
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol., 08 September 2022
Sec. Nutritional Immunology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.1007165
This article is part of the Research TopicImprove Metabolic and Autoimmune Diseases by Regulating Gut MicrobiotaView all 8 articles
Rheumatoid arthritis (RA) is a chronic autoimmune disease that primarily affects the joints. Microbial infection is considered a crucial inducer of RA. Alterations in the composition of intestinal bacteria in individuals with preclinical and established RA suggest a vital role of the gut microbiota in immune dysfunction characteristic of RA. However, the mechanisms by which gut dysbiosis contributes to RA are not fully understood. Furthermore, multiple therapies commonly used to treat RA may alter gut microbiota diversity, suggesting that modulating the gut microbiota may help prevent or treat RA. Hence, a better understanding of the changes in the gut microbiota that accompany RA should aid the development of novel therapeutic approaches. This mini-review discusses the impact of gut dysbiosis in the pathogenesis of RA, the selection of gut microbiota-related biomarkers for diagnosing RA, and provides examples of cross-modulation between the gut microbiota and some drugs commonly used to treat RA. Some suggestions and outlooks are also raised, which may help guide future research efforts.
Rheumatoid arthritis (RA) is a chronic, immune-mediated disease in which multiple immune cell types and signaling networks misfunction to elicit a maladaptive tissue repair process; this leads to organ damage, predominantly in the vascular system, lungs, and joints (1). RA affects ~1% of the population worldwide (2), and its pathogenesis may be linked to genetic and environmental factors (3, 4). At present, the specific pathogenesis of RA is not well understood.
Humans are one of the most complex microbial ecosystems on the planet, hosting over 100 trillion bacteria, mainly in the distal gut (5). Some gut bacteria species can induce autoimmunity in genetically predisposed animal models (6, 7). Dysbiosis of specific bacterial lineages and alterations in gut microbiota metabolism led to changes in the host immune profile that contribute to RA (8). It has been proposed that the mechanism by which gut microbiota imbalance leads to RA may be related to regulation of immune function by metabolites produced by gut microbes (9, 10). Immune T and B cells have position-specific phenotypes and functions in the mucosa, influenced by the microbiota (11). In turn, bacterial peptidoglycan components are found in the synovial tissue of RA patients, which may contribute to inflammation within the microenvironment of the joint (12, 13). Substantial data published in the past few years demonstrate that an altered composition of the gut microbiota in RA patients is one of the major factors triggering aberrant systemic immunity (14–16). Notably, different strains of gut bacteria can have profoundly different regulatory effects on immune system function. Some strains can stimulate an immune response, benefiting immunocompromised patients, while others can suppress the immune response, affecting immune regulation in RA patients (17–21). For example, segmented filamentous bacteria (SFB) have a unique ability to drive T helper 17 (Th17) cell accumulation in the small intestine’s lamina propria through SFB-derived antigens presented by dendritic cells (22–24). In contrast, the colonization of Bacteroides fragilis is associated with enhanced activity of regulatory T cells (Tregs), which may alleviate autoimmune disease (25, 26). Therefore, the relative abundance of different bacterial lineages may lead to changes in the host immune profile and drive inflammatory responses contributing to RA.
This mini-review discusses the role of the gut microbiota in the pathogenesis of RA, summarizes the diagnostic value of gut microbe-based biomarkers, and outlines mutual influences between the gut microbiota and some drugs used to treat RA. Some suggestions and outlooks are also raised to guide future research efforts.
Numerous studies highlighted a critical role of the gut microbiota in RA pathogenesis, through mechanisms including mainly production of proinflammatory metabolites, impairment of the intestinal mucosal barrier, and molecular mimicry of autoantigens.
The gastrointestinal tract hosts the majority of immune cells in the body, with constant interaction with the gut microbiota shaping their function and phenotypes. The gut microbiota mediates constant bidirectional communication with the host immune system in a delicate balance of inducing pathogenic infection or residing in the human body in a commensal state (27). The innate immune cells in the gut-associated lymphoid tissue comprise the first-line of defense against xenobiotics from the gastrointestinal tract. Disturbed gut microbiota can trigger the aberrant activation of innate immune cells, which leads to the upregulation of proinflammatory cytokine including interleukin-12 (IL-12), IL-23, and type I interferons, etc., as well as reduction of anti-inflammatory cytokines including transforming growth factor β and IL-10, etc. (28). Moreover, the adaptive lymphocytes are critical players in autoimmunity, and aberrant activation of T and B cells instigates RA. Gut pathogens with proinflammatory capacities can reshape the immune milieu through innate immune overactivation, followed by aberrant activation of the adaptive immune system. Microbial antigens can be presented to CD4+ T cells by dendritic cells and macrophages, leading to differentiation of inflammatory T cell subtypes. Th17 cells are a subset of proinflammatory CD4+ T cells characterized by production of interleukin-17 (IL-17) (23). Tregs are also derived from CD4+ T cells, show instead immunosuppressive actions, and may inhibit Th17 responses (29, 30). Studies have demonstrated that an increased Th17/Treg ratio is closely related to RA, and that the Th17/Treg balance is strongly regulated by gut microbiota and their metabolites (31, 32). Microbial antigens can also induce overactivation of B lymphocytes with the help of T follicular helper cells, differentiating into plasma cells and producing pathogenic autoantibodies. This may influence the pathogenesis of RA (28). Therefore, the dysbiosis gut microbiota, inflammatory factors, and immune responses are interrelated and jointly affect the development of RA (33) (Figure 1).
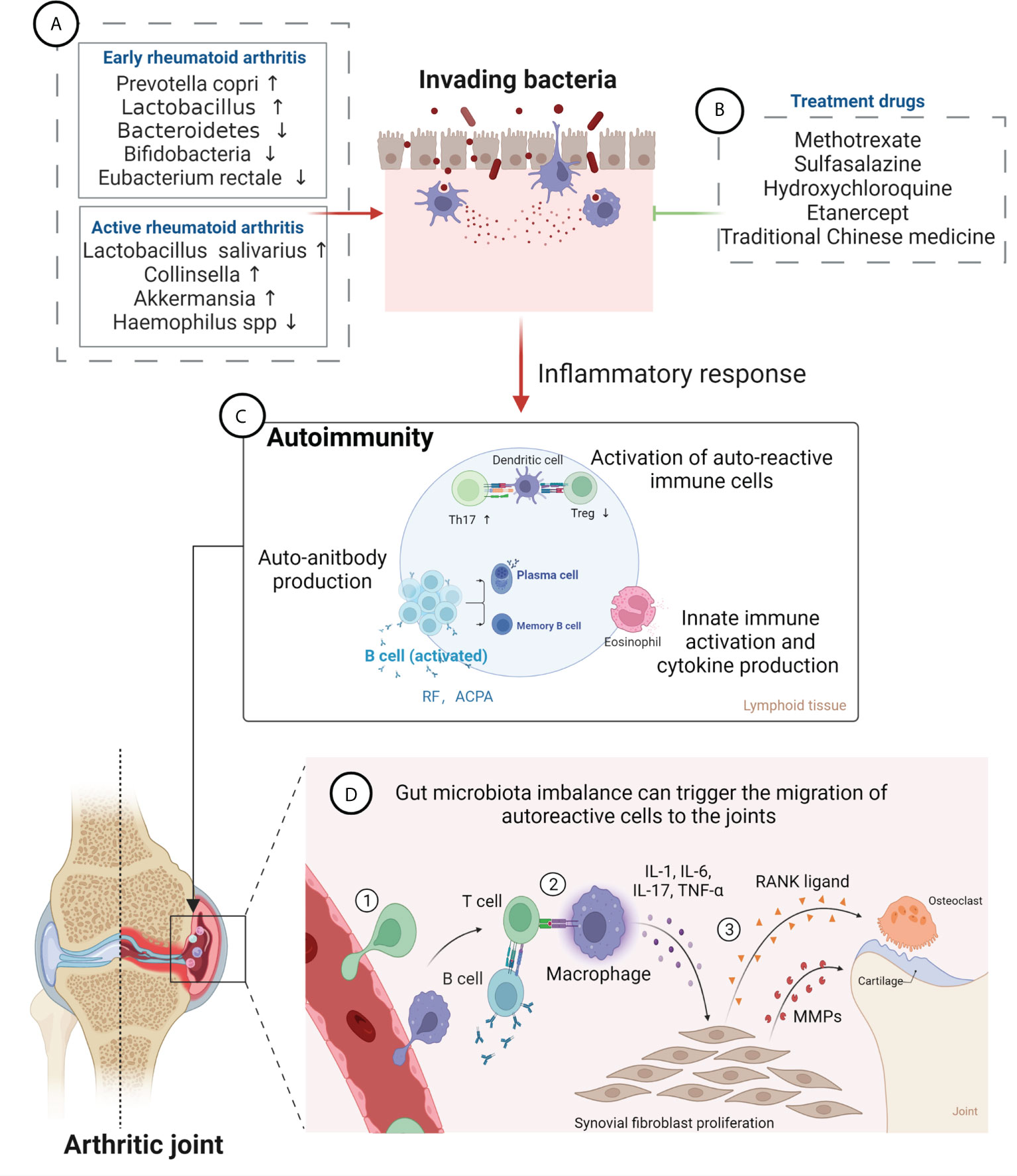
Figure 1 Gut microbiota in the pathogenesis of rheumatoid arthritis (RA) and effects of RA therapeutic drugs on the gut microbiota. (A) Changes in the composition of gut microbiota at different stages of RA. Levels of Prevotella copri and Lactobacillus are increased, while those of Bacteroidetes, Bifidobacteria and Eubacterium rectale are decreased, at an early stage; Abundance of Lactobacillus salivarius, Collinsella, and Akkermansia is increased, while that of Haemophilus spp. is decreased, in the active RA phase. (B) RA treatment drugs can improve gut microbiota imbalance and relieve disease symptoms, mainly including methotrexate, sulfasalazine, hydroxychloroquine, etanercept, and traditional Chinese medicine. (C) Gut microbiota can lead to damage of the epithelium and to the opening of the paracellular pathway and can cross the epithelium and get in contact with the immune cells beneath the epithelial layer, which leads to inflammation. Furthermore, bacterial antigens promote activation of autoreactive immune cells (B cell and T cell) in the lymphoid tissues, resulting in an imbalance between regulatory T cells (Tregs) and T helper 17 (Th17) cells, leading to expansion of inflammatory response. Activated B cells produce autoantibodies (anti-citrullinated protein antibody and rheumatoid factor). (D) Gut microbiota imbalance can trigger the migration of autoreactive cells to the joints, causing cartilage and bone damage. ① Bacterial antigens trigger promotes inflammation in synovial membrane, attracting leukocytes into the tissue. ② Autoreactive cells activate macrophages, resulting in inflammatory cytokine production. ③ Cytokines induce fibroblasts to produce MMPs (matrix metalloproteinases) and RANKL (receptor activator of nuclear factor κB ligand), which mediate destruction of bone and cartilage tissue, leading to the development of RA.
The intestinal mucosal barrier, formed and maintained by the intestinal epithelium, serves to isolate harmful substances in the intestinal lumen and prevent the invasion of pathogenic antigens. The gut integrity is impaired in RA patients, which leads to the translocation of microbes across the gut barrier into gut tissue and even circulation. Gut microbiota can promote the overactivation of innate and adaptive immunity in the local tissue, resulting in systemic immune dysregulation (28). In addition, disturbed gut microbiota can also trigger the migration of autoreactive cells to the joints, causing local joint inflammation (34). Autoreactive cells activate macrophages, resulting in inflammatory cytokine production. Further, cytokines such as tumor necrosis factor-alpha (TNF-α), IL-6, and IL-1 can induce fibroblasts to produce matrix metalloproteinases and receptor activator of nuclear factor κB ligand, which mediate destruction of the bone and cartilage tissue, leading to the development of RA (35) (Figure 1). A study has shown that Collinsella aerofaciens, a commensal gut bacterium found to be overrepresented in RA patients, reduces the expression of tight junction proteins in human intestinal epithelial cells and increases disease incidence in HLA-DQ8 transgenic mice subjected to collagen-induced arthritis (CIA) by disrupting the integrity of the intestinal mucosa (36). In contrast, Faecalibacterium prausnitzii, a prominent member of the human commensal gut microbiota whose abundance is reduced in patients with RA, was shown to sustain intestinal barrier function, maintain Th17/Treg balance, and exert significant anti-inflammatory effects (37). These findings indicate that changes in gut microbiota diversity may impair intestinal mucosal permeability, facilitating the onset of RA (38, 39). can trigger the migration of autoreactive cells to the joints, causing cartilage and bone damage.
Molecular mimicry is a mechanism by which pathogen-derived antigens that share sequence homology with self-peptides may lead to cross-activation of autoreactive T or B cells, triggering autoimmunity. Many identical peptides between human tissues and gut microbes bind to HLA-II alleles. The autoimmune candidates have been shown to be enriched in bacterial species belonging to the Firmicutes and Proteobacteria, which may have a higher disease impact in genetically susceptible individuals (40). N-acetylglucosamine-6-sulfatase and filamin A were identified as T- and B-cell-targeted autoantigens in more than 50% of RA patients (41). The HLA-DR-presented N-acetylglucosamine-6-sulfatase peptide has marked sequence homology with epitopes from sulfatase proteins of the Prevotella sp. and Parabacteroides sp., whereas the HLA-DR-presented filamin A peptide has homology with epitopes from proteins of the Prevotella sp. and Butyricimonas sp., another gut commensal (41). In turn, the presence of shared sequences between Collinsella and DRB1*0401 suggested that Collinsella may induce RA through molecular mimicry (36). These findings thus identify molecular mimicry as a plausible link between disrupted mucosal immune tolerance and systemic immunity in RA patients.
Gut microbiota composition can alter sex hormone levels to affect the occurrence of RA. Sex hormone deficiency increases intestinal permeability, thereby increasing the number of Th17 cells in peripheral blood and the levels of osteoclastogenic cytokines RANKL, IL-17 and TNF-α, promoting bone resorption (42). Clostridium can produce enzymes that catalyze the conversion of glucocorticoids into androgens, exerting immunomodulatory effects (43). Implying a strong connection between periodontitis and RA, Porphyromonas gingivalis, a major pathogen in periodontitis, promotes citrullination of proteins and production of anti-citrullinated protein antibodies, a hallmark of RA. This is mediated by production of peptidyl arginine deiminase (PAD) and leukotoxin A, which triggers in turn endogenous PAD activation in neutrophils (44). Besides, several metabolites including short-chain fatty acids, serotonin, as well as plant enzymes in the gut can also influence RA (45). However, despite major advances, the mechanisms linking dysbiosis of the gut microbiota and RA remain still incompletely characterized.
Early diagnosis of RA is critical to provide prompt treatment to slow down joint damage. Scher et al. (2013) reported that the abundance of Prevotella copri was increased, while that of Bacteroidetes was decreased, in patients with early RA; the presence of P. copri was correlated with a reduction in the abundance of other bacterial groups, including many beneficial microbes (46). Transplanting gut microbiota (with a high abundance of Prevotella) from patients with early RA into germ-free SKG mice can induce severe arthritis (47). In addition, increased number and species of intestinal Lactobacillus were observed in early RA patients than in healthy individuals (48). Studies have shown that the monocontamination of germ-free IL-1 receptor antagonist–deficient (IL1rn-/-) mice, which develop spontaneous arthritis due to excessive IL-1 signaling, with indigenous Lactobacillus bifidus resulted in rapid onset of arthritis that reached incidence rate and severity scores comparable to those recorded in non–germ-free mice (49). It was in turn reported that in early RA patients the fecal microbiota contained significantly less Bifidobacteria, B. fragilis, and Eubacterium rectale than in patients with fibromyalgia (50).
In the active period of RA, depletion of Haemophilus spp. in the patients’ gut, teeth, and saliva correlated negatively with RA severity and serum antibody levels. In contrast, Lactobacillus salivarius was present in increased amounts in cases of very active RA (16). Meanwhile, gut microbiota analysis revealed higher relative abundances of the genera Collinsella and Akkermansia in patients with active, compared to inactive, RA disease status (51). A study indicated that treatment with Collinsella exacerbated CIA in HLA-DQ8 transgenic mice (36). These observations reaffirm the impact of alterations in the gut microbiota on RA severity and suggest that changes in gut microbiota composition may serve as markers for the diagnosis of RA. However, further research is needed to conclusively identify reliable gut microbiota biomarkers for diagnosing RA (Figure 1).
The gut microbiota was found to predict drug response in RA (52). On the other hand, in recent years oral probiotics and fecal flora transplantation have shown promising results when used as adjuvant therapy for treating RA, by directly and indirectly modulating the gut microbiota. Indirect regulation of gut microbiota in RA patients and animal models is also exerted by disease-modifying anti-rheumatic drugs (DMARDs), traditional Chinese medicine (TCM) herbs and prescriptions, and by adjusting diet structure.
Probiotics, defined as “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host” (53), can reduce the abundance of pathogenic bacteria by competing for nutrition and colonization sites. At the same time, probiotics can alleviate RA symptoms by producing antibiotics and strengthening the intestinal barrier, with beneficial modulation of the immune function (54). Studies conducted in rats with adjuvant-induced arthritis (AIA) showed that oral administration of L. casei or L. acidophilus reduced arthritic inflammation, pannus formation, and cartilage destruction (55–57). More recently, it was reported that administration of L. casei to AIA rats significantly suppressed arthritis and protected against bone loss by reducing dysbiosis of the gut microbiota (58). However, supplementation with L. reuteri and L. rhamnosus GG did not significantly reduce RA disease activity (59, 60), suggesting that different Lactobacillus species may act differently on RA.
Fecal microbiota transplantation (FMT) refers to the introduction of gut microbiota obtained from the feces of a healthy donor into a patient’s gastrointestinal tract (61). Normalizing the gut microbiota through FMT may potentially improve RA symptoms. A case of a patient with refractory RA successfully treated with FMT indicated that FMT may have an excellent therapeutic effect on RA (62). However, clinical studies examining the efficacy of FMT in RA patients are so far scarce.
DMARDs may indirectly affect and remodel the structure and function of gut microbiota to regulate systemic immunity. Studies have demonstrated that microbial differences in the gastrointestinal tract of RA patients may partially determine the bioavailability and subsequent clinical outcome of methotrexate (63, 64). In turn, methotrexate treatment was shown to partially restore normal gut microbiota composition in RA patients (16, 65). Thus, the gut microbiota may be a predictor of clinical response to methotrexate, influencing the treatment response rate.
Another DMARD, i.e. sulfasalazine, is cleaved by the action of bacterial azoreductases in the large intestine into sulfapyridine and mesalazine (66, 67). Sulfapyridine affects the immune system and appears to normalize lymphocyte activity by regulating gut microbiota (68). A study in active RA patients showed that sulfasalazine therapy led to a substantial fall in fecal counts of Clostridium perfringens and Escherichia coli (69). Furthermore, sulfasalazine treatment significantly altered fecal microflora of RA patients by reducing total aerobic bacteria, Bacteroides, and Escherichia coli, and increasing the numbers of Bacillus (70). However, thorough characterization of the effects of sulfasalazine therapy on the gut microbiota in RA is still lacking.
Treatment with etanercept, a TNF-α antagonist, was shown to beneficially impact gut microbiota composition. In RA patients, etanercept treatment was associated with enrichment of Cyanobacteria, including members of the Nostocophycideae class and the Nostocales order (which were not represented among naïve patients), as well as with decreased abundance of Clostridiaceae and Deltaproteobacteria (71). In CIA mice, etanercept treatment led to decreased microbial community richness and diversity, increasing the abundance of Escherichia and Shigella and decreasing the abundance of Clostridium XIVa, Tannerella, and Lactobacillus (72). In contrast, hydroxychloroquine treatment in RA patients was associated with increased intestinal bacterial richness and diversity, suggestive of restoration of normal microbiota. In addition, the abundance of Faecalibacterium, found to be decreased prior to treatment, was positively correlated with the use of hydroxychloroquine (36, 71).
Numerous studies indicated that TCM-based therapies provide significant curative effects and elicit minor adverse reactions in RA (73). Notably, part of the beneficial effects of TCM on RA may be ascribed to regulation of the intestinal flora. Mei et al. (2021) showed that depletion of Clostridium celatum in RA patients could be reversed by treatment with the Huayu-Qiangshen-Tongbi formula (74). Administration of total glucosides of paeony to CIA rats corrected 78% of the taxonomic differences in microbial structure, while also increasing the relative abundance of certain forms of beneficial commensal bacteria (75). Similarly, most of the 19 types of bacteria found to be altered at the family level in CIA rats could be regulated by the Zushima tablet (76). It was also reported that Qingluo Tongbi decoction can effectively ameliorate arthritis in AIA rats at least partly by decreasing inflammatory responses regulated by the gut microbiota (77).
In addition to therapeutic drugs, dietary nutrients also affect the composition and function of the gut microbiota and may thus have an important impact on the prevention and treatment of RA. Dietary fiber, abundant in vegetarian diets, can improve gut microbiota composition in RA patients and reduce joint pain (78). Reducing the intake of carbohydrates can help improve the balance of intestinal flora and immune function (79, 80). The omega-3 polyunsaturated fatty acids can help to maintain the intestinal barrier integrity and interact with host immune cells (81). A low ratio of omega-3/omega-6 fatty acids has been shown to promote inflammation, increasing the risk of RA (82, 83). Increased sodium intake can also increase the risk of RA (84). Studies have confirmed that a high-salt diet may lead to dysbiosis of gut microbiota, which promotes the micro-inflammatory state and autoimmune processes (85, 86). Indeed, several clinical trials have demonstrated that RA severity can be alleviated through dietary interventions (87). Thus, a better understanding of the mutual influences between the gut microbiota and some drugs or dietary nutrients will help to achieve optimal therapeutic effects for RA.
After decades of research, the fundamental role of the gut microbiota in health and disease is now firmly established. It is thus widely recognized that the gut microbiota can affect almost all aspects of the host, and its dysregulation is associated with dysregulated immune tolerance and RA development. Indeed, changes in the gut microbiota can precede the onset of RA and are closely related to disease activity afterwards. Analysis of gut microbiota composition can also predict susceptibility to RA, and has become a useful method to predict and control RA incidence. Furthermore, the human gut microbiota and their enzymatic products can affect the bioavailability, clinical efficacy, and toxicity of a wide array of drugs through direct and indirect mechanisms. Conversely, various medicines and active ingredients modulate immune cell function by normalizing the composition of the gut microbiota. Although significant variations in some specific microbial communities have been detected in association with RA, further research is needed to clarify the role of the gut microbiota in RA and its impact on the mechanisms of action and efficacy of DMARDs. Notwithstanding, mounting evidence indicating that targeted modulation of the gut microbiota may alleviate RA suggests that personalized treatment approaches based on patient microbiome profiles may increase drug efficacy, lower toxicity risk, and improve clinical outcome.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
National Natural Science Foundation of China (31960178, 82160923, 81960863, 82160901, and 81960870); Construction Project of National TCM Clinical Research Base (2018 No. 131); Yunnan Provincial Fund for Medical Research Center: Clinical Evaluation and Basic Research on the Treatment of rheumatoid arthritis and gout by TCM (202102AA310006); Clinical Trial for the Treatment of Rheumatoid Arthritis with Warming yang and Smoothening Meridians (201507001-07, registration number: ChiCTR-INR-16010290); Clinical Cooperative Project of Chinese and Western Medicine for Major and Knotty Diseases; Yunnan Provincial Key Laboratory Construction Project Funding; Yunnan Provincial Key Laboratory of Chinese Medicine Rheumatology and Immunology; Yunnan Provincial Ten Thousands Program Famous Doctor Special; Yunnan Province Qingguo Wang Expert Workstation Construction Project (202005AF150017); Yunnan Applied Basic Research Projects- Union Foundation [2019FF002(-031)]; Applied Basic Research Programs of Science and Technology Commission Foundation of Yunnan Province (2019FA007); Key Laboratory of Traditional Chinese Medicine for Prevention and Treatment of Neuropsychiatric Diseases, Yunnan Provincial Department of Education; Scientific Research Projects for High-level Talents of Yunnan University of Chinese Medicine (2019YZG01); Young Top-Notch Talent in 10,000 Talent Program of Yunnan Province (YNWR-QNBJ-2019-235).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Weyand CM, Goronzy JJ. The immunology of rheumatoid arthritis. Nat Immunol (2021) 22(1):10–8. doi: 10.1038/s41590-020-00816-x
2. Firestein GS. Evolving concepts of rheumatoid arthritis. Nature (2003) 423(6937):356–61. doi: 10.1038/nature01661
3. McInnes IB, Schett G. The pathogenesis of rheumatoid arthritis. N Engl J Med (2011) 365(23):2205–19. doi: 10.1056/NEJMra1004965
4. Edilova MI, Akram A, Abdul-Sater AA. Innate immunity drives pathogenesis of rheumatoid arthritis. Biomed J (2021) 44(2):172–82. doi: 10.1016/j.bj.2020.06.010
5. Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol (2009) 9(5):313–23. doi: 10.1038/nri2515
6. Scher JU, Abramson SB. The microbiome and rheumatoid arthritis. Nat Rev Rheumatol (2011) 7(10):569–78. doi: 10.1038/nrrheum.2011.121
7. Gomez A, Luckey D, Yeoman CJ, Marietta EV, Berg Miller ME, Murray JA, et al. Loss of sex and age driven differences in the gut microbiome characterize arthritis-susceptible 0401 mice but not arthritis-resistant 0402 mice. PLoS One (2012) 7(4):e36095. doi: 10.1371/journal.pone.0036095
8. Wang Q, Zhang SX, Chang MJ, Qiao J, Wang CH, Li XF, et al. Characteristics of the gut microbiome and its relationship with peripheral CD4(+) T cell subpopulations and cytokines in rheumatoid arthritis. Front Microbiol (2022) 13:799602. doi: 10.3389/fmicb.2022.799602
9. He J, Chu Y, Li J, Meng Q, Liu Y, Jin J, et al. Intestinal butyrate-metabolizing species contribute to autoantibody production and bone erosion in rheumatoid arthritis. Sci Adv (2022) 8(6):eabm1511. doi: 10.1126/sciadv.abm1511
10. Yu D, Du J, Pu X, Zheng L, Chen S, Wang N, et al. The gut microbiome and metabolites are altered and interrelated in patients with rheumatoid arthritis. Front Cell Infect Microbiol (2021) 11:763507. doi: 10.3389/fcimb.2021.763507
11. Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature (2016) 535(7610):75–84. doi: 10.1038/nature18848
12. Schrijver IA, Melief MJ, Tak PP, Hazenberg MP, Laman JD. Antigen-presenting cells containing bacterial peptidoglycan in synovial tissues of rheumatoid arthritis patients coexpress costimulatory molecules and cytokines. Arthritis Rheum (2000) 43(10):2160–8. doi: 10.1002/1529-0131(200010)43:10<2160::Aid-anr3>3.0.Co;2-t
13. Pianta A, Arvikar S, Strle K, Drouin EE, Wang Q, Costello CE, et al. Evidence of the immune relevance of prevotella copri, a gut microbe, in patients with rheumatoid arthritis. Arthritis Rheumatol (2017) 69(5):964–75. doi: 10.1002/art.40003
14. Kinashi Y. Hase k. partners in leaky gut syndrome: Intestinal dysbiosis and autoimmunity. Front Immunol (2021) 12:673708. doi: 10.3389/fimmu.2021.673708
15. Chen B, Sun L, Zhang X. Integration of microbiome and epigenome to decipher the pathogenesis of autoimmune diseases. J Autoimmun (2017) 83:31–42. doi: 10.1016/j.jaut.2017.03.009
16. Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med (2015) 21(8):895–905. doi: 10.1038/nm.3914
17. Jung H, Jung SM, Rim YA, Park N, Nam Y, Lee J, et al. Arthritic role of porphyromonas gingivalis in collagen-induced arthritis mice. PLoS One (2017) 12(11):e0188698. doi: 10.1371/journal.pone.0188698
18. Ferreira-Halder CV, Faria AVS, Andrade SS. Action and function of faecalibacterium prausnitzii in health and disease. Best Pract Res Clin Gastroenterol (2017) 31(6):643–8. doi: 10.1016/j.bpg.2017.09.011
19. Iljazovic A, Roy U, Gálvez EJC, Lesker TR, Zhao B, Gronow A, et al. Perturbation of the gut microbiome by prevotella spp. enhances host susceptibility to mucosal inflammation. Mucosal Immunol (2021) 14(1):113–24. doi: 10.1038/s41385-020-0296-4
20. Amdekar S, Singh V, Kumar A, Sharma P, Singh R. Lactobacillus casei and lactobacillus acidophilus regulate inflammatory pathway and improve antioxidant status in collagen-induced arthritic rats. J Interferon Cytokine Res (2013) 33(1):1–8. doi: 10.1089/jir.2012.0034
21. Alpizar-Rodriguez D, Lesker TR, Gronow A, Gilbert B, Raemy E, Lamacchia C, et al. Prevotella copri in individuals at risk for rheumatoid arthritis. Ann Rheum Dis (2019) 78(5):590–3. doi: 10.1136/annrheumdis-2018-214514
22. Flannigan KL, Denning TL. Segmented filamentous bacteria-induced immune responses: A balancing act between host protection and autoimmunity. Immunology (2018) 154(4):537–46. doi: 10.1111/imm.12950
23. Wang Y, Yin Y, Chen X, Zhao Y, Wu Y, Li Y, et al. Induction of intestinal Th17 cells by flagellins from segmented filamentous bacteria. Front Immunol (2019) 10:2750. doi: 10.3389/fimmu.2019.02750
24. Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, et al. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell (2015) 163(2):367–80. doi: 10.1016/j.cell.2015.08.058
25. Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, et al. The toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science (2011) 332(6032):974–7. doi: 10.1126/science.1206095
26. Telesford KM, Yan W, Ochoa-Reparaz J, Pant A, Kircher C, Christy MA, et al. A commensal symbiotic factor derived from bacteroides fragilis promotes human CD39(+)Foxp3(+) T cells and treg function. Gut Microbes (2015) 6(4):234–42. doi: 10.1080/19490976.2015.1056973
27. Shin C, Kim YK. Autoimmunity in microbiome-mediated diseases and novel therapeutic approaches. Curr Opin Pharmacol (2019) 49:34–42. doi: 10.1016/j.coph.2019.04.018
28. Zhang X, Chen BD, Zhao LD, Li H. The gut microbiota: Emerging evidence in autoimmune diseases. Trends Mol Med (2020) 26(9):862–73. doi: 10.1016/j.molmed.2020.04.001
29. Lina C, Conghua W, Nan L, Ping Z. Combined treatment of etanercept and MTX reverses Th1/Th2, Th17/Treg imbalance in patients with rheumatoid arthritis. J Clin Immunol (2011) 31(4):596–605. doi: 10.1007/s10875-011-9542-6
30. Chen X, Oppenheim JJ. Th17 cells and tregs: Unlikely allies. J Leukoc Biol (2014) 95(5):723–31. doi: 10.1189/jlb.1213633
31. Kelly D, Mulder IE. Microbiome and immunological interactions. Nutr Rev (2012) 70 Suppl 1:S18–30. doi: 10.1111/j.1753-4887.2012.00498.x
32. Cheng H, Guan X, Chen D, Ma W. The Th17/Treg cell balance: A gut microbiota-modulated story. Microorganisms (2019) 7(12):583. doi: 10.3390/microorganisms7120583
33. Li Y, Zhang SX, Yin XF, Zhang MX, Qiao J, Xin XH, et al. The gut microbiota and its relevance to peripheral lymphocyte subpopulations and cytokines in patients with rheumatoid arthritis. J Immunol Res (2021) 2021:6665563. doi: 10.1155/2021/6665563
34. Brandl C, Bucci L, Schett G, Zaiss MM. Crossing the barriers: Revisiting the gut feeling in rheumatoid arthritis. Eur J Immunol (2021) 51(4):798–810. doi: 10.1002/eji.202048876
35. Pacifici R. T Cells, osteoblasts, and osteocytes: Interacting lineages key for the bone anabolic and catabolic activities of parathyroid hormone. Ann N Y Acad Sci (2016) 1364(1):11–24. doi: 10.1111/nyas.12969
36. Chen J, Wright K, Davis JM, Jeraldo P, Marietta EV, Murray J, et al. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med (2016) 8(1):43. doi: 10.1186/s13073-016-0299-7
37. Zhou L, Zhang M, Wang Y, Dorfman RG, Liu H, Yu T, et al. Faecalibacterium prausnitzii produces butyrate to maintain Th17/Treg balance and to ameliorate colorectal colitis by inhibiting histone deacetylase 1. Inflammation Bowel Dis (2018) 24(9):1926–40. doi: 10.1093/ibd/izy182
38. Tajik N, Frech M, Schulz O, Schälter F, Lucas S, Azizov V, et al. Targeting zonulin and intestinal epithelial barrier function to prevent onset of arthritis. Nat Commun (2020) 11(1):1995. doi: 10.1038/s41467-020-15831-7
39. Matei DE, Menon M, Alber DG, Smith AM, Nedjat-Shokouhi B, Fasano A, et al. Intestinal barrier dysfunction plays an integral role in arthritis pathology and can be targeted to ameliorate disease. Med (N Y) (2021) 2(7):864–83.e9. doi: 10.1016/j.medj.2021.04.013
40. Negi S, Singh H, Mukhopadhyay A. Gut bacterial peptides with autoimmunity potential as environmental trigger for late onset complex diseases: In-silico study. PloS One (2017) 12(7):e0180518. doi: 10.1371/journal.pone.0180518
41. Pianta A, Arvikar SL, Strle K, Drouin EE, Wang Q, Costello CE, et al. Two rheumatoid arthritis-specific autoantigens correlate microbial immunity with autoimmune responses in joints. J Clin Invest (2017) 127(8):2946–56. doi: 10.1172/jci93450
42. Li JY, Chassaing B, Tyagi AM, Vaccaro C, Luo T, Adams J, et al. Sex steroid deficiency-associated bone loss is microbiota dependent and prevented by probiotics. J Clin Invest (2016) 126(6):2049–63. doi: 10.1172/jci86062
43. Ridlon JM, Ikegawa S, Alves JM, Zhou B, Kobayashi A, Iida T, et al. Clostridium scindens: A human gut microbe with a high potential to convert glucocorticoids into androgens. J Lipid Res (2013) 54(9):2437–49. doi: 10.1194/jlr.M038869
44. Farquharson D, Butcher JP, Culshaw S. Periodontitis, porphyromonas, and the pathogenesis of rheumatoid arthritis. Mucosal Immunol (2012) 5(2):112–20. doi: 10.1038/mi.2011.66
45. Rosser EC, Piper CJM, Matei DE, Blair PA, Rendeiro AF, Orford M, et al. Microbiota-derived metabolites suppress arthritis by amplifying aryl-hydrocarbon receptor activation in regulatory b cells. Cell Metab (2020) 31(4):837–51.e10. doi: 10.1016/j.cmet.2020.03.003
46. Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, et al. Expansion of intestinal prevotella copri correlates with enhanced susceptibility to arthritis. Elife (2013) 2:e01202. doi: 10.7554/eLife.01202
47. Maeda Y, Kurakawa T, Umemoto E, Motooka D, Ito Y, Gotoh K, et al. Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheumatol (2016) 68(11):2646–61. doi: 10.1002/art.39783
48. Liu X, Zou Q, Zeng B, Fang Y, Wei H. Analysis of fecal lactobacillus community structure in patients with early rheumatoid arthritis. Curr Microbiol (2013) 67(2):170–6. doi: 10.1007/s00284-013-0338-1
49. Abdollahi-Roodsaz S, Joosten LA, Koenders MI, Devesa I, Roelofs MF, Radstake TR, et al. Stimulation of TLR2 and TLR4 differentially skews the balance of T cells in a mouse model of arthritis. J Clin Invest (2008) 118(1):205–16. doi: 10.1172/jci32639
50. Vaahtovuo J, Munukka E, Korkeamäki M, Luukkainen R, Toivanen P. Fecal microbiota in early rheumatoid arthritis. J Rheumatol (2008) 35(8):1500–5.
51. Chiang HI, Li JR, Liu CC, Liu PY, Chen HH, Chen YM, et al. An association of gut microbiota with different phenotypes in Chinese patients with rheumatoid arthritis. J Clin Med (2019) 8(11):1770. doi: 10.3390/jcm8111770
52. Onuora S. Gut microbiome could predict drug response in RA. Nat Rev Rheumatol (2021) 17(3):129. doi: 10.1038/s41584-021-00582-6
53. Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, et al. Expert consensus document. the international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol (2014) 11(8):506–14. doi: 10.1038/nrgastro.2014.66
54. Ferro M, Charneca S, Dourado E, Guerreiro CS, Fonseca JE. Probiotic supplementation for rheumatoid arthritis: A promising adjuvant therapy in the gut microbiome era. Front Pharmacol (2021) 12:711788. doi: 10.3389/fphar.2021.711788
55. So JS, Kwon HK, Lee CG, Yi HJ, Park JA, Lim SY, et al. Lactobacillus casei suppresses experimental arthritis by down-regulating T helper 1 effector functions. Mol Immunol (2008) 45(9):2690–9. doi: 10.1016/j.molimm.2007.12.010
56. Bungau SG, Behl T, Singh A, Sehgal A, Singh S, Chigurupati S, et al. Targeting probiotics in rheumatoid arthritis. Nutrients (2021) 13(10):3376. doi: 10.3390/nu13103376
57. Paul AK, Paul A, Jahan R, Jannat K, Bondhon TA, Hasan A, et al. Probiotics and amelioration of rheumatoid arthritis: Significant roles of lactobacillus casei and lactobacillus acidophilus. Microorganisms (2021) 9(5):1070. doi: 10.3390/microorganisms9051070
58. Pan H, Guo R, Ju Y, Wang Q, Zhu J, Xie Y, et al. A single bacterium restores the microbiome dysbiosis to protect bones from destruction in a rat model of rheumatoid arthritis. Microbiome (2019) 7(1):107. doi: 10.1186/s40168-019-0719-1
59. Hatakka K, Martio J, Korpela M, Herranen M, Poussa T, Laasanen T, et al. Effects of probiotic therapy on the activity and activation of mild rheumatoid arthritis–a pilot study. Scand J Rheumatol (2003) 32(4):211–5. doi: 10.1080/03009740310003695
60. Pineda Mde L, Thompson SF, Summers K, de Leon F, Pope J, Reid G. A randomized, double-blinded, placebo-controlled pilot study of probiotics in active rheumatoid arthritis. Med Sci Monit (2011) 17(6):Cr347–54. doi: 10.12659/msm.881808
61. Antushevich H. Fecal microbiota transplantation in disease therapy. Clin Chim Acta (2020) 503:90–8. doi: 10.1016/j.cca.2019.12.010
62. Zeng J, Peng L, Zheng W, Huang F, Zhang N, Wu D, et al. Fecal microbiota transplantation for rheumatoid arthritis: A case report. Clin Case Rep (2021) 9(2):906–9. doi: 10.1002/ccr3.3677
63. Yan H, Su R, Xue H, Gao C, Li X, Wang C. Pharmacomicrobiology of methotrexate in rheumatoid arthritis: Gut microbiome as predictor of therapeutic response. Front Immunol (2021) 12:789334. doi: 10.3389/fimmu.2021.789334
64. Zaharko DS, Bruckner H, Oliverio VT. Antibiotics alter methotrexate metabolism and excretion. Science (1969) 166(3907):887–8. doi: 10.1126/science.166.3907.887
65. Rodrigues GSP, Cayres LCF, Gonçalves FP, Takaoka NNC, Lengert AH, Tansini A, et al. Detection of increased relative expression units of bacteroides and prevotella, and decreased clostridium leptum in stool samples from Brazilian rheumatoid arthritis patients: A pilot study. Microorganisms (2019) 7(10):413. doi: 10.3390/microorganisms7100413
66. Krook A. Effect of metronidazole and sulfasalazine on the normal human faecal flora. Scand J Gastroenterol (1981) 16(5):587–92. doi: 10.3109/00365528109182016
67. Peppercorn MA, Goldman P. The role of intestinal bacteria in the metabolism of salicylazosulfapyridine. J Pharmacol Exp Ther (1972) 181(3):555–62.
68. Pullar T, Hunter JA, Capell HA. Which component of sulphasalazine is active in rheumatoid arthritis? Br Med J (Clin Res Ed) (1985) 290(6481):1535–8. doi: 10.1136/bmj.290.6481.1535
69. Neumann VC, Shinebaum R, Cooke EM, Wright V. Effects of sulphasalazine on faecal flora in patients with rheumatoid arthritis: A comparison with penicillamine. Br J Rheumatol (1987) 26(5):334–7. doi: 10.1093/rheumatology/26.5.334
70. Kanerud L, Scheynius A, Nord CE, Hafström I. Effect of sulphasalazine on gastrointestinal microflora and on mucosal heat shock protein expression in patients with rheumatoid arthritis. Br J Rheumatol (1994) 33(11):1039–48. doi: 10.1093/rheumatology/33.11.1039
71. Picchianti-Diamanti A, Panebianco C, Salemi S, Sorgi ML, Di Rosa R, Tropea A, et al. Analysis of gut microbiota in rheumatoid arthritis patients: Disease-related dysbiosis and modifications induced by etanercept. Int J Mol Sci (2018) 19(10):2938. doi: 10.3390/ijms19102938
72. Wang B, He Y, Tang J, Ou Q, Lin J. Alteration of the gut microbiota in tumor necrosis factor-α antagonist-treated collagen-induced arthritis mice. Int J Rheum Dis (2020) 23(4):472–9. doi: 10.1111/1756-185x.13802
73. Wang Y, Chen S, Du K, Liang C, Wang S, Owusu Boadi E, et al. Traditional herbal medicine: Therapeutic potential in rheumatoid arthritis. J Ethnopharmacol (2021) 279:114368. doi: 10.1016/j.jep.2021.114368
74. Mei L, Yang Z, Zhang X, Liu Z, Wang M, Wu X, et al. Sustained drug treatment alters the gut microbiota in rheumatoid arthritis. Front Immunol (2021) 12:704089. doi: 10.3389/fimmu.2021.704089
75. Peng J, Lu X, Xie K, Xu Y, He R, Guo L, et al. Dynamic alterations in the gut microbiota of collagen-induced arthritis rats following the prolonged administration of total glucosides of paeony. Front Cell Infect Microbiol (2019) 9:204. doi: 10.3389/fcimb.2019.00204
76. Shan J, Peng L, Qian W, Xie T, Kang A, Gao B, et al. Integrated serum and fecal metabolomics study of collagen-induced arthritis rats and the therapeutic effects of the zushima tablet. Front Pharmacol (2018) 9:891. doi: 10.3389/fphar.2018.00891
77. Huang Y, Li M, Zhou L, Xu D, Qian F, Zhang J, et al. Effects of qingluo tongbi decoction on gut flora of rats with adjuvant-induced arthritis and the underlying mechanism. Evid Based Complement Alternat Med (2019) 2019:6308021. doi: 10.1155/2019/6308021
78. Alwarith J, Kahleova H, Rembert E, Yonas W, Dort S, Calcagno M, et al. Nutrition interventions in rheumatoid arthritis: The potential use of plant-based diets. A Review Front Nutr (2019) 6:141. doi: 10.3389/fnut.2019.00141
79. DeChristopher LR, Uribarri J, Tucker KL. Intake of high-fructose corn syrup sweetened soft drinks, fruit drinks and apple juice is associated with prevalent arthritis in US adults, aged 20-30 years. Nutr Diabetes (2016) 6(3):e199. doi: 10.1038/nutd.2016.7
80. Wagnerberger S, Spruss A, Kanuri G, Volynets V, Stahl C, Bischoff SC, et al. Toll-like receptors 1-9 are elevated in livers with fructose-induced hepatic steatosis. Br J Nutr (2012) 107(12):1727–38. doi: 10.1017/s0007114511004983
81. Costantini L, Molinari R, Farinon B, Merendino N. Impact of omega-3 fatty acids on the gut microbiota. Int J Mol Sci (2017) 18(12):2645. doi: 10.3390/ijms18122645
82. Gioxari A, Kaliora AC, Marantidou F, Panagiotakos DP. Intake of ω-3 polyunsaturated fatty acids in patients with rheumatoid arthritis: A systematic review and meta-analysis. Nutrition (2018) 45:114–24.e4. doi: 10.1016/j.nut.2017.06.023
83. Di Giuseppe D, Wallin A, Bottai M, Askling J, Wolk A. Long-term intake of dietary long-chain n-3 polyunsaturated fatty acids and risk of rheumatoid arthritis: A prospective cohort study of women. Ann Rheum Dis (2014) 73(11):1949–53. doi: 10.1136/annrheumdis-2013-203338
84. Minamino H, Katsushima M, Hashimoto M, Fujita Y, Yoshida T, Ikeda K, et al. Urinary sodium-to-potassium ratio associates with hypertension and current disease activity in patients with rheumatoid arthritis: A cross-sectional study. Arthritis Res Ther (2021) 23(1):96. doi: 10.1186/s13075-021-02479-x
85. Wang C, Huang Z, Yu K, Ding R, Ye K, Dai C, et al. High-salt diet has a certain impact on protein digestion and gut microbiota: A sequencing and proteome combined study. Front Microbiol (2017) 8:1838. doi: 10.3389/fmicb.2017.01838
86. Yang YH, Istomine R, Alvarez F, Al-Aubodah TA, Shi XQ, Takano T, et al. Salt sensing by Serum/Glucocorticoid-regulated kinase 1 promotes Th17-like inflammatory adaptation of Foxp3(+) regulatory T cells. Cell Rep (2020) 30(5):1515–29.e4. doi: 10.1016/j.celrep.2020.01.002
Keywords: rheumatoid arthritis, gut microbiota, immune response, inflammation, drug treatment
Citation: Zhao T, Wei Y, Zhu Y, Xie Z, Hai Q, Li Z and Qin D (2022) Gut microbiota and rheumatoid arthritis: From pathogenesis to novel therapeutic opportunities. Front. Immunol. 13:1007165. doi: 10.3389/fimmu.2022.1007165
Received: 30 July 2022; Accepted: 17 August 2022;
Published: 08 September 2022.
Edited by:
Ying Yang, Yunnan University, ChinaReviewed by:
Xinchang Wang, Zhejiang Chinese Medical University, ChinaCopyright © 2022 Zhao, Wei, Zhu, Xie, Hai, Li and Qin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dongdong Qin, cWluZG9uZzEwOEAxNjMuY29t; Qingshan Hai, aGFpcWluZ3NoYW5AMTYzLmNvbQ==; Zhaofu Li, bHpmMDgxN0AxMjYuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.