- 1College of Food Science and Engineering, Ocean University of China, Qingdao, China
- 2Shanghai Engineering Research Center of Food Microbiology, School of Medical Instrument and Food Engineering, University of Shanghai for Science and Technology, Shanghai, China
Probiotics plays an important role in regulating gut microbiota and maintaining intestinal homeostasis. Extracellular vesicles (EVs) derived from probiotics have emerged as potential mediators of host immune response and anti-inflammatory effect. However, the anti-inflammatory effect and mechanism of probiotics derived EVs on inflammatory bowel disease (IBD) remains unclear. In this study, the effect of Lactobacillus plantarum Q7-derived extracellular vesicles (Q7-EVs) on gut microbiota and intestinal inflammation was investigated in C57BL/6J mice. The results showed that Q7-EVs alleviated DSS-induced colitis symptoms, including colon shortening, bleeding, and body weight loss. Consumption of Q7-EVs reduced the degree of histological damage. DSS-upregulated proinflammatory cytokine levels including IL-6, IL-1β, IL-2 and TNF-α were reduced significantly by Q7-EVs (p < 0.05). 16S rRNA sequencing results showed that Q7-EVs improved the dysregulation of gut microbiota and promoted the diversity of gut microbiota. It was observed that the pro-inflammatory bacteria (Proteobacteria) were reduced and the anti-inflammatory bacteria (Bifidobacteria and Muribaculaceae) were increased. These findings indicated that Q7-EVs might alleviate DSS-induced ulcerative colitis by regulating the gut microbiota.
Introduction
Inflammatory bowel disease (IBD), including ulcerative colitis (UC) and Crohn’s disease, are chronic inflammatory disorders of the gastrointestinal tract with a high incidence (1). Currently, the pathogenesis of IBD remains incompletely understood. The use of IBD drugs is severely limited by low efficacy, side effects and intolerance (2). It was reported that probiotics could alleviate IBD by maintaining normal gut microbiota, enhancing mucosal barrier function, and inhibiting exposure to inflammatory signals, which are expected to be a new treatment for IBD (3–5). The clinical effect and mechanism of probiotics on IBD are in progress (6).
Extracellular vesicles (EVs) are nano-scale membrane vesicles with phospholipid bilayer structures, which are secreted by almost all cells. At present, EVs have been isolated from animals (7, 8), plants (9) and microorganisms (10). Microorganism-derived EVs contain nucleic acids, proteins, and lipids, which play a crucial role in host metabolism and health (11). Microorganisms-host interactions mediated by EVs could result in various responses, which played potential roles as virulence factor delivery vehicles or inflammatory response modulators (12). For example, Escherichia coli Nissle 1917 derived EVs could mediate the anti-inflammatory and barrier protection effects in experimental colitis (13). EVs had been considered to be absent in Gram-positive bacteria due to their thick cell wall (11). Until 2009, Gram-positive bacteria-derived EVs was firstly confirmed (14). As Gram-positive bacteria, the functions of EVs derived from probiotics such as Lactobacillus and Bifidobacteria in immunity and intestinal diseases have attracted widespread attention. EVs derived from Bifidobacterium longum KACC 91563 alleviated food allergy through mast cell suppression (15). Lactobacillus rhamnosus GG derived EVs played a role in anti-proliferative on HepG2 cancer cells (16). Lactobacillus reuteri BBC3 derived EVs inhibited the inflammatory response mediated by activating macrophages, and played an important role in intestinal immune regulation (17). Lactobacillus plantarum derived EVs could be used as anti-inflammatory and immunomodulatory substances to improve inflammatory skin diseases and had the potential in treating antimicrobial-resistant pathogens (18, 19). Lactobacillus was considered to be the potential components involved with intestinal immune regulation (6). The effect of EVs derived from Lactobacillus on IBD has been rarely reported.
In our previous study, Lactobacillus plantarum Q7 (L. plantarum Q7) was isolated from the traditional Chinese fermented foods (20). It was observed that L. plantarum Q7 could exhibit activity against pathogenic bacteria such as Escherichia coli, Listeria monocytogenes and Staphylococcus aureus, which were associated with intestinal inflammation. The aim of this study was to explore the effect of L. plantarum Q7 derived EVs (Q7-EVs) on gut microbiota and ulcerative colitis in a DSS-induced colitis mouse model. It would provide a basis for probiotics to alleviate colitis from the perspective of EVs.
Materials and Methods
Isolation and Purification of Q7-EVs
L. plantarum Q7 (GenBank: CP019712-16) was isolated from traditional fermented yak yogurt made by herdsmen in Qinghai province, China (21). The strain was cultured in MRS medium at 37°C overnight. Q7-EVs were isolated from the culture supernatants using ultrafiltration combined with ultracentrifugation (22). In brief, the culture supernatants were centrifuged at 8000×g for 30 min to remove cell debris and impurities. After filtered by 0.22 μm filter, the supernatant was concentrated using centricon-plμs-70 100-kDa ultrafiltration tube (Millipore) and ultracentrifuged at 100000×g for 2 h (Hitachi, Ltd., Tokyo, Japan). The precipitate was aspirated gently by adding PBS and centrifuged at 100000×g for 1 h, then resuspended the pellet with PBS. Q7-EVs were collected and stored at -80°. The morphology and the size distribution of Q7-EVs were observed by Transmission electron microscope (H-7650, Hitachi, Ltd., Tokyo, Japan) and Dynamic light scattering (Nanotrac Wave II, Microtrac, Inc., Montgomeryville, USA).
Animals Model
4-5 weeks old SPF male C57BL/6J mice were purchased from the Laboratory Animal Breeding Center of Pengyue (Jinan, China). Mice were housed in an animal care facility and provided with sufficient food (Beijing Keao Xieli Feed Co., Ltd., Beijing, China) and water. Colitis was induced by 3.5% dextran sulfate sodium (DSS, Yeasen Biotech, Shanghai, CN) administration in the drinking water (23) and the mice were treated Q7-EVs by gavage (23, 24). The mice were acclimated for a week and randomly divided into four groups: control group (PBS, daily), DSS+PBS group (PBS, daily), DSS+10 μg Q7-EVs group (0.5 mg/kg body weight Q7-EVs, daily), DSS+20 μg Q7-EVs group (1 mg/kg body weight Q7-EVs, daily). The mice in control group were administered untreated drinking water for 18 days. The DSS group and treatment group were pre-garaged with PBS or Q7-EVs for 10 day and received 3.5% DSS in drinking water from 11th day to 18th day. All experimental processes were approved by Animal Ethics Committee of Ocean University of China (permission number: spxy20200720215).
Evaluation of Disease Activity Index (DAI) and Spleen Index
Body weight, stool characteristic and rectal bleeding were recorded every two days. DAI scores were calculated according to previously described methods (25–27). The details of each score were listed in Supplementary Materials (Table S1). The weight of spleen was determined, and the spleen index was defined as the ratio of spleen weight to body weight as previous report (28).
Histological Analysis
Colon tissue was fixed in 4% paraformaldehyde, dehydrated with gradient alcohol solution and embedded in paraffin. Paraffin-embed samples were cut into continuous longitudinal sections, and then stained by H&E. The histological scoring was measured according to the criteria listed in Table S2 (29).
Enzyme-Linked Immunosorbent Assay
According to the manuscript’s protocol, the expression level of IL-6, IL-1β, and TNF-α in the serum was determined by ELISA kit (Nanjing Jiancheng Biotechnology, Nanjing, China). Multiskan FC (Thermo Scientific, Waltham, MA, USA) was used to detect the absorbance of each sample at 450 nm.
RT-qPCR Analysis
The colon tissue was put into Trizol reagent and homogenized using a tissue grinder (Wuhan Sevelco Co., Ltd., Wuhan, China). 1 μg RNA was reverse transcribed via a high-capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, USA). Genes were quantitated using the StepOnePlus real-time PCR instrument (Applied Biosystems, Foster City, USA). The specific primers used were shown in Table S3. The 2-ΔΔCt method was utilized to normalize the expression results. The glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the housekeeper gene to normalize each mRNA.
16S rRNA Gene Sequencing
Fecal DNA extraction and high-throughput sequencing were performed by Personal Biotechnology Co., Ltd (Shanghai, China). Briefly, the Fast DNA SPIN extraction kit (MP Biomedicals, Santa Ana, CA, USA) was used to extract genomic DNA from mice feces. The highly variable V3-V4 region of the bacterial 16S rDNA gene was selected for amplification. The library was qualified based on the TruSeq Nano DNA Library Prep Kit from Illumina, and Illumina NovaSeq platform was used for paired-end sequencing. The filtered sequences were clustered as operational taxa (OTU) using the UCLUST algorithm with a similarity threshold of 97%. The OTU abundance and the six-level taxonomic classification from phylum to species were obtained for analysis.
Statistical Analyses
The experimental data were analyzed by GraphPad Prism version 7.0 and SPSS 25.0 statistical software. Comparisons of various anatomical measurements by two-way analysis of variance (ANOVA), followed by Fisher’s LSD post hoc tests. PCA and PCoA analyzed were used R language pack. The linear discriminant analysis (LDA) was applied to identify the differences of microbiota compositions between different groups by using an LDA score threshold of >3.5.
Results
Characterization of Q7-EVs
EVs were isolated from the supernatant of L. plantarum Q7 by ultrafiltration combined with ultracentrifugation (Figure 1A). The Q7-EVs were characterized by Transmission electron microscope and Dynamic light scattering. The results showed that Q7-EVs were double-layer membrane-enclosed nanoparticles with spherical morphology (Figure 1B). It was observed that Q7-EVs ranged from 70 nm to 500 nm in size (Figure 1C), the mean size was 185.5 ± 65.4 nm, which was consistent with the size of previous reports (30).
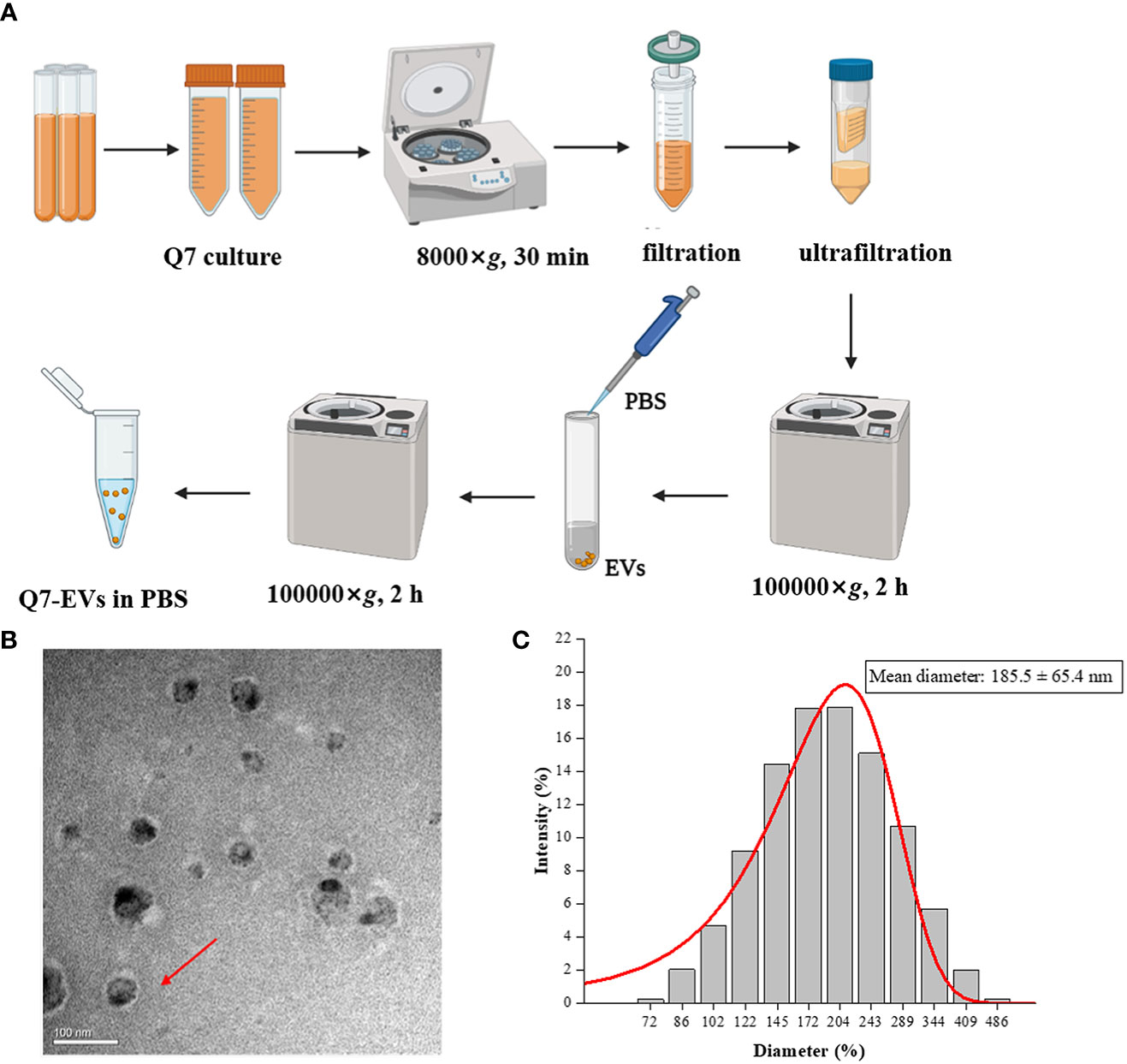
Figure 1 Characterization of Q7-EVs. (A) Isolation and purification procedures of Q7-EVs. (B) Transmission electron microscopy images of Q7-EVs. (C) Size distribution of Q7-EVs.
Effect of Q7-EVs on DSS-Induced Colitis
The effect of Q7-EVs on DSS-induced colitis was evaluated in C57BL/6J mice (Figure 2A) (31). The body weight, DAI, length of the colon, and colonic histomorphology were analyzed. Compared with the control group, the weight loss could be slowed by Q7-EVs intervention since the 6th day. The weight loss of 22.6% was observed in the DSS+PBS group, while the weight loss of 12.8% and 12.6% were found in the 10 μg and 20 μg Q7-EVs group on the 8th day, respectively. The weight loss could be slowed by Q7-EVs intervention with no dose correlation (Figure 2B). DAI scores of Q7-EVs (10 μg and 20 μg) treated mice were less than that of DSS+PBS group mice on the 8th day (p < 0.05) (Figure 2C). Colon length was regarded as a key indicator to assess the severity of acute colitis induced by DSS (31, 32). It was confirmed that Q7-EVs treatment improved the shortening of the colon significantly (p < 0.05) (Figures 2D, E). The colon length in the DSS+PBS group was 4.16 ± 0.24 cm. After treatment with Q7-EVs, the average length of the colon was observed to be 5.14 ± 0.07 and 5.19 ± 0.34 cm, respectively. The spleen is one of important immune organ of the body, which exerted a regulatory effect on the immunity. It was found that the Q7-EVs reduced the spleen index of DSS-induced colitis mouse model (Figure S1).
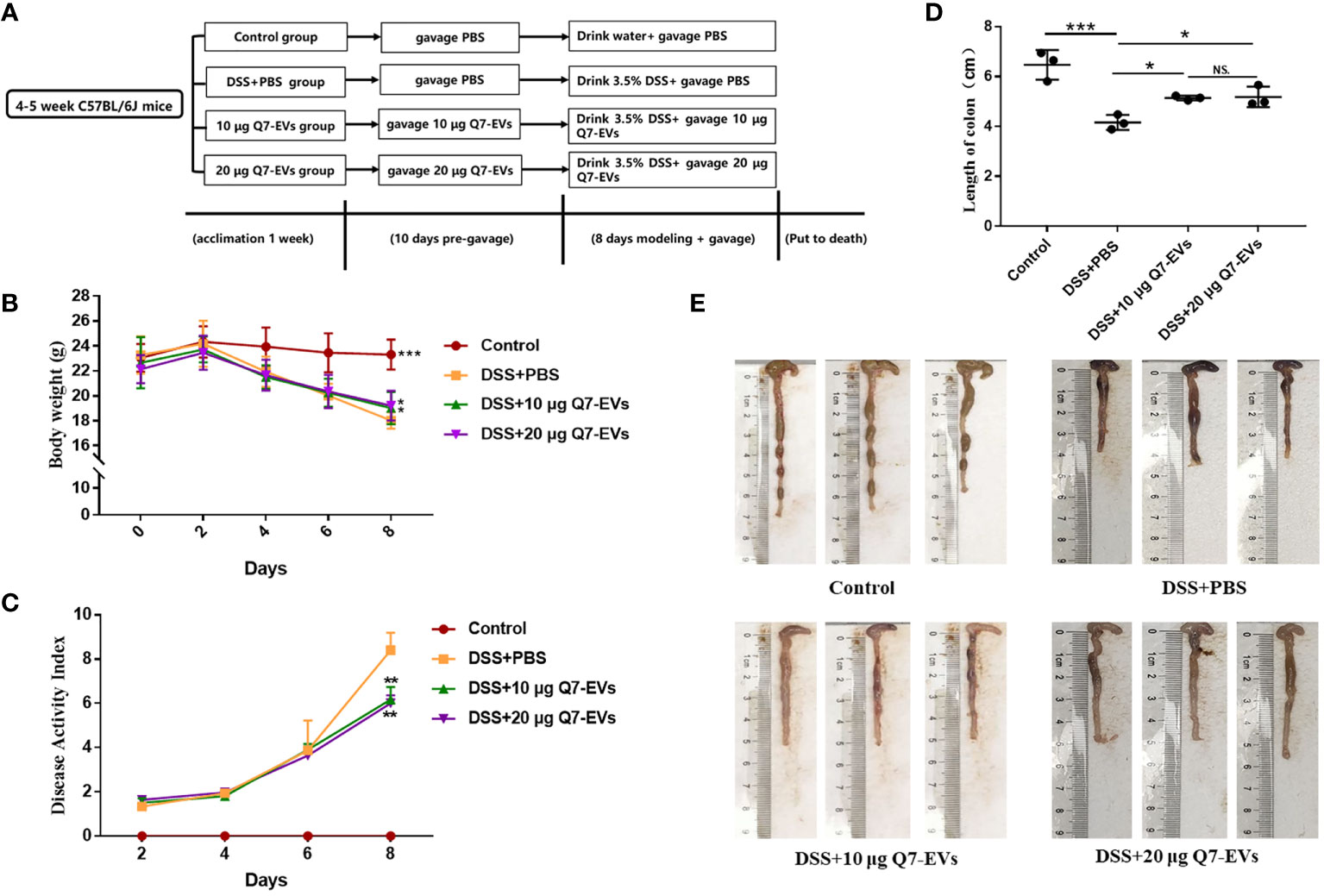
Figure 2 The effect of Q7-EVs on DSS-induced colitis in C57BL/6J mice. (A) Overall experimental design. (B) Body weight. (C) Disease activity index. (D, E) Representative photograph and statistical analysis of colon length. Values were means ± SD (*p < 0.05, **p < 0.01, ***p < 0.001, NS. represents no significant difference).
H&E staining showed that DSS treatment caused damage to the colon structure, while the pathological features were improved by oral Q7-EVs (Figure 3A). It suggested that Q7-EVs could reduce the colonic pathology score of mice and alleviate colitis in mice significantly (Figure 3B).
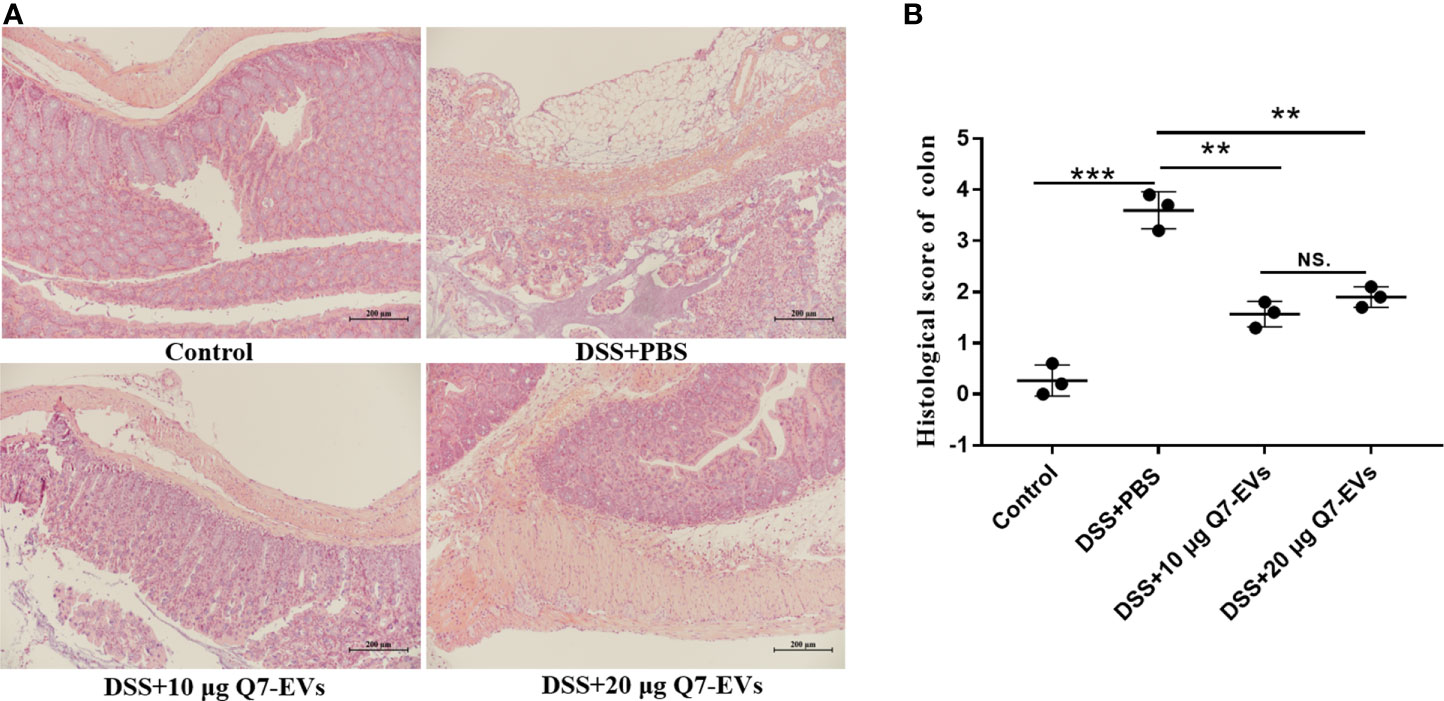
Figure 3 Pathophysiological analysis of colon sections. (A) Representative microscopy images of H&E-stained colonic sections. (B) Histological scoring of the colon section. Data were expressed as means ± S.D (**p < 0.01, ***p < 0.001, NS. represents no significant difference).
Effect of Q7-EVs on the Expression of Inflammatory Cytokines
It was observed that there was no significant difference in the effects of two doses of Q7-EVs (10 μg and 20 μg) on the body weight, colon length and colonic pathology score. Therefore, 10 μg Q7-EVs was selected in the following experiments. It was reported that the increase of pro-inflammatory cytokines was a hallmark of DSS-induced colitis (33). The level of inflammatory cytokines in colon tissues was detected (34). It was found that the expression levels of IL-6, IL-1β, TNF-α, and IL-2 were increased in the colon tissue of the DSS+PBS group mice. In contrast, these cytokines were significantly reduced in the colon under the intervention of Q7-EVs (Figure 4). A similar trend was found for the determination of IL-6, IL-1β, and TNF-α in serum (Figure S2).
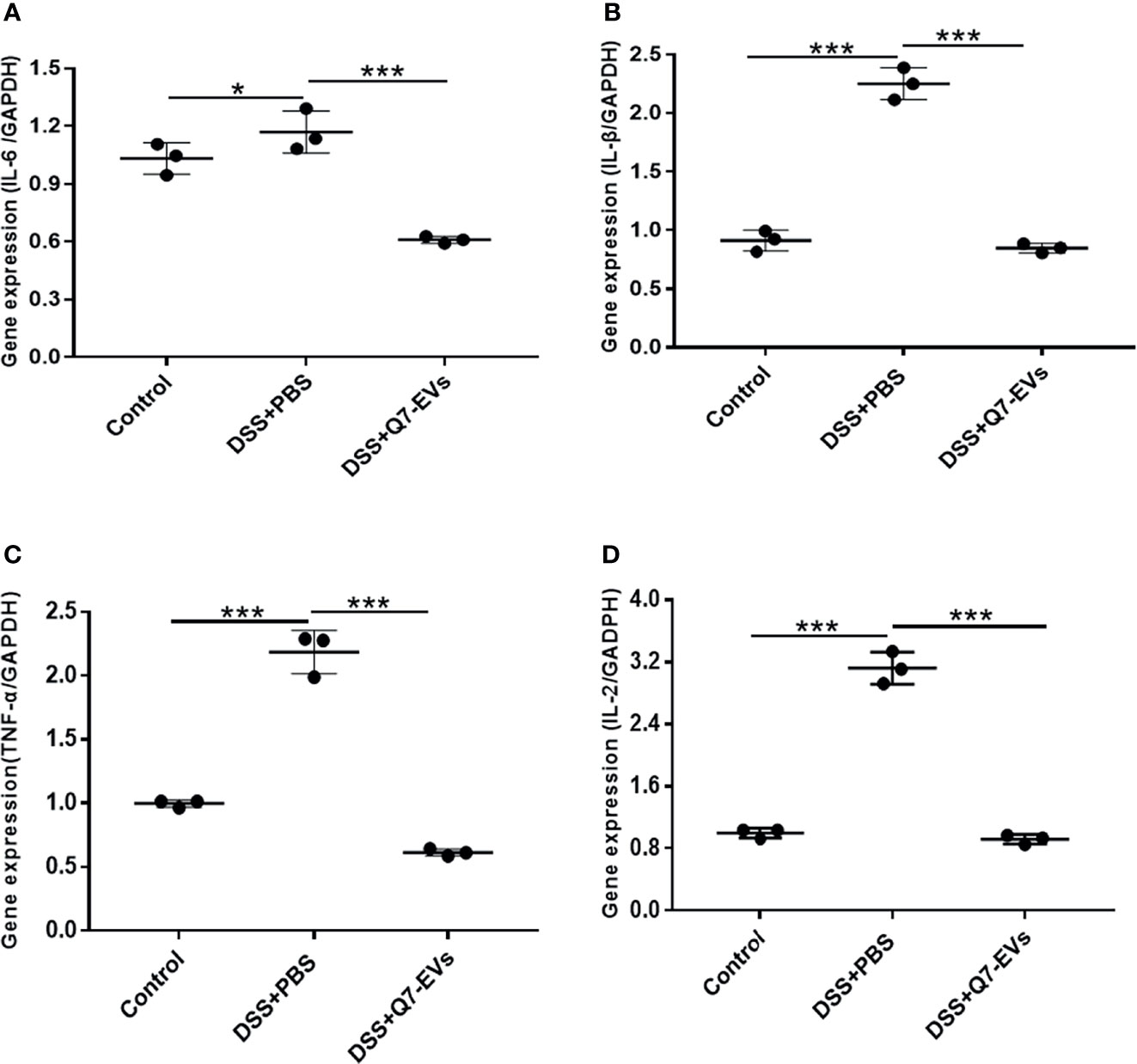
Figure 4 The effect of Q7-EVs on the mRNA Expression level in colon tissue. (A) IL-6, (B) TNE-α, (C) IL-β, (D) IL-2. Data were expressed as means ± S.D (*p < 0.05, ***p < 0.001).
Effect of Q7-EVs on Intestinal Microbiota Composition in Colitis Mice
It was found that microbial richness, evenness and diversity were significantly decreased in the DSS+PBS group in comparison with that of the control group, whereas Q7-EVs could restore the species diversity, richness and community evenness (Figures 5A–F).
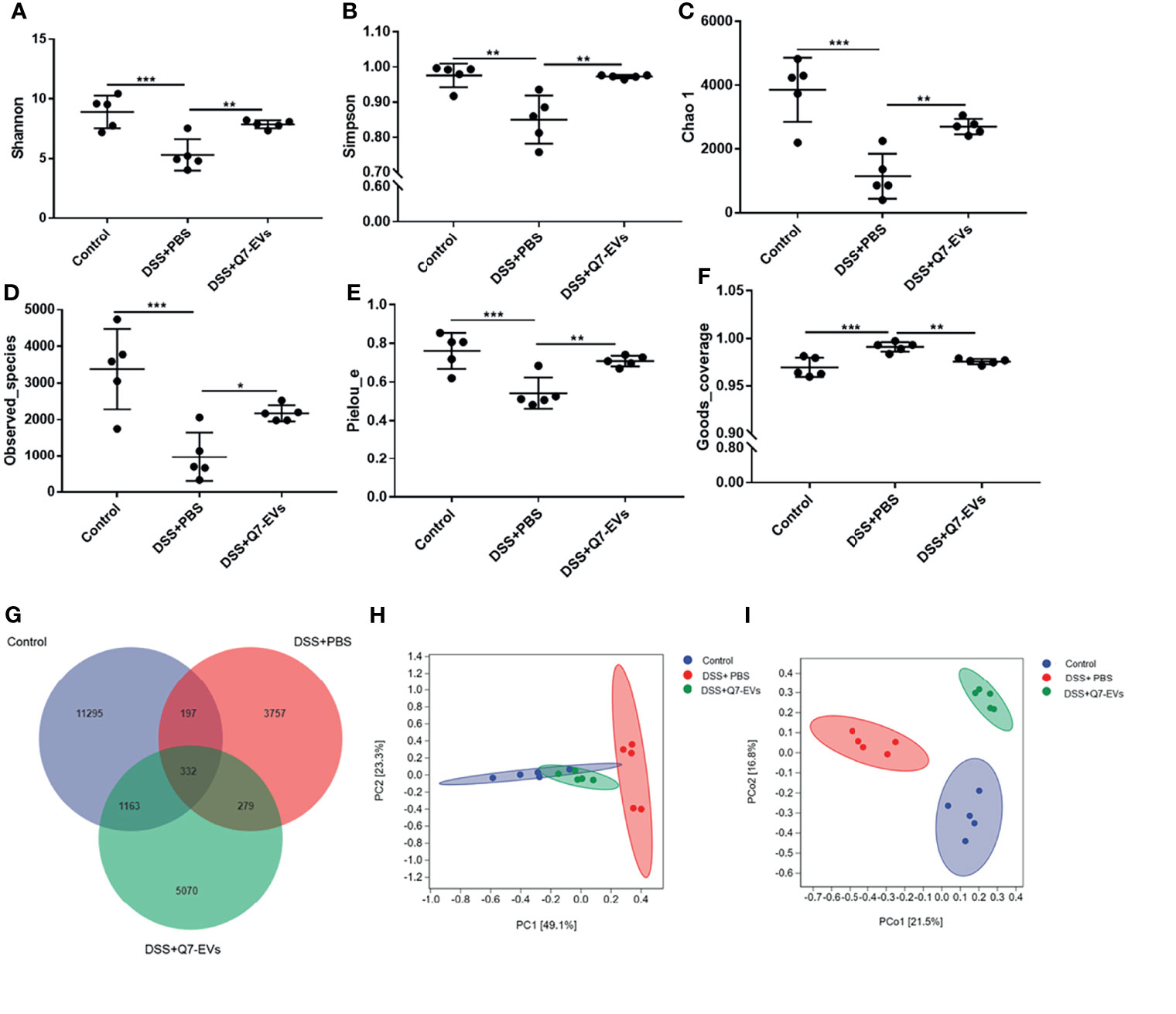
Figure 5 The effect of Q7-EVs on gut microbiota in colitis mice. Alpha diversity analysis included (A) Shannon index, (B) Simpson index, (C) Chao 1 index, (D) Observed_species, (E) Pielou_e, and (F) Goods_coverage. Comparative analysis of microbiota composition included (G) Venn diagram of OTU, (H) PCA analysis, and (I) PCoA analysis. Data were expressed as means ± S.D (*p < 0.05, **p < 0.01, ***p < 0.001).
Venn diagram was used to display the composition information of species in each group. It was observed that 332 OTUs overlapped in all three groups, 197 OTUs were present in both the control and DSS+PBS groups, 279 OTUs in the DSS+PBS and Q7-EVs groups, and 1163 OTUs in the control and DSS+Q7-EVs groups (Figure 5G). Compared with the DSS+PBS group, intestinal microbial composition in the DSS+Q7-EVs group was more similar to the control group. The results showed that the gut microbiota of the DSS+PBS group was significantly different from that of the control group, and Q7-EVs alleviated the shift of gut microbiota induced by DSS (Figures 5H, I). LEfSe analysis was demonstrated that 37 key phylotypes were determined as distinguished biomarkers on the genus (Figure 6A). The control group had 10 dominant microorganisms, the DSS+PBS group and the Q7-EVs group had 14 dominant microorganisms, respectively (Figure 6B).
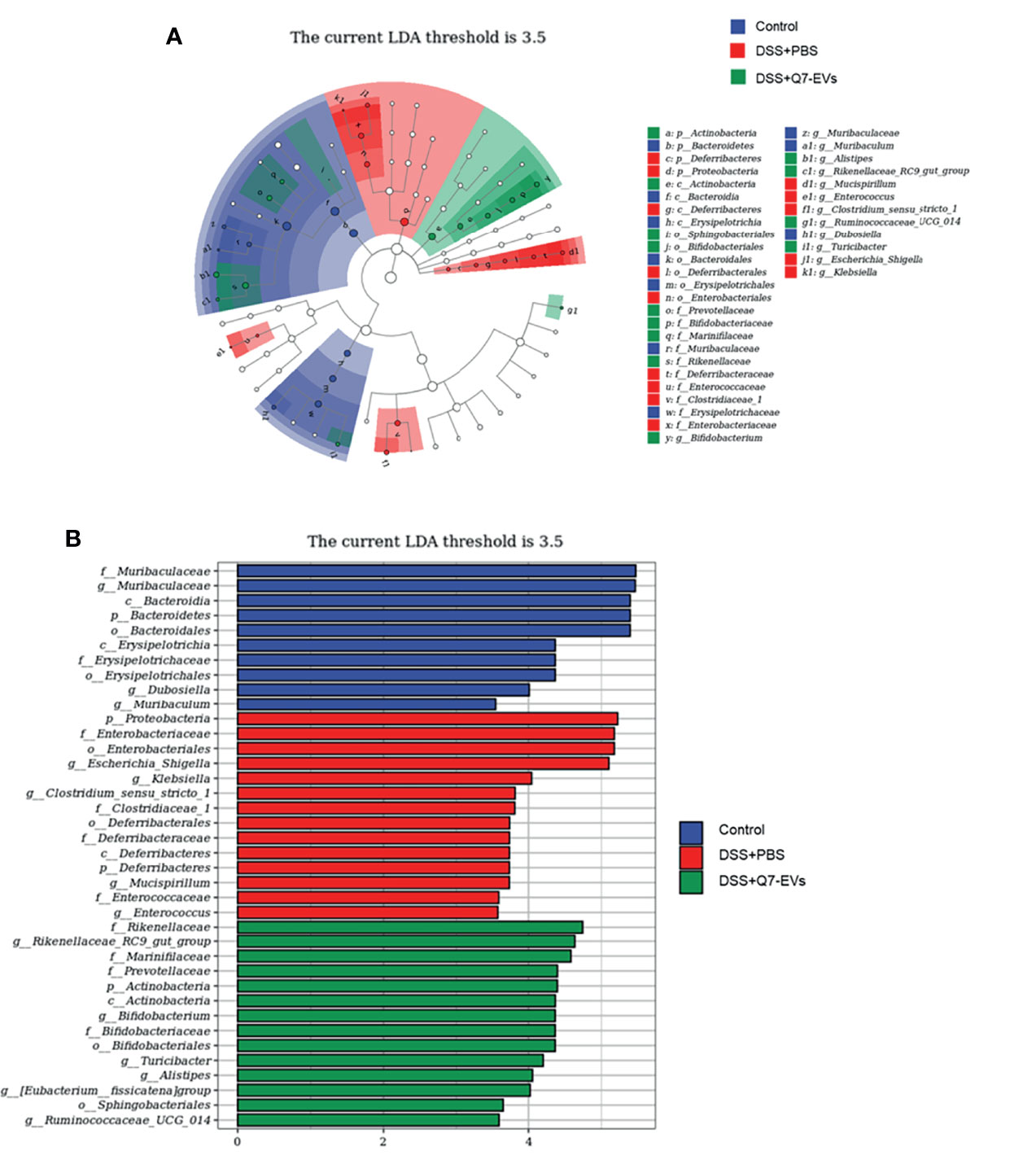
Figure 6 The effect of Q7-EVs on gut dominant microorganisms. (A) Cladogram displayed the taxonomic tree of differentially abundant taxa by LEfSe analysis on genus. (B) Distribution histogram of gut dominant microorganisms on genus.
The composition of the gut microbiota at the phylum level was shown in Figure 7A. Compared with the control group, Bacteroidetes, and Verrucomicrobia in the DSS+PBS group were significantly decreased, while Proteobacteria, Epsilonbacteraeota were increased significantly. These changes of gut microbiota were recovered in the Q7-EVs group. The abundance of Proteobacteria was decreased in the DSS+Q7-EVs group. The Firmicutes/Bacteroidetes (F/B) ratio in the DSS+PBS group was higher than that of the control group, and was restored after the intervention of Q7-EVs (Figure 7B), which could be used as a biomarker for intestinal inflammation (31). The difference of gut microbiota in each group at the genus level was shown in Figure 7C. Compared with the DSS+PBS group, Bifidobacterium, Rikenellaceae_RC9_gut_group, Akkermansia, Muribaculaceae, Lactobacillus and Alitipes in the DSS+Q7-EVs group were recovered to the level of the control group (Figure S3).
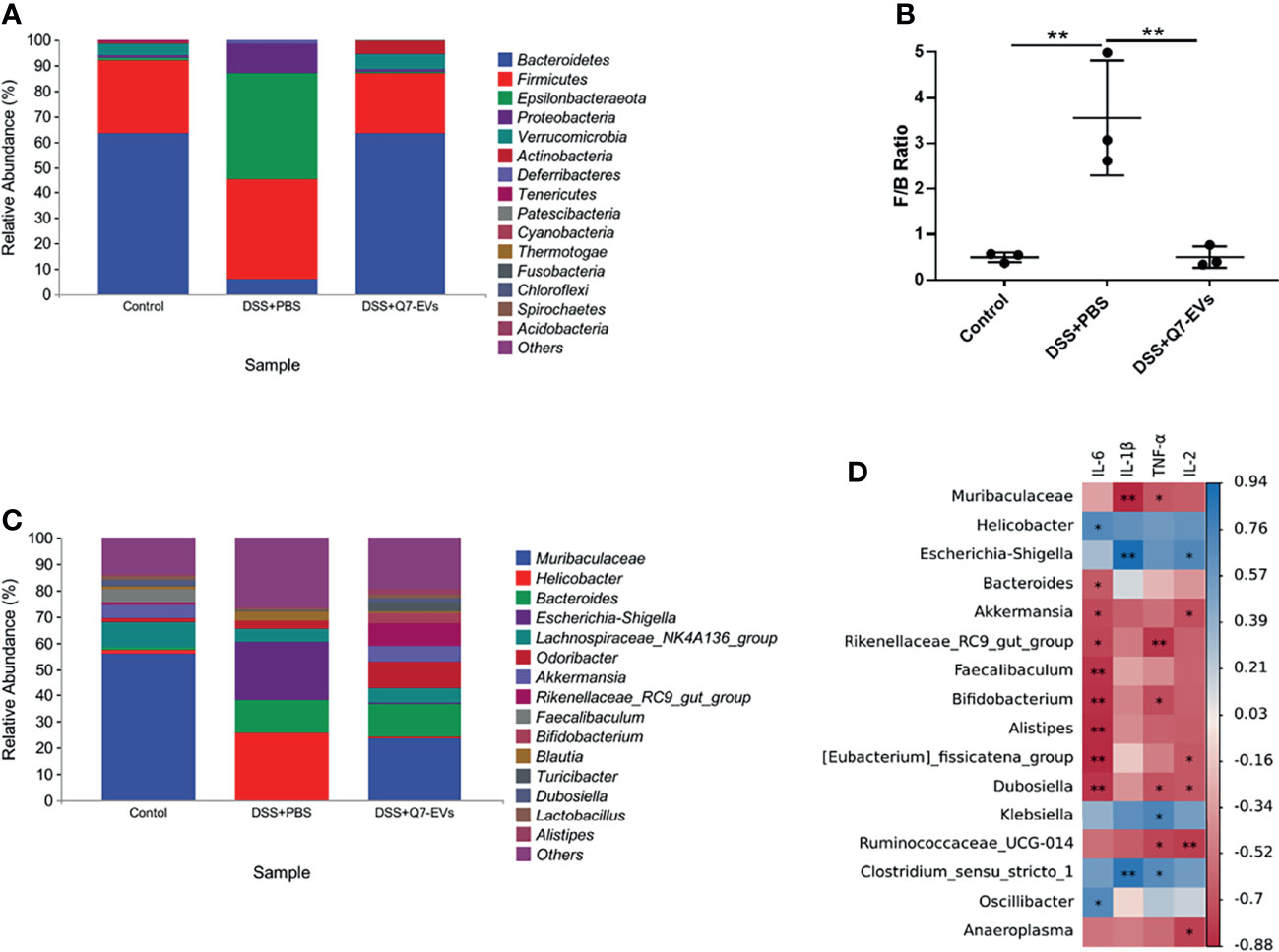
Figure 7 The effect of Q7-EVs on the community structures of gut microbiota and its relationship with inflammatory cytokines. (A) Microbial community bar plot by phylum. (B) The ratio of Firmicutes/Bacteroidetes. (C) Microbial community bar plot by genus. (D) Correlation matrix between the microbiota and inflammatory cytokines. Data were expressed as means ± S.D **p < 0.01.
Correlation analysis was conducted to determine the relationship between the gut microbiota and inflammatory cytokines. Proteobacteria (Helicobacter, Escherichia-shigella, Klebsiella, Oscillibacter) and Firmicutes (Clostridium_sensu_stricto_1) were positively correlated with inflammatory cytokines. Muribaculaceae, Bacteroides, Akkermansia, Rikenellaceae_RC9_gut_group, Faecalibaculum, Bifidobacterium, Alistipes, Eubacteriu_fissicatena_ group, Dubosiella, Ruminococcaceae_UCG-014 and Anaeroplasma were negatively correlated with inflammatory cytokines (Figure 7D). Combined with the LEfSe analysis, the dominant microorganisms in the DSS+Q7-EVs group contained more anti-inflammatory bacteria.
Discussion
UC is a common type of IBD with an increasing incidence in recent years (28). Probiotics can protect the intestinal mucosal barrier, inhibit intestinal inflammation, and maintain the balance of the gut microbiota structure (31), which may offer an alternative or adjuvant strategy to UC therapy (35). Probiotics-derived EVs has attracted increasing attention due to the potential for the treatment of multiple diseases, such as inflammation (17, 18, 22), bacterial infection (19) and obesity (36). L. plantarum Q7 was isolated from Chinese traditional food in our previous study (21). The potential probiotic character of L. plantarum Q7 had been investigated in the view of bacteriocin. In this study, the effect of L. plantarum Q7 on DSS-induced colitis was explored from the perspective of EVs.
It was found in this study that Q7-EVs could ameliorate DSS-induced colitis via regulating gut microbiota in C57BL/6J mice. The changes of inflammatory cytokine in mice serum and colon tissue were explored. Cytokines such as IL-6, IL-1β, IL-2 and TNF-α were involved in the inflammatory response (37–40). As expected, the Q7-EVs ameliorated the immune response and restored the expression of the cytokines. NF-κB was the main inflammatory pathway in the transcription of pro-inflammatory cytokines (such as IL-6, IL-1β, and TNF-α) (28). TLR4 and MyD88 played a critical role in the development of DSS-induced colitis (41). In our study, the gene expression of TLR4 and MyD88 in the colon was determined. The results showed that oral Q7-EVs down-regulated TLR4 and MyD88 gene expression compared with DSS-induced colitis mice (Figure S4). After binding to the ligand, MyD88-dependent signal transduction could lead to phosphorylation of NF-κB, thereby regulating the levels of transcription factors of IL-1β, IL-6, and TNF-α (24). It was speculated that Q7-EVs improved colitis by regulating the TLR4-MyD88-NF-κB pathway.
Gut microbiota played an important role in intestinal inflammation, which was related to the etiopathogenesis of UC. Different microbes participated in either pro-inflammatory or anti-inflammatory processes (42). It was reported that EVs could play a role in host-microbe responses and regulate the gut microflora (43). Proteobacteria was reported to be the main phylum of pathogenic bacteria, which promoted the production of excessive pro-inflammatory cytokines and related to IBD pathogenesis (24, 42). Akkermansia was a confirmed probiotic with anti-inflammatory effect by improving neutrophil infiltration and alleviating intestinal inflammation (44–46). Bifidobacteria could exert an anti-inflammatory effect through induction of intestinal IL-10 and protect from Th1-driven inflammation (42). Muribaculaceae could inhibit the CD8+ T cell activation to tolerate the immunity stimulation and present a negative correlation with inflammation status (47). In our study, the correlation between gut microbiota and inflammatory cytokines (IL-6, IL-1β, TNF-α and IL-2) was analyzed at the genus level. It was found that Q7-EVs increased the number of Akkermansia, Bifidobacteria, Muribaculaceae and Lactobacillus. On the other hand, Q7-EVs reduced the number of Proteobacteria, Deferribacteres and Epsilonbacteraeota. These results suggested that Q7-EVs might improve inflammation by regulating the gut microbiota in DSS-induced colitis mouse model. Lactobacillus and Bifidobacteria are common probiotics in the intestine, which help to inhibit harmful bacteria and improve gastrointestinal barrier function (48). Proteobacteria were considered as harmful bacteria in UC (49, 50). These findings indicated that Q7-EVs relieved DSS-induced colitis by increasing the beneficial bacteria and reducing the harmful bacteria in the intestine. Though it was confirmed that Q7-EVs could regulate the gut microbiota and alleviate DSS-induced colitis, the specific functional components and the mechanism of probiotic-derived EVs alleviating UC remained to be investigated in the future.
In conclusion, EVs derived from L. plantarum Q7 could regulate the intestinal microbiota and ameliorate DSS-induced colitis in C57BL/6J mice. L. plantarum Q7 might exert the probiotic function mediated by its extracellular vesicles. These findings provided a novel perspective to explore the function mechanism of probiotic and develop a potential therapeutic treatment of IBD.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/bioproject/, PRJNA778054.
Ethics Statement
The animal study was reviewed and approved by Animal Ethics Committee of Ocean University of China (permission number: spxy20200720215).
Author Contributions
Conceptualization, HY and LA. Methodology, HH, XZ, and TL. Formal analysis, HH, LT, and XL. Software, HH and QL Supervision, HY and LZ. Data curation, HH and YX. Writing-original draft preparation, HH, XZ, and LT. Writing-review and editing, HY, TL, and PG. Visualization, YB and YX. Project administration, HY. Funding acquisition, HY. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by National Natural Science Foundation of China (No. 32172180, 31771988) and Key Program of Natural Science Foundation of Shandong Province in China (No. ZR2020KC009).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.777147/full#supplementary-material
References
1. Guo F, Cai D, Li Y, Gu H, Qu H, Zong Q, et al. How Early-Life Gut Microbiota Alteration Sets Trajectories for Health and Inflammatory Bowel Disease? Front Nutr (2021) 8:760443. doi: 10.3389/fnut.2021.760443
2. Wang Z, Guo K, Gao P, Pu Q, Lin P, Qin S, et al. Microbial and Genetic-Based Framework Identifies Drug Targets in Inflammatory Bowel Disease. Theranostics (2021) 11(15):7491–506. doi: 10.7150/thno.59196
3. Santos Rocha C, Lakhdari O, Blottiere HM, Blugeon S, Sokol H, Bermudez-Humaran LG, et al. Anti-Inflammatory Properties of Dairy Lactobacilli. Inflammation Bowel Dis (2012) 18(4):657–66. doi: 10.1002/ibd.21834
4. Sánchez B, Delgado S, Blanco-M´ıguez A, Lourenc¸o A, Gueimonde M, Margolles A. Probiotics, Gut Microbiota, and Their Influence on Host Health and Disease. Mol Nutr Food Res (2017) 1(61):1600240. doi: 10.1002/mnfr.201600240
5. Hudson LE, Anderson SE, Corbett AH, Lamb TJ. Gleaning Insights From Fecal Microbiota Transplantation and Probiotic Studies for the Rational Design of Combination Microbial Therapies. Clin Microbiol Rev (2017) 30(1):191–231. doi: 10.1128/CMR.00049-16
6. Molina-Tijeras JA, Galvez J, Rodriguez-Cabezas ME. The Immunomodulatory Properties of Extracellular Vesicles Derived From Probiotics: A Novel Approach for the Management of Gastrointestinal Diseases. Nutrients (2019) 11(1038):1–15. doi: 10.3390/nu11051038
7. Sun Y, Shi H, Yin S, Ji C, Zhang X, Zhang B, et al. Human Mesenchymal Stem Cell Derived Exosomes Alleviate Type 2 Diabetes Mellitus by Reversing Peripheral Insulin Resistance and Relieving Beta-Cell Destruction. ACS Nano (2018) 12(8):7613–28. doi: 10.1021/acsnano.7b07643
8. Wu C, Liu Z. Proteomic Profiling of Sweat Exosome Suggests Its Involvement in Skin Immunity. J Invest Dermatol (2018) 138(1):89–97. doi: 10.1016/j.jid.2017.05.040
9. Zhuang X, Deng ZB, Mu J, Zhang L, Yan J, Miller D, et al. Ginger-Derived Nanoparticles Protect Against Alcohol-Induced Liver Damage. J Extracell Vesicles (2015) 4:28713. doi: 10.3402/jev.v4.28713
10. Briaud P, Carroll RK. Extracellular Vesicle Biogenesis and Functions in Gram-Positive Bacteria. Infect Immun (2020) 88(12):e00433. doi: 10.1128/IAI.00433-20
11. Liu Y, Alexeeva S, Defourny KA, Smid EJ, Abee T. Tiny But Mighty: Bacterial Membrane Vesicles in Food Biotechnological Applications. Curr Opin Biotechnol (2018) 49:179–84. doi: 10.1016/j.copbio.2017.09.001
12. Kim YS, Choi EJ, Lee WH, Choi SJ, Roh TY, Park J, et al. Extracellular Vesicles, Especially Derived From Gram-Negative Bacteria, in Indoor Dust Induce Neutrophilic Pulmonary Inflammation Associated With Both Th1 and Th17 Cell Responses. Clin Exp Allergy (2013) 43(4):443–54. doi: 10.1111/cea.12085
13. Fabrega MJ, Rodriguez-Nogales A, Garrido-Mesa J, Algieri F, Badia J, Gimenez R, et al. Intestinal Anti-Inflammatory Effects of Outer Membrane Vesicles From Escherichia Coli Nissle 1917 in DSS-Experimental Colitis in Mice. Front Microbiol (2017) 8:1274. doi: 10.3389/fmicb.2017.01274
14. Lee EY, Choi DY, Kim DK, Kim JW, Park JO, Kim S, et al. Gram-Positive Bacteria Produce Membrane Vesicles: Proteomics-Based Characterization of Staphylococcus Aureus-Derived Membrane Vesicles. Proteomics (2009) 9(24):5425–36. doi: 10.1002/pmic.200900338
15. Kim JH, Jeun EJ, Hong CP, Kim SH, Jang MS, Lee EJ, et al. Extracellular Vesicle-Derived Protein From Bifidobacterium Longum Alleviates Food Allergy Through Mast Cell Suppression. J Allergy Clin Immunol (2016) 137(2):507–16. doi: 10.1016/j.jaci.2015.08.016
16. Behzadi E, Mahmoodzadeh Hosseini H, Imani Fooladi AA. The Inhibitory Impacts of Lactobacillus Rhamnosus GG-Derived Extracellular Vesicles on the Growth of Hepatic Cancer Cells. Microb Pathog (2017) 110:1–6. doi: 10.1016/j.micpath.2017.06.016
17. Hu R, Lin H, Wang M, Zhao Y, Liu H, Min Y, et al. Lactobacillus Reuteri-Derived Extracellular Vesicles Maintain Intestinal Immune Homeostasis Against Lipopolysaccharide-Induced Inflammatory Responses in Broilers. J Anim Sci Biotechnol (2021) 12(1):25. doi: 10.1186/s40104-020-00532-4
18. Kim MH, Choi SJ, Choi HI, Choi JP, Park HK, Kim EK, et al. Lactobacillus Plantarum-Derived Extracellular Vesicles Protect Atopic Dermatitis Induced by Staphylococcus Aureus-Derived Extracellular Vesicles. Allergy Asthma Immunol Res (2018) 10(5):516–32. doi: 10.4168/aair.2018.10.5.516
19. Li M, Lee K, Hsu M, Nau G, Mylonakis E, Ramratnam B. Lactobacillus-Derived Extracellular Vesicles Enhance Host Immune Responses Against Vancomycin-Resistant Enterococci. BMC Microbiol (2017) 17(1):66. doi: 10.1186/s12866-017-0977-7
20. Bu Y, Liu Y, Li J, Liu T, Wang Y. Analyses of Plantaricin Q7 Synthesis by Lactobacillus Plantarum Q7 Based on Comparative Transcriptomics. Food Control (2021) 1):107909. doi: 10.1016/j.foodcont.2021.107909
21. Liu H, Zhang L, Yi H, Han X, Chi C. Identification and Characterization of Plantaricin Q7, a Novel Plantaricin Produced by Lactobacillus Plantarum Q7. LWT - Food Sci Technol (2016) 71:386–90. doi: 10.1016/j.lwt.2016.04.009
22. Kim W, Lee EJ, Bae IH, Myoung K, Kim ST, Park PJ, et al. Lactobacillus Plantarum-Derived Extracellular Vesicles Induce Anti-Inflammatory M2 Macrophage Polarization In Vitro. J Extracell Vesicles (2020) 9(1):1793514. doi: 10.1080/20013078.2020.1793514
23. Tong L, Hao H, Zhang Z, Lv Y, Liang X, Liu Q, et al. Milk-Derived Extracellular Vesicles Alleviate Ulcerative Colitis by Regulating the Gut Immunity and Reshaping the Gut Microbiota. Theranostics (2021) 11(17):8570–86. doi: 10.7150/thno.62046
24. Qu Y, Li X, Xu F, Zhao S, Wu X, Wang Y, et al. Kaempferol Alleviates Murine Experimental Colitis by Restoring Gut Microbiota and Inhibiting the LPS-TLR4-NF-kappaB Axis. Front Immunol (2021) 12:679897. doi: 10.3389/fimmu.2021.679897
25. Liu W, Zhang Y, Qiu B, Fan S, Ding H, Liu Z. Quinoa Whole Grain Diet Compromises the Changes of Gut Microbiota and Colonic Colitis Induced by Dextran Sulfate Sodium in C57BL/6 Mice. Sci Rep (2018) 8(1):14916. doi: 10.1038/s41598-018-33092-9
26. Wang Y, Xie Q, Zhang Y, Ma W, Ning K, Xiang JY, et al. Combination of Probiotics With Different Functions Alleviate DSS-Induced Colitis by Regulating Intestinal Microbiota, IL-10, and Barrier Function. Appl Microbiol Biotechnol (2020) 104(1):335–49. doi: 10.1007/s00253-019-10259-6
27. Kim JJ, Shajib MS, Manocha MM, Khan WI. Investigating Intestinal Inflammation in DSS-Induced Model of IBD. J Vis Exp (2012) 60:3678. doi: 10.3791/3678
28. Gao R, Shen Y, Shu W, Jin W, Bai F, Wang J, et al. Sturgeon Hydrolysates Alleviate DSS-Induced Colon Colitis in Mice by Modulating NF-Kappab, MAPK, and Microbiota Composition. Food Funct (2020) 11(8):6987–99. doi: 10.1039/c9fo02772f
29. Sann H, Erichsen J, Hessmann M, Pahl A, Hoffmeyer A. Efficacy of Drugs Used in the Treatment of IBD and Combinations Thereof in Acute DSS-Induced Colitis in Mice. Life Sci (2013) 92(12):708–18. doi: 10.1016/j.lfs.2013.01.028
30. Yanez-Mo M, Siljander PR, Andreu Z, Zavec AB, Borras FE, Buzas EI, et al. Biological Properties of Extracellular Vesicles and Their Physiological Functions. J Extracell Vesicles (2015) 4:27066. doi: 10.3402/jev.v4.27066
31. Rodriguez-Nogales A, Algieri F, Garrido-Mesa J, Vezza T, Utrilla MP, Chueca N, et al. Intestinal Anti-Inflammatory Effect of the Probiotic Saccharomyces Boulardii in DSS-Induced Colitis in Mice: Impact on microRNAs Expression and Gut Microbiota Composition. J Nutr Biochem (2018) 61:129–39. doi: 10.1016/j.jnutbio.2018.08.005
32. Axelsson LG, Landström E, Bylund-Fellenius AC. Experimental Colitis Induced by Dextran Sulphate Sodium in Mice: Beneficial Effects of Sulphasalazine and Olsalazine. Aliment Pharm Ther (1998) 12(9):925–34. doi: 10.1046/j.1365-2036.1998.00357.x
33. Yan F, Wang L, Shi Y, Cao H, Liu L, Washington MK, et al. Berberine Promotes Recovery of Colitis and Inhibits Inflammatory Responses in Colonic Macrophages and Epithelial Cells in DSS-Treated Mice. Am J Physiol Gastrointest Liver Physiol (2012) 302(5):504. doi: 10.1152/ajpgi.00312.2011
34. Li J, Zhong W, Wang W, Hu S, Yuan J, Zhang B, et al. Ginsenoside Metabolite Compound K Promotes Recovery of Dextran Sulfate Sodium-Induced Colitis and Inhibits Inflammatory Responses by Suppressing NF-kappaB Activation. PloS One (2014) 9(2):e87810. doi: 10.1371/journal.pone.0087810
35. Coqueiro AY, Raizel R, Bonvini A, Tirapegui J, Rogero MM. Probiotics for Inflammatory Bowel Diseases: A Promising Adjuvant Treatment. Int J Food Sci Nutr (2019) 70(1):20–9. doi: 10.1080/09637486.2018.1477123
36. Ashrafian F, Shahriary A, Behrouzi A, Moradi HR, Keshavarz Azizi Raftar S, Lari A, et al. Akkermansia Muciniphila-Derived Extracellular Vesicles as a Mucosal Delivery Vector for Amelioration of Obesity in Mice. Front Microbiol (2019) 10:2155. doi: 10.3389/fmicb.2019.02155
37. Jonas M, Neurath MF. Il-6 Signaling in Inflammatory Bowel Disease: Pathophysiological Role and Clinical Relevance. Inflammation Bowel Dis (2010) 8):1016–23. doi: 10.1002/ibd.20148
38. Puleston J, Cooper M, Murch S, Bid K, Makh S, Ashwood P, et al. A Distinct Subset of Chemokines Dominates the Mucosal Chemokine Response in Inflammatory Bowel Disease. Aliment Pharmacol Ther (2005) 21(2):109–20. doi: 10.1111/j.1365-2036.2004.02262.x
39. Ishiguro Y. Mucosal Proinflammatory Cytokine Production Correlates With Endoscopic Activity of Ulcerative Colitis. J Gastroenterol (1999) 34(1):66–74. doi: 10.1007/s005350050218
40. Hu T, Fan Y, Long X, Pan Y, Mu J, Tan F, et al. Protective Effect of Lactobacillus Plantarum YS3 on Dextran Sulfate Sodium-Induced Colitis in C57BL/6J Mice. J Food Biochem (2021) 45(2):e13632. doi: 10.1111/jfbc.13632
41. Chaniotou Z, Giannogonas P, Theoharis S, Teli T, Gay J, Savidge T, et al. Corticotropin-Releasing Factor Regulates TLR4 Expression in the Colon and Protects Mice From Colitis. Gastroenterology (2010) 139(6):2083–92. doi: 10.1053/j.gastro.2010.08.024
42. Mohamed SS, Abdeltawab NF, Wadie W, Ahmed LA, Ammar RM, Rabini S, et al. Effect of the Standard Herbal Preparation, STW5, Treatment on Dysbiosis Induced by Dextran Sodium Sulfate in Experimental Colitis. BMC Complement Med Ther (2021) 21(1):168. doi: 10.1186/s12906-021-03337-8
43. van Bergenhenegouwen J, Kraneveld AD, Rutten L, Kettelarij N, Garssen J, Vos AP. Extracellular Vesicles Modulate Host-Microbe Responses by Altering TLR2 Activity and Phagocytosis. PloS One (2014) 9(2):e89121. doi: 10.1371/journal.pone.0089121
44. Wang YY, Guo YL, Chen H, Wei H. CX W. Potential of Lactobacillus Plantarum ZDY2013 and Bifidobacterium Bifidum WBIN03 in Relieving Colitis by Gut Microbiota, Immune and Anti-Oxidative Stress. Can J Microbiol (2018) 64:327–37. doi: 10.1139/cjm-2017-0716
45. Liu Z, Liu F, Wang W, Sun C, Gao D, Ma J, et al. Study of the Alleviation Effects of a Combination of Lactobacillus Rhamnosus and Inulin on Mice With Colitis. Food Funct (2020) 11(5):3823–37. doi: 10.1039/c9fo02992c
46. Liu JH, Chen CY, Liu ZZ, Luo ZW, Rao SS, Jin L, et al. Extracellular Vesicles From Child Gut Microbiota Enter Into Bone to Preserve Bone Mass and Strength. Adv Sci (Weinh) (2021) 8(9):2004831. doi: 10.1002/advs.202004831
47. Shang L, Liu H, Yu H, Chen M, Yang T, Zeng X, et al. Core Altered Microorganisms in Colitis Mouse Model: A Comprehensive Time-Point and Fecal Microbiota Transplantation Analysis. Antibiotics (2021) 10:1–11. doi: 10.3390/antibiotics10060643
48. Azad MAK, Sarker M, Li T, Yin J. Probiotic Species in the Modulation of Gut Microbiota: An Overview. BioMed Res Int (2018) 2018:9478630. doi: 10.1155/2018/9478630
49. O’Shea CJ, O’Doherty JV, Callanan JJ, Doyle D, Thornton K, Sweeney T. The Effect of Algal Polysaccharides Laminarin and Fucoidan on Colonic Pathology, Cytokine Gene Expression and Enterobacteriaceae in a Dextran Sodium Sulfate-Challenged Porcine Model. J Nutr Sci (2016) 5:e15. doi: 10.1017/jns.2016.4
Keywords: Lactobacillus plantarum Q7, extracellular vesicles, gut microbiota, inflammatory cytokines, ulcerative colitis
Citation: Hao H, Zhang X, Tong L, Liu Q, Liang X, Bu Y, Gong P, Liu T, Zhang L, Xia Y, Ai L and Yi H (2021) Effect of Extracellular Vesicles Derived From Lactobacillus plantarum Q7 on Gut Microbiota and Ulcerative Colitis in Mice. Front. Immunol. 12:777147. doi: 10.3389/fimmu.2021.777147
Received: 15 September 2021; Accepted: 16 November 2021;
Published: 02 December 2021.
Edited by:
Christoph Mueller, University of Bern, SwitzerlandReviewed by:
Bailiang Li, Northeast Agricultural University, ChinaWei Chen, Hefei University of Technology, China
Waliul I. Khan, McMaster University, Canada
Copyright © 2021 Hao, Zhang, Tong, Liu, Liang, Bu, Gong, Liu, Zhang, Xia, Ai and Yi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huaxi Yi, eWloeEBvdWMuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
 Haining Hao
Haining Hao Xinyi Zhang1†
Xinyi Zhang1† Lingjun Tong
Lingjun Tong Tongjie Liu
Tongjie Liu Lanwei Zhang
Lanwei Zhang Yongjun Xia
Yongjun Xia Lianzhong Ai
Lianzhong Ai Huaxi Yi
Huaxi Yi