- 1Research Service, Veteran Administration Greater Los Angeles Healthcare System, Los Angeles, CA, United States
- 2Division of Rheumatology, Department of Medicine, University of California, Los Angeles, Los Angeles, CA, United States
- 3Department of Medicine, University of California, Los Angeles, Los Angeles, CA, United States
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease with widespread inflammation, immune dysregulation, and is associated with the generation of destructive anti-DNA autoantibodies. We have shown previously the immune modulatory properties of pCons peptide in the induction of both CD4+ and CD8+ regulatory T cells which can in turn suppress development of the autoimmune disease in (NZB/NZW) F1 (BWF1) mice, an established model of lupus. In the present study, we add novel protein information and further demonstrate the molecular and cellular phenotypes of pCons-induced CD4+ and CD8+ Treg subsets. Flow cytometry analyses revealed that pCons induced CD8+ Treg cells with the following cell surface molecules: CD25highCD28high and low subsets (shown earlier), CD62Lhigh, CD122low, PD1low, CTLA4low, CCR7low and 41BBhigh. Quantitative real-time PCR (qRT-PCR) gene expression analyses revealed that pCons-induced CD8+ Treg cells downregulated the following several genes: Regulator of G protein signaling (RGS2), RGS16, RGS17, BAX, GPT2, PDE3b, GADD45β and programmed cell death 1 (PD1). Further, we confirmed the down regulation of these genes by Western blot analyses at the protein level. To our translational significance, we showed herein that pCons significantly increased the percentage of CD8+FoxP3+ T cells and further increased the mean fluorescence intensity (MFI) of FoxP3 when healthy peripheral blood mononuclear cells (PBMCs) are treated with pCons (10 μg/ml, for 24-48 hours). In addition, we found that pCons reduced apoptosis in CD4+ and CD8+ T cells and B220+ B cells of BWF1 lupus mice. These data suggest that pCons stimulates cellular, immunological, and molecular changes in regulatory T cells which in turn protect against SLE autoimmunity.
Introduction
SLE is an autoimmune disease characterized by widespread inflammation, autoantibody production, and immune complex deposition. Regulatory T cells (Treg) are protective in many inflammatory and autoimmune diseases including SLE. The modulation of abnormal immune regulation is an object of intense investigation in autoimmune diseases. A therapeutic goal is to limit the number and activity of abnormal pathogenic cells and autoantibodies through restoration of immune system self-tolerance. One way to achieve that is by administrating peptides (such as pConsensus peptide, edratide and nucleosomal peptides) that induce regulatory T cells (1–11). Another approach used recently utilized nanoparticles for expanding regulatory T cells to treat autoimmune diseases including lupus (12–15). Whereas a decrease in the number and/or function of regulatory CD4+ T cells has been extensively studied in SLE (16–24), the role and characterization of the CD8+ Treg subset is less clear. Investigating the genes, regulatory networks, and signaling pathways that regulate the functional activity and survival of CD8+ Treg cells is important for development of therapies for restoring immune homeostasis in SLE and other autoimmune diseases. However, in order to rationally intervene to restore immune homeostasis, there is much that remains to be understood about the molecular phenotypes, mechanisms and pathways that govern the differentiation, expansion, maintenance, and regulatory function of CD8+ Treg. We have developed a unique model in which CD8+ regulatory T cells can be induced to suppress the development of autoimmune disease in an animal model of lupus, the (NZB/NZW) F1 (BWF1) mouse (3, 5, 25, 26). In this model, we have demonstrated that synthetic peptides (pCons) based on T cell determinants in the VH region of IgG which encode murine antibodies to DNA that bind to MHC Class I/II regions can activate CD8+ T cells in vitro, which can result in the suppression of co-cultured CD4+ T helper cells and B cell activities (26, 27). In addition, when pCons is administered in vivo, we can demonstrate the suppression of anti-DNA antibody production, and subsequent nephritis. However, the cellular and molecular phenotypes of pCons-induced CD8+ regulatory T cells are not yet completely clear. In this study, we have further provided novel information and defined the immunological and molecular phenotypes of pCons-induced CD8+ T regulatory cells and CD4+ regulatory T cells. We also showed that pCons treatment reduces apoptosis in CD4+ T cells, and CD8+ T cells and B220+ B cells. To the translational significance, we showed herein that pCons also induces CD8+FoxP3+ Treg cells in healthy human peripheral blood mononuclear cells (PBMCs).
Materials and Methods
Mice
NZB (H-2d/d), NZW (H-2z/z) and NZB/NZW F1 (H-2d/z) mice were purchased from the Jackson Laboratories (Bar Harbor, ME, USA) or bred at the University of California Los Angeles (UCLA). All mice were treated in accordance with the guidelines of the University of California Los Angeles Animal Research Committee, an Institution accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). Mice were housed in pathogen-free conditions according to the National Institutes of Health (NIH) guidelines for the use of experimental animals. Female mice were used for all experiments.
Subjects
We enrolled 6 healthy female donors (19-70 years of age) with no history of autoimmune disease. Subjects had regular menstrual cycles and were not taking any contraceptives or sex hormones. Written informed consent was obtained from each subject who participated in the study. The study was approved by the Institutional Review Board (# 11-000907) of the University of California Los Angeles.
Peptides
The pCons peptide used in this study and the MHC molecules they bind have been described earlier (26, 28). pCons (FIEWNKLRFRQGLEW), the artificial tolerizing peptide, contains T-cell determinants based on the J558 VH regions of several murine anti-dsDNA Ab from BWF1 mice (3, 5, 23, 26, 27, 29). Peptides were synthesized at Chiron Biochemicals (San Diego, CA, USA), purified to a single peak on high-performance liquid chromatography, and analyzed by mass spectroscopy for expected amino acid content.
Treatment of Mice
Ten- to twelve-week-old female BWF1 mice received a single i.v. dose of 1 mg of pCons, dissolved in saline, as reported previously (26, 27, 30) for tolerance induction. For immunophenotyping of regulatory T cells, female BWF1 mice were used and injected with pCons. Control mice received either a similar amount of pNeg (negative control peptide) or saline.
Cell Isolation, Preparation, Immunophenotyping, and Flow Cytometry
Spleen cells were isolated ~1 week after administration of the pCons peptide from tolerized, saline-treated, or naïve BWF1 mice. Single cell suspensions of splenocytes were prepared by passing cells through cell strainers (40µm) (Fisher). ACK lysing buffer, (Sigma, St Louis, MO, USA) was used to lyse red blood cells. Cells were washed and re-suspended in RPMI complete media. RPMI 1640-complete media was supplemented with L-glutamine (2 mM), penicillin (100 units/ml), streptomycin (0.1 mg/ml), 2-mercaptoethanol (Gibco) and 10% fetal bovine serum (FBS). FACS staining buffer was obtained from eBiosciences, BD Pharmingen, and/or BioLegend Inc. Cell subsets were further enriched following incubation with anti-CD4 (L3T4), anti-B (CD45R/B220), anti-CD8 (CD8a Ly-2), and microbeads from Miltenyi Biotech (Auburn, CA, USA). Purity of cells was determined to be more than 90% pure as assessed by flow cytometry (FACS). For immunophenotyping, isolated cells were washed with FACS buffer and 1–2 million cells were used for cell surface staining. Before staining, cells were incubated with rat anti-mouse CD16/CD32 (FC III/II receptor) Ab to block nonspecific binding.
For regulatory T cell immunophenotyping, spleen cells were stained with CD4 (L3T4), (RPA-T4), CD8 (Ly-2), CD25 (PC61), CD28 (37.51), CD62L (MEL-14), CD122 (TM-β1), PD1(RMP1), CCR7(4B12), GITR (DTA-1, AITR, TNFRSF18), CTLA-4 (UC10-4F10-11) and 4IBB(1AH2) antibodies for FACS analysis. Antibodies for cell surface staining and isotype controls were from BD Biosciences, BD Pharmingen, eBiosciences, or BioLegend. FoxP3 (PCH101) staining was performed with an eBiosciences intracellular kit (Cat #12-4776). Before intracellular FoxP3 staining, cells were stained with cell surface molecules (CD4, CD8, CD25, CD28, CD62L, CD122) as per manufacturer’s protocol. Cells were fixed and permeabilized, washed with permeabilization buffer, and then stained with anti-human FoxP3 (PCH101) antibody in 1X permeabilization buffer (eBiosciences), washed again with permeabilization buffer, and then the samples analyzed by FACS at the UCLA flow Core facility. Data was collected using an FACSCalibur (BD Biosciences) and analyzed with BD Cell Quest software (Becton-Dickinson, Mountain View, CA) or De Novo FCS Express Ver. 7 software (Ontario, Canada).
Human Peripheral Blood Mononuclear Cells (PBMCs) Isolation and Preparation
For human studies, peripheral blood mononuclear cells (PBMCs) were isolated on a density gradient (Histopaque-1077, Sigma-Aldrich, St. Louis, MO, USA) from blood samples of healthy volunteers. Lymphocytes were washed twice in RPMI complete media. Red blood cells (RBC) were lysed with RBC lysing solution (Sigma-Aldrich, St. Louis, MO, USA). After washing cells were stained with fluorochrome -labeled monoclonal antibodies (mAbs) and analyzed by FACS.
Western Blot Analysis
Western blot analyses were performed as described earlier (31). In brief, cell lysates were prepared from the CD8+ T cells of naïve and pCons-treated BWF1 mice. Cells were lysed with RIPA buffer (150 nM NaCl, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 10 mM Tris, pH 7.3) supplemented with Protease Arrest protease inhibitor cocktail solution (G Biosciences, Maryland Heights, MO, USA). Protein was measured from each sample using the Bradford assay (Bio-Rad Laboratories, Hercules, CA, USA) and an equal amount of protein was loaded in each well. The lysates were resolved on a 4–12% NuPage gel (Invitrogen, Carlsbad, CA, USA) under reducing conditions. Proteins were electro-transferred onto a polyvinylidene fluoride membrane (Invitrogen). The membranes were blocked with 3% BSA and immunoblotted with a protein-specific antibodies [GPT2 (ab80947), Abcam; PD1 (DO-1), sc-126 Santa Cruz Biotechnology, Inc; PD1 (ab58811) Abcam; GADD45b (K-12), sc-133606, Santa Cruz Biotechnologies, Inc; p53 (DO-1) sc-126, Santa Cruz Biotechnologies, Inc, Santa Cruz, CA, USA, (1: 200 - 1:1000 dilution range); Bax (1:1000 dilution) Cat # #2772, Cell Signaling Technology, Danvers, MA; PDE3b, H-300, sc-20793 (1:1000 dilution); RGS16 (H-100), sc-30218 (1:1000 dilution) or β-actin (1:100 000 dilution; Sigma, Inc]. Following washing, the membranes were incubated in secondary antibodies (1:2500 dilution; Santa Cruz Inc, Santa Cruz, CA, USA). All blocking, incubation and washing steps were performed in TBST (TBS and 0.1% Tween-20). Proteins were visualized using ECL (GE Healthcare, Buckinghamshire, UK).
RNA Isolation and Real-Time PCR
Total cellular RNA was isolated from purified cell subsets from saline-treated or pCons-tolerized BWF1 mice with TRIzol (Invitrogen, Carlsbad, CA, USA) as per manufacturer’s protocols. One-step-real time PCR was analyzed as described earlier (3, 5, 26, 29). Each experimental group consists of the pooled spleen cells of 3–4 mice from each group, naïve CD8+ T cells or tolerized CD8+ T cells. One-step RT-PCR was performed (Applied Biosystems, Foster City, CA, USA) using 100 ng of total RNA. Quantitative real-time reverse transcription was performed using TaqMan technology on an ABI Prism 7900 HT Sequence Detection System (Applied Biosystems). Primers and probes of regulator of G protein signaling genes (RGS2, RGS16, and RGS17), glutamic pyruvate transaminase 2 (GPT2), BAX (Bcl-2-associated X protein), programmed cell death-1 (PD1), growth arrest and DNA damage inducible 45 beta (GADD45β), and phosphodiesterase 3b (PDE3b), and GAPDH were obtained from Applied Biosystems (Foster City, CA, USA). The other oligonucleotide sequences used for the primers and TaqMan probes (Applied Biosystem, Foster City, CA) are described (3, 5, 26, 29). GAPDH was used as an endogenous control in each experimental set.
Measurement of Apoptosis
Assays were performed to measure apoptosis as described earlier (5, 26). In brief, splenocytes were obtained from both naïve and pCons-treated BWF1 mice. RBC were lysed, cells washed, and stained with fluorochrome-labeled specific antibodies [CD4 (PerCP), CD8 (PE), B220 (APC), and Annexin V (FITC)] and flow cytometry performed.
Statistical Analyses
Data was analyzed using GraphPad Prism 4.0 Software (San Diego, CA). Comparisons between the two groups were performed using paired one- or two-tailed Student’s t-test. Nonparametric testing among more than two groups was performed by one-way ANOVA. Results are expressed as mean ± SEM. p<0.05 was considered significant.
Results
pCons Treatment Alters Cell Surface Expression of CD25, CD122, and Increased Intracellular FoxP3 Expression in CD4+ T Cells and Further Modifies Cell Surface Expression of CD25, CD28, CTLA-4 and 41BB in CD8+ T Cells in BWF1 Mice
To explore the tolerogenic immune responses of pCons peptide in the present study, we determined the various cell surface expression markers by flow cytometry in both CD4+ and CD8+ T cells in pCons-treated and negative control peptide and or saline-treated BWF1 mice. We found that pCons treatment increased the cell surface expression of CD25 and CD122 in CD4+ T cells compared to naïve CD4+ T cells (Figure 1A Panels A, C, E, H, J, L). We also found that pCons treatment increased the intracellular FoxP3 expression in pCons-treated CD4+ T cells (Figure 1A Panels B, D, F, G, I, K). Next, we investigated the effect of pCons on CD8+ T cells. Our data demonstrate that pCons treatment modified the cell surface expression of CD25 (increased), CD28 (increased), CTLA-4 (decreased) and 41BB (no changed) in CD8+ T cells (Figure 1B Panels A–L). Gating strategy is shown in (Supplementary Figure 1A and Figure 1B Panels A–H). Previously, we have demonstrated that pCons treatment increased the FoxP3 expression in CD8+ T cells (3, 26). In this study, we re-validated our previous findings of FoxP3 and added novel information for additional cell surface phenotypes including FoxP3 with cumulative data of 4-6 experiments (Figure1A Panel K). Altogether, these data indicate that pCons treatment induced the various cell surface markers including intracellular FoxP3 in both CD4+ and CD8+T cells.
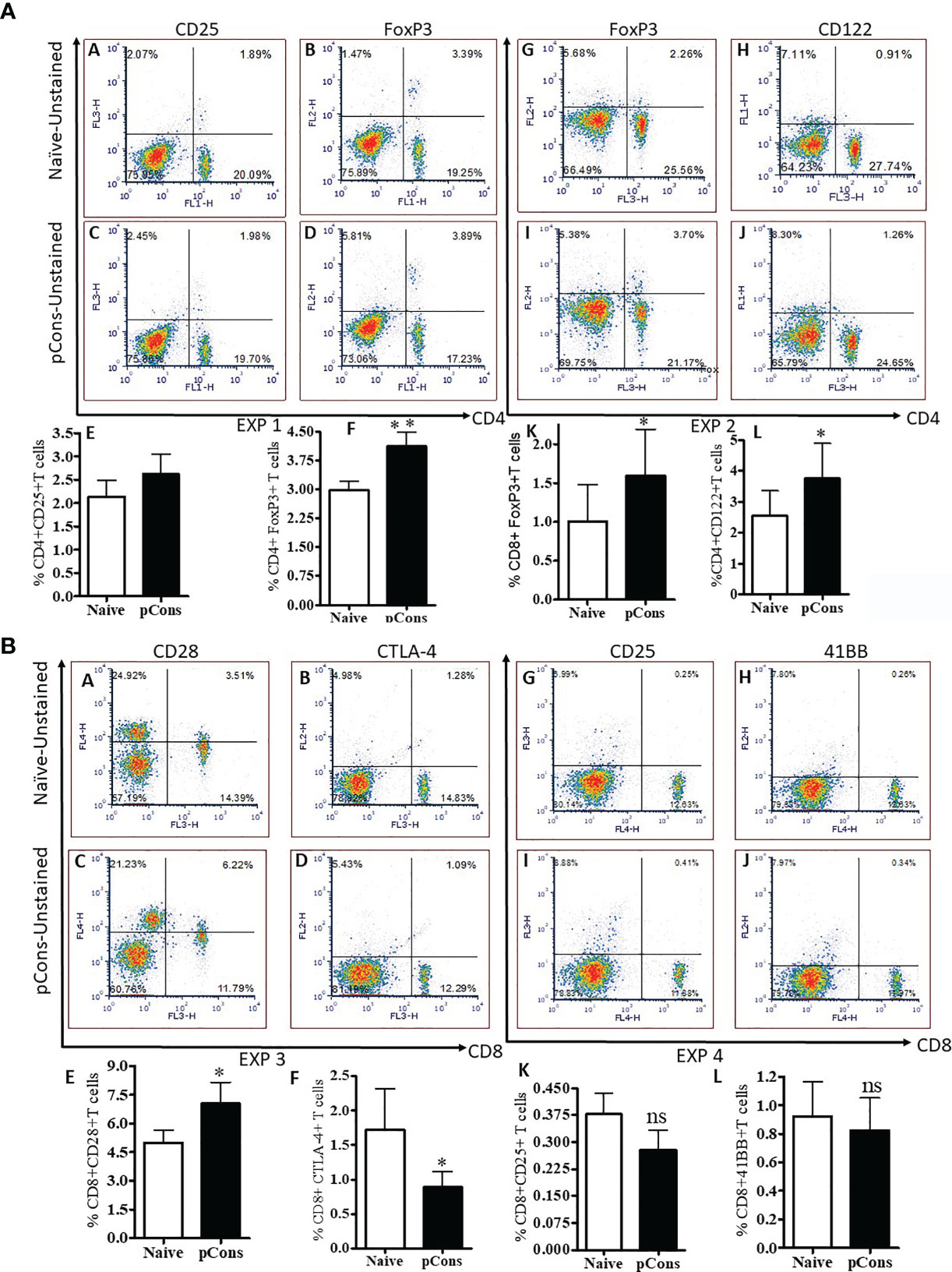
Figure 1 pCons treatment alters cell surface expression of CD25, CD122, and increased intracellular FoxP3 expression in CD4+ T cells and further modifies cell surface expression of CD25, CD28, CTLA-4 and 41BB in CD8+ T cells in BWF1 mice. Female 10-12-wk old BWF1 mice were treated with pCons (1 mg i.v.). After one-two week treatment, splenocytes were obtained both from naïve and pCons-treated BWF1 mice. RBC were lysed, cells washed, and stained with fluorochrome-labeled specific antibodies (CD4, CD8, CD25, CD28, CD122, CTLA-4, and 41BB). FACS analysis was performed on a FACSCalibur™ with cell Quest™ software (BD Biosciences, San Jose, CA) and analyzed using De Novo FCS Express software (Ontario, Canada). Intracellular FoxP3 expression was analyzed after cell fixation and permeabilization as per manufacturer’s protocol (eBiosciences, San Diego, CA, USA). (A) CD4+ T cells, and (B) CD8+ T cells data, two experiments each. (A) Exp 1. Panel (A) Naïve CD4+CD25+ T cells; (B) Naïve CD4+FoxP3+ T cells; (C) pCons CD4+CD25+ T cells; (D) pCons CD4+FoxP3+ T cells. (E) Cumulative data of CD4+CD25+T cells (4-5 experiments of two/three mice); (F) Cumulative data of CD4+FoxP3+ T cells (4-5 experiments of two/three mice). Exp 2. (G) Naïve CD4+FoxP3+ T cells; (H) Naïve CD4+CD122+ T cells; (I) pCons CD4+FoxP3+ T cells., (J) pCons CD4+CD122+ T cells. (K) Cumulative data (6 experiments) of CD8+FoxP3+ T cells. (L) Cumulative data of CD4+CD122+ T cells (4 experiments of two/three mice). (B) Exp 3. (A) Naïve CD8+CD28+ T cells; (B) Naïve CD8+CTLA-4+ T cells; (C) pCons CD8+CD28+ T cells; (D) pCons CD8+CTLA-4+ T cells. (E) Cumulative data of CD8+CD28+ T cells (5-6 experiments). (F) Cumulative data of CD8+CTLA-4+ T cells (5-6 experiments). Exp.4. (G) Naïve CD8+CD25+ T cells; (H) Naïve CD8+41BB+ T cells, (I) pCons CD8+CD25+ T cells; (J) pCons CD8+41BB+ T cells. (K) Cumulative (6-7 experiments data) of CD8+CD25+ T cells, (L) Cumulative (4 experiments data) of CD8+41BB+ T cells. Minimum 10,000 cells were gated, and only live cells were used for data analyses. Dead cells were excluded from the analyses. *p < 0.05. **p < 0. 001. ns, not significant.
pCons Treatment Induces and Modifies Cell Surface Expression of CD62L, CD122, and CCR7 in CD8+ T Cells in BWF1 Mice
Previously we showed that pCons treatment increased the number of regulatory CD4+ and CD8+ T cells and modulated their functions including the ability for the suppression of anti-DNA antibody in BWF1 mice (3, 5, 23, 26). In the present study, we were interested to see whether pCons treatment of BWF1 mice changes the cell surface expression of CD122, CD62L, PD1, and CCR7 in CD8+ T cells since these markers have been implicated in the regulatory phenotypes of CD8+ T cells (32–36). We found two-fold increase in cell surface expression of CD62L and further increase in percent expression of CD8+ CD62L+ T cells and a decrease in the CD122 and CCR7 mean fluorescence intensity in pCons-treated CD8+ T cells compared to negative control peptide or saline-treated CD8+ T cells (Figures 2A–E). PD1 cell surface expression was decreased in pCons-treated CD8+ T cells (Figure 2D). Further, Western blot analysis demonstrated that protein levels of CCR7 are decreased in CD8+ T cells after pCons tolerance (Figure 2F). Taken together, these data suggest that pCons treatment has differential immune-regulatory effects on CD8+ T cells and on CD62L, CD122, PD1, and CCR7.
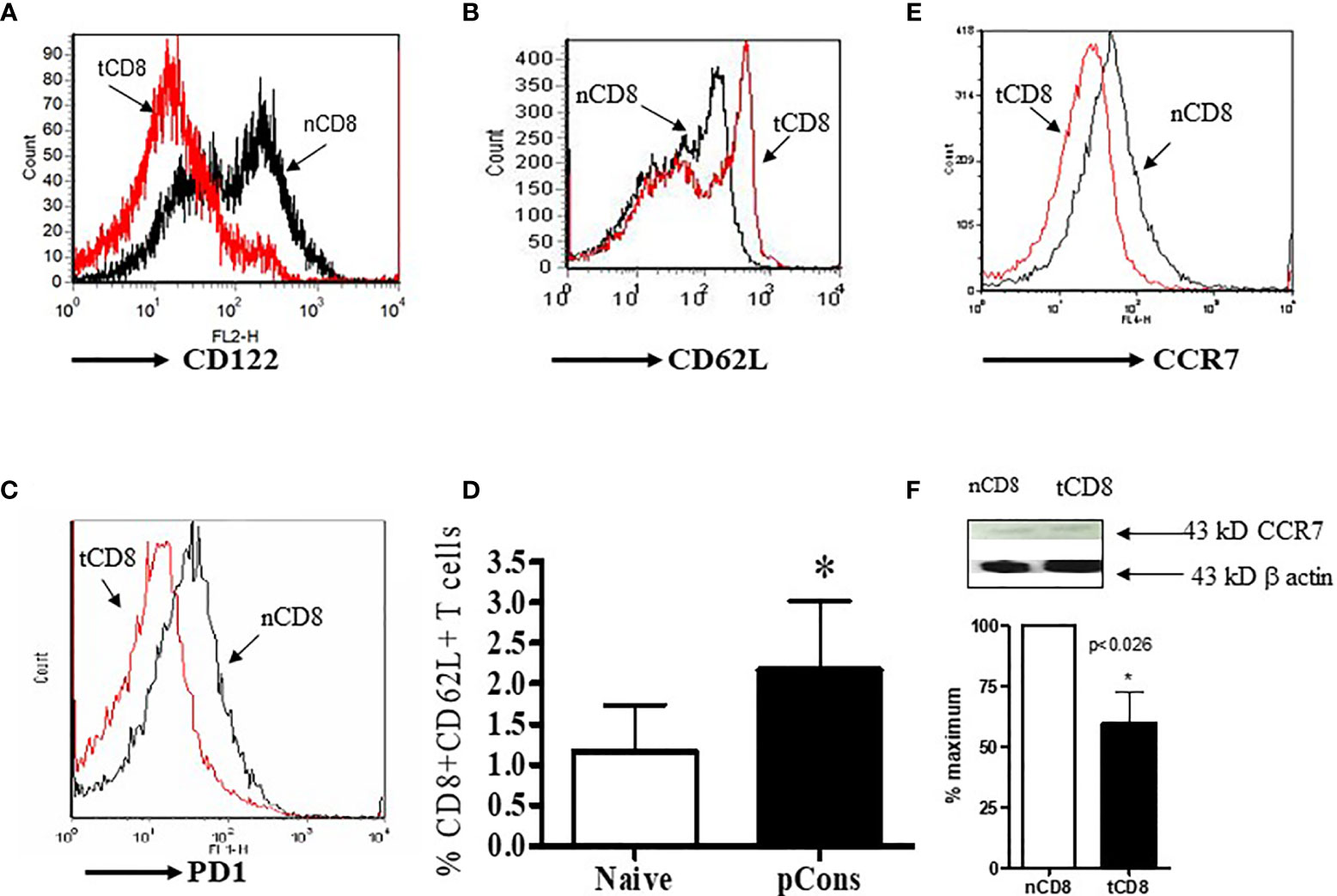
Figure 2 pCons treatment modified the cell surface expression of CD122, CD62L, CCR7, and PD1 in CD8+ T cells in BWF1 mice. Female 10-12-wk old BWF1 mice were treated with pCons (1 mg i.v.). After one-two week treatment, splenocytes were obtained both from naïve and pCons-treated BWF1 mice (2-3 mice in each group). RBC lysed, cells washed and stained with specific antibodies (CD8, CD62L, CD122, CCR7), and flow cytometry performed. (A) CD122 (PE) cell surface expression of naïve vs tolerized CD8+ T cells treated with pCons; (B) CD62L (FITC) cell surface expression of naïve vs tolerized CD8+ T cells pCons; (C) PD1 (PE) cell surface expression of naïve vs tolerized CD8+ (pCons); (D) Percent expression of naïve CD8+CD62L+ vs tolerized CD8+CD62L+ T cells (pCons); (E) CCR7 (APC) cell surface expression of naïve CD8+ vs tolerized CD8+ T cells (pCons-treated group). Minimum 10,000 cells were gated, and live cells were used for data analyses. Dead cells were excluded from the analyses. Data were analyzed with FCS Express Ver. 7 (De Novo, Ontario, Canada). *p < 0.05. (F) Naïve CD8+ and tolerized CD8+ T cells were obtained, lysed, and Western Blot analysis performed with CCR7 and β-actin antibodies. CCR7 value normalized to those of β−actin. *p < 0.05.
pCons Treatment Modifies Expression of Regulator of G-Protein Signaling Genes (RGS2, RGS16, RGS17), Interferon-Induced, and Apoptotic Genes in CD8+ T Cells
To determine whether there is cross-regulation of regulator of G protein signaling and interferon genes and whether pCons affect this cross-regulation in CD8+ T cells, we tested the expression of RGS and IFNs genes in pCons-treated CD8+ T cells. We have shown previously that pCons-induced CD8+ T regulatory cells are genetically reprogrammed following pCons induction (26, 29, 31). These pCons-induced CD8+ Treg cells display i) resist to apoptosis; ii) have immunosuppressive programs; and iii) traffic to sites of inflammation to inhibit the development of autoantibody formation. Our previous gene chip array analyses demonstrated that pCons-induced CD8+ Treg cells have upregulated genes including; interferon inducible 202b (Ifi202b), FoxP3, Bcl2, transformation related protein 53 (TP53) and interferon receptor IFNAR1 (29). In the present study, we showed herein that pCons treatment significantly downregulated and decreased (~2-5 fold) the expression of 6 genes: RGS2, RGS16, RGS17, BAX, GPT2, GADD45β (Figures 3A–D). We further confirmed the downregulation of GPT2, RGS16, GADD45β, and BAX proteins by Western blot analyses (Figures 3E–H). The downregulated BAX expression (37) in pCons-induced CD8+ T cells may contribute to the survival of these cells in vitro/vivo. Overall, these data indicate that pCons regulates RGS, IFNs, and apoptotic genes.
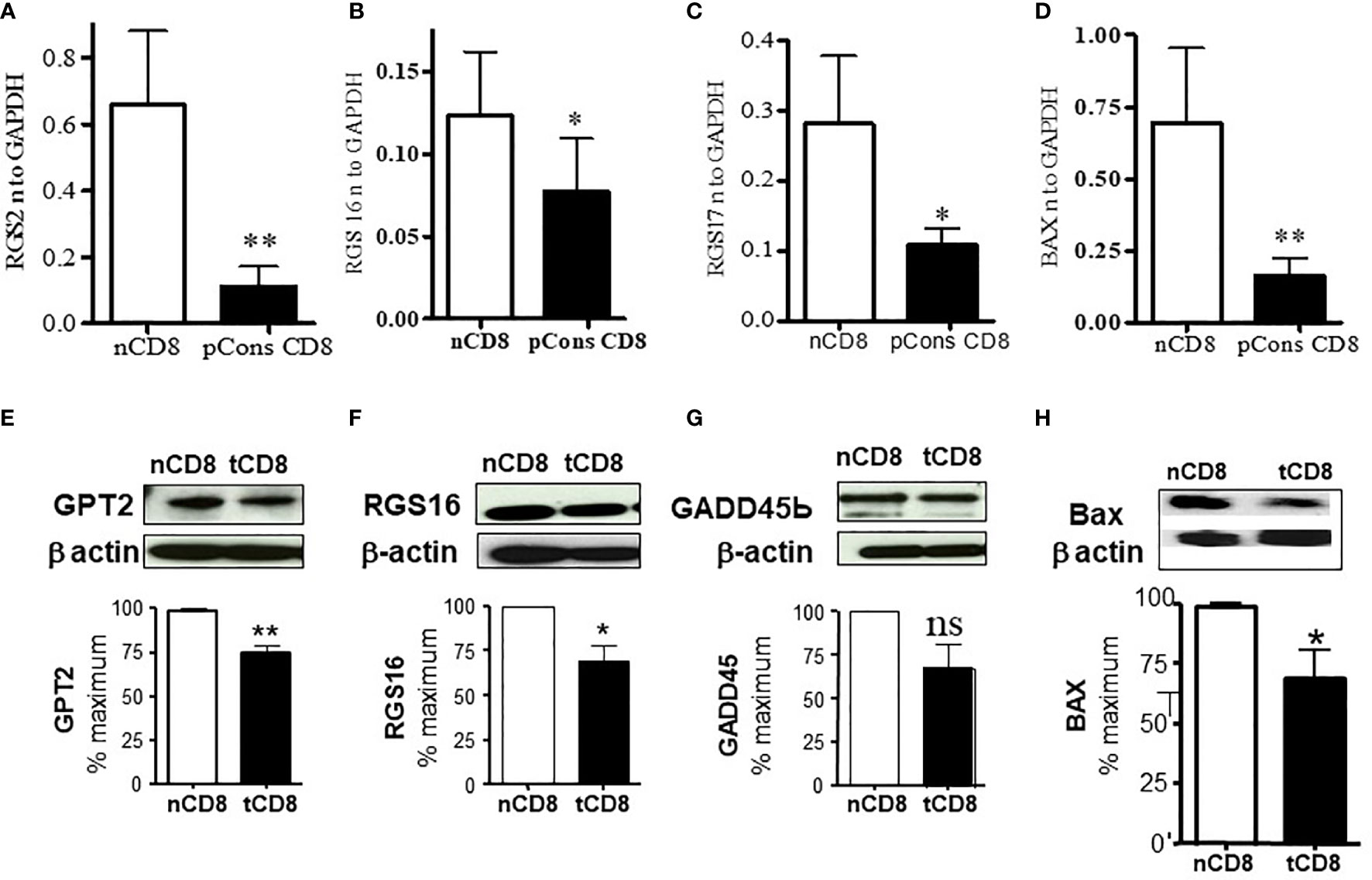
Figure 3 pCons treatment downregulated the expression of Regulator of G protein signaling (RGS2, RGS16, and RGS17) and Bax (Bcl2-associated X protein) in pCons-induced CD8+ T cells. Female 10-12-wk old BWF1 mice (n=3-5 mice) were treated with pCons (1 mg i.v.). After one-two week, splenocytes were obtained both from naïve and pCons-treated BWF1 mice. Naïve CD8+ T cells and pCons-treated CD8+ T cells were isolated from BWF1 mice spleen cells using microbeads from (Miltenyi Biotech (Auburn, CA, USA). Splenocytes were labelled with CD8-specific antibody, and naïve and pCons-treated CD8+ T cells sorted by FACS. Cells were lysed with RNA lysis solution (Trizol) and total cellular RNA obtained. Murine primers and probes (RGS2, RGS16, RGS17, BAX, and GAPDH) were obtained from Applied Biosystems (Foster City CA, USA). Real time PCR was performed with 100 ng of RNA with gene specific primers and probes comparing naïve and pCons-treated CD8+ T cells for each protein. (A) RGS2 normalized to GAPDH in naïve CD8+ T cells vs pCons-treated CD8+ T cells. (B) RGS16. (C) RGS17. (D) BAX. Data was normalized with GAPDH mRNA levels. (E–H). Western blot analyses. (E) GPT2 protein level normalized to β-actin in naïve CD8+ T cells vs pCons-treated CD8+ T cells. (F) RGS16. (G) GADD45β. (H) BAX. Data was normalized to β actin protein levels. *p < 0.05, **p < 0.001. ns, not significant.
pCons Treatment Reduces Apoptosis of CD4+ and CD8+ T Cells, and of B220+ B Cells in BWF1 Lupus Mice
Since earlier studies demonstrated that apoptosis affects immune tolerance, we were interested to investigate whether pCons influences apoptosis in various immune cell subsets including T cells which play an important role in lupus. To address this, we treated BWF1 lupus mice with pCons. After 1-2 weeks, splenocytes were obtained, stained with Annexin V FITC, and then stained with fluorochrome-labeled CD4+, CD8+ and B cells specific monoclonal antibodies, and analyzed by FACS. We found that pCons treatment significantly decreased (~5 fold) apoptosis in CD4+ T cells, CD8+ T cells (8-10-fold) and B220+B cells (~2.5-fold) compared to naïve or saline treated cells (Figures 4A–K). These data clearly demonstrate that pCons reduces apoptosis in both T and B cells.

Figure 4 pCons treatment alters and reduces apoptosis in both T and B cells of BWF1 mice. Female 10-12-wk old BWF1 mice were treated with pCons (1 mg i.v.). After one-two week treatment, splenocytes were obtained from both naïve and pCons-treated BWF1 mice (two/three mice in each group). RBC were lysed, cells washed, and stained with fluorochrome-labeled specific antibodies [CD4 (PerCP), CD8 (PE), B220 (APC), and Annexin V (FITC)] and flow cytometry performed. (A) Naïve unstained splenocytes; (B) Naïve CD8+ T cells; (C) Naïve CD4+ T cells; (D) Naïve B220+ B cells; (E) pCons-treated unstained splenocytes; (F) pCons-treated CD8+ T cells, (G) pCons-treated CD4+ T cells; (H) pCons-treated B220+ B cells; (I) Combined data of experiments (n=3-4) of naïve vs pCons-treated CD8+Annexin V+ T cells; (J) Combined data of experiments (n=3-4) of naïve vs pCons-treated CD4+Annexin V+ T cells; (K) Combined data of experiments (n=3-4) of naïve vs pCons-treated B220+Annexin V+ B cells. Minimum of 10,000 live cells were gated for data analyses. Dead cells were excluded from the analyses. Data was analyzed with FCS Express Ver. 7 (De Novo, Ontario, Canada). Statistical differences were determined by paired two-tailed Student’s t-test. *p < 0.05, ***p < 0.0001.
pCons Increases CD8+FoxP3+ T Cells in Healthy Human Subjects
Having examined the immunomodulatory properties of pCons in murine cells, we investigated whether pCons induces CD8+FoxP3+ T regulatory cells in healthy human subjects. To determine this, we isolated PBMCs from healthy subjects and cultured them with negative control peptide (pNeg) and pCons peptide (10 μg/ml) for 24-48 hours. After culture, cells were washed, stained with CD4, CD8, CD25 and FoxP3 fluorochrome-labeled monoclonal antibodies and analyzed by flow cytometry. As shown in Figure 5, pCons significantly increases the mean fluorescence intensity of cells expressing FoxP3 (Figure 5A) and the percentage of CD8+FoxP3+ T cells are significantly increased (~5 fold) (Figure 5B). These data suggest that pCons induces CD8+FoxP3+ Treg cells and has translational significance in humans.
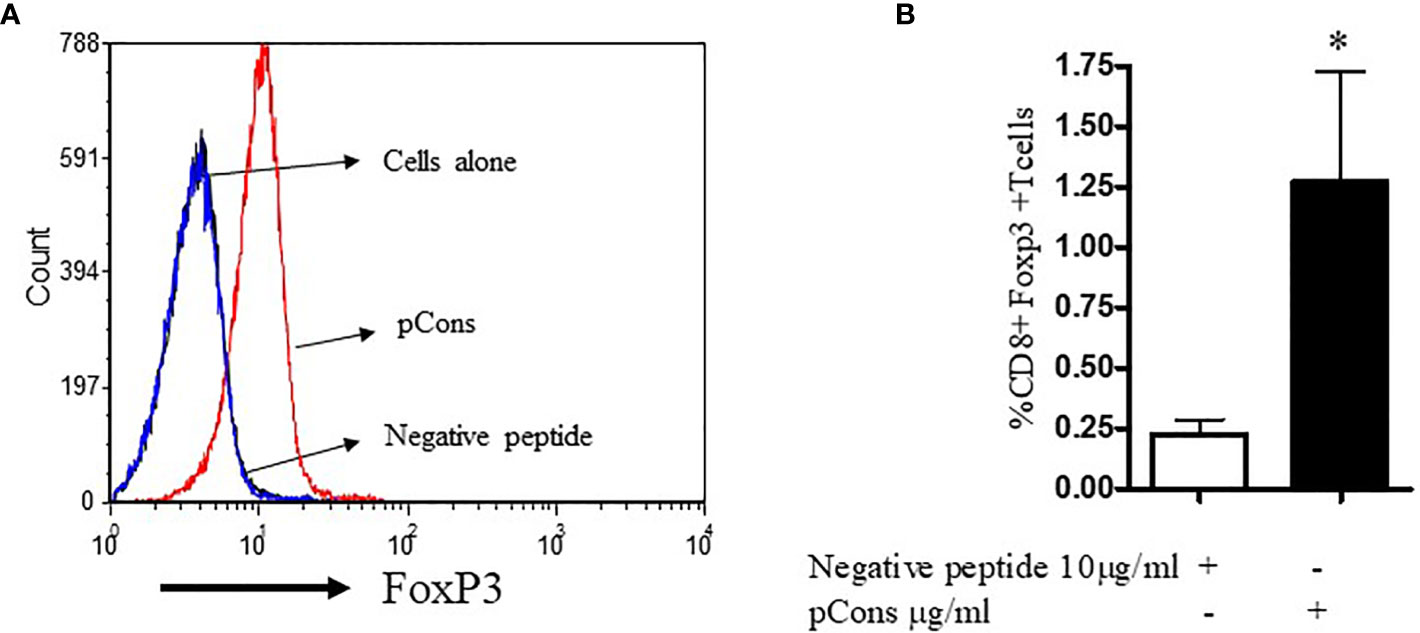
Figure 5 pCons treatment increases CD8+FoxP3+ T cells in healthy subjects. 10x106 peripheral blood mononuclear cells (PBMCs) were obtained from healthy volunteers (n=6). Cells were cultured with recombinant IL-2 (50 U/ml), negative control peptide A or pCons peptide (10 ug/ml) for 24-48 hours. Cells were stained with CD4, CD8, CD25 and FoxP3 fluorochrome-conjugated antibodies. FoxP3 intracellular staining was performed after cell surface staining. Cell were fixed and permeabilized as per manufacturer’s protocol. Minimum 10,000 cells were gated, and only live cells were used for data analyses. Dead cells were excluded from the analyses. (A) Mean fluorescence intensity (MFI) of FoxP3 among naïve, negative control peptide, and pCons-treated PBMCs of healthy subjects. (B) Percent expression of CD8+FoxP3+ T cells between negative control peptide and pCons-treated PBMCs of healthy controls. Dead cells were excluded from the analyses. Data were analyzed with FCS Express Ver. 7 (De Novo, Ontario, Canada) *p < 0.05.
Discussion
Regulatory T cells play a key role in the regulation of immune responses and maintaining immune homeostasis. Impairment in the development and function of regulatory T cells is a major contributing factor in the development of autoimmune diseases, including SLE (17, 19, 38–40). Thus, inducing and regulating the function of Treg is currently one of the prime goals not only in the study of autoimmune diseases, but graft versus host disease, organ transplantation, and neoplastic disease (41–43). In this study, we provided novel information for the immunological, cellular, and molecular phenotypes of regulatory T cells especially CD8+ Treg cells induced by the pCons treatment in BWF1 lupus mice. pCons-induced CD8+ T cells express high levels of CD62L, and low levels of CD122, PD1, CTLA4, and CCR7. We did not find major changes in the expression of 41BB. Further, pCons modulated the expression of CD25, CD28, CD122, and FoxP3 in both CD4+ and CD8+ Treg cells. The molecular phenotypes of suppressive CD8+ Treg cells include low level of regulator of G protein signaling (RGS2, RGS16, RGS17), BAX, GPT2, GADD45β, PDE3b, and PD1 (programmed cell death 1). The downregulation of these genes in pCons-tolerized CD8+ T cells was confirmed with Western blot analyses (Figure 3). Thus, our data revealed a molecular signature phenotype in CD8+ T cells induced by pCons peptide in BWF1 lupus mice that has clinical and functional importance in the immune tolerance and their immunoregulation. For example, L-selectin (CD62L) is a type-I transmembrane glycoprotein and adhesion molecule that plays an important role in T cell activation. A recent study revealed that CD62L expression on blood basophils may predict future response to standard induction therapy for patients with lupus nephritis (44). In addition, another study found that the expression levels of CD62L decreased on T cells during the inflammatory state and levels of CD8+CD62L+ T cells negatively correlated with disease severity (45). Previously glucocorticoids have been shown to increase the CD62L expression in patients with lupus (46). In agreement with our study, a recent study also showed an increase in expression of CD62L on CD8+ regulatory T cells in lupus mice (47). Thus, altogether, our finding of increased CD62L expression levels on CD8+ T cells after induction by pCons treatment in BWF1 lupus mice points to a therapeutic beneficial effect.
Recent evidence suggest that both CD8+CD122+ and CD8+C122- T cells are regulatory and can suppress autoimmunity (7, 48). Importantly, these cells express CD122 (IL-2Rβ), CD62Lhigh, PD1low and CCR7low (32, 49). Further, our data showed that pCons-induced CD8+ T cells in BWF1 mice have less cytotoxic-T lymphocyte-antigen-4 (CTLA-4) expression as compared to naïve CD8+ T cells. This is an important finding, since CTLA-4 (CD152) is an inhibitory cell-surface molecule that plays an important role in the promotion of anergy, immune regulation, and the prevention of autoimmunity. Abnormal function and susceptibility of CTLA-4 gene expression has been reported in SLE patients (50, 51). Further, it was demonstrated that CTLA-4 modulates regulatory and follicular helper T cells, thus controlling humoral immunity (52–54). CTLA-4 has been shown to downregulates CD80 and CD86 on antigen presenting cells (APC) (54). However, its precise mechanism of action has not been fully understood. Thus, our data supports the notion that pCons-induced CD8+ T cells are regulatory in nature and possess all the cellular and immunological phenotypes to induce immune tolerance.
Recent reports suggest that CCR7 was involved in the progression of lupus and its expression was increased in SLE patients (55, 56). Additionally, CCR7-CCL19 couples interaction of T helper, and B cells, and dendritic cell migration (56); thus CCR7 helps in immune complex deposition and autoantibody production. Our findings of reduction of cell surface expression of CCR7 in CD8+ T cells is important because CCR7, a G protein-coupled receptor may also help in production of TGFβ. We have previously shown that pCons-induced CD8+ T cells increase TGFβ mRNA and protein levels (3, 5, 26, 31); therefore, it is possible that TGFβ may be released via exosomes in CD8+ T cells in a CCR7-dependent manner. Exosomes are important in immunity (57), and we envision that they may play a role in CD8+ Treg-mediated suppression to establish and maintain self-tolerance. Earlier, we found that the mRNA of CCR7 was increased in pCons-tolerized CD8+ T cells (29). These differences may be due to cell surface trafficking and or differences in transcription/translational or “half-life” of CCR7 after pCons treatment in our model system. It is also conceivable that upregulated genes and cell surface receptors, e.g., IFNAR, IFI202b, may facilitate the expression, packaging, and release of TGFβ. In contrast, downregulation of CCR7 signaling may halt the inflammatory signals in pathogenic effectors CD4+ T cells, dendritic cells, antigen presenting cells (APCs), and B cells. Thus, CCR7 plays an important synergistic role in our model of immune tolerance. However, detailed mechanistic studies will be needed to address this issue.
Our findings that pCons-induced CD8+ T cells have decreased level of RGS proteins are important because reduction of RGS2 signaling increases Ca2+ mobilization and ERK1/2 activation in response to GPCR stimulation (58) which may be contributory to the observed CD8+ Treg expansion and maintenance. RGS proteins are potent GTPase-activating proteins (GAP) for heterotrimeric G protein (Gq, Gi, and Go family) alpha subunits acting as multifunctional inhibitors of signal transduction in many cells (59, 60), including “fine-tuning” GPCR signaling in lymphocytes (61). In particular, RGS2, also known as growth-inhibitory protein, plays a role in leukemogenesis (62). Thus, our finding that multiple RGS proteins are downregulated in tolerized CD8+ T cells (Figure 3) reinforces the positive effect on GPCR signaling pathways, together with reduced PDE3b, that may enhance cAMP signaling and plays an important role in the suppression of T cell function (63). The downregulation of PDE3b has been associated with enhanced insulin secretion, suggesting that secretion of other factors could also be positively modulated. Consistent with the notion that reversal of its cAMP-degrading activity is important for maintenance of CD8+ T regulatory cells. However, future studies will be required to address these possibilities.
In the current study, we found that GPT2 protein level was significantly decreased following pCons treatment in CD8+ T cells in BWF1 mice. GPT2 (glutamic-pyruvic transaminase 2) also known as alanine aminotransferase (ALT) plays an important role in gluconeogenesis and amino acid homeostasis, and is an hepatic enzyme/biomarker (64) that is upregulated in disease states. GPT2 has been shown to exacerbating autoimmune disease (65), and its levels were shown to be increased in NZB/NZW F1 mice (66). Increased serum alanine aminotransferases have been reported to be associated with anti-mitochondrial antibodies in SLE patients with autoimmune liver disease (67). Further Liu et al. found that liver injury including increased level of ALT correlates with biomarkers of autoimmunity and disease activity in patients with SLE (68). Thus, our finding that pCons treatment reduces GPT2 protein level has both clinical and translational significance in immune tolerance of our model and in SLE.
Previously, we demonstrated that pCons-induced CD8+ regulatory cells have upregulated genes including interferon inducible 202b (Ifi202b), FoxP3, Bcl2, transformation related protein 53 (TP53) and interferon receptor IFNAR1 (29). We showed previously utilizing gene silencing studies that these genes are important in the suppression of anti-DNA ab in the BWF1 lupus mice (3, 5, 26, 31). In the current study, we added novel information of candidate’s downregulated genes in CD8+ T cells. For example, the Bax gene has been implicated in lupus nephritis and in apoptosis (37) suggesting that apoptosis dysregulation in SLE was affected by polymorphic variants in apoptotic-related genes including Fas, FasL, Bcl2, and Bax. While high expression of FasL expression contributes to increased apoptosis and to the breakdown of immune tolerance favoring autoantibody production and inflammation, low expression of the Bax protein was found to be protective in the SLE patients (69). In general, apoptotic T cells and neutrophils are increased in SLE patients and have positive correlation with SLE disease activity index (70, 71). Earlier, we showed that apoptosis was decreased in pCons-induced CD8+ T cells (26). In this study, we also found decreased percent of annexin V+ T cells and B cells in pCons-treated BWF1 mice (Figure 4). We postulate that pCons treatment has direct effect on both T and B cells. In addition, pCons might have an indirect effect through CD8+ Tregs. This would require additional experiments to pinpoint the exact role. Thus, our finding of reduced Bax and annexin V in pCons-induced CD8+ T cells agrees with previous studies and suggests clinical significance. Similarly, GADD45β is a critical regulator of autoimmunity (72) that plays an important role in B cell apoptosis in response to Fas stimulation through activation of NF-κB (73). A recent report suggests that ablation of GADD45β ameliorates the inflammation and renal fibrosis caused by unilateral ureteral obstruction (UUO) in a chronic kidney disease mouse model (74). Previously it was shown that GADD45β was also induced in CD4+ T cells by inflammatory cytokines, such as IL-12 and IL-18 (75, 76). Furthermore, mRNA expression of GADD45β was associated with cytokine production and T helper cell differentiation (77, 78) and a genetic polymorphism study indicated a role for GADD45β in rheumatoid arthritis and lupus (79). Based on all these data, its plausible to hypothesize that some of the downregulated genes in pCons-induced CD8+ Treg cells may be regulated by specific miRNAs as these molecules have been identified with important roles in immune regulation (80). How these genes or their gene products cross-regulate in the overall suppression mechanism in our immune tolerance model has not been fully elucidated. Future detailed mechanistic studies are warranted to pinpoint the exact role.
In addition, we have shown herein that pCons peptide induces CD8+FoxP3+Treg cells in healthy human subjects suggesting translational significance. This finding is significant since patients with SLE have circulating T cells that can be activated by various peptides isolated from the variable regions of human anti-DNA antibodies (81, 82). Although, we were not able to study the modulation of CD8+FoxP3+Tregs in SLE patients with pCons, it may be possible that the pCons-modulation of CD8+ Tregs can be employed to reset the regulatory function of CD8+ regulatory T cells in lupus patients. Future study will be required to address this issue.
In summary, we found that pCons treatment promoted tolerogenic immune responses and modified the various cellular and molecular phenotypes in both CD8+ and CD4+ T regulatory cells in BWF1 mice. The data further demonstrate that CD8+FoxP3+ T cells can be modulated by pCons peptide in human cells indicating clinical and translational significance in SLE.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The animal study was reviewed and approved by the Chancellor Animal Review Committee (ARC), University of California, Los Angeles. The human study was reviewed and approved by the University of California, Los Angeles, Institutional Review Board (UCLA-IRB).
Author Contributions
RPS contributed to the experimental design, obtaining funding, conducting experiments, analyzing data, preparing figures, and writing of the manuscript. BHH contributed to funding and editing of the manuscript. DSB contributed to figure and manuscript editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the NIH grants AR54034, AI 083894, AI65645 to RPS; UCLA Senate Core Grant to BHH and RPS; UCLA Oppenheimer Clinical Seed Grant and American Autoimmune Related Disease Association grant to RPS.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.718359/full#supplementary-material
Supplementary Figure 1 | (A, B) Splenocytes were obtained both from naïve and pCons-treated BWF1 mice. RBC were lysed, cells washed, and stained with fluorochrome-labeled specific antibodies (CD4, CD8, CD25, CD28, CD122, CTLA-4, and 41BB). Live cell gating strategy is shown in (A) (panels A–D) for CD4+ T cells and (B) (panels E-H) for CD8+ T cells. Lymphocytes were first identified by a low forward scatter (FSC) and low side scatter (SSC) gate, and then further phenotyped for CD4 (CD4), CD8 (CD8a) and B (B220) cells, followed by gating for CD25, CD28, CTLA-4, 41BB, CD122. Intracellular FoxP3 expression was analyzed after cell fixation and permeabilization as per manufacturer’s protocol (eBiosciences, San Diego, CA, USA).
References
1. Filaci G, Bacilieri S, Fravega M, Monetti M, Contini P, Ghio M, et al. Impairment of CD8+ T Suppressor Cell Function in Patients With Active Systemic Lupus Erythematosus. J Immunol (2001) 166:6452–7. doi: 10.4049/jimmunol.166.10.6452
2. Karpouzas GA, La Cava A, Ebling FM, Singh RR, Hahn BH. Differences Between CD8+ T Cells in Lupus-Prone (NZB X NZW) F1 Mice and Healthy (BALB/c X NZW) F1 Mice may Influence Autoimmunity in the Lupus Model. Eur J Immunol (2004) 34:2489–99. doi: 10.1002/eji.200424978
3. Singh RP, La Cava A, Wong M, Ebling F, Hahn BH. CD8+ T Cell-Mediated Suppression of Autoimmunity in a Murine Lupus Model of Peptide-Induced Immune Tolerance Depends on Foxp3 Expression. J Immunol (2007) 178:7649–57. doi: 10.4049/jimmunol.178.12.7649
4. Singh RP, Hahn BH, La Cava A. Tuning Immune Suppression in Systemic Autoimmunity With Self-Derived Peptides. Inflammation Allergy Drug Targets (2008) 7:253–9. doi: 10.2174/187152808786848423
5. Singh RP, La Cava A, Hahn BH. Pconsensus Peptide Induces Tolerogenic CD8+ T Cells in Lupus-Prone (NZB X NZW)F1 Mice by Differentially Regulating Foxp3 and PD1 Molecules. J Immunol (2008) 180:2069–80. doi: 10.4049/jimmunol.180.4.2069
6. Skaggs BJ, Singh RP, Hahn BH. Induction of Immune Tolerance by Activation of CD8+ T Suppressor/Regulatory Cells in Lupus-Prone Mice. Hum Immunol (2008) 69:790–6. doi: 10.1016/j.humimm.2008.08.284
7. Suzuki M, Konya C, Goronzy JJ, Weyand CM. Inhibitory CD8+ T Cells in Autoimmune Disease. Hum Immunol (2008) 69:781–9. doi: 10.1016/j.humimm.2008.08.283
8. Kang HK, Michaels MA, Berner BR, Datta SK. Very Low-Dose Tolerance With Nucleosomal Peptides Controls Lupus and Induces Potent Regulatory T Cell Subsets. J Immunol (2005) 174:3247–55. doi: 10.4049/jimmunol.174.6.3247
9. Sharabi A, Azulai H, Sthoeger ZM, Mozes E. Clinical Amelioration of Murine Lupus by a Peptide Based on the Complementarity Determining Region-1 of an Autoantibody and by Cyclophosphamide: Similarities and Differences in the Mechanisms of Action. Immunology (2007) 121:248–57. doi: 10.1111/j.1365-2567.2007.02565.x
10. Sharabi A, Zinger H, Zborowsky M, Sthoeger ZM, Mozes E. A Peptide Based on the Complementarity-Determining Region 1 of an Autoantibody Ameliorates Lupus by Up-Regulating CD4+CD25+ Cells and TGF-Beta. Proc Natl Acad Sci USA (2006) 103:8810–5. doi: 10.1073/pnas.0603201103
11. Urowitz MB, Isenberg DA, Wallace DJ. Safety and Efficacy of Hcdr1 (Edratide) in Patients With Active Systemic Lupus Erythematosus: Results of Phase II Study. Lupus Sci Med (2015) 2:e000104. doi: 10.1136/lupus-2015-000104
12. Clemente-Casares X, Blanco J, Ambalavanan P, Yamanouchi J, Singha S, Fandos C, et al. Expanding Antigen-Specific Regulatory Networks to Treat Autoimmunity. Nature (2016) 530:434–40. doi: 10.1038/nature16962
13. Serra P, Santamaria P. Nanoparticle-Based Approaches to Immune Tolerance for the Treatment of Autoimmune Diseases. Eur J Immunol (2018) 48:751–6. doi: 10.1002/eji.201747059
14. Horwitz DA, Bickerton S, Koss M, Fahmy TM, La Cava A. Suppression of Murine Lupus by CD4+ and CD8+ Treg Cells Induced by T Cell-Targeted Nanoparticles Loaded With Interleukin-2 and Transforming Growth Factor Beta. Arthritis Rheumatol (2019) 71:632–40. doi: 10.1002/art.40773
15. Giang S, Horwitz DA, Bickerton S, La Cava A. Nanoparticles Engineered as Artificial Antigen-Presenting Cells Induce Human CD4(+) and CD8(+) Tregs That Are Functional in Humanized Mice. Front Immunol (2021) 12:628059. doi: 10.3389/fimmu.2021.628059
16. Crispin JC, Alcocer-Varela J, de Pablo P, Martinez A, Richaud-Patin Y, Alarcon-Segovia D. Immunoregulatory Defects in Patients With Systemic Lupus Erythematosus in Clinical Remission. Lupus (2003) 12:386–93. doi: 10.1191/0961203303lu368oa
17. Crispin JC, Martinez A, Alcocer-Varela J. Quantification of Regulatory T Cells in Patients With Systemic Lupus Erythematosus. J Autoimmun (2003) 21:273–6. doi: 10.1016/S0896-8411(03)00121-5
18. Liu MF, Wang CR, Fung LL, Wu CR. Decreased CD4+CD25+ T Cells in Peripheral Blood of Patients With Systemic Lupus Erythematosus. Scand J Immunol (2004) 59:198–202. doi: 10.1111/j.0300-9475.2004.01370.x
19. Miyara M, Amoura Z, Parizot C, Badoual C, Dorgham K, Trad S, et al. Global Natural Regulatory T Cell Depletion in Active Systemic Lupus Erythematosus. J Immunol (2005) 175:8392–400. doi: 10.4049/jimmunol.175.12.8392
20. Valencia X, Yarboro C, Illei G, Lipsky PE. Deficient CD4+CD25high T Regulatory Cell Function in Patients With Active Systemic Lupus Erythematosus. J Immunol (2007) 178:2579–88. doi: 10.4049/jimmunol.178.4.2579
21. Scalapino KJ, Tang Q, Bluestone JA, Bonyhadi ML, Daikh DI. Suppression of Disease in New Zealand Black/New Zealand White Lupus-Prone Mice by Adoptive Transfer of Ex Vivo Expanded Regulatory T Cells. J Immunol (2006) 177:1451–9. doi: 10.4049/jimmunol.177.3.1451
22. Horwitz DA, Gray JD, Zheng SG. The Potential of Human Regulatory T Cells Generated Ex Vivo as a Treatment for Lupus and Other Chronic Inflammatory Diseases. Arthritis Res (2002) 4:241–6. doi: 10.1186/ar414
23. La Cava A, Ebling FM, Hahn BH. Ig-Reactive CD4+CD25+ T Cells From Tolerized (New Zealand Black X New Zealand White)F1 Mice Suppress In Vitro Production of Antibodies to DNA. J Immunol (2004) 173:3542–8. doi: 10.4049/jimmunol.173.5.3542
24. Horwitz DA, Zheng SG, Gray JD, Wang JH, Ohtsuka K, Yamagiwa S. Regulatory T Cells Generated Ex Vivo as an Approach for the Therapy of Autoimmune Disease. Semin Immunol (2004) 16:135–43. doi: 10.1016/j.smim.2003.12.009
25. Dinesh RK, Skaggs BJ, La Cava A, Hahn BH, Singh RP. CD8+ Tregs in Lupus, Autoimmunity, and Beyond. Autoimmun Rev (2010) 9:560–8. doi: 10.1016/j.autrev.2010.03.006
26. Hahn BH, Singh RP, La Cava A, Ebling FM. Tolerogenic Treatment of Lupus Mice With Consensus Peptide Induces Foxp3-Expressing, Apoptosis-Resistant, TGFbeta-Secreting CD8+ T Cell Suppressors. J Immunol (2005) 175:7728–37. doi: 10.4049/jimmunol.175.11.7728
27. Hahn BH, Singh RR, Wong WK, Tsao BP, Bulpitt K, Ebling FM. Treatment With a Consensus Peptide Based on Amino Acid Sequences in Autoantibodies Prevents T Cell Activation by Autoantigens and Delays Disease Onset in Murine Lupus. Arthritis Rheum (2001) 44:432–41. doi: 10.1002/1529-0131(200102)44:2<432::AID-ANR62>3.0.CO;2-S
28. Hahn BHA, Anderson M, La Cava A. The Anti-DNA Ig Consensus Peptide pCONS Facilitates Regulatory T Cell Activity in SLE Patients. Arthritis Rheumatism (2007) 56:S546–547.
29. Singh RP, Dinesh R, Elashoff D, de Vos S, Rooney RJ, Patel D, et al. Distinct Gene Signature Revealed in White Blood Cells, CD4(+) and CD8(+) T Cells in (NZBx NZW) F1 Lupus Mice After Tolerization With Anti-DNA Ig Peptide. Genes Immun (2010) 11:294–309. doi: 10.1038/gene.2010.6
30. Hahn BH, Anderson M, Le E, La Cava A. Anti-DNA Ig Peptides Promote Treg Cell Activity in Systemic Lupus Erythematosus Patients. Arthritis Rheum (2008) 58:2488–97. doi: 10.1002/art.23609
31. Dinesh R, Hahn BH, La Cava A, Singh RP. Interferon-Inducible Gene 202b Controls CD8(+) T Cell-Mediated Suppression in Anti-DNA Ig Peptide-Treated (NZB X NZW) F1 Lupus Mice. Genes Immun (2011) 12:360–9. doi: 10.1038/gene.2011.4
32. Liu J, Chen D, Nie GD, Dai Z. CD8(+)CD122(+) T-Cells: A Newly Emerging Regulator With Central Memory Cell Phenotypes. Front Immunol (2015) 6:494. doi: 10.3389/fimmu.2015.00494
33. Dai Z, Zhang S, Xie Q, Wu S, Su J, Li S, et al. Natural CD8+CD122+ T Cells are More Potent in Suppression of Allograft Rejection Than CD4+CD25+ Regulatory T Cells. Am J Transplant (2014) 14:39–48. doi: 10.1111/ajt.12515
34. Smigiel KS, Richards E, Srivastava S, Thomas KR, Dudda JC, Klonowski KD, et al. CCR7 Provides Localized Access to IL-2 and Defines Homeostatically Distinct Regulatory T Cell Subsets. J Exp Med (2014) 211:121–36. doi: 10.1084/jem.20131142
35. Suzuki M, Jagger AL, Konya C, Shimojima Y, Pryshchep S, Goronzy JJ, et al. CD8+CD45RA+CCR7+FOXP3+ T Cells With Immunosuppressive Properties: A Novel Subset of Inducible Human Regulatory T Cells. J Immunol (2012) 189:2118–30. doi: 10.4049/jimmunol.1200122
36. Rifa’i M, Kawamoto Y, Nakashima I, Suzuki H. Essential Roles of CD8+CD122+ Regulatory T Cells in the Maintenance of T Cell Homeostasis. J Exp Med (2004) 200:1123–34. doi: 10.1084/jem.20040395
37. Badillo-Almaraz I, Daza L, Avalos-Diaz E, Herrera-Esparza R. Glomerular Expression of Fas Ligand and Bax mRNA in Lupus Nephritis. Autoimmunity (2001) 34:283–9. doi: 10.3109/08916930109014697
38. Buckner JH. Mechanisms of Impaired Regulation by CD4(+)CD25(+)FOXP3(+) Regulatory T Cells in Human Autoimmune Diseases. Nat Rev Immunol (2010) 10:849–59. doi: 10.1038/nri2889
39. Mellor-Pita S, Citores MJ, Castejon R, Tutor-Ureta P, Yebra-Bango M, Andreu JL, et al. Decrease of Regulatory T Cells in Patients With Systemic Lupus Erythematosus. Ann Rheum Dis (2006) 65:553–4. doi: 10.1136/ard.2005.044974
40. Singh RP, Bischoff DS. Sex Hormones and Gender Influence the Expression of Markers of Regulatory T Cells in SLE Patients. Front Immunol (2021) 12:619268. doi: 10.3389/fimmu.2021.619268
41. Flippe L, Bezie S, Anegon I, Guillonneau C. Future Prospects for CD8(+) Regulatory T Cells in Immune Tolerance. Immunol Rev (2019) 292:209–24. doi: 10.1111/imr.12812
42. Tsai YG, Lee CY, Lin TY, Lin CY. CD8(+) Treg Cells Associated With Decreasing Disease Activity After Intravenous Methylprednisolone Pulse Therapy in Lupus Nephritis With Heavy Proteinuria. PloS One (2014) 9:e81344. doi: 10.1371/journal.pone.0081344
43. Deng Q, Luo Y, Chang C, Wu H, Ding Y, Xiao R. The Emerging Epigenetic Role of CD8+T Cells in Autoimmune Diseases: A Systematic Review. Front Immunol (2019) 10:856. doi: 10.3389/fimmu.2019.00856
44. Halfon M, Bachelet D, Hanouna G, Dema B, Pellefigues C, Manchon P, et al. CD62L on Blood Basophils: A First Pre-Treatment Predictor of Remission in Severe Lupus Nephritis. Nephrol Dial Transplant (2020) 14:gfaa263. doi: 10.1093/ndt/gfaa263
45. Liu Y, Hoang TK, Wang T, He B, Tran DQ, Zhou J, et al. Circulating L-Selectin Expressing-T Cell Subsets Correlate With the Severity of Foxp3 Deficiency Autoimmune Disease. Int J Clin Exp Pathol (2016) 9:899–909.
46. Tan G-z, Mao Y-p, Zeng F-q. Glucocorticoids Treatment Upregulates the Expression of CD62L in CD4+CD25+T Cells in Peripheral Blood Monocytes of Patients With Systemic Lupus Erythematosus. Chin J Dermatol (2007) 40:362–4.
47. Indriyanti N, Soeroso J, Khotib J. T-Cell Activation Controlling Effects of Ethyl Acetate Fraction of Kalanchoe Pinnata (Lmk) Pers on Tmpd-Treated Lupus Mice. Int J Pharm Sci Res (2018) 9:475–82. doi: 10.13040/IJPSR.0975-8232.9(2).475-82
48. Suzuki H, Shi Z, Okuno Y, Isobe K. Are CD8+CD122+ Cells Regulatory T Cells or Memory T Cells? Hum Immunol (2008) 69:751–4. doi: 10.1016/j.humimm.2008.08.285
49. Okuno Y, Murakoshi A, Negita M, Akane K, Kojima S, Suzuki H. CD8+ CD122+ Regulatory T Cells Contain Clonally Expanded Cells With Identical CDR3 Sequences of the T-Cell Receptor Beta-Chain. Immunology (2013) 139:309–17. doi: 10.1111/imm.12067
50. Barreto M, Santos E, Ferreira R, Fesel C, Fontes MF, Pereira C, et al. Evidence for CTLA4 as a Susceptibility Gene for Systemic Lupus Erythematosus. Eur J Hum Genet (2004) 12:620–6. doi: 10.1038/sj.ejhg.5201214
51. Jury EC, Flores-Borja F, Kalsi HS, Lazarus M, Isenberg DA, Mauri C, et al. Abnormal CTLA-4 Function in T Cells From Patients With Systemic Lupus Erythematosus. Eur J Immunol (2010) 40:569–78. doi: 10.1002/eji.200939781
52. Sage PT, Paterson AM, Lovitch SB, Sharpe AH. The Coinhibitory Receptor CTLA-4 Controls B Cell Responses by Modulating T Follicular Helper, T Follicular Regulatory, and T Regulatory Cells. Immunity (2014) 41:1026–39. doi: 10.1016/j.immuni.2014.12.005
53. Wing K, Onishi Y, Prieto-Martin P, Yamaguchi T, Miyara M, Fehervari Z, et al. CTLA-4 Control Over Foxp3+ Regulatory T Cell Function. Science (2008) 322:271–5. doi: 10.1126/science.1160062
54. Wang CJ, Heuts F, Ovcinnikovs V, Wardzinski L, Bowers C, Schmidt EM, et al. CTLA-4 Controls Follicular Helper T-Cell Differentiation by Regulating the Strength of CD28 Engagement. Proc Natl Acad Sci USA (2015) 112:524–9. doi: 10.1073/pnas.1414576112
55. Yin S, Mao Y, Li X, Yue C, Zhou C, Huang L, et al. Hyperactivation and in Situ Recruitment of Inflammatory Vdelta2 T Cells Contributes to Disease Pathogenesis in Systemic Lupus Erythematosus. Sci Rep (2015) 5:14432. doi: 10.1038/srep14432
56. Clatworthy MR, Aronin CE, Mathews RJ, Morgan NY, Smith KG, Germain RN. Immune Complexes Stimulate CCR7-Dependent Dendritic Cell Migration to Lymph Nodes. Nat Med (2014) 20:1458–63. doi: 10.1038/nm.3709
57. Thery C, Zitvogel L, Amigorena S. Exosomes: Composition, Biogenesis and Function. Nat Rev Immunol (2002) 2:569–79. doi: 10.1038/nri855
58. Semplicini A, Lenzini L, Sartori M, Papparella I, Calo LA, Pagnin E, et al. Reduced Expression of Regulator of G-Protein Signaling 2 (RGS2) in Hypertensive Patients Increases Calcium Mobilization and ERK1/2 Phosphorylation Induced by Angiotensin II. J Hypertens (2006) 24:1115–24. doi: 10.1097/01.hjh.0000226202.80689.8f
59. Druey KM, Blumer KJ, Kang VH, Kehrl JH. Inhibition of G-Protein-Mediated MAP Kinase Activation by a New Mammalian Gene Family. Nature (1996) 379:742–6. doi: 10.1038/379742a0
60. Heximer SP, Blumer KJ. RGS Proteins: Swiss Army Knives in Seven-Transmembrane Domain Receptor Signaling Networks. Sci STKE (2007) 2007:pe2. doi: 10.1126/stke.3702007pe2
61. Kehrl JH. G-Protein-Coupled Receptor Signaling, RGS Proteins, and Lymphocyte Function. Crit Rev Immunol (2004) 24:409–23. doi: 10.1615/CritRevImmunol.v24.i6.20
62. Schwable J, Choudhary C, Thiede C, Tickenbrock L, Sargin B, Steur C, et al. RGS2 Is an Important Target Gene of Flt3-ITD Mutations in AML and Functions in Myeloid Differentiation and Leukemic Transformation. Blood (2005) 105:2107–14. doi: 10.1182/blood-2004-03-0940
63. Gavin MA, Rasmussen JP, Fontenot JD, Vasta V, Manganiello VC, Beavo JA, et al. Foxp3-Dependent Programme of Regulatory T-Cell Differentiation. Nature (2007) 445:771–5. doi: 10.1038/nature05543
64. Yang RZ, Blaileanu G, Hansen BC, Shuldiner AR, Gong DW. cDNA Cloning, Genomic Structure, Chromosomal Mapping, and Functional Expression of a Novel Human Alanine Aminotransferase. Genomics (2002) 79:445–50. doi: 10.1006/geno.2002.6722
65. Sundrud MS, Koralov SB, Feuerer M, Calado DP, Kozhaya AE, Rhule-Smith A, et al. Halofuginone Inhibits TH17 Cell Differentiation by Activating the Amino Acid Starvation Response. Science (2009) 324:1334–8. doi: 10.1126/science.1172638
66. Hsu TC, Huang CY, Chiang SY, Lai WX, Tsai CH, Tzang BS. Transglutaminase Inhibitor Cystamine Alleviates the Abnormality in Liver From NZB/W F1 Mice. Eur J Pharmacol (2008) 579:382–9. doi: 10.1016/j.ejphar.2007.10.059
67. Li CH, Xu PS, Wang CY, Zhang Y, Zou GL. The Presence of Anti-Mitochondrial Antibodies in Chinese Patients With Liver Involvement in Systemic Lupus Erythematosus. Rheumatol Int (2006) 26:697–703. doi: 10.1007/s00296-005-0034-y
68. Liu Y, Yu J, Oaks Z, Marchena-Mendez I, Francis L, Bonilla E, et al. Liver Injury Correlates With Biomarkers of Autoimmunity and Disease Activity and Represents an Organ System Involvement in Patients With Systemic Lupus Erythematosus. Clin Immunol (2015) 160:319–27. doi: 10.1016/j.clim.2015.07.001
69. Glesse N, Vianna P, Paim LMG, Matte MCC, Aguiar AKK, Palhano PL, et al. Evaluation of Polymorphic Variants in Apoptotic Genes and Their Role in Susceptibility and Clinical Progression to Systemic Lupus Erythematosus. Lupus (2017) 26:746–55. doi: 10.1177/0961203316678671
70. Yang X, Sun B, Wang H, Yin C, Wang X, Ji X. Increased Serum IL-10 in Lupus Patients Promotes Apoptosis of T Cell Subsets via the Caspase 8 Pathway Initiated by Fas Signaling. J BioMed Res (2015) 29:232–40. doi: 10.7555/JBR.29.20130037
71. Courtney PA, Crockard AD, Williamson K, Irvine AE, Kennedy RJ, Bell AL. Increased Apoptotic Peripheral Blood Neutrophils in Systemic Lupus Erythematosus: Relations With Disease Activity, Antibodies to Double Stranded DNA, and Neutropenia. Ann Rheum Dis (1999) 58:309–14. doi: 10.1136/ard.58.5.309
72. Liu L, Tran E, Zhao Y, Huang Y, Flavell R, Lu B. Gadd45 Beta and Gadd45 Gamma Are Critical for Regulating Autoimmunity. J Exp Med (2005) 202:1341–7. doi: 10.1084/jem.20051359
73. Zazzeroni F, Papa S, Algeciras-Schimnich A, Alvarez K, Melis T, Bubici C, et al. Gadd45 Beta Mediates the Protective Effects of CD40 Costimulation Against Fas-Induced Apoptosis. Blood (2003) 102:3270–9. doi: 10.1182/blood-2003-03-0689
74. Moon SJ, Kim JH, Choi YK, Lee CH, Hwang JH. Ablation of Gadd45beta Ameliorates the Inflammation and Renal Fibrosis Caused by Unilateral Ureteral Obstruction. J Cell Mol Med (2020) 24:8814–25. doi: 10.1111/jcmm.15519
75. Lu B, Ferrandino AF, Flavell RA. Gadd45beta Is Important for Perpetuating Cognate and Inflammatory Signals in T Cells. Nat Immunol (2004) 5:38–44. doi: 10.1038/ni1020
76. Yang J, Zhu H, Murphy TL, Ouyang W, Murphy KM. IL-18-Stimulated GADD45 Beta Required in Cytokine-Induced, But Not TCR-Induced, IFN-Gamma Production. Nat Immunol (2001) 2:157–64. doi: 10.1038/84264
77. Luo Y, Boyle DL, Hammaker D, Edgar M, Franzoso G, Firestein GS. Suppression of Collagen-Induced Arthritis in Growth Arrest and DNA Damage-Inducible Protein 45beta-Deficient Mice. Arthritis Rheum (2011) 63:2949–55. doi: 10.1002/art.30497
78. Du F, Wang L, Zhang Y, Jiang W, Sheng H, Cao Q, et al. Role of GADD45 Beta in the Regulation of Synovial Fluid T Cell Apoptosis in Rheumatoid Arthritis. Clin Immunol (2008) 128:238–47. doi: 10.1016/j.clim.2008.03.523
79. Li RN, Lin YZ, Pan YC, Lin CH, Tseng CC, Sung WY, et al. GADD45a and GADD45b Genes in Rheumatoid Arthritis and Systemic Lupus Erythematosus Patients. J Clin Med (2019) 8(6):801. doi: 10.3390/jcm8060801
80. Vinuesa CG, Rigby RJ, Yu D. Logic and Extent of miRNA-Mediated Control of Autoimmune Gene Expression. Int Rev Immunol (2009) 28:112–38. doi: 10.1080/08830180902934909
81. Kalsi JK, Ravirajan CT, Wiloch-Winska H, Blanco F, Longhurst CM, Williams W, et al. Analysis of Three New Idiotypes on Human Monoclonal Autoantibodies. Lupus (1995) 4:375–89. doi: 10.1177/096120339500400508
Keywords: pCons, regulatory T cells, systemic lupus erythematosus, anti-DNA Ab, immune tolerance, immune regulation
Citation: Singh RP, Hahn BH and Bischoff DS (2021) Cellular and Molecular Phenotypes of pConsensus Peptide (pCons) Induced CD8+ and CD4+ Regulatory T Cells in Lupus. Front. Immunol. 12:718359. doi: 10.3389/fimmu.2021.718359
Received: 31 May 2021; Accepted: 26 October 2021;
Published: 19 November 2021.
Edited by:
Ciriaco A Piccirillo, McGill University, CanadaReviewed by:
Bergithe Eikeland Oftedal, University of Bergen, NorwayAijing Liu, Second Hospital of Hebei Medical University, China
Copyright © 2021 Singh, Hahn and Bischoff. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ram P. Singh, cmFtLnNpbmdoQHZhLmdvdg==; cnBzaW5naDExQGhvdG1haWwuY29t
†Present Address: Ram P. Singh, Research Service, Veteran Administration Greater Los Angeles Healthcare System, Los Angeles, CA, United States
 Ram P. Singh
Ram P. Singh Bevra H. Hahn2,3
Bevra H. Hahn2,3 David S. Bischoff
David S. Bischoff