
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 22 November 2021
Sec. Comparative Immunology
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.733266
This article is part of the Research TopicImmunity to Emerging Pathogens in Poikilothermic VertebratesView all 9 articles
 Hajarooba Gnanagobal1,2
Hajarooba Gnanagobal1,2 Trung Cao1
Trung Cao1 Ahmed Hossain1
Ahmed Hossain1 My Dang1
My Dang1 Jennifer R. Hall3
Jennifer R. Hall3 Surendra Kumar1,4
Surendra Kumar1,4 Doan Van Cuong5
Doan Van Cuong5 Danny Boyce6
Danny Boyce6 Javier Santander1*
Javier Santander1*Renibacterium salmoninarum is a Gram-positive, intracellular pathogen that causes Bacterial Kidney Disease (BKD) in several fish species in freshwater and seawater. Lumpfish (Cyclopterus lumpus) is utilized as a cleaner fish to biocontrol sea lice infestation in Atlantic salmon (Salmo salar) farms. Atlantic salmon is susceptible to R. salmoninarum, and it can transfer the infection to other fish species. Although BKD outbreaks have not been reported in lumpfish, its susceptibility and immune response to R. salmoninarum is unknown. In this study, we evaluated the susceptibility and immune response of lumpfish to R. salmoninarum infection. Groups of lumpfish were intraperitoneally (i.p.) injected with either R. salmoninarum (1×107, 1×108, or 1×109 cells dose-1) or PBS (control). R. salmoninarum infection kinetics and mortality were followed for 98 days post-infection (dpi). Transcript expression levels of 33 immune-relevant genes were measured in head kidney (n = 6) of fish infected with 1×109 cells/dose and compared to the control at 28 and 98 dpi. Infected lumpfish displayed characteristic clinical signs of BKD. Lumpfish infected with high, medium, and low doses had a survival rate of 65%, 93%, and 95%, respectively. Mortality in the high-dose infected group stabilized after 50 dpi, but R. salmoninarum persisted in the fish tissues until 98 dpi. Cytokines (il1β, il8a, il8b), pattern recognition receptors (tlr5a), interferon-induced effectors (rsad2, mxa, mxb, mxc), and iron regulation (hamp) and acute phase reactant (saa5) related genes were up-regulated at 28 dpi. In contrast, cell-mediated adaptive immunity-related genes (cd4a, cd4b, ly6g6f, cd8a, cd74) were down-regulated at 28 dpi, revealing the immune suppressive nature of R. salmoninarum. However, significant upregulation of cd74 at 98 dpi suggests induction of cell-mediated immune response. This study showed that R. salmoninarum infected lumpfish in a similar fashion to salmonid fish species and caused a chronic infection, enhancing cell-mediated adaptive immune response.
Bacterial Kidney Disease (BKD) caused by Renibacterium salmoninarum is a chronic disease of wild and cultured fish, including Atlantic salmon (Salmo salar), chinook salmon (Oncorhynchus tshawytscha), rainbow trout (Oncorhynchus mykiss), Arctic char (Salvelinus alpinus L.), Pacific herring (Clupea pallasii pallasii), sablefish (Anoplopoma fimbria), flathead minnow (Pimephales promelas), North Pacific hake (Merluccius productus), ayu (Plecoglossus altivelis), eel (Anguilla anguilla) and bivalve molluscs, in both fresh and marine waters (1–7). R. salmoninarum has primarily adapted to infect and persist in salmonids (8). However, R. salmoninarum experimentally infected and caused mortality in non-salmonids including, sablefish and Pacific herring, shiner perch (Cymatogaster aggregate), common shiner (Notropis cornutus), and flathead minnow, and it caused mortalities events in minnow (Phoxinus phoxinus) and three-spined stickleback (Gasterosteus aculeatus) (2, 9–12).
R. salmoninarum is a Gram-positive, slow-growing, fastidious, and facultative intracellular pathogen (7, 13), which persistence within wild and farmed fish populations is high (2). R. salmoninarum is the only marine bacterial pathogen that has been documented of both horizontal (i.e. from fish to fish) and vertical (i.e. from parent to progeny) transmission (2). R. salmoninarum has caused substantial losses in the salmonid aquaculture industry, affecting up to 80% and 40% of the Pacific and Atlantic salmon stocks, respectively (8). The poor efficacy of antibiotics and vaccines in BKD prophylaxis has stymied the control of this pathogen (14, 15).
Lumpfish (Cyclopterus lumpus), a globiform teleost native to the North Atlantic, is used as an eco-friendly cleaner fish to biocontrol sea lice (e.g., Lepeophtheirus salmonis) infestations in the Atlantic salmon aquaculture (16). Lumpfish reduces the utilization of chemotherapeutants against sea lice in Atlantic salmon farms, consequently its annual demands have significantly increased in the North Atlantic (17). Lumpfish health is critical for its optimal performance and elimination of potential risk of disease transmission between lumpfish and salmon (18, 19). Pasteurella sp., Piscirickettsia salmonis, Vibrio anguillarum, Vibrio ordalii, Aeromonas salmonicida, Pseudomonas anguilliseptica, Moritella viscosa, and Tenacibaculum maritimum have been reported to be primary bacterial pathogens in lumpfish (17). Although R. salmoninarum outbreaks have not been reported in lumpfish, due to the broad host range of R. salmoninarum (i.e., salmonids, non-salmonids, bivalves and molluscs) and its horizontal transmission ability (20, 21), it is important to determine the susceptibility of lumpfish to R. salmoninarum and its potential risk for BKD. The risk of R. salmoninarum infection in lumpfish is significant because sea lice, like other blood-sucking ectoparasites, act as R. salmoninarum vector and could transfer R. salmoninarum from salmon to lumpfish and vice versa (22–24). R. salmoninarum transmission may occur as a result of the dynamic interplay between a susceptible host and virulent R. salmoninarum in an environmental context that facilitates such disease conditions (i.e., environmental stressors in the marine environment, high stocking densities in cultured conditions or parasitic infestations) (25, 26). For instance, horizontal transmission of R. salmoninarum between fish species like sockeye salmon (Oncorhynchus nerka) and chinook salmon (O. tshawytscha) has been reported (20, 21), and high biomass within sea cages and the free movement of seawater in and out of cages could increase the opportunity for disease transmission (27). Cleaner fish like lumpfish poses moderate risk of disease transmission to salmon (28). Transmission of amoebic parasite (Paramoeba perurans) from lumpfish to Atlantic salmon was demonstrated under controlled conditions (29). Though, the anticipated risk of infected lumpfish transmit bacterial disease to salmon is low, Atlantic salmon showed susceptibility to a lumpfish isolate of M. viscosa (28, 30). Thus, it could be possible that lumpfish act as an asymptomatic carrier and transmit disease threat to salmon (19). Several studies on the fish immune response to R. salmoninarum infection have been conducted in salmonids (31–34). However, the lumpfish susceptibility and immune response to R. salmoninarum infection is unknown. In addition, lumpfish is becoming an accessible model to study marine infectious diseases and teleost immunity (35).
Here, we evaluated the lumpfish susceptibility to a type strain of R. salmoninarum (ATCC 33209) and immune response at early and chronic infection stages. We determined that lumpfish is susceptible to R. salmoninarum, causing mortality and a chronic infection in the surviving individuals, similar to salmonid fish. The immune response profile of lumpfish head kidney at early and chronic infection stages showed that R. salmoninarum dysregulates the expression of transcripts with functional annotations related to pattern recognition, inflammation, cytokines, iron regulation, and cell-mediated adaptive immunity.
R. salmoninarum type strain [ATCC (American Type Culture Collection) 33209] was cultured in complex KDM2 broth [1.0% (w/v) peptone (Difco), 0.05% (w/v) yeast (Difco), 0.05% (w/v) L-cysteine HCl (Sigma-Aldrich, St. Louis, MO, USA), 10% (v/v) fetal bovine serum (Gibco, Thermofisher, CA, USA), 1.5% (v/v) nurse medium contained filter-sterilized supernatant from R. salmoninarum cultures] (36) at 15°C with aeration in an orbital shaker (180 rpm). When required, KDM2 broth was supplemented with 1.8% (w/v) agar (Difco), and cycloheximide (0.005% (w/v); Sigma-Aldrich), D-cycloserine (0.00125% (w/v); Sigma-Aldrich), polymyxin-B sulfate [0.0025% (w/v); Sigma-Aldrich], and oxolinic acid [0.00025% (w/v); Sigma-Aldrich] to make R. salmoninarum selective KDM2 plates (SKDM2) (37). Bacterial growth was monitored by spectrophotometry (Genova Nano, Jenway, UK), flow cytometry (BD FACS Aria II flow cytometer and BD FACS Diva v7.0 software, BD Biosciences, San Jose, CA, USA) and/or by colony forming units (CFU) plate counting (38). The purity and integrity of bacterial cells were evaluated and confirmed by Gram-staining (39) (Figure 1A) and PCR (40, 41).
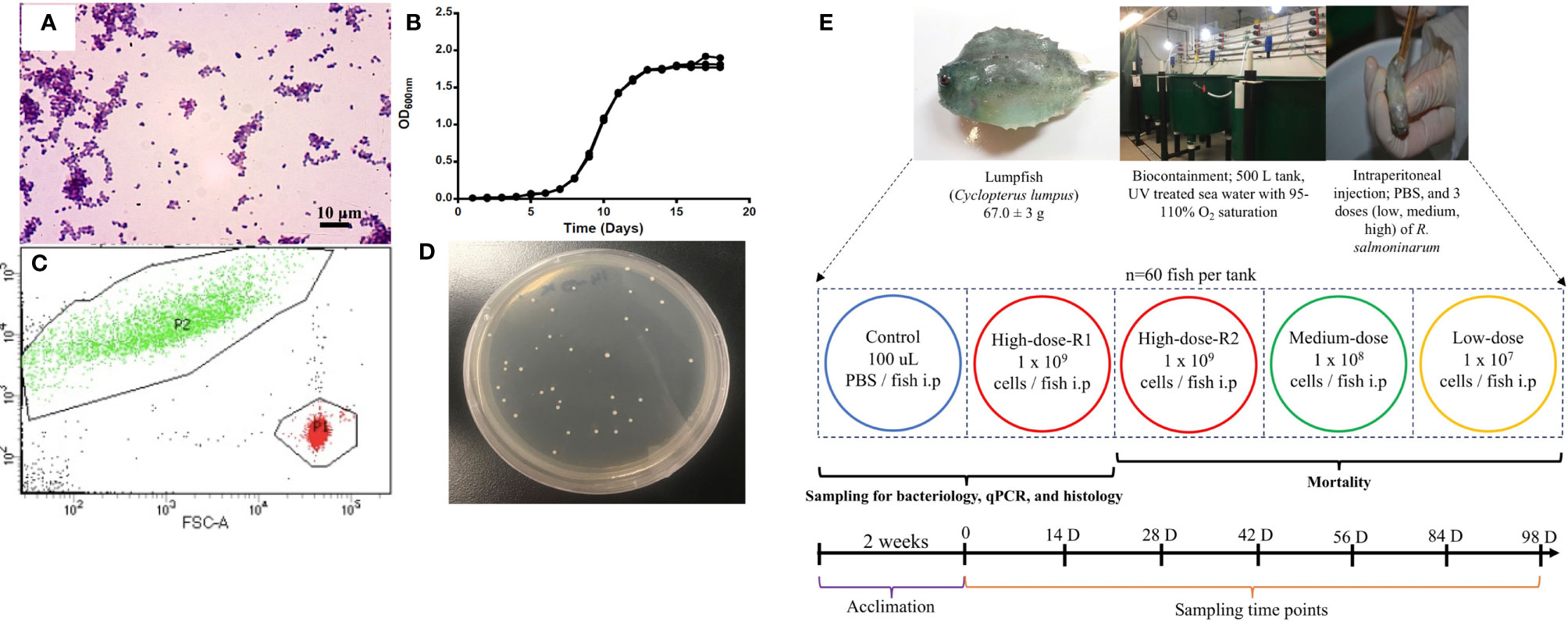
Figure 1 Renibacterium salmoninarum infection in lumpfish. (A) Characterization of R. salmoninarum by Gram staining; (B) R. salmoninarum growth curve in KDM2 broth; (C) Flow cytometric enumeration of R. salmoninarum. FSC: Forward Scatter; FITC: Green Fluorescence. In this plot of forward scatter versus fluorescence, green signals in the upper left-hand frame represent bacteria stained with the SYTO BC bacterial stain; red signals in the lower right-hand frame represent microsphere particles, which serve as the standard used to indicate sample volume. P1: Number of signals in the microsphere frame; P2: Number of signals in the bacterial frame. (D) R. salmoninarum colonies growing on a SKDM2 spread plate inoculated for quantitative culture of the bacterium from fish head kidney; (E) Experimental design for this study. 300 lumpfish (average weight: 67.0 ± 3 g) were divided in to 5 tanks (60 fish per tank) at the biocontainment facility. Fish from the control tank were intraperitoneally injected with 100 μl of PBS. Fish from the high (2 tanks for high dose; R1 and R2), medium, and low dose tanks were intraperitoneally injected with 100 μl of 109, 108, and 107 cells per fish of R. salmoninarum, respectively. The mock-infected tank (PBS control) and 3 experimental tanks [low, medium, and high (R2 tank) doses of R. salmoninarum] were monitored for mortality. Fish for sampling of spleen, liver and head kidney were collected from the mock-infected tank and high dose R1 tank. Sampling time points were 0, 14, 28, 42, 56, 84, and 98 days post infection, and 6 fish were sampled at each time point.
The bacterial infection inoculum was prepared as described previously (42), with modification for R. salmoninarum. Briefly, bacterial cells were cultured in 1 L of KDM2 at 15°C for 10 days and harvested at mid-logarithmic phase [Optical Density (O.D.) 600 nm = 0.8 ~1x108 CFU mL-1] (Figure 1B) by centrifugation at 6,000 rpm for 10 min at 4°C, and washed once with sterile phosphate-buffered saline (PBS, pH 7.0; 136 mM NaCl, 2.7 mM KCl, 10.1 mM Na2HPO4, 1.5 mM KH2PO4) (43). The bacterial pellet was resuspended in 100 ml of PBS and subjected to bacterial enumeration using a bacteria counting kit (Invitrogen) and flow cytometry according to manufacturers’ instructions. The number of bacterial cells in the inoculum was calculated by dividing the number of signals in the bacterial frame by the number of signals in the microsphere frame (Figure 1C). The bacterial cells suspension was normalized to 3x1010 cells ml-1 and serially diluted in PBS to the final infection doses of 1×109 cells dose-1 (high dose), 1×108 cells dose-1 (medium dose), and 1×107 cells dose-1 (low dose).
All animal protocols required for this research were reviewed and approved by the Institutional Animal Care Committee and the Biosafety Committee at Memorial University of Newfoundland (MUN) (https://www.mun.ca/research/about/acs/acc/) based on the guidelines of the Canadian Council on Animal Care (https://ccac.ca/). Experiments were conducted under protocols #18-01-JS, #18-03-JS, and biohazard license L-01.
Specific pathogen-free lumpfish (67.0 ± 3 g; mean ± SD) were produced and cultivated at the Joe Brown Aquatic Research Building (JBARB; Ocean Sciences Centre, St. John’s, NL, Canada). Infection studies were conducted in the aquatic level 3 (AQ3) biocontainment unit at the Cold-Ocean Deep-Sea Research Facility (CDRF; Ocean Sciences Centre, St. John’s, NL, Canada). Fish were distributed into five 500 L tanks (60 fish per tank) at a biomass of 25 kg m-3 and acclimated for 2 weeks at 10˚C before R. salmoninarum infection. Prior to and throughout the experimental study, fish were kept at optimal conditions (500 L tanks with flow-through (75 L min-1) filtered and UV-treated (8-10˚C) seawater, 95-110% air saturation, ambient photoperiod (12 h light:12 h dark). The fish were fed daily at a rate of 0.5% of their body weight per day with the commercial aquafeed Skretting - Europa 15 (55% crude protein, 15% crude lipid, and 1.5% crude fiber, 3% calcium, 2% phosphorus, 1% sodium, 5000 IU/kg vitamin A, 3000 IU kg-1 vitamin D, and 200 IU kg-1 vitamin E).
Lumpfish were intraperitoneally (i.p.) injected with 100 μl of 107, 108, or 109 cells of R. salmoninarum dose-1, similar to infection studies in salmonids and other fish species (44–47). A duplicate group of lumpfish i.p. injected with the 109 cells dose-1, was utilized for tissue sampling. Lumpfish i.p. injected with PBS were used as a control group (Figure 1E). Fish were monitored daily for mortality and clinical signs until 98 days post-injection (dpi) (Figure 1E). The survival rate was calculated according to Survival rate (%) = (Survivors at the end of the experiment/Initial individuals) × 100 (48).
Samples of spleen, liver, and head kidney were taken at 14, 28, 42, 56, 84, and 98 dpi from six lumpfish infected with 109 cells of R. salmoninarum dose-1 and PBS injected lumpfish groups (Figure 1E). Before sampling, lumpfish were netted and euthanized with an overdose of MS222 (400 mg L−1; Syndel Laboratories, Vancouver, BC, Canada). Each tissue was aseptically collected and consistently subsampled for bacteriology, histology and immune-relevant transcript expression analyses. For bacteriology analysis, 30-100 mg of tissue was individually placed into a sterile homogenizer bag (Nasco whirl-pak®, USA), kept on ice and processed soon after harvesting (< 1 h). For histology, tissue sections were fully submerged into 15 ml falcon tubes containing 10% neutral-buffered formalin. For transcript expression analyses, 50-100 mg of tissue was placed in a 1.5 mL RNase-free tube, flash-frozen using liquid nitrogen, and stored at -80 ˚C until RNA preparation.
To study R. salmoninarum kinetics in lumpfish tissues, bacterial loads per g of tissue of infected lumpfish (n = 6, from high dose infected group) were determined at 14, 28, 42, 56, 84, and 98 dpi according to previously described procedures for R. salmoninarum isolation from salmonid kidney (49) with modifications. Briefly, tissues were kept cold on ice after extraction and during all the procedures. Tissue samples were aseptically weighed in the sterile homogenizer bag, suspended in PBS peptone [PBS (pH 7.4); 0.1% peptone] in the ratio of 1 mL PBS peptone per 0.1 g of tissue, and mechanically homogenized. Tissue homogenates were then transferred into sterile 1.5 mL centrifuge tubes and centrifuged at 2500 x g for 20 minutes at 4°C. The absence of bacteria in the supernatant was confirmed by sub-culturing 10 μL on SKDM2 plates. The pellet was resuspended in PBS peptone at a ratio of 1:1 (w/v) (i.e., 0.1 g of tissue was resuspended in 100 μl of PBS peptone) and mixed using Vortex mixer (Corning, Life Sciences, USA). The suspension was serially diluted in PBS peptone (1:10), and either 10 μl of the tissue homogenate or 10 μL of the serial 10-fold dilution was spread onto SKDM2 agar plates (Figure 1D). The plates were sealed with paraffin film to prevent desiccation and incubated at 15°C for up to 4-8 weeks. In each sampling point, the R. salmoninarum recovered on SKDM2 agar plates from lumpfish tissues were pure, and the observed R. salmoninarum colonies showed a homogenous morphology (Figure 1D). Also, the inocula obtained from these colonies were confirmed as R. salmoninarum by Gram-staining (i.e., presence of pure, Gram-positive diplobacilli) and PCR (i.e., positive amplification with the R. salmoninarum specific primers (40, 41). R. salmoninarum loads (CFU g of tissue-1) were quantified by dividing the number of colonies by the weight of tissue plated (i.e., for a starting tissue weight of 0.1 g, 10 μl of the homogenate was spread onto SKDM2, then the tissue plated was equivalent to 0.01 g).
Tissue samples of spleen, liver, head kidney collected at 14, 28, 42 and 98 dpi from PBS-control and high dose R. salmoninarum infected lumpfish groups were analyzed for histopathology. Tissues were fixed in 10% PBS-buffered formalin for three days at room temperature. The formalin was then removed and the fixed tissues were preserved in PBS at 4°C until processing for paraffin embedded tissue block according to established procedures (50). Tissue sections of 5 μm thickness were stained with hematoxylin and eosin (Leica Biosystems) using established protocols (51, 52) and observed for histopathological changes under the light microscope (Olympus CX40, USA).
To study the lumpfish immune response to R. salmoninarum chronic infection, head kidney samples (n = 6 per group) extracted at 28 and 98 dpi from control (PBS-injected group) and infected lumpfish (109 cell dose-1) groups were selected for real-time quantitative polymerase chain reaction (qPCR) analyses. Approximately 80-100 mg of tissue was added to a 1.5 mL RNase-free centrifuge tube containing 500 µL of TRIzol reagent (Invitrogen) and homogenized using a motorized RNase-Free Pellet Pestle Grinder (Fisherbrand, Fisher Scientific, USA). Then, additional 500 µL of TRIzol was added, mixed by pipetting, and RNA extractions were completed following manufacturer’s instructions. Extracted RNA samples were then purified using RNeasy MinElute Cleanup Kit (QIAGEN, Mississauga, ON, Canada) following manufacturer’s instructions. RNA samples were treated with TURBO DNA-free™ Kit (Invitrogen) for complete digestion of DNA and removal of remaining DNase and divalent cations, such as magnesium and calcium. Purified RNA samples were quantified and verified for purity using a Genova Nano microvolume spectrophotometer (Jenway, UK), and RNA integrity was tested by 1% agarose gel electrophoresis (43). All RNA samples used in this study showed acceptable purity ratios (A260/230 > 1.8 and A260/280 > 2.0) and integrity (28S and 18S ribosomal RNA bands at a 2:1 ratio) (Supplementary Figure S1).
First-strand cDNA templates for qPCR were synthesized in 20 μL reactions from 1 mg purified RNA using SuperScript IV VILO Master Mix (Invitrogen) following the manufacturer’s instructions.
PCR amplifications were performed in 13 ml reactions using 1X Power SYBR Green PCR Master Mix (Applied Biosystems), 50 nM of both the forward and reverse primers and the indicated cDNA quantity (see below). Amplifications were performed using the QuantStudio 6 Flex Real-Time PCR system (384-well format) (Applied Biosystems). The real-time analysis program consisted of 1 cycle of 50°C for 2 min, 1 cycle of 95°C for 10 min, 40 cycles of 95°C for 15 sec, and 60°C for 1 min, with fluorescence detection at the end of each 60°C step and was followed by dissociation curve analysis.
For each gene that was subjected to qPCR analyses, a group of transcripts (with associated TRINITY IDs) were obtained from the NCBI Sequence Read Archive (SRA) under accession number SRP238224 (Supplementary File S1). To confirm the identity of a given transcript, determine its orientation and identify the coding sequence (CDS), a BLASTx search of the non-redundant (nr) protein sequences database using a translated nucleotide query was performed between June and July 2019. A database of all confirmed transcript sequences for a given gene was created using Vector NTI (Vector NTI Advance 11.5.4, Life Technologies). Next, for a given gene, multiple sequence alignments were performed for its corresponding transcripts using AlignX (Vector NTI Advance 11.5.4). These alignments were used to determine if the transcripts were identical, contained single nucleotide polymorphisms (SNPs)/sequencing errors or represented different gene paralogues/isoforms. In the case of gene paralogues/isoforms, these alignments were also helpful to determine their percentage identity and to identify regions where paralogue/isoform-specific qPCR primers could be designed.
Primers were designed using Primer3 (53–55). However, in the case of the gene paralogues/isoforms, some were custom-designed in paralogue/isoform-specific areas to ensure specificity. All primers are located in the CDS and in an area, which overlapped with that of the best BLASTx-identified sequence. In the case of gene paralogues/isoforms, primers were designed in an area with ≥ 3 bp difference between them to ensure specificity. The amplicon size range was between 90-160 bp. The sequences, amplicon sizes and efficiencies for all primer pairs used in the qPCR analyses are presented in Table 1.
Each primer pair was quality tested to ensure that a single product was amplified (dissociation curve analysis) and that there was no primer-dimer present in the no-template control. Amplicons were electrophoretically separated on 2% agarose gels and compared with a 1 kb plus ladder (Invitrogen) to verify that the correct size fragment was being amplified (43). Amplification efficiencies (56) were calculated for both control and immune-stimulated cDNA pools from head kidney samples. Standard curves were generated for both cDNA pools using a 5-point 1:3 dilution series starting with cDNA representing 10 ng of input total RNA. The reported efficiencies are an average of the two values (Table 1).
Expression levels of the genes of interest (GOIs) were normalized to expression levels of two endogenous gene controls. To select these endogenous controls, 5 genes [60S ribosomal protein L32 (rpl32), elongation factor 1-alpha (ef1a), eukaryotic translation initiation factor 3 subunit D (etif3d), polyadenylate-binding protein 1a (pabpc1a) and polyadenylate-binding protein 1b (pabpc1b)] were analyzed. Briefly, the fluorescence threshold cycle (CT) values of all 24 samples in the study were measured (in duplicate) for each of these transcripts using cDNA representing 4 ng of input total RNA and then analyzed using geNorm (57). geNorm M values for all of the candidate normalizers were < 0.3, suggesting stable expression; however, pabpc1b (geNorm M = 0.165) and etif3d (geNorm M = 0.168) were selected as the two endogenous controls as they were the most stably expressed.
For experimental qPCR, head kidney samples from the control and from the high dose R. salmoninarum infected fish at both 28 and 98 dpi were chosen to represent early (28 dpi) and chronic (98 dpi) infection stages of R. salmoninarum based on the survival and head kidney colonization data (i.e., fish showed mortality along with highest bacterial load at 28 dpi whereas fish mortality was stabilized even with the considerable amount of bacterial load at 98 dpi).
qPCR assays were designed for 33 transcripts with immune-relevant functional annotations (Table 1). These transcripts include pattern recognition receptors, cytokines, antimicrobial peptides, acute phase reactants, interferon regulators, interferon-induced effectors, humoral and cell-mediated adaptive immune response-related transcripts. An analysis of these transcripts-related innate and adaptive immunity would provide insight into host-pathogen interactions between lumpfish and R. salmoninarum at early and chronic infection stages.
The experimental qPCR analyses were conducted according to MIQE guidelines (58). cDNA representing 4 ng of input RNA was used as a template in the PCR reactions. All samples were analyzed on a single plate (3 GOIs and the two endogenous controls per plate; 33 GOIs over 11 plates). On each plate, for every sample, the GOIs and endogenous controls were tested in triplicate, and a no-template control was included. The relative quantity (RQ) of each transcript was determined using the QuantStudio Real-Time PCR Software (version 1.3) (Applied Biosystems) relative quantification study application, with normalization to both pabpc1b and etif3d transcript levels, and with amplification, efficiencies incorporated. For each GOI, the sample with the lowest normalized expression (mRNA) level was set as the calibrator sample (i.e., assigned an RQ value = 1) (Supplementary Table S1). Also, transcript expression levels were determined using the comparative 2-ΔΔCt method (59–61) (Supplementary Table S2). The levels of transcript expression data from the 2-ΔΔCt and the RQ data analysis methods were used in the main (Figures 4–6), and the supplementary (Supplementary Figures S3–S5) graphs, respectively.
All data are expressed as mean ± standard error (SE). Assumptions of normality and homoscedasticity were tested for the detected variances. Kaplan-Meier estimator was used to obtain survival fractions after the R. salmoninarum infection. The log‐rank test was used to compare the survival curve trends (p<0.0001), and a one-way ANOVA followed by Tukey’s multiple comparison post hoc test was used to determine significant differences between the survival of control and infected groups. Also, one-way ANOVA followed by the Holm-Sidak post hoc test was conducted to compare differences between tissues and within fish individuals at a single time point, whereas a non-parametric Kruskal-Wallis test was performed to compare the tissue bacterial loads between various time points per organ.
Transcript expression data were analyzed using a two-way ANOVA test, followed by the Sidak multiple comparisons post hoc test to identify significant differences between treatments (control and infected groups) at a single time point and for each treatment at different time points (i.e., 28 and 98 dpi). In all cases, p < 0.05 was considered statistically significant. All statistical analyses were performed using GraphPad Prism 8.0 (GraphPad Software, La Jolla California USA, www.graphpad.com).
BKD is a slowly progressing systemic infection depending on the virulence of the R. salmoninarum strain that correlates with their number of major soluble antigen (msa) gene copies (2, 62). In this study, we used R. salmoninarum type strain ATCC 33209, which has only two msa copies (63), and it is known to exhibit lower pathogenicity, cause low mortality and a chronic infection in salmonids (36, 45, 64). Lumpfish infected with R. salmoninarum ATCC 33209 displayed characteristic clinical signs of a chronic BKD infection (Figure 2A). Mortality began at 20 dpi, gradually increased and stabilized after 50 dpi in the high dose infected group (1x109 cells dose-1) (Figure 2B). In the medium (1x108 cells dose-1) and low (1x107 cells dose-1) dose groups, mortality began at 40 dpi and stabilized after 50 dpi as well (Figure 2B). External and internal BKD clinical signs and symptoms were observed in both dead and sampled fish. The clinical signs of R. salmoninarum infected lumpfish included hyper-pigmentation, lethargy, abdominal ascites, and hemorrhages in ventral sites. Examination of internal organs revealed splenomegaly, hydronephrosis, pale liver, pseudomembrane formation on internal organs, and ascites (Figure 1F). The survival rate for the high, medium, and low doses of R. salmoninarum groups was 65%, 93%, and 95%, respectively (Figure 2B). Cumulative number of fish mortalities (and mortality rate) observed during the experiment were 21 dead fish out of 60 total fish (35%), 4 dead out of 60 total fish (7%) and 3 dead out of 60 total fish (5%) for high, medium and low R. salmoninarum doses, respectively. The mortality data often considered the fish deaths from the tanks assigned for mortality observation (i.e., sampled fish were not considered in the analyses) (Figure 1E). Significantly lower survival (p<0.001) was observed in the high-dose R. salmoninarum infected group, whereas there were no significant differences in survival between PBS control, low and medium-dose fish groups.
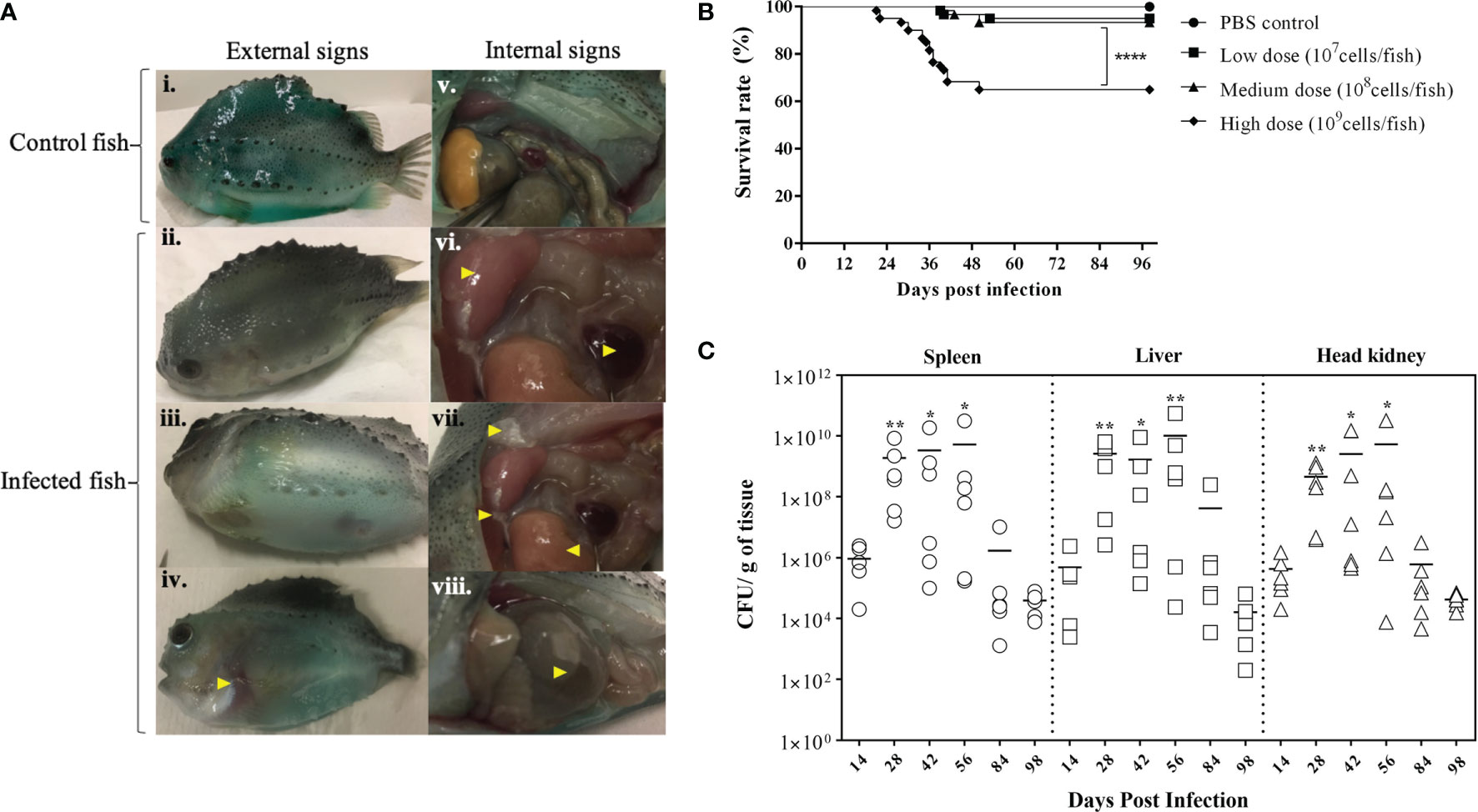
Figure 2 Bacterial Kidney Disease clinical signs, survival rates, and tissue colonization of Renibacterium salmoninarum infected lumpfish (A) Bacterial Kidney Disease signs and symptoms in lumpfish detected from 21 to 56 dpi. Pictures were randomly selected to visualize the external and internal signs compared to the control. Specific signs are indicated with yellow arrowheads. The external signs observed were (i) Control lumpfish; ii. Skin darkening; iii. Abdominal distension due to ascites; iv. Hemorrhages in ventral sites. The internal signs observed were v. Internal organs of control lumpfish; vi. Enlarged spleen and kidney; vii. Diffuse white membranous layer (pseudo membrane) on internal organs and pale liver; and viii. Accumulation of turbid fluid inside the abdominal sacs and cavities; (B) Percent survival of lumpfish exposed to experimental infection with high (1×109 cells/fish), medium (1×108 cells/fish) or low (1×107 cells/fish) doses of R. salmoninarum compared to a PBS control; **** denotes the significant differences between infected and control groups (p < 0.001); (C) Renibacterium salmoninarum tissue colonization in lumpfish. Bacterial loads in spleen, liver and head kidney of lumpfish (n = 6) infected with the high dose (1 x 109 cells/fish) of R. salmoninarum after 14, 28, 42, 56, 84, and 98 days post infection (dpi). Asterisks (*) represent significant differences (*p < 0.05, **p < 0.01) in the bacterial loads between time points (14, 28, 42, 56 and 84 dpi) per organ compared to the bacterial load at 98 dpi, as determined by the non-parametric Kruskal-Wallis test.
R. salmoninarum colonized all of the organs sampled in the high-dose infected lumpfish (Figure 2C; Supplementary Figure S2). Significantly higher bacterial loads were observed at 28, 42, and 56 dpi compared to 98 dpi (Figure 2C). A substantial decrease in the bacterial load was observed at 84 and 98 dpi. Tissue colonization results correlated with the mortality data (Figures 2B, C).
In contrast to the control fish, spleen, liver, and head kidney of high dose infected fish at 14, 28, and 42 dpi showed apparent histopathological damages (Figures 3B–D, G–I, L–N). Tissue damage was observed in all three organs at 14, 28, and 42 dpi (Figures 3C, G, M, N). Hemorrhages were observed in the spleen and liver at 14 and 42 dpi (Figures 3B, D, G, I). The liver sections showed increased vacuolations in hepatocytes at 28 and 42 dpi (Figures 3H, I). Melanomacrophage centers were observed in the spleen and liver at 42 dpi (Figures 3D, I). Head kidney sections showed congested glomerulus with diffuse thickening of the basement membrane at 14 and 42 dpi (Figures 3L, N). Tissue sections of control fish and high dose infected fish at 98 dpi seemed similar without any significant histopathological damages (Figures 3A, E, F, J, K, O).
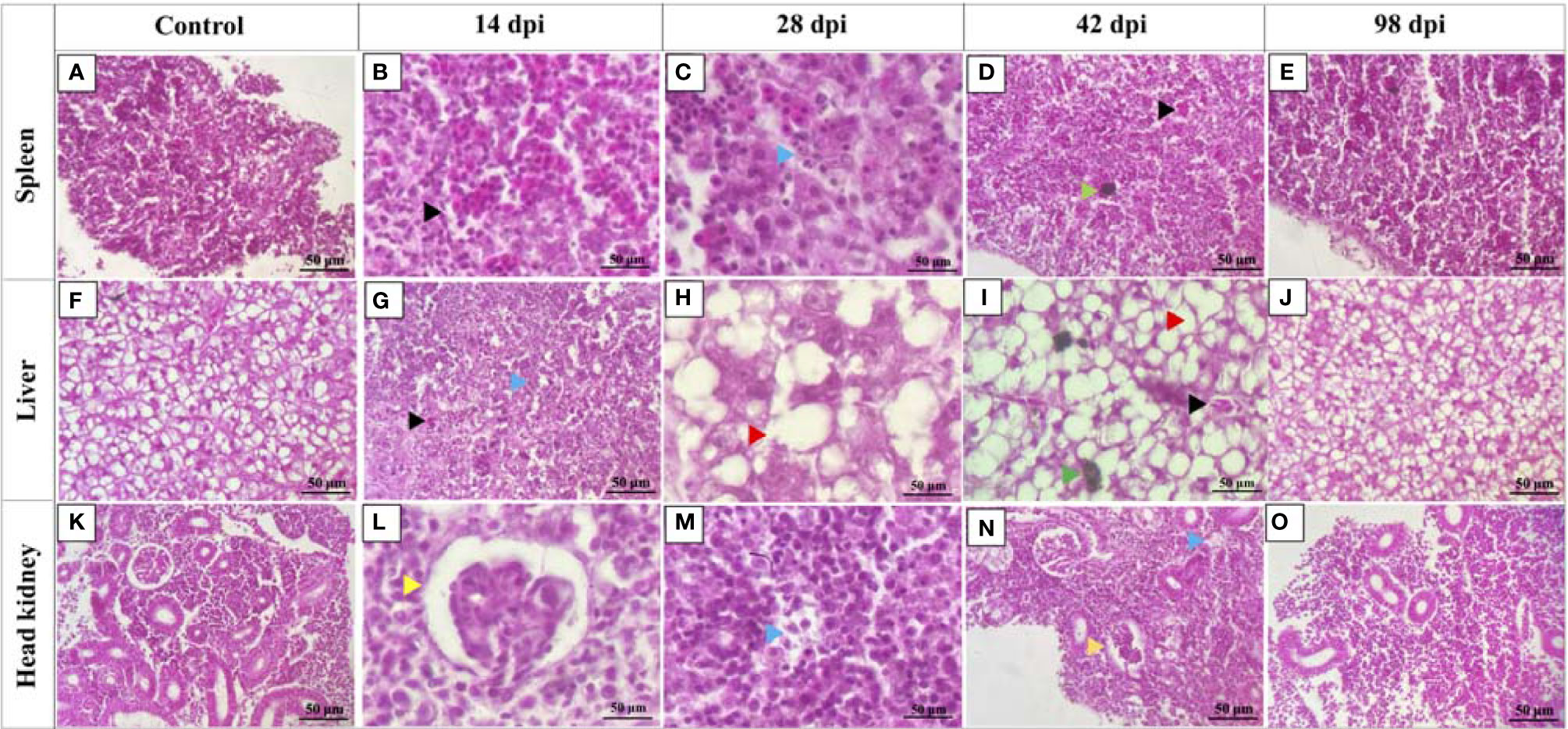
Figure 3 Histopathology changes in lumpfish tissues during Renibacterium salmoninarum infection. Lumpfish spleen, liver, and head kidney were collected from the high-dose (1 x 109 cells/fish) infected group at 14, 28, 42 and 98 days post infection (dpi) and from the control (PBS-mock infected) group and stained with haematoxylin and eosin (H & E). Histopathological changes in lumpfish spleen: (A) Spleen section from control fish; (B) Spleen section from infected fish at 14 dpi, showing hemorrhages (black arrowhead); (C) Spleen section from infected fish at 28 dpi, showing degenerations (blue arrowhead); (D) Spleen section from infected fish at 42 dpi, showing hemorrhage (black arrowhead) and melanomacrophage center [MMC] (green arrowhead); (E) Spleen section from infected fish at 98 dpi. Histopathological changes in lumpfish liver: (F) Liver section from control fish; (G) Liver section from infected fish at 14 dpi, showing hemorrhage (black arrowhead) and degeneration (blue arrowhead); (H) Liver section from infected fish at 28 dpi, showing increased vacuolations (red arrowhead); (I) Liver section from infected fish at 42 dpi, showing hemorrhage (black arrowhead), MMC (Green arrowhead) and vacuolation (red arrowhead); (J) Liver section from infected fish at 98 dpi. Histopathological changes in lumpfish head kidney: (K) Head kidney section from control fish; (L). Head kidney section from infected fish at 14 dpi, showing congested glomerulus (yellow arrowhead); (M). Head kidney section from infected fish at 28 dpi, showing degenerations (blue arrowhead); (N). Head kidney section from infected fish at 42 dpi, showing degeneration (blue arrowhead) and congested glomerulus (yellow arrowhead); (O). Head kidney section from infected fish at 98 dpi. Stain: H & E; Magnification: ✕ 400.
The immune response of lumpfish to R. salmoninarum infection was evaluated in head kidney at 28 dpi and 98 dpi in 109 cells dose-1 infected fish and compared to non-infected fish (PBS-control) at the same time points. Of the 33 genes (Table 1) that were evaluated, 12 genes were upregulated, and 4 genes were downregulated at both 28 and 98 dpi, whereas 17 genes were dissimilarly regulated.
Thirteen genes related to pattern recognition (Figures 4A–E) and cytokines (Figures 4F–M) were differentially regulated. toll-like receptor 3 (tlr3) and toll-like receptor 7 (tlrl7) were significantly downregulated in infected fish compared to the control fish at 28 dpi (Figures 4A, D). toll-like receptor 5a (tlr5a) expression was significantly upregulated at 28 dpi compared to the respective control group (Figure 4B). toll-like receptor 5b (tlr5b) and ATP-dependent RNA helicase lgp2 (lgp2) showed no significant differences in their expression levels between control and infected fish at 28 dpi and 98 dpi, respectively (Figures 4C, E).
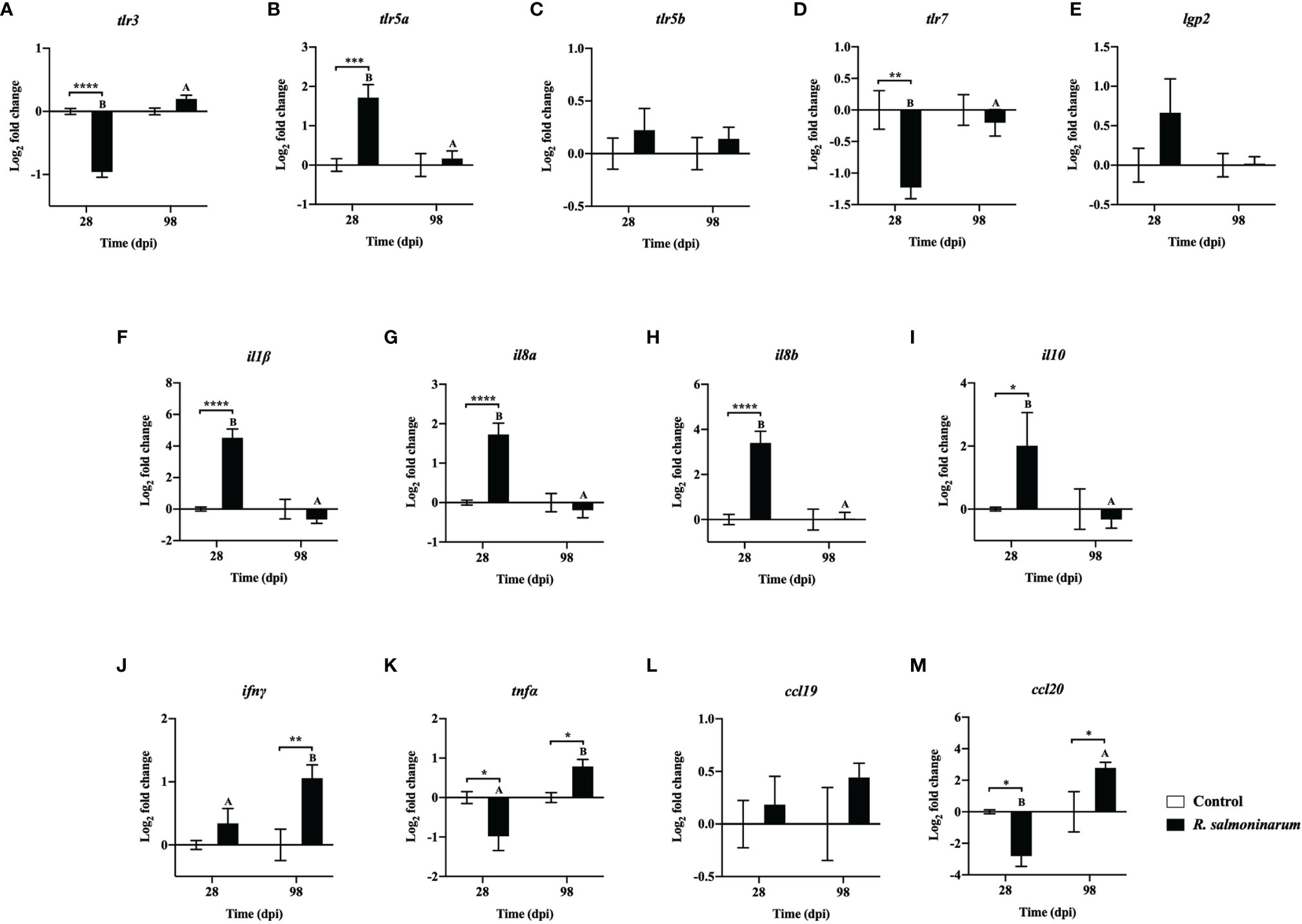
Figure 4 Expression of transcripts related to pattern recognition (A-E) and cytokines (F-M) in lumpfish head kidney in response to R. salmoninarum infection at 28 and 98 days post infection (dpi). Transcript expression levels in head kidney from control (PBS-mock infected group) and infected [high dose (1×109 cells/dose) of R. salmoninarum] lumpfish at 28 and 98 dpi were analyzed using qPCR. Relative expression was calculated using the 2−ΔΔCt method and log2 transformed; etif3d and pabpc1b were the endogenous control genes. A two-way ANOVA test, followed by the Sidak multiple comparisons post hoc test was used to identify significant differences between treatments (control and infected groups) at a single time point, and for a given treatment at different time points (28 and 98 dpi). Asterisks (*) represent significant differences between treatments at each time-point (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). Different letters represent significant differences between control (lower case) and infected (upper case) groups at 28 compared to 98 dpi. Each value is the mean ± S.E.M (n = 6).
Canonical proinflammatory cytokines encoding genes, including interleukin 1 beta (il1β), interleukin 8a (il8a), interleukin 8b (il8b), and the anti-inflammatory cytokine interleukin 10 (il10), showed significantly higher expression in infected fish at 28 dpi compared to the non-infected control fish (Figures 4F–I). The expression levels of il1β, il8a, il8b, and il10 in infected fish were not significantly different at 98 dpi compared to the control fish (Figures 4F–I).
R. salmoninarum infection significantly downregulated tumor necrosis factor alpha (tnfα) and C-C motif chemokine-like 20 (ccl20) expression at 28 dpi (Figures 4K, M). In contrast, interferon gamma (ifnγ), tnfα and ccl20 levels were significantly upregulated at 98 dpi compared to the respective non-infected fish group (Figures 4J, K, M).
Expression levels of 9 genes regulating the innate (Figures 5A–E) and inflammatory (Figures 5F–I) immune response were assessed. Gene expression levels of hepcidin antimicrobial peptide (hamp) and serum amyloid A 5 (saa5) were significantly upregulated in the head kidney of infected fish at 28 dpi compared to the respective non-infected control (Figures 5A, B).
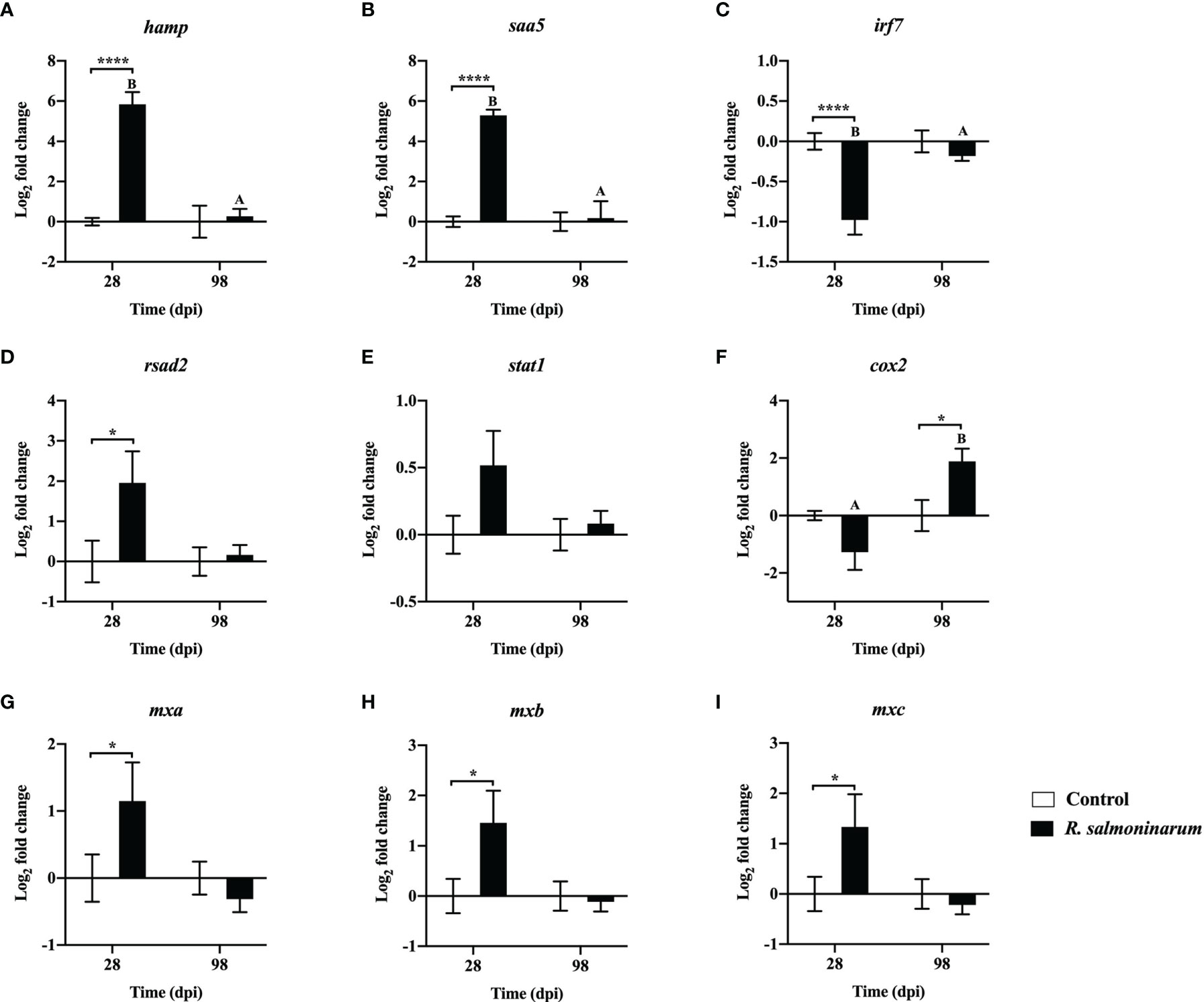
Figure 5 Expression of transcripts related to regulation of the innate (A–E) and inflammatory (F–I) immune response in lumpfish head kidney in response to R. salmoninarum infection at 28 and 98 days post infection (dpi). Transcript expression levels in head kidney from control (PBS-mock infected group) and infected [high dose (1×109 cells/dose) of R. salmoninarum] lumpfish at 28 and 98 dpi were analyzed using qPCR. Relative expression was calculated using the 2−ΔΔCt method and log2 transformed; etif3d and pabpc1b were the endogenous control genes. A two-way ANOVA test, followed by the Sidak multiple comparisons post hoc test was used to identify significant differences between treatments (control and infected groups) at a single time point, and for a given treatment at different time points (28 and 98 dpi). Asterisks (*) represent significant differences between treatments at each time-point (*p < 0.05, ****p < 0.0001). Different letters represent significant differences between control (lower case) and infected (upper case) groups at 28 compared to 98 dpi. Each value is the mean ± S.E.M (n = 6).
At 28 dpi, interferon regulatory factor 7 (irf7) was significantly downregulated (Figure 5C). Conversely, interferon-induced effectors such as radical S-adenosyl methionine domain-containing protein 2/viperin (rsad2) and three gene isoforms of interferon-induced GTP-binding protein (mxa, mxb and mxc) were significantly upregulated compared to the control fish at 28 dpi (Figures 5D, G–I). cyclooxygenase-2 (cox2) expression was significantly upregulated in infected fish at 98 dpi compared to the control (Figure 5F).
The expression levels of 11 genes playing putative roles in humoral (Figures 6A–F) and cellular-mediated adaptive immunity (Figures 6G–K) were assessed. Humoral (immunoglobulin heavy chain variable region a (igha), immunoglobulin delta heavy chain (ighδ), immunoglobulin mu heavy chain a (ighma), and immunoglobulin mu heavy chain b (ighmb)), and cellular-mediated (T-cell surface glycoprotein CD4a (cd4a), T-cell surface glycoprotein CD4b (cd4b), lymphocyte antigen 6 complex locus protein G6f (ly6g6f), T-cell surface glycoprotein CD8 alpha chain (cd8α), and HLA class II histocompatibility antigen gamma chain (cd74)) adaptive immunity-related genes showed significant downregulation at 28 dpi in the infected head kidney compared to the non-infected control (Figures 6A, C–E, G–K).
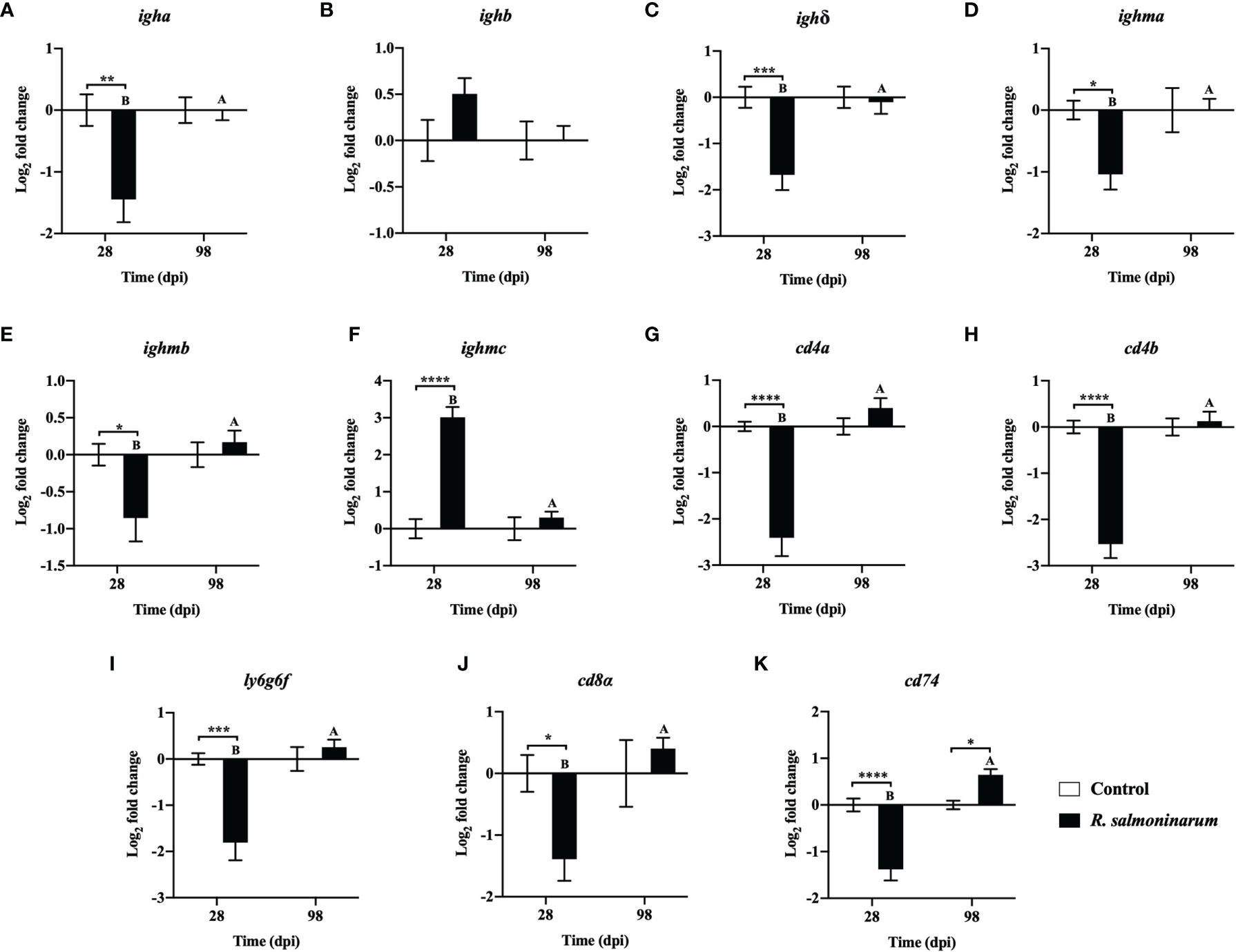
Figure 6 Expression of transcripts related to humoral (A–F) and cellular mediated (G–K) immunity in lumpfish head kidney in response to R. salmoninarum infection at 28 and 98 days post infection (dpi). Transcript expression levels in head kidney from control (PBS-mock infected group) and infected [high dose (1×109 cells/dose) of R. salmoninarum] lumpfish at 28 and 98 dpi were analyzed using qPCR. Relative expression was calculated using the 2−ΔΔCt method and log2 transformed; etif3d and pabpc1b were the endogenous control genes. A two-way ANOVA test, followed by the Sidak multiple comparisons post hoc test was used to identify significant differences between treatments (control and infected groups) at a single time point, and for a given treatment at different time points (28 and 98 dpi). Asterisks (*) represent significant differences between treatments at each time-point (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). Different letters represent significant differences between control (lower case) and infected (upper case) groups at 28 compared to 98 dpi. Each value is the mean ± S.E.M (n = 6).
At 98 dpi, only one adaptive immune-related gene, cd74, was significantly upregulated in infected fish compared to the control (Figure 6K). Expression of most of the genes related to humoral and cellular-mediated immunity in infected fish at 98 dpi was restored to similar levels observed in the control fish (Figures 6B, D, E–K).
The qPCR results were similar between the 2-ΔΔCt and the RQ data analysis methods. However, a few differences in the significance levels were detected for tlr3, tnfα, rsad2, ighma and cd8α expression at 28 or 98 dpi (Figures 4A, K; 5E; 6D, J and Supplementary Figures S3A, K; S4E; S5D, J).
As previously mentioned, lumpfish are in close contact with salmon when delousing sea lice in sea cage aquaculture (17, 19), and this interaction could result in the horizontal transmission of infectious disease agents between both species, including R. salmoninarum. Atlantic salmon is susceptible to R. salmoninarum, and it could transfer this pathogen to other fish species (20, 21). It is believed that lumpfish could act as a non-symptomatic carrier and transmit disease to cohabitating salmon (19). Haugland et al. (29) confirmed the experimental transmission of amoebic parasite from lumpfish to salmon (29). Also, Atlantic salmon susceptibility to a lumpfish isolate of M. viscosa reflecting the disease risk to salmon (30). Although BKD episodes have not been reported in lumpfish, its susceptibility and immune response to R. salmoninarum are unknown. Here, we examined the susceptibility of lumpfish to R. salmoninarum (ATCC 33209) type strain, which has been utilized for several infection studies in different fish species. Using similar R. salmoninarum infection doses like other studies, we also determined the infection kinetics and lumpfish molecular immune response at early and late chronic infection with R. salmoninarum. This study is the first report of R. salmoninarum experimental infection on lumpfish and provides immune-relevant information on how the lumpfish respond to R. salmoninarum.
For the R. salmoninarum infection kinetics studies, we selected the plate counting method in SKDM2 over typical methods for R. salmoninarum quantification (e.g., FAT, ELISA, and PCR) because it directly enumerates viable bacteria (7). Spleen and liver were also analyzed, in addition to the head kidney, to consider non-kidney R. salmoninarum infections, which have been well described in salmonids (65, 66). In lumpfish, R. salmoninarum infection becomes evident at 2 weeks post-infection, similar to chinook salmon (O. tshawytscha) i.p. infected with 1×106 R. salmoninarum cells dose-1 (67). In an antibody capture ELISA and western blot based analysis, Turaga et al. (1987) reported that levels of R. salmoninarum soluble antigens in infected coho salmon (Oncorhynchus kisutch) gradually increased during the course of infection, and peaked at 20 dpi and thereafter, fish mortality was observed (68). Although in the current study we did not measure R. salmoninarum soluble antigen levels, mortality of R. salmoninarum infected lumpfish started at 20 dpi, similar to coho salmon, i.p. infected with R. salmoninarum cells in the exponential phase of growth (O.D. 500 nm = 1.0) (68). This suggests that mortality could be initiated by the accumulation of R. salmoninarum MSA in the infected lumpfish. Because increased MSA levels correlated with the severity of infection and mortality (68, 69).
Lumpfish infected with a lethal dose of R. salmoninarum showed prominent BKD associated clinical signs at 14, 28, and 56 dpi (Figure 2A), similar to clinical signs described in other fish species (7). Bacterial loads in spleen, liver and head kidney at various time points indicated that R. salmoninarum established an infection in all infected individuals (Figure 2C). In contrast, carp (Cyprinus carpio L.), a non-salmonid-like lumpfish, showed resistance to R. salmoninarum infection (4.8×107 and 4.8×108 cells/dose), and no bacteria were recovered from head kidney after infection (47). These results indicate that the lumpfish is susceptible to R. salmoninarum and could be a potential vector for this pathogen.
Significantly higher tissue bacterial loads at 28, 42, and 56 dpi correlated with higher mortality (Figures 2B, C). However, after fish mortality ended (Figure 2B), R. salmoninarum remained in the internal tissues (Figure 2C and Supplementary Figures S2E, F), indicating a pattern of chronic infection. Arctic charr (15), chinook salmon (62), lamprey (70) and carp (47) cleared R. salmoninarum infection after 175, 115, 92 and 38 days, respectively. R. salmoninarum persisted in lumpfish tissues at least for 98 dpi, which is consistent with studies in chinook salmon where R. salmoninarum caused a chronic infection and persisted for up to 100 dpi (36). However, if the current study had been extended, it is possible that lumpfish could have cleared the R. salmoninarum after 98-100 dpi, as seen in the Arctic charr (15) or remains in other tissues like gonads (i.e., R. salmoninarum in ovarian fluid is an important source of infection for the eggs) to facilitate vertical transmission (7).
The lethal dose 50 (LD50) of R. salmoninarum ATCC 33209 in various salmonid hosts ranged from 1.4×105 to 2.94×108 CFU dose-1 (64). We could not determine the LD50 for R. salmoninarum ATCC 33209 in lumpfish because the fish infected with the highest dose (1×109 cells dose-1), similar to other studies, showed only 35% mortality. In contrast, mortality reached 100% within 15 days in Atlantic salmon infected with 108 cells dose-1 of highly virulent R. salmoninarum strains (71). The LD50 of R. salmoninarum type strain in lumpfish might be greater than 1×109 CFU dose-1, and although 109 cells dose-1 of R. salmoninarum ATCC 33209 was sufficient to invade, replicate, and establish an infection in lumpfish, its lethality was lower than in salmonid species (36, 72–75).
Differences in virulence between R. salmoninarum isolates from several geographical regions and fish hosts have been reported (64). Rhodes et al. (76) demonstrated the positive correlation between the functional msa gene copy number per bacterial cell and virulence (i.e., increased mortality) (76). The type strain R. salmoninarum ATCC 33209 used in this study has two msa gene copies, and both of these msa gene copies are essential for disease development and mortality (62, 63). Compared to other R. salmoninarum strains, R. salmoninarum ATCC 33209 has a reduced virulence. For example, this strain showed lower virulence in chinook and coho salmon compared to the other isolates, and it is not capable of causing BKD in rainbow trout (45, 64). Furthermore, R. salmoninarum type strain does not infect the carp (Epithelioma papillosum) cell line, even with a dose of 1×109 cells, in contrast to more virulent strains of R. salmoninarum (e.g., FT10) that are capable of invading and proliferate in these cells (45, 77). R. salmoninarum ATCC 33209 type strain was isolated in 1974, and it has been subjected to extensive laboratory passages, which may have contributed to its relatively reduced virulence (76). In the present study, R. salmoninarum ATCC 33209 was unable to kill all infected lumpfish even at a high dose, and this could be linked to the low virulence documented for R. salmoninarum ATCC 33209.
Histology observations in the sampled lumpfish infected with R. salmoninarum showed similarities with the histopathological characteristics of BKD in salmonids (Figure 3). For instance, glomerulopathy is related to antigen-antibody complexes deposition in the glomeruli, which causes thickening of the glomerular basement membrane (10, 78). In concordance with BKD histopathology, congested glomeruli were observed in the head kidney of infected lumpfish at 14 and 42 dpi (Figures 3L, N). Also, lysed and disrupted melanomacrophages resulting from the dispersal of pigments in tissues during BKD (79, 80) were observed in spleen and liver from infected lumpfish at 42 dpi (Figures 3D, I). No histopathological differences were observed at 98 dpi (Figure 3). The persistence of R. salmoninarum in lumpfish tissues at 98 dpi was indicative of a chronic infection, and the bacterium may remain dormant or controlled by the fish immune system (81). The lack of tissue inflammation and damage at 98 dpi could be explained by the known immune-suppressive nature of R. salmoninarum (8, 10, 82).
At 98 dpi, R. salmoninarum was isolated from spleen, liver and headkidney of the high dose infected lumpfish which showed no external, internal and histopathological disease signs (Figures 2C and 3). Similar to our results, M. viscosa was isolated from kidneys of non-symptomatic lumpfish at 27 days post bath challenge (30). This implies that lumpfish could be asymptomatic carriers for R. salmoninarum, and chronic infection could be a common strategy of marine bacterial pathogens.
The BKD-related histopathology observations in lumpfish coincided with the downregulation of immune-related genes in lumpfish head kidney after R. salmoninarum infection. For instance, we observed that R. salmoninarum influenced the expression of genes related to pathogen recognition, immune signalling, antibacterial activity, and humoral and cell-mediated immunity in lumpfish (Figures 4–6).
TLR5 is associated with flagellin detection (83). tlr5a was significantly upregulated at 28 dpi (Figure 4B). Increased expression of tlr5a in lumpfish upon exposure to Gram-positive, non-motile or non-flagellated bacteria like R. salmoninarum (84) is controversial. However, a similar upregulation of tlr5 in response to alive and formalin-killed R. salmoninarum has been reported (46, 85). Also, increased expression of tlr5a and tlr5b was reported in turbot (Scophthalmus maximus L.) mucosal tissues (i.e., intestine and gills) in response to the Gram-positive non-flagellated pathogen Streptococcus iniae (86). Therefore, the role of TLR5 beyond the recognition of flagellin, specifically in infection with non-flagellated bacteria in teleosts, warrants further investigation.
R. salmoninarum increased gene expression levels of the proinflammatory cytokine (il1β) and of the proinflammatory response related chemokines (il8a, il8b) at 28 dpi (Figures 4F–H) in lumpfish, which coincided with a canonical innate immune response. Simultaneously, il10, an anti-inflammatory mediator, was significantly upregulated at 28 dpi (Figure 4I). This pattern strongly suggests an R. salmoninarum induced immune suppression (8, 82). Similar to our results, IL-10 induction upon R. salmoninarum strain H-2 infection in Atlantic Salmon Kidney (ASK) cell line was observed by Bethke et al. (87) (87). IL10 counteracted the induced inflammatory immune responses (e.g., ILβ, IL8), and as a result, the pathogen could move forward in disease progression. However, as teleost fish IL-10 demonstrates immune suppressive function, il-10 expression upon pathogen infection could be the natural way of lumpfish to regulate its early innate immune responses (88). Thus, IL-10 upregulation might be seen from a host point of view in which host is trying to create a conducive environment to alleviate host-mediated pathology. For instance, IL-10 can promote tissue repair to overcome the tissue damage due to disease progression (89).
IL-1β was activated in fish leucocytes and macrophages and induced the expression of proinflammatory transcripts such as cox2 and tnfα (90–92). However, at 28 dpi, we observed that cox2 and tnfα were not upregulated even with high expression of il1β (Figures 4F, K, and 5C). IL-1β can also initiate an acute phase response and induce the synthesis of acute-phase proteins (APPs) such as serum amyloid A5 (SAA5) upon invasion of the pathogen (93, 94). We observed a significant upregulation of saa5 at 28 dpi in lumpfish (Figure 5B) indicating an inflammatory response to the infection (95).
TNF-α is associated with inflammation and chronic infections (96). TNF-α can either improve the phagocytic activity of fish leucocytes or support the intracellular survival of pathogens (97–100). In the current study, despite the high bacterial load in the fish tissues (Figure 2C), significant downregulation of tnfα at 28 dpi was observed in lumpfish (Figure 4K), which could affect the tnfα dependent killing pathways, thereby facilitating the infection and intracellular survival of R. salmoninarum (32). Also, this tnfα repression could reflect the immune-suppressive action of R. salmoninarum in lumpfish.
Reducing the availability of iron to bacteria as a means of nutritional immunity is one strategy used by vertebrates such as fish to control pathogens (101). On the other hand, intracellular bacteria compete for iron for their survival (102). HAMP is an antimicrobial peptide (AMP) that has anti-bacterial and immuno-modulatory functions and plays a role in iron homeostasis in fish (103). Here, we found that hamp was significantly upregulated at 28 dpi (Figure 5A). Similar to our results, increased expression of hamp in head kidney of Atlantic salmon has also been observed with R. salmoninarum infection (46, 85). Additionally, transferrin, an AMP encoding gene, which has a putative role in iron sequestration from bacteria, is upregulated in response to R. salmoninarum in salmonid hosts (67) and is involved in BKD resistance in coho salmon (O. kisutch) (104). Thus, hamp and transferrin in lumpfish might play an essential role in the BKD response.
IFN-γ is associated with adaptive immunity and has a role in both the early and late immune responses and in the host immune defense to intracellular bacteria (96, 102). ifnγ stimulation in lumpfish at 28 dpi was not significant in our study. In contrast, significant upregulation of ifnγ was reported in Atlantic salmon and chinook salmon infected with R. salmoninarum (46, 67). Interferon-induced effectors (rsad2, mxa, mxb and mxc) were significantly upregulated at 28 dpi (Figures 4J and 5D, G–I). Similar to our results, upregulation of rsad2 has also been observed in Atlantic salmon head kidney upon R. salmoninarum infection (46). In addition, increased expression of mx genes, mx1, mx2, and mx3 in rainbow trout macrophages (32) and mx1 in chinook salmon (67), after R. salmoninarum infection was also reported.
The immune-suppressive effects of R. salmoninarum were also observed in the adaptive immune response of lumpfish at 28 dpi. For instance, significant downregulation of humoral (igha, ighδ, ighma, ighmb) (Figures 6A, C–E) and cell-mediated (cd4a, cd4b, ly6g6f, cd8α, cd74) (Figures 6G–K) adaptive immune-related transcripts at 28 dpi, was observed. Mortality in lumpfish during the early time points could be attributed to this immune suppressive function of R. salmoninarum observed at 28 dpi. Significant downregulation of cd74 (an invariant polypeptide involved in major histocompatibility complex-II (MHC-II) formation and transport) (Figure 6K) in lumpfish head kidney at 28 dpi suggests that the T-cell responses could be modified towards an enhanced MHC-I and a reduced MHC-II dependent pathway, perhaps caused by an increased amount of MSA, similar to R. salmoninarum infection in rainbow trout (32, 105). This skewing towards the MHC-I pathway in lumpfish at the early stages of R. salmoninarum infection correlates with the BKD-dependent major histocompatibility-1 (mh1) induction observed in Atlantic salmon at 13 dpi (46). Further, Rozas-Serri et al. (106) demonstrated that the humoral and cell-mediated adaptive immune responses against R. salmoninarum in Atlantic salmon pre-smolts were significantly downregulated at the later stage of infection (55 dpi) (106), which agrees with our findings at 28 dpi. In contrast, most of the humoral-immune genes showed strong down-regulation at 28 dpi (Figures 6A, C–E), only the ighmc was significantly upregulated (Figure 6F). This observation at 28 dpi is controversial but in line with the triggered humoral response against R. salmoninarum in salmonids, which does not necessarily correlate with immune protection (7, 8, 106).
R. salmoninarum persisted in the lumpfish tissues for at least 98 dpi (Figure 2C), which correlates with the chronic nature of BKD (2, 107). Significant upregulation of the eicosanoid cox2 at 98 dpi (Figure 5F) could be related to the inflammatory response and supports the chronic persistence of R. salmoninarum in lumpfish tissues. tnfα was significantly upregulated at 98 dpi, which could be the result of MSA accumulation in infected lumpfish (32). Chronic stimulation of tnfα is known to assist the chronic inflammatory pathology of BKD and contributed to the host-mediated destruction of the kidney tissues in rainbow trout (32). In contrast, survivor lumpfish with considerable R. salmoninarum burden remaining in their internal tissues for at least 98 dpi (Figure 2C) did so in the absence of BKD clinical signs (Figures 3E, J, O) even with high expression of tnfα with respect to the control (Figure 4K). This immune pattern might be related to the chronic stage of R. salmoninarum infection. On the other hand, tnfα upregulation at 98 dpi (Figure 4K) could be linked to the low bacterial loads in lumpfish tissues at 98 dpi compared to 28 dpi (Figure 2C), because of the role of TNF-α in restricting the bacterial growth in infected macrophages and promoting macrophage survival in zebrafish (Danio rerio) infected Mycobacterium marinum (108).
Most of the downregulated adaptive immune genes (igha, ighδ, ighma, ighmb, cd4a, cd4b, ly6g6f, and cd8a) in infected lumpfish at 28 dpi returned to basal expression levels at 98 dpi (Figures 6A, C–E, G–J). Upregulation of cd74 at 98 dpi (Figure 6K) could induce MHC-II expression. Also, significant stimulation of ifnγ at 98 dpi (Figure 4J) could enhance antigen presentation through MHC-I, as was observed in rainbow trout (109). Thus, the interaction between this intracellular pathogen and teleost MHC-pathways warrants further investigation.
Based on gene expression results, R. salmoninarum could immune-suppressed lumpfish at the early infection stages (28 dpi). In contrast, at late stages (98 dpi), it seems that R. salmoninarum is partially controlled by the lumpfish immune system, which may be attributed to the induced cell-mediated immunity. It is not clear whether the R. salmoninarum will be cleared or if it will persist and be horizontally transmitted or vertically transferred to the next generation of lumpfish. On the other hand, the majority of the lumpfish (65%) survived R. salmoninarum infection and presented the bacteria in head kidney until 98 dpi. These observations suggest that lumpfish is susceptible to R. salmoninarum. Lumpfish susceptibility to high virulent strains of R. salmoninarum with multiple msa gene copies (i.e., msa gene copies ranged from two to five among 68 isolates) (110) and its transmission potential to other fish species warrants future research.
This study revealed that lumpfish is susceptible to R. salmoninarum ATCC 33209 i.p infection, exhibiting a chronic infection pattern. R. salmoninarum caused immune suppression and modulated the lumpfish immune response towards the MHC-I pathway at 28 dpi. Lumpfish seemed to trigger a cell-mediated immune response against R. salmoninarum at the chronic stage of infection. Although R. salmoninarum persisted for at least 98 dpi in lumpfish tissues, it is not known whether lumpfish is able to clear the infection or if R. salmoninarum will persist and use lumpfish as a vector during cohabitation with salmon. Lumpfish susceptibility to more virulent R. salmoninarum strains or different routes of infection warrants further investigation.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, SRP238224 and Supplementary Material (Supplementary File S1).
All animal protocols required for this research were reviewed and approved by the Institutional Animal Care Committee and the Biosafety Committee at Memorial University of Newfoundland (MUN) (https://www.mun.ca/research/about/acs/acc/) based on the guidelines of the Canadian Council on Animal Care (https://ccac.ca/). Experiments were conducted under protocols #18-01-JS, #18-03-JS, and biohazard license L-01.
Conceptualization: JS and HG. Methodology: JS, HG, TC, AH, MD, JH, SK, DC, and DB. Investigation: HG, TC, MD, AH, JH, SK, DC, DB, and JS. Writing original-draft: HG, JH, and JS. Resources: JS and DB. Writing - Review & Editing: HG, TC, AH, MD, JH, SK, DC, DB, and JS. Visualization: HG and JS. Supervision: JS. Funding acquisition: JS and DB. All authors contributed to the article and approved the submitted version.
This work was funded through grants from the Canada First - Ocean Frontier Institute to JS (sub-module J3), the Vietnam International Education Development (VIED) fellowship, and NSERC-Discovery grant (RGPIN-2018-05942).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the staff at the Cold-Ocean and Deep-Sea Research Facility (Steve Hill) and at the Dr. Joe Brown Aquaculture Research Building (staff members: Jessica Fry, Jennifer Monk, Denise Tucker and Kelsie Jeannot) for expert technical assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.733266/full#supplementary-material
1. Buller NB. Aquatic Animal Species and Organism Relationship. In: Bacteria and Fungi From Fish and Other Aquatic Animals : A Practical Identification Manual. London: CABI (2014). p. 368–73.
2. Evelyn TP. Bacterial Kidney Disease - BKD. In: Roberts V, R. J, Bromage NR, editors. Bacterial Disease of Fish. London: Blackwell Scientific (1993). p. 177–95.
3. Kent ML, Traxler GS, Kieser D, Richard J, Dawe SC, Shaw RW, et al. Survey of Salmonid Pathogens in Ocean-Caught Fishes in British Columbia, Canada. J Aquat Anim Health (1998) 10:211–9. doi: 10.1577/1548-8667(1998)010<0211:SOSPIO>2.0.CO;2
4. Starliper CE, Morrison P. Bacterial Pathogen Contagion Studies Among Freshwater Bivalves and Salmonid Fishes. J Shellfish Res (2000) 19:251–8.
5. Chambers EM, Nagel DA, Elloway EA, Addison KL, Barker GA, Verner-Jeffreys DW, et al. Polymerase Chain Reaction Detection of Renibacterium salmoninarum in Fish: Validation of a Modified Protocol. Aquaculture (2009) 287:35–9. doi: 10.1016/j.aquaculture.2008.10.031
6. Austin B, Austin DA. Aerobic Gram-Positive Rods and Cocci. In: Austin B, Austin D, editors. Bacterial Fish Pathogens. Germany: Springer (2016). doi: 10.1007/978-1-4020-6069-4
7. Pascho RJ, Elliott DG, Chase DM. Comparison of Traditional and Molecular Methods for Detection of Renibacterium Salmoninarum. In: Cunningham CO, editor. Molecular Diagnosis of Salmonid Diseases. Reviews: Methods and Technologies in Fish Biology and Fisheries, vol 3. Dordrecht: Springer (2002) p. 157–209. doi: 10.1007/978-94-017-2315-2_7
8. Wiens GD. Bacterial Kidney Disease (Renibacterium salmoninarum). In: Woo PTK, Bruno DW, editors. Fish Diseases and Disorders. Wallingford, UK: CABI (2011). p. 338–74.
9. Bell GR, Hoffmann RW, Brown LL. Pathology of Experimental Infections of the Sablefish, Anoplopoma fimbria (Pallas), With Renibacterium salmoninarum, the Agent of Bacterial Kidney Disease in Salmonids. J Fish Dis (1990) 13:355–67. doi: 10.1111/j.1365-2761.1990.tb00794.x
10. Eliott DG. Renibactrium salmoninarum. In: Woo PTK, Ciaprino R, editors. In Fish Viruses and Bacteria: Pathobiology and Protection. U.K: CABI (2017). p. 286–97.
11. Nagai T, Iida Y. Occurrence of Bacterial Kidney Disease in Cultured Ayu. Fish Pathol (2002) 37:77–81. doi: 10.3147/jsfp.37.77
12. Wallace IS, Munro LA, Kilburn R, Hall M, Black J, Raynard RS, et al. A Report on the Effectiveness of Cage and Farm-Level Fallowing for the Control of Bacterial Kidney Disease and Sleeping Disease on Large Cage-Based Trout Farms in Scotland. Scottish Mar Freshwater Sci Rep (2011) 2:36.
13. Fryer JL, Lannan CN. The History and Current Status of Renibacterium salmoninarum, the Causative Agent of Bacterial Kidney Disease in Pacific salmon. Fish Res (1993) . 17:15–33. doi: 10.1016/0165-7836(93)90004-Q
14. Boerlage AS, Stryhn H, Sanchez J, Hammell KL. Case Definition for Clinical and Subclinical Bacterial Kidney Disease (BKD) in Atlantic salmon (Salmo salar L.) in New Brunswick, Canada. J Fish Dis (2017) 40:395–409. doi: 10.1111/jfd.12521
15. Gudmundsdóttir S, Kristmundsson Á, Árnason ÍÖ. Experimental Challenges With Renibacterium salmoninarum in Arctic Charr Salvelinus alpinus. Dis Aquat Organ (2017) 124:21–30. doi: 10.3354/dao03107
16. Hvas M, Folkedal O, Imsland A, Oppedal F. Metabolic Rates, Swimming Capabilities, Thermal Niche and Stress Response of the Lumpfish, Cyclopterus lumpus. Biol Open (2018) 7:1–9. doi: 10.1242/bio.036079
17. Powell A, Treasurer JW, Pooley CL, Keay AJ, Lloyd R, Imsland AK, et al. Use of Lumpfish for Sea-Lice Control in Salmon Farming: Challenges and Opportunities. Rev Aquac (2018) 10:683–702. doi: 10.1111/raq.12194
18. Murray AG. A Modelling Framework for Assessing the Risk of Emerging Diseases Associated With the Use of Cleaner Fish to Control Parasitic Sea Lice on Salmon Farms. Transbound Emerg Dis (2016) 63:e270–7. doi: 10.1111/tbed.12273
19. Brooker AJ, Papadopoulou A, Gutierrez C, Rey S, Davie A, Migaud H. Sustainable Production and Use of Cleaner Fish for the Biological Control of Sea Lice: Recent Advances and Current Challenges. Vet Rec (2018) 183:383. doi: 10.1136/vr.104966
20. Bell GR, Higgs DA, Traxler GS. The Effect of Dietary Ascorbate, Zinc, and Manganese on the Development of Experimentally Induced Bacterial Kidney Disease in Sockeye salmon (Oncorhynchus nerka). Aquaculture (1984) 36:293–311. doi: 10.1016/0044-8486(84)90323-5
21. Murray C, Evelyn T, Beacham T, Bamer L, Ketcheson J, Prosperi-Porta L. Experimental Induction of Bacterial Kidney Disease in Chinook salmon by Immersion and Cohabitation Challenges. Dis Aquat Organ (1992) 12:91–6. doi: 10.3354/dao012091
22. Richards RH, Roberts RJ, Schlotfeldt H-J. Bacterial Diseases of the Bony Fish. In: Roberts RJ, Schlotfeldt H-J, editors. Basics of Fish Pathology. Hamburg, Germany: Paul Parey (1985). p. 174–207.
24. Scientific Committee on Animal Health and Animal Welfare. Bacterial Kidney Disease - BKD. In: Bacterial Disease of Fish UK: European Commission Health & Consumer protection (1999). p. 3–21.
25. Snieszko SF. Nutritional Fish Diseases. In: Fish Nutrition. New York: Academic Press (1972). p. 403–37. doi: 10.1016/b978-0-12-319650-7.50014-6
26. Lafferty KD, Harvell CD, Conrad JM, Friedman CS, Kent ML, Kuris AM, et al. Infectious Diseases Affect Marine Fisheries and Aquaculture Economics. Ann Rev Mar Sci (2015) 7:471–96. doi: 10.1146/annurev-marine-010814-015646
27. Conlon H, Imsland AKD. Lumpfish Habitat Development for Use in Salmon Farming. [Master’s Thesis]. [Akureyri (Bogir)]: University of Akureyri (2019).
28. Rimstad E, Basic D, Gulla S, Hjeltnes B, Mortensen S. Risk Assessment of Fish Health Associated With the Use of Cleaner Fish in Aquaculture. Opin panel Anim Heal Welf Nor Sci Comm Food Environ VKM Rep (2017) 32:45–7.
29. Haugland GT, Olsen A-B, Rønneseth A, Andersen L. Lumpfish (Cyclopterus lumpus L.) Develop Amoebic Gill Disease (AGD) After Experimental Challenge With Paramoeba perurans and can Transfer Amoebae to Atlantic salmon (Salmo salar L.). Aquaculture (2017) 478:48–55. doi: 10.1016/j.aquaculture.2016.04.001
30. Einarsdottir T, Sigurdardottir H, Bjornsdottir TS, Einarsdottir E. Moritella viscosa in Lumpfish (Cyclopterus lumpus) and Atlantic salmon (Salmo Salar). J Fish Dis (2018) 41:1751–8. doi: 10.1111/jfd.12884
31. Campos-Perez JJ, Ward M, Grabowski PS, Ellis AE, Secombes CJ. The Gills are an Important Site of iNOS Expression in Rainbow Trout Oncorhynchus mykiss After Challenge With the Gram-Positive Pathogen Renibacterium salmoninarum. Immunology (2000) 99:153–61. doi: 10.1046/j.1365-2567.2000.00914.x
32. Grayson TH, Cooper LF, Wrathmell AB, Roper J, Evenden AJ, Gilpin ML. Host Responses to Renibacterium salmoninarum and Specific Components of the Pathogen Reveal the Mechanisms of Immune Suppression and Activation. Immunology (2002) 106:273–83. doi: 10.1046/j.1365-2567.2002.01420.x
33. Jansson E, Hongslo T, Johannisson A, Pilström L, Timmusk S, Norrgren L. Bacterial Kidney Disease as a Model for Studies of Cell Mediated Immunity in Rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol (2003) 14:347–62. doi: 10.1006/fsim.2002.0442
34. Rhodes LD, Wallis S, Demlow SE. Genes Associated With an Effective Host Response by Chinook salmon to Renibacterium salmoninarum. Dev Comp Immunol (2009) 33:176–86. doi: 10.1016/j.dci.2008.08.006
35. Ellul RM, Walde C, Haugland GT, Wergeland H, Rønneseth A. Pathogenicity of Pasteurella sp. In Lumpsuckers (Cyclopterus lumpus L.). J Fish Dis (2019) 42:35–46. doi: 10.1111/jfd.12905
36. Purcell MK, Murray AL, Elz A, Park LK, Marcquenski SV, Winton JR, et al. Decreased Mortality of Lake Michigan Chinook Salmon After Bacterial Kidney Disease Challenge: Evidence for Pathogen-Driven Selection? J Aquat Anim Health (2008) 20:225–35. doi: 10.1577/H08-028.1
37. Austin B, Embley TM, Goodfellow M. Selective Isolation of Renibacterium salmoninarum. FEMS Microbiol Lett (1983) 17:111–4. doi: 10.1111/j.1574-6968.1983.tb00383.x
38. Valderrama K, Saravia M, Santander J. Phenotype of Aeromonas salmonicida Sp. salmonicida Cyclic Adenosine 3’,5’-Monophosphate Receptor Protein (Crp) Mutants and its Virulence in Rainbow trout (Oncorhynchus mykiss). J Fish Dis (2017) 40:1849–56. doi: 10.1111/jfd.12658
39. Leboffe MJ, Pierce BE. Microbiology: Laboratory Theory & Application. Ferguson D, editor. Englewood, CO: Morton Publishing Company (2015).
40. Brown LL, Iwama GK, Evelyn TPT, Nelson WS, Levine RP. Use of the Polymerase Chain Reaction (PCR) to Detect DNA From Renibacterium salmoninarum Within Individual Salmonid Eggs. Dis Aquat Organ (1994) 18:165–71. doi: 10.3354/dao018165
41. Brown LL, Evelyn TP, Iwama GK, Nelson WS, Levine RP. Bacterial Species Other Than Renibacterium salmoninarum Cross-React With Antisera Against R. salmoninarum But are Negative for the P57 Gene of R. salmoninarum as Detected by the Polymerase Chain Reaction (PCR). Dis Aquat Organ (1995) 21:227–31. doi: 10.3354/dao021227
42. Vasquez I, Cao T, Hossain A, Valderrama K, Gnanagobal H, Dang M, et al. Aeromonas salmonicida Infection Kinetics and Protective Immune Response to Vaccination in Sablefish (Anoplopoma fimbria). Fish Shellfish Immunol (2020) 104:557–66. doi: 10.1016/j.fsi.2020.06.005
43. Sambrook. J, Russel. W. Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press (2001).
44. Sakai M, Atsuta S, Kobayashi M. Susceptibility of Five Salmonid Fishes to Renibacterium salmoninarum. Fish Pathol (1991) 26:159–60. doi: 10.3147/jsfp.26.159
45. McIntosh D, Flaño E, Grayson TH, Gilpin ML, Austin B, Villena AJ. Production of Putative Virulence Factors by Renibacterium salmoninarum Grown in Cell Culture. Microbiology (1997) 143:3349–56. doi: 10.1099/00221287-143-10-3349
46. Eslamloo K, Caballero-Solares A, Inkpen SM, Emam M, Kumar S, Bouniot C, et al. Transcriptomic Profiling of the Adaptive and Innate Immune Responses of Atlantic salmon to Renibacterium salmoninarum Infection. Front Immunol (2020) 11:567838. doi: 10.3389/fimmu.2020.567838
47. Sakai M, Ogasawara K, Atsuta S, Kobayashi M. Comparative Sensitivity of Carp, Cyprinus carpio L. And Rainbow trout, Salmo gairdneri Richardson, to Renibacterium salmoninarum. J Fish Dis (1989) 12:367–72. doi: 10.1111/j.1365-2761.1989.tb00325.x
48. Adewoye SO, Ofawole O. Growth Performance and Survival Rate of Clarias Gariepinus Fed Lactobacillus acidophilus Supplemented Diets. IOSR J Agric Vet Sci (2013) 3:45–50. doi: 10.9790/2380-0364550
49. U.S Geological Survey. Culture of Renibacterium salmoninarum From Tissues and Ovarian Fluid. Western Fisheries Research Centre, Seattle, WA, USA: Standard Operating Procedure (SOP (2011) p. 1–6.
50. Chandler DE, Roberson RW. Bioimaging: Current Concepts in Light and Electron Microscopy. Sudbury, London: Johns and Barlett Publishers (2009). 345–50 p.
51. Luna LG. Manual of Histologic Staining Methods of the Armed Forces Institute of Pathology. New York: McGraw-Hill (1968).
52. Bernet D, Schmidt H, Meier W, Burkhardt-Holm P, Wahli T. Histopathology in Fish: Proposal for a Protocol to Assess Aquatic Pollution. J Fish Dis (1999) 22:25–34. doi: 10.1046/j.1365-2761.1999.00134.x
53. Kõressaar T, Remm M. Enhancements and Modifications of Primer Design Program Primer3. Bioinformatics (2007) 23:1289–91. doi: 10.1093/bioinformatics/btm091
54. Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3–new Capabilities and Interfaces. Nucleic Acids Res (2012) 40:e115. doi: 10.1093/nar/gks596
55. Kõressaar T, Lepamets M, Kaplinski L, Raime K, Andreson R, Remm M. Primer3_masker: Integrating Masking of Template Sequence With Primer Design Software. Bioinformatics (2018) 34:1937–8. doi: 10.1093/bioinformatics/bty036
56. Pfaffl MW. A New Mathematical Model for Relative Quantification in Real-Time RT-PCR. Nucleic Acids Res (2001) 29:e45. doi: 10.1093/nar/29.9.e45
57. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate Normalization of Real-Time Quantitative RT-PCR Data by Geometric Averaging of Multiple Internal Control Genes. Genome Biol (2002) 3:1–12. doi: 10.1186/gb-2002-3-7-research0034
58. Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin Chem (2009) 55:611–22. doi: 10.1373/clinchem.2008.112797
59. Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-ΔΔCT Method. Methods (2001) 25:402–8. doi: 10.1006/meth.2001.1262
60. Riedel G, Düker U, Fekete-Drimusz N, Manns M, Vondran F, Bock M. An Extended Δct-Method Facilitating Normalisation With Multiple Reference Genes Suited for Quantitative RT-PCR Analyses of Human Hepatocyte-Like Cells. PloS One (2014) 9:e93031. doi: 10.1371/journal.pone.0093031
61. Soto-Dávila M, Valderrama K, Inkpen SM, Hall JR, Rise ML, Santander J. Effects of Vitamin D2 (Ergocalciferol) and D3 (Cholecalciferol) on Atlantic salmon (Salmo salar) Primary Macrophage Immune Response to Aeromonas salmonicida subsp. salmonicida Infection. Front Immunol (2020) 10:3011. doi: 10.3389/fimmu.2019.03011
62. Coady AM, Murray AL, Elliott DG, Rhodes LD. Both Msa Genes in Renibacterium salmoninarum are Needed for Full Virulence in Bacterial Kidney Disease. Appl Environ Microbiol (2006) 72:2672–8. doi: 10.1128/AEM.72.4.2672-2678.2006
63. O’Farrell CL, Strom MS. Differential Expression of the Virulence-Associated Protein P57 and Characterization of its Duplicated Gene Msa in Virulent and Attenuated Strains of Renibacterium salmoninarum. Dis Aquat Organ (1999) 38:115–23. doi: 10.3354/dao038115
64. Starliper CE, Smith DR, Shatzer T. Virulence of Renibacterium salmoninarum to Salmonids. Dis Aquat Organ (1997) 9:37–41. doi: 10.1577/1548-8667(1997)009
65. Hoffmann R, Popp W, Van de Graaff S. Atypical BKD Predominantly Causing Ocular and Skin Lesions. Bull Eur Assoc Fish Pathol (1984) 4:7–9.
66. Bruno DW, Noguera PA, Poppe TT. A Colour Atlas of Salmonid Diseases. A Colour Atlas of Salmonid Diseases. Springer Netherlands (2013) 84–7. doi: 10.1007/978-94-007-2010-7
67. Metzger DC, Elliott DG, Wargo A, Park LK, Purcell MK. Pathological and Immunological Responses Associated With Differential Survival of Chinook salmon Following Renibacterium salmoninarum Challenge. Dis Aquat Organ (2010) 90:31–41. doi: 10.3354/dao02214
68. Turaga PSD, Wiens GD, Kaattari SL. Analysis of Renibacterium salmoninarum Antigen Production in Situ. Fish Pathol (1987) 22:209–14. doi: 10.3147/jsfp.22.209
69. Turaga P, Wiens G, Kaattari S. Bacterial Kidney Disease: The Potential Role of Soluble Protein Antigen(s). J Fish Biol (1987) 31:191–4. doi: 10.1111/j.1095-8649.1987.tb05312.x
70. Bell GR, Traxler GS. Resistance of the Pacific Lamprey, Lampetra tridentata (Gairdner), to Challenge by Renibacterium salmoninarum, the Causative Agent of Kidney Disease in Salmonids. J Fish Dis (1986) 9:277–9. doi: 10.1111/j.1365-2761.1986.tb01014.x
71. Daly JG, Griffiths SG, Kew AK, Moore AR, Olivier G. Characterization of Attenuated Renibacterium salmoninarum Strains and Their Use as Live Vaccines. Dis Aquat Organ (2001) 44:121–6. doi: 10.3354/dao044121
72. Withler RE, Evelyn TPT. Genetic Variation in Resistance to Bacterial Kidney Disease Within and Between Two Strains of Coho salmon From British Columbia. Trans Am Fish Soc (1990) 119:1003–9. doi: 10.1577/1548-8659(1990)119
73. Becham TD, Evelyn TPT. Population and Genetic Variation in Resistance of Chinook salmon to Vibriosis, Furunculosis, and Bacterial Kidney Disease. J Aquat Anim Health (1992) 4:153–67. doi: 10.1577/1548-8667(1992)004<0153:PAGVIR>2.3.CO;2
74. Gjedrem T, Gjøen HM. Genetic Variation in Susceptibility of Atlantic salmon, Salmo salar L., to Furunculosis, BKD and Cold Water Vibriosis. Aquac Res (1995) 26:129–34. doi: 10.1111/j.1365-2109.1995.tb00892.x
75. Purcell MK, Hard JJ, Neely KG, Park LK, Winton JR, Elliott DG. Genetic Variation in Bacterial Kidney Disease (BKD) Susceptibility in Lake Michigan Chinook salmon and its Progenitor Population From the Puget Sound. J Aquat Anim Health (2014) 26:9–18. doi: 10.1080/08997659.2013.860061
76. Rhodes LD, Coady AM, Deinhard RK. Identification of a Third Msa Gene in Renibacterium salmoninarum and the Associated Virulence Phenotype. Appl Environ Microbiol (2004) 70:6488–94. doi: 10.1128/AEM.70.11.6488-6494.2004
77. McIntosh D, Austin B. The Validity of Western Blotting for the Diagnosis of Bacterial Kidney Disease Based on the Detection of the P57 Antigen of Renibacterium salmoninarum. J Microbiol Methods (1996) 25:329–35. doi: 10.1016/0167-7012(96)00004-8
78. Elliott DG, Wiens GD, Hammell KL, Rhodes LD. Vaccination Against Bacterial Kidney Disease. Fish Vaccin (2014) p:255–72. doi: 10.1002/9781118806913.ch22
79. Bruno DW. Histopathology of Bacterial Kidney Disease in Laboratory Infected Rainbow trout, Salmo gairdneri Richardson, and Atlantic salmon, Salmo salar L., With Reference to Naturally Infected Fish. J Fish Dis (1986) 9:523–37. doi: 10.1111/j.1365-2761.1986.tb01049.x
80. Flano E, Kaattari SL, Razquin B, Villena AJ. Histopathology of the Thymus of Coho salmon Oncorhynchus kisutch Experimentally Infected With Renibacterium salmoninarum. Dis Aquat Org (1996) 26:11–8. doi: 10.3354/dao026011
81. Gutenberger SK, Duimstra JR, Rohovec JS FJ. Intracellular Survival of Renibacterium salmoninarum in Trout Mononuclear Phagocytes. Dis Aquat Org (1997) 28:93–106. doi: 10.3354/dao028093
82. Fredriksen Å, Endresen C, Wergeland HI. Immunosuppressive Effect of a Low Molecular Weight Surface Protein From Renibacterium salmoninarum on Lymphocytes From Atlantic salmon (Salmo salar L.). Fish Shellfish Immunol (1997) 7:273–82. doi: 10.1006/fsim.1997.0082
83. Pietretti D, Wiegertjes GF. Ligand Specificities of Toll-Like Receptors in Fish: Indications From Infection Studies. Dev Comp Immunol (2014) 43:205–22. doi: 10.1016/j.dci.2013.08.010
84. Fryer JL, Sanders JE. Bacterial Kidney Disease of Salmonid Fish. Annu Rev Microbiol (1981) 35:273–98. doi: 10.1146/annurev.mi.35.100181.001421
85. Eslamloo K, Kumar S, Caballero-Solares A, Gnanagobal H, Santander J, Rise ML. Profiling the Transcriptome Response of Atlantic salmon Head Kidney to Formalin-Killed Renibacterium salmoninarum. Fish Shellfish Immunol (2020) 98:937–49. doi: 10.1016/j.fsi.2019.11.057
86. Liu F, Su B, Fu Q, Shang M, Gao C, Tan F, et al. Identification, Characterization and Expression Analysis of TLR5 in the Mucosal Tissues of Turbot (Scophthalmus maximus L.) Following Bacterial Challenge. Fish Shellfish Immunol (2017) 68:272–9. doi: 10.1016/j.fsi.2017.07.021
87. Bethke J, Arias-Muñoz E, Yáñez A, Avendaño-Herrera R. Renibacterium salmoninarum Iron-Acquisition Mechanisms and ASK Cell Line Infection: Virulence and Immune Response. J Fish Dis (2019) 42:1283–91. doi: 10.1111/jfd.13051
88. Piazzon MC, Savelkoul HFJ, Pietretti D, Wiegertjes GF, Forlenza M. Carp Il10 has Anti-Inflammatory Activities on Phagocytes, Promotes Proliferation of Memory T Cells, and Regulates B Cell Differentiation and Antibody Secretion. J Immunol (2015) 194:187–99. doi: 10.4049/jimmunol.1402093
89. King A, Balaji S, Le LD, Crombleholme TM, Keswani SG. Regenerative Wound Healing: The Role of Interleukin-10. Adv Wound Care (2014) 3:315–23. doi: 10.1089/wound.2013.0461
90. Hong S, Zou J, Crampe M, Peddie S, Scapigliati G, Bols N, et al. The Production and Bioactivity of Rainbow trout (Oncorhynchus mykiss) Recombinant IL-1β. Vet Immunol Immunopathol (2001) 81(1–2):1–14. doi: 10.1016/S0165-2427(01)00328-2
91. Lu DQ, Bei JX, Feng LN, Zhang Y, Liu XC, Wang L, et al. Interleukin-1β Gene in Orange-Spotted grouper, Epinephelus Coioides: Molecular Cloning, Expression, Biological Activities and Signal Transduction. Mol Immunol (2008) 45(4):857–67. doi: 10.1016/j.molimm.2007.08.009
92. Hong S, Li R, Xu Q, Secombes CJ, Wang T. Two Types of TNF-α Exist in Teleost Fish: Phylogeny, Expression, and Bioactivity Analysis of Type-II TNF-α3 in Rainbow Trout Oncorhynchus mykiss. J Immunol (2013) 191(12):5959–72. doi:10.4049/jimmunol.1301584
93. Thorn CF, Whitehead AS. Differential Transcription of the Mouse Acute Phase Serum Amyloid A Genes in Response to Proinflammatory Cytokines. Amyloid (2002) 9:229–36. doi: 10.3109/13506120209114098
94. Bayne CJ, Gerwick L. The Acute Phase Response and Innate Immunity of Fish. Dev Comp Immunol (2001) 25:725–43. doi: 10.1016/S0145-305X(01)00033-7
95. Berliner JA, Navab M, Fogelman AM, Frank JS, Demer LL, Edwards PA, et al. Atherosclerosis: Basic Mechanisms. Oxidation, Inflammation, and Genetics. Circulation (1995) 91:2488–96. doi: 10.1161/01.cir.91.9.2488
96. Zou J, Secombes CJ. The Function of Fish Cytokines. Biol (Basel) (2016) 5:14-7. doi: 10.3390/biology5020023
97. Zou J, Peddie S, Scapigliati G, Zhang Y, Bols NC, Ellis AE, et al. Functional Characterisation of the Recombinant Tumor Necrosis Factors in Rainbow trout, Oncorhynchus mykiss. Dev Comp Immunol (2003) 27:813–22. doi: 10.1016/S0145-305X(03)00077-6
98. García-Castillo J, Chaves-Pozo E, Olivares P, Pelegrín P, Meseguer J, Mulero V. The Tumor Necrosis Factor Alpha of the Bony Fish Seabream Exhibits the In Vivo Proinflammatory and Proliferative Activities of its Mammalian Counterparts, Yet it Functions in a Species-Specific Manner. Cell Mol Life Sci (2004) 61:1331–40. doi: 10.1007/s00018-004-4068-1
99. Grayfer L, Walsh JG, Belosevic M. Characterization and Functional Analysis of Goldfish (Carassius auratus L.) Tumor Necrosis Factor-Alpha. Dev Comp Immunol (2008) 32:532–43. doi: 10.1016/j.dci.2007.09.009
100. Engele M, Stöβel E, Castiglione K, Schwerdtner N, Wagner M, Bölcskei P, et al. Induction of TNF in Human Alveolar Macrophages As a Potential Evasion Mechanism of Virulent Mycobacterium tuberculosis. J Immunol (2002) 168:1328–37. doi: 10.4049/jimmunol.168.3.1328
101. Skaar EP. The Battle for Iron Between Bacterial Pathogens and Their Vertebrate Hosts. PloS Pathog (2010) 6:1–2. doi: 10.1371/journal.ppat.1000949
102. Kaufman SHE. Immunity to Intracellular Bacteria. In: Paul WE, editor. Fundamental Immunology. Philadelphia: Lipincott-Raven Publishers (1999). p. 1335–71.
103. Valero Y, Chaves-Pozo E, Meseguer J, Esteban MA, Cuesta A. Biological Role of Fish Antimicrobial Peptides. Antimcrobial Pept: Prop Funct Role Immune Response (2013) . p:31–60. Available at: https://www.researchgate.net/publication/245745108_Biologial_Role_of_Fish_Antimicrobial_Peptides
104. Winter GW, Schreck CB, McIntyre JD. Resistance of Different Stocks and Transferrin Genotypes of Coho salmon, Oncorhynchus kisutch, and Steelhead trout, Salmo gairdneri, to Bacterial Kidney Disease and Vibriosis. Fish Bull (1980) 77:795–802.
105. Cresswell P. Assembly, Transport, and Function of MHC Class II Molecules. Annu Rev Immunol (1994) 12(1):259–91. doi: 10.1146/annurev.iy.12.040194.001355
106. Rozas-Serri M, Lobos C, Correa R, Ildefonso R, Vásquez J, Muñoz A, et al. Atlantic Salmon Pre-Smolt Survivors of Renibacterium salmoninarum Infection Show Inhibited Cell-Mediated Adaptive Immune Response and a Higher Risk of Death During the Late Stage of Infection at Lower Water Temperatures. Front Immunol (2020) 11:1378. doi: 10.3389/fimmu.2020.01378
107. Evenden AJ, Grayson TH, Gilpin ML, Munn CB. Renibacterium salmoninarum and Bacterial Kidney Disease - the Unfinished Jigsaw. Annu Rev Fish Dis (1993) 3:87–104. doi: 10.1016/0959-8030(93)90030-F
108. Clay H, Volkman HE, Ramakrishnan L. Tumor Necrosis Factor Signaling Mediates Resistance to Mycobacteria by Inhibiting Bacterial Growth and Macrophage Death. Immunity (2008) 29:283–94. doi: 10.1016/j.immuni.2008.06.011
109. Martin SAM, Zou J, Houlihan DF, Secombes CJ. Directional Responses Following Recombinant Cytokine Stimulation of Rainbow trout (Oncorhynchus mykiss) RTS-11 Macrophage Cells as Revealed by Transcriptome Profiling. BMC Genomics (2007) 8:150. doi: 10.1186/1471-2164-8-150
Keywords: Bacterial Kidney Disease (BKD), Gram-positive pathogen, Renibacterium salmoninarum, lumpfish, cell-mediated immunity
Citation: Gnanagobal H, Cao T, Hossain A, Dang M, Hall JR, Kumar S, Van Cuong D, Boyce D and Santander J (2021) Lumpfish (Cyclopterus lumpus) Is Susceptible to Renibacterium salmoninarum Infection and Induces Cell-Mediated Immunity in the Chronic Stage. Front. Immunol. 12:733266. doi: 10.3389/fimmu.2021.733266
Received: 30 June 2021; Accepted: 19 October 2021;
Published: 22 November 2021.
Edited by:
Eva-Stina Isabella Edholm, Arctic University of Norway, NorwayReviewed by:
Louise Jørgensen, University of Copenhagen, DenmarkCopyright © 2021 Gnanagobal, Cao, Hossain, Dang, Hall, Kumar, Van Cuong, Boyce and Santander. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Javier Santander, anNhbnRhbmRlckBtdW4uY2E=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.