
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 20 September 2021
Sec. Microbial Immunology
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.731845
This article is part of the Research TopicUnderstanding and Engineering Antibody-Superantigen InteractionsView all 5 articles
Superantigens are unconventional antigens which recognise immune receptors outside their usual recognition sites e.g. complementary determining regions (CDRs), to elicit a response within the target cell. T-cell superantigens crosslink T-cell receptors and MHC Class II molecules on antigen-presenting cells, leading to lymphocyte recruitment, induction of cytokine storms and T-cell anergy or apoptosis among many other effects. B-cell superantigens, on the other hand, bind immunoglobulins on B-cells, affecting opsonisation, IgG-mediated phagocytosis, and driving apoptosis. Here, through a review of the structural basis for recognition of immune receptors by superantigens, we show that their binding interfaces share specific physicochemical characteristics when compared with other protein-protein interaction complexes. Given that antibody-binding superantigens have been exploited extensively in industrial antibody purification, these observations could facilitate further protein engineering to optimize the use of superantigens in this and other areas of biotechnology.
Superantigens are unconventional antigens in the sense that they elicit a response by binding outside the complementary determining regions (CDRs) of their target immune receptor macromolecules (antibodies or T-cell receptors). At their initial description in 1989, superantigens were originally defined as proteins that hyper-stimulate T-cells via the crosslinking of T-cell receptors (TCRs) and MHC Class II molecules (1, 2). This definition required extension following the discovery of B-cell superantigens. B-cell superantigens can hyper-stimulate a large population of B-cells without necessarily having the ability to crosslink TCRs with MHC Class II receptors; they therefore have a different mechanism and specificity compared to T-cell superantigens (3). B-cell superantigens are commonly known to (i) stimulate a high proportion of B-cells, and (ii) bind outside of the CDRs (4). An extended definition of the term ‘superantigen’ was suggested to incorporate both functions, as a molecule which has antigen-receptor mediated interactions with over 5% of the lymphocyte pool (5). This functional definition is therefore based on the hyper-activity of the target receptor upon exposure, and we will use the term in this context here.
Here we review the current understanding of superantigens, how they directly interact with immune receptors of T and B-cells, what common features may be identified in recognition interfaces and how such insights could be adapted to facilitate further protein engineering of these versatile macromolecules for therapeutic, diagnostic, and biotechnological applications.
T-cell superantigens are typically microbial proteins. They were first identified from observation of the hyper-stimulation of T-cells by Staphylococcal Enterotoxin B (SEB). This phenomenon was caused by the crosslinking of T-cell receptors (TCRs) Vβ with MHC class II α1 on antigen presenting cells (APC) by SEB (1, 2). By crosslinking MHC Class II to TCR, small amounts of superantigens can stimulate extensive T-cell proliferation. In a normal adaptive immune response, only around 0.0001% of T-cells are activated. In contrast, superantigen exposure can activate up to 30% of the T-cell pool, leading to severe pathologies following infection (6, 7).
Enterotoxins produced by Staphylococcus aureus and Streptococcus pyogenes form a common family of T-cell superantigens. These enterotoxins are small (20-28 kDa), two domain proteins which are diverse in sequence (15-90%) (8). Despite this variation, enterotoxins and enterotoxin-like proteins from both Staphylococcus aureus and Streptococcus pyogenes are structurally similar (Figure 1), possessing a conserved Greek key motif at the N-terminus known as an oligonucleotide (OB)-fold (9). The C-terminus consists of a conserved β-domain capped by an α-helix (9) with the two β-folds separated by a cluster of α-helices. Due to their structural similarity, it has been suggested by others that the enterotoxins from Staphylococcus aureus and Streptococcus pyogenes shared a common ancestor (8).
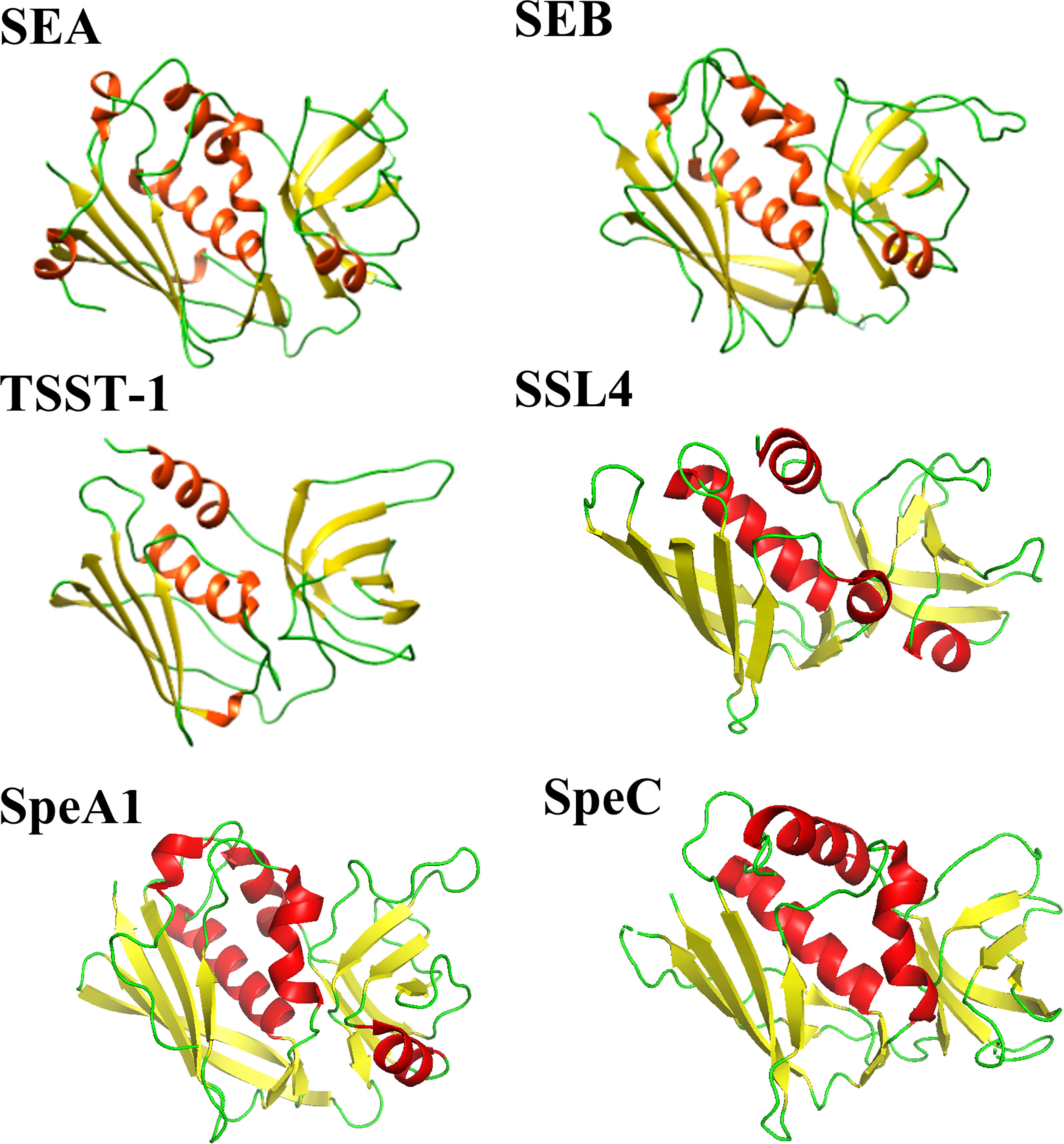
Figure 1 Comparison of selected Staphylococcal aureus and Streptococcus pyogenes enterotoxin and enterotoxin-like structures. Staphylococcal aureus enterotoxins include SEA (PDB code: 1ESF), SEB (PDB code: 1SE4) and TSST-1 (PDB code: 2QIL). The Staphylococcal aureus enterotoxin-like protein shown is SSL4 (PDB code: 4DXF). Two Streptococcal pyogenes enterotoxins are displayed: SpeA1 (PDB code: 1UUP) and SpeC (PDB code: 1KTK). The structures are shown as a ribbon plot with α-helices, β-strands and loops coloured in red, yellow, and green, respectively.
Enterotoxins are thermostable, can withstand extreme pH and are resistant to degradation by proteolytic enzymes such as pepsin and trypsin (10, 11). Some can retain activity after the cooking and digestive process to cause food poisoning (12): nearly 25% of food poisoning cases in the USA are attributed to Staphylococcal enterotoxins (13). In addition, T-cell superantigens also contribute to the development of systemic inflammatory response syndrome (SIRS) known as sepsis (14), toxic shock syndrome (15, 16), scarlet fever (17) and atopic dermatitis (18).
Observations of the cellular responses of T-cells to superantigens are inconsistent, depending on the type and maturity of the T-cell populations studied. T-cell superantigens can cause immature CD4+ and CD8+ T-cells to become depleted. Mature CD4+ and CD8+ T-cells on the other hand, proliferate and produce a cytokine storm (19–25) driving mature T-cells into a state of anergy (26). TCR activation upregulates Fyn signalling, preventing the protein tyrosine kinase ZAP-70 from associating with TCRs via CD3, thus inhibiting TCR signalling (27). The depletion of immature T-cells and anergy of mature T-cells would potentially allow a pathogen to evade the innate immune response, increasing pathogen survivability.
TCR binding to the MHC class II receptors on APCs results in a variety of responses that is dependent on the APC type; the principal pathways and components are summarized in Figure 2. During infection, neutrophils are recruited along with other effector cells through the release of cytokines (e.g. IFN-γ, IL-17, IL-12) and CXC chemokines produced primarily by CD4+ T-cells (28, 50–52). Counterintuitively, the recruitment of leukocytes increases the survivability of Staphylococcus aureus, due to the hyper-stimulation of T-cells, eventually leading to T-cell anergy and cell death. S. aureus is known to survive within neutrophils and macrophages in abscesses (29, 30).
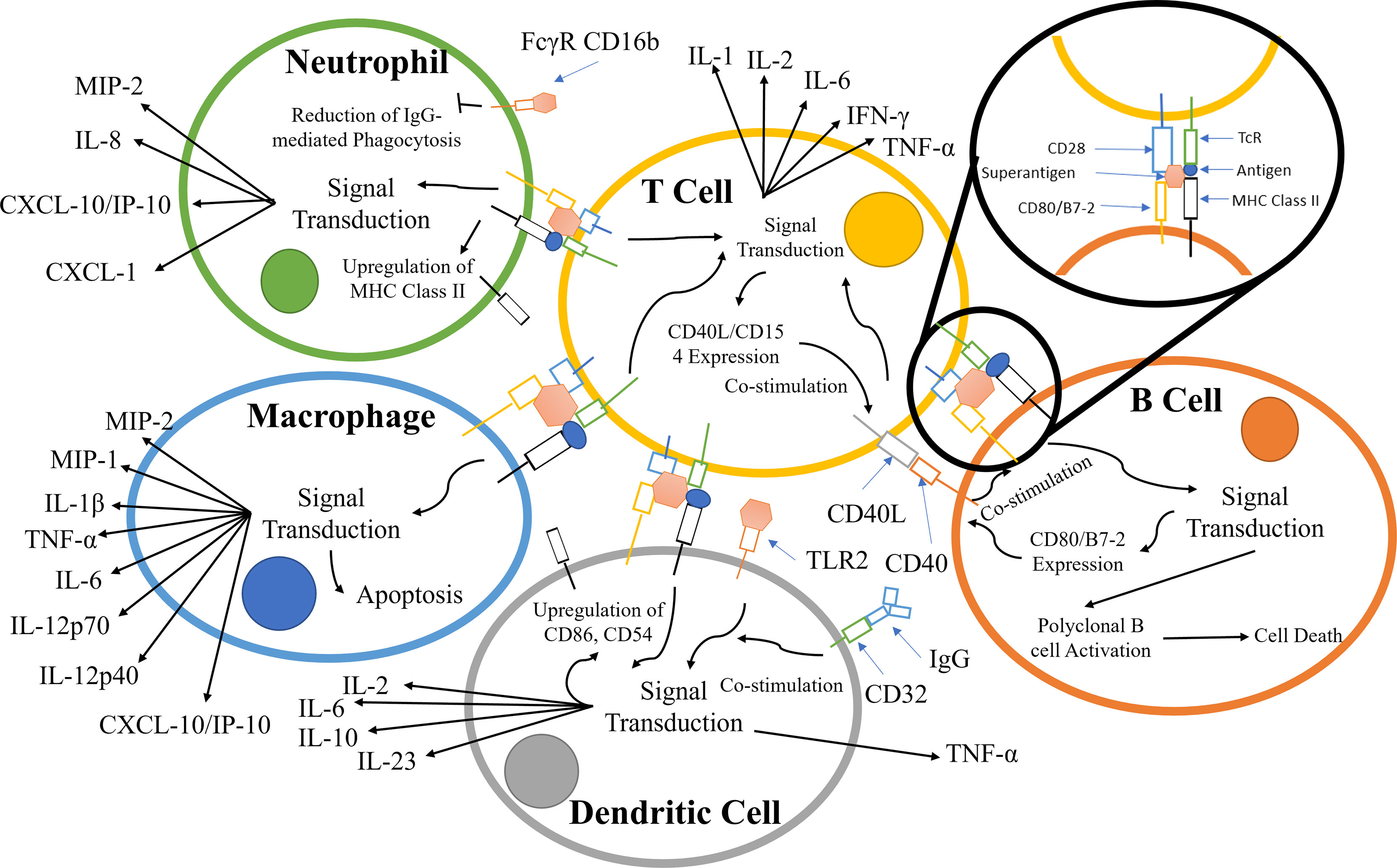
Figure 2 Principal components involved in the superantigen activation of T-cells, B-cells, macrophages, and neutrophils. The interactions displayed are based on material from references (28–49). The responses contribute to and escalate the hyper-activation of T-cells and subsequent cytokine storm.
Alongside TCR/MHC Class II activation, signalling pathways are co-stimulated by crosslinking CD28 on the T-cell with CD80/B7-2 on APCs (53–56). T-cell superantigens can also crosslink the α-subunit of laminin, LAMA2, with G-protein coupled receptor (GPCR), resulting in T-cell stimulation (57–59) (Figure 2).
T-cell superantigens first bind to MHC Class II receptors and accumulate on the surface of the APC before binding to the TCR (9). There are two possible binding sites on MHC Class II: a Zn-dependent high affinity site (Kd = 10-7 - 10-8 M) located on MHC Class II β chain, and a low affinity site (Kd = ~10-5 M) located on MHC Class II α chain (60). Most superantigens bind via the Zn-dependent binding site, forming a complex which is stable for more than 40 hours (61). The high affinity interface between SEH and MHC Class II α chain is shown in Figure 3A, showing a hydrophobic pocket surrounded by polar residues. In addition to H-bonds and salt bridges, a Zn ion contributes to the high binding affinity by stabilizing the complex through crosslinking H81 on the MHC Class II β-strand and H206, N208 on the β-strand 12 on SEH (62). This stabilization allows for the formation of 4 extra H-bonds due to the proximity of the chains where the removal of the Zn ion results in a decrease in binding for SEA, SED, SEE and SEH (63).
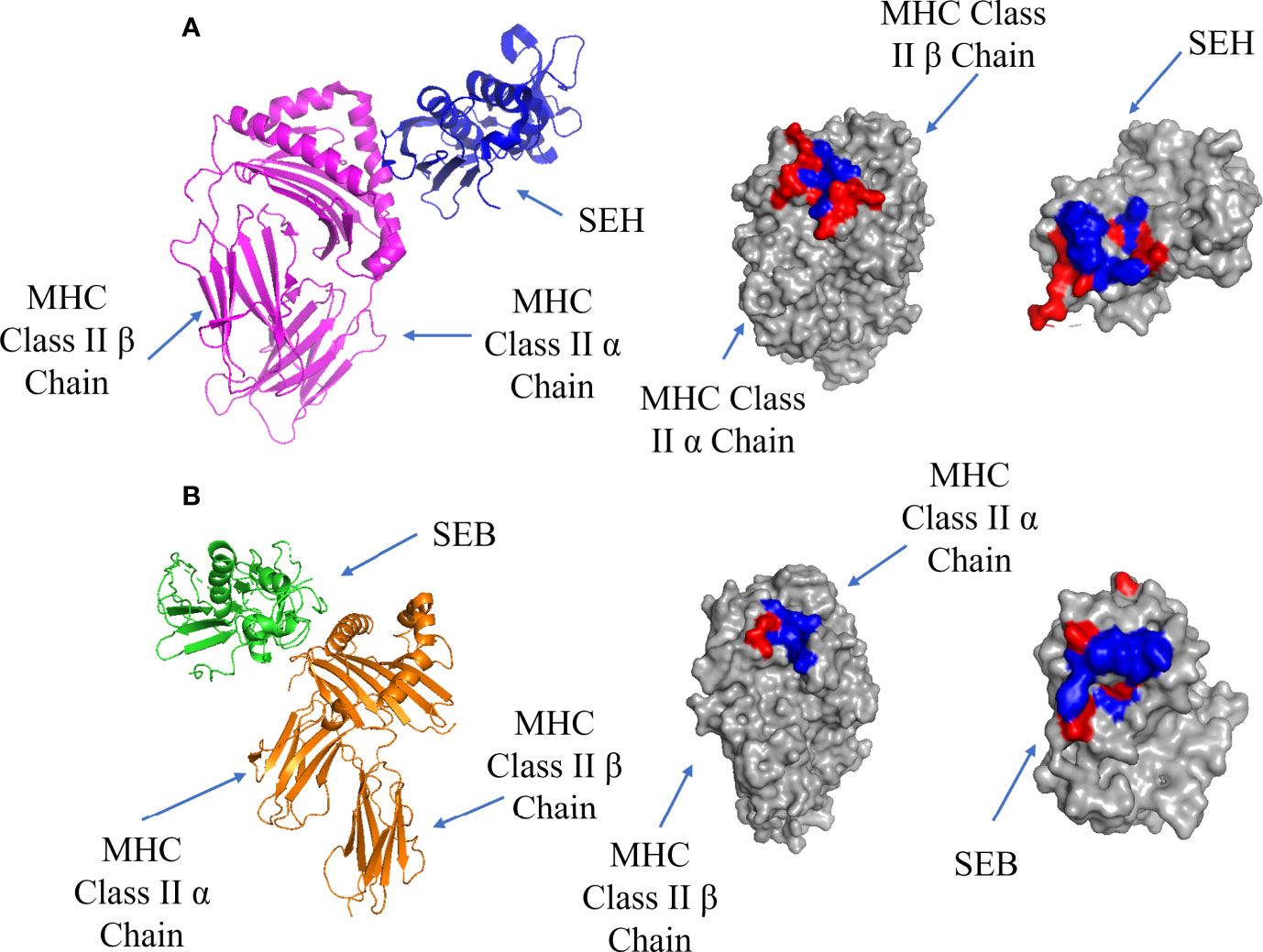
Figure 3 Binding of SEH and SEB superantigens to -MHC Class II. (A) Left panel: ribbon plot of SEH (blue) bound to the high affinity site on MHC Class II α Chain (purple) (PDB code: 2XN9). Middle Panel: MHC Class II showing polar (red) and hydrophobic residues (blue). Right Panel: SEH. (B). Left panel: ribbon plot of SEB (green) bound to the low affinity site on MHC Class II β Chain (orange) (PDB code: 1SEB). Middle panel: MHC Class II β Chain. Right panel: SEB.
The low affinity binding site is exemplified by a structure containing the enterotoxin SEB, which forms a complex with MHC class II (61): the low affinity interface is shown in Figure 3B. In addition to cross-interface bonds, there is a hydrophobic patch on SEB comprised of F44, L45 and F47 which inserts into a hydrophobic pocket on MHC Class II α chain.
The enterotoxin SEA can also bind to the low and high affinity sites to crosslink two MHC Class II molecules (61, 64, 65). Staphylococcal Enterotoxin H (SEH) was shown to bind the Zn-dependent high affinity site on MHC class II (62), as well as to TCR Vα instead of Vβ (66, 67). A list of T-cell superantigens and their site specificities has been previously summarized by Proft and Fraser (9). T-cell superantigen selectivity for the α or β chains of the MHC Class II complex is dependent on the presence of the Zn atom at the C-terminal β domain. Its absence leads to the binding of the α-chain of MHC Class II via a hydrophobic ridge on the N-terminal OB-domain (9).
Superantigens bind to the TCR after adhesion to MHC Class II; there are also two sites on the TCRs in all superantigen complexes studied to date. Some T-cell superantigens bind to the α chain [SEH (68)], although most recognize the β chain. Unlike complexes with MHC Class II, both TCR interfaces bind T-cell superantigens at low affinity (Kd = 10-4 – 10-6 M) (68, 69) and yet both are capable of mediating activation of a cytokine storm (8, 70–73). SEB binding to the TCR β chain is shown in Figure 4A where the interface is located at the TCR binding cleft between the N-terminal β-barrel and the second α-helix. It is characterised by several cross-interface bonds, with N23 playing a crucial role, and a nearby hydrophobic patch formed of V26, Y79 and Y80 on SEH packing against the CDR2 loop of TCR Vβ (74).
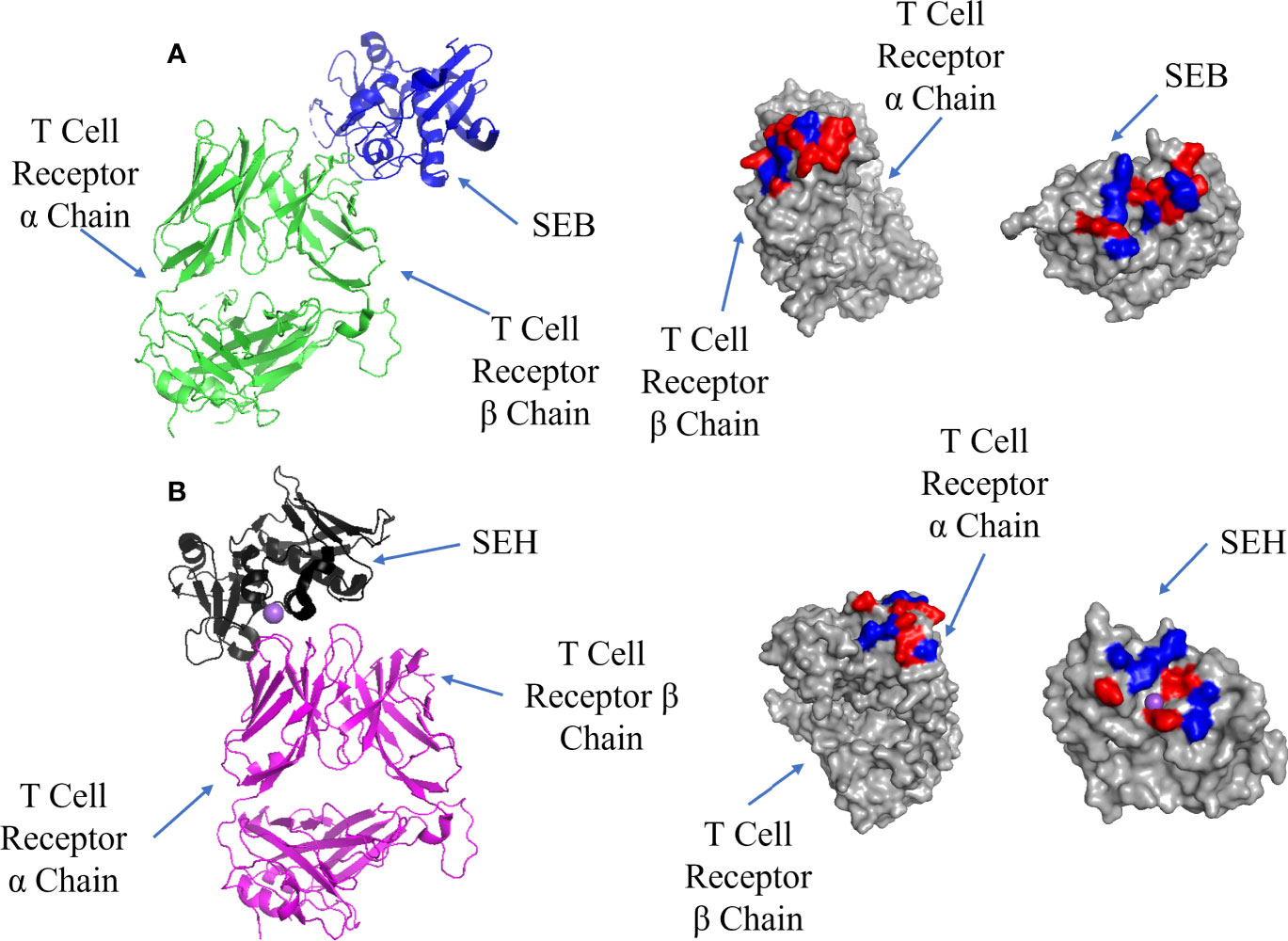
Figure 4 Binding of SEB and SEH superantigens to -TCRs. (A) Left panel: ribbon plot of SEB (blue) bound to TCR (green) (PDB code: 4C56). Middle panel: TCR showing polar (red) and hydrophobic residues (blue). Right panel: SEB. (B) Left panel: ribbon plot of SEH (black) bound to TCR (purple) (PDB code: 2XN9). Middle panel: TCR. Right panel: SEH.
SEH binding to the TCR α chain forms an interface comprising hydrophobic and hydrophilic patches, with a notable hydrophilic patch surrounding a Na ion (Figure 4B). Comparing this to the SEB-TCR β chain interface, there are 7 fewer H-bonds and 2 fewer salt bridges, although the binding affinities are similar (Kd = 10-4 – 10-6 M) (68). The lack of contacts between SEH and the TCR α chain may be bolstered by the presence of the Na ion. N16 found on the second α-helix and the hydrophobic patch (Y79 and Y80) on SEH are well conserved among T-cell superantigens whether they bind to the TCR α or β chains (67). The mutation N23A (equivalent to N16 in SEH) in SEC2 caused the loss of mitogenic activity (75) and the same mutation in SEB resulted in poorer proliferation of T-cells (76).
B-cell superantigens bind immunoglobulins outside the CDRs; proteins which would fit this definition of a superantigen were first described in the early 1990s (3). Binding to the Fab fragment drives B-cells into apoptosis by hyper-activation of B-cell receptors (BCRs). Considering that 20 to 50% of B-cells have BCRs on their surfaces (77), B-cell superantigens can elicit a potent immune response. However, B-cell superantigens are better known for their ability to bind Fc and their applications as affinity resins for antibody purification.
Staphylococcal Protein A (SpA), Streptococcal Protein G (SpG) and Peptostreptococcal Protein L (PpL) are B-cell superantigens located on the bacterial cell wall (78, 79). SpA was classified as a superantigen in 1995 due to its observed effect on B-cells (4). However, SpA was first isolated in 1940 and identified in 1964 due to its Fc binding ability (78, 80). It comprises a 42 kDa protein arranged into five homologous domains (E-D-A-B-C), each forming a three α-helix bundle fold (Figure 5A) (81, 82). The domains are linked by conserved, flexible linkers (82). Native SpA also includes region X, a 12 x 8-residue repeat sequence which binds peptidoglycan. All 5 A-E domains can bind both Fc and Fab fragments (83). The binding affinity for specific immunoglobulins depends on the isotype and species origin. In humans, SpA binds strongly to IgG1, IgG2, IgG4 and weakly to IgA1, IgA2 and IgM. Mutations R435H and F436Y on hIgG3 have been identified as the reason SpA cannot bind human IgG3 (84). Interestingly, mutations in CDR2 from the therapeutic antibodies Herceptin and Pertuzumab were shown to contribute to binding SpA (85).
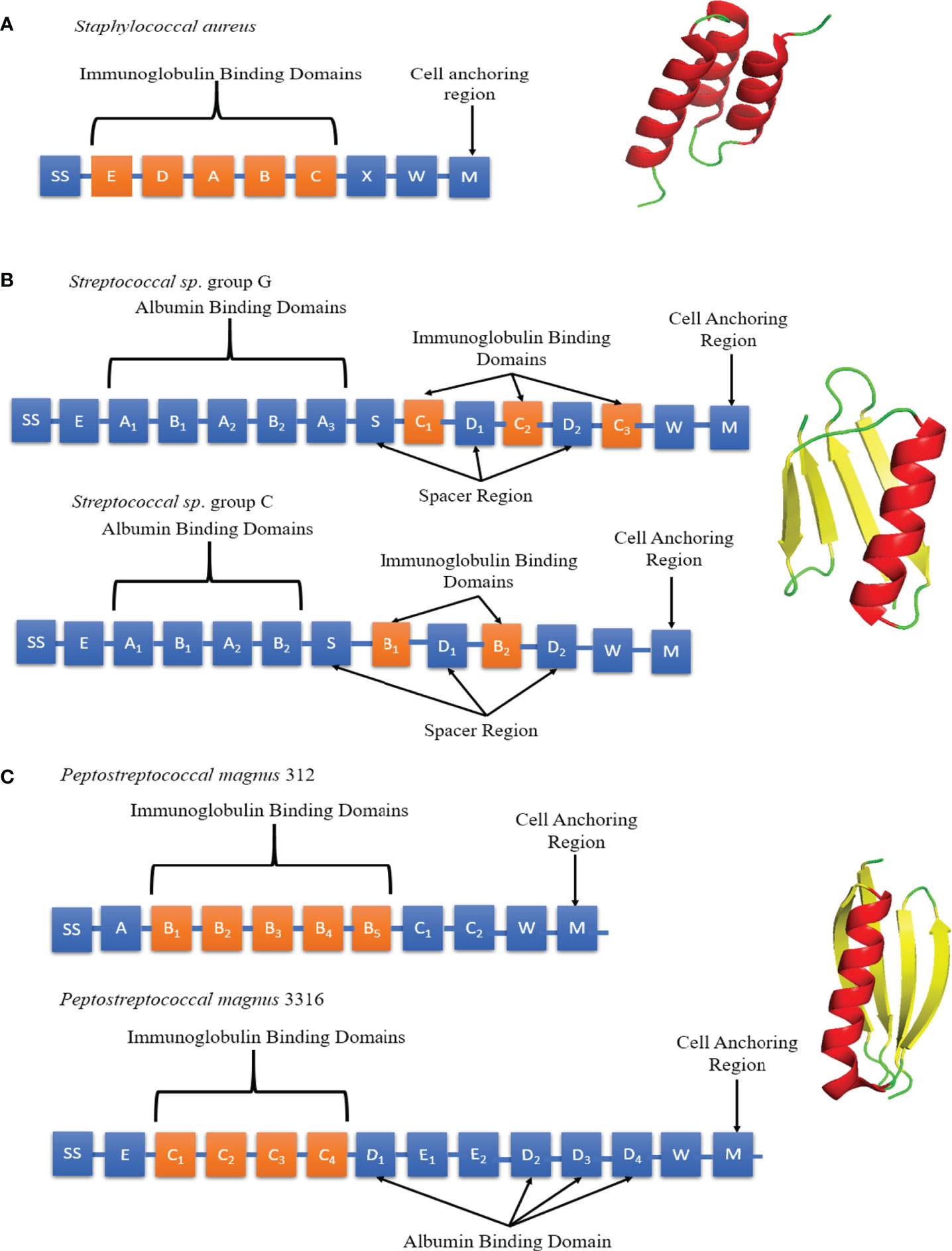
Figure 5 Schematic diagrams of SpA, SpG and PpL domain structures. (A) Left panel: Individual SpA domains including S (sorting peptide), Domains E-D-A-B-C, Region X and Region M. Right panel: SpA Domain C (PDB code: 4WWI) Each SpA immunoglobulin binding domains consists of 3 α-helices (red). (B) Left panel: Individual SpG domains including S (sorting peptide), Region E, Albumin Binding Domains A1-A2-A3, immunoglobulin binding domains B1-B2/C1-C2-C3 and Region W. Right panel: SpG Domain B1 (PDB code: 3GB1) Each SpG immunoglobulin binding domain consists of 1 α-helix (red) and 4 anti-parallel β-strands (yellow). (C) Left panel: Individual PpL domains including S (sorting peptide), Immunoglobulin Binding Domains B1-B2-B3-B4-B5/C1-C2-C3-C4, Albumin Binding Domains D1 to D4, Region W and M. Right panel: PpL Domain B1 (PDB code: 1HEZ). Each PpL immunoglobulin binding domain consists of 1 α-helix (red) and 4 anti-parallel β-strands (yellow).
SpG was first identified in 1984 by Björck and Kronvall (86) and subsequently described as a B-cell superantigen. The sequence of SpG differs depending on the Streptococcus strain of origin (Figure 5B). SpG from group C Streptococcus sp. contains 2 immunoglobulin binding domains (B1-B2) whereas group G has 3 (C1-C2-C3) (87–89). Between each immunoglobulin binding domain are ‘spacers’, known as D domains. All SpG immunoglobulin-binding domains can bind both the Fc and Fab fragments (83). SpG has provided an alternative to SpA in antibody manufacturing, due it its ability to bind some antibody isotypes not recognised by SpA. It can strongly bind to all four human IgG subclasses (IgG1, IgG2, IgG3 and IgG4).
PpL was shown to induce apoptosis in B-cells by binding to the VL region outside of the CDRs of BCRs, fulfilling the definition of a B-cell superantigen (90). It was first isolated in 1985 and characterised as an immunoglobulin-binding protein capable of binding to the variable light chain in 1988 (79, 91). Of the two most common strains of Peptostreptococcus magnus, strain 312 produces a 79 kDa, 5 domain (B1-B2-B3-B4-B5) protein whereas strain 3316 expresses a 106 kDa 4 domain (C1-C2-C3-C4) protein (Figure 5C) (92). PpL recognizes the light chain exclusively and cannot bind to the Fc region. This makes it highly suitable for affinity-purification of non-IgG antibodies (93, 94).
All three B-cell superantigens (SpA, SpG and PpL) share several common features; they form small, stable, multidomain structures with a ‘beads on a string’ type structure. Kim et al. compared antibody levels of IgG and VH3+ IgM in mice when infected with SpA mutants with one to 6 domains. The results showed that the optimal number of immunoglobulin binding domains to induce the largest B-cell response was 5 (95). This observation suggests that B-cell superantigens are driven by the need for multivalency of binding and the consequent improved cross-linking of BCRs. These results were corroborated by a similar study with PpL (92). Although SpG and PpL share no significant sequence homology (15%), their immunoglobulin binding domains have similar folds, forming a β-sheet packed against a single α-helix. A gene transfer event between Streptococcus aureus and Peptostreptococcus magnus has been proposed to explain a possible common evolutionary origin of SpG and PpL (96). All three B-cell superantigens also utilise regions W and M for crossing the cell membrane, featuring the common Gram-positive protein anchoring motif LPXTG (97, 98). SpG and PpL also contain albumin binding domains (99, 100), which are absent in SpA.
B-cell superantigens cross-link BCRs to activate BCR dependent signalling (101, 102). This initial signal transduction leads to the downregulation of BCRs, and an upregulation of several cluster of differentiation (CD) receptors (102), resulting in B-cell capping (summarized schematically in Figure 6). MHC Class II is also upregulated (102). The upregulation of these receptors leads to the activation of pro-apoptotic signals, such as Caspase 3, causing mitochondrial permeabilization and apoptosis (5, 101, 102). Recently it has been shown that SpA B-cell superantigen activity is dependent on the presence of the LPXTG anchoring motif as well as the ‘LysM domain’ between region X and the LPXTG motif (103). These observations imply SpA must be bound to peptidoglycan to cause B-cell stimulation.
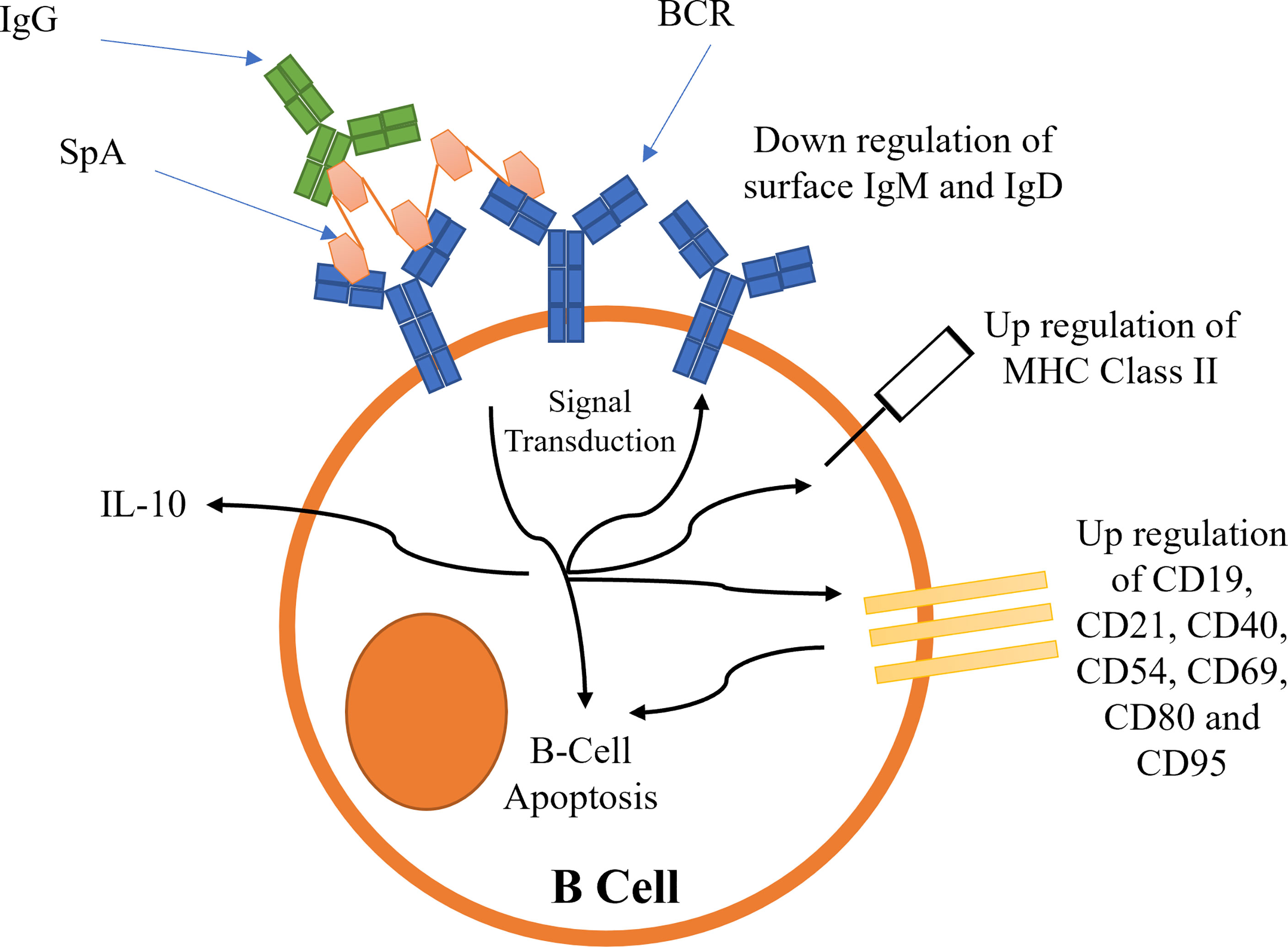
Figure 6 Proposed mechanisms for the activation of B-cell receptors by SpA. Activation leads to B-cell capping and B-cell apoptosis.
The precise functional role of microbial B-cell superantigens binding to Fc is obscure, although it has been shown recently that soluble IgG is a requirement for the successful activation of BCRs by SpA (104). The efficiency of BCR activation was dependent on the strength of Fc binding to each IgG subclass (104). The binding of SpA-IgG complex to BCR is predicted to increase the functional valency of the complex (104). SpA-IgG is thought to form a ‘lattice’ structure around the B-cells by crosslinking BCR Fab with IgG Fc and other BCR Fab regions promoting a sustained stimulation.
Other potential functions of the B-cell superantigen-immunoglobulin interaction are the blocking of immunoglobulin effector functions, opsonization and immunoglobulin-mediated phagocytosis, antibody-dependent cell mediated cytotoxicity (ADCC) and complement-dependent cytotoxicity (CDC) (105–107). Expression of B-cell superantigens ultimately leads to B-cell depletion and evasion of the immune system: in this sense, they can be considered as virulence factors (108–111).
SpA, SpG and PpL bind to BCRs at different sites on the Fab fragment, although the activation results in similar cellular responses. SpG binds to the CH1 domain (112), implying isotype dependent binding, whereas SpA binds to the VH3 family only. A comparison of the conservation of key residues between the seven VH families shows that, although many residues are conserved, there are several which are key and which, when mutated, result in the loss of binding for SpA (113), including in the VH-CDR2 (85). PpL domains only bind the κ light chain VL region and therefore lacks the ability to bind λ chains. The binding affinity of PpL differs between the families of κ light chain, specifically to FW1: it can bind to human Vκ I, III and IV, but not II (114, 115).
Several T-cell superantigens have the ability to bind BCRs, although generally in a weak and non-specific manner (4), and without a B-cell response. Exceptions have been noted, for example, SEA increased the survival of VH3 B-cells (116). SED has also been shown to increase survival of VH4 B-cells (117). However, the in vivo response is yet to be determined.
Recent research suggests that B-cell superantigens also enhance immune defences (118). Two superantigens have been identified from the commensal bacteria Lachnospiraceae sp: Immunoglobulin Binding Proteins A (IbpA) and B (IbpB). Both were observed to activate BCRs by binding VH3 leading to the increased secretion of IgA, although this was only shown in vitro.
The crystal structure of the SpA and IgM Fab complex is illustrated in Figure 7A, showing that the interface occurs at the VH domain (involving residues from β-strands B to E) of the Fab fragment and α-helices 2 and 3 of SpA (113). The interface is dominated by polar residues with three negatively charged residues from SpA and two positively charged residues from Fab forming electrostatic interactions (113). All SpA domains can bind to the Fab fragment (119), and each domain varies in its affinity towards VH3. The interacting residues form a predominantly hydrophilic interface forming several cross-interface bonds.
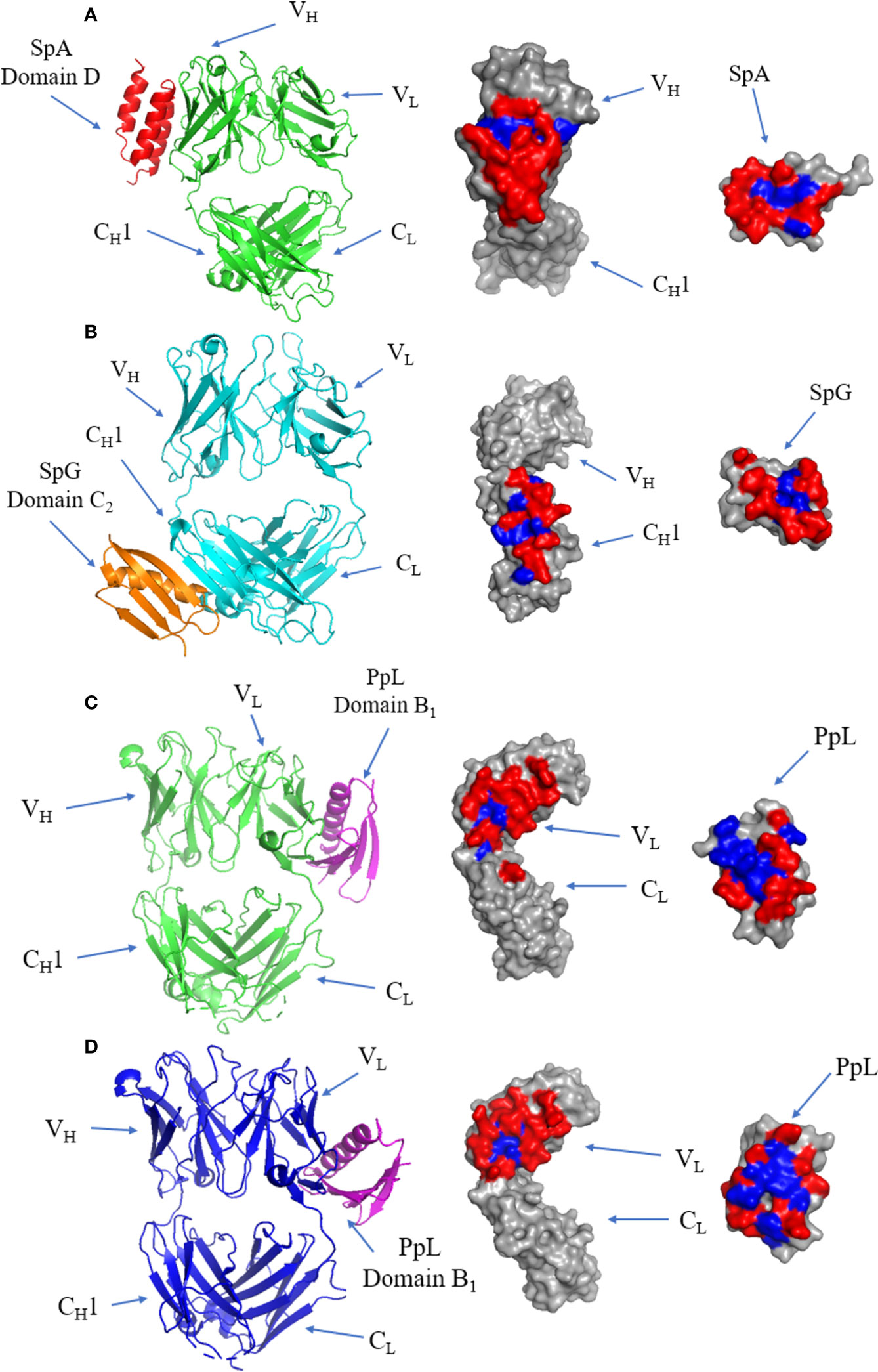
Figure 7 Binding of SpA, SpG and PpL to antibody Fab fragments: (A) Left panel: ribbon plot of SpA (Red) bound to VH3 domain of IgM Fab (Green) (PDB code: 1DEE). Middle panel: IgM Fab showing polar (red) and hydrophobic residues (blue). Right panel: SpA. (B) Left panel: ribbon plot of SpG (Orange) bound to CH1 domain of IgG Fab (Cyan) (PDB code: 1QKZ). Middle panel: IgG Fab. Right panel: SpG. (C) Left panel: ribbon plot of PpL Domain B1 (Purple) interface 1 bound to IgM Fab (Green) at the VL domain (PDB code: 1HEZ). Middle panel: IgM Fab. Left panel: PpL. (D) Left panel: ribbon plot of PpL Domain B1 (Purple) interface 2 bound to IgM Fab (Blue) at the VL domain (PDB code: 1HEZ). Middle panel: IgM Fab. Right panel: PpL.
SpG domain C2 co-crystalized with IgG Fab (Figure 7B): the interface forms an antiparallel alignment between the last β-strand of the CH1 domain and the second β-strand of SpG (120). The antiparallel complex also results in interactions occurring between first β-strand of CH1 and the C-terminal end of the α-helix of SpG (121). The interface is formed by mostly hydrophilic residues flanking a small hydrophobic patch.
The first of two binding sites of PpL to IgM Fab is shown in (Figure 7C). The majority of the interface occurs at framework region 1 (FR1) of the VL region, with several contacts occurring outside of the VL region: K107 between the VL and CL regions, E143 from the CL region and R24 on the β-strand of CDR-L1 of IgM Fab. The interface includes residues from the α-helix and second β-strand of PpL domain B1 (122). The interface has a high affinity (Kd: 110 nM) (123), forming a predominantly hydrophilic interaction characterised by 9 H-bonds, although several residues have been proposed as hotspots from in silico alanine scanning of the Fab and PpL (115, 123, 124). Interestingly, recent evidence showed distal FWR3 effects on the PpL binding site at the FW1 (125) adding to the considerations for the light chain pairing with the heavy chain (126, 127).
The second binding site is formed from 15 residues at β-strands A, B, C and D of the VL region of IgM Fab, and the α-helix and third β-strand of PpL (Figure 7D). Although the second binding site is slightly larger and composed of more cross-interface contacts, it has a lower binding affinity (3.4 µM) (115). The first and second binding sites of PpL share only one common residue from PpL (R52) but 10 out of 15 residues from IgM Fab.
The crystal structure of a single domain from SpA was determined in complex with IgG Fc: it showed that the protein-protein interface occurs between α-helix 1 and 2 of SpA domain B and CH2 and CH3 of the Fc (113) (Figure 8A). The residues forming the interface are generally hydrophilic (128). SpA residues Q9, Q10, D36 and D37, are conserved in the five immunoglobulin binding domains of SpA and are required for Fc binding (129). Mutating residue H435 in IgG eliminates SpA binding, as this residue is situated on the C-terminus of the CH3 region and protrudes into the CH2-CH3 cleft forming surface contacts with SpA (84).
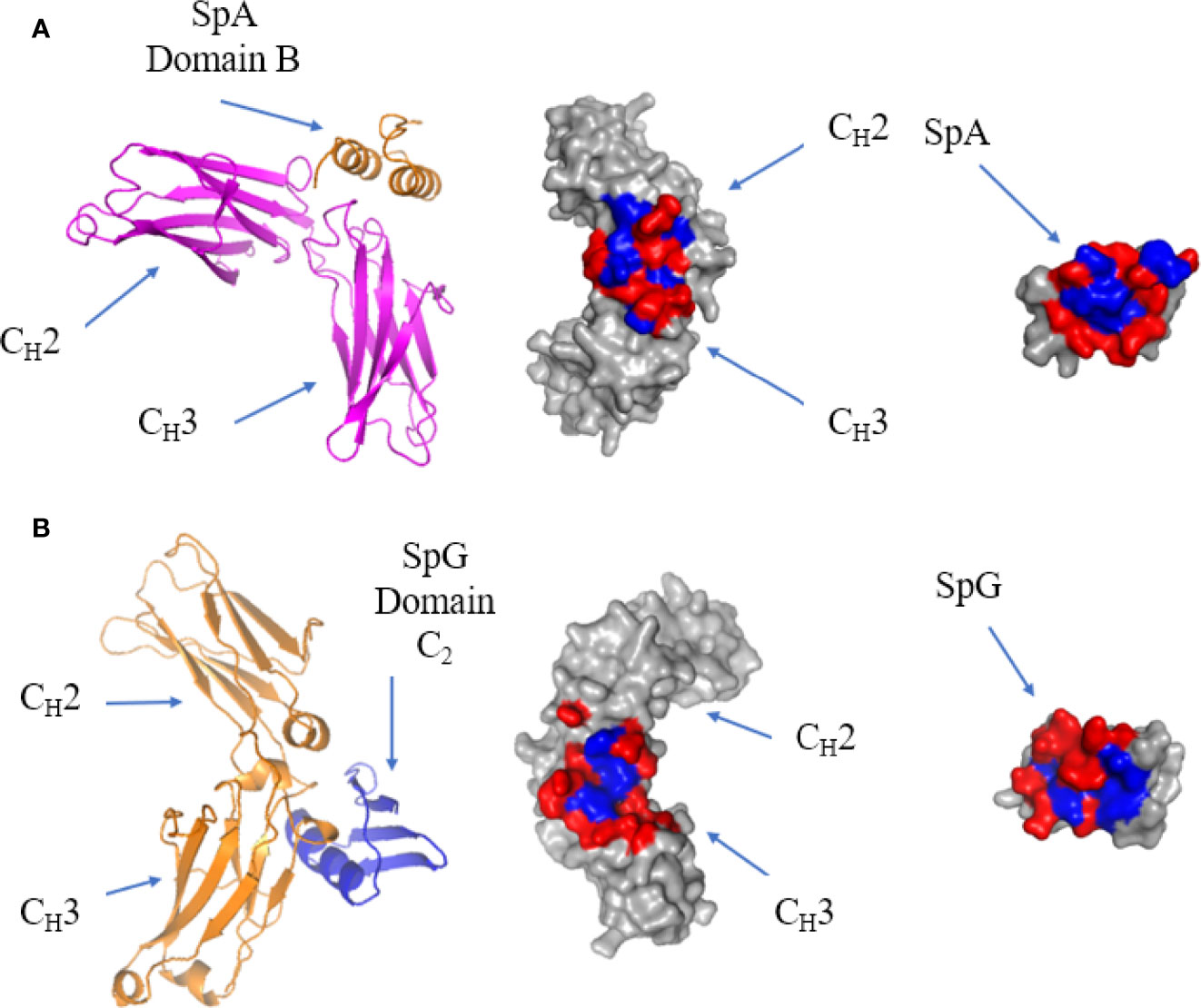
Figure 8 Binding of SpA and SpG superantigens to -IgG Fc. (A) Left panel: ribbon plot of SpA Domain B (Orange) bound to IgG Fc (Pink) at the CH2-CH3 interface (PDB code: 5U4Y). Middle panel: IgG Fc showing polar (red) and hydrophobic residues (blue). Right panel: SpA. (B) Left panel: ribbon plot of SpG Domain C2 (Blue) bound to IgG Fc (Orange) at the CH2-CH3 interface (PDB code: 1FCC). Middle panel: IgG Fc. Right panel: SpG.
The crystal structure of SpG C2 in complex with IgG Fc showed that it binds at the same site as SpA, with SpG binding to IgG Fc at the CH2-CH3 interface (Figure 8B). SpG fits within the CH2-CH3 cleft and binds through residues on the α-helix and third β-strand. As they recognise essentially the same site, SpA and SpG bind competitively to IgG Fc (130, 131). The strong binding affinity of SpG for IgG Fc is contributed by a hydrophobic pocket surrounded by hydrophilic residues. Comparing the binding sites of SpG for Fab and Fc, Fab binding uses β-strands 1 and 2 as well as the α-helix, whereas Fc binding uses β-strand 3 as well as a more prominent contribution of α-helix residues.
The structures of the complexes of T and B-cell superantigens with immune macromolecules were examined to compare the nature of the interfaces with all other structures of protein-protein complexes. A list was compiled from the Protein Data Bank, extracting specific data on hydrophobicity, number of hydrogen bonds, salt bridges, interface area, binding affinity, and charges at the interface. These values were then condensed onto a two-dimensional plot using t-distributed stochastic neighbor embedding, such that each point represents a complex (Figure 9).
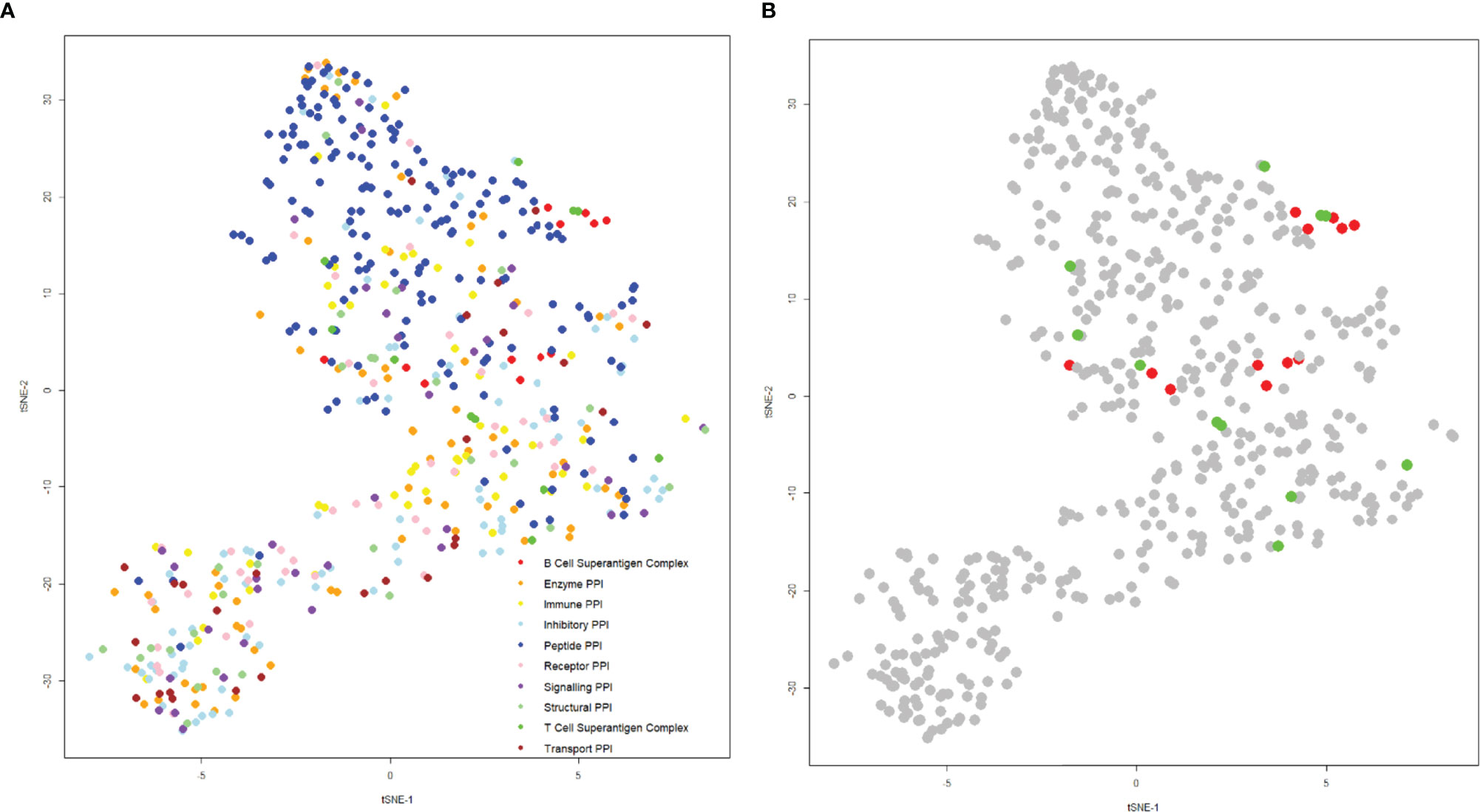
Figure 9 t-SNE plot of protein-protein interactions highlighting superantigen-antibody complexes. The position of each complex was determined using the following parameters: “Buried Surface Area (A2)”, “Buried Surface Area Hydrophobicity (A2)”, “Number of Interface Residues”, “H-Bonds”, “Salt Bridges”, Category”, “Total Positive Charge at Interface (A2)”, “Total Negative Charge at Interface (A2)” with a perplexity of 30. (A) Distribution of general protein-protein interactions categorized by function (T-cell superantigen complex, B-cell superantigen complex, enzyme PPI, immune PPI, inhibitory PPI, peptide PPI, receptor PPI, signaling PPI, structural PPI, toxin-antitoxin PPI and transport PPI). (B) The same plot as (A), but coloured for T-cell superantigen complexes (green) and B-cell superantigen complexes (red).
Complexes were categorized according to function: most were well dispersed by functional category, although peptide complexes tended to predominate in the upper half of the plot (Figure 8A). The superantigen complexes are grouped in the central and right side of the plot, indicating that their binding interfaces share some physicochemical characteristics (Figure 9B). An explanation for this phenomenon is that T-cell and B-cell superantigen interactions are transient-type complexes, as defined by Noreen and Thornton (132). Such complexes tend to be small and less hydrophobic than obligate, homo oligomeric complexes. Both T- and B-cell superantigen interfaces form interface areas less than 1000 Å2 and range from slightly to very hydrophilic.
The T-cell superantigens are located on the centre and right-hand side of the plot and are more scattered than the B-cell superantigen interfaces (Figure 9B). The interface areas of T-cell and B-cell superantigens have similar ranges: 436 – 944 Å2 and 517 – 714 Å2 respectively. Fractional hydrophobicity of the T-cell superantigen interfaces range from 18 – 49%, similar to those for B-cell superantigens (9 – 40%).
Although B-cell superantigens recognise different binding sites within the Fab molecule (VH, VL and CH1) they share very similar interface physicochemical properties, which align closely to those seen in peptide complexes.
The fact that superantigens are promiscuous and capable of recognition of different binding partners indicates there is scope for improving binding affinity and extending specificity for specific targets. The observation that B-cell superantigens, and more specifically superantigen-Fab complexes, are physicochemically similar may allow for development of engineering strategies which makes use of this facility. Nonetheless, just as we expanded the definition of superantigens in this review to include B-cell activation based on new findings, we are also aware of novel superantigen-like behaviours by non-proteins e.g., nickel (133) that may in time be included as superantigens in the future.
Superantigens have been employed in multiple applications, both clinically and industrially. Though many improvements have been made, there is room to engineer and expand their scope and applications. Understanding the biochemistry of the superantigen-antibody interfaces provides an information resource for the development of novel biotechnological and pharmaceutical applications.
Since the approval of the first therapeutic monoclonal antibody in 1986 (Muromonab-CD3), the use of antibody-based drugs has expanded significantly with technological developments such as scFv, antibody-drug conjugates (ADCs) and bispecifics. In 2019, 70% of all biopharmaceutical products sold were monoclonal antibodies in a market worth over $150 billion. Antibody-based drugs continue to increase their market share, with current estimates predicting global revenue to increase to over $300 billion by 2025 (134). The expansion of antibody-based drugs has therefore created a need for improved manufacturing and purification processes.
The most prominent industrial application of B-cell superantigens is their use as affinity resins for the purification of antibodies, allowing highly efficient separation of antibodies for clinical and research applications. Improvement of these affinity resins has allowed pharmaceutical companies to develop cost-effective antibody purification techniques, increasing the feasibility of large-scale manufacturing of antibodies, resulting in the expansion of the industry. There are, however, some limitations; 80% of the downstream processing cost occurs at the capture and purification phase (135), and there is no single resin which can bind all antibody isotypes from all species of interest. Furthermore, antibodies are eluted from the affinity resins at low pH values, frequently causing aggregation.
Some investigators have engineered superantigens to optimize their application in antibody purification (136, 137). For example “Domain Z” was developed in 1987- a mutant of Domain B with two mutations, A1V and G29A (138), which resulted in SpA losing the ability to bind VH3-Fab while retaining Fc affinity (139); this innovation allows for the selective purification of the Fc fragment after pepsin digestion. One such Z domain affinity resin is Cytiva’s mAb Select SuRe (140). When testing mutant N23T, the stability of the SpA Z domain resin increased (141). Recently a new SpA resin has been developed: AviPure. This resin is formed of two B domains with two cysteine and histidine residues at the C-terminus to the steric hindrance, increasing binding capacity and increasing its resistance to pHs extremes, while retaining high binding affinity (135). Affinity resins undergo cleaning in place (CIP) procedures commonly using 0.5 M NaOH; therefore, affinity resins with high alkaline stability are desirable. To address the issue with CIP procedures, SpA was shown to have higher alkaline stability with a single mutation at position 29, with G29W being the most stable (142). Two further mutations N23T and F30A to the SpA Z domain resulted in a higher alkaline resistance when compared to wild type (143). SpA Z domain was also engineered to include six glycine residues on the second loop, which resulted in an increase in the elution pH (143). Wild type SpA is less susceptible to extreme alkaline conditions, with a half-life of 16 h (141) compared to SpG, which has a half-life of under 10 mins (144). Asn residues were identified as the most susceptible to deamination: mutation of all three Asn residues of SpG (N8T, N35A and N37A) improved alkaline stability by 8-fold (145). SpG was further demonstrated to have higher alkaline stability when three other mutations Y3F, T16I and T1I were introduced (145). By increasing alkaline stability, the lifespan of affinity resins can be increased, lowering the overall cost of antibody production. It has also been recently demonstrated that adding an additional alkaline wash step after the antibody capture step results in a decrease in antibody aggregation, lower impurity levels and an increase in antibody yield (146).
With the development of new formats such as single chain variable fragments (scFvs), strong Fab binding is required. Unfortunately, SpA and SpG have lower binding affinities for the Fab fragment compared to Fc. PpL has the advantage over other B-cell superantigens of binding strongly to the Vκ of scFv, (Kd = 4.5nM) (147). The scFv structural arrangement consists of the VH and VL domains connected by a short linker. scFv molecules have the advantage of retaining the CDRs while being significantly smaller than whole antibodies. SpA can also bind scFv but at a lower binding affinity than PpL, whereas SpG is unable to bind scFv. The most significant downside for the use of PpL as an affinity resin is its inability to bind λ light chains. Therefore, in human antibody production, roughly 34% of the antibodies will not bind to the resin, suggesting that engineering PpL to bind λ light chains could be valuable.
Superantigens are used to detect IgG in serum (148), making use of their immunoglobulin binding specificity. On the contrary, superantigens recognized by IgGs allow for the detection of Staphylococcus aureus (149, 150) in disease states.
Engineering of superantigens to be specific to regions of TCRs or antibody V-region families or isotypes for the development of diagnostic kits could be applied to the quantification of disease-associated proteins e.g. IgE in allergy. The ability to specifically bind antibodies can also allow its development in easy-to-use, non-technical point-of-care testing home-use devices (151), as recently applied during the COVID-19 pandemic. Such superantigen-based diagnostics can be coupled with colorimetric, home-made devices [e.g. mobile spectrophotometers (152, 153)]. Given the increasing association of antibody VH families with certain diseases e.g. [VH5 in nickel allergy (85)], superantigens that can differentiate antibody VH families have clear potential in diagnostic kit development.
The role of superantigens in sepsis, a leading cause of death listed by the WHO, makes them an important target for toxic-shock syndrome (154). Several short peptide regions (~40 residues) from SEA and SPEA have been identified as causes of vasodilation (155), suggesting an application in the development of antihypertension drugs.
Superantigens can also be used as a target for an anti-Staphylococcus aureus vaccine. There have been several attempts at producing a vaccine against S. aureus, without success, although it has been shown that the use of anti-SpA antibodies leads to the promotion of opsonophagocytic clearance of Staphylococcus aureus (156, 157).
Superantigens have also shown promise in the treatment of cancer through a synergistic effect with antibodies in the recruitment of T-cells (158). The ability of SEB to hyper-stimulate and proliferate CAR T-cells led to a more effective antitumour response when used in combination (159). PpL has been shown to induce apoptosis in malignant κ+ B cell lymphomas in humans and mice (160), demonstrating the potential use of superantigens as anti-cancer drugs, particularly when sagaciously paired with a suitable Vκ light chain (94). A range of potential T-cell superantigen-based anticancer drugs have been recently reviewed (161), including SEB, demonstrating the ability to inhibit metastasis and tumour growth (162). Several Fab-superantigen fusion proteins show promising Phase I/II clinical trial results. A major drawback with using superantigens is their potential to elicit a toxic response. One way to prevent this is to reduce the over-stimulation of T-cells. SEA was split into two functionally inactive domains and attached to a scFv. When used in combination, the two SEA fragments reassemble, forming a functionally active superantigen and resulting in the selective activation of T-cells (163). Another way to avoid superantigen toxicity is to utilize superantigen-like proteins which, as mentioned previously, are very similar in structure and function, although they do not result in emesis. They have been shown to inhibit tumour growth by 30% without significant toxicity (164).
The importance of understanding superantigens goes beyond bacterial sepsis to viruses, where for example, SARS-CoV2 spike protein displays superantigen properties (165, 166) causing multisystem inflammatory syndrome in children through its unspecific activation of T-cells (167).
With the development of new clinical therapeutics, B-cell superantigen engineering presents an opportunity to develop novel applications, as well as improving current superantigen-based technology, such as purification resins. Structural information on B-cell superantigen interfaces has been useful in providing a basis for the engineering of binding characteristics. The application of protein engineering principles offers considerable scope for directed modification of superantigen binding properties and harnessing for applications in medicine and the pharmaceutical industry.
AD wrote the manuscript, with critical revisions from SG and JD. AD and JD prepared the figures. All authors contributed to the article and approved the submitted version.
AD is a PhD student funded by The University of Manchester, UK and the Agency for Science, Technology and Research (A*STAR), Singapore. This review was partially funded by the National Research Foundation (NRF) Singapore grant to Experimental Drug Development Centre (EDDC), ASTAR.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. White J, Herman A, Pullen AM, Kubo R, Kappler JW, Marrack P. The V Beta-Specific Superantigen Staphylococcal Enterotoxin B: Stimulation of Mature T Cells and Clonal Deletion in Neonatal Mice. Cell (1989) 56:27–35. doi: 10.1016/0092-8674(89)90980-x
2. Seth A, Stern LJ, Ottenhoff TH, Engel I, Owen MJ, Lamb JR, et al. Binary and Ternary Complexes Between T-Cell Receptor, Class II MHC and Superantigen In Vitro. Nature (1994) 369:324–7. doi: 10.1038/369324a0
3. Pascual V, Capra JD. B-Cell Superantigens? Curr Biol (1991) 1:315–7. doi: 10.1016/0960-9822(91)90097-g
4. Levinson AI, Kozlowski L, Zheng Y, Wheatley L. B-Cell Superantigens: Definition and Potential Impact on the Immune Response. J Clin Immunol (1995) 15:26S–36S. doi: 10.1007/BF01540891
5. Silverman GJ, Goodyear CS. Confounding B-Cell Defences: Lessons From a Staphylococcal Superantigen. Nat Rev Immunol (2006) 6:465–75. doi: 10.1038/nri1853
6. Choi Y, Lafferty JA, Clements JR, Todd JK, Gelfand EW, Kappler J, et al. Selective Expansion of T Cells Expressing V Beta 2 in Toxic Shock Syndrome. J Exp Med (1990) 172:981–4. doi: 10.1084/jem.172.3.981
7. Kotzin BL, Leung DY, Kappler J, Marrack P. Superantigens and Their Potential Role in Human Disease. Adv Immunol (1993) 54:99–166. doi: 10.1016/s0065-2776(08)60534-9
8. Proft T, Fraser JD. Bacterial Superantigens. Clin Exp Immunol (2003) 133:299–306. doi: 10.1046/j.1365-2249.2003.02203.x
9. Fraser JD, Proft T. The Bacterial Superantigen and Superantigen-Like Proteins. Immunol Rev (2008) 225:226–43. doi: 10.1111/j.1600-065X.2008.00681.x
10. Thomas D, Chou S, Dauwalder O, Lina G. Diversity in Staphylococcus Aureus Enterotoxins. Chem Immunol Allergy (2007) 93:24–41. doi: 10.1159/000100856
11. Li SJ, Hu DL, Maina EK, Shinagawa K, Omoe K, Nakane A. Superantigenic Activity of Toxic Shock Syndrome Toxin-1 Is Resistant to Heating and Digestive Enzymes. J Appl Microbiol (2011) 110:729–36. doi: 10.1111/j.1365-2672.2010.04927.x
12. Argudin MA, Mendoza MC, Rodicio MR. Food Poisoning and Staphylococcus Aureus Enterotoxins. Toxins (Basel) (2010) 2:1751–73. doi: 10.3390/toxins2071751
13. Herman A, Kappler JW, Marrack P, Pullen AM. Superantigens: Mechanism of T-Cell Stimulation and Role in Immune Responses. Annu Rev Immunol (1991) 9:745–72. doi: 10.1146/annurev.iy.09.040191.003525
14. Martin GS, Mannino DM, Eaton S, Moss M. The Epidemiology of Sepsis in the United States From 1979 Through 2000. N Engl J Med (2003) 348:1546–54. doi: 10.1056/NEJMoa022139
15. Bergdoll MS, Crass BA, Reiser RF, Robbins RN, Davis JP. A New Staphylococcal Enterotoxin, Enterotoxin F, Associated With Toxic-Shock-Syndrome Staphylococcus Aureus Isolates. Lancet (1981) 1:1017–21. doi: 10.1016/s0140-6736(81)92186-3
16. Schlievert PM, Shands KN, Dan BB, Schmid GP, Nishimura RD. Identification and Characterization of an Exotoxin From Staphylococcus Aureus Associated With Toxic-Shock Syndrome. J Infect Dis (1981) 143:509–16. doi: 10.1093/infdis/143.4.509
17. Stevens DL, Tanner MH, Winship J, Swarts R, Ries KM, Schlievert PM, et al. Severe Group A Streptococcal Infections Associated With a Toxic Shock-Like Syndrome and Scarlet Fever Toxin a. N Engl J Med (1989) 321:1–7. doi: 10.1056/NEJM198907063210101
18. Kim J, Kim BE, Ahn K, Leung DYM. Interactions Between Atopic Dermatitis and Staphylococcus Aureus Infection: Clinical Implications. Allergy Asthma Immunol Res (2019) 11:593–603. doi: 10.4168/aair.2019.11.5.593
19. Jenkinson EJ, Kingston R, Owen JJ. Newly Generated Thymocytes Are Not Refractory to Deletion When the Alpha/Beta Component of the T Cell Receptor is Engaged by the Superantigen Staphylococcal Enterotoxin B. Eur J Immunol (1990) 20:2517–20. doi: 10.1002/eji.1830201125
20. Coppola MA, Blackman MA. Bacterial Superantigens Reactivate Antigen-Specific CD8+ Memory T Cells. Int Immunol (1997) 9:1393–403. doi: 10.1093/intimm/9.9.1393
21. Murphy KM, Heimberger AB, Loh DY. Induction by Antigen of Intrathymic Apoptosis of CD4+CD8+TCRlo Thymocytes In Vivo. Science (1990) 250:1720–3. doi: 10.1126/science.2125367
22. Lin YS, Lei HY, Low TL, Shen CL, Chou LJ, Jan MS. In Vivo Induction of Apoptosis in Immature Thymocytes by Staphylococcal Enterotoxin B. J Immunol (1992) 149:1156–63.
23. Meilleur CE, Wardell CM, Mele TS, Dikeakos JD, Bennink JR, Mu HH, et al. Bacterial Superantigens Expand and Activate, Rather Than Delete or Incapacitate, Preexisting Antigen-Specific Memory CD8+ T Cells. J Infect Dis (2019) 219:1307–17. doi: 10.1093/infdis/jiy647
24. Xu R, Shears RK, Sharma R, Krishna M, Webb C, Ali R, et al. IL-35 is Critical in Suppressing Superantigenic Staphylococcus Aureus-Driven Inflammatory Th17 Responses in Human Nasopharynx-Associated Lymphoid Tissue. Mucosal Immunol (2020) 13:460–70. doi: 10.1038/s41385-019-0246-1
25. Florquin S, Amraoui Z, Abramowicz D, Goldman M. Systemic Release and Protective Role of IL-10 in Staphylococcal Enterotoxin B-Induced Shock in Mice. J Immunol (1994) 153:2618–23.
26. MacDonald HR, Baschieri S, Lees RK. Clonal Expansion Precedes Anergy and Death of V Beta 8+ Peripheral T Cells Responding to Staphylococcal Enterotoxin B In Vivo. Eur J Immunol (1991) 21:1963–6. doi: 10.1002/eji.1830210827
27. Watson AR, Janik DK, Lee WT. Superantigen-Induced CD4 Memory T Cell Anergy. I. Staphylococcal Enterotoxin B Induces Fyn-Mediated Negative Signaling. Cell Immunol (2012) 276:16–25. doi: 10.1016/j.cellimm.2012.02.003
28. Xu SX, Gilmore KJ, Szabo PA, Zeppa JJ, Baroja ML, Haeryfar SM, et al. Superantigens Subvert the Neutrophil Response to Promote Abscess Formation and Enhance Staphylococcus Aureus Survival In Vivo. Infect Immun (2014) 82:3588–98. doi: 10.1128/IAI.02110-14
29. Gresham HD, Lowrance JH, Caver TE, Wilson BS, Cheung AL, Lindberg FP. Survival of Staphylococcus Aureus Inside Neutrophils Contributes to Infection. J Immunol (2000) 164:3713–22. doi: 10.4049/jimmunol.164.7.3713
30. Pidwill GR, Gibson JF, Cole J, Renshaw SA, Foster SJ. The Role of Macrophages in Staphylococcus Aureus Infection. Front Immunol (2020) 11:620339. doi: 10.3389/fimmu.2020.620339
31. Zeng X, Moore TA, Newstead MW, Deng JC, Lukacs NW, Standiford TJ. IP-10 Mediates Selective Mononuclear Cell Accumulation and Activation in Response to Intrapulmonary Transgenic Expression and During Adenovirus-Induced Pulmonary Inflammation. J Interferon Cytokine Res (2005) 25:103–12. doi: 10.1089/jir.2005.25.103
32. Lei L, Altstaedt J, von der Ohe M, Proft T, Gross U, Rink L. Induction of Interleukin-8 in Human Neutrophils After MHC Class II Cross-Linking With Superantigens. J Leukoc Biol (2001) 70:80–6. doi: 10.1189/jlb.70.1.80
33. Gainet J, Dang PM, Chollet-Martin S, Brion M, Sixou M, Hakim J, et al. Neutrophil Dysfunctions, IL-8, and Soluble L-Selectin Plasma Levels in Rapidly Progressive Versus Adult and Localized Juvenile Periodontitis: Variations According to Disease Severity and Microbial Flora. J Immunol (1999) 163:5013–9.
34. Tuffs SW, James DBA, Bestebroer J, Richards AC, Goncheva MI, O’Shea M, et al. The Staphylococcus Aureus Superantigen SElX Is a Bifunctional Toxin That Inhibits Neutrophil Function. PloS Pathog (2017) 13:e1006461. doi: 10.1371/journal.ppat.1006461
35. Gonzalez CD, Ledo C, Giai C, Garofalo A, Gomez MI. The Sbi Protein Contributes to Staphylococcus Aureus Inflammatory Response During Systemic Infection. PloS One (2015) 10:e0131879. doi: 10.1371/journal.pone.0131879
36. Pliyev BK, Dimitrieva TV, Savchenko VG. Cytokine-Mediated Induction of MHC Class II in Human Neutrophils Is Dependent on NADPH Oxidase Activity. Eur J Cell Biol (2015) 94:67–70. doi: 10.1016/j.ejcb.2014.11.001
37. Noli Truant S, De Marzi MC, Sarratea MB, Antonoglou MB, Meo AP, Iannantuono Lopez LV, et al. Egc Superantigens Impair Monocytes/Macrophages Inducing Cell Death and Inefficient Activation. Front Immunol (2019) 10:3008. doi: 10.3389/fimmu.2019.03008
38. Chen CL, Wu YY, Lin CF, Kuo CF, Han CL, Wang S, et al. Streptococcal Pyrogenic Exotoxin B Inhibits Apoptotic Cell Clearance by Macrophages Through Protein S Cleavage. Sci Rep (2016) 6:26026. doi: 10.1038/srep26026
39. Watanabe I, Ichiki M, Shiratsuchi A, Nakanishi Y. TLR2-Mediated Survival of Staphylococcus Aureus in Macrophages: A Novel Bacterial Strategy Against Host Innate Immunity. J Immunol (2007) 178:4917–25. doi: 10.4049/jimmunol.178.8.4917
40. Nabavi N, Freeman GJ, Gault A, Godfrey D, Nadler LM, Glimcher LH. Signalling Through the MHC Class II Cytoplasmic Domain is Required for Antigen Presentation and Induces B7 Expression. Nature (1992) 360:266–8. doi: 10.1038/360266a0
41. Mourad W, Scholl P, Diaz A, Geha R, Chatila T. The Staphylococcal Toxic Shock Syndrome Toxin 1 Triggers B Cell Proliferation and Differentiation via Major Histocompatibility Complex-Unrestricted Cognate T/B Cell Interaction. J Exp Med (1989) 170:2011–22. doi: 10.1084/jem.170.6.2011
42. Bekeredjian-Ding I, Inamura S, Giese T, Moll H, Endres S, Sing A, et al. Staphylococcus Aureus Protein A Triggers T Cell-Independent B Cell Proliferation by Sensitizing B Cells for TLR2 Ligands. J Immunol (2007) 178:2803–12. doi: 10.4049/jimmunol.178.5.2803
43. Parcina M, Miranda-Garcia MA, Durlanik S, Ziegler S, Over B, Georg P, et al. Pathogen-Triggered Activation of Plasmacytoid Dendritic Cells Induces IL-10-Producing B Cells in Response to Staphylococcus Aureus. J Immunol (2013) 190:1591–602. doi: 10.4049/jimmunol.1201222
44. Narita K, Hu DL, Asano K, Nakane A. Interleukin-10 (IL-10) Produced by Mutant Toxic Shock Syndrome Toxin 1 Vaccine-Induced Memory T Cells Downregulates IL-17 Production and Abrogates the Protective Effect Against Staphylococcus Aureus Infection. Infect Immun (2019) 87:1–12. doi: 10.1128/IAI.00494-19
45. Keener AB, Thurlow LT, Kang S, Spidale NA, Clarke SH, Cunnion KM, et al. Staphylococcus Aureus Protein A Disrupts Immunity Mediated by Long-Lived Plasma Cells. J Immunol (2017) 198:1263–73. doi: 10.4049/jimmunol.1600093
46. Parcina M, Wendt C, Goetz F, Zawatzky R, Zahringer U, Heeg K, et al. Staphylococcus Aureus-Induced Plasmacytoid Dendritic Cell Activation Is Based on an IgG-Mediated Memory Response. J Immunol (2008) 181:3823–33. doi: 10.4049/jimmunol.181.6.3823
47. Bhardwaj N, Friedman SM, Cole BC, Nisanian AJ. Dendritic Cells are Potent Antigen-Presenting Cells for Microbial Superantigens. J Exp Med (1992) 175:267–73. doi: 10.1084/jem.175.1.267
48. Coutant KD, de Fraissinette AB, Cordier A, Ulrich P. Modulation of the Activity of Human Monocyte-Derived Dendritic Cells by Chemical Haptens, a Metal Allergen, and a Staphylococcal Superantigen. Toxicol Sci (1999) 52:189–98. doi: 10.1093/toxsci/52.2.189
49. Wu X, Xu F. Dendritic Cells During Staphylococcus Aureus Infection: Subsets and Roles. J Transl Med (2014) 12:358. doi: 10.1186/s12967-014-0358-z
50. Laan M, Cui ZH, Hoshino H, Lotvall J, Sjostrand M, Gruenert DC, et al. Neutrophil Recruitment by Human IL-17 via C-X-C Chemokine Release in the Airways. J Immunol (1999) 162:2347–52.
51. McLoughlin RM, Solinga RM, Rich J, Zaleski KJ, Cocchiaro JL, Risley A, et al. CD4+ T Cells and CXC Chemokines Modulate the Pathogenesis of Staphylococcus Aureus Wound Infections. Proc Natl Acad Sci USA (2006) 103:10408–13. doi: 10.1073/pnas.0508961103
52. McLoughlin RM, Lee JC, Kasper DL, Tzianabos AO. IFN-Gamma Regulated Chemokine Production Determines the Outcome of Staphylococcus Aureus Infection. J Immunol (2008) 181:1323–32. doi: 10.4049/jimmunol.181.2.1323
53. Arad G, Levy R, Nasie I, Hillman D, Rotfogel Z, Barash U, et al. Binding of Superantigen Toxins Into the CD28 Homodimer Interface is Essential for Induction of Cytokine Genes That Mediate Lethal Shock. PloS Biol (2011) 9:e1001149. doi: 10.1371/journal.pbio.1001149
54. Levy R, Rotfogel Z, Hillman D, Popugailo A, Arad G, Supper E, et al. Superantigens Hyperinduce Inflammatory Cytokines by Enhancing the B7-2/CD28 Costimulatory Receptor Interaction. Proc Natl Acad Sci USA (2016) 113:E6437–46. doi: 10.1073/pnas.1603321113
55. Kaempfer R, Popugailo A, Levy R, Arad G, Hillman D, Rotfogel Z. Bacterial Superantigen Toxins Induce a Lethal Cytokine Storm by Enhancing B7-2/CD28 Costimulatory Receptor Engagement, a Critical Immune Checkpoint. Receptors Clin Investig (2017) 4:1–9. doi: 10.14800/rci.1500
56. Popugailo A, Rotfogel Z, Supper E, Hillman D, Kaempfer R. Staphylococcal and Streptococcal Superantigens Trigger B7/CD28 Costimulatory Receptor Engagement to Hyperinduce Inflammatory Cytokines. Front Immunol (2019) 10:942. doi: 10.3389/fimmu.2019.00942
57. Yamasaki S, Tachibana M, Shinohara N, Iwashima M. Lck-Independent Triggering of T-Cell Antigen Receptor Signal Transduction by Staphylococcal Enterotoxins. J Biol Chem (1997) 272:14787–91. doi: 10.1074/jbc.272.23.14787
58. Bueno C, Lemke CD, Criado G, Baroja ML, Ferguson SS, Rahman AK, et al. Bacterial Superantigens Bypass Lck-Dependent T Cell Receptor Signaling by Activating a Galpha11-Dependent, PLC-Beta-Mediated Pathway. Immunity (2006) 25:67–78. doi: 10.1016/j.immuni.2006.04.012
59. Li Z, Zeppa JJ, Hancock MA, McCormick JK, Doherty TM, Hendy GN, et al. Staphylococcal Superantigens Use LAMA2 as a Coreceptor To Activate T Cells. J Immunol (2018) 200:1471–9. doi: 10.4049/jimmunol.1701212
60. Li Y, Li H, Dimasi N, McCormick JK, Martin R, Schuck P, et al. Crystal Structure of a Superantigen Bound to the High-Affinity, Zinc-Dependent Site on MHC Class II. Immunity (2001) 14:93–104. doi: 10.1016/s1074-7613(01)00092-9
61. Pless DD, Ruthel G, Reinke EK, Ulrich RG, Bavari S. Persistence of Zinc-Binding Bacterial Superantigens at the Surface of Antigen-Presenting Cells Contributes to the Extreme Potency of These Superantigens as T-Cell Activators. Infect Immun (2005) 73:5358–66. doi: 10.1128/IAI.73.9.5358-5366.2005
62. Petersson K, Hakansson M, Nilsson H, Forsberg G, Svensson LA, Liljas A, et al. Crystal Structure of a Superantigen Bound to MHC Class II Displays Zinc and Peptide Dependence. EMBO J (2001) 20:3306–12. doi: 10.1093/emboj/20.13.3306
63. Nilsson H, Bjork P, Dohlsten M, Antonsson P. Staphylococcal Enterotoxin H Displays Unique MHC Class II-Binding Properties. J Immunol (1999) 163:6686–93.
64. Hudson KR, Tiedemann RE, Urban RG, Lowe SC, Strominger JL, Fraser JD. Staphylococcal Enterotoxin A has Two Cooperative Binding Sites on Major Histocompatibility Complex Class II. J Exp Med (1995) 182:711–20. doi: 10.1084/jem.182.3.711
65. Tiedemann RE, Fraser JD. Cross-Linking of MHC Class II Molecules by Staphylococcal Enterotoxin A Is Essential for Antigen-Presenting Cell and T Cell Activation. J Immunol (1996) 157:3958–66.
66. Petersson K, Pettersson H, Skartved NJ, Walse B, Forsberg G. Staphylococcal Enterotoxin H Induces V Alpha-Specific Expansion of T Cells. J Immunol (2003) 170:4148–54. doi: 10.4049/jimmunol.170.8.4148
67. Saline M, Rodstrom KE, Fischer G, Orekhov VY, Karlsson BG, Lindkvist-Petersson K. The Structure of Superantigen Complexed With TCR and MHC Reveals Novel Insights Into Superantigenic T Cell Activation. Nat Commun (2010) 1:119. doi: 10.1038/ncomms1117
68. Pumphrey N, Vuidepot A, Jakobsen B, Forsberg G, Walse B, Lindkvist-Petersson K. Cutting Edge: Evidence of Direct TCR Alpha-Chain Interaction With Superantigen. J Immunol (2007) 179:2700–4. doi: 10.4049/jimmunol.179.5.2700
69. Krakauer T. Update on Staphylococcal Superantigen-Induced Signaling Pathways and Therapeutic Interventions. Toxins (Basel) (2013) 5:1629–54. doi: 10.3390/toxins5091629
70. Krakauer T. Differential Inhibitory Effects of Interleukin-10, Interleukin-4, and Dexamethasone on Staphylococcal Enterotoxin-Induced Cytokine Production and T Cell Activation. J Leukoc Biol (1995) 57:450–4. doi: 10.1002/jlb.57.3.450
71. Bohach GA, Fast DJ, Nelson RD, Schlievert PM. Staphylococcal and Streptococcal Pyrogenic Toxins Involved in Toxic Shock Syndrome and Related Illnesses. Crit Rev Microbiol (1990) 17:251–72. doi: 10.3109/10408419009105728
72. Marrack P, Kappler J. The Staphylococcal Enterotoxins and Their Relatives. Science (1990) 248:1066. doi: 10.1126/science.2343314
73. Miethke T, Wahl C, Heeg K, Echtenacher B, Krammer PH, Wagner H. T Cell-Mediated Lethal Shock Triggered in Mice by the Superantigen Staphylococcal Enterotoxin B: Critical Role of Tumor Necrosis Factor. J Exp Med (1992) 175:91–8. doi: 10.1084/jem.175.1.91
74. Rodstrom KE, Elbing K, Lindkvist-Petersson K. Structure of the Superantigen Staphylococcal Enterotoxin B in Complex With TCR and Peptide-MHC Demonstrates Absence of TCR-Peptide Contacts. J Immunol (2014) 193:1998–2004. doi: 10.4049/jimmunol.1401268
75. Leder L, Llera A, Lavoie PM, Lebedeva MI, Li H, Sekaly RP, et al. A Mutational Analysis of the Binding of Staphylococcal Enterotoxins B and C3 to the T Cell Receptor Beta Chain and Major Histocompatibility Complex Class II. J Exp Med (1998) 187:823–33. doi: 10.1084/jem.187.6.823
76. Kappler JW, Herman A, Clements J, Marrack P. Mutations Defining Functional Regions of the Superantigen Staphylococcal Enterotoxin B. J Exp Med (1992) 175:387–96. doi: 10.1084/jem.175.2.387
77. Silverman GJ, Sasano M, Wormsley SB. Age-Associated Changes in Binding of Human B Lymphocytes to a VH3-Restricted Unconventional Bacterial Antigen. J Immunol (1993) 151:5840–55.
78. Verwey WF. A Type-Specific Antigenic Protein Derived From the Staphylococcus. J Exp Med (1940) 71:635–44. doi: 10.1084/jem.71.5.635
79. Erntell M, Myhre EB, Kronvall G. Non-Immune IgG F(Ab’)2 Binding to Group C and G Streptococci is Mediated by Structures on Gamma Chains. Scand J Immunol (1985) 21:151–7. doi: 10.1111/j.1365-3083.1985.tb01414.x
80. Oeding P, Grov A, Myklestad B. Immunochemical Studies on Antigen Preparations From Staphylococcus Aureus. 2. Precipitating and Erythrocyte-Sensitizing Properties of Protein a (Antigen a) and Related Substances. Acta Pathol Microbiol Scand (1964) 62:117–27. doi: 10.1111/apm.1964.62.1.117
81. Myers JK, Oas TG. Preorganized Secondary Structure as an Important Determinant of Fast Protein Folding. Nat Struct Biol (2001) 8:552–8. doi: 10.1038/88626
82. Deis LN, Pemble C, Qi Y, Hagarman A, Richardson DC, Richardson JS, et al. Multiscale Conformational Heterogeneity in Staphylococcal Protein a: Possible Determinant of Functional Plasticity. Structure (2014) 22:1467–77. doi: 10.1016/j.str.2014.08.014
83. Moks T, Abrahmsen L, Nilsson B, Hellman U, Sjoquist J, Uhlen M. Staphylococcal Protein A Consists of Five IgG-Binding Domains. Eur J Biochem (1986) 156:637–43. doi: 10.1111/j.1432-1033.1986.tb09625.x
84. Jendeberg L, Nilsson P, Larsson A, Denker P, Uhlen M, Nilsson B, et al. Engineering of Fc(1) and Fc(3) From Human Immunoglobulin G to Analyse Subclass Specificity for Staphylococcal Protein a. J Immunol Methods (1997) 201:25–34. doi: 10.1016/s0022-1759(96)00215-3
85. Lua WH, Su CT, Yeo JY, Poh JJ, Ling WL, Phua SX, et al. Role of the IgE Variable Heavy Chain in FcepsilonRIalpha and Superantigen Binding in Allergy and Immunotherapy. J Allergy Clin Immunol (2019) 144:514–23.e5. doi: 10.1016/j.jaci.2019.03.028
86. Bjorck L, Kronvall G. Purification and Some Properties of Streptococcal Protein G, a Novel IgG-Binding Reagent. J Immunol (1984) 133:969–74.
87. Guss B, Eliasson M, Olsson A, Uhlen M, Frej AK, Jornvall H, et al. Structure of the IgG-Binding Regions of Streptococcal Protein G. EMBO J (1986) 5:1567–75. doi: 10.1002/j.1460-2075.1986.tb04398.x
88. Sjobring U, Bjorck L, Kastern W, Streptococcal protein G. Gene Structure and Protein Binding Properties. J Biol Chem (1991) 266:399–405. doi: 10.1016/S0021-9258(18)52448-0
89. Nilvebrant J, Hober S. The Albumin-Binding Domain as a Scaffold for Protein Engineering. Comput Struct Biotechnol J (2013) 6:e201303009. doi: 10.5936/csbj.201303009
90. Goodyear CS, Narita M, Silverman GJ. In Vivo VL-Targeted Activation-Induced Apoptotic Supraclonal Deletion by a Microbial B Cell Toxin. J Immunol (2004) 172:2870–710. doi: 10.4049/jimmunol.172.5.2870
91. Bjorck L, Protein L. A Novel Bacterial Cell Wall Protein With Affinity for Ig L Chains. J Immunol (1988) 140:1194–7.
92. Kastern W, Sjobring U, Bjorck L. Structure of Peptostreptococcal Protein L and Identification of a Repeated Immunoglobulin Light Chain-Binding Domain. J Biol Chem (1992) 267:12820–5. doi: 10.1016/S0021-9258(18)42349-6
93. Lua WH, Ling WL, Yeo JY, Poh JJ, Lane DP, Gan SK. The Effects of Antibody Engineering CH and CL in Trastuzumab and Pertuzumab Recombinant Models: Impact on Antibody Production and Antigen-Binding. Sci Rep (2018) 8:718. doi: 10.1038/s41598-017-18892-9
94. Ling WL, Lua WH, Gan SK. Sagacity in Antibody Humanization for Therapeutics, Diagnostics and Research Purposes: Considerations of Antibody Elements and Their Roles. Antib Ther (2020) 3:71–7910.
95. Kim HK, Falugi F, Missiakas DM, Schneewind O. Peptidoglycan-Linked Protein A Promotes T Cell-Dependent Antibody Expansion During Staphylococcus Aureus Infection. Proc Natl Acad Sci USA (2016) 113:5718–23. doi: 10.1073/pnas.1524267113
96. Goward CR, Scawen MD, Murphy JP, Atkinson T. Molecular Evolution of Bacterial Cell-Surface Proteins. Trends Biochem Sci (1993) 18:136–40. doi: 10.1016/0968-0004(93)90021-e
97. Schneewind O, Model P, Fischetti VA. Sorting of Protein A to the Staphylococcal Cell Wall. Cell (1992) 70:267–81. doi: 10.1016/0092-8674(92)90101-h
98. Navarre WW, Schneewind O. Surface Proteins of Gram-Positive Bacteria and Mechanisms of Their Targeting to the Cell Wall Envelope. Microbiol Mol Biol Rev (1999) 63:174–229. doi: 10.1128/MMBR.63.1.174-229.1999
99. Akerstrom B, Nielsen E, Bjorck L. Definition of IgG- and Albumin-Binding Regions of Streptococcal Protein G. J Biol Chem (1987) 262:13388–91. doi: 10.1016/S0021-9258(19)76438-2
100. Murphy JP, Duggleby CJ, Atkinson MA, Trowern AR, Atkinson T, Goward CR. The Functional Units of a Peptostreptococcal Protein L. Mol Microbiol (1994) 12:911–20. doi: 10.1111/j.1365-2958.1994.tb01079.x
101. Broker BM, Holtfreter S, Bekeredjian-Ding I. Immune Control of Staphylococcus Aureus - Regulation and Counter-Regulation of the Adaptive Immune Response. Int J Med Microbiol (2014) 304:204–14. doi: 10.1016/j.ijmm.2013.11.008
102. Goodyear CS, Silverman GJ. Death by a B Cell Superantigen: In Vivo VH-Targeted Apoptotic Supraclonal B Cell Deletion by a Staphylococcal Toxin. J Exp Med (2003) 197:1125–39. doi: 10.1084/jem.20020552
103. Shi M, Willing SE, Kim HK, Schneewind O, Missiakas D. Peptidoglycan Contribution to the B Cell Superantigen Activity of Staphylococcal Protein a. mBio (2021) 12:1–13. doi: 10.1128/mBio.00039-21
104. Ulloa-Morales AJ, Goodyear CS, Silverman GJ. Essential Domain-Dependent Roles Within Soluble IgG for In Vivo Superantigen Properties of Staphylococcal Protein A: Resolving the B-Cell Superantigen Paradox. Front Immunol (2018) 9:2011. doi: 10.3389/fimmu.2018.02011
105. Horstmann RD, Sievertsen HJ, Knobloch J, Fischetti VA. Antiphagocytic Activity of Streptococcal M Protein: Selective Binding of Complement Control Protein Factor H. Proc Natl Acad Sci USA (1988) 85:1657–61. doi: 10.1073/pnas.85.5.1657
106. Kihlberg BM, Collin M, Olsen A, Bjorck L. Protein H, an Antiphagocytic Surface Protein in Streptococcus Pyogenes. Infect Immun (1999) 67:1708–14. doi: 10.1128/IAI.67.4.1708-1714.1999
107. Tuffs SW, Haeryfar SMM, McCormick JK. Manipulation of Innate and Adaptive Immunity by Staphylococcal Superantigens. Pathogens (2018) 7:1–23. doi: 10.3390/pathogens7020053
108. Patella V, Casolaro V, Bjorck L, Marone G, Protein L. A Bacterial Ig-Binding Protein That Activates Human Basophils and Mast Cells. J Immunol (1990) 145:3054–61.
109. Genovese A, Bouvet JP, Florio G, Lamparter-Schummert B, Bjorck L, Marone G. Bacterial Immunoglobulin Superantigen Proteins A and L Activate Human Heart Mast Cells by Interacting With Immunoglobulin E. Infect Immun (2000) 68:5517–24. doi: 10.1128/iai.68.10.5517-5524.2000
110. Genovese A, Borgia G, Bjorck L, Petraroli A, de Paulis A, Piazza M, et al. Immunoglobulin Superantigen Protein L Induces IL-4 and IL-13 Secretion From Human Fc Epsilon RI+ Cells Through Interaction With the Kappa Light Chains of IgE. J Immunol (2003) 170:1854–61. doi: 10.4049/jimmunol.170.4.1854
111. Patel AH, Nowlan P, Weavers ED, Foster T. Virulence of Protein A-Deficient and Alpha-Toxin-Deficient Mutants of Staphylococcus Aureus Isolated by Allele Replacement. Infect Immun (1987) 55:3103–10. doi: 10.1128/IAI.55.12.3103-3110.1987
112. Derrick JP, Wigley DB. Crystal Structure of a Streptococcal Protein G Domain Bound to an Fab Fragment. Nature (1992) 359:752–4. doi: 10.1038/359752a0
113. Graille M, Stura EA, Corper AL, Sutton BJ, Taussig MJ, Charbonnier JB, et al. Crystal Structure of a Staphylococcus Aureus Protein A Domain Complexed With the Fab Fragment of a Human IgM Antibody: Structural Basis for Recognition of B-Cell Receptors and Superantigen Activity. Proc Natl Acad Sci USA (2000) 97:5399–404. doi: 10.1073/pnas.97.10.5399
114. Nilson BH, Solomon A, Bjorck L, Akerstrom B. Protein L From Peptostreptococcus Magnus Binds to the Kappa Light Chain Variable Domain. J Biol Chem (1992) 267:2234–9. doi: 10.1016/S0021-9258(18)45867-X
115. Housden NG, Harrison S, Housden HR, Thomas KA, Beckingham JA, Roberts SE, et al. Observation and Characterization of the Interaction Between a Single Immunoglobulin Binding Domain of Protein L and Two Equivalents of Human Kappa Light Chains. J Biol Chem (2004) 279:9370–8. doi: 10.1074/jbc.M312938200
116. Domiati-Saad R, Lipsky PE. Staphylococcal Enterotoxin A Induces Survival of VH3-Expressing Human B Cells by Binding to the VH Region With Low Affinity. J Immunol (1998) 161:1257–66.
117. Domiati-Saad R, Attrep JF, Brezinschek HP, Cherrie AH, Karp DR, Lipsky PE. Staphylococcal Enterotoxin D Functions as a Human B Cell Superantigen by Rescuing VH4-Expressing B Cells From Apoptosis. J Immunol (1996) 156:3608–20.
118. Bunker JJ, Drees C, Watson AR, Plunkett CH, Nagler CR, Schneewind O, et al. B Cell Superantigens in the Human Intestinal Microbiota. Sci Transl Med (2019) 11:1–27. doi: 10.1126/scitranslmed.aau9356
119. Jansson B, Uhlen M, Nygren PA. All Individual Domains of Staphylococcal Protein A Show Fab Binding. FEMS Immunol Med Microbiol (1998) 20:69–78. doi: 10.1111/j.1574-695X.1998.tb01112.x
120. Derrick JP, Maiden MC, Feavers IM. Crystal Structure of an Fab Fragment in Complex With a Meningococcal Serosubtype Antigen and a Protein G Domain. J Mol Biol (1999) 293:81–91. doi: 10.1006/jmbi.1999.3144
121. Derrick JP, Wigley DB. The Third IgG-Binding Domain From Streptococcal Protein G. An Analysis by X-Ray Crystallography of the Structure Alone and in a Complex With Fab. J Mol Biol (1994) 243:906–18. doi: 10.1006/jmbi.1994.1691
122. Graille M, Stura EA, Housden NG, Beckingham JA, Bottomley SP, Beale D, et al. Complex Between Peptostreptococcus Magnus Protein L and a Human Antibody Reveals Structural Convergence in the Interaction Modes of Fab Binding Proteins. Structure (2001) 9:679–87. doi: 10.1016/s0969-2126(01)00630-x
123. Beckingham JA, Housden NG, Muir NM, Bottomley SP, Gore MG. Studies on a Single Immunoglobulin-Binding Domain of Protein L From Peptostreptococcus Magnus: The Role of Tyrosine-53 in the Reaction With Human IgG. Biochem J (2001) 353:395–401. doi: 10.1042/0264-6021:3530395
124. Svensson HG, Wedemeyer WJ, Ekstrom JL, Callender DR, Kortemme T, Kim DE, et al. Contributions of Amino Acid Side Chains to the Kinetics and Thermodynamics of the Bivalent Binding of Protein L to Ig Kappa Light Chain. Biochemistry (2004) 43:2445–57. doi: 10.1021/bi034873s
125. Su CT, Ling WL, Lua WH, Poh JJ, Gan SK. The Role of Antibody Vkappa Framework 3 Region Towards Antigen Binding: Effects on Recombinant Production and Protein L Binding. Sci Rep (2017) 7:3766. doi: 10.1038/s41598-017-02756-3
126. Ling WL, Lua WH, Poh JJ, Yeo JY, Lane DP, Gan SK. Effect of VH-VL Families in Pertuzumab and Trastuzumab Recombinant Production, Her2 and FcgammaIIA Binding. Front Immunol (2018) 9:469. doi: 10.3389/fimmu.2018.00469
127. Ling WL, Su CT, Lua WH, Poh JJ, Ng YL, Wipat A, et al. Essentially Leading Antibody Production: An Investigation of Amino Acids, Myeloma, and Natural V-Region Signal Peptides in Producing Pertuzumab and Trastuzumab Variants. Front Immunol (2020) 11:604318. doi: 10.3389/fimmu.2020.604318
128. Salvalaglio M, Zamolo L, Busini V, Moscatelli D, Cavallotti C. Molecular Modeling of Protein A Affinity Chromatography. J Chromatogr A (2009) 1216:8678–86. doi: 10.1016/j.chroma.2009.04.035
129. Kim HK, Cheng AG, Kim HY, Missiakas DM, Schneewind O. Nontoxigenic Protein A Vaccine for Methicillin-Resistant Staphylococcus Aureus Infections in Mice. J Exp Med (2010) 207:1863–70. doi: 10.1084/jem.20092514
130. Stone GC, Sjobring U, Bjorck L, Sjoquist J, Barber CV, Nardella FA. The Fc Binding Site for Streptococcal Protein G Is in the C Gamma 2-C Gamma 3 Interface Region of IgG and is Related to the Sites That Bind Staphylococcal Protein A and Human Rheumatoid Factors. J Immunol (1989) 143:565–70.
131. Kato K, Lian LY, Barsukov IL, Derrick JP, Kim H, Tanaka R, et al. Model for the Complex Between Protein G and an Antibody Fc Fragment in Solution. Structure (1995) 3:79–85. doi: 10.1016/s0969-2126(01)00136-8
132. Nooren IM, Thornton JM. Diversity of Protein-Protein Interactions. EMBO J (2003) 22:3486–92. doi: 10.1093/emboj/cdg359
133. Chinh W-HL, Su T, Poh J-J, Ling W-L, Yeo JY, Gan SK. Molecular Insights of Nickel Binding to Therapeutic Antibodies as a Possible New Antibody Superantigen. Front Immunol (2021) 12:1–11. doi: 10.3389/fimmu.2021.676048
134. Grilo AL, Mantalaris A. The Increasingly Human and Profitable Monoclonal Antibody Market. Trends Biotechnol (2019) 37:9–16. doi: 10.1016/j.tibtech.2018.05.014
135. Kangwa M, Yelemane V, Ponnurangam A, Fernandez-Lahore M. An Engineered Staphylococcal Protein A Based Ligand: Production, Characterization and Potential Application for the Capture of Immunoglobulin and Fc-Fusion Proteins. Protein Expr Purif (2019) 155:27–34. doi: 10.1016/j.pep.2018.11.003
136. Inganas M. Comparison of Mechanisms of Interaction Between Protein A From Staphylococcus Aureus and Human Monoclonal IgG, IgA and IgM in Relation to the Classical FC Gamma and the Alternative F(Ab’)2 Epsilon Protein A Interactions. Scand J Immunol (1981) 13:343–52. doi: 10.1111/j.1365-3083.1981.tb00143.x
137. Ey PL, Prowse SJ, Jenkin CR. Isolation of Pure IgG1, IgG2a and IgG2b Immunoglobulins From Mouse Serum Using Protein A-Sepharose. Immunochemistry (1978) 15:429–36. doi: 10.1016/0161-5890(78)90070-6
138. Nilsson B, Moks T, Jansson B, Abrahmsen L, Elmblad A, Holmgren E, et al. A Synthetic IgG-Binding Domain Based on Staphylococcal Protein a. Protein Eng (1987) 1:107–13. doi: 10.1093/protein/1.2.107
139. Starovasnik MA, O’Connell MP, Fairbrother WJ, Kelley RF. Antibody Variable Region Binding by Staphylococcal Protein A: Thermodynamic Analysis and Location of the Fv Binding Site on E-Domain. Protein Sci (1999) 8:1423–31. doi: 10.1110/ps.8.7.1423
140. Seldon TA, Hughes KE, Munster DJ, Chin DY, Jones ML. Improved Protein-A Separation of V(H)3 Fab From Fc After Papain Digestion of Antibodies. J Biomol Tech (2011) 22:50–2.
141. Linhult M, Gulich S, Graslund T, Simon A, Karlsson M, Sjoberg A, et al. Improving the Tolerance of a Protein a Analogue to Repeated Alkaline Exposures Using a Bypass Mutagenesis Approach. Proteins (2004) 55:407–16. doi: 10.1002/prot.10616
142. Minakuchi K, Murata D, Okubo Y, Nakano Y, Yoshida S. Remarkable Alkaline Stability of an Engineered Protein A as Immunoglobulin Affinity Ligand: C Domain Having Only One Amino Acid Substitution. Protein Sci (2013) 22:1230–8. doi: 10.1002/pro.2310
143. Xia HF, Liang ZD, Wang SL, Wu PQ, Jin XH. Molecular Modification of Protein A to Improve the Elution pH and Alkali Resistance in Affinity Chromatography. Appl Biochem Biotechnol (2014) 172:4002–12. doi: 10.1007/s12010-014-0818-1
144. Gulich S, Linhult M, Stahl S, Hober S. Engineering Streptococcal Protein G for Increased Alkaline Stability. Protein Eng (2002) 15:835–42. doi: 10.1093/protein/15.10.835
145. Palmer B, Angus K, Taylor L, Warwicker J, Derrick JP. Design of Stability at Extreme Alkaline pH in Streptococcal Protein G. J Biotechnol (2008) 134:222–30. doi: 10.1016/j.jbiotec.2007.12.009
146. Imura Y, Tagawa T, Miyamoto Y, Nonoyama S, Sumichika H, Fujino Y, et al. Washing With Alkaline Solutions in Protein A Purification Improves Physicochemical Properties of Monoclonal Antibodies. Sci Rep (2021) 11:1827. doi: 10.1038/s41598-021-81366-6
147. Akerstrom B, Nilson BH, Hoogenboom HR, Bjorck L. On the Interaction Between Single Chain Fv Antibodies and Bacterial Immunoglobulin-Binding Proteins. J Immunol Methods (1994) 177:151–63. doi: 10.1016/0022-1759(94)90152-x
148. Nielsen K, Smith P, Yu W, Nicoletti P, Elzer P, Vigliocco A, et al. Enzyme Immunoassay for the Diagnosis of Brucellosis: Chimeric Protein A-Protein G as a Common Enzyme Labeled Detection Reagent for Sera for Different Animal Species. Vet Microbiol (2004) 101:123–9. doi: 10.1016/j.vetmic.2004.02.014
149. Urmann K, Reich P, Walter JG, Beckmann D, Segal E, Scheper T. Rapid and Label-Free Detection of Protein a by Aptamer-Tethered Porous Silicon Nanostructures. J Biotechnol (2017) 257:171–7. doi: 10.1016/j.jbiotec.2017.01.005
150. Yue H, Zhou Y, Wang P, Wang X, Wang Z, Wang L, et al. A Facile Label-Free Electrochemiluminescent Biosensor for Specific Detection of Staphylococcus Aureus Utilizing the Binding Between Immunoglobulin G and Protein a. Talanta (2016) 153:401–6. doi: 10.1016/j.talanta.2016.03.043
151. Ling WL, Ng YL, Wipat A, Lane DP, Gan SK. The Quantification of Antibody Elements and Receptors Subunit Expression Using qPCR: The Design of VH, VL, CH, CL, FcR Subunits Primers for a More Holistic View of the Immune System. J Immunol Methods (2020) 476:112683. doi: 10.1016/j.jim.2019.112683
152. Ng K, Liang W, Liew A, Yeo Y-H, Lua J, Qian W-H, et al. Republication – APD SpectBT: Arduino-Based Mobile Vis-Spectrophotometer. Sci Phone Apps Mobile Device (2019) 5:1–4. doi: 10.30943/2019/s23122019
153. Poh J-JG, Tan N, Gan S, Ken-En S. Spectrophotometer on-the-Go: The Development of a 2-in-1 UV–Vis Portable Arduino-Based Spectrophotometer. Sensors Actuators A: Phys (2021) 325:1–8. doi: 10.1016/j.sna.2021.112698
154. Salgado-Pabon W, Breshears L, Spaulding AR, Merriman JA, Stach CS, Horswill AR, et al. Superantigens are Critical for Staphylococcus Aureus Infective Endocarditis, Sepsis, and Acute Kidney Injury. mBio (2013) 4:1–9. doi: 10.1128/mBio.00494-13
155. Bashraheel SS, AlQahtani AD, Rashidi FB, Al-Sulaiti H, Domling A, Orie NN, et al. Studies on Vascular Response to Full Superantigens and Superantigen Derived Peptides: Possible Production of Novel Superantigen Variants With Less Vasodilation Effect for Tolerable Cancer Immunotherapy. BioMed Pharmacother (2019) 115:108905. doi: 10.1016/j.biopha.2019.108905
156. Yang Y, Qian M, Yi S, Liu S, Li B, Yu R, et al. Monoclonal Antibody Targeting Staphylococcus Aureus Surface Protein A (SasA) Protect Against Staphylococcus Aureus Sepsis and Peritonitis in Mice. PloS One (2016) 11:e0149460. doi: 10.1371/journal.pone.0149460
157. Varshney AK, Kuzmicheva GA, Lin J, Sunley KM, Bowling RA Jr, Kwan TY, et al. A Natural Human Monoclonal Antibody Targeting Staphylococcus Protein A Protects Against Staphylococcus Aureus Bacteremia. PloS One (2018) 13:e0190537. doi: 10.1371/journal.pone.0190537
158. Forsberg G, Ohlsson L, Brodin T, Bjork P, Lando PA, Shaw D, et al. Therapy of Human Non-Small-Cell Lung Carcinoma Using Antibody Targeting of a Modified Superantigen. Br J Cancer (2001) 85:129–36. doi: 10.1054/bjoc.2001.1891
159. von Scheidt B, Wang M, Oliver AJ, Chan JD, Jana MK, Ali AI, et al. Enterotoxins Can Support CAR T Cells Against Solid Tumors. Proc Natl Acad Sci USA (2019) 116:25229–35. doi: 10.1073/pnas.1904618116
160. Lorenzo D, Duarte A, Mundinano J, Berguer P, Nepomnaschy I, Piazzon I. A B-Cell Superantigen Induces the Apoptosis of Murine and Human Malignant B Cells. PloS One (2016) 11:e0162456. doi: 10.1371/journal.pone.0162456
161. Chen JY. Superantigens, Superantigen-Like Proteins and Superantigen Derivatives for Cancer Treatment. Eur Rev Med Pharmacol Sci (2021) 25:1622–30. doi: 10.26355/eurrev_202102_24873
162. Heidary MF, Mahmoodzadeh Hosseini H, Mehdizadeh Aghdam E, Nourani MR, Ranjbar R, Mirnejad R, et al. Overexpression of Metastatic Related MicroRNAs, Mir-335 and Mir-10b, by Staphylococcal Enterotoxin B in the Metastatic Breast Cancer Cell Line. Adv Pharm Bull (2015) 5:255–9. doi: 10.15171/apb.2015.035
163. Golob-Urbanc A, Rajcevic U, Strmsek Z, Jerala R. Design of Split Superantigen Fusion Proteins for Cancer Immunotherapy. J Biol Chem (2019) 294:6294–305. doi: 10.1074/jbc.RA118.006742
164. He Y, Sun Y, Ren Y, Qiao L, Guo R, Du J, et al. The T Cell Activating Properties and Antitumour Activity of Staphylococcal Enterotoxin-Like Q. Med Microbiol Immunol (2019) 208:781–92. doi: 10.1007/s00430-019-00614-9
165. Brown M, Bhardwaj N. Super(antigen) Target for SARS-CoV-2. Nat Rev Immunol (2021) 21:72. doi: 10.1038/s41577-021-00502-5
166. Cheng MH, Zhang S, Porritt RA, Noval Rivas M, Paschold L, Willscher E, et al. Superantigenic Character of an Insert Unique to SARS-CoV-2 Spike Supported by Skewed TCR Repertoire in Patients With Hyperinflammation. Proc Natl Acad Sci USA (2020) 117:25254–62. doi: 10.1073/pnas.2010722117
Keywords: superantigen, T-cell, B-cell, cytokine storm, interface, antibody purification
Citation: Deacy AM, Gan SK-E and Derrick JP (2021) Superantigen Recognition and Interactions: Functions, Mechanisms and Applications. Front. Immunol. 12:731845. doi: 10.3389/fimmu.2021.731845
Received: 28 June 2021; Accepted: 30 August 2021;
Published: 20 September 2021.
Edited by:
Ester Boix, Universitat Autònoma de Barcelona, SpainReviewed by:
Marco Pio La Manna, University of Palermo, ItalyCopyright © 2021 Deacy, Gan and Derrick. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jeremy P. Derrick, amVyZW15LmRlcnJpY2tAbWFuY2hlc3Rlci5hYy51aw==; Samuel Ken-En Gan, c2FtZ2FuQGFwZHNrZWcuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.