- 1State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-Products, Ningbo University, Ningbo, China
- 2Laboratory of Biochemistry and Molecular Biology, School of Marine Sciences, Ningbo University, Ningbo, China
- 3Key Laboratory of Applied Marine Biotechnology of Ministry of Education, Ningbo University, Ningbo, China
Akin to their mammalian counterparts, teleost fish possess a complex assortment of highly specialized immune cells that are capable of unleashing potent innate immune responses to eradicate or mitigate incoming pathogens, and also differentiate into memory lymphocytes to provide long-term protection. Investigations into specific roles and functions of fish immune cells depend on the precise separation of each cell type. Commonly used techniques, for example, density gradient centrifugation, rely on immune cells to have differing sizes or densities and thus fail to separate between similar cell types (e.g. T and B lymphocytes). Furthermore, a continuously growing database of teleost genomic information has revealed an inventory of cellular markers, indicating the possible presence of immune cell subsets in teleost fish. This further complicates the interpretation of results if subsets of immune cells are not properly separated. Consequently, monoclonal antibodies (mAbs) against specific cellular markers are required to precisely identify and separate novel subsets of immune cells in fish. In the field of fish immunology, mAbs are largely generated using the hybridoma technology, resulting in the development of mAbs against specific cellular markers in different fish species. Nevertheless, this technology suffers from being labour-intensive, time-consuming and most importantly, the inevitable loss of diversities of antibodies during the fusion of antibody-expressing B lymphocytes and myeloma cells. In light of this, the focus of this review is to discuss the potential applications of fluorescence-activated cell sorting and droplet-based microfluidics, two emerging technologies capable of screening and identifying antigen-specific B lymphocytes in a high-throughput manner, in promoting the development of valuable reagents for fish immunology studies. Our main goal is to encourage the incorporation of alternative technologies into the field of fish immunology to promote the production of specific antibodies in a high-throughput and cost-effective way, which could better allow for the precise separation of fish immune cells and also facilitate the identification of novel immune cell subsets in teleost fish.
Introduction
Teleost fishes represent a large and diverse group of vertebrates thriving in an extremely dynamic range of aquatic ecosystems that are populated with numerous parasitic, bacterial, and viral pathogens. Conceivably, a robust immune system has evolved to deal with these continuous assaults. A critical component in teleost immunity is specialized immune cells and it has been reported that immune cells isolated from fish are capable of preforming a range of effector responses that are indistinguishable from their mammalian counterparts (1–4). Investigations into functions and roles of teleost immune cells have revealed interesting discoveries, including the identification of a novel antibody isotype (i.e. IgT) that is unique to fish, and also B cells exhibiting phagocytic capabilities that later on are found to be evolutionarily conserved features (2, 5). These observations highlight the importance of using teleost fish as model organisms to study conserved and also divergent aspects of immunity across evolution.
The key first step in investigating cellular immunity is the precise separation of immune cells of interest. In the field of teleost immunology, this is commonly achieved via density gradient centrifugation. Generally, this technique involves layering a mixture of cells on gradient density medium (e.g. Ficoll®-Paque and Percoll) and after centrifugation, cells of higher buoyant density (i.e. erythrocytes and granulocytes) pellet to the bottom whereas mononuclear cells (i.e. macrophages/monocytes and lymphocytes) settle in an interphase layer (6–11). Afterwards, an added separation process can be applied to further purify cells of interest. For example, macrophages/monocytes can be further purified to eliminate lymphocytes based on different adherent properties (12–15). Of note, cell types of similar densities, such as T and B lymphocytes, are unlikely to be separated due to the nature of this technique. Furthermore, a growing number of reports demonstrated that subsets of immune cells with differential functions in immunity are also present in teleost fish (16–19). For example, similar to their mammalian counterparts, CD4 and CD8 molecules have been identified in a range of teleost species, indicating the presence of helper and cytotoxic T lymphocytes (16, 17). However, it is important to not assume that teleost immune cells will express the same surface markers as mammals do. Teleost immune cell subsets could also be determined by the expression of membrane-bound molecules that are unique to fish (e.g. novel immune type receptors) or evolutionally conserved (e.g. channel catfish leukocyte immune-type receptors) or secretions of cytokines (20–23). This further illustrates the complexity of cellular components in teleost immunity and a detailed characterization of immune cell subsets is required to further dissect specific roles of each cell type/subset, which can only be achieved when they are properly separated. Overall, density gradient centrifugation is a useful technique to separate cells exhibiting varying densities, but it fails to resolve cell types of similar physical characteristics.
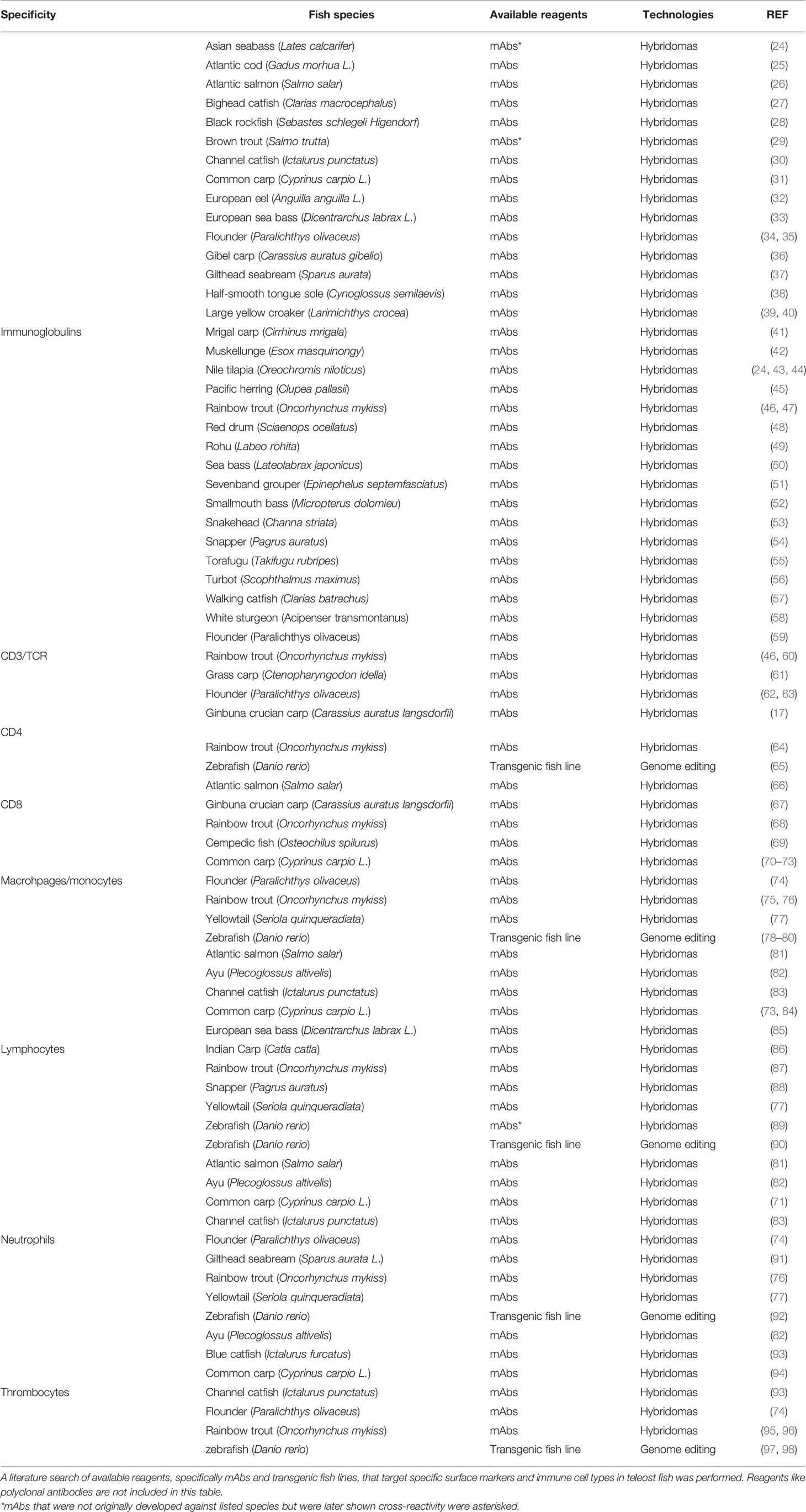
To characterize teleost immune cells with better resolution, alternative methods like antibody-based separation techniques are developed to isolate cells of interest. These techniques take advantage of monoclonal antibodies (mAbs) that are capable of recognizing epitopes with high specificity and thus, allowing to label cells of interest for precise separation. Indeed, mAbs targeting known immune cell markers in model organisms (e.g. mammals) have been successfully developed through various technologies. In teleost research, however, these valuable reagents are limited to a few species and largely generated through traditional hybridoma-based technology. A detailed summary of available reagents targeting well-characterized surface markers and immune cell types in teleost fish is presented in Table 1. The scope of this article is to review currently available technologies for the production of mAbs. Specifically, the hybridoma technology, fluorescence-activation cell sorting (FACS) and droplet-based microfluidics technologies with an emphasis on the latter two. Other methods, like phage display technology is not included in this review due to inherently different antibody-producing mechanisms, and readers interested in this specific technology are referred to excellent reviews elsewhere (99, 100). Our main goal is to provide alternative options for the development of mAbs in this field, which will be the basis of identifying and separating immune cell types and characterizing respective roles to further our understanding of teleost cellular immunity.
Hybridoma Technology
Antibodies, also known as immunoglobulins, are glycoproteins exclusively produced by B cells and come in two forms. Specifically, membrane-bound antibodies (e.g. B cell receptors) that normally expressed on the surface of naïve B cells to recognize foreign antigens, and soluble antibodies, for example, immunoglobulin G (IgG) that secreted by mature B cells after undergoing somatic hypermutations and class switching. Each species of antibodies specifically recognize epitopes of the same structure and this specificity of antibodies makes them valuable tools in therapeutics, diagnosis and research (101, 102). Currently, most of available antibodies on the market is IgG isotype since it exhibits the highest binding affinity to cognate antigens and thus, antibodies mentioned below are all referring to IgG isotype unless stated otherwise. It has been a daunting challenge to culture mature B lymphocytes in vitro for the continuous production of antibodies. Consequently, numerous resources and efforts have been invested to establish and optimize platforms for screening and culturing B lymphocytes secreting antibodies with desired specificity and affinity. Hybridoma technology represents the first platform for the continuous production of mAbs with known specificity and affinity since the invention in 1975 (103). This technology involves the fusion of splenic B cells isolated from immunized animals with an immortal B cell line (i.e. myeloma cells). These hybrid cells, called hybridomas, are immortal as myeloma cells while still maintain the ability of immunized splenic B cells to produce antibodies of interest. These hybridomas are further cloned and secreted mAbs are characterized in binding assays (e.g. ELISA) to select for hybridoma cell lines producing desired mAbs. Since each hybridoma cell line normally secrets a single species of antibodies, selected hybridoma cell lines could then serve as a continuous source for the production of mAbs with known specificity and affinity.
Currently, hybridoma technology remains the most popular method for generating mAbs and in the field of teleost immunology, majority of reported mAbs targeting surface markers and immune cell lineages were generated using this technology (Table 1). That being said, it is important to note that there are several inherent limitations pertaining to this technology. As mentioned above, generation of hybridomas requires the fusion of antigen-specific B cells and myeloma cells, which is commonly achieved using chemical reagents, like polyethylene glycol. This chemical reagent dehydrates lipid head groups on the cell membrane, resulting in an asymmetry of the membrane bilayer that favours the fusion of two adjacent cells. However, this fusion process is non-selective that often leads to the fusion of the same kind of cells and thus, result in the loss of antigen-specific B cells. Although variants of hybridoma technology have been developed to improve fusion efficiency, for example the B cell targeting method, repertoire of mAbs obtained using this technology is still biased since only B cells survive the fusion process could be selected (104). This largely reduces antibody diversity and consequently, selected mAbs are less likely the one with the optimal specificity and affinity. Furthermore, mAbs secreted by hybridomas are not always monospecific as assumed and gene expression of additional heavy and/or light chains was found within individual hybridomas (105). Collectively, these inherent limitations prevent the application of hybridoma technology in certain settings and with the advent of novel technologies, new platforms have emerged as alternative options for the production of mAbs, which are discussed below.
FACS and Droplet-BASED Microfluidics as Alternative Technologies for the Rapid Production of mAbs
A common theme in any mAbs discovery campaigns is to identify and isolate single B cells expressing and secreting mAbs with desired specificity and affinity. This posts a daunting task as the pool of antigen-specific B cells is highly heterogenous that each B cells normally secretes a single species of antibodies with unique properties (106). Although it is practically difficult, if not impossible, to screen all available B cells, various high-throughput technologies have been developed that allow a greater number of B cells to be screened, thus increasing the chance of finding optimal mAbs. FACS and droplet-based microfluidics, represent two high-throughput technologies for the in vitro screen of B cells of interest (Figure 1). Unlike traditional hybridoma technology that requires the immortalization of antibody-expressing B cells before any type of analysis, these two technologies offer unique opportunities to screen a repertoire of primary B cells that can be individually sorted in a high-throughput fashion. Sorted B cells are further analyzed, normally to obtain sequences of immunoglobulins heavy and light chain variable regions (VH and VL) since the binding specificity and affinity of immunoglobulins are largely determined by these two regions (106). These obtained sequences could then be subcloned and expressed for the production of recombinant mAbs. Using these two technologies coupled with single cell expression cloning, numerous success in the rapid identification and generation of rare mAbs species against a range of infectious agents has been reported in mammalian research (107–109). To date, very few teleost studies exploit these technologies for the generation of mAbs and therefore in this section, we will review these two technologies and also their potential applications in generating specific mAbs for teleost immunology studies.
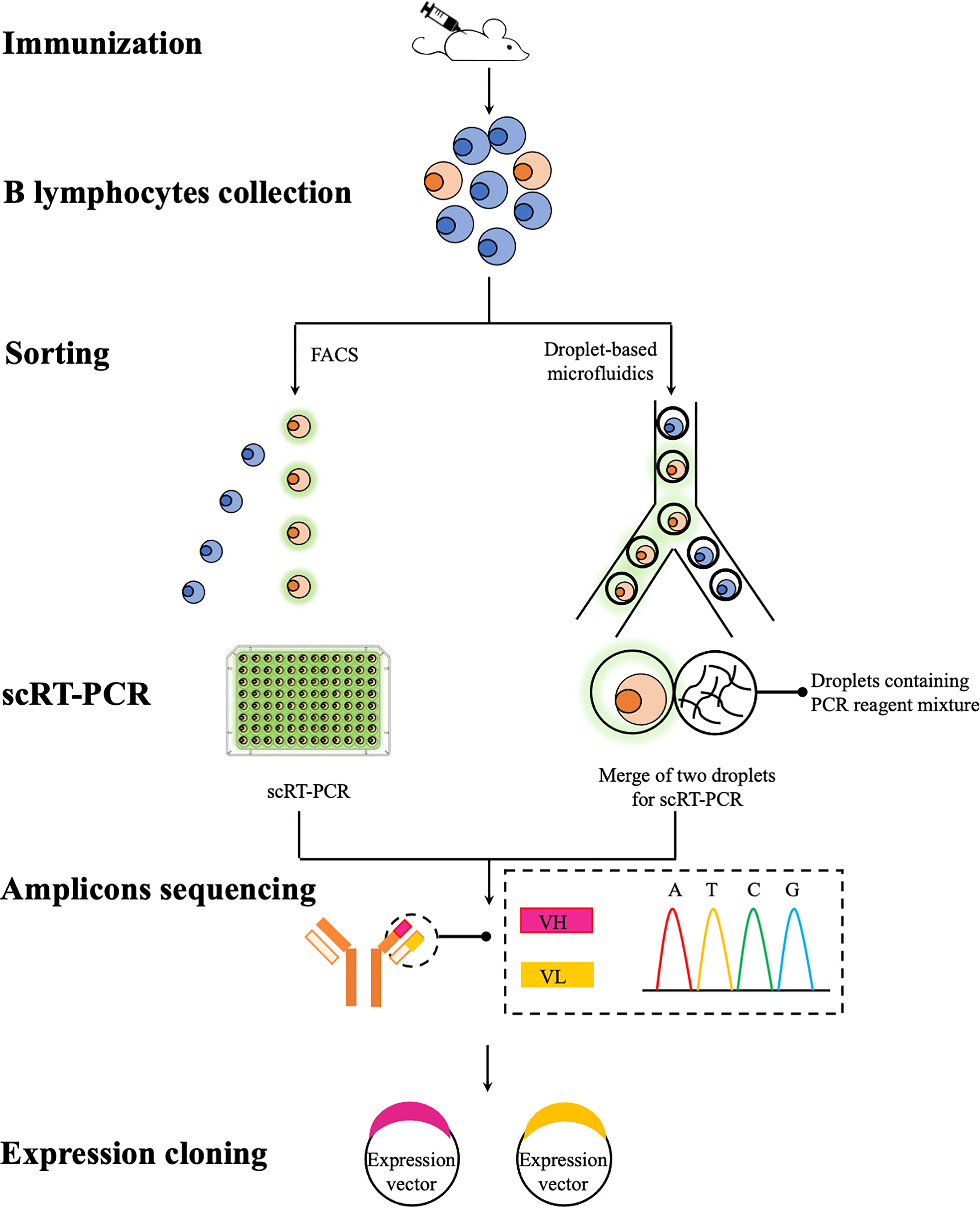
Figure 1 A schematic overview of recombinant mAbs production using FACS and droplet-based microfluidics. B lymphocytes (e.g. splenocytes and plasmablasts) are collected from immunized mice and then individually sorted via FACS or droplet-based microfluidics for antigen-specific lymphocytes. Paired VH and VL sequences are then amplified using scRT-PCR for subsequent sequencing (e.g. traditional Sanger sequencing and next-generation sequencing if bar-coded primers used during the scRT-PCR step) and subcloning. Expression vectors containing VH and VL sequences can be co-transfected into a range of expression systems for the production of recombinant mAbs.
FACS
Modern cell sorter (a type of flow cytometer capable of analyzing and also sorting cells of interest) is a multi-parameter instrument that allows the simultaneous measurement of optical and fluorescence characteristics of each single cell. Parameters, such as size, granularity and fluorescence features derived from fluorescent dyes bound to cells, are commonly used to differentiate and sort cells of interest. To date, a sophisticated cell sorter is capable of sorting up to thousands of cells per second while maintain reasonable viability. This makes it a valuable tool to efficiently discriminate B cells of interest from a large pool of highly heterogenous cells for further analysis at a single cell level.
Generally speaking, there are two strategies identifying B cells of interest using FACS for subsequent recombinant mAb production. One is antigen baiting that involves the use of fluorescently labelled antigens as a bait to sort antigen-specific B cells. This strategy is amenable to subsets of B cells expressing surface-bound antibodies (e.g. memory B cells) and often obtained recombinant mAbs exhibit desired specificity (110, 111). An alternative strategy relies on the fluorescent staining of defined surface markers, allowing to identify and isolate a broader spectrum of B cell subsets. This is particularly useful for identifying short lived plasmablasts that represent a transient population of antigen-specific B cells, normally observed 5~8 days after immunization (112–114). Of note, plasmablasts are unlikely to be identified using antigen baiting due to the low expression levels of surface-bound antibodies. Overall, these two FACS-based strategies allow an efficient and selective isolation of individual antigen-specific B cells for further analysis.
After FACS of antigen-specific B cells of interest, the next step is to obtain sequence information of paired VH and VL genes of each cell. This is often achieved through variants of polymerase chain reactions (PCR) coupled with different sequencing platforms. Single cell reverse transcription PCR (scRT-PCR) represents a major advance in single cell technology and allows to amplify genes of interest from a single cell (110). Sorted antigen-specific B cells are lysed in microtiter plates and after reverse transcription, cDNA templates of single cells are collected for amplification of VH and VL genes followed by Sanger sequencing. This combination of technologies was first employed to study autoantibody production during B cell development (115). Since then it has been adapted to antibody discovery campaigns and has led to the successful identification of neutralizing mAbs against a range of viral pathogens, including HIV and influenza virus (107, 108). However, this system is inherently low throughput since each sorted cell has to be processed and sequenced individually and practically, a few hundred VH and VL pairs at most could be identified using this system (116–118). This number is dwarfed by the enormous size of antibody repertoire that potentially, >1015 possible antibodies recognizing unique binding sites could be generated in mouse and human (119). More recently, a smart design of two-dimensional primer matrix largely improves the throughput of scRT-PCR; amplicons (e.g. paired VH and VL genes) from the same cell are bar-coded and thus, all amplicons could be pooled and analyzed using next generation sequencing (NGS) technology. This increased the number of analyzed B cells up to ~50,000 per experiment (120). Alternatively, the idea of linking VH and VL segments from single antigen-specific B cells was revisited and has been achieved via emulsion over-extension linkage RT-PCR. In this method, antigen-specific B cells are deposited by gravity into microwell arrays, in which individual cells are lysed and mRNAs are captured by poly(dT) beads in microwells. mRNAs-poly(dT) beads complexes are further washed and emulsified with primers, reverse transcriptase and polymerase to perform reverse transcription followed by linkage RT-PCR and Illumina sequencing (121). This method allows to screen >50,000 antigen-specific B cells per experiment with the potential to further scale up.
After obtaining sequencing information, paired VH and VL sequences originating from single antigen-specific B cells are then could be subcloned and expressed to produce recombinant mAbs. Numerous expression systems using eukaryotic and prokaryotic hosts have been developed (122). Each system is balanced by advantages and drawbacks and the choice of particular systems largely relies on applications of recombinant mAbs, which is detailed reviewed elsewhere (106). Overall, This FACS-based platform coupled with single cell sequencing and expression cloning has facilitated an efficient identification of antigen-specific B cells that leads to a rapid generation of recombinant mAbs. That being said, selecting paired VH and VL candidates for subsequent expression and validation of recombinant mAbs still presents a tedious task; furthermore, this platform primarily screens for antigen-specific B cells based on antibody binding specificity while other information regarding to antibodies, such as binding affinity and functional characteristics (e.g. neutralizing capacity), is not available until recombinant mAbs are generated. More recently, droplet-based microfluidics system has emerged as a cutting-edge platform and gradually established as a valuable tool for a range of applications, including single cell analysis with improved resolution, and will be discussed below.
Droplet-Based Microfluidics
Microfluidics is a technology that manipulates the flow of fluids through microchannels. At microscales, fluids behave very differently and give rise to unique features that are useful for a range of applications (123, 124). One branch of microfluidics is droplet-based microfluidics, which involves precise controls of two immiscible liquids (e.g. water and oil) through microchannels and leads to the formation of highly monodisperse aqueous droplets flowing in the carrier oil (Figure 2). These droplets, normally ranging from pico- to nanoliters, are functionally equivalent to individual wells in microtiter plates but with significantly reduced size, and biological entities (e.g. cells) encapsulated in each droplet are chemically and physically separated. Once formed, droplets could be further manipulated in a variety of ways (Figure 2). For example, addition of new reagents through droplets merge (125); static on/off-chip incubation of droplets (126, 127); detection of fluorescent signals in droplets and selective sorting of droplets of interest using electric field (128). The small volume of droplets combined with various ways of manipulation offers unique opportunities to study single cell biology in an unprecedent way. For example, secreted proteins are co-encapsulated with single cells inside droplets and quickly reach detectable concentrations due to the small droplet volume, resulting in a rapid detection of molecules/cells of interest. This is particularly useful in isolating plasmablasts (a subset of antibody-secreting B cells with low expression level of surface-bound antibodies) and overcomes limitations of aforementioned antigen baiting strategy. In addition, encapsulated cells could be lysed and intracellular molecules assayed within droplets; one example is the reverse transcription and amplification of genetic materials. This enables a more comprehensive analysis of single cells at biochemical and genetical levels. In antibody discovery campaigns, these features offer a unique opportunity to link genotype (e.g. sequencing information of VH and VL regions) with phenotype (e.g. properties of antibodies, such as specificity and affinity) in a one-stop manner. Indeed, various droplet-based microfluidics platforms have been developed and allows to characterize B cells of interest with improved resolution.
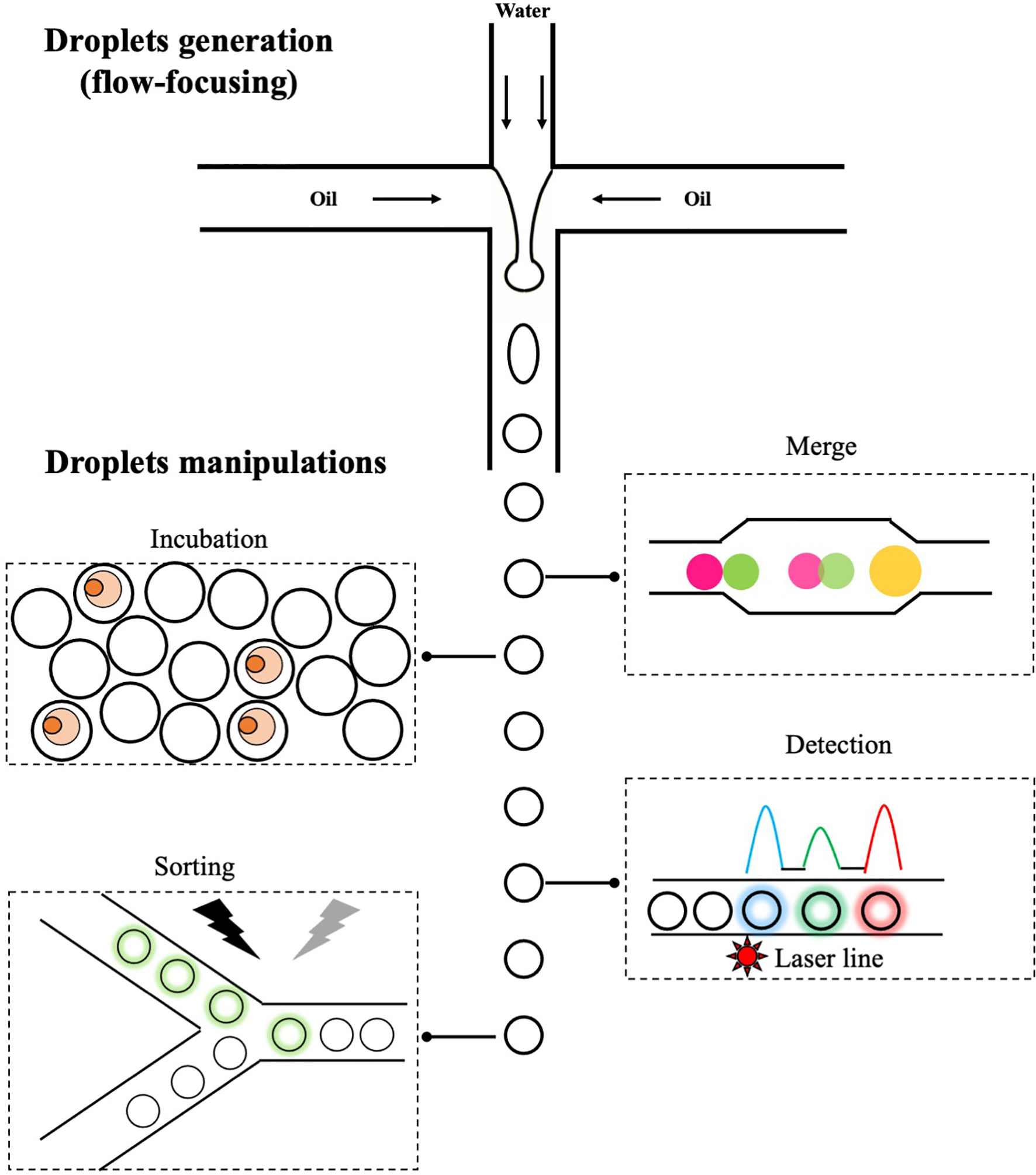
Figure 2 The generation and manipulations of droplets. One of popular methods to generate droplets in microfluidics is flow-focusing, in which the injected aqueous stream (water phase) is sheared by perpendicular oil streams pumped from two side channels. Once fluids meet, water in oil droplets are formed and those droplets can be further manipulated: merge of two droplets with different contents, on/off-chip incubation of droplets to allow reactions to occur, detection of fluorescent signals and sorting of droplets of interest via electric field.
A prototype of droplet-based microfluidics platform that enables a rapid detection of secreted antibodies was developed in 2013 and demonstrated as a powerful tool to sort antibody-secreting cells in a high-throughput way (129). Briefly, single cells are encapsulated with a capture bead (i.e. beads are opsonized with anti-IgG antibodies) and fluorescent probes (i.e. detection antibodies specifically targeting antigen-binding fragment (Fab) regions of IgGs). This is essentially a sandwich enzyme linked immunosorbent assay (ELISA) detecting secreted proteins from single cells and after 15min incubation, secreted antibodies are captured on the bead, together with localized fluorescent probes, and result in a clearly distinguishable increase in fluorescent intensity on beads and thus, droplets containing antibody-secreting cells can be sorted (Figure 3A). This represents a major advance in identifying antibody-secreting cells within a short time frame and could be potentially incorporated with other technologies to retrieve the genotype. However, this prototype fails to obtain useful information regarding to secreted antibodies, such as binding specificity and affinity, and therefore limit its applications in screening antigen-specific B cells.
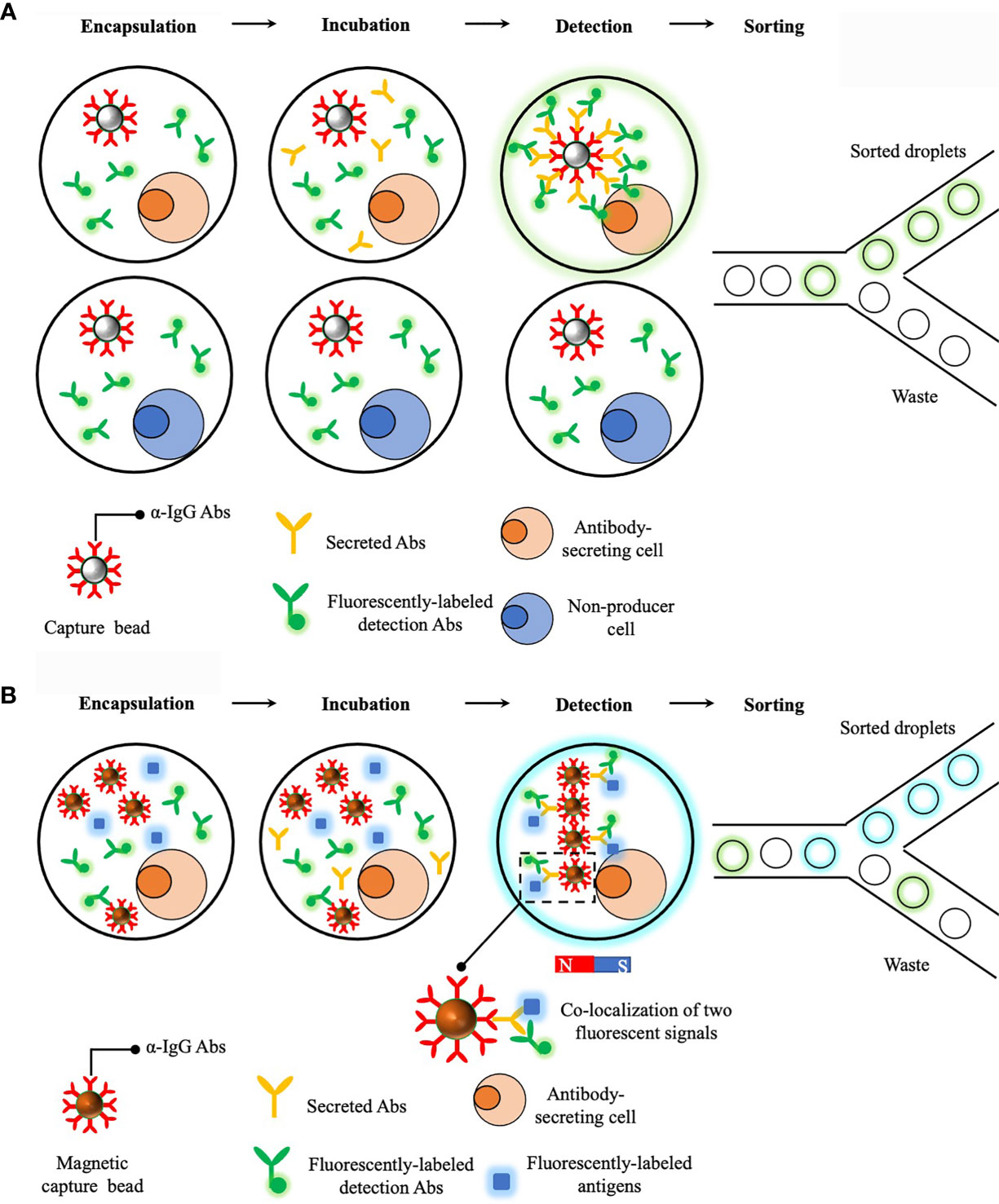
Figure 3 Droplet-based microfluidics platforms for single cell antibodies secretion measurements. (A) Sorting of antibody-secreting cells. Secreted antibodies were captured on the bead together with fluorescently-labeled detection antibodies. This leads to a localization of fluorescent signals on the bead and fluorescent droplets can then be sorted. (B) Dropmap platform for studying antibody secretion and affinity to cognate antigens. Compared to the previous platform, a major improvement of Dropmap is the encapsulation of magnetic beads and fluorescently-labeled antigens within each droplets. After antibody secretion, a magnetic field is applied to induce the formation of elongated and easily observable bead aggregates line, termed beadline. This helps amplify fluorescent signals and therefore, increase the sensitivity. Normally there are three scenarios after incubation, the first one is no fluorescent signal is observed and this indicates the cell is not able to secret antibodies; the second scenario is that only fluorescent signals from detection antibodies is localized on magnetic beads and this indicates the cell is able to secret antibodies but with poor specificity and affinity to cognate antigens; lastly, co-localization of two fluorescent signals is observed and this indicates cell is able to secret antibodies with desired specificity and affinity to cognate antigens.
An updated version of aforementioned prototype, termed DropMap, was further developed with features of determining antibody affinity to specific antigens (130). A major improvement of this updated platform is the co-encapsulation of single cells, antigens and detection antibodies that are differentially labeled, together with multiple magnetic capture beads (Figure 3B). Similar to the previous prototype, secreted antibodies are captured on beads, together with detection antibodies and potentially, antigens. Then a magnetic field is applied to induce the formation of an elongated and easily observable bead aggregates line, termed beadline, within each droplet. Consequently, binding specificity and affinity of secreted antibodies are determined by analysis of co-localization of fluorescent signals on the beadline and also the measurement of fluorescent intensity of antigens localized to the beadline, respectively. This DropMap platform allows a simultaneous measurement of antibody secretion rate, specificity and affinity, and this likely potentiates antibody screening process. Most recently, a novel platform, termed CelliGO, that integrates DropMap with NGS technology was established (131). In this platform, droplets containing single cells of interest (i.e. cells secreting antibodies with desired specificity and affinity) are sorted and merged with droplets containing PCR reagents. Paired VH and VL regions derived from single cells are amplified using bar-coded primers within droplets before NGS analysis. This platform seamlessly integrates multiple steps, ranging from sorting single cells, characterizing properties of secreted antibodies and obtaining sequencing information that are normally performed separately, into one device. Combined with NGS, this CelliGO platform represents a great advance in coupling phenotype with genotype of antigen-secreting cells and exhibits great potentials in discovering rare antigen-specific antibodies against difficult targets.
Although successful development of desired mAbs has been seen using droplet-based microfluidics, its applications have been largely limited to translational research so far using mammalian models due to the cost of this platform (e.g. expensive droplet-based microfluidics systems combined with the cost on regular maintenance and costly consumables). One example is to identify neutralizing mAbs against infectious agents, which have great therapeutic potentials (132, 133). Currently, it is economically impractical to apply this technology to generate mAbs reagents that are sorely for basic immunology research in non-model species. However, this still represents an alternative option and can be taken into account if specific mAbs reagents against difficulty targets are urgently required to address interesting hypotheses.
Potential Applications of FACS and Droplet-based Microfluidics Technologies for Teleost Immunology Research
Comparative immunology studies using non-model organisms like teleost fish have been expanding our understanding of immune systems and uncover interesting discoveries that are unique to fish or mechanistically conserved across evolution (2, 5). Most of these findings in teleost immunology studies are partially attributing to the availability of specific mAbs that allow the identification and isolation of different immune cell types. One example is the discovery of phagocytic B cells in rainbow trout, which would be impossible if no mAbs against rainbow trout IgM was available (2). In mammalian research, panels of mAbs targeting most of known immune-related molecules (e.g. surface markers) are commercially available. This enables precise isolations of different immune cell types, which is the key first step in further understanding specific roles of each cell type under different contexts and also how immune system operates in general. In comparison, these valuable reagents are mostly produced in-house and only limited to a few teleost species. Although teleost fish possess a variety of immune cells that are actively involved in innate and adaptive immunity, detailed functional characterizations of different immune cell types in ongoing immune responses is largely unavailable due to the lack of mAbs of high quality. To date, most of available mAbs in teleost immunology studies were generated using the hybridoma technology that requires lengthy immunization processes and labour-intensive cell fusion, cloning and screening procedures, which to some extent, discouraging investigators from generating these reagents. Currently, the advent in FACS and microfluidics technologies provide alternative options for a rapid identification of antigen-specific cells and thus, lead to the generation of mAbs in a more efficient way.
One of challenges associated with teleost fish for immunology studies is the presence of multiple copies of immune-related genes due to a teleost-specific whole genome duplication (WGD) events followed by a lineage-specific WGDs in several fish groups (e.g. salmonids and cyprinids) (134). This leads to an extensive expansion of genes encoding for immune receptors, cytokines and other components involved in immunity in teleost fish (135–144). For example, several isoforms of CD4 molecules differing slightly in extracellular domain structure have been identified in a range of fish species as opposed to only one copy of CD4 gene in human (145–148). This adds an additional layer of complexity in terms of reagents development, which requires mAbs with extreme specificity to differentiate isoforms that could potentially be identifiers of novel immune cell subsets. In this regard, FACS combined with scRT-PCR can potentially serve as a high-throughput platform to sort antigen-specific B cells via antigen-baiting for subsequent mAbs production. In addition to antigens of interest, antigens derived from other duplicated genes can also be differentially labeled with fluorochromes and serve as controls to exclude B cells exhibiting cross-reactivity during the sorting process, which will largely increase the chance of obtaining individual B cells with high specificity for subsequent scRT-PCR and expression cloning. In comparison to traditional hybridoma technology, this platform is more selective and avoids unnecessary waste (e.g. reagents and time) on non-producer/specific hybridomas and thus, improve the efficiency of developing mAb targeting closely-related isoforms in teleost fish. Furthermore, cytokine genes are also expanded in teleost fish and in the context of immunology, multiple copies of pro-/anti-inflammatory cytokine genes are present and responsible for shaping ongoing immune responses (149, 150). To date, teleost fish cytokines are mostly examined at transcript level and data regarding expression of cytokine molecules under resting state/upon stimulations is scarce due to the lack of mAb reagents (151). Besides, expression levels of cytokine molecules are relatively low under physiological conditions (151). These scenarios combined make it fairly difficult to generate mAbs reagents that are satisfying the requirement (mAbs exhibiting extremely high specificity and affinity) to allow sensitive and accurate detections of teleost cytokines. In this regard, droplet-based microfluidics combined with NGS is a better alternative for identifying antigen-specific B cells for subsequent sequencing and expression cloning. Properties of secreted antibodies derived from individual B cells can be parallelly analysed within droplets to retrieve optimal antigen-specific B cells. This platform provides better resolutions to allow a stringent selection of B cells based on both specificity and affinity of secreted antibodies and will largely reduce efforts and time spent in screening and validating generated mAbs afterwards and thus, increase the likelihood of obtaining desired mAbs against these difficulty targets.
Overall, the choice of particular technologies for mAb development is dependent on multiple factors, such as the applications/requirements of mAbs and availability of specialized equipment. There is no doubt that integration of novel technologies into our field will aid in the development of mAbs in a more efficient way, which is the basis of interesting discoveries in teleost immunology studies.
Conclusions
Teleost fishes represent the first species with an adaptive immunity and have been valuable model species in comparative immunology for understanding the evolution of immune systems. Knowledge obtained from teleost immunology studies have led to discoveries that were previously not aware of through studies of mammalian models (e.g. human and mouse). Despite their critical places in the evolutionary tree, teleost immunology studies are still at an early stage and in-depth characterizations of teleost immune cell types and their cytokine secretion profiles remain to be established primarily due to the lack of mAbs reagents of high quality, which largely prevents a further understanding of how immune systems operate in fish. Since the development of mAbs in this field largely relies on hybridoma technology, incorporation of available technologies, such as FACS and droplet-based microfluidics, into this process should be taken into account as this would largely increase the efficiency of mAbs production and eventually, lead to investigations on a broader spectrum of teleost species which may reveal novel immune cell types/subsets with distinct functions.
Author Contributions
CF, LN, and JC wrote and edited the manuscript. JZ provided the data and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by following grants: a Natural Science Foundation of Zhejiang Province (LZ18C190001); a Program for the Natural Science Foundation of China (31972821); a Natural Science Foundation of Ningbo (202003N4011; 20211JCGY020201) and a research fund of Ningbo University (422109623).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Dezfuli BS, Giari L. Mast Cells in the Gills and Intestines of Naturally Infected Fish: Evidence of Migration and Degranulation. J Fish Dis (2008) 31:845–52. doi: 10.1111/j.1365-2761.2008.00961.x
2. Li J, Barreda DR, Zhang Y-A, Boshra H, Gelman AE, LaPatra S, et al. B Lymphocytes From Early Vertebrates Have Potent Phagocytic and Microbicidal Abilities. Nat Immunol (2006) 7:1116–24. doi: 10.1038/ni1389
3. Shen L, Stuge TB, Bengtén E, Wilson M, Chinchar VG, Naftel JP, et al. Identification and Characterization of Clonal NK-Like Cells From Channel Catfish (Ictalurus Punctatus). Dev Comp Immunol (2004) 28:139–52. doi: 10.1016/S0145-305X(03)00119-8
4. Stuge TB, Wilson MR, Zhou H, Barker KS, Bengtén E, Chinchar G, et al. Development and Analysis of Various Clonal Alloantigen- Dependent Cytotoxic Cell Lines From Channel Catfish. J Immunol (2000) 164:2971–7. doi: 10.4049/jimmunol.164.6.2971
5. Zhang Y-A, Salinas I, Li J, Parra D, Bjork S, Xu Z, et al. IgT, a Primitive Immunoglobulin Class Specialized in Mucosal Immunity. Nat Immunol (2010) 11:827–35. doi: 10.1038/ni.1913
6. Braun-Nesje R, Bertheussen K, Kaplan G, Seljelid R. Salmonid Macrophages: Separation, In Vitro Culture and Characterization. J Fish Dis (1981) 4:141–51. doi: 10.1111/j.1365-2761.1981.tb01118.x
7. Rastgar S, Movahedinia A, Salamat N, Salati A, Zabihi E. Optimization of Macrophage Isolation From the Persian Sturgeon and the Caspian Kutum Fish: A Comparative Study. Cytotechnology (2018) 70:1643–54. doi: 10.1007/s10616-018-0256-y
8. Zhao M, Chi H, Sun L. Neutrophil Extracellular Traps of Cynoglossus Semilaevis: Production Characteristics and Antibacterial Effect. Front Immunol (2017) 8:290. doi: 10.3389/fimmu.2017.00290
9. Havixbeck JJ, Rieger AM, Wong ME, Hodgkinson JW, Barreda DR. Neutrophil Contributions to the Induction and Regulation of the Acute Inflammatory Response in Teleost Fish. J Leukoc Biol (2016) 99:241–52. doi: 10.1189/jlb.3HI0215-064R
10. Katzenback BA, Belosevic M. Isolation and Functional Characterization of Neutrophil-Like Cells, From Goldfish (Carassius Auratus L.) Kidney. Dev Comp Immunol (2009) 33:601–11. doi: 10.1016/j.dci.2008.10.011
11. Sørensen KK, Sveinbjørnsson B, Dalmo RA, Smedsrød B, Bertheussen K. Isolation, Cultivation and Characterization of Head Kidney Macrophages From Atlantic Cod, Gadus Morhua L. J Fish Dis (1997) 20:93–107. doi: 10.1046/j.1365-2761.1997.d01-112.x
12. Li Q, Ai Q, Mai K, Xu W, Zheng Y. A Comparative Study: In Vitro Effects of EPA and DHA on Immune Functions of Head-Kidney Macrophages Isolated From Large Yellow Croaker (Larmichthys Crocea). Fish Shellfish Immunol (2013) 35:933–40. doi: 10.1016/j.fsi.2013.07.004
13. He Y-Q, Chen J, Lu X-J, Shi Y-H. Characterization of P2X7R and Its Function in Themacrophages of Ayu, Plecoglossus Altivelis. PloS One (2013) 8:e57505. doi: 10.1371/journal.pone.0057505
14. Meng Z, Shao J, Xiang L. CpG Oligodeoxynucleotides Activate Grass Carp (Ctenopharyngodon Idellus) Macrophages. Dev Comp Immunol (2003) 27:313–21. doi: 10.1016/S0145-305X(02)00104-0
15. Yang D, Yang H, Cao Y, Jiang M, Zheng J, Peng B. Succinate Promotes Phagocytosis of Monocytes/Macrophages in Teleost Fish. Front Mol Biosci (2021) 8:644957. doi: 10.3389/fmolb.2021.644957
16. Nakanishi T, Toda H, Shibasaki Y, Somamoto T. Cytotoxic T Cells in Teleost Fish. Dev Comp Immunol (2011) 35:1317–23. doi: 10.1016/j.dci.2011.03.033
17. Toda H, Saito Y, Koike T, Takizawa F, Araki K, Yabu T, et al. Conservation of Characteristics and Functions of CD4 Positive Lymphocytes in a Teleost Fish. Dev Comp Immunol (2011) 35:650–60. doi: 10.1016/j.dci.2011.01.013
18. Wentzel AS, Janssen JJE, de Boer VCJ, van Veen WG, Forlenza M, Wiegertjes GF. Fish Macrophages Show Distinct Metabolic Signatures Upon Polarization. Front Immunol (2020) 11:152. doi: 10.3389/fimmu.2020.00152
19. Lu X-J, Chen J. Specific Function and Modulation of Teleost Monocytes/Macrophages: Polarization and Phagocytosis. Zool Res (2019) 40:146–50. doi: 10.24272/j.issn.2095-8137.2019.035
20. Hawke NA, Yoder JA, Litman GW. Expanding Our Understanding of Immunoglobulin, T-Cell Antigen Receptor, and Novel Immune-Type Receptor Genes: A Subset of the Immunoglobulin Gene Superfamily. Immunogenetics (1999) 50:124–33. doi: 10.1007/s002510050588
21. Litman GW, Hawke NA, Yoder JA. Novel Immune-Type Receptor Genes. Immunol Rev (2001) 181:250–9. doi: 10.1034/j.1600-065X.2001.1810121.x
22. Stafford JL, Bengtén E, Du Pasquier L, McIntosh RD, Quiniou SM, Clem LW, et al. A Novel Family of Diversified Immunoregulatory Receptors in Teleosts Is Homologous to Both Mammalian Fc Receptors and Molecules Encoded Within the Leukocyte Receptor Complex. Immunogenetics (2006) 58:758–73. doi: 10.1007/s00251-006-0134-1
23. Stafford JL, Bengtén E, Du Pasquier L, Miller NW, Wilson M. Channel Catfish Leukocyte Immune-Type Receptors Contain a Putative MHC Class I Binding Site. Immunogenetics (2007) 59:77–91. doi: 10.1007/s00251-006-0169-3
24. Soonthonsrima T, Wangman P, Chaivisuthangkura P, Pengsuk C, Sithigorngul P, Longyant S. Generation of Mouse Monoclonal Antibodies Specific to Tilapia Immunoglobulin Using Fish Immunoglobulin/BSA Complex for Monitoring of the Immune Response in Nile Tilapia Oreochromis Niloticus. Aquac Res (2019) 50:277–83. doi: 10.1111/are.13894
25. Israelson O, Petersson A, Bengtén E, Wiersma EJ, Anderson J, Gezelius G, et al. Immunoglobulin Concentration in Atlantic Cod, Gadus Morhua L., Serum and Cross-Reactivity Between Anti-Cod-Antibodies and Immunoglobulins From Other Species. J Fish Biol (1991) 39:265–78. doi: 10.1111/j.1095-8649.1991.tb04361.x
26. Pettersen FE, Fyllingen I, Kavlie A, Maaseide NP, Glette J, Endresen C, et al. Monoclonal Antibodies Reactive With Serum IgM and Leukocytes From Atlantic Salmon (Salmo salar L.). Fish Shellfish Immunol (1995) 5:275–87. doi: 10.1006/fsim.1995.0027
27. Bunnoy A, Na-Nakorn U, Srisapoome P. Development of a Monoclonal Antibody Specific to the IgM Heavy Chain of Bighead Catfish (Clarias Macrocephalus): A Biomolecular Tool for the Detection and Quantification of IgM Molecules and IgM + Cells in Clarias Catfish. Biomolecules (2020) 10:567. doi: 10.3390/biom10040567
28. Shin G, Lee H, Palaksha KJ, Kim Y, Lee E, Shin Y, et al. Production of Monoclonal Antibodies Against Serum Immunoglobulins of Black Rockfish (Sebastes Schlegeli Higendorf). J Vet Sci (2006) 7:293–5. doi: 10.4142/jvs.2006.7.3.293
29. Thuvander A, Fossum C, Lorenzen N. Monoclonal Antibodies to Salmonid Immunoglobulin: Characterization and Applicability in Immunoassays. Dev Comp Immunol (1990) 14:415–23. doi: 10.1016/0145-305X(90)90034-C
30. Edholm E-S, Hudgens ED, Tompkins D, Sahoo M, Burkhalter B, Miller NW, et al. Characterization of Anti-Channel Catfish IgL ς Monoclonal Antibodies. Vet Immunol Immunopathol (2010) 135:325–8. doi: 10.1016/j.vetimm.2010.01.004
31. Vesely T, Reschova S, Pokorova D, Hulova J, Nevorankova Z. Production of Monoclonal Antibodies Against Immunoglobulin Heavy Chain in Common Carp (Cyprinus Carpio L.). Vet Med (2012) 51:296–302. doi: 10.17221/5549-VETMED
32. van der Heijden MHT, Rooijakkers JBMA, Booms GHR, Rombout JHWM, Boon JH. Production, Characterisation and Applicability of Monoclonal Antibodies to European Eel (Anguilla Anguilla L., 1758) Immunoglobulin. Vet Immunol Immunopathol (1995) 45:151–64. doi: 10.1016/0165-2427(94)05335-P
33. Scapigliati G, Romano N, Picchietti S, Mazzini M, Mastrolia L, Scalia D, et al. Monoclonal Antibodies Against Sea Bass Dicentrarchus Labrax(L.) Immunoglobulins: Immunolocalisation of Immunoglobulin-Bearing Cells and Applicability in Immunoassays. Fish Shellfish Immunol (1996) 6:383–401. doi: 10.1006/fsim.1996.0038
34. Li Q, Zhan W, Xing J, Sheng X. Production, Characterisation and Applicability of Monoclonal Antibodies to Immunoglobulin of Japanese Flounder (Paralichthys Olivaceus). Fish Shellfish Immunol (2007) 23:982–90. doi: 10.1016/j.fsi.2007.03.008
35. Jang H-N, Woo J-K, Cho Y-H, Kyong S-B, Choi S-H. Characterization of Monoclonal Antibodies Against Heavy and Light Chains of Flounder (Paralichthys Olivaceus) Immunoglobulin. BMB Rep (2004) 37:314–9. doi: 10.5483/BMBRep.2004.37.3.314
36. Wu R, Shen J, Lai X, He T, Li Y. Development of Monoclonal Antibodies Against Serum Immunoglobulins From Gibel Carp (Carassius Auratus Gibelio) and Their Applications in Serodiagnosis of Inapparent Infection and Evaluation of Vaccination Strategies. Fish Shellfish Immunol (2020) 96:69–77. doi: 10.1016/j.fsi.2019.11.059
37. Navarro V, Quesada JA, Abad ME, Taverne N, Rombout JHWM. Immuno(cyto)chemical Characterisation of Monoclonal Antibodies to Gilthead Seabream (Sparus Aurata) Immunoglobulin. Fish Shellfish Immunol (1993) 3:167–77. doi: 10.1006/fsim.1993.1017
38. Yang S, Tang X, Sheng X, Xing J, Zhan W. Development of Monoclonal Antibodies Against IgM of Half-Smooth Tongue Sole (Cynoglossus Semilaevis) and Analysis of Phagocytosis of Fluorescence Microspheres by Migm+ Lymphocytes. Fish Shellfish Immunol (2017) 66:280–8. doi: 10.1016/j.fsi.2017.05.019
39. Huang Y, Yuan X, Mu P, Li Q, Ao J, Chen X. Development of Monoclonal Antibody Against IgM of Large Yellow Croaker (Larimichthys Crocea) and Characterization of IgM+ B Cells. Fish Shellfish Immunol (2019) 91:216–22. doi: 10.1016/j.fsi.2019.05.035
40. Fu Q, Wei Z, Chen Y, Xie J, Zhang X, He T, et al. Development of Monoclonal Antibody Against IgT of a Perciform Fish, Large Yellow Croaker (Larimichthys Crocea) and Characterization of IgT+ B Cells. Dev Comp Immunol (2021) 119:104027. doi: 10.1016/j.dci.2021.104027
41. Mohanty S, Makesh M, Rajendran KV, Suresh Babu PP, Anand D, Kumar S, et al. Production and Characterisation of Monoclonal Antibodies Against Immunoglobulins of Cirrhinus Mrigala (Hamilton 1822). Indian J Fish (2020) 67:55–61. doi: 10.21077/ijf.2019.67.2.84594-08
42. Faisal M, Standish IF, Vogelbein MA, Millard EV, Kaattari SL. Production of a Monoclonal Antibody Against of Muskellunge (Esox Masquinongy) IgM Heavy Chain and Its Use in Development of an Indirect ELISA for Titrating Circulating Antibodies Against VHSV-IVB. Fish Shellfish Immunol (2019) 88:464–71. doi: 10.1016/j.fsi.2019.03.002
43. Yin X, Mu L, Fu S, Wu L, Han K, Wu H, et al. Expression and Characterization of Nile Tilapia (Oreochromis Niloticus) Secretory and Membrane-Bound IgM in Response to Bacterial Infection. Aquaculture (2019) 508:214–22. doi: 10.1016/j.aquaculture.2019.03.058
44. Velázquez J, Rodríguez A, Aragón H, Haidar A, González M, Valdés R, et al. Monoclonal Antibody Against Nile Tilapia (Oreochromis Niloticus) IgM Heavy Chain: A Valuable Tool for Detection and Quantification of IgM and IgM+ Cells. Fish Shellfish Immunol (2021) 110:44–54. doi: 10.1016/j.fsi.2020.12.007
45. Purcell MK, Bromage ES, Silva J, Hansen JD, Badil SM, Woodson JC, et al. Production and Characterization of Monoclonal Antibodies to IgM of Pacific Herring (Clupea Pallasii). Fish Shellfish Immunol (2012) 33:552–8. doi: 10.1016/j.fsi.2012.06.006
46. Timmusk S, Jansson E, Pilstrom L. The Generation of Monoclonal Antibodies by Genetic Immunisation: Antibodies Against Trout TCR Alpha and IgL Isotypes. Fish Shellfish Immunol (2003) 14:187–206. doi: 10.1006/fsim.2002.0429
47. Sánchez C, López-Fierro P, Zapata A, Dominguez J. Characterisation of Monoclonal Antibodies Against Heavy and Light Chains of Trout Immunoglobulin. Fish Shellfish Immunol (1993) 3:237– 251. doi: 10.1006/fsim.1993.1024
48. MacDougal KC, Johnstone M-B, Burnett KG. Monoclonal Antibodies Reactive With the Immunoglobulin Heavy Chain in Sciaenid Fishes. Fish Shellfish Immunol (1995) 5:389–91. doi: 10.1006/fsim.1995.0038
49. Rathore G, Kumar G, Sood N, Kapoor D, Lakra WS. Development of Monoclonal Antibodies to Rohu [Labeo Rohita] Immunoglobulins for Use in Immunoassays. Fish Shellfish Immunol (2008) 25:761–74. doi: 10.1016/j.fsi.2008.02.014
50. Yang S, Tang X, Sheng X, Xing J, Zhan W. Development of Monoclonal Antibodies Against IgM of Sea Bass (Lateolabrax Japonicus) and Analysis of Phagocytosis by Migm+ Lymphocytes. Fish Shellfish Immunol (2018) 78:372–82. doi: 10.1016/j.fsi.2018.04.042
51. Kim S-W, Kim J-O, Kong K-H, Oh M-J, Kim W-S. Production of Monoclonal Antibodies to Immunoglobulin M of Sevenband Grouper (Epinephelus Septemfasciatus). Fish Pathol (2021) 34:111–5. doi: 10.7847/jfp.2021.34.1.111
52. Ottinger CA, Smith CR, Blazer VS, Iwanowicz LR, Vogelbein MA, Kaattari S. Production and Characterization of a Mouse Monoclonal Antibody Against Smallmouth Bass (Micropterus Dolomieu) IgM. Fish Shellfish Immunol (2021) 113:20–3. doi: 10.1016/j.fsi.2021.03.006
53. Sood N, Chaudhary DK, Rathore G, Singh A, Lakra WS. Monoclonal Antibodies to Snakehead, Channa Striata Immunoglobulins: Detection and Quantification of Immunoglobulin-Positive Cells in Blood and Lymphoid Organs. Fish Shellfish Immunol (2011) 30:569–75. doi: 10.1016/j.fsi.2010.12.004
54. Morrison RN, Hayball JD, Cook MT, Nowak BF. Anti-Immunoglobulin Binding and Activation of Snapper (Pagrus Auratus) Leucocytes. Dev Comp Immunol (2002) 26:247–55. doi: 10.1016/S0145-305X(01)00070-2
55. Miyadai T, Ootani M, Tahara D, Aoki M, Saitoh K. Monoclonal Antibodies Recognising Serum Immunoglobulins and Surface Immunoglobulin-Positive Cells of Puffer Fish, Torafugu (Takifugu Rubripes). Fish Shellfish Immunol (2004) 17:211–22. doi: 10.1016/j.fsi.2004.03.005
56. Estévez J, Leiro J, Santamarina MT, Domínguez J, Ubeira FM. Monoclonal Antibodies to Turbot (Scophthalmus Maximus) Immunoglobulins: Characterization and Applicability in Immunoassays. Vet Immunol Immunopathol (1994) 41:353–66. doi: 10.1016/0165-2427(94)90107-4
57. Sood N, Chaudhary DK, Singh A, Rathore G. Monoclonal Antibody to Serum Immunoglobulins of Clarias Batrachus and Its Application in Immunoassays. Gene (2012) 511:411–9. doi: 10.1016/j.gene.2012.09.044
58. Adkison MA, Basurco B, Hedrick RP. Humoral Immunoglobulins of the White Sturgeon, Acipenser Transmontanus: Partial Characterization of and Recognition With Monoclonal Antibodies. Dev Comp Immunol (1996) 20:285–98. doi: 10.1016/0145-305X(96)00015-8
59. Jung JW, Lee JS, Kim YR, Im SP, Kim SW, Lazarte JMS, et al. Development of a Monoclonal Antibody Against the CD3 Epsilon of Olive Flounder (Paralichthys Olivaceus) and Its Application in Evaluating Immune Response Related to CD3 Epsilon. Fish Shellfish Immunol (2017) 65:179–85. doi: 10.1016/j.fsi.2017.04.016
60. Boardman T, Warner C, Ramirez-Gomez F, Matrisciano J, Bromage E. Characterization of an Anti-Rainbow Trout (Oncorhynchus Mykiss) CD3 Epsilon Monoclonal Antibody. Vet Immunol Immunopathol (2012) 145:511–5. doi: 10.1016/j.vetimm.2011.11.017
61. Qin Y, Sun Z, Wang W, Xu J, Wang B, Jia Z, et al. Characterization of CD3γ/δ+ Cells in Grass Carp (Ctenopharyngodon Idella). Dev Comp Immunol (2021) 114:103791. doi: 10.1016/j.dci.2020.103791
62. Jung JW, Chun JH, Lee JS, Kim SW, Lee AR, Kim J, et al. Characterization of CD4-Positive Lymphocytes in the Antiviral Response of Olive Flounder (Paralichthys Oliveceus) to Nervous Necrosis Virus. Int J Mol Sci (2020) 21:4180. doi: 10.3390/ijms21114180
63. Xing J, Tian H, Tang X, Sheng X, Zhan W. Kinetics of T Lymphocyte Subsets and B Lymphocytes in Response to Immunostimulants in Flounder (Paralichthys Olivaceus): Implications for CD4+ T Lymphocyte Differentiation. Sci Rep (2020) 10:13827. doi: 10.1038/s41598-020-69542-6
64. Takizawa F, Magadan S, Parra D, Xu Z, Korytář T, Boudinot P, et al. Novel Teleost CD4-Bearing Cell Populations Provide Insights Into the Evolutionary Origins and Primordial Roles of CD4+ Lymphocytes and CD4+ Macrophages. J Immunol (2016) 196:4522–35. doi: 10.4049/jimmunol.1600222
65. Dee CT, Nagaraju RT, Athanasiadis EI, Gray C, Fernandez del Ama L, Johnston SA, et al. CD4-Transgenic Zebrafish Reveal Tissue-Resident Th2- and Regulatory T Cell–Like Populations and Diverse Mononuclear Phagocytes. J Immunol (2016) 197:3520–30. doi: 10.4049/jimmunol.1600959
66. Hetland DL, Jørgensen SM, Skjødt K, Dale OB, Falk K, Xu C, et al. In Situ Localisation of Major Histocompatibility Complex Class I and Class II and CD8 Positive Cells in Infectious Salmon Anaemia Virus (ISAV)-Infected Atlantic Salmon. Fish Shellfish Immunol (2010) 28:30–9. doi: 10.1016/j.fsi.2009.09.011
67. Toda H, Shibasaki Y, Koike T, Ohtani M, Takizawa F, Ototake M, et al. Alloantigen-Specific Killing Is Mediated by CD8-Positive T Cells in Fish. Dev Comp Immunol (2009) 33:646–52. doi: 10.1016/j.dci.2008.11.008
68. Takizawa F, Dijkstra JM, Kotterba P, Korytář T, Kock H, Köllner B, et al. The Expression of CD8α Discriminates Distinct T Cell Subsets in Teleost Fish. Dev Comp Immunol (2011) 35:752–63. doi: 10.1016/j.dci.2011.02.008
69. Dowding AJ, Maggs A, Scholes J. Diversity Amongst the Microglia in Growing and Regenerating Fish CNS: Immunohistochemical Characterization Using FL. 1, an Anti-Macrophage Monoclonal Antibody. Glia (1991) 4:345–64. doi: 10.1002/glia.440040403
70. Romano N, Picchietti S, Taverne-Thiele JJ, Taverne N, Abelli L, Mastrolia L, et al. Distribution of Macrophages During Fish Development: An Immunohistochemical Study in Carp (Cyprinus Carpio , L.). Anat Embryol (1998) 198:31–41. doi: 10.1007/s004290050162
71. Nakayasu C, Omori M, Hasegawa S, Kurata O, Okamoto N. Production of a Monoclonal Antibody for Carp (Cyprinus carpioL.) Phagocytic Cells and Separation of the Cells. Fish Shellfish Immunol (1998) 8:91–100. doi: 10.1006/fsim.1997.0125
72. Weyts FAA, Rombout JHWM, Flik G, Verburg-van kemenade BML. A Common Carp (Cyprinus carpio L.) Leucocyte Cell Line Shares Morphological and Functional Characteristics With Macrophages. Fish Shellfish Immunol (1997) 7:123–33. doi: 10.1006/fsim.1996.0069
73. Woo J-K, Jang H-N, Cho Y-H, Jang Y-S, Choi S-H. Production and Monoclonal Antibodies (Mabs) Against Surface Antigens on Israeli Carp Lymphocytes and Their Applications. Asian- Australas J Anim (2001) 14:1179–87. doi: 10.5713/ajas.2001.1179
74. Matsuyama T, Nakayasu C, Sakai T, Oseko N. Production and Characterization of Monoclonal Antibodies to Leukocytes and Serum Immunoglobulin in Japanese Flounder Paralichthys Olivaceus. Fish Sci (2009) 75:335–41. doi: 10.1007/s12562-008-0038-0
75. Köllner B, Blohm U, Kotterba G, Fischer U. A Monoclonal Antibody Recognising a Surface Marker on Rainbow Trout (Oncorhynchus Mykiss) Monocytes. Fish Shellfish Immunol (2001) 11:127–42. doi: 10.1006/fsim.2000.0300
76. Kuroda A, Okamoto N, Fukuda H. Characterization of Monoclonal Antibodies Against Antigens Shared With Neutrophils and Macrophages in Rainbow Trout Oncorhynchus Mykiss. Fish Pathol (2000) 35:205–13. doi: 10.3147/jsfp.35.205
77. Ishikawa T, Kawai K. Production of Monoclonal Antibodies Against Yellowtail Leucocyte. Bull Mar Sci Fish Kochi Univ (1998) 18:27–31
78. Ellett F, Pase L, Hayman JW, Andrianopoulos A, Lieschke GJ. Mpeg1 Promoter Transgenes Direct Macrophage-Lineage Expression in Zebrafish. Blood (2011) 117:e49–56. doi: 10.1182/blood-2010-10-314120
79. Hsu K, Traver D, Kutok JL, Hagen A, Liu T-X, Paw BH, et al. The Pu.1 Promoter Drives Myeloid Gene Expression in Zebrafish. Blood (2004) 104:1291–7. doi: 10.1182/blood-2003-09-3105
80. Hall C, Flores MV, Storm T, Crosier K, Crosier P. The Zebrafish Lysozyme C Promoter Drives Myeloid-Specific Expression in Transgenic Fish. BMC Dev Biol (2007) 7:42. doi: 10.1186/1471-213X-7-42
81. Pettersen EF, Bjerknes R, Wergeland HI. Studies of Atlantic Salmon (Salmo Salar L.) Blood, Spleen and Head Kidney Leucocytes Using Specific Monoclonal Antibodies, Immunohistochemistry and Flow Cytometry. Fish Shellfish Immunol (2000) 10:695–710. doi: 10.1006/fsim.2000.0284
82. Kato G, Takano T, Nishiki I, Kai W, Yasuike M, Nakamura Y, et al. Expressed Sequence Tag Analyses of Three Leukocyte Subpopulations in Ayu Plecoglossus Altivelis Altivelis, Separated by Monoclonal Antibodies. Mar Genomics (2015) 23:123–32. doi: 10.1016/j.margen.2015.06.011
83. Ainsworth AJ, Dexiang C, Greenway T. Characterization of Monoclonal Antibodies to Channel Catfish, Ictalurus Punctatus, Leucocytes. Vet Immunol Immunopathol (1990) 26:81–92. doi: 10.1016/0165-2427(90)90134-E
84. Koumans - van Diepen JCE. Characterisation of Fish Leucocytes: An Immunocytochemical and Functional Study in Carp (Cyprinus Carpio L.). Wageningen: Wageningen Agricultural University (1993).
85. Romano N, Abelli L, Mastrolia L, Scapigliati G. Immunocytochemical Detection and Cytomorphology of Lymphocyte Subpopulations in a Teleost Fish Dicentrarchus Labrax. Cell Tissue Res (1997) 289:163–71. doi: 10.1007/s004410050862
86. Chaudhary DK, Sood N, Pradhan PK, Agarwal NK, Rathore G. Production and Characterization of a Monoclonal Antibody Against Putative T Lymphocytes of Catla Catla. In Vitro Cell Dev Biol Anim (2012) 48:483–92. doi: 10.1007/s11626-012-9532-x
87. Jansson E, Grönvik K-O, Johannisson A, Näslund K, Westergren E, Pilström L. Monoclonal Antibodies to Lymphocytes of Rainbow Trout (Oncorhynchus Mykiss). Fish Shellfish Immunol (2003) 14:239–57. doi: 10.1006/fsim.2002.0434
88. Cook MT, Morrison RN, Wilkinson R, Nowak BF, Hayball PJ, Hayball JD. A Screen of Mammalian Antibodies on Snapper (Pagrus Auratus, Sparidae) Peripheral Blood Leukocytes Reveals Cross Reactivity of an Anti-Human CD3 Antibody With a Population of mIg– Cells. Dev Comp Immunol (2001) 25:553–9. doi: 10.1016/S0145-305X(01)00024-6
89. Miyazawa R, Matsuura Y, Shibasaki Y, Imamura S, Nakanishi T. Cross-Reactivity of Monoclonal Antibodies Against CD4-1 and CD8α of Ginbuna Crucian Carp With Lymphocytes of Zebrafish and Other Cyprinid Species. Dev Comp Immunol (2018) 80:15–23. doi: 10.1016/j.dci.2016.12.002
90. Langenau DM, Ferrando AA, Traver D, Kutok JL, Hezel J-PD, Kanki JP, et al. In Vivo Tracking of T Cell Development, Ablation, and Engraftment in Transgenic Zebrafish. PNAS (2004) 101:7369–74. doi: 10.1073/pnas.0402248101
91. Sepulcre MP, Pelegrín P, Mulero V, Meseguer J. Characterisation of Gilthead Seabream Acidophilic Granulocytes by a Monoclonal Antibody Unequivocally Points to Their Involvement in Fish Phagocytic Response. Cell Tissue Res (2002) 308:97–102. doi: 10.1007/s00441-002-0531-1
92. Renshaw SA, Loynes CA, Trushell DMI, Elworthy S, Ingham PW, Whyte MKB. A Transgenic Zebrafish Model of Neutrophilic Inflammation. Blood (2006) 108:3976–8. doi: 10.1182/blood-2006-05-024075
93. Passer BJ, Chen CH, Miller NW, Cooper MD. Catfish Thrombocytes Express an Integrin-Like CD41/CD61 Complex. Exp Cell Res (1997) 234:347–53. doi: 10.1006/excr.1997.3611
94. Rombout JHWM, Koumans-van Diepen JCE, Emmer PM, Taverne-Thiele JJ, Taverne N. Characterization of Carp Thrombocytes With Specific Monoclonal Antibodies. J Fish Biol (1996) 49:521–31. doi: 10.1111/j.1095-8649.1996.tb00047.x
95. Kollner B, Fischer U, Rombout J, Taverne-Thiele JJ, Hansen JD. Potential Involvement of Rainbow Trout Thrombocytes in Immune Functions: A Study Using a Panel of Monoclonal Antibodies and RT-PCR. Dev Comp Immunol (2004) 28:1049–62. doi: 10.1016/j.dci.2004.03.004
96. Slierendrecht WJ, Lorenzen N, Glamann J, Koch C, Rombout JHWM. Immunocytochemical Analysis of a Monoclonal Antibody Specific for Rainbow Trout (Oncorhynchus Mykiss) Granulocytes and Thrombocytes. Vet Immunol Immunopathol (1995) 46:349–60. doi: 10.1016/0165-2427(94)05362-V
97. Lin H-F, Traver D, Zhu H, Dooley K, Paw BH, Zon LI, et al. Analysis of Thrombocyte Development in CD41-GFP Transgenic Zebrafish. Blood (2005) 106:3803–10. doi: 10.1182/blood-2005-01-0179
98. Lin Q, Zhang Y, Zhou R, Zheng Y, Zhao L, Huang M, et al. Establishment of a Congenital Amegakaryocytic Thrombocytopenia Model and a Thrombocyte– Specific Reporter Line in Zebrafish. Leukemia (2017) 31:1206–16. doi: 10.1038/leu.2016.320
99. Alfaleh MA, Alsaab HO, Mahmoud AB, Alkayyal AA, Jones ML, Mahler SM, et al. Phage Display Derived Monoclonal Antibodies: From Bench to Bedside. Front Immunol (2020) 11:1986. doi: 10.3389/fimmu.2020.01986
100. Nagano K, Tsutsumi Y. Phage Display Technology as a Powerful Platform for Antibody Drug Discovery. Viruses (2021) 13:178. doi: 10.3390/v13020178
101. Jain M, Kamal N, Batra SK. Engineering Antibodies for Clinical Applications. Trends Biotechnol (2007) 25:307–16. doi: 10.1016/j.tibtech.2007.05.001
102. Mandell JW. Phosphorylation State-Specific Antibodies: Applications in Investigative and Diagnostic Pathology. Am J Pathol (2003) 163:1687–98. doi: 10.1016/S0002-9440(10)63525-0
103. Köhler G, Milstein C. Continuous Cultures of Fused Cells Secreting Antibody of Predefined Specificity. Nature (1975) 256:495–7. doi: 10.1038/256495a0
104. Lo MMS, Tsong TY, Conrad MK, Strittmatter SM, Hester LD, Snyder SH. Monoclonal Antibody Production by Receptor-Mediated Electrically Induced Cell Fusion. Nature (1984) 310:792–4. doi: 10.1038/310792a0
105. Bradbury ARM, Trinklein ND, Thie H, Wilkinson IC, Tandon AK, Anderson S, et al. When Monoclonal Antibodies Are Not Monospecific: Hybridomas Frequently Express Additional Functional Variable Regions. mAbs (2018) 10:539– 546. doi: 10.1080/19420862.2018.1445456
106. Basu K, Green EM, Cheng Y, Craik CS. Why Recombinant Antibodies — Benefits and Applications. Curr Opin Biotechnol (2019) 60:153–8. doi: 10.1016/j.copbio.2019.01.012
107. Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, et al. Rapid Cloning of High-Affinity Human Monoclonal Antibodies Against Influenza Virus. Nature (2008) 453:667–71. doi: 10.1038/nature06890
108. Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, et al. Broad Diversity of Neutralizing Antibodies Isolated From Memory B Cells in HIV-Infected Individuals. Nature (2009) 458:636–40. doi: 10.1038/nature07930
109. Wrammert J, Koutsonanos D, Li G-M, Edupuganti S, Sui J, Morrissey M, et al. Broadly Cross-Reactive Antibodies Dominate the Human B Cell Response Against 2009 Pandemic H1N1 Influenza Virus Infection. J Exp Med (2011) 208:181–93. doi: 10.1084/jem.20101352
110. Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H. Efficient Generation of Monoclonal Antibodies From Single Human B Cells by Single Cell RT-PCR and Expression Vector Cloning. J Immunol Methods (2008) 329:112–24. doi: 10.1016/j.jim.2007.09.017
111. Niro RD, Mesin L, Raki M, Zheng N-Y, Lund-Johansen F, Lundin KEA, et al. Rapid Generation of Rotavirus-Specific Human Monoclonal Antibodies From Small-Intestinal Mucosa. J Immunol (2010) 185:5377–83. doi: 10.4049/jimmunol.1001587
112. Bernasconi NL, Traggiai E, Lanzavecchia A. Maintenance of Serological Memory by Polyclonal Activation of Human Memory B Cells. Science (2002) 298:2199–202. doi: 10.1126/science.1076071
113. Sasaki S, Jaimes MC, Holmes TH, Dekker CL, Mahmood K, Kemble GW, et al. Comparison of the Influenza Virus-Specific Effector and Memory B-Cell Responses to Immunization of Children and Adults With Live Attenuated or Inactivated Influenza Virus Vaccines. J Virol (2007) 81:215–28. doi: 10.1128/JVI.01957-06
114. Brokstad KA, Cox RJ, Olofsson J, Jonsson R, Haaheim LR. Parenteral Influenza Vaccination Induces a Rapid Systemic and Local Immune Response. J Infect Dis (1995) 171:198–203. doi: 10.1093/infdis/171.1.198
115. Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant Autoantibody Production by Early Human B Cell Precursors. Science (2003) 301:1374–7. doi: 10.1126/science.1086907
116. Küppers R, Zhao M, Hansmann ML, Rajewsky K. Tracing B Cell Development in Human Germinal Centres by Molecular Analysis of Single Cells Picked From Histological Sections. EMBO J (1993) 12:4955–67. doi: 10.1002/j.1460-2075.1993.tb06189.x
117. Ehlich A, Martin V, Müller W, Rajewsky K. Analysis of the B-Cell Progenitor Compartment at The Level of Single Cells. Curr Biol (1994) 4:573–83. doi: 10.1016/S0960-9822(00)00129-9
118. Klein U, Rajewsky K, Küppers R. Human Immunoglobulin (Ig)M+IgD+ Peripheral Blood B Cells Expressing the CD27 Cell Surface Antigen Carry Somatically Mutated Variable Region Genes: CD27 as a General Marker for Somatically Mutated (Memory) B Cells. J Exp Med (1998) 188:1679–89. doi: 10.1084/jem.188.9.1679
119. Schroeder HW. Similarity and Divergence in the Development and Expression of the Mouse and Human Antibody Repertoires. Dev Comp Immunol (2006) 30:119–35. doi: 10.1016/j.dci.2005.06.006
120. Busse CE, Czogiel I, Braun P, Arndt PF, Wardemann H. Single-Cell Based High-Throughput Sequencing of Full-Length Immunoglobulin Heavy and Light Chain Genes. Eur J Immunol (2014) 44:597–603. doi: 10.1002/eji.201343917
121. DeKosky BJ, Ippolito GC, Deschner RP, Lavinder JJ, Wine Y, Rawlings BM, et al. High-Throughput Sequencing of the Paired Human Immunoglobulin Heavy and Light Chain Repertoire. Nat Biotechnol (2013) 31:166–9. doi: 10.1038/nbt.2492
122. Frenzel A, Hust M, Schirrmann T. Expression of Recombinant Antibodies. Front Immunol (2013) 4:217. doi: 10.3389/fimmu.2013.00217
123. Beebe DJ, Mensing GA, Walker GM. Physics and Applications of Microfluidics in Biology. Annu Rev BioMed Eng (2002) 4:261–86. doi: 10.1146/annurev.bioeng.4.112601.125916
124. Squires TM, Quake SR. Microfluidics: Fluid Physics at the Nanoliter Scale. Rev Mod Phys (2005) 77:977–1026. doi: 10.1103/RevModPhys.77.977
125. Mazutis L, Griffiths AD. Selective Droplet Coalescence Using Microfluidic Systems. Lab Chip (2012) 12:1800–6. doi: 10.1039/C2LC40121E
126. Shim J, Olguin LF, Whyte G, Scott D, Babtie A, Abell C, et al. Simultaneous Determination of Gene Expression and Enzymatic Activity in Individual Bacterial Cells in Microdroplet Compartments. J Am Chem Soc (2009) 131:15251–6. doi: 10.1021/ja904823z
127. Mazutis L, Araghi AF, Miller OJ, Baret J-C, Frenz L, Janoshazi A, et al. Droplet-Based Microfluidic Systems for High-Throughput Single DNA Molecule Isothermal Amplification and Analysis. Anal Chem (2009) 81:4813–21. doi: 10.1021/ac900403z
128. Ahn K, Kerbage C, Hunt TP, Westervelt RM, Link DR, Weitz DA. Dielectrophoretic Manipulation of Drops for High-Speed Microfluidic Sorting Devices. Appl Phys Lett (2006) 88:024104. doi: 10.1063/1.2164911
129. Mazutis L, Gilbert J, Ung WL, Weitz DA, Griffiths AD, Heyman JA. Single-Cell Analysis and Sorting Using Droplet-Based Microfluidics. Nat Protoc (2013) 8:870–91. doi: 10.1038/nprot.2013.046
130. Eyer K, Doineau RCL, Castrillon CE, Briseño-Roa L, Menrath V, Mottet G, et al. Single-Cell Deep Phenotyping of IgG-Secreting Cells for High- Resolution Immune Monitoring. Nat Biotechnol (2017) 35:977–82. doi: 10.1038/nbt.3964
131. Gérard A, Woolfe A, Mottet G, Reichen M, Castrillon C, Menrath V, et al. High-Throughput Single-Cell Activity-Based Screening and Sequencing of Antibodies Using Droplet Microfluidics. Nat Biotechnol (2020) 38:715–21. doi: 10.1038/s41587-020-0466-7
132. Adler AS, Mizrahi RA, Spindler MJ, Adams MS, Asensio MA, Edgar RC, et al. Rare, High-Affinity Anti-Pathogen Antibodies From Human Repertoires, Discovered Using Microfluidics and Molecular Genomics. mAbs (2017) 9:1282–96. doi: 10.1080/19420862.2017.1371383
133. Setliff I, Shiakolas AR, Pilewski KA, Murji AA, Mapengo RE, Janowska K, et al. High-Throughput Mapping of B Cell Receptor Sequences to Antigen Specificity. Cell (2019) 179:1636–46.e15. doi: 10.1016/j.cell.2019.11.003
134. Glasauer SMK, Neuhauss SCF. Whole-Genome Duplication in Teleost Fishes and Its Evolutionary Consequences. Mol Genet Genom (2014) 289:1045–60. doi: 10.1007/s00438-014-0889-2
135. Chang M, Nie P, Collet B, Secombes CJ, Zou J. Identification of an Additional Two-Cysteine Containing Type I Interferon in Rainbow Trout Oncorhynchus Mykiss Provides Evidence of a Major Gene Duplication Event Within This Gene Family in Teleosts. Immunogenetics (2009) 61:315–25. doi: 10.1007/s00251-009-0366-y
136. Harte A, Tian G, Xu Q, Secombes CJ, Wang T. Five Subfamilies of β-Defensin Genes Are Present in Salmonids: Evolutionary Insights and Expression Analysis in Atlantic Salmon Salmo Salar. Dev Comp Immunol (2020) 104:103560. doi: 10.1016/j.dci.2019.103560
137. Hong S, Wang T-Y, Secombes CJ, Wang T. Different Origins of Paralogues of Salmonid TNR1 and TNFR2: Characterisation and Expression Analysis of Four TNF Receptor Genes in Rainbow Trout Oncorhynchus Mykiss. Dev Comp Immunol (2019) 99:103403. doi: 10.1016/j.dci.2019.103403
138. Husain M, Bird S, Zwieten Rv, Secombes CJ, Wang T. Cloning of the IL-1β3 Gene and IL- 1β4 Pseudogene in Salmonids Uncovers a Second Type of IL-1β Gene in Teleost Fish. Dev Comp Immunol (2012) 38:431–46. doi: 10.1016/j.dci.2012.07.010
139. Liu F, Wang T, Petit J, Forlenza M, Chen X, Chen L, et al. Evolution of IFN Subgroups in Bony Fish - 2. Analysis of Subgroup Appearance and Expansion in Teleost Fish With a Focus on Salmonids. Fish Shellfish Immunol (2020) 98:564–73. doi: 10.1016/j.fsi.2020.01.039
140. Liu F, Wang T, Hu Y, Tian G, Secombes CJ, Wang T. Expansion of Fish CCL20_like Chemokines by Genome and Local Gene Duplication: Characterisation and Expression Analysis of 10 CCL20_like Chemokines in Rainbow Trout (Oncorhynchus Mykiss). Dev Comp Immunol (2020) 103:103502. doi: 10.1016/j.dci.2019.103502
141. Wang T, Bird S, Koussounadis A, Holland JW, Carrington A, Zou J, et al. Identification of a Novel IL-1 Cytokine Family Member in Teleost Fish. J Immunol (2009) 183:962–74. doi: 10.4049/jimmunol.0802953
142. Wang T, Liu F, Tian G, Secombes CJ, Wang T. Lineage/species-Specific Expansion of the Mx Gene Family in Teleosts: Differential Expression and Modulation of Nine Mx Genes in Rainbow Trout Oncorhynchus Mykiss. Fish Shellfish Immunol (2019) 90:413–30. doi: 10.1016/j.fsi.2019.04.303
143. Wang Y, Wang Q, Baoprasertkul P, Peatman E, Liu Z. Genomic Organization, Gene Duplication, and Expression Analysis of Interleukin-1beta in Channel Catfish (Ictalurus Punctatus). Mol Immunol (2006) 43:1653–64. doi: 10.1016/j.molimm.2005.09.024
144. Zou J, Mercier C, Koussounadis A, Secombes C. Discovery of Multiple Beta-Defensin Like Homologues in Teleost Fish. Mol Immunol (2007) 4:638–47. doi: 10.1016/j.molimm.2006.01.012
145. Edholm E-S, Stafford JL, Quiniou SM, Waldbieser G, Miller NW, Bengtén E, et al. Channel Catfish, Ictalurus Punctatus, CD4-Like Molecules. Dev Comp Immunol (2007) 31:172–87. doi: 10.1016/j.dci.2006.05.012
146. Laing KJ, Zou JJ, Purcell MK, Phillips R, Secombes CJ, Hansen JD. Evolution of the CD4 Family: Teleost Fish Possess Two Divergent Forms of CD4 in Addition to Lymphocyte Activation Gene-3. J Immunol (2006) 177:3939–51. doi: 10.4049/jimmunol.177.6.3939
147. Nonaka S, Somamoto T, Kato-Unoki Y, Ototake M, Nakanishi T, Nakao M. Molecular Cloning of CD4 From Ginbuna Crucian Carp Carassius Auratus Langsdorfii. Fish Sci (2008) 74:341–6. doi: 10.1111/j.1444-2906.2008.01530.x
148. Øvergård A-C, Nerland AH, Patel S. Cloning, Characterization, and Expression Pattern of Atlantic Halibut (Hippoglossus Hippoglossus L.) CD4-2, Lck, and ZAP-70. Fish Shellfish Immunol (2010) 29:987–97. doi: 10.1016/j.fsi.2010.08.011
149. Zou J, Secombes CJ. The Function of Fish Cytokines. Biology (2016) 5:23. doi: 10.3390/biology5020023
150. Wang T, Secombes CJ. The Cytokine Networks of Adaptive Immunity in Fish. Fish Shellfish Immunol (2013) 35:1703–18. doi: 10.1016/j.fsi.2013.08.030
Keywords: teleost fish, immune cells, monoclonal antibodies, fluorescence-activated cell sorting, droplet-based microfluidics
Citation: Fei C, Nie L, Zhang J and Chen J (2021) Potential Applications of Fluorescence-Activated Cell Sorting (FACS) and Droplet-Based Microfluidics in Promoting the Discovery of Specific Antibodies for Characterizations of Fish Immune Cells. Front. Immunol. 12:771231. doi: 10.3389/fimmu.2021.771231
Received: 06 September 2021; Accepted: 21 October 2021;
Published: 16 November 2021.
Edited by:
Jia Cai, Guangdong Ocean University, ChinaReviewed by:
Teruyuki Nakanishi, Goto Aquaculture Research Institute, JapanJianmin Ye, South China Normal University, China
Copyright © 2021 Fei, Nie, Zhang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiong Chen, amNoZW4xOTc1QDE2My5jb20=
 Chenjie Fei
Chenjie Fei Li Nie
Li Nie Jianhua Zhang
Jianhua Zhang Jiong Chen
Jiong Chen