- 1Division of Neonatology, Department of Pediatrics, Erasmus University Medical Center, Rotterdam, Netherlands
- 2Division of Pharmacology and Vascular Medicine, Department of Internal Medicine, Erasmus University Medical Center, Rotterdam, Netherlands
- 3Division of Experimental Cardiology, Department of Cardiology, Erasmus University Medical Center, Rotterdam, Netherlands
- 4Department of Pathology, Erasmus University Medical Center, Rotterdam, Netherlands
- 5The Tumor Immuno-Pathology (TIP) Laboratory, Erasmus University Medical Center, Rotterdam, Netherlands
- 6Department of Obstetrics and Gynecology, Erasmus University Medical Center, Rotterdam, Netherlands
- 7Department of Surgery, Erasmus University Medical Center, Rotterdam, Netherlands
Preeclampsia is a severe placenta-related pregnancy disorder that is generally divided into two subtypes named early-onset preeclampsia (onset <34 weeks of gestation), and late-onset preeclampsia (onset ≥34 weeks of gestation), with distinct pathophysiological origins. Both forms of preeclampsia have been associated with maternal systemic inflammation. However, alterations in the placental immune system have been less well characterized. Here, we studied immunological alterations in early- and late-onset preeclampsia placentas using a targeted expression profile approach. RNA was extracted from snap-frozen placenta samples (healthy n=13, early-onset preeclampsia n=13, and late-onset preeclampsia n=6). The expression of 730 immune-related genes from the Pan Cancer Immune Profiling Panel was measured, and the data were analyzed in the advanced analysis module of nSolver software (NanoString Technology). The results showed that early-onset preeclampsia placentas displayed reduced expression of complement, and toll-like receptor (TLR) associated genes, specifically TLR1 and TLR4. Mast cells and M2 macrophages were also decreased in early-onset preeclampsia compared to healthy placentas. The findings were confirmed by an immunohistochemistry approach using 20 healthy, 19 early-onset preeclampsia, and 10 late-onset preeclampsia placentas. We conclude that the placental innate immune system is altered in early-onset preeclampsia compared to uncomplicated pregnancies. The absence of these alterations in late-onset preeclampsia placentas indicates dissimilar immunological profiles. The study revealed distinct pathophysiological processes in early-onset and late-onset preeclampsia placentas and imply that a tailored treatment to each subtype is desirable.
1. Introduction
Placentation and subsequent development of the placenta are essential processes that determine an uneventful pregnancy and good fetal outcome. Problems of placental development and adaptation can result in impaired fetal growth and preeclampsia affecting 2-8% of all pregnancies (1). The serious complications of preeclampsia can ultimately progress into maternal and fetal death, but if prevented, can also have other short-term and long-term negative impacts on the health of both mother and infant (2, 3). Apart from symptom relief and the undesirable option to terminate pregnancy and deliver the placenta, often necessitating preterm delivery of the child, preeclampsia cannot be treated successfully yet.
Although the etiology is incompletely understood, preeclampsia is generally considered a condition of placental insufficiency, accompanied by an exaggeration of the normal pregnancy-induced maternal systemic inflammatory response (4). The immune system plays important roles in placental development; directing trophoblast invasion, maintaining feto-maternal tolerance, and fighting invading pathogens when necessary (5). In a healthy pregnancy, immune cells are present in and around the placenta throughout all trimesters of pregnancy and are involved in interactions between the mother and the placenta. The placental bed, the decidua, is mainly occupied by cells of the innate immune system, including uterine natural killer (NK) cells, macrophages, mast cells, and dendritic cells (DCs) (5, 6). Adaptive immune cells including regulatory T cells and B cells are present in lower numbers and may be involved in maintaining a stable and appropriate immune response during later stages of pregnancy (5, 7, 8). The placenta itself mostly contains fetal innate immune cells, particularly fetal macrophages named Hofbauer cells. Immunological perturbations might be related to placental insufficiency, and multiple studies have shown elevated circulating inflammatory markers in pregnant women with preeclampsia (9–13). Preeclamptic placentas were also shown to express increased levels of the antiangiogenic factors endoglin and soluble fms-like tyrosine kinase-1 (14–16), which can both be shed into the maternal circulation. An increase in the concentrations of maternally circulating endoglin and sFlt1, and a subsequent decrease in placental growth factor (PlGF) signaling were identified as the major causal factors for the development of preeclampsia (1, 2). However, the involvement of the placental immune system is less clear as studies on placental immune cell alterations in preeclampsia have reported contradictory results, and the exact roles of placental immune cells in preeclampsia remain to be elucidated (5, 6).
Hence, in contrast to the well-defined maternal systemic inflammatory state in preeclampsia, placental immune alterations have been less well characterized. Tumors and placentas display striking similarities in developmental processes, and these similarities are increasingly recognized to be useful for understanding processes in either tissue (17, 18). A methodology that has emerged in the field of oncology uses the expression of immune-related genes to investigate the immune responses and the involved immune cell types and pathways in various tissue types (www.nanostring.com). A high sensitivity of this technique allows the identification of non-abundant immune cells, even in diseases with an immune-suppressive microenvironment like pancreatic cancers (19–21). Therefore, this technique provides a novel opportunity to investigate the immune profile of the placentas of women with preeclampsia.
While most studies cluster preeclampsia as one condition, it is generally acknowledged that two entities, with separate pathophysiological origins, can be distinguished: early-onset preeclampsia (onset <34 weeks of gestation), thought to originate from deficient placentation, and late-onset preeclampsia (onset ≥34 weeks of gestation), believed to be predominantly driven by maternal cardiovascular risk factors and placental insufficiency (2). Distinct placental mRNA expression profiles between early-onset preeclampsia and late-onset preeclampsia (22) suggest that inconsistent immunological findings in the placenta might be explained by a lacking subtype definition of preeclampsia. The identification of placental immunological differences may lead to the discovery of unidentified pathophysiological processes and potential targets for future therapeutic interventions. Therefore, with the present study, we aimed to reveal placental immune alterations between early-onset preeclampsia, late-onset preeclampsia, and healthy placentas. This study investigated this for the first time in the placenta by measuring an immune profiling gene expression array that was developed to investigate changes in cancer tissue, studying transcriptional, cellular and pathway alterations.
2. Methods
2.1. Study Cohort
All patients who were admitted to the Erasmus MC in Rotterdam gave written approval to participate in this study, prior to giving birth. The participants were either patients with singleton term pregnancies undergoing elective caesarean section or patients with preeclampsia, diagnosed based on the new ISSHP 2018 criteria, i.e., new-onset hypertension after 20 weeks of gestation (systolic blood pressure ≥140 mmHg or diastolic blood pressure ≥90 mmHg), accompanied by maternal organ dysfunction (e.g. proteinuria as defined by urinary protein/creatinine ratio >30 mg/mmol) and/or uteroplacental dysfunction defined as fetal growth restriction (23). Early-onset preeclampsia was defined based on the onset of symptoms <34 weeks of gestation, while late-onset preeclampsia ≥34 weeks of gestation. Clinical data were retrieved from the digital medical files. The study was exempted from medical approval according to the Dutch Medical Research with Human Subjects Law (MEC-2016-418 and MEC-2017-418). The gene expression analysis was performed in 16 healthy, 13 early-onset preeclampsia and 6 late-onset preeclampsia placentas. Since formalin-fixed, paraffin-embedded (FFPE) material was not available for all samples included in the gene expression analysis, an additional independent cohort of patients (9 healthy, 9 early-onset preeclampsia, and 6 late-onset preeclampsia), was added to validate the results by immunohistochemistry. Immunohistochemistry was carried out in a final cohort of 20 healthy, 20 early-onset preeclampsia and 10 late-onset preeclampsia patients.
2.2. Tissue Preparation
Within 20 minutes after delivery of the placenta, biopsies were taken from areas without obvious infarcts/necrosis, snap-frozen and stored at -80°C. In a limited number of samples from the same population, one additional full-thickness slice (1 cm in thickness) was collected per placenta and fixed in formalin, then embedded in paraffin (FFPE). Since FFPE samples were not available from all patients included in the gene expression profiling measurements, the number of FFPE samples was expanded as described in the previous section to confirm the results by immunohistochemistry.
2.3. RNA Extraction and Gene Expression Array Measurements
For RNA extraction, frozen biopsies were homogenized in RLT lysis buffer (Qiagen, Venlo, the Netherlands) with β-mercaptoethanol. After proteinase K (Invitrogen, Breda, the Netherlands) treatment for ten minutes at 55°C, RNA was extracted with the Qiagen RNA easy mini kit (Venlo, the Netherlands) according to the manufacturer’s instructions, eluted in RNA free water, and stored at -80°C. RNA Quality Control (QC) was measured using the 2100 Bioanalyzer (Agilent, CA, USA). NanoString Technology is based on detecting the genes of interest using 100 base pairs, but a cut-off value of 300 base pairs is necessary to increase the accuracy of measuring all the genes. Therefore, the total RNA concentration was corrected to include fragments seized between 300 and 4000 nucleotides. A total of 200 ng RNA was hybridized to the PanCancer Immune Profiling Panel for 17 hours at 67°C, following the manufacturing procedure (NanoString, Seattle, USA). The nCounter FLEX platform was used and genes were counted by scanning 490 Fields-of-view (FOV).
2.3.1. Data Analysis
Data analysis was performed using the advanced analysis module (version 2.0) of nSolver™ software (version 4.0, NanoString Technology). The quality control of the measurements was done according to the general workflow used in the NanoString nCounter software analysis (24). Based on expression stability and minimum variance, thirty-three housekeeping genes (out of 40) were selected for normalization with the geNorm algorithm embedded in the advanced analysis module (Supplementary Table S1). The lower threshold of expression was calculated by multiplying the average expression of the negative controls by two. Genes that showed an expression count below the threshold in >80% of the samples were excluded from further analysis.
To identify differentially expressed genes in preeclampsia samples, healthy placenta samples were used as controls. Hence, data of the early-onset preeclampsia samples and that of the late-onset preeclampsia samples were compared to healthy control samples. In addition, to investigate the differences between the two groups of preeclampsia, data of late-onset preeclampsia were compared to that of early-onset preeclampsia. The normalized data were log2 transformed and the differentially expressed genes were identified using simplified negative binomial models, mixture negative binomial models, or log-linear models based on the convergence of each gene. The adjusted P-value was calculated using the Benjamini-Hoghberg method. Genes were considered differentially expressed when the adjusted P-value < 0.05 and the linear fold of change (FOC) > 1.5 in any direction.
2.3.2. Immune Cell Subset Analysis
Specific and unique cell type marker genes were used to define multiple immune cell subtypes (Supplementary Table S2). The marker genes were selected based on the default settings of the advanced analysis module of nSolver, combined with placenta-specific immune cell genes identified in single-cell RNA sequencing studies (25–27). By using marker genes to define the cell types we assume that the immune cell’s genes are expressed in that specific cell type only (mutually exclusive) and that the expression of the marker genes is equal between cells for all samples. As a quality check of the cell definition based on previous research, pairwise similarities between the marker genes of each cell type were considered sufficient if R ≥ 0.6 (Supplementary Figure S1) (28). Unfortunately, dendritic cell marker genes did not pass the quality control criteria, and therefore this cell type was not analyzed. The cell type marker genes were used to calculate a cell type score for each sample.
2.3.3. Pathway Analysis
Pathway alterations were analyzed by clustering genes into pathways using the default settings of the PanCancer Immune Profiling advanced analysis module in nSolver. The pathway scores were calculated for each sample as the square root of the average squared t-statistic of all genes in the corresponding pathway.
The database for annotation, visualization, and integrated discovery (DAVID) v6.8 (29–31) was used as an alternative method to functionally annotate and cluster genes. For this analysis, separate lists were used containing either the downregulated or upregulated genes with an unadjusted P-value < 0.05, comparing early-onset preeclampsia and late-onset preeclampsia to healthy placentas. Genes that were related to either TLR signaling, the complement pathway or chemotaxis-associated cytokines as identified in DAVID were used to perform unsupervised hierarchical heatmap clustering using Ward’s method and visualized using the ComplexHeatmap package in R (32).
2.4. Immunohistochemistry
2.4.1. M1/M2 Macrophage Staining
Sections of 4µm of FFPE samples were used for immunohistochemistry analysis. To visualize M1 and M2 macrophage subsets in placental tissue, the chromogenic duplex staining was done with CD68 and CD163 by an automated staining procedure using the Ventana Benchmark Discovery (Ventana Medical Systems Inc.). In brief, following deparaffinization and heat-induced antigen retrieval with CC1 (#950-500, Ventana) for 40 minutes at 95°C, the tissue samples were firstly incubated with CD68 for 60 minutes at 37˚C, followed by detection with anti-mouse HQ (#760-4814, Ventana) for 16 minutes and subsequently anti-HQ HRP (#760-4820), and visualized with Discovery Purple (#760-229, Ventana) for 32 minutes. An antibody denature step was performed using CC2 (#950-123, Ventana) for 20 minutes at 100˚C. Secondly, CD163 was incubated for 32 minutes at 37˚C, followed by detection with anti-mouse HQ and subsequently anti-HQ HRP, and visualized with Discovery Teal (#760-247, Ventana) for 32 minutes. Finally, all slides were counterstained with hematoxylin II (#760-2208) and bluing reagents (#760-2037) for 4 minutes. Antibody information and clonality can be found in Supplementary Table S3.
2.4.2. Mast Cell Staining
To visualize mast cells in placental tissue, sequential 4 µm thick (FFPE) sections were stained by conventional immunohistochemistry for tryptase, using the optiview universal DAB detection kit (#760-700, Ventana). In brief, following deparaffinization and heat-induced antigen retrieval with CC1 (#950-500, Ventana) for 64 minutes, the tissue samples were incubated with the antibody of interest (Supplementary Table S3) for 32 minutes at 37°C. Incubation was followed by hematoxylin II counterstain for 8 minutes and then a blue coloring reagent for 8 minutes according to the manufactures instructions (Ventana).
For analysis, the immunohistochemistry slides were scanned on a NanoZoomer 2.0-HT (Hamamatsu Photonics K.K.). In NDPview (version 2.5.19 Hamamatsu Photonics K.K.), 1 mm2 areas were selected in each slide, three in the central villus part of the placenta, and three covering the decidua. Only areas with open villus structures were selected, avoiding infarcted tissue and placental septa. The images were imported as separate JPEG images into ImageJ (version 1.53a Wayne Rasband, USA). The number of positive cells with visible nuclei was counted by hand. In the central areas of the placenta, only cells inside the villi were counted, excluding cells in the intervillous space. To normalize for variation in decidual thickness, the decidual area was drawn by hand, measured, and cell counts were reported as cells/mm2. For the combined CD68/CD163 staining, cells were counted as either double-positive cells or CD68 single-positive cells. These cells were predefined as M2, and M1 macrophages, respectively. All analyses were performed blinded by one researcher. Power analysis for immunohistochemistry revealed a requirement of n=20 per group to obtain statistical significance at α=0.05 and β=0.2. However, there was not enough late-onset preeclampsia material available, and one early-onset preeclampsia placenta had too many infarcts and was therefore excluded.
2.5. Statistical Tests
Unless stated otherwise, analyses were conducted and images were produced in Graphpad Prism 8 or R 4.1 using the ggplot2 package (33, 34). The cell type scores, pathway scores, and immunohistochemistry results were statistically tested between groups using a Kruskall-Wallis test with Tukey’s multiple comparisons test. A result was considered statistically significant if the P-value < 0.05.
3. Results
3.1. Clinical Characteristics
Out of the 36 mRNA samples which were measured by the gene expression array, four samples had to be excluded from further analysis. One sample was retrospectively excluded due to misclassification as late-onset preeclampsia while the patient suffered from pregnancy-induced hypertension. Three healthy control samples were excluded because these did not meet the quality control criteria after gene expression measurement. The binding density in these samples, reflecting the total percentage of detected genes, was <0.2 compared to >0.7 in the 32 included samples, this was also reflected by significantly lower counts of positive controls and housekeeping genes (Supplementary Figure S2). The remaining study population included samples of patients with early-onset preeclampsia (n=13), late-onset preeclampsia (n=6), and uncomplicated pregnancies (n=13) (Table 1).
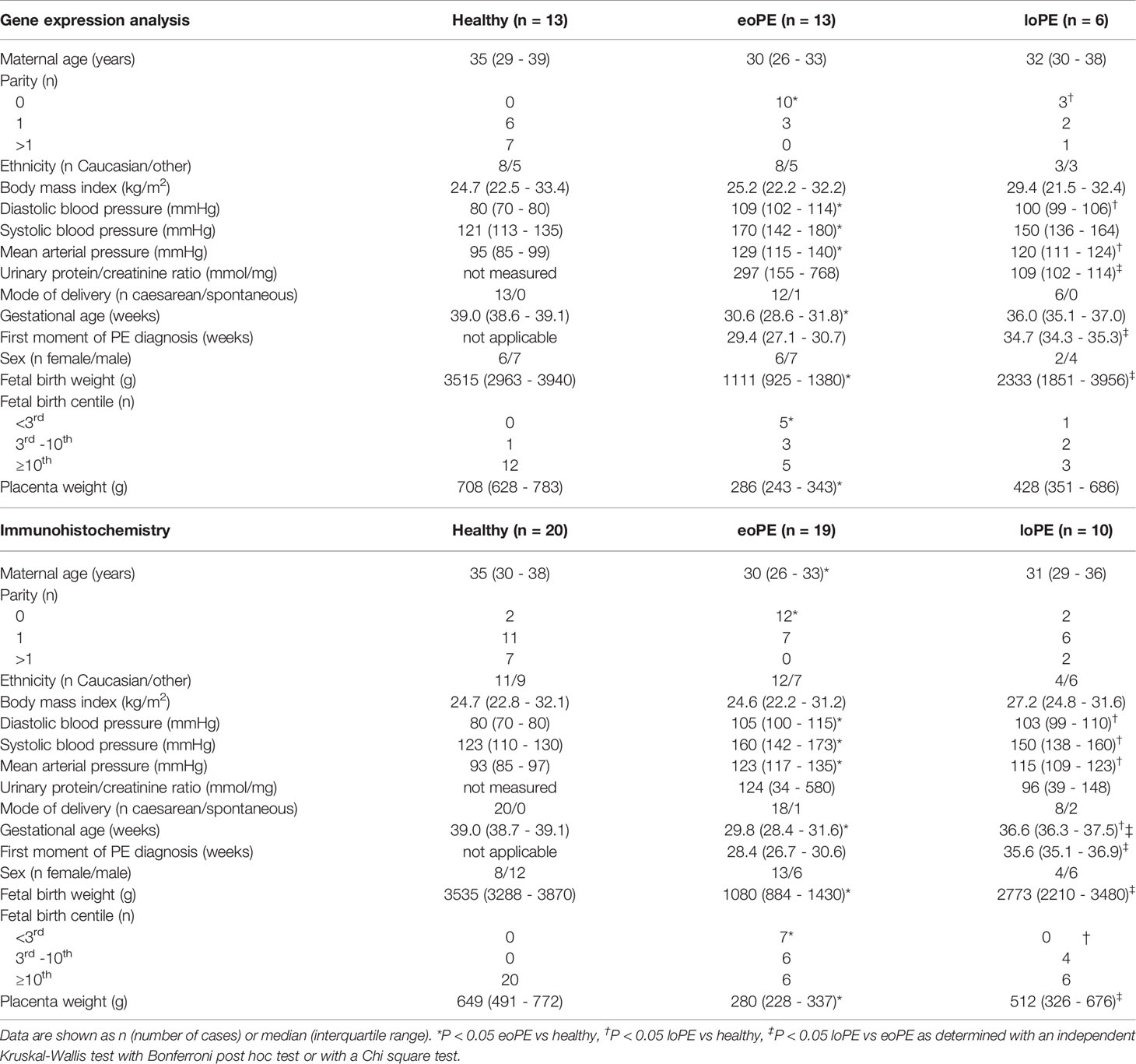
Table 1 Clinical characteristics of the healthy, early-onset preeclampsia (eoPE) and late-onset preeclampsia (loPE) patients included in the gene expression analysis and immunohistochemistry studies.
3.2. More Immune-Related Gene Expression Alterations in Early-Onset, Than in Late-Onset Preeclampsia Placentas
Analysis of 730 individual genes revealed 33 downregulated, and 15 upregulated genes in early-onset preeclampsia compared to healthy controls (Figure 1A). Late-onset preeclampsia placentas did not display any immune-related gene expression differences compared to healthy controls after correction for multiple testing (Figure 1B). However, eight downregulated and six upregulated genes were found in late-onset compared to early-onset preeclampsia placentas (Figure 1C).
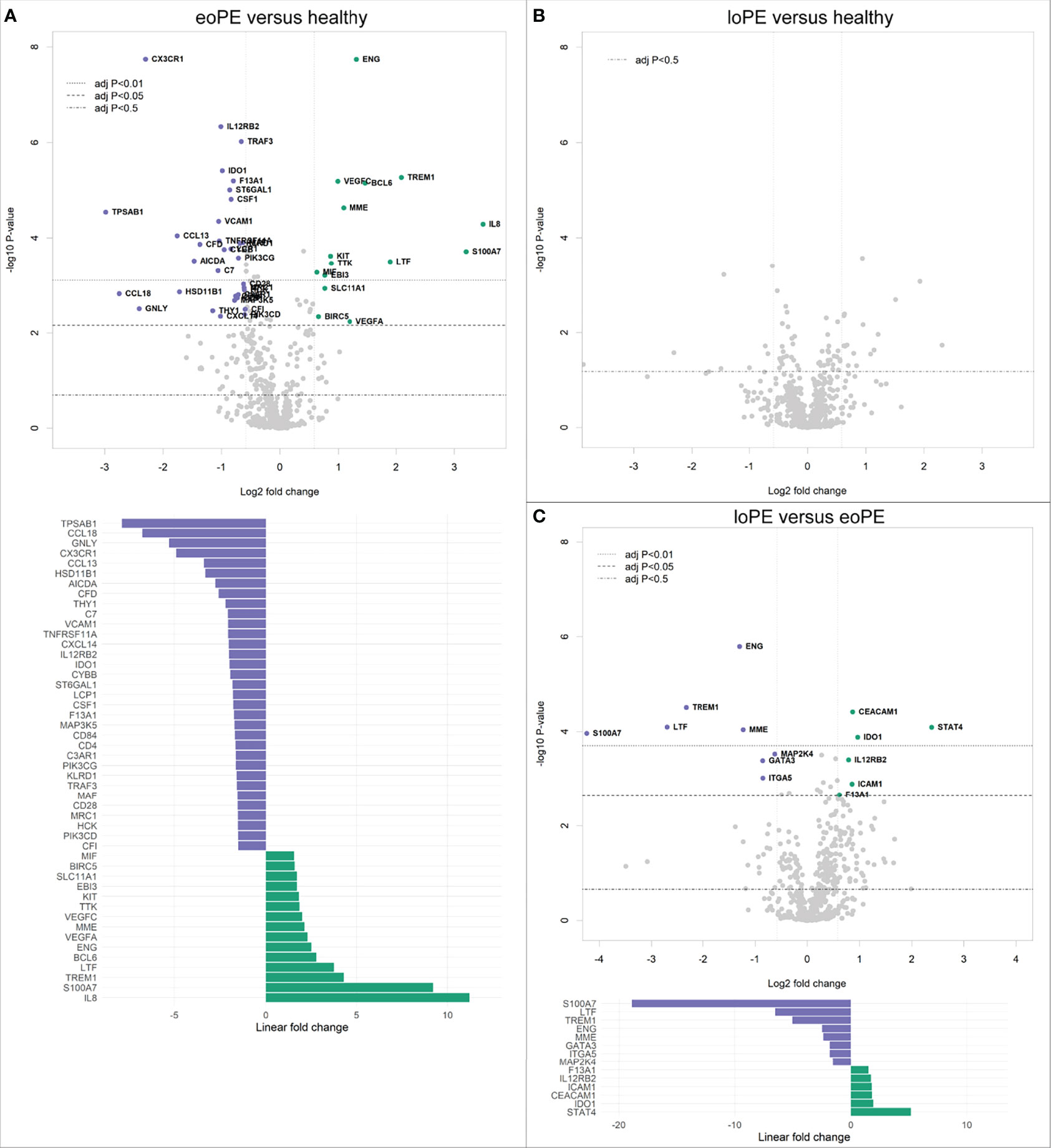
Figure 1 Gene expression is altered in early-onset preeclampsia (eoPE) compared to healthy and late-onset preeclampsia (loPE) placentas. Differential gene expression analyses in early-onset preeclampsia compared to healthy (A), late-onset preeclampsia compared to healthy (B), and late-onset compared to early-onset preeclampsia placentas (C). Points with annotation in the volcano plots depict genes with an adjusted P-value < 0.05 and FOC > 1.5. The bottom graphs in panels (A, C) display the linear fold changes of the differentially expressed genes. The annotations represent the official gene names.
3.3. Decreased Toll-Like Receptor (TLR) and Complement Signaling in Early-Onset Preeclampsia Placentas
Pathway analysis of the gene expression data, using multiple predefined pathways (Supplementary Table S4), revealed decreased expression of TLR-associated genes in early-onset preeclampsia compared to healthy placentas (P=0.005), mainly due to reduced expression of TLR4 and TLR1 (Figure 2A). Additionally, complement-associated genes were reduced in early-onset preeclampsia compared to healthy placentas (P=0.007, Figure 2B), including complement 7 (C7), complement C1q A chain (C1QA), complement 2 (C2), and complement C1r (C1R). None of the predefined pathways were statistically significantly altered in late-onset preeclampsia.
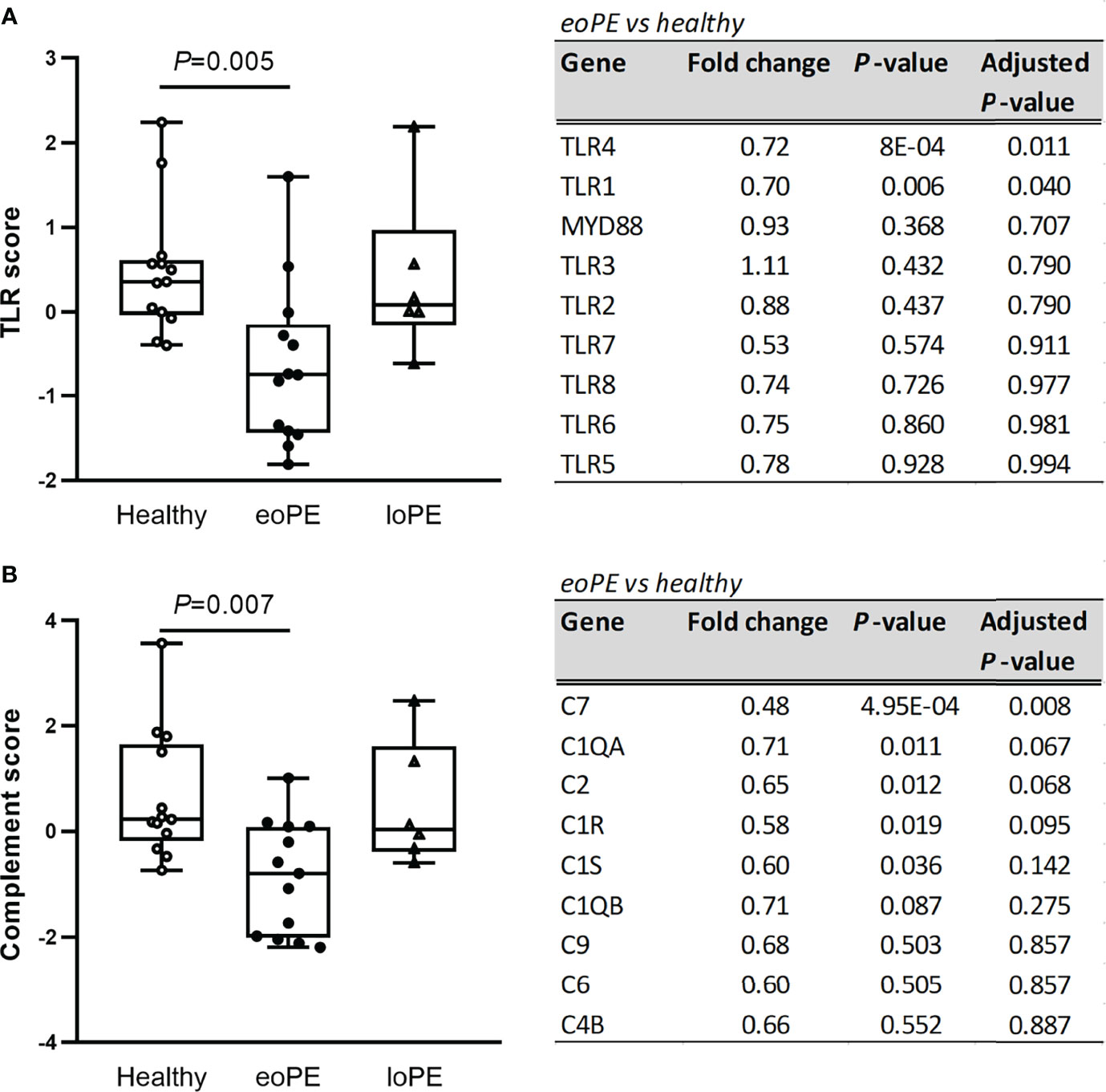
Figure 2 TLR- and complement-associated gene expression are diminished in early-onset preeclampsia (eoPE) placentas, but not in late-onset preeclampsia (loPE). The TLR score (A) and complement score (B) were calculated in each group using the genes displayed in the adjacent tables. Statistical significance was determined using the Kruskal-Wallis test followed by Tukey’s multiple comparisons tests.
DAVID functional clustering and pathway analysis was performed using the genes with an unadjusted P-value < 0.05 to investigate further interactions. In agreement with the gene expression findings, TLR signaling and complement activation were found to be reduced in early-onset preeclampsia compared to healthy placentas (Supplementary Table S5). In addition, “responses against non-self tissue” and “chemotaxis-associated chemokines” were identified as downregulated pathways in early-onset preeclampsia placentas. Genes that were upregulated in early-onset preeclampsia did not result in any significant pathway interactions. The pathways “inflammatory signaling” and “increased chemotaxis” networks were found to be significantly upregulated in late-onset preeclampsia compared to healthy placentas (Supplementary Table S5). No significant pathway interactions were obtained concerning the downregulated genes in late-onset preeclampsia. To highlight the genes which were involved in the most significant pathways, hierarchical heatmap clustering was performed and shown in Figure 3.
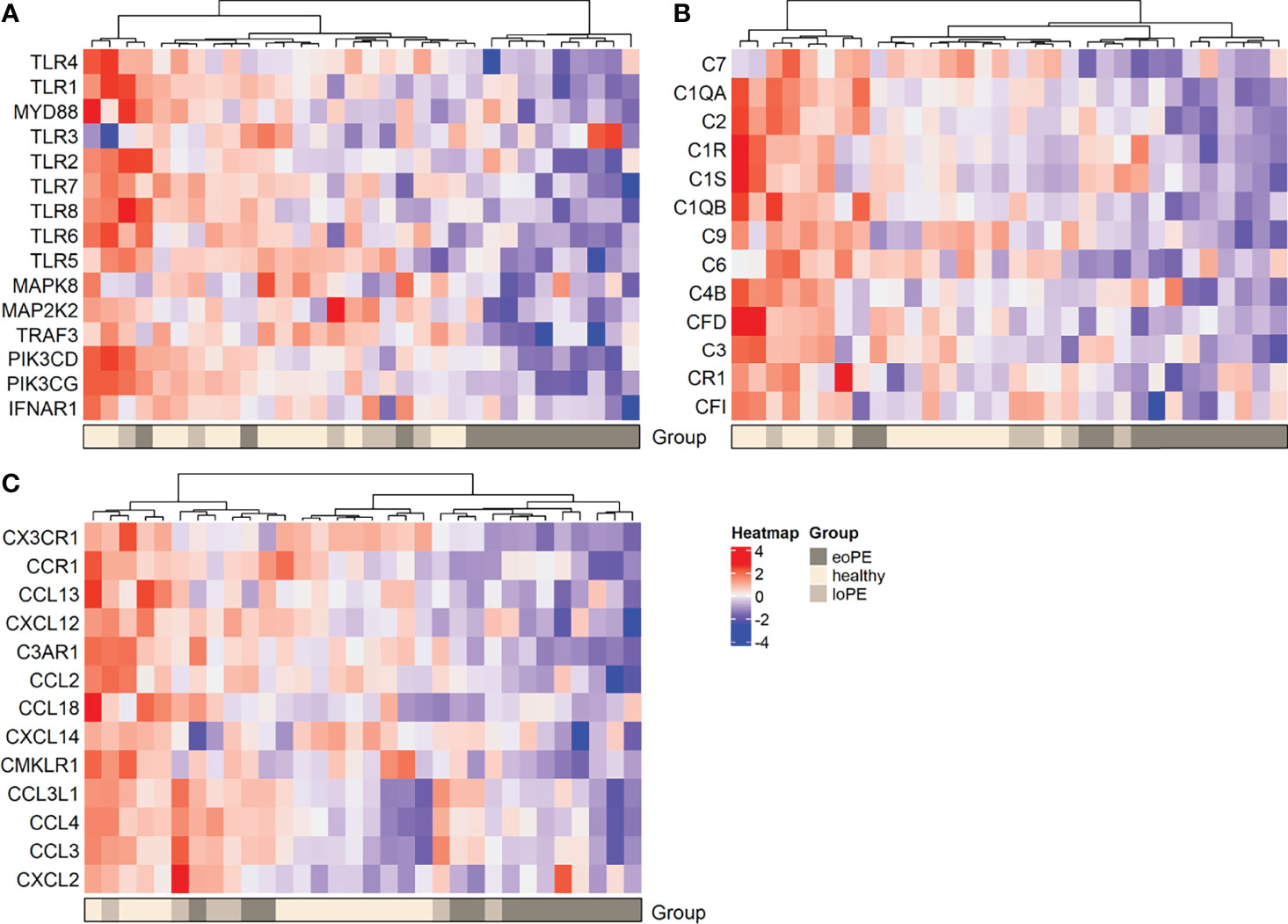
Figure 3 Heatmap hierarchical clustering displays lower expression of genes associated with the TLR pathway (A), complement activation (B), and chemokines (C) in early-onset preeclampsia placentas. Red depicts increased and blue depicts decreased mRNA expression scaled to the gene average. eoPE, early-onset preeclampsia; loPE, late-onset preeclampsia.
3.4. Reduced Innate Immune Cell Populations in Placentas From Early-Onset Preeclampsia Patients
Using the gene expression analysis, we identified multiple immune cell subtypes in the placenta (Supplementary Table S2). The genes that define NK CD56dim, regulatory T cell, Th1 cell, and CD8 T cell signals were expressed below the threshold of expression and therefore not identified nor compared between the samples. M2 macrophages (CD136+MRC1+C1QA+C1QB+, P=0.007) and mast cells (TPSAB1, P=0.007) were decreased in early-onset preeclampsia samples, while M1 macrophages (CD86+MSR1+) were not altered based on gene expression (Figures 4A–C). Although the expression of the B cell marker gene CD19 was increased in late-onset preeclampsia versus early-onset preeclampsia (P=0.015), it did not differ in either condition when compared to healthy placentas. All other investigated cell types did not statistically significantly differ between our groups.
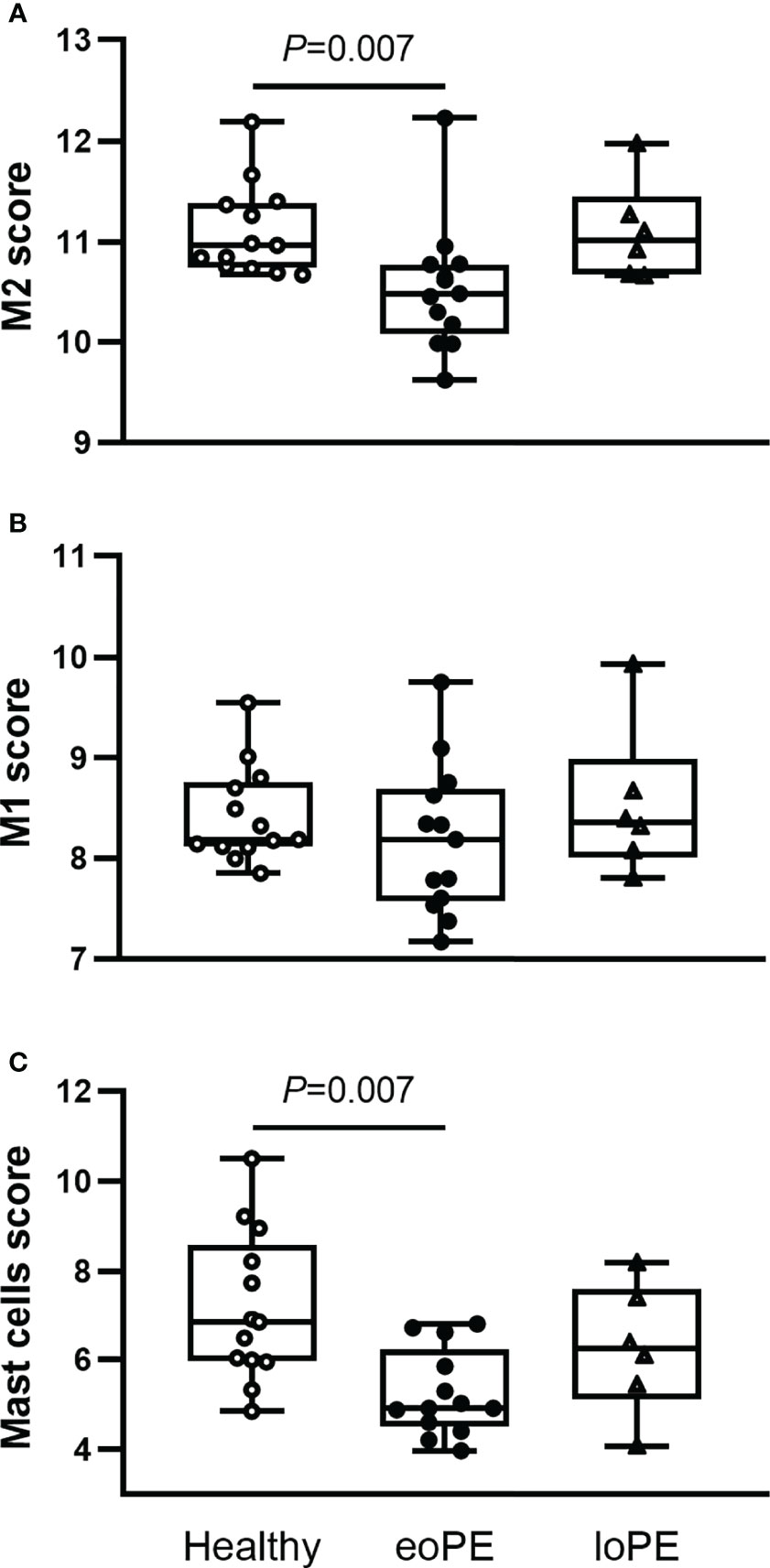
Figure 4 Mast cells and M2 macrophages are less abundant in early-onset preeclampsia (eoPE) placentas. Cell type scores were calculated based on the gene expression of CD163, MRC1, C1QA and C1QB for M2 macrophages (A), CD86 and MSR1 for M1 macrophages (B), TPSAB1 for mast cells (C). The lines represent median and interquartile ranges. Statistical significance was determined using the Kruskal-Wallis test followed by Tukey’s multiple comparisons tests. eoPE, early-onset preeclampsia; loPE, late-onset preeclampsia.
To verify these findings, immunohistochemistry was performed in a partially independent cohort as described in the Methods and defined in Table 1. Examples of the M1/M2 and mast cell immunohistochemistry images are shown in Figure 5, and the quantification of the immunohistochemistry is displayed in Figure 6. M2 macrophages were more abundant inside the villi of healthy compared to early-onset preeclampsia placentas (P=0.0003, Figure 6A), but not different in the decidua. M1 macrophages were not altered between the various groups (Figure 6B). While a few mast cells were detected inside the villi of the healthy placentas, they were rarely detected in the villi of early-onset preeclampsia placentas (P=0.02, Figure 6C). These early-onset preeclampsia placentas displayed a trend towards reduced mast cell numbers in the decidua as well (P=0.09, Figure 6C).
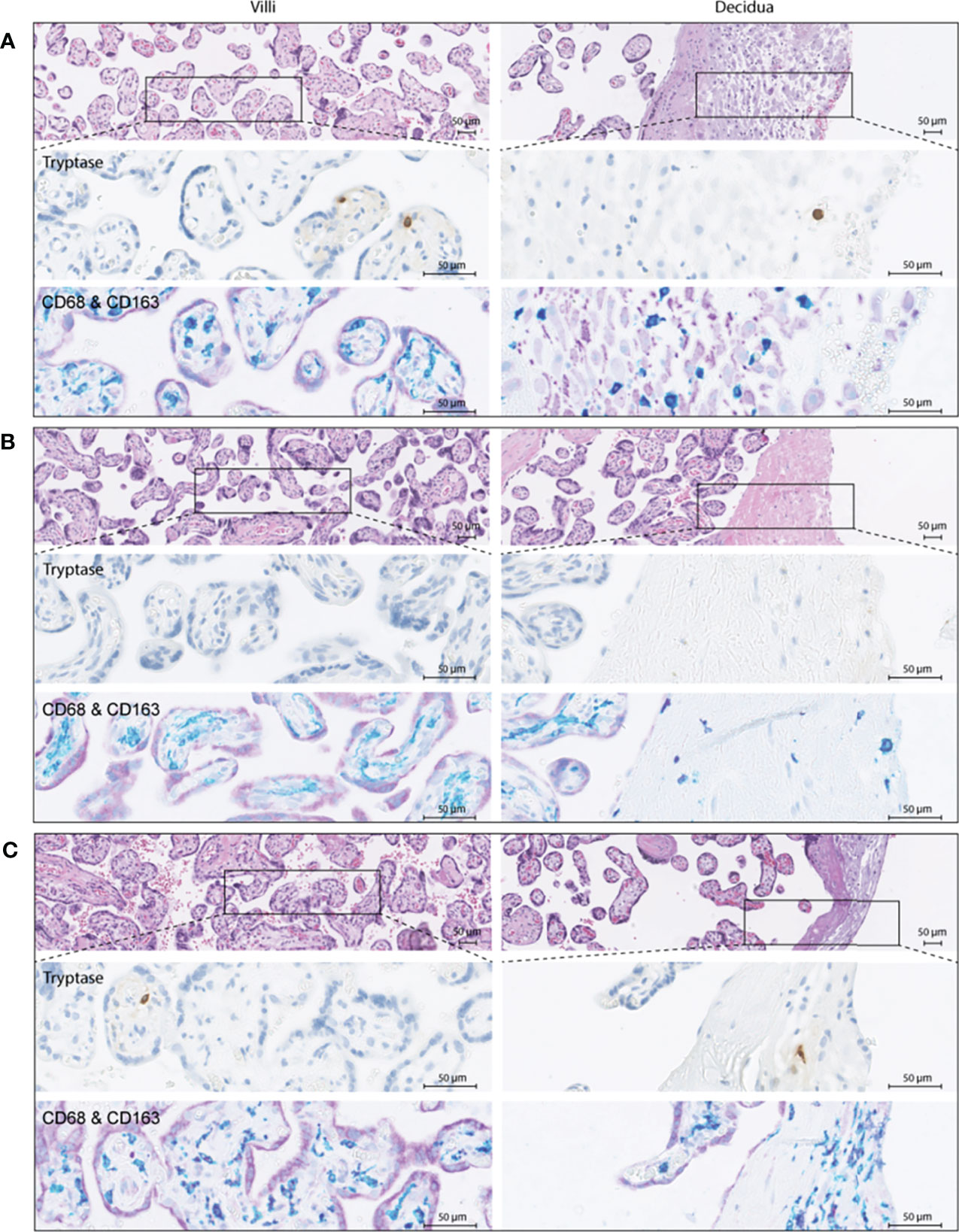
Figure 5 Immunohistochemistry examples of healthy (A), early-onset preeclampsia (eoPE, B) and late-onset preeclampsia (loPE, C) placentas. In each panel the top images were stained for hematoxylin and eosin, the middle images for tryptase to visualize mast cells, and the bottom images for CD68 (teal) & CD163 (purple) to visualize macrophages. CD68+CD163+ (dark purple) cells were defined as M2 macrophages, CD68+CD163- (teal) cells as M1 macrophages. The left panels display the placental villi, and the right panels display the decidua.
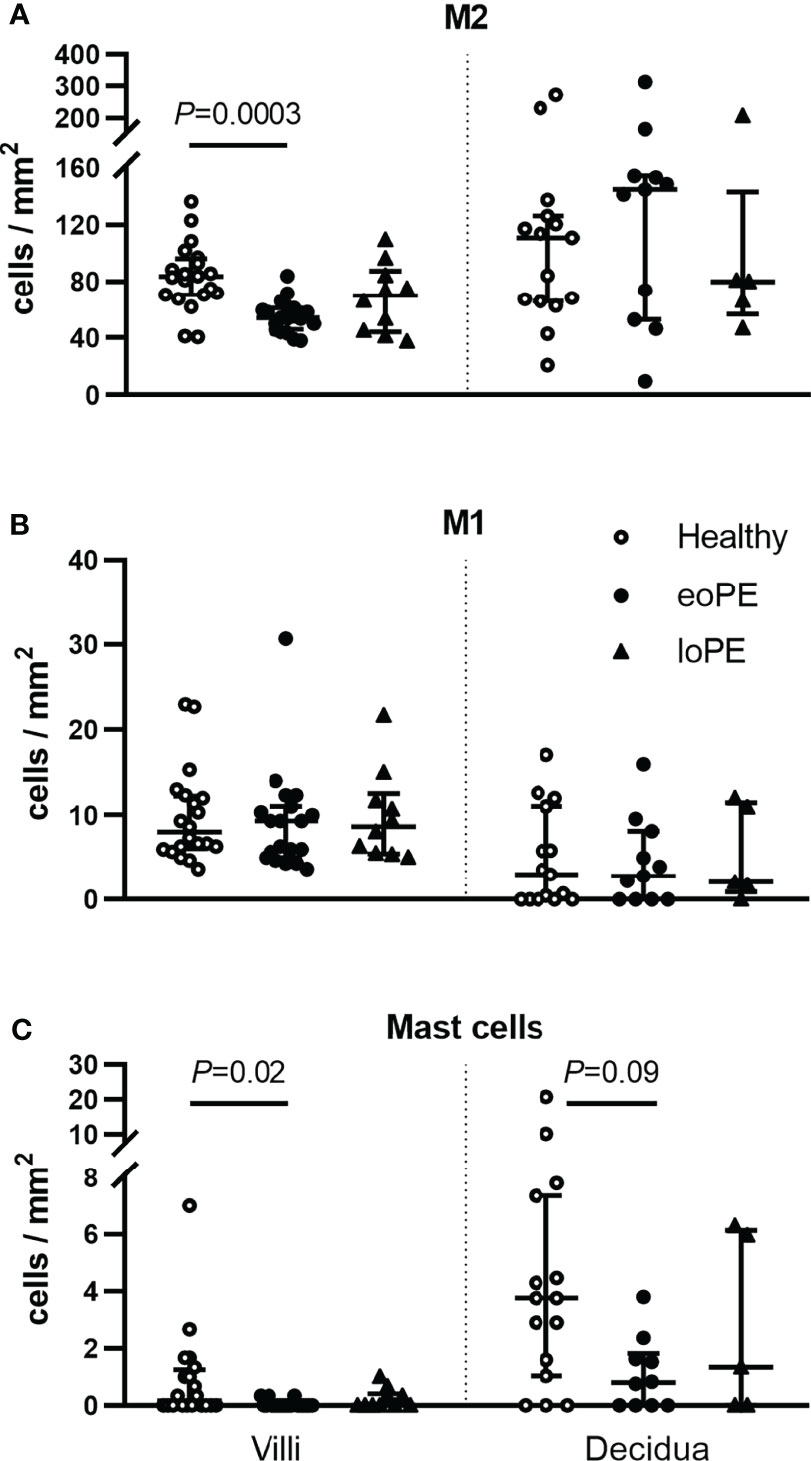
Figure 6 Mast cells and M2 macrophages are less abundant in the villi of early-onset preeclampsia (eoPE) placentas. M2 macrophages (CD68+CD163+, A), M1 macrophages (CD68+CD163-, B) and mast cells (tryptase, C) were stained by immunohistochemistry and quantified in the villi and in the decidual area. The decidua was missing in 5 healthy, 9 early-onset preeclampsia and 5 late-onset preeclampsia slides. The lines represent median and interquartile ranges. Statistical significance was determined using the Kruskal-Wallis test followed by Tukey’s multiple comparisons tests. eoPE, early-onset preeclampsia; loPE, late-onset preeclampsia.
4. Discussion
Placental immunological processes are important for the development of the placenta and fetus, and the maintenance of a tolerogenic environment. Disturbances in these processes could lead or contribute to preeclampsia, a condition that has been associated with a maternal systemic inflammatory state. However, placental immune alterations have been less well characterized. Data from the present study suggest that in contrast to the maternal systemic inflammatory state, placental innate immune factors are compromised in early-onset preeclampsia. Absence of these placental immune alterations in late-onset preeclampsia, suggests that both subtypes have dissimilar placental immune profiles, confirming distinct pathophysiological processes. It has been shown that placental dysfunction contributes to the maternal (inflammatory) syndrome in both subtypes. Nevertheless, the results of this study indicate that a maternal systemic inflammatory state does not necessarily translate to an inflammatory placenta.
Findings from the current study agree with the distinct gene expression profiles that have been identified between placentas from women with early-onset preeclampsia and late-onset preeclampsia (22, 35), and further extends this knowledge by identifying specific immune alterations. The placentas from patients with early-onset preeclampsia displayed increased placental endoglin expression as shown by the differential gene expression analysis, and in agreement with previous studies in women with severe preeclampsia (15, 16). However, earlier studies have also shown that placental expression of endoglin was increased in early-onset preeclampsia versus late-onset preeclampsia, and that elevated levels of circulating endoglin were more pronounced in preterm versus term preeclampsia (36, 37). Therefore, endoglin seems to have a more significant role in early-onset preeclampsia than in late-onset preeclampsia. Another gene that was differentially expressed between early-onset preeclampsia and late-onset preeclampsia was indoleamine 2,3-dioxygenase (IDO)1. The decreased IDO1 expression in early-onset preeclampsia is in agreement with previous studies (38–41), and based on the current study, seems to be specific to early-onset preeclampsia. IDO1 is the first enzyme of the kynurenine pathway of which we recently reviewed its placental functions and alterations in preeclampsia (42). The kynurenine pathway provides an important source of de novo NAD+ synthesis, and is involved in vascular development and functioning, anti- and pro-oxidative processes, and immune regulation. However, the function of the kynurenine pathway is mainly determined by tryptophan transporters (38), and by the spatial localization of IDO1 (42). The spatial distribution was not preserved in this study, therefore, it impossible to draw any further conclusions on the differential roles of IDO1 in preeclampsia.
Early-onset preeclampsia placentas displayed a decreased expression of TLR4, in agreement with earlier research (43), while studies in mild preeclampsia have reported either increased or unaltered TLR4 expression (44–49), similar to our late-onset preeclampsia patients. A role for reduced TLR4 signaling in early-onset preeclampsia was also suggested by a higher prevalence of allelic TLR4 gene variants with attenuated function in women with a history of early-onset preeclampsia (50). TLR4 can recognize several other pathogen-associated molecular patterns, in addition to its classical anti-microbial function (51, 52). The absence of visible infections, and the expression of TLR4 in many placental cells including trophoblasts (data not shown), suggests that TLR4 has other roles in the placenta than microbial defense only. In contrast to a decreased placental TLR4 expression as identified in the present study, previous studies have shown that the expression of TLR4 was elevated in the circulating innate immune cells of women with preeclampsia (53, 54). This suggests that the regulation of TLR4 is differently altered in preeclampsia in the fetal compared to the maternal compartment.
The present study confirmed that C7 is a potential gene candidate to discriminate early-onset from late-onset preeclampsia (22), and suggests a generally compromised complement system in early-onset preeclampsia placentas. Downregulation of C7 in the placentas of women with preeclampsia, but not in pregnancies with fetal growth restriction only, suggests this is specific to preeclampsia (55). While under normal conditions the complement system plays a role in regulating tolerance, apoptosis, angiogenesis, inflammation and clearance of free fetal DNA (56), locally decreased C1q production leads to decreased trophoblast invasion (57). Moreover, pregnant C1q deficient mice display a preeclampsia-like phenotype (58). Hence, insufficient production of complement factors can lead to impaired placental development and might precede early-onset preeclampsia.
A lower abundance of mast cells in the villi of early-onset preeclampsia placentas is in agreement with previous research (59), and seems specific to early-onset preeclampsia (60, 61). Mast cells are a source of angiogenic factors (62), and their depletion results in aberrant spiral artery remodeling and eventually fetal growth restriction (63). An increased mast cell number is related to a higher microvascular density in diabetic placentas (64), which was also associated with increased CX3CR1 expression (65). CX3CR1, the most statistically significant downregulated gene in the present study, is a transmembrane protein on immune cells, involved in cell adhesion, migration, and angiogenesis through interaction with its ligand CX3CL1 (fractalkine), and activation of hypoxia-inducible factor 1α (HIF1α) and vascular endothelial growth factor (VEGF) signaling (66). Hence, reductions in mast cells and CX3CR1 expression might contribute to impaired villous vascular development in early-onset preeclampsia placentas. However, CX3CR1 and the mast cell marker TPSAB1 did not correlate in the current gene expression analysis (data not shown), suggesting that CX3CR1 is expressed by other placental (immune) cells as well.
This study presents for the first time a decrease of M2 macrophages, presumably Hofbauer cells, in the villi of early-onset preeclampsia placentas. Reduced M2 macrophage numbers have been reported in fetal growth restriction and pregnancy-induced hypertension (67, 68). Since this already occurred in the first trimester (68), M2 alterations seemed to precede pathological placental development. While at the beginning of pregnancy M2 macrophages play a role in trophoblast invasion and spiral artery remodeling, decidual M2 macrophages were not altered in the current study. However, loss of M2 macrophages inside the villi, might be related to impaired clearance of apoptotic bodies, and insufficient protection of the fetus against intrauterine infection (69, 70). Earlier studies have shown that total macrophage numbers were not altered in preeclampsia (presumably late-onset based on the gestational age) and fetal growth restriction, but increased in the case of a decreased fetal growth rate (71, 72), suggesting that the lower M2 numbers in early-onset preeclampsia are not related to the co-occurrence of fetal growth restriction. Additionally, T cell infiltration was increased in preeclampsia and fetal growth restriction (71), although CD8 T cell genes were expressed below the detection limit in our study.
Recently, pyroptosis was identified as one of the key events that can contribute to systemic sterile inflammation specifically in patients with early-onset preeclampsia (73). Pyroptosis can trigger inflammatory responses independent of TLR4 and can cause membrane rupture and consequent release of pro-inflammatory cytokines like IL-1β, IL-18, and IL-33 (73, 74). Pyroptosis has been documented mainly in cells of the myeloid lineage such as macrophages, but recent work has demonstrated that pyroptosis occurs in primary human trophoblasts in the placenta from women with early-onset preeclampsia (75). Since pyroptosis was not investigated in the present study, its link to the currently identified alterations remains subject for future studies.
The current study has a few limitations. Firstly, an influence of the lower gestational age in women with early-onset preeclampsia cannot be ruled out. However, concurrent results between the present study and data from Nevalainen et al. (22) and Than et al. (35), who did use gestational age-matched controls, and absence of a correlation between gestational age and our most important findings within the early-onset preeclampsia group (data not shown), argue the results are unrelated to pregnancy duration. Secondly, as part of clinical care to improve fetal long maturation, twelve out of the thirteen early-onset preeclampsia patients in the gene expression analysis cohort received one or two dosages of the corticosteroid betamethasone within 48 hours of each other. Additional analyses reassured that the results were not confounded by betamethasone (Supplementary Figure S3). Thirdly, it should be acknowledged that the lower number of late-onset preeclampsia samples may have resulted in power loss for this group, and therefore potentially contributed to the absence of alterations. For example, the immunohistochemistry results revealed large variations in M2 macrophage and mast cell numbers in both late-onset preeclampsia and healthy samples, but the power to detect a statistically significant difference compared to early-onset preeclampsia was probably not achieved in late-onset preeclampsia, because of the lower number of included samples. Lastly, in the current study mRNA from whole placental tissue was used that included fetal and maternal blood, and some decidual tissue. This makes it impossible to discern which tissue compartments contributed to the identified differences. Moreover, rather than causal, certain immune components might have been downregulated in response to a maternal systemic inflammatory condition, although this generally described pro-inflammatory state may not be preeclampsia-specific but confounded by an increased maternal body mass index (76), which did not differ between our groups. Studying the spatial alterations of the placental immune compartments in both early-onset preeclampsia and late-onset preeclampsia will provide substantial information to understand the disease. In addition, changes in the placental microenvironment should be connected to those in the circulation, especially in early-onset preeclampsia, to investigate whether those women truly experience systemic inflammation.
In conclusion, the present study identified alterations in the innate immune system in placentas of women with early-onset preeclampsia, reflected by downregulated TLR and complement pathways, and a lower abundance of mast cells and M2 macrophages in the fetal villi. The identification of placental immune alterations in early-onset preeclampsia but not in late-onset preeclampsia asserts distinct pathophysiological processes, underlining the need to investigate them separately. These data provide a basis to direct future research into the placental innate immune system, and to discriminate between early-onset and late-onset preeclampsia, to eventually interfere and tailor treatment to these distinct pathological pregnancy conditions.
Data Availability Statement
The data presented in the study are deposited in the Gene Expression Omnibus (GEO) repository, accession number GSE190639, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE190639.
Ethics Statement
The study was reviewed by the Medische Ethische Toetsings Commisie Erasmus MC and exempted from medical approval according to the Dutch Medical Research with Human Subjects Law (MEC-2016-418 and MEC-2017-418). The participants provided their written informed consent to participate in this study.
Author Contributions
The study was designed by MB, IR, and DM. Data collection was performed by MB, EH, TB, and JD, whilst data analysis was done by MB and DM. MB, EH, RV, BR, and CE contributed to the data interpretation. MB wrote the manuscript and designed the figures. All authors contributed to the article and approved the submitted version.
Funding
This work was partially supported by the Support Casper Foundation (www.supportcasper.org).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Dr. S. Schoenmakers for his help with collecting the placentas.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.780043/full#supplementary-material
References
1. Rana S, Lemoine E, Granger JP, Karumanchi SA. Preeclampsia: Pathophysiology, Challenges, and Perspectives. Circ Res (2019) 124(7):1094–112. doi: 10.1161/CIRCRESAHA.118.313276
2. Steegers EAP, von Dadelszen P, Duvekot JJ, Pijnenborg R. Pre-Eclampsia. Lancet (2010) 376(9741):631–44. doi: 10.1016/S0140-6736(10)60279-6
3. Bellamy L, Casas JP, Hingorani AD, Williams DJ. Pre-Eclampsia and Risk of Cardiovascular Disease and Cancer in Later Life: Systematic Review and Meta-Analysis. BMJ (2007) 335(7627):974. doi: 10.1136/bmj.39335.385301.BE
4. Redman CWG, Sargent IL. Pre-Eclampsia, the Placenta and the Maternal Systemic Inflammatory Response—A Review. Placenta (2003) 24:S21–S7. doi: 10.1053/plac.2002.0930
5. Bulmer JN, Williams PJ, Lash GE. Immune Cells in the Placental Bed. Int J Dev Biol (2010) 54(2-3):281–94. doi: 10.1387/ijdb.082763jb
6. Faas MM, De Vos P. Innate Immune Cells in the Placental Bed in Healthy Pregnancy and Preeclampsia. Placenta (2018) 69:125–33. doi: 10.1016/j.placenta.2018.04.012
7. Tong M, Abrahams VM. Immunology of the Placenta. Obstet Gynecol Clin North Am (2020) 47(1):49–63. doi: 10.1016/j.ogc.2019.10.006
8. Wienke J, Brouwers L, van der Burg LM, Mokry M, Scholman RC, Nikkels PG, et al. Human Tregs at the Materno-Fetal Interface Show Site-Specific Adaptation Reminiscent of Tumor Tregs. JCI Insight (2020) 5(18):e137926. doi: 10.1172/jci.insight.137926
9. Ferguson KK, Meeker JD, McElrath TF, Mukherjee B, Cantonwine DE. Repeated Measures of Inflammation and Oxidative Stress Biomarkers in Preeclamptic and Normotensive Pregnancies. Am J Obstet Gynecol (2017) 216(5):527.e1–.e9. doi: 10.1016/j.ajog.2016.12.174
10. Szarka A, Rigo J Jr., Lazar L, Beko G, Molvarec A. Circulating Cytokines, Chemokines and Adhesion Molecules in Normal Pregnancy and Preeclampsia Determined by Multiplex Suspension Array. BMC Immunol (2010) 11:59. doi: 10.1186/1471-2172-11-59
11. Xie C, Yao MZ, Liu JB, Xiong LK. A Meta-Analysis of Tumor Necrosis Factor-Alpha, Interleukin-6, and Interleukin-10 in Preeclampsia. Cytokine (2011) 56(3):550–9. doi: 10.1016/j.cyto.2011.09.021
12. Kalinderis M, Papanikolaou A, Kalinderi K, Ioannidou E, Giannoulis C, Karagiannis V, et al. Elevated Serum Levels of Interleukin-6, Interleukin-1β and Human Chorionic Gonadotropin in Pre-Eclampsia. Am J Reprod Immunol (2011) 66(6):468–75. doi: 10.1111/j.1600-0897.2011.01019.x
13. Tosun M, Celik H, Avci B, Yavuz E, Alper T, Malatyalioğlu E. Maternal and Umbilical Serum Levels of Interleukin-6, Interleukin-8, and Tumor Necrosis Factor-α in Normal Pregnancies and in Pregnancies Complicated by Preeclampsia. J Matern Fetal Neonatal Med (2010) 23(8):880–6. doi: 10.3109/14767051003774942
14. Ahmad S, Ahmed A. Elevated Placental Soluble Vascular Endothelial Growth Factor Receptor-1 Inhibits Angiogenesis in Preeclampsia. Circ Res (2004) 95(9):884–91. doi: 10.1161/01.RES.0000147365.86159.f5
15. Toft JH, Toft JH, Lian IA, Tarca AL, Erez O, Espinoza J, et al. Whole-Genome Microarray and Targeted Analysis of Angiogenesis-Regulating Gene Expression (ENG, FLT1, VEGF, PlGF) in Placentas From Pre-Eclamptic and Small-for-Gestational-Age Pregnancies. J Matern Fetal Neonatal Med (2008) 21(4):267–73. doi: 10.1080/14767050801924118
16. Taki A, Abe M, Komaki M, Oku K, Iseki S, Mizutani S, et al. Expression of Angiogenesis-Related Factors and Inflammatory Cytokines in Placenta and Umbilical Vessels in Pregnancies With Preeclampsia and Chorioamnionitis/Funisitis. Congenital Anomalies (2012) 52(2):97–103. doi: 10.1111/j.1741-4520.2012.00359.x
17. Macklin PS, McAuliffe J, Pugh CW, Yamamoto A. Hypoxia and HIF Pathway in Cancer and the Placenta. Placenta (2017) 56:8–13. doi: 10.1016/j.placenta.2017.03.010
18. Costanzo V, Bardelli A, Siena S, Abrignani S. Exploring the Links Between Cancer and Placenta Development. Open Biol (2018) 8(6):180081. doi: 10.1098/rsob.180081
19. Lau SP, van Montfoort N, Kinderman P, Lukkes M, Klaase L, van Nimwegen M, et al. Dendritic Cell Vaccination and CD40-Agonist Combination Therapy Licenses T Cell-Dependent Antitumor Immunity in a Pancreatic Carcinoma Murine Model. J Immunother Cancer (2020) 8(2):e000772. doi: 10.1136/jitc-2020-000772
20. van der Sijde F, Li Y, Schraauwen R, de Koning W, van Eijck CHJ, Mustafa DAM. RNA From Stabilized Whole Blood Enables More Comprehensive Immune Gene Expression Profiling Compared to RNA From Peripheral Blood Mononuclear Cells. PloS One (2020) 15(6):e0235413. doi: 10.1371/journal.pone.0235413
21. Groeneveldt C, Kinderman P, van den Wollenberg DJM, van den Oever RL, Middelburg J, Mustafa DAM, et al. Preconditioning of the Tumor Microenvironment With Oncolytic Reovirus Converts CD3-Bispecific Antibody Treatment Into Effective Immunotherapy. J Immunother Cancer (2020) 8(2):e001191. doi: 10.1136/jitc-2020-001191
22. Nevalainen J, Skarp S, Savolainen E-R, Ryynänen M, Järvenpää J. Intrauterine Growth Restriction and Placental Gene Expression in Severe Preeclampsia, Comparing Early-Onset and Late-Onset Forms. J Perinatal Med (2017) 45(7):869–77. doi: 10.1515/jpm-2016-0406
23. Brown MA, Magee LA, Kenny LC, Karumanchi SA, McCarthy FP, Saito S, et al. Hypertensive Disorders of Pregnancy: ISSHP Classification, Diagnosis, and Management Recommendations for International Practice. Hypertension (2018) 72(1):24–43. doi: 10.1161/HYPERTENSIONAHA.117.10803
24. Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, et al. Direct Multiplexed Measurement of Gene Expression With Color-Coded Probe Pairs. Nat Biotechnol (2008) 26(3):317–25. doi: 10.1038/nbt1385
25. Tsang JCH, Vong JSL, Ji L, Poon LCY, Jiang P, Lui KO, et al. Integrative Single-Cell and Cell-Free Plasma RNA Transcriptomics Elucidates Placental Cellular Dynamics. Proc Natl Acad Sci (2017) 114(37):E7786–E95. doi: 10.1073/pnas.1710470114
26. Vento-Tormo R, Efremova M, Botting RA, Turco MY, Vento-Tormo M, Meyer KB, et al. Single-Cell Reconstruction of the Early Maternal–Fetal Interface in Humans. Nature (2018) 563(7731):347–53. doi: 10.1038/s41586-018-0698-6
27. Pique-Regi R, Romero R, Tarca AL, Sendler ED, Xu Y, Garcia-Flores V, et al. Single Cell Transcriptional Signatures of the Human Placenta in Term and Preterm Parturition. Elife (2019) 8:e52004. doi: 10.7554/eLife.52004
28. de Koning W, Latifi D, Li Y, van Eijck CHJ, Stubbs AP, Mustafa DAM. Identification, Validation, and Utilization of Immune Cells in Pancreatic Ductal Adenocarcinoma Based on Marker Genes. Front Immunol (2021) 12:649061. doi: 10.3389/fimmu.2021.649061
29. DAVID-Bioinformatics-Resources. Functional Annotation Tool . Available at: https://david.ncifcrf.gov/.
30. Huang da W, Sherman BT, Lempicki RA. Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat Protoc (2009) 4(1):44–57. doi: 10.1038/nprot.2008.211
31. Huang da W, Sherman BT, Lempicki RA. Bioinformatics Enrichment Tools: Paths Toward the Comprehensive Functional Analysis of Large Gene Lists. Nucleic Acids Res (2009) 37(1):1–13. doi: 10.1093/nar/gkn923
32. Gu Z, Eils R, Schlesner M. Complex Heatmaps Reveal Patterns and Correlations in Multidimensional Genomic Data. Bioinformatics (2016) 32(18):2847–9. doi: 10.1093/bioinformatics/btw313
33. R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing (2021).
35. Than NG, Romero R, Tarca AL, Kekesi KA, Xu Y, Xu Z, et al. Integrated Systems Biology Approach Identifies Novel Maternal and Placental Pathways of Preeclampsia. Front Immunol (2018) 9(1661). doi: 10.3389/fimmu.2018.01661
36. Levine RJ, Lam C, Qian C, Yu KF, Maynard SE, Sachs BP, et al. Soluble Endoglin and Other Circulating Antiangiogenic Factors in Preeclampsia. N Engl J Med (2006) 355(10):992–1005. doi: 10.1056/NEJMoa055352
37. Romero R, Nien JK, Espinoza J, Todem D, Fu W, Chung H, et al. A Longitudinal Study of Angiogenic (Placental Growth Factor) and Anti-Angiogenic (Soluble Endoglin and Soluble Vascular Endothelial Growth Factor Receptor-1) Factors in Normal Pregnancy and Patients Destined to Develop Preeclampsia and Deliver a Small for Gestational Age Neonate. J Matern Fetal Neonatal Med (2008) 21(1):9–23. doi: 10.1080/14767050701830480
38. Broekhuizen M, Klein T, Hitzerd E, de Rijke YB, Schoenmakers S, Sedlmayr P, et al. L-Tryptophan-Induced Vasodilation Is Enhanced in Preeclampsia: Studies on Its Uptake and Metabolism in the Human Placenta. Hypertension (2020) 76(1):184–94. doi: 10.1161/HYPERTENSIONAHA.120.14970
39. Iwahashi N, Yamamoto M, Nanjo S, Toujima S, Minami S, Ino K. Downregulation of Indoleamine 2, 3-Dioxygenase Expression in the Villous Stromal Endothelial Cells of Placentas With Preeclampsia. J Reprod Immunol (2017) 119:54–60. doi: 10.1016/j.jri.2017.01.003
40. Zardoya-Laguardia P, Blaschitz A, Hirschmugl B, Lang I, Herzog SA, Nikitina L, et al. Endothelial Indoleamine 2,3-Dioxygenase-1 Regulates the Placental Vascular Tone and is Deficient in Intrauterine Growth Restriction and Pre-Eclampsia. Sci Rep (2018) 8(1):5488. doi: 10.1038/s41598-018-23896-0
41. Kudo Y, Boyd CA, Sargent IL, Redman CW. Decreased Tryptophan Catabolism by Placental Indoleamine 2,3-Dioxygenase in Preeclampsia. Am J Obstet Gynecol (2003) 188(3):719–26. doi: 10.1067/mob.2003.156
42. Broekhuizen M, Danser AHJ, Reiss IKM, Merkus D. The Function of the Kynurenine Pathway in the Placenta: A Novel Pharmacotherapeutic Target? Int J Environ Res Public Health (2021) 18(21):11545. doi: 10.3390/ijerph182111545
43. Kulikova GV, Nizyaeva NV, Nagovitsina MN, Lyapin VM, Loginova NS, Kan NE, et al. Specific Features of TLR4 Expression in Structural Elements of Placenta in Patients With Preeclampsia. Bull Exp Biol Med (2016) 160(5):718–21. doi: 10.1007/s10517-016-3259-8
44. Zhang L, Yang H. Expression and Localization of TLR4 and its Negative Regulator Tollip in the Placenta of Early-Onset and Late-Onset Preeclampsia. Hypertens Pregnancy (2012) 31(2):218–27. doi: 10.3109/10641955.2011.642434
45. Bernardi FC, Felisberto F, Vuolo F, Petronilho F, Souza DR, Luciano TF, et al. Oxidative Damage, Inflammation, and Toll-Like Receptor 4 Pathway Are Increased in Preeclamptic Patients: A Case-Control Study. Oxid Med Cell Longev (2012) 2012:636419. doi: 10.1155/2012/636419
46. Kim YM, Romero R, Oh SY, Kim CJ, Kilburn BA, Armant DR, et al. Toll-Like Receptor 4: A Potential Link Between “Danger Signals,” the Innate Immune System, and Preeclampsia? Am J Obstet Gynecol (2005) 193(3, Supplement):921.e1–.e8. doi: 10.1016/j.ajog.2005.07.076
47. Pineda A, Verdin-Terán SL, Camacho A, Moreno-Fierros L. Expression of Toll-Like Receptor TLR-2, TLR-3, TLR-4 and TLR-9 Is Increased in Placentas From Patients With Preeclampsia. Arch Med Res (2011) 42(5):382–91. doi: 10.1016/j.arcmed.2011.08.003
48. Parthiban PS, Mahendra J, Logaranjani A, Shanmugam S, Balakrishnan A, Junaid M, et al. Association Between Specific Periodontal Pathogens, Toll-Like Receptor-4, and Nuclear Factor-κb Expression in Placental Tissues of Pre-Eclamptic Women With Periodontitis. J Invest Clin Dent (2018) 9(1):e12265. doi: 10.1111/jicd.12265
49. Dabagh-Gorjani F, Anvari F, Zolghadri J, Kamali-Sarvestani E, Gharesi-Fard B. Differences in the Expression of TLRs and Inflammatory Cytokines in Pre-Eclamptic Compared With Healthy Pregnant Women. Iran J Immunol (2014) 11(4):233–45.
50. van Rijn BB, Franx A, Steegers EA, de Groot CJ, Bertina RM, Pasterkamp G, et al. Maternal TLR4 and NOD2 Gene Variants, Pro-Inflammatory Phenotype and Susceptibility to Early-Onset Preeclampsia and HELLP Syndrome. PloS One (2008) 3(4):e1865. doi: 10.1371/journal.pone.0001865
51. Beutler B, Jiang Z, Georgel P, Crozat K, Croker B, Rutschmann S, et al. Genetic Analysis of Host Resistance: Toll-Like Receptor Signaling and Immunity at Large. Annu Rev Immunol (2006) 24:353–89. doi: 10.1146/annurev.immunol.24.021605.090552
52. Lu Y-C, Yeh W-C, Ohashi PS. LPS/TLR4 Signal Transduction Pathway. Cytokine (2008) 42(2):145–51. doi: 10.1016/j.cyto.2008.01.006
53. Panda B, Panda A, Ueda I, Abrahams VM, Norwitz ER, Stanic AK, et al. Dendritic Cells in the Circulation of Women With Preeclampsia Demonstrate a Pro-Inflammatory Bias Secondary to Dysregulation of TLR Receptors. J Reprod Immunol (2012) 94(2):210–5. doi: 10.1016/j.jri.2012.01.008
54. Xie F, Hu Y, Turvey SE, Magee LA, Brunham RM, Choi KC, et al. Toll-Like Receptors 2 and 4 and the Cryopyrin Inflammasome in Normal Pregnancy and Pre-Eclampsia. BJOG: Int J Obstet Gynaecol (2010) 117(1):99–108. doi: 10.1111/j.1471-0528.2009.02428.x
55. Nishizawa H, Ota S, Suzuki M, Kato T, Sekiya T, Kurahashi H, et al. Comparative Gene Expression Profiling of Placentas From Patients With Severe Pre-Eclampsia and Unexplained Fetal Growth Restriction. Reprod Biol Endocrinol (2011) 9(1):107. doi: 10.1186/1477-7827-9-107
56. Teirila L, Heikkinen-Eloranta J, Kotimaa J, Meri S, Lokki AI. Regulation of the Complement System and Immunological Tolerance in Pregnancy. Semin Immunol (2019) 45:101337. doi: 10.1016/j.smim.2019.101337
57. Agostinis C, Bulla R, Tripodo C, Gismondi A, Stabile H, Bossi F, et al. An Alternative Role of C1q in Cell Migration and Tissue Remodeling: Contribution to Trophoblast Invasion and Placental Development. J Immunol (2010) 185(7):4420–9. doi: 10.4049/jimmunol.0903215
58. Singh J, Ahmed A, Girardi G. Role of Complement Component C1q in the Onset of Preeclampsia in Mice. Hypertension (2011) 58(4):716–24. doi: 10.1161/HYPERTENSIONAHA.111.175919
59. Mitani R, Maeda K, Fukui R, Endo S, Saijo Y, Shinohara K, et al. Production of Human Mast Cell Chymase in Human Myometrium and Placenta in Cases of Normal Pregnancy and Preeclampsia. Eur J Obstet Gynecol Reprod Biol (2002) 101(2):155–60. doi: 10.1016/S0301-2115(01)00546-2
60. Szewczyk G, Pyzlak M, Klimkiewicz J, Smiertka W, Miedzinska-Maciejewska M, Szukiewicz D. Mast Cells and Histamine: Do They Influence Placental Vascular Network and Development in Preeclampsia? Mediators Inflamm (2012) 2012:307189. doi: 10.1155/2012/307189
61. Szukiewicz D, Szukiewicz A, Maslinska D, Gujski M, Poppe P, Mazurek-Kantor J. Mast Cell Number, Histamine Concentration and Placental Vascular Response to Histamine in Preeclampsia. Inflamm Res (1999) 48(1):39–40. doi: 10.1007/s000110050390
62. Mukai K, Tsai M, Saito H, Galli SJ. Mast Cells as Sources of Cytokines, Chemokines, and Growth Factors. Immunol Rev (2018) 282(1):121–50. doi: 10.1111/imr.12634
63. Meyer N, Woidacki K, Knöfler M, Meinhardt G, Nowak D, Velicky P, et al. Chymase-Producing Cells of the Innate Immune System Are Required for Decidual Vascular Remodeling and Fetal Growth. Sci Rep (2017) 7(1):45106. doi: 10.1038/srep45106
64. Szukiewicz D, Szukiewicz A, Maslinska D, Markowski MW. Placental Mast Cells (MC) and Histamine (HA) in Pregnancy Complicated by Diabetes Class C - Relation to the Development of Villous Microvessels. Placenta (1999) 20:503–10. doi: 10.1016/S0143-4004(99)80039-3
65. Szukiewicz D, Kochanowski J, Pyzlak M, Szewczyk G, Stangret A, Mittal TK. Fractalkine (CX3CL1) and its Receptor CX3CR1 may Contribute to Increased Angiogenesis in Diabetic Placenta. Mediators Inflamm (2013) 2013:437576. doi: 10.1155/2013/437576
66. Ryu J, Lee CW, Hong KH, Shin JA, Lim SH, Park CS, et al. Activation of Fractalkine/CX3CR1 by Vascular Endothelial Cells Induces Angiogenesis Through VEGF-A/KDR and Reverses Hindlimb Ischaemia. Cardiovasc Res (2008) 78(2):333–40. doi: 10.1093/cvr/cvm067
67. Bezemer RE, Schoots MH, Timmer A, Scherjon SA, Erwich J, van Goor H, et al. Altered Levels of Decidual Immune Cell Subsets in Fetal Growth Restriction, Stillbirth, and Placental Pathology. Front Immunol (2020) 11:1898. doi: 10.3389/fimmu.2020.01898
68. Prins JR, Faas MM, Melgert BN, Huitema S, Timmer A, Hylkema MN, et al. Altered Expression of Immune-Associated Genes in First-Trimester Human Decidua of Pregnancies Later Complicated With Hypertension or Foetal Growth Restriction. Placenta (2012) 33(5):453–5. doi: 10.1016/j.placenta.2012.02.010
69. Co EC, Gormley M, Kapidzic M, Rosen DB, Scott MA, Stolp HAR, et al. Maternal Decidual Macrophages Inhibit NK Cell Killing of Invasive Cytotrophoblasts During Human Pregnancy. Biol Reprod (2013) 88(6):155–. doi: 10.1095/biolreprod.112.099465
70. Faas MM, de Vos P. Uterine NK Cells and Macrophages in Pregnancy. Placenta (2017) 56:44–52. doi: 10.1016/j.placenta.2017.03.001
71. Lager S, Sovio U, Eddershaw E, van der Linden MW, Yazar C, Cook E, et al. Abnormal Placental CD8+ T-Cell Infiltration is a Feature of Fetal Growth Restriction and Pre-Eclampsia. J Physiol (2020) 598(23):5555–71. doi: 10.1113/JP279532
72. Sharps MC, Baker BC, Guevara T, Bischof H, Jones RL, Greenwood SL, et al. Increased Placental Macrophages and a Pro-Inflammatory Profile in Placentas and Maternal Serum in Infants With a Decreased Growth Rate in the Third Trimester of Pregnancy. Am J Reprod Immunol (2020) 84(3):e13267. doi: 10.1111/aji.13267
73. Banerjee S, Huang Z, Wang Z, Nakashima A, Saito S, Sharma S, et al. Etiological Value of Sterile Inflammation in Preeclampsia: Is It a Non-Infectious Pregnancy Complication? Front Cell Infect Microbiol (2021) 11(760). doi: 10.3389/fcimb.2021.694298
74. Bortoluci KR, Medzhitov R. Control of Infection by Pyroptosis and Autophagy: Role of TLR and NLR. Cell Mol Life Sci (2010) 67(10):1643–51. doi: 10.1007/s00018-010-0335-5
75. Cheng S-B, Nakashima A, Huber WJ, Davis S, Banerjee S, Huang Z, et al. Pyroptosis is a Critical Inflammatory Pathway in the Placenta From Early Onset Preeclampsia and in Human Trophoblasts Exposed to Hypoxia and Endoplasmic Reticulum Stressors. Cell Death Dis (2019) 10(12):927. doi: 10.1038/s41419-019-2162-4
Keywords: placenta, preeclampsia, immune system, toll like receptor, complement, macrophage, mast cell
Citation: Broekhuizen M, Hitzerd E, van den Bosch TPP, Dumas J, Verdijk RM, van Rijn BB, Danser AHJ, van Eijck CHJ, Reiss IKM and Mustafa DAM (2021) The Placental Innate Immune System Is Altered in Early-Onset Preeclampsia, but Not in Late-Onset Preeclampsia. Front. Immunol. 12:780043. doi: 10.3389/fimmu.2021.780043
Received: 20 September 2021; Accepted: 29 November 2021;
Published: 21 December 2021.
Edited by:
Kenneth Beaman, Rosalind Franklin University of Medicine and Science, United StatesReviewed by:
Umida Ganieva, Rosalind Franklin University of Medicine and Science, United StatesSurendra Sharma, Women & Infants Hospital of Rhode Island, United States
Copyright © 2021 Broekhuizen, Hitzerd, van den Bosch, Dumas, Verdijk, van Rijn, Danser, van Eijck, Reiss and Mustafa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dana A. M. Mustafa, ZC5tdXN0YWZhQGVyYXNtdXNtYy5ubA==
†These authors share last authorship
 Michelle Broekhuizen
Michelle Broekhuizen Emilie Hitzerd
Emilie Hitzerd Thierry P. P. van den Bosch
Thierry P. P. van den Bosch Jasper Dumas
Jasper Dumas Robert M. Verdijk4
Robert M. Verdijk4 Bas B. van Rijn
Bas B. van Rijn A. H. Jan Danser
A. H. Jan Danser Casper H. J. van Eijck
Casper H. J. van Eijck Irwin K. M. Reiss
Irwin K. M. Reiss Dana A. M. Mustafa
Dana A. M. Mustafa