- 1Department of Orthopedic Surgery, The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 2Orthopedic Research Institute of Zhejiang University, Hangzhou, China
- 3Key Laboratory of Motor System Disease Research and Precision Therapy of Zhejiang Province, The Second Affiliated Hospital, Zhejiang University, Hangzhou, China
- 4State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, National Clinical Research Center for Infectious Diseases, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, Department of Infectious Diseases, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, China
- 5Joint Surgery, Ningbo 6th Hospital, Ningbo, China
Objectives: To find out the genetic association between IL6 and autoimmune arthritis.
Methods: We performed a two-sample Mendelian randomization (MR) study using multiple genome-wide association studies (GWAS) datasets. Furthermore, a sex-stratified MR study was performed to identify sexual dimorphism in the association between IL6 and autoimmune arthritis. Then, LocusZoom plots were displayed based on the IL6R gene region to present evidence of genetic colocalization between diseases.
Results: The MR result denoted a genetic association between the increased level of IL-6 signaling and risk of RA (β=0.325, 95%CI 0.088, 0.561, p=7.08E-03) and AS (β=1.240, 95%CI 0.495, 1.980, p=1.1E-03). Accordingly, sIL6R was found to have negatively correlation with the onset of RA (β=-0.020, 95%CI -0.0320, -0.008, p=1.18E-03) and AS (β=-0.125, 95%CI -0.177, -0.073, p=2.29E-06). However, no genetic association between IL6/sIL6R and PsA was detected. The gender-stratified MR analysis showed that IL6 was associated with AS in the male population, with RA in the female population, and with PsA in the male population. Additionally, ADAR, a gene identified by a sensitive test, could be the reason for the nonsignificant association between IL6 and PsA in a pooled population.
Conclusion: Our findings showed that the overactive IL6 signal pathway led to autoimmune arthritis, especially in RA and AS. Sexual difference was also observed in IL6-intermediate susceptibility to autoimmune arthritis.
Highlights
1. Genetic association was identified between IL6-signaling and autoimmune arthritis.
2. IL6 was correlated with male AS, male PsA, and female RA development in a sex-dependent manner.
Introduction
Interleukin-6 (IL-6), as a proinflammatory cytokine, plays major roles in the inflammation process and the development of the immune system (1, 2). However, abnormal immune response could lead to autoimmune arthritis including rheumatoid arthritis (RA), ankylosing spondylitis (AS), and psoriatic arthritis (PsA), which places a great burden on patients by causing joint destruction and even ending up with disability (3, 4). As previously reported, elevating levels of IL-6 have been detected in pathological sites among RA, AS, and PsA patients, and the serum level of IL-6 is associated with the autoimmune arthritis pathogenesis (5–7). Hence, IL-6 blockade seems to be a therapeutic target for alleviating the process of autoimmune arthritis. Anti-IL-6 signaling drugs such as tocilizumab and siltuximab have been approved for RA and systemic juvenile idiopathic arthritis therapy by the Food and Drug Administration (FDA) (4). However, treating AS or PsA with anti-IL-6 therapy could only relatively alleviate the progression of diseases in some reports (8, 9).
It is known that genetic susceptibility accounts for 30% in terms of the risk of autoimmune disease (10). By using information from genome-wide association studies (GWAS), Mendelian randomization (MR), a newly introduced approach, has the potential to evaluate the causal relationship between exposures and phenotype (11, 12). Briefly, MR uses genetic variants as the instrumental variable to interpret a different outcome without bias caused by reverse causation or confounding factors (13, 14). Previous MR studies have shown that the genetic level of serum sIL6R is negatively associated with the risk of developing RA (15). Li and his colleges found that the IL-6 gene -174G/C variant is associated with the risk of RA using MR meta-analysis (16). In order to evaluate the casual association between IL-6 and other autoimmune arthritis, large-scale MR study should be carried out to verify the exact correlation and find out potential therapeutic strategy.
In this study, up-to-date GWAS level summary data for IL-6 signaling and circulating sIL6R were utilized in the MR analysis. The results presented a genetic association between IL6 and different types of autoimmune arthritis. Additionally, we further discovered the underlying sexual dimorphism in these autoimmune diseases based on the MR results.
Method
Genetic Instrumental Variables
Two sets of IL6-related genetic instruments were based on recent MR reports. Genetic variants that represented for IL6 signaling were obtained from a large-scale GWAS meta-analysis assessing chronic inflammation, which included 204,402 European individuals (17). Based on prior MR reports from Kappelmann et al. and Georgakis et al. (18, 19), who investigated the association between IL6 signaling on specific depressive symptom outcomes and cardiovascular outcomes, respectively, IL6 signaling-related SNPs were used in our study (Table S1). According to Kappelmann et al.’s definition, “IL6 signaling” referred to IL-6R genetic instruments and weighted by the level of CRP.
Plasma sIL6R is a decoy receptor that is able to suppress IL6 signaling by forming an inhibitory complex with sgp130 (20). To further identify the role of IL6 signaling in autoimmune arthritis, genetic IV SNPs associated with the level of sIL6R were generated from a European ancestry GWAS (n=3,301) (21). A total of 34 SNPs utilized in this study were based on Rosa et al.’s study (Table S1) (15). Linkage disequilibrium levels of SNPs chosen above from two datasets were tested using the LDmatrix Tool (https://ldlink.nci.nih.gov/?tab=ldmatrix, CEU; r2 < 0.1). When IV SNPs could not be found in autoimmune arthritis summary statistics, the LDproxy Tool was used to identify potential proxy SNPs (CEU; r2>0.8).
Summary Statistics for Autoimmune Arthritis
Publicly available meta-analysis datasets for autoimmune arthritis (RA, AS, PsA) were derived from some different studies. GWAS summary statistics of 14,361 RA cases and 43,923 controls of European ancestry were obtained from a large-scale GWAS (22). The summary of the GWAS statistic for AS and PsA was obtained from FinnGen, a large-scale project combining genotype data from Finnish biobanks and digital health record data from Finnish health registries. We selected datasets containing 541 AS cases, 74,589 controls and 562 PsA cases, and 93,959 controls of Finnish ancestry. Another set of autoimmune arthritis meta-analysis summary statistic was acquired from UK Biobank (UKBB) for replication study, encompassing 361,141 of European ancestry with 4,017 RA cases, 3,154 AS cases, and 712 PsA cases.
Moreover, summary GWAS results of these three types of autoimmune arthritis in different genders were extracted from UKBB (with 194,174 female and 167,020 male) for further sex-stratified MR analysis to avoid potential sexual bias, due to the fact that gender differences had been widely reported in autoimmune diseases.
Statistical Analysis
To evaluate casual effect, an MR study was carried out with four modes: MR-Egger mode, weighted median mode, and inverse variance-weighted (IVW) mode with the “TwoSampleMR” R-package (23). With the minimum weighted average variance, the IVW method was mainly selected for analysis (24). Because the MR analysis is based on the hypothesis that genetic exposure influences the outcome directly, it is not suitable to perform the analysis in the presence of pleiotropy. Thus, we conducted several approaches to detect potential pleiotropy (25), Firstly, MR-Egger regression was performed to capture the horizontal pleiotropy (26). Additionally, we applied MR pleiotropy residual sum and outlier test (MR-PRESSO) to detect potential horizontal pleiotropy using the MR-PRESSO global test. For any detected pleiotropic SNP, the MR-PRESSO outlier test was performed to remove these SNPs and rectify the horizontal pleiotropy (27).
Heterogeneity test was performed using Cochran’s Q statistics analysis to identify whether the MR results were biased by the heterogenic factors. Leave-one-out sensitivity analysis was performed to identify whether the estimate was driven by an SNP. These two methods were also applied through the “TwoSampleMR” R-package. The estimated effect represented the log odds ratio (OR) between IL6R and autoimmune arthritis risk. All the analyses with P<0.05 were considered statistically significant.
Results
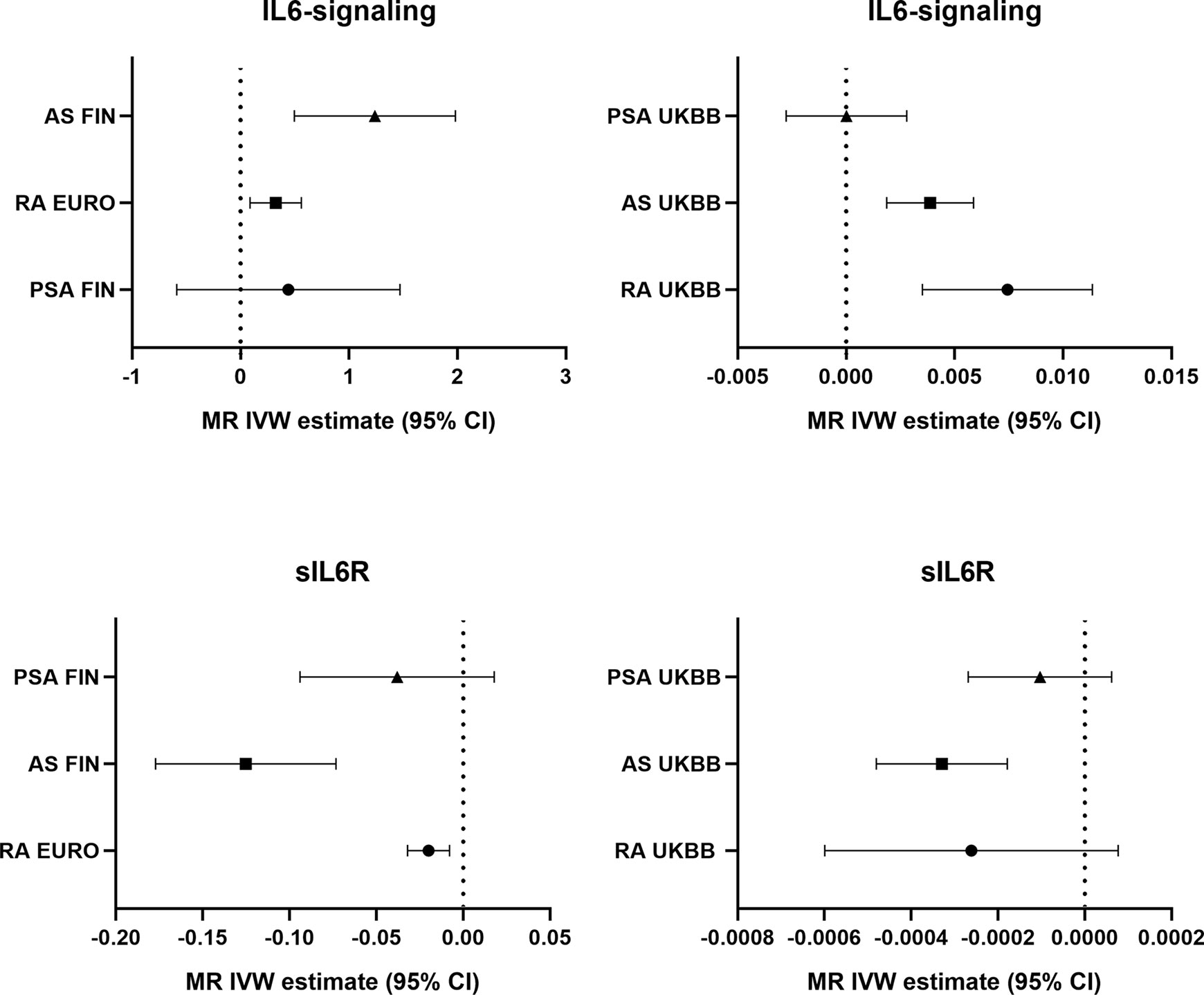
Potential genetic association between the levels of IL6-signaling, sIL6R, and autoimmune arthritis were detected by using two-sample MR analysis (Figure 1). Summarized results of associations are shown in Table 1, and a forest plot is shown in Figure 2.
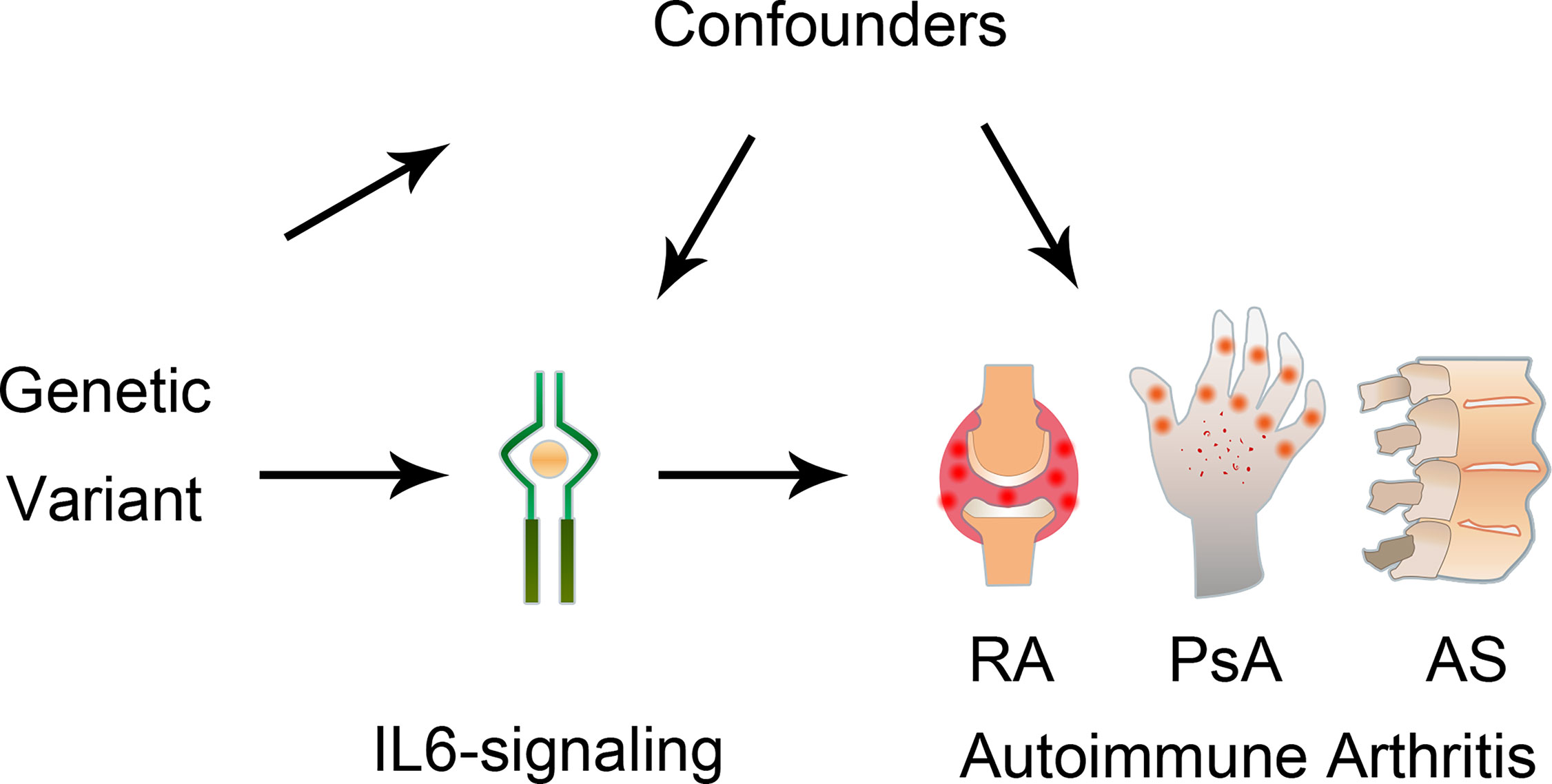
Figure 1 Graphical diagram of the Mendelian randomization analysis between IL6 and risk of autoimmune arthritis. RA, rheumatoid arthritis; AS, ankylosing spondylitis; PsA, psoriatic arthritis.
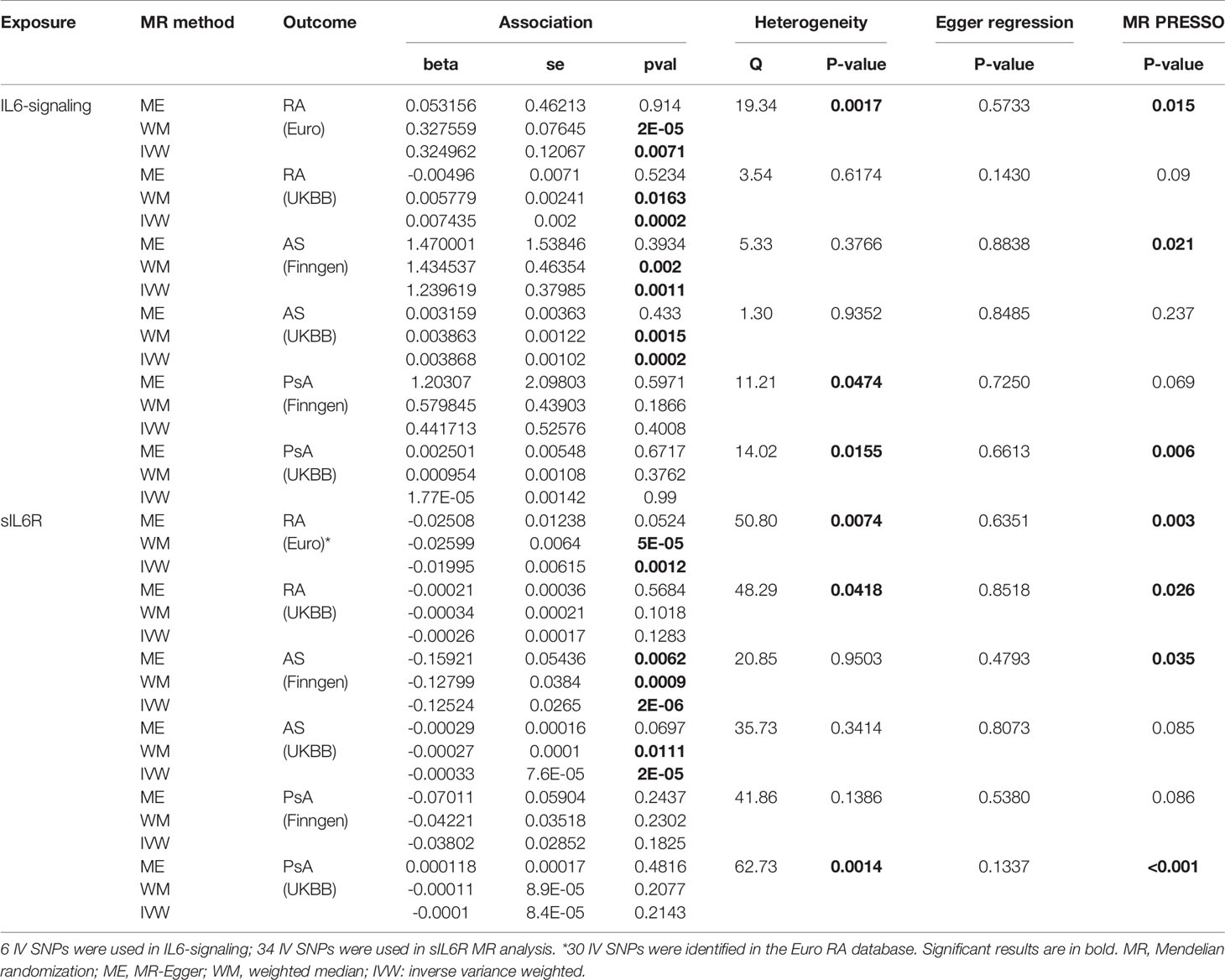
Table 1 MR estimates of the causal effect of IL6-signaling and sIL6R level on the risk of autoimmune arthritis.
Finding for Upregulated IL6-Signaling and Levels of sIL6R in Autoimmune Arthritis
Casual effects of upregulated IL6-signaling have been found to be positively associated with RA (IVW mode effect=0.325, 95%CI 0.088 to 0.561, p=7.08E-03) and AS (IVW mode effect=1.240, 95%CI 0.495 to 1.980, p=1.1E-03). As the results of MR present, the elevated concentration of sIL6R, which inhibits IL6-signaling, was negatively associated with the risk of RA (IVW mode effect=-0.020, 95%CI -0.0320 to -0.008, p=1.18E-03) and AS (IVW mode effect=-0.125, 95%CI -0.177 to -0.073, p=2.29E-06). Replicated MR analysis confirmed the connection between IL6 and risk of RA (IVW mode effect=0.003, 95%CI 0.007 to 0.011, p=1.98E-04) or AS (IVW mode effect=0.0039, 95%CI 0.0019 to 0.0059, p=1.51E-04). The negative association between sIL6R and AS was validated through replicated MR analysis (IVW mode effect=-0.00033, 95%CI -0.00048 to -0.00018, p=-0.00018). However, no genetic association was found between RA and sIL6R in the replicated MR analysis. Similar with the MR analysis results in PsA, no genetic effects were found between the risk of PsA and IL6-signaling (Table 1).
Pleiotropy test using MR-Egger did not denote any pleiotropic SNP in the MR study. However, the horizontal pleiotropy was detected among the MR results by using MRPRESSO (Table 1). After removing the pleiotropic SNPs by the MRPRESSO outlier test, the corrected MR estimates are listed in Table S2.
Furthermore, we carried out Cochran’s Q statistics test to evaluate the MR results. There was no sign of heterogenetic effects between the genetic effects and risk of AS (Table 1). However, heterogeneity was found among the MR estimate concerning the association between IL6 and two other autoimmune arthritis diseases (Table 1). Thus, leave-one-out sensitivity analysis was performed to identify the important SNP and the possible source of heterogeneity (Figure S1). Rs2228145, a well-reported IL6 receptor genetic variant, was confirmed to strongly influence the effects of IL6-signaling on RA or AS (28) (Figure S1). In addition, rs4129267, an IL6R-related SNP in LD with rs2228145(r2 = 1) identified by Ferreira and her colleges (29), was found to make a great contribution to getting RA. On the other hand, rs12059682 had a biased role of IL6 in the onset of PsA (Figure S1).
Gender-Stratified MR Analysis for IL6-Signaling and Levels of sIL6R in Autoimmune Arthritis
Sexual dimorphic effects on autoimmune arthritis were widely demonstrated previously. In order to further identify the potential heterogeneity in order to further identify among MR analyses. We performed sex-stratified MR analysis to judge whether a sex-specific genetic relationship existed between the IL6 and the risk of autoimmune arthritis (Figure 3). The UKBB GWAS datasets were used, which include publicly available sex-specific summary data. We found casual association between IL6 and RA in women rather than in men. Conversely, the male population seemed easier to suffer from AS when exposed to elevating IL6-signaling levels (Table 2). Consistent with the casual estimate in AS, men could be at higher risk of getting PsA than women. However, it is intriguing that upregulation of IL6-signaling even negatively associated with the risk of PsA (Table 2). Next, as shown by MRPRESSO, the pleiotropy in the MR estimate was found only in PsA. After removing pleiotropic SNP from MR analysis, there was no casual association between IL6 and PSA in the female population (corrected estimate listed in Table S2).
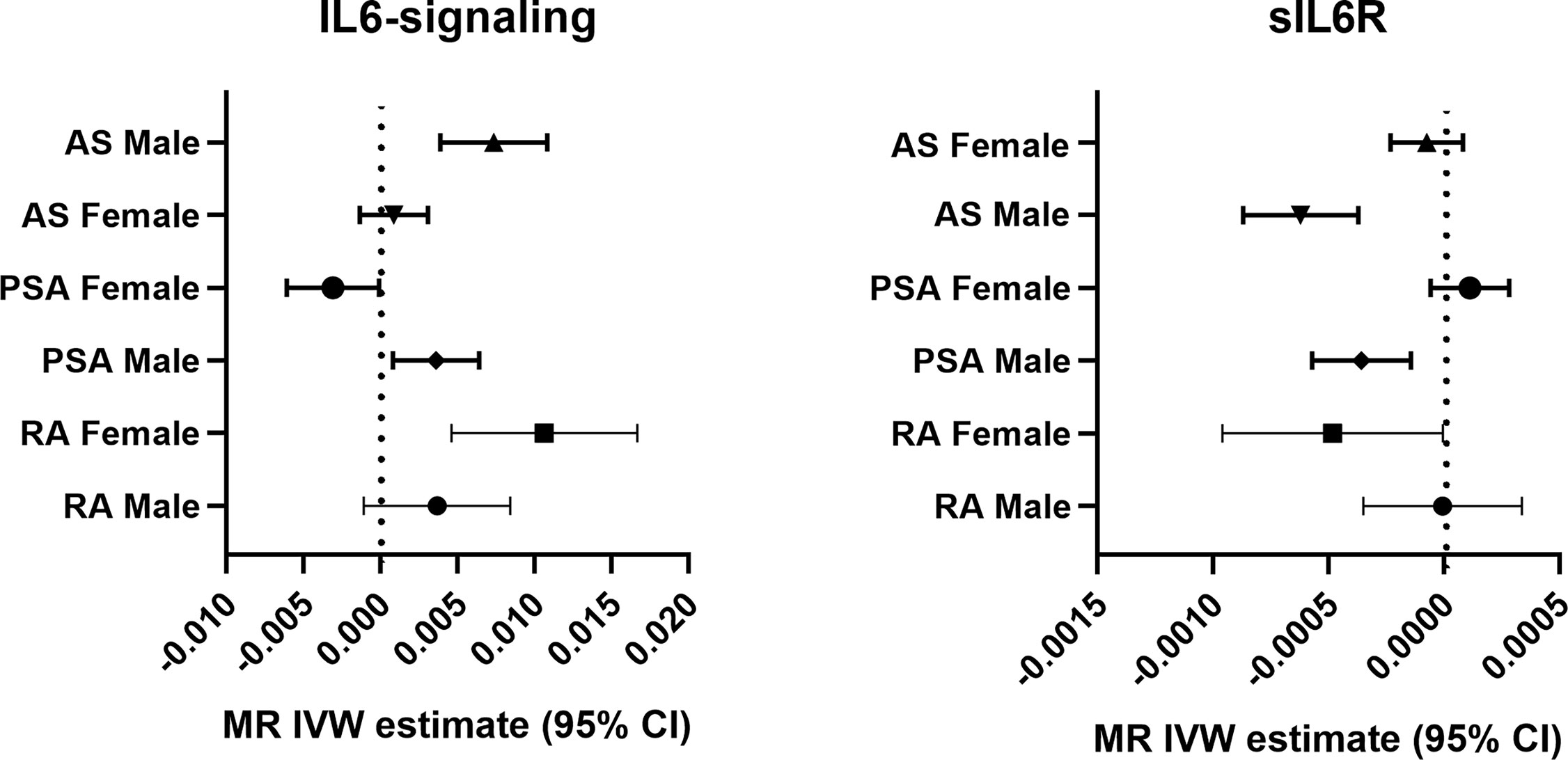
Figure 3 Forest plot of the causal association between IL6-signaling/sIL6R and autoimmune arthritis by sex.
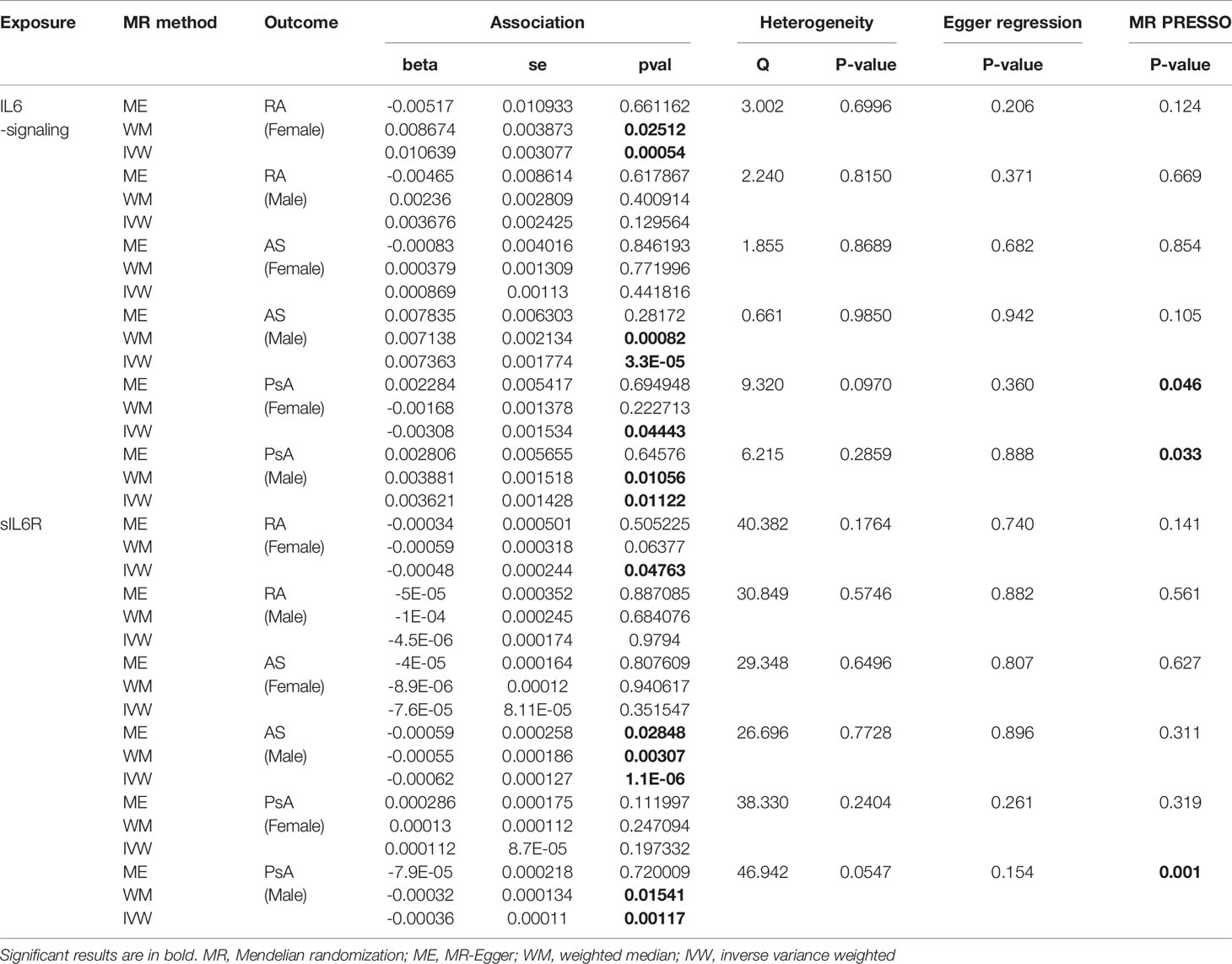
Table 2 Gender-stratified MR estimates of the causal effect of IL6-signaling and sIL6R level on the risk of autoimmune arthritis.
Next, we apply the leave-one-out sensitive test to evaluate whether some potential SNP creates bias on the gender effects on the MR estimate. No SNP was discovered to create bias in the genetic effect between IL-6 exposure and RA or AS (Table 2). Accordingly, Rs2228145 was also verified as the key SNP in the development of PsA in men (Figure S2). Because rs12059682 was identified as the potential bias SNP among overall PsA populations, we found that it posed gender difference in the effect contribution to the onset of PsA (Figure S2). When leaving it out, upregulating IL6-signaling was also found to be not associated with the risk of PsA.
To deeply identify the role of rs12059682 in the PsA among different genders, firstly, the genetic variant was mapped as the variation of ADAR (adenosine deaminase acting on RNA). Single-tissue eQTL evaluation was carried out using GTEX. Rs12059682 T>C variation would lead to a higher transcriptional level of ADAR in the adrenal gland, EBV-transformed lymphocytes, lung esophagus mucosa, and fibroblast (Figure S3 and Table S3). Based on Shallev et al.’s work (30), decreased A-to-I RNA editing could lead to the accumulation of (double-stranded RNA) dsRNA, which played an important role in psoriasis pathogenesis. Variation of rs12059682 was found riskier in women. Therefore, we brought out the hypothesis that a lower level of ADAR promoted the development of PsA in females.
Thus, we browsed the GEO database for sex-stratified PsA gene expression profile. As shown by Figure 4A, the transcriptional level of ADAR was lower in female PsA patients than healthy donors (GSE61281, p=0.0371) (31), while there was no statistically significant difference between male PsA patients and the control group (p=0.3333). In addition, the ADAR level was found to be correlated with RA (32). The rs12059682 T>C mutation could result in a higher level of ADAR and increased the risk of developing RA based on the GWAS datasets. Moreover, the level of ADAR was found to be higher in female RA patients compared to age-matched male patients (Figure 4B, GSE74143) (32).
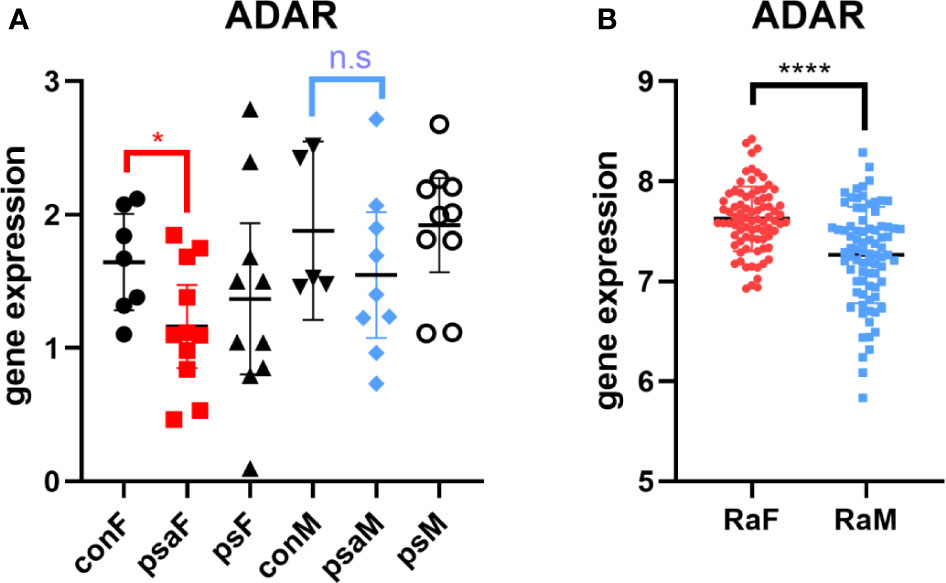
Figure 4 Expression of ADAR in RA or PsA from GEO datasets GSE61281 (A) and GSE74143 (B). con, control; psa, psoriatic arthritis; ps, psoriasis; RA, rheumatoid arthritis; F, female; M, male. Data from all bar graphs are presented as mean ± SEM. (*P < 0.05, ****P < 0.0001, n.s, not significant).
Regional Visualization of Autoimmune Arthritis GWAS Datasets for Further Analysis
As shown in Figure 5, LocusZoom plots were displayed based on the IL6R gene region (1q21.3) to present evidence of genetic colocalization between diseases. However, no genome-wide level association signals were found in this region. At first, we browsed the IL6R gene region to identify the top SNPs with a p-value lower than 0.01 (MAF >0.01). If the candidate SNP could not be annotated to the RS number and substituted by proxy SNP, then the SNP in this region with the next lowest p-value was chosen for analysis.
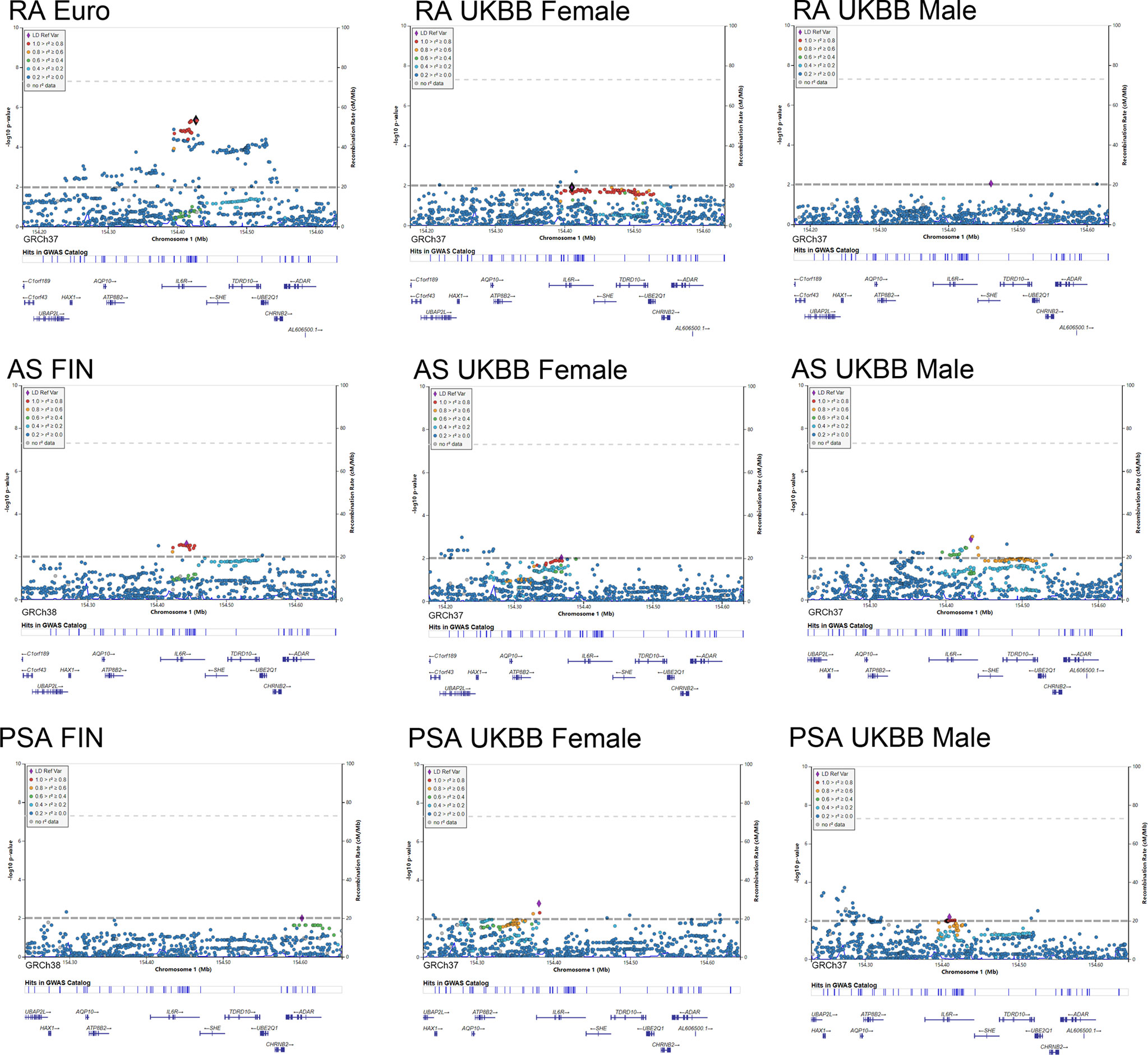
Figure 5 Regional plot of the genetic variants at 1q21.3 and their association with autoimmune arthritis.
By using the Finngen GWAS dataset for AS, rs4845372 (1:154442920_C/A, P=2.48 × 10^-3) was identified as the potential SNP accounting for IL6’s effects in the onset of AS. After LD evaluation, this genetic variance was found to be in high linkage with rs2228145 (r2 = 0.882). Next, a sex-stratified plot using UKBB GWAS data presented rs6695045 (1:154432957_A/G, P=1.47 × 10^-3) as the top influencer in the IL6R region with the male population, which was also an SNP that was relatively in linkage with rs2228145 (r2 = 0.438). On the other hand, in the female population, none of the candidate SNPs (p<0.01) was detected in the IL6R gene region. When looking for other than the IL6R gene region, there were no other signals having eQTL data or a literature-based relationship with IL6R.
In addition, our regional plot further emphasized rs2228145 (1:154426970_A/C, P=4.50 × 10^-6) as the lead SNP, putting great genetic predisposition to RA (Figure 5). As for further sex-specific analysis, no SNP in the IL6R region met our criteria when analyzing male GWAS data. Rather, a weak signal (1:154410686_C/CT p=0.012) was identified in women with RA. The C to T variance of proxy SNP rs55800510 (R2 = 0.9666) was associated with the risk of RA (Table S3).
As for the regional plotting result in male PsA patients, we found that rs12730036 (1:154416969_C/T p=9.37 × 10^-3) was in high linkage with rs2228145 (r2 = 0.941). This result denoted consistency with the MR analysis, which resulted in a higher risk of PsA in male by upregulating IL6 signaling. Interestingly, the lowest-p value-polymorphism rs12083537 G>A, which was able to activate the IL6-related pathway, presented to be a protective genetic variance for women against PsA (Table S3).
Discussion
In the present MR study, by applying two sets of genetic IVs, our findings showed corroborating evidence that the overactive IL6 signal pathway led to autoimmune arthritis, especially in RA and AS. Genetic predisposition to a higher IL6 level was also genetically associated with the risk of PsA in the male population. Consistent with a previous study, elevating level of IL6 was found in patients with autoimmune arthritis, which played a great role in the pathogenesis of joint lesions (6, 7, 33). As Rosa and her colleague presented, by using sIL6R genetical IV and PheWAS analysis, the genetically determined sIL6R was negatively associated with the risk of RA (15). We extended the result and complemented the evidence from a regional plot and MR analysis. rs2228145, a genetic variant that accounted for 51% variance of sIL6R levels (34), was identified as the common susceptibility locus for autoimmune arthritis. Additionally, the strategy of targeting the IL6 axis by using novel biological medicine to treat RA had been well validated (35). However, anti-IL6 therapy against AS or PsA was still not introduced clinically.
We observed that sexual differences influenced IL6-intermediate susceptibility to autoimmune arthritis. Sex differences were reported in the level of IL-6 among monocytes (36). Consistently, gender differences in the prevalence of RA were well described based on some large epidemiological studies (37). In the previous study, hormonal factors and sex chromosomes were thought to cause the sex difference in autoimmune disease (38, 39). Here, we present the evidence that IL6 acted differently during the onset of RA, which could be another possible explanation for gender difference in RA (40). Likewise, AS is another typical autoimmune arthritis where the sex ratio is 3:1 (M/F) (41). Our MR analysis confirmed the casual association between IL6 and the development of AS in a gender-dependent manner. However, Gracey and his colleagues found that there was no sex dimorphism in the level of IL-6 among AS patients (42). Additionally, another observational study identified a higher level of IL6 in female patients with syndesmophytes than those without them. No significant change of the IL6 level was found in the pooled populations or male patients with syndesmophytes (43). Here, by using the summary dependent t-test, we found that the IL6 level was higher in male than in female patients without syndesmophytes (3.46 ± 2.86 vs 1.47 ± 1.1) (p=0.024). We presumed that the result of the IL6 level was confounded by the progression of diseases. However, the underlying mechanism is still unclear and further study is needed.
As for the onset of PSA, unlike the equal gender ratio observed in epidemiology, we found that a genetically elevating level of IL6 could increase the risk of PsA for men and decrease the risk for women. A cross-sectional analysis reported by Eder et al., who specifically compared the sexual difference in PsA, presented the phenotype that male PsA patients developed more severe axial and joint damage than female (44). Contrarily, some researchers reported that female patients were suffering from RA with more severe joint damages (45, 46). Here we identified a genetic variant rs12059682, located in the ADAR1 gene region, causing heterogeneity in MR analysis. A recent study introduced the role of the IL6R-STAT3-ADAR1 axis in the oncogenicity of multiple myeloma (47). It should be noted that enrichment of ADAR SNP loci was identified in GWAS signals for autoimmune diseases (48). Corresponding to the evidence from the rna-seq dataset, rs12059682 C>T mutation could lead to a lower level of ADAR in the lymphocyte and fibroblast. The decreased level of DNA editing, regulated by the ADAR (49), was correlated with a higher risk of PsA due to the accumulation of dsRNA (30). Furthermore, the impaired post-translational modification of dsRNA and unwinding of dsRNA structures led to adaptive immune activation through producing IFNβ (50). On the other hand, Vlachogiannis et al. observed elevated adenosine-to-inosine RNA editing in the progress of RA (32). Consistently, a higher transcriptional level of ADAR was observed in female RA patients. As Vlachogiannis et al. suggested, more single-stranded RNA (ssRNA) was produced by adenosine-to-inosine RNA editing, which resulted in the binding of ssRNA-binding proteins and increase in the expression of these pro-inflammatory genes (such as HuR). Therefore, the opposite effect of ADAR could be the reason for a different role of IL6 in these two autoimmune arthritis diseases. Elsewise, this SNP could partially account for the sexual difference in RA or PsA susceptibility. However, further sex-stratified GWAS research and pathological studies should be carried out to validate these hypotheses and undermine the potential mechanism.
This MR study was performed by using two sets of IL6-related IV and multiple GWAS datasets, which aimed to ensure the consistency of the results. However, there were still some limitations. Firstly, the sex-stratified MR analysis was mainly based on the UKBB GWAS data. Some genetic variants were different from Finland and UK populations, which was caused by “population bottlenecks” (51). Gender-stratified GWAS datasets from Finland should be collected to replicate the findings, although the allele frequency of major SNP was similar between the two ethnic groups. Moreover, our study identified rs12059682 as the potential source of bias. Other GWAS focused on the level of ADRA editing that should be performed to identify the casual association between ADRA and autoimmune arthritis. Finally, however, the genetic pleiotropy was rectified by the MRPRESSO in the pooled or sex-stratified MR under a sensitive test. Other potential pathway for the associations cannot be fully ruled out.
To summarize, we identified the genetic association between IL6-signaling and onset of autoimmune arthritis. These results further validated IL6 inhibition as the therapeutic strategies for RA. Rather, the genetic role of IL6 in a sex-dependent manner was discovered in the development of male AS, male PsA, and female RA. Those who have started randomized controlled trials of IL6 inhibition on autoimmune arthritis should take gender differences into account. In the end, the hypothesis that ADAR reduces the effectiveness of the IL6 inhibitor in PsA populations, supported by the statistical findings, should be further investigated through pathophysiological studies.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author Contributions
SY, JH, and WW designed this study. Study conduct: JH and WW. Data collection: ZQ, CL, XJ, and JW. Data analysis: CF, YZ, XJ, and GZ. Drafting the manuscript: JH, ZQ, and XJ. Revising the manuscript content: JM, CJ, YS, and CZ. Approving the final version of the manuscript: all authors. SY takes responsibility for the integrity of the data. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Natural Science Foundation of China under Grant Nos. 81772360 and 81800782, Zhejiang Province Medical and Health Project No. 2020391395, and Yinzhou District Agriculture and Social Development Science and Technology Project No. 2017YZQ104.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We want to acknowledge the participants and investigators of FinnGen study and Neale Lab (http://www.nealelab.is/uk-biobank). The authors thank all investigators for sharing these data.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.707617/full#supplementary-material
Supplementary Figure S1 | Forest plots and “leave-one-out” plot for MR analyses of the causal effect of IL6 on autoimmune arthritis in pooled population.
Supplementary Figure S2 | Forest plots and “leave-one-out” plot for MR analyses of the causal effect of IL6 on autoimmune arthritis in female or male population.
Supplementary Figure S3 | Violin plot and multiple tissues expression level of rs12059682.
References
1. Choy E, Rose-John S. Interleukin-6 as a Multifunctional Regulator: Inflammation, Immune Response, and Fibrosis. J Scleroderma Rel Disord (2017) 2(2_suppl):S1–5. doi: 10.5301/jsrd.5000265
2. Tanaka T, Narazaki M, Kishimoto T. IL-6 in Inflammation, Immunity, and Disease. Cold Spring Harb Perspect Biol (2014) 6(10):a016295. doi: 10.1101/cshperspect.a016295
3. Hirano T. Interleukin 6 in Autoimmune and Inflammatory Diseases: A Personal Memoir. Proc Jpn Acad Ser B Phys Biol Sci (2010) 86(7):717–30. doi: 10.2183/pjab.86.717
4. Lai JH, Luo SF, Ho LJ. Targeting the CD40-CD154 Signaling Pathway for Treatment of Autoimmune Arthritis. Cells (2019) 8(8):927. doi: 10.3390/cells8080927
5. McInnes IB, Schett G. Cytokines in the Pathogenesis of Rheumatoid Arthritis. Nat Rev Immunol (2007) 7(6):429–42. doi: 10.1038/nri2094
6. Liu W, Wu YH, Zhang L, Liu XY, Xue B, Wang Y, et al. Elevated Serum Levels of IL-6 and IL-17 may Associate With the Development of Ankylosing Spondylitis. Int J Clin Exp Med (2015) 8(10):17362–76.
7. Ogata A, Kumanogoh A, Tanaka T. Pathological Role of Interleukin-6 in Psoriatic Arthritis. Arthritis (2012) 2012:713618. doi: 10.1155/2012/713618
8. Mease PJ, Gottlieb AB, Berman A, Drescher E, Xing J, Wong R, et al. The Efficacy and Safety of Clazakizumab, an Anti-Interleukin-6 Monoclonal Antibody, in a Phase IIb Study of Adults With Active Psoriatic Arthritis. Arthritis Rheumatol (2016) 68(9):2163–73. doi: 10.1002/art.39700
9. Merashli M, De Marco G, Podgorski M, McGonagle D, Marzo-Ortega H. Evidence of Response to IL-6 Inhibition in Some Cases of Refractory Spondyloarthritis-Associated Peripheral Synovitis. Ann Rheum Dis (2016) 75(7):1418–20. doi: 10.1136/annrheumdis-2016-209275
10. Azizov V, Dietel K, Steffen F, Dürholz K, Meidenbauer J, Lucas S, et al. Ethanol Consumption Inhibits TFH Cell Responses and the Development of Autoimmune Arthritis. Nat Commun (2020) 11(1):1998. doi: 10.1038/s41467-020-15855-z
11. Porcu E, Rueger S, Lepik K, eQTLGen Consortium, BIOS Consortium, Santoni F, et al. Mendelian Randomization Integrating GWAS and eQTL Data Reveals Genetic Determinants of Complex and Clinical Traits. Nat Commun (2019) 10(1):3300. doi: 10.1038/s41467-019-10936-0
12. Marson A, Housley WJ, Hafler DA. Genetic Basis of Autoimmunity. J Clin Invest (2015) 125(6):2234–41. doi: 10.1172/JCI78086
13. Burgess S, Butterworth A, Thompson SG. Mendelian Randomization Analysis With Multiple Genetic Variants Using Summarized Data. Genet Epidemiol (2013) 37(7):658–65. doi: 10.1002/gepi.21758
14. Sekula P, Del Greco MF, Pattaro C, Köttgen A. Mendelian Randomization as an Approach to Assess Causality Using Observational Data. J Am Soc Nephrol (2016) 27(11):3253–65. doi: 10.1681/ASN.2016010098
15. Rosa M, Chignon A, Li Z, Boulanger M, Arsenault BJ, Bossé Y, et al. A Mendelian Randomization Study of IL6 Signaling in Cardiovascular Diseases, Immune-Related Disorders and Longevity. NPJ Genom Med (2019) 4:23. doi: 10.1038/s41525-019-0097-4
16. Li B, Xiao Y, Xing D, Ma XL, Liu J. Circulating Interleukin-6 and Rheumatoid Arthritis: A Mendelian Randomization Meta-Analysis. Med (Baltimore) (2016) 95(23):e3855. doi: 10.1097/MD.0000000000003855
17. Ligthart S, Vaez A, Vosa U, Stathopoulou MG, de Vries PS, Prins BP, et al. Genome Analyses of >200,000 Individuals Identify 58 Loci for Chronic Inflammation and Highlight Pathways That Link Inflammation and Complex Disorders. Am J Hum Genet (2018) 103(5):691–706. doi: 10.1016/j.ajhg.2018.09.009
18. Kappelmann N, Arloth J, Georgakis MK, Czamara D, Rost N, Ligthart S, et al. Dissecting the Association Between Inflammation, Metabolic Dysregulation, and Specific Depressive Symptoms: A Genetic Correlation and 2-Sample Mendelian Randomization Study. JAMA Psychiatry (2021) 78(2):161–70. doi: 10.1001/jamapsychiatry.2020.3436
19. Georgakis MK, Malik R, Gill D, Franceschini N, Sudlow C, Dichgans M, et al. Interleukin-6 Signaling Effects on Ischemic Stroke and Other Cardiovascular Outcomes: A Mendelian Randomization Study. Circ Genom Precis Med (2020) 13(3):e002872.doi: 10.1161/CIRCGEN.119.002872
20. Jostock T, Müllberg J, Ozbek S, Atreya R, Blinn G, Voltz N, et al. Soluble Gp130 is the Natural Inhibitor of Soluble Interleukin-6 Receptor Transsignaling Responses. Eur J Biochem (2001) 268(1):160–7. doi: 10.1046/j.1432-1327.2001.01867.x
21. Sun BB, Maranville JC, Peters JE, Stacey D, Staley JR, Blackshaw J, et al. Genomic Atlas of the Human Plasma Proteome. Nature (2018) 558(7708):73–9. doi: 10.1038/s41586-018-0175-2
22. Okada Y, Wu D, Trynka G, Raj T, Terao C, Ikari K, et al. Genetics of Rheumatoid Arthritis Contributes to Biology and Drug Discovery. Nature (2014) 506(7488):376–81. doi: 10.1038/nature12873
23. Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-Base Platform Supports Systematic Causal Inference Across the Human Phenome. Elife (2018) 7:e34408. doi: 10.7554/eLife.34408
24. Lee CH, Cook S, Lee JS, Han B. Comparison of Two Meta-Analysis Methods: Inverse-Variance-Weighted Average and Weighted Sum of Z-Scores. Genomics Inform (2016) 14(4):173–80. doi: 10.5808/GI.2016.14.4.173
25. Bucur IG, Claassen T, Heskes T. Inferring the Direction of a Causal Link and Estimating its Effect via a Bayesian Mendelian Randomization Approach. Stat Methods Med Res (2020) 29(4):1081–111. doi: 10.1177/0962280219851817
26. Hemani G, Bowden J, Davey Smith G. Evaluating the Potential Role of Pleiotropy in Mendelian Randomization Studies. Hum Mol Genet (2018) 27(R2):R195–208. doi: 10.1093/hmg/ddy163
27. Verbanck M, Chen CY, Neale B, Do R. Detection of Widespread Horizontal Pleiotropy in Causal Relationships Inferred From Mendelian Randomization Between Complex Traits and Diseases. Nat Genet (2018) 50(5):693–8. doi: 10.1038/s41588-018-0099-7
28. Garbers C, Monhasery N, Aparicio-Siegmund S, Lokau J, Baran P, Nowell MA, et al. The Interleukin-6 Receptor Asp358Ala Single Nucleotide Polymorphism Rs2228145 Confers Increased Proteolytic Conversion Rates by ADAM Proteases. Biochim Biophys Acta (2014) 1842(9):1485–94. doi: 10.1016/j.bbadis.2014.05.018
29. Ferreira MA, Matheson MC, Duffy DL, Marks GB, Hui J, Souëf PL, et al. Identification of IL6R and Chromosome 11q13.5 as Risk Loci for Asthma. Lancet (2011) 378(9795):1006–14. doi: 10.1016/S0140-6736(11)60874-X
30. Shallev L, Kopel E, Feiglin A, Leichner GS, Avni D, Sidi Y, et al. Decreased A-To-I RNA Editing as a Source of Keratinocytes’ dsRNA in Psoriasis. Rna (2018) 24(6):828–40. doi: 10.1261/rna.064659.117
31. Pollock RA, Abji F, Liang K, Chandran V, Pellett FJ, Virtanen C, et al. Gene Expression Differences Between Psoriasis Patients With and Without Inflammatory Arthritis. J Invest Dermatol (2015) 135(2):620–3. doi: 10.1038/jid.2014.414
32. Vlachogiannis NI, Gatsiou A, Silvestris DA, Stamatelopoulos K, Tektonidou MG, Gallo A, et al. Increased Adenosine-to-Inosine RNA Editing in Rheumatoid Arthritis. J Autoimmun (2020) 106:102329. doi: 10.1016/j.jaut.2019.102329
33. Hashizume M, Mihara M. The Roles of Interleukin-6 in the Pathogenesis of Rheumatoid Arthritis. Arthritis (2011) 2011:765624. doi: 10.1155/2011/765624
34. van Dongen J, Jansen R, Smit D, Hottenga JJ, Mbarek H, Willemsen G, et al. The Contribution of the Functional IL6R Polymorphism Rs2228145, eQTLs and Other Genome-Wide SNPs to the Heritability of Plasma sIL-6R Levels. Behav Genet (2014) 44(4):368–82. doi: 10.1007/s10519-014-9656-8
35. Ogata A, Kato Y, Higa S, Yoshizaki K. IL-6 Inhibitor for the Treatment of Rheumatoid Arthritis: A Comprehensive Review. Mod Rheumatol (2019) 29(2):258–67. doi: 10.1080/14397595.2018.1546357
36. O’Connor MF, Motivala SJ, Valladares EM, Olmstead R, Irwin MR. Sex Differences in Monocyte Expression of IL-6: Role of Autonomic Mechanisms. Am J Physiol Regul Integr Comp Physiol (2007) 293(1):R145–51. doi: 10.1152/ajpregu.00752.2006
37. Linos A, Worthington JW, O’Fallon WM, Kurland LT. The Epidemiology of Rheumatoid Arthritis in Rochester, Minnesota: A Study of Incidence, Prevalence, and Mortality. Am J Epidemiol (1980) 111(1):87–98. doi: 10.1093/oxfordjournals.aje.a112878
38. Rubtsov AV, Rubtsova K, Kappler JW, Marrack P. Genetic and Hormonal Factors in Female-Biased Autoimmunity. Autoimmun Rev (2010) 9(7):494–8. doi: 10.1016/j.autrev.2010.02.008
39. Jirholt J, Lindqvist AB, Holmdahl R. The Genetics of Rheumatoid Arthritis and the Need for Animal Models to Find and Understand the Underlying Genes. Arthritis Res (2001) 3(2):87–97. doi: 10.1186/ar145
40. van Vollenhoven RF. Sex Differences in Rheumatoid Arthritis: More Than Meets the Eye. BMC Med (2009) 7:12. doi: 10.1186/1741-7015-7-12
41. Will R, Edmunds L, Elswood J, Calin A. Is There Sexual Inequality in Ankylosing-Spondylitis - a Study of 498 Women and 1202 Men. J Rheumatol (1990) 17(12):1649–52.
42. Gracey E, Yao Y, Green B, Qaiyum Z, Baglaenko Y, Lin A, et al. Sexual Dimorphism in the Th17 Signature of Ankylosing Spondylitis. Arthritis Rheumatol (2016) 68(3):679–89. doi: 10.1002/art.39464
43. Huang WN, Tso TK, Kuo YC, Tsay GJ. Distinct Impacts of Syndesmophyte Formation on Male and Female Patients With Ankylosing Spondylitis. Int J Rheum Dis (2012) 15(2):163–8. doi: 10.1111/j.1756-185X.2011.01687.x
44. Eder L, Thavaneswaran A, Chandran V, Gladman DD. Gender Difference in Disease Expression, Radiographic Damage and Disability Among Patients With Psoriatic Arthritis. Ann Rheum Dis (2013) 72(4):578–82. doi: 10.1136/annrheumdis-2012-201357
45. Jawaheer D, Maranian P, Park G, Lahiff M, Amjadi SS, Paulus HE. Disease Progression and Treatment Responses in a Prospective DMARD-Naive Seropositive Early Rheumatoid Arthritis Cohort: Does Gender Matter? J Rheumatol (2010) 37(12):2475–85. doi: 10.3899/jrheum.091432
46. Kuiper S, van Gestel AM, Swinkels HL, de Boo TM, da Silva JA, van Riel PL. Influence of Sex, Age, and Menopausal State on the Course of Early Rheumatoid Arthritis. J Rheumatol (2001) 28(8):1809–16.
47. Teoh PJ, Chung TH, Chng PYZ, Toh SHM, Chng WJ. IL6R-STAT3-ADAR1 (P150) Interplay Promotes Oncogenicity in Multiple Myeloma With 1q21 Amplification. Haematologica (2020) 105(5):1391–404. doi: 10.3324/haematol.2019.221176
48. Zhang H, Fu Q, Shi X, Pan Z, Yang W, Huang Z, et al. Human A-To-I RNA Editing SNP Loci Are Enriched in GWAS Signals for Autoimmune Diseases and Under Balancing Selection. Genome Biol (2020) 21(1):288. doi: 10.1186/s13059-020-02205-x
49. Qu L, Yi Z, Zhu S, Wang C, Cao Z, Zhou Z, et al. Programmable RNA Editing by Recruiting Endogenous ADAR Using Engineered RNAs. Nat Biotechnol (2019) 37(9):1059–69. doi: 10.1038/s41587-019-0178-z
50. Zhang LJ. Type1 Interferons Potential Initiating Factors Linking Skin Wounds With Psoriasis Pathogenesis. Front Immunol (2019) 10:1440. doi: 10.3389/fimmu.2019.01440
Keywords: Mendelian randomization, IL6, sIL6R, autoimmune arthritis, ADAR
Citation: Hong J, Qu Z, Ji X, Li C, Zhang G, Jin C, Wang J, Zhang Y, Shen Y, Meng J, Zhou C, Fang C, Wang W and Yan S (2021) Genetic Associations Between IL-6 and the Development of Autoimmune Arthritis Are Gender-Specific. Front. Immunol. 12:707617. doi: 10.3389/fimmu.2021.707617
Received: 10 May 2021; Accepted: 11 August 2021;
Published: 03 September 2021.
Edited by:
James Cheng-Chung Wei, Chung Shan Medical University Hospital, TaiwanReviewed by:
Zhiming Lin, Third Affiliated Hospital of Sun Yat-sen University, ChinaKaterina Theodoropoulou, Centre Hospitalier Universitaire Vaudois (CHUV), Switzerland
Copyright © 2021 Hong, Qu, Ji, Li, Zhang, Jin, Wang, Zhang, Shen, Meng, Zhou, Fang, Wang and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shigui Yan, zrjwsj@zju.edu.cn; Wei Wang, sunny01@zju.edu.cn
†These authors have contributed equally to this work
 Jianqiao Hong
Jianqiao Hong