
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 25 January 2022
Sec. Microbial Immunology
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.802760
We have successfully designed and constructed a RAEV vector system with regulated-delayed attenuation in vivo attributes that synthesizes Ichthyophthirius multifiliis (Ich) protective antigen IAG52B to enable vaccination of fish susceptible to edwardsiellosis and white spot disease. The first feature of this vaccine delivery system is an Edwardsiella piscicida strain carrying genomic deletions of asdA. AsdA is an enzyme necessary for the synthesis of diaminopimelic acid (DAP), which is an essential component of the peptidoglycan layer of the cell wall of Gram-negative bacteria. asdA mutant strains have obligate growth requirements for DAP in the medium or a plasmid vector with the wild-type asdA gene enabling synthesis of DAP. This balanced-lethal plasmid vector-host system in E. piscicida enables as a second feature the synthesis of recombinant antigens to induce protective immunity against fish pathogens. Recombinant protective antigen IAG52B from the fish pathogen I. multifiliis was synthesized by RAEV strains harboring the AsdA+ plasmid pG8R8029. The third feature of this vaccine strain is a regulated-delayed attenuation in vivo phenotype that is based on the replacement of an arabinose-regulated araC ParaBAD cassette for the promoters of the fur and crp genes of E. piscicida such that the expression of these genes is dependent on arabinose provided during growth. Thus, following colonization, the Fur and Crp proteins stop being synthesized due to the lack of arabinose and attenuation is progressively achieved in vivo to prevent generation of diseases symptoms. Our vaccine strain χ16022 with the genotype ΔasdA10 ΔPfur170::TT araC ParaBAD fur ΔPcrp68::TT araC ParaBAD crp contains the AsdA+ plasmid, pG8R8029, which encodes the IAG52B antigen. Vaccine strain χ16022(pG8R8029) is attenuated and induces systemic and mucosal IgM titer against E. piscicida and Ich in zebrafish. In addition, transcript levels of tnf-α, il-1β, il-6 and il-8 were significantly increased in different tissues of vaccinated zebrafish compared to unimmunized fish. Zebrafish vaccinated with χ16022(pG8R8029) showed 60% survival upon intracoelomic (i.c.) challenge with a lethal dose of virulent E. piscicida strain J118. Our RAEV system could be used as a generalized vaccine-vector system to protect teleost fish against multiple bacterial, viral and parasitic infectious diseases.
Edwardsiella piscicida is a Gram-negative, facultative anaerobic bacterium of the family Enterobacteriaceae. It is the causative agent of edwardsiellos in many economically important species of fish including catfish (Ictalurus furtatus), tilapia (Tilapia nilotica), European eel (Anguilla anguilla) and Indian major carp (Catla catla) (1–3). Vaccination would be an effective method to prevent and control Edwardsiella outbreaks. Live-recombinant attenuated bacterial vaccines must be fully attenuated to prevent disease symptoms and highly immunogenic. Conventional in-frame gene deletion can over attenuate the strain, making it more susceptible to host immune defenses and limit its ability to colonize internal tissue (4). To overcome these problems, our lab has previously demonstrated the ability to effectively attenuate E. piscicida through the use of regulated-delayed attenuation systems (5, 6). The promoters of the fur and crp genes were replaced by a tightly-regulated araC ParaBAD cassette such that expression was arabinose dependent during growth in vitro. This feature enables the strain to phenotypically mimic the virulent of wild-type E. piscicida strain at the time of vaccination and enhance the tissue colonization. Inside host tissues, absence of arabinose prevents the synthesis of the Fur and Crp proteins, allowing attenuation to gradually manifest and prevent disease symptoms (4). The safety and control of these strains can be further enhanced by replacing the promoters of several virulence factors with externally-regulated promoters. Additionally, these recombinant attenuated Edwardsiella vaccines (RAEVs) are capable of stimulating both innate and adaptive immune responses in fish (5, 6). Zebrafish vaccinated with an RAEV showed up-regulation in the transcription of genes for several pro-inflammatory cytokines, including TNF-α and IL-1β (5, 7). Additionally, immunized fish had significantly higher levels of serum IgM and increased levels of protection against challenge with the wild-type strain (5). These attributes suggest that these RAEV strains would be optimal delivery systems for heterologous recombinant proteins.
The ability of an organism to act as a recombinant protein producer is contingent on its ability to effectively retain the expression vector. To ensure that the vector is not lost, a selective pressure is created so that only organisms harboring the vector with the selective marker are able to grow. In laboratory settings, antibiotics typically serve as the selective agent. However, vectors with antibiotic selection markers may not be retained in vivo when the selective pressure is removed. Thus, vectors used in live-attenuated vaccines should have a selective marker that is applicable in host tissue. Diaminopimelic acid (DAP) is a necessary precursor for peptidoglycan synthesis in Gram-negative bacteria (8). Aspartate ß-semialdehyde dehydrogenase is an enzyme required for the production of DAP and encoded for by the asdA gene (9, 10). Gram-negative bacteria lacking asdA are unable to synthesize DAP and undergo lysis if they cannot obtain it from their surroundings (11–14). DAP is not synthesized nor metabolized in mammalian tissue and the nutritional requirements of fish suggests they also lack biological pathways that utilize DAP as a substrate (15, 16). This makes asdA an appropriate selection marker for live vaccines and is the basis for the balanced-lethal system, in which asdA deficient bacteria are dependent on a vector containing the cloned wild-type asdA gene for survival (17). This system is utilized in many Recombinant Attenuated Salmonella Vaccines (RASVs) to deliver protective antigens against multiple pathogens through antigen secretion or surface display (18, 19). Additionally, E. ictaluri and E. tarda ΔasdA mutants are capable of expressing heterologous antigens harbored on AsdA+ vectors (14, 20). These findings suggest that an RAEV possessing an asdA deletion could also function in a same manner.
Ichthyopthirius multifiliis is a ciliated protozoan and the etiological agent of white spot disease in fish. Infection by I. multifiliis has been shown to elicit various primary and secondary immune responses in catfish that promote parasite clearance (21–24). These adaptive immune responses in fish suggests that vaccination would be a safe and effective method to prevent disease. Surface immobilization antigens (IAGs) expressed by I. multifiliis are immuno-dominant and have become candidate proteins for vaccine development (23). Channel catfish and rainbow trout immunized with DNA vaccines encoding IAG52B produced specific antibodies, but showed no significant increase in the level of protection in challenge studies, suggesting that naked DNA alone is insufficient to induce a strong enough immune response to confer protection (25, 26).
The purpose of this study was to construct an antibiotic-sensitive and highly immunogenic Recombinant Attenuated Edwardsiella Vaccine (RAEV) vector system with regulated-delayed attenuation in vivo attributes. The asdA gene was deleted and an optimal balanced-lethal system was developed by using diverse recombinant AsdA+ plasmids containing different promoter sequences and origins of replication to generate as a platform for the heterologous antigen delivery system. The I. multifiliis gene encoding the surface immobilization antigen 52B (IAG52B) was codon optimized to enable high-level expression in Edwardsiella. In addition, a codon specifying incorporation of glutamine in Edwardsiella was specified to replace the nonsense codon used to specify glutamine in I. multifiliis. This modified sequence was then inserted into an AsdA+ expression vector and fused with the β-lactamase type II signal sequence for better secretion of the cloned gene products into the periplasm. The strain was then evaluated for its ability to elicit innate and adaptive immune responses towards both E. piscicida and I. multifiliis in immunized zebrafish.
All bacterial strains and plasmids used in this study are listed in Table 1. Bacterial strains were grown on Luria-Bertani (LB) agar or in LB broth. Where necessary, media was supplemented with 15% agar, 10% sucrose, colistin sulfate (Col) (12.5 µg/ml), chloramphenicol (Cm) (25 µg/ml), diaminopimelic acid (DAP) 50 µg/ml. Growth of bacteria was determined by spectrophotometrically and/or by plating following serial dilution. Oligonucleotides were from IDT (Coralville, IA). New England BioLabs restriction endonucleases and T4 ligase were used for cloning. For all PCR reactions, GoTaq DNA polymerase (Promega, catalog# M3008) was used. For plasmid DNA isolation and purification of gel fragments and PCR products, Qiagen products (Hilden, Germany) were used.
Adult zebrafish, Danio rerio (2 ± 0.5 cm and 0.4 ± 0.05 g) deemed clinically healthy were acclimated to laboratory conditions for two weeks after purchase from Aquatic Research Organisms, Hampton, NH, U.S.A. Conditioned reverse osmosis (RO) water was used in the zebrafish cultivation system. Water temperature was maintained at 26°C with a conductivity between 300-400 µS and pH between 7.0 and 7.4 by adding instant sea salt and sodium bicarbonate. Water was changed every day. A 14/10 h light/dark cycle was utilized, with lights turned on at 7:00 am and off at 9:00 pm. Fish were fed commercial zebrafish feed, GP 500-800 Micron Weaning Diet (Brine shrimp direct) two times per day. Zebrafish were anaesthetized by immersion in 100 ng/ml of tricaine methanesulphonate (MS-222) (Tricaine-S, Syndel, USA). Before manipulations, fish were euthanized for at least 10 min with Tris-buffered MS-222 at 20 mg/lit.
Publicly available AsdA protein sequences were retrieved from NCBI GenBank database. E. piscicida (CP001135), E. hoshinae (WP_024522689), E. ictaluri (WP_015872886), E. anguillarum (WP_034163973.1), E. coli (AP_004358), S. Gallinarum (WP_000799940), Salmonella Paratyphi A (ATF61156.1), E. tarda (WP_109728620), A. hydrophila (ABK39477.1), A. salmonicida (WP_005310917), Vibrio (WP_001263690), S. Typhimurium (AKH09169). S. flexneri (YP_690789), and Yersiniaceae (WP_120132887.1) sequences were used to construct the unrooted phylogenetic tree of AsdA by the neighbor-joining method of MEGA6 program (33). The three-dimensional (3D) structures of the E. piscicida, E. ictaluri and S. Typhimurium AsdA protein was predicted by using Phyre2 web portal (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index).
To develop antibiotic-sensitive strains of live-attenuated recombinant bacterial vaccines, a balanced-lethal host-vector system was constructed by deletion of the aspartate β-semialdehyde dehydrogenase (asdA) gene. The ΔasdA10 defined deletion mutation encompasses a 1100 base pair deletion including the ATG start codon but not does not include the last four bases, “CTAG”, specifying the stop codon for the gene. Primers asdA-1F-XbaI and asdA-2R (Table 2) were designed to amplify the upstream of the asdA gene flanking region (432 bp). The downstream asdA gene flanking region (583 bp) was amplified by primers asdA-3F and asdA-4R-KpnI (Table 2). A XbaI site was included in asdA-1F-XbaI, and a KpnI site was included in asdA-4R-KpnI. The flanking regions were amplified from E. piscicida J118 genomic DNA. The two PCR fragments were joined by overlapping PCR with primers asdA-1F-XbaI and asdA-4R-KpnI, and the products were cloned into the XbaI/KpnI site of the suicide vector pRE112 (29). The resulting plasmid was designated pG8R8000. To construct the E. piscicida ΔasdA10 mutant, the suicide plasmid was transferred from Escherichia coli χ7213 to E. piscicida wild-type strain J118 through conjugation. LB agar plates containing Col, Cm, and DAP were used to isolate strains with single-crossover plasmid insertions. A sacB-based sucrose sensitivity counter-selection system was used to select for bacteria that had lost the suicide vector after a second homologous recombination (i.e., allelic exchange) (29). The colonies were screened for growth in the presence of DAP, as well as for CmS, and Colr. The ΔasdA-deletion mutant was confirmed by PCR and DNA sequencing.
To construct the E. piscicida ΔasdA10 strain (χ16000) with a regulated-delayed attenuation phenotype, ΔPfur and ΔPcrp deletion insertion mutations were added sequentially to χ16000 by using suicide plasmids pG8R8024 and pG8R8009 as described previously (5, 6). The resultant strain had a genotype ΔasdA10 ΔPfur170::TT araC ParaBAD fur ΔPcrp68::TT araC ParaBAD crp and numbered as χ16022.
A series of different AsdA+ plasmid vectors were constructed with pUC ori and pBR ori containing the E. piscicida asdA gene with modifications of the asdA promoter, SD sequence and start codon. The E. piscicida asdA gene was amplified with its wild-type promoter and Shine-Dalgarno (SD) sequence, or with only the SD asdA sequence and also with modification of the start codon from ATG to GTG or the asdA gene without its SD sequence by using primers listed in Table 2. Forward and reverse primers were tagged with restriction enzyme sites for XbaI and KpnI. Fragments of the pYA3341 (pUC ori) (30) and pYA3342 (pBR ori) plasmids (30) minus the S. Typhimurium asdA gene were amplified by PCR with the primer pair P42F-KpnI and p42R-XbaI (Table 2). After gel purification, fragments were ligated with T4 DNA ligase and transformed into the E. piscicida ΔasdA strain χ16000 and plated on LB agar plates. The recombinant plasmids were confirmed by restriction digestion with XbaI and KpnI and sequencing. The resulting plasmids were named pG8R8011, pG8R8012, pG8R8013, pG8R8014, pG8R8015, pG8R8016, pG8R8017 and pG8R8018 (Table 1).
The growth of the E. piscicida ΔasdA mutant strain harboring different AsdA+ vectors was analyzed and compared to the mutant strain without a plasmid in LB both in presence and absence of DAP. A series of plasmids were constructed with differing copy numbers, presence or absence of the promoter for the asdA gene and with either ATG or GTG start codons as described in Table 1. Standing overnight 30°C cultures (OD600 ~ 0.6) of E. piscicida strains were diluted 1:100 into prewarmed LB or LB plus DAP broth and incubated at 30°C with shaking at 180 RPM. The OD600 was measured every 60 min. The growth curves were calculated using the automated growth curve device Bioscreen C (Growth Curves USA, Piscataway, NJ).
To determine the LD50 of E. piscicida strains i.e. J118, χ16000, χ16010, χ16012 and χ16012(pYA3493) ten-fold serial dilutions of E. piscicida fresh cultures were made in sterile BSG, and the concentration of bacteria was determined by the spread-plate method. Fish were i.c. injected in a dose of 10 µl of BSG containing different concentrations of CFU/fish (Table 3) with 10 fish in each group (two replicate tanks, 5 fish in each tank). Mortality was documented daily over a 15-day period, and the Reed and Muench method was used to calculate the LD50 values (34).
Zebrafish were inoculated with ~5 x 103 cells of χ16022, carrying the Ich IAG52B antigen gene, in a dose of 10 µl of BSG by i.c. injection. The spleen and kidneys were collected at days 3 and 5 after inoculation. Tissues were homogenized in 200 µL of BSG. A 10-fold serial dilution of each sample was used and plated on LB agar plates containing 0.2% arabinose and 10 µg/mL of colistin sulfate. The plates were incubated at 30˚C for 48 h and then the colonies were counted to determine the bacterial load in each organ. The results obtained from five fish (n = 5) in each time point were plotted with the bars indicating standard error.
The immune protection mediated by χ16022 against wild-type E. piscicida challenge was evaluated following i.c. vaccination, with a dose of 1 x 104 cells/fish using 15 fish/group. Fish were given a booster dose of 1 x 104 cells/fish after 2 weeks. Control fish were injected with 10 µL of BSG. Two weeks after receiving the booster dose, fish were challenged by i.c injection of virulent, wild-type E. piscicida J118 at a dose of 1 x 105 cells/fish. Mortalities were recorded for 14 days and represented as percent survival. The experiment was repeated once using 15 fish/group. Since the results from the two independent experiments were similar, the data were pooled together and represented as 30 fish total.
The periplasmic fraction was prepared by following the method described by Kang et al., 2002 (30). E. piscicida and E. coli cells harboring the pG8R8029 plasmid were grown in LB broth at 30˚C or 37˚C up to OD600 of 0.8 and centrifuged at 6000 g for 10 min. The supernatant fluid was saved for analysis of secreted proteins. Equal volumes of periplasmic, cytoplasmic, outer membrane, supernatant fractions and total lysate samples were separated by SDS-PAGE for western blot analysis.
Plasmid encoding IAG52B (pG8R8029) and control plasmid (pYA3493) were electroporated into E. piscicida or E. ictaluri or E. coli cells. Strains were grown in LB broth and 0.2% arabinose was added when necessary. Bacterial cells were grown at 30°C or 37°C with aeration (180 rpm) to an optical density at 600 nm (OD600) of 0.8. For the western blot analysis, 1 ml of bacterial culture was centrifuged, suspended in 100 µl of phosphate buffered saline (PBS; pH 7.4) and mixed with 100 µl of 2X SDS loading buffer. Protein samples were boiled for 10 minutes, and then 10 µl samples were loaded onto a 12% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) gels and electrophoresed. Samples were transferred onto nitrocellulose membranes. Membranes were blocked overnight at 4°C using fat-free milk powder dissolved in phosphate buffered saline (PBS) (5%, wt/vol) supplemented with 0.05% Tween 20 (PBS-T). The membranes were incubated with a primary rabbit polyclonal anti-Ich antibody (35). Membranes were washed with PBS-T three times, and then incubated with an alkaline phosphatase-conjugated anti-rabbit immunoglobulin G (IgG) (Sigma) diluted 1:10,000 in blocking buffer at room temperature for 1 h. A mixture of nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate (NBT-BCIP) (Amaresco), chromo-genic substrates for alkaline phosphatase was used to develop color. The reaction was stopped after 10 min by washing with several large volumes of deionized water.
Total RNA was extracted from zebrafish tissue samples (i.e., gill, kidney, intestine, and spleen) using TRIzol®(Ambion) following the standard protocol (Invitrogen). The total RNA concentration was measured by a UV-spectrophotometer (NanoDrop2000c, Thermo), and the relative purity was analyzed by the ratio of the absorbance value at 260 nm (A260) divided by the absorbance value at 280 nm, with a value of ~2.0 being considered highly pure. 3 µg of total RNA was then treated with 1 U of Thermo Scientific™ DNase I, RNase-free (FEREN0521) to remove any residual DNA from the sample. Reverse transcription was carried out using oligo-dT primer and Thermo Scientific™ RevertAid™ Premium First Strand cDNA Synthesis Kit (Catalog #FERK1622). cDNA synthesis was confirmed by PCR amplification of the ß-actin gene, keeping DNase-treated RNA as a negative control. Until further analysis, the synthesized cDNA was stored at -80˚C.
To evaluate the ability of χ16022 specifying the Ich IAG52B antigen to stimulate an immune response, the expression of the genes encoding IL-1β, IL-6, IL-8, and TNF-α in zebrafish tissues was analyzed by qRT-PCR. A 10 µL reaction mixture was prepared consisting of 5 µL of 2 x PowerUp™ SYBR™ Green Master Mix (Thermofisher Catalog # A25742), 3.5 µL of PCR grade H2O, 0.25 µL of FW and RV primers (2.5 mM each), and 1.0 µL of cDNA. The reaction was carried out in a Quantstudio 3 thermocycler (Applied Biosystems) in three separate wells with conditions of initial denaturation at 95°C for 10 min followed by 45 cycles with denaturation at 94°C for 10 s, annealing at 58°C for 10 s and extension at 72°C for 10 s. The β-actin gene was used for the internal normalization and reaction mixtures without the cDNA template served as the negative controls. PCR efficiencies were determined by analyzing cDNA serial dilutions. Since the efficiencies were almost 100%, the 2−ΔΔCT method (36) could be used to calculate the relative gene expression compared to the B-actin gene reference. The relative expression ratios were obtained by normalizing expression of the target gene, as determined by mean crossing point (cp) deviation by that of B-actin. The single band amplification and correct size was confirmed by ethidium bromide-stained 1% agarose gels using 8 µL of the qRT-PCR product. The qRT-PCR data was expressed as a mean of three individual experiments with standard error. To determine the significant difference between control and treated groups, Student’s t-test was conducted using Microsoft Excel 2010 with a significance level of P < 0.05.
ELISA was used to assay antibodies in gill, skin and serum to E. piscicida LPS and Ich membrane protein. Samples were prepared as described previously (5). Polystyrene 96-well flat-bottom microtiter plates (Dynatech Laboratories Inc., Chantilly, Va.) were coated with E. piscicida LPS or Ich membrane protein (100 ng/well), in sodium carbonate-bicarbonate coating buffer (pH 9.6) 100 μl volumes in each well. The coated plates were incubated for an overnight at 4°C. Free binding sites were blocked with 5% bovine serum albumin (BSA) for 1 h at room temperature. After washing, 100-μl volume of diluted zebrafish anti-serum/mucus sample was added to individual wells in duplicate and incubated for 2 h at 37°C. The plates were treated with mouse anti-zebrafish IgM monoclonal antibody (Aquatic Diagnostics Ltd) for 1 h at room temperature. Plates were then incubated with biotinylated goat anti-mouse IgG (Southern Biotechnology Associates, Birmingham, AL) for 1 h at room temperature. After incubation of wells with a streptavidin-alkaline phosphatase conjugate (Southern Biotechnology) for 1 h at 37°C, p-nitrophenyl phosphate (PNPP, Thermo Fisher Scientific) was added for color development. The optical density (OD) units were read at 405 nm using an automated ELISA plate reader (model EL311SX; Biotek, Winooski, VT).
Statistical analysis was performed by GraphPad Prism 6 (Graph Pad Software, Inc., San Diego, CA, USA). Survival data was analyzed with the log-rank (Mantel-Cox) test. Differences between the groups were analyzed by two-way ANOVA, where asterisks (*) indicate a significant difference (*P < 0.05, **P < 0.01, ***P < 0.001).
The E. piscicida EIB202 (J118) asdA open reading frame consisted of 1104 base pairs (bp) that encoded a putative 368 amino acid (aa) residue protein with an estimated molecular mass of 40 kilodaltons (kDa). To explore the evolutionary development history of bacterial asdA genes, a phylogenetic tree was constructed based on the amino acid sequences (Figure 1A). The amino acid sequence of E. piscicida AsdA shares a high percentage of sequence identity with E. anguillarum (98.64%), E. ictaluri (97.28%), E. tarda (94.02%) and E. hoshinae (92.92%). These sequence identities were reflected in the phylogenetic tree. E. piscicida and E. anguillarum form a cluster together with a high bootstrap value. E. ictaluri forms a separate but close cluster to E. piscicida. E. hoshinae and E. tarda fall in same cluster and were separated from other Edwardsiella species. Salmonella Typhimurium, Salmonella Gallinarum and Salmonella Paratyphi A shared 82.83% of sequence identity with E. piscicida.
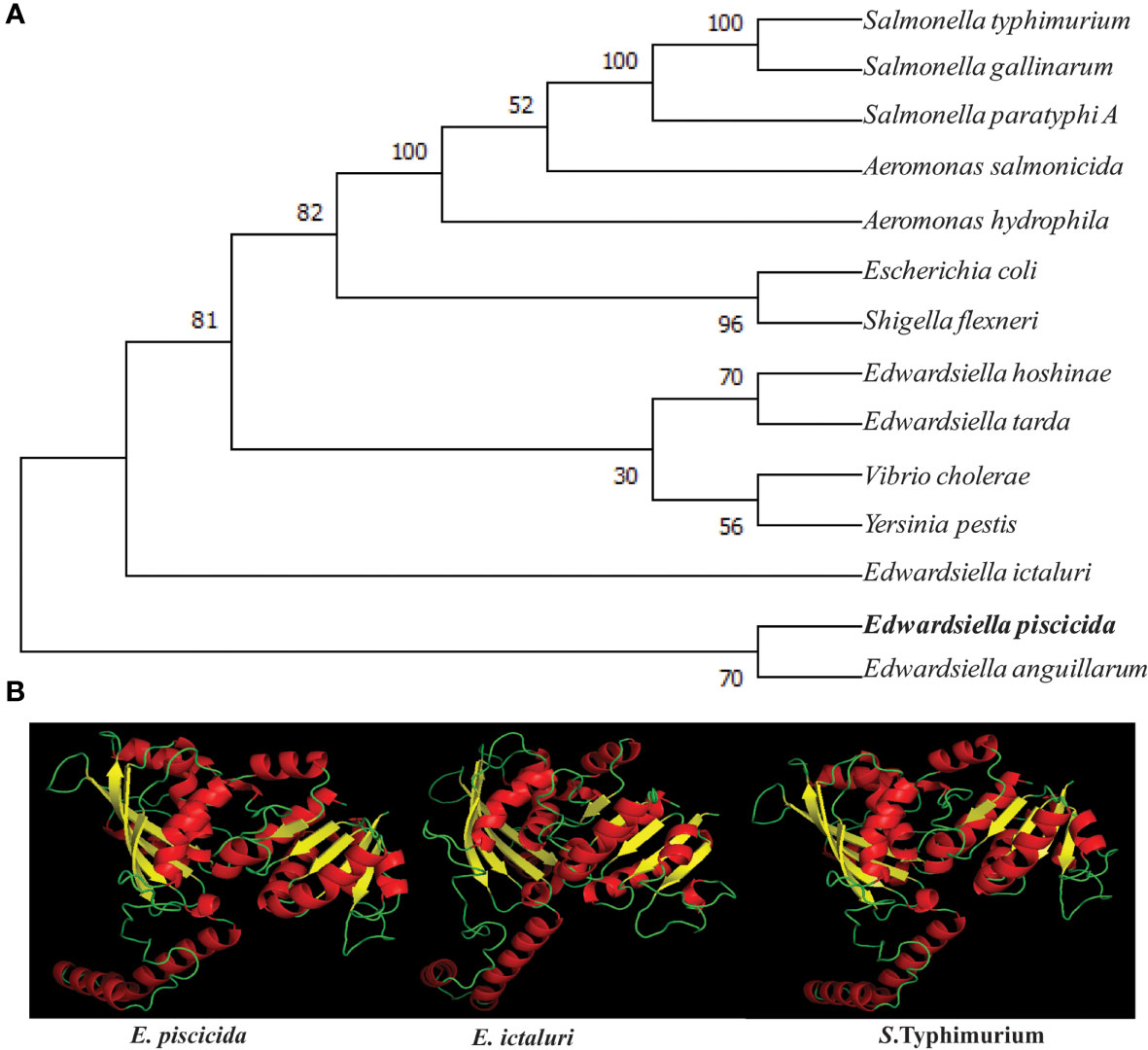
Figure 1 Phylogenetic and structural similarity of E piscicida AsdA with other bacteria. (A) Amino acid sequences of AsdA of various bacteria were retrieved from the GenBank database and the unrooted phylogenetic tree of AsdA was constructed by the neighbor-joining method within the MEGAX program. (B) 3D structure of E. piscicida, E. ictaluri and S. Typhimurium AsdA proteins were predicted by using (PS)2: Protein Structure Prediction Server Version 3.0 web portal.
The 3D structure of the E. piscicida, E. ictaluri and S. Typhimurium AsdA were predicted by using the Phyre2 web portal. Our results revealed that they share high sequence homology and with similar 3D structures. Each protein showed 16 α-helices and 9 β-sheets that were connected to each other by loops (Figure 1B).
To develop antibiotic-sensitive strains of live-attenuated recombinant bacterial vaccines, we used a balanced-lethal host-vector system to delete the aspartate β-semialdehyde dehydrogenase (asdA) gene. The ΔasdA10 defined deletion mutation encompasses a 1100 base pair deletion including the ATG start codon but does not include the last four bases, “CTAG”, of the gene. The upper panel of Figure 2A illustrates the chromosomal structures of the wild-type and mutant strains. Construction of the E. piscicida ΔasdA strain was done through suicide plasmid mediated homologous recombination, using suicide vector pG8R8000, a pR112 (Cm) based suicide vector (Table 1), by allelic replacement in the parent strain J118. The genotype was confirmed via PCR (Figure 1B) and sequencing. Agarose gel analysis and sequencing of PCR product confirmed that ΔasdA was 1100 bp smaller than the wild-type strain. The resultant E. piscicida ΔasdA mutant was named χ16000 (Table 1). The phenotype was verified by growth of ΔasdA mutant (χ16000;) in presence or absence of DAP (Figures 2C, D). The ΔasdA mutant could not survive in LB agar plates without being supplemented with DAP, but in the presence of DAP, the ΔasdA mutant strain grew like the wild-type strain. These results confirm that the E. piscicida asdA gene is functional and might work in a similar manner to that observed in E. ictaluri (14) and Salmonella (17).
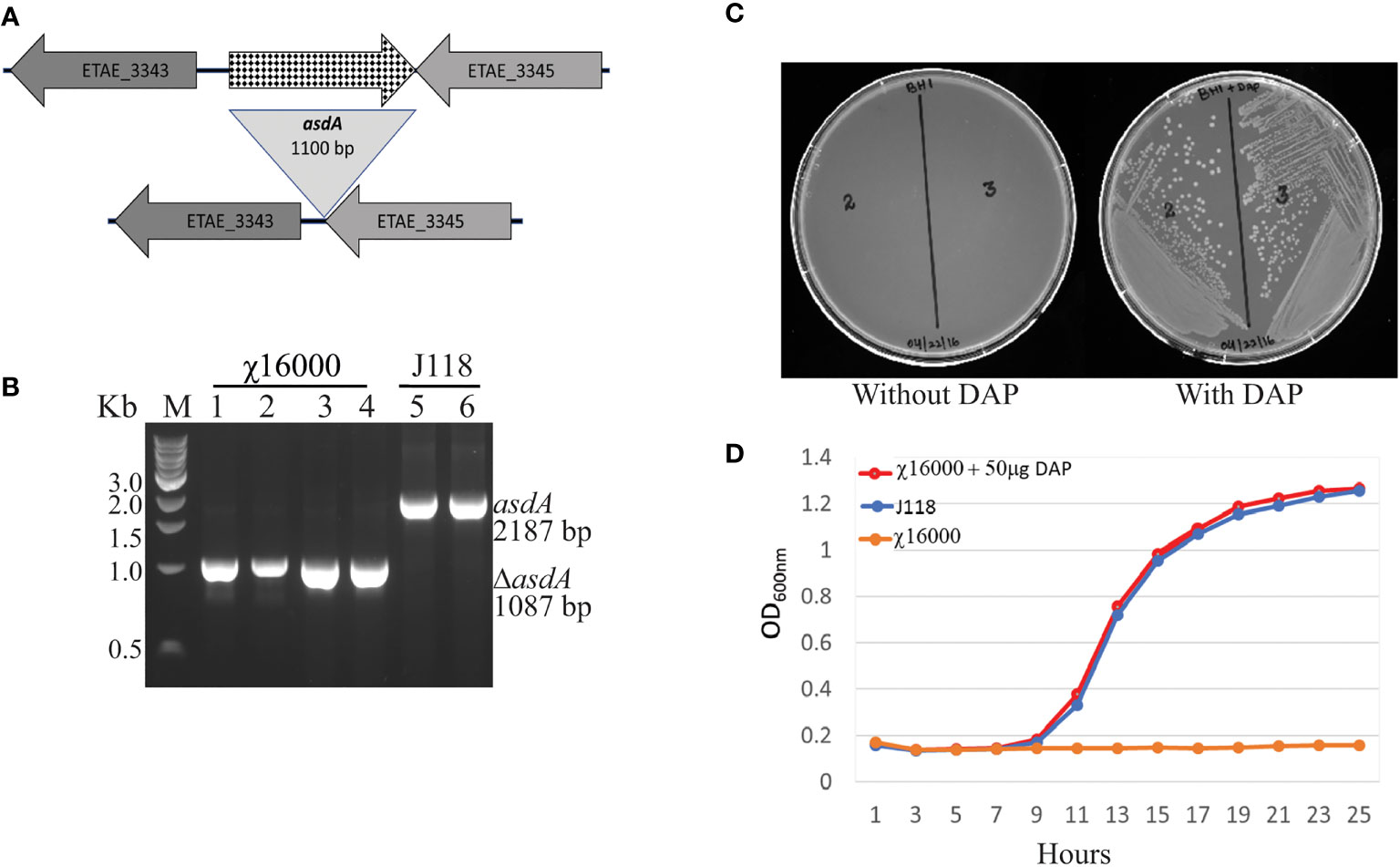
Figure 2 (E) piscicida asdA (χ16000) genotype and phenotype verification. (A) Deletion map of ΔasdA. (B) Genotype verification of E. piscicida ΔasdA (χ16000) by PCR. (C) Phenotypic verification of ΔasdA (χ16000) in the presence or absence of DAP in LB agar plates. (D) Growth curves for J118 and χ16000 in presence or absence of DAP.
Deletion of either fur or crp attenuated E. piscicida (5, 6) and E. ictaluri (37, 38). Strains were constructed with deletion insertion mutations that conferred a phenotype of regulated-delayed attenuation in vivo (5). The principle of regulated-delayed attenuation in vivo is based on the replacement of the fur and crp gene promoters with a tightly regulated araC ParaBAD cassette such that the expression of these genes is dependent on arabinose supplied during growth. Thus, following internal tissue colonization, Fur and Crp protein synthesis is terminated due to the absence of arabinose and attenuation is steadily manifested in vivo to prevent disease symptoms. The promoters, including all sequences that interact with activator or repressor proteins, for the fur and crp genes were deleted, and the improved araC ParaBAD cassette was substituted in E. piscicida ΔasdA strain (χ16000) to yield a strain with genotype ΔasdA10; ΔPfur170::TT araC ParaBAD fur; ΔPcrp68::TT araC ParaBAD crp (P stands for promoter and TT for transcription terminator), and the strain was designated χ16022.
Gram-negative bacteria with asdA mutants have an obligate requirement for diaminopimelic acid (DAP), which is a crucial component of the peptidoglycan layer (4). In surroundings deprived of DAP, i.e., animal tissues, they will undergo lysis. Deletion of the asdA gene has previously been used to develop antibiotic-sensitive strains of live-attenuated recombinant bacterial vaccines (14, 19). Introduction of an AsdA+ plasmid into a ΔasdA mutant makes the bacterial strain plasmid dependent (17). This dependence on the AsdA+ plasmid vector creates a balanced-lethal complementation between the bacterial strain and the recombinant plasmid.
Eight different AsdA+ plasmid vectors were constructed with pUC ori or pBR ori containing the E. piscicida asdA gene with modifications of the asdA promoter, SD sequence and start codon (Figures 3A, B). Plasmids pG8R8011 to pG8R8014 contain the pUC ori whereas plasmids pG8R8015 to pG8R8018 have the pBR ori. Plasmid pG8R8011 and pG8R8015 contain the wild-type asdA gene with the wild-type promoter, which includes the 118 bp upstream sequence from the start codon (ATG). Plasmid pG8R8012 and pG8R8016 contain E. piscicida asdA ORF with Shine-Delgrano (SD) sequence “AGGA”. pG8R8013 and pG8R8017 contain only the wild-type asdA ORF sequence. Plasmid pG8R8014 and pG8R8018 encode asdA sequence with the start codon “GTG” and (SD) sequence “AGGA”. Each of these plasmids were individually incorporated into the ΔadsA strain χ16000, and growth curves determined (Figure 3C). All of these E. piscicida AsdA+ plasmids were able to complement the asdA gene of χ16000. The strain complemented with pG8R8018, which has the pBR ori and encodes the asdA sequence with the start codon “GTG” and (SD) sequence “AGGA” grew similar to the wild-type. χ16000 complemented with pG8R8014 grew slightly slower compared to other complemented strains. pG8R8014 and pG8R8018 have similar features other than the origin of replication.
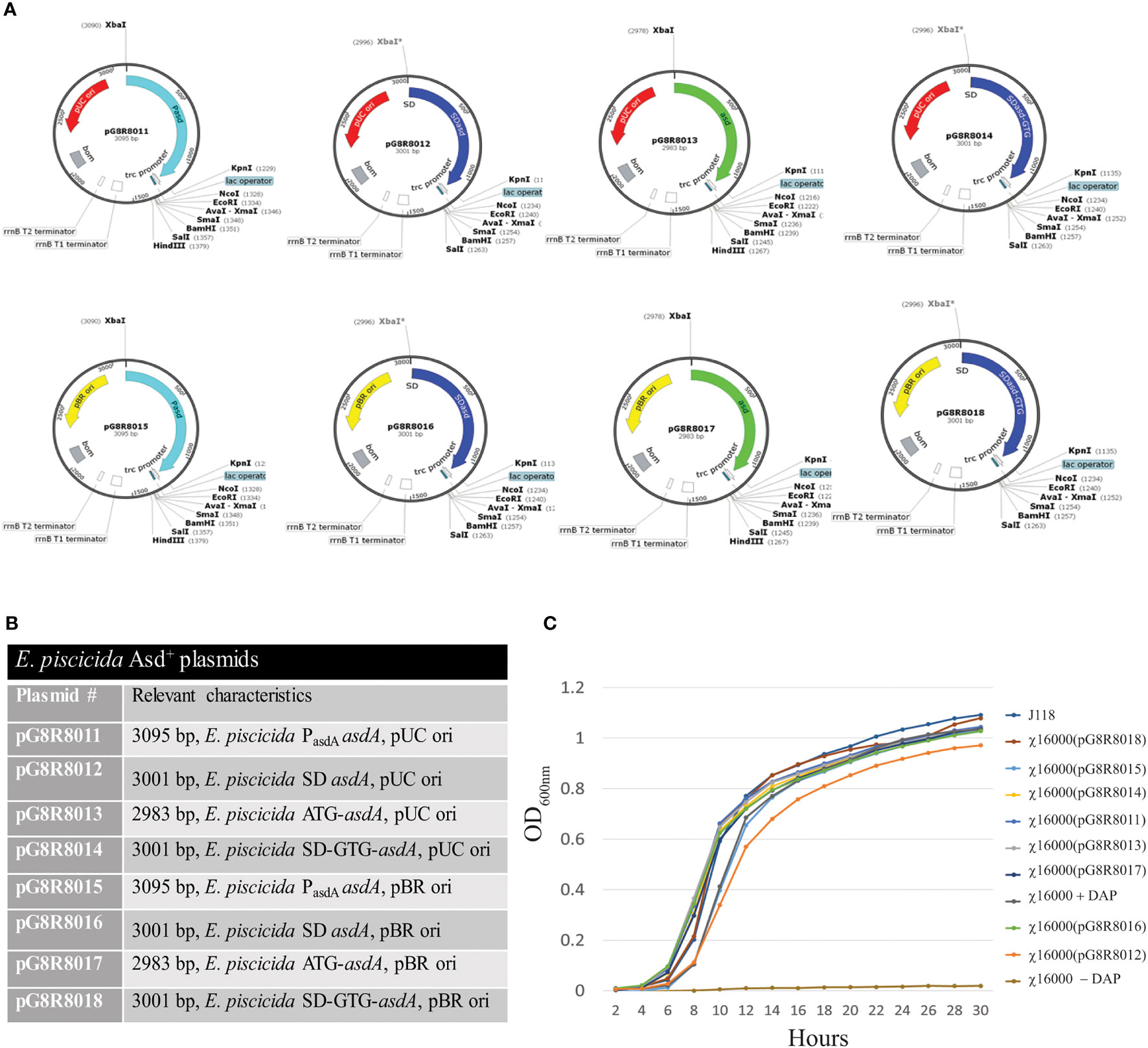
Figure 3 Complementation of ΔasdA mutant strains with E. piscicida AsdA+ Plasmids. (A) E. piscicida AsdA+ Plasmid maps (pG8R8011-pG8R8018) showing origin of replication, orientation of asdA gene, presence or absence of the promoter for the asdA gene and with either ATG or GTG start codons, Ptrc promoter and multiple cloning sites. (B) Characteristic of E. piscicida AsdA+ plasmids or various strategies to construct AsdA+ plasmids via modification of the plasmid copy number (pUC ori and pBR ori), promoter, SD sequence and start codon. (C) Growth of ΔasdA (χ16000) mutant strains complemented with different E. piscicida AsdA+ plasmids (pG8R8011 - pG8R8018).
The E. piscicida AsdA enzyme shares 97% and 81% sequence identity with E. ictaluri and Salmonella, respectivelly. Therefore, compared the ability of the asdA gene from E. ictaluri and Salmonella to complement the E. piscicida ΔasdA mutant (Figure 4). χ16000 complemented with pEZ142 showed a similar growth curve to wild-type J118. A high (pUC ori), medium (pBR & p15A ori) and low-copy (pSC101 ori) number Salmonella AsdA+ plasmids were used (Table 1) to complement E. piscicida ΔasdA mutants. All of these plasmids efficiently complemented the ΔasdA chromosomal mutation; however, the growth of these strains was slower than those complemented by plasmids with the E. piscicida AsdA+ encoding sequences. A slower growth of χ16000(pYA3337) was observed compared to other strains. The slower growth might be due to the low-copy number plasmid pYA3337, which may not produce a sufficient amount of AsdA enzyme needed for the growth of the bacteria.
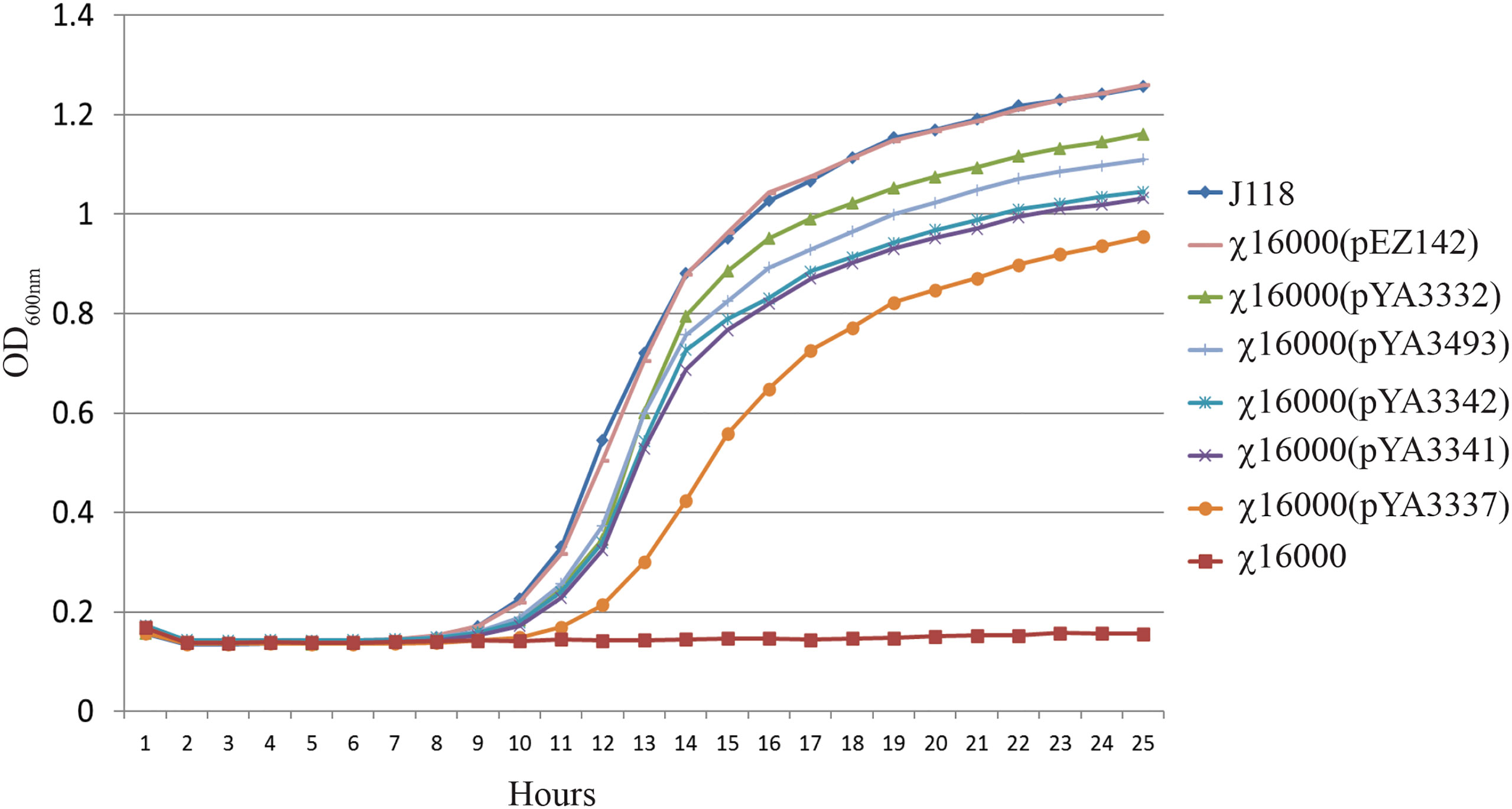
Figure 4 Growth of E. piscicida ΔasdA (χ16000) strains complemented with asdA gene from E. ictaluri and Salmonella. Growth of E. piscicida ΔasdA (χ16000) strains complemented with Salmonella AsdA+ plasmids with different copy numbers (high, medium and low) and modified asdA gene promoter.
IAG52B ORF consisted of 1383 base pairs and encoded a protein of 460 aa. Different domains of IAG52B were predicted by the InterPro web portal (https://www.ebi.ac.uk/interpro/). IAG52B contains a C-terminal signal IP of 20 amino acids, a middle non-cytoplasmic domain of 422 amino acids and a N-terminal transmembrane region of 18 amino acids. Transgene expression of i-antigen genes imposes a challenge in heterologous systems such as bacteria, because Ich, like other hymenostome ciliates, uses a nonstandard genetic code (39–41) in which the stop codons UAA and UGA encode glutamine. This problem was addressed by synthesizing and inserting i-antigen gene constructs with altered codons that were optimized for expression in E. coli and E. piscicida (Figure 5A). The composition of the IAG52B amino acid is depicted in Figure 5B. The secondary structure of IAG52B (Figure 5C) was predicted by Protean software, and the 3D structure (Figure 5D) was predicted by Phyre2 web portal. Our results indicate that the IAG52B protein contains 20 β-sheets and 7 α-helices and is connected by loops.
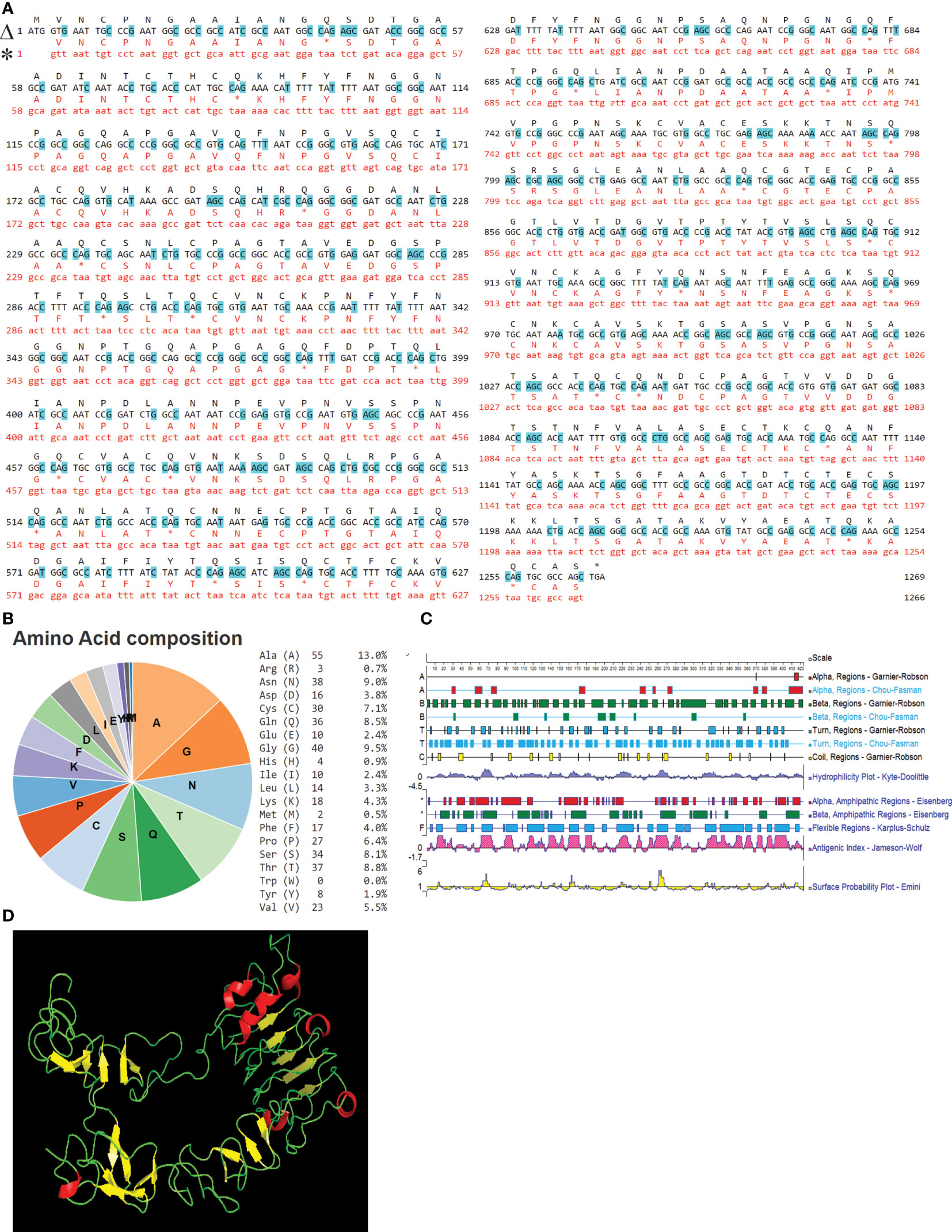
Figure 5 IAG52B sequence analysis. (A) Alignment of nucleotide and amino acid sequences of the original and codon-optimized IAG52B gene. Line marked with Δ is the codon optimized sequence and * is the original IAG52B sequence. Changed nucleotide sequences are highlighted with blue. (B) Pie chart and table illustrates the amino acid composition of IAG52B. (C) The secondary structures of IAG52B protein were predicted by Protean software by using Gramier-Robson and Chou-Fasman methods. Lines 1, 3, 5, and 7 are the Gramier -Robson methods; the red represents the alpha helix, the green represents the beta fold, the blue represents the turn, the yellow represents the random coil. Lines 2, 4, 6 are for the Chou -Fasman method; the red represents the alpha helix, the green represents the beta fold, the blue for the turn, without random coil prediction. (D) The 3D structure of IAG52B protein was predicted using the Phyre2 web portal.
The codon-optimized sequence of IAG52B non-cytoplasmic domain of 1269 bp was PCR amplified and cloned into the pYA3493 (Figure 6A) at the EcoRI-BamHI site. The IAG52B gene was fused into the same reading frame of bla SS under the Ptrc promoter. The resultant plasmid was named as pG8R8029 (Figure 6B). Both pYA3493 and pG8R8029 were introduced into the ΔasdA strains: E. piscicida (χ16000), E. ictaluri (J111) and E. coli (χ6212). The synthesis of IAG52B was analyzed by western blotting. As shown in Figure 6C, synthesis of IAG52B was detected in E. piscicida and E. coli, but no signal was noticed in E. ictaluri and cells harboring the control plasmid pYA3493.
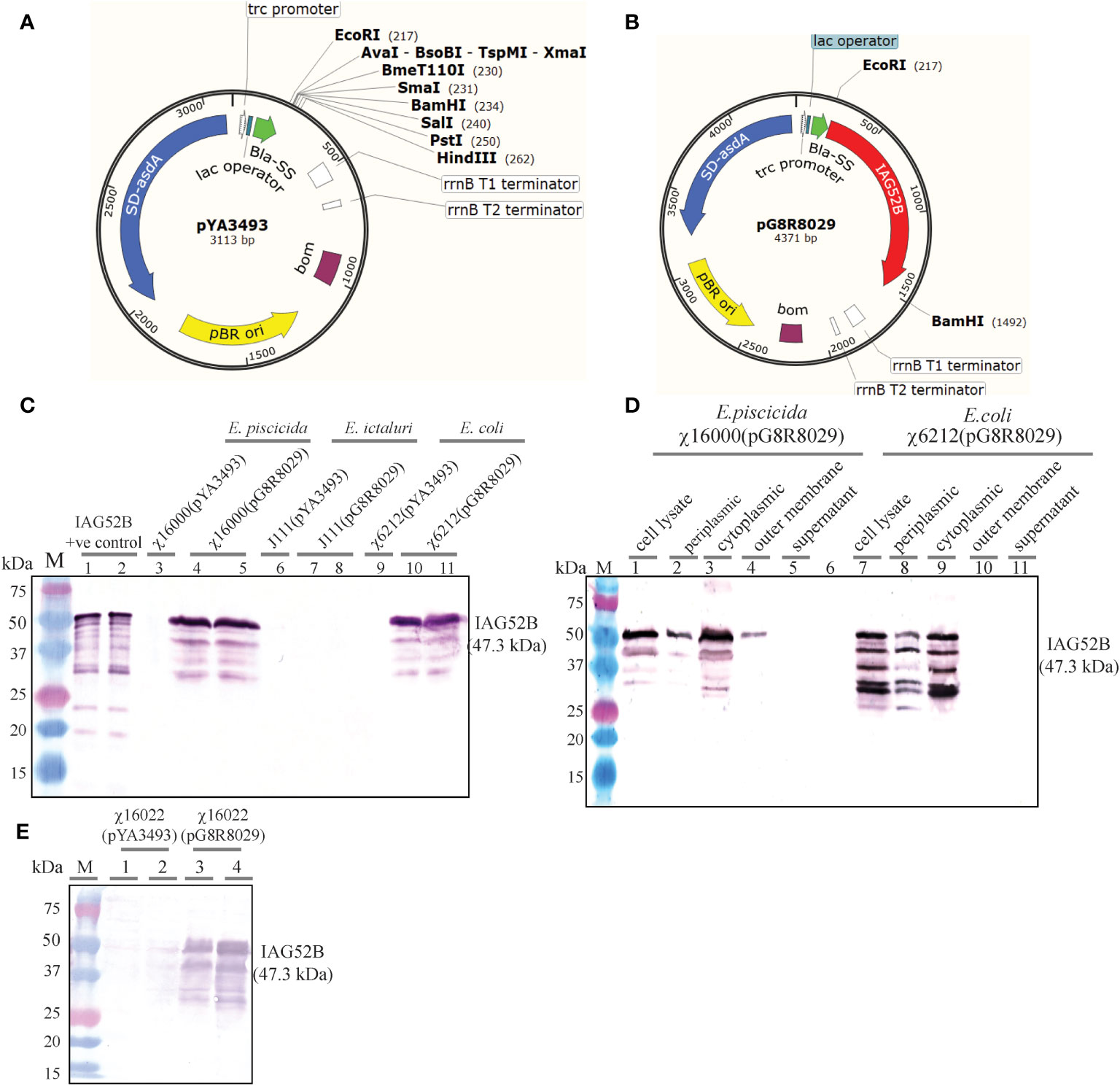
Figure 6 (A) Vector map of pYA3493 (pBR ori), which is a periplasmic secretion AsdA+ vector. A DNA fragment encoding the β-lactamase signal sequence (bla SS) and 12 amino acid residues of the N-terminus of mature-lactamase was positioned under the control of the Ptrc promoter. Map of pYA3493 shows the unique restriction enzyme sites in the multiple cloning site. (B) Vector map of recombinant plasmid pG8R8029. Codon-optimized gene of IAG52B non-cytoplasmic domain of 1269 bp was PCR amplified and cloned into the pYA3493 at EcoRI-BamHI sites. The IAG52B gene was fused into the same reading frame of bla SS and under the control of the Ptrc promoter. (C) Synthesis of IAG52B antigen in E. piscicida, E. ictaluri and E. coli asdA mutant strains. pYA3493 (vector control) or pG8R8029 (encoding IAG52B) were electroporated into E. piscicida (χ16000) or E. ictaluri (J111) or E. coli (χ6212). IAG52B antigen synthesis was analyzed in these cells by western blotting using an anti-IAG52B antibody. (D) Subcellular location of synthesized IAG52B in E. piscicida and E. coli. Western blot showing IAG52B synthesis in whole cell lysate, periplasmic, cytoplasmic, outer membrane and supernatant fraction of χ16000 and χ6212 harboring pG8R8029. (E) Analysis of IAG52B antigen synthesis in E. piscicida vaccine strain χ16022. pYA3493 (vector control) or pG8R8029 (encoding IAG52B) was electroporated into χ16022, IAG52B antigen synthesis was analyzed by western blotting by using anti-IAG52B antibody.
E. piscicida strain χ16000 and E. coli strain χ6212 harboring pG8R8029 were analyzed for subcellular localization of IAG52B. As expected, IAG52B was detectable in cytoplasm, periplasm and the outer membrane in both χ16000(pG8R8029) and χ6212(pG8R8029) (Figure 6D). A high amount of protein was detected in the cytoplasmic fraction but only half of the IAG52B was secreted into the periplasmic fraction in both E. piscicida and E. coli. Interestingly, some of i-antigen associated with the outer membrane in E. piscicida was detected; this suggests that some antigen may be incorporated into outer membrane vesicle (OMVs) that are very immunogenic and excellent means for antigen delivery. Next, the synthesis of IAG52B in the vaccine delivery strain χ16022 was analyzed. Large amounts of IAG52B synthesis were detected in χ16022(pG8R8029) and no signal was observed in the vector control χ16022(pYA3493) (Figure 6E). This result confirms that IAG52B is synthesized in the E. piscicida vaccine strain χ16022(pG8R8029). A similar subcellular localization pattern was observed in χ16000(pG8R8029).
To examine the stability of plasmids pYA3493 and pG8R8029 in E. piscicida χ16022, strains χ16022(pYA3493) and χ16022(pG8R8029) were cultured with daily passage of 1:1,000 dilutions for five consecutive days in LB broth containing DAP and arabinose. Hundred colonies were screened daily for DAP sensitivity in LB agar plates in the presence or absence of DAP. All the colonies were DAP independent, indicating that pYA3493 and pG8R8029 are very stable in the χ16022 vaccine strain. IAG52B synthesis was analyzed by western blotting from cells obtained from the last day of culture of the stability test, and there was a detectable signal, which suggests both pG8R8029 in the vaccine strain and antigen expression are stable.
The virulence of the E. piscicida vaccine and wild-type strains was determined by LD50 values in zebrafish injected intracoelomically with different concentrations of bacteria. Our result showed that the vaccine strain χ16022 harboring the control plasmid pYA3493 had significantly higher LD50 levels compare to the wild-type strain (Table 3). The LD50 of the wild-type strain J118 was 1.1 × 104 CFU, while the LD50 of the χ16022(pYA3493) increased up to 2×105 CFU, which is more than a 10-fold increase (Table 3). As expected, the LD50 of the χ16022(pYA3429) was similar to the LD50 of χ16022(pYA3493) (data not shown). This result indicates that the E. piscicida crp gene product contributes to the virulence and pathogenicity of E. piscicida. The LD50 of χ16012 was 2 x 104 CFU, which was similar to the wild-type strain J118. This result confirms that ΔPfur170::TT araC ParaBAD fur deletion insertion mutation may not affect the virulence of E. piscicida by i.c. injection. As expected, the LD50 of the χ16022(pYA3429) was similar to the LD50 of χ16022(pYA3493) (data not shown).
To examine the ability of χ16022(pG8R8029) to colonize and disseminate into fish tissues, zebrafish were immunized with the vaccine strain χ16022(pG8R8029) by i.c. injection. The recovery of the vaccine strain from the kidney and spleen was determined at days 3 or 5 post-vaccination. A significant number of χ16022(pG8R8029) was recovered in both the kidney and spleen at days 3 or 5 of post-vaccination. However, the number of bacteria was higher at day 3 compared to day 5 (Figure 7). This result indicates that the vaccine strain successfully colonized and disseminated into different tissues of fish.
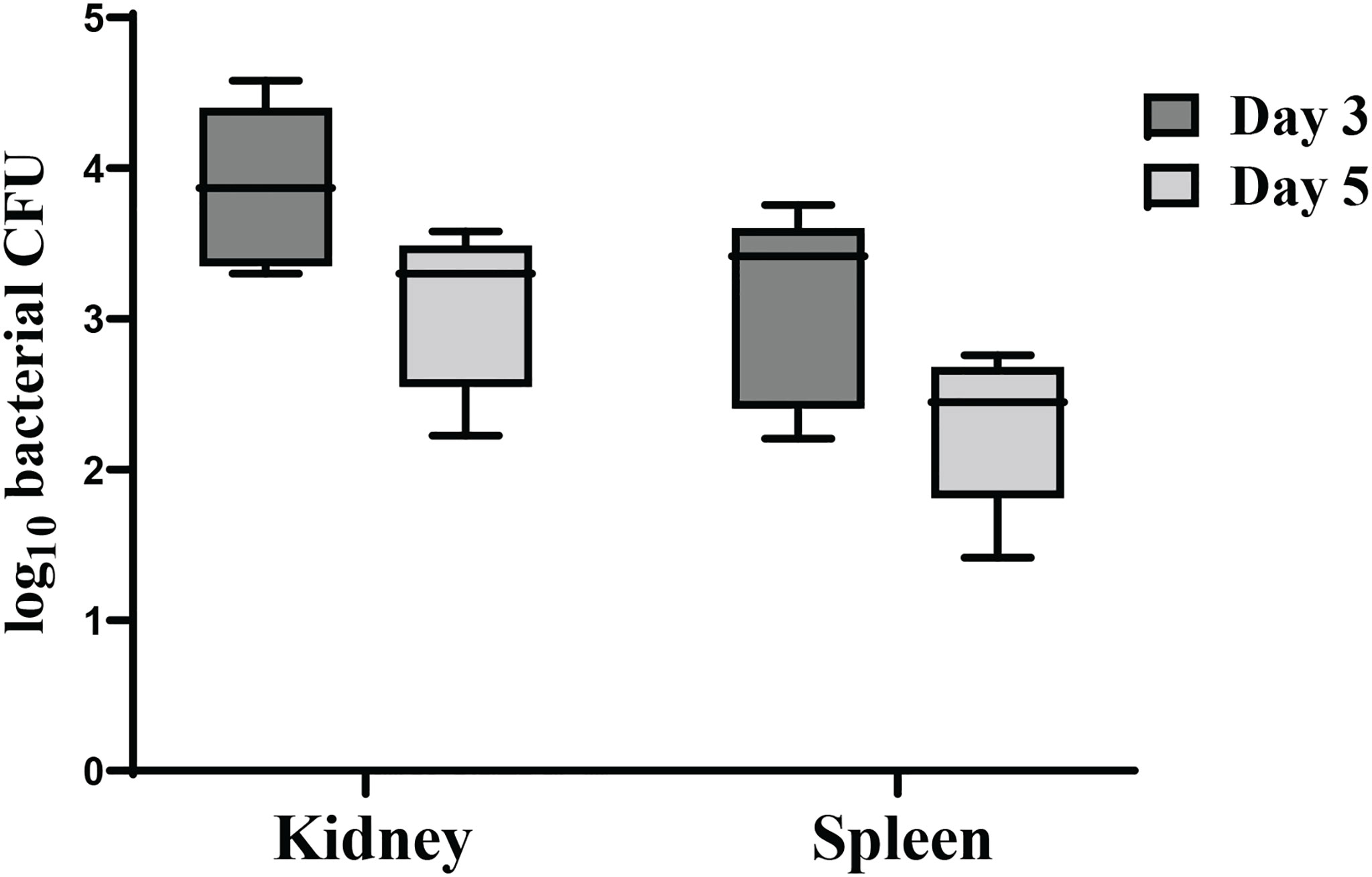
Figure 7 Dissemination and colonization of χ16022(pG8R8029) in zebrafish tissues. (A) Zebrafish were vaccinated with χ16022(pG8R8029) by i.c. injection. Kidney and spleen were collected from immunized fish at 3- and 5-days post-vaccination (five fish in each group). The tissues were homogenized in 200 μL of BSG and plated on LB agar plates supplemented with 0.2% agarose and 10 μg/mL of colistin. The plates were incubated at 30°C for 48 h and the colonies were counted. The data was a combination of three independent assays.
To investigate the efficiency of the vaccine strain in modulating immune responses of zebrafish, the expression of genes associated with the immune response including tumor necrosis factor-α (tnf-α), interleukin 1β (il-1β), interleukin-6 (il-6) and interleukin 8 (il-8) in the gills, kidney, intestine and spleen were assessed by qRT-PCR. All of these cytokine genes were significantly upregulated in the tissues of vaccinated fish compared to unvaccinated fish. Furthermore, their expression was higher at day 3 compared to day 5 and 7 in all tested tissues of the vaccinated fish. A significant increase in tnf-α gene expression was detected in kidney and spleen (Figure 8A). il-1β gene expression levels were significantly increased in all organs after 3 days post-vaccination. No changes were found in the expression levels of il-1β at day 5 and 7 in vaccinated fish compared to the control group (Figure 8B). The expression of il-6 and il-8 patterns were similar to il-1β, showing significant upregulation in all tissues at day 3, whereas, the changes at day 5 and day 7 were insignificant compared to control tissues (Figures 8C, D).
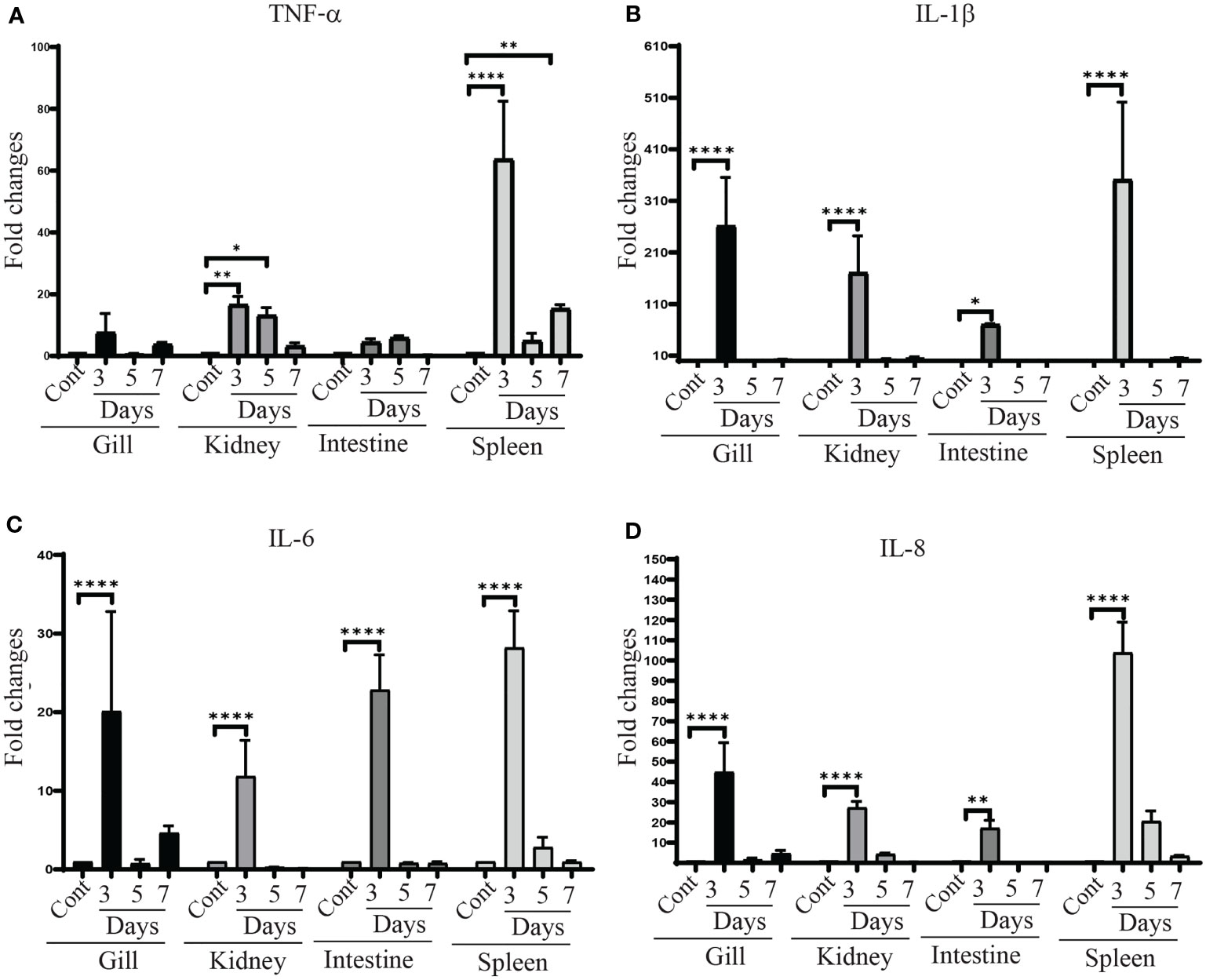
Figure 8 tnf-α, il-1β, il–6 and il-8 gene expression in χ16022(pG8R8029) vaccinated and control zebrafish. Zebrafish were vaccinated with χ16022(pG8R8029) by i.c. injection. At three, five and seven days of post-immunization, total RNA was extracted from the gills, kidney, intestine and spleen and cDNA was prepared. The qRT-PCR assay was conducted to analyze the expression of tnf-α, il-1β, il-6 and il-8 genes using β-actin as an internal control. The results are expressed as mean ± standard error (bars) from three separate experiments. Differences between uninfected (control) and infected groups were analyzed by two-way ANOVA, where asterisks (*) indicate significant difference (*P < 0.05, **P < 0.01, ****P < 0.0001) with respect to the control group.
Serum and mucosal immunoglobulin M (IgM) responses to E. piscicida lipopolysaccharide (LPS) and Ich membrane protein were measured by ELISA at 4 weeks (before challenge) and 6 weeks (2 weeks after challenge) post-vaccination of χ16022(pG8R8029). The LPS and Ich specific IgM titers were induced in both serum and mucosal tissues of vaccinated fish and compared to unvaccinated control fish. There was a significant upregulation of anti-LPS IgM titers in the serum at both 4- and 6-weeks post-vaccination compared to control group. The anti-Ich IgM titer was significantly increased only at 6 weeks of post-vaccination (Figure 9).
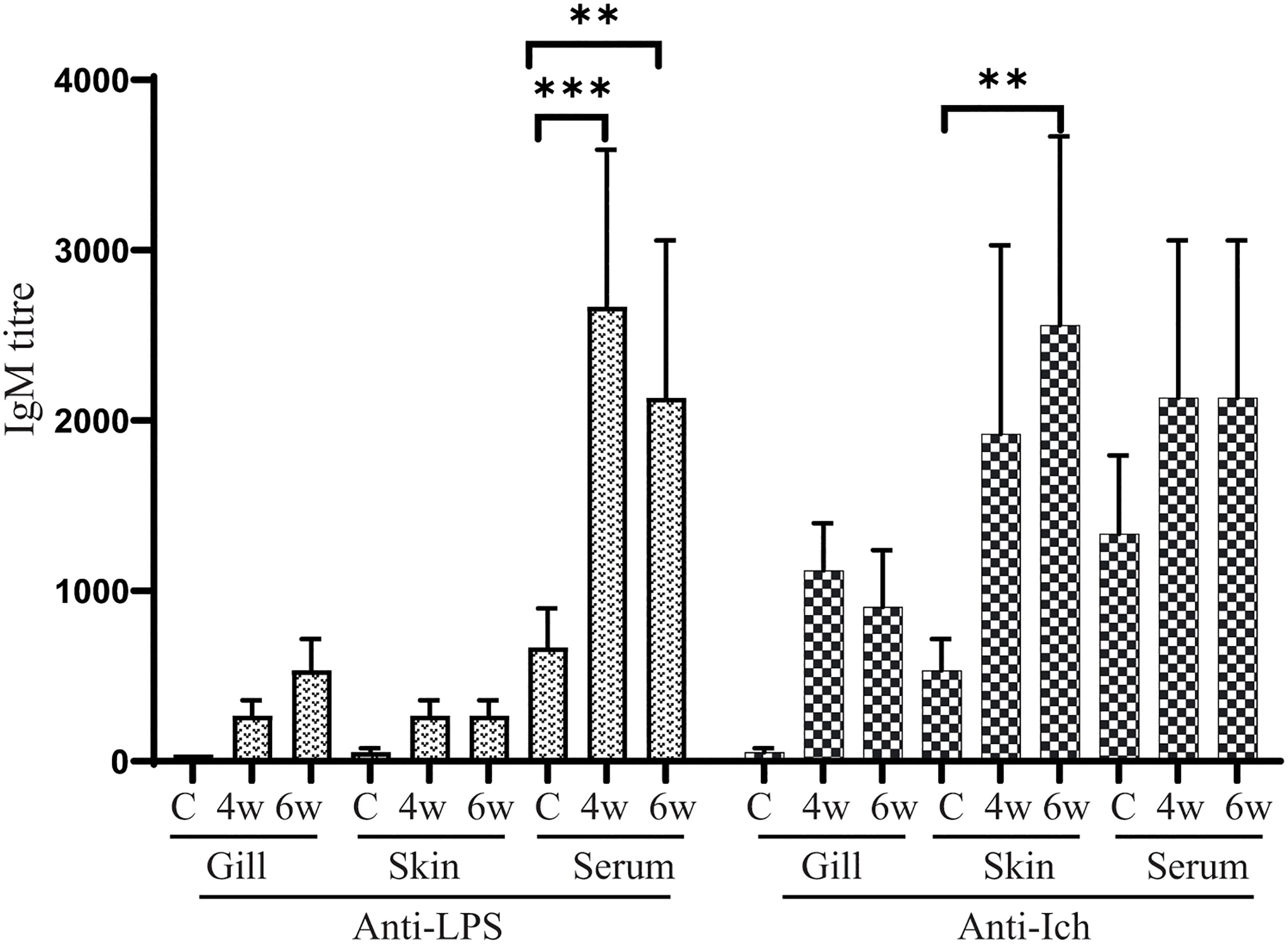
Figure 9 Anti-LPS and anti-Ich antibody responses in zebrafish. Serum and mucosal immunoglobulin M (IgM) responses to E. piscicida lipopolysaccharide (LPS) and Ich IAG52B membrane protein were measured by ELISA at 4 weeks (4w) post-vaccination with χ16022(pG8R8029) and 2 weeks after J118 challenge (6w). The results are expressed as mean ± standard error (bars) from three separate experiments. Differences between treated and control groups were analyzed by two-way ANOVA, where asterisks (*) indicate significant difference (**P < 0.01, ***P < 0.001).
In order to investigate the protective efficacy of χ16022(pG8R8029) in zebrafish against E. piscicida infection, the vaccinated and control fish were i.c. challenged with live virulent E. piscicida (J118) 1 x 105 cells/fish (10 × LD50) at 4 weeks of post-vaccination. The mortality in fish started within 48 hours post E. piscicida infection (confirmed by isolating J118 from the kidney of dead fish). The vaccinated fish showed typical symptoms of E. piscicida infection such as hemorrhages of the skin and in internal organs and swelling of abdomen. Significantly greater survival was observed in the χ16022(pG8R8029)-vaccinated group (60%) and in the vector control χ16022(pY3493) group (55%) in comparison to that of the control unvaccinated fish (0%) over a period of 15 days post-challenge (Figure 10) against virulent E. piscicida strain J118.
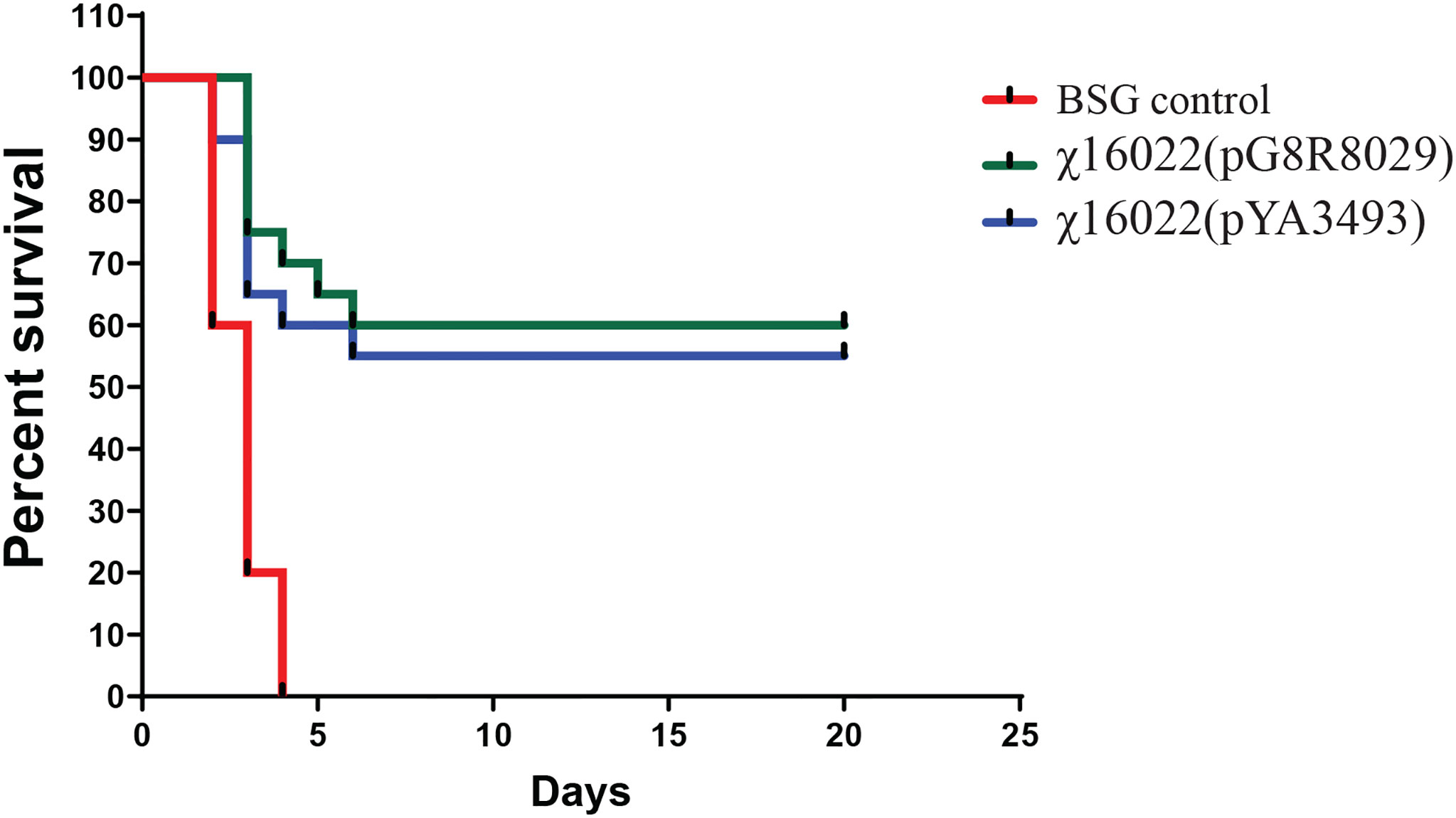
Figure 10 Survival of χ16022(pG8R8029), χ16022(pYA3493) vaccinated and BSG control fish after wild-type E. piscicida challenge. Control and 4 weeks post-vaccination zebrafish were i.c. challenged with 1 × 105 CFU/dose (10 × LD50) of wild-type E. piscicida. Mortality was recorded daily and represented as percent survival.
The advantage of using live bacterial vaccine vectors is their capability to invade and colonize internal tissues after mucosal delivery, which is critically important for inducing immune protection (32). Bacterial vaccine vectors have been used to produce protection against self as well as heterologous antigens (42–44). Our lab has developed a strategy of regulated-delayed attenuation in Salmonella, which makes the vaccine vectors safe and immunogenic for delivery of heterologous antigens to mice (45–47). We recently established the regulated-delayed attenuation phenotype in E. piscicida by using the arabinose-regulated promoter (5, 6). In this study, the promoters of both fur and crp genes were replaced with a tightly regulated araC ParaBAD cassette such that expression of these genes is dependent on arabinose which is provided during growth in vitro. Fur and Crp proteins cease to be synthesized due to the absence of arabinose and attenuation is progressively established in vivo to preclude induction of disease symptoms. Our study indicated that the E. piscicida strain carrying these two deletion insertion mutations is attenuated in zebrafish by i.c. injection.
To eliminate the need for an antibiotic-resistance selection marker for maintenance of plasmids in bacterial cells, our lab developed the balanced-lethal vector–host systems in Salmonella and E. ictaluri to ensure maintenance of the plasmid vectors in vivo and in vitro, without reliance on antibiotic-resistance markers (12, 14, 17). To develop a balanced-lethal system in E. piscicida, the asdA gene was characterized, whose product is essential for the synthesis of DAP, a crucial element of bacterial cell wall peptidoglycan. E. piscicida ΔasdA strain χ16000 can only survive when DAP is supplied in the growth media or contains a plasmid with the wild-type asdA gene. Since DAP is not present in in vivo environments, the ΔasdA strain will undergo lysis unless a plasmid with the asdA gene is present within the bacteria, establishing a powerful selection marker for maintenance of the plasmids within the RAEV, as observed previously in Salmonella and E. ictaluri (12, 14, 17). The ideal balanced-lethal system should have similar level of virulence as the wild-type strain. The amount of AsdA synthesis by the Asd+ vector in RASV affects the growth and immunogenicity of the strain (30). E. piscicida AsdA shares a high percentage of sequence identity and structural identity with, E. ictaluri (97.28%) and Salmonella (82.83%). As expected, plasmids containing the wild-type asdA gene from E. ictaluri and Salmonella were able to complement and establish the balanced-lethal system in E. piscicida ΔasdA strains. The pYA3493 AsdA+ vector system has been successfully used in live recombinant Salmonella and E. ictaluri vaccines (14, 30, 31, 48). Therefore, the pYA3493 AsdA+ vector system was used to deliver the antigens by RAEV. Vaccine efficacy depends on successful delivery of protein antigens for presentation in an optimal form and it has been observed that secreted proteins are immunogenic and interact with antigen-presenting cells because of their subcellular locations (49). β-Lactamase is a well-characterized periplasmic secreted protein in Gram-negative bacteria and translocation depends on β-Lactamase signal sequence (bla SS) composed of 23 amino acid (aa) N-terminal residues (50, 51). The pYA3493 plasmid contains the Ptrc promoter region, β-lactamase signal sequence (bla SS), and a multicloning sites. It was designed for secretion of recombinant antigens to the periplasm from the cytoplasm (48).
Infection by I. multifiliis has been shown to elicit both primary and secondary immune responses in catfish that promote parasite clearance (21–24). This adaptive immune response in fish suggests that vaccination would be a safe and effective method to prevent white spot disease. Surface immobilization antigens (IAGs) expressed by I. multifiliis are immuno-dominant and have become candidate proteins for vaccine development (23). Channel catfish and rainbow trout immunized with DNA vaccines encoding IAG52B produced specific antibodies, but showed no significant increase in the level of protection in challenge studies, suggesting that naked DNA alone is insufficient to induce a strong enough immune response to confer protection (25, 26). We expect that the Ich antigen delivered by RAEV should solve this problem. Sera from immunized fish immobilize I. multifiliis theronts in vitro demonstrating a role for i-antigens in protective immunity (52). Cytokines play an important role in immune surveillance against bacterial infection in fish (53–55). It was observed that, RAEV-Ich strain induced cytokine tnf-α, il-1β, il-6 and il-8 gene expression in different tissues of zebrafish. Activation of proinflammatory cytokines is involved in regulating immunoglobulin synthesis in teleosts (5, 7, 56). Zebrafish receiving booster doses of a live attenuated Vibrio anguillarum vaccine at 2 weeks after primary vaccination were better protected against vibriosis in comparison to fish that received a single vaccination (57). In the current study, we delivered booster immunizations at 2 weeks after primary vaccination to ensure long-term protective immunity. We did not detect signs of disease in vaccinated fish during the immunization periods. Our study demonstrates that RAEV-Ich induced a mucosal and serum anti-IAG52B IgM titer in zebrafish. As expected, RAEV-Ich vaccine strain also induced E. piscicida protective immunity in the immunized zebrafish. Immunized fish showed elevated levels of E. piscicida anti-LPS IgM in both mucus and serum. The immune protection mediated by RAEV-Ich vaccine strain χ16022(pG8R8029) and vector-control strain χ16022(pYA3493) against E. piscicida infection were evaluated. The survival rate was higher than 60% and 55% in zebrafish immunized with χ16022(pG8R8029) and χ16022(pYA3493) respectively. This confirms that RAEV confers immune protection against the virulent E. piscicida.
In summary, we have invented and developed an innovative antibiotic-sensitive Recombinant Attenuated Edwardsiella Vaccine (RAEV) vector system with in vivo display of regulated-delayed attenuation. The bla SS-IAG52B fusion protein was synthesized and secreted into the RAEV periplasm and delivered it to fish. RAEV-Ich induced different levels of immune responses and gave significant protection to the fish against E. piscicida infection. RAEV-Ich also induces anti- IAG52B IgM in zebrafish suggesting this vaccine strain may protect fish against white-spot diseases. This multidisciplinary approach using cutting-edge technologies will address the sustainability challenges of aquaculture increased food security.
Publicly available datasets were analyzed in this study. This data can be found here: https://www.ncbi.nlm.nih.gov/genbank/CP001135.1, AAK94941.1.
The animal study was reviewed and approved by University of Florida IACUC.
BS and RC contributed to conception funding acquisition and design of the study. BS wrote the first draft of the manuscript and contributed for the Methodology, Investigation, data analysis. CP wrote the sections of the manuscript, editing and methodology. All authors contributed to manuscript revision, read, and approved the submitted version.
This work was supported by the grant of U.S. Department of Agriculture (USDA) - National Institute of Food and Agriculture – USDA-NIFA Grant No. 2018-67015-28286.
RC is a co-founder and part owner of Curtiss Healthcare, Inc., which is involved in developing vaccines against infectious diseases of farm animals.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank RCF and H. W. Dickerson, Department of Infectious Diseases, The University of Georgia for I. multifiliis reagents.
1. Bujan N, Toranzo A, Magarinos B. Edwardsiella Piscicida: A Significant Bacterial Pathogen of Cultured Fish. Dis Aquat Organisms (2018) 131:59–71. doi: 10.3354/dao03281
2. Leung KY, Wang Q, Yang Z, Siame BA. Edwardsiella Piscicida: A Versatile Emerging Pathogen of Fish. Virulence (2019) 10(1):555–67. doi: 10.1080/21505594.2019.1621648
3. Park SB, Aoki T, Jung TS. Pathogenesis of and Strategies for Preventing Edwardsiella Tarda Infection in Fish. Vet Res (2012) 43(1):67. doi: 10.1186/1297-9716-43-67
4. Wang S, Kong Q, Curtiss R. New Technologies in Developing Recombinant Attenuated Salmonella Vaccine Vectors. Microb Pathog (2013) 58:17–28. doi: 10.1016/j.micpath.2012.10.006
5. Swain B, Powell CT, Curtiss R 3rd. Pathogenicity and Immunogenicity of Edwardsiella Piscicida Ferric Uptake Regulator (Fur) Mutations in Zebrafish. Fish Shellfish Immunol (2020) 107(Pt B):497–510. doi: 10.1016/j.fsi.2020.10.029
6. Zhou P, Han X, Ye X, Zheng F, Yan T, Xie Q, et al. Phenotype, Virulence and Immunogenicity of Edwardsiella Piscicida Cyclic AMP Receptor Protein (Crp) Mutants in Catfish Host. Microorganisms (2020) 8(4):517. doi: 10.3390/microorganisms8040517
7. Swain B, Powell CT, Curtiss R. Virulence, Immunogenicity and Live Vaccine Potential of aroA and phoP Mutants of Edwardsiella Piscicida in Zebrafish. Microb Pathog (2022) 162:105355. doi: 10.1016/j.micpath.2021.105355
8. Schleifer KH, Kandler O. Peptidoglycan Types of Bacterial Cell Walls and Their Taxonomic Implications. Bacteriol Rev (1972) 36(4):407–77. doi: 10.1128/br.36.4.407-477.1972
9. Pavelka MS Jr, Jacobs WR Jr. Biosynthesis of Diaminopimelate, the Precursor of Lysine and a Component of Peptidoglycan, Is an Essential Function of Mycobacterium Smegmatis. J Bacteriol (1996) 178(22):6496–507. doi: 10.1128/jb.178.22.6496-6507.1996
10. Viola RE. The Central Enzymes of the Aspartate Family of Amino Acid Biosynthesis. Accounts Chem Res (2001) 34(5):339–49. doi: 10.1021/ar000057q
11. Harb OS, Abu Kwaik Y. Identification of the Aspartate-Beta-Semialdehyde Dehydrogenase Gene of Legionella Pneumophila and Characterization of a Null Mutant. Infect Immun (1998) 66(5):1898–903. doi: 10.1128/IAI.66.5.1898-1903.1998
12. Galán JE, Nakayama K, Curtiss R 3rd. Cloning and Characterization of the Asd Gene of Salmonella Typhimurium: Use in Stable Maintenance of Recombinant Plasmids in Salmonella Vaccine Strains. Gene (1990) 94(1):29–35. doi: 10.1016/0378-1119(90)90464-3
13. Cardineau GA, Curtiss R 3rd. Nucleotide Sequence of the Asd Gene of Streptococcus Mutans. Identification of the Promoter Region and Evidence for Attenuator-Like Sequences Preceding the Structural Gene. J Biol Chem (1987) 262(7):3344–53. doi: 10.1016/S0021-9258(18)61509-1
14. Santander J, Xin W, Yang Z, Curtiss R. The Aspartate-Semialdehyde Dehydrogenase of Edwardsiella Ictaluri and Its Use as Balanced-Lethal System in Fish Vaccinology. PloS One (2010) 5(12):e15944. doi: 10.1371/journal.pone.0015944
15. Halver JE, Delong DC, Mertz ET. Nutrition of Salmonoid Fishes. V. Classification of Essential Amino Acids for Chinook Salmon. J Nutr (1957) 63(1):95–105. doi: 10.1093/jn/63.1.95
16. Mazid MA, Tanaka Y, Katayama T, Simpson KL, Chichester CO. Matabolism of Amino Acids in Aquatic Animals-III Indispensable Amino Acids for Tilapia Zillii. Nippom Suisan Gakkaishi (1978) 44(7):739–42. doi: 10.2331/suisan.44.739
17. Nakayama K, Kelly SM, Curtiss R. Construction of an ASD+ Expression-Cloning Vector: Stable Maintenance and High Level Expression of Cloned Genes in a Salmonella Vaccine Strain. Bio/Technology (1988) 6(6):693–7. doi: 10.1038/nbt0688-693
18. Curtiss R, Zhang X, Wanda S, HoYoung K, Konjufca V, Li Y, et al. Induction of Host Immune Responses Using Salmonella-Vectored Vaccines. In: Virulence Mechanisms of Bacterial Pathogens, Fourth Edition. American Society of Microbiology (2006). pp. 297–313.
19. Yan Y, Mu W, Zhang L, Guan L, Liu Q, Zhang Y. Asd-Based Balanced-Lethal System in Attenuated Edwardsiella Tarda to Express a Heterologous Antigen for a Multivalent Bacterial Vaccine. Fish Shellfish Immunol (2013) 34(5):1188–94. doi: 10.1016/j.fsi.2013.01.027
20. Clark-Curtiss JE, Curtiss R. Salmonella Vaccines: Conduits for Protective Antigens. J Immunol (2018) 200(1):39–48. doi: 10.4049/jimmunol.1600608
21. Zhao F, Li YW, Pan HJ, Shi CB, Luo XC, Li AX, et al. Expression Profiles of Toll-Like Receptors in Channel Catfish (Ictalurus Punctatus) After Infection With Ichthyophthirius Multifiliis. Fish Shellfish Immunol (2013) 35(3):993–7. doi: 10.1016/j.fsi.2013.05.023
22. Goven BA, Dawe DL, Gratzek JB. Protection of Channel Catfish, Ictalurus Punctatus Rafmesque, Against Ichthyophthirius Multifiliis Fouquet by Immunization. J Fish Biol (1980) 17:311–6. doi: 10.1111/j.1095-8649.1980.tb02764.x
23. Dickerson HW, Findly RC. Immunity to Ichthyophthirius Infections in Fish: A Synopsis. Dev Comp Immunol (2014) 43(2):290–9. doi: 10.1016/j.dci.2013.06.004
24. Maki JL, Dickerson HW. Systemic and Cutaneous Mucus Antibody Responses of Channel Catfish Immunized Against the Protozoan Parasite Ichthyophthirius Multifiliis. Clin Diagn Lab Immunol (2003) 10(5):876–81. doi: 10.1128/CDLI.10.5.876-881.2003
25. von Gersdorff Jørgensen L, Sigh J, Kania PW, Holten-Andersen L, Buchmann K, Clark T, et al. Approaches Towards DNA Vaccination Against a Skin Ciliate Parasite in Fish. PloS One (2012) 7(11):e48129. doi: 10.1371/journal.pone.0048129
26. Lin Y, Cheng G, Wang X, Clark TG. The Use of Synthetic Genes for the Expression of Ciliate Proteins in Heterologous Systems. Gene (2002) 288(1):85–94. doi: 10.1016/S0378-1119(02)00433-X
27. Roland K, Curtiss R 3rd, Sizemore D. Construction and Evaluation of a Delta Cya Delta Crp Salmonella Typhimurium Strain Expressing Avian Pathogenic Escherichia Coli O78 LPS as a Vaccine to Prevent Airsacculitis in Chickens. Avian Dis (1999) 43(3):429–41. doi: 10.2307/1592640
28. Wang Q, Yang M, Xiao J, Wu H, Wang X, Lv Y, et al. Genome Sequence of the Versatile Fish Pathogen Edwardsiella Tarda Provides Insights Into Its Adaptation to Broad Host Ranges and Intracellular Niches. PloS One (2009) 4(10):e7646. doi: 10.1371/journal.pone.0007646
29. Edwards RA, Keller LH, Schifferli DM. Improved Allelic Exchange Vectors and Their Use to Analyze 987P Fimbria Gene Expression. Gene (1998) 207(2):149–57. doi: 10.1016/S0378-1119(97)00619-7
30. Kang HY, Srinivasan J, Curtiss R 3rd. Immune Responses to Recombinant Pneumococcal PspA Antigen Delivered by Live Attenuated Salmonella Enterica Serovar Typhimurium Vaccine. Infect Immun (2002) 70(4):1739–49. doi: 10.1128/IAI.70.4.1739-1749.2002
31. Branger CG, Fetherston JD, Perry RD, Curtiss R 3rd. Oral Vaccination With Different Antigens From Yersinia Pestis KIM Delivered by Live Attenuated Salmonella Typhimurium Elicits a Protective Immune Response Against Plague. Adv Exp Med Biol (2007) 603:387–99. doi: 10.1007/978-0-387-72124-8_36
32. Torres-Escobar A, Juárez-Rodríguez MD, Branger CG, Curtiss R 3rd. Evaluation of the Humoral Immune Response in Mice Orally Vaccinated With Live Recombinant Attenuated Salmonella Enterica Delivering a Secreted Form of Yersinia Pestis PsaA. Vaccine (2010) 28(36):5810–6. doi: 10.1016/j.vaccine.2010.06.070
33. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol (2013) 30(12):2725–9. doi: 10.1093/molbev/mst197
34. Reed LJ, Muench H. A Simple Method of Estimating Fifty Per Cent Endpoints12. Am J Epidemiol (1938) 27(3):493–7. doi: 10.1093/oxfordjournals.aje.a118408
35. Wang X, Dickerson HW. Surface Immobilization Antigen of the Parasitic Ciliate Ichthyophthirius Multifiliis Elicits Protective Immunity in Channel Catfish (Ictalurus Punctatus). Clin Vaccine Immunol (2002) 9(1):176–81. doi: 10.1128/CDLI.9.1.176-181.2002
36. Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2–ΔΔct Method. Methods (2001) 25(4):402–8. doi: 10.1006/meth.2001.1262
37. Santander J, Golden G, Wanda S-Y, Curtiss R 3rd. Fur-Regulated Iron Uptake System of Edwardsiella Ictaluri and Its Influence on Pathogenesis and Immunogenicity in the Catfish Host. Infect Immun (2012) 80(8):2689–703. doi: 10.1128/IAI.00013-12
38. Santander J, Mitra A, Curtiss R 3rd. Phenotype, Virulence and Immunogenicity of Edwardsiella Ictaluri Cyclic Adenosine 3',5'-Monophosphate Receptor Protein (Crp) Mutants in Catfish Host. Fish Shellfish Immunol (2011) 31(6):1142–53. doi: 10.1016/j.fsi.2011.10.009
39. Helftenbein E. Nucleotide Sequence of a Macronuclear DNA Molecule Coding for Alpha-Tubulin From the Ciliate Stylonychia Lemnae. Special Codon Usage: TAA Is Not a Translation Termination Codon. Nucleic Acids Res (1985) 13(2):415–33. doi: 10.1093/nar/13.2.415
40. Horowitz S, Gorovsky MA. An Unusual Genetic Code in Nuclear Genes of Tetrahymena. Proc Natl Acad Sci (1985) 82(8):2452–5. doi: 10.1073/pnas.82.8.2452
41. Preer JR, Preer LB, Rudman BM, Barnett AJ. Deviation From the Universal Code Shown by the Gene for Surface Protein 51a in Paramecium. Nature (1985) 314(6007):188–90. doi: 10.1038/314188a0
42. Juárez-Rodríguez MD, Arteaga-Cortés LT, Kader R, Curtiss R 3rd, Clark-Curtiss JE. Live Attenuated Salmonella Vaccines Against Mycobacterium Tuberculosis With Antigen Delivery via the Type III Secretion System. Infect Immun (2012) 80(2):798–814. doi: 10.1128/IAI.05525-11
43. Su H, Liu Q, Bian X, Wang S, Curtiss R, Kong Q. Synthesis and Delivery of Streptococcus Pneumoniae Capsular Polysaccharides by Recombinant Attenuated Salmonella Vaccines. Proc Natl Acad Sci (2021) 118(2):e2013350118. doi: 10.1073/pnas.2013350118
44. Kong W, Wang X, Fields E, Okon B, Jenkins MC, Wilkins G, et al. Mucosal Delivery of a Self-Destructing Salmonella-Based Vaccine Inducing Immunity Against Eimeria. Avian Dis (2020) 64(3):254–68. doi: 10.1637/aviandiseases-D-19-00159
45. Shi H, Wang S, Roland KL, Gunn BM, Curtiss R 3rd. Immunogenicity of a Live Recombinant Salmonella Enterica Serovar Typhimurium Vaccine Expressing pspA in Neonates and Infant Mice Born From Naive and Immunized Mothers. Clin Vaccine Immunol CVI (2010) 17(3):363–71. doi: 10.1128/CVI.00413-09
46. Li Y, Wang S, Scarpellini G, Gunn B, Xin W, Wanda S-Y, et al. Evaluation of New Generation Salmonella Enterica Serovar Typhimurium Vaccines With Regulated Delayed Attenuation to Induce Immune Responses Against PspA. Proc Natl Acad Sci USA (2009) 106(2):593–8. doi: 10.1073/pnas.0811697106
47. Shi H, Santander J, Brenneman KE, Wanda SY, Wang S, Senechal P, et al. Live Recombinant Salmonella Typhi Vaccines Constructed to Investigate the Role of rpoS in Eliciting Immunity to a Heterologous Antigen. PloS One (2010) 5(6):e11142. doi: 10.1371/journal.pone.0011142
48. Xin W, Wanda SY, Li Y, Wang S, Mo H, Curtiss R 3rd. Analysis of Type II Secretion of Recombinant Pneumococcal PspA and PspC in a Salmonella Enterica Serovar Typhimurium Vaccine With Regulated Delayed Antigen Synthesis. Infect Immun (2008) 76(7):3241–54. doi: 10.1128/IAI.01623-07
49. Spreng S, Dietrich G, Weidinger G. Rational Design of Salmonella-Based Vaccination Strategies. Methods (San Diego Calif) (2006) 38(2):133–43. doi: 10.1016/j.ymeth.2005.09.012
50. Kochi SK, Killeen KP, Ryan US. Advances in the Development of Bacterial Vector Technology. Expert Rev Vaccines (2003) 2(1):31–43. doi: 10.1586/14760584.2.1.31
51. Kadonaga JT, Plückthun A, Knowles JR. Signal Sequence Mutants of Beta-Lactamase. J Biol Chem (1985) 260(30):16192–9. doi: 10.1016/S0021-9258(17)36220-8
52. Hines RS, Spira DT. Ichthyophthiriasis in the Mirror Carp Cyprinus Carpio (L.) V. Acquired Immunity. J Fish Biol (1974) 6(4):373–8. doi: 10.1111/j.1095-8649.1974.tb04554.x
53. Swain B, Basu M, Samanta M. Molecular Cloning and Characterization of Nucleotide Binding and Oligomerization Domain-1 (NOD1) Receptor in the Indian Major Carp, Rohu (Labeo Rohita), and Analysis of Its Inductive Expression and Down-Stream Signalling Molecules Following Ligands Exposure and Gram-Negative Bacterial Infections. Fish Shellfish Immunol (2012) 32(5):899–908. doi: 10.1016/j.fsi.2012.02.018
54. Swain B, Basu M, Sahoo BR, Maiti NK, Routray P, Eknath AE, et al. Molecular Characterization of Nucleotide Binding and Oligomerization Domain (NOD)-2, Analysis of Its Inductive Expression and Down-Stream Signaling Following Ligands Exposure and Bacterial Infection in Rohu (Labeo Rohita). Dev Comp Immunol (2012) 36(1):93–103. doi: 10.1016/j.dci.2011.06.018
55. Swain B, Basu M, Samanta M. NOD1 and NOD2 Receptors in Mrigal (Cirrhinus Mrigala): Inductive Expression and Downstream Signalling in Ligand Stimulation and Bacterial Infections. J Biosci (2013) 38(3):533–48. doi: 10.1007/s12038-013-9330-y
56. Zou J, Secombes CJ. The Function of Fish Cytokines. Biol (Basel) (2016) 5(2):23. doi: 10.3390/biology5020023
Keywords: fish vaccine, immunity, pathogenicity, E. piscicida, Ichthyophthirius multifiliis (Ich), innate & adaptive immune response
Citation: Swain B, Powell CT and Curtiss R III (2022) Construction and Evaluation of Recombinant Attenuated Edwardsiella piscicida Vaccine (RAEV) Vector System Encoding Ichthyophthirius multifiliis (Ich) Antigen IAG52B. Front. Immunol. 12:802760. doi: 10.3389/fimmu.2021.802760
Received: 27 October 2021; Accepted: 21 December 2021;
Published: 25 January 2022.
Edited by:
Maryam Dadar, Razi Vaccine and Serum Research Institute, IranReviewed by:
Ozan Ozdemir, Mississippi State University, United StatesCopyright © 2022 Swain, Powell and Curtiss. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Banikalyan Swain, c3dhaW5iYW5pa2FseWFuQHlhaG9vLmNvbQ==; c3dhaW5iQHVmbC5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.