
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 27 September 2022
Sec. Parasite Immunology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.1013159
 Zhaohai Wen1†
Zhaohai Wen1† Yue Zhang1†
Yue Zhang1† Jiajun Feng1
Jiajun Feng1 Kalibixiati Aimulajiang1,2
Kalibixiati Aimulajiang1,2 Muhammad Tahir Aleem1
Muhammad Tahir Aleem1 Mingmin Lu1
Mingmin Lu1 Lixin Xu1
Lixin Xu1 Xiaokai Song1
Xiaokai Song1 Xiangrui Li1
Xiangrui Li1 Ruofeng Yan1*
Ruofeng Yan1*Haemonchus contortus is an important parasitic nematode of ruminants. Previous studies showed that H. contortus escape the immunity through complex mechanisms, including releasing excretory/secretory proteins (ESPs) to modulate the host immune response. However, the detailed mechanism through which H. contortus excretory/secretory proteins (HcESPs) promote immune evasion remains unknown. In the present study, we demonstrated that HcESPs inhibit the adaptive immune response of goats including downregulation of immune cell antigen presentation, upregulation of immune checkpoint molecules, activation of the STAT3/PD-L1 pathway, and activation of immunosuppressive regulatory T (Treg) cells. Furthermore, HcESPs reversed the LPS-induced upregulation of pro-inflammatory mediators in PBMCs by inhibiting the TLR4/NF-κB/MAPKs/NLRP3 signaling pathway. Our study provides a better understanding of the evasion mechanisms for H. contortus, which could be helpful in providing an alternative way to prevent the infection of this parasite.
Parasites are grossly neglected pathogens that infect humans, animals, and crops (1–4). Particularly, parasitic nematodes cause severe morbidity and mortality worldwide (1–4). Haemonchus contortus is one of the most distributed parasites for ruminants, causing enormous losses to the global livestock industry and negatively impacting animal welfare (5). Unlike viruses and bacteria, parasitic nematodes establish a long-term infection in the host, usually with only a mild inflammatory response. This is attributed to the release of complex excretory/secretory protein (ESP) mixtures into host tissues that interfere with host signaling mechanisms and immune homeostasis (6). However, the detailed mechanism of the role of nematode ESP in immune evasion remains to be further investigated.
Previous studies in our laboratory found that the H. contortus ESPs (HcESPs) inhibited the proliferation and nitric oxide (NO) release from PBMCs in vitro (7). However, we know little about the molecular mechanisms, including the related signaling pathways that HcESPs employ to suppress the host innate and adaptive immunity.
Immune checkpoints are inhibitory pathways in the immune system, regulated by the interaction of receptors expressed on immune cells with their ligands (8). LAG-3 (Lymphocyte-activation gene 3), PD-1 (programmed death 1), PD-L1 (programmed death ligand 1), and CTLA-4 (cytotoxic lymphocyte-associated antigen 4) are immune checkpoint molecules that cause immune cell dysfunction and are negative regulators of the immune response (9, 10). Studies have shown that many pathogens (including parasites, bacteria, and viruses) can use the PD-1 pathway to evade host adaptive immunity (11–14). During Trypanosoma cruzi infection, blockade of CTLA-4 resulted in increased host resistance to T. cruzi infection, which was associated with increased production of IFN-γ, TNF-α, and NO (15). Butler NS et al. reported that therapeutic blockade of LAG-3 and PD-L1 rapidly cleared blood-stage Plasmodium vivax in mice (16). However, the role of immune checkpoint molecules in H. contortus infection has not been reported.
Pattern recognition receptor (PRR) recognition of pathogen-associated molecular patterns (PAMP) is critical in initiating innate immune responses in animals. The immune system can detect invading pathogens through toll-like receptors (TLRs) and instruct the subsequent immune response to clear the infection (17). TLRs recognize PAMPs of various pathogens, such as parasites, bacteria, viruses, and fungi. TLRs initiate complex signaling cascades within cells, such as promoting the release of inflammatory mediators, promoting the formation of inflammasomes, and thus activating various innate immune responses. The innate immune response is the host’s first line of defense against pathogen invasion, and TLRs are a vital component of the immune defense; therefore, many pathogens have evolved strategies to manipulate the TLRs signaling pathway to suppress the host response (16, 18–22). However, nothing is known about the effect of HcESPs on goat PRR.
In this study, we analyzed the effects of HcESPs on goats in antigen presentation, expression of immune checkpoint molecules, expression of Tregs cell marker molecules, and STAT3/PD-L1 signaling pathway in PBMCs to investigate the molecular mechanisms by which HcESPs regulate adaptive immune responses. Moreover, we also analyzed the effect of HcESPs on the transcriptional levels of PRR genes in PBMCs. Furthermore, we analyzed the anti-inflammatory effects of different concentrations of HcESPs and its impact on inflammatory pathways such as nuclear factor-kappa B (NF-κB) and Mitogen-activated protein kinases (MAPKs) by establishing an inflammation model of PBMCs using lipopolysaccharides (LPS). Our study identified the molecular mechanisms involved in the modulation of immune cell function caused by HcESPs and the associated signaling pathways, which provides new insights to understand the strategies employed by H. contortus to survive in the host.
The 3- to 6-month-old local goats were housed in the experimental animal center of the college of veterinary medicine, Nanjing Agricultural University, and provided with sterile hay, corn feed, and free access to water. H. contortus (Nanjing strain) were maintained by successive passages of goats in vivo.
Adult H. contortus (females and males) and HcESPs were collected as described in previous studies (7, 23). Briefly, each goat was orally infected with 8000 infective third-stage larvae (iL3s). The excretion of fecal egg was checked since 21 days post infection (DPI), and adult worms were collected from the fourth stomach of goats at 35 DPI. About 300 to 500 worms were collected from each goat. The worms were washed with sterile phosphate-buffered solution (PBS), and cultured in RPMI 1640 medium (100 worms/mL) containing 1% penicillin and 1% streptomycin at 37°C in a 5% CO2 incubator for 24 h. HcESPs in the culture supernatant was collected, desalted, and concentrated using ultrafiltration tubes (3 kDa-cut-off). The concentration of HcESPs was determined by the BCA method and confirmed by SDS-PAGE (Supplementary material Figure S1). Total of 48 mg of HcESPs were collected by incubation of 400 adult worms for 24 h. The possible concentration of HcESPs in infected goat was estimated from 34 to 86 µg/mL. However, differences in host body weight and worm burden might affect the assessment of physiological concentrations of HcESPs.
Goat PBMCs were isolated from peripheral blood using the standard Ficoll-Hypaque (GE Healthcare, Little Chalfont, UK) gradient centrifugation method described previously (24). PBMCs were washed three times with PBS, and the cell density was adjusted to 1 × 106 cells/mL. Then, the cell viability was detected with Trypan blue dye (25).
Cell proliferation was assayed with the Cell Counting Kit-8 (CCK-8, Beyotime) with reference to previous studies (26). Briefly, freshly isolated PBMCs (concentration of 1×106/mL) were homogenously added to 96-well cell culture plates at 100 μL per well. Subsequently, HcESPs at different concentrations (10, 20, 40, 80 μg/mL) were added to co-incubate with PBMCs, and a control group was established (equal volume of RPMI 1640 medium instead of HcESP). The cell culture plates were incubated in a 5% CO2 cell incubator at 37°C for 24 h. Then, 10 μL of CCK-8 solution was added to each well, and incubated for another 2 h, the absorbance values at OD450 were measured by a microplate reader.
PBMCs (concentration of 1×106/mL) were homogenously added to 6-well cell culture plates at 2 mL per well. Different concentrations (5, 10, 20, 40, 80 μg/mL) of HcESPs were added and co-incubated with PBMCs, and a control group was established by adding an equal volume of RPMI 1640 medium. The cells were incubated for 0, 6, 12, 18, and 24 h at 37°C in a 5% CO2 incubator. Subsequently, the cells were collected for RNA extraction.
Total RNA was extracted from PBMCs by the TRIzol method (Vazyme Biotech, Nanjing, Jiangsu, China) according to the manufacturer’s protocol. The cDNA was synthesized by the HiScript III 1st Strand cDNA Synthesis Kit (Vazyme Biotech, Nanjing, Jiangsu, China). The reaction system contains 3.4 μL ddH2O, 0.2 μL forward and reverses primers, 5 μL 2×ChamQ SYBR qPCR Master Mix (Vazyme Biotech co.,ltd), and 1 μL cDNA. The qPCR reaction was performed under the following conditions: one cycle at 95°C/30 s; 40 cycles at 95°C/10 s, 60°C/30 s; and one cycle at 95°C/15 s, 60°C/60 s, and 95°C/15 s. The β-actin was used as an internal reference gene (27). Primer sequences for Arg1, iNOs, CLA-DQB1, CLA-DQB3, PD-L1, PD-1, LAG-3, CTLA-4, IL-10, TGF-β, Foxp3, and PRRs were shown in the Supplementary material Table S1. The data were analyzed using the comparative Ct (2−ΔΔ Ct) method, based on cycle thresholds (Ct), which were obtained by ABI Prism 7500 software (Applied Biosystems, Foster City, CA, USA).
The effect of HcESPs on the antigen uptake of PBMCs was determined by detecting FITC-dextran internalization. FITC-dextran was used as a fluorescent antigen, and FITC fluorescence was observed in PBMCs after phagocytosis of FITC-dextran. Therefore, the phagocytic function of PBMCs can be assessed by the fluorescence intensity. Briefly, different concentrations (10, 20, 40, 80 μg/mL) of HcESPs were co-incubated with PBMCs for 24 h at 37°C in a 5% CO2 incubator, and a control group was established by adding an equal volume of RPMI 1640 medium. The PBMCs were then incubated with 100 ng/mL FITC-dextran for 1 h. The cells were subsequently washed 3 times with PBS and placed under a fluorescent microscope for observation. The phagocytosis was assessed by the intensity of FITC-dextran in PBMCs.
Freshly isolated PBMCs (concentration of 1×106/mL) were homogenously added to 6-well cell culture plates at 2 mL per well. Subsequently, HcESPs at different concentrations (10, 20, 40, 80 μg/mL) were added and co-incubated with PBMCs, and a control group was established by adding an equal volume of RPMI 1640 medium. The cells were incubated for 24 h at 37°C in a 5% CO2 incubator. Subsequently, the cells were collected for total protein extraction.
Western blot assays were performed with reference to the methods of previous studies (28). Briefly, total protein was obtained by lysing PBMCs using RIPA Lysis Buffer (Beyotime, Shanghai, China), and the concentration of protein samples was determined by the BCA kit (Beyotime, Shanghai, China). The proteins (30µg) were transferred onto PVDF membranes (GE Healthcare Life Science, Beijing) by 10% SDS-PAGE. To block the non-specific binding sites, the membranes were incubated with 5% BSA for 2h at room temperature. The membranes were washed 5 times with TBST and incubated with primary antibody at 4°C overnight. Washed with TBST, the membranes were incubated with goat anti-rabbit IgG (H+L) HRP secondary antibody for 2 h at room temperature. The expression of β-actin was used as an internal quantitative control. The expression of the target proteins was detected by chemiluminescence. Image J was used to analyze the relative protein expression levels.
Source antibodies: TGF-β1 (1:500; Santa Cruz Biotechnology, Inc., Dallas, TX, USA); IL-10, PD-L1 p705-STAT3 (1:1000; Affinity Biosciences, Jiangsu province, China); β-actin (ABclonal, China).
To investigate the effect of HcESPs on inflammation and inflammation-related signaling pathways, we used Lipopolysaccharides (LPS) to treat PBMCs to establish an inflammation model. Briefly, freshly isolated PBMCs (concentration of 1×106/mL) were homogenously added to 6-well cell culture plates at 2 mL per well. Then, the PBMCs were stimulated using LPS (1 µg/mL), and series concentrations (0, 10, 20, 40, 80 μg/mL) of HcESPs were co-incubated with PBMCs for 24 h at 37°C in a 5% CO2 incubator. At the same time, a blank control group (RPMI 1640 medium instead of LPS and HcESP) was set up.
Group settings were consistent with the description in section 2.7 (“Inflammation modeling”). The detailed procedure for qPCR assays is consistent with the description in Section 2.3. The primers TNF-α, IL-6, IL-1β, TLR4, myD88, ASC, NLRP3, and IL-18 for qPCR are listed in Supplementary material Table S1.
Group settings were consistent with the description in section 2.7 (“Inflammation modeling”). The detailed procedure for western blot assays is consistent with the description in Section 2.5. Source antibodies: IL-1β, TNF-α, TGF-β1, TLR4 (1:500; Santa Cruz Biotechnology, Inc., Dallas, TX, USA); IL-6, IL-10, p705-STAT3, NLRP3 (1:1000; Affinity Biosciences, Jiangsu province, China); p38, p-p38, ERK, p-ERK (Cell Signaling Technology, USA); myD88, JNK, p-JNK, p65, p-p65 (Proteintech Group, Wuhan, China); β-actin (ABclonal, China).
Statistical analysis was performed using IBM SPSS Statistics 21 software and GraphPad Premier 6.0 software package (GraphPad Prism). The differences between the groups were statistically calculated by one-way analysis of variance (ANOVA). Data are presented as the mean ± standard error of the mean (SEM), and considered significant at *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns: non-significant.
The proliferation of PBMCs incubated with various doses (10, 20, 40, and 80 μg/ml) of HcESPs was considerably restrained when compared with the control group (Supplementary material: Figure S2).
The transcript levels of specific phenotypic markers of M1 (inducible nitric oxide synthase (iNOS)) and M2 (arginase-1(Arg1)) macrophages were assessed by qPCR assays. As shown in Figure 1A, HcESPs significantly down-regulated the transcript levels of iNOs in PBMCs, while significantly up-regulating the transcript levels of Arg1 (Figure 1B). This suggests that HcESPs may have contributed to the polarization of macrophages toward M2 macrophages.
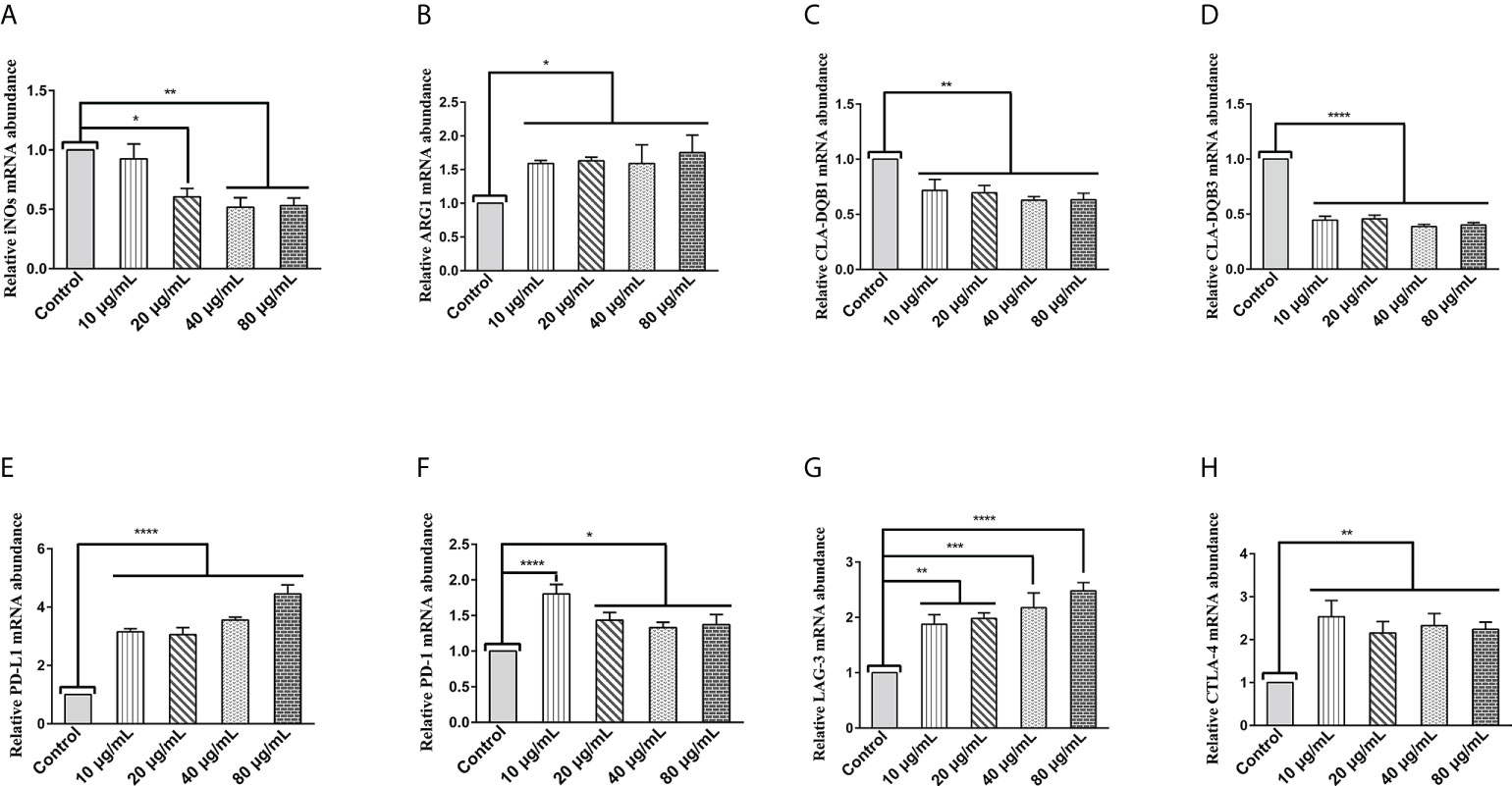
Figure 1 HcESPs upregulated the transcript levels of Arg1 and immune checkpoint molecules in PBMCs, while downregulated the transcript levels of iNOs and MHC II in PBMCs. Cells were incubated with different concentrations (10, 20, 40, 80 μg/mL) of HcESPs or an equal volume of RPMI 1640 medium (control) at 37°C and 5% CO2 for 24 h. (A, B) The transcript levels of iNOs and Arg1 were detected by qPCR assay; (C, D) The transcript levels of MHC II (CLA-DQB1 and CLA-DQB3) were detected by qPCR assay; (E-H) The transcript levels of immune checkpoints molecules (PD-L1, PD-1, LAG-3, CTLA-4) were detected by qPCR assay; Data are presented as the mean ± SEM from three independent experiments. * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001 vs the control group.
Extracellular peptides (e.g., bacteria and helminths) are presented to CD4+- T cells via major histocompatibility complex (MHC) class II molecules (29). MHC class II molecules play an essential role in fighting against parasitic infections due to their known antigen presentation function (30, 31). Therefore, we evaluated the effect of HcESPs on the transcript levels of MHC class II molecules (CLA-DQBI, CLA-DQB3) in PBMCs by qPCR assays. As shown in Figures 1C, D, HcESPs significantly downregulated the mRNA expression levels of antigen-presenting molecules (CLA-DQBI, CLA-DQB3) in PBMCs, suggesting that HcESPs affected the antigen-presenting function in PBMCs. In addition, HcESPs significantly inhibited the ability of PBMCs to take up FITC-dextran (Supplementary material: Figure S3), suggesting that HcESPs inhibited the phagocytosis in PBMCs.
LAG-3, PD-1, CTLA-4, and PD-L1 are suppressive immune checkpoints impair lymphocyte function (9, 32, 33). Similarly, the expression of these immune checkpoint molecules plays a vital role in the parasite’s evasion of the host immune system (34–38). Therefore, in this study, the effect of HcESPs on the transcript levels of suppressive immune checkpoint molecules in PBMCs was assessed by qPCR assays. Interestingly, different concentrations of HcESPs significantly upregulated the mRNA levels of PD-L1, PD-1, LAG-3, and CTLA-4 in PBMCs (Figures 1E–H), suggesting that HcESPs impaired T-cell function and created an “immunosuppressive microenvironment”. Interestingly, the transcriptional levels of PD-1 in the HcESPs groups of 20, 40, and 80 μg/mL showed a down-regulation trend compared with the HcESPs group of 10 μg/mL. This suggested that the transcriptional regulation of PD-1 in PBMCs was diminished when the concentration of HcESPs reached 20 μg/mL. This may be related to negative feedback regulation of PD-1 transcription in PBMCs. The precise regulatory mechanism needs to be further elucidated by subsequent studies.
Regulatory T (Treg) cells are specialized immunosuppressive CD4+ T lymphocytes that play a critical role in parasite immune escape (39, 40). The transcription factor Forkhead box protein 3 (Foxp3) is a marker molecule of Treg cells (41, 42). The qPCR results showed that different concentrations of HcESPs treatment significantly upregulated the transcript levels of IL-10, TGF-β, and Foxp3 in PBMCs (Figures 2A–C). Moreover, western blot results showed that HcESPs significantly promoted the protein expression levels of IL-10 and TGF-β in PBMCs (Figures 2D–F). These results suggest that HcESPs promote the proliferation and differentiation of Treg cells in PBMCs.
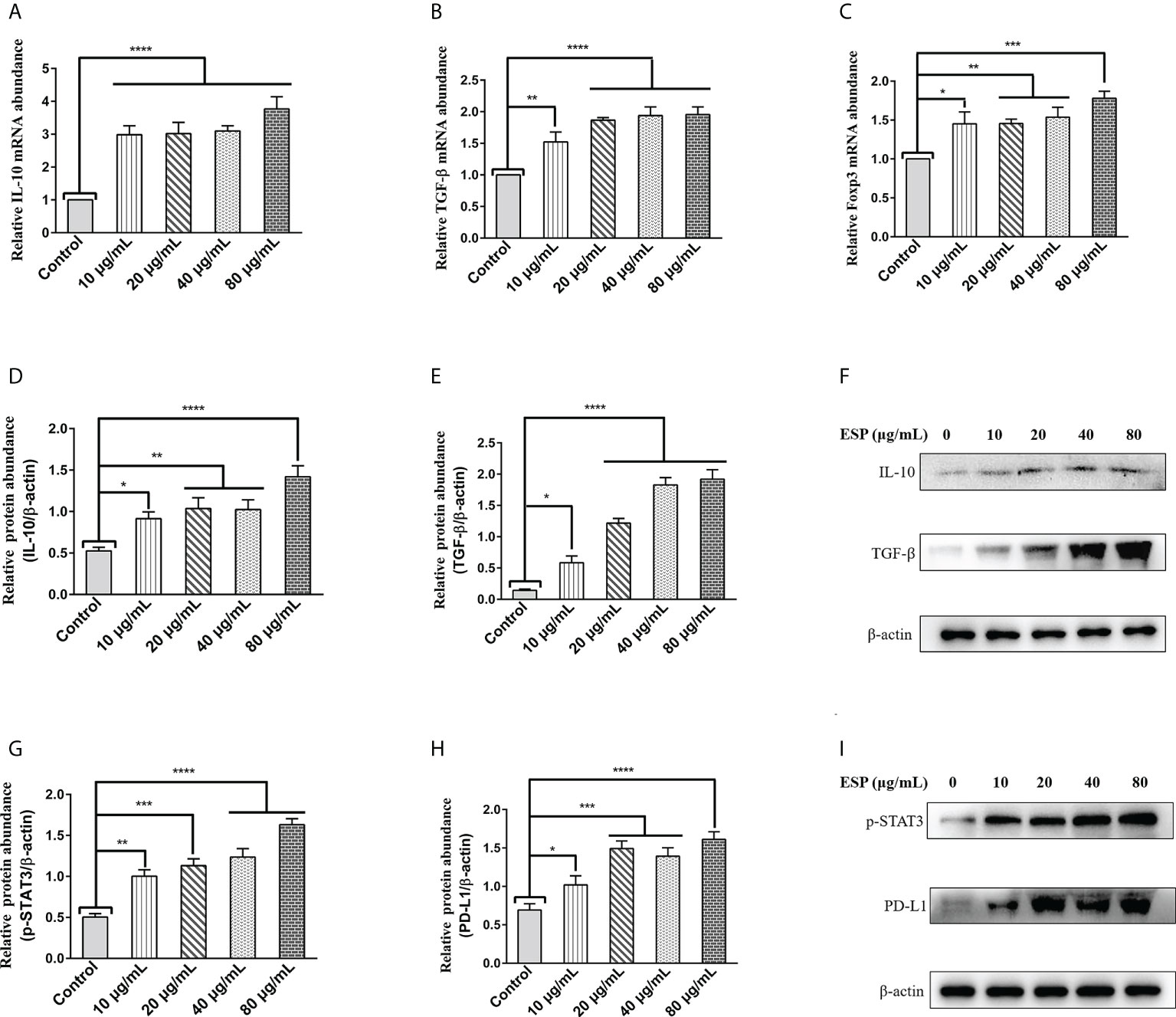
Figure 2 HcESPs promoted the expression of Treg cell markers and activated the STAT3/PD-L1 pathway. Cells were incubated with different concentrations (10, 20, 40, 80 μg/mL) of HcESPs, and an equal volume of RPMI 1640 medium (control) at 37°C and 5% CO2 for 24 h. (A–C): The transcript levels of IL-10, TGF-β, and Foxp3 were detected by qPCR assays; (D–F): Detection of relative protein expression levels of IL-10 and TGF-β by Western blot; (G–I): Detection of relative protein expression levels of p-STAT3, PD-L1 by Western blot; Data are presented as the mean ± SEM from three independent experiments. * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001 vs the control group.
A growing body of evidence suggests that STAT3 activation is a critical molecular hub for immune evasion by bacteria, tumors, etc. (43–45). However, is it possible that worm ESPs activates STAT3/PD-L1 pathway like tumor cells, bacteria, and other pathogens, which has not been reported yet? Therefore, we analyzed the effect of HcESPs treatment on the STAT3/PD-L1 pathway in PBMCs by western blot assays. Interestingly, western blot results (Figures 2G-I) showed that different concentrations of HcESPs treatment significantly upregulated the protein expression levels of p-STAT3, and PD-L1, suggesting that HcESPs activated the STAT3/PD-L1 pathway in PBMCs.
The above results suggest that HcESPs inhibit the adaptive immune response and create a “microenvironment” of immunosuppression. We next analyzed the effect of HcESP on PRR in PBMCs. We analyzed the changes in mRNA levels of TLR1, TLR2, TLR3, TLR4, TLR5, TLR6, TLR7, TLR8, TLR9, TLR10, C-type lectin MCL (CLEC4D), macrophage-inducible C-type lectin (CLEC4E), a significant decrease in C-terminal caspase recruitment domain [CARD] domain-containing protein 4 (NLRC4), and MDA5 by qPCR assays when different concentrations of HcESPs were co-incubated with PBMCs for different times. The results are shown in Figure 3. HcESPs significantly down-regulated the transcript levels of TLR2, TLR4, and TLR5 in PBMCs and showed a dose-dependent relationship (Figures 3B–D) while up-regulating the transcript level of TLR1 in PBMCs and showed a dose-dependent relationship (Figure 3A). However, the effect of HcESPs on the transcript levels of TLR3, TLR6, TLR7, TLR8, TLR9, TLR10, CLEC4D, CLEC4E, MDA5, and NLRC4 in PBMC showed turbulent changes over time such as up-regulation or down-regulation (Supplementary material: Figure S4).
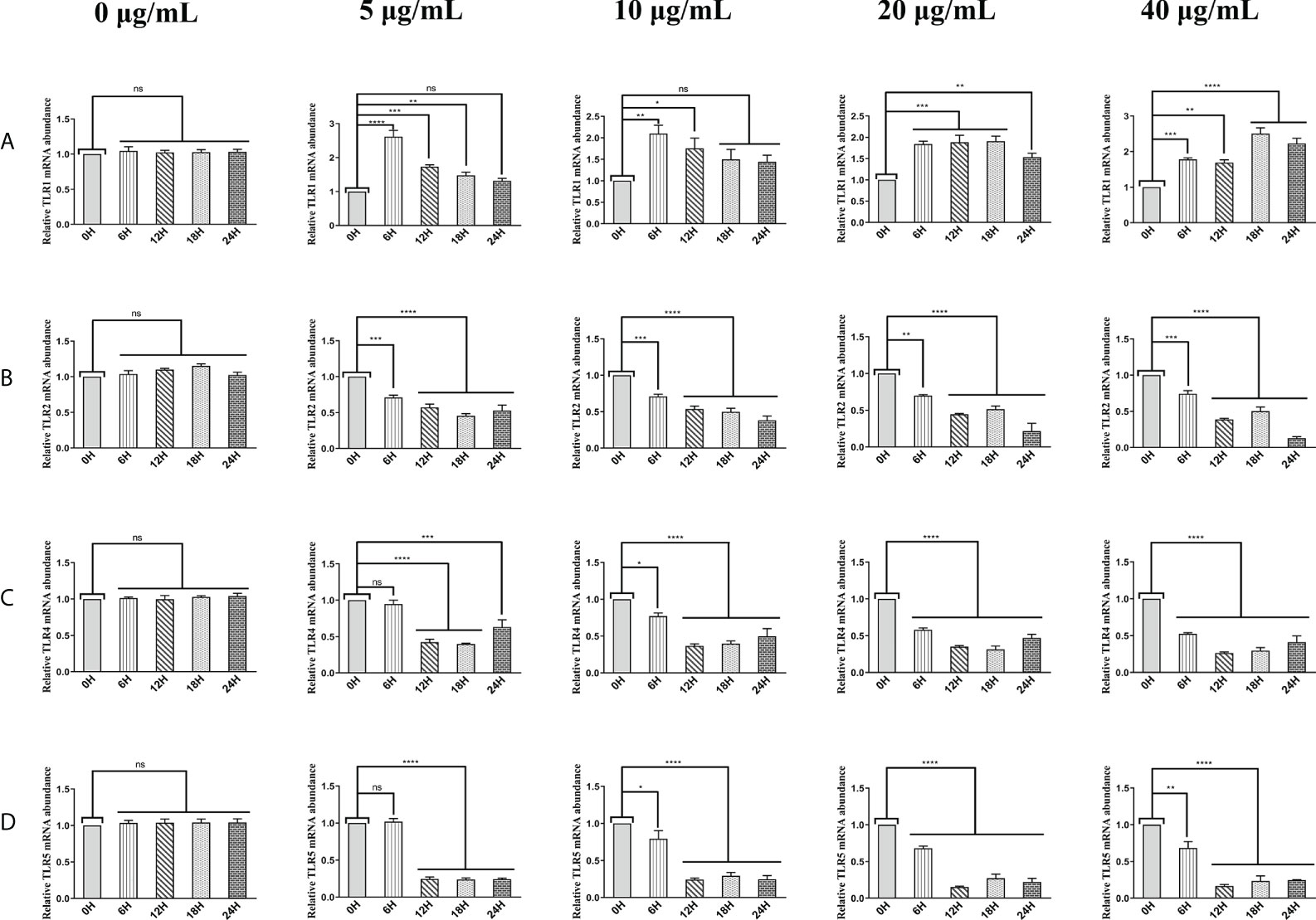
Figure 3 HcESPs down-regulated the transcript level of TLR2, TLR4, and TLR5 while up-regulated the transcript level of TLR1 in PBMCs. Cells were incubated with different concentrations (0, 5, 10, 20, 40 μg/mL) of HcESPs at 37°C and 5% CO2 for 0, 6, 12, 18, and 24 hours, respectively. (A–D): The transcript levels of TLR1, TLR2, TLR4, and TLR5 were detected by qPCR assays; Data are presented as the mean ± SEM from three independent experiments. Asterisks indicate significant differences at *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; ns: non-significant.
HcESPs inhibit the expression of TLR2, TLR4, and TLR5, which may be one of the mechanisms by which HcESPs suppress the host inflammatory response. It is well known that TLR4/NF-κB is a classical pro-inflammatory pathway, and activation of the TLR4/NF-κB signaling pathway promotes the release of inflammatory mediators such as TNF-α, IL-6, and IL-1β. Therefore, we next analyzed the effect of HcESPs on TLR4-related inflammatory pathways.
As shown in Figures 4A–C, LPS upregulated the transcript levels of TNF-α, IL-6, and IL-1β in PBMCs. However, HcESPs treatment significantly reversed the LPS-induced upregulation of the transcript levels of pro-inflammatory mediators (TNF-α, IL-6, IL-1β) (Figures 4A–C). Consistent with the qPCR results, western blot results (Figures 4D–G) showed that HcESPs significantly reversed the upregulation of pro-inflammatory mediator (TNF-α, IL-6, IL-1β) protein expression levels caused by LPS. These results suggest that HcESPs can attenuate the inflammatory response of PBMCs due to LPS.
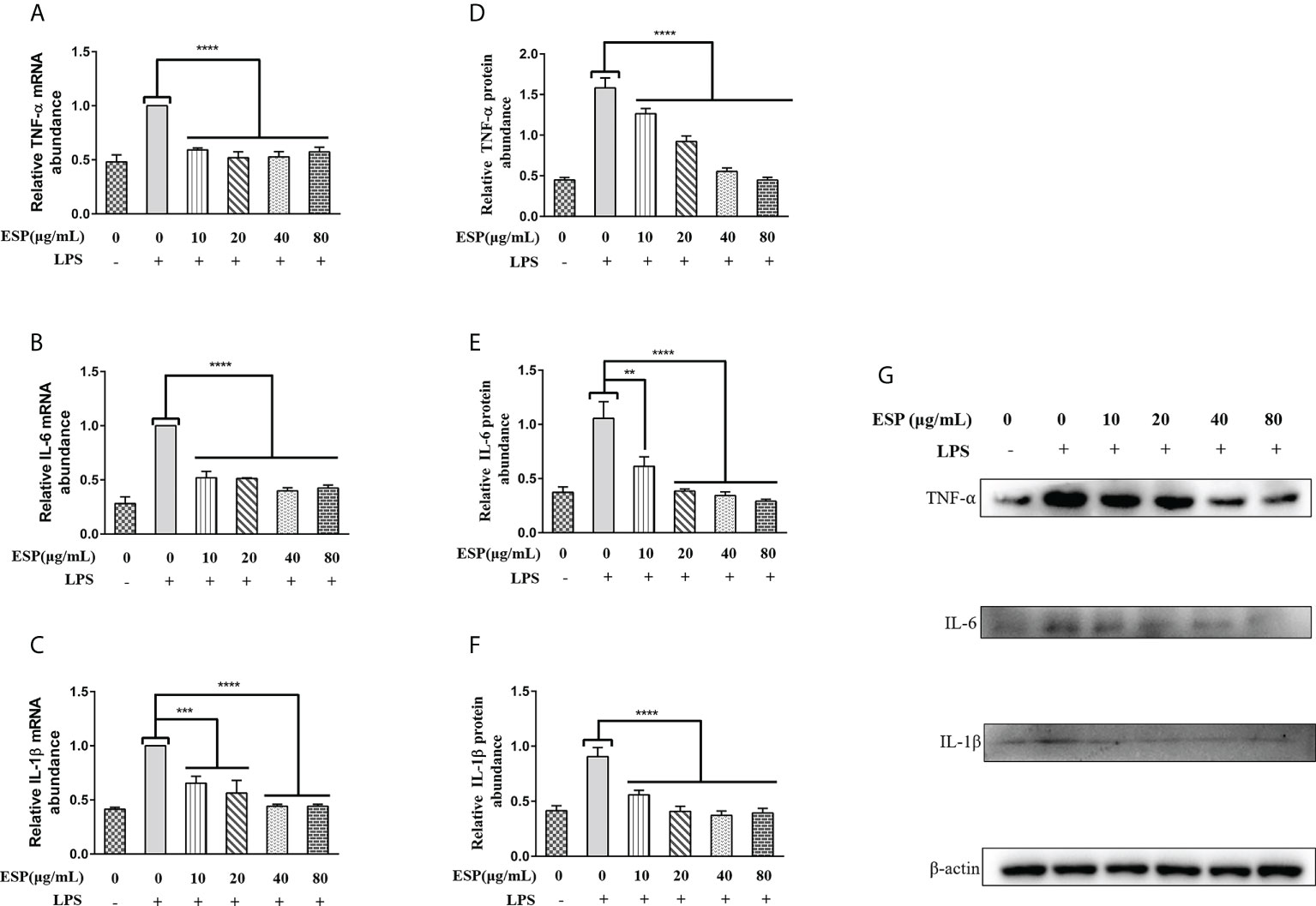
Figure 4 HcESPs reversed the LPS-induced upregulation of inflammatory mediators in PBMCs. Cells were stimulated with LPS (1 μg/mL) to establish an inflammation model, followed by incubation with different concentrations (0, 10, 20, 40, 80 μg/mL) of HcESPs at 37°C and 5% CO2 for 24 h. A blank control group was set up (equal volumes of RPMI 1640 were used instead of LPS and HcESPs). (A–C): Detection of relative transcript levels of TNF-α, IL-6, and IL-1β in PBMCs by qPCR assays; (D–G): Detection of relative protein expression levels of TNF-α, IL-6, and IL-1β by Western blot; Data are presented as the mean ± SEM from three independent experiments. Asterisks indicate significant differences at **P < 0.01, ***P < 0.001, and ****P < 0.0001.
The mRNA level of TNF-αremains constant irrespective of the increase in the concentration of HcESP, but the relative protein abundance gradually decreases. This may indicate that the regulation of TNF-α transcription in PBMCs by HcESPs saturates when HcESPs reaches 10 µg/mL, and the transcriptional regulation of TNF-α is not enhanced with its increasing concentration. However, the regulation of post-transcriptional translation of TNF-α by HcESPs in PBMCs showed a dose-dependent relationship with increasing concentration. The precise regulatory mechanism needs to be further studied.
We examined the effects of HcESPs on the transcriptional expression of key proteins of the TLR4/myD88/NF-κB signaling pathway by qPCR and western blot assays. The qPCR results (Figures 5A, B) showed that HcESPs significantly reversed the LPS-induced upregulation of mRNA levels of TLR4 and myD88. Consistent with the expected results, HcESPs treatment significantly down-regulated the protein expression levels of TLR4, myD88, and p-p65 in PBMCs compared with the LPS-treated group (Figures 5C–F). This suggests that HcESPs inhibit the TLR4/myD88/NF-κB signaling pathway in PBMCs.
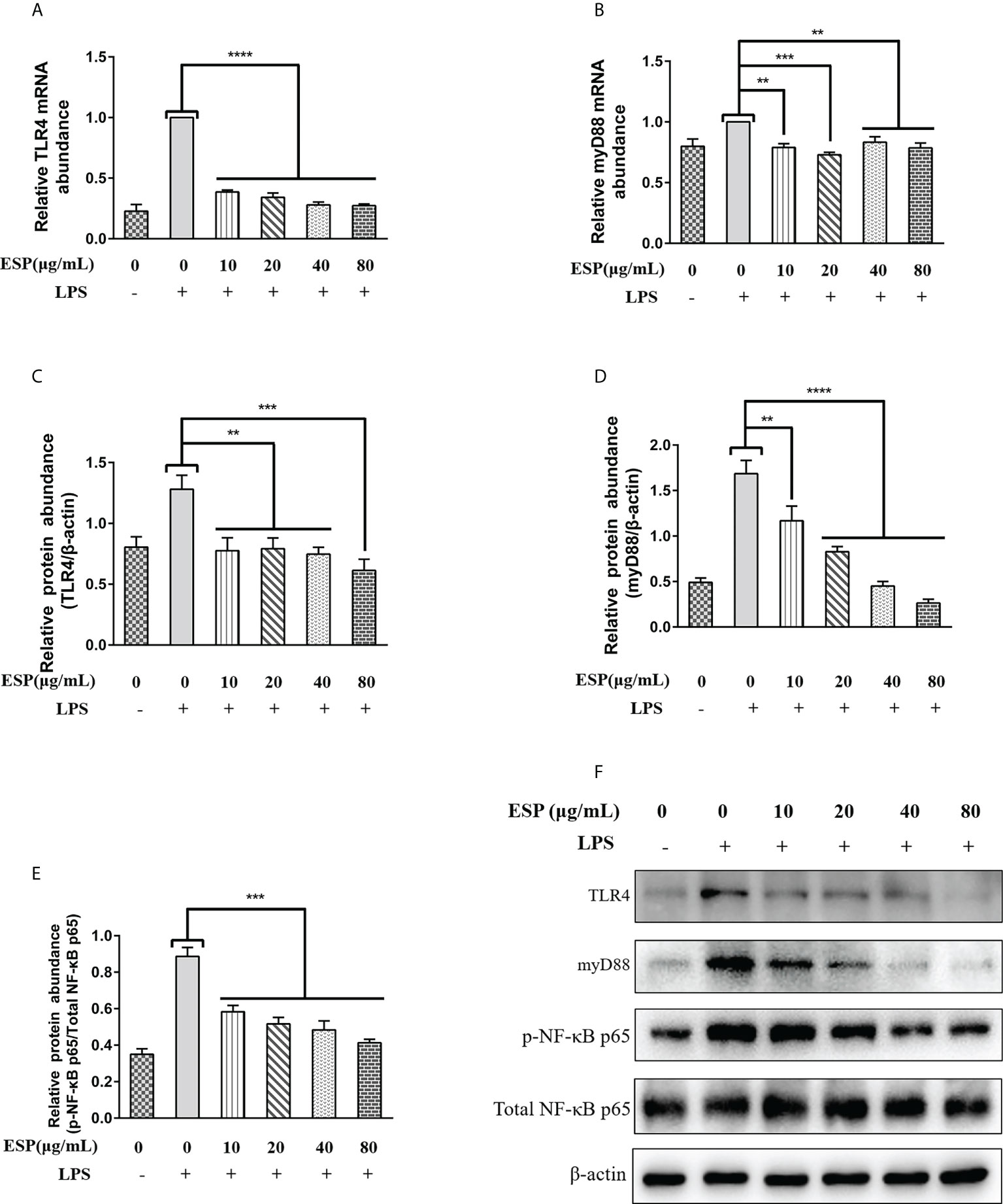
Figure 5 HcESPs reversed LPS-induced activation of the TLR4/NF-κB signaling pathway. Cells were stimulated with LPS (1 μg/mL) to establish an inflammation model, followed by incubation with different concentrations (0, 10, 20, 40, 80 μg/mL) of HcESPs at 37°C and 5% CO2 for 24 h. A blank control group was set up (equal volumes of RPMI 1640 were used instead of LPS and HcESPs). (A, B): Detection of relative transcript levels of TLR4 and myD88 in PBMCs by qPCR assays; (C–F): Detection of relative protein expression levels of TLR4, myD88, p65, and p-p65 by Western blot; Data are presented as the mean ± SEM from three independent experiments. Asterisks indicate significant differences at **P < 0.01, ***P < 0.001, and ****P < 0.0001.
The down-regulation of myD88 transcript levels showed no dose-dependent relationship with the concentration of HcESPs, while the downregulation of myD88 protein levels showed a dose-dependent relationship with the concentration of HcESPs. This suggests that the transcriptional regulation of myD88 in PBMCs may be saturated when the HcESPs reaches 10 μg/mL.
It is well known that activation of the MAPKs signaling pathway promotes inflammatory responses, which in turn promotes the release of pro-inflammatory mediators and plays a vital role in host immune defense. Therefore, we analyzed the effect of HcESPs treatment on the expression levels of key proteins of MAPKs signaling pathway in PBMCs using western blot. Interestingly, HcESPs treatment significantly reversed the LPS-induced upregulation of p-p38, p-JNK, and p-ERK protein phosphorylation levels in PBMCs (Figures 6A–D), indicating that HcESPs ameliorated LPS-induced activation of the MAPKs signaling pathway.
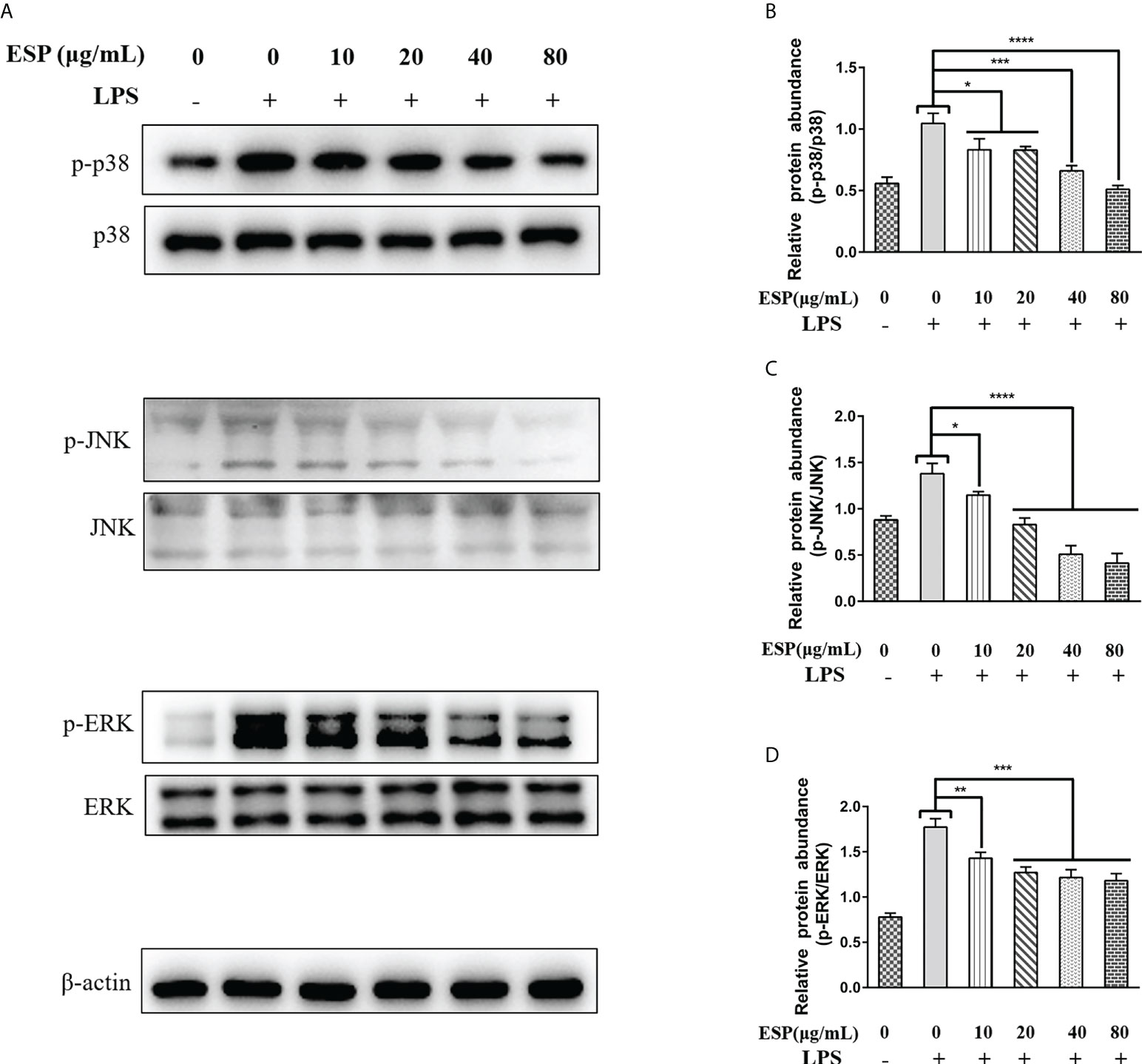
Figure 6 HcESPs inhibited the activation of the MAPKs signaling pathway. Cells were stimulated with LPS (1 μg/Ml) to establish an inflammation model, followed by incubation with different concentrations (10, 20, 40, 80 μg/mL) of HcESPs or equal volumes of RPMI 1640 medium at 37°C and 5% CO2 for 24 h. A blank control group was set up (RPMI 1640 instead of LPS and HcESPs). (A): Detection of protein expression levels of p38, p-p38, JNK, p-JNK, ERK, and p-ERK by Western blot; (B–D): Statistics of the p-p38, p-JNK, and p-ERK; Data are presented as the mean ± SEM from three independent experiments. Asterisks indicate significant differences at *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Activation of the Nod-like receptor pyrin domain containing protein-3 (NLRP3) signaling pathway contributes to the release of pro-inflammatory factors that help the host resist pathogen invasion and play a key role in innate immunity. However, little is known about whether HcESPs regulate the NLRP3 signaling pathway in PBMCs. Therefore, we next analyzed the mRNA levels of ASC, NLRP3, and IL-18 in PBMCs by qPCR assays, and the protein levels of NLRP3 were detected by western blot assays. The results (Figures 7A–C) showed that HcESPs significantly downregulated the transcript levels of ASC, NLRP3, and IL-18 in PBMCs compared with the LPS-treated group. Consistent with the qPCR results, HcESPs significantly down-regulated the protein expression level of NLRP3 in PBMCs compared with the LPS-treated group (Figure 7D). This suggests that HcESPs inhibit the activation of the NLRP3 signaling pathway in PBMCs.
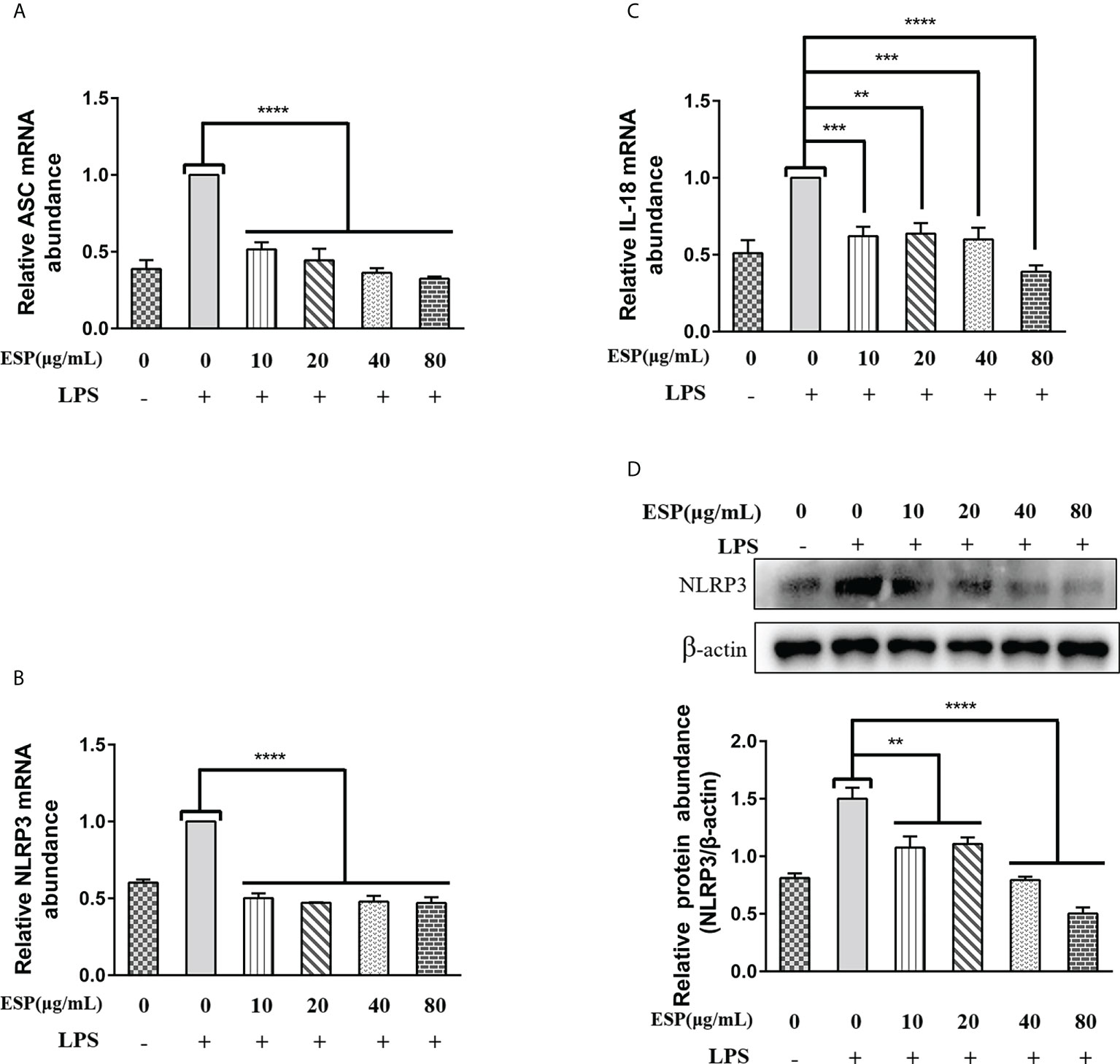
Figure 7 HcESPs inhibited the NLRP3 signaling pathway in PBMCs. Cells were stimulated with LPS (1 μg/mL) to establish an inflammation model, and incubated with different concentrations (10, 20, 40, 80 μg/mL) of HcESPs or equal volume of RPMI 1640 medium at 37°C and 5% CO2 for 24 h. A blank control group was set up (equal volume of RPMI 1640 instead of LPS and HcESPs). (A–C): Detection of relative transcript levels of ASC, NLRP3, and IL-18 in PBMCs by qPCR assays; (D): Detection of relative protein expression of NLRP3 by Western blot; Data are presented as the mean ± SEM from three independent experiments. Asterisks indicate significant differences at **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Many studies have reported that the ESPs of worms play a crucial role in aiding worm infection and parasitism (46–48). Previous studies in our laboratory have shown that a single component protein from HcESPs has a significant protective effect as a vaccine candidate (49–52). This may be because the antibodies blocked the immunosuppressive molecules in HcESPs, which reactivated the host immune function. Interestingly, passive immunization with IgG against a single component protein from HcESPs also achieved a significant protective effect against H. contortus infection (anti-H. contortus α/β-hydrolase domain protein IgG: 54% reduction in fecal egg counts and 74.2% reduction in worm burden; anti-H. contortus adhesion-regulating molecule 1 IgG: 48.9% reduction in fecal egg counts and 58.6% reduction in worm burden; anti-H. contortus transthyretin domain–containing protein IgG: 63.7% reduction in fecal egg counts and 66.4% reduction in worm burden) (49, 50, 52).
In adaptive immunity, antigen-presenting cells (APC) present extracellular pathogen antigen molecules to CD4-T cells via MHC-II molecules (53). Thus, the peptide encoded by the MHC gene constitutes the first line of defense of the host against pathogens. In the present study, we found that HcESPs significantly downregulated the mRNA levels of MHC-II (CLA-DQB1, CLA-DQB3) in PBMCs. The release of ESPs during nematode infection resulted in a decrease in the antigen presentation efficiency of APC, which may be one of the factors limiting vaccine efficacy. A growing number of studies have shown that APC-expressed MHC molecules are one of the key factors influencing immunotherapy (54–56).
Macrophages are essential in the immune response and can be divided into two types: classically activated macrophages with pro-inflammatory potency (M1, mainly expressing iNOs as a marker) and alternatively activated macrophages with anti-inflammatory function (M2, mainly expressing Arg1 as a marker) (57, 58). It has been shown that the anti-inflammatory microenvironment created by M2 macrophages contributes to tumor and parasite immune evasion (59–62). In the present study, we found that HcESPs significantly promoted the transcription of Arg1 in PBMCs and inhibited the transcription of iNOs in PBMCs, which may be the result of HcESPs promoting the polarization of macrophages to M2 macrophages in PBMCs.
Immune checkpoint blockade therapy is effective in oncology, and chronic viral infections. Development of inhibitors targeting PD-L1, PD-1, CTLA-4, and LAG-3 showed great potentialities in oncology immunotherapy (9, 63–65). In our study, results showed that HcESPs significantly upregulated the transcriptional levels of PD-L1, PD-1, CTLA-4, and LAG-3 in PBMCs and activated the STAT3/PD-L1 signaling pathway, which may be one of the mechanisms by which HcESPs promote immune evasion in H. contortus. Activation of immune checkpoint molecules suppressed T cell function, which may explain the downregulation of PBMCs activity due to HcESPs (7). Therefore, in subsequent studies, it is necessary to investigate the efficacy of targeted blocked immune checkpoint molecular therapies for H. contortus infection. The helminth parasite fights immune rejection through complex evasion mechanisms, including activation of host immunosuppressive Treg cells (66). Studies showed that Treg cells suppressed inflammatory reactions by secreting IL-10 and TGF-β, and contributed to parasite immune evasion (47, 67). In our study, we found that H. contortus ESPs activated goat Treg cells and promoted the secretion of IL-10 and TGF-β, which was similar with previous studies (47, 67).
The PRR-related signaling pathway is a critical link between innate and adaptive immunity and is crucial for worm invasion, survival, pathogenesis, and elimination (68, 69). TLRs can be expressed on the surface of various immune cells, such as lymphocytes, macrophages, dendritic cells, and eosinophils (70, 71). During helminth infection, various glycolipids and glycoproteins from helminths can bind to TLRs on the surface of immune cells, activating or modulating TLRs-mediated immune responses (72, 73). ESPs play an essential role in worm parasitism by interfering with TLRs-mediated immune responses. It was demonstrated that ESPs from Trichinella spiralis down-regulated TLR2\TLR4 mRNA expression in macrophages, suppressing the pro-inflammatory response induced by LPS (74). Recent studies have shown that ESPs of Echinococcus granulosus downregulated TLR2/TLR4 expression in goat PBMCs (75). Interestingly, we found that different concentrations of HcESPs co-incubated with PBMCs at different times all significantly down-regulated the mRNA levels of TLR2, TLR4, and TLR5, while up-regulating the mRNA of TLR1. Similar to the previous findings (74, 75), HcESP down-regulated TLR2 and TLR4 mRNA expression in PBMCs, which may be one of the mechanisms by which HcESPs suppress host inflammatory responses.
During helminth infection, TLR4 activation induced eosinophil infiltration and increased secretion of mucosal secretory immunoglobulin A (SIgA) antibodies in the infected area, facilitating worm expulsion (76). NF-κB is one of the core downstream signals of TLRs/MyD88, and its activation promotes the release of inflammatory mediators (TNF-α, IL-6, IL-1β) (77). Interestingly, our study found that HcESPs reversed the LPS-induced upregulation of the expression levels of inflammatory mediators. This suggests that HcESPs might have anti-inflammatory effects. We hypothesized that the downregulation of pro-inflammatory mediators in PBMCs by HcESPs might be related to the suppression of the TLR4/myD88/NF-κB signaling pathway. Consistent with the predicted results, HcESPs inhibited the expression of TLR4, myD88, and p-NF-κB P65, suggesting that HcESPs downregulate LPS-induced inflammatory responses by suppressing the TLR4/myD88/NF-κB signaling pathway.
NF-κB/MAPKs/NLRP3 is a classical signaling pathway of inflammation that regulates the release of pro-inflammatory mediators and plays a vital role in fighting against pathogen invasion. Key molecules in the NF-κB, MAPKs, and NLRP3 signaling pathways are essential targets for anti-inflammatory drug research (78–80). The activation of the p38 MAPK signaling pathway was found to promote the phosphorylation and degradation of IκBα, which activates the NF-κB signaling pathway (81). In the present study, we found that HcESPs inhibited LPS-induced activation of MAPKs (JNK, ERK, p38) signaling pathway and had the best inhibitory effect on the JNK pathway. In addition, HcESPs also inhibited the LPS-induced activation of the NLRP3 signaling pathway. These results are consistent with the downregulation of inflammatory mediator expression levels. Inhibition of TLR4/MAPKs/NF-κB/NLRP3 signaling pathway by HcESPs may be a strategy for H. contortus evasion of the host innate immunity.
In this study, we analyzed the mechanisms involved in suppressing the host immune system by HcESPs. However, due to limitations in the availability of goat antibodies, we only analyzed the transcriptional level of iNOs, Arg1, immune checkpoint molecules (PD-1, LAG-3, CTLA-4), MHC-II, Foxp3, pattern recognition receptors, ASC and IL-18. The changes in protein levels deserve further study.
In the present study, we demonstrate several ways that HcESPs help the parasite to escape the host immunity (Figure 8), including (1) inhibition of antigen presentation, (2) upregulation of immune checkpoint molecules, (3) promotion of Tregs cell proliferation, (4) activation of STAT3/PD-L1 pathway, (5) down-regulated the expressions of TLR2, TLR4, and TLR5 and (6) downregulated the expression of pro-inflammatory cytokines by inhibiting the TLR4/NF-κB/MAPKs/NLRP3 signaling pathway.
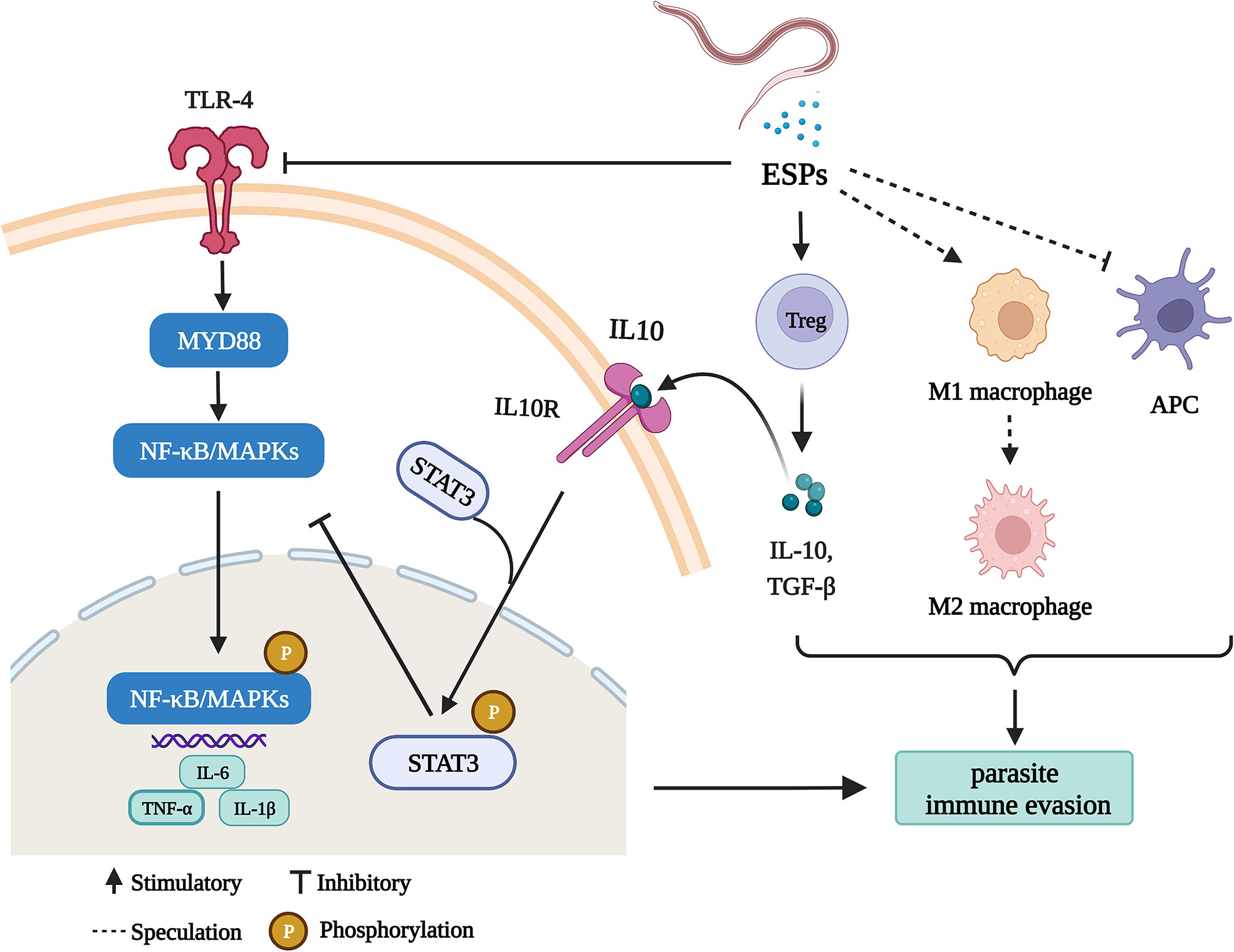
Figure 8 HcESPs suppress goat immune function: a possible mechanism of immune evasion in Haemonchus contortus (Charting with BioRender.com software). HcESPs suppress adaptive immunity by upregualtion of Treg cells, inhibiting the antigen presenting cells, and promoting the shift of macrophages from pro-inflammatory type (M1) to anti-inflammatory type (M2). In addition, HcESP suppresses innate immunity by inhibiting TLR4//NF-κB pathway and activating IL-10/STAT3 pathway, which result in downregulated inflammatory responses.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
The animal study was reviewed and approved by the Science and Technology Agency of Jiangsu Province. The approval ID is SYXK (SU) 2010-0005.
Data curation, ZW and YZ. Formal analysis, ZW, JF, YZ, and KA. Funding acquisition, RY. Investigation, JF. Methodology, ZW and KA. Project administration, RY. Resources, RY. Software, ZW and KA. Supervision, ML, XS, LX, XL, and RY. Visualization, ZW and MA. Writing – original draft, ZW. Writing – review and editing, ZW and RY. All authors contributed to the article and approved the submitted version.
This research was funded by the National Natural Science Foundation of China (31872464) and State Key Laboratory of Pathogenesis, Prevention and Treatment of High Incidence Diseases in Central Asia Fund (SKL-HIDCA-2022-20).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.1013159/full#supplementary-material
1. Hotez PJ, Alvarado M, Basáñez MG, Bolliger I, Bourne R, Boussinesq M, et al. The global burden of disease study 2010: interpretation and implications for the neglected tropical diseases. PloS Negl Trop Dis (2014) 8:e2865. doi: 10.1371/journal.pntd.0002865
2. Pullan RL, Smith JL, Jasrasaria R, Brooker SJ. Global numbers of infection and disease burden of soil transmitted helminth infections in 2010. Parasites Vectors (2014) 7:37. doi: 10.1186/1756-3305-7-37
3. Chitwood DJ. Research on plant-parasitic nematode biology conducted by the united states department of agriculture-agricultural research service. Pest Manage Sci (2003) 59:748–53. doi: 10.1002/ps.684
4. Rashid M, Rashid MI, Akbar H, Ahmad L, Hassan MA, Ashraf K, et al. A systematic review on modelling approaches for economic losses studies caused by parasites and their associated diseases in cattle. Parasitology (2019) 146:129–41. doi: 10.1017/S0031182018001282
5. Sallé G, Doyle SR, Cortet J, Cabaret J, Berriman M, Holroyd N, et al. The global diversity of haemonchus contortus is shaped by human intervention and climate. Nat Commun (2019) 10:4811. doi: 10.1038/s41467-019-12695-4
6. Maizels RM, Smits HH, McSorley HJ. Modulation of host immunity by helminths: The expanding repertoire of parasite effector molecules. Immunity (2018) 49:801–18. doi: 10.1016/j.immuni.2018.10.016
7. Gadahi JA, Yongqian B, Ehsan M, Zhang ZC, Wang S, Yan RF, et al. Haemonchus contortus excretory and secretory proteins (HcESPs) suppress functions of goat PBMCs in vitro. Oncotarget (2016) 7:35670–9. doi: 10.18632/oncotarget.9589
8. Ruffo E, Wu RC, Bruno TC, Workman CJ, Vignali DAA. Lymphocyte-activation gene 3 (LAG3): The next immune checkpoint receptor. Semin Immunol (2019) 42:101305. doi: 10.1016/j.smim.2019.101305
9. Tawbi HA, Schadendorf D, Lipson EJ, Ascierto PA, Matamala L, Castillo Gutiérrez E, et al. Relatlimab and nivolumab versus nivolumab in untreated advanced melanoma. New Engl J Med (2022) 386:24–34. doi: 10.1056/NEJMoa2109970
10. Zhao SJ, Muyayalo KP, Luo J, Huang D, Mor G, Liao AH. Next generation of immune checkpoint molecules in maternal-fetal immunity. Immunol Rev (2022) 308:40–54. doi: 10.1111/imr.13073
11. Furtado R, Chorro L, Zimmerman N, Guillen E, Spaulding E, Chin SS, et al. Blockade of LAG-3 in PD-L1-Deficient mice enhances clearance of blood stage malaria independent of humoral responses. Front Immunol (2020) 11:576743. doi: 10.3389/fimmu.2020.576743
12. Day CL, Kaufmann DE, Kiepiela P, Brown JA, Moodley ES, Reddy S, et al. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature (2006) 443:350–4. doi: 10.1038/nature05115
13. Horne-Debets JM, Faleiro R, Karunarathne DS, Liu XQ, Lineburg KE, Poh CM, et al. PD-1 dependent exhaustion of CD8+ T cells drives chronic malaria. Cell Rep (2013) 5:1204–13. doi: 10.1016/j.celrep.2013.11.002
14. Huang X, Venet F, Wang YL, Lepape A, Yuan Z, Chen Y, et al. PD-1 expression by macrophages plays a pathologic role in altering microbial clearance and the innate inflammatory response to sepsis. Proc Natl Acad Sci United States America (2009) 106:6303–8. doi: 10.1073/pnas.0809422106
15. Martins GA, Tadokoro CE, Silva RB, Silva JS, Rizzo LV. CTLA-4 blockage increases resistance to infection with the intracellular protozoan trypanosoma cruzi. J Immunol (2004) 172:4893–901. doi: 10.4049/jimmunol.172.8.4893
16. Pathak SK, Basu S, Basu KK, Banerjee A, Pathak S, Bhattacharyya A, et al. Corrigendum: Direct extracellular interaction between the early secreted antigen ESAT-6 of mycobacterium tuberculosis and TLR2 inhibits TLR signaling in macrophages. Nat Immunol (2015) 16:326. doi: 10.1038/ni0315-326b
17. Udgata A, Dolasia K, Ghosh S, Mukhopadhyay S. Dribbling through the host defence: targeting the TLRs by pathogens. Crit Rev Microbiol (2019) 45:354–68. doi: 10.1080/1040841X.2019.1608904
18. Udgata A, Qureshi R, Mukhopadhyay S. Transduction of functionally contrasting signals by two mycobacterial PPE proteins downstream of TLR2 receptors. J Immunol (2016) 197:1776–87. doi: 10.4049/jimmunol.1501816
19. Hornef MW, Wick MJ, Rhen M, Normark S. Bacterial strategies for overcoming host innate and adaptive immune responses. Nat Immunol (2002) 3:1033–40. doi: 10.1038/ni1102-1033
20. Panda SK, Kumar S, Tupperwar NC, Vaidya T, George A, Rath S, et al. Chitohexaose activates macrophages by alternate pathway through TLR4 and blocks endotoxemia. PloS Pathog (2012) 8:e1002717. doi: 10.1371/journal.ppat.1002717
21. Abend JR, Ramalingam D, Kieffer-Kwon P, Uldrick TS, Yarchoan R, Ziegelbauer JM. Kaposi's sarcoma-associated herpesvirus microRNAs target IRAK1 and MYD88, two components of the toll-like receptor/interleukin-1R signaling cascade, to reduce inflammatory-cytokine expression. J Virol (2012) 86:11663–74. doi: 10.1128/JVI.01147-12
22. Bowie AG, Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol (2008) 8:911–22. doi: 10.1038/nri2436
23. Lu M, Tian X, Yang Z, Wang W, Tian AL, Li C, et al. Proteomic analysis revealed T cell hyporesponsiveness induced by haemonchus contortus excretory and secretory proteins. Veterinary Res (2020) 51:65. doi: 10.1186/s13567-020-00790-0
24. Naqvi MA, Memon MA, Jamil T, Naqvi SZ, Aimulajiang K, Gadahi JA, et al. Galectin domain containing protein from haemonchus contortus modulates the immune functions of goat PBMCs and regulates CD4+ T-helper cells in vitro. Biomolecules (2020) 10:116. doi: 10.3390/biom10010116
25. Wen Z, Xie X, Aleem MT, Aimulajiang K, Chen C, Liang M, et al. In vitro characterization of haemonchus contortus trehalose-6-phosphate phosphatase and its immunomodulatory effects on peripheral blood mononuclear cells (PBMCs). Parasit Vectors (2021) 14:611. doi: 10.1186/s13071-021-05115-4
26. Wen Z, Zhang Z, Aimulajiang K, Aleem MT, Feng J, Liang M, et al. Histidine acid phosphatase domain-containing protein from haemonchus contortus is a stimulatory antigen for the Th1 immune response of goat PBMCs. Parasites Vectors (2022) 15:282. doi: 10.1186/s13071-022-05411-7
27. Wen Z, Aleem MT, Aimulajiang K, Chen C, Liang M, Song X, et al. The GT1-TPS structural domain protein from haemonchus contortus could be suppressive antigen of goat PBMCs. Front Immunol (2021) 12:787091. doi: 10.3389/fimmu.2021.787091
28. Yan S, Xie N, Aleem MT, Ji X, Zhang C, Cao X, et al. Overexpression of angiotensin-converting enzyme 2 contributes to the amelioration of streptococcus uberis-induced inflammatory injury in mammary epithelial cells. Veterinary Microbiol (2022) 268:109398. doi: 10.1016/j.vetmic.2022.109398
29. Neefjes J, Jongsma ML, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol (2011) 11:823–36. doi: 10.1038/nri3084
30. Montero BK, Wasimuddin, Schwensow N, Gillingham MAF, Ratovonamana YR, Rakotondranary SJ, et al. Evidence of MHC class I and II influencing viral and helminth infection via the microbiome in a non-human primate. PloS Pathog (2021) 17:e1009675. doi: 10.1371/journal.ppat.1009675
31. Trowsdale J, Knight JC. Major histocompatibility complex genomics and human disease. Annu Rev Genomics Hum Genet (2013) 14:301–23. doi: 10.1146/annurev-genom-091212-153455
32. Tsai HI, Wu Y, Liu X, Xu Z, Liu L, Wang C, et al. Engineered small extracellular vesicles as a FGL1/PD-L1 dual-targeting delivery system for alleviating immune rejection. Adv Sci (Weinheim Baden-Wurttemberg Germany) (2022) 9:e2102634. doi: 10.1002/advs.202102634
33. Dai E, Zhu Z, Wahed S, Qu Z, Storkus WJ, Guo ZS. Epigenetic modulation of antitumor immunity for improved cancer immunotherapy. Mol Cancer (2021) 20:171. doi: 10.1186/s12943-021-01464-x
34. Lanza R, Russell DW, Nagy A. Engineering universal cells that evade immune detection. Nat Rev Immunol (2019) 19:723–33. doi: 10.1038/s41577-019-0200-1
35. Dey NS, Senaratne S, Somaratne V, Madarasinghe NP, Seneviratne B, Forrester S, et al. Early reduction in PD-L1 expression predicts faster treatment response in human cutaneous leishmaniasis. J Clin Invest (2021) 131:e142765. doi: 10.1172/JCI142765
36. da Fonseca-Martins AM, de Souza Lima-Gomes P, Antunes MM, de Moura RG, Covre LP, Calôba C, et al. Leishmania parasites drive PD-L1 expression in mice and human neutrophils with suppressor capacity. Front Immunol (2021) 12:598943. doi: 10.3389/fimmu.2021.598943
37. Butler NS, Moebius J, Pewe LL, Traore B, Doumbo OK, Tygrett LT, et al. Therapeutic blockade of PD-L1 and LAG-3 rapidly clears established blood-stage plasmodium infection. Nat Immunol (2011) 13:188–95. doi: 10.1038/ni.2180
38. Kurup SP, Obeng-Adjei N, Anthony SM, Traore B, Doumbo OK, Butler NS, et al. Regulatory T cells impede acute and long-term immunity to blood-stage malaria through CTLA-4. Nat Med (2017) 23:1220–5. doi: 10.1038/nm.4395
39. Wu D, Li H, Liu M, Qin J, Sun Y. The Ube2m-Rbx1 neddylation-Cullin-RING-Ligase proteins are essential for the maintenance of regulatory T cell fitness. Nat Commun (2022) 13:3021. doi: 10.1038/s41467-022-30707-8
40. Hodge SH, McSorley HJ. A good day for helminths: how parasite-derived GDH suppresses inflammatory responses. EMBO Rep (2022) 23:e55054. doi: 10.15252/embr.202255054
41. Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Sci (New York NY) (2003) 299:1057–61. doi: 10.1126/science.1079490
42. Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol (2003) 4:330–6. doi: 10.1038/ni904
43. Jia L, Wang Y, Wang CY. circFAT1 promotes cancer stemness and immune evasion by promoting STAT3 activation. Adv Sci (Weinheim Baden-Wurttemberg Germany) (2021) 8:2003376. doi: 10.1002/advs.202003376
44. Zhang L, Kuca K, You L, Zhao Y, Musilek K, Nepovimova E, et al. Signal transducer and activator of transcription 3 signaling in tumor immune evasion. Pharmacol Ther (2022) 230:107969. doi: 10.1016/j.pharmthera.2021.107969
45. Siddiqui KF, Amir M, Gurram RK, Khan N, Arora A, Rajagopal K, et al. Latency-associated protein Acr1 impairs dendritic cell maturation and functionality: a possible mechanism of immune evasion by mycobacterium tuberculosis. J Infect Dis (2014) 209:1436–45. doi: 10.1093/infdis/jit595
46. Drurey C, Lindholm HT, Coakley G, Poveda MC, Löser S, Doolan R, et al. Intestinal epithelial tuft cell induction is negated by a murine helminth and its secreted products. J Exp Med (2022) 219:e20211140. doi: 10.1084/jem.20211140
47. Lothstein KE, Gause WC. Mining helminths for novel therapeutics. Trends Mol Med (2021) 27:345–64. doi: 10.1016/j.molmed.2020.12.010
48. Boyett D, Hsieh MH. Wormholes in host defense: how helminths manipulate host tissues to survive and reproduce. PloS Pathog (2014) 10:e1004014. doi: 10.1371/journal.ppat.1004014
49. Lu M, Tian X, Zhang Y, Wang W, Tian AL, Aimulajiang K, et al. Protection studies of an excretory-secretory protein HcABHD against haemonchus contortus infection. Veterinary Res (2021) 52:3. doi: 10.1186/s13567-020-00871-0
50. Lu M, Tian X, Wang W, Zhang Y, Aimulajiang K, Tian AL, et al. The excretory-secretory antigen HcADRM1 to generate protective immunity against haemonchus contortus. Parasitology (2021) 148:1497–508. doi: 10.1017/S0031182021001141
51. Wang Q, Sun X, Huang X, Huang J, Hasan MW, Yan R, et al. Nanoparticles of Chitosan/Poly(D,L-Lactide-Co-Glycolide) enhanced the immune responses of haemonchus contortus HCA59 antigen in model mice. Int J Nanomed (2021) 16:3125–39. doi: 10.2147/IJN.S301851
52. Tian X, Lu M, Jia C, Bu Y, Aimulajiang K, Zhang Y, et al. Haemonchus contortus transthyretin domain - containing protein (HcTTR): A promising vaccine candidate against haemonchus contortus infection. Veterinary Parasitol (2020) 279:109045. doi: 10.1016/j.vetpar.2020.109045
53. Cobb BA, Wang Q, Tzianabos AO, Kasper DL. Polysaccharide processing and presentation by the MHCII pathway. Cell (2004) 117:677–87. doi: 10.1016/j.cell.2004.05.001
54. Ochoa R, Lunardelli VAS, Rosa DS, Laio A, Cossio P. Multiple-allele MHC class II epitope engineering by a molecular dynamics-based evolution protocol. Front Immunol (2022) 13:862851. doi: 10.3389/fimmu.2022.862851
55. Badrinath S, Dellacherie MO, Li A, Zheng S, Zhang X, Sobral M, et al. A vaccine targeting resistant tumours by dual T cell plus NK cell attack. Nature (2022) 606:992–8. doi: 10.1038/s41586-022-04772-4
56. Hinke DM, Andersen TK, Gopalakrishnan RP, Skullerud LM, Werninghaus IC, Grødeland G, et al. Antigen bivalency of antigen-presenting cell-targeted vaccines increases b cell responses. Cell Rep (2022) 39:110901. doi: 10.1016/j.celrep.2022.110901
57. Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature (2013) 496:445–55. doi: 10.1038/nature12034
58. Varotto-Boccazzi I, Epis S, Arnoldi I, Corbett Y, Gabrieli P, Paroni M, et al. Boosting immunity to treat parasitic infections: Asaia bacteria expressing a protein from wolbachia determine M1 macrophage activation and killing of leishmania protozoans. Pharmacol Res (2020) 161:105288. doi: 10.1016/j.phrs.2020.105288
59. Sharma A, Sharma P, Ganga L, Satoeya N, Mishra S, Vishwakarma AL, et al. Infective larvae of brugia malayi induce polarization of host macrophages that helps in immune evasion. Front Immunol (2018) 9:194. doi: 10.3389/fimmu.2018.00194
60. Cai J, Huang L, Tang H, Xu H, Wang L, Zheng M, et al. Macrophage migration inhibitory factor of thelazia callipaeda induces M2-like macrophage polarization through TLR4-mediated activation of the PI3K-akt pathway. FASEB J (2021) 35:e21866. doi: 10.1096/fj.202100676R
61. Sun R, Zhang Z, Bao R, Guo X, Gu Y, Yang W, et al. Loss of SIRT5 promotes bile acid-induced immunosuppressive microenvironment and hepatocarcinogenesis. J Hepatol (2022) (77):453–66. doi: 10.1016/j.jhep.2022.02.030
62. Noe JT, Rendon BE, Geller AE, Conroy LR, Morrissey SM, Young LEA, et al. Lactate supports a metabolic-epigenetic link in macrophage polarization. Sci Adv (2021) 7:eabi8602. doi: 10.1126/sciadv.abi8602
63. O'Donnell JS, Teng MWL, Smyth MJ. Cancer immunoediting and resistance to T cell-based immunotherapy. Nat Rev Clin Oncol (2019) 16:151–67. doi: 10.1038/s41571-018-0142-8
64. Chen H, Pan Y, Zhou Q, Liang C, Wong CC, Zhou Y, et al. METTL3 inhibits anti-tumor immunity by targeting m(6)A-BHLHE41-CXCL1/CXCR2 axis to promote colorectal cancer. Gastroenterology (2022) 11:1–17. doi: 10.1053/j.gastro.2022.06.024
65. Xie J, El Rami F, Zhou K, Simonetta F, Chen Z, Zheng X, et al. Multiparameter longitudinal imaging of immune cell activity in chimeric antigen receptor T cell and checkpoint blockade therapies. ACS Cent Sci (2022) 8:590–602. doi: 10.1021/acscentsci.2c00142
66. Johnston CJC, Smyth DJ, Kodali RB, White MPJ, Harcus Y, Filbey KJ, et al. A structurally distinct TGF-β mimic from an intestinal helminth parasite potently induces regulatory T cells. Nat Commun (2017) 8:1741. doi: 10.1038/s41467-017-01886-6
67. Kalia I, Anand R, Quadiri A, Bhattacharya S, Sahoo B, Singh AP. Plasmodium berghei-released factor, PbTIP, modulates the host innate immune responses. Front Immunol (2021) 12:699887. doi: 10.3389/fimmu.2021.699887
68. Lee WS, Kim DS, Kim JH, Heo Y, Yang H, Go EJ, et al. Intratumoral immunotherapy using a TLR2/3 agonist, l-pampo, induces robust antitumor immune responses and enhances immune checkpoint blockade. J Immunother Cancer (2022) 10:e004799. doi: 10.1136/jitc-2022-004799
69. Yan C, Wu J, Xu N, Li J, Zhou QY, Yang HM, et al. TLR4 deficiency exacerbates biliary injuries and peribiliary fibrosis caused by clonorchis sinensis in a resistant mouse strain. Front Cell Infect Microbiol (2020) 10:526997. doi: 10.3389/fcimb.2020.526997
70. Lanzavecchia A, Sallusto F. Toll-like receptors and innate immunity in b-cell activation and antibody responses. Curr Opin Immunol (2007) 19:268–74. doi: 10.1016/j.coi.2007.04.002
71. Kawasaki T, Kawai T. Toll-like receptor signaling pathways. Front Immunol (2014) 5:461. doi: 10.3389/fimmu.2014.00461
72. Onguru D, Liang Y, Griffith Q, Nikolajczyk B, Mwinzi P, Ganley-Leal L. Human schistosomiasis is associated with endotoxemia and toll-like receptor 2- and 4-bearing b cells. Am J Trop Med Hygiene (2011) 84:321–4. doi: 10.4269/ajtmh.2011.10-0397
73. Ludwig-Portugall I, Layland LE. TLRs, treg, and b cells, an interplay of regulation during helminth infection. Front Immunol (2012) 3:8. doi: 10.3389/fimmu.2012.00008
74. Han C, Yu J, Zhang Z, Zhai P, Zhang Y, Meng S, et al. Immunomodulatory effects of trichinella spiralis excretory-secretory antigens on macrophages. Exp Parasitol (2019) 196:68–72. doi: 10.1016/j.exppara.2018.10.001
75. Soleymani N, Taran F, Nazemshirazi M, Naghibi A, Torabi M, Borji H, et al. Dysregulation of ovine toll-like receptors 2 and 4 expression by hydatid cyst-derived antigens. Iranian J Parasitol (2021) 16:219–28. doi: 10.18502/ijpa.v16i2.6271
76. Nogueira DS, de Oliveira LM, Amorim CCO, Gazzinelli-Guimarães AC, Barbosa FS, Oliveira FMS, et al. Eosinophils mediate SIgA production triggered by TLR2 and TLR4 to control ascaris suum infection in mice. PloS Pathog (2021) 17:e1010067. doi: 10.1371/journal.ppat.1010067
77. Lawrence T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harbor Perspect Biol (2009) 1:a001651. doi: 10.1101/cshperspect.a001651
78. Fang L, Wu HM, Ding PS, Liu RY. TLR2 mediates phagocytosis and autophagy through JNK signaling pathway in staphylococcus aureus-stimulated RAW264.7 cells. Cell Signal (2014) 26:806–14. doi: 10.1016/j.cellsig.2013.12.016
79. Jahan S, Kumar D, Chaturvedi S, Rashid M, Wahajuddin M, Khan YA, et al. Therapeutic targeting of NLRP3 inflammasomes by natural products and pharmaceuticals: A novel mechanistic approach for inflammatory diseases. Curr Med Chem (2017) 24:1645–70. doi: 10.2174/0929867324666170227121619
80. Zhou H, Luan W, Wang Y, Song Y, Xu H, Tang X, et al. The combination of cloxacillin, thioridazine and tetracycline protects mice against staphylococcus aureus peritonitis by inhibiting α-hemolysin-induced MAPK/NF-κB/NLRP3 activation. Int J Biol Macromol (2022) 198:1–10. doi: 10.1016/j.ijbiomac.2021.12.112
Keywords: Haemonchus contortus, HcESPs, PBMCs, TLR4/NF-κB/MAPKs signaling pathway, immune evasion, pattern recognition receptor
Citation: Wen Z, Zhang Y, Feng J, Aimulajiang K, Aleem MT, Lu M, Xu L, Song X, Li X and Yan R (2022) Excretory/secretory proteins inhibit host immune responses by downregulating the TLR4/NF-κB/MAPKs signaling pathway: A possible mechanism of immune evasion in parasitic nematode Haemonchus contortus. Front. Immunol. 13:1013159. doi: 10.3389/fimmu.2022.1013159
Received: 06 August 2022; Accepted: 13 September 2022;
Published: 27 September 2022.
Edited by:
Tiago W. P. Mineo, Federal University of Uberlandia, BrazilReviewed by:
Adler Ray Dillman, University of California, Riverside, United StatesCopyright © 2022 Wen, Zhang, Feng, Aimulajiang, Aleem, Lu, Xu, Song, Li and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruofeng Yan, eWFucnVvZmVuZ0BuamF1LmVkdS5jbg==
†These authors contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.