- 1Diabetes Research Group, Division of Infection and Immunity, School of Medicine, Cardiff University, Cardiff, United Kingdom
- 2Department of Biological and Chemical Sciences, School of Natural and Social Sciences, Roberts Wesleyan College, Rochester, NY, United States
- 3Wellcome Centre for Human Genetics, University of Oxford, Oxford, United Kingdom
In the past few years, huge advances have been made in techniques to analyse cells at an individual level using RNA sequencing, and many of these have precipitated exciting discoveries in the immunology of type 1 diabetes (T1D). This review will cover the first papers to use scRNAseq to characterise human lymphocyte phenotypes in T1D in the peripheral blood, pancreatic lymph nodes and islets. These have revealed specific genes such as IL-32 that are differentially expressed in islet –specific T cells in T1D. scRNAseq has also revealed wider gene expression patterns that are involved in T1D and can predict its development even predating autoantibody production. Single cell sequencing of TCRs has revealed V genes and CDR3 motifs that are commonly used to target islet autoantigens, although truly public TCRs remain elusive. Little is known about BCR repertoires in T1D, but scRNAseq approaches have revealed that insulin binding BCRs commonly use specific J genes, share motifs between donors and frequently demonstrate poly-reactivity. This review will also summarise new developments in scRNAseq technology, the insights they have given into other diseases and how they could be leveraged to advance research in the type 1 diabetes field to identify novel biomarkers and targets for immunotherapy.
Introduction
It is widely accepted that in T1D “there remains a paucity of robust and accepted biomarkers that can effectively inform on the activity of T cells during the natural history of the disease or in response to treatment” (1). Furthermore, the phenotype and roles of autoreactive B cells in T1D have received less attention than T cells (2–4). Whilst flow and mass cytometry approaches have enabled many insights into cell phenotypes and antigen specificity in type 1 diabetes [reviewed (5)], they allow detection of a relatively small number of markers, limiting the potential to discover truly novel biomarkers. In turn this limits the ability to monitor the natural history of diabetes development and patient responses to immunotherapy. In addition, although a number of immunomodulatory agents are in clinical trials for type 1 diabetes, these are generally non-specific in their actions (for example targeting CD3 or CD20) (6), and there remains a need to identify and target pathways that are perturbed specifically in islet-antigen specific lymphocytes.
Traditional RNA sequencing involves taking all cells of interest, and combining their RNA in a single sample before sequencing. In contrast, single cell RNA sequencing isolates individual cells, either through sorting into wells, or using droplet based technology (7). Transcripts from each cell are barcoded (a unique molecular identifier is also added to each transcript to circumvent any amplification bias), before being combined for sequencing. This allows quantification of the expression of every gene in every individual cell, so that cell phenotypes and heterogeneity can be fully elucidated. Of particular interest to immunologists are scRNAseq methods that allow sequencing across the V(D)J region of TCRs and BCRs. This allows capture of the paired TCRα and β chains (or paired heavy and light chains of BCRs) which is key to determining antigen specificity (8, 9) and being able to reconstitute the receptor in a cell line or to express it as a secreted antibody. A single cell sequencing approach also avoids much of the bias of bulk RNAseq of receptors (10).
There are a variety of methods used for scRNAseq [reviewed (7)] although the 10x Genomics platform has come to dominate the field, due to the relatively large number of cells that can be sampled and options of combining, for example, protein expression and V(D)J sequencing with standard gene expression (GEX) data (11, 12). In parallel, there has been an explosion in techniques to deal with the vast quantity of data generated, perform quality control and extract meaningful findings (13).
However, scRNAseq also comes with a number of caveats. Firstly it is technically challenging and poor sample preparation can lead to doublet formation in a similar manner to that seen in flow cytometry, but in addition scRNAseq samples are susceptible to contamination with RNA from dying cells and the downstream clustering algorithms can also produce seemingly novel cell populations which are in fact artefacts (14). Secondly, the high cost can make it somewhat inaccessible and limit sample numbers and sizes. Lastly, it requires stringent statistical analysis to avoid type 1 errors, preferably backed up by follow up experiments to verify findings (13).
Nevertheless, scRNAseq offers an exciting opportunity to identify novel biomarkers that could be indicative of diabetes progression in at risk individuals, and allow real-time monitoring of clinical trials through tracking expression of specific immuno-receptor sequences and cell phenotypes. Furthermore, it has the potential to discover novel targets for immunotherapy of type 1 diabetes, through the identification of genes that are differentially expressed in islet-antigen specific lymphocytes.
Using scRNAseq to Identify Biomarkers for Progression to Type 1 Diabetes and Phenotypes in T1D
scRNAseq’s potential is demonstrated in a paper by Kallionpää et al. They revealed that high IL-32 expression in PBMCs was strongly associated with seroconversion and progression to T1D, contributed mainly by activated, highly differentiated, T cells and NK cells. Interestingly insulin (INS), glucagon (GCG), and REG1A were found to be upregulated in T1D and AAB+ individuals in the bulk RNAseq of PBMC but not in scRNAseq (15). These genes are normally associated with the pancreas, but expressed at the mRNA level in whole blood and lymph nodes at much lower levels (www.genecards.org). For insulin in particular this wider expression is thought to be involved in peripheral maintenance of tolerance (16).
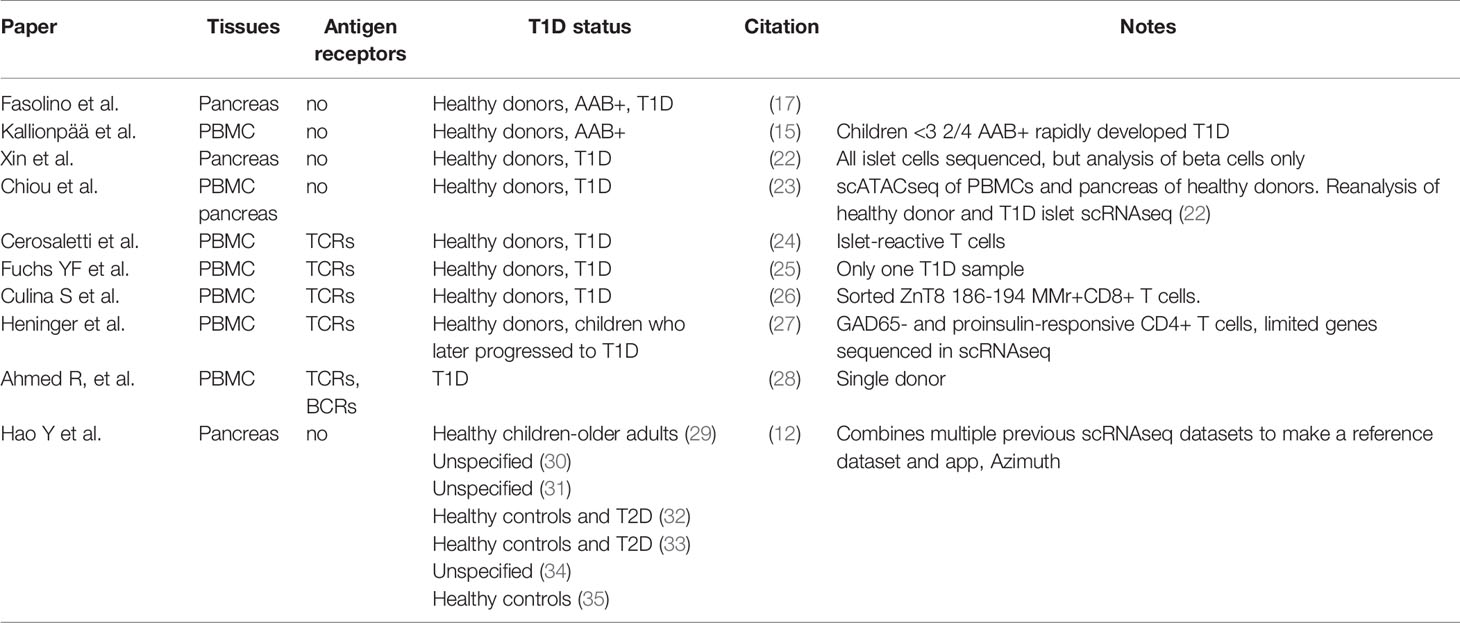
We can also glean insight into the immunology of T1D from scRNAseq studies of the pancreas, as in T1D these will include infiltrating immune cells. For example, the Vahedi group performed scRNAseq of human pancreatic islet cells and found particular enrichment of antigen-presenting cells and macrophages in T1D (17). In a strong replication of Kallionpää et al.’s findings, an analysis of differentially expressed genes (DEG) in immune cells between healthy and type 1 diabetes pancreas samples identified REG1B, REG1A, INS and REG3A and IL-32 as highly differentially expressed (17). As with INS, GCG and REG1A, REG1B and REG3A are highly expressed in the pancreas but at lower levels in the blood and lymph nodes. Furthermore REG genes are reported to be upregulated in the pancreas not only in people that have T1D, but also those who are autoantibody positive (18). They are upregulated in inflammatory conditions and are thought to be important in the survival of beta cells in T1D (18). An alternative explanation for the association of these RNA transcripts with immune cells is that RNA transcripts from dying beta cells are contaminating other cell types during the scRNAseq process (19). A similar scRNAseq analysis of the NOD mouse pancreas has also been conducted (20) and scRNAseq has been used to characterise hESCs differentiating into beta cells (21). Studies using scRNAseq to investigate the human pancreas and T1D are summarised in Table 1.
Closely related to scRNAseq is scATACseq, whereby the DNA from individual cell nuclei is analysed to identify open or accessible chromatin regions and hence predict which genes are being expressed in each cell. Recently, Chiou et al. combined scATACseq with bulk ATACseq and scRNAseq approaches to link cis-regulatory elements (CREs e.g. gene promoters and enhancers) in peripheral blood cells and pancreatic cells with GWAS of diabetes risk (23). As would be expected this identified many CREs used in T cells and beta cells that had genetic variants associated with T1D susceptibility. For example CREs that controlled CTLA4 and CCR7 expression in T cells had variants associated with T1D. Importantly, this paper also identified CREs used in pancreatic cells that had polymorphisms associated with T1D risk, particularly those used in acinar and ductal cells. They were further able to map the T1D risk allele of rs7795896 to a CRE used in ductal cells. The risk variant was associated with decreased CFTR expression in ductal cells. Mutations in CFTR itself cause cystic fibrosis, frequently associated with pancreatic exocrine and endocrine abnormalities, but this is this first demonstration of a role for it, and may other genes expressed in the exocrine pancreas, in T1D pathogenesis. This paper also produced a reference map of single-cell chromatin accessibility from T1D-relevant cells from healthy donors (i.e. lymphoid, myeloid and pancreatic endocrine and non-endocrine cells). Interestingly scRNAseq of the human pancreas also identified multiple changes in gene expression in ductal cells in T1D (17). In particular expression of MHC Class II pathway and interferon alpha and beta pathway genes were increased. Other developments in the field of epigenetics of T1D and the interplay with environmental triggers [reviewed (36–38)] have also started to yield evidence of pathogenic roles for molecules such as BACH2, IL23A, IL6R and IL6ST in T cell function in T1D (39). It will be of great interest to see how our understanding of epigenetics in T1D develops at the single cell level.
scRNAseq of TCRs
Methods to Identify Antigen Specific T Cells
As discussed above, there are many advantages of single cell sequencing TCRs over bulk TCR repertoire sequencing. Before the advent of large scale scRNAseq, many people in the type 1 diabetes field appreciated the importance of sequencing immunoreceptors on a single cell basis and linking this to antigen specificity and affinity (40–42). As of 2017 there were 1655 clonotypes of known specificity for T1D autoantigens (41), a number which has increased substantially with the advent of higher throughput scRNAseq.
These have been identified through a numbers of methods. HLA class I or class II multimers may be used to select antigen specific T cells. This has the advantage of being able to select cells from the peripheral blood but is limited by HLA restriction and to known epitopes or mimotopes (43). In addition non-specific binding may yield false-positive TCRs. Alternatively peripheral blood T cells can be stimulated in vitro with islet peptide pools and selected on the basis of upregulation of activation markers, allowing wider specificities and HLA compatibilities, but with the risk of bystander activation again resulting in false negatives. A third approach is to sample T cells directly from the pLN or pancreas, where islet-specific T cells will be massively enriched. These cells can then either be stimulated in vitro with peptide pools the TCRs re-expressed ex vivo to determine specificity. Alternatively, TCR sequences can be compared to those in the literature known to be islet antigen specific.
Diabetes Autoantigen- Specific Paired TCRs in the Peripheral Blood
Eugster and colleagues performed an heroic effort to sequence paired TCRs from 1650 T cells that either bound a GAD tetramer or responded to GAD in vitro, by sorting single cells from the peripheral blood and performing plate based scRNAseq (44). GAD specific TCRs were highly heterogenous both within and between donors, with no shared TCRs between donors, although individual TCRα or TCRβ chains were often shared. Moreover, there was limited overlap between the TCRs identified by tetramer binding and T cell activation methods, indicating that epitope recognition and MHC usage by GAD specific TCRs was likely to be broad (44).
Cerosaletti et al. performed scRNAseq of islet-reactive TCRs from the peripheral blood (identified by ex vivo response to stimulation with islet-peptide pools). They found that T cells from T1D had higher numbers of identical CDR3, which had arisen by clonal expansion (i.e. T cells with identical TCRα and TCRβ chains, that have arisen by division of a parent cell), rather than convergent recombination (24). By re-expressing the TCRs in cell lines it was found that many of these TCRs in people with T1D were IGRP specific (24). It was further shown that donors with T1D had large clonal expansions of IGRP-reactive T cells in the peripheral blood and frequently used a specific shared TCRα chain, which was paired with different beta chains in each donor (45). Preferential usage of TRAJ53 and TRAV29 and TCRα chains bearing the motif SGGSNYKLTF were identified in single cell TCR sequencing of people with T1D. When a bulk sequencing approach was taken, a particular TCRα chain bearing this motif was highly enriched in the memory CD8+ T cells of autoantibody positive people and those with T1D compared to controls. Clones bearing the motif were also shown to directly kill IGRP- peptide bearing cells (45). T cell clones bearing both IGRP (24, 45) and hybrid insulin peptide- responsive TCRs are persistent over time (46). However, others have examined TCR repertoires in children progressing to diabetes and shared TCRs were not seen either between children or within the same child over time, indicating high diversity in the peripheral blood at this age (27).
Diabetes Autoantigen- Specific Paired TCRs in the Pancreas and Pancreatic Lymph Nodes
Early work examined T cells from the pancreatic lymph nodes (pLN) of people with T1D and found high clonal expansions (47). Additionally there were many T cells that shared a TCRβ but had divergent TCRα. Many clonally expanded CD4+ T cells recognised insulin A1-15 in the context of DR4 (47). Pathiraja et al. grew out CD4+T cells from the pancreatic islets of a donor with T1D using anti CD3 and cytokine stimulation. Over 25% of these clones had TCRs that responded to proinsulin peptides restricted by HLA-DQ8 or the HLA-DQ8 transdimer and 30% of clones used TRBV5–1*01 (48). Whilst it is difficult to make direct comparisons to frequencies of islet-reactive T cells in the peripheral blood (26), it is clear that in the peripheral blood frequencies are much lower [around 0.01-0.05% of T cells in people with T1D (3, 44, 49)]. Most T cells isolated from the pancreas had unique clonotypes, whilst the majority of in vivo clonally expanded T cells were specific for proinsulin (48). It has also been found that ZnT8- reactive T cells were present at similar frequencies in the blood of healthy controls and people with T1D, but were enriched in the pancreas of the latter (26). Single cell sequencing of TCRs found a public ZnT8 specific CDR3B in the peripheral blood, and enriched in the pancreas of people with T1D, although the full TCRβ had divergent sequences due to different gene usages. ZnT8 reactive T cells also showed a bias towards TRBV19 and TRAV12-2 usage (26).
Seay et al. also found sharing of CDR3s between donors in the pancreas, with a TCRβ with homology to a known GAD reactive TCR found in 7/18 T1D donors (50). Furthermore a shared CDR3β chain was found in all people with T1D in the conventional T cell compartment, whilst in healthy controls it was predominantly in the Treg compartment (50). Interestingly TCR sequencing of GAD-responsive CD4+IL-13+ T cells from patients who had received injected GAD Alum found that they often used a highly public TCRβ (TCRα sequencing was not available) (51).
Direct capture of pancreatic T cells by the Nakayama group enabled single cell TCR sequencing and confirmed infiltration of proinsulin specific cells into the pancreas in T1D. Of the hundreds of TCRs sequenced, most were present only on a single cell, indicating a diversity of response even years after diagnosis. Clonal expansions were more likely in CD8+ T cells and these clones were found in multiple islets from the same donor, indicating in vivo migration. Furthermore, across three donors it was noted that whilst there were no identical TCRs, there were identical TCRα sequences and TCR subunits (52). When the TCRs were re-expressed, the B9-23 reactive TCRs isolated from the pancreas induced much higher IL-2 secretion compared to control B9-23 TCRs isolated from peripheral blood (52) which may indicate the former have a higher affinity for B9-23. Moreover, only the pancreas-derived TCRs were capable of a response to whole proinsulin presented by APCs (52).
Recently the Nakayama group has reconstructed individual TCRs from the pancreas of people with T1D. TCRs were selected for re-expression on the basis of clonal expansion or V gene usage previously associated with proinsulin C19-A3 specificity, and were found to recognise epitopes across preproinsulin and presented by a variety of MHC class II (53). Many TCRs recognised peptides in the region of B9-23, but others, (many from clonally expanded cells) recognised peptide right across from the signal peptide to the A chain. Furthermore, these TCRs recognised peptides in the context of diverse MHCII, although a preference was shown for DQ (53). Even with these constraints of the selection criteria in this study, this single cell approach showed a diversity of peptide and MHCII specificity that would have been missed using tetramers.
New Avenues for scRNAseq of TCRs
Taken together, the evidence suggests that T cells with TCRs with higher affinity for diabetes autoantigens are more likely to be found in the pancreas than in the peripheral circulation. This represents a major challenge in T1D research as in other autoimmune diseases it is relatively straightforward to obtain samples from the site of autoimmune attack (54). For example in psoriatic arthritis, extraction of viable T cells directly from the affected joints enabled sequencing of paired TCR receptors and scRNAseq profiling of cells phenotypes (55). Even in the pancreas, clonal expansion is modest and whilst CDR3 sequences specific for many diabetes autoantigens are shared between donors, there is not yet evidence of truly public TCRs with identical TCRα and β chains. However, more widespread use of the VDJdb repository (56), IEDB (57) and the JDRF/nPOD CloneSearch might allow enriched motifs to become apparent across different experiments, although this would still be limited by HLA restriction. To further complicate the picture, scRNAseq has demonstrated that islet antigen –reactive T cells (24) and HIP reactive T cells in particular (58) sometimes express two TCRα chains, which are known to contribute to autoimmunity (59, 60).
Phenotypes of Antigen-Specific T Cells: Combining TCR Sequencing With Gene Expression
Combining TCR sequencing (or selection based on autoantigen reactivity), with scRNAseq has the potential to give further insights into T cell function. This has not always been straightforward to demonstrate, for example analysis of IGRP- specific T cells from the peripheral blood did not show a distinctive gene expression (GEX) pattern in response to stimulation (25). Similarly scRNAseq of ZnT8 reactive cells from the peripheral blood of people with T1D showed similar GEX profiles to healthy controls, indicating that these peripheral T cells may not be playing a driving role in T1D, although T1D patients had higher expression of aryl hydrocarbon receptor (AHR) and aurora kinase A (AURKA) and lower expression of RORA (26).
The approach was more successful for Heninger et al, who demonstrated that proinflammatory responses to diabetes autoantigens were dominant in children who progressed to autoantibody positivity, whilst regulatory T cell responses were seen in those who didn’t (27). An algorithm based on gene expression in response to autoantigens enabled identification of which children would later progress to autoantibody positivity. As this group developed autoantibodies the GEX profiles of their CD4+ T cells changed towards increased expression of Th1 genes (27). These findings suggest that biomarkers of T1D susceptibility may allow identification of at risk children prior to seroconversion (61).
In addition, Cerosaletti et al. used islet peptide pools to stimulate T cells from the peripheral blood in vitro and characterised those that activated by scRNAseq. They did not observe a significant level of differentially expressed genes between healthy controls to those from people with T1D. However, when they focussed on cells from people with T1D that were highly clonally expanded (termed T1D-E cells), they found that these cells did have a unique transcriptional profile compared to islet reactive T cells from healthy controls or those from people with T1D that were not clonally expanded. T1D-E cells preferentially expressed genes associated with T cell activation and leukocyte differentiation (24). These experiments demonstrate how focussing on antigen specificity can enhance findings from scRNAseq.
scRNAseq of BCRs
Early Work to Determine Antibody Sequences
Early interest in autoantibodies in T1D, before the advent of scRNAseq, focussed on isolating GAD-specific B cells from people with T1D (62) and sequencing the BCRs from clones, which provided evidence that GAD autoantibodies have frequently undergone somatic maturation and are therefore from antigen-experienced B cells (63, 64). Similarly IA-2 specific antibodies sequenced from B cells from people with T1D also show evidence of somatic mutation (65–67). Anti-insulin antibodies have been sequenced from people with T1D, but may have arisen in response to injected insulin rather than endogenous insulin (68, 69) [reviewed (70)]. BCR sequencing combined with phenotyping of B cells has given great insight into B cell response in other autoimmune diseases (71) and in response to vaccinations (72) and in B cell lymphoma (73). Yet without the equivalent of a tetramer approach to identifying autoreactive B cells, phenotyping and characterisation of the BCR has lagged behind T cell research in T1D.
Identifying Islet-Reactive BCRs in the Periphery, Pancreatic Lymph Nodes and Pancreas
Smith et al. developed an approach to isolate insulin reactive B cells from the peripheral blood of people with T1D, by flow cytometric sorting B cells that bound insulin conjugated to fluorescent tags. The authors were then able to sequence BCRs from individual cells. They demonstrated that insulin binding BCRs preferentially used JH6 gene segments which have previously been associated with autoreactivity and were biased towards use of positively charged amino acids in the CDR3 region (74). When re-expressed as antibodies, BCRs from anergic naive IgD+, IgM− B cells demonstrated binding at levels thought to induce anergic B cell responses, whilst those from naïve B cells bound weakly and would likely be ignorant of insulin under physiological conditions (74).
scRNAseq has been used to characterise a novel lymphocyte population that express both TCRs and BCRs (28). It is suggested that these “dual expressors” (DE) are increased in frequency in type 1 diabetes and that in people with type 1 diabetes there is a public BCR which can stimulate insulin-reactive CD4+ T cells. However, this work remains controversial as others have been unable to replicate the enrichment of DE in T1D nor the specific public BCR sequence (75). This highlights the importance of good quality control at every step of scRNAseq experiments.
Isolation of CD19+IgG+ B cells from pancreatic lymph nodes from autoantibody positive donors and single cell sequencing of their BCRs demonstrated that no clonally expanded B cells were identified in the pLN. Antibodies were reconstructed from BCR sequencing, although very few of these were found to be specific for IA2 (none were specific for GAD and insulin was not tested) (76). Seay et al. also sorted and single cell sequenced the BCRs from pancreatic LNs. They found an enrichment of insulin binding motifs in pLN from people with T1D compared to controls (50). They also observed sequence overlap with autoreactive BCRs cloned from precursor (early immature) B cells from healthy donors previously published by Wardemann et al. (77). Wardemann et al. observed that not only are many BCRs from healthy donor precursor B cells insulin reactive, they are often also polyreactive to other autoantigens for example dsDNA, ssDNA or nuclear proteins (77). This polyreactivity has also been noted for both IgM and IgG insulin antibodies (68). Similarly Smith et al. demonstrated that all of their high affinity insulin binding BCRs were also reactive to LPS and chromatin (74). Polyreactive antibodies have been postulated to play a key role in the healthy immune system but are also implicated in a variety of autoimmune diseases (78, 79). It therefore appears that autoreactive B cells in T1D may span a wide range of phenotypes and the antibodies produced may often be polyreactive, however the limited number of studies make it difficult to draw firm conclusions.
Future Perspectives on scRNAseq in Type 1 Diabetes
New Single Cell Methods and Analysis Tools
scRNAseq is beginning to give fascinating insights into type 1 diabetes and new approaches may yield further discoveries. The first of these is spatial transcriptomics (80). In this technique, indexed oligos capture RNA from either fresh-frozen or formalin-fixed, paraffin-embedded tissue sections. This allows determination of gene expression on a level that is fast approaching single cell resolution. It has already been used to give insights into cell interactions in other diseases such as rheumatoid arthritis, where infiltrating leukocytes interact with target cells (81, 82). Spatial transcriptomics therefore has great potential to unravel lymphocyte interactions with beta cells in the pancreas and to give insight into different patterns of immune cell infiltration (2). In both type 1 (23) and type 2 diabetes (83) scATACseq has recently been used to link GWAS to epigenetic regulation of gene expression. New methodologies enabling combination of ATACseq, and CITEseq with scRNAseq in the same experiment will also contribute to the field (84). New analysis tools such as CellPhoneDB give the ability to map interactions between subsets of cells, based on DEG in scRNAseq datasets, which would allow identification of novel interactions between immune cells and beta cells in the pancreas (85, 86). This may become increasingly important as we begin to understand the role of beta cell stress and signalling in type 1 diabetes (6) as well as the involvement of other pancreatic cells in diabetes development (23). CellPhoneDB has been used to identify crosstalk between T cells and epithelial cells in ulcerative colitis (87) whilst in rheumatoid arthritis scRNAseq has revealed interaction pathways between B cells, fibroblasts and monocytes (88). Additionally, recent work from the Satija lab has brought together previously published scRNAseq datasets of pancreatic cells, including immune cells from healthy pancreatic samples (12), which will facilitate this type of analysis. This would be further enhanced were there a unified repository for T1D scRNAseq datasets, similar to those for COVID-19 (89).
Technological and Analytical Approaches to Enhance Immunoreceptor Sequencing
We have seen how combining GEX with V(D)J sequencing has increased insights into T1D. The recent development of DNA barcoded multimers will allow now the determination of T cell antigen specificity in scRNAseq experiments (90, 91), whilst conjugation of whole proteins or large folded protein fragments to DNA barcodes will facilitate identification of antigen specific B cells (92).
Computational approaches to determine the likely interaction of an immunoreceptor with target antigen also have the potential to revolutionise the search for antigen specific TCRs and BCRs. Approaches such as tcrdist (93), GLIPH (94) and immune receptor network generation for BCRs (95) enable BCR and TCR sequences to be mapped and visualised, and those that differ by only one or two amino acids are assumed to target the same antigens. NetTCR (96) and TCRex (97) use neural networks and machine learning algorithms to cluster TCRs predicited to bind the same epitope. Recent advances such as ICON and TCRAI leverage scRNAseq technology along with oligo labelled dextramers. They utilise the paired TCRα and TCRβ transcripts to build libraries of antigen specific receptors, with a neural network to predict antigen specificity of TCRs. However, many of these approaches have been validated using viral or tumour antigens with well-defined epitopes. As we have seen in the sections above, whilst there definitely are peptide sequences from diabetes autoantigens that are widely recognised, the immune response also targets diverse sequences in different individuals. Furthermore, auto-antigenic TCRs tend to bind pMHC with lower affinity than TCRs targeting pathogens (98, 99) as high affinity self-reactive TCRs are generally deleted in the thymus. It is not clear how this lower affinity and lack of public TCRs may impact upon the usefulness of computational approaches for T1D.
Biomarkers in Clinical Trials
In 2019, it was demonstrated that teplizumab could delay progression to T1D in high risk individuals (100). Further work confirmed a correlation between fold change in C-peptide and change in frequency of CD8+KLRG1+TIGIT+ T cells (101). scRNAseq of T cells from the clinical trials of teplizumab and other immunotherapies in T1D could offer an amazing opportunity to identify all biomarkers predictive of successful treatment. For example, scRNAseq studies have shown a variety of phenotypic markers induced in vitro with anti-CD3 antibodies in human PBMC, including a variety of interleukin receptors and markers of regulation and exhaustion including FOXP3, CTLA4, TNFRSF18, LAG3 and PDCD1 (102). In contrast, anti-CD3/CD28 stimulation of PBMC analysed with scRNAseq and CITEseq, showed phenotypes strongly associated with activation (although memory subsets also upregulated senescence) (103).
In the future, a deeper understanding of TCRs and BCRs has the potential to better quantify the risk of progression in autoantibody positive people. Monitoring the abundance and phenotypes of lymphocytes bearing specific CDR3 sequences or using specific V genes may also prove useful in monitoring immunotherapies, particularly antigen specific immunotherapies, where phenotypic changes in whole lymphocyte populations may not be so obvious (1, 104–106). In addition, BCRs also have the potential to be used in CAR-Treg cell immunotherapy as has been demonstrated in the NOD mouse (107).
A Computational Approach to Move Beyond scRNAseq
scRNAseq has demonstrated its great potential to identify novel biomarkers both in T1D and other autoimmune diseases. However, it is both technically challenging and expensive. Therefore it is crucial that researchers should be able to translate findings from scRNAseq into more accessible diagnostic and monitoring tests, for example using standardised flow cytometry or qPCR panels as is starting to happen in cancer research (108, 109). Similarly in IBD, a machine learning approach allowed identification of a CD8+ T cell signature that could predict prognosis. These biomarkers were then developed into a commercially available whole blood qPCR test to facilitate personalised therapy (110).
In T1D, recent advances in computational analysis are beginning to allow discrimination of changes in cell subsets from bulk RNAseq. Mehdi et al. identified a peripheral blood transcriptomic signature that predicted autoantibody development (111). Of the DEG identified, many were associated with the ubiquitin-proteasome pathway, DC and T cell function and were potentially targets of drugs approved for other conditions (111). Xhonneux et al. (112), demonstrated from transcriptomics of whole blood that they could undertake “digital cytometry”, by mapping groups of genes back to cell types. Children who developed autoantibodies against insulin first, had a signature of increased NK cells and CD4+ memory T cells. In contrast, those who first developed autoantibodies to GAD had a reduced percentage of CD4+ memory T cells and NK cells, but increased activated NK cells. Harmonizome (113) was used to identify a G protein–coupled receptor, GPR171, predicted to control the immune signature found in IAA+ children (112). Adding gene expression information to predictive models, increased their accuracy in predicting later T1D development in children under 18 months (112).
Discussion
The first papers to analyse lymphocytes from type 1 diabetes using scRNAseq have provided fascinating insights into phenotypes involved in driving the disease and identified new potential targets for immunotherapy, such as IL-32 (15, 17). scRNAseq of TCRs involved in T1D has revealed that autoantigen specific TCRs have a wide range of targets and that whilst single chains or CDR3s are often shared between donors, it is rare to see TCRs with both chains identical in multiple donors; hence public TCRs remain elusive. In the peripheral blood, diabetes autoantigen reactive cells do not always have distinct phenotypes in healthy donors compared to those with T1D (25, 26), and enrichment of islet reactive cells is much more pronounced in the pancreas and pancreatic lymph nodes. Combining TCR sequencing with T cell phenotyping has led to a deeper understanding of islet antigen-specific cells in the peripheral blood (24, 27). A key challenge, for which scRNAseq is ideally suited, will be to develop methods to identify which T cells in the periphery are truly involved in beta cell destruction, and which are simply able to bind islet antigen multimers but are not capable of either trafficking to the islets or contributing to beta cell killing. Looking to the future, it is clear that combining antigen specificity with scRNA phenotyping and new computational approaches, such as those that can give insight into interactions between islet cells and infiltrating lymphocytes, have the potential to revolutionise the field.
Relatively few papers have tackled single cell sequencing (or indeed bulk sequencing) of BCRs repertoires in T1D, but those available suggest that these BCRs have unique properties and are often polyreactive (50, 74, 77). New approaches to identify islet-antigen specific B cells with scRNAseq (92) will therefore have much to contribute to our knowledge of how islet autoantibodies develop and are involved in disease progression.
scRNAseq is ideally suited to identifying subtle phenotypic differences between cohorts and has demonstrated promise in identifying differentially expressed genes in people that will later progress to autoantibody positivity and T1D (27). Developing this approach will be key to identifying at-risk individuals and matching them to a novel immunotherapy that is appropriate for their stage and phenotype of disease (100, 101, 114, 115). Furthermore, new analytical approaches will enable scRNAseq findings to be translated into new immunotherapies and biomarkers to monitor effectiveness of those already in clinical trials.
Author Contributions
SH wrote the first draft of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
SH is funded by the Diabetes Research and Wellness Foundation Professor David Matthews Non-Clinical Research Fellowship 2020. DT, TT, and CD are funded by Diabetes UK, JDRF, Diabetes Research and Wellness Foundation, Wellcome Trust ISSF 204824/Z/16/Z IND-B-SS003 and the Association of Physicians of GB and Ireland. Open access publication fees were provided by the Wellcome Trust via an open access funding grant to Cardiff University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
AAB+, Autoantibody positive; DE cell, dual expressor cell reported to express both a TCR and a BCR’; DEG, differentially expressed genes; GEX, gene expression; scRNAseq, single cell RNA sequencing.
References
1. Ahmed S, Cerosaletti K, James E, Long SA, Mannering S, Speake C, et al. Standardizing T-Cell Biomarkers in Type 1 Diabetes: Challenges and Recent Advances. Diabetes (2019) 68:1366–79. doi: 10.2337/db19-0119
2. Leete P, Oram RA, McDonald TJ, Shields BM, Ziller C, TIGI study team, et al. Studies of Insulin and Proinsulin in Pancreas and Serum Support the Existence of Aetiopathological Endotypes of Type 1 Diabetes Associated With Age at Diagnosis. Diabetologia (2020) 63:1258–67. doi: 10.1007/s00125-020-05115-6
3. Hanna SJ, Powell WE, Long AE, Robinson EJS, Davies J, Megson C, et al. Slow Progressors to Type 1 Diabetes Lose Islet Autoantibodies Over Time, Have Few Islet Antigen-Specific CD8+ T Cells and Exhibit a Distinct CD95hi B Cell Phenotype. Diabetologia (2020) 63:1174–85. doi: 10.1007/s00125-020-05114-7
4. Powell WE, Hanna SJ, Hocter CN, Robinson E, Davies J, Dunseath GJ, et al. Loss of CXCR3 Expression on Memory B Cells in Individuals With Long-Standing Type 1 Diabetes. Diabetologia (2018) 61:1794–803. doi: 10.1007/s00125-018-4651-x
5. Rahman AH, Homann D. Mass Cytometry and Type 1 Diabetes Research in the Age of Single-Cell Data Science. Curr Opin Endocrinol Diabetes Obes (2020) 27:231–9. doi: 10.1097/MED.0000000000000549
6. von Scholten BJ, Kreiner FF, Gough SCL, von Herrath M. Current and Future Therapies for Type 1 Diabetes. Diabetologia (2021) 64:1037–48. doi: 10.1007/s00125-021-05398-3
7. Ding J, Adiconis X, Simmons SK, Kowalczyk MS, Hession CC, Marjanovic ND, et al. Systematic Comparison of Single-Cell and Single-Nucleus RNA-Sequencing Methods. Nat Biotechnol (2020) 38:737–46. doi: 10.1038/s41587-020-0465-8
8. Efremova M, Vento-Tormo R, Park J-E, Teichmann SA, James KR. Immunology in the Era of Single-Cell Technologies. Annu Rev Immunol (2020) 38:727–57. doi: 10.1146/annurev-immunol-090419-020340
9. Carter JA, Preall JB, Grigaityte K, Goldfless SJ, Jeffery E, Briggs AW, et al. Single T Cell Sequencing Demonstrates the Functional Role of αβ TCR Pairing in Cell Lineage and Antigen Specificity. Front Immunol (2019) 10:1516. doi: 10.3389/fimmu.2019.01516
10. Barennes P, Quiniou V, Shugay M, Egorov ES, Davydov AN, Chudakov DM, et al. Benchmarking of T Cell Receptor Repertoire Profiling Methods Reveals Large Systematic Biases. Nat Biotechnol (2021) 39:236–45. doi: 10.1038/s41587-020-0656-3
11. See P, Lum J, Chen J. Ginhoux F. A Single-Cell Sequencing Guide for Immunologists. Front Immunol (2018) 9:2425. doi: 10.3389/fimmu.2018.02425
12. Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, et al. Integrated Analysis of Multimodal Single-Cell Data. Cell (2021) 184:3573–87.e29. doi: 10.1016/j.cell.2021.04.048
13. Andrews TS, Kiselev VY, McCarthy D, Hemberg M. Tutorial: Guidelines for the Computational Analysis of Single-Cell RNA Sequencing Data. Nat Protoc (2021) 16:1–9. doi: 10.1038/s41596-020-00409-w
14. Sun B, Bugarin-Estrada E, Overend LE, Walker CE, Tucci FA, Bashford-Rogers RJM. Double-Jeopardy: scRNA-Seq Doublet/Multiplet Detection Using Multi-Omic Profiling. Cell Rep Methods (2021) 1:100008. doi: 10.1016/j.crmeth.2021.100008
15. Kallionpää H, Somani J, Tuomela S, Ullah U, de Albuquerque R, Lönnberg T, et al. Early Detection of Peripheral Blood Cell Signature in Children Developing β-Cell Autoimmunity at a Young Age. Diabetes (2019) 68:2024–34. doi: 10.2337/db19-0287
16. Thayer TC, Pearson JA, De Leenheer E, Hanna SJ, Boldison J, Davies J, et al. Peripheral Proinsulin Expression Controls Low-Avidity Proinsulin-Reactive CD8 T Cells in Type 1 Diabetes. Diabetes (2016) 65:3429–39. doi: 10.2337/db15-1649
17. Fasolino M, Schwartz GW, Golson ML, Wang YJ, Morgan A, Liu C, et al. Multiomics Single-Cell Analysis of Human Pancreatic Islets Reveals Novel Cellular States in Health and Type 1 Diabetes. Genomics (2021). doi: 10.1101/2021.01.28.428598
18. Nyalwidhe JO, Grzesik WJ, Burch TC, Semeraro ML, Waseem T, Gerling IC, et al. Comparative Quantitative Proteomic Analysis of Disease Stratified Laser Captured Microdissected Human Islets Identifies Proteins and Pathways Potentially Related to Type 1 Diabetes. PloS One (2017) 12:e0183908. doi: 10.1371/journal.pone.0183908
19. AlRashidi FT, Gillespie KM. Biomarkers in Islet Cell Transplantation for Type 1 Diabetes. Curr Diabetes Rep (2018) 18:94. doi: 10.1007/s11892-018-1059-4
20. Zakharov PN, Hu H, Wan X, Unanue ER. Single-Cell RNA Sequencing of Murine Islets Shows High Cellular Complexity at All Stages of Autoimmune Diabetes. J Exp Med (2020) 217:e20192362. doi: 10.1084/jem.20192362
21. Sharon N, Vanderhooft J, Straubhaar J, Mueller J, Chawla R, Zhou Q, et al. Wnt Signaling Separates the Progenitor and Endocrine Compartments During Pancreas Development. Cell Rep (2019) 27:2281–91.e5. doi: 10.1016/j.celrep.2019.04.083
22. Xin Y, Dominguez Gutierrez G, Okamoto H, Kim J, Lee A-H, Adler C, et al. Pseudotime Ordering of Single Human β-Cells Reveals States of Insulin Production and Unfolded Protein Response. Diabetes (2018) 67:1783–94. doi: 10.2337/db18-0365
23. Chiou J, Geusz RJ, Okino M-L, Han JY, Miller M, Melton R, et al. Interpreting Type 1 Diabetes Risk With Genetics and Single-Cell Epigenomics. Nature (2021) 594:398–402. doi: 10.1038/s41586-021-03552-w
24. Cerosaletti K, Barahmand-pour-Whitman F, Yang J, DeBerg HA, Dufort MJ, Murray SA, et al. Single-Cell RNA Sequencing Reveals Expanded Clones of Islet Antigen-Reactive CD4 + T Cells in Peripheral Blood of Subjects With Type 1 Diabetes. JI (2017) 199:323–35. doi: 10.4049/jimmunol.1700172
25. Fuchs YF, Sharma V, Eugster A, Kraus G, Morgenstern R, Dahl A, et al. Gene Expression-Based Identification of Antigen-Responsive CD8+ T Cells on a Single-Cell Level. Front Immunol (2019) 10:2568. doi: 10.3389/fimmu.2019.02568
26. Culina S, Lalanne AI, Afonso G, Cerosaletti K, Pinto S, Sebastiani G, et al. Islet-Reactive CD8 + T Cell Frequencies in the Pancreas, But Not in Blood, Distinguish Type 1 Diabetic Patients From Healthy Donors. Sci Immunol (2018) 3:eaao4013. doi: 10.1126/sciimmunol.aao4013
27. Heninger A-K, Eugster A, Kuehn D, Buettner F, Kuhn M, Lindner A, et al. A Divergent Population of Autoantigen-Responsive CD4 + T Cells in Infants Prior to β Cell Autoimmunity. Sci Transl Med (2017) 9:eaaf8848. doi: 10.1126/scitranslmed.aaf8848
28. Ahmed R, Omidian Z, Giwa A, Cornwell B, Majety N, Bell DR, et al. A Public BCR Present in a Unique Dual-Receptor-Expressing Lymphocyte From Type 1 Diabetes Patients Encodes a Potent T Cell Autoantigen. Cell (2019) 177:1583–99.e16. doi: 10.1016/j.cell.2019.05.007
29. Enge M, Arda HE, Mignardi M, Beausang J, Bottino R, Kim SK, et al. Single-Cell Analysis of Human Pancreas Reveals Transcriptional Signatures of Aging and Somatic Mutation Patterns. Cell (2017) 171:321–30.e14. doi: 10.1016/j.cell.2017.09.004
30. Grün D, Muraro MJ, Boisset J-C, Wiebrands K, Lyubimova A, Dharmadhikari G, et al. De Novo Prediction of Stem Cell Identity Using Single-Cell Transcriptome Data. Cell Stem Cell (2016) 19:266–77. doi: 10.1016/j.stem.2016.05.010
31. Muraro MJ, Dharmadhikari G, Grün D, Groen N, Dielen T, Jansen E, et al. A Single-Cell Transcriptome Atlas of the Human Pancreas. Cell Syst (2016) 3:385–94.e3. doi: 10.1016/j.cels.2016.09.002
32. Segerstolpe Å, Palasantza A, Eliasson P, Andersson E-M, Andréasson A-C, Sun X, et al. Single-Cell Transcriptome Profiling of Human Pancreatic Islets in Health and Type 2 Diabetes. Cell Metab (2016) 24:593–607. doi: 10.1016/j.cmet.2016.08.020
33. Lawlor N, George J, Bolisetty M, Kursawe R, Sun L, Sivakamasundari V, et al. Single-Cell Transcriptomes Identify Human Islet Cell Signatures and Reveal Cell-Type–Specific Expression Changes in Type 2 Diabetes. Genome Res (2017) 27:208–22. doi: 10.1101/gr.212720.116
34. Baron M, Veres A, Wolock SL, Faust AL, Gaujoux R, Vetere A, et al. A Single-Cell Transcriptomic Map of the Human and Mouse Pancreas Reveals Inter- and Intra-Cell Population Structure. Cell Syst (2016) 3:346–60.e4. doi: 10.1016/j.cels.2016.08.011
35. Xin Y, Adler C, Kim J, Ding Y, Ni M, Wei Y, et al. Single-Cell RNA Sequencing and Analysis of Human Pancreatic Islets. JoVE (2019) 149. doi: 10.3791/59866
36. Zhang J, Chen L-M, Zou Y, Zhang S, Xiong F, Wang C-Y. Implication of Epigenetic Factors in the Pathogenesis of Type 1 Diabetes. Chin Med J (2021) 134:1031–42. doi: 10.1097/CM9.0000000000001450
37. Esposito S, Toni G, Tascini G, Santi E, Berioli MG, Principi N. Environmental Factors Associated With Type 1 Diabetes. Front Endocrinol (2019) 10:592. doi: 10.3389/fendo.2019.00592
38. Xie Z, Chang C, Huang G, Zhou Z. The Role of Epigenetics in Type 1 Diabetes. In: Chang C, Lu Q, editors. Epigenetics in Allergy and Autoimmunity Advances in Experimental Medicine and Biology. Singapore: Springer Singapore (2020). p. 223–57. doi: 10.1007/978-981-15-3449-2_9
39. Robertson CC, Inshaw JRJ, Onengut-Gumuscu S, Chen W-M, Santa Cruz DF, Yang H, et al. Fine-Mapping, Trans-Ancestral and Genomic Analyses Identify Causal Variants, Cells, Genes and Drug Targets for Type 1 Diabetes. Nat Genet (2021) 53:962–71. doi: 10.1038/s41588-021-00880-5
40. Clark M, Kroger CJ, Ke Q, Tisch RM. The Role of T Cell Receptor Signaling in the Development of Type 1 Diabetes. Front Immunol (2021) 11:615371. doi: 10.3389/fimmu.2020.615371
41. Jacobsen LM, Posgai A, Seay HR, Haller MJ. Brusko TM. T Cell Receptor Profiling in Type 1 Diabetes. Curr Diabetes Rep (2017) 17:118. doi: 10.1007/s11892-017-0946-4
42. Wang Y, Sosinowski T, Novikov A, Crawford F, White J, Jin N, et al. How C-Terminal Additions to Insulin B-Chain Fragments Create Superagonists for T Cells in Mouse and Human Type 1 Diabetes. Sci Immunol (2019) 4:eaav7517. doi: 10.1126/sciimmunol.aav7517
43. James EA, Mallone R, Kent SC. DiLorenzo TP. T-Cell Epitopes and Neo-Epitopes in Type 1 Diabetes: A Comprehensive Update and Reappraisal. Diabetes (2020) 69:1311–35. doi: 10.2337/dbi19-0022
44. Eugster A, Lindner A, Catani M, Heninger A-K, Dahl A, Klemroth S, et al. High Diversity in the TCR Repertoire of GAD65 Autoantigen-Specific Human CD4 + T Cells. JI (2015) 194:2531–8. doi: 10.4049/jimmunol.1403031
45. Fuchs YF, Eugster A, Dietz S, Sebelefsky C, Kühn D, Wilhelm C, et al. CD8+ T Cells Specific for the Islet Autoantigen IGRP Are Restricted in Their T Cell Receptor Chain Usage. Sci Rep (2017) 7:44661. doi: 10.1038/srep44661
46. Baker RL, Rihanek M, Hohenstein AC, Nakayama M, Michels A, Gottlieb PA, et al. Hybrid Insulin Peptides Are Autoantigens in Type 1 Diabetes. Diabetes (2019) 68:1830–40. doi: 10.2337/db19-0128
47. Kent SC, Chen Y, Bregoli L, Clemmings SM, Kenyon NS, Ricordi C, et al. Expanded T Cells From Pancreatic Lymph Nodes of Type 1 Diabetic Subjects Recognize an Insulin Epitope. Nature (2005) 435:224–8. doi: 10.1038/nature03625
48. Pathiraja V, Kuehlich JP, Campbell PD, Krishnamurthy B, Loudovaris T, Coates PTH, et al. Proinsulin-Specific, HLA-DQ8, and HLA-DQ8-Transdimer–Restricted CD4 + T Cells Infiltrate Islets in Type 1 Diabetes. Diabetes (2015) 64:172–82. doi: 10.2337/db14-0858
49. Skowera A, Ladell K, McLaren JE, Dolton G, Matthews KK, Gostick E, et al. β-Cell–Specific CD8 T Cell Phenotype in Type 1 Diabetes Reflects Chronic Autoantigen Exposure. Diabetes (2015) 64:916–25. doi: 10.2337/db14-0332
50. Seay HR, Yusko E, Rothweiler SJ, Zhang L, Posgai AL, Campbell-Thompson M, et al. Tissue Distribution and Clonal Diversity of the T and B Cell Repertoire in Type 1 Diabetes. JCI Insight (2016) 1:e88242. doi: 10.1172/jci.insight.88242
51. Arif S, Gomez-Tourino I, Kamra Y, Pujol-Autonell I, Hanton E, Tree T, et al. GAD-Alum Immunotherapy in Type 1 Diabetes Expands Bifunctional Th1/Th2 Autoreactive CD4 T Cells. Diabetologia (2020) 63:1186–98. doi: 10.1007/s00125-020-05130-7
52. Michels AW, Landry LG, McDaniel KA, Yu L, Campbell-Thompson M, Kwok WW, et al. Islet-Derived CD4 T Cells Targeting Proinsulin in Human Autoimmune Diabetes. Diabetes (2017) 66:722–34. doi: 10.2337/db16-1025
53. Landry LG, Anderson AM, Russ HA, Yu L, Kent SC, Atkinson MA, et al. Proinsulin-Reactive CD4 T Cells in the Islets of Type 1 Diabetes Organ Donors. Front Endocrinol (2021) 12:622647. doi: 10.3389/fendo.2021.622647
54. Zhao M, Jiang J, Zhao M, Chang C, Wu H, Lu Q. The Application of Single-Cell RNA Sequencing in Studies of Autoimmune Diseases: A Comprehensive Review. Clinic Rev Allerg Immunol (2021) 60:68–86. doi: 10.1007/s12016-020-08813-6
55. Penkava F, Velasco-Herrera MDC, Young MD, Yager N, Nwosu LN, Pratt AG, et al. Single-Cell Sequencing Reveals Clonal Expansions of Pro-Inflammatory Synovial CD8 T Cells Expressing Tissue-Homing Receptors in Psoriatic Arthritis. Nat Commun (2020) 11:4767. doi: 10.1038/s41467-020-18513-6
56. Bagaev DV, Vroomans RMA, Samir J, Stervbo U, Rius C, Dolton G, et al. VDJdb in 2019: Database Extension, New Analysis Infrastructure and a T-Cell Receptor Motif Compendium. Nucleic Acids Res (2020) 48:D1057–62. doi: 10.1093/nar/gkz874
57. Vita R, Mahajan S, Overton JA, Dhanda SK, Martini S, Cantrell JR, et al. The Immune Epitope Database (IEDB): 2018 Update. Nucleic Acids Res (2019) 47:D339–43. doi: 10.1093/nar/gky1006
58. Wiles TA, Hohenstein A, Landry LG, Dang M, Powell R, Guyer P, et al. Characterization of Human CD4 T Cells Specific for a C-Peptide/C-Peptide Hybrid Insulin Peptide. Front Immunol (2021) 12:668680. doi: 10.3389/fimmu.2021.668680
59. Ji Q, Perchellet A, Goverman JM. Viral Infection Triggers Central Nervous System Autoimmunity via Activation of CD8+ T Cells Expressing Dual TCRs. Nat Immunol (2010) 11:628–34. doi: 10.1038/ni.1888
60. Schuldt NJ, Auger JL, Spanier JA, Martinov T, Breed ER, Fife BT, et al. Cutting Edge: Dual Tcrα Expression Poses an Autoimmune Hazard by Limiting Regulatory T Cell Generation. JI (2017) 199:33–8. doi: 10.4049/jimmunol.1700406
61. Schallenberg S, Kretschmer K. New Insight Into Type 1 Diabetes Development: Resolving Early Diabetogenic CD4+ T Cell Responses That Precede Seroconversion. Ann Transl Med (2018) 6:58–8. doi: 10.21037/atm.2017.12.14
62. Richter W, Endl J, Eiermann TH, Brandt M, Kientsch-Engel R, Thivolet C, et al. Human Monoclonal Islet Cell Antibodies From a Patient With Insulin-Dependent Diabetes Mellitus Reveal Glutamate Decarboxylase as the Target Antigen. Proc Natl Acad Sci (1992) 89:8467–71. doi: 10.1073/pnas.89.18.8467
63. Madec AM, Rousset F, Ho S, Robert F, Thivolet C, Orgiazzi J, et al. Four IgG Anti-Islet Human Monoclonal Antibodies Isolated From a Type 1 Diabetes Patient Recognize Distinct Epitopes of Glutamic Acid Decarboxylase 65 and Are Somatically Mutated. J Immunol (1996) 156:3541–9.
64. Jury KM, Loeffler D, Eiermann TH, Ziegler B, Boehm BO, Richter W. Evidence for Somatic Mutation and Affinity Maturation of Diabetes Associated Human Autoantibodies to Glutamate Decarboxylase. J Autoimmun (1996) 9:371–7. doi: 10.1006/jaut.1996.0050
65. Kolm-Litty V, Berlo S, Bonifacio E, Bearzatto M, Engel AM, Christie M, et al. Human Monoclonal Antibodies Isolated From Type I Diabetes Patients Define Multiple Epitopes in the Protein Tyrosine Phosphatase-Like IA-2 Antigen. J Immunol (2000) 165:4676–84. doi: 10.4049/jimmunol.165.8.4676
66. Ananieva-Jordanova R, Evans M, Nakamatsu T, Premawardhana LDKE, Sanders J, Powell M, et al. Isolation and Characterisation of a Human Monoclonal Autoantibody to the Islet Cell Autoantigen IA-2. J Autoimmun (2005) 24:337–45. doi: 10.1016/j.jaut.2005.03.003
67. Weenink SM, Lo J, Stephenson CR, McKinney PA, Ananieva-Jordanova R, Rees Smith B, et al. Autoantibodies and Associated T-Cell Responses to Determinants Within the 831–860 Region of the Autoantigen IA-2 in Type 1 Diabetes. J Autoimmun (2009) 33:147–54. doi: 10.1016/j.jaut.2009.04.002
68. Ikematsu H, Ichiyoshi Y, Schettino EW, Nakamura M, Casali P. VH and V Kappa Segment Structure of Anti-Insulin IgG Autoantibodies in Patients With Insulin-Dependent Diabetes Mellitus. Evidence for Somatic Selection. J Immunol (1994) 152:1430–41.
69. Ichiyoshi Y, Zhou M, Casali P. A Human Anti-Insulin IgG Autoantibody Apparently Arises Through Clonal Selection From an Insulin-Specific “Germ-Line” Natural Antibody Template. Analysis by V Gene Segment Reassortment and Site-Directed Mutagenesis. J Immunol (1995) 154:226–38.
70. Potter KN, Wilkin TJ. The Molecular Specificity of Insulin Autoantibodies. Diabetes Metab Res Rev (2000) 16:338–53. doi: 10.1002/1520-7560(2000)9999:9999<::aid-dmrr145>3.0.co;2-l
71. Bashford-Rogers RJM, Bergamaschi L, McKinney EF, Pombal DC, Mescia F, Lee JC, et al. Analysis of the B Cell Receptor Repertoire in Six Immune-Mediated Diseases. Nature (2019) 574:122–6. doi: 10.1038/s41586-019-1595-3
72. Horns F, Dekker CL, Quake SR. Memory B Cell Activation, Broad Anti-Influenza Antibodies, and Bystander Activation Revealed by Single-Cell Transcriptomics. Cell Rep (2020) 30:905–13.e6. doi: 10.1016/j.celrep.2019.12.063
73. Milpied P, Cervera-Marzal I, Mollichella M-L, Tesson B, Brisou G, Traverse-Glehen A, et al. Human Germinal Center Transcriptional Programs Are De-Synchronized in B Cell Lymphoma. Nat Immunol (2018) 19:1013–24. doi: 10.1038/s41590-018-0181-4
74. Smith MJ, Packard TA, O’Neill SK, Henry Dunand CJ, Huang M, Fitzgerald-Miller L, et al. Loss of Anergic B Cells in Prediabetic and New-Onset Type 1 Diabetic Patients. Diabetes (2015) 64:1703–12. doi: 10.2337/db13-1798
75. Japp AS, Meng W, Rosenfeld AM, Perry DJ, Thirawatananond P, Bacher RL, et al. TCR+/BCR+ Dual-Expressing Cells and Their Associated Public BCR Clonotype Are Not Enriched in Type 1 Diabetes. Cell (2021) 184:827–39.e14. doi: 10.1016/j.cell.2020.11.035
76. Catani M, Walther D, Christie MR, McLaughlin KA, Bonifacio E, Eugster A. Isolation of Human Monoclonal Autoantibodies Derived From Pancreatic Lymph Node and Peripheral Blood B Cells of Islet Autoantibody-Positive Patients. Diabetologia (2016) 59:294–8. doi: 10.1007/s00125-015-3792-4
77. Wardemann H. Predominant Autoantibody Production by Early Human B Cell Precursors. Science (2003) 301:1374–7. doi: 10.1126/science.1086907
78. Avrameas S, Alexopoulos H, Moutsopoulos HM. Natural Autoantibodies: An Undersugn Hero of the Immune System and Autoimmune Disorders—A Point of View. Front Immunol (2018) 9:1320. doi: 10.3389/fimmu.2018.01320
79. Zhou Z-H, Tzioufas AG, Notkins AL. Properties and Function of Polyreactive Antibodies and Polyreactive Antigen-Binding B Cells. J Autoimmun (2007) 29:219–28. doi: 10.1016/j.jaut.2007.07.015
80. Ståhl PL, Salmén F, Vickovic S, Lundmark A, Navarro JF, Magnusson J, et al. Visualization and Analysis of Gene Expression in Tissue Sections by Spatial Transcriptomics. Science (2016) 353:78–82. doi: 10.1126/science.aaf2403
81. Carlberg K, Korotkova M, Larsson L, Catrina AI, Ståhl PL, Malmström V. Exploring Inflammatory Signatures in Arthritic Joint Biopsies With Spatial Transcriptomics. Sci Rep (2019) 9:18975. doi: 10.1038/s41598-019-55441-y
82. Vickovic S, Schapiro D, Carlberg K, Lötstedt B, Larsson L, Korotkova M, et al. Three-Dimensional Spatial Transcriptomics Uncovers Cell Type Dynamics in the Rheumatoid Arthritis Synovium. Genomics (2020). doi: 10.1101/2020.12.10.420463
83. Rai V, Quang DX, Erdos MR, Cusanovich DA, Daza RM, Narisu N, et al. Single-Cell ATAC-Seq in Human Pancreatic Islets and Deep Learning Upscaling of Rare Cells Reveals Cell-Specific Type 2 Diabetes Regulatory Signatures. Mol Metab (2020) 32:109–21. doi: 10.1016/j.molmet.2019.12.006
84. Mimitou EP, Lareau CA, Chen KY, Zorzetto-Fernandes AL, Hao Y, Takeshima Y, et al. Scalable, Multimodal Profiling of Chromatin Accessibility, Gene Expression and Protein Levels in Single Cells. Nat Biotechnol (2021). doi: 10.1038/s41587-021-00927-2
85. Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: Inferring Cell–Cell Communication From Combined Expression of Multi-Subunit Ligand–Receptor Complexes. Nat Protoc (2020) 15:1484–506. doi: 10.1038/s41596-020-0292-x
86. Hao S, Yan K-K, Ding L, Qian C, Chi H, Yu J. Network Approaches for Dissecting the Immune System. iScience (2020) 23:101354. doi: 10.1016/j.isci.2020.101354
87. Corridoni D, Antanaviciute A, Gupta T, Fawkner-Corbett D, Aulicino A, Jagielowicz M, et al. Single-Cell Atlas of Colonic CD8+ T Cells in Ulcerative Colitis. Nat Med (2020) 26:1480–90. doi: 10.1038/s41591-020-1003-4
88. Palla G, Ferrero E. Latent Factor Modeling of scRNA-Seq Data Uncovers Dysregulated Pathways in Autoimmune Disease Patients. iScience (2020) 23:101451. doi: 10.1016/j.isci.2020.101451
89. Chan Zuckerberg Initiative Single-Cell COVID-19 Consortia, Ballestar E, Farber DL, Glover S, Horwitz B, Meyer K, et al. Single Cell Profiling of COVID-19 Patients: An International Data Resource From Multiple Tissues. MedRxiv (2020). doi: 10.1101/2020.11.20.20227355
90. Bentzen AK, Marquard AM, Lyngaa R, Saini SK, Ramskov S, Donia M, et al. Large-Scale Detection of Antigen-Specific T Cells Using Peptide-MHC-I Multimers Labeled With DNA Barcodes. Nat Biotechnol (2016) 34:1037–45. doi: 10.1038/nbt.3662
91. Minervina AA, Pogorelyy MV, Kirk AM, Allen EK, Allison KJ, Lin C-Y, et al. Convergent Epitope-Specific T Cell Responses After SARS-CoV-2 Infection and Vaccination. MedRxiv (2021). doi: 10.1101/2021.07.12.21260227
92. Setliff I, Shiakolas AR, Pilewski KA, Murji AA, Mapengo RE, Janowska K, et al. High-Throughput Mapping of B Cell Receptor Sequences to Antigen Specificity. Cell (2019) 179:1636–46.e15. doi: 10.1016/j.cell.2019.11.003
93. Dash P, Fiore-Gartland AJ, Hertz T, Wang GC, Sharma S, Souquette A, et al. Quantifiable Predictive Features Define Epitope-Specific T Cell Receptor Repertoires. Nature (2017) 547:89–93. doi: 10.1038/nature22383
94. Glanville J, Huang H, Nau A, Hatton O, Wagar LE, Rubelt F, et al. Identifying Specificity Groups in the T Cell Receptor Repertoire. Nature (2017) 547:94–8. doi: 10.1038/nature22976
95. Bashford-Rogers RJM, Palser AL, Huntly BJ, Rance R, Vassiliou GS, Follows GA, et al. Network Properties Derived From Deep Sequencing of Human B-Cell Receptor Repertoires Delineate B-Cell Populations. Genome Res (2013) 23:1874–84. doi: 10.1101/gr.154815.113
96. Montemurro A, Schuster V, Povlsen HR, Bentzen AK, Jurtz V, Chronister WD, et al. NetTCR-2.0 Enables Accurate Prediction of TCR-Peptide Binding by Using Paired TCRα and β Sequence Data. Commun Biol (2021) 4:1060. doi: 10.1038/s42003-021-02610-3
97. Gielis S, Moris P, Bittremieux W, De Neuter N, Ogunjimi B, Laukens K, et al. Detection of Enriched T Cell Epitope Specificity in Full T Cell Receptor Sequence Repertoires. Front Immunol (2019) 10:2820. doi: 10.3389/fimmu.2019.02820
98. Bridgeman JS, Sewell AK, Miles JJ, Price DA, Cole DK. Structural and Biophysical Determinants of αβ T-Cell Antigen Recognition: T-Cell Antigen Recognition. Immunology (2012) 135:9–18. doi: 10.1111/j.1365-2567.2011.03515.x
99. Cole DK, Bulek AM, Dolton G, Schauenberg AJ, Szomolay B, Rittase W, et al. Hotspot Autoimmune T Cell Receptor Binding Underlies Pathogen and Insulin Peptide Cross-Reactivity. J Clin Invest (2016) 126:2191–204. doi: 10.1172/JCI85679
100. Herold KC, Bundy BN, Long SA, Bluestone JA, DiMeglio LA, Dufort MJ, et al. An Anti-CD3 Antibody, Teplizumab, in Relatives at Risk for Type 1 Diabetes. N Engl J Med (2019) 381:603–13. doi: 10.1056/NEJMoa1902226
101. Sims EK, Bundy BN, Stier K, Serti E, Lim N, Long SA, et al. Teplizumab Improves and Stabilizes Beta Cell Function in Antibody-Positive High-Risk Individuals. Sci Transl Med (2021) 13:eabc8980. doi: 10.1126/scitranslmed.abc8980
102. Sousa IG, Simi KCR, do Almo MM, Bezerra MAG, Doose G, Raiol T, et al. Gene Expression Profile of Human T Cells Following a Single Stimulation of Peripheral Blood Mononuclear Cells With Anti-CD3 Antibodies. BMC Genomics (2019) 20:593. doi: 10.1186/s12864-019-5967-8
103. Lawlor N, Nehar-Belaid D, Grassmann JDS, Stoeckius M, Smibert P, Stitzel ML, et al. Single Cell Analysis of Blood Mononuclear Cells Stimulated Through Either LPS or Anti-CD3 and Anti-Cd28. Front Immunol (2021) 12:636720. doi: 10.3389/fimmu.2021.636720
104. Alhadj Ali M, Liu Y-F, Arif S, Tatovic D, Shariff H, Gibson VB, et al. Metabolic and Immune Effects of Immunotherapy With Proinsulin Peptide in Human New-Onset Type 1 Diabetes. Sci Transl Med (2017) 9:eaaf7779. doi: 10.1126/scitranslmed.aaf7779
105. Casas R, Dietrich F, Barcenilla H, Tavira B, Wahlberg J, Achenbach P, et al. Glutamic Acid Decarboxylase Injection Into Lymph Nodes: Beta Cell Function and Immune Responses in Recent Onset Type 1 Diabetes Patients. Front Immunol (2020) 11:564921. doi: 10.3389/fimmu.2020.564921
106. Assfalg R, Knoop J, Hoffman KL, Pfirrmann M, Zapardiel-Gonzalo JM, Hofelich A, et al. Oral Insulin Immunotherapy in Children at Risk for Type 1 Diabetes in a Randomised Controlled Trial. Diabetologia (2021) 64:1079–92. doi: 10.1007/s00125-020-05376-1
107. Tenspolde M, Zimmermann K, Weber LC, Hapke M, Lieber M, Dywicki J, et al. Regulatory T Cells Engineered With a Novel Insulin-Specific Chimeric Antigen Receptor as a Candidate Immunotherapy for Type 1 Diabetes. J Autoimmun (2019) 103:102289. doi: 10.1016/j.jaut.2019.05.017
108. Zheng l, Li l, Xie j, Jin h, Zhu n. Six Novel Biomarkers for Diagnosis and Prognosis of Esophageal Squamous Cell Carcinoma: Validated by scRNA-Seq and qPCR. J Cancer (2021) 12:899–911. doi: 10.7150/jca.50443
109. Liu J, Xu T, Jin Y, Huang B, Zhang Y. Progress and Clinical Application of Single-Cell Transcriptional Sequencing Technology in Cancer Research. Front Oncol (2021) 10:593085. doi: 10.3389/fonc.2020.593085
110. Biasci D, Lee JC, Noor NM, Pombal DR, Hou M, Lewis N, et al. A Blood-Based Prognostic Biomarker in IBD. Gut (2019) 68:1386–95. doi: 10.1136/gutjnl-2019-318343
111. Mehdi AM, Hamilton-Williams EE, Cristino A, Ziegler A, Bonifacio E, Le Cao K-A, et al. A Peripheral Blood Transcriptomic Signature Predicts Autoantibody Development in Infants at Risk of Type 1 Diabetes. JCI Insight (2018) 3:e98212. doi: 10.1172/jci.insight.98212
112. Xhonneux L-P, Knight O, Lernmark Å, Bonifacio E, Hagopian WA, Rewers MJ, et al. Transcriptional Networks in at-Risk Individuals Identify Signatures of Type 1 Diabetes Progression. Sci Transl Med (2021) 13:eabd5666. doi: 10.1126/scitranslmed.abd5666
113. Rouillard AD, Gundersen GW, Fernandez NF, Wang Z, Monteiro CD, McDermott MG, et al. The Harmonizome: A Collection of Processed Datasets Gathered to Serve and Mine Knowledge About Genes and Proteins. Database (2016) 2016:baw100. doi: 10.1093/database/baw100
114. Jacobsen LM, Bundy BN, Greco MN, Schatz DA, Atkinson MA, Brusko TM, et al. Comparing Beta Cell Preservation Across Clinical Trials in Recent-Onset Type 1 Diabetes. Diabetes Technol Ther (2020) 22:948–53. doi: 10.1089/dia.2020.0305
115. Sosenko JM, Skyler JS, Herold KC, Schatz DA, Haller MJ, Pugliese A, et al. Slowed Metabolic Decline After 1 Year of Oral Insulin Treatment Among Individuals at High Risk for Type 1 Diabetes in the Diabetes Prevention Trial–Type 1 (DPT-1) and TrialNet Oral Insulin Prevention Trials. Diabetes (2020) 69:1827–32. doi: 10.2337/db20-0166
Keywords: type 1 diabetes, scRNAseq, immunology, lymphocytes, TCR - T cell receptor, BCR - B cell receptor
Citation: Hanna SJ, Tatovic D, Thayer TC and Dayan CM (2021) Insights From Single Cell RNA Sequencing Into the Immunology of Type 1 Diabetes- Cell Phenotypes and Antigen Specificity. Front. Immunol. 12:751701. doi: 10.3389/fimmu.2021.751701
Received: 01 August 2021; Accepted: 14 September 2021;
Published: 01 October 2021.
Edited by:
Anne Cooke, University of Cambridge, United KingdomReviewed by:
Aaron Michels, University of Colorado, United StatesStefania Canè, University of Verona, Italy
Copyright © 2021 Hanna, Tatovic, Thayer and Dayan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stephanie J. Hanna, SGFubmFTSkBjZi5hYy51aw==
 Stephanie J. Hanna
Stephanie J. Hanna Danijela Tatovic1
Danijela Tatovic1