- 1Laboratory for Immune Homeostasis, RIKEN Center for Integrative Medical Sciences, Yokohama, Japan
- 2Graduate School of Medical Life Science, Yokohama City University, Yokohama, Japan
- 3Cell Signal Unit, Okinawa Institute of Science and Technology Graduate University, Okinawa, Japan
Development of lymphocytes is precisely regulated by various mechanisms. In addition to transcriptional rates, post-transcriptional regulation of mRNA abundance contributes to differentiation of lymphocytes. mRNA decay is a post-transcriptional mechanism controlling mRNA abundance. The carbon catabolite repression 4 (CCR4)-negative on TATA-less (NOT) complex controls mRNA longevity by catalyzing mRNA deadenylation, which is the rate-limiting step in the mRNA decay pathway. mRNA decay, regulated by the CCR4-NOT complex, is required for differentiation of pro-B to pre-B cells and V(D)J recombination in pro-B cells. In this process, it is likely that the RNA-binding proteins, ZFP36 ring finger protein like 1 and 2, recruit the CCR4-NOT complex to specific target mRNAs, thereby inducing cell quiescence of pro-B cells. A recent study showed that the CCR4-NOT complex participates in positive selection of thymocytes. Mechanistically, the CCR4-NOT deadenylase complex inhibits abnormal apoptosis by reducing the expression level of mRNAs encoding pro-apoptotic proteins, which are otherwise up-regulated during positive selection. We discuss mechanisms regulating CCR4-NOT complex-dependent mRNA decay in lymphocyte development and selection.
Introduction
Pleiotropic mechanisms control cytoplasmic mRNA abundance. Besides transcriptional regulation, post-transcriptional mechanisms are critical for controlling the level of cytoplasmic mRNA. mRNA decay is a post-transcriptional mechanism for reducing mRNA abundance. One major role of mRNA decay systems is to control homeostatic turnover and quality of mRNA. In addition, mRNA decay pathways actively regulate mRNA abundance, which is necessary to maintain and alter mRNA quantity in response to physiological signals. Many studies have suggested that active regulation of mRNA decay mechanisms must be critical for immune regulation and homeostasis (1–14). In this review, we focus on functions of the mRNA decay system regulated by the carbon catabolite repression 4 (CCR4)-negative on TATA-less (NOT) deadenylase complex in early lymphocyte development.
mRNA Decay Mechanisms
Functionally, mRNA decay comprises two classes. The first is the mRNA decay system required for RNA surveillance to prevent generation of potentially toxic proteins (15). Nonsense-mediated mRNA decay (NMD) is an mRNA quality control pathway that degrades aberrant mRNAs with premature termination codons (16–18). In addition, No-go decay and No-stop decay pathways lead to mRNA decay in cases of ribosome stalling due to accidental blockades of translation and failure of termination, respectively (19).
The second class is the mRNA decay system that actively regulates amounts of mRNA encoding functional proteins. Exonuclease and endonuclease mRNA decay pathways mainly contribute to this “active” mRNA decay (14, 16, 20). In addition, some recent studies have proposed involvement of the NMD mechanism in regulation of mRNAs encoding full-length proteins during embryonic development and tissue-specific cell differentiation (18, 21).
Initiation Of The mRNA Decay Pathway By Deadenylation
The exonuclease pathway of mRNA decay is initiated by removing polyA tails from mRNAs (16, 20). Following deadenylation of polyA tails, the 5’ cap structure of deadenylated mRNAs is removed by recruiting the decapping complex (Dcp1/Dcp2). Then 5’-3’ exoribonuclease 1 and 2 degrade decapped mRNAs from their 5′ ends.
Deadenylation of mRNA is the rate-limiting step for the exonuclease pathway. At present, three deadenylases have been reported: carbon catabolite repressor 4-negative on the TATA (CCR4-NOT) complex, polyA nuclease 2 (Pan2)-Pan3, and polyA-specific ribonuclease (PARN) (16, 20). A previous study suggested that cytoplasmic deadenylase activity of the CCR4-NOT complex predominated (22). In contrast, PAN2/3 trims relatively long tails of polyA (above 150 nt) and exerts minimal influence on the transcriptome.
In humans and mice, The CCR4-NOT complex is composed of eight protein subunits (23–25). CCR4-NOT transcription complex subunit 1 (CNOT1) serves as a scaffold to assemble the other subunits and recruits RNA-binding proteins (26–28). Two subunits, CNOT6/6L and CNOT7/8, have deadenylase activity (29–32). Other CNOT subunits (CNOT2, 3, 9,10, and 11) lack deadenylase activity and may regulate catalytic functions of the complex (6, 33, 34). Individual deletion of CNOT2, CNOT3 and CNOT10 destabilized the complex and caused degradation of other subunits (6, 33, 34). Thus, these CNOT subunits evidently also contribute to the integrity of the whole CCR4-NOT complex. In addition, CNOT2 and CNOT3 form a heterodimer that recruits RNA-binding proteins to the CCR4-NOT complex (35, 36). Moreover, in vitro reconstitution experiments showed that the CNOT2-CNOT3 heterodimer maximizes the deadenylase activity and poly (A) selectivity of the CCR4-NOT complex (37).
Ablation of genes encoding individual subunits of the CCR4-NOT complex revealed its roles in various physiological functions. For instance, CNOT3 is required for postnatal liver functions (38), pancreatic β cell function and identity (39), maintaining cardiac homeostasis (40), and bone resorption (41). CNOT7, a catalytic subunit, has non-redundant functions in spermatogenesis (42, 43). In addition, some studies showed a requirement of deadenylation induced by the CCR4-NOT complex in early lymphocyte development (3, 4, 6).
Functions Of The Ccr4-Not Complex In Lymphocyte Development
Early B cell differentiation has been widely studied (44, 45). Pro-B cells derived from common progenitor cells differentiate into pre-B cells and subsequently immature B cells. In the transition of pro-B cells to pre-B cells, generation of the immunoglobulin (Ig) μ heavy chain assembled from variable (VH), diversity (DH), and joining (JH) gene segments in pro-B cells is essential. Together with surrogate light chains, Igμ heavy chains form a precursor B cell receptor (pre-BCR). Pre-BCR signaling terminates the V(D)J recombination and induces rapid proliferation. After quiescence is re-established by later signaling from pre-BCR, recombination of light chains occurs for further differentiation of pre-B cells into immature B cells.
In B cell development, CNOT3 protein is up-regulated during differentiation from pro-B cells to pre-B cells (3). Two studies reported a requirement for CNOT3 in B cell development (3, 4). Inoue et al. (3) showed that B cell-specific deletion of Cnot3 in mice with Mbl1-Cre deleter resulted in a severe reduction of pre-B cell differentiation thereafter. Moreover, rearrangements resulting from joining of distal variable gene segments of the Ig heavy chain (Igh) gene (VH) to its diversity and joining (DH JH) gene segments were impaired, although the proximal VH to DH JH rearrangement and DH to JH rearrangement were not affected. Mechanistically, the CCR4-NOT complex mediates deadenylation of Trp53 mRNA coding p53 protein, reducing its transcript level in pro-B cells. Cnot3 deletion caused up-regulation of p53, thereby causing an increase in the expression level of pro-apoptotic genes regulated by p53. Interestingly, deletion of the p53 gene partially rescued the defect of pro-B cell differentiation due to the CNOT3 deficiency, but it did not rescue the failure of the distal VH to DH JH rearrangement. Thus, CCR4-NOT complex-mediated RNA decay ensures gene rearrangement in pro-B cells through p53-independent mechanisms and prevents abnormal apoptosis in both p53-dependent and independent mechanisms during the pro-B cell to pre-B cell transition (Figure 1).
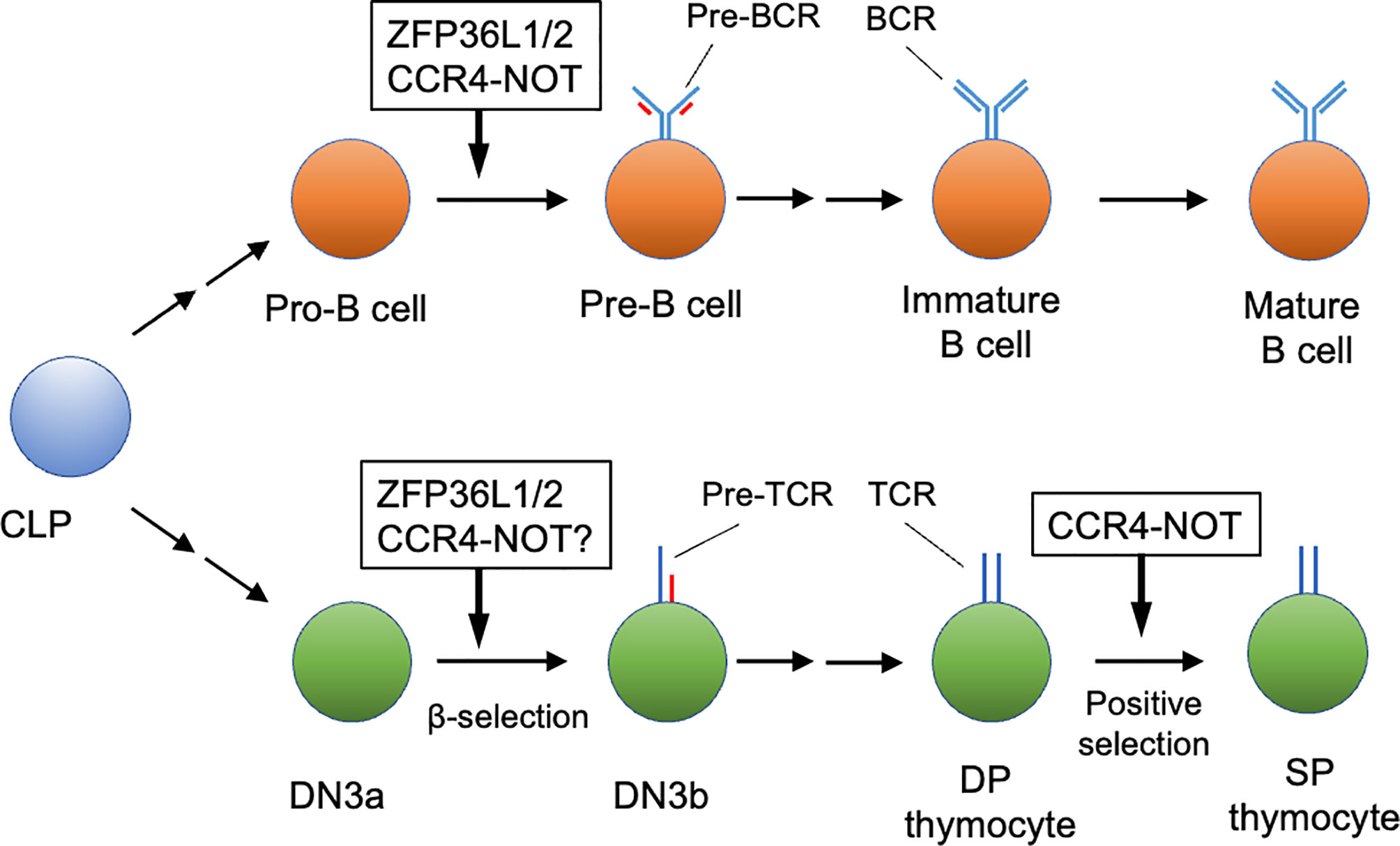
Figure 1 Early lymphocyte development regulated by the CCR4-NOT complex. The CCR4-NOT complex is required for differentiation of pro-B cells to pre-B cells and positive selection of DP thymocytes. ZFP36L1 and ZFP36L2 (ZFP36L1/2) regulate the pro-B cell to pre-B cell transition and β-selection of the DN3 thymocyte stage. Involvement of the CCR4-NOT complex in β selection has not been verified yet. Some differentiation stages were omitted for simplicity. CLP, common lymphoid progenitor; DN, CD4 CD8 double negative thymocyte; DP, CD4 CD8 double positive thymocyte; SP, CD4 or CD8 single positive thymocytes.
Yang et al. also reported a defect in the transition from pro-B cells to pre-B cells due to deletion of the Cnot3 gene (4). Because Cnot3-deletion throughout the body causes embryonic lethality, the tamoxifen-inducible Cre-driver mouse strain in addition to Mbl1-Cre mice was used. Even though both mouse lines showed pro-B cell arrest, the effect was more severe in the Cnot3 deletion in Mbl1-Cre. This difference seems to be due to genotoxic stress from excess nuclear accumulation of CRE protein, driven by the Mbl1-promoter. Notably, CNOT3 interacts with Early B-cell factor 1 (EBF1), which is critical for establishing B cell lineages. Mutation of a histidine residue in the DNA-binding domain (DBD) of EBF1 abolished its binding to CNOT3. Moreover, transduction of DBD mutant protein rescued the differentiation defect of EBF1-deficient B progenitors less efficiently in vitro compared to wild-type protein, suggesting that the interaction between CNOT3 and EBF1 is critical for B cell differentiation. Thus, the CCR4-NOT complex may function in both mRNA decay and transcription in pro-B cell differentiation. However, it is still not clear whether these two events are coupled and whether they influence each other.
Early development of conventional T cells occurs in the thymus (46). Briefly, T cell progenitors from bone marrow differentiate into CD4–CD8– (DN) thymocytes. In the DN thymocyte stage, recombination of TCRβ chain genes and their selection occurs. DN thymocytes then differentiate into CD4+CD8+ (DP) thymocytes. After completing TCRα chain recombination, DP thymocytes undergo positive selection to test the quality and specificity of the TCR αβ complex. Positively selected DP thymocytes differentiate into CD4 or CD8 single-positive (SP) cells. In the process of positive selection, DP and SP thymocytes are further separated by expression levels of surface CD3 and CD69 (47). Before positive selection, DP thymocytes express low levels of CD69 and CD3. During positive selection, CD3 and CD69 expression are up-regulated by TCR signaling. After completion, the expression level of CD3 persists, but surface CD69 expression is down-regulated.
A recent study revealed the activity of the CCR4-NOT complex in positive selection (Figure 1). In thymic T cell development, some protein subunits of the CCR4-NOT complex (CNOT1, 2, 3 and 6) are up-regulated in the transition from DN thymocytes to DP thymocytes (6). T-cell-specific deletion of the murine Cnot3 gene by the CD4-Cre deleter caused a severe developmental defect of CD4 and CD8SP thymocytes in the thymus. Specifically, the Cnot3 deletion caused a severe reduction of CD69hiCD3hi cells and CD69loCD3hi cells, but did not influence pre-selected CD69loCD3int thymocytes, suggesting that the CCR4-NOT complex is required during positive selection. mRNAs encoding pro-apoptotic molecules, DAB2-interacting protein (DAB2IP) (48) and BCL2-binding component 3 (BBC3) (49), were up-regulated and their polyA tails were elongated in Cnot3-deficient thymocytes during the course of positive selection. Moreover, transduction of anti-apoptotic Bcl-2 protein into Cnot3-deficient bone marrow progenitor cells rescued the developmental defect of thymocytes caused by Cnot3-deletion. Thus, by trimming their polyA tails, the CCR4-NOT complex promotes degradation of mRNAs encoding these pro-apoptotic proteins, which are up-regulated during positive selection. This regulation by the CCR4-NOT complex is necessary to prevent abnormal apoptosis during positive selection. Interestingly, up-regulation of Dab2ip resulting from Cnot3 deletion occurred in the CD69loCD3int and CD69hiCD3hi stages, but not in the CD69loCD3hi stage. In contrast, Bbc3 was up-regulated from the CD69hiCD3hi stage in the absence of CNOT3. Thus, the CCR4-NOT complex may prevent thymocyte apoptosis during two distinct stages of positive selection via two different mechanisms.
Upstream Events Leading To Ccr4-Not-Mediated Rna Degradation
Several mechanisms reportedly trigger RNA decay by recruiting the CCR4-NOT complex to target mRNAs (Figure 2). ZFP36 family proteins, including ZFP36, ZFP36 ring finger protein-like (ZFP36L) 1 and ZFP36L2, bind to specific sequences in 3’ untranslated regions (UTRs) of mRNAs (50, 51). The CCR4-NOT complex is recruited by ZFP36L1 and ZFP36L2 (28), and initiates deadenlyation of mRNAs bound to ZFP36L1 and ZFP36L2, thereby promoting decay of targeted mRNAs. B cell-specific deletion of both Zfp36l1 and Zfp36l2 genes caused a severe reduction of cellularity from the pre-B cell stage onward (5). Furthermore, Igμ chain expression in pro-B and early pre-B cells was reduced in these mutant mice. Thus, ZFP36L1 or ZFP36L2 is required for differentiation of pro-B cells into pre-B cells and recombination of Igh in early B cells (Figure 1). Notably, phenotypes of these mutant mice were quite similar to those of B-cell-specific Cnot3-deficient mouse lines. Transcriptome analysis of late pre-B cells in Zfp36l1 and Zfp36l2 doubly-deficient mice showed an increase in the expression level of several cell-cycle related genes. Consistently, cell cycle analysis showed that pro-B cells in S phase were significantly increased and that those in G0 phase were severely reduced by depletion of ZFP36L1 and ZFP36L2. Progression of V(D)J recombination requires cellular quiescence because expression of RAG2 protein is restricted to G0 and G1 phase (52–55). Thus, ZFP36L1 and ZFP36L2 redundantly suppress cell cycle progression in early B cell stages via degradation of mRNAs encoding cell cycle-related genes, which may be required to promote V(D)J recombination. Putative target mRNAs of ZFP36L1 were upregulated in Cnot3-deficient pro-B cells (3). In addition, as described, CNOT3 deficiency resulted in failure of V(D)J recombination in pro-B cells in a p53-independent manner (3). Overall, these findings suggest that ZFP36L1 or ZFP36L2 recruits the CCR4-NOT complex and leads to degradation of mRNAs encoding cell cycle-promoting genes, thereby regulating cell cycle entry and exit to promote progression of V(D)J recombination. Because Zfp36l1 and Zfp36l2 genes are expressed throughout B cell development, there may be up-stream events activating ZFP36L1 and ZFP36L2, or promoting recruitment to their target mRNAs.
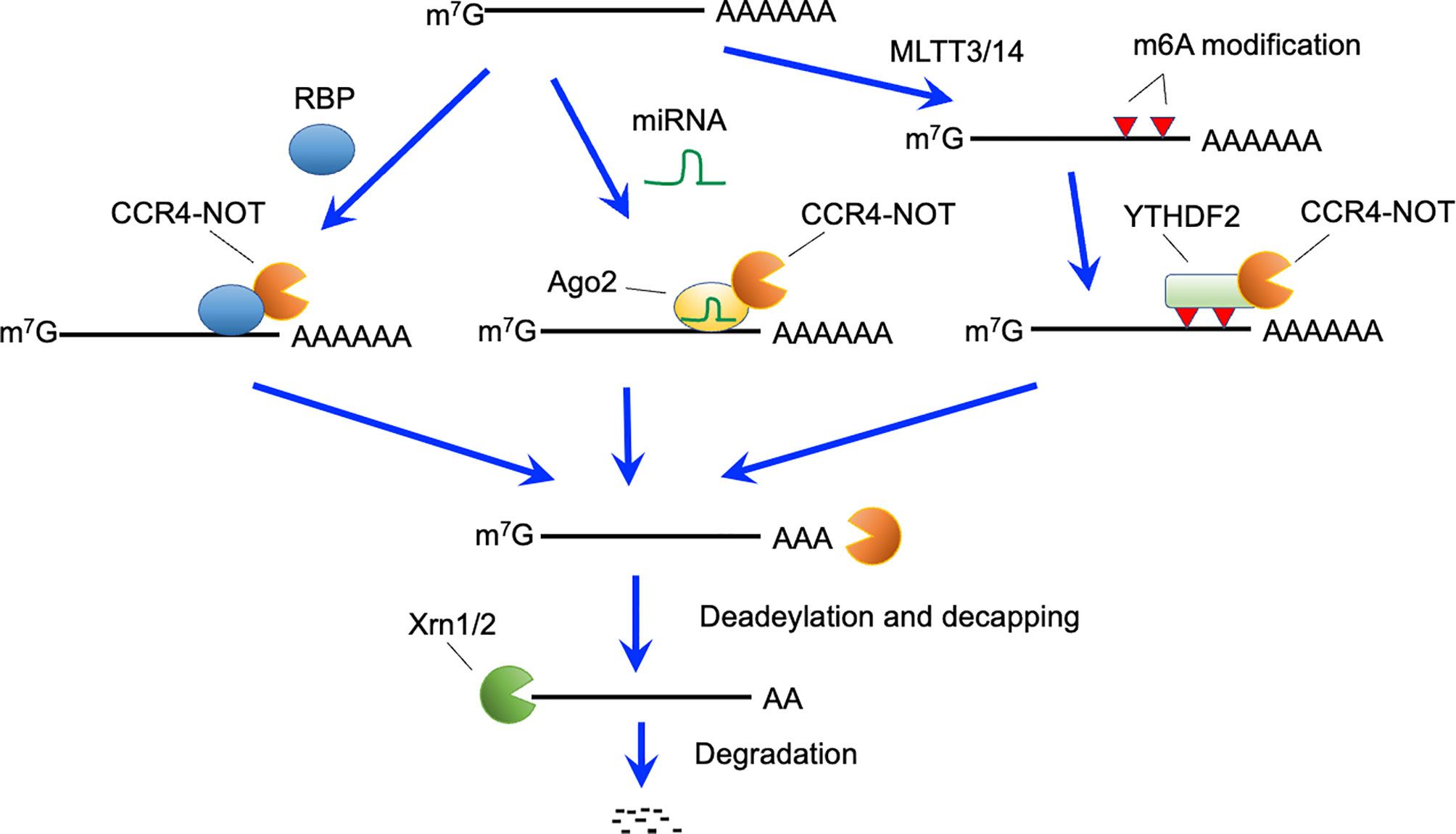
Figure 2 Active RNA decay pathways mediated by the CCR4-NOT complex. The CCR4-NOT complex can be recruited by RNA binding proteins (RBP), miRNA-Ago2 complex, and YTHDF2 bound to N6-methyladeonsine, which is generated by the METTL3 and METTL14 methyltransferase complexes. The CCR4-NOT complex deadenylates polyA tails of recruited mRNAs. After deadenylation, 5’-decapping enzymes are recruited and eliminate the cap structure. Finally, 5’-exonucleases (Xrn1 and 2) causes degradation of target mRNAs.
In addition to B cell development, ZFP36L1 and ZFP36L2 regulate early T cell differentiation. As in B cells, Zfp36l1 and Zfp36l2 genes are expressed throughout thymocyte development. Mice in which both ZFP36L1 and ZFP36L2 were depleted in early T cell progenitors developed T cell acute lymphoblastic leukemia (56). Importantly, V(D)J recombination of TCRβ gene was defective in DN thymocytes of these doubly-deficient mice. Thus, DN thymocytes in mutant mice by-pass the β-selection checkpoint without expression of TCRβ and are converted into T lymphoblasts. Mechanistically, the ZFP36L1- and ZFP36L2-dependent RNA decay pathway appeared to inhibit up-regulation of genes involved in DNA damage-response and cell proliferation (57). This suppression appeared crucial to induce cellular quiescence required for V(D)J recombination in DN thymocytes. Considering that V(D)J recombination occurs in both DN thymocytes and pro-B cell stages, functions of ZFP36L1 and ZFP36L2 may be similar in these two cell types (Figure 1). Although this may imply a requirement for CCR4-NOT complex-dependent RNA decay in the DN stages, involvement of the CCR4-NOT complex in V(D)J recombination of DN thymocytes has not been addressed yet.
Roquin family proteins, ROQUIN-1 and ROQUIN-2, can also recruit the CCR4-NOT complex in the 3’-UTRs of target mRNAs (58). T cell-specific deletion of both Rc3h1 and Rc3h2 (encoding ROQUIN-1 and ROQUIN-2, respectively) genes resulted in enhanced helper T cell activation and follicular helper T cell differentiation with spontaneous inflammation (10). Puzzlingly, these phenotypes have not been observed in T-cell-specific Cnot3-deficient mice. However, because T-cell-specific deletion of Cnot3 severely impaired mature T cell development in the thymus, the phenotype that characterized the later T cell stage needs to be clarified. Alternatively, other mRNA decay systems may function downstream of these ROQUIN family proteins.
Among epigenetic modifications of mRNA, the N6-methyl adenosine (m6A) modification in RNA is intensively studied (59, 60). The m6A modification is recognized by the YT521-B homology domain-containing family 2 that recruits the CCR4-NOT complex (61) (Figure 2). Thus, m6A modification can lead to CCR4-NOT complex-mediated RNA decay. The N6-methylation of adenosine is catalyzed by methyl transferase (METTL) 3 and METTL14 complexes (62). CD4-Cre-mediated deletion of the Mettl3 gene suppressed homeostatic proliferation of peripheral CD4-positive T cells under lymphopenic conditions (63). Socs mRNA degradation induced by interleukin 7-signaling, which is required for proliferation and differentiation of naïve T cells, was impaired in these mice. Notably, again, the link between m6A modification and CCR4-NOT complex-mediated mRNA decay is still obscure in T cells because of the severe reduction of mature naive T cells in T cell-specific Cnot3-deficient mice.
miRNAs form complexes with Argonaute family proteins on their target sequences (64, 65). The miRNA- Argonaute family complex recruits the CCR4-NOT complex or the PAN2-PAN3 complex, leading to degradation of target mRNAs (64, 65). Conditional depletion of Dicer, an enzyme critical for miRNA generation (66) in early B cells inhibited the transition of pro-B to pre-B cells, suggesting a requirement of miRNA for this differentiation stage (67). This phenotype is similar to that in mice deficient for CNOT3. However, the Dicer deficiency did not impair the V(D)J recombination reaction, whereas generation of an antibody repertoire was disturbed (67). Thus, the miRNA-CCR4-NOT complex axis in early B cells still needs to be clarified.
Although deletion of the Dicer gene in early thymocytes by the Lck-Cre deleter caused a severe reduction in cellularity of thymocytes, the percentage of each thymocyte fraction was not affected (68). Moreover, Dicer deletion by CD4-Cre showed normal thymocyte number and percentages of thymocyte subsets (69). Overall, miRNA-dependent mRNA decay through recruitment of the CCR4-NOT complex might not be essential for early thymocyte development and selection.
Concluding Remarks
Rapid, simultaneous and dynamic transcription of several mRNAs occurs during cell differentiation for generation of functional proteins. In addition, protein abundance must be precisely regulated to avoid deleterious cellular consequences. Regulation of mRNA levels should be an efficient way to control protein expression levels because one mRNA molecule generates about 3000 protein molecules on average in mammalian cells (70). However, once proteins are produced, RNA decay mechanisms should be less effective than direct protein degradation for regulating protein levels. Accordingly, it is likely that RNA decay-dependent regulation of protein concentrations by the CCR4-NOT complex is most effective when cells receive signals initiating production of large numbers of mRNA transcripts, such as during lymphocyte development.
It is likely that ZFP36L family proteins recruit the CCR4-NOT complex to suppress cell-cycle-related genes and DNA damage-responsive genes during early lymphocyte differentiation. In contrast, upstream events of CCR4-NOT complex-dependent regulation of thymic positive selection remain unknown. Moreover, possible upstream mechanisms of the CCR4-NOT complex, i.e., ROQUIN family, m6A modification, are necessary to regulate differentiation and functions of lymphocytes in later differentiation stages. Therefore, it is also important to address whether CCR4-NOT complex-induced RNA decay controls these lymphocyte differentiation and functions.
Finally, given that recent studies on human disease progression and onset by dysregulation of RNA decay systems (71–74), understanding RNA decay mechanisms would be beneficial for developing therapies and preventive measures against such diseases.
Author Contributions
TA wrote the first draft of the manuscript and TY critically reviewed it. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science, Japan (20H03441) (TA), and CREST from Japan Science and Technology Agency (JPMJCR2011) (TA), and funding from the Okinawa Institute of Science and Technology Graduate University, Japan (TY).
References
1. Matsushita K, Takeuchi O, Standley DM, Kumagai Y, Kawagoe T, Miyake T, et al. Zc3h12a Is an RNase Essential for Controlling Immune Responses by Regulating mRNA Decay. Nature (2009) 458(7242):1185–90. doi: 10.1038/nature07924
2. Yu D, Tan AH, Hu X, Athanasopoulos V, Simpson N, Silva DG, et al. Roquin Represses Autoimmunity by Limiting Inducible T-Cell Co-Stimulator Messenger RNA. Nature (2007) 450(7167):299–303. doi: 10.1038/nature06253
3. Inoue T, Morita M, Hijikata A, Fukuda-Yuzawa Y, Adachi S, Isono K, et al. CNOT3 Contributes to Early B Cell Development by Controlling Igh Rearrangement and p53 mRNA Stability. J Exp Med (2015) 212(9):1465–79. doi: 10.1084/jem.20150384
4. Yang CY, Ramamoorthy S, Boller S, Rosenbaum M, Rodriguez Gil A, Mittler G, et al. Interaction of CCR4-NOT With EBF1 Regulates Gene-Specific Transcription and mRNA Stability in B Lymphopoiesis. Genes Dev (2016) 30(20):2310–24. doi: 10.1101/gad.285452.116
5. Galloway A, Saveliev A, Lukasiak S, Hodson DJ, Bolland D, Balmanno K, et al. RNA-Binding Proteins ZFP36L1 and ZFP36L2 Promote Cell Quiescence. Science (2016) 352(6284):453–9. doi: 10.1126/science.aad5978
6. Ito-Kureha T, Miyao T, Nishijima S, Suzuki T, Koizumi SI, Villar-Briones A, et al. The CCR4-NOT Deadenylase Complex Safeguards Thymic Positive Selection by Down-Regulating Aberrant Pro-Apoptotic Gene Expression. Nat Commun (2020) 11(1):6169. doi: 10.1038/s41467-020-19975-4
7. Vinuesa CG, Cook MC, Angelucci C, Athanasopoulos V, Rui L, Hill KM, et al. A RING-type Ubiquitin Ligase Family Member Required to Repress Follicular Helper T Cells and Autoimmunity. Nature (2005) 435(7041):452–8. doi: 10.1038/nature03555
8. Lee SK, Silva DG, Martin JL, Pratama A, Hu X, Chang PP, et al. Interferon-Gamma Excess Leads to Pathogenic Accumulation of Follicular Helper T Cells and Germinal Centers. Immunity (2012) 37(5):880–92. doi: 10.1016/j.immuni.2012.10.010
9. Linterman MA, Rigby RJ, Wong RK, Yu D, Brink R, Cannons JL, et al. Follicular Helper T Cells Are Required for Systemic Autoimmunity. J Exp Med (2009) 206(3):561–76. doi: 10.1084/jem.20081886
10. Vogel KU, Edelmann SL, Jeltsch KM, Bertossi A, Heger K, Heinz GA, et al. Roquin Paralogs 1 and 2 Redundantly Repress the Icos and Ox40 Costimulator mRNAs and Control Follicular Helper T Cell Differentiation. Immunity (2013) 38(4):655–68. doi: 10.1016/j.immuni.2012.12.004
11. Essig K, Hu D, Guimaraes JC, Alterauge D, Edelmann S, Raj T, et al. Roquin Suppresses the PI3K-mTOR Signaling Pathway to Inhibit T Helper Cell Differentiation and Conversion of Treg to Tfr Cells. Immunity (2017) 47(6):1067–82 e12. doi: 10.1016/j.immuni.2017.11.008
12. Taylor GA, Carballo E, Lee DM, Lai WS, Thompson MJ, Patel DD, et al. A Pathogenetic Role for TNF Alpha in the Syndrome of Cachexia, Arthritis, and Autoimmunity Resulting From Tristetraprolin (TTP) Deficiency. Immunity (1996) 4(5):445–54. doi: 10.1016/S1074-7613(00)80411-2
13. Molle C, Zhang T, Ysebrant de Lendonck L, Gueydan C, Andrianne M, Sherer F, et al. Tristetraprolin Regulation of Interleukin 23 mRNA Stability Prevents a Spontaneous Inflammatory Disease. J Exp Med (2013) 210(9):1675–84. doi: 10.1084/jem.20120707
14. Akiyama T, Suzuki T, Yamamoto T. RNA Decay Machinery Safeguards Immune Cell Development and Immunological Responses. Trends Immunol (2021) 42(5):447–60. doi: 10.1016/j.it.2021.03.008
15. Wolin SL, Maquat LE. Cellular RNA Surveillance in Health and Disease. Science (2019) 366(6467):822–7. doi: 10.1126/science.aax2957
16. Labno A, Tomecki R, Dziembowski A. Cytoplasmic RNA Decay Pathways - Enzymes and Mechanisms. Biochim Biophys Acta (2016) 1863(12):3125–47. doi: 10.1016/j.bbamcr.2016.09.023
17. Yi Z, Sanjeev M, Singh G. The Branched Nature of the Nonsense-Mediated Mrna Decay Pathway. Trends Genet (2021) 37(2):143–59. doi: 10.1016/j.tig.2020.08.010
18. Nasif S, Contu L, Muhlemann O. Beyond Quality Control: The Role of Nonsense-Mediated mRNA Decay (NMD) in Regulating Gene Expression. Semin Cell Dev Biol (2018) 75:78–87. doi: 10.1016/j.semcdb.2017.08.053
19. Powers KT, Szeto JA, Schaffitzel C. New Insights Into No-Go, Non-Stop and Nonsense-Mediated mRNA Decay Complexes. Curr Opin Struct Biol (2020) 65:110–8. doi: 10.1016/j.sbi.2020.06.011
20. Schoenberg DR, Maquat LE. Regulation of Cytoplasmic mRNA Decay. Nat Rev Genet (2012) 13(4):246–59. doi: 10.1038/nrg3160
21. Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense Surveillance Regulates Expression of Diverse Classes of Mammalian Transcripts and Mutes Genomic Noise. Nat Genet (2004) 36(10):1073–8. doi: 10.1038/ng1429
22. Yi H, Park J, Ha M, Lim J, Chang H, Kim VN. Pabp Cooperates With the CCR4-NOT Complex to Promote Mrna Deadenylation and Block Precocious Decay. Mol Cell (2018) 70(6):1081–8.e5. doi: 10.1016/j.molcel.2018.05.009
23. Collart MA. The Ccr4-Not Complex Is a Key Regulator of Eukaryotic Gene Expression. Wiley Interdiscip Rev RNA (2016) 7(4):438–54. doi: 10.1002/wrna.1332
24. Inada T, Makino S. Novel Roles of the Multi-Functional CCR4-NOT Complex in Post-Transcriptional Regulation. Front Genet (2014) 5:135. doi: 10.3389/fgene.2014.00135
25. Shirai YT, Suzuki T, Morita M, Takahashi A, Yamamoto T. Multifunctional Roles of the Mammalian CCR4-NOT Complex in Physiological Phenomena. Front Genet (2014) 5:286. doi: 10.3389/fgene.2014.00286
26. Sandler H, Kreth J, Timmers HT, Stoecklin G. Not1 Mediates Recruitment of the Deadenylase Caf1 to mRNAs Targeted for Degradation by Tristetraprolin. Nucleic Acids Res (2011) 39(10):4373–86. doi: 10.1093/nar/gkr011
27. Petit AP, Wohlbold L, Bawankar P, Huntzinger E, Schmidt S, Izaurralde E, et al. The Structural Basis for the Interaction Between the CAF1 Nuclease and the NOT1 Scaffold of the Human CCR4-NOT Deadenylase Complex. Nucleic Acids Res (2012) 40(21):11058–72. doi: 10.1093/nar/gks883
28. Fabian MR, Frank F, Rouya C, Siddiqui N, Lai WS, Karetnikov A, et al. Structural Basis for the Recruitment of the Human CCR4-NOT Deadenylase Complex by Tristetraprolin. Nat Struct Mol Biol (2013) 20(6):735–+. doi: 10.1038/nsmb.2572
29. Chen J, Chiang YC, Denis CL. CCR4, a 3’-5’ Poly(a) RNA and ssDNA Exonuclease, Is the Catalytic Component of the Cytoplasmic Deadenylase. EMBO J (2002) 21(6):1414–26. doi: 10.1093/emboj/21.6.1414
30. Viswanathan P, Ohn T, Chiang YC, Chen J, Denis CL. Mouse CAF1 can Function as a Processive Deadenylase/3’-5’-Exonuclease In Vitro But in Yeast the Deadenylase Function of CAF1 Is Not Required for mRNA Poly(a) Removal. J Biol Chem (2004) 279(23):23988–95. doi: 10.1074/jbc.M402803200
31. Bianchin C, Mauxion F, Sentis S, Seraphin B, Corbo L. Conservation of the Deadenylase Activity of Proteins of the Caf1 Family in Human. RNA (2005) 11(4):487–94. doi: 10.1261/rna.7135305
32. Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CY, et al. Concerted Action of Poly(a) Nucleases and Decapping Enzyme in Mammalian mRNA Turnover. Nat Struct Mol Biol (2005) 12(12):1054–63. doi: 10.1038/nsmb1016
33. Ito K, Inoue T, Yokoyama K, Morita M, Suzuki T, Yamamoto T. CNOT2 Depletion Disrupts and Inhibits the CCR4-NOT Deadenylase Complex and Induces Apoptotic Cell Death. Genes Cells (2011) 16(4):368–79. doi: 10.1111/j.1365-2443.2011.01492.x
34. Farber V, Erben E, Sharma S, Stoecklin G, Clayton C. Trypanosome CNOT10 Is Essential for the Integrity of the NOT Deadenylase Complex and for Degradation of Many Mrnas. Nucleic Acids Res (2013) 41(2):1211–22. doi: 10.1093/nar/gks1133
35. Bhandari D, Raisch T, Weichenrieder O, Jonas S, Izaurralde E. Structural Basis for the Nanos-Mediated Recruitment of the CCR4-NOT Complex and Translational Repression. Genes Dev (2014) 28(8):888–901. doi: 10.1101/gad.237289.113
36. Raisch T, Bhandari D, Sabath K, Helms S, Valkov E, Weichenrieder O, et al. Distinct Modes of Recruitment of the CCR4-NOT Complex by Drosophila and Vertebrate Nanos. EMBO J (2016) 35(9):974–90. doi: 10.15252/embj.201593634
37. Raisch T, Chang CT, Levdansky Y, Muthukumar S, Raunser S, Valkov E. Reconstitution of Recombinant Human CCR4-NOT Reveals Molecular Insights Into Regulated Deadenylation. Nat Commun (2019) 10(1):3173. doi: 10.1038/s41467-019-11094-z
38. Suzuki T, Kikuguchi C, Nishijima S, Nagashima T, Takahashi A, Okada M, et al. Postnatal Liver Functional Maturation Requires Cnot Complex-Mediated Decay of mRNAs Encoding Cell Cycle and Immature Liver Genes. Development (2019) 146(4):dev168146. doi: 10.1242/dev.168146
39. Mostafa D, Yanagiya A, Georgiadou E, Wu Y, Stylianides T, Rutter GA, et al. Loss of Beta-Cell Identity and Diabetic Phenotype in Mice Caused by Disruption of CNOT3-dependent mRNA Deadenylation. Commun Biol (2020) 3(1):476. doi: 10.1038/s42003-020-01201-y
40. Yamaguchi T, Suzuki T, Sato T, Takahashi A, Watanabe H, Kadowaki A, et al. The CCR4-NOT Deadenylase Complex Controls Atg7-dependent Cell Death and Heart Function. Sci Signal (2018) 11(516). doi: 10.1126/scisignal.aan3638
41. Watanabe C, Morita M, Hayata T, Nakamoto T, Kikuguchi C, Li X, et al. Stability of mRNA Influences Osteoporotic Bone Mass Via CNOT3. Proc Natl Acad Sci USA (2014) 111(7):2692–7. doi: 10.1073/pnas.1316932111
42. Nakamura T, Yao R, Ogawa T, Suzuki T, Ito C, Tsunekawa N, et al. Oligo-Astheno-Teratozoospermia in Mice Lacking Cnot7, a Regulator of Retinoid X Receptor Beta. Nat Genet (2004) 36(5):528–33. doi: 10.1038/ng1344
43. Berthet C, Morera AM, Asensio MJ, Chauvin MA, Morel AP, Dijoud F, et al. CCR4-Associated Factor CAF1 is an Essential Factor for Spermatogenesis. Mol Cell Biol (2004) 24(13):5808–20. doi: 10.1128/MCB.24.13.5808-5820.2004
44. Melchers F. Checkpoints That Control B Cell Development. J Clin Invest (2015) 125(6):2203–10. doi: 10.1172/JCI78083
45. Clark MR, Mandal M, Ochiai K, Singh H. Orchestrating B Cell Lymphopoiesis Through Interplay of IL-7 Receptor and Pre-B Cell Receptor Signalling. Nat Rev Immunol (2014) 14(2):69–80. doi: 10.1038/nri3570
46. Rothenberg EV. Programming for T-lymphocyte Fates: Modularity and Mechanisms. Genes Dev (2019) 33(17-18):1117–35. doi: 10.1101/gad.327163.119
47. Hare KJ, Jenkinson EJ, Anderson G. CD69 Expression Discriminates MHC-dependent and -Independent Stages of Thymocyte Positive Selection. J Immunol (1999) 162(7):3978–83.
48. Xie D, Gore C, Zhou J, Pong RC, Zhang H, Yu L, et al. DAB2IP Coordinates Both PI3K-Akt and ASK1 Pathways for Cell Survival and Apoptosis. Proc Natl Acad Sci USA (2009) 106(47):19878–83. doi: 10.1073/pnas.0908458106
49. Yu J, Zhang L. PUMA, a Potent Killer With or Without P53. Oncogene (2008) 27(Suppl 1):S71–83. doi: 10.1038/onc.2009.45
50. Adachi S, Homoto M, Tanaka R, Hioki Y, Murakami H, Suga H, et al. ZFP36L1 and ZFP36L2 Control LDLR mRNA Stability Via the ERK-RSK Pathway. Nucleic Acids Res (2014) 42(15):10037–49. doi: 10.1093/nar/gku652
51. Otsuka H, Fukao A, Tomohiro T, Adachi S, Suzuki T, Takahashi A, et al. ARE-Binding Protein ZFP36L1 Interacts With CNOT1 to Directly Repress Translation Via a Deadenylation-Independent Mechanism. Biochimie (2020) 174:49–56. doi: 10.1016/j.biochi.2020.04.010
52. Li Z, Dordai DI, Lee J, Desiderio S. A Conserved Degradation Signal Regulates RAG-2 Accumulation During Cell Division and Links V(D)J Recombination to the Cell Cycle. Immunity (1996) 5(6):575–89. doi: 10.1016/S1074-7613(00)80272-1
53. Zhang L, Reynolds TL, Shan X, Desiderio S. Coupling of V(D)J Recombination to the Cell Cycle Suppresses Genomic Instability and Lymphoid Tumorigenesis. Immunity (2011) 34(2):163–74. doi: 10.1016/j.immuni.2011.02.003
54. Johnson K, Chaumeil J, Micsinai M, Wang JM, Ramsey LB, Baracho GV, et al. IL-7 Functionally Segregates the Pro-B Cell Stage by Regulating Transcription of Recombination Mediators Across Cell Cycle. J Immunol (2012) 188(12):6084–92. doi: 10.4049/jimmunol.1200368
55. Bendall SC, Davis KL, Amir el AD, Tadmor MD, Simonds EF, Chen TJ, et al. Single-Cell Trajectory Detection Uncovers Progression and Regulatory Coordination in Human B Cell Development. Cell (2014) 157(3):714–25. doi: 10.1016/j.cell.2014.04.005
56. Hodson DJ, Janas ML, Galloway A, Bell SE, Andrews S, Li CM, et al. Deletion of the RNA-binding Proteins ZFP36L1 and ZFP36L2 Leads to Perturbed Thymic Development and T Lymphoblastic Leukemia. Nat Immunol (2010) 11(8):717–24. doi: 10.1038/ni.1901
57. Vogel KU, Bell LS, Galloway A, Ahlfors H, Turner M. The RNA-Binding Proteins Zfp36l1 and Zfp36l2 Enforce the Thymic Beta-Selection Checkpoint by Limiting DNA Damage Response Signaling and Cell Cycle Progression. J Immunol (2016) 197(7):2673–85. doi: 10.4049/jimmunol.1600854
58. Leppek K, Schott J, Reitter S, Poetz F, Hammond MC, Stoecklin G. Roquin Promotes Constitutive mRNA Decay Via a Conserved Class of Stem-Loop Recognition Motifs. Cell (2013) 153(4):869–81. doi: 10.1016/j.cell.2013.04.016
59. Shulman Z, Stern-Ginossar N. The RNA Modification N(6)-methyladenosine as a Novel Regulator of the Immune System. Nat Immunol (2020) 21(5):501–12. doi: 10.1038/s41590-020-0650-4
60. Yang C, Hu Y, Zhou B, Bao Y, Li Z, Gong C, et al. The Role of M(6)a Modification in Physiology and Disease. Cell Death Dis (2020) 11(11):960. doi: 10.1038/s41419-020-03143-z
61. Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, et al. YTHDF2 Destabilizes M(6)a-Containing RNA Through Direct Recruitment of the CCR4-NOT Deadenylase Complex. Nat Commun (2016) 7:12626. doi: 10.1038/ncomms12626
62. Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 Complex Mediates Mammalian Nuclear RNA N6-Adenosine Methylation. Nat Chem Biol (2014) 10(2):93–5. doi: 10.1038/nchembio.1432
63. Li HB, Tong J, Zhu S, Batista PJ, Duffy EE, Zhao J, et al. M(6)a mRNA Methylation Controls T Cell Homeostasis by Targeting the IL-7/STAT5/SOCS Pathways. Nature (2017) 548(7667):338–42. doi: 10.1038/nature23450
64. Iwakawa HO, Tomari Y. The Functions of MicroRNAs: mRNA Decay and Translational Repression. Trends Cell Biol (2015) 25(11):651–65. doi: 10.1016/j.tcb.2015.07.011
65. Jonas S, Izaurralde E. Towards a Molecular Understanding of microRNA-mediated Gene Silencing. Nat Rev Genet (2015) 16(7):421–33. doi: 10.1038/nrg3965
66. Ha M, Kim VN. Regulation of microRNA Biogenesis. Nat Rev Mol Cell Biol (2014) 15(8):509–24. doi: 10.1038/nrm3838
67. Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, et al. Dicer Ablation Affects Antibody Diversity and Cell Survival in the B Lymphocyte Lineage. Cell (2008) 132(5):860–74. doi: 10.1016/j.cell.2008.02.020
68. Cobb BS, Nesterova TB, Thompson E, Hertweck A, O’Connor E, Godwin J, et al. T Cell Lineage Choice and Differentiation in the Absence of the RNase III Enzyme Dicer. J Exp Med (2005) 201(9):1367–73. doi: 10.1084/jem.20050572
69. Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T Cell Differentiation in the Absence of Dicer. J Exp Med (2005) 202(2):261–9. doi: 10.1084/jem.20050678
70. Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, et al. Global Quantification of Mammalian Gene Expression Control. Nature (2011) 473(7347):337–42. doi: 10.1038/nature10098
71. De Keersmaecker K, Atak ZK, Li N, Vicente C, Patchett S, Girardi T, et al. Exome Sequencing Identifies Mutation in CNOT3 and Ribosomal Genes RPL5 and RPL10 in T-Cell Acute Lymphoblastic Leukemia. Nat Genet (2013) 45(2):186–90. doi: 10.1038/ng.2508
72. Tavernier SJ, Athanasopoulos V, Verloo P, Behrens G, Staal J, Bogaert DJ, et al. A Human Immune Dysregulation Syndrome Characterized by Severe Hyperinflammation With a Homozygous Nonsense Roquin-1 Mutation. Nat Commun (2019) 10(1):4779. doi: 10.1038/s41467-019-12704-6
73. Kakiuchi N, Yoshida K, Uchino M, Kihara T, Akaki K, Inoue Y, et al. Frequent Mutations That Converge on the NFKBIZ Pathway in Ulcerative Colitis. Nature (2020) 577(7789):260–5. doi: 10.1038/s41586-019-1856-1
Keywords: mRNA decay, CCR4-NOT complex, lymphocyte development, Apoptosis, VDJ recombination
Citation: Akiyama T and Yamamoto T (2021) Regulation of Early Lymphocyte Development via mRNA Decay Catalyzed by the CCR4-NOT Complex. Front. Immunol. 12:715675. doi: 10.3389/fimmu.2021.715675
Received: 27 May 2021; Accepted: 05 July 2021;
Published: 19 July 2021.
Edited by:
Osamu Takeuchi, Kyoto University, JapanReviewed by:
Xiaocui He, La Jolla Institute for Immunology (LJI), United StatesRyuya Fukunaga, Johns Hopkins University, United States
Copyright © 2021 Akiyama and Yamamoto. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Taishin Akiyama, dGFpc2hpbi5ha2l5YW1hQHJpa2VuLmpw
 Taishin Akiyama
Taishin Akiyama Tadashi Yamamoto3
Tadashi Yamamoto3