
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 08 December 2022
Sec. Viral Immunology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.1006395
This article is part of the Research TopicA year in review: Coronavirus infection in Humans and Viral ImmunologyView all 21 articles
 Ruoxuan Xiao1,2
Ruoxuan Xiao1,2 Ao Zhang1,2*
Ao Zhang1,2*The coronavirus disease 2019 (COVID-19) pandemic caused by the infection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has cast a notorious damage to the public health and global economy. The Stimulator of Interferon Genes (STING) is a crucial element of the host antiviral pathway and plays a pivotal but complex role in the infection and development of COVID-19. Herein, we discussed the antagonistic mechanism of viral proteins to the STING pathway as well as its activation induced by host cells. Specifically, we highlighted that the persistent activation of STING by SARS-CoV-2 led to abnormal inflammation, and STING inhibitors could reduce the excessive inflammation. In addition, we also emphasized that STING agonists possessed antiviral potency against diverse coronavirus and showed adjuvant efficacy in SARS-CoV-2 vaccines by inducing IFN responses.
Coronavirus disease 2019 (COVID-19) has rapidly spread across the world since the end of 2019, resulting in over 500 million confirmed infected cases and over 6 million deaths so far (https://www.who.int/). The corresponding virus of COVID-19 was identified as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus, which contains a positive single-stranded RNA genome (1, 2). COVID-19 has caused disastrous damage to the public health and the economic development of the world, and a few treatment options and vaccines have been developed to reduce it (3, 4). Remdesivir, a broad-spectrum antiviral drug, is the first drug approved by FDA for COVID-19 treatment (5). Another small molecule antiviral agent, Paxlovid (nirmatrelvir/ritonavir), was approved for the treatment of adult patients with mild-to-moderate COVID-19 (6). Meanwhile, oral or intravenous administration of dexamethasone was reported to reduce the 28-day mortality in patients hospitalized with COVID-19 (7). Additionally, various therapeutic monoclonal antibodies have been applied to treat COVID-19, such as Regkirona (regdanvimab) and REGEN-COV (casirivimab and imdevimab) (8–10). Combination of interleukin-6 receptor blocker (tocilizumab or sarilumab) and JAK inhibitor baricitinib is strong recommended for patients with severe or critical COVID-19 (11). Moreover, a number of vaccines has been developed to prevent the infection of SARS-CoV-2, such as BNT162b2 and mRNA-1273, demonstrating appreciable efficacy in phase III clinical trials (12, 13). Nevertheless, available drugs and vaccines are insufficient to combat the continuous emergence of viral variants, and the excessive inflammation induced by existent treatments should be concerned. Therefore, it is urgent to develop novel prophylactic and therapeutical measures to prevent SARS-CoV-2 from continuous infection, mutation, and transmission.
SARS-CoV-2 belongs to the β-coronavirus genus, which includes SARS-CoV, Middle East respiratory syndrome (MERS)-CoV and bat coronavirus HKU4 and so on (14–17). The virus genome consists of 14 open reading frames (ORFs) that encode 16 nonstructural proteins (nsp), structural proteins (spike protein S, membrane protein M, envelope protein E, and nucleocapsid protein N) and 9 accessory proteins (ORF3a, ORF3b, ORF6, ORF7a, ORF7b, ORF8, ORF9b, ORF9c and ORF10) (Figure 1A) (1, 18). The life cycle of SARS-CoV-2 is displayed in Figure 1B, and the interaction between the S protein of SARS-CoV-2 and the ACE2 on host cells is essential for the infection, thus ACE2 and S protein are important targets for treatment of COVID-19 (18, 19).
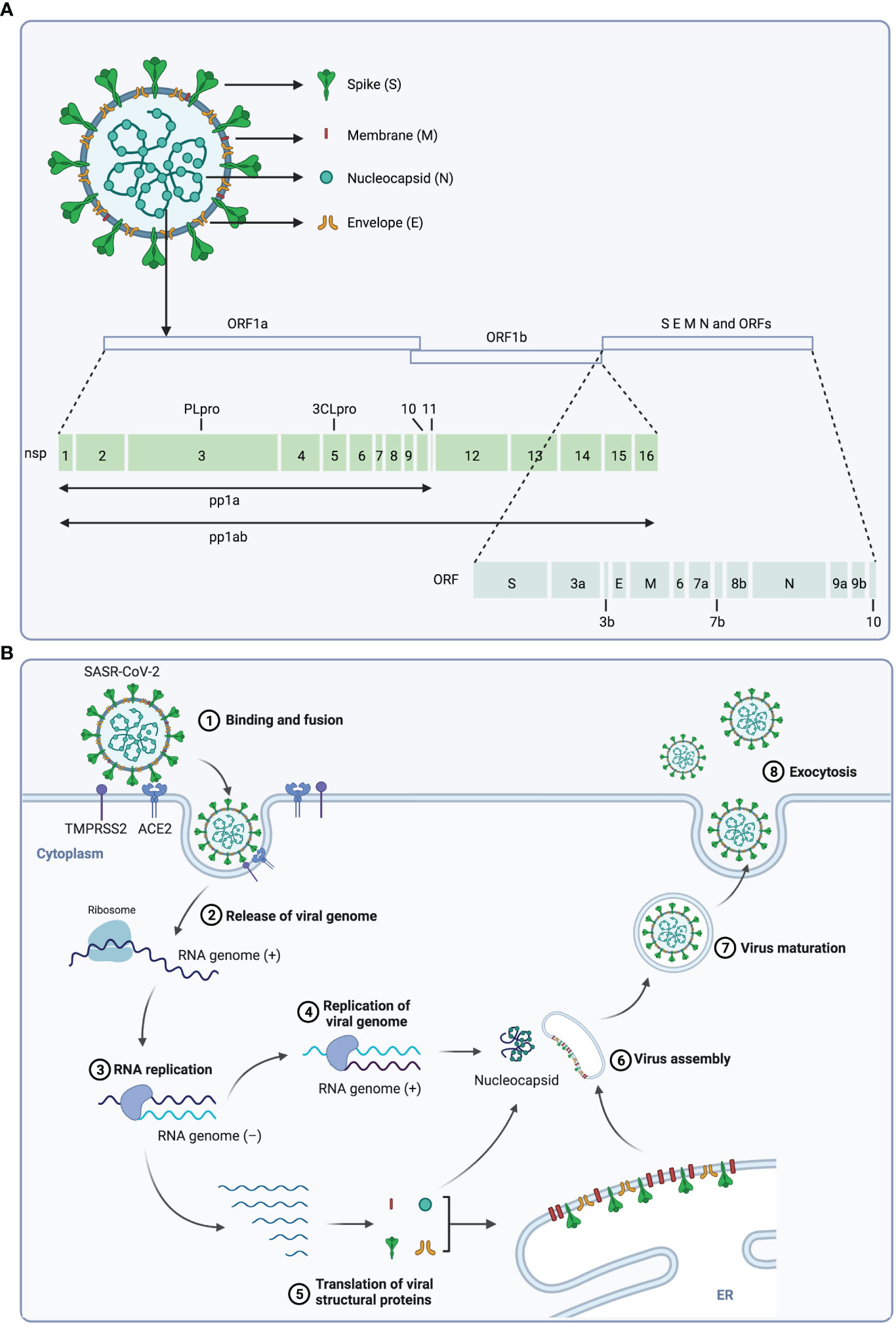
Figure 1 The structure and life cycle of SARS-CoV-2 virus. (A) The structure and genome of SARS-CoV-2 virus. The SARS-CoV-2 virus is composed of four structural proteins (spike protein S, membrane protein M, envelope protein E, and nucleocapsid protein N) and a single-stranded RNA genome. The virus genome contains 14 ORFs encoding 16 nonstructural proteins, 4 structural proteins and 9 accessory proteins respectively. (B) Scheme of the SARS-CoV-2 replication cycle. At the initial step of infection of this virus, the S1 subunit of the S protein interacts with the receptor angiotensin-converting enzyme 2 (ACE2) of host cells, and the S2 subunit is cleaved by TMPRSS2, a serine protease on the host cell surface, to promote uptake and fusion. Subsequently, the viral RNA is released into the cytoplasm of the host cell, and the ORF1a and ORF1b at the 5’-end are translated to polyproteins (pp1a and pp1ab), which is then cleaved by viral proteases 3CLpro and PLpro to 16 nonstructural proteins (nsps). These nsps form the replication and transcription complex to synthesize progeny viral genomic RNA. In parallel, ORFs at 3’-end are translated to structural proteins, and the S, M and E proteins are translocated to the ER-to-Golgi compartment, where they are assembled with N-encapsulated genomic RNA and then secreted out of cell through exocytosis.
The innate immune system is the first line of defense against evading pathogens (20). It recognizes pathogen/damage associated molecular patterns (PAMPs/DAMPs) by pattern recognition receptors (PPRs), including Toll-like receptors (TLRs), Nod-like receptors (NLRs), RIG-I-like receptors (RLRs) and the DNA sensor cyclic guanosine monophosphate (GMP)-adenosine monophosphate (AMP) synthase (cGAS)-stimulator of interferon genes (STING) signaling pathway. Among them, the cGAS-STING pathway plays an important role in innate immune response to pathogen infection. Mechanistically, the double stranded DNA (dsDNA) of pathogens is accumulated in cytoplasm and activates cGAS to generate 2′3′-cyclic GMP-AMP (2′3′-cGAMP), which binds to and activates STING (21–24). The bound STING is translocated from endoplasmic reticulum (ER) to Golgi, where it recruits the kinase TANK-binding kinase 1 (TBK1) and stimulate IκB kinase (IKK), causing phosphorylation of interferon regulatory factor 3 (IRF3) and nuclear factor-κB (NF-κB) (25, 26). Subsequently, the transcription of type I interferons (IFNs) and other inflammatory genes was triggered, which mediate immune response to eliminate pathogens (27–29). In addition to dsDNA from pathogens, endogenous DNA including chromosomal DNA and mitochondrial DNA can also trigger the cGAS-STING signaling pathway. Normally, chromatin is strictly compartmentalized in the nucleus to prevent cGAS-STING activation, while chromosome mis-segregation during cell mitosis leads to the generation of micronuclei, induing the aberrant recognition by cGAS (30, 31). Similarly, abnormal packaging of mitochondrial DNA (mtDNA) facilitates the escape of mtDNA into cytosol, which induces the activation of cGAS-STING (32).
As an RNA virus, SARS-CoV-2 is primarily recognized by RLRs in the host cells (33). Interestingly, increasing evidences demonstrated that the cGAS-STING pathway, a key DNA sensor, restricted the infection of RNA virus, and the proteins of RNA virus could antagonize the cGAS-STING signalling (34). For example, Sting-/- mice were more sensitive to vesicular stomatitis virus (VSV), a negative-stranded virus, and the production of type I IFNs was decreased in Sting-/- mice (35). Besides, the papain-like protease (PLpro) of SARS-CoV was reported to disrupt the STING-tumor necrosis factor receptor-associated factor 3 (TRAF3)-TBK1 complex by directly binding to it, and the dimerization and ubiquitination of STING were blocked by the PLpro of SARS-CoV and human coronavirus (HCoV) (36, 37). Hence, as an RNA virus, how SARS-CoV-2 interacts with STING pathway is worthy of further exploration.
SARS-CoV-2 infection has a double-edged effect on the STING signaling, dependent on the stage of disease procession and the infected tissues. Initially, Rui and colleagues hypothesized that SARS-CoV-2 might antagonize the innate immune pathway due to the antiviral function of STING (38). They investigated the effect of SARS-CoV-2 proteins on STING and RLR-mediated immune response, and found that both ORF3a and 3CL of the virus could inhibit STING and the downstream NF-κB signaling, but not IRF3 signaling, and this process was independent of cGAS. Further, it was found that ORF3a directly interacted with both the N-terminal and the C-terminal fragment of STING and suppressed the nuclear accumulation of p65, which then inhibited STING-mediated NF-κB signaling. While viral 3CL, through its enzymatic activity, inhibited the NF-κB pathway by suppressing the K63-ubiquitination of STING. In addition, the polymorphisms of STING from different species, including human, mouse and chicken could be inhibited by 3CL and ORF3a. However, bat STING, the natural host of SARS-CoV-2, was found defective to produce type-I interferon (IFN) and thus showed compromised anti-viral potency (39, 40). These results suggest that STING might be involved in the transmission of the virus. This is the first study supporting that SARS-CoV-2 can suppress STING signaling to escape from innate immune response.
Similarly, Han and co-workers reported that the ORF9b of SARS-CoV-2 suppressed the induction of type I and III interferons through multiple innate immune signaling, including RLR, TLR and STING (41). The antiviral activity of the host cell against SARS-CoV-2 depends on the production of type I and III IFNs, which is impaired in the serum of COVID-19 patients. However, the ORF9b of SARS-CoV-1 has been reported to inhibit IFNs response (2, 42–44). Based on these findings, they explored the effect of ORF9b on IFNs response and found that ORF9b of SARS-CoV-2 antagonized type I and III IFN responses induced by SeV and suppressed the activation of RLR, TLR and the cGAS-STING pathway. Mechanistic studies indicated that ORF9b directly interacted with RIG-I, MDA-5, MAVS, TBK1, TRIF and STING, and suppressed the phosphorylation of TBK1 and IRF3 along with IRF3 nuclear translocation. Furthermore, overexpression of viral ORF9b facilitated VSV infection, suggesting that ORF9b is closely implicated with the pathogenesis of COVID-19.
Recently, the ORF10 of SARS-CoV-2 has also been found to antagonize STING-dependent interferon response. Han et al. screened 29 SARS-CoV-2 viral proteins, and found ORF10 could suppress the activation of the cGAS-STING pathway by interacting with STING directly. As a result, the STING-TBK1 interaction was impeded, and the translocation of STING was blocked, leading the immune evasion of SARS-CoV-2 (45).
Taken together, various components of SARS-CoV-2 could inhibit the STING pathway and subsequent interferon response, leading to virus escape from innate immunity (Table 1).
Intriguingly, contrary to the aforementioned findings that SARS-CoV-2 antagonizes the STING signaling, many other studies suggest that infection with SARS-CoV-2 could trigger STING signaling. Transcriptome data showed that at the time of diagnosis, the content of STING protein was increased in the blood of patients with mild or moderate symptoms, whereas there is no significant change in the severe patients (46). Furthermore, STING expression was found to be elevated only in moderate patients at a few days after diagnosis (46). These data indicate that activation of STING might be associated with the severity and stage of COVID-19.
More detailed mechanistic studies indicate that cell fusion and formation of syncytia and micronuclei play crucial roles in SARS-CoV-2-induced cGAS-STING signaling (47, 48). Previous study has demonstrated that cell fusion mediated by the interaction of S protein with host ACE2 results in the formation of syncytia, presenting as a single cell containing several nuclei (49). Further, Ren and co-workers explored molecular events after syncytium formation in the well-established syncytia model. The results showed that both S protein and SARS-CoV-2 induced syncytium formation in HeLa-ACE2 cells and then led to production of micronuclei. Eventually, DNA damage and genome instability of the micronuclei promoted the activation of the cGAS-STING pathway as well as the downstream IFN response (50).
Furthermore, more studies were conducted to elucidate the mechanism of innate immune activation caused by SARS-CoV-2 infection. In addition to the activation of cGAS-STING induced by DNA from micronuclei, Zhou and co-workers further demonstrated that syncytia formation caused cytoplasmic chromatin by disrupting the actin cytoskeleton and nuclear lamin A/C, which are important factors for maintaining nuclear morphology. Meanwhile, they found that STING agonists (diABZI and SR-717) exhibited antiviral activity against SARS-CoV-2 (51). Meanwhile, cleavage of S protein by host proteases was found essential for cell fusion and IFN response (52).
The generation of syncytia provides a possible mechanism for delayed IFN response in COVID-19 patients, indicating that the production of type I IFN is inhibited at early stage of SARS-CoV-2 infection, but then substantially activated at the late stage (44, 53).
Although STING activation presents antiviral potential, the sustained STING signaling results in excessive amounts of type I IFNs. Indeed, patients with severe COVID-19 exhibited robust type I interferon response, which was associated with acute respiratory distress syndrome, lung injury and poor clinical outcome (1, 54–57). Therefore, over-activation of STING pathway may lead to hyperinflammation and related syndromes in COVID-19 patients.
By profiling the transcriptome and secreted cytokines of SARS-CoV-2-infected lung epithelial cells, Neufeldt and co-workers found that the NF-κB and pro-inflammatory pathway was up-regulated in infected lung epithelial cells, while the antiviral IFN response was not enhanced (58). These observations were consistent with the clinical features of severe patients (2, 59). Subsequent studies further proved that the hyper-inflammatory response is attributed to the activation of NF-κB but not IRF3, and is mediated by cGAS-STING pathway rather the RLR and TLR pathways. This imbalanced immune response recruited macrophage and neutrophils to cause cell death and lung pathology (58, 60). Finally, VS-X4 and H151, the established STING inhibitors (61), suppressed the upregulation of inflammatory cytokines and alleviated the abnormal immune response, which may protect COVID-19 patients from further suffering of this disease (62, 63).
Different from other studies on lung epithelial cells or tumor cell models, Domizio and colleagues focus on skin manifestations in SARS-CoV-2 infection (64). They found that cGAS-STING signaling and subsequent type I IFN production were initiated in endothelial cells and perivascular macrophages around injured vessels. As a result, the production of type I IFN in endothelial cells promoted cell death and tissue damage. Accordingly, administration of the STING inhibitor H151 reduced type I IFN response and related lung pathology in mice infected with SARS-CoV-2 (19).
Since viral proteins suppress the STING pathway in the early stage of infection, while the micronucleus and DNA damage caused by cell fusion in the host activate the STING signaling to suppress viral infection, treatment with STING agonists in the early stage of COVID-19 provides be a potential antiviral strategy.
Recently, Li and co-workers found that SASR-CoV-2 infection induced delay of IFN response to evade innate immunity, which could be controlled by type I IFN treatment (65). Subsequently, they screened 75 agonists targeting diverse PRR pathway and identified cyclic dinucleotides (CDNs), the endogenous stimulator of STING (66), showing antiviral activity against SARS-CoV-2. One of the potent STING agonists diABZI was then tested in subsequent studies due to its significant potency and higher bioavailability. As expected, diABZI elicited potent and transient innate signaling and prevented SARS-CoV-2 infection in primary human respiratory epithelial cells as well as the lung of mice. A single intranasal delivery of diZBAI protected mice from lethality induced by SARS-CoV-2 and its South African variant B.1.351, thus supporting the therapeutic potential of diABZI against diverse trains of SARS-CoV-2 (65).
Similarly, the diABZI analogue, diABZI-4, was proved as well to prevent SARS-CoV-2 replication in ACE2-A549 cells and in 3D-cultured embryonic stem cell–derived induced alveolar type II (iAT2) cells (67). Intranasal administration of diABZI-4 before or after virus infection reduced weight loss and death in K18-ACE2 mice without pathological damage in lung tissues. Furthermore, diABZI induced transient pro-inflammatory cytokines production and promoted the activation of myeloid cells, T cells and NK cells, without pathological damage and excessive inflammation in lung tissue (67).
Taken together, STING agonists could effectively activate the antiviral response and prevent SARS-CoV-2 infection in vivo and in vitro. The activation is transient, but can prevent lung tissue damage from abnormal inflammation. Therefore, STING agonists may provide alternative strategy for the treatment of COVID-19 in the early stage (Table 2).
Vaccines have made great contribution to COVID-19 prevention (70). Of which subunit vaccines are the most used due to their excellent efficacy and safety. The spike protein of SARS-Cov-2 and its receptor binding domain (RBD) have been considered as the two main antigens in COVID-19 vaccine, because their corresponding epitope domains could induce the production of neutralizing antibodies (71, 72). However, the poor immunogenicity of the highly purified S protein and RBD limits the development of effective SARS-CoV-2 vaccine. Therefore, additional adjuvants are necessary to elicit robust and durable immune response. Aluminum salts (Alum) are the most commonly used adjuvant, however, poor antibody immune response and predominant Th2 response restrict their use for various antigens (73, 74). With the development of innate immunity research, PAMPs/DAMPs has attracted attention as potential vaccine adjuvants, and STING agonists has been employed as adjuvants in multiple pre-clinical vaccines (75). Mechanistically, activation of STING could maturate DCs and prime T cells, leading to subsequent humoral immunity to control virus (76, 77). Neutralizing antibodies produced by humoral immunity contributes to the virus clearance potency of vaccines, and STING agonists were reported to increase antibody titers and trigger potent humoral immune response (78).Additionally, STING agonists induced the formation of germinal center (GC), where B cells were primed and differenced into memory B cells to achieve long-lasting profection towards virus (79, 80). Therefore, STING agonists are promising adjuvant for constructing effective SARS-CoV-2 vaccines.
cGAMP, the natural ligand of STING, has been widely investigated as adjuvant in vaccine development. cGAMP and S protein-loaded HIV-derived virus-like particles (VLPs) was reported to induce more potential antibody response compared with S protein-loaded VLPs. The virus neutralizing capacity of resulting antibodies was improved as well (81). An intranasal subunit vaccine accompanied by liposomal cGAMP and lyophilized S protein was reported to trigger robust neutralizing antibodies and comprehensive immune response in lung, spleen and nasal compartments (82). Additionally, a ternary adjuvant system consisting of Alum, cGAMP and TLR3 agonist poly(I:C) was used to construct a S1 protein vaccine, and the ternary adjuvant showed potent adjuvant effect on inducing humoral and cellular immunity without apparent biological toxicity in immunized mice (83).
In addition to cGAMP, a few novel STING agonists were also used as adjuvants in SARS-CoV-2 vaccine. For instance, Wu and co-workers synthesized the analogue CDGSF by modifying cyclic di-GMP (CDG) with one phosphorothioate and one fluorine moieties (84). The fluorine modification enhanced the liposolubility and stability of CDG (85), and increased the expression of CD86 in macrophage and bone marrow-derived dendritic cells (BMDC). As adjuvant, CDGSF significantly improved the S protein-specific IFN-γ secretion and IgG titers, more potent than classical adjuvant Alum, thus highlighting the adjuvant potential of CDGSF in SARS-CoV-2 vaccine preparation.
CF501 is a new STING agonist and was found to show potent adjuvant efficacy in pan-sarbecovirus vaccine (86). Compared with Alum- or cGAMP-adjuvanted RBD-Fc-based vaccines, intramuscular injection of CF501-adjuvanted RBD-Fc vaccine (CF501/RBD-Fc) triggered stronger humoral and cellular immune responses against various variants of SARS-CoV-2, SARS-CoV and SARSs-CoVs from bats in mice, rabbit and rhesus macaques models. Further, CF501/RBD-Fc induced long-term protective immunity against SARS-CoV-2 challenge in both macaques and hACE-transgenic mice. Moreover, CF501 transiently triggered innate immunity without obvious lesion in the tissue of CF501/RBD-Fc-immunized mice, suggesting the good safety profile of CF501 as adjuvant (86).
It is well established that the STING pathway elucidates a double-edged effect on COVID-19. At the early stage of infection, STING signaling is suppressed by vital proteins containing 3CL, ORF3a and ORF9b, resulting in the impaired innate immune response (38, 41). In contrast, the fusion of S protein of the virus with the ACE2 receptor in host cells leads to syncytia formation, resulting in formation of micronucleus and DNA damage, and consequently triggering the STING signaling and antiviral response (50, 51). These may account for the observed activation delay of type I IFN response in COVID-19 patients (57). Therefore, treatment with STING agonists could effectively activate innate immune response to inhibit virus infection and replication. However, excessive and sustained activation of STING signaling leads to accumulation of inflammatory factors, resulting in abnormal lung inflammation and poor clinical outcomes (Figure 2) (52, 58, 64). Hence, trials of STING agonists and inhibitors in the treatment of COVID-19 should be cautiously evaluated in context. Timing and duration are critical factors, at the early state of infection, STING agonists may be used to recuse the deficiency of IFNs production. Instead, the inhibitors of STING could be applied to suppress the excessive inflammatory response and to alleviate the tissue injury caused by the disease. Since type I IFN response is the bridge between STING pathway and COVID-19 progression, we speculate that the content of interferons in the patients might be a bio-marker for the use of STING regulators in clinical.
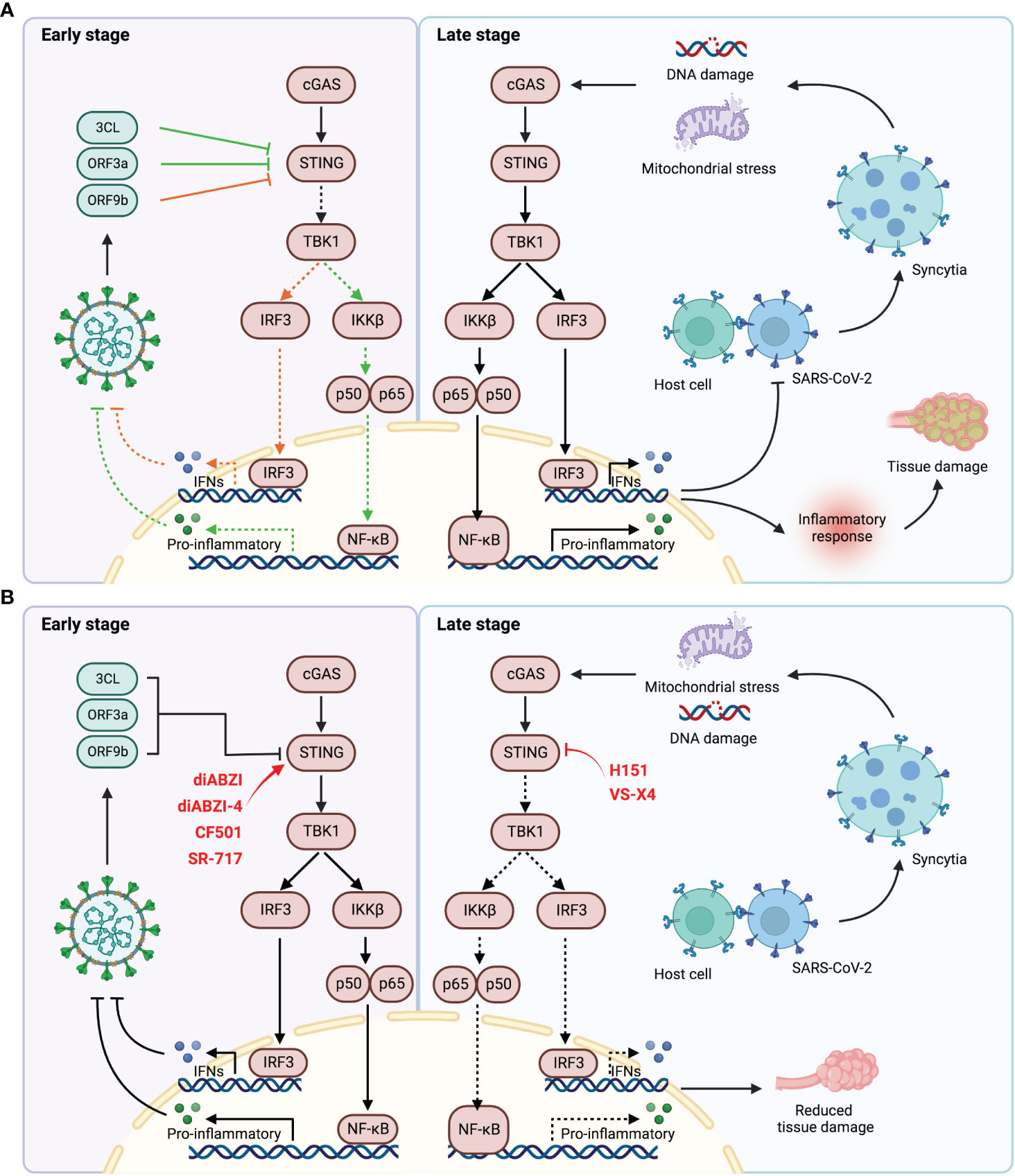
Figure 2 The relationship of SARS-CoV-2 infection and STING pathway. (A) At the early stage of SARS-CoV-2 infection, the viral proteins inhibited the activation of STING pathway by direct interaction with STING (left). During the late stage of infection, the host cells fused with virus through the interaction of ACE2 and S protein to form syncytia, which contains a large number of micronuclei, mediating DNA damage and thus activating STING signaling. Durable and excessive STING activation lead to abnormal inflammatory response, resulting in tissue damage and poor prognosis (right). (B) STING agonists could be used to activate STING signaling at the early stage of infection to elicit anti-viral response (left). STING inhibitors could attenuate tissue damage by suppressing excessive STING activation and aberrant inflammatory response (right).
XR collated the literature and wrote the review, and ZA established that the structure of the review and revised the article.
We thank financial supports from the Major Projects for Shanghai Zhangjiang National Independent Innovation of China (ZJ2021-ZD-007)), the National Natural Science Foundation of China (82273767), China National Innovation of Science and Technology-2030 (Program of Brain Science and Brain-Inspired Intelligence Technology, 2021ZD0204004). The figures were created with biorender.com.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Kim D, Lee JY, Yang JS, Kim JW, Kim VN, Chang H. The architecture of SARS-CoV-2 transcriptome. Cell (2020) 181:914–21.e910. doi: 10.1016/j.cell.2020.04.011
2. Blanco-Melo D, Nilsson-Payant BE, Liu WC, Uhl S, Hoagland D, Møller R, et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell (2020) 181:1036–1045.e1039. doi: 10.1016/j.cell.2020.04.026
3. Burki TK. The role of antiviral treatment in the COVID-19 pandemic. Lancet Respir Med (2022) 10:e18. doi: 10.1016/S2213-2600(22)00011-X
4. Kumari M, Lu RM, Li MC, Huang JL, Hsu FF, Ko SH, et al. A critical overview of current progress for COVID-19: development of vaccines, antiviral drugs, and therapeutic antibodies. J BioMed Sci (2022) 29:68. doi: 10.1186/s12929-022-00852-9
5. Malin JJ, Suárez I, Priesner V, Fätkenheuer G, Rybniker J. Remdesivir against COVID-19 and other viral diseases. Clin Microbiol Rev 34 (2020) 34, e00162-20. doi: 10.1128/CMR.00162-20
6. Reis S, Metzendorf MI, Kuehn R, Popp M, Gagyor I, Kranke P, et al. Nirmatrelvir combined with ritonavir for preventing and treating COVID-19. Cochrane Database Syst Rev 9 Cd015395 (2022) 9, CD015395. doi: 10.1002/14651858.CD015395.pub2
7. Horby P, Lim WS, Emberson JR, Mafham M, Bell JL, Linsell L, et al. Dexamethasone in hospitalized patients with covid-19. N Engl J Med (2021) 384:693–704. doi: 10.1056/NEJMoa2021436
9. Deeks ED. Casirivimab/Imdevimab: First approval. Drugs (2021) 81:2047–55. doi: 10.1007/s40265-021-01620-z
10. van de Veerdonk FL, Giamarellos-Bourboulis E, Pickkers P, Derde L, Leavis H, van Crevel R, et al. A guide to immunotherapy for COVID-19. Nat Med (2022) 28:39–50. doi: 10.1038/s41591-021-01643-9
11. Baricitinib in patients admitted to hospital with COVID-19 (RECOVERY): a randomised, controlled, open-label, platform trial and updated meta-analysis. Lancet (2022) 400:359–68. doi: 10.1016/S0140-6736(22)01109-6
12. Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, et al. Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N Engl J Med (2020) 383:2603–15. doi: 10.1056/NEJMoa2034577
13. Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med (2021) 384:403–16. doi: 10.1056/NEJMoa2035389
14. Woo PCY, Lau SKP, Chu CM, Chan KH, Tsoi HW, Huang Y, et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol (2005) 79:884–95. doi: 10.1128/JVI.79.2.884-895.2005
15. Drosten C, Günther S, Preiser W, van der Werf S, Brodt HR, Becker S, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med (2003) 348:1967–76. doi: 10.1056/NEJMoa030747
16. Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med (2012) 367:1814–20. doi: 10.1056/NEJMoa1211721
17. Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, et al. A new coronavirus associated with human respiratory disease in China. Nature (2020) 579:265–9. doi: 10.1038/s41586-020-2008-3
18. V'Kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat Rev Microbiol (2021) 19:155–70. doi: 10.1038/s41579-020-00468-6
19. Cevik M, Kuppalli K, Kindrachuk J, Peiris M. Virology, transmission, and pathogenesis of SARS-CoV-2. Bmj (2020) 371:m3862. doi: 10.1136/bmj.m3862
20. Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell (2006) 124:783–801. doi: 10.1016/j.cell.2006.02.015
21. Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature (2008) 455:674–8. doi: 10.1038/nature07317
22. Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science (2013) 339:786–91. doi: 10.1126/science.1232458
23. Wu J, Sun L, Chen X, Du F, Shi H, Chen C, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science (2013) 339:826–30. doi: 10.1126/science.1229963
24. Sun W, Li Y, Chen L, Chen H, You F, Zhou X, et al. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci U.S.A. (2009) 106:8653–8. doi: 10.1073/pnas.0900850106
25. Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity (2008) 29:538–50. doi: 10.1016/j.immuni.2008.09.003
26. Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U.S.A. (2009) 106:20842–6. doi: 10.1073/pnas.0911267106
27. Zhang X, Bai XC, Chen ZJ. Structures and mechanisms in the cGAS-STING innate immunity pathway. Immunity (2020) 53:43–53. doi: 10.1016/j.immuni.2020.05.013
28. Ritchie C, Carozza JA, Li L. Biochemistry, cell biology, and pathophysiology of the innate immune cGAS-cGAMP-STING pathway. Annu Rev Biochem (2022) 91:599–628. doi: 10.1146/annurev-biochem-040320-101629
29. Liu S, Cai X, Wu J, Cong Q, Chen X, Li , et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science (2015) 347:aaa2630. doi: 10.1126/science.aaa2630
30. Mackenzie KJ, Carroll P, Martin CA, Murina O, Fluteau A, Simpson DJ, et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature (2017) 548:461–5. doi: 10.1038/nature23449
31. Harding SM, Benci JL, Irianto J, Discher DE, Minn AJ, Greenberg RA. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature (2017) 548:466–70. doi: 10.1038/nature23470
32. West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature (2015) 520:553–7. doi: 10.1038/nature14156
33. Yoneyama M, Fujita T. RNA Recognition and signal transduction by RIG-i-like receptors. Immunol Rev (2009) 227:54–65. doi: 10.1111/j.1600-065X.2008.00727.x
34. Ma Z, Damania B. The cGAS-STING defense pathway and its counteraction by viruses. Cell Host Microbe (2016) 19:150–8. doi: 10.1016/j.chom.2016.01.010
35. Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature (2009) 461:788–92. doi: 10.1038/nature08476
36. Chen X, Yang X, Zheng Y, Yang Y, Xing Y, Chen Z. SARS coronavirus papain-like protease inhibits the type I interferon signaling pathway through interaction with the STING-TRAF3-TBK1 complex. Protein Cell (2014) 5:369–81. doi: 10.1007/s13238-014-0026-3
37. Sun L, Xing Y, Chen X, Zheng Y, Yang Y, Nichols DB, et al. Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. PloS One (2012) 7:e30802. doi: 10.1371/journal.pone.0030802
38. Rui Y, Su J, Shen S, Hu Y, Huang D, Zheng W, et al. Unique and complementary suppression of cGAS-STING and RNA sensing- triggered innate immune responses by SARS-CoV-2 proteins. Signal Transduct Target Ther (2021) 6:123. doi: 10.1038/s41392-021-00515-5
39. Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature (2020) 579:270–3. doi: 10.1038/s41586-020-2012-7
40. Xie J, Li Y, Shen X, Goh G, Zhu Y, Cui J, et al. Dampened STING-dependent interferon activation in bats. Cell Host Microbe (2018) 23:297–301.e294. doi: 10.1016/j.chom.2018.01.006
41. Han L, Zhuang MW, Deng J, Zheng Y, Zhang J, Nan ML, et al. SARS-CoV-2 ORF9b antagonizes type I and III interferons by targeting multiple components of the RIG-I/MDA-5-MAVS, TLR3-TRIF, and cGAS-STING signaling pathways. J Med Virol (2021) 93:5376–89. doi: 10.1002/jmv.27050
42. Hung IF, Lung KC, Tso EY, Liu R, Chung TW, Chu MY, et al. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet (2020) 395:1695–704. doi: 10.1016/S0140-6736(20)31042-4
43. Vanderheiden A, Ralfs P, Chirkova T, Upadhyay AA, Zimmerman MG, Bedoya S, et al. Type I and type III interferons restrict SARS-CoV-2 infection of human airway epithelial cultures. J Virol 94 (2020) 94, e00985-20. doi: 10.1128/JVI.00985-20
44. Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science (2020) 369:718–24. doi: 10.1126/science.abc6027
45. Han L, Zheng Y, Deng J, Nan ML, Xiao Y, Zhuang MW, et al. SARS-CoV-2 ORF10 antagonizes STING-dependent interferon activation and autophagy. J Med Virol (2022) 94:5174–88. doi: 10.1002/jmv.27965
46. Yang CA, Huang YL, Chiang BL. Innate immune response analysis in COVID-19 and kawasaki disease reveals MIS-c predictors. J Formos Med Assoc (2022) 121:623–32. doi: 10.1016/j.jfma.2021.06.009
47. Buchrieser J, Dufloo J, Hubert M, Monel B, Planas D, Rajah MM, et al. Syncytia formation by SARS-CoV-2-infected cells. EMBO J (2020) 39:e106267. doi: 10.15252/embj.2020106267
48. Rockx B, Kuiken T, Herfst S, Bestebroer T, Lamers MM, Oude Munnink , et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science (2020) 368:1012–5. doi: 10.1126/science.abb7314
49. Buchrieser J, Dufloo J, Hubert M, Monel B, Planas D, Rajah MM, et al. Syncytia formation by SARS-CoV-2-infected cells. EMBO J (2021) 40:e107405. doi: 10.15252/embj.2020107405
50. Ren H, Ma C, Peng H, Zhang B, Zhou L, Su Y, et al. Micronucleus production, activation of DNA damage response and cGAS-STING signaling in syncytia induced by SARS-CoV-2 infection. Biol Direct (2021) 16:20. doi: 10.1186/s13062-021-00305-7
51. Zhou Z, Zhang X, Lei X, Xiao X, Jiao T, Ma R, et al. Sensing of cytoplasmic chromatin by cGAS activates innate immune response in SARS-CoV-2 infection. Signal Transduct Target Ther (2021) 6:382. doi: 10.1038/s41392-021-00800-3
52. Liu X, Wei L, Xu F, Zhao F, Huang Y, Fan Z, et al. SARS-CoV-2 spike protein-induced cell fusion activates the cGAS-STING pathway and the interferon response. Sci Signal (2022) 15:eabg8744. doi: 10.1126/scisignal.abg8744
53. Yang D, Chu H, Hou Y, Chai Y, Shuai H, Lee AC, et al. Attenuated interferon and proinflammatory response in SARS-CoV-2-Infected human dendritic cells is associated with viral antagonism of STAT1 phosphorylation. J Infect Dis (2020) 222:734–45. doi: 10.1093/infdis/jiaa356
54. Ramasamy S, Subbian S. Critical determinants of cytokine storm and type I interferon response in COVID-19 pathogenesis. Clin Microbiol Rev 34 (2021) 34, e00299-20. doi: 10.1128/CMR.00163-21
55. Channappanavar R, Fehr AR, Vijay R, Mack M, Zhao J, Meyerholz DK, et al. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-Infected mice. Cell Host Microbe (2016) 19:181–93. doi: 10.1016/j.chom.2016.01.007
56. Channappanavar R, Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin immunopathol (2017) 39:529–39. doi: 10.1007/s00281-017-0629-x
57. Lee JS, Shin EC. The type I interferon response in COVID-19: implications for treatment. Nat Rev Immunol (2020) 20:585–6. doi: 10.1038/s41577-020-00429-3
58. Neufeldt CJ, Cerikan B, Cortese M, Frankish J, Lee JY, Plociennikowska A, et al. SARS-CoV-2 infection induces a pro-inflammatory cytokine response through cGAS-STING and NF-κB. Commun Biol (2022) 5:45. doi: 10.1038/s42003-021-02983-5
59. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in wuhan, China. Lancet (2020) 395:497–506. doi: 10.1016/S0140-6736(20)30183-5
60. Dunphy G, Flannery SM, Almine JF, Connolly DJ, Paulus C, Jønsson KL, et al. Non-canonical activation of the DNA sensing adaptor STING by ATM and IFI16 mediates NF-κB signaling after nuclear DNA damage. Mol Cell (2018) 71:745–760.e745. doi: 10.1016/j.molcel.2018.07.034
61. Haag SM, Gulen MF, Reymond L, Gibelin A, Abrami L, Decout A, et al. Targeting STING with covalent small-molecule inhibitors. Nature (2018) 559:269–73. doi: 10.1038/s41586-018-0287-8
62. Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson , et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet (2020) 395:1033–4. doi: 10.1016/S0140-6736(20)30628-0
63. Hoagland DA, Møller R, Uhl SA, Oishi K, Frere J, Golynker I, et al. Leveraging the antiviral type I interferon system as a first line of defense against SARS-CoV-2 pathogenicity. Immunity (2021) 54:557–570.e555. doi: 10.1016/j.immuni.2021.01.017
64. Domizio JD, Gulen MF, Saidoune F, Thacker VV, Yatim A, Sharma K, et al. The cGAS-STING pathway drives type I IFN immunopathology in COVID-19. Nature (2022) 603:145–51. doi: 10.1038/s41586-022-04421-w
65. Li M, Ferretti M, Ying B, Descamps H, Lee E, Dittmar M, et al. Pharmacological activation of STING blocks SARS-CoV-2 infection. Sci Immunol (2021) 6:eabi9007. doi: 10.1126/sciimmunol.abi9007
66. Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature (2011) 478:515–8. doi: 10.1038/nature10429
67. Humphries F, Shmuel-Galia L, Jiang Z, Wilson R, Landis P, Ng SL, et al. A diamidobenzimidazole STING agonist protects against SARS-CoV-2 infection. Sci Immunol (2021) 6:eabi9002. doi: 10.1126/sciimmunol.abi9002
68. Zhu Q, Zhang Y, Wang L, Yao X, Wu D, Cheng J, et al. Inhibition of coronavirus infection by a synthetic STING agonist in primary human airway system. Antiviral Res (2021) 187:105015. doi: 10.1016/j.antiviral.2021.105015
69. Liu W, Reyes HM, Yang JF, Li Y, Stewart KM, Basil MC, et al. Activation of STING signaling pathway effectively blocks human coronavirus infection. J Virol (2021) 95:e00490-21. doi: 10.1128/JVI.00490-21
70. Altmann DM, Boyton RJ. COVID-19 vaccination: The road ahead. Science (2022) 375:1127–32. doi: 10.1126/science.abn1755
71. Dai L, Gao GF. Viral targets for vaccines against COVID-19. Nat Rev Immunol (2021) 21:73–82. doi: 10.1038/s41577-020-00480-0
72. Wang Y, Wang L, Cao H, Liu C. SARS-CoV-2 S1 is superior to the RBD as a COVID-19 subunit vaccine antigen. J Med Virol (2021) 93:892–8. doi: 10.1002/jmv.26320
73. Reed SG, Orr MT, Fox CB. Key roles of adjuvants in modern vaccines. Nat Med (2013) 19:1597–608. doi: 10.1038/nm.3409
74. Arunachalam PS, Walls AC, Golden N, Atyeo C, Fischinger S, Li C, et al. Adjuvanting a subunit COVID-19 vaccine to induce protective immunity. Nature (2021) 594:253–8. doi: 10.1038/s41586-021-03530-2
75. Van Herck S, Feng B, Tang L. Delivery of STING agonists for adjuvanting subunit vaccines. Adv Drug Delivery Rev (2021) 179:114020. doi: 10.1016/j.addr.2021.114020
76. Gutjahr A, Papagno L, Nicoli F, Kanuma T, Kuse N, Cabral-Piccin MP, et al. The STING ligand cGAMP potentiates the efficacy of vaccine-induced CD8+ T cells. JCI Insight (2019) 4:e125107. doi: 10.1172/jci.insight.125107
77. Wang J, Li P, Yu Y, Fu Y, Jiang H, Lu M, et al. Pulmonary surfactant-biomimetic nanoparticles potentiate heterosubtypic influenza immunity. Science (2020) 367:eaau0810. doi: 10.1126/science.aau0810
78. Gray PM, Forrest G, Wisniewski T, Porter G, Freed DC, DeMartino JA, et al. Evidence for cyclic diguanylate as a vaccine adjuvant with novel immunostimulatory activities. Cell Immunol (2012) 278:113–9. doi: 10.1016/j.cellimm.2012.07.006
79. Hanson MC, Crespo MP, Abraham W, Moynihan KD, Szeto GL, Chen SH, et al. Nanoparticulate STING agonists are potent lymph node-targeted vaccine adjuvants. J Clin Invest (2015) 125:2532–46. doi: 10.1172/JCI79915
80. Zhang R, Wang C, Guan Y, Wei X, Sha M, Yi M, et al. Manganese salts function as potent adjuvants. Cell Mol Immunol (2021) 18:1222–34. doi: 10.1038/s41423-021-00669-w
81. Chauveau L, Bridgeman A, Tan TK, Beveridge R, Frost JN, Rijal P, et al. Inclusion of cGAMP within virus-like particle vaccines enhances their immunogenicity. EMBO Rep (2021) 22:e52447. doi: 10.15252/embr.202152447
82. An X, Martinez-Paniagua M, Rezvan A, Sefat SR, Fathi M, Singh S, et al. Single-dose intranasal vaccination elicits systemic and mucosal immunity against SARS-CoV-2. iScience (2021) 24:103037. doi: 10.1016/j.isci.2021.103037
83. Zhang RY, Yin XG, Zhou SH, Zhang HW, Lu J, He CB, et al. A protein vaccine with alum/c-GAMP/poly(I:C) rapidly boosts robust immunity against SARS-CoV-2 and variants of concern. Chem Commun (Camb) (2022) 58:3925–8. doi: 10.1039/D2CC00271J
84. Wu JJ, Zhao L, Han BB, Hu HG, Zhang BD, Li WH, et al. A novel STING agonist for cancer immunotherapy and a SARS-CoV-2 vaccine adjuvant. Chem Commun (Camb) (2021) 57:504–7. doi: 10.1039/D0CC06959K
85. Cavaliere A, Probst KC, Westwell AD, Slusarczyk M. Fluorinated nucleosides as an important class of anticancer and antiviral agents. Future Med Chem (2017) 9:1809–33. doi: 10.4155/fmc-2017-0095
Keywords: COVID-19, SARS-CoV-2, STING, vaccine, IFNs
Citation: Xiao R and Zhang A (2022) Involvement of the STING signaling in COVID-19. Front. Immunol. 13:1006395. doi: 10.3389/fimmu.2022.1006395
Received: 29 July 2022; Accepted: 24 November 2022;
Published: 08 December 2022.
Edited by:
Pei-Hui Wang, Shandong University, ChinaReviewed by:
Jianwei Wang, Chinese Academy of Medical Sciences and Peking Union Medical College, ChinaCopyright © 2022 Xiao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ao Zhang, YW82OTE5emhhbmdAc2p0dS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.