- 1Department of Radiotherapy, The Second Hospital of Jilin University, Changchun, China
- 2Department of Thyroid Surgery, The Second Hospital of Jilin University, Changchun, China
- 3Department of Hepatobiliary and Pancreatic Surgery, The Second Hospital of Jilin University, Changchun, China
- 4Department of Pathology, The Second Hospital of Jilin University, Changchun, China
- 5Department of Biobank, The China-Japan Union Hospital of Jilin University, Changchun, China
- 6Department of Breast Surgery, The Affiliated Hospital Changchun University of Chinese Medicine, Changchun, China
- 7Department of Orthopedics, The China-Japan Union Hospital of Jilin University, Changchun, China
- 8Department of Pharmacy, The Second Hospital of Jilin University, Changchun, China
- 9Jilin Provincial Key Laboratory on Molecular and Chemical Genetic, The Second Hospital of Jilin University, Changchun, China
Several C2 domain-containing proteins play key roles in tumorigenesis, signal transduction, and mediating protein–protein interactions. Tandem C2 domains nuclear protein (TC2N) is a tandem C2 domain-containing protein that is differentially expressed in several types of cancers and is closely associated with tumorigenesis and tumor progression. Notably, TC2N has been identified as an oncogene in lung and gastric cancer but as a tumor suppressor gene in breast cancer. Recently, a large number of tumor-associated antigens (TAAs), such as heat shock proteins, alpha-fetoprotein, and carcinoembryonic antigen, have been identified in a variety of malignant tumors. Differences in the expression levels of TAAs between cancer cells and normal cells have led to these antigens being investigated as diagnostic and prognostic biomarkers and as novel targets in cancer treatment. In this review, we summarize the clinical characteristics of TC2N-positive cancers and potential mechanisms of action of TC2N in the occurrence and development of specific cancers. This article provides an exploration of TC2N as a potential target for the diagnosis and treatment of different types of cancers.
1 Introduction
Cancer is an important public health concern worldwide and continues to be of great interest to the scientific community. It is one of the leading causes of death, with approximately 14 million new cases and 8.2 million cancer-related deaths occurring in 2018 (1). This disease is considered the biggest barrier to improving life expectancy in countries in the 21st century (2). Annually, over 4 million new cancer patients and over 2 million cancer-related mortalities are reported in China. Despite the availability of multiple treatment modalities such as surgery, chemotherapy, radiation therapy, and targeted therapy, the 3- and 5-year cancer-specific survival rates remain poor (3–7). While overall cancer related mortalities have decreased (8), it is notable that the reduction in mortality is largely due to early detection and prevention rather than development of better treatment options (9–12).
Most cancers are asymptomatic in the early stages of development (13, 14) largely because of their ability to evade immune surveillance (15, 16). Immune evasion is thought to be driven by two major mechanisms. First, owing to altered antigen presentation or receptor library editing, the immune system is unable to detect tumor populations. Second, the initially effective immune response may become ineffective owing to the presence of an immunosuppressive tumor microenvironment (17–19). Therefore, it is important to explore mechanisms of cancer development to identify new markers for diagnosis and prognosis and to develop effective and novel treatment methods. Developments in both fronts will have substantial implications for improving survival rates of cancer patients.
Numerous studies have shown that certain genes, such as oncogenes and tumor suppressor genes, are risk factors for many types of cancer (20–24). When activated, oncogenes stimulate tumor growth whereas tumor suppressor genes prevent tumor growth and development. In mouse models, where oncogene expression is driven by tissue-specific promoters, tumors appear at high frequency but disappear when the inductive stimulus is turned off (25–27), suggesting that oncogenes are the Achilles’ heel of cancer (28). Tumor suppressor genes play a critical role in controlling the cell cycle assuring proper proliferation and differentiation (29). Therefore, identifying these genes is crucial because targeting them may prevent or treat different types of cancers.
The C2 domain was initially thought to be a protein structural domain of calcium-dependent protein kinase C (30–32). Further studies confirmed that the function of the C2 domain was not only calcium-dependent phospholipid binding, but also involved in cellular signal transduction and protein-protein interactions (33). Several proteins that contain a structural domain called the C2 domain have been linked to the regulation of tumorigenesis. For example, a C2 domain-containing protein, DOC2B, plays a tumor-suppressive role in cervical cancer by inhibiting cell proliferation, migration, and invasion (34). Conversely, another C2 domain-containing protein, myoferlin, plays a tumor-enhancing role by promoting metastasis in patients with triple-negative breast cancer (35). Gene encoding tandem C2 domains nuclear protein (TC2N)—a putative C2 domain-containing protein—has recently been shown to function both as an oncogene and a tumor suppressor gene (36–38). TC2N is located on human chromosome 14q32, belongs to the carboxyl-terminal type (C-type) tandem C2 protein family, and contains two C-terminal C2 domains (C2A and C2B) (39). TC2N is also an immune system gene similar to IFI27, CASS4, and SMARCD3 (40). Given its tumorigenesis properties and its association with the immune system, it has been proposed as a potential target for the detection and treatment of various cancers. In this review, we summarize recent progress in understanding the role and underlying mechanisms of TC2N in the occurrence and development of cancer, with a focus on lung cancer, breast cancer, and gastric cancer.
2 TC2N in Cancers
TC2N expression is upregulated in different types of cancers, including lung, breast, and gastric cancers. The relevant clinicopathological features and the molecular mechanisms of TC2N in these cancers are summarized in Table 1 and detailed in the rest of this section.
2.1 Lung Cancer
2.1.1 Functional Characteristics and Clinical Features of TC2N in Lung Cancer
TC2N is overexpressed in cancerous lung tissues and cell lines compared with that in normal lung tissues and a human bronchial epithelial cell line, respectively. Hao XL et al. (36) showed that upregulation of TC2N was significantly correlated with advanced TNM stage and high histological grade of disease. Additionally, high TC2N expression levels were associated with poor clinical outcomes and significantly short overall survival. Hence, TC2N expression has been proposed as an independent prognostic factor affecting patient survival. Mechanistically, overexpression of TC2N significantly inhibited apoptosis, promoted cell proliferation, and increased migration and invasion of tumor cells in vitro; in contrast, knockdown of TC2N promoted apoptosis and inhibited proliferation of lung cancer cells (41). Furthermore, knockdown of TC2N in tumor tissues resulted in an increase in apoptotic cells, supporting the hypothesis that TC2N overexpression promotes tumorigenesis and growth of lung cancer tumors in vivo.
In summary, TC2N is a potential novel oncogene in lung cancer, whose expression levels are correlated with cancer progression and patient survival. TC2N stimulates cell proliferation, migration, and invasion and reduces apoptosis of lung cancer cells in vitro and in vivo.
2.1.2 Signaling Pathways Influenced by TC2N in Lung Cancer
2.1.2.1 TC2N Inhibits p53 Signaling Pathway in Lung Cancer
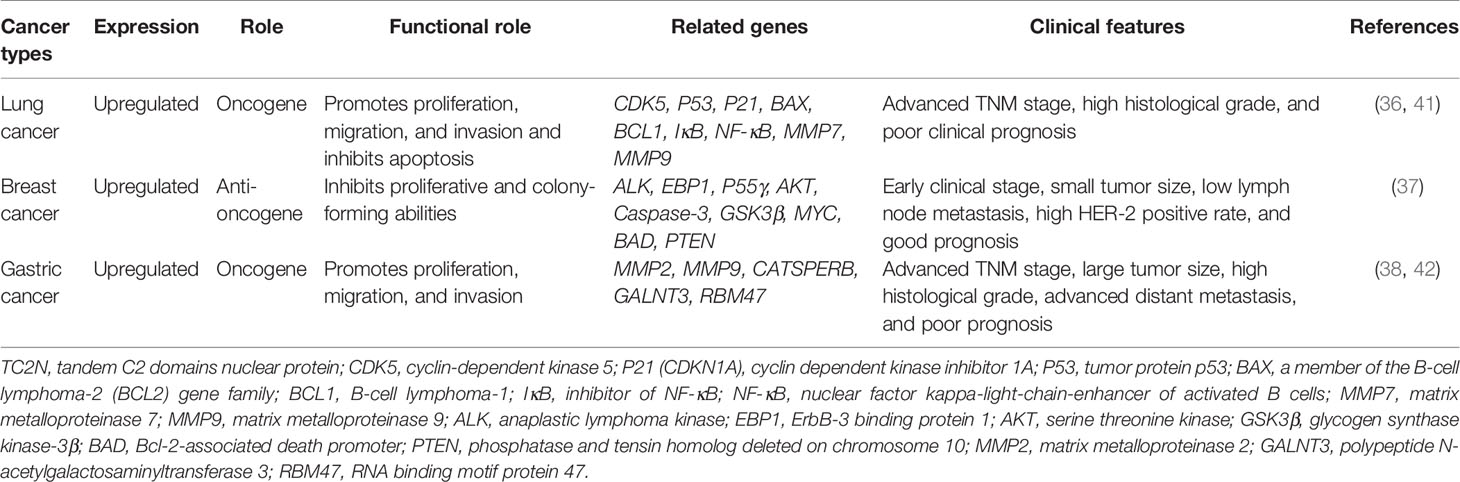
Hao XL et al. (36) proposed that the regulation of cell proliferation, cell cycle, and apoptosis by TC2N is dependent on the p53 signaling pathway. TP53, which encodes p53, was initially classified as an oncogene due to its ability to transform cells (43–46). However, the identification of growth-inhibiting and temperature-sensitive mutants of p53 in sporadic cancer samples and familial cancers has shown that p53 is in fact a tumor suppressor protein (47–53). p53 functions as the major regulator of central signaling and cell fate decision pathways (54). It is a nuclear transcription factor composed of 393 amino acids with four major functional domains: a transcriptional, a DNA binding, a tetramerization, and a regulatory domain (55). It modifies the expression of multiple genes involved in a variety of biological processes, including cell cycle, apoptosis, senescence, differentiation, and DNA repair (56–66). Moreover, p53 has been associated with diverse biological processes, such as regeneration (67), metabolism (68, 69), interaction with viruses (70), prevention of liver pathologies (71, 72), forming a barrier to stem cell formation (73, 74), endocrinology circuits (75) and serving as the guardian of the tissue hierarchy (76). p53 activity is largely controlled by post-translational modifications, such as phosphorylation (77). CDK5 is a protein kinase that phosphorylates p53 at Ser-15, Ser-33 and Ser-46 (78, 79) and binding of CDK5 to p53 induces activation of the p53 signaling pathway (77). Overexpression of TC2N interferes with CDK5-p53 interaction in the nucleus and induces significant CDK5 degradation by increasing the ubiquitination of CDK5 (Figure 1). Therefore, an increase in TC2N levels suppresses CDK5-induced p53 phosphorylation and p53 pathway activation. The expression of other key players in the p53 signaling pathway, such as P21, BAX, and BCL-2, is also downregulated by TCN2 (36). When cells experience stress or undergo uncontrolled division and proliferation, p53 is activated (56, 80). Under these conditions, p53 induces p21 expression, causing cell cycle arrest (81, 82). Furthermore, p53 triggers programmed cell death by triggering apoptosis-related genes, including bax, a pro-apoptotic member of the bcl-2 family, when a DNA damage cannot be repaired (83). Hence, reduction of the downstream players in the p53 pathway by TCN2 promotes proliferation and prevents apoptosis.
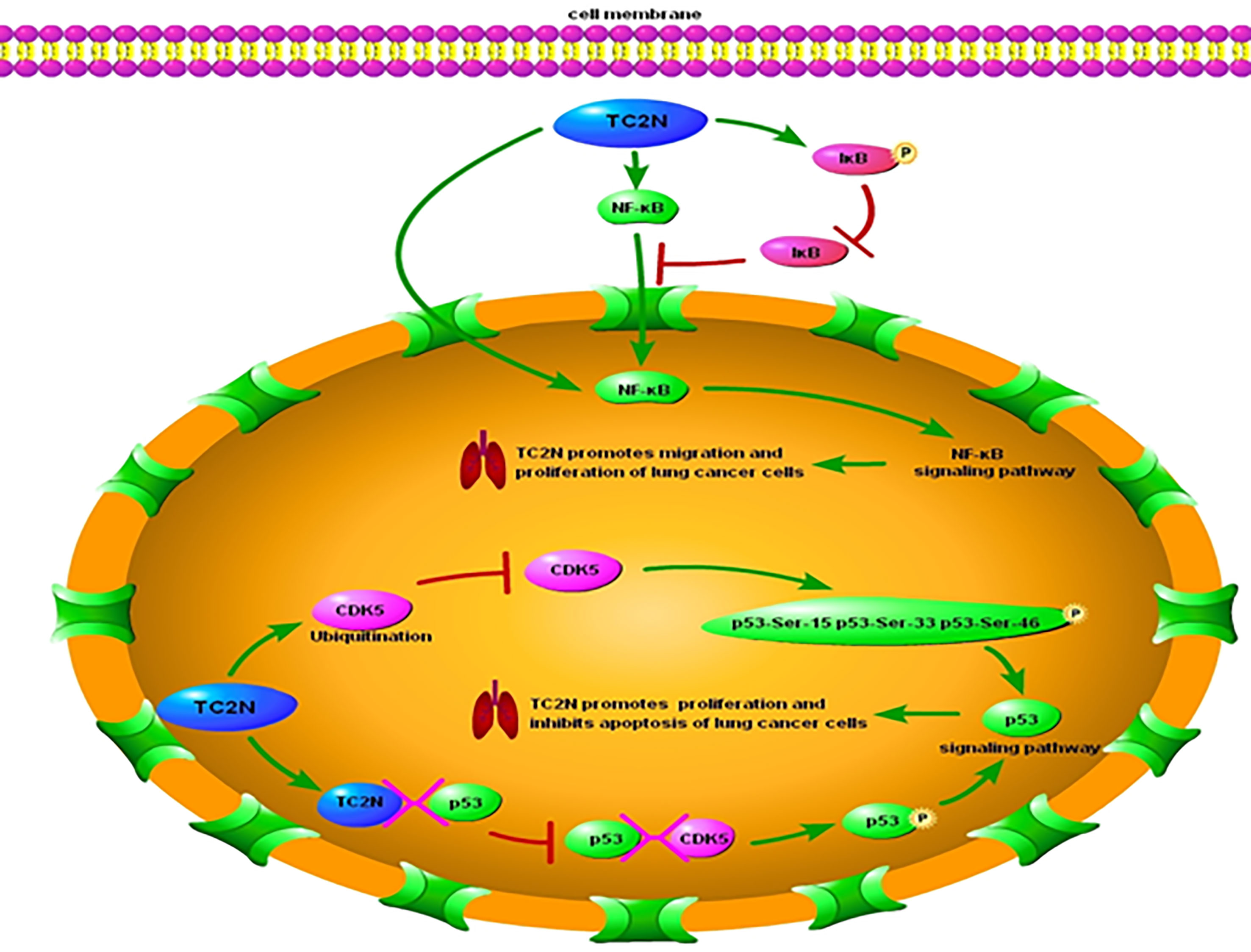
Figure 1 Underlying molecular mechanisms of TC2N in p53 and NF-κB signaling pathways in lung cancer. TC2N, tandem C2 domain nuclear protein; CDK5, cyclin-dependent kinase 5; P53, tumor protein p53; IκB, inhibitor of NF-κB; NF-κB, nuclear factor kappa-light-chain-enhancer of activated B cells.
2.1.2.2 TC2N Promotes NF-κB Signaling Pathway in Lung Cancer
In addition to suppressing the p53 pathway, TCN2 was observed to affect another key signaling pathway in lung cancer cells (41). Over 30 years ago, Sen et al. (84) identified a protein that bound to a specific, conserved DNA sequence in the nucleus of activated B lymphocytes. The protein was named after the identified cell type and the gene it affected: nuclear factor binding near the k light-chain gene in B cells (NF-kB) (85). Since its discovery, NF-κB has been found to be involved in several key processes such as immune regulation, inflammation, cell survival, stress response, embryogenesis, differentiation, proliferation, and cell death (86–96) and it functions primarily by orchestrating the expression of many functionally diverse genes (85, 89, 93, 97, 98). Due to its extensive physiological effects, dysregulation of NF-κB can lead to severe consequences (99, 100), including cancer, neurodegenerative diseases, autoimmune diseases, cardiovascular diseases, and diabetes (85, 99–105).
Most lymphatic or solid tumors, including lung cancer, present with increased NF-κB levels (106). NF-κB in the nucleus is an indicator of active NF-κB signaling, and its levels correlate with the transcription of its target genes (107). Typically, NF-κB levels in the nucleus and its activity are regulated by inhibitor of NF-κB (IκB). IκB acts as a gatekeeper, limiting NF-κB migration into the nucleus by masking its nuclear localization domains (108, 109). Additionally, it prevents activation of NF-κB target genes by masking the DNA-binding domains of NF-κB (108), thereby leading to interruption of the NF-κB signaling pathway. Notably, overexpression of NF-κB—both in the nucleus and cytoplasm of lung cancer cells—correlated with increased expression level of TC2N in these cells (41). Hao XL et al. (41) proposed that this increase in NF-κB expression level is a direct consequence of TC2N overexpression in these cells (Figure 1). Overexpression of TC2N enhanced the phosphorylation of IκB but decreased the total IκB protein levels, leading to increased nuclear translocation of NF-κB and subsequent activation of the signaling pathway (41). Additionally, TC2N modulates this process through other downstream proteins in the pathway such as MMP7 and MMP9 (41).
2.2 Breast Cancer
2.2.1 Functional Characteristics and Clinical Features of TC2N in Breast Cancer
Similar to that in lung cancer tissues, the expression of TC2N was markedly upregulated in breast cancer tissues compared with that in adjacent non-cancerous tissues (37). However, unlike that in lung cancer, upregulated TC2N was associated with good prognosis and overall survival. It positively correlated with the early clinical stage of disease, small tumor size, low lymph node metastasis, and high human epidermal growth factor receptor 2 (HER-2) positivity rate. Additionally, upregulated TC2N inhibited the proliferative and colony-forming abilities of breast cells both in vitro and in vivo. In summary, in contrast to its role in lung cancer cells, TC2N is a potential tumor suppressor in breast cancer.
2.2.2 Signaling Pathways Influenced by TC2N in Breast Cancer
2.2.2.1 TC2N Inhibits PI3K/AKT Signaling Pathway in Breast Cancer
To explain the tumor suppressive function of TC2N in breast cancer cells, Hao XL et al. (37) proposed that upregulation of TC2N represses the Phosphoinositide 3-kinases/serine-threonine kinase (PI3K/AKT) signaling pathway, which is typically constitutively active in some human cancers (110, 111). PI3K/AKT is a growth-regulating cellular pathway and it is well established that PI3K/AKT signaling enhances tumor cell survival, proliferation, and motility in different tumor types (112–119). PI3Ks form a family of kinases that are expressed in almost all mammalian cells and play essential roles in survival, migration, cell cycle progression, and cell growth (120). PI3K phosphorylates phosphatidylinositol to form inositol lipid, which functions as a second messenger in the human body (121). Similarly, AKT is involved in various physiological processes and is a key regulatory protein for cell growth, survival, metabolism, and proliferation (116, 122–124). The pathogenesis of a variety of human cancers is associated with aberrant regulation of the PI3K/AKT pathway (125–127).
Anaplastic Lymphoma Kinase (ALK) is an activator of the PI3K/AKT signaling pathway, and it induces phosphorylation of the p55γ subunit of PI3K in cancer cells, rather than the usual p85 subunit that is phosphorylated (128, 129). It has been shown that the interaction between ALK and p55γ is crucial for ALK-induced p55γ phosphorylation (128) and subsequent activation of the PI3K/AKT signaling pathway. Another key regulatory step in the activation of PI3K/AKT signaling is phosphorylation of AKT by ErbB-3 binding protein 1 (EBP1). TC2N targets both these key steps to inhibit the PI3K/AKT signaling pathway. TC2N forms a complex with ALK, which prevents the ALK-p55γ interaction and therefore inhibits downstream AKT phosphorylation and consequently the PI3K-AKT signaling pathway (Figure 2). Additionally, TC2N inhibits the interaction of EBP1 with AKT, which is necessary for phosphorylation of AKT (130, 131) and subsequent PI3K/AKT signaling. Upregulation of TC2N has also been shown to activate AKT inhibitors such as caspase-3 and block AKT inhibitors such as GSK3β, MYC, BAD, and PTEN.
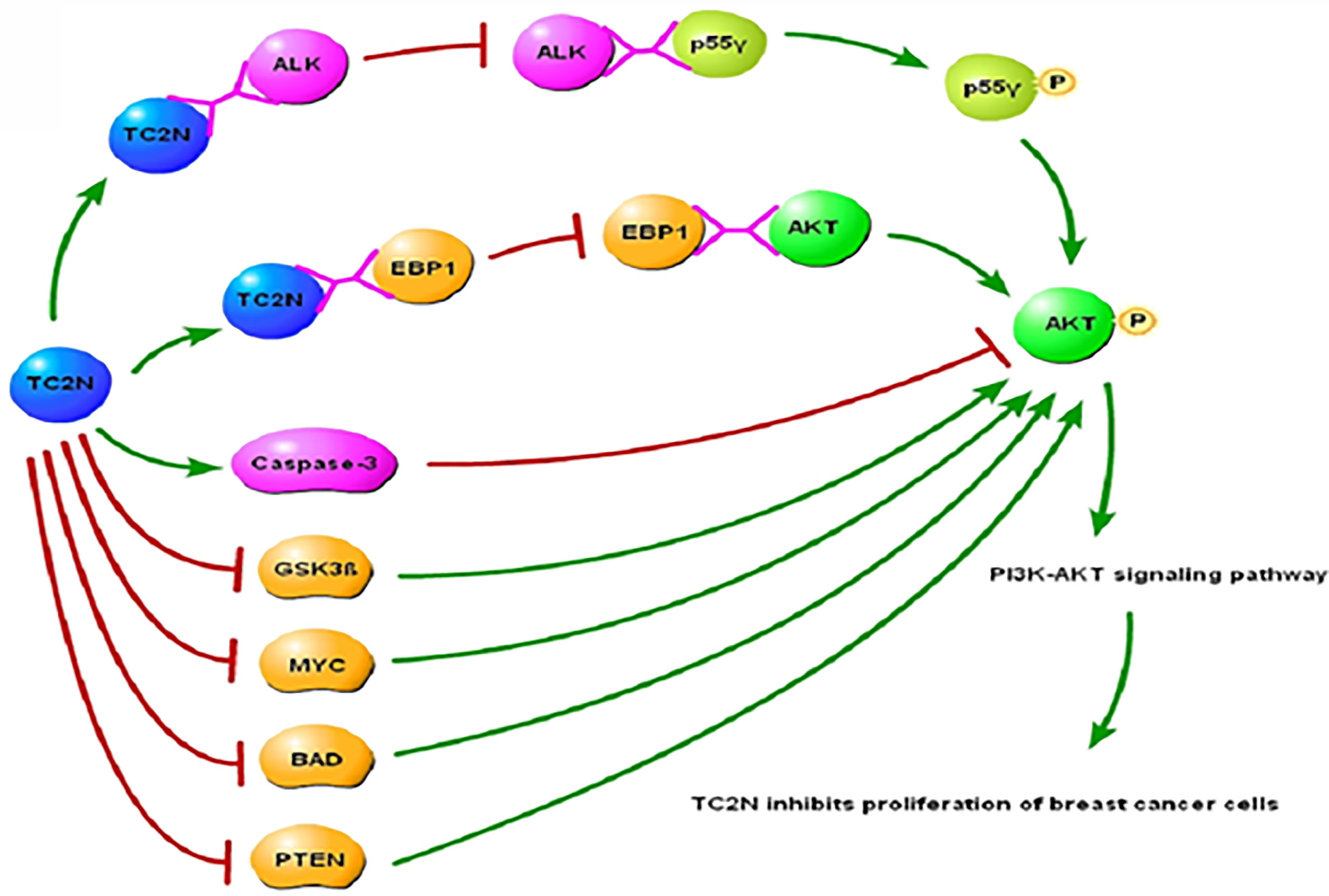
Figure 2 Underlying molecular mechanisms of TC2N in PI3K-AKT signaling pathway in breast cancer. TC2N, tandem C2 domain nuclear protein; ALK, anaplastic lymphoma kinase; EBP1, ErbB-3 binding protein 1; AKT, serine threonine kinase; GSK3β, glycogen synthase kinase-3β; BAD, Bcl-2-associated death promoter; PTEN, phosphatase and tensin homolog deleted on chromosome 10.
2.3 Gastric Cancer
2.3.1 Functional Characteristics and Clinical Features of TC2N in Gastric Cancer
Similar to lung and breast cancer cells, TC2N is overexpressed in a variety of gastric cancer cell lines and tumor samples compared to normal cells and tissues (38, 42). High TC2N levels were significantly correlated with poorly differentiated histological classification, large tumor size, advanced TNM stage, and advanced distant metastasis. Furthermore, patients with high TC2N expression showed poorer prognosis regardless of TNM stage compared to patients with low TC2N expression. In vitro, TC2N knockdown significantly inhibited the proliferation of gastric cancer cells, while TC2N overexpression promoted the growth of these cells. Similar results were observed in vivo where downregulation of TC2N inhibited the migration and invasion of gastric cancer cells, whereas overexpression had the opposite effect. Thus, similar to lung cancer, TC2N potentially functions as an oncogene in gastric cancer.
2.3.2 Signaling Pathway of TC2N in Gastric Cancer
The mechanism of action of TC2N in gastric cancer remains unclear. Shen L et al. (42) suggested that TC2N might partially affect the migration and invasion ability of gastric cancer by regulating the expression levels of MMP2 and MMP9. MMP2 and MMP9 are known to be involved in cell invasion and tumor metastasis (132). TC2N expression also showed strong positive correlation with the expression of CATSPERB and other cancer related genes such as GALNT3 and RBM47 (38). However, the detailed molecular mechanism by which TC2N promotes gastric cancer progression needs further evaluation.
3 Conclusion and Future Perspectives
High-throughput gene expression profiling facilitates the simultaneous measurement of the expression levels of thousands of genes. A key application of gene expression profiling in cancer is to identify differences in gene expression patterns between tumor and control samples (133). Advances in technology and the declining costs of DNA sequencing have spurred global efforts to discover differentially expressed genes in various cancers. From one such effort, TC2N was found to be widely upregulated in several human cancers, including lung, breast, and gastric cancers. TC2N levels were correlated with multiple clinicopathological features and prognosis, such as TNM stage, histological grade, tumor size, overall survival, lymph node metastasis, and distant metastasis. In support of its involvement in tumorigenesis and tumor progression, in vitro and in vivo experiments have shown that TC2N affects proliferation, apoptosis, migration, invasion of tumor cells and tumor growth in many cancers. The underlying molecular mechanisms of TC2N in several cancers have also been preliminarily explored and suggest that TC2N modulates several key signaling pathways that influence carcinogenesis and cancer progression, including p53, NF-κB, and PI3K/AKT signaling pathways.
Although TC2N is a potential therapeutic target, several questions remain to be addressed. First, the molecular mechanism of TC2N in different types of cancers is not completely understood. For example, while preliminary data suggest that in gastric cancer TC2N modulates the expression of several cancer related genes, the specific pathway affected by TC2N is unclear. Furthermore, while the function of TC2N in lung, breast and gastric cancer have been studied to some extent, its potential role in other cancers, such as cancers associated with the urinary and reproductive systems remain unexplored. Second, while TC2N is upregulated in tumor tissues of some specific cancers, it is not known if TC2N is also upregulated in body fluids such as plasma and urine. The identification of diagnostic biomarkers is a promising avenue for early cancer diagnosis. If TC2N is detectable in plasma or urine, it may facilitate early detection and prognosis assessment using simple and non-invasive tests. Third, it is unknown if TC2N is a tumor-associated antigen and requires further evaluation. Fourth, TC2N is an immune system associated gene, but whether it can serve as a target in personalized immunotherapies remains to be seen. Therefore, more attention should be paid to the clinical value of TC2N in cancer diagnosis and treatment.
In summary, TC2N has been shown to have oncogenic or tumor-suppressive functions in different types of cancers, and could potentially serve as a cancer-specific molecular biomarker for early diagnosis, treatment, and prognosis assessment. While some progress has been made in the mechanistic analysis of TC2N, several questions remain unanswered. Future work needs to focus on understanding the precise molecular mechanism of TC2N in carcinogenesis and tumor progression to explore the potential clinical application of TC2N.
Author Contributions
HL and TW contributed to the conception and design of the review. HL wrote the first draft of the manuscript. BL revised the manuscript. All authors contributed to the article and approved the submitted version
Funding
This work was supported by National Key R&D Program of China (Grant #2018YFC1311600), Jilin Province Financial and Health Project (Research on the Differential Expression of Cyclic RNA in Cervical Cancer HeLa Cells and the Mechanism of Radiation Resistance under Radiation Induction); Jilin Provincial Department of Finance (Consortium of Medical Consortium for Diagnosis and Treatment of Difficult Women’s Tumors and Precision Radiotherapy Training Base Construction Project); Jilin Province Medical and Health Talent Special Project (2019SCZT010); National Key Clinical Specialty Capacity Building Project (Application of Non-coplanar 3D Printing and Intertissue Interpolation Technology in Improving the Diagnosis and Treatment Ability of Recurrent and Refractory Cervical Cancer).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
TC2N, tandem C2 domains nuclear protein; TAAs, tumor-associated antigens; CDK5, cyclin-dependent kinase 5; P21(CDKN1A) , cyclin dependent kinase inhibitor 1A; P53, tumor protein p53; BAX, a member of the B-cell lymphoma-2(BCL2) gene family; BCL1, B-cell lymphoma-1; IκB, inhibitor of NF-κB; NF-κB, nuclear factor kappa-light-chain-enhancer of activated B cells; MMP7, matrix metalloproteinase 7; MMP9, matrix metalloproteinase 9; ALK, anaplastic lymphoma kinase; EBP1, ErbB-3 binding protein 1; AKT, serine threonine kinase; GSK3β, glycogen synthase kinase-3β; PTEN, phosphatase and tensin homolog deleted on Chromosome 10; MMP2, matrix metalloproteinase 2; GALNT3, polypeptide N-acetylgalactosaminyltransferase 3; RBM47, RNA binding motif protein 47.
References
1. Galván Morales MA, Barrera Rodríguez R, Santiago Cruz JR, Teran LM. Overview of New Treatments With Immunotherapy for Breast Cancer and a Proposal of a Combination Therapy. Molecules (2020) 25(23):5686. doi: 10.3390/molecules25235686
2. Fitzgerald J, Higgins D, Mazo Vargas C, Watson W, Mooney C, Rahman A, et al. Future of Biomarker Evaluation in the Realm of Artificial Intelligence Algorithms: Application in Improved Therapeutic Stratification of Patients With Breast and Prostate Cancer. J Clin Pathol (2021) 74(7):429–34. doi: 10.1136/jclinpath-2020-207351
3. Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, et al. Cancer Treatment and Survivorship Statistics, 2016. CA: Cancer J Clin (2016) 66(4):271–89. doi: 10.3322/caac.21349
4. Nakashima L. Evolution of Cancer Treatment and Evolving Challenges. Healthcare Manage Forum (2018) 31(1):26–8. doi: 10.1177/0840470417722568
5. Kim Y, Kim DH. CpG Island Hypermethylation as a Biomarker for the Early Detection of Lung Cancer. Methods Mol Biol (2015) 1238:141–71. doi: 10.1007/978-1-4939-1804-1_8
6. Koinis F, Kotsakis A, Georgoulias V. Small Cell Lung Cancer (SCLC): No Treatment Advances in Recent Years. Trans Lung Cancer Res (2016) 5(1):39–50. doi: 10.3978/j.issn.2218-6751.2016.01.03
7. Vijayvergia N, Shah PC, Denlinger CS. Survivorship in Non-Small Cell Lung Cancer: Challenges Faced and Steps Forward. J Natl Compr Canc Netw (2015) 13(9):1151–61. doi: 10.6004/jnccn.2015.0140
8. Henley SJ, Ward EM, Scott S, Ma J, Anderson RN, Firth AU, et al. Annual Report to the Nation on the Status of Cancer, Part I: National Cancer Statistics. Cancer (2020) 126(10):2225–49. doi: 10.1002/cncr.32802
9. Chabner BA, Roberts TG Jr. Timeline: Chemotherapy and the War on Cancer. Nat Rev Cancer (2005) 5(1):65–72. doi: 10.1038/nrc1529
10. Etzioni R, Urban N, Ramsey S, McIntosh M, Schwartz S, Reid B, et al. The Case for Early Detection. Nat Rev Cancer (2003) 3(4):243–52. doi: 10.1038/nrc1041
11. Huff CA, Matsui W, Smith BD, Jones RJ. The Paradox of Response and Survival in Cancer Therapeutics. Blood (2006) 107(2):431–4. doi: 10.1182/blood-2005-06-2517
12. Buskwofie A, David-West G, Clare CA. A Review of Cervical Cancer: Incidence and Disparities. J Natl Med Assoc (2020) 112(2):229–32. doi: 10.1016/j.jnma.2020.03.002
13. Smith JS, Lindsay L, Hoots B, Keys J, Franceschi S, Winer R, et al. Human Papillomavirus Type Distribution in Invasive Cervical Cancer and High-Grade Cervical Lesions: A Meta-Analysis Update. Int J Cancer (2007) 121(3):621–32. doi: 10.1002/ijc.22527
14. Zheng X, Li X, Wang X. Extracellular Vesicle-Based Liquid Biopsy Holds Great Promise for the Management of Ovarian Cancer. Biochim Biophys Acta Rev Cancer (2020) 1874(1):188395. doi: 10.1016/j.bbcan.2020.188395
15. Zitvogel L, Galluzzi L, Smyth MJ, Kroemer G. Mechanism of Action of Conventional and Targeted Anticancer Therapies: Reinstating Immunosurveillance. Immunity (2013) 39(1):74–88. doi: 10.1016/j.immuni.2013.06.014
16. Quigley DA, Kristensen V. Predicting Prognosis and Therapeutic Response From Interactions Between Lymphocytes and Tumor Cells. Mol Oncol (2015) 9(10):2054–62. doi: 10.1016/j.molonc.2015.10.003
17. Menon H, Ramapriyan R, Cushman TR, Verma V, Kim HH, Schoenhals JE, et al. Role of Radiation Therapy in Modulation of the Tumor Stroma and Microenvironment. Front Immunol (2019) 10:193. doi: 10.3389/fimmu.2019.00193
18. Durgin JS, Weiner DM, Wysocka M, Rook AH. The Immunopathogenesis and Immunotherapy of Cutaneous T Cell Lymphoma: Pathways and Targets for Immune Restoration and Tumor Eradication. J Am Acad Dermatol (2021) 84(3):587–95. doi: 10.1016/j.jaad.2020.12.027
19. Vesely MD, Kershaw MH, Schreiber RD, Smyth MJ. Natural Innate and Adaptive Immunity to Cancer. Annu Rev Immunol (2011) 29:235–71. doi: 10.1146/annurev-immunol-031210-101324
20. Vysotskaia V, Kaseniit KE, Bucheit L, Ready K, Price K, Johansen Taber K. Clinical Utility of Hereditary Cancer Panel Testing: Impact of PALB2, ATM, CHEK2, NBN, BRIP1, RAD51C, and RAD51D Results on Patient Management and Adherence to Provider Recommendations. Cancer (2020) 126(3):549–58. doi: 10.1002/cncr.32572
21. Shareefi G, Turkistani AN, Alsayyah A, Kussaibi H, Abdel Hadi M, Alkharsah KR. Pathway-Affecting Single Nucleotide Polymorphisms (SNPs) in RPS6KA1 and MBIP Genes Are Associated With Breast Cancer Risk. Asian Pac J Cancer Prev (2020) 21(7):2163–8. doi: 10.31557/apjcp.2020.21.7.2163
22. Xiong Z, Wu J, Sun Y, Bai M, Niu F, Jin T. Variants in Multiple Genes Are Associated With Esophageal Cancer Risk in a Chinese Han Population: A Case-Control Study. J Gene Med (2020) 22(12):e3266. doi: 10.1002/jgm.3266
23. Khan RT, Siddique A, Shahid N, Khokher S, Fatima W. Breast Cancer Risk Associated With Genes Encoding DNA Repair MRN Complex: A Study From Punjab, Pakistan. Breast Cancer (2018) 25(3):350–5. doi: 10.1007/s12282-018-0837-9
24. Elek Z, Kovács Z, Keszler G, Szabó M, Csanky E, Luo J, et al. High Throughput Multiplex SNP-Analysis in Chronic Obstructive Pulmonary Disease and Lung Cancer. Curr Mol Med (2020) 20(3):185–93. doi: 10.2174/1566524019666191017123446
25. Chin L, Tam A, Pomerantz J, Wong M, Holash J, Bardeesy N, et al. Essential Role for Oncogenic Ras in Tumour Maintenance. Nature (1999) 400(6743):468–72. doi: 10.1038/22788
26. Huettner CS, Zhang P, Van Etten RA, Tenen DG. Reversibility of Acute B-Cell Leukaemia Induced by BCR-Abl1. Nat Genet (2000) 24(1):57–60. doi: 10.1038/71691
27. Boxer RB, Jang JW, Sintasath L, Chodosh LA. Lack of Sustained Regression of C-MYC-Induced Mammary Adenocarcinomas Following Brief or Prolonged MYC Inactivation. Cancer Cell (2004) 6(6):577–86. doi: 10.1016/j.ccr.2004.10.013
28. Weinstein IB. Cancer. Addiction to Oncogenes–the Achilles Heal of Cancer. Sci (New York NY) (2002) 297(5578):63–4. doi: 10.1126/science.1073096
29. Witek Ł, Janikowski T, Gabriel I, Bodzek P, Olejek A. Analysis of microRNA Regulating Cell Cycle-Related Tumor Suppressor Genes in Endometrial Cancer Patients. Hum Cell (2021) 34(2):564–9. doi: 10.1007/s13577-020-00451-6
30. Duncan RR, Shipston MJ, Chow RH. Double C2 Protein. A Review. Biochimie (2000) 82(5):421–6. doi: 10.1016/s0300-9084(00)00214-5
31. Corbalan-Garcia S, Gómez-Fernández JC. Signaling Through C2 Domains: More Than One Lipid Target. Biochim Biophys Acta (2014) 1838(6):1536–47. doi: 10.1016/j.bbamem.2014.01.008
32. Cho W, Stahelin RV. Membrane Binding and Subcellular Targeting of C2 Domains. Biochim Biophys Acta (2006) 1761(8):838–49. doi: 10.1016/j.bbalip.2006.06.014
33. Farah CA, Sossin WS. The Role of C2 Domains in PKC Signaling. Adv Exp Med Biol (2012) 740:663–83. doi: 10.1007/978-94-007-2888-2_29
34. Kabekkodu SP, Bhat S, Radhakrishnan R, Aithal A, Mascarenhas R, Pandey D, et al. DNA Promoter Methylation-Dependent Transcription of the Double C2-Like Domain β (DOC2B) Gene Regulates Tumor Growth in Human Cervical Cancer. J Biol Chem (2014) 289(15):10637–49. doi: 10.1074/jbc.M113.491506
35. Blomme A, Costanza B, de Tullio P, Thiry M, Van Simaeys G, Boutry S, et al. Myoferlin Regulates Cellular Lipid Metabolism and Promotes Metastases in Triple-Negative Breast Cancer. Oncogene (2017) 36(15):2116–30. doi: 10.1038/onc.2016.369
36. Hao XL, Han F, Zhang N, Chen HQ, Jiang X, Yin L, et al. TC2N, a Novel Oncogene, Accelerates Tumor Progression by Suppressing P53 Signaling Pathway in Lung Cancer. Cell Death Differ (2019) 26(7):1235–50. doi: 10.1038/s41418-018-0202-8
37. Hao XL, Gao LY, Deng XJ, Han F, Chen HQ, Jiang X, et al. Identification of TC2N as a Novel Promising Suppressor of PI3K-AKT Signaling in Breast Cancer. Cell Death Dis (2019) 10(6):424. doi: 10.1038/s41419-019-1663-5
38. Xu J, Ou X, Li J, Cai Q, Sun K, Ye J, et al. Overexpression of TC2N Is Associated With Poor Prognosis in Gastric Cancer. J Cancer (2021) 12(3):807–17. doi: 10.7150/jca.50653
39. Fukuda M, Mikoshiba K. Tac2-N, An Atypical C-Type Tandem C2 Protein Localized in the Nucleus. FEBS Lett (2001) 503(2-3):217–8. doi: 10.1016/s0014-5793(01)02738-7
40. Prokop JW, Hartog NL, Chesla D, Faber W, Love CP, Karam R, et al. High-Density Blood Transcriptomics Reveals Precision Immune Signatures of SARS-CoV-2 Infection in Hospitalized Individuals. Front Immunol (2021) 12:694243. doi: 10.3389/fimmu.2021.694243
41. Hao X, Gao LY, Zhang N, Chen H, Jiang X, Liu W, et al. Tac2-N Acts as a Novel Oncogene and Promotes Tumor Metastasis via Activation of NF-κb Signaling in Lung Cancer. J Exp Clin Cancer Res CR (2019) 38(1):319. doi: 10.1186/s13046-019-1316-7
42. Shen L, Zhang P, Wang J, Ji P. Tac2-N Serves an Oncogenic Role and Promotes Drug Resistance in Human Gastric Cancer Cells. Exp Ther Med (2020) 20(5):113. doi: 10.3892/etm.2020.9241
43. Eliyahu D, Raz A, Gruss P, Givol D, Oren M. Participation of P53 Cellular Tumour Antigen in Transformation of Normal Embryonic Cells. Nature (1984) 312(5995):646–9. doi: 10.1038/312646a0
44. Jenkins JR, Rudge K, Currie GA. Cellular Immortalization by a cDNA Clone Encoding the Transformation-Associated Phosphoprotein P53. Nature (1984) 312(5995):651–4. doi: 10.1038/312651a0
45. Parada LF, Land H, Weinberg RA, Wolf D, Rotter V. Cooperation Between Gene Encoding P53 Tumour Antigen and Ras in Cellular Transformation. Nature (1984) 312(5995):649–51. doi: 10.1038/312649a0
46. Wolf D, Harris N, Rotter V. Reconstitution of P53 Expression in a Nonproducer Ab-MuLV-Transformed Cell Line by Transfection of a Functional P53 Gene. Cell (1984) 38(1):119–26. doi: 10.1016/0092-8674(84)90532-4
47. Baker SJ, Fearon ER, Nigro JM, Hamilton SR, Preisinger AC, Jessup JM, et al. Chromosome 17 Deletions and P53 Gene Mutations in Colorectal Carcinomas. Sci (New York NY) (1989) 244(4901):217–21. doi: 10.1126/science.2649981
48. Eliyahu D, Michalovitz D, Eliyahu S, Pinhasi-Kimhi O, Oren M. Wild-Type P53 can Inhibit Oncogene-Mediated Focus Formation. Proc Natl Acad Sci USA (1989) 86(22):8763–7. doi: 10.1073/pnas.86.22.8763
49. Finlay CA, Hinds PW, Levine AJ. The P53 Proto-Oncogene can Act as a Suppressor of Transformation. Cell (1989) 57(7):1083–93. doi: 10.1016/0092-8674(89)90045-7
50. Malkin D, Li FP, Strong LC, Fraumeni JF Jr, Nelson CE, Kim DH, et al. Germ Line P53 Mutations in a Familial Syndrome of Breast Cancer, Sarcomas, and Other Neoplasms. Sci (New York NY) (1990) 250(4985):1233–8. doi: 10.1126/science.1978757
51. Michalovitz D, Halevy O, Oren M. Conditional Inhibition of Transformation and of Cell Proliferation by a Temperature-Sensitive Mutant of P53. Cell (1990) 62(4):671–80. doi: 10.1016/0092-8674(90)90113-s
52. Srivastava S, Zou ZQ, Pirollo K, Blattner W, Chang EH. Germ-Line Transmission of a Mutated P53 Gene in a Cancer-Prone Family With Li-Fraumeni Syndrome. Nature (1990) 348(6303):747–9. doi: 10.1038/348747a0
53. Hollstein M, Sidransky D, Vogelstein B, Harris CC. P53 Mutations in Human Cancers. Sci (New York NY) (1991) 253(5015):49–53. doi: 10.1126/science.1905840
54. Stein Y, Rotter V, Aloni-Grinstein R. Gain-Of-Function Mutant P53: All the Roads Lead to Tumorigenesis. Int J Mol Sci (2019) 20(24):6197. doi: 10.3390/ijms20246197
55. May P, May E. Twenty Years of P53 Research: Structural and Functional Aspects of the P53 Protein. Oncogene (1999) 18(53):7621–36. doi: 10.1038/sj.onc.1203285
56. Harris SL, Levine AJ. The P53 Pathway: Positive and Negative Feedback Loops. Oncogene (2005) 24(17):2899–908. doi: 10.1038/sj.onc.1208615
57. Molchadsky A, Rivlin N, Brosh R, Rotter V, Sarig R. P53 Is Balancing Development, Differentiation and De-Differentiation to Assure Cancer Prevention. Carcinogenesis (2010) 31(9):1501–8. doi: 10.1093/carcin/bgq101
58. Bieging KT, Mello SS, Attardi LD. Unravelling Mechanisms of P53-Mediated Tumour Suppression. Nat Rev Cancer (2014) 14(5):359–70. doi: 10.1038/nrc3711
59. Luo Q, Beaver JM, Liu Y, Zhang Z. Dynamics of P53: A Master Decider of Cell Fate. Genes (2017) 8(2):178. doi: 10.3390/genes8020066
60. Wynford-Thomas D. P53: Guardian of Cellular Senescence. J Pathol (1996) 180(2):118–21. doi: 10.1002/(sici)1096-9896(199610)180:2<118::Aid-path673>3.0.Co;2-i
61. Gao Y, Ferguson DO, Xie W, Manis JP, Sekiguchi J, Frank KM, et al. Interplay of P53 and DNA-Repair Protein XRCC4 in Tumorigenesis, Genomic Stability and Development. Nature (2000) 404(6780):897–900. doi: 10.1038/35009138
62. Ryan KM. P53 and Autophagy in Cancer: Guardian of the Genome Meets Guardian of the Proteome. Eur J Cancer (Oxford Engl 1990) (2011) 47(1):44–50. doi: 10.1016/j.ejca.2010.10.020
63. Wang SJ, Ou Y, Jiang L, Gu W. Ferroptosis: A Missing Puzzle Piece in the P53 Blueprint? Mol Cell Oncol (2016) 3(3):e1046581. doi: 10.1080/23723556.2015.1046581
64. Lane D, Levine A. P53 Research: The Past Thirty Years and the Next Thirty Years. Cold Spring Harbor Perspect Biol (2010) 2(12):a000893. doi: 10.1101/cshperspect.a000893
65. Levav-Cohen Y, Goldberg Z, Tan KH, Alsheich-Bartok O, Zuckerman V, Haupt S, et al. The P53-Mdm2 Loop: A Critical Juncture of Stress Response. Sub-Cell Biochem (2014) 85:161–86. doi: 10.1007/978-94-017-9211-0_9
66. Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of P53. Cell (2009) 137(3):413–31. doi: 10.1016/j.cell.2009.04.037
67. Charni M, Aloni-Grinstein R, Molchadsky A, Rotter V. P53 on the Crossroad Between Regeneration and Cancer. Cell Death Differ (2017) 24(1):8–14. doi: 10.1038/cdd.2016.117
68. Labuschagne CF, Zani F, Vousden KH. Control of Metabolism by P53 - Cancer and Beyond. Biochim Biophys Acta Rev Cancer (2018) 1870(1):32–42. doi: 10.1016/j.bbcan.2018.06.001
69. Goldstein I, Rotter V. Regulation of Lipid Metabolism by P53 - Fighting Two Villains With One Sword. Trends Endocrinol Metab: TEM (2012) 23(11):567–75. doi: 10.1016/j.tem.2012.06.007
70. Aloni-Grinstein R, Charni-Natan M, Solomon H, Rotter V. P53 and the Viral Connection: Back Into the Future (‡). Cancers (2018) 10(6):178. doi: 10.3390/cancers10060178
71. Charni M, Rivlin N, Molchadsky A, Aloni-Grinstein R, Rotter V. P53 in Liver Pathologies-Taking the Good With the Bad. J Mol Med (Berlin Germany) (2014) 92(12):1229–34. doi: 10.1007/s00109-014-1223-5
72. Yan Z, Miao X, Zhang B, Xie J. P53 as a Double-Edged Sword in the Progression of non-Alcoholic Fatty Liver Disease. Life Sci (2018) 215:64–72. doi: 10.1016/j.lfs.2018.10.051
73. Aloni-Grinstein R, Shetzer Y, Kaufman T, Rotter V. P53: The Barrier to Cancer Stem Cell Formation. FEBS Lett (2014) 588(16):2580–9. doi: 10.1016/j.febslet.2014.02.011
74. Molchadsky A, Rotter V. P53 and its Mutants on the Slippery Road From Stemness to Carcinogenesis. Carcinogenesis (2017) 38(4):347–58. doi: 10.1093/carcin/bgw092
75. Charni-Natan M, Aloni-Grinstein R, Osher E, Rotter V. Liver and Steroid Hormones-Can a Touch of P53 Make a Difference? Front Endocrinol (2019) 10:374. doi: 10.3389/fendo.2019.00374
76. Koifman G, Aloni-Grinstein R, Rotter V. P53 Balances Between Tissue Hierarchy and Anarchy. J Mol Cell Biol (2019) 11(7):553–63. doi: 10.1093/jmcb/mjz022
77. Liu G, Chen X. Regulation of the P53 Transcriptional Activity. J Cell Biochem (2006) 97(3):448–58. doi: 10.1002/jcb.20700
78. Zhang J, Krishnamurthy PK, Johnson GV. Cdk5 Phosphorylates P53 and Regulates its Activity. J Neurochem (2002) 81(2):307–13. doi: 10.1046/j.1471-4159.2002.00824.x
79. Lee JH, Kim HS, Lee SJ, Kim KT. Stabilization and Activation of P53 Induced by Cdk5 Contributes to Neuronal Cell Death. J Cell Sci (2007) 120(Pt 13):2259–71. doi: 10.1242/jcs.03468
80. Vogelstein B, Lane D, Levine AJ. Surfing the P53 Network. Nature (2000) 408(6810):307–10. doi: 10.1038/35042675
81. Georgakilas AG, Martin OA, Bonner WM. P21: A Two-Faced Genome Guardian. Trends Mol Med (2017) 23(4):310–9. doi: 10.1016/j.molmed.2017.02.001
82. Faria MH, Patrocínio RM, Moraes Filho MO, Rabenhorst SH. Immunoexpression of Tumor Suppressor Genes P53, P21 WAF1/CIP1 and P27 KIP1 in Humam Astrocystic Tumors. Arq Neuro Psiquiatr (2007) 65(4b):1114–22. doi: 10.1590/s0004-282x2007000700004
83. Gottlieb TM, Oren M. P53 and Apoptosis. Semin Cancer Biol (1998) 8(5):359–68. doi: 10.1006/scbi.1998.0098
84. Sen R, Baltimore D. Multiple Nuclear Factors Interact With the Immunoglobulin Enhancer Sequences. Cell (1986) 46(5):705–16. doi: 10.1016/0092-8674(86)90346-6
85. Zhang Q, Lenardo MJ, Baltimore D. 30 Years of NF-κb: A Blossoming of Relevance to Human Pathobiology. Cell (2017) 168(1-2):37–57. doi: 10.1016/j.cell.2016.12.012
86. Tong L, Tergaonkar V. Rho Protein GTPases and Their Interactions With Nfκb: Crossroads of Inflammation and Matrix Biology. Biosci Rep (2014) 34(3):283–95. doi: 10.1042/bsr20140021
87. Perkins ND. The Diverse and Complex Roles of NF-κb Subunits in Cancer. Nat Rev Cancer (2012) 12(2):121–32. doi: 10.1038/nrc3204
88. Irelan JT, Murphy TJ, DeJesus PD, Teo H, Xu D, Gomez-Ferreria MA, et al. A Role for IkappaB Kinase 2 in Bipolar Spindle Assembly. Proc Natl Acad Sci USA (2007) 104(43):16940–5. doi: 10.1073/pnas.0706493104
89. Beg AA, Baltimore D. An Essential Role for NF-kappaB in Preventing TNF-Alpha-Induced Cell Death. Sci (New York NY) (1996) 274(5288):782–4. doi: 10.1126/science.274.5288.782
90. Liu ZG, Hsu H, Goeddel DV, Karin M. Dissection of TNF Receptor 1 Effector Functions: JNK Activation Is Not Linked to Apoptosis While NF-kappaB Activation Prevents Cell Death. Cell (1996) 87(3):565–76. doi: 10.1016/s0092-8674(00)81375-6
91. Van Antwerp DJ, Martin SJ, Kafri T, Green DR, Verma IM. Suppression of TNF-Alpha-Induced Apoptosis by NF-kappaB. Sci (New York NY) (1996) 274(5288):787–9. doi: 10.1126/science.274.5288.787
92. Wang CY, Mayo MW, Baldwin AS Jr. TNF- and Cancer Therapy-Induced Apoptosis: Potentiation by Inhibition of NF-kappaB. Sci (New York NY) (1996) 274(5288):784–7. doi: 10.1126/science.274.5288.784
93. Joyce D, Albanese C, Steer J, Fu M, Bouzahzah B, Pestell RG. NF-kappaB and Cell-Cycle Regulation: The Cyclin Connection. Cytokine Growth Factor Rev (2001) 12(1):73–90. doi: 10.1016/s1359-6101(00)00018-6
94. Huang S, Pettaway CA, Uehara H, Bucana CD, Fidler IJ. Blockade of NF-kappaB Activity in Human Prostate Cancer Cells Is Associated With Suppression of Angiogenesis, Invasion, and Metastasis. Oncogene (2001) 20(31):4188–97. doi: 10.1038/sj.onc.1204535
95. Pasparakis M, Luedde T, Schmidt-Supprian M. Dissection of the NF-kappaB Signalling Cascade in Transgenic and Knockout Mice. Cell Death Differ (2006) 13(5):861–72. doi: 10.1038/sj.cdd.4401870
96. Gerondakis S, Grossmann M, Nakamura Y, Pohl T, Grumont R. Genetic Approaches in Mice to Understand Rel/NF-kappaB and IkappaB Function: Transgenics and Knockouts. Oncogene (1999) 18(49):6888–95. doi: 10.1038/sj.onc.1203236
97. Grilli M, Chiu JJ, Lenardo MJ. NF-Kappa B and Rel: Participants in a Multiform Transcriptional Regulatory System. Int Rev Cytol (1993) 143:1–62. doi: 10.1016/s0074-7696(08)61873-2
98. Barnes PJ, Karin M. Nuclear Factor-Kappab: A Pivotal Transcription Factor in Chronic Inflammatory Diseases. New Engl J Med (1997) 336(15):1066–71. doi: 10.1056/nejm199704103361506
99. Courtois G, Gilmore TD. Mutations in the NF-kappaB Signaling Pathway: Implications for Human Disease. Oncogene (2006) 25(51):6831–43. doi: 10.1038/sj.onc.1209939
100. Kadhim HS, Al-Jeboori TI, Tawfik MS. Possible Role of Nuclear Factor kappaB Detected by in Situ Hybridization in the Pathogenesis of Transitional Cell Carcinoma of the Bladder. Le J Med Libanais Lebanese Med J (2006) 54(4):196–9.
101. Taniguchi K, Karin M. NF-κb, Inflammation, Immunity and Cancer: Coming of Age. Nat Rev Immunol (2018) 18(5):309–24. doi: 10.1038/nri.2017.142
102. Hayden MS, Ghosh S. Shared Principles in NF-kappaB Signaling. Cell (2008) 132(3):344–62. doi: 10.1016/j.cell.2008.01.020
103. Vallabhapurapu S, Karin M. Regulation and Function of NF-kappaB Transcription Factors in the Immune System. Annu Rev Immunol (2009) 27:693–733. doi: 10.1146/annurev.immunol.021908.132641
104. Baker RG, Hayden MS, Ghosh S. NF-κb, Inflammation, and Metabolic Disease. Cell Metab (2011) 13(1):11–22. doi: 10.1016/j.cmet.2010.12.008
105. Kumar A, Takada Y, Boriek AM, Aggarwal BB. Nuclear Factor-Kappab: Its Role in Health and Disease. J Mol Med (Berlin Germany) (2004) 82(7):434–48. doi: 10.1007/s00109-004-0555-y
106. Karin M, Cao Y, Greten FR, Li ZW. NF-kappaB in Cancer: From Innocent Bystander to Major Culprit. Nat Rev Cancer (2002) 2(4):301–10. doi: 10.1038/nrc780
107. Chaturvedi MM, Sung B, Yadav VR, Kannappan R, Aggarwal BB. NF-κb Addiction and its Role in Cancer: 'One Size Does Not Fit All'. Oncogene (2011) 30(14):1615–30. doi: 10.1038/onc.2010.566
108. DiDonato JA, Mercurio F, Karin M. NF-κb and the Link Between Inflammation and Cancer. Immunol Rev (2012) 246(1):379–400. doi: 10.1111/j.1600-065X.2012.01099.x
109. Hayden MS, Ghosh S. NF-κb, the First Quarter-Century: Remarkable Progress and Outstanding Questions. Genes Dev (2012) 26(3):203–34. doi: 10.1101/gad.183434.111
110. Uddin S, Hussain AR, Al-Hussein KA, Manogaran PS, Wickrema A, Gutierrez MI, et al. Inhibition of Phosphatidylinositol 3'-Kinase/AKT Signaling Promotes Apoptosis of Primary Effusion Lymphoma Cells. Clin Cancer Res Off J Am Assoc Cancer Res (2005) 11(8):3102–8. doi: 10.1158/1078-0432.Ccr-04-1857
111. Koul D, Shen R, Kim YW, Kondo Y, Lu Y, Bankson J, et al. Cellular and In Vivo Activity of a Novel PI3K Inhibitor, PX-866, Against Human Glioblastoma. Neuro-oncology (2010) 12(6):559–69. doi: 10.1093/neuonc/nop058
112. Yu JS, Cui W. Proliferation, Survival and Metabolism: The Role of PI3K/AKT/mTOR Signalling in Pluripotency and Cell Fate Determination. Dev (Cambridge England) (2016) 143(17):3050–60. doi: 10.1242/dev.137075
113. Wee P, Wang Z. Epidermal Growth Factor Receptor Cell Proliferation Signaling Pathways. Cancers (2017) 9(5):52. doi: 10.3390/cancers9050052
114. Porta C, Paglino C, Mosca A. Targeting PI3K/Akt/mTOR Signaling in Cancer. Front Oncol (2014) 4:64. doi: 10.3389/fonc.2014.00064
115. Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B. The Emerging Mechanisms of Isoform-Specific PI3K Signalling. Nat Rev Mol Cell Biol (2010) 11(5):329–41. doi: 10.1038/nrm2882
116. Manning BD, Cantley LC. AKT/PKB Signaling: Navigating Downstream. Cell (2007) 129(7):1261–74. doi: 10.1016/j.cell.2007.06.009
117. Engelman JA. Targeting PI3K Signalling in Cancer: Opportunities, Challenges and Limitations. Nat Rev Cancer (2009) 9(8):550–62. doi: 10.1038/nrc2664
118. Fruman DA, Rommel C. PI3K and Cancer: Lessons, Challenges and Opportunities. Nat Rev Drug Discovery (2014) 13(2):140–56. doi: 10.1038/nrd4204
119. Bader AG, Kang S, Zhao L, Vogt PK. Oncogenic PI3K Deregulates Transcription and Translation. Nat Rev Cancer (2005) 5(12):921–9. doi: 10.1038/nrc1753
120. Fruman DA, Chiu H, Hopkins BD, Bagrodia S, Cantley LC, Abraham RT. The PI3K Pathway in Human Disease. Cell (2017) 170(4):605–35. doi: 10.1016/j.cell.2017.07.029
121. Eto K, Iwatsuki M, Watanabe M, Ida S, Ishimoto T, Iwagami S, et al. The microRNA-21/PTEN Pathway Regulates the Sensitivity of HER2-Positive Gastric Cancer Cells to Trastuzumab. Ann Surg Oncol (2014) 21(1):343–50. doi: 10.1245/s10434-013-3325-7
122. Kennedy SG, Wagner AJ, Conzen SD, Jordán J, Bellacosa A, Tsichlis PN, et al. The PI 3-Kinase/Akt Signaling Pathway Delivers an Anti-Apoptotic Signal. Genes Dev (1997) 11(6):701–13. doi: 10.1101/gad.11.6.701
123. Vivanco I, Sawyers CL. The Phosphatidylinositol 3-Kinase AKT Pathway in Human Cancer. Nat Rev Cancer (2002) 2(7):489–501. doi: 10.1038/nrc839
124. Sanidas I, Polytarchou C, Hatziapostolou M, Ezell SA, Kottakis F, Hu L, et al. Phosphoproteomics Screen Reveals Akt Isoform-Specific Signals Linking RNA Processing to Lung Cancer. Mol Cell (2014) 53(4):577–90. doi: 10.1016/j.molcel.2013.12.018
125. Shayesteh L, Lu Y, Kuo WL, Baldocchi R, Godfrey T, Collins C, et al. PIK3CA Is Implicated as an Oncogene in Ovarian Cancer. Nat Genet (1999) 21(1):99–102. doi: 10.1038/5042
126. Sun M, Wang G, Paciga JE, Feldman RI, Yuan ZQ, Ma XL, et al. AKT1/PKBalpha Kinase Is Frequently Elevated in Human Cancers and its Constitutive Activation Is Required for Oncogenic Transformation in NIH3T3 Cells. Am J Pathol (2001) 159(2):431–7. doi: 10.1016/s0002-9440(10)61714-2
127. Bellacosa A, Kumar CC, Di Cristofano A, Testa JR. Activation of AKT Kinases in Cancer: Implications for Therapeutic Targeting. Adv Cancer Res (2005) 94:29–86. doi: 10.1016/s0065-230x(05)94002-5
128. Seo M, Lee S, Kim JH, Lee WH, Hu G, Elledge SJ, et al. RNAi-Based Functional Selection Identifies Novel Cell Migration Determinants Dependent on PI3K and AKT Pathways. Nat Commun (2014) 5:5217. doi: 10.1038/ncomms6217
129. Seo M, Kim JH, Suk K. Role of the P55-Gamma Subunit of PI3K in ALK-Induced Cell Migration: RNAi-Based Selection of Cell Migration Regulators. Cell Adhes Migr (2017) 11(3):205–10. doi: 10.1080/19336918.2016.1202385
130. Kim CK, Lee SB, Nguyen TL, Lee KH, Um SH, Kim J, et al. Long Isoform of ErbB3 Binding Protein, P48, Mediates Protein Kinase B/Akt-Dependent HDM2 Stabilization and Nuclear Localization. Exp Cell Res (2012) 318(2):136–43. doi: 10.1016/j.yexcr.2011.08.013
131. Liu Z, Ahn JY, Liu X, Ye K. Ebp1 Isoforms Distinctively Regulate Cell Survival and Differentiation. Proc Natl Acad Sci USA (2006) 103(29):10917–22. doi: 10.1073/pnas.0602923103
132. Wang X, Yang B, She Y, Ye Y. The lncRNA TP73-AS1 Promotes Ovarian Cancer Cell Proliferation and Metastasis via Modulation of MMP2 and MMP9. J Cell Biochem (2018) 119(9):7790–9. doi: 10.1002/jcb.27158
Keywords: TC2N, tumor-associated antigens (TAAs), cancer, signal pathway, molecular mechanism, functional characterization, clinical feature
Citation: Li H, Fang H, Chang L, Qiu S, Ren X, Cao L, Bian J, Wang Z, Guo Y, Lv J, Sun Z, Wang T and Li B (2021) TC2N: A Novel Vital Oncogene or Tumor Suppressor Gene In Cancers. Front. Immunol. 12:764749. doi: 10.3389/fimmu.2021.764749
Received: 25 August 2021; Accepted: 29 October 2021;
Published: 02 December 2021.
Edited by:
Xiangqian Guo, Henan University, ChinaReviewed by:
Chengqi Xu, Huazhong University of Science and Technology, ChinaJiateng Zhong, Xinxiang Medical University, China
Shuangyu Lv, Henan University, China
Copyright © 2021 Li, Fang, Chang, Qiu, Ren, Cao, Bian, Wang, Guo, Lv, Sun, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tiejun Wang, ZHJ3YW5ndGllanVuQHllYWgubmV0; Bingjin Li, bGliaW5namluQGpsdS5lZHUuY24=
 Hanyang Li
Hanyang Li He Fang3
He Fang3 Tiejun Wang
Tiejun Wang Bingjin Li
Bingjin Li