- 1School of Medical Sciences, The University of Sydney, Sydney, NSW, Australia
- 2Brain and Mind Centre, The University of Sydney, Sydney, NSW, Australia
- 3School of Psychology, The University of Sydney, Sydney, NSW, Australia
Behavioral variant frontotemporal dementia (bvFTD) is a younger onset form of neurodegeneration initiated in the frontal and/or temporal lobes with a slow clinical onset but rapid progression. bvFTD is highly complex biologically with different pathological signatures and genetic variants that can exhibit a spectrum of overlapping clinical manifestations. Although the role of innate immunity has been extensively investigated in bvFTD, the involvement of adaptive immunity in bvFTD pathogenesis is poorly understood. We analyzed blood serum proteomics to identify proteins that are associated with autoimmune disease in bvFTD. Eleven proteins (increased: ATP5B, CALML5, COLEC11, FCGBP, PLEK, PLXND1; decreased: APOB, ATP8B1, FAM20C, LOXL3, TIMD4) were significantly altered in bvFTD with autoimmune disease compared to those without autoimmune disease. The majority of these proteins were enriched for glycoprotein-associated proteins and pathways, suggesting that the glycome is targeted in bvFTD with autoimmune disease.
Introduction
Behavioral variant frontotemporal dementia (bvFTD) is a non-Alzheimer’s younger onset neurodegenerative disease with a slow and subtle onset and rapid progression (1, 2). Affected individuals exhibit marked behavioral disturbances (1, 2). bvFTD is biologically complex with different pathological signatures and genetic variants that impact on similar cell types and networks in the brain (1, 2). Therefore, deciphering the precise disease mechanism(s) that give rise to various degenerative proteinopathies in the same neuronal systems in bvFTD remains a significant challenge. As indicated in other neurodegenerative diseases, the immune system and inflammation are involved, with innate immunity extensively investigated in bvFTD. There is evidence of a significantly altered glial landscape in diseased brain regions (3–6), a consistent association of bvFTD with the HLA immune loci (7, 8) and disease-causative genes that are associated with inflammation in bvFTD (i.e. C9ORF72, PGRN, TREM2), as reviewed in detail elsewhere (9).
Recently, we utilized a discovery proteomics approach to assess serum changes in patients with bvFTD demonstrating significant peripheral changes in calcium ion binding and innate immune pathway proteins (10). In contrast, the involvement of adaptive immunity in bvFTD is poorly understood biologically and remains to be explored. Evidence supporting a role for autoimmunity in bvFTD includes bvFTD-associated genetic variants linked to autoimmune conditions (11, 12), an overrepresentation of autoimmune disease in bvFTD, specifically non-thyroid autoimmune conditions linked to clinical and genetic bvFTD variants (13–15), and the presence of autoantibodies in individuals with bvFTD (16–19).
Autoimmunity is suggested to be an integral part of neurodegeneration (20, 21) with considerable evidence of immune system upregulation in neurodegenerative diseases, implying that the body’s immune system attacks cells of the CNS in a similar way to other autoimmune diseases (22). Indeed, international retrospective studies have demonstrated a significantly higher (80%) risk of dementia in middle-aged individuals with autoimmune diseases (23) and individuals admitted to hospital with an autoimmune disease are 20% more likely to have a subsequent admission for dementia (24).
In this study we investigated alterations in adaptive immunity, firstly by determining the prevalence of autoimmune disease in our patient cohort, and then applying our discovery proteomics approach to determine any blood serum changes in bvFTD patients with and without autoimmune disease.
Materials and Methods
Patient Information
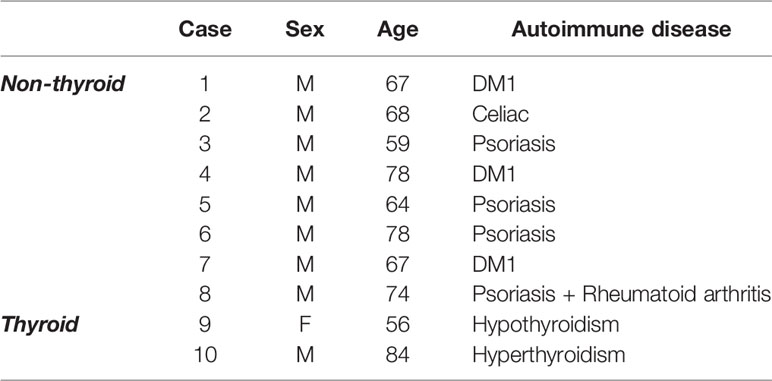
The proteomics dataset generated in our previous study (10) was re-analyzed specifically for changes in proteins associated with autoimmune disease. bvFTD patients were from FRONTIER, the frontotemporal dementia clinical research group at the University of Sydney Brain and Mind Centre, and from the ForeFront FTD and motor neuron disease clinic at the University of Sydney Brain and Mind Centre. Each patient in the cohort previously underwent neurological examination including a comprehensive cognitive assessment and structural brain MRI, and met current consensus diagnostic criteria for bvFTD (25). In the present study, the bvFTD cohort specifically were screened for prevalence of a panel of pre-determined autoimmune diseases that were collated from the Australian Society of Clinical Immunology and Allergy (ASCIA) (26) and adapted from previously published studies investigating autoimmune disease in international bvFTD cohorts (13, 14). We compared bvFTD patients with autoimmune disease (N=10) (Table 1) to those without autoimmune disease (N=62). Human research ethics approval was granted by the University of New South Wales (approval number: HC12573). All information on the bvFTD cohort and materials and methods relating to proteomics were reported in our previous study (10).
Blood Sampling and Proteomics Analysis
The current data is derived from our previous proteomics analysis (10). Briefly, blood samples (9 mL) were collected in tubes (BD Vacutainer SST II Advance Tube #367958), and serum prepared by centrifugation at 3,500 rpm for 10 min at 4°C, which was then aliquoted and stored at −80°C until use. A comprehensive analysis of bvFTD serum proteins using proteomics based on the advanced liquid chromatography-tandem mass spectrometry (LC–MS–MS) technology was then undertaken. Briefly, protein depletion method was used in which 96% of 14 high-abundant proteins (e.g. albumin, IgG) were removed (27) using a 4.6 mm × 100 mm Multiple Affinity Removal System column (MARS, Agilent, Santa Clara, CA, USA) based on the depletion method (27) and following the manufacturer’s instructions, allowing for greater accuracy in identifying less abundant proteins. A total of 40 μl of serum was diluted with 120 μl of buffer A and passed through a 0.22 μm filter and centrifuged at 16,000g. The supernatant was injected into a MARS column and the flow through collected. The column was washed with buffer B, to elute the bound proteins, before re-equilibrating with buffer A prior to the next sample. Collected fractions were buffer exchanged into 100 mM TEAB. Following this, we performed nano-capillary liquid chromatography-tandem mass spectrometry (LC-MS-MS) using a Dionex Ultimate 3000 HPLC system (Thermo Fisher Scientific, Waltham, MA, USA) coupled to an in-house fritless nano 75 μm × 30 cm column packed with Repro-Sil Pur 120 C18 stationary phase (1.9 μm, Dr, Maisch GmbH, Germany). Separated compounds were analyzed with an Orbitrap Fusion Tribrid Mass Spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) and a synchronous precursor selection MS3 method (28) was used for data collection. Proteome Discoverer 2.2 (Thermo Fisher Scientific, Waltham, MA, USA) was used to analyze the MS data and the raw mass spectrometry data was processed using MaxQuant (29). Proteomics data are available from the corresponding author upon request.
Western Blotting
Serum (equal volumes) were heated with sample buffer (3.2% SDS, 32% glycerol, 0.16% bromophenol blue, 100 mM Tris-HCl, pH 6.8, 8% 2-mercaptoethanol), electrophoresed on Criterion Stain-free 4-20% SDS-PAGE gels (Bio-Rad) and transferred onto nitrocellulose membranes at 100 volts for 30 min. The membranes were blocked with TBS containing 5% nonfat dry milk and probed with anti-LOXL3 antibody (mouse monoclonal, 1:1000, Santa Cruz, sc377216) overnight at 4°C. The membranes were then washed three times in TBS containing 0.1% Tween 20 and incubated with horseradish peroxidase-conjugated secondary antibody for 2 h at room temperature. Protein bands were detected using enhanced chemiluminescence and Gel Doc System (Bio-Rad). The blots were stripped and probed for housekeeper proteins transferrin. The signal intensity was quantified using Image Lab (Bio-Rad) and NIH ImageJ software (v1.45s).
Gene Ontology Analysis
Two gene ontology software programs, Bioprofiling (30) (www.bioprofiling.de, 16 Dec 2019) and STRING (31) v11 (16 Dec 2019), were used to interpret and predict function or pathway on a set of proteins identified by the proteomics analysis. The proteins that were significantly altered were inputted separately into each of the programs following their instructions.
Statistical Analysis
Statistical analysis on proteomics data was performed as previously described (10). Briefly, protein peak intensities were log2 transformed and any missing values were imputed using the k nearest neighbor algorithm (impute. knn function from the impute package in R). Following imputation, protein intensities were normalized across batches using the RUV-III (Removing Unwanted Variation-III) algorithm (32). Default parameters from the RUVIII function were used. After normalization, any proteins that were originally missing were removed, and samples with replicates averaged. Linear models were fitted using the R/Bioconductor software package limma (33). A design matrix which included age and sex as covariates was used and tested for significance of disease status; neither age nor sex had any effect on protein levels. The Benjamini–Hochberg method was used to control for multiple testing, and proteins with an adjusted P < 0.05 were considered to be statistically significant. For western blotting data, statistical analysis was performed using SPSS Statistics software (IBM, Chicago, Illinois), using a univariate analysis (general linear model), with age and sex as covariates, and statistical significance set at P < 0.05.
Results
Analysis of Serum Proteins Altered in bvFTD With Autoimmune Disease
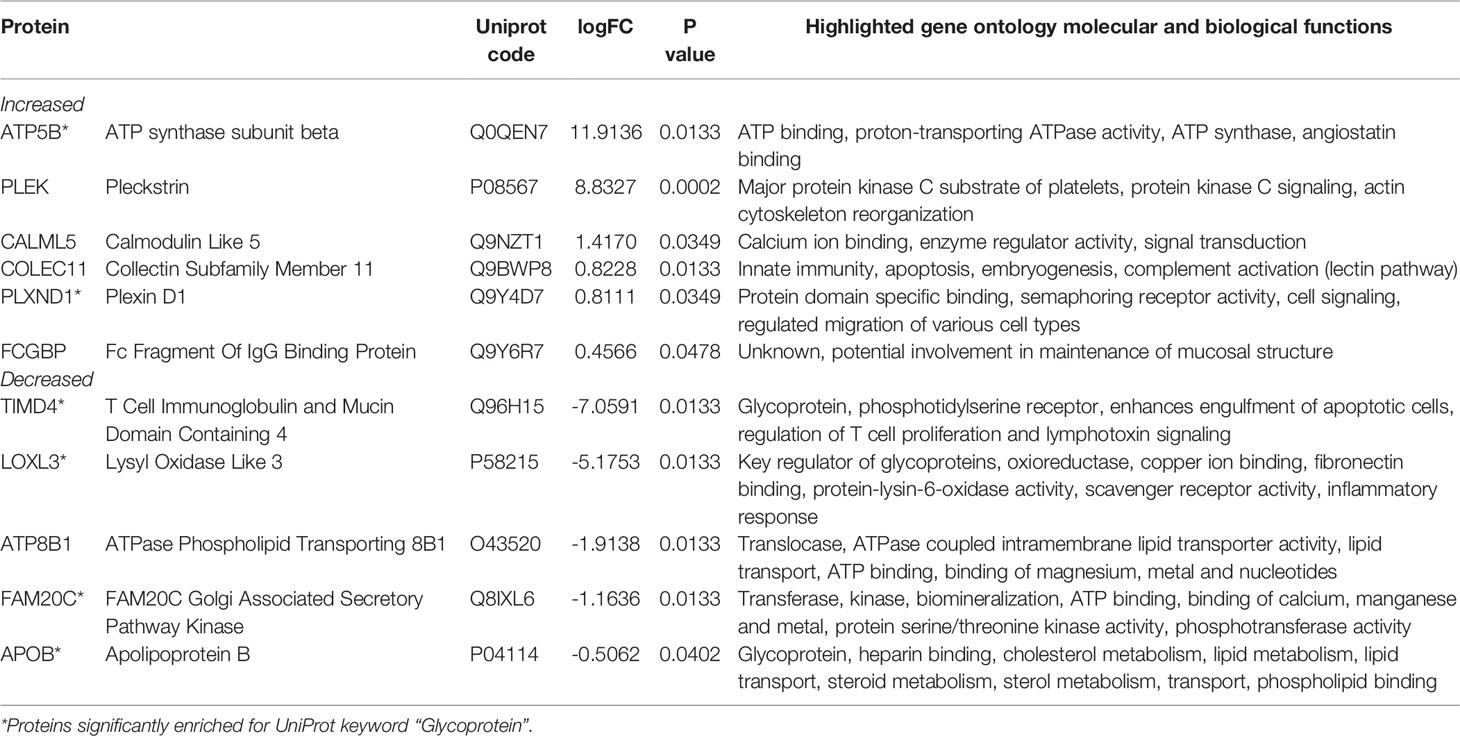
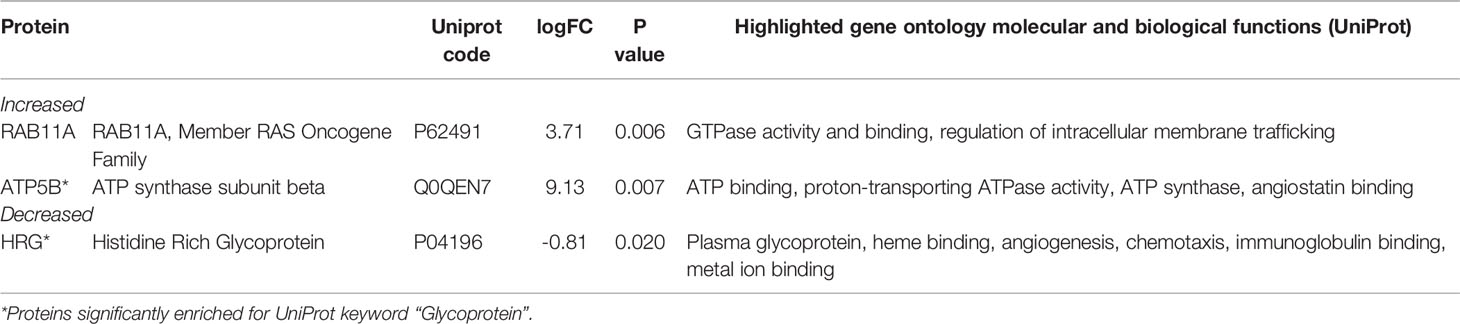
We analyzed serum proteins in bvFTD patients with autoimmune disease (bvFTD-autoimmune; N=10) (Table 1) and bvFTD patients without autoimmune disease (bvFTD-nonautoimmune; N=62) to identify proteins that are associated with autoimmune disease in bvFTD. We found that 11 proteins were significantly altered in bvFTD-autoimmune compared to bvFTD-nonautoimmune covarying for age and sex. Six proteins were significantly increased in bvFTD-autoimmune – ATP5B, CALML5, COLEC11, FCGBP, PLEK, PLXND1; and 5 proteins were significantly decreased – APOB, ATP8B1, FAM20C, LOXL3, TIMD4 (Table 2). Although measuring proteins in serum by western blotting is difficult, because of the low sensitivity of interfering proteins, we were able to validate the decrease in LOXL3 in bvFTD-autoimmune by this method (Figure 1). Also, we separated non-thyroid autoimmune disease (bvFTD-nonthyroid, N=8) (Table 1) from the bvFTD-autoimmune group and compared them to bvFTD-nonautoimmune, and found that 3 proteins were significantly altered; increased – ATP5B, RAB11A; decreased – HRG (Table 3).
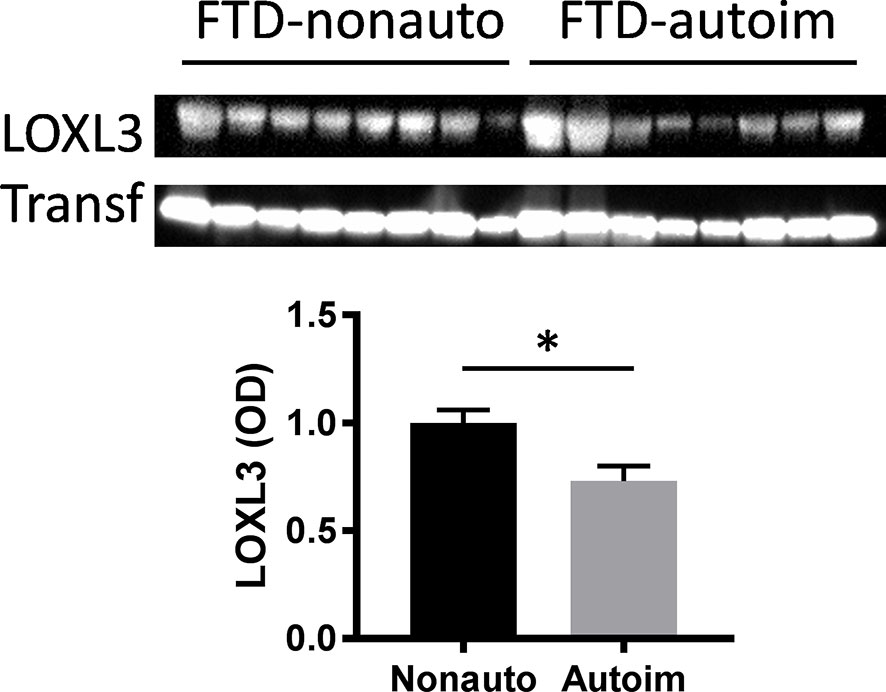
Figure 1 Validation of LOXL3 alteration in bvFTD-autoimmune compared to bvFTD-nonautoimmune serum by western blotting; normalized to the housekeeper protein transferrin (Transf) and optical density (OD) measurements of the bands. Data represent mean and SE as error bars, *P < 0.05.
Predicting Dysregulated Pathways in bvFTD With Autoimmune Disease
We then used two gene ontology software programs, STRING and Bioprofiling, to identify or predict functions/pathways possibly altered in bvFTD with autoimmune disease. The 11 proteins altered in bvFTD-autoimmune compared to bvFTD-nonautoimmune were assessed using Bioprofiling. Only one prominent pathway was generated: “Extracellular region” with 5 hits (APOB, FAM20C, FCGBP, LOXL3, PLEK). This same pathway was generated in STRING with the same 5 protein hits (APOB, FAM20C, FCGBP, LOXL3, PLEK). Interestingly, 6 of the 11 altered proteins in bvFTD-autoimmune (APOB, ATP5B, FAM20C, LOXL3, PLXND1, TIMD4) and 2 of the 3 altered proteins in bvFTD-nonthyroid (ATP5B and HRG) (Figure 2) were enriched for the Uniprot keyword “Glycoprotein” (KW-0325) and “Immune system process” (GO:0002376).
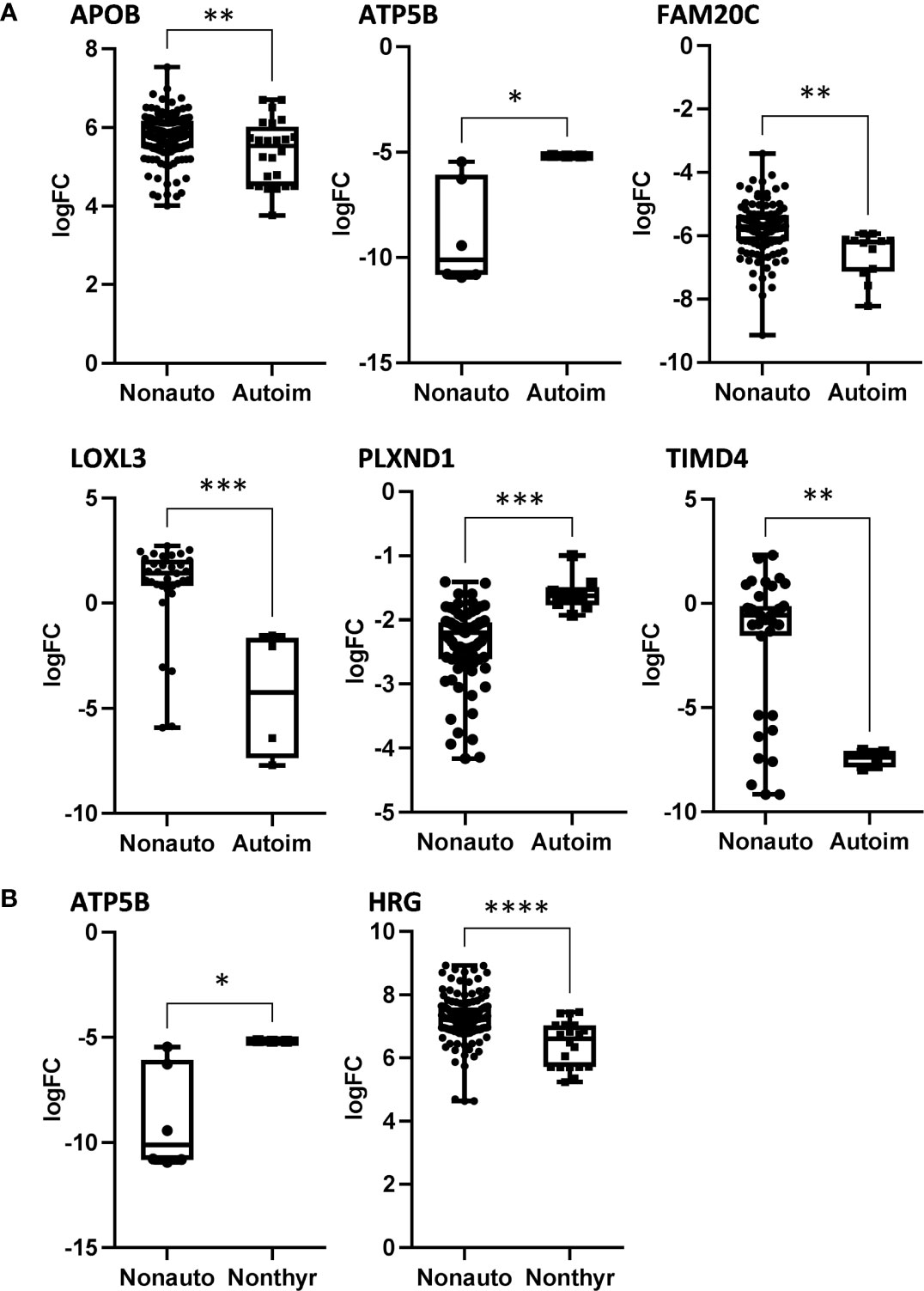
Figure 2 The significantly altered proteins in bvFTD-autoimmune (A) and bvFTD-nonthyroid (B) compared to bvFTD-nonautoimmune serum that were enriched for the Uniprot keyword “Glycoprotein”. FC, fold change. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Discussion
This study sought to investigate blood serum changes in the context of adaptive immunity in an Australian clinical bvFTD cohort with and without the presence of autoimmune disease. We analyzed our proteomics dataset to determine any blood serum changes in bvFTD with autoimmune disease compared to bvFTD without autoimmune disease. In addition, we determined if there was any change in blood serum in bvFTD with non-thyroid autoimmune disease, given previously published studies reported an increased prevalence specifically of non-thyroid autoimmune conditions in international FTD cohorts (13, 14).
Ten bvFTD individuals (14%) within the cohort were identified to have an autoimmune disease, the majority of which had non-thyroid autoimmune conditions (N=8). In Australia, autoimmune disease occurs in 5% of the population (26) and while our cohort is limited in number, the increased prevalence of autoimmune disease does support previous published findings of an overrepresentation of autoimmune disease in international FTD cohorts (13–15). Interestingly, of these individuals with autoimmune disease, nine were male (90%) and of these bvFTD males with autoimmune disease, all except one had non-thyroid autoimmune conditions. Autoimmune diseases are typically more prevalent in females than males (34, 35) and while analysis of any differences in sex could not be determined given more males than females were identified as having an autoimmune disease in this limited cohort, which is a limitation of the present study, the heightened presence of autoimmune disease in bvFTD males could potentially represent specific adaptive immune alterations driven by the combination of both male sex and autoimmune disease in bvFTD. There is significant evidence demonstrating clear differences between male and female immunity (35, 36) thus potentially a heightened chronic inflammatory environment such as that in neurodegeneration, could further exacerbate sex-related adaptive and peripheral immune responses or vice versa. Further investigation of potential sex-related differences specifically in adaptive immunity within bvFTD is required.
Proteomics analysis of blood serum identified 11 proteins that were significantly altered in bvFTD-autoimmune compared to bvFTD-nonautoimmune participants. While each of the 11 proteins are involved in diverse molecular and biological functions (Table 2), gene ontology analysis of these altered proteins identified ‘Extracellular region’ (GO:0005576) as being a significantly enriched pathway in bvFTD-autoimmune serum. Whereas in bvFTD-nonthyroid individuals, only 3 proteins were significantly altered, limiting the ability to perform gene ontology analysis. Of particular interest in the present study is that the majority of the significantly altered proteins were enriched for the Uniprot annotated keyword “Glycoprotein” (KW-0325) and the pathway “Immune system process” (GO:0002376). Glycoproteins play central and diverse roles in inflammatory processes and in the pathophysiology of chronic low-grade inflammatory conditions including diabetes type II, cardiovascular disease and cancer (37, 38). The process of glycosylation, is one of the most essential post-translational modifications (39) and plays a critical role in regulating functional immune responses via complex receptor-glycan motif interactions (40, 41). Specifically, protein glycosylation is identified to have a central role in the biochemical stabilization of 3D protein structure, protein folding, protein trafficking on cell membranes as antigen (42). The majority of glycoproteins are localized to the cell surface and are involved in cell adhesion, signal transduction, and structural maintenance of cells and tissues (42).
Glycosylation can be influenced by multiple factors including the type of cell, its activation state, environmental factors, age of the cell and inflammatory mediators such as cytokines (43). Each of these factors can be altered in the setting of disease, therefore glycoprotein expression or in general the ‘glycome’ could represent the overall health status of an individual (38, 44). Indeed, alterations in the human glycome have been associated with cancer and various autoimmune diseases (37, 38). Specifically, glycan processing contributes to the pathogenesis of autoimmune diseases, where abnormal glycosylation of one or more glycoproteins can occur (38). In the adaptive immune system, glycans have important roles in B and T cell differentiation and alterations in glycosylation can modulate inflammatory responses. Importantly, immunoglobulins are themselves glycoproteins, whose biological functions are modulated by their glycosylation patterns. Immunoglobulin glycosylation patterns that are identified to skew the immune system toward a pro- or anti-inflammatory direction are involved in the pathophysiology of autoimmune disease as reviewed in detail elsewhere (38, 45).
In the CNS, glycosylation is vital for maintaining normal brain functions and various glycan-rich molecules within the brain are involved in neural functions, including neuronal development, migration and regeneration (42, 46, 47). In addition, inflammation itself can induce glycan modifications that alter protein folding by masking sites for protease cleavage. This prevents proteolysis and extends the circulating half-life of serum proteins in addition to altering their structure, therefore redirecting the protein to different cell membrane receptors and altering its downstream cellular effects (48, 49). Given inflammation is considered a pathological hallmark of neurodegeneration, it is important to consider what affect this inflammatory response could have on adaptive immunity, particularly given adaptive immunity also serves to support the function of innate immunity and the dialogue between the two is critical and constant. In support of neurodegeneration driving these immune processes, variation in CSF glycan expression has been detected in multiple neurodegenerative diseases, including Alzheimer’s disease (AD), Parkinson’s disease, Huntington’s disease and amyotrophic lateral sclerosis (46, 50–56). Our data adds bvFTD to this list of CNS diseases.
There have been few investigations on the glycome in bvFTD. In AD affected brain regions, there are marked differences in the levels of soluble protein glycans compared to controls, particularly O-GlycNAcytylated and N-O-glycosylated proteins. Of note, individuals with FTD-tau Pick’s disease demonstrate similar lower levels of protein O-GlcNAcylation compared to AD, although they do not have the other AD variations in their glycome. As previously suggested, the glycome of each neurodegenerative disease is likely to differ and should be examined independently (57) as the glycome changes and adapts to maintain optimal function (42, 46, 47).
When comparing both neurodegeneration and autoimmunity, it is important to distinguish that the type of inflammatory/immune response involved in both is different. Autoimmune disease involves adaptive immunity, whereas neurodegenerative disease primarily involves innate immune responses. However, neurodegeneration has been suggested to exist on the same disease spectrum as autoimmunity, despite having different etiologies (20, 21). While the innate immune system has been the focus of investigation in FTD, adaptive immunity has yet to be fully examined, despite evidence to suggest a link between FTD and autoimmunity.
To date, the association between autoimmune disease and FTD has been unified by underlying TDP-43 pathology, and extends to GRN carriers (13). Furthermore, GWAS studies have identified novel risk loci that strongly implicate immune pathways in the pathogenesis of TDP-43 specifically (8), and an enrichment of FTD-associated genetic variants is observed in multiple autoimmune disorders (11). While there has been limited investigation into the glycome of sporadic bvFTD specifically, it is important to note that the GRN gene is itself a lysosomal glycoprotein critical for proper lysosomal function. Furthermore, GRN mutations have also been linked to autoimmunity with multiple studies reporting prominent upregulation of serum GRN levels in individuals with various autoimmune diseases (12, 58–62). In addition, antibodies to GRN have been demonstrated in individuals with histories of particular autoimmune conditions (63). This suggests that bvFTD individuals with GRN mutations (not assessed in the present study) in combination with autoimmune disease could have enhanced dysregulation of the glycome. Another bvFTD susceptibility gene triggering receptor expressed on myeloid cells 2 (TREM2) located in 6p21.1 MHC/HLA region of the genome is also a cell surface transmembrane glycoprotein (64, 65). Variations in TREM2 are implicated in autoimmunity and increased risk of autoimmune disease (66, 67).
Given the relationship between these FTD disease causative and susceptibility genes, their involvement in both innate and adaptive immunity, warrants further investigation of the link between glycoproteins and the various FTD subtypes with and without coinciding autoimmune disease. The genetic and pathological attributes of our bvFTD cohort was beyond the scope of the present study, however, in future to determine any specific glycoprotein changes that may be specifically linked to bvFTD and FTD overall further investigation in larger independent cohorts with genetic and pathologically confirmed individuals, with and without autoimmune disease is required.
Despite the need for further investigation and validation, what makes the findings of glycoprotein-associated alterations in bvFTD-autoimmune serum within our cohort particularly interesting, is the clinical utility of assays that are able to measure inflammatory glycoproteins. Unfortunately, sensitive and specific biomarkers of disease for FTD remain elusive which impedes the ability to make accurate diagnosis of the underlying disease subtype during life and also prevents the ability to track disease progression, both of these are critical features required to inform clinical trials. However, in the context of glycoproteins, newly established diagnostic and prognostic tests are able to utilize information from measuring the amount or structure of attached glycans to proteins which could be unique to both the individual and the disease.
In summary, discovery proteomics and gene ontology analysis in serums from bvFTD individuals with (14%) versus without autoimmune disease identified numerous glycoprotein-associated serum proteins and pathways that were significantly altered. While an altered glycome is not a new concept in terms of neurodegeneration or autoimmunity, this study provides evidence to suggest that the glycome is particularly affected in individuals with both bvFTD and autoimmune disease. This implies a unique adaptive immune profile specific to bvFTD in the setting of autoimmune disease, either driven by autoimmunity or potentially disease pathogenesis. Future follow up investigation utilizing quantitative glycoproteomics is required in larger independent genetically and pathologically confirmed FTD cohorts to confirm these findings and to further explore potential sex differences that may be associated with bvFTD and autoimmunity. The use of secreted and cell surface glycomes to reflect overall cellular status is routinely assessed (38) and quantified, not only to understand disease mechanisms, but also to improve diagnosis, prognosis and risk prediction (37, 68, 69). Further insights into the structure and function of the glycome in bvFTD could offer an approach for therapeutic development and the ability to fine tune immunological responses and inflammation to optimize the performance of therapeutics specifically targeting bvFTD, as has been suggested for other diseases (38).
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by University of New South Wales Human Research Ethics. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
WSK designed and supervised the project, analyzed the data, and wrote the manuscript. FB conducted the project, analyzed the data, and wrote the manuscript. JSK conducted the project, analyzed the data, and wrote the manuscript. GMH analyzed the data and revised the manuscript. JJK analyzed the data and revised the manuscript. OP and JRH recruited the patients and performed the neurological examinations, and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by funding to ForeFront, a collaborative research group dedicated to the study of frontotemporal dementia and motor neuron disease, from the National Health and Medical Research Council of Australia (NHMRC) program grant (#1037746). GMH is a NHMRC Senior Leadership Fellow (#1176607) and OP is a NHMRC Senior Research Fellow (#1103258).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank A. Connolly and B. Crossett for proteomics analysis and Kitty Lo for statistical analysis.
References
1. Hogan DB, Jette N, Fiest KM, Roberts JI, Pearson D, Smith EE, et al. The Prevalence and Incidence of Frontotemporal Dementia: A Systematic Review. Can J Neurol Sci (2016) 43(Suppl 1):S96–S109. doi: 10.1017/cjn.2016.25
2. Coyle-Gilchrist IT, Dick KM, Patterson K, Vazquez Rodriquez P, Wehmann E, Wilcox A, et al. Prevalence, Characteristics, and Survival of Frontotemporal Lobar Degeneration Syndromes. Neurology (2016) 86(18):1736–43. doi: 10.1212/WNL.0000000000002638
3. Lant SB, Robinson AC, Thompson JC, Rollinson S, Pickering-Brown S, Snowden JS, et al. Patterns of Microglial Cell Activation in Frontotemporal Lobar Degeneration. Neuropathol Appl Neurobiol (2014) 40(6):686–96. doi: 10.1111/nan.12092
4. Bellucci A, Bugiani O, Ghetti B, Spillantini MG. Presence of Reactive Microglia and Neuroinflammatory Mediators in a Case of Frontotemporal Dementia With P301S Mutation. Neurodegener Dis (2011) 8(4):221–9. doi: 10.1159/000322228
5. Sakae N, Roemer SF, Bieniek KF, Murray ME, Baker MC, Kasanuki K, et al. Microglia in Frontotemporal Lobar Degeneration With Progranulin or C9ORF72 Mutations. Ann Clin Transl Neurol (2019) 6(9):1782–96. doi: 10.1002/acn3.50875
6. Taipa R, Brochado P, Robinson A, Reis I, Costa P, Mann DM, et al. Patterns of Microglial Cell Activation in Alzheimer Disease and Frontotemporal Lobar Degeneration. Neurodegener Dis (2017) 17(4-5):145–54. doi: 10.1159/000457127
7. Ferrari R, Hernandez DG, Nalls MA, Rohrer JD, Ramasamy A, Kwok JB, et al. Frontotemporal Dementia and Its Subtypes: A Genome-Wide Association Study. Lancet Neurol (2014) 13(7):686–99. doi: 10.1016/S1474-4422(14)70065-1
8. Pottier C, Ren Y, Perkerson RB 3rd, Baker M, Jenkins GD, van Blitterswijk M, et al. Genome-Wide Analyses as Part of the International FTLD-TDP Whole-Genome Sequencing Consortium Reveals Novel Disease Risk Factors and Increases Support for Immune Dysfunction in FTLD. Acta Neuropathol (2019) 137(6):879–99. doi: 10.1007/s00401-019-01962-9
9. Bright F, Werry EL, Dobson-Stone C, Piguet O, Ittner LM, Halliday GM, et al. Neuroinflammation in Frontotemporal Dementia. Nat Rev Neurol (2019) 15(9):540–55. doi: 10.1038/s41582-019-0231-z
10. Katzeff JS, Bright F, Lo K, Kril JJ, Connolly A, Crossett B, et al. Altered Serum Protein Levels in Frontotemporal Dementia and Amyotrophic Lateral Sclerosis Indicate Calcium and Immunity Dysregulation. Sci Rep (2020) 10(1):13741. doi: 10.1038/s41598-020-70687-7
11. Broce I, Karch CM, Wen N, Fan CC, Wang Y, Tan CH, et al. Immune-Related Genetic Enrichment in Frontotemporal Dementia: An Analysis of Genome-Wide Association Studies. PloS Med (2018) 15(1):e1002487. doi: 10.1371/journal.pmed.1002487
12. Jian J, Li G, Hettinghouse A, Liu C. Progranulin: A Key Player in Autoimmune Diseases. Cytokine (2018) 101:48–55. doi: 10.1016/j.cyto.2016.08.007
13. Miller ZA, Rankin KP, Graff-Radford NR, Takada LT, Sturm VE, Cleveland CM, et al. TDP-43 Frontotemporal Lobar Degeneration and Autoimmune Disease. J Neurol Neurosurg Psychiatry (2013) 84(9):956–62. doi: 10.1136/jnnp-2012-304644
14. Miller ZA, Sturm VE, Camsari GB, Karydas A, Yokoyama JS, Grinberg LT, et al. Increased Prevalence of Autoimmune Disease Within C9 and FTD/MND Cohorts: Completing the Picture. Neurol Neuroimmunol Neuroinflamm (2016) 3(6):e301. doi: 10.1212/NXI.0000000000000301
15. Katisko K, Solje E, Koivisto AM, Kruger J, Kinnunen T, Hartikainen P, et al. Prevalence of Immunological Diseases in a Finnish Frontotemporal Lobar Degeneration Cohort With the C9orf72 Repeat Expansion Carriers and Non-Carriers. J Neuroimmunol (2018) 321:29–35. doi: 10.1016/j.jneuroim.2018.05.011
16. Gelpi E, Hoftberger R, Graus F, Ling H, Holton JL, Dawson T, et al. Neuropathological Criteria of Anti-IgLON5-Related Tauopathy. Acta Neuropathol (2016) 132(4):531–43. doi: 10.1007/s00401-016-1591-8
17. Borroni B, Stanic J, Verpelli C, Mellone M, Bonomi E, Alberici A, et al. Anti-AMPA GluA3 Antibodies in Frontotemporal Dementia: A New Molecular Target. Sci Rep (2017) 7(1):6723. doi: 10.1038/s41598-017-06117-y
18. Palese F, Bonomi E, Nuzzo T, Benussi A, Mellone M, Zianni E, et al. Anti-GluA3 Antibodies in Frontotemporal Dementia: Effects on Glutamatergic Neurotransmission and Synaptic Failure. Neurobiol Aging (2020) 86:143–55. doi: 10.1016/j.neurobiolaging.2019.10.015
19. Cavazzana I, Alberici A, Bonomi E, Ottaviani R, Kumar R, Archetti S, et al. Antinuclear Antibodies in Frontotemporal Dementia: The Tip’s of Autoimmunity Iceberg? J Neuroimmunol (2018) 325:61–3. doi: 10.1016/j.jneuroim.2018.10.006
20. de Haan P, Klein HC, t Hart BA. Autoimmune Aspects of Neurodegenerative and Psychiatric Diseases: A Template for Innovative Therapy. Front Psychiatry (2017) 8:46. doi: 10.3389/fpsyt.2017.00046
21. Schwartz M, Butovsky O, Kipnis J. Does Inflammation in an Autoimmune Disease Differ From Inflammation in Neurodegenerative Diseases? Possible Implications for Therapy. J Neuroimmun Pharmacol (2006) 1(1):4–10. doi: 10.1007/s11481-005-9010-2
22. Geschwind MD, Haman A, Miller BL. Rapidly Progressive Dementia. Neurol Clin (2007) 25(3):783–807, vii. doi: 10.1016/j.ncl.2007.04.001
23. Lin TM, Chen WS, Sheu JJ, Chen YH, Chen JH, Chang CC. Autoimmune Rheumatic Diseases Increase Dementia Risk in Middle-Aged Patients: A Nationwide Cohort Study. PloS One (2018) 13(1):e0186475. doi: 10.1371/journal.pone.0186475
24. Wotton CJ, Goldacre MJ. Associations Between Specific Autoimmune Diseases and Subsequent Dementia: Retrospective Record-Linkage Cohort Study, UK. J Epidemiol Community Health (2017) 71(6):576–83. doi: 10.1136/jech-2016-207809
25. Rascovsky K, Hodges JR, Knopman D, Mendez MF, Kramer JH, Neuhaus J, et al. Sensitivity of Revised Diagnostic Criteria for the Behavioural Variant of Frontotemporal Dementia. Brain (2011) 134(Pt 9):2456–77. doi: 10.1093/brain/awr179
26. ASCIA. Fast Facts Autoimmune Diseases. Australasian Society of Clinical Immunology and Allergy (2019). Available at: https://www.allergy.org.au/patients/fast-facts/autoimmune-diseases.
27. Zolotarjova N, Mrozinski P, Chen H, Martosella J. Combination of Affinity Depletion of Abundant Proteins and Reversed-Phase Fractionation in Proteomic Analysis of Human Plasma/Serum. J Chromatogr A (2008) 1189(1-2):332–8. doi: 10.1016/j.chroma.2007.11.082
28. Ting L, Rad R, Gygi SP, Haas W. MS3 Eliminates Ratio Distortion in Isobaric Multiplexed Quantitative Proteomics. Nat Methods (2011) 8(11):937–40. doi: 10.1038/nmeth.1714
29. Cox J, Mann M. MaxQuant Enables High Peptide Identification Rates, Individualized P.P.B.-Range Mass Accuracies and Proteome-Wide Protein Quantification. Nat Biotechnol (2008) 26(12):1367–72. doi: 10.1038/nbt.1511
30. Antonov AV. BioProfiling.de: Analytical Web Portal for High-Throughput Cell Biology. Nucleic Acids Res (2011) 39(Web Server issue):W323–7. doi: 10.1093/nar/gkr372
31. Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING V11: Protein-Protein Association Networks With Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res (2019) 47(D1):D607–D13. doi: 10.1093/nar/gky1131
32. Molania R, Gagnon-Bartsch JA, Dobrovic A, Speed TP. A New Normalization for Nanostring Ncounter Gene Expression Data. Nucleic Acids Res (2019) 47(12):6073–83. doi: 10.1093/nar/gkz433
33. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res (2015) 43(7):e47. doi: 10.1093/nar/gkv007
34. Desai MK, Brinton RD. Autoimmune Disease in Women: Endocrine Transition and Risk Across the Lifespan. Front Endocrinol (Lausanne) (2019) 10:265. doi: 10.3389/fendo.2019.00265
35. Ngo ST, Steyn FJ, McCombe PA. Gender Differences in Autoimmune Disease. Front Neuroendocrinol (2014) 35(3):347–69. doi: 10.1016/j.yfrne.2014.04.004
36. Klein SL, Flanagan KL. Sex Differences in Immune Responses. Nat Rev Immunol (2016) 16(10):626–38. doi: 10.1038/nri.2016.90
37. Connelly MA, Gruppen EG, Otvos JD, Dullaart RPF. Inflammatory Glycoproteins in Cardiometabolic Disorders, Autoimmune Diseases and Cancer. Clin Chim Acta (2016) 459:177–86. doi: 10.1016/j.cca.2016.06.012
38. Reily C, Stewart TJ, Renfrow MB, Novak J. Glycosylation in Health and Disease. Nat Rev Nephrol (2019) 15(6):346–66. doi: 10.1038/s41581-019-0129-4
39. Ohtsubo K, Marth JD. Glycosylation in Cellular Mechanisms of Health and Disease. Cell (2006) 126(5):855–67. doi: 10.1016/j.cell.2006.08.019
40. Wahl A, van den Akker E, Klaric L, Stambuk J, Benedetti E, Plomp R, et al. Genome-Wide Association Study on Immunoglobulin G Glycosylation Patterns. Front Immunol (2018) 9:277. doi: 10.3389/fimmu.2018.00277
41. Plomp R, Ruhaak LR, Uh HW, Reiding KR, Selman M, Houwing-Duistermaat JJ, et al. Subclass-Specific IgG Glycosylation is Associated With Markers of Inflammation and Metabolic Health. Sci Rep (2017) 7(1):12325. doi: 10.1038/s41598-017-12495-0
42. Varki A. Biological Roles of Glycans. Glycobiology (2017) 27(1):3–49. doi: 10.1093/glycob/cww086
43. Maverakis E, Kim K, Shimoda M, Gershwin ME, Patel F, Wilken R, et al. Glycans in the Immune System and The Altered Glycan Theory of Autoimmunity: A Critical Review. J Autoimmun (2015) 57:1–13. doi: 10.1016/j.jaut.2014.12.002
44. Merleev AA, Park D, Xie Y, Kailemia MJ, Xu G, Ruhaak LR, et al. A Site-Specific Map of the Human Plasma Glycome and Its Age and Gender-Associated Alterations. Sci Rep (2020) 10(1):17505. doi: 10.1038/s41598-020-73588-x
45. Goulabchand R, Vincent T, Batteux F, Eliaou JF, Guilpain P. Impact of Autoantibody Glycosylation in Autoimmune Diseases. Autoimmun Rev (2014) 13(7):742–50. doi: 10.1016/j.autrev.2014.02.005
46. Iqbal S, Ghanimi Fard M, Everest-Dass A, Packer NH, Parker LM. Understanding Cellular Glycan Surfaces in the Central Nervous System. Biochem Soc Trans (2019) 47(1):89–100. doi: 10.1042/BST20180330
47. Scott H, Panin VM. N-Glycosylation in Regulation of the Nervous System. Adv Neurobiol (2014) 9:367–94. doi: 10.1007/978-1-4939-1154-7_17
48. Schnaar RL. Glycobiology Simplified: Diverse Roles of Glycan Recognition in Inflammation. J Leukoc Biol (2016) 99(6):825–38. doi: 10.1189/jlb.3RI0116-021R
49. van Kooyk Y, Rabinovich GA. Protein-Glycan Interactions in the Control of Innate and Adaptive Immune Responses. Nat Immunol (2008) 9(6):593–601. doi: 10.1038/ni.f.203
50. Varadi C, Nehez K, Hornyak O, Viskolcz B, Bones J. Serum N-Glycosylation in Parkinson’s Disease: A Novel Approach for Potential Alterations. Molecules (2019) 24(12):2220. doi: 10.3390/molecules24122220
51. Akasaka-Manya K, Kawamura M, Tsumoto H, Saito Y, Tachida Y, Kitazume S, et al. Excess APP O-Glycosylation by GalNAc-T6 Decreases Abeta Production. J Biochem (2017) 161(1):99–111. doi: 10.1093/jb/mvw056
52. Gaunitz S, Tjernberg LO, Schedin-Weiss S. The N-Glycan Profile in Cortex and Hippocampus is Altered in Alzheimer Disease. J Neurochem (2020) 00:1–13. doi: 10.1111/jnc.15202
53. Zhang Q, Ma C, Chin LS, Li L. Integrative Glycoproteomics Reveals Protein N-Glycosylation Aberrations and Glycoproteomic Network Alterations in Alzheimer’s Disease. Sci Adv (2020) 6(40):eabc5802. doi: 10.1126/sciadv.abc5802
54. Moll T, Shaw PJ, Cooper-Knock J. Disrupted Glycosylation of Lipids and Proteins is a Cause of Neurodegeneration. Brain (2020) 143(5):1332–40. doi: 10.1093/brain/awz358
55. Edri-Brami M, Rosental B, Hayoun D, Welt M, Rosen H, Wirguin I, et al. Glycans in Sera of Amyotrophic Lateral Sclerosis Patients and Their Role in Killing Neuronal Cells. PloS One (2012) 7(5):e35772. doi: 10.1371/journal.pone.0035772
56. Bras IC, Konig A, Outeiro TF. Glycation in Huntington’s Disease: A Possible Modifier and Target for Intervention. J Huntingtons Dis (2019) 8(3):245–56. doi: 10.3233/JHD-190366
57. Frenkel-Pinter M, Shmueli MD, Raz C, Yanku M, Zilberzwige S, Gazit E, et al. Interplay Between Protein Glycosylation Pathways in Alzheimer’s Disease. Sci Adv (2017) 3(9):e1601576. doi: 10.1126/sciadv.1601576
58. Tang W, Lu Y, Tian QY, Zhang Y, Guo FJ, Liu GY, et al. The Growth Factor Progranulin Binds to TNF Receptors and Is Therapeutic Against Inflammatory Arthritis in Mice. Science (2011) 332(6028):478–84. doi: 10.1126/science.1199214
59. Zhao YP, Tian QY, Liu CJ. Progranulin Deficiency Exaggerates, Whereas Progranulin-Derived Atsttrin Attenuates, Severity of Dermatitis in Mice. FEBS Lett (2013) 587(12):1805–10. doi: 10.1016/j.febslet.2013.04.037
60. Zhou B, Li H, Liu J, Xu L, Guo Q, Sun H, et al. Progranulin Induces Adipose Insulin Resistance and Autophagic Imbalance via TNFR1 in Mice. J Mol Endocrinol (2015) 55(3):231–43. doi: 10.1530/JME-15-0075
61. Matsubara T, Mita A, Minami K, Hosooka T, Kitazawa S, Takahashi K, et al. PGRN is a Key Adipokine Mediating High Fat Diet-Induced Insulin Resistance and Obesity Through IL-6 in Adipose Tissue. Cell Metab (2012) 15(1):38–50. doi: 10.1016/j.cmet.2011.12.002
62. Wei F, Zhang Y, Zhao W, Yu X, Liu CJ. Progranulin Facilitates Conversion and Function of Regulatory T Cells Under Inflammatory Conditions. PloS One (2014) 9(11):e112110. doi: 10.1371/journal.pone.0112110
63. Thurner L, Preuss KD, Fadle N, Regitz E, Klemm P, Zaks M, et al. Progranulin Antibodies in Autoimmune Diseases. J Autoimmun (2013) 42:29–38. doi: 10.1016/j.jaut.2012.10.003
64. Bouchon A, Dietrich J, Colonna M. Cutting Edge: Inflammatory Responses can be Triggered by TREM-1, a Novel Receptor Expressed on Neutrophils and Monocytes. J Immunol (2000) 164(10):4991–5. doi: 10.4049/jimmunol.164.10.4991
65. Allcock RJ, Barrow AD, Forbes S, Beck S, Trowsdale J. The Human TREM Gene Cluster at 6p21.1 Encodes Both Activating and Inhibitory Single IgV Domain Receptors and Includes Nkp44. Eur J Immunol (2003) 33(2):567–77. doi: 10.1002/immu.200310033
66. Matzaraki V, Kumar V, Wijmenga C, Zhernakova A. The MHC Locus and Genetic Susceptibility to Autoimmune and Infectious Diseases. Genome Biol (2017) 18(1):76. doi: 10.1186/s13059-017-1207-1
67. Gough SC, Simmonds MJ. The HLA Region and Autoimmune Disease: Associations and Mechanisms of Action. Curr Genomics (2007) 8(7):453–65. doi: 10.2174/138920207783591690
68. Hu M, Lan Y, Lu A, Ma X, Zhang L. Glycan-Based Biomarkers for Diagnosis of Cancers and Other Diseases: Past, Present, and Future. Prog Mol Biol Transl Sci (2019) 162:1–24. doi: 10.1016/bs.pmbts.2018.12.002
Keywords: frontotemporal dementia, autoimmune disease, proteomics, serum, thyroid, glycoprotein, glycome, biomarker
Citation: Bright F, Katzeff JS, Hodges JR, Piguet O, Kril JJ, Halliday GM and Kim WS (2021) Glycoprotein Pathways Altered in Frontotemporal Dementia With Autoimmune Disease. Front. Immunol. 12:736260. doi: 10.3389/fimmu.2021.736260
Received: 05 July 2021; Accepted: 16 August 2021;
Published: 01 September 2021.
Edited by:
Li-Tung Huang, Kaohsiung Chang Gung Memorial Hospital, TaiwanReviewed by:
Jordi A. Matias-Guiu, Hospital Clínico San Carlos, SpainVita Golubovskaya, University of Oklahoma Health Sciences Center, United States
Copyright © 2021 Bright, Katzeff, Hodges, Piguet, Kril, Halliday and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Woojin Scott Kim, d29vamluLmtpbUBzeWRuZXkuZWR1LmF1
†These authors have contributed equally to this work
 Fiona Bright
Fiona Bright Jared S. Katzeff
Jared S. Katzeff John R. Hodges1,2
John R. Hodges1,2 Olivier Piguet
Olivier Piguet Glenda M. Halliday
Glenda M. Halliday Woojin Scott Kim
Woojin Scott Kim