
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 05 November 2021
Sec. Viral Immunology
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.744242
 Fuxing Lou1†
Fuxing Lou1† Maochen Li1*†
Maochen Li1*† Zehan Pang1†
Zehan Pang1† Lin Jiang1†
Lin Jiang1† Lin Guan1†
Lin Guan1† Lili Tian1
Lili Tian1 Jiaming Hu2
Jiaming Hu2 Junfen Fan3*
Junfen Fan3* Huahao Fan1*
Huahao Fan1*The global pandemic of the coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), places a heavy burden on global public health. Four SARS-CoV-2 variants of concern including B.1.1.7, B.1.351, B.1.617.2, and P.1, and two variants of interest including C.37 and B.1.621 have been reported to have potential immune escape, and one or more mutations endow them with worrisome epidemiologic, immunologic, or pathogenic characteristics. This review introduces the latest research progress on SARS-CoV-2 variants of interest and concern, key mutation sites, and their effects on virus infectivity, mortality, and immune escape. Moreover, we compared the effects of various clinical SARS-CoV-2 vaccines and convalescent sera on epidemic variants, and evaluated the neutralizing capability of several antibodies on epidemic variants. In the end, SARS-CoV-2 evolution strategies in different transmission stages, the impact of different vaccination strategies on SARS-CoV-2 immune escape, antibody therapy strategies and COVID-19 epidemic control prospects are discussed. This review will provide a systematic and comprehensive understanding of the secret of SARS-CoV-2 variants of interest/concern and immune escape.
The coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), broke out in Wuhan, China in December 2019. Since then this epidemic has been spread and intensified internationally and defined as a Public Health Emergency of International Concern (PHEIC) by the World Health Organization (WHO) on January 30, 2020. As of September 26, 2021, 231,614,338 cases of COVID-19 have been confirmed, including 4,744,918 deaths caused by SARS-CoV-2-induced inflammatory infections or other complications [https://coronavirus.jhu.edu/map.html].
SARS-CoV-2 is a single-stranded RNA virus, typical symptoms of infected patients are fever, cough, chest discomfort, and respiratory distress syndrome (RDS) often occurs in severe cases (1). The inherent error-prone characteristics of viral RNA-dependent RNA polymerase (RdRp) result in the random introduction of mutations in the viral genome during replication. Although the virus encodes an exonuclease (ExoN, nsp14) with a proofreading function, it cannot eliminate the occurrence of viral mutations (2, 3). With continued spreading and viral replication, chronic infection will increase the possibility of virus adaptive mutation (4, 5). The multiple emerging mutations of SARS-CoV-2 variants confer worrisome epidemiologic, immunologic, or pathogenic characteristics (6). WHO developed a Variant Classification scheme, and the categories of variants of concern (VOC) and variants of interest (VOI) were proposed. VOC refers to the SARS-CoV-2 variants that can increase the transmissibility or cause detrimental change in COVID-19 epidemiology by strongly impairing the effectiveness of vaccines and neutralizing antibodies (Table 1), or can cause more serious disease conditions. VOI means the SARS-CoV-2 variants that harbor mutations which are predictable or known to affect viral characteristics, such as infectivity, disease severity, immune escape, or showing a sudden risk to global public health security [https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/]. At present, there are mainly four kinds of VOC: B.1.1.7 (Alpha, originated in the United Kingdom), B.1.351 (Beta, originated in South Africa), P.1 (Gamma, originated in Brazil), and B.1.617.2 (Delta, originated in India) (Figures 1, 2). VOI mainly includes C.37 (Lambda, first detected in Peru) and B.1.621 (Mu, first detected in Colombia) (Figures 3, 4) [https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/].
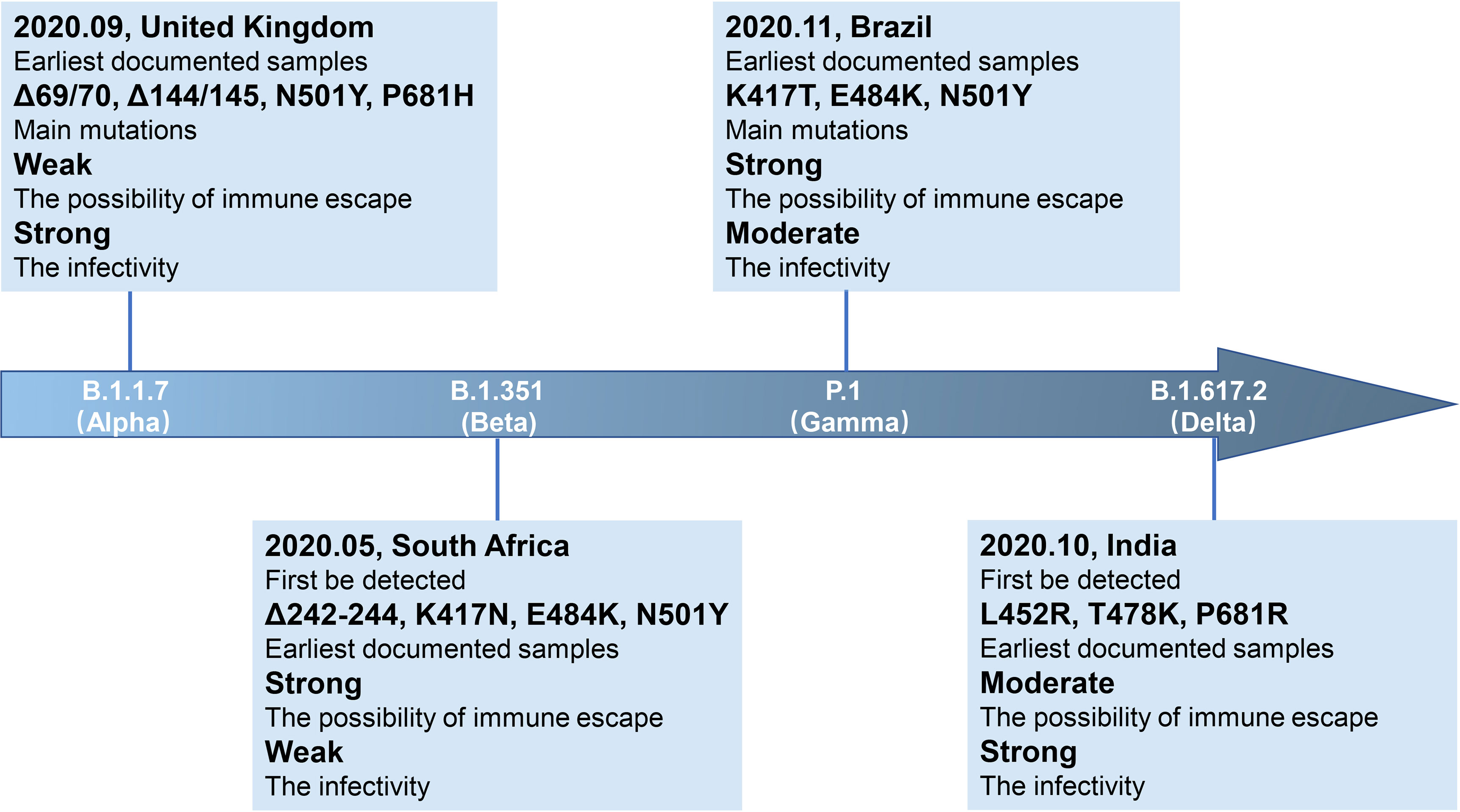
Figure 1 Brief information of four VOC variants. Four VOC variants (B.1.1.7, B.1.351, P.1, and B.1.617.2) are marked in the arrow according to the date of designation, and their related brief information (e.g., the time and location of earliest documented samples, infectivity, main mutations, immune escape ability) are displayed in the corresponding location.
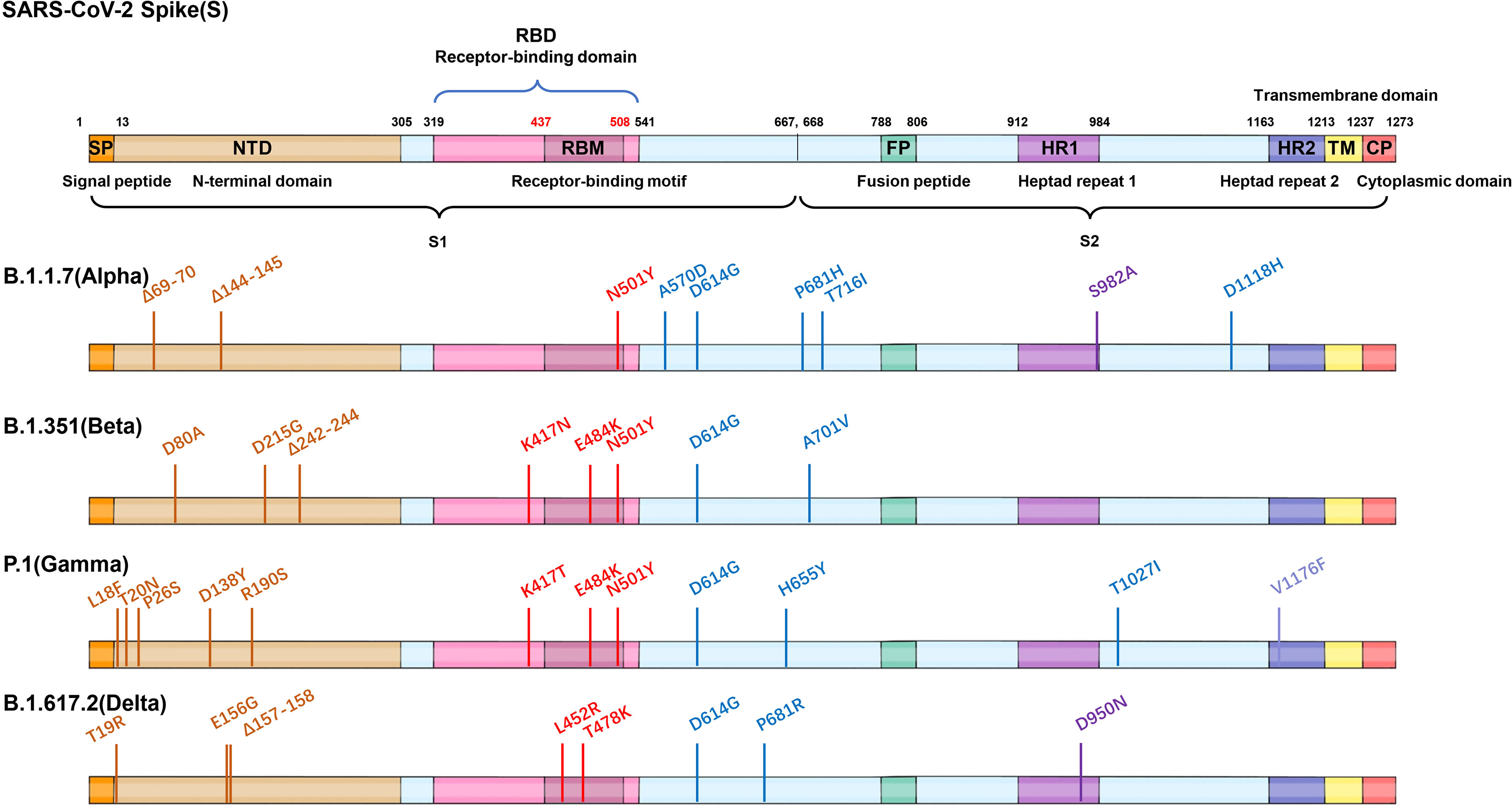
Figure 2 Mutations of four VOC variants. Schematic showing the locations of amino acid substitutions of four VOCs (B.1.1.7, B.1.351, P.1, and B.1.617.2) in spike protein. The RBD region is shown in modena, the NTD region is shown in shallow orange.
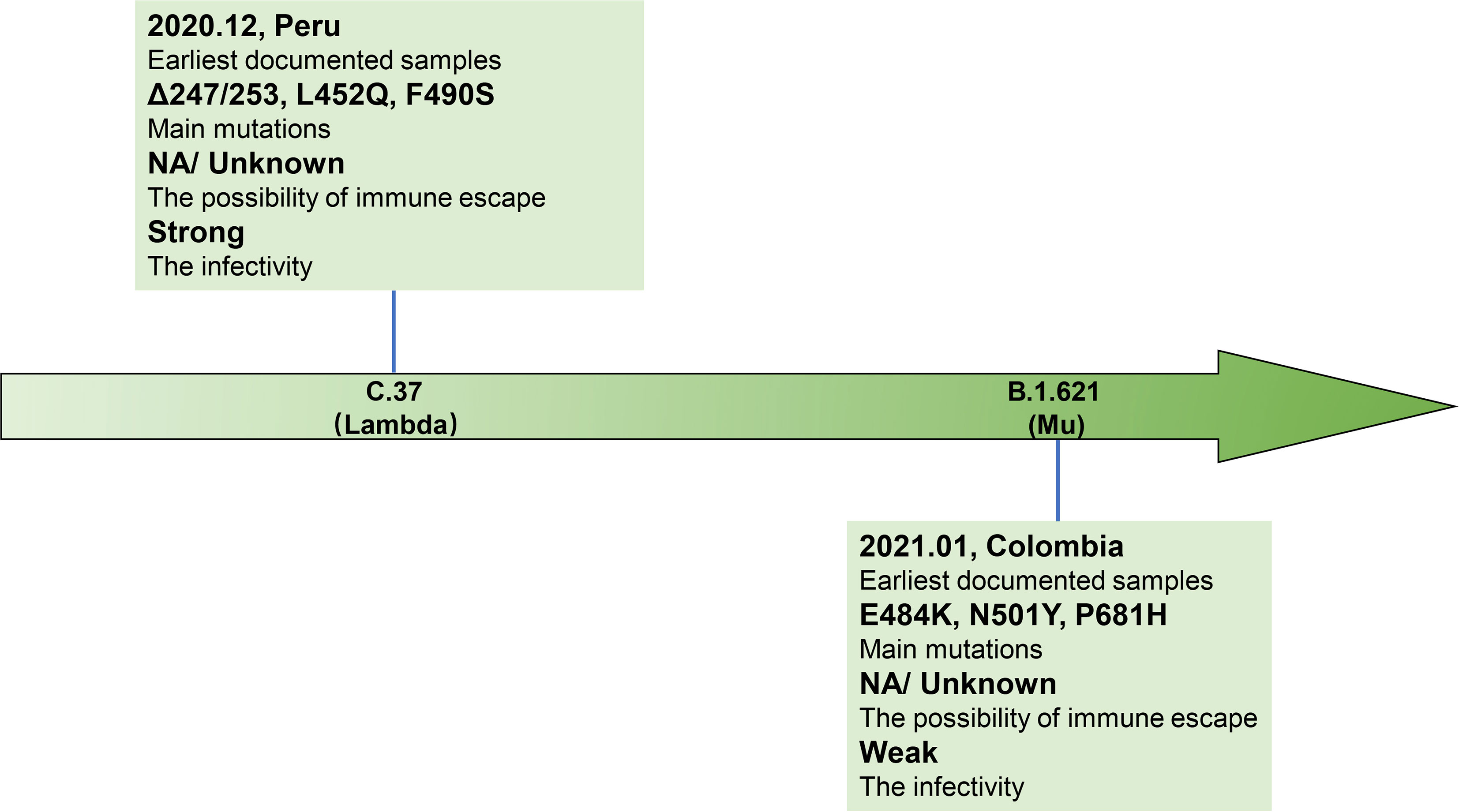
Figure 3 Brief information of two VOI variants. Two VOI variants (C.37 and B.1.621) are marked in the arrow according to the date of designation, and their related brief information (e.g., the time and location of earliest documented samples, infectivity, main mutations, immune escape ability) are displayed in the corresponding location.
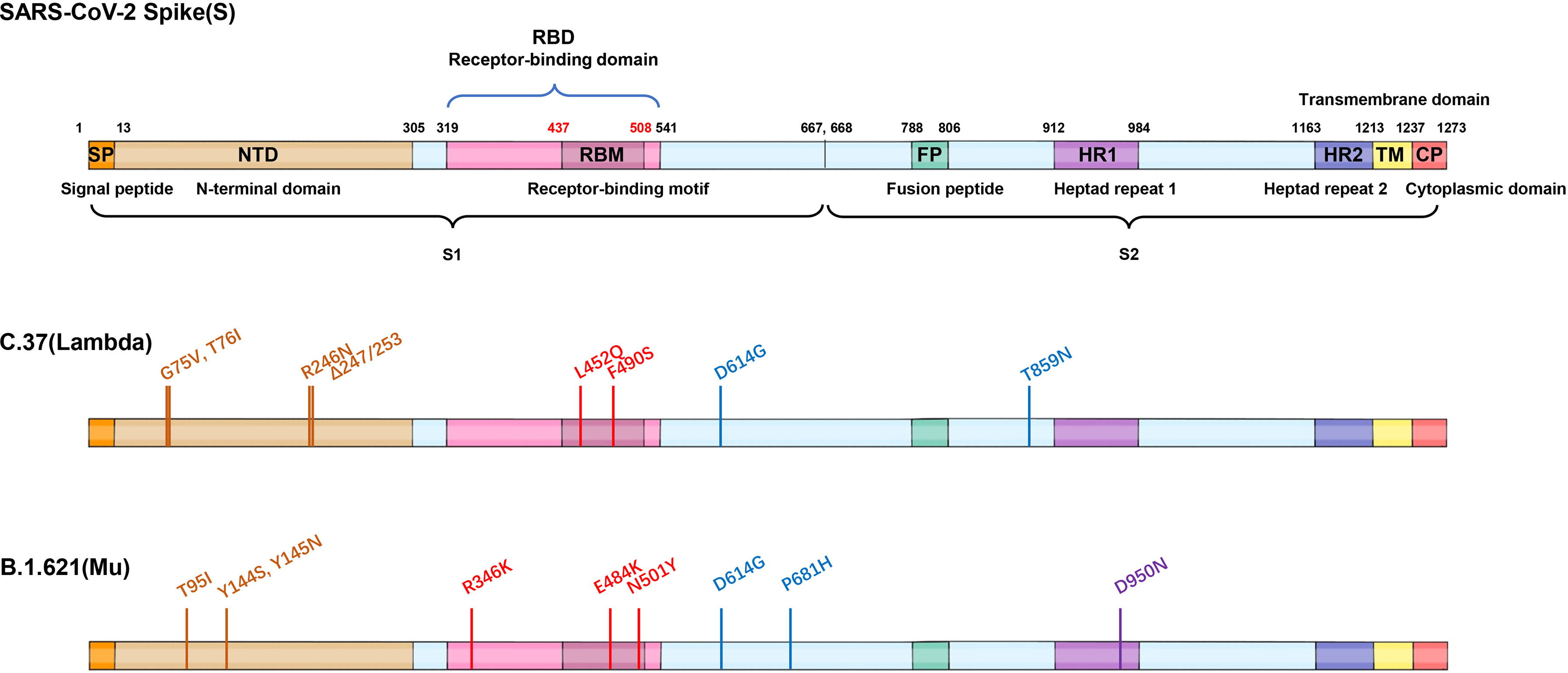
Figure 4 Mutations of two VOI variants. Schematic showing the locations of amino acid substitutions of two VOI variants (C.37 and B.1.621) in spike protein. The RBD region is shown in modena, the NTD region is shown in shallow orange.
Variants Under Monitoring (VUM) means a SARS-CoV-2 variant with specific mutations that affect virus characteristics, and may pose a potential threat in the future, but there is no definitive evidence of phenotypic or epidemiological impact at present [https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/]. Currently designated VUMs include B.1.427/B.1.429 (first detected in California, USA), R.1 (first detected in several countries in January 2021), B.1.466.2 (first detected in Indonesia), B.1.1.318 (detected in multiple countries in January 2021), B.1.1.519 (detected in several countries in January 2021), C.36.3 (detected in several countries in January 2021), B.1.214.2 (detected in several countries in November 2020), B.1.1.523 (detected in several countries in May 2020), B.1.619 (detected in several countries in May 2020), B.1.620 (detected in several countries in November 2020), C.1.2 (first detected in South Africa), B.1.525 (Eta, detected in several countries in December 2020), B.1.526 (Iota, first detected in the United States), and B.1.617.1 (Kappa, first detected in India) [https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/]. Some former VOIs but no longer designated as VUMs include P.2 (first detected in Brazil) and P.3 (first detected in the Philippines) (Supplementary Figures 1, 2) [Variants: distribution of cases data, 20 May 2021-GOV.UK (www.gov.uk)].
In this article, we provide a systematic and comprehensive summary of the key genetic variants of SARS-CoV-2 and elucidate the impacts of pivotal mutations on viral transmissibility, infectivity, and immune escape of vaccines and antibodies.
B.1.1.7 is the earliest prevalent variant, which was originally identified in the United Kingdom in September 2020 (18), and has a significant transmission superiority with a higher reproduction number (R) than non-VOC lineages (19). On December 18, 2020, it was designated by the Public Health England (PHE) as a VOC lineage [https://researchportal.phe.gov.uk/en/]. This variant with 10 mutations in spike (S) protein (20, 21) can be divided into three subgroups. The major strain contains Δ69-70, and the other two subgroups lack this deletion, which indicated that Δ69-70 obtained selective advantages in the variation process of SARS-CoV-2 (22). As of January 13, 2021, there were 76 confirmed cases of B.1.1.7 in 12 states in the United States (23), and this variant has been circulating in 174 countries so far, indicating that B.1.1.7 is highly contagious [https://cov-lineages.org/]. According to the survey of PHE, B.1.1.7 would lead to a 30%-50% increase in secondary attack rate (18). In some studies, convalescent plasma and vaccine sera were applied in B.1.1.7 neutralization assays, and no widespread immune escape was observed (22). Pseudovirus neutralization assay showed that the neutralization of BNT162b2-immune sera against B.1.1.7 was largely preserved, indicating that the variants have difficulty escaping from vaccine-mediated immune protection (20). Wang et al. evaluated the neutralization effect of two vaccines BBIBP-CorV (Sinopharm) and CoronaVac (Sinovac) developed in China against B.1.1.7. The results showed that compared with wildtype (WT), the BBIBP-CorV-immune serum remained neutralizing potency to B.1.1.7, but the geometric mean titers (GMTs) of CoronaVac-immune serum against B.1.1.7 decreased significantly (24). The above studies suggested that B.1.1.7 did not pose a great threat to the protective efficacy of COVID-19 vaccines. However, B.1.1.7 variant infection was related to higher virus titer in nasopharyngeal swabs, which accounted for the increased mortality (25). Therefore, the necessity of continuous SARS-CoV-2 sequence surveillance should be highlighted.
B.1.351 was discovered in South Africa in May 2020, and then expanded rapidly to become the dominant lineage in South Africa. It was related to the sharp increase of infected cases nationwide in mid-December, which strongly indicated that this variant had a selective advantage (26). Through the analysis of virus sequencing, it was found that B.1.351 also has three subgroups: 501Y.V2-1, 501Y.V2-2, and 501Y.V2-3. Compared with the receptor binding domain (RBD) and N terminal domain (NTD) sequences of the other two subgroups, 501Y.V2-3 contains R246I mutation and lacks L18F mutation, implying the evolution of SARS-CoV-2. The RBD of B.1.351 harbors three notable mutation sites: K417N, E484K and N501Y, and the combined effect of these three mutations could enhance the affinity of viral spike protein to ACE2 (27). A previous study showed that 21 of 44 convalescent plasma samples lost neutralizing activity against B.1.351 (28). Pseudovirus neutralization assay confirmed that 12 of 17 monoclonal antibodies were ineffective against B.1.351 (29). Compared with WT, the 50% plaque reduction neutralization titer (PRNT50) of 14 convalescent serum against live B.1.351 virus decreased by 3.2- to 41.9-fold (30). The impaired efficacy of vaccine-immune sera including mRNA-1273 (Modena) and BNT162b2 (Pfizer) against 501Y.V2 was demonstrated (11), and 20 of 25 BBIBP-CorV (Sinopharm) vaccinated serum samples showed complete or partial neutralization loss against B.1.351, and the CoronaVac vaccinated sera showed a significant decrease of GMTs against B.1.351, accompanied by the complete or partial neutralization loss of most samples (24). The efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against symptomatic infection caused by B.1.351 was only 10.4%, which directly led to the suspension of the ChAdOx1 nCoV-19 vaccine in South Africa (12). The neutralizing activity against B.1.351 of three potent monoclonal antibodies (2-15, LY-CoV555, and REGN10933) approved for emergency use also decreased significantly (11). These studies raised concerns about the efficacy of vaccines and antibodies against B.1.351. Interestingly, a study carried out in South Africa showed that the convalescent serum from B.1.351-infected patients retained the neutralization activity against the virus in the first-wave epidemic with only a 2.3-fold decrease (30). However, the serum obtained from the first wave of the epidemic infection could not effectively neutralize B.1.351 (30). These findings suggested that the sera from individuals infected with B.1.351 possess cross-neutralization activity to other variants, and the antibodies elicited by the variants with stronger immune escape ability may have more extensive neutralization ability (Figure 5). On the other hand, in the presence of cross-neutralization activity, the neutralization efficacy of antibodies induced by original strains against other strains still decreased to some extent. Therefore, the existence of the cross-neutralization provided an idea for vaccine optimization. Universal vaccines using variants in the epidemic as seed strains against multiple variants could quickly and effectively restrain the spread of the epidemic (21).
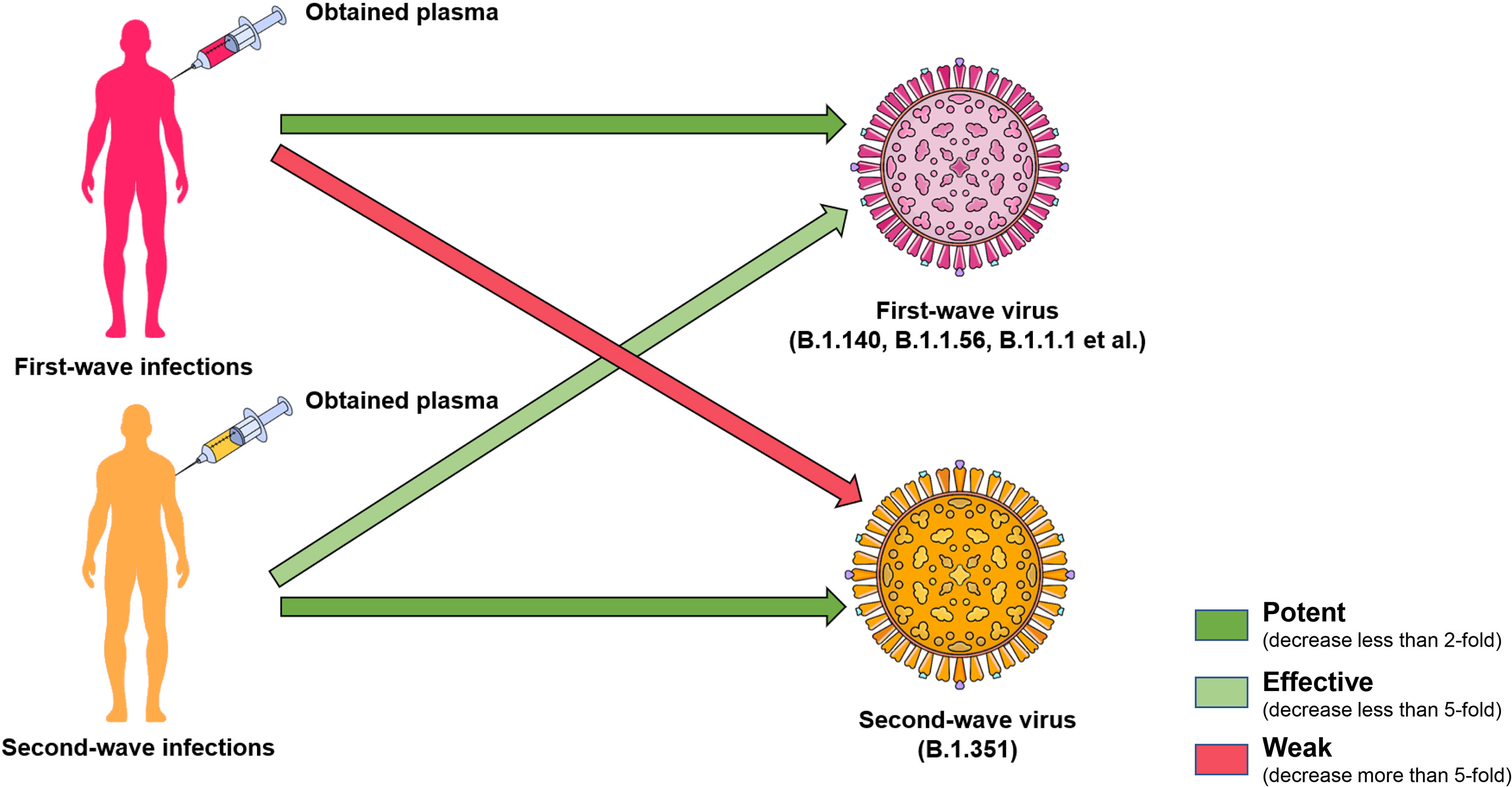
Figure 5 Cross neutralization of convalescent plasma. The cross neutralization of convalescent serum has been noticed by the neutralization activity test. The convalescent serum from B.1.351-infected patients retained the neutralization against the virus in the first-wave epidemic in South Africa. However, the serum obtained from the first-wave of epidemic infection could not effectively neutralize B.1.351. The green arrow indicates that the neutralization still remains, and the light green means a worse neutralization effect. Pink indicates a significant decrease in neutralization efficacy.
In November 2020, a second wave of COVID-19 epidemic broke out in Brazil, causing 76% of the population to be infected (31), which was similar to the epidemic in South Africa. After virus sequence analysis, the variant was named P.1. The RBDs of P.1 and B.1.351 contain mutations in site 417, 484, and 501 residues, except that P.1 harbors K417T, B.1.351 harbors K417N. In addition, the L18F mutation located in the NTD was shown to be related to immune escape potentiality (32). Because of its strong infectivity, P.1 was designated as VOC by WHO on January 11, 2021 (20, 33). The transmissibility of P.1 could be 1.7-fold to 2.4-fold higher than that of non-P.1 lineages, and the convalescent serum from non-P.1 SARS-CoV-2 infected patients could only provide 54%-79% protection against the infection of P.1 (16). According to data from WHO, a variant named P.2 with E484K mutation was originally sequenced in Brazil in April 2020 [https://www.who.int]. P.2 variant showed significant resistance to the vaccinated serum, as the neutralization efficacy of BNT162b2 (Pfizer) against the Brazilian/Japanese P.2 strain decreased 5.8-fold and mRNA-1273 (Moderna) decreased 2.9-fold. Similarly, the vaccine neutralizing activity against the Brazilian/Japanese P.1 strain also decreased significantly (6.7-fold for BNT162b2 and 4.5-fold for mRNA-1273) (17). Few studies have been published on P.2 since it did not cause large-scale outbreaks in Brazil and other countries, but further analysis of the sequence of P.1 and P.2 may reveal the evolution of the virus. Another SARS-CoV-2 variant named P.3 was reported in the Philippines in March 2021. This lineage has several notable mutations in the S protein, including E484K, N501Y, and P681H (Supplementary Figures 1, 2). P.3 as well as P.1 belong to B.1.1.28 lineage (https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/).
With increased infectivity and virulence, a new variant with an L452R mutation in S protein appeared in California in March 2020, which was named B.1.427/429. Since October 2020, a peak of the COVID-19 epidemic occurred in southern California. According to the sequence analysis of local strains, it was found that most of the variants came from clade 20C (34), which first emerged in Europe and then mutated in Britain and other places, becoming the most widely distributed variant B.1.1.7. From September 2020 to January 2021, 2172 nasal/nasopharyngeal swabs from 44 counties in California were sequenced and it was found that the B.1.427/429 positive sample ratio increased from 0% to more than 50% (34). There are four missense mutations in the S protein of B.1.427/B.1.429, including L452R, S13I, W152C, and D614G, among which the L452R mutation is located in the RBD region (Supplementary Figures 1, 2). It was found that the virus shedding amount of this variant in vivo was 2-fold higher than that of WT, and the production of pseudovirus containing L452R in cell culture and lung tissue also increased, but was lower than that of B.1.1.7, B.1.351, and P.1, which showed that this variant had higher infectious ability (35). Furthermore, B.1.427/B.1.429 had a certain immune escape ability, which was demonstrated by the significantly reduced neutralization ability of plasma from Pfizer/BioNTech BNT162b2 or Moderna mRNA-1273 vaccinated participants and impaired effects of convalescent serum from patients against B.1.427/B.1.429 (36). Worryingly, more mutations may be accumulated on the basis of B.1.427/B.1.429 in the future, which would further increase the possibility of immune escape (37).
A previous VOI, currently designated VUM variant B.1.526 was first identified in the New York region in November 2020, and began to spread at an alarming rate (38). The most significant mutations in the spike of this lineage are L5F, T95I, D253G or S477N, and D614G (39) (Supplementary Figure 2). Preliminary data analyzed by the New York City Department of Health and Mental Hygiene (DOHMH) suggested that the B.1.526 variant was not associated with an increased risk of breakthrough infection or reinfection after vaccination and with more serious disease conditions (38). The antibodies elicited by infection and vaccines (Pfizer BNT162b2 and Moderna mRNA-1273) could retain the complete neutralization titer against B.1.526 harboring S477N, but the neutralization was 3.5-fold lower against B.1.526 harboring E484K than that of D614G strain (40). In addition, the titer of E484K neutralized by REGN10933 monoclonal antibody decreased by 12-fold, but the neutralization activity of its combined cocktail with REGN10987 against B.1.526 was completely retained (40). Other studies also evaluated the resistance of B.1.526 variant to neutralizing antibodies and ACE2 blocking antibodies induced by the mRNA-1273 vaccine within 7 months. These results suggest that current vaccines still retained neutralization ability for B.1.526 and the variant did not show widespread immune escape (41).
B.1.525, also known as 20A/S: 484K, was discovered and expanded rapidly in many countries in December 2020 (https://covariants.org/variants/20A.S.484K). Phylogenetic analysis showed that the lineage originated from Nigeria (42). Although it did not circulate all over the world, it was defined as the lineage of international significance (43) and was classified as VUM for the possibility of increasing infectivity, virulence and reducing the effectiveness of the vaccine by notable mutations carried by the variant (https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/). Eight mutations (D614G, Q677H, E484K, F888L, A67V, Δ69/70, Δ144/145, and Q52R) in the Spike protein of B.1.525 were identified (Supplementary Figure 2). Q677H harbored by B.1.525 can regulate the transmissibility (42). E484K exists in B.1.351, P.1, and P.2 variants and is related to immune escape. Δ69/70 and Δ144/145 were detected in B.1.1.7, Δ69/70 was shown to have a selective advantage (42), and 144/145 site was verified to be a binding epitope of multiple antibodies targeting NTD (36). The average viral load of the upper respiratory tract between this variant and B.1.1.7 infected patients was similar (43). At the same time, B.1.525 was resistant to neutralization of convalescent serum, vaccine-elicited serum, and monoclonal antibodies (44).
As of September 26, 2021, India’s confirmed cases in has reached 33,652,745, becoming the country with the second largest cumulative number of confirmed cases in the world, which was propelled by variant B.1.617 [https://coronavirus.jhu.edu/map.html]. Through genome sequencing of local confirmed cases, it was found that the local epidemic variant B.1.617 harbored E484K, L452R, and P681R mutations, all of which have been present in other epidemic strains. E484K was detected in B.1.351 while the 681 residue was detected as H681 in B.1.1.7. This showed that the B.1.617 may obtain the characteristics of both B.1.1.7 and B.1.351, and it was considered as one of the most concerned epidemic variants (45). With the spread of the local epidemic in India, B.1.617.1, B.1.617.2, and B.1.617.3 strains emerged successively. B.1.617.2, named Delta, was first sequenced in India in October 2020 and then spread globally. It posed a greater threat to global public health security than the other two lineages and was recognized as VOC by WHO on May 11, 2021 [https://www.who.int/publications/]. At present, at least 163 countries were under the shadow of B.1.617.2 [https://outbreak.info/situation-reports]. Under the background of the prevalence of B.1.1.7, B.1.617.2 strain also appeared in the United Kingdom, and gradually increased or even became a dominant strain, indicating that this variant had a significant competitive advantage (46). In May 2021, a local outbreak caused by Delta occurred in Guangzhou, China, where the variant spread four generations in only 10 days, indicating its remarkable infectivity. A research team collected the clinical information of 159 Delta infection cases in Guangzhou, China and analyzed their clinical characteristics and viral dynamics. Compared with the WT strain, the Delta strain harbored a significantly shorter median incubation period (4 days vs. 6 days), and was featured with a higher viral load (median Ct 20.6 vs. 34.0). Moreover, patients with Delta infection were associated with a shorter time of the deterioration to critical illness, a higher risk of critical status and longer period for RNA-negative conversion than WT (47). These clinical data confirmed that Delta carried unique properties that require continuous monitoring and follow-up.
B.1.617.2 with Δ156-157, G158R, L452R, T478K and other mutations is considered to pose great challenges to the effectiveness of vaccines and neutralizing antibodies (48). Compared with the WT, the neutralizing antibody titers (NAbTs) of sera from BNT162b2 recipients against B.1.617.2 reduced by 5.8-fold (49), and the neutralization titer of serum from Pfizer Comirnaty vaccine had a 3-fold decrease against B.1.617.2 compared with B.1.1.7 (14). Serum from participants injected with a single dose of AstraZeneca vaccine almost completely lost neutralizing activity against B.1.617.2. The effective dose 50% (ED50) analysis showed that compared with B.1.1.7, the neutralization titer of patients sera in 6 and 12 months after infection against B.1.617.2 decreased by 4- to 6-fold, respectively (14). In addition, a recent study held by PHE showed that even after two doses of vaccine, recipients could still be infected by B.1.617.2. Among the four clinically approved antibodies Bamlanivimab, Etesevimab, Casirivimab, and Imdevimab, Bamlanivimab lost its neutralization activity against B.1.617.2, which is considered to be caused by the mutation at site L452, while the other three antibodies remained neutralizing activity (14). Therefore, it is still necessary to monitor the efficacy of the vaccines and antibodies against circulating variants.
Recently, a new SARS-CoV-2 variant C.37 had infected more than 80% of the population in Peru (14). C.37 has a similar mutation as B.1.617.2 in 452 site (L452Q) as well as a mutation F490S in the antibody-binding epitopes of RBD, which may reduce the neutralization of partial RBD antibodies. WHO named the variant as Lambda and designated it as a variant of interest in June 2021 [https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/]. Given C.37 may have higher contagious and immune escape ability, it is necessary to take continuous surveillance for the Lambda variant.
The SARS-CoV-2 Mu variant (B.1.621), a new variant of interest classified by WHO on August 30, 2021, has been detected in at least 50 countries, predominantly in Colombia [https://outbreak.info/situation-reports]. In the context of Gamma’s dominance, Mu transcended Gamma within only five months, propelling the epidemic in Colombia, implying the conspicuous infectivity. The Mu variant was first detected on August 1, 2021 with nine mutations (D614G, P681H, R346K, N501Y, E484K, T95I, D950N, Y145N, Y144S) in the Spike protein (Figure 4). The distinctive substitutions distributed in NTD (T95I, Y145N, Y144S), RBD (R346K, N501Y, E484K) and furin cleavage site (P681H) were associated with increased viral infectivity and immune escape ability. Pseudovirus neutralization assay showed that the neutralization titers of BNT162b2-vaccinated sera against Mu strain were reduced by 7.6-fold compared with WT, showing significantly more resistant than other VOCs (2.6-fold against Alpha; 8.2-fold against Beta; 4.1-fold against Gamma; 4.0-fold against Delta) and VOIs (3.4-fold against Lambda), and a similar situation also existed in convalescent sera (50).
SARS-CoV-2 variants appeared frequently all over the world, which aroused great global attention. According to previous studies, climatic factors will change the speed of virus transmission to a certain extent and affect the spread of the epidemic (51). However, with highly contagious characteristics of SARS-CoV-2, it seems that the regulatory role of climate in virus spread is attenuated. If effective control measures are not taken in time, large-scale epidemic may still break out in hot and humid climates (52), which is proved by the emergence of B.1.351 and B.1.617 strains.
Spike protein of SARS-CoV-2 contains N-terminal domain (NTD), receptor binding domain (RBD), and other regions (such as fusion peptide region) (1). The RBD and NTD regions are mainly located on the S1 subunit, while the fusion peptides and other regions are located on the S2 subunit. Based on the sequence analysis of the main epidemic strains (B.1.1.7, B.1.351, P.1, B.1.427/429, and B.1.617), it was found that the main mutations located in the RBD region are K417N/T, N439K, L452R, E484K/Q, and N501Y. The mutations located in the NTD region are mainly as follows: L18F, T20N, P26S, Δ69-70, D80A, D111D, D138Y, G142D, Δ144, W152G, R190S, D215G, and Δ242-244; other regions are A570D, D614G, H655Y, P681H/R, A701V, T716I, S982A, and T1027I (22, 26, 29, 45, 53, 54) (Figures 2, 4).
The N439K mutation located in the receptor binding motif (RBM) was first found in Scotland in March 2020 and spread widely in European countries (4). N439K is thought to enhance the binding of RBD to ACE2 (55) and escape the neutralization of some monoclonal and polyclonal antibodies in the convalescent serum (4). Another amino acid mutation in the RBM region, Y453F, can also enhance the affinity of the virus to ACE2 (55). It is worth noting that the virus in all patients infected with SARS-CoV-2 is associated with minks harboring Y453F strain (56).
The serum analysis of nearly 650 recovered patients with SARS-CoV-2 infection showed that 90% of the neutralizing antibodies targeted the RBD region (57), which may be due to the lack of glycan shielding in the amino acids in the RBD region compared with other regions (58). K417N/T was detected in B.1.351 variant and P.1 variant, and 417 residue was a potential key site for immune escape (4, 28). VH3-53/66 encodes a class of common and effective neutralizing antibodies, and K417N mutation could reduce the affinity of these antibodies to S protein (28, 58, 59). Different from N501Y and E484K, which belong to the same RBD region, no evidence for positive selection of K417N was demonstrated (26). K417N mutation both in B.1.351 and P.1 damaged the affinity between them (4, 55, 60), whereas the E484K and N501Y mutations increased the interaction between S protein and ACE2 (5). Furthermore, the effect of virus mutation on neutralizing antibodies may have a cumulative effect, the more mutation sites, the lower neutralizing response of antibodies (26). Thus, timely and effective prevention and control measures should be taken to restrain the spread of the epidemic and thus reduce the emergence of new variants.
L452R was first found in a novel lineage in California named CAL.20C (34), and then detected in B.1.617 [https://www.gisaid.org/], which made the conformation of the S protein more stable (54), leading to the increased affinity of the virus and ACE2 (2). L452 residue did not directly interact with ACE2, but the L452R mutation could affect the structural stability of the region where S protein interacts with ACE2 and facilitate SARS-CoV-2 to enter into human respiratory organs (37), which accounted for the prevalence of B.1.427/429 in North America and B.1.617 in India. In addition, the mutation in this site seemed to have a positive effect on immune escape, as the mutation of L452 residue may induce the conformational change of RBD, thus reducing the binding ability of several monoclonal neutralizing antibodies (2, 61–63) and diminishing the neutralization activity of convalescent sera (37, 61). Single L452R mutation could reduce or abolish the neutralizing activity of clinical-stage monoclonal antibodies such as regdanvimab (CT-P59), etesevimab (LY-CoV016), and bamlanivimab (LY-CoV555) (36). L452R has also been confirmed to reduce the neutralizing activity of some antibodies, which do not directly bind to the ACE2-binding epitopes, and it is considered to be a moderate immune escape site for these antibodies (64). The presence of L452R mutation in multiple lineages and regions indicated that this mutation has a positive selection, which may be due to the selective pressure of RBD-specific neutralizing antibodies (36).
All three variants B.1.1.7/B.1.351/P.1 contain N501Y mutation [https://cov-lineages.org/], and this mutation could enhance the affinity of virus S protein with ACE2, especially with the side chains of residues Y41 and K353 of ACE2 (18, 20, 22, 55, 65–67). In addition, the N501Y mutation enabled the virus to infect BALB/c mice, which expanded its host range (67). The neutralization ability of the serum inoculated with Pfizer BNT162b2 vaccine against pseudovirus with N501Y was almost the same as that of the pseudovirus without the mutation (20, 68). However, in the presence of E484K, N501Y, and K417N, the neutralization activity of sera from Moderna mRNA-1273 or Pfizer BNT162b2 vaccinated individuals decreased to a certain extent (63). These findings indicated that a single N501Y mutation is not essential for immune escape, but the accumulation of such key sites will eventually promote immune escape.
The mutation of 484 site exists in the form of K484 in B.1.351 and P.1 while Q484 in B.1.617 (https://cov-lineages.org/) (45). 484 site may also be one of the important immune dominant epitopes. As one of the most important amino acid sites in S protein, E484 mutates to K, Q, or P, the antibody neutralization titer decreases more than 10-fold (69). E484K has been shown to reduce the neutralization of convalescent serum and some antibodies (69). In addition, further mutation of E484K on B.1.1.7 will further reduce the serum neutralizing response of BNT162b2 vaccines (8). Some antibodies from IGHV3-53 and IGHV3-66 genes target the E484 residue of SRAS-CoV-2 S protein, and the mutation at position 484 has a negative effect on the neutralization of these antibodies (28, 70). The 484 site mutation also reduced the neutralizing activity of a variety of monoclonal antibodies in clinical stage, including REGN10933 and LY-CoV-555 (2). Some studies have confirmed that the mutation of E484K could eliminate the key interaction between epitope antibodies against 484 and Arg50 or Arg96, resulting in decreased antibody neutralization efficiency (71, 72). In contrast, Brii-196, COV2-2130, P2C-1F11, and H014 still preserved high neutralization ability, which may be due to their broad-spectrum antigen-binding epitopes (33, 53, 73). The above results provided a novel approach for the future antibody cocktail therapy, which means that the antibody cocktail against different immune epitopes could improve the overall neutralization activity. Although the Indian variant did not contain mutations at sites 417 and 501, given that India currently has the second-largest number of infected people in the world, the E484K mutation may occur on the main epidemic strain B.1.617.2 in India. And it will further improve the affinity to ACE2 and immune escape ability of B.1.617.2, thus improving its infectivity. Antibody neutralizing response was mainly affected by a few dominant epitope mutations in RBD. E484 position, the targeting site of antibodies such as heavy chain germline IGHV3-53 and IGHV3-66, has the greatest influence on antibody binding and neutralization in RBD region (54, 62, 69, 74–76). E484K can escape not only the neutralization of monoclonal antibodies C121, C144 (77) and the serum from convalescent patients (72), but also the neutralization of the combination of REGN10989 and REGN10934 monoclonal antibody cocktail (78).
The deletion of fragments in the NTD region of SARS-CoV-2 was repeatedly observed in the process of evolution, and included sites being considered to be related to immune escape such as L18F and R246I mutations (32, 33, 72).
B.1.1.7 harbored two mutations in the NTD region, Δ69-70 and Δ144. As mentioned earlier, strains with Δ69-70 occupied the dominant position in the middle and later stages of the epidemic in the UK, indicating that these mutations are positive selective. Δ69-70 was speculated to change the conformation of the exposed NTD loop and increase the infectivity of the virus (79). Δ144 significantly reduced the neutralization of most antibodies targeting NTD against B.1.1.7 variants, indicating that the 144 site was one of the neutralizing epitopes for antibodies targeting NTD (33).
B.1.351 and P.1 contain multiple mutations in the NTD region (B.1.351: D80A, D215G, Δ242-244, P.1: L18F, T20N, P26S, D138Y, R190S) [https://cov-lineages.org/]. However, the main targeting site of NTD for antibody against B.1.351 was 242-244 residues, whose deletion reduced the neutralization ability of many kinds of potent antibodies targeting NTD, including 4A8 monoclonal antibody, by more than 1000-fold (33).
The NTD of B.1.427/B.1.429 contains S13I and W152C mutations. This variant achieves neutralization escape through an indirect strategy (36). The S13I mutation could extinguish the integrity of the NTD vulnerable sites by destroying the C15/C136 disulfide bond (36).
The neutralizing potency of the antibody targeting NTD region was poor, greatly reducing the antibody types available for clinical use. However, the mechanisms that determine the antibody failure remain unclear. Present studies have suggested that antibodies targeting NTD may play the role by: (1) blocking the fusion of virus and cell membrane; (2) promoting antibody-mediated cytotoxicity (ADCC); (3) interfering with other coreceptors, such as DC-SIGN and L-SIGN (8, 32, 80). Further studies found that there was a repetitive deletion region (RDR) in the NTD region, which contains most of the immune epitopes of NTD, and the mutation in the RDR region could lead to a decrease in the neutralization ability of antibodies targeting NTD (36). However, Δ144 and Δ242-244 are located in the RDR2 and RDR4 region, respectively, which eliminate the binding of 4A8 targeting RDR2 and RDR4 (33). The existence of the RDR region could explain how antibodies targeting NTD cannot effectively neutralize several mainstream variants. The above analysis showed that multiple immune escape mutations exist in NTD, thereby this region is also in a stage of immune pressure similar to RBD. When studying the antigen drift of new variants, neutralizing antibodies targeting NTD epitopes should be considered. NTD specific antibodies could be divided into two types: highly effective antivirus and low efficacy but polysaccharide-dependent neutralizing activity (dominant epitopes are RBD neutralizing epitopes, subdominant epitopes are neutralizing epitopes other than RBD) (81).
D614G mutation is an important mutation in SARS-CoV-2 and has become the dominant mutation site in all circulating SARS-CoV-2 variants (82, 83). Compared with D614, G614 could increase the viral load in the upper respiratory tract of patients but not in the lungs and may be conducive to the virus spread (84). Interestingly, G614 seemed to be more sensitive to neutralization by increasing the percentage of 1-RBD “up” conformation (85). In addition, neutralizing antibody titration (NT50) assay confirmed that no significant difference in neutralizing antibody titer between serum from the hamster infected with D614 strain and G614 strain (86). However, D614G mutation could not decrease the neutralization potency of most antibodies, indicating that this mutation is not the main immune escape site (29).
P681H/R mutations appeared in both B.1.1.7 and B.1.617 variants, and were proximal to the furin cleavage site (33), which could accelerate virus spread by increasing the membrane fusion rate (53). At present, the effect of P681H/R mutation on the affinity between virus and ACE2, and the neutralization potency of antibody are not clear. It is still necessary to carry out related tests and closely monitor the newly emerged mutations.
Residue 769 is located on the exposed S2 loop. In an immunocompromised SARS-CoV-2 infected person who was treated with convalescent serum, the variant harboring Δ69-70 and D769H was detected, and D769H was thought to be associated with immune escape (53).
As no effective drugs are available for SARS-CoV-2, vaccination becomes an important strategy in preventing and controlling the epidemic. The purpose of vaccination is to induce an immune response similar to natural infection and to produce associated immune cells and antibodies, and the antibody response caused by the vaccine is stronger than that caused by natural infection (36). Neutralization assay confirmed that the neutralization effect of three RBD mutations N439K, Y453F, and N501Y on convalescent serum was greater than that of vaccinated serum, which indicated that mRNA vaccine was more resistant to single RBD mutation than natural infection (9).
Since the outbreak of the SARS-CoV-2 epidemic, the world has accelerated the development process of vaccines (87). At present, there are seven kinds of potential SARS-CoV-2 vaccines: inactivated vaccines, live attenuated vaccines, DNA vaccines, mRNA vaccines, viral vectored vaccines, protein subunit vaccines, and virus-like particle vaccines. As of 26 September 2021, a total of 6,078,264,761 doses of vaccine [https://coronavirus.jhu.edu/map.html] have been administered worldwide. However, the emerging SARS-CoV-2 variants aroused great concern as a variety of vaccine sera showed weakened neutralization effect against variants (Table 2).
Inactivated vaccine is the most classical vaccine form, easy to prepare and can cause effective immune response (93). In clinical phase I and phase II trials, BBIBP-CorV vaccine produced extensively high titer of neutralization antibodies (94). At present, three inactivated vaccines independently developed by China have been put into use. However, it may be difficult to protect the variants since the seed strain of inactivated vaccine is from the Wuhan virus. Wang et al. evaluated the neutralization effect of two inactivated vaccines BBIBP-CorV and CoronaVac against B.1.1.7 and B.1.351 by SARS-CoV-2 pseudoviruses. The results showed that BBIBP-CorV could still retain a neutralization effect against B.1.1.7, but 20 of 25 BBIBP-CorV serum samples were ineffective or partially lost activity against B.1.351. In addition, the neutralization effects of CoronaVac against B.1.1.7 and B.1.351 variants were impaired than that of the WT strain. In addition, no neutralization antibody was detected in 6 of 25 BBIBP-CorV serum samples and 4 of 34 CoronaVac serum samples (24). This result indicated that antibody response heterogeneity exists between individuals after vaccination. Therefore, prevention and control measures such as wearing masks and keeping social distance should still be maintained in risky areas. Yadav PD et al. investigated the inactivated vaccine BBV152 and found that compared with WT strain, the neutralization activity of BBV152 vaccine against B.1.617 decreased by about 2-fold, but it was able to potently neutralize B.1.1.7 (89). All of the above findings indicated that the mutation should be monitored continuously, and new seed strains of SARS-CoV-2 could be considered to update the existing inactivated vaccine (21, 95).
The vectored vaccines are made by using viruses or bacteria as vectors and inserting genes encoding effective immunogens of pathogens into the vector. Usually, most of the marketed vector vaccines targeting SARS-CoV-2 employ adenoviruses vectors (e.g., vaccines from ConSino and AstraZeneca), which can induce both innate immunity and adaptive immunity (96).
The neutralizing activity of the AZD1222 vaccine against B.1.1.7 and B.1.351 strains was evaluated, and the results showed that widespread immune escape was not observed in B.1.1.7, while greater resistance to B.1.351 was observed in both the pseudovirus and the live-virus neutralization assays (12, 22). Therefore, it is still necessary to update the immunogen according to the virus mutation. Real-world research has confirmed that the protective effectiveness of AstraZeneca vaccine against B.1.617.2 strain is 59.8%, while that of B.1.1.7 strain is 66.1%, suggesting that B.1.617.2 has stronger immune escape ability than B.1.1.7 (97). The ChAdOx1 nCoV-19 vaccine was still effective against the B.1.1.7 variant in clinic, but it had a poor protective effect on mild-to-moderate diseases caused by the B.1.351 variant (12, 98, 99).
Protein subunit vaccines use specific protein regions of pathogens to exert immunogenicity. Such vaccines only hold the necessary antigens related to infection and have fewer side effects on the body (100). At present, NVX-CoV2372 vaccines comprise S protein in full-length for vaccine antigen component in clinical use, ZF2001 vaccines apply the dimer form of RBD as vaccine antigen component, while Pfizer and Moderna utilize the trimeric RBD (101, 102).
Huang et al. evaluated the neutralization effects of BBIBP-CorV and ZF2001 on B.1.351. The results showed that although the antibody titer of these vaccinated sera against B.1.351 decreased slightly compared with WT, both of them could effectively neutralize B.1.351 (91). NVX-CoV 2373 (Novavax) protein vaccines provided 95.6% protection efficacy against WT virus, 85.6% efficacy against the B.1.1.7 variant and 60.0% efficacy against the B.1.351 variant (99). Similarly, single-dose vaccination of JNJ-78436735 (Johnson/Janssen) remained 72% protective effect on moderate-to-severe COVID-19 patients in the United States, but in South Africa, where B.1.351 strain had been widely prevalent, it only had a 57% protective effect on moderate-to-severe SARS-CoV-2 infection (103). These results suggest that the protein subunit vaccines have a better neutralization effect on WT and B.1.1.7 strains, and the effect on highly mutated variants including B.1.351 should be monitored continuously.
Nucleic acid vaccines can be divided into RNA vaccines and DNA vaccines. At present, most nucleic acid vaccines for SARS-CoV-2 are mRNA vaccines. These kind of vaccines have been confirmed to be durable, effective and safe in animal experiments, and could simultaneously induce both T and B cell immune responses in the body (104). Supasa P et al. evaluated the neutralizing response of serum from BNT162b2 vaccines against the B.1.1.7 variant, the neutralization activity decreased by 3.3-fold compared to the WT strain, and no immune escape was observed (22). Real-world studies in Qatar showed that the protective efficacy of BNT162b2 vaccines against B.1.1.7 variant was estimated to be 87.0%, and the efficacy against B.1.351 variant was estimated to be 72.1%. Fortunately, its protection against severe disease was still above 90% (105). Similar to the vector vaccines, the neutralization activity of mRNA vaccines against B.1.351 variant was significantly reduced (11, 88, 106). In a study conducted in Israel, BNT162b2 vaccine could still effectively neutralize most of the variants (e.g., B.1.427/B.1.429, B.1.526) (88, 107). Accordingly, the neutralization potency assay using pseudoviruses found that compared with WT (D614), the average neutralization potency of BNT162B2 vaccinated sera decreased by 2.8-fold for B.1.427/B.1.429, 3.2-fold for B.1.351, 1.2-fold for B.1.1.7 and 1.7-fold for P.1, respectively (36). In addition, mixed use of mRNA vaccines seemed to be an effective method. The novel mRNA vaccine mRNA-1273.211, which is a mixture of mRNA-1273 and vaccine encoding B.1.351 S protein, has stronger neutralization efficacy against B.1.351 and P.1 than the monovalent vaccine in animal experiments, and is still valid for B.1.427/429 (108). Compared with mRNA vaccines, DNA vaccines have higher stability and can be stored for a long time (109), as of September 26, 2021, 11 DNA vaccines have been approved for clinical trials [https://www.who.int/emergencies/diseases/novel-coronavirus-2019/covid-19-vaccines]. Momin T et al. reported the results of phase I clinical trial of ZyCoV-D vaccine, a DNA candidate vaccine composed of plasmid DNA carrying RBD gene of Spike and signal peptide gene. The study revealed that this vaccine had good safety, but the serum titer was less than convalescent serum (110). In addition, another DNA candidate vaccine INO-4800, based on full-length of spike protein, showed higher efficiency by the inoculation method of electroporation, and induced neutralizing antibodies in all vaccinated cases (111). Compared with mRNA vaccines, the titers of neutralizing antibody induced by DNA vaccines are still low (112, 113). Thus, it is necessary to improve the efficiency of DNA vaccines and optimize the inoculation method.
The prevalence of variants triggers significant challenges in vaccines design. Compared with conventional subunit vaccines with non-granular antigens, polyvalent nanoparticles usually significantly enhance neutralizing antibody responses, and polyvalent RBD nanoparticles using two-component protein nanoparticles I53-50 have strong antigenic effects, eliciting strong antibody responses against multiple epitopes (114). Compared with the vaccine based on soluble S protein, the new nanoparticle vaccine produced 10-fold more neutralizing antibodies in mice (114). Another study also developed a nano-vaccine supplemented with 3M-052 adjuvant to induce protective immunity against SARS-CoV-2 and a variety of β-coronaviruses (115).
Antibodies are important tools for the treatment of infectious diseases (116). Most of the antibodies used in the treatment of COVID-19 target RBD or NTD, as these two regions have higher immunogenicity (117). The neutralizing antibodies targeting SARS-CoV-2 RBD can be classified into four categories (Figure 6) (54). Among these categories, the immune superiority of RBM epitopes directly contacting with ACE2 are more obvious than other epitopes (56). The mechanism of monoclonal antibodies targeting NTD is still not clear (54, 118).
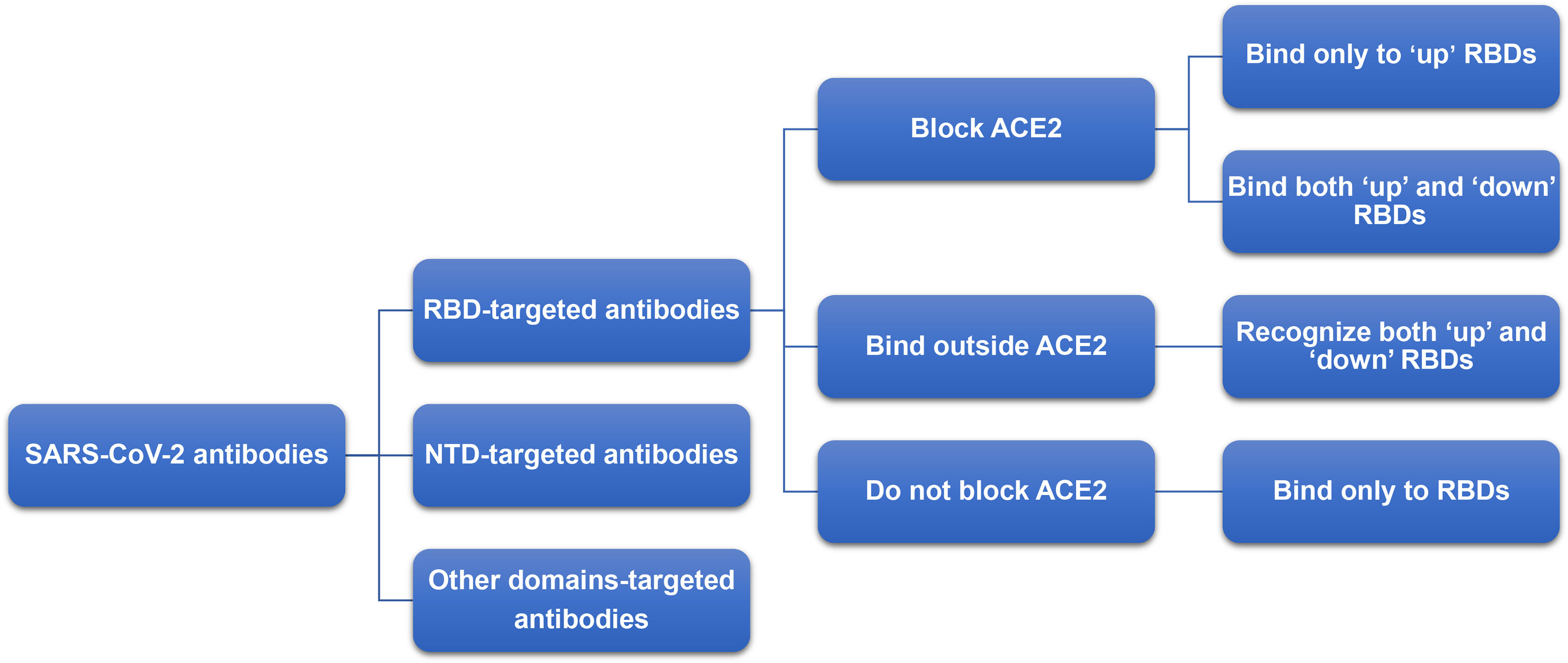
Figure 6 Classification of neutralizing antibody targeting RBD. The neutralizing antibodies targeting SARS-CoV-2 RBD can be classified into four categories: (1) block ACE2 and bind only to ‘up’ RBDs; (2) block ACE2 and can bind to both ‘up’ and ‘down’ RBDs and can contact adjacent RBDs; (3) bind outside the ACE2 sites and recognize both ‘up’ and ‘down’ RBDs; (4) bind to the external residue sites of ACE2 and bind only to ‘up’ RBDs.
Both antibody therapy and convalescent serum therapy can lead to higher immune pressure (118). Constantly subjected to immune pressure caused by the same class of neutralizing epitopes leads to the emergence of novel mutations in these neutralizing epitopes, providing the possibility of antibody response escape (Table 3). However, the simultaneous use of two antibodies targeting different epitopes will help to alleviate this situation, because viruses with simultaneous double-site mutations are not easy to survive under such conditions (73, 119). In addition, due to the key role of S2 subunit in membrane fusion (120), and the high conservation of S2 sequence, antibodies or vaccines targeting fusion peptide (FP) can be designed to reduce mutations caused by immune pressure and enhance the persistence and effectiveness of antibodies and vaccines. Antibodies targeting FP can play a role by preventing protease-mediated cleavage of S2 site. In view of the fact that RBD has a high mutation entropy, which increases the possibility of vaccine-induced immune escape, these conservative targets in S2 may be considered for future vaccine design (121, 122).
At present, most of the monoclonal antibodies (mAbs) used in clinical (e.g., REGN10933, LY-CoV-555) still maintain high neutralization efficacy against B.1.1.7 (11), but most of the mAbs showed a remarkable decrease against B.1.351 (11, 29). mAbs therapy can produce selective pressure, which would increase the possibility of virus escaping from targeted antigen mutations. The combination of two or more mAbs that target non-overlapping epitopes could reduce the escape possibility (123). Moreover, due to the key roles of L452R, LY-CoV-555 completely lost the neutralization activity to B.1.427/B.1.429 (36). Other studies showed that the cocktail therapy was less affected by B.1.1.7 (22). Therefore, cocktail therapy may become an effective method for the treatment of SARS-CoV-2 variants infection (73, 119). The non-overlapping epitopes and lack of binding competition are the critical factors in mAbs cocktail selection (73). According to this principle, some studies have designed the combination of CoV2-06 and CoV2-14 as cocktail antibodies. The targeted epitopes of CoV2-06 and CoV2-14 were mutated at the same time, which was not conducive to virus survival. Therefore, the epitopes of CoV2-06 and CoV2-14 can be considered in future vaccine design so as to reduce the immune escape of variants (73). Similarly, COVID-19 IgGs can be obtained from the isolated convalescent serum. These IgGs can target different epitopes of S protein to play effective neutralizing roles (124).
In addition, some mAbs have cross neutralization activity such as COVA1-16, which can maintain neutralization activity against a variety of variants (e.g., B.1.1.7, B.1.351) by binding to highly conserved epitopes of S protein (125). The infection of B.1.1.7 or B.1.351 can also cause low levels of cross neutralization antibodies (30, 36).
Notably, Sinopharm announced that they have isolated a potent monoclonal antibody 2B11 with higher neutralization activity from the convalescent sera. The IC50 of antibody 2B11 against Delta variant is 5 ng/mL, which is a promising alternative treatment for COVID-19 (126).
Continuous surveillance of viral genome sequence data and the effectiveness of vaccines would help to better understand the drivers of SARS-CoV-2 transmission and evolution, providing a basis for vaccine development and update (123). The same point mutation (e.g., D614G, N501Y) sequenced in different virus strains began to spread globally, suggesting that these mutations have certain evolutionary advantages (118). Two important determinants of variant spread are occurrence frequency in individuals and transmission possibility. Immune escape mutations in a single host may be relatively rare, at least in early infections. However, potential host adaptive mutations can be observed even in the absence of vaccine or antibody therapy selection pressure. It is suggested that transmission-enhancing and/or immune-escape SARS-CoV-2 variants are unlikely to arise frequently, but could spread rapidly if they are successfully transmitted (65). We speculate that early mutations of the virus such as B.1.1.7 mainly enhance its transmission ability, while in the later stage of the epidemic, immune escape strains start to emerge with the increased immune pressure.
The number of people with acquired immunity against COVID-19 continues to increase after natural infection or vaccination, but due to unequal interventions and access to vaccines, the virus would subject to greater immune pressure and require repeated immunization rounds to deal with the continuous arising of virus variants (127). Postponement of the second dose could help more people get vaccination when the vaccine production dose is not enough, but the proposal to change the vaccine scheme to a single dose may accelerate the evolution of virus strains. The antibody titer produced by only one dose of vaccination would not be sufficient for virus infection prevention and virus clearance, which is likely to contribute to the production of vaccine-resistant strains (118).
The emergence of new variants emphasizes the need for continued vigilance. Since vaccine-induced herd immunity increases the probability of immune escape, it is difficult to determine which variants or sequences should be selected to update the vaccine sequence. B.1.351 is the variant of greatest concern, with the strong resistance to vaccine serum and antibodies. Therefore, it is believed that the development of vaccine constructs using B.1.351 is the top priority (5). As COVID-19 continues to spread, more virus variants with the ability to escape the neutralization of antibodies would appear. The protection of these variants can be ensured by the combination of two or more potent neutralizing antibodies against different epitopes (128). In addition, using antibody cocktails to resist virus mutation seems to be a sensible strategy. However, it must be recognized that the use of mAbs for long-term treatment or prevention, especially in chronic infected individuals who are immunocompromised, may lead to the emergence of neutralization resistance mutations. In order to avoid selective pressure and immune escape, it is suggested that antibody therapy might consist of a combination of antibodies targeting non-overlapping or highly conserved epitopes (129). Considering the important role of site 484 for antibody binding and neutralization, it is a good strategy to identify monoclonal antibodies from E484K infected individuals (59).
The global circulation of multiple SARS-CoV-2 variants undermines confidence in whether the current vaccines will provide long-term protection. The vaccines elicited antibody responses against RBD in a manner similar to natural infection (63), suggesting that vaccines could reduce the severity of disease caused by natural infection more or less. In addition, the response of T cells against spikes cannot be disturbed by mutations and could still prevent severe diseases caused by variants (59). Moreover, under the continuous emergence of SARS-CoV-2 mutations, the body’s immune system is also constantly improving its ability to deal with the evolution. The memory B cells did not decrease after 6.2 months of effective vaccination, but continued to evolve and were involved in preventing reinfection. These results strongly suggest that vaccinated individuals can respond quickly and effectively to the virus upon exposure (130).
New variants will continue to emerge (123), and intensive surveillance systems are needed to monitor the arising of new variants. Moreover, breakthrough infections among vaccinees urgently need to be elucidated. The second or even third generation vaccines targeting virus variants, as well as the more extensive development of immunogens targeting ACE2-RBD-independent surfaces, are worthy of further study (59).
HF, JF, and ML designed the research. FL, ML, ZP, LJ, LG, JF, and HF read and analyzed the papers. LT and JH participated in discussion. FL, ML, JF, and HF wrote and revised the manuscript. All authors contributed to the article and approved the submitted version.
This manuscript was funded by grants from National Key Research and Development Program of China (grant No. 2020YFA0712102), Fundamental Research Funds for Central Universities (grant No. BUCTZY2022), and H&H Global Research and Technology Center (grant No. H2021028).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.744242/full#supplementary-material
Supplementary Figure 1 | Brief information of six former VOI variants. Six former VOC variants (B.1.427/429, P.2, P.3, B.1.525, B.1.526, and B.1.617.1) are marked in the arrow according to the date of designation, and their related brief information (e.g., the time and location of earliest documented samples, infectivity, main mutations, immune escape ability) are displayed in the corresponding location.
Supplementary Figure 2 | Mutations of six former VOI variants. Schematic showing the locations of amino acid substitutions of six former VOI variants (B.1.427/429, P.2, P.3, B.1.525, B.1.526, and B.1.617.1) in spike protein. The RBD region is shown in modena, the NTD region is shown in shallow orange.
1. Hu B, Guo H, Zhou P, Shi ZL. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol (2021) 19(3):141–54. doi: 10.1038/s41579-020-00459-7
2. Liu Z, VanBlargan LA, Bloyet LM, Rothlauf PW, Chen RE, Stumpf S, et al. Identification of SARS-CoV-2 Spike Mutations That Attenuate Monoclonal and Serum Antibody Neutralization. Cell Host Microbe (2021) 29(3):477–88.e4. doi: 10.1016/j.chom.2021.01.014
3. Smith EC, Blanc H, Surdel MC, Vignuzzi M, Denison MR. Coronaviruses Lacking Exoribonuclease Activity are Susceptible to Lethal Mutagenesis: Evidence for Proofreading and Potential Therapeutics. PloS Pathog (2013) 9(8):e1003565. doi: 10.1371/journal.ppat.1003565
4. Thomson EC, Rosen LE, Shepherd JG, Spreafico R, da Silva Filipe A, Wojcechowskyj JA, et al. Circulating SARS-CoV-2 Spike N439K Variants Maintain Fitness While Evading Antibody-Mediated Immunity. Cell (2021) 184(5):1171–87.e20. doi: 10.1016/j.cell.2021.01.037
5. Dejnirattisai W, Zhou D, Supasa P, Liu C, Mentzer AJ, Ginn HM, et al. Antibody Evasion by the P.1 Strain of SARS-CoV-2. Cell (2021) 184(11):2939–54.e9. doi: 10.1016/j.cell.2021.03.055
6. Neuzil KM. Interplay Between Emerging SARS-CoV-2 Variants and Pandemic Control. N Engl J Med (2021) 384(20):1952–4. doi: 10.1056/NEJMe2103931
7. Davies NG, Abbott S, Barnard RC, Jarvis CI, Kucharski AJ, Munday JD, et al. Estimated Transmissibility and Impact of SARS-CoV-2 Lineage B.1.1.7 in England. Science (2021) 372(6538):eabg3055. doi: 10.1126/science.abg3055
8. Collier DA, De Marco A, Ferreira I, Meng B, Datir RP, Walls AC, et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA Vaccine-Elicited Antibodies. Nature (2021) 593(7857):136–41. doi: 10.1038/s41586-021-03412-7
9. Shen X, Tang H, McDanal C, Wagh K, Fischer W, Theiler J, et al. SARS-CoV-2 Variant B.1.1.7 is Susceptible to Neutralizing Antibodies Elicited by Ancestral Spike Vaccines. Cell Host Microbe (2021) 29(4):529–39.e3. doi: 10.1016/j.chom.2021.03.002
10. Mahase E. Covid-19: What New Variants are Emerging and How are They Being Investigated. BMJ (2021) 372:n158. doi: 10.1136/bmj.n158
11. Wang P, Nair MS, Liu L, Iketani S, Luo Y, Guo Y, et al. Antibody Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. Nature (2021) 593(7857):130–5. doi: 10.1038/s41586-021-03398-2
12. Madhi SA, Baillie V, Cutland CL, Voysey M, Koen AL, Fairlie L, et al. Efficacy of the ChAdOx1 Ncov-19 Covid-19 Vaccine Against the B.1.351 Variant. N Engl J Med (2021) 384(20):1885–98. doi: 10.1056/NEJMoa2102214
13. Campbell F, Archer B, Laurenson-Schafer H, Jinnai Y, Konings F, Batra N, et al. Increased Transmissibility and Global Spread of SARS-CoV-2 Variants of Concern as at June 2021. Euro Surveill (2021) 26(24):2100509. doi: 10.2807/1560-7917.ES.2021.26.24.2100509
14. Planas D, Veyer D, Baidaliuk A, Staropoli I, Guivel-Benhassine F, Rajah MM, et al. Reduced Sensitivity of SARS-CoV-2 Variant Delta to Antibody Neutralization. Nature (2021) 596(7871):276–80. doi: 10.1038/s41586-021-03777-9
15. Liu C, Ginn HM, Dejnirattisai W, Supasa P, Wang B, Tuekprakhon A, et al. Reduced Neutralization of SARS-CoV-2 B.1.617 by Vaccine and Convalescent Serum. Cell (2021) 184(16):4220–36.e13. doi: 10.1016/j.cell.2021.06.020
16. Faria NR, Mellan TA, Whittaker C, Claro IM, Candido D, Mishra S, et al. Genomics and Epidemiology of the P.1 SARS-CoV-2 Lineage in Manaus, Brazil. Science (2021) 372(6544):815–21. doi: 10.1126/science.abh2644
17. Garcia-Beltran WF, Lam EC, St Denis K, Nitido AD, Garcia ZH, Hauser BM, et al. Multiple SARS-CoV-2 Variants Escape Neutralization by Vaccine-Induced Humoral Immunity. Cell (2021) 184(9):2372–83.e9. doi: 10.1016/j.cell.2021.03.013
18. Alpert T, Brito AF, Lasek-Nesselquist E, Rothman J, Valesano AL, MacKay MJ, et al. Early Introductions and Transmission of SARS-CoV-2 Variant B.1.1.7 in the United States. Cell (2021) 184(10):2595–604.e13. doi: 10.1016/j.cell.2021.03.061
19. Volz E, Mishra S, Chand M, Barrett JC, Johnson R, Geidelberg L, et al. Assessing Transmissibility of SARS-CoV-2 Lineage B.1.1.7 in England. Nature (2021) 593(7858):266–9. doi: 10.1038/s41586-021-03470-x
20. Muik A, Wallisch AK, Sänger B, Swanson KA, Mühl J, Chen W, et al. Neutralization of SARS-CoV-2 Lineage B.1.1.7 Pseudovirus by BNT162b2 Vaccine-Elicited Human Sera. Science (2021) 371(6534):1152–3. doi: 10.1126/science.abg6105
21. Li M, Lou F, Fan H. SARS-CoV-2 Variants: A New Challenge to Convalescent Serum and mRNA Vaccine Neutralization Efficiency. Signal Transduct Target Ther (2021) 6(1):151. doi: 10.1038/s41392-021-00592-6
22. Supasa P, Zhou D, Dejnirattisai W, Liu C, Mentzer AJ, Ginn HM, et al. Reduced Neutralization of SARS-CoV-2 B.1.1.7 Variant by Convalescent and Vaccine Sera. Cell (2021) 184(8):2201–11.e7. doi: 10.1016/j.cell.2021.02.033
23. Galloway SE, Paul P, MacCannell DR, Johansson MA, Brooks JT, MacNeil A, et al. Emergence of SARS-CoV-2 B.1.1.7 Lineage - United States, December 29, 2020-January 12, 2021. MMWR Morb Mortal Wkly Rep (2021) 70(3):95–9. doi: 10.15585/mmwr.mm7003e2
24. Wang GL, Wang ZY, Duan LJ, Meng QC, Jiang MD, Cao J, et al. Susceptibility of Circulating SARS-CoV-2 Variants to Neutralization. N Engl J Med (2021) 384(24):2354–6. doi: 10.1056/NEJMc2103022
25. Davies NG, Jarvis CI. CMMID COVID-19 Working Group, Et al. Increased Mortality in Community-Tested Cases of SARS-CoV-2 Lineage B.1.1.7. Nature (2021) 593(7858):270–4. doi: 10.1038/s41586-021-03426-1
26. Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, et al. Detection of a SARS-CoV-2 Variant of Concern in South Africa. Nature (2021) 592(7854):438–43. doi: 10.1038/s41586-021-03402-9
27. Ramanathan M, Ferguson ID, Miao W, Khavari PA. SARS-CoV-2 B.1.1.7 and B.1.351 Spike Variants Bind Human ACE2 With Increased Affinity. Lancet Infect Dis (2021) 21(8):1070. doi: 10.1016/S1473-3099(21)00262-0
28. Wibmer CK, Ayres F, Hermanus T, Madzivhandila M, Kgagudi P, Oosthuysen B, et al. SARS-CoV-2 501y.V2 Escapes Neutralization by South African COVID-19 Donor Plasma. Nat Med (2021) 27(4):622–5. doi: 10.1038/s41591-021-01285-x
29. Li Q, Nie J, Wu J, Zhang L, Ding R, Wang H, et al. SARS-CoV-2 501y.V2 Variants Lack Higher Infectivity But do Have Immune Escape. Cell (2021) 184(9):2362–71.e9. doi: 10.1016/j.cell.2021.02.042
30. Cele S, Gazy I, Jackson L, Hwa SH, Tegally H, Lustig G, et al. Escape of SARS-CoV-2 501y.V2 From Neutralization by Convalescent Plasma. Nature (2021) 593(7857):142–6. doi: 10.1038/s41586-021-03471-w
31. Sabino EC, Buss LF, Carvalho M, Prete CA Jr, Crispim M, Fraiji NA, et al. Resurgence of COVID-19 in Manaus, Brazil, Despite High Seroprevalence. Lancet (2021) 397(10273):452–5. doi: 10.1016/S0140-6736(21)00183-5
32. McCallum M, De Marco A, Lempp FA, Tortorici MA, Pinto D, Walls AC, et al. N-Terminal Domain Antigenic Mapping Reveals a Site of Vulnerability for SARS-CoV-2. Cell (2021) 184(9):2332–47.e16. doi: 10.1016/j.cell.2021.03.028
33. McCarthy KR, Rennick LJ, Nambulli S, Robinson-McCarthy LR, Bain WG, Haidar G, et al. Recurrent Deletions in the SARS-CoV-2 Spike Glycoprotein Drive Antibody Escape. Science (2021) 371(6534):1139–42. doi: 10.1126/science.abf6950
34. Zhang W, Davis BD, Chen SS, Sincuir Martinez JM, Plummer JT, Vail E. Emergence of a Novel SARS-CoV-2 Variant in Southern California. JAMA (2021) 325(13):1324–6. doi: 10.1001/jama.2021.1612
35. Aleem A, Samad ABA, Amy KS. Emerging Variants of SARS-CoV-2 And Novel Therapeutics Against Coronavirus (COVID-19). Treasure Island (FL: StatPearls Publishing (2021).
36. McCallum M, Bassi J, De Marco A, Chen A, Walls AC, Di Iulio J, et al. SARS-CoV-2 Immune Evasion by the B.1.427/B.1.429 Variant of Concern. Science (2021) 373(6555):648–54. doi: 10.1126/science.abi7994
37. Deng X, Garcia-Knight MA, Khalid MM, Servellita V, Wang C, Morris MK, et al. Transmission, Infectivity, and Neutralization of a Spike L452R SARS-CoV-2 Variant. Cell (2021) 184(13):3426–37.e8. doi: 10.1016/j.cell.2021.04.025
38. Thompson CN, Hughes S, Ngai S, Baumgartner J, Wang JC, McGibbon E, et al. Rapid Emergence and Epidemiologic Characteristics of the SARS-CoV-2 B.1.526 Variant - New York City, New York, January 1-April 5, 2021. MMWR Morb Mortal Wkly Rep (2021) 70(19):712–6. doi: 10.15585/mmwr.mm7019e1
39. West AP Jr, Wertheim JO, Wang JC, Vasylyeva TI, Havens JL, Chowdhury MA, et al. Detection and Characterization of the SARS-CoV-2 Lineage B.1.526 in New York. Nat Commun (2021) 12(1):4886. doi: 10.1038/s41467-021-25168-4
40. Zhou H, Dcosta BM, Samanovic MI, Mulligan MJ, Landau NR, Tada T. B.1.526 SARS-CoV-2 Variants Identified in New York City are Neutralized by Vaccine-Elicited and Therapeutic Monoclonal Antibodies. mBio (2021) 12(4):e0138621. doi: 10.1128/mBio.01386-21
41. Pegu A, O'Connell SE, Schmidt SD, O'Dell S, Talana CA, Lai L, et al. Durability of mRNA-1273 Vaccine-Induced Antibodies Against SARS-CoV-2 Variants. Science (2021) 373(6561):1372–7. doi: 10.1126/science.abj4176
42. Pereira F, Tosta S, Lima MM, Reboredo de Oliveira da Silva L, Nardy VB, Gómez M, et al. Genomic Surveillance Activities Unveil the Introduction of the SARS-CoV-2 B.1.525 Variant of Interest in Brazil: Case Report. J Med Virol (2021) 93(9):5523–6. doi: 10.1002/jmv.27086
43. Ozer EA, Simons LM, Adewumi OM, Fowotade AA, Omoruyi EC, Adeniji JA, et al. High Prevalence of SARS-CoV-2 B.1.1.7 (UK Variant) and the Novel B.1.5.2.5 Lineage in Oyo State, Nigeria. medRxiv (2021). doi: 10.1101/2021.04.09.21255206
44. Janik E, Niemcewicz M, Podogrocki M, Majsterek I, Bijak M. The Emerging Concern and Interest SARS-CoV-2 Variants. Pathogens (2021) 10(6):633. doi: 10.3390/pathogens10060633
45. Cherian S, Potdar V, Jadhav S, Yadav P, Gupta N, Das M, et al. SARS-CoV-2 Spike Mutations, L452R, T478K, E484Q and P681R, in the Second Wave of COVID-19 in Maharashtra, India. Microorganisms (2021) 9(7):1542. doi: 10.3390/microorganisms9071542
46. Challen R, Dyson L, Overton CE, Guzman-Rincon LM, Hill EM, Stage HB, et al. Early Epidemiological Signatures of Novel SARS-CoV-2 Variants: Establishment of B.1.617.2 in England. MedRxiv: The Preprint Server Health Sci (2021). doi: 10.1101/2021.06.05.21258365
47. Wang Y, Chen R, Hu F, Lan Y, Yang Z, Zhan C, et al. Transmission, Viral Kinetics and Clinical Characteristics of the Emergent SARS-CoV-2 Delta VOC in Guangzhou, China. EClinicalMedicine (2021) 40:101129. doi: 10.1016/j.eclinm.2021.101129
48. Li M, Lou F, Fan H. SARS-CoV-2 Variants of Concern Delta: A Great Challenge to Prevention and Control of COVID-19. Signal Transduct Target Ther (2021) 6(1):349. doi: 10.1038/s41392-021-00767-1
49. Wall EC, Wu M, Harvey R, Kelly G, Warchal S, Sawyer C, et al. Neutralising Antibody Activity Against SARS-CoV-2 VOCs B.1.617.2 and B.1.351 by BNT162b2 Vaccination. Lancet (2021) 397(10292):2331–3. doi: 10.1016/S0140-6736(21)01290-3
50. Uriu K, Kimura I, Shirakawa K, Takaori-Kondo A, Nakada T, Kaneda A, et al. Ineffective Neutralization of the SARS-CoV-2 Mu Variant by Convalescent and Vaccine Sera. BioRxiv Preprint Server Biol (2021). doi: 10.1101/2021.09.06.459005
51. Qasim Bukhari YJ. Will Coronavirus Pandemic Diminish by Summer. BioRxiv Preprint Server Biol (2021). doi: 10.2139/ssrn.3556998
52. Baker RE, Yang W, Vecchi GA, Metcalf C, Grenfell BT. Susceptible Supply Limits the Role of Climate in the Early SARS-CoV-2 Pandemic. Science (2020) 369(6501):315–9. doi: 10.1126/science.abc2535
53. Kemp SA, Collier DA, Datir RP, Ferreira I, Gayed S, Jahun A, et al. SARS-CoV-2 Evolution During Treatment of Chronic Infection. Nature (2021) 592(7853):277–82. doi: 10.1038/s41586-021-03291-y
54. Barnes CO, Jette CA, Abernathy ME, Dam KA, Esswein SR, Gristick HB, et al. SARS-CoV-2 Neutralizing Antibody Structures Inform Therapeutic Strategies. Nature (2020) 588(7839):682–7. doi: 10.1038/s41586-020-2852-1
55. Starr TN, Greaney AJ, Hilton SK, Ellis D, Crawford K, Dingens AS, et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell (2020) 182(5):1295–310.e20. doi: 10.1016/j.cell.2020.08.012
56. ECDC. Detection of New SARS-CoV-2 Variants Related to Mink (2020). Available at: https://www.ecdc.europa.eu/sites/default/files/ocuments/RRA-SARS-CoV-2-in-mink-12-nov-2020.Pdf.
57. Piccoli L, Park YJ, Tortorici MA, Czudnochowski N, Walls AC, Beltramello M, et al. Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Cell (2020) 183(4):1024–42.e21. doi: 10.1016/j.cell.2020.09.037
58. Watanabe Y, Berndsen ZT, Raghwani J, Seabright GE, Allen JD, Pybus OG, et al. Vulnerabilities in Coronavirus Glycan Shields Despite Extensive Glycosylation. Nat Commun (2020) 11(1):2688. doi: 10.1101/2020.02.20.957472
59. Zhou D, Dejnirattisai W, Supasa P, Liu C, Mentzer AJ, Ginn HM, et al. Evidence of Escape of SARS-CoV-2 Variant B.1.351 From Natural and Vaccine-Induced Sera. Cell (2021) 184(9):2348–61.e6. doi: 10.1016/j.cell.2021.02.037
60. Starr TN, Greaney AJ, Addetia A, Hannon WW, Choudhary MC, Dingens AS, et al. Prospective Mapping of Viral Mutations That Escape Antibodies Used to Treat COVID-19. Science (2021) 371(6531):850–4. doi: 10.1126/science.abf9302
61. Li Q, Wu J, Nie J, Zhang L, Hao H, Liu S, et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell (2020) 182(5):1284–94.e9. doi: 10.1016/j.cell.2020.07.012
62. Greaney AJ, Starr TN, Gilchuk P, Zost SJ, Binshtein E, Loes AN, et al. Complete Mapping of Mutations to the SARS-CoV-2 Spike Receptor-Binding Domain That Escape Antibody Recognition. Cell Host Microbe (2021) 29(1):44–57.e9. doi: 10.1016/j.chom.2020.11.007
63. Wang Z, Schmidt F, Weisblum Y, Muecksch F, Barnes CO, Finkin S, et al. mRNA Vaccine-Elicited Antibodies to SARS-CoV-2 and Circulating Variants. Nature (2021) 592(7855):616–22. doi: 10.1038/s41586-021-03324-6
64. Greaney AJ, Starr TN, Barnes CO, Weisblum Y, Schmidt F, Caskey M, et al. Mapping Mutations to the SARS-CoV-2 RBD That Escape Binding by Different Classes of Antibodies. Nat Commun (2021) 12(1):4196. doi: 10.1038/s41467-021-24435-8
65. Lythgoe KA, Hall M, Ferretti L, de Cesare M, MacIntyre-Cockett G, Trebes A, et al. SARS-CoV-2 Within-Host Diversity and Transmission. Science (2021) 372(6539):eabg0821. doi: 10.1126/science.abg0821
66. Saad-Roy CM, Morris SE, Metcalf C, Mina MJ, Baker RE, Farrar J, et al. Epidemiological and Evolutionary Considerations of SARS-CoV-2 Vaccine Dosing Regimes. Science (2021) 372(6540):363–70. doi: 10.1126/science.abg8663
67. Gu H, Chen Q, Yang G, He L, Fan H, Deng YQ, et al. Adaptation of SARS-CoV-2 in BALB/c Mice for Testing Vaccine Efficacy. Science (2020) 369(6511):1603–7. doi: 10.1126/science.abc4730
68. Xie X, Zou J, Fontes-Garfias CR, Xia H, Swanson KA, Cutler M, et al. Neutralization of N501Y Mutant SARS-CoV-2 by BNT162b2 Vaccine-Elicited Sera. bioRxiv (2021). doi: 10.1101/2021.01.07.425740
69. Greaney AJ, Loes AN, Crawford K, Starr TN, Malone KD, Chu HY, et al. Comprehensive Mapping of Mutations in the SARS-CoV-2 Receptor-Binding Domain That Affect Recognition by Polyclonal Human Plasma Antibodies. Cell Host Microbe (2021) 29(3):463–76.e6. doi: 10.1016/j.chom.2021.02.003
70. Yuan M, Huang D, Lee CD, Wu NC, Jackson AM, Zhu X, et al. Structural and Functional Ramifications of Antigenic Drift in Recent SARS-CoV-2 Variants. Science (2021) 373(6556):818–23. doi: 10.1126/science.abh1139
71. Wang L, Zhou T, Zhang Y, Yang ES, Schramm CA, Shi W, et al. Ultrapotent Antibodies Against Diverse and Highly Transmissible SARS-CoV-2 Variants. Science (2021) 373(6556):eabh1766. doi: 10.1126/science.abh1766
72. Andreano E, Piccini G, Licastro D, Casalino L, Johnson NV, Paciello I, et al. SARS-CoV-2 Escape From a Highly Neutralizing COVID-19 Convalescent Plasma. Proc Natl Acad Sci U S A (2021) 118(36):e2103154118. doi: 10.1073/pnas.2103154118
73. Ku Z, Xie X, Davidson E, Ye X, Su H, Menachery VD, et al. Molecular Determinants and Mechanism for Antibody Cocktail Preventing SARS-CoV-2 Escape. Nat Commun (2021) 12(1):469. doi: 10.1038/s41467-020-20789-7
74. Robbiani DF, Gaebler C, Muecksch F, Lorenzi J, Wang Z, Cho A, et al. Convergent Antibody Responses to SARS-CoV-2 in Convalescent Individuals. Nature (2020) 584(7821):437–42. doi: 10.1038/s41586-020-2456-9
75. Yuan M, Liu H, Wu NC, Lee CD, Zhu X, Zhao F, et al. Structural Basis of a Shared Antibody Response to SARS-CoV-2. Science (2020) 369(6507):1119–23. doi: 10.1126/science.abd2321
76. Zost SJ, Gilchuk P, Chen RE, Case JB, Reidy JX, Trivette A, et al. Rapid Isolation and Profiling of a Diverse Panel of Human Monoclonal Antibodies Targeting the SARS-CoV-2 Spike Protein. Nat Med (2020) 26(9):1422–7. doi: 10.1038/s41591-020-0998-x
77. Weisblum Y, Schmidt F, Zhang F, DaSilva J, Poston D, Lorenzi J, et al. Escape From Neutralizing Antibodies by SARS-CoV-2 Spike Protein Variants. Elife (2020) 9:e61312. doi: 10.7554/eLife.61312
78. Baum A, Fulton BO, Wloga E, Copin R, Pascal KE, Russo V, et al. Antibody Cocktail to SARS-CoV-2 Spike Protein Prevents Rapid Mutational Escape Seen With Individual Antibodies. Science (2020) 369(6506):1014–8. doi: 10.1126/science.abd0831
79. Meng B, Kemp SA, Papa G, Datir R, Ferreira I, Marelli S, et al. Recurrent Emergence of SARS-CoV-2 Spike Deletion H69/V70 and Its Role in the Alpha Variant B.1.1.7. Cell Rep (2021) 35(13):109292. doi: 10.1016/j.celrep.2021.109292
80. Soh WT, Liu Y, Nakayama EE, Ono C, Torii S, Nakagami H, et al. The N-Terminal Domain of Spike Glycoprotein Mediates SARS-CoV-2 Infection by Associating With L-SIGN and DC-SIGN. BioRxiv: Preprint Server Biol (2020). doi: 10.1101/2020.11.05.369264
81. Graham C, Seow J, Huettner I, Khan H, Kouphou N, Acors S, et al. Neutralization Potency of Monoclonal Antibodies Recognizing Dominant and Subdominant Epitopes on SARS-CoV-2 Spike is Impacted by the B.1.1.7 Variant. Immunity (2021) 54(6):1276–89.e6. doi: 10.1016/j.immuni.2021.03.023
82. Yurkovetskiy L, Wang X, Pascal KE, Tomkins-Tinch C, Nyalile TP, Wang Y, et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell (2020) 183(3):739–51.e8. doi: 10.1016/j.cell.2020.09.032
83. Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, et al. Tracking Changes in SARS-CoV-2 Spike: Evidence That D614G Increases Infectivity of the COVID-19 Virus. Cell (2020) 182(4):812–27.e19. doi: 10.1016/j.cell.2020.06.043
84. Plante JA, Liu Y, Liu J, Xia H, Johnson BA, Lokugamage KG, et al. Spike Mutation D614G Alters SARS-CoV-2 Fitness. Nature (2021) 592(7852):116–21. doi: 10.1038/s41586-020-2895-3
85. Weissman D, Alameh MG, de Silva T, Collini P, Hornsby H, Brown R, et al. D614G Spike Mutation Increases SARS CoV-2 Susceptibility to Neutralization. Cell Host Microbe (2021) 29(1):23–31.e4. doi: 10.1016/j.chom.2020.11.012
86. Stauft CB, Lien CZ, Selvaraj P, Liu S, Wang TT. The G614 Pandemic SARS-CoV-2 Variant is Not More Pathogenic Than the Original D614 Form in Adult Syrian Hamsters. Virology (2021) 556:96–100. doi: 10.1016/j.virol.2021.01.005
87. Stern PL. Key Steps in Vaccine Development. Ann Allergy Asthma Immunol (2020) 125(1):17–27. doi: 10.1016/j.anai.2020.01.025
88. Liu Y, Liu J, Xia H, Zhang X, Fontes-Garfias CR, Swanson KA, et al. Neutralizing Activity of BNT162b2-Elicited Serum. N Engl J Med (2021) 384(15):1466–8. doi: 10.1056/NEJMc2102017
89. Yadav PD, Sapkal GN, Abraham P, Ella R, Deshpande G, Patil DY, et al. Neutralization of Variant Under Investigation B.1.617 With Sera of BBV152 Vaccinees. Clin Infect Dis (2021). doi: 10.1093/cid/ciab411
90. Di Caro A, Cunha F, Petrosillo N, Beeching NJ, Ergonul O, Petersen E, et al. Severe Acute Respiratory Syndrome Coronavirus 2 Escape Mutants and Protective Immunity From Natural Infections or Immunizations. Clin Microbiol Infect (2021) 27:823-6. doi: 10.1016/j.cmi.2021.03.011
91. Baoying Huang LD, Hui Wang ZH, Xiaoming Yang WT, Gao GF. Neutralization of SARS-CoV-2 VOC 501y.V2 by Human Antisera Elicited by Both Inactivated BBIBP-CorV and Recombinant Dimeric RBD ZF2001 Vaccines. BioRxiv Preprint Server Biol (2020). doi: 10.1101/2021.02.01.429069
92. Wu K, Werner AP, Koch M, Choi A, Narayanan E, Stewart-Jones G, et al. Serum Neutralizing Activity Elicited by mRNA-1273 Vaccine. N Engl J Med (2021) 384(15):1468–70. doi: 10.1056/NEJMc2102179
93. Murdin AD, Barreto L, Plotkin S. Inactivated Poliovirus Vaccine: Past and Present Experience. Vaccine (1996) 14(8):735–46. doi: 10.1016/0264-410X(95)00211-I
94. Xia S, Zhang Y, Wang Y, Wang H, Yang Y, Gao GF, et al. Safety and Immunogenicity of an Inactivated SARS-CoV-2 Vaccine, BBIBP-CorV: A Randomised, Double-Blind, Placebo-Controlled, Phase 1/2 Trial. Lancet Infect Dis (2021) 21(1):39–51. doi: 10.1016/S1473-3099(20)30831-8
95. Wang L, Wang L, Zhuang H. Profiling and Characterization of SARS-CoV-2 Mutants' Infectivity and Antigenicity. Signal Transduct Target Ther (2020) 5(1):185. doi: 10.1038/s41392-020-00302-8
96. Tatsis N, Ertl HC. Adenoviruses as Vaccine Vectors. Mol Ther (2004) 10(4):616–29. doi: 10.1016/j.ymthe.2004.07.013
97. Lopez Bernal J, Andrews N, Gower C, Gallagher E, Simmons R, Thelwall S, et al. Effectiveness of Covid-19 Vaccines Against the B.1.617.2 (Delta) Variant. N Engl J Med (2021) 385(7):585–94. doi: 10.1056/NEJMoa2108891
98. Emary K, Golubchik T, Aley PK, Ariani CV, Angus B, Bibi S, et al. Efficacy of ChAdOx1 Ncov-19 (AZD1222) Vaccine Against SARS-CoV-2 Variant of Concern 202012/01 (B.1.1.7): An Exploratory Analysis of a Randomised Controlled Trial. Lancet (2021) 397(10282):1351–62. doi: 10.1016/S0140-6736(21)00628-0
99. Mahase E. Covid-19: Novavax Vaccine Efficacy is 86% Against UK Variant and 60% Against South African Variant. BMJ (2021) 372:n296. doi: 10.1136/bmj.n296
100. Pollet J, Chen WH, Strych U. Recombinant Protein Vaccines, a Proven Approach Against Coronavirus Pandemics. Adv Drug Deliv Rev (2021) 170:71–82. doi: 10.1016/j.addr.2021.01.001
101. Keech C, Albert G, Cho I, Robertson A, Reed P, Neal S, et al. Phase 1-2 Trial of a SARS-CoV-2 Recombinant Spike Protein Nanoparticle Vaccine. N Engl J Med (2020) 383(24):2320–32. doi: 10.1056/NEJMoa2026920
102. Yang S, Li Y, Dai L, Wang J, He P, Li C, et al. Safety and Immunogenicity of a Recombinant Tandem-Repeat Dimeric RBD-Based Protein Subunit Vaccine (ZF2001) Against COVID-19 in Adults: Two Randomised, Double-Blind, Placebo-Controlled, Phase 1 and 2 Trials. Lancet Infect Dis (2021) 21(8):1107–19. doi: 10.1016/S1473-3099(21)00127-4
103. Mahase E. Covid-19: Where Are We on Vaccines and Variants. BMJ (2021) 372:n597. doi: 10.1136/bmj.n597
104. Maruggi G, Zhang C, Li J, Ulmer JB, Yu D. mRNA as a Transformative Technology for Vaccine Development to Control Infectious Diseases. Mol Ther (2019) 27(4):757–72. doi: 10.1016/j.ymthe.2019.01.020
105. Abu-Raddad LJ, Chemaitelly H, Butt AA. National Study Group for COVID-19 Vaccination. Effectiveness of the BNT162b2 Covid-19 Vaccine Against the B.1.1.7 and B.1.351 Variants. N Engl J Med (2021) 385(2):187–9. doi: 10.1056/NEJMc2104974
106. Doria-Rose N, Suthar MS, Makowski M, O'Connell S, McDermott AB, Flach B, et al. Antibody Persistence Through 6 Months After the Second Dose of mRNA-1273 Vaccine for Covid-19. N Engl J Med (2021) 384(23):2259–61. doi: 10.1056/NEJMc2103916
107. Dagan N, Barda N, Kepten E, Miron O, Perchik S, Katz MA, et al. BNT162b2 mRNA Covid-19 Vaccine in a Nationwide Mass Vaccination Setting. N Engl J Med (2021) 384(15):1412–23. doi: 10.1056/NEJMoa2101765
108. Wu K, Choi A, Koch M, Elbashir S, Ma L, Lee D, et al. Variant SARS-CoV-2 mRNA Vaccines Confer Broad Neutralization as Primary or Booster Series in Mice. bioRxiv (2021). doi: 10.1101/2021.04.13.439482
109. Karpiński TM, Ożarowski M, Seremak-Mrozikiewicz A, Wolski H, Wlodkowic D. The 2020 Race Towards SARS-CoV-2 Specific Vaccines. Theranostics (2021) 11(4):1690–702. doi: 10.7150/thno.53691
110. Momin T, Kansagra K, Patel H, Sharma S, Sharma B, Patel J, et al. Safety and Immunogenicity of a DNA SARS-CoV-2 Vaccine (ZyCoV-D): Results of an Open-Label, Non-Randomized Phase I Part of Phase I/II Clinical Study by Intradermal Route in Healthy Subjects in India. EClinicalMedicine (2021) 38:101020. doi: 10.1016/j.eclinm.2021.101020
111. Tebas P, Yang S, Boyer JD, Reuschel EL, Patel A, Christensen-Quick A, et al. Safety and Immunogenicity of INO-4800 DNA Vaccine Against SARS-CoV-2: A Preliminary Report of an Open-Label, Phase 1 Clinical Trial. EClinicalMedicine (2021) 31:100689. doi: 10.1016/j.eclinm.2020.100689
112. Anderson EJ, Rouphael NG, Widge AT, Jackson LA, Roberts PC, Makhene M, et al. Safety and Immunogenicity of SARS-CoV-2 mRNA-1273 Vaccine in Older Adults. N Engl J Med (2020) 383(25):2427–38. doi: 10.1056/NEJMoa2028436
113. Walsh EE, Frenck RW Jr, Falsey AR, Kitchin N, Absalon J, Gurtman A, et al. Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates. N Engl J Med (2020) 383(25):2439–50. doi: 10.1056/NEJMoa2027906
114. Walls AC, Fiala B, Schäfer A, Wrenn S, Pham MN, Murphy M, et al. Elicitation of Potent Neutralizing Antibody Responses by Designed Protein Nanoparticle Vaccines for SARS-CoV-2. Cell (2020) 183(5):1367–82.e17. doi: 10.1016/j.cell.2020.10.043
115. Saunders KO, Lee E, Parks R, Martinez DR, Li D, Chen H, et al. Neutralizing Antibody Vaccine for Pandemic and Pre-Emergent Coronaviruses. Nature (2021) 594(7864):553–9. doi: 10.1038/s41586-021-03594-0
116. Hansen J, Baum A, Pascal KE, Russo V, Giordano S, Wloga E, et al. Studies in Humanized Mice and Convalescent Humans Yield a SARS-CoV-2 Antibody Cocktail. Science (2020) 369(6506):1010–4. doi: 10.1126/science.abd0827
117. Liu L, Wang P, Nair MS, Yu J, Rapp M, Wang Q, et al. Potent Neutralizing Antibodies Against Multiple Epitopes on SARS-CoV-2 Spike. Nature (2020) 584(7821):450–6. doi: 10.1038/s41586-020-2571-7
118. Kupferschmidt K. New Mutations Raise Specter of 'Immune Escape’. Science (2021) 371(6527):329–30. doi: 10.1126/science.371.6527.329
119. Xiang Y, Nambulli S, Xiao Z, Liu H, Sang Z, Duprex WP, et al. Versatile and Multivalent Nanobodies Efficiently Neutralize SARS-CoV-2. Science (2020) 370(6523):1479–84. doi: 10.1126/science.abe4747
120. Zhu Y, Feng F, Hu G, Wang Y, Yu Y, Zhu Y, et al. A Genome-Wide CRISPR Screen Identifies Host Factors That Regulate SARS-CoV-2 Entry. Nat Commun (2021) 12(1):961. doi: 10.1038/s41467-021-21213-4
121. Garrett ME, Galloway J, Chu HY, Itell HL, Stoddard CI, Wolf CR, et al. High-Resolution Profiling of Pathways of Escape for SARS-CoV-2 Spike-Binding Antibodies. Cell (2021) 184(11):2927–38.e11. doi: 10.1016/j.cell.2021.04.045
122. Cohen J. Vaccines That can Protect Against Many Coronaviruses Could Prevent Another Pandemic. Sci (New York NY) (2021) 372(6539):227–31. doi: 10.1126/science.372.6539.227
123. Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, et al. SARS-CoV-2 Variants, Spike Mutations and Immune Escape. Nat Rev Microbiol (2021) 19(7):409–24. doi: 10.1038/s41579-021-00573-0
124. Barnes CO, West AP Jr, Huey-Tubman KE, Hoffmann M, Sharaf NG, Hoffman PR, et al. Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell (2020) 182(4):828–42.e16. doi: 10.1016/j.cell.2020.06.025
125. Liu H, Wu NC, Yuan M, Bangaru S, Torres JL, Caniels TG, et al. Cross-Neutralization of a SARS-CoV-2 Antibody to a Functionally Conserved Site Is Mediated by Avidity. Immunity (2020) 53(6):1272–80.e5. doi: 10.1016/j.immuni.2020.10.023
126. Pan Y, Du J, Liu J, Wu H, Gui F, Zhang N, et al. Screening of Potent Neutralizing Antibodies Against SARS-CoV-2 Using Convalescent Patients-Derived Phage-Display Libraries. Cell Discov (2021) 7(1):57. doi: 10.1038/s41421-021-00295-w
127. Di Caro A, Cunha F, Petrosillo N, Beeching NJ, Ergonul O, Petersen E, et al. Severe Acute Respiratory Syndrome Coronavirus 2 Escape Mutants and Protective Immunity From Natural Infections or Immunizations. Clin Microbiol Infect (2021) 27(6):823–6. doi: 10.1016/j.cmi.2021.03.011
128. Yao H, Sun Y, Deng YQ, Wang N, Tan Y, Zhang NN, et al. Rational Development of a Human Antibody Cocktail That Deploys Multiple Functions to Confer Pan-SARS-CoVs Protection. Cell Res (2021) 31(1):25–36. doi: 10.1038/s41422-020-00444-y
129. Chen EC, Gilchuk P, Zost SJ, Suryadevara N, Winkler ES, Cabel CR, et al. Convergent Antibody Responses to the SARS-CoV-2 Spike Protein in Convalescent and Vaccinated Individuals. Cell Rep (2021) 36(8):109604. doi: 10.1016/j.celrep.2021.109604
Keywords: SARS-CoV-2 variants, immune escape, vaccine, neutralizing antibody, variants of interest, variants of concern
Citation: Lou F, Li M, Pang Z, Jiang L, Guan L, Tian L, Hu J, Fan J and Fan H (2021) Understanding the Secret of SARS-CoV-2 Variants of Concern/Interest and Immune Escape. Front. Immunol. 12:744242. doi: 10.3389/fimmu.2021.744242
Received: 20 July 2021; Accepted: 29 September 2021;
Published: 05 November 2021.
Edited by:
Paul Roesch, University of Bayreuth, GermanyCopyright © 2021 Lou, Li, Pang, Jiang, Guan, Tian, Hu, Fan and Fan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huahao Fan, ZmFuaHVhaGFvQG1haWwuYnVjdC5lZHUuY24=; Junfen Fan, ZmFuanVuZmVuQHh3aG9zcC5vcmc=; Maochen Li, bGltYW9jaGVuQG1haWwuYnVjdC5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.