- 1Laboratory of Neuro-oncology, Tianjin Neurological Institute, Tianjin Medical University General Hospital, Key Laboratory of Post-Neuro Injury Neuro-repair and Regeneration in Central Nervous System, Ministry of Education, Tianjin, China
- 2Department of Neuro-Oncology and Neurosurgery, Tianjin Medical University Cancer Institute and Hospital, National Clinical Research Center for Cancer, Key Laboratory of Cancer Prevention and Therapy of Tianjin, Tianjin’s Clinical Research Center for Cancer, Tianjin, China
- 3Beijing Neurosurgical Institute, Department of Neurosurgery, Beijing Tiantan Hospital, Capital Medical University, Beijing, China
- 4Department of Neurosurgery, Affiliated Hospital of Hebei University, Baoding, China
- 5Key Laboratory of Precise Diagnosis and Treatment of Glioma in Hebei Province, Baoding, China
- 6Department of Neurosurgery, Tianjin Medical University General Hospital, Tianjin, China
- 7Department of Pathology, Affiliated Hospital of Hebei University, Baoding, China
- 8Department of Pathology, Hebei University School of Basic Medical Sciences, Baoding, China
Background: Immunotherapy, especially checkpoint inhibitors targeting PD-1 or PD-L1, has revolutionized cancer therapy. However, PD-1/PD-L1 inhibitors have not been investigated thoroughly in glioblastoma (GBM). Studies have shown that polymerase 1 and transcript release factor (PTRF/Cavin-1) has an immune-suppressive function in GBM. Thus, the relationship between PTRF and PD-L1 and their role in immune suppression requires further investigation in GBM.
Methods: We used public databases and bioinformatics analysis to investigate the relationship between PTRF and PD-L1. We next confirmed the predicted relationship between PTRF and PD-L1 in primary GBM cell lines by using different experimental approaches. RIP-Seq, RIP, ChIP, and qRT-PCR were conducted to explore the molecular mechanism of PTRF in immunosuppression.
Results: We found that PTRF stabilizes lncRNA NEAT1 to induce NF-κB and PD-L1 and promotes immune evasion in GBM. PTRF was found to correlate with immunosuppression in the public GBM databases. PTRF increased the level of PD-L1 in primary cell lines from GBM patients. We carried out RIP-Seq of GBM cells and found that PTRF interacts with lncRNA NEAT1 and stabilizes its mRNA. PTRF also promoted the activity of NF-κB by suppressing UBXN1 expression via NEAT1 and enhanced the transcription of PD-L1 through NF-κB activation. Finally, PTRF promoted immune evasion in GBM cells by regulating PD-1 binding and PD-L1 mediated T cell cytotoxicity.
Conclusions: In summary, our study identified the PTRF-NEAT1-PD-L1 axis as a novel immune therapeutic target in GBM.
Introduction
Glioblastoma (GBM) is the most aggressive primary brain tumor in adults (1). Standard treatment regimens, including surgery, temozolomide chemotherapy, and radiation therapy, fail to improve patient survival (2). Immunotherapy, particularly immune checkpoint blockade with anti-programmed death 1 (PD-1)/PD-1 ligand (PD-L1) antibodies, has radically changed the treatment of multiple tumors (3–5). However, clinical studies investigating the use of anti-PD-1/PD-L1 in GBM remain are limited (6).
PD-L1 is an important immune checkpoint molecule on cancer cells which plays a critical role in immune surveillance and immune evasion. The majority of GBM patients are resistant to anti-PD-1/PD-L1 therapy, and only a minority of patients respond to this immunotherapy (7). The response to PD-1/PD-L1 blockade is correlated with PD-L1 expression in tumor cells (8, 9). Previous studies demonstrated that PD-L1 protein levels are associated with glioma grades, with elevated levels on tumor cells facilitating immune evasion in glioma patients (10, 11). Thus, one possible mechanism of immune evasion in GBM is the upregulation of PD-L1 which requires an in-depth investigation. Recent studies show that the expression of PD-L1 in tumors is regulated by signaling pathways, transcription factors and epigenetic factors, such as the Wnt/β-catenin pathway (12), STAT3 (13), and NF-κB (14). To develop an effective strategy to enhance antitumor immunity, the underlying mechanisms of PD-L1 regulation in GBM need to be elucidated.
Polymerase 1 and Transcript Release Factor (PTRF, also known as Cavin-1) is upregulated in GBM with mesenchymal phenotype compared with other subtypes of GBM (15). The prognosis of GBM patients with high PTRF expression is also worse (15). Previous studies found that PTRF protects vascular endothelial cells from cytotoxic T cell-mediated lysis (16). Further, in the challenge phase of asthma, loss of PTRF led to intense airway inflammation and potent type 2 immune responses (17). In GBM, bioinformatics analysis showed a negative correlation between PTRF expression and cytotoxic lymphocytes (18). We also demonstrated that PTRF overexpression increased tumor proliferation and decreased tumor-infiltrating CD8+ T cells mediated anti-tumor immunity (19). However, the mechanism by which PTRF influences immunosuppression, particularly PD-L1 expression, in GBM needs further research.
Here, we show that PTRF correlates with immunosuppression and increases the expression of PD-L1 in GBM. Further we found that PTRF interacts with lncRNA NEAT1 and maintains the mRNA stability of NEAT1, increasing H3K27me3 level on the UBXN1 promoter, which results in the binding of NF-κB to the PD-L1 promoter. These findings highlight the role of the PTRF-NEAT1-PD-L1 axis in immune evasion in GBM; this immune-suppressive axis may be targeted as a novel immunotherapeutic strategy.
Materials and Methods
Cell Culture, Lentiviruses, and Chemicals
N9 cell lines derived from GBM patients, were established by Professor Fan of Beijing Normal University. TBD0220 cell lines derived from GBM patients, were established by Professor Kang of Tianjin Medical University (20, 21). N9 and TBD0220 cells were cultured in DMEM/F12 supplemented with 1% Penicillin-Streptomycin and 10% FBS. Cells were maintained at 37°C in a humidified 5% CO2 atmosphere. All GBM cells, except in vivo cultures, were maintained for less than eight generations. Lentiviruses encoding the PTRF-EGFP fusion protein (GOSL87854) and luciferase (GCNL84615) were purchased from GENECHEM (Shanghai, China). Small interfering RNAs targeting PTRF were synthesized by GenePharma (Shanghai, China). Sequences of siRNAs were as follows: PTRF siRNA#1 5’-GCCGCAACUUUAAAGUCAUGAUCUA; PTRF siRNA#2 5’-AGGAGUCCCGCGCAGAGCGUAUCAA. Small interfering RNAs targeting NEAT1 were synthesized by GenePharma (Shanghai, China). Sequences of siRNAs were as follows: NEAT1 siRNA 5’- GAACUUUACUUCGUUAGAUTT. We treated cells with Actinomycin D (Selleck, S7418) at a final concentration of 1 μM.
Western Blot Analysis
The protein was extracted GBM cells by RIPA protein lysis buffer (Solarbio, R0010), with a freshly added mixture of protease inhibitor cocktail and PMSF. Then, 20 µg of protein was run on an SDS-PAGE gel, separated by electrophoresis and transferred to PVDF membranes. The membrane was first blocked by 5% BSA for 1-2 hours at room temperature, followed by incubation with primary antibodies at 4°C overnight. After washing three times with PBST, the membrane was blocked with secondary antibodies (Promega, 1:10000). Protein bands were detected was performed using G:BOXF3 (Syngene, UK) with Western HRP Substrate (Millipore, WBLUF0500). The antibodies used for western blots were: PD-L1 (Abcam, ab58810, 1:500), NF-κB (Cell Signaling Technology, 8242, 1:1000), Phospho-NF-κB (Cell Signaling Technology, 3033, 1:1000), UBXN1 (Proteintech, 16135-1-AP, 1:1000), GAPDH (Proteintech, 60004-1-Ig, 1:1000).
Quantitative RT-PCR
Cells were lysed in TRIzol Reagent and mixed with chloroform, then centrifuged at 12000 rpm for 10 minutes at 4°C. The upper aqueous phase was transferred to a clean tube and an equal volume of isopropanol was added. The samples were mixed and stored at −20°C overnight. The concentration of total RNA was measured by NanoDrop 2000 (Thermo Scientific, USA). cDNA was synthesized from 1 µg of RNA using a reverse transcription kit (Promega, USA), according to the manufacturer’s protocol. qRT-PCR was performed using the DNA Engine Opticon 2 Two-Color qRT-PCR detection system (Bio-Rad Laboratories, USA) and the results were normalized to the GAPDH. The following primers were used:
PD-L1-F: AGGAGTACCTTGGCTTTGCC
PD-L1-R: GCCTTGCTCAGCCACAATTC
NEAT1-1F: CCAGTTTTCCGAGAACCAAA
NEAT1-1R: ATGCTGATCTGCTGCGTATG
NEAT1-2F: CTAGAGGCTCGCATTGTGTG
NEAT1-2R: GCCCACACGAAACCTTACAT
UBXN1-F: AGTGAAGAGGAAAGACAGGAA
UBXN1-R: TTTGTCCCTCTCGATCTTTTC
TNFα-F: CCCAGGCAGTCAGATCATCTTC
TNFα-R: GGTTTGCTACAACATGGGCTACA
IL1β-F: CCACAGACCTTCCAGGAGAATG
IL1β-R: GTGCAGTTCAGTGATCGTACAGG
IL8-F: GAGTGATTGAGAGTGGACCACACT
IL8-R: AGACAGAGCTCTCTTCCATCAGAAA
GAPDH-F: TGCACCACCAACTGCTTAGC
GAPDH-R: GGCATGGACTGTGGTCATGAG
Flow Cytometry
For the detection of cell-surface PD-L1, cells were trypsinized and collected into a flow tube. The cells were incubated with 100 μL PBS by adding PE anti-human PD-L1 Antibody (329705, BioLegend; the dilution ratio is 1: 100) at room temperature for 1 hour. Then, the cells were analyzed via flow cytometer after washing with PBS.
RNA-Binding Protein Immunoprecipitation
RIP experiment was performed using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, No.17-701) according to the manufacturer’s instructions. For high-throughput sequencing (RIP-Seq), the libraries were prepared following the manufacturer’s instructions and applied to the Illumina HiSeq X Ten system by ABlife. Inc (Wuhan, China). To further validate the RIP-Seq results, the RNA fraction, precipitated by RIP, was analyzed via RT-qPCR. The data presented in the study are deposited in the SRA repository, accession number PRJNA777377
Chromatin Immunoprecipitation
ChIP experiment was performed using the Magna ChIP Chromatin Immunoprecipitation Kit (Millipore, No.17-10085) according to the manufacturer’s instructions. The antibodies against H3K27me3 and NF-κB were purchased from Cell Signaling Technology (Denver, MA, USA). The primers used for UBXN1 have been described. The primers used for UBXN1 and PDL1 were as follows:
UBXN1-F: GGCAGGGGAAATGATGGATA
UBXN1-R: TAACCTGCCCACTCCATTAC
PD-L1-1F: ACTGAAAGCTTCCGCCGATT
PD-L1-1R: GAGGAACAACGCTCCCTACC
PD-L1-2F: GGGTGGCAGAATATCAGGGAC
PD-L1-2R: CGTGGATTCTGTGACTTCCTC
PDL1-3F: ACCTGTAAACTGTATTGCCACA
PDL1-3R: TGGTGACTGTAAGTTTGGGTGA
PDL1-4F: AGGGTAGAAACAGGTGGGAA
PDL1-4R: GAAAGCAGTGTTCAGGGTCTAC
Luciferase Reporter Assay
The expression vectors encoding PGL3-Basic-PD-L1WT and PGL3-Basic-PD-L1MUT were synthesized by Integrated Biotech Solutions (Shanghai, China). Briefly, the GBM cells were transfected with 0.5 μg of PGL3-Basic empty vector, PGL3-Basic-PD-L1WT and PGL3-Basic-PD-L1MUT by Lipofectamine 3000 (Invitrogen) separately. After incubation for 48 hours, luciferase activity was measured using the luciferase assay system (E1501, Promega) according to the manufacturer’s protocol.
In Situ Hybridization Histochemistry
The glioma tissues were surgically resected from patients at the Beijing Tiantan Hospital. The ISH experiments for NEAT1 were performed using the RNA ISH kit (Boster, China) according to the manufacturer’s instructions. The probes were designed and synthesized by Boster (Wuhan, China).
In Vivo Intracranial Patient-Derived Xenografts Model
Female BALB/C nude mice (4 weeks old) were used for intracranial GBM Patient-derived xenografts, and TBD0220 cells were used to construct the intracranial model. The cells were injected into the mice under the guidance of a stereotactic instrument at coordinates relative to bregma. Bioluminescence imaging was used to detect intracranial tumor growth by In Vivo Imaging System (IVIS) Spectrum. On day 21, the mice were sacrificed, and the brain tissues were removed. The brain tissue was then fixed in 10% formalin for 24 h, embedded in paraffin, and sectioned into 8 μm slices. All experiments were approved by the Animal Ethical and Welfare Committee of Tianjin Medical University. Kaplan-Meier survival curves were plotted to analyze the survival.
HE and Immunohistochemical Staining
The glioma tissues were surgically resected from patients at the Beijing Tiantan Hospital. The tissues were embedded in paraffin, sectioned, and stained with HE. IHC staining was performed as described previously. Staining intensity was quantified by a pathologist. Antibodies include Ki-67 (ZSGB-BIO, ZM-0167, 1: 100), Phospho-NF-κB (Cell Signaling Technology, 3033, 1:400), UBXN1 (Proteintech, 16135-1-AP, 1:200), PTRF (Proteintech, 18892-1-AP, 1:200), PD-L1 (Cell Signaling Technology, 13684, 1:400), CD8 (Proteintech, 66868-1-lg, 1:200).
PD-L1/PD-1–Binding Assay
Confocal laser scanning microscopy (CLSM) and flow cytometry were used to study the PD-L1/PD-1–binding in GBM cells. Cells were trypsinized and collected into a flow tube. Control and PTRF overexpressing TBD0220 cells were incubated with 5 μg/mL recombinant human PD-1 FC chimera protein (1086-PD-050, R&D Systems) at room temperature for 1 hour. After incubation, the cells were washed three times with PBS and incubated with anti-Human IgG (H+L) Cross-Adsorbed Secondary Antibody (Alexa Fluor 488) (A-11013, Thermo Fisher) for 1 hour at room temperature. For CLSM analysis, the cells were fixed with paraformaldehyde for 15 minutes at room temperature and further stained with DAPI for detecting the cell nucleus. CLSM images were observed by FV-1200 laser scanning confocal microscope (Olympus Corporation, Japan). For flow cytometry analysis, the cells were trypsinized and collected by centrifugation, then resuspended in PBS. The data were analyzed by FlowJo v10.6.2.
T Cell Cytotoxicity Assay
Control and PTRF overexpressing TBD0220 cells were separately transfected with siRNA for NEAT1 in DMEM/F12 medium. Human peripheral blood mononuclear cells (PBMC; CBP-002, StemEry) were maintained in DMEM/F12 medium. The PBMC cells were activated with 100 ng/mL CD3 antibody (317325, BioLegend), 100 ng/mL CD28 antibody (302933, BioLegend), and 10 ng/mL IL2 (589102, BioLegend) for 24 h. Then, activated PBMC cells were incubated with TBD0220 cells at 10:1 ratio in the presence of caspase-3/7 green detection reagent (C10723, Thermo Fisher, 1:1000) at 37°C for 12 hours according to the manufacturer’s protocol. The cell nuclei were visualized by staining with DAPI. All the images were observed using FV-1200 laser scanning confocal microscope (Olympus Corporation, Japan).
Clinical Information And Transcriptome Data of Glioma Patients
The clinical and RNA sequencing data sets of glioma patients in TCGA database were downloaded from the University of California Santa Cruz (UCSC) Xena Functional Genomics Explorer (https://xenabrowser.net/). CGGA-693 RNA sequencing data sets and Rembrandt microarray data were obtained from the Chinese Glioma Genome Atlas (CGGA) (http://www.cgga.org.cn/). All the RNA sequencing data were transformed by log2 (normalized count + 1).
PTRF-correlated genes were obtained by a filtering condition: Pearson correlation coefficients between the expression of PTRF and other genes ≥ 0.3 and p-value ≤ 0.05. GO-BP analysis of PTRF-correlated genes was performed by the cluster Profiler v3.18.0 R package and visualized by bubble charts. Tumor purity of glioma data in TCGA, CGGA, and Rembrandt databases was measured by estimate v1.0.13 R package. The univariant and multivariant cox analyses were employed by survival v3.1.12 R package and displayed as forest maps. The PNP score of each patient was calculated by the formula below, and overall survival data of patients with PNP score-high or low were shown as survival curves.
β-value (PTRF) × expression data (PTRF) + β-value (NEAT1) × expression data (NEAT1) + β-value (PD-L1) × expression data (PD-L1)
Statistical Analysis
All data visualization was performed using Microsoft Excel, GraphPad Prism 8 software, and R. Two-tail unpaired Student’s t-test was used to compare two experimental groups, and one-way or two-way ANOVA was used when comparing three or more experimental groups. All data represent the mean ± SD. A p-value less than 0.05 was considered statistically significant, ns = not significant. Significance was defined as *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Results
PTRF Increases PD-L1 Levels and Correlates With Immunosuppression in GBM
In this study, considering the immune-regulation function of PTRF, we assessed the relationships between PTRF and immune responses in GBM by examining GBM cases in public databases. We acquired the transcriptome data of glioma in CGGA, TCGA, and Rembrandt databases, and calculated the correlation between PTRF and other genes. The PTRF co-expression gene sets (r>0.3) were filtered out and interrogated by gene ontology (GO) enrichment analysis. GO-biological process (GO-BP) results show that the functions associated with T cell activation was significantly enriched (Figure 1A and Supplemental Figure 1A). Elevated expression of PTRF was associated with low tumor purity and had a positive correlation with the expressions of genes, involved in tumor immunity (Figure 1B, Supplemental Figure 1B). The correlation between PTRF and the representative genes of immune cells in TCGA, CGGA, and Rembrandt databases, were calculated and visualized as scatter maps (Figure 1C and Supplemental Figure 1C). We found that the expression of PTRF positively correlated with PD-L1 (correlation coefficient of 0.56), suggesting that PTRF may regulate PD-L1. To investigate the regulation of PD-L1 by PTRF, we stably overexpressed PTRF in primary GBM cell lines (N9 and TBD0220) by transfecting lentivirus. We conducted immunoblotting analysis of cultured N9 and TBD0220 cells and found that PTRF overexpression promoted the increase of PD-L1 protein levels in N9 PTRF and TBD0220 PTRF cells compared with their respective control cells (Figure 1D and Supplemental Figure 1D). Real-time quantitative PCR (RT-qPCR) determined that the mRNA level of PD-L1 was increased in N9 PTRF and TBD0220 PTRF cells compared with the control cells (Figure 1E).
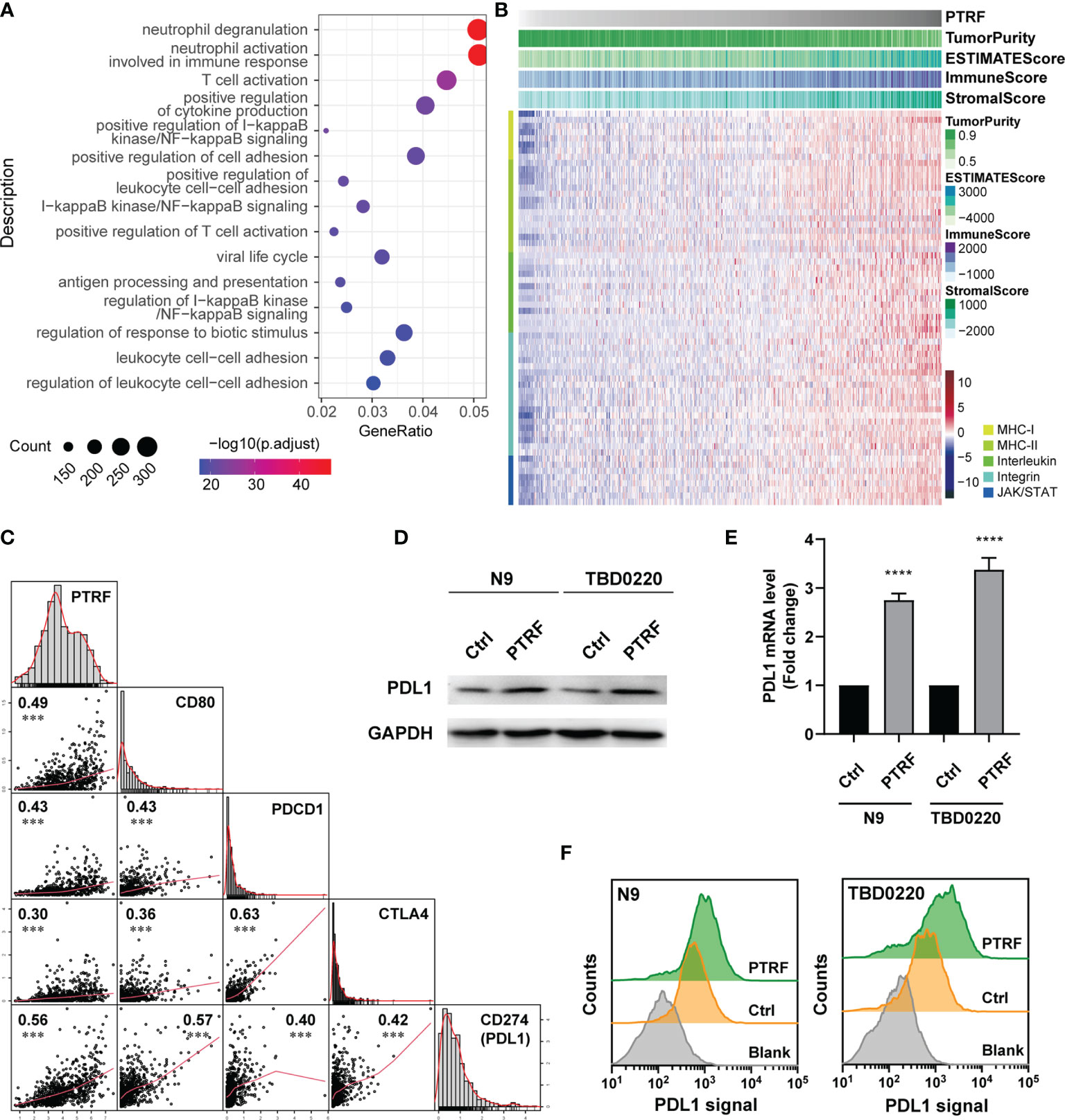
Figure 1 PTRF increases PD-L1 levels and correlates with immunosuppression in GBM. (A) Gene ontology enrichment analysis of genes that are positively associated with PTRF in the CGGA GBM database. (B) Heatmap showing the relationship between PTRF, low tumor purity, and genes regulating immunity in the CGGA GBM database. (C) The correlation between PTRF, CD80, PDCD1, CTLA4, and PD-L1 levels in the CGGA GBM database, measured by Pearson’s correlation test. (D) Western blot analysis of PD-L1 expression in primary GBM cells (N9 and TBD0220; both control and PTRF overexpression cells). (E) Relative mRNA level of PD-L1 in control and PTRF overexpressing cells. (F) Flow cytometry analysis of PD-L1 in control and PTRF overexpressing cells. ****p ≤ 0.001.
We further employed flow cytometry to interrogate the PD-L1 level changes on the cell membranes of N9 and TBD0220 cells overexpressed PTRF. We confirmed that PD-L1 protein levels were higher in the PTRF overexpressing cells compared with the control cells (Figure 1F). We verified the effect of PTRF on PD-L1 by PTRF knock-down in N9 and TBD0220 cells. The mRNA and protein level of PD-L1 was significantly decreased by PTRF knockdown in N9 and TBD0220 cells compared with their respective control cells (Supplemental Figures 1E, F). These results show that PTRF regulates the expression of PD-L1 at the transcription level in GBM cells.
PTRF Maintains mRNA Stability of lncRNA NEAT1
PTRF is a novel non-canonical RNA-binding protein (RBP) (22) and participates in the transcription regulation of PD-L1 in GBM. To investigate the potential mechanism of PTRF as an RBP affecting the expression of PD-L1, we performed RBP immunoprecipitation-coupled high-throughput sequencing (RIP-Seq) by Illumina HiSeq X Ten system in N9 cells with PTRF-Flag over-expression (Figure 2A). The experiments were biologically repeated twice and the clean reads were mapped to the human reference genome (GRCh38) using TopHat2. The reads were analyzed for PTRF-associated RNA sequences after normalization by three computational methods: (a) CIMS, (b) Piranha, and (c) ABlife. Based on screening with two replicates by three different computations, we identified ten long noncoding RNAs (lncRNAs) as potential PTRF-associated RNA molecules (Figure 2B).
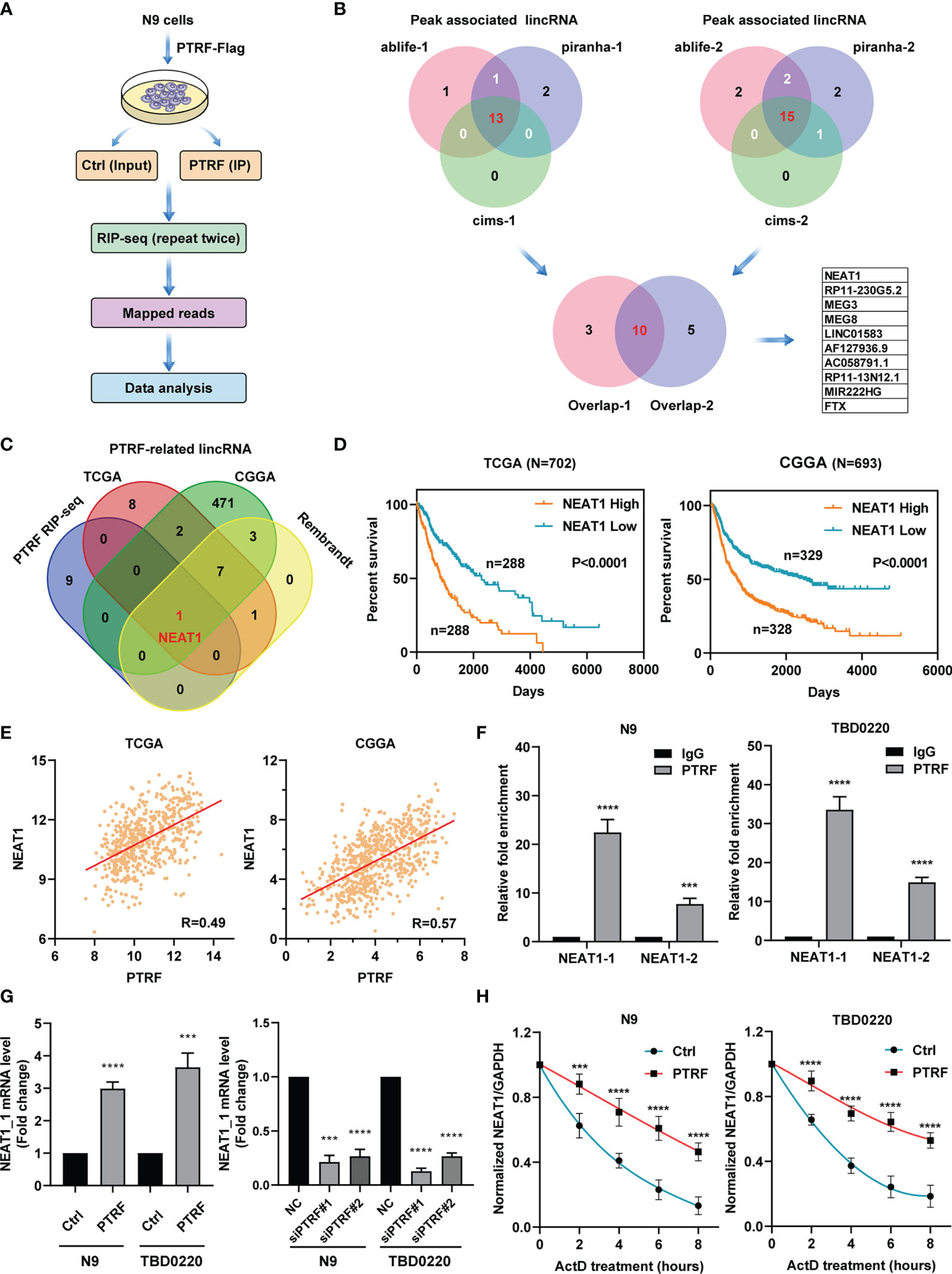
Figure 2 PTRF interacts with LncRNA NEAT1 and maintains its mRNA stability. (A) Schematic of RIP-seq analysis in primary GBM cells with PTRF overexpression. (B) Venn diagrams showing the number of peaks associated with lncRNA that overlap between two repeated experiments by three computational methods: (a) CIMS, (b) Piranha, and (c) ABlife. Ten lncRNAs were identified as potential PTRF-associated RNA molecules. (C) Venn diagrams of results for potential lncRNAs between PTRF RIP-Seq, TCGA, CGGA, and Rembrandt database. (D) The Kaplan–Meier curves of patients with high or low lncRNA NEAT1 expression (the TCGA database and CGGA database). (E) The correlation between PTRF and NEAT1 levels in the TCGA and CGGA GBM database. (F) RIP-qPCR verification of RIP-Seq results with PTRF antibodies for the enrichment of NEAT1 in N9 and TBD0220 cells. (G) The relative level of NEAT1 in control and PTRF overexpressing cells (left). The level of NEAT1 after PTRF knockdown in N9 and TBD0220 cells (right). (H) RT-qPCR analysis of NEAT1, treated with actinomycin D (ActD) at the indicated time points in N9 and TBD0220 cells. ***p ≤ 0.001 and ****p ≤ 0.001.
To clarify the function of PTRF as RBP, we performed the co-expression analysis to identify PTRF-related lncRNAs (r > 0.3) in TCGA, CGGA, and Rembrandt glioma databases. We overlapped the lncRNAs data from RIP-Seq results and public GBM databases. The cross‐comparison of the four lists revealed that nuclear enriched abundant transcript 1 (NEAT1) is the lncRNA that interacts with PTRF (Figure 2C). Patients with a high level of NEAT1 had shorter overall survival times compared with those with low NEAT1 expression (Figure 2D and Supplemental Figure 2A). Further, we found a significant correlation between PTRF and NEAT1 in TCGA (r = 0.49), CGGA (r = 0.57), and Rembrandt (r = 0.49) databases (Figure 2E and Supplemental Figure 2B). RBP immunoprecipitation–coupled RT-qPCR was performed in N9 and TBD0220 cells in which the cell lysates were followed by IP with the antibody against PTRF. Given that two transcripts of NEAT1, NEAT1_1 (3.7 kb) and NEAT1_2 (23 kb), we designed two pairs of primers for PCR amplification, respectively. Quantitative PCR analysis showed the enrichment of NEAT1_1 and NEAT1_2 in the PTRF group, compared with the IgG group, validating the RIP-Seq results. The enrichment of NEAT1_1 was significantly higher than that of NEAT1_2 in RBP immunoprecipitation (Figure 2F).
To verify the effect of PTRF on NEAT1, we conducted quantitative PCR analysis to determine that the mRNA level of NEAT1_1 was increased in N9 PTRF and TBD0220 PTRF cells than control cells. The mRNA level of NEAT1_1 was significantly decreased by PTRF knockdown in N9 and TBD0220 cells (Figure 2G). The effect of PTRF on NEAT1_2 is consistent with the results of NEAT1_1 (Supplemental Figures 2C, D). Considering the pivotal role of RBP in post-transcriptional regulation, we investigated whether PTRF regulated the stability of NEAT1 with actinomycin D (ActD) blocking mRNA synthesis. We conducted a quantitative PCR–based time curve (0–8 h) analysis of NEAT1, which showed that PTRF resulted in significantly lower degradation of NEAT1 level (Figure 2H and Supplemental Figure 2E).
PTRF Suppresses UBXN1 Expression and Promotes the Activity of NF-κB via NEAT1
The genes that positively correlated with PTRF were found to be closely related to the NF-κB signaling pathway (Figure 1A). Since, NF-κB plays a crucial role in the immune response and PD-L1 regulation, we explored the role of NEAT1 in the regulation of NF-κB in a PTRF-dependent manner. We previously demonstrated that the PRC2 complex promoted NF-κB activity via the trimethylation of H3K27 (H3K27me3) in the UBXN1 promoter (23, 24). Based on the interaction between NEAT1 and EZH2, we designed siRNA to knock-down NEAT1 expression. The level of UBXN1 was significantly decreased by PTRF overexpression in N9 and TBD0220 cells, and this decrease was reversed by NEAT1 knockdown (Figure 3A). Further, the level of UBXN1 was significantly increased in PTRF knockdown cells compared with the control cells (Figure 3B). We next performed chromatin immunoprecipitation (ChIP)-qPCR analysis to evaluate the chromatin state of the UBXN1 promoter and found that the enrichment levels of H3K27me3 were increased at the UBXN1 promoter region in cells overexpressing PTRF (Figure 3C). Further, knockdown of PTRF or NEAT1 reduced H3K27me3 enrichment at the UBXN1 promoter region in N9 and TBD0220 cells (Figure 3D).
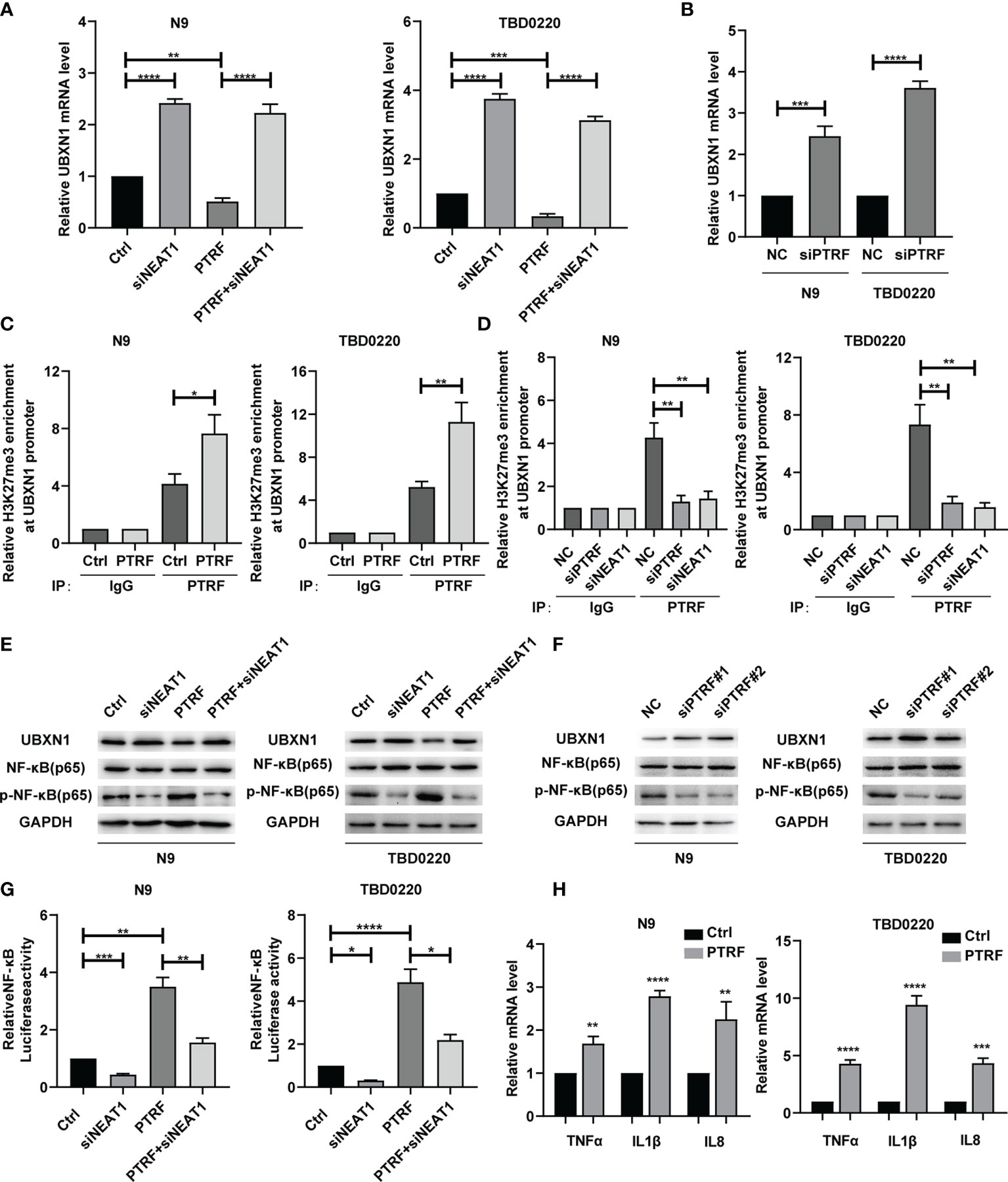
Figure 3 (A) RT-qPCR analysis of UBXN1 in N9 and TBD0220 cells with or without PTRF overexpression, as well as treatment with NEAT1 knockdown. (B) The mRNA level of UBXN1 after PTRF knockdown in N9 and TBD0220 cells. (C) ChIP-PCR analysis of the ability of H3K27me3 to bind to the UBXN1 promoter using H3K27me3 antibody in N9 and TBD0220 cells with or without PTRF overexpression. (D) ChIP-PCR analysis of the ability of H3K27me3 to bind to the UBXN1 promoter normalized to IgG with PTRF or NEAT1 knockdown in N9 and TBD0220 cells, respectively. (E) Western blot analysis of UBXN1, NF-κB, and P-NF-κB expression in primary GBM cells (N9 and TBD0220) with or without PTRF overexpression, as well as treatment with NEAT1 knockdown respectively. (F) Western blot analysis of UBXN1, NF-κB, and P-NF-κB expression with PTRF knockdown in N9 and TBD0220 cells. (G) The Luciferase activity of NF-κB in N9 and TBD0220 cells with or without PTRF overexpression, as well as treatment with NEAT1 knockdown respectively. (H) The mRNA level of NF-κB downstream target genes, including TNFα, IL-1β, and IL-8 in N9 and TBD0220 cells with PTRF overexpression. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, and ****p ≤ 0.001.
We next investigated how PTRF regulates the activity of NF-κB. PTRF overexpression decreased the proteins level of UBXN1 (Figure 3E and Supplemental Figure 3A) and increased the nuclear levels of p-NF-κB in N9 and TBD0220 cells, which were reversed by knocking down NEAT1 (Figure 3E and Supplemental Figure 3B). In contrast, knockdown of PTRF increased the UBXN1 proteins level (Figure 3F and Supplemental Figure 3C) and decreased the nuclear levels of p-NF-κB (Figure 3F). In addition, we cultured N9 and TBD0220 cells with lentivirus vector expressing a NF-κB luciferase construct. Luciferase reporter assay showed that the activity of NF-κB was significantly up-regulated by overexpression of PTRF in N9 and TBD0220 cells and the effect was reversed by NEAT1 knockdown (Figure 3G). Knockdown of PTRF decreased the luciferase activity of NF-κB reporter (Supplemental Figure 3D). We also detected the expression of NF-κB target genes, confirming that NF-κB was activated by PTRF overexpression in N9 and TBD0220 cells (Figure 3H). PTRF knockdown produced the opposite effects (Supplemental Figure 3E). These results suggest that PTRF suppresses UBXN1 expression and promotes the activity of NF-κB via NEAT1.
PTRF Contributes to GBM Proliferation Through the UBXN1/NF-κB Axis by Regulating NEAT1 In Vivo
To further verify the role of the PTRF/NEAT1/NF-κB axis in vivo, we established an intracranial GBM patient-derived xenografts model in BALB/c nude mice. We transfected TBD0220 and TBD0220 PTRF cells with NEAT1 siRNA respectively. The bioluminescence imaging showed that PTRF overexpression resulted in a significant increase in tumor growth. NEAT1 knockdown reduced the growth of the tumor and attenuated the tumor growth induced by PTRF (Figures 4A, B). The mice bearing tumors, derived from the PTRF overexpressing cells, showed poor survival compared with the control group by Kaplan–Meier survival curves. NEAT1 knockdown group had a longer survival time compared with their respective control groups (Figure 4C). Similar results could be found in Hematoxylin-Eosin (H&E) staining that NEAT1 knockdown partly reverse the promotion of GBM proliferation induced by PTRF (Figure 4D). The expression of NEAT1 was increased in the PTRF overexpressing group compared with the control group (Figure 4E). Overexpression of PTRF resulted in a significant decrease in UBXN1 and an increase in Ki-67 and p-NF-κB (Figure 4E). NEAT1 knockdown significantly increased the level of UBXN1 and decreased the level of Ki-67 and p-NF-κB (Figure 4E). These data suggest that PTRF regulates NEAT1 and promotes GBM proliferation through the UBXN1/NF-κB axis.
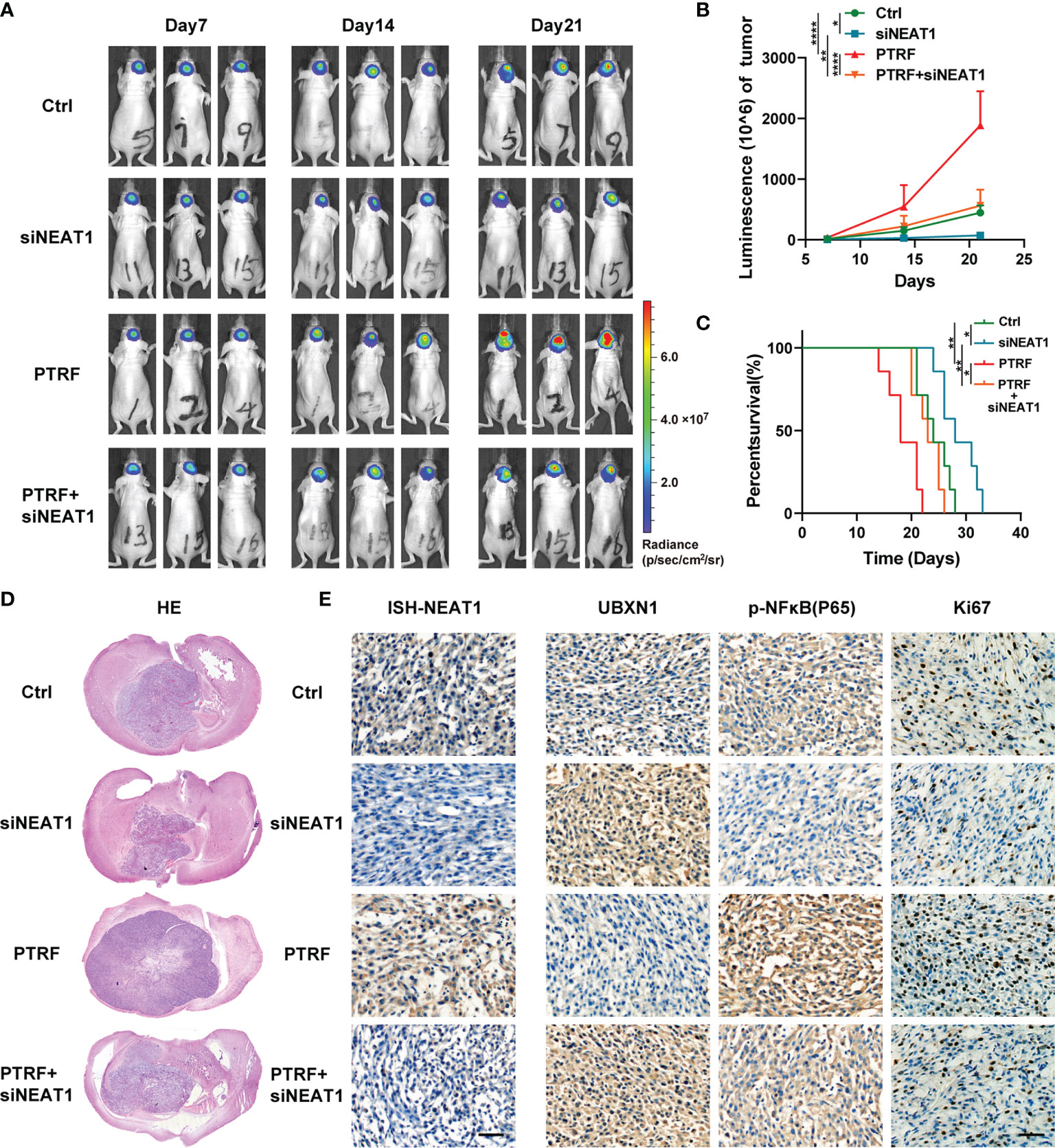
Figure 4 PTRF contributes to GBM proliferation through the UBXN1/NF-κB axis by regulating NEAT1 In Vivo. (A) Representative tumor bioluminescence images of mice at 7, 14, and 21 days post tumor implantation. (B) Tumor growth curves using quantification of bioluminescence imaging signal intensities for mice bearing GBM with or without PTRF overexpression, as well as treatment with NEAT1 knockdown respectively. (C) Kaplan-Meier survival curve of nude mice implanted with TBD0220 cells with or without PTRF overexpression, as well as treatment with NEAT1 knockdown respectively. (D) Representative images of H&E staining of tumors from the nude mice. (E) ISH staining of NEAT1 in tumors (left). IHC staining of UBXN1, P-NF-κB, and Ki-67 in tumors (right). Scale bar, 50 μm. *p ≤ 0.05, **p ≤ 0.01, and ****p ≤ 0.001.
The NF-κB Binds to the PD-L1 Promoter Region in Response to PTRF Activation and Enhances PD-L1 Transcription
To identify the binding sequence of NF-κB in the promoter region of PD-L1, we analyzed the PD-L1 promoter and found four putative binding sites for NF-κB in the PD-L1 promoter region (25). Four pairs of primers with CHIP-qPCR for PD-L1 promoter were designed respectively. The sonicated chromatin from PTRF overexpression in N9 and TBD0220 cells was immunoprecipitated with an anti-NF-κB antibody, and only one fragment was amplified in N9 PTRF and TBD0220 PTRF cells compared with control cells by PCR, indicating (–111)-GGAAAGTCCA- (–102) as a consensus sequence recognized by NF-κB (Figure 5A). We constructed a luciferase reporter driven by the PD-L1 promoter that contains the WT or a mutated potential NF-κB-binding sequence (Figure 5B). We transfected the luciferase reporter construct into N9 and TBD0220 cells and measured luciferase activity. PTRF overexpression in N9 and TBD0220 cells significantly increased the activity of the WT but not of mutated promoter, indicating that PTRF increased the binding of NF-κB to PD-L1. The knockdown of PTRF reduced the luciferase activity driven by the PD-L1 promoter in N9 and TBD0220 cells (Figure 5C). A similar inhibitory effect was also observed by NEAT1 knockdown in N9 and TBD0220 cells (Figure 5D). These results demonstrate that NF-κB binds to the PD-L1 promoter region and enhances the PD-L1 transcription in response to PTRF activation in GBM cells, and this regulation is mediated via one of the NF-kB binding sites (-111 to -102 nt region).
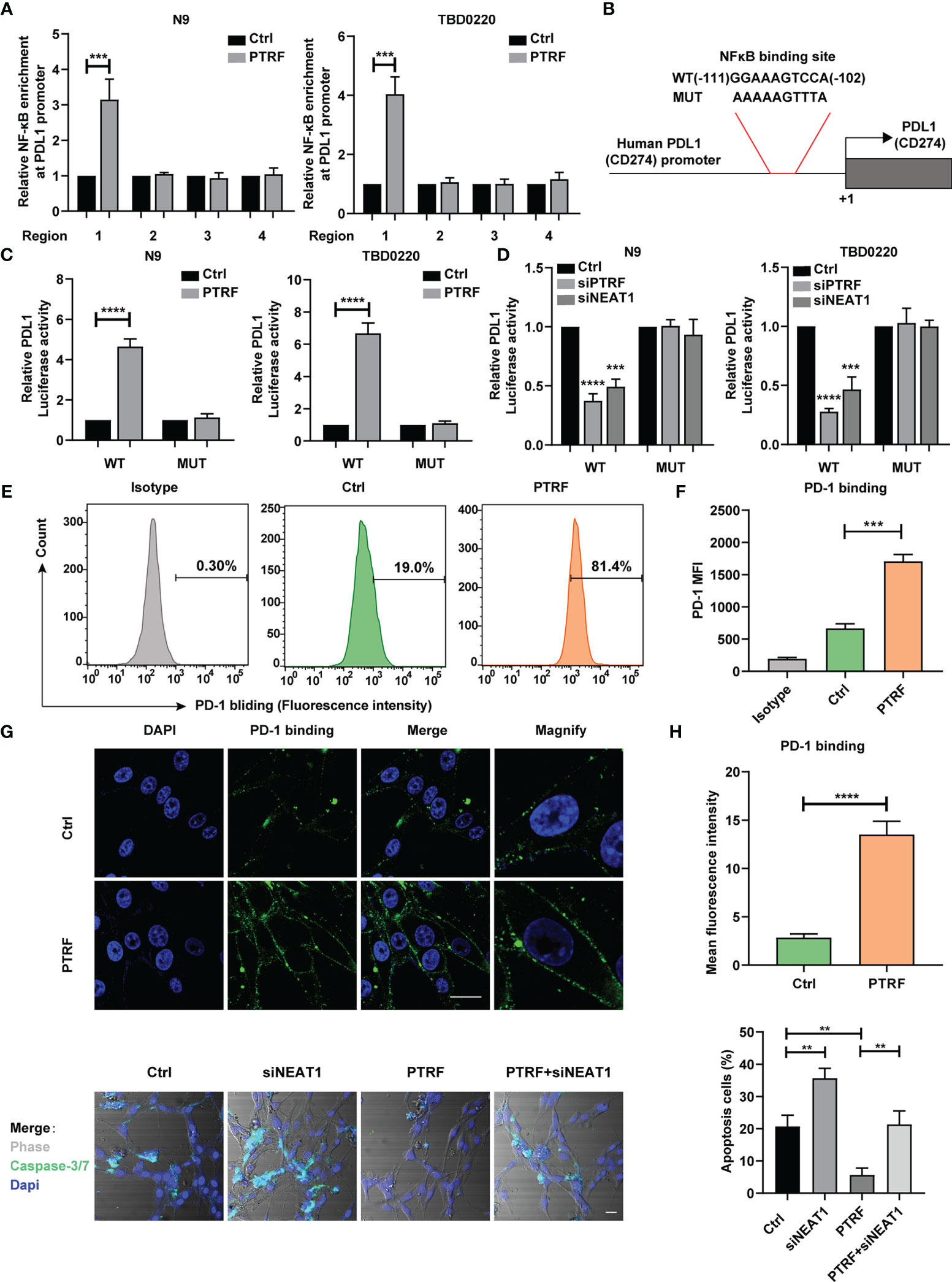
Figure 5 PTRF enhances PD-L1 transcription via NF-κB and promotes immune evasion of GBM cells through PD-L1. (A) ChIP-PCR analysis of the ability of NF-κB to bind to the PD-L1 promoter in N9 and TBD0220 cells with or without PTRF overexpression. Results are normalized to IgG. (B) Schematic illustration of the proximal region of the human PD-L1 promoter. (C) N9 and TBD0220 cells (with the expression of empty vector or PTRF) were transfected with a luciferase reporter vector containing WT or mutated NF-κB sequence of PD-L1 promoter. (D) N9 and TBD0220 cells with PTRF or NEAT1 knockdown were transfected with a luciferase reporter vector containing WT or mutated NF-κB sequence of PD-L1 promoter. (E) Flow cytometry analysis of PD-1 binding in TBD0220 cells with or without PTRF overexpression. (F) Quantification of fluorescence intensity in (E). The y axis represents the mean fluorescence intensity of PD-1. (G) Immunofluorescence staining of control and PTRF overexpressing cells. PD-1 (green), Nuclei (blue), scale bar, 20 μm. (H) Quantification of fluorescence intensity in (G). (I) T cell killing assay of TBD0220 cells with or without PTRF overexpression, as well as treatment with NEAT1 knockdown respectively. caspase3/7 cleavage (green), Nuclei (blue), scale bar, 20 μm. **p ≤ 0.01, ***p ≤ 0.001, and ****p ≤ 0.001.
PTRF Promotes Immune Evasion of GBM Cells by Affecting PD-1 Binding and PD-L1–Mediated T Cell Toxicity
Previous studies have shown that elevated PD-L1 expression in tumor cells mediates T cell tolerance and activates anti-tumor immunosuppression (10, 12). To determine whether PTRF regulation on PD-L1 may affect its interaction with PD-1, we incubated a recombinant protein containing human PD-1 and Fc fragment of IgG with TBD0220 cells. We found that PTRF overexpression increased PD-1 binding on the TBD0220 cell surface compared with the control cells (Figures 5E, F). Immunofluorescence assays revealed that overexpression of PTRF was accompanied by high expression of PD-1 (Figures 5G, H), indicating that PTRF significantly increased PD-1 binding on the GBM cell surface, which is consistent with the flow cytometry results. Furthermore, PTRF overexpression decreased the sensitivity of GBM cells to T cell-mediated cytotoxicity. The knockdown of NEAT1 sensitized GBM cells to T cell-mediated cell death and attenuated the immune evasion induced by PTRF in GBM (Figure 5I). These results demonstrate that PTRF alters PD-1 binding and T cell-mediated cytotoxicity through regulation of PD-L1 expression in GBM.
To further assess relationships between PTRF, NEAT1, and PD-L1 in GBM, we chose tissue samples of GBM patients with high or low PTRF expression. GBM patients with high PTRF-expression had high levels of NEAT1 and p-NF-κB, showing that PTRF significantly correlates with NEAT1 and p-NF-κB (Figure 6A). Furthermore, high levels of PTRF positively correlated with PD-L1 expression and inversely correlated with CD8+ T cell infiltration in GBM (Figure 6A). These results suggest a role of PTRF in PD-L1 regulation and immune evasion in GBM.
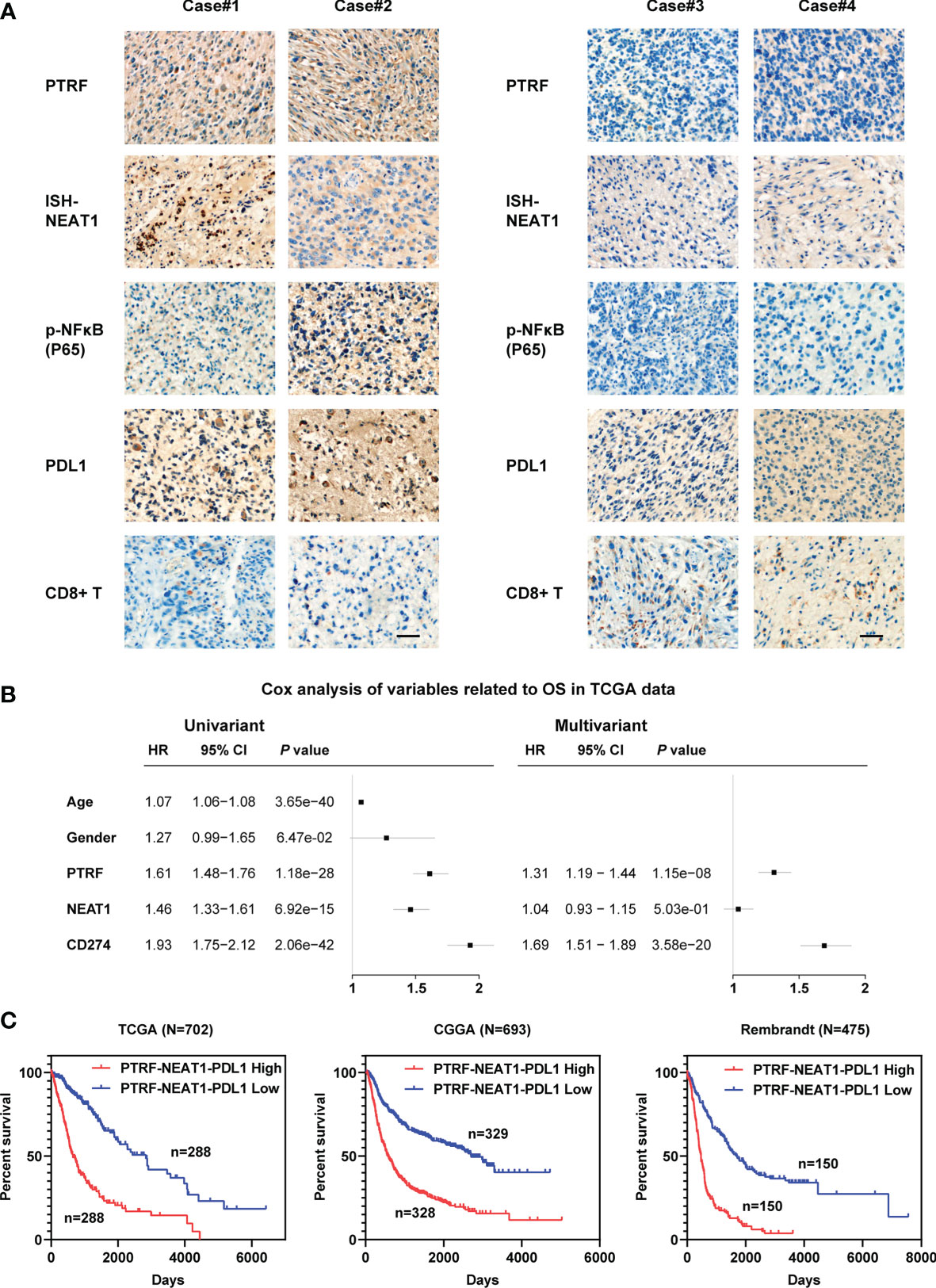
Figure 6 The relationships between PTRF, NEAT1, and PD-L1 in GBM. (A) IHC or ISH staining of PTRF, NEAT1, P-NF-Kb, PD-L1, and CD8 in four representative GBM specimens. Scale bar, 50 μm. (B) Univariate and multivariate Cox analyses of PTRF, NEAT1, and PD-L1 expression in the TCGA databases. (C) The Kaplan–Meier curves for patients with different expressions of PTRF, NEAT1, and PD-L1.
To evaluate the prognostic relevance of PTRF, NEAT1, and PD-L1 expression, we analyzed glioma data from TCGA, CGGA, and Rembrandt databases. As shown in Figure 6B and Supplemental Figure 4, high expressions of PTRF, NEAT1, and PD-L1 are independent poor prognostic markers. Next, we integrated the expression levels of these genes and evaluated the suggestive effect of the PNP geneset (PTRF-NEAT1-PD-L1) on the prognosis of GBM patients. We show that a higher level of PNP indicates a shorter survival time in patients with glioma in TCGA, CGGA, and Rembrandt databases (Figure 6C).
Discussion
In this study, we show that PTRF induces the stability of lncRNA NEAT1 and promotes the activity of NF-κB, leading to PD-L1 mediated immune evasion in GBM (Figure 7). According to previous studies, PTRF plays different roles in different types of tumors. PTRF inhibits tumor metastasis in prostate cancer and tumorigenesis in colorectal cancer (26, 27). In contrast, PTRF promotes the progression of pancreatic cancer and proliferation of rhabdomyosarcoma (28, 29). The tumor-promoting effect of PTRF in gliomas is widely recognized. Previously, we showed that the expression of PTRF positively correlates with the WHO grade of glioma, and the prognosis of glioma patients with high PTRF expression is worse (15). We further found a reduction in the number of CD8+ T cells and CD8+ T cells mediated cytotoxic activity in tumor tissues with PTRF overexpression (19). In this study, we demonstrated that PTRF increases the expression of PD-L1 and promotes immune evasion in GBM.
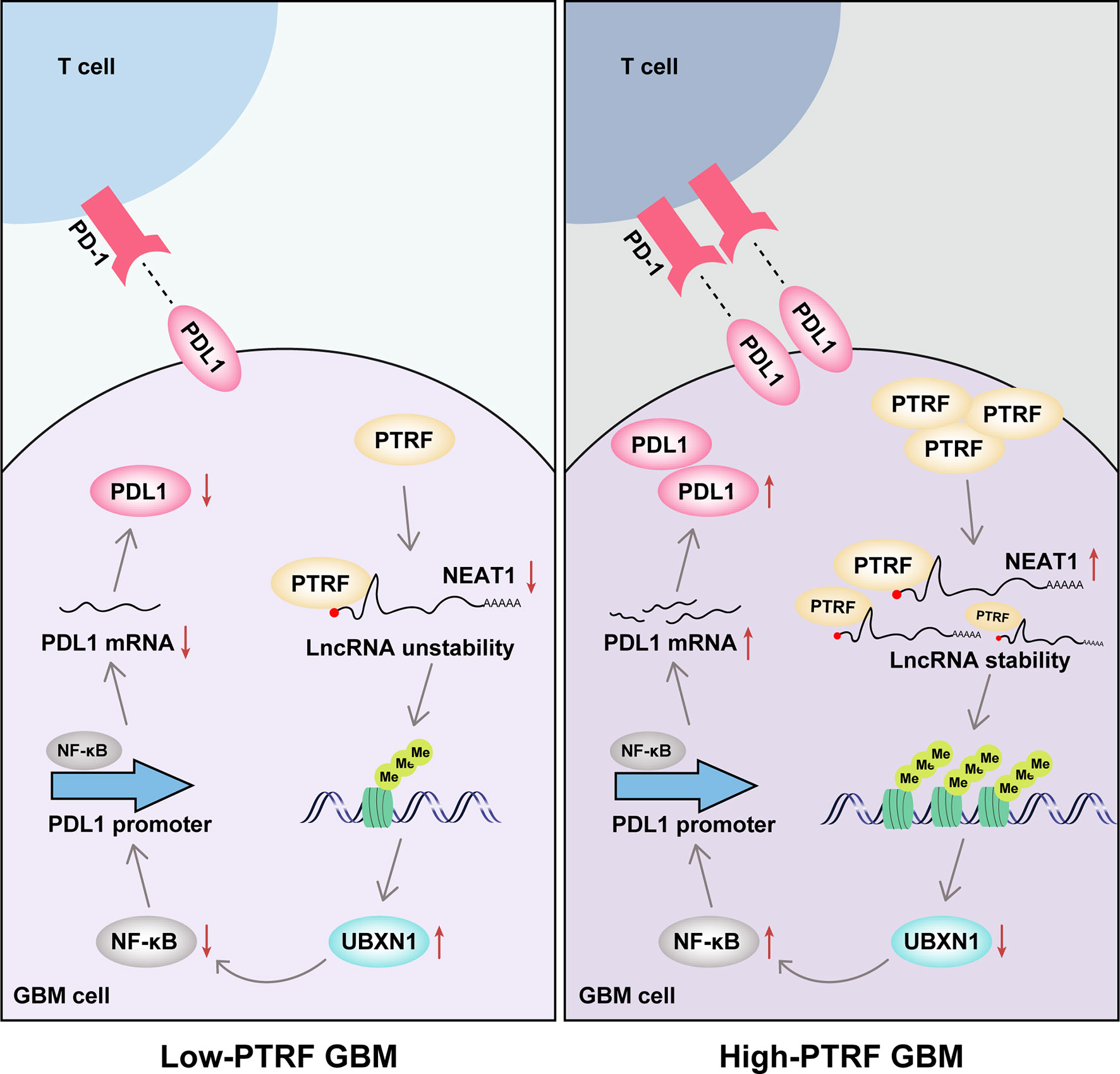
Figure 7 The mechanistic model showing how PTRF/Cavin-1 stabilizes lncRNA NEAT1 to maintain the activity of NF-κB and promote immune evasion via PD-L1 in glioblastoma.
Recently studies have shown that lncRNAs play a crucial roles in regulating immunity, including the activation and function of macrophages and T cells (30). One such lncRNA is NEAT1. Previous studies have found that NEAT1-knockout mice have abnormalities in their Treg and Th cell differentiation (31). NEAT1 has also been found to target STAT3 and regulate the differentiation of Th17 cells in rheumatoid arthritis (32). Imamura et al. found that NEAT1 activate the transcription of IL-8 by removing SFPQ from the IL-8 promoter (33). Knockdown of NEAT1 also limits the apoptosis of CD8+ T cells through the miR-155/Tim-3 pathway (34). Although NEAT1 has been studied in immune response, the relationship between NEAT1 and PD-L1 remains unclear. In this study, we found that NEAT1, induced by PTRF, mediates PD-L1 regulation and promotes immune evasion in GBM.
UBXN1 is a UBX domain-containing protein which inhibits NF-κB activation. The knockdown of UBXN1 decreases the stability of IκBα and enhances NF-κB signaling (35). In addition, UBXN1 is the negative regulator of NF-κB signaling and interacts with cIAPs through the UBA domain (36). YTHDF2 was also shown to accelerate UBXN1 mRNA degradation via METTL3−mediated m6A, promoting the activation of NF−κB (37). Our previous study found that UBXN1 is also a target for EZH2 mediated H3K27me3, and is regulated by EGFR and lncRNA PRADX (23, 24). In addition, we also detected that lncRNA NEAT1 binds to EZH2 and mediates the trimethylation of H3K27 in their promoters (38), which regulated by EGFR pathway activity. From the discussion above, we further demonstrated that PTRF maintains the stability of NEAT1 to suppresses UBXN1 expression and activate NF-κB activity.
Although the expression of PD-L1 is regulated by multiple mechanisms, transcriptional regulation plays an important part in its regulation in cancer. MYC was shown to directly bind to the promoter of PD-L1 and enhanced the expression of PD-L1, regulating the anti-tumor response in mouse tumors and human tumor cells (39). Interferon regulatory factor-1 (IRF-1), a transcription factor, regulates the constitutive and IFN-γ-mediated PD-L1 expression in human lung cancer cell (40). In classical Hodgkin lymphoma, AP-1-responsive enhancer in the PD-L1 gene binds to the PD-L1 enhancer and increases the activity of PD-L1 promoter (41). Recently, MUC1-C was found to activate PD-L1 transcription by recruiting MYC and NF-κB to the PD-L1 promoter region in triple-negative breast cancer (42). Our study identified another novel mechanism of PD-L1 regulation in cancer—PTRF induces the binding of NF-κB to the PD-L1 gene promoter region and enhances PD-L1 transcription in GBM.
Data Availability Statement
The data presented in the study are deposited in the SRA repository, accession number PRJNA777377.
Ethics Statement
The animal study was reviewed and approved by the Animal Ethical and Welfare Committee (Protocol # TMUaMEC 2018034). Written informed consent was not obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author Contributions
Developed the study concept and designed the study: YT and QW. Conducted the experiments and acquired the data: KY, XC, XL, YW, JZ, SY, CX, EY, MX, BH, CF, YT and QW. Analyzed the data, generated the figures, and wrote the paper: CK, KY and XC. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81772667 and 82002657), the Tianjin Key R&D Plan of Tianjin Science and Technology Plan Project (No. 20YFZCSY00360), the Central Government Guided Local Science and Technology Development fund Project of Hebei Province (No. 216Z7711G), the Hebei Natural Science Foundation Precision Medicine Joint Project (No. H2020201206), the Key Scientific Research Project in Colleges and Universities of Hebei Province (No. ZD2021308), The National Key Research and Development Programs of China (No. 2018YFA0209700), and the Science and Technology Project of Tianjin Municipal Health Commission (No. TJWJ2021QN007 and TJWJ2021QN003).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.802795/full#supplementary-material
References
1. Ostrom QT, Patil N, Cioffi G, Waite K, Kruchko C, Barnholtz-Sloan JS. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2013-2017. Neuro Oncol (2020) 22(12 Suppl 2):iv1–96. doi: 10.1093/neuonc/noaa200
2. Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting New Advances in Neuro-Oncology: The Avenue to a Cure for Malignant Glioma. CA: Cancer J Clin (2010) 60(3):166–93. doi: 10.3322/caac.20069
3. Xu F, Jin T, Zhu Y, Dai C. Immune Checkpoint Therapy in Liver Cancer. J Exp Clin Cancer Res (2018) 37(1):110. doi: 10.1186/s13046-018-0777-4
4. Reck M, Rodriguez-Abreu D, Robinson AG, Hui R, Csoszi T, Fulop A, et al. Pembrolizumab Versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med (2016) 375(19):1823–33. doi: 10.1056/NEJMoa1606774
5. Ferris RL, Blumenschein G Jr., Fayette J, Guigay J, Colevas AD, Licitra L, et al. Nivolumab for Recurrent Squamous-Cell Carcinoma of the Head and Neck. N Engl J Med (2016) 375(19):1856–67. doi: 10.1056/NEJMoa1602252
6. Chuntova P, Chow F, Watchmaker PB, Galvez M, Heimberger AB, Newell EW, et al. Unique Challenges for Glioblastoma Immunotherapy-Discussions Across Neuro-Oncology and Non-Neuro-Oncology Experts in Cancer Immunology. Meeting Report From the 2019 SNO Immuno-Oncology Think Tank. Neuro Oncol (2021) 23(3):356–75. doi: 10.1093/neuonc/noaa277
7. Sharma P, Allison JP. The Future of Immune Checkpoint Therapy. Science (2015) 348(6230):56–61. doi: 10.1126/science.aaa8172
8. Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, et al. Predictive Correlates of Response to the Anti-PD-L1 Antibody MPDL3280A in Cancer Patients. Nature (2014) 515(7528):563–7. doi: 10.1038/nature14011
9. Iwai Y, Ishida M, Tanaka Y, Okazaki T, Honjo T, Minato N. Involvement of PD-L1 on Tumor Cells in the Escape From Host Immune System and Tumor Immunotherapy by PD-L1 Blockade. Proc Natl Acad Sci USA (2002) 99(19):12293–7. doi: 10.1073/pnas.192461099
10. Garber ST, Hashimoto Y, Weathers SP, Xiu J, Gatalica Z, Verhaak RG, et al. Immune Checkpoint Blockade as a Potential Therapeutic Target: Surveying CNS Malignancies. Neuro Oncol (2016) 18(10):1357–66. doi: 10.1093/neuonc/now132
11. Berghoff AS, Kiesel B, Widhalm G, Rajky O, Ricken G, Wohrer A, et al. Programmed Death Ligand 1 Expression and Tumor-Infiltrating Lymphocytes in Glioblastoma. Neuro Oncol (2015) 17(8):1064–75. doi: 10.1093/neuonc/nou307
12. Du L, Lee JH, Jiang H, Wang C, Wang S, Zheng Z, et al. Beta-Catenin Induces Transcriptional Expression of PD-L1 to Promote Glioblastoma Immune Evasion. J Exp Med (2020) 217(11):e20191115. doi: 10.1084/jem.20191115
13. Xiang Z, Zhou Z, Song S, Li J, Ji J, Yan R, et al. Dexamethasone Suppresses Immune Evasion by Inducing GR/STAT3 Mediated Downregulation of PD-L1 and IDO1 Pathways. Oncogene (2021) 40(31):5002–12. doi: 10.1038/s41388-021-01897-0
14. Peng J, Hamanishi J, Matsumura N, Abiko K, Murat K, Baba T, et al. Chemotherapy Induces Programmed Cell Death-Ligand 1 Overexpression via the Nuclear Factor-KappaB to Foster an Immunosuppressive Tumor Microenvironment in Ovarian Cancer. Cancer Res (2015) 75(23):5034–45. doi: 10.1158/0008-5472.CAN-14-3098
15. Huang K, Fang C, Yi K, Liu X, Qi H, Tan Y, et al. The Role of PTRF/Cavin1 as a Biomarker in Both Glioma and Serum Exosomes. Theranostics (2018) 8(6):1540–57. doi: 10.7150/thno.22952
16. Thommen DS, Schuster H, Keller M, Kapoor S, Weinzierl AO, Chennakesava CS, et al. Two Preferentially Expressed Proteins Protect Vascular Endothelial Cells From an Attack by Peptide-Specific CTL. J Immunol (2012) 188(11):5283–92. doi: 10.4049/jimmunol.1101506
17. Ni Y, Hao J, Hou X, Du W, Yu Y, Chen T, et al. Dephosphorylated Polymerase I and Transcript Release Factor Prevents Allergic Asthma Exacerbations by Limiting IL-33 Release. Front Immunol (2018) 9:1422. doi: 10.3389/fimmu.2018.01422
18. Guo Q, Guan GF, Cheng W, Zou CY, Zhu C, Cheng P, et al. Integrated Profiling Identifies Caveolae-Associated Protein 1 as a Prognostic Biomarker of Malignancy in Glioblastoma Patients. CNS Neurosci Ther (2019) 25(3):343–54. doi: 10.1111/cns.13072
19. Yi K, Zhan Q, Wang Q, Tan Y, Fang C, Wang Y, et al. PTRF/cavin-1 Remodels Phospholipid Metabolism to Promote Tumor Proliferation and Suppress Immune Responses in Glioblastoma by Stabilizing Cpla2. Neuro Oncol (2021) 23(3):387–99. doi: 10.1093/neuonc/noaa255
20. Li X, Qian X, Wang B, Xia Y, Zheng Y, Du L, et al. Programmable Base Editing of Mutated TERT Promoter Inhibits Brain Tumour Growth. Nat Cell Biol (2020) 22(3):282–8. doi: 10.1038/s41556-020-0471-6
21. Zhang Y, Li J, Yi K, Feng J, Cong Z, Wang Z, et al. Elevated Signature of a Gene Module Coexpressed With CDC20 Marks Genomic Instability in Glioma. Proc Natl Acad Sci USA (2019) 116(14):6975–84. doi: 10.1073/pnas.1814060116
22. Wang Z, Tang W, Yuan J, Qiang B, Han W, Peng X. Integrated Analysis of RNA-Binding Proteins in Glioma. Cancers (Basel) (2020) 12(4):892. doi: 10.3390/cancers12040892
23. Li Y, Liu X, Cui X, Tan Y, Wang Q, Wang Y, et al. LncRNA PRADX-Mediated Recruitment of PRC2/DDX5 Complex Suppresses UBXN1 Expression and Activates NF-KappaB Activity, Promoting Tumorigenesis. Theranostics (2021) 11(9):4516–30. doi: 10.7150/thno.54549
24. Huang K, Yang C, Wang QX, Li YS, Fang C, Tan YL, et al. The CRISPR/Cas9 System Targeting EGFR Exon 17 Abrogates NF-KappaB Activation via Epigenetic Modulation of UBXN1 in EGFRwt/vIII Glioma Cells. Cancer Lett (2017) 388:269–80. doi: 10.1016/j.canlet.2016.12.011
25. Huang G, Wen Q, Zhao Y, Gao Q, Bai Y. NF-KappaB Plays a Key Role in Inducing CD274 Expression in Human Monocytes After Lipopolysaccharide Treatment. PloS One (2013) 8(4):e61602. doi: 10.1371/journal.pone.0061602
26. Wang F, Zheng Y, Orange M, Yang C, Yang B, Liu J, et al. PTRF Suppresses the Progression of Colorectal Cancers. Oncotarget (2017) 8(30):48650–9. doi: 10.18632/oncotarget.9424
27. Moon H, Lee CS, Inder KL, Sharma S, Choi E, Black DM, et al. PTRF/cavin-1 Neutralizes Non-Caveolar Caveolin-1 Microdomains in Prostate Cancer. Oncogene (2014) 33(27):3561–70. doi: 10.1038/onc.2013.315
28. Faggi F, Chiarelli N, Colombi M, Mitola S, Ronca R, Madaro L, et al. Cavin-1 and Caveolin-1 Are Both Required to Support Cell Proliferation, Migration and Anchorage-Independent Cell Growth in Rhabdomyosarcoma. Lab Invest; J Tech Methods Pathol (2015) 95(6):585–602. doi: 10.1038/labinvest.2015.45
29. Liu L, Xu HX, Wang WQ, Wu CT, Chen T, Qin Y, et al. Cavin-1 Is Essential for the Tumor-Promoting Effect of Caveolin-1 and Enhances Its Prognostic Potency in Pancreatic Cancer. Oncogene (2014) 33(21):2728–36. doi: 10.1038/onc.2013.223
30. Chen W, Liu S, Wang F. Potential Impact and Mechanism of Long Non-Coding RNAs on Cancer and Associated T Cells. J Cancer (2021) 12(16):4873–82. doi: 10.7150/jca.58859
31. Gast M, Rauch BH, Haghikia A, Nakagawa S, Haas J, Stroux A, et al. Long Noncoding RNA NEAT1 Modulates Immune Cell Functions and Is Suppressed in Early Onset Myocardial Infarction Patients. Cardiovasc Res (2019) 115(13):1886–906. doi: 10.1093/cvr/cvz085
32. Shui X, Chen S, Lin J, Kong J, Zhou C, Wu J. Knockdown of lncRNA NEAT1 Inhibits Th17/CD4(+) T Cell Differentiation Through Reducing the STAT3 Protein Level. J Cell Physiol (2019) 234(12):22477–84. doi: 10.1002/jcp.28811
33. Imamura K, Imamachi N, Akizuki G, Kumakura M, Kawaguchi A, Nagata K, et al. Long Noncoding RNA NEAT1-Dependent SFPQ Relocation From Promoter Region to Paraspeckle Mediates IL8 Expression Upon Immune Stimuli. Mol Cell (2014) 53(3):393–406. doi: 10.1016/j.molcel.2014.01.009
34. Yan K, Fu Y, Zhu N, Wang Z, Hong JL, Li Y, et al. Repression of lncRNA NEAT1 Enhances the Antitumor Activity of CD8(+)T Cells Against Hepatocellular Carcinoma via Regulating miR-155/Tim-3. Int J Biochem Cell Biol (2019) 110:1–8. doi: 10.1016/j.biocel.2019.01.019
35. Hu Y, O'Boyle K, Auer J, Raju S, You F, Wang P, et al. Multiple UBXN Family Members Inhibit Retrovirus and Lentivirus Production and Canonical NFkappaBeta Signaling by Stabilizing IkappaBalpha. PloS Pathog (2017) 13(2):e1006187. doi: 10.1371/journal.ppat.1006187
36. Wang YB, Tan B, Mu R, Chang Y, Wu M, Tu HQ, et al. Ubiquitin-Associated Domain-Containing Ubiquitin Regulatory X (UBX) Protein UBXN1 Is a Negative Regulator of Nuclear Factor KappaB (NF-Kappab) Signaling. J Biol Chem (2015) 290(16):10395–405. doi: 10.1074/jbc.M114.631689
37. Chai RC, Chang YZ, Chang X, Pang B, An SY, Zhang KN, et al. YTHDF2 Facilitates UBXN1 mRNA Decay by Recognizing METTL3-Mediated M(6)A Modification to Activate NF-KappaB and Promote the Malignant Progression of Glioma. J Hematol Oncol (2021) 14(1):109. doi: 10.1186/s13045-021-01124-z
38. Chen Q, Cai J, Wang Q, Wang Y, Liu M, Yang J, et al. Long Noncoding RNA NEAT1, Regulated by the EGFR Pathway, Contributes to Glioblastoma Progression Through the WNT/beta-Catenin Pathway by Scaffolding Ezh2. Clin Cancer Res (2018) 24(3):684–95. doi: 10.1158/1078-0432.CCR-17-0605
39. Casey SC, Tong L, Li Y, Do R, Walz S, Fitzgerald KN, et al. MYC Regulates the Antitumor Immune Response Through CD47 and PD-L1. Science (2016) 352(6282):227–31. doi: 10.1126/science.aac9935
40. Lee SJ, Jang BC, Lee SW, Yang YI, Suh SI, Park YM, et al. Interferon Regulatory Factor-1 Is Prerequisite to the Constitutive Expression and IFN-Gamma-Induced Upregulation of B7-H1 (Cd274). FEBS Lett (2006) 580(3):755–62. doi: 10.1016/j.febslet.2005.12.093
41. Green MR, Rodig S, Juszczynski P, Ouyang J, Sinha P, O'Donnell E, et al. Constitutive AP-1 Activity and EBV Infection Induce PD-L1 in Hodgkin Lymphomas and Posttransplant Lymphoproliferative Disorders: Implications for Targeted Therapy. Clin Cancer Res (2012) 18(6):1611–8. doi: 10.1158/1078-0432.CCR-11-1942
Keywords: glioblastoma, PTRF, lncRNA NEAT1, PDL1, immunosuppression
Citation: Yi K, Cui X, Liu X, Wang Y, Zhao J, Yang S, Xu C, Yang E, Xiao M, Hong B, Fang C, Kang C, Tan Y and Wang Q (2022) PTRF/Cavin-1 as a Novel RNA-Binding Protein Expedites the NF-κB/PD-L1 Axis by Stabilizing lncRNA NEAT1, Contributing to Tumorigenesis and Immune Evasion in Glioblastoma. Front. Immunol. 12:802795. doi: 10.3389/fimmu.2021.802795
Received: 27 October 2021; Accepted: 14 December 2021;
Published: 06 January 2022.
Edited by:
Junxia Zhang, First Affiliated Hospital, Nanjing Medical University, ChinaReviewed by:
Xiuping Zhou, Xuzhou Medical University, ChinaXudong Zhao, Sichuan University, China
Ailiang Zeng, University of Texas MD Anderson Cancer Center, United States
Copyright © 2022 Yi, Cui, Liu, Wang, Zhao, Yang, Xu, Yang, Xiao, Hong, Fang, Kang, Tan and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qixue Wang, cWl4dWVfd2FuZ0B0bXUuZWR1LmNu; Yanli Tan, dGFueWFubGk1NTM2QDEyNi5jb20=
†These authors have contributed equally to this work and share first authorship
 Kaikai Yi
Kaikai Yi Xiaoteng Cui
Xiaoteng Cui Xing Liu
Xing Liu Yunfei Wang
Yunfei Wang Jixing Zhao1
Jixing Zhao1 Chuan Fang
Chuan Fang Yanli Tan
Yanli Tan Qixue Wang
Qixue Wang