- 1Department of Medical Oncology, Shuguang Hospital, Shanghai University of Traditional Chinese Medicine, Shanghai, China
- 2Academy of Integrative Medicine, Shanghai University of Traditional Chinese Medicine, Shanghai, China
Introduction: Dysregulation of the Hippo signaling pathway has been implicated in multiple pathologies, including cancer, and YAP1 is the major effector of the pathway. In this study, we assessed the role of YAP1 in prognostic value, immunomodulation, and drug response from a pan-cancer perspective.
Methods: We compared YAP1 expression between normal and cancerous tissues and among different pathologic stages survival analysis and gene set enrichment analysis were performed. Additionally, we performed correlation analyses of YAP1 expression with RNA modification-related gene expression, tumor mutation burden (TMB), microsatellite instability (MSI), immune checkpoint regulator expression, and infiltration of immune cells. Correlations between YAP1 expression and IC50s (half-maximal inhibitory concentrations) of drugs in the CellMiner database were calculated.
Results: We found that YAP1 was aberrantly expressed in various cancer types and regulated by its DNA methylation and post-transcriptional modifications, particularly m6A methylation. High expression of YAP1 was associated with poor survival outcomes in ACC, BLCA, LGG, LUAD, and PAAD. YAP1 expression was negatively correlated with the infiltration of CD8+ T lymphocytes, CD4+ Th1 cells, T follicular helper cells, NKT cells, and activated NK cells, and positively correlated with the infiltration of myeloid-derived suppressor cells (MDSCs) and cancer-associated fibroblasts (CAFs) in pan-cancer. Higher YAP1 expression showed upregulation of TGF-β signaling, Hedgehog signaling, and KRAS signaling. IC50s of FDA-approved chemotherapeutic drugs capable of inhibiting DNA synthesis, including teniposide, dacarbazine, and doxorubicin, as well as inhibitors of hypoxia-inducible factor, MCL-1, ribonucleotide reductase, and FASN in clinical trials were negatively correlated with YAP1 expression.
Discussion: In conclusion, YAP1 is aberrantly expressed in various cancer types and regulated by its DNA methylation and post-transcriptional modifications. High expression of YAP1 is associated with poor survival outcomes in certain cancer types. YAP1 may promote tumor progression through immunosuppression, particularly by suppressing the infiltration of CD8+ T lymphocytes, CD4+ Th1 cells, T follicular helper cells, NKT cells, and activated NK cells, as well as recruiting MDSCs and CAFs in pan-cancer. The tumor-promoting activity of YAP1 is attributed to the activation of TGF-β, Hedgehog, and KRAS signaling pathways. AZD2858 and varlitinib might be effective in cancer patients with high YAP1 expression.
Introduction
The Hippo signaling pathway is an evolutionarily conserved pathway with a biological role in cell fate determination, organ size control, and tissue regeneration in most tissues and organs (1–4). Dysregulation of this pathway has been implicated in a variety of pathologies and has received extensive attention over the past two decades (5). In cancer research, the activated Hippo pathway is considered a tumor suppressor pathway due to its role in inhibiting cell proliferation and promoting apoptosis (6–8).
Growth factors, glucose, hypoxia, cell polarity, and mechanical cues from cell–cell or cell–extracellular matrix attachment regulate the Hippo pathway (9). The core components of the Hippo pathway in mammals consist of a kinase cascade, MST1/2 and LATS1/2, and the main effector YAP1/TAZ, a transcriptional coactivator without DNA-binding domains. The major binding partners of YAP1/TAZ are TEAD1–4. Other transcriptional factors, including AP1, PITX2, ZEB1, MYC, E2F, and SMADs, have also been reported to cooperate with the YAP1/TAZ–TEAD complex (10–15). In addition, RUNX2, TP73, and FOXO1 also directly bind to YAP1/TAZ (16). YAP1 overexpression has been reported to be oncogenic in bile duct, breast, colon, lung, and liver cancers (17–23). Inhibition of the Hippo pathway or overexpression of YAP1 may lead to the nuclear translocation of YAP1, which then binds to transcription factors to promote the expression of tumor-promoting genes. Of note, YAP1 also functions as a tumor corepressor to repress the expression of downstream genes, including the cell-cycle kinase inhibitor p27, by recruiting the NuRD (nucleosome remodeling deacetylase) complex, YY1, or EZH2, a polycomb repressive complex member (24, 25). Thus, the role of transcriptional cofactor or transcriptional corepressor of YAP1 is largely context-dependent. Therefore, better defining the role of YAP1 in each cancer type will be a key challenge for future studies about target identification and cancer therapy. We investigated the potential role of YAP1 in survival predication from a pan-cancer perspective.
Tumor-infiltrating immune cells are a major component of the ecosystem in tumor microenvironment (TME) and regulate tumor progression (26). Recently, the Hippo signaling pathway is emerging as an important pathway to affect immune function in cancer (27). YAP1 has been reported to affect the activity of B cells, Tregs, macrophages, and myeloid-derived suppressor cells (MDSCs) in several cancer types (28). However, the role of YAP1 in different cancer types and its mechanisms in immune regulation remain to be investigated. In this study, we analyzed the correlation between YAP1 expression and infiltration of various immune cells in 33 cancer types.
Strategies to inhibit YAP1 activity include the following (1): Disrupting the YAP1–TEAD binding or blocking the transcriptional activity of the YAP1–TEAD complex. Carbonic anhydrase 3 (CA3) and verteporfin act by disrupting the YAP1–TEAD binding (29–31). The TDU domain of vestigial like family member 4 (VGLL4), a natural antagonist of YAP1, competes with YAP1 to bind TEADs (32). Narciclasine and peptide17 competes with TEAD4 for binding to YAP1 (33, 34). In addition, K-975, a TEAD inhibitor, inhibits YAP1/TAZ–TEAD interaction (35) (2). Targeting downstream targets of YAP1/TAZ (BCL-xL, FOXM1, and TG2). A37, celecoxib, TP-0903, cyclic peptide RA-V (deoxybouvardin), navitoclax, thiostrepton, and NC-9 fall into this category (36–43). Of these drugs, only verteporfin has been approved by Food and Drug Administration (FDA). However, YAP1 may act in a TEAD-independent manner (44). The efficacy and selectivity of other drugs are not satisfactory. New drugs are needed to inhibit YAP1 activity. In this study, we calculated the correlation between YAP1 expression and IC50s of drugs with FDA approval or in clinical trials, which is a simple way to roughly assess the drug sensitivity.
In this study, we analyzed YAP1 expression in 33 cancer types to reveal its role in predicating prognosis, modulating TME, and drug response to chemotherapeutic and targeted drugs that were FDA-approved or are in clinical trials.
Methods and materials
Data collection
Transcriptional RNA-sequence data [cohort: TCGA Pan-Cancer (PANCAN), batch effects normalized mRNA data], genome-wide DNA methylation levels (DNA Methylation 450K), and clinical characteristics (curated clinical data) of samples related to 33 cancer types were downloaded from UCSC Xena (https://xenabrowser.net/datapages/), which is derived from TCGA resources (45).
Survival analysis
Survival information, including overall survival (OS), progression-free interval (PFI), disease-free interval (DFI), and disease-specific survival (DSS), was also downloaded from the UCSC Xena database. The Kaplan–Meier model and univariate Cox regression were then used to assess the prognostic value of YAP1. Bivariate YAP1 expression levels were used to perform Kaplan–Meier curves analysis using the optimal cut point from the survminer R package (0.4.9) and survival R package (version 3.3.1). p-values of the Kaplan–Meier method and hazard ratio (HR) with a 95% confidence interval (95% CI) were calculated for each cancer type and presented as forest plots using the forestplot R package (version 2.0.1).
Infiltration of immune cells
The proportions of 22 immune cell types and immune scores of all samples of 33 cancer types were downloaded from the supplementary data of the published paper (46). Infiltration of immune cells was performed with the CIBERSORT program, a method that uses gene expression profiles of complex tissues to calculate cell composition (47). The Xcell, TIMER, EPIC, quanTIseq, and MCP-counter programs were also used. TIMER provides the coefficients of six immune infiltrating cells indicating the relative abundance of immune cells (48). The EPIC program estimates the proportions of immune and cancer cells by separating the reference gene expression profiles of major non-malignant cell types (49). The ESTIMATE program was performed to infer tumor purity and immune cell admixture (immune score) from expression data (50). Pearson correlation coefficients between YAP1 expression and infiltration of immune cells or immune score were calculated using the ggpubr R package (version 0.4.0), and scatter plots were visualized using ggplot2 (version 3.3.6).
Gene set enrichment analysis
For each cancer type, samples with YAP1 expression above the median level were grouped as high YAP1 expression, and the others were grouped as low YAP1 expression to compare the difference in their hallmark gene sets. Fifty hallmark gene sets were downloaded from the Molecular Signatures Database (MSigDB, https://www.gsea-msigdb.org/gsea/index.jsp), and gene set enrichment analysis (GSEA) was performed using GSEA software (version 4.2.3). One thousand times was set as the number of permutations and phenotypes was set as permutation type. Normalized enrichment score (NES) and nominal p value for each biological process were calculated for each cancer type. Hallmark gene sets with nominal p value < 0.05 were presented as a bubble plot using the ggplot2 R package.
Correlation of YAP1 expression with tumor mutation burden and microsatellite instability
Tumor mutation burden (TMB) andmicrosatellite instability (MSI) levels for each sample in pan-cancer were downloaded from the supplementary data of the published paper (46). Pearson correlation coefficients between YAP1 expression and TMB or MSI levels for each cancer type were calculated using ggpubr R package and displayed as radar charts using fmsb R package (version 0.7.3).
Azoxymethane/dextran sulfate sodium-induced colorectal cancer model and immunohistochemical staining of YAP1
CRC was induced in C57BL/6J mice by azoxymethane (AOM)/dextran sulfate sodium (DSS) as previously described (51). Colon sections of each mouse were collected for IHC staining and YAP1 was stained as previously described (51).
Assessment of drug sensitivity
IC50s (half-maximal inhibitory concentrations) of drugs and gene expression of cancer cell lines were downloaded from the CellMiner database (https://discover.nci.nih.gov/cellminer/home.do). Only drugs with FDA approval or in clinical trials were included in further analyses. Pearson correlation coefficients between YAP1 expression and the IC50 z score of each drug were calculated using the ggpubr R package and represented as bubble plots or scatter plots using the ggplot2 R package.
Cell viability assay
Cells were seeded at a density of 1 × 104 cells/well in 96-well plates. When cells reached 60% confluence, drugs were added into the wells and incubated for 24 h or 48 h. Medium containing 10% Cell Counting Kit-8 (CCK-8) reagent (Dojindo, Japan) was then added into the cells and incubated for another 1.5 h at 37°C. The light absorbance was measured at 450 nm on the microplate reader (Bio-Rad, USA). Each group was performed in sextuplicate. Verteporfin, AZD-2858 were purchased from TargetMol, USA, and varlitinib was purchased from MCE, USA
Western blot assay
Cellular proteins were extracted with protein extraction reagent (Beyotime Biotechnology) and quantified by BCA protein assay (Beyotime Biotechnology). A total of 12 μg of protein per sample was added to SDS-PAGE gels for electrophoresis (100 V, 2 h), followed by constant flow membrane transfer (ice bath, 210 mA, 2 h). The transferred polyvinylidene fluoride (PVDF) membranes were blocked with 5% BSA-containing Tris-buffered saline with Tween (TBST) for 2 h at room temperature, and incubated with primary antibodies overnight at 4°C. Then, membranes were washed three times with TBST and incubated with secondary antibody at room temperature for 2 h. Membranes were examined with a gel imager (ECL, Millipore, USA). Antibodies of Vimentin, E-Cadherin, smad2, ERK, p-ERK, CREB, and GAPDH were purchased from Cell Signaling Technology (CST); YAP1, p-YAP1, and PD-L1 were purchased from Proteintech. All primary antibodies were used at 1:1,000 dilution. Secondary antibodies were purchased from Beyotime Biotechnology (1:2,000 dilution).
Reverse transcription-quantitative polymerase chain reaction
Primers were designed using the Primer-Blast tool at NCBI (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) and synthesized by Sangon Biological Engineering Co., Ltd. (Shanghai, China). The sequences of the primers are listed in Supplementary Table S1. Total cellular RNA was extracted with RNA Trizol (Beyotime Biotechnology). Reverse transcription of RNA was performed using HiScript III RT SuperMix (Vazyme). The SYBR qPCR Mix (Vazyme) was used in a 10-µl reaction mixture that included 1 µl of cDNA template, 0.4 µl of each 0.5 µM primer, 3.6 µl of ddH2O, and 5 µl of 2× SYBR buffer. The reaction was performed with 1 cycle of 30 s at 95°C and 50 cycles of 10 s at 95°C, 30 s at 60°C, and 15 s at 60°C. Actin was used as the reference mRNA. The qPCR reaction was performed in triplicate.
Statistical analysis
Differences in YAP1 mRNA expression between normal and cancer tissues or between the two pathological stages of each cancer type were tested by Wilcox test. The differences in the proportions of the four pathological stages between groups with high or low YAP1 expression were compared by chi-squared test. Differences in OS, PFI, DFI, and DSS between those two subgroups were compared using the Kaplan–Meier method and log-rank rest. The HRs were calculated by univariate Cox regression. All p values were two-sided, and p < 0.05 was considered statistically significant.
Results
YAP1 is aberrantly expressed in various cancer tissues
We compared YAP1 mRNA expression between normal and primary cancer tissues in 23 cancer types. For the other 10 cancer types, RNA-sequence data from paired normal tissues were not available. YAP1 was differently expressed in 14 of the 23 cancer types with statistical significance. Among them, YAP1 expression was upregulated in cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), and thyroid carcinoma (THCA), and was downregulated in bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), head and neck squamous cell carcinoma (HNSC), kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), pheochromocytoma and paraganglioma (PCPG), prostate adenocarcinoma (PRAD), and uterine corpus endometrial carcinoma (UCEC) (Figure 1A). IHC staining also identified that YAP1 was upregulated in colon cancer tissues versus normal colonic mucosa tissues (52). YAP1 has been reported to be upregulated in CHOL and THCA (17, 18). YAP1 does not contain DNA-binding sequences; thus, the binding partners are important for its function. YAP1 binds to TEADs to facilitate the expression of tumor-promoting genes, and YAP1 may switch to bind to TP73 to promote apoptosis of cancer cells (19). Therefore, we also analyzed the expression of TEADs and TP73 in pan-cancer. Consistent with YAP1 expression, TEAD2 was upregulated not only in CHOL, COAD, and THCA, but also in BRCA, HNSC, liver hepatocellular carcinoma (LIHC), LUAD, LUSC, and UCEC (Figure 1B). Expression of TEAD1/2/4 is shown in Supplementary Figure S1. TP73 was also upregulated in most cancer types, including CHOL, COAD, and THCA (Figure 1C). Xia et al. reported that high levels of YAP1 expression were positively correlated with TEAD4 gene expression, and their co-expression was a prognostic marker for poor ovarian cancer survival (53). High expressions of YAP1 and TEADs and their target genes were associated with low OS in patients with non-metastatic human gastric carcinomas (54). Strano et al. reported that physical interaction with YAP1 protein enhanced transcriptional activity of TP73 (55). High expression of YAP1, TEAD4, and TP73 was significantly associated with high grade, advanced stage, supraglottic location of tumors, nodal metastases, and recurrence of human laryngeal cancer. In addition, high expression of all proteins was significantly associated with poor overall and disease-free survival (56). The tumor-promoting or tumor-suppressing role of the YAP1–TP73 complex, as well as the binding preference mechanism of YAP1 remains to be revealed.
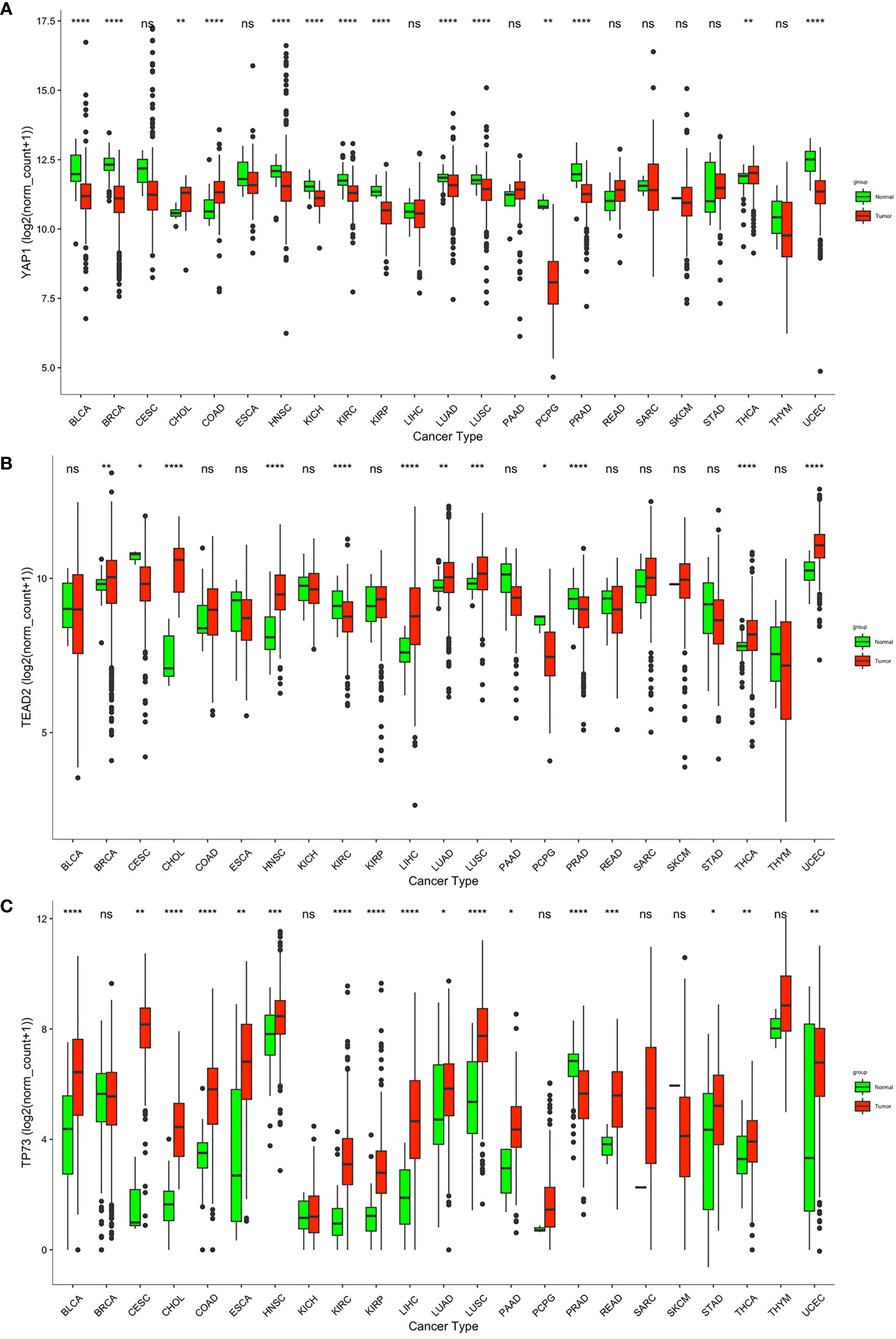
Figure 1 Transcriptional level of YAP1, TEAD2, and TP53 in normal and cancer tissues according to RNA-sequence data from TCGA. ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05, and ns p > 0.05.
YAP1 expression correlates with its DNA methylation and RNA modification
To seek the potential regulation of YAP1 expression by DNA methylation and post-transcriptional RNA modifications, we performed correlation analyses. DNA methylation levels of eight CpG sites in the 5’UTR of YAP1 were included. In general, YAP1 expression was negatively correlated with its DNA methylation level in most cancer types, suggesting that DNA demethylation in the 5’UTR of YAP1 may promote its expression (Figure 2A). RNA modifications play pivotal roles in RNA stability and translation efficiency. We found that a wide range of RNA modification-related genes were positively correlated with YAP1 expression. Genes responsible for reading, writing, and erasing the modifications on m1A, m5C, and m6A were broadly associated with YAP1 expression in all cancer types, particularly in lymphoid neoplasm diffuse large B-cell lymphoma (DLBC), rectum adenocarcinoma (READ), skin cutaneous melanoma (SKCM), uveal melanoma (UVM), and testicular germ cell tumors (TGCT) (Figure 2B). Among the 37 regulators, RNA m6A methylation readers and writers, including ZC3H13, LRPPPRC, YTHDC2, YTHDF3, and KIAA1429, were positively correlated with YAP1 expression in more than 20 of the 33 cancer types with statistical significance, suggesting the potential role of RNA m6A methylation in facilitating YAP1 mRNA stability.
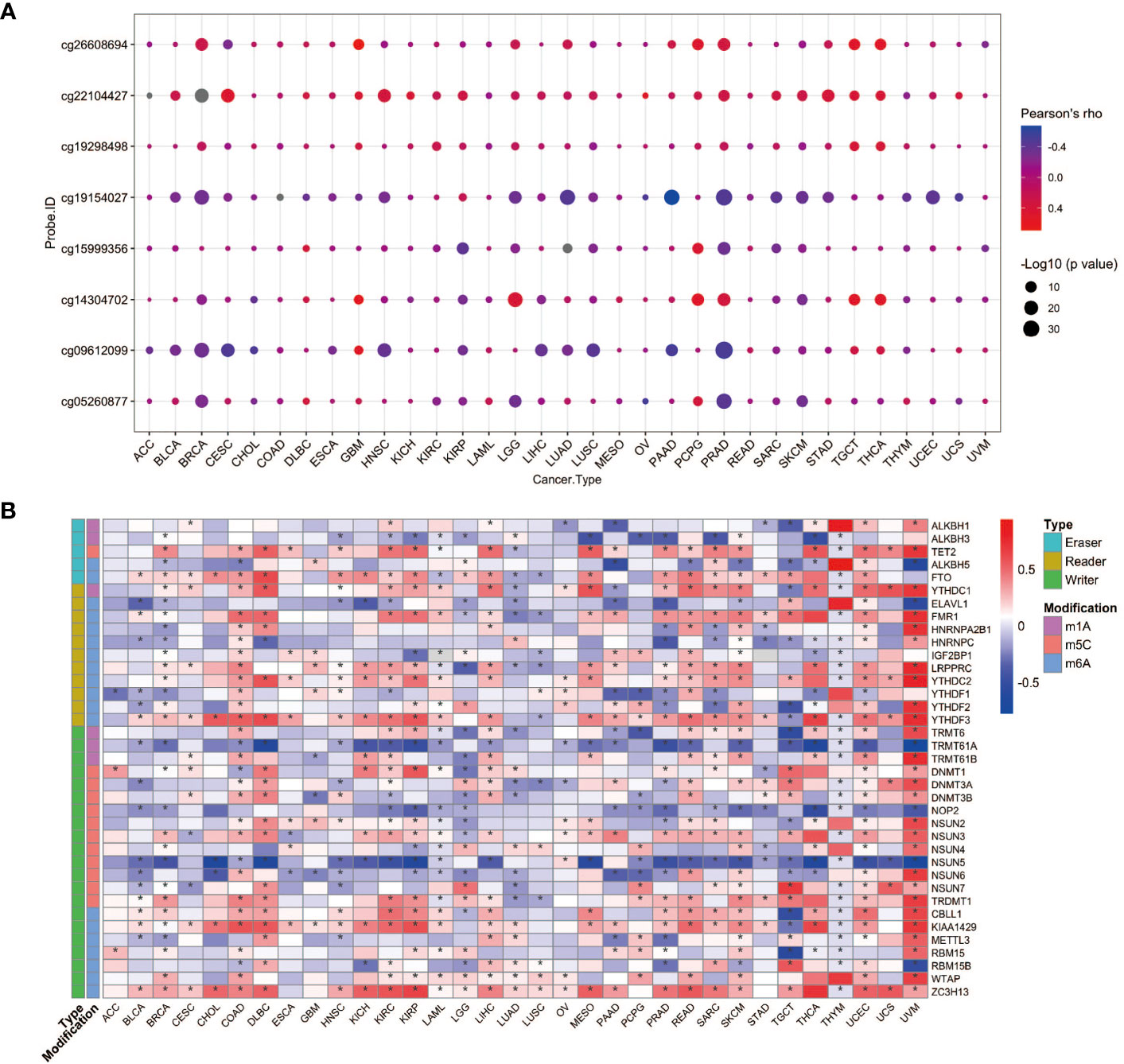
Figure 2 YAP1 expression correlates with its DNA methylation and RNA modification. (A) Pearson correlation between YAP1 expression and its DNA methylation in 33 cancer types. (B) Pearson correlation between YAP1 expression and RNA modification regulators. *p < 0.05.
Clinical prognostic value of YAP1 in pan-cancer
To assess the role of YAP1 in predicating clinical outcomes of patients in 33 cancer types, we analyzed OS, DSS, DFI, and PFI using the Kaplan–Meier method (log-rank test) and univariate Cox regression. High YAP1 expression was a risk factor for the OS in seven cancer types, PFI in eight cancer types, DFI in six cancer types, and DSS in eight cancer types. In general, high YAP1 expression was risky in adrenocortical carcinoma (ACC), BLCA, COAD, brain lower grade glioma (LGG), LUAD, and pancreatic adenocarcinoma (PAAD), but it is a protective factor in esophageal carcinoma (ESCA), KIRC, PRAD, and mesothelioma (MESO) (Figures 3A, B). Of note, high YAP1 expression was a risk factor for all four prognostic survival indicators of ACC and BLCA (Figure 2A). Therefore, the role of YAP1 is largely context-dependent. In ACC, COAD, and TGCT, YAP1 expression was higher in the late pathologic stage (AJCC pathologic stages III and IV) than that in the early stage (AJCC pathologic stages I and II) (Figure 3C). In BLCA, PAAD, and TGCT, a higher frequency of patients with more severe stages was observed in patients with higher YAP1 expression compared with those with lower YAP1 expression (Figure 3D). Those results demonstrate that YAP1 may promote cancer progression in ACC, COAD, PAAD, and TGCT. Among them, the tumor-promoting role of YAP1 in COAD and PAAD has been determined experimentally (57, 58).
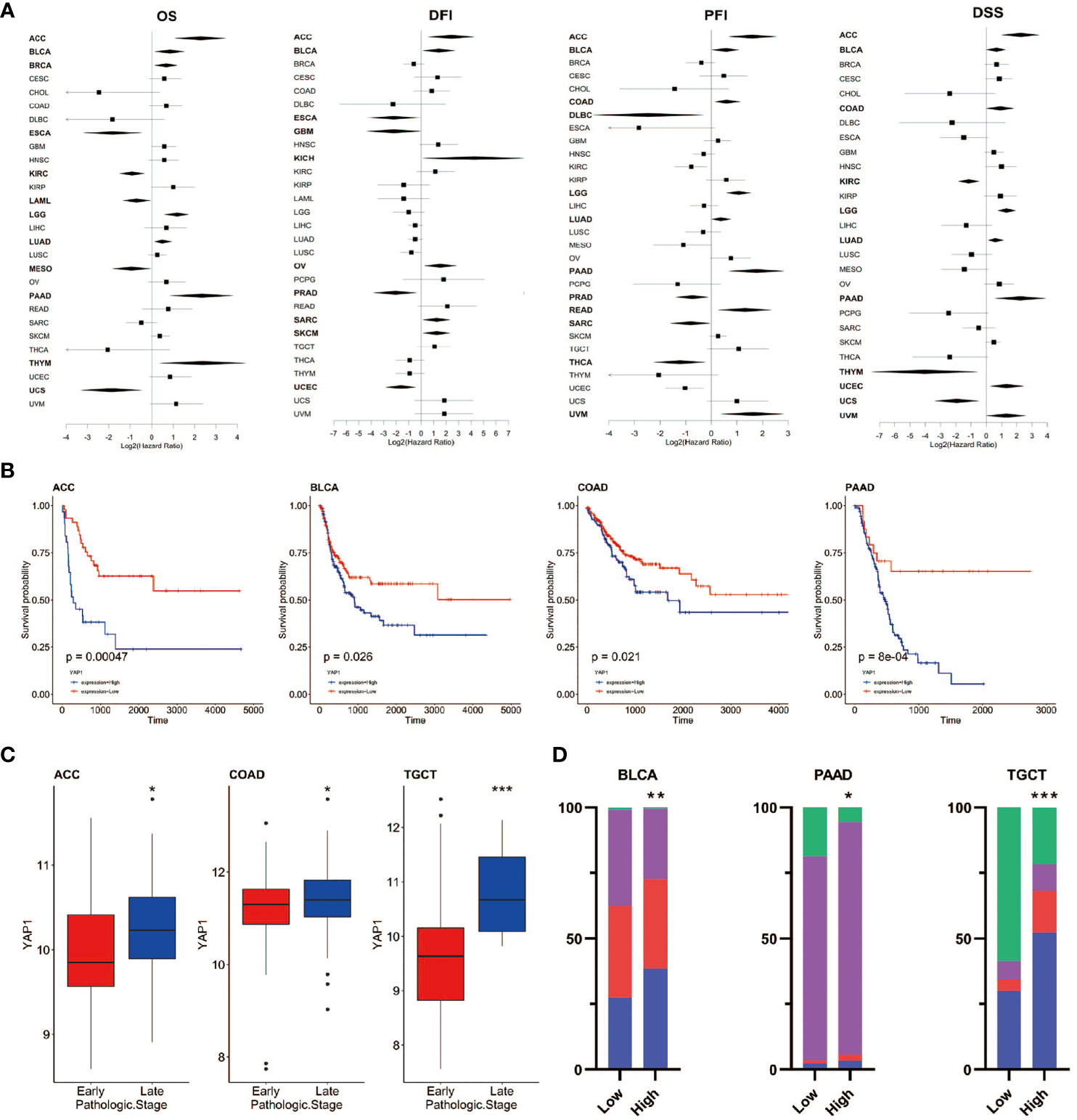
Figure 3 Prognostic value of YAP1 expression in pan-cancer. (A) Survival outcomes of cancer patients with high or low YAP1 expression. Bold lines: p < 0.05. (B) Survival curves of PFI in ACC, BLCA, COAD, and PAAD. (C)YAP1 expression between patients with the early or late pathologic stages of cancer. (D) Distribution of pathologic stages in BLCA, PAAD, and TGCT. ***p < 0.001, **p < 0.01, and *p < 0.05.
YAP1 induces immunosuppressive TME
TME contains tumor cells, immune cells, and stromal cells, which play pivotal roles in cancer initiation, progression, and drug response. We analyzed the correlation between YAP1 expression and infiltration of 19 cell types in tumor tissues using CIBERSORT, Xcell, TIMER, EPIC, quanTIseq, and MCP-counter programs. We found that YAP1 expression was associated with infiltration of cells in both innate and adaptive immune system in pan-cancer. In general, YAP1 expression was negatively correlated with the infiltration of CD8+ T cells, T follicular helper cells, γδ T cells, activated natural killer (NK) T cells, CD4+ Th1 cells, and myeloid dendritic cells in most cancer types, but positively correlated with the infiltration of MDSCs, cancer-associated fibroblasts (CAFs), and neutrophil cells (Figure 4A). These results were consistent using different tools, except for the filtration of CD8+ T cells calculated by TIMER and other programs. EPIC and TIMER programs showed an opposite correlation between YAP1 expression and infiltration of CD4+ T cells in a few cancer types. The correlation of YAP1 expression with infiltration of regulatory T cells (Tregs), plasma B cells, NK cells, and macrophage was contradictory when using different programs (Figure 4A). Thus, experiments are needed to confirm the results. Notably, CD8+ T cells are a key subset of MHC class I-restricted T cells and are one of the major mediators of adaptive immunity. High expression of YAP1 may inhibit the infiltration of CD8+ T cells in 21 of the 33 cancer types (BLCA, BRCA, COAD, HNSC, KIRC, KIRP, LGG, LIHC, LUAD, LUSC, OV, MESO, PCPG, PRAD, SARC, SKCM, TGCT, THCA, THYM, UCEC, and UCS), indicating its immunosuppressive role in TME (Figure 4B). Immune score predicts the level of infiltrating immune cells. YAP1 expression was negatively with the immune score in 12 cancer types (HNSC, KIRC, KIRP, LIHC, MESO, OV, SARC, SKCM, TGCT, THCA, THYM, and UCEC). However, it was positively correlated with the immune score in BRCA, DLBC, GBM, LGG, PAAD, PCPG and PRAD (Figure 4C). IL-6, CSF-1, CSF-2, CSF-3, and CXCL5, which are capable of recruiting MDSCs, were downregulated in CRC cells when YAP1 was inhibited by verteporfin (Figure 8C).
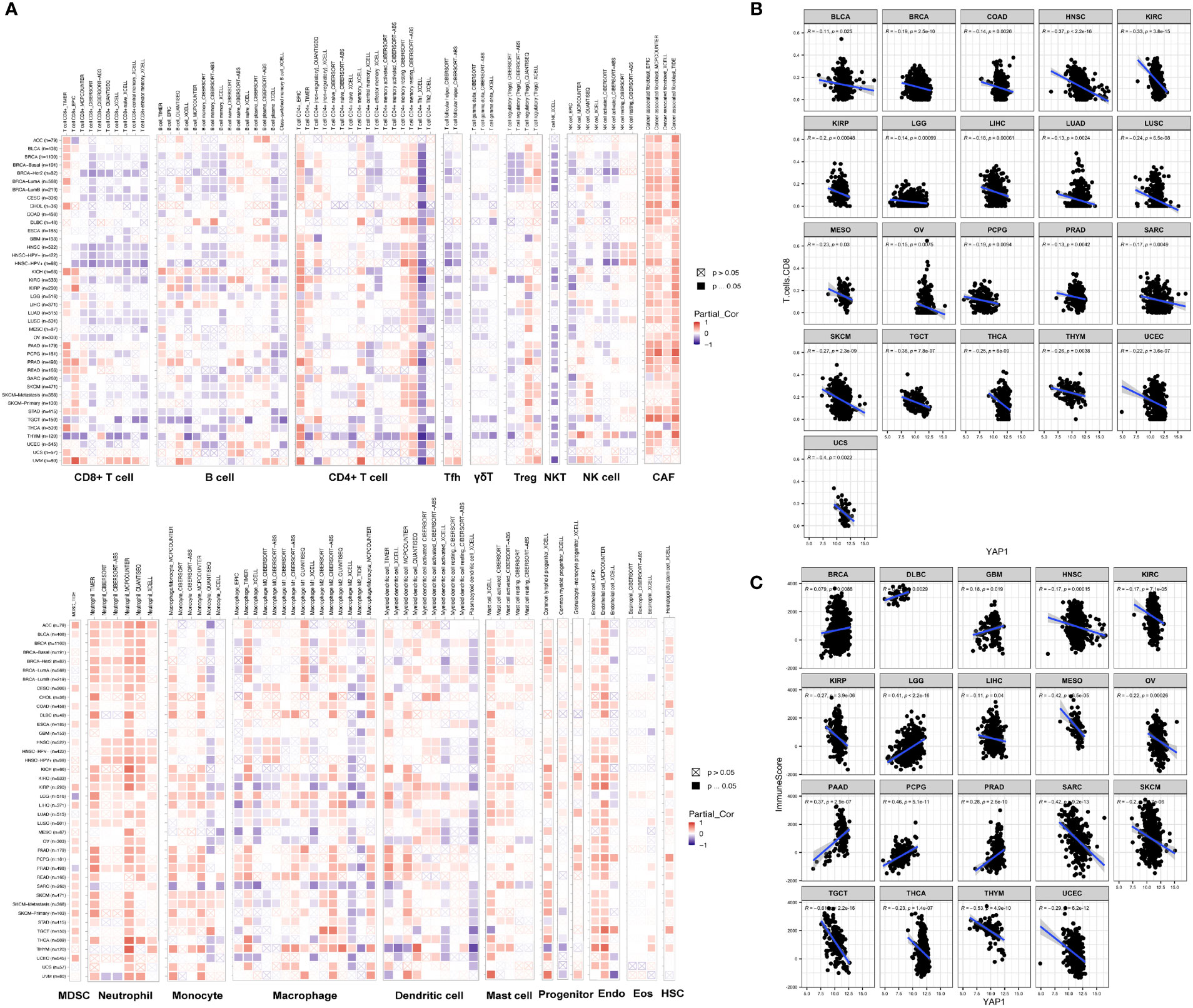
Figure 4 Correlation of YAP1 expression with immune cell infiltration. (A) Pearson correlation between YAP1 expression and infiltration of 22 immune cell types using different programs. Positive correlation is in red and negative correlation is in blue. *p < 0.05. (B) Pearson correlation between YAP1 expression and infiltration of CD8+ T cells in pan-cancer. Only statistically significant cancer types were shown. (C) Pearson correlation between YAP1 expression and immune score. Only statistically significant cancer types were shown. Tfh, T follicular helper cells. Treg, regulatory T cells; NKT, natural killer T cells; NK, natural killer cells; CAF, cancer-associated fibroblast; MDSC, myeloid-derived suppressor cell; Endo, endothelial cell; Eos, eosinophil; HSC, hematopoietic stem cell. γδ T cells, gamma-delta T cells.
Correlations between YAP1 and immune checkpoints, TMB, and MSI
Immune checkpoints are regulators of the immune system. They consist of a group of programmed death receptors and their ligands expressed on immune cells. Tumor cells can evade immune destruction by upregulating immune checkpoints (59). Therefore, we performed the Pearson correlation analyses to reveal the relationship between YAP1 expression and 46 immune checkpoint regulators. We found that YAP1 expression was positively correlated with most immune checkpoint genes and immune cell marker genes in all 33 cancer types. CTLA-4, TIM-3 (HAVCR2), and PD-1 (PDCD1) are key checkpoint regulators that suppress immune response. They all positively correlated with YAP1 expression in LGG, PAAD, and PRAD, but negatively correlated with YAP1 expression in MESO, SARC, TGCT, and UCEC (Figure 5A). We confirmed that YAP1 inhibitor verteporfin greatly reduced PD-L1 expression in CRC cell lines (Figure 8B). In most cancer types, CD274 (PD-L1), NRP1, and TNFSF15 were positively correlated with YAP1 expression. In PAAD, PRAD and PCPG, YAP1 expression was highly correlated with the expression of most immune checkpoint genes, suggesting that YAP1 may help cancer cells to evade immune destruction (Figure 5A). TMB refers to the total number of mutations per million bases (60). It is considered a promising biomarker of immune response, as tumors with high mutations are associated with high tumor neo-antigen burden, making them immunogenic, and therefore being more responsive to immunotherapy (61–64). MSI is a condition of genetic susceptibility to mutation due to impaired DNA mismatch repair (65). It is also used as a major predictive marker for the efficacy of immune checkpoint blockade therapies (66). We found that YAP1 expression was positively correlated with TMB in TGCT and was negatively correlated in PCPG, KIRP, and COAD (Figure 5B). In addition, YAP1 expression was positively correlated with MSI in GBM, HNSC, and TGCT and was negatively correlated with MSI in DLBC, PRAD, THCA, and UCS (Figure 5C). Our results suggest that YAP1 may predict the efficacy of immune checkpoint inhibitors in TGCT, GBM, and HNSC.
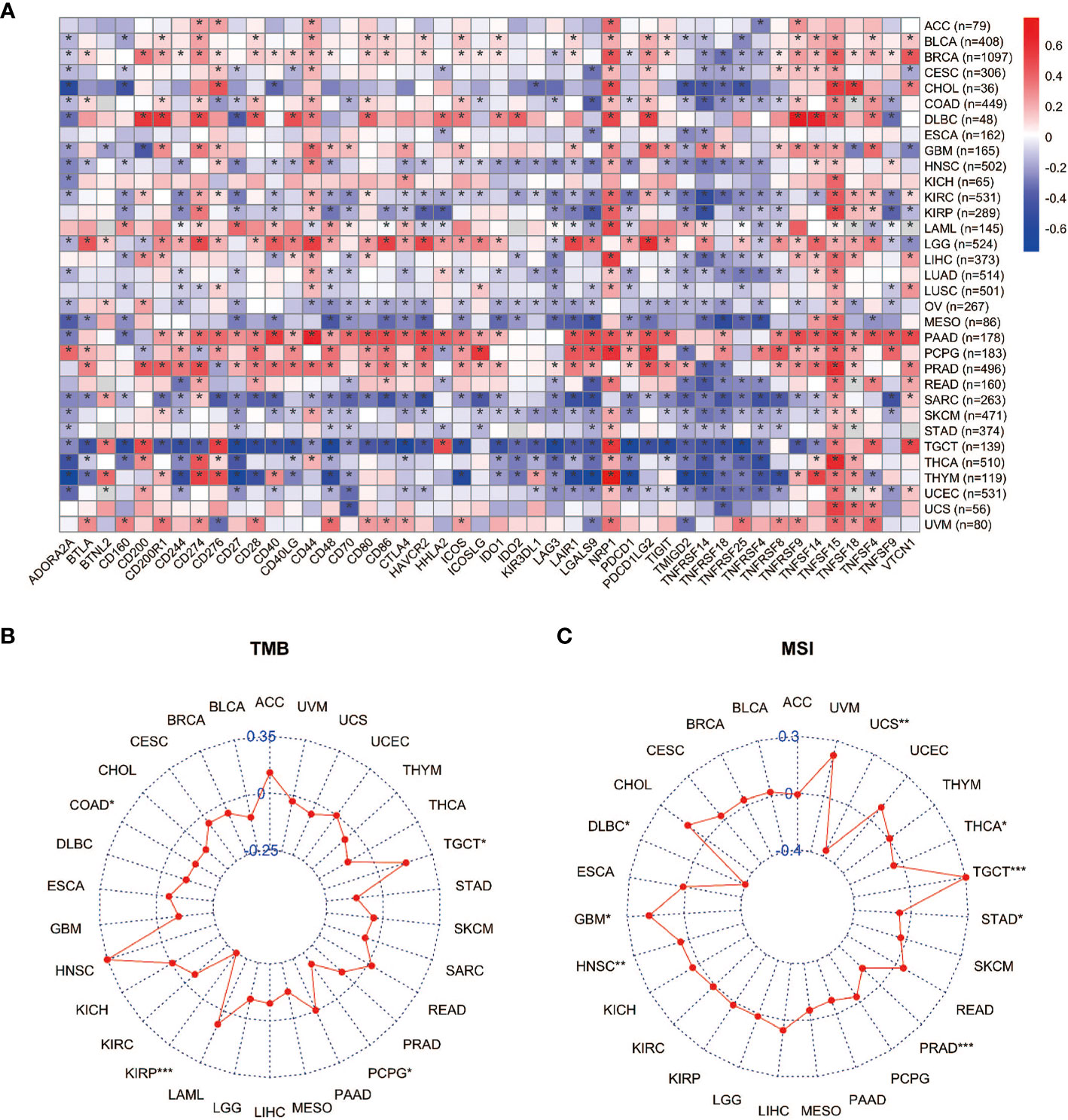
Figure 5 Correlation between expression of YAP1 and immune checkpoints, TMB, and MSI. (A) Heat map of correlation between YAP1 expression and 46 immune checkpoint regulators. *p < 0.05. (B, C) Correlation between YAP1 expression, TMB, and MSI. ***p < 0.001, **p < 0.01, and *p < 0.05.
YAP1-associated cancer hallmarks
Hallmark gene sets summarize and represent specific well-defined biological states or processes and display coherent expression. We subdivided patients into two groups for each cancer type based on YAP1 expression above or below the median level, and analyzed the differences in 50 hallmark gene sets across 33 cancer types. More than 23 hallmarks were aberrantly upregulated in LAML, LGG, and PCPG patients with high YAP1 expression versus those with low YAP1 expression. Various hallmark gene sets were upregulated in pan-cancer. TGF-β signaling pathway was upregulated in 23 cancer types, mitotic spindle process in 16 cancer types, Hedgehog signaling pathway in 15 cancer types, KRAS signaling in 8 cancer types, epithelial mesenchymal transition (EMT) in 10 cancer types, and angiogenesis in 7 cancer types, indicating that YAP1 is involved in promoting cell proliferation, cancer cell stemness, invasiveness, and migration processes (Figure 6). We then confirmed these results on CRC cell lines (Lovo and SW620). As expected, YAP1 was upregulated in AOM/DSS-induced colitis-associated cancer in C57BL/6J mice (Figure 8A). The YAP1 inhibitor verteporfin significantly reduced their cell viability (Figure 8B). Verteporfin also reduced expression of p-ERK (KRAS signaling pathway) and smad2 (TGF-β signaling pathway), and downregulated Wnt target genes AXIN2, BIRC5, CCND1, and CD44 (Figures 8C, D). In addition, upregulation of E-cadherin (epithelial maker) and downregulation of vimentin (mesenchymal marker) were observed in verteporfin treatment, suggesting a role of YAP1 in promoting EMT (Figure 8D). Of note, those results were based on enrolled cancerous samples. The hallmark difference between samples with higher and lower YAP1 expression is less than that between the samples with or without YAP1 function. Thus, the YAP1-associated hallmarks included but were not limited to those mentioned above
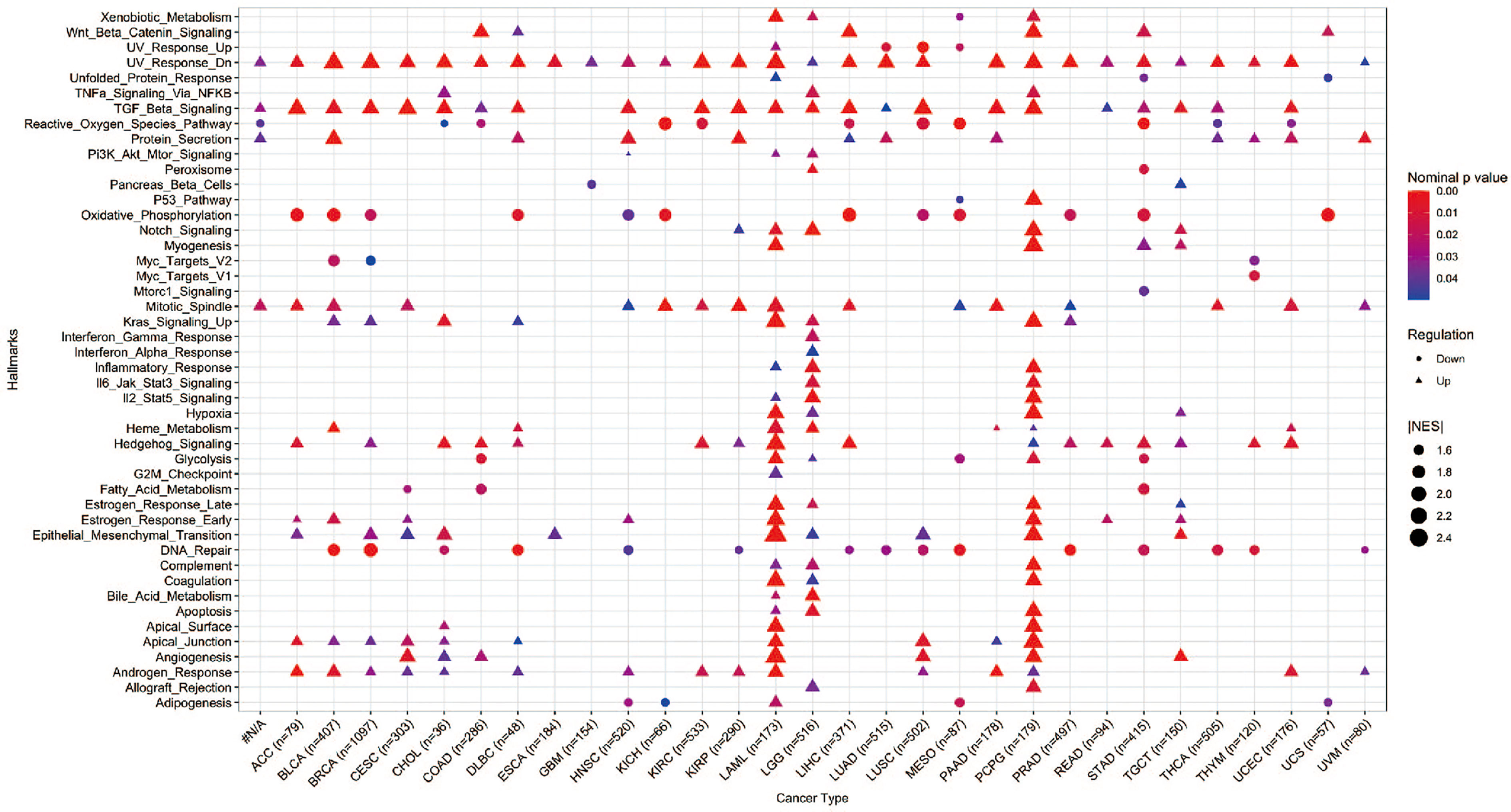
Figure 6 GSEA of hallmarks in cancer patients with high YAP1 expression versus those with low YAP1 expression. Only statistically significant hallmarks (p < 0.05) were shown.
Potential drugs for YAP1
We further screened potential drugs that may be effective in cancer patients with high YAP1 expression. We downloaded the z scores of IC50s of 24,360 drugs in 60 cancer cell lines. Only 218 FDA-approved drugs and 574 drugs in clinical trials were included for correlation analysis. Of the 218 FDA-approved drugs, chemotherapeutic agents that inhibit DNA synthesis (teniposide, dacarbazine, doxorubicin, triethylenemelamine, nitrogen mustard, etoposide, and thiotepa) topped the list, which might be effective in treating cancer patients with high YAP1 expression. Interestingly, an antipsychotic medication, fluphenazine, may also have an effect. Inhibitors of hypoxia-inducible factor (IDF-11774), MCL-1 (S63845, AZD-5991, pyridoclax, and S-64315), ribonucleotide reductase (imexon), FASN (JNJ-54302833), WNT signaling (CCT251545), STAT/STAT3 (CPD-401), and CHK (rabusertib) were among the top 10 drugs in clinical trial that may work in patients with high YAP1 expression, whereas inhibitors of EGFR (TAS6417), Bruton’s agammaglobulinemia tyrosine kinase (BTK) (spebrutinib), SYK (entospletinib), RET (Blu667), VEGFR2 (ENMD-2076, P-529), and αvβ3 integrin (MK-0429, cilengitide) may not work (Figure 7). We then treated Lovo and SW620 cell lines with AZD2858 and varlitinib alone or in combination with verteporfin. These results showed that the inhibitory rate of combined treatment versus verteporfin is smaller than that of AZD2858 or varlitinib alone versus control, indicating that the role of AZD2858 and varlitinib in cytotoxicity was partially dependent on YAP1 activity (Figure 8E). In addition, the combination of varlitinib and verteporfin was more effective than either single agent. KRAS is an effector molecule responsible for signal transduction from ligand-bound EGFR to the nucleus. Varlitinib is a targeted drug against EGFR. Dual inhibition of EGFR and YAP1 obtained better therapeutic outcomes, suggesting that the KRAS pathway was involved in YAP1 activity, as we analyzed in silico (Figure 6).
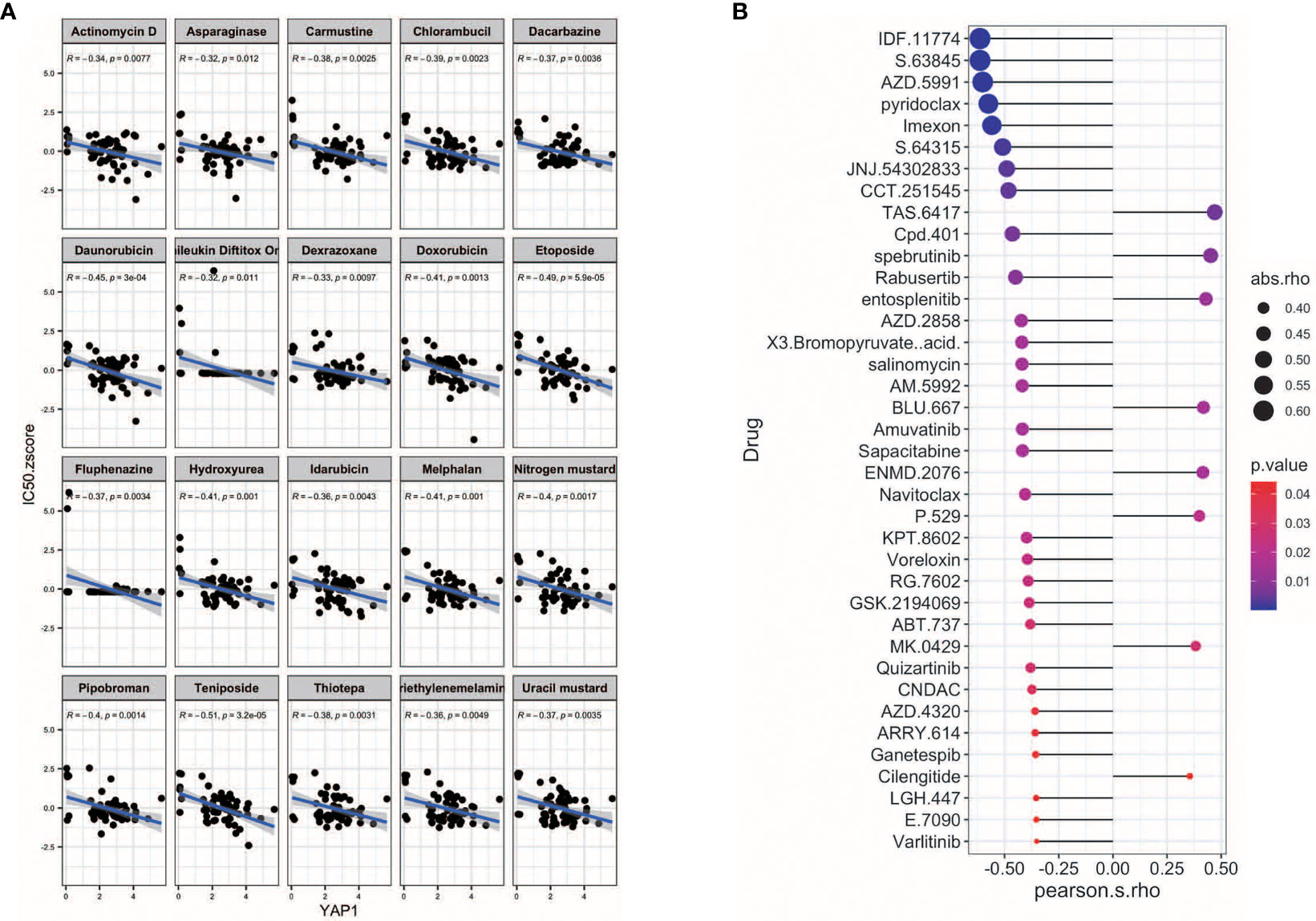
Figure 7 Potential drugs for YAP1. (A) Correlation of YAP1 expression with and IC50 z score of FDA-approved drugs (top 25). (B) Correlation of YAP1 expression and IC50 of drugs in clinical trial.
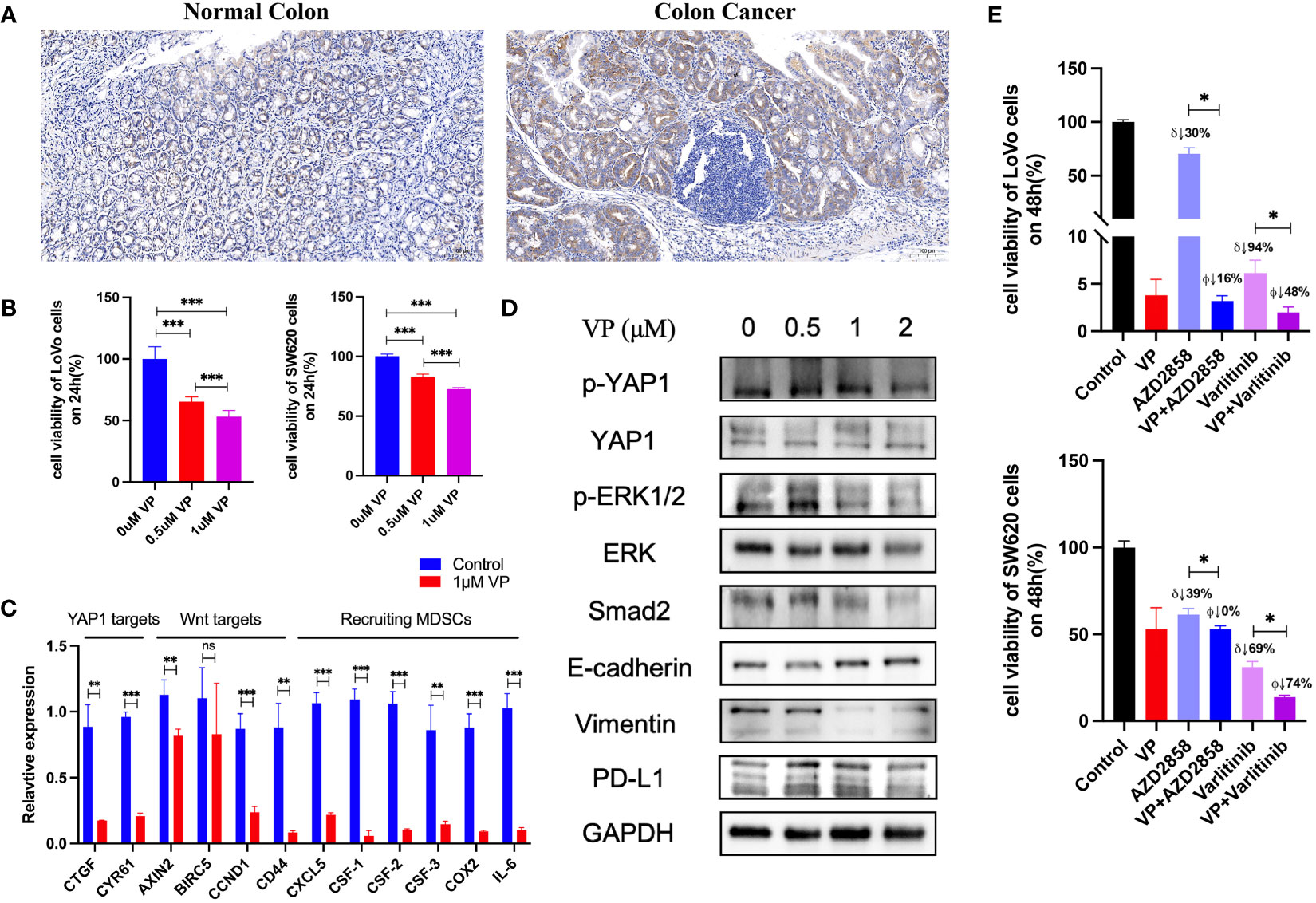
Figure 8 Experiments to confirm the activities of YAP1. (A) IHC staining of YAP1 in normal colon tissue and AOM/DSS-induced colitis-associated CRC in C57BL/6J mice. (B) Effect of YAP1 inhibitor verteporfin (VP) on the viability of CRC cells (24 h). (C) Regulation of YAP1 inhibitor on Wnt target genes and chemokines that recruit MDSCs’ infiltration in LoVo cell line. (D) YAP1 is involved in KRAS and TGF-β signaling pathways as well as EMT processes in Lovo cell line. (E) Effect of AZD2858 and varlitinib in combination with YAP1 inhibitor on the viability of CRC cells. δ: versus control group, ϕ versus verteporfin group. VP: 1 μM; AZD2858: 1 μM; varlitinib: 5 mM. ns (no significance), *p < 0.05, **p < 0.01, and ***p < 0.001.
Discussion
YAP1 is a transcriptional co-activator and a major effector of the Hippo signaling pathway. Emerging work indicates that YAP1 is widely activated in human malignancies and is essential for cancer initiation, progression, and drug resistance in most solid tumors. High expression of YAP1 has been reported to promote the excessive cell proliferation in multiple tissues, including liver, gastrointestinal tissue, skin, and heart (67–70). TEAD-dependent YAP1 function has also been linked to invasive and metastatic behavior of tumor cells (71). Compelling evidence also showed that YAP1 can alter the TME by recruiting immunosuppressive cell types, suppressing cytotoxic T-cell function or promoting the polarization of tumor-associated macrophages towards the pro-tumor M2 phenotype (72–74). In this study, we analyzed YAP1 expression in pan-cancer and evaluated its role in prognostic value, immunomodulation, and drug response.
According to the TCGA database, YAP1 expression was elevated in CHOL, COAD, and THCA, but decreased in 11 of the 23 cancer types, namely, BLCA, BRCA, HNSC, KICH, KIRC, KIRP, LUAD, LUSC, PCPG, PRAD, and UCEC (Figure 1). However, YAP1 has been reported to be oncogenic in BLCA (75, 76), BRCA (77), HNSC (78), KICH (79), KIRC (80), KIRP (81), LUAD (82), LUSC (83), PRAD (84), and UCEC (85). PCPG is a rare adrenal tumor. The role of YAP1 in PCPG remains to be investigated. The downregulation of YAP1 in those caners might be due to case number limitation, and in addition to YAP1 expression level, YAP1 activity also depends on its cellular location. High YAP1 expression is a risk factor for survival outcomes in ACC, BLCA, LGG, LUAD, PAAD, and COAD (Figure 3). Moreover, in ACC, BLCA, COAD, and TGCT, YAP1 was associated with more severe pathologic stages (Figure 3). Among them, the prognostic value of YAP1 on BLCA (84), LUAD (86), PAAD (87) and COAD (88) has been reported.
According to our result, high expression of YAP1 predicted poor survival outcomes in patients with LUAD, COAD, and PAAD, and was associated with more severe pathologic stages in patients with COAD, PAAD, and TGCT (Figure 3). PAAD has been reported to be characterized by immunosuppressive TME. In this study, YAP1 in PAAD was positively correlated with a wide range of immune checkpoint regulators. YAP1 expression was positively correlated with levels of TMB and MSI in TGCT. In addition, important immune cells, including activated CD4+ memory T cells and T follicular helper cells, were positively correlated with YAP1 expression in LUAD. Therefore, we would like to discuss in more detail the role of YAP1 in these cancer types.
Elevated expression of the gene signature for YAP1/TAZ activity is associated with poor prognosis in patients with non-small-cell lung cancer (NSCLC) (89, 90), which is consistent with the results of LUAD in our study (Figure 3). Our GSEA results suggested that high YAP1 expression was associated with reduced activities of DNA repair processes (Figure 6), and defective DNA repair contributes to individual susceptibility to lung cancer development (91). We also showed that TGF-β signaling was upregulated in LUAD with high YAP1 expression (Figure 6). TGF-β is the most potent inducer of EMT in NSCLC and is pivotal to the development of tumor-promoting microenvironment in the lung cancer tissues (92). In our findings, YAP1 may inhibit infiltration of CD8+ T cells, CD4+ Th1 cells, T follicular helper cells, and NKT cells, but increase infiltration of CAFs, which promote the development of LUAD. Zhang et al. have reported that YAP1 activation was not sufficient to trigger NSCLC formation, but promoted its progression to higher grades (89, 93). The role of YAP1 in immune cell infiltration in LUAD will be further experimentally confirmed in our future studies.
High level of YAP activity has been found to be prognostic for poor outcome in four datasets of CRC patients (94). In our study, YAP1 expression was associated with severe pathologic stage and poor survival outcome in COAD (Figure 3). YAP1/TAZ is required for the formation and growth of intestinal tumors (95). Among the enriched hallmarks, higher YAP1 expression was associated with upregulated Wnt β-catenin signaling, TGF-β signaling via NFKB, and Hedgehog signaling (Figure 6). Overactivation of the Wnt signaling pathway is the most oncogenic pathway in CRC. In addition, β-catenin-driven cancers require the YAP1 transcriptional complex for tumorigenesis (96), suggesting the network between the two signaling pathways. Barry et al. reported that forced YAP1 overexpression in the gut epithelium was not sufficient to form tumor (95). The lack of Wnt signaling pathway might be the reason. In COAD, YAP1 expression was positively correlated with the infiltration of MDSCs and CAFs, and was negatively correlated with the infiltration of CD8+ T cells, CD4+ Th1 cells, activated NK cells, and NKT cells (Figure 4). Mechanically, YAP1 promotes MDSC induction by inhibiting PTEN expression, resulting in upregulation of COX-2, p-AKT, and p-p65 in CRC-derived cells, which leads to secretion of the cytokine granulocyte-macrophage colony-stimulating factor (97). YAP1-dependent matrix remodeling is required for the generation and maintenance of CAFs (98). Mechanisms of YAP1 in recruiting CAFs and suppressing CD4+ Th1 cells, T follicular helper cells, activated NK cells, and NKT cells are yet to be unraveled.
PAAD is a very aggressive neoplasia that seems to arise from pancreatic exocrine cells. Pancreas-specific YAP1 knockout halted tumor progression (99). Activation of YAP1 in acinar cells upregulated JAK-STAT3 signaling and promoted the development of pancreatic cancer (100). In this study, YAP1 expression was higher in PAAD patients with more severe pathological stages (Figure 3). Mechanically, YAP1 and the ZEB1 complex activates ITGA3 to promote its metastasis (101). PAAD is characterized by immunosuppressive TME. In our results, YAP1 expression was positively correlated with MDSC infiltration and negatively correlated with NKT cells. Mielgo et al. reported that YAP1 recruited MDSCs to suppress T-cell function and generate an immunosuppressive microenvironment (102). The mechanism remains to be clarified.
Testicular cancer is a very common malignancy in young men. Although testicular cancer has a high cure rate, patients have a high long-term risk of secondary malignant tumors. Ji et al. have reported that different immune status in TME may be responsible for the different survival outcomes of TGCT patients (103). Moreover, YAP1 inhibition enhanced the chemosensitivity of cisplatin in TGCT (104). Studies about the YAP1 in immunomodulation in TGCT remain absent.
In this study, from a pan-cancer perspective, high expression of YAP1 was associated with upregulation of TGF-β signaling, KRAS signaling, Hedgehog signaling, EMT, and androgen response in most cancer types (Figure 6). There was a close interplay between YAP1 and TGF-β signaling. YAP1 activation promoted TGF-β expression, which was involved in the biological processes of endothelial-to-mesenchymal transition (EMT) and fibrosis (105–107) in the liver, lung, and kidney. Their interactions in other organ tissues need to be investigated. We confirmed that YAP1 was able to upregulate smad2 in TGF-β signaling in CRC cell lines (Figure 8). KRAS and YAP1 converged on the transcription factor FOS and activated a transcriptional program involved in regulating EMT (108). However, another study found limited overlap of gene expression between KRAS G12V and YAP1 S127A-driven tumors (109). The interplay between YAP1 and KRAS signaling remains largely unknown. In this study, YAP1 inhibitor reduced the KRAS pathway activity in CRC (Figure 8). Hedgehog signaling activation could upregulate YAP1 expression and induce osteosarcoma development (110), or aid in generation of liver (111). The cooperation between Hedgehog and YAP1 signaling in tumor formation and progression remains to be uncovered. YAP1 mRNA was upregulated in androgen-insensitive prostate cancer cells (112), but the mechanism of their interaction is largely unknown. Furthermore, in our findings, the YAP1 inhibitor did reduce EMT markers in CRC.
Emerging evidence demonstrated the role of YAP1 in modulating TME. In this study, we found that YAP1 may suppress the infiltration of CD8+ T lymphocytes, γδ T cells, T follicular helper cells, NKT cells, and activated NK cells. Moreover, YAP1 may recruit the CAFs and MDSCs to tumor site to suppress the immune response. Among them, the relationship between YAP1 and infiltration of CD8+ T lymphocytes, MDSCs, and CAFs has been reported in several cancer types. However, the mechanisms are incomplete. The role of YAP1 in the infiltration and activities of T follicular helper cells, NKT cells, and activated NK cells remains to be investigated. Stampouloglou et al. reported that YAP1 overexpression in T cells reduced their activation, differentiation, and function, which translated in vivo into an impairment of T-cell infiltration and tumor repression (113). Another study confirmed that YAP1 attenuated CD8+ T cell-mediated anti-tumor response (114). Mechanically, YAP1 overexpression in cancer cells could upregulate PD-L1 expression and impede the activities of CD8+ T cells in melanoma (115). Another mechanism is that YAP1/TEAD directly upregulates CXCL5 in cancer cells to recruit CXCR2-expressing MDSCs, leading to decreased infiltration of CD8+ T cells (74). In addition, IL-6 and CSF1-3 induced by YAP1 in PAAD stimulated the accumulation of MDSCs (116), and upregulation of COX2 by YAP1 in human granulosa cells promoted the recruitment of MDSCs (117). The role of YAP1 in CD8+ T cells and MDSCs in other cancer types remains to be unraveled. In this study, we confirmed the results by experiments on CRC cells and found that the YAP1 inhibitor verteporfin decreased the expression of PD-L1, CXCL5, COX-2, IL-6, and CSF1-3 (Figure 8). Mechanotransduction-mediated YAP1 activation establishes a feed-forward self-reinforcing loop that contributes to maintenance of the CAF phenotype and promotes breast cancer invasion (98). Mechanistically, active YAP1 promotes the expression of ANLN and DIAPH3 and stabilizes actomyosin proteins, which is required for the generation and maintenance of CAFs (98). Another mechanism is that high expression of YAP1 in the tumor stromal cells converts normal fibroblasts into CAFs in the TME of prostate cancer (118). The function of YAP1 in CAFs of other cancer types remains largely unknown. In this study, high expression of YAP1 was associated with TGF-β signaling in pan-cancer. TGF-β-associated extracellular matrix genes link CAFs to immune evasion and immunotherapy failure (57). Therefore, YAP1 may be involved in the function of CAF through TGF-β signaling. We found that YAP1 inhibitor verteporfin reduced smad2 expression in CRC cell lines, suggesting a downregulation of TGF-β signaling (Figure 8). The role of YAP1 in the infiltration and activity of T follicular helper cells, NKT cells, and activated NK cells remains to be investigated. Another interesting result in this study is that YAP1 expression was negatively correlated with infiltration of CD8+ T cells, but positively correlated with infiltration of resting CD4+ memory T cells in most cancer types. This might not be due to the direct relationship between these two cells. YAP1 expression was also negatively correlated with infiltration of activated CD4+ memory T cells, suggesting that YAP1 may impede the activation of memory CD4+ T cells. CD4+ T cells are required for survival of CD8+ T cells during both primary and memory recall responses (119). Therefore, the possible relationship is that YAP1 may impair the activation of memory CD4+ T cells, and impede the survival of CD8 T cells. Further experimental studies on CD4+ T cells will be conducted in the near future to verify the role of YAP1 in memory CD4+ T-cell activation.
Further analyses showed that YAP1 expression was positively correlated to a wide range of immune checkpoints, especially in PAAD, PCPG, and PRAD, suggesting that YAP1 is a potential new drug target for anti-cancer immunotherapy in these cancer types. In PAAD, PRAD, and LGG, YAP1 expression was positively correlated with the expression of CTLA-4, TIM-3 (HAVCR2), and PD-1 (PDCD1), which are key checkpoint regulators that suppress immune reaction. In most cancer types, CD274 (PD-L1), NRP1, and TNFSF15 were positively correlated with YAP1 expression. YAP1 has been reported to induce PD-L1 expression (115). Apart from PD-L1, YAP1 might also regulate the expression of other immune checkpoints. The relationship between these immune checkpoints and YAP1 and the role of their interactions in TME regulation remain to be investigated. These checkpoint regulators may play key roles in YAP1-induced immunosuppressive TME. In PAAD, PRAD, and PCPG, YAP1 expression was highly correlated with the expression of most immune checkpoint genes in our result. Among them, PAAD has been reported to be characterized by the immunosuppressive microenvironment, suggesting a role for YAP1 in PAAD development (102).
Methylation of DNA cytosine bases leads to the inaccessibility of DNA regulatory elements to their transcription factors through a number of mechanisms, leading to the gene transcription shutdown. We found that YAP1 expression was negatively regulated by its DNA methylation in the 5’UTR. Cellular RNAs are naturally decorated with a variety of chemical modifications, which affect the mRNA stability and translation. In our study, extensive modification “effectors,” including enzymes of “writers” and “erasers” that alter the modification level and binding proteins of “readers” that recognize the chemical marks, were positively correlated with YAP1 mRNA level. Further research should be conducted to study whether aberrant DNA methylation and RNA modifications of YAP1 are involved in cancer development and how they work.
FDA-approved chemotherapeutic drugs that are capable of inhibiting DNA synthesis, including teniposide, dacarbazine, doxorubicin, and triethylenemelamine, and inhibitors of hypoxia-inducible factor, MCL-1, ribonucleotide reductase, and FASN in clinical trials are potential drugs to treat cancer patients with high YAP1 expression. Among them, the cytotoxicity effect of AZD2858 and varlitinib was partially attributed to YAP1 activity.
Conclusion
YAP1 was aberrantly expressed in various cancer types and regulated by its DNA methylation and post-transcriptional modifications. High expression of YAP1 was associated with poor survival outcomes in ACC, BLCA, LGG, LUAD, and PAAD. YAP1 may promote tumor progression through immunosuppression, particularly by suppressing the infiltration of CD8+ T lymphocytes, CD4+ Th1 cells, T follicular helper cells, NKT cells, and activated NK cells, as well as recruiting MDSCs and CAFs in pan-cancer. The YAP1-promoting tumor activity is probably attributed to the activation of TGF-β, Hedgehog, or KRAS signaling pathways. AZD2858 and varlitinib in clinical trials might be effective in cancer patients with high YAP1 expression.
Data availability statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Author contributions
XH contributed to designing the article structure, writing—original draft, editing the figures and funding acquisition. YRZ contributed to performing experiments, editing the figures, and writing—review. HY contributed to performing experiments. YYZ and XS contributed to formal analysis. QL contributed to funding acquisition. YW contributed to supervision, funding acquisition, and project administration. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (81904130, 82122075, and 82074232), the Project of Shanghai Science and Technology Committee (19YF1449900), the Shanghai Frontier Research Base of Disease and Syndrome Biology of Inflammatory Cancer Transformation (2021KJ03–12), the “Shu Gguang” project supported by Shanghai Municipal Education Commission and Shanghai Education Development Foundation (21SG43), the Clinical Research Plan of SHDC (SHDC2020CR4043), and the Shanghai Youth Talent Support Program.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.1012173/full#supplementary-material
Supplementary Figure 1 | Transcriptional level of TEAD1, TEAD3 and TEAD4 in normal and cancer tissues according to RNA-sequence data from TCGA. ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05, ns p > 0.05.
Glossary

References
1. Yildirim E, Bora G, Onel T, Talas N, Yaba A. Cell fate determination and hippo signaling pathway in preimplantation mouse embryo. Cell Tissue Res (2021) 386(3):423–44. doi: 10.1007/s00441-021-03530-8
2. Davis JR, Tapon N. Hippo signalling during development. Development (2019) 146(18). doi: 10.1242/dev.167106
3. Wang J, Liu S, Heallen T, Martin JF. The hippo pathway in the heart: Pivotal roles in development, disease, and regeneration. Nat Rev Cardiol (2018) 15(11):672–84. doi: 10.1038/s41569-018-0063-3
4. Boopathy GTK, Hong W. Role of hippo pathway-Yap/Taz signaling in angiogenesis. Front Cell Dev Biol (2019) 7:49. doi: 10.3389/fcell.2019.00049
5. Calses PC, Crawford JJ, Lill JR, Dey A. Hippo pathway in cancer: Aberrant regulation and therapeutic opportunities. Trends Cancer (2019) 5(5):297–307. doi: 10.1016/j.trecan.2019.04.001
6. Mo JS, Park HW, Guan KL. The hippo signaling pathway in stem cell biology and cancer. EMBO Rep (2014) 15(6):642–56. doi: 10.15252/embr.201438638
7. Pan D. The hippo signaling pathway in development and cancer. Dev Cell (2010) 19(4):491–505. doi: 10.1016/j.devcel.2010.09.011
8. Zhao B, Lei QY, Guan KL. The hippo-yap pathway: New connections between regulation of organ size and cancer. Curr Opin Cell Biol (2008) 20(6):638–46. doi: 10.1016/j.ceb.2008.10.001
9. Luo J, Li P. Context-dependent transcriptional regulations of Yap/Taz in stem cell and differentiation. Stem Cell Res Ther (2022) 13(1):10. doi: 10.1186/s13287-021-02686-y
10. Beyer TA, Weiss A, Khomchuk Y, Huang K, Ogunjimi AA, Varelas X, et al. Switch enhancers interpret tgf-beta and hippo signaling to control cell fate in human embryonic stem cells. Cell Rep (2013) 5(6):1611–24. doi: 10.1016/j.celrep.2013.11.021
11. Croci O, De Fazio S, Biagioni F, Donato E, Caganova M, Curti L, et al. Transcriptional integration of mitogenic and mechanical signals by myc and yap. Genes Dev (2017) 31(20):2017–22. doi: 10.1101/gad.301184.117
12. Kapoor A, Yao W, Ying H, Hua S, Liewen A, Wang Q, et al. Yap1 activation enables bypass of oncogenic kras addiction in pancreatic cancer. Cell (2014) 158(1):185–97. doi: 10.1016/j.cell.2014.06.003
13. Lehmann W, Mossmann D, Kleemann J, Mock K, Meisinger C, Brummer T, et al. Zeb1 turns into a transcriptional activator by interacting with Yap1 in aggressive cancer types. Nat Commun (2016) 7:10498. doi: 10.1038/ncomms10498
14. Tao G, Kahr PC, Morikawa Y, Zhang M, Rahmani M, Heallen TR, et al. Pitx2 promotes heart repair by activating the antioxidant response after cardiac injury. Nature (2016) 534(7605):119–23. doi: 10.1038/nature17959
15. Zanconato F, Forcato M, Battilana G, Azzolin L, Quaranta E, Bodega B, et al. Genome-wide association between Yap/Taz/Tead and ap-1 at enhancers drives oncogenic growth. Nat Cell Biol (2015) 17(9):1218–27. doi: 10.1038/ncb3216
16. Totaro A, Panciera T, Piccolo S. Yap/Taz upstream signals and downstream responses. Nat Cell Biol (2018) 20(8):888–99. doi: 10.1038/s41556-018-0142-z
17. Pei T, Li Y, Wang J, Wang H, Liang Y, Shi H, et al. Yap is a critical oncogene in human cholangiocarcinoma. Oncotarget (2015) 6(19):17206–20. doi: 10.18632/oncotarget.4043
18. Huang CY, Han Z, Li X, Xie HH, Zhu SS. Inhibition of thyroid carcinoma cells with Yap1 protein interference and its mechanism. Eur Rev Med Pharmacol Sci (2018) 22(8):2351–5. doi: 10.26355/eurrev_201804_14826
19. Sinclear CK, Maruyama J, Nagashima S, Arimoto-Matsuzaki K, Kuleape JA, Iwasa H, et al. Protein kinase calpha activation switches Yap1 from tead-mediated signaling to P73-mediated signaling. Cancer Sci (2022) 113(4):1305–20. doi: 10.1111/cas.15285
20. Muhammad JS, Guimei M, Jayakumar MN, Shafarin J, Janeeh AS, AbuJabal R, et al. Estrogen-induced hypomethylation and overexpression of Yap1 facilitate breast cancer cell growth and survival. Neoplasia (2021) 23(1):68–79. doi: 10.1016/j.neo.2020.11.002
21. Bai Z, Wu Q, Zhang C, Chen J, Cao L. Effects of Yap1 and Sfrp2 overexpression on the biological behavior of colorectal cancer cells and their molecular mechanisms. J Gastrointest Oncol (2021) 12(4):1601–12. doi: 10.21037/jgo-21-418
22. Dai Y, Liu S, Zhang WQ, Yang YL, Hang P, Wang H, et al. Yap1 regulates Abcg2 and cancer cell side population in human lung cancer cells. Oncotarget (2017) 8(3):4096–109. doi: 10.18632/oncotarget.13686
23. He Q, Lin ZY, Wang ZH, Huang WJ, Tian D, Liu M, et al. Six4 promotes hepatocellular carcinoma metastasis through upregulating Yap1 and c-met. Oncogene (2020) 39(50):7279–95. doi: 10.1038/s41388-020-01500-y
24. Hoxha S, Shepard A, Troutman S, Diao H, Doherty JR, Janiszewska M, et al. Yap-mediated recruitment of Yy1 and Ezh2 represses transcription of key cell-cycle regulators. Cancer Res (2020) 80(12):2512–22. doi: 10.1158/0008-5472.CAN-19-2415
25. Kim M, Kim T, Johnson RL, Lim DS. Transcriptional Co-repressor function of the hippo pathway transducers yap and taz. Cell Rep (2015) 11(2):270–82. doi: 10.1016/j.celrep.2015.03.015
26. Chen YP, Yin JH, Li WF, Li HJ, Chen DP, Zhang CJ, et al. Single-cell transcriptomics reveals regulators underlying immune cell diversity and immune subtypes associated with prognosis in nasopharyngeal carcinoma. Cell Res (2020) 30(11):1024–42. doi: 10.1038/s41422-020-0374-x
27. Zhang Y, Zhang H, Zhao B. Hippo signaling in the immune system. Trends Biochem Sci (2018) 43(2):77–80. doi: 10.1016/j.tibs.2017.11.009
28. Matthaios D, Tolia M, Mauri D, Kamposioras K, Karamouzis M. Yap/Hippo pathway and cancer immunity: It takes two to tango. Biomedicines (2021) 9(12):1949. doi: 10.3390/biomedicines9121949
29. Song S, Xie M, Scott AW, Jin J, Ma L, Dong X, et al. A novel Yap1 inhibitor targets csc-enriched radiation-resistant cells and exerts strong antitumor activity in esophageal adenocarcinoma. Mol Cancer Ther (2018) 17(2):443–54. doi: 10.1158/1535-7163.MCT-17-0560
30. Barsyte-Lovejoy D, Li F, Oudhoff MJ, Tatlock JH, Dong A, Zeng H, et al. (R)-Pfi-2 is a potent and selective inhibitor of Setd7 methyltransferase activity in cells. Proc Natl Acad Sci U.S.A. (2014) 111(35):12853–8. doi: 10.1073/pnas.1407358111
31. Scott LJ, Goa KL. Verteporfin. Drugs Aging (2000) 16(2):139–46. doi: 10.2165/00002512-200016020-00005
32. Jiao S, Wang H, Shi Z, Dong A, Zhang W, Song X, et al. A peptide mimicking Vgll4 function acts as a yap antagonist therapy against gastric cancer. Cancer Cell (2014) 25(2):166–80. doi: 10.1016/j.ccr.2014.01.010
33. Kawamoto R, Nakano N, Ishikawa H, Tashiro E, Nagano W, Sano K, et al. Narciclasine is a novel yap inhibitor that disturbs interaction between yap and Tead4. BBA Adv (2021) 1:100008. doi: 10.1016/j.bbadva.2021.100008
34. Zhang Z, Lin Z, Zhou Z, Shen HC, Yan SF, Mayweg AV, et al. Structure-based design and synthesis of potent cyclic peptides inhibiting the yap-tead protein-protein interaction. ACS Med Chem Lett (2014) 5(9):993–8. doi: 10.1021/ml500160m
35. Kaneda A, Seike T, Danjo T, Nakajima T, Otsubo N, Yamaguchi D, et al. The novel potent tead inhibitor, K-975, inhibits Yap1/Taz-tead protein-protein interactions and exerts an anti-tumor effect on malignant pleural mesothelioma. Am J Cancer Res (2020) 10(12):4399–415.
36. Yu J, Alharbi A, Shan H, Hao Y, Snetsinger B, Rauh MJ, et al. Taz induces lung cancer stem cell properties and tumorigenesis by up-regulating Aldh1a1. Oncotarget (2017) 8(24):38426–43. doi: 10.18632/oncotarget.16430
37. Guerrant W, Kota S, Troutman S, Mandati V, Fallahi M, Stemmer-Rachamimov A, et al. Yap mediates tumorigenesis in neurofibromatosis type 2 by promoting cell survival and proliferation through a cox-2-Egfr signaling axis. Cancer Res (2016) 76(12):3507–19. doi: 10.1158/0008-5472.CAN-15-1144
38. Ghiso E, Migliore C, Ciciriello V, Morando E, Petrelli A, Corso S, et al. Yap-dependent axl overexpression mediates resistance to egfr inhibitors in nsclc. Neoplasia (2017) 19(12):1012–21. doi: 10.1016/j.neo.2017.10.003
39. Ji X, Song L, Sheng L, Gao A, Zhao Y, Han S, et al. Cyclopeptide Ra-V inhibits organ enlargement and tumorigenesis induced by yap activation. Cancers (Basel) (2018) 10(11):449. doi: 10.3390/cancers10110449
40. Lin L, Sabnis AJ, Chan E, Olivas V, Cade L, Pazarentzos E, et al. The hippo effector yap promotes resistance to raf- and mek-targeted cancer therapies. Nat Genet (2015) 47(3):250–6. doi: 10.1038/ng.3218
41. Eisinger-Mathason TS, Mucaj V, Biju KM, Nakazawa MS, Gohil M, Cash TP, et al. Deregulation of the hippo pathway in soft-tissue sarcoma promotes Foxm1 expression and tumorigenesis. Proc Natl Acad Sci U.S.A. (2015) 112(26):E3402–11. doi: 10.1073/pnas.1420005112
42. Fisher ML, Kerr C, Adhikary G, Grun D, Xu W, Keillor JW, et al. Transglutaminase interaction with Alpha6/Beta4-integrin stimulates Yap1-dependent Deltanp63alpha stabilization and leads to enhanced cancer stem cell survival and tumor formation. Cancer Res (2016) 76(24):7265–76. doi: 10.1158/0008-5472.CAN-16-2032
43. Liu CY, Pobbati AV, Huang ZY, Cui L, Hong WJ. Transglutaminase 2 is a direct target gene of Yap/Taz-letter. Cancer Res (2017) 77(17):4734–5. doi: 10.1158/0008-5472.Can-16-3535
44. Han D, Lee SM, Kwon M, Noh H, Lee JH, Yoon Y, et al. Yap enhances Fgf2-dependent neural stem cell proliferation by induction of fgf receptor expression. Stem Cells Dev (2020) 29(18):1240–6. doi: 10.1089/scd.2019.0281
45. Tomczak K, Czerwinska P, Wiznerowicz M. The cancer genome atlas (Tcga): An immeasurable source of knowledge. Contemp Oncol (Pozn) (2015) 19(1A):A68–77. doi: 10.5114/wo.2014.47136
46. Li N, Zhan X. Machine learning identifies pan-cancer landscape of Nrf2 oxidative stress response pathway-related genes. Oxid Med Cell Longev (2022) 2022:8450087. doi: 10.1155/2022/8450087
47. Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods (2015) 12(5):453–7. doi: 10.1038/nmeth.3337
48. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. Timer: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res (2017) 77(21):e108–e10. doi: 10.1158/0008-5472.CAN-17-0307
49. Racle J, de Jonge K, Baumgaertner P, Speiser DE, Gfeller D. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. Elife (2017) 6:e26476. doi: 10.7554/eLife.26476
50. Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun (2013) 4:2612. doi: 10.1038/ncomms3612
51. Zhou Y, Feng Y, Cen R, Hou X, Yu H, Sun J, et al. San-Wu-Huang-Qin decoction attenuates tumorigenesis and mucosal barrier impairment in the Aom/Dss model by targeting gut microbiome. Phytomedicine (2022) 98:153966. doi: 10.1016/j.phymed.2022.153966
52. Shao Q, Xu J, Deng R, Wei W, Zhou B, Yue C, et al. The expressions of Yap1, beta-catenin and survivin in colon cancer tissues and their clinical significance. Int J Clin Exp Pathol (2018) 11(12):6032–8.
53. Xia Y, Chang T, Wang Y, Liu Y, Li W, Li M, et al. YAP promotes ovarian cancer cell tumorigenesis and is indicative of a poor prognosis for ovarian cancer patients. PloS One (2014) 9(3):e91770. doi: 10.1371/journal.pone.0091770
54. Giraud J, Molina-Castro S, Seeneevassen L, Sifré E, Izotte J, Tiffon C, et al. Verteporfin targeting YAP1/TAZ-TEAD transcriptional activity inhibits the tumorigenic properties of gastric cancer stem cells. Int J Cancer (2020) 146(8):2255–67. doi: 10.1002/ijc.32667
55. Tsinias G, Nikou S, Mastronikolis N, Bravou V, Papadaki H. Expression and prognostic significance of YAP, TAZ, TEAD4 and p73 in human laryngeal cancer. Histol Histopathol (2020) 35(9):983–95. doi: 10.14670/HH-18-228
56. Strano S, Munarriz E, Rossi M, Castagnoli L, Shaul Y, Sacchi A, et al. Physical interaction with yes-associated protein enhances p73 transcriptional activity. J Biol Chem (2001) 276(18):15164–73. doi: 10.1074/jbc.M010484200
57. Sun Z, Ou C, Liu J, Chen C, Zhou Q, Yang S, et al. Yap1-induced Malat1 promotes epithelial-mesenchymal transition and angiogenesis by sponging mir-126-5p in colorectal cancer. Oncogene (2019) 38(14):2627–44. doi: 10.1038/s41388-018-0628-y
58. Huang SS, Li YQ, Hu JH, Li L, Liu Z, Guo H, et al. Lncrna Pwar6 regulates proliferation and migration by epigenetically silencing Yap1 in tumorigenesis of pancreatic ductal adenocarcinoma. J Cell Mol Med (2021) 25(9):4275–86. doi: 10.1111/jcmm.16480
59. Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer (2012) 12(4):252–64. doi: 10.1038/nrc3239
60. Chan TA, Yarchoan M, Jaffee E, Swanton C, Quezada SA, Stenzinger A, et al. Development of tumor mutation burden as an immunotherapy biomarker: Utility for the oncology clinic. Ann Oncol (2019) 30(1):44–56. doi: 10.1093/annonc/mdy495
61. Brown SD, Warren RL, Gibb EA, Martin SD, Spinelli JJ, Nelson BH, et al. Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome Res (2014) 24(5):743–50. doi: 10.1101/gr.165985.113
62. Choucair K, Morand S, Stanbery L, Edelman G, Dworkin L, Nemunaitis J. Tmb: A promising immune-response biomarker, and potential spearhead in advancing targeted therapy trials. Cancer Gene Ther (2020) 27(12):841–53. doi: 10.1038/s41417-020-0174-y
63. McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science (2016) 351(6280):1463–9. doi: 10.1126/science.aaf1490
64. Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science (2015) 348(6230):69–74. doi: 10.1126/science.aaa4971
65. Cohen R, Rousseau B, Vidal J, Colle R, Diaz LA, Andre T. Immune checkpoint inhibition in colorectal cancer: Microsatellite instability and beyond. Target Oncol (2020) 15(1):11–24. doi: 10.1007/s11523-019-00690-0
66. Picard E, Verschoor CP, Ma GW, Pawelec G. Relationships between immune landscapes, genetic subtypes and responses to immunotherapy in colorectal cancer. Front Immunol (2020) 11:369. doi: 10.3389/fimmu.2020.00369
67. Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, et al. Yap1 increases organ size and expands undifferentiated progenitor cells. Curr Biol (2007) 17(23):2094. doi: 10.1016/j.cub.2007.11.016
68. Dong JX, Feldmann G, Huang JB, Wu S, Zhang NL, Comerford SA, et al. Elucidation of a universal size-control mechanism in drosophila and mammals. Cell (2007) 130(6):1120–33. doi: 10.1016/j.cell.2007.07.019
69. Heallen T, Zhang M, Wang J, Bonilla-Claudio M, Klysik E, Johnson RL, et al. Hippo pathway inhibits wnt signaling to restrain cardiomyocyte proliferation and heart size. Science (2011) 332(6028):458–61. doi: 10.1126/science.1199010
70. Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, Rodriguez JR, Zhou DW, et al. Yap1 acts downstream of alpha-catenin to control epidermal proliferation. Cell (2011) 144(5):782–95. doi: 10.1016/j.cell.2011.02.031
71. Lamar JM, Stern P, Liu H, Schindler JW, Jiang ZG, Hynes RO. The hippo pathway target, yap, promotes metastasis through its tead-interaction domain. P Natl Acad Sci USA (2012) 109(37):E2441–E50. doi: 10.1073/pnas.1212021109
72. Austenaa L, Natoli G. A shortcut for early macrophage recruitment into tumors by activated oncogenes. Gene Dev (2017) 31(3):223–5. doi: 10.1101/gad.296905.117
73. Sarkar S, Bristow CA, Dey P, Rai K, Perets R, Ramirez-Cardenas A, et al. Prkci promotes immune suppression in ovarian cancer. Gene Dev (2017) 31(11):1109–21. doi: 10.1101/gad.296640.117
74. Wang GC, Lu X, Dey P, Deng PN, Wu CC, Jiang S, et al. Targeting yap-dependent mdsc infiltration impairs tumor progression. Cancer Discovery (2016) 6(1):80–95. doi: 10.1158/2159-8290.Cd-15-0224
75. Chen XX, Li S, Yu Z, Tan WL. Yes-associated protein 1 promotes bladder cancer invasion by regulating epithelial-mesenchymal transition. Int J Clin Exp Patho (2019) 12(3):1070–+.
76. Li S, Yu Z, Chen SS, Li F, Lei CY, Chen XX, et al. The Yap1 oncogene contributes to bladder cancer cell proliferation and migration by regulating the H19 long noncoding rna. Urol Oncol-Semin Ori (2015) 33(10):427.e1-10. doi: 10.1016/j.urolonc.2015.06.003
77. Qadir J, Riaz SK, Taj K, Sattar N, Sahar NE, Khan JS, et al. Increased Yap1 expression is significantly associated with breast cancer progression, metastasis and poor survival. Future Oncol (2021) 17(21):2725–34. doi: 10.2217/fon-2020-1080
78. Omori H, Sato K, Nakano T, Wakasaki T, Toh S, Taguchi K, et al. Stress-triggered Yap1/Sox2 activation transcriptionally reprograms head and neck squamous cell carcinoma for the acquisition of stemness. J Cancer Res Clin (2019) 145(10):2433–44. doi: 10.1007/s00432-019-02995-z
79. Xiao JT, Shi Q, Li WG, Mu XY, Peng JT, Li MZ, et al. Arrdc1 and Arrdc3 act as tumor suppressors in renal cell carcinoma by facilitating Yap1 degradation. Am J Cancer Res (2018) 8(1):132–+.
80. Rybarczyk A, Klacz J, Wronska A, Matuszewski M, Kmieci Z, Wierzbicki PM. Overexpression of the Yap1 oncogene in clear cell renal cell carcinoma is associated with poor outcome. Oncol Rep (2017) 38(1):427–39. doi: 10.3892/or.2017.5642
81. Sourbier C, Liao PJ, Ricketts CJ, Wei D, Yang Y, Baranes SM, et al. Targeting loss of the hippo signaling pathway in Nf2-deficient papillary kidney cancers. Oncotarget (2018) 9(12):10723–33. doi: 10.18632/oncotarget.24112
82. Pu X, Wang J, Li W, Fan W, Wang L, Mao Y, et al. Copb2 promotes cell proliferation and tumorigenesis through up-regulating Yap1 expression in lung adenocarcinoma cells. BioMed Pharmacother (2018) 103:373–80. doi: 10.1016/j.biopha.2018.04.006
83. Huang H, Zhang W, Pan Y, Gao Y, Deng L, Li F, et al. Yap suppresses lung squamous cell carcinoma progression Via deregulation of the Dnp63-Gpx2 axis and ros accumulation. Cancer Res (2017) 77(21):5769–81. doi: 10.1158/0008-5472.CAN-17-0449
84. Cheng SY, Prieto-Dominguez N, Yang S, Connelly ZM, StPierre S, Rushing B, et al. The expression of Yap1 is increased in high-grade prostatic adenocarcinoma but is reduced in neuroendocrine prostate cancer. Prostate Cancer P D (2020) 23(4):661–9. doi: 10.1038/s41391-020-0229-z
85. Wang CY, Zhu T, Zhao ZX, Chen B, Chen TT, Dong Q, et al. Pan-cancer multi-omics analyses reveal crosstalk between the hippo and immune signaling pathways in the tumor microenvironment. FEBS Lett (2022) 596(4):449–64. doi: 10.1002/1873-3468.14249
86. Chen J, Zhang K, Zhi YR, Wu Y, Chen BA, Bai JY, et al. Tumor-derived exosomal mir-19b-3p facilitates M2 macrophage polarization and exosomal Linc00273 secretion to promote lung adenocarcinoma metastasis Via hippo pathway. Clin Transl Med (2021) 11(9):e478. doi: 10.1002/ctm2.478
87. Li C, Peng ZZ, Wang YZ, Lam G, Nissen N, Tang J, et al. Epithelial cell transforming 2 is regulated by yes-associated protein 1 and mediates pancreatic cancer progression and metastasis. Am J Physiol-Gastr L (2021) 320(3):G380–G95. doi: 10.1152/ajpgi.00185.2020
88. Wang LJ, Shi SJ, Guo ZY, Zhang X, Han SX, Yang AG, et al. Overexpression of yap and taz is an independent predictor of prognosis in colorectal cancer and related to the proliferation and metastasis of colon cancer cells. PloS One (2013) 8(6):e65539. doi: 10.1371/journal.pone.0065539
89. Lau AN, Curtis SJ, Fillmore CM, Rowbotham SP, Mohseni M, Wagner DE, et al. Tumor-propagating cells and Yap/Taz activity contribute to lung tumor progression and metastasis (Vol 33, pg 468, 2014). EMBO J (2014) 33(13):1502–. doi: 10.15252/embj.201488991
90. Noguchi S, Saito A, Horie M, Mikami Y, Suzuki HI, Morishita Y, et al. An integrative analysis of the tumorigenic role of taz in human non-small cell lung cancer. Clin Cancer Res (2014) 20(17):4660–72. doi: 10.1158/1078-0432.Ccr-13-3328
91. Li WZ, Zhang M, Huang CJ, Meng JL, Yin XX, Sun GY. Genetic variants of DNA repair pathway genes on lung cancer risk. Pathol Res Pract (2019) 215(10):152548. doi: 10.1016/j.prp.2019.152548
92. Saito A, Horie M, Nagase T. Tgf-beta signaling in lung health and disease. Int J Mol Sci (2018) 19(8):2460. doi: 10.3390/ijms19082460
93. Zhang WJ, Gao YJ, Li FM, Tong XY, Ren Y, Han XK, et al. Yap promotes malignant progression of Lkb1-deficient lung adenocarcinoma through downstream regulation of survivin. Cancer Res (2015) 75(21):4450–7. doi: 10.1158/0008-5472.Can-14-3396
94. Lee KW, Lee SS, Kim SB, Sohn BH, Lee HS, Jang HJ, et al. Significant association of oncogene Yap1 with poor prognosis and cetuximab resistance in colorectal cancer patients. Clin Cancer Res (2015) 21(2):357–64. doi: 10.1158/1078-0432.Ccr-14-1374
95. Barry ER, Morikawa T, Butler BL, Shrestha K, de la Rosa R, Yan KS, et al. Restriction of intestinal stem cell expansion and the regenerative response by yap. Nature (2013) 493(7430):106–+. doi: 10.1038/nature11693
96. Rosenbluh J, Nijhawan D, Cox AG, Li XN, Neal JT, Schafer EJ, et al. Beta-Catenin-Driven cancers require a Yap1 transcriptional complex for survival and tumorigenesis. Cell (2013) 153(1):267–70. doi: 10.1016/j.cell.2013.03.007
97. Yang R, Cai TT, Wu XJ, Liu YN, He J, Zhang XS, et al. Tumour Yap1 and pten expression correlates with tumour-associated myeloid suppressor cell expansion and reduced survival in colorectal cancer. Immunology (2018) 155(2):263–72. doi: 10.1111/imm.12949
98. Calvo F, Ege N, Grande-Garcia A, Hooper S, Jenkins RP, Chaudhry SI, et al. Mechanotransduction and yap-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat Cell Biol (2013) 15(6):637–+. doi: 10.1038/ncb2756
99. Zhang WY, Nandakumar N, Shi YH, Manzano M, Smith A, Graham G, et al. Downstream of mutant kras, the transcription regulator yap is essential for neoplastic progression to pancreatic ductal adenocarcinoma. Sci Signal (2014) 7(324):ra42. doi: 10.1126/scisignal.2005049
100. Gruber R, Panayiotou R, Nye E, Spencer-Dene B, Stamp G, Behrens A. Yap1 and taz control pancreatic cancer initiation in mice by direct up-regulation of jak-Stat3 signaling. Gastroenterology (2016) 151(3):526–39. doi: 10.1053/j.gastro.2016.05.006
101. Liu M, Zhang Y, Yang J, Zhan H, Zhou Z, Jiang Y, et al. Zinc-dependent regulation of Zeb1 and Yap1 coactivation promotes epithelial-mesenchymal transition plasticity and metastasis in pancreatic cancer. Gastroenterology (2021) 160(5):1771–83.e1. doi: 10.1053/j.gastro.2020.12.077
102. Mielgo A, Schmid MC. Impact of tumour associated macrophages in pancreatic cancer. Bmb Rep (2013) 46(3):131–8. doi: 10.5483/BMBRep.2013.46.3.036
103. Ji C, Wang Y, Wang Y, Luan J, Yao L, Wang Y, et al. Immune-related genes play an important role in the prognosis of patients with testicular germ cell tumor. Ann Transl Med (2020) 8(14):866. doi: 10.21037/atm-20-654
104. He K, Li Z, Ye K, Zhou Y, Yan M, Qi H, et al. Novel sequential therapy with metformin enhances the effects of cisplatin in testicular germ cell tumours Via Yap1 signalling. Cancer Cell Int (2022) 22(1):113. doi: 10.1186/s12935-022-02534-w
105. Savorani C, Malinverno M, Seccia R, Maderna C, Giannotta M, Terreran L, et al. A dual role of yap in driving tgf beta-mediated endothelial-to-Mesenchymal transition. J Cell Sci (2021) 134(15):jcs251371. doi: 10.1242/jcs.251371
106. Oh SH, Swiderska-Syn M, Jewell ML, Premont RT, Diehl AM. Liver regeneration requires Yap1-tgf beta-dependent epithelial-mesenchymal transition in hepatocytes. J Hepatol (2018) 69(2):359–67. doi: 10.1016/j.jhep.2018.05.008
107. Ding H, Xu YY, Jiang N. Upregulation of mir-101a suppresses chronic renal fibrosis by regulating Kdm3a Via blockade of the yap-Tgf-Beta-Smad signaling pathway. Mol Ther-Nucl Acids (2020) 19:1276–89. doi: 10.1016/j.omtn.2020.01.002
108. Shao DD, Xue W, Krall EB, Bhutkar A, Piccioni F, Wang XX, et al. Kras and Yap1 converge to regulate emt and tumor survival. Cell (2014) 158(1):171–84. doi: 10.1016/j.cell.2014.06.004
109. Mohamed AD, Shah N, Hettmer S, Vargesson N, Wackerhage H. Analysis of the relationship between the kras G12v oncogene and the hippo effector Yap1 in embryonal rhabdomyosarcoma. Sci Rep-Uk (2018) 8:15674. doi: 10.1038/s41598-018-33852-7
110. Chan LH, Wang W, Yeung W, Deng Y, Yuan P, Mak KK. Hedgehog signaling induces osteosarcoma development through Yap1 and H19 overexpression. Oncogene (2014) 33(40):4857–66. doi: 10.1038/onc.2013.433
111. Swiderska-Syn M, Xie GH, Michelotti GA, Jewell ML, Premont RT, Syn WK, et al. Hedgehog regulates yes-associated protein 1 in regenerating mouse liver. Hepatology (2016) 64(1):232–44. doi: 10.1002/hep.28542
112. Zhang L, Yang SP, Chen XC, Stauffer S, Yu F, Lele SM, et al. The hippo pathway effector yap regulates motility, invasion, and castration-resistant growth of prostate cancer cells. Mol Cell Biol (2015) 35(8):1350–62. doi: 10.1128/Mcb.00102-15
113. Stampouloglou E, Cheng N, Federico A, Slaby E, Monti S, Szeto GL, et al. Yap suppresses T-cell function and infiltration in the tumor microenvironment. PloS Biol (2020) 18(1):e3000591. doi: 10.1371/journal.pbio.3000591
114. Lebid A, Chung L, Pardoll DM, Pan F. Yap attenuates Cd8 T cell-mediated anti-tumor response. Front Immunol (2020) 11:580. doi: 10.3389/fimmu.2020.00580
115. Kim MH, Kim CG, Kim SK, Shin SJ, Choe EA, Park SH, et al. Yap-induced pd-L1 expression drives immune evasion in brafi-resistant melanoma. Cancer Immunol Res (2018) 6(3):255–66. doi: 10.1158/2326-6066.Cir-17-0320
116. Murakami S, Shahbazian D, Surana R, Zhang W, Chen H, Graham GT, et al. Yes-associated protein mediates immune reprogramming in pancreatic ductal adenocarcinoma. Oncogene (2017) 36(9):1232–44. doi: 10.1038/onc.2016.288
117. Cheng JC, Chang HM, Liu PP, Leung PCK. Sphingosine-1-Phosphate induces cox-2 expression and Pge2 production in human granulosa cells through a S1p(1/3)-mediated yap signaling. Cell Signal (2016) 28(6):643–51. doi: 10.1016/j.cellsig.2016.03.006
118. Shen TY, Li Y, Zhu SM, Yu JP, Zhang BY, Chen XR, et al. Yap1 plays a key role of the conversion of normal fibroblasts into cancer-associated fibroblasts that contribute to prostate cancer progression. J Exp Clin Canc Res (2020) 39(1):36. doi: 10.1186/s13046-020-1542-z
Keywords: YAP1, pan-cancer, immune, survival, drug response
Citation: Hu X, Zhang Y, Yu H, Zhao Y, Sun X, Li Q and Wang Y (2022) The role of YAP1 in survival prediction, immune modulation, and drug response: A pan-cancer perspective. Front. Immunol. 13:1012173. doi: 10.3389/fimmu.2022.1012173
Received: 05 August 2022; Accepted: 31 October 2022;
Published: 21 November 2022.
Edited by:
Jongdae Lee, Guangzhou Medical University, ChinaReviewed by:
Hong Sook Kim, Sungkyunkwan University, South KoreaNor Hazwani Ahmad, Universiti Sains Malaysia (USM), Malaysia
Copyright © 2022 Hu, Zhang, Yu, Zhao, Sun, Li and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Wang, eWFud2FuZ0BzaHV0Y20uZWR1LmNu
†These authors have contributed equally to this work
 Xueqing Hu
Xueqing Hu Yingru Zhang
Yingru Zhang Hao Yu1
Hao Yu1 Yiyang Zhao
Yiyang Zhao Xiaoting Sun
Xiaoting Sun Qi Li
Qi Li Yan Wang
Yan Wang