- 1Department of Clinical Laboratory, The Second Hospital, Cheeloo College of Medicine, Shandong University, Jinan, China
- 2Shandong Engineering & Technology Research Center for Tumor Marker Detection , Jinan, China
- 3Shandong Provincial Clinical Medicine Research Center for Clinical Laboratory, Jinan, China
- 4School of Public Health, Shandong First Medical University & Shandong Academy of Medical Sciences, Taian, China
- 5Department of Respiratory and Critical Care Medicine, Qilu Hospital, Cheeloo College of Medicine, Shandong University, Jinan, China
As a non-classic major histocompatibility complex (MHC) class I molecule, human leukocyte antigen G (HLA-G) is expressed in fetal-maternal interface and immunoprivileged site only in healthy condition, and in pathological conditions such as cancer, it can be de novo expressed. It is now widely accepted that HLA-G is a key molecule in the process of immune escape of cancer cells, which is ubiquitously expressed in the tumor environment. This raises the possibility that it may play an adverse role in tumor immunity. The expression level of HLA-G has been demonstrated to be highly correlated with clinical parameters in many tumors, and its potential significance in the diagnosis and prognosis of cancer has been postulated. However, because HLA-G itself has up to seven different subtypes, and for some subtypes, detected antibodies are few or absent, it is hard to evaluate the actual expression of HLA-G in tumors. In the present work, we described (a) the structure and three main forms of HLA-G, (b) summarized the mechanism of HLA-G in the immune escape of tumor cells, (c) discussed the potential role of HLA-G as a tumor marker, and reviewed (d) the methods for detecting and quantifying HLA-G.
Introduction
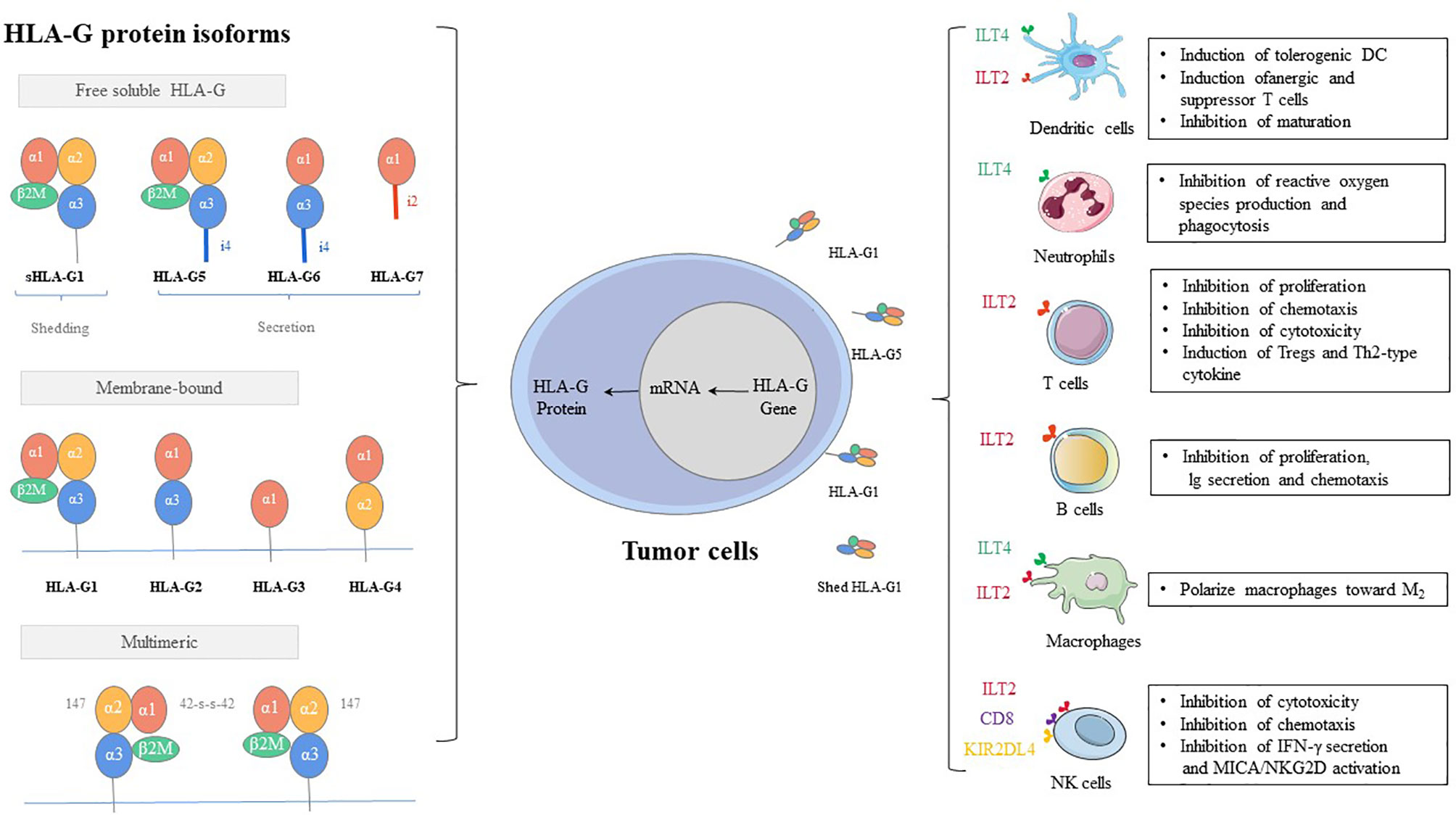
As early as 1983, human leukocyte antigen G (HLA-G) is first observed on the cytotrophoblast at the fetal-maternal interface (1). As a class of major histocompatibility complex (MHC) I molecules, HLA-G showed low polymorphic in the coding region, while several polymoprphism have been describe in non-coding region of the locus (3’UTR, and 5’URR regions) (2). The exons and introns of the HLA-G gene are the same as those of classic MHC class molecules, consisting of eight exons and seven introns (3, 4). However, HLA-G shows only limited genetic polymorphism. The main reason can be attributed to that the terminator of HLA-G is located in the second codon of exon 6, and thus most of exon 6 and all of exons 7 and 8 cannot be translated into protein (5, 6). HLA-G can exist in a variety of structures, which can not only be expressed on the cell surface but also exist in the form of secretion (7, 8). There are seven isoforms, which encode four membrane-bound (HLA-G1, -G2, -G3, and -G4) and three soluble (HLA-G5, -G6, and -G7) protein isoforms (9). Each HLA-G subtype contains one to three spherical domains (α1, α2, and α3) encoded by exon 2, exon 3, and exon 4 (Figure 1).
HLA-G is initially considered to be useful for fetal establishment and maintenance of maternal-fetal immune tolerance (10, 11). Therefore, initial studies have mainly focused on its role in regulating maternal immune cell responses and protecting the fetus from natural killer (NK) cell-mediated lysis (12–14). Fetal and tumor development are similar and characterized by rapid tissue proliferation, which is associated with the expressions of anti-apoptotic factors and telomerase (14–17). It is worth noting that since immunoregulatory sites can be shared between tumor development and placenta formation, both placenta and tumor are protected by the immune system and resistant to induction by the immune microenvironment. Therefore, the research focus of HLA-G has gradually shifted to tumors (18).
In the malignant environment where tumors occur, the expression of HLA-G in melanoma is first reported in 1998 (19), and then the abnormal expression of HLA-G is observed in a variety of malignant tumors, such as lung cancer (20–28), gastric cancer (29–37), ovarian cancer (OC) (38–47), breast cancer (48–54), and hematopoietic tumor (55–58). With the deepening of research, the expression of HLA-G in solid malignant tumors and its potential clinical relevance have attracted increasing attention (59). Abnormal expression of HLA-G plays a variety of roles in the progression of malignant tumors, such as inducing apoptosis, inhibiting immune cytolysis and cytotoxicity, and chemotaxis of regulatory cells and damage of different immune effector cells through receptor binding and/or trogocytosis (60).
In addition, HLA-G is secreted in a variety of body fluids, as either free soluble HLA-G (sHLA-G) or part of extracellular vesicles (EVs), and it has been extensively studied as tumor markers. Among them, sHLA-G is derived from the secretion of sHLA-G homotypes, such as HLA-G5, HLA-G6, and HLA-G7, and the shedding of membrane-bound HLA-G homotypes, such as HLA-G1, cleaved by proteolytic hydrolysis (61). Soluble isoforms can be detected in saliva (62, 63), ascitic fluid (47), plasma (26, 33, 48, 49, 56, 64–69), thymus (70), seminal plasma (71), cerebrospinal fluid (CSF) (72, 73), human first trimester and term placentas in situ and in vitro (74), and cell culture supernatant (75, 76). High levels of sHLA-G are correlated with tumor histological type, lymph node metastasis, and patient survival, which can be used as a tumor marker to provide a basis for early diagnosis, differentiation, and prognosis (21). However, it remains largely unclear whether HLA-G-bearing EVs are produced by tumor cells, while there is a functional association between the HLA-G-bearing EVs and various tumors, such as melanoma, breast cancer, and kidney cancer (77). In summary, sHLA-G and HLA-G-bearing EVs may provide unpredictable diagnostic opportunities to monitor tumor status and progression.
HLA-G: A Key Immune Evasion Molecule in Tumors
In cancer, abnormal expression of HLA-G is considered a key strategy of tumor cells to evade immune surveillance, which is strongly supported by the high incidence of tumors in patients treated with immunosuppressive agents after organ and stem cell transplantation (78). The continued construction of tumor phenotypes is thought to be a result of immune-mediated tumor recognition, a phenomenon known as immune editing of cancer. Three stages define the process of immune editing: elimination (immune surveillance), balance (duration/dormancy), and escape (progression) (79, 80). These three stages integrate the immune system’s ability to protect the host from cancer and promote cancer development (81). HLA-G is involved in all three stages, and it is highly necessary to understand the role of HLA-G in tumor immune escape to better develop effective anti-tumor strategies. In the present work, we summarized the main mechanisms of HLA-G-mediated immunosuppression in three aspects as follows:
1. Inhibitory receptors of HLA-G, such as KIR2DL4/CD158d, ILT-2/CD85j, ILT-4/CD85d, CD8, and CD160, can directly exert the immunosuppressive effect by HLA-G. These inhibitory receptors can express in all monocytes, as well as B cells, T cells, and NK cells. Especially, ILT2 receptors are present in subgroups of dendritic cells (DCs) and myeloid-derived suppressive cells (MDSCs) (82–85). ILT4 receptors are mainly expressed in DCs, neutrophils, monocytes, and MDSCs (86–88). KIR2DL4 receptors are mainly expressed in decidual NK cells (89). The mechanisms by which these receptors participate in immunosuppressive effects induced by HLA-G include inhibition of differentiation, cytokine secretion and chemotaxis, immune cell proliferation, cytotoxicity, and induction of MDSCs or M2-type macrophages and regulatory cells (90, 91) (Figure 1).
2. MDSCs, regulatory T cells (Tregs), and tolerogenic DCs can participate in immune escape regulated by HLA-G via an indirect immunosuppressive way. The sub-population of tolerogenic DCs, DC-10s, expresses a high level of HLA-G and induces adaptive type 1 Tregs (Tr1) through the HLA-G/ILT4 signaling pathway (92). On the other hand, tolerogenic DCs produce CD8+CD28+ and CD4+CD25+CTLA-4+ Tregs under the induction of HLA-G, which further strengthens the ability of immune escape of tumor cells (93).
3. HLA-G uses dynamic transfer mechanisms between cells, such as trogocytosis (membrane-bound HLA-G) and EVs (membrane-bound and sHLA-G) (77, 94–96) (Figure 2). Trogocytosis is the process of transporting secretory vesicles or other membrane vesicles from the cell through the cell membrane (97). Activated T cells and NK cells obtain membrane fragments containing functional HLA-G from HLA-G+ or tumor cells through the process of exocytosis. HLA-G-modified cells can immediately reverse immune effector functions to tolerogenic function (98, 99). The generic term EVs are phospholipid bilayer-enclosed vesicles, which are highly heterogeneous in size, molecular content, and membrane composition depending on the state, micro-environment, and the cell of origin (100, 101). According to biogenesis, EVs are characterized by apoptotic bodies (AB) (>500 nm), microvesicles (100-1,000 nm), and exosomes (70-150 nm) (102). In the tumor state, especially in the established acidic microenvironment, EVs can directly fuse with cancer cells, or carry out the transportation and exchange of biologically active substances through endocytosis, phagocytosis, and micropinocytosis, thereby contributing to the intercellular signaling mechanism, providing the tumor with oxygen, metabolites, nutrients and other soluble factors, and making tumor development possible (103).
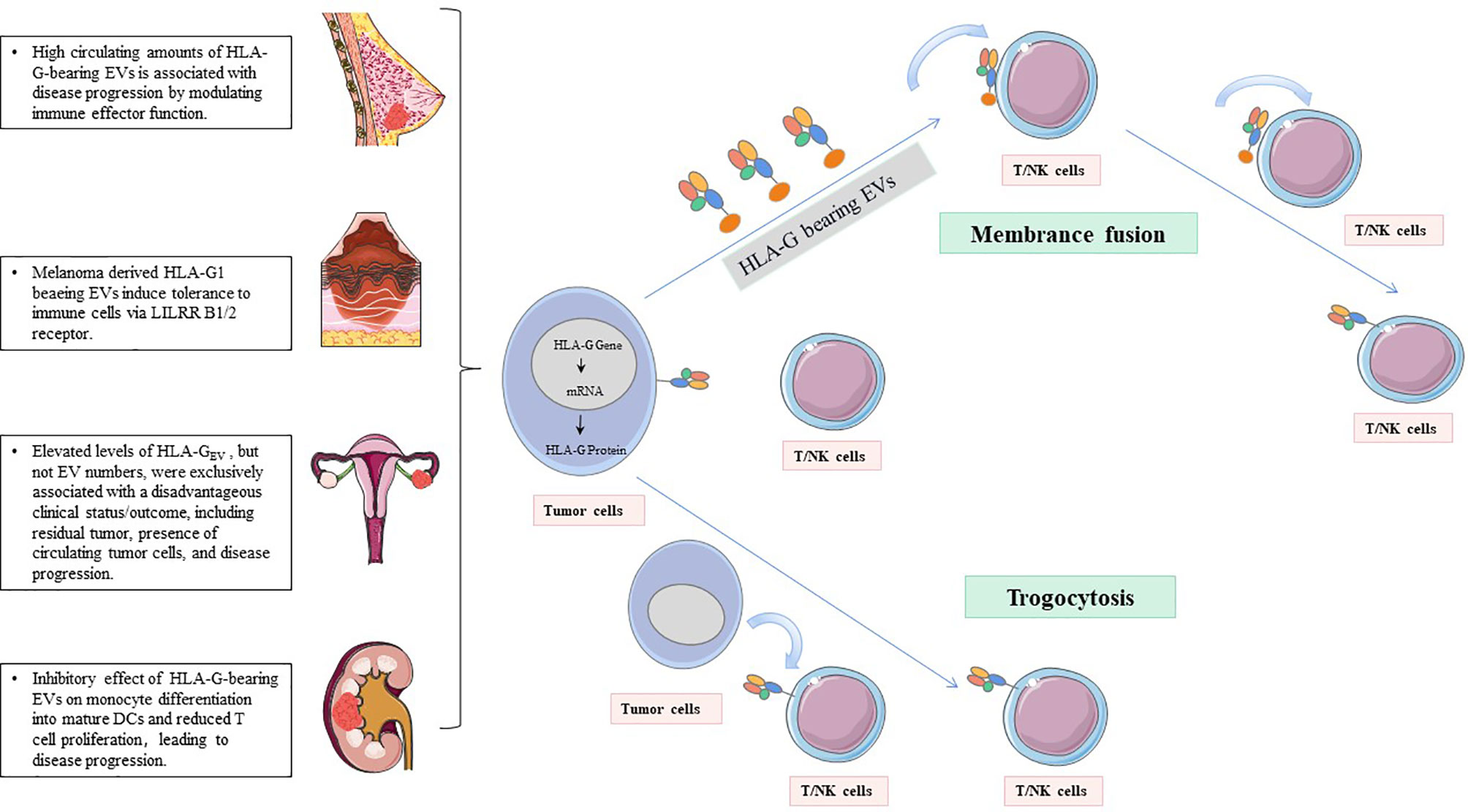
Figure 2 HLA-G-bearing EVs with potential immunological and clinical relevance. NK cells and T cells acquire HLA-G from HLA-G+tumor cells or HLA-G+immune cells via the process of trogocytosis and/or EVs.
Elevated HLA-G in Tumor Patient Tissues as Tumor Indicators
Over the years, many studies have reported that HLA-G is preferentially detected in the primary tumor site and metastatic tumor site, while it is rarely detected in the tumor spontaneous regression site, adjacent tissues, or healthy tissues (104–106). Immunohistochemical (IHC) staining is usually used to detect the expression frequency of HLA-G in tissues, which is combined with clinical results for analysis. In most cancers, the expression of HLA-G is related to the patient’s poor clinical outcome (25, 107, 108).
In liver tissue, HLA-G is detected in the primary site of hepatocellular carcinoma, while its expression is low in benign lesions represented by liver cirrhosis (109). In thyroid tissue, the cell staining efficiency of HLA-G antigen in follicular adenocarcinoma and thyroid cancer is significantly higher compared with the normal thyroid and goiter (110). In ovarian tissue, HLA-G is always co-localized with CA125 protein, indicating that OC cells express HLA-G, while normal cells do not (39). In cervical cancer lesions, the expression of HLA G is an important predictor of CIN I and age, which is not affected by other variables. In addition, HLA-G interacts with immunosuppression induced by human papillomavirus infection, leading to more serious clinical outcomes observed in patients with CIN III and invasive cervical cancer (111, 112).
In oral squamous cell carcinoma (OSCC), according to IHC and reverse transcription-polymerase chain reaction (RT-PCR) results, the higher the TNM stage, the higher the protein expression level of HLA-G, and the histological grade and lymph node metastasis are positively correlated with the expression of HLA-G (107). These results indicate that HLA-G is related to the malignant transformation of tumors, supporting that HLA-G is an indicator of early diagnosis and dynamic monitor. The expression of HLA-G in tissues may also be an important indicator for the prognosis of cancer patients. In 201 colon cancer patients, IHC staining shows that the survival time of patients with HLA-G-positive tumors is significantly shorter compared with those carrying HLA-G-negative tumors (113). In multivariate analysis, HLA-G shows the potential as an independent prognostic factor.
In a joint analysis of HLA class I, HLA-E, and HLA-G to predict the prognosis of colorectal cancer (CRC), three tumor immune phenotypes are generated by comprehensively analyzing the expressions of all markers, resulting in strong immune system tumor recognition, intermediate immune system tumor recognition, and poor immune system tumor recognition. These immune phenotypes represent important and independent clinical prognostic characteristics of colon cancer (108). In nasopharyngeal carcinoma (NPC), HLA-G is positively correlated with CD68+ macrophages and IL-10 expression, indicating that HLA-G may regulate immune escape in NPC (114). Among 522 NPC specimens, the expression of HLA-G at the protein level is detected in 79.2% of cases. In addition, the high expression of HLA-G predicts the low survival rate of NPC patients (114). Moreover, HLA-G is suggested to be an independent predictor of cancers, such as esophageal squamous cell carcinoma (115), gastric cancer (37, 116), breast cancer (52, 117, 118), and OC (38, 44, 119). Finally, the differential expressions of HLA-G can also help predict and diagnose different subtypes. Significant differences between groups have been observed between low-grade glioma and high-grade glioma tissues (120). Besides, the expression of HLA-G in the non-luminal subtypes of invasive ductal carcinoma of the breast is significantly higher compared with the luminal subtypes (121).
sHLA-G in the Blood of Tumor Patients as Circulating Tumor Markers
In the serum of healthy people, the content of HLAG is 20 ng/mL and significantly lower compared with cancer patients. sHLA-G is produced and secreted mainly by immune cells and tumors (122). For example, in acute leukemia, the level of sHLA-G in T cells and monocytes in the serum is detected by enzyme-linked immunosorbent assay (ELISA), which is averagely five times higher compared with healthy controls. Moreover, sHLA-G can be secreted in vitro by DCs, lymphocytes, plasma cells, and monocytes/macrophages (56), and these secreted sHLA-G molecules cause anti-tumor reactions locally in the tumor or along with the circulatory system to the whole body. Next, we described the current status of sHLA-G in the serum of tumor patients as tumor markers from the aspects of diagnosis, prognosis, and identification (Table 1).
In terms of diagnosis, sHLA-G is abnormally expressed in the plasma in breast ductal carcinoma (131), head and neck squamous cell carcinoma (129), gastric cancer (33), CRC (125), and papillary thyroid carcinoma (65), which is considered to be a preoperative diagnosis of cancer histopathology potential marker of aggressiveness. It is worth noting that in gastric cancer, researchers have found that sHLA-G in combination with common serum tumor markers, such as CA72-4, CA125, and CA19-1, can improve the clinical screening of gastric cancer compared with sHLA-G alone (132). In patients with breast cancer, the expression of HLA-G is negatively correlated with proliferation factor (133). In addition, the concentration of sHLA-G in plasma helps predict and diagnose cancers of different subtypes. Compared with patients with simple lobular carcinoma and simple ductal carcinoma, patients with mixed lobular lesions and breast ducts have a more significant increase in sHLA-G (131). ELISA also shows that the plasma levels of sHLA-G in patients with liver cancer are higher compared with the control group (134). These results support that sHLA-G can be used for tumor early diagnosis.
In terms of prognosis, the high expression of sHLA-G in patients with non-small cell lung cancer is significantly related to poor overall survival (OS), especially in patients with advanced cancer (135). An interesting phenomenon has been found in the Tunisian population that patients with HLA-G*01:04:01 alleles have elevated plasma levels of sHLA-G, while patients who do not carry HLA G*0105N alleles (cannot encode HLA-G1 protein) express a dramatically reduced level of sHLA-G in plasma (22). In CRC, the prognostic value of plasma sHLA-G is mainly reflected by predicting the risk of liver metastasis in patients with stage II and III CRC. Hauben and colleagues have found that the sHLA-G levels are associated with shorter liver metastasis-free survival (LMFS) in patients with stage II CRC and longer LMFS in patients with stage III CRC. The possible reason can be attributed to the fact that the stage III patients receive chemotherapy before the sample collection, and the tumor cells are damaged (64). Therefore, the differential level of plasma sHLA-G is a predictive biomarker of treatment response to advanced CRC, and it is also a potential prognostic marker.
sHLA-G in Body Fluids of Tumor Patients as Tumor Markers
The source of sHLA-G is related to the inflammatory factors present in the cancer microenvironment and the reduction of NK cells and memory T cells (136, 137). Researchers have revealed that the expression of HLA-G in ascites, saliva, and bronchial lavage fluid can be useful for tumor diagnosis and prognosis (Table 2).
Ascites
Results obtained from Ullah’s group have shown that sHLA-G in ascites is mainly expressed by ascites cells, tumor cells, and stromal cells (39). Moreover, Sun et al. have found that the level of sHLA-G in malignant ascites induced by various solid tumors is significantly higher compared with benign ascites. Especially for ascites caused by gynecological tumors and gastrointestinal tumors, the levels of related tumor markers CEA and CA199 are also elevated in malignant ascites, while its specificity and sensitivity are significantly lower compared with sHLA-G (139). The level of sHLA-G in malignant ascites caused by OC and breast cancer is significantly higher compared with benign ascites. In addition, the specificity, sensitivity, and AUC are greatly improved when the critical value of 13 ng/mL is achieved. Besides, in OC, sHLA-G1/G5 is negatively correlated with CD3-/CD56+ subgroups and CD4+ CD45RO+ memory cells, suggesting that sHLA-G plays a role in the tumor microenvironment by up-regulating T-reg cells and down-regulating NK cells (39). In cytology-negative malignant ascites, sHLA-G also has outstanding diagnostic performance. In 32 cases of cytology-negative ascites, the positive rate of sHLA-G is 75%, which is significantly higher compared with CEA and CA199 (50). Collectively, sHLA-G can be used as an independent indicator for early diagnosis of malignant ascites, and it is helpful to screen for malignant ascites when cytology is negative.
Saliva
Saliva is a body fluid that can be collected easily, quickly, and non-invasively (140). Biomolecules in saliva have been used as promising markers for early diagnosis, detection, and treatment in OSCC (141, 142), breast cancer (143, 144), OC (145), and other tumors. In OSCC, saliva is the body fluid that has the closest contact with oral tumors. Researchers have collected the saliva samples from 22 OSCC patients. Compared with non-metastatic OSCC, the HLA-G level of metastatic OSCC is dramatically elevated and correlated with poor OS (63). The sHLA-G level in CRC patients is also significantly higher compared with healthy controls, especially in patients with stage III-IV tumor (62). In the bronchial lavage fluid of patients with different histological types of lung cancer, the level of sHLA-G is significantly correlated with lower Karnofsky index in metastatic tumors and can be used as a prognostic marker for lung cancer (24). The exudate of OC patients is collected before, during, or after chemotherapy. It is found that the expression of HLA-G is decreased after chemotherapy, and the decreased expression of HLA-G predicts the improvement of patient survival rate. This may be related to the preferential sensitivity of HLA-G-expressing cells (47). Therefore, researchers believe that saliva sHLA-G can be used as a diagnostic and prognostic biomarker for multiple cancers, and it is a less invasive alternative to venipuncture. However, more studies are needed to confirm the significance of saliva sHLA-G as a tumor indicator.
HLA-G Derived From EVs as a Tumor Marker
EVs are composed of growth factors, biologically active lipids, genetic information, and antibodies/ligands/receptors, and they are resistant to RNase, enduing their great potential as a tumor biomarker (146–148). Secreted HLA-G can exist in the form of free sHLA-G or be secreted by EVs and found in various body fluids, such as plasma, ascites, and pleural exudate (96) (Figure 2). Recent studies have found that tumor cells, cytotrophoblast cells, and mesenchymal stem cells can secrete HLA-G-carrying EVs, playing a role in regulating the tumor microenvironment and immunosuppressive function (149). Therefore, HLA-G can be shedding from the cell surface by metalloproteases or released from various cells, incorporated into EVs, and serve as a promising tumor indicator (77, 104, 150).
Riteau et al. have isolated an HLA-G-positive cell line from primary and metastatic lymph node melanomas and named it Fon. For the first time, this melanoma cell line is found to secrete exosomes containing HLA-G1. They speculate that the immune tolerance produced by melanoma-derived HLA-G exosomes may be a method for tumors to regulate host immunity. So far, the specific mechanism of this method is still unclear (150). In breast cancer patients receiving neoadjuvant chemotherapy (NACT), the relationship between exosomes carrying HLA-G and the prognosis of the disease has been evaluated. Before NACT, the sHLA-GEV levels are correlated with circulating stem cell-like tumor cells. The total amount of sHLA-GEV is significantly increased after NACT, and it is related to the disease process, while the total sHLA-G level has nothing to do with the clinical prognosis (48). In addition, HLA-G-bearing EVs released by renal cancer cells damage the differentiation of monocytes into DCs and inhibit the maturation process of DCs. These findings suggest that HLA-G-mediated tumor immune escape mechanisms can spread to HLA-G-negative tumor cells through the EV pathway (151, 152). In epithelial ovarian cancer (EOC), the levels of HLA-GEV are increased by seven times compared with healthy controls, and the elevated HLA-GEV can serve as independent 3-year and 10-year progression-free survival (PFS) prognostic factors. It is worth noting that all patients with high levels of HLA-GEV experience disease progression within approximately 5 years after the initial diagnosis. Schwich’s group has also shown that HLA-GEV serves as an independent risk assessment marker for disease progression of EOC (40).
HLA-G Detection and Quantification
Based on the current understanding of HLA-G and its various forms that have multiple immune tolerance regulating functions in malignant tumors, HLA-G is generally recognized as a biomarker that can be used to monitor the disease state and progression in cancer patients (153). However, due to the diversity of HLA-G structures, the standardization of HLA-G detection methods has been a topic of discussion from the past to the present (154). HLA-G test results vary greatly in different locations of HLA-G acquisition, between different tumors, or the same tumor in different laboratory test results.
Detection of HLA-G in Tissues
The status of HLA-G in tumor tissues is usually detected by IHC. Although IHC is an experimental technique that has been widely used, there is controversy about the use of such a method to detect HLA-G. Because HLA-G has multiple subtypes, and each antibody recognizes only specific epitopes, leading to different staining results (52, 54, 124, 155). In addition, the experimental procedures of IHC also differ greatly, such as the type of antibody used and its dilution, incubation time, and staining evaluation criteria. The differences in treatment history, tumor subtypes, and individual tumor microenvironment between different patients will also affect the evaluation of HLA-G expression levels (156–158).
In 81 patients with colon cancer, Swets and colleagues have used three monoclonal antibodies (4H84, MEM-G/1, and MEM-G/2) to evaluate the expression of HLA-G. In primary tumors, the positive staining rates of HLA-G using monoclonal antibody 4H84, MEM-G/1, and MEM-G/2 are 29%, 6%, and 10%, respectively. They have found that different epitopes of HLA-G detected by different monoclonal antibodies are differentially expressed in CRC tissues (159–161). Although it has been confirmed that 4H84 and MEM-G/1 can recognize all subtypes of HLA-G, the reason for this difference in expression is generally believed to be a cross-reaction with HLA-I (160, 161). For example, 4H84 has been shown to cross-react with the existence of β2M free classic HLA class I molecules on activated leukocytes. This may lead to an overestimation of HLA-G expression in pathological tissues recognized by leukocyte infiltration (such as CRC), leading to differences among studies (162). Therefore, it is recommended to use a variety of different monoclonal antibodies to detect HLA-G.
In addition, whether these antibodies can block the binding of HLA-G to its receptors (ILT2, ILT4, and KIR2DL4) is also a challenge (163). Antibodies targeting the α3 domain and β2M of HLA-G can block the binding of HLA-G to ILT2 and ILT4. The antibody targets the α2 domain and can block the binding of HLA-G to KIR2DL4. It has been confirmed that MEM-G/1 blocks the binding site of HLA-G2 and ILT4, while MEM-G/9 and G233 can bind to HLA-G1, depriving the binding site of ILT2. Besides, antibody 87G can block the interaction between HLA-G1 and its receptor (164). Therefore, to block the interaction of HLA-G with all its receptors, it is necessary to develop an antibody mixture that can recognize all HLA-G subtypes and block the binding of HLA-G to ILT2, ILT4, and KIR2DL4, thereby reducing HLA-G cross-reaction with its receptors.
Detection of sHLA-G in Fluids
sHLA-G is also expressed and released by cancer cells, which is a potential biomarker in the body fluids of cancer patients (62). In particular, sHLA-G in the supernatant of IVF embryos has been considered as an independent factor predicting pregnancy outcome (165–167). Therefore, as early as 2004, in the Wet Workshop for the quantification of sHLA-G, the standardization of sHLA-G detection and quantification methods has been discussed, including sensitivity, standard reagents, and antibody specificity (168). At present, ELISA is mainly employed to quantify the level of sHLA-G in body fluids. The monoclonal antibody MEM-G/9 is used to simultaneously detect the shedding of HLA-G1 and the secreted HLA-G5. MEM-G/9 specifically captures β2M-related HLA-G1 and HLA-G5 as well as polyclonal anti-human β2M antibodies (169). In this seminar, HLA-G-expressing EVs are also discovered. This structure cannot be detected by the combination of MEM-G/9 and anti-β2 but can be detected by an antibody combination of mAb 5A6G7 and W6/32. Given the often-antagonistic composition and complex structure of EVs, the various components carried by EVs may drive the functional activity of EVs expressing HLA-G and eliminate or enhance the immune tolerance function of HLA-G.
ExoQuick™ precipitation is now usually used to extract EVs from samples and to quantify the number of EV particles and the number of vesicular-bound HLA-G (HLA-GEV) (48). The results show that in OC, poor clinical status and presence of CTC, and PFS are associated with elevated HLA-GEV levels (40). In breast cancer patients receiving NACT, high levels of vesicular sHLA-G are also associated with disease progression (48). Given these, future work should focus on the standardization process. Before being applied to routine clinical practice, a larger research cohort, prospective research, and internationally recommended standardized testing methods are needed to verify the application of HLA-G in diseases. Importantly, it is necessary to develop more specific antibodies against HLA-G subtypes, explore new undiscovered HLA-G subtypes, and make full use of existing experimental techniques to evaluate the role of various subtypes in various tumors.
Conclusions
After more than 30 years of research, it has been shown that compared with other tumor markers, HLA-G has unique characteristics as follows: 1) it participates in the immune tolerance network in healthy individuals and tumor patients; 2) it is barely expressed in normal tissues and frequently identified in tumor cells. Given these characteristics of HLA-G, one of the current research goals is to serve as a biomarker for diagnosis, prognosis, and clinical testing. However, many factors affect the interpretation of the clinical significance of HLA-G, such as the number of patients included in the study, different clinical parameters of patients (such as medical history, disease stage, and treatment plan), and differences in testing protocols. To better explain the structural diversity of HLA-G, sHLA-G, HLA-GEV, and their expression and clinical significance in tumors, future efforts should be devoted to studies focusing on multi-center and large sample analysis, and the establishment of standardized and feasible HLA-G detection methods.
Author Contributions
PL, NW and YZ wrote and edited the manuscript. CW and LD edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by grant from the National Natural Science Foundation of China (82002228 to PL, 82072368 to LD), the Key Research and Development Program of Shandong Province (2019GSF108091 to LD), Taishan Scholars Program of Shandong Province awarded to CXW and LD.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Ellis SA, Sargent IL, Redman CW, McMichael AJ. Evidence for a Novel HLA Antigen Found on Human Extravillous Trophoblast and a Choriocarcinoma Cell Line. Immunology (1986) 59(4):595–601.
2. Heinrichs H, Orr HT. HLA Non-A,B,C Class I Genes: Their Structure and Expression. Immunol Res (1990) 9(4):265–74. doi: 10.1007/BF02935526
3. Geraghty DE, Koller BH, Orr HT. A Human Major Histocompatibility Complex Class I Gene That Encodes a Protein With a Shortened Cytoplasmic Segment. Proc Natl Acad Sci USA (1987) 84(24):9145–9. doi: 10.1073/pnas.84.24.9145
4. Shiroishi M, Kuroki K, Ose T, Rasubala L, Shiratori I, Arase H, et al. Efficient Leukocyte Ig-Like Receptor Signaling and Crystal Structure of Disulfide-Linked HLA-G Dimer. J Biol Chem (2006) 281(15):10439–47. doi: 10.1074/jbc.M512305200
5. Ishitani A, Geraghty DE. Alternative Splicing of HLA-G Transcripts Yields Proteins With Primary Structures Resembling Both Class I and Class II Antigens. Proc Natl Acad Sci USA (1992) 89(9):3947–51. doi: 10.1073/pnas.89.9.3947
6. Xu HH, Yan WH, Lin A. The Role of HLA-G in Human Papillomavirus Infections and Cervical Carcinogenesis. Front Immunol (2020) 11:1349. doi: 10.3389/fimmu.2020.01349
7. Frumento G, Franchello S, Palmisano GL, Nicotra MR, Giacomini P, Loke YW, et al. Melanomas and Melanoma Cell Lines do Not Express HLA-G, and the Expression Cannot be Induced by GammaIFN Treatment. Tissue Antigens (2000) 56(1):30–7. doi: 10.1034/j.1399-0039.2000.560104.x
8. HoWangYin KY, Loustau M, Wu J, Alegre E, Daouya M, Caumartin J, et al. Multimeric Structures of HLA-G Isoforms Function Through Differential Binding to LILRB Receptors. Cell Mol Life Sci CMLS (2012) 69(23):4041–9. doi: 10.1007/s00018-012-1069-3
9. Paul P, Cabestre FA, Ibrahim EC, Lefebvre S, Khalil-Daher I, Vazeux G, et al. Identification of HLA-G7 as a New Splice Variant of the HLA-G mRNA and Expression of Soluble HLA-G5, -G6, and -G7 Transcripts in Human Transfected Cells. Hum Immunol (2000) 61(11):1138–49. doi: 10.1016/S0198-8859(00)00197-X
10. Carosella ED, Moreau P, Le Maoult J, Le Discorde M, Dausset J, Rouas-Freiss N. HLA-G Molecules: From Maternal-Fetal Tolerance to Tissue Acceptance. Adv Immunol (2003) 81:199–252. doi: 10.1016/S0065-2776(03)81006-4
11. Rebmann V, da Silva Nardi F, Wagner B, Horn PA. HLA-G as a Tolerogenic Molecule in Transplantation and Pregnancy. J Immunol Res (2014) 2014:297073. doi: 10.1155/2014/297073
12. Newmark CS, Gentry L, Simpson M, Jones T. MMPI Criteria for Diagnosing Schizophrenia. J Pers Assess (1978) 42(4):366–73. doi: 10.1207/s15327752jpa4204_6
13. Jurisicova A, Casper RF, MacLusky NJ, Mills GB, Librach CL. HLA-G Expression During Preimplantation Human Embryo Development. Proc Natl Acad Sci USA (1996) 93(1):161–5. doi: 10.1073/pnas.93.1.161
14. McMaster MT, Librach CL, Zhou Y, Lim KH, Janatpour MJ, DeMars R, et al. Human Placental HLA-G Expression Is Restricted to Differentiated Cytotrophoblasts. J Immunol (1995) 154(8):3771–8.
15. Lehner R, Bobak J, Kim NW, Shroyer AL, Shroyer KR. Localization of Telomerase hTERT Protein and Survivin in Placenta: Relation to Placental Development and Hydatidiform Mole. Obstet Gynecol (2001) 97(6):965–70. doi: 10.1097/00006250-200106000-00018
16. Li F, Ambrosini G, Chu EY, Plescia J, Tognin S, Marchisio PC, et al. Control of Apoptosis and Mitotic Spindle Checkpoint by Survivin. Nature (1998) 396(6711):580–4. doi: 10.1038/25141
17. Kyo S, Takakura M, Kohama T, Inoue M. Telomerase Activity in Human Endometrium. Cancer Res (1997) 57(4):610–4.
18. Loustau M, Anna F, Dréan R, Lecomte M, Langlade-Demoyen P, Caumartin J. HLA-G Neo-Expression on Tumors. Front Immunol (2020) 11:1685. doi: 10.3389/fimmu.2020.01685
19. Paul P, Rouas-Freiss N, Khalil-Daher I, Moreau P, Riteau B, Le Gal FA, et al. HLA-G Expression in Melanoma: A Way for Tumor Cells to Escape From Immunosurveillance. Proc Natl Acad Sci USA (1998) 95(8):4510–5. doi: 10.1073/pnas.95.8.4510
20. Fu J, Mao J, Wang C. The microRNA-152/Human Leukocyte Antigen-G Axis Affects Proliferation and Immune Escape of Non-Small Cell Lung Cancer Cells. J Int Med Res (2020) 48(11):300060520970758. doi: 10.1177/0300060520970758
21. Kowal A, Wiśniewski A, Kuśnierczyk P, Jankowska R. Human Leukocyte Antigen (HLA)-G Gene Polymorphism in Patients With Non-Small Cell Lung Cancer. Thorac Cancer (2015) 6(5):613–9. doi: 10.1111/1759-7714.12232
22. Ben Amor A, Beauchemin K, Faucher MC, Hamzaoui A, Hamzaoui K, Roger M. Human Leukocyte Antigen G Polymorphism and Expression Are Associated With an Increased Risk of Non-Small-Cell Lung Cancer and Advanced Disease Stage. PloS One (2016) 11(8):e0161210. doi: 10.1371/journal.pone.0161210
23. Lin A, Zhu CC, Chen HX, Chen BF, Zhang X, Zhang JG, et al. Clinical Relevance and Functional Implications for Human Leucocyte Antigen-G Expression in Non-Small-Cell Lung Cancer. J Cell Mol Med (2010) 14(9):2318–29. doi: 10.1111/j.1582-4934.2009.00858.x
24. Montilla D, Pérez M, Borges L, Bianchi G, Cova JA. Soluble Human Leukocyte Antigen-G in the Bronchoalveolar Lavage of Lung Cancer Patients. Archivos Bronconeumologia (2016) 52(8):420–4. doi: 10.1016/j.arbr.2016.06.018
25. Yie SM, Yang H, Ye SR, Li K, Dong DD, Lin XM. Expression of Human Leucocyte Antigen G (HLA-G) Is Associated With Prognosis in Non-Small Cell Lung Cancer. Lung Cancer (Amsterdam Netherlands) (2007) 58(2):267–74. doi: 10.1016/j.lungcan.2007.06.011
26. Schütt P, Schütt B, Switala M, Bauer S, Stamatis G, Opalka B, et al. Prognostic Relevance of Soluble Human Leukocyte Antigen-G and Total Human Leukocyte Antigen Class I Molecules in Lung Cancer Patients. Hum Immunol (2010) 71(5):489–95. doi: 10.1016/j.humimm.2010.02.015
27. Yan WH, Liu D, Lu HY, Li YY, Zhang X, Lin A. Significance of Tumour Cell HLA-G5/-G6 Isoform Expression in Discrimination for Adenocarcinoma From Squamous Cell Carcinoma in Lung Cancer Patients. J Cell Mol Med (2015) 19(4):778–85. doi: 10.1111/jcmm.12400
28. Urosevic M, Kurrer MO, Kamarashev J, Mueller B, Weder W, Burg G, et al. Human Leukocyte Antigen G Up-Regulation in Lung Cancer Associates With High-Grade Histology, Human Leukocyte Antigen Class I Loss and Interleukin-10 Production. Am J Pathol (2001) 159(3):817–24. doi: 10.1016/S0002-9440(10)61756-7
29. Murdaca G, Calamaro P, Lantieri F, Pigozzi S, Mastracci L, Grillo F, et al. HLA-G Expression in Gastric Carcinoma: Clinicopathological Correlations and Prognostic Impact. Virchows Archiv an Int J Pathol (2018) 473(4):425–33. doi: 10.1007/s00428-018-2379-0
30. Xu DP, Shi WW, Zhang TT, Lv HY, Li JB, Lin A, et al. Elevation of HLA-G-Expressing DC-10 Cells in Patients With Gastric Cancer. Hum Immunol (2016) 77(9):800–4. doi: 10.1016/j.humimm.2016.01.003
31. Ishigami S, Natsugoe S, Miyazono F, Nakajo A, Tokuda K, Matsumoto M, et al. HLA-G Expression in Gastric Cancer. Anticancer Res (2006) 26(3b):2467–72.
32. Khorrami S, Rahimi R, Mohammadpour H, Bahrami S, Yari F, Poustchi H, et al. Association of HLA-G*01:01:02:01/G*01:04:01 Polymorphism With Gastric Adenocarcinoma. Hum Immunol (2016) 77(2):153–7. doi: 10.1016/j.humimm.2015.11.005
33. Pan YQ, Ruan YY, Peng JB, Han QY, Zhang X, Lin A, et al. Diagnostic Significance of Soluble Human Leukocyte Antigen-G for Gastric Cancer. Hum Immunol (2016) 77(4):317–24. doi: 10.1016/j.humimm.2016.01.009
34. Zhang Y, Liu Y, Lu N, Shan NN, Zheng GX, Zhao SM, et al. Expression of the Genes Encoding Human Leucocyte Antigens-A, -B, -DP, -DQ and -G in Gastric Cancer Patients. J Int Med Res (2010) 38(3):949–56. doi: 10.1177/147323001003800321
35. Yie SM, Yang H, Ye SR, Li K, Dong DD, Lin XM. Expression of Human Leukocyte Antigen G (HLA-G) Correlates With Poor Prognosis in Gastric Carcinoma. Ann Surg Oncol (2007) 14(10):2721–9. doi: 10.1245/s10434-007-9464-y
36. Du L, Xiao X, Wang C, Zhang X, Zheng N, Wang L, et al. Human Leukocyte Antigen-G Is Closely Associated With Tumor Immune Escape in Gastric Cancer by Increasing Local Regulatory T Cells. Cancer Sci (2011) 102(7):1272–80. doi: 10.1111/j.1349-7006.2011.01951.x
37. Peng Y, Xiao J, Li W, Li S, Xie B, He J, et al. Prognostic and Clinicopathological Value of Human Leukocyte Antigen G in Gastrointestinal Cancers: A Meta-Analysis. Front Oncol (2021) 11:642902. doi: 10.3389/fonc.2021.642902
38. Schwich E, Rebmann V, Michita RT, Rohn H, Voncken JW, Horn PA, et al. HLA-G 3' Untranslated Region Variants +3187g/G, +3196g/G and +3035T Define Diametrical Clinical Status and Disease Outcome in Epithelial Ovarian Cancer. Sci Rep (2019) 9(1):5407. doi: 10.1038/s41598-019-41900-z
39. Ullah M, Azazzen D, Kaci R, Benabbou N, Pujade Lauraine E, Pocard M, et al. High Expression of HLA-G in Ovarian Carcinomatosis: The Role of Interleukin-1β. Neoplasia (New York NY) (2019) 21(3):331–42. doi: 10.1016/j.neo.2019.01.001
40. Schwich E, Rebmann V, Horn PA, Celik AA, Bade-Döding C, Kimmig R, et al. Vesicular-Bound HLA-G as a Predictive Marker for Disease Progression in Epithelial Ovarian Cancer. Cancers (2019) 11(8):1106. doi: 10.3390/cancers11081106
41. Babay W, Ben Yahia H, Boujelbene N, Zidi N, Laaribi AB, Kacem D, et al. Clinicopathologic Significance of HLA-G and HLA-E Molecules in Tunisian Patients With Ovarian Carcinoma. Hum Immunol (2018) 79(6):463–70. doi: 10.1016/j.humimm.2018.02.012
42. Zhang X, Han QY, Li JB, Ruan YY, Yan WH, Lin A. Lesion HLA-G5/-G6 Isoforms Expression in Patients With Ovarian Cancer. Hum Immunol (2016) 77(9):780–4. doi: 10.1016/j.humimm.2015.12.003
43. Jung YW, Kim YT, Kim SW, Kim S, Kim JH, Cho NH, et al. Correlation of Human Leukocyte Antigen-G (HLA-G) Expression and Disease Progression in Epithelial Ovarian Cancer. Reprod Sci (Thousand Oaks Calif) (2009) 16(11):1103–11. doi: 10.1177/1933719109342131
44. Lin A, Zhang X, Xu HH, Xu DP, Ruan YY, Yan WH. HLA-G Expression Is Associated With Metastasis and Poor Survival in the Balb/c Nu/Nu Murine Tumor Model With Ovarian Cancer. Int J Cancer (2012) 131(1):150–7. doi: 10.1002/ijc.26375
45. Mach P, Blecharz P, Basta P, Marianowski P, Skret-Magierlo J, Kojs Z, et al. Differences in the Soluble HLA-G Blood Serum Concentration Levels in Patients With Ovarian Cancer and Ovarian and Deep Endometriosis. Am J Reprod Immunol (New York NY 1989) (2010) 63(5):387–95. doi: 10.1111/j.1600-0897.2009.00806.x
46. Rutten MJ, Dijk F, Savci-Heijink CD, Buist MR, Kenter GG, van de Vijver MJ, et al. HLA-G Expression Is an Independent Predictor for Improved Survival in High Grade Ovarian Carcinomas. J Immunol Res (2014) 2014:274584. doi: 10.1155/2014/274584
47. Davidson B, Elstrand MB, McMaster MT, Berner A, Kurman RJ, Risberg B, et al. HLA-G Expression in Effusions Is a Possible Marker of Tumor Susceptibility to Chemotherapy in Ovarian Carcinoma. Gynecologic Oncol (2005) 96(1):42–7. doi: 10.1016/j.ygyno.2004.09.049
48. König L, Kasimir-Bauer S, Hoffmann O, Bittner AK, Wagner B, Manvailer LF, et al. The Prognostic Impact of Soluble and Vesicular HLA-G and Its Relationship to Circulating Tumor Cells in Neoadjuvant Treated Breast Cancer Patients. Hum Immunol (2016) 77(9):791–9. doi: 10.1016/j.humimm.2016.01.002
49. Chen HX, Lin A, Shen CJ, Zhen R, Chen BG, Zhang X, et al. Upregulation of Human Leukocyte Antigen-G Expression and Its Clinical Significance in Ductal Breast Cancer. Hum Immunol (2010) 71(9):892–8. doi: 10.1016/j.humimm.2010.06.009
50. Singer G, Rebmann V, Chen YC, Liu HT, Ali SZ, Reinsberg J, et al. HLA-G Is a Potential Tumor Marker in Malignant Ascites. Clin Cancer Res an Off J Am Assoc Cancer Res (2003) 9(12):4460–4.
51. Kleinberg L, Flørenes VA, Skrede M, Dong HP, Nielsen S, McMaster MT, et al. Expression of HLA-G in Malignant Mesothelioma and Clinically Aggressive Breast Carcinoma. Virchows Archiv an Int J Pathol (2006) 449(1):31–9. doi: 10.1007/s00428-005-0144-7
52. He X, Dong DD, Yie SM, Yang H, Cao M, Ye SR, et al. HLA-G Expression in Human Breast Cancer: Implications for Diagnosis and Prognosis, and Effect on Allocytotoxic Lymphocyte Response After Hormone Treatment In Vitro. Ann Surg Oncol (2010) 17(5):1459–69. doi: 10.1245/s10434-009-0891-9
53. de Kruijf EM, Sajet A, van Nes JG, Natanov R, Putter H, Smit VT, et al. HLA-E and HLA-G Expression in Classical HLA Class I-Negative Tumors Is of Prognostic Value for Clinical Outcome of Early Breast Cancer Patients. J Immunol (Baltimore Md 1950) (2010) 185(12):7452–9. doi: 10.4049/jimmunol.1002629
54. Ramos CS, Gonçalves AS, Marinho LC, Gomes Avelino MA, Saddi VA, Lopes AC, et al. Analysis of HLA-G Gene Polymorphism and Protein Expression in Invasive Breast Ductal Carcinoma. Hum Immunol (2014) 75(7):667–72. doi: 10.1016/j.humimm.2014.04.005
55. Tawfeek GA, Alhassanin S. HLA-G Gene Polymorphism in Egyptian Patients With Non-Hodgkin Lymphoma and Its Clinical Outcome. Immunol Invest (2018) 47(3):315–25. doi: 10.1080/08820139.2018.1430826
56. Gros F, Sebti Y, de Guibert S, Branger B, Bernard M, Fauchet R, et al. Soluble HLA-G Molecules Increase During Acute Leukemia, Especially in Subtypes Affecting Monocytic and Lymphoid Lineages. Neoplasia (New York NY) (2006) 8(3):223–30. doi: 10.1593/neo.05703
57. Urosevic M, Kamarashev J, Burg G, Dummer R. Primary Cutaneous CD8+ and CD56+ T-Cell Lymphomas Express HLA-G and Killer-Cell Inhibitory Ligand, Ilt2. Blood (2004) 103(5):1796–8. doi: 10.1182/blood-2003-10-3372
58. Chen J, Shen B, Jiang Y, Jun L, Zhu M, Chen B, et al. Analysis of Immunoglobulin-Like Transcripts (ILTs) in Lymphocytes With sHLA-G and IL10 From SLE Patients. Clin Exp Med (2013) 13(2):135–42. doi: 10.1007/s10238-012-0185-6
59. Negrini S, Fenoglio D, Parodi A, Kalli F, Battaglia F, Nasi G, et al. Phenotypic Alterations Involved in CD8+ Treg Impairment in Systemic Sclerosis. Front Immunol (2017) 8:18. doi: 10.3389/fimmu.2017.00018
60. Lin A, Yan WH. Human Leukocyte Antigen-G (HLA-G) Expression in Cancers: Roles in Immune Evasion, Metastasis and Target for Therapy. Mol Med (Cambridge Mass) (2015) 21(1):782–91. doi: 10.2119/molmed.2015.00083
61. Jacobsen DP, Lekva T, Moe K, Fjeldstad HES, Johnsen GM, Sugulle M, et al. Pregnancy and Postpartum Levels of Circulating Maternal sHLA-G in Preeclampsia. J Reprod Immunol (2021) 143:103249. doi: 10.1016/j.jri.2020.103249
62. Lázaro-Sánchez AD, Salces-Ortiz P, Velásquez LI, Orozco-Beltrán D, Díaz-Fernández N, Juárez-Marroquí A. HLA-G as a New Tumor Biomarker: Detection of Soluble Isoforms of HLA-G in the Serum and Saliva of Patients With Colorectal Cancer. Clin Trans Oncol Off Publ Fed Spanish Oncol Societies Natl Cancer Institute Mexico (2020) 22(7):1166–71. doi: 10.1007/s12094-019-02244-2
63. Gonçalves AS, Arantes DA, Bernardes VF, Jaeger F, Silva JM, Silva TA, et al. Immunosuppressive Mediators of Oral Squamous Cell Carcinoma in Tumour Samples and Saliva. Hum Immunol (2015) 76(1):52–8. doi: 10.1016/j.humimm.2014.11.002
64. Kirana C, Ruszkiewicz A, Stubbs RS, Hardingham JE, Hewett PJ, Maddern GJ, et al. Soluble HLA-G Is a Differential Prognostic Marker in Sequential Colorectal Cancer Disease Stages. Int J Cancer (2017) 140(11):2577–86. doi: 10.1002/ijc.30667
65. Bertol BC, de Araújo JNG, Sadissou IA, Sonon P, Dias FC, Bortolin RH, et al. Plasma Levels of Soluble HLA-G and Cytokines in Papillary Thyroid Carcinoma Before and After Thyroidectomy. Int J Clin Pract (2020) 74(10):e13585. doi: 10.1111/ijcp.13585
66. Babay W, Boujelbene N, Ben Yahia H, Bortolotti D, Zemni I, Ouzari HI, et al. Prognostic Significance of High Circulating sHLA-G in Ovarian Carcinoma. Hla (2021) 98(4):357–65. doi: 10.1111/tan.14374
67. Dardano A, Rizzo R, Polini A, Stignani M, Tognini S, Pasqualetti G, et al. Soluble Human Leukocyte Antigen-G and Its Insertion/Deletion Polymorphism in Papillary Thyroid Carcinoma: Novel Potential Biomarkers of Disease? J Clin Endocrinol Metab (2012) 97(11):4080–6. doi: 10.1210/jc.2012-2231
68. Zheng J, Xu C, Chu D, Zhang X, Li J, Ji G, et al. Human Leukocyte Antigen G Is Associated With Esophageal Squamous Cell Carcinoma Progression and Poor Prognosis. Immunol Lett (2014) 161(1):13–9. doi: 10.1016/j.imlet.2014.04.007
69. Xu YF, Lu Y, Cheng H, Jiang J, Xu J, Long J, et al. High Expression of Human Leukocyte Antigen-G Is Associated With a Poor Prognosis in Patients With PDAC. Curr Mol Med (2015) 15(4):360–7. doi: 10.2174/1566524015666150401102218
70. Mallet V, Blaschitz A, Crisa L, Schmitt C, Fournel S, King A, et al. HLA-G in the Human Thymus: A Subpopulation of Medullary Epithelial But Not CD83(+) Dendritic Cells Expresses HLA-G as a Membrane-Bound and Soluble Protein. Int Immunol (1999) 11(6):889–98. doi: 10.1093/intimm/11.6.889
71. Larsen MH, Bzorek M, Pass MB, Larsen LG, Nielsen MW, Svendsen SG, et al. Human Leukocyte Antigen-G in the Male Reproductive System and in Seminal Plasma. Mol Hum Reprod (2011) 17(12):727–38. doi: 10.1093/molehr/gar052
72. Fainardi E, Rizzo R, Melchiorri L, Vaghi L, Castellazzi M, Marzola A, et al. Presence of Detectable Levels of Soluble HLA-G Molecules in CSF of Relapsing-Remitting Multiple Sclerosis: Relationship With CSF Soluble HLA-I and IL-10 Concentrations and MRI Findings. J Neuroimmunol (2003) 142(1-2):149–58. doi: 10.1016/S0165-5728(03)00266-2
73. Fainardi E, Rizzo R, Melchiorri L, Stignani M, Castellazzi M, Tamborino C, et al. CSF Levels of Soluble HLA-G and Fas Molecules Are Inversely Associated to MRI Evidence of Disease Activity in Patients With Relapsing-Remitting Multiple Sclerosis. Multiple Sclerosis (Houndmills Basingstoke England) (2008) 14(4):446–54. doi: 10.1177/1352458507085137
74. Blaschitz A, Juch H, Volz A, Hutter H, Daxboeck C, Desoye G, et al. The Soluble Pool of HLA-G Produced by Human Trophoblasts Does Not Include Detectable Levels of the Intron 4-Containing HLA-G5 and HLA-G6 Isoforms. Mol Hum Reprod (2005) 11(10):699–710. doi: 10.1093/molehr/gah185
75. Fuzzi B, Rizzo R, Criscuoli L, Noci I, Melchiorri L, Scarselli B, et al. HLA-G Expression in Early Embryos Is a Fundamental Prerequisite for the Obtainment of Pregnancy. Eur J Immunol (2002) 32(2):311–5. doi: 10.1002/1521-4141(200202)32:2<311::AID-IMMU311>3.0.CO;2-8
76. Noci I, Fuzzi B, Rizzo R, Melchiorri L, Criscuoli L, Dabizzi S, et al. Embryonic Soluble HLA-G as a Marker of Developmental Potential in Embryos. Hum Reprod (Oxford England) (2005) 20(1):138–46. doi: 10.1093/humrep/deh572
77. Rebmann V, König L, Nardi Fda S, Wagner B, Manvailer LF, Horn PA. The Potential of HLA-G-Bearing Extracellular Vesicles as a Future Element in HLA-G Immune Biology. Front Immunol (2016) 7:173. doi: 10.3389/fimmu.2016.00173
78. Carosella ED, Horuzsko A. HLA-G and Cancer. Semin Cancer Biol (2007) 17(6):411–2. doi: 10.1016/j.semcancer.2007.06.014
79. Kim R, Emi M, Tanabe K. Cancer Immunoediting From Immune Surveillance to Immune Escape. Immunology (2007) 121(1):1–14. doi: 10.1016/B978-012372551-6/50066-3
80. Rodríguez JA. HLA-Mediated Tumor Escape Mechanisms That may Impair Immunotherapy Clinical Outcomes via T-Cell Activation. Oncol Lett (2017) 14(4):4415–27. doi: 10.3892/ol.2017.6784
81. Dunn GP, Old LJ, Schreiber RD. The Three Es of Cancer Immunoediting. Annu Rev Immunol (2004) 22:329–60. doi: 10.1146/annurev.immunol.22.012703.104803
82. Naji A, Menier C, Maki G, Carosella ED, Rouas-Freiss N. Neoplastic B-Cell Growth Is Impaired by HLA-G/ILT2 Interaction. Leukemia (2012) 26(8):1889–92. doi: 10.1038/leu.2012.62
83. Własiuk P, Putowski M, Giannopoulos K. PD1/PD1L Pathway, HLA-G and T Regulatory Cells as New Markers of Immunosuppression in Cancers. Postepy Higieny i Medycyny Doswiadczalnej (Online) (2016) 70(0):1044–58. doi: 10.5604/17322693.1220994
84. Persson G, Jørgensen N, Nilsson LL, Andersen LHJ, Hviid TVF. A Role for Both HLA-F and HLA-G in Reproduction and During Pregnancy? Hum Immunol (2020) 81(4):127–33. doi: 10.1016/j.humimm.2019.09.006
85. Morel E, Bellón T. HLA Class I Molecules Regulate IFN-Gamma Production Induced in NK Cells by Target Cells, Viral Products, or Immature Dendritic Cells Through the Inhibitory Receptor ILT2/CD85j. J Immunol (Baltimore Md 1950) (2008) 181(4):2368–81. doi: 10.4049/jimmunol.181.4.2368
86. Liang S, Ristich V, Arase H, Dausset J, Carosella ED, Horuzsko A. Modulation of Dendritic Cell Differentiation by HLA-G and ILT4 Requires the IL-6–STAT3 Signaling Pathway. Proc Natl Acad Sci USA (2008) 105(24):8357–62. doi: 10.1073/pnas.0803341105
87. Baudhuin J, Migraine J, Faivre V, Loumagne L, Lukaszewicz AC, Payen D, et al. Exocytosis Acts as a Modulator of the ILT4-Mediated Inhibition of Neutrophil Functions. Proc Natl Acad Sci USA (2013) 110(44):17957–62. doi: 10.1073/pnas.1221535110
88. LeMaoult J, Zafaranloo K, Le Danff C, Carosella ED. HLA-G Up-Regulates ILT2, ILT3, ILT4, and KIR2DL4 in Antigen Presenting Cells, NK Cells, and T Cells. FASEB J Off Publ Fed Am Societies Exp Biol (2005) 19(6):662–4. doi: 10.1096/fj.04-1617fje
89. Bai Y, Liang J, Liu W, Wang F, Li C. Possible Roles of HLA-G Regulating Immune Cells in Pregnancy and Endometrial Diseases via KIR2DL4. J Reprod Immunol (2020) 142:103176. doi: 10.1016/j.jri.2020.103176
90. Lee CL, Guo Y, So KH, Vijayan M, Guo Y, Wong VH, et al. Soluble Human Leukocyte Antigen G5 Polarizes Differentiation of Macrophages Toward a Decidual Macrophage-Like Phenotype. Hum Reprod (Oxford England) (2015) 30(10):2263–74. doi: 10.1093/humrep/dev196
91. Morandi F, Rouas-Freiss N, Pistoia V. The Emerging Role of Soluble HLA-G in the Control of Chemotaxis. Cytokine Growth Factor Rev (2014) 25(3):327–35. doi: 10.1016/j.cytogfr.2014.04.004
92. Gregori S, Tomasoni D, Pacciani V, Scirpoli M, Battaglia M, Magnani CF, et al. Differentiation of Type 1 T Regulatory Cells (Tr1) by Tolerogenic DC-10 Requires the IL-10-Dependent ILT4/HLA-G Pathway. Blood (2010) 116(6):935–44. doi: 10.1182/blood-2009-07-234872
93. Fainardi E, Castellazzi M, Stignani M, Morandi F, Sana G, Gonzalez R, et al. Emerging Topics and New Perspectives on HLA-G. Cell Mol Life Sci CMLS (2011) 68(3):433–51. doi: 10.1007/s00018-010-0584-3
94. Carosella ED, Rouas-Freiss N, Tronik-Le Roux D, Moreau P, LeMaoult J. HLA-G: An Immune Checkpoint Molecule. Adv Immunol (2015) 127:33–144. doi: 10.1016/bs.ai.2015.04.001
95. Lin A, Yan WH. Intercellular Transfer of HLA-G: Its Potential in Cancer Immunology. Clin Trans Immunol (2019) 8(9):e1077. doi: 10.1002/cti2.1077
96. Alegre E, Rebmann V, Lemaoult J, Rodriguez C, Horn PA, Díaz-Lagares A, et al. In Vivo Identification of an HLA-G Complex as Ubiquitinated Protein Circulating in Exosomes. Eur J Immunol (2013) 43(7):1933–9. doi: 10.1002/eji.201343318
97. Brown R, Kabani K, Favaloro J, Yang S, Ho PJ, Gibson J, et al. CD86+ or HLA-G+ can be Transferred via Trogocytosis From Myeloma Cells to T Cells and Are Associated With Poor Prognosis. Blood (2012) 120(10):2055–63. doi: 10.1182/blood-2012-03-416792
98. LeMaoult J, Caumartin J, Daouya M, Switala M, Rebmann V, Arnulf B, et al. Trogocytic Intercellular Membrane Exchanges Among Hematological Tumors. J Hematol Oncol (2015) 8:24. doi: 10.1186/s13045-015-0114-8
99. Tilburgs T, Evans JH, Crespo ÂC, Strominger JL. The HLA-G Cycle Provides for Both NK Tolerance and Immunity at the Maternal-Fetal Interface. Proc Natl Acad Sci USA (2015) 112(43):13312–7. doi: 10.1073/pnas.1517724112
100. Han L, Lam EW, Sun Y. Extracellular Vesicles in the Tumor Microenvironment: Old Stories, But New Tales. Mol Cancer (2019) 18(1):59. doi: 10.1186/s12943-019-0980-8
101. Simon T, Jackson E, Giamas G. Breaking Through the Glioblastoma Micro-Environment via Extracellular Vesicles. Oncogene (2020) 39(23):4477–90. doi: 10.1038/s41388-020-1308-2
102. Colombo M, Raposo G, Théry C. Biogenesis, Secretion, and Intercellular Interactions of Exosomes and Other Extracellular Vesicles. Annu Rev Cell Dev Biol (2014) 30:255–89. doi: 10.1146/annurev-cellbio-101512-122326
103. Wu T, Dai Y. Tumor Microenvironment and Therapeutic Response. Cancer Lett (2017) 387:61–8. doi: 10.1016/j.canlet.2016.01.043
104. Curigliano G, Criscitiello C, Gelao L, Goldhirsch A. Molecular Pathways: Human Leukocyte Antigen G (HLA-G). Clin Cancer Res an Off J Am Assoc Cancer Res (2013) 19(20):5564–71. doi: 10.1158/1078-0432.CCR-12-3697
105. Amiot L, Vu N, Samson M. Biology of the Immunomodulatory Molecule HLA-G in Human Liver Diseases. J Hepatol (2015) 62(6):1430–7. doi: 10.1016/j.jhep.2015.03.007
106. Khodabandeh Shahraki P, Zare Y, Azarpira N, Hosseinzadeh M, Farjadian S. Prognostic Value of HLA-G in Malignant Liver and Pancreas Lesions. Iranian J Immunol IJI (2018) 15(1):28–37.
107. Shen X, Wang P, Dai P, Jin B, Tong Y, Lin H, et al. Correlation Between Human Leukocyte Antigen-G Expression and Clinical Parameters in Oral Squamous Cell Carcinoma. Indian J Cancer (2018) 55(4):340–3. doi: 10.4103/ijc.IJC_602_17
108. Zeestraten EC, Reimers MS, Saadatmand S, Goossens-Beumer IJ, Dekker JW, Liefers GJ, et al. HLA-E and HLA-G Predicts Prognosis in Colon Cancer Patients. Br J Cancer (2014) 110(2):459–68. doi: 10.1038/bjc.2013.696
109. Wang Y, Ye Z, Meng XQ, Zheng SS. Expression of HLA-G in Patients With Hepatocellular Carcinoma. Hepatobil Pancreatic Dis Int HBPD Int (2011) 10(2):158–63. doi: 10.1016/S1499-3872(11)60025-8
110. de Figueiredo Feitosa NL, Crispim JC, Zanetti BR, Magalhães PK, Soares CP, Soares EG, et al. HLA-G Is Differentially Expressed in Thyroid Tissues. Thyroid Off J Am Thyroid Assoc (2014) 24(3):585–92. doi: 10.1089/thy.2013.0246
111. Ferguson R, Ramanakumar AV, Koushik A, Coutlée F, Franco E, Roger M. Human Leukocyte Antigen G Polymorphism Is Associated With an Increased Risk of Invasive Cancer of the Uterine Cervix. Int J Cancer (2012) 131(3):E312–9. doi: 10.1002/ijc.27356
112. Rodríguez JA, Galeano L, Palacios DM, Gómez C, Serrano ML, Bravo MM, et al. Altered HLA Class I and HLA-G Expression Is Associated With IL-10 Expression in Patients With Cervical Cancer. Pathobiol J Immunopathol Mol Cell Biol (2012) 79(2):72–83. doi: 10.1159/000334089
113. Cai Z, Wang L, Han Y, Gao W, Wei X, Gong R, et al. Immunoglobulin−like Transcript 4 and Human Leukocyte Antigen−G Interaction Promotes the Progression of Human Colorectal Cancer. Int J Oncol (2019) 54(6):1943–54. doi: 10.3892/ijo.2019.4761
114. Cai MB, Han HQ, Bei JX, Liu CC, Lei JJ, Cui Q, et al. Expression of Human Leukocyte Antigen G Is Associated With Prognosis in Nasopharyngeal Carcinoma. Int J Biol Sci (2012) 8(6):891–900. doi: 10.7150/ijbs.4383
115. Lin A, Zhang X, Zhou WJ, Ruan YY, Xu DP, Wang Q, et al. Human Leukocyte Antigen-G Expression Is Associated With a Poor Prognosis in Patients With Esophageal Squamous Cell Carcinoma. Int J Cancer (2011) 129(6):1382–90. doi: 10.1002/ijc.25807
116. Wan R, Wang ZW, Li H, Peng XD, Liu GY, Ou JM, et al. Human Leukocyte Antigen-G Inhibits the Anti-Tumor Effect of Natural Killer Cells via Immunoglobulin-Like Transcript 2 in Gastric Cancer. Cell Physiol Biochem Int J Exp Cell Physiol Biochem Pharmacol (2017) 44(5):1828–41. doi: 10.1159/000485819
117. Vangangelt KMH, van Pelt GW, Engels CC, Putter H, Liefers GJ, Smit V, et al. Prognostic Value of Tumor-Stroma Ratio Combined With the Immune Status of Tumors in Invasive Breast Carcinoma. Breast Cancer Res Treat (2018) 168(3):601–12. doi: 10.1007/s10549-017-4617-6
118. Ishibashi K, Kumai T, Ohkuri T, Kosaka A, Nagato T, Hirata Y, et al. Epigenetic Modification Augments the Immunogenicity of Human Leukocyte Antigen G Serving as a Tumor Antigen for T Cell-Based Immunotherapy. Oncoimmunology (2016) 5(6):e1169356. doi: 10.1080/2162402X.2016.1169356
119. Andersson E, Poschke I, Villabona L, Carlson JW, Lundqvist A, Kiessling R, et al. Non-Classical HLA-Class I Expression in Serous Ovarian Carcinoma: Correlation With the HLA-Genotype, Tumor Infiltrating Immune Cells and Prognosis. Oncoimmunology (2016) 5(1):e1052213. doi: 10.1080/2162402X.2015.1052213
120. Fan X, Wang Y, Zhang C, Liu X, Qian Z, Jiang T. Human Leukocyte Antigen-G Overexpression Predicts Poor Clinical Outcomes in Low-Grade Gliomas. J Neuroimmunol (2016) 294:27–31. doi: 10.1016/j.jneuroim.2016.03.015
121. Dong DD, Yie SM, Li K, Li F, Xu Y, Xu G, et al. Importance of HLA-G Expression and Tumor Infiltrating Lymphocytes in Molecular Subtypes of Breast Cancer. Hum Immunol (2012) 73(10):998–1004. doi: 10.1016/j.humimm.2012.07.321
122. Rouas-Freiss N, Moreau P, Ferrone S, Carosella ED. HLA-G Proteins in Cancer: Do They Provide Tumor Cells With an Escape Mechanism? Cancer Res (2005) 65(22):10139–44. doi: 10.1158/0008-5472.CAN-05-0097
123. Ben Yahia H, Babay W, Bortolotti D, Boujelbene N, Laaribi AB, Zidi N, et al. Increased Plasmatic Soluble HLA-G Levels in Endometrial Cancer. Mol Immunol (2018) 99:82–6. doi: 10.1016/j.molimm.2018.04.007
124. Jeong S, Park S, Park BW, Park Y, Kwon OJ, Kim HS. Human Leukocyte Antigen-G (HLA-G) Polymorphism and Expression in Breast Cancer Patients. PloS One (2014) 9(5):e98284. doi: 10.1371/journal.pone.0098284
125. Zhu CB, Wang CX, Zhang X, Zhang J, Li W. Serum sHLA-G Levels: A Useful Indicator in Distinguishing Colorectal Cancer From Benign Colorectal Diseases. Int J Cancer (2011) 128(3):617–22. doi: 10.1002/ijc.25372
126. Tan B, Guo J, Wang L, Wang L, Gan X, Chen B. Expression and Change of miR-199b-5p, s HLA-G in Thyroid Carcinoma. Exp Mol Pathol (2021) 120:104643. doi: 10.1016/j.yexmp.2021.104643
127. Wang Z, Zhao L, Liu L, Liu X. Human Leucocyte Antigen-G 14-Bp InDel Polymorphism and Oral Squamous Cell Carcinoma Risk in Chinese Han Population: A Case-Control Study. Int J Immunogenet (2018). doi: 10.1111/iji.12390
128. Agnihotri V, Gupta A, Kumar R, Upadhyay AD, Dwivedi S, Kumar L, et al. Promising Link of HLA-G Polymorphism, Tobacco Consumption and Risk of Head and Neck Squamous Cell Carcinoma (HNSCC) in North Indian Population. Hum Immunol (2017) 78(2):172–8. doi: 10.1016/j.humimm.2016.12.007
129. Agnihotri V, Gupta A, Kumar L, Dey S. Serum sHLA-G: Significant Diagnostic Biomarker With Respect to Therapy and Immunosuppressive Mediators in Head and Neck Squamous Cell Carcinoma. Sci Rep (2020) 10(1):3806. doi: 10.1038/s41598-020-60811-y
130. Chen Y, Gao XJ, Deng YC, Zhang HX. Relationship Between HLA-G Gene Polymorphism and the Susceptibility of Esophageal Cancer in Kazakh and Han Nationality in Xinjiang. Biomarkers Biochem Indic Exposure Response Susceptibility to Chemicals (2012) 17(1):9–15. doi: 10.3109/1354750X.2011.633242
131. Provatopoulou X, Kalogera E, Sagkriotis A, Zagouri F, Nonni A, Zografos GC, et al. Soluble Human Leukocyte Antigen-G Expression in Patients With Ductal and Lobular Breast Malignancy. Anticancer Res (2012) 32(3):1021–6.
132. Farjadian S, Tabebordbar M, Mokhtari M, Safaei A, Malekzadeh M, Ghaderi A. HLA-G Expression in Tumor Tissues and Soluble HLA-G Plasma Levels in Patients With Gastrointestinal Cancer. Asian Pacific J Cancer Prev APJCP (2018) 19(10):2731–5. doi: 10.22034/APJCP.2018.19.10.2731
133. Wuerfel FM, Huebner H, Häberle L, Gass P, Hein A, Jud SM, et al. HLA-G and HLA-F Protein Isoform Expression in Breast Cancer Patients Receiving Neoadjuvant Treatment. Sci Rep (2020) 10(1):15750. doi: 10.1038/s41598-020-72837-3
134. Park Y, Park Y, Lim HS, Kim YS, Hong DJ, Kim HS. Soluble Human Leukocyte Antigen-G Expression in Hepatitis B Virus Infection and Hepatocellular Carcinoma. Tissue Antigens (2012) 79(2):97–103. doi: 10.1111/j.1399-0039.2011.01814.x
135. Cai MY, Xu YF, Qiu SJ, Ju MJ, Gao Q, Li YW, et al. Human Leukocyte Antigen-G Protein Expression Is an Unfavorable Prognostic Predictor of Hepatocellular Carcinoma Following Curative Resection. Clin Cancer Res an Off J Am Assoc Cancer Res (2009) 15(14):4686–93. doi: 10.1158/1078-0432.CCR-09-0463
136. Fu B, Zhou Y, Ni X, Tong X, Xu X, Dong Z, et al. Natural Killer Cells Promote Fetal Development Through the Secretion of Growth-Promoting Factors. Immunity (2017) 47(6):1100–13.e6. doi: 10.1016/j.immuni.2017.11.018
137. Anna F, Bole-Richard E, LeMaoult J, Escande M, Lecomte M, Certoux JM, et al. First Immunotherapeutic CAR-T Cells Against the Immune Checkpoint Protein HLA-G. J Immunother Cancer (2021) 9(3):e001998. doi: 10.1136/jitc-2020-001998
138. Chen QY, Chen YX, Han QY, Zhang JG, Zhou WJ, Zhang X, et al. Prognostic Significance of Immune Checkpoints HLA-G/ILT-2/4 and PD-L1 in Colorectal Cancer. Front Immunol (2021) 12:679090. doi: 10.3389/fimmu.2021.679090
139. Sun J, Chang YX, Niu CY. Evaluation of Ascitic Soluble Human Leukocyte Antigen-G for Distinguishing Malignant Ascites From Benign Ascites. Tumour Biol J Int Soc Oncodevelopmental Biol Med (2017) 39(11):1010428317726840. doi: 10.1177/1010428317726840
140. Grande MA, Belstrøm D, Damgaard C, Holmstrup P, Thangaraj SS, Nielsen CH, et al. Complement Split Product C3c in Saliva as Biomarker for Periodontitis and Response to Periodontal Treatment. J Periodontal Res (2021) 56(1):27–33. doi: 10.1111/jre.12788
141. Cristaldi M, Mauceri R, Di Fede O, Giuliana G, Campisi G, Panzarella V. Salivary Biomarkers for Oral Squamous Cell Carcinoma Diagnosis and Follow-Up: Current Status and Perspectives. Front Physiol (2019) 10:1476. doi: 10.3389/fphys.2019.01476
142. Song X, Yang X, Narayanan R, Shankar V, Ethiraj S, Wang X, et al. Oral Squamous Cell Carcinoma Diagnosed From Saliva Metabolic Profiling. Proc Natl Acad Sci USA (2020) 117(28):16167–73. doi: 10.1073/pnas.2001395117
143. Bentata M, Morgenstern G, Nevo Y, Kay G, Granit Mizrahi A, Temper M, et al. Splicing Factor Transcript Abundance in Saliva as a Diagnostic Tool for Breast Cancer. Genes (2020) 11(8):880. doi: 10.3390/genes11080880
144. Delmonico L, Bravo M, Silvestre RT, Ornellas MH, De Azevedo CM, Alves G. Proteomic Profile of Saliva and Plasma From Women With Impalpable Breast Lesions. Oncol Lett (2016) 12(3):2145–52. doi: 10.3892/ol.2016.4828
145. Zermeño-Nava JJ, Martínez-Martínez MU, Rámirez-de-Ávila AL, Hernández-Arteaga AC, García-Valdivieso MG, Hernández-Cedillo A, et al. Determination of Sialic Acid in Saliva by Means of Surface-Enhanced Raman Spectroscopy as a Marker in Adnexal Mass Patients: Ovarian Cancer vs Benign Cases. J Ovarian Res (2018) 11(1):61. doi: 10.1186/s13048-018-0433-9
146. ELA S, Mäger I, Breakefield XO, Wood MJ. Extracellular Vesicles: Biology and Emerging Therapeutic Opportunities. Nat Rev Drug Discovery (2013) 12(5):347–57. doi: 10.1038/nrd3978
147. Mittelbrunn M, Gutiérrez-Vázquez C, Villarroya-Beltri C, González S, Sánchez-Cabo F, González M, et al. Unidirectional Transfer of microRNA-Loaded Exosomes From T Cells to Antigen-Presenting Cells. Nat Commun (2011) 2:282. doi: 10.1038/ncomms1285
148. Ludwig AK, Giebel B. Exosomes: Small Vesicles Participating in Intercellular Communication. Int J Biochem Cell Biol (2012) 44(1):11–5. doi: 10.1016/j.biocel.2011.10.005
149. Yáñez-Mó M, Siljander PR, Andreu Z, Zavec AB, Borràs FE, Buzas EI, et al. Biological Properties of Extracellular Vesicles and Their Physiological Functions. J Extracell Vesicles (2015) 4:27066. doi: 10.3402/jev.v4.27066
150. Riteau B, Faure F, Menier C, Viel S, Carosella ED, Amigorena S, et al. Exosomes Bearing HLA-G Are Released by Melanoma Cells. Hum Immunol (2003) 64(11):1064–72. doi: 10.1016/j.humimm.2003.08.344
151. García M, Palma MB, Verine J, Miriuka S, Inda AM, Errecalde AL, et al. The Immune-Checkpoint HLA-G/ILT4 Is Involved in the Regulation of VEGF Expression in Clear Cell Renal Cell Carcinoma. BMC Cancer (2020) 20(1):624. doi: 10.1186/s12885-020-07113-8
152. Grange C, Tapparo M, Tritta S, Deregibus MC, Battaglia A, Gontero P, et al. Role of HLA-G and Extracellular Vesicles in Renal Cancer Stem Cell-Induced Inhibition of Dendritic Cell Differentiation. BMC Cancer (2015) 15:1009. doi: 10.1186/s12885-015-2025-z
153. González A, Rebmann V, LeMaoult J, Horn PA, Carosella ED, Alegre E. The Immunosuppressive Molecule HLA-G and Its Clinical Implications. Crit Rev Clin Lab Sci (2012) 49(3):63–84. doi: 10.3109/10408363.2012.677947
154. Nardi Fda S, König L, Wagner B, Giebel B, Santos Manvailer LF, Rebmann V. Soluble Monomers, Dimers and HLA-G-Expressing Extracellular Vesicles: The Three Dimensions of Structural Complexity to Use HLA-G as a Clinical Biomarker. Hla (2016) 88(3):77–86. doi: 10.1111/tan.12844
155. da Silva GB, Silva TG, Duarte RA, Neto NL, Carrara HH, Donadi EA, et al. Expression of the Classical and Nonclassical HLA Molecules in Breast Cancer. Int J Breast Cancer (2013) 2013:250435. doi: 10.1155/2013/250435
156. Lin A, Yan WH. Heterogeneity of HLA-G Expression in Cancers: Facing the Challenges. Front Immunol (2018) 9:2164. doi: 10.3389/fimmu.2018.02164
157. Anastassiou G, Rebmann V, Wagner S, Bornfeld N, Grosse-Wilde H. Expression of Classic and Nonclassic HLA Class I Antigens in Uveal Melanoma. Invest Ophthalmol Visual Sci (2003) 44(5):2016–9. doi: 10.1167/iovs.02-0810
158. Goldman-Wohl D, Ariel I, Greenfield C, Hochner-Celnikier D, Lavy Y, Yagel S. A Study of Human Leukocyte Antigen G Expression in Hydatidiform Moles. Am J Obstet Gynecol (2001) 185(2):476–80. doi: 10.1067/mob.2001.115994
159. Swets M, König MH, Zaalberg A, Dekker-Ensink NG, Gelderblom H, van de Velde CJ, et al. HLA-G and Classical HLA Class I Expression in Primary Colorectal Cancer and Associated Liver Metastases. Hum Immunol (2016) 77(9):773–9. doi: 10.1016/j.humimm.2016.03.001
160. Swets M, Wouters A, Krijgsman D, van Vlierberghe RLP, Boot A, van Eendenburg JD, et al. HLA-G Protein Expression in Colorectal Cancer Evaluated by Immunohistochemistry and Western Blot Analysis: Its Expression Characteristics Remain Enigmatic. Clin Immunol (Orlando Fla) (2018) 194:80–6. doi: 10.1016/j.clim.2018.07.005
161. Poláková K, Kuba D, Russ G. The 4h84 Monoclonal Antibody Detecting Beta2m Free Nonclassical HLA-G Molecules Also Binds to Free Heavy Chains of Classical HLA Class I Antigens Present on Activated Lymphocytes. Hum Immunol (2004) 65(2):157–62. doi: 10.1016/j.humimm.2003.10.005
162. Apps R, Gardner L, Moffett A. A Critical Look at HLA-G. Trends Immunol (2008) 29(7):313–21. doi: 10.1016/j.it.2008.02.012
163. Attia JVD, Dessens CE, van de Water R, Houvast RD, Kuppen PJK, Krijgsman D. The Molecular and Functional Characteristics of HLA-G and the Interaction With Its Receptors: Where to Intervene for Cancer Immunotherapy? Int J Mol Sci (2020) 21(22):8678. doi: 10.3390/ijms21228678
164. Furukawa A, Meguro M, Yamazaki R, Watanabe H, Takahashi A, Kuroki K, et al. Evaluation of the Reactivity and Receptor Competition of HLA-G Isoforms Toward Available Antibodies: Implications of Structural Characteristics of HLA-G Isoforms. Int J Mol Sci (2019) 20(23):5947. doi: 10.3390/ijms20235947
165. Rebmann V, Switala M, Eue I, Grosse-Wilde H. Soluble HLA-G Is an Independent Factor for the Prediction of Pregnancy Outcome After ART: A German Multi-Centre Study. Hum Reprod (Oxford England) (2010) 25(7):1691–8. doi: 10.1093/humrep/deq120
166. Rebmann V, Switala M, Eue I, Schwahn E, Merzenich M, Grosse-Wilde H. Rapid Evaluation of Soluble HLA-G Levels in Supernatants of In Vitro Fertilized Embryos. Hum Immunol (2007) 68(4):251–8. doi: 10.1016/j.humimm.2006.11.003
167. Rizzo R, Vercammen M, van de Velde H, Horn PA, Rebmann V. The Importance of HLA-G Expression in Embryos, Trophoblast Cells, and Embryonic Stem Cells. Cell Mol Life Sci CMLS (2011) 68(3):341–52. doi: 10.1007/s00018-010-0578-1
168. Rebmann V, Lemaoult J, Rouas-Freiss N, Carosella ED, Grosse-Wilde H. Report of the Wet Workshop for Quantification of Soluble HLA-G in Essen, 2004. Hum Immunol (2005) 66(8):853–63. doi: 10.1016/j.humimm.2005.05.003
Keywords: HLA-G, tumor, immune escape, extracellular vesicles, biomarker
Citation: Li P, Wang N, Zhang Y, Wang C and Du L (2021) HLA-G/sHLA-G and HLA-G-Bearing Extracellular Vesicles in Cancers: Potential Role as Biomarkers. Front. Immunol. 12:791535. doi: 10.3389/fimmu.2021.791535
Received: 08 October 2021; Accepted: 25 October 2021;
Published: 11 November 2021.
Edited by:
Wei-Hua Yan, Wenzhou Medical University, ChinaReviewed by:
Giuseppe Murdaca, University of Genoa, ItalySilvia Gregori, San Raffaele Telethon Institute for Gene Therapy (SR-Tiget), Italy
Copyright © 2021 Li, Wang, Zhang, Wang and Du. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuanxin Wang, Y3h3YW5nQHNkdS5lZHUuY24=; Lutao Du, bHV0YW9kdUBzZHUuZWR1LmNu
†These authors have contributed equally to this work
 Peilong Li
Peilong Li Nan Wang
Nan Wang Yi Zhang5
Yi Zhang5 Chuanxin Wang
Chuanxin Wang Lutao Du
Lutao Du