- 1Radiotherapy & OncoImmunology Laboratory, Department of Radiation Oncology, Radboud Institute for Molecular Life Sciences, Radboud University Medical Center, Nijmegen, Netherlands
- 2Hubrecht Institute, Royal Netherlands Academy of Arts and Sciences (KNAW), Utrecht, Netherlands
- 3Copenhagen Center for Glycomics, Departments of Cellular and Molecular Medicine, Faculty of Health Sciences, University of Copenhagen, Copenhagen, Denmark
Sialic acid-binding immunoglobulin-like lectins (Siglecs) are a family of receptors that recognize sialoglycans – sialic acid containing glycans that are abundantly present on cell membranes. Siglecs are expressed on most immune cells and can modulate their activity and function. The majority of Siglecs contains immune inhibitory motifs comparable to the immune checkpoint receptor PD-1. In the tumor microenvironment (TME), signaling through the Siglec-sialoglycan axis appears to be enhanced through multiple mechanisms favoring tumor immune evasion similar to the PD-1/PD-L1 signaling pathway. Siglec expression on tumor-infiltrating immune cells appears increased in the immune suppressive microenvironment. At the same time, enhanced Siglec ligand expression has been reported for several tumor types as a result of aberrant glycosylation, glycan modifications, and the increased expression of sialoglycans on proteins and lipids. Siglec signaling has been identified as important regulator of anti-tumor immunity in the TME, but the key factors contributing to Siglec activation by tumor-associated sialoglycans are diverse and poorly defined. Among others, Siglec activation and signaling are co-determined by their expression levels, cell surface distribution, and their binding preferences for cis- and trans-ligands in the TME. Siglec binding preference are co-determined by the nature of the proteins/lipids to which the sialoglycans are attached and the multivalency of the interaction. Here, we review the current understanding and emerging conditions and factors involved in Siglec signaling in the TME and identify current knowledge gaps that exist in the field.
Introduction
Humans express 14 members of the Sialic acid-binding immunoglobulin-like lectins (Siglecs) that are divided in two subfamilies based on their sequence similarity and evolutionary conservation. Siglec-1 (also known as sialoadhesin and CD169), Siglec-2 (also known as CD22) Siglec-4 and Siglec-15 have clear orthologous in mammalian species (1–4). The CD33-related Siglecs (Siglec-3 also known as CD33, Siglec-5 to -11, -14 and -16) evolved more rapidly among species. These transmembrane receptors are mainly present on immune cells, but also on other cell types such as trophoblasts, myelin-forming cells, and stromal cells (1, 3, 4). Siglecs bind their ligands via an extracellular N-terminal V-set domain. Intracellularly, most Siglecs (Siglec-3, -5 to -9, and -11) harbor a combination of a membrane proximal ITIM (immunoreceptor tyrosine-based inhibition motif) domain and a membrane distal ITIM-like domain (1–3). Upon Siglec activation, this ITIM domain is phosphorylated by Src family kinases. Phosphorylation subsequently leads to recruitment of SH2-domain containing phosphatases SHP-1 and/or SHP-2, which dephosphorylate downstream components of immune stimulatory pathways, thus inhibiting cellular activation. Interestingly, three Siglecs (Siglec-14, -15 and -16) do not contain ITIM domains, but have a positively charged residue in their transmembrane domain that enables them to complex with ITAM (immunoreceptor tyrosine-based activation motif) containing adaptor proteins, such as DAP10 or DAP12 (2, 3, 5). This leads to recruitment of protein kinases that can phosphorylate downstream targets, eventually triggering downstream signaling pathways (1–3, 5). Besides ITIM- and ITAM-containing Siglecs, Siglec-1 does not contain known intracellular signaling motifs.
The ligands for the Siglecs are constituted by glycans, highly diverse biomolecules composed of various monosaccharides that are linked to membrane bound and secreted glycoproteins, glycolipids, and as recently suggested also small noncoding glycoRNAs (3, 4, 6–9). Siglecs selectively recognize glycans that contain one or more negatively charged sialic acid residues, so called sialoglycans (1, 6). Sialoglycans are present on virtually every human cell, but are aberrantly expressed on tumor cells where they contribute to tumor growth and progression, including metastasis (10). Interestingly, Siglec expression has been reported to be enhanced and/or induced on other cells within the TME, including immune cells (11–16). Concomitantly, the combination of altered Siglec expression on immune cells and aberrant sialoglycan expression on tumor cells, could possibly lead to strong Siglec activation and resulting in immune cell inhibition within the TME (Figure 1). The enhanced sialoglycans expression within the TME is clearly described in literature, however, the evidence that this directly translates into more Siglec ligand expression and Siglec signaling remains scarce.

Figure 1 Schematic overview of changes in the Siglec-sialoglycan axis upon tumor formation. Immune cells circulating in healthy tissue express Siglecs and cells in this environment express ‘normal’ sialic acid (red diamonds) types and levels. Upon tumor development, immune cells infiltrate the TME, such as tumor-associated macrophages, T cells, and myeloid-derived suppressor cells (and many other types). These immune cells display changed and partially enhanced Siglec expression levels and can interact with tumor-associated sialoglycans that are formed and often increased as result of altered glycosylation in cancer cells. Siglec-sialoglycan interactions in the TME can lead to functional modulation of infiltrating immune cells and the anti-tumor immune response.
The structure and signaling motifs of Siglec receptors show great resemblance to the well-known inhibitory receptor PD-1 (3, 17) that is used as target in immunotherapy of cancer. Moreover, Siglecs are often co-expressed on T cells with other inhibitory receptors, such as PD-1, TIM-3 and LAG-3 (11). Therefore, the immune inhibitory Siglecs are currently envisioned as potential immune checkpoint receptors that can be targeted in cancer (17–20). Despite Siglecs emerging as an attractive target for cancer immunotherapy, the exact cell and glycobiological conditions that trigger, regulate and control Siglec signaling in the TME remain largely elusive. Here, we will review the current knowledge on factors (co-)determining Siglec signaling and discuss knowledge gaps regarding this Siglec-sialoglycan signaling axis focusing on the TME.
Siglec Expression Within the TME
Siglec family members are expressed by most cell subsets of the human immune system and by several tissues and cell types outside of the immune system (1–4). Some Siglecs have immune cell type specific expression and can therefore be used as differentiation marker such as Siglec-1 that is found mainly on macrophages, Siglec-2 that is specifically expressed by B cells, Siglec-3 that marks cells from the myeloid lineage including microglia cells, or Siglec-8 that is found predominantly on eosinophils (3, 8). In the TME, focus has been mainly on the inhibitory Siglecs-7, -9, and -10 as well as the activating Siglec-15. Siglec-7 and -9 are abundantly present on natural killer (NK) cells with Siglec-7 being a pan-marker for human NK cells and Siglec-9 being present on a population of CD56dim CD16+ NK cells (21). Strong interactions of these Siglecs with synthetic and natural multivalent sialoglycan ligands appears to be sufficient to inhibit NK cell activation (22). Accordingly, blocking Siglec-7 and -9 interactions with sialoglycans using monoclonal antibodies increased killing of several tumor cell lines (K562 and HeLa) by peripheral blood NK cells (21). Another example of a Siglec-mediated immune modulatory effect was observed in T cells. Only a small percentage (1-3%) of peripheral blood CD8+ T cells express Siglec-7 and/or Siglec-9 (11, 23). Remarkably, Siglec-9 expression on CD8+ T cells present in peripheral blood and tumor tissue of non-small cell lung cancer (NSCLC) patients is upregulated up to 25% and 40% respectively (11). Similarly, tumor-infiltrating CD8+ T cells in melanoma patients have also been found to display enhanced levels of Siglec-9 (13). Functionally, Fab fragments targeting Siglec-9 could modulate activation and IL-2 production of staphylococcal enterotoxin B stimulated tumor-infiltrating lymphocytes of primary NSCLC patients (11). Siglec-9 antibodies could increase cytotoxicity of healthy donor CD8+ T cells against anti-CD3-loaded P815 tumor cells and enhance IFNγ and TNFα production (13). Besides NK and T cells, immune modulation of Siglec-9 expressing myeloid cells has also been observed. For instance, Rodriguez et al. showed that pancreatic ductal adenocarcinoma-derived sialic acid can cause monocytes to differentiate to macrophages, which was prevented using anti-Siglec-7 and anti-Siglec-9 blocking antibodies (24).
Siglec-15 has emerged as an interesting target for cancer immunotherapy (25). Siglec-15 contains a positively charged residue in its transmembrane domain and is therefore regarded as an activating Siglec (26). Siglec-15 expression was observed on tumor-associated macrophages (TAMs) in the TME of human lung and rectal adenocarcinoma and human hepatocellular carcinoma (27). The interaction between Siglec-15 expressing monocytic THP-1 cells and sialyl-Tn expressing H157 human lung carcinoma cells enhanced TGF-β production. Next to its expression on myeloid cells, Siglec-15 was observed on stromal cells and even on tumor cells (28). Siglec-15 expression on tumor cells was verified by others on several types of cancer, such as lymphoma, gastric cancer and acute myeloid leukemia (29–31). Similarly, Siglec-8 has been reported to be expressed by breast cancer cells (32). Next to that, several Siglecs were found to be expressed on hematological cancers (33–35). Siglec-6 expression was observed on acute myeloid leukemia cell lines, primary acute myeloid leukemia blasts, transformed B-cells in chronic lymphocytic leukemia (33). Siglec-2 expression on acute lymphoid leukemia has also been documented and Siglec-3 is found on blasts in nearly all acute myeloid leukemia patients (34, 35). Besides Siglec expression on immune cells and on tumor cells, Siglecs might also be expressed within the stromal compartment, as Siglec-11 expression was found on human and chimpanzee ovarian stromal cells (36). However, the influence of the stromal compartment on the Siglec-sialoglycan signaling axis remains largely unexplored.
Regulation of Siglec Expression
How Siglec expression is being triggered and regulated is still largely unknown, including the factors contributing to the enhanced expression in the TME (12). Siglec-10 was found to be upregulated on CD52+ CD4+ T cells upon CD3/CD28 activation (37, 38). Therefore, one factor contributing to enhanced Siglec expression is cellular activation, but this was not investigated within the TME context. Another way tumor cells influence Siglec-10 expression on immune cells has been studied by Li et al. (39). They showed that extracellular vesicles (EVs) isolated from ascites from epithelial ovarian cancer patients could induce Siglec-10 expression on Jurkat T cells. Functionally, treatment of Jurkat T cells with tumor cell derived EVs inhibited PMA/ionomycin-induced protein kinase C activity and impaired phosphorylation of tyrosine kinase zeta-chain-associated protein kinase 70 after activation with anti-CD3, but direct evidence for Siglec signaling is lacking.
Siglec-15 expression was found to be downregulated by IFN-γ on monocyte-derived macrophages and RAW264.7 cells and to be dependent on macrophage-colony stimulating factor (28). Interestingly, also Siglec-1 expression was shown to be influenced by cytokines, as it could be induced by IFN-α and IFN-γ (40). Moreover, a few studies have observed that immune suppressive cytokines can influence Siglec expression, which might explain how the immune suppressive TME affects Siglec expression on immune cells. For instance, De Saint Jean et al. (2017) reported that TGF-β1 can enhance Siglec-1, but not Siglec-5, -7, -9, -10 and -14, expression on monocyte-derived dendritic cells (41). Nagase et al. did not observe increased Siglec-1 expression on the monocytic THP-1 cell line by treatment with TGF-β, but rather after stimulation with IL-1β and TNF-α (42). Another study showed that high-expressing Siglec-1 peripheral blood mononuclear cells (PBMC) derived macrophages (CD14+ monocytes differentiated with 10% human AB serum) downregulate Siglec-1 expression upon treatment with tumor culture supernatant from HepG2 cells and upon treatment with recombinant human TGF-β, but not with recombinant human TNF-α or IL-10 (43). Lastly, Calzada-Wack et al. showed that IL-10 treatment of in vitro cultured blood monocytes resulted in decreased Siglec-3 expression (44). Furthermore, it has been shown that Siglec-expressing cells can also be recruited to the TME, for example the TME of gliomas can contain many myeloid-derived suppressor cells (MDSCs) that express Siglecs (45). These data show that both monocytic MDSCs and polymorphonuclear MDSCs from the glioma TME express Siglec-3, -5, 7 and -9. Potentially, TME characteristics such as metabolic shift, hypoxic areas and changes in stromal compartment could all affect Siglec expression and signaling in the TME.
One component that might influence Siglec expression in general and in the TME in particular is the dynamics of their recruitment to the membrane, internalization, recycling, and degradation. Siglec-2 has been shown to be constitutively endocytosed, and several Siglec receptors (Siglec-1, Siglec-2, Siglec-3) have been demonstrated to be internalized after ligand binding or antibody crosslinking, and it was shown that Siglec-2 internalization was controlled by tyrosine residues Tyr (843) or Tyr (863) in its intracellular ITIM motif (46–52). Phosphorylation of the ITIM motifs can induce an intracellular pathway that eventually leads to Siglec degradation. Suppressor of cytokine signaling 3 and Cbl, a RING finger-containing E3 ligase, can bind the phosphorylated ITIM of Siglec-3 (51, 53). This leads to ubiquitination and proteasomal degradation of Siglec-3.
Regarding Siglec recruitment to the membrane, a recent study by Chen et al. showed that N-glycosylation of Siglec-15 affects its localization (54). Treatment with a variety of lysosome inhibitors showed that Siglec-15 was degraded in a lysosome-dependent manner in Siglec-15 overexpressing HEK293T cells. Moreover, inhibition of glycosylation using tunicamycin diminished transportation of Siglec-15 to the cell membrane and promoted lysosomal degradation of the receptor. Interestingly, this process was regulated by glucose uptake. As we will touch upon later, differences in glucose uptake are observed within the hypoxic TME, so how this affects Siglec expression is an interesting topic for future studies (55).
A few studies have observed soluble Siglec receptors, indicating that receptor shedding could contribute to Siglec expression as well. Ito et al. for instance showed presence of the ectodomain of Siglec-9 in the secretome of serum-free conditioned medium from stem cells derived from human exfoliated deciduous teeth (56). Moreover, soluble Siglec-8 has been observed in serum from patients with different forms of eosinophilia (57). However, the contribution and effect of Siglec receptor shedding in the TME remains to our knowledge largely unstudied.
In summary, recent studies support the notion that Siglec expression on immune cells in the TME is enhanced compared to normal physiology. These changes in Siglec expression may be the result of altered cellular signaling and activation, secreted factors such as EVs and cytokines in the TME, as well as altered expression, glycosylation, internalization, and degradation dynamics of Siglecs themselves. In addition, to upregulation of expression, the enhanced presence of Siglec expressing cells could also be due to preferential recruitment to the TME. More detailed investigation is required to understand how Siglec expression and function is controlled under steady state conditions and within the TME, including the transcription factors and epigenetic mechanisms involved.
Sialoglycan Ligands for Siglecs in the TME
Both healthy cells and cancer cells generate structurally highly diverse sialoglycans that are displayed on the cell surface and on secreted glycoproteins and glycolipids. Early studies already reported changes in sialoglycan expression during cancer development and found sialoglycans on multiple distinct cell types within the TME, including tumor cells and immune cells (6, 10, 15, 16, 58–60). Sialic acids were described to play a role in tumor progression, for instance by masking antigens on tumor cells to prevent immune cell recognition, by avoiding complement activation, hindering physical interactions with immune cells, and by functioning as ligand for immunosuppressive Siglecs (10). As reviewed extensively elsewhere, there are many different ways employed by tumors to change their glycosylation patterns including sialic acid levels (15, 61–65). In addition to tumor cells, Siglec ligands have been observed in the stromal compartment of the TME. For instance, Siglec-7 and -9 ligands were identified on human bone marrow-derived mesenchymal stromal cells (66). Interestingly, Siglec-9 ligands were higher expressed on cancer-associated fibroblasts than on normal fibroblasts. In mice, Siglec-E ligands were found on mesenchymal stromal cells in the presence of tumor conditioned medium and on aortic endothelial cells after lipopolysaccharide (LPS) treatment (67). Siglec-9 ligands were also expressed on human aorta and HUV-EC-C endothelial cells at high glucose levels and Siglec-10 was shown to bind the endothelial protein VAP-1 (67, 68). However, expression of Siglec ligands on stromal cells needs further confirmation in vivo.
Printed arrays of synthetic glycans and cell-based arrays have revealed that the individual Siglec family members have unique and partially overlapping binding specificities for distinct ‘healthy’ or cancer-associated sialoglycan structures (3, 8, 9, 69–71). However, the binding epitome for all the human Siglecs and ligand dynamics in the TME is of tremendous structural diversity and has not been fully mapped (Figure 2). Arguably, the glycosylation changes in the TME that result in specific Siglec binding or potentially abrogate binding events largely determine signaling of inhibitory and activating Siglecs.
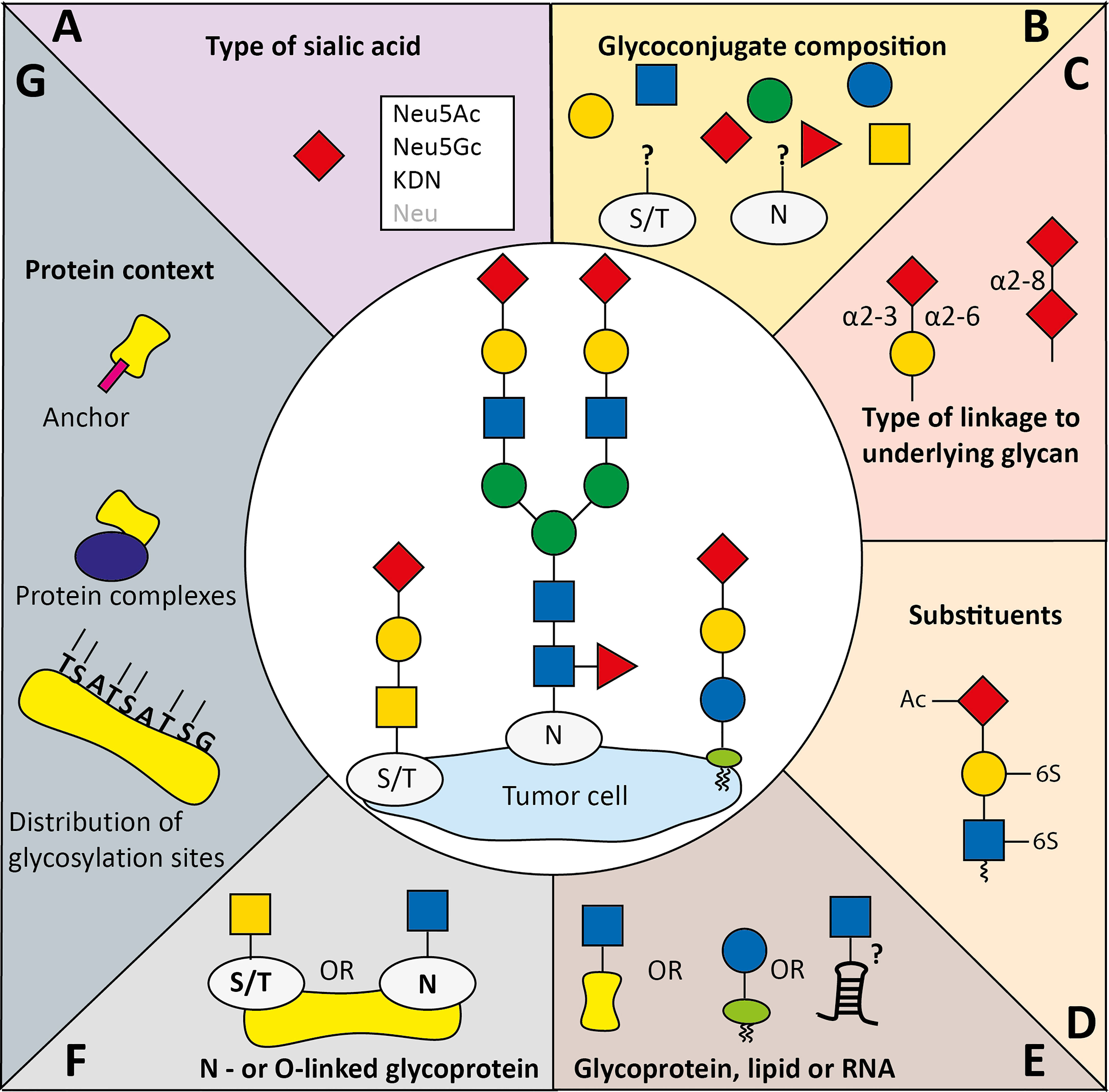
Figure 2 Variables implicated in co-determining Siglec specificity for sialoglycans. Siglecs display specific binding preferences for sialoglycans which is among others determined by (A) the type of sialic acid, (B) the glycan composition, and (C) the type of glycosidic linkage of sialic acid to the penultimate glycan. Siglec binding preferences to sialoglycans can be (co-)determined by (D) glycan modifications to the sialic acids or other glycans (Ac: acetylation, 6-S: 6-O-sulfation), (E) the glycan carrier backbone which can be lipids, proteins, and potentially RNA, (F) the type of protein glycosylation (O-linked to the oxygen atom in serine or threonine, or N-linked to the nitrogen atom in asparagine). Besides, (G) the protein backbone, including distribution of glycosylation sites, complexing with other proteins and specific (glycosylated) anchors can provide specific binding context for Siglecs.
It is important to note that the increased sialylation within the TME is mostly demonstrated with plant lectin staining and gene expression data of the enzymes involved in the sialylation pathway. Recombinant Siglec proteins are used to show whether these changes in sialoglycans also lead to more Siglec ligand binding, however, data demonstrating how this translates into more Siglec signaling is lacking or mostly indirect.
Many structural aspects of sialoglycans can contribute to Siglec binding. Here, we will summarize the current knowledge on factors implicated in Siglec preferences for specific sialoglycans and identify factors regulating expression of these sialoglycans within the TME (Figure 2).
Siglec Binding Preferences for Sialic Acids
All functionally expressed human Siglecs contain a conserved arginine residue in the V-set domain that forms a salt bridge with the carboxylate group of sialic acids and is essential for sialic acid recognition (1, 4). Sequence diversity between the Siglecs further co-determines their glycan fine binding specificities. The term sialic acids refers to a large family of related sugar derivatives that share the same 9-carbon backbone which can undergo extensive natural modification (6, 72). The four core sialic acids are N-acetylneuraminic acid (Neu5Ac), N-glycolylneuraminic acid (Neu5Gc), 3-deoxy-D-glycero-D-galacto-nonulosonic acid (KDN), and neuraminic acid (Neu) which can be further altered by additional modifications (Figures 2A–D) (6). Generally human Siglecs prefer Neu5Ac, the most abundant sialic acid in human, but some including human Siglec-2 recognize both Neu5Ac and Neu5Gc with similar affinity (73–76).
Next to the sialic acid core structure, Siglecs feature specific preference for how the sialic acid is linked (α2-3, α2-6, or α2-8 linkages) to underlying glycans. For instance, Siglec-2 has clear preference for α2-6-linkages, Siglec-9 prefers α2-3-linked sialic acid, and Siglec-7 seems to have preference for both α2-3 and α2-8 linkages (3, 9, 69, 71, 77) (Figure 2C). Altered expression of sialyltransferase enzymes during cancer development could change the sialic acid linkage types towards one or the other. For instance, ST6GalNAc-I has been associated with cancer, which leads to α2-6 linked sialyl Tn antigen that has been shown to be recognized by Siglec-7 (9, 78, 79). Siglec-15 has also been suggested to bind Sialyl Tn, although a recent study could not confirm this interaction (9, 26, 80).
Modification of Monosaccharides
Besides the type of the sialic acid core, Siglec binding to its ligands can be influenced by modification of monosaccharides within the sialoglycan (Figure 2D) (3, 70). For instance, galactose 6-O-sulfation has been identified as critical component for Siglec-3, -5, -8 and -14 binding and 6-O-acetylation was shown to be important for Siglec-9 binding (9, 81–83). Accordingly, expression of the sulfotransferase CHST1 in HEK293 cells that installs a 6-O-sulfo group to galactose induced binding of recombinant Siglec-3, -8, and -15 and largely enhanced Siglec-7 binding (9, 82). In line with this, several studies showed that Siglec-7 binds the carbohydrate sialyl 6-sulfo Lewisx that is expressed by non-transformed colonic cells, rather than sialyl Lewisx, which is expressed by tumor cells (23, 84, 85). This could result in decreased Siglec activation and concomitant enhanced inflammation (23). It was demonstrated that Siglec-7 on macrophages exerts an immunosuppressive effect upon ligand binding. Besides sulfation, other modifications have also been described to affect Siglec binding. For instance, a study that investigated binding of recombinant Siglec Fc chimeras to biotinylated polyacrylamide probes that were conjugated to sialylated glycans found that fucosylation of sialoglycans can reduce Siglec binding (86). Furthermore, sialic acid O-acetylation has been shown to generally abrogate Siglec binding to sialoglycans (86, 87). Using CRISPR-Cas9-mediated knockouts of Sialic acid acetylesterase and Sialic acid acetyltransferase, a recent study by Grabenstein et al. showed that Siglec-7, -9, -10 and -11 binding to HCT 116 colon cancer cells and A549 lung cancer cells was decreased upon knock out of the Sialic acid acetylesterase (88). Furthermore, knock out of Sialic acid acetylesterase resulted in enhanced NK cell-mediated cytotoxicity in colon and lung cancer cells.
These studies suggest that glycan modifications such as sulfation and acetylation and the regulation of expression and activity of glycan modifying enzymes in the TME can have a strong impact on Siglec interactions with sialoglycans. Further studies into the presence of these modification in the TME compared to healthy tissue and effects on Siglec signaling could reveal a major role for glycan modifications in the tumor immune response.
Influence of Protein/Lipid Context on Siglec Binding
Sialoglycans are attached to different backbones, such as proteins/peptides (either N-linked to asparagine or O-linked to serine or threonine) or lipids (89, 90). Recently Flynn et al. provided evidence that sialoglycans can be attached to conserved small noncoding RNAs (7) (Figures 2E, F). Cell surface glycoRNAs were proposed to specifically interact with Siglec-11 and -14, but not with Siglec-5 which shares the same V-set domain sequence with Siglec-14 (7, 91). Siglecs can harbor a very distinct binding preferences for N- and O-linked sialoglycans which is presumably mediated by the variable Siglec C-C’ loop (3, 4, 9, 59, 70, 71). Ligand interactions with Siglec-7, and probably the other Siglecs, induced conformational changes in the sialic acid binding site and in the C-C’ loop resulting in further interactions with the glycosphingolipid core region itself, indicating that the lipid context is of importance for Siglec-7 binding (92, 93). Furthermore, Siglec-7 binds the two linked sialic acid residues (Neu5Acα2,8Neu5Acα) and most of the other sugar moieties of a GT1b analog, demonstrating that not only the sialic acid monomer is important for binding, but the underlying glycan as well (93). More Siglec preferences have extensively been reviewed elsewhere (3, 8). Thus, Siglec binding is determined by both the context of the protein or lipid backbone and the structure of the accompanied glycans.
The hypothesis that the protein context is important for Siglec binding can be demonstrated by two specific ligands, CD24 and CD52, that were identified for Siglec-10 and which are highly similar in structure (94–96). Both are relatively small glycoproteins, consisting of 31-35 (CD24) or 12 amino acids (CD52) (97–99). They both harbor a glycosylphosphatidylinositol anchor (GPI anchor) and can be released from the cell membrane by the action of phospholipases. Interestingly, the GPI anchor, consisting of a phospholipid tail, a glycan core and a phosphoetanolamine linker to which the protein is attached, contains a glycolipid that can be sialylated as well (100–102). Therefore, CD24 and CD52 are glycoproteins attached to a glycolipid, which makes their context highly interesting for future studies. CD24 and CD52 have been shown to complex with HMGB1, adding even an additional layer of complexity, as binding of Siglec-10 to CD52 has even been shown to be enhanced by HGMB1 (95, 103, 104). Therefore, the context of the glycoprotein can cause binding by third proteins that influence Siglec binding. Both CD24 and CD52 have been shown to inhibit macrophage and T cell immune cell function through Siglec-10 binding (94, 96, 103). Studies showed that the interaction affects a diverse set of processes, such as phagocytosis by TAMs, Lck and ZAP-70 phosphorylation in CD4+ T cells and cytokine secretion by activated T cells.
In summary, the protein/lipid context of a Siglec ligand can be of importance for Siglec binding. At least, we hypothesize that the protein backbone can provide specific binding context for Siglecs (Figure 2G) and multivalency for high avidity binding can be created by dense sialoglycan presentation on a protein backbone or via dense glycan clusters at the cell membrane, which we will touch upon later in more detail.
Regulation of Siglec Ligands
A remaining knowledge gap is how expression of specific Siglec ligands is regulated within the TME. Multiple factors might play a role here. For instance, the hypoxic environment could influence glycan composition. Human cancer-associated gangliosides have been observed to incorporate more Neu5Gc into their glycans than non-transformed cells (105). Gangliosides are a special class of glycosphingolipids – glycans attached to a ceramide anchor - that contain at least one sialic acid residue (106). Due to a deletion in the Cmah gene, humans are incapable of synthesizing Neu5Gc and are dependent on dietary Neu5Gc (107–109). The increase in ganglioside associated Neu5Gc was suggested to be associated with the hypoxic environment of tumors. Potentially, Siglec binding or loss of binding due to Neu5Gc incorporation can alter signaling in the TME.
Interestingly, hypoxia was shown to induce transcription of the sialic acid transporter Sialin (SLC17A5), which mediates transport of external sialic acid into the cell (110). Moreover, hypoxia was reported to influence expression of sialyltransferases (111, 112). Therefore, Siglec binding could be influenced by the hypoxic environment by adjustment of the main type of sialic acid that is incorporated in sialoglycans, but future studies should elucidate how this exactly affects Siglec activation. Besides gene expression, also enzyme activity of for example specific sialyltransferases could influence incorporation of the type of sialic acid, although additional research is required to further study enzyme activity within the TME and its influence on Siglec activation.
The shift in metabolism that is accompanied with malignant transformation adds an additional layer to regulation of glycosylation in the TME (113). HIF-1α is a key player in regulating cellular energetics within the hypoxic TME as it causes enhanced glucose uptake, which is not only required to generate ATP via various biosynthetic pathways, but it is also an important glycosylation precursor (55, 114, 115). HIF-1α suppresses the tricarboxylic acid cycle, ultimately preventing generation of UDP-N-Acetylglucosamine (UDP-GlcNAc), which is required for N-glycan branching (116, 117). Besides, hypoxia also influences sialylation, as it activates biosynthesis of CMP-NeuAc nucleotides through conversion of UDP-GlcNAc to ManNAc by GNE (118). This eventually results in enhanced overall cell surface sialylation. Overall, the changed metabolism that is observed in the TME is intertwined with aberrant glycosylation patterns, and future research should elucidate how this specifically affects Siglec signaling within this hypoxic TME.
Tumor cells have been demonstrated to release EVs able to modulate immune cell activation in the TME (119). Recently, the presence of Siglec ligands on EVs has been shown, which could be an additional manner in which tumor cells influence Siglec ligand expression within the TME (Figure 3A) (120). For example, Dusoswa et al. showed that isolated EVs (including exosomes and small membrane budded vesicles) of glioblastoma cell lines specifically express ligands for Siglec-9 (120). Moreover, exosomes derived from ovarian ascites fluid from cancer patients express GD3 on their surface (121). Shenoy et al. reported that expression of GD3 on liposomes inhibits T cell activation in a sialic acid-dependent way. GD3 has been reported to be a ligand of Siglec-7, which is expressed on subsets of T cells, and might be activated by GD3 positive exosomes (122–125). These data imply that T cell inhibition by GD3 can at least in part be Siglec-mediated, but this finding needs further experimental confirmation.
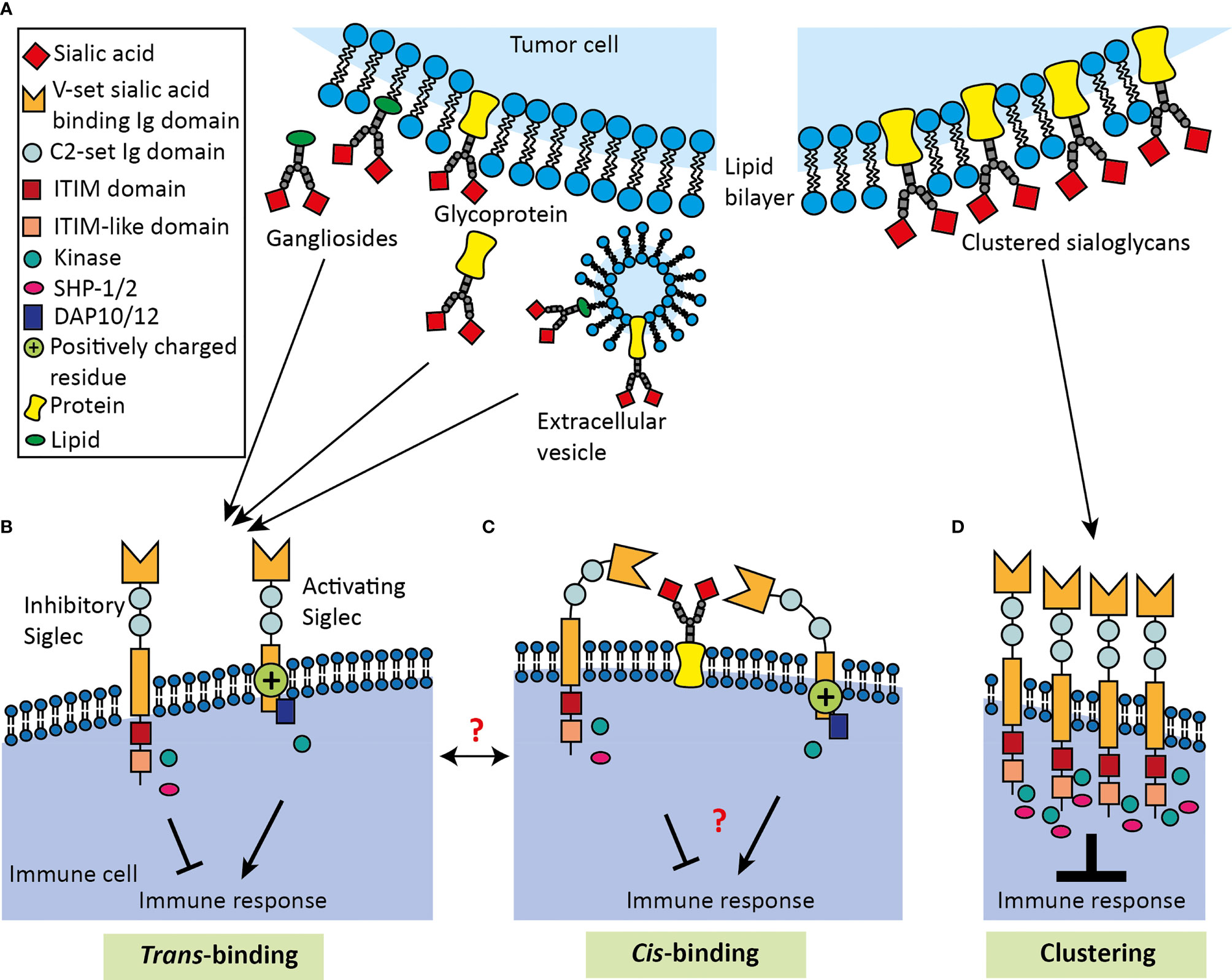
Figure 3 Schematic overview of Siglec binding to cis- and trans-ligands. (A) Tumor derived Siglec trans-ligands comprise glycolipids and glycoproteins, which can either be membrane-bound, secreted or present on EVs. Inhibitory and activating Siglecs can bind ligands either in (B) trans-fashion or in (C) cis-fashion. Siglecs bind their ligand using the V-set sialic acid-binding Ig domain, which is positioned away from the membrane by a variable number of C2-set Ig domains. Siglecs can bear ITIM and ITIM-like domains, which lead to inhibition of the immune response. In contrast, activating Siglecs can carry a positively charged residue in the transmembrane domain, which can associate with DAP10/12 activating transmembrane adaptor proteins to activate the immune response. Cis-ligand interactions probably contribute to immune homeostasis, but remain largely unknown and the dynamics of Siglecs binding in trans or cis mode requires more investigation. (D) Multivalent ligands are capable of clustering Siglecs to induce signaling, causing an intracellular accumulation of signaling molecules that eventually results in enhanced immune modulation.
Another interaction with exosomes has been reported for Siglec-1 that is expressed on macrophages and dendritic cells. Siglec-1 is an atypical Siglec, as it has no intracellular signaling motif and has 16 C2 Ig domains that extend the V-set domain far away from the cell membrane (1). Although Siglec-1 does not have intracellular motifs and is mostly described in uptake of α2-3-sialylated proteins or particles, multiple studies have shown that Siglec-1+ macrophages exert immunosuppressive effects (43, 46, 126, 127). Functionally, Siglec-1 expression on macrophages has been described to be important for antigen transfer to dendritic cells or T cells (43, 46, 127). An in vivo mouse study demonstrated that Siglec-1+ macrophages in tumor-draining lymph nodes bind tumor-derived EVs to physically block further dissemination and subsequent lymphocyte activation (128). Black et al. reported that Siglec-1 also binds a special type of EVs, apoptotic bodies, derived from the EL4 lymphoma cell line (129). Importantly, they showed in vivo that siglec-1-/- mice had an increased cytotoxic T cell response to apoptotic vesicles displaying ovalbumin protein than wild type C57BL/6. Thus, the data show that Siglec-1 activation could be enhanced by tumor cells secreting EVs displaying Siglec-1 ligands. However, it remains unknown whether Siglec-1 ligands are actively incorporated into membranes of EVs, or whether this is a passive process. Moreover, many factors regarding Siglec signaling induced by tumor-derived EVs remain unknown, such as the strength and kinetics of the activation and how these factors compare to Siglec activation induced by ligands on tumor cells. Furthermore, systemic transport of tumor-derived EVs and the resulting effect of these on Siglec activation outside the TME also requires further research, as well as the comparison between EVs derived from tumors and from healthy tissue.
In summary, Siglec ligand expression within the TME can be affected by various factors, such as hypoxia, its associated change in metabolism and the secretion of Siglec ligand-containing EVs. However, additional research is required to elucidate the contribution of these factors and possible others that have yet to be determined.
Siglec Clustering and Multivalent Ligands
Immune inhibitory Siglec signaling requires a few steps that are similar to induction of signaling by PD-1. First of all, PD-1 receptors have been described to require clustering to initiate signaling and literature now supports the idea that Siglecs require clustering as well (Figure 3D) (3, 130). Inhibitory Siglecs harbor ITIM domains like the PD-1 receptors to which kinases get recruited upon clustering and activation to phosphorylate the ITIM domains (1, 131). Research has previously shown that intracellular kinases can locally accumulate at receptor clusters, which might also be the case for Siglec receptors (132). Lastly, SHP-1/2 phosphatases are recruited intracellularly to the clusters and these will cause dephosphorylation of downstream molecules to inhibit immune cell activation (133). Localization of the clusters of PD-1 receptors to T cell receptors (TCRs) enhances efficiency of PD-1 mediated suppression, and this might also be true for Siglecs and their downstream targets (130). Here, we will discuss the current evidence for Siglec clustering and that multivalent ligands can be bound by clusters of Siglec receptors.
Siglec Clustering
It has been observed that Siglecs can form foci (such as Siglec-E on neutrophils) or accumulate at cell-cell contact sites, like the Siglec-9 accumulation on neutrophils that was observed at contact sites with LS180 and A549 carcinoma cells (60, 134). Furthermore, several studies used antibodies to cross-link Siglecs on the cell membrane to induce a functional effect (135–138). A typical example that demonstrates the relevance of Siglec clustering is given by Siglec-2. Siglec-2 binds cis-ligands resulting in cluster formation that inhibits B cell receptor signaling, which we will discuss in more detail in the section Siglec binding to cis-ligands (139–142). Another example was given by Ikehara et al., who transfected Jurkat T cells with Siglec-7 or -9 and observed clustering and partial co-localization of these receptors with CD3 of the TCR (125). Subsequently, they found that Siglec expression leads to reduced phosphorylation of molecules downstream of the TCR following TCR engagement.
Multivalent Siglec Ligands
The monovalent binding affinity of a Siglec for a sialoglycan ligand is relatively low (Kd of 100-300 μM) (4, 8, 59). Multivalent interactions, the simultaneous binding between clustered receptors and multimeric ligands, increases the binding affinity between Siglecs and sialoglycans as has been shown for Siglec-2 clusters (143–149). For instance, lipid rafts that contain many gangliosides can be seen as a multivalent Siglec ligand. Accordingly, Nicoll et al. reported clustering of unmasked Siglec-7 on NK cells at the contact site with ganglioside GD3-synthase transfected P815 cells (150).
Multivalent Siglec ligands can be formed at the cell surface through clustering of individual sialoglycoproteins or lipids or are formed by densely glycosylated protein such as the mucins (151–155). Mucins are a family of large secreted and membrane-bound glycoproteins consisting for up to 60% of tandem repeat domains formed by repeating serine, threonine and proline sequences (156, 157). Both serine and threonine are acceptors of O-GalNAc-type glycosylation resulting in dense glycosylation of mucins that can account for up to 80% of their total mass. Gel-forming mucins such as MUC2 and MUC5AC form a barrier at the epithelial surfaces and house the microbiome, and membrane-bound mucins have many functions in cell signaling and cellular interactions (157).
CD43 is another example of multivalent Siglec ligand (158, 159). Wisnovsky et al. discovered CD43 as a multivalent ligand for Siglec-7 on the K562 chronic myeloid leukemia cell line using a genome-wide CRISPR screen (158). CD43, or sialophorin, is a mucin-type protein harboring a heavily O-glycosylated extracellular domain (160, 161). This study showed that CD43 can relocalize Siglec-7 on NK cells to the immunological synapse with the K562 cells. In parallel, Yoshimura et al. identified CD43 on K562 cells as Siglec-7 binding partner using biochemical techniques (159).
In summary, binding of Siglecs to their ligands can be strengthened by multivalent interactions and in line with this, several multivalent ligands for Siglecs have been identified.
Multivalent Siglec Ligands Present in the TME
Mucins are overexpressed and aberrantly glycosylated in many types of cancer (162, 163). Several studies have reported Siglec binding to (cancer-associated) mucins suggesting that they can mediate communication between the epithelial surface and the immune system and that they could alter the anti-tumor immune response (151–154). For instance, MUC1 that is overexpressed in many cancer types has been reported to interact with Siglec-1 and Siglec-4, mediating adhesion between Siglec-4 expressing Schwann cells and MUC1 expressing pancreatic cells (152, 154). Beatson et al. reported that the cancer-associated sialyl T glycoform of MUC1 binds with Siglec-9 on myeloid cells and that this interaction can induce a tumor-associated macrophage phenotype (151, 164). Sialyl T-MUC1 instructed macrophages secrete soluble factors (IL-6, M-CSF, PAI-1) associated with tumor progression and display enhanced levels of the inhibitory receptor PD-L1 (151, 164). Moreover, these mucins enabled tumor-associated macrophages to inhibit T cell proliferation. Recently, the interaction of the human Siglecs with a large panel of recombinant mucins and mucin-like proteins decorated with defined O-GalNAc-type glycans was dissected (9). Although Siglec-9 interactions with sialyl T-MUC1 were not confirmed, this study revealed Siglec-4, -7, and -15 interactions with mucins that were largely determined by the mucin O-glycan pattern and type.
Another multivalent Siglec ligand that has been found on tumor cells or secreted in the TME is LGALS3BP (Mac-2 binding protein) (165, 166). LGALS3BP is a heavily N-glycosylated protein and Siglec-5, -9, and -10 have been shown to bind it (166). Importantly, recombinant LGALS3BP was demonstrated to inhibit neutrophil activation and siRNA-mediated reduction of LGALS3BP expression in HT-29 colon cancer cells increased neutrophil-induced apoptosis of the HT-29 cells (166). Interestingly, Koths et al. have shown before that LGALS3BP can form multimeric complexes (167). These data suggest a role for complex formation of Siglec ligands to offer a multivalent Siglec ligand to activate Siglec receptors.
Altogether, Siglecs can accumulate at the cell surface to induce a functional effect. Multivalent ligands increase binding affinity between Siglecs and sialoglycans. Classic example of tumor associated multivalent Siglec ligands are mucins. Smaller Siglec ligands, such as CD24 for Siglec-10, however, have also been identified in the TME that are less likely to act as multivalent ligand (106–108). Potentially, clustering of such ligands within the cell membrane could result in local presentation of Siglec ligands in a multivalent manner. In the same way, glycolipids could possibly organize in lipid rafts to activate Siglecs on the same cell (cis) as well as on opposing cells (trans) (59).
It would be interesting to study whether there is a critical amount of specific interactions and/or Siglec clustering that is required to trigger Siglec signaling. Also, comparison of the strength and kinetics of Siglec activation by ligands that either offer a multivalent or monovalent ligand would contribute to the current knowledge on Siglec activation by multivalent ligands and would aid in the rational development of Siglec targeting therapeutics. We hypothesize that the multivalency of Siglec ligands can have multiple functions, for example to induce Siglec clustering and thereby have a functional effect or to outcompete cis-ligands with a lower affinity. Nevertheless, more research is required to study these hypotheses and elucidate whether and how monovalent ligands can activate Siglecs.
The Function of Siglec Binding to Cis-Ligands
In order for Siglecs to interact with trans-ligands (Figure 3B), for instance on tumor cells, Siglecs have to be available for binding. Cells expressing Siglecs also express themselves sialoglycan ligands on their membrane that can interact in cis thereby ‘masking’ the V-set binding site (59) (Figure 3C). This has been shown for most Siglecs and potentially lowers the threshold for Siglec signaling by trans-interactions (45, 150, 168–172). Naturally, masking interactions are overcome by the dynamic on/off binding of Siglecs with their ligands, in particular for lower (monovalent) affinity ligands, that enables binding to higher affinity ligands (1, 4). Noteworthy, masking can also be abolished by sialidases or experimentally by treatment with sialylation inhibitors (17, 173). Furthermore, the C2-set domain repeats allows orientation of the V-set domain away from cis-ligands which likely contributes to recognition of trans-ligands. The 16 C2 Ig domains that orient the V-set domain of Siglec-1 into the extracellular space were suggested to enable Siglec-1 to mediate intercellular interactions. Still, interactions of Siglec-1 with cis-ligands has been observed (168, 174). However, it remains unknown whether cis-ligands only mask Siglec-1 or also activate the receptor. Variation in the C2-set repeat numbers between the Siglecs possibly determines the sensitivity of individual family members for the cis- and trans-ligands and also endogenous membrane and secreted sialidases (e.g. NEU1, NEU3).
Glycosylation changes in the cell have also been suggested to regulate Siglec binding dynamics for cis- and trans-ligands (169, 175). Masking/unmasking dynamics under physiological conditions and particularly in the TME and consequences for signaling and immune regulation is likely of key importance in Siglec biology.
Effect of Siglecs Binding to Cis-Ligands
One of the main knowledge gaps regarding cis-ligands is whether Siglecs can only be activated by trans- ligands or also by cis-ligands. The best studied Siglec receptor in terms of cis-interactions is Siglec-2, which is a receptor intracellularly harboring multiple ITIMs, an ITIM-like domain and a Grb2 binding motif (1, 176). Siglec-2 and B cell receptor (BCR) are present on the membrane as clusters that partially overlap (139). Co-clustering was found to be highly important for Siglec-2-mediated inhibition of BCR signaling as increased clustering of Siglec-2 with the BCR inhibits BCR signaling (139, 177). Siglec-2 is scavenged away from the BCR by cis-ligands on neighboring sialylated Siglec-2 receptors present on the same cell membrane leading to enhanced BCR activity (139–142). Collins et al. demonstrated that Siglec-2 on B cells can redistribute to the sites of cell contact with other lymphocytes, despite the presence of cis-ligands (178). This indicates that clustered patterns of Siglecs have to be tightly regulated and are affected by both cis- and trans-ligands (140, 141).
Human Siglec-2 has a high affinity for Neu5Ac as sialic acid and a 6-O-sulfate on the GlcNAc in the underlying glycan (175, 179). When human B cells are activated, the 6-O-sulfotransferases needed for the Siglec-2 ligands are downregulated. This results in a decrease in interactions between Siglec-2 and cis-ligands and therefore Siglec-2 becomes available to inhibit the BCR. Accordingly, in vivo mouse studies established that mice that lack the α2-6 sialyltransferase St6gal1 and mice with a mutation of the critical Arginine in the V-set domain of Siglec-2 both showed enhanced BCR inhibition. Murine Siglec-2 on B cells has a similar mechanism to inhibit BCR signaling. Murine Siglec-2 has a preference for Neu5Gc, rather than Neu5Ac (180). It is unmasked by downregulation of the hydroxylase that is responsible for the conversion of Neu5Ac to Neu5Gc, eventually resulting in a reduction of Siglec-2-cis-interactions and enhanced BCR inhibition (175, 181). Nevertheless, despite the ability of cis-ligands to prevent Siglec-2 to inhibit BCR signaling, Siglec-2 is always able to bind trans-ligands on an opposing cell (178, 182, 183). This interaction causes Siglec-2 to translocate to the immunological synapse, where it can inhibit BCR signaling (184, 185).
A recent study by Ballet et al. (2021) has further established Siglec-2 signaling on B cells induced by cis-ligands (186). Here, Siglec-2 was shown to associate with β7 integrin in a sialic acid-dependent manner. Next, recruitment of SHP-1 to Siglec-2 was demonstrated to inhibit β7 integrin endocytosis and restrain β7 integrin phosphorylation at the cell surface. The β7 integrin complexes with the α4 integrin, and this complex is involved in homing B lymphocytes to gut-associated lymphoid tissue, where B cells are being activated. Indeed, the Siglec-2-SHP-1 axis was shown to enhance B cell homing to the gut-associated lymphoid tissue.
Another study on Siglec signaling induced by cis-ligands has recently been performed by Delaveris et al. (2021) (187). Applying synthetic lipid-tethered glycopolypeptides that inserted into cell membranes in combination with Förster resonance energy transfer analysis revealed binding between Siglec-9 and glycopolypeptides in cis. Strikingly, Siglec-9 binding to ligands in cis reduced LPS-mediated MAPK signaling, cytokine secretion and phagocytosis in macrophages, while the same ligand used in trans did not. Of note, these studies on Siglec interactions with cis-ligands have not been performed within the context of the TME. Activation of Siglecs by cis-ligands in the TME therefore remains to be elucidated.
Relevance of Cis-Ligands in the TME
Although it has been established that Siglecs are masked by cis-interactions, for many Siglecs the biological role of these interactions is poorly understood. Varki and Angata (2006) proposed that cis-interactions set a threshold for recognition of trans-ligands with higher affinity or to monitor sialylation on the own cell surface (188). Cis-interactions could have a gatekeeper function blocking insubstantial interactions and signaling enabling sensing of high affinity or avidity Siglec ligands that lead to a biologically relevant signaling outcome. Cis-interactions, on the other hand, may exert baseline signaling which in case of the inhibitory Siglec members may support a resting state or return to a resting state after activation. Understanding the physiological relevance of Siglec cis-interactions and possible signaling requires further investigation. Likewise, the relevance of Siglec cis-interactions in the TME is largely unknown.
It is conceivable that tumor-associated trans-ligands outcompete cis-ligands either due to higher affinity or avidity for instance. The malignant transformation could potentially guide expression of specific glycan/glycoprotein or higher expression levels and clustering of trans-ligands. Altered biosynthesis, or degradation of cis-ligands on Siglec-expressing immune cells upon infiltration into the TME could further increase Siglec activation on immune cells upon encounter with tumor cells. For instance, it has been reported that T cell activation can result in a loss of sialylation on core 1-O glycans (189–191). Similarly, IL-2 stimulation of NK the cell line NK-92 reduced gene expression of the sialyltransferases ST8SIA1, ST6GAL1, and ST3GAL1, but increased overall α2-6-linked sialic acids and poly sialic acid likely due to an increase in the expression of poly sialic acid carrier molecule NCAM/CD56 (192). Occurrence of such alterations in glycosyltransferase and sialyltransferase expression in the TME, but also sialidase expression or changes in nucleotide sugar metabolism could thus alter cis-ligand expression. Together with the observed changes in immune cell phenotype like the enhanced Siglec expression on T cells and macrophages in response to factors in the TME, this would enable strong trans-interactions with sialoglycans on tumor cells and modulate the immune cell function (11, 28, 94). Clearly, multiple aspects need further investigation to understand how Siglec cis-interactions in the TME are altered and the biological effects thereof.
Challenges for the Study of Trans- and Cis- Siglec Interaction Dynamics
The dynamics of Siglec binding to trans- and cis-ligands is emerging as a key event in Siglec biology determining their clustering, signaling and biological effects. Studying these dynamics, however, is challenging. To study cis-binding-mediated Siglec activation, studies have installed lipid-conjugated glycopolypeptides into cell membranes (187, 193, 194). Similarly, to study trans-binding-mediated Siglec activation, Siglec-ligand containing liposomes have been used (195–197). However, addition of Siglec ligands in such manners requires careful interpretation, as this can lead to overexpression of unnatural sialic acids that can be recognized by Siglecs either in cis or trans and does not represent a natural situation. Besides, novel tools have been developed to remove sialic acids from the cell membrane to assess Siglec binding in either cis or trans manner. Sialic acid removal from the cell membrane can be established using sialidase or sialic acid mimetics that can enter the cell to inhibit sialyltransferases and thereby prevent transfer of sialic acid onto glycan chains (4, 173).
Nevertheless, altering cellular sialylation by different means can co-affect other factors. For instance, a recent study by Edgar et al. has provided an example that abrogation of sialic acid expression can result in Siglec-independent effects (198). They have shown that the costimulatory molecule CD28 on T cells can bind to sialoglycans in cis and in trans. The interactions of CD28 with cis-ligands limited binding of CD28 to its trans protein ligands CD80 and CD86 on antigen-presenting cells thereby negatively affecting co-stimulation. This example illustrates that also the function of non-Siglec receptors can be affected by abrogation of sialic acid expression. Moreover, removal of sialic acids will expose galactose, which can be recognized by galectins, which can modulate the immune response and affect tumor development (199). Similar mechanisms may be in place for other sialoglycan binding receptors, such as factor H and selectins (6). Furthermore, removal of the negatively charged sialic acid molecules could affect total charge of the cell and might therefore affect cellular interactions. Indeed, it was demonstrated that diminished sialic acid expression caused enhanced tumor cell killing by cytotoxic CD8+ T cells, which was explained at least in part by increased clustering of the T cells with tumor cells (200).
Additionally, studies have genetically manipulated the glycosylation machinery to modify sialylation, for example via genetic glycoengineering with inducible sialyltransferase expression to tune Siglec ligand expression (11, 201–203). However, this might also affect other processes, as Kohnz et al. have found that knockdown of cytidine monophosphate N-acetylneuraminic acid synthase, an enzyme required for sialic acid activation to be incorporated in glycans, affected levels of more than 200 other gene transcripts including oncogenes (204).
Overall, results have to be carefully interpreted when interrupting Siglec ligands, as these type of experiments are likely to have various side effects.
Concluding Remarks
The Siglec-sialoglycan axis emerges as important regulator of immune cell-tumor cell interactions in the TME that co-determines the outcome of tumor immunity (17). Qualitative and quantitative changes in Siglec ligand expression as well as Siglec receptors on immune cells in the TME have been linked to immune evasion. However, unraveling the biological effects and consequences of Siglec signaling in the TME remains challenging for several reasons. First, healthy cells and cancer cells can produce a highly diverse Siglec interactome that is formed by the sialoglycan structures, their modifications, their display on particular glycoproteins and glycolipids as well as multivalent higher order binding patterns created by protein glycosylation density and ligand clustering. Deducing the specific and biological meaningful cis- and trans-interactions of each of the individual Siglec family member is currently ongoing aided by recent technological advances with recombinant (multimeric) Siglec probes, proximity labeling approaches, cell-based glycan arrays, and genome-wide CRISPR screens (9, 22, 158, 173, 205).
Second, the expression patterns and membrane organization of most Siglecs on immune cells in the periphery and the TME (and other tissues) are not well understood. Siglec-2 is the best studied Siglec in that context and shows the importance of cis-interactions in tuning B cell receptor activation. Siglec-2 interactions with high avidity ligands in trans and downregulation of cis-ligands result in clustering around the B cell receptor and potent inhibitory signaling sufficient to block B cell activation (139–142). The membrane organization and interaction partners, recruitment to the membrane, internalization and recycling kinetics of the other Siglecs are less well understood, but should move more into focus as they likely determine their individual signaling modes.
Third, still relatively little is known regarding the signaling of Siglecs and the downstream molecular targets and effects. The immune inhibitory Siglecs that recruit SHP-1 or SHP-2 after ITIM phosphorylation are best described, but the molecular and cellular consequences of their signaling are not fully understood. Likewise, signaling of the activating Siglecs-14, -15, and -16 via adaptor proteins needs further dissection as well as other potential signaling residues (Grb2, Fyn kinase sites). Next to understanding the consequences of Siglec signaling and their integration with other immune signaling pathways, systems to determine whether ligand binding equals signaling are required. Studies with Siglec-Fc chimeras inform on binding, but this does not necessarily correlate with signaling (22). Suematsu et al. developed a reporter system for direct measurement of Siglec activation using a receptor consisting of the extracellular Siglec domains and the transmembrane and intracellular domains of the CD3ζ chain (206). Such cell-based assays could allow direct measurement of Siglec activation in response to a ligand, measure effects of signaling kinetics with different (multivalent) ligands, and shed light into signaling through trans- and cis-ligand binding dynamics.
Finally, further insight into general aspects of Siglec biology is needed to advance our currently limited understanding of inhibiting and activating Siglec signaling in the TME and the role that hypoxia, altered metabolism and cytokines play in regulating cis/trans-Siglec ligand and Siglec expression that guide cell-cell communication in the TME. The identification of Siglecs as important immune checkpoints in the TME implies that blocking monoclonal antibodies or other strategies to abolish Siglec-sialic acid interactions and signaling can boost anti-tumor immunity (16, 17, 28, 94). Further research into the diverse Siglec signaling modes in the TME may turn out to be highly awarding to uncover the impact of therapeutic Siglec targeting on the efficacy of cancer immunotherapy.
Author Contributions
EH designed and wrote the review, CB edited the review and LC and GA designed, supervised and edited the review. All authors contributed to and approved the submitted version.
Funding
This work was supported by a Radboudumc grant awarded to EH and grants from the Dutch Cancer Foundation awarded to GA, K.C.M. Santegoets and P. Wesseling (KWF 11266) and GA, LC and DG (KWF 2021-1 13373).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Varki A, Schnaar RL, Crocker PR. I-Type Lectins. In: Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, et al., editors. Essentials of Glycobiology p . Cold Spring Harbor (NY: Cold Spring Harbor Laboratory Press (2015). p. 453–67.
2. Macauley MS, Crocker PR, Paulson JC. Siglec-Mediated Regulation of Immune Cell Function in Disease. Nat Rev Immunol (2014) 14:10. doi: 10.1038/nri3737
3. Duan S, Paulson JC. Siglecs as Immune Cell Checkpoints in Disease. Annu Rev Immunol (2020) 38:365–95. doi: 10.1146/annurev-immunol-102419-035900
4. Büll C, Heise T, Adema GJ, Boltje TJ. Sialic Acid Mimetics to Target the Sialic Acid-Siglec Axis. Trends Biochem Sci (2016) 41:6. doi: 10.1016/j.tibs.2016.03.007
5. Angata T. Siglecs That Associate With DAP12. Adv Exp Med Biol (2020) 1204:215–30. doi: 10.1007/978-981-15-1580-4_9
6. Varki A, Schnaar RL, Schauer R. Sialic Acids and Other Nonulosonic Acids. In: Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, et al., editors. Essentials of Glycobiology Cold Spring Harbor (NY: Cold Spring Harbor Laboratory Press (2015). p. 179–95.
7. Flynn RA, Pedram K, Malaker SA, Batista PJ, Smith BAH, Johnson AG, et al. Small RNAs Are Modified With N-Glycans and Displayed on the Surface of Living Cells. Cell (2021) 184:12. doi: 10.1016/j.cell.2021.04.023
9. Büll C, Nason R, Sun L, Van Coillie J, Madriz Sørensen D, Moons SJ, et al. Probing the Binding Specificities of Human Siglecs by Cell-Based Glycan Arrays. Proc Natl Acad Sci USA (2021) 118:17. doi: 10.1073/pnas.2026102118
10. Büll C, den Brok MH, Adema GJ. Sweet Escape: Sialic Acids in Tumor Immune Evasion. Biochim Biophys Acta (2014) 1846:1. doi: 10.1016/j.bbcan.2014.07.005
11. Stanczak MA, Siddiqui SS, Trefny MP, Thommen DS, Boligan KF, von Gunten S, et al. Self-Associated Molecular Patterns Mediate Cancer Immune Evasion by Engaging Siglecs on T Cells. J Clin Invest (2018) 128:11. doi: 10.1172/JCI120612
12. Chen Z, Yu M, Guo L, Zhang B, Liu S, Zhang W, et al. Tumor Derived SIGLEC Family Genes May Play Roles in Tumor Genesis, Progression, and Immune Microenvironment Regulation. Front Oncol (2020) 10:586820. doi: 10.3389/fonc.2020.586820
13. Haas Q, Boligan KF, Jandus C, Schneider C, Simillion C, Stanczak MA, et al. Siglec-9 Regulates an Effector Memory CD8(+) T-Cell Subset That Congregates in the Melanoma Tumor Microenvironment. Cancer Immunol Res (2019) 7:5. doi: 10.1158/2326-6066.Cir-18-0505
14. Cassetta L, Fragkogianni S, Sims AH, Swierczak A, Forrester LM, Zhang H, et al. Human Tumor-Associated Macrophage and Monocyte Transcriptional Landscapes Reveal Cancer-Specific Reprogramming, Biomarkers, and Therapeutic Targets. Cancer Cell (2019) 35:4. doi: 10.1016/j.ccell.2019.02.009
15. Büll C, Stoel MA, den Brok MH, Adema GJ. Sialic Acids Sweeten a Tumor’s Life. Cancer Res (2014) 74:12. doi: 10.1158/0008-5472.Can-14-0728
16. Pearce OM, Laubli H. Sialic Acids in Cancer Biology and Immunity. Glycobiology (2016) 26:2. doi: 10.1093/glycob/cwv097
17. van de Wall S, Santegoets KCM, van Houtum EJH, Büll C, Adema GJ. Sialoglycans and Siglecs Can Shape the Tumor Immune Microenvironment. Trends Immunol (2020) 41:4. doi: 10.1016/j.it.2020.02.001
18. Murugesan G, Weigle B, Crocker PR. Siglec and Anti-Siglec Therapies. Curr Opin Chem Biol (2021) 62:34–42. doi: 10.1016/j.cbpa.2021.01.001
19. Bärenwaldt A, Läubli H. The Sialoglycan-Siglec Glyco-Immune Checkpoint - a Target for Improving Innate and Adaptive Anti-Cancer Immunity. Expert Opin Ther Targets (2019) 23:10. doi: 10.1080/14728222.2019.1667977
20. Wielgat P, Rogowski K, Niemirowicz-Laskowska K, Car H. Sialic Acid-Siglec Axis as Molecular Checkpoints Targeting of Immune System: Smart Players in Pathology and Conventional Therapy. Int J Mol Sci (2020) 21:12. doi: 10.3390/ijms21124361
21. Jandus C, Boligan KF, Chijioke O, Liu H, Dahlhaus M, Démoulins T, et al. Interactions Between Siglec-7/9 Receptors and Ligands Influence NK Cell-Dependent Tumor Immunosurveillance. J Clin Invest (2014) 124:4. doi: 10.1172/jci65899
22. Hudak JE, Canham SM, Bertozzi CR. Glycocalyx Engineering Reveals a Siglec-Based Mechanism for NK Cell Immunoevasion. Nat Chem Biol (2014) 10:1. doi: 10.1038/nchembio.1388
23. Miyazaki K, Sakuma K, Kawamura YI, Izawa M, Ohmori K, Mitsuki M, et al. Colonic Epithelial Cells Express Specific Ligands for Mucosal Macrophage Immunosuppressive Receptors Siglec-7 and -9. J Immunol (2012) 188:9. doi: 10.4049/jimmunol.1100605
24. Rodriguez E, Boelaars K, Brown K, Eveline Li RJ, Kruijssen L, Bruijns SCM, et al. Sialic Acids in Pancreatic Cancer Cells Drive Tumour-Associated Macrophage Differentiation via the Siglec Receptors Siglec-7 and Siglec-9. Nat Commun (2021) 12:1. doi: 10.1038/s41467-021-21550-4
25. Sun J, Lu Q, Sanmamed MF, Wang J. Siglec-15 as an Emerging Target for Next-Generation Cancer Immunotherapy. Clin Cancer Res (2021) 27:3. doi: 10.1158/1078-0432.Ccr-19-2925
26. Angata T, Tabuchi Y, Nakamura K, Nakamura M. Siglec-15: An Immune System Siglec Conserved Throughout Vertebrate Evolution. Glycobiology (2007) 17:8. doi: 10.1093/glycob/cwm049
27. Takamiya R, Ohtsubo K, Takamatsu S, Taniguchi N, Angata T. The Interaction Between Siglec-15 and Tumor-Associated Sialyl-Tn Antigen Enhances TGF-Beta Secretion From Monocytes/Macrophages Through the DAP12-Syk Pathway. Glycobiology (2013) 23:2. doi: 10.1093/glycob/cws139
28. Wang J, Sun J, Liu LN, Flies DB, Nie X, Toki M, et al. Siglec-15 as an Immune Suppressor and Potential Target for Normalization Cancer Immunotherapy. Nat Med (2019) 25:4. doi: 10.1038/s41591-019-0374-x
29. Fudaba H, Momii Y, Hirakawa T, Onishi K, Asou D, Matsushita W, et al. Sialic Acid-Binding Immunoglobulin-Like Lectin-15 Expression on Peritumoral Macrophages Is a Favorable Prognostic Factor for Primary Central Nervous System Lymphoma Patients. Sci Rep (2021) 11:1. doi: 10.1038/s41598-020-79742-9
30. Quirino MWL, Pereira MC, Deodato de Souza MF, Pitta IDR, Da Silva Filho AF, Albuquerque MSS, et al. Immunopositivity for Siglec-15 in Gastric Cancer and Its Association With Clinical and Pathological Parameters. Eur J Histochem (2021) 65:1. doi: 10.4081/ejh.2021.3174
31. Cao H, Neerincx A, de Bono B, Lakner U, Huntington C, Elvin J, et al. Sialic Acid-Binding Immunoglobulin-Like Lectin (Sigelac)-15 Is a Rapidly Internalised Cell-Surface Antigen Expressed by Acute Myeloid Leukaemia Cells. Br J Haematol (2021) 193:5. doi: 10.1111/bjh.17496
32. Trebo A, Ditsch N, Degenhardt T, Kuhn C, Rahmeh M, Schmoeckel E, et al. First Evidence for a Role of Siglec-8 in Breast Cancer. Int J Mol Sci (2021) 22:4. doi: 10.3390/ijms22042000
33. Jetani H, Navarro-Bailón A, Maucher M, Frenz S, Verbruggen CM, Yeguas A, et al. Siglec-6 Is a Novel Target for CAR T-Cell Therapy in Acute Myeloid Leukemia (AML). Blood (2021) 138:1830–42. doi: 10.1182/blood.2020009192
34. Raponi S, De Propris MS, Intoppa S, Milani ML, Vitale A, Elia L, et al. Flow Cytometric Study of Potential Target Antigens (CD19, CD20, CD22, CD33) for Antibody-Based Immunotherapy in Acute Lymphoblastic Leukemia: Analysis of 552 Cases. Leuk Lymphoma (2011) 52:6. doi: 10.3109/10428194.2011.559668
35. Piccaluga PP, Arpinati M, Candoni A, Laterza C, Paolini S, Gazzola A, et al. Surface Antigens Analysis Reveals Significant Expression of Candidate Targets for Immunotherapy in Adult Acute Lymphoid Leukemia. Leuk Lymphoma (2011) 52:2. doi: 10.3109/10428194.2010.529206
36. Wang X, Chow R, Deng L, Anderson D, Weidner N, Godwin AK, et al. Expression of Siglec-11 by Human and Chimpanzee Ovarian Stromal Cells, With Uniquely Human Ligands: Implications for Human Ovarian Physiology and Pathology. Glycobiology (2011) 21:8. doi: 10.1093/glycob/cwr039
37. Bandala-Sanchez E, Bediaga NG, Naselli G, Neale AM, Harrison LC. Siglec-10 Expression Is Up-Regulated in Activated Human CD4(+) T Cells. Hum Immunol (2020) 81:2–3. doi: 10.1016/j.humimm.2020.01.009
38. Zhang P, Lu X, Tao K, Shi L, Li W, Wang G, et al. Siglec-10 Is Associated With Survival and Natural Killer Cell Dysfunction in Hepatocellular Carcinoma. J Surg Res (2015) 194:1. doi: 10.1016/j.jss.2014.09.035
39. Li Y, Zhou J, Zhuo Q, Zhang J, Xie J, Han S, et al. Malignant Ascite-Derived Extracellular Vesicles Inhibit T Cell Activity by Upregulating Siglec-10 Expression. Cancer Manag Res (2019) 11:7123–34. doi: 10.2147/cmar.S210568
40. Rempel H, Calosing C, Sun B, Pulliam L. Sialoadhesin Expressed on IFN-Induced Monocytes Binds HIV-1 and Enhances Infectivity. PloS One (2008) 3:4. doi: 10.1371/journal.pone.0001967
41. De Saint Jean A, Lucht F, Bourlet T, Delézay O. Transforming Growth Factor Beta 1 Up-Regulates CD169 (Sialoadhesin) Expression on Monocyte-Derived Dendritic Cells: Role in HIV Sexual Transmission. Aids (2014) 28:16. doi: 10.1097/qad.0000000000000431
42. Nagase R, Kajitani N, Shikata K, Ogawa D, Kodera R, Okada S, et al. Phenotypic Change of Macrophages in the Progression of Diabetic Nephropathy; Sialoadhesin-Positive Activated Macrophages Are Increased in Diabetic Kidney. Clin Exp Nephrol (2012) 16:5. doi: 10.1007/s10157-012-0625-3
43. Zhang Y, Li JQ, Jiang ZZ, Li L, Wu Y, Zheng L. CD169 Identifies an Anti-Tumour Macrophage Subpopulation in Human Hepatocellular Carcinoma. J Pathol (2016) 239:2. doi: 10.1002/path.4720
44. Calzada-Wack JC, Frankenberger M, Ziegler-Heitbrock HW. Interleukin-10 Drives Human Monocytes to CD16 Positive Macrophages. J Inflammation (1996) 46:2.
45. Santegoets KCM, Gielen PR, Büll C, Schulte BM, Kers-Rebel ED, Küsters B, et al. Expression Profiling of Immune Inhibitory Siglecs and Their Ligands in Patients With Glioma. Cancer Immunol Immunother (2019) 68:6. doi: 10.1007/s00262-019-02332-w
46. Edgar LJ, Kawasaki N, Nycholat CM, Paulson JC. Targeted Delivery of Antigen to Activated CD169(+) Macrophages Induces Bias for Expansion of CD8(+) T Cells. Cell Chem Biol (2019) 26:1. doi: 10.1016/j.chembiol.2018.10.006
47. Veninga H, Borg EG, Vreeman K, Taylor PR, Kalay H, van Kooyk Y, et al. Antigen Targeting Reveals Splenic CD169+ Macrophages as Promoters of Germinal Center B-Cell Responses. Eur J Immunol (2015) 45:3. doi: 10.1002/eji.201444983
48. van Dinther D, Lopez Venegas M, Veninga H, Olesek K, Hoogterp L, Revet M, et al. Activation of CD8(+) T Cell Responses After Melanoma Antigen Targeting to CD169(+) Antigen Presenting Cells in Mice and Humans. Cancers (Basel) (2019) 11:2. doi: 10.3390/cancers11020183
49. van Dinther D, Veninga H, Revet M, Hoogterp L, Olesek K, Grabowska J, et al. Comparison of Protein and Peptide Targeting for the Development of a CD169-Based Vaccination Strategy Against Melanoma. Front Immunol (2018) 9:1997. doi: 10.3389/fimmu.2018.01997
50. John B, Herrin BR, Raman C, Wang YN, Bobbitt KR, Brody BA, et al. The B Cell Coreceptor CD22 Associates With AP50, a Clathrin-Coated Pit Adapter Protein, via Tyrosine-Dependent Interaction. J Immunol (2003) 170:7. doi: 10.4049/jimmunol.170.7.3534
51. Orr SJ, Morgan NM, Elliott J, Burrows JF, Scott CJ, McVicar DW, et al. CD33 Responses Are Blocked by SOCS3 Through Accelerated Proteasomal-Mediated Turnover. Blood (2007) 109:3. doi: 10.1182/blood-2006-05-023556
52. Shan D, Press OW. Constitutive Endocytosis and Degradation of CD22 by Human B Cells. J Immunol (1995) 154:9.
53. Walter RB, Hausermann P, Raden BW, Teckchandani AM, Kamikura DM, Bernstein ID, et al. Phosphorylated ITIMs Enable Ubiquitylation of an Inhibitory Cell Surface Receptor. Traffic (2008) 9:2. doi: 10.1111/j.1600-0854.2007.00682.x
54. Chen X, Dang X, Song J, Wang G, Liu C, Cui L, et al. N-Glycosylation of Siglec-15 Decreases Its Lysosome-Dependent Degradation and Promotes Its Transportation to the Cell Membrane. Biochem Biophys Res Commun (2020) 533:1. doi: 10.1016/j.bbrc.2020.08.111
55. Chen C, Pore N, Behrooz A, Ismail-Beigi F, Maity A. Regulation of Glut1 mRNA by Hypoxia-Inducible Factor-1. Interaction Between H-Ras and Hypoxia. J Biol Chem (2001) 276:12. doi: 10.1074/jbc.M010144200
56. Ito T, Ishigami M, Matsushita Y, Hirata M, Matsubara K, Ishikawa T, et al. Secreted Ectodomain of SIGLEC-9 and MCP-1 Synergistically Improve Acute Liver Failure in Rats by Altering Macrophage Polarity. Sci Rep (2017) 7:44043. doi: 10.1038/srep44043
57. Na HJ, Hamilton RG, Klion AD, Bochner BS. Biomarkers of Eosinophil Involvement in Allergic and Eosinophilic Diseases: Review of Phenotypic and Serum Markers Including a Novel Assay to Quantify Levels of Soluble Siglec-8. J Immunol Methods (2012) 383:1–2. doi: 10.1016/j.jim.2012.05.017
58. Gianchecchi E, Arena A, Fierabracci A. Sialic Acid-Siglec Axis in Human Immune Regulation, Involvement in Autoimmunity and Cancer and Potential Therapeutic Treatments. Int J Mol Sci (2021) 22:11. doi: 10.3390/ijms22115774
59. Crocker PR, Paulson JC, Varki A. Siglecs and Their Roles in the Immune System. Nat Rev Immunol (2007) 7:4. doi: 10.1038/nri2056
60. Läubli H, Pearce OM, Schwarz F, Siddiqui SS, Deng L, Stanczak MA, et al. Engagement of Myelomonocytic Siglecs by Tumor-Associated Ligands Modulates the Innate Immune Response to Cancer. Proc Natl Acad Sci USA (2014) 111:39. doi: 10.1073/pnas.1409580111
61. Boligan KF, Mesa C, Fernandez LE, von Gunten S. Cancer Intelligence Acquired (CIA): Tumor Glycosylation and Sialylation Codes Dismantling Antitumor Defense. Cell Mol Life Sci (2015) 72:7. doi: 10.1007/s00018-014-1799-5
62. Pinho SS, Reis CA. Glycosylation in Cancer: Mechanisms and Clinical Implications. Nat Rev Cancer (2015) 15:9. doi: 10.1038/nrc3982
63. Bagdonaite I, Pallesen EMH, Nielsen MI, Bennett EP, Wandall HH. Mucin-Type O-GalNAc Glycosylation in Health and Disease. Adv Exp Med Biol (2021) 1325:25–60. doi: 10.1007/978-3-030-70115-4_2
64. Varki A, Kannagi R, Toole B, Stanley P. Glycosylation Changes in Cancer. In: Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, et al., editors. Essentials of Glycobiology. Cold Spring Harbor (NY: Cold Spring Harbor Laboratory Press Copyright 2015-2017 by The Consortium of Glycobiology Editors, La Jolla, California. All rights reserved (2015). p. 597–609.
65. Stowell SR, Ju T, Cummings RD. Protein Glycosylation in Cancer. Annu Rev Pathol (2015) 10:473–510. doi: 10.1146/annurev-pathol-012414-040438
66. Egan H, Treacy O, Lynch K, Leonard NA, O’Malley G, De Veirman K, et al. Stromal Cell Sialylation Suppresses T Cells in Inflammatory Tumour Microenvironments: A New Tumour Stromal Cell Immune Checkpoint? bioRxiv [Preprint] (2021). doi: 10.1101/2021.06.18.447879
67. Liu H, Zheng Y, Zhang Y, Li J, Fernandes SM, Zeng D, et al. Immunosuppressive Siglec-E Ligands on Mouse Aorta Are Up-Regulated by LPS via NF-κb Pathway. BioMed Pharmacother (2020) 122:109760. doi: 10.1016/j.biopha.2019.109760
68. Kivi E, Elima K, Aalto K, Nymalm Y, Auvinen K, Koivunen E, et al. Human Siglec-10 can Bind to Vascular Adhesion Protein-1 and Serves as Its Substrate. Blood (2009) 114:26. doi: 10.1182/blood-2009-04-219253
69. Blixt O, Collins BE, van den Nieuwenhof IM, Crocker PR, Paulson JC. Sialoside Specificity of the Siglec Family Assessed Using Novel Multivalent Probes: Identification of Potent Inhibitors of Myelin-Associated Glycoprotein. J Biol Chem (2003) 278:33. doi: 10.1074/jbc.M304331200
70. Gao C, Hanes MS, Byrd-Leotis LA, Wei M, Jia N, Kardish RJ, et al. Unique Binding Specificities of Proteins Toward Isomeric Asparagine-Linked Glycans. Cell Chem Biol (2019) 26:4. doi: 10.1016/j.chembiol.2019.01.002
71. Narimatsu Y, Joshi HJ, Nason R, Van Coillie J, Karlsson R, Sun L, et al. An Atlas of Human Glycosylation Pathways Enables Display of the Human Glycome by Gene Engineered Cells. Mol Cell (2019) 75:2. doi: 10.1016/j.molcel.2019.05.017
72. Schauer R, Kamerling JP. Exploration of the Sialic Acid World. Adv Carbohydr Chem Biochem (2018) 75:1–213. doi: 10.1016/bs.accb.2018.09.001
73. Di Carluccio C, Forgione RE, Montefiori M, Civera M, Sattin S, Smaldone G, et al. Behavior of Glycolylated Sialoglycans in the Binding Pockets of Murine and Human CD22. iScience (2021) 24:1. doi: 10.1016/j.isci.2020.101998
74. Brinkman-Van der Linden EC, Sjoberg ER, Juneja LR, Crocker PR, Varki N, Varki A. Loss of N-Glycolylneuraminic Acid in Human Evolution. Implications for Sialic Acid Recognition by Siglecs. J Biol Chem (2000) 275:12. doi: 10.1074/jbc.275.12.8633
75. Sonnenburg JL, Altheide TK, Varki A. A Uniquely Human Consequence of Domain-Specific Functional Adaptation in a Sialic Acid-Binding Receptor. Glycobiology (2004) 14:4. doi: 10.1093/glycob/cwh039
76. Angata T. Possible Influences of Endogenous and Exogenous Ligands on the Evolution of Human Siglecs. Front Immunol (2018) 9:2885. doi: 10.3389/fimmu.2018.02885
77. Avril T, North SJ, Haslam SM, Willison HJ, Crocker PR. Probing the Cis Interactions of the Inhibitory Receptor Siglec-7 With Alpha2,8-Disialylated Ligands on Natural Killer Cells and Other Leukocytes Using Glycan-Specific Antibodies and by Analysis of Alpha2,8-Sialyltransferase Gene Expression. J Leukoc Biol (2006) 80:4. doi: 10.1189/jlb.1005559
78. Marcos NT, Pinho S, Grandela C, Cruz A, Samyn-Petit B, Harduin-Lepers A, et al. Role of the Human ST6GalNAc-I and ST6GalNAc-II in the Synthesis of the Cancer-Associated Sialyl-Tn Antigen. Cancer Res (2004) 64:19. doi: 10.1158/0008-5472.Can-04-1921
79. Schjoldager KT, Narimatsu Y, Joshi HJ, Clausen H. Global View of Human Protein Glycosylation Pathways and Functions. Nat Rev Mol Cell Biol (2020) 21:12. doi: 10.1038/s41580-020-00294-x
80. Murugesan G, Correia VG, Palma AS, Chai W, Li C, Feizi T, et al. Siglec-15 Recognition of Sialoglycans on Tumor Cell Lines can Occur Independently of Sialyl Tn Antigen Expression. Glycobiology (2020) 31:1. doi: 10.1093/glycob/cwaa048
81. Pröpster JM, Yang F, Rabbani S, Ernst B, Allain FH, Schubert M. Structural Basis for Sulfation-Dependent Self-Glycan Recognition by the Human Immune-Inhibitory Receptor Siglec-8. Proc Natl Acad Sci USA (2016) 113:29. doi: 10.1073/pnas.1602214113
82. Jung J, Enterina JR, Bui DT, Mozaneh F, Lin P-H, Nitin, et al. Carbohydrate Sulfation as a Mechanism for Fine-Tuning Siglec Ligands. bioRxiv [Preprint] (2021). doi: 10.1101/2021.06.27.450109
83. Yu H, Gonzalez-Gil A, Wei Y, Fernandes SM, Porell RN, Vajn K, et al. Siglec-8 and Siglec-9 Binding Specificities and Endogenous Airway Ligand Distributions and Properties. Glycobiology (2017) 27:7. doi: 10.1093/glycob/cwx026
84. Izawa M, Kumamoto K, Mitsuoka C, Kanamori A, Ohmori K, Ishida H, et al. Expression of Sialyl 6-Sulfo Lewis X Is Inversely Correlated With Conventional Sialyl Lewis X Expression in Human Colorectal Cancer. Cancer Res (2000) 60:5.
85. Yusa A, Miyazaki K, Kimura N, Izawa M, Kannagi R. Epigenetic Silencing of the Sulfate Transporter Gene DTDST Induces Sialyl Lewisx Expression and Accelerates Proliferation of Colon Cancer Cells. Cancer Res (2010) 70:10. doi: 10.1158/0008-5472.Can-09-2383
86. Brinkman-Van der Linden EC, Varki A. New Aspects of Siglec Binding Specificities, Including the Significance of Fucosylation and of the Sialyl-Tn Epitope. Sialic Acid-Binding Immunoglobulin Superfamily Lectins. J Biol Chem (2000) 275:12. doi: 10.1074/jbc.275.12.8625
87. Padler-Karavani V, Hurtado-Ziola N, Chang YC, Sonnenburg JL, Ronaghy A, Yu H, et al. Rapid Evolution of Binding Specificities and Expression Patterns of Inhibitory CD33-Related Siglecs in Primates. FASEB J (2014) 28:3. doi: 10.1096/fj.13-241497
88. Grabenstein S, Barnard KN, Anim M, Armoo A, Weichert WS, Bertozzi CR, et al. Deacetylated Sialic Acids Modulates Immune Mediated Cytotoxicity via the Sialic Acid-Siglec Pathway. Glycobiology (2021) 31:1279–94. doi: 10.1093/glycob/cwab068
89. Varki A, Esko JD, Colley KJ. Cellular Organization of Glycosylation. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, et al., editors. Essentials of Glycobiology. Cold Spring Harbor (NY: Cold Spring Harbor Laboratory Press (2009).
90. Varki A, Sharon N. Historical Background and Overview. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, et al., editors. Essentials of Glycobiology. Cold Spring Harbor (NY: Cold Spring Harbor Laboratory Press (2009).
91. Angata T, Hayakawa T, Yamanaka M, Varki A, Nakamura M. Discovery of Siglec-14, a Novel Sialic Acid Receptor Undergoing Concerted Evolution With Siglec-5 in Primates. FASEB J (2006) 20:12. doi: 10.1096/fj.06-5800com
92. Lopez PHH, Schnaar RL. Gangliosides in Cell Recognition and Membrane Protein Regulation. Curr Opin Struct Biol (2009) 19:5. doi: 10.1016/j.sbi.2009.06.001
93. Attrill H, Imamura A, Sharma RS, Kiso M, Crocker PR, van Aalten DM. Siglec-7 Undergoes a Major Conformational Change When Complexed With the Alpha(2,8)-Disialylganglioside GT1b. J Biol Chem (2006) 281:43. doi: 10.1074/jbc.M601714200
94. Barkal AA, Brewer RE, Markovic M, Kowarsky M, Barkal SA, Zaro BW, et al. CD24 Signalling Through Macrophage Siglec-10 Is a Target for Cancer Immunotherapy. Nature (2019) 572:7769. doi: 10.1038/s41586-019-1456-0
95. Chen GY, Tang J, Zheng P, Liu Y. CD24 and Siglec-10 Selectively Repress Tissue Damage-Induced Immune Responses. Science (2009) 323:5922. doi: 10.1126/science.1168988
96. Bandala-Sanchez E, Zhang Y, Reinwald S, Dromey JA, Lee BH, Qian J, et al. T Cell Regulation Mediated by Interaction of Soluble CD52 With the Inhibitory Receptor Siglec-10. Nat Immunol (2013) 14:7. doi: 10.1038/ni.2610
97. Kay R, Rosten PM, Humphries RK. CD24, a Signal Transducer Modulating B Cell Activation Responses, Is a Very Short Peptide With a Glycosyl Phosphatidylinositol Membrane Anchor. J Immunol (1991) 147:4.
98. Xia MQ, Hale G, Lifely MR, Ferguson MAJ, Campbell D, Packman L, et al. Structure of the CAMPATH-1 Antigen, a Glycosylphosphatidylinositol-Anchored Glycoprotein Which Is an Exceptionally Good Target for Complement Lysis. Biochem J (1993) 293:3. doi: 10.1042/bj2930633
99. Xia MQ, Tone M, Packman L, Hale G, Waldmann H. Characterization of the CAMPATH-1 (CDw52) Antigen: Biochemical Analysis and cDNA Cloning Reveal an Unusually Small Peptide Backbone. Eur J Immunol (1991) 21:7. doi: 10.1002/eji.1830210714
100. Paulick MG, Bertozzi CR. The Glycosylphosphatidylinositol Anchor: A Complex Membrane-Anchoring Structure for Proteins. Biochemistry (2008) 47:27. doi: 10.1021/bi8006324
101. Stahl N, Baldwin MA, Hecker R, Pan KM, Burlingame AL, Prusiner SB. Glycosylinositol Phospholipid Anchors of the Scrapie and Cellular Prion Proteins Contain Sialic Acid. Biochemistry (1992) 31:21. doi: 10.1021/bi00136a600
102. Kobayashi A, Hirata T, Nishikaze T, Ninomiya A, Maki Y, Takada Y, et al. α2,3 Linkage of Sialic Acid to a GPI Anchor and an Unpredicted GPI Attachment Site in Human Prion Protein. J Biol Chem (2020) 295:22. doi: 10.1074/jbc.RA120.013444
103. Bandala-Sanchez E, GB N, Goddard-Borger ED, Ngui K, Naselli G, Stone NL, et al. CD52 Glycan Binds the Proinflammatory B Box of HMGB1 to Engage the Siglec-10 Receptor and Suppress Human T Cell Function. Proc Natl Acad Sci USA (2018) 115:30. doi: 10.1073/pnas.1722056115
104. Lotze MT, Tracey KJ. High-Mobility Group Box 1 Protein (HMGB1): Nuclear Weapon in the Immune Arsenal. Nat Rev Immunol (2005) 5:4. doi: 10.1038/nri1594
105. Labrada M, Dorvignit D, Hevia G, Rodriguez-Zhurbenko N, Hernandez AM, Vazquez AM, et al. GM3(Neu5Gc) Ganglioside: An Evolution Fixed Neoantigen for Cancer Immunotherapy. Semin Oncol (2018) 45:1–2. doi: 10.1053/j.seminoncol.2018.04.003
106. Ledeen RW, Kopitz J, Abad-Rodriguez J, Gabius HJ. Glycan Chains of Gangliosides: Functional Ligands for Tissue Lectins (Siglecs/Galectins). Prog Mol Biol Transl Sci (2018) 156:289–324. doi: 10.1016/bs.pmbts.2017.12.004
107. Chou HH, Takematsu H, Diaz S, Iber J, Nickerson E, Wright KL, et al. A Mutation in Human CMP-Sialic Acid Hydroxylase Occurred After the Homo-Pan Divergence. Proc Natl Acad Sci USA (1998) 95:20. doi: 10.1073/pnas.95.20.11751
108. Irie A, Koyama S, Kozutsumi Y, Kawasaki T, Suzuki A. The Molecular Basis for the Absence of N-Glycolylneuraminic Acid in Humans. J Biol Chem (1998) 273:25. doi: 10.1074/jbc.273.25.15866
109. Hayakawa T, Satta Y, Gagneux P, Varki A, Takahata N. Alu-Mediated Inactivation of the Human CMP- N-Acetylneuraminic Acid Hydroxylase Gene. Proc Natl Acad Sci USA (2001) 98:20. doi: 10.1073/pnas.191268198
110. Yin J, Hashimoto A, Izawa M, Miyazaki K, Chen GY, Takematsu H, et al. Hypoxic Culture Induces Expression of Sialin, a Sialic Acid Transporter, and Cancer-Associated Gangliosides Containing Non-Human Sialic Acid on Human Cancer Cells. Cancer Res (2006) 66:6. doi: 10.1158/0008-5472.Can-05-2615
111. Albuquerque APB, Balmaña M, Mereiter S, Pinto F, Reis CA, Beltrão EIC. Hypoxia and Serum Deprivation Induces Glycan Alterations in Triple Negative Breast Cancer Cells. Biol Chem (2018) 399:7. doi: 10.1515/hsz-2018-0121
112. Jones RB, Dorsett KA, Hjelmeland AB, Bellis SL. The ST6Gal-I Sialyltransferase Protects Tumor Cells Against Hypoxia by Enhancing HIF-1α Signaling. J Biol Chem (2018) 293:15. doi: 10.1074/jbc.RA117.001194
113. Peixoto A, Relvas-Santos M, Azevedo R, Santos LL, Ferreira JA. Protein Glycosylation and Tumor Microenvironment Alterations Driving Cancer Hallmarks. Front Oncol (2019) 9:380. doi: 10.3389/fonc.2019.00380
114. Masson N, Ratcliffe PJ. Hypoxia Signaling Pathways in Cancer Metabolism: The Importance of Co-Selecting Interconnected Physiological Pathways. Cancer Metab (2014) 2:1. doi: 10.1186/2049-3002-2-3
115. Freeze HH, Hart GW, Schnaar RL. Glycosylation Precursors. In: Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, et al., editors. Essentials of Glycobiology. Cold Spring Harbor (NY: Cold Spring Harbor Laboratory Press (2015). p. 51–63.
116. Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-Mediated Expression of Pyruvate Dehydrogenase Kinase: A Metabolic Switch Required for Cellular Adaptation to Hypoxia. Cell Metab (2006) 3:3. doi: 10.1016/j.cmet.2006.02.002
117. Sasai K, Ikeda Y, Fujii T, Tsuda T, Taniguchi N. UDP-GlcNAc Concentration Is an Important Factor in the Biosynthesis of Beta1,6-Branched Oligosaccharides: Regulation Based on the Kinetic Properties of N-Acetylglucosaminyltransferase V. Glycobiology (2002) 12:2. doi: 10.1093/glycob/12.2.119
118. Keppler OT, Hinderlich S, Langner J, Schwartz-Albiez R, Reutter W, Pawlita M. UDP-GlcNAc 2-Epimerase: A Regulator of Cell Surface Sialylation. Science (1999) 284:5418. doi: 10.1126/science.284.5418.1372
119. Robbins PD, Morelli AE. Regulation of Immune Responses by Extracellular Vesicles. Nat Rev Immunol (2014) 14:3. doi: 10.1038/nri3622
120. Dusoswa SA, Horrevorts SK, Ambrosini M, Kalay H, Paauw NJ, Nieuwland R, et al. Glycan Modification of Glioblastoma-Derived Extracellular Vesicles Enhances Receptor-Mediated Targeting of Dendritic Cells. J Extracell Vesicles (2019) 8:1. doi: 10.1080/20013078.2019.1648995
121. Shenoy GN, Loyall J, Berenson CS, Kelleher RJ Jr, Iyer V, Balu-Iyer SV, et al. Sialic Acid-Dependent Inhibition of T Cells by Exosomal Ganglioside GD3 in Ovarian Tumor Microenvironments. J Immunol (2018) 201:12. doi: 10.4049/jimmunol.1801041
122. Ito A, Handa K, Withers DA, Satoh M, Hakomori S. Binding Specificity of Siglec7 to Disialogangliosides of Renal Cell Carcinoma: Possible Role of Disialogangliosides in Tumor Progression. FEBS Lett (2001) 504:1–2. doi: 10.1016/s0014-5793(01)02734-x
123. Hashimoto N, Ito S, Tsuchida A, Bhuiyan RH, Okajima T, Yamamoto A, et al. The Ceramide Moiety of Disialoganglioside (GD3) Is Essential for GD3 Recognition by the Sialic Acid-Binding Lectin SIGLEC7 on the Cell Surface. J Biol Chem (2019) 294:28. doi: 10.1074/jbc.RA118.007083
124. Nicoll G, Ni J, Liu D, Klenerman P, Munday J, Dubock S, et al. Identification and Characterization of a Novel Siglec, Siglec-7, Expressed by Human Natural Killer Cells and Monocytes. J Biol Chem (1999) 274:48. doi: 10.1074/jbc.274.48.34089
125. Ikehara Y, Ikehara SK, Paulson JC. Negative Regulation of T Cell Receptor Signaling by Siglec-7 (P70/AIRM) and Siglec-9. J Biol Chem (2004) 279:41. doi: 10.1074/jbc.M403538200
126. Asano K, Nabeyama A, Miyake Y, Qiu CH, Kurita A, Tomura M, et al. CD169-Positive Macrophages Dominate Antitumor Immunity by Crosspresenting Dead Cell-Associated Antigens. Immunity (2011) 34:1. doi: 10.1016/j.immuni.2010.12.011
127. van Dinther D, Veninga H, Iborra S, Borg EGF, Hoogterp L, Olesek K, et al. Functional CD169 on Macrophages Mediates Interaction With Dendritic Cells for CD8(+) T Cell Cross-Priming. Cell Rep (2018) 22:6. doi: 10.1016/j.celrep.2018.01.021
128. Pucci F, Garris C, Lai CP, Newton A, Pfirschke C, Engblom C, et al. SCS Macrophages Suppress Melanoma by Restricting Tumor-Derived Vesicle-B Cell Interactions. Science (2016) 352:6282. doi: 10.1126/science.aaf1328
129. Black LV, Saunderson SC, Coutinho FP, Muhsin-Sharafaldine MR, Damani TT, Dunn AC, et al. The CD169 Sialoadhesin Molecule Mediates Cytotoxic T-Cell Responses to Tumour Apoptotic Vesicles. Immunol Cell Biol (2016) 94:5. doi: 10.1038/icb.2015.111
130. Yokosuka T, Takamatsu M, Kobayashi-Imanishi W, Hashimoto-Tane A, Azuma M, Saito T. Programmed Cell Death 1 Forms Negative Costimulatory Microclusters That Directly Inhibit T Cell Receptor Signaling by Recruiting Phosphatase SHP2. J Exp Med (2012) 209:6. doi: 10.1084/jem.20112741
131. Boussiotis VA, Chatterjee P, Li L. Biochemical Signaling of PD-1 on T Cells and Its Functional Implications. Cancer J (2014) 20:4. doi: 10.1097/ppo.0000000000000059
132. Wilson BS, Pfeiffer JR, Oliver JM. Observing FcepsilonRI Signaling From the Inside of the Mast Cell Membrane. J Cell Biol (2000) 149:5. doi: 10.1083/jcb.149.5.1131
133. Bolland S, Ravetch JV. Inhibitory Pathways Triggered by ITIM-Containing Receptors. Adv Immunol (1999) 72:2084–94. doi: 10.1016/s0065-2776(08)60019-x
134. McMillan SJ, Sharma RS, McKenzie EJ, Richards HE, Zhang J, Prescott A, et al. Siglec-E Is a Negative Regulator of Acute Pulmonary Neutrophil Inflammation and Suppresses CD11b Beta2-Integrin-Dependent Signaling. Blood (2013) 121:11. doi: 10.1182/blood-2012-08-449983
135. Lajaunias F, Dayer JM, Chizzolini C. Constitutive Repressor Activity of CD33 on Human Monocytes Requires Sialic Acid Recognition and Phosphoinositide 3-Kinase-Mediated Intracellular Signaling. Eur J Immunol (2005) 35:1. doi: 10.1002/eji.200425273
136. Vitale C, Romagnani C, Falco M, Ponte M, Vitale M, Moretta A, et al. Engagement of P75/AIRM1 or CD33 Inhibits the Proliferation of Normal or Leukemic Myeloid Cells. Proc Natl Acad Sci USA (1999) 96:26. doi: 10.1073/pnas.96.26.15091
137. Varchetta S, Brunetta E, Roberto A, Mikulak J, Hudspeth KL, Mondelli MU, et al. Engagement of Siglec-7 Receptor Induces a Pro-Inflammatory Response Selectively in Monocytes. PloS One (2012) 7:9. doi: 10.1371/journal.pone.0045821
138. von Gunten S, Yousefi S, Seitz M, Jakob SM, Schaffner T, Seger R, et al. Siglec-9 Transduces Apoptotic and Nonapoptotic Death Signals Into Neutrophils Depending on the Proinflammatory Cytokine Environment. Blood (2005) 106:4. doi: 10.1182/blood-2004-10-4112
139. Gasparrini F, Feest C, Bruckbauer A, Mattila PK, Muller J, Nitschke L, et al. Nanoscale Organization and Dynamics of the Siglec CD22 Cooperate With the Cytoskeleton in Restraining BCR Signalling. EMBO J (2016) 35:3. doi: 10.15252/embj.201593027
140. Grewal PK, Boton M, Ramirez K, Collins BE, Saito A, Green RS, et al. ST6Gal-I Restrains CD22-Dependent Antigen Receptor Endocytosis and Shp-1 Recruitment in Normal and Pathogenic Immune Signaling. Mol Cell Biol (2006) 26:13. doi: 10.1128/mcb.00308-06
141. Collins BE, Smith BA, Bengtson P, Paulson JC. Ablation of CD22 in Ligand-Deficient Mice Restores B Cell Receptor Signaling. Nat Immunol (2006) 7:2. doi: 10.1038/ni1283
142. Han S, Collins BE, Bengtson P, Paulson JC. Homomultimeric Complexes of CD22 in B Cells Revealed by Protein-Glycan Cross-Linking. Nat Chem Biol (2005) 1:2. doi: 10.1038/nchembio713
143. Mammen M, Choi SK, Whitesides GM. Polyvalent Interactions in Biological Systems: Implications for Design and Use of Multivalent Ligands and Inhibitors. Angew Chem Int Ed Engl (1998) 37:20. doi: 10.1002/(sici)1521-3773(19981102)37:20<2754::Aid-anie2754>3.0.Co;2-3
144. Cohen M, Varki A. Modulation of Glycan Recognition by Clustered Saccharide Patches. Int Rev Cell Mol Biol (2014) 308:75–125. doi: 10.1016/b978-0-12-800097-7.00003-8
145. Powell LD, Varki A. The Oligosaccharide Binding Specificities of CD22 Beta, a Sialic Acid-Specific Lectin of B Cells. J Biol Chem (1994) 269:14. doi: 10.1016/S0021-9258(17)34106-6
146. Powell LD, Jain RK, Matta KL, Sabesan S, Varki A. Characterization of Sialyloligosaccharide Binding by Recombinant Soluble and Native Cell-Associated CD22. Evidence for a Minimal Structural Recognition Motif and the Potential Importance of Multisite Binding. J Biol Chem (1995) 270:13. doi: 10.1074/jbc.270.13.7523
147. van Rossenberg SM, Sliedregt LA, Autar R, Piperi C, van der Merwe AP, van Berkel TJ, et al. A Structure-Function Study of Ligand Recognition by CD22beta. J Biol Chem (2001) 276:16. doi: 10.1074/jbc.M009276200
148. O’Reilly MK, Paulson JC. Multivalent Ligands for Siglecs. Methods Enzymol (2010) 478:343–63. doi: 10.1016/s0076-6879(10)78017-4
149. Peng W, Paulson JC. CD22 Ligands on a Natural N-Glycan Scaffold Efficiently Deliver Toxins to B-Lymphoma Cells. J Am Chem Soc (2017) 139:36. doi: 10.1021/jacs.7b03208
150. Nicoll G, Avril T, Lock K, Furukawa K, Bovin N, Crocker PR. Ganglioside GD3 Expression on Target Cells can Modulate NK Cell Cytotoxicity via Siglec-7-Dependent and -Independent Mechanisms. Eur J Immunol (2003) 33:6. doi: 10.1002/eji.200323693
151. Beatson R, Tajadura-Ortega V, Achkova D, Picco G, Tsourouktsoglou TD, Klausing S, et al. The Mucin MUC1 Modulates the Tumor Immunological Microenvironment Through Engagement of the Lectin Siglec-9. Nat Immunol (2016) 17:11. doi: 10.1038/ni.3552
152. Swanson BJ, McDermott KM, Singh PK, Eggers JP, Crocker PR, Hollingsworth MA. MUC1 Is a Counter-Receptor for Myelin-Associated Glycoprotein (Siglec-4a) and Their Interaction Contributes to Adhesion in Pancreatic Cancer Perineural Invasion. Cancer Res (2007) 67:21. doi: 10.1158/0008-5472.Can-06-2483
153. Tanida S, Akita K, Ishida A, Mori Y, Toda M, Inoue M, et al. Binding of the Sialic Acid-Binding Lectin, Siglec-9, to the Membrane Mucin, MUC1, Induces Recruitment of Beta-Catenin and Subsequent Cell Growth. J Biol Chem (2013) 288:44. doi: 10.1074/jbc.M113.471318
154. Nath D, Hartnell A, Happerfield L, Miles DW, Burchell J, Taylor-Papadimitriou J, et al. Macrophage-Tumour Cell Interactions: Identification of MUC1 on Breast Cancer Cells as a Potential Counter-Receptor for the Macrophage-Restricted Receptor, Sialoadhesin. Immunology (1999) 98:2. doi: 10.1046/j.1365-2567.1999.00827.x
155. Nason R, Büll C, Konstantinidi A, Sun L, Ye Z, Halim A, et al. Display of the Human Mucinome With Defined O-Glycans by Gene Engineered Cells. Nat Commun (2021) 12:1. doi: 10.1038/s41467-021-24366-4
156. Bansil R, Turner BS. Mucin Structure, Aggregation, Physiological Functions and Biomedical Applications. Curr Opin Colloid Interface Sci (2006) 11:2. doi: 10.1016/j.cocis.2005.11.001
157. Hansson GC. Mucins and the Microbiome. Annu Rev Biochem (2020) 89:769–93. doi: 10.1146/annurev-biochem-011520-105053
158. Wisnovsky S, Möckl L, Malaker SA, Pedram K, Hess GT, Riley NM, et al. Genome-Wide CRISPR Screens Reveal a Specific Ligand for the Glycan-Binding Immune Checkpoint Receptor Siglec-7. Proc Natl Acad Sci USA (2021) 118:5. doi: 10.1073/pnas.2015024118
159. Yoshimura A, Asahina Y, Chang LY, Angata T, Tanaka H, Kitajima K, et al. Identification and Functional Characterization of a Siglec-7 Counter-Receptor on K562 Cells. J Biol Chem (2021) 296:100477. doi: 10.1016/j.jbc.2021.100477
160. Rosenstein Y, Santana A, Pedraza-Alva G. CD43, a Molecule With Multiple Functions. Immunol Res (1999) 20:2. doi: 10.1007/bf02786465
161. Tuccillo FM, Palmieri C, Fiume G, de Laurentiis A, Schiavone M, Falcone C, et al. Cancer-Associated CD43 Glycoforms as Target of Immunotherapy. Mol Cancer Ther (2014) 13:3. doi: 10.1158/1535-7163.Mct-13-0651
162. Kufe DW. Mucins in Cancer: Function, Prognosis and Therapy. Nat Rev Cancer (2009) 9:12. doi: 10.1038/nrc2761
163. Chugh S, Gnanapragassam VS, Jain M, Rachagani S, Ponnusamy MP, Batra SK. Pathobiological Implications of Mucin Glycans in Cancer: Sweet Poison and Novel Targets. Biochim Biophys Acta (2015) 1856:2. doi: 10.1016/j.bbcan.2015.08.003
164. Beatson R, Graham R, Grundland Freile F, Cozzetto D, Kannambath S, Pfeifer E, et al. Cancer-Associated Hypersialylated MUC1 Drives the Differentiation of Human Monocytes Into Macrophages With a Pathogenic Phenotype. Commun Biol (2020) 3:1. doi: 10.1038/s42003-020-01359-5
165. Grassadonia A, Tinari N, Iurisci I, Piccolo E, Cumashi A, Innominato P, et al. 90k (Mac-2 BP) and Galectins in Tumor Progression and Metastasis. Glycoconj J (2002) 19:7–9. doi: 10.1023/B:GLYC.0000014085.00706.d4
166. Laubli H, Alisson-Silva F, Stanczak MA, Siddiqui SS, Deng L, Verhagen A, et al. Lectin Galactoside-Binding Soluble 3 Binding Protein (LGALS3BP) Is a Tumor-Associated Immunomodulatory Ligand for CD33-Related Siglecs. J Biol Chem (2014) 289:48. doi: 10.1074/jbc.M114.593129
167. Koths K, Taylor E, Halenbeck R, Casipit C, Wang A. Cloning and Characterization of a Human Mac-2-Binding Protein, a New Member of the Superfamily Defined by the Macrophage Scavenger Receptor Cysteine-Rich Domain. J Biol Chem (1993) 268:19. doi: 10.1016/S0021-9258(19)85233-X
168. Nakamura K, Yamaji T, Crocker PR, Suzuki A, Hashimoto Y. Lymph Node Macrophages, But Not Spleen Macrophages, Express High Levels of Unmasked Sialoadhesin: Implication for the Adhesive Properties of Macrophages In Vivo. Glycobiology (2002) 12:3. doi: 10.1093/glycob/12.3.209
169. Razi N, Varki A. Masking and Unmasking of the Sialic Acid-Binding Lectin Activity of CD22 (Siglec-2) on B Lymphocytes. Proc Natl Acad Sci USA (1998) 95:13. doi: 10.1073/pnas.95.13.7469
170. Perez-Oliva AB, Martinez-Esparza M, Vicente-Fernandez JJ, Corral-San Miguel R, Garcia-Penarrubia P, Hernandez-Caselles T. Epitope Mapping, Expression and Post-Translational Modifications of Two Isoforms of CD33 (CD33M and CD33m) on Lymphoid and Myeloid Human Cells. Glycobiology (2011) 21:6. doi: 10.1093/glycob/cwq220
171. Kawasaki Y, Ito A, Withers DA, Taima T, Kakoi N, Saito S, et al. Ganglioside DSGb5, Preferred Ligand for Siglec-7, Inhibits NK Cell Cytotoxicity Against Renal Cell Carcinoma Cells. Glycobiology (2010) 20:11. doi: 10.1093/glycob/cwq116
172. Tateno H, Crocker PR, Paulson JC. Mouse Siglec-F and Human Siglec-8 Are Functionally Convergent Paralogs That Are Selectively Expressed on Eosinophils and Recognize 6’-Sulfo-Sialyl Lewis X as a Preferred Glycan Ligand. Glycobiology (2005) 15:11. doi: 10.1093/glycob/cwi097
173. Läubli H, Kawanishi K, George Vazhappilly C, Matar R, Merheb M, Sarwar Siddiqui S. Tools to Study and Target the Siglec-Sialic Acid Axis in Cancer. FEBS J (2020) 288:6206–25. doi: 10.1111/febs.15647
174. Ducreux J, Crocker PR, Vanbever R. Analysis of Sialoadhesin Expression on Mouse Alveolar Macrophages. Immunol Lett (2009) 124:2. doi: 10.1016/j.imlet.2009.04.006
175. Macauley MS, Kawasaki N, Peng W, Wang SH, He Y, Arlian BM, et al. Unmasking of CD22 Co-Receptor on Germinal Center B-Cells Occurs by Alternative Mechanisms in Mouse and Man. J Biol Chem (2015) 290:50. doi: 10.1074/jbc.M115.691337
176. Chen J, Wang H, Xu WP, Wei SS, Li HJ, Mei YQ, et al. Besides an ITIM/SHP-1-Dependent Pathway, CD22 Collaborates With Grb2 and Plasma Membrane Calcium-ATPase in an ITIM/SHP-1-Independent Pathway of Attenuation of Ca2+i Signal in B Cells. Oncotarget (2016) 7:35. doi: 10.18632/oncotarget.9794
177. Bednar KJ, Shanina E, Ballet R, Connors EP, Duan S, Juan J, et al. Human CD22 Inhibits Murine B Cell Receptor Activation in a Human CD22 Transgenic Mouse Model. J Immunol (2017) 199:9. doi: 10.4049/jimmunol.1700898
178. Collins BE, Blixt O, DeSieno AR, Bovin N, Marth JD, Paulson JC. Masking of CD22 by Cis Ligands Does Not Prevent Redistribution of CD22 to Sites of Cell Contact. Proc Natl Acad Sci USA (2004) 101:16. doi: 10.1073/pnas.0400851101
179. Kimura N, Ohmori K, Miyazaki K, Izawa M, Matsuzaki Y, Yasuda Y, et al. Human B-Lymphocytes Express Alpha2-6-Sialylated 6-Sulfo-N-Acetyllactosamine Serving as a Preferred Ligand for CD22/Siglec-2. J Biol Chem (2007) 282:44. doi: 10.1074/jbc.M702341200
180. Kelm S, Schauer R, Manuguerra J-C, Gross H-J, Crocker PR. Modifications of Cell Surface Sialic Acids Modulate Cell Adhesion Mediated by Sialoadhesin and CD22. Glycoconjugate J (1994) 11:6. doi: 10.1007/BF00731309
181. Naito Y, Takematsu H, Koyama S, Miyake S, Yamamoto H, Fujinawa R, et al. Germinal Center Marker GL7 Probes Activation-Dependent Repression of N-Glycolylneuraminic Acid, a Sialic Acid Species Involved in the Negative Modulation of B-Cell Activation. Mol Cell Biol (2007) 27:8. doi: 10.1128/mcb.02047-06
182. Macauley MS, Paulson JC. Siglecs Induce Tolerance to Cell Surface Antigens by BIM-Dependent Deletion of the Antigen-Reactive B Cells. J Immunol (2014) 193:9. doi: 10.4049/jimmunol.1401723
183. Lanoue A, Batista FD, Stewart M, Neuberger MS. Interaction of CD22 With Alpha2,6-Linked Sialoglycoconjugates: Innate Recognition of Self to Dampen B Cell Autoreactivity? Eur J Immunol (2002) 32:2. doi: 10.1002/1521-4141(200202)32:2<348::Aid-immu348>3.0.Co;2-5
184. Müller J, Obermeier I, Wöhner M, Brandl C, Mrotzek S, Angermüller S, et al. CD22 Ligand-Binding and Signaling Domains Reciprocally Regulate B-Cell Ca2+ Signaling. Proc Natl Acad Sci USA (2013) 110:30. doi: 10.1073/pnas.1304888110
185. Hennet T, Chui D, Paulson JC, Marth JD. Immune Regulation by the ST6Gal Sialyltransferase. Proc Natl Acad Sci USA (1998) 95:8. doi: 10.1073/pnas.95.8.4504
186. Ballet R, Brennan M, Brandl C, Feng N, Berri J, Cheng J, et al. A CD22-Shp1 Phosphatase Axis Controls Integrin β(7) Display and B Cell Function in Mucosal Immunity. Nat Immunol (2021) 22:3. doi: 10.1038/s41590-021-00862-z
187. Delaveris CS, Chiu SH, Riley NM, Bertozzi CR. Modulation of Immune Cell Reactivity With Cis-Binding Siglec Agonists. Proc Natl Acad Sci USA (2021) 118:3. doi: 10.1073/pnas.2012408118
188. Varki A, Angata T. Siglecs–the Major Subfamily of I-Type Lectins. Glycobiology (2006) 16:1. doi: 10.1093/glycob/cwj008
189. Chervenak R, Cohen JJ. Peanut Lectin Binding as a Marker for Activated T-Lineage Lymphocytes. Thymus (1982) 4:2.
190. Galvan M, Murali-Krishna K, Ming LL, Baum L, Ahmed R. Alterations in Cell Surface Carbohydrates on T Cells From Virally Infected Mice can Distinguish Effector/Memory CD8+ T Cells From Naive Cells. J Immunol (1998) 161:2.
191. Piller F, Piller V, Fox RI, Fukuda M. Human T-Lymphocyte Activation Is Associated With Changes in O-Glycan Biosynthesis. J Biol Chem (1988) 263:29. doi: 10.1016/S0021-9258(18)68157-8
192. Rosenstock P, Bork K, Massa C, Selke P, Seliger B, Horstkorte R. Sialylation of Human Natural Killer (NK) Cells Is Regulated by IL-2. J Clin Med (2020) 9:6. doi: 10.3390/jcm9061816
193. Wang X, Lang S, Tian Y, Zhang J, Yan X, Fang Z, et al. Glycoengineering of Natural Killer Cells With CD22 Ligands for Enhanced Anticancer Immunotherapy. ACS Cent Sci (2020) 6:3. doi: 10.1021/acscentsci.9b00956
194. Hong S, Yu C, Rodrigues E, Shi Y, Chen H, Wang P, et al. Modulation of Siglec-7 Signaling Via In Situ-Created High-Affinity Cis-Ligands. ACS Cent Sci (2021) 7:8. doi: 10.1021/acscentsci.1c00064
195. Nycholat CM, Duan S, Knuplez E, Worth C, Elich M, Yao A, et al. A Sulfonamide Sialoside Analogue for Targeting Siglec-8 and -F on Immune Cells. J Am Chem Soc (2019) 141:36. doi: 10.1021/jacs.9b05769
196. Duan S, Arlian BM, Nycholat CM, Wei Y, Tateno H, Smith SA, et al. Nanoparticles Displaying Allergen and Siglec-8 Ligands Suppress IgE-Fcϵri-Mediated Anaphylaxis and Desensitize Mast Cells to Subsequent Antigen Challenge. J Immunol (2021) 206:10. doi: 10.4049/jimmunol.1901212
197. Bednar KJ, Nycholat CM, Rao TS, Paulson JC, Fung-Leung WP, Macauley MS. Exploiting CD22 To Selectively Tolerize Autoantibody Producing B-Cells in Rheumatoid Arthritis. ACS Chem Biol (2019) 14:4. doi: 10.1021/acschembio.8b01018
198. Edgar LJ, Thompson AJ, Vartabedian VF, Kikuchi C, Woehl JL, Teijaro JR, et al. Sialic Acid Ligands of CD28 Suppress Costimulation of T Cells. ACS Cent Sci (2021) 7:9. doi: 10.1021/acscentsci.1c00525
199. Yang RY, Rabinovich GA, Liu FT. Galectins: Structure, Function and Therapeutic Potential. Expert Rev Mol Med (2008) 10:e17. doi: 10.1017/s1462399408000719
200. Büll C, Boltje TJ, Balneger N, Weischer SM, Wassink M, van Gemst JJ, et al. Sialic Acid Blockade Suppresses Tumor Growth by Enhancing T-Cell-Mediated Tumor Immunity. Cancer Res (2018) 78:13. doi: 10.1158/0008-5472.Can-17-3376
201. Perdicchio M, Cornelissen LA, Streng-Ouwehand I, Engels S, Verstege MI, Boon L, et al. Tumor Sialylation Impedes T Cell Mediated Anti-Tumor Responses While Promoting Tumor Associated-Regulatory T Cells. Oncotarget (2016) 7:8. doi: 10.18632/oncotarget.6822
202. Cornelissen LAM, Blanas A, van der Horst JC, Kruijssen L, Zaal A, O’Toole T, et al. Disruption of Sialic Acid Metabolism Drives Tumor Growth by Augmenting CD8(+) T Cell Apoptosis. Int J Cancer (2019) 144:9. doi: 10.1002/ijc.32084
203. Narimatsu Y, Büll C, Chen YH, Wandall HH, Yang Z, Clausen H. Genetic Glycoengineering in Mammalian Cells. J Biol Chem (2021) 296:100448. doi: 10.1016/j.jbc.2021.100448
204. Kohnz RA, Roberts LS, DeTomaso D, Bideyan L, Yan P, Bandyopadhyay S, et al. Protein Sialylation Regulates a Gene Expression Signature That Promotes Breast Cancer Cell Pathogenicity. ACS Chem Biol (2016) 11:8. doi: 10.1021/acschembio.6b00433
205. Wu G, Nagala M, Crocker PR. Identification of Lectin Counter-Receptors on Cell Membranes by Proximity Labeling. Glycobiology (2017) 27:9. doi: 10.1093/glycob/cwx063
Keywords: Siglecs, sialic acids, sialoglycans, tumor microenvironment, immune checkpoint, cancer, immunotherapy
Citation: van Houtum EJH, Büll C, Cornelissen LAM and Adema GJ (2021) Siglec Signaling in the Tumor Microenvironment. Front. Immunol. 12:790317. doi: 10.3389/fimmu.2021.790317
Received: 06 October 2021; Accepted: 23 November 2021;
Published: 13 December 2021.
Edited by:
Heinz Laubli, University Hospital of Basel, SwitzerlandReviewed by:
Matthew S. Macauley, University of Alberta, CanadaLubor Borsig, University of Zurich, Switzerland
Copyright © 2021 van Houtum, Büll, Cornelissen and Adema. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gosse J. Adema, Z29zc2UuYWRlbWFAcmFkYm91ZHVtYy5ubA==
 Eline J. H. van Houtum
Eline J. H. van Houtum Christian Büll
Christian Büll Lenneke A. M. Cornelissen
Lenneke A. M. Cornelissen Gosse J. Adema
Gosse J. Adema