- 1Department of Rheumatology and Immunology, West China Hospital, Sichuan University, Chengdu, China
- 2Division of Arthritis and Rheumatic Diseases, Oregon Health & Science University, Portland, OR, United States
- 3Section of Rheumatology, Veterans Affairs (VA) Portland Healthcare System, Portland, OR, United States
Besides its contribution to the development of rheumatic diseases, the gut microbiota interact with anti-rheumatic drugs. The intestinal microbiota can directly metabolize many drugs and indirectly change drug metabolism through a complex multi-dimensional interaction with the host, thus affecting individual response to drug therapy and adverse effects. The focus of the current review is to address recent advances and important progress in our understanding of how the gut microbiota interact with anti-rheumatic drugs and provide perspectives on promoting precision treatment, drug discovery, and better therapy for rheumatic diseases.
Introduction
Although much of the mechanism is yet to be learnt, current evidence indicates that microbes might be vital environmental factors in initiating and propagating the onset of autoimmune rheumatic diseases. For a long time, specific infectious microorganisms have been suspected to trigger rheumatic diseases in genetically susceptible individuals—for example, Mycobacterium tuberculosis was once postulated to cause rheumatoid arthritis (RA), leading to the gold salt remedy to treat the communicable disease (1). This concept was abandoned because of the lack of evidence to prove this was the case. Other bacteria, such as Porphyromonas gingivalis (P. gingivalis) and Proteus mirabilis (P. mirabilis), were also considered to be candidate pathogens to cause RA (2–8) as well as Klebsiella pneumoniae, Salmonella, and Yersinia as pathogens for spondyloarthritis (9, 10). However, none of these was proven to directly cause these diseases.
Findings of recent studies rather suggest that dysbacteriosis in microbiome contributes to a range of chronic conditions, such as inflammatory bowel disease (IBD) (11), diabetes (12), multiple sclerosis (13), autism (14), various cancers (15–17), and rheumatic diseases (18).
Dysbacteriosis is the alteration of bacterial composition from healthy status to disease, and this has been well documented in several studies in patients with RA. Vaahtovuo et al. (19) found that, compared with fibromyalgia, patients with early RA had significantly fewer bifidobacteria and bacteria of the Bacteroides-Porphyromonas-Prevotella group, Bacteroides fragilis subgroup, and Eubacterium rectale–Clostridium coccoides group. Scher et al. (20) reported an expansion of Prevotellaceae but a reduction of Bacteroidaceae in new-onset-RA patients. The disbacteriosis in RA patients was further confirmed and expanded in oral microbiota (21). Interestingly, medically treated arthritic animals in preclinical models and RA patients in clinical remission can restore their gut flora composition (21, 22). These findings highlight the importance of gut microbiota ecological balance to the wellbeing of the host and imply that certain bacteria are beneficial to the host by countering the potential harmful bacteria. On the other hand, these raise the points as to how medications affect the community of microbiota and vice versa. The term pharmacomicrobiotics has been introduced for studies investigating the effect of microbiome variations on drug disposition, action, and toxicity (23). This review will focus on our current understanding of the interaction between disease-modifying anti-rheumatic drugs (DMARD) and the gut microbiota.
How Does Microbiota Influence Rheumatic Diseases?
It is assumed that bacteria first colonize the body of most infants soon after birth. However, several studies reported that bacterial DNA were found in the placenta (24, 25), meconium (26), and amniotic fluid (27). This has put forth an idea that mothers are transferring bacteria to the fetus in the womb before birth to establish a fetal–maternal microbiome relationship. Interestingly, activated memory CD4+ T cells developed in fetal circulation (28). The important question raised by these findings is that the placenta is not a totally sterile environment as we thought; the fetus may harbor microbes, which possibly shape our immune system during the earliest days of life even before birth.
The enormous and diverse community of gut microbiota constitutes a distinct network that is vital to make the immune system work functionally, but how the dynamics of microbiome shape autoimmune diseases is unclear. Firstly, the alien microorganisms become the fruitful sources of antigenic variation to interact with immune cells to maintain homeostasis (29). Approximately 70–80% of the immune cells of the body populate in our gastrointestinal tract. As a result of the coevolution, microorganisms and intestinal immune cells form a bidirectional relationship. Multiple autoimmune and inflammatory diseases, such as RA, are classically considered T cell-mediated disorders (30). The microbiota and its metabolite-associated signals are responsible for the activation, polarization, and function of CD4+ T cells, including T-bet+ T helper type 1 (Th1), GATA3+ Th2, retinoic receptor-associated orphan receptor (ROR)-γt+ Th17, and FOXP3+ T regulatory (Treg) cells (31). A landmark study highlighted that segmented filamentous bacteria (SFB) alone is sufficient to induce the differentiation of lamina propria Th17 cells in mice (32). Notably, mono-colonization with SFB in germ-free mice rapidly induces the onset of autoimmune arthritis and reinstated the lamina propria Th17 cell compartment (33). However, SFB is not found to be colonized in humans. Additionally, a similar sequence between specific microbial peptides and host autoantigens, resulting in the production of cross-reactive T cells targeting both parts, has long been recognized as the molecular mimicry that may be another potential mechanism for the involvement of the microbiome in rheumatic disease (34–36).
Aside from T cells, microbial exposures can activate B cells and induce immunoglobulin (37–40). Antinuclear antibodies (ANA) are a hallmark feature of systemic lupus erythematosus (SLE) (41). Lymphotoxin-deficient mice show the development of ANA by 3 months of age, including anti-U1-ribonucleoprotein, anti-Sm, anti-Scl70/topoisomerase-I, anti-centromere protein B, anti-SSA/Ro52, and anti-Jo1 antibodies. Treating lymphotoxin-deficient mice with antibiotics or in a germ-free condition (42) reduced the prevalence of ANA compared to their littermates (43). Antibodies directed against dsDNA have been associated with SLE disease severity. A recent study reported that serum anti-Ruminococcus gnavus strain-restricted antibodies correlated directly with SLE disease activity index (SLEDAI) score and anti-native DNA levels but inversely correlated with C3 and C4 in lupus patients (44). Fecal transplantation from lupus mice to germ-free recipients resulted in enhanced intestinal immune response and upregulated expression of antibody titers against dsDNA (45). Oral gavage of Roseburia intestinalis into a mouse model of spontaneous antiphospholipid syndrome in NZW × BXSB F1 triggered the development of anti-human β2 glycoprotein protein I antibodies and thrombotic events (46).
Taking these data together, the above-mentioned studies indicate that commensal microbiota play a role in impacting on the physiological state of the immune cell subsets and are prone to increase the susceptibility to autoimmune responses and reprogramming of immune cells. It must be emphasized that the autoimmunity-triggering effect of these bacteria is the result of a defective counter-mechanism from other commensal microbiota. Prebiotics and probiotics are intended to impact dysbacteriosis and restore the balance between harmful and beneficial bacteria.
Dialogue Between the Gut Microbiota and the Rheumatic Therapeutics
Since the long-term use of anti-rheumatic drugs and the individual response of patients may vary greatly, the ultimate aim of rheumatologists is to maximize clinical outcomes and minimize their side effects. Most anti-rheumatic drugs are orally administrated and under the process of commensal microorganisms that alter the bioavailability of the drug in the intestine directly or indirectly. With the advent of pharmacomicrobiomics, it has drawn a growing interest to profile interactions between drugs and gut bacteria (47–49).
The metabolism of drugs by trillions of gut microbiota is multidimensional—for example, the prodrug sulfasalazine (SSZ) can hardly be absorbed in the upper intestine, and the majority of this agent is metabolized by the bacterial enzyme azoreductase into its active components sulfapyridine and 5‐aminosalicylic acid (ASA) functionally targeting colon sites (50). The intestinal microbiota harbor diverse β-glucuronidase enzymes that manipulate the pharmacokinetics of oral drugs. Bacteria such as Clostridium, Peptostreptococcus, and Staphylococcus are able to secrete β-glucuronidases (51), which release glucuronic acid (GlcA) sugars from complex carbohydrates. Some chemicals, like nonsteroidal anti-inflammatory drugs (NSAIDs), are conjugated to GlcA (52, 53). Targeting luminal bacterial β-D-glucuronidase can reduce NSAID-related intestinal mucosal injury through halting the hydrolysis of NSAID glucuronides (54).
Aside from the mechanism of microbial enzymes to affect their biotransformation, a recent study emphasized that drug-metabolizing microbial proteins can contribute to the in vivo drug metabolism of gnotobiotic mice and provide evidence that metagenomics and genomics sequence data can explain the capacity of both isolated gut bacteria and complete communities to convert specific drugs (55). It was found that, at 7 h after oral gavage of dexamethasone to germ-free mice or gnotobiotic mice mono-colonized with Clostridium scindens (C. scindens), dexamethasone was significantly reduced and androgen metabolite increased in gnotobiotic mice mono-colonized with C. scindens in the caecum. This phenomenon was also found in prednisone, prednisolone, cortisone, and cortisol and demonstrated that C. scindens metabolizes endogenous steroid hormones.
A recent study screened 1,197 drugs against 40 representative gut bacterial strains and found that 24% of the drugs affected the in vitro growth of bacteria (56). Therefore, the mechanistic understanding of gut microbiota and drugs is still complex; however, manipulating the microbiota in order to promote a better response needs to be further investigated.
Interaction Between Microbes and Cortisone
Since compound E was introduced by Philip Hench to treat RA successfully (57), glucocorticoids become the strong, broad-spectrum anti-inflammatory and immunosuppressive approach in the art of healing in a range of inflammatory rheumatic diseases (58–60). However, the exact mechanisms of how this first-line therapy impacts on anti-inflammatory pathways are still obscure, and the long-term safety of glucocorticoids is still challenging in rheumatic diseases (59, 61). Recently, a number of corpus have highlighted dysbiotic gut microbiota in SLE (44, 62–64); a reduction of species richness diversity was noted in patients with lupus, with reductions in taxonomic complexity most pronouncedly related to SLEDAI (44). Glucocorticoids are a mainstay therapy to manage flares and remission in SLE (65). The study by Mukherji and co-workers showed that oral gavage with prednisone appeared to have the most significant proportion of Bacteroidetes and Firmicutes than the control group in MRL/lpr mice. In total, thirty-three bacterial taxa were significantly changed in the prednisone treatment group, and Rikenella, Mucispirillum, Oscillospira, and Bilophila were of relatively lower abundance at the genera level; Prevotella and Anaerostipes were enriched as well (66). Additionally, this study also identified that glucocorticoids downregulated Mucispirillum, which positively correlated with SLEDAI, and it was previously reported to degrade colonic mucin in the intestines (67). Oscillospira, Rikenella, and Bilophila were positively associated with anti-dsDNA.
Meanwhile, another study screened the gut microbiota in glucocorticoid therapy among patients with SLE. Generally, the observed diversity of bacterial communities was similar between healthy controls and SLE patients with glucocorticoid therapy but statistically different between healthy controls and SLE patients without glucocorticoid therapy. SLE patients treated with glucocorticoids restored the ratio of Firmicutes to Bacteroidetes and increased a group of core bacteria genera, including Lactococcus, Streptococcus, and Bifidobacterium, which were reduced in the SLE without glucocorticoids. SLE treated with glucocorticoids reduced activity-related glycan metabolism via increasing the abundance of Bacteroides in lupus (68).
Taken together, these findings suggest that glucocorticoid therapy has the potential ability to modulate the gut microbiota composition of lupus through some bacteria-based corticosterone synthesis pattern which is still far from clear. Meanwhile, they open up many novel questions and further emphasize the need for novel, more effective treatments for SLE that minimize or eliminate the need for glucocorticoids. Low-dose glucocorticoids are commonly used in combination with other DMARDs to treat RA. How the gut microbiota are affected by glucocorticoids in RA is a subject that is of interest to investigate. Numerous studies have highlighted gut dysbiosis during the different phases of RA, although the variability of results could be subjected to the analysis technology, geographic factors, and clinical progression.
Perturbed microbiome can be normalized after a combination of DMARDs, which may include corticosteroids in RA patients (21). However, it is difficult to dissect how corticosteroids contribute to the collective effects of DMARDs in combination.
Interaction Between Microbes and Methotrexate
Although originally designed as an anti-cancer therapy, methotrexate (MTX) is now the cornerstone drug for the treatment of various rheumatic diseases and the first-line anchor drug for the treatment of RA over decades (69–71). The possible pharmacological and anti-inflammatory mechanism of this drug is to antagonize folate-dependent processes to suppress the synthesis of purines and pyrimidines, inhibit nuclear factor-κB, Janus kinase signal transducer, and STAT signaling pathway, and promote adenosine signaling (72).
Gastrointestinal side effect is commonly induced by MTX therapy due to the intestinal barrier damage (73–75). Dietary restriction dramatically increased the survival rate of mice exposed to lethal doses of MTX administration. Dietary restriction may suppress intestinal inflammation by upregulating protective intestinal bacteria (Lactobacillus genus). However, ablating the gut microbiota through applying broad-spectrum antibiotics eliminates the beneficial effect achieved by dietary restriction. Moreover, administration of probiotic with Lactobacillus rhamnosus GG partially mimicked the rescue effect of a dietary restriction (76). Another study indicated that the number of Bacteroides fragilis in feces was dramatically decreased in low-dose-MTX-treated mice, while gavage with B. fragilis could profoundly ameliorate the MTX-induced inflammatory process (77). In a pharmacokinetic study, a low dose (10 mg/kg) of MTX altered the microbial profile that induced a higher abundance of Firmicutes over Bacteroidetes and the reverse at high dose (100 mg/kg) in Sprague–Dawley rats. The relative abundance of Firmicutes was positively correlated with 2,4-diamino-N-10-methylpteroic acid, which is the MTX degradation produced via the excretion of the intestinal bacterial enzyme carboxypeptidase glutamate 2 after MTX treatment at 48 h (78). The microbiota composition also changed after monotherapy with MTX, with a lower abundance of Enterobacteriales compared with non-treated patients with RA (79). Zhang et al. reported patients with restored RA-related gut and oral microbiome abundance of microbial linkage groups (MLGs) similar to the normal situation after MTX treatment. Enriched gut and oral MLGs also negatively correlated with clinical parameters such as C-reactive protein, anti-citrullinated protein antibodies, and rheumatoid factor (21).
It is well known that MTX response varies among patients with RA, that is, around half of patients fail to achieve an adequately clinical response after MTX therapy (69). A recent study analyzed the gut microbiomes of drug‐naïve, new‐onset-RA patients and observed that the overall bacterial diversity is distinct between MTX responders and non-responders. These non-responders had significantly enriched communities than the responders. A further study revealed a significant increase of MAP-kinase signaling, DNA replication, fatty acid degradation, and ABC transporters in non-responders, as well as a significant decrease of lipopolysaccharide and folate biosynthesis. These data suggest that the human gut microbiota was able to metabolize oral MTX (80). Furthermore, the baseline abundance of gut microbiome features is of great value in predicting treatment outcomes in response to MTX. Notably, a microbiome‐based model by machine learning techniques could suggest a possible future clinical response of the gut microbiome on MTX metabolism (80).
For MTX working as the folate competitive antagonist, folate has been given as an additional medication to reduce the adverse events of MTX, like intestinal toxicity and liver function abnormalities (81). Huang et al. reported that leucovorin supplementation not only ameliorated MTX-induced intestinal damage but also remodeled the MTX-induced composition of the bacterial community alternation and increased the abundance of Bifidobacterium. Oral gavage of Bifidobacterium longum exerts a trophic effect on the intestinal mucosa to ameliorated MTX-induced intestinal damage (82).
The impact of MTX on human gut microbiota has been directly tested using a humanized mouse model (83). Germ-free mice were colonized by stool samples from a healthy human donor. MTX significantly altered the gut microbiota as soon as day 1 of MTX administration, and it lasted for 4 days. A high dose of MTX (50-mg/kg dose for cancer treatment) significantly decreased the Bacteroidetes phylum, while low-dose MTX (1 mg/kg—dose for arthritis treatment) showed the same trend but with a moderate effect. Interestingly, the route of administration of MTX (oral vs. intraperitoneal injection) and rescue with folic acid did not significantly affect the overall effect of MTX. The perturbed growth of Bacteriodetes by MTX is confirmed in culture. These findings are reflected in RA patients. Thus, new-onset-RA patients who were responsive to MTX showed a significant decrease in Bacteriodetes relative to those who were not responsive to MTX (80).
Interestingly, the microbiota from MTX-treated and MTX-responsive RA patients was able to transfer immunosuppressive effects in gnotobiotic mice. The recipient mice showed a decrease of multiple immune cells, including activated T cells, Th1 cells, B cells, and myeloid cells in the spleen (83). Furthermore, a reduction of activated T cells, Th17 cells, and myeloid cells was also observed in the intestinal mucosa (83). The immunosuppressive effects by MTX-exposed microbiota may be attributed to the different abundance of one phylum (Proteobacteria), 26 genera, and 41 amplicon sequence variants (83). These results suggest that the effects of MTX on microbiota can contribute to the immunosuppressive therapeutic effect of MTX in the hosts. Further investigations are required to delineate how the altered immune cell populations, especially those in the intestinal mucosa, will affect the community of gut flora. The interaction of MTX with the gut microbiota and the effects on host immune activation are illustrated in Figure 1.
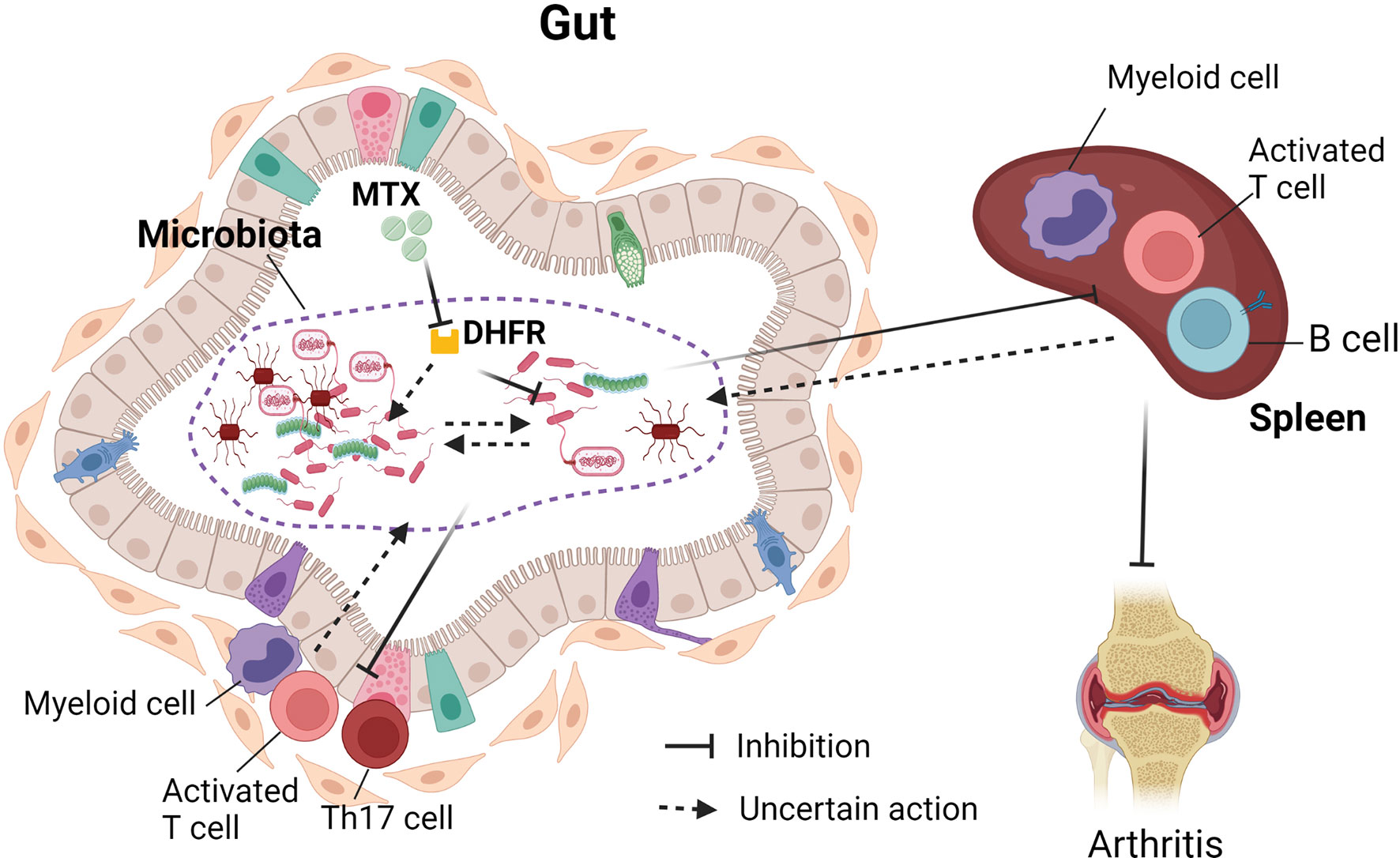
Figure 1 Interaction between gut microbiota and methotrexate (MTX). MTX inhibits bacterial dehydrofolate reductase and affects the growth of bacteria, bacterial transcriptome, and metabolome. There are MTX-sensitive and MTX-resistant bacteria in the human gut. Therefore, MTX treatment affects the community of gut microbiota. The change of gut microbiota post-MTX treatment can suppress immune cells in the periphery and in the intestinal mucosa. It is not clear whether MTX will also affect interactions between gut bacteria and whether the change of immune cellular components will, in turn, affect the gut microbiota.
Interaction Between Microbes and Sulfasalazine
As Streptococcus found in milk was thought as the bacteriological evidence to provoke RA (84), SSZ has been initiated to treat this “infective polyarthritis” since the 1940s (85). Designed as a compound, the most amount of SSZ reaches the colon and is broken into 5‐ASA and sulfapyridine by gut organisms. Although the pharmacological mechanism of action is obscure, SSZ has anti-inflammatory and antibacterial properties to exert beneficial effects on RA, ankylosing spondylitis (AS), and IBD (86, 87).
The administration of antibiotics on germ-free rats showed unchanged SSZ in their caecum and feces. When germ-free rats are infected with four specific bacteria normally found in the intestinal tract of rodents, the rats regain the ability to metabolize SSZ as the conventional rats. These findings suggest that intestinal bacterial metabolism is essential for activating SSZ transformation (88).
Probiotics are “friendly” live microorganisms (bacteria or yeasts) taken as food supplement that promotes favorable benefits for the host by improving the intestinal microbial balance. Co-administration of probiotic strains such as Lactobacillus acidophilus, Bifidobacterium lactis, and Streptococcus salivarius with SSZ modulates azoreductase activity and SSZ metabolism in the colon (89). However, some clinical studies did not claim that the short-term co-administration of probiotics could exert beneficial effects on patients with RA (90) or IBD by changing the metabolism of SSZ. These studies suggest that certain bacterial species possess diazoreductase activity by producing more extensive metabolism of SSZ.
An earlier study showed a significant reduction of the total counts of aerobic bacteria Escherichia coli and Bacteroides after SSZ therapy and a high frequency of Bacillus, but there had been very limited effects on the upper jejunal gastrointestinal flora in patients with RA (91). Treatment with SSZ could alleviate the inflammation and regulate bacterial composition with increasing short-chain fatty acid-producing bacteria (Lachnospiraceae—Blautia), lactic acid-producing bacteria (Lactococcus), Mycoplasma, and decreasing proportions of Enterococcus and Proteobacteria in 2,4,6-trinitrobenzene sulfonic acid-induced colitis in rats (92).
These results indicate that, in addition to the conversion of SSZ to active drug bacteria in the gut, changes in microbiota composition by SSZ will also contribute to the anti-inflammatory effects of SSZ.
Interaction Between Microbes and Hydroxychloroquine
Hydroxychloroquine (HCQ) was initially used to prevent and treat malaria and then employed, for its anti-inflammatory properties, to successfully treat various rheumatic diseases, such as SLE, RA, and other inflammatory rheumatic diseases (65, 93, 94).
Previous studies investigated HCQ influence on gut microbiota in Q-fever endocarditis patients. These patients, treated with doxycycline and HCQ, presented significantly lower amounts of Bacteroidetes and Lactobacillus compared with the controls (95). Systemic rheumatologic conditions are prone to develop more cardiovascular events compared to the general population (96). HCQ administration was reported as a potentially beneficial therapy for K/BxN mice with high-fat diet (HFD) in a mouse model of RA that develops atherosclerosis. HCQ could alleviate the HFD-induced dyslipidemia and atherosclerosis as well as profoundly restored abnormal gut microbiota with a higher abundance of Akkermansia and Parabacteroides and a lower abundance of Clostridium sensu stricto cluster (97). The total glucosides of paeony (TGP) is a traditional Chinese herb medication which has been approved for a variety of rheumatic disease for its anti-inflammatory and immunomodulatory functions (98, 99). The TGP + HCQ group had increased richness of microbiota and had significant changes of Bacteroidetes and Firmicutes in NOD mice with Sjögren’s syndrome. The proportion of Lactobacillus and Incertae of phylum Firmicutes and Desulfovibrio of phylum Proteobacteria was significantly increased, and the abundance of Bacteroides and Alloprevotella of phylum Bacteroidetes and Pseudoflavonifractor of phylum Firmicutes was significantly decreased in the TGP + HCQ group compared with the control group. The abundance of Akkermansia of phylum Verrucomicrobia was significantly decreased in the TGP and TGP + HCQ groups compared with the HCQ group. However, most of these studies do not test the interaction of HCQ alone with gut microbiota alone. Recently, a paper observed that oral gavage of high dose (100 mg/kg) HCQ for 2 weeks significantly increased the relative abundance of phylum Bacteroidetes, whereas it decreased that of Firmicutes without changing the intestinal integrity and the immunological responses in mice (100).
Investigations into the direct effects of HCQ on gut microbiota will be required to delineate whether HCQ directly impacts bacteria growth or indirectly via the immune system of the host.
Biological Disease-Modifying Anti-rheumatic Drugs
Bioengineered fusion proteins and therapeutic monoclonal antibodies used to treat rheumatic diseases are collectively called disease-modifying anti-rheumatic drugs (bDMARDs). These include agents that inhibit tumor necrosis factor (TNF), interleukin (IL)-1, 6, 17, and 23, T cell co-stimulation, B cell growth factors, and B cell-depleting monoclonal antibody. The targets of these bDMARDs are clearly defined.
TNF inhibitors (TNFi) are the most effective treatments for RA, spondyloarthritis, and IBD after the failure of traditional therapy (101). Ample evidence indicates that TNFi therapy induces mucosal healing and restores gut microbiota dysbiosis in clinical and experimental models (102, 103). However, the interaction between gut microbiota in patients with rheumatic disorders and TNFi is not thoroughly investigated. Etanercept (ETN) therapy showed major intestinal composition changes compared with treatment-naïve RA patients who possessed more abundant Lactobacillus as reported before (21, 104). Patients under treatment with ETN present enriched Cyanobacteria, while Deltaproteobacteria and Clostridiaceae were decreased than in treatment-naïve patients (79). Cyanobacteria produce a source of novel bioactive secondary metabolites that may help to modulate the immune system and result in attenuating RA (105, 106).
The gut microbiota is always considered as a vital environmental factor in triggering AS (107). ETN therapy markedly reduced the incidence, arthritis progression, and inflammatory cytokines, such as TNF and IL-17A, in the serum, recovered intestinal barrier function as well as restored the gut microbiota composition similar to that in naïve mice in a proteoglycan‐induced AS model (108). A recent study observed that TNFi treatment had better improvement in AS nonsmokers than in AS smokers. The relative abundance of the microbiota is more prone to be increased in AS nonsmokers after treatment with TNFi for 6 months. In addition, some bacteria, including Actinomyces, Agathobacter, Bilophila, Klebsiella, Lachnospiraceae_NK4A136, Ruminococcaceae-UCG- 002, and Ruminococcaceae_UCG-005, were sensitive to TNFi treatment in AS nonsmokers, while Bacteroides, Faecalibacterium, Lachnoclostridium, Parabacteroides, Blautia, Butyricicoccus, and Escherichia-Shigella were not. This suggests that these bacteria were tolerant to TNFi treatment (109). Since TNFi do not work for all patients, one challenge to clinicians is to investigate the biomarker that can predict the clinical response to TNFi. Another recent study in patients with spondyloarthritis treated by TNFi (most of which are ETN) revealed no significant modification of a particular taxa after 3 months of treatment. It should be noted that the responder patients showed only few mild differences in microbiota composition at order level than in non-responder patients. Interestingly, a higher proportion of the Burkholderiales order before TNFi treatment was strongly correlated with the responding patients after 3 months of treatment, suggesting that certain intestinal bacteria can possibly predict the clinical response as a biomarker for TNFi efficacy in patients with spondyloarthritis (110). In Crohn’s disease, infliximab non-responders had a higher abundance of baseline Blautia, Faecalibacterium, Roseburia, and Negativibacillus genera, while a higher abundance of baseline Hungatella, Ruminococcus gnavus, and Parvimonas was found in infliximab responders (111). Clearly, more studies including a large number of patients are required to replicate the findings in these studies before profiling of microbiota as a biomarker for predicting response to TNFi can be applied in clinical practice.
Concluding Remarks
Over a decade of intensive work on the biological activity of gut microbiota spurs inspired enthusiasm to explore the involvement of our resident bacteria in immune processes of the host. There is ample evidence highlighting that gut microbiota interact extensively with anti-rheumatic drugs. In addition to the well-known effect of bacteria on the conversion of inactive prodrugs to active drug, we now learned that DMARDs, such as MTX, can directly affect the growth of gut flora. Furthermore, alteration of the gut microbiota may also contribute to the immunosuppressive effects of MTX. Clearly, further studies are required to identify microbiota which can mediate immune suppression in the host. The other clinically relevant aspect of the interaction of microbiota with DMRADs is towards personalized medicine. Identifying unique individual gut microbial signature may help clinicians to choose a most likely responsive drug for the patient and one devoid of adverse effects.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
The work of LZ is supported by the Scientific Application and Foundation Project of Science and Technology, Department of Sichuan Province (no. 2020YJ0021), and the Scientific Research Project of Health Commission of Sichuan Province (no. 20PJ050). The work of CQC is supported by an innovative award from the American College of Rheumatology Research Foundation and by a VA Merit Review grant (I01BX005195).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank John Rutledge for proofreading the manuscript.
References
1. Sigler JW, Bluhm GB, Duncan H, Sharp JT, Ensign DC, McCrum WR. Gold Salts in the Treatment of Rheumatoid Arthritis. A Double-Blind Study. Ann Intern Med (1974) 80(1):21–6. doi: 10.7326/0003-4819-80-1-21
2. Martinez-Martinez RE, Abud-Mendoza C, Patino-Marin N, Rizo-Rodriguez JC, Little JW, Loyola-Rodriguez JP. Detection of Periodontal Bacterial DNA in Serum and Synovial Fluid in Refractory Rheumatoid Arthritis Patients. J Clin Periodontol (2009) 36(12):1004–10. doi: 10.1111/j.1600-051X.2009.01496.x
3. Mikuls TR, Payne JB, Reinhardt RA, Thiele GM, Maziarz E, Cannella AC, et al. Antibody Responses to Porphyromonas Gingivalis (P. Gingivalis) in Subjects With Rheumatoid Arthritis and Periodontitis. Int Immunopharmacol (2009) 9(1):38–42. doi: 10.1016/j.intimp.2008.09.008
4. de Pablo P, Dietrich T, McAlindon TE. Association of Periodontal Disease and Tooth Loss With Rheumatoid Arthritis in the US Population. J Rheumatol (2008) 35(1):70–6.
5. Dissick A, Redman RS, Jones M, Rangan BV, Reimold A, Griffiths GR, et al. Association of Periodontitis With Rheumatoid Arthritis: A Pilot Study. J Periodontol (2010) 81(2):223–30. doi: 10.1902/jop.2009.090309
6. Maresz KJ, Hellvard A, Sroka A, Adamowicz K, Bielecka E, Koziel J, et al. Porphyromonas Gingivalis Facilitates the Development and Progression of Destructive Arthritis Through Its Unique Bacterial Peptidylarginine Deiminase (PAD). PloS Pathog (2013) 9(9):e1003627. doi: 10.1371/journal.ppat.1003627
7. Puntis D, Malik S, Saravanan V, Rynne M, Heycock C, Hamilton J, et al. Urinary Tract Infections in Patients With Rheumatoid Arthritis. Clin Rheumatol (2013) 32(3):355–60. doi: 10.1007/s10067-012-2129-7
8. Newkirk MM, Goldbach-Mansky R, Senior BW, Klippel J, Schumacher HR Jr, El-Gabalawy HS. Elevated Levels of IgM and IgA Antibodies to Proteus Mirabilis and IgM Antibodies to Escherichia Coli Are Associated With Early Rheumatoid Factor (RF)-Positive Rheumatoid Arthritis. Rheumatol (Oxford) (2005) 44(11):1433–41. doi: 10.1093/rheumatology/kei036
9. Zhang L, Zhang YJ, Chen J, Huang XL, Fang GS, Yang LJ, et al. The Association of HLA-B27 and Klebsiella Pneumoniae in Ankylosing Spondylitis: A Systematic Review. Microb Pathog (2018) 117:49–54. doi: 10.1016/j.micpath.2018.02.020
10. Fendler C, Laitko S, Sorensen H, Gripenberg-Lerche C, Groh A, Uksila J, et al. Frequency of Triggering Bacteria in Patients With Reactive Arthritis and Undifferentiated Oligoarthritis and the Relative Importance of the Tests Used for Diagnosis. Ann Rheum Dis (2001) 60(4):337–43. doi: 10.1136/ard.60.4.337
11. Franzosa EA, Sirota-Madi A, Avila-Pacheco J, Fornelos N, Haiser HJ, Reinker S, et al. Author Correction: Gut Microbiome Structure and Metabolic Activity in Inflammatory Bowel Disease. Nat Microbiol (2019) 4(5):898. doi: 10.1038/s41564-019-0442-5
12. Gurung M, Li Z, You H, Rodrigues R, Jump DB, Morgun A, et al. Role of Gut Microbiota in Type 2 Diabetes Pathophysiology. EBioMedicine (2020) 51:102590. doi: 10.1016/j.ebiom.2019.11.051
13. Mielcarz DW, Kasper LH. The Gut Microbiome in Multiple Sclerosis. Curr Treat Options Neurol (2015) 17(4):344. doi: 10.1007/s11940-015-0344-7
14. Strati F, Cavalieri D, Albanese D, De Felice C, Donati C, Hayek J, et al. New Evidences on the Altered Gut Microbiota in Autism Spectrum Disorders. Microbiome (2017) 5(1):24. doi: 10.1186/s40168-017-0242-1
15. Wong SH, Yu J. Gut Microbiota in Colorectal Cancer: Mechanisms of Action and Clinical Applications. Nat Rev Gastroenterol Hepatol (2019) 16(11):690–704. doi: 10.1038/s41575-019-0209-8
16. Louis P, Hold GL, Flint HJ. The Gut Microbiota, Bacterial Metabolites and Colorectal Cancer. Nat Rev Microbiol (2014) 12(10):661–72. doi: 10.1038/nrmicro3344
17. Gunjur A. Cancer and the Microbiome. Lancet Oncol (2020) 21(7):888. doi: 10.1016/S1470-2045(20)30351-X
18. Scher JU, Abramson SB. The Microbiome and Rheumatoid Arthritis. Nat Rev Rheumatol (2011) 7(10):569–78. doi: 10.1038/nrrheum.2011.121
19. Vaahtovuo J, Munukka E, Korkeamaki M, Luukkainen R, Toivanen P. Fecal Microbiota in Early Rheumatoid Arthritis. J Rheumatol (2008) 35(8):1500–5.
20. Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, et al. Expansion of Intestinal Prevotella Copri Correlates With Enhanced Susceptibility to Arthritis. Elife (2013) 2:e01202. doi: 10.7554/eLife.01202
21. Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, et al. The Oral and Gut Microbiomes Are Perturbed in Rheumatoid Arthritis and Partly Normalized After Treatment. Nat Med (2015) 21(8):895–905. doi: 10.1038/nm.3914
22. Zhang L, Song P, Zhang X, Metea C, Schleisman M, Karstens L, et al. Alpha-Glucosidase Inhibitors Alter Gut Microbiota and Ameliorate Collagen-Induced Arthritis. Front Pharmacol (2019) 10:1684. doi: 10.3389/fphar.2019.01684
23. Rizkallah MSR, Aziz RK. The Human Microbiome Project, Personalized Medicine and the Birth of Pharmacomicrobiomics. Curr Pharmacogenom Person Med (2010) 8:182–93. doi: 10.2174/187569210792246326
24. Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The Placenta Harbors a Unique Microbiome. Sci Transl Med (2014) 6(237):237ra65. doi: 10.1126/scitranslmed.3008599
25. Stout MJ, Conlon B, Landeau M, Lee I, Bower C, Zhao Q, et al. Identification of Intracellular Bacteria in the Basal Plate of the Human Placenta in Term and Preterm Gestations. Am J Obstet Gynecol (2013) 208(3):226.e1–7. doi: 10.1016/j.ajog.2013.01.018
26. Jimenez E, Marin ML, Martin R, Odriozola JM, Olivares M, Xaus J, et al. Is Meconium From Healthy Newborns Actually Sterile? Res Microbiol (2008) 159(3):187–93. doi: 10.1016/j.resmic.2007.12.007
27. Jimenez E, Fernandez L, Marin ML, Martin R, Odriozola JM, Nueno-Palop C, et al. Isolation of Commensal Bacteria From Umbilical Cord Blood of Healthy Neonates Born by Cesarean Section. Curr Microbiol (2005) 51(4):270–4. doi: 10.1007/s00284-005-0020-3
28. Zhang X, Mozeleski B, Lemoine S, Deriaud E, Lim A, Zhivaki D, et al. CD4 T Cells With Effector Memory Phenotype and Function Develop in the Sterile Environment of the Fetus. Sci Transl Med (2014) 6(238):238ra72. doi: 10.1126/scitranslmed.3008748
29. Hooper LV, Littman DR, Macpherson AJ. Interactions Between the Microbiota and the Immune System. Science (2012) 336(6086):1268–73. doi: 10.1126/science.1223490
30. Smolen JS, Aletaha D, Barton A, Burmester GR, Emery P, Firestein GS, et al. Rheumatoid Arthritis. Nat Rev Dis Primers (2018) 4:18001. doi: 10.1038/nrdp.2018.1
31. Brown EM, Kenny DJ, Xavier RJ. Gut Microbiota Regulation of T Cells During Inflammation and Autoimmunity. Annu Rev Immunol (2019) 37:599–624. doi: 10.1146/annurev-immunol-042718-041841
32. Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of Intestinal Th17 Cells by Segmented Filamentous Bacteria. Cell (2009) 139(3):485–98. doi: 10.1016/j.cell.2009.09.033
33. Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, et al. Gut-Residing Segmented Filamentous Bacteria Drive Autoimmune Arthritis via T Helper 17 Cells. Immunity (2010) 32(6):815–27. doi: 10.1016/j.immuni.2010.06.001
34. Zhang X, Chen BD, Zhao LD, Li H. The Gut Microbiota: Emerging Evidence in Autoimmune Diseases. Trends Mol Med (2020) 26(9):862–73. doi: 10.1016/j.molmed.2020.04.001
35. Greiling TM, Dehner C, Chen X, Hughes K, Iniguez AJ, Boccitto M, et al. Commensal Orthologs of the Human Autoantigen Ro60 as Triggers of Autoimmunity in Lupus. Sci Transl Med (2018) 10(434):eaan2306. doi: 10.1126/scitranslmed.aan2306
36. Chen BD, Jia XM, Xu JY, Zhao LD, Ji JY, Wu BX, et al. An Autoimmunogenic and Proinflammatory Profile Defined by the Gut Microbiota of Patients With Untreated Systemic Lupus Erythematosus. Arthritis Rheumatol (2021) 73(2):232–43. doi: 10.1002/art.41511
37. Lindner C, Wahl B, Fohse L, Suerbaum S, Macpherson AJ, Prinz I, et al. Age, Microbiota, and T Cells Shape Diverse Individual IgA Repertoires in the Intestine. J Exp Med (2012) 209(2):365–77. doi: 10.1084/jem.20111980
38. Hapfelmeier S, Lawson MA, Slack E, Kirundi JK, Stoel M, Heikenwalder M, et al. Reversible Microbial Colonization of Germ-Free Mice Reveals the Dynamics of IgA Immune Responses. Science (2010) 328(5986):1705–9. doi: 10.1126/science.1188454
39. Lindner C, Thomsen I, Wahl B, Ugur M, Sethi MK, Friedrichsen M, et al. Diversification of Memory B Cells Drives the Continuous Adaptation of Secretory Antibodies to Gut Microbiota. Nat Immunol (2015) 16(8):880–8. doi: 10.1038/ni.3213
40. Li H, Limenitakis JP, Greiff V, Yilmaz B, Scharen O, Urbaniak C, et al. Mucosal or Systemic Microbiota Exposures Shape the B Cell Repertoire. Nature (2020) 584(7820):274–8. doi: 10.1038/s41586-020-2564-6
41. Ward MM, Deodhar A, Akl EA, Lui A, Ermann J, Gensler LS, et al. American College of Rheumatology/Spondylitis Association of America/Spondyloarthritis Research and Treatment Network 2015 Recommendations for the Treatment of Ankylosing Spondylitis and Nonradiographic Axial Spondyloarthritis. Arthritis Rheumatol (2016) 68(2):282–98. doi: 10.1002/art.39298
42. Skoldstam L, Hagfors L, Johansson G. An Experimental Study of a Mediterranean Diet Intervention for Patients With Rheumatoid Arthritis. Ann Rheum Dis (2003) 62(3):208–14. doi: 10.1136/ard.62.3.208
43. Van Praet JT, Donovan E, Vanassche I, Drennan MB, Windels F, Dendooven A, et al. Commensal Microbiota Influence Systemic Autoimmune Responses. EMBO J (2015) 34(4):466–74. doi: 10.15252/embj.201489966
44. Azzouz D, Omarbekova A, Heguy A, Schwudke D, Gisch N, Rovin BH, et al. Lupus Nephritis Is Linked to Disease-Activity Associated Expansions and Immunity to a Gut Commensal. Ann Rheum Dis (2019) 78(7):947–56. doi: 10.1136/annrheumdis-2018-214856
45. Ma Y, Xu X, Li M, Cai J, Wei Q, Niu H. Gut Microbiota Promote the Inflammatory Response in the Pathogenesis of Systemic Lupus Erythematosus. Mol Med (2019) 25(1):35. doi: 10.1186/s10020-019-0102-5
46. Ruff WE, Dehner C, Kim WJ, Pagovich O, Aguiar CL, Yu AT, et al. Pathogenic Autoreactive T and B Cells Cross-React With Mimotopes Expressed by a Common Human Gut Commensal to Trigger Autoimmunity. Cell Host Microbe (2019) 26(1):100–13.e8. doi: 10.1016/j.chom.2019.05.003
47. Scher JU, Nayak RR, Ubeda C, Turnbaugh PJ, Abramson SB. Pharmacomicrobiomics in Inflammatory Arthritis: Gut Microbiome as Modulator of Therapeutic Response. Nat Rev Rheumatol (2020) 16(5):282–92. doi: 10.1038/s41584-020-0395-3
48. Saad R, Rizkallah MR, Aziz RK. Gut Pharmacomicrobiomics: The Tip of an Iceberg of Complex Interactions Between Drugs and Gut-Associated Microbes. Gut Pathog (2012) 4(1):16. doi: 10.1186/1757-4749-4-16
49. Doestzada M, Vila AV, Zhernakova A, Koonen DPY, Weersma RK, Touw DJ, et al. Pharmacomicrobiomics: A Novel Route Towards Personalized Medicine? Protein Cell (2018) 9(5):432–45. doi: 10.1007/s13238-018-0547-2
50. Smedegard G, Bjork J. Sulphasalazine: Mechanism of Action in Rheumatoid Arthritis. Br J Rheumatol (1995) 34(Suppl 2):7–15. doi: 10.1093/rheumatology/XXXIV.suppl_2.7
51. Gadelle D, Raibaud P, Sacquet E. Beta-Glucuronidase Activities of Intestinal Bacteria Determined Both In Vitro and In Vivo in Gnotobiotic Rats. Appl Environ Microbiol (1985) 49(3):682–5. doi: 10.1128/aem.49.3.682-685.1985
52. Boelsterli UA, Redinbo MR, Saitta KS. Multiple NSAID-Induced Hits Injure the Small Intestine: Underlying Mechanisms and Novel Strategies. Toxicol Sci (2013) 131(2):654–67. doi: 10.1093/toxsci/kfs310
53. Little MS, Pellock SJ, Walton WG, Tripathy A, Redinbo MR. Structural Basis for the Regulation of Beta-Glucuronidase Expression by Human Gut Enterobacteriaceae. Proc Natl Acad Sci USA (2018) 115(2):E152–61. doi: 10.1073/pnas.1716241115
54. LoGuidice A, Wallace BD, Bendel L, Redinbo MR, Boelsterli UA. Pharmacologic Targeting of Bacterial Beta-Glucuronidase Alleviates Nonsteroidal Anti-Inflammatory Drug-Induced Enteropathy in Mice. J Pharmacol Exp Ther (2012) 341(2):447–54. doi: 10.1124/jpet.111.191122
55. Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R, Goodman AL. Mapping Human Microbiome Drug Metabolism by Gut Bacteria and Their Genes. Nature (2019) 570(7762):462–7. doi: 10.1038/s41586-019-1291-3
56. Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE, et al. Extensive Impact of Non-Antibiotic Drugs on Human Gut Bacteria. Nature (2018) 555(7698):623–8. doi: 10.1038/nature25979
57. Hench PS, Kendall EC, Slocumb CH, Polley HF. The Effect of a Hormone of the Adrenal Cortex (17-Hydroxy-11-Dehydrocorticosterone; Compound E) and of Pituitary Adrenocorticotropic Hormone on Rheumatoid Arthritis. Proc Staff Meet Mayo Clin (1949) 24(8):181–97.
58. Vandewalle J, Luypaert A, De Bosscher K, Libert C. Therapeutic Mechanisms of Glucocorticoids. Trends Endocrinol Metab (2018) 29(1):42–54. doi: 10.1016/j.tem.2017.10.010
59. Hardy RS, Raza K, Cooper MS. Therapeutic Glucocorticoids: Mechanisms of Actions in Rheumatic Diseases. Nat Rev Rheumatol (2020) 16(3):133–44. doi: 10.1038/s41584-020-0371-y
60. Rhen T, Cidlowski JA. Antiinflammatory Action of Glucocorticoids–New Mechanisms for Old Drugs. N Engl J Med (2005) 353(16):1711–23. doi: 10.1056/NEJMra050541
61. Ugarte A, Danza A, Ruiz-Irastorza G. Glucocorticoids and Antimalarials in Systemic Lupus Erythematosus: An Update and Future Directions. Curr Opin Rheumatol (2018) 30(5):482–9. doi: 10.1097/BOR.0000000000000527
62. Choi SC, Brown J, Gong M, Ge Y, Zadeh M, Li W, et al. Gut Microbiota Dysbiosis and Altered Tryptophan Catabolism Contribute to Autoimmunity in Lupus-Susceptible Mice. Sci Transl Med (2020) 12(551):eaax2220. doi: 10.1126/scitranslmed.aax2220
63. Luo XM, Edwards MR, Mu Q, Yu Y, Vieson MD, Reilly CM, et al. Gut Microbiota in Human Systemic Lupus Erythematosus and a Mouse Model of Lupus. Appl Environ Microbiol (2018) 84(4):e02288–17. doi: 10.1128/AEM.02288-17
64. Johnson BM, Gaudreau MC, Gudi R, Brown R, Gilkeson G, Vasu C. Gut Microbiota Differently Contributes to Intestinal Immune Phenotype and Systemic Autoimmune Progression in Female and Male Lupus-Prone Mice. J Autoimmun (2020) 108:102420. doi: 10.1016/j.jaut.2020.102420
65. Fanouriakis A, Kostopoulou M, Alunno A, Aringer M, Bajema I, Boletis JN, et al. 2019 Update of the EULAR Recommendations for the Management of Systemic Lupus Erythematosus. Ann Rheum Dis (2019) 78(6):736–45. doi: 10.1136/annrheumdis-2019-215089
66. He Z, Kong X, Shao T, Zhang Y, Wen C. Alterations of the Gut Microbiota Associated With Promoting Efficacy of Prednisone by Bromofuranone in MRL/lpr Mice. Front Microbiol (2019) 10:978. doi: 10.3389/fmicb.2019.00978
67. Cai W, Ran Y, Li Y, Wang B, Zhou L. Intestinal Microbiome and Permeability in Patients With Autoimmune Hepatitis. Best Pract Res Clin Gastroenterol (2017) 31(6):669–73. doi: 10.1016/j.bpg.2017.09.013
68. Guo M, Wang H, Xu S, Zhuang Y, An J, Su C, et al. Alteration in Gut Microbiota Is Associated With Dysregulation of Cytokines and Glucocorticoid Therapy in Systemic Lupus Erythematosus. Gut Microbes (2020) 11(6):1758–73. doi: 10.1080/19490976.2020.1768644
69. Brown PM, Pratt AG, Isaacs JD. Mechanism of Action of Methotrexate in Rheumatoid Arthritis, and the Search for Biomarkers. Nat Rev Rheumatol (2016) 12(12):731–42. doi: 10.1038/nrrheum.2016.175
70. Bleyer WA. The Clinical Pharmacology of Methotrexate: New Applications of an Old Drug. Cancer (1978) 41(1):36–51. doi: 10.1002/1097-0142(197801)41:1<36::AID-CNCR2820410108>3.0.CO;2-I
71. Chan ES, Cronstein BN. Methotrexate–How Does It Really Work? Nat Rev Rheumatol (2010) 6(3):175–8. doi: 10.1038/nrrheum.2010.5
72. Cronstein BN, Aune TM. Methotrexate and Its Mechanisms of Action in Inflammatory Arthritis. Nat Rev Rheumatol (2020) 16(3):145–54. doi: 10.1038/s41584-020-0373-9
73. de Araujo AA, Borba PB, de Souza FH, Nogueira AC, Saldanha TS, Araujo TE, et al. In a Methotrexate-Induced Model of Intestinal Mucositis, Olmesartan Reduced Inflammation and Induced Enteropathy Characterized by Severe Diarrhea, Weight Loss, and Reduced Sucrose Activity. Biol Pharm Bull (2015) 38(5):746–52. doi: 10.1248/bpb.b14-00847
74. Aparicio-Domingo P, Romera-Hernandez M, Karrich JJ, Cornelissen F, Papazian N, Lindenbergh-Kortleve DJ, et al. Type 3 Innate Lymphoid Cells Maintain Intestinal Epithelial Stem Cells After Tissue Damage. J Exp Med (2015) 212(11):1783–91. doi: 10.1084/jem.20150318
75. Lee YS, Kim TY, Kim Y, Lee SH, Kim S, Kang SW, et al. Microbiota-Derived Lactate Accelerates Intestinal Stem-Cell-Mediated Epithelial Development. Cell Host Microbe (2018) 24(6):833–46.e6. doi: 10.1016/j.chom.2018.11.002
76. Tang D, Zeng T, Wang Y, Cui H, Wu J, Zou B, et al. Dietary Restriction Increases Protective Gut Bacteria to Rescue Lethal Methotrexate-Induced Intestinal Toxicity. Gut Microbes (2020) 12(1):1714401. doi: 10.1080/19490976.2020.1714401
77. Zhou B, Xia X, Wang P, Chen S, Yu C, Huang R, et al. Induction and Amelioration of Methotrexate-Induced Gastrointestinal Toxicity Are Related to Immune Response and Gut Microbiota. EBioMedicine (2018) 33:122–33. doi: 10.1016/j.ebiom.2018.06.029
78. Letertre MPM, Munjoma N, Wolfer K, Pechlivanis A, McDonald JAK, Hardwick RN, et al. A Two-Way Interaction Between Methotrexate and the Gut Microbiota of Male Sprague-Dawley Rats. J Proteome Res (2020) 19(8):3326–39. doi: 10.1021/acs.jproteome.0c00230
79. Picchianti-Diamanti A, Panebianco C, Salemi S, Sorgi ML, Di Rosa R, Tropea A, et al. Analysis of Gut Microbiota in Rheumatoid Arthritis Patients: Disease-Related Dysbiosis and Modifications Induced by Etanercept. Int J Mol Sci (2018) 19(10):2938. doi: 10.3390/ijms19102938
80. Artacho A, Isaac S, Nayak R, Flor-Duro A, Alexander M, Koo I, et al. The Pre-Treatment Gut Microbiome Is Associated With Lack of Response to Methotrexate in New Onset Rheumatoid Arthritis. Arthritis Rheumatol (2021) 73(6):931–42. doi: 10.1002/art.41622
81. Liu L, Liu S, Wang C, Guan W, Zhang Y, Hu W, et al. Folate Supplementation for Methotrexate Therapy in Patients With Rheumatoid Arthritis: A Systematic Review. J Clin Rheumatol (2019) 25(5):197–202. doi: 10.1097/RHU.0000000000000810
82. Huang X, Fang Q, Rao T, Zhou L, Zeng X, Tan Z, et al. Leucovorin Ameliorated Methotrexate Induced Intestinal Toxicity via Modulation of the Gut Microbiota. Toxicol Appl Pharmacol (2020) 391:114900. doi: 10.1016/j.taap.2020.114900
83. Nayak RR, Alexander M, Deshpande I, Stapleton-Gray K, Rimal B, Patterson AD, et al. Methotrexate Impacts Conserved Pathways in Diverse Human Gut Bacteria Leading to Decreased Host Immune Activation. Cell Host Microbe (2021) 29(3):362–77 e11. doi: 10.1016/j.chom.2020.12.008
84. Svartz N. The Primary Cause of Rheumatoid Arthritis Is an Infection–The Infectious Agent Exists in Milk. Acta Med Scand (1972) 192(3):231–9. doi: 10.1111/j.0954-6820.1972.tb04807.x
85. Rains CP, Noble S, Faulds D. Sulfasalazine. A Review of Its Pharmacological Properties and Therapeutic Efficacy in the Treatment of Rheumatoid Arthritis. Drugs (1995) 50(1):137–56. doi: 10.2165/00003495-199550010-00009
86. Sammaritano LR, Bermas BL, Chakravarty EE, Chambers C, Clowse MEB, Lockshin MD, et al. 2020 American College of Rheumatology Guideline for the Management of Reproductive Health in Rheumatic and Musculoskeletal Diseases. Arthritis Rheumatol (2020) 72(4):529–56. doi: 10.1002/art.41191
87. Figueroa V, Rodriguez MS, Lanari C, Lamb CA. Nuclear Action of FGF Members in Endocrine-Related Tissues and Cancer: Interplay With Steroid Receptor Pathways. Steroids (2019) 152:108492. doi: 10.1016/j.steroids.2019.108492
88. Peppercorn MA, Goldman P. The Role of Intestinal Bacteria in the Metabolism of Salicylazosulfapyridine. J Pharmacol Exp Ther (1972) 181(3):555–62.
89. Lee HJ, Zhang H, Orlovich DA, Fawcett JP. The Influence of Probiotic Treatment on Sulfasalazine Metabolism in Rat. Xenobiotica (2012) 42(8):791–7. doi: 10.3109/00498254.2012.660508
90. Lee HJ, Waller RD, Stebbings S, Highton J, Orlovich DA, Schmierer D, et al. The Effects of an Orally Administered Probiotic on Sulfasalazine Metabolism in Individuals With Rheumatoid Arthritis: A Preliminary Study. Int J Rheum Dis (2010) 13(1):48–54. doi: 10.1111/j.1756-185X.2009.01449.x
91. Kanerud L, Scheynius A, Nord CE, Hafstrom I. Effect of Sulphasalazine on Gastrointestinal Microflora and on Mucosal Heat Shock Protein Expression in Patients With Rheumatoid Arthritis. Br J Rheumatol (1994) 33(11):1039–48. doi: 10.1093/rheumatology/33.11.1039
92. Zheng H, Chen M, Li Y, Wang Y, Wei L, Liao Z, et al. Modulation of Gut Microbiome Composition and Function in Experimental Colitis Treated With Sulfasalazine. Front Microbiol (2017) 8:1703. doi: 10.3389/fmicb.2017.01703
93. Fanouriakis A, Kostopoulou M, Cheema K, Anders HJ, Aringer M, Bajema I, et al. 2019 Update of the Joint European League Against Rheumatism and European Renal Association-European Dialysis and Transplant Association (EULAR/ERA-EDTA) Recommendations for the Management of Lupus Nephritis. Ann Rheum Dis (2020) 79(6):713–23. doi: 10.1136/annrheumdis-2020-216924
94. Schrezenmeier E, Dorner T. Mechanisms of Action of Hydroxychloroquine and Chloroquine: Implications for Rheumatology. Nat Rev Rheumatol (2020) 16(3):155–66. doi: 10.1038/s41584-020-0372-x
95. Angelakis E, Million M, Kankoe S, Lagier JC, Armougom F, Giorgi R, et al. Abnormal Weight Gain and Gut Microbiota Modifications Are Side Effects of Long-Term Doxycycline and Hydroxychloroquine Treatment. Antimicrob Agents Chemother (2014) 58(6):3342–7. doi: 10.1128/AAC.02437-14
96. Kurmann RD, Mankad R. Atherosclerotic Vascular Disease in the Autoimmune Rheumatologic Woman. Clin Cardiol (2018) 41(2):258–63. doi: 10.1002/clc.22916
97. Shi N, Zhang S, Silverman G, Li M, Cai J, Niu H. Protective Effect of Hydroxychloroquine on Rheumatoid Arthritis-Associated Atherosclerosis. Anim Model Exp Med (2019) 2(2):98–106. doi: 10.1002/ame2.12065
98. Zhang W, Dai SM. Mechanisms Involved in the Therapeutic Effects of Paeonia Lactiflora Pallas in Rheumatoid Arthritis. Int Immunopharmacol (2012) 14(1):27–31. doi: 10.1016/j.intimp.2012.06.001
99. Feng Z, Zhang BQ, Zhu YM, Yu BB, Fu L, Zhou LL, et al. The Effectiveness and Safety of Total Glucosides of Paeony in Primary Sjogren’s Syndrome: A Systematic Review and Meta-Analysis. Front Pharmacol (2019) 10:550. doi: 10.3389/fphar.2019.00550
100. Pan ZY, Chang YX, Han N, Hou FY, Lee BJY, Zhi FC, et al. Short-Term High-Dose Gavage of Hydroxychloroquine Changes Gut Microbiota But Not the Intestinal Integrity and Immunological Responses in Mice. Life Sci (2021) 264:118450. doi: 10.1016/j.lfs.2020.118450
101. Croft M, Siegel RM. Beyond TNF: TNF Superfamily Cytokines as Targets for the Treatment of Rheumatic Diseases. Nat Rev Rheumatol (2017) 13(4):217–33. doi: 10.1038/nrrheum.2017.22
102. Sakurai T, Nishiyama H, Sakai K, De Velasco MA, Nagai T, Komeda Y, et al. Mucosal Microbiota and Gene Expression Are Associated With Long-Term Remission After Discontinuation of Adalimumab in Ulcerative Colitis. Sci Rep (2020) 10(1):19186. doi: 10.1038/s41598-020-76175-2
103. Jones-Hall YL, Kozik A, Nakatsu C. Ablation of Tumor Necrosis Factor Is Associated With Decreased Inflammation and Alterations of the Microbiota in a Mouse Model of Inflammatory Bowel Disease. PloS One (2015) 10(3):e0119441. doi: 10.1371/journal.pone.0119441
104. Liu X, Zou Q, Zeng B, Fang Y, Wei H. Analysis of Fecal Lactobacillus Community Structure in Patients With Early Rheumatoid Arthritis. Curr Microbiol (2013) 67(2):170–6. doi: 10.1007/s00284-013-0338-1
105. Singh RK, Tiwari SP, Rai AK, Mohapatra TM. Cyanobacteria: An Emerging Source for Drug Discovery. J Antibiot (Tokyo) (2011) 64(6):401–12. doi: 10.1038/ja.2011.21
106. Singh R, Parihar P, Singh M, Bajguz A, Kumar J, Singh S, et al. Uncovering Potential Applications of Cyanobacteria and Algal Metabolites in Biology, Agriculture and Medicine: Current Status and Future Prospects. Front Microbiol (2017) 8:515. doi: 10.3389/fmicb.2017.00515
107. Scher JU, Littman DR, Abramson SB. Microbiome in Inflammatory Arthritis and Human Rheumatic Diseases. Arthritis Rheumatol (2016) 68(1):35–45. doi: 10.1002/art.39259
108. Liu B, Yang L, Cui Z, Zheng J, Huang J, Zhao Q, et al. Anti-TNF-Alpha Therapy Alters the Gut Microbiota in Proteoglycan-Induced Ankylosing Spondylitis in Mice. Microbiologyopen (2019) 8(12):e927. doi: 10.1002/mbo3.927
109. Zhang F, Ma C, Zhang B, Bi L. Dynamic Changes in Gut Microbiota Under the Influence of Smoking and TNF-Alpha-Blocker in Patients With Ankylosing Spondylitis. Clin Rheumatol (2020) 39(9):2653–61. doi: 10.1007/s10067-020-05032-4
110. Bazin T, Hooks KB, Barnetche T, Truchetet ME, Enaud R, Richez C, et al. Microbiota Composition May Predict Anti-Tnf Alpha Response in Spondyloarthritis Patients: An Exploratory Study. Sci Rep (2018) 8(1):5446. doi: 10.1038/s41598-018-23571-4
111. Dovrolis N, Michalopoulos G, Theodoropoulos GE, Arvanitidis K, Kolios G, Sechi LA, et al. The Interplay Between Mucosal Microbiota Composition and Host Gene-Expression Is Linked With Infliximab Response in Inflammatory Bowel Diseases. Microorganisms (2020) 8(3):438. doi: 10.3390/microorganisms8030438
Keywords: microbiota, disease modifying anti-rheumatic drugs, personalized medicine, biomarkers, prediction and opportunity
Citation: Zhang L and Chu C-Q (2021) Gut Microbiota–Medication Interaction in Rheumatic Diseases. Front. Immunol. 12:796865. doi: 10.3389/fimmu.2021.796865
Received: 17 October 2021; Accepted: 15 November 2021;
Published: 03 December 2021.
Edited by:
Xia Li, Dalian Medical University, ChinaCopyright © 2021 Zhang and Chu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cong-Qiu Chu, Y2h1Y0BvaHN1LmVkdQ==
 Lingshu Zhang
Lingshu Zhang Cong-Qiu Chu
Cong-Qiu Chu