
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 21 December 2021
Sec. Inflammation
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.780779
This article is part of the Research Topic Epigenetics of the Immune Component of Inflammation View all 43 articles
 Huan Yang1,2,3,4†
Huan Yang1,2,3,4† Jie He1,3,4†
Jie He1,3,4† Shuai Huang1,3,4†
Shuai Huang1,3,4† Hongbing Yang2
Hongbing Yang2 Qingjie Yi2,5
Qingjie Yi2,5 Yuelan Tao1,3,4
Yuelan Tao1,3,4 Miaomiao Chen1,3,4
Miaomiao Chen1,3,4 Xuemei Zhang1,3,4*
Xuemei Zhang1,3,4* Hongbo Qi1,3,4*
Hongbo Qi1,3,4*Many studies have confirmed that extrachromosomal circular DNAs (eccDNAs/ecDNAs) exist in tumor and normal cells independently of the chromosome and are essential for oncogene plasticity and drug resistance. Studies have confirmed that there are many eccDNAs/ecDNAs in maternal plasma derived from the fetus. Fetal growth restriction (FGR) is a pregnancy-related disease associated with high newborn morbidity and mortality. However, the characteristics and nature of eccDNAs/ecDNAs in FGR are poorly understood. This study aims to deconstruct the properties and potential functions of eccDNAs/ecDNAs in FGR. We performed circle-seq to identify the expression profile of eccDNAs/ecDNAs, analyzed by bioinformatics, and verified by real-time Polymerase Chain Reaction (PCR) combined with southern blot in FGR compared with the normal groups. A total of 45,131 eccDNAs/ecDNAs (including 2,118 unique ones) were identified, which had significantly higher abundance in FRG group than in normal group, and was bimodal in length, peaking at ~146bp and ~340bp, respectively. Gestational age may be one independent factor affecting the production of eccDNAs/ecDNAs, most of which come from genomic regions with high gene density, with a 4~12bp repeat around the junction, and their origin had a certain genetic preference. In addition, some of the host-genes overlapped with non-coding RNAs (ncRNAs) partially or even completely. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis revealed that host-genes on the differentially expressed eccDNAs/ecDNAs (DEEECs/DEECs) were mainly enriched in immune-related functions and pathways. The presence of some ecDNAs were verified, and whose variability were consistent with the circle-seq results. We identified and characterized eccDNAs/ecDNAs in placentas with FGR, and elucidated the formation mechanisms and the networks with ncRNAs, which provide a new vision for the screening of new biomarkers and therapeutic targets for FGR.
Extrachromosomal circular DNAs(eccDNAs/ecDNAs) was first described by Alix Bassel more than 50 years ago (1), which widely exist in eukaryotic cells such as yeast, nematodes, ciliates, plants, and mammals (2–5), and has homology with chromosomal DNA. They have extensive heterogeneity in length, number, and origin due to different types and genetic backgrounds of cells and tissues (6). EccDNAs/ecDNAs are tissue-specific in mammals, which may lead to senescence and participate in gene compensation and intercellular communication. According to the size of the circular chromosome, we usually refer to the small circular DNA entities as eccDNAs (2, 7) and the large entity as ecDNAs (8–10). The latter typically contain oncogenes or drug resistance genes, which leads to oncogene amplification and is a powerful driving force for tumor heterogeneity (9, 11, 12). In addition, Dutta et al. found that eccDNAs can also produce small regulatory RNAs to regulate gene expression in a promoter-independent manner (13). Dennis Lo and his colleagues have identified and characterized fetal-derived eccDNAs in maternal plasma with a high abundance, indicating that it can be used as a potential serological marker for non-invasive prenatal diagnosis (14). However, the characteristics of eccDNAs/ecDNAs in the placenta are unknown, and the effect of cyclization on gene remodeling mediates pregnancy-related diseases is unclear.
Fetal growth restriction (FGR) refers to the failure of the fetus to reach its due growth potential in the maternal uterus due to various adverse reasons, which is a severe complication that endangers the safety of perinatal babies (15). The diagnostic criteria are that the fetal weight is lower than the 10th percentile for the same gestational age (16). Birth weight lower than the 3rd percentile is regarded as severe FGR, which is also directly related to severe adverse pregnancy outcomes of the fetus (17, 18). The pathogenesis of FGR is complex, with placental dysgenesis being an essential pathogenetic foundation (19). Therefore, we are intrigued by the nature and origin of eccDNAs/ecDNAs derived from the placenta with FGR.
In this study, we used Circle-Seq to extract, sequence, and locate extracellular cyclization elements in the placenta from the normal and severe FGR groups. To facilitate subsequent analysis, we divided the circular entities into eccDNAs and ecDNAs with the length of 100kb as the boundary. This study found that: (1) EccDNAs/ecDNAs is abundant and significant individual heterogeneity in the placenta, with higher abundance in the FGR group. (2) The formation of eccDNAs/ecDNAs is not a random incident and has some genetic preference, with microhomologous repeats as its main feature. (3) EccDNAs as a potential driver of FGR through immune signaling pathways and a complex network with non-coding RNAs (ncRNAs). We have validated by real-time Polymerase Chain Reaction (PCR) and Southern blot that the relevant ecDNAs are indeed present in the placenta. Our results provide an unprecedented view of these enigmatic molecules in the placenta during gestation, but one that remains naive.
This study was approved by the Ethics Committee of the First Affiliated Hospital of Chongqing Medical University (No: 2019-136), Chongqing, China. Informed written consent was obtained from all patients. We recruited three pregnant women with Severe FGR and three healthy pregnant women in this study, whose detailed clinical characteristics of patients are shown in Supplementary Table 1. Severe FGR was defined as an estimated fetal weight below the 3rd percentile for gestational age and confirmed after birth (17, 18). All participants were singleton pregnancy and underwent cesarean section without any major pregnancy complications, such as premature labor or preeclampsia, gestational diabetes mellitus, renal disease, premature rupture of membranes, etc. Placental tissues were collected immediately after delivery, then snap-frozen in liquid nitrogen and stored at -80°C for preservation.
Total genomic DNA was extracted from the placenta tissue samples according to the Cetyl trimethyl ammonium bromide (CATB), and its concentrations were measured using a NanoDrop Microvolume Spectrophotometer (Thermo Scientific). Qualified DNA samples (OD260/280 values between 1.8 and 2.0) were used for further experiments.
For each sample, the remaining linear genomic DNA was removed by exonuclease V (New England Biolabs) following the manufacturer’s instructions to enrich circular DNA. After rolling circle amplification (RCA) of circular DNA as previously described (20), the resultant circular DNA was digested with MspI (New England Biolabs) according to the manufacturer’s instructions. Then the next step is to implement library sequencing.
We used 1ug amplified Circular DNA for paired-end (PE) library construction using the NEBNext DNA Library Prep Master Mix set for Illumina (New England Biolabs). PE high-throughput sequencing (150 cycles) was performed according to the manufacturer’s protocol (Illumina), and 150bp (PE150) on both ends of the library was sequenced by Illumina Novaseq 6000(Genedenovo Bio, Guangzhou, China).
Reads obtained from the sequencing machines included raw reads containing adapters or low-quality bases, which would affect the following assembly and analysis. fastp software (21) was used to get high-quality clean reads according to four stringent filtering standards: (1) Removing reads containing adapters; (2) Removing reads containing more than 10% of unknown nucleotides; (3) Removing reads that are all A bases;(4) Removing low quality reads containing more than 50% of low quality (Q-value ≤ 20) bases.
The circular DNA analysis software Circle-map was used for eccDNAs/ecDNAs detection (20). The basic principle of Circle-map was used for discordant reads pair across the circular DNA interface to locate the position of the circular DNA interface. Then, the soft clipped read was used to determine the exact position of the circular DNA interface. However, both discordant reads pair and soft-clipped read can quantify the subsequent eccDNAs/ecDNAs. In addition, the software will also comprehensively consider the sequencing reads coverage of the eccDNAs/ecDNAs interval and the change of sequencing depth compared with the peripheral interval (theoretically, the sequencing depth of the eccDNAs/ecDNAs interval will increase) to judge the reliability of eccDNAs/ecDNAs. Circular DNA with a reserved length of < 100kb is defined as eccDNA, and the length of circular DNA between 100kb and 10M was identified as ecDNA for the subsequent analysis, respectively. The detected split reads in at least one sample are greater than or equal to one.
The software Burrows-Wheeler Aligner(BWA) (22) was used to align the clean reads to Ensembl release 98(Human genes, GRch38.p13). Picard Tools was used to sort the results and mark the repeated sequences, and BEDTools (23) was used to make genome coverage statistics.
To analyze eccDNAs/ecDNAs distribution in the whole-genome chromosome region, we split each chromosome into 50kb windows, counted the number of Circular DNA in each window, and showed it with a Manhattan chart. We mapped the sequenced base sequences with the whole-genome sequences and referred to the matching motifs as overlapping motifs. According to the source region of circular DNA, we annotated eccDNAs/ecDNAs. We divided the genome into 5 ‘ UTR, coding-exon, Exon plus, introns, 3’ UTR, Genic, Genic_ up/down 2000bp, CpG Island, CpG Island_ up/down 2000bp, transposons, Alu regions, and then annotated eccDNAs/ecDNAs according to which region it belongs. If eccDNAs/ecDNAs come from coding gene regions (including Exon, intron, gene_up2000 bp, and gene_down2000 bp), we referred to such coding genes as eccDNAs/ecDNAs-related genes. We annotated the coding genes related to eccDNAs/ecDNAs. The common and unique eccDNAs/ecDNAs-related genes was compared among samples, as long as eccDNAs/ecDNAs is detected in any sample in a certain treatment group in the genomic region where a certain gene is located, the gene will be judged as the eccDNAs/ecDNAs -related gene.
BEDTools combined with an in-house algorithm were used to determine the median percent GC of each identified eccDNAs/ecDNAs and their up-stream and down-stream regions with the same length as themself.
For all detected eccDNAs/ecDNAs, the sum of split reads and discordant reads pair of each eccDNAs/ecDNAs was regarded as the number of effective interface positions Tags supporting eccDNAs/ecDNAs, and based on which Tags per million (TPM) value of each eccDNAs/ecDNAs was calculated using the following formula: TPM= T*1000000/M (T: the number of influential interface positions Tags supporting an eccDNAs/ecDNAs; M: the total number of effective interface positions Tags of all eccDNAs/ecDNAs).
Based on the sequencing information, we used R to carry out Principal Component Analysis (PCA), using dimension reduction to study the distance relationship between samples. We also took the expression of each eccDNAs/ecDNAs in any two samples, calculated their Pearson correlation coefficient, and then visually displayed their correlation in the form of a heat map.
The input data of eccDNAs/ecDNAs differential expression analysis were eccDNAs/ecDNAs tag count data, which was analyzed by edgeR software (24). The analysis was mainly divided into three parts: (1) normalized the tag count; (2) Calculated the probability of hypothesis testing (p-value) according to the model; (3) Multiple hypothesis testing and correction for obtaining FDR value (error detection rate). Based on the differential analysis results, we selected eccDNAs/ecDNAs with P-value<0.05 and |log2FC|>1 as the differentially expressed eccDNAs/ecDNAs (DEEECs/DEECs), for which Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of related host-genes were consequently carried out.
We performed GO and KEGG pathway enrichment analysis by using the “Clusterprofiler” R package to investigate the biological characteristics of these DEEECs/DEECs related host-genes.
To explore the motif patterns on both sides of the eccDNAs/ecDNAs junction, we located the base composition of the up-stream and down-stream 20bp at the start and end positions of each eccDNAs/ecDNAs. The 20bp up-stream and down-stream of these eccDNAs/ecDNAs genome coordinates were output to BED file, and then the function “BEDTools getFasta” was used to extract all relevant sequences (23), which were further analyzed by R (14, 25).
To investigate the relationship of circular DNA with transposon elements (TEs) and ncRNAs, we downloaded annotations for TEs, lncRNAs, miRNAs, snoRNAs from Ensembl release 98 (Human genes, GRCh38.p13), and circRNAs from circBase (26), respectively. Then, the coordinates of TEs and ncRNAs were compared with that of eccDNAs/ecDNAs using the R/Bioconductor GenomicRanges and the SplicingTypesAnno packages.
Outward PCR oligonucleotides were designed on NCBI and Primer5 and the DEEECs/DEECs were targeted to design primers that can produce products across-junction (Supplementary Table 2). Total genomic DNA from 6 normal and 6 FGR placentas (screening criteria as above) was obtained by CTAB. Linear genomic DNA was digested with restriction exonuclease V. The remaining looped elements were PCR amplified using specific primers across the junction, and the product was loaded on a 2% agarose gel for Southern bolt electrophoresis.
SPSS 26.0 statistical software was used for statistical description and analysis with a=0.05 as the test level. Statistical descriptions were expressed as mean ± standard (x̄ ± s) deviation. The statistically significant differences using the independent-sample t-test. P < 0.05 was considered statistically significant.
Purification, enrichment, and detection of eccDNAs/ecDNAs from placental samples were performed in four steps (Figure 1). A total of 41,247 eccDNAs and 3,884 ecDNAs were found in 6 samples, of which the number of unique eccDNAs and ecDNAs was 1,981 and 207, respectively (Table 1). We discovered that these circular elements were highly dynamic, and the result of Veen analysis in the normal and the FRG groups revealed only 188 eccDNAs and 52 ecDNAs overlaps (Figures 2A, B). We found that circular DNA can derive from any chromosome, and surprisingly chromosome 17 contains 193 unique eccDNAs and 67 unique ecDNAs, which was more than any other chromosome (Figure 3A and Supplementary Table 3). The result may be related to the fact that chromosome 17 carries more coding genes than other chromosomes, especially those associated with DNA damage repair (e.g., TP53) (11, 27), consistent with the results previously detected in normal human muscles and blood (20). We found that the length of eccDNAs in the placenta varies greatly but concentrates upon 100 ~ 400bp with two peaks at ~146bp and ~340bp, respectively (Figures 3B, C). The length of eccDNAs around the mononucleosome or polynucleosome, suggested that DNA wrap around one or more nucleosomes may contribute to extrachromosomal circular DNA formation (7, 28), in line with previous reports on the characteristics of eccDNAs in maternal plasma derived from fetuses (14). In addition, we also found that many AA/AT/TT dinucleotide repeats were around the junction of cyclic elements (Supplementary Data 7A, B). CpG islands were periodically acid-interrupted by AA/AT/TT dinucleotides, which were characteristic of sequences that preferentially assemble into nucleosomes (29).
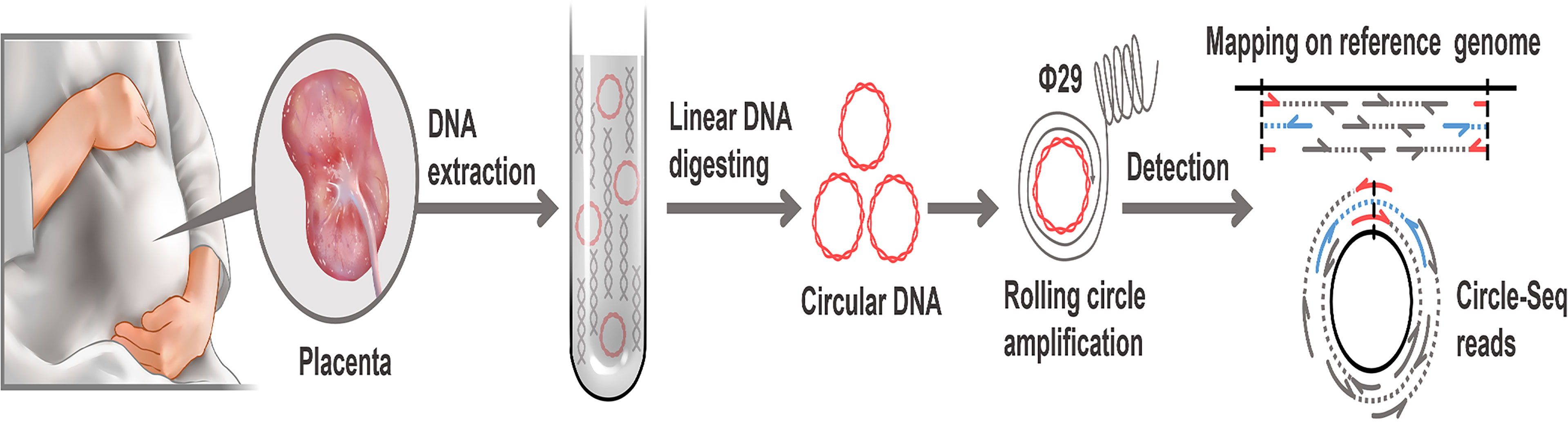
Figure 1 Workflow of eccDNAs/ecDNAs identification in placentas. Placentas were collected after elective deliveries. Total DNA from the placenta including linear and circular was extracted. linear DNA was digested off using exonuclease V. After the RCA of eccDNAs/ecDNAs. Then paired-end sequencing, localization and detection of eccDNAs/ecDNAs with paired-end reads (concordant reads, gray; discordant reads, blue. soft-clipped, split, red). Sequencing data were processed using bioinformatics for identification.
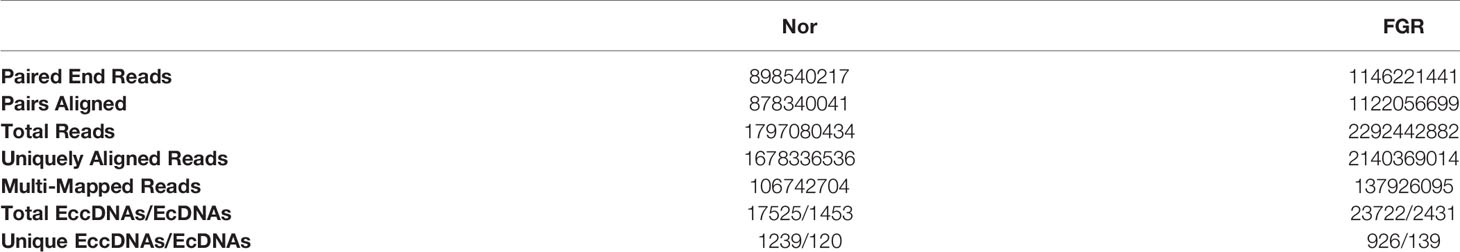
Table 1 Summary of eccDNAs and ecDNAs in placental tissues sequencing and mapping to the Human genome.
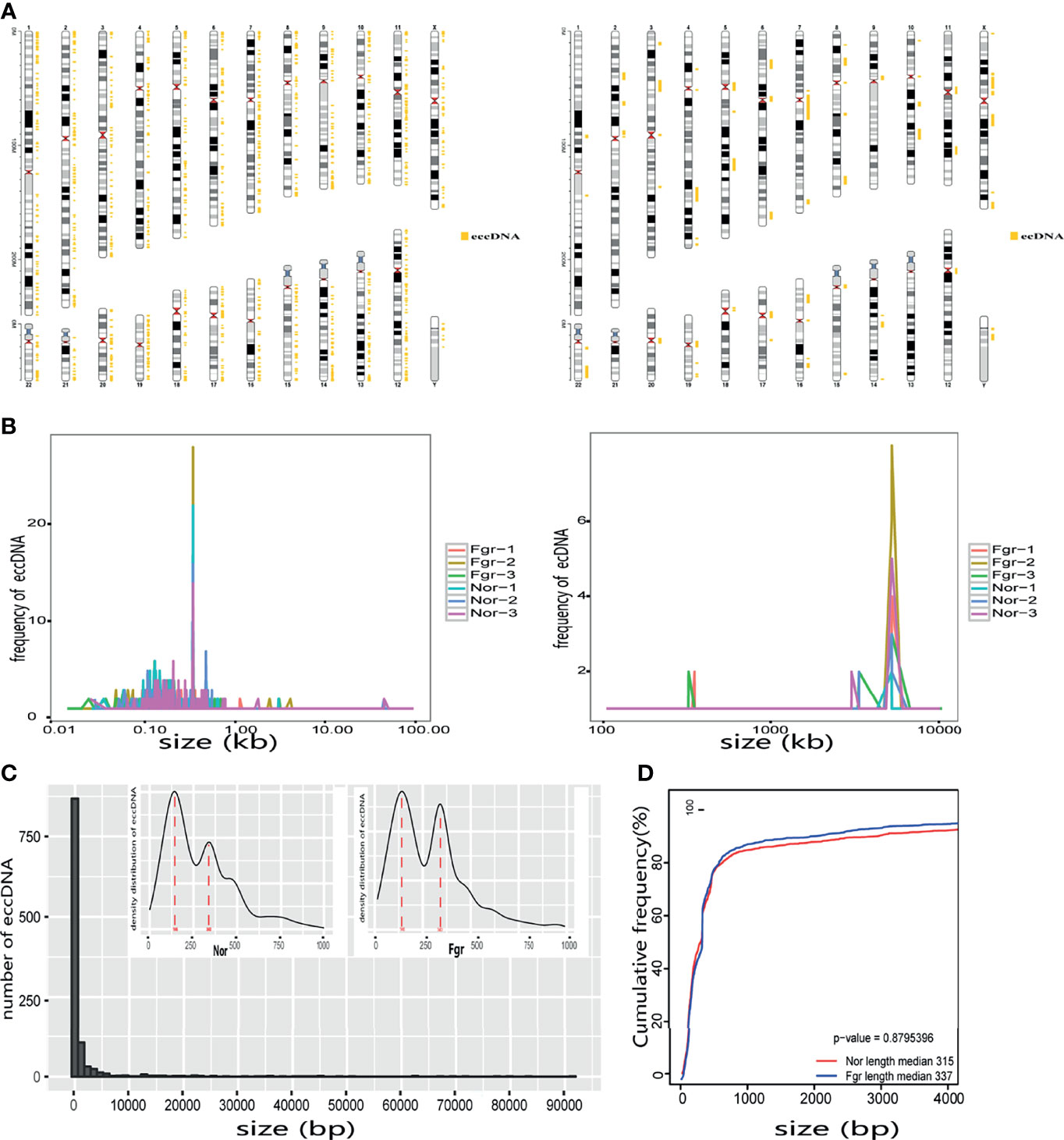
Figure 2 Expression profiles of eccDNAs/ecDNAs between FGR and normal groups. (A) Some eccDNAs/ecDNAs are shared between FGR and normal groups. (B) Hierarchical clustering of eccDNAs/ecDNAs profiles from six samples. (C) Heatmap showing the DEEECs/DEECs between FGR and normal groups.
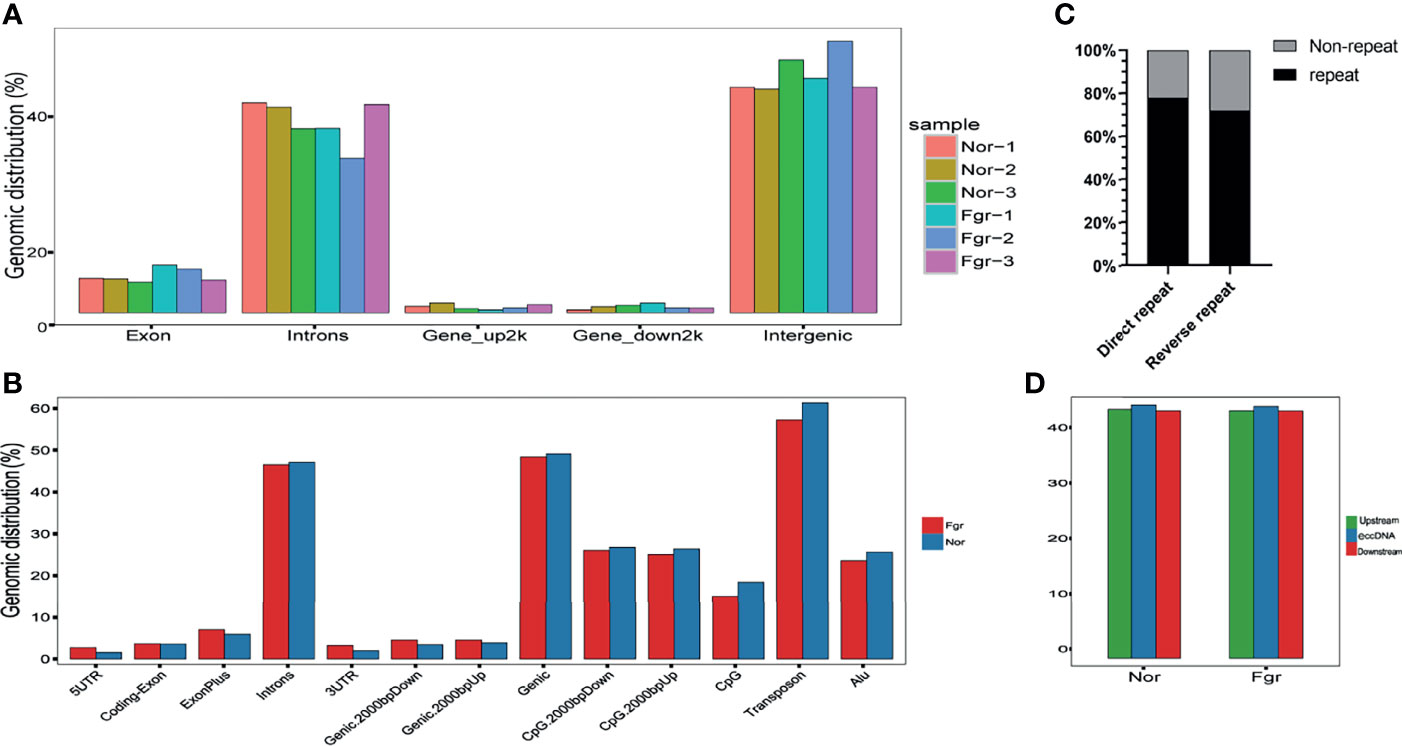
Figure 3 Size distribution of placental-derived eccDNAs/ecDNAs and their location on different chromosomes. (A) The distribution of eccDNAs/ecDNAs in different chromosomes. (B) Size profiling of total eccDNAs/ecDNAs in the placentas. (C) The length density distribution of eccDNAs of FGR and normal groups. There are three peaks at 146 bp, and 340 bp, respectively. (D) Cumulative frequency plot of eccDNAs in FGR and normal placentas. Sizes were capped at 4000bp for both groups of molecules.
To understand the origin of circular DNA in the genome and whether its characteristics affect its formation, we mapped the total population of eccDNAs/ecDNAs detected onto the whole-genome chromosome region, showing that 50.5% of eccDNAs and 50.8% of ecDNAs overlapped with gene regions (Figure 4A and Supplementary Figure 1A). Within the overlapping genomic motifs, they were concentrated in gene regions, CpG islands, introns, and TEs, respectively (Figure 4B and Supplementary Figure 1B), in a pattern similar to the observation in mice and non-pregnant human muscle (7, 30). The genes overlapped on those circular elements were highly dynamic, of which 52% (1030/1981) of eccDNAs covered partial gene fragments and 42% (87/207) of ecDNAs covered more than one complete gene (Supplementary Data 1A, B). Among these, 1030 unique eccDNAs partially overlapped with 939 genes, while the 87 unique ecDNAs overlapped with 3316 genes (Supplementary Data 9A, B). What is exciting is the large number of ecDNAs carried multiple genomes, forming super ecDNA [e.g., ecDNA(chr10:62267596-65472354) carrying 121 genes]. In addition, we found a preference for some genes being circular, with 5.9% (55/939) of host-genes derived from eccDNAs localized to more than three eccDNAs, while 0.81% (27/3316) of host-genes derived from ecDNAs had a similar profile. There were even many super genes positioned on circular DNAs, e.g., PTPRN2, which formed 11 different eccDNAs and was found in all 6 samples (Supplementary Data 9A, B). Notably, those circular elements from the X chromosome (41/92) tend to come from the gene region rather than the intergenic region compared with the Y chromosome (2/44) (Supplementary Data 1A, B). Therefore, eccDNAs/ecDNAs from the placenta is not completely random and shows a certain bias. Regrettably, we did not find a statistically significant difference in the genomic distribution of eccDNAs/ecDNAs derived between the normal and FGR groups (Figure 3).
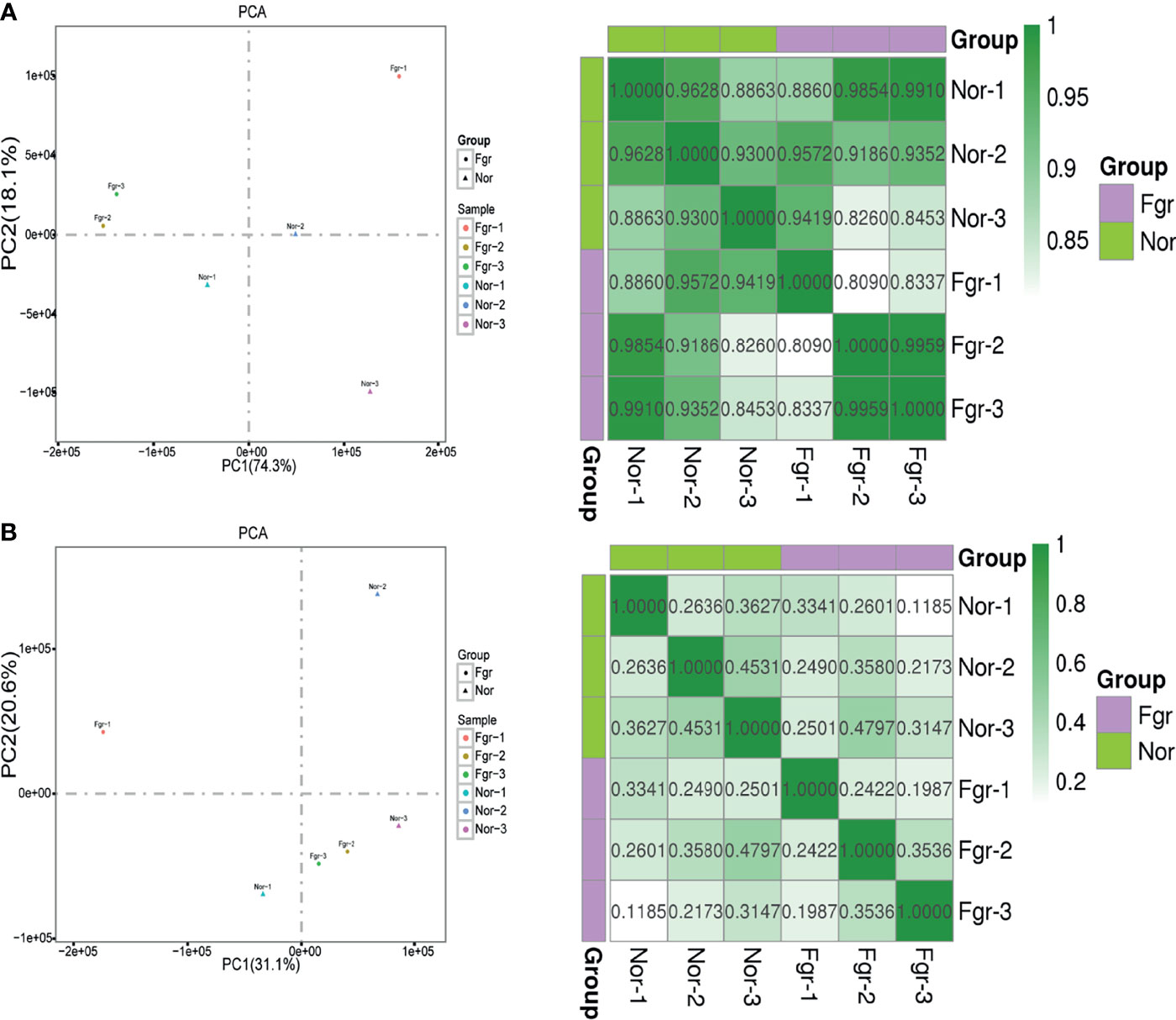
Figure 4 Properties of the loci that release eccDNAs. (A) The distribution of eccDNAs of each sample on whole-genome chromosome region. (B) Distribution of eccDNAs of FGR and normal groups in the indicated genomic region. (C) Percentage of eccDNAs with (black) or without (gray) 4~12bp direct or reverse repeats flanking junction locus at the genomic source. (D) Median percent GC content of eccDNAs and the genomic sequences of equal length upstream or downstream of the eccDNAs source loci.
We performed a BLAST analysis of 20bp upstream and downstream of the eccDNAs/ecDNAs junctions locus. Interestingly, we found that 77.7% of the eccDNAs had a direct repeat motif of 4~12bp upstream and downstream of the junction, and 71.5% of eccDNAs junction had a reverse repeating motif of 4~12bp (Figure 4C). Similar results were obtained for ecDNAs (Supplementary Figure 1C). The presence of “double-repeated trinucleotide sequences” on both sides of the junction was also found by using R (Supplementary Data 7A, B). These results suggest that microhomologous recombination may be an essential potential trigger for the promotion of double-stranded DNA cyclization.
Next, we evaluated whether pathological factors affect circular DNA in placental tissue during pregnancy. The results showed that the total number of eccDNAs of FGR group (7907 ± 1514; n=3) was slightly higher than those in the normal group (5842 ± 479.4; n=3) (P=0.087) (Supplementary Data 2A). The content of total ecDNAs also increased in the FGR group (810.3 ± 187.9; n=3) compared with the normal group (484.3 ± 56.59; n=3) (P=0.045) (Supplementary Data 2B). No significant difference in the number of unique eccDNAs and ecDNAs were found between the two groups (unique eccDNAs: FGR group (353.7 ± 53.80; n=3) vs normal group (459.3 ± 67.16; n=3) (P=0.100) (Supplementary Data 3A); unique ecDNAs: FGR group [64.33 ± 9.074; n=3) vs normal group (54.3 ± 4.619; n= 3) (P= 0.164) (Supplementary Data 3B)]. To clarify whether pathological factors affect the length of circular elements, we compared the length distribution of eccDNAs between the two groups, and there was no significant difference between the two groups (P=0.879) (Figure 3D). PCA and Pearson analysis was carried out between groups using R to investigate whether other relevant factors influenced eccDNAs/ecDNAs, and results showed that FGR1 was significantly away from the shared features within the group (Figures 5A, B).
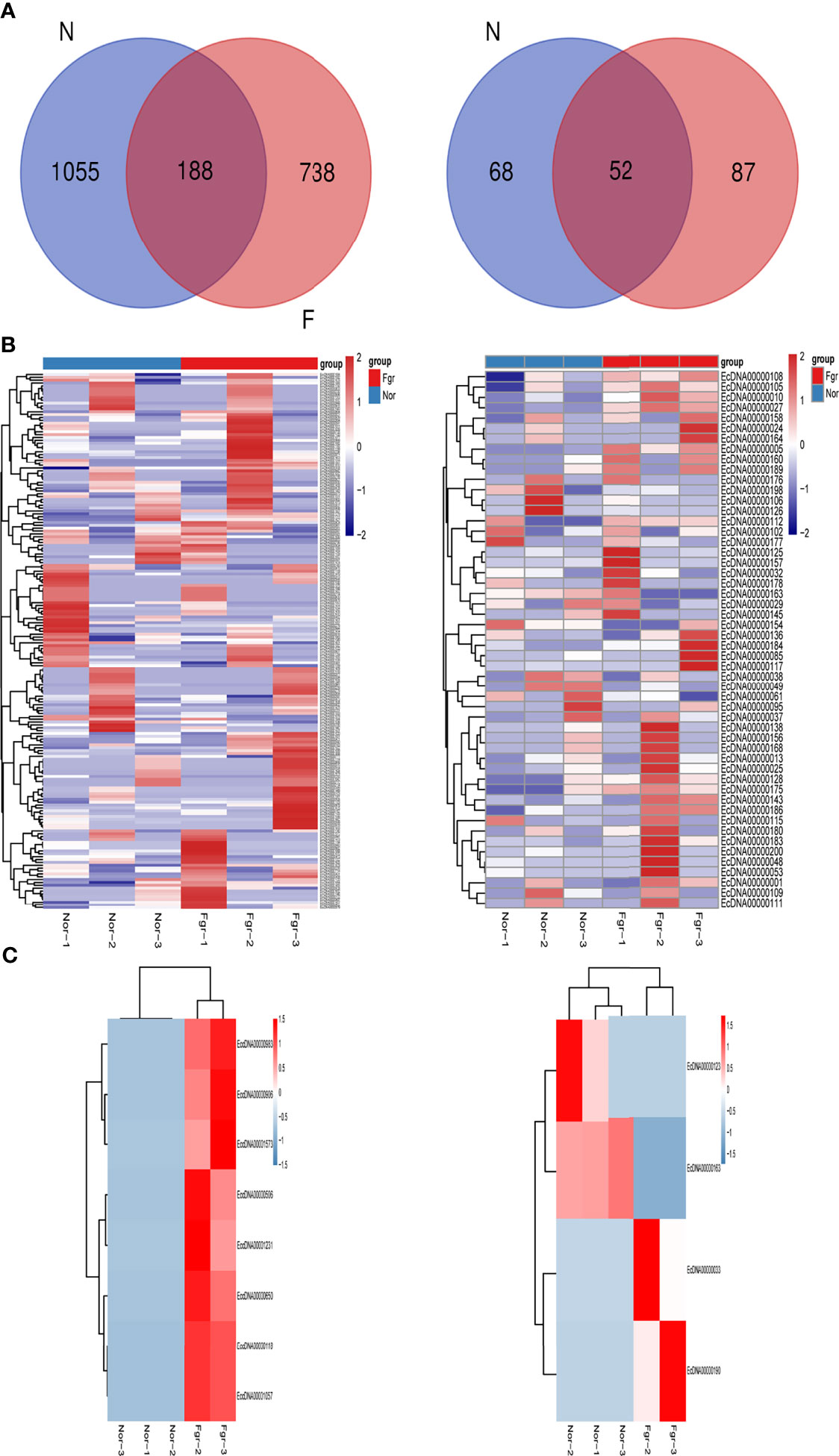
Figure 5 Correlation analysis between samples. (A) Principal component analysis and pearson correlation coefficient base on profile of eccDNAs of six samples. (B) Principal component analysis and pearson correlation coefficient base on profile of ecDNAs of six samples.
To investigate the potential biological functions of circular DNA in the placenta for disease pathogenesis, we performed a differential analysis of eccDNAs/ecDNAs between groups. The “ edgeR” package was used with the cut-off criteria of adjusted P-value<0.05 and |log2 FC|≥1. Results found that only one differential eccDNA (Chr2: 90394483-90399164) and one differential ecDNA (Chr17: 23204862-26614801), and their host-gene all annotated at the intergenic region, urging us to further re-analyze the sample information. Combined with the results of PCA, we excluded FGR1 and re-analyzed, which generated 8 DEEECs and 4 DEECs (Figure 2C). The results were compared with the reference genome using BWA software (22). We performed GO and KEGG pathway enrichment analysis by using the “Clusterprofiler” R package. The most enriched GO terms of host-genes on DEEECs were showed (Figure 6A and Table 2). The top 3 enriched terms of biological process were “lymphocyte-mediated immunity (GO:0002449, adjusted P = 0.001), regulation of immune response (GO:0050776, adjusted P = 0.002), and regulation of immune system process (GO:0002682, adjusted P = 0.012). The top 3 enriched terms of cellular component were “immunoglobulin complex, circulating (GO:0042571, adjusted P = 0.019), immunoglobulin complex (GO: GO:0019814, adjusted P = 0.029), and plasma membrane part (GO: GO:0044459, adjusted P = 0.029)”. The top 3 enriched terms of molecular function were “HLA-B specific inhibitory MHC class I receptor activity (GO: GO:0030109, adjusted P = 0.01), immunoglobulin receptor binding (GO: GO:0034987, adjusted P = 0.01), and formate-tetrahydrofolate ligase activity (GO:0004329, adjusted P =0.01)”. This host-gene significantly enriched GO terms on eccDNAs, which helps us further understand their role in FGR. Meanwhile, KEGG pathway analysis was further performed (Figure 6B and Table 3), in which host-genes were significantly enriched in “Antigen processing and presentation (ko04612, P = 3.85E-08), Natural killer cell-mediated cytotoxicity (ko04650, P = 4.43E-07), and Graft-versus-host disease (ko05332, adjusted P = 0.00025)”. We also analyzed the biological function of DEECs in the same way (Figures 6C, D). Interestingly, most of these different circular elements were in the FGR group.
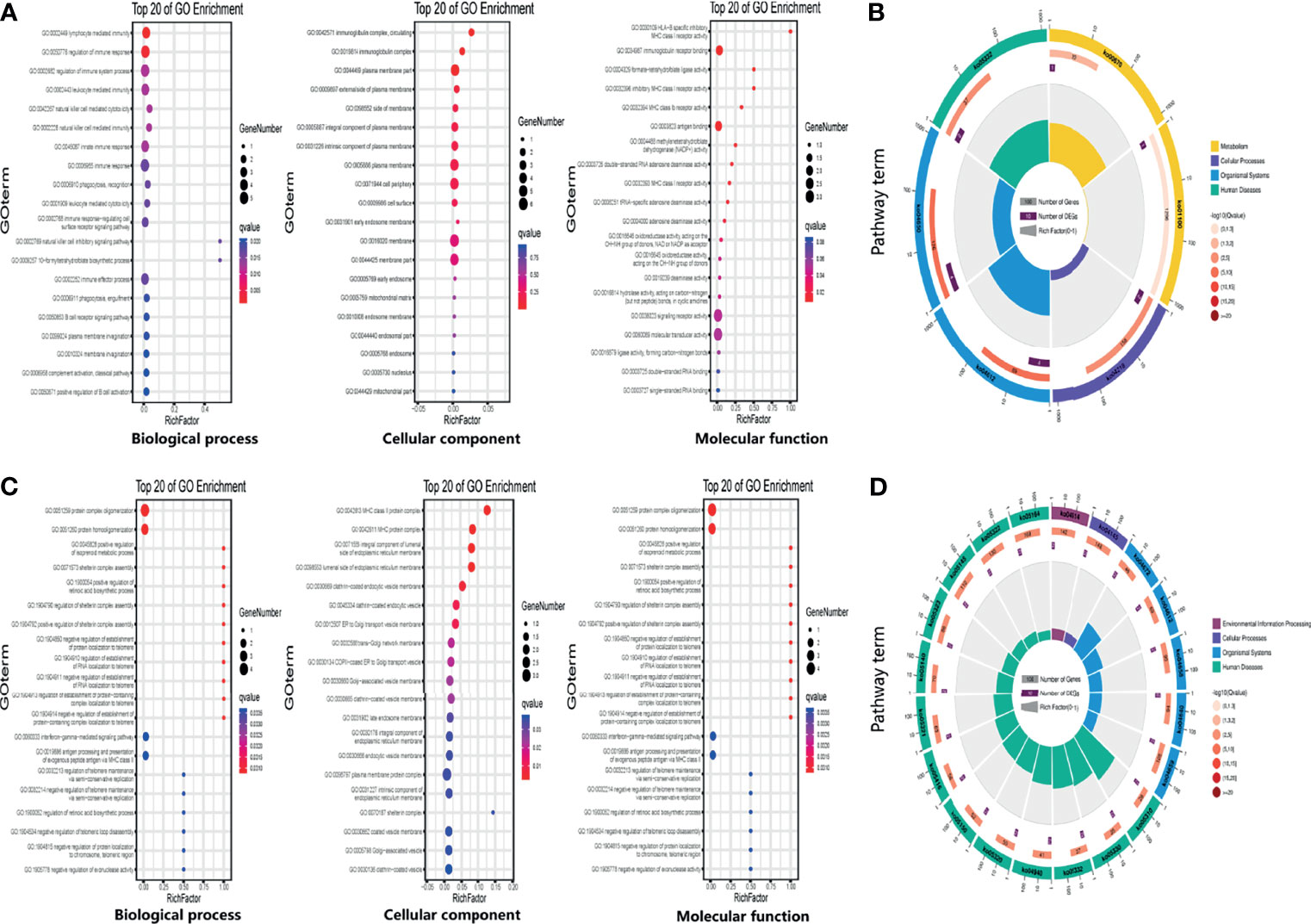
Figure 6 The enrichment map for differentially expressed host-genes on DEEECs/DEECs generated using GO and KEGG pathway analyses. (A) GO enrichment analyses of host-genes on DEEECs. (B) KEGG enrichment analyses of host-genes on DEEECs. (C) GO enrichment analyses of host-genes on DEECs. (D) KEGG enrichment analyses of host-genes on DEECs.
Studies have shown that ncRNAs play a significant role in FGR (31). We compared the genomic coordinates of eccDNAs/ecDNAs with ncRNAs using R/Bioconductor GenomicRanges and SplicingTypesAnno software package. Results revealed that 12.6% (250/1981) of the eccDNAs overlapped with the coordinates of 199 unique lncRNAs, and 0.05% (1/1981) of the eccDNAs overlapped with the coordinates of 22 unique tRNAs (Supplementary Data 11A). The same analysis uncovered that 40.6% (84/207) of ecDNAs overlapped with the coordinates of 1288 unique lncRNAs, 20.8% (43/207) of ecDNAs overlapped coordinates of 184 unique miRNAs, and 14.9% (31/207) of ecDNAs overlapped coordinates of 57 unique snoRNAs (Supplementary Data 11B). The results indicate that the function of these cyclic elements may be associated with lncRNA, tRNA, miRNA, snoRNA, etc., and some studies have demonstrated that under the conditions of stress, the expression of ncRNAs may change and participate in the progression of FGR (32, 33).
Lanciano et al. reported that eccDNAs were related to TEs activity (25, 34). In addition, it was recently found that the Ty1 subfamily of long terminal repeat sequence (LTR) retrotransposons was overexpressed in yeast eccDNAs (35). We further analyzed the correlation of TEs with eccDNAs/ecDNAs and found a high overlapping of 61.38% (Figure 3B and Supplementary Table 4). TEs is a movable DNA sequence (36) and contains a large number of repeating elements, which may contribute to the generation of eccDNAs/ecDNAs and drive tumor evolution and genetic heterogeneity (8). The high overlapping of ecDNAs/ecDNAs in the placenta annotated with the TEs is worthy to investigate.
Iparraguirre et al. proposed that circular DNA potentially forms transcripts analogous to circRNAs (37). We mapped all sequences of eccDNAs/ecDNAs to the database downloaded from circBase (26). 11,148 circRNAs were found to partially even all overlap with the circular DNA, among which the coordinates of ecDNA (Chr 6:57345464-60256901) could highly overlap with multiple circRNAs [e.g., hsa_circ_0076892: Chr6:57317623-60430572 (93.5%) and hsa_circ_0076899: Chr6: 57324200-60425439 (93.9%)] (Supplementary Table 5). Moreover, the vast majority of ecDNAs coordinates were are completely overlapped with their corresponding circRNAs. In light of this, we also attach a high priority to the possibility that eccDNAs/ecDNAs influences disease pathogenesis through the direct transcript form of circRNAs.
It has been confirmed that ecDNAs carry complete genes in tumors, which is one reason for promoting disease progression and leading to poor prognosis expression (38). Therefore, we validated whether some ecDNAs were present in 6 normal and 6 FGR placentas (including 3 normal and 3 FGR samples for Circle-seq). After DNA was extracted, PCR amplification was performed by using cross-junction reverse complementary primers (7), e.g. (F: GGGGCCACCTATGTGCAATG R: TGGGAACCAGGAATGAGGGA) (product 275bp) (F: AAGACAGTATGGCGATTCCT R: CATTGTTGGACATTGGGTTG) (product184 bp) (Supplementary Table 2), and then further verified by Southern blotting. Encouragingly, we detected the presence of ecDNA (Chr 6: 32482948-32590591) and ecDNA (Chr 8: 71825887-73468945) in placentas (Figure 7 and Supplementary Figure 2), whose variability was consistent with the sequencing results, stimulating our curiosity about the possible role of eccDNAs/ecDNAs in FGR.
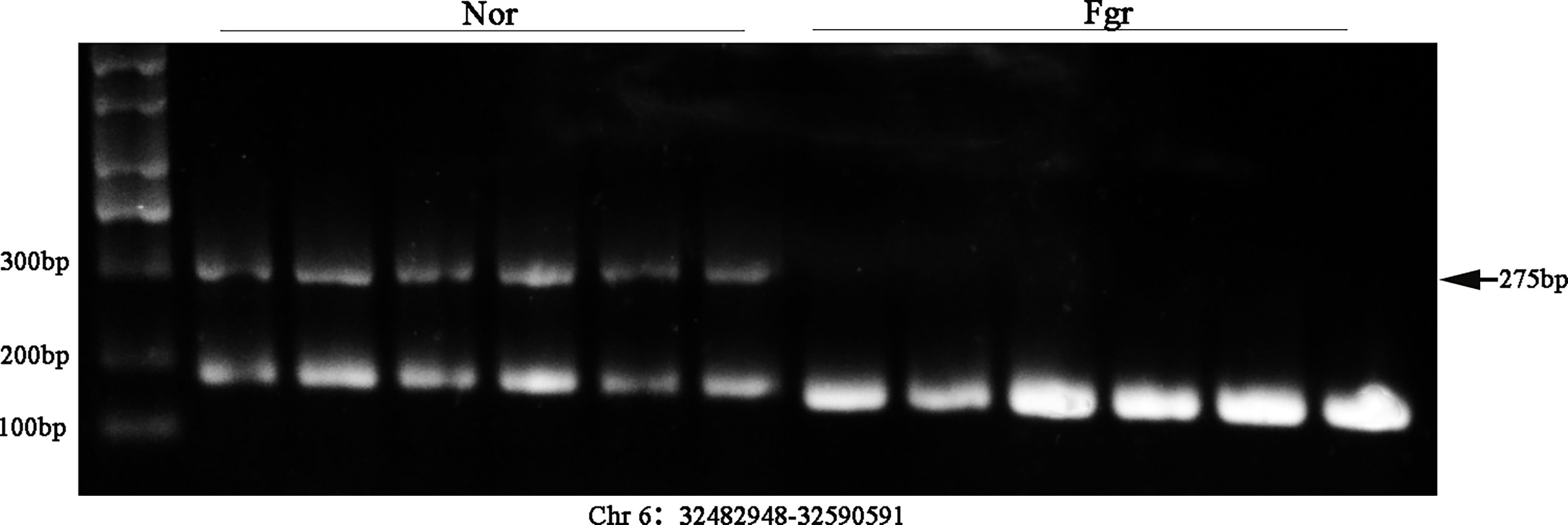
Figure 7 Validation of ecDNA (Chr 6: 32482948-32590591) from 12 placentas (including 6 FGR and 6 normal) using Southern blot.
This study aimed to determine the origin and characteristics of placental-derived eccDNAs/ecDNAs and further explore their possible effects on pregnancy. We identified that eccDNAs/ecDNAs were abundant in the placenta with significant heterogeneity, and shared the same characteristics in length, GC content, genomic distribution, and repetitive sequence, which similar to previous reports of humans and mice (7, 20, 28). This study is the first report to compare the eccDNAs/ecDNAs in the normal human placenta and FGR human placenta, which is significant for further analysis on pathogeny and mechanism of FGR.
Although we did not observe any difference in the length, GC content (Figure 3D and Supplementary Figure 1D), genomic distribution, and repetitive sequence between the FGR and normal groups, it was surprising that the amount of eccDNAs/ecDNAs in the FGR placenta was significantly higher than that in the normal group. Abnormal apoptosis of trophoblast cells can cause invasion dysfunction of placental trophoblast cells, and shallow implantation of placental trophoblast cells is a potential pathological mechanism of FGR, thus leading to the occurrence of FGR (39). Given that the production of eccDNAs/ecDNAs is regulated to some extent by apoptosis (40), we speculate that apoptosis plays a critical part in facilitating the DNA elements detached from chromosomes and cyclization in FGR. In addition, placental dysplasia is an important pathological basis of FGR, in which excessive oxidative stress affects the proliferation and apoptosis of trophoblasts (41). The increased level of oxidative stress may aggravate the damage of chromosomes, which will not only promote base oxidation but also lead to single-strand breaks (SSB) and double-strand breaks (DSB) (42). Thereby, they promote the formation of circular elements, which may also be why there are more eccDNAs/ecDNAs in the placenta associated with FGR.
To understand their potential formation mechanisms, we explored the nucleotide modes around the junction locus of the eccDNAs/ecDNAs. Shibata et al. have reported that there are microhomologous base modes around the junction of eccDNAs, and microhomology may promote cyclization of double-terminal DNA through homologous recombination (4, 22). In addition, Sun and his colleagues reported the presence of reverse repeat motif in Arabidopsis (25). In parallel to this, Dennis Lo et al. proposed a new view on the formation of eccDNAs/ecDNAs, where the presence of “double-repeated trinucleotide sequences” on both sides of the junction might be a critical element in the production of those circular elements (14). Repeated deoxyribonucleotide elements usually represent DNA breakpoints and susceptibility sites for R-loop-out events (2, 20, 28). Our statistical analysis found not only direct repeating motifs around the junction of eccDNAs/ecDNAs, but also many reverse repeating motifs, suggesting that microhomology repeats may be the critical factors mediating the formation of eccDNAs/ecDNAs.
The human genome contains many other types of repeated sequences, e.g., TEs, the insertion mutation factor of the eukaryotic genome, affecting the evolution and adaptation of species and individual physiology or disease (43). The proportion of transposons in the human genome is approximately 44% (44, 45). Surprisingly, our results revealed that more than half of the circular entities overlapped TEs in placenta (Supplementary Table 4). Furthermore, studies have reported that excessive loss, generation, or insertion of TEs into other DNA sequences on chromosomes of mice during gestation can lead to placental and fetal capillary dysfunction, affecting fetal mice growth and increased embryonic mortality (46, 47). Consequently, a higher percentage of TEs forming circular DNA may also significantly contribute to disease progression.
Additionally, we found that gestational age may also contribute to the formation of these circular entities. Using PCA and Pearson analysis, we discovered that FGR1 deviated from the characteristics of the FGR group. In yeast, the abundance of extrachromosomal rDNA circles (ERCs) is highly related to age (48). Due to the absence of centromeres in circular DNA, including ERCs, eccDNAs/ecDNAs accumulate rapidly in aging yeast through a highly asymmetric mitotic separation process (49). The abundance of eccDNAs in rat lymphocytes and human lung fibroblasts also underwent age-related dynamic procedures (50). However, asymmetric inheritance alone is not enough to promote the accumulation of circular DNA, and the active replication origin carried on circular DNA is an important reason to encourage the production of more copies of circular DNA (49). When elements of circular DNA are neither produced quickly nor have an active origin for replication, they tend to decline with age. Although our findings are very innovative and clinically significant, some limitations still need to be explored further. Since the normal placental tissue matching the gestational week of the FGR group is difficult to obtain, we selected placental tissue delivered at full-term pregnancy as a control. The gestational age might play a role in the differences found. We need more data on placenta with different gestational age to help us explore the effect of gestational weeks for circular elements.
Shibata and colleagues had reported that eccDNAs/ecDNAs originate from the microdeletions of chromosomes (7). Data from the Thousand Genomes Project describe the length of chromosome microdeletions in germline cells peaking at 100bp and 350bp, concentrated in gene regions, with a high frequency of short repeat sequences on both sides microdeletions (51), consistent with our results. Moreover, the origin of eccDNAs is related to the nucleosomes (7), and the characteristics of dinucleosomes are fully reflected in the data of the placenta.
Current studies have demonstrated that eccDNAs/ecDNAs is predominantly derived from genetically dense chromosomal regions. In addition, our data have revealed a genetic preference for the formation of circular DNA, e.g., PTPRN2 forms 11 unique eccDNAs. PTRRN2 mediates dysregulation of innate immune pathways. The maternal-fetal interface is in an environment of altered immune homeostasis during pregnancy. Karli S et al. reported that altered methylation of the PTPRN2 leads to T cell dysfunction and increases reactive oxygen species (ROS) levels, resulting in an increased risk of FGR (52, 53). If so, whether gene cyclization has a similar or opposite effect to methylation makes for fascinating speculation.
To further investigate the possibility of eccDNAs/ecDNAs driving disease progression through its host-genes, we performed a differential analysis between groups. And after GO and KEGG clustering analysis of the host-genes of DEEECs/DEECs, we found an interesting phenomenon: the host-genes of DEEECs mainly cluster in immune-related cellular components, biological processes, and functions. EccDNAs/ecDNAs arise from many functional hotspots, one of which is the recombination of genes encoding T-cell receptor variable regions and immunoglobulin Flight chains (54, 55). Furthermore, the activation of homotypic conversion of B-cell-derived immunoglobulin heavy chains also contributes to the production of eccDNA (56). Studies have demonstrated that altered cytokine patterns at the maternal-fetal interface during pregnancy are an essential mechanism for FGR. Such as increased or decreased populations of dNK cells, macrophages, T cells, dendritic cells, B cells, and NKT cells, which affect placental trophoblast proliferation, apoptosis, vascular endothelial cell activity, etc., thus influencing the pathogenesis of FGR (57–59). We found many host-genes on DEEECs/DEECs have been confirmed to be associated with FGR. Such as the host-gene KIR2DL1 on eccDNA (Chr19: 54775137-54839510), which plays a critical role in the pathogenesis of FGR by binding to HLA-C2, and can increase the secretion of cytokines and thus enhances trophoblast invasion (60, 61). As described above, we conjecture that the imbalance of the maternal-fetal interface immune defenses drives the production of more eccDNAs/ecDNAs, and host-genes of eccDNAs/ecDNAs activation of immune-related pathways and functions exacerbates FGR in turn.
Presently, the exact function of eccDNAs/ecDNAs is not fully understood. Several reports have described its possible role in disease: (1) Unequal replication inheritance of circular DNA leads to heterogeneity of the disease (9, 38, 62). (2) Due to the cyclizing, topological structure change and the sequence rearrange of those elements (e.g., enhancers are connected to the promoter topological structure) result in enhanced gene transcription (38, 63, 64). (3) eccDNAs/ecDNAs can produce miRNA or siRNA, which interact with mRNA to inhibit translation function (13). (4) The cyclization of DNA does not directly affect the expression of the gene itself. It affects the gene expression that circular DNA is inserted into linear chromosomes to form chromosome recombination (including tumor suppressor gene loci, proto-oncogene loci, or other sites) (62, 65). (5) The integration of DNA from different chromosomes or between humans and viruses affects the expression of genes, such as human papillomavirus from cervical cancer (66) and Torque Teno virus gene in pigeon circular DNA genome (67). Here we have specifically explored other potential functions of circular DNA, such as the relationship with ncRNAs. The results show that the annotations of lncRNAs, tRNAs, circRNAs, miRNAs, and snoRNAs partially even completely overlap with these circles, especially circRNAs.
In mammalian genomes, 4%~9% of the sequences that produce transcripts are lncRNAs. lncRNAs are extensively involved in important regulatory processes such as chromosome silencing, genome imprinting, chromatin modification, transcriptional activation, transcriptional interference, and intranuclear transport (68).In addition, numerous studies have reported the critical role of lncRNAs in pregnancy (69–71). Some meaningful data were also found in our research, e.g., lncRNA-PVT1 overlapped with eccDNA (Chr8: 128144301-128144456), which could significantly promote apoptosis and inhibits proliferation, migration, and invasion of trophoblast cells. Additionally, PVT1 was also proved cross-talking with many miRNAs and genes involved in maintaining the physiological placenta (72, 73). Thus, in addition to a vital oncogene, PVT1 is also a key regulator in gestational mediating the development of the placenta.
The primary function of tRNAs is to decode mRNA sequences and transport specific amino acids from aminoacyl-tRNA synthetases (AARs) to ribosomes (74). Besides their typical role in protein translation, tRNAs are actively involved in other biology areas, such as primers for retroviral genome reverse transcription (75). Dutta A et al. found that small fragments of tRNA and tRNA halves also can regulate gene expression, protein translation, reverse transcription transposon activity, transgenerational epigenetic changes and responses to environmental stress during pregnancy, which can influence fetal growth and alter maternal immune activation (76). We found that eccDNA overlapped 22 species of tRNAs with high abundance coverage, which make it worthwhile to explore whether the epigenetic modification of tRNAs altered by eccDNAs can influence disease pathogenesis in the future.
Over the past decade, considerable researches have been conducted on ncRNAs in pregnancy-related diseases, of which miRNAs play a central role. Dysregulation of miRNAs from the placenta was proved to influence placental progression and function and these miRNAs to export to the mother and fetus, thus directly affecting maternal and fetal physiology (77–79). Our data showed that many miRNAs (e.g., miR-191, miR-224, miR-374a, miR-193b, miR-365a, miR-4287, miR-590, miR-664b, miR-3622b, etc.) overlapped with the genomic loci of ecDNAs. Considering that they can be involved in the onset and progression of FGR by affecting different cellular pathways and functions including oxidative stress, angiogenesis, immunity, inflammation, etc. (80–83), it is reasonable to believe that ecDNAs mediating FGR progression through interaction with miRNAs is also with high potential. Further, circRNAs, as miRNA sponges, can influence the expression of downstream genes (84, 85), which may affect the invasion and apoptosis of trophoblast cells by acting on certain target miRNAs in the human placenta, thus mediating the onset of pregnancy-related diseases (86, 87). Since this study showed some of the ecDNAs almost completely overlapping with circRNAs, we speculate that some circRNAs are not formed by backsplicing of precursor-mRNAs, but by direct transcription of circular DNA.
SnoRNA comes from a conserved family of nuclear-derived RNAs involved in ribosomal subunit maturation and rRNA processing (88), in addition to snoRNA that increases lncRNA stability and maintains subcellular localization through the formation of Sno-lncRNA (89). Mohandas N et al. described that the snoRNA mediated rRNA maturation defects is a distinctive feature of Diamond-Blackfan anemia (DBA), which leads to cell cycle arrest and increased apoptosis by producing nucleolar stress, affecting P53 activity and activation of its downstream targets, and increasing ROS (90). The locus of some ecDNAs in the genome highly overlapped with the small nucleolar RNA H/ACA box (SNORA) and small nucleolar RNA C/D/box (SNORD), and U3, etc. in the snoRNA family. Studies have demonstrated that the SNORA/SNORD family promotes immune imbalance and aging by increasing the immune system’s dendritic cells (DCs) to affect antigen presentation and energy production pathways (91). Our next step is to investigate whether ecDNAs can mediate the progress of FGR by modifying the expression of snoRNAs.
Cell-free DNA (cfDNA) has been shown great promise in the diagnosis, prognosis, and monitoring of many illnesses (e.g., carcinoma) (92, 93), among which cfDNA analysis for examination of trisomy during pregnancy is widely used with high accuracy (94). Recently, Dennis Lo and colleagues characterized maternal plasma containing high amounts of fetal-derived eccDNAs (14). Given the fact that eccDNAs/ecDNAs are more stable than linear DNA, as well as that the large quantity of eccDNAs/ecDNAs was found derived from FGR placentas in our data, and that some genetics preferred to form circular DNA, these findings may help us to develop a new tool for non-invasive prenatal diagnosis that is superior to cfDNA. Besides, the monitoring of eccDNAs in current studies on cancer-related genes can be used as a therapeutic efficacy target (95). Similarly, the tumor-like nature of trophoblasts (plentiful of genes affecting the proliferation, migration, apoptosis, and immune microenvironment) is closely involved in pregnancy-related diseases. Therefore, establishing of eccDNAs/ecDNAs as a biomarker for pregnancy complications has important clinical value.
Considering that most of the discussions above on eccDNAs/ecDNAs function are still not fully confirmed, Liu, X et al. developed a biotechnology-based on CRISPR-C that can facilitate the artificial release of eccDNA from cells (96). We can verify the effect of altered eccDNAs/ecDNAs abundance and types on gene expression and cellular function by performing CRISPR-C intervention at the cellular level in the following study. In view of this, we may venture to imagine whether targeted interventions in circular DNA also have therapeutic potential, a question that certainly deserves further investigation.
In conclusion, we described the landscapes of eccDNAs/ecDNAs in placental tissues of normal and FGR pregnancies by the circle-seq technique and investigated their characteristics and formation mechanisms in detail (Figure 8). Our study also uncovers eccDNAs as a potential driver of FGR through immune signaling pathways. Numerous genomic annotations of ncRNAs overlapped highly with those of eccDNAs/ecDNAs, suggesting the existence of a complex network and underlying biological functions between them. Additionally, the greater stability of eccDNAs/ecDNAs than linear DNA confers excellent biomarker properties, therefore whether their targeted interventions have therapeutic potential is of great benefit for clinical. However, these ideas have high innovation, and the function of these mysterious molecules needs to be further investigated.
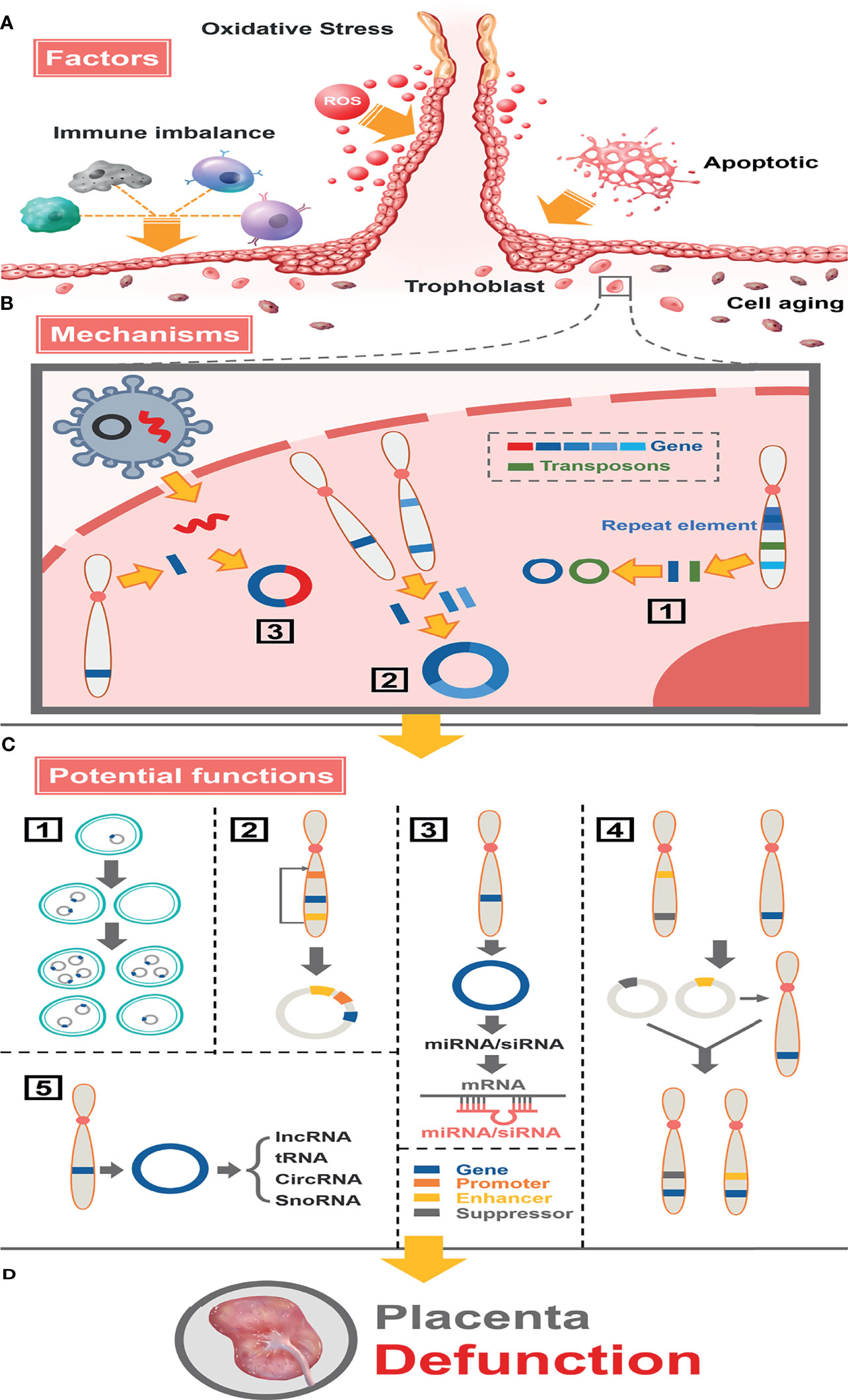
Figure 8 Full landscape of extrachromosomal circular DNA in fetal growth restriction. (A) Factors promoting the formation of eccDNAs/ecDNAs in the placenta. Oxidative stress, apoptosis, aging, and immune imbalance at the maternal-fetal interface (such dNK cells, macrophage T cells, B cells, etc.) deteriorate chromosome fragmentation and deletion of gene fragments in trophoblast. (B) EccDNAs/ecDNAs of different ontogenetic origins in the placenta and potential mechanisms of formation. (1) Different DNA fragments shed from the same chromosome form multiple cycles by microhomologous recombination. (2) Different DNA fragments shed from different chromosomes are integrated into a cycle by microhomologous recombination.(3) Virus-derived DNA fragments integrate with human chromosomally shed DNA fragments forming cycles. (C) Potential functions of eccDNAs/ecDNAs in the placenta. (1) Circular DNA lacks centromere, which inherits genes unequally into offspring cells promoting disease heterogeneity. (2) Effect of altered topology due to cyclization, or promotion of (enhancer linked to promoter topology) gene transcriptional enhancement by sequence rearrangement on the cycle. (3) miRNA or siRNA is produced by eccDNA and interacts with mRNA to suppress gene expression. (4) Cycles carrying enhancers or repressors are chimerized into the genomes of different linear DNAs, forming chromosomal recombination that enhances or represses gene expression. (5) Influence genes expression through network and cross-talk with ncRNAs. (D) Placental dysfunction leading to FGR.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: SRR17054686 and PRJNA784191.
The studies involving human participants were reviewed and approved by The Ethics Committee of the First Affiliated Hospital of Chongqing Medical University. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
HY, JH, and SH designed the research and wrote the manuscript. HY and JH conducted the raw data analysis. HY conducted experiments. HBY supervised part of the study. YT and MC collected the placenta samples. QY performed the statistical analysis. XZ and HQ conceptualized, supervised, and funded this study. All authors contributed to the article and approved the submitted version.
This work was supported by The National Natural Science Foundation of China, Grant/Award Numbers: 81771613, 82171679.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors are grateful to all the pregnant women who participated in our study and the support of the First Affiliated Hospital of Chongqing Medical University, Chongqing, China. we also appreciate the support from “111 program” of the Ministry of Education of P.R.C and the State Administration of Foreign Experts Affairs of P.R.C and the cooperation of the Obstetrics, Nurse and Midwifery staff of the First Affiliated Hospital of Chongqing Medical University, Chongqing, China.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.780779/full#supplementary-material
eccDNAs/ecDNAs, Extrachromosomal circular DNAs; FGR, Fetal growth restriction; PCR, Real-time Polymerase Chain Reaction; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; DEEECs/DEECs, Differentially expressed eccDNAs/ecDNAs; ncRNAs, Non-coding RNAs; Nor, Normal; TEs, Transposon elements; CATB, Cetyl trimethyl ammonium bromide; RCA, Rolling circle amplification; PE Paired-end; BWA, Burrows-Wheeler Aligner; TPM, Tags per million; PCA, Principal Component Analysis; SSB, Single-strand breaks; DSB, Double-strand breaks; LTR, Long terminal repeat sequence; ERCs, Extrachromosomal rDNA circles; ROS, Reactive oxygen species; AARs, AminoacyltRNA synthetases; DBA, Diamond-Blackfan anemia; SNORA, Small nucleolar RNA H/ACA box; SNORD, Small nucleolar RNA C/D/box; DCs, Dendritic cells; cfDNA, Cell-free DNA.
1. Gaubatz J. Extrachromosomal Circular DNAs and Genomic Sequence Plasticity in Eukaryotic Cells. Mutat Res (1990) 237:271–92. doi: 10.1016/0921-8734(90)90009-g
2. Møller H, Parsons L, Jørgensen T, Botstein D, Regenberg B. Extrachromosomal Circular DNA Is Common in Yeast. Proc Natl Acad Sci USA (2015) 112(24):E3114–22. doi: 10.1073/pnas.1508825112
3. Møller H, Bojsen R, Tachibana C, Parsons L, Botstein D, Regenberg B. Genome-Wide Purification of Extrachromosomal Circular DNA From Eukaryotic Cells. J Vis Exp: JoVE (2016) 110):e54239. doi: 10.3791/54239
4. Kumar P, Dillon LW, Shibata Y, Jazaeri AA, Jones DR, Dutta A. Normal and Cancerous Tissues Release Extrachromosomal Circular DNA (eccDNA) Into the Circulation. Mol Cancer Res (2017) 15(9):1197–205. doi: 10.1158/1541-7786
5. Yerlici V, Lu M, Hoge C, Miller R, Neme R, Khurana J, et al. Programmed Genome Rearrangements in Oxytricha Produce Transcriptionally Active Extrachromosomal Circular DNA. Nucleic Acids Res (2019) 47(18):9741–60. doi: 10.1093/nar/gkz725
6. Cohen S, Segal D. Extrachromosomal Circular DNA in Eukaryotes: Possible Involvement in the Plasticity of Tandem Repeats. Cytogenet Genome Res (2009) 124:327–38. doi: 10.1159/000218136
7. Shibata Y, Kumar P, Layer R, Willcox S, Gagan JR, Griffith JD, et al. Extrachromosomal microDNAs and Chromosomal Microdeletions in Normal Tissues. Science (2012) 336(6077):82–6. doi: 10.1126/science.1213307
8. Turner KM, Deshpande V, Beyter D, Koga T, Rusert J, Lee C, et al. Extrachromosomal Oncogene Amplification Drives Tumour Evolution and Genetic Heterogeneity. Nature (2017) 543(7643):122–5. doi: 10.1038/nature21356
9. Nikolaev S, Santoni F, Garieri M, Makrythanasis P, Falconnet E, Guipponi M, et al. Extrachromosomal Driver Mutations in Glioblastoma and Low-Grade Glioma. Nat Commun (2014) 5:5690. doi: 10.1038/ncomms6690
10. Xu K, Ding L, Chang TC, Shao Y, Chiang J, Mulder H, et al. Structure and Evolution of Double Minutes in Diagnosis and Relapse Brain Tumors. Acta Neuropathol (2019) 137(1):123–37. doi: 10.1007/s00401-018-1912-1
11. Rausch T, Jones D, Zapatka M, Stütz A, Zichner T, Weischenfeldt J, et al. Genome Sequencing of Pediatric Medulloblastoma Links Catastrophic DNA Rearrangements With TP53 Mutations. Cell (2012) 148:59–71. doi: 10.1016/j.cell.2011.12.013
12. deCarvalho A, Kim H, Poisson L, Winn M, Mueller C, Cherba D, et al. Discordant Inheritance of Chromosomal and Extrachromosomal DNA Elements Contributes to Dynamic Disease Evolution in Glioblastoma. Nat Genet (2018) 50(5):708–17. doi: 10.1038/s41588-018-0105-0
13. Paulsen T, Shibata Y, Kumar P, Dillon L, Dutta A. Small Extrachromosomal Circular DNAs, microDNA, Produce Short Regulatory RNAs That Suppress Gene Expression Independent of Canonical Promoters. Nucleic Acids Res (2019) 47(9):4586–96. doi: 10.1093/nar/gkz155
14. Sin STK, Jiang P, Deng J, Ji L, Cheng SH, Dutta A, et al. Identification and Characterization of Extrachromosomal Circular DNA in Maternal Plasma. Proc Natl Acad Sci USA (2020) 117(3):1658–65. doi: 10.1073/pnas.1914949117
15. Mook-Kanamori D, Steegers E, Eilers P, Raat H, Hofman A, Jaddoe V. Risk Factors and Outcomes Associated With First-Trimester Fetal Growth Restriction. JAMA (2010) 303(6):527–34. doi: 10.1001/jama.2010.78
16. Henry G, William G. ACOG Practice Bulletin No. 204: Fetal Growth Restriction. Obstet Gynecol (2019) 133(2):e97–e109. doi: 10.1097/aog.0000000000003070
17. Gordijn S, Beune I, Thilaganathan B, Papageorghiou A, Baschat A, Baker P, et al. Consensus Definition of Fetal Growth Restriction: A Delphi Procedure. Ultrasound Obstetrics Gynecol: Off J Int Soc Ultrasound ObstetGynecol (2016) 48(3):333–9. doi: 10.1002/uog.15884
18. Beune I, Bloomfield F, Ganzevoort W, Embleton N, Rozance P, van Wassenaer-Leemhuis A, et al. Consensus Based Definition of Growth Restriction in the Newborn. J Pediatr (2018) 196:71–6.e1. doi: 10.1016/j.jpeds.2017.12.059
19. Gurugubelli Krishna R, Vishnu Bhat B. Molecular Mechanisms of Intrauterine Growth Restriction. J Matern-Fetal Neonatal Med: Off J Eur Assoc Perinatal Med Fed Asia Oceania Perinatal Soc Int Soc Perinatal Obstet (2018) 31(19):2634–40. doi: 10.1080/14767058.2017.1347922
20. Moller HD, Mohiyuddin M, Prada-Luengo I, Sailani MR, Halling JF, Plomgaard P, et al. Circular DNA Elements of Chromosomal Origin are Common in Healthy Human Somatic Tissue. Nat Commun (2018) 9(1):1069. doi: 10.1038/s41467-018-03369-8
21. Chen S, Zhou Y, Chen Y, Gu J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics (2018) 34(17):i884–i90. doi: 10.1093/bioinformatics/bty560
22. Li H, Durbin R. Fast and Accurate Long-Read Alignment With Burrows-Wheeler Transform. Bioinf (Oxf Engl) (2010) 26(5):589–95. doi: 10.1093/bioinformatics/btp698
23. Quinlan AR, Hall IM. BEDTools: A Flexible Suite of Utilities for Comparing Genomic Features. Bioinformatics (2010) 26(6):841–2. doi: 10.1093/bioinformatics/btq033
24. Robinson MD, McCarthy DJ, Smyth GK. Edger: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics (2010) 26(1):139–40. doi: 10.1093/bioinformatics/btp616
25. Wang K, Tian H, Wang L, Wang L, Tan Y, Zhang Z, et al. Deciphering Extrachromosomal Circular DNA in Arabidopsis. Comput Struct Biotechnol J (2021) 19:1176–83. doi: 10.1016/j.csbj.2021.01.043
26. Glažar P, Papavasileiou P, Rajewsky N. Circbase: A Database for Circular RNAs. RNA (New York NY) (2014) 20(11):1666–70. doi: 10.1261/rna.043687.113
27. McCabe M, Bäcklund L, Leong H, Ichimura K, Collins V. Chromosome 17 Alterations Identify Good-Risk and Poor-Risk Tumors Independently of Clinical Factors in Medulloblastoma. Neuro-oncology (2011) 13(4):376–83. doi: 10.1093/neuonc/noq192
28. Dillon LW, Kumar P, Shibata Y, Wang YH, Willcox S, Griffith JD, et al. Production of Extrachromosomal MicroDNAs Is Linked to Mismatch Repair Pathways and Transcriptional Activity. Cell Rep (2015) 11(11):1749–59. doi: 10.1016/j.celrep.2015.05.020
29. Segal E, Fondufe-Mittendorf Y, Chen L, Thåström A, Field Y, Moore I, et al. A Genomic Code for Nucleosome Positioning. Nature (2006) 442(7104):772–8. doi: 10.1038/nature04979
30. Zhu J, Zhang F, Du M, Zhang P, Fu S, Wang L. Molecular Characterization of Cell-Free eccDNAs in Human Plasma. Sci Rep (2017) 7(1):10968. doi: 10.1038/s41598-017-11368-w
31. Goyal D, Limesand SW, Goyal R. Epigenetic Responses and the Developmental Origins of Health and Disease. J Endocrinol (2019) 242(1):T105–t19. doi: 10.1530/joe-19-0009
32. Monteiro LJ, Peñailillo R, Sánchez M, Acuña-Gallardo S, Mönckeberg M, Ong J, et al. The Role of Long Non-Coding RNAs in Trophoblast Regulation in Preeclampsia and Intrauterine Growth Restriction. Genes (2021) 12(7):970. doi: 10.3390/genes12070970
33. Torrent M, Chalancon G, de Groot NS, Wuster A, Madan Babu M. Cells Alter Their tRNA Abundance to Selectively Regulate Protein Synthesis During Stress Conditions. Sci Signal (2018) 11(546):eaat6409. doi: 10.1126/scisignal.aat6409
34. Lanciano S, Carpentier MC, Llauro C, Jobet E, Robakowska-Hyzorek D, Lasserre E, et al. Sequencing the Extrachromosomal Circular Mobilome Reveals Retrotransposon Activity in Plants. PloS Genet (2017) 13(2):e1006630. doi: 10.1371/journal.pgen.1006630
35. Møller H, Larsen C, Parsons L, Hansen A, Regenberg B, Mourier T. Formation of Extrachromosomal Circular DNA From Long Terminal Repeats of Retrotransposons in Saccharomyces Cerevisiae. G3 (Bethesda Md) (2015) 6(2):453–62. doi: 10.1534/g3.115.025858
36. Conrad D, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, et al. Origins and Functional Impact of Copy Number Variation in the Human Genome. Nature (2010) 464(7289):704–12. doi: 10.1038/nature08516
37. Iparraguirre L, Prada-Luengo I, Regenberg B, Otaegui D. To Be or Not to Be: Circular RNAs or mRNAs From Circular DNAs? Front Genet (2019) 10:940. doi: 10.3389/fgene.2019.00940
38. Wu S, Turner KM, Nguyen N, Raviram R, Erb M, Santini J, et al. Circular ecDNA Promotes Accessible Chromatin and High Oncogene Expression. Nature (2019) 575(7784):699–703. doi: 10.1038/s41586-019-1763-5
39. Paules C, Dantas A, Miranda J, Crovetto F, Eixarch E, Rodriguez-Sureda V, et al. Premature Placental Aging in Term Small-for-Gestational-Age and Growth-Restricted Fetuses. Ultrasound Obstet Gynecol: Off J Int Soc Ultrasound Obstetrics Gynecol (2019) 53(5):615–22. doi: 10.1002/uog.20103
40. Mehanna P, Gagne V, Lajoie M, Spinella JF, St-Onge P, Sinnett D, et al. Characterization of the microDNA Through the Response to Chemotherapeutics in Lymphoblastoid Cell Lines. PloS One (2017) 12(9):e0184365. doi: 10.1371/journal.pone.0184365
41. Sun C, Groom KM, Oyston C, Chamley LW, Clark AR, James JL. The Placenta in Fetal Growth Restriction: What is Going Wrong? Placenta (2020) 96:10–8. doi: 10.1016/j.placenta.2020.05.003
42. Fishel M, Vasko M, Kelley M. DNA Repair in Neurons: So If They Don’t Divide What’s to Repair? Mutat Res (2007) 614:24–36. doi: 10.1016/j.mrfmmm.2006.06.007
43. Lanciano S, Cristofari G. Measuring and Interpreting Transposable Element Expression. Nat Rev Genet (2020) 21(12):721–36. doi: 10.1038/s41576-020-0251-y
44. Bourque G, Burns KH, Gehring M, Gorbunova V, Seluanov A, Hammell M, et al. Ten Things You Should Know About Transposable Elements. Genome Biol (2018) 19(1):199. doi: 10.1186/s13059-018-1577-z
45. Burns KH, Boeke JD. Human Transposon Tectonics. Cell (2012) 149(4):740–52. doi: 10.1016/j.cell.2012.04.019
46. Sekita Y, Wagatsuma H, Nakamura K, Ono R, Kagami M, Wakisaka N, et al. Role of Retrotransposon-Derived Imprinted Gene, Rtl1, in the Feto-Maternal Interface of Mouse Placenta. Nat Genet (2008) 40(2):243–8. doi: 10.1038/ng.2007.51
47. Ono R, Nakamura K, Inoue K, Naruse M, Usami T, Wakisaka-Saito N, et al. Deletion of Peg10, an Imprinted Gene Acquired From a Retrotransposon, Causes Early Embryonic Lethality. Nat Genet (2006) 38(1):101–6. doi: 10.1038/ng1699
48. Borghouts C, Benguria A, Wawryn J, Jazwinski S. Rtg2 Protein Links Metabolism and Genome Stability in Yeast Longevity. Genetics (2004) 166(2):765–77. doi: 10.1534/genetics.166.2.765
49. Cruz C, Della Rosa M, Krueger C, Gao Q, Horkai D, King M, et al. Tri-Methylation of Histone H3 Lysine 4 Facilitates Gene Expression in Ageing Cells. eLife (2018) 7:e34081. doi: 10.7554/eLife.34081
50. Kunisada T, Yamagishi H, Ogita Z, Kirakawa T, Mitsui Y. Appearance of Extrachromosomal Circular DNAs During In Vivo and In Vitro Ageing of Mammalian Cells. Mech Ageing Dev (1985) 29(1):89–99. doi: 10.1016/0047-6374(85)90050-8
51. Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, Alkan C, et al. Mapping Copy Number Variation by Population-Scale Genome Sequencing. Nature (2011) 470(7332):59–65. doi: 10.1038/nature09708
52. Krishna RG, Vishnu Bhat B, Bobby Z, Papa D, Badhe B, Kalidoss VK, et al. Identification of Differentially Methylated Candidate Genes and Their Biological Significance in IUGR Neonates by Methylation EPIC Array. J Matern-Fetal Neonatal Med: Off J Eur Assoc Perinatal Med Fed Asia Oceania Perinatal Soc Int Soc Perinatal Obstet (2020) 2(24):1–9. doi: 10.1080/14767058.2020.1727881
53. Chen PY, Chu A, Liao WW, Rubbi L, Janzen C, Hsu FM, et al. Prenatal Growth Patterns and Birthweight Are Associated With Differential DNA Methylation and Gene Expression of Cardiometabolic Risk Genes in Human Placentas: A Discovery-Based Approach. Reprod Sci (Thousand Oaks Calif) (2018) 25(4):523–39. doi: 10.1177/1933719117716779
54. Fujimoto S, Yamagishi H. Isolation of an Excision Product of T-Cell Receptor Alpha-Chain Gene Rearrangements. Nature (1987) 327(6119):242–3. doi: 10.1038/327242a0
55. McCormack WT, Tjoelker LW, Carlson LM, Petryniak B, Barth CF, Humphries EH, et al. Chicken IgL Gene Rearrangement Involves Deletion of a Circular Episome and Addition of Single Nonrandom Nucleotides to Both Coding Segments. Cell (1989) 56(5):785–91. doi: 10.1016/0092-8674(89)90683-1
56. von Schwedler U, Jäck HM, Wabl M. Circular DNA Is a Product of the Immunoglobulin Class Switch Rearrangement. Nature (1990) 345(6274):452–6. doi: 10.1038/345452a0
57. Fu B, Zhou Y, Ni X, Tong X, Xu X, Dong Z, et al. Natural Killer Cells Promote Fetal Development Through the Secretion of Growth-Promoting Factors. Immunity (2017) 47(6):1100–13.e6. doi: 10.1016/j.immuni.2017.11.018
58. Yang F, Zheng Q, Jin L. Dynamic Function and Composition Changes of Immune Cells During Normal and Pathological Pregnancy at the Maternal-Fetal Interface. Front Immunol (2019) 10:2317. doi: 10.3389/fimmu.2019.02317
59. Robertson SA, Care AS, Moldenhauer LM. Regulatory T Cells in Embryo Implantation and the Immune Response to Pregnancy. J Clin Invest (2018) 128(10):4224–35. doi: 10.1172/jci122182
60. Xiong S, Sharkey AM, Kennedy PR, Gardner L, Farrell LE, Chazara O, et al. Maternal Uterine NK Cell-Activating Receptor KIR2DS1 Enhances Placentation. J Clin Invest (2013) 123(10):4264–72. doi: 10.1172/jci68991
61. Moffett A, Hiby SE, Sharkey AM. The Role of the Maternal Immune System in the Regulation of Human Birthweight. Philos Trans R Soc London Ser B Biol Sci (2015) 370(1663):20140071. doi: 10.1098/rstb.2014.0071
62. Koche RP, Rodriguez-Fos E, Helmsauer K, Burkert M, MacArthur IC, Maag J, et al. Extrachromosomal Circular DNA Drives Oncogenic Genome Remodeling in Neuroblastoma. Nat Genet (2020) 52(1):29–34. doi: 10.1038/s41588-019-0547-z
63. Morton A, Dogan-Artun N, Faber Z, MacLeod G, Bartels C, Piazza M, et al. Functional Enhancers Shape Extrachromosomal Oncogene Amplifications. Cell (2019) 179(6):1330–41.e13. doi: 10.1016/j.cell.2019.10.039
64. Beroukhim R, Zhang X, Meyerson M. Copy Number Alterations Unmasked as Enhancer Hijackers. Nat Genet (2016) 49(1):5–6. doi: 10.1038/ng.3754
65. Storlazzi C, Lonoce A, Guastadisegni M, Trombetta D, D’Addabbo P, Daniele G, et al. Gene Amplification as Double Minutes or Homogeneously Staining Regions in Solid Tumors: Origin and Structure. Genome Res (2010) 20(9):1198–206. doi: 10.1101/gr.106252.110
66. Deshpande V, Luebeck J, Nguyen N, Bakhtiari M, Turner K, Schwab R, et al. Exploring the Landscape of Focal Amplifications in Cancer Using AmpliconArchitect. Nat Commun (2019) 10(1):392. doi: 10.1038/s41467-018-08200-y
67. Møller H, Ramos-Madrigal J, Prada-Luengo I, Gilbert M, Regenberg B. Near-Random Distribution of Chromosome-Derived Circular DNA in the Condensed Genome of Pigeons and the Larger, More Repeat-Rich Human Genome. Genome Biol Evol (2020) 12(1):3762–77. doi: 10.1093/gbe/evz281
68. Kopp F, Mendell JT. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell (2018) 172(3):393–407. doi: 10.1016/j.cell.2018.01.011
69. Medina-Bastidas D, Guzmán-Huerta M, Borboa-Olivares H, Ruiz-Cruz C, Parra-Hernández S, Flores-Pliego A, et al. Placental Microarray Profiling Reveals Common mRNA and lncRNA Expression Patterns in Preeclampsia and Intrauterine Growth Restriction. Int J Mol Sci (2020) 21(10):3597. doi: 10.3390/ijms21103597
70. Apicella C, Ruano CSM, Méhats C, Miralles F, Vaiman D. The Role of Epigenetics in Placental Development and the Etiology of Preeclampsia. Int J Mol Sci (2019) 20(11):2837. doi: 10.3390/ijms20112837
71. Song X, Luo X, Gao Q, Wang Y, Gao Q, Long W. Dysregulation of LncRNAs in Placenta and Pathogenesis of Preeclampsia. Curr Drug Targets (2017) 18(10):1165–70. doi: 10.2174/1389450118666170404160000
72. Lipka A, Paukszto L, Majewska M, Jastrzebski JP, Myszczynski K, Panasiewicz G, et al. Identification of Differentially Expressed Placental Transcripts During Multiple Gestations in the Eurasian Beaver (Castor Fiber L.). Reprod Fertil Dev (2017) 29(10):2073–84. doi: 10.1071/rd16186
73. Matthews JC, Beveridge MJ, Malandro MS, Rothstein JD, Campbell-Thompson M, Verlander JW, et al. Activity and Protein Localization of Multiple Glutamate Transporters in Gestation Day 14 vs. Day 20 Rat Placenta. Am J Physiol (1998) 274(3):C603–14. doi: 10.1152/ajpcell.1998.274.3.C603
74. Schimmel P. The Emerging Complexity of the tRNA World: Mammalian tRNAs Beyond Protein Synthesis. Nat Rev Mol Cell Biol (2018) 19(1):45–58. doi: 10.1038/nrm.2017.77
75. Nunes A, Ribeiro DR, Marques M, Santos MAS, Ribeiro D, Soares AR. Emerging Roles of tRNAs in RNA Virus Infections. Trends Biochem Sci (2020) 45(9):794–805. doi: 10.1016/j.tibs.2020.05.007
76. Su Z, Frost EL, Lammert CR, Przanowska RK, Lukens JR, Dutta A. tRNA-Derived Fragments and microRNAs in the Maternal-Fetal Interface of a Mouse Maternal-Immune-Activation Autism Model. RNA Biol (2020) 17(8):1183–95. doi: 10.1080/15476286.2020.1721047
77. Ali A, Bouma GJ, Anthony RV, Winger QA. The Role of LIN28-Let-7-ARID3B Pathway in Placental Development. Int J Mol Sci (2020) 21(10):3637. doi: 10.3390/ijms21103637
78. Addo KA, Palakodety N, Hartwell HJ, Tingare A, Fry RC. Placental microRNAs: Responders to Environmental Chemicals and Mediators of Pathophysiology of the Human Placenta. Toxicol Rep (2020) 7:1046–56. doi: 10.1016/j.toxrep.2020.08.002
79. Kotlabova K, Doucha J, Hromadnikova I. Placental-Specific microRNA in Maternal Circulation–Identification of Appropriate Pregnancy-Associated microRNAs With Diagnostic Potential. J Reprod Immunol (2011) 89(2):185–91. doi: 10.1016/j.jri.2011.02.006
80. Li H, Ouyang Y, Sadovsky E, Parks WT, Chu T, Sadovsky Y. Unique microRNA Signals in Plasma Exosomes From Pregnancies Complicated by Preeclampsia. Hypertension (2020) 75(3):762–71. doi: 10.1161/hypertensionaha.119.14081
81. Kim SH, MacIntyre DA, Binkhamis R, Cook J, Sykes L, Bennett PR, et al. Maternal Plasma miRNAs as Potential Biomarkers for Detecting Risk of Small-for-Gestational-Age Births. EBioMedicine (2020) 62:103145. doi: 10.1016/j.ebiom.2020.103145
82. Mouillet JF, Chu T, Hubel CA, Nelson DM, Parks WT, Sadovsky Y. The Levels of Hypoxia-Regulated microRNAs in Plasma of Pregnant Women With Fetal Growth Restriction. Placenta (2010) 31(9):781–4. doi: 10.1016/j.placenta.2010.07.001
83. Awamleh Z, Gloor GB, Han VKM. Placental microRNAs in Pregnancies With Early Onset Intrauterine Growth Restriction and Preeclampsia: Potential Impact on Gene Expression and Pathophysiology. BMC Med Genomics (2019) 12(1):91. doi: 10.1186/s12920-019-0548-x
84. Geng HH, Li R, Su YM, Xiao J, Pan M, Cai XX, et al. The Circular RNA Cdr1as Promotes Myocardial Infarction by Mediating the Regulation of miR-7a on Its Target Genes Expression. PloS One (2016) 11(3):e0151753. doi: 10.1371/journal.pone.0151753
85. Liu Q, Zhang X, Hu X, Dai L, Fu X, Zhang J, et al. Circular RNA Related to the Chondrocyte ECM Regulates MMP13 Expression by Functioning as a MiR-136 ‘Sponge’ in Human Cartilage Degradation. Sci Rep (2016) 6:22572. doi: 10.1038/srep22572
86. Maass PG, Glažar P, Memczak S, Dittmar G, Hollfinger I, Schreyer L, et al. A Map of Human Circular RNAs in Clinically Relevant Tissues. J Mol Med (Berlin Germany) (2017) 95(11):1179–89. doi: 10.1007/s00109-017-1582-9
87. Qian Y, Lu Y, Rui C, Qian Y, Cai M, Jia R. Potential Significance of Circular RNA in Human Placental Tissue for Patients With Preeclampsia. Cell Physiol Biochem: Int J Exp Cell Physiol Biochem Pharmacol (2016) 39(4):1380–90. doi: 10.1159/000447842
88. Esteller M. Non-Coding RNAs in Human Disease. Nat Rev Genet (2011) 12(12):861–74. doi: 10.1038/nrg3074
89. Yin QF, Yang L, Zhang Y, Xiang JF, Wu YW, Carmichael GG, et al. Long Noncoding RNAs With snoRNA Ends. Mol Cell (2012) 48(2):219–30. doi: 10.1016/j.molcel.2012.07.033
90. Da Costa L, Leblanc T, Mohandas N. Diamond-Blackfan Anemia. Blood (2020) 136(11):1262–73. doi: 10.1182/blood.2019000947
91. Rahmatpanah F, Agrawal S, Scarfone VM, Kapadia S, Mercola D, Agrawal A. Transcriptional Profiling of Age-Associated Gene Expression Changes in Human Circulatory CD1c+ Myeloid Dendritic Cell Subset. J Gerontol Ser A Biol Sci Med Sci (2019) 74(1):9–15. doi: 10.1093/gerona/gly106
92. Muinelo-Romay L, Casas-Arozamena C, Abal M. Liquid Biopsy in Endometrial Cancer: New Opportunities for Personalized Oncology. Int J Mol Sci (2018) 19(8):2311. doi: 10.3390/ijms19082311
93. Khakoo S, Georgiou A, Gerlinger M, Cunningham D, Starling N. Circulating Tumour DNA, a Promising Biomarker for the Management of Colorectal Cancer. Crit Rev Oncol/Hematol (2018) 122:72–82. doi: 10.1016/j.critrevonc.2017.12.002
94. Norton ME, Jacobsson B, Swamy GK, Laurent LC, Ranzini AC, Brar H, et al. Cell-Free DNA Analysis for Noninvasive Examination of Trisomy. N Engl J Med (2015) 372(17):1589–97. doi: 10.1056/NEJMoa1407349
95. Paulsen T, Kumar P, Koseoglu MM, Dutta A. Discoveries of Extrachromosomal Circles of DNA in Normal and Tumor Cells. Trends Genet: TIG (2018) 34(4):270–8. doi: 10.1016/j.tig.2017.12.010
Keywords: eccDNA, ecDNA, fetal growth restriction, placenta, Circle-seq, immunity
Citation: Yang H, He J, Huang S, Yang H, Yi Q, Tao Y, Chen M, Zhang X and Qi H (2021) Identification and Characterization of Extrachromosomal Circular DNA in Human Placentas With Fetal Growth Restriction. Front. Immunol. 12:780779. doi: 10.3389/fimmu.2021.780779
Received: 21 September 2021; Accepted: 29 November 2021;
Published: 21 December 2021.
Edited by:
Cheong-Meng Chong, University of Macau, ChinaReviewed by:
Xing-Hua Gao, The First Affiliated Hospital of China Medical University, ChinaCopyright © 2021 Yang, He, Huang, Yang, Yi, Tao, Chen, Zhang and Qi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuemei Zhang, emhhbmd4dWVtZWk0MDlAMTYzLmNvbQ==; Hongbo Qi, cWlob25nYm9jeUBnbWFpbC5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.