattern recognition receptors (PRRs) to sense invading viruses (5) (6). The retinoic acid-inducible gene-I (RIG-I)-like receptors (RLRs) are the key intracellular PRRs that can detect viral RNA (7), including RIG-I, melanoma differentiation-associated protein 5 (MDA5) and laboratory of genetics and physiology 2 (LGP2) (8). Upon RNA virus recognition, RIG-I and MDA5 undergo conformational changes and then recruit and interact with a mitochondrial adaptor protein (MAVS) (9). MAVS aggregation transmits a signal to downstream signaling molecules, activating the TANK-binding kinase 1 (TBK1) and the inhibitor of nuclear factor kappa-B kinase epsilon (IKKϵ). These kinases, in turn, activate interferon regulatory factor 3 (IRF3) and nuclear factor-kappa B (NF-κB), leading to the production of interferon (IFN) and proinflammatory cytokine (10).
LGP2, also called DExH-Box helicase 58 (DHX58), is the third member of RLRs (11). Owing to the lack of N-terminal caspase activation and recruitment domains (CARD), it is widely believed to regulate RIG-I and MDA5 via different mechanisms (12–14). LGP2 was initially identified as a negative regulator of RIG-I and MDA5 signaling. For instance, LGP2 interacts with RIG-I via its C-terminal regulatory domain (RD) to block RIG-I self-association and signaling (15). In contrast, LGP2-deficient mice have a positive regulatory role in RIG-I- and MDA5-mediated antiviral responses (16). So far, RLR orthologs have been cloned and characterized from various fish species (17). However, the molecular mechanism of LGP2 regulation in RIG-I and MDA5 remains inconsistent in teleost. For instance, black carp LGP2 associates with RIG-I/MDA5 to activate IFN signaling and the host antiviral innate response (18). Nonetheless, grass carp LGP2 inhibits IFN-β promoter stimulator 1 and IRF3 activation via RIG-I and MDA5 (19). Hence, it is important to determine the role of fish LGP2 in RIG-I/MDA5-mediated antiviral response.
Common carp (Cyprinus carpio L.), an important freshwater aquaculture species worldwide, is facing disease outbreak problems from bacterial, parasitic, and viral pathogens (20, 21). Particularly, SVCV poses a serious threat to common carp health (22, 23). Fish mucosal tissues, such as gills and gut, act as a protective barrier where pathogens against pathogen invasion in the aquatic environment (24). Hence, in this study, RNA-seq technology was utilized to describe differentially expressed genes (DEGs) in the gills, foregut and hindgut of common carp challenged by SVCV. Our results demonstrated that SVCV infection elicited a strong immune response in the mucosal tissues. CcMDA5 and CcLGP2 were selected for further research among these DEGs. Their antiviral activities and regulatory mechanisms were specifically investigated. The study emphasized the fish mucosal immune response to SVCV and will provide new insights into the potential role of fish LGP2 in the MDA5 regulation.
Material and methods
Animal samples and viral challenge
Common carp (C. carpio L.) with a body weight of 150−200 g were obtained from a fish farm in Jinan and adapted for at least one week before being used in experiments. SVCV were kindly gifted from Yuguang Qin (Shandong Freshwater Fisheries Research Institute, China) and quantified by our laboratory according to previous research (25). The fishes were randomly divided into SVCV-infected and control groups. Carps in the infected group were intraperitoneally injected with 500 μL of SVCV (106 TCID50/mL), and the control group received an equal volume of Medium 199 (Gibco, Grand Island, NY, USA). Three fishes were randomly selected from the control and infected groups and immersed in a 100 mg/L-Tricaine Methane Sulfonate solution. Additionally, carp gills, foregut and hindgut samples were collected at 72 h post SVCV or Medium 199 injection. The samples were promptly frozen in liquid nitrogen and stored at -80°C until use.
RNA isolation, library construction and sequencing
Total RNA was isolated using TRNpure Total RNA Kit (Nobelab, Beijing, China) following the manufacturer’s protocol. The quantity and purity of total RNA were determined using NanoDrop 2000 (Thermo Fisher, Waltham, MA, USA) and Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA), respectively. RNA purification, reverse transcription, library construction and sequencing were performed at Shanghai Majorbio Bio-pharm Biotechnology Co., Ltd. (Shanghai, China) according to the manufacturer’s instructions (Illumina, San Diego, CA, USA). The transcriptome library was prepared following the TruSeqTM RNA sample preparation Kit from Illumina (San Diego, CA, USA) using 1 μg of total RNA. The messenger RNA was isolated using oligo(dT) beads and fragmented by fragmentation buffer. The double-stranded complementary DNA (cDNA) was synthesized using a SuperScript double-stranded cDNA synthesis kit (Invitrogen, Carlsbad, CA, USA) with random hexamer primers (Illumina, San Diego, CA, USA) and subjected to end-repair, phosphorylation, and ‘A’ base addition per Illumina’s library construction protocol. Libraries were size selected for cDNA target fragments of 300 bp on 2% Low Range Ultra Agarose followed by PCR amplification including 15 PCR cycles, using Phusion DNA polymerase (NEB, Ipswich, MA, USA). After being quantified by TBS380, the paired-end RNA-seq sequencing library was sequenced with the Illumina NovaSeq 6000 sequencer (2 × 150 bp read length).
Identification and functional enrichment analysis of DEGs
Raw reads were first quality checked by Sickl (https://github.com/najoshi/sickle) and SeqPrep (https://github.com/jstjohn/SeqPrep) to eliminate adaptor sequence, low-quality sequence and reads containing polyN. The trimmed reads were assessed by fastp v0.19.5, and the high-quality clean reads were aligned with the reference genome of C. carpio (GCF_000951615.1) using TopHat Version v2.1.1. The clean data were analyzed on the Majorbio Cloud Platform as previously described (26). Briefly, HISAT2 software was used to map the clean reads to the common carp genome. The mapped reads were assembled using StringTie software. To identify DEGs between the control and SVCV groups (three replicates in each group), TPM (transcripts per million) were used to assess the gene expression. DEGs analysis between two groups was done using DESeq2 software. A false discovery rate (FDR) < 0.05 and |log2 (Fold Change, FC)| ≥1 were used as the threshold for the selection of the DEGs. Lastly, functional-enrichment analysis including Gene Ontology (GO, http://www.geneontology.org) and Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.jp/kegg/) were performed to identify DEGs that were significantly enriched in GO terms and metabolic pathways at P-adjust ≤ 0.05 when compared to the whole-transcriptome background. GO functional enrichment and KEGG pathway analysis were performed using Goatools (https://github.com/tanghaibao/Goatools) and KOBAS (http://kobas.cbi.pku.edu.cn/home.do).
Quantitative polymerase chain reaction
Total RNA was extracted from tissues and cells using TRNpure Total RNA Kit (Nobelab, Beijing, China), and 2 μg of total RNA was reverse-transcribed using ReScriptTM II RT SuperMix for qPCR (+gDNA Eraser) (Nobelab, Beijing, China) following the manufacturer’s instructions. Subsequently, the gene expression was detected using 2 × SYBR qPCR Mix (Nobelab, Beijing, China) and performed on a LightCycler 96 instrument (Roche, Switzerland). The qPCR was performed as follows: 95°C for 120 s followed by 40 cycles of 95°C for 10 s and 60°C for 30 s. The relative expression of genes was analyzed using the 2−ΔΔCT method. The primers used in this study were listed in Table 1.
Cell culture and transfection
Epithelioma papulosum cyprini (EPC) cell line is derived from fathead minnow (Pimephales promelas), which is widely used in fish disease research. 293T cell line is a derivative of human embryonic kidney 293 cells commonly used to detect interactions between proteins. EPC and 293T cells were generously donated by Yuguang Qin (Shandong Freshwater Fisheries Research Institute, China) and Guangxun Meng (Institute Pasteur of Shanghai, Chinese Academy of Sciences, China), respectively. EPC cells were cultured in Medium 199 (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, Grand Island, NY, USA) and cultivated at 25°C without CO2. 293T cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Grand Island, NY, USA) supplemented with 10% FBS (Gibco, Grand Island, NY, USA), 100 U/mL penicillin and 100 μg/mL streptomycin (Life Technologies, Grand Island, NY, USA) and maintained at 37°C with 5% CO2. Target genes were transfected into 293T and EPC cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) and jetPRIME reagent (Polyplus, Illkirch, France), respectively.
Gene cloning and plasmid construction
Plasmid enhanced green fluorescent protein (pEGFP)-N1 plasmid was purchased from Clontech (Mountain View, CA, USA). The plasmids of pmCherry-N1, pFUGW-2FLAG and pCMV-HA were generously donated by Guangxun Meng. To construct the pEGFP-CcMDA5 and pEGFP-CcLGP2 fusion vector, the whole open reading frame of CcMDA5 (GenBank accession number: KM374815.1) and CcLGP2 (GenBank accession number: KM374816.1) were produced using PCR and digested with the corresponding restriction enzymes (HindIII/BamHI for CcMDA5 and KpnI/SmaI for CcLGP2). The digested sequence was ligated into pEGFP-N1 using T4 DNA ligase (Life Technologies, Carlsbad, CA, USA). The other recombinant plasmids of CcMDA5 and CcLGP2 used in this study were cloned with the same method. The ifn-1 promoter luciferase reporter plasmid was constructed as described previously (27).
Confocal microscopy
EPC cells were seeded onto coverslips in a 24-well plate overnight before being transfected with EGFP-CcMDA5 and mCherry-CcLGP2 recombinant plasmids as depicted in the figures. The cells were rinsed with PBS and fixed with 4% PFA (paraformaldehyde) for 30 min at room temperature. Images were obtained using a laser scanning confocal microscope (Leica SP8, Wetzlar, Germany) after staining with 4’,6-diamidino-2-phenylindole (DAPI, Sigma Aldrich, St. Loius, MO, USA) for 10 min at room temperature.
Luciferase activity assays
293T cells in a 96-well plate were co-transfected with a luciferase reporter plasmid (ifn-1-Luc), pRL-TK Renilla luciferase plasmid, target gene recombinant plasmid and the control vectors. After 48 h of transfection, cells were lysed using Dual-Glo® Reagent (Promega, Madison, WI, USA). Firefly luminescence was measured after 10 min of incubation at room temperature. Dual-Glo® Stop & Glo® Reagent (Promega, Madison, WI, USA) were further combined in an equivalent volume. Ten minutes later, the activity of Renilla luciferase was determined. The Firefly luciferase/Renilla luciferase ratio was calculated following the manufacturer’s instructions.
Crystal violet staining
EPC cells were cultivated in a 24-well plate and transfected with 0.5 μg of CcMDA5/CcLGP2 recombinant plasmid or the empty vector. After transfection for 24 h, the cells were infected with SVCV (106 TCID50/mL, 30 μL) for twenty-four hours. Then the cells were washed with PBS, fixed with 4% PFA and dyed with 1% crystal violet for 30 min at room temperature.
Coimmunoprecipitation and immunoblot analysis
Immunoprecipitation and immunoblotting were performed as described previously (28). Briefly, 293T cells seeded in 6-well plates were collected after 48-h transfection with the indicated plasmids. The cells were washed with PBS and lysed using lysis buffer (50 mM Tris-HCl, 15 mM ethylenediamine tetraacetic acid with pH 8.0, 150 mM NaCl, 1 mM NaF, 1 mM Na3VO4, and 1% Nonidet P-40) with protease inhibitor cocktail (Roche, Switzerland) at 4°C. After being centrifugated for 10 min at 12,000 × g, the supernatant was collected and transferred into a fresh tube. The remaining supernatants were immunoprecipitated using anti-FLAG conjugated agarose (Sigma-Aldrich, St. Louis, MO, USA) at 4°C overnight with constant agitation. The immunoprecipitated proteins were collected on the following day. After washing four times with lysis buffer, the complex was resuspended with 2 × sodium dodecyl sulfate (SDS) loading buffer (as the IP sample) and heated at 95°C. Subsequently, the whole-cell lysates and IP samples were separated by 10% SDS-polyacrylamide gel electrophoresis and transferred into a polyvinylidene fluoride membrane (Millipore, Bedford, MA, USA). The remaining steps of immunoblot were the same as previously stated (28).
Statistical analysis
Statistical analysis was performed using GraphPad Prism 7.0 software. All data were presented as mean ± standard deviations (SD). Comparisons in experiments between two groups were assessed by t-test and a one-way analysis of variance was used in more than two groups. P < 0.05 was considered statistically significant.
Results
RNA Sequencing
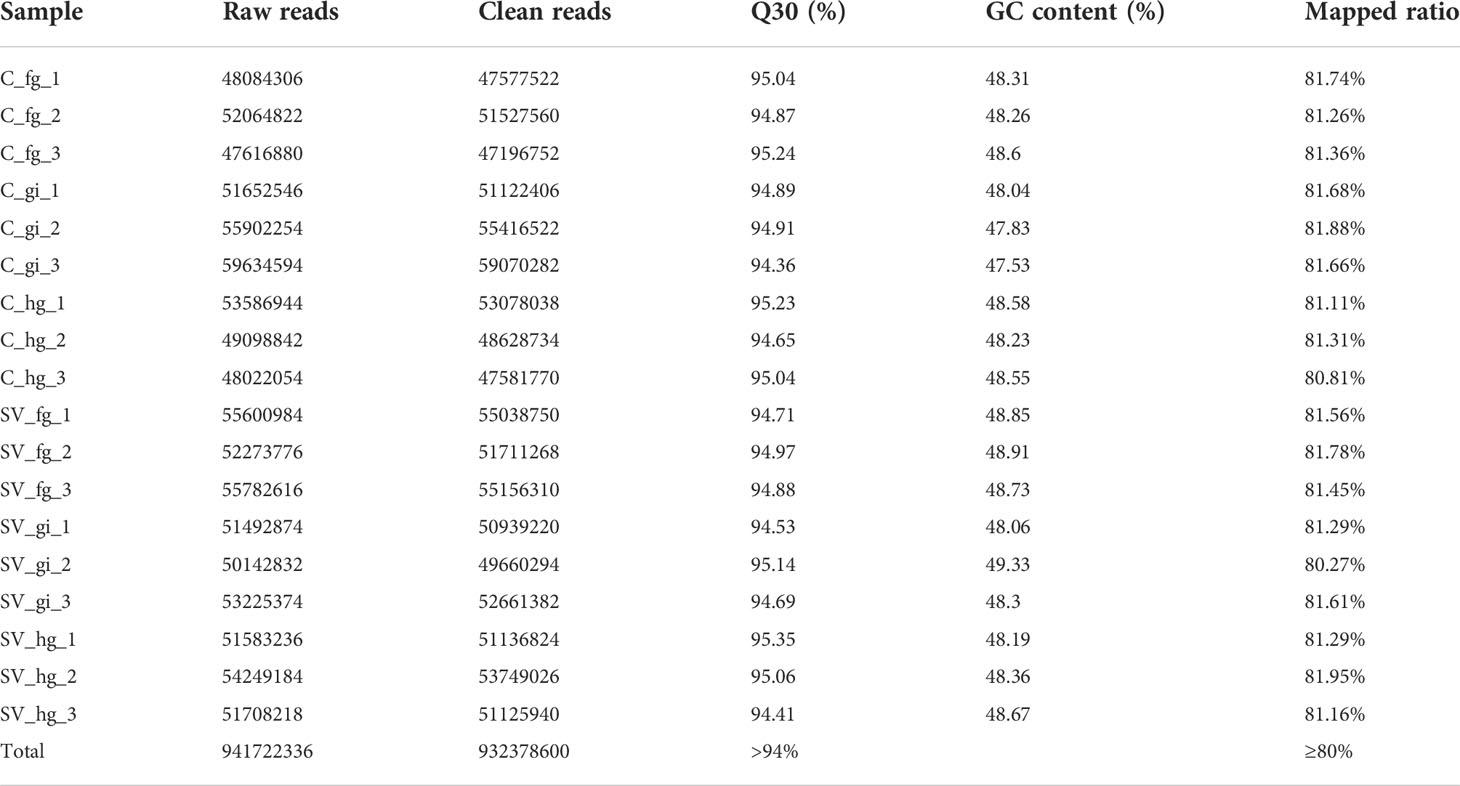
A total of 18 samples (gills, foregut and hindgut tissues from the control and SVCV-infected group) were sequenced to screen for the critical genes involved in the immune response to SVCV infection. After Illumina sequencing, a total of 941,722,336 reads were identified. Furthermore, 932,378,600 clean reads were selected after quality control by filtering low-quality sequences. The benchmark for sequencing quality Q30 > 90%. Among the 18 samples, more than 80% of the clean reads were successfully mapped to the reference genome (Table 2). These results indicate that the transcriptome data were of high quality and the obtained unigenes were reliable and suitable for annotation analysis.
Identification of DEGs
To study the genetic variation in response to SVCV, DEGs were screened by comparing transcriptome data. As presented in the volcanic plots (Figure 1A), a total of 2,645 DEGs were identified between the control and SVCV-infected groups. Among the 2,645 DEGs, 417 up-regulated and 160 down-regulated genes were detected in the gills, 815 up-regulated and 239 down-regulated genes were found in the foregut and 701 up-regulated and 313 down-regulated genes were identified in the hindgut, respectively. The DEGs induced by SVCV in the gills, foregut and hindgut are depicted in the Venn diagram. As presented in Figures 1B, C, 217 up-regulated and 7 down-regulated DEGs were common in the gills, foregut and hindgut.
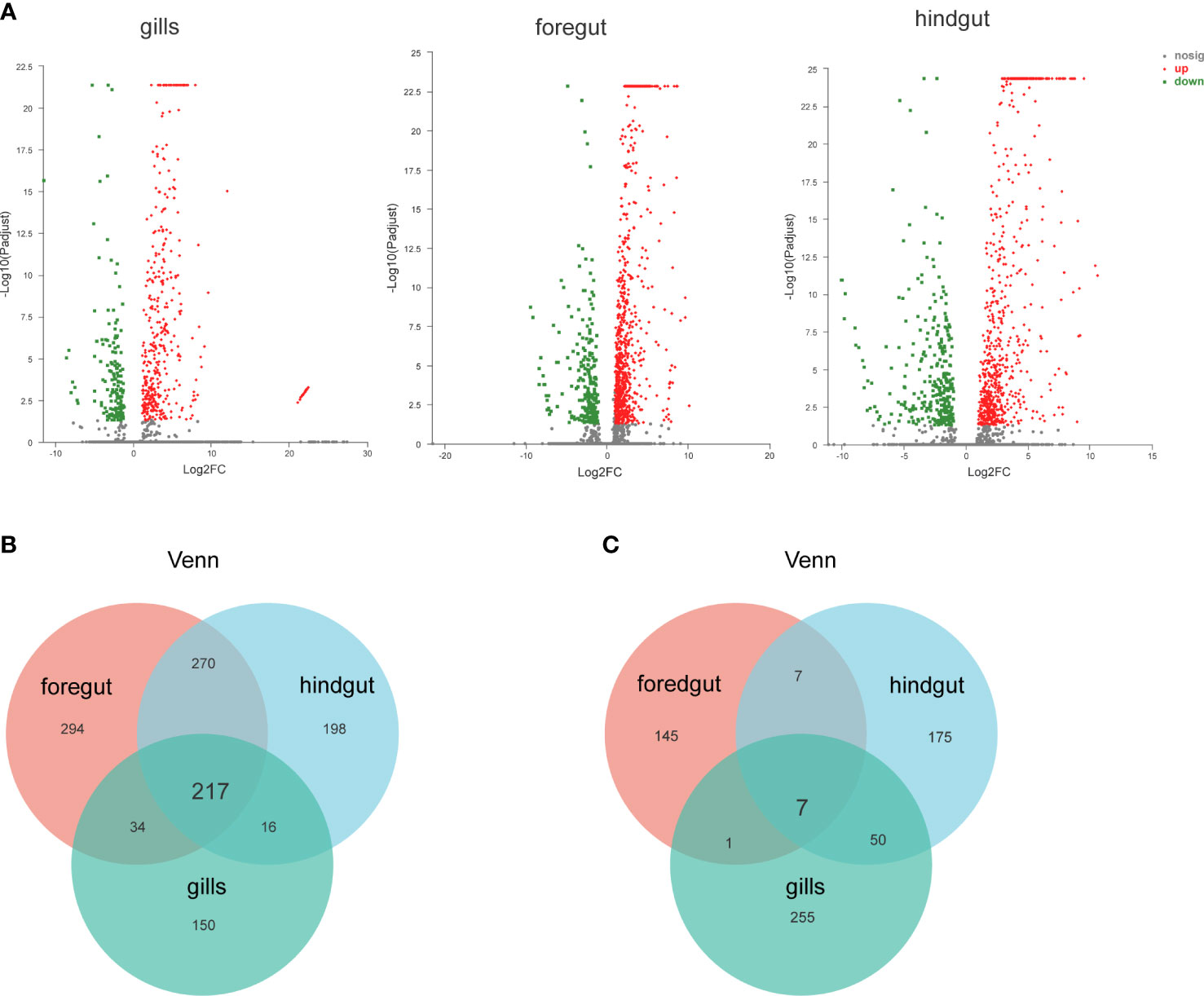
Figure 1 Identification and analysis of differentially expressed genes (DEGs). (A) Volcano plot of DEGs from the transcriptome in carp gills, foregut and hindgut between the control and spring viremia of carp virus (SVCV)-infected groups. (B) Venn diagrams representing the number of transcripts co-upregulated DEGs for each sample. (C) Venn diagrams representing the number of transcripts co-downregulated DEGs for each sample.
GO and KEGG enrichment analyses of upregulated DEGs
GO and KEGG function enrichment analyses were used to better understand the biological roles of DEGs observed in the gills, foregut and hindgut. GO enrichment analysis revealed that the identified upregulated DEGs were significantly enriched in terms associated with innate immune response and host responses to the virus, such as defense response to the virus (biological process) and transferase activity, transferring pentosyl groups (molecular function) (Figure 2A). Furthermore, to determine the putative biochemical pathways of important upregulated DEGs, we identified the top 20 pathways using KEGG enrichment analysis. As depicted in Figure 2B, the upregulated DEGs were also significantly enriched in infectious diseases and pathways associated with innate immunity, such as Influenza A (22 genes), RIG-I-like signaling pathway (11 genes), Cytosolic DNA-sensing pathway (10 genes), PPAR (Peroxisome proliferator-activated receptors) signaling pathway (10 genes), Toll-like receptor signaling pathway (7 genes) and NF-κB signaling pathway (6 genes). The GO and KEGG function enrichment analyses were performed to analyze the respective identified upregulated DEGs in the gills, foregut and hindgut (Supplementary Figure 1). Furthermore, we analyzed the upregulated DEGs enriched in infectious diseases and pathways associated with innate immunity and constructed a heatmap. It was revealed that many RLR signaling pathway molecules, including LGP2, MDA5, IRF3 and IRF7, were significantly upregulated (Figure 2C). RLR signaling pathway has been reported to play pivotal roles in the antiviral innate immune response to RNA virus. Considering the controversial roles of LGP2 regulating MDA5 in various viral infections, we selected CcMDA5 and CcLGP2 for further analysis.
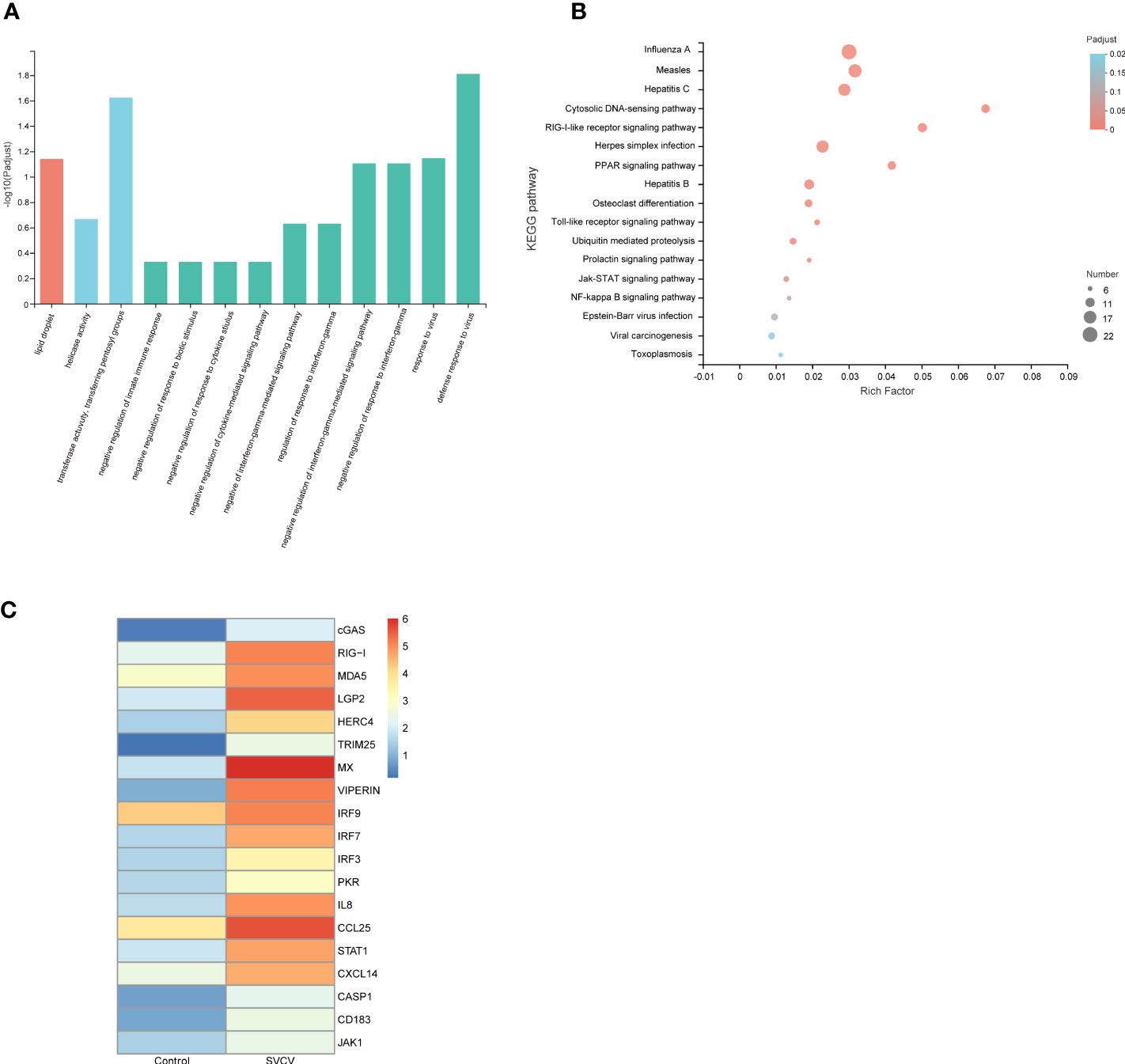
Figure 2 Function analysis of DEGs. (A) The gene ontology (GO) enrichment analysis of co-upregulated DEGs in gills, foregut and hindgut between the control and SVCV-infected groups. The X-axis indicates the GO terms, and the Y-axis indicates the degree of enrichment in this term. (B) The Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of co-upregulated DEGs in gills, foregut and hindgut. The X-axis indicates the rich factors of corresponding pathways, and the Y-axis indicates the pathway names. The different colors of the dots represent the q-value, and the number of DEGs in each pathway is represented by the size of the dots. (C) A heatmap plot of DEGs involved in infectious diseases and immune-related pathways that are enriched in the control group versus the SVCV-infected group.
The validation of DEGs
To verify the reliability of the transcriptome data, ten genes were randomly selected, and qPCR was performed in the foregut. These genes comprised tripartite motif-containing protein 21 (TRIM21), MDA5, LGP2, cyclic GMP-AMP synthase (cGAS), interferon-induced protein 44 (IFI44), ras-related GTP-binding protein (RAS), golgi membrane protein 2 (GLOM2), heat shock factor protein (HSP), forkhead-box (FOX) and TLC domain-containing protein 4-B (TLCD4B). As depicted in Figure 3, the expression of TRIM21, MDA5, cGAS, LGP2, and IFI44 was increased and the expression of RAS, GLOM2, HSP, FOX and TLCD4B was decreased in the foregut following SVCV infection. Consequently, the confirmed expression profile of DEGs was consistent with obtained RNA-seq analysis.
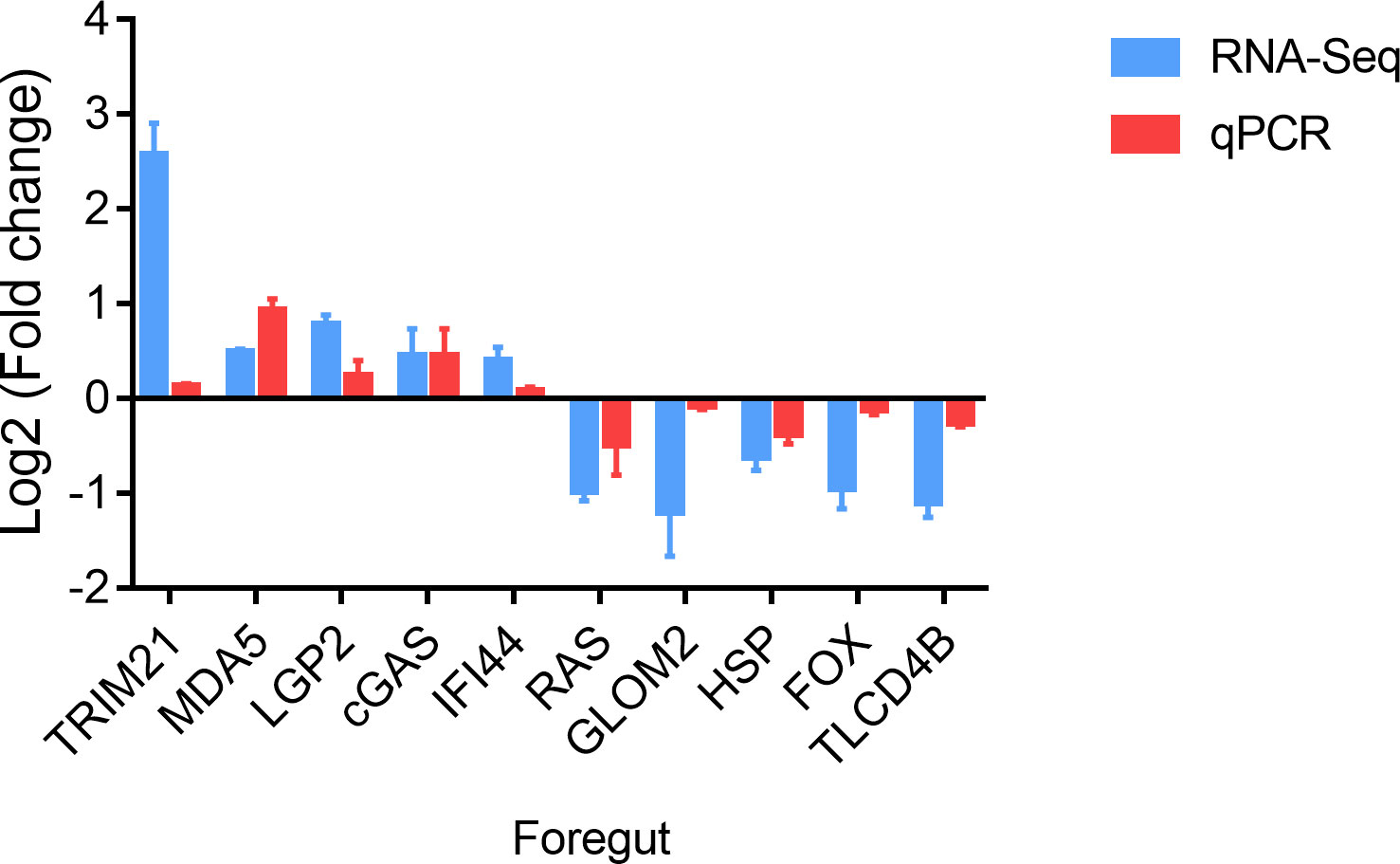
Figure 3 Validation of DEGs in the foregut. The mRNA levels of DEGs were normalized to the level of the S11 gene.
CcMDA5-mediated signaling inhibits SVCV infection
To describe the potential role of CcMDA5, we overexpressed CcMDA5 in EPC cells and assessed the expression of the downstream antiviral molecules. As illustrated in Figure 4A, Western blot results revealed that CcMDA5 overexpression induced phosphorylation of TBK1 (p-TBK1) and IRF3 (p-IRF3) in EPC cells. Additionally, overexpression of CcMDA5 caused a marked upregulation of interferon-1 (ifn-1), myxovirus resistance (mx), viperin and interferon-stimulated gene 15 (isg15) (324.25-fold, 72.07-fold, 129.99-fold and 84.15-fold, respectively), compared with the control cells (Figure 4B). To identify the role of CcMDA5 in virus proliferation, the mRNA expression of the svcv-g gene and virus-induced cytopathic effects (CPEs) were explored. Compared with the control cells, CcMDA5 significantly decreased the transcriptional level of the svcv-g gene and CPEs (Figures 4C, D). These results revealed that CcMDA5 activates IFN signaling and inhibit virus proliferation.
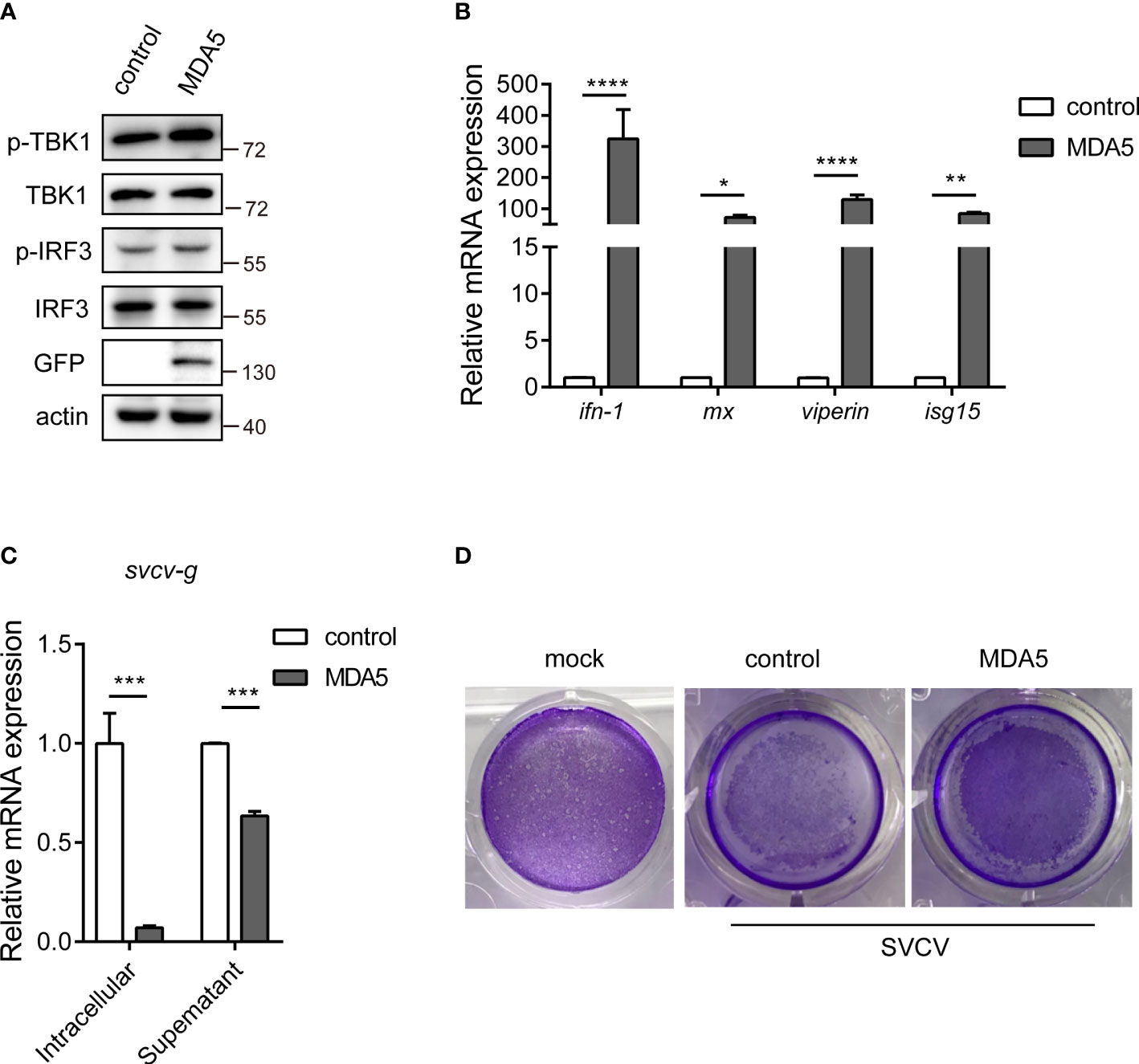
Figure 4 The effect of CcMDA5 in the antiviral immune responses. (A, B) Empty vector or GFP-tagged CcMDA5 plasmid was transfected into epithelioma papulosum cyprini (EPC) cells for 24 h. (A) Western blot was performed to test the phosphorylation of TBK1 and IRF3. β-Actin was used as an internal reference. (B) Total RNA was extracted to detect interferon-1 (ifn-1), myxovirus resistance (mx), viperin, and interferon-stimulated gene 15 (isg15) using qPCR. Data are represented as fold induction relative to the transcriptional level in control, which was set to 1. Mean ± SD (n = 3). (C, D) EPC cells were transfected with the above-mentioned plasmids. At 24 h post-transfection, the cells were stimulated with SVCV or Medium 199 and incubated at 25°C for 24 h. (C) Cell samples and supernatants were collected. The mRNA expression of the svcv-g gene was detected using qPCR. (D) The cell monolayers were stained with crystal violet, and CPEs were observed. The mock group represented the cells without treatment. Each experiment was conducted thrice. *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001.
CcLGP2 positively regulates CcMDA5 inducing ifn-1 expression
Previous studies have revealed that teleost LGP2 exhibited different antiviral activities against viral infection. To describe the potential role of LGP2 in common carp, we overexpressed CcLGP2 in EPC cells. As illustrated in Figures 5A, B, the phosphorylation of TBK1 and IRF3 and the transcription of ifn-1, mx, viperin and isg15 were significantly enhanced after overexpression of CcLGP2. Additionally, CcLGP2 also reduced the replication of the svcv-g gene and CPEs (Figures 5C, D).
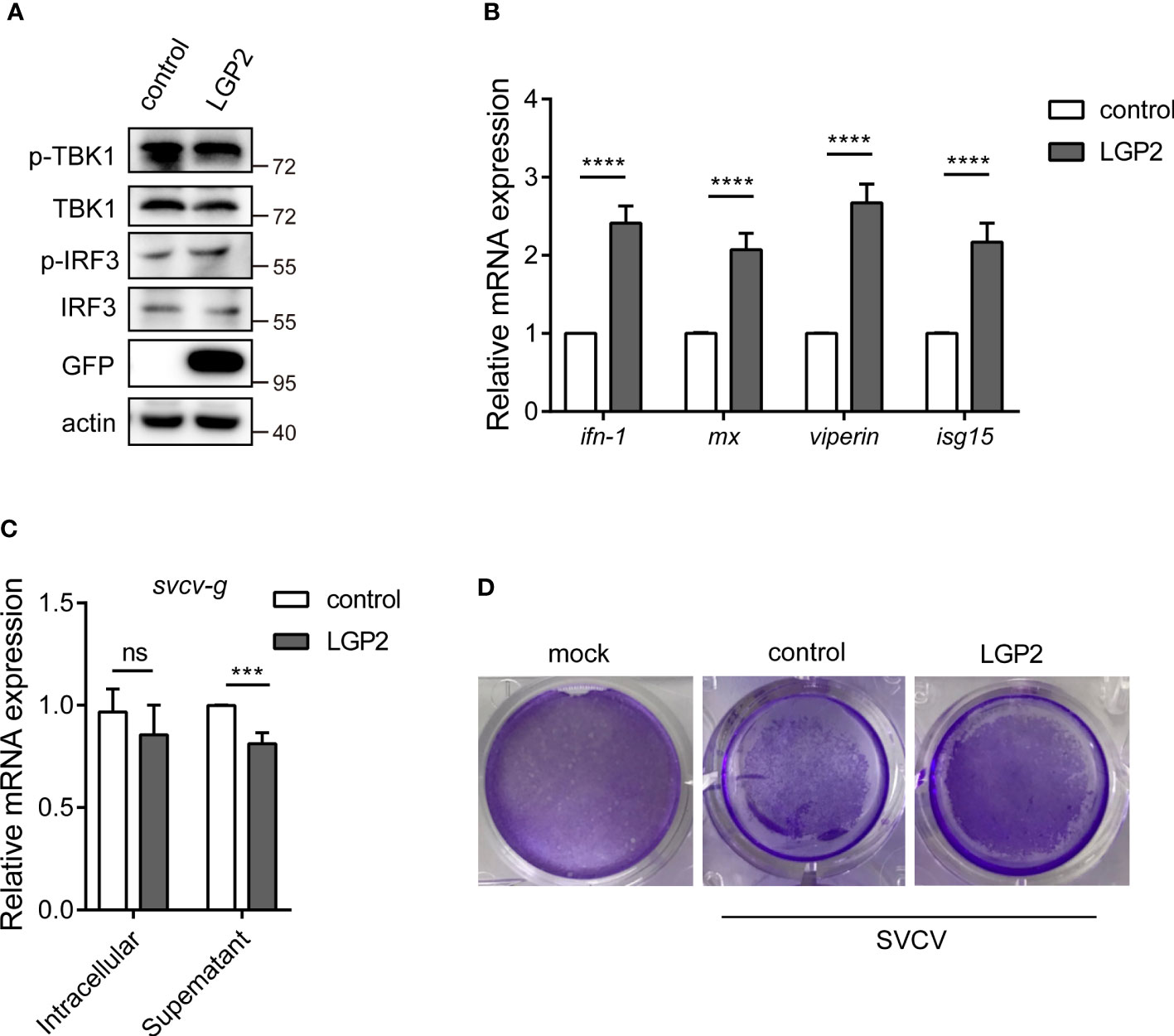
Figure 5 CcLGP2 enhances the IFN activity and inhibits viral replication. Empty vector or GFP-tagged CcLGP2 plasmid were transfected into EPC cells for 24 h. Further, the protein and RNA were extracted to detect downstream signaling molecules by Western blot (A) and qPCR (B). (C, D) EPC cells were transfected with the above-mentioned plasmids. Twenty-four hours post-transfection, the cells were infected with SVCV and incubated at 25°C for 24 h. (C) Cell samples and supernatants were collected, and the expression of svcv-g was detected using qPCR. (D) The cell monolayers were stained with crystal violet, and CPEs were observed. Each experiment was conducted thrice. ***P < 0.001, ****P < 0.0001 and ns indicates no significant difference.
To further explore the effect of CcLGP2 on the CcMDA5-mediated signaling pathway, qPCR and reporter assay were performed in EPC and 293T cells, respectively. As illustrated in Figure 6A, in comparison with the CcMDA5 group, the expression of ifn-1 was significantly up-regulated following co-transfection of CcLGP2 and CcMDA5. Additionally, CcLGP2 significantly enhanced CcMDA5-induced ifn-1 promoter activation in 293T cells (Figure 6B). These results revealed that CcLGP2 acts as the positive regulator in the MDA5-mediated ifn-1 activation.
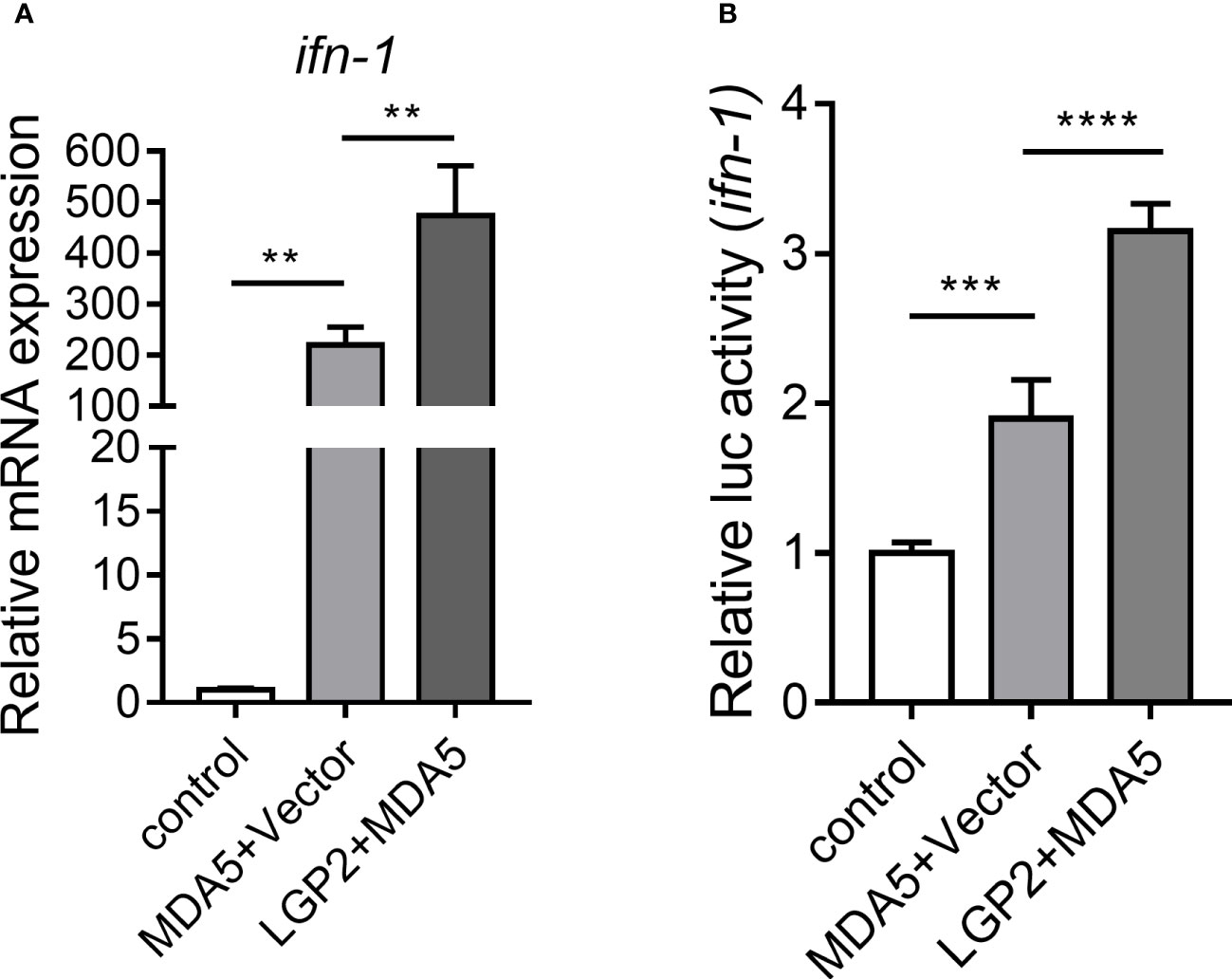
Figure 6 CcLGP2 activates the CcMDA5-inducing ifn-1. (A) Empty vector or FLAG-tagged CcMDA5 plasmid was co-transfected with GFP-tagged CcLGP2 plasmid into EPC cells for 24 h. Total RNA was extracted to detect ifn-1. Data are represented as fold induction relative to the transcriptional level in control, which was set to 1. (B) 293T cells were co-transfected with expression vectors for Firefly luciferase reporter gene with ifn-1 promoter, Renilla luciferase and target gene for 48 h. The relative activity of ifn-1 was the ratio of Firefly fluorescence to Renilla fluorescence. Mean ± SD (n = 3). Each experiment was conducted thrice. **P < 0.01, ***P < 0.001 and ****P < 0.0001.
Association between CcLGP2 and CcMDA5
To clarify the regulatory mechanism between CcLGP2 and CcMDA5, confocal microscopy and coimmunoprecipitation experiments were conducted. It was observed that CcLGP2 colocalized with CcMDA5 in the EPC cells cytoplasm (Figure 7A and Supplementary Figure 2). The CO-IP assay further verified their association, indicating an interaction between CcLGP2 and CcMDA5 (Figure 7B). Additionally, we investigated the domain of CcMDA5 that interacted with CcLGP2. As depicted in Figures 7C, D, different truncations of CcMDA5 were generated, and the CO-IP assay revealed that CcLGP2 was associated with the full-length of CcMDA5, albeit not with other truncations, suggesting that CcLGP2 bound with the C-terminal RD of CcMDA5. Overall, these observations indicate that CcLGP2 interacts with CcMDA5 via the RD.
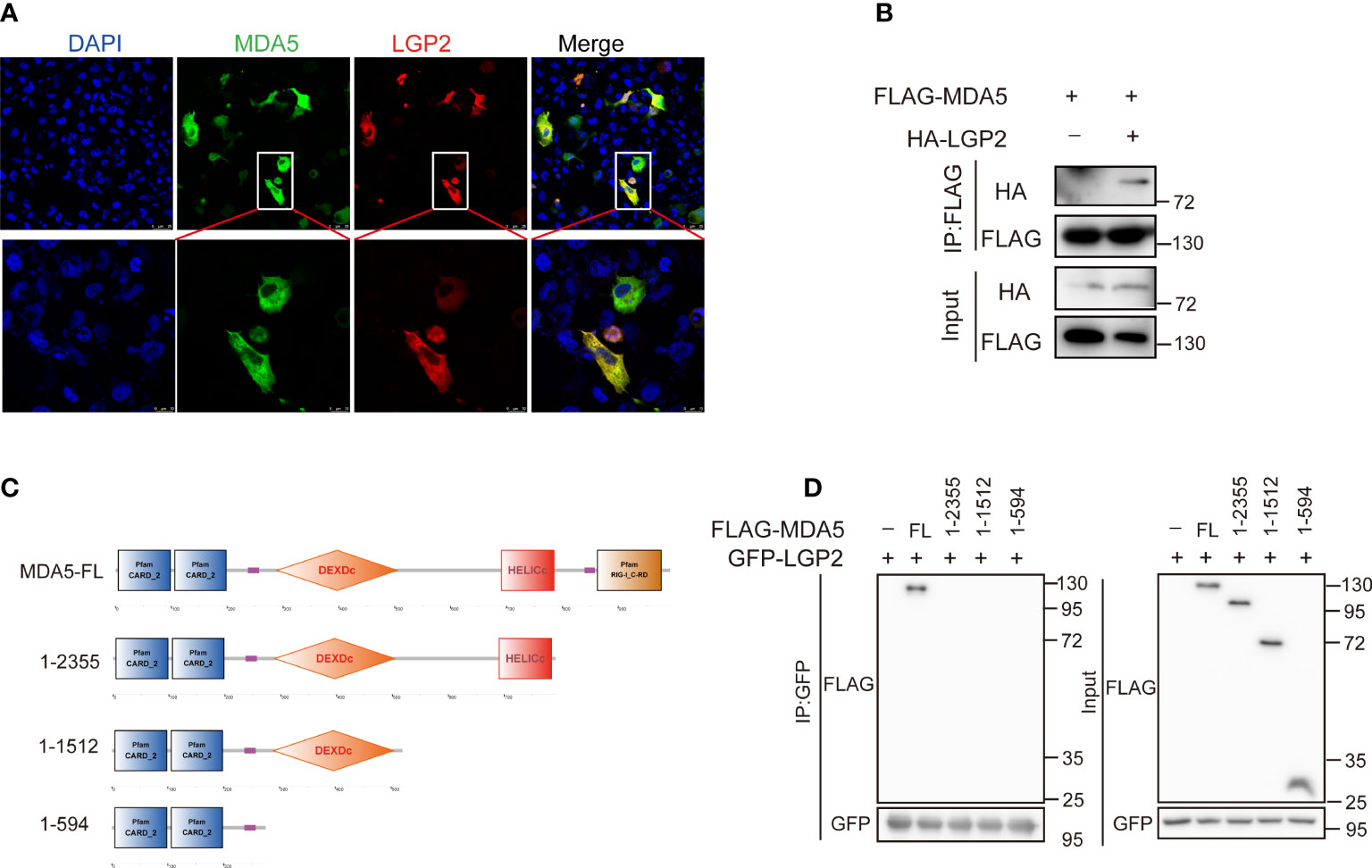
Figure 7 CcLGP2 protein interacts with CcMDA5 through the RD domain. (A) EPC cells were transfected for 24 h with plasmids expressing GFP-tagged CcMDA5 and mCherry-tagged CcLGP2. After staining with DAPI, the coverslip was observed under a laser scanning confocal microscope. (B) As indicated in the figure, FLAG-tagged CcMDA5 and HA-tagged CcLGP2 were separately transfected into 293T cells. The cell lysates were immunoprecipitated with anti-FLAG antibodies and further immunoblotted with anti-FLAG and anti-HA antibodies. The control group was transfected with FLAG-tagged CcMDA5 and empty vector pCMV-HA. (C) Schematic diagram of full-length and mutant CcMDA5. (D) GFP-tagged CcLGP2 and different truncations of CcMDA5 were transfected into 293T cells for 48 h as indicated in the figure. The cell lysates were immunoprecipitated with anti-GFP antibodies and further immunoblotted with anti-FLAG and anti-GFP antibodies. All the experiments were repeated at least thrice.
Discussion
Teleost mucosal tissues serve as the first line of defense in innate immunity and play an essential role against pathogen infection (29, 30). Previous studies have demonstrated that the microbial composition of mucosal tissues is changed, and the mucosal immune response is also activated during pathogen stimulation (31). Although studies have illustrated the changes in microbial composition following pathogen infection and environment changes (31, 32), the antiviral mechanism of mucosal tissues remains elusive. Therefore, in this study, we selected carp mucosal immune tissues (gills, foregut and hindgut) for RNA-Seq analysis to explore the changes in the host immune system following SVCV infection. Following Illumina sequencing, 932,378,600 high-quality reads and 2,645 DEGs were identified.
KEGG analysis revealed that most of the co-upregulated DEGs were involved in pathogenic infection-related pathways, such as Influenza A, Cytosolic DNA-sensing pathway, RIG-I-like signaling pathway, PPAR signaling pathway and Toll-like receptor signaling pathway. As reported in previous studies, these pathways play essential roles in antiviral immune response (33–35). In transcriptome profiling, there were several genes identified as significantly up-regulated genes, including RIG-I, MDA5, LGP2, cGAS, IRF3 and IRF7. cGAS is the major sensor for detecting DNA and RNA viruses (36). Mice with cGAS deficiency have a higher mortality rate and viral titer (37). In zebrafish, cGAS knockdown inhibits Edwardsiella tarda-induced production of immunoglobulin (Ig)Z in gills and skin (38). Therefore, cGAS plays a crucial part in mucosal immunity against SVCV in common carp. IRFs are a family of transcription factors, playing important and diverse roles in viral infections by activating IFNs (39). IRF3 and IRF7 are the most important regulators of type I IFN against viral infection (40, 41). Hence, the initiation of transcription of IRF3 and IRF7 regulates IFN signaling against SVCV infection. RIG-I, MDA5 and LGP2, the members of RIG-I-like receptors, have been reported to be involved in the mucosal immune response against viral invasion (42). Additionally, our previous study has indicated a potential role of RIG-I in SVCV infection (26). Further studies are required to unravel the biological roles of MDA5 and LGP2 in SVCV infection.
Previous studies have demonstrated that MDA5, together with RIG-I, are cytoplasmic sensors of double-stranded RNAs (43, 44). MDA5 recruits the adaptor MAVS after interacting with viral RNA to initiate the production of IFN and antiviral molecules (45). The role of fish MDA5 in the induction of IFN has been well-elucidated during the last few decades (43, 46, 47). For instance, zebrafish MDA5 forms a complex with MAVS and then induces the IFN-I promoter activity (48). MDA5 from orange-spotted grouper activates the IFN promoter activity and interferon-stimulated response element (ISRE) activity and increases the transcription of IRF3, IRF7 and tumor necrosis factor receptor-associated factor 6 (TRAF6) (49). Consistent with these results, CcMDA5 overexpression not only significantly induced the phosphorylation of TBK1 and IRF3 but also promoted the expression of ifn-1 and ISGs. IFNs are a group of vital cytokines, which can induce a wide range of ISGs to limit virus replication and spread (50) (51). For instance, gibel carp IFNα remarkably inhibits cyprinid herpesvirus 2 (CyHV-2) propagation by activating signal transducer and activator of transcription 1 (STAT1), MX and protein kinase R (PKR) (52). Similarly, in EPC cells, CcMDA5 can reduce the expression level of the svcv-g gene and inhibit SVCV replication by inducing the expression of ifn-1, mx, viperin and isg15. MDA5-mediated antiviral signaling pathways are conserved in mammals and teleost.
LGP2, the third member of the RLR family, possesses the DExD/H box RNA helicase domain that is important for inducing IFN-β response (11). However, LGP2 lacks the CARD domain that is required for signaling, which is responsible for the controversial roles of LGP2 in RLR-mediated signaling (53). To date, numerous pieces of evidence have illustrated the positive regulatory mechanism between the LGP2 and MDA5 (54). For instance, mammalian LGP2 can amplify the MDA5-mediated signaling pathway by augmenting the ability of MDA5 to form stable filaments (54, 55). In teleost, LGP2 plays a positive role in viral infection in Japanese flounder (53) and miiuy croaker (56). In line with these results, CcLGP2 overexpression significantly upregulated TBK1, IRF3, ifn-1, mx, viprein and isg15, and promoted MDA5-mediated ifn-1 activation. In contrast, LGP2, in mandarin fish (57), grouper (58) and rainbow trout (59) functions as a negative regulator in antiviral response by decreasing the transcription of IRF3, NF-κB and IFN-β. Recent studies have revealed that LGP2 is a bifunctional protein and yields a biphasic effect on MDA5 signaling (60, 61). Hence, the role of fish LGP2 in antiviral response may be associated with fish and virus species.
Conclusively, the transcriptome profiling in the common carp gills, foregut and hindgut after SVCV infection was elaborated. Two key genes, CcMDA5 and CcLGP2, were identified post SVCV infection. CcLGP2 acted as a positive regulator in MDA5-induced ifn-1 activation. Collectively, the study will provide a better understanding for further investigation into the innate immune mechanism in common carp fight against viral infection.
Data availability statement
The data presented in the study are deposited in National Center for Biotechnology Information (NCBI) Short Read Archive (SRA) under accession number PRJNA873930. (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA873930).
Ethics statement
The animal study was reviewed and approved by Animal Experimental Ethics Committee of Shandong Normal University.
Author contributions
SS and GY conceived and designed the experiments. RL, YN and performed the experiments and analyzed the data. YQ and HL helped with the experiments. SS and RL wrote the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.1019872/full#supplementary-material
References
1. Wang Y, Zhang H, Lu Y, Wang F, Liu L, Liu J, et al. Comparative transcriptome analysis of zebrafish (Danio rerio) brain and spleen infected with spring viremia of carp virus (Svcv). Fish Shellfish Immunol (2017) 69:35–45. doi: 10.1016/j.fsi.2017.07.055
2. Ashraf U, Lu Y, Lin L, Yuan J, Wang M, Liu X. Spring viraemia of carp virus: Recent advances. J Gen Virol (2016) 97(5):1037–51. doi: 10.1099/jgv.0.000436
3. Kumari SG, Makkouk KM, Attar N, Ghulam W, Lesemann DE. First report of chickpea chlorotic dwarf virus infecting spring chickpea in Syria. Plant Dis (2004) 88(4):424. doi: 10.1094/PDIS.2004.88.4.424C
4. Shao L, Zhao J. Isolation of a highly pathogenic spring viraemia of carp virus strain from grass carp (Ctenopharyngodon idella) in late summer, China, 2016. Virus Res (2017) 238:183–92. doi: 10.1016/j.virusres.2017.06.025
5. Gong XY, Zhang QM, Gui JF, Zhang YB. Svcv infection triggers fish ifn response through rlr signaling pathway. Fish Shellfish Immunol (2019) 86:1058–63. doi: 10.1016/j.fsi.2018.12.063
6. Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell (2006) 124(4):783–801. doi: 10.1016/j.cell.2006.02.015
7. Kawai T, Akira S. Toll-like receptor and rig-I-Like receptor signaling. Ann New York Acad Sci (2008) 1143:1–20. doi: 10.1196/annals.1443.020
8. Rehwinkel J, Gack MU. Rig-I-Like receptors: Their regulation and roles in rna sensing. Nat Rev Immunol (2020) 20(9):537–51. doi: 10.1038/s41577-020-0288-3
9. Fan X, Jin T. Structures of rig-I-Like receptors and insights into viral rna sensing. In: Jin T, Yin Q, editors. Structural immunology. Singapore: Springer Singapore (2019). p. 157–88.
10. Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. Mavs forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell (2011) 146(3):448–61. doi: 10.1016/j.cell.2011.06.041
11. Bamming D, Horvath CM. Regulation of signal transduction by enzymatically inactive antiviral rna helicase proteins Mda5, rig-I, and Lgp2. J Biol Chem (2009) 284(15):9700–12. doi: 10.1074/jbc.M807365200
12. Loo YM, Gale M Jr. Immune signaling by rig-I-Like receptors. Immunity (2011) 34(5):680–92. doi: 10.1016/j.immuni.2011.05.003
13. Sanchez David RY, Combredet C, Najburg V, Millot GA, Beauclair G, Schwikowski B, et al. Lgp2 binds to pact to regulate rig-i- and Mda5-mediated antiviral responses. Sci Signaling (2019) 12(601):eaar3993. doi: 10.1126/scisignal.aar3993
14. Li S, Yang J, Zhu Y, Wang H, Ji X, Luo J, et al. Analysis of porcine rig-I like receptors revealed the positive regulation of rig-I and Mda5 by Lgp2. Front Immunol (2021) 12:609543. doi: 10.3389/fimmu.2021.609543
15. Malur M, Gale M Jr., Krug RM. Lgp2 downregulates interferon production during infection with seasonal human influenza a viruses that activate interferon regulatory factor 3. J Virol (2012) 86(19):10733–8. doi: 10.1128/JVI.00510-12
16. Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, et al. Lgp2 is a positive regulator of rig-i- and Mda5-mediated antiviral responses. Proc Natl Acad Sci United States America (2010) 107(4):1512–7. doi: 10.1073/pnas.0912986107
17. Chen SN, Zou PF, Nie P. Retinoic acid-inducible gene I (Rig-I)-Like receptors (Rlrs) in fish: Current knowledge and future perspectives. Immunology (2017) 151(1):16–25. doi: 10.1111/imm.12714
18. Liu J, Li J, Xiao J, Chen H, Lu L, Wang X, et al. The antiviral signaling mediated by black carp Mda5 is positively regulated by Lgp2. Fish Shellfish Immunol (2017) 66:360–71. doi: 10.1016/j.fsi.2017.05.035
19. Rao Y, Wan Q, Yang C, Su J. Grass carp laboratory of genetics and physiology 2 serves as a negative regulator in retinoic acid-inducible gene I- and melanoma differentiation-associated gene 5-mediated antiviral signaling in resting state and early stage of grass carp reovirus infection. Front Immunol (2017) 8:352. doi: 10.3389/fimmu.2017.00352
20. Bostock J, McAndrew B, Richards R, Jauncey K, Telfer T, Lorenzen K, et al. Aquaculture: Global status and trends. Philos Trans R Soc Lond B Biol Sci (2010) 365(1554):2897–912. doi: 10.1098/rstb.2010.0170
21. Xu P, Zhang X, Wang X, Li J, Liu G, Kuang Y, et al. Genome sequence and genetic diversity of the common carp, cyprinus carpio. Nat Genet (2014) 46(11):1212–9. doi: 10.1038/ng.3098
22. Salinas I. The mucosal immune system of teleost fish. Biology (2015) 4(3):525–39. doi: 10.3390/biology4030525
23. Ahne W, Bjorklund HV, Essbauer S, Fijan N, Kurath G, Winton JR. Spring viremia of carp (Svc). Dis Aquat Org (2002) 52(3):261–72. doi: 10.3354/dao052261
24. Salinas I, Zhang YA, Sunyer JO. Mucosal immunoglobulins and b cells of teleost fish. Dev Comp Immunol (2011) 35(12):1346–65. doi: 10.1016/j.dci.2011.11.009
25. Liu R, Liu X, Song M, Qi Y, Li H, Yang G, et al. Cyprinus carpio trif participates in the innate immune response by inducing nf-κb and ifn activation and promoting apoptosis. Front Immunol (2021) 12:725150(3324). doi: 10.3389/fimmu.2021.725150
26. Liu R, Li H, Liu X, Liang B, Qi Y, Meng F, et al. Trim25 inhibits spring viraemia of carp virus replication by positively regulating rig-I signaling pathway in common carp (Cyprinus carpio l.). Fish Shellfish Immunol (2022) 127:306–17. doi: 10.1016/j.fsi.2022.06.033
27. Shan S, Liu R, Feng H, Meng F, Aizaz M, Yang G. Identification and functional characterization of a fish-specific Tlr19 in common carp (Cyprinus carpio l.) that recruits trif as an adaptor and induces ifn expression during the immune response. Vet Res (2021) 52(1):88. doi: 10.1186/s13567-021-00957-3
28. Liu R, Qi Y, Feng H, Niu Y, Zhang F, Yang G, et al. Fish-specific toll-like receptor 14 (Tlr14) from Asian swamp eel (Monopterus albus) is involved in immune response to bacterial infection. Fish Shellfish Immunol (2022) 124:313–23. doi: 10.1016/j.fsi.2022.04.010
29. Perez-Lopez A, Behnsen J, Nuccio SP, Raffatellu M. Mucosal immunity to pathogenic intestinal bacteria. Nat Rev Immunol (2016) 16(3):135–48. doi: 10.1038/nri.2015.17
30. Lowrey L, Woodhams DC, Tacchi L, Salinas I. Topographical mapping of the rainbow trout (Oncorhynchus mykiss) microbiome reveals a diverse bacterial community with antifungal properties in the skin. Appl Environ Microbiol (2015) 81(19):6915–25. doi: 10.1128/AEM.01826-15
31. Zhang X, Ding L, Yu Y, Kong W, Yin Y, Huang Z, et al. The change of teleost skin commensal microbiota is associated with skin mucosal transcriptomic responses during parasitic infection by ichthyophthirius multifillis. Front Immunol (2018) 9:2972. doi: 10.3389/fimmu.2018.02972
32. Meng KF, Ding LG, Wu S, Wu ZB, Cheng GF, Zhai X, et al. Interactions between commensal microbiota and mucosal immunity in teleost fish during viral infection with svcv. Front Immunol (2021) 12:654758. doi: 10.3389/fimmu.2021.654758
33. Dai Z, Li J, Hu C, Wang F, Wang B, Shi X, et al. Transcriptome data analysis of grass carp (Ctenopharyngodon idella) infected by reovirus provides insights into two immune-related genes. Fish Shellfish Immunol (2017) 64:68–77. doi: 10.1016/j.fsi.2017.03.008
34. Lin Y, Wang B, Wang N, Ouyang G, Cao H. Transcriptome analysis of rare minnow (Gobiocypris rarus) infected by the grass carp reovirus. Fish Shellfish Immunol (2019) 89:337–44. doi: 10.1016/j.fsi.2019.04.013
35. Wang H, Tang X, Sheng X, Xing J, Chi H, Zhan W. Transcriptome analysis reveals temperature-dependent early immune response in flounder (Paralichthys olivaceus) after hirame novirhabdovirus (Hirrv) infection. Fish Shellfish Immunol (2020) 107(Pt A):367–78. doi: 10.1016/j.fsi.2020.10.020
36. Li X, Shu C, Yi G, Chaton CT, Shelton CL, Diao J, et al. Cyclic gmp-amp synthase is activated by double-stranded DNA-induced oligomerization. Immunity (2013) 39(6):1019–31. doi: 10.1016/j.immuni.2013.10.019
37. Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, et al. Pan-viral specificity of ifn-induced genes reveals new roles for cgas in innate immunity. Nature (2014) 505(7485):691–5. doi: 10.1038/nature12862
38. Liu ZF, Ji JF, Jiang XF, Shao T, Fan DD, Jiang XH, et al. Characterization of cgas homologs in innate and adaptive mucosal immunities in zebrafish gives evolutionary insights into cgas-sting pathway. FASEB J: Off Publ Fed Am Soc Exp Biol (2020) 34(6):7786–809. doi: 10.1096/fj.201902833R
39. Harada H, Taniguchi T, Tanaka N. The role of interferon regulatory factors in the interferon system and cell growth control. Biochimie (1998) 80(8-9):641–50. doi: 10.1016/s0300-9084(99)80017-0
40. Clement JF, Bibeau-Poirier A, Gravel SP, Grandvaux N, Bonneil E, Thibault P, et al. Phosphorylation of irf-3 on ser 339 generates a hyperactive form of irf-3 through regulation of dimerization and cbp association. J Virol (2008) 82(8):3984–96. doi: 10.1128/JVI.02526-07
41. Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, et al. Irf-7 is the master regulator of type-I interferon-dependent immune responses. Nature (2005) 434(7034):772–7. doi: 10.1038/nature03464
42. Zhang L, Cao M, Li Q, Yan X, Xue T, Song L, et al. Genome-wide identification of nod-like receptors and their expression profiling in mucosal tissues of turbot (Scophthalmus maximus l.) upon bacteria challenge. Mol Immunol (2021) 134:48–61. doi: 10.1016/j.molimm.2021.02.003
43. Su J, Huang T, Dong J, Heng J, Zhang R, Peng L. Molecular cloning and immune responsive expression of Mda5 gene, a pivotal member of the rlr gene family from grass carp ctenopharyngodon idella. Fish Shellfish Immunol (2010) 28(4):712–8. doi: 10.1016/j.fsi.2010.01.009
44. Jia P, Jia K, Chen L, Le Y, Jin Y, Zhang J, et al. Identification and characterization of the melanoma differentiation - associated gene 5 in Sea perch, lateolabrax japonicus. Dev Comp Immunol (2016) 61:161–8. doi: 10.1016/j.dci.2016.03.029
45. Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, et al. Ips-1, an adaptor triggering rig-i- and Mda5-mediated type I interferon induction. Nat Immunol (2005) 6(10):981–8. doi: 10.1038/ni1243
46. Ohtani M, Hikima J, Kondo H, Hirono I, Jung TS, Aoki T. Characterization and antiviral function of a cytosolic sensor gene, Mda5, in Japanese flounder, paralichthys olivaceus. Dev Comp Immunol (2011) 35(5):554–62. doi: 10.1016/j.dci.2010.12.013
47. Bhat A, Paria A, Deepika A, Sreedharan K, Makesh M, Bedekar MK, et al. Molecular cloning, characterisation and expression analysis of melanoma differentiation associated gene 5 (Mda5) of green chromide, etroplus suratensis. Gene (2015) 557(2):172–81. doi: 10.1016/j.gene.2014.12.028
48. Zou PF, Chang MX, Xue NN, Liu XQ, Li JH, Fu JP, et al. Melanoma differentiation-associated gene 5 in zebrafish provoking higher interferon-promoter activity through signalling enhancing of its shorter splicing variant. Immunology (2014) 141(2):192–202. doi: 10.1111/imm.12179
49. Huang Y, Yu Y, Yang Y, Yang M, Zhou L, Huang X, et al. Antiviral function of grouper Mda5 against iridovirus and nodavirus. Fish Shellfish Immunol (2016) 54:188–96. doi: 10.1016/j.fsi.2016.04.001
50. Basler CF, Garcia-Sastre A. Viruses and the type I interferon antiviral system: Induction and evasion. Int Rev Immunol (2002) 21(4-5):305–37. doi: 10.1080/08830180213277
51. Altmann SM, Mellon MT, Distel DL, Kim CH. Molecular and functional analysis of an interferon gene from the zebrafish, danio rerio. J Virol (2003) 77(3):1992–2002. doi: 10.1128/jvi.77.3.1992-2002.2003
52. Zhou Y, Jiang N, Fan Y, Zhou Y, Xu C, Liu W, et al. Identification, expression profiles and antiviral activities of a type I ifn from gibel carp carassius auratus gibelio. Fish Shellfish Immunol (2019) 91:78–86. doi: 10.1016/j.fsi.2019.04.063
53. Ohtani M, Hikima J, Kondo H, Hirono I, Jung TS, Aoki T. Evolutional conservation of molecular structure and antiviral function of a viral rna receptor, Lgp2, in Japanese flounder, paralichthys olivaceus. J Immunol (2010) 185(12):7507–17. doi: 10.4049/jimmunol.1001850
54. Bruns AM, Leser GP, Lamb RA, Horvath CM. The innate immune sensor Lgp2 activates antiviral signaling by regulating Mda5-rna interaction and filament assembly. Mol Cell (2014) 55(5):771–81. doi: 10.1016/j.molcel.2014.07.003
55. Komuro A, Horvath CM. Rna- and virus-independent inhibition of antiviral signaling by rna helicase Lgp2. J Virol (2006) 80(24):12332–42. doi: 10.1128/JVI.01325-06
56. Han J, Wang Y, Chu Q, Xu T. The evolution and functional characterization of miiuy croaker cytosolic gene Lgp2 involved in immune response. Fish Shellfish Immunol (2016) 58:193–202. doi: 10.1016/j.fsi.2016.09.025
57. Gu T, Lu L, An C, Chen B, Wei W, Wu X, et al. Mda5 and Lgp2 acts as a key regulator though activating nf-kappab and Irf3 in rlrs signaling of mandarinfish. Fish Shellfish Immunol (2019) 86:1114–22. doi: 10.1016/j.fsi.2018.12.054
58. Yu Y, Huang Y, Yang Y, Wang S, Yang M, Huang X, et al. Negative regulation of the antiviral response by grouper Lgp2 against fish viruses. Fish Shellfish Immunol (2016) 56:358–66. doi: 10.1016/j.fsi.2016.07.015
59. Chang M, Collet B, Nie P, Lester K, Campbell S, Secombes CJ, et al. Expression and functional characterization of the rig-I-Like receptors Mda5 and Lgp2 in rainbow trout (Oncorhynchus mykiss). J Virol (2011) 85(16):8403–12. doi: 10.1128/JVI.00445-10
60. Zhang QM, Zhao X, Li Z, Wu M, Gui JF, Zhang YB. Alternative splicing transcripts of zebrafish Lgp2 gene differentially contribute to ifn antiviral response. J Immunol (2018) 200(2):688–703. doi: 10.4049/jimmunol.1701388
Keywords: Cyprinus carpio L., spring viremia of carp virus (SVCV), transcriptome analysis, MDA5, LGP2
Citation: Liu R, Niu Y, Qi Y, Li H, Yang G and Shan S (2022) Transcriptome analysis identifies LGP2 as an MDA5-mediated signaling activator following spring viremia of carp virus infection in common carp (Cyprinus carpio L.). Front. Immunol. 13:1019872. doi: 10.3389/fimmu.2022.1019872
Received: 15 August 2022; Accepted: 03 October 2022;
Published: 18 October 2022.
Edited by:
Tor Gjøen, University of Oslo, NorwayReviewed by:
Min Feng, South China Agricultural University, ChinaJude Juventus Aweya, Jimei University, China
Copyright © 2022 Liu, Niu, Qi, Li, Yang and Shan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shijuan Shan, c2hhbnNqQHNkbnUuZWR1LmNu; Guiwen Yang, eWFuZ2d3QHNkbnUuZWR1LmNu
†These authors have contributed equally to this work
 Rongrong Liu
Rongrong Liu Yan Niu
Yan Niu Yue Qi
Yue Qi Hua Li
Hua Li Guiwen Yang
Guiwen Yang Shijuan Shan
Shijuan Shan