
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 24 November 2021
Sec. Cytokines and Soluble Mediators in Immunity
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.790379
This article is part of the Research Topic Harnessing the Complexity of Normal and Pathological Hematopoietic Supportive Niches View all 13 articles
The journey of a hematopoietic stem cell (HSC) involves the passage through successive anatomical sites where HSCs are in direct contact with their surrounding microenvironment, also known as niche. These spatial and temporal cellular interactions throughout development are required for the acquisition of stem cell properties, and for maintaining the HSC pool through balancing self-renewal, quiescence and lineage commitment. Understanding the context and consequences of these interactions will be imperative for our understanding of HSC biology and will lead to the improvement of in vitro production of HSCs for clinical purposes. The aorta-gonad-mesonephros (AGM) region is in this light of particular interest since this is the cradle of HSC emergence during the embryonic development of all vertebrate species. In this review, we will focus on the developmental origin of HSCs and will discuss the novel technological approaches and recent progress made to identify the cellular composition of the HSC supportive niche and the underlying molecular events occurring in the AGM region.
The origin of the hematopoietic system lies within the early developing embryo (1–5). Successive waves give rise to hematopoietic stem and progenitor cells (HSPCs) with various lineage potentials and self-renewal capacities. While initially a pool of short-lived differentiated cells is formed to sustain the fast-growing embryo, multilineage and self-renewing hematopoietic stem cells (HSCs) are then produced to support long-term hematopoiesis. HSCs are first detected in the aorta-gonad-mesonephros (AGM) region prior to colonize mainly the fetal liver (and also the placenta) where they mature and expand through self-renewal. Shortly before birth, HSCs emigrate from the fetal liver and home to the bone marrow (BM) where they form the pool of adult HSCs that will participate to the replenishment of all blood lineages for the remaining life of the organism. For long, stem cells were considered as independent entities able to self-regulate their own behavior. Four decades ago, Schofield was the first to postulate that stem cells are not complete autonomous entities, as they require external signals from the local microenvironment or niche to regulate their behavior and fate decisions to either remain quiescent, to self-renew or to differentiate in response to the need of the organism (6). HSCs are in this regard unique as their formation requires sequential interactions with distinct anatomical sites throughout development and in adults, e.g. the AGM, fetal liver and BM. While HSC niches are well documented in the adult [reviewed in (7–9)], this is far from being the case during ontogeny.
How embryonic and fetal niches exactly support and contribute to the development of HSCs and the hematopoietic system is of great interest both for our fundamental knowledge and for the clinic to advance the therapeutic application of hematopoietic (stem) cells. The increased number of diseases and disorders treated at least in part by HSC transplantations and the difficulties to find HSCs with the best donor-patient compatibility is a major issue. Decades of efforts to develop culture conditions, either to expand HSCs ex vivo or to generate new HSCs in vitro, hold great promise but success remains limited (10–13). The low production of HSC-like cells with limited multilineage and/or self-renewal properties remains a major barrier to a successful use of HSCs for transplantation and gene modification. The reconstitution of a complete microenvironment or at least some of its key components to support the generation, maintenance and/or expansion of HSCs in vitro will be necessary to overcome this barrier. However important clues on how HSC production is supported by successive niches in vivo is missing. In this review, we will focus on the developmental origin of HSPCs and will discuss the novel technological approaches and recent progress made to identify the cellular composition, the importance of cell cross-talk and the underlying molecular events involved between the HSCs and the supportive microenvironment in the embryonic aorta, the physiological cradle of the first adult-type HSCs.
In mammals, the first hematopoietic cells are formed independently of HSCs. This first or primitive wave generates nucleated erythrocytes that emerge with and in close proximity to endothelial cells (14). They derive from mesodermal derivatives in blood islands of polyclonal origin, in the extra-embryonic yolk sac (YS), around mouse embryonic day (E)7.25 (15–17). Beside primitive erythrocytes, a first wave of primitive megakaryocyte and macrophage progenitors also arise in the YS blood islands (18–20). A portion of these “primitive” macrophages will persist throughout adulthood and give rise to microglia, the tissue-resident macrophages in the adult brain and central nervous system (21). Multipotent erythro-myeloid progenitors (EMPs) will then produce erythrocytes, macrophages, granulocytes and megakaryocytes (and possibly also few B lymphocytes and NK cells) through a second hematopoietic wave (also referred to as transient definitive wave or EMP wave) in the YS vascular plexus, starting at ∼E8 - E8.25 (18, 22–26). The YS does not provide a competent niche for the differentiation of EMPs in mature cells, which instead occurs in the fetal liver (23). Some macrophages reside in tissues, also referred to as tissue-resident macrophages, of embryos and adults where they act as immune sentinels involved in tissue homeostasis (27, 28). The YS and the developing dorsal aorta in the para-aortic splanchnopleura (which will give rise to the AGM region) also generate lymphoid progenitors at ∼E8.5 - E9.5, independently of HSCs, that are responsible for the initial immunity during development and the persistence of some immune cells (i.e. B-cells) into adulthood (29–33). The importance of these long thought short-life “primitive” hematopoietic cells, of which in fact some subsist and play important roles in late fetuses and adults, reinforces the need for a better identification of the niche-derived signals regulating their production. The 3rd or definitive hematopoietic wave leads to the formation of HSCs that can be detected as early as E10.5, in the aorta of the AGM region and in the extra-embryonic vitelline and umbilical arteries (34–36). Slightly later, HSCs are also found in other highly vascularized tissues such as the YS, and then the placenta and the fetal liver where they expand before colonizing their final destination, the BM (37–40). The impact of the microenvironment on HSC behavior is well illustrated in the fetal liver. During birth, the fetal liver undergoes dramatic changes in hemodynamic forces when the umbilical inlets are ligated. These changes trigger the transformation of arterial endothelial cells lining the portal vessels into venous endothelial cells, characterized by the loss of arterial markers (Neuropilin-1 and Ephrin-B2), acquisition of the venous marker (Eph Receptor B4) and the loss of Nestin+NG2+ pericytes through apoptosis (41). The latter cells are critical niche components and are probably the main cause of the emigration of HSCs from the fetal liver and their homing to the BM.
Hemogenic endothelial (HE) cells are a small subset of endothelial cells (1–3% in distinct tissues), which have either an arterial or venous identity (28, 42). HE cells can transdifferentiate into hematopoietic cells through a so-called endothelial to hematopoietic transition (EHT), a highly conserved process across vertebrate species (17, 43–49). HE cells can give rise to different types of hematopoietic cells, suggesting that different types of HE cells exist (50, 51). In the floor of the dorsal aorta, HE cells give rise to both EMPs and the first adult-type HSCs (17, 43–47, 49, 52). Most HSPCs are generated from HE cells with arterial characteristics, e.g. in the AGM, vitelline and umbilical vessels and the vascular labyrinth of the placenta and YS (53, 54). However, EMPs can also be produced by venous HE cells in the plexus of the YS (55, 56) and from HE cells in the heart that have not yet acquired an arterial-venous specification (57). Macrophages can also directly derive from HE cells via EHT in the placenta (58). The mere presence of HE cells does not guarantee that an EHT event occurs, indicating that the EHT process is differentially steered by signals from the niche, depending on their precise anatomical location and surroundings. For example, HE cells in the aorta that are not exposed to hemodynamic forces from blood flow do not give rise to HSCs, while EMP emergence is unaffected (59–63). How precisely HE cells acquire their hemogenic potential and how this potential leads to the formation of different hematopoietic cells through an EHT event remains to date largely elusive. Nevertheless, there are strong evidences that spatial and temporal signals from the microenvironment play a major role in hemogenic specification (to acquire a hemogenic potential), EHT, IAHC formation and HSPC production. As described in this review, new single cell technologies will shed new light on these processes to better understand how the establishment of the hematopoietic system is regulated.
After EHT, hematopoietic cells are organized in intra-aortic hematopoietic clusters (IAHCs) that remain transiently attached to the inner side of the vessels in most species (44) (Figure 1). IAHC-like structures have also been observed in non/less-hematopoietic sites such as the somites or the cerebrovascular sinusoids in the head (64). However, the detection of HSCs (which circulate through the blood circulation) and the presence of IAHCs in a tissue do not prove that EHT occurs in situ, as shown in the mouse embryonic head (65, 66). The ultimate proof that the de novo formation occurs in a tissue was provided for the aorta by performing confocal imaging on zebrafish embryos in vivo and on thick mouse embryo slices ex vivo (43, 67–69). The continued existence of HE cells beyond embryonic stages, leading to the formation of multipotent progenitors (MPP-3) and few HSCs in the sinusoids of the BM in fetal/neonate chicken and mouse was also reported and imaged, which could be referred to as a 4th hematopoietic wave (70).
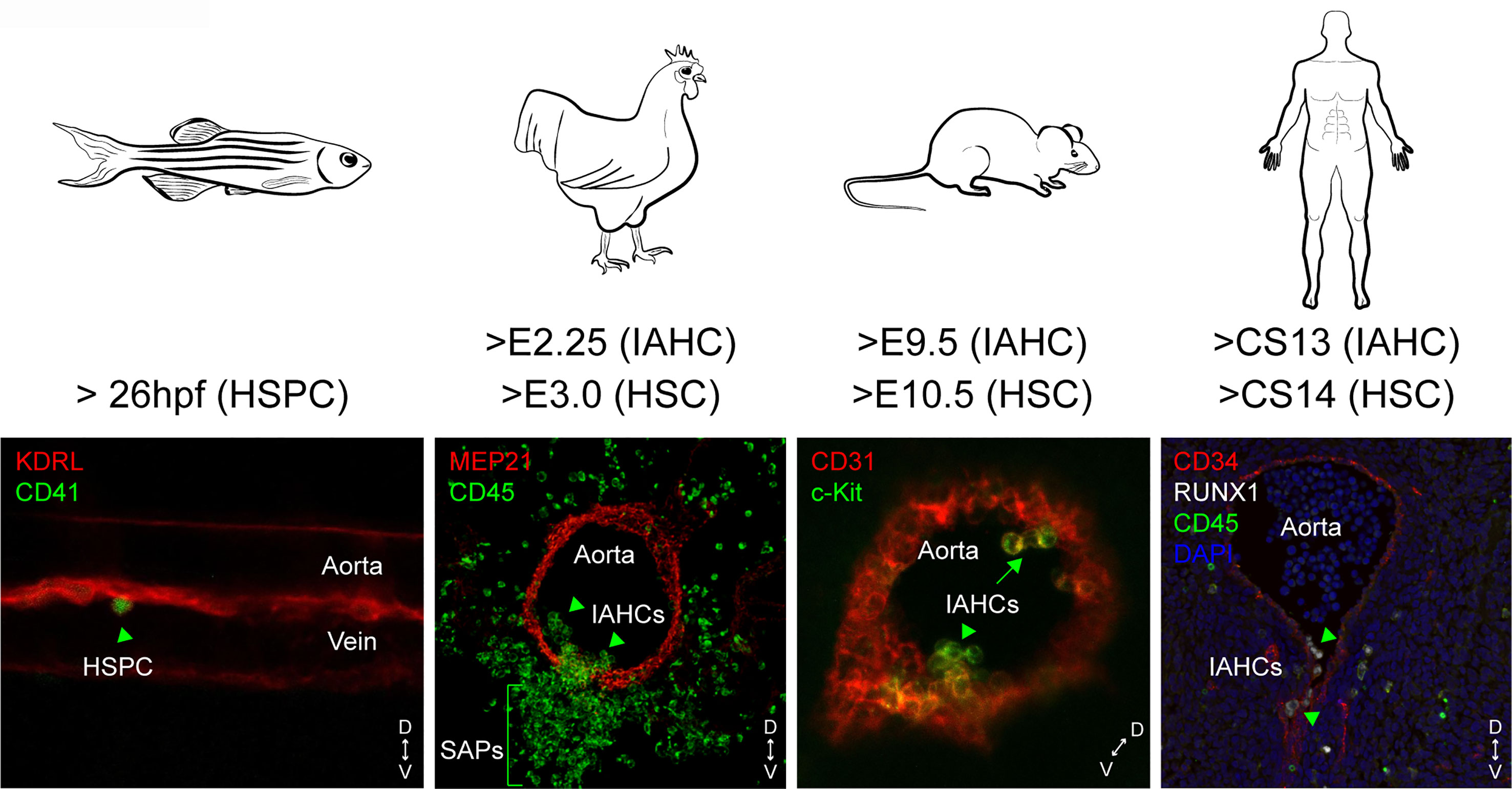
Figure 1 Spatial and temporal locations of IAHCs and HSPCs in the aorta of zebrafish, chicken, mouse and human embryos. The endothelial to hematopoietic transition (EHT) leads to the production of single HSPCs in the aorta of zebrafish embryos, or clusters (intra-aortic hematopoietic clusters or IAHCs). IAHCs emerge exclusively in the ventral side of the aorta in chicken and human embryos (arrow heads). In contrast, IAHCs also emerge in the dorsal side of the aorta in mouse embryos (arrows). In chicken, IAHC cells ingress underneath the ventral aortic endothelium to form sub-aortic patches (SAPs). The starting time of IAHCs and HSPCs/HSCs detected is indicated for each embryo species. D, dorsal; V, ventral.
Extensive research has explored the formation of the first HSCs during development, with a particular focus on IAHCs. IAHC cells express similar hematopoietic and endothelial markers as HSCs (67, 71, 72), and both are absent in Runx1 knock-out embryos (72, 73), suggesting that HSCs are likely part of IAHCs. However, IAHCs appear earlier than HSCs in the aorta (E9.5 versus E10.5), suggesting that some IAHC cells will become HSCs through gradual specification and maturation (Figure 1). Indeed, it was found that IAHCs are mainly composed out of HSC precursors (pre-HSCs type I and type II) that progressively mature into functional HSCs (72, 74–76). Accordingly, IAHCs contain very few HSCs and committed progenitors (that are mainly present in the blood circulation and transiting from the YS to the fetal liver). The process of maturation begins in the aorta but mainly takes place after migration of the cells into the fetal liver and placenta in mammals or in the caudal hematopoietic tissue (CHT) in zebrafish embryos (77). Limiting dilution transplantations and statistical analyses suggest that the pool of adult HSCs in the fetal liver is formed by the pool of IAHC cells (76). This finding raises the question what the contribution of HSCs found in the YS and placenta (39) and the pre-HSCs found in vitelline and umbilical arteries (54) is. A transient production of lympho-myeloid-biased progenitors and lymphoid cells prior to pre-HSC production was also reported in IAHCs (78–80). Although the connections between these different cell types remain unclear, it underlines the heterogeneity and complexity of IAHC composition at different time points and location during development. Of note, various differentiation potentials of IAHC cells have been revealed ex vivo, in presence of supportive stromal cell lines (OP-9) and/or cytokines, which might not reflect the true fate of these cells in vivo. The EHT is orchestrated by hematopoietic transcription factors primarily driven by RUNX1 (46, 81, 82), although it was reported that Runx1 deficiency does not preclude the formation of pre-HSCs type I but most likely blocks their maturation into pre-HSCs type II (83). Ectopic expression of Runx1 is sufficient to induce EMPs from non-HE cells (i.e. in YS, aorta, heart), but only between E7.5 to E8.5 of mouse development (82). Therefore, more complex events, such as the initiation of the blood circulation and the proper arterial–venous specialization of endothelial cells in the vascular niche microenvironment, might contribute to temporal restriction of EMPs and (pre-)HSCs appearance in IAHCs.
IAHCs originate from single HE cells that undergo 1 or 2 divisions to form a monoclonal core of IAHCs. Neighboring HE cells are then recruited into IAHCs that become thereby polyclonal while the cellularity increases (84, 85). Intra-cardiac injection of Dll4 blocking antibodies results in the enhanced recruitment of HE cells into IAHCs, suggesting that Dll4-Notch signaling in HE cells regulates IAHC cellularity (84). Unusually large IAHCs were also observed in Svep1-/- embryos that did not originate from ectopic proliferation of IAHC cells (86). Svep1 is expressed and secreted by mesenchymal cells surrounding the aorta, highlighting the importance of microenvironmental factors (Notch independent) in determining IAHC size (86). Size and composition might also be determined by an increased proliferation during pre-HSC expansion and a decrease when cells start to acquire HSC identity (87). While slowly cycling cells are located at the base of IAHCs, more proliferating cells preferentially locate at the more apical part of IAHCs (87). Thus, IAHC size is determined by recruitment of neighboring HE cells, controlled by a combination of direct signaling between HE cells and factors derived from the microenvironment, and proliferation of the different cell types within IAHCs. Such local regulation within and between IAHCs is an important concept to further explore since HSC activity is affected by the increased cellularity in IAHCs, as shown in Svep1-/- embryos (86).
The mechanism of HSC emergence is highly conserved and regulated both in space and time in between species, with few species-specific differences most likely due to anatomical constrains (1) (Figure 1). The EHT is polarized and restricted to the ventral side of the aorta in chicken, zebrafish and human embryos (4, 43, 88–91). After EHT, cells form IAHCs that remain transiently attached to the endothelium before detaching and leaving via the circulation to colonize the fetal liver in human and mouse. In chicken, the entire floor of the aorta becomes hemogenic, forming IAHC cells that ingress, at least in part, in the mesenchyme underneath the ventral endothelium to form sub-aortic patches, a site considered as the mammalian fetal liver equivalent (89) (Figure 1). In zebrafish, HSPCs bud off as single cells from the floor of the dorsal aorta into the sub-aortic space, where they transiently reside and roughly half of them divide before entering the circulation via the posterior cardinal vein (43, 68, 91) (Figure 1). Half of these HSPCs are considered as HSCs, the rest being possibly committed myeloid progenitors (92). Although “true” HSCs exist in chicken (89), it remains unknown whether they emerge as pre-HSCs and whether maturation and/or expansion occurs in the sub-aortic patches.
HSPCs start to emerge at 26 hours post fertilization (hpf) with a peak at 40 hpf, in the aorta of zebrafish embryos. IAHCs are found between E2.25 and E5.5 (with a peak at E3) in the anterior portion of the chicken aorta, from days 27 to 42 in the middle portion of the human aorta, and between E9.5 and E14.5 (with a peak at E10.5) in the middle portion of the mouse aorta (4, 34, 43, 68, 88–90, 93, 94). In contrast to other species, the mouse embryo has the particularity to produce IAHCs also in the dorsal part of the aorta (95, 96) (Figure 1). RNA-sequencing (RNA-seq) comparative analyses performed on whole IAHCs isolated from the ventral or dorsal part of the aorta revealed a strong similarity at the molecular level at both E10.5 and E11.5 (97). However, dorsal IAHCs are less numerous and have a lower HSC potential (four times less) compared to ventral IAHCs, as shown by limiting dilution transplantations of the subdissected parts of the aorta (ventral [AoV] versus dorsal [AoD]) (95, 98). Using a dissociation-reaggregation culture system that recapitulates HSC development ex vivo (99), it was shown that the AoD tissue induces a higher HSC production in the AoV tissue isolated at E10.5 but not at E11.5. In contrast, the AoV induces the production of HSCs in AoD at E11.5 but not at E10.5 (98). Such experiments, although performed ex vivo, reveal that the ventral and dorsal aortic microenvironment have reciprocal effects on HSC development, depending of the developmental time, and most likely on the differential release of factors by the two regions (i.e. SCF, Shh, BMP) and the capacity of IAHC cells to respond to specific signals by expressing the right level of receptors at the right time point (75, 98, 100–102). The lack of hemogenic potential of the dorsal aortic endothelium of most other vertebrate species might also be explained by the different origin of the endothelial cells populating the ventral and the dorsal part of the aorta. In zebrafish embryos, the dorsal endothelium does not originate from the splanchnopleura (lateral plate mesoderm) but from the paraxial mesoderm (103). In the avian embryo, the dorsal endothelium that derives from the splanchnopleura is progressively replaced by paraxial mesoderm-derived endothelial cells (as the ventral endothelium), which corresponds to the end of aortic hematopoiesis (104–106).
Various interacting signals from unique niche populations present in the different anatomical sites, as well as biomechanical forces, form an intricate signaling network that regulates the formation of HSPCs [for reviews (2, 9, 107–109)]. These signaling events are far from fully understood and many questions remain such as the exact nature (timing and duration) of signaling interactions between the niche and HSPCs and how these interactions contribute to determine different cell fates in endothelial/HE cells and IAHCs. Single-cell (sc) qPCR analyses paved the way for the (single-cell) genomic techniques to explore HSPC development. Although sc-qPCR offers high sensitivity and specificity, the quality of the data relies on high cell numbers and carefully selected and tested primer panels. Such approach identified important players that specify early blood formation (110) or revealed that HE cells are molecularly specified toward a hematopoietic fate two days prior the emergence of the first HSCs (111). Bulk RNA-seq, scRNA-seq and microarray analyses have then been performed to analyze the intrinsic regulation (e.g. by transcription factors) of HSPC formation by sequencing phenotypically enriched populations for arterial endothelial cells, HE cells, cells undergoing EHT, pre-HSCs and/or HSCs sorted from mouse, chicken, human and/or zebrafish embryos (78, 86, 97, 112–123). Overall, these studies highlighted important features. Among them are (i) the molecular heterogeneity of the HE, pre-HSC and HSC populations, (ii) the gene regulatory networks and trajectories involved during HSPC formation, (iii) new surface markers for a better localization and isolation of these rare embryonic cells, (iv) the identification of specific cell-cell-interactions and (v) important novel niche secreted factors. Single-cell transcriptomics of whole mouse and human embryos or organs collected at early time points of development also provided information on early mesoderm specification and important regulators of the early hematopoietic development (124–126). Several signaling molecules and pathways critical for HSC emergence or maturation have been identified by using knock-out/knock-down approaches, large drug screening (in zebrafish) or by performing mouse tissue explant or dissociation-reaggregation cultures in presence of either growth factors or stromal cell lines. Among others are Wnt, Notch, vitamin-A derived retinoic acid signals, BMP4, cytokines such as the interleukin-3 (IL-3) and stem cell factor (SCF), the catecholamines produced by the sympathetic nervous system, pro-inflammatory signals, the blood shear stress, chemokine such as Cxcl12 (SDF1), hyaluronan and extracellular matrix compounds (9, 75, 100, 109, 127–129). Overall, these approaches hardly link a regulator to a specific cell type (e.g. due to the limited purity of the cell populations tested) or to an anatomical location, particularly for the soluble factors.
In the avian model, dissection procedures that prevent the migration of the sub-aortic mesenchyme, also abolish Runx1 expression in HE cells. Subsequently, the formation of IAHCs is inhibited, proving the supportive role of mesenchymal cells in hematopoiesis in the aorta (130). Notch expression also tightly controls aortic hematopoiesis at specific time points of chicken, zebrafish and mouse development (84, 130, 131). Obtaining a global picture of all the molecular players expressed by the surroundings of the aorta during HSC emergence is still not achieved. In an attempt to identify putative molecules secreted by the HSC supportive microenvironment, several groups have compared cell lines derived from embryonic, fetal and postnatal mouse blood-forming tissues where HSCs emerge, expand or are maintained in vivo (e.g. from AGM sub-regions (aorta-mesonephros [AM] and urogenital ridges [UG]), the embryonic liver [EL] or the BM) (132, 133). Such cell lines have been tested and characterized for their competency to maintain/expand mouse and human HSPCs at different levels in vitro. Macro-array-based gene expression analyses of HSC-supportive (UG26-1B6 and EL08-1D2) versus less/non-supportive (UG15-1B7, AM20-1B4, EL28-1B3, and AM30-3F4) stromal cell lines revealed an up-regulation of fibroblast growth factor-7 (FGF-7), cathepsin K, thrombospondin 2 (TSP2), pleiotrophin (PTN), and IGFBP-3 and -4 in the supportive cell lines (134). This study demonstrated that ‘niche’ cells are not necessarily in direct contact nor need to be in contact with HSCs to fulfil their support capacity since secreted factors from the microenvironment are sufficient to maintain the HSC stemness properties (134). Using a similar approach, bulk transcriptome comparative analyses of AGM (UG26.1B6 [supportive] vs UG26.3B5 [less-supportive]), fetal liver (AFT024 [supportive] vs BFC012 [non-supportive]) and BM (BMC9 [supportive] vs BMC10 [less-supportive]) cell lines established the genetic signature of the sequential embryonic, fetal and adult HSC niches (135) (Figure 2A). Through comprehensive transcriptomic meta-analyses, 481 mRNAs and 17 micro-RNAs were found organized in modular networks and involved in critical signaling pathways. Beside known HSC regulators, this study also identified unexpected ones such as Pax9 and Ccdc80 that were functionally validated using morpholino injections in the zebrafish model. While these studies used solely in vitro cell lines, they opened the way for a better identification/characterization of the molecular landscapes of the sequential supportive HSC microenvironments.
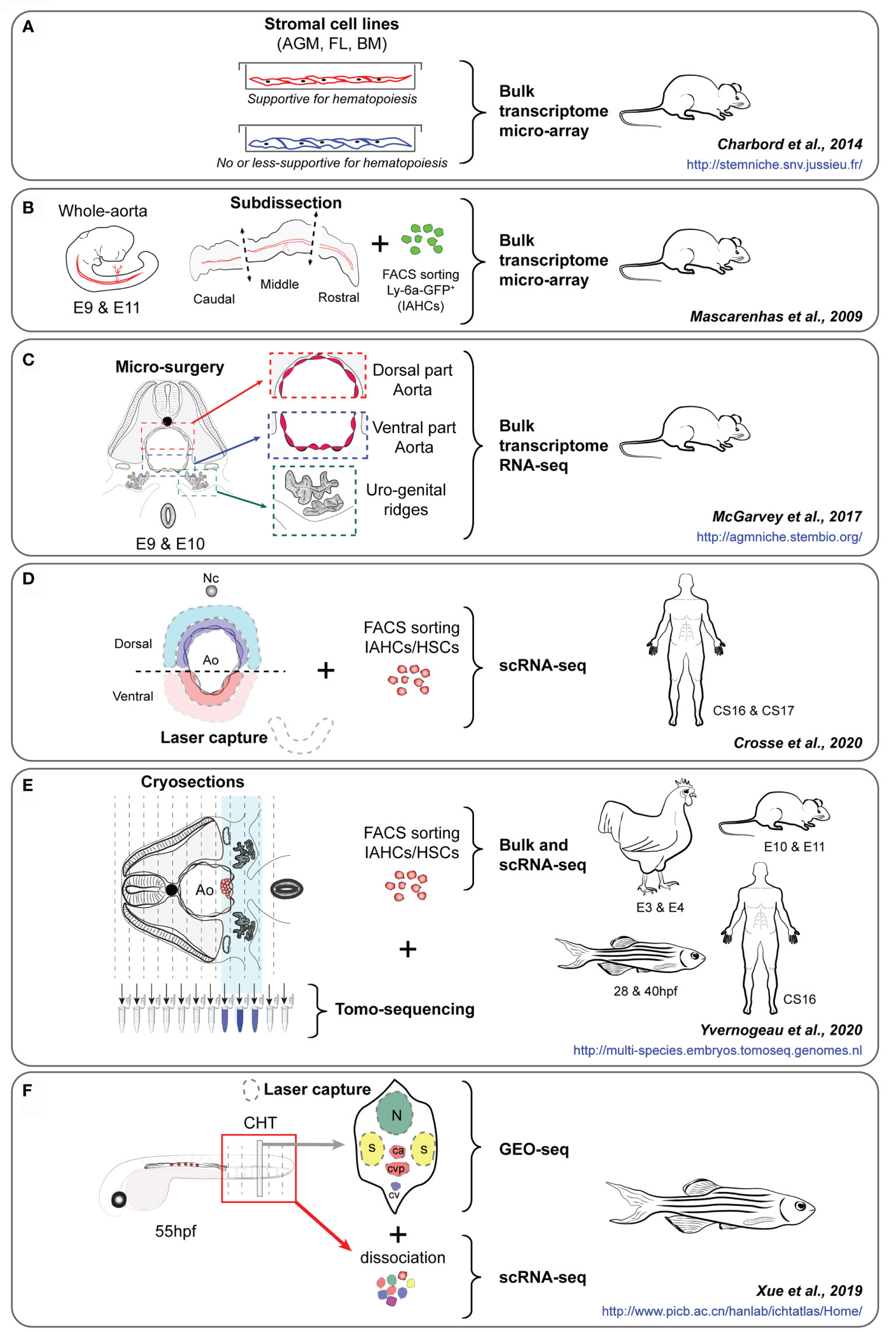
Figure 2 (A–F) Experimental approaches of key studies to identify the HSPC supportive landscape of the embryonic aorta, i.e. the studies of Charbord et al. (135) (A), Mascarenhas et al. (93) (B), McGarvey et al. (136) (C), Crosse et al. (117) (D), Yvernogeau et al. (86) (E) and Xue et al. (137) (F). The embryo species and stages, the type of cells/tissues analyzed, and the experimental approach used for each study are indicated. The interactives website resources provided in each study are also indicated (in blue). AGM, Aorta-Gonad-Mesonephros; FL, Fetal liver; BM, Bone marrow; E, Embryonic day; IAHCs, Intra-Aortic Hematopoietic Clusters, HSCs, Hematopoietic stem cells; Nc, Notochord; Ao, Aorta; BM, Bone marrow; CS, Carnegie stage; CHT, Caudal hematopoietic tissue; hpf, hour post-fertilization.
Although very informative, studies performed on stromal cell lines that are often clonal do not recapitulate the complexity of the HSC microenvironment. Moreover, critical in vivo components (i.e. blood flow/shear stress, circulating cells and growth factors) as well as the spatial three-dimensional organization of the aortic surrounding tissues are missing. It is not certain that all the in vivo features need to be reproduced in vitro, since HSC development can occur ex vivo when AGMs are cultured after dissociation-reaggregation or as explant for few days (35, 99). Several labs explored the aortic microenvironment by dissecting intact hematopoietic organs or by dissecting defined sub-regions, based on their HSC activity (i.e. emergence, expansion). In mice, the middle third of the aorta was identified as the HSC-containing region compared to the most anterior or posterior third regions that were devoid of HSCs (93) (Figure 2B). A micro-array transcriptomic analyses performed on these different regions collected at E9 and E11 (before and during the acquisition of an HSC potential, respectively) identified p57Kip2 and IgF2 as important hematopoietic regulators (93). Using a more precise microsurgery, the dorsal and ventral parts of the aorta and the urogenital ridges (UGRs) were isolated from E9.5 to E10.5 since the polarity along the dorsal to ventral axis of the embryo was demonstrated as a clear demarcation of the supportive HSC niche (136) (Figure 2C). Bulk RNA-sequencing on these different tissues allowed to identify critical signaling pathways and several secreted molecules, including Bmper as a ventrally polarized new regulator of HSC development in the AGM region. The use of human embryos at early stages when IAHCs appear is challenging due to the difficulty to collect intact embryos at the time of IAHC emergence in the aorta [around Carnegie stage (CS) 13 (post-fertilization age of 30-33 days) (4)] Different layers of cells surrounding the ventral and dorsal sides of the aorta of human embryos were captured by laser dissection and sequenced (LMO-seq) (117) (Figure 2D). In parallel, scRNA-seq data obtained from sorted human CD34+ IAHC cells were used to explore the cross-talk (e.g. involving secreted factors, cell surface receptors) occurring between IAHCs and the surroundings of the aorta. This approach allowed to highlight the cardiac epidermal growth factor (EGF) and its major receptor, endothelin 1 (expressed and secreted by endothelial cells), as a potent enhancer of HSC generation in human embryos (117).
A major drawback in single cell and bulk sequencing experiments is the lack of spatial information, which is essential to understand the interactions between niche and HSC cells. To perform RNA-seq while keeping spatial information, tomography-sequencing (tomo-seq) was developed (138). Embryonic thick slices or complete embryos are sequentially cryosectioned along the axis of the embryo, e.g. following a transversal or longitudinal orientation, and RNA from each slice is collected for sequencing. RNA expression profiles can then be visualized along the embryo axis. Genes expressed highly or solely in specific tissues/areas can be used for anatomical or orientation confirmation, e.g. Shh expression to locate the notochord, or Mpl (chicken), Gata2 (mouse) or Cxcl12 (zebrafish) expression for the aortic region containing IAHCs/HSPCs (86) (Figure 2E). Genes expressed in tissues or regions of interest can then be readily filtered out. By using this technique, thick transversal embryo slices and/or embryo trunks were collected from the 4 main species used to study developmental hematopoiesis, i.e. zebrafish, chicken, mouse and human, at two different developmental time points (beginning and time of HSPC production) (86). Genes specifically expressed in the ventral mesenchyme located underneath the aorta, and more expressed in this region than in the rest of the tissue sample, were identified. To further enhance the usability of these data, transcriptomic datasets of sorted mouse (97) and chicken (86) IAHC cells were generated and compared to the tomo-seq datasets to identify genes and pathways potentially involved in the cross-talk between IAHCs and the ventral aortic microenvironment. Known ligands and corresponding receptors in the aortic ventral microenvironment and IAHC cells were identified, validating this experimental approach and analysis. These were known to be involved in EHT and HSPC survival, attachment, maturation, and/or expansion (e.g. integrin, WNT, BMP, FGF, NOTCH, catecholamines), in inflammation, extracellular matrix organization, cytoskeleton rearrangement, in various cellular processes (protein phosphorylation, intracellular protein transduction) and specific pathways (Pi3K-AKT, MAPK, ERK, RAP1, and RAS). Molecules with unknown hematopoietic role were also identified and functionally validated in vitro and in vivo as important and conserved HSPC regulators in mouse, chicken and zebrafish embryos. These included (1): the adrenomedullin (ADM), a hypotensive and vasodilator agent and its receptor RAMP2, which regulates HSPC emergence in the aorta, and (2) SVEP1 (Sushi, Von Willebrand Factor Type A, EGF and Pentraxin Domain Containing 1), a secreted extracellular factor critical for proper lymphangiogenesis (139), that was shown to also regulate IAHC cellularity and HSPC production in the aorta (86).
The cellular and molecular mechanisms underlying HSC and multipotent progenitor expansion remain poorly understood. Zebrafish embryos have been used to generate a “3D transcriptional atlas” to characterize the spatiotemporal transcriptome during HSPC expansion in the CHT region (137) (Figure 2F). In this study, multi-dimensional RNA-seq approaches were used, including bulk and scRNA-seq on HSPCs isolated at 6 different time points (between 28 hpf and 3 mpf), and in vivo GEO-seq performed on the CHT region where six regions were collected on embryo sections by laser capture microdissection at 55 hpf. These regions included the neural tube (in the dorsal region), the muscles (left and right regions), and the caudal artery, caudal vein and caudal vein plexus (in the middle, intermediate and ventral regions, respectively). Such approach combined with functional validation allowed to reconstruct the panoramic transcriptome landscape (temporal and spatial) of the zebrafish CHT, and highlighted the integrin signaling protein Smchd1 as critical for HSPC expansion. Single-cell and spatial transcriptomics recently provided a spatio-temporal transcriptome map of the mouse fetal liver and thereby identified transcriptionally heterogeneous HSPC subsets, as well as HSC ‘pocket-like’ units composed of niche cells (i.e. hepatoblasts, stromal cells, endothelial cells, and macrophages), where macrophages and growth factors (MDK, PTN, and IGFBP5) played an important role in HSPC expansion (140).
The ability to visualize HSCs in their native environment has been paramount for our current knowledge regarding HSC dynamics and behavior in vivo (43, 48, 67, 68, 89, 141, 142). However, immunostainings, in situ hybridizations and the use of transgenic reporters only allow for the simultaneous visualization of a handful of genes. Ideally, one would like to image HSCs embedded in their niche in detail, or even follow them by time-lapse imaging until they display the desired behavior, capture the entire transcriptome and map this back to the imaging data. This combination would permit the precise identification of the different (sub)types of cells, as well as their transcriptomic state. Such approach would be especially powerful to e.g. elucidate the exact composition of small and large IAHCs, the heterogeneity of various endothelium (hemogenic and non-hemogenic) and the direct interactions of HSCs with their successive embryonic, fetal and adult niches. Although laser micro-dissection allows for the isolation of relatively small sections of tissue after imaging that can be processed for transcriptomic analysis (137), current technological advances are heading towards a more precise capture of the transcriptome of a full slide. Visium spatial gene expression by 10X Genomics permits such capture of the whole transcriptome from a tissue section. Prior capture, there is an optional step that allows for the visualization of proteins of interest by immunofluorescence to gain a deeper understanding of tissue organization or the localization of cells of interest (Figure 3A). This first version of Visium has 5.000 uniquely barcoded spots on 4 separate capture areas on each slide. Each spot will capture a range of about 1-10 cells, depending on the tissue thickness and tissue architecture (e.g. cell size). This capture grid will undoubtedly become smaller in future versions to reduce the number of cells captured per spot. While such approach will definitely add to our understanding of how HSCs are embedded and interacting with their microenvironment, the financial burden to systematically study HSCs in their native environment will be extremely high.
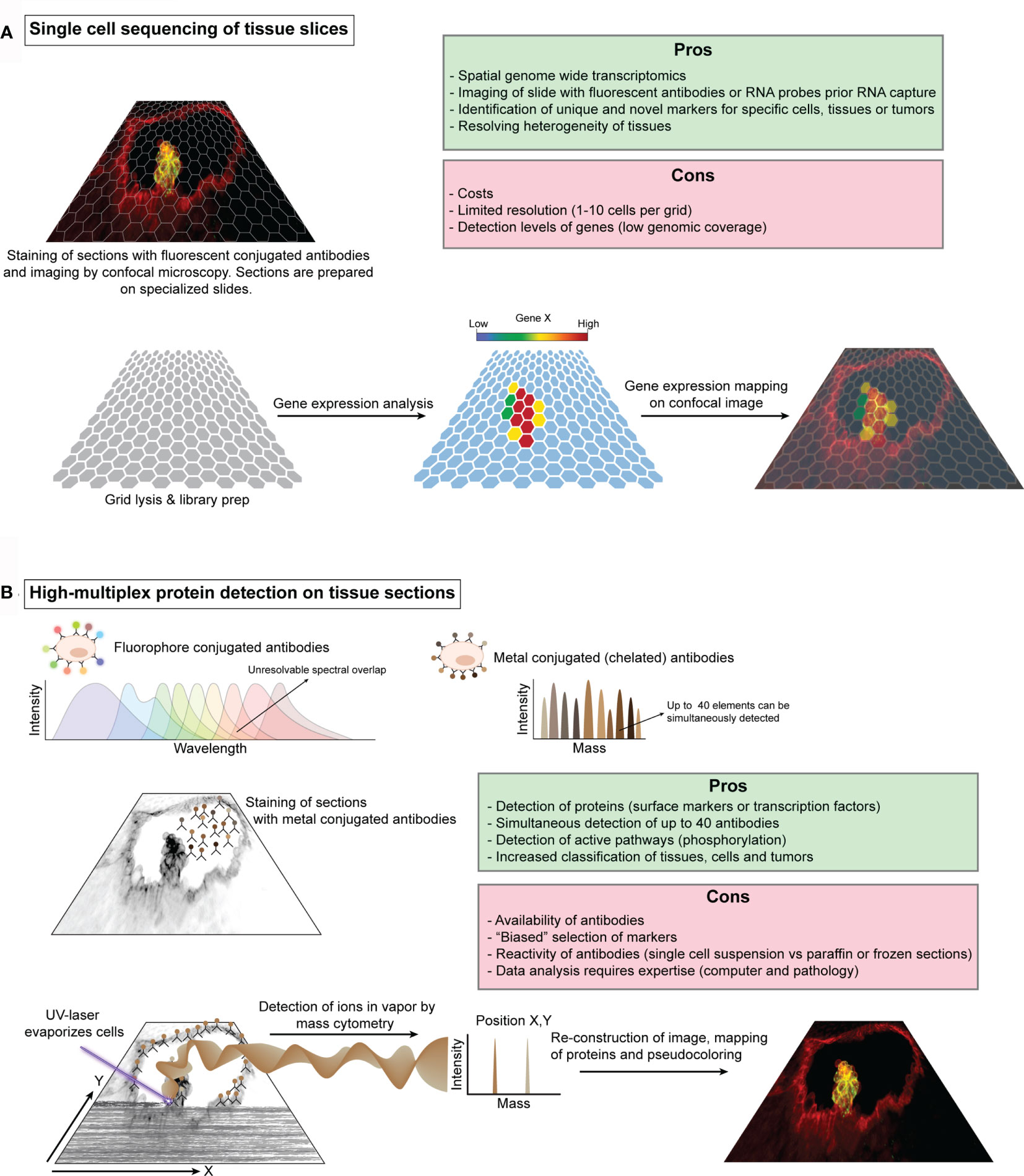
Figure 3 Schematic illustration of single cell sequencing of tissue slices and high-multiplex protein detection on tissue sections. (A) Schematic representation of 10X Genomics Visium workflow. Fresh-frozen or paraffin embedded tissue sections are prepared on specialized slides that contain several capture areas or grid. Each grid position has a known unique barcode, which is used for reconstructing the tissue after sequencing. These tissues can be stained with fluorescent antibodies or with one of the principle tissue stains, e.g. hematoxylin & eosin, and subsequently imaged by microscopy prior RNA capture. Cells are then permeabilized and the RNA content is captured for barcoding and library preparation. The library is sequenced by standard procedures and the data can be visualized in relation to the imaged tissues. (B). The use of multiple fluorescent proteins and quantum dots to simultaneously label different cell components is limited to the ability to resolve spectral overlap between the different fluorochromes. To increase the number of labels, classic fluorescent proteins have been replaced by specific elements or isotopes such as noble and post-transition metals, rare-earth elements and halogens, which can be detected by mass cytometry. Antibodies against specific proteins (e.g. receptors or transcription factors) can be tagged with these isotopes and used for staining of tissue sections and an image of the general morphology can be taken. A UV-laser evaporates the cells in a typical confocal scanning movement (X,Y) and fumes of the cells are transported by an inert gas into the mass cytometer for measurement. The measured isotopes and the corresponding X,Y coordinates can then be used to reconstruct an image which can be superimposed on top of the microscope images.
Whilst much attention goes out to transcriptomic approaches (122), recent advances in protein-based techniques should not be overlooked. Measuring proteins present in or on the cell by fluorescence-based flow cytometry has proven to be a rapid and powerful tool for isolating, sub-typing and phenotyping the cells of the immune system, including HSCs (143, 144). Multiplexing classic fluorescent and quantum dots labeled antibodies have stretched the limit of this technique up to 17 parameters (145, 146). Further expansion of fluorescent based cytometry seems unlikely due to the limitations to resolve the spectral overlap. Replacing fluorescent proteins or quantum dots with element isotopes (chelated antibody tags) dramatically reduced the cross-talk between channels and enabled the simultaneous measurement of up to 40 parameters, which is referred to as cytometry by time-of-flight (CyTOF) (147, 148). Distinct isotopes can be used to label different antibody panels that include surface markers, transcription factors as well as signaling molecules (phosphoproteins). By using CyTOF with about 31 different isotopes, functional and hierarchical maps of the immune/hematopoietic system have been drawn and show that hematopoiesis is a continuum rather than a collection of defined subsets (149–151). CyTOF was also instrumental in identifying a pro-inflammatory subset of macrophages that is involved in the development of HSCs (152). In addition, integrating CyTOF with scRNA-seq could provide additional discriminatory power for further sub-setting or functional analysis between distinct subsets of cells like HSCs and progenitors (153). Besides the precise immunophenotyping of cells in suspension, recent advances have enabled CyTOF of tissue sections or cells cultured on a slide. Aerosols of evaporated cells by laser ablation are transported to the CyTOF mass cytometry by an inert gas for detection (154). Data for each cell is then mapped based on the laser ablation coordinates to reassemble the original tissue architecture (Figure 3B). While the selection and availability of antibodies used is crucial to the success of this technique, it provides a new powerful way to study for example sections of the embryonic aorta to elucidate the composition of the vascular aortic, fetal liver and BM niches.
Limitless access to different types of blood producing cells manufactured in vitro is the “holy grail” of regenerative medicine. This would, among others, combat the current shortage of donor HSCs by providing a readily accessible source to all blood groups (red blood cells) (155, 156) or functional T-cells that can be engineered for anti-cancer therapies (157, 158). The crux of the matter is that these in vitro produced cells should faithfully mimic their in vivo counterparts. In vitro production of hematopoietic progenitors and mature blood cells (e.g. red blood cells, platelets, megakaryocytes, T-cells) from pluripotent stem cells or somatic cells, through reprogramming or transgene free protocols, is achievable (159). Reprogramming by (transient) expression of transcription factors is also a very promising strategy to generate HSC-like cells in vitro since a decade (11, 160–166). However, these HSC-like cells are produced at a very low yield and remain limited in their capacities to self-renew and/or to replenish all blood lineages, which is an absolute requirement for therapeutic use. Moreover, the association of the reprogramming factors with the development of leukemia remains an underlying risk. Indirect reprogramming (transgene free) through co-culturing pluripotent stem or progenitor cells with supportive cells that mimic the microenvironment in combination with chemical manipulation has therefore become a more favorable option. In its simplest form, this would be co-culturing pluripotent stem cells (e.g. iPSCs, ESCs) or somatic cells with a supportive cell line (e.g. OP9-cells) and/or a cocktail of growth factors, hormones and/or cytokines. Under these conditions, the formation of HE cells and some hematopoietic progenitors was successfully obtained, while HSCs are not or very rarely produced (167–173). As discussed above, the formation of HSCs in vivo requires a chain of events involving a complex sequence of both cell intrinsic and extrinsic factors that are difficult to recapitulate in a “simple” in vitro setting. Furthermore, one of the important open standing questions is whether it is necessary to first mimic the AGM-like microenvironment, to ensure HE formation and pre-HSC production in vitro, and second to mimic the fetal liver microenvironment to support pre-HSC maturation and HSC expansion. Comparing the transcriptomes of in vitro-generated HSC-like cells to fetal liver HSCs is an interesting approach to identify transcription factors and molecular pathways that could improve the in vitro production of HSCs (174). However, in vitro-generated HSCs that faithfully mimic the functionality of bona fide HSCs might have a different transcriptional landscape.
Recent advances in the ex utero culture of post-implantation mouse embryos, enabling the development until the hindlimb formation stage (E11) (175), or in 3D-culturing systems (176, 177) might offer a more sophisticated way of producing transplantable HSCs in vitro, as these systems recapitulate key aspects of developmental processes or organs. Disaggregation-reaggregation assays in the 1950’s showed that a suspension of chicken mesonephric cells could self-organize into the structural pattern of the original tissue, now generally referred to as organoids (178, 179). To date, organoid technology is mainly used to model the development of organs “in a dish”, including various diseases affecting these organs. The AGM is a complex region composed of a myriad of cells, including endothelial cells, HE cells, mesenchymal cells and various immune cells and it would be a challenging task to mimic the precise organization and timing of influx of supportive immune cells, like macrophages, within organoids. Hence, reports of successful HSC production by organoid technology are scarce, although there are some 3D iPSC-derived organoid-like culture systems that produce hematopoietic progenitors (180). The succeeding step of organoid cultures are techniques to mimic the first days of embryonic development in a dish. Pluripotent stem cells or iPSCs in a round bottom plate or in a hanging drop will aggregate into embryoid bodies (EBs) to form the three embryonic germ layers. Providing these EBs with a tailored cocktail of growth factors and cytokines at the right time will result in the production of endothelial cells, hematopoietic progenitors, erythrocytes, macrophages, neutrophils and mast cells (181–186). Until recently, this approach did not yield any transplantable HSCs. However, by extending the EB culture and optimizing the cocktail of cytokines and growth factors, which includes BMP4, VEGF, IGF1, SCF, FLT3, TPO, IL-1, -3, -6 and G-CSF, the formation of multilineage HSCs that produced myeloid, lymphoid and erythrocytes in sequential transplanted recipients was achieved (187). The success of generating HSCs from EBs might result from the extension of the EB culture time, allowing germ-layer specification and lineage commitment, or alternatively from the maturation of pre-HSCs. Reconstitution of sub-lethally irradiated recipients by these EB-derived HSCs required the injection of 400.000 cells from dissociated EBs per mouse, indicating that the production of HSCs in EBs is extremely low. One potential explanation for the low yield might be the restricted number of cells receiving the required spatial and temporal signals. This might be due to a sub-optimal organization of the different germ layers in these EBs. Interestingly, stimulating EBs with a pulse of a WNT/β-catenin signaling agonist results in the break of symmetry and subsequently leads to an anteroposterior axial organization with a bilateral symmetry similar to vertebrate embryos (188, 189). These so-called gastruloids display key features of mammalian development after implantation although tissue organization is often limited. Improvements in culturing conditions, such as embedding in Matrigel or fusion of a pulsed and non-pulsed EB, enhanced tissue organization and led to the formation of somite-like and neural structures (190–192). Single cell transcriptomics of gastruloids of different “developmental stages” and culturing methods showed different mesodermal derived populations with expression of endothelial and early blood markers like Etv2, Kdr, Cdh5, Kit, Gata2, Runx1, Cd34 and Itga2b (190, 191, 193). This suggests that gastruloids produce a hemangioblast-like cell type that might differentiate into endothelial, HE and/or even early HSPCs. Conformingly, detailed analysis of endothelial and blood markers in gastruloids revealed the formation of a vascular plexus, the production of blood progenitors and erythroid-like populations (193). The presence of blood vessel like structures and expression of markers like Kit, Itga2b and Runx1 is suggestive for the presence of HE cells and/or pre-HSCs, although additional functional assays are needed to confirm whether gastruloids can produce such cell types. Gastruloids are thus a promising and exciting new tool that does not need to use animals (respect of the 3R rules) and might proof valuable in understanding how extrinsic signals derived from the microenvironment instruct HE cells to undergo EHT and maybe produce (pre-)HSCs. However, the limited time when gastruloids can be cultured (equivalent to a mouse embryo around E8.5-E9.5) might preclude the formation of (pre-)HSCs.
An increasing number of studies based on RNA-sequencing and/or spatial transcriptomics performed in different embryo species confirmed the complexity of the aortic niche. The development of new molecular approaches and the increasing power of scRNA-seq technologies now offers the possibility to go one step further in the study of HSC regulation by the surrounding microenvironment and to generate high throughput datasets with limited material. The main challenge will be to manage and integrate all RNA-seq, spatial transcriptomic datasets in a comprehensive manner to obtain a global/real picture of what is happening in the AGM region when pre-HSCs/HSCs are generated, and to identify the fine tuning of all the regulators that evolve both in time and space. Most datasets are freely accessible and several labs have invested in creating interactive websites, which makes the exploration of these data relatively easy and allows to interrogate for the expression of any mRNA and miR of interest in supportive/non-supportive cell lines (135), gene expression in different sub-dissected regions of the AGM in mouse (136) and in multiple species for comparison and conservation (86), as well as in the CHT, the HSPC expansion niche in zebrafish embryos (137) (Figure 2). The number of molecules and pathways identified to be involved in hemogenic specification, EHT, IAHC formation and pre-HSC maturation within the aorta are continuously increasing. However, the mechanism by which most of these molecules interact and/or interfere, directly or indirectly, to regulate HSPC fate remains to be elucidated.
The evolution and combination of sc-genomic and multiomic techniques (e.g. Scifi-seq, ASAP-seq, ECCITE-seq, Visium) and the efforts made to increase cell throughput with lower costs or the detection of rare cell populations will continue to pave the way for a better understanding of HSPC production and its fine-tuned regulation by the supportive niche. The integration of transcriptomics, proteomics, and epigenetic changes at single-cell resolution and functional validations in vitro or in vivo will be essential to understand HSPC development in physiological condition with the goal to improve cell-replacement therapy, but also in immune and blood disease conditions, e.g. in the case of childhood leukemia that originate in utero.
BW and LY contributed equally to the review. All authors contributed to the article and approved the submitted version.
This work was supported by a European Research Council Grant (ERC, project number 220-H75001EU/HSCOrigin-309361), a TOP subsidy from NWO/ZonMw (912.15.017), a KWF grant (Dutch Cancer Society, project 13549) and the UMC Utrecht “Regenerative Medicine & Stem Cells” priority research program.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Klaus A, Robin C. Embryonic Hematopoiesis Under Microscopic Observation. Dev Biol (2017) 428(2):318–27. doi: 10.1016/j.ydbio.2017.03.008
2. Dzierzak E, Bigas A. Blood Development: Hematopoietic Stem Cell Dependence and Independence. Cell Stem Cell (2018) 22(5):639–51. doi: 10.1016/j.stem.2018.04.015
3. Ottersbach K. Endothelial-To-Haematopoietic Transition: An Update on the Process of Making Blood. Biochem Soc Trans (2019) 47(2):591–601. doi: 10.1042/BST20180320
4. Ivanovs A, Rybtsov S, Ng ES, Stanley EG, Elefanty AG, Medvinsky A. Human Haematopoietic Stem Cell Development: From the Embryo to the Dish. Development (2017) 144(13):2323–37. doi: 10.1242/dev.134866
5. Clements WK, Traver D. Signalling Pathways That Control Vertebrate Haematopoietic Stem Cell Specification. Nat Rev Immunol (2013) 13(5):336–48. doi: 10.1038/nri3443
6. Schofield R. The Relationship Between the Spleen Colony-Forming Cell and the Haemopoietic Stem Cell. Blood Cells (1978) 4(1-2):7–25.
7. Comazzetto S, Shen B, Morrison SJ. Niches That Regulate Stem Cells and Hematopoiesis in Adult Bone Marrow. Dev Cell (2021) 56(13):1848–60. doi: 10.1016/j.devcel.2021.05.018
8. Pinho S, Frenette PS. Haematopoietic Stem Cell Activity and Interactions With the Niche. Nat Rev Mol Cell Biol (2019) 20(5):303–20. doi: 10.1038/s41580-019-0103-9
9. Gao X, Xu C, Asada N, Frenette PS. The Hematopoietic Stem Cell Niche: From Embryo to Adult. Development (2018) 145(2):1–12. doi: 10.1242/dev.139691
10. Freire AG, Butler JM. Blood Making: Learning What to Put Into the Dish. F1000Res (2020) 9:1–11. doi: 10.12688/f1000research.21245.1
11. Barcia Duran JG, Lis R, Rafii S. Haematopoietic Stem Cell Reprogramming and the Hope for a Universal Blood Product. FEBS Lett (2019) 593(23):3253–65. doi: 10.1002/1873-3468.13681
12. Zimran E, Papa L, Hoffman R. Ex Vivo Expansion of Hematopoietic Stem Cells: Finally Transitioning From the Lab to the Clinic. Blood Rev (2021) 50:100853. doi: 10.1016/j.blre.2021.100853
13. Wilkinson AC, Igarashi KJ, Nakauchi H. Haematopoietic Stem Cell Self-Renewal In Vivo and Ex Vivo. Nat Rev Genet (2020) 21(9):541–54. doi: 10.1038/s41576-020-0241-0
14. Moore MA, Metcalf D. Ontogeny of the Haemopoietic System: Yolk Sac Origin of In Vivo and In Vitro Colony Forming Cells in the Developing Mouse Embryo. Br J Haematol (1970) 18(3):279–96. doi: 10.1111/j.1365-2141.1970.tb01443.x
15. Furuta C, Ema H, Takayanagi S, Ogaeri T, Okamura D, Matsui Y, et al. Discordant Developmental Waves of Angioblasts and Hemangioblasts in the Early Gastrulating Mouse Embryo. Development (2006) 133(14):2771–9. doi: 10.1242/dev.02440
16. Ueno H, Weissman IL. Clonal Analysis of Mouse Development Reveals a Polyclonal Origin for Yolk Sac Blood Islands. Dev Cell (2006) 11(4):519–33. doi: 10.1016/j.devcel.2006.08.001
17. Padron-Barthe L, Temino S, Villa del Campo C, Carramolino L, Isern J, Torres M. Clonal Analysis Identifies Hemogenic Endothelium as the Source of the Blood-Endothelial Common Lineage in the Mouse Embryo. Blood (2014) 124(16):2523–32. doi: 10.1182/blood-2013-12-545939
18. Hoeffel G, Ginhoux F. Fetal Monocytes and the Origins of Tissue-Resident Macrophages. Cell Immunol (2018) 330:5–15. doi: 10.1016/j.cellimm.2018.01.001
19. Palis J, Robertson S, Kennedy M, Wall C, Keller G. Development of Erythroid and Myeloid Progenitors in the Yolk Sac and Embryo Proper of the Mouse. Development (1999) 126(22):5073–84. doi: 10.1242/dev.126.22.5073
20. Tober J, Koniski A, McGrath KE, Vemishetti R, Emerson R, de Mesy-Bentley KK, et al. The Megakaryocyte Lineage Originates From Hemangioblast Precursors and is an Integral Component Both of Primitive and of Definitive Hematopoiesis. Blood (2007) 109(4):1433–41. doi: 10.1182/blood-2006-06-031898
21. Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, et al. Fate Mapping Analysis Reveals That Adult Microglia Derive From Primitive Macrophages. Science (2010) 330(6005):841–5. doi: 10.1126/science.1194637
22. Iturri L, Freyer L, Biton A, Dardenne P, Lallemand Y, Gomez Perdiguero E. Megakaryocyte Production is Sustained by Direct Differentiation From Erythromyeloid Progenitors in the Yolk Sac Until Midgestation. Immunity (2021) 54(7):1433–46.e5. doi: 10.1016/j.immuni.2021.04.026
23. Palis J. Hematopoietic Stem Cell-Independent Hematopoiesis: Emergence of Erythroid, Megakaryocyte, and Myeloid Potential in the Mammalian Embryo. FEBS Lett (2016) 590(22):3965–74. doi: 10.1002/1873-3468.12459
24. Hoeffel G, Chen J, Lavin Y, Low D, Almeida FF, See P, et al. C-Myb(+) Erythro-Myeloid Progenitor-Derived Fetal Monocytes Give Rise to Adult Tissue-Resident Macrophages. Immunity (2015) 42(4):665–78. doi: 10.1016/j.immuni.2015.03.011
25. Dege C, Fegan KH, Creamer JP, Berrien-Elliott MM, Luff SA, Kim D, et al. Potently Cytotoxic Natural Killer Cells Initially Emerge From Erythro-Myeloid Progenitors During Mammalian Development. Dev Cell (2020) 53(2):229–39.e7. doi: 10.1016/j.devcel.2020.02.016
26. McGrath KE, Frame JM, Fegan KH, Bowen JR, Conway SJ, Catherman SC, et al. Distinct Sources of Hematopoietic Progenitors Emerge Before HSCs and Provide Functional Blood Cells in the Mammalian Embryo. Cell Rep (2015) 11(12):1892–904. doi: 10.1016/j.celrep.2015.05.036
27. Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, et al. Tissue-Resident Macrophages Originate From Yolk-Sac-Derived Erythro-Myeloid Progenitors. Nature (2015) 518(7540):547–51. doi: 10.1038/nature13989
28. Wu Y, Hirschi KK. Regulation of Hemogenic Endothelial Cell Development and Function. Annu Rev Physiol (2021) 83:17–37. doi: 10.1146/annurev-physiol-021119-034352
29. Godin IE, Garcia-Porrero JA, Coutinho A, Dieterlen-Lievre F, Marcos MA. Para-Aortic Splanchnopleura From Early Mouse Embryos Contains B1a Cell Progenitors. Nature (1993) 364(6432):67–70. doi: 10.1038/364067a0
30. Yoshimoto M, Montecino-Rodriguez E, Ferkowicz MJ, Porayette P, Shelley WC, Conway SJ, et al. Embryonic Day 9 Yolk Sac and Intra-Embryonic Hemogenic Endothelium Independently Generate a B-1 and Marginal Zone Progenitor Lacking B-2 Potential. Proc Natl Acad Sci USA (2011) 108(4):1468–73. doi: 10.1073/pnas.1015841108
31. Boiers C, Carrelha J, Lutteropp M, Luc S, Green JC, Azzoni E, et al. Lymphomyeloid Contribution of an Immune-Restricted Progenitor Emerging Prior to Definitive Hematopoietic Stem Cells. Cell Stem Cell (2013) 13(5):535–48. doi: 10.1016/j.stem.2013.08.012
32. Kobayashi M, Shelley WC, Seo W, Vemula S, Lin Y, Liu Y, et al. Functional B-1 Progenitor Cells are Present in the Hematopoietic Stem Cell-Deficient Embryo and Depend on Cbfbeta for Their Development. Proc Natl Acad Sci USA (2014) 111(33):12151–6. doi: 10.1073/pnas.1407370111
33. Godin I, Dieterlen-Lievre F, Cumano A. Emergence of Multipotent Hemopoietic Cells in the Yolk Sac and Paraaortic Splanchnopleura in Mouse Embryos, Beginning at 8.5 Days Postcoitus. Proc Natl Acad Sci USA (1995) 92(3):773–7. doi: 10.1073/pnas.92.3.773
34. Muller AM, Medvinsky A, Strouboulis J, Grosveld F, Dzierzak E. Development of Hematopoietic Stem Cell Activity in the Mouse Embryo. Immunity (1994) 1(4):291–301. doi: 10.1016/1074-7613(94)90081-7
35. Medvinsky A, Dzierzak E. Definitive Hematopoiesis Is Autonomously Initiated by the AGM Region. Cell (1996) 86(6):897–906. doi: 10.1016/S0092-8674(00)80165-8
36. de Bruijn MF, Speck NA, Peeters MC, Dzierzak E. Definitive Hematopoietic Stem Cells First Develop Within the Major Arterial Regions of the Mouse Embryo. EMBO J (2000) 19(11):2465–74. doi: 10.1093/emboj/19.11.2465
37. Christensen JL, Wright DE, Wagers AJ, Weissman IL. Circulation and Chemotaxis of Fetal Hematopoietic Stem Cells. PLoS Biol (2004) 2(3):E75. doi: 10.1371/journal.pbio.0020075
38. Ottersbach K, Dzierzak E. The Murine Placenta Contains Hematopoietic Stem Cells Within the Vascular Labyrinth Region. Dev Cell (2005) 8(3):377–87. doi: 10.1016/j.devcel.2005.02.001
39. Kumaravelu P, Hook L, Morrison AM, Ure J, Zhao S, Zuyev S, et al. Quantitative Developmental Anatomy of Definitive Haematopoietic Stem Cells/Long-Term Repopulating Units (HSC/RUs): Role of the Aorta-Gonad-Mesonephros (AGM) Region and the Yolk Sac in Colonisation of the Mouse Embryonic Liver. Development (2002) 129(21):4891–9. doi: 10.1242/dev.129.21.4891
40. Gekas C, Dieterlen-Lievre F, Orkin SH, Mikkola HK. The Placenta is a Niche for Hematopoietic Stem Cells. Dev Cell (2005) 8(3):365–75. doi: 10.1016/j.devcel.2004.12.016
41. Khan JA, Mendelson A, Kunisaki Y, Birbrair A, Kou Y, Arnal-Estape A, et al. Fetal Liver Hematopoietic Stem Cell Niches Associate With Portal Vessels. Science (2016) 351(6269):176–80. doi: 10.1126/science.aad0084
42. Gritz E, Hirschi KK. Specification and Function of Hemogenic Endothelium During Embryogenesis. Cell Mol Life Sci (2016) 73(8):1547–67. doi: 10.1007/s00018-016-2134-0
43. Kissa K, Herbomel P. Blood Stem Cells Emerge From Aortic Endothelium by a Novel Type of Cell Transition. Nature (2010) 464(7285):112–5. doi: 10.1038/nature08761
44. Jaffredo T, Gautier R, Eichmann A, Dieterlen-Lievre F. Intraaortic Hemopoietic Cells are Derived From Endothelial Cells During Ontogeny. Development (1998) 125(22):4575–83. doi: 10.1242/dev.125.22.4575
45. Zovein AC, Hofmann JJ, Lynch M, French WJ, Turlo KA, Yang Y, et al. Fate Tracing Reveals the Endothelial Origin of Hematopoietic Stem Cells. Cell Stem Cell (2008) 3(6):625–36. doi: 10.1016/j.stem.2008.09.018
46. Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, Speck NA. Runx1 Is Required for the Endothelial to Haematopoietic Cell Transition But Not Thereafter. Nature (2009) 457(7231):887–91. doi: 10.1038/nature07619
47. Eilken HM, Nishikawa S, Schroeder T. Continuous Single-Cell Imaging of Blood Generation From Haemogenic Endothelium. Nature (2009) 457(7231):896–900. doi: 10.1038/nature07760
48. Lancrin C, Sroczynska P, Stephenson C, Allen T, Kouskoff V, Lacaud G. The Haemangioblast Generates Haematopoietic Cells Through a Haemogenic Endothelium Stage. Nature (2009) 457(7231):892–5. doi: 10.1038/nature07679
49. Oberlin E, El Hafny B, Petit-Cocault L, Souyri M. Definitive Human and Mouse Hematopoiesis Originates From the Embryonic Endothelium: A New Class of HSCs Based on VE-Cadherin Expression. Int J Dev Biol (2010) 54(6-7):1165–73. doi: 10.1387/ijdb.103121eo
50. Chen MJ, Li Y, De Obaldia ME, Yang Q, Yzaguirre AD, Yamada-Inagawa T, et al. Erythroid/myeloid Progenitors and Hematopoietic Stem Cells Originate From Distinct Populations of Endothelial Cells. Cell Stem Cell (2011) 9(6):541–52. doi: 10.1016/j.stem.2011.10.003
51. Dignum T, Varnum-Finney B, Srivatsan SR, Dozono S, Waltner O, Heck AM, et al. Multipotent Progenitors and Hematopoietic Stem Cells Arise Independently From Hemogenic Endothelium in the Mouse Embryo. Cell Rep (2021) 36(11):109675. doi: 10.1016/j.celrep.2021.109675
52. de Bruijn MF, Ma X, Robin C, Ottersbach K, Sanchez MJ, Dzierzak E. Hematopoietic Stem Cells Localize to the Endothelial Cell Layer in the Midgestation Mouse Aorta. Immunity (2002) 16(5):673–83. doi: 10.1016/S1074-7613(02)00313-8
53. Li W, Ferkowicz MJ, Johnson SA, Shelley WC, Yoder MC. Endothelial Cells in the Early Murine Yolk Sac Give Rise to CD41-Expressing Hematopoietic Cells. Stem Cells Dev (2005) 14(1):44–54. doi: 10.1089/scd.2005.14.44
54. Gordon-Keylock S, Sobiesiak M, Rybtsov S, Moore K, Medvinsky A. Mouse Extraembryonic Arterial Vessels Harbor Precursors Capable of Maturing Into Definitive HSCs. Blood (2013) 122(14):2338–45. doi: 10.1182/blood-2012-12-470971
55. Kasaai B, Caolo V, Peacock HM, Lehoux S, Gomez-Perdiguero E, Luttun A, et al. Erythro-Myeloid Progenitors can Differentiate From Endothelial Cells and Modulate Embryonic Vascular Remodeling. Sci Rep (2017) 7:43817. doi: 10.1038/srep43817
56. Frame JM, Fegan KH, Conway SJ, McGrath KE, Palis J. Definitive Hematopoiesis in the Yolk Sac Emerges From Wnt-Responsive Hemogenic Endothelium Independently of Circulation and Arterial Identity. Stem Cells (2016) 34(2):431–44. doi: 10.1002/stem.2213
57. Nakano H, Liu X, Arshi A, Nakashima Y, van Handel B, Sasidharan R, et al. Haemogenic Endocardium Contributes to Transient Definitive Haematopoiesis. Nat Commun (2013) 4:1564. doi: 10.1038/ncomms2569
58. Liang G, Zhou C, Jiang X, Zhang Y, Huang B, Gao S, et al. De Novo Generation of Macrophage From Placenta-Derived Hemogenic Endothelium. Dev Cell (2021) 56(14):2121–33.e6. doi: 10.1016/j.devcel.2021.06.005
59. Lux CT, Yoshimoto M, McGrath K, Conway SJ, Palis J, Yoder MC. All Primitive and Definitive Hematopoietic Progenitor Cells Emerging Before E10 in the Mouse Embryo are Products of the Yolk Sac. Blood (2008) 111(7):3435–8. doi: 10.1182/blood-2007-08-107086
60. Adamo L, Naveiras O, Wenzel PL, McKinney-Freeman S, Mack PJ, Gracia-Sancho J, et al. Biomechanical Forces Promote Embryonic Haematopoiesis. Nature (2009) 459(7250):1131–5. doi: 10.1038/nature08073
61. North TE, Goessling W, Peeters M, Li P, Ceol C, Lord AM, et al. Hematopoietic Stem Cell Development Is Dependent on Blood Flow. Cell (2009) 137(4):736–48. doi: 10.1016/j.cell.2009.04.023
62. Lundin V, Sugden WW, Theodore LN, Sousa PM, Han A, Chou S, et al. YAP Regulates Hematopoietic Stem Cell Formation in Response to the Biomechanical Forces of Blood Flow. Dev Cell (2020) 52(4):446–60.e5. doi: 10.1016/j.devcel.2020.01.006
63. Wang L, Zhang P, Wei Y, Gao Y, Patient R, Liu F. A Blood Flow-Dependent Klf2a-NO Signaling Cascade is Required for Stabilization of Hematopoietic Stem Cell Programming in Zebrafish Embryos. Blood (2011) 118(15):4102–10. doi: 10.1182/blood-2011-05-353235
64. Yzaguirre AD, Speck NA. Insights Into Blood Cell Formation From Hemogenic Endothelium in Lesser-Known Anatomic Sites. Dev Dyn (2016) 245(10):1011–28. doi: 10.1002/dvdy.24430
65. Li Z, Lan Y, He W, Chen D, Wang J, Zhou F, et al. Mouse Embryonic Head as a Site for Hematopoietic Stem Cell Development. Cell Stem Cell (2012) 11(5):663–75. doi: 10.1016/j.stem.2012.07.004
66. Iizuka K, Yokomizo T, Watanabe N, Tanaka Y, Osato M, Takaku T, et al. Lack of Phenotypical and Morphological Evidences of Endothelial to Hematopoietic Transition in the Murine Embryonic Head During Hematopoietic Stem Cell Emergence. PLoS One (2016) 11(5):e0156427. doi: 10.1371/journal.pone.0156427
67. Boisset JC, van Cappellen W, Andrieu-Soler C, Galjart N, Dzierzak E, Robin C. In Vivo Imaging of Haematopoietic Cells Emerging From the Mouse Aortic Endothelium. Nature (2010) 464(7285):116–20. doi: 10.1038/nature08764
68. Bertrand JY, Chi NC, Santoso B, Teng S, Stainier DY, Traver D. Haematopoietic Stem Cells Derive Directly From Aortic Endothelium During Development. Nature (2010) 464(7285):108–11. doi: 10.1038/nature08738
69. Lam EY, Hall CJ, Crosier PS, Crosier KE, Flores MV. Live Imaging of Runx1 Expression in the Dorsal Aorta Tracks the Emergence of Blood Progenitors From Endothelial Cells. Blood (2010) 116(6):909–14. doi: 10.1182/blood-2010-01-264382
70. Yvernogeau L, Gautier R, Petit L, Khoury H, Relaix F, Ribes V, et al. In Vivo Generation of Haematopoietic Stem/Progenitor Cells From Bone Marrow-Derived Haemogenic Endothelium. Nat Cell Biol (2019) 21(11):1334–45. doi: 10.1038/s41556-019-0410-6
71. Brachtendorf G, Kuhn A, Samulowitz U, Knorr R, Gustafsson E, Potocnik AJ, et al. Early Expression of Endomucin on Endothelium of the Mouse Embryo and on Putative Hematopoietic Clusters in the Dorsal Aorta. Dev Dyn (2001) 222(3):410–9. doi: 10.1002/dvdy.1199
72. Boisset JC, Clapes T, Klaus A, Papazian N, Onderwater J, Mommaas-Kienhuis M, et al. Progressive Maturation Toward Hematopoietic Stem Cells in the Mouse Embryo Aorta. Blood (2015) 125(3):465–9. doi: 10.1182/blood-2014-07-588954
73. Cai Z, de Bruijn M, Ma X, Dortland B, Luteijn T, Downing RJ, et al. Haploinsufficiency of AML1 Affects the Temporal and Spatial Generation of Hematopoietic Stem Cells in the Mouse Embryo. Immunity (2000) 13(4):423–31. doi: 10.1016/S1074-7613(00)00042-X
74. Rybtsov S, Sobiesiak M, Taoudi S, Souilhol C, Senserrich J, Liakhovitskaia A, et al. Hierarchical Organization and Early Hematopoietic Specification of the Developing HSC Lineage in the AGM Region. J Exp Med (2011) 208(6):1305–15. doi: 10.1084/jem.20102419
75. Rybtsov S, Batsivari A, Bilotkach K, Paruzina D, Senserrich J, Nerushev O, et al. Tracing the Origin of the HSC Hierarchy Reveals an SCF-Dependent, IL-3-Independent CD43(-) Embryonic Precursor. Stem Cell Rep (2014) 3(3):489–501. doi: 10.1016/j.stemcr.2014.07.009
76. Rybtsov S, Ivanovs A, Zhao S, Medvinsky A. Concealed Expansion of Immature Precursors Underpins Acute Burst of Adult HSC Activity in Foetal Liver. Development (2016) 143(8):1284–9. doi: 10.1242/dev.131193
77. Chen MJ, Lummertz da Rocha E, Cahan P, Kubaczka C, Hunter P, Sousa P, et al. Transcriptome Dynamics of Hematopoietic Stem Cell Formation Revealed Using a Combinatorial Runx1 and Ly6a Reporter System. Stem Cell Rep (2020) 14(5):956–71. doi: 10.1016/j.stemcr.2020.03.020
78. Zhu Q, Gao P, Tober J, Bennett L, Chen C, Uzun Y, et al. Developmental Trajectory of Prehematopoietic Stem Cell Formation From Endothelium. Blood (2020) 136(7):845–56. doi: 10.1182/blood.2020004801
79. Hadland BK, Varnum-Finney B, Mandal PK, Rossi DJ, Poulos MG, Butler JM, et al. A Common Origin for B-1a and B-2 Lymphocytes in Clonal Pre- Hematopoietic Stem Cells. Stem Cell Rep (2017) 8(6):1563–72. doi: 10.1016/j.stemcr.2017.04.007
80. Kobayashi M, Tarnawsky SP, Wei H, Mishra A, Azevedo Portilho N, Wenzel P, et al. Hemogenic Endothelial Cells Can Transition to Hematopoietic Stem Cells Through a B-1 Lymphocyte-Biased State During Maturation in the Mouse Embryo. Stem Cell Rep (2019) 13(1):21–30. doi: 10.1016/j.stemcr.2019.05.025
81. Yzaguirre AD, de Bruijn MF, Speck NA. The Role of Runx1 in Embryonic Blood Cell Formation. Adv Exp Med Biol (2017) 962:47–64. doi: 10.1007/978-981-10-3233-2_4
82. Yzaguirre AD, Howell ED, Li Y, Liu Z, Speck NA. Runx1 is Sufficient for Blood Cell Formation From non-Hemogenic Endothelial Cells In Vivo Only During Early Embryogenesis. Development (2018) 145(2):1–7. doi: 10.1242/dev.158162
83. Liakhovitskaia A, Rybtsov S, Smith T, Batsivari A, Rybtsova N, Rode C, et al. Runx1 Is Required for Progression of CD41+ Embryonic Precursors Into HSCs But Not Prior to This. Development (2014) 141(17):3319–23. doi: 10.1242/dev.110841
84. Porcheri C, Golan O, Calero-Nieto FJ, Thambyrajah R, Ruiz-Herguido C, Wang X, et al. Notch Ligand Dll4 Impairs Cell Recruitment to Aortic Clusters and Limits Blood Stem Cell Generation. EMBO J (2020) 39(8):e104270. doi: 10.15252/embj.2019104270
85. Zape JP, Lizama CO, Cautivo KM, Zovein AC. Cell Cycle Dynamics and Complement Expression Distinguishes Mature Haematopoietic Subsets Arising From Hemogenic Endothelium. Cell Cycle (2017) 16(19):1835–47. doi: 10.1080/15384101.2017.1361569
86. Yvernogeau L, Klaus A, Maas J, Morin-Poulard I, Weijts B, Schulte-Merker S, et al. Multispecies RNA Tomography Reveals Regulators of Hematopoietic Stem Cell Birth in the Embryonic Aorta. Blood (2020) 136(7):831–44. doi: 10.1182/blood.2019004446
87. Batsivari A, Rybtsov S, Souilhol C, Binagui-Casas A, Hills D, Zhao S, et al. Understanding Hematopoietic Stem Cell Development Through Functional Correlation of Their Proliferative Status With the Intra-Aortic Cluster Architecture. Stem Cell Rep (2017) 8(6):1549–62. doi: 10.1016/j.stemcr.2017.04.003
88. Yokomizo T, Dzierzak E. Three-Dimensional Cartography of Hematopoietic Clusters in the Vasculature of Whole Mouse Embryos. Development (2010) 137(21):3651–61. doi: 10.1242/dev.051094
89. Yvernogeau L, Robin C. Restricted Intra-Embryonic Origin of Bona Fide Hematopoietic Stem Cells in the Chicken. Development (2017) 144(13):2352–63. doi: 10.1242/dev.151613
90. Tavian M, Coulombel L, Luton D, Clemente HS, Dieterlen-Lievre F, Peault B. Aorta-Associated CD34+ Hematopoietic Cells in the Early Human Embryo. Blood (1996) 87(1):67–72. doi: 10.1182/blood.V87.1.67.67
91. Bonkhofer F, Rispoli R, Pinheiro P, Krecsmarik M, Schneider-Swales J, Tsang IHC, et al. Blood Stem Cell-Forming Haemogenic Endothelium in Zebrafish Derives From Arterial Endothelium. Nat Commun (2019) 10(1):3577. doi: 10.1038/s41467-019-11423-2
92. Henninger J, Santoso B, Hans S, Durand E, Moore J, Mosimann C, et al. Clonal Fate Mapping Quantifies the Number of Haematopoietic Stem Cells That Arise During Development. Nat Cell Biol (2017) 19(1):17–27. doi: 10.1038/ncb3444
93. Mascarenhas MI, Parker A, Dzierzak E, Ottersbach K. Identification of Novel Regulators of Hematopoietic Stem Cell Development Through Refinement of Stem Cell Localization and Expression Profiling. Blood (2009) 114(21):4645–53. doi: 10.1182/blood-2009-06-230037
94. Tavian M, Peault B. Embryonic Development of the Human Hematopoietic System. Int J Dev Biol (2005) 49(2-3):243–50. doi: 10.1387/ijdb.041957mt
95. Taoudi S, Medvinsky A. Functional Identification of the Hematopoietic Stem Cell Niche in the Ventral Domain of the Embryonic Dorsal Aorta. Proc Natl Acad Sci USA (2007) 104(22):9399–403. doi: 10.1073/pnas.0700984104
96. Taylor E, Taoudi S, Medvinsky A. Hematopoietic Stem Cell Activity in the Aorta-Gonad-Mesonephros Region Enhances After Mid-Day 11 of Mouse Development. Int J Dev Biol (2010) 54(6-7):1055–60. doi: 10.1387/ijdb.103152et
97. Baron CS, Kester L, Klaus A, Boisset JC, Thambyrajah R, Yvernogeau L, et al. Single-Cell Transcriptomics Reveal the Dynamic of Haematopoietic Stem Cell Production in the Aorta. Nat Commun (2018) 9(1):2517. doi: 10.1038/s41467-018-04893-3
98. Souilhol C, Gonneau C, Lendinez JG, Batsivari A, Rybtsov S, Wilson H, et al. Inductive Interactions Mediated by Interplay of Asymmetric Signalling Underlie Development of Adult Haematopoietic Stem Cells. Nat Commun (2016) 7:10784. doi: 10.1038/ncomms10784
99. Taoudi S, Gonneau C, Moore K, Sheridan JM, Blackburn CC, Taylor E, et al. Extensive Hematopoietic Stem Cell Generation in the AGM Region via Maturation of VE-Cadherin+CD45+ Pre-Definitive HSCs. Cell Stem Cell (2008) 3(1):99–108. doi: 10.1016/j.stem.2008.06.004
100. Durand C, Robin C, Bollerot K, Baron MH, Ottersbach K, Dzierzak E. Embryonic Stromal Clones Reveal Developmental Regulators of Definitive Hematopoietic Stem Cells. Proc Natl Acad Sci USA (2007) 104(52):20838–43. doi: 10.1073/pnas.0706923105
101. Crisan M, Solaimani Kartalaei P, Neagu A, Karkanpouna S, Yamada-Inagawa T, Purini C, et al. BMP and Hedgehog Regulate Distinct AGM Hematopoietic Stem Cells Ex Vivo. Stem Cell Rep (2016) 6(3):383–95. doi: 10.1016/j.stemcr.2016.01.016
102. Peeters M, Ottersbach K, Bollerot K, Orelio C, de Bruijn M, Wijgerde M, et al. Ventral Embryonic Tissues and Hedgehog Proteins Induce Early AGM Hematopoietic Stem Cell Development. Development (2009) 136(15):2613–21. doi: 10.1242/dev.034728
103. Sahai-Hernandez P, Pouget C, Svoboda O, Traver D. Dermomyotome-Derived Endothelial Cells Migrate to the Dorsal Aorta to Support Hematopoietic Stem Cell Emergence. bioRxiv (2020). doi: 10.1101/2020.05.14.096305
104. Ambler CA, Nowicki JL, Burke AC, Bautch VL. Assembly of Trunk and Limb Blood Vessels Involves Extensive Migration and Vasculogenesis of Somite-Derived Angioblasts. Dev Biol (2001) 234(2):352–64. doi: 10.1006/dbio.2001.0267
105. Pardanaud L, Luton D, Prigent M, Bourcheix LM, Catala M, Dieterlen-Lievre F. Two Distinct Endothelial Lineages in Ontogeny, One of Them Related to Hemopoiesis. Development (1996) 122(5):1363–71. doi: 10.1242/dev.122.5.1363
106. Pouget C, Gautier R, Teillet MA, Jaffredo T. Somite-Derived Cells Replace Ventral Aortic Hemangioblasts and Provide Aortic Smooth Muscle Cells of the Trunk. Development (2006) 133(6):1013–22. doi: 10.1242/dev.02269
107. Horton PD, Dumbali SP, Bhanu KR, Diaz MF, Wenzel PL. Biomechanical Regulation of Hematopoietic Stem Cells in the Developing Embryo. Curr Tissue Microenviron Rep (2021) 2(1):1–15. doi: 10.1007/s43152-020-00027-4
108. Espin-Palazon R, Weijts B, Mulero V, Traver D. Proinflammatory Signals as Fuel for the Fire of Hematopoietic Stem Cell Emergence. Trends Cell Biol (2018) 28(1):58–66. doi: 10.1016/j.tcb.2017.08.003
109. Bigas A, Guiu J, Gama-Norton L. Notch and Wnt Signaling in the Emergence of Hematopoietic Stem Cells. Blood Cells Mol Dis (2013) 51(4):264–70. doi: 10.1016/j.bcmd.2013.07.005
110. Moignard V, Woodhouse S, Haghverdi L, Lilly AJ, Tanaka Y, Wilkinson AC, et al. Decoding the Regulatory Network of Early Blood Development From Single-Cell Gene Expression Measurements. Nat Biotechnol (2015) 33(3):269–76. doi: 10.1038/nbt.3154
111. Swiers G, Baumann C, O'Rourke J, Giannoulatou E, Taylor S, Joshi A, et al. Early Dynamic Fate Changes in Haemogenic Endothelium Characterized at the Single-Cell Level. Nat Commun (2013) 4:2924. doi: 10.1038/ncomms3924
112. Blokzijl-Franke S, Ponsioen B, Schulte-Merker S, Herbomel P, Kissa K, Choorapoikayil S, et al. Phosphatidylinositol-3 Kinase Signaling Controls Survival and Stemness of Hematopoietic Stem and Progenitor Cells. Oncogene (2021) 40(15):2741–55. doi: 10.1038/s41388-021-01733-5
113. Zhou J, Xu J, Zhang L, Liu S, Ma Y, Wen X, et al. Combined Single-Cell Profiling of lncRNAs and Functional Screening Reveals That H19 Is Pivotal for Embryonic Hematopoietic Stem Cell Development. Cell Stem Cell (2019) 24(2):285–98.e5. doi: 10.1016/j.stem.2018.11.023
114. Hu Y, Huang Y, Yi Y, Wang H, Liu B, Yu J, et al. Single-Cell RNA Sequencing Highlights Transcription Activity of Autophagy-Related Genes During Hematopoietic Stem Cell Formation in Mouse Embryos. Autophagy (2017) 13(4):770–1. doi: 10.1080/15548627.2016.1278093
115. Sugiyama D, Joshi A, Kulkeaw K, Tan KS, Yokoo-Inoue T, Mizuochi-Yanagi C, et al. A Transcriptional Switch Point During Hematopoietic Stem and Progenitor Cell Ontogeny. Stem Cells Dev (2017) 26(5):314–27. doi: 10.1089/scd.2016.0194
116. Zhou F, Li X, Wang W, Zhu P, Zhou J, He W, et al. Tracing Haematopoietic Stem Cell Formation at Single-Cell Resolution. Nature (2016) 533(7604):487–92. doi: 10.1038/nature17997
117. Crosse EI, Gordon-Keylock S, Rybtsov S, Binagui-Casas A, Felchle H, Nnadi NC, et al. Multi-Layered Spatial Transcriptomics Identify Secretory Factors Promoting Human Hematopoietic Stem Cell Development. Cell Stem Cell (2020) 27(5):822–39.e8. doi: 10.1016/j.stem.2020.08.004
118. Vink CS, Calero-Nieto FJ, Wang X, Maglitto A, Mariani SA, Jawaid W, et al. Iterative Single-Cell Analyses Define the Transcriptome of the First Functional Hematopoietic Stem Cells. Cell Rep (2020) 31(6):107627. doi: 10.1016/j.celrep.2020.107627
119. Hou S, Li Z, Zheng X, Gao Y, Dong J, Ni Y, et al. Embryonic Endothelial Evolution Towards First Hematopoietic Stem Cells Revealed by Single-Cell Transcriptomic and Functional Analyses. Cell Res (2020) 30(5):376–92. doi: 10.1038/s41422-020-0300-2
120. Bergiers I, Andrews T, Vargel Bolukbasi O, Buness A, Janosz E, Lopez-Anguita N, et al. Single-Cell Transcriptomics Reveals a New Dynamical Function of Transcription Factors During Embryonic Hematopoiesis. Elife (2018) 7:1–38. doi: 10.7554/eLife.29312
121. Zeng Y, He J, Bai Z, Li Z, Gong Y, Liu C, et al. Tracing the First Hematopoietic Stem Cell Generation in Human Embryo by Single-Cell RNA Sequencing. Cell Res (2019) 29(11):881–94. doi: 10.1038/s41422-019-0228-6
122. Karlsson G, Sommarin MNE, Boiers C. Defining the Emerging Blood System During Development at Single-Cell Resolution. Front Cell Dev Biol (2021) 9:660350. doi: 10.3389/fcell.2021.660350
123. Hadland B, Varnum-Finney B, Dozono S, Dignum T, Nourigat-McKay C, Jackson DL, et al. Engineering a Niche Supporting Haematopoietic Stem Cell Development Using Integrated Single Cell Transcriptomics. bioRxiv (2021). doi: 10.1101/2021.01.25.427999
124. Ibarra-Soria X, Jawaid W, Pijuan-Sala B, Ladopoulos V, Scialdone A, Jorg DJ, et al. Defining Murine Organogenesis at Single-Cell Resolution Reveals a Role for the Leukotriene Pathway in Regulating Blood Progenitor Formation. Nat Cell Biol (2018) 20(2):127–34. doi: 10.1038/s41556-017-0013-z
125. Pijuan-Sala B, Griffiths JA, Guibentif C, Hiscock TW, Jawaid W, Calero-Nieto FJ, et al. A Single-Cell Molecular Map of Mouse Gastrulation and Early Organogenesis. Nature (2019) 566(7745):490–5. doi: 10.1038/s41586-019-0933-9
126. Cao J, O'Day DR, Pliner HA, Kingsley PD, Deng M, Daza RM, et al. A Human Cell Atlas of Fetal Gene Expression. Science (2020) 370(6518):1–42. doi: 10.1126/science.aba7721
127. Chanda B, Ditadi A, Iscove NN, Keller G. Retinoic Acid Signaling Is Essential for Embryonic Hematopoietic Stem Cell Development. Cell (2013) 155(1):215–27. doi: 10.1016/j.cell.2013.08.055
128. Fitch SR, Kimber GM, Wilson NK, Parker A, Mirshekar-Syahkal B, Gottgens B, et al. Signaling From the Sympathetic Nervous System Regulates Hematopoietic Stem Cell Emergence During Embryogenesis. Cell Stem Cell (2012) 11(4):554–66. doi: 10.1016/j.stem.2012.07.002
129. Robin C, Ottersbach K, Durand C, Peeters M, Vanes L, Tybulewicz V, et al. An Unexpected Role for IL-3 in the Embryonic Development of Hematopoietic Stem Cells. Dev Cell (2006) 11(2):171–80. doi: 10.1016/j.devcel.2006.07.002
130. Richard C, Drevon C, Canto PY, Villain G, Bollerot K, Lempereur A, et al. Endothelio-Mesenchymal Interaction Controls Runx1 Expression and Modulates the Notch Pathway to Initiate Aortic Hematopoiesis. Dev Cell (2013) 24(6):600–11. doi: 10.1016/j.devcel.2013.02.011
131. Guo Y, Cheng BYL, Wang D, Ma ACH, He BL, Man TK, et al. Function of Arl4aa in the Initiation of Hematopoiesis in Zebrafish by Maintaining Golgi Complex Integrity in Hemogenic Endothelium. Stem Cell Rep (2020) 14(4):575–89. doi: 10.1016/j.stemcr.2020.02.012
132. Oostendorp RA, Harvey KN, Kusadasi N, de Bruijn MF, Saris C, Ploemacher RE, et al. Stromal Cell Lines From Mouse Aorta-Gonads-Mesonephros Subregions are Potent Supporters of Hematopoietic Stem Cell Activity. Blood (2002) 99(4):1183–9. doi: 10.1182/blood.V99.4.1183
133. Charbord P, Oostendorp R, Pang W, Herault O, Noel F, Tsuji T, et al. Comparative Study of Stromal Cell Lines Derived From Embryonic, Fetal, and Postnatal Mouse Blood-Forming Tissues. Exp Hematol (2002) 30(10):1202–10. doi: 10.1016/S0301-472X(02)00895-0
134. Oostendorp RA, Robin C, Steinhoff C, Marz S, Brauer R, Nuber UA, et al. Long-Term Maintenance of Hematopoietic Stem Cells Does Not Require Contact With Embryo-Derived Stromal Cells in Cocultures. Stem Cells (2005) 23(6):842–51. doi: 10.1634/stemcells.2004-0120
135. Charbord P, Pouget C, Binder H, Dumont F, Stik G, Levy P, et al. A Systems Biology Approach for Defining the Molecular Framework of the Hematopoietic Stem Cell Niche. Cell Stem Cell (2014) 15(3):376–91. doi: 10.1016/j.stem.2014.06.005
136. McGarvey AC, Rybtsov S, Souilhol C, Tamagno S, Rice R, Hills D, et al. A Molecular Roadmap of the AGM Region Reveals BMPER as a Novel Regulator of HSC Maturation. J Exp Med (2017) 214(12):3731–51. doi: 10.1084/jem.20162012
137. Xue Y, Liu D, Cui G, Ding Y, Ai D, Gao S, et al. A 3d Atlas of Hematopoietic Stem and Progenitor Cell Expansion by Multi-Dimensional RNA-Seq Analysis. Cell Rep (2019) 27(5):1567–78.e5. doi: 10.1016/j.celrep.2019.04.030
138. Junker JP, Noel ES, Guryev V, Peterson KA, Shah G, Huisken J, et al. Genome-Wide RNA Tomography in the Zebrafish Embryo. Cell (2014) 159(3):662–75. doi: 10.1016/j.cell.2014.09.038
139. Karpanen T, Padberg Y, van de Pavert SA, Dierkes C, Morooka N, Peterson-Maduro J, et al. An Evolutionarily Conserved Role for Polydom/Svep1 During Lymphatic Vessel Formation. Circ Res (2017) 120(8):1263–75. doi: 10.1161/CIRCRESAHA.116.308813
140. Gao S, Shi Q, Zhang Y, Liang G, Kang Z, Huang B, et al. Identification of HSC/MPP Expansion Units in Fetal Liver by Single-Cell Spatiotemporal Transcriptomics. Cell Res (2021). doi: 10.1038/s41422-021-00540-7
141. Kissa K, Murayama E, Zapata A, Cortes A, Perret E, Machu C, et al. Live Imaging of Emerging Hematopoietic Stem Cells and Early Thymus Colonization. Blood (2008) 111(3):1147–56. doi: 10.1182/blood-2007-07-099499
142. Upadhaya S, Krichevsky O, Akhmetzyanova I, Sawai CM, Fooksman DR, Reizis B. Intravital Imaging Reveals Motility of Adult Hematopoietic Stem Cells in the Bone Marrow Niche. Cell Stem Cell (2020) 27(2):336–45.e4. doi: 10.1016/j.stem.2020.06.003
143. Majeti R, Park CY, Weissman IL. Identification of a Hierarchy of Multipotent Hematopoietic Progenitors in Human Cord Blood. Cell Stem Cell (2007) 1(6):635–45. doi: 10.1016/j.stem.2007.10.001
144. Tarnok A, Ulrich H, Bocsi J. Phenotypes of Stem Cells From Diverse Origin. Cytometry A (2010) 77(1):6–10. doi: 10.1002/cyto.a.20844
145. Perfetto SP, Chattopadhyay PK, Roederer M. Seventeen-Colour Flow Cytometry: Unravelling the Immune System. Nat Rev Immunol (2004) 4(8):648–55. doi: 10.1038/nri1416
146. Chattopadhyay PK, Price DA, Harper TF, Betts MR, Yu J, Gostick E, et al. Quantum Dot Semiconductor Nanocrystals for Immunophenotyping by Polychromatic Flow Cytometry. Nat Med (2006) 12(8):972–7. doi: 10.1038/nm1371
147. Bandura DR, Baranov VI, Ornatsky OI, Antonov A, Kinach R, Lou X, et al. Mass Cytometry: Technique for Real Time Single Cell Multitarget Immunoassay Based on Inductively Coupled Plasma Time-of-Flight Mass Spectrometry. Anal Chem (2009) 81(16):6813–22. doi: 10.1021/ac901049w
148. Lou X, Zhang G, Herrera I, Kinach R, Ornatsky O, Baranov V, et al. Polymer-Based Elemental Tags for Sensitive Bioassays. Angew Chem Int Ed Engl (2007) 46(32):6111–4. doi: 10.1002/anie.200700796
149. Bendall SC, Simonds EF, Qiu P, Amir el AD, Krutzik PO, Finck R, et al. Single-Cell Mass Cytometry of Differential Immune and Drug Responses Across a Human Hematopoietic Continuum. Science (2011) 332(6030):687–96. doi: 10.1126/science.1198704
150. Fragiadakis GK, Baca QJ, Gherardini PF, Ganio EA, Gaudilliere DK, Tingle M, et al. Mapping the Fetomaternal Peripheral Immune System at Term Pregnancy. J Immunol (2016) 197(11):4482–92. doi: 10.4049/jimmunol.1601195
151. Spitzer MH, Gherardini PF, Fragiadakis GK, Bhattacharya N, Yuan RT, Hotson AN, et al. IMMUNOLOGY. An Interactive Reference Framework for Modeling a Dynamic Immune System. Science (2015) 349(6244):1259425. doi: 10.1126/science.1259425
152. Mariani SA, Li Z, Rice S, Krieg C, Fragkogianni S, Robinson M, et al. Pro-Inflammatory Aorta-Associated Macrophages Are Involved in Embryonic Development of Hematopoietic Stem Cells. Immunity (2019) 50(6):1439–52.e5. doi: 10.1016/j.immuni.2019.05.003
153. Ha MK, Kwon SJ, Choi JS, Nguyen NT, Song J, Lee Y, et al. Mass Cytometry and Single-Cell RNA-Seq Profiling of the Heterogeneity in Human Peripheral Blood Mononuclear Cells Interacting With Silver Nanoparticles. Small (2020) 16(21):e1907674. doi: 10.1002/smll.201907674
154. Giesen C, Wang HA, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, et al. Highly Multiplexed Imaging of Tumor Tissues With Subcellular Resolution by Mass Cytometry. Nat Methods (2014) 11(4):417–22. doi: 10.1038/nmeth.2869
155. Giarratana MC, Rouard H, Dumont A, Kiger L, Safeukui I, Le Pennec PY, et al. Proof of Principle for Transfusion of In Vitro-Generated Red Blood Cells. Blood (2011) 118(19):5071–9. doi: 10.1182/blood-2011-06-362038
156. Giarratana MC, Kobari L, Lapillonne H, Chalmers D, Kiger L, Cynober T, et al. Ex Vivo Generation of Fully Mature Human Red Blood Cells From Hematopoietic Stem Cells. Nat Biotechnol (2005) 23(1):69–74. doi: 10.1038/nbt1047
157. Timmermans F, Velghe I, Vanwalleghem L, De Smedt M, Van Coppernolle S, Taghon T, et al. Generation of T Cells From Human Embryonic Stem Cell-Derived Hematopoietic Zones. J Immunol (2009) 182(11):6879–88. doi: 10.4049/jimmunol.0803670
158. June CH, Sadelain M. Chimeric Antigen Receptor Therapy. N Engl J Med (2018) 379(1):64–73. doi: 10.1056/NEJMra1706169
159. Batta K, Menegatti S, Garcia-Alegria E, Florkowska M, Lacaud G, Kouskoff V. Concise Review: Recent Advances in the In Vitro Derivation of Blood Cell Populations. Stem Cells Transl Med (2016) 5(10):1330–7. doi: 10.5966/sctm.2016-0039
160. Pereira CF, Chang B, Qiu J, Niu X, Papatsenko D, Hendry CE, et al. Induction of a Hemogenic Program in Mouse Fibroblasts. Cell Stem Cell (2013) 13(2):205–18. doi: 10.1016/j.stem.2013.05.024
161. Sandler VM, Lis R, Liu Y, Kedem A, James D, Elemento O, et al. Reprogramming Human Endothelial Cells to Haematopoietic Cells Requires Vascular Induction. Nature (2014) 511(7509):312–8. doi: 10.1038/nature13547
162. Riddell J, Gazit R, Garrison BS, Guo G, Saadatpour A, Mandal PK, et al. Reprogramming Committed Murine Blood Cells to Induced Hematopoietic Stem Cells With Defined Factors. Cell (2014) 157(3):549–64. doi: 10.1016/j.cell.2014.04.006
163. Sugimura R, Jha DK, Han A, Soria-Valles C, da Rocha EL, Lu YF, et al. Haematopoietic Stem and Progenitor Cells From Human Pluripotent Stem Cells. Nature (2017) 545(7655):432–8. doi: 10.1038/nature22370
164. Doulatov S, Vo LT, Chou SS, Kim PG, Arora N, Li H, et al. Induction of Multipotential Hematopoietic Progenitors From Human Pluripotent Stem Cells via Respecification of Lineage-Restricted Precursors. Cell Stem Cell (2013) 13(4):459–70. doi: 10.1016/j.stem.2013.09.002
165. Lis R, Karrasch CC, Poulos MG, Kunar B, Redmond D, Duran JGB, et al. Conversion of Adult Endothelium to Immunocompetent Haematopoietic Stem Cells. Nature (2017) 545(7655):439–45. doi: 10.1038/nature22326
166. Daniel MG, Rapp K, Schaniel C, Moore KA. Induction of Developmental Hematopoiesis Mediated by Transcription Factors and the Hematopoietic Microenvironment. Ann N Y Acad Sci (2020) 1466(1):59–72. doi: 10.1111/nyas.14246
167. Chadwick K, Wang L, Li L, Menendez P, Murdoch B, Rouleau A, et al. Cytokines and BMP-4 Promote Hematopoietic Differentiation of Human Embryonic Stem Cells. Blood (2003) 102(3):906–15. doi: 10.1182/blood-2003-03-0832
168. Fujimoto TT, Kohata S, Suzuki H, Miyazaki H, Fujimura K. Production of Functional Platelets by Differentiated Embryonic Stem (ES) Cells In Vitro. Blood (2003) 102(12):4044–51. doi: 10.1182/blood-2003-06-1773
169. Kennedy M, Awong G, Sturgeon CM, Ditadi A, LaMotte-Mohs R, Zuniga-Pflucker JC, et al. T Lymphocyte Potential Marks the Emergence of Definitive Hematopoietic Progenitors in Human Pluripotent Stem Cell Differentiation Cultures. Cell Rep (2012) 2(6):1722–35. doi: 10.1016/j.celrep.2012.11.003
170. Choi KD, Vodyanik MA, Togarrati PP, Suknuntha K, Kumar A, Samarjeet F, et al. Identification of the Hemogenic Endothelial Progenitor and its Direct Precursor in Human Pluripotent Stem Cell Differentiation Cultures. Cell Rep (2012) 2(3):553–67. doi: 10.1016/j.celrep.2012.08.002
171. Uchida N, Haro-Mora JJ, Fujita A, Lee DY, Winkler T, Hsieh MM, et al. Efficient Generation of Beta-Globin-Expressing Erythroid Cells Using Stromal Cell-Derived Induced Pluripotent Stem Cells From Patients With Sickle Cell Disease. Stem Cells (2017) 35(3):586–96. doi: 10.1002/stem.2517
172. Ruiz JP, Chen G, Haro Mora JJ, Keyvanfar K, Liu C, Zou J, et al. Robust Generation of Erythroid and Multilineage Hematopoietic Progenitors From Human iPSCs Using a Scalable Monolayer Culture System. Stem Cell Res (2019) 41:101600. doi: 10.1016/j.scr.2019.101600
173. Zambidis ET, Peault B, Park TS, Bunz F, Civin CI. Hematopoietic Differentiation of Human Embryonic Stem Cells Progresses Through Sequential Hematoendothelial, Primitive, and Definitive Stages Resembling Human Yolk Sac Development. Blood (2005) 106(3):860–70. doi: 10.1182/blood-2004-11-4522
174. Fidanza A, Stumpf PS, Ramachandran P, Tamagno S, Babtie A, Lopez-Yrigoyen M, et al. Single-Cell Analyses and Machine Learning Define Hematopoietic Progenitor and HSC-Like Cells Derived From Human PSCs. Blood (2020) 136(25):2893–904. doi: 10.1182/blood.2020006229
175. Aguilera-Castrejon A, Oldak B, Shani T, Ghanem N, Itzkovich C, Slomovich S, et al. Ex Utero Mouse Embryogenesis From Pre-Gastrulation to Late Organogenesis. Nature (2021) 593(7857):119–24. doi: 10.1038/s41586-021-03416-3
176. Chatterjee C, Schertl P, Frommer M, Ludwig-Husemann A, Mohra A, Dilger N, et al. Rebuilding the Hematopoietic Stem Cell Niche: Recent Developments and Future Prospects. Acta Biomater (2021) 132:129–48. doi: 10.1016/j.actbio.2021.03.061
177. Congrains A, Bianco J, Rosa RG, Mancuso RI, Saad STO. 3d Scaffolds to Model the Hematopoietic Stem Cell Niche: Applications and Perspectives. Materials (Basel) (2021) 14(3):1–18. doi: 10.3390/ma14030569
178. Moscona A, Moscona H. The Dissociation and Aggregation of Cells From Organ Rudiments of the Early Chick Embryo. J Anat (1952) 86(3):287–301.
179. Clevers H. Modeling Development and Disease With Organoids. Cell (2016) 165(7):1586–97. doi: 10.1016/j.cell.2016.05.082
180. Ackermann M, Haake K, Kempf H, Kaschutnig P, Weiss AC, Nguyen AHH, et al. A 3d iPSC-Differentiation Model Identifies Interleukin-3 as a Regulator of Early Human Hematopoietic Specification. Haematologica (2021) 106(5):1354–67. doi: 10.3324/haematol.2019.228064
181. Keller G, Kennedy M, Papayannopoulou T, Wiles MV. Hematopoietic Commitment During Embryonic Stem Cell Differentiation in Culture. Mol Cell Biol (1993) 13(1):473–86. doi: 10.1128/MCB.13.1.473
182. Nakano T, Kodama H, Honjo T. In Vitro Development of Primitive and Definitive Erythrocytes From Different Precursors. Science (1996) 272(5262):722–4. doi: 10.1126/science.272.5262.722
183. Subramanian A, Guo B, Marsden MD, Galic Z, Kitchen S, Kacena A, et al. Macrophage Differentiation From Embryoid Bodies Derived From Human Embryonic Stem Cells. J Stem Cells (2009) 4(1):29–45.
184. Vittet D, Prandini MH, Berthier R, Schweitzer A, Martin-Sisteron H, Uzan G, et al. Embryonic Stem Cells Differentiate In Vitro to Endothelial Cells Through Successive Maturation Steps. Blood (1996) 88(9):3424–31. doi: 10.1182/blood.V88.9.3424.bloodjournal8893424
185. Wiles MV, Keller G. Multiple Hematopoietic Lineages Develop From Embryonic Stem (ES) Cells in Culture. Development (1991) 111(2):259–67. doi: 10.1242/dev.111.2.259
186. Yamaguchi T, Tashiro K, Tanaka S, Katayama S, Ishida W, Fukuda K, et al. Two-Step Differentiation of Mast Cells From Induced Pluripotent Stem Cells. Stem Cells Dev (2013) 22(5):726–34. doi: 10.1089/scd.2012.0339
187. Guyonneau-Harmand BLH L, Birebent B, Desterke C, Chevallier N, Garçon L, Lapillonne H, et al. Transgene-Free Hematopoietic Stem and Progenitor Cells From Human Induced Pluripotent Stem Cells. bioRxiv (2017). doi: 10.1101/177691
188. Turner DA, Girgin M, Alonso-Crisostomo L, Trivedi V, Baillie-Johnson P, Glodowski CR, et al. Anteroposterior Polarity and Elongation in the Absence of Extra-Embryonic Tissues and of Spatially Localised Signalling in Gastruloids: Mammalian Embryonic Organoids. Development (2017) 144(21):3894–906. doi: 10.1242/dev.150391
189. van den Brink SC, Baillie-Johnson P, Balayo T, Hadjantonakis AK, Nowotschin S, Turner DA, et al. Symmetry Breaking, Germ Layer Specification and Axial Organisation in Aggregates of Mouse Embryonic Stem Cells. Development (2014) 141(22):4231–42. doi: 10.1242/dev.113001
190. Veenvliet JV, Bolondi A, Kretzmer H, Haut L, Scholze-Wittler M, Schifferl D, et al. Mouse Embryonic Stem Cells Self-Organize Into Trunk-Like Structures With Neural Tube and Somites. Science (2020) 370(6522):1–7. doi: 10.1126/science.aba4937
191. van den Brink SC, Alemany A, van Batenburg V, Moris N, Blotenburg M, Vivie J, et al. Single-Cell and Spatial Transcriptomics Reveal Somitogenesis in Gastruloids. Nature (2020) 582(7812):405–9. doi: 10.1038/s41586-020-2024-3
192. Xu PF, Borges RM, Fillatre J, de Oliveira-Melo M, Cheng T, Thisse B, et al. Construction of a Mammalian Embryo Model From Stem Cells Organized by a Morphogen Signalling Centre. Nat Commun (2021) 12(1):3277. doi: 10.1038/s41467-021-23653-4
Keywords: hematopoietic stem cells, aorta-gonad-mesonephros, microenvironment, niche, single cell RNA sequencing, tomography sequencing, embryo, hemogenic endothelium
Citation: Weijts B, Yvernogeau L and Robin C (2021) Recent Advances in Developmental Hematopoiesis: Diving Deeper With New Technologies. Front. Immunol. 12:790379. doi: 10.3389/fimmu.2021.790379
Received: 06 October 2021; Accepted: 28 October 2021;
Published: 24 November 2021.
Edited by:
Dominique Bonnet, Francis Crick Institute, United KingdomReviewed by:
Mihaela Crisan, University of Edinburgh, United KingdomCopyright © 2021 Weijts, Yvernogeau and Robin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Catherine Robin, Yy5yb2JpbkBodWJyZWNodC5ldQ==
†Present address: Laurent Yvernogeau, Sorbonne Université, IBPS, CNRS UMR7622, Inserm U1156, Laboratoire de Biologie du Développement, Paris, France
‡These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.