
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 21 October 2021
Sec. Immunological Tolerance and Regulation
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.772017
This article is part of the Research TopicImmunopathology of Type 1 DiabetesView all 21 articles
Type 1 diabetes (T1d) results from a sustained autoreactive T and B cell response towards insulin-producing β cells in the islets of Langerhans. The autoreactive nature of the condition has led to many investigations addressing the genetic or cellular changes in primary lymphoid tissues that impairs central tolerance- a key process in the deletion of autoreactive T and B cells during their development. For T cells, these studies have largely focused on medullary thymic epithelial cells (mTECs) critical for the effective negative selection of autoreactive T cells in the thymus. Recently, a new cellular player that impacts positively or negatively on the deletion of autoreactive T cells during their development has come to light, thymic B cells. Normally a small population within the thymus of mouse and man, thymic B cells expand in T1d as well as other autoimmune conditions, reside in thymic ectopic germinal centres and secrete autoantibodies that bind selective mTECs precipitating mTEC death. In this review we will discuss the ontogeny, characteristics and functionality of thymic B cells in healthy and autoimmune settings. Furthermore, we explore how in silico approaches may help decipher the complex cellular interplay of thymic B cells with other cells within the thymic microenvironment leading to new avenues for therapeutic intervention.
Type 1 diabetes (T1d) is an autoimmune condition characterised by the destruction of the insulin producing beta (β) cells in the islets of Langerhans by co-operative interaction between the innate and adaptive immune systems; the final assault being perpetuated by CD8+ cytotoxic T cells (1–3). The non-obese diabetic (NOD) mouse that spontaneously develops T1d in a manner thought to be similar to man, has been an important resource in analysing the complexity of the T1d immunopathology (4). In line with the important role that the thymus plays in purging thymocytes with autoreactive T cell receptors (TcRs) (5), T1d progression is linked to defects in central tolerance increasing the output of islet-reactive T cells (6, 7), although definitive understanding of why negative selection of islet-reactive T cells occurs remains elusive.
Studies in NOD mice deficient in B cells have revealed an initially unappreciated role for B cells not only in helping the CD4+ T cell response to islet antigens (8–10), but also promoting the survival of islet-reactive CD8+ CTL in situ (11). This important role for B cells in the T1d process has been translated to man, where T1d progression risk is determined by the number of serum antibodies to islet antigens (12) and evidence that T1d severity is characterised by increased infiltration of islets letion of B cells in people newly diagnosed with T1d results in transient remission of the condition (14).
Recently, we reported a new role for B cells in the T1d process- mediators of breakdown in thymic central tolerance (15). We showed that thymic B cells, normally a minor constituent of a healthy thymus in both man and mouse (16, 17), rapidly increase following the initiation of islet infiltration with immune cells (termed insulitis) but prior to overt clinical manifestation of T1d. Further, we showed that thymic B cells reside in putative thymic germinal centres, undergo in situ class switching and differentiation into plasma cells. Autoantibodies from these plasma cells targeted specific medullary thymic epithelial cells (mTECs) for antibody-mediated apoptosis, leading to increased output of T cells that bypassed negative selection thereby enhancing T1d progression. Our finding was reminiscent of the immunopathology of the autoimmune conditions of myasthenia gravis (18) and systemic lupus erythematosus (19, 20), suggesting that thymic B cell abnormalities may be a common link between certain autoimmune conditions.
Analysis of thymic B cells is challenging; thymic involution as we age necessitates human thymic studies to be largely restricted to foetal or paediatric tissue which, quite rightly, is ethically sensitive to procure. Although it could be argued the availability of murine thymi negates issues of thymic cellularity during the involution process, researchers have an ethical responsibility to minimise the size of murine cohorts undergoing experimental procedures. Systems biology, which incorporates the development of computational models that can recapitulate the complexity of the immune response in defined tissues- offers an attractive approach to evaluate the molecules and signal pathways that contribute to the thymic B cell-mediated progression to T1d.
Examples of existing simulation studies in biology are numerous: granuloma formation (21–23), breast cancer metastasis (24) and lymphoid tissue formation (25, 26). There has also been a considerable body of work on modelling epithelial tissues (27, 28). Following this precedent it will be instructive to create a computer simulation of the early thymic events in T1d development.
This review is, therefore, structured in two parts; first we will discuss our current understanding of the ontogeny, phenotype and function of thymic B cells in health and disease, and secondly, we review the development and challenges of the systems biology approach of generating a computational model of thymic autoimmunity.
Thymic B cells were initially discovered during immunohistochemical studies of human thymic tissue (17) and subsequently in mice (29, 30).Thymic B cells, a minor population of cells within the thymic cellular pool, are present from embryonic age through to adulthood, the number of cells remaining stable throughout life, although reports of the ageing murine thymus suggest increasing numbers of thymic B cells are a characteristic of the thymic involution (6, 7, 31). Since the discovery of thymic B cells, the origin of the cells- in situ development versus recirculation from the periphery- has been debated. For example, Sato et al., demonstrated that peripheral B cells preferentially migrate to the thymus in response to increasing levels of intrathymic B lymphocyte chemoattractant CXCL13, although it was noted this occurred in aged mice (32). In contrast, parabiosis models argue against peripheral B cell migration to the thymus (33). Our adoptive transfer studies in NOD mice, revealed splenic B cells had minimum capacity to migrate to the thymus even when transferred at the post-insulitic, pre-diabetic phase when thymic B cell numbers rapidly increase (Davis and Green, unpublished observations). Interestingly, B cells isolated from the thymus almost exclusively migrated back to this organ following adoptive transfer into recipient mice. These findings related to peripheral B cell thymic migration, align with others, and suggest that thymic B cells originate from in situ development (34, 35). Further support for this hypothesis was provided by studies in NOD-RAG2p-GFP reporter mice (15), that enable monitoring of recombination activating gene (RAG) activity in developing B and T cells (36). Comparison of thymic B cell RAG activity in NOD mice revealed a significant increase in rearrangement of the B cell receptor (BcR) in the post-insulitic, pre-diabetic phase compared to pre-insulitic phase (15). Interestingly, the level of thymic B cell RAG activity was similar between NOD mice in the pre-insulitic phase to the levels seen for control, age-matched mice, and in this latter strain thymic B cell RAG activity remained at a constant level as mice aged. Although it could be speculated that increased RAG activity in thymic B cells may represent aberrant re-ignition of RAG genes in peripheral B cells that migrate to the thymus and potentially undergo receptor editing of the BcR, studies by Gay et al., demonstrating that peripheral RAG- B cells were unable to reactivate RAG following a series of mitogenic and antigenic B cell stimulations, argue against this possibility (37). Similar findings looking at the ability of immunisation to re-ignite RAG activity supports the hypothesis that RAG expression in B cells is restricted to their developmental stages (38, 39). Thus, the thymus of mice per se can support B cell development, but in the context of T1d, there is an acceleration in B cell development specifically during the post-insulitis, prediabetic phase, and this coincides with a rapid increase in thymic B cell numbers (15).
If thymic B cells develop in situ, what are the progenitor cells and signal pathways involved? Although it is well established that the thymus is seeded with common lymphoid progenitor cells (CLP) that have T, B and NK cell potential (40), it has been challenging to map the thymic B cell development pathway assuming it originates from the CLP population. Several studies have identified B lineage-committing transcription factors and cells within the thymus that have a phenotype akin to B cell committed precursors in their natural developmental habitat, the bone marrow (34). In addition, McKenna et al., demonstrated that addition of bone marrow isolated pro-B cells into thymic organ cultures stimulated with Fms-related receptor tyrosine kinase 3 (FLT3) and IL-7 induced immature B cell development (41). Interestingly, similar culture of pro-B cells with cell-lines derived from bone marrow or thymic stroma cultured did not induce complete B cell development in the presence of FLT3 and IL-7. However, these studies are based on B cell committed progenitors, that is pro-B cells, and it has been far more challenging to identify within the thymus the cell(s) that lie upstream of the pro-B cell that have been identified in the bone marrow (42, 43).
As well as speculation on the definitive progenitor from which intrathymic B cells develop, there also been controversy in the signal pathways that lead to intrathymic B cell development. For example, it has been proposed that intrathymic B cell development may be a default pathway resulting from perturbation of the T cell developmental pathway due to impaired CD3 or T cell receptor β (TcRβ) signalling (44) or inappropriate Notch signalling (45). However, Feyerabend et al., using the cre-lox system to delete Notch in the DN1 population, a population that has both B and T cell potential, did not divert development down the B cell developmental pathway (46). More understanding is required as to both the progenitor and signal pathways that enable the development of thymic B cells. Such studies will be invaluable in determining whether increased thymic B cell development characteristic of T1d progression in NOD mice relates to perturbation of these signal pathways.
B cells can be divided into several classifications: follicular (FO), marginal zone (MZ), peritoneal (B1a) and regulatory (Bregs). Each subgroup of B cells can be identified by their surface markers, and each has particular functions to play in the immune response. Although Bregs have been identified in the thymus (47) Bregs will be discussed elsewhere in this Special Edition and will not be considered here. MZ B cells have received particular attention in the autoimmune setting, due to an aberrant increase in both their numbers and location in murine autoimmune models (48–50) as well as in humans living with certain autoimmune conditions (51, 52). Innate-like MZ B cells express high levels of IgM and low levels of IgD alongside coexpression of CD21 and CD35 (53). They can function to remove apoptotic cell debris impeding autoimmunity (54), or conversely, they may promote autoimmunity via their polyreactive receptors (48). Furthermore, weak signalling through the BcR (55, 56) or perturbation of the negative regulators of BcR signalling- including FcγRIIB- promotes expansion of MZ B cells that are more efficient at presenting antigen to T cells (57), including potentially autoreactive T cells. In light of the strong link between mutations in FcγRIIB and T1d in both mouse and man, we assessed whether thymic B cells in NOD mice had a MZ or FO phenotype (15). Our data showed that although MZ-like cells are detectable in the thymus of both NOD and control mice, there are significantly fewer in the NOD mouse thymus compared to control animals. It is intriguing that MZ-like cells are present in the thymus of mice, and it will be interesting to see if they function here as surveillance cells against infection or participate in the removal of apoptotic thymocytes.
Nevertheless, our data overwhelmingly ascribes thymic B cells to have a FO phenotype in the NOD mouse, a finding that is in line with several studies in mice and man (33). In NOD mice, CD5, which has been described as a marker for B1a cells in the peritoneal cavity, is also expressed on the thymic FO B cells, although not as extensively as seen in non-NOD strains (29). Thymic FO B cells, in comparison to splenic FO B cells, express much higher levels of MHC class I and II, as well as costimulatory molecule CD40, signalling through which may regulate expression of CD5 (58). Although the majority of thymic B cells in the NOD mice expressed an IgM+IgD+ BcR, class-switched B cells expressing IgM-IgD-IgG+ BcRs were readily detectable, although notably control mice also had these class-switched cells too. In contrast to this shared phenotype of NOD thymic B cells with non-NOD strains of mice, the NOD thymus harbours class-switched unusual IgM-IgD+IgA+ and IgM-IgD+IgG+ FO B cells where such cells were largely absent in control mice (15). Interestingly, human peripheral B cells with this unusual expression of IgD+ in the absence of IgM has recently been described in people living with T1d (59). In people with T1d, these IgD+ B cells express polyreactive receptors and can interact with insulin. Although we established that insulin-reactive B cells reside in the thymus of NOD mice, similar numbers of insulin-reactive B cells were present in control, non-autoimmune mice (15). Further, our studies focused on the thymic B cell population in its entirety, not this specific subgroup of B cells. Thus, the antigen specificity of these unique thymic IgD+IgG+ B cells in NOD mice is yet to be resolved, but potentially harbours an autoreactive BcR repertoire.
B cell activation takes place in specialised germinal centres (GCs) within B cell follicles of secondary lymphoid organs. GCs tend to form in response to infection, although small transient GCs have been seen in non-inflammatory conditions. GC formation requires cross-talk between stromal cells and immune cells, and are integral for the somatic hypermutation, class switching and differentiation of activated B cells into plasma cells and memory B cells reviewed by (60). Aside from conventional GCs, B cell activation can also take place in extrafollicular structures and ectopic GCs. These ectopic GCs occur in non-lymphoid tissue, and result from the remodelling of the tissue stromal cell network in response to inflammation (61). Ectopic GCs are of particular importance in autoimmunity, and several conditions have reported the presence of these structures in the target tissue (62–64), including in the islets of T1d murine models (65), and the presence of ectopic GCs correlates with disease severity. Like conventional GCs, ectopic GCs are compartmentalised into B and T cell areas (64), and several cytokines have been attributed to their localised formation including IL-22 (66) and IL-23 (67). Interferon gamma (IFNγ) seems to be one of the most critical players in ectopic GCs formation (68–70). Despite the remarkable knowledge we now have on conventional and ectopic GCs, little is known about GCs that can form in thymic tissue despite being a characteristic of autoimmune conditions like myasthenia gravis (18, 71), systemic lupus erythematosus (19) and T1d (15). Consistent among the autoimmune conditions where thymic GC occur, the GCs form at the cortical-medullary junction, and in NOD mice, such structures only materialise at the post-insulitic, pre-diabetic phase. Furthermore, the thymus becomes enriched with IL-21 as thymic GCs form, a cytokine that is critical for regulating GC maintenance and promotion of B cell differentiation and proliferation (72). Expression of activation-induced cytidine deaminase (AID) increases, suggesting active somatic hypermutation/class-switching is ongoing. IL-21 has proven a particularly interesting cytokine in promoting T1d both in man (73) and NOD mice (74). At the heart of the link between IL-21 and T1d are the T follicular helper (Tfh) cells (73) and in children at risk of T1d progression, circulating Tfh cell numbers peak around onset of clinical symptoms (75). In man it has been described that Tfh cells within conventional GCs may be identifiable from their recirculating counterpart on the basis of expression of the master transcription factor for Tfh cells- Bcl-6; whereas GC residing Tfh cells express Bcl-6, recirculating Tfh cells may not (76, 77). Nevertheless, circulating Tfh cells are uniquely gifted at entering inflamed tissue, and participating in ectopic GC formation (78) and in vitro, can promote B cell class switching retaining characteristics of GC Tfh cells (79, 80). As expected, considering their essential role in GC/ectopic GCs, Tfh cells are enhanced in the thymus of NOD mice at an age when thymic GC formation occurs. These Tfh cells express Bcl-6 and IL-21, as well as other known markers of Tfh cells (15). This suggests that intrathymic Tfh cells may derive from in situ CD4+ T cells, unless circulating Tfh cells that migrate to the thymus take on the phenotype of GC Tfh cells with respect to Bcl-6 expression.
Certain questions remain about the nature of the thymic GCs in T1d; do they contain follicular dendritic cell structures and what is the source of the inflammation to push thymic GC formation? GCs are populated with specialised follicular dendritic cells (FDC) that act as a depot for antigen presentation within the GC (81). However, we have yet to confirm such cells exist in our thymic GCs (Pinto and Green, unpublished observations). Interestingly, it has been postulated that the type of FDC present in a GC/ectopic GC may be unique to the tissue and type of inflammation (67, 82) and markers of conventional GCs may not be present on thymic FDCs. In terms of inflammation, it could be speculated that viral infections linked to T1d development in man may also infect the thymus (83) or endogenous retroviral infection (84) could induce thymic GC formation. Alternatively thymic GC formation may simply be a reflection of an accelerated ageing of the thymus (85). Indeed, others, have documented accelerated thymic involution in NOD mice (6). It will be important to determine if people who develop T1d also have thymic GCs, and potentially accelerated ageing of this tissue.
The immunohistochemical evidence that thymic B cells form follicles at the cortico-medullary junction, and can form rosette-like structures around T cells in the medulla (86, 87) led to early speculation that thymic B cells were involved in negative selection of autoreactive T cells. Subsequent studies from different groups have substantiated that hypothesis (discussed below), although whether thymic B cells positively or negatively contribute to negative selection seems dependent on the autoimmune nature of the mammal studied. Before we discuss the evidence for the role of thymic B cells in negative selection, let’s first consider their antigen specificity.
We earlier touched on the finding that thymic B cells in NOD mice or control animals expressed receptors for insulin i.e. they were autoreactive. In normal B cell development and maturation, efficiency in removing B cells with autoreactive BcRs is high, with approximately 20% of circulating B cells bearing autoreactive BcRs (88). Interestingly, the removal of B cell with autoreactive BcRs occurs in two stages; initially at the immature B cell stage in the bone marrow and subsequently during the transition of immature B cells to mature B cells following their recent egress from the bone marrow, with the early immature to immature B cell development stage exhibiting the largest removal of self-reactive B cells from the repertoire (88). Thus the earliest stages of B cell development in the bone marrow the repertoire of B cells has a high level of self-reactivity. To determine if the thymic B cell repertoire, similar to bone marrow-derived B cells, had self-reactivity, Rother et al. performed comparative sequencing studies of single cell sorted paediatric thymic B cells versus foetal bone marrow B cells (89). Such studies demonstrated that thymic B cells had a greater specificity for self-peptide autoantigens than similar sequenced foetal bone marrow B cells, these latter cells being more specific for dsDNA. Furthermore, thymic B cells had polyreactivity, recognising multiple autoantigens, including insulin. This prevalence of thymic B cells to harbour an autoreactive BcR has been shown by others (90). It can be envisaged that autoreactive thymic B cells may participate in negative selection by presenting ‘free’ autoreactive antigens- either trafficked to the thymus or captured from dying medullary thymic epithelial cells. Indeed, thymic B cells have been shown to efficiently present antigens their BcR is specific for to developing T cells (91) promoting efficient deletion of autoreactive T cells during T cell development (92–95). However, capture of autoantigens by the thymic B cell autoreactive BcR is not the sole way thymic B cells contribute to negative selection. Yamano et al., using a transgenic autoimmune regulator (AIRE) gene locus encoding a chimeric influenza haemagglutinin protein and human CD2 promoter demonstrated that 50% of thymic B cells express AIRE. In contrast, splenic or bone marrow B cells in the transgenic animal did not (94). Interestingly these AIRE+ thymic B cells resided in the medulla, and comparative studies of tissue restricted antigen (TRA) expression between AIRE+ thymic B cells and AIRE+ medullary thymic epithelial cells found there was no overlap between the two cell types in the TRAs presented. This suggests that thymic B cells may work in concert with medullary thymic epithelial cells to negatively select a greater range of autoreactive T cells. The work of Yamano builds on early reports that thymic B cells deleted superantigen specific T cells and that B cell specific expression of myelin oligodendrocyte glycoprotein enhanced negative selection of MOG-reactive transgenic T cells (16).
This ability of thymic B cells to participate in negative selection is suggested to be linked to their intrathymic class-switching activity (96), autoreactive thymocytes enabling selection and expansion of their cognate autoreactive thymic B cell counterpart. Perera et al., used an AID reporter mouse in a parabiotic model to show that self-antigen can drive class-switching of thymic B cells in situ, and the class-switched cells predominantly expressed IgG2b and IgA BcRs (96), and on an autoimmune background, class-switching of autoreactive thymic B cells numbers was enhanced. Further, they showed that impeding class switching of thymic B cells impaired their ability to negatively selective autoreactive T cells.
Others have documented a role for thymic B cells in the development of thymic T regulatory cells (Treg), and animals with expanded thymic B cell compartments have a correlating expanded thymic Treg compartment too (97, 98).
It would appear that there is strong evidence that thymic B cells are important in central tolerance. However, this positive role for thymic B cells in central tolerance is contrasted by the evidence that thymic B cells are key players in mediating tissue damage in myasthenia gravis (18, 63, 71, 99) and more recently systemic lupus erythematosus (19, 100) and T1d (15). Myasthenia gravis is the most documented condition where thymic B cells participate negatively in the autoimmune outcome. In myasthenia gravis, the thymus is the key source for pathogenic acetylcholine receptor antibodies that target this receptor on muscles leading to chronic muscle weakness. The thymus in myasthenia gravis patients have medullary thymic epithelial hyperplasia (18) with autoreactive thymic B cells secreting antibodies to acetylcholine receptors expressed on the medullary thymic epithelial cells triggering their demise via Complement-mediated attack (101).
In animal models of systemic lupus erythematosus, thymic B cells have a distinct transcriptome compared to thymic B cells from non-autoimmune prone, with increased prevalence of genes related to B cell survival (19). These increased thymic B cells were shown to promote expansion of the Tfh cells which in turn could enhance the systemic autoantibody response.
Our own studies in NOD mice, suggest that in T1d, thymic B cells may act in a manner similar to that seen in myasthenia gravis. Enhancement of thymic B cell numbers, the formation of thymic germinal centres results in a significant increase in intrathymic antibody levels in contrast to non-autoimmune prone mice. These antibodies are predominantly of the IgG1 and IgA subclass and were unique to the thymic compartment (15). Furthermore, in situ binding of these IgG antibodies to thymic medullary epithelial cells correlated with enhanced apoptosis of these antibody-selected cells. We have yet to establish that the antigen recognised by the autoantibodies is insulin, however, we showed that loss of certain medullary thymic epithelial cells resulted in decreased negative selection of autoreactive T cells, and enhanced survival of insulin-reactive thymocytes (15).
These contrasting roles for thymic B cells in negative selection are intriguing. Whether thymic B cells harbour pro-negative selection and anti-negative selection subpopulations, with the latter population having an advantage over the former in the autoimmune setting remains to be established. Alternatively, it may be that as an autoimmune process is ongoing, pro-negative selection thymic B cells switch to an anti-negative selection functionality. The potential mechanisms that promote these divergent properties of thymic B cells is shown in Figure 1. Studies that address these hypotheses will be important in understanding the relationship between thymic B cells and central thymic tolerance.
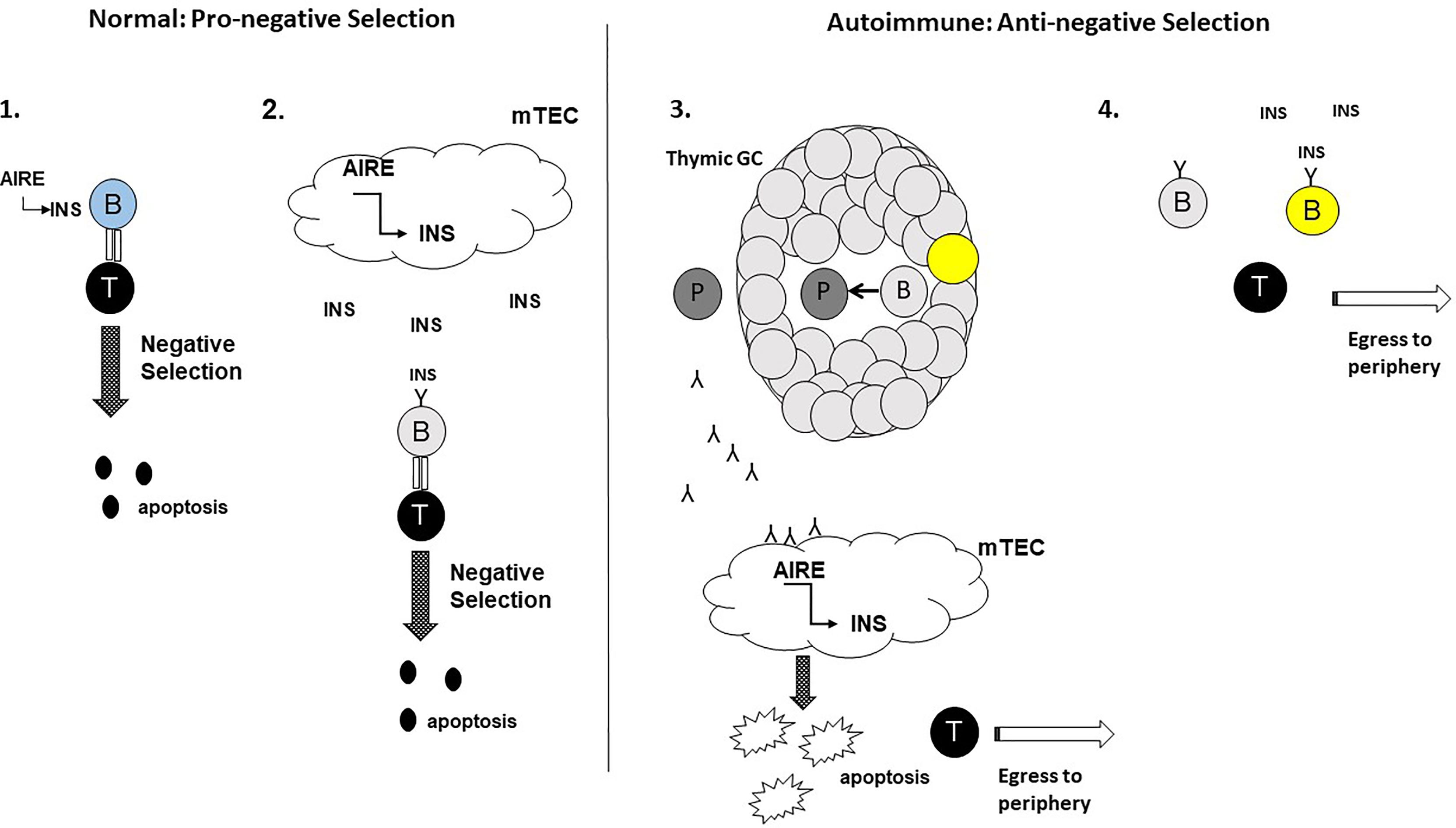
Figure 1 Potential roles for thymic B cells in the thymic negative selection process: a hypothesis. In the normal setting, thymic B cells enhance the negative selection of autoreactive T cells. 1. Aire+ Thymic B cells (blue) express and present self-antigens e.g. insulin (INS) to autoreactive T cells with high affinity TcRs for the self-antigens leading to T cell apoptosis. 2. Self-antigens e.g. INS secreted by Aire+ mTECs are acquired by thymic B cells expressing self-reactive BcRs (light grey). Internalisation, processing and presentation of the self-antigen to autoreactive T cells leads to T cell apoptosis. In the autoimmune setting, thymic B cells impede the negative selection of autoreactive T cells. 3. The emergence of thymic GCs results in thymic B cells receiving signals to develop into plasma cells (dark grey) secreting autoantibodies for self-antigens expressed by mTECs. Binding of the autoantibodies to mTECs leads to mTEC apoptosis, leading to decreased negative selection of autoreactive T cells and increased egress of the T cells to the peripheral tissues. 4. Somatic hypermutated (yellow) thymic B cells egressing from thymic GCs, may outcompete ‘normal’ thymic B cells for the binding of self-antigen in the thymic milieu impeding negative selection. For this scenario, the sm thymic B cells would either fail to adequately present the self-antigens to autoreactive T cells to support negative selection.
Many challenges face immunologists studying the thymus; early involution of thymic tissue meaning most human studies are based on procured tissue from foetuses, neonates or young children, and acquiring such tissue is not readily accessible due to, among other issues, ethical reasons. Studies of the thymus, particularly the ageing thymus and its many intricacies in cellular cross-talk, requires large cohorts of mice, again rearing questions of ethics. Recent years have seen a drive towards using computational algorithms and in silico models to recapitulate the dynamic environments of immune tissues, and assess the role of candidate molecules in a particular pathway. In this last part of the review, we will discuss the potential of developing in silico computational models to identify therapeutic avenues for manipulating thymic B cells in autoimmune disorders like T1d.
Systems biology is the integration of wet-lab experimentation and computational research in order to understand complex biological systems. Computational biology provides tools for the theoretical exploration of biology, permitting scientists to address critical questions directly (102). Simulation is one facet of computational biology that is finding increasing usage (103–106).
The use of simulation would bypass the necessary ethical, financial and practical issues surrounding acquisition of thymic tissue. In addition, the very nature of the system lends itself very precisely to Agent Based Modelling and Simulation (ABMS); consisting as it does of very many heterogeneous individuals of different types e.g. B cells, T cells, thymic medullary epithelial cells. These cell populations can interact with each other in specific ways under specific conditions. The cells will also possess some concept of location (the medulla or the cortico-medullary junction) and state (e.g. some stage of the cellular life cycle). Use of agent-based modelling will permit us to investigate how manipulating cell behaviours will give rise to altered system-wide (‘emergent’) behaviour. The results will be easily interpretable and so simple to put into words accessible to a wider audience.
A simulation is typically either mathematical or computational. Mathematical models are generally based on series of differential equations (107) and cells are modelled as populations rather than as individuals (108). Such models are often seen as opaque to non-mathematicians (109) and the model is typically difficult to extend should further cell types need to be considered. A further perceived disadvantage of mathematical models is that the resulting differential equations tend be complicated to solve exactly and often require solving via numerical methods which entails potentially unacceptable approximations to the model.
On the other hand, computational simulation methodologies such as Agent Based Modelling (ABM) are typically conceptually much simpler (110, 111). Cellular populations are modelled as sets of individuals (112) that share similar behaviours e.g. a population of Single Positive T cells. In this way more descriptive explanations of experimental observations may be proposed from simulation outcomes. Additionally agent-based models are better suited than other modelling techniques to capturing the system-wide, or emergent, behaviour of the system (113). Here, emergent behaviour is understood to be the system behaviour arising from the combined behaviours of the component entities e.g. cells.
An ABM will represent some abstraction of the system created jointly by a scientist, expert in the system of interest, and a software developer. The model should aim to include all factors e.g. cells and signalling molecules, generally held to be essential to system function. It is also important to include any potential roles for the biological environment in the model.
A number of short tutorials on the creation of agent based models can be found in the literature e.g. Bandini et al., (114). Generally, the principal steps in developing an agent-based simulation are:
i. Identify the agents that are important to what you want to model:
For example in a model of negative selection events in the thymic medulla, we might wish to model the behaviours of Single Positive T cells, B cells and medullary thymic epithelial cells (mTECs).
ii. Identify the environment in which your agents exist:
We might for instance break the thymus into three distinct environments: the cortex, the cortico-medullary junction (CMJ) and the medulla; placing relevant cells in each.
iii. Identify mechanisms whereby the agents interact with each other and with their environment:
We might consider that cells behave differently in different environments. For example, SP T cells will not tend to remain in the cortex, but will rather migrate across the CMJ to the medulla where they will interact with mTECs, via recognition of the insulin fragments presented by the mTECs, to facilitate negative selection. Aggressive B cells will also be able to interact directly with mTECs, also via recognition of insulin presented by the mTECs.
iv. Consider how best to implement the model as computer code and any assumptions and simplifications necessary in achieving this:
It will be very difficult to exactly replicate precise biological behaviour as computer code. For example, cells are unlikely to be of regular shapes e.g. circular, so the geometry of the system will not be exactly reproducible in silico and appropriate approximations will be necessary. In the case of thymic B cells it will be necessary to decide how we will differentiate between those B cells which serve to enhance the negative selective of autoreactive T cells and those that prevent the negative selection of autoaggressive T cells.
Despite the benefits of employing simulation as an investigative tool, it is wise to be mindful of the potential pitfalls in the use of the technique.
As an experiment, a simulation must be reproducible. It is therefore of paramount importance that simulation design be comprehensively documented. The documentation must incorporate all assumptions and design decisions to make them available to the scientific community to assess. The CoSMoS simulation design protocol (115) provides guidance in documentation and development of simulations, in which the user can have confidence.
The first challenge in the design of agent-based models is to correctly capture the relevant entities (cells, signalling molecules etc.) of the system and their behaviours in the model. If a key component of the system (or its behaviour) is omitted from the model then the model will be unfit for the purpose for which it was designed. As an example, two different types of behaviour in the B cell population have been noted. Some cells appear to be essentially quiescent, though may play an active role in sustaining negative selection, whereas others adopt a more aggressive role, apoptosing mTECs and thus triggering the breakdown of negative selection and hence central tolerance. Effective communication between the software developer and the expert biologist will help to mitigate this problem.
Also, simulation outcomes will be determined by the choice of values for the required simulation parameters. Model parameters may represent quantities such as the duration of a particular cell cycle stage or the concentration of cytokine that brings about progression to the next differentiation state. Such values will, in all likelihood, not be available from experiment and must be estimated. Chosen parameter values will ultimately impact on overall system behaviour and simulation output. Calibration is the process of adjusting parameter values so as to align in silico simulation results with observed in vivo behaviours. The issues surrounding simulation parameterization are discussed briefly below, but are addressed more fully in the relevant literature:
Optimization of parameter values is the subject of numerous widely used techniques such as the Latin Hypercube (116). This technique is based on the random sampling of the entire parameter space and is time and resource intensive to perform for a typical biological system.
The stability of simulation performance with parameter perturbation can be assessed using two different types of analysis. Robustness analysis is used to gauge the impact of simulation parameters on the simulation’s ability to execute stably i.e. which parameter values might cause the simulation to crash (117). Sensitivity Analysis can be used to investigate the effects of individual parameters or combinations of parameters on simulation outputs via the systematic variation of parameter values and observation of the effect on simulation performance (117). Sensitivity analyses are further discussed in (118, 119).
Recent research has aimed to refine techniques that facilitate calibration of simulations involving large numbers of parameters in a less resource intensive manner than the Latin Hypercube method described above. These novel techniques include the use of Machine Learning and multi-objective calibration (120, 121).
Although the scope and particularly the calibration of computational simulations is potentially limited by the scale of the computational resources available, there is a trend towards easier access to powerful high performance computer clusters which makes these concerns less relevant.
Despite the challenges entailed in employing an ABMS approach to biological simulation, ABMS remains a highly descriptive and powerful methodology for elucidation of cellular and molecular mechanisms of disease and many of the challenges posed by the use of the technique, particularly those relating to parameterization, are becoming more easily addressed using recent developments.
Commonalities in autoimmune conditions offer insights into novel therapies that may target multiple autoimmune conditions. Evidence that abnormality in the thymic B cell compartment is a shared characteristic between several autoimmune conditions highlights the need for further studies into this enigmatic cell. Nevertheless, increasing ethical considerations and availability of thymic tissue for analysis makes studies of thymic B cells somewhat challenging, particularly in man. Computational models of the dynamic thymic environment may offer a new approach to address the role of thymic B cells in health and disease, leading to candidate molecules or signal pathways as novel therapeutic targets in autoimmune conditions, including T1d.
DC and EAG wrote the thymic B cell section. RG wrote the computational modelling section. All authors contributed to the article and approved the submitted version.
This work was supported by a Diabetes UK Project Grant 18/0005804 awarded to EAG, and a Daphne Jackson Trust Fellowship awarded to RG. The University of York and the Medical Research Council funded the Fellowship.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Kaufman DL, Clare-Salzler M, Tian J, Forsthuber T, Ting GS, Robinson P, et al. Spontaneous Loss of T-Cell Tolerance to Glutamic Acid Decarboxylase in Murine Insulin-Dependent Diabetes. Nature (1993) 366(6450):69–72. doi: 10.1038/366069a0
2. Lieberman SM, DiLorenzo TP. A Comprehensive Guide to Antibody and T-Cell Responses in Type 1 Diabetes. Tissue Antigens (2003) 62(5):359–77. doi: 10.1034/j.1399-0039.2003.00152.x
3. Tisch R, Yang XD, Singer SM, Liblau RS, Fugger L, McDevitt HO. Immune Response to Glutamic Acid Decarboxylase Correlates With Insulitis in non-Obese Diabetic Mice. Nature (1993) 366(6450):72–5. doi: 10.1038/366072a0
4. Kachapati K, Adams D, Bednar K, Ridgway WM. The non-Obese Diabetic (NOD) Mouse as a Model of Human Type 1 Diabetes. Methods Mol Biol (2012) 933:3–16. doi: 10.1007/978-1-62703-068-7_1
5. Takahama Y, Ohigashi I, Baik S, Anderson G. Generation of Diversity in Thymic Epithelial Cells. Nat Rev Immunol (2017) 17(5):295–305. doi: 10.1038/nri.2017.12
6. sO’Reilly LA, Healey D, Simpson E, Chandler P, Lund T, Ritter MA, et al. Studies on the Thymus of non-Obese Diabetic (NOD) Mice: Effect of Transgene Expression. Immunology (1994) 82(2):275–86.
7. Parish NM, Chandler P, Quartey-Papafio R, Simpson E, Cooke A. The Effect of Bone Marrow and Thymus Chimerism Between non-Obese Diabetic (NOD) and NOD-E Transgenic Mice, on the Expression and Prevention of Diabetes. Eur J Immunol (1993) 23(10):2667–75. doi: 10.1002/eji.1830231042
8. Noorchashm H, Lieu YK, Noorchashm N, Rostami SY, Greeley SA, Schlachterman A, et al. I-Ag7-Mediated Antigen Presentation by B Lymphocytes is Critical in Overcoming a Checkpoint in T Cell Tolerance to Islet Beta Cells of Nonobese Diabetic Mice. J Immunol (1999) 163(2):743–50.
9. Serreze DV, Fleming SA, Chapman HD, Richard SD, Leiter EH, Tisch RM. B Lymphocytes are Critical Antigen-Presenting Cells for the Initiation of T Cell-Mediated Autoimmune Diabetes in Nonobese Diabetic Mice. J Immunol (1998) 161(8):3912–8.
10. Serreze DV, Gallichan WS, Snider DP, Croitoru K, Rosenthal KL, Leiter EH, et al. MHC Class I-Mediated Antigen Presentation and Induction of CD8(+) Cytotoxic T-Cell Responses in Autoimmune Diabetes-Prone NOD Mice. Diabetes (1996) 45(7):902–8. doi: 10.2337/diab.45.7.902
11. Brodie GM, Wallberg M, Santamaria P, Wong FS, Green EA. B-Cells Promote Intra-Islet CD8+ Cytotoxic T-Cell Survival to Enhance Type 1 Diabetes. Diabetes (2008) 57(4):909–17. doi: 10.2337/db07-1256
12. Parikka V, Nanto-Salonen K, Saarinen M, Simell T, Ilonen J, Hyoty H, et al. Early Seroconversion and Rapidly Increasing Autoantibody Concentrations Predict Prepubertal Manifestation of Type 1 Diabetes in Children at Genetic Risk. Diabetologia (2012) 55(7):1926–36. doi: 10.1007/s00125-012-2523-3
13. Leete P, Willcox A, Krogvold L, Dahl-Jorgensen K, Foulis AK, Richardson SJ, et al. Differential Insulitic Profiles Determine the Extent of Beta-Cell Destruction and the Age at Onset of Type 1 Diabetes. Diabetes (2016) 65(5):1362–9. doi: 10.2337/db15-1615
14. Pescovitz MD, Greenbaum CJ, Krause-Steinrauf H, Becker DJ, Gitelman SE, Goland R, et al. Rituximab, B-Lymphocyte Depletion, and Preservation of Beta-Cell Function. N Engl J Med (2009) 361(22):2143–52. doi: 10.1056/NEJMoa0904452
15. Pinto AI, Smith J, Kissack MR, Hogg KG, Green EA. Thymic B Cell-Mediated Attack of Thymic Stroma Precedes Type 1 Diabetes Development. Front Immunol (2018) 9:1281. doi: 10.3389/fimmu.2018.01281
16. Inaba M, Inaba K, Hosono M, Kumamoto T, Ishida T, Muramatsu S, et al. Distinct Mechanisms of Neonatal Tolerance Induced by Dendritic Cells and Thymic B Cells. J Exp Med (1991) 173(3):549–59. doi: 10.1084/jem.173.3.549
17. Isaacson PG, Norton AJ, Addis BJ. The Human Thymus Contains a Novel Population of B Lymphocytes. Lancet (1987) 2(8574):1488–91. doi: 10.1016/S0140-6736(87)92622-5
18. Christensson B, Biberfeld P, Matell G. B-Cell Compartment in the Thymus of Patients With Myasthenia Gravis and Control Subjects. Ann N Y Acad Sci (1988) 540:293–7. doi: 10.1111/j.1749-6632.1988.tb27079.x
19. Hidalgo Y, Nunez S, Fuenzalida MJ, Flores-Santibanez F, Saez PJ, Dorner J, et al. Thymic B Cells Promote Germinal Center-Like Structures and the Expansion of Follicular Helper T Cells in Lupus-Prone Mice. Front Immunol (2020) 11:696. doi: 10.3389/fimmu.2020.00696
20. Mackay IR, Masel M, Burnet FM. Thymic Abnormality in Systemic Lupus Erythematosus. Australas Ann Med (1964) 13:5–14. doi: 10.1111/imj.1964.13.1.5
21. Kirschner D, Pienaar E, Marino S, Linderman JJ. A Review of Computational and Mathematical Modeling Contributions to Our Understanding of Mycobacterium Tuberculosis Within-Host Infection and Treatment. Curr Opin Syst Biol (2017) 3:170–85. doi: 10.1016/j.coisb.2017.05.014
22. Segovia-Juarez JL, Ganguli S, Kirschner D. Identifying Control Mechanisms of Granuloma Formation During M. Tuberculosis Infection Using an Agent-Based Model. J Theor Biol (2004) 231(3):357–76. doi: 10.1016/j.jtbi.2004.06.031
23. Warsinske HC, DiFazio RM, Linderman JJ, Flynn JL, Kirschner DE. Identifying Mechanisms Driving Formation of Granuloma-Associated Fibrosis During Mycobacterium Tuberculosis Infection. J Theor Biol (2017) 429:1–17. doi: 10.1016/j.jtbi.2017.06.017
24. Tang J, Enderling H, Becker-Weimann S, Pham C, Polyzos A, Chen CY, et al. Phenotypic Transition Maps of 3D Breast Acini Obtained by Imaging-Guided Agent-Based Modeling. Integr Biol (Camb) (2011) 3(4):408–21. doi: 10.1039/c0ib00092b
25. Butler JA, Cosgrove J, Alden K, Timmis J, Coles MC. Model-Driven Experimentation: A New Approach to Understand Mechanisms of Tertiary Lymphoid Tissue Formation, Function, and Therapeutic Resolution. Front Immunol (2017) 7:658. doi: 10.3389/fimmu.2016.00658
26. Cosgrove J, Novkovic M, Albrecht S, Pikor NB, Zhou Z, Onder L, et al. B Cell Zone Reticular Cell Microenvironments Shape CXCL13 Gradient Formation. Nat Commun (2020) 11(1):3677. doi: 10.1038/s41467-020-17135-2
27. Walker DC, Hill G, Wood SM, Smallwood RH, Southgate J. Agent-Based Computational Modeling of Wounded Epithelial Cell Monolayers. IEEE Trans Nanobiosci (2004) 3(3):153–63. doi: 10.1109/TNB.2004.833680
28. Walker DC, Southgate J, Hill G, Holcombe M, Hose DR, Wood SM, et al. The Epitheliome: Agent-Based Modelling of the Social Behaviour of Cells. Biosystems (2004) 76(1-3):89–100. doi: 10.1016/j.biosystems.2004.05.025
29. Miyama-Inaba M, Kuma S, Inaba K, Ogata H, Iwai H, Yasumizu R, et al. Unusual Phenotype of B Cells in the Thymus of Normal Mice. J Exp Med (1988) 168(2):811–6. doi: 10.1084/jem.168.2.811
30. Nango K, Inaba M, Inaba K, Adachi Y, Than S, Ishida T, et al. Ontogeny of Thymic B Cells in Normal Mice. Cell Immunol (1991) 133(1):109–15. doi: 10.1016/0008-8749(91)90183-C
31. Bredenkamp N, Nowell CS, Blackburn CC. Regeneration of the Aged Thymus by a Single Transcription Factor. Development (2014) 141(8):1627–37. doi: 10.1242/dev.103614
32. Sato T, Ishikawa S, Akadegawa K, Ito T, Yurino H, Kitabatake M, et al. Aberrant B1 Cell Migration Into the Thymus Results in Activation of CD4 T Cells Through its Potent Antigen-Presenting Activity in the Development of Murine Lupus. Eur J Immunol (2004) 34(12):3346–58. doi: 10.1002/eji.200425373
33. Perera J, Huang H. The Development and Function of Thymic B Cells. Cell Mol Life Sci (2015) 72(14):2657–63. doi: 10.1007/s00018-015-1895-1
34. Akashi K, Richie LI, Miyamoto T, Carr WH, Weissman IL. B Lymphopoiesis in the Thymus. J Immunol (2000) 164(10):5221–6. doi: 10.4049/jimmunol.164.10.5221
35. Than S, Inaba M, Inaba K, Fukuba Y, Adachi Y, Ikehara S. Origin of Thymic and Peritoneal Ly-1 B Cells. Eur J Immunol (1992) 22(5):1299–303. doi: 10.1002/eji.1830220527
36. McCaughtry TM, Wilken MS, Hogquist KA. Thymic Emigration Revisited. J Exp Med (2007) 204(11):2513–20. doi: 10.1084/jem.20070601
37. Gay D, Saunders T, Camper S, Weigert M. Receptor Editing: An Approach by Autoreactive B Cells to Escape Tolerance. J Exp Med (1993) 177(4):999–1008. doi: 10.1084/jem.177.4.999
38. Yu W, Nagaoka H, Jankovic M, Misulovin Z, Suh H, Rolink A, et al. Continued RAG Expression in Late Stages of B Cell Development and No Apparent Re-Induction After Immunization. Nature (1999) 400(6745):682–7. doi: 10.1038/23287
39. Yu W, Nagaoka H, Misulovin Z, Meffre E, Suh H, Jankovic M, et al. RAG Expression in B Cells in Secondary Lymphoid Tissues. Cold Spring Harbor Symp Quant Biol (1999) 64:207–10. doi: 10.1101/sqb.1999.64.207
40. Benz C, Bleul CC. A Multipotent Precursor in the Thymus Maps to the Branching Point of the T Versus B Lineage Decision. J Exp Med (2005) 202(1):21–31. doi: 10.1084/jem.20050146
41. McKenna HJ, Morrissey PJ. Flt3 Ligand Plus IL-7 Supports the Expansion of Murine Thymic B Cell Progenitors That can Mature Intrathymically. J Immunol (1998) 160(10):4801–9.
42. Inlay MA, Bhattacharya D, Sahoo D, Serwold T, Seita J, Karsunky H, et al. Ly6d Marks the Earliest Stage of B-Cell Specification and Identifies the Branchpoint Between B-Cell and T-Cell Development. Genes Dev (2009) 23(20):2376–81. doi: 10.1101/gad.1836009
43. Mansson R, Zandi S, Welinder E, Tsapogas P, Sakaguchi N, Bryder D, et al. Single-Cell Analysis of the Common Lymphoid Progenitor Compartment Reveals Functional and Molecular Heterogeneity. Blood (2010) 115(13):2601–9. doi: 10.1182/blood-2009-08-236398
44. Ceredig R. The Ontogeny of B Cells in the Thymus of Normal, CD3 Epsilon Knockout (KO), RAG-2 KO and IL-7 Transgenic Mice. Int Immunol (2002) 14(1):87–99. doi: 10.1093/intimm/14.1.87
45. He Y, Pear WS. Notch Signalling in B Cells. Semin Cell Dev Biol (2003) 14(2):135–42. doi: 10.1016/s1084-9521(02)00182.
46. Feyerabend TB, Terszowski G, Tietz A, Blum C, Luche H, Gossler A, et al. Deletion of Notch1 Converts Pro-T Cells to Dendritic Cells and Promotes Thymic B Cells by Cell-Extrinsic and Cell-Intrinsic Mechanisms. Immunity (2009) 30(1):67–79. doi: 10.1016/j.immuni.2008.10.016
47. Xing C, Ma N, Xiao H, Wang X, Zheng M, Han G, et al. Critical Role for Thymic CD19+CD5+CD1dhiIL-10+ Regulatory B Cells in Immune Homeostasis. J Leukoc Biol (2015) 97(3):547–56. doi: 10.1189/jlb.3A0414-213RR
48. Carnrot C, Prokopec KE, Rasbo K, Karlsson MC, Kleinau S. Marginal Zone B Cells are Naturally Reactive to Collagen Type II and are Involved in the Initiation of the Immune Response in Collagen-Induced Arthritis. Cell Mol Immunol (2011) 8(4):296–304. doi: 10.1038/cmi.2011.2
49. Rolf J, Motta V, Duarte N, Lundholm M, Berntman E, Bergman ML, et al. The Enlarged Population of Marginal Zone/CD1d(high) B Lymphocytes in Nonobese Diabetic Mice Maps to Diabetes Susceptibility Region Idd11. J Immunol (2005) 174(8):4821–7. doi: 10.4049/jimmunol.174.8.4821
50. Wellmann U, Werner A, Winkler TH. Altered Selection Processes of B Lymphocytes in Autoimmune NZB/W Mice, Despite Intact Central Tolerance Against DNA. Eur J Immunol (2001) 31(9):2800–10. doi: 10.1002/1521-4141(200109)31:9<2800::AID-IMMU2800>3.0.CO;2-E
51. Daridon C, Pers JO, Devauchelle V, Martins-Carvalho C, Hutin P, Pennec YL, et al. Identification of Transitional Type II B Cells in the Salivary Glands of Patients With Sjogren’s Syndrome. Arthritis Rheum (2006) 54(7):2280–8. doi: 10.1002/art.21936
52. Segundo C, Rodriguez C, Garcia-Poley A, Aguilar M, Gavilan I, Bellas C, et al. Thyroid-Infiltrating B Lymphocytes in Graves’ Disease are Related to Marginal Zone and Memory B Cell Compartments. Thyroid (2001) 11(6):525–30. doi: 10.1089/105072501750302813
53. Palm AE, Kleinau S. Marginal Zone B Cells: From Housekeeping Function to Autoimmunity? J Autoimmun (2021) 119:102627. doi: 10.1016/j.jaut.2021.102627
54. Ferguson AR, Youd ME, Corley RB. Marginal Zone B Cells Transport and Deposit IgM-Containing Immune Complexes Onto Follicular Dendritic Cells. Int Immunol (2004) 16(10):1411–22. doi: 10.1093/intimm/dxh142
55. Hammad H, Vanderkerken M, Pouliot P, Deswarte K, Toussaint W, Vergote K, et al. Transitional B Cells Commit to Marginal Zone B Cell Fate by Taok3-Mediated Surface Expression of ADAM10. Nat Immunol (2017) 18(3):313–20. doi: 10.1038/ni.3657
56. Hampel F, Ehrenberg S, Hojer C, Draeseke A, Marschall-Schroter G, Kuhn R, et al. CD19-Independent Instruction of Murine Marginal Zone B-Cell Development by Constitutive Notch2 Signaling. Blood (2011) 118(24):6321–31. doi: 10.1182/blood-2010-12-325944
57. Palm AK, Friedrich HC, Mezger A, Salomonsson M, Myers LK, Kleinau S. Function and Regulation of Self-Reactive Marginal Zone B Cells in Autoimmune Arthritis. Cell Mol Immunol (2015) 12(4):493–504. doi: 10.1038/cmi.2015.37
58. Wortis HH, Teutsch M, Higer M, Zheng J, Parker DC. B-Cell Activation by Crosslinking of Surface IgM or Ligation of CD40 Involves Alternative Signal Pathways and Results in Different B-Cell Phenotypes. Proc Natl Acad Sci U S A (1995) 92(8):3348–52. doi: 10.1073/pnas.92.8.3348
59. Smith MJ, Packard TA, O’Neill SK, Dunand CJH, Huang M, Fitzgerald-Miller L, et al. Loss of Anergic B Cells in Prediabetic and New-Onset Type 1 Diabetic Patients. Diabetes (2015) 64(5):1703–12. doi: 10.2337/db13-1798
60. Elsner RA, Shlomchik MJ. Germinal Center and Extrafollicular B Cell Responses in Vaccination, Immunity, and Autoimmunity. Immunity (2020) 53(6):1136–50. doi: 10.1016/j.immuni.2020.11.006
61. Silva-Cayetano A, Linterman MA. Stromal Cell Control of Conventional and Ectopic Germinal Centre Reactions. Curr Opin Immunol (2020) 64:26–33. doi: 10.1016/j.coi.2020.03.007
62. Carrillo-Ballesteros FJ, Palafox-Sanchez CA, Franco-Topete RA, Munoz-Valle JF, Orozco-Barocio G, Martinez-Bonilla GE, et al. Expression of BAFF and BAFF Receptors in Primary Sjogren’s Syndrome Patients With Ectopic Germinal Center-Like Structures. Clin Exp Med (2020) 20(4):615–26. doi: 10.1007/s10238-020-00637-0
63. Zhang X, Liu S, Chang T, Xu J, Zhang C, Tian F, et al. Intrathymic Tfh/B Cells Interaction Leads to Ectopic GCs Formation and Anti-AChR Antibody Production: Central Role in Triggering MG Occurrence. Mol Neurobiol (2016) 53(1):120–31. doi: 10.1007/s12035-014-8985-1
64. Zuckerman NS, Howard WA, Bismuth J, Gibson K, Edelman H, Berrih-Aknin S, et al. Ectopic GC in the Thymus of Myasthenia Gravis Patients Show Characteristics of Normal GC. Eur J Immunol (2010) 40(4):1150–61. doi: 10.1002/eji.200939914
65. Kendall PL, Yu G, Woodward EJ, Thomas JW. Tertiary Lymphoid Structures in the Pancreas Promote Selection of B Lymphocytes in Autoimmune Diabetes. J Immunol (2007) 178(9):5643–51. doi: 10.4049/jimmunol.178.9.5643
66. Barone F, Nayar S, Campos J, Cloake T, Withers DR, Toellner KM, et al. IL-22 Regulates Lymphoid Chemokine Production and Assembly of Tertiary Lymphoid Organs. Proc Natl Acad Sci U S A (2015) 112(35):11024–9. doi: 10.1073/pnas.1503315112
67. Canete JD, Celis R, Yeremenko N, Sanmarti R, van Duivenvoorde L, Ramirez J, et al. Ectopic Lymphoid Neogenesis is Strongly Associated With Activation of the IL-23 Pathway in Rheumatoid Synovitis. Arthritis Res Ther (2015) 17:173. doi: 10.1186/s13075-015-0688-0
68. Domeier PP, Chodisetti SB, Soni C, Schell SL, Elias MJ, Wong EB, et al. IFN-Gamma Receptor and STAT1 Signaling in B Cells are Central to Spontaneous Germinal Center Formation and Autoimmunity. J Exp Med (2016) 213(5):715–32. doi: 10.1084/jem.20151722
69. Jackson SW, Jacobs HM, Arkatkar T, Dam EM, Scharping NE, Kolhatkar NS, et al. B Cell IFN-Gamma Receptor Signaling Promotes Autoimmune Germinal Centers via Cell-Intrinsic Induction of BCL-6. J Exp Med (2016) 213(5):733–50. doi: 10.1084/jem.20151724
70. Lee SK, Silva DG, Martin JL, Pratama A, Hu X, Chang PP, et al. Interferon-Gamma Excess Leads to Pathogenic Accumulation of Follicular Helper T Cells and Germinal Centers. Immunity (2012) 37(5):880–92. doi: 10.1016/j.immuni.2012.10.010
71. Leprince C, Cohen-Kaminsky S, Berrih-Aknin S, Vernet-Der Garabedian B, Treton D, Galanaud P, et al. Thymic B Cells From Myasthenia Gravis Patients are Activated B Cells. Phenotypic and Functional Analysis. J Immunol (1990) 145(7):2115–22.
72. Zotos D, Coquet JM, Zhang Y, Light A, D’Costa K, Kallies A, et al. IL-21 Regulates Germinal Center B Cell Differentiation and Proliferation Through a B Cell-Intrinsic Mechanism. J Exp Med (2010) 207(2):365–78. doi: 10.1084/jem.20091777
73. Ferreira RC, Simons HZ, Thompson WS, Cutler AJ, Dopico XC, Smyth DJ, et al. IL-21 Production by CD4(+) Effector T Cells and Frequency of Circulating Follicular Helper T Cells are Increased in Type 1 Diabetes Patients. Diabetologia (2015) 58(4):781–90. doi: 10.1007/s00125-015-3509-8
74. Sutherland AP, Van Belle T, Wurster AL, Suto A, Michaud M, Zhang D, et al. Interleukin-21 is Required for the Development of Type 1 Diabetes in NOD Mice. Diabetes (2009) 58(5):1144–55. doi: 10.2337/db08-0882
75. Viisanen T, Ihantola EL, Nanto-Salonen K, Hyoty H, Nurminen N, Selvenius J, et al. Circulating CXCR5+PD-1+ICOS+ Follicular T Helper Cells Are Increased Close to the Diagnosis of Type 1 Diabetes in Children With Multiple Autoantibodies. Diabetes (2017) 66(2):437–47. doi: 10.2337/db16-0714
76. He J, Tsai LM, Leong YA, Hu X, Ma CS, Chevalier N, et al. Circulating Precursor CCR7(lo)PD-1(Hi) CXCR5(+) CD4(+) T Cells Indicate Tfh Cell Activity and Promote Antibody Responses Upon Antigen Reexposure. Immunity (2013) 39(4):770–81. doi: 10.1016/j.immuni.2013.09.007
77. Morita R, Schmitt N, Bentebibel SE, Ranganathan R, Bourdery L, Zurawski G, et al. Human Blood CXCR5(+)CD4(+) T Cells are Counterparts of T Follicular Cells and Contain Specific Subsets That Differentially Support Antibody Secretion. Immunity (2011) 34(1):108–21. doi: 10.1016/j.immuni.2010.12.012
78. Rao DA. T Cells That Help B Cells in Chronically Inflamed Tissues. Front Immunol (2018) 9:1924. doi: 10.3389/fimmu.2018.01924
79. Hill DL, Pierson W, Bolland DJ, Mkindi C, Carr EJ, Wang J, et al. The Adjuvant GLA-SE Promotes Human Tfh Cell Expansion and Emergence of Public TCRbeta Clonotypes. J Exp Med (2019) 216(8):1857–73. doi: 10.1084/jem.20190301
80. Vella LA, Buggert M, Manne S, Herati RS, Sayin I, Kuri-Cervantes L, et al. T Follicular Helper Cells in Human Efferent Lymph Retain Lymphoid Characteristics. J Clin Invest (2019) 129(8):3185–200. doi: 10.1172/JCI125628
81. Barone F, Gardner DH, Nayar S, Steinthal N, Buckley CD, Luther SA. Stromal Fibroblasts in Tertiary Lymphoid Structures: A Novel Target in Chronic Inflammation. Front Immunol (2016) 7:477. doi: 10.3389/fimmu.2016.00477
82. Rangel-Moreno J, Carragher DM, de la Luz Garcia-Hernandez M, Hwang JY, Kusser K, Hartson L, et al. The Development of Inducible Bronchus-Associated Lymphoid Tissue Depends on IL-17. Nat Immunol (2011) 12(7):639–46. doi: 10.1038/ni.2053
83. Michaux H, Martens H, Jaidane H, Halouani A, Hober D, Geenen V. How Does Thymus Infection by Coxsackievirus Contribute to the Pathogenesis of Type 1 Diabetes? Front Immunol (2015) 6:338. doi: 10.3389/fimmu.2015.00338
84. Tugnet N, Rylance P, Roden D, Trela M, Nelson P. Human Endogenous Retroviruses (HERVs) and Autoimmune Rheumatic Disease: Is There a Link? Open Rheumatol J (2013) 7:13–21. doi: 10.2174/1874312901307010013
85. Domeier PP, Schell SL, Rahman ZS. Spontaneous Germinal Centers and Autoimmunity. Autoimmunity (2017) 50(1):4–18. doi: 10.1080/08916934.2017.1280671
86. Spencer J, Choy M, Hussell T, Papadaki L, Kington JP, Isaacson PG. Properties of Human Thymic B-Cells. Immunology (1992) 75(4):596–600.
87. Weerkamp F, de Haas EF, Naber BA, Comans-Bitter WM, Bogers AJ, van Dongen JJ, et al. Age-Related Changes in the Cellular Composition of the Thymus in Children. J Allergy Clin Immunol (2005) 115(4):834–40. doi: 10.1016/j.jaci.2004.10.031
88. Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant Autoantibody Production by Early Human B Cell Precursors. Science (2003) 301(5638):1374–7. doi: 10.1126/science.1086907
89. Rother MB, Schreurs MWJ, Kroek R, Bartol SJW, van Dongen JJM, van Zelm MC. The Human Thymus Is Enriched for Autoreactive B Cells. J Immunol (2016) 197(2):441–8. doi: 10.4049/jimmunol.1501992
90. Tonnelle C, D’Ercole C, Depraetere V, Metras D, Boubli L, Fougereau M. Human Thymic B Cells Largely Overexpress the VH4 Ig Gene Family. A Possible Role in the Control of Tolerance in Situ? Int Immunol (1997) 9(3):407–14. doi: 10.1093/intimm/9.3.407
91. Roche PA, Furuta K. The Ins and Outs of MHC Class II-Mediated Antigen Processing and Presentation. Nat Rev Immunol (2015) 15(4):203–16. doi: 10.1038/nri3818
92. Dunn-Walters DK, Howe CJ, Isaacson PG, Spencer J. Location and Sequence of Rearranged Immunoglobulin Genes in Human Thymus. Eur J Immunol (1995) 25(2):513–9. doi: 10.1002/eji.1830250231
93. Frommer F, Waisman A. B Cells Participate in Thymic Negative Selection of Murine Auto-Reactive CD4(+) T Cells. PloS One (2010) 5(10):e15372. doi: 10.1371/journal.pone.0015372
94. Yamano T, Nedjic J, Hinterberger M, Steinert M, Koser S, Pinto S, et al. Thymic B Cells Are Licensed to Present Self Antigens for Central T Cell Tolerance Induction. Immunity (2015) 42(6):1048–61. doi: 10.1016/j.immuni.2015.05.013
95. Yamano T, Steinert M, Klein L. Thymic B Cells and Central T Cell Tolerance. Front Immunol (2015) 6:376. doi: 10.3389/fimmu.2015.00376
96. Perera J, Zheng Z, Li S, Gudjonson H, Kalinina O, Benichou JIC, et al. Self-Antigen-Driven Thymic B Cell Class Switching Promotes T Cell Central Tolerance. Cell Rep (2016) 17(2):387–98. doi: 10.1016/j.celrep.2016.09.011
97. Walters S, Webster KE, Sutherland A, Gardam S, Groom J, Liuwantara D, et al. Increased CD4+Foxp3+ T Cells in BAFF-Transgenic Mice Suppress T Cell Effector Responses. J Immunol (2009) 182(2):793–801. doi: 10.4049/jimmunol.182.2.793
98. Walters SN, Webster KE, Daley S, Grey ST. A Role for Intrathymic B Cells in the Generation of Natural Regulatory T Cells. J Immunol (2014) 193(1):170–6. doi: 10.4049/jimmunol.1302519
99. Thangarajh M, Masterman T, Helgeland L, Rot U, Jonsson MV, Eide GE, et al. The Thymus is a Source of B-Cell-Survival Factors-APRIL and BAFF-In Myasthenia Gravis. J Neuroimmunol (2006) 178(1-2):161–6. doi: 10.1016/j.jneuroim.2006.05.023
100. Xing C, Zhu G, Xiao H, Fang Y, Liu X, Han G, et al. B Cells Regulate Thymic CD8(+)T Cell Differentiation in Lupus-Prone Mice. Oncotarget (2017) 8(52):89486–99. doi: 10.18632/oncotarget.19002
101. Leite MI, Jones M, Strobel P, Marx A, Gold R, Niks E, et al. Myasthenia Gravis Thymus: Complement Vulnerability of Epithelial and Myoid Cells, Complement Attack on Them, and Correlations With Autoantibody Status. Am J Pathol (2007) 171(3):893–905. doi: 10.2353/ajpath.2007.070240
102. Kitano H. Computational Systems Biology. Nature (2002) 420(6912):206–10. doi: 10.1038/nature01254
103. An G, Mi Q, Dutta-Moscato J, Vodovotz Y. Agent-Based Models in Translational Systems Biology. Wiley Interdiscip Rev Syst Biol Med (2009) 1(2):159–71. doi: 10.1002/wsbm.45
104. Bauer AL, Beauchemin CA, Perelson AS. Agent-Based Modeling of Host-Pathogen Systems: The Successes and Challenges. Inf Sci (N Y) (2009) 179(10):1379–89. doi: 10.1016/j.ins.2008.11.012
105. Forrest S, Beauchemin C. Computer Immunology. Immunol Rev (2007) 216:176–97. doi: 10.1111/j.1600-065X.2007.00499.x
106. Vodovotz Y, An G. Agent-Based Models of Inflammation in Translational Systems Biology: A Decade Later. Wiley Interdiscip Rev Syst Biol Med (2019) 11(6):e1460. doi: 10.1002/wsbm.1460
107. Perelson AS. Modelling Viral and Immune System Dynamics. Nat Rev Immunol (2002) 2(1):28–36. doi: 10.1038/nri700
108. Kleinstein SH, Seiden PE. Simulating the Immune System. Comput Sci Eng (2000) 2:69–77. doi: 10.1109/5992.852392
109. Seiden PE, Celada F. A Model for Simulating Cognate Recognition and Response in the Immune System. J Theor Biol (1992) 158(3):329–57. doi: 10.1016/S0022-5193(05)80737-4
110. Bonabeau E. Agent-Based Modelling: Methods and Techniques for Simulating Human Systems. ProcNatlAcadSci (2002) 99:7280–7. doi: 10.1073/pnas.082080899
111. Kam N, Cohen IR, Harel D. “The Immune System as a Reactive System: Modeling T Cell Activation With Statecharts”. In: Symposium on Human Centric Computing Languages and Environments (HCC). Stresa, Italy: IEEE (2001). p. 15–22.
112. Macal CM, North MJ. Tutorial on Agent-Based Modeling and Simulation. J Simulation (2010) 4:151–62. doi: 10.1057/jos.2010.3
113. Ghorbani A, Dechesne F, Dignum V, Jonker C. Enhancing ABM Into an Inevitable Tool for Policy Analysis. Policy Complex Syst (2014) 1(1). doi: 10.18278/jpcs.1.1.3
114. Bandini S, Manzoni S, Vizzari G. Agent Based Modeling and Simulation: An Informatics Perspective. J Artif Soc Soc Simulation (2009) 12(4):4. doi: 10.1007/978-1-0716-0368-0_12
115. Stepney S, Polack FAC, Alden K, Andrews PS, Bown JL, Droop A, et al. Engineering Simulations as Scientific Instruments: A Pattern Language. Berlin: Springer (2018).
116. McKay MD, Beckman RJ, Conover WJ. A Comparison of Three Methods for Selecting Values of Input Variables in the Analysis of Output From Computer Code. Technometrics (1979) 21(2):239–45. doi: 10.1080/00401706.1979.10489755
117. Alden K, Timmis J, Andrews PS, Veiga-Fernandes H, Coles MC. Pairing Experimentation and Computational Modeling to Understand the Role of Tissue Inducer Cells in the Development of Lymphoid Organs. Front Immunol (2012) 3:172. doi: 10.3389/fimmu.2012.00172
118. Marino S, Hogue IB, Ray CJ, Kirschner DE. A Methodology for Performing Global Uncertainty and Sensitivity Analysis in Systems Biology. J Theor Biol (2008) 254(1):178–96. doi: 10.1016/j.jtbi.2008.04.011
119. Renardy M, Hult C, Evans S, Linderman JJ, Kirschner DE. Global Sensitivity Analysis of Biological Multi-Scale Models. Curr Opin BioMed Eng (2019) 11:109–16. doi: 10.1016/j.cobme.2019.09.012
120. Alden K, Cosgrove J, Coles MC, Timmis J. Using Emulation to Engineer and Understand Simulations of Biological Systems. IEEE/ACM Trans Comput Biol Bioinform (2020) 17(1):302–15. doi: 10.1109/TCBB.2018.2843339
Keywords: type 1 diabetes, thymic B cells, autoimmunity, computational modelling, negative selection
Citation: Greaves RB, Chen D and Green EA (2021) Thymic B Cells as a New Player in the Type 1 Diabetes Response. Front. Immunol. 12:772017. doi: 10.3389/fimmu.2021.772017
Received: 07 September 2021; Accepted: 01 October 2021;
Published: 21 October 2021.
Edited by:
Ludger Klein, Ludwig Maximilian University of Munich, GermanyReviewed by:
Tomoyoshi Yamano, Kanazawa University, JapanCopyright © 2021 Greaves, Chen and Green. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: E. Allison Green, YWxsaXNvbi5ncmVlbkB5b3JrLmFjLnVr
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.