
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 08 December 2022
Sec. Molecular Innate Immunity
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.1016165
 Matija Rijavec1,2*†‡
Matija Rijavec1,2*†‡ Aleš Maver3†
Aleš Maver3† Paul J. Turner4
Paul J. Turner4 Keli Hočevar3
Keli Hočevar3 Mitja Košnik1,5
Mitja Košnik1,5 Amnah Yamani6,7
Amnah Yamani6,7 Simon P. Hogan6,7
Simon P. Hogan6,7 Adnan Custovic4
Adnan Custovic4 Borut Peterlin3
Borut Peterlin3 Peter Korošec1,8
Peter Korošec1,8Background: Anaphylaxis is an acute life-threatening allergic reaction and a concern at a global level; therefore, further progress in understanding the underlying mechanisms and more effective strategies for diagnosis, prevention and management are needed.
Objective: We sought to identify the global architecture of blood transcriptomic features of anaphylaxis by integrating expression data from human patients and mouse model of anaphylaxis.
Methods: Bulk RNA-sequencings of peripheral whole blood were performed in: i) 14 emergency department (ED) patients with acute anaphylaxis, predominantly to Hymenoptera venom, ii) 11 patients with peanut allergy undergoing double-blind, placebo-controlled food challenge (DBPCFC) to peanut, iii) murine model of IgE-mediated anaphylaxis. Integrative characterisation of differential gene expression, immune cell-type-specific gene expression profiles, and functional and pathway analysis was undertaken.
Results: 1023 genes were commonly and significantly dysregulated during anaphylaxis in ED and DBPCFC patients; of those genes, 29 were also dysregulated in the mouse model. Cell-type-specific gene expression profiles showed a rapid downregulation of blood basophil and upregulation of neutrophil signature in ED and DBPCFC patients and the mouse model, but no consistent and/or significant differences were found for other blood cells. Functional and pathway analysis demonstrated that human and mouse blood transcriptomic signatures of anaphylaxis follow trajectories of upregulation of cell movement, migration and neuroinflammatory signalling, and downregulation of lipid activating nuclear receptors signalling.
Conclusion: Our study highlights the matched and extensive blood transcriptomic changes and suggests the involvement of discrete cellular components and upregulation of migration and neuroinflammatory pathways during anaphylaxis.
Anaphylaxis is an acute systemic hypersensitivity reaction that is rapid in onset and usually associated with potentially life-threatening airway, breathing, and/or circulatory compromise, often with skin and mucosal changes (1). It is mostly triggered by food, drugs, or insect stings (1, 2). Anaphylaxis is increasingly a concern at a global level; therefore, further progress in understanding the underlying mechanisms and more effective strategies for diagnosis, prevention and management are needed (1–4).
Although mast cell activation via IgE-mediated pathways is postulated to have a pivotal role in anaphylaxis, there has been steady progress in understanding that other pathways, effector cells and mediators potentially contribute to symptoms (1), with evidence for a potential role involving complement, basophils, neutrophils, monocytes/macrophages, platelets, and mediators, such as platelet-activating factor (PAF), cysteinyl leukotrienes (CysLTs), tryptase, and cytokines/chemokines (1, 5–9).
Transcriptomic data relating to anaphylaxis in human subjects, indicate that innate immune pathways (10) and immune cells, such as neutrophils, naïve CD4+ T cells, and macrophages are involved in the pathogenesis of human anaphylaxis (11). However, these data are not accomplished or integrated by transcriptomic analysis from mouse models of anaphylaxis, which have been developed to investigate the functions of antibodies, Fc receptors, and effector cells in anaphylaxis (1, 12).
To address this evidence gap, we integrated acute transcriptomic changes (bulk RNA-sequencing) of peripheral whole blood observed during anaphylaxis (i) in patients presenting to the hospital emergency department (ED) with acute anaphylaxis triggered mostly by insect stings, (ii) in peanut-allergic patients undergoing double-blind placebo-controlled food challenge (DBPCFC), and (iii) in a study of iIL9Tg mouse model using the passive antigen-induced anaphylaxis model which mimics food-induced IgE-mediated anaphylaxis (12, 13). Integrated data revealed 1023 genes commonly and significantly dysregulated during anaphylaxis in ED and DBPCFC patients; of those genes, 29 were also dysregulated in the mouse model; and common changes in blood basophils and neutrophils, upregulation of cell movement, migration, and neuroinflammatory signalling, and downregulation of lipid activating nuclear receptors signalling.
We recruited 15 patients (8 female patients; age, 20-79 years) presenting with an acute episode of anaphylaxis to the ED of University Hospital Golnik, Slovenia (6). Reaction severity was graded according to the Mueller criteria (14). We collected blood samples at presentation to the ED and 7-14 days and/or 1 month after the anaphylactic episode (Table 1).
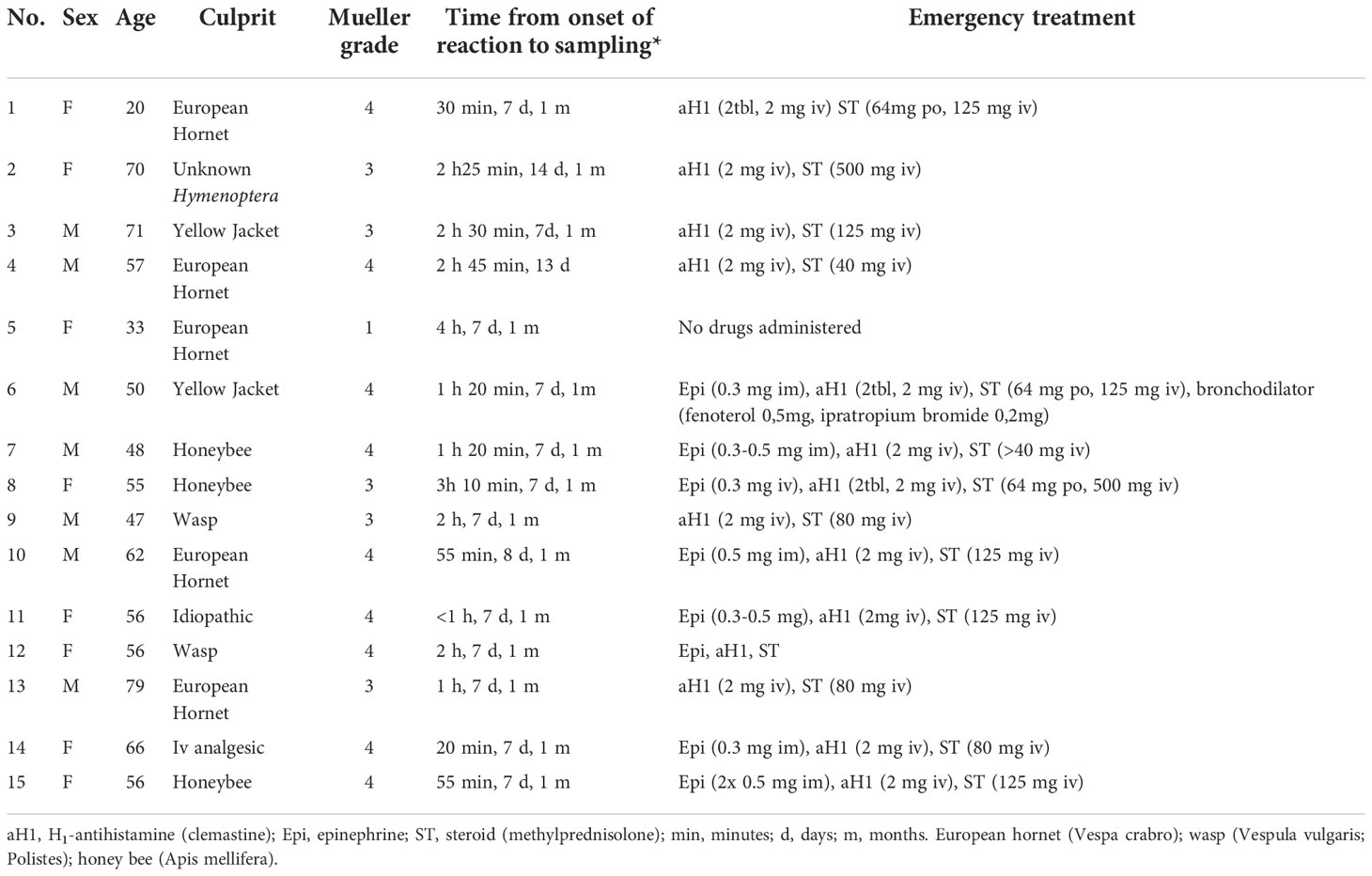
Table 1 Patients with acute anaphylactic reactions recruited from the hospital emergency department.
We recruited 11 patients with peanut allergy in whom allergy was confirmed by DBPCFCs as described elsewhere (15). Blood samples were collected prior to the challenge, at the onset of objective symptoms, and 2-4 hours after the challenge (Table 2).
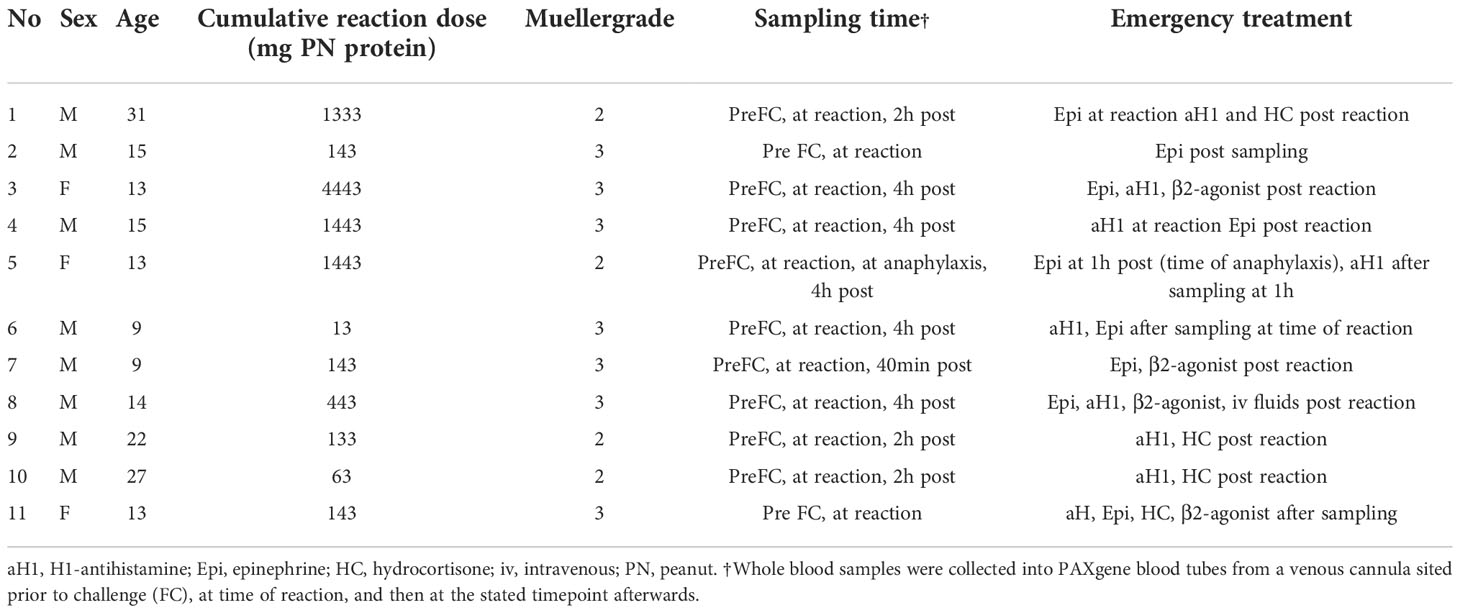
Table 2 Patients with peanut allergy undergoing double-blind placebo-controlled food challenge to peanut.
For passive oral antigen-induced anaphylaxis, iIL9Tg mice were sensitised and challenged as previously described (12). In iIL9Tg mice, murine IL-9 is under the control of the intestine-specific promoter of the rat fatty acid–binding protein (iFABPp) gene. This promoter has been extensively used to direct the expression of genes, specifically in enterocytes of the small intestine (16). A consequence of constitutive expression of IL-9 in the SI is a significant increase in mast cell levels predominantly localised to intraepithelial, intercryptic, and lamina propria regions of the SI. We have previously demonstrated that administration of antigen-specific IgE (anti-TNP-IgE) and subsequently challenged with a single oral dose of antigen [TNP-ovalbumin (OVA)] induces an acute allergic reaction characterised by gastrointestinal (GI) and systemic symptoms (17). Briefly, mice were injected intravenously (i.v.) with 10 μg of anti-TNP-IgE (200 μL of saline) with or without IL-4C (recombinant, IL-4–neutralising, anti–IL-4 monoclonal antibody [mAb] complex, 1:5 weight) (anti–IL-4 mAb, clone BVD4-1D11.2 obtained from Fred Finkelman, CCHMC) (17). Twenty-four hours later, mice were held in the supine position and orally gavaged with 250 μL of TNP-Ovalbumin (Ova; 50 mg) in saline (17). Before the intragastric (i.g.) challenge, mice were deprived of food for 3-4 hours. Challenges were performed with i.g. feeding needles (01-290-2B; Fisher Scientific Co., Pittsburgh, PA). For passive anaphylaxis, iIL9Tg mice were injected i.v. with 10 μg of anti-IgE [EM-95; obtained from Fred Finkelman, CCHMC (18)] in 100 μl saline and anaphylaxis assessment performed (12). Hypothermia (significant loss of body temperature) was used as an indication of anaphylaxis. Rectal temperature was measured with a rectal probe and a digital thermocouple thermometer (Model BAT-12; Physitemp Instruments, Clifton, NJ, USA). Temperature was taken prior to the challenge and then every 15 min for 30 or 60 minutes to identify maximum temperature change as previously described (12). Control mice were defined as mice that received anti-TNP IgE and subsequently received saline via o.g. and did not experience shock (hypothermia) response 30 minutes following challenge (FC). Mice that experienced a mild to moderate reaction received anti-TNP IgE and subsequently received o.g. OVA-TNP and experienced a 1.5 - 1.9˚C temperature loss within 30 minutes of oral FC. Mice that experienced a severe reaction received anti-TNP IgE and IL-4C and o.g. OVA-TNP and experienced a ≥ 3.7°C temperature loss within 30 minutes of the FC. Peripheral whole blood was drawn from iIL9Tg mice 2 hours following the intragastric challenge.
This study was conducted in accordance with the amended Declaration of Helsinki. Ethical approval was obtained from the Slovenian National Medical Ethics Committee (ED study) and the London Central Research Ethics Committee (DBPCFC study). All subjects provided written informed consent.
All mice were maintained and bred in a barrier facility, and animals were handled under approved Institutional Animal Care and Use Committee protocols at the University of Michigan animal facility.
Whole blood samples were collected into Tempus™ Blood RNA tubes (Thermo Fisher Scientific, Waltham, MA, USA) or PAXgene blood tubes (Qiagen, Hilden, Germany) containing RNA stabilising reagent. Total RNA was isolated using QuickGene blood cell kit (RB-S) and Fujifilm QuickGene-810 automated system (Fujifilm Life Sciences, Tokyo, Japan) or PAXgene Blood miRNA Kit and fully automated QIAcube system (Qiagen), according to the manufacturer’s isolation protocol. The purity of isolated RNA was assessed using the NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific), and integrity was determined on Agilent 2100 Bioanalyzer using RNA 6000 Nano LabChip kit (Agilent Technologies, Santa Clara, CA, USA). All samples used for library preparation had RNA integrity values greater than 7.0. Extracted RNA samples were stored at -80°C before further processing.
We performed library preparation using the TrueSeq stranded total RNA sample preparation kit (Illumina, San Diego, CA, USA), according to the manufacturer’s protocol. After Ribo-Zero rRNA depletion, the remaining RNA was purified, fragmented, and primed for first strand cDNA synthesis with reverse transcriptase (SuperScript II) and random primers, followed by second strand cDNA synthesis performed in the presence of dUTP. Blunt-ended double-strand DNA was 3’ adenylated, and multiple indexing adapters (T-tailed) were ligated to the ends of the ds cDNA. Fragments with adapter molecules on both ends were selectively enriched with 15 cycles of PCR reaction. 44 libraries were normalised to the final concentration of 10 nM and then pooled in equimolar concentrations. Sequencing was performed on the Illumina Hiseq 2000 sequencing system in 2×100 sequencing cycles using pair-end sequencing mode.
After initial read quality filtering and demultiplexing using Bcl2Fastq v1.8., alignments to the human reference genome (hg19) and mouse genome (mm10) were performed bowtie and tophat alignment softwares. Transcript abundances were then estimated using read counts function in Subread package for R (19), followed by intersample normalisation by variance modelling at the observational level (voom) approach implemented in limma Bioconductor package (20). Differences in gene expression were estimated using linear modelling and significance estimation procedures in limma. Principal component analysis was performed using the built-in function in R 4.0.3. Prior to further calculations, we removed the low-expressed genes by filtering out the genes that displayed counts per million (cpm) values below 1 in over 95% of the samples tested. This enabled removal of genes characterised by the low expression across all the samples, while still retaining potential biologically relevant genes that displayed low expression in a subset of samples, but a higher expression level in others.
The significance estimations were controlled for multiple testing using the false discovery rate approach (FDR) implemented in limma, and genes with FDR values of 0.1 or less were deemed dysregulated (differentially expressed).For comparison of human and mouse transcriptional patterns, we performed the conversion of the gene identifiers using orthology data in the Ensembl database, using biomaRt package for R.
In order to gain insights into biological mechanisms during anaphylaxis and to determine whether examined gene sets show statistically significant differences between stages, we performed Gene Set Enrichment Analysis (GSEA). Normalised sample expression levels (normalised RPKM values) of genes were used in the analysis. Gene sets collections (hallmark - H, curated gene sets – C2, ontology gene sets - C5, immunologic signature gene sets - C7) were obtained from the Molecular Signature Database (MSigDB) complied by Broad Institute: http://www.broadinstitute.org/gsea/index.jsp. To assess the statistical significance of the enrichment score, we used 1000 permutations.
We generated the gene signatures of immune cells based on the data from published immune cell expression profiling studies. Briefly, we analysed the publicly available immune cell transcriptome data for humans (GSE3982) (21) and mice (GSE37448) (22). For each immune cell type characterised in these studies, we selected genes that performed best at differentiating a specific immune cell type from others characterised in the study (23, 24). For each cell type investigated in the published immune cell profiling datasets, we performed a moderated t-test comparison (limma) of gene expression of the selected cell type and compared it to expression profiles of other cell types analysed in the study. We performed this iteratively across all the cell types investigated in the published datasets. Using this approach, we identified genes that were specifically over-expressed and were therefore considered as characteristic of a specific immune cell type versus other cell types. Only genes that attained a highly significant upregulation were included in each signature (P-value < 10-12). GSEA analysis was then used to detect the enrichment of gene sets belonging to each immune signature set. The list of genes included in human and mouse immune cell signatures are provided in the Supplementary Materials (Supplementary Tables 11, 12).
The functional analyses were generated through the use of QIAGEN’s Ingenuity® Pathway Analysis (IPA®, QIAGEN Redwood City, www.qiagen.com/ingenuity). DEGs with corresponding expression values were uploaded into the IPA software (Qiagen). The core analysis function included in the software was used to interpret the DEG data, which included diseases and functions, canonical pathways, and upstream transcriptional regulators.
Raw sequencing and read-count level data were submitted to GEO repository (GSE215184).
Table 1 shows detailed information on demographic and clinical characteristics, emergency treatment, and sampling data of 15 ED patients. The reaction was caused by a Hymenoptera insect sting in 13 (87%) patients. One patient had a Mueller grade I, 5 a Mueller grade III, and 9 a Mueller grade IV reaction. The median time from the onset of symptoms to peripheral blood sample collection was 1 hour 20 minutes (range 20 minutes to 4 hours). Convalescent samples were collected from all patients median 7 days (range 7 to 14 days) and 14 patients one month after the anaphylactic episode.
In 11 peanut-allergic individuals, peripheral blood samples were collected prior to DBPCFC. Four patients had a Mueller grade II and 7 a Mueller grade III reaction. According to Mueller grading, the reaction severity was significantly lower in the DBPCFC group compared to the ED group (Median 3 vs 4; P = 0.002; Mann-Whitney U test). Following the objective reaction, further blood samples were obtained from all patients, and in 9, 2-4 hours after reaction. One patient (patient 5) experienced anaphylaxis 1 hour after an initial non-anaphylaxis reaction and therefore had 2 post-reaction samples collected (Table 2).
Individual transcriptomic analysis was ascertained from the blood of 8 mice with severe IL-4C amplified anaphylaxis within 30 minutes of oral challenge, 8 with mild to moderate anaphylaxis within 30 minutes of the challenge, and 7 control mice, that did not experience shock (hypothermia) response 30 minutes following challenge for baseline comparison (17).
1023 genes were commonly and significantly dysregulated during anaphylaxis in ED and DBPCFC patients; of those genes, 29 were also dysregulated in the mouse model.
We compared gene expression signatures during the anaphylaxis episode between different provoking agents (insect stings and peanut) and human (ED, Peanut-DBPCFC studies) or murine model of anaphylaxis. Therefore, we first compared differentially expressed genes (DEGs) between all three study groups, and found a high correlation in transcriptional response during human anaphylaxis, between ED and DBPCFC (r = 0.61, P < 10-16; Figure 1A). While only limited correlation was found between human and mouse model (MOUSE and ED (r = 0.19, P < 10-16); MOUSE and DBPCFC (r = 0.17, P < 10-16; Figures 1B, C). Detailed analysis revealed that in the emergency department (ED) study, no differences in RNA expression when comparing the convalescent samples 7 days and 1 month after the acute allergic reaction was detected. On the other hand, significant differences in RNA expression profiles were observed when comparing samples after acute allergic reaction with either 7 days and 1 month after the episode. Therefore, in all downstream analyses, RNA expression after the acute allergic reaction was compared with merged convalescent samples after 7 days and 1 month after the episode, as 3224 differentially expressed gene (DEGs) were found during anaphylaxis in comparison to convalescent state.
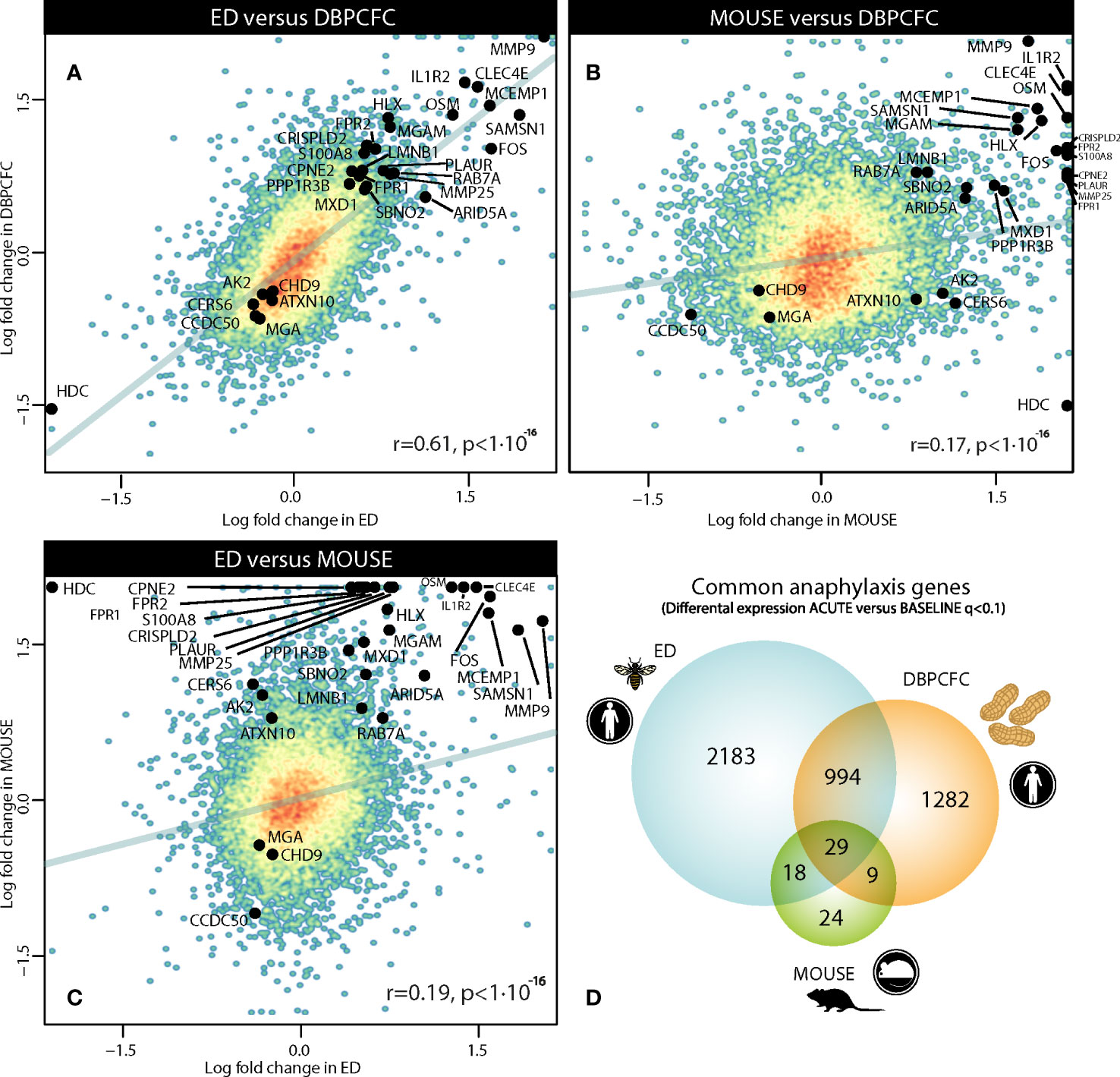
Figure 1 Comparison of gene dysregulation during the anaphylactic episode between human and mouse model of anaphylaxis: (A) between ED and DBPCFC study, (B) between MOUSE and DBPCFC study, and (C) between ED and MOUSE study. Overall 3224, 2314 and 80 genes were differentially expressed in ED, DBPCFC and mouse study group, respectively with (D) the overlap of 1023 DEGs for both human situations of anaphylaxis and 29 DEGs for all three study groups. Genes (n=29) differentially expressed in human and mouse model during anaphylaxis are highlighted. ACUTE, acute anaphylactic episode; ED, Emergency Department; DBPCFC, double-blind placebo-controlled food challenge.
When analysing the transcriptional changes in the double-blind placebo-controlled food challenge (DBPCFC) study group, only modest changes in gene expression were detected at the cessation of the challenge in comparison to before, whereas most profound changes were detected when comparing after the challenge with before DBPCFC, where 2314 DEGs were detected. Therefore, for all subsequent analyses, RNA expression after the challenge was compared with the status before the challenge.
In the Mouse study group, we compared RNA expression levels in samples with anaphylaxis (mild and severe) in comparison to mouse samples without anaphylaxis and found 80 DEGs between the two groups. Only modest changes in RNA expression levels were observed between mice with mild and severe anaphylaxis.
To identify the key genes dysregulated during the anaphylactic episode we then analysed which genes are differentially expressed in all three study groups; 1023 genes were commonly and significantly dysregulated during anaphylaxis in ED and DBPCFC patients; of those genes, 29 were also dysregulated in the mouse model (q < 0.1; Figure 1D). Top DEGs found only in human anaphylaxis are presented in Table 3, while top overlapping DEGs regulated in anaphylaxis are presented in Table 4. Details in Supplementary Tables 1–5. DEGs in peripheral whole blood might reflect alterations in transcriptional regulation or immune cell type composition, both reflecting the pathophysiological mechanism of anaphylaxis (11). Among the six key upregulated drivers of acute peanut allergic reactions recently identified by Watson et al. (11), five, specifically, KLHL2, PPP1R3D, PADI4, IL1R2, and ECHDC3, were also found to be upregulated (all q<0.05) in both human ED and DBPCFC study. No dysregulation of the LTB4R gene was found in our study.
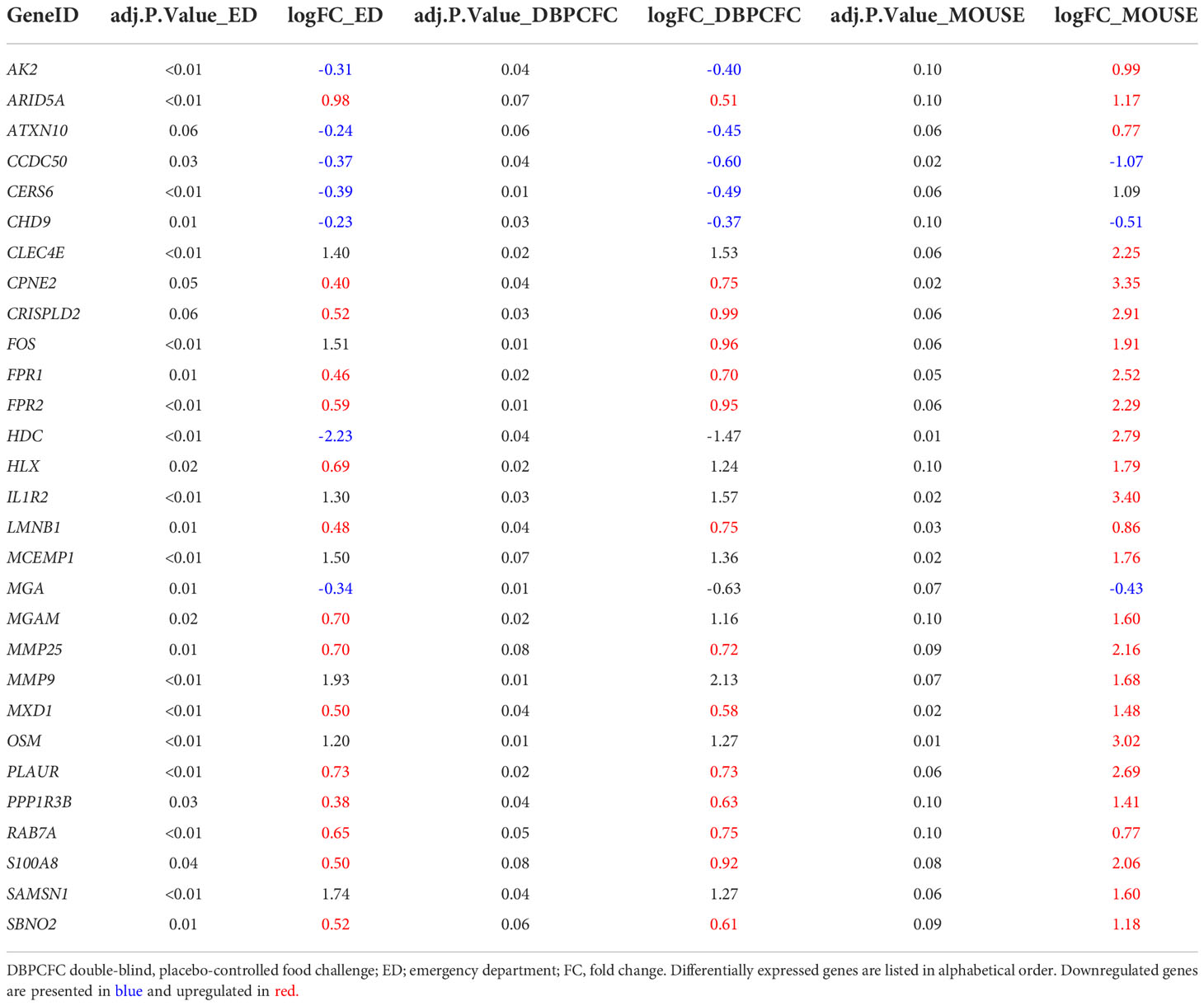
Table 4 Differentially expressed genes (q < 0.1) in all three study groups (human (ED and DBPCFC) and mouse study groups).
Anaphylaxis involves complex interactions, recruitment, and activities of several different immune cell types. To identify key blood immune cell types participating in anaphylaxis comparative analysis with expression signatures of basophils, B cells, dendritic cells, eosinophils, macrophages, monocytes, neutrophils, NK cells, T cells (CD4+ and CD8+, Th1 and Th2), were obtained from Molecular Signature Database (MSigDB). Immune cell-type-specific gene expression profiles identified major changes in basophil and neutrophil expression signatures for all three studies: ED, DBPCFC and mouse study of anaphylaxis (Figure 2). Eosinophil signatures were different only for ED and mouse studies, but not DBPCFC study. No consistent differences were observed in the expression signatures of other immune cells (Details in Supplementary Tables 6, 7).
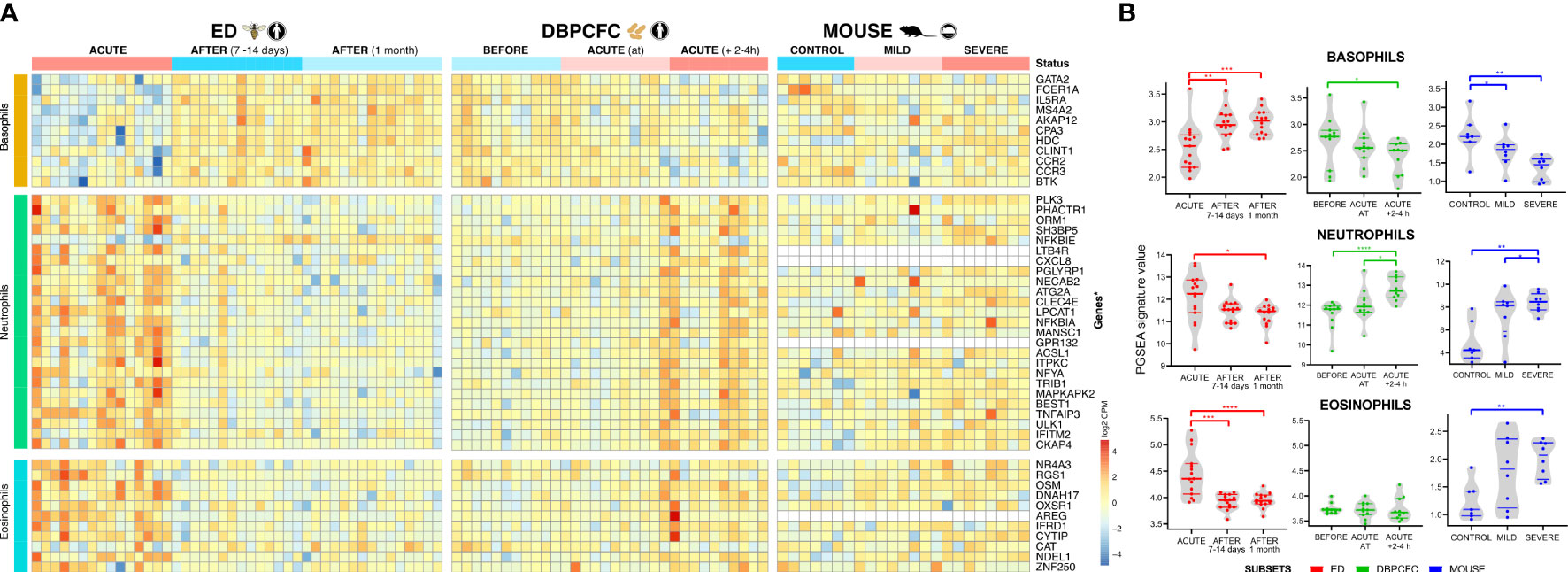
Figure 2 Immune cell-specific profile changes in peripheral blood during anaphylaxis. (A) Cell-specific gene signatures of basophils, neutrophils and eosinophils are differentially expressed during anaphylaxis. (B) Changes in cell profile in peripheral blood in different time points. No consistent differences were observed in expression signatures of B cells, dendritic cells, macrophages, monocytes, NK cells, T cells (CD4+ and CD8+, Th1 and Th2) (Details in Supplementary Tables 6, 7). ACUTE, acute anaphylactic episode; ED, Emergency Department; DBPCFC, double-blind placebo-controlled food challenge. *P <0.05, **P <0.01, ***P <0.001, and ***P <0.0001 by the Mann-Whitney test.
During anaphylaxis, we observed a significant decrease in basophil gene signature and an increase in neutrophil specific gene signature, and those changes were evident in all three study groups (Figures 2A, B). In the DBPCFC study, trends in basophil and neutrophil changes already become evident at the onset of objective anaphylactic symptoms and then achieved significance at post sampling (Figure 2B). Eosinophil specific gene signature was increased only in the ED study, but not under the controlled setting of the DBPCFC study, while in the mouse study group it was increased in mice with severe anaphylaxis (Figure 2). The changes in gene expression signatures of basophils and neutrophils might represent either a change in the number of these cells (reflecting movement and/or migration of these cells) and/or changes in distinct basophil and neutrophil subsets with specific/different functional consequences.
To illustrate interactions, biological context and bio-function among the top DEGs during anaphylaxis, a network and pathway analysis was performed and the top terms enriched in all three study groups and each category are presented in Figure 3.
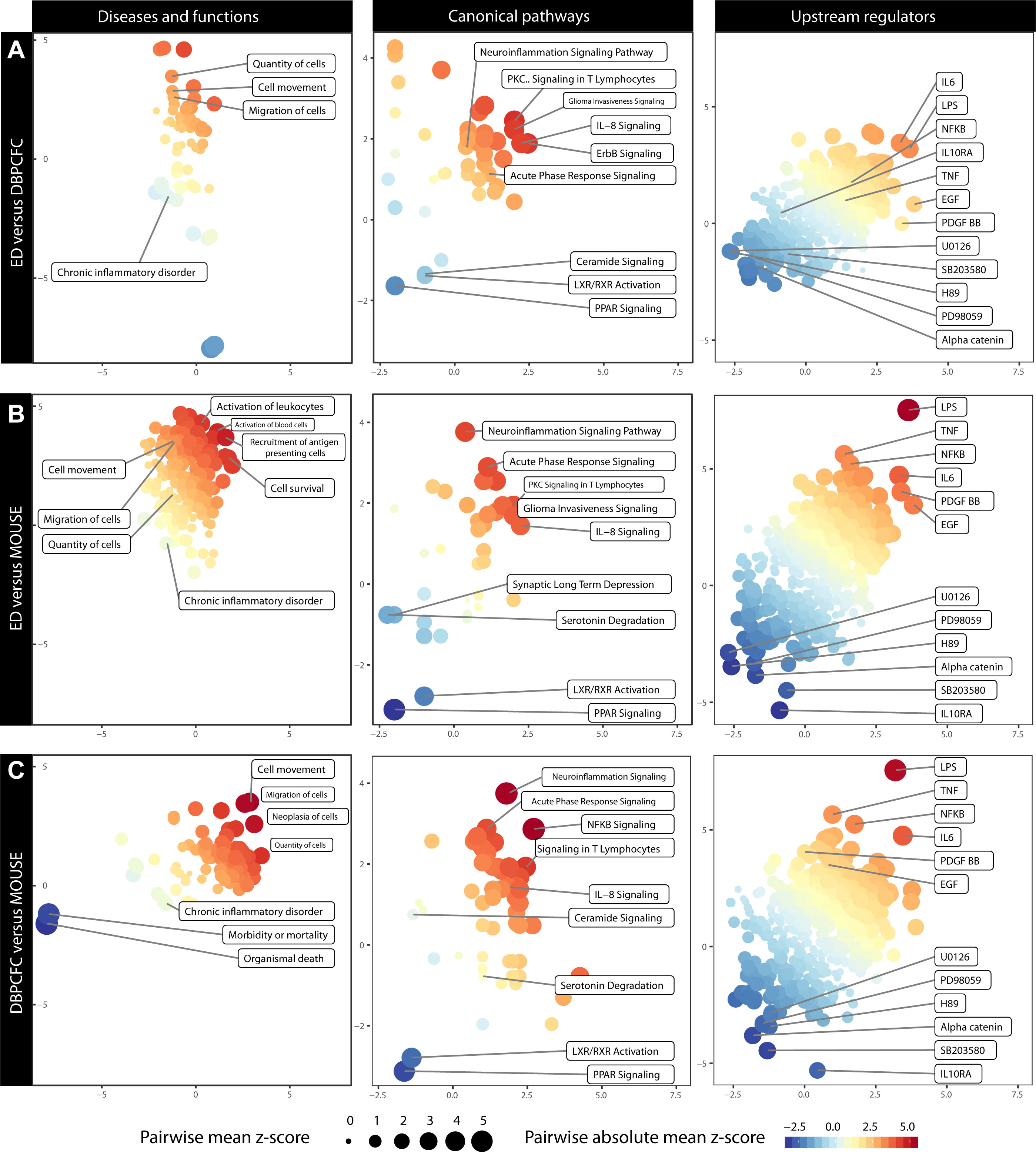
Figure 3 Comparison of altered Diseases and functions, Canonical pathways and Upstream regulators during anaphylaxis between different study groups: (A) ED and DBPCFC, (B) ED and MOUSE, (C) DBPCFC and MOUSE. The plot is based on the z score calculations obtained for a given set of genes using the IPA analysis. The positive values indicate an overall upregulation (red colour) of the genes in a set and negative values indicate a downregulation (blue colour). The colour of the points corresponds to a mean of z scores obtained for the pair of exposures in each panel, and the size of the points corresponds to an absolute average of z scores. The larger and more intensely coloured points, therefore, indicate a more pronounced and analogous dysregulation of a gene set in a given pair of exposures. Details in Supplementary Table 8 (Diseases and functions), Supplementary Table 9 (Canonical pathways) and Supplementary Table 10 (Upstream regulators).
Enrichment of DEGs shows that increased cell movement and migration involving different immune cells represent the most important bio-function in peripheral blood during anaphylaxis. Therefore, as also demonstrated with immune cell-type-specific gene expression profiles, migration and activation of different immune cells – most notably basophils and neutrophils – represent key mechanisms in human and murine anaphylaxis.
Diseases and functions analysis showed that the upregulated DEGs were significantly enriched in biological functions related to cell movement (z score = 2.58, P = 1.19×10-15; z score = 2.04, P = 9.58×10-7; z score = 2.68, P = 2.64×10-10 for ED, DBPCFC and MOUSE study groups, respectively), the quantity of cells and migration of cells (z score = 2.06, P = 9.54×10-17; z score = 1.27, P = 1.04×10-5 z score = 2.49, P = 6.52×10-10 for ED, DBPCFC and MOUSE study groups, respectively), while the downregulated DEGs were enriched in functions related to chronic inflammatory disorders (z score = -2.05, P = 3.84×10-14; z score = -0.42, P = 3.57×10-5; z score = -0.90, P = 4.31×10-12 for ED, DBPCFC and MOUSE study groups, respectively). Details in Supplementary Table 8.
The most significantly enriched canonical pathways of the upregulated DEGs were acute phase response (z scores were 1.98, 2.18, and 1.13, for ED, DBPCFC and MOUSE study groups, respectively), IL-8 signalling (z scores were 2.98, 1.21, and 1.34, for ED, DBPCFC and MOUSE study groups, respectively), signalling in T lymphocytes, and neuroinflammatory signalling pathway. For the downregulated DEGs, the most significantly enriched pathways were for lipid-activated nuclear receptors of transcription factors (PPAR (z scores were -1.34, -0.78, and -2.24, for ED, DBPCFC and MOUSE study groups, respectively) and LXR/RXR [z scores were -0.73, -1.39, and -2.12, for ED, DBPCFC and MOUSE study groups, respectively)] signalling. Further details are in Supplementary Table 9.
Upstream regulator analysis revealed that LPS-like response (z scores were 3.30, 3.41, and 4.87, for ED, DBPCFC and MOUSE study groups, respectively), TNF (z scores were 2.58, 2.16, and 4.01, for ED, DBPCFC and MOUSE study groups, respectively), IL-6, NF-κB (z scores were 1.15, 2.40, and 2.00, for ED, DBPCFC and MOUSE study groups, respectively), EGF, and platelet-derived growth factor (PDGF-BB; z scores were 3.60, 1.98, and 2.58, for ED, DBPCFC and MOUSE study groups, respectively)) were the most significantly upregulated pathways during anaphylaxis in all three study groups. While alpha-catenin (z scores were -2.11, -2.61, and -2.60, for ED, DBPCFC and MOUSE study groups, respectively) and inhibitors of mitogen-activated protein kinases (OU126, PD98059, H89, SB2003580) being among the most downregulated upstream regulators during anaphylaxis. Further details are in Supplementary Table 10.
Herein, we have integrated analysis of ED insect sting and controlled DBPCFC peanut human studies with mouse study of anaphylaxis to identify the global architecture of blood transcriptome features of anaphylaxis. The iIL9Tg mouse model permits the induction of an IgE-mediated food allergic reaction following a single oral dose of antigen, as in human anaphylaxis (12, 13). Our study highlights the extensive blood transcriptomic changes, which included dysregulation of 1023 genes in human anaphylaxis and 29 genes that were dysregulated in both human and mouse anaphylaxis, the involvement of blood basophils and neutrophils, and upregulation of cell movement, migration, neuroinflammatory signalling, and downregulation of lipid activating nuclear receptors signalling during anaphylaxis. Notably, the extent and correlations of the transcriptomic changes in human anaphylaxis (1023 genes; correlation coefficient r = 0.61) versus that induced in both human and mouse anaphylaxis (29 genes; correlation coefficients r = 0.17 and 0.19) differ significantly.Among the six key drivers of acute peanut allergic reactions previously identified by Watson et al. (11), five of six, specifically PADI4, IL1R2, PPP1R3D, KLHL2, and ECHDC3, but not LTB4R, were found to be dysregulated in ED and DBPCFC studies, supporting the importance of those genes in human anaphylaxis and the relevance of previous results (11). However, of those 6 genes, only IL1R2 was dysregulated in the mouse study. Importantly, IL1R2 was recently identified as one of the genes associated with reaction severity in peanut-allergic children (25). IL-1R2 is expressed by neutrophils, B cells, monocytes, and macrophages, and it is a decoy receptor that neutralises IL-1β, limiting the activation of the pro-inflammatory IL-1 pathway, and influencing the production of different interleukins and NF-κB signalling (11, 26). OSM, oncostatin M, and IL-6 family cytokines are potent neutrophil recruiters (27). OSM stimulates endothelial cell proliferation and migration (28) and is implicated in endothelial signalling (29) and activation (30). Besides, OSM upregulates matrix metalloproteinase-9 (MMP9) through the MAPK/ERK pathway. MMP9 is an important pro-inflammatory protein, which is involved in activation of mast cells (31) and tissue remodelling (10) and was also identified in our study. The most important source of MMP9 is reported to be activated neutrophils (10, 31). CCDC50, and SBNO2 are involved in NF-κB signalling, ARID5A, FOS, MGA, MXD1, and S100A8 in cellular proliferation, differentiation and apoptosis, and CD177, CPNE2, CST7, FPR1, FPR2, HLX, HRH4, IL5RA, MCEMP1, ORM1, PTGDR2, and SBNO2 in immune system regulation and signalling (32).
Multiple lines of evidence suggest that, besides mast cells viewed as key players, discrete cells may contribute to the pathology of anaphylaxis (1, 6, 10, 11, 33). By employing immune cell-type-specific gene expression profiles analysis (23, 24), we showed a rapid change in basophils and neutrophil signatures during anaphylaxis. However, the importance of basophils and neutrophils in the pathophysiology of anaphylaxis is currently unsettled (1). Nevertheless, a rapid and significant decrease in basophil gene signature during anaphylaxis supports speculations that basophil migrate from the circulation rapidly upon allergen challenge (6, 34), possibly to sites of inflammation (7, 35, 36) where they might have an effector role (34). Alternatively this migration might have a protection role by limiting basophil activation in circulation (34). Basophil migration might be mediated by the chemokine CCL2, levels of which are increased during anaphylaxis (6, 7) and which can induce selective migration of human basophils (7, 37).
Recent data suggests that neutrophils are not only important in murine anaphylaxis (1, 8, 38) but might be also important in human anaphylaxis (1, 8). This is supported by our findings of a rapid increase in neutrophil gene signature in both human ED insect sting and controlled DBPCFC peanut studies and consistent with a previous study in peanut-allergic children in which allergen challenge (but not placebo) upregulates neutrophil gene signatures (11). Human and murine neutrophils express several activating FcµRs and human neutrophils also FcϵRI, can release histamine (although in much smaller amounts than mast cells), PAF (which can cause bronchoconstriction, increased vascular permeability, chemotaxis, and degranulation of eosinophils and neutrophils) and CysLTs after stimulation with immune complexes (1, 8). In murine models of anaphylaxis, neutrophil depletion can ameliorate anaphylaxis. A potential contribution of neutrophils during anaphylaxis in humans is supported by increased serum levels of PAF (9) and myeloperoxidase (MPO), the major enzyme stored in neutrophils (39). Recently, increased neutrophil activation markers were also demonstrated during drug, food, and insect-induced anaphylaxis (15, 40).
Aside from basophils and neutrophils, we also observed increased eosinophil gene signatures, but only in patients of the ED study and mice with severe anaphylaxis. Eosinophils express FcϵRI as well as several other receptors (including IL-5 receptor and CCR5) on their surface; their engagement induces eosinophil activation and the release of several mediators including cationic proteins (eosinophil peroxidase, major basic protein, eosinophil cationic protein, eosinophil-derived neurotoxin), lipid mediators (LTC4, PAF), and a variety of cytokines (IL-1, IL-3, IL-4, IL-5, IL-13, GM-CSF, TGF-α/β, TNF-α, chemokines (CCL3, CCL5, CCL11), and neuromodulators (Substance P, vasoactive intestinal peptide) (1, 41–43) – which could sustain or augment the immune response. However, no previous data did show that eosinophils may play a role in anaphylaxis.
Integrated data-driven functional and pathway analysis using IPA disease and function, canonic pathways, and upstream regulator analysis identified cell movement, migration, and neuroinflammatory and lipid activating nuclear receptors signalling as the major blood-related pathways of anaphylaxis. Upregulation of cell movement and migration is consistent with the rapid change in basophils and neutrophil signatures during anaphylaxis and emphasises the potential importance of cellular interactions in anaphylaxis (1, 6, 10, 11, 33). Neurogenic inflammation may be a key driver of some allergic symptoms (44), including rhinitis, conjunctivitis, coughing, bronchoconstriction, airway mucus secretion, dysphagia, altered gastrointestinal motility, and urticaria/angioedema (44). Allergen-induced activation of mast cells could lead to alterations in the function of afferent neurons like intrapulmonary airway C-fiber (45), neurons within the CNS (46, 47), and somatosensory itch fiber in the skin (48), sensory nerve excitability (49) and neuroplasticity (50), increases in synaptic efficacy in autonomic ganglia (51, 52) and stimulation of enteric neurons (53, 54) – an effect which might be more prominent in atopic individuals compared to non-atopic controls (44). Of note, a recent neuronal receptor-based controlling mechanism of anaphylaxis involving the glutamatergic receptor mGluR7was described (55). These data would therefore be consistent with our hypothesis that upregulation of neuroinflammatory signalling pathways might impact neuromodulation in anaphylaxis.
Lipid-activating nuclear receptors signalling, specifically lipid-activated nuclear receptors of transcription factors PPAR and LXR/RXR, was found to be highly downregulated. Ligand-activated transcription factors play important roles in cellular processes such as differentiation, proliferation, survival, apoptosis, and motility in a variety of biological contexts, including inflammation and immune responses (56). LXR and PPAR can antagonise inflammatory pathways through transrepression (57). PPARs can modulate the intensity, duration, and outcomes of inflammatory responses through anti-inflammatory effects (58). PPAR activation also reduces the production of multiple pro-inflammatory mediators, including TNF-α, IL-1α, IL-6, and IL-8. PPAR activation in endothelial cells inhibits endothelial inflammation, improves endothelial function, and inhibits cell proliferation and migration (59). Downregulation/inhibition of these effects thus results in vascular dysregulation. Lipid-activated nuclear receptors also shape macrophage and dendritic cell function (60), and studies in mice have shown that depletion of monocytes/macrophages can reduce anaphylaxis (1).
There was also upregulation of NFκB signalling and lipopolysaccharide (LPS) pathway, upregulation of inflammatory cytokines IL-6, IL-8, TNF-α, and IL-1β, and angiogenic platelet-derived growth factor (PDGF). These cytokines induce the inactivation of cadherin, which mediates cell adhesion and lead to vascular leakage by increased capillary permeability and target several immune cells with the FcγR receptor (involved in acute inflammatory responses) (61). LPS modulates immune responses by interacting with toll-like receptor 4 (TLR4, and can induce both Th1 and Th2 responses (62). PDGF is synthesised and released by platelets upon activation and is increased in children with anaphylaxis; rapid IgE desensitisation, in turn, downregulates PDGF (63).
There are some important limitations to our study. The first and most significant limitation is that we did not perform follow-up functional in-vitro experiments and analyses. However, it is important to note that recent studies corroborate possible contributing mechanisms of many of our major discovery genes (ARG1, CPA3, FOS, GATA2, HDC, IL18R1, IL18RAP, MMP9, OSM, and PER1) and related them to mast cells or basophils, or other contributing factors. Thus, Do et al. (25) recently identified ARG1 as a major hub in the network of the genetic and epigenetic architecture of reaction severity in peanut allergy. Dwyer et al. (64) demonstrated that shared mast cell and basophil gene signature networks include ten-fold higher expression of CPA3 and GATA2 than other immune cells. Further, the transcription factor GATA2 regulates HDC gene expression in mast cells and is required for IgE/mast cell-mediated anaphylaxis (65), and conditional deletions of HDC confirm the role of histamine in IgE-mediated anaphylaxis (66). Inhibition of FOS expression attenuates IgE−mediated mast cell activation and allergic inflammation (67). Genetic variations at the IL-18 locus (IL18R1 and IL18RAP) were associated with exercise-induced anaphylaxis (68), while MMP9 and OSM provide evidence for the involvement of innate immune pathways (40). OSM is also involved in hyper-IgE syndrome (69). Finally, increased expression of PER1 inhibits mTORC1 signalling, thus reducing Th1 differentiation and shifting the T-helper subset balance towards Th2 (70). Another important limitation is that due to the relatively small number of included subjects, additional subanalysis by stratifying patients according to reaction severity and trigger could not be performed. Further, bulk RNA-sequencing data was based on a mixed cell population from peripheral blood. Therefore, variations in the immune cell composition of the samples may impact the transcriptional profile. However, to our knowledge, this is the first report of integrated transcriptomic analysis of human anaphylaxis (ED patients with acute reaction, predominantly to Hymenoptera venom and patients with peanut allergy undergoing DBPCFC to peanut) and iIL9T mouse model, which mimics food-induced IgE-mediated anaphylaxis. Additionally, it was recently proposed that Hymenoptera venom and medications such as quinolone antibiotics can cause non–IgE-mediated anaphylaxis through the activation of Mas-related G protein-coupled receptor X2 (MRGPRX2) on mast cells (71–73). For Hymenoptera venom, this novel and intriguing concept is based on a venom constituent mastoparan, which can directly, through MRGPRX2, activate mast cells (74–76). Thus, direct activation of mast cells by mastoparan via this receptor (71, 74) may also affect transcriptomic changes observed in our ED patients. However, a broader assessment will be required to replicate these findings (71, 74) and confirm this hypothesis.
In summary, integrative blood transcriptomic analysis in human and murine model of anaphylaxis demonstrated extensive gene dysregulation, mainly involved in NF-κB, MAPK/ERK and endothelial signalling, inflammation, cellular proliferation, differentiation, apoptosis, and immune system regulation and signalling. Importantly, we confirmed, in both human and mouse studies, the rapid changes in blood basophils and neutrophils signatures during anaphylaxis. For the first time, we have identified the upregulation of cell movement, migration, and neuroinflammatory signalling during anaphylaxis. These findings highlight the involvement of distinct effector cells, and complex signalling changes, which reflect cellular movement and interactions, as well as possible implications of the nervous system. Further studies of those pathways might be important for more effective strategies for preventing this disorder and might, in future, also provide novel options for treating anaphylaxis.
The data presented in this study are deposited in the GEO repository, accession number GSE215184.
This study was conducted in accordance with the amended Declaration of Helsinki. Ethical approval was obtained from the Slovenian National Medical Ethics Committee (ED study) and the London Central Research Ethics Committee (DBPCFC study). All subjects provided written informed consent. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin. All mice were maintained and bred in a barrier facility, and animals were handled under approved Institutional Animal Care and Use Committee protocols at the University of Michigan animal facility.
MR, AM, PK conceived the idea and designed the experiments; MR, AM, PT, KH, MK, AY, SH, AC, BP, PK acquired clinical data, performed experiments, data analysis and interpretation. MR, PK wrote the manuscript. All authors read, revised and approved the final version of the manuscript.
Slovenian Research Agency (P3-0360), NIHR Biomedical Research Centre based at Imperial College Healthcare NHS Trust and Imperial College London, and National Institutes of Health (AI138177 and AI112626).
We thank all the patients, as well as the clinical and laboratory staff for their assistance.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.1016165/full#supplementary-material
1. Reber LL, Hernandez JD, Galli SJ. The pathophysiology of anaphylaxis. J Allergy Clin Immunol (2017) 140:335–48. doi: 10.1016/j.jaci.2017.06.003
2. Yu JE, Lin RY. The epidemiology of anaphylaxis. Clin Rev Allergy Immunol (2018) 54:366–74. doi: 10.1007/s12016-015-8503-x
3. Turner PJ, Gowland MH, Sharma V, Ierodiakonou D, Harper N, Garcez T, et al. Increase in anaphylaxis-related hospitalisations but no increase in fatalities: An analysis of united kingdom national anaphylaxis data, 1992-2012. J Allergy Clin Immunol (2015) 135:956–63. doi: 10.1016/j.jaci.2014.10.021
4. LoVerde D, Iweala OI, Eginli A, Krishnaswamy G. Anaphylaxis. Chest (2018) 153:339–48. doi: 10.1016/j.chest.2017.07.033
5. Lyons JJ, Chovanec J, O’Connell MP, Liu Y, Šelb J, Zanotti R, et al. Heritable risk for severe anaphylaxis associated with increased α-tryptase–encoding germline copy number at TPSAB1. J Allergy Clin Immunol (2021) 147:622–32. doi: 10.1016/j.jaci.2020.06.035
6. Korosec P, Turner PJ, Silar M, Kopac P, Kosnik M, Gibbs BF, et al. Basophils, high-affinity IgE receptors, and CCL2 in human anaphylaxis. J Allergy Clin Immunol (2017) 140:750–8. doi: 10.1016/j.jaci.2016.12.989
7. Vantur R, Rihar M, Koren A, Rijavec M, Kopac P, Bidovec-Stojkovic U, et al. Chemokines during anaphylaxis: The importance of CCL2 and CCL2-dependent chemotactic activity for basophils. Clin Transl Allergy (2020) 10:1–11. doi: 10.1186/s13601-020-00367-2
8. Jonsson F, Mancardi DA, Albanesi M, Bruhns P. Neutrophils in local and systemic antibody-dependent inflammatory and anaphylactic reactions. J Leukoc Biol (2013) 94:643–56. doi: 10.1189/jlb.1212623
9. Vadas P, Gold M, Perelman B, Liss GM, Lack G, Blyth T, et al. Platelet-activating factor, PAF acetylhydrolase, and severe anaphylaxis. N Engl J Med (2008) 358:28–35. doi: 10.1056/NEJMoa070030
10. Stone SF, Bosco A, Jones A, Cotterell CL, Van Eeden PE, Arendts G, et al. Genomic responses during acute human anaphylaxis are characterised by upregulation of innate inflammatory gene networks. PloS One (2014) 9:1–10. doi: 10.1371/journal.pone.0101409
11. Watson CT, Cohain AT, Griffin RS, Chun Y, Grishin A, Hacyznska H, et al. Integrative transcriptomic analysis reveals key drivers of acute peanut allergic reactions. Nat Commun (2017) 8:1943. doi: 10.1038/s41467-017-02188-7
12. Yamani A, Wu D, Waggoner L, Noah T, Koleske AJ, Finkelman F, et al. The vascular endothelial specific IL-4 receptor alpha-ABL1 kinase signaling axis regulates the severity of IgE-mediated anaphylactic reactions. J Allergy Clin Immunol (2018) 142:1159–72. doi: 10.1016/j.jaci.2017.08.046
13. Ganesan V, Sharma A, Tomar S, Schuler CF, Hogan SP. IL-4 receptor alpha signaling alters oral food challenge and immunotherapy outcomes in mice. J Allergy Clin Immunol (2022) S0091-6749(22). doi: 10.1016/j.jaci.2022.07.011
14. Mueller UR. Clinical presentation and pathogenesis. In: Mueller UR, editor. Insect sting allergy. Stuttgart: Gustav Fischer (1990). p. 33–65.
15. Muñoz-Cano R, Pascal M, Bartra J, Picado C, Valero A, Kim DK, et al. Distinct transcriptome profiles differentiate nonsteroidal anti-inflammatory drug-dependent from nonsteroidal anti-inflammatory drug-independent food-induced anaphylaxis. J Allergy Clin Immunol (2016) 137:137–46. doi: 10.1016/j.jaci.2015.05.042
16. Sweetser DA, Birkenmeier EH, Hoppe PC, McKeel DW, Gordon JI. Mechanisms underlying generation of gradients in gene expression within the intestine: an analysis using transgenic mice containing fatty acid binding protein-human growth hormone fusion genes. Genes Dev (1988) 2:1318–32. doi: 10.1101/gad.2.10.1318
17. Ahrens R, Osterfeld H, Wu D, Chen CY, Arumugam M, Groschwitz K, et al. Intestinal mast cell levels control severity of oral antigen-induced anaphylaxis in mice. Am J Pathol (2012) 180:1535–46. doi: 10.1016/j.ajpath.2011.12.036
18. Baniyash M, Eshhar Z. Inhibition of IgE binding to mast cells and basophils by monoclonal antibodies to murine IgE. Eur J Immunol (1984) 14:799–807. doi: 10.1002/eji.1830140907
19. Liao Y, Smyth GK, Shi W. The r package rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res (2019) 47:e47. doi: 10.1093/nar/gkz114
20. Law CW, Chen Y, Shi W, Smyth GK. Voom: Precision weights unlock linear model analysis tools for RNA-seq read counts (2014). Genome Biol (2014) 15:R29. doi: 10.1186/gb-2014-15-2-r29
21. Jeffrey KL, Brummer T, Rolph MS, Liu SM, Callejas NA, Grumont RJ, et al. Positive regulation of immune cell function and inflammatory responses by phosphatase PAC-1. Nat Immunol (2006) 7:274–83. doi: 10.1038/ni1310
22. Elpek KG, Cremasco V, Shen H, Harvey CJ, Wucherpfennig KW, Goldstein DR, et al. The tumor microenvironment shapes lineage, transcriptional, and functional diversity of infiltrating myeloid cells. Cancer Immunol Res (2014) 2:655–67. doi: 10.1158/2326-6066.CIR-13-0209
23. Shen-Orr SS, Tibshirani R, Khatri P, Bodian DL, Staedtler F, Perry NM, et al. Cell type–specific gene expression differences in complex tissues. Nat Methods (2010) 7:287–9. doi: 10.1038/nmeth.1439
24. Shen-Orr SS, Gaujoux R. Computational deconvolution: Extracting cell type-specific information from heterogeneous samples. Curr Opin Immunol (2013) 25:571–8. doi: 10.1016/j.coi.2013.09.015
25. Do AN, Watson CT, Cohain AT, Griffin RS, Grishin A, Wood RA, et al. Dual transcriptomic and epigenomic study of reaction severity in peanut-allergic children. J Allergy Clin Immunol (2020) 145:1219–30. doi: 10.1016/j.jaci.2019.10.040
26. Peters VA, Joesting JJ, Freund GG. IL-1 receptor 2 (IL-1R2) and its role in immune regulation. Brain Behav Immun (2013) 32:1–8. doi: 10.1016/j.bbi.2012.11.006
27. Kerfoot SM, Raharjo E, Ho M, Kaur J, Serirom S, McCafferty D-M, et al. Exclusive neutrophil recruitment with oncostatin m in a human system. Am J Pathol (2001) 159:1531–9. doi: 10.1016/S0002-9440(10)62538-2
28. Nagata T, Kai H, Shibata R, Koga M, Yoshimura A, Imaizumi T. Oncostatin m, an interleukin-6 family cytokine, upregulates matrix metalloproteinase-9 through the mitogen-activated protein kinase kinase-extracellular signal-regulated kinase pathway in cultured smooth muscle cells. Arterioscler Thromb Vasc Biol (2003) 23:588–93. doi: 10.1161/01.ATV.0000060891.31516.24
29. Setiadi H, Yago T, Liu Z, McEver RP. Endothelial signaling by neutrophil-released oncostatin m enhances p-selectin–dependent inflammation and thrombosis. Blood Adv (2019) 3:168–83. doi: 10.1182/bloodadvances.2018026294
30. Van Keulen D, Pouwer MG, Pasterkamp G, Van Gool AJ, Gelpke MDS, Princen HMG, et al. Inflammatory cytokine oncostatinMinduces endothelial activation in macro- and microvascular endothelial cells and in APOE &z.txt 3Leiden.CETP mice. PloS One (2018) 13:1–20. doi: 10.1371/journal.pone.0204911
31. Xu L, Cai Z, Yang F, Chen M. Activation-induced upregulation of MMP9 in mast cells is a positive feedback mediator for mast cell activation. Mol Med Rep (2017) 15:1759–64. doi: 10.3892/mmr.2017.6215
32. Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, et al. The GeneCards suite: From gene data mining to disease genome sequence analyses. Curr Protoc Bioinforma (2016) 54:1.30.1–1.30.33. doi: 10.1002/cpbi.5
33. Brown SGA, Stone SF, Fatovich DM, Burrows SA, Holdgate A, Celenza A, et al. Anaphylaxis: Clinical patterns, mediator release, and severity. J Allergy Clin Immunol (2013) 132:1141–9. doi: 10.1016/j.jaci.2013.06.015
34. Korošec P, Gibbs BF, Rijavec M, Custovic A, Turner PJ. Important and specific role for basophils in acute allergic reactions. Clin Exp Allergy (2018) 48:502–12. doi: 10.1111/cea.13117
35. Gauvreau GM, Lee JM, Watson RM, Irani A-MA, Sschwartz LB, O’Byrne PM. Increased numbers of both airway basophils and mast cells in sputum after allergen inhalation challenge of atopic asthmatics. Am J Respir Crit Care Med (2000) 161:1473–8. doi: 10.1164/ajrccm.161.5.9908090
36. Guo CB, Liu MC, Galli SJ, Bochner BS, Kagey-Sobotka A, Lichtenstein LM. Identification of IgE-bearing cells in the late-phase response to antigen in the lung as basophils. Am J Respir Cell Mol Biol (1994) 10:384–90. doi: 10.1165/ajrcmb.10.4.7510984
37. Iikura M, Ebisawa M, Yamaguchi M, Tachimoto H, Ohta K, Yamamoto K, et al. Transendothelial migration of human basophils. J Immunol (2004) 173:5189–95. doi: 10.4049/jimmunol.173.8.5189
38. Jönsson F, Mancardi D, Kita Y, Karasuyama H, Iannascoli B, Van Rooijen N, et al. Mouse and human neutrophils induce anaphylaxis. J Clin Invest (2011) 121:1484–96. doi: 10.1172/JCI45232
39. Francis A, Bosio E, Stone SF, Fatovich DM, Arendts G, Nagree Y, et al. Neutrophil activation during acute human anaphylaxis: analysis of MPO and sCD62L. Clin Exp Allergy (2017) 47:361–70. doi: 10.1111/cea.12868
40. Francis A, Bosio E, Stone SF, Fatovich DM, Arendts G, MacDonald SPJ, et al. Markers involved in innate immunity and neutrophil activation are elevated during acute human anaphylaxis: Validation of a microarray study. J Innate Immun (2019) 11:63–73. doi: 10.1159/000492301
41. Davis BP, Rothenberg ME. Inflammatory and effector Cells/Cell migration. In: Pediatric allergy: Principles and practice, 3rd ed. Elsevier Inc (2015). p. 41–53.e4. doi: 10.1016/B978-0-323-29875-9.00005-7
42. Bochner BS. Systemic activation of basophils and eosinophils: Markers and consequences. J Allergy Clin Immunol (2000) 106(5 SUPPL):292–302. doi: 10.1067/mai.2000.110164
43. Ogawa Y, Grant JA. Mediators of anaphylaxis. Immunol Allergy Clin North Am (2007) 27:249–60. doi: 10.1016/j.iac.2007.03.013
44. Undem BJ, Taylor-Clark T. Mechanisms underlying the neuronal-based symptoms of allergy. J Allergy Clin Immunol (2014) 133:1521–34. doi: 10.1016/j.jaci.2013.11.027
45. Potenzieri C, Meeker S, Undem BJ. Activation of mouse bronchopulmonary c-fibres by serotonin and allergen-ovalbumin challenge. J Physiol (2012) 590:5449–59. doi: 10.1113/jphysiol.2012.237115
46. Kreis ME, Müller M, Zittel TT, Glatzle J, Grundy D. Mediators of neuronal activation in the rat brainstem following intestinal anaphylaxis. Neurosci Lett (2000) 289:45–8. doi: 10.1016/S0304-3940(00)01265-9
47. Chen CY, Bonham AC, Joad JP, Gershwin LJ, Schelegle ES, Plopper CG, et al. Extended allergen exposure in asthmatic monkeys induces neuroplasticity in nucleus tractus solitarius. J Allergy Clin Immunol (2001) 108:557–62. doi: 10.1067/mai.2001.118132
48. Omori Y, Andoh T, Shirakawa H, Ishida H, Hachiga T, Kuraishi Y. Itch-related responses of dorsal horn neurons to cutaneous allergic stimulation in mice. Neuroreport (2009) 20:478–81. doi: 10.1097/WNR.0b013e3283293725
49. Jiang W, Kreis ME, Eastwood C, Kirkup AJ, Humphrey PPA, Grundy D. 5-HT(3) and histamine H(1) receptors mediate afferent nerve sensitivity to intestinal anaphylaxis in rats. Gastroenterology (2000) 119:1267–75. doi: 10.1053/gast.2000.19461
50. Zhang G, Lin RL, Wiggers M, Snow DM, Lee LY. Altered expression of TRPV1 and sensitivity to capsaicin in pulmonary myelinated afferents following chronic airway inflammation in the rat. J Physiol (2008) 586:5771–86. doi: 10.1113/jphysiol.2008.161042
51. Weinreich D, Undem BJ. Immunological regulation of synaptic transmission in isolated guinea pig autonomic ganglia. J Clin Invest (1987) 79:1529–32. doi: 10.1172/JCI112984
52. Undent BJ, Myers AC, Weinreich D. Antigen-induced modulation of autonomic and sensory neurons in vitro. Int Arch Allergy Appl Immunol (1991) 94:319–24. doi: 10.1159/000235394
53. Schemann M, Michel K, Ceregrzyn M, Zeller F, Seidl S, Bischoff SC. Human mast cell mediator cocktail excites neurons in human and guinea-pig enteric nervous system. Neurogastroenterol Motil (2005) 17:281–9. doi: 10.1111/j.1365-2982.2004.00591.x
54. Liu S, Hu HZ, Gao N, Gao C, Wang G, Wang X, et al. Neuroimmune interactions in guinea pig stomach and small intestine. Am J Physiol Gastrointest Liver Physiol (2003) 284:G154–64. doi: 10.1152/ajpgi.00241.2002
55. Rogoz K, Aresh B, Freitag FB, Pettersson H, Magnúsdóttir EI, Larsson Ingwall L, et al. Identification of a neuronal receptor controlling anaphylaxis. Cell Rep (2016) 14:370–9. doi: 10.1016/j.celrep.2015.12.033
56. Daynes RA, Jones DC. Emerging roles of PPARs in inflammation and immunity. Nat Rev Immunol (2002) 2:748–59. doi: 10.1038/nri912
57. Hong C, Tontonoz P. Coordination of inflammation and metabolism by PPAR and LXR nuclear receptors. Curr Opin Genet Dev (2008) 18:461–7. doi: 10.1016/j.gde.2008.07.016
58. Le Menn G, Neels J. Regulation of immune cell function by PPARs and the connection with metabolic and neurodegenerative diseases. Int J Mol Sci (2018) 19:1575. doi: 10.3390/ijms19061575
59. Duan SZ, Usher MG, Mortensen RM. Peroxisome proliferator-activated receptor-γ–mediated effects in the vasculature. Circ Res (2008) 102:283–94. doi: 10.1161/CIRCRESAHA.107.164384
60. Kiss M, Czimmerer Z, Nagy L. The role of lipid-activated nuclear receptors in shaping macrophage and dendritic cell function: From physiology to pathology. J Allergy Clin Immunol (2013) 132:264–86. doi: 10.1016/j.jaci.2013.05.044
61. Jimenez-Rodriguez T, Garcia-Neuer M, Alenazy LA, Castells M. Anaphylaxis in the 21st century: phenotypes, endotypes, and biomarkers. J Asthma Allergy (2018) 11:121–42. doi: 10.2147/JAA.S159411
62. Eisenbarth SC, Piggott DA, Huleatt JW, Visintin I, Herrick CA, Bottomly K. Lipopolysaccharide-enhanced, toll-like receptor 4-dependent T helper cell type 2 responses to inhaled antigen. J Exp Med (2002) 196:1645–51. doi: 10.1084/jem.20021340
63. Poza-Guedes P, Barrios Y, Fuentes V, Franco A, Sánchez-Machín I, Alonso E, et al. Downregulation of angiogenesis factors, VEGF and PDGF, after rapid IgE desensitisation and oral immunotherapy in children with food allergy. BioMed Res Int (2014) 2014:372567. doi: 10.1155/2014/372567
64. Dwyer DF, Barrett NA, Austen KF. Expression profiling of constitutive mast cells reveals a unique identity within the immune system. Nat Immunol (2016) 17:878–87. doi: 10.1038/ni.3445
65. Li Y, Liu B, Harmacek L, Long Z, Liang J, Lukin K, et al. The transcription factors GATA2 and microphthalmia-associated transcription factor regulate hdc gene expression in mast cells and are required for IgE/mast cell-mediated anaphylaxis. J Allergy Clin Immunol (2018) 142:1173–84. doi: 10.1016/j.jaci.2017.10.043
66. Morin F, Singh N, Mdzomba JB, Dumas A, Pernet V, Vallières L. Conditional deletions of hdc confirm roles of histamine in anaphylaxis and circadian activity but not in autoimmune encephalomyelitis. J Immunol (2021) 206:2029–37. doi: 10.4049/jimmunol.2000719
67. Wang HN, Ji K, Zhang LN, Xie CC, Li WY, Zhao ZF, et al. Inhibition of c-fos expression attenuates IgE-mediated mast cell activation and allergic inflammation by counteracting an inhibitory AP1/Egr1/IL-4 axis. J Transl Med (2021) 19:261. doi: 10.1186/s12967-021-02932-0
68. Gao X, Wen L, Li H, Wang R, Yin J. Genetic variation at the interleukin-18 locus is associated with wheat-dependent exercise-induced anaphylaxis in the han Chinese population. Gene (2020) 737:144462. doi: 10.1016/j.gene.2020.144462
69. Béziat V, Tavernier SJ, Chen YH, Ma CS, Materna M, Laurence A, et al. Dominant-negative mutations in human IL6ST underlie hyper-IgE syndrome. J Exp Med (2020) 217:e20191804. doi: 10.1084/jem.20191804
70. Capelle CM, Chen A, Zeng N, Baron A, Grzyb K, Arns T, et al. Stress hormone signalling inhibits Th1 polarisation in a CD4 T-cell-intrinsic manner via mTORC1 and the circadian gene PER1. Immunology (2022) 165:428–44. doi: 10.1111/imm.13448
71. Giavina-Bianchi P, Gonçalves DG, Zanandréa A, Borges de Castro R, Garro LS, Kalil J, et al. Anaphylaxis to quinolones in mastocytosis: Hypothesis on the mechanism. J Allergy Clin Immunol Pract (2019) 7:2089–90. doi: 10.1016/j.jaip.2019.01.059
72. Weiler CR. Mastocytosis, quinolones, MRGPRX2, and anaphylaxis. J Allergy Clin Immunol Pract (2019) 7:2091–2. doi: 10.1016/j.jaip.2019.02.015
73. Kelso JM. MRGPRX2 signaling and skin test results. J Allergy Clin Immunol Pract (2020) 8:426. doi: 10.1016/j.jaip.2019.09.038
74. Arifuzzaman M, Mobley YR, Choi HW, Bist P, Salinas CA, Brown ZD, et al. MRGPR-mediated activation of local mast cells clears cutaneous bacterial infection and protects against reinfection. Sci Adv (2019) 5:eaav0216. doi: 10.1126/sciadv.aav0216
75. Roy S, Chompunud Na Ayudhya C, Thapaliya M, Deepak V, Ali H. Multifaceted MRGPRX2: New insight into the role of mast cells in health and disease. J Allergy Clin Immunol (2021) 148:293–308. doi: 10.1016/j.jaci.2021.03.049
Keywords: transcriptome analysis, anaphylaxis, basophils, neutrophils, cell movement, migration, neuroinflammatory signaling, lipid activating nuclear receptors signaling
Citation: Rijavec M, Maver A, Turner PJ, Hočevar K, Košnik M, Yamani A, Hogan S P, Custovic A, Peterlin B and Korošec P (2022) Integrative transcriptomic analysis in human and mouse model of anaphylaxis identifies gene signatures associated with cell movement, migration and neuroinflammatory signalling. Front. Immunol. 13:1016165. doi: 10.3389/fimmu.2022.1016165
Received: 11 August 2022; Accepted: 10 November 2022;
Published: 08 December 2022.
Edited by:
Philipp Starkl, Medical University of Vienna, AustriaReviewed by:
Jelle Folkerts, Erasmus Medical Center, NetherlandsCopyright © 2022 Rijavec, Maver, Turner, Hočevar, Košnik, Yamani, Hogan, Custovic, Peterlin and Korošec. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Matija Rijavec, bWF0aWphLnJpamF2ZWNAa2xpbmlrYS1nb2xuaWsuc2k=
†These authors have contributed equally to this work and share first authorship
‡ORCID: Matija Rijavec, orcid.org/0000-0002-2596-4952
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.