
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol., 06 September 2021
Sec. Microbial Immunology
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.728742
This article is part of the Research TopicStrategies Played by Immune Cells and Mycobacteria in the Battle between Antimicrobial Activity and Bacterial SurvivalView all 13 articles
Autophagy is critically involved in host defense pathways through targeting and elimination of numerous pathogens via autophagic machinery. Nontuberculous mycobacteria (NTMs) are ubiquitous microbes, have become increasingly prevalent, and are emerging as clinically important strains due to drug-resistant issues. Compared to Mycobacterium tuberculosis (Mtb), the causal pathogen for human tuberculosis, the roles of autophagy remain largely uncharacterized in the context of a variety of NTM infections. Compelling evidence suggests that host autophagy activation plays an essential role in the enhancement of antimicrobial immune responses and controlling pathological inflammation against various NTM infections. As similar to Mtb, it is believed that NTM bacteria evolve multiple strategies to manipulate and hijack host autophagy pathways. Despite this, we are just beginning to understand the molecular mechanisms underlying the crosstalk between pathogen and the host autophagy system in a battle with NTM bacteria. In this review, we will explore the function of autophagy, which is involved in shaping host–pathogen interaction and disease outcomes during NTM infections. These efforts will lead to the development of autophagy-based host-directed therapeutics against NTM infection.
Around 200 species of nontuberculous mycobacteria (NTMs) have been identified as the causal pathogens of pulmonary and ulcerative human diseases in both immunocompromised and immunocompetent subjects. The Mycobacterium avium complex (MAC) group including M. intracellulare, M. avium subsp. hominissuis, and M. intracellulare subsp. chimaera are the most common causes of NTM pulmonary diseases (NTM-PD), which are more emerging (1–3). Mycobacteroides abscessus (Mabc) is another frequently encountered pathogen that causes NTM-PD (4–6). The prevalence and incidence of NTM infections are increasing worldwide, and the risk of antibiotics resistance is often challenging and complex in the treatment of NTM diseases (7). Despite this, we have a lack of understanding of the virulence factors and host–pathogen interactions in terms of NTM infection.
Autophagy is an intracellular process for the maintenance of homeostasis upon stress conditions through lysosomal degradation of cytoplasmic cargos (8, 9). During a variety of infections, autophagy plays a cell-autonomous and/or a non-autonomous function to protect the hosts from infectious hazards and harmful inflammation (10). Recent studies highlighted multiple layered crosstalks of autophagy with several important processes, including innate immunity, immunometabolism, and mitochondrial function, to prevent harmful inflammation and to augment host protective function (10, 11). Therefore, autophagy-activating strategies are becoming promising not only for the development of host-directed therapeutics but also for the design of potential vaccines against mycobacterial infection (3). However, intracellular pathogens are able to develop sophisticated strategies of exploitation and subvert autophagy in order to enhance their survival in the host cells (12–14). Compared with Mycobacterium tuberculosis (Mtb), an extensively studied pathogen, much less is known about the function of autophagy pathways against NTM infection. In addition, the individual picture of NTM interaction with host autophagy machinery and how each NTM escapes from host autophagic responses remain uncharacterized. In this review, we focus on the recent progress of our understanding of autophagy functions in the context of host defense against NTM infections.
NTM bacteria are diverse species that grow in the environment and are opportunistic pathogens that cause a wide spectrum of diseases in humans. The prevalence, morbidity, and mortality of NTM diseases are increasing worldwide, particularly in developed countries, associated with several predisposing factors such as aging, immunosuppressive therapy or conditions, and relevant comorbidity with chronic pulmonary diseases (15–21). NTM-PD is common in immunocompetent persons, whereas immunocompromised patients primarily suffer from disseminated diseases (16, 18). The most important human pathogens causing NTM-PD include MAC, Mabc, and Mycobacterium kansasii. In addition, the infections caused by NTMs also vary by geographic distribution (22, 23). NTM-PD also can be organized into clinical phenotypes (24). For example, “Lady Windermere’s syndrome” usually occurs in elderly females with a fibronodular radiographic pattern of NTM-PD (25, 26). Besides NTM-PD, NTM causes extrapulmonary diseases, including skin and soft-tissue infections, musculoskeletal infections, lymphadenitis, and disseminated disease (27, 28). Importantly, NTM treatment is often toxic and difficult because of intrinsic multidrug resistance, limited treatment options, and lengthy duration (24, 29, 30).
After infection, NTMs are found in different types of cells but extensively studied in macrophages as the primary host cells where a vast number of NTMs are able to arrest phagosomal maturation and persist, form biofilms, and even replicate (31–34). Thus, innate immune signaling activated by numerous pathogen-associated molecular patterns may contribute to host immune defense against NTM infection (32). Although the exact nature of host protective factors is uncertain, it has been long thought that T helper 1 (Th1) responses induced by interferon (IFN)-γ and interleukin (IL)-12 are crucial in the defense against NTM infection (31, 35). In addition, several genetic factors, including cystic fibrosis transmembrane conductance regulator mutations, vitamin D receptors, and polymorphisms of solute carrier 11A1 (or natural resistance-associated macrophage protein 1), are associated with NTM-PD (35, 36), although these are not specific to NTM infections. Moreover, anti-tumor necrosis factor (TNF)-α therapy during autoimmune diseases may lead to the increased risk of NTM diseases as well as tuberculosis, suggesting that TNF-α is also crucial for host defense against NTM infection (37, 38). Recent studies highlight the function of autophagy and apoptosis as another key factor for controlling mycobacteria (1, 3). In this review, we primarily discuss the current understanding of host cell autophagy in terms of host defense and controlling immunopathology during NTM infection.
Although this session covers a general understanding of the autophagy/xenophagy pathways and their interaction with intracellular Mtb, much uncertainty remains on the specific function of autophagy in the context of each NTM infection. In Mtb infection, there are at least three types of autophagy pathways participating in the antibacterial host defense (39). Xenophagy involves the cytoplasmic escape of Mtb through the ESAT-6 secretion (ESX)-1 system, thereby being subjected to ubiquitination system and recognized by selective cargo receptors, i.e., p62 and NDP52, for lysosomal degradation (40). Although the ESX-1 system is required for early autophagy induction, it functions in a late inhibition of autophagy flux in human primary dendritic cells (41). Xenophagy involves the core autophagy-related genes (Atg), including ULK1, Atg14, Beclin-1, and Atg5-12, which are important in the initiation of autophagosome formation and elongation step of autophagy (42). Another type of noncanonical autophagy, LC3-associated phagocytosis (LAP), involves Rubicon, NADPH oxidase 2, Beclin-1, and Atg5-12, which is also crucial for combating intracellular Mtb, which resists this process through its own effector, the LCP protein CpsA (43). In recent years, we have made considerable progress in revealing the signaling pathways that regulate xenophagy against Mtb infection. The cytosolic DNA sensor cyclic GMP-AMP synthase (cGAS)-STING signaling pathway is critically required to recognize cytosolic Mtb DNA to induce autophagy (44). A recent study showed that xenophagy could be triggered by the direct ubiquitination of Mtb surface protein Rv1468c, which contains a eukaryotic-like ubiquitin-associated domain (45). In addition, xenophagic clearance of Mtb is mediated by various E3 ubiquitin ligases, including PARK2 (46), Smurf1 via K48-linked ubiquitination (47), and TRIM16 through interaction with galectin-3 (48). Moreover, the lysosomal damage recognized by galectin-8 and -9 signaling promotes autophagy and antimicrobial responses against Mtb infection (49). However, it is largely unclear whether these or other signaling pathways are involved in regulating autophagy defense against NTM bacteria, which may operate different strategies compared to Mtb to survive within host cells.
So far, numerous autophagy-activating agents/drugs have been reported to enhance the activation of autophagy and phagosomal maturation through colocalization of bacterial phagosomes with autophagosomes/lysosomes (50–52). Accumulating evidence suggests that a wide range of antimicrobial strategies can be applied to promote antimicrobial activities for infectious diseases through autophagy modulation (50–53). These strategies include multiple biological pathways such as targeting selective autophagy through adaptors, regulation of posttranslational modification of key proteins, modulation of inflammatory responses, etc. (10, 52, 53).
Earlier studies showed that both Mycolicibacterium smegmatis and Mycolicibacterium fortuitum exhibit strong autophagy induction, whereas M. kansasii induces less induction of autophagy in macrophages (54). In addition, autophagy induced by M. smegmatis is independent of mTOR activity, and lipid components of M. smegmatis activate mTOR signaling (54). Because mTOR inhibition by rapamycin results in decreasing intracellular bacterial burden (55), simultaneous activation of both autophagy and mTOR signaling might be another immune escaping strategy manipulated by bacteria. Mabc smooth (Mabc S) variant exhibits pathogenesis mainly through the suppression of phagosomal maturation and induction of phagosome-cytosol communications. However, the Mabc rough (Mabc R) variant enhances autophagy and apoptosis and can form extracellular cords, thereby evading phagocytosis (56, 57). However, it remains largely unknown how various NTM microbes induce or suppress host cell autophagy in different tissues/cells and whether it exerts to modify host defensive system during infection. In the next session, we discuss the recent advances and perspectives on the roles of host cell autophagy in the context of infection with each NTM pathogen and explore in brief the potential autophagy-activating strategies against NTM infections.
M. avium complex (MAC), among other NTMs, is the most commonly isolated species in the world (58). M. avium infection leads to the increase in numerous microRNAs, including miR-125a-5p that is required for autophagy activation and suppression of intracellular survival of M. avium in macrophages (59). MiR-125a-5p-mediated autophagy activation is induced by targeting of signal transducers and activators of transcription 3 (STAT3) in macrophages (59).
Alpha-1-antitrypsin (AAT) deficiency is closely related to the increased risk of emphysema and bronchiectasis (60), which are important predisposing factors for NTM-PD (36). Previous studies reported that AAT treatment results in the control of intracellular growth of M. abscessus and M. intracellulare in human macrophages (61, 62). Interestingly, human primary monocyte-derived macrophage culture with plasma obtained from patients with post-AAT infusion significantly increases the autophagosome formation during M. intracellulare infection (61). These studies may provide potential clinical significance because AAT-based, autophagy-related, adjunctive therapy could be beneficial for treatment of NTM-PD patients who have underlying diseases such as bronchiectasis along with AAT deficiency.
Mabc is the rapidly growing NTM strain and unique in the characteristics for survival inside macrophages (56). Mabc is classified into two morphotypes, i.e., S and R forms, depending on the presence of glycopeptidolipids (GPL) (56, 63–65). Mabc ESX-4 locus that encodes an ESX-4 type VII secretion system is crucial for the growth and survival within host cells through blockade of phagosomal acidification and the ability to damage phagosomes (66).
Mabc S variants reside within more intact phagosomes and are surrounded by an electron translucent zone (ETZ), whereas Mabc R variants possess a loose phagosomal membrane and lack ETZ (63). Thus, it is thought that Mabc S strains are capable of successful phagosome-cytosol communication and are more resistant to phagosomal acidification. In addition, the nature of Mabc S to favor phagosome-cytosol communication is associated with less induction of autophagy and apoptosis than those by Mabc R morphotype (63). In accordance with this, Mabc S infection of macrophages upregulates the LC3-II and p62 levels in a time-dependent manner, suggesting that Mabc S inhibits autophagic flux (67). Earlier studies suggest that the use of antibiotics such as azithromycin aggravates the impairment of autophagy during Mabc infection, thus predisposing patients with cystic fibrosis to NTM infections. Mechanistically, long-term use of macrolide drug azithromycin results in an inhibition of intracellular clearance of Mabc in human macrophages, at least due to defective autophagy and prevention of lysosomal acidification of NTM bacteria (68). Furthermore, the virulent clinical strain UC22 of Mabc, the R variant, robustly inhibits autophagic flux, thereby escaping from the clearance by host defense (69).
However, recent reports showed that treatment of the autophagy inhibitor and activator (chloroquine and rapamycin, respectively) does not affect antimycobacterial effects against Mabc R and S infection in neutrophils (70). These data suggest that autophagy is not critically involved in neutrophil antimicrobial pathways against Mabc infection. Future studies are warranted to discover the exact roles and mechanisms by which autophagy activation regulates the virulence or protective responses in different cell types and tissues during infection with Mabc and their related strains.
M. marinum is a natural pathogen of ectotherms to cause systemic tuberculosis-like disease and is widely used as a model organism of Mtb (71–73). M. marinum usually grows at 25 to 35°C and causes extrapulmonary infections at cooler surfaces like skin in humans (71, 73). The genomes of Mtb and M. marinum are closely related at a high degree of homology and share amino acid identity averages of 85% (72, 74).
A microscopic imaging approach through zebrafish injection of mycobacteria has lighted on tracking an in vivo autophagic process related to Mtb and NTM infectious diseases (75). Earlier studies reported that M. marinum, a model NTM for tuberculosis-like disease in zebrafish, can induce autophagosome formation via the ESX-1 secretion system but simultaneously actively block the autophagic flux to escape from xenophagic degradation during infection (76, 77). In M. marinum–infected macrophages, phagosomal escape and bacterial ubiquitination are followed by targeting the lysosome-like organelle through the autophagy-independent pathway, which does not involve atg5 or LC3 association (78). Another study showed that M. marinum mimG, an orthologue of Mtb Rv3242c that contains phosphoribosyltransferase, enhances intracellular bacterial survival and virulence in zebrafish. M. marinum mimG-induced pathogenesis is at least partly mediated due to the inhibition of autophagy in macrophages (79). Future studies are warranted to identify other bacterial effectors that alter host cell autophagy to exert immune evasion during infection. These efforts will facilitate the presentation of attractive targets for potential host-directed drug therapies during NTM infection.
M. marinum is targeted by selective autophagy through autophagic adaptors optineurin and p62/SQSTM1 for bacterial clearance (80). DNA damage regulated autophagy modulator 1 (DRAM1), a critical regulator of autophagy and cell death, is activated by Toll-like receptor signaling and plays an essential function in selective autophagic defense against M. marinum infection (81). The selective autophagy activation by DRAM1 is mediated through cytosolic DNA sensor STING and the adaptor p62/SQSTM1 (81). Indeed, DRAM1 functions through autophagic targeting and phagosomal maturation of M. marinum, thereby restricting bacteria during the early phase of infection. Dissemination of M. marinum infection is associated with defective autophagy and gasdermin Eb-mediated pyroptotic cell death in dram1 mutant zebrafish larvae (82). However, it remains to be characterized whether DRAM1 plays a crucial role in host defense to other NTM strains through activation of autophagy and prevention of cell death.
Moreover, M. marinum infection of microglial cells induces autophagy that can limit the intracellular replication of M. marinum (83). Notably, rapamycin-induced autophagy activation inhibits the intracellular survival of M. marinum, suggesting the role of autophagy in microglial defense against M. marinum (83). Because M. marinum is genetically closely related to Mtb in a high degree of homology (74), autophagy activation may provide a new strategy for the treatment of tubercular meningitis.
Drosophila melanogaster is another model host for M. marinum and is widely used for innate immune defense and xenophagy during mycobacterial infection (46, 84–86). In the Drosophila model, autophagy-related gene Atg2 is required to inhibit intracellular mycobacterial growth and lipid droplets in phagocytes without changing bulk autophagy during M. marinum infection (85). By using atg7 mutant Drosophila, autophagy activation in vivo was found to contribute to antibiotic-mediated antimicrobial effects during M. marinum infection (87). Using unicellular eukaryote Dictyostelium discoideum, another model host for M. marinum, transcriptome analysis identified that M. marinum induces transcriptional activation of autophagy genes and endosomal sorting complexes required for transport (ESCRT) (88). Another study showed that the Mycobacterium-containing vacuole (MCV) damage induced by the ESX-1 system of M. marinum is recognized and repaired by the ESCRT component Tsg101, thereby leading to the containment of M. marinum in an intact compartment (89). In this process, autophagy and ESCRT pathways function in separate membrane repair processes in parallel for the restriction of mycobacterial growth in the cytosols during M. marinum infection (89). However, it is yet to be elucidated how ESCRT components are recruited to vacuolar damage sites in D. discoideum and whether ubiquitination system is involved in the ESCRT recruitment for membrane repair.
Nonpathogenic M. smegmatis is well known to induce autophagy in macrophages through the upregulation of several autophagy-related genes and TLR2 activation. However, autophagy targeting of M. smegmatis is not dependent on membrane damage and ubiquitination of bacteria (90). In addition, a high dose of rapamycin treatment leads to antimicrobial activities to M. smegmatis, presumably due to autophagy-independent modality, because bacterial growth is also inhibited in autophagy-deficient macrophages (91). Thus, there might be an alternative mechanism by which the autophagy pathway is functional in the recognition of mycobacteria to enhance phagosomal maturation and antimicrobial responses during infection.
M. smegmatis infection of PC12 and C17.2 cells induces neural differentiation through an autophagy-independent pathway via IFN-γ and PI3K-Akt signaling pathways (92). Vitamin D3, known as a protective factor for human tuberculosis, increases intracellular M. smegmatis clearance and restricts host cell cytotoxicity (93). Because vitamin D3 induces the activation of antibacterial autophagy and cathelicidin to inhibit intracellular Mtb survival (94), vitamin D3-mediated M. smegmatis clearance is presumably mediated through autophagy and antimicrobial proteins. Future studies are warranted to clarify the roles of autophagy in antimicrobial host defense against M. smegmatis infection.
Buruli ulcer, the third most common mycobacterial disease and destructive necrotizing skin infection caused by M. ulcerans, is common in West and Central Africa and becoming increasingly common in southeastern Australia (95, 96). Several studies have highlighted the genetic susceptibility of M. ulcerans infection in the context of autophagy. Recent genetic studies showed the protective effect of the minor allele G of ATG16L1 (rs2241880) from the ulcer phenotype in Buruli ulcer (97, 98). In addition, several autophagy genes, including PRKN, NOD2, and ATG16L1, are related to susceptibility to severe Buruli ulcer (98). Importantly, the missense variant T300A (rs2241880) of the ATG16L1 gene is associated with the development of Buruli ulcer (98). A mechanistic study showed that knock-in mice (Atg16L1T316A) harboring the human ATG16L1 variant (T300A) functions in a decrease in bacterial autophagy, thereby protective to Citrobacter rodentium infection through type I interferon response, similar to hypomorphic ATG16L1 mice (99). However, it is still unclear whether a certain allele of ATG16L1 (T300A) functions in the suppression of autophagy against M. ulcerans to confer host protection against Buruli ulcer.
A proteomics study showed that mycolactone, the potent exotoxin of M. ulcerans, significantly increases autophagosome formation and protein ubiquitination (100). These data strongly suggest that toxin affects host cell homeostasis, although the molecular mechanisms underlying these phenomena have not been elucidated. A genome-wide association study (GWAS) identified two variants in LncRNA genes (rs9814705 and rs76647377) in association with Buruli ulcer (97), suggesting the potential roles for LncRNAs in the pathogenesis of Buruli ulcer. Given the findings that the expression of long intergenic noncoding RNA erythroid prosurvival (lincRNA-EPS) is downregulated in primary monocytes from patients with active pulmonary tuberculosis and silencing of lincRNA-EPS enhances autophagy in macrophages during bacillus Calmette-Guérin (BCG) infection (101, 102), the LncRNA variant may play a role in the autophagy activation to modulate antimicrobial responses during Buruli ulcer further. Future studies are warranted to determine the exact role of autophagy and its related function of the identified variants of LncRNAs in Buruli ulcer. Such an effort will facilitate the development of new strategies against Buruli ulcer.
M. terrae is a member of the M. terrae complex and slow-growing NTM and can cause antibiotic-resistant debilitating diseases, including tenosynovitis and pulmonary disease (103). Interestingly, IL-17A and IL-17F are capable of activating autophagosome formation and autophagic flux, thereby restricting the intracellular growth of M. terrae in RAW264.7 cells (104). The autophagic responses during several NTM infections are summarized in Table 1.
Because different mycobacterial species have distinct virulence mechanisms for their pathogenesis, numerous NTMs and Mtb may possess differential activities and strategies to regulate host autophagy. Although there remain many gaps in the knowledge to address differential regulation of various NTMs as well as Mtb in the host defensive pathways, a recent finding reported differential immune and autophagic responses induced by Mtb and four different NTMs (Mabc, M. smegmatis, M. intracellulare, and M. avium) in human THP-1 cells (105). Compared to the autophagy-inducing activities by M. smegmatis and Mabc, the levels of autophagy induction are less in the infection with MAC and Mtb (105). Another study in RAW264.7 cells also has shown that autophagy induction by mycobacteria differs in magnitude among several species, including Mtb, BCG, and NTM (M. smegmatis, M. foruitum, and M. kansasii); and autophagy induction was minimal with the M. kansasii infection (54). Although a study suggested that long incubation of M. kansasii with Rapamycin could reduce the growth rate of bacteria (91), the exact role of autophagy in the regulation of M. kansasii infection is yet to be identified. A better understanding of the differential activities that regulate host autophagy pathways could offer a new insight for controlling a variety of mycobacterial infections.
Several reports have highlighted the antimicrobial roles of autophagy-activating agents against NTM. Recent studies showed that trehalose treatment results in the activation of the xenophagic flux to inhibit intracellular bacterial survival of various NTM strains as well as Mtb. Importantly, trehalose-mediated autophagy promotes the eradication of intracellular Mtb or NTMs, even in the status with co-infection with human immunodeficiency virus (HIV) (106). Trehalose-induced autophagy is mediated through the activation of TFEB, the key transcriptional factor for autophagy and lysosomal biogenesis (107), in macrophages (106). In addition, the autophagy induction by trehalose is dependent on lysosomal calcium release via MCOLN1 (106). These findings are corroborative with our recent data showing that the activation of nuclear receptor peroxisome proliferator-activated receptor α (PPARα) by gemfibrozil suppresses in vitro and in vivo bacterial growth of Mabc through TFEB activation (67). During Mabc infection, PPARα activation promotes nuclear translocation of TFEB and colocalization of bacterial phagosomes with lysosomes in macrophages (67).
Autophagy activation by vitamin D treatment induces autophagy to facilitate antimicrobial function through CAMP production in macrophages infected with M. marinum (108). In addition, the blockade of glycolysis by inhibitors such as 2-deoxyglucose (2-DG) prior to infection inhibits the proliferation of M. marinum in macrophages and zebrafishes (109). Thiostrepton (TSR) is an antibiotic harboring a quinaldic acid (QA) moiety that targets bacterial ribosome and induces ER stress-mediated autophagy to promote antimicrobial host defense during M. marinum infection (110). Similarly, rifampicin and amikacin also have antimicrobial activities against M. marinum in Drosophila melanogaster through the activation of the autophagic flux (87). Ohmyungsamycins, the cyclic peptides harboring autophagy activity, have antimicrobial activities against M. marinum in D. melanogaster (84). Moreover, autophagy activation by rapamycin exhibits a defense against M. marinum in microglial cells (83). These data strongly suggest that several antibiotics exhibit both direct antimicrobial and indirect host-targeting ability to enhance their effects to eliminate intracellular NTM strains. Future studies are warranted to clarify the dual mode of actions mediated by several drugs that possess potential host defense activities.
Through selective targeting intracellular pathogens, the autophagy pathway functions in the activation of antimicrobial responses, regulation of immunologic balance, and anti-inflammatory effects during infection (42, 111). Recent studies showed that the NTM-PD patients with Mabc or Mycobacteroides abscessus subsp. massiliense have pathological inflammatory responses in their peripheral blood mononuclear cells (112). In addition, resveratrol, an agonist of sirtuin 1 and 3 (113, 114), exerts a beneficial role through controlling excessive inflammation and mitochondrial homeostasis upon Mabc infection in vivo (115). Combined with resveratrol-induced antibacterial autophagy effects during Mtb infection (116), these data strongly suggest that autophagy-activating agents provide potential candidates for host-directed therapeutics during NTM infection. Antimicrobial responses of autophagy-activating exogenous agents against various NTM infections are summarized in Table 2.
Additionally, BCG vaccination currently in use for immunization against Mtb could be exploited against NTM (117, 118). However, BCG vaccine interference by NTM mycobacterial species is thought to be a potential cause of its reduced efficacy against Mtb (119). In addition, several vaccine candidates with autophagy activation as a major element have been tested against Mtb in animal models (120, 121), but there are currently no recommended vaccine protocols established to study the vaccine efficacy against NTM infections. Using autophagy-related strategies to develop effective vaccinations against NTM could be a huge advance in the fight against NTM infections (3). A schematic representation of the autophagy process during several NTM infections is shown in Figure 1.
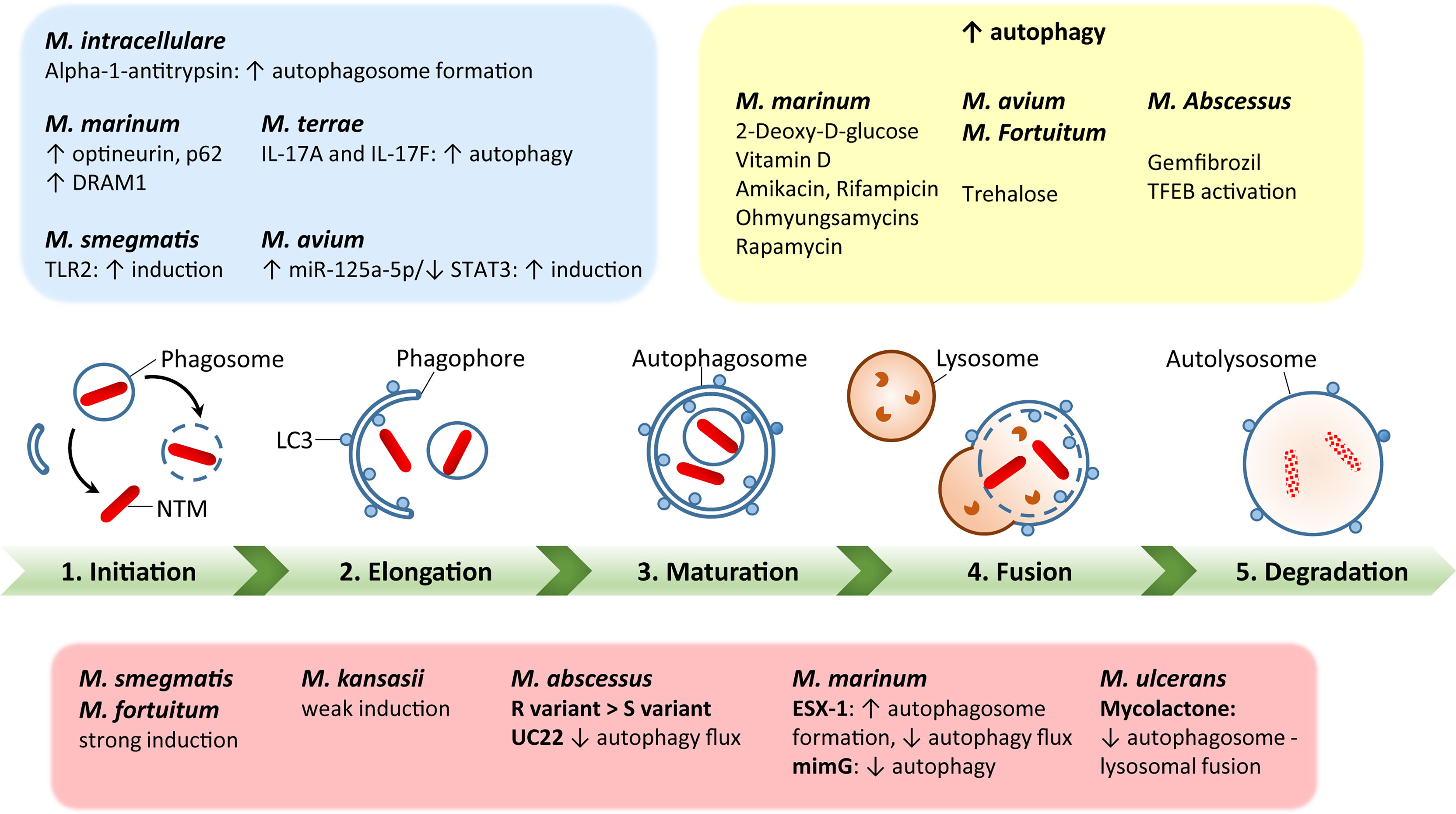
Figure 1 A schematic diagram of autophagy regulation by each nontuberculous mycobacterial infection. Green bar depicts the steps of autophagy, including initiation, elongation, maturation, fusion with lysosomes, and degradation. The red box summarizes each NTM response to regulate host autophagy processes. The blue box represents host factors to regulate autophagy during NTM infection. The yellow box shows the effects of autophagy-regulating agents for modulation of autophagy in the context of each NTM infection. The detailed mechanisms have been described in the text.
Compared with the autophagy activation against Mtb infection, it remains to be largely uncharacterized in the roles for host autophagy/xenophagy in the context of infection caused by a variety of NTM bacteria. However, autophagy modulation seems to be a potential pathway to provide novel adjunctive therapeutics based on autophagy against various NTM infections. Future studies are warranted to understand differential roles for autophagy to regulate a complex layer of host–pathogen interaction during NTM infection.
Current knowledge is very limited on how various NTMs circumvent the autophagy process during infection. Future studies are warranted to elucidate the mechanisms by which each NTM strain induces and/or manipulates the host autophagy signaling pathway during pulmonary or extrapulmonary manifestation. Such an effort to understand autophagy functions upon NTM infection will advance the development of potential host-directed therapeutics against NTM infection.
E-KJ: designed. E-KJ, PS, and ISK: wrote and reviewed the manuscript. PS: summarized the tables. ISK: drew the figure. All authors contributed to the article and approved the submitted version.
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2017R1A5A2015385) and a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI 20C0017).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We are indebted to current and past members of our Medical Research Center (i-MRC) for discussions and investigations that contributed to this article. We apologize to colleagues whose work and publications could not be cited due to space limitations.
1. Kilinc G, Saris A, Ottenhoff THM, Haks MC. Host-Directed Therapy to Combat Mycobacterial Infections. Immunol Rev (2021) 301(1):62–83. doi: 10.1111/imr.12951
2. Crilly NP, Ayeh SK, Karakousis PC. The New Frontier of Host-Directed Therapies for Mycobacterium Avium Complex. Front Immunol (2020) 11:623119. doi: 10.3389/fimmu.2020.623119
3. Strong EJ, Lee S. Targeting Autophagy as a Strategy for Developing New Vaccines and Host-Directed Therapeutics Against Mycobacteria. Front Microbiol (2020) 11:614313. doi: 10.3389/fmicb.2020.614313
4. Johansen MD, Herrmann JL, Kremer L. Non-Tuberculous Mycobacteria and the Rise of Mycobacterium Abscessus. Nat Rev Microbiol (2020) 18(7):392–407. doi: 10.1038/s41579-020-0331-1
5. van Ingen J, Wagner D, Gallagher J, Morimoto K, Lange C, Haworth CS, et al. Poor Adherence to Management Guidelines in Nontuberculous Mycobacterial Pulmonary Diseases. Eur Respir J (2017) 49(2):1601855. doi: 10.1183/13993003.01855-2016
6. Park IK, Olivier KN. Nontuberculous Mycobacteria in Cystic Fibrosis and non-Cystic Fibrosis Bronchiectasis. Semin Respir Crit Care Med (2015) 36(2):217–24. doi: 10.1055/s-0035-1546751
7. Falkinham JO 3rd. Challenges of NTM Drug Development. Front Microbiol (2018) 9:1613. doi: 10.3389/fmicb.2018.01613
8. Deretic V, Saitoh T, Akira S. Autophagy in Infection, Inflammation and Immunity. Nat Rev Immunol (2013) 13(10):722–37. doi: 10.1038/nri3532
9. Mizushima N, Levine B. Autophagy in Mammalian Development and Differentiation. Nat Cell Biol (2010) 12(9):823–30. doi: 10.1038/ncb0910-823
10. Deretic V. Autophagy in Inflammation, Infection, and Immunometabolism. Immunity (2021) 54(3):437–53. doi: 10.1016/j.immuni.2021.01.018
11. Rao L, Eissa NT. Autophagy in Pulmonary Innate Immunity. J Innate Immun (2020) 12(1):21–30. doi: 10.1159/000497414
12. Pareja ME, Colombo MI. Autophagic Clearance of Bacterial Pathogens: Molecular Recognition of Intracellular Microorganisms. Front Cell Infect Microbiol (2013) 3:54. doi: 10.3389/fcimb.2013.00054
13. Jo EK, Yuk JM, Shin DM, Sasakawa C. Roles of Autophagy in Elimination of Intracellular Bacterial Pathogens. Front Immunol (2013) 4:97. doi: 10.3389/fimmu.2013.00097
14. Ogawa M, Yoshikawa Y, Mimuro H, Hain T, Chakraborty T, Sasakawa C. Autophagy Targeting of Listeria Monocytogenes and the Bacterial Countermeasure. Autophagy (2011) 7(3):310–4. doi: 10.4161/auto.7.3.14581
15. Pereira AC, Ramos B, Reis AC, Cunha MV. Non-Tuberculous Mycobacteria: Molecular and Physiological Bases of Virulence and Adaptation to Ecological Niches. Microorganisms (2020) 8(9):1380. doi: 10.3390/microorganisms8091380
16. Swenson C, Zerbe CS, Fennelly K. Host Variability in NTM Disease: Implications for Research Needs. Front Microbiol (2018) 9:2901. doi: 10.3389/fmicb.2018.02901
17. Jeon D. Infection Source and Epidemiology of Nontuberculous Mycobacterial Lung Disease. Tuberc Respir Dis (Seoul) (2019) 82(2):94–101. doi: 10.4046/trd.2018.0026
18. Koh WJ. Nontuberculous Mycobacteria-Overview. Microbiol Spectr (2017) 5(1). doi: 10.1128/microbiolspec.TNMI7-0024-2016
19. Wassilew N, Hoffmann H, Andrejak C, Lange C. Pulmonary Disease Caused by Non-Tuberculous Mycobacteria. Respiration (2016) 91(5):386–402. doi: 10.1159/000445906
20. Kwon YS, Koh WJ. Diagnosis and Treatment of Nontuberculous Mycobacterial Lung Disease. J Korean Med Sci (2016) 31(5):649–59. doi: 10.3346/jkms.2016.31.5.649
21. Honda JR, Knight V, Chan ED. Pathogenesis and Risk Factors for Nontuberculous Mycobacterial Lung Disease. Clin Chest Med (2015) 36(1):1–11. doi: 10.1016/j.ccm.2014.10.001
22. Davidson RM. A Closer Look at the Genomic Variation of Geographically Diverse Mycobacterium Abscessus Clones That Cause Human Infection and Disease. Front Microbiol (2018) 9:2988. doi: 10.3389/fmicb.2018.02988
23. Zweijpfenning SMH, Ingen JV, Hoefsloot W. Geographic Distribution of Nontuberculous Mycobacteria Isolated From Clinical Specimens: A Systematic Review. Semin Respir Crit Care Med (2018) 39(3):336–42. doi: 10.1055/s-0038-1660864
24. Holt MR, Miles JJ, Inder WJ, Thomson RM. Exploring Immunomodulation by Endocrine Changes in Lady Windermere Syndrome. Clin Exp Immunol (2019) 196(1):28–38. doi: 10.1111/cei.13265
25. Kartalija M, Ovrutsky AR, Bryan CL, Pott GB, Fantuzzi G, Thomas J, et al. Patients With Nontuberculous Mycobacterial Lung Disease Exhibit Unique Body and Immune Phenotypes. Am J Respir Crit Care Med (2013) 187(2):197–205. doi: 10.1164/rccm.201206-1035OC
26. Kim RD, Greenberg DE, Ehrmantraut ME, Guide SV, Ding L, Shea Y, et al. Pulmonary Nontuberculous Mycobacterial Disease: Prospective Study of a Distinct Preexisting Syndrome. Am J Respir Crit Care Med (2008) 178(10):1066–74. doi: 10.1164/rccm.200805-686OC
27. Wi YM. Treatment of Extrapulmonary Nontuberculous Mycobacterial Diseases. Infect Chemother (2019) 51(3):245–55. doi: 10.3947/ic.2019.51.3.245
28. Piersimoni C, Scarparo C. Extrapulmonary Infections Associated With Nontuberculous Mycobacteria in Immunocompetent Persons. Emerg Infect Dis (2009) 15(9):1351–8; quiz 544. doi: 10.3201/eid1509.081259
29. Nasiri MJ, Haeili M, Ghazi M, Goudarzi H, Pormohammad A, Imani Fooladi AA, et al. New Insights in to the Intrinsic and Acquired Drug Resistance Mechanisms in Mycobacteria. Front Microbiol (2017) 8:681. doi: 10.3389/fmicb.2017.00681
30. Griffith DE, Aksamit TR. Therapy of Refractory Nontuberculous Mycobacterial Lung Disease. Curr Opin Infect Dis (2012) 25(2):218–27. doi: 10.1097/QCO.0b013e3283511a64
31. Abe Y, Fukushima K, Hosono Y, Matsumoto Y, Motooka D, Ose N, et al. Host Immune Response and Novel Diagnostic Approach to NTM Infections. Int J Mol Sci (2020) 21(12):4351. doi: 10.3390/ijms21124351
32. Prasla Z, Sutliff RL, Sadikot RT. Macrophage Signaling Pathways in Pulmonary Nontuberculous Mycobacteria Infections. Am J Respir Cell Mol Biol (2020) 63(2):144–51. doi: 10.1165/rcmb.2019-0241TR
33. Sousa S, Borges V, Joao I, Gomes JP, Jordao L. Nontuberculous Mycobacteria Persistence in a Cell Model Mimicking Alveolar Macrophages. Microorganisms (2019) 7(5):113. doi: 10.3390/microorganisms7050113
34. Sousa S, Bandeira M, Carvalho PA, Duarte A, Jordao L. Nontuberculous Mycobacteria Pathogenesis and Biofilm Assembly. Int J Mycobacteriol (2015) 4(1):36–43. doi: 10.1016/j.ijmyco.2014.11.065
35. Sexton P, Harrison AC. Susceptibility to Nontuberculous Mycobacterial Lung Disease. Eur Respir J (2008) 31(6):1322–33. doi: 10.1183/09031936.00140007
36. Chan ED, Iseman MD. Underlying Host Risk Factors for Nontuberculous Mycobacterial Lung Disease. Semin Respir Crit Care Med (2013) 34(1):110–23. doi: 10.1055/s-0033-1333573
37. Brode SK, Jamieson FB, Ng R, Campitelli MA, Kwong JC, Paterson JM, et al. Increased Risk of Mycobacterial Infections Associated With Anti-Rheumatic Medications. Thorax (2015) 70(7):677–82. doi: 10.1136/thoraxjnl-2014-206470
38. Yoo JW, Jo KW, Kang BH, Kim MY, Yoo B, Lee CK, et al. Mycobacterial Diseases Developed During Anti-Tumour Necrosis Factor-Alpha Therapy. Eur Respir J (2014) 44(5):1289–95. doi: 10.1183/09031936.00063514
39. Silwal P, Paik S, Kim JK, Yoshimori T, Jo EK. Regulatory Mechanisms of Autophagy-Targeted Antimicrobial Therapeutics Against Mycobacterial Infection. Front Cell Infect Microbiol (2021) 11:633360. doi: 10.3389/fcimb.2021.633360
40. Watson RO, Manzanillo PS, Cox JS. Extracellular M. Tuberculosis DNA Targets Bacteria for Autophagy by Activating the Host DNA-Sensing Pathway. Cell (2012) 150(4):803–15. doi: 10.1016/j.cell.2012.06.040
41. Romagnoli A, Etna MP, Giacomini E, Pardini M, Remoli ME, Corazzari M, et al. ESX-1 Dependent Impairment of Autophagic Flux by Mycobacterium Tuberculosis in Human Dendritic Cells. Autophagy (2012) 8(9):1357–70. doi: 10.4161/auto.20881
42. Bah A, Vergne I. Macrophage Autophagy and Bacterial Infections. Front Immunol (2017) 8:1483. doi: 10.3389/fimmu.2017.01483
43. Koster S, Upadhyay S, Chandra P, Papavinasasundaram K, Yang G, Hassan A, et al. Mycobacterium Tuberculosis is Protected From NADPH Oxidase and LC3-Associated Phagocytosis by the LCP Protein CpsA. Proc Natl Acad Sci U S A (2017) 114(41):E8711–E20. doi: 10.1073/pnas.1707792114
44. Watson RO, Bell SL, MacDuff DA, Kimmey JM, Diner EJ, Olivas J, et al. The Cytosolic Sensor cGAS Detects Mycobacterium Tuberculosis DNA to Induce Type I Interferons and Activate Autophagy. Cell Host Microbe (2015) 17(6):811–9. doi: 10.1016/j.chom.2015.05.004
45. Chai Q, Wang X, Qiang L, Zhang Y, Ge P, Lu Z, et al. A Mycobacterium Tuberculosis Surface Protein Recruits Ubiquitin to Trigger Host Xenophagy. Nat Commun (2019) 10(1):1973. doi: 10.1038/s41467-019-09955-8
46. Manzanillo PS, Ayres JS, Watson RO, Collins AC, Souza G, Rae CS, et al. The Ubiquitin Ligase Parkin Mediates Resistance to Intracellular Pathogens. Nature (2013) 501(7468):512–6. doi: 10.1038/nature12566
47. Franco LH, Nair VR, Scharn CR, Xavier RJ, Torrealba JR, Shiloh MU, et al. The Ubiquitin Ligase Smurf1 Functions in Selective Autophagy of Mycobacterium Tuberculosis and Anti-Tuberculous Host Defense. Cell Host Microbe (2017) 21(1):59–72. doi: 10.1016/j.chom.2016.11.002
48. Chauhan S, Kumar S, Jain A, Ponpuak M, Mudd MH, Kimura T, et al. TRIMs and Galectins Globally Cooperate and TRIM16 and Galectin-3 Co-Direct Autophagy in Endomembrane Damage Homeostasis. Dev Cell (2016) 39(1):13–27. doi: 10.1016/j.devcel.2016.08.003
49. Jia J, Abudu YP, Claude-Taupin A, Gu Y, Kumar S, Choi SW, et al. Galectins Control mTOR in Response to Endomembrane Damage. Mol Cell (2018) 70(1):120–35.e8. doi: 10.1016/j.molcel.2018.03.009
50. Maphasa RE, Meyer M, Dube A. The Macrophage Response to Mycobacterium Tuberculosis and Opportunities for Autophagy Inducing Nanomedicines for Tuberculosis Therapy. Front Cell Infect Microbiol (2020) 10:618414. doi: 10.3389/fcimb.2020.618414
51. Kim YS, Silwal P, Kim SY, Yoshimori T, Jo EK. Autophagy-Activating Strategies to Promote Innate Defense Against Mycobacteria. Exp Mol Med (2019) 51(12):1–10. doi: 10.1038/s12276-019-0290-7
52. Paik S, Kim JK, Chung C, Jo EK. Autophagy: A New Strategy for Host-Directed Therapy of Tuberculosis. Virulence (2019) 10(1):448–59. doi: 10.1080/21505594.2018.1536598
53. Rubinsztein DC, Bento CF, Deretic V. Therapeutic Targeting of Autophagy in Neurodegenerative and Infectious Diseases. J Exp Med (2015) 212(7):979–90. doi: 10.1084/jem.20150956
54. Zullo AJ, Lee S. Mycobacterial Induction of Autophagy Varies by Species and Occurs Independently of Mammalian Target of Rapamycin Inhibition. J Biol Chem (2012) 287(16):12668–78. doi: 10.1074/jbc.M111.320135
55. Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy Is a Defense Mechanism Inhibiting BCG and Mycobacterium Tuberculosis Survival in Infected Macrophages. Cell (2004) 119(6):753–66. doi: 10.1016/j.cell.2004.11.038
56. Viljoen A, Herrmann JL, Onajole OK, Stec J, Kozikowski AP, Kremer L. Controlling Extra- and Intramacrophagic Mycobacterium Abscessus by Targeting Mycolic Acid Transport. Front Cell Infect Microbiol (2017) 7:388. doi: 10.3389/fcimb.2017.00388
57. Bernut A, Herrmann JL, Kissa K, Dubremetz JF, Gaillard JL, Lutfalla G, et al. Mycobacterium Abscessus Cording Prevents Phagocytosis and Promotes Abscess Formation. Proc Natl Acad Sci U S A (2014) 111(10):E943–52. doi: 10.1073/pnas.1321390111
58. Daley CL, Winthrop KL. Mycobacterium Avium Complex: Addressing Gaps in Diagnosis and Management. J Infect Dis (2020) 222(Suppl 4):S199–211. doi: 10.1093/infdis/jiaa354
59. Wang Y, Chen C, Xu XD, Li H, Cheng MH, Liu J, et al. Levels of miR-125a-5p are Altered in Mycobacterium Avium-Infected Macrophages and Associate With the Triggering of an Autophagic Response. Microbes Infect (2020) 22(1):31–9. doi: 10.1016/j.micinf.2019.07.002
60. Parr DG, Guest PG, Reynolds JH, Dowson LJ, Stockley RA. Prevalence and Impact of Bronchiectasis in Alpha1-Antitrypsin Deficiency. Am J Respir Crit Care Med (2007) 176(12):1215–21. doi: 10.1164/rccm.200703-489OC
61. Bai X, Bai A, Honda JR, Eichstaedt C, Musheyev A, Feng Z, et al. Alpha-1-Antitrypsin Enhances Primary Human Macrophage Immunity Against Non-Tuberculous Mycobacteria. Front Immunol (2019) 10:1417. doi: 10.3389/fimmu.2019.01417
62. Chan ED, Kaminska AM, Gill W, Chmura K, Feldman NE, Bai X, et al. Alpha-1-Antitrypsin (AAT) Anomalies are Associated With Lung Disease Due to Rapidly Growing Mycobacteria and AAT Inhibits Mycobacterium Abscessus Infection of Macrophages. Scand J Infect Dis (2007) 39(8):690–6. doi: 10.1080/00365540701225744
63. Roux AL, Viljoen A, Bah A, Simeone R, Bernut A, Laencina L, et al. The Distinct Fate of Smooth and Rough Mycobacterium Abscessus Variants Inside Macrophages. Open Biol (2016) 6(11):1601. doi: 10.1098/rsob.160185
64. Awuh JA, Flo TH. Molecular Basis of Mycobacterial Survival in Macrophages. Cell Mol Life Sci (2017) 74(9):1625–48. doi: 10.1007/s00018-016-2422-8
65. Howard ST, Rhoades E, Recht J, Pang X, Alsup A, Kolter R, et al. Spontaneous Reversion of Mycobacterium Abscessus From a Smooth to a Rough Morphotype Is Associated With Reduced Expression of Glycopeptidolipid and Reacquisition of an Invasive Phenotype. Microbiol (Reading) (2006) 152(Pt 6):1581–90. doi: 10.1099/mic.0.28625-0
66. Laencina L, Dubois V, Le Moigne V, Viljoen A, Majlessi L, Pritchard J, et al. Identification of Genes Required for Mycobacterium Abscessus Growth In Vivo With a Prominent Role of the ESX-4 Locus. Proc Natl Acad Sci U S A (2018) 115(5):E1002–E11. doi: 10.1073/pnas.1713195115
67. Kim YS, Kim JK, Hanh BTB, Kim SY, Kim HJ, Kim YJ, et al. The Peroxisome Proliferator-Activated Receptor Alpha- Agonist Gemfibrozil Promotes Defense Against Mycobacterium Abscessus Infections. Cells (2020) 9(3):648. doi: 10.3390/cells9030648
68. Renna M, Schaffner C, Brown K, Shang S, Tamayo MH, Hegyi K, et al. Azithromycin Blocks Autophagy and may Predispose Cystic Fibrosis Patients to Mycobacterial Infection. J Clin Invest (2011) 121(9):3554–63. doi: 10.1172/JCI46095
69. Kim SW, Subhadra B, Whang J, Back YW, Bae HS, Kim HJ, et al. Clinical Mycobacterium Abscessus Strain Inhibits Autophagy Flux and Promotes Its Growth in Murine Macrophages. Pathog Dis (2017) 75(8). doi: 10.1093/femspd/ftx107
70. Pohl K, Grimm XA, Caceres SM, Poch KR, Rysavy N, Saavedra M, et al. Mycobacterium Abscessus Clearance by Neutrophils Is Independent of Autophagy. Infect Immun (2020) 88(8):e00024–20. doi: 10.1128/IAI.00024-20
71. Aubry A, Mougari F, Reibel F, Cambau E. Mycobacterium Marinum. Microbiol Spectr (2017) 5(2). doi: 10.1128/microbiolspec.TNMI7-0038-2016
72. Stinear TP, Seemann T, Harrison PF, Jenkin GA, Davies JK, Johnson PD, et al. Insights From the Complete Genome Sequence of Mycobacterium Marinum on the Evolution of Mycobacterium Tuberculosis. Genome Res (2008) 18(5):729–41. doi: 10.1101/gr.075069.107
73. Clark HF, Shepard CC. Effect of Environmental Temperatures on Infection With Mycobacterium Marinum (Balnei) of Mice and a Number of Poikilothermic Species. J Bacteriol (1963) 86:1057–69. doi: 10.1128/jb.86.5.1057-1069.1963
74. Tobin DM, Ramakrishnan L. Comparative Pathogenesis of Mycobacterium Marinum and Mycobacterium Tuberculosis. Cell Microbiol (2008) 10(5):1027–39. doi: 10.1111/j.1462-5822.2008.01133.x
75. Hosseini R, Lamers GE, Hodzic Z, Meijer AH, Schaaf MJ, Spaink HP. Correlative Light and Electron Microscopy Imaging of Autophagy in a Zebrafish Infection Model. Autophagy (2014) 10(10):1844–57. doi: 10.4161/auto.29992
76. Cardenal-Munoz E, Arafah S, Lopez-Jimenez AT, Kicka S, Falaise A, Bach F, et al. Mycobacterium Marinum Antagonistically Induces an Autophagic Response While Repressing the Autophagic Flux in a TORC1- and ESX-1-Dependent Manner. PLoS Pathog (2017) 13(4):e1006344. doi: 10.1371/journal.ppat.1006344
77. Lerena MC, Colombo MI. Mycobacterium Marinum Induces a Marked LC3 Recruitment to its Containing Phagosome That Depends on a Functional ESX-1 Secretion System. Cell Microbiol (2011) 13(6):814–35. doi: 10.1111/j.1462-5822.2011.01581.x
78. Collins CA, De Maziere A, van Dijk S, Carlsson F, Klumperman J, Brown EJ. Atg5-Independent Sequestration of Ubiquitinated Mycobacteria. PLoS Pathog (2009) 5(5):e1000430. doi: 10.1371/journal.ppat.1000430
79. Mohanty S, Jagannathan L, Ganguli G, Padhi A, Roy D, Alaridah N, et al. A Mycobacterial Phosphoribosyltransferase Promotes Bacillary Survival by Inhibiting Oxidative Stress and Autophagy Pathways in Macrophages and Zebrafish. J Biol Chem (2015) 290(21):13321–43. doi: 10.1074/jbc.M114.598482
80. Zhang R, Varela M, Vallentgoed W, Forn-Cuni G, van der Vaart M, Meijer AH. The Selective Autophagy Receptors Optineurin and P62 are Both Required for Zebrafish Host Resistance to Mycobacterial Infection. PLoS Pathog (2019) 15(2):e1007329. doi: 10.1371/journal.ppat.1007329
81. van der Vaart M, Korbee CJ, Lamers GE, Tengeler AC, Hosseini R, Haks MC, et al. The DNA Damage-Regulated Autophagy Modulator DRAM1 Links Mycobacterial Recognition via TLR-MYD88 to Autophagic Defense [Corrected]. Cell Host Microbe (2014) 15(6):753–67. doi: 10.1016/j.chom.2014.05.005
82. Zhang R, Varela M, Forn-Cuni G, Torraca V, van der Vaart M, Meijer AH. Deficiency in the Autophagy Modulator Dram1 Exacerbates Pyroptotic Cell Death of Mycobacteria-Infected Macrophages. Cell Death Dis (2020) 11(4):277. doi: 10.1038/s41419-020-2477-1
83. Chen Z, Shao XY, Wang C, Hua MH, Wang CN, Wang X, et al. Mycobacterium Marinum Infection in Zebrafish and Microglia Imitates the Early Stage of Tuberculous Meningitis. J Mol Neurosci (2018) 64(2):321–30. doi: 10.1007/s12031-018-1026-1
84. Kim TS, Shin YH, Lee HM, Kim JK, Choe JH, Jang JC, et al. Ohmyungsamycins Promote Antimicrobial Responses Through Autophagy Activation via AMP-Activated Protein Kinase Pathway. Sci Rep (2017) 7(1):3431. doi: 10.1038/s41598-017-03477-3
85. Pean CB, Schiebler M, Tan SW, Sharrock JA, Kierdorf K, Brown KP, et al. Regulation of Phagocyte Triglyceride by a STAT-ATG2 Pathway Controls Mycobacterial Infection. Nat Commun (2017) 8:14642. doi: 10.1038/ncomms14642
86. Floto RA, Sarkar S, Perlstein EO, Kampmann B, Schreiber SL, Rubinsztein DC. Small Molecule Enhancers of Rapamycin-Induced TOR Inhibition Promote Autophagy, Reduce Toxicity in Huntington's Disease Models and Enhance Killing of Mycobacteria by Macrophages. Autophagy (2007) 3(6):620–2. doi: 10.4161/auto.4898
87. Kim JJ, Lee HM, Shin DM, Kim W, Yuk JM, Jin HS, et al. Host Cell Autophagy Activated by Antibiotics Is Required for Their Effective Antimycobacterial Drug Action. Cell Host Microbe (2012) 11(5):457–68. doi: 10.1016/j.chom.2012.03.008
88. Kjellin J, Pranting M, Bach F, Vaid R, Edelbroek B, Li Z, et al. Investigation of the Host Transcriptional Response to Intracellular Bacterial Infection Using Dictyostelium Discoideum as a Host Model. BMC Genomics (2019) 20(1):961. doi: 10.1186/s12864-019-6269-x
89. Lopez-Jimenez AT, Cardenal-Munoz E, Leuba F, Gerstenmaier L, Barisch C, Hagedorn M, et al. The ESCRT and Autophagy Machineries Cooperate to Repair ESX-1-Dependent Damage at the Mycobacterium-Containing Vacuole But Have Opposite Impact on Containing the Infection. PLoS Pathog (2018) 14(12):e1007501. doi: 10.1371/journal.ppat.1007501
90. Bah A, Lacarriere C, Vergne I. Autophagy-Related Proteins Target Ubiquitin-Free Mycobacterial Compartment to Promote Killing in Macrophages. Front Cell Infect Microbiol (2016) 6:53. doi: 10.3389/fcimb.2016.00053
91. Zullo AJ, Jurcic Smith KL, Lee S. Mammalian Target of Rapamycin Inhibition and Mycobacterial Survival Are Uncoupled in Murine Macrophages. BMC Biochem (2014) 15:4. doi: 10.1186/1471-2091-15-4
92. Feng X, Lu J, He Z, Wang Y, Qi F, Pi R, et al. Mycobacterium Smegmatis Induces Neurite Outgrowth and Differentiation in an Autophagy-Independent Manner in PC12 and C17.2 Cells. Front Cell Infect Microbiol (2018) 8:201. doi: 10.3389/fcimb.2018.00201
93. Gough ME, Graviss EA, May EE. The Dynamic Immunomodulatory Effects of Vitamin D3 During Mycobacterium Infection. Innate Immun (2017) 23(6):506–23. doi: 10.1177/1753425917719143
94. Yuk JM, Shin DM, Lee HM, Yang CS, Jin HS, Kim KK, et al. Vitamin D3 Induces Autophagy in Human Monocytes/Macrophages via Cathelicidin. Cell Host Microbe (2009) 6(3):231–43. doi: 10.1016/j.chom.2009.08.004
95. Buultjens AH, Vandelannoote K, Meehan CJ, Eddyani M, de Jong BC, Fyfe JAM, et al. Comparative Genomics Shows That Mycobacterium Ulcerans Migration and Expansion Preceded the Rise of Buruli Ulcer in Southeastern Australia. Appl Environ Microbiol (2018) 84(8):e02612–17. doi: 10.1128/AEM.02612-17
96. Capela C, Sopoh GE, Houezo JG, Fiodessihoue R, Dossou AD, Costa P, et al. Clinical Epidemiology of Buruli Ulcer From Benin (2005-2013): Effect of Time-Delay to Diagnosis on Clinical Forms and Severe Phenotypes. PLoS Negl Trop Dis (2015) 9(9):e0004005. doi: 10.1371/journal.pntd.0004005
97. Manry J, Vincent QB, Johnson C, Chrabieh M, Lorenzo L, Theodorou I, et al. Genome-Wide Association Study of Buruli Ulcer in Rural Benin Highlights Role of Two LncRNAs and the Autophagy Pathway. Commun Biol (2020) 3(1):177. doi: 10.1038/s42003-020-0920-6
98. Capela C, Dossou AD, Silva-Gomes R, Sopoh GE, Makoutode M, Menino JF, et al. Genetic Variation in Autophagy-Related Genes Influences the Risk and Phenotype of Buruli Ulcer. PLoS Negl Trop Dis (2016) 10(4):e0004671. doi: 10.1371/journal.pntd.0004671
99. Martin PK, Marchiando A, Xu R, Rudensky E, Yeung F, Schuster SL, et al. Autophagy Proteins Suppress Protective Type I Interferon Signalling in Response to the Murine Gut Microbiota. Nat Microbiol (2018) 3(10):1131–41. doi: 10.1038/s41564-018-0229-0
100. Gama JB, Ohlmeier S, Martins TG, Fraga AG, Sampaio-Marques B, Carvalho MA, et al. Proteomic Analysis of the Action of the Mycobacterium Ulcerans Toxin Mycolactone: Targeting Host Cells Cytoskeleton and Collagen. PLoS Negl Trop Dis (2014) 8(8):e3066. doi: 10.1371/journal.pntd.0003066
101. Manry J. Human Genetics of Buruli Ulcer. Hum Genet (2020) 139(6-7):847–53. doi: 10.1007/s00439-020-02163-1
102. Ke Z, Lu J, Zhu J, Yang Z, Jin Z, Yuan L. Down-Regulation of lincRNA-EPS Regulates Apoptosis and Autophagy in BCG-Infected RAW264.7 Macrophages via JNK/MAPK Signaling Pathway. Infect Genet Evol (2020) 77:104077. doi: 10.1016/j.meegid.2019.104077
103. Smith DS, Lindholm-Levy P, Huitt GA, Heifets LB, Cook JL. Mycobacterium Terrae: Case Reports, Literature Review, and In Vitro Antibiotic Susceptibility Testing. Clin Infect Dis (2000) 30(3):444–53. doi: 10.1086/313693
104. Orosz L, Papanicolaou EG, Seprenyi G, Megyeri K. IL-17A and IL-17F Induce Autophagy in RAW 264.7 Macrophages. BioMed Pharmacother (2016) 77:129–34. doi: 10.1016/j.biopha.2015.12.020
105. Feng Z, Bai X, Wang T, Garcia C, Bai A, Li L, et al. Differential Responses by Human Macrophages to Infection With Mycobacterium Tuberculosis and Non-Tuberculous Mycobacteria. Front Microbiol (2020) 11:116. doi: 10.3389/fmicb.2020.00116
106. Sharma V, Makhdoomi M, Singh L, Kumar P, Khan N, Singh S, et al. Trehalose Limits Opportunistic Mycobacterial Survival During HIV Co-Infection by Reversing HIV-Mediated Autophagy Block. Autophagy (2021) 17(2):476–95. doi: 10.1080/15548627.2020.1725374
107. Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, et al. TFEB Links Autophagy to Lysosomal Biogenesis. Science (2011) 332(6036):1429–33. doi: 10.1126/science.1204592
108. Sato E, Imafuku S, Ishii K, Itoh R, Chou B, Soejima T, et al. Vitamin D-Dependent Cathelicidin Inhibits Mycobacterium Marinum Infection in Human Monocytic Cells. J Dermatol Sci (2013) 70(3):166–72. doi: 10.1016/j.jdermsci.2013.01.011
109. Kan Y, Meng L, Xie L, Liu L, Dong W, Feng J, et al. Temporal Modulation of Host Aerobic Glycolysis Determines the Outcome of Mycobacterium Marinum Infection. Fish Shellfish Immunol (2020) 96:78–85. doi: 10.1016/j.fsi.2019.11.051
110. Zheng Q, Wang Q, Wang S, Wu J, Gao Q, Liu W. Thiopeptide Antibiotics Exhibit a Dual Mode of Action Against Intracellular Pathogens by Affecting Both Host and Microbe. Chem Biol (2015) 22(8):1002–7. doi: 10.1016/j.chembiol.2015.06.019
111. Sharma V, Verma S, Seranova E, Sarkar S, Kumar D. Selective Autophagy and Xenophagy in Infection and Disease. Front Cell Dev Biol (2018) 6:147. doi: 10.3389/fcell.2018.00147
112. Kim HJ, Kim IS, Lee SG, Kim YJ, Silwal P, Kim JY, et al. MiR-144-3p is Associated With Pathological Inflammation in Patients Infected With Mycobacteroides Abscessus. Exp Mol Med (2021) 53(1):136–49. doi: 10.1038/s12276-020-00552-0
113. Bagul PK, Katare PB, Bugga P, Dinda AK, Banerjee SK. SIRT-3 Modulation by Resveratrol Improves Mitochondrial Oxidative Phosphorylation in Diabetic Heart Through Deacetylation of TFAM. Cells (2018) 7(12):235. doi: 10.3390/cells7120235
114. Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, et al. Small Molecule Activators of Sirtuins Extend Saccharomyces Cerevisiae Lifespan. Nature (2003) 425(6954):191–6. doi: 10.1038/nature01960
115. Kim YJ, Lee SH, Jeon SM, Silwal P, Seo JY, Hanh BTB, et al. Sirtuin 3 is Essential for Host Defense Against Mycobacterium Abscessus Infection Through Regulation of Mitochondrial Homeostasis. Virulence (2020) 11(1):1225–39. doi: 10.1080/21505594.2020.1809961
116. Kim TS, Jin YB, Kim YS, Kim S, Kim JK, Lee HM, et al. SIRT3 Promotes Antimycobacterial Defenses by Coordinating Mitochondrial and Autophagic Functions. Autophagy (2019) 15(8):1356–75. doi: 10.1080/15548627.2019.1582743
117. Rampacci E, Stefanetti V, Passamonti F, Henao-Tamayo M. Preclinical Models of Nontuberculous Mycobacteria Infection for Early Drug Discovery and Vaccine Research. Pathogens (2020) 9(8):641. doi: 10.3390/pathogens9080641
118. Kwon BE, Ahn JH, Park EK, Jeong H, Lee HJ, Jung YJ, et al. B Cell-Based Vaccine Transduced With ESAT6-Expressing Vaccinia Virus and Presenting Alpha-Galactosylceramide Is a Novel Vaccine Candidate Against ESAT6-Expressing Mycobacterial Diseases. Front Immunol (2019) 10:2542. doi: 10.3389/fimmu.2019.02542
119. Verma D, Chan ED, Ordway DJ. Non-Tuberculous Mycobacteria Interference With BCG-Current Controversies and Future Directions. Vaccines (Basel) (2020) 8(4):688. doi: 10.3390/vaccines8040688
120. Hu D, Wu J, Zhang R, Chen L, Chen Z, Wang X, et al. Autophagy-Targeted Vaccine of LC3-LpqH DNA and its Protective Immunity in a Murine Model of Tuberculosis. Vaccine (2014) 32(20):2308–14. doi: 10.1016/j.vaccine.2014.02.069
Keywords: autophagy, nontuberculous mycobacteria, host defense, innate immunity, infection
Citation: Silwal P, Kim IS and Jo E-K (2021) Autophagy and Host Defense in Nontuberculous Mycobacterial Infection. Front. Immunol. 12:728742. doi: 10.3389/fimmu.2021.728742
Received: 22 June 2021; Accepted: 16 August 2021;
Published: 06 September 2021.
Edited by:
Malcolm Scott Duthie, HDT Biotech Corporation, United StatesReviewed by:
Gian Maria Fimia, Sapienza University of Rome, ItalyCopyright © 2021 Silwal, Kim and Jo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eun-Kyeong Jo, aGF5b3VuZ2pAY251LmFjLmty
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.