
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 16 December 2021
Sec. Nutritional Immunology
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.788638
This article is part of the Research Topic Immune Pathogenesis of Intestinal Inflammatory Diseases View all 24 articles
 Yuheng Luo1,2,3*†
Yuheng Luo1,2,3*† Cong Lan1,2,3†
Cong Lan1,2,3† Kunhong Xie1,2,3†
Kunhong Xie1,2,3† Hua Li1,2,3†
Hua Li1,2,3† Estelle Devillard4
Estelle Devillard4 Jun He1,2,3
Jun He1,2,3 Li Liu5
Li Liu5 Jingyi Cai1,2,3
Jingyi Cai1,2,3 Gang Tian1,2,3
Gang Tian1,2,3 Aimin Wu1,2,3
Aimin Wu1,2,3 Zhihua Ren6
Zhihua Ren6 Daiwen Chen1,2,3
Daiwen Chen1,2,3 Bing Yu1,2,3
Bing Yu1,2,3 Zhiqing Huang1,2,3
Zhiqing Huang1,2,3 Ping Zheng1,2,3
Ping Zheng1,2,3 Xiangbing Mao1,2,3
Xiangbing Mao1,2,3 Jie Yu1,2,3
Jie Yu1,2,3 Junqiu Luo1,2,3
Junqiu Luo1,2,3 Hui Yan1,2,3
Hui Yan1,2,3 Quyuan Wang1,2,3
Quyuan Wang1,2,3 Huifen Wang1,2,3
Huifen Wang1,2,3 Jiayong Tang1,2,3
Jiayong Tang1,2,3Intestinal inflammation is a major threat to the health and growth of young animals such as piglets. As a next-generation probiotics, limited studies have shown that Akkermansia muciniphila could alleviate inflammation of intestinal epithelial cells (IECs). In this study, a TNF-α-induced inflammatory model of IPEC-J2 cells, the intestinal porcine enterocytes, was built to evaluate the effects of active or inactive A. muciniphila on the inflammation of IECs. The viability of IPEC-J2 cells was the highest when treated with active (108 copies/mL) or inactive (109 copies/mL) A. muciniphila for 7.5 h (P < 0.01). Treated with 20 ng/mL of TNF-α and followed by a treatment of A. muciniphila, the mRNA level of proinflammatory cytokines (IL-8, IL-1β, IL-6 and TNF-α) was remarkably reduced (P < 0.05) along with the increased mRNA level of tight junction proteins (ZO-1 and Occludin, P < 0.05). Flow cytometry analysis showed that active or inactive A. muciniphila significantly suppressed the rate of the early and total apoptotic of the inflammatory IPEC-J2 cells (P < 0.05). According to results of transcriptome sequencing, active and inactive A. muciniphila may decline cell apoptosis by down-regulating the expression of key genes in calcium signaling pathway, or up-regulating the expression of key genes in cell cycle signaling pathway. And the bacterium may alleviate the inflammation of IECs by down-regulating the expression of PI3K upstream receptor genes. Our results indicate that A. muciniphila may be a promising NGP targeting intestinal inflammation.
The gastrointestinal epithelium represents the direct contact surface between the body and external environment. Its selective permeability allows the appropriate absorption of nutrients and water, while prevents the invasion of noxious molecules and microorganisms in the lumen (1). The cellular architecture of the intestines features a continuous monolayer of intestinal epithelial cells (IECs) that represent the first defensive barrier against environmental and microbial attacks by carrying out several critical innate immune functions, and many factors such as pathogens can cause the inflammation or apoptosis of IECs (2). Normal intestinal functions are closely linked to the balance of proliferation and apoptosis of IECs. An obvious example for the dysfunction of IECs is the intestinal inflammation caused by social, environmental or dietary stress in young animals, which leads to a growth defect (3). Antibiotics, used as growth promoters, are also recognized as effective anti-inflammatory additives for animals (4). However, the increasing number of antibiotic-resistant bacterial strains in animal gut accelerates research to develop alternative methods for the prevention and therapy of intestinal inflammation in young animals. Amongst those alternatives, probiotics, could modulate inflammation, by regulating for instance the production of cytokines and IgA (5, 6).
Compared with the first-generation probiotics (FGPs), the next-generation probiotics (NGPs) which are anaerobic gut commensal microbes that have been shown to suppress mucosal inflammation (7, 8). Of all NGPs, Akkermansia muciniphila, a newly isolated Gram-negative bacterium (9), has been shown to have beneficial effect on type II diabetes, obesity and colorectal cancer (10–14). It is also reported that A. muciniphila enhances the thickness of the mucus layer in obese mice induced by high-fat diet and the integrity of IECs in several in vitro studies (10, 15), and may contribute to the epithelial homeostasis and cell fate (16). Today, these beneficial effects were mainly observed using active A. muciniphila. Some special structures of A. muciniphila, such as a specific protein Amuc_1100 isolated from the outer membrane or A. muciniphila-derived extracellular vesicles (AmEVs), can also modulate the intestinal barrier and immune responses of host (17, 18). These limited results indicate that A. muciniphila compounds may alleviate the inflammation, suggesting that having active cells might not be necessary to obtain an effect on inflammation.
The aim of the current study was to assess the effects of active or inactive A. muciniphila in an in vitro inflammatory model based on IPEC-J2 cells which is closed to human physiology and has been widely used as an in vitro model in physiological, pathological and nutritional studies (19). A specific focus was put on monitoring the expression of inflammatory cytokines and apoptosis, as well as the transcriptome of the cells. These data were then discussed to provide new thoughts about the application of A. muciniphila as a NGP in animal feed or human food.
A. muciniphila (strain DSM 22959) was cultured in brain heart infusion broth (BHI) (Qingdao Rishui Bio·Technology Co., Ltd, Qingdao, China) (9). The medium was prepared with double distilled water (ddH2O) under a complete anaerobic condition with high-purity carbon dioxide. Then each of 10 mL medium was allocated in a 20 mL tubes and immediately sealed with butyl rubber stopper and aluminum crimp cap followed by an autoclave at 121°C for 20 min. All tubes inoculated with A. muciniphila were incubated at 37°C. To establish the growth curve of A. muciniphila, each of 500 µL culture was firstly collected at 6, 12, 18, 24, 30, 36, 42 and 48 h and then centrifuged at 10,000 × g for 15 min, respectively. Bacterial growth was measured by qPCR. Genomic DNA was extracted using a described method (20). Specific primers, AM1 (5’-CAGCACGTGAAGGTGGGGAC-3’) and AM2 (5’-CCTTGCGGTTGGCTTCAGAT-3’) (21), were used to amplify partial 16s rRNA gene of A. muciniphila. The real-time PCR was performed on a CFX96TM Real-Time System (Bio-Rad, USA) in triplicate, and the 10 µL reaction solution included 5 µL SYBR Premix EX Taq (TaKaRa, Dalian, China), 1 µL each of the primers (10 µM), 1 µL DNA template and 3 µL ddH2O. The PCR reaction was performed as follows: a pre-denaturation at 95°C for 10min, a total of 40 cycles of 95°C for 15 s, 60°C for 40 s and 72°C for 30 s, as well as a final extension at 72°C for 5 min.
The process for the preparation of active and inactive A. muciniphila cells is shown in Figure 1. In detail, 108 copies/mL was collected and centrifuged at 5,000 × g for 5 min. The pellet containing the cells of A. muciniphila was washed with 1 × anaerobic phosphate-buffered saline (PBS) (Solarbio, Beijing, China) and centrifuged for three times and then resuspended into 1 mL cell culture medium (descripted below) to obtain the concentrated active A. muciniphila (different concentrations from 106 to 109 copies/mL). Aliquots of those diluted cells were then autoclaved at 120°C for 30 min to obtain inactive A. muciniphila cells. For the following survival test, active or inactive A. muciniphila was inoculated and cultured in BHI medium, respectively. Then, the OD600 value of culture containing the two types of bacteria were tested at 3 h, 6 h, 18 h. To investigate whether autoclave caused damage to the integrity of the bacterium, the prepared cells (active and inactive) of A. muciniphila were collected and fixed for 2 h in a solution with 4% glutaraldehyde away from light. The specimens were dehydrated in absolute alcohol and critical-point dried with carbon dioxide. Finally, each sample were glued onto a sample holder using a carbon adhesive tab and sputter-coated with 10 nm platinum, and observed with a field emission scanning electron microscope (JEOL 7500 F) at 15 kV.
To investigate the role of A. muciniphila on inflammatory IECs, we used tumor necrosis factor-α (TNF-α) as an inducer of inflammatory response on IPEC-J2 cells, the intestinal porcine enterocytes isolated from the jejunum of a neonatal, as described previously (22).
IPEC-J2 cells used in the current study were donated by Dr. Per Torp Sangild from University of Copenhagen (Denmark) and cultured in a complete medium containing Dulbecco’s Modified Eagle Medium: F-12 (DMEM-F12) (HyClone, USA), 10% (v/v) fetal bovine serum (FBS) (HyClone, USA) which is more suitable in this experiment (19, 23, 24) and 1% (v/v) Penicillin-Streptomycin (Solarbio, Beijing, China). 1×105 cells/well were seeded and grown at 37°C in a CO2 incubator (5% v/v). When the cell fusion rate reaches 80%, the IPEC-J2 cells were treated with DMEM-F12 and starved for 12 h. To construct an inflammatory response model, the prepared IPEC-J2 cells were treated with 0, 10, 20, 40, 80 and 120 ng/mL of Recombinant Susscrofa TNF-α (Raybiotech, Inc Cat.# 230-00875) for 48 hours, respectively. Next, the mRNA level of Interleukin-1β (IL-1β), Interleukin-6 (IL-6) and TNF-α was determined using real-time PCR to assess whether the inflammatory model was built successfully, combining observed morphology of the cells with an inverted microscope (NiKon Ts100, C-W 10× (F.O.V. 22mm), Japan).
To determine the cell viability of IPEC-J2 cells, an Enhanced Cell Counting Kit-8 (CCK-8) (Biyuntian, Shanghai, China) was used. In brief, the activated IPEC-J2 cells were seeded in a 96-well plate (Corning, New York City, USA) at a concentration of 1×105 cells per well for 48 h. The prepared living (active) and inactive A. muciniphila at 106, 107, 108 and 109 copies/mL were then inoculated into the cells in triplicate and incubated at 37°C for 2.5, 5, 7.5 and 10 h, respectively. At the end of the incubation, 10 µL CCK-8 solution was added to each well and continuously incubated for 2 h. The absorbance for each well was then measured at a wavelength of 450 nm using the Absorbance Microplate Reader (Molecular Devices, SpectraMax 190, USA). The optimum concentration of A. muciniphila to apply on IPEC was then confirmed via the results of cell viability of IPEC-J2 cells (results are shown below).
To investigate the protection of active and inactive A. muciniphila on IPEC-J2 cells, a total of 6 treatments were included. IPEC-J2 Cells in the negative control (CON) were treated with PBS, while cells in the positive control (TNF) were challenged with Recombinant Susscrofa TNF-α. The remaining 4 treatments included the following: IPEC-J2 cells with the optimum concentration (determined as described above) of active A. muciniphila (A), IPEC-J2 cells with the optimum concentration (determined as described above) of inactive A. muciniphila (IA), IPEC-J2 cells challenged with TNF-α and then incubated with active A. muciniphila (TNF+A), IPEC-J2 cells challenged with TNF-α and then incubated with inactive A. muciniphila (TNF+IA). Of these treatments, A and IA were designed to investigate the effect of both active and inactive A. muciniphila on normal IECs, TNF+A and TNF+IA aimed to assess the possible remission of inflammatory IECs by A. muciniphila.
In this study, the relative expression of four genes encoding inflammation related cytokines and two tight junction proteins were selected as the markers of inflammation. The specific primers for IL-1β, IL-6 and TNF-α were designed with Primer Primer5 software, while the primers for IL-8 (22), zonula occludens-1 (ZO-1) (25), Occludin (26) and the three house-keeping genes, β-actin (27), GAPDH (28) and TBP (29) were referred to existing researches (Table 1). The total RNA of the collected IPEC-J2 cells was extracted using a TRIzol reagent (Invitrogen, Carlsbad, California, USA). Then, each RNA sample was reverse-transcribed into cDNA using PrimeScript RT reagent kit (TaKaRa, Dalian, China) after detection of RNA purity and integrity by a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Massachusetts, USA) and 1.5% agarose gel electrophoresis. Real-time PCR was also performed on a CFX96TM Real-Time System (Bio-Rad, USA) in triplicate. The 25 μL reaction solution consisted of 12.5 μL SYBR Premix EX Taq (TaKaRa, Dalian, China), 0.5 μL of each primer (10 μM), 2 μL cDNA template and 9.5 μL ddH2O. Procedures of the PCR reaction included a pre-denaturation at 95°C for 30 s followed by 40 cycles of 95°C for 5 s and 60°C for 34 s. The data of targeting genes were then normalized with house-keeping genes, and the relative expression of each gene was calculated using the 2-△△Ct method (30).
The apoptosis of IPEC-J2 cells was detected using an Annexin V-Fluorescein isothiocyanate isomer/Propidium Iodide (Annexin V-FITC/PI) kit (Beyotime, Shanghai, China) according to the manufacturer’s instructions. Shortly, the medium and cells in each well were collected at the end of the incubation, and then treated with 2 mL 0.25% ethylenediaminetetraacetic acid (EDTA)-free pancreatin (Beyotime, China) for 2 min. The solution was then collected into a 10 mL centrifugal tube and centrifuged at 1,000 × g for 5 min. Cells were then resuspended in 100 μL 1 × Binding Buffer to reach a final concentration of 1 × 105 cells/mL. Next, 5 μL FITC Annexin V and 5 μL PI were added into the tube respectively, and the mixture was incubated at room temperature for 15 min away from light. Finally, 400 μL 1 × Binding Buffer was added to each tube and cell apoptosis was analyzed using a flow cytometry (FACSVerse, BD, USANIK) within 1 h.
Total RNA of each sample was extracted using TRIzol (Invitrogen, USA), and the quality of extracted RNA was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). Approximate 1μg RNA of each sample was used for the preparation of library. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumia). Then, the prepared RNA samples were sequenced on an Illumina Novaseq platform and 150 bp paired-end reads were generated. All the downstream analyses were based on the clean data with high quality. FeatureCounts v1.5.0-p3 was used to count the reads numbers mapped to each gene. The FPKM (Fragments per Kilobase Million) of each gene was calculated based on the length of the gene and reads count mapped to this gene. Principal component analysis (PCA) was performed and visualized using the DESeq (2012) R package.
Differential expression genes (DEGs) were filtered according to the following parameters: P value < 0.05, and log2 fold change > 1 or < –1 in each pairwise comparison. The differential expression genes in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were annotated using KOBAS 3.0 (http://kobas.cbi.pku.edu.cn/kobas3/genelist/) and visualized using the ggplot2 R package. The DEGs were annotated using STRING (https://cn.stringdb.org) and the protein interaction network was visualized using Cytoscape (3.7.1). All sequences were uploaded to NCBI database with an accession number (PRJNA752872).
Data were analyzed using a one-way ANOVA with Holm-Sidak’s multiple comparison (e.g., the optimum concentration of Recombinant Susscrofa TNF-α) or Tukey’s multiple comparison (e.g., the mRNA level of genes and the rate of apoptosis). The multiple t-test was used when comparing only two groups (e.g., the bacteria viability test and the processing time of TNF-α on the IPEC-J2 cells). The viability of IPEC-J2 cells to active or inactive A. muciniphila was analyzed using a two-way ANOVA with Sidak’s multiple comparison. Data were shown as mean ± standard error of the mean (SEM). Differences were considered to be significant when P < 0.05. Statistical analysis was performed using a GraphPad Prism 8 software (GraphPad Software, La Jolla, USA).
First, the appropriate concentration of Recombinant Susscrofa TNF-α to be applied on IPEC-J2 was determined. Cells were treated with TNF-α at 10, 20, 40, 80 and 120 ng/mL after 48 h, the expression of IL-1β, IL-6 and TNF-α were measured. The highest response of IL-1β and TNF-α was obtained with the concentration of 20 ng/mL of TNF-α (P < 0.001, Figures 2A, C). IL-6 was only increased with the concentration of 80 ng/mL TNF-α (Figure 2B). Taking these data together, the dose of 20 ng/mL TNF-α was chosen for further studies. To confirm the optimal processing time, the IPEC-J2 cells were then treated with 20 ng/mL TNF-α for 12, 24, 36, and 48 h. In the TNF-α-treated cells, the expression of the cytokines was increased when compared to the control at 48h for IL-1β (P < 0.05, Figure 2D), 12 and 24 h for IL-6 (P < 0.01 or 0.05, Figure 2E), as well as 12, 24, 36 and 48h for TNF-α (P < 0.01 or 0.05, Figure 2F). Meanwhile, the bright-field images (Figure 2G) of inverted microscope showed that the integrity of those TNF-α-challenged cells was destroyed with obviously increased gap and the volume of nucleus compared to the control. Particularly, the IPEC-J2 cells treated with 20 ng/mL for 48 h showed the highest degree of cell destruction and the most cellular fragments, which are typical characteristics of apoptosis. Taking these data together, the incubation of 48 h with 20 ng/mL TNF-α was chosen to test the effects of A. muciniphila on stressed IPEC-J2 cells.
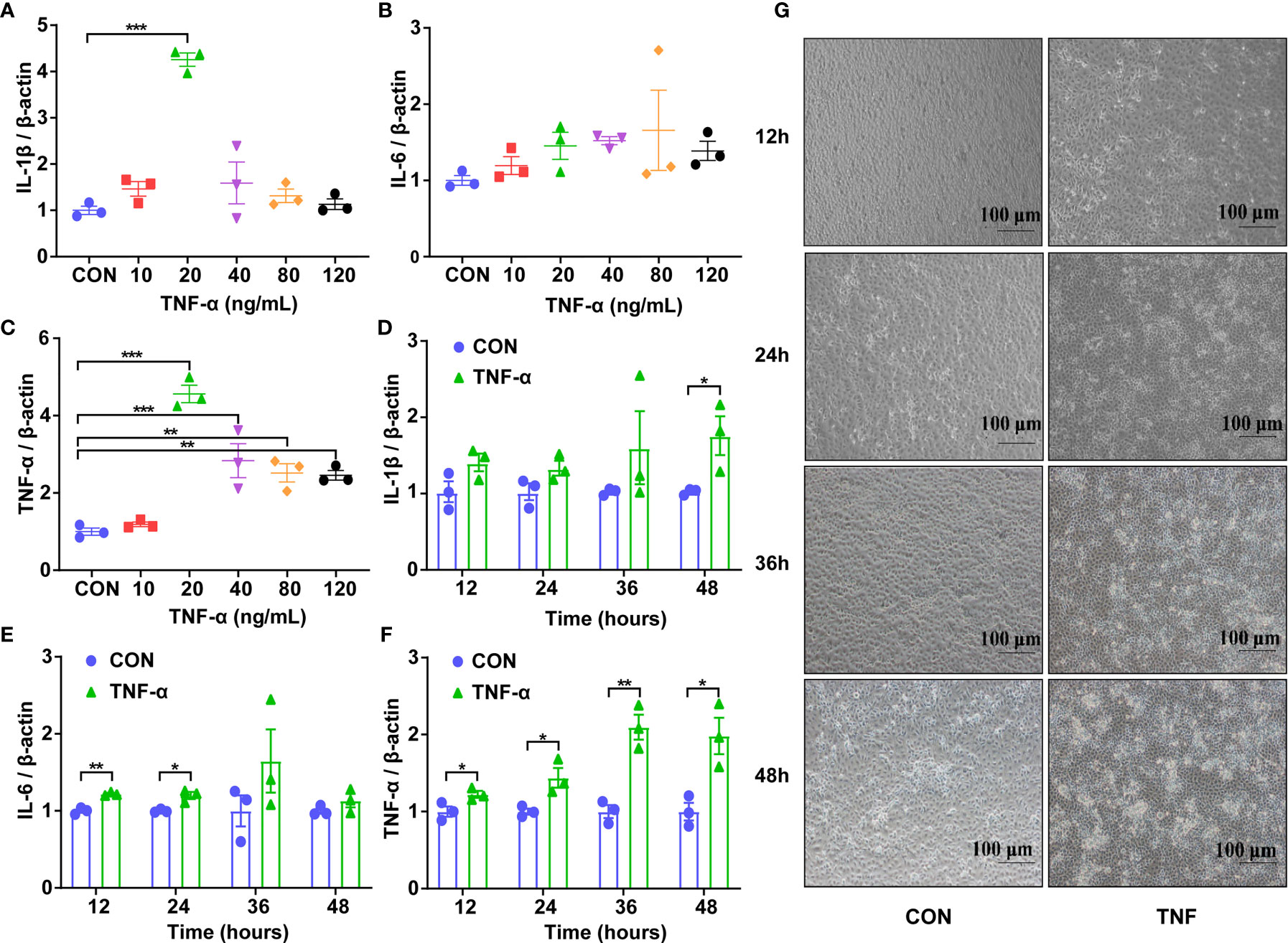
Figure 2 Expression of genes encoding inflammatory cytokines and the representative morphology of IPEC-J2 cells in different groups. (A) The mRNA level of IL-1β of cells challenged with Recombinant Susscrofa TNF-α with different concentrations (0, 10, 20, 40, 80, 120 ng/mL) for 48 h. (B) The mRNA level of IL-6 of cells challenged with TNF-α with different concentrations (0, 10, 20, 40, 80, 120 ng/mL) for 48 h. (C) The mRNA level of TNF-α of cells challenged with TNF-α with different concentrations (0, 10, 20, 40, 80, 120 ng/mL) for 48 h. (D) The mRNA level of IL-1β of cells challenged with 20 ng/mL TNF-α for 12, 24, 36 and 48 h. (E) The mRNA level of IL-6 of cells challenged with 20 ng/mL TNF-α for 12, 24, 36 and 48 h. (F) The mRNA level of TNF-α of cells challenged with 20-ng/mL TNF-α for 12, 24, 36 and 48 h. Data are represented as mean ± SEM (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001. CON, cells without TNF-α treatment. Results represented one of the three independent experiments. (G) The representative images of IPEC-J2 cells challenged with 20 ng/mL Recombinant Susscrofa TNF-α photographed by an inverted microscope under bright field at 12, 24, 36 and 48 h, respectively. The scale bar is 100 μm.
According to the growth curve based on copy numbers (Figure 3A), the growth of A. muciniphila in BHI medium reached a plateau at 48 h. To verify whether A. muciniphila was effectively inactivated by the autoclave treatment, we detected the OD value of the bacterium (pure cells without medium) during 18 hours. The non-autoclaved A. muciniphila showed a growth at 6 and 18 h (P < 0.001, Figure 3B), whereas the autoclaved cells did not grow during the same period (P > 0.05, Figure 3B).
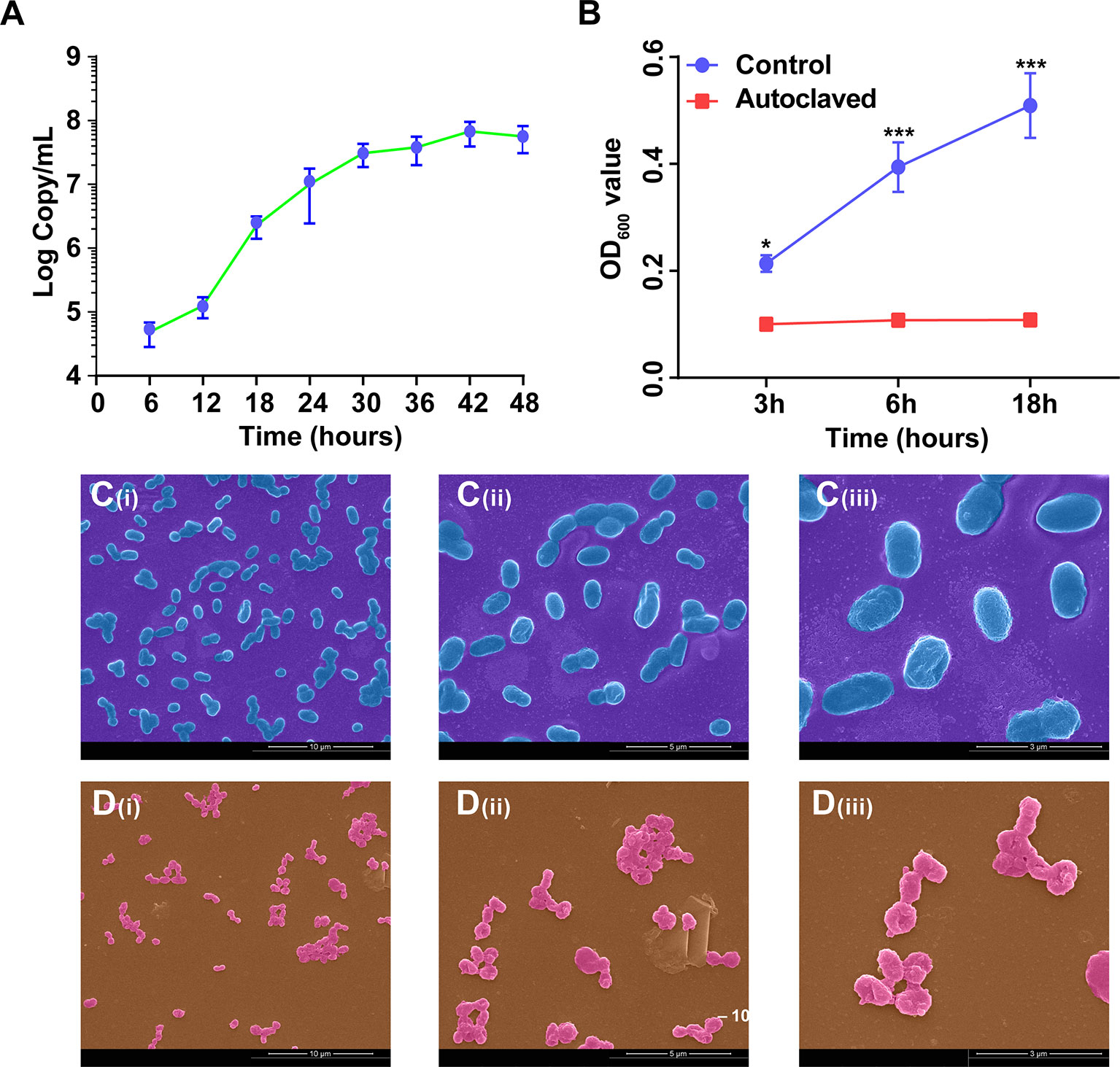
Figure 3 The growth curve and cellular morphology of A. muciniphila. (A) The growth curve of A. muciniphila cultured in BHI medium. (B) The growth curve of A. muciniphila with (Autoclaved) or without (Control) autoclaving in BHI medium within 18 hours. OD600 value. (C) SEM images of active A. muciniphila. (i) Bar, 10 µm. magnification times, × 5,000. (ii) Bar, 5 µm. magnification times, × 10,000. (iii) Bar, 3 µm. magnification times, × 20,000. (D) SEM images of inactive A. muciniphila. (i) Bar, 10 µm. magnification times, × 5,000. (ii) Bar, 5 µm. magnification times, × 10,000. (iii) Bar, 3 µm. magnification times, × 20,000. Data are expressed as mean ± SEM (n = 3). *P < 0.05, ***P < 0.001. Results represented one of the two independent experiments.
According to the profiles of scanning electron microscope (Figures 3C, D), the cellular morphology of active and inactive A. muciniphila showed less different. Specifically, the cells of active A. muciniphila were oval-shaped, owned complete and smooth cell walls, and the typical cell of autoclaved A. muciniphila showed irregular shape, sticked together, partially collapsed, accidented and even broken.
To determine the viability of IPEC-J2 cells in presence of A. muciniphila, the IPEC-J2 cells were co-cultured with active or autoclaved A. muciniphila applied at 106 to 109 copies/mL for 2.5, 5, 7.5 and 10 hours. Compared to non-treated IPEC-J2 cells, the viability of IPEC-J2 cells co-cultured with active A. muciniphila was increased for all times and all concentrations of the bacteria (P < 0.05 or 0.01 or 0.001, Figure 4A), except for the concentration of 109 of active A. muciniphila for 7.5 and 10 h which led to a significant decrease in viability of IPEC-J2 cells (P < 0.001, Figure 4A). When treated with inactive A. muciniphila, IPEC-J2 showed an increased viability for a concentration of 109 of inactive A. muciniphila for 7.5 and 10 h (P < 0.05, Figure 4B). From these results, the optimum concentration of active and inactive A. muciniphila should be 108 and 109 copies/mL respectively, and the incubation time with IPEC-J2 cells should be 7.5 h.
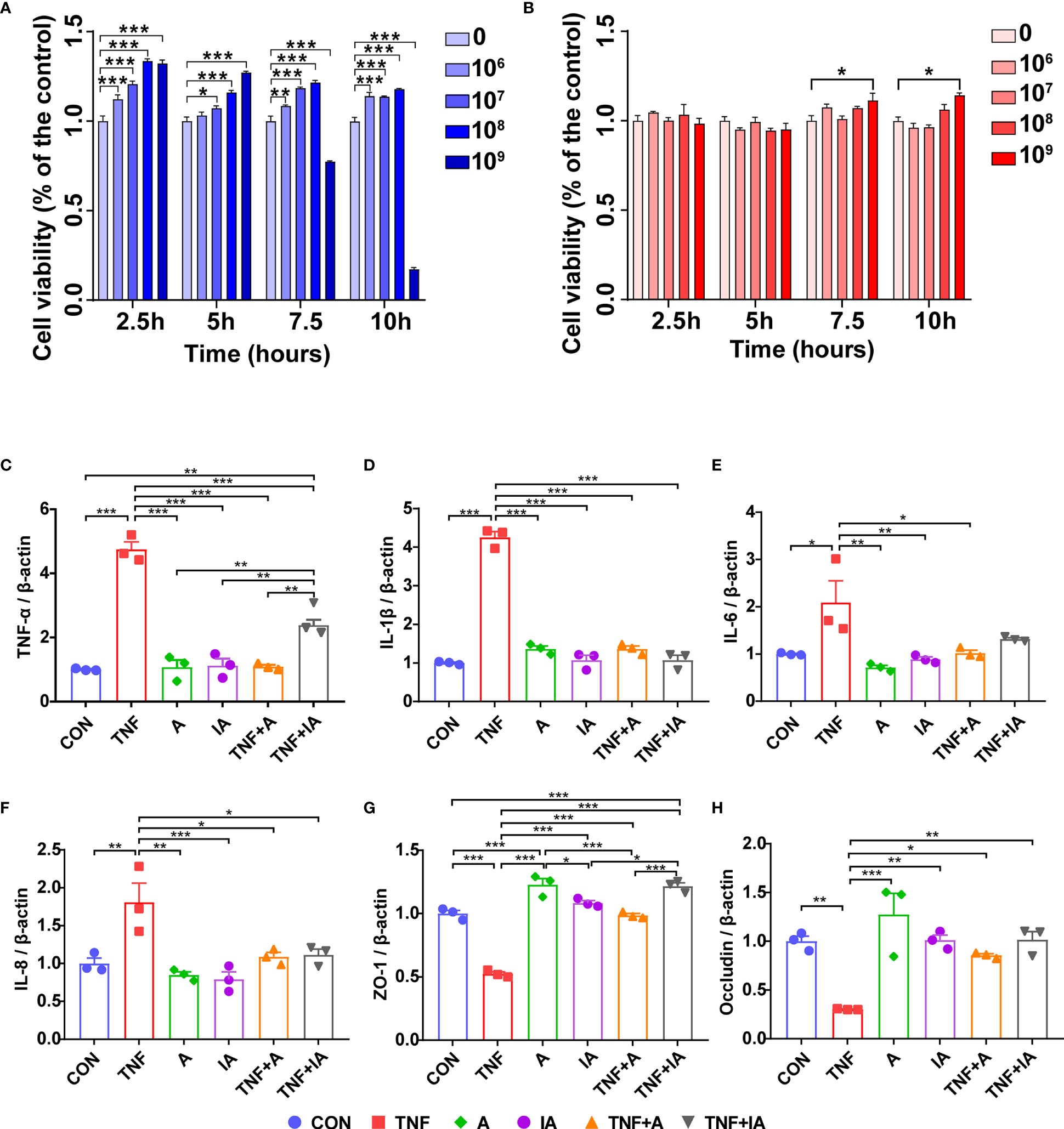
Figure 4 The cell viability of IPEC-J2 cells treated with active or inactive A. muciniphila. and mRNA level of inflammation related cytokines and tight junctions of the IPEC-J2 cells in different groups. (A) The viability of IPEC-J2 cells co-cultured with living (active) A. muciniphila at 0, 106, 107, 108 and 109 copies/mL for 2.5, 5, 7.5 and 10 h, respectively. (B) The viability of IPEC-J2cells co-cultured with inactive (autoclaved) A. muciniphila at 0, 106, 107, 108 and 109 copies/mL for 2.5, 5, 7.5 and 10 h, respectively. (C) The mRNA level of TNF-α of IPEC-J2 cells in different groups. (D) The mRNA level of IL-1β of IPEC-J2 cells in different groups. (E) The mRNA level of IL-6 of cells in different groups. (F) The mRNA level of IL-8 of IPEC-J2 cells in different groups. (G) The mRNA level of Occludin of IPEC-J2 cells in different groups. (H) The mRNA level of Occludin of IPEC-J2 cells in different groups. CON, cells treated with PBS for 48 h. TNF, cells challenged with 20 ng/mL Recombinant Susscrofa TNF-α for 48 hours. A, cells co-cultured with 108 copies/mL active A. muciniphila for 48 h. IA, cells co-cultured with109 copies/mL inactive A. muciniphila for 48 h. TNF+A, cells challenged with 20 ng/mL TNF-α for 40.5 h and then co-cultured with 108 copies/mL active A. muciniphila for 7.5 h. TNF+IA, cells challenged with 20 ng/mL TNF-α for 40.5 h and then co-cultured with 109 copies/mL inactive A. muciniphila for 7.5 h. *P < 0.05, **P < 0.01, ***P < 0.001. Data are expressed as mean ± SEM (n = 3). Results represented one of the three independent experiments.
In order to assess the effects of A. muciniphila on IECs, the normal or TNF-α challenged IPEC-J2 cells were co-cultured with active or inactive A. muciniphila. According to the results of real-time PCR, the expression of TNF-α (P < 0.001, Figure 4C), IL-1β (P < 0.001, Figure 4D), IL-6 (P < 0.05, Figure 4E) and IL-8 (P < 0.001, Figure 4F) showed increased in the cells in group TNF, while the expression of ZO-1 (Figure 4G) and Occludin (Figure 4H) in these cells was decreased compared to CON group (P < 0.01 or 0.001). Comparing with cells in group TNF, the mRNA levels of TNF-α (Figure 4C), IL-1β (Figure 4D) and IL-8 (Figure 4F) of the cells in groups A, IA, TNF+A and TNF+IA, as well as the mRNA level of IL-6 (Figure 4E) of cells in groups A, IA and TNF+A were reduced (P < 0.05 or 0.01 or 0.001). The expression of ZO-1 (Figure 4G) and Occludin (Figure 4H) of the cells co-cultured with both active and inactive A. muciniphila was higher than cells in TNF group (P < 0.05 or 0.01 or 0.001).
The degree of cellular damage can be reflected by the rate of apoptosis. In the current study, an annexin V-FITC/PI assay with flow cytometry was performed to investigate whether A. muciniphila could attenuate the apoptosis induced by TNF-α in IPEC-J2 cells. Results showed that compared with CON cells, the early-stage apoptosis (Figures 5A, B) of the cells in groups TNF (P < 0.001) was increased (Figures 5A, B). While the early-stage apoptosis of the cells in groups A, IA, TNF+A and TNF+IA was decreased compared to those cells in TNF group (P < 0.01 or 0.001).
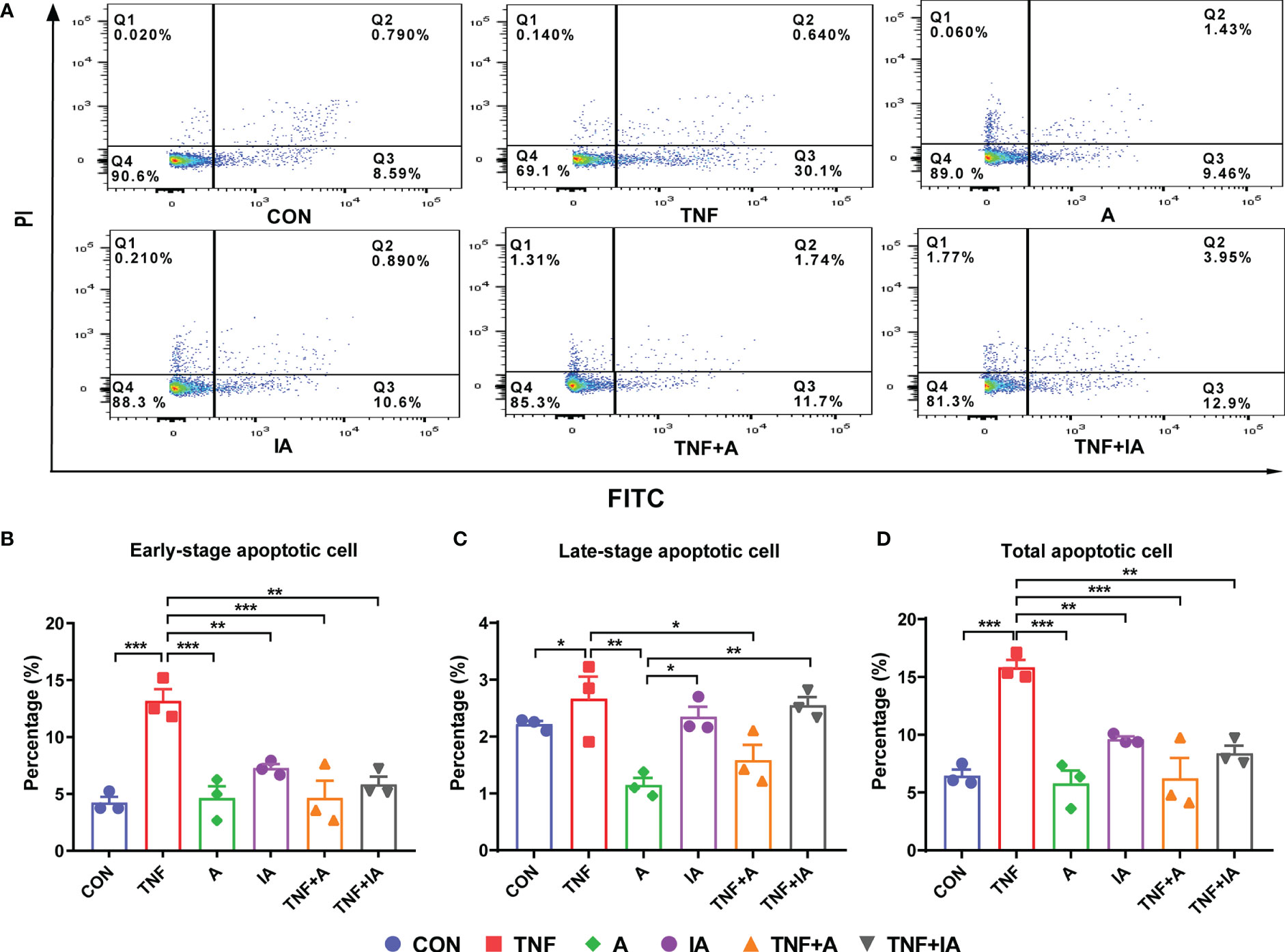
Figure 5 The apoptosis of IPEC-J2cells in different groups. (A) The profile of flow cytometry with an annexin V-FITC/PI kit of the IPEC-J2 cells in different groups. Each of the frames is divided into four quadrants: Q1, necrotic cells. Q2, cells in the late-stage of apoptosis. Q3, cells in the early-stage of apoptosis. Q4, normal cells. (B) The rate of IPEC-J2 cells in the early stage of apoptosis in different groups. (C) The rate of IPEC-J2 cells in the late stage of apoptosis in different groups. (D) The rate of total apoptosis of IPEC-J2 cells in different groups. Data are represented as mean± SEM (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001. CON, cells treated with PBS for 48 h. TNF, cells challenged with 20 ng/mL Recombinant Susscrofa TNF-α for 48 hours. A, cells co-cultured with 108 copies/mL active A. muciniphila for 48 h. IA, cells co-cultured with 109 copies/mL inactive A. muciniphila for 48 h. TNF+A, cells challenged with 20 ng/mL TNF-α for 40.5 h and then co-cultured with 108 copies/mL active A. muciniphila for 7.5 h. TNF+IA, cells challenged with 20 ng/mL TNF-α for 40.5 h and then co-cultured with 109 copies/mL inactive A. muciniphila for 7.5 h.
Comparing with cells in CON group, the treatment of TNF-α increased the total apoptosis rate of the IPEC-J2 cells (P < 0.001, Figure 5). However, the treatment of active (TNF+A) A. muciniphila and group A and IA was found to reduce the total apoptosis rate of the TNF-challenged cells compared to TNF group (P < 0.01 or 0.001, Figure 5D). Meanwhile, the late-stage apoptosis of cells in group IA and TNF+IA was higher than those in other group (P < 0.05 or 0.01, Figures 5A, C).
As shown above, we found that both live and inactive A. muciniphila probably alleviated the inflammatory injury of IPEC-J2 induced by TNF-α, but the underlying pathway remains unclear. In order to find possible mechanism, a transcriptome analysis was performed. After removing low-quality reads, the average number of reads in each library was over 45 million. All filtered reads were aligned with the reference genome of Sus scrofa by Hisat2 software, and the transcripts identified in each sample (FPKM) were analyzed. The PCA targeting the top 20,000 genes with the highest expression level from the total 9 samples from the three groups revealed three corresponding independent clusters (Figure 6A). Further, a total of 197 DEGs were unique in TNF+A vs TNF, and 151 DEGs were only found in TNF+IA vs TNF (Figure 6B). The KEGG function of the filtered DEGs was then annotated using a KOBAS 3.0 (http://kobas.cbi.pku.edu.cn/kobas3/genelist/), and the first 20 paths with the lowest p-value were selected to generate bubble chart (Figures 6C, D). When comparing groups TNF+A and TNF, the different pathways involved in inflammatory response were identified as calcium signaling pathway and PI3K-Akt signaling pathway that also showed different between groups TNF+IA and TNF. The cell cycle pathway associated with cell proliferation and cycle also showed different between TNF+IA and TNF. According to the visualized protein interaction network (Figure 7A, B), the expression of proteins CACNA1S (Calcium channel, voltage-dependent, L type, alpha 1S subunit), CACNA1G (Calcium channel, voltage-dependent, T type, alpha 1G subunit), P2RX1 (Purinergic Receptor P2X 1) and P2RX2 (Purinergic Receptor P2X 2) in calcium signaling pathway, as well as the expression of proteins IL2RA (Interleukin-2 receptor subunit alpha), EPOR (Erythropoietin receptor) and PRLR (Prolactin Receptor) in PI3K-Akt signaling pathway were decreased (P < 0.05), and only the expression of ATP2B2 (ATPase plasma membrane calcium transporter) showed increased (P < 0.05) in TNF+A group compared to group TNF. Comparing with group TNF, the expression of proteins EPOR and IL2R (Interleukin-2 receptor) in PI3K-Akt signaling pathway and CDKN1C (Cyclin-dependent kinase inhibitor 1c) in cell cycle pathway showed decreased (P < 0.05), while the expression of proteins PCNA (Proliferating Cell Nuclear Antigen), CCNE2 (Cyclin E2), CDC23 (Cell division cycle 23), ORC1 (Origin Recognition Complex 1) and MCM3 (Mini-chromosome maintenance 3) in cell cycle pathway showed raised (P < 0.05).
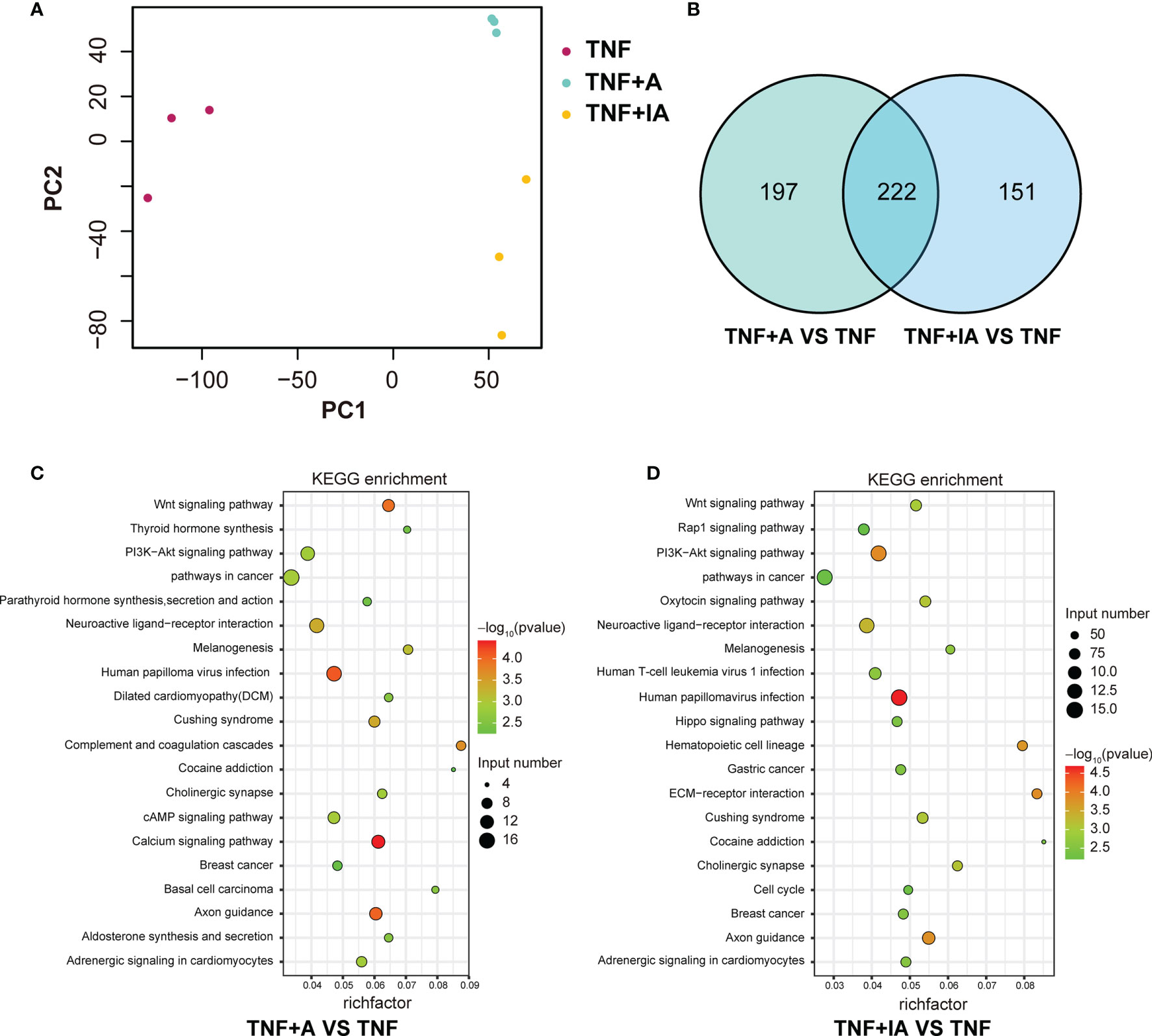
Figure 6 Comparison of gene expression abundance and KEGG enrichment in T, and TNF+A and TNF+IA groups. (A) The principal component analysis based on the RNA-Seq data. (B) Venn diagrams of differentially expressed transcripts. (C) The top 20 KEGG pathways enriched in TNF+A group compared to group TNF. (D) The top 20 KEGG pathways enriched in TNF+IA group compared to group TNF. TNF, cells challenged with 20 ng/mL Recombinant Susscrofa TNF-α for 48 hours. TNF+A, cells challenged with 20 ng/mL TNF-α for 40.5 h and then co-cultured with 108 copies/mL active A. muciniphila for 7.5 h. TNF+IA, cells challenged with 20 ng/mL TNF-α for 40.5 h and then co-cultured with 109 copies/mL inactive A. muciniphila for 7.5 h.
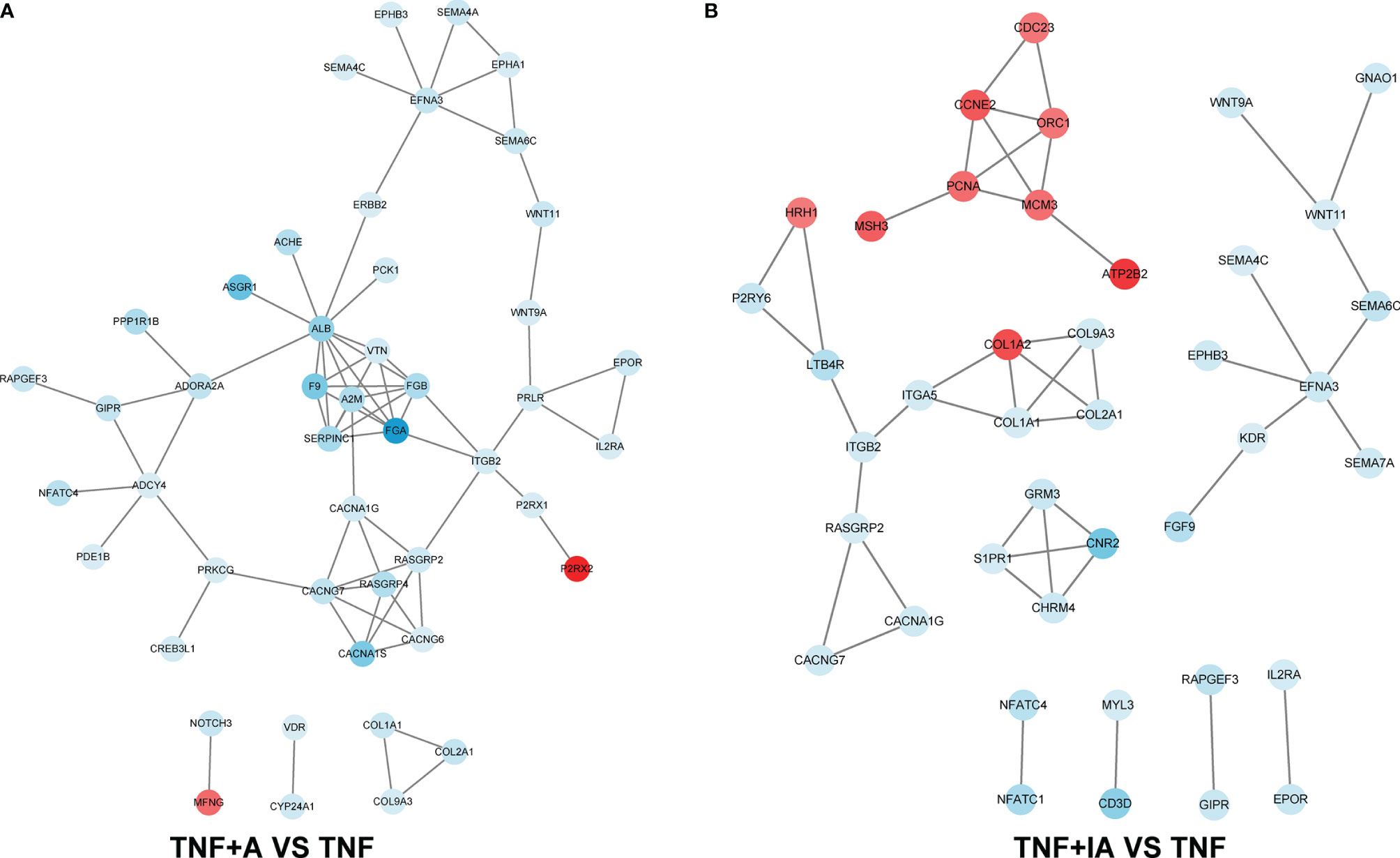
Figure 7 The protein-protein interaction network based on the KEGG annotation. (A) Change of protein abundance in TNF+A group compared to group TNF. (B) Change of protein abundance in TNF+IA group compared to group TNF. TNF, cells challenged with 20 ng/mL Recombinant Susscrofa TNF-α for 48 hours. TNF+A, cells challenged with 20 ng/mL TNF-α for 40.5 h and then co-cultured with 108 copies/mL active A. muciniphila for 7.5 h. TNF+IA, cells challenged with 20 ng/mL TNF-α for 40.5 h and then co-cultured with 109 copies/mL inactive A. muciniphila for 7.5 h.
Besides human beings, A. muciniphila is also found in the gut of other mammals such as guinea pigs, swines and rabbits (31–33). The effects of A. muciniphila on obesity and metabolic disorders have been widely discussed. However, its effect on the intestinal inflammation is controversial. In the gut of patients with inflammatory bowel disease (IBD) (34) or those with Crohn’s disease below 16 years of age (35), the abundant of A. muciniphila is reported to be reduced, indicating a positive role of this bacterium on gut health. But in two other studies, commensal A. muciniphila is observed to exacerbate the gut inflammation in Salmonella typhimurium-infected gnotobiotic mice (36) and was sufficient for promoting intestinal inflammation in germ-free Il10-/-mice (37). Another particularity of this bacterium is that some of its affects have been attributed to cellular components, such as proteins, which led us to think that similar effect could also be observed with inactivated cells. In the present study, we investigated the effect of A. muciniphila as alive or inactivated cultures on the IPEC-J2 cells with inflammation induced by Recombinant Susscrofa TNF-α to explore whether A. muciniphila benefit the health of IECs. To the best of our knowledge, the current study is the first to evaluate the role of both active and inactive A. muciniphila in an inflammatory model of IPEC-J2 cells.
Inflammatory response of IECs generally results in the occurrence of proinflammatory cytokines such as IL-1β, IL-6 and TNF-α (38, 39). The level of these cytokines is also widely used in clinical diagnosis as typical biomarkers of inflammation (40). In the current study, the mRNA level of TNF-α and IL-1β was remarkably increased in those cells treated with 20 ng/mL of Recombinant Susscrofa TNF-α for 48 hours, accompanied by visible lesion of cells under microscope. These results show that an inflammatory model of IPEC-J2 cells had been successfully induced by the recombinant TNF-α.
The potential to prevent or cure a certain disease is the main characteristic of NGPs different from FGPs, which makes most of the NGPs be classified to live biotherapeutics products (LBPs) (41). A. muciniphila is regarded as one of the most promising LBPs against metabolic diseases like obesity and diabetes. On the other hand, A. muciniphila may also play a role in the maintenance of intestinal homeostasis and improvement of intestinal inflammation. In patients with IBD, the relative abundance of A. muciniphila in the intestinal mucosa is decreased (42). Oral gavage of A. muciniphila is increases the level of 2-arachidonoylglycerol and in turn reduces the inflammatory responses in mice receiving a high-fat diet (10) and relieve dextran sulfate sodium (DSS)-induced colitis via its extracellular vesicles (43). In the current study, we not only showed the prevention of A. muciniphila as a LBP on the inflammation of IECs, but also showed that inactive A. muciniphila cells could be an available candidate against the inflammation of IECs. Whether firstly treated with 20 ng/mL of TNF-α for 40.5 h and followed by a treatment of active or inactive A. muciniphila for 7.5 h the mRNA level of proinflammatory cytokines was remarkably reduced along with the increased mRNA level of TJs. The epithelial permeability largely depends on adherin junctions and TJs. An increased intestinal permeability always contributes to the severity of some diseases, such as IBD and irritable bowel syndrome (44). The expression of genes encoding TJs is usually decreased when the function of IECs is impaired in case of inflammation (45–47). As a negative correlation has been found between TJs and the permeability of IPEC-J2 cells treated with TNF-α (45), the relative expression of TJs genes can be regarded as a marker for the damage of IECs. Our results thus showed a probable repair of A. muciniphila to the inflammatory damage of IECs, which was confirmed by the following analysis of flow cytometry. Apoptosis is a classical program of cell death, regulated by many factors such as TNF-α, IL-1β and caspase-2 (48–52). TNF-α has previously been reported to increase the apoptosis of IECs and cause the injury of intestinal barrier, which give rise to the occurrence of IBD. It was well grounded to hypothesize that A. muciniphila protecting the IECs from TNF-α-induced inflammation might be associated with the suppression of apoptosis. According to our results, the supplement of both active and inactive A. muciniphila after the treatment of TNF-α indeed reduced the early and total apoptosis of the IPEC-J2 cells. Similar results were also found in mice that the intestinal colonization of A. muciniphila strongly altered the expression of genes involved in the death of IECs at transcriptional level (16) and decreased the apoptosis of hepatocytes (53).
Limited to the descriptive results of the current study, the underlying mechanism of the positive effect of inactive A. muciniphila on the inflammatory IECs cannot be explained in detail. It has been shown that some cellular components, such as AmEVs, could increase the expression of TJs in Caco-2 cells and ultimately improve the integrity of intestinal barrier in mice with high-fat diet-induced diabetes (17). But this hypothesis cannot explain our results, as we used only cells but not total culture (and thus not extracellular metabolites) which were then autoclaved. The effects of A. muciniphila we observed is therefore might be due to a cellular component which is resistant to autoclave treatment. Although the profiles of scanning electron microscope showed that the integrity of inactive A. muciniphila cells were partially destroyed, some components of cell wall or membrane might still be biologically active, for instance, autoclave may not affect the structure of polysaccharides. Similarly, the specific membrane protein of A. muciniphila (Amuc_1100) has previously been reported to improve the intestinal barrier and can blunt colitis in mice (54, 55) even when the protein is pasteurized. Another clinical study shows that pasteurized 1010 of A. muciniphila is safe for human and can reduce the level of multiple inflammatory markers in serum (56), which proves the positive effect of inactive A. muciniphila on the systemic inflammation.
We further used transcriptome analysis to scan the potential signaling pathway of A. muciniphila in alleviating the TNF-α induced inflammation of IECs. We found that the treatment of active A. muciniphila down-regulated the expression of some key genes in calcium signaling pathway, such as CACNA1S, P2RX1 and P2RX2. It has been reported that under oxidative stress and other conditions, the increase of intracellular Ca2+ concentration can induce cell apoptosis and autophagy (57–59), while its decrease is associated with the remission of inflammation (60). Therefore, our results suggest that the apoptosis of TNF-α-challenged IECs may be inhibited by active A. muciniphila by reducing the influx of Ca2+ and increasing the outflow of Ca2+. In addition, although the treatment of inactive A. muciniphila was also found to reduce apoptosis of TNF-α-challenged IECs, the underlying mechanism may be different from that of active bacterium. We found that the expression of key genes involved in cell cycle, such as PCNA, CCNE2, CDC23, ORC1 and MCM3 in TNF-α-challenged IECs was up-regulated by the treatment of inactive A. muciniphila. All these genes play an important role in maintaining the growth and survival, the stability of genome and the life span of cells (61–70). At the same time, the expression of CDKN1C was down-regulated by inactive A. muciniphila, which may normalize the cell cycle (71–73). Another noteworthy path was PI3K-Akt signaling pathway. Studies have shown that PI3K-Akt signaling pathway is involved in the occurrence of ulcerative colitis and can be activated by intestinal inflammation (74, 75), and its inhibition can down-regulate the expression of nuclear transcription factors-κB (NF-κB) to reduce the expression of proinflammatory cytokines (76, 77). We found that the treatment of both active and inactive A. muciniphila down-regulated the expression of upstream receptors (IL2RA, EPOR, PRLR and/or PIK3R5) in PI3K-Akt signaling pathway, which may directly decline the expression of cytokines like TNF- α, IL-8, IL-1β and IL-6 in these TNF-α-challenged cells.
Our study suggests that active and autoclaved inactive A. muciniphila improved the TNF-α-induced inflammation of IECs by reducing the expression of pro-inflammatory cytokines and increasing the expression of TJs, which might be associated with the decreased apoptosis. Although both active and inactive A. muciniphila showed significant remission of inflammation and apoptosis of TNF-α-challenged IECs, the underlying mechanism may be different. Active A. muciniphila may decline cell apoptosis by down-regulating the expression of key genes in calcium signaling pathway, while inactive A. muciniphila may reduce cell apoptosis by up-regulating the expression of key genes in cell cycle signaling pathway. Both live and inactive A. muciniphila may alleviate the inflammation of IECs by down-regulating the expression of PI3K upstream receptor genes. The specific effect of A. muciniphila on inflammatory IECs indicates that it may be a promising candidate of NGP for piglets or other animals with similar physiology and anatomy to cure inflammatory. However, further in vivo researches are still needed to explore the underlying mechanism of active and inactive A. muciniphila on the inflammation of IECs.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA752872.
YL designed the experiment and wrote the manuscript. CL and KX finished the experiment and the real-time PCR analysis. HL conducted transcriptome analysis and helped finish the experiment. ED and JH helped design the experiment. LL provided the strain and helped design the experiment. JC, GT, AW, ZH, and DC helped finish the laboratory analysis. BY, ZH, PZ, XM, JY, and JL helped revise the manuscript. HY, QW, and HW helped collect the references and purchase reagents. JT helped analyze real-time PCR data. All authors contributed to the article and approved the submitted version.
This work was supported by Adisseo and the National Natural Science Foundation of China (NSFC, grant number 31872369 and 32072743).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Suzuki T. Regulation of Intestinal Epithelial Permeability by Tight Junctions. Cell Mol Life Sci (2013) 70:631–59. doi: 10.1007/s00018-012-1070-x
2. Patankar JV, Becker C. Cell Death in the Gut Epithelium and Implications for Chronic Inflammation. Nat Rev Gastroenterol Hepatol (2020) 17:543–56. doi: 10.1038/s41575-020-0326-4
3. Lallès J-P, Boudry G, Favier C, Le Floc’h N, Luron I, Montagne L, et al. Gut Function and Dysfunction in Young Pigs: Physiology. Anim Res (2004) 53:301–16. doi: 10.1051/animres:2004018
4. Niewold TA. The Nonantibiotic Anti-Inflammatory Effect of Antimicrobial Growth Promoters, the Real Mode of Action? A Hypothesis. Poult Sci (2007) 86:605–9. doi: 10.1093/ps/86.4.605
5. Maassen CB, van Holten-Neelen C, Balk F, den Bak-Glashouwer MJ, Leer RJ, Laman JD, et al. Strain-Dependent Induction of Cytokine Profiles in the Gut by Orally Administered Lactobacillus Strains. Vaccine (2000) 18:2613–23. doi: 10.1016/S0264-410X(99)00378-3
6. Gotteland M, Cruchet S, Verbeke S. Effect of Lactobacillus Ingestion on the Gastrointestinal Mucosal Barrier Alterations Induced by Indometacin in Humans. Aliment Pharmacol Ther (2001) 15:11–7. doi: 10.1046/j.1365-2036.2001.00898.x
7. Ott SJ, Musfeldt M, Wenderoth DF, Hampe J, Brant O, Fölsch UR, et al. Reduction in Diversity of the Colonic Mucosa Associated Bacterial Microflora in Patients With Active Inflammatory Bowel Disease. Gut (2004) 53:685–93. doi: 10.1136/gut.2003.025403
8. Tlaskalová-Hogenová H, Stepánková R, Hudcovic T, Tucková L, Cukrowska B, Lodinová-Zádníková R, et al. Commensal Bacteria (Normal Microflora), Mucosal Immunity and Chronic Inflammatory and Autoimmune Diseases. Immunol Lett (2004) 93:97–108. doi: 10.1016/j.imlet.2004.02.005
9. Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia Muciniphila Gen. Nov., Sp. Nov., a Human Intestinal Mucin-Degrading Bacterium. Int J Syst Evol Microbiol (2004) 54:1469–76. doi: 10.1099/ijs.0.02873-0
10. Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, et al. Cross-Talk Between Akkermansia Muciniphila and Intestinal Epithelium Controls Diet-Induced Obesity. Proc Natl Acad Sci USA (2013) 110:9066–71. doi: 10.1073/pnas.1219451110
11. Ijaz MU, Ahmad MI, Hussain M, Khan IA, Zhao D, Li C. Meat Protein in High-Fat Diet Induces Adipogensis and Dyslipidemia by Altering Gut Microbiota and Endocannabinoid Dysregulation in the Adipose Tissue of Mice. J Agric Food Chem (2020) 68:3933–46. doi: 10.1021/acs.jafc.0c00017
12. Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A Metagenome-Wide Association Study of Gut Microbiota in Type 2 Diabetes. Nature (2012) 490:55–60. doi: 10.1038/nature11450
13. Seo K-H, Kim D-H, Yokoyama WH, Kim H. Synbiotic Effect of Whole Grape Seed Flour and Newly Isolated Kefir Lactic Acid Bacteria on Intestinal Microbiota of Diet-Induced Obese Mice. J Agric Food Chem (2020) 68:13131–7. doi: 10.1021/acs.jafc.0c01240
14. Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL, Ryan EP. Stool Microbiome and Metabolome Differences Between Colorectal Cancer Patients and Healthy Adults. PLoS One (2013) 8:e70803. doi: 10.1371/journal.pone.0070803
15. Reunanen J, Kainulainen V, Huuskonen L, Ottman N, Belzer C, Huhtinen H, et al. Akkermansia Muciniphila Adheres to Enterocytes and Strengthens the Integrity of the Epithelial Cell Layer. Appl Environ Microbiol (2015) 81:3655–62. doi: 10.1128/AEM.04050-14
16. Derrien M, van Baarlen P, Hooiveld G, Norin E, Müller M, de Vos WM. Modulation of Mucosal Immune Response, Tolerance, and Proliferation in Mice Colonized by the Mucin-Degrader Akkermansia Muciniphila. Front Microbiol (2011) 2:166. doi: 10.3389/fmicb.2011.00166
17. Chelakkot C, Choi Y, Kim D-K, Park HT, Ghim J, Kwon Y, et al. Akkermansia Muciniphila-Derived Extracellular Vesicles Influence Gut Permeability Through the Regulation of Tight Junctions. Exp Mol Med (2018) 50:e450. doi: 10.1038/emm.2017.282
18. Ottman N, Reunanen J, Meijerink M, Pietilä TE, Kainulainen V, Klievink J, et al. Pili-Like Proteins of Akkermansia Muciniphila Modulate Host Immune Responses and Gut Barrier Function. PLoS One (2017) 12:e0173004. doi: 10.1371/journal.pone.0173004
19. Vergauwen H. The Impact of Food Bioactives on Health: In Vitro and Ex Vivo Models: The IPEC-J2 Cell Line. Cham (CH): Springer (2015). p. 10.
20. Wilson K. Preparation of Genomic DNA From Bacteria. Curr Protoc Mol Biol (2001) 56:2.4.1–5. doi: 10.1002/0471142727.mb0204s56
21. Collado MC, Derrien M, Isolauri E, de Vos WM, Salminen S. Intestinal Integrity and Akkermansia Muciniphila, A Mucin-Degrading Member of the Intestinal Microbiota Present in Infants, Adults, and the Elderly. Appl Environ Microbiol (2007) 73:7767–70. doi: 10.1128/AEM.01477-07
22. Yang JW, Tian G, Chen DW, Yao Y, He J, Zheng P, et al. Involvement of PKA Signalling in Anti-Inflammatory Effects of Chitosan Oligosaccharides in IPEC-J2 Porcine Epithelial Cells. J Anim Physiol Anim Nutr (Berl) (2018) 102:252–9. doi: 10.1111/jpn.12686
23. Geens MM, Niewold TA. Optimizing Culture Conditions of a Porcine Epithelial Cell Line IPEC-J2 Through a Histological and Physiological Characterization. Cytotechnology (2011) 63:415–23. doi: 10.1007/s10616-011-9362-9
24. Vergauwen H, Tambuyzer B, Jennes K, Degroote J, Wang W, de Smet S, et al. Trolox and Ascorbic Acid Reduce Direct and Indirect Oxidative Stress in the IPEC-J2 Cells, an In Vitro Model for the Porcine Gastrointestinal Tract. PLoS One (2015) 10:e0120485. doi: 10.1371/journal.pone.0120485
25. Pu J, Chen D, Tian G, He J, Zheng P, Mao X, et al. Effects of Benzoic Acid, Bacillus Coagulans and Oregano Oil Combined Supplementation on Growth Performance, Immune Status and Intestinal Barrier Integrity of Weaned Piglets. Anim Nutr (2020) 6:152–9. doi: 10.1016/j.aninu.2020.02.004
26. Mao X, Gu C, Hu H, Tang J, Chen D, Yu B, et al. Dietary Lactobacillus Rhamnosus GG Supplementation Improves the Mucosal Barrier Function in the Intestine of Weaned Piglets Challenged by Porcine Rotavirus. PLoS One (2016) 11:e0146312. doi: 10.1371/journal.pone.0146312
27. Su G, Xie K, Chen D, Yu B, Huang Z, Luo Y, et al. Differential Expression, Molecular Cloning, and Characterization of Porcine Beta Defensin 114. J Anim Sci Biotechnol (2019) 10:60. doi: 10.1186/s40104-019-0367-0
28. Chen J, Li Y, Yu B, Chen D, Mao X, Zheng P, et al. Dietary Chlorogenic Acid Improves Growth Performance of Weaned Pigs Through Maintaining Antioxidant Capacity and Intestinal Digestion and Absorption Function. J Anim Sci (2018) 96:1108–18. doi: 10.1093/jas/skx078
29. Martino A, Cabiati M, Campan M, Prescimone T, Minocci D, Caselli C, et al. Selection of Reference Genes for Normalization of Real-Time PCR Data in Minipig Heart Failure Model and Evaluation of TNF-α mRNA Expression. J Biotechnol (2011) 153:92–9. doi: 10.1016/j.jbiotec.2011.04.002
30. Ginzinger DG. Gene Quantification Using Real-Time Quantitative PCR. Exp Hematol (2002) 30:503–12. doi: 10.1016/S0301-472X(02)00806-8
31. Hildebrand F, Ebersbach T, Nielsen HB, Li X, Sonne SB, Bertalan M, et al. A Comparative Analysis of the Intestinal Metagenomes Present in Guinea Pigs (Cavia Porcellus) and Humans (Homo Sapiens). BMC Genomics (2012) 13:514. doi: 10.1186/1471-2164-13-514
32. McCormack UM, Curião T, Buzoianu SG, Prieto ML, Ryan T, Varley P, et al. Exploring a Possible Link Between the Intestinal Microbiota and Feed Efficiency in Pigs. Appl Environ Microbiol (2017) 83:e00380–17. doi: 10.1128/AEM.00380-17
33. Fang S, Chen X, Zhou L, Wang C, Chen Q, Lin R, et al. Faecal Microbiota and Functional Capacity Associated With Weaning Weight in Meat Rabbits. Microb Biotechnol (2019) 12:1441–52. doi: 10.1111/1751-7915.13485
34. Derrien M, Belzer C, de Vos WM. Akkermansia Muciniphila and Its Role in Regulating Host Functions. Microb Pathog (2017) 106:171–81. doi: 10.1016/j.micpath.2016.02.005
35. Lopez-Siles M, Enrich-Capó N, Aldeguer X, Sabat-Mir M, Duncan SH, Garcia-Gil LJ, et al. Alterations in the Abundance and Co-Occurrence of Akkermansia Muciniphila and Faecalibacterium Prausnitzii in the Colonic Mucosa of Inflammatory Bowel Disease Subjects. Front Cell Infect Microbiol (2018) 8:281. doi: 10.3389/fcimb.2018.00281
36. Ganesh BP, Klopfleisch R, Loh G, Blaut M. Commensal Akkermansia Muciniphila Exacerbates Gut Inflammation in Salmonella Typhimurium-Infected Gnotobiotic Mice. PLoS One (2013) 8:e74963. doi: 10.1371/journal.pone.0074963
37. Seregin SS, Golovchenko N, Schaf B, Chen J, Pudlo NA, Mitchell J, et al. NLRP6 Protects Il10-/- Mice From Colitis by Limiting Colonization of Akkermansia Muciniphila. Cell Rep (2017) 19:733–45. doi: 10.1016/j.celrep.2017.03.080
38. Akira S, Hirano T, Taga T, Kishimoto T. Biology of Multifunctional Cytokines: IL 6 and Related Molecules (IL 1 and TNF). FASEB J (1990) 4:2860–7. doi: 10.1096/fasebj.4.11.2199284
39. Shanahan F. Inflammatory Bowel Disease: Immunodiagnostics, Immunotherapeutics, and Ecotherapeutics. Gastroenterology (2001) 120:622–35. doi: 10.1053/gast.2001.22122
40. Stevens C, Walz G, Singaram C, Lipman ML, Zanker B, Muggia A, et al. Tumor Necrosis Factor-Alpha, Interleukin-1 Beta, and Interleukin-6 Expression in Inflammatory Bowel Disease. Dig Dis Sci (1992) 37:818–26. doi: 10.1007/bf01300378
41. O’Toole PW, Marchesi JR, Hill C. Next-Generation Probiotics: The Spectrum From Probiotics to Live Biotherapeutics. Nat Microbiol (2017) 2:17057. doi: 10.1038/nmicrobiol.2017.57
42. Png CW, Lindén SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, et al. Mucolytic Bacteria With Increased Prevalence in IBD Mucosa Augment In Vitro Utilization of Mucin by Other Bacteria. Am J Gastroenterol (2010) 105:2420–8. doi: 10.1038/ajg.2010.281
43. Kang C-S, Ban M, Choi E-J, Moon H-G, Jeon J-S, Kim D-K, et al. Extracellular Vesicles Derived From Gut Microbiota, Especially Akkermansia Muciniphila, Protect the Progression of Dextran Sulfate Sodium-Induced Colitis. PLoS One (2013) 8:e76520. doi: 10.1371/journal.pone.0076520
44. Vancamelbeke M, Vermeire S. The Intestinal Barrier: A Fundamental Role in Health and Disease. Expert Rev Gastroenterol Hepatol (2017) 11:821–34. doi: 10.1080/17474124.2017.1343143
45. Bruewer M, Luegering A, Kucharzik T, Parkos CA, Madara JL, Hopkins AM, et al. Proinflammatory Cytokines Disrupt Epithelial Barrier Function by Apoptosis-Independent Mechanisms. J Immunol (2003) 171:6164–72. doi: 10.4049/jimmunol.171.11.6164
46. Watson AJ, Hughes KR. TNF-α-Induced Intestinal Epithelial Cell Shedding: Implications for Intestinal Barrier Function. Ann NY Acad Sci (2012) 1258:1–8. doi: 10.1111/j.1749-6632.2012.06523.x
47. Lechuga S, Ivanov AI. Disruption of the Epithelial Barrier During Intestinal Inflammation: Quest for New Molecules and Mechanisms. Biochim Biophys Acta Mol Cell Res (2017) 1864:1183–94. doi: 10.1016/j.bbamcr.2017.03.007
48. Liao Y-C, Liang W-G, Chen F-W, Hsu J-H, Yang J-J, Chang M-S. IL-19 Induces Production of IL-6 and TNF-Alpha and Results in Cell Apoptosis Through TNF-Alpha. J Immunol (2002) 169:4288–97. doi: 10.4049/jimmunol.169.8.4288
49. Miao EA, Rajan JV, Aderem A. Caspase-1-Induced Pyroptotic Cell Death. Immunol Rev (2011) 243:206–14. doi: 10.1111/j.1600-065X.2011.01044.x
50. Miwa K, Asano M, Horai R, Iwakura Y, Nagata S, Suda T. Caspase 1-Independent IL-1beta Release and Inflammation Induced by the Apoptosis Inducer Fas Ligand. Nat Med (1998) 4:1287–92. doi: 10.1038/3276
51. Rath PC, Aggarwal BB. TNF-Induced Signaling in Apoptosis. J Clin Immunol (1999) 19:350–64. doi: 10.1023/a:1020546615229
52. Wallach D, Varfolomeev EE, Malinin NL, Goltsev YV, Kovalenko AV, Boldin MP. Tumor Necrosis Factor Receptor and Fas Signaling Mechanisms. Annu Rev Immunol (1999) 17:331–67. doi: 10.1146/annurev.immunol.17.1.331
53. Wu W, Lv L, Shi D, Ye J, Fang D, Guo F, et al. Protective Effect of Akkermansia Muciniphila Against Immune-Mediated Liver Injury in a Mouse Model. Front Microbiol (2017) 8:1804. doi: 10.3389/fmicb.2017.01804
54. Plovier H, Everard A, Druart C, Depommier C, van Hul M, Geurts L, et al. A Purified Membrane Protein From Akkermansia Muciniphila or the Pasteurized Bacterium Improves Metabolism in Obese and Diabetic Mice. Nat Med (2017) 23:107–13. doi: 10.1038/nm.4236
55. Wang L, Tang L, Feng Y, Zhao S, Han M, Zhang C, et al. A Purified Membrane Protein From Akkermansia Muciniphila or the Pasteurised Bacterium Blunts Colitis Associated Tumourigenesis by Modulation of CD8+ T Cells in Mice. Gut (2020) 69:1988–97. doi: 10.1136/gutjnl-2019-320105
56. Depommier C, Everard A, Druart C, Plovier H, van Hul M, Vieira-Silva S, et al. Supplementation With Akkermansia Muciniphila in Overweight and Obese Human Volunteers: A Proof-of-Concept Exploratory Study. Nat Med (2019) 25:1096–103. doi: 10.1038/s41591-019-0495-2
57. Ferrari D, Pinton P, Szabadkai G, Chami M, Campanella M, Pozzan T, et al. Endoplasmic Reticulum, Bcl-2 and Ca2+ Handling in Apoptosis. Cell Calcium (2002) 32:413–20. doi: 10.1016/s0143416002002014
58. Giorgi C, Bonora M, Sorrentino G, Missiroli S, Poletti F, Suski JM, et al. P53 at the Endoplasmic Reticulum Regulates Apoptosis in a Ca2+-Dependent Manner. Proc Natl Acad Sci USA (2015) 112:1779–84. doi: 10.1073/pnas.1410723112
59. Rasola A, Bernardi P. Mitochondrial Permeability Transition in Ca(2+)-Dependent Apoptosis and Necrosis. Cell Calcium (2011) 50:222–33. doi: 10.1016/j.ceca.2011.04.007
60. Liu X, Wang N, Zhu Y, Yang Y, Chen X, Fan S, et al. Inhibition of Extracellular Calcium Influx Results in Enhanced IL-12 Production in LPS-Treated Murine Macrophages by Downregulation of the Camkkβ-AMPK-SIRT1 Signaling Pathway. Mediators Inflamm (2016) 2016:6152713. doi: 10.1155/2016/6152713
61. Ables ET, Drummond-Barbosa D. Cyclin E Controls Drosophila Female Germline Stem Cell Maintenance Independently of Its Role in Proliferation by Modulating Responsiveness to Niche Signals. Development (2013) 140:530–40. doi: 10.1242/dev.088583
62. Chesnokov IN. Multiple Functions of the Origin Recognition Complex. Int Rev Cytol (2007) 256:69–109. doi: 10.1016/S0074-7696(07)56003-1
63. Choe KN, Moldovan G-L. Forging Ahead Through Darkness: PCNA, Still the Principal Conductor at the Replication Fork. Mol Cell (2017) 65:380–92. doi: 10.1016/j.molcel.2016.12.020
64. Harkness TAA. Activating the Anaphase Promoting Complex to Enhance Genomic Stability and Prolong Lifespan. Int J Mol Sci (2018) 19:1888. doi: 10.3390/ijms19071888
65. Huygelen V, de Vos M, Willemen S, Fransen E, Casteleyn C, van Cruchten S, et al. Age-Related Differences in Mucosal Barrier Function and Morphology of the Small Intestine in Low and Normal Birth Weight Piglets. J Anim Sci (2014) 92:3398–406. doi: 10.2527/jas.2014-7742
66. Izumi M, Mizuno T, Yanagi K-I, Sugimura K, Okumura K, Imamoto N, et al. The Mcm2-7-Interacting Domain of Human Mini-Chromosome Maintenance 10 (Mcm10) Protein Is Important for Stable Chromatin Association and Origin Firing. J Biol Chem (2017) 292:13008–21. doi: 10.1074/jbc.M117.779371
67. Parker DJ, Iyer A, Shah S, Moran A, Hjelmeland AB, Basu MK, et al. A New Mitochondrial Pool of Cyclin E, Regulated by Drp1, Is Linked to Cell-Density-Dependent Cell Proliferation. J Cell Sci (2015) 128:4171–82. doi: 10.1242/jcs.172429
68. Sivakumar S, Gorbsky GJ. Spatiotemporal Regulation of the Anaphase-Promoting Complex in Mitosis. Nat Rev Mol Cell Biol (2015) 16:82–94. doi: 10.1038/nrm3934
69. Slaymaker IM, Fu Y, Toso DB, Ranatunga N, Brewster A, Forsburg SL, et al. Mini-Chromosome Maintenance Complexes Form a Filament to Remodel DNA Structure and Topology. Nucleic Acids Res (2013) 41:3446–56. doi: 10.1093/nar/gkt022
70. Wang S-C. PCNA: A Silent Housekeeper or a Potential Therapeutic Target? Trends Pharmacol Sci (2014) 35:178–86. doi: 10.1016/j.tips.2014.02.004
71. Evans-Anderson HJ, Alfieri CM, Yutzey KE. Regulation of Cardiomyocyte Proliferation and Myocardial Growth During Development by FOXO Transcription Factors. Circ Res (2008) 102:686–94. doi: 10.1161/CIRCRESAHA.107.163428
72. Roskoski R. Cyclin-Dependent Protein Kinase Inhibitors Including Palbociclib as Anticancer Drugs. Pharmacol Res (2016) 107:249–75. doi: 10.1016/j.phrs.2016.03.012
73. Stampone E, Caldarelli I, Zullo A, Bencivenga D, Mancini FP, Della Ragione F, et al. Genetic and Epigenetic Control of CDKN1C Expression: Importance in Cell Commitment and Differentiation, Tissue Homeostasis and Human Diseases. Int J Mol Sci (2018) 19:1055. doi: 10.3390/ijms19041055
74. Huang XL, Xu J, Zhang XH, Qiu BY, Peng L, Zhang M, et al. PI3K/Akt Signaling Pathway Is Involved in the Pathogenesis of Ulcerative Colitis. Inflamm Res (2011) 60:727–34. doi: 10.1007/s00011-011-0325-6
75. Lee G, Goretsky T, Managlia E, Dirisina R, Singh AP, Brown JB, et al. Phosphoinositide 3-Kinase Signaling Mediates Beta-Catenin Activation in Intestinal Epithelial Stem and Progenitor Cells in Colitis. Gastroenterology (2010) 139:869–81, 881.e1–9. doi: 10.1053/j.gastro.2010.05.037
76. Dai C, Zheng CQ, Meng FJ, Zhou Z, Sang LX, Jiang M. VSL3 Probiotics Exerts the Anti-Inflammatory Activity via PI3k/Akt and NF-κb Pathway in Rat Model of DSS-Induced Colitis. Mol Cell Biochem (2013) 374:1–11. doi: 10.1007/s11010-012-1488-3
77. Schwanke RC, Marcon R, Meotti FC, Bento AF, Dutra RC, Pizzollatti MG, et al. Oral Administration of the Flavonoid Myricitrin Prevents Dextran Sulfate Sodium-Induced Experimental Colitis in Mice Through Modulation of PI3K/Akt Signaling Pathway. Mol Nutr Food Res (2013) 57:1938–49. doi: 10.1002/mnfr.201300134
Keywords: Akkermansia muciniphila, inflammation, intestinal epithelial cells, TNF-α, apoptosis
Citation: Luo Y, Lan C, Xie K, Li H, Devillard E, He J, Liu L, Cai J, Tian G, Wu A, Ren Z, Chen D, Yu B, Huang Z, Zheng P, Mao X, Yu J, Luo J, Yan H, Wang Q, Wang H and Tang J (2021) Active or Autoclaved Akkermansia muciniphila Relieves TNF-α-Induced Inflammation in Intestinal Epithelial Cells Through Distinct Pathways. Front. Immunol. 12:788638. doi: 10.3389/fimmu.2021.788638
Received: 03 October 2021; Accepted: 30 November 2021;
Published: 16 December 2021.
Edited by:
Xia Xiong, Institute of Subtropical Agriculture (CAS), ChinaReviewed by:
Xiangfeng Kong, Institute of Subtropical Agriculture (CAS), ChinaCopyright © 2021 Luo, Lan, Xie, Li, Devillard, He, Liu, Cai, Tian, Wu, Ren, Chen, Yu, Huang, Zheng, Mao, Yu, Luo, Yan, Wang, Wang and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuheng Luo, bHVvbHVvMjEyQDEyNi5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.