- Max Perutz Labs, University of Vienna, Vienna Biocenter (VBC), Vienna, Austria
Regulated changes in mRNA stability are critical drivers of gene expression adaptations to immunological cues. mRNA stability is controlled mainly by RNA-binding proteins (RBPs) which can directly cleave mRNA but more often act as adaptors for the recruitment of the RNA-degradation machinery. One of the most prominent RBPs with regulatory roles in the immune system is tristetraprolin (TTP). TTP targets mainly inflammation-associated mRNAs for degradation and is indispensable for the resolution of inflammation as well as the maintenance of immune homeostasis. Recent advances in the transcriptome-wide knowledge of mRNA expression and decay rates together with TTP binding sites in the target mRNAs revealed important limitations in our understanding of molecular mechanisms of TTP action. Such orthogonal analyses lead to the discovery that TTP binding destabilizes some bound mRNAs but not others in the same cell. Moreover, comparisons of various immune cells indicated that an mRNA can be destabilized by TTP in one cell type while it remains stable in a different cell linage despite the presence of TTP. The action of TTP extends from mRNA destabilization to inhibition of translation in a subset of targets. This article will discuss these unexpected context-dependent functions and their implications for the regulation of immune responses. Attention will be also payed to new insights into the role of TTP in physiology and tissue homeostasis.
Introduction
It is now well accepted that regulation of mRNA stability by RNA-binding proteins (RBPs) is indispensable for healthy immune responses. RBPs orchestrate the immune system by modulating gene expression through mRNA destabilization or stabilization, or by controlling translation (1–3). Although this basic knowledge is established, many important questions remain unresolved. These include mechanistic explanations of the phenotype caused by an RBP deletion in mice and the selective functions of RBPs in specific cell types despite ubiquitous expression. Improved models of the molecular mechanisms of RBP action are needed to answer the open questions. These models will likely abandon the linear schemes in which RBP binding to a target mRNA inevitably results in a canonical consequence, e.g. mRNA decay. The aim of this review is to provide a framework for updated models of RBP action in immune responses.
The history of mRNA decay research, both at the level of mechanisms and functions, is tightly connected to the immune system. The first evidence that selective mRNA degradation is driven by a cis-acting element was reported for the mRNA encoding the granulocyte/monocyte growth factor GM-CSF (4). This study established that an adenylate-uridylate-rich element (AU-rich element; ARE) in the 3’ untranslated region (3’ UTR) of the GM-CSF mRNA (encoded by the CSF2 gene) confers mRNA instability if introduced into the 3’ UTR of a stable mRNA. The autonomous effect of AREs on mRNA stability has been subsequently documented for many other mRNAs. The key role of ARE-dependent mRNA decay in vivo was revealed by the deletion of the ARE in the mouse Tnf gene which resulted in a spontaneous development of gut and joint inflammation (5). However, genome sequencing and transcriptome-wide mRNA stability assays indicated that the initial model of an autonomous function of 3’ UTR-located AREs in mRNA destabilization was too simple. Approximately 20% of human genes contain AREs in their 3’ UTRs, yet most of the corresponding mRNAs are stable (6, 7). The medium half-life of mRNA in human HepG3 cells is approximately 10 h with mRNAs of metabolic genes having on average the highest half-lives (6). In comparison, inflammation-associated mRNAs belong to those with the shortest average half-lives. For illustration, the decay rate of TNF mRNA is in the range of 20 – 40 min, depending on the cell type and stimulus (8, 9). Although inflammation-induced mRNAs are enriched in AREs, it is now accepted that the presence of an ARE is not sufficient to destabilize the mRNA. Hence, new and more comprehensive models of regulation of mRNA decay by cis-acting elements are needed.
Mechanisms of RBP-Driven Changes in mRNA Stability
mRNA-destabilizing RBPs bind and facilitate the target mRNA degradation in two ways, depending on the properties of the particular RBP (1, 2, 10, 11). One class of RBPs possesses an endonuclease activity which allows the RBP to cleave the target mRNA and generate ends devoid of the 5’ m7G cap and the 3’ poly(A) tail. These unprotected ends serve as substrates for exonucleases which process the mRNA in 3’ – 5’ direction via the exosome and 5’ - 3’ direction via XRN1 (12, 13). The best characterized endonucleolytic RBP relevant for the immune system is Regnase-1 (gene name Zc3h12a) which destabilizes mRNAs of transcription factors and cytokines such as Icos, Ox40, c-Rel, IL-2 and IL-6 (14). Regnase-1-deficient mice show severe systemic inflammation associated with T and B cell activation. The phenotype is largely recapitulated by a T cell-specific deletion (14). The second class of RBPs destabilize the target mRNA by promoting the recruitment of the CCR4-NOT deadenylase and the DCP1/DCP2-containing decapping complexes (15, 16). A number of RBPs in this class are known to regulate the immune system. Tristetraprolin (TTP), as one of the most prominent members, will be described in detail below. Other well characterized members are Roquin-1 (gene name Rc3h1), Roquin-2 (Rc3h2), Zfp36l1, Zfp36l2 and Auf1. The Roquin proteins redundantly target the mRNAs of Icos and Ox40 to control T cell activation. Deletion of both Roquin-1 and Roquin-2 genes specifically in CD4 T cells results in an autoimmune phenotype resembling systemic lupus erythematosus while deletion of the single genes remains without severe consequences (17). The proteins Zfp36l1 and Zfp36l2 are members of the TTP family but, in contrast to TTP, have more pleotropic functions as demonstrated by embryonic or postnatal lethality of the respective knockouts in mice (18, 19). Zfp36l1 and Zfp36l2 are involved in the regulation of immune system in multiple ways. They control the expression of proliferative cell cycle regulators during B and T cell development: double deletion of Zfp36l1 and Zfp36l2 in T cells results in lymphopenia and malignant transformation of immature CD8 T cells while similar deletion in pro-B cells causes a block in B cell development owing to a failure in entering quiescence hence genome safeguarding prior to VDJ recombination (20, 21). The protein Auf1 exhibits anti-inflammatory functions by promoting the degradation of cytokine mRNAs as revealed by the hypersensitivity of Auf1-deficient mice to endotoxic shock (22). Auf1 has been subsequently found to regulate many other processes in addition to immune responses including telomere maintenance and muscle regeneration (23, 24).
mRNA-stabilizing RBPs are less well understood and their functions are more pleiotropic as compared to the destabilizing RBPs such as TTP. The general opinion is that mRNA-stabilizing RBPs act by preventing the destabilizing proteins from binding to the target. As a consequence, the target mRNAs are more stable and/or more efficiently translated. mRNA-stabilizing RBPs regulating immune responses include HuR (gene name Elavl1) and Arid5a. Deletion of HuR or Arid5a in mice resulted in increased resistance to experimental autoimmune encephalomyelitis (25, 26). Furthermore, HuR is required for antibody production by B cells (27). HuR is involved in regulation of other processes including liver metabolism, cell proliferation and cancer (28–30).
RNA Binding of RBPs Controlling mRNA Stability
RBPs bind to RNA through interactions of their RNA-binding domains with specific sequences or defined structural elements in the target mRNA. The most frequent RNA-binding domain in the immunoregulatory RBPs is the C3H1 (Cys-Cys-Cys-His) zinc finger domain present for example in TTP, Zfp36l1 and Zfp36l2 (2, 31). Regnase and Roquin contain a C3H1 zinc finger and an additional RNA-binding domain: a PIN domain and a ROQ domain, respectively (32, 33). Auf1 and HuR bind to RNA through the RNA recognition motif (RRM) domains which occur in 2 or 3 repeats in these proteins (34, 35). Arid5a interacts with RNA via an ARID domain which is known to recognize DNA in other ARID domain-containing proteins (26, 36).
The target site in the RNA is defined by the RNA-binding domain. The C3H1 zinc finger present in TTP, Zfp36l1 and Zfp36l2 binds preferentially to AREs with the core sequence UAUUUAU although divergent target sites have been identified as well (9, 21, 37). A preference for AU-rich sequences shows also the ARID domain of Arid5a (26). The RRM domain of Auf1 recognizes U- and GU-rich stretches and, albeit less frequently, AREs (38). The RRM motif of HuR prefers U-rich sequences (9, 39, 40). The preference of these binding domains for AREs or U-rich sequences reflects the unstructured nature of such sequences: AREs and U-rich sequences in general do not adopt a secondary structure. In contrast, Regnase and Roquin bind to RNAs exhibiting stem-loop folds with the loop part formed by three bases with a pyrimidine–purine–pyrimidine sequence while the stem is more variable both in length (5 – 8 bases in each half of the stem) and sequence (41, 42).
Transcriptome-wide binding assays revealed that most of these RBPs bind frequently to 3’ UTR and, unexpectedly, introns (9, 21, 38–40). Binding to introns regulates splicing in case of HuR (40, 43). However, it appears that functional interactions are largely confined to elements located in the 3’ UTRs as intronic binding in general does not result in changes in stability or splicing of the transcript.
The TTP Protein Family: Evolutionary Conserved RBPs With Diverse Functions From Yeast to Mammals
TTP contains an RNA-binding domain formed by a characteristic tandem C3H1 zinc finger in the middle part and protein-protein interaction domains at the N- and C-termini (31). The tandem zinc finger and the overall domain structure are conserved in similar RBPs from yeast to plants and mammals hence these RBPs constitute the TTP protein family (44). Interestingly, no TTP protein members are found in birds despite their presence in reptiles (44). Although all these proteins facilitate mRNA degradation their functions in cells and/or organisms are diverse. For example, the yeast TTP family member Cth2 regulates mRNA stability upon iron deficiency while the Xenopus TTP proteins act during embryonic development and the C. elegans homologues are required for meiosis and oocyte production (45–47). Humans contain three TTP family members (Zfp36, Zfp36l1 and Zfp36l2) and mice express the Zfp36l3 member in addition. Much of what we now know about the functions of the TTP protein family has been learned from knockouts in mice carried out by the Blackshear laboratory. Deletion of Zfp36l1 (also known as BRF1 and TIS11b) is embryonic lethal because of failure in umbilical circulation resulting from absent fusion of the allantois with the chorion (18). Mice lacking Zfp36l2 (also known as BRF2 and TIS11D) die within a few weeks after birth due to a marked deficiency in hematopoiesis (19). Zfp36l3 is a paternally imprinted X chromosome gene which is likely involved in regulation of iron metabolism in the placenta; Zfp36l3 deletion results in decreased neonatal survival rates without obvious morphological aberrances in surviving offspring (48).
TTP: a TTP Family Member With Unique Selectivity for the Regulation of Immune Responses
TTP (Zfp36) is an outstanding member of the TTP family as its function is remarkably specific and related to the regulation of immune responses. TTP knockout in mice results in systemic inflammation characterized by arthritis, dermatitis, conjunctivitis and cachexia (49). This so called TTP deficiency syndrome develops within approximately 8 weeks of birth and progressively worsens leading to death of most animals at around 6-8 months of age. TTP-deficient mice do not show any developmental abnormalities or health defects at birth; the mice are not fertile presumably owing to their poor health (49). The TTP deficiency syndrome was shown to be dependent on TNF signaling and mechanistically explained by increased stability of Tnf mRNA (49, 50). Subsequent studies established that the inflammatory disease of TTP-deficient mice is caused, albeit to variable extent, by increased stability of other cytokine and chemokine mRNAs as well, notably Il23, Ccl3, Il1a and Il1b mRNAs (51–53).
Given this multiple evidence for its indispensable role in the immune system, it comes with no surprise that TTP has become one of the best studied RBPs. However, many important questions remain open. For example, it is not well understood which cell types drive the inflammatory disease in TTP-deficient mice. Mice bearing LysM-Cre-mediated TTP deletion in the myeloid compartment are healthy which is unexpected given that myeloid cells are cells with arguably the highest TTP expression (54, 55). Although these mice exhibit lethal hypersensitivity to endotoxic shock, the absence of a spontaneous inflammation suggests that deletion of TTP in myeloid cells alone is not sufficient to cause the TTP deficiency syndrome. Similarly, mice with CD11c-Cre-driven deletion of TTP in dendritic cells remain without a spontaneous phenotype (56). Surprisingly, systemic inflammation arises upon deletion of TTP in keratinocytes (56). The inflammatory disease in these mice develops from psoriasis-like focal skin lesions containing neutrophilic infiltrates, indicating that persistent local inflammation can become systemic with time. The model of keratinocyte-specific TTP deletion suggests that TTP expression is particularly critical in barrier tissues, i.e. tissues constantly exposed to environmental cues. However, it is remarkable that the full-body TTP knockout mice remain without pathology in the intestinal or lung epithelium, i.e. the most prominent mucosal barriers. The absence of mucosal inflammation in TTP-deficient mice suggests that TTP has more complex roles in these tissues. Such functional complexity is supported by findings showing that the lack of intestinal pathology in TTP knockout mice is associated with a local expansion of regulatory T cells (57). Moreover, Villin-Cre-driven TTP deletion in intestinal epithelial cells increases the resistance against dextran sulfate-induced colitis suggesting, that the lack of TTP might enhance the robustness of the intestinal barrier (58). Although the mechanism is yet to be determined, the improved mucosal barrier might be caused by accelerated tissue regeneration since these mice exhibit higher numbers of Goblet cells. These findings suggest that the absence of TTP augments proliferation signals that are commonly associated with inflammatory conditions. In agreement, skin inflammation caused by TTP deficiency in keratinocytes promotes tumorigenesis that appears to be causally associated with overproduction of the growth factor amphiregulin (59). Consistently, amphiregulin mRNA is a TTP target. However, TTP deficiency can cause increased cell numbers also by means of decreased apoptosis as shown for TTP-deficient neutrophils: neutrophils devoid of TTP express higher levels of the TTP target Mcl1 mRNA which codes for an anti-apoptotic factor particularly relevant for neutrophils (60). Interestingly, this effect pertains only to immunostimulated (e.g. pathogen-engaged) neutrophils, not to the circulating dormant neutrophil pool.
Cumulatively, the available animal models of TTP deficiency clearly indicate that the major function of TTP is to control the immune response. Although TTP restricts cell numbers in some cases, this function is also largely related to control of inflammation: (i) by ameliorating inflammation TTP prevents the expression of inflammation-associated growth factors or anti-apoptotic proteins, (ii) TTP directly targets the mRNAs of several inflammation-associated growth or anti-apoptotic factors. More studies directly investigating cells from tissues are needed to complete our understanding of TTP effects in vivo.
mRNA Destabilization by TTP
TTP promotes mRNA decay through the recruitment of the CCR4-NOT deadenylase and the DCP1/DCP2 decapping complexes to the bound target. The N- and C-termini of TTP represent the protein-protein interaction domains in this process. The CCR4-NOT deadenylase complex interacts with the N- and C-terminal domains with the CNOT1 subunit being directly involved in binding to TTP (61–64). The DCP1 and DCP2 decapping protein complexes interact with the N-terminal TTP domain (62). Following decapping and deadenylation, the target mRNA is degraded through the 5′-3′ exonuclease Xrn1 and the 3′-5′ exonuclease of the exosome, respectively. The identification of these interactions suggested that TTP-mediated mRNA degradation is governed by a protein recruitment cascade. However, this model does not explain why many TTP-bound RNAs (including mRNAs and introns) are stable as shown by more recent studies (9, 65–67). The surprising findings of these studies delineate that the process of mRNA destabilization by TTP is more complex and dependent on yet unidentified regulatory mechanisms.
TTP Binding to RNA
Both zinc finger domains are required for interaction of TTP with RNA as mutation of either of them abrogates RNA binding (68). Moreover, mutation of the first zinc finger in the TTP locus in mice phenocopied the complete TTP deletion (69). This was a significant finding as it definitively proved that the function of TTP is entirely dependent on its RNA binding activity. Initial characterization of the motif recognized by TTP focused on the TNF mRNA, the first known TTP target: the motif is a 9-mer with the sequence UUAUUUAUU which is repeated several times in the TNF 3’ UTR (37, 70). Subsequent analysis of RNAs enriched in RNA immunoprecipitation assays suggested that TTP binds to AREs also in other target mRNAs (71). A precise genome-wide mapping of target sequences was generated by several CLIP-Seq (cross-linking immunoprecipitation-high-throughput sequencing) studies employing immune cells. Although these nucleotide resolution analyses confirmed the preference of TTP for the UAUUUAU sequence, they also provided several unexpected findings (9, 65–67). The studies showed that TTP binds also to sites that were divergent from the canonical TTP binding sequence as visualized in the searchable TTP Atlas (https://ttp-atlas.univie.ac.at) (9). Moreover, TTP binding was not limited to 3’ UTRs but was detected at sites located in 5’ UTRs, coding sequences and introns as well (Figure 1). Particularly striking was the high incidence of TTP binding to introns. Although the number of identified intronic binding sites was dependent on the CLIP-Seq method, the peak finding algorithm and experimental cell system, the studies convincingly established that TTP interacts with pre-mRNA in addition to mRNA. This finding implies that TTP can engage RNA interactions in the nucleus. The biological significance of the intronic binding remains to be determined as no effects on splicing or stability of the intron-bound RNA has so far been observed (9). Given the high frequency of TTP binding to introns it is possible that introns act as sponge to titrate away TTP molecules. This mechanism was reported for circular RNAs that function as sponge molecules for micro RNAs (72, 73). Similar to intronic binding, it is currently unclear whether interactions of TTP with 5’ UTRs or coding sequences entail changes in RNA processing.
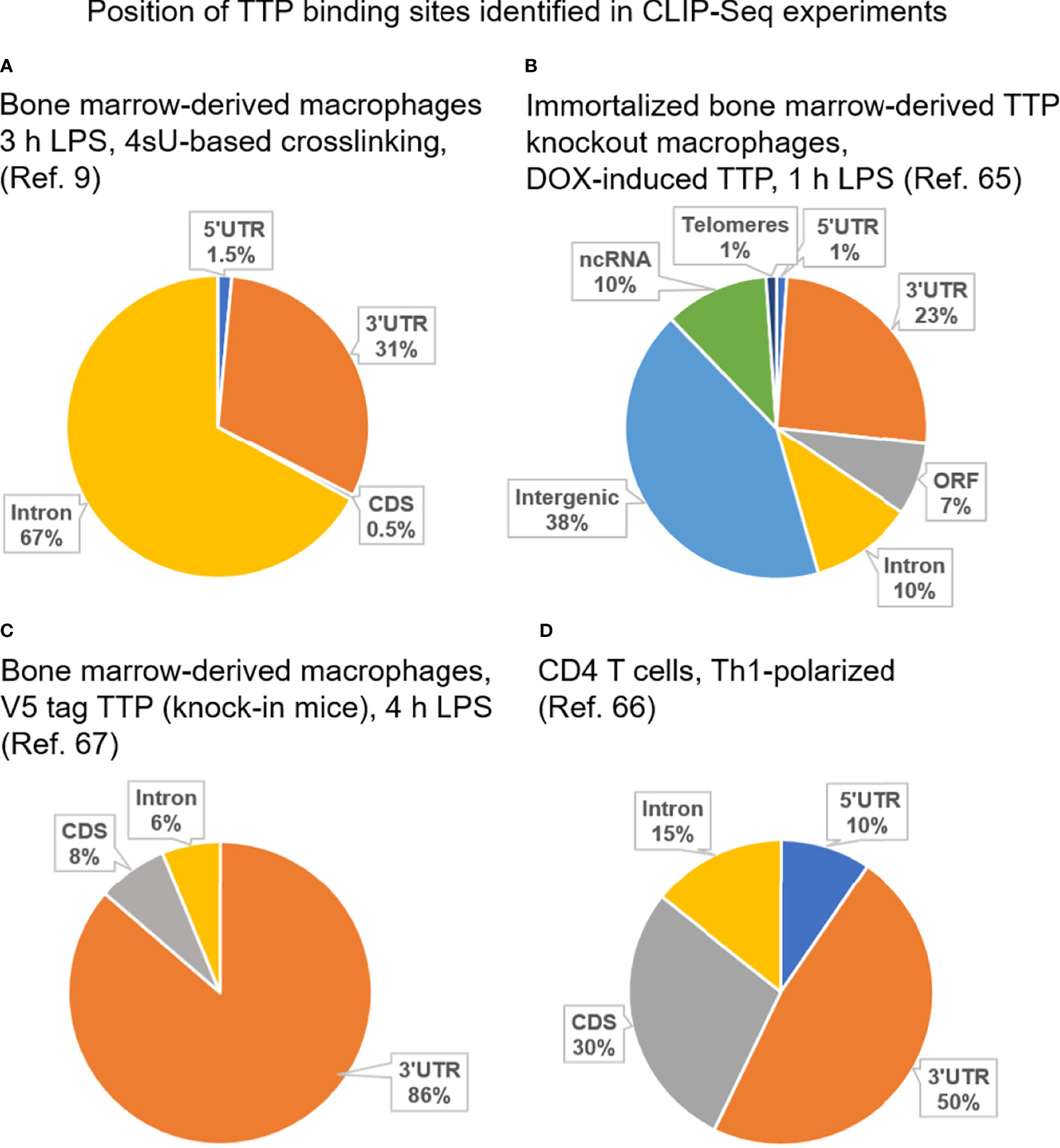
Figure 1 Position of TTP binding sites identified in reported CLIP-Seq experiments employing immune cells. (A) CLIP-Seq experiment carried out using bone marrow-derived macrophages isolated from wild type mice (i.e. expressing solely endogenous TTP). Cells were stimulated for 3 h with LPS prior to CLIP-Seq which was based on thiouridine (4sU)-mediated crosslinking allowing crosslinking with 365 nm UV, i.e. mild conditions [(9)]. (B) CLIP-Seq experiment performed using immortalized bone marrow-derived macrophages isolated from TTP knockout mice and engineered to express doxycycline-inducible TTP. Cells were treated with doxycycline and stimulated for 1 h with LPS prior to CLIP-Seq [(65)]. (C) CLIP-Seq experiment carried out using bone marrow-derived macrophages isolated from mice expressing V5-tagged TTP from the endogenous locus (knock-in mice). Cells were stimulated for 4 h with LPS prior to CLIP-Seq [(67)]. (D) CLIP-Seq experiment carried out using CD4+ T cells from wild type mice [(i.e. expressing solely endogenous TTP)]. CD4+ T cells were polarized under Th1 conditions prior to CLIP-Seq [(66)]. 3’ UTR, 3’ untranslated region; 5’ UTR, 5’ untranslated region; CDS, coding sequence; ORF, open reading frame; ncRNA, non-coding RNA; DOX, doxycycline.
The CLIP-Seq data show that functional TTP bindings sites are located in 3' UTR. Remarkably, binding of TTP to 3’ UTR does not always cause destabilization of the target mRNA, as revealed in recent studies. This enigmatic and probably significant property of TTP is discussed further below.
Regulation of TTP
TTP function is regulated in multiple ways with many of them remaining poorly understood. Moreover, it is likely that some key regulatory events are still not known. Comprehensive knowledge about the regulation of TTP is critical for our understanding of the remarkably selective function of TTP in the immune system and for the control of inflammation in general. TTP is regulated at the level of transcription, mRNA stability, protein stability and by posttranslational modifications (31, 74–76). As far as we can say, all these mechanisms are critical for the appropriate extent, timing and selectivity of TTP-driven mRNA degradation. They act in concert to allow the immune system launching an efficient but not exaggerated inflammatory response.
TTP mRNA levels are low under steady state conditions but dramatically induced in response to inflammatory stimuli which are mostly associated with stress signaling. The increase in TTP mRNA levels is achieved mostly by transcriptional induction and to some extent also through mRNA stabilization. As an immediate early gene, TTP is transcriptionally activated rapidly after stimulation. The activation signals include growth factors, cytokines such as TNF, IL-4, IL-10, or IFN-γ, and bacterial products e.g. LPS (50, 77–80). The transcription factors involved in the transcriptional upregulation were characterized in few instances: IFNs, IL-10 and IL-4 drive TTP expression through STAT1, STAT3 and STAT6, respectively (78–80). STAT1 employs a GAS (Gamma interferon activation site) element which is conserved in the TTP promoter in mice and humans (78). This GAS element is likely involved also in response to IL-4 and IL-10. The activating signals often synergize to achieve maximal induction of TTP (78, 79).
Stabilization of TTP mRNA by p38 MAPK signaling contributes to induction of TTP expression (81). TTP mRNA contains AREs which interact with TTP protein suggesting that autoregulation is the mechanism underlying the low TTP mRNA stability. TTP mRNA is indeed moderately more stable in TTP knock-in mice expressing the zinc finger-inactivated mutant (69).
A central aspect of the regulation of TTP levels in cells is the control of TTP protein stability. TTP is continuously degraded in a proteasome-dependent way which appears to proceed without ubiquitination and is likely to involve the intrinsically unfolded N- and/or C-terminal domains (82, 83). The mechanism of this important process is not resolved and its elucidation would significantly advance our understanding of protein degradation in general. TTP protein stability increases by orders of magnitude upon phosphorylation of S52 and S178 (in mouse coordinates) (82, 84). Phosphorylation of these two residues is brought about by MK2, a kinase that is activated by p38 MAPK. Although the p38 MAPK/MK2-driven phosphorylation of S52 and S178 increases TTP protein stability and thereby positively regulates TTP levels, it inhibits the mRNA-destabilization activity of TTP. This phosphorylation-dependent TTP inhibition probably results from a combination of several processes: (i) S52 and S178 phosphorylation causes association of TTP with 14-3-3 proteins thereby preventing relocation of TTP to stress granules and processing bodies, (ii) 14-3-3 protein binding promotes export of TTP from the nucleus, (iii) S52 and S178 phosphorylation decreases association of TTP with the CCR4-NOT deadenylase, and (iv) MK2-dependent TTP phosphorylation diminishes TTP binding to RNA (85–88). Although the mechanistic details of the function of S52 and S178 phosphorylation are not fully understood, the biological consequences have been convincingly revealed by generation of double knock-in mice bearing S52A and S178A mutations in the TTP locus (89). These mice are unable to express high TTP protein levels, consistent with a rapid TTP protein degradation. Nevertheless, the mice are protected against LPS-induced systemic inflammation indicating that the S52A/S178A mutant acts as hyperactive TTP in vivo. The double knock-in mouse confirmed the previously proposed model of TTP function according to which p38 MAPK leads to accumulation of inactive (i.e. phosphorylated) TTP in the initial phase of inflammation. Later, i.e. in the resolution phase of inflammation, the gradual decrease of p38 MAPK activity releases TTP from its inhibited state thereby facilitating degradation of TTP target mRNAs (9, 55). In parallel, the diminishing phosphorylation accelerates proteasomal degradation of TTP rendering the cells responsive to a new inflammatory stimulus.
TTP contains more than 30 phosphorylation sites out of which only S52 and S178 have been functionally annotated in cells and animals (90). A recent quest for a better understanding of TTP phosphorylation has revealed MK2-dependent phosphorylation of T84, S85, T250, and S316 out of which the phosphorylation of S316 is the most robust one (91). Notably, S316 phosphorylation in not involved in regulation of TTP protein stability; instead, it appears to regulate interactions of TTP with the translation inhibition proteins (91). It will be exciting to see the progress in functional characterization of other phosphorylation sites as they likely impinge on TTP in unexpected ways.
To Degrade or Not to Degrade the Bound mRNA?
Transcriptome-wide mRNA stability studies coupled to CLIP-Seq analyses revealed that, surprisingly, TTP does not always cause degradation of the bound target. This has been convincingly demonstrated by employing bone marrow-derived macrophages expressing solely endogenous TTP (9). The study showed that 71% of mRNAs bound by TTP in their 3’ UTR are stable. A similar conclusion was drawn from CLIP-Seq and mRNA stability assays in HEK293 cells overexpressing TTP (92). These observations were supported by other CLIP-Seq studies employing primary cells (i.e. cells not overexpressing TTP) although the evidence was indirect as it was based on differential expression analysis (RNA-Seq) but not on transcriptome-wide mRNA stability assessments (66, 67). These unexpected results indicate that a more complex model of TTP action needs to be developed. The model will probably involve proteins acting in cis with TTP which prevent recruitment of the RNA degradation machinery to stable transcripts or facilitate such recruitment to unstable transcripts (Figure 2). This new concept could also involve yet uncharacterized TTP phosphorylation events; in this scenario a particular phosphorylation (activating or inactivating) would occur only on certain target mRNAs and/or subcellular locations. The advanced concept of TTP function might also consider a recently reported hypothesis that TTP stabilizes mRNA under certain circumstances: The dramatic induction of TTP following an inflammatory stimulus was proposed to generate a pool of free TTP that sequesters the RNA degradation machinery thereby preventing mRNA decay, but a direct evidence for this hypothesis was not provided (93).
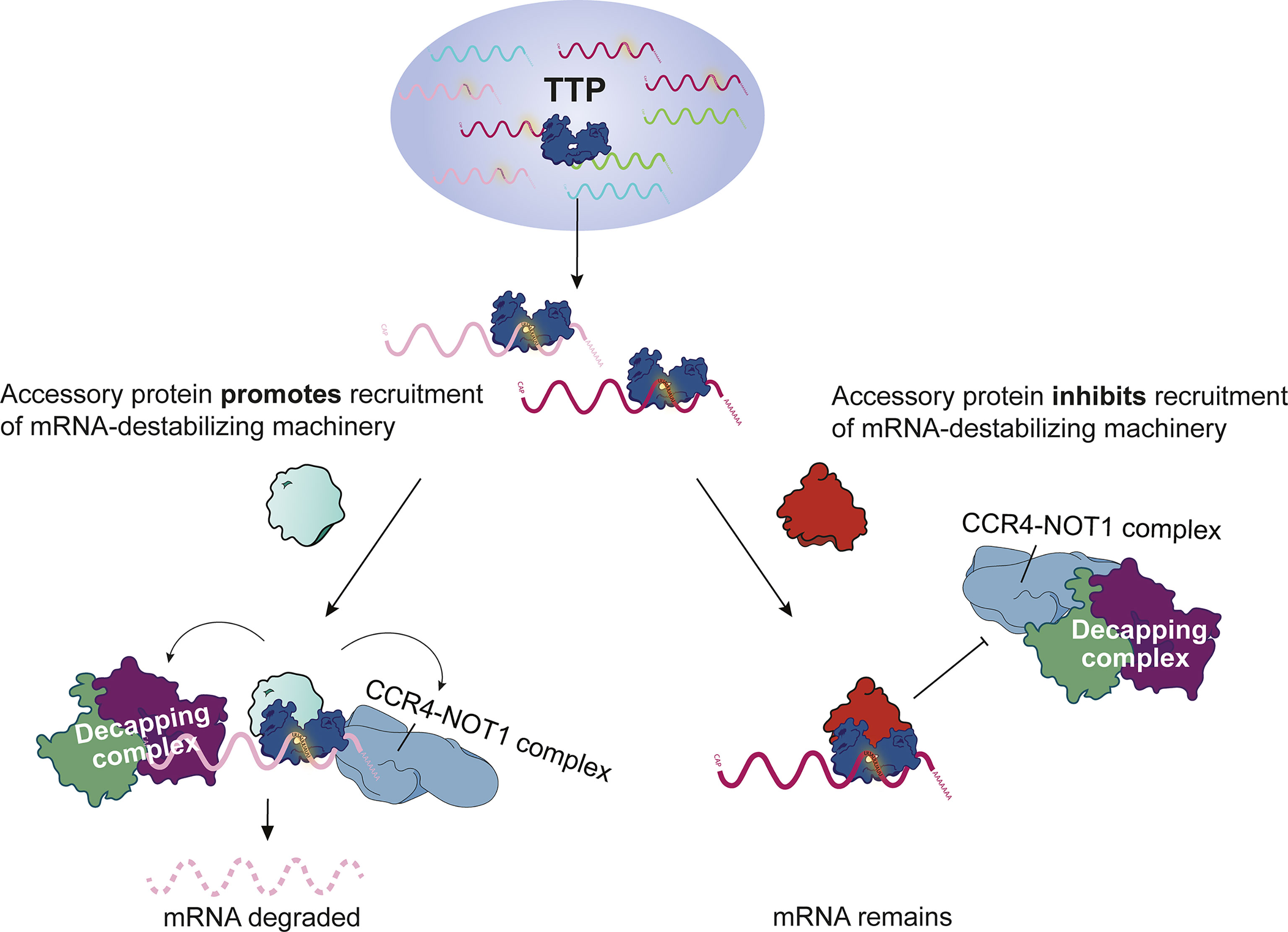
Figure 2 Model of functional and silent binding of TTP to mRNA. Model of possible mechanisms explaining how mRNA-bound TTP destabilizes some mRNAs but not others.
Related to the question of whether TTP destabilizes or not a specific subset of bound mRNAs are ribosome profiling data showing that TTP can affect mRNA stability but also inhibit translation (65–67). Particularly conclusive were studies employing primary macrophages and T cells, i.e. experimental systems expressing solely endogenous and naturally regulated TTP (66, 67). All these studies revealed transcript-selective effects of TTP on translation: some TTP target mRNAs were less abundant at polysomes while others were not depleted from polysomes. Negative regulation of translation has been also implicated in control of inflammatory gene expression in tumor-associated macrophages (94). All these data are consistent with an updated model of TTP function in which cis-acting and transcript-specific RBPs determine the final consequence of TTP binding to the target mRNA (Figure 2). This updated model is supported by the finding that TTP binds to the cytoplasmic poly(A)-binding protein and that such interactions are required for inhibition of translation by TTP in primary macrophages (67). The model will become more complex once data on tissue/cell type-specific functions of TTP are included. First data on biologically relevant cell type-specific effects of TTP have only recently become available: The mRNA coding for the IL-1β cytokine (Il1b mRNA) is destabilized by TTP in bone marrow-derived dendritic cells but not in bone marrow-derived macrophages despite strong binding to TTP (53). The regulation of Il1b expression by TTP is important in vivo as TTP-deficient mice show higher Il1b mRNA levels in several tissues. Moreover, genetic inactivation of IL-1 signaling in TTP-deficient animals ameliorates the TTP deficiency syndrome (53).
The extent of cell type-specific regulation of TTP activity can be indirectly estimated from a number of RNA-seq studies comparing mRNA levels in wild-type versus TTP knockout cells. For example, the levels of Tnf mRNA, the bona fide TTP target, are comparable in wild-type and TTP-deficient T cells, suggesting that TTP does not target Tnf mRNA for degradation in T cells in contrast to most other cell types (66). Similarly, Il6 mRNA, which is known to be bound and destabilized by TTP in bone marrow-derived macrophages (BMDMs) (55), is more highly expressed also in TTP-deficient dendritic cells (upon 3 h or 6 h LPS stimulation) and T cells (upon 4 h activation) but not in peritoneal neutrophils (53, 60). Reported RNA-seq expression data for selected TTP targets in primary immune cells (BMDMs, BMDCs, peritoneal neutrophils and T cells) from wild-type and TTP-deficient mice are summarized in Table 1. These data convincingly visualize that a comparison of mRNA levels does not provide a definitive information about the destabilization of a particular mRNA by TTP since indirect (possibly cell type-specific) effects of transcription can mask differences in mRNA stability. A good example is Il6 mRNA in T cells: while a short (4 h) activation results in 50% higher Il6 mRNA levels in TTP-deficient T cells as compared to controls, consistent with Il6 mRNA being destabilized by TTP, a 3-day activation causes TTP-deficient T cells to express 15% less Il6 mRNA than the control cells [Table 1 and (66)]. Thus, mRNA stability assays combined, whenever possible, with TTP binding analyses are required when defining a TTP target in a given cell type. A combination of transcriptome-wide mRNA stability and TTP binding assays has been so far reported only for BMDMs so that a comprehensive TTP target collection (searchable at https://ttp-atlas.univie.ac.at/) is available only for this cell type (9). Future studies should include transcriptome-wide mRNA stability analyses in other cell types.
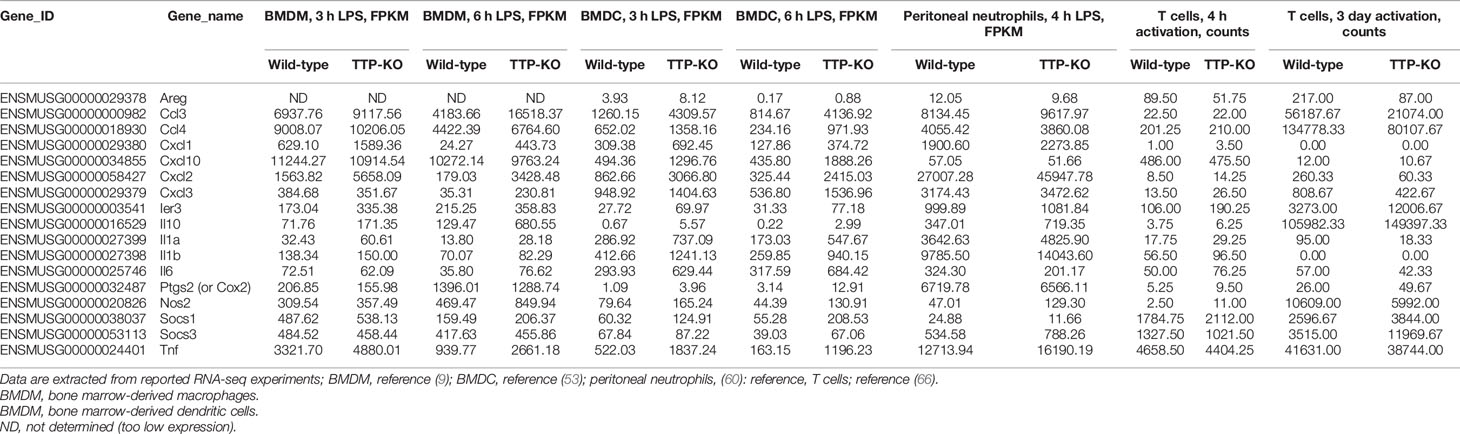
Table 1 Expression data (FPKMs or counts) of selected TTP targets in different immune cell types from wild type and TTP-KO mice.
In summary, these findings implicate that cell type-specific RBPs act together with TTP to stabilize or destabilize select TTP targets. Comprehensive biochemical studies including reconstitution assays are needed to precisely determine the underlying mechanisms. The results of these studies will be relevant for the entire TTP family, as the functional versus silent binding to target mRNA is important also for Zfp36l1 and Zfp36l2 (21, 95).
Outlook
Despite more than 25 years of research, TTP continues to represent an important and fruitful model for studies on RBPs in general and on the regulation of immune responses by RBPs in particular. Technological progress in recent years and advanced animal models were instrumental for the identification of novel regulatory facets and functional consequences of TTP which fundamentally improved our understating of physiological and pathological inflammation. Most of these new findings remain mechanistically poorly defined and represent challenging topics for future research. This will include analyses of TTP-containing protein complexes and yet uncharacterized phosphorylation sites which will help addressing the mechanism of functional versus silent binding of TTP to RNA. An underexplored area are tissue- and cell type-specific functions of TTP in vivo to answer the still incompletely understood phenotype of TTP-deficient mice. Finally, an attractive avenue is the exploitation of TTP and mRNA decay in therapy of inflammatory diseases and cancer.
Author Contributions
PK designed the concept and wrote the manuscript. AB wrote the manuscript and prepared figures. JF prepared figures. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Austrian Science Fund (FWF) grants P33000-B, P31848-B and W1261 to PK.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Turner M, Diaz-Munoz MD. RNA-Binding Proteins Control Gene Expression and Cell Fate in the Immune System. Nat Immunol (2018) 19:120–9. doi: 10.1038/s41590-017-0028-4
2. Akira S, Maeda K. Control of RNA Stability in Immunity. Annu Rev Immunol (2021) 39:481–509. doi: 10.1146/annurev-immunol-101819-075147
3. Carpenter S, Ricci EP, Mercier BC, Moore MJ, Fitzgerald KA. Post-Transcriptional Regulation of Gene Expression in Innate Immunity. Nat Rev (2014) 14:361–76. doi: 10.1038/nri3682
4. Shaw G, Kamen R. A Conserved AU Sequence From the 3’ Untranslated Region of GM-CSF mRNA Mediates Selective mRNA Degradation. Cell (1986) 46:659–67. doi: 10.1016/0092-8674(86)90341-7
5. Kontoyiannis D, Pasparakis M, Pizarro TT, Cominelli F, Kollias G. Impaired on/Off Regulation of TNF Biosynthesis in Mice Lacking TNF AU-Rich Elements: Implications for Joint and Gut-Associated Immunopathologies. Immunity (1999) 10:387–98. doi: 10.1016/S1074-7613(00)80038-2
6. Yang E, van Nimwegen E, Zavolan M, Rajewsky N, Schroeder M, Magnasco M, et al. Decay Rates of Human mRNAs: Correlation With Functional Characteristics and Sequence Attributes. Genome Res (2003) 13:1863–72. doi: 10.1101/gr.1272403
7. Bakheet T, Hitti E, Al-Saif M, Moghrabi WN, Khabar KSA. The AU-Rich Element Landscape Across Human Transcriptome Reveals a Large Proportion in Introns and Regulation by ELAVL1/HuR. Biochim Biophys Acta Gene Regul Mech (2018) 1861:167–77. doi: 10.1016/j.bbagrm.2017.12.006
8. Hao S, Baltimore D. The Stability of mRNA Influences the Temporal Order of the Induction of Genes Encoding Inflammatory Molecules. Nat Immunol (2009) 10:281–8. doi: 10.1038/ni.1699
9. Sedlyarov V, Fallmann J, Ebner F, Huemer J, Sneezum L, Ivin M, et al. Tristetraprolin Binding Site Atlas in the Macrophage Transcriptome Reveals a Switch for Inflammation Resolution. Mol Syst Biol (2016) 12:868. doi: 10.15252/msb.20156628
10. Diaz-Munoz MD, Turner M. Uncovering the Role of RNA-Binding Proteins in Gene Expression in the Immune System. Front Immunol (2018) 9:1094. doi: 10.3389/fimmu.2018.01094
11. Yoshinaga M, Takeuchi O. RNA Binding Proteins in the Control of Autoimmune Diseases. Immunol Med (2019) 42:53–64. doi: 10.1080/25785826.2019.1655192
12. Labno A, Tomecki R, Dziembowski A. Cytoplasmic RNA Decay Pathways - Enzymes and Mechanisms. Biochim Biophys Acta (2016) 1863:3125–47. doi: 10.1016/j.bbamcr.2016.09.023
13. Zinder JC, Lima CD. Targeting RNA for Processing or Destruction by the Eukaryotic RNA Exosome and its Cofactors. Genes Dev (2017) 31:88–100. doi: 10.1101/gad.294769.116
14. Uehata T, Iwasaki H, Vandenbon A, Matsushita K, Hernandez-Cuellar E, Kuniyoshi K, et al. Malt1-Induced Cleavage of Regnase-1 in CD4(+) Helper T Cells Regulates Immune Activation. Cell (2013) 153:1036–49. doi: 10.1016/j.cell.2013.04.034
15. Collart MA. The Ccr4-Not Complex is a Key Regulator of Eukaryotic Gene Expression. Wiley Interdiscip Rev RNA (2016) 7:438–54. doi: 10.1002/wrna.1332
16. Grudzien-Nogalska E, Kiledjian M. New Insights Into Decapping Enzymes and Selective mRNA Decay. Wiley Interdiscip Rev RNA (2017) 8:e1379. doi: 10.1002/wrna.1379
17. Vogel KU, Edelmann SL, Jeltsch KM, Bertossi A, Heger K, Heinz GA, et al. Roquin Paralogs 1 and 2 Redundantly Repress the Icos and Ox40 Costimulator mRNAs and Control Follicular Helper T Cell Differentiation. Immunity (2013) 38:655–68. doi: 10.1016/j.immuni.2012.12.004
18. Stumpo DJ, Byrd NA, Phillips RS, Ghosh S, Maronpot RR, Castranio T, et al. Chorioallantoic Fusion Defects and Embryonic Lethality Resulting From Disruption of Zfp36L1, a Gene Encoding a CCCH Tandem Zinc Finger Protein of the Tristetraprolin Family. Mol Cell Biol (2004) 24:6445–55. doi: 10.1128/MCB.24.14.6445-6455.2004
19. Stumpo DJ, Broxmeyer HE, Ward T, Cooper S, Hangoc G, Chung YJ, et al. Targeted Disruption of Zfp36l2, Encoding a CCCH Tandem Zinc Finger RNA-Binding Protein, Results in Defective Hematopoiesis. Blood (2009) 114:2401–10. doi: 10.1182/blood-2009-04-214619
20. Hodson DJ, Janas ML, Galloway A, Bell SE, Andrews S, Li CM, et al. Deletion of the RNA-Binding Proteins ZFP36L1 and ZFP36L2 Leads to Perturbed Thymic Development and T Lymphoblastic Leukemia. Nat Immunol (2010) 11:717–24. doi: 10.1038/ni.1901
21. Galloway A, Saveliev A, Lukasiak S, Hodson DJ, Bolland D, Balmanno K, et al. RNA-Binding Proteins ZFP36L1 and ZFP36L2 Promote Cell Quiescence. Science (2016) 352:453–9. doi: 10.1126/science.aad5978
22. Lu JY, Sadri N, Schneider RJ. Endotoxic Shock in AUF1 Knockout Mice Mediated by Failure to Degrade Proinflammatory Cytokine mRNAs. Genes Dev (2006) 20:3174–84. doi: 10.1101/gad.1467606
23. Abbadi D, Yang M, Chenette DM, Andrews JJ, Schneider RJ. Muscle Development and Regeneration Controlled by AUF1-Mediated Stage-Specific Degradation of Fate-Determining Checkpoint mRNAs. Proc Natl Acad Sci USA (2019) 116:11285–90. doi: 10.1073/pnas.1901165116
24. Pont AR, Sadri N, Hsiao SJ, Smith S, Schneider RJ. mRNA Decay Factor AUF1 Maintains Normal Aging, Telomere Maintenance, and Suppression of Senescence by Activation of Telomerase Transcription. Mol Cell (2012) 47:5–15. doi: 10.1016/j.molcel.2012.04.019
25. Chen J, Cascio J, Magee JD, Techasintana P, Gubin MM, Dahm GM, et al. Posttranscriptional Gene Regulation of IL-17 by the RNA-Binding Protein HuR Is Required for Initiation of Experimental Autoimmune Encephalomyelitis. J Immunol (2013) 191:5441–50. doi: 10.4049/jimmunol.1301188
26. Masuda K, Ripley B, Nishimura R, Mino T, Takeuchi O, Shioi G, et al. Arid5a Controls IL-6 mRNA Stability, Which Contributes to Elevation of IL-6 Level In Vivo. Proc Natl Acad Sci U.S.A. (2013) 110:9409–14. doi: 10.1073/pnas.1307419110
27. Diaz-Munoz MD, Bell SE, Fairfax K, Monzon-Casanova E, Cunningham AF, Gonzalez-Porta M, et al. The RNA-Binding Protein HuR Is Essential for the B Cell Antibody Response. Nat Immunol (2015) 16:415–25. doi: 10.1038/ni.3115
28. Zhang Z, Zong C, Jiang M, Hu H, Cheng X, Ni J, et al. Hepatic HuR Modulates Lipid Homeostasis in Response to High-Fat Diet. Nat Commun (2020) 11:3067. doi: 10.1038/s41467-020-16918-x
29. Katsanou V, Milatos S, Yiakouvaki A, Sgantzis N, Kotsoni A, Alexiou M, et al. The RNA-Binding Protein Elavl1/HuR is Essential for Placental Branching Morphogenesis and Embryonic Development. Mol Cell Biol (2009) 29:2762–76. doi: 10.1128/MCB.01393-08
30. Lang M, Berry D, Passecker K, Mesteri I, Bhuju S, Ebner F, et al. HuR Small-Molecule Inhibitor Elicits Differential Effects in Adenomatosis Polyposis and Colorectal Carcinogenesis. Cancer Res (2017) 77:2424–38. doi: 10.1158/0008-5472.CAN-15-1726
31. Wells ML, Perera L, Blackshear PJ. An Ancient Family of RNA-Binding Proteins: Still Important! Trends Biochem Sci (2017) 42(4):285–96. doi: 10.1016/j.tibs.2016.12.003
32. Tan D, Zhou M, Kiledjian M, Tong L. The ROQ Domain of Roquin Recognizes mRNA Constitutive-Decay Element and Double-Stranded RNA. Nat Struct Mol Biol (2014) 21:679–85. doi: 10.1038/nsmb.2857
33. Yokogawa M, Tsushima T, Noda NN, Kumeta H, Enokizono Y, Yamashita K, et al. Structural Basis for the Regulation of Enzymatic Activity of Regnase-1 by Domain-Domain Interactions. Sci Rep (2016) 6:22324. doi: 10.1038/srep22324
34. Wagner BJ, DeMaria CT, Sun Y, Wilson GM, Brewer G. Structure and Genomic Organization of the Human AUF1 Gene: Alternative pre-mRNA Splicing Generates Four Protein Isoforms. Genomics (1998) 48:195–202. doi: 10.1006/geno.1997.5142
35. Pabis M, Popowicz GM, Stehle R, Fernandez-Ramos D, Asami S, Warner L, et al. HuR Biological Function Involves RRM3-Mediated Dimerization and RNA Binding by All Three RRMs. Nucleic Acids Res (2019) 47:1011–29. doi: 10.1093/nar/gky1138
36. Iwahara J, Clubb RT. Solution Structure of the DNA Binding Domain From Dead Ringer, a Sequence-Specific AT-Rich Interaction Domain (ARID). EMBO J (1999) 18:6084–94. doi: 10.1093/emboj/18.21.6084
37. Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS, Blackshear PJ. Evidence That Tristetraprolin Binds to AU-Rich Elements and Promotes the Deadenylation and Destabilization of Tumor Necrosis Factor Alpha mRNA. Mol Cell Biol (1999) 19:4311–23. doi: 10.1128/MCB.19.6.4311
38. Yoon JH, De S, Srikantan S, Abdelmohsen K, Grammatikakis I, Kim J, et al. PAR-CLIP Analysis Uncovers AUF1 Impact on Target RNA Fate and Genome Integrity. Nat Commun (2014) 5:5248. doi: 10.1038/ncomms6248
39. Lebedeva S, Jens M, Theil K, Schwanhausser B, Selbach M, Landthaler M, et al. Transcriptome-Wide Analysis of Regulatory Interactions of the RNA-Binding Protein HuR. Mol Cell (2011) 43:340–52. doi: 10.1016/j.molcel.2011.06.008
40. Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, et al. Integrative Regulatory Mapping Indicates That the RNA-Binding Protein HuR Couples pre-mRNA Processing and mRNA Stability. Mol Cell (2011) 43:327–39. doi: 10.1016/j.molcel.2011.06.007
41. Leppek K, Schott J, Reitter S, Poetz F, Hammond MC, Stoecklin G. Roquin Promotes Constitutive mRNA Decay via a Conserved Class of Stem-Loop Recognition Motifs. Cell (2013) 153:869–81. doi: 10.1016/j.cell.2013.04.016
42. Mino T, Murakawa Y, Fukao A, Vandenbon A, Wessels HH, Ori D, et al. Regnase-1 and Roquin Regulate a Common Element in Inflammatory mRNAs by Spatiotemporally Distinct Mechanisms. Cell (2015) 161:1058–73. doi: 10.1016/j.cell.2015.04.029
43. Chang SH, Elemento O, Zhang J, Zhuang ZW, Simons M, Hla T. ELAVL1 Regulates Alternative Splicing of Eif4e Transporter to Promote Postnatal Angiogenesis. Proc Natl Acad Sci USA (2014) 111:18309–14. doi: 10.1073/pnas.1412172111
44. Blackshear PJ, Perera L. Phylogenetic Distribution and Evolution of the Linked RNA-Binding and NOT1-Binding Domains in the Tristetraprolin Family of Tandem CCCH Zinc Finger Proteins. J Interferon Cytokine Res (2014) 34:297–306. doi: 10.1089/jir.2013.0150
45. Puig S, Askeland E, Thiele DJ. Coordinated Remodeling of Cellular Metabolism During Iron Deficiency Through Targeted mRNA Degradation. Cell (2005) 120:99–110. doi: 10.1016/j.cell.2004.11.032
46. Treguer K, Faucheux C, Veschambre P, Fedou S, Theze N, Thiebaud P. Comparative Functional Analysis of ZFP36 Genes During Xenopus Development. PloS One (2013) 8:e54550. doi: 10.1371/journal.pone.0054550
47. Kaymak E, Ryder SP. RNA Recognition by the Caenorhabditis Elegans Oocyte Maturation Determinant OMA-1. J Biol Chem (2013) 288:30463–72. doi: 10.1074/jbc.M113.496547
48. Stumpo DJ, Trempus CS, Tucker CJ, Huang W, Li L, Kluckman K, et al. Deficiency of the Placenta- and Yolk Sac-Specific Tristetraprolin Family Member ZFP36L3 Identifies Likely mRNA Targets and an Unexpected Link to Placental Iron Metabolism. Development (2016) 143:1424–33. doi: 10.1242/dev.130369
49. Taylor GA, Carballo E, Lee DM, Lai WS, Thompson MJ, Patel DD, et al. A Pathogenetic Role for TNF Alpha in the Syndrome of Cachexia, Arthritis, and Autoimmunity Resulting From Tristetraprolin (TTP) Deficiency. Immunity (1996) 4:445–54. doi: 10.1016/S1074-7613(00)80411-2
50. Carballo E, Lai WS, Blackshear PJ. Feedback Inhibition of Macrophage Tumor Necrosis Factor-Alpha Production by Tristetraprolin. Science (1998) 281:1001–5. doi: 10.1126/science.281.5379.1001
51. Molle C, Zhang T, Ysebrant de Lendonck L, Gueydan C, Andrianne M, Sherer F, et al. Tristetraprolin Regulation of Interleukin 23 mRNA Stability Prevents a Spontaneous Inflammatory Disease. J Exp Med (2013) 210:1675–84. doi: 10.1084/jem.20120707
52. Kang JG, Amar MJ, Remaley AT, Kwon J, Blackshear PJ, Wang PY, et al. Zinc Finger Protein Tristetraprolin Interacts With CCL3 mRNA and Regulates Tissue Inflammation. J Immunol (2011) 187:2696–701. doi: 10.4049/jimmunol.1101149
53. Sneezum L, Eislmayr K, Dworak H, Sedlyarov V, Le Heron A, Ebner F, et al. Context-Dependent IL-1 mRNA-Destabilization by TTP Prevents Dysregulation of Immune Homeostasis Under Steady State Conditions. Front Immunol (2020) 11:1398. doi: 10.3389/fimmu.2020.01398
54. Qiu LQ, Stumpo DJ, Blackshear PJ. Myeloid-Specific Tristetraprolin Deficiency in Mice Results in Extreme Lipopolysaccharide Sensitivity in an Otherwise Minimal Phenotype. J Immunol (2012) 188:5150–9. doi: 10.4049/jimmunol.1103700
55. Kratochvill F, Machacek C, Vogl C, Ebner F, Sedlyarov V, Gruber AR, et al. Tristetraprolin-Driven Regulatory Circuit Controls Quality and Timing of mRNA Decay in Inflammation. Mol Syst Biol (2011) 7:560. doi: 10.1038/msb.2011.93
56. Andrianne M, Assabban A, La C, Mogilenko D, Salle DS, Fleury S, et al. Tristetraprolin Expression by Keratinocytes Controls Local and Systemic Inflammation. JCI Insight (2017) 2(11):e92979. doi: 10.1172/jci.insight.92979
57. La C, de Toeuf B, Bindels LB, Van Maele L, Assabban A, Melchior M, et al. The RNA-Binding Protein Tristetraprolin Regulates RALDH2 Expression by Intestinal Dendritic Cells and Controls Local Treg Homeostasis. Mucosal Immunol (2021) 14(1):80–91. doi: 10.1038/s41385-020-0302-x
58. Eshelman MA, Matthews SM, Schleicher EM, Fleeman RM, Kawasawa YI, Stumpo DJ, et al. Tristetraprolin Targets Nos2 Expression in the Colonic Epithelium. Sci Rep (2019) 9:14413. doi: 10.1038/s41598-019-50957-9
59. Assabban A, Dubois-Vedrenne I, Van Maele L, Salcedo R, Snyder BL, Zhou L, et al. Tristetraprolin Expression by Keratinocytes Protects Against Skin Carcinogenesis. JCI Insight (2021) 6. doi: 10.1172/jci.insight.140669
60. Ebner F, Sedlyarov V, Tasciyan S, Ivin M, Kratochvill F, Gratz N, et al. The RNA-Binding Protein Tristetraprolin Schedules Apoptosis of Pathogen-Engaged Neutrophils During Bacterial Infection. J Clin Invest (2017) 127:2051–65. doi: 10.1172/JCI80631
61. Lai WS, Kennington EA, Blackshear PJ. Tristetraprolin and its Family Members can Promote the Cell-Free Deadenylation of AU-Rich Element-Containing mRNAs by Poly(A) Ribonuclease. Mol Cell Biol (2003) 23:3798–812. doi: 10.1128/MCB.23.11.3798-3812.2003
62. Lykke-Andersen J, Wagner E. Recruitment and Activation of mRNA Decay Enzymes by Two ARE-Mediated Decay Activation Domains in the Proteins TTP and BRF-1. Genes Dev (2005) 19:351–61. doi: 10.1101/gad.1282305
63. Sandler H, Kreth J, Timmers HT, Stoecklin G. Not1 Mediates Recruitment of the Deadenylase Caf1 to mRNAs Targeted for Degradation by Tristetraprolin. Nucleic Acids Res (2011) 39:4373–86. doi: 10.1093/nar/gkr011
64. Fabian MR, Frank F, Rouya C, Siddiqui N, Lai WS, Karetnikov A, et al. Structural Basis for the Recruitment of the Human CCR4-NOT Deadenylase Complex by Tristetraprolin. Nat Struct Mol Biol (2013) 20:735–9. doi: 10.1038/nsmb.2572
65. Tiedje C, Diaz-Munoz MD, Trulley P, Ahlfors H, Laass K, Blackshear PJ, et al. The RNA-Binding Protein TTP is a Global Post-Transcriptional Regulator of Feedback Control in Inflammation. Nucleic Acids Res (2016) 44:7418–40. doi: 10.1093/nar/gkw474
66. Moore MJ, Blachere NE, Fak JJ, Park CY, Sawicka K, Parveen S, et al. ZFP36 RNA-Binding Proteins Restrain T Cell Activation and Anti-Viral Immunity. eLife (2018) 7. doi: 10.7554/eLife.33057
67. Zhang X, Chen X, Liu Q, Zhang S, Hu W. Translation Repression via Modulation of the Cytoplasmic Poly(A)-Binding Protein in the Inflammatory Response. eLife (2017) 6. doi: 10.7554/eLife.27786
68. Lai WS, Kennington EA, Blackshear PJ. Interactions of CCCH Zinc Finger Proteins With mRNA: non-Binding Tristetraprolin Mutants Exert an Inhibitory Effect on Degradation of AU-Rich Element-Containing mRNAs. J Biol Chem (2002) 277:9606–13. doi: 10.1074/jbc.M110395200
69. Lai WS, Stumpo DJ, Qiu L, Faccio R, Blackshear PJ. A Knock-In Tristetraprolin (TTP) Zinc Finger Point Mutation in Mice: Comparison With Complete TTP Deficiency. Mol Cell Biol (2018) 38(4):e00488–17. doi: 10.1128/MCB.00488-17
70. Brewer BY, Malicka J, Blackshear PJ, Wilson GM. RNA Sequence Elements Required for High Affinity Binding by the Zinc Finger Domain of Tristetraprolin: Conformational Changes Coupled to the Bipartite Nature of Au-Rich MRNA-Destabilizing Motifs. J Biol Chem (2004) 279:27870–7. doi: 10.1074/jbc.M402551200
71. Stoecklin G, Tenenbaum SA, Mayo T, Chittur SV, George AD, Baroni TE, et al. Genome-Wide Analysis Identifies Interleukin-10 mRNA as Target of Tristetraprolin. J Biol Chem (2008) 283:11689–99. doi: 10.1074/jbc.M709657200
72. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs Are a Large Class of Animal RNAs With Regulatory Potency. Nature (2013) 495(7441):333–8. doi: 10.1038/nature11928
73. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA Circles Function as Efficient microRNA Sponges. Nature (2013) 495(7441):384–8. doi: 10.1038/nature11993
74. Sandler H, Stoecklin G. Control of mRNA Decay by Phosphorylation of Tristetraprolin. Biochem Soc Trans (2008) 36:491–6. doi: 10.1042/BST0360491
75. Kovarik P, Ebner F, Sedlyarov V. Posttranscriptional Regulation of Cytokine Expression. Cytokine (2017) 89:21–6. doi: 10.1016/j.cyto.2015.11.007
76. O’Neil JD, Ammit AJ, Clark AR. MAPK P38 Regulates Inflammatory Gene Expression via Tristetraprolin: Doing Good by Stealth. Int J Biochem Cell Biol (2018) 94:6–9. doi: 10.1016/j.biocel.2017.11.003
77. Lai WS, Thompson MJ, Taylor GA, Liu Y, Blackshear PJ. Promoter Analysis of Zfp-36, the Mitogen-Inducible Gene Encoding the Zinc Finger Protein Tristetraprolin. J Biol Chem (1995) 270:25266–72. doi: 10.1074/jbc.270.42.25266
78. Sauer I, Schaljo B, Vogl C, Gattermeier I, Kolbe T, Muller M, et al. Interferons Limit Inflammatory Responses by Induction of Tristetraprolin. Blood (2006) 107:4790–7. doi: 10.1182/blood-2005-07-3058
79. Schaljo B, Kratochvill F, Gratz N, Sadzak I, Sauer I, Hammer M, et al. Tristetraprolin is Required for Full Anti-Inflammatory Response of Murine Macrophages to IL-10. J Immunol (2009) 183:1197–206. doi: 10.4049/jimmunol.0803883
80. Suzuki K, Nakajima H, Ikeda K, Maezawa Y, Suto A, Takatori H, et al. IL-4-Stat6 Signaling Induces Tristetraprolin Expression and Inhibits TNF-Alpha Production in Mast Cells. J Exp Med (2003) 198:1717–27. doi: 10.1084/jem.20031701
81. Tchen CR, Brook M, Saklatvala J, Clark AR. The Stability of Tristetraprolin mRNA is Regulated by Mitogen-Activated Protein Kinase P38 and by Tristetraprolin Itself. J Biol Chem (2004) 279:32393–400. doi: 10.1074/jbc.M402059200
82. Brook M, Tchen CR, Santalucia T, McIlrath J, Arthur JS, Saklatvala J, et al. Posttranslational Regulation of Tristetraprolin Subcellular Localization and Protein Stability by P38 Mitogen-Activated Protein Kinase and Extracellular Signal-Regulated Kinase Pathways. Mol Cell Biol (2006) 26:2408–18. doi: 10.1128/MCB.26.6.2408-2418.2006
83. Ngoc LV, Wauquier C, Soin R, Bousbata S, Twyffels L, Kruys V, et al. Rapid Proteasomal Degradation of Posttranscriptional Regulators of the TIS11/tristetraprolin Family is Induced by an Intrinsically Unstructured Region Independently of Ubiquitination. Mol Cell Biol (2014) 34:4315–28. doi: 10.1128/MCB.00643-14
84. Hitti E, Iakovleva T, Brook M, Deppenmeier S, Gruber AD, Radzioch D, et al. Mitogen-Activated Protein Kinase-Activated Protein Kinase 2 Regulates Tumor Necrosis Factor mRNA Stability and Translation Mainly by Altering Tristetraprolin Expression, Stability, and Binding to Adenine/Uridine-Rich Element. Mol Cell Biol (2006) 26:2399–407. doi: 10.1128/MCB.26.6.2399-2407.2006
85. Clement SL, Scheckel C, Stoecklin G, Lykke-Andersen J. Phosphorylation of Tristetraprolin by MK2 Impairs AU-Rich Element mRNA Decay by Preventing Deadenylase Recruitment. Mol Cell Biol (2011) 31:256–66. doi: 10.1128/MCB.00717-10
86. Stoecklin G, Stubbs T, Kedersha N, Wax S, Rigby WF, Blackwell TK, et al. MK2-Induced Tristetraprolin:14-3-3 Complexes Prevent Stress Granule Association and ARE-mRNA Decay. EMBO J (2004) 23:1313–24. doi: 10.1038/sj.emboj.7600163
87. Johnson BA, Blackwell TK. Multiple Tristetraprolin Sequence Domains Required to Induce Apoptosis and Modulate Responses to TNFalpha Through Distinct Pathways. Oncogene (2002) 21:4237–46. doi: 10.1038/sj.onc.1205526
88. Tiedje C, Ronkina N, Tehrani M, Dhamija S, Laass K, Holtmann H, et al. The P38/MK2-Driven Exchange Between Tristetraprolin and HuR Regulates AU-Rich Element-Dependent Translation. PloS Genet (2012) 8:e1002977. doi: 10.1371/journal.pgen.1002977
89. Ross EA, Smallie T, Ding Q, O’Neil JD, Cunliffe HE, Tang T, et al. Dominant Suppression of Inflammation via Targeted Mutation of the mRNA Destabilizing Protein Tristetraprolin. J Immunol (2015) 195:265–76. doi: 10.4049/jimmunol.1402826
90. Clark AR, Dean JL. The Control of Inflammation via the Phosphorylation and Dephosphorylation of Tristetraprolin: A Tale of Two Phosphatases. Biochem Soc Trans (2016) 44:1321–37. doi: 10.1042/BST20160166
91. Ronkina N, Shushakova N, Tiedje C, Yakovleva T, Tollenaere MAX, Scott A, et al. The Role of TTP Phosphorylation in the Regulation of Inflammatory Cytokine Production by MK2/3. J Immunol (2019) 203:2291–300. doi: 10.4049/jimmunol.1801221
92. Mukherjee N, Jacobs NC, Hafner M, Kennington EA, Nusbaum JD, Tuschl T, et al. Global Target mRNA Specification and Regulation by the RNA-Binding Protein ZFP36. Genome Biol (2014) 15:R12. doi: 10.1186/gb-2014-15-1-r12
93. Mahmoud L, Moghrabi W, Khabar KSA, Hitti EG. Bi-Phased Regulation of the Post-Transcriptional Inflammatory Response by Tristetraprolin Levels. RNA Biol (2019) 16:309–19. doi: 10.1080/15476286.2019.1572437
94. Kratochvill F, Gratz N, Qualls JE, Van De Velde LA, Chi H, Kovarik P, et al. Tristetraprolin Limits Inflammatory Cytokine Production in Tumor-Associated Macrophages in an mRNA Decay-Independent Manner. Cancer Res (2015) 75:3054–64. doi: 10.1158/0008-5472.CAN-15-0205
Keywords: tristetraprolin (TTP), zinc finger protein 36 (Zfp36), RNA binding protein, mRNA stability/decay, inflammation, immune system, immune homeostasis
Citation: Kovarik P, Bestehorn A and Fesselet J (2021) Conceptual Advances in Control of Inflammation by the RNA-Binding Protein Tristetraprolin. Front. Immunol. 12:751313. doi: 10.3389/fimmu.2021.751313
Received: 31 July 2021; Accepted: 01 September 2021;
Published: 17 September 2021.
Edited by:
Manuel Daniel Díaz-Muñoz, U1043 Centre de Physiopathologie de Toulouse Purpan (INSERM), FranceReviewed by:
Matthias Gaestel, Hannover Medical School, GermanyHeiko Mühl, Goethe University Frankfurt, Germany
Copyright © 2021 Kovarik, Bestehorn and Fesselet. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pavel Kovarik, cGF2ZWwua292YXJpa0B1bml2aWUuYWMuYXQ=
 Pavel Kovarik
Pavel Kovarik Annika Bestehorn
Annika Bestehorn Jeanne Fesselet
Jeanne Fesselet