
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 29 October 2021
Sec. Autoimmune and Autoinflammatory Disorders
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.761890
This article is part of the Research Topic Recent Advances in Immune-Stromal Interactions in Autoimmune & Autoinflammatory Skin Diseases View all 6 articles
Inflammatory skin diseases are induced by disorders of the host defense system of the skin, which is composed of a barrier, innate and acquired immunity, as well as the cutaneous microbiome. These disorders are characterized by recurrent cutaneous lesions and intense itch, which seriously affecting life quality of people across all ages and ethnicities. To elucidate molecular factors for typical inflammatory skin diseases (such as psoriasis and atopic dermatitis), transcriptomic profiling assays have been largely performed. Additionally, single-cell RNA sequencing (scRNA-seq) as well as spatial transcriptomic profiling have revealed multiple potential translational targets and offered guides to improve diagnosis and treatment strategies for inflammatory skin diseases. High-throughput transcriptomics data has shown unprecedented power to disclose the complex pathophysiology of inflammatory skin diseases. Here, we will summarize discoveries from transcriptomics data and discuss how to maximize the transcriptomics data to propel the development of diagnostic biomarkers and therapeutic targets in inflammatory skin diseases.
The skin is the outmost layer of the body. It not only acts as the first line to defense against various biotic and abiotic stresses, but also play crucial roles in water homeostatic and thermoregulation. Dysregulation of the host defense system of the skin is usually accompanied by immune-mediated inflammation and abnormal keratinocyte differentiation that subsequently induce inflammatory skin diseases. Psoriasis and atopic dermatitis (AD) are the most common chronic inflammatory skin diseases (1, 2). The pathogenesis of these two inflammatory skin diseases is complex, with disease progression driven by a combination of multiple factors, including environmental factors, genetic factors in skin barriers, dysbiosis of skin resident microbiomes, and immune system defects (3–5). Various cutaneous cellular changes, like T-lymphocyte infiltration, vascular hyperplasia can be found in infected skin of psoriasis (3, 4). AD is clinically characterized by chronic, pruritic eczematous skin lesions (5). Different subsets of TH cells, for example, TH17 and TH2/TH22 for psoriasis and AD, respectively, trigger different arrays of cytokines (6). Although both psoriasis and AD can occur at any age, AD prefers to affect infants, especially those at 3-6 months old (5), while the peak onset of psoriasis is in adolescence and early adulthood (7).
Over the past decade, high-throughput RNA-sequencing (RNA-seq) has proven to be an indispensable tool for transcriptome-wide analysis of transcriptional variations. Various types of RNAs, including messenger RNAs (mRNAs), microRNAs (miRNAs), long noncoding RNAs (lncRNAs) and circular RNAs (circRNAs), can be captured by RNA-seq (8). Therefore, broad applications of RNA-seq open a window for deep understanding of multiple aspects of molecular biology in inflammatory skin diseases, such as mRNA splicing and non-coding RNAs regulation (9, 10). Among main RNA-seq technologies, short-read sequencing of cDNA comprises the majority of current available RNA-seq data of inflammatory skin disease studies. Questions that when and where transcription occurs and how transcription is regulated are intensively studied. Recently, newly developed single-cell and spatial transcriptomic sequencing are emerging as powerful techniques for mapping and quantifying transcriptional activity at single-cell and spatial resolution. These powerful techniques have been widely and successfully used in humans, animals and plants to resolve intercellular transcriptomics heterogeneity at single-cell level (11–13). Using scRNA-seq, exciting insights about the unique characteristics of skin-resident innate lymphoid cells, key inflammatory pathways as well as inflammatory fibroblasts have been reported (14–16).
In this review, we will summarize perspectives from the big data view to characterize the current understanding of inflammatory skin diseases, specially focusing on common and bias induced in transcriptional and post-transcriptional levels in psoriasis and AD (Table 1). These high-throughput transcriptomics studies will help to provide insightful knowledge for a more comprehensive understanding of molecular mechanisms underlying disease occurrence and pave the way to the identification of therapeutic targets for more specific and safer modulation of inflammatory skin diseases.
The rapid development of computational algorithms has largely expanded our understanding of molecular mechanisms of inflammatory skin diseases. After obtaining sequencing reads derived from human skin samples, quality control should be performed first (Figure 1). Computational tools, such as FastQC, NGSQC (37), fastp (38), FASTX-Toolkit, and Trimmomatic (39), can be used to evaluate the quality of sequencing bases and trim low-quality bases and reads. Next, quality-controlled RNA-seq reads are mapped to the human reference genome to determine where they are from by using such aligners as BWA (40), Bowtie2 (41), STAR (42), TopHat2 (43), and HISAT2 (44). Alignment results should be examined to filter low-quality read alignments by utilizing Picard (https://broadinstitute.github.io/picard/), RSeQC (45), or Qualimap (46). Then, reads mapped to genomic regions of specific genes were calculated to determine transcriptional abundance, this step could be realized by HTSeq-count (47), Kallisto (48), featureCounts (49), Cuffilinks (50), StringTie (51), RSEM (52), and Sailfish (53). Advanced analysis of RNA-seq data includes differential expression analysis, ncRNA analysis, alternative splicing (AS), RNA editing, and alternative polyadenylation (APA). The differential analysis is designed to statistically compare the same genes between different conditions to determine functional gene sets that participate in the development of pathological or physiological conditions. Frequently used computational tools that perform differential analysis are DESeq2 (54), edgeR (55), baySeq (56), EBseq (57), NOISeq (58), and SAMseq (59). Noncoding RNAs constitute the major component of human genome and exert important regulatory roles in a variety of physiological and pathological processes. The identification and quantification of noncoding RNAs could be performed based on the reference annotation or do novo transcriptome assembly (60). AS is the major contribution to transcriptional diversity in humans, which plays crucial roles in the pathological process of diseases. Computational algorithms, such as Suppa2 (61), rMATS (62), CuffDiff2 (63), MISO (64), DEXSeq (65), and spliceR (66), were designed to identify and quantify AS events and the alternative usage of gene exons. RNA editing is a post-transcriptional event that alters single bases at RNA levels, which has been demonstrated to modulate the AS process, RNA expression, and RNA translation. Multiple tools have been developed to identify RNA editing sites, such as RNAEditor (67), REDItools (68), JACUSA (69), and RES-Scanner (70). APA event is the alternative processing of mRNA 3’ end that generates mRNAs with diverse lengths of 3’ UTR.APA is emerging to play important roles in regulating RNA metabolism, which could be detected by DaPars (71), APAtrap (72), and PHMM (73). We will summarize major findings from analyzing large-scale transcriptomics data in inflammatory skin diseases.
Keratinocytes (KCs) were previously thought of passive bystanders in the inflammatory process. Nowadays, growing evidence has shown that KCs are actively involved in the initiation and maintenance of skin inflammation, by secreting chemokines, cytokines and antimicrobial peptides, which further amplifies the inflammatory process through chemoattraction and activation of skin immune cells (17).
In psoriasis, IL-17 activates epidermal KCs via IL-17R to produce proinflammatory cytokines and chemokines, such as IL-1, IL-6, CXCL1 (C-X-C motif), and CCL20 (C-C motif ligand 20) to induce and propagate psoriatic inflammation (17), while IL-23 drives a local skin inflammatory loop through IL-23 triggered production of IL-22, which in turn helps the maintenance of TH17 cells (18). Therefore, the interplay between IL-23-IL-17 axis and KCs is believed as the central of IL-17-mediated inflammatory loop in psoriasis. Thousands of differentially expressed genes (DEGs) by using large-scale transcriptome analysis were identified in psoriasis. As expected, IL-17, IL-1/IL-36, IL-12/IL-23, and TNF-α pathways were revealed to be the top enriched pathways (19, 21, 24, 74). To further reveal the cellular and molecular mechanisms behind this, IL-17 receptor-A (IL-17RA) antagonist brodalumab, was applied in moderate and severe psoriasis over 12 weeks. This treatment-induced global gene expression profiles revealed a rapid and extensive suppression of IL-17-dependent genes in KCs as well as specific KC-derived inflammatory mediators such as IL-8, IL-9, and IL-20 (24). Compared to human monoclonal antibody ustekinumab, this suppression occurs earlier and to a greater extent, suggesting that application of IL-17 antagonists producing a greater modulation of the synergistic/additive gene set of psoriasis. Similarly, IL17-RA, IL-12/IL-23, or TNF inhibition treatments, were found to mainly affect the expression of IL-17 dependent genes, which further confirmed the hypothesis that IL-17 -stimulated KCs were key drivers of psoriasis (19, 74). Page et al. reported that transcriptional dysregulation in moderate-to-severe psoriasis is dominated in IL-17 related genes in KCs. Down-regulation of IL-17A, IL-17F and IL-12B were detected as early as 2 weeks post-treatment with PF-06700841, suggesting the requirement of TYK2 and Janus kinase 1 for psoriasis signaling transduction (20). IL-36, another important mediator of psoriasis, was thought to regulate psoriasis by activating MAPKs and NF-kB pathways (75). To identify novel factors in psoriasis, RNA-seq analysis was applied in time-dependent IL-1B, IL-36A, IL-36B or IL-36G treated KC samples. This work found that early and later IL-1B-specific responses are all replicated by those of IL-36 treatments. Besides, Type I and II interferon genes exhibited a time-dependent response pattern: inducted at 8 h following no response or repression at 24 h, suggesting a fine-scale characterization of time and cytokine-specific response patterns after IL-1B and IL-36 treatment (76). Early studies showed that the inflammatory loop of AD initiates once skin-resident DCs or other immune cells are stimulated by external agents, with a consequence of releasing epithelial type 2 inflammatory cytokines, such as TSLP, IL-33, and IL-25 (33–35). Meanwhile, KC responses can also be directly induced by external agents to affect subsequent inflammatory events (18, 77). To explore molecular mechanisms behind this, RNA-seq by using biopsy specimens have been largely performed (22, 23, 29, 78). The first study of lesional AD RNA-seq, reported that TREM-1 and IL-36 were novel factors of AD (23). Recently, Tsoi and colleagues showed that dysregulated genes accompanying the transition from nonlesional to acute and chronic AD quantitatively different. Enrichment of DEGs in TNF, TH1, TH2, and TH17 pathways was observed during the whole process of disease progression in nonlesional, acute, and chronic AD. And the most heightened inflammatory response in chronic AD samples. Besides, 42 significant dysregulated genes involving in epidermal differentiation (e.g., IL-10, KRT16, S100A8, and S100A9), antimicrobial, immunomodulatory chemokines (CXCL1, CXCL6), and negative regulation of T-cell activation (FOXK1) were found in chronic versus acute AD (22). These findings provide novel insights and highlight underappreciated pathways in AD pathogenesis that may amenable to be future therapeutic targets. Medical treatment of moderate-to-severe AD-like oral JAK/SYK inhibitor ASN002 induces rapid and sustained improvements in TH2, TH22/TH17, and TH1 pathways, as well as epidermal barrier abnormalities (78). AD in pediatric and Asian populations trigger strong TH2 and TH17 responses, whereas European American adults mainly induce TH2/TH22 activation, suggesting diverse oral therapeutics in treatment with moderate-to-severe AD in different region patients need to be carefully chosen. Very recently, study exploring moderate-to-severe AD before and after systemic treatment characterized a “core” signature of AD by dysregulation of genes related to keratinocyte differentiation and IL-31/IL-1 itch signaling. Additionally, a dynamic signature reflects progressive immune response dominated by type 2 cytokines, with an additional role of TH17 and natural killer cell signaling (29).
The skin can be mainly divided into three layers: epidermis (consists of four strata of keratinocytes), dermis (mainly consists of fibroblasts and immune cells), and subcutaneous layer (consists of subcutaneous fat and connective tissue) (79). As a complex and dynamic system, skin barriers consist of four parts: microbiome barrier (skin microbiome), chemical barrier (stratum corneum), physiological barrier (mainly by epidermis), and immune barrier (epidermis and dermis) (80).
Stratum corneum and tight junctions (TJs) in granular cells are two main constituents of the physiological barrier (81). IL-17 was found to downregulate filaggrin expression and impaired TJs of the skin (82). Though the role of Langerhans cells (LCs) in psoriasis pathogenesis is still controversial, it seems like that inflammation-associated LCs in impaired psoriasis barrier gain an enhanced capacity to promote polarization of naïve T cells into TH17, and subsequently induce TH17 responses (83).
As showed by Swindell. et al, a large number of psoriasis-induced DEGs were also differentially expressed in other immune defective diseases and epidermal differentiation complex (EDC), reflecting intrinsic immune defects in psoriatic KCs may contribute to compromising barrier function (84). To achieve a more comprehensive understanding of molecular pathogenesis, Zhang et al., re-analysis the gene expression profiles of 175 pairs of lesional and non-lesional skin samples from 5 previously published datasets. In this work, they showed that the most enriched biological processes of DEGs in psoriasis samples were immune responses and epidermal differentiation/development (85). In another study, 127 DEGs were screened and the most enriched GOs were keratinization by in-depth bioinformatic analyses of datasets deposited in GEO (GSE13355 and GSE14905) (86). Taking all these together, physical barrier is speculated to be critical for the development of psoriasis.
As defects in either stratum or tight junction triggers TH2 response, the crucial role of skin barriers in the development of AD was well accepted (87). Key epidermal genes, such as filaggrin (FLG), loricrin (LOR), and KRT10, which are also known as EDC genes, were frequently dysregulated in AD (88–90). Downregulation of these EDC genes that function as a physical barrier or antimicrobe factors were proposed to contribute to AD pathogenesis. Evidence supporting this is that mutation of the epidermal structure protein FLG causes approximately 20-40% of AD (5). Besides epidermal disturbances in structural proteins, proteases and their inhibitors are commonly the top dysregulated genes in AD transcriptomic profiles (29). This might be due to the maintenance of skin barrier function largely relies on homeostatic conditions between cellular motility and excessive tissue destruction. Kallikrein-related peptidase (KLK) genes, such as KLK5, KLK14, and lipid metabolism gene ELOVL1, were down-regulated in AD skin (29), while protease inhibitor SPINK5 (serine protease inhibitor of kazal type 5) and human aquaporin 3 (AQP3) were up-regulated, which function in elevating inflammatory responses and epidermal water loss, respectively (29, 91).
Since all itch stimuli sensed in the cutaneous are ultimately transmitted through nerves to the brain, the crucial role of the nervous system is thought as key factor to understand the mechanisms underlying itch in inflammatory skin diseases. Neuropeptides like substance P (SP) and calcitonin Gene-Related Peptides (CGRPs) were reported to be important in the enhancement of itch in psoriasis (92, 93). TRP cation channels are a superfamily of nonselective calcium-permeable cation channels, that in coupling with pruritogenic CGRPs (94, 95). TRPs such as TRPV1, TRPM8, TRPV3 and TRPC4 were reported to be significantly elevated in pruritic skin with psoriasis (30, 96). Meanwhile, various immune cells, such as T cells or mast cells, secrete diverse cytokines that directly or indirectly aggravate itch by increasing inflammatory responses. Examples are the well-known TH17 and TH12 cytokines IL-17, IL-22 and IL-23 (97, 98). Besides, nociceptors were also found to interact with dermal dendritic cells (DDCs) and regulate IL-23/IL-17 pathway via TRPV1 and Nav1.8 (99). In addition, gene transcription of IL-31 was elevated in pruritic lesions of psoriasis (30). While application Janus Kinase-Signal Transducer and Activator of Transcription (JAK-STAT) inhibitor down-regulating mRNA expression levels of IL-22, IL-23, and IL-31 to effectively attenuate itch in psoriatic patients (100–102). These studies highlighted the crosstalk between peripheral nerves and immune system in the development of psoriasis.
Similar to psoriasis, chronic itch is a well-defined symptom of AD. Individual proprioceptors are defined to respond to specific pruritogens, such as non-histaminergic to induce chronic itch in AD (103). Recently, the role of IgE-basophil-leukotrienes (LT) were revealed in the activation AD-caused acute itch flares (104). Likely, TH2 lymphocytes, eosinophils, neutrophils and mast cells work together to amplify inflammatory and itch pathways via releasing cytokines and neurogenic peptides (105, 106). Some AD-associated pro-inflammatory TH2 cytokines, such as IL-4 and IL-13 can directly activate sensory neurons through JAK signaling pathways (27). Additionally, IL-31 and TSLP also directly interact with cation channel TRPV1+TRPA1+ neurons to trigger robust itch behaviors in AD (107, 108). RNA-seq analysis showed that serine proteases KLK7 was the most abundant and differentially expressed KLKs in both human AD and murine AD-like skin. Surprisingly, KLK7 promotes AD-associated itch independently with skin inflammation (36).
Comparative analysis of global gene regulation in paired itchy, lesional and noitchy, and nonlesional skin biopsies from AD and psoriasis identified nearly 2,000 DEGs. Among them, up-regulation of phospholipase A2 IVD, voltage-gated sodium channel 1.7, and TRPV1 were positively correlated with itch intensity in both diseases. While upregulation of TRPV2, TRPA1, protease-activated receptor 2, protease-activated receptor 4, and IL-10 were only detected in AD, and TRPM8, TRPV3, phospholipase C and IL-36 were psoriatic specific. This “itchscriptome” extended our understanding of the molecular mechanism of sensory nerves in inflammatory skin diseases and provide potential targets for itch treatment (30).
Due to the complexity of skin surface microenvironments, skin located microbiota is quite diverse. Surveys of discrete skin sites, that were selected from microbial infections sites, demonstrated that skin physiology (moist, dry, and sebaceous) is the result of organized skin microbiota communities (109). Skin microbes metabolized host proteins and lipids to release bioactive molecules. These skin microbe-released antimicrobial peptides or metabolites can be directly coupled by host skin-resident dendritic cells (DCs) and trigger host immune responses. Meanwhile, these molecules released from skin microbiota promote host to secret cytokines (such as IL-1, IL-17) and further enhance cell microbicidal function (109). Dysbiosis of skin microbiota therefore is highly associated with inflammatory skin disease.
To reveal the role of microbiota in inflammatory skin diseases, large-scale analysis using 16S and RNA-sequencing have been performed and presented distinct microbiome compositions in AD and psoriasis skin. Unlike AD, wherein only, S. aureus was identified as the dominant microbe, multiple organisms including Corynebacterium and Finegoldia were identified in psoriasis skin. Corynebacterium was considered to negatively regulate interferon signaling to affect the initiation of psoriasis (110), while colonization or infection of bacterium S. aureus has been frequently reported in AD (111). Using 16S and RNA-sequencing of AD samples, a significant increase of S. aureus and loss of anerobic species, accompanying with host response by altering expression of genes related to barrier function, metabolic reprogramming, antimicrobial defense, and TH2 signaling was detected (110).
Besides protein-coding RNAs, the human genome also transcribes a large number of noncoding RNAs that have been classified into multiple families based on their size and biogenesis (112). These noncoding RNAs have been revealed to play pivotal roles in human complex diseases including cancer, autoimmunity diseases, and hereditary diseases (113–115). And deep mining of noncoding RNA transcriptomic data will provide the opportunity to discover novel biomarkers and therapeutic targets in inflammatory skin diseases (Table 2) (8).
miRNAs are small (approximately 22 nucleotides) and evolutionarily conserved noncoding RNAs that regulate gene expression at the post-transcriptionally level. To date, more than 2,500 human miRNAs have been reported, and their expression levels vary significantly depending on tissue and cell types (141). Genetical evidences form both gain and loss of function approaches exhibited that miRNAs functions to regulate gene expression in at least two aspects: prepress target mRNAs and buffer posttranscriptional genetic noise (142).
Evidence is rapidly accumulating for the role of miRNAs in the pathogenesis of inflammatory skin disorders. miR-203 was the first reported keratinocyte-derived miRNA, which was shown to target SOC3, NR1H3/LXR-α and PRAR-ϒ (116–118). miR-203 was reported to be upregulated in serum but downregulated in urine, suggesting as a potential biomarker for children AD (119). Some miRNAs, such as miR-146a and miR-155, were elevated in both psoriasis and AD (120, 143). miR-146a was found to negatively regulate keratinocyte proliferation and inflammation pathways by targeting CCL5, TRAF6, IRAK1, and CARD10 (120–122), while miR-155 was demonstrated to be involved in keratinocyte proliferation, inflammation, and tight junction disruption, by directly targeting CTL4 in T cells (123–125). In psoriasis, miR155 was also reported to regulate GATA3 downstream IL-37 mediated inflammatory responses (125). Several miRNAs have been demonstrated to be efficiency diagnostic biomarkers for inflammatory skin diseases, such as miR-223, miR-143, and miR-369-3p in psoriasis (126, 130). In addition, miR-143 suppresses IL-13 activity and inflammatory responses via directly targeting IL-13Rα1 (127). In serum of AD patients,miR-614, miR-223, and miR-151a were significantly up-regulated, Collectively these studies suggested miRNAs as new diagnostic biomarkers for inflammatory skin diseases (128, 132, 133).
LncRNAs are RNA transcripts that are over 200 nucleotides long and commonly recognized to have limited potential to encode any identifiable peptide products (144). LncRNAs can be classified into sense, antisense transcripts, long intergenic noncoding RNAs (lincRNAs), and long intronic RNAs (lncRNAs) based on their genome location (145), and can also be classified as cis- and trans-acting lncRNAs according to their function (146). Expression of lncRNA is usually tissue-restricted, development-regulated and vary largely under different disease conditions. Many studies revealed that lncRNAs may serve as scaffolds to form ribonucleoprotien (RNP) complexes or as decoys for proteins and miRNAs (147, 148).
With the widespread applications of next-generation sequencing (NGS) technologies, huge numerous of human lncRNAs have been identified, producing a plenty of lncRNA resources in different contexts, such as LNCipedia (149) and NONCODE (150). Recently, an enrichment of dysregulated lncRNAs of typical inflammatory skin diseases has been reported (151). These dysregulated lncRNAs were mainly involved in epidermal differentiation, apoptosis and immune responses pathways (134, 140, 152). For example, lncRNA-H19 regulate Dsg1 expression and consequently regulates keratinocyte differentiation through directly binding to miR-130b-3p (135). Similarly, Qiao et al. reported that the expression level of lncRNA-MSX2P1 positively correlated with S100A7. And up-regulation of lncRNA-MSX2P1 promoted the IL-22-stimulated KCs by inhibiting miR-6731-5p, suggesting a network module of lncRNA-MSX2P1-miR-6731-5p-S100A7 (136). By using weighted gene co-expression network analysis (WGCNA), 67 miRNA-lncRNA co-expression pairs have been found (153). Studies of lncRNA SPRR2C revealed that it competed with STAT1 and S1000A7 to counteract miR-330-mediated suppression of STAT1 and S100A7 (139). Psoriatic down-regulated lncRNA MEG3 was found to activate apoptosis through miR-21-suppressed caspase-8 (137). Alternatively, psoriatic up-regulated lncRNA PRINS targets the anti-apoptotic G1P3 in KCs, therefore diminishing the sensitivity of KCs to spontaneous apoptosis through G1P3 (138). Recently, several lncRNAs that were previously annotated as noncoding RNAs are reported to encode micropeptides or small proteins (140, 154, 155). These lncRNA-encoded micropeptides were shown to be key regulators of vital cell functions, such as muscle development, cancer development, and inflammatory skin disease development. Niu et al. found that lncRNA MIR155HG could encode a micropeptide to regulate antigen presentation and suppress autoimmune inflammation in psoriasis, unlike well-known lncRNA/miRNA module, this study, for the first time, illustrated a new mechanism of lncRNA in inflammatory diseases (140).
Applications of NGS-based methods revolutionize our understanding of dysregulated genes at not only the transcriptome level but also post-transcriptional levels. It is known that human immature transcripts go through extensive post-transcriptional regulation to generate mature functional transcripts (156). Major post-transcriptional events such as AS and RNA editing are well studied in human complex diseases, including cancer at immune diseases (157, 158). Here, we will discuss the important roles of post-transcriptional events in the progression of inflammatory skin diseases.
AS is a post-transcriptional process by which pre-mRNA transcripts are spliced in different ways. Nearly all human protein-coding genes undergo one or more forms of alternative splicing to generate various functional mRNA and protein products from a single gene. This process largely contributes to the complexity in the transcriptome and lead to protein diversity (8). AS needs the formation of the splicing complex to exert inclusion or skipping of exons, alternative 5’ splice-site selection, intron retention, exclusive splicing of adjacent exons, and switching between alternative splice sites (159–161). The formation of the splicing complex is a series of interplays between small nuclear ribonucleoprotein particles (snRNPs, U1, U2, U4/6, and U5), small nuclear RNAs (snRNAs), and overs 150 additional associated proteins which not directly bound to the snRNPs (162). With the growing applications of RNA-seq, more roles of AS events are unveiled. For example, cancer cells can generate cancer type-specific and subtype-specific alterations in the splicing process and contribute to cancer progression as well as cancer immune responses (163). Similarly, Shimizu et al. reported that the ST2 gene encoded both membrane-bound ST2L and soluble ST2 (sST2) by AS, wherein ST2L promoted TH2 activity with a result of dysregulation of TH1/TH2 immune balance and severe AD diseases (164). Human ACT1 undergoes AS in SNP-D10N region with a result of expressed ACT1 isoforms ACT-D19N and ACT-D10N. ACT1-D19N is fully responsive to IL-17 through interacting with Hsp90, while ACT-D10N loses this ability. Although these two isoforms are equally expressed in ACT1D10N/D10 fibroblasts, ACT1D10N/D10 T cell expressed predominantly ACT-D10N, leading to a dysregulated hyperactive TH17 response with elevated IL-17A and IL-22 expression in ACT1D10N/D10 T cells and consequently severe psoriasis (165). Kyong et al. reported that ESRP1-mediated AS of Rho GTP exchange factor ARHGEF11 was essential for epithelial tight junction (TJ) integrity in the dysregulation of skin barriers (166). Together, these studies not only illustrated the vital role of AS events in the development of inflammatory skin diseases but also revealed a potential application of mining AS events to deep our understanding of inflammatory skin diseases.
However, to our best knowledge, comparing with a large application of transcriptome analysis in the identification of novel AS events in cancer, most of the AS investigations in inflammatory skin diseases were low-throughput experiment-based, suggesting the potential applications of identification AS events through high-throughput transcriptomics in discovering novel factors in inflammatory skin diseases.
RNA editing is a post-transcriptional event that modifies single-base changes on RNA nucleotides without altering their genomic DNA (158). Thus, impaired RNA editing activity can lead to increased modulation of alternative splicing, missense codon changes, and modifications of noncoding RNAs (158, 167, 168) RNA editing has been reported as an import process that contributes to proteomic diversity in human diseases (169). In 2011, Cailin E. Joyce reported a low frequency of RNA editing in normal and psoriasis skin (170). This work was later confirmed by the Shoshana Greenberger group, as they reported that psoriasis patients demonstrated a global A-to-I RNA editing reduction in psoriatic lesions, which may account for the accumulation of double-stranded RNA (dsRNA). This process, in turn, stimulates the production of IFNs and is instrumental in triggering the initiation and progression of diseases (171). Besides global alteration, RNA editing changes were also detected in IGFBP7, COPA, and FLNA genes sites, suggesting a link of autoimmune diseases to a reduction in global RNA editing (171). These studies together suggested that RNA-editing mediated post-transcriptional regulation may be involved in the process of inflammatory skin diseases.
Single-cell RNA sequencing (scRNA-seq) technology is emerging as a powerful tool for characterizing heterogeneity between and within tissue/cell types. It enables more rapid identification of novel cell types, cell states, lineages as well as circuitry (172, 173). Together with spatial transcriptomics, these revolutionized techniques have been widely and successfully used in both mammalian and plant kingdoms (174, 175) and prompted our understanding of multiple complex diseases, such as inflammatory skin diseases (176). The generation of scRNA-seq data mainly includes sample collection, tissue dissociation, cell sorting, library construction, and sequencing (Figure 2). The raw scRNA-seq data was first mapped to reference genome by utilizing tools like Cell Ranger, SEQC (177), and zUMIs (178) to generate an expression matrix of all detected genes in all cells. The data also needs to be checked to remove doublet cells. Scrublet (179), scds (180), DoubleFinder (181) were designed to identify doublets. Next, data normalization, integration, dimensionality reduction, and clustering should be performed, which could be realized by different functions implemented in Seurat (182). Cell clusters are further assigned “real” cell names by using computational tools, such as SingleR (183), CellAssign (184), AUCell (185). These cell names should be manually checked and will be used in the following advanced analysis. Most physiological and pathological processes are accompanied by transcriptional dynamics, which could be influenced by the pseudo-temporal ordering of single cells by using scRNA-seq data. Most commonly used expression trajectory inference tools include Monocle (186), Slingshot (187), PAGA (188), and DPT (189). Cell-cell interactions mediated by ligand-receptor complexes are critical in diverse biological processes. Investigation of context-dependent crosstalk of different cell types enables a deep understanding of specific physiological and pathological processes. ScRNA-seq data could also be used to infer cell-cell communications by using such computational tools as CellPhoneDB (190), CellChat (191), and NicheNet (192). Spatial transcriptomics techniques can assay cells in their native tissue context, which enables spatial characterization of transcriptional activities. Multiple computational tools have been developed to analyze spatial transcriptomics data, such as Seurat, BayesSpace (193), Giotto (194), stLearn (195). Integrative analysis of scRNA-seq and spatial transcriptomics data will help to precisely decode intercellular communications in specific tissue locations.
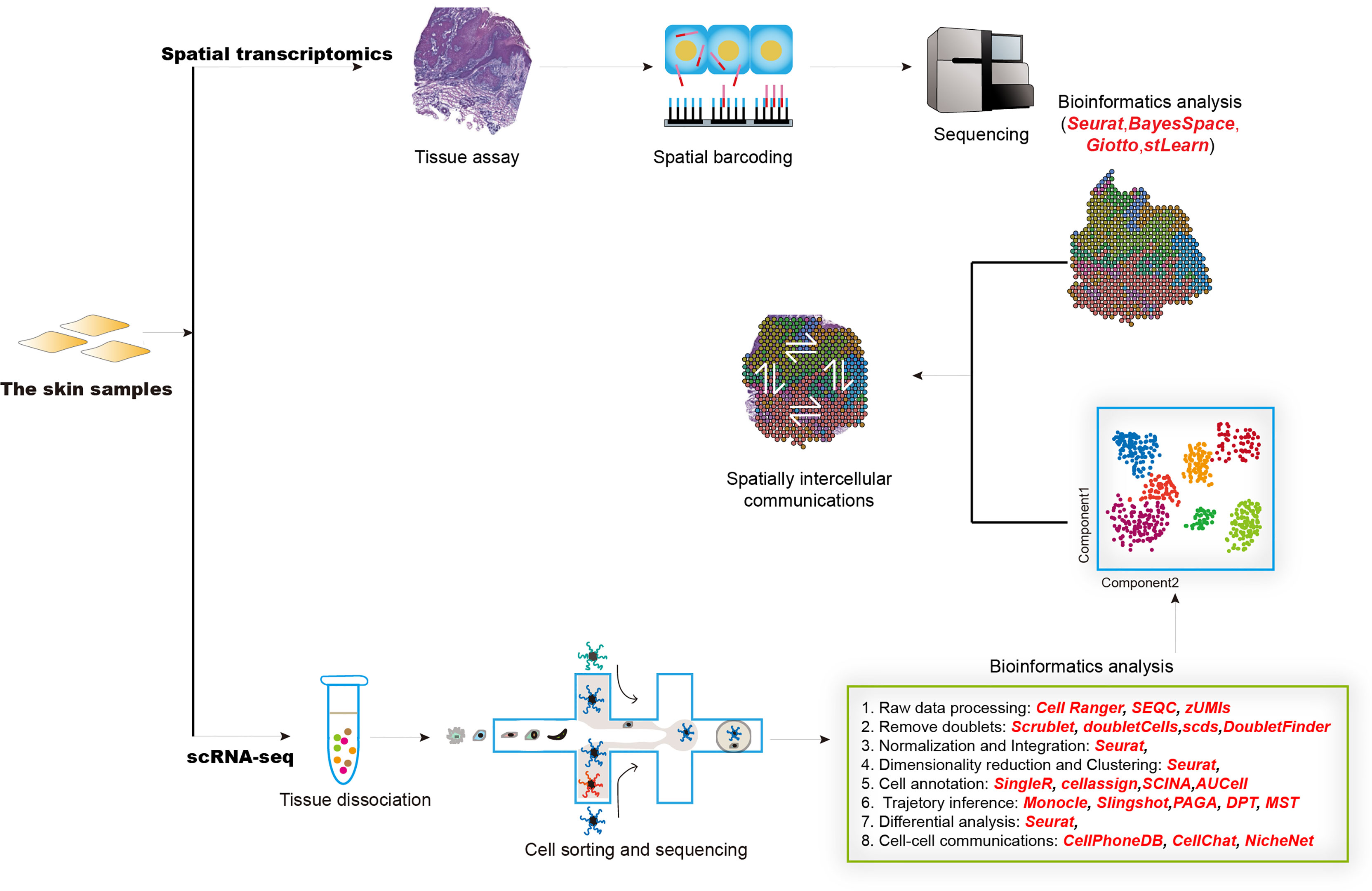
Figure 2 The overall design of experimental and computational analysis for single-cell transcriptomics and spatial transcriptomics studies in inflammatory diseases.
Here, we will summarize newly reported discoveries about inflammatory skin disease at single-cell resolution.
In 2018, Cheng et al. reported the first single-cell transcriptomics study of human epidermis from multiple anatomic sites and psoriasis-like skins. Intriguingly, high levels of previous reported inflammatory transcripts such as S100 transcripts and IFI27 levels in normal scalp were detected, suggesting a cause for the inflammation that often occurs at this site. While in foreskin keratinocytes, upregulation of proliferation-related transcripts was detected. In the psoriatic epidermis, enrichment of channel cells and mitotic subfraction, enhancement of inflammatory transcripts like S100A7, S100A8, S100A9, IFI27, PI3 as well as CD1C+CD301A+ myeloid dendritic cell population were detected. This study provided a critical step toward epidermal development, differentiation, and inflammation (196). Psoriasis is known as an IL-17-driven inflammatory skin disease, in which autoantigen-induced CD8+ T cells have been identified as pathogenic drivers. By using scRNA-seq, a total of 11 transcriptionally diverse CD8+ T cell subsets in psoriatic and healthy skin were identified, including 2 non-exhausted Tc17 cell subsets. Besides, CXCL13, which achieved greater accuracy than IL-17A, was thought as a novel biomarker of psoriasis severity. This study uncovered the diverse landscape of CD8+ T cells in psoriatic and healthy skin (197). To uncover the expression of key phenotypic features of cells in both high fidelity and high throughput, Hughes et al. developed a massive parallel scRNA-seq, also called Seq-Well S3 (Second-Strand Synthesis) protocol to chart the transcriptional landscape of five human inflammatory skin diseases, including acne, alopecia areata, granuloma annulare, leprosy, and psoriasis (198). Over-representation of Tregs, dysfunctional NR4A1-expressing T cells, and senescent SESN3+ T cells were detected in psoriasis. Besides, IRF4+ cDC2 cluster that displays an elevated expression of CCL17, CCL22, and a population of fibroblasts that expressed CCL19 and BAFF were reported in psoriasis biopsies. Notably, a large population of shared signals among cell types and states in the tested five inflammatory diseases were identified, suggesting potentially common outputs between these diseases (198). He et al. reported the first scRNA-seq analysis of healthy, lesional, and nonlesional skin from AD patients. In this work, they detected a high expression of TH2 (IL-13) and TH22 (IL-22) T cells in AD. And identified a novel COL6A5+COL18A1+ subpopulation inflammatory fibroblasts. These COL6A5+COL18A1+ subpopulation fibroblasts expressed CCL2 and CCL9 cytokines and were unique to lesional AD. Another unique subpopulation to AD lesions identified in this work is LAMP3+ DC, which expressed the CCL19 receptor CCR7. These findings together revealed a potential role of fibroblast in cross-talk with DCs and T-cells (21). Rojahn et al. characterized the pathogenesis of AD on both transcriptomic and proteomic levels, by using suction blistering captured epidermal and biopsies samples. Comparing transcriptional profiles of key inflammatory pathways (such as TH2 pathways) were detected, but suction blistering was superior in cell-specific resolution for high-abundance transcripts (i.e. KRT1/KRT10, KRT16/KRT6A, S100A8/S100A9) (14). An elevated level of AD-typical cytokines such as IL-13 and IL-22 in TH2 and TH22 cells, as well as antimicrobial cytokines like IL-26 are which expressed in proliferating T cells and natural killer T cells, were detected. Gao et al. , evaluated the intrinsic and intercellular alterations of healthy donors and patients with psoriasis. They revealed that the evolutionally conserved epidermal keratinocytes and dermal mesenchymal cells could self-transform into immune active states via intensively evoking expression of major histocompatibility complex (MHC) genes during psoriasis. They uncovered the immunoregulatory axis from skin resident cells to immune cells (199). To generate the human skin cell atlas, single-cell technology combined with immunostaining in situ of human skin biopsies in early prenatal life, adulthood, and typical inflammatory skin diseases were characterized (176). In total, 34 cell states were identified in healthy human skin, with dynamic changes across embryonic, adult life, and upon perturbation during inflammatory skin diseases. In the view of the skin immune system, the dominant cells are lymphocytes and macrophages in first-trimester embryonic skin and clonal expansion of disease-associated lymphocytes in inflammatory diseases. In adult skin, two inferred trajectories for keratinocyte differentiation and the presence of endothelial cells were detected. Besides, augmented migratory DC signature was detected during the development of human thymus and in disease states. Taken together, this study revealed the dynamic nature of cutaneous homeostasis across the fetal development and immune-mediated inflammatory disorders (176).
The best strategy to characterize the pathological process and develop therapeutic targets of inflammatory skin diseases is to comparatively measure every key gene. However, it will take years to portray a large spectrum of genes by using traditional molecular techniques. The RNA-seq and single-cell transcriptomics technologies offer a great opportunity to extensively identify abnormalities in the pathological progression of inflammatory skin diseases. In this review, we discussed how transcriptomics data expedites significant findings in inflammatory skin diseases.
SL conceived and supervised this project. JW, ZF, TL, WH, and YW collected the data and literatures. ZF, SL, and WH draw the plots. SL and JW wrote the manuscript with comments from all listed authors. All authors contributed to the article and approved the submitted version.
This study was supported by Shanghai General Hospital Startup Funding (02.06.01.20.06 and 02.06.02.21.01).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
2. Wer S, Novak N. Atopic Dermatitis. Lancet (2016) 387:11091122. doi: 10.1016/S0140-6736(15)00149-X
3. Griffiths CE, Barker JN. Pathogenesis and Clinical Features of Psoriasis. Lancet (2007) 370:263–71. doi: 10.1016/S0140-6736(07)61128-3
4. Nickoloff BJ. Cracking the Cytokine Code in Psoriasis. Nat Med (2007) 13:242–4. doi: 10.1038/nm0307-242
5. Langan SM, Irvine AD, Weidinger S. Atopic Dermatitis. Lancet (2020) 396:345–60. doi: 10.1016/S0140-6736(20)31286-1
6. Guttman-Yassky E, Krueger JG. Atopic Dermatitis and Psoriasis: Two Different Immune Diseases or One Spectrum? Curr Opin Immunol (2017) 48:68–73. doi: 10.1016/j.coi.2017.08.008
7. Noda S, Krueger JG, Guttman-Yassky E. The Translational Revolution and Use of Biologics in Patients With Inflammatory Skin Diseases. J Allergy Clin Immunol (2015) 135:324–36. doi: 10.1016/j.jaci.2014.11.015
8. Xiang Y, Ye Y, Zhang Z, Han L. Maximizing the Utility of Cancer Transcriptomic Data. Trends Cancer (2018) 4:823–37. doi: 10.1016/j.trecan.2018.09.009
9. Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of Transcription in Human Cells. Nature (2012) 489:101–8. doi: 10.1038/nature11233
10. Morris KV, Mattick JS. The Rise of Regulatory RNA. Nat Rev Genet (2014) 15:423–37. doi: 10.1038/nrg3722
11. Hammond TR, Dufort C, Dissing-Olesen L, Giera S, Young A, Wysoker A, et al. Single-Cell RNA Sequencing of Microglia Throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell-State Changes. Immunity (2019) 50:253–71.e6. doi: 10.1016/j.immuni.2018.11.004
12. Suvà ML, Tirosh I. Single-Cell RNA Sequencing in Cancer: Lessons Learned and Emerging Challenges. Mol Cell (2019) 75:7–12. doi: 10.1016/j.molcel.2019.05.003
13. Zhang TQ, Xu ZG, Shang GD, Wang JW. A Single-Cell RNA Sequencing Profiles the Developmental Landscape of Arabidopsis Root. Mol Plant (2019) 12:648–60. doi: 10.1016/j.molp.2019.04.004
14. Rojahn TB, Vorstandlechner V, Krausgruber T, Bauer WM, Alkon N, Bangert C, et al. Single-Cell Transcriptomics Combined With Interstitial Fluid Proteomics Defines Cell Type–Specific Immune Regulation in Atopic Dermatitis. J Allergy Clin Immunol (2020) 146:1056–69. doi: 10.1016/j.jaci.2020.03.041
15. He H, Suryawanshi H, Morozov P, Gay-Mimbrera J, Del Duca E, Kim HJ, et al. Single-Cell Transcriptome Analysis of Human Skin Identifies Novel Fibroblast Subpopulation and Enrichment of Immune Subsets in Atopic Dermatitis. J Allergy Clin Immunol (2020) 145:1615–28. doi: 10.1016/j.jaci.2020.01.042
16. Hughes TK, Wadsworth 2MH, Gierahn TM, Do T, Weiss D, Andrade PR, et al. Second-Strand Synthesis-Based Massively Parallel scRNA-Seq Reveals Cellular States and Molecular Features of Human Inflammatory Skin Pathologies. Immunity (2020) 53:878–94.e7. doi: 10.1016/j.immuni.2020.09.015
17. Lowes MA, Suárez-Fariñas M, Krueger JG. Immunology of Psoriasis. Annu Rev Immunol (2014) 32:227–55. doi: 10.1146/annurev-immunol-032713-120225
18. Teunissen MBM, Munneke JM, Bernink JH, Spuls PI, Res PCM, Te Velde A, et al. Composition of Innate Lymphoid Cell Subsets in the Human Skin: Enrichment of NCR + ILC3 in Lesional Skin and Blood of Psoriasis Patients. J Invest Dermatol (2014) 134:2351–60. doi: 10.1038/jid.2014.146
19. Zaba LC, Suárez-Fariñas M, Fuentes-Duculan J, Nograles KE, Guttman-Yassky E, Cardinale I, et al. Effective Treatment of Psoriasis With Etanercept Is Linked to Suppression of IL-17 Signaling, Not Immediate Response TNF Genes. J Allergy Clin Immunol (2009) 124:1022–395. doi: 10.1016/j.jaci.2009.08.046
20. Page KM, Suarez-Farinas M, Suprun M, Zhang W, Garcet S, Fuentes-Duculan J, et al. Molecular and Cellular Responses to the TYK2/JAK1 Inhibitor PF-06700841 Reveal Reduction of Skin Inflammation in Plaque Psoriasis. J Invest Dermatol (2020) 140:1546–55.e4. doi: 10.1016/j.jid.2019.11.027
21. Chiricozzi A, Guttman-Yassky E, Suárez-Farĩas M, Nograles KE, Tian S, Cardinale I, et al. Integrative Responses to IL-17 and TNF-α in Human Keratinocytes Account for Key Inflammatory Pathogenic Circuits in Psoriasis. J Invest Dermatol (2011) 131:677–87. doi: 10.1038/jid.2010.340
22. Tsoi LC, Rodriguez E, Stölzl D, Wehkamp U, Sun J, Gerdes S, et al. Progression of Acute-to-Chronic Atopic Dermatitis Is Associated With Quantitative Rather Than Qualitative Changes in Cytokine Responses. J Allergy Clin Immunol (2020) 145:1406–15. doi: 10.1016/j.jaci.2019.11.047
23. RNA Sequencing Atopic Dermatitis Transcriptome Profiling Provides Insights Into Novel Disease Mechanisms With Potential Therapeutic Implications. Elsevier Enhanced Reader. Available at: https://reader.elsevier.com/reader/sd/pii/S0091674915003437?token=BAFDDE6514541DC5D3B583E90010F95B09AA0F504EA52C5FF8FDE5A477F04C0E1D2C02D825B8C6A15CBBBD245500FA10 (Accessed March 22, 2021).
24. Tomalin LE, Russell CB, Garcet S, Ewald DA, Klekotka P, Nirula A, et al. Short-Term Transcriptional Response to IL-17 Receptor-A Antagonism in the Treatment of Psoriasis. J Allergy Clin Immunol (2020) 145:922–32. doi: 10.1016/j.jaci.2019.10.041
25. Pasquali L, Srivastava A, Meisgen F, Das Mahapatra K, Xia P, Xu Landén N, et al. The Keratinocyte Transcriptome in Psoriasis: Pathways Related to Immune Responses, Cell Cycle and Keratinization. Acta Derm Venereol (2019) 99:196–205. doi: 10.2340/00015555-3066
26. Vu YH, Hashimoto-Hachiya A, Takemura M, Yumine A, Mitamura Y, Nakahara T, et al. IL-24 Negatively Regulates Keratinocyte Differentiation Induced by Tapinarof, an Aryl Hydrocarbon Receptor Modulator: Implication in the Treatment of Atopic Dermatitis. Int J Mol Sci (2020) 21:9412. doi: 10.3390/ijms21249412
27. Oetjen LK, Mack MR, Feng J, Whelan TM, Niu H, Guo CJ, et al. Sensory Neurons Co-Opt Classical Immune Signaling Pathways to Mediate Chronic Itch. Cell (2017) 171:217–228.e13. doi: 10.1016/j.cell.2017.08.006
28. Beck LA, Thaçi D, Hamilton JD, Graham NM, Bieber T, Rocklin R, et al. Dupilumab Treatment in Adults With Moderate-to-Severe Atopic Dermatitis. N Engl J Med (2014) 371:130–9. doi: 10.1056/NEJMoa1314768
29. Möbus L, Rodriguez E, Harder I, Stölzl D, Boraczynski N, Gerdes S, et al. Atopic Dermatitis Displays Stable and Dynamic Skin Transcriptome Signatures. J Allergy Clin Immunol (2021) 147:213–23. doi: 10.1016/j.jaci.2020.06.012
30. Nattkemper LA, Tey HL, Valdes-Rodriguez R, Lee H, Mollanazar NK, Albornoz C, et al. The Genetics of Chronic Itch: Gene Expression in the Skin of Patients With Atopic Dermatitis and Psoriasis With Severe Itch. J Invest Dermatol (2018) 138:1311–7. doi: 10.1016/j.jid.2017.12.029
31. Oldhoff JM, Darsow U, Werfel T, Katzer K, Wulf A, Laifaoui J, et al. Anti-IL-5 Recombinant Humanized Monoclonal Antibody (Mepolizumab) for the Treatment of Atopic Dermatitis. Allergy (2005) 60:693–6. doi: 10.1111/j.1398-9995.2005.00791.x
32. Paller AS, Kabashima K, Bieber T. Therapeutic Pipeline for Atopic Dermatitis: End of the Drought? J Allergy Clin Immunol (2017) 140:633–43. doi: 10.1016/j.jaci.2017.07.006
33. Van Dyken SJ, Mohapatra A, Nussbaum JC, Molofsky AB, Thornton EE, Ziegler SF, et al. Chitin Activates Parallel Immune Modules That Direct Distinct Inflammatory Responses via Innate Lymphoid Type 2 and γδ T Cells. Immunity (2014) 40:414–24. doi: 10.1016/j.immuni.2014.02.003
34. Lambrecht BN, Hammad H. The Immunology of the Allergy Epidemic and the Hygiene Hypothesis. Nat Immunol (2017) 18:1076–83. doi: 10.1038/ni.3829
35. Nowarski R, Jackson R, Flavell RA. The Stromal Intervention: Regulation of Immunity and Inflammation at the Epithelial-Mesenchymal Barrier. Cell (2017) 168:362–75. doi: 10.1016/j.cell.2016.11.040
36. Guo CJ, Mack MR, Oetjen LK, Trier AM, Council ML, Pavel AB, et al. Kallikrein 7 Promotes Atopic Dermatitis-Associated Itch Independently of Skin Inflammation. J Invest Dermatol (2020) 140:1244–52.e4. doi: 10.1016/j.jid.2019.10.022
37. Patel RK, Jain M. NGS QC Toolkit: A Toolkit for Quality Control of Next Generation Sequencing Data. PloS One (2012) 7:e30619. doi: 10.1371/journal.pone.0030619
38. Chen S, Zhou Y, Chen Y, Gu J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics (2018) 34:i884i890. doi: 10.1093/bioinformatics/bty560
39. Bolger AM, Lohse M, Usadel B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics (2014) 30:2114–20. doi: 10.1093/bioinformatics/btu170
40. Li H, Durbin R. Fast and Accurate Long-Read Alignment With Burrows-Wheeler Transform. Bioinformatics (2010) 26:589–95. doi: 10.1093/bioinformatics/btp698
41. Langmead B, Salzberg SL. Fast Gapped-Read Alignment With Bowtie 2. Nat Methods (2012) 9:357360. doi: 10.1038/nmeth.1923
42. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics (2013) 29:15–21. doi: 10.1093/bioinformatics/bts635
43. Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential Gene and Transcript Expression Analysis of RNA-Seq Experiments With TopHat and Cufflinks. Nat Protoc (2012) 7:562–78. doi: 10.1038/nprot.2012.016
44. Kim D, Langmead B, Salzberg SL. HISAT: A Fast Spliced Aligner With Low Memory Requirements. Nat Methods (2015) 12:357–60. doi: 10.1038/nmeth.3317
45. Wang L, Wang S, Li W. RSeQC: Quality Control of RNA-Seq Experiments. Bioinformatics (2012) 28:2184–5. doi: 10.1093/bioinformatics/bts356
46. Okonechnikov K, Conesa A, Garcı F. Genome Analysis Qualimap 2?: Advanced Multi-Sample Quality Control for High-Throughput Sequencing Data. Bioinformatics (2015) 32:292294. doi: 10.1093/bioinformatics/btv566
47. Anders S, Pyl PT, Huber W. HTSeq–A Python Framework to Work With High-Throughput Sequencing Data. Bioinformatics (2015) 31:166–9. doi: 10.1093/bioinformatics/btu638
48. Bray NL, Pimentel H, Melsted P, Pachter L. Near-Optimal Probabilistic RNA-Seq Quantification. Nat Biotechnol (2016) 34:525–7. doi: 10.1038/nbt.3519
49. Liao Y, Smyth GK, Shi W. FeatureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics (2014) 30:923–30. doi: 10.1093/bioinformatics/btt656
50. Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript Assembly and Quantification by RNA-Seq Reveals Unannotated Transcripts and Isoform Switching During Cell Differentiation. Nat Biotechnol (2010) 28:511–5. doi: 10.1038/nbt.1621
51. Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie Enables Improved Reconstruction of a Transcriptome From RNA-Seq Reads. Nat Biotechnol (2015) 33:290–5. doi: 10.1038/nbt.3122
52. Parrish N, Hormozdiari F, Eskin E. RSEM: Accurate Transcript Quantification From RNA-Seq Data With or Without a Reference Genome. BMC Bioinf (2014) 12:21–40. doi: 10.1186/1471-2105-12-323
53. Patro R, Mount SM, Kingsford C. Sailfish Enables Alignment-Free Isoform Quantification From RNA-Seq Reads Using Lightweight Algorithms. Nat Biotechnol (2014) 32:462–4. doi: 10.1038/nbt.2862
54. Love MI, Huber W, Anders S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data With Deseq2. Genome Biol (2014) 15:550. doi: 10.1186/s13059-014-0550-8
55. Robinson MD, McCarthy DJ, Smyth GK. Edger: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics (2010) 26:139–40. doi: 10.1093/bioinformatics/btp616
56. Hardcastle TJ, Kelly KA. BaySeq: Empirical Bayesian Methods for Identifying Differential Expression in Sequence Count Data. BMC Bioinf (2010) 11:422. doi: 10.1186/1471-2105-11-422
57. Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BMG, et al. EBSeq: An Empirical Bayes Hierarchical Model for Inference in RNA-Seq Experiments. Bioinformatics (2013) 29:1035–43. doi: 10.1093/bioinformatics/btt087
58. Tarazona S, Furió-Tarí P, Turrà D, Di Pietro A, Nueda MJ, Ferrer A, et al. Data Quality Aware Analysis of Differential Expression in RNA-Seq With NOISeq R/Bioc Package. Nucleic Acids Res (2015) 43:e140. doi: 10.1093/nar/gkv711
59. Li J, Tibshirani R. Finding Consistent Patterns: A Nonparametric Approach for Identifying Differential Expression in RNA-Seq Data. Stat Methods Med Res (2013) 22:519–36. doi: 10.1177/0962280211428386
60. Li S, Hu Z, Zhao Y, Huang S, He X. Transcriptome-Wide Analysis Reveals the Landscape of Aberrant Alternative Splicing Events in Liver Cancer. Hepatology (2019) 69:359–75. doi: 10.1002/hep.30158
61. Trincado JL, Entizne JC, Hysenaj G, Singh B, Skalic M, Elliott DJ, et al. SUPPA2: Fast, Accurate, and Uncertainty-Aware Differential Splicing Analysis Across Multiple Conditions. Genome Biol (2018) 19:40. doi: 10.1186/s13059-018-1417-1
62. Shen S, Park JW, Lu Z, Lin L, Henry MD, Wu YN, et al. rMATS: Robust and Flexible Detection of Differential Alternative Splicing From Replicate RNA-Seq Data. Proc Natl Acad Sci USA (2014) 111:E5593–601. doi: 10.1073/pnas.1419161111
63. Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential Analysis of Gene Regulation at Transcript Resolution With RNA-Seq. Nat Biotechnol (2013) 31:46–53. doi: 10.1038/nbt.2450
64. Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and Design of RNA Sequencing Experiments for Identifying Isoform Regulation. Nat Methods (2010) 7:1009–15. doi: 10.1038/nmeth.1528
65. Anders S, Reyes A, Huber W. Detecting Differential Usage of Exons From RNA-Seq Data. Genome Res (2012) 22:2008–17. doi: 10.1101/gr.133744.111
66. WJ K, V-S. Splicer: Classification of Alternative Splicing and Prediction of Coding Potential From RNA-Seq Data. BMC Bioinf (2014) 15:81. doi: 10.1186/1471-2105-15-81
67. John D, Weirick T, Dimmeler S, Uchida S. RNAEditor : Easy Detection of RNA Editing Events and the Introduction of Editing Islands. Brief Bioinform (2017) 18:993–1001. doi: 10.1093/bib/bbw087
68. Lo Giudice C, Tangaro MA, Pesole G, Picardi E. Investigating RNA Editing in Deep Transcriptome Datasets With REDItools and REDIportal. Nat Protoc (2020) 15:10981131. doi: 10.1038/s41596-019-0279-7
69. Piechotta M, Wyler E, Ohler U, Landthaler M, Dieterich C. JACUSA: Site-Specific Identification of RNA Editing Events From Replicate Sequencing Data. BMC Bioinf (2017) 18:1–15. doi: 10.1186/s12859-016-1432-8
70. Wang Z, Lian J, Li Q, Zhang P, Zhou Y, Zhan X, et al. RES-Scanner: A Software Package for Genome-Wide Identification of RNA-Editing Sites. Gigascience (2016) 5:1–9. doi: 10.1186/s13742-016-0143-4
71. Xia Z, Donehower LA, Cooper TA, Neilson JR, Wheeler DA, Wagner EJ, et al. Dynamic Analyses of Alternative Polyadenylation From RNA-Seq Reveal a 3’2-UTR Landscape Across Seven Tumour Types. Nat Commun (2014) 5:5274. doi: 10.1038/ncomms6274
72. Ye C, Long Y, Ji G, Li QQ, Wu X. APAtrap: Identification and Quantification of Alternative Polyadenylation Sites From RNA-Seq Data. Bioinformatics (2018) 34:1841–9. doi: 10.1093/bioinformatics/bty029
73. Lu J, Bushel PR. Dynamic Expression of 3 ′ UTRs Revealed by Poisson Hidden Markov Modeling of RNA-Seq: Implications in Gene Expression Pro Fi Ling. Gene (2013) 527:616–23. doi: 10.1016/j.gene.2013.06.052
74. Gaffen SL, Jain R, Garg AV, Cua DJ. The IL-23-IL-17 Immune Axis: From Mechanisms to Therapeutic Testing. Nat Rev Immunol (2014) 14:585–600. doi: 10.1038/nri3707
75. Towne J, Sims J. IL-36 in Psoriasis. Curr Opin Pharmacol (2012) 12:486–90. doi: 10.1016/j.coph.2012.02.009
76. Swindell WR, Beamer MA, Sarkar MK, Loftus S, Fullmer J, Xing X, et al. RNA-Seq Analysis of IL-1B and IL-36 Responses in Epidermal Keratinocytes Identifies a Shared MyD88-Dependent Gene Signature. Front Immunol (2018) 9:80. doi: 10.3389/fimmu.2018.00080
77. Nakajima S, Igyártó BZ, Honda T, Egawa G, Otsuka A, Hara-Chikuma M, et al. Langerhans Cells Are Critical in Epicutaneous Sensitization With Protein Antigen via Thymic Stromal Lymphopoietin Receptor Signaling. J Allergy Clin Immunol (2012) 129:1048–55.e6. doi: 10.1016/j.jaci.2012.01.063
78. Pavel AB, Song T, Kim HJ, Del Duca E, Krueger JG, Dubin C, et al. Oral Janus Kinase/SYK Inhibition (ASN002) Suppresses Inflammation and Improves Epidermal Barrier Markers in Patients With Atopic Dermatitis. J Allergy Clin Immunol (2019) 144:1011–24. doi: 10.1016/j.jaci.2019.07.013
79. Chambers ES, Vukmanovic-Stejic M. Skin Barrier Immunity and Ageing. Immunology (2020) 160:116–25. doi: 10.1111/imm.13152
80. Eyerich S, Eyerich K, Traidl-Hoffmann C, Biedermann T. Cutaneous Barriers and Skin Immunity: Differentiating A Connected Network. Trends Immunol (2018) 39:315–27. doi: 10.1016/j.it.2018.02.004
81. Dainichi T, Kitoh A, Otsuka A, Nakajima S, Nomura T, Kaplan DH, et al. The Epithelial Immune Microenvironment (EIME) in Atopic Dermatitis and Psoriasis. Nat Immunol (2018) 19:1286–98. doi: 10.1038/s41590-018-0256-2
82. Gutowska-Owsiak D, Schaupp AL, Salimi M, Selvakumar TA, McPherson T, Taylor S, et al. IL-17 Downregulates Filaggrin and Affects Keratinocyte Expression of Genes Associated With Cellular Adhesion. Exp Dermatol (2012) 21:104–10. doi: 10.1111/j.1600-0625.2011.01412.x
83. Kirby B. Langerhans Cells in Psoriasis: Getting to the Core of the Disease. Br J Dermatol (2018) 178:1240. doi: 10.1111/bjd.16596
84. Swindell WR, Sarkar MK, Liang Y, Xing X, Baliwag J, Elder JT, et al. RNA-Seq Identifies a Diminished Differentiation Gene Signature in Primary Monolayer Keratinocytes Grown From Lesional and Uninvolved Psoriatic Skin. Sci Rep (2017) 7:18045. doi: 10.1038/s41598-017-18404-9
85. Zhang Y-J, Sun Y-Z, Gao X-H, Qi R-Q. Integrated Bioinformatic Analysis of Differentially Expressed Genes and Signaling Pathways in Plaque Psoriasis. Mol Med Rep (2019) 20:225–35. doi: 10.3892/mmr.2019.10241
86. Luo Y, Luo Y, Chang J, Xiao Z, Zhou B. Identification of Candidate Biomarkers and Pathways Associated With Psoriasis Using Bioinformatics Analysis. Hereditas (2020) 157:30. doi: 10.1186/s41065-020-00141-1
87. Egawa G, Kabashima K. Multifactorial Skin Barrier Deficiency and Atopic Dermatitis: Essential Topics to Prevent the Atopic March. J Allergy Clin Immunol (2016) 138:350–8.e1. doi: 10.1016/j.jaci.2016.06.002
88. Rodríguez E, Baurecht H, Herberich E, Wagenpfeil S, Brown SJ, Cordell HJ, et al. Meta-Analysis of Filaggrin Polymorphisms in Eczema and Asthma: Robust Risk Factors in Atopic Disease. J Allergy Clin Immunol (2009) 123:1361–70.e7. doi: 10.1016/j.jaci.2009.03.036
89. Catunda R, Rekhi U, Clark D, Levin L, Febbraio M. Loricrin Downregulation and Epithelial-Related Disorders: A Systematic Review. J der Dtsch Dermatologischen Gesellschaft = J Ger Soc Dermatol JDDG (2019) 17:1227–38. doi: 10.1111/ddg.14001
90. Goleva E, Berdyshev E, Leung DYM. Epithelial Barrier Repair and Prevention of Allergy. J Clin Invest (2019) 129:1463–74. doi: 10.1172/JCI124608
91. Lee A-Y. Molecular Sciences The Role of MicroRNAs in Epidermal Barrier. Int J Mol Sci (2020) 21:5781. doi: 10.3390/ijms21165781
92. Saraceno R, Kleyn CE, Terenghi G, Griffiths CEM. The Role of Neuropeptides in Psoriasis. Br J Dermatol (2006) 155:876–82. doi: 10.1111/j.1365-2133.2006.07518.x
93. Komiya E, Tominaga M, Kamata Y, Suga Y, Takamori K. Molecular and Cellular Mechanisms of Itch in Psoriasis. Int J Mol Sci (2020) 21:8406. doi: 10.3390/ijms21218406
94. Kittaka H, Tominaga M. The Molecular and Cellular Mechanisms of Itch and the Involvement of TRP Channels in the Peripheral Sensory Nervous System and Skin. Allergol Int (2017) 66:22–30. doi: 10.1016/j.alit.2016.10.003
95. Sun S, Dong X. Trp Channels and Itch. Semin Immunopathol (2016) 38:293–307. doi: 10.1007/s00281-015-0530-4
96. Lee SH, Tonello R, Choi Y, Jung SJ, Berta T. Sensory Neuron-Expressed TRPC4 Is a Target for the Relief of Psoriasiform Itch and Skin Inflammation in Mice. J Invest Dermatol (2020) 140:2221–9.e6. doi: 10.1016/j.jid.2020.03.959
97. Moynes DM, Vanner SJ, Lomax AE. Participation of Interleukin 17A in Neuroimmune Interactions. Brain Behav Immun (2014) 41:1–9. doi: 10.1016/j.bbi.2014.03.004
98. Barry DM, Liu X-T, Liu B, Liu X-Y, Gao F, Zeng X, et al. Exploration of Sensory and Spinal Neurons Expressing Gastrin-Releasing Peptide in Itch and Pain Related Behaviors. Nat Commun (2020) 11:1397. doi: 10.1038/s41467-020-15230-y
99. Riol-Blanco L, Ordovas-Montanes J, Perro M, Naval E, Thiriot A, Alvarez D, et al. Nociceptive Sensory Neurons Drive Interleukin-23-Mediated Psoriasiform Skin Inflammation. Nature (2014) 510:157–61. doi: 10.1038/nature13199
100. Bushmakin AG, Mamolo C, Cappelleri JC, Stewart M. The Relationship Between Pruritus and the Clinical Signs of Psoriasis in Patients Receiving Tofacitinib. J Dermatol Treat (2015) 26:19–22. doi: 10.3109/09546634.2013.861891
101. Feldman SR, Thaçi D, Gooderham M, Augustin M, de la Cruz C, Mallbris L, et al. Tofacitinib Improves Pruritus and Health-Related Quality of Life Up to 52 Weeks: Results From 2 Randomized Phase III Trials in Patients With Moderate to Severe Plaque Psoriasis. J Am Acad Dermatol (2016) 75:1162–1170.e3. doi: 10.1016/j.jaad.2016.07.040
102. Hashimoto T, Sakai K, Sanders KM, Yosipovitch G, Akiyama T. Antipruritic Effects of Janus Kinase Inhibitor Tofacitinib in a Mouse Model of Psoriasis. Acta Derm Venereol (2019) 99:298–303. doi: 10.2340/00015555-3086
103. Paus R, Schmelz M, Bíró T, Steinhoff M. Frontiers in Pruritus Research: Scratching the Brain for More Effective Itch Therapy. J Clin Invest (2006) 116:1174–86. doi: 10.1172/JCI28553
104. Wang F, Trier AM, Li F, Kim S, Chen Z, Chai JN, et al. A Basophil-Neuronal Axis Promotes Itch. Cell (2021) 184:422–40.e17. doi: 10.1016/j.cell.2020.12.033
105. Steinhoff M, Buddenkotte J, Lerner EA. Role of Mast Cells and Basophils in Pruritus. Immunol Rev (2018) 282:248–64. doi: 10.1111/imr.12635
106. Walsh CM, Hill RZ, Schwendinger-Schreck J, Deguine J, Brock EC, Kucirek N, et al. Neutrophils Promote CXCR3-Dependent Itch in the Development of Atopic Dermatitis. Elife (2019) 8:e48448. doi: 10.7554/eLife.48448
107. Cevikbas F, Wang X, Akiyama T, Kempkes C, Savinko T, Antal A, et al. A Sensory Neuron-Expressed IL-31 Receptor Mediates T Helper Cell-Dependent Itch: Involvement of TRPV1 and TRPA1. J Allergy Clin Immunol (2014) 133:448–60. doi: 10.1016/j.jaci.2013.10.048
108. Wilson SR, Thé L, Batia LM, Beattie K, Katibah GE, McClain SP, et al. The Epithelial Cell-Derived Atopic Dermatitis Cytokine TSLP Activates Neurons to Induce Itch. Cell (2013) 155:285–95. doi: 10.1016/j.cell.2013.08.057
109. Belkaid Y, Segre JA. Dialogue Between Skin Microbiota and Immunity. Science (2014) 346:954–9. doi: 10.1126/science.1260144
110. Fyhrquist N, Muirhead G, Prast-Nielsen S, Jeanmougin M, Olah P, Skoog T, et al. Microbe-Host Interplay in Atopic Dermatitis and Psoriasis. Nat Commun (2019) 10:4703. doi: 10.1038/s41467-019-12253-y
111. Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, et al. Temporal Shifts in the Skin Microbiome Associated With Disease Flares and Treatment in Children With Atopic Dermatitis. Genome Res (2012) 22:850–9. doi: 10.1101/gr.131029.111
112. Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The Landscape of Long Noncoding RNAs in the Human Transcriptome. Nat Genet (2015) 47:199–208. doi: 10.1038/ng.3192
113. Anastasiadou E, Jacob LS, Slack FJ. Non-Coding RNA Networks in Cancer. Nat Rev Cancer (2018) 18:5–18. doi: 10.1038/nrc.2017.99
114. Chen YG, Satpathy AT, Chang HY. Gene Regulation in the Immune System by Long Noncoding RNAs. Nat Immunol (2017) 18:962–72. doi: 10.1038/ni.3771
115. Allou L, Balzano S, Magg A, Quinodoz M, Royer-Bertrand B, Schöpflin R, et al. Non-Coding Deletions Identify Maenli lncRNA as a Limb-Specific En1 Regulator. Nature (2021) 592:9398. doi: 10.1038/s41586-021-03208-9
116. Sonkoly E, Wei T, Janson PCJ, Sääf A, Lundeberg L, Tengvall-Linder M, et al. MicroRNAs: Novel Regulators Involved in the Pathogenesis of Psoriasis? PloS One (2007) 2:e610. doi: 10.1371/journal.pone.0000610
117. Xu Y, Ji Y, Lan X, Gao X, Chen H-D, Geng L. Mir−203 Contributes to IL−17−induced VEGF Secretion by Targeting SOCS3 in Keratinocytes. Mol Med Rep (2017) 16:8989–96. doi: 10.3892/mmr.2017.7759
118. Xiao Y, Wang H, Wang C, Zeng B, Tang X, Zhang Y, et al. miR-203 Promotes HaCaT Cell Overproliferation Through Targeting LXR-α and PPAR-γ. Cell Cycle (2020) 19:1928–40. doi: 10.1080/15384101.2020.1783934
119. Lv Y, Qi R, Xu J, Di Z, Zheng H, Huo W, et al. Profiling of Serum and Urinary microRNAs in Children With Atopic Dermatitis. PloS One (2014) 9:e115448. doi: 10.1371/journal.pone.0115448
120. Rebane A, Runnel T, Aab A, Maslovskaja J, Rückert B, Zimmermann M, et al. MicroRNA-146a Alleviates Chronic Skin Inflammation in Atopic Dermatitis Through Suppression of Innate Immune Responses in Keratinocytes. J Allergy Clin Immunol (2014) 134:836–47.e11. doi: 10.1016/j.jaci.2014.05.022
121. Taganov KD, Boldin MP, Chang K-J, Baltimore D. NF-kappaB-Dependent Induction of microRNA miR-146, an Inhibitor Targeted to Signaling Proteins of Innate Immune Responses. Proc Natl Acad Sci USA (2006) 103:12481–6. doi: 10.1073/pnas.0605298103
122. Crone SG, Jacobsen A, Federspiel B, Bardram L, Krogh A, Lund AH, et al. microRNA-146a Inhibits G Protein-Coupled Receptor-Mediated Activation of NF-κb by Targeting CARD10 and COPS8 in Gastric Cancer. Mol Cancer (2012) 11:71. doi: 10.1186/1476-4598-11-71
123. Sonkoly E, Janson P, Majuri M-L, Savinko T, Fyhrquist N, Eidsmo L, et al. MiR-155 Is Overexpressed in Patients With Atopic Dermatitis and Modulates T-Cell Proliferative Responses by Targeting Cytotoxic T Lymphocyte-Associated Antigen 4. J Allergy Clin Immunol (2010) 126:520–81. doi: 10.1016/j.jaci.2010.05.045
124. Wang X, Chen Y, Yuan W, Yao L, Wang S, Jia Z, et al. MicroRNA-155-5p Is a Key Regulator of Allergic Inflammation, Modulating the Epithelial Barrier by Targeting Pkiα. Cell Death Dis (2019) 10:884. doi: 10.1038/s41419-019-2124-x
125. Wang H, Zhang Y, Luomei J, Huang P, Zhou R, Peng Y. The miR-155/GATA3/IL37 Axis Modulates the Production of Proinflammatory Cytokines Upon TNF-α Stimulation to Affect Psoriasis Development. Exp Dermatol (2020) 29:647658. doi: 10.1111/exd.14117
126. Løvendorf MB, Zibert JR, Gyldenløve M, Røpke MA, Skov L. MicroRNA-223 and miR-143 Are Important Systemic Biomarkers for Disease Activity in Psoriasis. J Dermatol Sci (2014) 75:133–9. doi: 10.1016/j.jdermsci.2014.05.005
127. Yu X, Wang M, Li L, Zhang L, Chan MTV, Wu WKK. MicroRNAs in Atopic Dermatitis: A Systematic Review. J Cell Mol Med (2020) 24:5966–72. doi: 10.1111/jcmm.15208
128. Jia H-Z, Liu S-L, Zou Y-F, Chen X-F, Yu L, Wan J, et al. MicroRNA-223 Is Involved in the Pathogenesis of Atopic Dermatitis by Affecting Histamine-N-Methyltransferase. Cell Mol Biol (Noisy-le-grand) (2018) 64:103–7. doi: 10.14715/cmb/2018.64.3.17
129. Wang R, Wang F-F, Cao H-W, Yang J-Y. MiR-223 Regulates Proliferation and Apoptosis of IL-22-Stimulated HaCat Human Keratinocyte Cell Lines via the PTEN/Akt Pathway. Life Sci (2019) 230:28–34. doi: 10.1016/j.lfs.2019.05.045
130. Scalavino V, Liso M, Cavalcanti E, Gigante I, Lippolis A, Mastronardi M, et al. miR-369-3p Modulates Inducible Nitric Oxide Synthase and Is Involved in Regulation of Chronic Inflammatory Response. Sci Rep (2020) 10:15942. doi: 10.1038/s41598-020-72991-8
131. Guo S, Zhang W, Wei C, Wang L, Zhu G, Shi Q, et al. Serum and Skin Levels of miR-369-3p in Patients With Psoriasis and Their Correlation With Disease Severity. Eur J Dermatol (2013) 23:608–13. doi: 10.1684/ejd.2013.2148
132. Bélanger É, Madore A-M, Boucher-Lafleur A-M, Simon M-M, Kwan T, Pastinen T, et al. Eosinophil microRNAs Play a Regulatory Role in Allergic Diseases Included in the Atopic March. Int J Mol Sci (2020) 21:9011. doi: 10.3390/ijms21239011
133. Chen X-F, Zhang L-J, Zhang J, Dou X, Shao Y, Jia X-J, et al. MiR-151a Is Involved in the Pathogenesis of Atopic Dermatitis by Regulating Interleukin-12 Receptor β2. Exp Dermatol (2018) 27:427–32. doi: 10.1111/exd.13276
134. Tsoi LC, Iyer MK, Stuart PE, Swindell WR, Gudjonsson JE, Tejasvi T, et al. Analysis of Long Non-Coding RNAs Highlights Tissue-Specific Expression Patterns and Epigenetic Profiles in Normal and Psoriatic Skin. Genome Biol (2015) 16:24. doi: 10.1186/s13059-014-0570-4
135. Li CX, Li HG, Huang LT, Kong YW, Chen FY, Liang JY, et al. H19 lncRNA Regulates Keratinocyte Differentiation by Targeting miR-130b-3p. Cell Death Dis (2017) 8:e3174. doi: 10.1038/cddis.2017.516
136. Qiao M, Li R, Zhao X, Yan J, Sun Q. Up-Regulated lncRNA-MSX2P1 Promotes the Growth of IL-22-Stimulated Keratinocytes by Inhibiting miR-6731-5p and Activating S100A7. Exp Cell Res (2018) 363:243–54. doi: 10.1016/j.yexcr.2018.01.014
137. Jia H-Y, Zhang K, Lu W-J, Xu G-W, Zhang J-F, Tang Z-L. LncRNA MEG3 Influences the Proliferation and Apoptosis of Psoriasis Epidermal Cells by Targeting miR-21/Caspase-8. BMC Mol Cell Biol (2019) 20:46. doi: 10.1186/s12860-019-0229-9
138. Szegedi K, Sonkoly E, Nagy N, Németh IB, Bata-Csörgo Z, Kemény L, et al. The Anti-Apoptotic Protein G1P3 Is Overexpressed in Psoriasis and Regulated by the Non-Coding RNA, PRINS. Exp Dermatol (2010) 19:269–78. doi: 10.1111/j.1600-0625.2010.01066.x
139. Luo M, Huang P, Pan Y, Zhu Z, Zhou R, Yang Z, et al. Weighted Gene Coexpression Network and Experimental Analyses Identify lncRNA SPRR2C as a Regulator of the IL-22-Stimulated HaCaT Cell Phenotype Through the miR-330/STAT1/S100A7 Axis. Cell Death Dis (2021) 12:86. doi: 10.1038/s41419-020-03305-z
140. Niu L, Lou F, Sun Y, Sun L, Cai X, Liu Z, et al. A Micropeptide Encoded by lncRNA MIR155HG Suppresses Autoimmune Inflammation via Modulating Antigen Presentation. Sci Adv (2020) 6:eaaz2059. doi: 10.1126/sciadv.aaz2059
141. Lewis BP, Burge CB, Bartel DP. Conserved Seed Pairing, Often Flanked by Adenosines, Indicates That Thousands of Human Genes Are microRNA Targets. Cell (2005) 120:15–20. doi: 10.1016/j.cell.2004.12.035
142. Hornstein E, Shomron N. Canalization of Development by microRNAs. Nat Genet (2006) 38 Suppl:S20–4. doi: 10.1038/ng1803
143. El-Komy M, Amin I, El-Hawary MS, Saadi D, Shaker O. Upregulation of the miRNA-155, miRNA-210, and miRNA-20b in Psoriasis Patients and Their Relation to IL-17. Int J Immunopathol Pharmacol (2020) 34:2058738420933742. doi: 10.1177/2058738420933742
144. Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative Annotation of Human Large Intergenic Noncoding RNAs Reveals Global Properties and Specific Subclasses. Genes Dev (2011) 25:1915–27. doi: 10.1101/gad.17446611
145. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE V7 Catalog of Human Long Noncoding RNAs: Analysis of Their Gene Structure, Evolution, and Expression. Genome Res (2012) 22:1775–89. doi: 10.1101/gr.132159.111
146. Ponting CP, Oliver PL, Reik W. Evolution and Functions of Long Noncoding RNAs. Cell (2009) 136:629–41. doi: 10.1016/j.cell.2009.02.006
147. Kopp F, Mendell JT. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell (2018) 172:393–407. doi: 10.1016/j.cell.2018.01.011
148. Long Y, Wang X, Youmans DT, Cech TR. How do lncRNAs Regulate Transcription? Sci Adv (2017) 3:eaao2110. doi: 10.1126/sciadv.aao2110
149. Volders P-J, Anckaert J, Verheggen K, Nuytens J, Martens L, Mestdagh P, et al. LNCipedia 5: Towards a Reference Set of Human Long Non-Coding RNAs. Nucleic Acids Res (2019) 47:D135–9. doi: 10.1093/nar/gky1031
150. Zhao L, Wang J, Li Y, Song T, Wu Y, Fang S, et al. NONCODEV6: An Updated Database Dedicated to Long Non-Coding RNA Annotation in Both Animals and Plants. Nucleic Acids Res (2021) 49:D165–71. doi: 10.1093/nar/gkaa1046
151. Ghafouri-Fard S, Eghtedarian R, Taheri M, Rakhshan A. The Eminent Roles of ncRNAs in the Pathogenesis of Psoriasis. Non-coding RNA Res (2020) 5:99–108. doi: 10.1016/j.ncrna.2020.06.002
152. Yan J, Song J, Qiao M, Zhao X, Li R, Jiao J, et al. Long Noncoding RNA Expression Profile and Functional Analysis in Psoriasis. Mol Med Rep (2019) 19:3421–30. doi: 10.3892/mmr.2019.9993
153. Li H, Yang C, Zhang J, Zhong W, Zhu L, Chen Y. Identification of Potential Key mRNAs and LncRNAs for Psoriasis by Bioinformatic Analysis Using Weighted Gene Co-Expression Network Analysis. Mol Genet Genomics (2020) 295:741–9. doi: 10.1007/s00438-020-01654-0
154. Matsumoto A, Pasut A, Matsumoto M, Yamashita R, Fung J, Monteleone E, et al. MTORC1 and Muscle Regeneration Are Regulated by the LINC00961-Encoded SPAR Polypeptide. Nature (2017) 541:228–32. doi: 10.1038/nature21034
155. Wu P, Mo Y, Peng M, Tang T, Zhong Y, Deng X, et al. Emerging Role of Tumor-Related Functional Peptides Encoded by lncRNA and circRNA. Mol Cancer (2020) 19:22. doi: 10.1186/s12943-020-1147-3
156. de Klerk E, ‘t Hoen PAC. Alternative mRNA Transcription, Processing, and Translation: Insights From RNA Sequencing. Trends Genet (2015) 31:128–39. doi: 10.1016/j.tig.2015.01.001
157. Li Y, Sun N, Lu Z, Sun S, Huang J, Chen Z, et al. Prognostic Alternative mRNA Splicing Signature in Non-Small Cell Lung Cancer. Cancer Lett (2017) 393:40–51. doi: 10.1016/j.canlet.2017.02.016
158. Xu X, Wang Y, Liang H. The Role of A-To-I RNA Editing in Cancer Development. Curr Opin Genet Dev (2018) 48:51–6. doi: 10.1016/j.gde.2017.10.009
159. Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep Surveying of Alternative Splicing Complexity in the Human Transcriptome by High-Throughput Sequencing. Nat Genet (2008) 40:1413–5. doi: 10.1038/ng.259
160. Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, et al. Alternative Isoform Regulation in Human Tissue Transcriptomes. Nature (2008) 456:470–6. doi: 10.1038/nature07509
161. Wahl MC, Will CL, Lührmann R. The Spliceosome: Design Principles of a Dynamic RNP Machine. Cell (2009) 136:701–18. doi: 10.1016/j.cell.2009.02.009
162. Ule J, Blencowe BJ. Alternative Splicing Regulatory Networks: Functions, Mechanisms, and Evolution. Mol Cell (2019) 76:329–45. doi: 10.1016/j.molcel.2019.09.017
163. Bonnal SC, López-Oreja I, Valcárcel J. Roles and Mechanisms of Alternative Splicing in Cancer - Implications for Care. Nat Rev Clin Oncol (2020) 17:457–74. doi: 10.1038/s41571-020-0350-x
164. Shimizu M, Matsuda A, Yanagisawa K, Hirota T, Akahoshi M, Inomata N, et al. Functional SNPs in the Distal Promoter of the ST2 Gene Are Associated With Atopic Dermatitis. Hum Mol Genet (2005) 14:2919–27. doi: 10.1093/hmg/ddi323
165. Wu L, Wang C, Boisson B, Misra S, Rayman P, Finke JH, et al. The Differential Regulation of Human ACT1 Isoforms by Hsp90 in IL-17 Signaling. J Immunol (2014) 193:1590–9. doi: 10.4049/jimmunol.1400715
166. Lee SK, Cieply B, Yang Y, Peart N, Glaser C, Chan P, et al. Esrp1-Regulated Splicing of Arhgef11 Isoforms Is Required for Epithelial Tight Junction Integrity. Cell Rep (2018) 25:2417–2430.e5. doi: 10.1016/j.celrep.2018.10.097
167. Wang Y, Xu X, Yu S, Jeong KJ, Zhou Z, Han L, et al. Systematic Characterization of A-To-I RNA Editing Hotspots in microRNAs Across Human Cancers. Genome Res (2017) 27:1112–25. doi: 10.1101/gr.219741.116
168. Gong J, Liu C, Liu W, Xiang Y, Diao L, Guo A-Y, et al. LNCediting: A Database for Functional Effects of RNA Editing in lncRNAs. Nucleic Acids Res (2017) 45:D79–84. doi: 10.1093/nar/gkw835
169. Peng X, Xu X, Wang Y, Hawke DH, Yu S, Han L, et al. A-To-I RNA Editing Contributes to Proteomic Diversity in Cancer. Cancer Cell (2018) 33:817–28.e7. doi: 10.1016/j.ccell.2018.03.026
170. Joyce CE, Zhou X, Xia J, Ryan C, Thrash B, Menter A, et al. Deep Sequencing of Small RNAs From Human Skin Reveals Major Alterations in the Psoriasis Mirnaome. Hum Mol Genet (2011) 20:4025–40. doi: 10.1093/hmg/ddr331
171. Shallev L, Kopel E, Feiglin A, Leichner GS, Avni D, Sidi Y, et al. Decreased A-To-I RNA Editing as a Source of Keratinocytes’ dsRNA in Psoriasis. Rna (2018) 24:828–40. doi: 10.1261/rna.064659.117
172. Haque A, Engel J, Teichmann SA, Lönnberg T. A Practical Guide to Single-Cell RNA-Sequencing for Biomedical Research and Clinical Applications. Genome Med (2017) 9:75. doi: 10.1186/s13073-017-0467-4
173. Tanay A, Regev A. Scaling Single-Cell Genomics From Phenomenology to Mechanism. Nature (2017) 541:331–8. doi: 10.1038/nature21350
174. Baccin C, Al-Sabah J, Velten L, Helbling PM, Grünschläger F, Hernández-Malmierca P, et al. Combined Single-Cell and Spatial Transcriptomics Reveal the Molecular, Cellular and Spatial Bone Marrow Niche Organization. Nat Cell Biol (2020) 22:38–48. doi: 10.1038/s41556-019-0439-6
175. Shaw R, Tian X, Xu J. Single-Cell Transcriptome Analysis in Plants: Advances and Challenges. Mol Plant (2021) 14:115–26. doi: 10.1016/j.molp.2020.10.012
176. Reynolds G, Vegh P, Fletcher J, Poyner EFM, Stephenson E, Goh I, et al. Developmental Cell Programs Are Co-Opted in Inflammatory Skin Disease. Science (2021) 371:eaba6500. doi: 10.1126/science.aba6500
177. Azizi E, Carr AJ, Plitas G, Mazutis L, Rudensky AY, Pe D, et al. Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell (2018) 174:1293–308.e36. doi: 10.1016/j.cell.2018.05.060
178. Parekh S, Ziegenhain C, Vieth B, Enard W, Hellmann I. zUMIs - A Fast and Flexible Pipeline to Process RNA Sequencing Data With UMIs. Gigascience (2018) 7:1–9. doi: 10.1093/gigascience/giy059
179. Wolock SL, Lopez R, Klein AM. Scrublet: Computational Identification of Cell Doublets in Single-Cell Transcriptomic Data. Cell Syst (2019) 8:281–91.e9. doi: 10.1016/j.cels.2018.11.005
180. Bais AS, Kostka D. Scds: Computational Annotation of Doublets in Single-Cell RNA Sequencing Data. Bioinformatics (2020) 36:1150–8. doi: 10.1093/bioinformatics/btz698
181. McGinnis CS, Murrow LM, Gartner ZJ. DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst (2019) 8:329–37.e4. doi: 10.1016/j.cels.2019.03.003
182. Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, et al. Integrated Analysis of Multimodal Single-Cell Data. Cell (2021) 184:3573–87.e29. doi: 10.1016/j.cell.2021.04.048
183. Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, et al. Reference-Based Analysis of Lung Single-Cell Sequencing Reveals a Transitional Profibrotic Macrophage. Nat Immunol (2019) 20:163–72. doi: 10.1038/s41590-018-0276-y
184. Zhang AW, O’Flanagan C, Chavez EA, Lim JLP, Ceglia N, McPherson A, et al. Probabilistic Cell-Type Assignment of Single-Cell RNA-Seq for Tumor Microenvironment Profiling. Nat Methods (2019) 16:1007–15. doi: 10.1038/s41592-019-0529-1
185. Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, et al. SCENIC: Single-Cell Regulatory Network Inference and Clustering. Nat Methods (2017) 14:1083–6. doi: 10.1038/nmeth.4463
186. Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, et al. The Dynamics and Regulators of Cell Fate Decisions Are Revealed by Pseudotemporal Ordering of Single Cells. Nat Biotechnol (2014) 32:381–6. doi: 10.1038/nbt.2859
187. Metallurgy R. Slingshot: Cell Lineage and Pseudotime Inference for Single-Cell Transcriptomics. BMC Genomics (2018) 19:477. doi: 10.1186/s12864-018-4772-0
188. Wolf FA, Hamey FK, Plass M, Solana J, Dahlin JS, Göttgens B, et al. PAGA: Graph Abstraction Reconciles Clustering With Trajectory Inference Through a Topology Preserving Map of Single Cells. Genome Biol (2019) 20:1–9. doi: 10.1186/s13059-019-1663-x
189. Haghverdi L, Büttner M, Wolf FA, Buettner F, Theis FJ. Diffusion Pseudotime Robustly Reconstructs Lineage Branching. Nat Methods (2016) 13:845–8. doi: 10.1038/nmeth.3971
190. Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: Inferring Cell–Cell Communication From Combined Expression of Multi-Subunit Ligand–Receptor Complexes. Nat Protoc (2020) 15:1484–506. doi: 10.1038/s41596-020-0292-x
191. Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, et al. Inference and Analysis of Cell-Cell Communication Using CellChat. Nat Commun (2021) 12:1–20. doi: 10.1038/s41467-021-21246-9
192. Browaeys R, Saelens W, Saeys Y. NicheNet: Modeling Intercellular Communication by Linking Ligands to Target Genes. Nat Methods (2020) 17:159–62. doi: 10.1038/s41592-019-0667-5
193. Zhao E, Stone MR, Ren X, Guenthoer J, Smythe KS, Pulliam T, et al. Spatial Transcriptomics at Subspot Resolution With BayesSpace. Nat Biotechnol (2021). doi: 10.1038/s41587-021-00935-2
194. Dries R, Zhu Q, Dong R, Eng CHL, Li H, Liu K, et al. Giotto: A Toolbox for Integrative Analysis and Visualization of Spatial Expression Data. Genome Biol (2021) 22:1–31. doi: 10.1186/s13059-021-02286-2
195. Pham D, Tan X, Xu J, Grice LF, Lam PY, Raghubar A, et al. Stlearn: Integrating Spatial Location, Tissue Morphology and Gene Expression to Find Cell Types, Cell-Cell Interactions and Spatial Trajectories Within Undissociated Tissues. bioRxiv (2020). doi: 10.1101/2020.05.31.125658
196. Cheng JB, Sedgewick AJ, Finnegan AI, Harirchian P, Lee J, Kwon S, et al. Transcriptional Programming of Normal and Inflamed Human Epidermis at Single-Cell Resolution. Cell Rep (2018) 25:871–83. doi: 10.1016/j.celrep.2018.09.006
197. Liu J, Chang HW, Huang ZM, Nakamura M, Sekhon S, Ahn R, et al. Single-Cell RNA Sequencing of Psoriatic Skin Identifies Pathogenic Tc17 Cell Subsets and Reveals Distinctions Between CD8+ T Cells in Autoimmunity and Cancer. J Allergy Clin Immunol (2021) 147:23702380. doi: 10.1016/j.jaci.2020.11.028
198. Hughes TK, Wadsworth MH, Gierahn TM, Do T, Weiss D, Andrade PR, et al. Second-Strand Synthesis-Based Massively Parallel scRNA-Seq Reveals Cellular States and Molecular Features of Human Inflammatory Skin Pathologies. Immunity (2020) 53:878–94.e7. doi: 10.1016/j.immuni.2020.09.015
Keywords: transcriptomics, inflammatory skin diseases, RNA-Seq, bioinformatics, atopic dermatitis, psoriasis
Citation: Wu J, Fang Z, Liu T, Hu W, Wu Y and Li S (2021) Maximizing the Utility of Transcriptomics Data in Inflammatory Skin Diseases. Front. Immunol. 12:761890. doi: 10.3389/fimmu.2021.761890
Received: 20 August 2021; Accepted: 15 October 2021;
Published: 29 October 2021.
Edited by:
Jillian M. Richmond, University of Massachusetts Medical School, United StatesReviewed by:
Chunjie Jiang, University of Pennsylvania, United StatesCopyright © 2021 Wu, Fang, Liu, Hu, Wu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shengli Li, c2hlbmdsaS5saUBzaHNtdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.