
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 02 August 2021
Sec. Mucosal Immunity
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.712915
 Gilda Varricchi1,2,3*†
Gilda Varricchi1,2,3*† Remo Poto1,2†
Remo Poto1,2† Gianluca Ianiro4
Gianluca Ianiro4 Alessandra Punziano1,2
Alessandra Punziano1,2 Gianni Marone1,2,3
Gianni Marone1,2,3 Antonio Gasbarrini4
Antonio Gasbarrini4 Giuseppe Spadaro1,2
Giuseppe Spadaro1,2Common variable immunodeficiency (CVID) is the most common symptomatic primary antibody immunodeficiency, characterized by reduced serum levels of IgG, IgA, and/or IgM. The vast majority of CVID patients have polygenic inheritance. Immune dysfunction in CVID can frequently involve the gastrointestinal tract and lung. Few studies have started to investigate the gut microbiota profile in CVID patients. Overall, the results suggest that in CVID patients there is a reduction of alpha and beta diversity compared to controls. In addition, these patients can exhibit increased plasma levels of lipopolysaccharide (LPS) and markers (sCD14 and sCD25) of systemic immune cell activation. CVID patients with enteropathy exhibit decreased IgA expression in duodenal tissue. Mouse models for CVID unsatisfactorily recapitulate the polygenic causes of human CVID. The molecular pathways by which gut microbiota contribute to systemic inflammation and possibly tumorigenesis in CVID patients remain poorly understood. Several fundamental questions concerning the relationships between gut microbiota and the development of chronic inflammatory conditions, autoimmune disorders or cancer in CVID patients remain unanswered. Moreover, it is unknown whether it is possible to modify the microbiome and the outcome of CVID patients through specific therapeutic interventions.
Elie Metchnikoff, a founder of modern Immunology, suggested that indigenous microbiota provide several pivotal functions for health and disease (1). However, only in recent years, due to the growing access to DNA sequencing technology, important mechanistic insights have been clarified (2). The human gut microbiota consists of 10-100 trillion symbiotic microbes (e.g., bacteria, yeast, and viruses) within each individual (3), whereas the human microbiome encodes over 3 million genes these cells harbor (4). New culture-independent techniques based on high-throughput or next-generation sequencing (NGS) technologies have revolutionized the knowledge of the gut microbiota (5). These approaches have allowed comprehensive analysis of the gut microbiota composition without the limitations of classic culture methods. The most common omics quantification is sequencing of the 16S ribosomal RNA subunit. Such sequencing is relatively simple, but can miss potentially relevant pathobions (6). The whole-community shotgun sequencing (WCS) has been proposed to be a more accurate technique (7). In addition, new omics technologies (e.g., metabolomics and proteomics) go beyond species abundance and have enabled more comprehensive insights on microbiota functions in health and disease (8, 9).
Gut microbiome composition is a dynamic process changing through life. Colonization of the gut begins before delivery (10). The most important contribution to the genesis of the microbiome is vertical transmission of maternal microbiota and the colonization process during delivery (10). Important factors that can affect microbiome composition are type of delivery (vaginal versus caesarean) and feeding (formula versus breast). Early colonization of the gut microbiome has major effects on its future composition (11). Diet, antibiotics and the environment in early life have an essential role in determining gut microbiota composition in adults (12). For instance, diet can remarkably shape the gut microbiome, making results difficult to interpret in the absence of dietary control.
The gut microbiota plays an important role in educating and modulating the host innate and adaptive immune system (13, 14). The gut microbiome also maintains the intestinal epithelial barrier homeostasis, defense against pathogens (15) and harvests energy from food (16). Germ-free mice, lacking a gut microbial flora, show defects in multiple immune cell populations, such as TH2 cells, ILCs and have few IgA-producing intestinal plasma cells, and generally, greater susceptibility to infections (17, 18). Intestinal IgA maturation occurs in response to bacterial colonization of the intestine (19). IgA deficiency causes imbalance of the gut microbiota, resulting in activation of the systemic immune system (20). These findings suggest a close link between gut microbiota, and the local and systemic immune system (21, 22).
Dysbiosis, any change in diversity of gut microbiome, is characterized by loss of beneficial microbes (symbionts) and expansion of potentially pathological organisms (pathobionts) (23). The human gut microbiota is mainly composed of two bacterial phyla: Bacteroidetes and Firmicutes, which are obligate anaerobic bacteria and constitute approximately 90% of microbial community. Additional phyla include Actinobacteria, Proteobacteria, Verrucomicrobia and Fusobacteria. Quantitative and qualitative changes of the gut microbiome are important contributors not only to gastrointestinal disorders but also to systemic metabolic (e.g., obesity, diabetes) and inflammatory diseases (24). There is also evidence for a potential role in health and disease of the viral (virome) and fungal community (mycobiome) (25).
The aim of this review was to provide an overview of current results and many outstanding questions related to the study of microbiome in common variable immunodeficiency (CVID). We will outline the factors (e.g., decreased secretory IgA, recurrent infections, antibiotics) that shape gut microbiota composition in patients with CVID, and the potential clinical interventions (e.g., diet, probiotics, prebiotics, drugs and fecal microbiota transplantation) to re-establish and/or to promote a “healthier” microbial community.
Common variable immunodeficiency (CVID) is the most common symptomatic primary antibody immunodeficiency (PID) in adulthood with a prevalence of approximately 1/25,000 (26). CVID is characterized by low serum levels of IgG, IgA, and/or IgM and impaired antibody synthesis in response to vaccines and pathogens (27, 28). CVID may present with a wide spectrum of clinical manifestations including increased susceptibility to infections, inflammatory and autoimmune diseases, solid and hematological malignancies (29, 30). The age of onset of CVID varies between 20 and 40 years. The variable age of onset and the heterogeneity of clinical manifestations, which can involve various segments of the respiratory and gastrointestinal tract, is consistent with different phenotypes of this PID.
CVID has complex pathogenic mechanisms resulting from intrinsic or extrinsic defects in B cell differentiation and/or from an impaired cross-talk between B and T cells (31–33). The majority of CVID patients lack a monogenic basis and the disease has probably polygenic inheritance (34). Monogenic disorders involving mutations in genes necessary for B-cell functions represent 2-10% of all CVID patients in different cohorts (35–40).
With the advent of intravenous (i.v.) or subcutaneous (s.c.) immunoglobulin replacement therapy (IgRT), CVID patients are relatively free from life-threatening infections and have a more prolonged survival than several years ago (26). However, IgRT has no proven efficacy in the prevention/treatment of immune dysregulation-related complications (41, 42). It has been estimated that the vast majority (68 to 83%) of CVID patients develop non-infectious complications such as autoimmunity, granulomatous and lymphoproliferative diseases, or malignancies, causing significant morbidity and mortality (29, 42).
It has been suggested that environmental factors (e.g., microbial dysbiosis), via epigenetic mechanisms, play a role in inflammatory and immune dysregulation in CVID (43).
Immune dysfunction in CVID can involve different sections of the gastrointestinal tract and cause manifestations including bloating, diarrhea, protein-energy malnutrition and malabsorption (44, 45). Gastrointestinal involvement with malabsorption (=∼ 6%), often complicated by nutritional deficiency requiring total parenteral nutrition, is associated with increased mortality and remains a major clinical challenge (42). Moreover, protein-losing enteropathy (PLE) can cause therapeutic failure of the IgRT (46, 47), because circulating IgG, as well as other plasma proteins, are lost through epithelial exudation in the setting of mucosal inflammation and damage (48). Gastrointestinal disorders in CVID can be classified into five groups: infectious (Giardia lamblia, Cryptosporidium parvum, Campylobacter jejuni, Salmonella species, Clostridioides difficile, Cytomegalovirus, Norovirus), small intestinal bacterial overgrowth (SIBO), inflammatory (celiac disease and celiac-like villous atrophy, microscopic colitis, ulcerative colitis, Crohn disease), malignant (gastrointestinal cancers, lymphoma), and autoimmune disorders (pernicious anemia, autoimmune thrombocytopenia and hepatitis, primary biliary cholangitis) (44, 49, 50).
The gastrointestinal tracts involved in CVID display a wide spectrum of histologic patterns. The intestinal mucosa can show increased intraepithelial lymphocytes, villous blunting, crypt distortion, overexpression of apoptosis, paucity of plasma cells and nodular lymphoid hyperplasia, reflecting a mucosal response to gut antigens (51, 52). Among this broad spectrum of gastrointestinal manifestations, CVID-related enteropathy (E-CVID) is characterized by villous atrophy, malabsorption and diarrhea (48, 53).
In the last decade, few studies have started to investigate the gut microbiota profile in CVID patients. Gut microbial imbalance of patients with CVID mainly includes changes in microbial diversity, decrease in symbiotic beneficial bacteria, and increase in pathobionts (22, 54–56). The term alpha diversity was introduced to describe the mean microbial species diversity in the gut (43). Higher species diversity leads to higher resilience in the gut ecosystem. In other words, a healthy, resilient gut microbiome relies on high richness and biodiversity (57).
Jørgensen and colleagues performed a 16S ribosomal RNA-based profiling of stool samples in 44 patients with CVID, 45 patients with inflammatory bowel disease (IBD) and 263 healthy controls (22). Alpha diversity of the gut microbiota was reduced in patients with CVID and IBD. CVID patients were also different in beta diversity compared to controls and IBD. Patients with very low serum IgA had reduced alpha diversity. Increased plasma levels of lipopolysaccharide (LPS) were associated with reduced alpha diversity. Two biomarkers [i.e., soluble CD14 (sCD14) and sCD25] of increased microbial translocation and immune cell activation were increased in CVID patients. LPS and sCD25 levels were strong determinants of reduced alpha diversity in a subgroup of patients with inflammatory and autoimmune phenotype. In particular, they found that certain Firmicutes of Clostridia class (Lachnospiraceae Dorea and Lachnospiraceae Roseburia families), Bacilli and certain Proteobacteria of Gammaproteobacteria class were increased in CVID. Other Firmicutes of Clostridia class (Christensenellaceae and Lachnospiraceae Blautia families), Actinobacteria (Bifidobacteriaceae family) and other Proteobacteria of Deltaproteobacteria class (Desulfovibrionales order) were reduced in CVID. Overall, these results, including the decrease of beneficial taxa (e.g., Bifidobacteriaceae family) and the increase of the detrimental classes Bacilli and Gammaproteobacteria, emphasize a link between CVID, alterations of gut microbiota, and systemic inflammation.
Shulzhenko et al. examined the duodenal microbiome in 7 patients with E-CVID and 8 patients without enteropathy (54). Both groups had very low levels of serum IgA, but those with enteropathy showed decreased mRNA levels of both IgA subclasses in the duodenal mucosa. IGHA1 mRNA was approximately 10 times higher than IGHA2 and =∼ 100 times higher than for IgG genes suggesting that IgA in general and IgA1 in particular is the dominant gut Ig immunoglobulin in CVID small intestine. These data were extended by showing reduced IgA expression, by immunohistochemistry, in the duodenal tissues of CVID patients with enteropathy compared to those without enteropathy. The authors, using transkingdom network analysis of the duodenal microbiome found Acinetobacter baumannii as a candidate microbe driving CVID enteropathy. The authors also found that A. baumannii activated the monocyte-derived THP-1 cell line, through the expression of interferon (IFN) type I (IFNB1) and CXCL9, with the induction of the Th1-driven inflammation found in CVID enteropathy (54, 58). In addition, type I and type II IFNs were responsible for the shift to pro-inflammatory metabolism in the small intestine. The authors suggested that A. baumannii was responsible for villous atrophy and malabsorption in the absence of mucosal IgA. In conclusion, this study suggests that patients with CVID and enteropathy exhibit decreased duodenal IgA expression compared to their counterparts without enteropathy. Moreover, A. baumannii exhibited some of the criteria one might expect of an enteropathy-inducing pathobiont.
Using 16S rRNA sequencing, Fiedorová et al. analyzed the bacterial and fungal gut microbiota (mycobiota) in 27 CVID patients and 28 matched healthy controls including 16 case-control pairs living in the same household (55). The alpha diversity of the CVID and control subjects gut community was evaluated in terms of a number of observed OTUs (richness), Shannon index and Chao1 index. All measured alpha diversity indices were lower in the CVID cohort than in controls. In particular, CVID patients with severe phenotype were associated with lower alpha diversity indices. In this study, alterations of alpha diversity were not associated with inflammatory and autoimmune disorders. Interestingly, the authors did not find any significant taxonomic differences of fungal microbiota between patients and controls, suggesting that gut mycobiota may not affect the CVID phenotype. More recently, the same group characterized the fecal microbiota and stool metabolome in a cohort of 6 CVID patients without gastroenterological symptoms and their healthy housemates (56). CVID fecal microbiome showed increased bacterial diversity and differences in several bacterial species compared to controls. Moreover, CVID patients showed in the stool samples decreased levels of adenosine and inosine, two metabolites of purine metabolism, which modulate several aspects of the immune response (59, 60).
The relationships between microbiota-specific systemic IgG and mucosal IgA were examined by Fadlallah et al. (61). These authors demonstrated that serum IgG directed against commensal microbiota are present in healthy subjects and are increased in patients with selective IgA deficiency (SIgAd). Serum IgG and secretory IgA target the same bacteria, and each individual targets a diverse microbiota repertoire. Hence, it is possible to hypothesize that IgRT with IgG preparations from IgA-deficient patients could offer better protection against gut microbial translocations in patients with CVID, especially for treating gastrointestinal-related inflammation (61). In this study, the authors also found that plasma levels of IL-6 and sCD14, two markers of systemic inflammation, were increased in CVID patients compared to healthy controls.
A fundamental question is whether microbiomes in other niches (e.g., oral or respiratory tract), besides gut, can control the immune dysregulation in CVID patients. A recent study highlighted the possible involvement of oral microbiota in CVID (62). The authors showed an increase of oral bacterial load, alpha diversity, and abundance of bacteria from the Prevotellaceae family in CVID patients with low serum IgA compared to controls. In addition, decreased serum IgA and expansion of Prevotellaceae bacteria were associated with lung disease (62).
The molecular pathways by which gut microbiota contribute to systemic inflammation in patients with CVID are of great importance but remain poorly understood. CVID patients may have increased intestinal permeability caused by a variety of direct and indirect microbial and immunological mechanisms associated to CVID-related enteropathy (44, 48, 63). This condition provides an impairment of the gut barrier, predominantly caused by disruption of intercellular tight junctions. Increased intestinal permeability leads to microbiota-host interactions involving a multitude of bacterial products (e.g., lipopolysaccharides, peptidoglycan, flagellin, RNA, and DNA) released into the portal circulation and influences microbial sensing and systemic immune response through recognition of microbe-associated molecular patterns by the innate immune system (64, 65). On the other hand, gut microbial dysbiosis and infectious triggers may further exacerbate gut leakiness causing increased microbial translocation and inflammation.
Jørgensen and collaborators carefully examined the role of gut microbiota to different aspects of systemic inflammation. In a first study, they confirmed that CVID patients have reduced gut microbial diversity compared to healthy donors (66). Eight-week administration of rifaximin, the oral non-absorbable intestinal antibiotic, did not modify circulating markers (LPS, sCD14, and sCD25) of inflammation in plasma of CVID patients. Rifaximin treatment was associated with a significant change in microbial alpha diversity (Chao1, Shannon index and OTUs). These results support the hypothesis that systemic inflammation in CVID is not influenced by rifaximin-sensitive bacteria. In another study, the same group highlighted the importance of the gut microbiota-dependent metabolite trimethylamine N-oxide (TMAO) in CVID. They found that CVID patients had significantly elevated plasma concentrations of TMAO and trimethylamine (TMA) compared to healthy controls. Increased circulating levels of TMAO correlated with raised LPS and increased inflammatory markers such as TNF-α and IL-12 (67). Plasma TMAO concentrations correlated positively with gut abundance of Gammaprobacteria. The authors concluded that TMAO could be a link between gut microbial dysbiosis and systemic inflammation.
In conclusion, there is evidence that the composition of the gut microbiota in patients with CVID is significantly different from healthy individuals (22, 55, 56, 63, 68). Whether changes in the intestinal microbiota observed in CVID are cause or effect of disease still needs to be determined.
Diarrhea is a common gastrointestinal symptom in CVID patients (69–72). Gut-microbial diversity, evaluated by the 16S rRNA gene sequencing, body mass index (BMI), spleen size, and lymphocyte phenotypes were examined in 46 CVID patients reporting ≥ 6 days/months of diarrhea (63). As expected, BMI was lower and malabsorption more common in patients with diarrhea vs. non-diarrhea patients. Patients with diarrhea had reduced naïve CD4+ T cell counts. Alpha diversity was lower in 15 CVID patients with diarrhea compared to 12 healthy donors. The colonic microbiota of CVID patients with diarrhea differed from that of patients without diarrhea in beta diversity. Alpha diversity was lower in CVID patients compared to controls. There was no difference in alpha diversity between patients with and without diarrhea. Very few CVID patients with diarrhea tested positive for viruses (norovirus, adenovirus, cytomegalovirus, sapovirus). Only one patient was positive for Clostridioides difficile.
Figure 1 schematically illustrates some of the structural features and the cells of the innate and adaptive immune system in human colonic mucosa in normal subjects and in CVID patients.
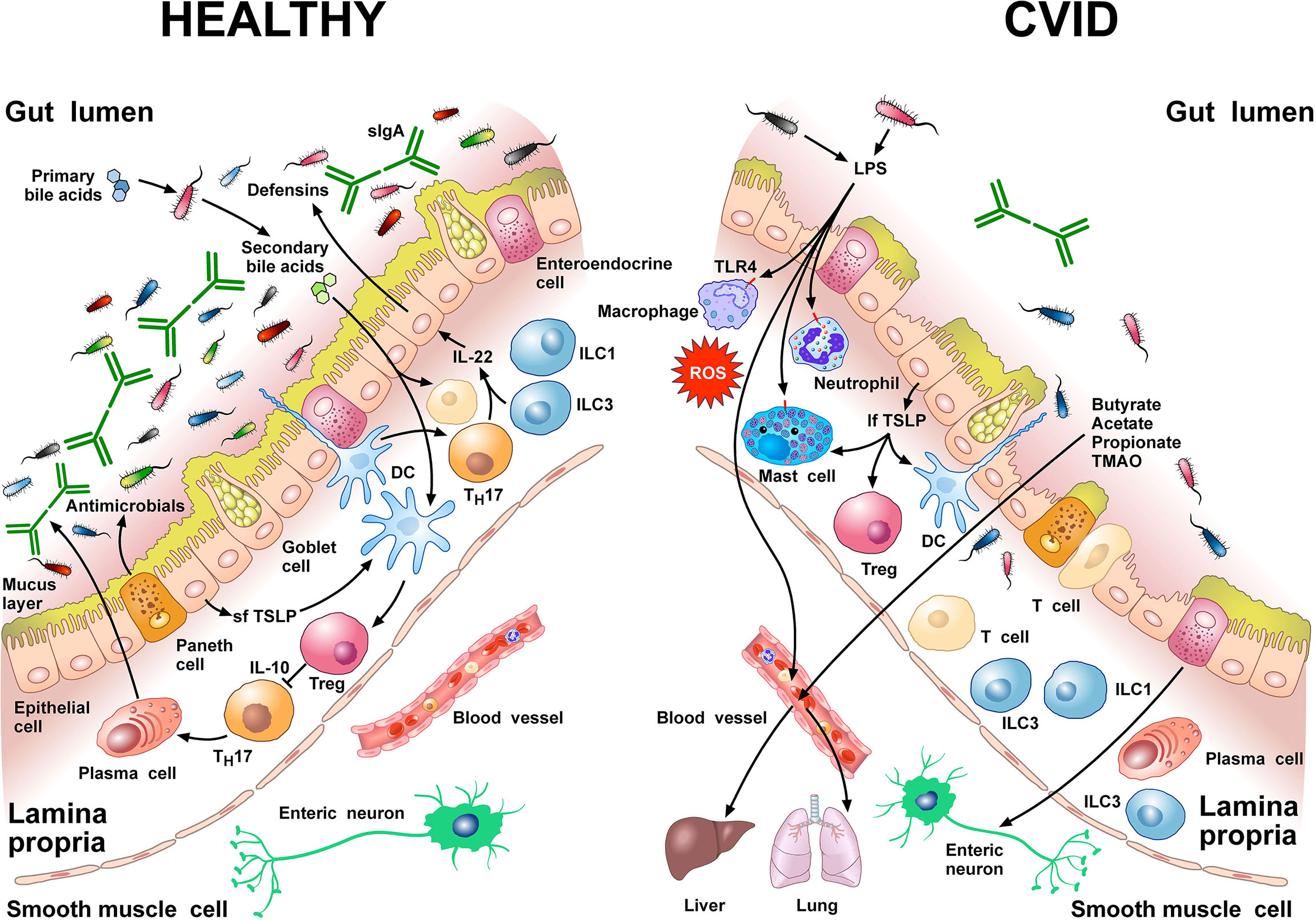
Figure 1 Left side Gut homeostasis and healthy gut are maintained by the interplay between physical barrier (intact mucus layer and epithelial cells) and several cells of the innate and adaptive immune system. In normal subjects four major phyla dominate the gut microbiome: Bacteriodetes, Firmicutes, Proteobacteria, and Actinobacteria. Dendritic cells (DCs) are pivotal for sensing bacterial products and activating antigen-specific CD4+ T cell differentiation. DCs also promote TH17 immunity, Foxp3+ Treg induction, and IgA production by plasma cells (73). Approximately 5% of primary bile acids transit to the colon and can be metabolized by commensal gut flora (74, 75). These secondary bile acids promote Treg generation and modulate the production of cytokines from DCs (76–78). Several other immune cells (e.g., macrophages, mast cells, neutrophils, ILC3, TH1 cells, plasma cells) are sentinels in the mucosal system (79). Neutrophils exert anti-bacterial effects through the release of their stored and newly synthesized mediators and the formation of neutrophils extracellular traps NETs (80). TH17 cells produce IL-22 that promotes secretion of anti-microbial peptides such as β-defensins by epithelial cells. ILC1 and ILC3 are present in human intestinal mucosa (81). ILC3 produces IL-22, which plays a role in containing the commensal flora (82) and protecting epithelial cells (83). Paneth cells produce several molecules with antimicrobial activity (α-defensin, REG3, ANG4, sPLA2) as well as cytokines that can recruit immune cells (84). The short thymic stromal lymphopoietin isoform (sfTSLP), constitutively expressed by human epithelial cells, is crucial in preserving immune tolerance in the gut (85–87). Right side In patients with CVID, alpha and beta diversity of the gut microbiota is reduced compared to healthy donors (22, 66). Repeated or chronic infections damage the intestinal epithelium (44). The disruption of the gut barrier integrity and the reduction of secretory IgA (54) increase microbial translocation (88) and the permeability of pathogen-associated molecular patterns (PAMPs) such as LPS (22, 66, 89). LPS activates TLR4 on human macrophages (90), neutrophils (91), and mast cells (92, 93) to release pro-inflammatory mediators and ROS. The numbers of ILC3 and ILC1 are abnormally high in the inflamed intestinal mucosa (94). The long TSLP isoform (lfTSLP), induced by several components of gut microbiota, exerts pro-inflammatory effects and contributes to intestinal damage (85–87). The passage of bacteria-derived products (e.g., LPS, butyrate, acetate propionate, TMAO) into the circulation is one of the means of communication between the gut microbiota and the lung or the liver (2, 89, 95, 96). The three most common short chain fatty acids (SCFAs) (butyrate, acetate, and propionate) can also exert immunomodulatory/anti-inflammatory roles (97, 98).
CD19 deficiency is a risk factor for monogenic CVID in humans (99). A mouse model of CVID (CD19−/− mice) is characterized by intestinal malabsorption and defects in lipid metabolism and transport (100). CD19−/− mice develop defective maturation, proliferation, and selection of B cells in the intestinal germinal center, thus resulting in impaired B cell memory and insufficient synthesis of high-affinity antibodies (101) (102, 103). CD19−/− mice had a B cell deficiency in gut-associated lymphoid tissue (GALT) resulting in a dramatic decrease of fecal IgA, IgG, and IgM compared to wild-type (WT) mice. As a consequence, CD19−/− mice cannot bind intestinal bacteria with IgA with subsequent expansion of fecal anaerobic bacteria. These mice develop chronic intestinal malabsorption and altered microbiota composition characterized by outgrowth of anaerobic bacteria within the order Bacteroidales (e.g., Rikenellaceae and Lachnospiraceae), in addition to specific species [Staphylococcus spp., Sutterella spp., segmented filamentous bacteria (SFB), an undescribed species of alphaproteobacteria, and Bilophila spp]. Intestinal malabsorption was associated with intestinal mast cell activation. In this study, treatment with metronidazole and the subsequent reduction in the severity of malabsorption highlights the hypothesis that expansion of anaerobic bacteria was driving the malabsorption in CD19−/− mice. In order to determine whether the intestinal malabsorption was due to dietary gluten exposure, the authors applied a strict gluten-free diet (GFD). Five weeks after initial GFD-exposure the intestinal mucosa histologically showed a reduction of malabsorption. The authors suggested that malabsorption in CD19−/− mice was, at least in part, not only microbiota-dependent but also gluten-sensitive. Of note, gluten sensitivity in patients with CVID is still a matter of debate (100). The results of the previous study suggest that gut microbiota could be an important co-factor in CD19−/− mice. Modification of gluten antigens by microbial transglutaminase may enhance the immunogenicity of gluten peptides, unleashing the inflammatory response (100). In this scenario, the mucosal IgA seems to play an intriguing role as modulators of the gut microbiota (104). In fact, secretory IgA regulates the composition and functions of the gut microbiota by promoting symbiosis between bacteria (105), whereas commensal bacteria induce local IgA response. In addition, IgA binds to members of the commensal gut microbiota and affects colonization levels (106). IgA-coated species include members of the Proteobacteria phylum (e.g., Enterobacteriaceae) and Firmicutes phylum (e.g., Lactobacilli), which were relatively increased in the CVID cohort studied by Fiedorová and colleagues. Hence, reduced secretory IgA in CVID patients with enteropathy can lead to microbial dysbiosis.
The use of CD19-/- mice as a clinically relevant model for CVID has been critically examined by Jorgensen and collaborators (107). These authors emphasized that this mouse model does not reflect the polygenic causes of the majority of CVID patients. Therefore, animal models should be carefully considered when studying the interplay between microbiota and immunodeficiency.
A recent cross-sectional study on a cohort of 27 CVID individuals reported that symptoms of small intestinal bacterial overgrowth (SIBO) were found in the majority (> 60%) of patients (72). This disorder is characterized by the presence of a pathological concentration of bacteria in the jejunal aspirate equal to or greater than 105 colony-forming units (CFU) per ml. Recognition of this syndrome is made possible by the non-invasive hydrogen tests, such as glucose and lactulose breath tests (108). Symptoms traditionally linked to SIBO include bloating, diarrhea, and abdominal pain/discomfort (72). Steatorrhoea is present in more severe cases. SIBO can contribute to or worsen the nutritional status of CVID patients. In fact, bacterial overgrowth can compromise the absorption of proteins, fats, carbohydrates, vitamins, and other micronutrients. For example, B-12 deficiency is caused by the consumption of cobalamin by anaerobes, malabsorption of the vitamin due to competitive binding with cobalamin from bacterially generated metabolites of cobalamin at the ileal receptor, and in more severe overgrowth, mucosal injury involving the binding site (109). The excess of bacteria, in addition to competing for nutrient intake, can also produce toxic metabolites (e.g., bile acids, hydroxylated fatty acids, and other organic acids) and cause direct damage to the enterocytes of the small intestine, leading to increased intestinal permeability (110). Deconjugation of bile acids by intestinal bacteria may result in malabsorption of fat and fat-soluble vitamins with consequent steatorrhoea and fat-soluble vitamin deficiencies. Bacterial synthesis of folic acid may be responsible for the unusual combination of high folate and B12 deficiency (72).
There is compelling evidence of bidirectional gut-lung axis in CVID. Several studies have demonstrated that respiratory infections, mostly prevalent in CVID, are associated with a change in the composition of the gut microbiota (111–117). In particular, many respiratory infections, common in CVID patients, are often accompanied by gastrointestinal symptoms (112, 118). In animal models, bacterial infections or intratracheal instillation of LPS led to alterations in the intestinal microbiota (118, 119). On the other side, repeated and/or chronic gastrointestinal infections can damage the intestinal epithelium resulting in an increased microbial translocation (88). Soluble microbial components (e.g., LPS, peptidoglycans) and metabolites (e.g., butyrate, acetate, propionate, TMAO) transported via the circulation are one means of communication between the gut microbiota and the lungs (2, 89, 95, 96). Immune cells can also migrate from the intestine to the respiratory tract via the circulation (120). Finally, the gut can modulate immunological and non-immunological responses in the lungs via host-derived inflammatory mediators (121). Further studies are needed to ascertain the pathophysiological relevance of the routes of communications within the gut-lung axis in CVID.
The microbiota-gut-liver axis might be a key player in the pathogenesis of liver involvement in CVID (44), as the passage of bacteria-derived products into the portal circulation can activate specific receptors on cells of innate immunity (e.g., TLR4 receptors on macrophages and monocytes), leading to liver inflammation (96).
The gut microbiota is now considered a “metabolic organ” that not only facilitates harvesting of nutrients and energy from the ingested food but also produces several metabolites (e.g., bile acids) that signal through their cognate receptors to modulate host metabolism (122) and immune system (123). One class of such metabolites, primary bile acids, produced in the liver, is metabolized to secondary bile acids in the intestine by the gut microbiota (74, 75, 122). Secondary bile acids activate the plasma membrane G protein-coupled bile acid receptor 1 (GPBAR1 or TGR5) and the nuclear Farnesoid-X-Receptor (FXR) (74, 123). GPBAR1 and FXR, highly expressed by intestinal epithelial cells, are also present in immune cells, intestinal muscle cells and neurons (123).
The interaction between the bile and the microbiota is bidirectional. Bile acids can modulate the microbiota through the engagement of FXR (124). GPBAR1 and FXR are expressed by macrophages, DCs, NK cells (125–127), and T helper cells (122). The activation of these receptors inhibits several aspects of the inflammatory pathways (74, 123).
It is well established that Foxp3+ Treg cells play a fundamental role in maintaining immune homeostasis in the intestinal lamina propria (79, 128, 129). Bile acid metabolites inhibited the differentiation of TH17 cells by binding to the transcription factor retinoid-related orphan receptor-γt (RORγt) (77). These authors also found that the bile acid metabolite isoallolithocholic acid increased the differentiation of Treg cells through the overexpression of Foxp3. More recently, it has been demonstrated that secondary bile acids increased Foxp3 induction acting on DCs to diminish their immunosuppressive properties (76). Ablating the FXR receptor in DCs enhanced the generation of Treg cells in vitro and in vivo. Collectively, these studies indicate that secondary bile acids are potent inducers of Tregs, suggesting that microbial metabolism of endogenous steroids contributes to immunological balance in the gut.
These results have translational relevance. Song and coworkers have demonstrated that both dietary and microbial factors modulate bile acid composition and colonic Foxp3+ Tregs (130). Importantly, genetic deletion of bile acid metabolic pathways in gut symbionts decreased Treg cell population.
Further studies should investigate possible alterations of bile acid metabolites and host immunological homeostasis in CVID patients.
Several studies reported an increased risk of malignancies in CVID (131–134). Gastric cancer is the leading cause of death in Italian CVID patients (135). The pathophysiological pathways underlying the relationships between CVID and the increased prevalence of cancers in CVID are of paramount importance but not fully understood. It is well established that chronic low-grade inflammation is a driving force of tumor initiation and progression (136, 137). Moreover, inflammatory stimuli such as LPS, activate immune cells (e.g., macrophages, neutrophils) present in tumor microenvironment (90, 91) and play a role in the switch between dormancy and proliferation of metastatic cells (138, 139). CVID patients exhibit immunological evidence of local and systemic inflammation (22, 61, 66, 67). Future studies should evaluate whether the gut microbiome could be used to identify CVID patients who may develop cancer.
The link between Helicobacter pylori (H. pylori) and gastric cancer represents a prototype of interactions between chronic low-grade inflammation caused by a pathobiont and cancer (140). H. pylori is a pathogen recognized to play a role in gastric tumorigenesis. H. pylori infection in CVID patients appears to facilitate the progression to gastric atrophy and cancer with higher rates compared with the general population, and this could be explained by impaired mucosal immune response to pathogens (141). In particular, low mucosal IgA levels (with bactericidal activity against H. pylori) and hypochlorhydria may enhance H. pylori colonization and mucosal inflammation, thus promoting gastric carcinogenesis (142). H. pylori may not be the only microbe implicated in gastric cancer development. Several studies demonstrate differences in the microbiota of individuals with atrophic gastritis and gastric cancer, suggesting that the alteration of gastric microbiome modulates gastric cancer initiation and progression (143–148). H. pylori colonization gradually fades in the final steps of gastric carcinogenesis with H. pylori frequently absent/undetectable at the cancer stage (143, 149, 150). Of note, progression to gastric cancer in some patients, can occur after H. pylori eradication (135). The latter findings highlight the involvement of additional factors (e.g., other components of the gastric microbiota) in the carcinogenesis process. The latter observation is clinically relevant because follow-up strategies targeting gastric cancer secondary prevention cannot rely exclusively on H. pylori identification but also on implementation of upper endoscopies in CVID patients. Further studies appear necessary to identify an altered gastric microbiome in CVID patients at risk of developing gastric cancer.
Despite adequate i.v. or s.c. IgRT, recurrent respiratory and gastrointestinal infections are the commonest clinical features in CVID (26, 151). Consequently, a significant percentage of CVID patients are frequently treated with antibiotics to control acute infections or as prophylaxis to reduce infection frequency (152). Antibiotics have also detrimental effects inducing profound alterations of the gut microbiota or dysbiosis and development of bacterial resistance (153). Interestingly, some antibiotics might also have beneficial effects on gut microbiota. For example, rifaximin demonstrated to have eubiotic effect on gut microbiota by increasing the abundance of some bacterial species considered to be beneficial in patients affected by different gastrointestinal and liver diseases (154). Early studies reported that rifaximin reduced plasma endotoxin levels in patients with cirrhosis (155, 156). By contrast, rifaximin had no effect on circulating markers of systemic inflammation (sCD14, sCD25 or LPS), whereas decreased microbial alpha diversity in patients with CVID (66). Moreover, none of the ten major bacteria hitherto indicated to differentiate CVID patients and healthy controls, the CVID specific dysbiosis index (22), was significantly modified by rifaximin. The CVID specific dysbiosis index correlates with circulating biomarkers of systemic inflammation and gut leakage. Hence, it is possible to hypothesize that the lack of CVID specific dysbiosis index modification could explain the absence of an anti-inflammatory effect of rifaximin.
The term “gut resistome” describes the collection of genes or genetic material of gut microbiota that confers antimicrobial resistance (157, 158). Microbial genes conferring resistance to antibiotics can be transferred to gut pathogens in a process named horizontal gene transfer (HGT), thus inducing disease in the host (159). Hence, gut microbiota can be considered as a potential reservoir of antibiotic resistance particularly following recurrent antibiotic therapy (160). Antibiotic-induced resistances can be found even years after the drug administration (161–164). These observations are relevant because prolonged or recurrent antibiotic treatments are frequently administered to CVID patients (152). Antibiotic therapies can also induce impairment in an otherwise healthy gut microbiota, thus contributing to Clostridioides difficile infection (CDI) (165, 166). Several antibiotics (i.e., clindamycin, cephalosporins, penicillins, and fluoroquinolones) have been associated with the development of CDI (167). Drug-related factors (e.g., antibiotic class, duration, dose and route of administration of therapy) and host-related factors can profoundly influence the microbial composition. This post-antibiotic dysbiosis can be expressed by loss of diversity, loss of crucial taxa, shifts of metabolic pathways, and reduced colonization resistance against invading pathogens. For these reasons targeted therapies with narrow-spectrum antibiotics and shorter treatment courses are advisable in CVID patients.
The human microbiological ecosystem changes in response to a variety of environmental stimuli, making difficult to identify specific microbiome signatures in human diseases. Assuming that human microbiome is easily shaped, perhaps we could modulate it to favor a desired outcome (168). The emerging field of microbiome engineering is in its infancy and faces several challenges (169). Figure 2 provides examples of treatments that can modify the microbiome.
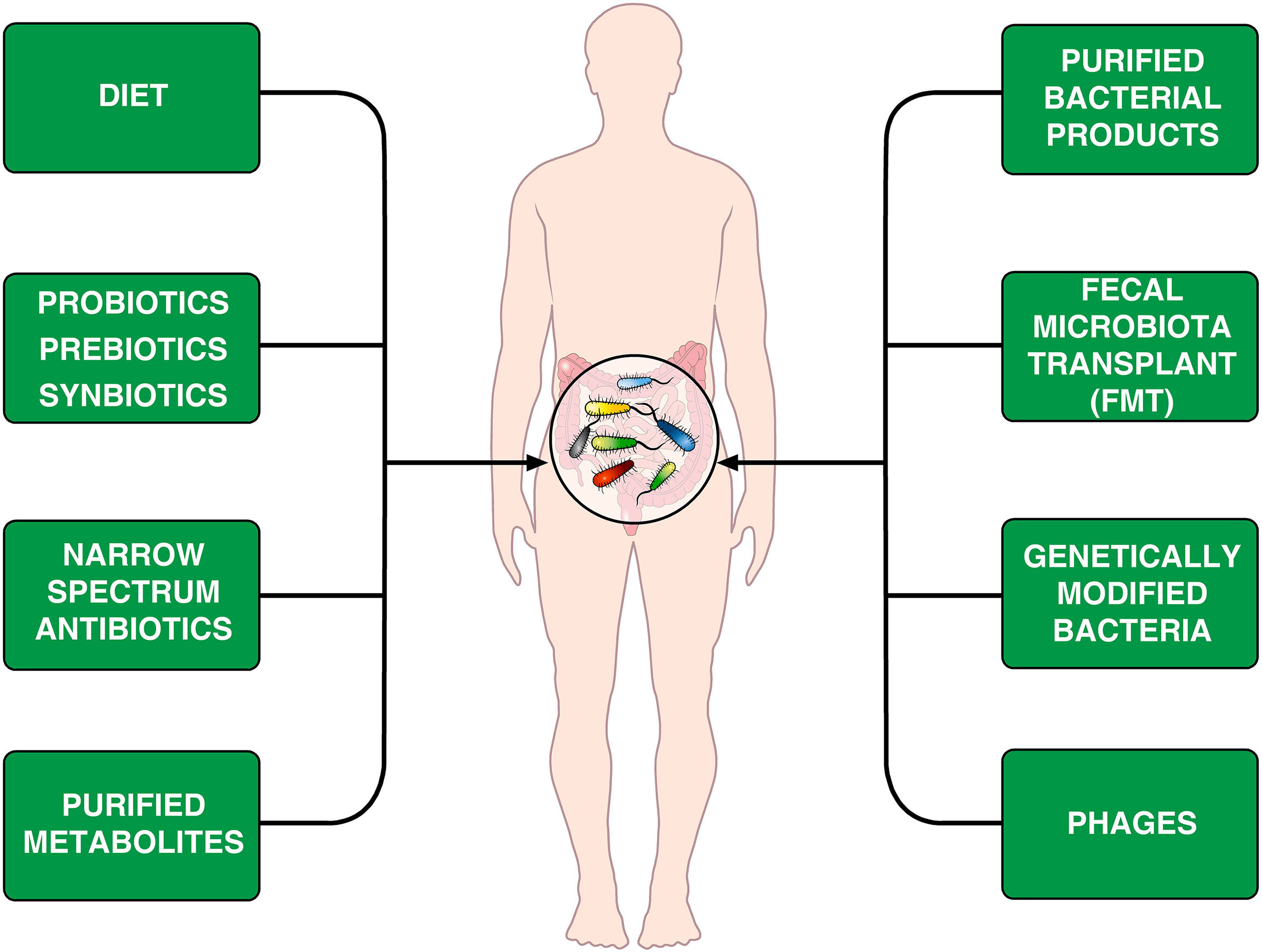
Figure 2 Schematic representation of theoretically therapeutic approaches that can modify the gut microbiome in CVID patients. Diet is an important environmental factor that shapes the microbiota composition (170–172) and can induce Foxp3+ Treg cells in the intestine (173). Another approach is the administration of prebiotics, probiotics or synbiotics (174, 175). The different synbiotics, doses, and regimens makes it difficult to perform controlled clinical trials (176). Rifaximin, which is beneficial in patients with certain gastrointestinal diseases (154), decreases alpha diversity in CVID patients without improving markers of systemic inflammation (66). It has been suggested that the administration of specific bacterial products can selectively modulate colonic immune cells in CVID patients (177). Fecal microbiota transplant (FMT), initially developed to treat recurrent C. difficile colitis (178, 179) is theoretically an appealing therapeutic tool. Genetically modified bacteria used to treat experimental colitis (180) and phages (181, 182) could theoretically be used for treating microbiome-associated pathologies in CVID patients.
Despite increasing evidence involving gut microbiota in CVID (54, 63, 66, 67), the therapeutic modulation of the gut microbiome has not been investigated yet as a potential treatment for these patients. Elie Metchnikoff first noted that not all microbes are dangerous and envisioned that oral administration of lactic acid-producing bacteria with food (e.g., soured milk) might improve health and longevity (1). Therefore, Metchnikoff was acclaimed as a founding father of contemporary probiotics. There is some evidence that probiotics exert immunomodulatory effects and are considered a promising alternative for the prevention and treatment of certain inflammatory disorders (183). Recent evidence indicates that probiotic-derived metabolites promote differentiation of Foxp3+ Tregs (184). The latter observation deserves attention because CVID patients have lower numbers of Treg cell compared to healthy controls in their peripheral blood (185, 186).
Dietary contributions to health and chronic conditions, such as obesity, cardiovascular and gastrointestinal disorders, and cancer are of universal importance. Gut microbiome has been implicated as one of several potentially causal human-environment interactions in these disorders (187–189). Host diet plays a fundamental role in shaping the gut microbiota (170–172, 188, 189). Dietary antigens can induce Foxp3+ Treg cells in the intestine (173), essential regulators of immunological homeostasis in the colon (190). Diet can modulate the population of colonic Treg cells in mice (130). The relationships between different habitual diets (e.g., healthy plant-based foods, animal-based foods, and butyrate-producer foods), microbiome composition and peripheral and colonic Treg cells in CVID are presently unknown and should be investigated.
Fecal microbiota transplant (FMT), which transfers an entire microbiome from a healthy donor to a recipient, is a therapeutic tool with several potential applications but numerous specific caveats. Transplant can be performed via an oral capsule containing fecal extracts (191–193), colonoscopy-guided insertion (193, 194), or enema (195, 196). Healthy donors vary widely in microbiome composition, and donor composition can affect success rates (197, 198). FMT involves risks, and regulatory bodies have recently released safety alerts regarding the risk for transmission of infectious agents via FMT (199). Specific guidelines have been released to offer FMT with high levels of safety at the time of COVID-19 pandemic (200).
Another approach is to administer a formulation of compounds and spores that promote growth of desirable bacteria in the gut. These are referred to as probiotics, although the term refers to formulations containing live microorganisms (201). The term prebiotics refers to compounds (e.g., oligosaccharides and fibers) promoting the growth of specific bacteria (174, 175). The combination of probiotics and prebiotics is called a synbiotic (202). The latter strategy is appealing because one could incorporate any combination of microorganisms to construct a personalized symbiotic (203). The variety of possible synbiotics different formulations, doses, and regimens makes it difficult to make comparisons. The activity of taxonomically similar or even identical strains manufactured by different producers varies. Therefore, the therapeutic properties of a formulation cannot be extrapolated from one preparation to another (203, 204). Moreover, it is not always evident whether gut microbiome can be actually altered by synbiotic regimens. Finally, synbiotics themselves can be associated with adverse events and their safety profile is often overlooked (205, 206). Probiotics, prebiotics, and synbiotics have shown some efficacy in certain experimental (203) and clinical studies (176). However, there is no published evidence addressing these issues in CVID patients.
Several bacterial genera have been engineered to produce microorganisms with modified genetic payloads. Steidler et al. developed a genetically modified strain of Lactococcus lactis to deliver IL-10 to treat colitis in a mouse model (180). It has been postulated that synthetic bacteria could become in the future a tool for microbiome engineering (207). No successful interventions that utilize engineered microbes to improve patient outcomes have been described.
Phage therapy is another technology for microbiome engineering. The goal is to design phages, infecting and killing undesired bacteria. It has been suggested that phages could serve as a tool for treating microbiome-associated pathologies (181). This approach has been preliminary used since the early twentieth century as a treatment for certain infections (208). Phage therapy has been reported to be successful in a patient infected with multidrug-resistant A. baumanii (209). Interestingly, Shulzhenko et al. found A. baumanii the microbe driving enteropathy in CVID patients (54).
Although phage therapy is a potential application for microbiome engineering (182), this approach poses several challenges. Phages are live organisms and they could theoretically evolve and cause unknown effects on a microbial community (210).
In the past decade, several studies have started to identify quantitative and qualitative gut microbiota alterations in CVID patients. It is unclear whether the disruption of eubiotic microbiota observed in CVID is primary or secondary to environmental exposures, antibiotic therapies or diet. Evidence for a role of gut microbiota in the development of local and systemic complications in CVID is still limited. Developments in microbiome field will allow as to shift from a statistical to a casual association research. CVID is not a single disease, but rather a heterogeneous syndrome. Therefore, specific and detailed analyses of the composition (e.g., bacteria, yeast, and viruses) of gut microbiota in different phenotypes of CVID are urgently needed. A common omics quantification of microbiota is sequencing of the 16S ribosomal RNA subunit (22, 54, 63). This is a relatively simple and convenient way to identify organisms in a microbiome, but can miss potentially relevant pathobionts (6, 168). Whole-community shotgun sequencing (WCS) is more accurate to identify individual species and genes (7, 211, 212). We anticipate that advances in sample and sequencing will be critical for further advances in the field. Table 1 summarizes some of the outstanding pathophysiological questions that should be addressed to better highlight the complex interactions between specific gut microbial species and different phenotypes of CVID.
Experimental and clinical studies aimed at modulating gut microbiota in CVID patients are in their infancy. A better knowledge of quantitative and qualitative alterations in pathobionts in these patients is a prerequisite to hypothesize applications from the emerging field of microbiome engineering. A preliminary approach has shown that systemic inflammation in CVID is not modified by the elimination of rifaximin-sensitive bacteria (66). The administration of probiotics, prebiotics or their combination (symbiotics) could represent another strategy to modify the microbiome in CVID patients. FMT, initially developed to treat recurrent C.difficile colitis, represents the most radical means of engineering a microbiome (178).
Recently, Geva-Zatorsky and collaborators have devised a sensitive screen that entailed monocolonization of mice with a large number of species of human gut symbionts followed by extensive phenotyping and transcriptomic of immune cells (177). Individual microbes induced colonic TH17 cells, Treg cells and DCs or decreased ILCs. These results indicate that future efforts should be devoted to identify bacterial products underlying specific immunomodulatory activity of therapeutic interest. The advantage of the latter approach, using microbial products dosed as a drug, would yield host responses more reproducible overusing whole bacteria to modify gut microbiota.
Table 2 illustrates some of the outstanding questions concerning the therapeutic corrections of microbiome in CVID patients (213). Further advances in this field might have the potential to revolutionize the approach to prevention and treatment of different phenotypes of CVID.
GV, RP, and GS conceived and wrote the manuscript and prepared figures. GI, AP, GM, and AG contributed to the modification and revision of the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported in part by grants from the CISI-Lab Project (University of Naples Federico II), TIMING Project and Campania Bioscience (Regione Campania).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Fabrizio Fiorbianco for the outstanding medical illustrations, Dr. Gjada Criscuolo for critical reading of the manuscript and the administrative staff (Dr. Roberto Bifulco and Dr. Anna Ferraro), without whom it would not be possible to work as a team.
1. Neyen C. Foundational Article: Mechnikov I, 1909: Intestinal Bacteriotherapy. J Leukoc Biol (2021) 109:519–33. doi: 10.1002/JLB.5RI1020-638
2. Wypych TP, Wickramasinghe LC, Marsland BJ. The Influence of the Microbiome on Respiratory Health. Nat Immunol (2019) 20:1279–90. doi: 10.1038/s41590-019-0451-9
3. Kamada N, Seo SU, Chen GY, Nunez G. Role of the Gut Microbiota in Immunity and Inflammatory Disease. Nat Rev Immunol (2013) 13:321–35. doi: 10.1038/nri3430
4. Ursell LK, Metcalf JL, Parfrey LW, Knight R. Defining the Human Microbiome. Nutr Rev (2012) 70(Suppl 1):S38–44. doi: 10.1111/j.1753-4887.2012.00493.x
5. Kaeberlein T, Lewis K, Epstein SS. Isolating “Uncultivable” Microorganisms in Pure Culture in a Simulated Natural Environment. Science (2002) 296:1127–9. doi: 10.1126/science.1070633
6. Janda JM, Abbott SL. 16s rRNA Gene Sequencing for Bacterial Identification in the Diagnostic Laboratory: Pluses, Perils, and Pitfalls. J Clin Microbiol (2007) 45:2761–4. doi: 10.1128/JCM.01228-07
7. Laudadio I, Fulci V, Palone F, Stronati L, Cucchiara S, Carissimi C. Quantitative Assessment of Shotgun Metagenomics and 16S Rdna Amplicon Sequencing in the Study of Human Gut Microbiome. OMICS (2018) 22:248–54. doi: 10.1089/omi.2018.0013
8. Segal JP, Mullish BH, Quraishi MN, Acharjee A, Williams HRT, Iqbal T, et al. The Application of Omics Techniques to Understand the Role of the Gut Microbiota in Inflammatory Bowel Disease. Therap Adv Gastroenterol (2019) 12:1756284818822250. doi: 10.1177/1756284818822250
9. Malla MA, Dubey A, Kumar A, Yadav S, Hashem A. And Abd Allah, E.F. Exploring the Human Microbiome: The Potential Future Role of Next-Generation Sequencing in Disease Diagnosis and Treatment. Front Immunol (2018) 9:2868. doi: 10.3389/fimmu.2018.02868
10. Rodriguez JM, Murphy K, Stanton C, Ross RP, Kober OI, Juge N, et al. The Composition of the Gut Microbiota Throughout Life, With an Emphasis on Early Life. Microb Ecol Health Dis (2015) 26:26050. doi: 10.3402/mehd.v26.26050
11. Gritz EC, Bhandari V. The Human Neonatal Gut Microbiome: A Brief Review. Front Pediatr (2015) 3:17. doi: 10.3389/fped.2015.00060
12. Zmora N, Suez J, Elinav E. You Are What You Eat: Diet, Health and the Gut Microbiota. Nat Rev Gastroenterol Hepatol (2019) 16:35–56. doi: 10.1038/s41575-018-0061-2
13. Rooks MG, Garrett WS. Gut Microbiota, Metabolites and Host Immunity. Nat Rev Immunol (2016) 16:341–52. doi: 10.1038/nri.2016.42
14. Schirmer M, Smeekens SP, Vlamakis H, Jaeger M, Oosting M, Franzosa EA, et al. Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell (2016) 167:1125–36.e1128. doi: 10.1016/j.cell.2016.10.020
15. Libertucci J, Young VB. The Role of the Microbiota in Infectious Diseases. Nat Microbiol (2019) 4:35–45. doi: 10.1038/s41564-018-0278-4
16. Tremaroli V, Kovatcheva-Datchary P, Backhed F. A Role for the Gut Microbiota in Energy Harvesting? Gut (2010) 59:1589–90. doi: 10.1136/gut.2010.223594
17. Surana NK, Kasper DL. Deciphering the Tete-a-Tete Between the Microbiota and the Immune System. J Clin Invest (2014) 124:4197–203. doi: 10.1172/JCI72332
18. Hepworth MR, Monticelli LA, Fung TC, Ziegler CG, Grunberg S, Sinha R, et al. Innate Lymphoid Cells Regulate CD4+ T-Cell Responses to Intestinal Commensal Bacteria. Nature (2013) 498:113–7. doi: 10.1038/nature12240
19. Suzuki K, Fagarasan S. How Host-Bacterial Interactions Lead to IgA Synthesis in the Gut. Trends Immunol (2008) 29:523–31. doi: 10.1016/j.it.2008.08.001
20. Kawamoto S, Maruya M, Kato LM, Suda W, Atarashi K, Doi Y, et al. Foxp3(+) T Cells Regulate Immunoglobulin a Selection and Facilitate Diversification of Bacterial Species Responsible for Immune Homeostasis. Immunity (2014) 41:152–65. doi: 10.1016/j.immuni.2014.05.016
21. Wei M, Shinkura R, Doi Y, Maruya M, Fagarasan S, Honjo T. Mice Carrying a Knock-in Mutation of Aicda Resulting in a Defect in Somatic Hypermutation Have Impaired Gut Homeostasis and Compromised Mucosal Defense. Nat Immunol (2011) 12:264–70. doi: 10.1038/ni.1991
22. Jorgensen SF, Troseid M, Kummen M, Anmarkrud JA, Michelsen AE, Osnes LT, et al. Altered Gut Microbiota Profile in Common Variable Immunodeficiency Associates With Levels of Lipopolysaccharide and Markers of Systemic Immune Activation. Mucosal Immunol (2016) 9:1455–65. doi: 10.1038/mi.2016.18
23. Mullish BH, Quraishi MN, Segal JP, Ianiro G, Iqbal TH. The Gut Microbiome: What Every Gastroenterologist Needs to Know. Frontline Gastroenterol (2021) 12:118–27. doi: 10.1136/flgastro-2019-101376
24. Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A Core Gut Microbiome in Obese and Lean Twins. Nature (2009) 457:480–4. doi: 10.1038/nature07540
25. Huseyin CE, O’Toole PW, Cotter PD, Scanlan PD. Forgotten Fungi-the Gut Mycobiome in Human Health and Disease. FEMS Microbiol Rev (2017) 41:479–511. doi: 10.1093/femsre/fuw047
26. Cunningham-Rundles C, Bodian C. Common Variable Immunodeficiency: Clinical and Immunological Features of 248 Patients. Clin Immunol (1999) 92:34–48. doi: 10.1006/clim.1999.4725
27. Bonilla FA, Barlan I, Chapel H, Costa-Carvalho BT, Cunningham-Rundles C, de la Morena MT, et al. International Consensus Document (Icon): Common Variable Immunodeficiency Disorders. J Allergy Clin Immunol Pract (2016) 4:38–59. doi: 10.1016/j.jaip.2015.07.025
28. Registry_ClinicalCriteria, E. (2014). Available at: https://esid.org/content/download/13053/372959/file/ESIDRegistry_ClinicalCriteria2014.pdf.
29. Chapel H, Lucas M, Lee M, Bjorkander J, Webster D, Grimbacher B, et al. Common Variable Immunodeficiency Disorders: Division Into Distinct Clinical Phenotypes. Blood (2008) 112:277–86. doi: 10.1182/blood-2007-11-124545
30. Jolles S. The Variable in Common Variable Immunodeficiency: A Disease of Complex Phenotypes. J Allergy Clin Immunol Pract (2013) 1:545–56. doi: 10.1016/j.jaip.2013.09.015
31. Ochtrop ML, Goldacker S, May AM, Rizzi M, Draeger R, Hauschke D, et al. T and B Lymphocyte Abnormalities in Bone Marrow Biopsies of Common Variable Immunodeficiency. Blood (2011) 118:309–18. doi: 10.1182/blood-2010-11-321695
32. Taubenheim N, von Hornung M, Durandy A, Warnatz K, Corcoran L, Peter HH, et al. Defined Blocks in Terminal Plasma Cell Differentiation of Common Variable Immunodeficiency Patients. J Immunol (2005) 175:5498–503. doi: 10.4049/jimmunol.175.8.5498
33. Le Saos-Patrinos C, Loizon S, Blanco P, Viallard JF, Duluc D. Functions of Tfh Cells in Common Variable Immunodeficiency. Front Immunol (2020) 11:6. doi: 10.3389/fimmu.2020.00006
34. Orange JS, Glessner JT, Resnick E, Sullivan KE, Lucas M, Ferry B, et al. Genome-Wide Association Identifies Diverse Causes of Common Variable Immunodeficiency. J Allergy Clin Immunol (2011) 127:1360–7.e1366. doi: 10.1016/j.jaci.2011.02.039
35. Kienzler AK, Hargreaves CE, Patel SY. The Role of Genomics in Common Variable Immunodeficiency Disorders. Clin Exp Immunol (2017) 188:326–32. doi: 10.1111/cei.12947
36. van Zelm MC, Reisli I, van der Burg M, Castano D, van Noesel CJ, van Tol MJ, et al. An Antibody-Deficiency Syndrome Due to Mutations in the CD19 Gene. N Engl J Med (2006) 354:1901–12. doi: 10.1056/NEJMoa051568
37. Kuijpers TW, Bende RJ, Baars PA, Grummels A, Derks IA, Dolman KM, et al. CD20 Deficiency in Humans Results in Impaired T Cell-Independent Antibody Responses. J Clin Invest (2010) 120:214–22. doi: 10.1172/JCI40231
38. Bossaller L, Burger J, Draeger R, Grimbacher B, Knoth R, Plebani A, et al. ICOS Deficiency Is Associated With a Severe Reduction of CXCR5+CD4 Germinal Center Th Cells. J Immunol (2006) 177:4927–32. doi: 10.4049/jimmunol.177.7.4927
39. Li J, Jorgensen SF, Maggadottir SM, Bakay M, Warnatz K, Glessner J, et al. Association of CLEC16A With Human Common Variable Immunodeficiency Disorder and Role in Murine B Cells. Nat Commun (2015) 6:6804. doi: 10.1038/ncomms7804
40. Thaventhiran JED, Lango Allen H, Burren OS, Rae W, Greene D, Staples E, et al. Whole-Genome Sequencing of a Sporadic Primary Immunodeficiency Cohort. Nature (2020) 583:90–5. doi: 10.1038/s41586-020-2265-1
41. Perez EE, Orange JS, Bonilla F, Chinen J, Chinn IK, Dorsey M, et al. Update on the Use of Immunoglobulin in Human Disease: A Review of Evidence. J Allergy Clin Immunol (2017) 139:S1–46. doi: 10.1016/j.jaci.2016.09.023
42. Resnick ES, Moshier EL, Godbold JH, Cunningham-Rundles C. Morbidity and Mortality in Common Variable Immune Deficiency Over 4 Decades. Blood (2012) 119:1650–7. doi: 10.1182/blood-2011-09-377945
43. Berbers RM, Nierkens S, van Laar JM, Bogaert D, Leavis HL. Microbial Dysbiosis in Common Variable Immune Deficiencies: Evidence, Causes, and Consequences. Trends Immunol (2017) 38:206–16. doi: 10.1016/j.it.2016.11.008
44. Pecoraro A, Crescenzi L, Varricchi G, Marone G, Spadaro G. Heterogeneity of Liver Disease in Common Variable Immunodeficiency Disorders. Front Immunol (2020) 11:338. doi: 10.3389/fimmu.2020.00338
45. Varricchi G, Pecoraro A, Crescenzi L, Marone G, Travaglino A, D’Armiento FP, et al. Gastroduodenal Disorders in Patients With CVID Undergoing Immunoglobulin Therapy. Curr Pharm Biotechnol (2018) 19:734–41. doi: 10.2174/1389201019666181010170630
46. Jolles S, Orange JS, Gardulf A, Stein MR, Shapiro R, Borte M, et al. Current Treatment Options With Immunoglobulin G for the Individualization of Care in Patients With Primary Immunodeficiency Disease. Clin Exp Immunol (2015) 179:146–60. doi: 10.1111/cei.12485
47. Bonilla FA. Intravenous and Subcutaneous Immunoglobulin G Replacement Therapy. Allergy Asthma Proc (2016) 37:426–31. doi: 10.2500/aap.2016.37.3987
48. Uzzan M, Ko HM, Mehandru S, Cunningham-Rundles C. Gastrointestinal Disorders Associated With Common Variable Immune Deficiency (CVID) and Chronic Granulomatous Disease (Cgd). Curr Gastroenterol Rep (2016) 18:17. doi: 10.1007/s11894-016-0491-3
49. Pecoraro A, Nappi L, Crescenzi L, D’Armiento FP, Genovese A, Spadaro G. Chronic Diarrhea in Common Variable Immunodeficiency: A Case Series and Review of the Literature. J Clin Immunol (2018) 38:67–76. doi: 10.1007/s10875-017-0461-z
50. Agarwal S, Mayer L. Pathogenesis and Treatment of Gastrointestinal Disease in Antibody Deficiency Syndromes. J Allergy Clin Immunol (2009) 124:658–64. doi: 10.1016/j.jaci.2009.06.018
51. Teahon K, Webster AD, Price AB, Weston J, Bjarnason I. Studies on the Enteropathy Associated With Primary Hypogammaglobulinaemia. Gut (1994) 35:1244–9. doi: 10.1136/gut.35.9.1244
52. Daniels JA, Lederman HM, Maitra A, Montgomery EA. Gastrointestinal Tract Pathology in Patients With Common Variable Immunodeficiency (CVID): A Clinicopathologic Study and Review. Am J Surg Pathol (2007) 31:1800–12. doi: 10.1097/PAS.0b013e3180cab60c
53. Malamut G, Verkarre V, Suarez F, Viallard JF, Lascaux AS, Cosnes J, et al. The Enteropathy Associated With Common Variable Immunodeficiency: The Delineated Frontiers With Celiac Disease. Am J Gastroenterol (2010) 105:2262–75. doi: 10.1038/ajg.2010.214
54. Shulzhenko N, Dong X, Vyshenska D, Greer RL, Gurung M, Vasquez-Perez S, et al. CVID Enteropathy is Characterized by Exceeding Low Mucosal IgA Levels and Interferon-Driven Inflammation Possibly Related to the Presence of a Pathobiont. Clin Immunol (2018) 197:139–53. doi: 10.1016/j.clim.2018.09.008
55. Fiedorova K, Radvansky M, Bosak J, Grombirikova H, Nemcova E, Kralickova P, et al. Bacterial But Not Fungal Gut Microbiota Alterations are Associated With Common Variable Immunodeficiency (Cvid) Phenotype. Front Immunol (2019) 10:1914. doi: 10.3389/fimmu.2019.01914
56. Bosak J, Lexa M, Fiedorova K, Gadara DC, Micenkova L, Spacil Z, et al. Patients With Common Variable Immunodeficiency (Cvid) Show Higher Gut Bacterial Diversity and Levels of Low-Abundance Genes Than the Healthy Housemates. Front Immunol (2021) 12:671239. doi: 10.3389/fimmu.2021.671239
57. Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, Stability and Resilience of the Human Gut Microbiota. Nature (2012) 489:220–30. doi: 10.1038/nature11550
58. Cols M, Rahman A, Maglione PJ, Garcia-Carmona Y, Simchoni N, Ko HM, et al. Expansion of Inflammatory Innate Lymphoid Cells in Patients With Common Variable Immune Deficiency. J Allergy Clin Immunol (2016) 137:1206–15.e1206. doi: 10.1016/j.jaci.2015.09.013
59. Welihinda AA, Kaur M, Greene K, Zhai Y, Amento EP. The Adenosine Metabolite Inosine is a Functional Agonist of the Adenosine A2A Receptor With a Unique Signaling Bias. Cell Signal (2016) 28:552–60. doi: 10.1016/j.cellsig.2016.02.010
60. He B, Hoang TK, Wang T, Ferris M, Taylor CM, Tian X, et al. Resetting Microbiota by Lactobacillus Reuteri Inhibits T Reg Deficiency-Induced Autoimmunity Via Adenosine A2A Receptors. J Exp Med (2017) 214:107–23. doi: 10.1084/jem.20160961
61. Fadlallah J, Sterlin D, Fieschi C, Parizot C, Dorgham K, El Kafsi H, et al. Synergistic Convergence of Microbiota-Specific Systemic IgG and Secretory Iga. J Allergy Clin Immunol (2019) 143:1575–85.e1574. doi: 10.1016/j.jaci.2018.09.036
62. Berbers RM, Mohamed Hoesein FAA, Ellerbroek PM, van Montfrans JM, Dalm V, van Hagen PM, et al. Low IgA Associated With Oropharyngeal Microbiota Changes and Lung Disease in Primary Antibody Deficiency. Front Immunol (2020) 11:1245. doi: 10.3389/fimmu.2020.01245
63. van Schewick CM, Noltner C, Abel S, Burns SO, Workman S, Symes A, et al. Altered Microbiota, Impaired Quality of Life, Malabsorption, Infection, and Inflammation in CVID Patients With Diarrhoea. Front Immunol (2020) 11:1654. doi: 10.3389/fimmu.2020.01654
64. Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, et al. Microbial Translocation Is a Cause of Systemic Immune Activation in Chronic HIV Infection. Nat Med (2006) 12:1365–71. doi: 10.1038/nm1511
65. Sandler NG, Douek DC. Microbial Translocation in HIV Infection: Causes, Consequences and Treatment Opportunities. Nat Rev Microbiol (2012) 10:655–66. doi: 10.1038/nrmicro2848
66. Jorgensen SF, Macpherson ME, Bjornetro T, Holm K, Kummen M, Rashidi A, et al. Rifaximin Alters Gut Microbiota Profile, But Does Not Affect Systemic Inflammation - A Randomized Controlled Trial in Common Variable Immunodeficiency. Sci Rep (2019) 9:167. doi: 10.1038/s41598-018-35367-7
67. Macpherson ME, Hov JR, Ueland T, Dahl TB, Kummen M, Otterdal K, et al. Gut Microbiota-Dependent Trimethylamine N-Oxide Associates With Inflammation in Common Variable Immunodeficiency. Front Immunol (2020) 11:574500. doi: 10.3389/fimmu.2020.574500
68. Franco-Esquivias AP, Pena CG, Torres-Lozano C, Vaca-Paniagua F, Diaz-Velasquez C, Ortega-Cisneros M, et al. Gut Microbiota in Mexican Patients With Common Variable Immunodeficiency. Gac Med Mex (2019) 155:447–52. doi: 10.24875/GMM.M20000330
69. Quinti I, Soresina A, Spadaro G, Martino S, Donnanno S, Agostini C, et al. Long-Term Follow-Up and Outcome of a Large Cohort of Patients With Common Variable Immunodeficiency. J Clin Immunol (2007) 27:308–16. doi: 10.1007/s10875-007-9075-1
70. Oksenhendler E, Gerard L, Fieschi C, Malphettes M, Mouillot G, Jaussaud R, et al. Infections in 252 Patients With Common Variable Immunodeficiency. Clin Infect Dis (2008) 46:1547–54. doi: 10.1086/587669
71. Fine KD, Seidel RH, Do K. The Prevalence, Anatomic Distribution, and Diagnosis of Colonic Causes of Chronic Diarrhea. Gastrointest Endosc (2000) 51:318–26. doi: 10.1016/S0016-5107(00)70362-2
72. Baniadam L, Arshi S, Nabavi M, Yousefi A, Fallahpour M, Shokri S, et al. Can Concurrent Lower Gastrointestinal Manifestations Help the Timely Diagnosis of Small Intestinal Bacterial Overgrowth in CVID Patients? Eur Ann Allergy Clin Immunol (2021) 53:18–22. doi: 10.23822/EurAnnACI.1764-1489.137
73. Zhang Z, Li J, Zheng W, Zhao G, Zhang H, Wang X, et al. Peripheral Lymphoid Volume Expansion and Maintenance are Controlled by Gut Microbiota Via RALDH+ Dendritic Cells. Immunity (2016) 44:330–42. doi: 10.1016/j.immuni.2016.01.004
74. Wahlstrom A, Sayin SI, Marschall HU, Backhed F. Intestinal Crosstalk Between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab (2016) 24:41–50. doi: 10.1016/j.cmet.2016.05.005
75. de Aguiar Vallim TQ, Tarling EJ, Edwards PA. Pleiotropic Roles of Bile Acids in Metabolism. Cell Metab (2013) 17:657–69. doi: 10.1016/j.cmet.2013.03.013
76. Campbell C, McKenney PT, Konstantinovsky D, Isaeva OI, Schizas M, Verter J, et al. Bacterial Metabolism of Bile Acids Promotes Generation of Peripheral Regulatory T Cells. Nature (2020) 581:475–9. doi: 10.1038/s41586-020-2193-0
77. Hang S, Paik D, Yao L, Kim E, Trinath J, Lu J, et al. Bile Acid Metabolites Control TH17 and Treg Cell Differentiation. Nature (2019) 576:143–8. doi: 10.1038/s41586-019-1785-z
78. Pandiyan P, Bhaskaran N, Zou M, Schneider E, Jayaraman S, Huehn J. Microbiome Dependent Regulation of Tregs and Th17 Cells in Mucosa. Front Immunol (2019) 10:426. doi: 10.3389/fimmu.2019.00426
79. Tanoue T, Atarashi K, Honda K. Development and Maintenance of Intestinal Regulatory T Cells. Nat Rev Immunol (2016) 16:295–309. doi: 10.1038/nri.2016.36
80. Cristinziano L, Poto R, Criscuolo G, Ferrara AL, Galdiero MR, Modestino L, et al. Il-33 and Superantigenic Activation of Human Lung Mast Cells Induce the Release of Angiogenic and Lymphangiogenic Factors. Cells (2021) 10:145–64. doi: 10.3390/cells10010145
81. Simoni Y, Fehlings M, Kloverpris HN, McGovern N, Koo SL, Loh CY, et al. Human Innate Lymphoid Cell Subsets Possess Tissue-Type Based Heterogeneity in Phenotype and Frequency. Immunity (2017) 46:148–61. doi: 10.1016/j.immuni.2016.11.005
82. Sonnenberg GF, Monticelli LA, Alenghat T, Fung TC, Hutnick NA, Kunisawa J, et al. Innate Lymphoid Cells Promote Anatomical Containment of Lymphoid-Resident Commensal Bacteria. Science (2012) 336:1321–5. doi: 10.1126/science.1222551
83. Hanash AM, Dudakov JA, Hua G, O’Connor MH, Young LF, Singer NV, et al. Interleukin-22 Protects Intestinal Stem Cells From Immune-Mediated Tissue Damage and Regulates Sensitivity to Graft Versus Host Disease. Immunity (2012) 37:339–50. doi: 10.1016/j.immuni.2012.05.028
84. Speakman EA, Dambuza IM, Salazar F, Brown GD. T Cell Antifungal Immunity and the Role of C-Type Lectin Receptors. Trends Immunol (2020) 41:61–76. doi: 10.1016/j.it.2019.11.007
85. Biancheri P, Di Sabatino A, Rescigno M, Giuffrida P, Fornasa G, Tsilingiri K, et al. Abnormal Thymic Stromal Lymphopoietin Expression in the Duodenal Mucosa of Patients With Coeliac Disease. Gut (2016) 65:1670–80. doi: 10.1136/gutjnl-2014-308876
86. Varricchi G, Pecoraro A, Marone G, Criscuolo G, Spadaro G, Thymic Stromal Lymphopoietin Isoforms A. Inflammatory Disorders, and Cancer. Front Immunol (2018) 9:1595. doi: 10.3389/fimmu.2018.01595
87. Fornasa G, Tsilingiri K, Caprioli F, Botti F, Mapelli M, Meller S, et al. Dichotomy of Short and Long Thymic Stromal Lymphopoietin Isoforms in Inflammatory Disorders of the Bowel and Skin. J Allergy Clin Immunol (2015) 136:413–22. doi: 10.1016/j.jaci.2015.04.011
88. Hand TW. The Role of the Microbiota in Shaping Infectious Immunity. Trends Immunol (2016) 37:647–58. doi: 10.1016/j.it.2016.08.007
89. Perreau M, Vigano S, Bellanger F, Pellaton C, Buss G, Comte D, et al. Exhaustion of Bacteria-Specific CD4 T Cells and Microbial Translocation in Common Variable Immunodeficiency Disorders. J Exp Med (2014) 211:2033–45. doi: 10.1084/jem.20140039
90. Staiano RI, Loffredo S, Borriello F, Iannotti FA, Piscitelli F, Orlando P, et al. Human Lung-Resident Macrophages Express CB1 and CB2 Receptors Whose Activation Inhibits the Release of Angiogenic and Lymphangiogenic Factors. J Leukoc Biol (2016) 99:531–40. doi: 10.1189/jlb.3HI1214-584R
91. Braile M, Cristinziano L, Marcella S, Varricchi G, Marone G, Modestino L, et al. LPS-Mediated Neutrophil VEGF-A Release Is Modulated by Cannabinoid Receptor Activation. J Leukoc Biol (2021) 109:621–31. doi: 10.1002/JLB.3A0520-187R
92. Marshall JS. Mast-Cell Responses to Pathogens. Nat Rev Immunol (2004) 4:787–99. doi: 10.1038/nri1460
93. Varricchi G, Loffredo S, Borriello F, Pecoraro A, Rivellese F, Genovese A, et al. Superantigenic Activation of Human Cardiac Mast Cells. Int J Mol Sci (2019) 20:1828–44. doi: 10.3390/ijms20081828
94. Bernink JH, Krabbendam L, Germar K, de Jong E, Gronke K, Kofoed-Nielsen M, et al. Interleukin-12 and -23 Control Plasticity of CD127(+) Group 1 and Group 3 Innate Lymphoid Cells in the Intestinal Lamina Propria. Immunity (2015) 43:146–60. doi: 10.1016/j.immuni.2015.06.019
95. Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites Produced by Commensal Bacteria Promote Peripheral Regulatory T-cell Generation. Nature (2013) 504:451–5. doi: 10.1038/nature12726
96. Li B, Selmi C, Tang R, Gershwin ME, Ma X. The Microbiome and Autoimmunity: A Paradigm From the Gut-Liver Axis. Cell Mol Immunol (2018) 15:595–609. doi: 10.1038/cmi.2018.7
97. Qiu Y, Yang J, Wang L, Yang X, Gao K, Zhu C, et al. Dietary Resveratrol Attenuation of Intestinal Inflammation and Oxidative Damage is Linked to the Alteration of Gut Microbiota and Butyrate in Piglets Challenged With Deoxynivalenol. J Anim Sci Biotechnol (2021) 12:71. doi: 10.1186/s40104-021-00596-w
98. Bayazid AB, Jang YA, Kim YM, Kim JG, Lim BO. Neuroprotective Effects of Sodium Butyrate Through Suppressing Neuroinflammation and Modulating Antioxidant Enzymes. Neurochem Res (2021). doi: 10.1007/s11064-021-03369-z
99. Durandy A, Kracker S, Fischer A. Primary Antibody Deficiencies. Nat Rev Immunol (2013) 13:519–33. doi: 10.1038/nri3466
100. Mohammed AD, Khan MAW, Chatzistamou I, Chamseddine D, Williams-Kang K, Perry M, et al. Gut Antibody Deficiency in a Mouse Model of CVID Results in Spontaneous Development of a Gluten-Sensitive Enteropathy. Front Immunol (2019) 10:2484. doi: 10.3389/fimmu.2019.02484
101. Wang Y, Carter RH. CD19 Regulates B Cell Maturation, Proliferation, and Positive Selection in the FDC Zone of Murine Splenic Germinal Centers. Immunity (2005) 22:749–61. doi: 10.1016/j.immuni.2005.04.012
102. Fehr T, Rickert RC, Odermatt B, Roes J, Rajewsky K, Hengartner H, et al. Antiviral Protection and Germinal Center Formation, But Impaired B Cell Memory in the Absence of CD19. J Exp Med (1998) 188:145–55. doi: 10.1084/jem.188.1.145
103. Salzer U, Grimbacher B. Monogenetic Defects in Common Variable Immunodeficiency: What Can We Learn About Terminal B Cell Differentiation? Curr Opin Rheumatol (2006) 18:377–82. doi: 10.1097/01.bor.0000231906.12172.6e
104. Okai S, Usui F, Ohta M, Mori H, Kurokawa K, Matsumoto S, et al. Intestinal IgA as a Modulator of the Gut Microbiota. Gut Microbes (2017) 8:486–92. doi: 10.1080/19490976.2017.1310357
105. Nakajima A, Vogelzang A, Maruya M, Miyajima M, Murata M, Son A, et al. Iga Regulates the Composition and Metabolic Function of Gut Microbiota by Promoting Symbiosis Between Bacteria. J Exp Med (2018) 215:2019–34. doi: 10.1084/jem.20180427
106. Pabst O, Slack E. Iga and the Intestinal Microbiota: The Importance of Being Specific. Mucosal Immunol (2020) 13:12–21. doi: 10.1038/s41385-019-0227-4
107. Jorgensen SF, Fevang B. And Aukrust, P. Commentary: Gut Antibody Deficiency in a Mouse Model of CVID Results in Spontaneous Development of a Gluten-Sensitive Enteropathy. Front Immunol (2020) 11:1921–23. doi: 10.3389/fimmu.2020.01921
108. Gasbarrini A, Corazza GR, Gasbarrini G, Montalto M, Di Stefano M, Basilisco G, et al. Methodology and Indications of H2-Breath Testing in Gastrointestinal Diseases: The Rome Consensus Conference. Aliment Pharmacol Ther (2009) 29(Suppl 1):1–49. doi: 10.1111/j.1365-2036.2009.03951.x
109. Quigley EMM, Murray JA, Pimentel M. Aga Clinical Practice Update on Small Intestinal Bacterial Overgrowth: Expert Review. Gastroenterology (2020) 159:1526–32. doi: 10.1053/j.gastro.2020.06.090
110. Lauritano EC, Valenza V, Sparano L, Scarpellini E, Gabrielli M, Cazzato A, et al. Small Intestinal Bacterial Overgrowth and Intestinal Permeability. Scand J Gastroenterol (2010) 45:1131–2. doi: 10.3109/00365521.2010.485325
111. Groves HT, Higham SL, Moffatt MF, Cox MJ, Tregoning JS. Respiratory Viral Infection Alters the Gut Microbiota by Inducing Inappetence. mBio (2020) 11:e03236-19. doi: 10.1128/mBio.03236-19
112. Wang J, Li F, Wei H, Lian ZX, Sun R, Tian Z. Respiratory Influenza Virus Infection Induces Intestinal Immune Injury Via Microbiota-Mediated Th17 Cell-Dependent Inflammation. J Exp Med (2014) 211:2397–410. doi: 10.1084/jem.20140625
113. Deriu E, Boxx GM, He X, Pan C, Benavidez SD, Cen L, et al. Influenza Virus Affects Intestinal Microbiota and Secondary Salmonella Infection in the Gut Through Type I Interferons. PloS Pathog (2016) 12:e1005572. doi: 10.1371/journal.ppat.1005572
114. Bartley JM, Zhou X, Kuchel GA, Weinstock GM. And Haynes, L. Impact of Age, Caloric Restriction, and Influenza Infection on Mouse Gut Microbiome: An Exploratory Study of the Role of Age-Related Microbiome Changes on Influenza Responses. Front Immunol (2017) 8:1164. doi: 10.3389/fimmu.2017.01164
115. Groves HT, Cuthbertson L, James P, Moffatt MF, Cox MJ, Tregoning JS. Respiratory Disease Following Viral Lung Infection Alters the Murine Gut Microbiota. Front Immunol (2018) 9:182. doi: 10.3389/fimmu.2018.00182
116. Yildiz S, Mazel-Sanchez B, Kandasamy M, Manicassamy B, Schmolke M. Influenza A Virus Infection Impacts Systemic Microbiota Dynamics and Causes Quantitative Enteric Dysbiosis. Microbiome (2018) 6:9. doi: 10.1186/s40168-017-0386-z
117. Hanada S, Pirzadeh M, Carver KY, Deng JC. Respiratory Viral Infection-Induced Microbiome Alterations and Secondary Bacterial Pneumonia. Front Immunol (2018) 9:2640. doi: 10.3389/fimmu.2018.02640
118. Samuelson DR, Charles TP, de la Rua NM, Taylor CM, Blanchard EE, Luo M, et al. Analysis of the Intestinal Microbial Community and Inferred Functional Capacities During the Host Response to Pneumocystis Pneumonia. Exp Lung Res (2016) 42:425–39. doi: 10.1080/01902148.2016.1258442
119. Sze MA, Tsuruta M, Yang SW, Oh Y, Man SF, Hogg JC, et al. Changes in the Bacterial Microbiota in Gut, Blood, and Lungs Following Acute LPS Instillation Into Mice Lungs. PloS One (2014) 9:e111228. doi: 10.1371/journal.pone.0111228
120. Huang Y, Mao K, Chen X, Sun MA, Kawabe T, Li W, et al. S1P-Dependent Interorgan Trafficking of Group 2 Innate Lymphoid Cells Supports Host Defense. Science (2018) 359:114–9. doi: 10.1126/science.aam5809
121. Young RP, Hopkins RJ, Marsland B. The Gut-Liver-Lung Axis. Modulation of the Innate Immune Response and Its Possible Role in Chronic Obstructive Pulmonary Disease. Am J Respir Cell Mol Biol (2016) 54:161–9. doi: 10.1165/rcmb.2015-0250PS
122. Fiorucci S, Distrutti E, Carino A, Zampella A, Biagioli M. Bile Acids and Their Receptors in Metabolic Disorders. Prog Lipid Res (2021) 82:101094. doi: 10.1016/j.plipres.2021.101094
123. Fiorucci S, Biagioli M, Zampella A, Distrutti E. Bile Acids Activated Receptors Regulate Innate Immunity. Front Immunol (2018) 9:1853. doi: 10.3389/fimmu.2018.01853
124. Parseus A, Sommer N, Sommer F, Caesar R, Molinaro A, Stahlman M, et al. Microbiota-Induced Obesity Requires Farnesoid X Receptor. Gut (2017) 66:429–37. doi: 10.1136/gutjnl-2015-310283
125. Kawamata Y, Fujii R, Hosoya M, Harada M, Yoshida H, Miwa M, et al. A G Protein-Coupled Receptor Responsive to Bile Acids. J Biol Chem (2003) 278:9435–40. doi: 10.1074/jbc.M209706200
126. Vavassori P, Mencarelli A, Renga B, Distrutti E, Fiorucci S. The Bile Acid Receptor FXR is a Modulator of Intestinal Innate Immunity. J Immunol (2009) 183:6251–61. doi: 10.4049/jimmunol.0803978
127. Mencarelli A, Renga B, Migliorati M, Cipriani S, Distrutti E, Santucci L, et al. The Bile Acid Sensor Farnesoid X Receptor is a Modulator of Liver Immunity in a Rodent Model of Acute Hepatitis. J Immunol (2009) 183:6657–66. doi: 10.4049/jimmunol.0901347
128. Yang BH, Hagemann S, Mamareli P, Lauer U, Hoffmann U, Beckstette M, et al. Foxp3(+) T Cells Expressing RORgammat Represent a Stable Regulatory T-cell Effector Lineage With Enhanced Suppressive Capacity During Intestinal Inflammation. Mucosal Immunol (2016) 9:444–57. doi: 10.1038/mi.2015.74
129. DiSpirito JR, Zemmour D, Ramanan D, Cho J, Zilionis R, Klein AM, et al. Molecular Diversification of Regulatory T Cells in Nonlymphoid Tissues. Sci Immunol (2018) 3:eaat5861. doi: 10.1126/sciimmunol.aat5861
130. Song X, Sun X, Oh SF, Wu M, Zhang Y, Zheng W, et al. Microbial Bile Acid Metabolites Modulate Gut RORgamma(+) Regulatory T Cell Homeostasis. Nature (2020) 577:410–5. doi: 10.1038/s41586-019-1865-0
131. Leone P, Vacca A, Dammacco F, Racanelli V. Common Variable Immunodeficiency and Gastric Malignancies. Int J Mol Sci (2018) 19:451–63. doi: 10.3390/ijms19020451
132. Mellemkjaer L, Hammarstrom L, Andersen V, Yuen J, Heilmann C, Barington T, et al. Cancer Risk Among Patients With IgA Deficiency or Common Variable Immunodeficiency and Their Relatives: A Combined Danish and Swedish Study. Clin Exp Immunol (2002) 130:495–500. doi: 10.1046/j.1365-2249.2002.02004.x
133. Vajdic CM, Mao L, van Leeuwen MT, Kirkpatrick P, Grulich AE, Riminton S. Are Antibody Deficiency Disorders Associated With a Narrower Range of Cancers Than Other Forms of Immunodeficiency? Blood (2010) 116:1228–34. doi: 10.1182/blood-2010-03-272351
134. Kiaee F, Azizi G, Rafiemanesh H, Zainaldain H, Sadaat Rizvi F, Alizadeh M, et al. Malignancy in Common Variable Immunodeficiency: A Systematic Review and Meta-Analysis. Expert Rev Clin Immunol (2019) 15:1105–13. doi: 10.1080/1744666X.2019.1658523
135. Pulvirenti F, Pecoraro A, Cinetto F, Milito C, Valente M, Santangeli E, et al. Gastric Cancer is the Leading Cause of Death in Italian Adult Patients With Common Variable Immunodeficiency. Front Immunol (2018) 9:2546. doi: 10.3389/fimmu.2018.02546
136. Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell (2011) 144:646–74. doi: 10.1016/j.cell.2011.02.013
137. Cristinziano L, Modestino L, Antonelli A, Marone G, Simon HU, Varricchi G, et al. Neutrophil Extracellular Traps in Cancer. Semin Canc Biol (2021).
138. Albrengues J, Shields MA, Ng D, Park CG, Ambrico A, Poindexter ME, et al. Neutrophil Extracellular Traps Produced During Inflammation Awaken Dormant Cancer Cells in Mice. Science (2018) 361:eaao4227. doi: 10.1126/science.aao4227
139. Barkan D, El Touny LH, Michalowski AM, Smith JA, Chu I, Davis AS, et al. Metastatic Growth From Dormant Cells Induced by a col-I-enriched Fibrotic Environment. Cancer Res (2010) 70:5706–16. doi: 10.1158/0008-5472.CAN-09-2356
140. Gullo I, Costa C, Silva SL, Ferreira C, Motta A, Silva SP, et al. The Dysfunctional Immune System in Common Variable Immunodeficiency Increases the Susceptibility to Gastric Cancer. Cells (2020) 9:1498–518. doi: 10.3390/cells9061498
141. Rajilic-Stojanovic M, Figueiredo C, Smet A, Hansen R, Kupcinskas J, Rokkas T, et al. Systematic Review: Gastric Microbiota in Health and Disease. Aliment Pharmacol Ther (2020) 51:582–602. doi: 10.1111/apt.15650
142. Quiding-Jarbrink M, Sundstrom P, Lundgren A, Hansson M, Backstrom M, Johansson C, et al. Decreased IgA Antibody Production in the Stomach of Gastric Adenocarcinoma Patients. Clin Immunol (2009) 131:463–71. doi: 10.1016/j.clim.2009.01.010
143. Eun CS, Kim BK, Han DS, Kim SY, Kim KM, Choi BY, et al. Differences in Gastric Mucosal Microbiota Profiling in Patients With Chronic Gastritis, Intestinal Metaplasia, and Gastric Cancer Using Pyrosequencing Methods. Helicobacter (2014) 19:407–16. doi: 10.1111/hel.12145
144. Wang Z, Gao X, Zeng R, Wu Q, Sun H, Wu W, et al. Changes of the Gastric Mucosal Microbiome Associated With Histological Stages of Gastric Carcinogenesis. Front Microbiol (2020) 11:997. doi: 10.3389/fmicb.2020.00997
145. Aviles-Jimenez F, Vazquez-Jimenez F, Medrano-Guzman R, Mantilla A. And Torres, J. Stomach Microbiota Composition Varies Between Patients With non-Atrophic Gastritis and Patients With Intestinal Type of Gastric Cancer. Sci Rep (2014) 4:4202. doi: 10.1038/srep04202
146. Wang L, Zhou J, Xin Y, Geng C, Tian Z, Yu X, et al. Bacterial Overgrowth and Diversification of Microbiota in Gastric Cancer. Eur J Gastroenterol Hepatol (2016) 28:261–6. doi: 10.1097/MEG.0000000000000542
147. Coker OO, Dai Z, Nie Y, Zhao G, Cao L, Nakatsu G, et al. Mucosal Microbiome Dysbiosis in Gastric Carcinogenesis. Gut (2018) 67:1024–32. doi: 10.1136/gutjnl-2017-314281
148. Ianiro G, Molina-Infante J, Gasbarrini A. Gastric Microbiota. Helicobacter (2015) 20(Suppl 1):68–71. doi: 10.1111/hel.12260
149. Ferreira RM, Pereira-Marques J, Pinto-Ribeiro I, Costa JL, Carneiro F, Machado JC, et al. Gastric Microbial Community Profiling Reveals a Dysbiotic Cancer-Associated Microbiota. Gut (2018) 67:226–36. doi: 10.1136/gutjnl-2017-314205
150. Yu G, Torres J, Hu N, Medrano-Guzman R, Herrera-Goepfert R, Humphrys MS, et al. Molecular Characterization of the Human Stomach Microbiota in Gastric Cancer Patients. Front Cell Infect Microbiol (2017) 7:302. doi: 10.3389/fcimb.2017.00302
151. Gathmann B, Mahlaoui N, Gerard L, Oksenhendler E, Warnatz K, Schulze I, et al. Clinical Picture and Treatment of 2212 Patients With Common Variable Immunodeficiency. J Allergy Clin Immunol (2014) 134:116–26. CEREDIH. doi: 10.1016/j.jaci.2013.12.1077
152. Sperlich JM, Grimbacher B, Workman S, Haque T, Seneviratne SL, Burns SO, et al. Respiratory Infections and Antibiotic Usage in Common Variable Immunodeficiency. J Allergy Clin Immunol Pract (2018) 6:159–68.e153. doi: 10.1016/j.jaip.2017.05.024
153. Blaser MJ. Antibiotic Use and its Consequences for the Normal Microbiome. Science (2016) 352:544–5. doi: 10.1126/science.aad9358
154. Rizzatti G, Ianiro G, Gasbarrini A. Antibiotic and Modulation of Microbiota: A New Paradigm? J Clin Gastroenterol (2018) 52(Suppl 1):S74–7. Proceedings from the 9th Probiotics, Prebiotics and New Foods, Nutraceuticals and Botanicals for Nutrition & Human and Microbiota Health Meeting, held in Rome, Italy from September 10 to 12, 2017. doi: 10.1097/MCG.0000000000001069
155. Bajaj JS, Heuman DM, Sanyal AJ, Hylemon PB, Sterling RK, Stravitz RT, et al. Modulation of the Metabiome by Rifaximin in Patients With Cirrhosis and Minimal Hepatic Encephalopathy. PloS One (2013) 8:e60042. doi: 10.1371/journal.pone.0060042
156. Kalambokis GN, Tsianos EV. Rifaximin Reduces Endotoxemia and Improves Liver Function and Disease Severity in Patients With Decompensated Cirrhosis. Hepatology (2012) 55:655–6. doi: 10.1002/hep.24751
157. Rolain JM. Food and Human Gut as Reservoirs of Transferable Antibiotic Resistance Encoding Genes. Front Microbiol (2013) 4:173. doi: 10.3389/fmicb.2013.00173
158. Modi SR, Collins JJ, Relman DA. Antibiotics and the Gut Microbiota. J Clin Invest (2014) 124:4212–8. doi: 10.1172/JCI72333
159. Emamalipour M, Seidi K, Zununi Vahed S, Jahanban-Esfahlan A, Jaymand M, Majdi H, et al. Horizontal Gene Transfer: From Evolutionary Flexibility to Disease Progression. Front Cell Dev Biol (2020) 8:229. doi: 10.3389/fcell.2020.00229
160. Palleja A, Mikkelsen KH, Forslund SK, Kashani A, Allin KH, Nielsen T, et al. Recovery of Gut Microbiota of Healthy Adults Following Antibiotic Exposure. Nat Microbiol (2018) 3:1255–65. doi: 10.1038/s41564-018-0257-9
161. De La Cochetiere MF, Durand T, Lepage P, Bourreille A, Galmiche JP, Dore J. Resilience of the Dominant Human Fecal Microbiota Upon Short-Course Antibiotic Challenge. J Clin Microbiol (2005) 43:5588–92. doi: 10.1128/JCM.43.11.5588-5592.2005
162. Jernberg C, Lofmark S, Edlund C, Jansson JK. Long-Term Ecological Impacts of Antibiotic Administration on the Human Intestinal Microbiota. ISME J (2007) 1:56–66. doi: 10.1038/ismej.2007.3
163. Dethlefsen L, Huse S, Sogin ML, Relman DA. The Pervasive Effects of an Antibiotic on the Human Gut Microbiota, as Revealed by Deep 16S rRNA Sequencing. PloS Biol (2008) 6:e280. doi: 10.1371/journal.pbio.0060280
164. Dethlefsen L, Relman DA. Incomplete Recovery and Individualized Responses of the Human Distal Gut Microbiota to Repeated Antibiotic Perturbation. Proc Natl Acad Sci USA (2011) 108(Suppl 1):4554–61. doi: 10.1073/pnas.1000087107
165. Sartelli M, Di Bella S, McFarland LV, Khanna S, Furuya-Kanamori L, Abuzeid N, et al. 2019 Update of the WSES Guidelines for Management of Clostridioides (Clostridium) Difficile Infection in Surgical Patients. World J Emerg Surg (2019) 14:8. doi: 10.1186/s13017-019-0228-3
166. Becattini S, Taur Y, Pamer EG. Antibiotic-Induced Changes in the Intestinal Microbiota and Disease. Trends Mol Med (2016) 22:458–78. doi: 10.1016/j.molmed.2016.04.003
167. Leffler DA, Lamont JT. Clostridium Difficile Infection. N Engl J Med (2015) 373:287–8. doi: 10.1056/NEJMc1506004
168. Khan S, Hauptman R, Kelly L. Engineering the Microbiome to Prevent Adverse Events: Challenges and Opportunities. Annu Rev Pharmacol Toxicol (2021) 61:159–79. doi: 10.1146/annurev-pharmtox-031620-031509
169. Lawson CE, Harcombe WR, Hatzenpichler R, Lindemann SR, Loffler FE, O’Malley MA, et al. Common Principles and Best Practices for Engineering Microbiomes. Nat Rev Microbiol (2019) 17:725–41. doi: 10.1038/s41579-019-0255-9
170. Zmora N, Zilberman-Schapira G, Suez J, Mor U, Dori-Bachash M, Bashiardes S, et al. Personalized Gut Mucosal Colonization Resistance to Empiric Probiotics Is Associated With Unique Host and Microbiome Features. Cell (2018) 174:1388–405.e1321. doi: 10.1016/j.cell.2018.08.041
171. Sheflin AM, Melby CL, Carbonero F, Weir TL. Linking Dietary Patterns With Gut Microbial Composition and Function. Gut Microbes (2017) 8:113–29. doi: 10.1080/19490976.2016.1270809
172. Wypych TP, Marsland BJ. Diet Hypotheses in Light of the Microbiota Revolution: New Perspectives. Nutrients (2017) 9:537–49. doi: 10.3390/nu9060537
173. Kim KS, Hong SW, Han D, Yi J, Jung J, Yang BG, et al. Dietary Antigens Limit Mucosal Immunity by Inducing Regulatory T Cells in the Small Intestine. Science (2016) 351:858–63. doi: 10.1126/science.aac5560
174. Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJ, et al. Expert Consensus Document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) Consensus Statement on the Definition and Scope of Prebiotics. Nat Rev Gastroenterol Hepatol (2017) 14:491–502. doi: 10.1038/nrgastro.2017.75
175. Simpson HL, Campbell BJ. Review Article: Dietary Fibre-Microbiota Interactions. Aliment Pharmacol Ther (2015) 42:158–79. doi: 10.1111/apt.13248
176. Markowiak P, Slizewska K. Effects of Probiotics, Prebiotics, and Synbiotics on Human Health. Nutrients (2017) 9:1021–50. doi: 10.3390/nu9091021
177. Geva-Zatorsky N, Sefik E, Kua L, Pasman L, Tan TG, Ortiz-Lopez A, et al. Mining the Human Gut Microbiota for Immunomodulatory Organisms. Cell (2017) 168:928–943 e911. doi: 10.1016/j.cell.2017.01.022
178. Millan B, Laffin M, Madsen K. Fecal Microbiota Transplantation: Beyond Clostridium Difficile. Curr Infect Dis Rep (2017) 19:31. doi: 10.1007/s11908-017-0586-5
179. Ianiro G, Murri R, Sciume GD, Impagnatiello M, Masucci L, Ford AC, et al. Incidence of Bloodstream Infections, Length of Hospital Stay, and Survival in Patients With Recurrent Clostridioides Difficile Infection Treated With Fecal Microbiota Transplantation or Antibiotics: A Prospective Cohort Study. Ann Intern Med (2019) 171:695–702. doi: 10.7326/M18-3635
180. Steidler L, Hans W, Schotte L, Neirynck S, Obermeier F, Falk W, et al. Treatment of Murine Colitis by Lactococcus Lactis Secreting Interleukin-10. Science (2000) 289:1352–5. doi: 10.1126/science.289.5483.1352
181. Paule A, Frezza D, Edeas M. Microbiota and Phage Therapy: Future Challenges in Medicine. Med Sci (Basel) (2018) 6:86–95. doi: 10.3390/medsci6040086
182. Schmitt FCF, Brenner T, Uhle F, Loesch S, Hackert T, Ulrich A, et al. Gut Microbiome Patterns Correlate With Higher Postoperative Complication Rates After Pancreatic Surgery. BMC Microbiol (2019) 19:42. doi: 10.1186/s12866-019-1399-5
183. Suez J, Zmora N, Segal E, Elinav E. The Pros, Cons, and Many Unknowns of Probiotics. Nat Med (2019) 25:716–29. doi: 10.1038/s41591-019-0439-x
184. Kang HJ, Kim GC, Lee CG, Park S, Sharma G, Verma R, et al. Probiotics-Derived Metabolite Ameliorates Skin Allergy by Promoting Differentiation of FOXP3(+) Regulatory T Cells. J Allergy Clin Immunol (2021) 147:1517–21. doi: 10.1016/j.jaci.2020.11.040
185. Genre J, Errante PR, Kokron CM, Toledo-Barros M, Camara NO, Rizzo LV. Reduced Frequency of CD4(+)CD25(HIGH)FOXP3(+) Cells and Diminished FOXP3 Expression in Patients With Common Variable Immunodeficiency: A Link to Autoimmunity? Clin Immunol (2009) 132:215–21. doi: 10.1016/j.clim.2009.03.519
186. Arandi N, Mirshafiey A, Jeddi-Tehrani M, Abolhassani H, Sadeghi B, Mirminachi B, et al. Evaluation of CD4+CD25+FOXP3+ Regulatory T Cells Function in Patients With Common Variable Immunodeficiency. Cell Immunol (2013) 281:129–33. doi: 10.1016/j.cellimm.2013.03.003
187. Brown JM, Hazen SL. Microbial Modulation of Cardiovascular Disease. Nat Rev Microbiol (2018) 16:171–81. doi: 10.1038/nrmicro.2017.149
188. Wang DD, Nguyen LH, Li Y, Yan Y, Ma W, Rinott E, et al. The Gut Microbiome Modulates the Protective Association Between a Mediterranean Diet and Cardiometabolic Disease Risk. Nat Med (2021) 27:333–43. doi: 10.1038/s41591-020-01223-3
189. Asnicar F, Berry SE, Valdes AM, Nguyen LH, Piccinno G, Drew DA, et al. Microbiome Connections With Host Metabolism and Habitual Diet From 1,098 Deeply Phenotyped Individuals. Nat Med (2021) 27:321–32. doi: 10.1038/s41591-020-01183-8
190. Sefik E, Geva-Zatorsky N, Oh S, Konnikova L, Zemmour D, McGuire AM, et al. Mucosal IMMUNOLOGY. Individual Intestinal Symbionts Induce a Distinct Population of RORgamma(+) Regulatory T Cells. Science (2015) 349:993–7. doi: 10.1126/science.aaa9420
191. Youngster I, Russell GH, Pindar C, Ziv-Baran T, Sauk J, Hohmann EL. Oral, Capsulized, Frozen Fecal Microbiota Transplantation for Relapsing Clostridium Difficile Infection. JAMA (2014) 312:1772–8. doi: 10.1001/jama.2014.13875
192. Hirsch BE, Saraiya N, Poeth K, Schwartz RM, Epstein ME, Honig G. Effectiveness of Fecal-Derived Microbiota Transfer Using Orally Administered Capsules for Recurrent Clostridium Difficile Infection. BMC Infect Dis (2015) 15:191. doi: 10.1186/s12879-015-0930-z
193. Kao D, Roach B, Silva M, Beck P, Rioux K, Kaplan GG, et al. Effect of Oral Capsule- vs Colonoscopy-Delivered Fecal Microbiota Transplantation on Recurrent Clostridium Difficile Infection: A Randomized Clinical Trial. JAMA (2017) 318:1985–93. doi: 10.1001/jama.2017.17077
194. Allegretti JR, Korzenik JR, Hamilton MJ. Fecal Microbiota Transplantation Via Colonoscopy for Recurrent C. Difficile Infection. J Vis Exp (2014) (94):52154. doi: 10.3791/52154
195. Silverman MS, Davis I, Pillai DR. Success of Self-Administered Home Fecal Transplantation for Chronic Clostridium Difficile Infection. Clin Gastroenterol Hepatol (2010) 8:471–3. doi: 10.1016/j.cgh.2010.01.007
196. Tariq R, Pardi DS, Bartlett MG, Khanna S. Low Cure Rates in Controlled Trials of Fecal Microbiota Transplantation for Recurrent Clostridium Difficile Infection: A Systematic Review and Meta-Analysis. Clin Infect Dis (2019) 68:1351–8. doi: 10.1093/cid/ciy721
197. Wilson BC, Vatanen T, Cutfield WS, O’Sullivan JM. The Super-Donor Phenomenon in Fecal Microbiota Transplantation. Front Cell Infect Microbiol (2019) 9:2. doi: 10.3389/fcimb.2019.00002
198. de Groot P, Scheithauer T, Bakker GJ, Prodan A, Levin E, Khan MT, et al. Donor Metabolic Characteristics Drive Effects of Faecal Microbiota Transplantation on Recipient Insulin Sensitivity, Energy Expenditure and Intestinal Transit Time. Gut (2020) 69:502–12. doi: 10.1136/gutjnl-2019-318320
199. Marcella C, Cui B, Kelly CR, Ianiro G, Cammarota G, Zhang F. Systematic Review: The Global Incidence of Faecal Microbiota Transplantation-Related Adverse Events From 2000 to 2020. Aliment Pharmacol Ther (2021) 53:33–42. doi: 10.1111/apt.16148
200. Ianiro G, Mullish BH, Kelly CR, Kassam Z, Kuijper EJ, Ng SC, et al. Reorganisation of Faecal Microbiota Transplant Services During the COVID-19 Pandemic. Gut (2020) 69:1555–63. doi: 10.1136/gutjnl-2020-321829
201. Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, et al. Expert Consensus Document. The International Scientific Association for Probiotics and Prebiotics Consensus Statement on the Scope and Appropriate Use of the Term Probiotic. Nat Rev Gastroenterol Hepatol (2014) 11:506–14. doi: 10.1038/nrgastro.2014.66
202. Schrezenmeir J, de Vrese M. Probiotics, Prebiotics, and Synbiotics–Approaching a Definition. Am J Clin Nutr (2001) 73:361S–4S. doi: 10.1093/ajcn/73.2.361s
203. Biagioli M, Capobianco D, Carino A, Marchiano S, Fiorucci C, Ricci P, et al. Divergent Effectiveness of Multispecies Probiotic Preparations on Intestinal Microbiota Structure Depends on Metabolic Properties. Nutrients (2019) 11:325-42. doi: 10.3390/nu11020325
204. Chapman CM, Gibson GR, Rowland I. Health Benefits of Probiotics: Are Mixtures More Effective Than Single Strains? Eur J Nutr (2011) 50:1–17. doi: 10.1007/s00394-010-0166-z
205. Didari T, Solki S, Mozaffari S, Nikfar S, Abdollahi M. A Systematic Review of the Safety of Probiotics. Expert Opin Drug Saf (2014) 13:227–39. doi: 10.1517/14740338.2014.872627
206. Quin C, Estaki M, Vollman DM, Barnett JA, Gill SK. And Gibson, D.L. Probiotic Supplementation and Associated Infant Gut Microbiome and Health: A Cautionary Retrospective Clinical Comparison. Sci Rep (2018) 8:8283. doi: 10.1038/s41598-018-26423-3
207. Ronda C, Chen SP, Cabral V, Yaung SJ, Wang HH. Metagenomic Engineering of the Mammalian Gut Microbiome In Situ. Nat Methods (2019) 16:167–70. doi: 10.1038/s41592-018-0301-y
208. Abedon ST, Kuhl SJ, Blasdel BG, Kutter EM. Phage Treatment of Human Infections. Bacteriophage (2011) 1:66–85. doi: 10.4161/bact.1.2.15845
209. Schooley RT, Biswas B, Gill JJ, Hernandez-Morales A, Lancaster J, Lessor L, et al. Development and Use of Personalized Bacteriophage-Based Therapeutic Cocktails To Treat a Patient With a Disseminated Resistant Acinetobacter Baumannii Infection. Antimicrob Agents Chemother (2017) 61:e00954-17. doi: 10.1128/AAC.00954-17
210. Henein A. What are the Limitations on the Wider Therapeutic Use of Phage? Bacteriophage (2013) 3:e24872. doi: 10.4161/bact.24872
211. Jovel J, Patterson J, Wang W, Hotte N, O’Keefe S, Mitchel T, et al. Characterization of the Gut Microbiome Using 16S or Shotgun Metagenomics. Front Microbiol (2016) 7:459. doi: 10.3389/fmicb.2016.00459
212. Quince C, Walker AW, Simpson JT, Loman NJ, Segata N. Shotgun Metagenomics, From Sampling to Analysis. Nat Biotechnol (2017) 35:833–44. doi: 10.1038/nbt.3935
Keywords: common variable immunodeficiency, fecal microbiota transplantation, inflammation, mucosal immunology, microbiota, probiotics
Citation: Varricchi G, Poto R, Ianiro G, Punziano A, Marone G, Gasbarrini A and Spadaro G (2021) Gut Microbiome and Common Variable Immunodeficiency: Few Certainties and Many Outstanding Questions. Front. Immunol. 12:712915. doi: 10.3389/fimmu.2021.712915
Received: 21 May 2021; Accepted: 12 July 2021;
Published: 02 August 2021.
Edited by:
Carmen Fernández, Stockholm University, SwedenReviewed by:
Leszek Rudzki, Medical University of Bialystok, PolandCopyright © 2021 Varricchi, Poto, Ianiro, Punziano, Marone, Gasbarrini and Spadaro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gilda Varricchi, Z2lsZGFuZXRAZ21haWwuY29t
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.