
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 16 December 2021
Sec. Microbial Immunology
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.760475
This article is part of the Research TopicCurrent Trends in Exploiting Molecular Signaling in Bacteria-Host CrosstalkView all 30 articles
 Maryam Ali Mohammadie Kojour1
Maryam Ali Mohammadie Kojour1 Tariku Tesfaye Edosa1,2
Tariku Tesfaye Edosa1,2 Ho Am Jang1
Ho Am Jang1 Maryam Keshavarz1,3
Maryam Keshavarz1,3 Yong Hun Jo1*
Yong Hun Jo1* Yeon Soo Han1*
Yeon Soo Han1*The dimeric cytokine ligand Spätzle (Spz) is responsible for Toll pathway activation and antimicrobial peptide (AMP) production upon pathogen challenge in Tenebrio molitor. Here, we indicated that TmSpz5 has a functional role in response to bacterial infections. We showed that the highest expression of TmSpz5 is induced by Candida albicans. However, TmSpz5 knockdown reduced larval survival against Escherichia coli and Staphylococcus aureus. To evaluate the molecular mechanism underlying the observed survival differences, the role of TmSpz5 in AMP production was examined by RNA interference and microbial injection. T. molitor AMPs that are active against Gram-negative and -positive bacteria, including Tmtenecins, Tmattacins, Tmcoleoptericins, Tmtaumatin-like-proteins, and Tmcecropin-2, were significantly downregulated by TmSpz-5 RNAi in the Malpighian tubules (MTs) following a challenge with E. coli and S. aureus. However, upon infection with C. albicans the mRNA levels of most AMPs in the dsTmSpz5-injected group were similar to those in the control groups. Likewise, the expression of the transcription factors NF-κB, TmDorX2, and TmRelish were noticeably suppressed in the MTs of TmSpz5-silenced larvae. Moreover, E. coli-infected TmSpz5 knockdown larvae showed decreased antimicrobial activity in the MTs and hindgut compared with the control group. These results demonstrate that TmSpz5 has a defined role in T. molitor innate immunity by regulating AMP expression in MTs in response to E. coli.
Insects have been the largest and most diverse class over millions of years of evolution and have adapted to survive in a vast range of ecological territories (1–3). Owing to their exposure to various pathogen sources including bacteria, fungi, parasites, and viruses, they have evolved several multifunctional defense mechanisms, making them an exceptional model for immunity studies (4, 5). Unlike mammals, insects do not have an adaptive immunity (2). However, they do possess a functional innate immune system, involving both humoral and cellular immune responses (6). Cellular immunity, mediated by hemocytes (insect blood cells), involves nodulation (7), encapsulation (8), and phagocytosis (9). Humoral immune response, on the other hand, is mainly mediated by fat bodies (the equivalent of the mammalian liver) and soluble plasma proteins. The production of antimicrobial peptides (AMPs) is the main determinant of humoral immunity (6). Following invader recognition, AMP production is stimulated by the activation of two major signaling pathways, the immune deficiency (Imd) and Toll pathways (6, 10).
The Toll signaling pathway was initially identified as a dorso-ventral axis establishment regulator during embryonic development in Drosophila melanogaster (11). Since then, extensive molecular and mapping studies have provided insight into the roles of the Toll pathway and its components in the Drosophila immune system. The key activator of the Toll transmembrane-associated receptor is the endogenous cytokine-like polypeptide Spätzle (Spz) (12). Invader detection by peptidoglycan recognition proteins (PGRPs) or β-1,3-glucan recognition protein (βGRP)/Gram-negative-binding proteins (GNBPs) in Drosophila leads to a proteolytic cascade that eventually results in Spätzle cleavage and activation. Spätzle can then bind to the Toll receptor and activate downstream signaling pathways, leading to AMP production (10, 13, 14). In addition to its role in D. melanogaster, Spätzle has been shown to have significant roles in different species and taxa, including mosquitoes (15–17), Manduca sexta (18), Bombyx mori (19), shrimp (20), and Tenebrio molitor (21).
Comprehensive biochemical studies of innate immunity claim for a relatively large insect model to enable the collection of sufficient hemolymph samples. Thus, in the last two decades, T. molitor has become a common model for biochemical and molecular studies on innate immunity pathways and their components (22).
Toll signaling in T. molitor is activated when PGRP-SA and GNBP1 recognize meso-diaminopimelic acid (DAP)-type peptidoglycan (PGN) of Gram-negative bacteria and some Bacillus species, and the lysine-type peptidoglycan of Gram-positive bacteria (21, 23–26). However, in Drosophila, the PGRP-SA/GNBP1 complex solely recognizes Gram-positive bacterial and fungal infections, whereas Gram-negative bacteria can be sensed by the alternative receptors PGRP-LC and PGRP-LE and triggers the Imd signaling pathway (6, 27–31). Following recognition, a proteolytic cascade activation, including modular serine protease (MSP), Spz-processing enzyme (SPE)-activating enzyme (SAE), and SPE, leads to the cleavage of Spätzle zymogen, and eventually, mature Spätzle recruitments to the Toll receptor (32, 33). Upon Spz–Toll association in T. molitor, an intracellular cascade is activated, resulting in the engagement of myeloid differentiation factor 88 (MyD88), tube, pelle, pellino, and tumor necrosis factor receptor associated factor (TRAF). This ultimately leads to the binding of cactin to cactus, a dorsal-related immunity factor (Dif) and dorsal inhibitor (34, 35). These transcription factors translocate to the nucleus (36, 37), where they bind to NF-κB-response elements and induce AMP genes transcription (38–43). We have identified nine Spätzle genes (TmSpz-like, -1b, -3, -4, -5, -6, -7, -7a, and -7b) in T. molitor. However, the functional importance of these isoforms is poorly understood. To date, only two T. molitor Spätzle genes (TmSpz4 and TmSpz6) have been functionally characterized (44, 45). In this study, we focused on the immunological significance of TmSpz5 against microbial infection (Supplementary Table 1).
T. molitor larvae were reared under dark conditions at 26 ± 1°C and 60 ± 5% relative humidity in an environmental chamber established in the laboratory. Larvae were fed an artificial diet consisting of 1.1 g sorbic acid, 1.1 ml propionic acid, 20 g bean powder, 10 g brewer’s yeast powder, and 200 g wheat bran in 4,400 ml distilled water. The feed was autoclaved at 121°C for 15 min and fed to healthy 10th–12th instar larvae for all experiments.
The Gram-negative bacterium Escherichia coli (strain K12), Gram-positive bacterium Staphylococcus aureus (strain RN4220), and fungus Candida albicans (strain AUMC 13529) were used as pathogenic invaders. E. coli and S. aureus were cultured in Luria–Bertani (LB) broth, and C. albicans was cultured in Sabouraud’s dextrose broth overnight at 37°C. The microorganisms were harvested and washed twice in phosphate-buffered saline (PBS; pH 7.0) and then centrifuged at 3,500 × g for 15 min. Subsequently, the samples were suspended in PBS, and concentrations were measured at 600 nm (OD600) by spectrophotometry (Eppendorf, Hamburg, Germany). E. coli and S. aureus were diluted to 1 × 106 cells/µl, and C. albicans was diluted to 5 × 104 cells/µl for immune challenge studies.
The TmSpz5 gene sequence (accession number: MW916536) was obtained from the T. molitor RNAseq analysis (unpublished) and NCBI Expressed Sequence Tag (EST) database. The Tribolium castaneum Spz5 amino acid sequence (accession number: XP_008193940.1) was used as the query for identification by local-tblastn searches. The full-length open reading frame (ORF) and deduced amino acid sequences of TmSpz5 were analyzed using BLASTp (NCBI; https://blast.ncbi.nlm.nih.gov/Blast.cgi). The domain architectures of the protein sequences were retrieved using InterProScan (https://www.ebi.ac.uk/interpro/search/sequence-search). Signal peptides were predicted using the SignalP 5.0 server (http://www.cbs.dtu.dk/services/SignalP/).
A multiple-sequence alignment of the TmSpz5 amino acid sequence with representative Spätzle amino acid sequences from other insects (retrieved from GenBank) was generated using ClustalX 2.1 (46). Estimation of the percent identity and phylogenetic analyses were performed using ClustalX 2.1 (pim as the output file) and MEGA version 7.0 (47), respectively. Evolutionary relationships were inferred using the neighbor-joining method (48), and the bootstrap consensus tree was inferred from 1,000 replicates. Several protein sequences were used to generate the phylogenetic tree, including those of TcSpz5, Tribolium castaneum spätzle 5 isoform X1 (XP_008193940.1); TcSpz5, Tribolium castaneum spätzle 5 isoform X2 (XP_015836109.1); AtSpz5like, Aethina tumida spätzle 5-like (XP_019879590.1); SoSpz5like, Sitophilus oryzae spätzle 5-like (XP_030767938.1); MsSpz5, M. sexta spätzli 5 (XP_037299529.1); BmSpz5, B. mori spätzli 5 (XP_004924790.1); AaSpz5like, Anopheles albimanus spätzle 5-like (XP_035790066.1); DmeSpz5, D. melanogaster spätzle5 (NP_647753.1); DmaSpz5, Drosophila mauritiana spätzle 5 (XP_033160799.1); ArSpz5, Athalia rosae spätzli 5 (XP_012261687.1); PgSpz5, Pseudomyrmex gracilis spätzle 5 isoform X3 (XP_020284715.1); and PvSpz4, Penaeus vannamei spätzle 4 (ANJ04742.1).
The protocols for the developmental stage- and tissue-specific analyses have been reported previously (44, 45, 49). Briefly, total RNA was isolated from different developmental stages (eggs, young larvae (instars 10–12), late larvae (instars 14-15), pre-pupae, 1- to 7-day-old pupae, and 1- to 5-day-old adults) and tissues [integument, gut, fat bodies, Malpighian tubule (MT), hemocytes of last instar larvae and 5-day-old adults, and ovary and testis of 5-day-old adults] of T. molitor.
To analyze the induction of TmSpz5, suspensions containing 1 × 106 cells/µl of E. coli and S. aureus and 5 × 104 cells/μl of C. albicans were injected into T. molitor larvae at instars 10–12 (n = 20). PBS-injected T. molitor larvae were used as the control group. Samples were collected at 3, 6, 9, 12, and 24 h post-microbial challenge.
Total RNA was isolated using the Clear-S Total RNA Extraction Kit (Invirustech Co., Gwangju, South Korea) according to the manufacturer’s instructions. Then, 2 μg of total RNA was used as the template to synthesize cDNA using the Oligo (dT)12–18 primers under the following reaction conditions: 72°C for 5 min, 42°C for 1 h, and 94°C for 5 min. The MyGenie96 Thermal Block (Bioneer, Daejeon, Korea) and AccuPower® RT PreMix (Bioneer) were used according to the manufacturer’s instructions. cDNA was stored at -20°C until further use.
Relative quantitative PCR (qRT-PCR) was performed using AccuPower® 2X GreenStar qPCR Master Mix (Bioneer) with synthesized cDNAs and specific primers (TmSpz5_qPCR_Fw and TmSpz5_qPCR_Rv), as depicted in Table 1, with an initial denaturation of 95°C for 20 s, followed by 40 cycles at 95°C for 5 s and 60°C for 20 s. T. molitor ribosomal protein L27a (TmL27a) was used as an internal control, and the results were analyzed using the 2-ΔΔCt method (50). The results are presented as means ± standard error (SE) of three biological replicates.
To synthesize the double-stranded RNA (dsRNA) of the TmSpz5 gene, primers containing the T7 promoter sequence at their 5′ ends were designed using SnapDragon-Long dsRNA Design (Table 1). PCR was performed using AccuPower® Pfu PCR PreMix with the TmSpz5_Fw and TmSpz5_Rv primers (Table 1) and according to the developmental expression pattern of TmSpz5, cDNA synthesized from pre-pupae (whole bodies) as a template under the following cycling conditions: an initial denaturation step at 94°C for 2 min followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 53°C for 30 s, and extension at 72°C for 30 s, with a final extension step at 72°C for 5 min. PCR products were purified using the AccuPrep PCR Purification Kit (Bioneer), and dsRNA was synthesized using the AmpliScribe T7-Flash Transcription Kit (Epicentre Biotechnologies, Madison, WI, USA) according to the manufacturer’s instructions. After synthesis, the dsRNA was purified by precipitation with 5 M ammonium acetate and 80% ethanol, followed by quantification using an Epoch spectrophotometer (BioTek Instruments, Inc., Winooski, VT, USA). The dsRNA for enhanced green fluorescent protein (dsEGFP) was synthesized for use as a control and was stored at -20°C until use.
To study the importance of TmSpz5 in the T. molitor immune response, dsTmSpz5 (1 µg/µl) was first injected into early-instar larvae (instars 10–12; n = 30) using disposable needles mounted onto a micro-applicator (Picospritzer III Micro Dispense System; Parker Hannifin, Hollis, NH, USA). An equal amount of dsEGFP was injected in the larvae at the same stage as the negative control. The efficiency of TmSpz5 knockdown was evaluated by qRT-PCR, and over 86% knockdown was achieved at 4 days postinjection. The TmSpz5-silenced and dsEGFP-injected larval groups were challenged with E. coli (106 cells/µl), S. aureus (106 cells/µl), or C. albicans (5 × 104 cells/µl) in triplicate experiments. The challenged larvae were maintained for 10 days, and the number of surviving larvae was recorded. The survival rates of TmSpz5-silenced larvae were compared with those of the control larvae. Relevant analysis was performed using Kaplan–Meier plots (51).
To evaluate the functional properties of TmSpz5 in the regulation of AMP gene expression in response to pathogens, RNAi was used for TmSpz5 gene silencing, followed by the injection of larvae with E. coli, S. aureus, or C. albicans. dsEGFP and PBS were used as the negative and injection controls, respectively. At 24 h postinjection, the hemocytes, fat body, gut, and MTs were dissected, total RNA was extracted from each tissue, and cDNA was synthesized as described above. Next, qRT-PCR was performed with specific primers (Table 1) to analyze the temporal expression patterns of 14 AMP genes: TmTenecin-1, -2, -3, and -4 (TmTene1, 2, 3, and 4), TmAttacin-1a, -1b, and -2 (TmAtt1a, 1b and 2), TmDefensin (TmDef), TmDefensin-like (TmDef-like), TmColeoptericin-A and -B (TmColeA and B), TmCecropin-2 (TmCec-2), and TmThaumatin like protein-1 and -2 (TmTLP1 and 2).
To study the effects of dsTmSpz5 on the expression of NF-κB genes, including TmDorsal isoform X2 (TmDorX2) and TmRelish (TmRel), TmSpz5 was silenced in early-instar larvae and E. coli, S. aureus, and C. albicans were injected at 4 days post-double-strand treatment. At 24 h after pathogen injection, the MTs, hemocytes, gut, and fat bodies were dissected. Total RNA extraction and cDNA synthesis were performed as described above.
The AMPs and NF-κB gene expression patterns led us to perform colony-forming unit (CFU) assay to assess the in vitro AMP activity against Gram-negative bacteria. Therefore, TmSpz5 dsRNA-treated young instar larvae of T. molitor were injected with E. coli (106 cells). At 48 h post–pathogen injection, the hemolymph, midgut, hindgut, and Malpighian tubules were isolated in 100 μl 1× PBS. PBS and dsEGFP were injected as uninfected and dsRNA control groups, respectively. Tissue samples were homogenized and centrifuged at 15,000 rpm at 4°C for 10 min, and then the supernatants were boiled at 100°C for 10 min and centrifuged again at 15,000 rpm at 4°C for 10 min. Consequently, the protein content of extracted peptides has been measured by an Epoch machine and 50 ng of tissue samples was assayed with 106 cells of E. coli in 1× PBS at 37°C for 2 h (52). Eventually, 2,000-fold serial dilutions were performed, and 100 μl of the resulting mixture was plated onto LB agar, followed by incubation at 37°C for 16 h. The colony numbers of assayed plates were then counted.
Statistical analyses were performed using SAS 9.4 (SAS Institute, Inc., Cary, NC, USA), and cumulative survival was analyzed by Tukey’s multiple-comparison test, with a significance level of p < 0.05. Fold change in expression of the AMP genes compared to the levels of the internal control (TmL27a) and external control (PBS) was calculated using the 2-ΔΔCt method.
To acquire the full-length cDNA sequence of TmSpz5, a local blast search of the T. molitor RNAseq database was performed using the T. castaneum Spätzle5 protein sequence as the query. The TmSpz5 full-length ORF consisted of 1,062 bp, encoding a polypeptide of 353 amino acid residues (Figure 1). As determined using InterProScan, the TmSpz5 amino acid sequence contained a cystine-knot domain at the C-terminus (which binds to the Toll receptor), one cleavage site predicted to be processed by SPE, and a predicted signal peptide at the N-terminus (Figure 1). Additionally, the conserved domains in TmSpz5 were compared at the amino acid level using ClustalX 2.1 and multiple-sequence alignment. TmSpz5 sequences were conserved at the protein level among insect species (Figure 2A). A phylogenetic analysis illustrated that TmSpätzle5 in the order Coleoptera formed a group with other isoforms of Spätzle5 from T. castaneum (Figure 2B).
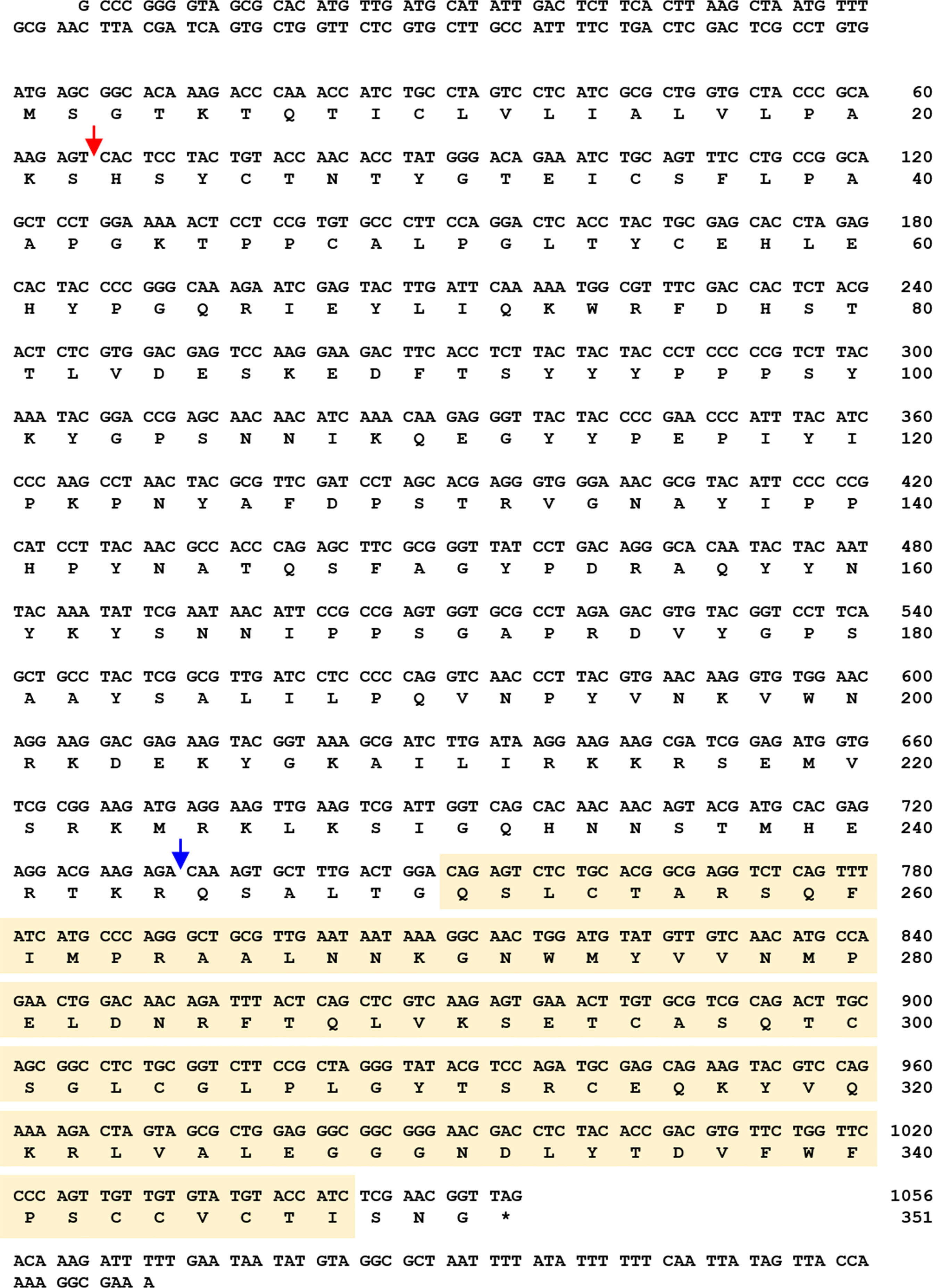
Figure 1 Nucleotide and deduced amino acid sequences of T. molitor Spätzle5 (TmSpz5). The TmSpz5 full-length open reading frame (ORF) consisted of 1,062 bp, encoding a polypeptide of 353 amino acid residues. The cystine-knot domain is shown in a yellow box, and the signal peptide region and cleavage site are indicated by red and blue arrows, respectively. The stop codon is shown with an asterisk.
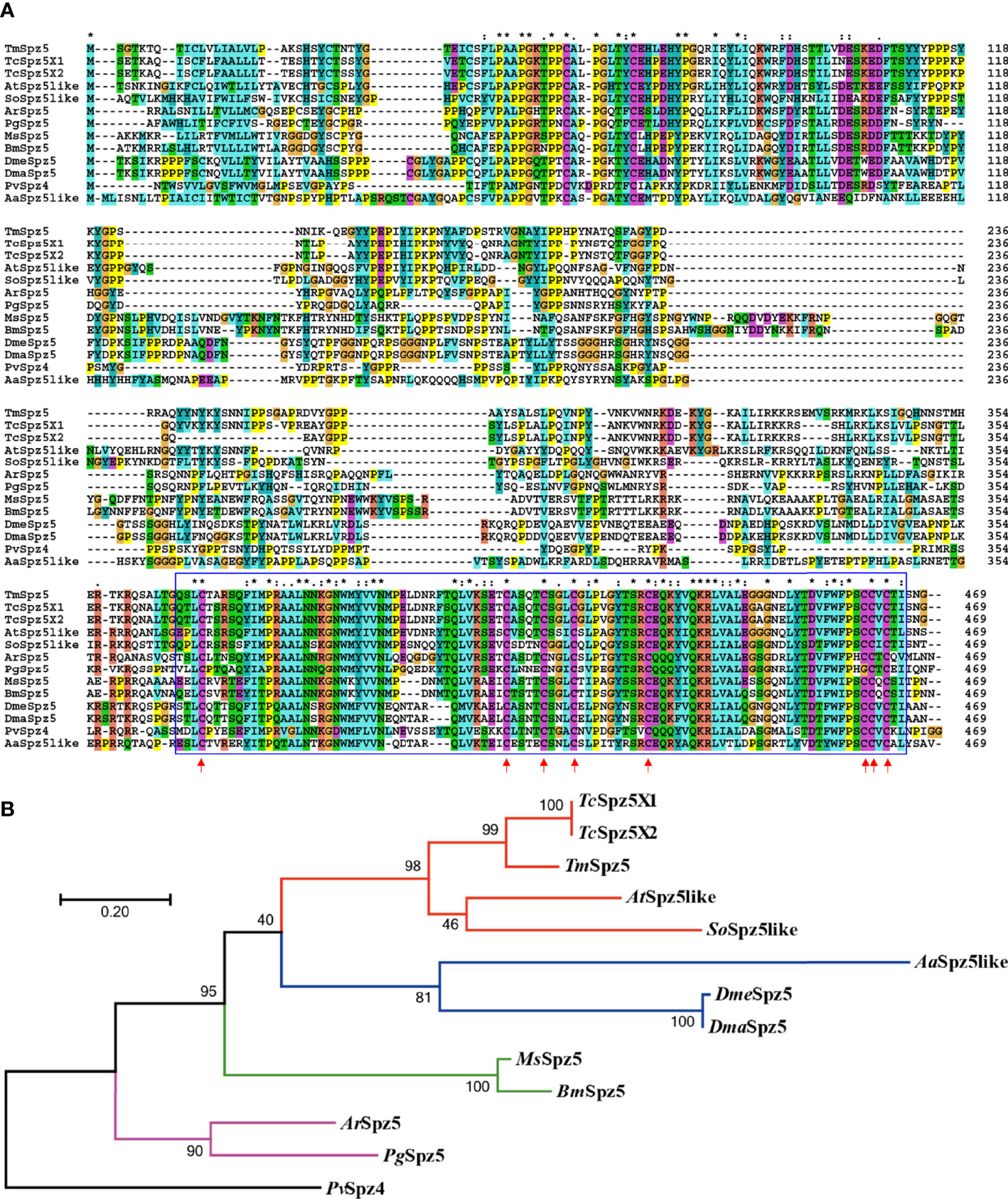
Figure 2 Multiple-sequence alignment (A) and phylogenetic analysis (B) of T. molitor Spätzle5 (TmSpz5). A domain analysis was performed using ClustalX2, and the phylogenetic tree was constructed using MEGA7 with the maximum likelihood method and 1,000 bootstrap replicates (where numbers at nodes indicate bootstrap support). The representative Spätzle 5 protein sequences showed high homology at the conserved domains marked in blue boxes, and the red arrows indicate conserved cystine-knot domain scores between groups. A neighbor-joining (NJ) tree was constructed based on the protein sequences of TcSpz5, Tribolium castaneum spätzle 5 isoform X1 (XP_008193940.1); TcSpz5, Tribolium castaneum spätzle 5 isoform X2 (XP_015836109.1); AtSpz5like, A. tumida spätzle 5-like (XP_019879590.1); SoSpz5like, S. oryzae spätzle 5-like (XP_030767938.1); MsSpz5, M. sexta spätzle 5 (XP_037299529.1); BmSpz5, B. mori spätzle 5 (XP_004924790.1); AaSpz5like, A. albimanus spätzle 5-like (XP_035790066.1); DmeSpz5, D. melanogaster spaetzle5 (NP_647753.1); DmaSpz5, D. mauritiana spätzle 5 (XP_033160799.1); ArSpz5, A. rosae spätzle 5 (XP_012261687.1); PgSpz5, P. gracilis spätzle 5 isoform X3 (XP_020284715.1); and PvSpz4, P. vannamei spätzle 4 (ANJ04742.1) which was used as the outgroup. Colored lines indicate different insect orders; red: Coleopteran, green: Lepidopteran, blue: Dipteran, purple: Hymenopteran. PvSpz4, illustrated black, belongs to the Crustacean class.
TmSpz5 mRNA expression patterns were evaluated by qRT-PCR at different developmental stages and in various tissues in larvae and adults. TmSpz5 was observed at essentially all developmental stages (Figure 3A). However, the highest expression levels were seen in embryos and pupae. The mRNA levels decreased at the larval stage, and in late larvae, it shows the lowest expression. We observed fluctuations in the expression pattern during pupal stages with a plateau phase in late pupae. Overall, increased TmSpz5 mRNA levels were observed during molting and each ecdysis, with a gradual fall across each individual stage.
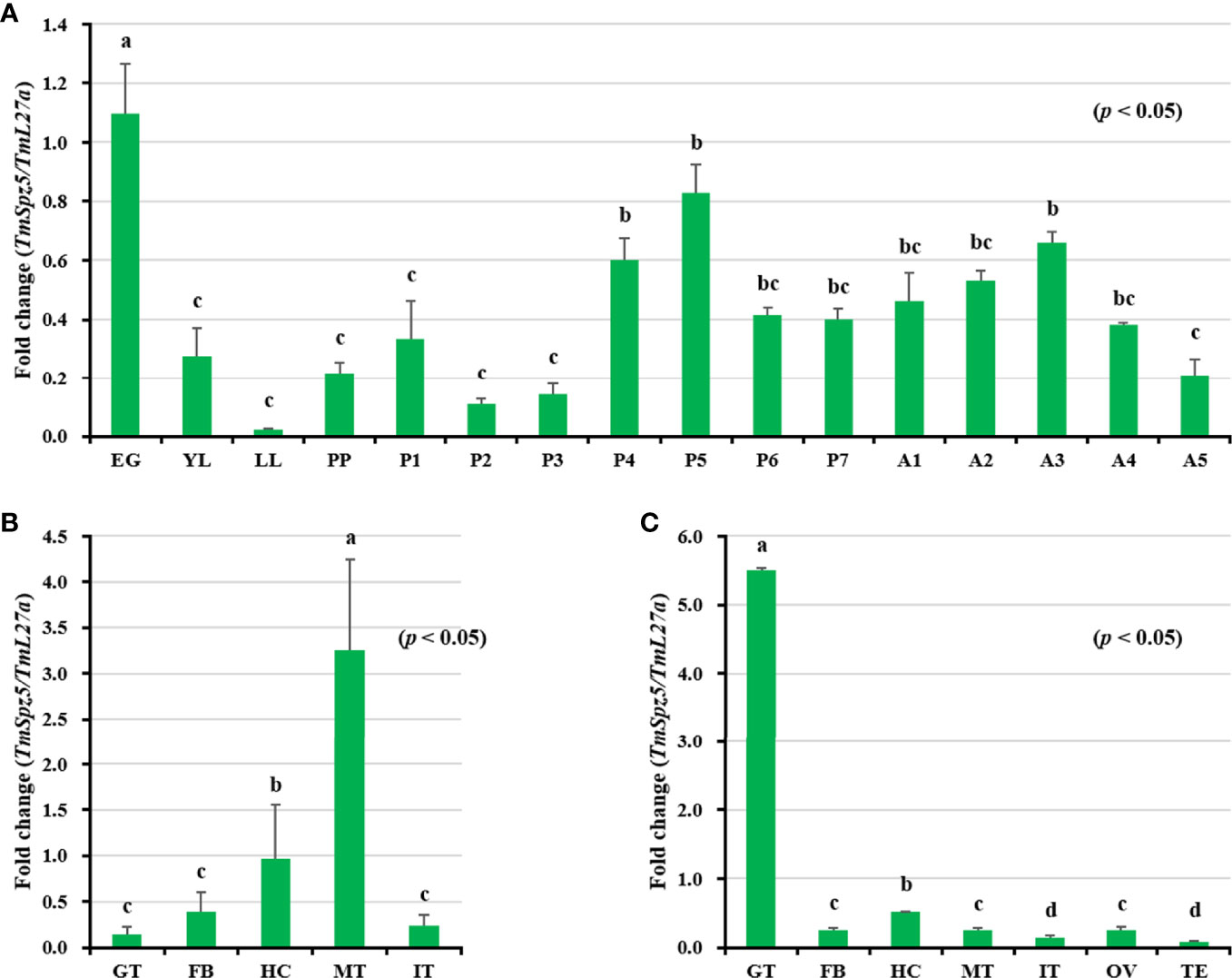
Figure 3 Developmental stage- and tissue-specific expression patterns of TmSpz5 measured by qRT-PCR. (A) Relative TmSpz5 mRNA levels in eggs (EG), young larvae (YL), late-instar larvae (LL), pre-pupae (PP), 1- to 7-day-old pupae (P1–P7), and 1- to 5-day-old adults (A1–A5) are illustrated. Expression levels were the highest in the eggs and the pupae. The mRNA levels decreased in the larval stage and were lowest in the late larval stage. Increases in TmSpz5 transcript levels were detected during molting and in each ecdysis with a gradual decrease across each individual stage. TmSpz5 tissue expression patterns in late instar larvae (B) and adults (C) were also examined. Total RNA was extracted from different tissues, including the integument (IT), Malpighian tubule (MT), gut (GT), hemocytes (HC), and fat bodies (FB) of late instar larvae and the integument (IT), Malpighian tubule (MT), gut (GT), hemocytes (HC), fat bodies (FB), ovary (OV), and testis (TE) of 5-day-old adults. Total RNA was isolated from 20 mealworms and T. molitor 60S ribosomal protein 27a (TmL27a) primers were used as internal control (N = 3). One-way ANOVA and Tukey’s multiple-range test at a 95% confidence level were used for comparisons. Bars with the same letter are not significantly different by Tukey’s multiple-range test (p < 0.05).
With respect to tissue expression patterns (Figures 3B, C), TmSpz5 expression levels were highest in MTs, followed by (in decreasing order) the hemocytes, fat bodies, integument, and gut in larvae. Contrarily, in adults, the mRNA expression of TmSpz5 was low in MTs and highest in the gut.
TmSpz5 expression in immune-challenged T. molitor larvae was examined after E. coli, S. aureus, and C. albicans injections (Figure 4), using PBS injection as the control. Four tissues, including the fat bodies (Figure 4A), hemocytes (Figure 4B), gut (Figure 4C), and MTs (Figure 4D), were collected at 3, 6, 9, 12, and 24 h post-pathogen injection for total RNA extraction. TmSpz5 expression was considerably upregulated in response to bacterial and fungal infections. TmSpz5 expression varied in tissue- and time-dependent manners. The highest expression levels were seen in the gut at 12 and 24 h and in the fat bodies at 9 and 24 h after infection (in that order), in response to all three pathogens. Of note, in the fat bodies, the expression of TmSpz5 was lowest at 12 h, possibly due to fluctuations in mRNA expression after infection as also reported earlier (35, 44, 49, 53). In MTs, there was also a noticeable upregulation in response to C. albicans and E. coli at 3 h post injection and in response to S. aureus at 9 h postinjection. C. albicans also induced TmSpz5 expression in the hemocytes at 12 h postinjection.
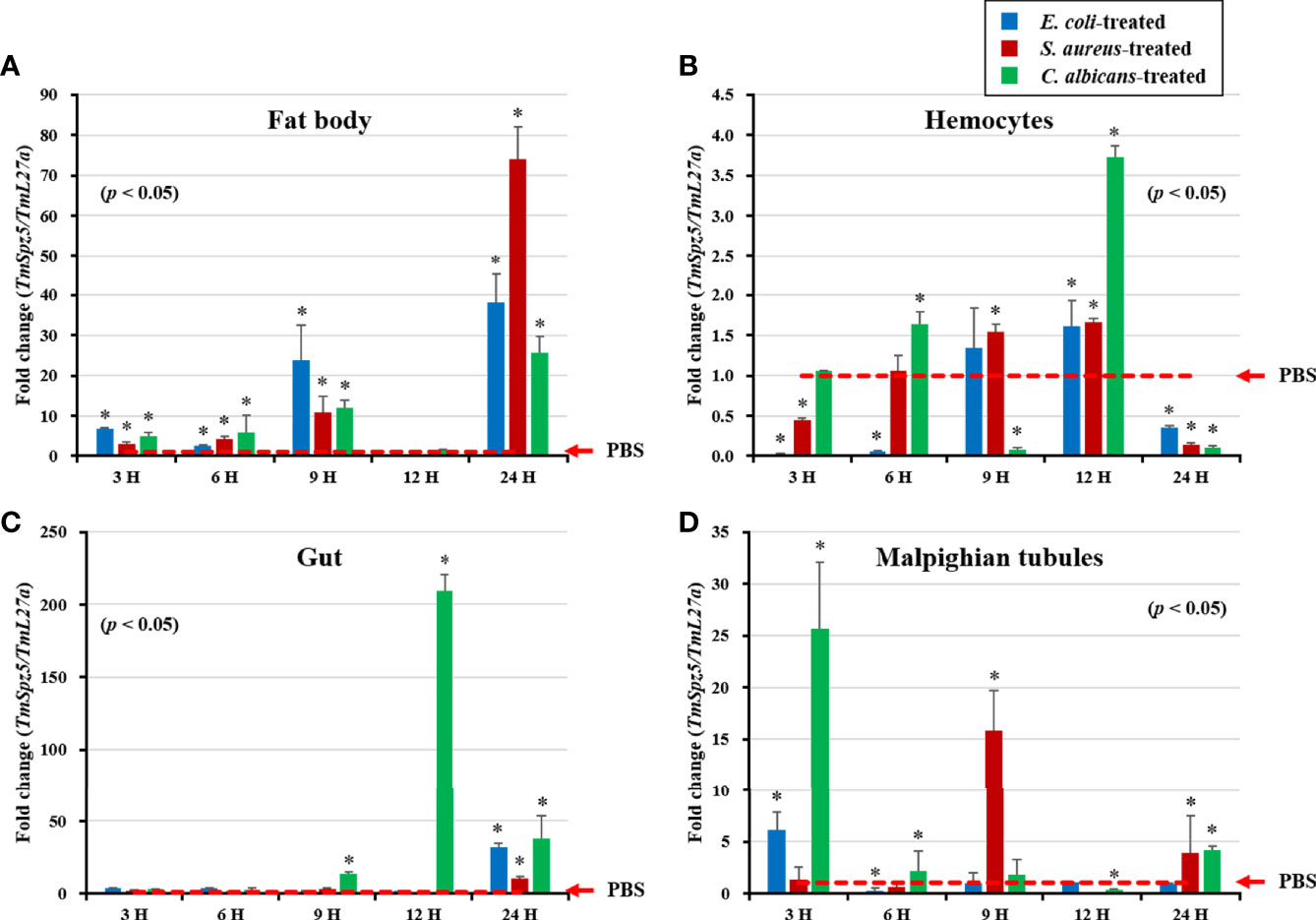
Figure 4 Temporal expression patterns of TmSpz5 in immune-challenged T. molitor larvae. Levels of TmSpz5 mRNA in the fat bodies (A), hemocytes (B), gut (C), and Malpighian tubules (D) were examined by qRT-PCR 3, 6, 9, 12, and 24 h after infection with E. coli (106 cells/µl), S. aureus (106 cells/µl), and C. albicans (5 × 104 cells/µl). TmSpz5 expression was highly induced in the presence of C. albicans and S. aureus in various tissues. PBS was used as an injection control, and T. molitor 60S ribosomal protein 27a (TmL27a) primers were used as internal control (n = 3). Asterisks indicate significant differences between infected and PBS-injected larval groups by Student’s t-test (p < 0.05). Vertical bars indicate means ± SD (n = 20).
Considering our observation that TmSpz5 expression is induced by different pathogens, we further examined the survival rate of TmSpz5-silenced larvae using the RNAi technique. TmSpz5 mRNA levels were decreased by 86% 4 days after dsTmSpz5 injection (Figure 5A), confirming the efficiency of the RNAi.
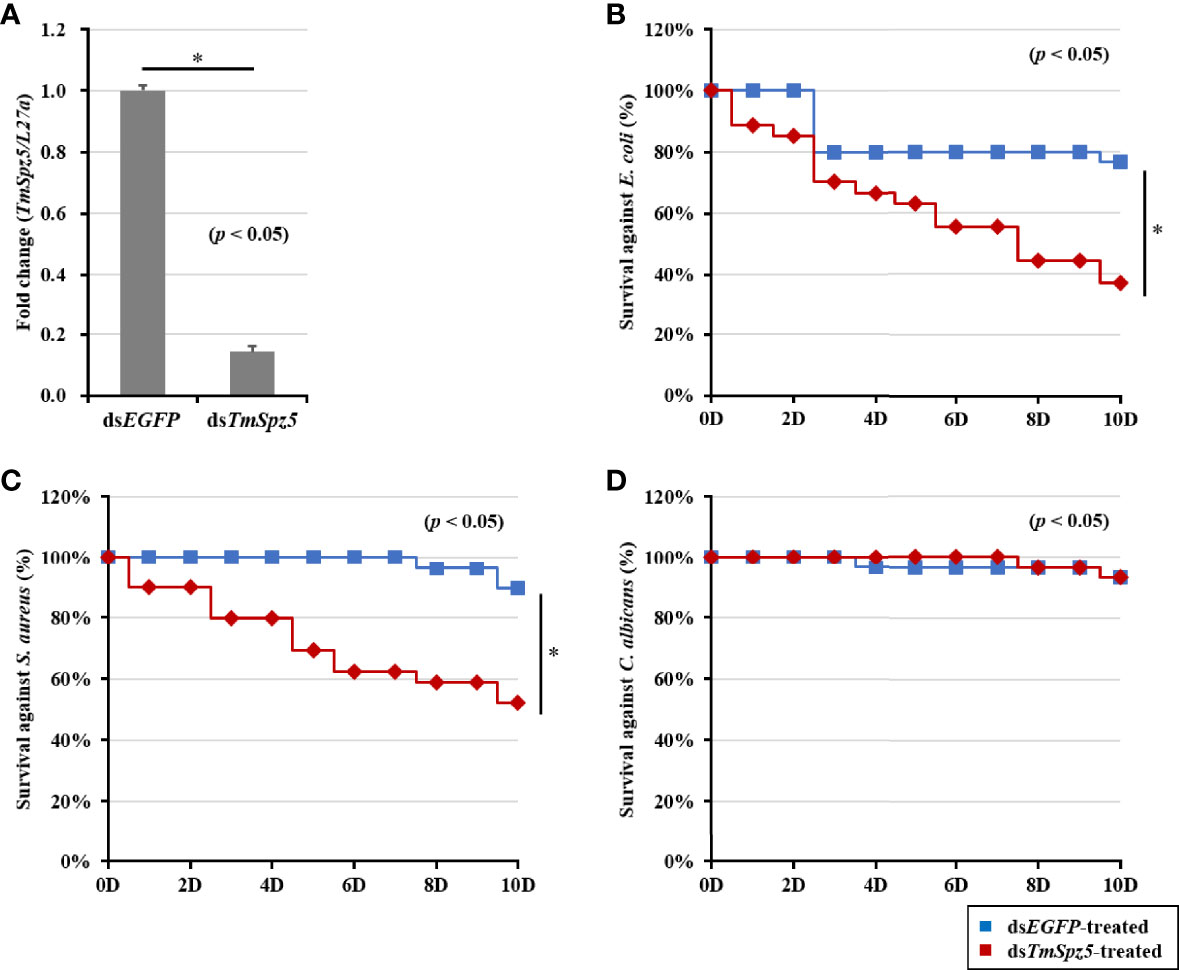
Figure 5 Effect of TmSpz5 gene silencing on the survival of T. molitor larvae. The silencing efficiency of dsTmSpz5 was measured by qRT-PCR at 4 days postinjection (A). TmSpz5-silenced larvae were injected with E. coli (B), S. aureus (C), and C. albicans (D), and survival rates were studied over 10 days post-pathogen injection (n = 10 per group). Larval survival rates at 10 days post-microbial injection were 33% after E. coli injection, 58% after S. aureus injection, and 90% after C. albicans injection compared with the levels in the dsEGFP-injected control group. The data are reported as averages of three biologically independent replicates. Asterisks indicate significant differences between dsTmSpz5- and dsEGFP-injected groups. The survival analysis was performed using Kaplan–Meier plots (log-rank chi-squared test; *p < 0.05).
Subsequent to confirmation of RNAi efficiency, pathogens of interest were injected. Survival rates of TmSpz5-silenced larvae were then evaluated over 10 days following microbial infection. dsEGFP was used as the control group for dsTmSpz5. PBS-injected larvae showed no statistically significant differences in survival between the dsTmSpz5 and dsEGFP groups (data not shown). dsTmSpz5 larvae showed considerable reductions in survival in response to E. coli and S. aureus (survival rates of approximately 33% and 58%, respectively) (Figures 5B, C). Interestingly, C. albicans-injected larvae showed similar survival rates to those of the PBS group (Figure 5D).
The survival analysis indicated that TmSpz5 gene silencing accelerated the vulnerability of larvae challenged with E. coli and S. aureus, but not C. albicans. We further evaluated the induction of AMPs following challenge with E. coli, S. aureus, and C. albicans in TmSpz5-silenced T. molitor larvae. In particular, we knocked down TmSpz5 and evaluated the levels of 14 AMP genes 24 h after the microbial challenge.
According to the results of the survival analysis, we expected TmSpz5 silencing to lead to AMP downregulation in response to E. coli and S. aureus. Our data illustrated that following confirmation of the TmSpz5 knockdown efficiency (Supplementary Figure 1), 10 out of 14 AMP genes were significantly downregulated in the MTs of TmSpz5-silenced larvae after E. coli and S. aureus injections but not after fungal infection. In particular, the E. coli challenge resulted in reductions in the levels of TmTene1, TmTene2, TmTene3, TmTene4, TmColeA, TmColeB, TmAtt1a, TmAtt1b, TmAtt2, TmTLP1, and TmTLP2 and the S. aureus challenge resulted in substantial reductions in the levels of TmTene2, TmTene4, TmColeA, TmColeB, TmAtt1a, TmAtt1b, TmAtt2, TmTLP1, and TmTLP2 (Figure 6). In the gut, silencing of TmSpz5 suppressed the E. coli-induced upregulation of TmColeA, TmAtt1a, and TmAtt1b as well as the S. aureus-induced regulation of TmTene2, TmTene4, TmColeB, TmAtt1a, and TmAtt1b (Figures 7B, D, H–K). In the hemocytes, TmTene1, TmDef, and TmAtt2 were downregulated in response to E. coli and TmDef and TmAtt2 were downregulated in response to S. aureus (Figures 8A, E, L). Moreover, in response to C.albicans, mRNA levels of TmTen3 and TmCec2 were downregulated (Figures 8C, G). In the fat bodies, only the levels of TmTene4, TmDef, and TmTLP1 were reduced in response to E. coli infection (Figures 9D, E, M). Surprisingly, dsTmSpz5 elevated the mRNA levels of some AMPs in response to pathogens in all dissected tissues, particularly the levels of the Cecropin, Attacin, and Tencin families in the gut, fat bodies, and hemocytes (Figures 7A, C, E–G, 8B, D, F, and 9A–C, F–L, N). Finally, mRNA levels of almost all AMPs did not differ between the dsTmSpz5 group and the control group in response to C. albicans.
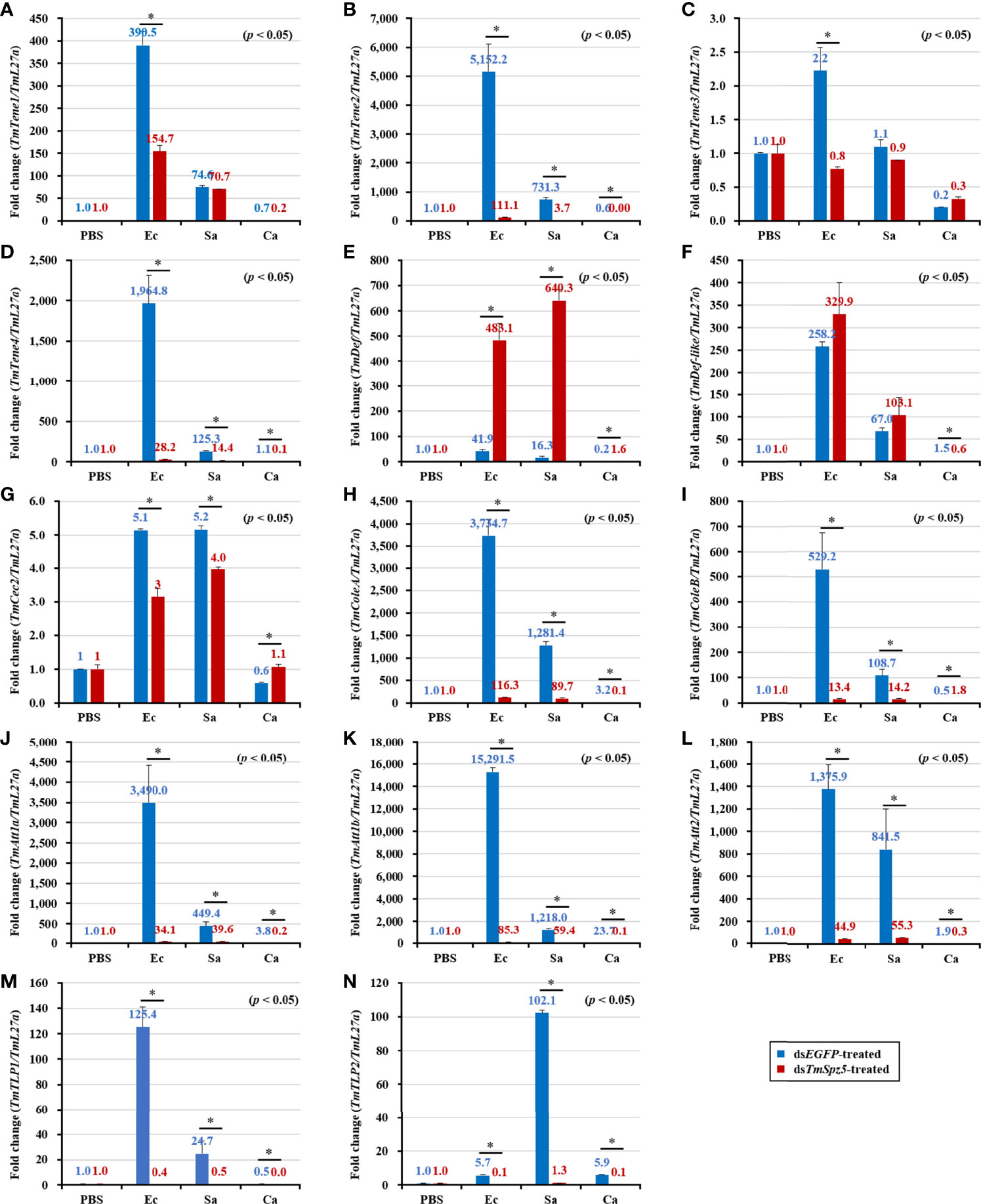
Figure 6 Induction of 14 AMP genes in the Malpighian tubules of TmSpz5-silenced T. molitor larvae infected with E. coli (Ec), S. aureus (Sa), and C. albicans (Ca) using PBS as control. At 24 h after microbial injection, AMP genes, including TmTene1 (A), TmTene2 (B), TmTene3 (C), TmTene4 (D), TmDef (E), TmDef-like (F), TmCec2 (G), TmColeA (H), TmColeB (I), TmAtt1a (J), TmAtt1b (K), TmAtt2 (L), TmTLP1 (M), and TmTLP2 (N) were examined by qPCR using dsEGFP as a knockdown control and T. molitor ribosomal protein (TmL27a) as an internal control. All experiments were performed in triplicate. Asterisks indicate significant differences between dsTmSpz5- and dsEGFP-treated groups determined by Student’s t-test (p < 0.05).
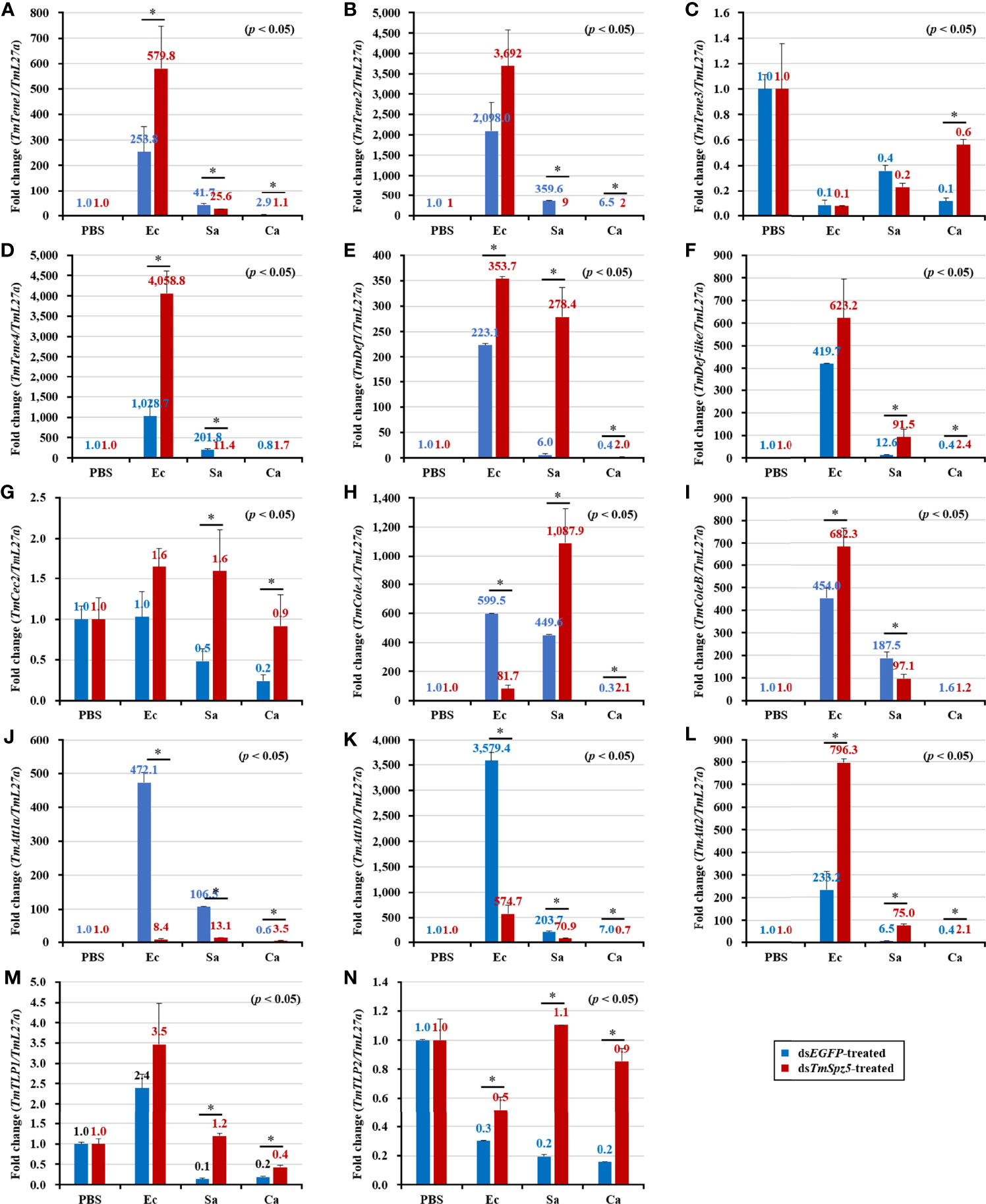
Figure 7 Effect of TmSpz5 gene silencing on antimicrobial peptide (AMP) gene expression levels in the T. molitor gut. Four days after TmSpz5 RNAi treatment, pathogens including E. coli (Ec), S. aureus (Sa), and C. albicans (Ca) and PBS as control were injected. Levels of the AMP genes, including TmTene1 (A), TmTene2 (B), TmTene3 (C), TmTene4 (D), TmDef (E), TmDef-like (F), TmCec2 (G), TmColeA (H), TmColeB (I), TmAtt1a (J), TmAtt1b (K), TmAtt2 (L), TmTLP1 (M), and TmTLP2 (N) were evaluated by qRT-PCR at 24 h post-microbial injection. dsEGFP was injected as a negative control, and TmL27a expression was evaluated as an internal control. All experiments were performed in triplicate. Asterisks indicate significant differences between dsTmSpz5- and dsEGFP-treated groups when compared using Student’s t-test (p < 0.05).

Figure 8 The mRNA expression levels of 14 antimicrobial peptide (AMP) genes after TmSpz5 gene silencing in the hemocytes of T. molitor. Four days after TmSpz5 dsRNA treatment, E. coli (Ec), S. aureus (Sa), and C. albicans (Ca) and PBS as a control were injected. At 24 h after injecting the microbes, the expression levels of TmTene1 (A), TmTene2 (B), TmTene3 (C), TmTene4 (D), TmDef (E), TmDef-like (F), TmCec2 (G), TmColeA (H), TmColeB (I), TmAtt1a (J), TmAtt1b (K), TmAtt2 (L), TmTLP1 (M), and TmTLP2 (N) were evaluated by qRT-PCR. dsEGFP was injected as a negative control, and TmL27a expression was measured as an internal control. All experiments were performed in triplicate. Asterisks indicate significant differences between dsTmSpz5- and dsEGFP-treated groups when compared using Student’s t-test (p < 0.05).
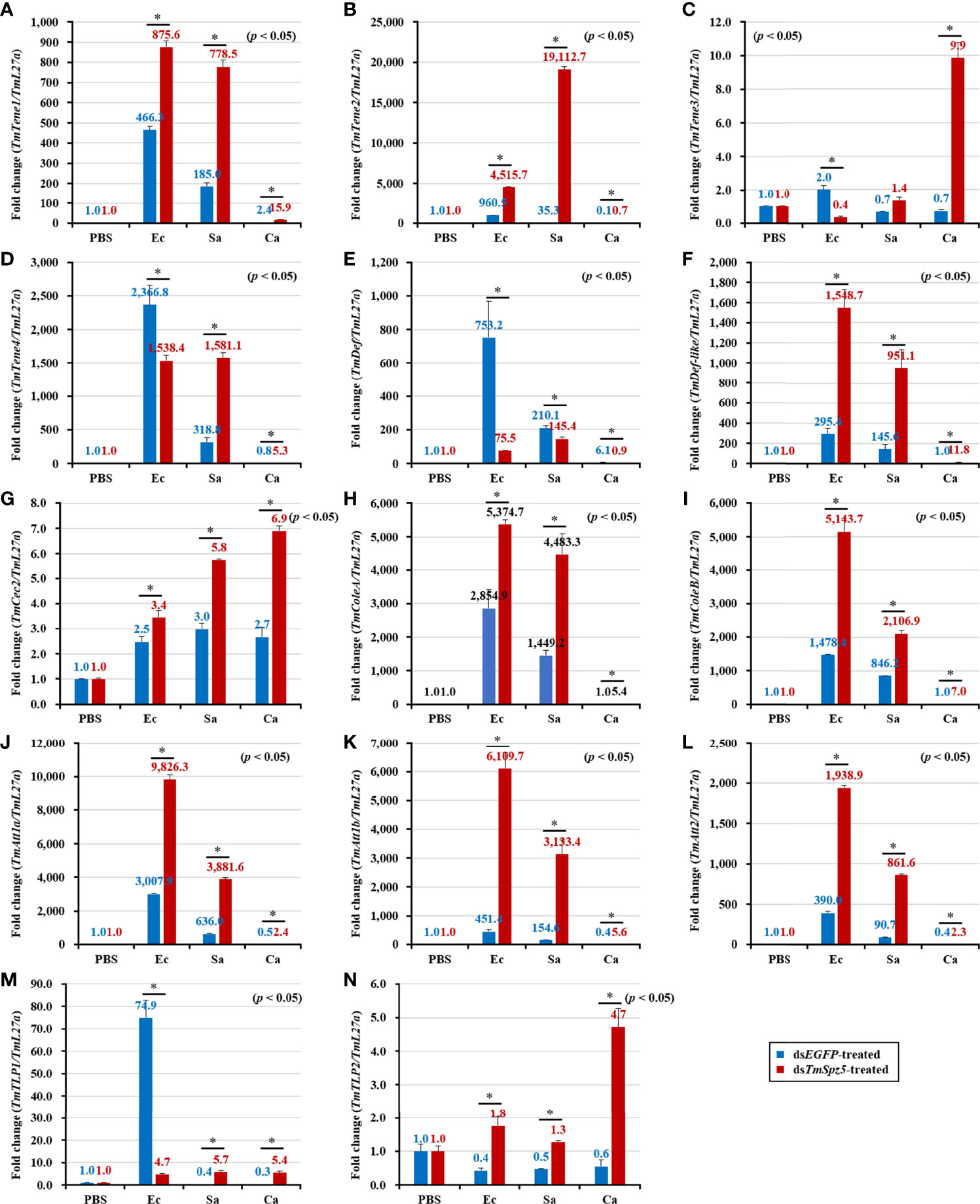
Figure 9 Antimicrobial peptide expression patterns in the fat bodies of dsTmSpz5-treated T. molitor larvae in response to microbial challenge. At 24 h post-infection by E. coli (Ec), S. aureus (Sa), and C. albicans (Ca), the mRNA levels of TmTene1 (A), TmTene2 (B), TmTene3 (C), TmTene4 (D), TmDef (E), TmDef-like (F), TmCec2 (G), TmColeA (H), TmColeB (I), TmAtt1a (J), TmAtt1b (K), TmAtt2 (L), TmTLP1 (M), and TmTLP2 (N) were evaluated by qRT-PCR. PBS was administered to the non-infected control group. dsEGFP was injected as a negative control, and TmL27a expression was measured as an internal control. All experiments were performed in triplicate. Asterisks indicate significant differences between dsTmSpz5- and dsEGFP-treated groups determined using Student’s t-test (p < 0.05).
Following the same protocol used to evaluate the expression of AMP genes following knockdown, the NF-κB pathway genes TmDorX2 and TmRelish were examined (Figure 10). dsTmSpz5 considerably depleted TmDorX2 expression levels in MTs following E. coli and S. aureus infection (Figure 10A). A less substantial reduction in TmRelish expression was observed in MTs (Figure 10B). Moreover, following the microbial challenge, TmDorX2 was upregulated in the fat bodies and gut and TmRelish was upregulated in the fat bodies.

Figure 10 Effect of Tmspz5 gene silencing on NF-κB gene expression patterns. dsTmspz5-treated T. molitor larvae were infected with E. coli, S. aureus and C. albicans and 24 h post-pathogen injection, mRNA levels of NF-κB genes, including TmDorX2 (A) and TmRel (B), were measured by RT-qPCR. EGFP dsRNA was assessed as a negative control and T. molitor ribosomal protein (TmL27a) was used as an internal control. All experiments were performed in triplicate. Asterisks indicate significant differences between dsTmSpz5- and dsEGFP-treated groups determined using Student’s t-test (p < 0.05).
The AMP assay result clearly demonstrated that E. coli and S. aureus infection in dsTmSpz5-treated larvae induced AMP expression significantly in Malpighian tubules and partially in the gut. Thus, we examined whether this suppression would affect bacterial growth in the hemolymph, MTs, midgut, and hindgut by CFU assay. Following dsEGFP and dsTmSpz5 injection, larvae were exposed to E. coli, and the aforementioned tissues were dissected 48 h postinfection and cultured with E. coli on LB agar plates. Elevated antimicrobial activity was observed in all dissected tissues in E. coli-injected larvae compared with the PBS-injected group (Figures 11A, C, E, G). Moreover, it was found that E. coli growth was hindered in the dsEGFP-injected gut, compared to the dsTmSpz5-injected larvae in the MTs, hindgut, and midgut (in decreasing order) (Figures 11D, F, H). In contrast, in the hemolymph, no significant difference in proliferation inhibition was observed between the dsEGFP- and dsTmSpz5-injected larvae (Figure 11B). These results imply that the effect of TmSpz5 knockdown on AMP gene depletion in MTs causes suppressed antimicrobial activity against Gram-negative bacteria. Additionally, while antimicrobial activity in hemolymph remained indifferent, downregulation of AMP genes in MTs subsequent to TmSpz5 knockdown exhibits reduced antimicrobial activity in the hindgut.
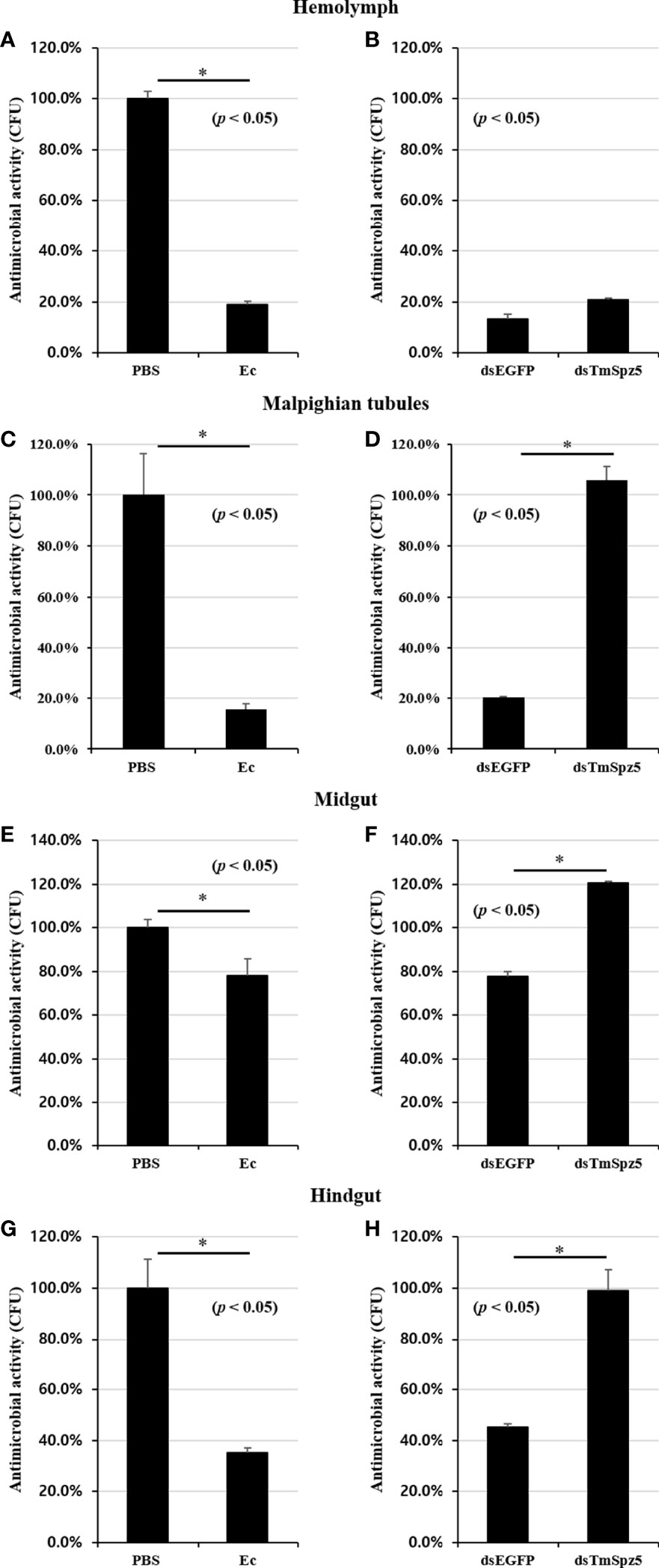
Figure 11 Antimicrobial activity against E. coli in TmSpz5-silenced larvae hemolymph, Malpighian tubules, midgut, and hindgut by CFU assay. (A, C, E, G) Antimicrobial activity evoked by E. coli (Ec) (106 cells/μl) elicitation. PBS-injected T. molitor was used as a negative control (Cont). E. coli-injected T. molitor hemolymph and Malpighian tubules had higher antimicrobial activity compared with control group (A, C). (B, D, F, H) E. coli (106 cells/μl) was injected into dsTmSpz5-treated T. molitor larvae. dsEGFP-treated larvae were used as a negative control. The result shows that the antimicrobial activity was decreased by treatment of dsSpz5 compared with the dsEGFP-treated group majorly in Malpighian tubules (D), hindgut (H), and midgut (F) in a depleting manner. E. coli proliferation remained indifferent in the dsTmSpz5-treated group compared with dsEGFP-treated larvae in hemolymph (B). Asterisks indicate significant differences between dsTmSpz5- and dsEGFP-injected groups.
Drosophila is one of the most potent genetic model systems for characterization of the Toll and Imd signaling pathways (24, 54, 55). Nevertheless, the focus on this model limits our understanding of the biochemical mechanisms of the Toll proteolytic cascade. For instance, the activation protocol (i.e., developmental factors or infection) influences pathway activity, making it difficult to comprehensively characterize the underlying mechanisms (12, 31).
T. molitor Toll signaling activation by Gram-positive bacteria or fungi, its compartments, and its relevant AMPs have been well elucidated (24). Surprisingly, similar to Lys-type PGN, TmPGRP-SA can recognize polymeric DAP-type PGN of Gram-negative bacteria, subsequently leading to activation of a three-step proteolytic cascade and the production of mature Spätzle (53, 56).
During Drosophila developmental stages, expression of the Spätzle gene is regulated by hormonal alteration. Radio-immunoassays have illustrated that ecdysone activity is high during prepupal and pupal stages (57). Likewise, cross talk between the steroid hormone 20-hydroxyecdysone (20E) and immune-regulatory genes in Drosophila has been reported (57, 58). Additionally, Drosophila MTs do not undergo histological alterations during pupal metamorphosis and therefore play an important role in innate immunity during this process (59, 60). Our developmental stage- and tissue-specific gene expression data revealed that TmSpz5 levels are high during the embryonic stage as well as at each ecdysis, consequently increasing susceptibility to possible attacks, showing that TmSpz5 contributes to both insect dorso-ventral axis formation during development and immune responses, respectively (11). The fact that TmSpz5 expression is the highest in larval MTs and in the adult gut supports its important role in epithelial defense organs.
Toll signaling is activated upon the recognition of Gram-positive bacteria and fungi by the cleavage of the cytokine-like polypeptide Spätzle (2, 6, 27). We detected a high and early expression of TmSpz5 following C. albicans and S. aureus challenge in descending order in all dissected tissues. The observed TmSpz5 expression in response to E. coli infection in MTs provides evidence for cross talk between the Toll and Imd signaling pathways. The unexpected results of the survival analysis demonstrated the importance of TmSpz5 in T. molitor immunity against E. coli and S. aureus, but not C. albicans. Consistently, TmSpz5 silencing leads to T. molitor vulnerability toward E. coli and S. aureus by decreasing AMP production in the presence of pathogens. Our results were predominantly consistent with those of previous studies on AMP production after treatment with dsTmSpz5. In Drosophila, attacin, diptericin, cecropin, and drosocin are active against Gram-negative bacteria, and metchnikowin and defensin act against Gram-positive bacteria (59, 61–65). In this study, TmCecropin-2 was also induced by Gram-positive bacteria and fungi. Surprisingly, the elevated mRNA levels of some AMPs in various tissues may suggest that there are alternative mechanisms to regulate gene expression. As it has been demonstrated previously, different T. molitor Späzle RNAi treatments (TmSpz4, TmSpz6, TmSpzlike) resulted in an increased expression of AMPs following microbial challenges (44, 45, 49). Additionally, the monomeric DAP-type peptidoglycan of Gram-negative bacteria activates TmIMD protein which triggers the expression of nine AMP genes (66). Likely, results of this study propose a possibility that the effect of TmSpz5 RNAi leads to the overexpression of other Späzle genes with a similar function. Overexpression of some AMP genes, mostly in hemolymph and fat bodies, maintains homeostasis. Moreover, other signaling pathways such as Imd can trigger an elevated expression of AMPs (35). Since the Imd signaling pathway has not been fully clarified, further studies regarding possible synergistic effects on induction of different AMPs are required to have a crystal understanding of Toll and Imd pathway association with regulation of AMP genes. Furthermore, the lack of change in mRNA levels of most AMPs in the dsTmSpz5 group in response to C. albicans appeared to be inconsistent with the induction data.
NF-κB family members in Drosophila, activated by the Toll and Imd pathways, regulate the expression of AMP genes (27). The Toll signaling pathway mediates activation of the transcription factors Dorsal and Dif and is predominantly actuated by the detection of Gram-positive bacteria and fungi (6, 40, 67, 68). In contrast, Gram-negative bacteria activate the Imd pathway, which triggers the NF-κB transcription factor Relish (6, 43, 69). In agreement with the AMP expression results, TmDorX2 was significantly suppressed in the MTs of TmSpz5-silenced larvae following challenges with E. coli and S. aureus, indicating that TmSpz5 is involved in regulating the expression of TmDorX2.
With respect to antimicrobial activity, AMPs extracted from all tissues except the hemolymph effectively inhibited E. coli growth. The effective inhibition of bacterial proliferation in the MTs and hindgut were consistent with the AMP mRNA expression and NF-κB results, suggesting that TmSpz5 acts as an immune component in the MTs and subsequently the hindgut. Further investigations are needed to verify these results.
Drosophila fat bodies are considered as the insect equivalent of the mammalian liver and are the main AMP-producing tissues, allowing an effective response to infection (6). Epithelial cells in the gut, MTs, genital tract, and trachea play important roles in systemic immunity by mediating the local response to invaders (70, 71). These epithelial tissues constitute the first line of defense toward possible invaders, and if pathogens invade these barriers, cellular and humoral immunity is induced (60, 71). Insect MTs form by hindgut–midgut joint invagination, and thus its secretions and hemolymph waste products are constantly transported to the hindgut (72). Thereupon, Drosophila MTs have osmoregulatory activity function as detoxification compartments in the hemolymph, acting as major innate immune organs (73). They are able to recognize pathogens and induce the production of high levels of AMPs (59, 60). They do not endure metamorphosis caused by ecdysone induction and are conveyed from larvae to adults. PGRP-LC expression is elevated by MT ecdysone production and has a marked effect on boosting host immunity (60). Additionally, PGRP-LE and PGRP-SC1 are immune elements predominantly functioning in the posterior midgut and anterior hindgut (74).
Our results show that MTs are critical immune organs in T. molitor, as has been observed in Drosophila. The radical shrinkage of the expression of almost all AMP genes and TmDorX2 in the MTs of TmSpz5-silenced larvae following E. coli infection suggests that DAP-type PGN is recognized by PRRs and the relevant proteolytic cascade leads to the activation of mature TmSpz5. Consequently, activated TmSpz5 binds to the Toll receptor and positively regulates the expression of the NF-κB response elements and AMP genes. In contrast to the lack of change in AMP expression in the gut of TmSpz5 knockdown larvae, the CFU results not only demonstrate the pivotal role of TmSpz5 in antibacterial activity of MT AMPs but also show that these AMPs act as hindgut disinfectants (Figure 12).
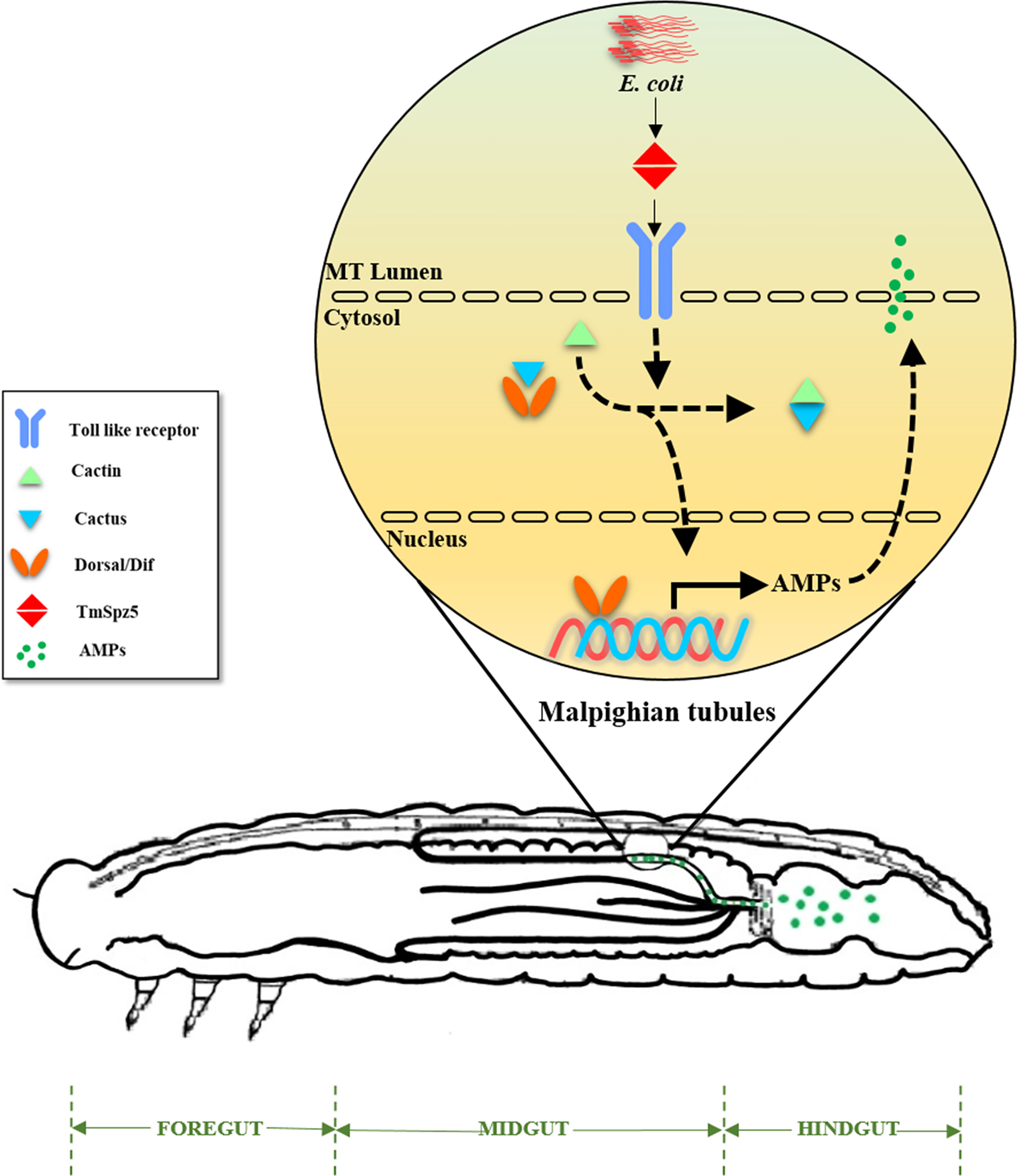
Figure 12 A schematic summary of TmSpz5 positive regulation in antimicrobial peptide production in Malpighian tubules (MTs) of bacterial infected larvae. 10 AMP-encoding genes including TmTen-1, TmTen-2, TmTen-4, TmColA, TmColB, TmAtt-1a, TmAtt-1b, TmAtt-2, TmTLP-1, and TmTLP-2 are positively regulated by TmSpz5 upon bacterial infections in MTs and produced peptides along with rest of tubules content, eventually flow to hindgut.
Our molecular analyses deepen our current knowledge of T. molitor immunity. Notably, the role of MTs in the innate immunity of T. molitor against the Gram-negative bacteria, E. coli, supports the results of previous studies, showing that polymeric DAP-type PG can be sensed by PGRP-SA, and Toll pathway activation leads to TmSpz5 cleavage and AMP production. A comprehensive understanding of these proteolytic cascades could provide a basis for the development of diagnostic kits and novel clinical trials for innate immune system-related diseases.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
YH, YJ, and MA conceived and designed the experiments. MA performed the experiments. YH contributed reagents/materials/analysis tools. MA, YJ, and HJ analyzed the data. MA wrote the first draft of the manuscript. YJ, MK, TE, HJ, and YH revised and edited the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (Grant No. 2018R1A2A2A05023367) and by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (IPET) through the Export Promotion Technology Development Program (Grant no. 617077‐5), funded by the Ministry of Agriculture, Food, and Rural Affairs (MAFRA).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.760475/full#supplementary-material
1. Coope G. Several Million Years of Stability Among Insect Species Because of, or in Spite of, Ice Age Climatic Instability? Philos Trans R Soc Lond Ser B Biol Sci (2004) 359(1442):209–14. doi: 10.1098/rstb.2003.1393
2. Ferrandon D, Imler J-L, Hoffmann JA. Sensing Infection in Drosophila: Toll and Beyond. Semin Immunol (2004) 16(1):43–53. Elsevier. doi: 10.1016/j.smim.2003.10.008
3. Grimaldi D, Engel MS, Engel MS, Engel MS. Evolution of the Insects. New York: Cambridge University Press (2005).
4. Janeway CA Jr, Medzhitov R. Innate Immune Recognition. Annu Rev Immunol (2002) 20(1):197–216. doi: 10.1146/annurev.immunol.20.083001.084359
5. Medzhitov R, Janeway CA. Decoding the Patterns of Self and Nonself by the Innate Immune System. Science (2002) 296(5566):298–300. doi: 10.1126/science.1068883
6. Hoffmann JA. The Immune Response of Drosophila. Nature (2003) 426(6962):33–8. doi: 10.1038/nature02021
7. Miller JS, Nguyen T, Stanley-Samuelson DW. Eicosanoids Mediate Insect Nodulation Responses to Bacterial Infections. Proc Natl Acad Sci (1994) 91(26):12418–22. doi: 10.1073/pnas.91.26.12418
8. Pech LL, Strand MR. Granular Cells Are Required for Encapsulation of Foreign Targets by Insect Haemocytes. J Cell Sci (1996) 109(8):2053–60. doi: 10.1242/jcs.109.8.2053
9. Leonard C, Ratcliffe NA, Rowley AF. The Role of Prophenoloxidase Activation in Non-Self Recognition and Phagocytosis by Insect Blood Cells. J Insect Physiol (1985) 31(10):789–99. doi: 10.1016/0022-1910(85)90072-1
10. Tauszig S, Jouanguy E, Hoffmann JA, Imler J-L. Toll-Related Receptors and the Control of Antimicrobial Peptide Expression in Drosophila. Proc Natl Acad Sci (2000) 97(19):10520–5. doi: 10.1073/pnas.180130797
11. Belvin MP, Anderson KV. A Conserved Signaling Pathway: The Drosophila Toll-Dorsal Pathway. Annu Rev Cell Dev Biol (1996) 12(1):393–416. doi: 10.1146/annurev.cellbio.12.1.393
12. Weber AN, Tauszig-Delamasure S, Hoffmann JA, Lelièvre E, Gascan H, Ray KP, et al. Binding of the Drosophila Cytokine Spätzle to Toll Is Direct and Establishes Signaling. Nat Immunol (2003) 4(8):794–800. doi: 10.1038/ni955
13. Shia AK, Glittenberg M, Thompson G, Weber AN, Reichhart J-M, Ligoxygakis P. Toll-Dependent Antimicrobial Responses in Drosophila Larval Fat Body Require Spätzle Secreted by Haemocytes. J Cell Sci (2009) 122(24):4505–15. doi: 10.1242/jcs.049155
14. Ming M, Obata F, Kuranaga E, Miura M. Persephone/Spätzle Pathogen Sensors Mediate the Activation of Toll Receptor Signaling in Response to Endogenous Danger Signals in Apoptosis-Deficient Drosophila. J Biol Chem (2014) 289(11):7558–68. doi: 10.1074/jbc.M113.543884
15. Richman AM, Dimopoulos G, Seeley D, Kafatos FC. Plasmodium Activates the Innate Immune Response of Anopheles Gambiae Mosquitoes. EMBO J (1997) 16(20):6114–9. doi: 10.1093/emboj/16.20.6114
16. Christophides GK, Zdobnov E, Barillas-Mury C, Birney E, Blandin S, Blass C, et al. Immunity-Related Genes and Gene Families in Anopheles Gambiae. Science (2002) 298(5591):159–65. doi: 10.1126/science.1077136
17. Cui C, Wang Y, Liu J, Zhao J, Sun P, Wang S. A Fungal Pathogen Deploys a Small Silencing RNA That Attenuates Mosquito Immunity and Facilitates Infection. Nat Commun (2019) 10(1):1–10. doi: 10.1038/s41467-019-12323-1
18. An C, Jiang H, Kanost MR. Proteolytic Activation and Function of the Cytokine Spätzle in the Innate Immune Response of a Lepidopteran Insect, Manduca Sexta. FEBS J (2010) 277(1):148–62. doi: 10.1111/j.1742-4658.2009.07465.x
19. Yu B, Sang Q, Pan G, Li C, Zhou Z. A Toll-Spätzle Pathway in the Immune Response of Bombyx Mori. Insects (2020) 11(9):586. doi: 10.3390/insects11090586
20. Huerlimann R, Wade NM, Gordon L, Montenegro JD, Goodall J, McWilliam S, et al. De Novo Assembly, Characterization, Functional Annotation and Expression Patterns of the Black Tiger Shrimp (Penaeus Monodon) Transcriptome. Sci Rep (2018) 8(1):1–14. doi: 10.1038/s41598-018-31148-4
21. Roh K-B, Kim C-H, Lee H, Kwon H-M, Park J-W, Ryu J-H, et al. Proteolytic Cascade for the Activation of the Insect Toll Pathway Induced by the Fungal Cell Wall Component. J Biol Chem (2009) 284(29):19474–81. doi: 10.1074/jbc.M109.007419
22. Park JB, Choi WH, Kim SH, Jin HJ, Han YS, Kim NJ. Developmental Characteristics of Tenebrio Molitor Larvae (Coleoptera: Tenebrionidae) in Different Instars. Int J Ind Entomol (2014) 28(1):5–9. doi: 10.7852/ijie.2014.28.1.5
24. Ali Mohammadie Kojour M, Han YS, Jo YH. An Overview of Insect Innate Immunity. Entomol Res (2020) 50(6):282–91. doi: 10.1111/1748-5967.12437
25. Keshavarz M, Jo YH, Edosa TT, Bae YM, Han YS. TmPGRP-SA Regulates Antimicrobial Response to Bacteria and Fungi in the Fat Body and Gut of Tenebrio Molitor. Int J Mol Sci (2020) 21(6):2113. doi: 10.3390/ijms21062113
26. Keshavarz M, Jo YH, Edosa TT, Han YS. Tenebrio Molitor PGRP-LE Plays a Critical Role in Gut Antimicrobial Peptide Production in Response to Escherichia Coli. Front Physiol (2020) 11:320. doi: 10.3389/fphys.2020.00320
27. Hoffmann JA. Innate Immunity of Insects. Curr Opin Immunol (1995) 7(1):4–10. doi: 10.1016/0952-7915(95)80022-0
28. Choe KM, Werner T, Stoven S, Hultmark D, Anderson KV. Requirement for a Peptidoglycan Recognition Protein (PGRP) in Relish Activation and Antibacterial Immune Responses in Drosophila. Science (2002) 296(5566):359–62. doi: 10.1126/science.1070216
29. Gottar M, Gobert V, Michel T, Belvin M, Duyk G, Hoffmann JA, et al. The Drosophila Immune Response Against Gram-Negative Bacteria Is Mediated by a Peptidoglycan Recognition Protein. Nature (2002) 416(6881):640–4. doi: 10.1038/nature734
30. Rämet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RAB. Functional Genomic Analysis of Phagocytosis and Identification of a Drosophila Receptor for E. Coli. Nature (2002) 416(6881):644–8. doi: 10.1038/nature735
31. Gobert V, Gottar M, Matskevich AA, Rutschmann S, Royet J, Belvin M, et al. Dual Activation of the Drosophila Toll Pathway by Two Pattern Recognition Receptors. Science (2003) 302(5653):2126–30. doi: 10.1126/science.1085432
32. Yang YT, Lee MR, Lee SJ, Kim S, Nai YS, Kim JS. Tenebrio Molitor Gram-Negative-Binding Protein 3 (TmGNBP3) Is Essential for Inducing Downstream Antifungal Tenecin 1 Gene Expression Against Infection With Beauveria Bassiana JEF-007. Insect Sci (2018) 25(6):969–77. doi: 10.1111/1744-7917.12482
33. Vigneron A, Jehan C, Rigaud T, Moret Y. Immune Defenses of a Beneficial Pest: The Mealworm Beetle, Tenebrio Molitor. Front Physiol (2019) 10:138. doi: 10.3389/fphys.2019.00138
34. Patnaik BB, Patnaik HH, Seo GW, Jo YH, Lee YS, Lee BL, et al. Gene Structure, cDNA Characterization and RNAi-Based Functional Analysis of a Myeloid Differentiation Factor 88 Homolog in Tenebrio Molitor Larvae Exposed to Staphylococcus Aureus Infection. Dev Comp Immunol (2014) 46(2):208–21. doi: 10.1016/j.dci.2014.04.009
35. Jo YH, Kim YJ, Park KB, Seong JH, Kim SG, Park S, et al. TmCactin Plays an Important Role in Gram-Negative and -Positive Bacterial Infection by Regulating Expression of 7 AMP Genes in Tenebrio Molitor. Sci Rep (2017) 7:46459. doi: 10.1038/srep46459
36. Keshavarz M, Jo YH, Park KB, Ko HJ, Edosa TT, Lee YS, et al. TmDorX2 Positively Regulates Antimicrobial Peptides in Tenebrio Molitor Gut, Fat Body, and Hemocytes in Response to Bacterial and Fungal Infection. Sci Rep (2019) 9(1):16878. doi: 10.1038/s41598-019-53497-4
37. Keshavarz M, Jo YH, Patnaik BB, Park KB, Ko HJ, Kim CE, et al. TmRelish is Required for Regulating the Antimicrobial Responses to Escherichia Coli and Staphylococcus Aureus in Tenebrio Molitor. Sci Rep (2020) 10(1):4258. doi: 10.1038/s41598-020-61157-1
38. Jang HA, Park KB, Kim BB, Ali Mohammadie Kojour M, Bae YM, Baliarsingh S, et al. In Silico Identification and Expression Analyses of Defensin Genes in the Mealworm Beetle Tenebrio Molitor. Entomol Res (2020) 50(12):575–85 doi: 10.1111/1748-5967.12468
39. Jo YH, Park S, Park KB, Noh MY, Cho JH, Ko HJ, et al. In Silico Identification, Characterization and Expression Analysis of Attacin Gene Family in Response to Bacterial and Fungal Pathogens in Tenebrio Molitor. Entomol Res (2018) 48(1):45–54. doi: 10.1111/1748-5967.12287
40. Keshavarz M, Jo YH, Park KB, Ko HJ, Edosa TT, Lee YS, et al. Tm DorX2 Positively Regulates Antimicrobial Peptides in Tenebrio Molitor Gut, Fat Body, and Hemocytes in Response to Bacterial and Fungal Infection. Sci Rep (2019) 9(1):1–19. doi: 10.1038/s41598-019-53497-4
41. Ali Mohammadie Kojour M, Jang HA, Edosa TT, Keshavarz M, Kim BB, Bae YM, et al. Identification, In Silico Characterization, and Expression Analysis of Tenebrio Molitor Cecropin-2. Entomol Res (2020) 51(2):74–82. doi: 10.1111/1748-5967.12476
42. Jang HA, Park KB, Kim BB, Ali Mohammadie Kojour M, Bae YM, Baliarsingh S, et al. Bacterial But Not Fungal Challenge Up-Regulates the Transcription of Coleoptericin Genes in Tenebrio Molitor. Entomol Res (2020) 50(9):440–9. doi: 10.1111/1748-5967.12465
43. Keshavarz M, Jo YH, Patnaik BB, Park KB, Ko HJ, Kim CE, et al. Tm Relish Is Required for Regulating the Antimicrobial Responses to Escherichia Coli and Staphylococcus Aureus in Tenebrio Molitor. Sci Rep (2020) 10(1):1–18. doi: 10.1038/s41598-020-61157-1
44. Edosa TT, Jo YH, Keshavarz M, Bae YM, Kim DH, Lee YS, et al. TmSpz4 Plays an Important Role in Regulating the Production of Antimicrobial Peptides in Response to Escherichia Coli and Candida Albicans Infections. Int J Mol Sci (2020) 21(5):1878. doi: 10.3390/ijms21051878
45. Edosa TT, Jo YH, Keshavarz M, Bae YM, Kim DH, Lee YS, et al. TmSpz6 Is Essential for Regulating the Immune Response to Escherichia Coli and Staphylococcus Aureus Infection in Tenebrio Molitor. Insects (2020) 11(2):105–19. doi: 10.3390/insects11020105
46. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X Version 2.0. Bioinformatics (2007) 23(21):2947–8. doi: 10.1093/bioinformatics/btm404
47. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis Across Computing Platforms. Mol Biol Evol (2018) 35(6):1547–9. doi: 10.1093/molbev/msy096
48. Saitou N, Nei M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol Biol Evol (1987) 4(4):406–25.
49. Jang HA, Patnaik BB, Ali Mohammadie Kojour M, Kim BB, Bae YM, Park KB, et al. TmSpz-Like Plays a Fundamental Role in Response to E. Coli But Not S. Aureus or C. Albican Infection in Tenebrio Molitor via Regulation of Antimicrobial Peptide Production. Int J Mol Sci (2021) 22(19):10888. doi: 10.3390/ijms221910888
50. Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (2001) 25(4):402–8. doi: 10.1006/meth.2001.1262
51. Goel MK, Khanna P, Kishore J. Understanding Survival Analysis: Kaplan-Meier Estimate. Int J Ayurveda Res (2010) 1(4):274. doi: 10.4103/0974-7788.76794
52. Moon HJ, Lee SY, Kurata S, Natori S, Lee BL. Purification and Molecular Cloning of cDNA for an Inducible Antibacterial Protein From Larvae of the Coleopteran, Tenebrio Molitor. J Biochem (1994) 116(1):53–8. doi: 10.1093/oxfordjournals.jbchem.a124502
53. Park S, Jo YH, Park KB, Ko HJ, Kim CE, Bae YM, et al. TmToll-7 Plays a Crucial Role in Innate Immune Responses Against Gram-Negative Bacteria by Regulating 5 AMP Genes in Tenebrio Molitor. Front Immunol (2019) 10:310. doi: 10.3389/fimmu.2019.00310
54. Anthoney N, Foldi I, Hidalgo A. Toll and Toll-Like Receptor Signalling in Development. Development (2018) 145(9):dev156018. doi: 10.1242/dev.156018
55. Harnish JM, Link N, Yamamoto S. Drosophila as a Model for Infectious Diseases. Int J Mol Sci (2021) 22(5):2724. doi: 10.3390/ijms22052724
56. Kim CH, Kim SJ, Kan H, Kwon HM, Roh KB, Jiang R, et al. A Three-Step Proteolytic Cascade Mediates the Activation of the Peptidoglycan-Induced Toll Pathway in an Insect. J Biol Chem (2008) 283(12):7599–607. doi: 10.1074/jbc.M710216200
57. Borst DW, Bollenbacher WE, O’Connor JD, King DS, Fristrom JW. Ecdysone Levels During Metamorphosis of Drosophila Melanogaster. Dev Biol (1974) 39(1):308–16. doi: 10.1016/S0012-1606(74)80032-1
58. Meister M, Richards G. Ecdysone and Insect Immunity: The Maturation of the Inducibility of the Diptericin Gene in Drosophila Larvae. Insect Biochem Mol Biol (1996) 26(2):155–60. doi: 10.1016/0965-1748(95)00076-3
59. Verma P, Tapadia MG. Immune Response and Anti-Microbial Peptides Expression in Malpighian Tubules of Drosophila Melanogaster Is Under Developmental Regulation. PloS One (2012) 7(7):e40714. doi: 10.1371/journal.pone.0040714
60. Zheng W, Rus F, Hernandez A, Kang P, Goldman W, Silverman N, et al. Dehydration Triggers Ecdysone-Mediated Recognition-Protein Priming and Elevated Anti-Bacterial Immune Responses in Drosophila Malpighian Tubule Renal Cells. BMC Biol (2018) 16(1):60. doi: 10.1186/s12915-018-0532-5
61. Bulet P, Hetru C, Dimarcq JL, Hoffmann D. Antimicrobial Peptides in Insects; Structure and Function. Dev Comp Immunol (1999) 23(4-5):329–44. doi: 10.1016/s0145-305x(99)00015-4
62. Ekengren S, Hultmark D. Drosophila Cecropin as an Antifungal Agent. Insect Biochem Mol Biol (1999) 29(11):965–72. doi: 10.1016/s0965-1748(99)00071-5
63. Tzou P, Ohresser S, Ferrandon D, Capovilla M, Reichhart JM, Lemaitre B, et al. Tissue-Specific Inducible Expression of Antimicrobial Peptide Genes in Drosophila Surface Epithelia. Immunity (2000) 13(5):737–48. doi: 10.1016/s1074-7613(00)00072-8
64. Imler JL, Bulet P. Antimicrobial Peptides in Drosophila: Structures, Activities and Gene Regulation. Chem Immunol Allergy (2005) 86:1–21. doi: 10.1159/000086648
65. Lemaitre B, Hoffmann J. The Host Defense of Drosophila Melanogaster. Annu Rev Immunol (2007) 25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615
66. Jo YH, Patnaik BB, Hwang J, Park KB, Ko HJ, Kim CE, et al. Regulation of the Expression of Nine Antimicrobial Peptide Genes by TmIMD Confers Resistance Against Gram-Negative Bacteria. Sci Rep (2019) 9(1):10138. doi: 10.1038/s41598-019-46222-8
67. Meng X, Khanuja BS, Ip YT. Toll Receptor-Mediated Drosophila Immune Response Requires Dif, an NF-kappaB Factor. Genes Dev (1999) 13(7):792–7. doi: 10.1101/gad.13.7.792
68. Rutschmann S, Jung AC, Hetru C, Reichhart JM, Hoffmann JA, Ferrandon D. The Rel Protein DIF Mediates the Antifungal But Not the Antibacterial Host Defense in Drosophila. Immunity (2000) 12(5):569–80. doi: 10.1016/S1074-7613(00)80208-3
69. Kaneko T, Yano T, Aggarwal K, Lim JH, Ueda K, Oshima Y, et al. PGRP-LC and PGRP-LE Have Essential Yet Distinct Functions in the Drosophila Immune Response to Monomeric DAP-Type Peptidoglycan. Nat Immunol (2006) 7(7):715–23. doi: 10.1038/ni1356
70. Brey PT, Lee WJ, Yamakawa M, Koizumi Y, Perrot S, François M, et al. Role of the Integument in Insect Immunity: Epicuticular Abrasion and Induction of Cecropin Synthesis in Cuticular Epithelial Cells. Proc Natl Acad Sci USA (1993) 90(13):6275–9. doi: 10.1073/pnas.90.13.6275
71. Huttner KM, Bevins CL. Antimicrobial Peptides as Mediators of Epithelial Host Defense. Pediatr Res (1999) 45(6):785–94. doi: 10.1203/00006450-199906000-00001
72. McGavin GC. Essential Entomology: An Order-by-Order Introduction. New York: Oxford University Press (2001).
73. McGettigan J, McLennan RKJ, Broderick KE, Kean L, Allan AK, Cabrero P, et al. Insect Renal Tubules Constitute a Cell-Autonomous Immune System That Protects the Organism Against Bacterial Infection. Insect Biochem Mol Biol (2005) 35(7):741–54. doi: 10.1016/j.ibmb.2005.02.017
Keywords: Tenebrio molitor, innate immune response, spätzle 5, antimicrobial peptides, NF-κB, Toll signaling pathway, Malpighian tubules
Citation: Ali Mohammadie Kojour M, Edosa TT, Jang HA, Keshavarz M, Jo YH and Han YS (2021) Critical Roles of Spätzle5 in Antimicrobial Peptide Production Against Escherichia coli in Tenebrio molitor Malpighian Tubules. Front. Immunol. 12:760475. doi: 10.3389/fimmu.2021.760475
Received: 18 August 2021; Accepted: 22 November 2021;
Published: 16 December 2021.
Edited by:
Carmen Mariana Chifiriuc, University of Bucharest, RomaniaReviewed by:
Fengliang Jin, South China Agricultural University, ChinaCopyright © 2021 Ali Mohammadie Kojour, Edosa, Jang, Keshavarz, Jo and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yeon Soo Han, aGFueXNAam51LmFjLmty; Yong Hun Jo, eWh1bjEyMjhAam51LmFjLmty
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.