- 1Cancer Center, Department of Radiation Oncology, Zhejiang Provincial People’s Hospital, Affiliated People’s Hospital, Hangzhou Medical College, Hangzhou, China
- 2Surgical Intensive Care Unit, First Affiliated Hospital, Zhejiang University, Hangzhou, China
- 3Department of Breast and Thyroid Surgery, Taizhou Hospital of Zhejiang Province, Taizhou, China
- 4Cancer Center, Department of Breast Surgery, Zhejiang Provincial People’s Hospital, People’s Hospital of Hangzhou Medical College, Hangzhou, China
- 5Department of Surgical Oncology, Sir Run Run Shaw Hospital, Zhejiang University, Hangzhou, China
- 6Department of Medical Oncology, First Affiliated Hospital, Zhejiang University, Hangzhou, China
NRF2 is an important regulatory transcription factor involved in tumor immunity and tumorigenesis. In this study, we firstly identified that FKBP4/NR3C1 axis was a novel negative regulator of NRF2 in human breast cancer (BC) cells. The effect of FKBP4 appeared to be at protein level of NRF2 since it could not suppress the expression of NRF2 at mRNA level. Bioinformatics analysis and in vitro experiments further demonstrated that FKBP4 regulated NRF2 via regulating nuclear translocation of NR3C1. We then reported that naringenin, a flavonoid, widely distributed in citrus and tomato, could suppress autophagy and proliferation of BC cells through FKBP4/NR3C1/NRF2 signaling pathway in vitro and in vivo. Naringenin was also found to promote dendritic cell (DC) differentiation and maturation through FKBP4/NR3C1/NRF2 axis. Therefore, our study found that naringenin could induce inhibition of autophagy and cell proliferation in BC cells and enhance DC differentiation and maturation, at least in part, though regulation of FKBP4/NR3C1/NRF2 signaling pathway. Identification of FKBP4/NR3C1/NRF2 axis would provide insights for novel anti-tumor strategy against BC among tumor microenvironment.
Introduction
Breast cancer (BC) is a leading cause of cancer-related deaths in women aged 40 years and younger (1). Early detection and comprehensive treatments, which consist of surgery, radiation, chemotherapy, endocrine therapy and targeted therapy, have dramatically improved the prognosis of BC patients. In recent years, immunotherapy in BC showed promising future. Cancer vaccines, bispecific antibodies, and immune checkpoint inhibitors are verified to have potential applied value in BC immunotherapy (2). For instance, adaptive immune checkpoint therapies by targeting cytotoxic T-lymphocyte antigen-4 (CTLA-4), proNR3C1ammed cell death-1 (PD-1), and ligand partner for PD-1 (PD-L1) for BC have been used in clinical trial (3, 4). Nevertheless, a portion of BC patients still cannot benefit from the above-mentioned immunotherapy strategies (5). Therefore, unraveling the potential molecular mechanisms of the immune system in BC cells is essential to further understand and improve immune related anti-tumor effects.
The transcription factor nuclear factor erythroid 2-related factor 2 (NRF2), also known as NFE2L2, is a Cap’n’collar, leucine zipper transcription factor comprised of seven Neh domains, and is regarded as a significant orchestrator of the cellular antioxidant response (6). Recently, NRF2 has been found to be involved in innate immunity and cytokine secretion orchestrated by dendritic cell (DC) (7). Additionally, several studies revealed the suppressive function of NRF2 in tumorigenesis, namely, BC, gastric cancer, leukemia, prostate cancer, colorectal cancer, melanomas and so forth (8–13). The previous study of our team has summarized the biological crosstalk between innate Immunity and BC (14), and we are wondering whether and how NRF2 performs its anti-tumor and pro-immunity effects by connection with novel molecular chaperones in BC.
As one of the most extensively studied proteins among the 18 identified human FK506-binding proteins (FKBPs), FK506-binding protein 4 (FKBP4), also known as FKBP52, has been reported to exhibit multiple functions which involve binding to different cellular receptors or targets in various kinds of cancers, such as lung cancer (15), prostate cancer (16), and glioblastoma (17). For example, FKBP4 has been demonstrated to interact with heat shock protein 90 (Hsp90) to affect steroid hormone receptor function in BC (18). In terms of immune regulation, phytanoyl-CoA alpha-hydroxylase (PAHX) has been regarded as a specific target of FKBP4 for studying the cellular signaling pathway in the presence of immunosuppressant drugs (19). Our previous work found that FKBP4 interacted with non-coding RNAs and mRNAs during the occurrence and development of BC, thus playing a role in promoting cancer (20, 21). Nevertheless, current immunologic mechanism of FKBP4 is still in its infancy for BC, and it is necessary to explore more new detailed contents of its regulation of innate and adaptive immunity functions during the occurrence and development of BC.
The NR3C1 (nuclear receptor subfamily 3, group C, member 1/glucocorticoid receptor) normally resides in the cell cytoplasm, where the NR3C1 protein translocates to the nucleus when bound to glucocorticoids, and thus is involved in growth, reproduction, metabolism, immune and inflammatory reactions, and cardiovascular functions and tumor cellular proliferation and differentiation (22). Researches on NR3C1 and BC have also been conducted in recent years, e.g., high levels of NR3C1 expression and high concentrations of cortisol have been shown to have an anti-proliferative effect in cancerous breast tissue (23). Some studies have preliminarily found that NR3C1 is associated with FKBPs, but the specific mechanisms remain unclear in BC (24).
In this study, we showed that naringenin, a flavonoid shown to have anti-tumor effects in various carcinomas in other studies (25), suppressed both autophagy and proliferation via new-found FKBP4/NR3C1/NRF2 signaling pathway in luminal A and basal-like subtype of BC cells. Mechanically, these effects relied on downregulation of FKBP4, which post-transcriptionally upregulated NRF2 through intensive nuclear translocation of NR3C1. Additionally, naringenin could promote differentiation and maturation of DC among tumor microenvironment through regulating FKBP4/NR3C1/NRF2 signaling pathway. Identification of FKBP4/NR3C1 axis as the novel NRF2 regulator would provide in-depth insights for immunological anti-tumor strategy to overcome BC.
Materials and Methods
Cell Culture
MCF10A, MCF7, T47D, BT549, MDA231, and SKBR3 cells were obtained from the American Type Culture Collection (ATCC). MCF10A was cultured in Mammary Epithelial Basal medium. MCF7 was cultured in Eagle’s Minimum Essential medium. T47D and SKBR3 were cultured in Dulbecco’s modified Eagle’s medium. BT549 was cultured in Roswell Park Memorial Institute (RPMI) medium. MDA231 was cultured in Leibovitz’s L15 medium. Growth media were supplemented with 10% fetal calf serum and penicillin/streptomycin (100 units per ml). All human cell lines were cultured at 37°C in a humidified incubator supplied with 5% CO2.
Antibodies and Reagents
Antibodies were used in the following dilutions: NRF2 (1:1,000, Proteintech, #16396-1-AP), FKBP4 (1:1,000, Proteintech, #10655-1-AP), NR3C1 (1:1,000, Proteintech, #24050-1-AP), P62 (1:1,000, MBL, #PM045), Histone (1:1,000, CST, #3638), GAPDH (1:1,000, Proteintech, #60004-1-Ig), Flag (1:1,000, Sigma, #F3165), HA (1:1,000, Biolegend, #901514), Secondary antibody goat anti-mouse (1:2,500, HuaBio, #HA1006), secondary antibody goat anti-rabbit (1:2,500, HuaBio, #HA1001), Anti-RABBIT IgG (H&L) (GOAT) Antibody Rhodamine Conjugated (1:200, MULTISCIENCES, #RK-611-1002), Anti-RABBIT IgG (H&L) (GOAT) Antibody ATTO 488 Conjugated (1:200, MULTISCIENCES, #RK-611-152-122S), Anti-Mouse CD11c, PE (1:20, MULTISCIENCES, #AM011C04), Anti-Mouse CD86 (B7-2), APC (AM08605). Naringenin was purchased from APExBIO (#N1370), and resolved in DMSO at 10 mM. BC cells were treated by medium with naringenin for different points in time.
Gene Silence
The shRNA against NRF2 was purchased from RIBBIO (Guangzhou, China), and shRNA against NR3C1 was purchased from Shanghai Generay Biotech Co., Ltd. The shRNA targeting sequences of NRF2 (26) and NR3C1 (27) were from published articles, lentiviral particles were produced as follows. In brief, HEK293T packaging cells were transfected with 800 ng pLKO.1 DNA in combination with the packaging plasmids 200 ng lenti-VSV-G, 400 ng lenti-RRE, and 140 ng lenti-REV. Virus containing supernatant was harvested at 36 and 48 h after transfection and filtered through a 0.45 μM syringe filter with the addition of 10 μM DEAE. Supernatants were used to infect target cells in another 12 h period.
Western Blotting and co-IP
Knockdown and overexpression efficiencies and biochemical responses were analyzed by western blotting. Cells were lysed in RIPA lysis buffer (EMD Millipore Corp.), supplemented with protease inhibitor and phosphatase inhibitor cocktail tablets (Roche). Separated proteins were transferred to nitrocellulose filter membranes and blocked in 5% milk in Tris-buffered saline, with 0.05% Tween-20. Immunodetection was done with various primary antibodies. Appropriate horseradish peroxidase-conjugated secondary antibodies were used and signals were visualized with Bio-Rad chemiluminescence by Bio-Rad ChemiDoc™ MP Imaging System. Cells for co-IP were lysed in lysis buffer (50 mM Tris, pH 7.5, with 150 mM NaCl, 0.5% NP-40, and protease inhibitor and phosphatase inhibitor cocktail tablets (Roche) at 4°C for 30 min. After sonication and centrifugation, cell lysates were incubated with beads (Sigma) at 4°C overnight on a rotator. After six washes with wash buffer (20 mM Tris, pH 7.5, 100 mM NaCl, 0.05% Tween-20, 0.1 mM EDTA), 50 μl of elution buffer (100 mM glycine-HCl, pH 2.5) was added to resuspend the beads, and the eluted proteins were obtained by centrifugation, followed by SDS-PAGE and immunoblotting analysis.
Quantitative RT-qPCR
Total RNA was extracted from cells by using TRIzol (Invitrogen). Reverse transcription was carried out with a 40-μl volume by using a PrimeScript™ RT Master Mix kit (TaKaRa) according to the manufacturer’s instructions. Quantitative real-time PCR (qPCR) was carried out on an Applied Biosystems Fast 7500 machine by using a TB NR3C1een® Premix Ex Taq™ II kit (TaKaRa) and the following primer sets were used for qPCR analysis: NRF2, 5′-GAGAGCCCAGTCTTCATTGC-3′ (forward) and 5′-TTGGCTTCTGGACTTGGAAC-3′ (reverse), FKBP4, 5′- CATTGCCATAGCCACCATGAA-3′ (forward) and 5′-TCCAGTGCAACCTCCACGATA-3′ (reverse), NR3C1, 5′-AGTGGTTGAAAATCTCCTTAACTATTGCT-3′ (forward) and 5′-GGTATCTGATTGGTGATGATTTCAGCTA-3′ (reverse), GAPDH, 5′-ATGACATCAAGAAGGTGGTG-3′ (forward) and 5′-CATACCAGGAAATGAGCTTG-3′ (reverse) as a control. The qPCR assay was carried out with a 15-μl volume consisting of 7.5 μl of a 2× TB NR3C1een mix solution, 0.3 μl of 10 μM of each oligonucleotide primer, 0.3 μl of ROX Reference Dye II and 2 μl of the cDNA template. Target fragment amplification was carried out as follows: 95°C for 30 s, followed by 40 cycles consisting of 95°C for 5 s and 60°C for 34 s. Melting-curve analysis was carried out at 90°C for 15 s and then at 60°C for 1 min and 95°C for 15 s.
Plasmids
FKBP4-HA, NR3C1-HA, NRF2-Flag, and the control plasmid were constructed by Shanghai Generay Biotech Co., Ltd.
Immunofluorescence
T47D or BT549 cells grown on coverslips were fixed for 15 min with 4% paraformaldehyde in PBS, permeabilized for 10 min in 0.1% Triton X-100 in PBS, and blocked using 5% BSA for 1 h. The cells were then incubated with primary antibodies at 4°C overnight. After a rinse with PBS, the cells were incubated with fluorescent-conjugated secondary antibodies for 1 h at 37°C. The nuclei were counterstained with 4, 6-diamidino-2-phenylindole (DAPI; Sigma–Aldrich). Images were captured using a ZEISS laser scanning confocal microscope (LSM710; Zeiss). ZEISS ZEN Microscope software was used for acquisition.
Autophagy Flux Monitoring
To evaluate the formation of fluorescent LC3B puncta, p−mCherry−C1−EGFP−hLC3B (LC3B) was used to monitor autophagy flux, 48 h after LC3B co−transfection with siRNAs, the cells were washed with 1× PBS and immediately analyzed via confocal microscopy (magnification, 500×). The nuclei were counterstained with 4, 6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich). Images were captured using a ZEISS laser scanning confocal microscope (LSM710; Zeiss). ZEISS ZEN Microscope software was used for acquisition.
Nuclear and Cytoplasmic Fractionation
T47D or BT549 cells were transfected with the indicated siRNAs for 72 h, then cells were harvested, and the nuclear and cytoplasmic fractions were separated using Thermo Fisher Scientific NE-PER Nuclear and Cytoplasmic Extraction Reagents (78833) according to the manufacturer’s protocol.
Transient Transfection
Breast cancer cells cultured in 12-well tissue culture plates were transiently transfected with plasmids using Lipofectamine® 2000 Reagent (Invitrogen) or siRNAs using Lipofectamine® RNAiMAX Reagent (Invitrogen) as instructed by the manufacturer. The siRNA targeting FKBP4, NR3C1, and NRF2, and also negative control siRNA were purchased from RIBBIO (Guangzhou, China). Seventy two hours later, the whole-cell extract was prepared for RT-qPCR or western blot analysis.
Cell Proliferation Assay
Cell proliferation was analyzed using a Cell Counting Kit-8 (CCK-8) (DOJINDO). All cells were seeded into 96-well plates at a density of 5,000 cells/well in a 100 µl volume and incubated at 37°C under 5% CO2 for 24, 48, and 72 h, followed by the addition of 10 μl of CCK-8 solution. The absorbance in each well was measured after 1 h incubation using a microculture plate reader at a test wavelength of 450 nm. Three replicate wells were set up in each group, and three independent experiments were performed.
Colony Formation Assay
Five hundred cells per well of breast cancer were seeded in a 6-well plate for colony formation assay. Two weeks after, they were fixed with 4% paraformaldehyde and stained with Crystal Violet. Colonies were quantified using ImageJ software.
Generation of Bone Marrow Cells
The DC cells were purchased from Wuhan Procell Life Science&Technology Co., Ltd. The cells were cultured in RPMI medium with 10% FBS, penicillin/streptomycin (100 units per ml), GMCSF (20 ng/ml) (Signalway Antibody, #AP73338), and IL4 (10 ng/ml) (Signalway Antibody, #AP73338).
Flow Cytometry Analysis
Single cell suspensions were surface-labeled using the mAbs mentioned above for 30 min at 4°C. After using Flow cytometry Staining buffer (MULTI SCIENCES, China), cells were analyzed with a BD FACSCalibur Flow Cytometer (Becton Dickinson). Cytometry data was analyzed using FlowJo software version 10 (CD11c gating was firstly used to screen for DC cells, and then CD86 gating was finally focused).
In Vivo Tumor Xenograft Assays
Six-week-old male BALB/c nude mice were purchased from Vital River Laboratory Animal Technology Co, Ltd (Beijing, P.R. China). Animal experimental procedures were approved by the Medical Ethics Committee of Zhejiang Provincial People’s Hospital. The six-week-old male mice were randomized into different groups. T47D shControl and T47D shNRF2 cells (5 × 106 cells/mice) were implanted subcutaneously into the flank of nude mice. After tumor formation, naringenin (50 mg/kg) was administered orally to its respective animal treatment groups for 28 days. Tumor volume (mm3) was measured every three days and calculated by the formula (length × width × width)/2. When the tumors had reached a volume of approximately 600 mm3, the mice were euthanized.
Bioinformatics Analysis
The localizations of FKBP4, NR3C1, and NRF2 protein were generated by PROTTER database (28) (https://wlab.ethz.ch/protter/start/). The immunofluorescence stainings of the subcellular distribution of FKBP4, NR3C1, and NRF2 protein were generated by from the HPA database (https://www.proteinatlas.org/). The expression, correlation, and prognostic module of the Breast Cancer Gene-Expression Miner v4.7 database (bc-GenExMiner v4.7) (29) (bcgenex.centregauducheau.fr) were used to evaluate the expression, correlation and prognostic merit of FKBP4, NR3C1, and NRF2 in human breast cancer. The lymphocyte, immune subtype, and molecular subtype module of the TISIDB database (30) (http://cis.hku.hk/TISIDB/), the gene, survival and scna module of the TIMER database (31) (https://cistrome.shinyapps.io/timer/) were used to evaluate relation between DC and immunological merit of FKBP4, NR3C1, and NRF2 in human breast cancer. Transcription factors in NRF2 promoter were predicted by PROMO (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3). Protein to protein interacting network was analyzed by STRING (https://string-db.org/).
Statistics
Two-tailed Student’s t-test was used in this study. Data shown was mean ± SD from at least three independent experiments. Statistical probability was expressed as *p <0.05, **p <0.01, and ***p <0.001.
Results
Negative Correlation of FKBP4 and NRF2 in Breast Cancer
Firstly, the FKBP4 and NRF2 protein topology both revealed intracellular membrane (cytosol and nucleoplasm) localization (Figures S1A–D), we also observed that FKBP4 and NRF2 colocalized with the nuclear marker in different cells by immunofluorescence assay of the HPA database, suggesting the subcellular localization of FKBP4 and NRF2 in nuclei (Figures S1E, F). Then we used bc-GenExMiner v4.7 database, an online public tool focused on BC, to explore the clinicopathological characteristics of FKBP4 and NRF2. Upregulated FKBP4 was found significantly related to luminal A, luminal B, HER2-positive and basal-like subtype of BC patients than the normal group (Figure 1A), and downregulated NRF2 was significantly related to luminal A, luminal B, HER2-positive and basal-like subtype patients than the normal group (Figure 1B), and the same results for TISIDB database (Figure S2). Furthermore, we validated a significant negative association between FKBP4 and NRF2 both in luminal A and basal-like subtype of BC patients (Figures 1C, D), but not in luminal B and HER2-positive subtype of BC patients (Figure S3). The prognostic merits of FKBP4 and NRF2 in luminal A and basal-like subtype of BC patients were further analyzed by using bc-GenExMiner v4.7, the Kaplan–Meier curve showed that increased levels of FKBP4 and decreased levels of NRF2 were strongly correlated with worse survival both in luminal A and basal-like subtype of BC patients (Figure S4).
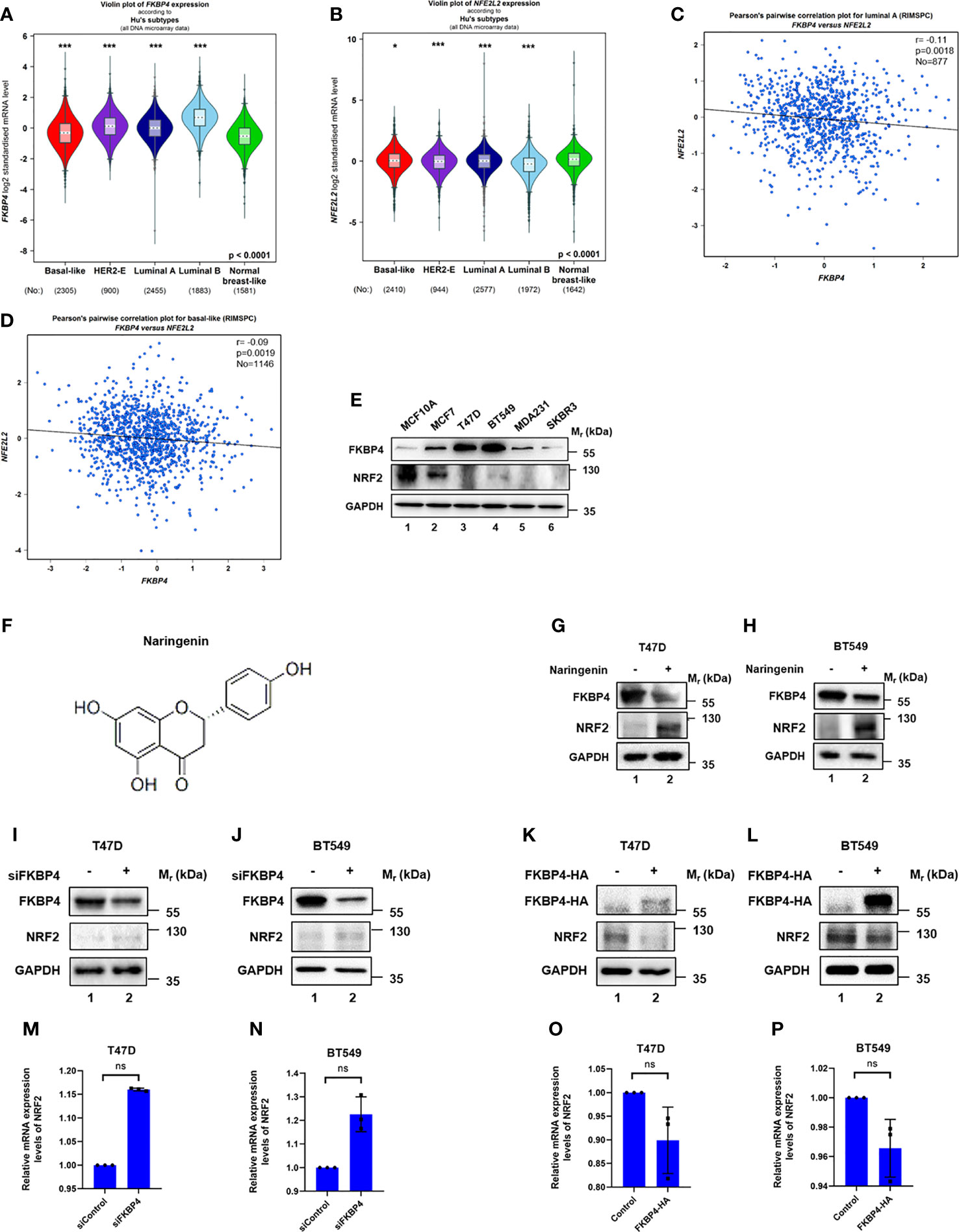
Figure 1 FKBP4 negatively regulates NRF2 at protein level. (A) A violin plot indicated upregulated FKBP4 in luminal A, luminal B, HER2-positive and basal-like subtype of BC patients than the normal group. (B) A violin plot indicated downregulated NRF2 in luminal A, luminal B, HER2-positive and basal-like subtype of BC patients than the normal group. (C) Pearson’s pairwise correlation plot of FKBP4 and NRF2 in luminal A subtype of BC patients, r = −0.11, p = 0.0018, No = 877. (D) Pearson’s pairwise correlation plot of FKBP4 and NRF2 in basal-like subtype of BC patients, r = −0.09, p = 0.0019, No = 1146. (E) Representative western blot analysis results of FKBP4, NRF2, and endogenous control GAPDH. Western blot analysis showed a negatively correlated expression of FKBP4 and NRF2 in BC and normal breast cells. (F) Chemical structure of naringenin. (G, H) Representative western blot analysis results of FKBP4, NRF2, and endogenous control GAPDH. Western blot analysis showed decreased FKBP4 expression and increased NRF2 expression in T47D and BT549 cells treated by 100 nM naringenin for 48 h (I, J) Representative western blot analysis results of FKBP4, NRF2, and endogenous control GAPDH. Western blot analysis showed silencing FKBP4 resulted in upregulation of NRF2 in T47D and BT549 cells. (K, L) Representative western blot analysis results of FKBP4-HA, NRF2, and endogenous control GAPDH. Western blot analysis showed overexpressing FKBP4 resulted in downregulation of NRF2 in T47D and BT549 cells. (M, N) RT-qPCR showed silencing FKBP4 resulted in no significant changes of NRF2 in T47D and BT549 cells (n = 3 independent biological replicates). (O, P) RT-qPCR showed overexpressing FKBP4 resulted in no significant changes of NRF2 in T47D and BT549 cells (n = 3 independent biological replicates). *p <0.05, ***p <0.001, ns, nonsense.
We also tested five BC cell lines (MCF7, T47D, BT549, MDA231, and SKBR3) and also normal breast cell line (MCF10A) for expression levels of FKBP4 and NRF2. As shown in Figure 1E, expression of FKBP4 and NRF2 were negatively correlated in BC cell lines, especially in T47D (representing luminal A subtype of BC) and BT549 (representing basal-like subtype of BC) cell lines. Since naringenin (shown in Figure 1F) has been reportedly involved in immunity regulation (32), we wondered its effect of pharmacologically modulating NRF2 expression. As shown in Figures 1G, H, naringenin significantly decreased FKBP4 expression and increased NRF2 expression in T47D and BT549 cells.
FKBP4 Negatively Regulates NRF2 at Protein Level
To confirm the specific interaction between FKBP4 and NRF2 at the molecular level, we used siRNA specifically targeting FKBP4 in T47D and BT549 cells, which led to upregulation of NRF2 at protein level (Figures 1I, J). In addition, we transfected FKBP4-HA plasmid in T47D and BT549 cells, which resulted in downregulation of endogenous NRF2 (Figures 1K, L). These results clearly indicated that FKBP4 had a role in negatively regulating NRF2 protein expression. However, we found no evidence that silencing or overexpressing FKBP4 could lead to changes of NRF2 at mRNA level in T47D and BT549 cells (Figures 1M–P).
Negative Correlation of FKBP4 and NR3C1 in Breast Cancer
We then continued to figure out potential factors involved in the FKBP4/NRF2 axis. Firstly, we used PROMO (33, 34), a virtual laboratory for the identification of putative transcription factors (TFs) binding sites in DNA sequences, to find predicted TFs binding to NRF2. After inputting NRF2 promoter sequence, namely, 1,000 bases upstream, CDS exons, and 150 bases downstream in PROMO (Table 1), 17 TFs were shown in order of frequency: NR3C1, HNF, GATA3, GCF, PXR1, RXRA, TFIID, P53, PAX5, TFII-I, IRF2, FOXP3, XBP1, cMyc, ER, CEBPB, and YY1 (Figure 2A). Besides FKBP4 was shown to connect to NR3C1 with a highest score in protein to protein interacting network on STRING database (35) (Figure 2B) (Table 2), we wondered whether NR3C1 was involved in FKBP4 associated NRF2 dysregulation. The NR3C1 protein topology also revealed intracellular membrane (cytosol and nucleoplasm) localization (Figures S5A, B), immunofluorescence assay of the HPA database suggested the subcellular localization of NR3C1 in nuclei (Figure S5C).
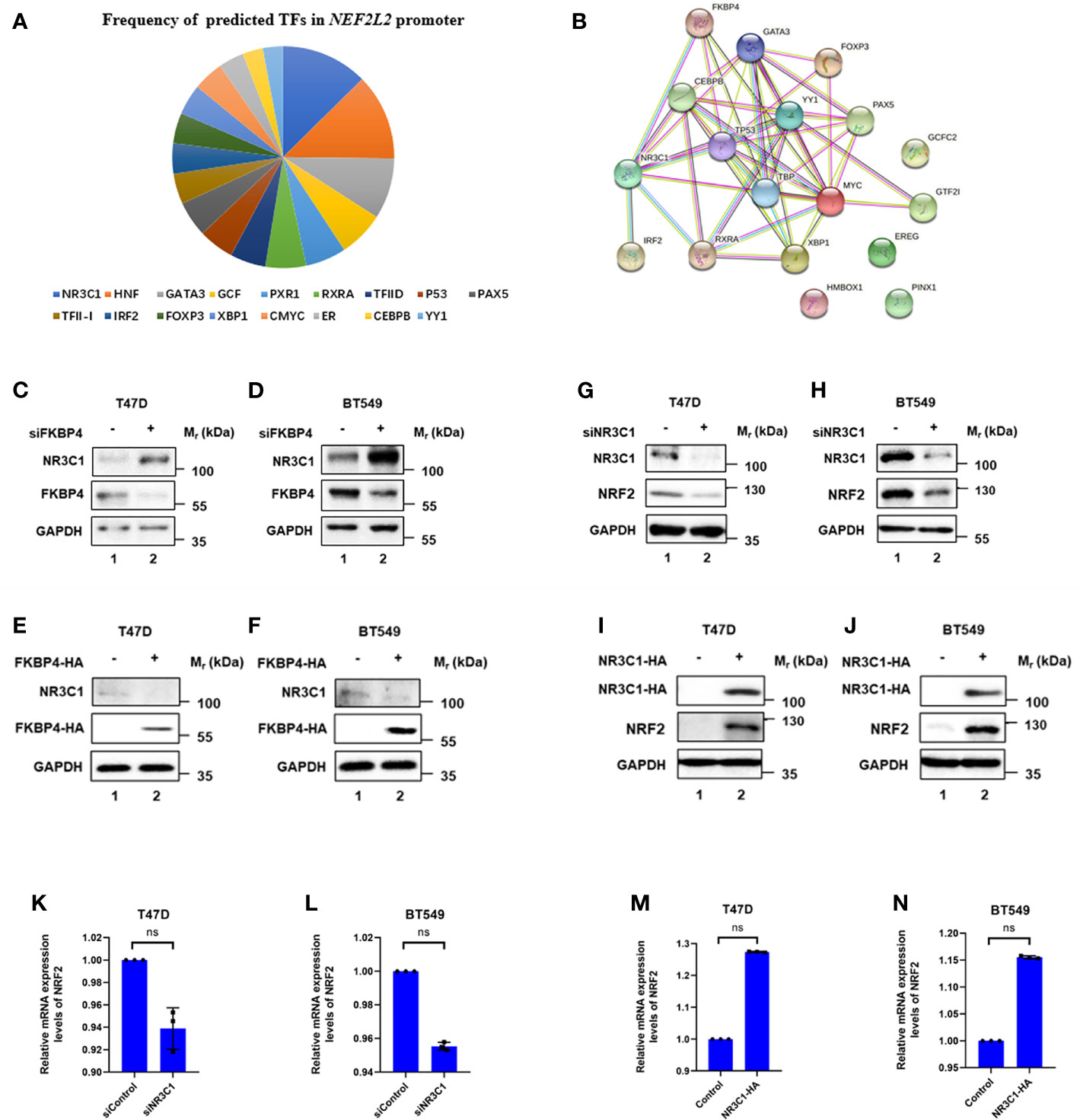
Figure 2 FKBP4 regulates NRF2 via NR3C1. (A) Pie chart showed frequency of 17 predicted transcription factors in NRF2 promoter. (B) Protein to protein interacting network of FKBP4 and 17 putative NRF2 transcription factors. (C–F) Representative western blot analysis results of FKBP4, NR3C1, and endogenous control GAPDH. Western blot analysis showed silencing FKBP4 resulted in upregulation of NR3C1, overexpressing FKBP4 resulted in downregulation of NR3C1 in T47D and BT549 cells. (G–J) Representative western blot analysis results of NR3C1, NRF2, and endogenous control GAPDH. Western blot analysis showed silencing NR3C1 resulted in downregulation of NRF2, overexpressing NR3C1 resulted in upregulation of NRF2 in T47D and BT549 cells. (K–N) RT-qPCR showed silencing NR3C1 resulted in no significant changes of NRF2, overexpressing NR3C1 resulted in no significant changes of NRF2 in T47D and BT549 cells (n = 3 independent biological replicates). ns, nonsense.
We firstly used siRNA specifically targeting FKBP4 in T47D and BT549 cells, and found it led to upregulation of NR3C1 at protein level (Figures 2C, D). In addition, we transfected FKBP4-HA plasmid in T47D and BT549 cells, which resulted in downregulation of endogenous NR3C1 (Figures 2E, F). These results clearly indicated that FKBP4 had a role in negatively regulating NR3C1 protein expression. Bioinformatics results also suggested a significant negative association between FKBP4 and NR3C1 both in luminal A and basal-like subtype of BC patients (Figures S6A, B). Furthermore, we found naringenin could upregulate NR3C1 protein expression in T47D and BT549 cells (Figure S7).
NR3C1 Positively Regulates NRF2 at Protein Level
We further confirmed the specific interaction between NR3C1 and NRF2 at the molecular level. We firstly used siRNA specifically targeting NR3C1 in T47D and BT549 cells, which led to downregulation of NRF2 at protein level (Figures 2G, H). In addition, we transfected NR3C1-HA plasmid in T47D and BT549 cells, which resulted in upregulation of endogenous NRF2 (Figures 2I, J). These results clearly indicated that NR3C1 had a role in positively regulating NRF2 protein expression. Given NR3C1 was demonstrated to regulate NRF2 at protein level, we wondered whether it affected the mRNA level of NRF2. We then found silencing of NR3C1 could not lead to downregulation of NRF2 at mRNA level in T47D and BT549 cells (Figures 2K, L). Whereas overexpressed NR3C1 neither resulted in upregulation of NRF2 at mRNA level in T47D and BT549 cells (Figures 2M, N).
Meanwhile, we verified a significant positive association between NR3C1 and NRF2 both in luminal A and basal-like subtype of BC patients (Figures S6C, D). Downregulated NR3C1 was significantly related to four molecular subtype patients than the normal group in two databases (Figure S8) and was strongly correlated with worse survival both in luminal A and basal-like subtype of BC patients (Figure S9). Hence, our results implied that FKBP4 might downregulate NRF2 by inhibiting NR3C1 at protein level.
FKBP4 Binds to NR3C1 and Regulates Nuclear Translocation of NR3C1
As FKBP4 regulates its cellular targets via protein–protein interaction (36, 37), we tested whether FKBP4 bound to NR3C1. We performed co-IP and western blot assays using BT549 and T47D cells, and results showed that FKBP4 and NR3C1 bound with each other (Figures 3A–D). We further observed that nuclear accumulations of NR3C1 were enhanced by siRNA targeting FKBP4 or inhibited by FKBP4-HA plasmid in BT549 and T47D cells by western blot analysis (Figures 3E–H). Silencing FKBP4 mediated promotion of NR3C1 nuclear accumulations were also observed in BT549 and T47D cells by immunofluorescence microscopy (Figures 3I, J).
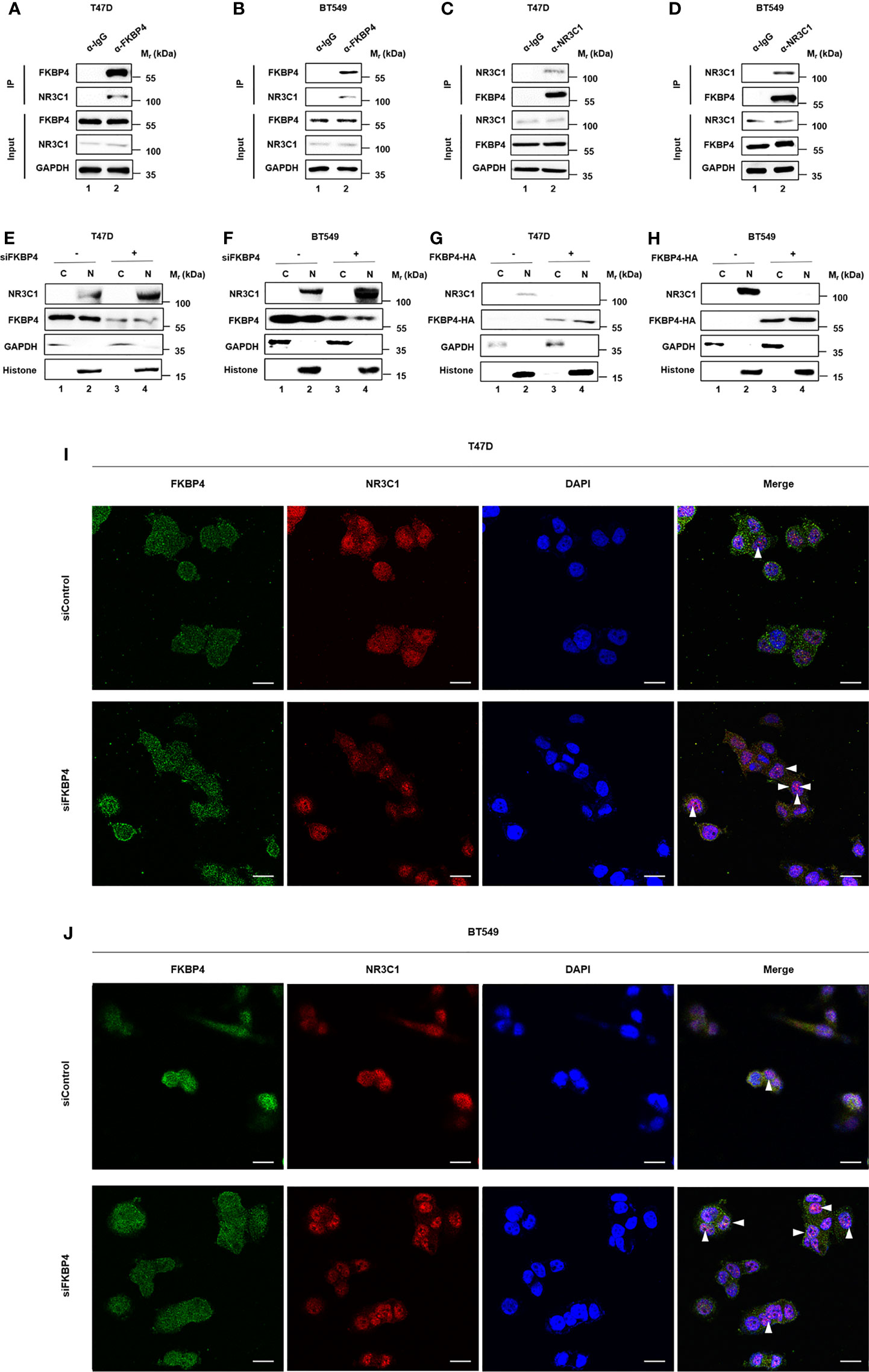
Figure 3 FKBP4 binds to NR3C1 and FKBP4 inhibition triggers nuclear translocation of NR3C1. (A–D) co-IP and western blot assays using antibodies as indicated for binding between endogenous FKBP4 and NR3C1 in T47D and BT549 cells. (E–H) T47D and BT549 cells were transfected with siRNAs or plasmids. Cells were harvested, and the nuclear and cytoplasmic fractions were separated. Proteins were analyzed by immunoblotting using anti-NR3C1, anti-FKBP4, anti- GAPDH, or anti-histone antibody. (I, J) T47D and BT549 cells were transfected with FKBP4 siRNAs. Immunofluorescent staining was carried out using anti-NR3C1 and anti-FKBP4. DAPI staining was performed to show the nuclei (n = 3 independent biological replicates). The arrowheads show the NR3C1 nuclear localization under FKBP4 inhibition. Scale bar = 20 μm.
These observations were consistent with the finding that FKBP4 inhibited NRF2 through interacting with NR3C1 at protein level.
FKBP4/NR3C1/NRF2 Signaling Pathway Involved in Naringenin-Restrained Autophagy
Since naringenin had been used to inhibit autophagy (38), we doubted whether naringenin related FKBP4/NR3C1/NRF2 axis was also involved in autophagy. Western blotting showed that FKBP4 overexpression decreased the expression of autophagy associated molecule P62 in T47D and BT549 cells (Figures 4A, B). Similarly, silencing NRF2 in T47D and BT549 cells led to the downregulation of P62 (Figures 4C, D). Meanwhile, western blotting showed that silencing FKBP4 increased the expression of P62 in T47D and BT549 cells (Figures 4E, F), and NRF2 overexpression led to the upregulation of P62 (Figures 4G, H). As shown in Figure S10, NR3C1 had the same effects as NRF2 on P62. Autophagy flux was also observed increased in T47D and BT549 cells silencing NRF2 (Figures 4I, J).
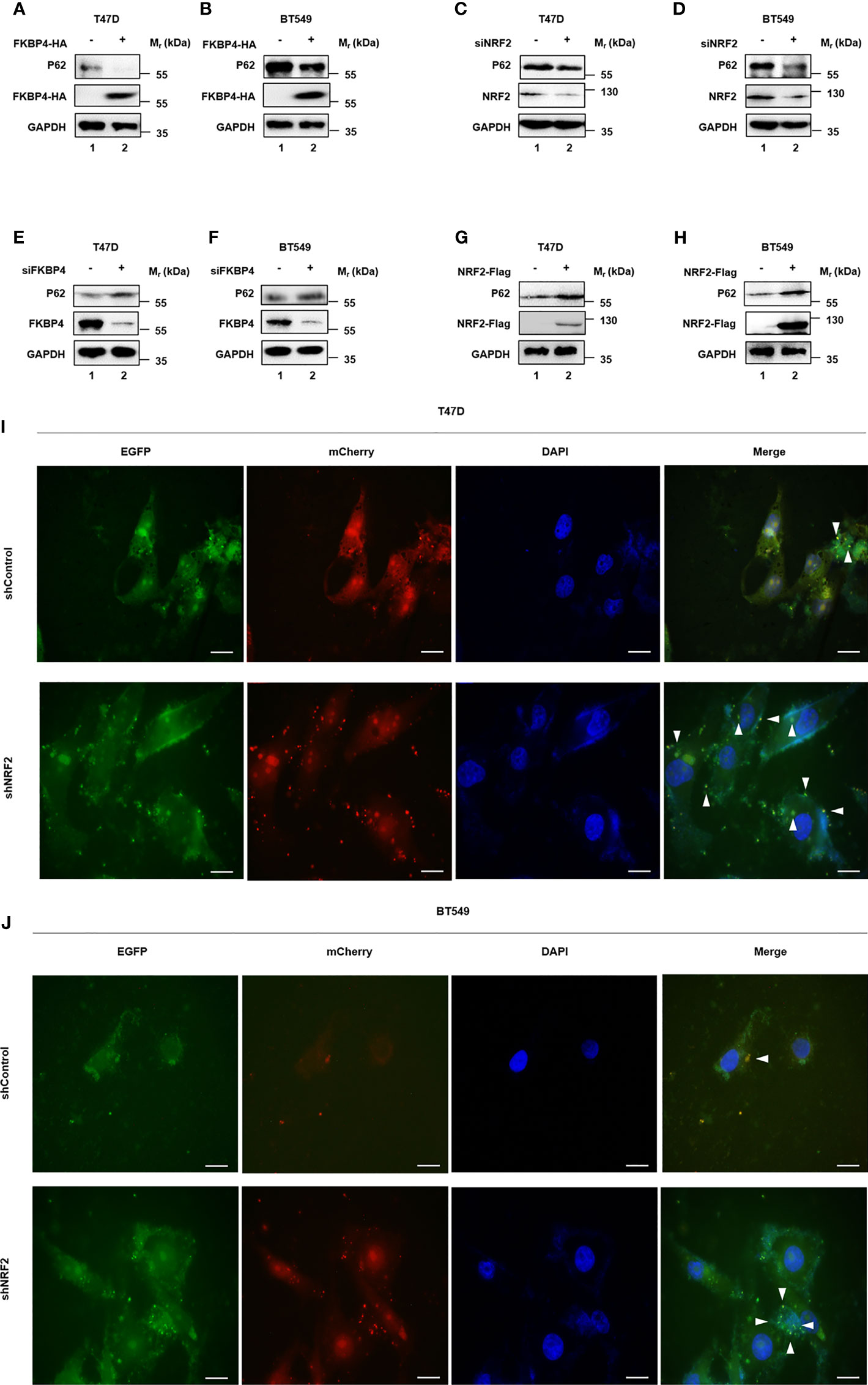
Figure 4 FKBP4/NR3C1/NRF2 axis is involved in naringenin-restrained autophagy. (A, B) Representative western blot analysis results of P62, FKBP4-HA, and endogenous control GAPDH. Western blot analysis showed overexpressing FKBP4 resulted in downregulation of P62 in T47D and BT549 cells. (C, D) Representative western blot analysis results of P62, NRF2, and endogenous control GAPDH. Western blot analysis showed silencing NRF2 resulted in downregulation of P62 in T47D and BT549 cells. (E, F) Representative western blot analysis results of P62, FKBP4, and endogenous control GAPDH. Western blot analysis showed silencing FKBP4 resulted in upregulation of P62 in T47D and BT549 cells. (G, H) Representative western blot analysis results of P62, NRF2-Flag, and endogenous control GAPDH. Western blot analysis showed overexpressing NRF2 resulted in upregulation of P62 in T47D and BT549 cells. (I, J) Measurement of autophagy flux in T47D and BT549 cells co−transfected with p−mCherry−C1−EGFP−hLC3B and shControl or shNRF2. Scale bar = 20 μm.
All together, these results suggested that naringenin enhanced BC cell autophagy partially owing to FKBP4/NR3C1/NRF2 axis.
FKBP4/NR3C1/NRF2 Signaling Pathway Involved in Naringenin-Restrained Cell Proliferation
While naringenin is well-known to inhibit cell proliferation (39), we doubted whether naringenin related FKBP4/NR3C1/NRF2 axis was also involved in cell proliferation. Cell viability assay showed that silencing FKBP4 or overexpressing NR3C1 prevented cell proliferation of both T47D and BT549 cells at 72 h (Figures 5A–D), while overexpressing FKBP4 or silencing NR3C1 promoted cell proliferation of both T47D and BT549 cells at 72 h (Figures 5E–H). Additionally, knockdown of NRF2 significantly prevented naringenin-restrained cell proliferation of T47D and BT549 cells during 72 h (Figures 5I–L). These findings were further confirmed by cell photography (Figure S11) and colony formation assay (Figure S12).
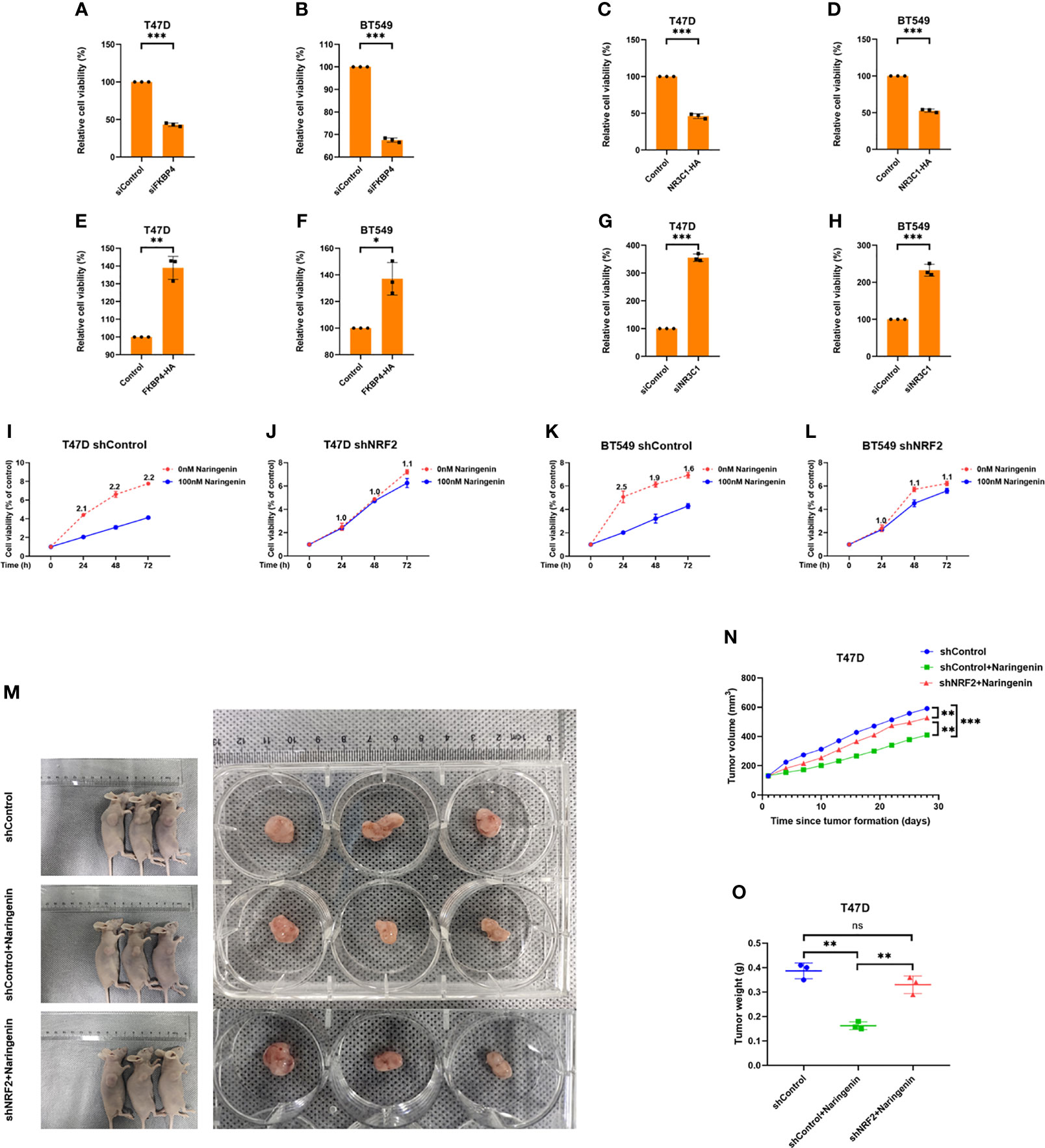
Figure 5 FKBP4/NR3C1/NRF2 axis is involved in naringenin-restrained cell proliferation. (A, B) Bar chart showed silencing FKBP4 resulted in decreased cell viability of T47D and BT549 cells at 72 h (n = 3 independent biological replicates). (C, D) Bar chart showed overexpressing NR3C1 resulted in decreased cell viability of T47D and BT549 cells at 72 h (n = 3 independent biological replicates). (E, F) Bar chart showed overexpressing FKBP4 resulted in increased cell viability of T47D and BT549 cells at 72 h (n = 3 independent biological replicates). (G, H) Bar chart showed silencing NR3C1 resulted in increased cell viability of T47D and BT549 cells at 72 h (n = 3 independent biological replicates). (I–L) Line chart showed knockdown of NRF2 by shRNA attenuated naringenin-restrained cell proliferation in T47D and BT549 cells treated by 100 nM naringenin, numeric representation at each dot is cell viability fold change of 0 and 100 nM naringenin treatment (n = 3 independent biological replicates). (M–O) The macroscopic appearance, volume and weight of subcutaneous tumors in mice (n = 3/group) transplanted with T47D cells treated with shControl, shControl + Naringenin, and shNRF2 + Naringenin. *p <0.05, **p <0.01, ***p <0.001, ns, nonsense.
T47D cells treated with silenced NRF2 were used to generate subcutaneous xenograft models in nude mice. Results showed that the volume and weight of tumors were significantly reduced in the naringenin treated group than in the control group, whereas those in the naringenin + shNRF2 group were significantly greater than in the naringenin treated group (Figures 5M–O).
Thus, in vivo and in vitro experiments indicated that naringenin inhibited BC proliferation partially owing to the FKBP4/NR3C1/NRF2 signaling pathway.
FKBP4/NR3C1/NRF2 Signaling Pathway Involved in Naringenin-Induced DC Differentiation and Maturation
Since naringenin had been found connected with the immune system (40), we investigated the correlations between FKBP4/NR3C1/NRF2 signaling pathway and various immune signatures using TISIDB database and TIMER database. Figures 6A–F showed that associations between FKBP4, NR3C1, and NRF2 expression and immune subtypes across human cancers, FKBP4 and NR3C1 were most closely associated with immune subtypes of BC. Similar to the finding of Liang et al. that naringenin induced more DCs infiltration into tumor (41), we firstly mined the relationship of FKBP4/NR3C1/NRF2 axis and DC infiltration level and abundance, namely, FKBP4 expression was significantly negatively correlated with DC infiltration level and abundance, while NR3C1 and NRF2 expression were significantly positively correlated with DC infiltration level and abundance in two databases (Figures 6G–L). However, FKBP4/NR3C1/NRF2 signaling pathway did not affect the relationship between DC infiltration and prognosis of BC, nor did their mutations in DC cells affect DC infiltration level in BC (Figure S13).
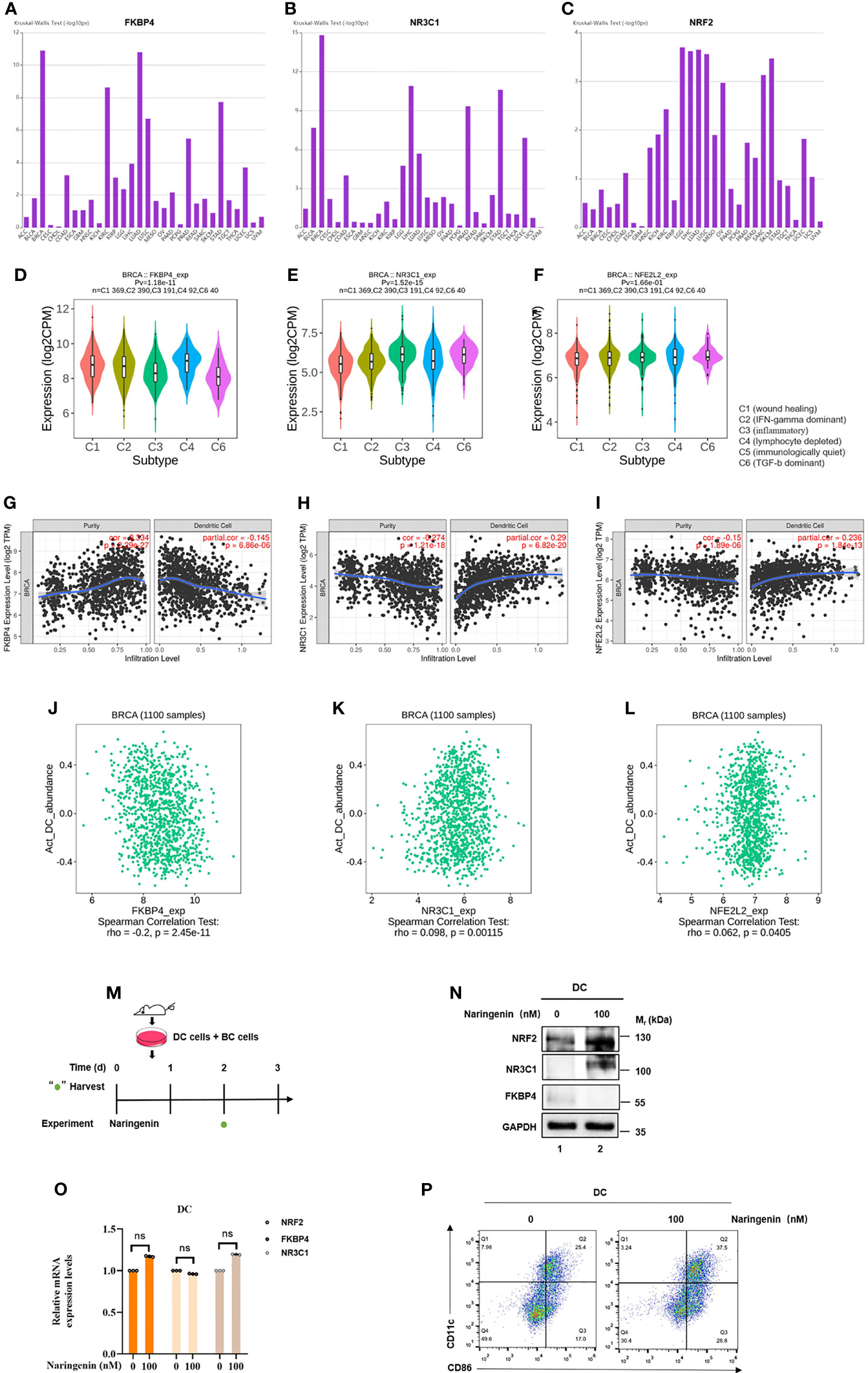
Figure 6 FKBP4/NR3C1/NRF2 signaling pathway involved in naringenin-induced DC differentiation and maturation. (A–C) Distribution of FKBP4, NR3C1, and NRF2 expression across immune subtypes in pan-cancer analysis using TISIDB database. (D–F) Distribution of FKBP4, NR3C1, and NRF2 expression across immune subtypes only in breast cancer using TISIDB database. (G–I) Spearman correlations between expression of FKBP4, NR3C1 or NRF2 and infiltration level of DCs in breast cancer using TIMER database. (J–L) Spearman correlations between expression of FKBP4, NR3C1 or NRF2 and abundance of DCs in breast cancer using TISIDB database. (M) Schema of mice DC cells co-cultured with BC cells treated by naringenin. (N) Western blot analysis of NR3C1, FKBP4, and NRF2 protein expression in DCs co-cultured with BC cells for 3 days. (O) RT-qPCR analysis of NR3C1, FKBP4, and NRF2 mRNA expression in DCs co-cultured with BC cells for 3 days. (P) DC cells were treated with naringenin for 72 h and analyzed for maturation by flow cytometry analysis. ns, nonsense.
Next, to confirm if FKBP4/NR3C1/NRF2 axis was truly regulated in DC, we co-cultured BC cells and bone marrow cells of mice with GMCSF + IL4, simulated as DC (Figure 6M). After naringenin treatment for 72 h, we found that FKBP4 was decreased while NR3C1 and NRF2 were increased at protein but not mRNA level (Figures 6N, O). Flow cytometry analysis results showed that the positive expression percentages of CD11c and CD86 of DC were significantly increased in the naringenin group in contrast to those in the control group (Figure 6P). These data suggested that FKBP4/NR3C1/NRF2 signaling pathway was partially involved in naringenin-induced DC differentiation and maturation.
Taken together, we demonstrated that naringenin mediated anti-autophagy, anti-proliferation of BC cells, and pro-DC differentiation and maturation in a new-found FKBP4/NR3C1/NRF2 dependent way (Figure 7).
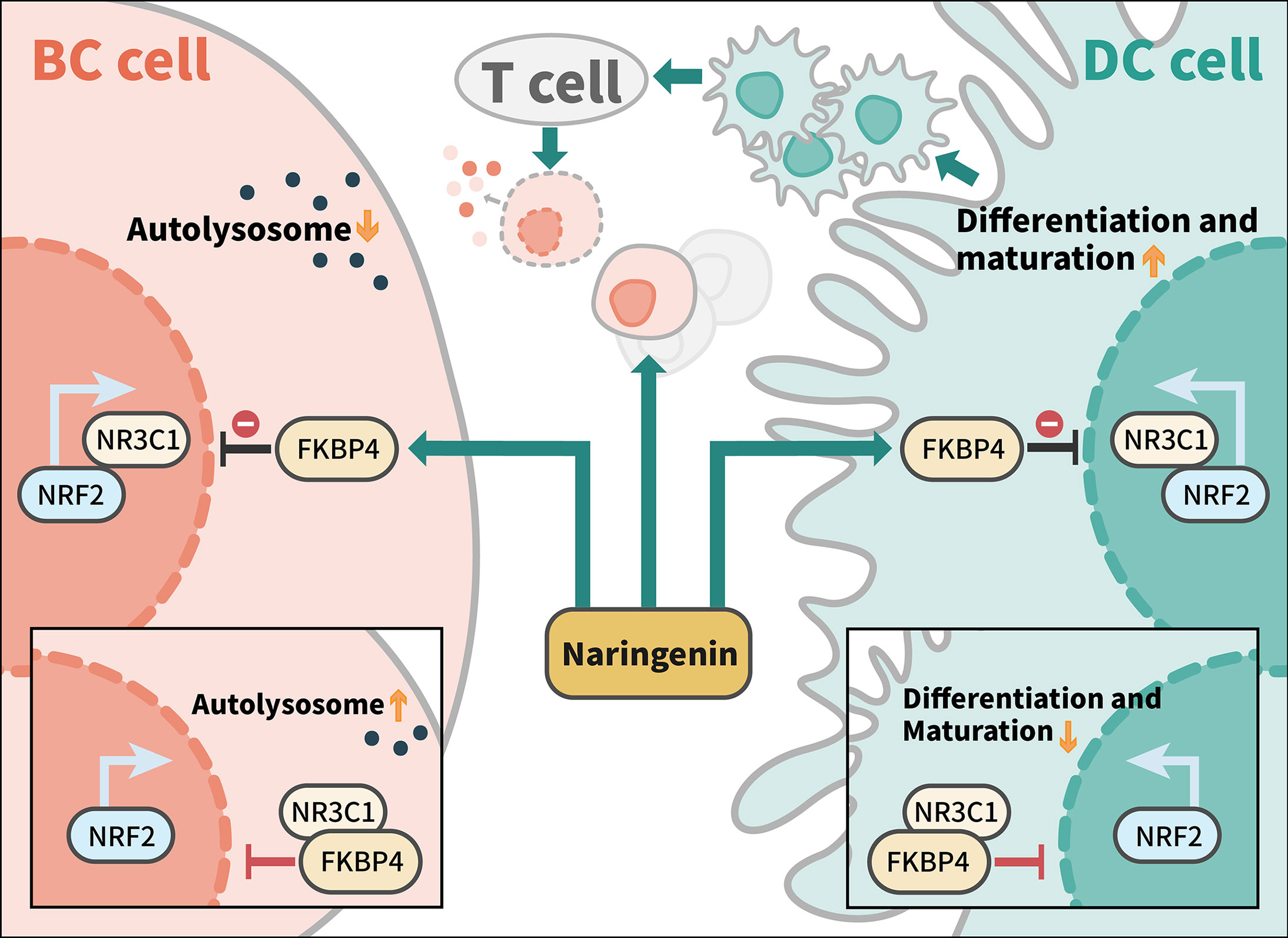
Figure 7 Model of naringenin-mediated inhibition of autophagy and cell proliferation and promotion of DC differentiation and maturation through FKBP4/NR3C1/NRF2 signaling pathway. Naringenin inhibits FKBP4 protein, thus releasing and inducing NR3C1 protein to bind to NRF2 protein, leading to autophagy and proliferation inhibition of BC cells, as well as differentiation and maturation promotion of DC. Among tumor microenvironment, naringenin might promote T cells to kill breast cancer cells by enhancing the antigen presentation function of DC cells.
Discussion
Endogenous host-derived regulators of NRF2 have been becoming the hotspot on related researches. For instance, when specific thiol residues are modified by electrophiles, NRF2 is released from the E3 ligase adaptor Kelch-like ECH-associated protein 1 (Keap1) related complex and translocates into the nucleus wherein it performs its functions (42). Yu et al. defined the Bax-binding protein MOAP-1 as a negative regulator of NRF2 via dissociation of p62 bodies (43). TRIM25 expression was found positively associated with NRF2 expression in hepatocellular carcinoma, mechanistically, TRIM25 directly targeted Keap1 by ubiquitination and degradation, leading to NRF2 activation (44). The current study firstly suggested that FKBP4/NR3C1 axis might be a novel endogenous negative regulator of NRF2 by altering its post-transcriptional activity in both BC cells and DCs. Thus, FKBP4 and NR3C1 could serve as notable therapeutic molecules against NRF2-dependent tumorigenesis, autoinflammation, and autoimmunity in the future.
FK506-binding protein family in human genomes has included 18 FKBPs up to date, which could target on various pathways in embryonic development, stress response, cardiac function, cancer tumorigenesis, and neuronal function (45). In colorectal cancer, silencing FKBP3 has been found to attenuate oxaliplatin resistance by regulation of the phosphatase and tensin homolog (PTEN)/AKT axis (46). In Alzheimer’s disease, FKBP12 and amyloid precursor protein (APP) interplay has been suspected to affect Aβ peptides expression (47). Although FKBP4 has been demonstrated to connect mammalian target of naringenin complex 2 (mTORC2) and phosphoinositide-3-kinase (PI3K) to enhance cell proliferation of BC, for the first time, we found that FKBP4 played a carcinogenic role by downregulating NRF2 in BC cells and co-cultured DCs.
Currently, there is no research of regulation mechanisms on NR3C1 and NRF2, but only a few studies suggest that NR3C1 is involved in the innate immune response (48, 49). In our study, we demonstrated FKBP4 post-transcriptionally downregulated NRF2 through binding to NR3C1, thus inhibiting nuclear translocation of NR3C1, but this effect was found relatively weak because shNR3C1 could not completely inhibit FKBP4 from regulating NRF2 (data not shown). Therefore, although we speculated that some of the predicted TFs were also involved in FKBP4/NRF2 pathway, subsequent mechanism studies need to be further improved to verify our conjecture.
As for the interaction of FKBP and autophagy, FKBP5 has been reported to change phosphorylation of Beclin1 by binding to Beclin1, thus triggering autophagic pathways (50). FKBP8 was found to recruit lipidated LC3A to induce mitochondrial autophagy via N-terminal LC3-interacting region motif of FKBP8 (51), while the regulation of autophagy by FKBP4 in tumor cells has not been reported yet. Meanwhile, NRF2 was also involved in autophagy, e.g., Moscat et al. demonstrated that protein kinase C (PKC) λ/ι loss in hepatocytes could promote autophagy and oxidative phosphorylation, through which NRF2 drove cancer cell through cell-autonomous and non-autonomous mechanisms (52). In the current study, we first demonstrated that inhibiting FKBP4 could weaken autophagy by increasing NRF2 expression of BC cells, thus autophagy plays an oncogenic role in this context. Actually, autophagy plays a dual role depending on the different types of context of BC (53). For instance, inhibition of autophagy along with HER2 inhibition is critical for promoting BC regression, but autophagy stimulation could transform the effectiveness of HER2 treatments (54), Meanwhile small-molecule RL71 triggered excessive autophagic cell death as a potential therapeutic strategy in triple-negative BC (55), implying that autophagy could function as a tumor suppressive mechanism. Additionally, autophagy seems to exert a dual role in luminal breast cancer, and also triple-negative BC (56).
Takabe et al. demonstrated that high NRF2 tumors were associated with high infiltration of DCs by conducting in silico analyses in 5,443 breast cancer patients from several large patient cohorts (57). Besides, it is likely that different methods mediate the transfer of cargos to DCs through NRF2 related signalings, e.g., the project that our team is currently working on has found that extracellular vesicles secreted by naringenin treated BC cells induced DC differentiation and maturation on NRF2 dependent way (data not shown). Further experimentation will be required to determine the relative contribution of these different mechanisms of tumor DNA delivery to DCs.
Recently, a growing body of evidence has demonstrated that the pharmacological agents, e.g., Ezetimibe ketone, Desfluoro-ezetimibe, NK-252, Bardoxolone, and TBHQ could induce activation of NRF2 (58–61). Here we proposed naringenin, a citrus flavonoid shown to have cytotoxic and antiproliferative effects on various cancer cell types (62) and regulates immunological pathways (63), to be another kind of potential NRF2 conditioning agent. With the development of immunotherapies such as cancer vaccine, immune checkpoint inhibitors, oncolytic virus, and chimeric antigen receptor T cell (CAR-T) therapy (64), a combination of NRF2-targeting agonists and immunotherapies may provide multiple feasible approaches to new BC treatment strategies.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The animal study was reviewed and approved by the Animal Welfare Ethics Committee of Zhejiang Provincial People’s Hospital.
Author Contributions
HX designed the experiment. HX and CZ performed most of experiments. BL, BX, XL, CC, and ZL performed the western blot assay and the real-time PCR experiments. YuJ, ZW, MY, and YoJ contributed to bioinformatics analysis. HX, ZC, and BL analyzed the data and wrote the paper. HX, LW, JZ and XM conducted the study supervision. All authors contributed to the article and approved the submitted version.
Funding
The work was supported by the National Natural Science Foundation of China (No. 81972453, No. 81972597, No. 82000212 and No. 82102814), Zhejiang Provincial Natural Science Foundation of China under grants (No. LY19H160055, LY19H160059, LY20H160026, LQ21H160022 and LQ22H160053). The work was sponsored by Zhejiang Provincial Medical and Health Science and Technology Project (No. 2018ZD028 and No. 2021RC003), Zhejiang Provincial People’s Hospital Scientific Research Foundation for The Excellent Youth (ZRY2020B007).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the Core Facilities, Department of Radiation Oncology, Zhejiang Provincial People’s Hospital for the technical support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.745111/full#supplementary-material
References
1. Ganz PA, Goodwin PJ. Breast Cancer Survivorship: Where Are We Today? Adv Exp Med Biol (2015) 862:1–8. doi: 10.1007/978-3-319-16366-6_1
2. Yu LY, Tang J, Zhang CM, Zeng WJ, Yan H, Li MP, et al. New Immunotherapy Strategies in Breast Cancer. Int J Environ Res Public Health (2017) 14:1–18. doi: 10.3390/ijerph14010068
3. Vonderheide RH, LoRusso PM, Khalil M, Gartner EM, Khaira D, Soulieres D, et al. Tremelimumab in Combination With Exemestane in Patients With Advanced Breast Cancer and Treatment-Associated Modulation of Inducible Costimulator Expression on Patient T Cells. Clin Cancer Res (2010) 16:3485–94. doi: 10.1158/1078-0432.CCR-10-0505
4. Lum LG, Thakur A, Al-Kadhimi Z, Colvin GA, Cummings FJ, Legare RD, et al. Targeted T-Cell Therapy in Stage IV Breast Cancer: A Phase I Clinical Trial. Clin Cancer Res (2015) 21:2305–14. doi: 10.1158/1078-0432.CCR-14-2280
5. De Iuliis F, Salerno G, Taglieri L, Scarpa S. Are Pharmacogenomic Biomarkers an Effective Tool to Predict Taxane Toxicity and Outcome in Breast Cancer Patients? Literature Review. Cancer Chemother Pharmacol (2015) 76:679–90. doi: 10.1007/s00280-015-2818-4
6. Rojo de la Vega M, Chapman E, Zhang DD. NRF2 and the Hallmarks of Cancer. Cancer Cell (2018) 34:21–43. doi: 10.1016/j.ccell.2018.03.022
7. Wei HJ, Gupta A, Kao WM, Almudallal O, Letterio JJ, Pareek TK. Nrf2-Mediated Metabolic Reprogramming of Tolerogenic Dendritic Cells is Protective Against Aplastic Anemia. J Autoimmun (2018) 94:33–44. doi: 10.1016/j.jaut.2018.07.005
8. Lien EC, Lyssiotis CA, Juvekar A, Hu H, Asara JM, Cantley LC, et al. Glutathione Biosynthesis Is a Metabolic Vulnerability in PI(3)K/Akt-Driven Breast Cancer. Nat Cell Biol (2016) 18:572–8. doi: 10.1038/ncb3341
9. Hu Z, Wang X, Li D, Cao L, Cui H, Xu G. UFBP1, a Key Component in Ufmylation, Enhances Drug Sensitivity by Promoting Proteasomal Degradation of Oxidative Stress-Response Transcription Factor Nrf2. Oncogene (2021) 40:647–62. doi: 10.1038/s41388-020-01551-1
10. Liu P, Ma D, Wang P, Pan C, Fang Q, Wang J. Nrf2 Overexpression Increases Risk of High Tumor Mutation Burden in Acute Myeloid Leukemia by Inhibiting MSH2. Cell Death Dis (2021) 12:20. doi: 10.1038/s41419-020-03331-x
11. Ghosh S, Dutta N, Banerjee P, Gajbhiye RL, Sareng HR, Kapse P, et al. Induction of Monoamine Oxidase A-Mediated Oxidative Stress and Impairment of NRF2-Antioxidant Defence Response by Polyphenol-Rich Fraction of Bergenia Ligulata Sensitizes Prostate Cancer Cells In Vitro and In Vivo. Free Radical Biol Med (2021) 172:136–51. doi: 10.1016/j.freeradbiomed.2021.05.037
12. Taha-Mehlitz S, Bianco G, Coto-Llerena M, Kancherla V, Bantug GR, Gallon J, et al. Adenylosuccinate Lyase is Oncogenic in Colorectal Cancer by Causing Mitochondrial Dysfunction and Independent Activation of NRF2 and mTOR-MYC-Axis. Theranostics (2021) 11:4011–29. doi: 10.7150/thno.50051
13. Nishida M, Yamashita N, Ogawa T, Koseki K, Warabi E, Ohue T, et al. Mitochondrial Reactive Oxygen Species Trigger Metformin-Dependent Antitumor Immunity via Activation of Nrf2/mTORC1/p62 Axis in Tumor-Infiltrating CD8T Lymphocytes. J Immunother Cancer (2021) 9. doi: 10.1136/jitc-2021-002954
14. Wang S, Zhou L, Ling L, Meng X, Chu F, Zhang S, et al. The Crosstalk Between Hippo-YAP Pathway and Innate Immunity. Front Immunol (2020) 11:323. doi: 10.3389/fimmu.2020.00323
15. Meng W, Meng J, Jiang H, Feng X, Wei D, Ding Q. FKBP4 Accelerates Malignant Progression of Non-Small-Cell Lung Cancer by Activating the Akt/mTOR Signaling Pathway. Analytical Cell Pathol (Amsterdam) (2020) 2020:6021602. doi: 10.1155/2020/6021602
16. Maeda K, Habara M, Kawaguchi M, Matsumoto H, Hanaki S, Masaki T, et al. FKBP51 and FKBP52 Regulate Androgen Receptor Dimerization and Proliferation in Prostate Cancer Cells. Mol Oncol (2021). doi: 10.1002/1878-0261.13030
17. Ott M, Litzenburger UM, Rauschenbach KJ, Bunse L, Ochs K, Sahm F, et al. Suppression of TDO-Mediated Tryptophan Catabolism in Glioblastoma Cells by a Steroid-Responsive FKBP52-Dependent Pathway. Glia (2015) 63:78–90. doi: 10.1002/glia.22734
18. Ebong IO, Beilsten-Edmands V, Patel NA, Morgner N, Robinson CV. The Interchange of Immunophilins Leads to Parallel Pathways and Different Intermediates in the Assembly of Hsp90 Glucocorticoid Receptor Complexes. Cell Discov (2016) 2:16002. doi: 10.1038/celldisc.2016.2
19. Chambraud B, Radanyi C, Camonis JH, Rajkowski K, Schumacher M, Baulieu EE. Immunophilins, Refsum Disease, and Lupus Nephritis: The Peroxisomal Enzyme Phytanoyl-COA Alpha-Hydroxylase Is a New FKBP-Associated Protein. Proc Natl Acad Sci USA (1999) 96:2104–9. doi: 10.1073/pnas.96.5.2104
20. Xiong H, Chen Z, Chen W, Li Q, Lin B, Jia Y. FKBP-Related ncRNA-mRNA Axis in Breast Cancer. Genomics (2020) 112:4595–607. doi: 10.1016/j.ygeno.2020.08.017
21. Xiong H, Chen Z, Zheng W, Sun J, Fu Q, Teng R, et al. FKBP4 is a Malignant Indicator in Luminal A Subtype of Breast Cancer. J Cancer (2020) 11:1727–36. doi: 10.7150/jca.40982
22. Argentieri MA, Nagarajan S, Seddighzadeh B, Baccarelli AA, Shields AE. Epigenetic Pathways in Human Disease: The Impact of DNA Methylation on Stress-Related Pathogenesis and Current Challenges in Biomarker Development. EBioMedicine (2017) 18:327–50. doi: 10.1016/j.ebiom.2017.03.044
23. Vilasco M, Communal L, Mourra N, Courtin A, Forgez P, Gompel A. Glucocorticoid Receptor and Breast Cancer. Breast Cancer Res Treat (2011) 130:1–10. doi: 10.1007/s10549-011-1689-6
24. Ruiz-Conca M, Gardela J, Martínez CA, Wright D, López-Bejar M, Rodríguez-Martínez H, et al. Natural Mating Differentially Triggers Expression of Glucocorticoid Receptor (NR3C1)-Related Genes in the Preovulatory Porcine Female Reproductive Tract. Int J Mol Sci (2020) 21:4437–53. doi: 10.3390/ijms21124437
25. Salehi B, Fokou PVT, Sharifi-Rad M, Zucca P, Pezzani R, Martins N, et al. The Therapeutic Potential of Naringenin: A Review of Clinical Trials. Pharmaceut (Basel Switzerland) (2019) 12:11–28. doi: 10.3390/ph12010011
26. Olagnier D, Brandtoft AM, Gunderstofte C, Villadsen NL, Krapp C, Thielke AL, et al. Nrf2 Negatively Regulates STING Indicating a Link Between Antiviral Sensing and Metabolic Reprogramming. Nat Commun (2018) 9:3506. doi: 10.1038/s41467-018-05861-7
27. Zhang Z, Zhou C, Li X, Barnes SD, Deng S, Hoover E, et al. Loss of CHD1 Promotes Heterogeneous Mechanisms of Resistance to AR-Targeted Therapy via Chromatin Dysregulation. Cancer Cell (2020) 37:584–98.e11. doi: 10.1016/j.ccell.2020.03.001
28. Omasits U, Ahrens CH, Müller S, Wollscheid B. Protter: Interactive Protein Feature Visualization and Integration With Experimental Proteomic Data. Bioinf (Oxford England) (2014) 30:884–6. doi: 10.1093/bioinformatics/btt607
29. Jézéquel P, Gouraud W, Ben Azzouz F, Guérin-Charbonnel C, Juin PP, Lasla H, et al. Bc-GenExMiner 4.5: New Mining Module Computes Breast Cancer Differential Gene Expression Analyses. Database: The Journal of Biological Databases and Curation. Database (Oxford) (2021) 7:1–10. doi: 10.1093/database/baab007
30. Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW, Wu WC, et al. TISIDB: An Integrated Repository Portal for Tumor-Immune System Interactions. Bioinf (Oxford England) (2019) 35:4200–2. doi: 10.1093/bioinformatics/btz210
31. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res (2017) 77:e108–10. doi: 10.1158/0008-5472.CAN-17-0307
32. Zhang L, He H, Zhang M, Wu Y, Xu X, Yang M, et al. Assessing the Effect and Related Mechanism of Naringenin on the Proliferation, Osteogenic Differentiation and Endothelial Differentiation of Human Periodontal Ligament Stem Cells. Biochem Biophys Res Commun (2021) 534:337–42. doi: 10.1016/j.bbrc.2020.11.081
33. Messeguer X, Escudero R, Farre D, Nunez O, Martinez J, Alba MM. PROMO: Detection of Known Transcription Regulatory Elements Using Species-Tailored Searches. Bioinf (Oxford England) (2002) 18:333–4. doi: 10.1093/bioinformatics/18.2.333
34. Farre D, Roset R, Huerta M, Adsuara JE, Rosello L, Alba MM, et al. Identification of Patterns in Biological Sequences at the ALGGEN Server: PROMO and MALGEN. Nucleic Acids Res (2003) 31:3651–3. doi: 10.1093/nar/gkg605
35. Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING V11: Protein-Protein Association Networks With Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res (2019) 47:D607–13. doi: 10.1093/nar/gky1131
36. Assimon VA, Southworth DR, Gestwicki JE. Specific Binding of Tetratricopeptide Repeat Proteins to Heat Shock Protein 70 (Hsp70) and Heat Shock Protein 90 (Hsp90) Is Regulated by Affinity and Phosphorylation. Biochemistry (2015) 54:7120–31. doi: 10.1021/acs.biochem.5b00801
37. Kimmins S, MacRae TH. Maturation of Steroid Receptors: An Example of Functional Cooperation Among Molecular Chaperones and Their Associated Proteins. Cell Stress Chaperones (2000) 5:76–86. doi: 10.1379/1466-1268(2000)005<0076:MOSRAE>2.0.CO;2
38. Li Q, Liu S, Yang G, Li M, Qiao P, Xue Q. Naringenin Inhibits Autophagy and Epithelial-Mesenchymal Transition of Human Lens Epithelial Cells by Regulating the Smad2/3 Pathway. Drug Dev Res (2021). doi: 10.1002/ddr.21868
39. Pateliya B, Burade V, Goswami S. Combining Naringenin and Metformin With Doxorubicin Enhances Anticancer Activity Against Triple-Negative Breast Cancer In Vitro and In Vivo. Eur J Pharmacol (2021) 891:173725. doi: 10.1016/j.ejphar.2020.173725
40. Miles EA, Calder PC. Effects of Citrus Fruit Juices and Their Bioactive Components on Inflammation and Immunity: A Narrative Review. Front Immunol (2021) 12:712608. doi: 10.3389/fimmu.2021.712608
41. Wang L, Zeng W, Wang L, Wang Z, Yin X, Qin Y, et al. Naringenin Enhances the Antitumor Effect of Therapeutic Vaccines by Promoting Antigen Cross-Presentation. J Immunol (Baltimore Md (2020) 1950) 204:622–31. doi: 10.4049/jimmunol.1900278
42. Saha S, Buttari B, Panieri E, Profumo E, Saso L. An Overview of Nrf2 Signaling Pathway and Its Role in Inflammation. Molecules (Basel Switzerland) (2020) 25:5474–504. doi: 10.3390/molecules25225474
43. Tan CT, Chang HC, Zhou Q, Yu C, Fu NY, Sabapathy K, et al. MOAP-1-Mediated Dissociation of P62/SQSTM1 Bodies Releases Keap1 and Suppresses Nrf2 Signaling. EMBO Rep (2021) 22:e50854. doi: 10.15252/embr.202050854
44. Liu Y, Tao S, Liao L, Li Y, Li H, Li Z, et al. TRIM25 Promotes the Cell Survival and Growth of Hepatocellular Carcinoma Through Targeting Keap1-Nrf2 Pathway. Nat Commun (2020) 11:348. doi: 10.1038/s41467-019-14190-2
45. Ghartey-Kwansah G, Li Z, Feng R, Wang L, Zhou X, Chen FZ, et al. Comparative Analysis of FKBP Family Protein: Evaluation, Structure, and Function in Mammals and Drosophila Melanogaster. BMC Dev Biol (2018) 18:7. doi: 10.1186/s12861-018-0167-3
46. Tong J, Shen Y, Chen X, Wang R, Hu Y, Zhang X, et al. FKBP3 Mediates Oxaliplatin Resistance in Colorectal Cancer Cells by Regulating HDAC2 Expression. Oncol Rep (2019) 42(4):1404–12. doi: 10.3892/or.2019.7259
47. Blair LJ, Baker JD, Sabbagh JJ, Dickey CA. The Emerging Role of Peptidyl-Prolyl Isomerase Chaperones in Tau Oligomerization, Amyloid Processing, and Alzheimer’s Disease. J Neurochem (2015) 133:1–13. doi: 10.1111/jnc.13033
48. Abdelmegeid MK, Vailati-Riboni M, Alharthi A, Batistel F, Loor JJ. Supplemental Methionine, Choline, or Taurine Alter In Vitro Gene Network Expression of Polymorphonuclear Leukocytes From Neonatal Holstein Calves. J Dairy Sci (2017) 100:3155–65. doi: 10.3168/jds.2016-12025
49. Smith AJ, Li Q, Wietgrefe SW, Schacker TW, Reilly CS, Haase AT. Host Genes Associated With HIV-1 Replication in Lymphatic Tissue. J Immunol (Baltimore Md: 1950) (2010) 185:5417–24. doi: 10.4049/jimmunol.1002197
50. Gassen NC, Hartmann J, Zschocke J, Stepan J, Hafner K, Zellner A, et al. Association of FKBP51 With Priming of Autophagy Pathways and Mediation of Antidepressant Treatment Response: Evidence in Cells, Mice, and Humans. PloS Med (2014) 11:e1001755. doi: 10.1371/journal.pmed.1001755
51. Bhujabal Z, Birgisdottir Å B, Sjøttem E, Brenne HB, Øvervatn A, Habisov S, et al. FKBP8 Recruits LC3A to Mediate Parkin-Independent Mitophagy. EMBO Rep (2017) 18:947–61. doi: 10.15252/embr.201643147
52. Kudo Y, Sugimoto M, Arias E, Kasashima H, Cordes T, Linares JF, et al. Pkcλ/ι Loss Induces Autophagy, Oxidative Phosphorylation, and NRF2 to Promote Liver Cancer Progression. Cancer Cell (2020) 38:247–62.e11. doi: 10.1016/j.ccell.2020.05.018
53. Han Y, Fan S, Qin T, Yang J, Sun Y, Lu Y, et al. Role of Autophagy in Breast Cancer and Breast Cancer Stem Cells (Review). Int J Oncol (2018) 52:1057–70. doi: 10.3892/ijo.2018.4270
54. Zambrano J, Yeh ES. Autophagy and Apoptotic Crosstalk: Mechanism of Therapeutic Resistance in HER2-Positive Breast Cancer. Breast Cancer: Basic Clin Res (2016) 10:13–23. doi: 10.4137/BCBCR.S32791
55. Gao J, Fan M, Peng S, Zhang M, Xiang G, Li X, et al. Small-Molecule RL71-Triggered Excessive Autophagic Cell Death as a Potential Therapeutic Strategy in Triple-Negative Breast Cancer. Cell Death Dis (2017) 8:e3049. doi: 10.1038/cddis.2017.444
56. Romero MA, Bayraktar Ekmekcigil O, Bagca BG, Avci CB, Sabitaliyevich UY, Zhenisovna TG, et al. Role of Autophagy in Breast Cancer Development and Progression: Opposite Sides of the Same Coin. Adv Exp Med Biol (2019) 1152:65–73. doi: 10.1007/978-3-030-20301-6_5
57. Oshi M, Angarita FA, Tokumaru Y, Yan L, Matsuyama R, Endo I, et al. High Expression of NRF2 Is Associated With Increased Tumor-Infiltrating Lymphocytes and Cancer Immunity in ER-Positive/HER2-Negative Breast Cancer. Cancers (Basel) (2020) 12:3856–70. doi: 10.3390/cancers12123856
58. Lee DH, Han DH, Nam KT, Park JS, Kim SH, Lee M, et al. Ezetimibe, an NPC1L1 Inhibitor, is a Potent Nrf2 Activator That Protects Mice From Diet-Induced Nonalcoholic Steatohepatitis. Free Radical Biol Med (2016) 99:520–32. doi: 10.1016/j.freeradbiomed.2016.09.009
59. Shimozono R, Asaoka Y, Yoshizawa Y, Aoki T, Noda H, Yamada M, et al. Nrf2 Activators Attenuate the Progression of Nonalcoholic Steatohepatitis-Related Fibrosis in a Dietary Rat Model. Mol Pharmacol (2013) 84:62–70. doi: 10.1124/mol.112.084269
60. Reisman SA, Chertow GM, Hebbar S, Vaziri ND, Ward KW, Meyer CJ. Bardoxolone Methyl Decreases Megalin and Activates Nrf2 in the Kidney. J Am Soc Nephrol: JASN (2012) 23:1663–73. doi: 10.1681/ASN.2012050457
61. Shi X, Li Y, Hu J, Yu B. Tert-Butylhydroquinone Attenuates the Ethanol-Induced Apoptosis of and Activates the Nrf2 Antioxidant Defense Pathway in H9c2 Cardiomyocytes. Int J Mol Med (2016) 38:123–30. doi: 10.3892/ijmm.2016.2605
62. Hermawan A, Ikawati M, Jenie RI, Khumaira A, Putri H, Nurhayati IP, et al. Identification of Potential Therapeutic Target of Naringenin in Breast Cancer Stem Cells Inhibition by Bioinformatics and In Vitro Studies. Saudi Pharm J: SPJ: Off Publ Saudi Pharm Soc (2021) 29:12–26. doi: 10.1016/j.jsps.2020.12.002
63. Zhong J, Yu R, Zhou Q, Liu P, Liu Z, Bian Y. Naringenin Prevents TNF-α-Induced Gut-Vascular Barrier Disruption Associated With Inhibiting the NF-κb-Mediated MLCK/p-MLC and NLRP3 Pathways. Food Funct (2021) 12:2715–25. doi: 10.1039/D1FO00155H
Keywords: FKBP4, NRF2, NR3C1, autophagy, Dendritic cell, Breast cancer
Citation: Xiong H, Chen Z, Lin B, Xie B, Liu X, Chen C, Li Z, Jia Y, Wu Z, Yang M, Jia Y, Wang L, Zhou J and Meng X (2022) Naringenin Regulates FKBP4/NR3C1/NRF2 Axis in Autophagy and Proliferation of Breast Cancer and Differentiation and Maturation of Dendritic Cell. Front. Immunol. 12:745111. doi: 10.3389/fimmu.2021.745111
Received: 21 July 2021; Accepted: 21 October 2021;
Published: 11 January 2022.
Edited by:
Mohammad Hojjat-Farsangi, Karolinska Institutet (KI), SwedenReviewed by:
Zhou Xunian, MD Anderson Cancer Center, United StatesBlanca Estela Garcia-Perez, Instituto Politécnico Nacional (IPN), Mexico
Daniela Laura Papademetrio, University of Buenos Aires, Argentina
Copyright © 2022 Xiong, Chen, Lin, Xie, Liu, Chen, Li, Jia, Wu, Yang, Jia, Wang, Zhou and Meng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hanchu Xiong, MTE3MTgyNjZAemp1LmVkdS5jbg==; Xuli Meng, bXhsbWFpbEAxMjYuY29t
†These authors have contributed equally to this work
 Hanchu Xiong
Hanchu Xiong Zihan Chen2†
Zihan Chen2† Cong Chen
Cong Chen Zhaoqing Li
Zhaoqing Li Linbo Wang
Linbo Wang Jichun Zhou
Jichun Zhou