- 1Vlaams Instituut voor Biotechnologie (VIB)-UGent Center for Medical Biotechnology, VIB, Ghent, Belgium
- 2Department of Biochemistry and Microbiology, Ghent University, Ghent, Belgium
- 3Section Virology, Division Infectious Diseases & Immunology, Department of Biomolecular Health Sciences, Utrecht University, Utrecht, Netherlands
Neuraminidase of influenza A and B viruses plays a critical role in the virus life cycle and is an important target of the host immune system. Here, we highlight the current understanding of influenza neuraminidase structure, function, antigenicity, immunogenicity, and immune protective potential. Neuraminidase inhibiting antibodies have been recognized as correlates of protection against disease caused by natural or experimental influenza A virus infection in humans. In the past years, we have witnessed an increasing interest in the use of influenza neuraminidase to improve the protective potential of currently used influenza vaccines. A number of well-characterized influenza neuraminidase-specific monoclonal antibodies have been described recently, most of which can protect in experimental challenge models by inhibiting the neuraminidase activity or by Fc receptor-dependent mechanisms. The relative instability of the neuraminidase poses a challenge for protein-based antigen design. We critically review the different solutions that have been proposed to solve this problem, ranging from the inclusion of stabilizing heterologous tetramerizing zippers to the introduction of inter-protomer stabilizing mutations. Computationally engineered neuraminidase antigens have been generated that offer broad, within subtype protection in animal challenge models. We also provide an overview of modern vaccine technology platforms that are compatible with the induction of robust neuraminidase-specific immune responses. In the near future, we will likely see the implementation of influenza vaccines that confront the influenza virus with a double punch: targeting both the hemagglutinin and the neuraminidase.
1 Introduction
Influenza A and B viruses (IAV and IBV) cause acute respiratory illness and are widespread in the human population, with seasonal appearance in moderate climate zones and year-round manifestation in the tropics (1, 2). The use of licensed influenza vaccines is considered one of the best measures to prevent human influenza. These vaccines are vital in the efforts to alleviate the burden of influenza illness and deaths and are especially recommended for individuals who have an increased risk of developing complications due to age or underlying disease (3, 4). The effectiveness of currently licensed influenza vaccines however leaves considerable room for improvement. Depending on the IAV subtype and the antigenic match between the influenza strains that are represented in the vaccine and the strains that circulate in the population, the vaccines prevent 10 to 60% of laboratory-confirmed medically attended influenza (5). The composition of seasonal influenza vaccines is reconsidered every year for each hemisphere in an attempt to keep pace with the antigenic drift of the viral hemagglutinin (HA), the major envelope protein on the influenza virions and the principal protective antigen in currently used influenza vaccines. These annual updates come with a risk of suboptimal predictions leading to a mismatch between the vaccine- and circulating influenza virus strains. There is a pressing need for more effective influenza vaccines that can elicit stronger and potentially broader protection against influenza. In the past decade, there has been a renewed interest in the exploration of influenza neuraminidase (NA) as a protective antigen component in influenza vaccines. Here, we review some of the seminal findings on NA structure and function, its immune-protective potential, as well as the current efforts to implement NA in next-generation influenza vaccines that aim for eliciting an immune response with increased magnitude and breadth.
2 Neuraminidase: Structure and Function
2.1 NA Structure
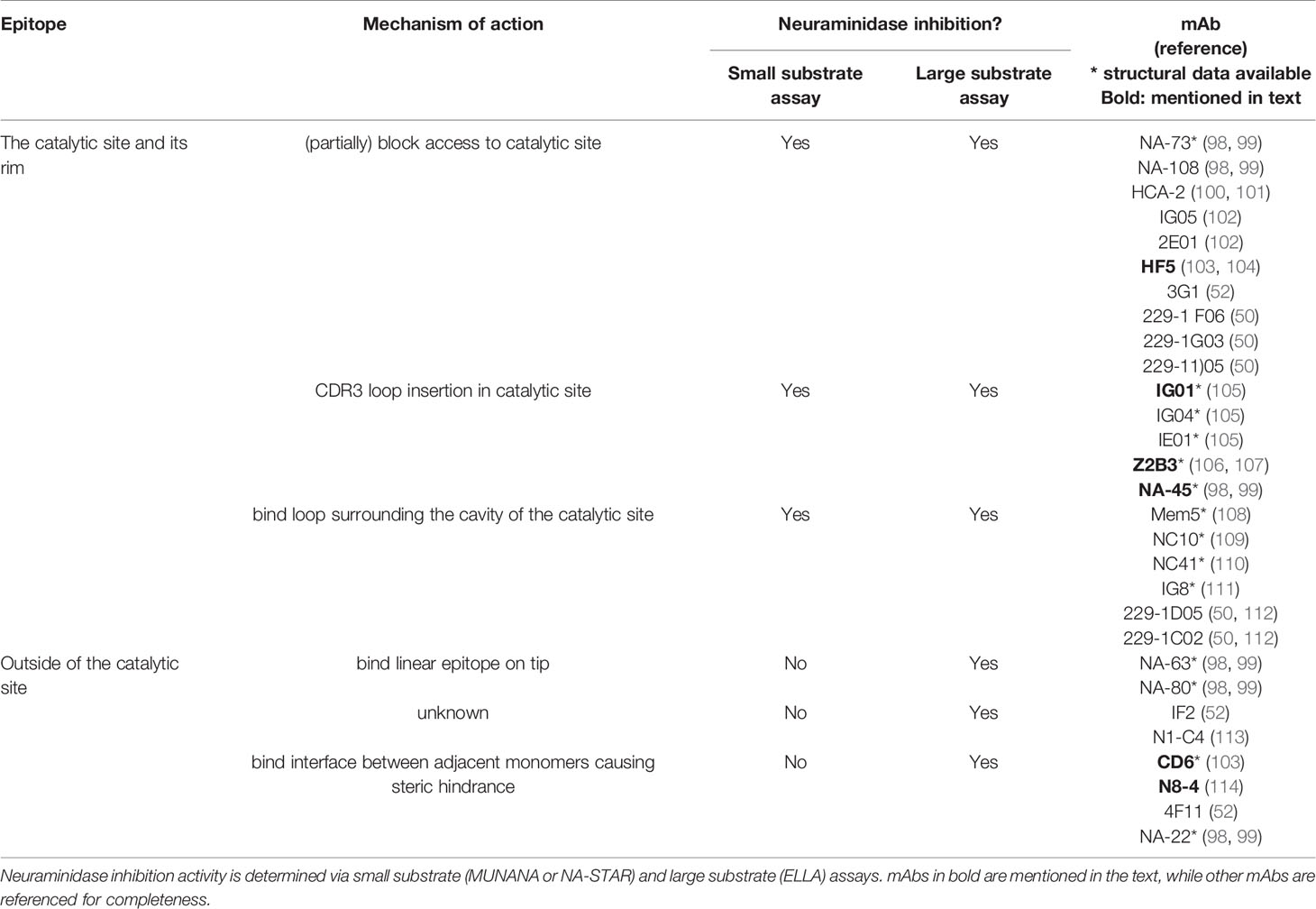
NA is one of the three membrane proteins expressed on IAV and IBV particles, next to HA and matrix protein 2 (M2). Label-free protein quantification of purified influenza A and B virions revealed that the NA : HA ratio ranges from 0.1 to 0.2 (6). NA is a homotetrameric type II membrane protein with a mushroom-like shape. Each protomer comprises approximately 470 amino acid residues and consists of a cytoplasmic tail, a transmembrane domain (TMD), a stalk and a head domain (Figure 1).
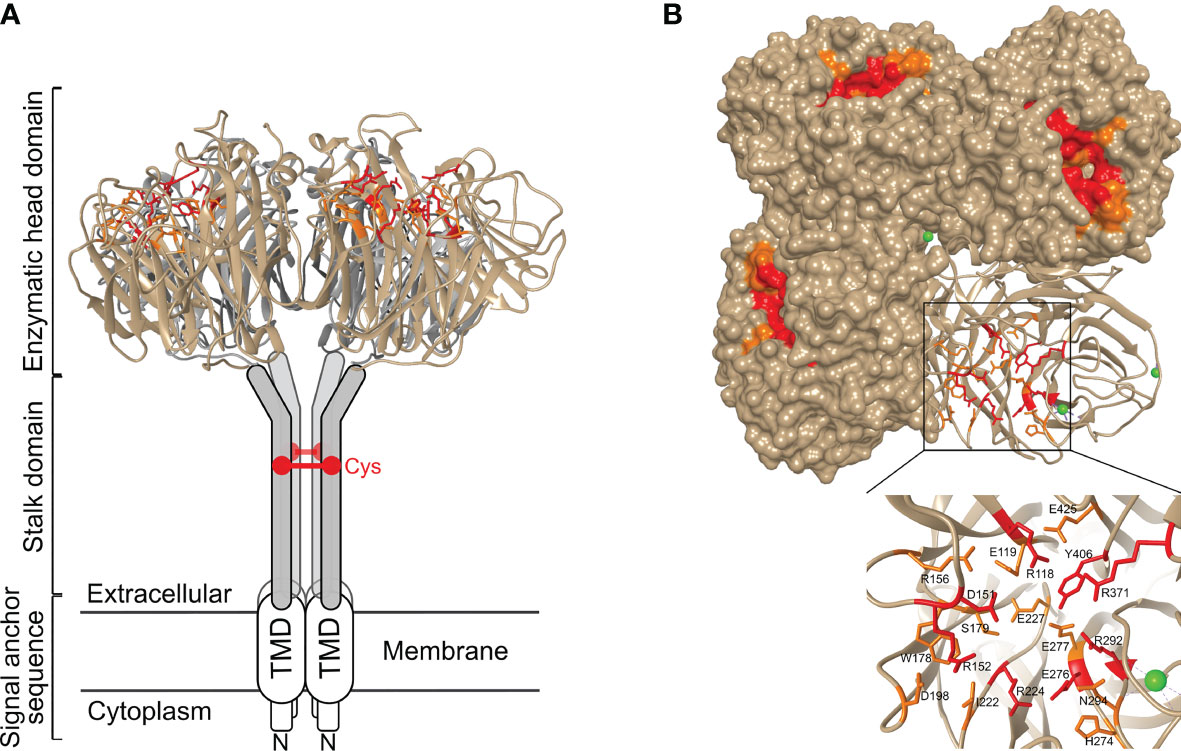
Figure 1 Structure of neuraminidase and its catalytic site. (A) Side view and (B) top view of N1 NA (PDB 6Q23). NA is a homotetrameric type II membrane protein consisting of a head domain, stalk domain, and a transmembrane domain (TMD) and cytoplasmic tail that together form the signal anchor sequence. In general, NA stalk domains contain a cysteine residue (Cys) involved in intermolecular disulphide bond formation. The inset in panel (B) shows the catalytic site with the residues that interact with the sialic acid-containing substrate depicted in red, and the residues that stabilize the catalytic site labelled in orange. Ca2+ ions are shown as green spheres.
The cytoplasmic tail of NA consists of 7 highly conserved amino acid residues. Alanine-scanning mutagenesis of the cytoplasmic tail of NA of A/WSN/33 virus indicated a role in virus budding. Notably bud release rather than bud formation was affected in the tested NA cytoplasmic tail mutants (7). The cytoplasmic tail is also important for the association of NA with lipid rafts. Lipid rafts on the apical site of polarized cells are sites where newly formed influenza virions initiate assembly and budding (8). Together with the cytoplasmic tail, the TMD plays a role in the transport of newly expressed NA to the apical plasma membrane (9). The sequence of the TMD is moderately conserved across IAV subtypes and predicted to form an alpha helix. The TMD ensures the membrane anchoring of NA and serves as a translocation signal. In addition, this domain is reported to be an important stabilizing factor for the tetrameric NA formation (10, 11). Substitution of hydrophilic residues in the TMD by alanine reduced or even abolished the interaction between the four TMDs of the NA tetramer (10).
The stalk domain connects the TMD with the catalytic head domain. There is no crystal structure available of the stalk domain. The NA stalk varies in length within and across NA subtypes, carries multiple predicted N-glycosylation sites and, with few exceptions, contains at least one cysteine residue that can form an intermolecular disulphide bond with a neighbouring NA molecule (12). Glycosylation of the stalk region may contribute to NA stability, whereas inter-stalk disulphide bond formation is important for the tetramer formation. Whereas the length of the NA stalk of human influenza viruses seems to be relatively constant, the NA stalk of avian NA subtypes, in particular N1, N2, N3, N5, N6, and N7, tolerate deletions (13). The stalk length affects NA enzymatic activity, presumably by modulating the accessibility to the sialic acid-containing substrates (14).
Crystal structures of the catalytic head domain of at least one representative NA from N1 to N9 and from influenza B NA have been resolved (15–22). The NA head domain is characterized by a six-bladed propeller that is folded around the catalytic site and which is typical for all known sialidases (23). Each blade is made up of four antiparallel β-sheets that are stabilized by disulphide bonds and connected by loops of variable length (12). Each monomer harbours a catalytic site, oriented towards the lateral side of the NA head, that is highly conserved over the different IAV subtypes (Figure 1). Remarkably, unlike the tetramer, influenza NA monomers and dimers show very little if any enzymatic activity (24). It is not known why tetramer formation is essential for NA to be active. Possibly, this is linked to calcium binding by tetrameric NA, which contributes to NA activity and stability. NA can bind up to nine Ca2+ ions in the case of the 2009 H1N1 pandemic (H1N1pdm09) virus derived NA (16, 25–28).
Despite the significant primary sequence variation between IAV subtype NAs, the catalytic site residues in N1–N9 NAs are highly conserved. Among these, residues R118, D151, R152, R224, E276, R292, R371, and Y406 (N2 numbering) directly contact the substrate, while residues E119, R156, W178, S179, D198, I222, E227, H274, E277, N294, and E425 play a key role in stabilizing the catalytic site residues (19) (Figure 1). Phylogenetic analysis indicates that IAV NAs fall into two distinct groups. In group 1 NAs, a cavity adjacent to the catalytic site is observed which is absent in group 2 NAs (29). This cavity is created by the so-called 150-loop that consists of residue 147-152 and is flexible in group 1 NAs such that it can adopt an open and a closed conformation. Group 2 NAs on the other hand lack this second cavity as the formation of a salt bridge between D147 and H150 stabilizes the 150-loop, which is absent in group 1 NAs due to the presence of a G147 there (29, 30). The catalytic site and its adjacent 150-cavity are further explored as targets for NA inhibitors (31). Additionally, most avian influenza NAs, but not NAs of human viruses, have a functional second sialic acid binding site (2SBS) or hemadsorption site next to the catalytic site (32). The 2SBS consists of three loops with residues that facilitate binding to sialic acid in its so-called chair conformation. The catalytic site, in contrast, binds sialic acid in its twisted boat conformation (16). Studies comparing human and avian NA catalytic properties show that the presence of a functional 2SBS in avian NA increases NA activity against multivalent substrates (33, 34).
2.2 NA Function
HA and NA exert different functions in the influenza virus life cycle. HA is vital in the entry process, by mediating binding to sialic acids on host cell glycoproteins or -lipids, which results in virion uptake into endocytic vesicles, and the subsequent fusion of the host cell and virus membrane through a pH-induced conformational change (35). NA, on the other hand, catalyses the removal of the terminal sialic acids and thus functions as a receptor-destroying enzyme. NA activity is involved in multiple steps of the virus life cycle (Figure 2). During viral entry NA cleaves decoy receptors present in the mucus that lines the epithelial cells of the respiratory tract, allowing the infection of underlying epithelial cells (36–38). In line herewith, inhibition of NA activity was shown to result in severely decreased infection of differentiated primary human airway epithelium cells (39). NA activity was also reported to stimulate HA-mediated membrane fusion (40). The best-known function of NA in the influenza virus replication cycle is its critical role in the release of newly formed virions from the infected cell and in prevention of HA-mediated virion aggregation by removing sialic acid from the viral and host cell membrane (41).
Figure 2 Important role for NA in the virus life cycle. NA contributes to virus motility, allowing the virus to move through the mucus layer and to reach functional receptors at the cell surface. NA also plays an essential role at the end of the virus life cycle by removing sialic acids from the cell surface thereby allowing efficient release of virions and preventing virion aggregation.
Given these partly opposing functions, a functional balance between HA and NA is critical for viral replication. If HA binding is much stronger than NA activity, for example, virions may be trapped by decoy receptors present in mucus and eventually be removed by mucociliary clearance before reaching the underlying epithelial cells. On the contrary, if NA activity dominates, the virion will quickly move through the mucus, but will more likely fail to stably bind to an epithelial cell and enter it. HA and NA can be found on the virion surface in a patch-wise distribution, which contributes together with the HA-NA balance to virus motility and entry. In filamentous influenza virions, NA is mainly localized at one pole of the filamentous virus, and the few NA molecules present along the side of the virus tend to cluster in patches as well (42). Such polarized viruses seem to move by Brownian ratchet-like diffusion in a mucus-rich extracellular environment, in which filamentous particles exhibit directed mobility away from their NA-rich pole and toward the HA crowded part of the virus (42). HA binding to sialic acid is reversible due to the low affinity. This reversible binding likely allows NA patches to catalyse the removal of the sialic acid when the HA is no longer bound thereby initiating movement of the virion through mucus and over the cell surface. This NA-driven virion motility presumably allows the virus to find and to dock to specific spots on the cell surface that trigger entry into the host cell (38, 43, 44). Although detailed mechanistic insights into the importance of the HA-NA balance are still lacking, it is clear that besides HA also NA contributes to IAV pathogenicity. For example, truncation of the NA stalk, which has been associated with adaptation of IAV from aquatic birds to poultry, resulted in increased virulence in poultry and mice (13, 45–49).
3 Antibodies Directed Against NA as a Correlate of Protection
Protection against disease caused by influenza virus infection has traditionally been correlated with the presence of hemagglutination-inhibiting (HAI) antibodies in the blood. Such antibodies can neutralize influenza viruses in vitro, by preventing the binding of the virus to its sialic acid-containing receptor on the target cell. Therefore, in order to induce such antibodies, current influenza vaccines are standardized for their HA content. Natural infection, however, elicits an immune response against both HA and NA (50). In mice it has been shown that NA inhibiting (NAI) antibodies are associated with a reduction in pulmonary virus titer. More recently, by using a guinea pig model, monoclonal NAI antibodies were demonstrated to reduce airborne transmission of human IAVs, both when the antibodies were administered post infection to the infected animals or to the exposed recipients (51). This observation is in line with the contribution of NA to the release and spread of newly formed viruses after infection (50, 52, 53). Further, a combination of HA and NA provides even enhanced protection against influenza compared to HA alone (54–57). Studies comparing protection in mice induced by conventional inactivated influenza vaccines with or without supplementation with recombinant NA showed that for protection against homosubtypic influenza virus infection anti-HA antibodies sufficed. However, when challenged with an influenza virus with a mismatched HA supplementation of the vaccine with NA was required to reach a clear reduction in pulmonary virus titer (58, 59). In a ferret study it was found that whereas vaccination with HA reduced viral titers, vaccination with NA particularly decreased the clinical effects of infection, with optimal protection being achieved by a combination of the two antigens (54).
Evidence of NA-based protection in human has also been observed during the 1968 Hong-Kong pandemic. This pandemic was caused by a H3N2 virus with the same N2 as the previously H2N2 circulating virus. Individuals with pre-existing antibodies against the NA of H2N2 were less likely to be infected with the newly emerged virus (60–63). In humans, NA-inhibiting antibodies in serum correlate with a reduced virus load in nasal wash (62). Couch et al. first showed that serum N2-specific antibodies, elicited by vaccination with H1N2 (H1 HA from A/equine/Prague, N2 NA from A/Aichi/2/68 (H3N2)) in humans who initially lacked anti-HA antibodies, correlate with a reduction of viral shedding after challenge of the subjects with A/Aichi/2/68 (H3N2) virus (62). Three decades later, during the 2009 H1N1 pandemic season, it was demonstrated that HAI and NAI antibodies in serum independently correlated with immunity against infection and infection-associated illness. Moreover, in H1N1pdm09-infected individuals, NAI antibodies in serum independently predicted reduced illness (64). This correlation of NAI antibodies with protection is also supported by more recent studies in humans. The presence of NAI antibodies in serum was associated with a reduction of PCR-confirmed influenza infection for both H3N2 and H1N1pdm09 virus (65, 66). Memoli et al. divided the participants in their controlled human challenge study with H1N1pdm09 virus in a group based on HAI or NAI titer and showed that subjects with a high (> 1:40) NAI titer at baseline not only presented with reduced symptoms and virus shedding duration, but also with a reduction in the number of symptoms and the symptom severity. In contrast, HAI titers correlated only with a reduction in the number of symptoms and virus shedding duration but not with symptom severity (66). These results highlight that anti-NA immunity can enhance protection against influenza virus infection.
4 Evolution and Antigenic Drift of NA
HA and NA are prime targets of the host’s immune response to influenza virus infection (67). Combined with the relatively high mutation rate of the replicating influenza virus RNA genome, human influenza viruses with antigenically altered HA and NA emerge that have a selective advantage over previously circulating strains because they are less likely to be recognized by antibodies that prevail in the population. This phenomenon of antigenic changes is called antigenic drift, and is at the basis for the frequent updating of human influenza vaccines. The selection of seasonal vaccine strains is based on three types of data: epidemiological information, HA and NA gene sequence phylogeny and serological analysis using an HA inhibition assay. Today, the main focus of genetic and antigenic surveillance is thus on HA since licensed influenza vaccine formulations are standardized for the amount of HA (68). The antigenic drift of HA has been extensively studied (69). In a seminal paper, Smith et al., discerned a pattern of eleven antigenic HA clusters in human H3N2 viruses that circulated between 1968 and 2003 (70). Later, two clusters were added to the antigenic map when HAI data was added up to 2011 showing that the human H3N2 virus continued to evolve antigenically (71). Interestingly, the antigenic change between the HA clusters is the result of a limited set of amino acid changes confined to positions near the receptor binding site of HA (72, 73). In contrast to HA, antigenic drift of NA is not routinely examined. In the next paragraphs, we discuss the genetic evolution of NA and the limited studies on NA antigenic drift.
4.1 Genetic Evolution
Several studies addressed the genetic evolution of HA and NA using IAV and IBV strains, which revealed that their evolution differs and is often asynchronous (74–78). Although the general topology of the NA and HA phylogenetic trees is similar with the typical ‘ladder-like’ gradual evolution and rapid replacement of old strains by newer ones, NA evolved slower and more gradually at the nucleotide level than HA. Looking at 40 years of evolution of human H3N2 viruses, Westgeest et al. showed that NA had fewer nucleotide substitutions over this time span compared with the HA head HA1, the most variable part of the HA gene (78). Nevertheless, it was concluded by Bhatt et al. (74) that with almost all adaptive evolution in NA being concentrated in residues on the surface of NA, the adaptation rate is higher for surface NA residues than for HA1. The observation that adaptive evolution in NA occurs almost exclusively in solvent-accessible surface residues indicates an important role for antibody-mediated immune responses in NA evolution. In Figure 3, the IAV N1, IAV N2 and IBV NA structural conservation for the viruses that were used in seasonal influenza vaccines from 1970 to 2021 are depicted. Overall, a relatively high conservation of the head domain residues is observed for each NA (sub)type. As expected, the catalytic site is for all NA subtypes highly conserved. However, the surface residues do show variation, especially for IAV N2, which is in agreement with a higher rate of adaptation for NA in H3N2 than H1N1 viruses (74).
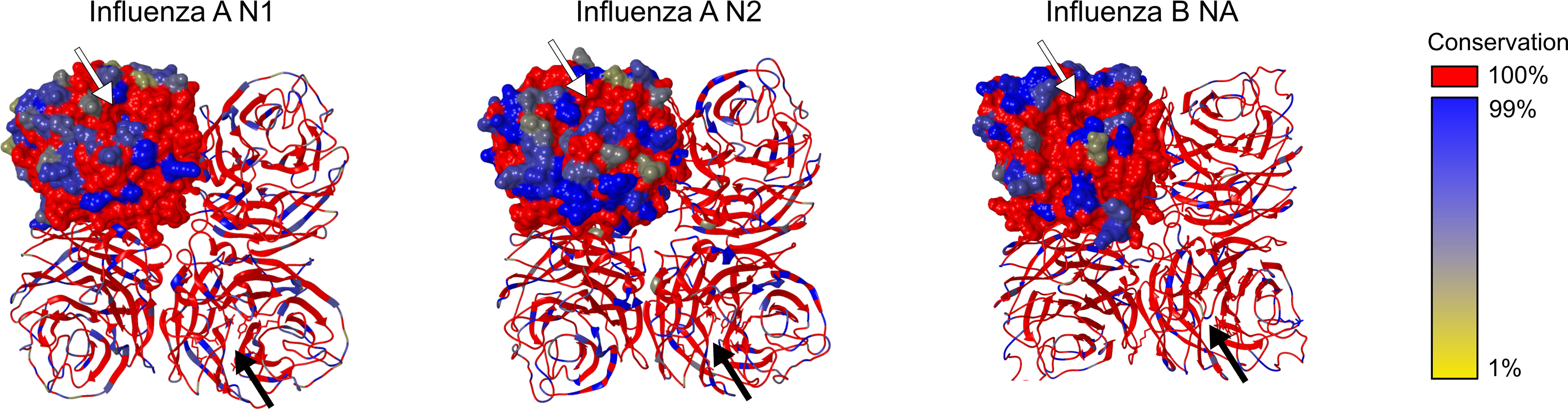
Figure 3 NA structural conservation. Structural alignments of all seasonal influenza vaccine included influenza strains in the period 1970-2021.The sequence conservation of Influenza A N1, Influenza A N2 and Influenza B NA amino acid residues was visualized using MUSCLE and shown on a crystal structure of the respective NA subtype using the render by conservation function in CHIMERA. Residues conserved in all sequences of a specific subtype NA are shown in red. Further distinction of conservation is indicated from dark blue (99% conservation) till yellow (1% conservation). Structure representations based on N1: PDB 4B7Q, N2: PDB 4GZX, and IBV NA: PDB 4CPL. Arrows point towards the catalytic site.
4.2 Antigenic Drift
The analysis of NA antigenic drift usually relies on the determination of NAI titers of polyclonal ferret antisera raised against a particular influenza virus strain (e.g. a vaccine strain) in an enzyme-linked lectin assay (ELLA) (79, 80). The antigenic relatedness of the NAs from two viruses can be assessed by comparing NAI titers, calculating percent relatedness (81) or by performing antigenic cartography (76) to generate an antigenic map (80). On such a map, based on the quantification of the raw data of NAI assays, the antigenic distances between influenza viruses or sera are visualized. Importantly, the reactivity of antisera of ferrets that have experienced a single experimental virus infection is quite different from human sera which have an antibody repertoire that has typically been shaped by multiple prior infections and vaccinations (80, 82). The antigenic changes identified this way based on sera from ferret that were infected once with a particular influenza virus strain may therefore not always accurately reflect the response of an (adult) human individual (83) although such discrepancies have so far only been demonstrated for HA-specific responses (84–86).
Only a few studies have investigated the antigenic drift of NA using large NAI data sets and by constructing antigenic maps. In 2011, Sandbulte et al. reported on the characterization of the antigenic drift of NA in human H1N1 and H3N2 viruses that were recommended for influenza vaccine implementation over a period of 15 years in the USA (76). Similarly, Gao et al. examined the antigenic drift of NA sequences of H1N1pdm09 viruses (80). Overall, the antigenic differences between NAs of human H1N1 viruses occurred in one direction, meaning that antisera raised against older strains reacted weakly with more recent NAs whereas reactivity between antisera against more recent strains and older NAs was high (76, 80, 87). Both raw NAI data and the antigenic map obtained by Sandbulte et al. show that the NA of pre-2009 seasonal human H1N1 was in antigenic stasis for over a decade despite its genetic evolution during that time. For human H3N2 viruses the antigenic drift of NA was also not proportional to the number of amino acid changes induced. Collectively, these data show that there is discordance between the antigenic and genetic evolution of NA. Indeed, For N2 it was found that a single point mutation (E329K) was responsible for the abrupt NA antigenic drift between 2006 and 2007 (76). Similarly, for H1N1pdm09 few substitutions appeared to be largely responsible for the observed antigenic changes (80). The presumed important role in antigenic drift of K432E (80) was later confirmed using recombinant NA proteins that only differed at this position (87). Epistatic interactions as well as by biophysical constraints may also play a role in NA antigenic drift as was shown for the evolution of an antigenic site in H3N2 NA (88, 89). Furthermore, changes in HA receptor binding, e.g. as a result of antigenic drift, may in turn select for substitutions in NA that affect enzymatic activity to restore a functional balance in HA and NA, while additionally affecting NA antigenicity (90).
Bidirectional IAV transmission between humans and swine represents a serious public health challenge. In the last decade, an increasing number of zoonotic infections with IAV from swine has been reported. For example, after two distinct human-to-swine H3N2 spill overs in 1998 and 2002, N2-98 and N2-02 lineage viruses have circulated in swine in the USA and antigenically evolved over the next 20 years. After IAV infection, pigs typically produce broadly cross-reactive NAI antibodies to the N2 protein, but no NAI cross-reactivity between the N2-98 and N2-02 lineages was observed (91). The antigenic distance between swine and human N2 antigens increased over time to the extent that there is by now very little antigenic similarity with the human seasonal H3N2 NA that these viruses were derived from, nor with the NA of currently circulating human H3N2 viruses. This suggest that there will be little to no cross-reactive NA mediated immunity in both the swine and human population which may impact the occurrence of future spill overs (91).
5 Neuraminidase-Specific Monoclonal Antibodies: Mechanisms of Action and Epitopes
Using large panels of monoclonal antibodies (mAbs) or recombinant NA proteins, several studies have set out to identify antigenic sites on NA (12, 52, 92–96). NA activity and inhibition thereof can be evaluated with two types of assays. The so-called 4-methylumbelli-feryl N-acetyl-α-D-neuraminic acid (MUNANA, a fluorogenic NA substrate) or NA-STAR (a chemiluminescent NA substrate) assays allow to quantify the hydrolysis of a small soluble substrate by NA. The small molecule NA inhibitor oseltamivir, which binds precisely inside the catalytic site, inhibits NA activity in these types of assays (97). Only a mAb that binds inside or in very close proximity to the catalytic site will be able to prevent cleavage of the small substrate whereas mAbs that bind distal to the catalytic site are less likely to have an effect in such assays.
ELLA assays use larger substrates compared to MUNANA and NA-STAR assays, such as the glycoprotein fetuin that contains sialylated N- and O-linked glycans. A mAb binding to the NA head domain, even outside of the catalytic site, may still sterically block access of fetuin to the catalytic site and prevent cleavage of the substrate, while cleavage of small soluble substrates may not be affected. Comparing the inhibition profile of NAI mAbs based on the outcome of both types of assays will thus give an indication on their possible binding site. Table 1 provides an overview of different NA-specific mAbs and their impact on NA activity as determined with a small molecule substrate or in the ELLA. Overall, two different antigenic regions have been described that characterize NAI antibodies: (i) the catalytic site and its rim and (ii) outside of the catalytic site, including the interface between two adjacent monomers. Additionally, several NA-specific mAbs have been described that lack detectable NAI activity in either assay, yet can protect in vivo against influenza A virus challenge. These mAbs rely on Fc effector functions and will be discussed separately (Figure 4).
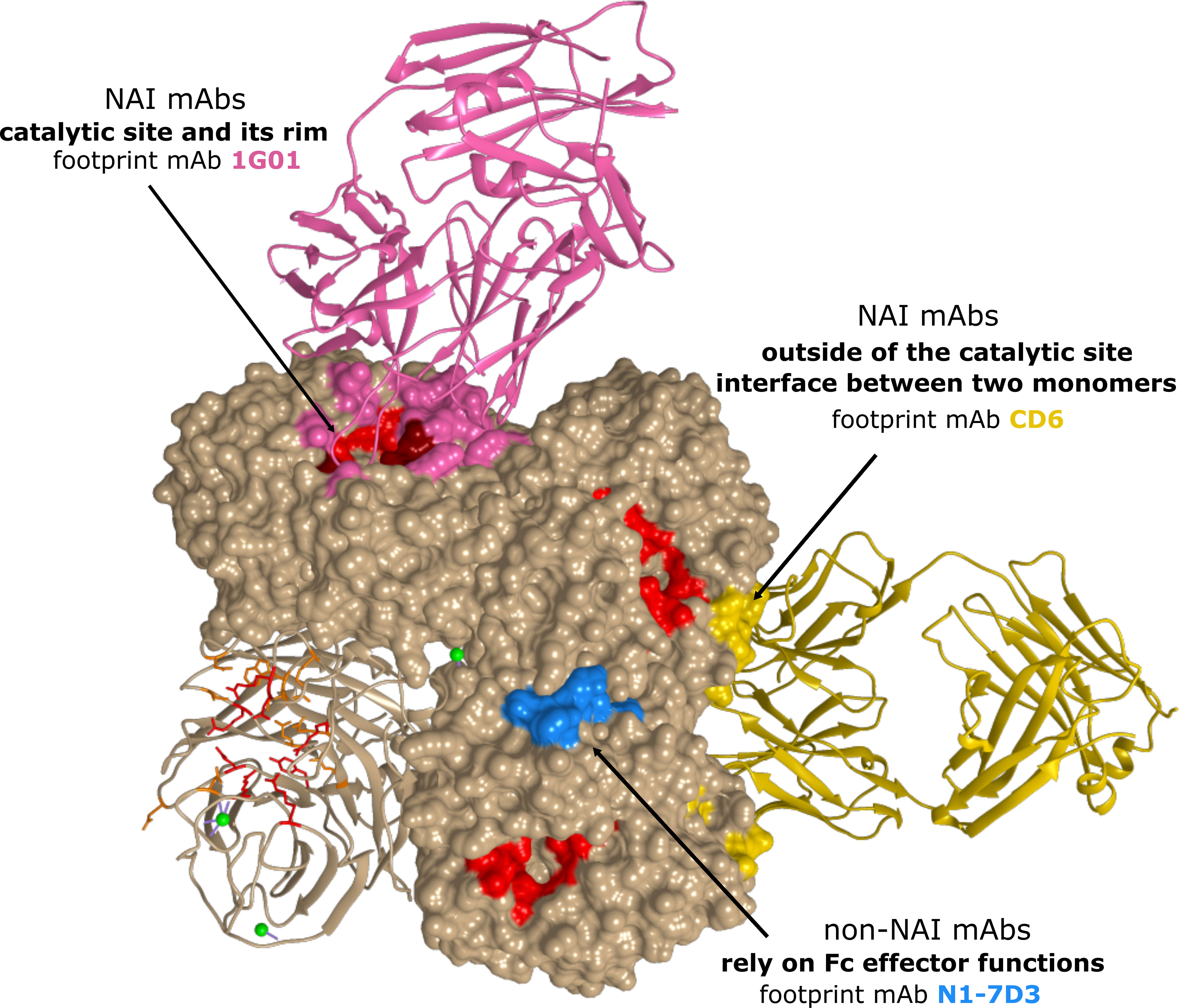
Figure 4 Antigenic regions of N1 NA-specific mAbs. The A/California/04/2009 (H1N1) tetramer is depicted with one protomer in ribbon representation. The catalytic residues are depicted in red. For each region, a representative antibody footprint is shown: 1G01 (105) in pink (PDB 6Q23) with antibody contact residues in NA that overlap with the catalytic site in dark red; CD6 (103) in gold (PDB 4QNP) and N1-7D3 (113) in light blue.
5.1 NAI mAbs That Target the Catalytic Site and Its Rim
Early antibody characterization and vaccination studies have shown that immunity to NA can be very broad within, but usually not across, subtypes (105). Nevertheless, mAbs that directly bind into the highly conserved catalytic site of NA (115) and inhibit enzymatic activity in small molecule assays are expected to exert a degree of heterosubtypic NAI. Recently, Stadlbauer et al. reported on a human mAb, 1G01, which binds to and inhibits the activity of several group 1 and group 2 NAs as well as NAs from both influenza B virus antigenic lineages in vitro. MAb 1G01 was shown to inhibit NA activity in a NA-Star assay and to occupy the catalytic site via a long CDR H3 loop by co-crystal structure analysis (Figure 4) (105). A similar mechanism of action was described for mAb NA-45 where the CDR H3 loop adopts a protruding conformation with a tip that inserts into the NA catalytic site (98). This type of substrate mimicry is unique among all structurally characterized NA antibodies so far but has also been reported for anti-HA antibodies that target the receptor-binding site (116, 117). Interestingly, the number of residues in the antibody footprint might be important for the cross-reactivity of mAbs. For example, the binding footprint of 1G01 (105) and Z2B3 (106, 107) significantly overlaps. However, the 1G01 footprint includes more catalytic and framework residues which explains the broader cross-reactivity of 1G01 compared to Z2B3 (107). It is important to note that the footprint of mAbs that bind into the highly conserved catalytic site of NA often overlaps with the rim of the catalytic site. The rim is less conserved and can tolerate amino acid substitution without NA losing enzymatic activity.
Next to conventional mAbs, other antibody moieties that target the NA catalytic site have been described. The first NA-specific single domain antibodies or VHHs were described by Harmsen et al. with several candidate cross-NA binders and some VHHs that could affect NA activity (118). The isolation and characterization of a set of alpaca-derived H5N1 NA-specific VHHs (N1-VHH) with NA-inhibitory activity was also described. Two monovalent candidates N1-3-VHHm and N1-5-VHHm could inhibit NA activity with N1-3-VHHm also inhibiting oseltamivir resistant H5N1 virus NA. Bivalency of these constructs enhanced their NA inhibitory capacities and resulted in VHH constructs that could protect mice against H5N1 challenge (119).
5.2 mAbs that Bind to Epitopes Outside the Catalytic Site
A large proportion of the reported NAI mAbs only display NAI activity in the ELLA, but not in assays with small molecule substrates, indicating they do not contact the catalytic site directly (99). Binding of these mAbs likely results in steric hindrance and restricts access of large glycoconjugate substrates to the catalytic site (52). The NAI activity in ELLA assays has been associated with the capacity to block virus egress from infected cells (98, 99). An example of such a mAb is CD6, which effectively inhibited H1N1pdm09 viruses both in vivo and in vitro (103). Interestingly, CD6 more efficiently inhibited NA as an intact IgG compared to Fab and Fab2 molecules of CD6, demonstrating that CD6 inhibits NA activity through steric hindrance and not structural distortion. Crystallization studies revealed a quaternary epitope that spans the lateral faces of two neighbouring N1 monomers (Figure 4). Such a quaternary epitope present in intact NA dimers had been proposed earlier by Saito et al. for the N8-4 mAb but until then had never been structurally defined (114). For HA, quaternary epitope-specific mAbs that span two HA monomers that are efficient in neutralizing virus in vitro have also been described (120, 121). Some studies showed a correlation between the relative distance of epitopes to the NA catalytic site and the in vitro properties of mAbs. For example, mAb HF5 which has contact residues that surround the catalytic site pocket, showed more efficient NAI activity compared to other mAbs with an epitope that is located more laterally on the NA head (92, 107). Additionally, murine anti-N9 mAbs that inhibit NA enzymatic activity with a small molecule substrate in vitro, provided superior in vivo protection compared with mAbs that inhibited NA activity only in the ELLA assay (93).
5.3 mAbs That Depend on Fc Effector Functions for Protection
It is becoming increasingly clear that Fc-mediated effector functions have an important role in providing in vivo protection against influenza virus challenge (122). The fragment crystallisable (Fc) region of an antibody can interact with Fc receptors and thereby mediate indirect effector functions such as antibody-dependent cellular cytotoxicity (ADCC), antibody-dependent cellular phagocytosis (ADCP) and complement-dependent cytotoxicity (CDC) (123). Using Fc receptor knockout mice and DA265 mutant mAbs that are unable to bind Fc receptors, it was demonstrated that broadly reactive HA-stalk antibodies depend on Fc-Fcγ receptor interactions in vivo (122, 124, 125). The contribution of Fcγ receptor engagement for in vivo protection by some NA-specific mAbs was demonstrated by Job et al. (126). The mouse monoclonal antibody N1-7D3 binds to a conserved linear epitope near the carboxy-terminus of N1 NA and shows no NAI activity (Figure 4) (113). However, a recombinant mouse-human chimeric version comprised of the variable domains of mouse N1-7D3 and the constant regions of human IgG1 could engage activating Fcγ receptors and protected FcγR-humanized mice against challenge with H1N1pdm09 virus (126). In addition, mAbs with no detectable NAI activity reported by Yasuhara et al., could protect mice against a lethal influenza virus infection. Interestingly, a N297Q mutant version of these mAbs that lacks Fcγ receptor-binding activity failed to protect, thereby supporting the crucial role for Fcγ effector functions in protection by non-NAI mAbs (127). In line with this, grafting of the non-NAI N1-7-VHH on a mouse IgG2a Fc could protect mice against an otherwise lethal H5N1 challenge (119).
Several studies have demonstrated that the Fc-mediated effector functions might also be crucial for in vivo protection by some anti-NA mAbs that possess weak NAI activity (99, 127). Thus, while for potent N9 NAI mAbs Fc effector functions were not needed, the ability to interact with Fc receptors was required for weakly NAI mAbs to protect against challenge (99). Likewise, a broadly cross-reactive mAb with low NAI activity required FcγR interactions to mediate protection, while a strain-specific mAbs with high NAI did not (124). Likely, the sum of the NAI activity and the FcγR-mediated effector functions determines the potency of an anti-NA antibody in vivo. An anti-NA mAb with high NAI activity can reduce virus spread in vivo mainly through its NAI activity, whereas an anti-NA mAb without NAI activity can suppress virus replication mainly through FcγR-mediated effector cell activation (127).
6 NA-Based Influenza Vaccines
6.1 NA Immunogenicity in Seasonal Influenza Vaccines
Despite the recognized importance of NA-reactive antibodies in protection against influenza, the current commercially available influenza vaccines do not consistently elicit these antibodies (50, 128). NA immunogenicity of these vaccines varies widely between manufacturers (65, 128–130) and is poor compared to natural infection. Whereas following natural infection the number of NA-reactive B cells were equal to or exceeded HA-reactive B cells, current vaccines rarely induced NA-reactive B cells (50). Furthermore, broadly cross-reactive NAI antibodies elicited by natural infection were unable to bind multiple commercially available inactivated vaccines, indicating that the vaccines lack the NA epitopes targeted by these antibodies (50).
The poor NA immunogenicity of the licensed influenza vaccines can be attributed to a few factors. As mentioned, current vaccines are optimized and standardized specifically for inducing high HAI antibody titers. As a result, the immunogenicity of the NA is not guaranteed. Quantity and quality of the NA antigen varies between manufacturers and vaccine batches (131, 132). Virus inactivation procedures may also affect NA immunogenicity. While treatment with EDTA or formalin did not compromise the immunogenicity of recombinant NA proteins (133), native NA in a viral particle may respond differently to these conditions. The stability and immunogenicity of NA also varies between strains (132). Nevertheless, in case vaccination does elicit NA antibodies, these can be long lived in healthy human subjects (134, 135). Better consideration of the NA component of current vaccines could therefore mean an important step forward in effectivity of the current vaccines. This could be achieved by also considering NA in selection of the vaccine strains, optimizing the manufacturing process to keep NA antigenically intact, and also standardizing the amounts of immunogenic NA in vaccine preparations.
6.2 Next-Generation NA-Based Vaccines
Next-generation seasonal and universal influenza vaccines call for an improved NA-directed immune response. Various options for rational design of the NA antigen and mode of presentation to the immune system are in development aiming to boost the breadth and magnitude of the NA-specific response (Figure 5). The following section summarizes promising ideas and the rationale behind them.
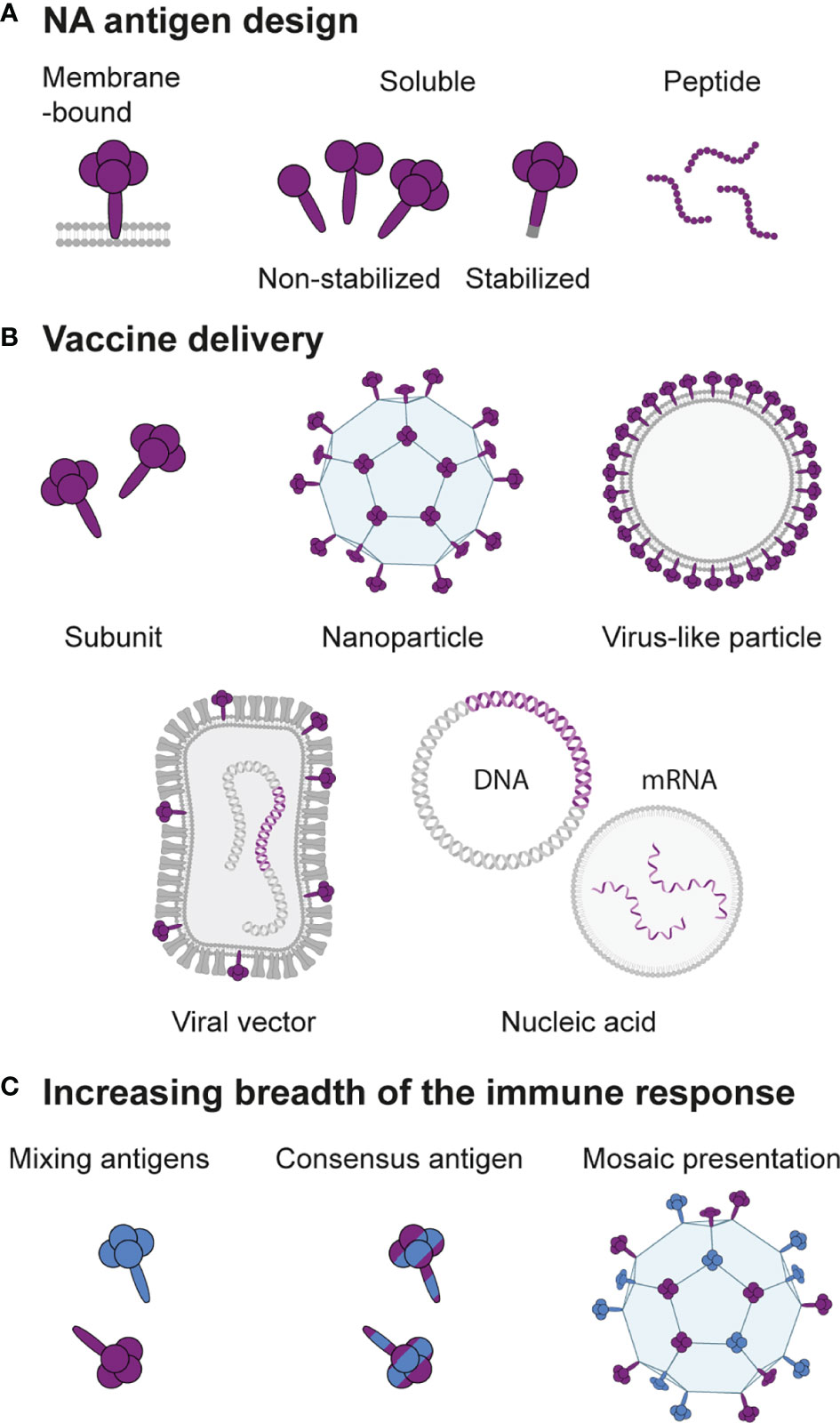
Figure 5 Design of next-generation NA-based vaccines. (A) The NA antigen can be presented in the native membrane-bound form, as a soluble protein that lacks the transmembrane domain with or without modifications to retain or stabilize the tetrameric structure, or as peptides containing an epitope of interest. (B) Depending on the antigen design, various methods for vaccine delivery are possible. Soluble NA can be administered as a subunit vaccine or coupled to a nanoparticle carrier. Membrane-bound NA can be presented on a virus-like particle or on the cell surface when encoded as RNA or DNA delivered directly or by a viral vector. (C) Strategies to increase the breadth of the immune response include mixing of NAs from different strains, computational design of consensus NAs, or hetero-multivalent mosaic presentation of NAs from different strains on a single particle.
6.2.1 Enhancing NA Immunogenicity Within Existing Influenza Virus Vaccines
In addition to optimizing the manufacturing conditions of the current seasonal vaccines, extra steps can be taken to improve the immunogenicity of NA while using the same influenza virion-based vaccine technology as a basis. For example, by re-structuring parts of the viral genome, a higher amount of NA can be incorporated into the viral particles (136, 137). Swapping the packaging signals of HA and NA that contain the viral RNA promoters resulted in virions with more NA, although at the expense of HA. Immunization with this virus resulted in increased levels of NAI and Fc-dependent antibodies directed against NA and induced protection against lethal challenge with an NA-matched IAV strain. HA-directed IgG titers were however significantly lower for the recombinant virus compared to the wild-type (136). Since glycosylation of the NA head domain was also found to affect NA incorporation in the virion (138), altering glycosylation sites could be explored as an alternative strategy to incorporate higher levels of NA in an IAV vaccine strain. Such modifications may, however, also affect NA antigenicity and immunogenicity. Extending the NA stalk so that NA surpassed HA in length was also shown to enhance NA immunogenicity of an inactivated virion-based vaccine in mice. Specifically, increased levels of antibodies with ADCC activity were induced. The authors hypothesized that their extended NA stalk domain method exposes NA epitopes to the immune system that were otherwise hidden (139).
6.2.2 Recombinant Protein Design
An alternative solution to increase the immune response to NA is to add extra NA to vaccine formulations. Johansson et al. supplemented inactivated vaccines with soluble NA purified from viral particles. This approach induced higher titers of NAI antibodies without compromising on HAI antibodies (58, 59). Recombinant soluble NA proteins (Figure 5A) are also attractive vaccine antigens to supplement inactivated virus vaccines or as a component of a multi-antigen subunit vaccine. Important in the design of such antigens is that a correctly folded tetrameric structure is essential for optimal immunogenicity (133, 140). Often enzymatic activity is measured as a proxy for a correct conformation of the NA antigen. While this is a reliable proxy for the presence of intact antigenic structure, NA activity in itself is not required to induce protective NA-specific antibodies in mice (133, 141, 142). Early on, recombinant NA was produced by replacing the membrane anchor sequence with a signal sequence derived from a type I membrane protein. This method typically results in a mix of tetrameric, dimeric, and some monomeric protein (24). To produce pure and stable tetrameric NA, ectodomains are better fused to heterologous tetramerization domains. Multiple tetramerization domains have successfully been used to produce immune-protective recombinant NA: VASP, GCN4, and Tetrabrachion (54, 143, 144). Tetrabrachion-stabilized NA proteins appear most stable and enzymatically active, presumably due to the parallel orientation of these domains (145, 146). In addition, a crystal structure of a tetrabrachion-stabilized N9 showed that the head domain maintained its native structure as found on virions (147). Whether immunization with these constructs also results in better quality antibody responses is yet to be determined. Interestingly, a recent study showed that the combined choice of tetramerization domain and inclusion of the stalk domain into recombinant NA profoundly impacts activity and immunogenicity. Thorough evaluation of individual recombinant NAs may therefore be required to determine the optimal antigen design strategy (142).
Mutations in the NA stalk (148) or the interface between protomers (149) can also enhance the stability and immunogenicity of recombinant NA. Cysteine mutations in the stalk led to more efficient dimer formation of recombinant N1, resulting in enhanced enzymatic activity and immune protection. The cysteine-stabilized dimers were however still outperformed by a VASP-stabilized tetrameric NA (148). It was only recently recognized that recombinant NA proteins can adopt an open conformation in addition to the closed conformation that is found on the surface of influenza virions. Structure-guided stabilization of the closed conformation was performed that improved thermal stability. More importantly, it also enhanced the affinity to protective antibodies elicited by viral infection, indicating that these antigens may elicit antibody responses to vulnerable quaternary epitopes more efficiently (149).
To broaden the protection elicited by a recombinant protein antigen, computational methods can be used to design constructs based on a consensus sequence of varying strains (Figure 5C). After similar techniques have shown promise for the induction of more broadly reactive HA responses (150), recent studies engineered NA constructs that combined sequences of various human, swine, and avian H1N1 and H5N1 strains (151, 152). The resulting antigens were immunogenic and induced antibodies against a broader range of viruses than wild-type NA antigens, including NAI antibodies against strains not included in the antigen design (151). Immunization or passive transfer of the sera of immunized mice offered protection against homologous and heterologous viral challenge (151, 152). While the broadened immune response may come at the cost of a lower magnitude compared to immunizations with a matched NA antigen (152), consensus NA antigens are promising candidates for inclusion in multi-antigen universal vaccine formulations.
6.2.3 Multivalent Presentation
The immunogenicity of soluble protein antigens can be substantially improved with multivalent presentation. Multivalent nanoparticles better mimic viruses in terms of size, shape, antigen valency, and repetitive organization, resulting in enhanced uptake by antigen presenting cells and stronger activation of B cell receptors (153). Such nanoparticle designs may constitute virus-like particles (VLPs) containing membrane-anchored NAs or protein nanoparticles to which soluble NAs are coupled (Figure 5B).
NA can self-assemble into VLPs, either when expressed alone or in combination with other IAV structural proteins (154–157). VLPs that display N1 alone or in combination with H5 and M2e induced high titers of NAI antibodies (154, 157–159). Vaccination with N1 VLPs derived from H1N1pdm09 provided cross-protection against lethal challenge with heterologous (H5N1) and even heterosubtypic (H3N2) IAV (159). However, in a more recent study N1 VLPs were not able to cross-protect against a historical H3N2 strain (160). N2 VLPs derived from a more recent H3N2 virus did protect against challenge with the distant H3N2. In addition, bivalent vaccination combining the N1 and N2 VLPs induced strong anti-N1 and -N2 antibody responses that exceeded the antibody levels induced by either one of the VLPs individually (160). Protective efficacy of NA-based VLP vaccination has also been demonstrated in ferrets. In these animals, VLPs containing H5N1-derived NA and matrix protein M1 induced high serum NAI antibody titers and protection against lethal homologous challenge. Incorporation of additional H5 or H3 into the VLPs further reduced clinical symptoms of the ferrets after H5N1 challenge. VLPs composed of H3/N2/M1 were however unable to cross-protect against the heterosubtypic H5N1 challenge (154). Efforts to further boost both humoral and cellular immunity of VLP vaccines currently focuses on attachment of adjuvants directly onto the VLPs (161–163).
In a different strategy based on multivalent presentation, nanoparticles consisting of little more than IAV structural proteins were generated (164). A dense core was constructed out of M2e protein and decorated with M2e-NA fusion proteins by chemical cross-linking. These double-layered nanoparticles greatly improved immunogenicity and (cross-)protection over soluble M2e-NA fusion proteins. M2e-NA nanoparticles were generated using NA from H5N1 and H3N2. Mice immunized with the M2e-N1 nanoparticles were fully protected against mortality following challenge with the homologous strain, as well as H1N1 and H3N2. Immunization with the M2e-N2 nanoparticles fully protected against mortality following challenge with the homologous strain and an H9N2 virus and conferred 60% protection against H1N1 (164).
Self-assembling protein nanoparticles are highly ordered, monodisperse carrier platforms that are increasingly used in experimental vaccines. The geometry of these nanoparticles is versatile and can be readily adapted for optimization to a specific antigen (165). While these carriers have not yet been used to generate NA-based nanoparticle vaccines, examples of their application for other viral glycoproteins illustrate their promise. HA nanoparticles, for example, elicited more than 10-fold higher antibody titers compared to the commercial inactivated vaccine, including antibodies directed against conserved vulnerable epitopes (166). The respiratory syncytial virus fusion protein similarly induced 10-fold higher neutralizing antibody titers when presented on a two-component protein scaffold (167). Self-assembling protein scaffolds are also well suited for presenting viral glycoproteins from multiple strains together on mosaic nanoparticles (Figure 5C). It was hypothesized that presentation of these diverse antigens alongside each other gives an avidity benefit for cross-reactive B cell receptors, resulting in a broader antibody response. This strategy was applied using the HA receptor binding domains (RBDs) of two H1N1 strains. Mosaic nanoparticles displaying two distinct H1 RBDs were found to induce broader antibody responses than a mixture of homotypic nanoparticles displaying the same set of RBDs (168). These results indicate that mosaic nanoparticles may enhance activation of B cells specific for the otherwise subdominant cross-reactive epitopes, which could be an interesting strategy to evaluate for NA.
6.2.4 Epitope-Based Vaccines
Epitope-based vaccine design aims to precisely direct the immune response towards conserved vulnerable B or T cell epitopes by presenting peptide epitopes (Figure 5A) on an immunogenic carrier or in a multi-epitope construct. In doing so, these techniques have potential for eliciting broadly protective immunity. The potential of an universally conserved linear B cell epitope near the NA catalytic site consisting of residues 222 through 228 or 230 (NA222) as an vaccine antigen was recently studied (169, 170). Kim et al. incorporated the epitope into the HA head domain of a H1N1 virus, creating a chimeric virus that was inactivated prior to immunizations. The NA222 chimeric virus, but not the inactivated wild-type virus, induced a strong Th1-type antibody response directed to this epitope. The chimeric virus protected against heterosubtypic challenge with H3N2 and H9N2 viruses and at 6 days post-challenge the mice expressed high levels of mucosal IgG and IgA specific for the epitope (169). Zeigler et al. developed a self-assembling protein nanoparticle containing the NA222 epitope in addition to two universal CD4 T cell epitopes that mediate high-affinity, long-lived antibody responses. The NA222 nanoparticle induced high IgG titers and conferred approximately 50% survival in otherwise lethal H1N1 and IBV challenge, whereas nanoparticles with HA or M2e epitopes were 70-75% protective with similar IgG titers. It was suggested that the NA222 epitope was less protective due to limited antibody accessibility (170). Alternative novel B and T cell epitopes may be identified using in silico predictions and be combined into a multi-epitope construct. Further in vivo studies are needed, however, to evaluate the protective potential of such vaccine candidates (171).
6.2.5 Viral Vector-Based Vaccines
Antigens delivered in the form of genetic information will be expressed in the natural environment of the cell, which helps to ensure the correct antigen generation and processing that is vital for a potent immune response. Vectored vaccines deliver the genetic material encoding the antigen of interest by incorporation into unrelated live attenuated or replication incompetent viruses (Figure 5B). A large number of viral vectors are available that each differ in their ability to stimulate different arms of the immune system, but also in more practical characteristics including genomic stability, accepted insertion size, and safety profile (172).
Experimental NA-based vector vaccines have been described among others for pox virus (173, 174) or parainfluenza virus (175) vectors. Modified Vaccinia Ankara (MVA), a safe and commonly used live poxviral vector, that expressed NA (MVA-NA) of H1N1pdm09 was analyzed for its protective efficacy. The MVA-NA vaccine induced high NAI titers and a potent cellular response characterized by particularly strong activation of CD8 T cells. Immunization conferred partial protection against replication of a homologous virus (173). A raccoonpox vector expressing the NA of avian H5N1 conferred full protection to mice against an otherwise lethal challenge when administered via the intranasal (IN) route. Interestingly, depletion of CD4 and CD8 T cells strongly reduced protection, suggesting an important role for cellular immunity in the protection provided by this vector (174). In contrast, parainfluenza 5 (PIV5) expressing the NA of H1N1pdm09 or avian H5N1 induced weak T cell responses and was dependent on antibodies for protection. PIV5 expressing the H1N1pdm09 NA conferred partial cross-protection against H5N1 and H3N2 challenge, whereas PIV5 expressing the H5N1 NA protected against H1N1, but not H3N2 (175). Other NA-expressing viral vectors that have been applied with varied success in a veterinary setting are Newcastle disease virus, infectious laryngotracheitis virus and alphavirus replicons (176–179).
6.2.6 Nucleic Acid-Based Vaccines
Nucleic acid-based vaccines directly deliver the genetic information encoding the antigen without the need for a viral vector (Figure 5B). In doing so, these methods exploit the benefits of in vivo antigen expression while eliminating the risks for reduced efficacy due to anti-vector immunity and safety concerns associated with viral vector-based vaccines. Production of this type of vaccines is rapid and scalable, which could be a critical advantage over other vaccine platforms in the event of a new emerging strain. Recent advances in stability and delivery of RNA and DNA vaccine formulations have led to improved immunogenicity, resulting in a renewed interest in using these technologies for emerging infectious diseases (172, 180, 181). mRNA vaccine technology was particularly accelerated by the current SARS-CoV-2 pandemic with the development of two highly effective vaccines based on this technology (182, 183).
Several studies reported the protective potential of NA antigens delivered by vaccination with plasmid DNA. An early study comparing the ability of various IAV antigens delivered by plasmid DNA to induce a protective immune response showed that only immunization with HA or NA, but not other internal IAV antigens, protected mice against homologous H1N1 challenge (184). In a follow-up study DNA vaccination with N2 induced full protection against challenge with homologous and drifted H3N2 strains, but was not effective in protecting against H1N1 challenge (185). Plasmid DNA encoding N1 from H1N1 conferred full protection against homologous challenge and 40-50% protection against H5N1 (186). Efficacy of a H5 DNA vaccine against challenge with a distant H5N1 strain was boosted from 75% to 100% with the addition of a N1-encoding plasmid (187).
NA-based mRNA vaccines have demonstrated high potency in some studies, but lower in another (188–190). Lipid encapsulated mRNA vaccines encoding various antigens of H1N1pdm09, separate or in a combination vaccine, elicited a strong humoral and cellular response in mice. The NA component of the vaccine was found to be the only one eliciting high NAI titers and protecting against a highly lethal dose of a matched challenge virus. A vaccine dose as low as 0.05 μg was sufficient to elicit a protective immune response. The NA mRNA vaccine however provided only limited or no protection against heterosubtypic challenge while the other more conserved vaccine components were fully protective (190) Similarly, in a recent study vaccination with mRNA encoding NA of H1N1pdm09 induced high NAI titers and protected against mortality from challenge with pre-pandemic H1N1 and H5N1. Serum antibodies from vaccinated mice however did not cross-react with H3N2 or influenza B virus (189) In an earlier study the immunogenicity of a N1-mRNA vaccine was considerably lower. High dose vaccination induced only 40% protection against a matched challenge in mice. Supplementing a H1-mRNA vaccine with the N1-mRNA however resulted in significantly reduced morbidity over the H1-only vaccine (188). The potential of mRNA vaccines might be boosted further by the use of self-replicating RNAs, which may induce high expression levels after low dose vaccination (191, 192).
7 Outlook
Various studies mentioned in this review describe NA-based vaccine candidates with impressive protective efficacy against homologous and heterologous challenge strains, although most candidates have yet to be tested in models other than mice. In view of the potential of NA to induce protective immunity, efforts to improve vaccine efficacy against influenza should not only focus on HA, but also on NA. Improving the NA component of current vaccines with respect to antigenic match and immunogenicity, would likely improve the efficacy of these vaccines in the short term. NA additionally should be considered in the development of next-generation vaccines, besides the largely HA-focused approaches.
While studying the protective efficacy of isolated NA-based vaccine candidates is informative, NA particularly has potential as a component in a multi-antigen vaccine. Vaccines targeting both HA and NA provide better and broader protection, as evidenced by reduced disease and transmission when compared to HA- or NA-only vaccines. Given that the antigenic drift of HA and NA is discordant (76) a seasonal vaccine combining both antigens would be less likely to be mismatched with circulating strains for both antigens, compared to a HA-focussed vaccine.
Most vaccine candidates described here induce a protective immune response against homologous and in some cases intra-subtypic heterologous NA, but not heterosubtypic NA. The application of strategies aimed at increasing the breadth of the immune response is vital to improve protection against drifted or new emerging strains. The induction of broadly protective heterologous immune responses may be enhanced by computational design of consensus antigens. Protective responses against different NA subtypes may be achieved by simple mixing of NA antigens, though the breadth may in such vaccines still be limited to the strains used for immunization like in the current seasonal vaccines. Additional measures may be required to ensure that the elicited immune response surpasses the immunization strains. To direct the immune responses more towards conserved epitopes the option of combining NAs from multiple strains onto heteromultivalent mosaic nanoparticles could be explored.
The recent market application of mRNA vaccines targeting SARS-CoV-2 is likely to pave the road for the clinical use of this vaccine platform for novel influenza vaccines. Prior to the emergence of SARS-CoV-2 it was already recognized that mRNA vaccines would be suitable specifically in an outbreak setting mostly due to the capacity for rapid development, in addition to the low dose requirement and high potency (180). mRNA vaccines encoding HA of potential pandemic strains have already demonstrated safety and immunogenicity in ferrets, non-human primates and humans (193) and clinical trials for seasonal HA-based mRNA vaccines are underway (Clinical Trials Identifiers NCT04956575 and NCT04969276). Addition of NA-encoding mRNA to such formulations is likely to improve the magnitude and breadth of protection and should be advocated.
Author Contributions
SC, MP, MB, XS, and CH wrote the manuscript. SC, MP, and MB created the figures. MB made Table 1. All authors contributed to the article and approved the submitted version.
Funding
This work was supported in part by the ENDFLU project that has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 874650. The present work was also a part of the research program of the Netherlands Centre for One Health (www.ncoh.nl) and was financially supported through the One Health Investment Fund from the Faculty of Veterinary Medicine of the Utrecht University.
Conflict of Interest
XS declares to receive funding from Sanofi Pasteur for research related to influenza vaccine development.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Paget J, Marquet R, Meijer A, van der Velden K. Influenza Activity in Europe During Eight Seasons (1999-2007): An Evaluation of the Indicators Used to Measure Activity and an Assessment of the Timing, Length and Course of Peak Activity (Spread) Across Europe. BMC Infect Dis (2007) 7(1):1–7. doi: 10.1186/1471-2334-7-141
2. Ng S, Gordon A. Influenza Burden and Transmission in the Tropics. Curr Epidemiol Rep (2015) 2(2):89–100. doi: 10.1007/s40471-015-0038-4
3. Demicheli V, Jefferson T, Di Pietrantonj C, Ferroni E, Thorning S, Thomas RE, et al. Vaccines for Preventing Influenza in the Elderly. Cochrane Database Syst Rev (2018) 2(2):CD004876. doi: 10.1002/14651858.CD004876.pub4
4. Rodrigues BS, David C, Costa J, Ferreira JJ, Pinto FJ, Caldeira D. Influenza Vaccination in Patients With Heart Failure: A Systematic Review and Meta-Analysis of Observational Studies. Heart (2020) 106(5):350–7. doi: 10.1136/heartjnl-2019-315193
5. Belongia EA, Kieke BA, Donahue JG, Greenlee RT, Balish A, Foust A, et al. Effectiveness of Inactivated Influenza Vaccines Varied Substantially With Antigenic Match From the 2004-2005 Season to the 2006-2007 Season. J Infect Dis (2009) 199(2):159–67. doi: 10.1086/595861
6. Hutchinson EC, Charles PD, Hester SS, Thomas B, Trudgian D, Martinez-Alonso M, et al. Conserved and Host-Specific Features of Influenza Virion Architecture. Nat Commun (2014) 5:4816. doi: 10.1038/ncomms5816
7. Barman S, Adhikary L, Chakrabarti AK, Bernas C, Kawaoka Y, Nayak DP. Role of Transmembrane Domain and Cytoplasmic Tail Amino Acid Sequences of Influenza A Virus Neuraminidase in Raft Association and Virus Budding. J Virol (2004) 78(10):5258–69. doi: 10.1128/jvi.78.10.5258-5269.2004
8. Rossman JS, Lamb RA. Influenza Virus Assembly and Budding. Virology (2011) 411(2):229–36. doi: 10.1016/j.virol.2010.12.003
9. Kundu A, Avalos RT, Sanderson CM, Nayak DP. Transmembrane Domain of Influenza Virus Neuraminidase, A Type II Protein, Possesses an Apical Sorting Signal in Polarized MDCK Cells. J Virol (1996) 70(9):6508–15. doi: 10.1128/jvi.70.9.6508-6515.1996
10. Nordholm J, Da Silva DV, Damjanovic J, Dou D, Daniels R. Polar Residues and Their Positional Context Dictate the Transmembrane Domain Interactions of Influenza a Neuraminidases. J Biol Chem (2013) 288(15):10652–60. doi: 10.1074/jbc.M112.440230
11. Da Silva DV, Nordholm J, Madjo U, Pfeiffer A, Daniels R. Assembly of Subtype 1 Influenza Neuraminidase Is Driven by Both the Transmembrane and Head Domains. J Biol Chem (2013) 288(1):644–53. doi: 10.1074/jbc.M112.424150
12. Air GM. Influenza Neuraminidase. Influenza Other Respi Viruses (2012) 6(4):245–5. doi: 10.1111/j.1750-2659.2011.00304.x
13. Li J, Dohna H, Cardona CJ, Miller J, Carpenter TE. Emergence and Genetic Variation of Neuraminidase Stalk Deletions in Avian Influenza Viruses. PloS One (2011) 6(2):e14722. doi: 10.1371/journal.pone.0014722
14. Baigent SJ, McCauley JW. Glycosylation of Haemagglutinin and Stalk-Length of Neuraminidase Combine to Regulate the Growth of Avian Influenza Viruses in Tissue Culture. Virus Res (2001) 79(1–2):177–85. doi: 10.1016/S0168-1702(01)00272-6
15. Wang M, Qi J, Liu Y, Vavricka C, Wu Y, Li Q, et al. Influenza A Virus N5 Neuraminidase Has an Extended 150-Cavity. J Virol (2011) 85(16):8431–5. doi: 10.1128/JVI.00638-11
16. Sun X, Li Q, Wu Y, Wang M, Liu Y, Qi J, et al. Structure of Influenza Virus N7: The Last Piece of the Neuraminidase “Jigsaw” Puzzle. J Virol (2014) 88(16):9197–207. doi: 10.1128/jvi.00805-14
17. Li Q, Qi J, Wu Y, Kiyota H, Tanaka K, Suhara Ys, et al. Functional and Structural Analysis of Influenza Virus Neuraminidase N3 Offers Further Insight Into the Mechanisms of Oseltamivir Resistance. J Virol (2013) 87(18):10016–24. doi: 10.1128/JVI.01129-13
18. Xu X, Zhu X, Dwek RA, Stevens J, Wilson IA. Structural Characterization of the 1918 Influenza Virus H1N1 Neuraminidase. J Virol (2008) 82(21):10493–501. doi: 10.1128/JVI.00959-08
19. Russell RJ, Haire LF, Stevens DJ, Collins PJ, Lin YP, Blackburn GM, et al. The Structure of H5N1 Avian Influenza Neuraminidase Suggests New Opportunities for Drug Design. Nature (2006) 443(7107):45–9. doi: 10.1038/nature05114
20. Varghese J, Colman P. Three-Dimensional Structure of the Neuraminidase of Influenza Virus A/Tokyo/3/67 at 2.2 A Resolution. J Mol Biol (1991) 221(2):473–86. doi: 10.1016/0022-2836(91)80068-6
21. Tulip WR, Varghese JN, Baker AT, van Donkelaar A, Laver WG, Webster RG, et al. Refined Atomic Structures of N9 Subtype Influenza Virus Neuraminidase and Escape Mutants. J Mol Biol (1991) 221(2):487–97. doi: 10.1016/0022-2836(91)80069-7
22. Burmeister WP, Ruigrok RWH, Cusack S. The 2.2 Å Resolution Crystal Structure of Influenza B Neuraminidase and Its Complex With Sialic Acid. EMBO J (1992) 11(1):49–5. doi: 10.1002/j.1460-2075.1992.tb05026.x
23. Roggentin P, Rothe B, Kaper JB, Galen J, Lawrisuk L, Vimr ER, et al. Conserved Sequences in Bacterial and Viral Sialidases. Glycoconj J (1989) 6(3):349–53. doi: 10.1007/BF01047853
24. Deroo T, Jou WM, Fiers W. Recombinant Neuraminidase Vaccine Protects Against Lethal Influenza. Vaccine (1996) 14(6):561–9. doi: 10.1016/0264-410X(95)00157-V
25. Chong AKJ, Pegg MS, von Itzstein M. Influenza Virus Sialidase: Effect of Calcium on Steady-State Kinetic Parameters. Biochim Biophys Acta (BBA)/Protein Struct Mol (1991) 1077(1):65–71. doi: 10.1016/0167-4838(91)90526-6
26. Wang H, Dou D, Östbye H, Revol R, Daniels R. Structural Restrictions for Influenza Neuraminidase Activity Promote Adaptation and Diversification. Nat Microbiol (2019) 4(12):2565–77. doi: 10.1038/s41564-019-0537-z
27. Burmeister WP, Cusack S, Ruigrok RWH. Calcium Is Needed for the Thermostability of Influenza B Virus Neuraminidase. J Gen Virol (1994) 75(2):381–8. doi: 10.1099/0022-1317-75-2-381
28. Dimmock NJ. Dependence of the Activity of an Influenza Virus Neuraminidase Upon Ca2+. J Gen Virol (1971) 13(3):481–3. doi: 10.1099/0022-1317-13-3-481
29. Li L, Li Y, Zhang L, Hou T. Theoretical Studies on the Susceptibility of Oseltamivir Against Variants of 2009 A/H1N1 Influenza Neuraminidase. J Chem Inf Model (2012) 52(10):2715–29. doi: 10.1021/ci300375k
30. Amaro RE, Swift RV, Votapka L, Li WW, Walker RC, Bush RM. Mechanism of 150-Cavity Formation in Influenza Neuraminidase. Nat Commun (2011) 2(1):388. doi: 10.1038/ncomms1390
31. Wu Y, Qin G, Gao F, Liu Y, Vavricka CJ, Qi J, et al. Induced Opening of Influenza Virus Neuraminidase N2 150-Loop Suggests an Important Role in Inhibitor Binding. Sci Rep (2013) 3(1):1–8. doi: 10.1038/srep01551
32. Du W, de Vries E, van Kuppeveld FJM, Matrosovich M, de Haan CAM. Second Sialic Acid-Binding Site of Influenza A Virus Neuraminidase: Binding Receptors for Efficient Release. FEBS J (2021) 288(19):5598–612. doi: 10.1111/febs.15668
33. Du W, Guo H, Nijman VS, Doedt J, van der Vries E, van der Lee J, et al. The 2nd Sialic Acid-Binding Site of Influenza a Virus Neuraminidase Is an Important Determinant of the Hemagglutinin-Neuraminidase-Receptor Balance. PloS Pathog (2019) 15(6):e1007860. doi: 10.1371/journal.ppat.1007860
34. Uhlendorff J, Matrosovich T, Klenk H, Matrosovich M. Functional Significance of the Hemadsorption Activity of Influenza Virus Neuraminidase and Its Alteration in Pandemic Viruses. Arch Virol (2009) 154(6):945–57. doi: 10.1007/S00705-009-0393-X
35. Benton DJ, Gamblin SJ, Rosenthal PB, Skehel JJ. Structural Transitions in Influenza Haemagglutinin at Membrane Fusion PH. Nature (2020) 583(7814):150–3. doi: 10.1038/s41586-020-2333-6
36. Cohen M, Zhang XQ, Senaati HP, Chen HW, Varki NM, Schooley RT, et al. Influenza A Penetrates Host Mucus by Cleaving Sialic Acids With Neuraminidase. Virol J (2013) 10(1):1–13. doi: 10.1186/1743-422X-10-321
37. Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk HD. Human and Avian Influenza Viruses Target Different Cell Types in Cultures of Human Airway Epithelium. Proc Natl Acad Sci USA (2004) 101(13):4620–4. doi: 10.1073/pnas.0308001101
38. de Vries E, Du W, Guo H, de Haan CAM. Influenza A Virus Hemagglutinin–Neuraminidase–Receptor Balance: Preserving Virus Motility. Trends Microbiol (2020) 28(1):57–67. doi: 10.1016/j.tim.2019.08.010
39. Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk H-D. Neuraminidase Is Important for the Initiation of Influenza Virus Infection in Human Airway Epithelium. J Virol (2004) 78(22):12665. doi: 10.1128/JVI.78.22.12665-12667.2004
40. Su B, Wurtzer S, Rameix-Welti MA, Dwyer D, van der Werf S, Naffakh N, et al. Enhancement of the Influenza a Hemagglutinin (HA)-Mediated Cell-Cell Fusion and Virus Entry by the Viral Neuraminidase (Na). PloS One (2009) 4(12):e8495. doi: 10.1371/journal.pone.0008495
41. Palese P, Compans RW. Inhibition of Influenza Virus Replication in Tissue Culture by 2 Deoxy 2,3 Dehydro N Trifluoroacetylneuraminic Acid (FANA): Mechanism of Action. J Gen Virol (1976) 33(1):159–63. doi: 10.1099/0022-1317-33-1-159
42. Vahey MD, Fletcher DA. Influenza a Virus Surface Proteins Are Organized to Help Penetrate Host Mucus. Elife (2019) 8:e43764. doi: 10.7554/eLife.43764
43. Fei Y, Sun YS, Li Y, Yu H, Lau K, Landry JP, et al. Characterization of Receptor Binding Profiles of Influenza a Viruses Using an Ellipsometry-Based Label-Free Glycan Microarray Assay Platform. Biomolecules (2015) 5(3):1480–98. doi: 10.3390/biom5031480
44. Guo H, Rabouw H, Slomp A, Dai M, van der Vegt F, van Lent JWM, et al. Kinetic Analysis of the Influenza A Virus HA/NA Balance Reveals Contribution of NA to Virus-Receptor Binding and NA-Dependent Rolling on Receptor-Containing Surfaces. PloS Pathog (2018) 14(8):e1007233. doi: 10.1371/journal.ppat.1007233
45. Park S, Il Kim J, Lee I, Bae JY, Yoo K, Nam M, et al. Adaptive Mutations of Neuraminidase Stalk Truncation and Deglycosylation Confer Enhanced Pathogenicity of Influenza A Viruses. Sci Rep (2017) 7(1):10928. doi: 10.1038/s41598-017-11348-0
46. Hossain MJ, Hickman D, Perez DR. Evidence of Expanded Host Range and Mammalian-Associated Genetic Changes in a Duck H9N2 Influenza Virus Following Adaptation in Quail and Chickens. PloS One (2008) 3(9):e3170. doi: 10.1371/journal.pone.0003170
47. Matrosovich M, Zhou N, Kawaoka Y, Webster R. The Surface Glycoproteins of H5 Influenza Viruses Isolated From Humans, Chickens, and Wild Aquatic Birds Have Distinguishable Properties. J Virol (1999) 73(2):1146–55. doi: 10.1128/jvi.73.2.1146-1155.1999
48. Munier S, Larcher T, Cormier-Aline F, Soubieux D, Su B, Guigand L, et al. A Genetically Engineered Waterfowl Influenza Virus With a Deletion in the Stalk of the Neuraminidase Has Increased Virulence for Chickens. J Virol (2010) 84(2):940–52. doi: 10.1128/jvi.01581-09
49. Zhou H, Yu Z, Hu Y, Tu J, Zou W, Peng Y, et al. The Special Neuraminidase Stalk-Motif Responsible for Increased Virulence and Pathogenesis of H5N1 Influenza A Virus. PloS One (2009) 4(7):e6277. doi: 10.1371/journal.pone.0006277
50. Chen YQ, Wohlbold TJ, Zheng NY, Huang M, Huang Y, Neu KE, et al. Influenza Infection in Humans Induces Broadly Cross-Reactive and Protective Neuraminidase-Reactive Antibodies. Cell (2018) 173(2):417–29. doi: 10.1016/j.cell.2018.03.030
51. Tan J, O’Dell G, Hernandez M, Sordillo E, Kahn Z, Kriti D, et al. Human Anti-Neuraminidase Antibodies Reduce Airborne Transmission of Clinical Influenza Virus Isolates in the Guinea Pig Model. J Virol (2021) JVI0142121. doi: 10.1128/JVI.01421-21
52. Wohlbold TJ, Podolsky KA, Chromikova V, Kirkpatrick E, Falconieri V, Meade P, et al. Broadly Protective Murine Monoclonal Antibodies Against Influenza B Virus Target Highly Conserved Neuraminidase Epitopes. Nat Microbiol (2017) 2(10):1415–24. doi: 10.1038/s41564-017-0011-8
53. Xiong FF, Liu XY, Gao FX, Luo J, Duan P, Tan WS, et al. Protective Efficacy of Anti-Neuraminidase Monoclonal Antibodies Against H7N9 Influenza Virus Infection. Emerg Microbes Infect (2020) 9(1):78–87. doi: 10.1080/22221751.2019.1708214
54. Bosch BJ, Bodewes R, Vries RP, de; Kreijtz JHCM, Bartelink W, Amerongen Gv, et al. Recombinant Soluble, Multimeric HA and NA Exhibit Distinctive Types of Protection Against Pandemic Swine-Origin 2009 A(H1N1) Influenza Virus Infection in Ferrets. J Virol (2010) 84(19):10366. doi: 10.1128/JVI.01035-10
55. Chen Z, Matsuo K, Asanuma H, Takahashi H, Iwasaki T, Suzuki Y, et al. Enhanced Protection Against a Lethal Influenza Virus Challenge by Immunization With Both Hemagglutinin- and Neuraminidase-Expressing DNAs. Vaccine (1999) 17(7–8):653–9. doi: 10.1016/S0264-410X(98)00247-3
56. Johansson BE. Immunization With Influenza A Virus Hemagglutinin and Neuraminidase Produced in Recombinant Baculovirus Results in a Balanced and Broadened Immune Response Superior to Conventional Vaccine. Vaccine (1999) 17(15–16):2073–80. doi: 10.1016/S0264-410X(98)00413-7
57. Johansson BE, Kilbourne ED. Dissociation of Influenza Virus Hemagglutinin and Neuraminidase Eliminates Their Intravirionic Antigenic Competition. J Virol (1993) 67(10):5721–3. doi: 10.1128/jvi.67.10.5721-5723.1993
58. Johansson BE, Matthews JT, Kilbourne ED. Supplementation of Conventional Influenza A Vaccine With Purified Viral Neuraminidase Results in a Balanced and Broadened Immune Response. Vaccine (1998) 16(9–10):1009–15. doi: 10.1016/S0264-410X(97)00279-X
59. Johansson BE, Pokorny BA, Tiso VA. Supplementation of Conventional Trivalent Influenza Vaccine With Purified Viral N1 and N2 Neuraminidases Induces a Balanced Immune Response Without Antigenic Competition. Vaccine (2002) 20(11–12):1670–4. doi: 10.1016/S0264-410X(01)00490-X
60. Monto AS, Kendal AP. Effect of Neuraminidase Antibody on Hong Kong Influenza. Lancet (1973) 301(7804):623–5. doi: 10.1016/S0140-6736(73)92196-X
61. Schulman JL, Khakpour M, Kilbourne ED. Protective Effects of Specific Immunity to Viral Neuraminidase on Influenza Virus Infection of Mice. J Virol (1968) 2(8):778–86. doi: 10.1128/jvi.2.8.778-786.1968
62. Couch RB, Kasel JA, Gerin JL, Schulman JL, Kilbourne ED. Induction of Partial Immunity to Influenza by a Neuraminidase Specific Vaccine. J Infect Dis (1974) 129(4):411–20. doi: 10.1093/infdis/129.4.411
63. Dowdle WR, Coleman MT, Mostow SR, Kaye HS, Schoenbaum SC. Inactivated Influenza Vaccines. 2. Laboratory Indices of Protection. Postgrad Med J (1973) 49(569):159–63. doi: 10.1136/pgmj.49.569.159
64. Couch RB, Atmar RL, Franco LM, Quarles JM, Wells J, Arden N, et al. Antibody Correlates and Predictors of Immunity to Naturally Occurring Influenza in Humans and the Importance of Antibody to the Neuraminidase. J Infect Dis (2013) 207(6):974–81. doi: 10.1093/infdis/jis935
65. Monto AS, Petrie JG, Cross RT, Johnson E, Liu M, Zhong W, et al. Antibody to Influenza Virus Neuraminidase: An Independent Correlate of Protection. J Infect Dis (2015) 212(8):1191–9. doi: 10.1093/INFDIS/JIV195
66. Memoli MJ, Shaw PA, Han A, Czajkowski L, Reed S, Athota R, et al. Evaluation of Antihemagglutinin and Antineuraminidase Antibodies as Correlates of Protection in an Influenza A/H1N1 Virus Healthy Human Challenge Model. MBio (2016) 7(2):e00417-16. doi: 10.1128/mBio.00417-16
67. Kosik I, Yewdell JW. Influenza Hemagglutinin and Neuraminidase: Yin-Yang Proteins Coevolving to Thwart Immunity. Viruses (2019) 11(4):346. doi: 10.3390/v11040346
68. Fiore AE, Uyeki TM, Broder K, Finelli L, Euler GL, Singleton JA, et al. Prevention and Control of Influenza With Vaccines: Recommendations of the Advisory Committee on Immunization Practices (ACIP), 2010. Morb Mortal Wkly Rep (2010) 6(59):RR–8.
69. Heaton NS, Sachs D, Chen CJ, Hai R, Palese P. Genome-Wide Mutagenesis of Influenza Virus Reveals Unique Plasticity of the Hemagglutinin and NS1 Proteins. Proc Natl Acad Sci USA (2013) 110(50):20248–53. doi: 10.1073/pnas.1320524110
70. Smith DJ, Lapedes AS, De Jong JC, Bestebroer TM, Rimmelzwaan GF, Osterhaus ADME, et al. Mapping the Antigenic and Genetic Evolution of Influenza Virus. Science (2004) 305(5682):371–6. doi: 10.1126/science.1097211
71. De Jong JC, Donker GA, Meijer A, van der Hoek W, Rimmelzwaan GF, Osterhaus ADME. Het Influenzaseizoen 2010/2011 in Nederland: Het Nieuwe A(H1N1)-Virus Van 2009 Blijft Actief. Ned Tijdschr Voor Med Microbiol (2011) 19(4):21–7.
72. Koel BF, Burke DF, Bestebroer TM, van der Vliet S, Zondag GCM, Vervaet G, et al. Substitutions Near the Receptor Binding Site Determine Major Antigenic Change During Influenza Virus Evolution. Science (2013) 342(6161):976–9. doi: 10.1126/science.1244730
73. Doud MB, Hensley SE, Bloom JD. Complete Mapping of Viral Escape From Neutralizing Antibodies. PloS Pathog (2017) 13(3):e1006271. doi: 10.1371/journal.ppat.1006271
74. Bhatt S, Holmes EC, Pybus OG. The Genomic Rate of Molecular Adaptation of the Human Influenza A Virus. Mol Biol Evol (2011) 28(9):2443–51. doi: 10.1093/molbev/msr044
75. Kilbourne ED, Johansson BE, Grajower B. Independent and Disparate Evolution in Nature of Influenza A Virus Hemagglutinin and Neuraminidase Glycoproteins. Proc Natl Acad Sci USA (1990) 87(2):786–90. doi: 10.1073/pnas.87.2.786
76. Sandbulte MR, Westgeest KB, Gao J, Xu X, Klimov AI, Russell CA, et al. Discordant Antigenic Drift of Neuraminidase and Hemagglutinin in H1N1 and H3N2 Influenza Viruses. Proc Natl Acad Sci USA (2011) 108(51):20748–53. doi: 10.1073/pnas.1113801108
77. Laver WG, Air GM, Webster RG, Markoff LJ. Amino Acid Sequence Changes in Antigenic Variants of Type A Influenza Virus N2 Neuraminidase. Virology (1982) 122(2):450–60. doi: 10.1016/0042-6822(82)90244-6
78. Westgeest KB, de Graaf M, Fourment M, Bestebroer TM, van Beek R, Spronken MIJ, et al. Genetic Evolution of the Neuraminidase of Influenza a (H3N2) Viruses From 1968 to 2009 and Its Correspondence to Haemagglutinin Evolution. J Gen Virol (2012) 93(Pt 9):1996–2007. doi: 10.1099/vir.0.043059-0
79. Kilbourne ED, Laver WG, Schulman JL, Webster RG. Antiviral Activity of Antiserum Specific for an Influenza Virus Neuraminidase. J Virol (1968) 2(4):281–8. doi: 10.1128/jvi.2.4.281-288.1968
80. Gao J, Couzens L, Burke DF, Wan H, Wilson P, Memoli MJ, et al. Antigenic Drift of the Influenza A(H1N1)Pdm09 Virus Neuraminidase Results in Reduced Effectiveness of A/California/7/2009 (H1N1pdm09)-Specific Antibodies. MBio (2019) 10(2):e00307–19. doi: 10.1128/MBIO.00307-19
81. Archetti I, Horsfall FL. Persistent Antigenic Variation of Influenza A Viruses After Incomplete Neutralization in Ovo With Heterologous Immune Serum. J Exp Med (1950) 92(5):441–62. doi: 10.1084/jem.92.5.441
82. Xie H, Wan XF, Ye Z, Plant EP, Zhao Y, Xu Y, et al. H3N2 Mismatch of 2014-15 Northern Hemisphere Influenza Vaccines and Head-To-Head Comparison Between Human and Ferret Antisera Derived Antigenic Maps. Sci Rep (2015) 5:15279. doi: 10.1038/srep15279
83. Linderman SL, Chambers BS, Zost SJ, Parkhousea K, Li Y, Herrmann C, et al. Potential Antigenic Explanation for Atypical H1n1 Infections Among Middle-Aged Adults During the 2013-2014 Influenza Season. Proc Natl Acad Sci USA (2014) 5:15279. doi: 10.1073/pnas.1409171111
84. Li GM, Chiu C, Wrammert J, McCausland M, Andrews SF, Zheng NY, et al. Pandemic H1N1 Influenza Vaccine Induces a Recall Response in Humans That Favors Broadly Cross-Reactive Memory B Cells. Proc Natl Acad Sci USA (2012) 109(23):9047–52. doi: 10.1073/pnas.1118979109
85. Chen Z, Wang W, Zhou H, Suguitan AL, Shambaugh C, Kim L, et al. Generation of Live Attenuated Novel Influenza Virus A/California/7/09 (H1N1) Vaccines With High Yield in Embryonated Chicken Eggs. J Virol (2010) 84(1):44–51. doi: 10.1128/jvi.02106-09
86. Lee MS, Yang CF. Cross-Reactive H1N1 Antibody Responses to a Live Attenuated Influenza Vaccine in Children: Implication for Selection of Vaccine Strains. J Infect Dis (2003) 188(9):1362–6. doi: 10.1086/379045
87. Dai M, Du W, Martínez-Romero C, Leenders T, Wennekes T, Rimmelzwaan GF, et al. Analysis of the Evolution of Pandemic Influenza a(H1N1) Virus Neuraminidase Reveals Entanglement of Different Phenotypic Characteristics. MBio (2021) 12(3):e00287–21. doi: 10.1128/mBio.00287-21
88. Lässig M, Mustonen V, Walczak AM. Predicting Evolution. Nat Ecol Evol (2017) 1(3):77. doi: 10.1038/s41559-017-0077
89. Wang Y, Lei R, Nourmohammed A, Wu NC. Antigenic Evolution of Human Influenza H3N2 Neuraminidase Is Constrained by Charge Balancing. bioRxiv (2021) 2021.07.10.451918. doi: 10.1101/2021.07.10.451918
90. Hensley SE, Das SR, Gibbs JS, Bailey AL, Schmidt LM, Bennink JR, et al. Influenza a Virus Hemagglutinin Antibody Escape Promotes Neuraminidase Antigenic Variation and Drug Resistance. PloS One (2011) 6(2):e15190. doi: 10.1371/journal.pone.0015190
91. Kaplan BS, Anderson TK, Chang J, Santos J, Perez D, Lewis N, et al. Evolution and Antigenic Advancement of N2 Neuraminidase of Swine Influenza A Viruses Circulating in the United States Following Two Separate Introductions From Human Seasonal Viruses. J Virol (2021) 95(20):e0063221. doi: 10.1128/JVI.00632-21
92. Wan H, Gao J, Xu K, Chen H, Couzens LK, Rivers KH, et al. Molecular Basis for Broad Neuraminidase Immunity: Conserved Epitopes in Seasonal and Pandemic H1N1 as Well as H5N1 Influenza Viruses. J Virol (2013) 87(16):9290–300. doi: 10.1128/jvi.01203-13
93. Wan H, Qi L, Gao J, Couzens LK, Jiang L, Gao Y, et al. Comparison of the Efficacy of N9 Neuraminidase-Specific Monoclonal Antibodies Against Influenza A(H7N9) Virus Infection. J Virol (2018) 92(4):e01588–17. doi: 10.1128/jvi.01588-17
94. Wilson JR, Guo Z, Reber A, Kamal RP, Music N, Gansebom S, et al. An Influenza A Virus (H7N9) Anti-Neuraminidase Monoclonal Antibody With Prophylactic and Therapeutic Activity In Vivo. Antiviral Res (2016) 135:48–55. doi: 10.1016/j.antiviral.2016.10.001
95. Guo Z, Wilson JR, York IA, Stevens J. Biosensor-Based Epitope Mapping of Antibodies Targeting the Hemagglutinin and Neuraminidase of Influenza A Virus. J Immunol Methods (2018) 461:23–9. doi: 10.1016/j.jim.2018.07.007
96. Eichelberger M, Wan H. Influenza Neuraminidase as a Vaccine Antigen. Curr Top Microbiol Immunol (2015) 386:275–99. doi: 10.1007/82_2014_398
97. Wang B, Wang K, Meng P, Hu Y, Yang F, Liu K, et al. Design, Synthesis, and Evaluation of Carboxyl-Modified Oseltamivir Derivatives With Improved Lipophilicity as Neuraminidase Inhibitors. Bioorg Med Chem Lett (2018) 28(21):3477–82. doi: 10.1016/j.bmcl.2018.09.014
98. Zhu X, Turner HL, Lang S, McBride R, Bangaru S, Gilchuk IM, et al. Structural Basis of Protection Against H7N9 Influenza Virus by Human Anti-N9 Neuraminidase Antibodies. Cell Host Microbe (2019) 26(6):729–38. doi: 10.1016/j.chom.2019.10.002
99. Gilchuk IM, Bangaru S, Gilchuk P, Irving RP, Kose N, Bombardi RG, et al. Influenza H7N9 Virus Neuraminidase-Specific Human Monoclonal Antibodies Inhibit Viral Egress and Protect From Lethal Influenza Infection in Mice. Cell Host Microbe (2019) 26(6):715–28. doi: 10.1016/j.chom.2019.10.003
100. Doyle TM, Hashem AM, Li C, Van Domselaar G, Larocque L, Wang J, et al. Universal Anti-Neuraminidase Antibody Inhibiting All Influenza A Subtypes. Antiviral Res (2013) 100(2):567–74. doi: 10.1016/j.antiviral.2013.09.018
101. Doyle TM, Li C, Bucher DJ, Hashem AM, Van Domselaar G, Wang K, et al. A Monoclonal Antibody Targeting a Highly Conserved Epitope in Influenza B Neuraminidase Provides Protection Against Drug Resistant Strains. Biochem Biophys Res Commun (2013) 441(1):226–9. doi: 10.1016/j.bbrc.2013.10.041
102. Madsen A, Dai YN, McMahon M, Schmitz AJ, Turner JS, Tan J, et al. Human Antibodies Targeting Influenza B Virus Neuraminidase Active Site Are Broadly Protective. Immunity (2020) 53(4):852–863.e. doi: 10.1016/j.immuni.2020.08.015
103. Wan H, Yang H, Shore DA, Garten RJ, Couzens L, Gao J, et al. Structural Characterization of a Protective Epitope Spanning A(H1N1)Pdm09 Influenza Virus Neuraminidase Monomers. Nat Commun (2015) 6(1):1–10. doi: 10.1038/ncomms7114
104. Jiang L, Fantoni G, Couzens L, Gao J, Plant E, Ye Z. Comparative Efficacy of Monoclonal Antibodies That Bind to Different Epitopes of the 2009 Pandemic H1N1 Influenza Virus Neuraminidase. J Virol (2016) 90(1):117–28. doi: 10.1128/JVI.01756-15
105. Stadlbauer D, Zhu X, McMahon M, Turner JS, Wohlbold TJ, Schmitz AJ, et al. Broadly Protective Human Antibodies That Target the Active Site of Influenza Virus Neuraminidase. Science (2019) 366(6464):499–504. doi: 10.1126/science.aay0678
106. Rijal P, Wang BB, Tan TK, Schimanski L, Janesch P, Dong T, et al. Broadly Inhibiting Antineuraminidase Monoclonal Antibodies Induced by Trivalent Influenza Vaccine and H7N9 Infection in Humans. J Virol (2020) 94(4):e01182–19. doi: 10.1128/jvi.01182-19
107. Jiang H, Peng W, Qi J, Chai Y, Song H, Bi Y, et al. Structure-Based Modification of an Anti-Neuraminidase Human Antibody Restores Protection Efficacy Against the Drifted Influenza Virus. MBio (2020) 11(5):e02315–20. doi: 10.1128/mBio.02315-20
108. Gulati U, Hwang C-C, Venkatramani L, Gulati S, Stray SJ, Lee JT, et al. Antibody Epitopes on the Neuraminidase of a Recent H3N2 Influenza Virus (A/Memphis/31/98). J Virol (2002) 76(23):12274–80. doi: 10.1128/jvi.76.23.12274-12280.2002?
109. Malby RL, Tulip WR, Harley VR, McKimm-Breschkin JL, Laver WG, Webster RG, et al. The Structure of a Complex Between the NC10 Antibody and Influenza Virus Neuraminidase and Comparison With the Overlapping Binding Site of the NC41 Antibody. Structure (1994) 2(8):733–46. doi: 10.1016/S0969-2126(00)00074-5
110. Tulip WR, Varghese JN, Laver WG, Webster RG, Colman PM. Refined Crystal Structure of the Influenza Virus N9 Neuraminidase-NC41 Fab Complex. J Mol Biol (1992) 227(1):122–48. doi: 10.1016/0022-2836(91)80069-7
111. Wan Z, Ye J, Sang J, Shao H, Qian K, Jin W, et al. Identification of Amino Acids in H9N2 Influenza Virus Neuraminidase That Are Critical for the Binding of Two Mouse Monoclonal Antibodies. Vet Microbiol (2016) 187:58–63. doi: 10.1016/j.vetmic.2016.03.011
112. Kirkpatrick Roubidoux E, McMahon M, Carreño JM, Capuano C, Jiang K, Simon V, et al. Identification and Characterization of Novel Antibody Epitopes on the N2 Neuraminidase. mSphere (2021) 6(1):e00958–20. doi: 10.1128/msphere.00958-20
113. Job ER, Schotsaert M, Ibañez LI, Smet A, Ysenbaert T, Roose K, et al. Antibodies Directed Toward Neuraminidase N1 Control Disease in a Mouse Model of Influenza. J Virol (2018) 92(4):e01584–17. doi: 10.1128/jvi.01584-17
114. Saito T, Taylor G, Laver WG, Kawaoka Y, Webster RG. Antigenicity of the N8 Influenza A Virus Neuraminidase: Existence of an Epitope at the Subunit Interface of the Neuraminidase. J Virol (1994) 68(3):1790–6. doi: 10.1128/jvi.68.3.1790-1796.1994
115. Colman PM, Hoyne PA, Lawrence MC. Sequence and Structure Alignment of Paramyxovirus Hemagglutinin-Neuraminidase With Influenza Virus Neuraminidase. J Virol (1993) 67(6):2972–80. doi: 10.1128/jvi.67.6.2972-2980.1993
116. Schmidt AG, Therkelsen MD, Stewart S, Kepler TB, Liao HX, Moody MA, et al. Viral Receptor-Binding Site Antibodies With Diverse Germline Origins. Cell (2015) 161(5):1026–34. doi: 10.1016/j.cell.2015.04.028
117. Lee PS, Ohshima N, Stanfield RL, Yu W, Iba Y, Okuno Y, et al. Receptor Mimicry by Antibody F045-092 Facilitates Universal Binding to the H3 Subtype of Influenza Virus. Nat Commun (2014) 5:3614. doi: 10.1038/ncomms4614
118. Harmsen M, Blokker J, Pritz-Verschuren S, Bartelink W, van der Burg H, Koch G. Isolation of Panels of Llama Single-Domain Antibody Fragments Binding All Nine Neuraminidase Subtypes of Influenza A Virus. Antibodies (2013) 2(2):168–92. doi: 10.3390/antib2020168
119. Cardoso FM, Ibanez LI, Van den Hoecke S, De Baets S, Smet A, Roose K, et al. Single-Domain Antibodies Targeting Neuraminidase Protect Against an H5N1 Influenza Virus Challenge. J Virol (2014) 88(15):8278–96. doi: 10.1128/jvi.03178-13
120. Barbey-Martin C, Gigant B, Bizebard T, Calder LJ, Wharton SA, Skehel JJ, et al. An Antibody That Prevents the Hemagglutinin Low PH Fusogenic Transition. Virology (2002) 294(1):70–4. doi: 10.1006/viro.2001.1320
121. Iba Y, Fujii Y, Ohshima N, Sumida T, Kubota-Koketsu R, Ikeda M, et al. Conserved Neutralizing Epitope at Globular Head of Hemagglutinin in H3N2 Influenza Viruses. J Virol (2014) 88(13):7130–44. doi: 10.1128/jvi.00420-14
122. Dilillo DJ, Tan GS, Palese P, Ravetch JV. Broadly Neutralizing Hemagglutinin Stalk-Specific Antibodies Require Fcγr Interactions for Protection Against Influenza Virus In Vivo. Nat Med (2014) 20(2):143–51. doi: 10.1038/nm.3443
123. Jegaskanda S, Vanderven HA, Wheatley AK, Kent SJ. Fc or Not Fc; That Is the Question: Antibody Fc-Receptor Interactions Are Key to Universal Influenza Vaccine Design. Hum Vaccines Immunother (2017) 13(6):1–9. doi: 10.1080/21645515.2017.1290018
124. DiLillo DJ, Palese P, Wilson PC, Ravetch JV. Broadly Neutralizing Anti-Influenza Antibodies Require Fc Receptor Engagement for In Vivo Protection. J Clin Invest (2016) 126(2):605–10. doi: 10.1172/JCI84428
125. Henry Dunand CJ, Leon PE, Huang M, Choi A, Chromikova V, Ho IY, et al. Both Neutralizing and Non-Neutralizing Human H7N9 Influenza Vaccine-Induced Monoclonal Antibodies Confer Protection. Cell Host Microbe (2016) 19(6):800–13. doi: 10.1016/j.chom.2016.05.014
126. Job ER, Ysenbaert T, Smet A, Van Hecke A, Meuris L, Kleanthous H, et al. Fcγ Receptors Contribute to the Antiviral Properties of Influenza Virus Neuraminidase-Specific Antibodies. MBio (2019) 10(5):e01667–19. doi: 10.1128/mBio.01395-19
127. Yasuhara A, Yamayoshi S, Kiso M, Sakai-Tagawa Y, Koga M, Adachi E, et al. Antigenic Drift Originating From Changes to the Lateral Surface of the Neuraminidase Head of Influenza A Virus. Nat Microbiol (2019) 4(6):1024–34. doi: 10.1038/s41564-019-0401-1
128. Ito H, Nishimura H, Kisu T, Hagiwara H, Watanabe O, Kadji FMN, et al. Low Response in Eliciting Neuraminidase Inhibition Activity of Sera Among Recipients of a Split, Monovalent Pandemic Influenza Vaccine During the 2009 Pandemic. PloS One (2020) 15(5):e0233001. doi: 10.1371/JOURNAL.PONE.0233001
129. Powers DC, Kilbourne ED, Johansson BE. Neuraminidase-Specific Antibody Responses to Inactivated Influenza Virus Vaccine in Young and Elderly Adults. Clin Diagn Lab Immunol (1996) 3(5):511–6. doi: 10.1128/CDLI.3.5.511-516.1996
130. Couch RB, Atmar RL, Keitel WA, Quarles JM, Wells J, Arden N, et al. Randomized Comparative Study of the Serum Antihemagglutinin and Antineuraminidase Antibody Responses to Six Licensed Trivalent Influenza Vaccines. Vaccine (2012) 31(1):190–5. doi: 10.1016/J.VACCINE.2012.10.065
131. Getie-Kebtie M, Sultana I, Eichelberger M, Alterman M. Label-Free Mass Spectrometry-Based Quantification of Hemagglutinin and Neuraminidase in Influenza Virus Preparations and Vaccines. Influenza Other Respi Viruses (2013) 7(4):521–30. doi: 10.1111/IRV.12001
132. Sultana I, Yang K, Getie-Kebtie M, Couzens L, Markoff L, Alterman M, et al. Stability of Neuraminidase in Inactivated Influenza Vaccines. Vaccine (2014) 32(19):2225–30. doi: 10.1016/J.VACCINE.2014.01.078
133. McMahon M, Strohmeier S, Rajendran M, Capuano C, Ellebedy AH, Wilson PC, et al. Correctly Folded - But Not Necessarily Functional - Influenza Virus Neuraminidase Is Required to Induce Protective Antibody Responses in Mice. Vaccine (2020) 38(45):7129–37. doi: 10.1016/J.VACCINE.2020.08.067
134. Petrie J, Ohmit S, Johnson E, Truscon R, Monto A. Persistence of Antibodies to Influenza Hemagglutinin and Neuraminidase Following One or Two Years of Influenza Vaccination. J Infect Dis (2015) 212(12):1914–22. doi: 10.1093/INFDIS/JIV313
135. Kilbourne E, Couch R, Kasel J, Keitel W, Cate T, Quarles J, et al. Purified Influenza A Virus N2 Neuraminidase Vaccine Is Immunogenic and Non-Toxic in Humans. Vaccine (1995) 13(18):1799–803. doi: 10.1016/0264-410X(95)00127-M
136. Zheng A, Sun W, Xiong X, Freyn AW, Peukes J, Strohmeier S, et al. Enhancing Neuraminidase Immunogenicity of Influenza A Viruses by Rewiring RNA Packaging Signals. J Virol (2020) 94(16):e00742–20. doi: 10.1128/JVI.00742-20
137. Gao J, Wan H, Li X, Martinez MR, Klenow L, Gao Y, et al. Balancing the Influenza Neuraminidase and Hemagglutinin Responses by Exchanging the Vaccine Virus Backbone. PloS Pathog (2021) 17(4):e1009171. doi: 10.1371/JOURNAL.PPAT.1009171
138. Östbye H, Gao J, Martinez MR, Wang H, Gier JW de, Daniels R. N-Linked Glycan Sites on the Influenza A Virus Neuraminidase Head Domain Are Required for Efficient Viral Incorporation and Replication. J Virol (2020) 94(19):e00874–20. doi: 10.1128/JVI.00874-20
139. Broecker F, Zheng A, Suntronwong N, Sun W, Bailey MJ, Krammer F, et al. Extending the Stalk Enhances Immunogenicity of the Influenza Virus Neuraminidase. J Virol (2019) 93(18):e00840–19. doi: 10.1128/JVI.00840-19
140. Deng X, Wang Q, Liu M, Zheng Q, Wu F, Huang J. Tetrameric Neuraminidase of Influenza A Virus Is Required to Induce Protective Antibody Responses in Mice. Front Microbiol (2021) 12:729914. doi: 10.3389/FMICB.2021.729914
141. Sultana I, Gao J, Markoff L, Eichelberger MC. Influenza Neuraminidase-Inhibiting Antibodies Are Induced in the Presence of Zanamivir. Vaccine (2011) 29(14):2601–6. doi: 10.1016/J.VACCINE.2011.01.047
142. Gao J, Klenow L, Parsons L, Malik T, Phue J-N, Gao Z, et al. Design of the Recombinant Influenza Neuraminidase Antigen Is Crucial for Protective Efficacy. J Virol (2021), JVI0116021. doi: 10.1101/2021.04.29.442077
143. Schotsaert M, Ysenbaert T, Smet A, Schepens B, Vanderschaeghe D, Stegalkina S, et al. Long-Lasting Cross-Protection Against Influenza A by Neuraminidase and M2e-Based Immunization Strategies. Sci Rep (2016) 6:24402. doi: 10.1038/SREP24402
144. Liu WC, Lin CY, Tsou YT, Jan JT, Wu SC. Cross-Reactive Neuraminidase-Inhibiting Antibodies Elicited by Immunization With Recombinant Neuraminidase Proteins of H5N1 and Pandemic H1N1 Influenza A Viruses. J Virol (2015) 89(14):7224–34. doi: 10.1128/JVI.00585-15
145. Schmidt PM, Attwood RM, Mohr PG, Barrett SA, McKimm-Breschkin JLA. Generic System for the Expression and Purification of Soluble and Stable Influenza Neuraminidase. PloS One (2011) 6(2):e16284. doi: 10.1371/JOURNAL.PONE.0016284
146. Dai M, Guo H, Dortmans JCFM, Dekkers J, Nordholm J, Daniels R, et al. Identification of Residues That Affect Oligomerization and/or Enzymatic Activity of Influenza Virus H5N1 Neuraminidase Proteins. J Virol (2016) 90(20):9457. doi: 10.1128/JVI.01346-16
147. Streltsov VA, Schmidt PM, McKimm-Breschkin JL. Structure of an Influenza A Virus N9 Neuraminidase With a Tetrabrachion-Domain Stalk. Acta Crystallogr Sect F Struct Biol Commun (2019) 75(Pt 2):89–97. doi: 10.1107/S2053230X18017892
148. Strohmeier S, Carreño JM, Brito RN, Krammer F. Introduction of Cysteines in the Stalk Domain of Recombinant Influenza Virus N1 Neuraminidase Enhances Protein Stability and Immunogenicity in Mice. Vaccines (2021) 9(4):404. doi: 10.3390/VACCINES9040404
149. Ellis D, Lederhofer J, Acton OJ, Tsybovsky Y, Kephart S, Yap C, et al. Structure-Based Design of Stabilized Recombinant Influenza Neuraminidase Tetramers. bioRxiv (2021), 2021.05.17.444468. doi: 10.1101/2021.05.17.444468
150. Bullard BL, Weaver EA. Strategies Targeting Hemagglutinin as a Universal Influenza Vaccine. Vaccines (2021) 9(3):257. doi: 10.3390/VACCINES9030257
151. Job ER, Ysenbaert T, Smet A, Christopoulou I, Strugnell T, Oloo EO, et al. Broadened Immunity Against Influenza by Vaccination With Computationally Designed Influenza Virus N1 Neuraminidase Constructs. NPJ Vaccines (2018) 3:55. doi: 10.1038/S41541-018-0093-1
152. Skarlupka AL, Bebin-Blackwell A-G, Sumner SF, Ross TM. Universal Influenza Virus Neuraminidase Vaccine Elicits Protective Immune Responses Against Human Seasonal and Pre-Pandemic Strains. J Virol (2021) 95(17):e0075921. doi: 10.1128/JVI.00759-21
153. Bachmann MF, Jennings GT. Vaccine Delivery: A Matter of Size, Geometry, Kinetics and Molecular Patterns. Nat Rev Immunol (2010) 10(11):787–96. doi: 10.1038/NRI2868
154. Smith GE, Sun X, Bai Y, Liu YV, Massare MJ, Pearce MB, et al. Neuraminidase-Based Recombinant Virus-Like Particles Protect Against Lethal Avian Influenza A(H5N1) Virus Infection in Ferrets. Virology (2017) 509:90–7. doi: 10.1016/J.VIROL.2017.06.006
155. Lai JCC, Chan WWL, Kien F, Nicholls JM, Peiris JSM, Garcia J-M. Formation of Virus-Like Particles From Human Cell Lines Exclusively Expressing Influenza Neuraminidase. J Gen Virol (2010) 91(9):2322–30. doi: 10.1099/VIR.0.019935-0
156. Tao P, Luo M, Zhu D, Qu S, Yang Z, Gao M, et al. Virus-Like Particle Vaccine Comprised of the HA, NA, and M1 Proteins of an Avian Isolated H5N1 Influenza Virus Induces Protective Immunity Against Homologous and Heterologous Strains in Mice. Viral Immunol (2009) 22(4):273–81. doi: 10.1089/VIM.2009.0017
157. Wu C-Y, Yeh Y-C, Chan J-T, Yang Y-C, Yang J-R, Liu M-T, et al. A VLP Vaccine Induces Broad-Spectrum Cross-Protective Antibody Immunity Against H5N1 and H1N1 Subtypes of Influenza A Virus. PloS One (2012) 7(8):e42363. doi: 10.1371/JOURNAL.PONE.0042363
158. Easterbrook JD, Schwartzman LM, Gao J, Kash JC, Morens DM, Couzens L, et al. Protection Against a Lethal H5N1 Influenza Challenge by Intranasal Immunization With Virus-Like Particles Containing 2009 Pandemic H1N1 Neuraminidase in Mice. Virology (2012) 432(1):39–44. doi: 10.1016/J.VIROL.2012.06.003
159. Kim KH, Lee YT, Park S, Jung YJ, Lee Y, Ko EJ, et al. Neuraminidase Expressing Virus-Like Particle Vaccine Provides Effective Cross Protection Against Influenza Virus. Virology (2019) 535:179–88. doi: 10.1016/J.VIROL.2019.07.008
160. Menne Z, Pliasas VC, Compans RW, Glover S, Kyriakis CS, Skountzou I. Bivalent Vaccination With NA1 and NA2 Neuraminidase Virus-Like Particles Is Protective Against Challenge With H1N1 and H3N2 Influenza A Viruses in a Murine Model. Virology (2021) 562:197–208. doi: 10.1016/J.VIROL.2021.08.001
161. Liu J, Ren Z, Wang H, Zhao Y, Wilker PR, Yu Z, et al. Influenza Virus-Like Particles Composed of Conserved Influenza Proteins and GPI-Anchored CCL28/GM-CSF Fusion Proteins Enhance Protective Immunity Against Homologous and Heterologous Viruses. Int Immunopharmacol (2018) 63:119–28. doi: 10.1016/J.INTIMP.2018.07.011
162. Skountzou I, Quan F-S, Gangadhara S, Ye L, Vzorov A, Selvaraj P, et al. Incorporation of Glycosylphosphatidylinositol-Anchored Granulocyte- MacrophageColony-Stimulating Factor or CD40 Ligand Enhances Immunogenicity of Chimeric Simian Immunodeficiency Virus-Like Particles. J Virol (2007) 81(3):1083–94. doi: 10.1128/JVI.01692-06
163. Patel JM, Kim MC, Vartabedian VF, Lee YN, He S, Song JM, et al. Protein Transfer-Mediated Surface Engineering to Adjuvantate Virus-Like Nanoparticles for Enhanced Anti-Viral Immune Responses. Nanomedicine (2015) 11(5):1097–107. doi: 10.1016/J.NANO.2015.02.008
164. Wang Y, Deng L, Gonzalez GX, Luthra L, Dong C, Ma Y, et al. Double-Layered M2e-NA Protein Nanoparticle Immunization Induces Broad Cross-Protection Against Different Influenza Viruses in Mice. Adv Healthc Mater (2020) 9(2):1901176. doi: 10.1002/ADHM.201901176
165. Ueda G, Antanasijevic A, Fallas JA, Sheffler W, Copps J, Ellis D, et al. Tailored Design of Protein Nanoparticle Scaffolds for Multivalent Presentation of Viral Glycoprotein Antigens. Elife (2020) 9:1–30. doi: 10.7554/ELIFE.57659
166. Kanekiyo M, Wei C-J, Yassine HM, McTamney PM, Boyington JC, Whittle JRR, et al. Self-Assembling Influenza Nanoparticle Vaccines Elicit Broadly Neutralizing H1N1 Antibodies. Nature (2013) 499(7456):102–6. doi: 10.1038/NATURE12202
167. Marcandalli J, Fiala B, Ols S, Perotti M, Schueren W devd, Snijder J, et al. Induction of Potent Neutralizing Antibody Responses by a Designed Protein Nanoparticle Vaccine for Respiratory Syncytial Virus. Cell (2019) 176(6):1420–31. doi: 10.1016/J.CELL.2019.01.046
168. Kanekiyo M, Joyce MG, Gillespie RA, Gallagher JR, Andrews SF, Yassine HM, et al. Mosaic Nanoparticle Display of Diverse Influenza Virus Hemagglutinins Elicits Broad B Cell Responses. Nat Immunol (2019) 20(3):362–72. doi: 10.1038/S41590-018-0305-X
169. Kim KH, Jung YJ, Lee Y, Park BR, Oh J, Lee YN, et al. Cross Protection by Inactivated Recombinant Influenza Viruses Containing Chimeric Hemagglutinin Conjugates With a Conserved Neuraminidase or M2 Ectodomain Epitope. Virology (2020) 550:51–60. doi: 10.1016/J.VIROL.2020.08.003
170. Zeigler DF, Gage E, Clegg CH. Epitope-Targeting Platform for Broadly Protective Influenza Vaccines. PloS One (2021) 16(5):e0252170. doi: 10.1371/JOURNAL.PONE.0252170
171. Behbahani M, Moradi M, Mohabatkar H. In Silico Design of a Multi-Epitope Peptide Construct as a Potential Vaccine Candidate for Influenza A Based on Neuraminidase Protein. Silico Pharmacol (2021) 9(1):36. doi: 10.1007/S40203-021-00095-W
172. Rauch S, Jasny E, Schmidt KE, Petsch B. New Vaccine Technologies to Combat Outbreak Situations. Front Immunol (2018) 9:1963. doi: 10.3389/FIMMU.2018.01963
173. Hessel A, Schwendinger M, Fritz D, Coulibaly S, Holzer GW, Sabarth N, et al. A Pandemic Influenza H1N1 Live Vaccine Based on Modified Vaccinia Ankara Is Highly Immunogenic and Protects Mice in Active and Passive Immunizations. PloS One (2010) 5(8):e12217. doi: 10.1371/JOURNAL.PONE.0012217
174. Kingstad-Bakke B, Kamlangdee A, Osorio JE. Mucosal Administration of Raccoonpox Virus Expressing Highly Pathogenic Avian H5N1 Influenza Neuraminidase Is Highly Protective Against H5N1 and Seasonal Influenza Virus Challenge. Vaccine (2015) 33(39):5155–62. doi: 10.1016/J.VACCINE.2015.08.005
175. Mooney AJ, Gabbard JD, Li Z, Dlugolenski DA, Johnson SK, Tripp RA, et al. Vaccination With Recombinant Parainfluenza Virus 5 Expressing Neuraminidase Protects Against Homologous and Heterologous Influenza Virus Challenge. J Virol (2017) 91(23):e01579–17. doi: 10.1128/JVI.01579-17
176. Sylte M, Hubby B, Suarez D. Influenza Neuraminidase Antibodies Provide Partial Protection for Chickens Against High Pathogenic Avian Influenza Infection. Vaccine (2007) 25(19):3763–72. doi: 10.1016/J.VACCINE.2007.02.011
177. Ramp K, Veits J, Deckers D, Rudolf M, Grund C, Mettenleiter TC, et al. Coexpression of Avian Influenza Virus H5 and N1 by Recombinant Newcastle Disease Virus and the Impact on Immune Response in Chickens. Avian Dis (2011) 55(3):413–21. doi: 10.1637/9652-011111-REG.1
178. Nayak B, Kumar S, DiNapoli J, Paldurai A, Perez D, Collins P, et al. Contributions of the Avian Influenza Virus HA, NA, and M2 Surface Proteins to the Induction of Neutralizing Antibodies and Protective Immunity. J Virol (2010) 84(5):2408–20. doi: 10.1128/JVI.02135-09
179. Pavlova S, Veits J, Keil G, Mettenleiter T, Fuchs W. Protection of Chickens Against H5N1 Highly Pathogenic Avian Influenza Virus Infection by Live Vaccination With Infectious Laryngotracheitis Virus Recombinants Expressing H5 Hemagglutinin and N1 Neuraminidase. Vaccine (2009) 27(5):773–85. doi: 10.1016/J.VACCINE.2008.11.033
180. Pardi N, Hogan MJ, Porter FW, Weissman D. MRNA Vaccines — a New Era in Vaccinology. Nat Rev Drug Discovery (2018) 17(4):261–79. doi: 10.1038/NRD.2017.243
181. Xu Z, Patel A, Tursi NJ, Zhu X, Muthumani K, Kulp DW, et al. Harnessing Recent Advances in Synthetic DNA and Electroporation Technologies for Rapid Vaccine Development Against COVID-19 and Other Emerging Infectious Diseases. Front Med Technol (2020) 0:571030. doi: 10.3389/FMEDT.2020.571030
182. Baden LR, Sahly HM El, Essink B, Kotloff K, Frey S, Novak R, et al. Efficacy and Safety of the MRNA-1273 SARS-CoV-2 Vaccine. N Eng J Med (2021) 384(5):403–16. doi: 10.1056/NEJMOA2035389
183. Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, et al. Safety and Efficacy of the BNT162b2 MRNA Covid-19 Vaccine. N Eng J Med (2020) 383(27):2603–15. doi: 10.1056/NEJMOA2034577
184. Chen Z, Sahashi Y, Matsuo K, Asanuma H, Takahashi H, Iwasaki T, et al. Comparison of the Ability of Viral Protein-Expressing Plasmid DNAs to Protect Against Influenza. Vaccine (1998) 16(16):1544–9. doi: 10.1016/S0264-410X(98)00043-7
185. Chen Z, Kadowaki SE, Hagiwara Y, Yoshikawa T, Matsuo K, Kurata T, et al. Cross-Protection Against a Lethal Influenza Virus Infection by DNA Vaccine to Neuraminidase. Vaccine (2000) 18(28):3214–22. doi: 10.1016/S0264-410X(00)00149-3
186. Sandbulte MR, Jimenez GS, Boon ACM, Smith LR, Treanor JJ, Webby RJ. Cross-Reactive Neuraminidase Antibodies Afford Partial Protection Against H5N1 in Mice and Are Present in Unexposed Humans. PloS Med (2007) 4(2):e59. doi: 10.1371/JOURNAL.PMED.0040059
187. Torrieri-Dramard L, Lambrecht B, Ferreira HL, Van Den Berg T, Klatzmann D, Bellier B. Intranasal DNA Vaccination Induces Potent Mucosal and Systemic Immune Responses and Cross-Protective Immunity Against Influenza Viruses. Mol Ther (2011) 19(3):602–11. doi: 10.1038/MT.2010.222
188. Petsch B, Schnee M, Vogel AB, Lange E, Hoffmann B, Voss D, et al. Protective Efficacy of In Vitro Synthesized, Specific MRNA Vaccines Against Influenza A Virus Infection. Nat Biotechnol (2012) 30(12):1210–6. doi: 10.1038/NBT.2436
189. Freyn AW, Pine M, Rosado VC, Benz M, Muramatsu H, Beattie M, et al. Antigen Modifications Improve Nucleoside-Modified MRNA-Based Influenza Virus Vaccines in Mice. Mol Ther - Methods Clin Dev (2021) 22:84–95. doi: 10.1016/J.OMTM.2021.06.003
190. Freyn AW, Silva JR da, Rosado VC, Bliss CM, Pine M, Mui BL, et al. A Multi-Targeting, Nucleoside-Modified MRNA Influenza Virus Vaccine Provides Broad Protection in Mice. Mol Ther (2020) 28(7):1569–84. doi: 10.1016/J.YMTHE.2020.04.018
191. Bloom K, Berg Fvd, Arbuthnot P. Self-Amplifying RNA Vaccines for Infectious Diseases. Gene Ther (2021) 28(3–4):117–29. doi: 10.1038/S41434-020-00204-Y
192. Blakney AK, Ip S, Geall AJ. An Update on Self-Amplifying MRNA Vaccine Development. Vaccines (2021) 9(2):97. doi: 10.3390/VACCINES9020097
Keywords: influenza, neuraminidase, antigenic drift, monoclonal antibodies, correlate of protection, vaccines
Citation: Creytens S, Pascha MN, Ballegeer M, Saelens X and de Haan CAM (2021) Influenza Neuraminidase Characteristics and Potential as a Vaccine Target. Front. Immunol. 12:786617. doi: 10.3389/fimmu.2021.786617
Received: 30 September 2021; Accepted: 29 October 2021;
Published: 16 November 2021.
Edited by:
Corey Patrick Mallett, GlaxoSmithKline (USA), United StatesReviewed by:
Irina V. Kiseleva, Institute of Experimental Medicine (RAS), RussiaMarion Russier, Max Planck Institute of Biochemistry, Germany
Copyright © 2021 Creytens, Pascha, Ballegeer, Saelens and de Haan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cornelis A. M. de Haan, Qy5BLk0uZGVIYWFuQHV1Lm5s; Xavier Saelens, eGF2aWVyLnNhZWxlbnNAdmliLXVnZW50LmJl
†These authors share first authorship
 Sarah Creytens1,2†
Sarah Creytens1,2† Mirte N. Pascha
Mirte N. Pascha Marlies Ballegeer
Marlies Ballegeer Xavier Saelens
Xavier Saelens Cornelis A. M. de Haan
Cornelis A. M. de Haan