- 1Centre for Inflammation and Tissue Repair, University College London (UCL) Respiratory, London, United Kingdom
- 2London School of Hygiene and Tropical Medicine, Infectious and Tropical Disease, Department of Infection Biology, London, United Kingdom
- 3Cellular and Molecular Immunology Unit, Centre for Research and Development of Medical Diagnostic Laboratories (CMDL), Faculty of Associated Medical Sciences, Khon Kaen University, Khon Kaen, Thailand
- 4Department of Microbiology, Faculty of Medicine Siriraj Hospital, Mahidol University, Bangkok-Noi, Bangkok, Thailand
- 5School of Pharmacy, University College London, London, United Kingdom
- 6Department of Medical Technology, Faculty of Associated Medical Sciences, Chiang Mai University, Chiang Mai, Thailand
Antibody therapy may be an alternative treatment option for infections caused by the multi-drug resistant (MDR) bacterium Acinetobacter baumannii. As A. baumannii has multiple capsular serotypes, a universal antibody therapy would need to target conserved protein antigens rather than the capsular polysaccharides. We have immunized mice with single or multiple A. baumannii strains to induce antibody responses to protein antigens, and then assessed whether these responses provide cross-protection against a collection of genetically diverse clinical A. baumannii isolates. Immunized mice developed antibody responses to multiple protein antigens. Flow cytometry IgG binding assays and immunoblots demonstrated improved recognition of both homologous and heterologous clinical strains in sera from mice immunized with multiple strains compared to a single strain. The capsule partially inhibited bacterial recognition by IgG and the promotion of phagocytosis by human neutrophils. However, after immunization with multiple strains, serum antibodies to protein antigens promoted neutrophil phagocytosis of heterologous A. baumannii strains. In an infection model, mice immunized with multiple strains had lower bacterial counts in the spleen and liver following challenge with a heterologous strain. These data demonstrate that antibodies targeting protein antigens can improve immune recognition and protection against diverse A. baumannii strains, providing support for their use as an antibody therapy.
Introduction
Antibiotic resistant Acinetobacter baumannii infections have spread rapidly across hospitals, particularly intensive care units, increasingly causing nosocomial infections that are associated with a mortality rate that can be greater than 50% (1–5). Clinical isolates of A. baumannii are often highly resistant to multiple antibiotics, including carbapenems, making them difficult to treat (5). Current treatment options use complex antibiotic combinations to overcome the antimicrobial resistance (AMR) profiles, but extremely multidrug resistant isolates with limited treatment options are now being identified in patients (6). This is compounded as currently, there are no licensed vaccines to counteract the growing threat of A. baumannii infections. As a consequence, A. baumannii is the top priority pathogen in recent WHO reports on AMR, and developing alternative methods of treating or preventing A. baumannii infections is a global imperative (1). A. baumannii seems to be a particularly prevalent pathogen in Asian countries: For example, in Thailand, infections with multi-drug resistant A. baumannii were the dominant cause of death due to AMR infections in 2010, estimated to cause over 25,000 deaths (7).
A potential adjunct treatment option for severe A. baumannii infections is monoclonal antibody therapy that specifically targets surface A. baumannii antigens, thereby improving immune recognition and clearance of the bacteria by the host (6, 8). Proof of principle studies have shown that whole A. baumannii cell preparations inactivated using either formalin (9, 10), antibiotic treatment (11), lipopolysaccharide removal (12), or by targeted gene deletion (13), induce antibody responses to A. baumannii that are partially protective against subsequent infections in mice with the homologous strain. Similarly, outer membrane complexes (OMCs) (14, 15), vesicles (OMVs) (16) and at least fifteen recombinant proteins (17–33) have been shown to elicit partially protective anti-A. baumannii antibody responses. However, the extensive genetic and antigenic heterogeneity in A. baumannii strains poses a significant challenge in the development of cross-protective antibody responses. Although anti-capsular antibody protects against A. baumannii infections in both mice (34) and rat (35) infection models, over 100 unique capsule loci (KL types) have been described for A. baumannii clinical strains (36). For example, a recent evaluation of 191 isolates collected from three tertiary hospitals in Thailand reported high capsule type diversity with a total of 24 KL types identified, and the three KL types with the highest prevalence KL10, KL6 and KL47 only accounting for 15.7%, 15.2% and 11.0% of the isolates evaluated respectively (37). This diversity inhibits using A. baumannii capsular antigen as the target for a monoclonal antibody therapy. Targeting conserved sub-capsular antigens offer an attractive alternative to targeting the capsular antigen. However, the relatively low abundance of protein antigens compared to capsular antigen and their sub-capsular localization suggests antibody to protein antigens are likely to be less protective than antibody to capsule (38). Furthermore, due to the extensive genetic diversity amongst A. baumannii strains, only approximately 2,000 genes are common to almost all strains, representing a minority of the over 20,000+ genes found in the A. baumannii pan-genome (39). Furthermore, only a small proportion of the core genes are predicted to be expressed on the surface of bacterial cells and therefore potentially accessible to therapeutic antibodies. Despite these problems, antibodies raised by vaccinating with either whole cells (9, 13, 14) or protein antigens (17, 20, 21, 29, 30, 40) have been shown to recognize and protect against infection with heterologous A. baumannii strains, indicating an anti-protein approach is feasible. However, the KL types and degree of genomic diversity between the strains used in these studies was unclear making the clinical potential of the demonstrated cross-protective immunity hard to assess.
Here we have investigated whether antibody responses developed in response to exposure to live A. baumannii are able to recognize heterologous strains, including those with a different KL type. To assess cross-reactivity between antisera and clinically relevant strains, we have tested mouse antisera raised against a single A. baumannii strain or three different strains against nine Thai clinical A. baumannii isolates. As antibody mediated immunity to bacterial pathogens can be mediated by improving bacterial agglutination, membrane attack complex (MAC)-dependent bacterial lysis or complement- or Fcγ receptor-dependent phagocytosis, we have used in vitro assays to evaluate these functions in different antisera. The protective efficacy of immunization with a selected combination of strains was tested in a mouse model of infection. Our hypothesis was that when compared to immunization with a single strain, sequential immunization with different clinical isolates would select for antibody responses to immunogenic, conserved A. baumannii protein antigens and thereby induce improved cross-protective antibody responses.
Materials and Methods
Bacterial Strains and Culture Conditions
The nine clinical A. baumannii isolates were obtained from patients admitted to Songklanagarind and Siriraj Hospitals, Thailand (37) (Table 1). AB5075 wild-type and unencapsulated AB5075Δwza were obtained from the Manoil lab A. baumannii mutant library (https://www.gs.washington.edu/labs/manoil/baumanniii.htm). Bacteria were cultured at 37°C on LB plates or in LB broth to an optical density at 600nm of 0.8 (approximately 109 CFU/ml) and stored at -80°C in 10% glycerol as single use aliquots.
Bacteremia Mouse Model of A. baumannii Infection
All in vivo experiments using mice were performed according to United Kingdom national guidelines for animal use and care. Experiments performed at UCL were approved by the UCL Biological Services Ethical Committee and the UK Home Office (P64714548). Experiments used 5–6-week-old outbred female CD1 mice obtained from Charles River Laboratories. A. baumannii associated bacteremia mouse model was established by the intraperitoneal route by testing bacterial doses ranging from 106 to 1010 CFU/mouse in the presence or absence of varying concentration of mucin. There was rapid clearance of bacteria at all doses when injected in the absence of mucin. A minimum lethal dose of 1.0 x 109 CFU/mouse was determined for the AB1 strain. With this dose, significant target organ CFU could be recovered consistently at 24 hours post-inoculation. Mice rapidly developed fatal infection at later time points. To evaluate the protective efficacy of the AB3879 and multi strain antisera, the AB1 stock was washed and resuspended in appropriate volume of 5% Mucin in PBS to inject a dose of 6.0 x 109 CFU/per mouse. Groups of mice injected with 5% Mucin/PBS served as a control. At 24 hours post infection, mice were sacrificed, and the lungs, liver and spleen harvested, homogenized and the bacterial counts obtained by plating serial dilutions on LB agar plates.
Anti-A. baumannii Mouse Sera
The majority of experiments were performed using pooled serum obtained from mice immunized with live sub-lethal doses (106 CFU/mouse/vaccination) of A. baumannii via the intraperitoneal route. Homologous sera namely anti-AB3879, anti-AB5075WT and anti-AB5075Δwza were generated following three immunizations, 14 days apart with the same A. baumannii isolate. Multi strain antisera was generated by sequential immunization with AB3879, AB1492-09 and AB1615-09 isolates respectively. Sera from non-immunized mice were used as controls.
Whole Cell ELISA
Antibodies specific to antigens in different A. baumannii strains were measured by whole cell ELISA. Briefly, A. baumannii were grown to late log-phase (OD600nm = 0.8, approximately 109 CFU/ml), washed, and resuspended in PBS. 96-well plates (Brand plates, IMMUNOGrade) were coated with 50 µl of this bacterial suspension, overnight at 4°C, inactivated using 50 µl of 4% paraformaldehyde for 10 minutes, then blocked with 5% non-fat milk/0.05% Tween 20/PBS (blocking buffer) for 3 hours at room temperature. Sera were diluted to a 1:100 dilution in blocking buffer before addition, and binding to bacterial antigens detected with goat anti-mouse IgG conjugated to Horse-radish peroxidase (Invitrogen) and detected using the TMB substrate.
Immunoblot Detection of Bacterial Lysate Recognition by IgG
A. baumannii bacterial lysate were obtained by resuspending bacteria in 1X lithium dodecyl sulfate (LDS) sample buffer (NuPAGE LDS Sample buffer (4X), Invitrogen) with 50mM dithiothreitol [NuPAGE sample reducing agent (10X)], boiled at 95°C for 10 min, separated on a gradient 4-12% SDS-polyacrylamide gel (NuPAGE Bis-Tris gels), and transferred to nitrocellulose membranes using the iBlot 2 Dry Blotting System (ThermoFisher Scientific). Nitrocellulose membranes were blocked with 10% non-fat milk/0.05% Tween 20/PBS (blocking buffer) for 2 hours then incubated with mouse sera at a 1:1000 dilution overnight at 4°C, washed and then probed using an anti-mouse IgG conjugated with horseradish peroxidase. Protein bands were visualized using the chemiluminescent ECL reagent (Amersham) on the ImageQuant™ LAS 4000.
Flow-Cytometry Detection of Bacterial Surface Recognition by IgG
Total IgG binding to A. baumannii was assessed using flow cytometry. 106 CFU of A. baumannii was incubated with mouse antisera in triplicate for 1 hour at 37°C, followed by goat anti-mouse IgG-allophycocyanin (APC) (Invitrogen) for 1 hour at room temperature. Samples were analyzed using a BD FACSVerse and data processed using FlowJo software for Windows (version 10). Markers for identifying IgG positive bacteria was set using bacteria incubated with PBS and then incubated with the secondary antibody. The proportion of IgG positive bacteria was gated on the single-cell population of unstained and bacteria labelled with secondary antibody only. Two independent experiments were conducted using stocks cultured on separate days and stored as single-use aliquots. A positive correlation between the two independent experiments was observed (Spearman correlation: R=0.7157, p-value=0.0359).
Genomic Analysis of the Capsule Locus
Capsule KL loci were previously determined using Kaptive and an A. baumannii KL database (37, 41).
Growth Inhibition Assays
102 CFU of A. baumannii was incubated with 1:100 dilution of mouse antisera in triplicate and bacterial growth (OD600nm) at 37°C in 5% C02 was monitored every hour over a 24-hour period using the Spark multimode microplate reader (TECAN).
Neutrophil Opsonophagocytosis Assays
Phagocytosis was investigated using an established flow cytometry assay (42), neutrophils extracted from fresh human blood from healthy adult donors and fluorescent A. baumannii labeled with 6-carboxyfluorescein succinimidyl ester (FAMSE, Molecular Probes). Bacteria was opsonized with mouse antisera and 5% heat-inactivated human sera for 30 minutes at 37°C followed by the addition of 105 neutrophils to a final multiplicity of infection (MOI) of approximately 1:100. A minimum of 5000 cells were analyzed by flowcytometry using a BD FACSVerse and data processed using FlowJo software for Windows (version 10). To identify the percentage of neutrophils associated with bacteria, neutrophils that had not been incubated with bacteria were used to identify the negative control. The proportion of bacteria positive neutrophils was gated on the single-cell population of neutrophils not incubated with bacteria. To combine the percentage of bacteria associated neutrophils and the intensity of association, a fluorescence index was determined by multiplying the percentage of positive neutrophils by the geometric median fluorescence of the positive population. All sera were tested in parallel on the same day, at the same concentration with the same batch of bacteria and neutrophils for each independent experiment.
Bacterial Cell Size Measurement
A. baumannii strains were cultured in LB broth to an OD600nm of 0.4, pelleted and resuspended in 300 µL sterile PBS. 10 µL bacteria solution was mixed with 2 µL 2000 kDa FITC-Dextran (Sigma) at 10 mg/ml before spotting 10 µL onto a glass microscope slide and sealed with a coverslip. Slides were imaged using a Zeiss LSM 880 confocal microscope with Plan-Apochromat 63x/1.4 Oil DIC M27 using standard confocal mode driven by ZEN Black 2.3 software (Carl Zeiss). A minimum of 1,000 individual bacteria were imaged using a 490 nm laser with collection of light at 520 nm. To measure bacterial cell length, ImageJ 1.53a software was used to first convert images to 8-bit with auto-threshold, before identifying singlet bacterial particles within a defined circularity and size range.
Statistical Analysis
Statistical analyses were conducted using GraphPad Prism version 8 (GraphPad, USA). Data are presented as means, and the error bars represent standard deviations. Parametric data were analyzed using Student’s T test and nonparametric data using the Mann-Whitney U test. Doubling times from growth curves were calculated using the log of exponential growth equation in GraphPad version 8 (https://www.graphpad.com/guides/prism/latest/curve-fitting/reg_log-of-exponential-growth.html). The optical density values obtained from bacterial growth at the exponential phase (6-16 hours) were log transformed and the non-linear regression of exponential growth with a log population equation used to calculate doubling time in the presence of respective anti-A. baumannii sera. The doubling time ratio was calculated by dividing doubling time in respective sera over doubling time in sera from non-immunized mice (DT sera/DT control).
Results
Immunization With A. baumannii Induces Antibody Responses That Recognize Multiple Conserved Proteins
Nine A. baumannii strains were isolated in 2016 from clinical specimens obtained from two tertiary hospitals in Thailand, Siriraj Hospital, Bangkok and Songklanagarind Hospital, Songklanagarind Hospital, Hat Yai, Songkhla Province. These strains subsequently underwent whole-genome sequencing and sequence typing (37). The nine isolates tested had a range of multilocus sequence typing (MLST) sequence types (ST) and KL types including five ST2 (representing global clone 2), three ST215 and one ST164 strains and three KL47, two KL10, two KL52, one KL6 and one KL2 capsular loci strains (Table 1). None of the isolates share both the capsule loci and MLST (Institute Pasteur Scheme) type. Two additional strains, the AB5075 (ST2, KL25) wild-type and the unencapsulated AB5075Δwza (strain name: tnab1_kr121203p06q153, genotype: wza-153::T26) strain were obtained from the Manoil lab A. baumannii mutant library (https://www.gs.washington.edu/labs/manoil/baumanniii.htm).
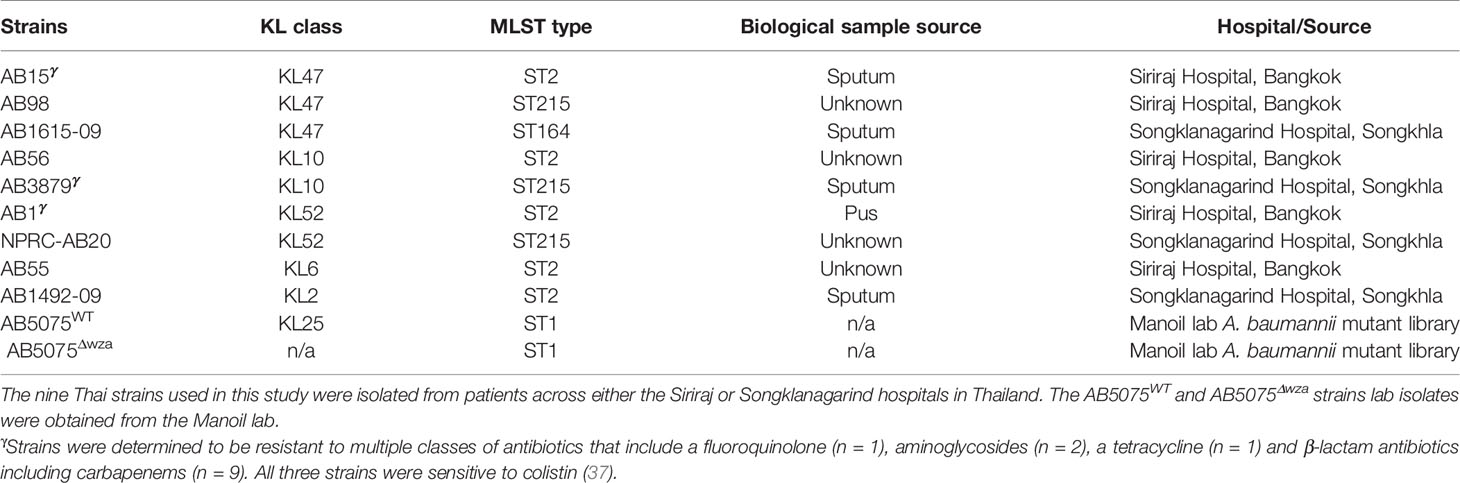
Groups of mice were immunized with either AB3879 (KL10 type) or sequentially with the AB3879, AB1492-09 (KL2) then AB1615-09 (KL47) strains (multi strain group) with both groups receiving the same total bacterial CFU. Sera was obtained 2 weeks after the third injection, pooled and used for whole cell ELISAs and to probe immunoblots of all nine A. baumannii strains. Whole cell ELISAs showed both antisera recognized all nine strains to varying degrees, with higher antibody levels to all strains observed with the multi strain antisera (Figures 1A, B). The immunoblots showed both antisera recognized multiple bands of similar size for all the probed A. baumannii strains (Figures 1C, D). More immunoreactive bands were visible in immunoblots probed with multi strain antisera (Figure 1D, black arrows), although one band above 70 KDa was weaker compared to the results for lysates probed with AB3879 antisera (Figure 1C, black arrow). These data demonstrate that mouse antisera generated by inoculation of live A. baumannii can recognize different A. baumannii strains expressing different KL types and from different STs, and that at least some of the antibody responses target multiple protein antigens that are likely to be conserved between A. baumannii strains.
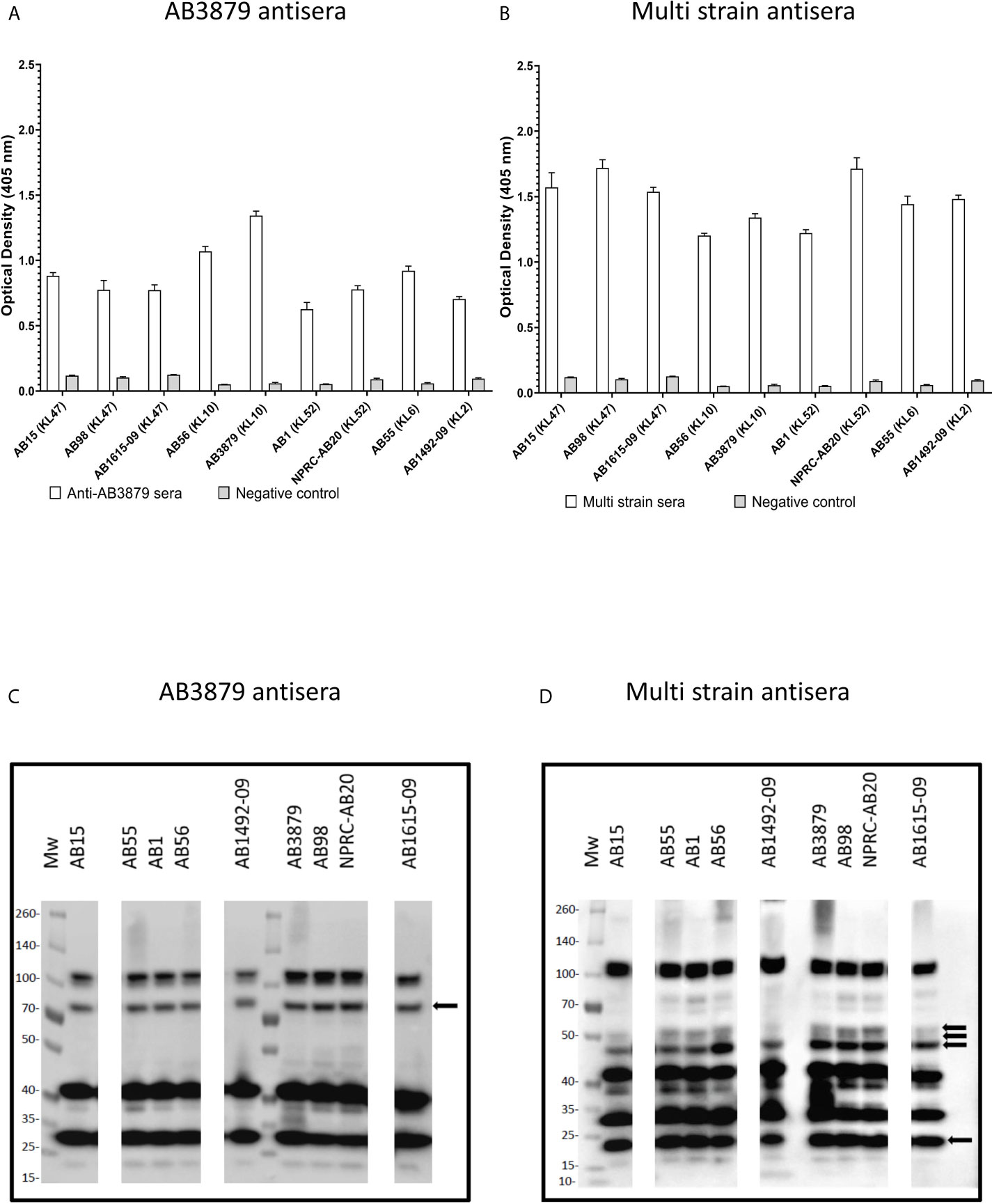
Figure 1 Whole cell ELISA and immunoblot detection of the reactivity of mouse antisera with A. baumannii isolates. Whole cell ELISAs of IgG binding to nine A. baumannii isolates using either AB3879 (A) or multi strain antisera (B). Whole cell lysates from the nine diverse A. baumannii isolates were resolved on a reduced 4-12% gradient gel, probed with mouse antisera and anti-mouse IgG-HRP. Each lane represents a clinical A. baumannii isolate indicated above. Immunoreactive proteins detected when blot was probed with either AB3879 (C) or multi strain antisera (D). Black arrows represent bands unique to immunoblots probed with either AB3879 or multi strain sera. Bars represent mean values for each group and the error bars indicate standard deviations (SDs) (n = 3). Representative data from two independent experiments is shown.
The Effects of the Capsule on Antibody Binding to A. baumannii in Sera From Immunized Mice
Previous authors have demonstrated that the A. baumannii polysaccharide capsule inhibits access of antibody to underlying protein antigens (18). In addition, antisera raised in mice to the wild-type A. baumannii strains is also likely to contain a mixture of IgG to capsular and protein antigens. To assess whether the A. baumannii capsule was a target antigen to IgG in the antisera and/or inhibited anti-protein antigen recognition, we used a flow cytometry IgG binding assay to measure IgG deposition on live A. baumannii incubated in antisera. Initially, we measured IgG deposition on the AB5075 wild-type (KL25, ST1) and unencapsulated AB5075Δwza strains using antisera raised by immunizing mice with either of these two strains. This confirmed that mouse antisera raised against encapsulated A. baumannii contained IgG that recognized the capsule, with increased IgG deposition on the AB5075 wild-type strain when incubated in antisera raised against the encapsulated AB5075 wild-type compared to the unencapsulated AB5075Δwza strain (Figure 2A, left panel and B). The results also indicated there was significant IgG in both AB5075WT and AB5075Δwza antisera against protein antigens, with marked IgG deposition occurring on the AB5075Δwza unencapsulated strains (Figure 2A, right panel and B). When incubated in antisera raised against the AB5075Δwza unencapsulated strain, there was reduced IgG deposition on the AB5075 strain compared to the AB5075Δwza strain demonstrating the capsule impaired recognition of A. baumannii by IgG to protein antigens (Figure 2A, 2nd row). When incubated in AB3879 and multi strain antisera there were high levels of IgG binding to the unencapsulated AB5075Δwza strains, confirming these sera contained a significant anti-protein antigen IgG response (Figures 2A, B). However, there was markedly reduced IgG binding to the encapsulated AB5075 wild-type strain in AB3879 and multi strain antisera, confirming the capsule impaired recognition of protein antigens. There was a small degree of residual IgG binding to the encapsulated AB5075 wild-type strain in the multi strain antisera, demonstrating that some recognition mediated by antiprotein IgG occurred in this serum even against an encapsulated strain.
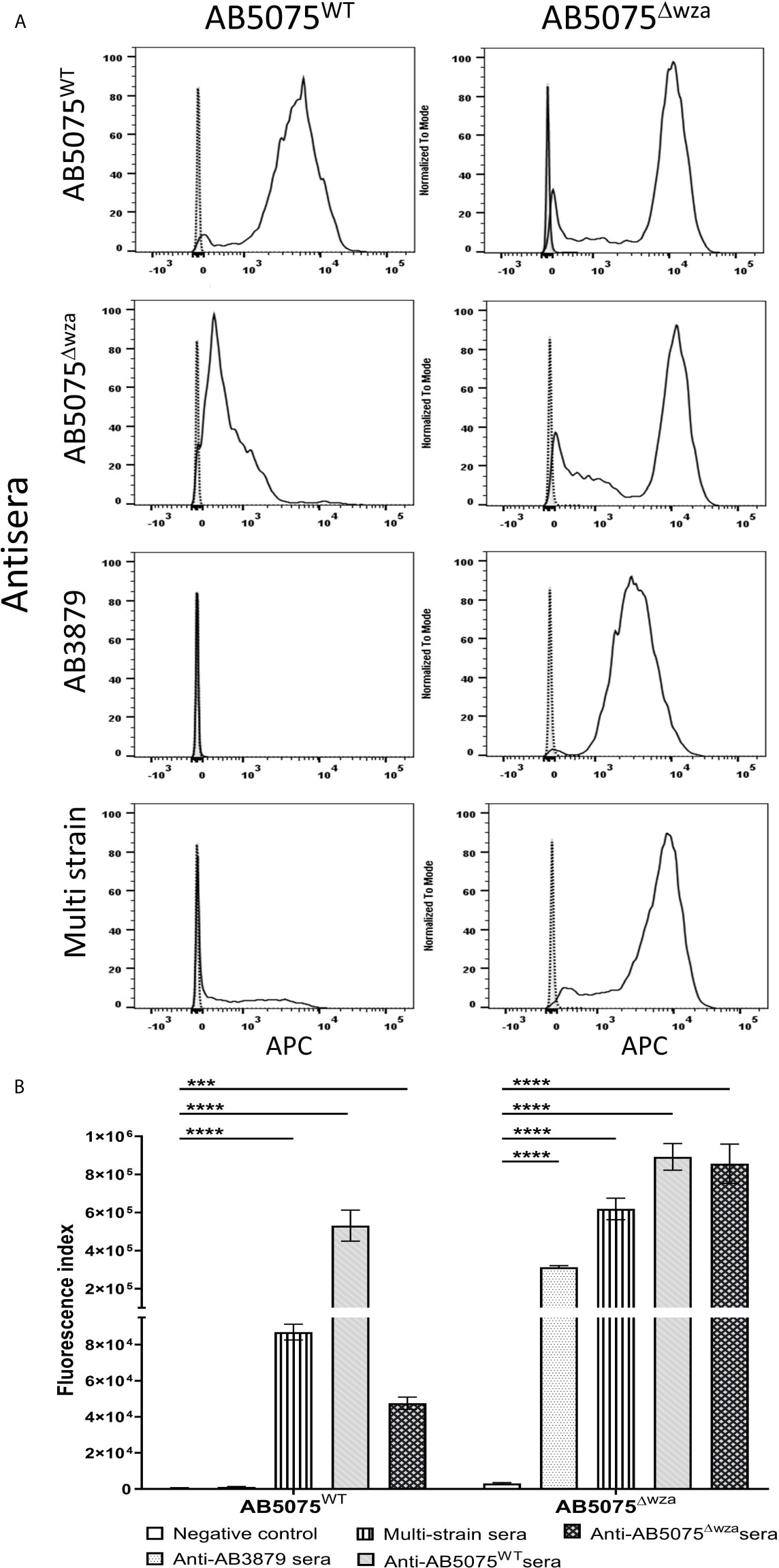
Figure 2 Flow cytometry detection of IgG binding to wild type and un-encapsulated AB5075 isolates by mouse antisera. Representative histograms showing the recognition of 1x106 CFU of AB5075 wild-type strain (left panel) and un-encapsulated AB5075Δwza strain (right panel) to either (A) AB5075WT, AB5075Δwza, AB3879 or multi strain antisera. (B) Bar graph presenting the proportion of AB5075WT and AB5075Δwza bacteria bound by IgG in AB3879, multi strain, AB5075WT or AB5075Δwza antisera. The Y axis shows the fluorescence index calculated by multiplying the percentage of IgG positive bacteria to the median fluorescent intensity of the positive population that was detected in each isolate. Bars represent mean values for each condition/strain and the error bars indicate standard deviations (SDs) (n = 3). T-test was used for statistical analysis ***p-value < 0.001, ****p-value =< 0.0001. Representative data from two independent experiments is shown.
IgG Binding to Diverse A. baumannii Strains Incubated in Antisera From Immunized Mice
To further characterize whether capsular and/or protein antigens were targets for IgG in the AB3879 and multi strain antisera, the IgG deposition assay was repeated with all nine Thai clinical isolates. For the AB3879 antisera, the highest levels of IgG binding occurred to the homologous AB3879 strain, and to strain AB56 that shares the same capsule locus (KL10) (Figures 3A, C). In addition, after incubation in AB3879 antisera, there was significant IgG binding to two of the KL47 capsule strains (AB98 and AB1615-09), one of which shared the same ST type to AB3879 (ST215). There was minimal IgG binding to the remaining strains (Figures 3A, C). These data show that although the sera to AB3879 contains antibodies to multiple conserved protein antigens, IgG deposition in this antiserum correlated with strain KL type except for the KL47 strains. Genomic analysis of the capsule loci of the Thai clinical isolates showed significant differences between the KL47 and KL10 capsule loci, suggesting that preservation of capsule structure between these KL types is unlikely to explain the cross-reactivity observed (Figure 4).
Figure 3 IgG recognition of a panel of nine A. baumannii isolates by mouse antisera generated against immunization with one strain or sequentially with three strains. (A) Mouse AB3879 antisera and (B) multi strain (AB3879+AB1492-09+AB1615-09) antisera IgG binding to the surface of A. baumannii isolates as measured by flow cytometry. The nine isolates are grouped in order of their respective capsule serotypes and are indicated in brackets on the X-axis. Round, square and triangle isolates belong to the ST2, ST215 and ST164 MLST type respectively. The Y axis shows the proportion of IgG positive bacteria multiplied by the median fluorescent intensity (MFI) of IgG positive population, in each of the nine A. baumannii isolates. (C) Representative histograms showing IgG binding of 1x106 CFU of the nine A. baumannii isolates to either AB3897 antisera (left panel) or multi strain antisera (right panel). Grey histogram represents bacteria labelled with 2nd antibody only, black-dotted line represents bacteria incubated with sera from non-immunized mice and black solid line represents bacteria incubated with the respective sera (AB3879 antisera left panel and multi strain antisera right panel). The nine isolates and their respective KL class denoting the capsule serotypes are indicated alongside respective histograms. Dot plots represent triplicate values with lines indicating the mean values and the error bars indicate standard deviations (SDs) (n = 3). Representative data from two independent experiments is shown.

Figure 4 Organization of the five KL loci represented in the A. baumannii strains. A comparison of the CPS biosynthesis genes clusters and their organization in the five KL loci represented in the A. baumannii strains. CPS export genes are in black, genes in dark blue are involved in the synthesis of common sugar substrates, genes in green are involved in the synthesis of complex sugars, genes in yellow encode glycosyltransferases, genes in red encode initiating transferases and genes in brown encode Acyl or acetyl transferases.
In contrast to the data for AB3879 antisera, incubation in multi strain antisera (raised against KL10, KL2 and KL47 strains) resulted in IgG binding to eight of the nine strains investigated (Figures 3B, C). The highest binding was to AB3879 (KL10) and AB1615-09 (KL47) strains included in the vaccination schedule, as well as the KL10 strain, AB56 (similar to the results for AB3878 antisera), to two other KL47 strains and the AB1492-09 (KL2) strain (included in the vaccination schedule). In addition, there were also lower levels of IgG binding detectable to the heterologous strains AB1 (KL52) and AB55 (KL6) (Figures 3B, C) indicating that anti-protein responses in multi strain antisera allows immune recognition of a wider range of strains than in the AB3879 antisera.
Effects of Antisera on Growth of Encapsulated AB5075WT and Unencapsulated AB5075ΔwzaA. baumannii Strains
Agglutination by specific antisera can inhibit bacterial growth in broth culture (43). To evaluate whether anti-capsular and/or anti-protein antibodies could inhibit A. baumannii growth, we used antisera raised against the encapsulated AB5075 wild-type and the unencapsulated AB5075Δwza strain to measure growth inhibition in both AB5075WT and AB5075Δwza strains. Growth of the encapsulated AB5075 wild-type strain was inhibited by antisera raised against the encapsulated AB5075 strain but not by antisera raised against the unencapsulated AB5075Δwza or AB3879 or the multi strain antisera (Figure 5). Reduced growth of the unencapsulated AB5075Δwza strain was only observed when approaching the stationary phase after incubation with antisera raised against AB5075WT, AB5075Δwza and the multi strain group (Supplementary Figure 1), suggesting late phase growth could be inhibited to a small degree by anti-protein antibodies. These data indicate growth inhibition was largely dependent on capsule specific antibodies.
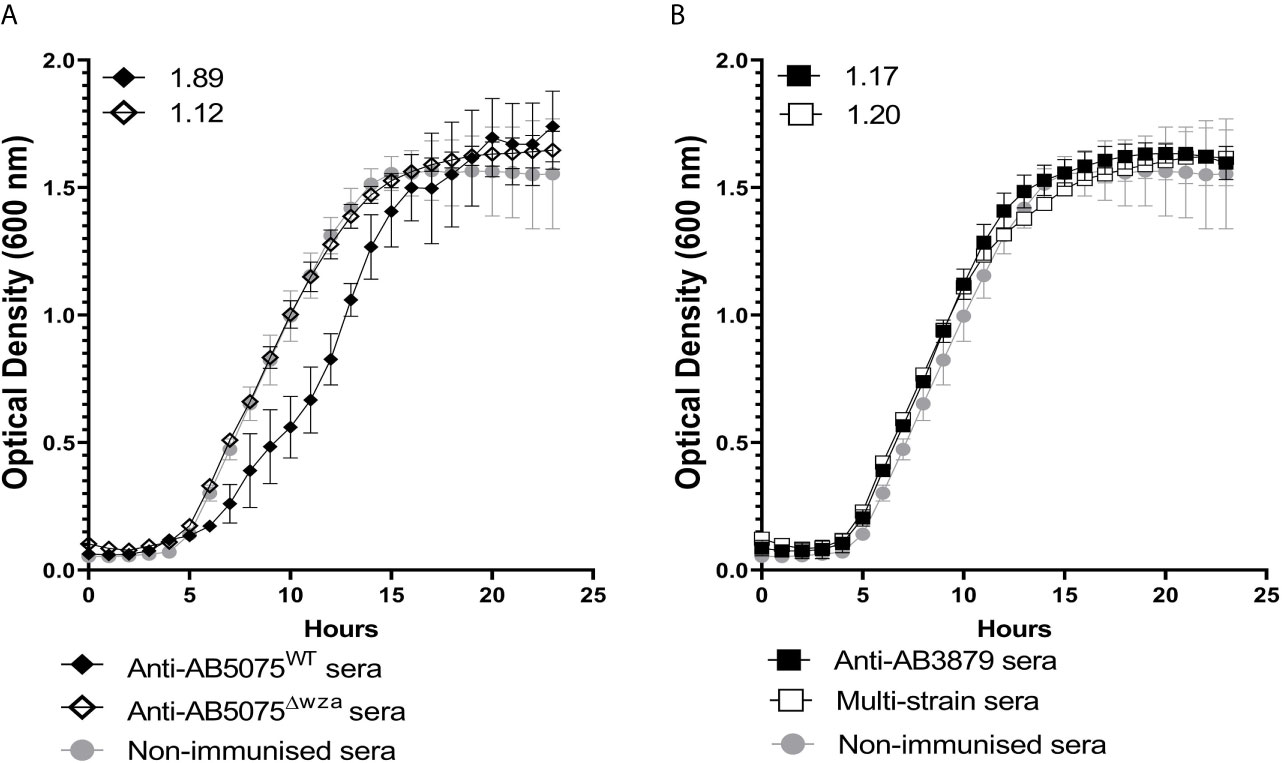
Figure 5 Growth inhibitory activity of wild type AB5075 isolates by mouse antisera. 102 CFU of encapsulated AB5075WTstrain was incubated with either, AB5075WT antisera (black line, solid diamond), AB5075Δwza antisera (black line, open diamond) and sera from non-immunized mice (grey line, solid circle) (A) or AB3879 antisera (black line, solid square), multi strain antisera (black line, open square), and sera from non-immunized mice (grey line, solid circle) (B). Numbers on the top left corner represent the ratios of doubling time for respective antisera. Ratios of doubling time in test sera/control sera were calculated with a ratio of > 1.2 indicating delayed growth in the antisera. Line graphs indicate the mean value, and the error bars indicate standard deviations (SDs) (n=6).
Growth Inhibitory Activity of AB3879 and Multi Strain Antisera Against Thai A. baumannii Strains
To further characterize the inhibitory activity in the AB3879 and multi strain antisera, the growth inhibition assay was repeated with all nine Thai clinical isolates. When compared to sera from non-immunized mice, AB3879 antisera inhibited the growth of both KL10 strains AB3879 and (to a lesser extent) AB56, with an increased doubling time and significantly reduced the total CFU reached in culture for both strains (Figure 6 and Table 2). Multi strain antisera (raised against KL10, KL2 and KL47 type strains) inhibited growth and increased the doubling times for all five KL10 or KL47 strains, and significantly reduced the total CFU reached for the AB98 (KL47), AB1615-09 (KL47) and AB3879 (KL10) strains (Figure 6 and Table 2). Overall, all the strains showing significant growth inhibition in A. baumannii antisera were the homologous KL types to strains used to generate the antisera and showed high levels of IgG binding in the flow cytometry assay (Figure 3), suggesting this effect was largely dependent on anti-capsular antibodies. However, growth was not inhibited in antisera for every strain showing high levels of IgG binding (e.g., AB98 and AB1615-09 in AB3879 antisera) or for all KL types used to generate the antisera (e.g., the KL2 strain AB1492-09 in multi strain antisera) (Supplementary Figure 2).
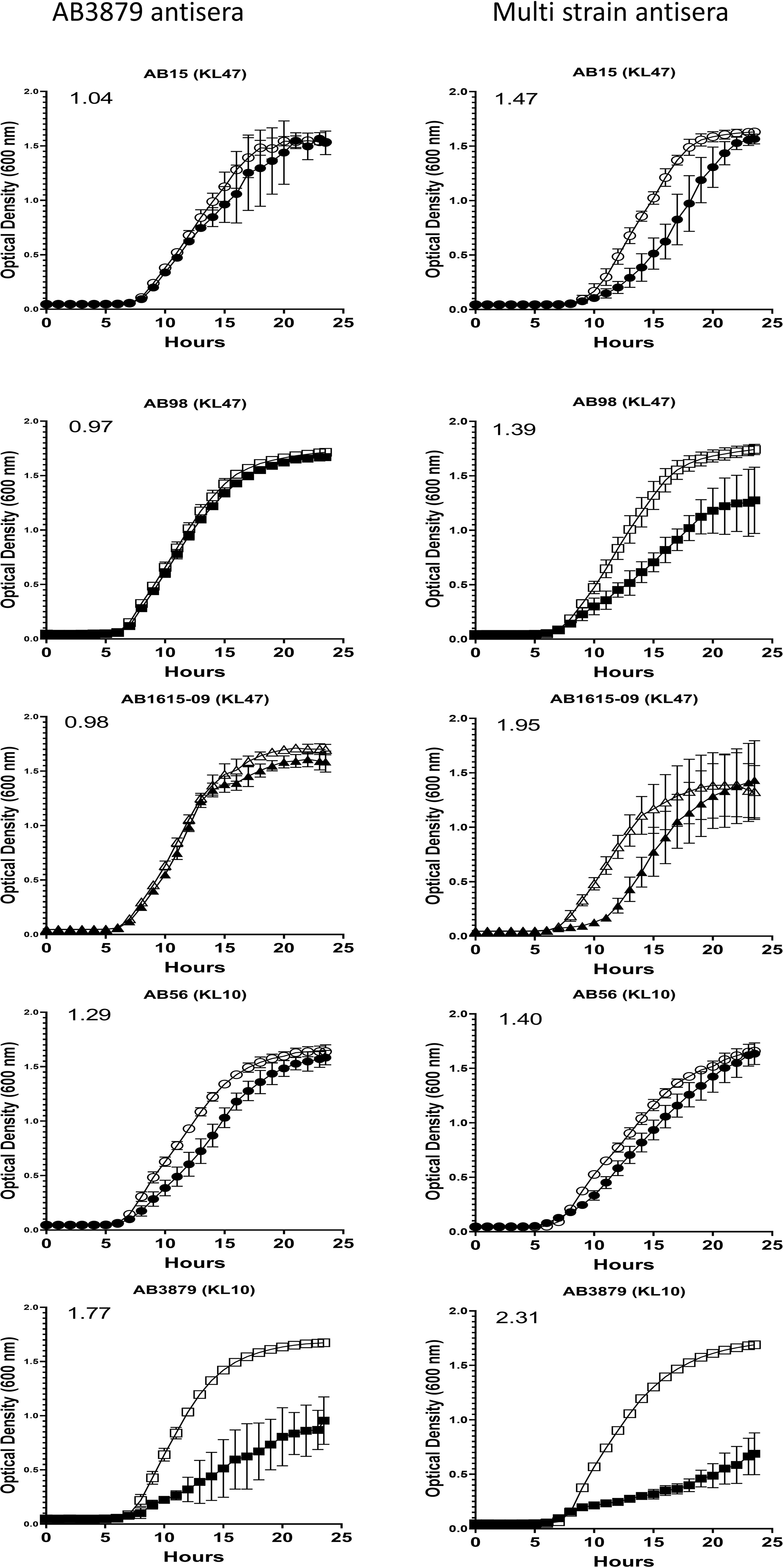
Figure 6 Growth inhibitory activity on a panel of five A. baumannii isolates by mouse antisera. 102 CFU of a panel of A. baumannii isolates were incubated in triplicate with either AB3879 antisera (left panel) or multi strain antisera (right panel) and the OD600nm measured every 30 minutes for 24 hours. Sera from non-immunized mice was included as a control (open symbols). Solid symbols represent respective anti-A. baumannii antisera. The five isolates are grouped by their capsule serotype and indicated at the top of each graph in brackets. Round, square and triangle symbols represent ST2, ST215, and ST164 MSLT types respectively. Numbers on the top left-hand corner represent the ratios of doubling time for respective antisera. Ratios of doubling time in test sera/control sera were calculated with a ratio of > 1.2 indicating delayed growth in the antisera. Line graphs indicate the mean value, and the error bars indicate standard deviations (SDs) (n = 6).
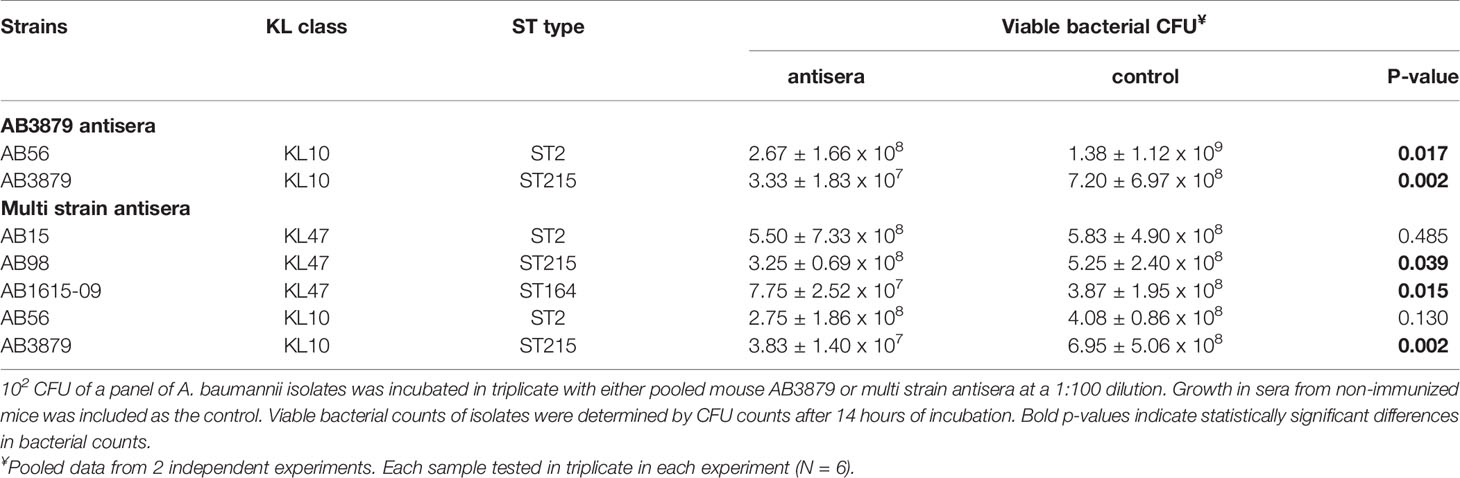
Table 2 Total CFU counts to measure the growth inhibitory effects of AB3879 antisera and multi strain antisera.
The Effects of the Capsule on Promotion of A. baumannii Neutrophil Phagocytosis by Antisera
To assess whether the A. baumannii capsule inhibited neutrophil phagocytosis following opsonization with antisera, we measured phagocytosis using a flow cytometry assay of the encapsulated AB5075 wild-type and the unencapsulated AB5075Δwza strains using antisera raised by immunizing mice with either of these two strains. As expected, the encapsulated AB5075 strain was considerably less susceptible to neutrophil phagocytosis compared to the unencapsulated AB5075Δwza strain following opsonization with all antibodies, including sera from non-immunized mice (Figure 7). Although IgG deposition on the AB5075 wild-type strain was observed with antisera raised against AB5075WT, AB5075Δwza and the multi strain group (Figure 2), only incubation in AB5075WT and to a lesser extent multi strain antiserum, increased neutrophil phagocytosis of this strain (Figures 7A, B). In contrast, the unencapsulated AB5075Δwza strain showed significant increases in susceptibility to neutrophil phagocytosis following opsonization with all antisera tested. Cumulatively, these data confirm the A. baumannii capsule inhibits promotion of neutrophil phagocytosis by antisera, but despite this, anti-protein antibodies in the multi strain antisera can promote a modest improvement of the phagocytosis of a heterologous encapsulated strain.
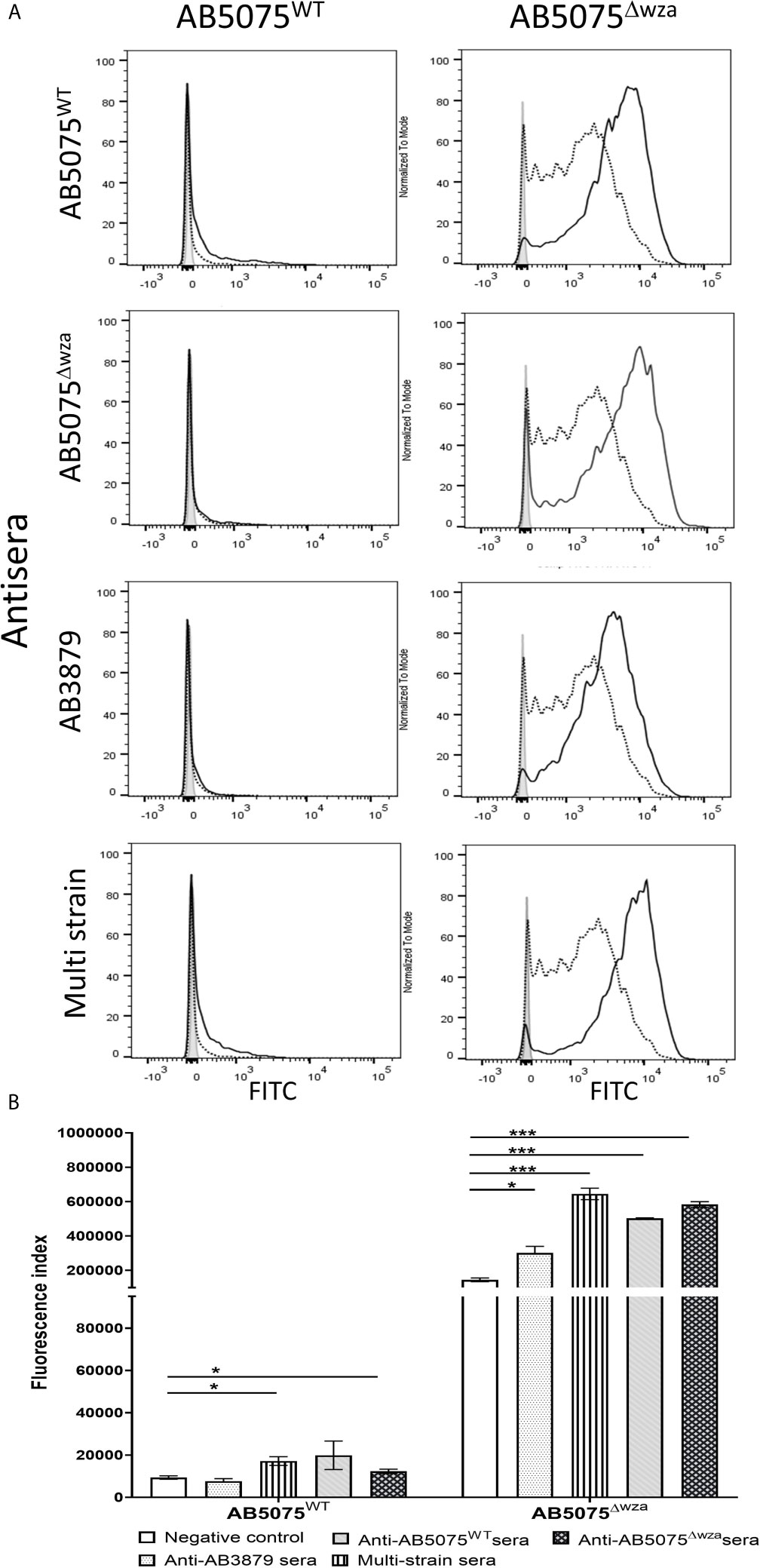
Figure 7 Neutrophil opsonophagocytosis of wild type and un-encapsulated AB5075 isolates by mouse antisera. (A) FAMSE labelled AB5075WT (left panel) and AB5075Δwza (right panel) were opsonized with either AB5075WT, AB5075Δwza, AB3879 or multi strain antisera and incubated with healthy human neutrophils at Bacteria: Neutrophil MOI of 100:1. Sera from non-immunized mice was included as a control. Each graph shows a grey histogram that represents neutrophils incubated with no sera, black-dotted line represents neutrophils with bacteria opsonized with sera from non-immunized mice and black solid line represents neutrophils with bacteria opsonized with the respective sera indicated on the left. (B) Bar graph showing fluorescence index calculated by multiplying the percentage of IgG positive bacteria to the median fluorescent intensity of the positive population that was detected in each isolate, on the Y axis. The X-axis shows either AB5075WT (left) or AB5075Δwza (right) opsonized with either AB3879, multi strain, AB5075WT or AB5075Δwza antisera as indicated in the legend. Bars represent mean values for each condition/strain tested in triplicate and the error bars indicate standard deviations (SDs) (n=3). T-test was used for statistical analysis *p-value < 0.05, ***p-value < 0.001 Representative data from two independent experiments is shown.
Effects of Incubation in Antisera on Phagocytosis of Thai A. baumannii Strains
To further characterize the ability of AB3879 and multi strain antisera, to promote neutrophil phagocytosis of A. baumannii strains, we repeated the flow cytometry neutrophil phagocytosis assay with all nine Thai clinical isolates. Susceptibility to neutrophil phagocytosis when incubated in naïve mouse sera varied markedly between A. baumannii strains even within KL type (e.g., KL47 and KL52 strains) (Figure 8). Compared to control sera, the AB3879 antisera enhanced phagocytosis of the homologous AB3879 (KL10) strain to the highest degree, while multi strain sera also enhanced phagocytosis of the homologous AB3879 (KL10) and AB1615-09 (KL47) strains to the highest degree. Higher levels of phagocytosis of the heterologous AB56 (KL10) and AB1 (KL52) strains were also detected when incubated in either antisera, and for the NPRC-AB20 (KL52) strain as well in AB3879 antisera (Figure 8A). Overall, the neutrophil uptake data showed three broad patterns of results. The first pattern consisted of strains that were susceptible to neutrophil phagocytosis and in which antisera had little additional effect (e.g., strains AB15, AB98, and AB1492-09). The second pattern were strains that were relatively resistant to neutrophil phagocytosis in naïve sera for which addition of antisera significantly improved phagocytosis (e.g., AB1615-09, AB56, AB3879, AB1, and NPRC-AB20). Importantly, these included strains in which phagocytosis was increased by A. baumannii antisera that was raised against heterologous KL types (e.g., AB1 and NPRC-AB20), demonstrating a potential effect of anti-protein rather than anti-capsular antibodies. The last pattern identified was a single strain that was highly resistant to neutrophil phagocytosis under all conditions (AB55).
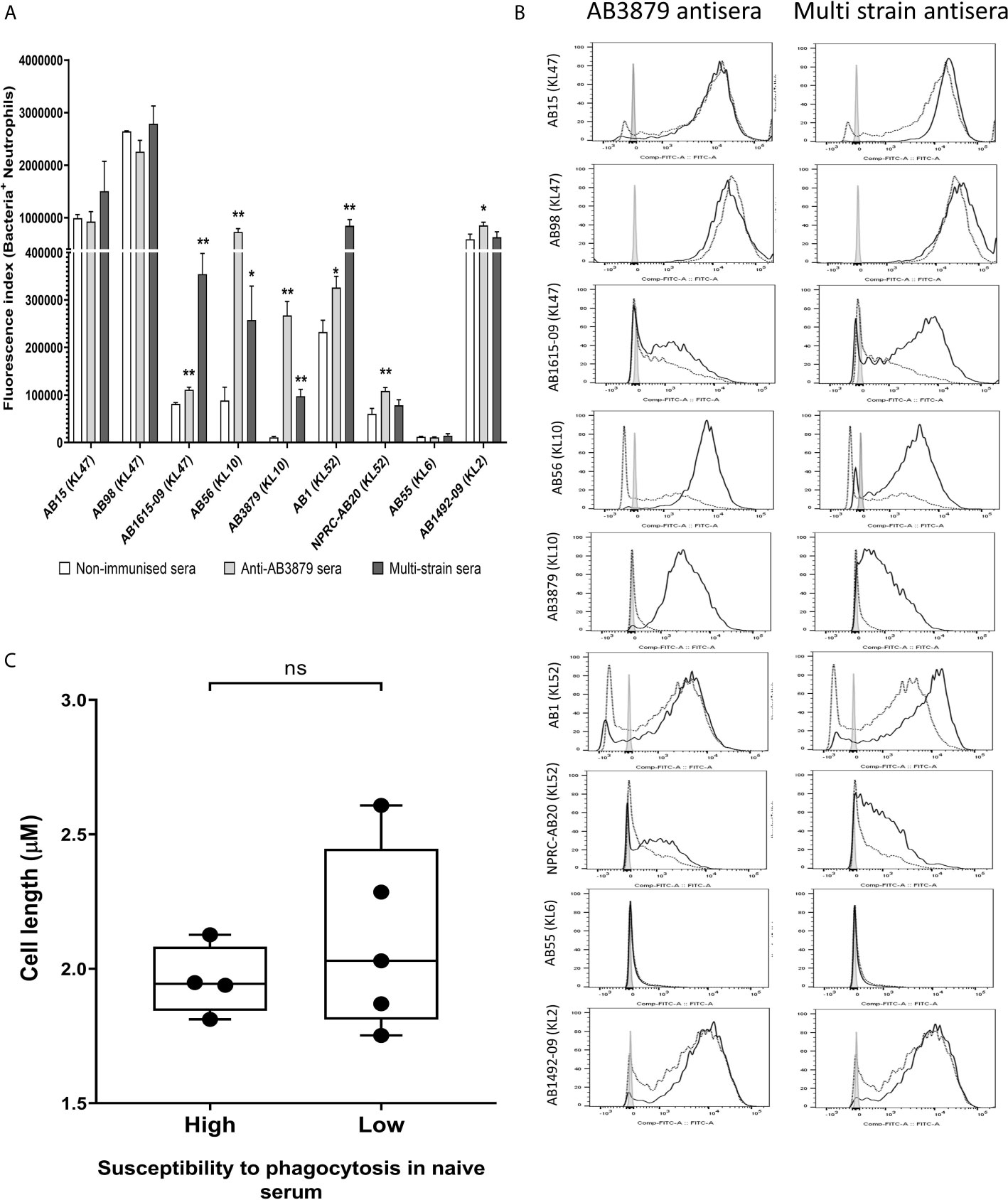
Figure 8 Opsonophagocytosis by human neutrophils of a panel of nine A. baumannii isolates by mouse antisera generated following immunization with one strain or sequentially with three strains. Nine FAMSE labelled A. baumannii isolates were opsonized with either AB3879 antisera, multi strain antisera or sera from non-immunized mice and incubated with healthy human neutrophils at Bacteria: Neutrophil MOI of 100:1. (A) Shows the fluorescence index calculated by multiplying the proportion (%) of FAMSE+ve neutrophils and the median fluorescence intensity of the positive population [mean (± standard deviation)] on the Y axis and the nine isolates with their capsule serotypes indicated in brackets are on the X-axis. Phagocytosis indices in bacteria incubated with non-immune sera, anti-AB3879 sera and multi strain sera for each isolate is represented in the white, grey and dark grey bars respectively. Bars represent mean values for each condition/strain tested in triplicate and the error bars indicate standard deviations (SDs) (n = 3). T-test was used for statistical analysis *p-value < 0.05, **p-value < 0.001, ***p-value < 0.0001, ns: p-value= not significant. Representative data from two independent experiments is shown. (B) Representative histograms showing phagocytosis of FAMSE labelled A. baumannii isolates incubated with either AB3897 antisera (left panel) and multi strain antisera (right panel). Grey histograms represent no sera (neutrophils only), black-dotted line represents addition of neutrophils to bacteria incubated with sera from non-immunized mice and black solid line represents addition of neutrophils to bacteria incubated with the respective mouse antisera. The nine isolates and their respective KL class denoting the capsule serotypes are indicated alongside respective histograms. (C) Comparison of the capsule size in bacterial isolates that show low (N = 5) versus high (N = 4) susceptibility to neutrophils phagocytosis. Box and whisker plots represent mean values for each group and the error bars indicate the minimum and maximum value. Mann-Whitney U-test was used for statistical analysis *p-value < 0.05, **p-value < 0.01, ns: p-value=not significant.
We observed that the A. baumannii capsule inhibited promotion of phagocytosis and the unencapsulated AB5075Δwza strain was significantly more susceptible to neutrophil phagocytosis compared to encapsulated AB5075WT strain, even in naïve sera (Figure 7). To further investigate the marked difference in susceptibility to neutrophil phagocytosis observed in the Thai clinical strains, we compared the bacterial cell length, as a proxy measure of capsule thickness, between the strains that showed low or no susceptibility to phagocytosis in naïve sera (AB1615-09, AB56, AB3879, NPRC-AB20 and AB55) and those strains that were highly susceptible to phagocytosis in naïve sera (AB15, AB98, AB1 and AB1492-09). No differences were observed suggesting that the capsule size between these two groups is unlikely to explain the difference in neutrophil phagocytosis (Figure 8C).
Sequential Immunization Partially Protects Against Bacteremia in Mouse Models of Heterologous A. baumannii Infection
We assessed whether the increased bacterial IgG binding and neutrophil phagocytosis observed in vitro with the multi strain antisera to heterologous strains, resulted in increased protection in a mouse model of A. baumannii bacteremia. Groups of 6 mice were immunized three times with either the single strains AB3879 (KL10) or AB1 (KL52) or sequentially with AB3879, AB1492-09 and AB1615-09 (identical to multi strain antisera group) and then challenged with the AB1 strain (Figures 9A). Mice immunized with the homologous strain AB1 had significantly lower bacterial CFU in the spleen, liver, lungs, and kidneys compared to control group (Figures 9B–E). Similar findings were observed in mice immunized with AB3879 followed by challenge with the homologous strain (Supplementary Figure 3), confirming homologous vaccination protected against subsequent infection.
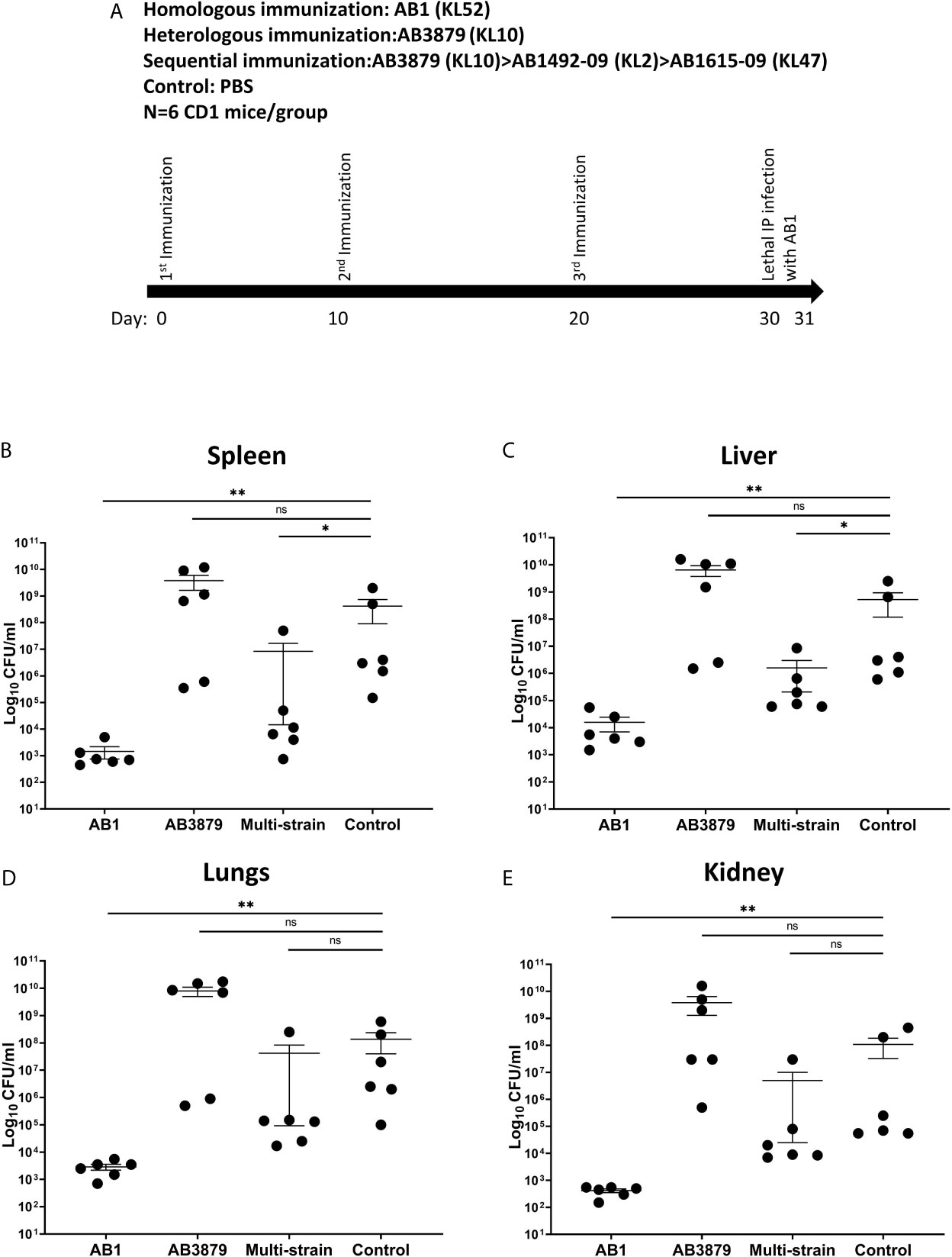
Figure 9 Bacterial load in the spleen, liver, lungs and kidneys from mice following lethal challenge with heterologous AB1 isolates. (A) CD1 mice were immunized three times with 106 CFU of either AB3879, AB1, sequentially with AB3879, AB1492-09 & AB1615-09, or PBS followed by IP infection with 6.0 x 109 CFU/mouse of AB1 isolate in a lethal bacteremia model of infection. The bacterial burden in the (B) spleen, (C) liver, (D) lungs and (E) kidney were determined 24 hpi. Dot plots represent individual values with lines indicating the mean and the error bars indicate standard deviations (SDs) (n = 6/group) Mann-Whitney U-test was used for statistical analysis *p-value < 0.05, **p-value < 0.01, ns: p-value=not significant.
In contrast, the bacterial burden of mice immunized with the AB3879 strain and challenged with the AB1 strain, did not differ from the control group in all organs tested (Figures 9B–E). However, mice immunized sequentially with three strains, all heterologous to the AB1 challenge strain, also had reduced bacterial CFU in the spleen and liver compared to control group (Figures 9B, C). These data demonstrate that sequential pre-exposure to heterologous A. baumannii strains partially protected mice from a lethal injection, indicating a protective effect of anti-protein antibodies in this serum.
Discussion
The use of monoclonal antibodies for the treatment of multi-drug resistant bacteria is currently undergoing evaluation in clinical trials for multi-antibiotic resistant bacterial pathogens such as Pseudomonas aeruginosa and methicillin-resistance Staphylococcus aureus. These monoclonal antibodies target either the capsular polysaccharide, secreted protein toxins or surface membrane proteins such as type III secretion systems involved in host-pathogens interactions (44). Although the A. baumannii polysaccharide capsule is highly immunogenic and induces protective antibodies (34, 35, 45), targeting the capsule faces significant challenges. These include a wide range of described KL types (capsule serotypes), and limited knowledge of which of these, are the predominant cause of clinical cases of A. baumannii infections, which may also vary in different geographical locations (37, 46). For example, a total of 24 serotypes were identified among a large number of A. baumannii clinical isolates from Thailand (37). In contrast, a study from Vietnam found a total of 4 KL serotypes (KL2, KL49, KL58, and KL32) (46) but these together represented only 15% of the isolates in the Thai study. Similar contradictory data have been obtained using antibodies to different A. baumannii capsular polysaccharides with the K1 specific monoclonal antibody 13D6 recognizing 13% of the isolates tested (35), whereas C8, targeting the HUMC1 capsular polysaccharide, and anti-KL22 polyclonal sera recognized 60% of the clinical strains tested (34, 45). This variation in the proportion of strains recognized by anti-capsular antibody is likely to reflect differences between geographical sources in the predominant KL types, and perhaps for the anti-KL22 data, partial cross recognition of surface proteins found in the capsular material. These data suggest that a monoclonal therapy targeting A. baumannii KL types could be ineffective due to KL type diversity and would also have to be tailored to the KL types prevalent in each specific geographical region.
Developing monoclonal antibodies that recognize conserved sub-capsular protein antigens could overcome some of the challenges faced by targeting the capsular polysaccharide. Identifying which conserved proteins are potential vaccine candidates or targets of antibody therapy has been addressed using reverse vaccinology approaches for many pathogens (47). However, there are few data on acquired protective immunity to A. baumannii to aid in protein target antigen identification. To begin to address this, we have generated anti-A. baumannii sera by immunization against a single strain or sequential immunization with 3 strains, an approach that has been exploited to induce broadly reactive antibody responses in HIV (48), Influenza (49) and Neisseria meningitidis (50). Our results demonstrated that polyclonal mouse antisera generated following immunization with live A. baumannii contained a mixture of antibodies that target the polysaccharide capsule and protein targets. KL-type specific anti-capsular IgG responses predominated but using immunoblots and assays with an unencapsulated or heterologous KL type strains, we have identified additional anti-protein antigen responses. Previous data using sequential vaccination with outer membrane vesicles from three heterologous N. meningitidis strains improved antibody responses against genetically diverse strains and boosted the antibody titers to the highly conserved surface protein A (NspA) (50). Similarly, our data show that compared to immunization with a single strain, sequential immunization with multiple strains elicited antibodies that recognized more immunoreactive protein bands on immunoblot, boosted IgG recognition of both homologous and heterologous isolates, and resulted in enhanced phagocytosis of some heterologous isolates. Furthermore, prior immunization with three heterologous strains reduced target organ CFU in mice challenged with another A. baumannii strain. These data provide proof of principle that antibody to protein antigens can provide cross-protective immunity, although the ability of the capsule to inhibit antisera dependent phagocytosis remains a significant challenge.
We have used in vitro assays to help clarify mechanisms by which antisera might protect against A. baumannii. Table 3 summarizes and allows cross-comparisons of the results from these assays. There was a lack of concordance between the whole-cell ELISA and immunoblot results, which recognized proteins in all strains, and the surface IgG binding data which showed variable reactivity to the same strains. These results suggest that the flow cytometry assay was dominated by anti-capsular antibody responses, although it clearly identified an anti-protein antigen response in some antisera/strain conditions. This was expected as the flow cytometry assay measures IgG binding to the surface of the whole bacterium and our data confirms previous data that the A. baumannii capsule blocks access to subcapsular protein antigens (18), whereas whole-cell ELISAs measure antibody responses to all bacterial antigens. However, there was heterologous recognition of some strains, especially in sera obtained from mice vaccinated with three different strains indicating IgG binding to heterologous A. baumannii strains through anti-protein antibody responses. The functional consequences of the anti-protein responses were investigated using growth inhibition in antisera and by flow cytometry neutrophil uptake assay. Inhibition of A. baumannii growth when cultured in broth in the presence of antisera was only caused by KL type specific anticapsular antibody. Susceptibility to neutrophil phagocytosis varied markedly between the Thai A. baumannii strains, an observation that needs further investigation to identify the underlying causes and whether strains that are less susceptible to neutrophil phagocytosis are more likely to cause invasive clinical infections (as has been shown for Streptococcus pneumoniae) (51). Although the capsule impaired both phagocytosis and antibody access to underlying protein antigens, our data showed improved phagocytosis of selected heterologous strains when incubated in sera from mice vaccinated with A. baumannii strains especially the multi strain antisera. The effects on neutrophil phagocytosis mirrored to an extent the IgG binding data, although with some exceptions (Table 3). Several studies using neutrophil depleted mice have demonstrated the important role of neutrophils in mediating resistance to both pneumonia (52) and bacteremia (53) following A. baumannii infection, suggesting the in vitro data could translate into in vivo protection against infection. Indeed, in mice previously immunized with the three different A. baumannii strains used to make the multi strain antisera, there was partial reductions in target organ CFU when infected with the heterologous AB1 strain in a bacteremia model, indicating some protective efficacy of the anti-protein antigen response. Protection is likely to be mediated by antibody (43), but this will require experimental confirmation using passive transfer of immune sera and/or B cell and T cell depletion experiments, as cell mediated immunity could also contribute to the protection observed. However, this is beyond the scope of the present work. A. baumannii also commonly causes pneumonia and infections on indwelling devices, urinary tract infections or soft tissue infections and additional suitable in vivo models will be required to comprehensively evaluate the efficacy of an antibody therapy for the treatment of all A. baumannii infections (54, 55). These are likely to require a degree of immunosuppression to allow A. baumannii to establish an infection at the target site. In summary, the in vitro data suggest that flow cytometry measurement of IgG binding and of neutrophil phagocytosis provide data that can correlate to in vivo protection against bacteremia by a heterologous strain, and hence could be used for rapid assessment of the potential protective efficacy of a particular antisera or antibody to A. baumannii.
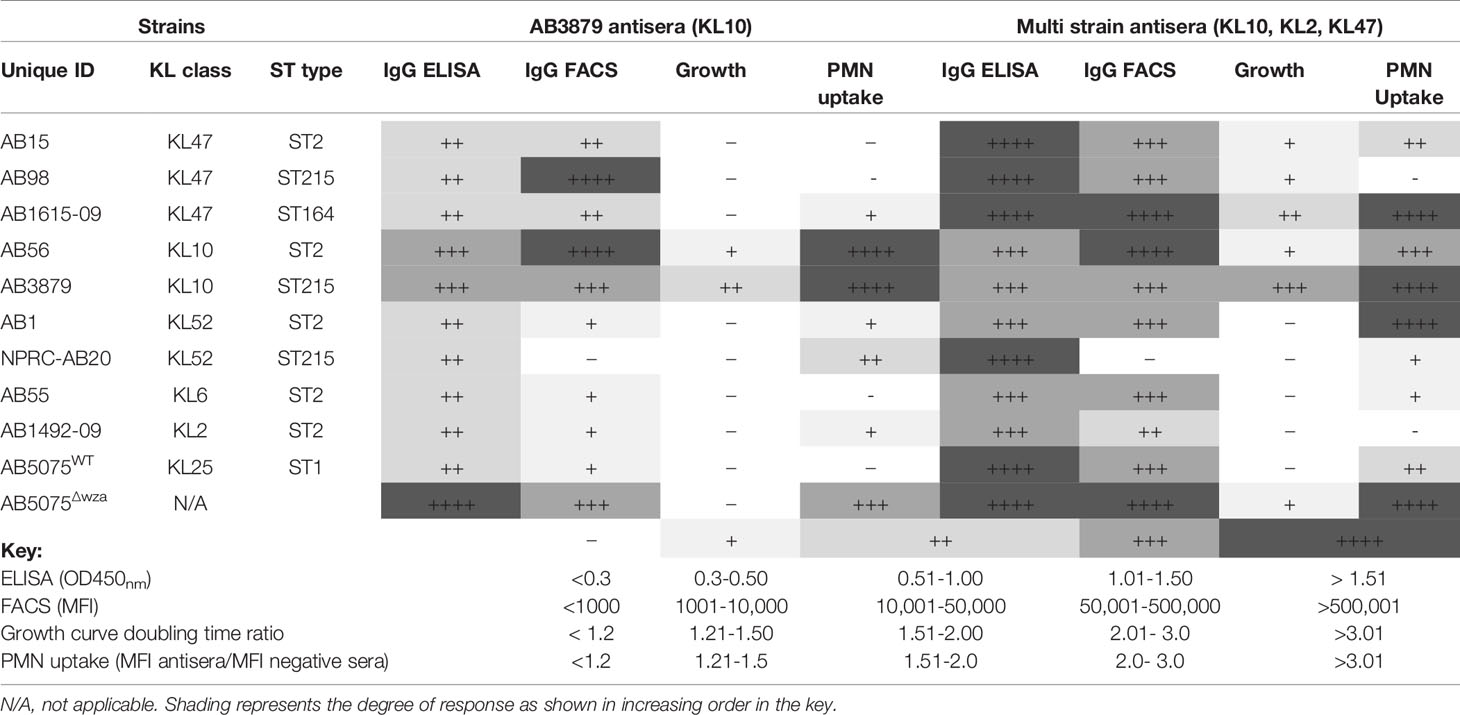
Table 3 Summary of the in vitro assays measuring AB3879 and multi strain antisera effect on A. baumannii strains.
Successive immunization of mice with different A. baumannii strains may have strengthened IgG recognition of heterologous strains compared to immunization with a single strain by amplifying recall responses to conserved protein antigens. A detailed characterization of the protein targets recognized by the protective multi strain antisera is underway and will help lead to the identification of potential monoclonal antibody targets for enhancing neutrophil clearance of an A. baumannii infection. Although we evaluated total IgG in this study, future work will also evaluate IgG isotypes when characterizing protective antibody responses to specific A. baumannii proteins, to help guide the development of monoclonal antibodies that elicit specific effector functions (56). However, a key challenge to monoclonal antibody development, is the inhibitory effect of A. baumannii capsule to both antibody binding and neutrophil phagocytosis, as well as the genetic variation between A. baumannii strains which makes targeting a single antigen unlikely to protect against all strains. An effective monoclonal antibody therapy may have to target several A. baumannii protein antigens to help overcome these challenges.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics Statement
The animal study was reviewed and approved by UCL Biological Services Ethical Committee and the UK Home Office (P64714548).
Author Contributions
JB led the design and setup of the project. PK and PT provided bacterial strains. GK, SW, and YC performed the experiments. RS performed the bioinformatics analysis. GK, SW, YC, and CK analyzed the data. GK and JB wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was undertaken at UCLH/UCL who received a proportion of funding from the Department of Health’s NIHR Biomedical Research Centre’s funding scheme and was also supported by an MRC DPFS MR/S004394/1 to RS, BW, GL and JB. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.705533/full#supplementary-material
Supplementary Figure 1 | Growth inhibitory activity of unencapsulated AB5075Δwza isolates by mouse antisera. 102 CFU of un-encapsulated AB5075Δwza strain was incubated in triplicate with either, AB5075WT antisera (black line, solid diamond), AB5075Δwza antisera (black line, open diamond) and sera from non-immunized mice (grey line, solid circle) (A) or AB3879 antisera (black line, solid square), multi strain antisera (black line, open square), and sera from non-immunized mice (grey line, solid circle) (B) and the OD600nm measured every 30 minutes over 24 hours.
Supplementary Figure 2 | Growth inhibitory activity on a panel of four A. baumannii isolates by mouse antisera. 102 CFU of a panel of A. baumannii isolates were incubated in triplicate with either AB3879 antisera (left panel) or multi strain antisera (right panel) and the OD600nm measured every 30 minutes for 24 hours. Sera from non-immunized mice was included as a control (open symbols). Solid symbols represent respective anti-A. baumannii antisera. Data from two independent experiments is shown. The five isolates are grouped by their capsule serotype and indicated at the top of each graph in brackets. Round, square and triangle symbols represent ST2, ST215, and ST164 MSLT types respectively.
Supplementary Figure 3 | Bacterial load in the spleen, liver and lungs from mice following lethal challenge with homologous AB3879 strain. CD1 mice were immunized three times with 106 CFU of the AB3879 or PBS followed by IP infection with 2.0 x 107 CFU/mouse of the homologous AB3879 strain in a bacteremia model of infection. The bacterial burden in the spleen, liver and lungs were determined 24 hpi. Dot plots represent individual values with lines indicating the mean and the error bars indicate standard deviations (SDs) (n=15/group) Mann-Whitney U-test was used for statistical analysis *p-value < 0.05, **p-value< 0.001, ***p-value < 0.0001, ns: p-value= not significant.
References
1. WHO. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotic. (2017).
2. Allegranzi B, Bagheri Nejad S, Combescure C, Graafmans W, Attar H, Donaldson L, et al. Burden of Endemic Health-Care-Associated Infection in Developing Countries: Systematic Review and Meta-Analysis. Lancet (2011) 377(9761):228–41. doi: 10.1016/S0140-6736(10)61458-4
3. ECDC. Carbapenem-Resistant Acinetobacter Baumannii in Healthcare Settings. Stockholm: European Centre for Disease Prevention and Control. (2016).
4. WHO. Report on the Burden of Endemic Health Care-Associated Infection Worldwide. A Systematic Review of the Literature. (2011).
5. Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, et al. Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet Infect Dis (2018) 18(3):318–27. doi: 10.1016/S1473-3099(17)30753-3
6. Isler B, Doi Y, Bonomo RA, Paterson DL. New Treatment Options Against Carbapenem-Resistant Acinetobacter Baumannii Infections. Antimicrob Agents Chemother (2019) 63(1):1110–18. doi: 10.1128/AAC.01110-18
7. Lim C, Takahashi E, Hongsuwan M, Wuthiekanun V, Thamlikitkul V, Hinjoy S, et al. Epidemiology and Burden of Multidrug-Resistant Bacterial Infection in a Developing Country. Elife (2016) 5:18082. doi: 10.7554/eLife.18082
8. McConnell MJ. Where are We With Monoclonal Antibodies for Multidrug-Resistant Infections? Drug Discov Today (2019) 24(5):1132–8. doi: 10.1016/j.drudis.2019.03.002
9. McConnell MJ, Pachon J. Active and Passive Immunization Against Acinetobacter Baumannii Using an Inactivated Whole Cell Vaccine. Vaccine (2010) 29(1):1–5. doi: 10.1016/j.vaccine.2010.10.052
10. KuoLee R, Harris G, Yan H, Xu HH, Conlan WJ, Patel GB, et al. Intranasal Immunization Protects Against Acinetobacter Baumannii-Associated Pneumonia in Mice. Vaccine (2015) 33(1):260–7. doi: 10.1016/j.vaccine.2014.02.083
11. Shu MH, MatRahim N, NorAmdan N, Pang SP, Hashim SH, Phoon WH, et al. An Inactivated Antibiotic-Exposed Whole-Cell Vaccine Enhances Bactericidal Activities Against Multidrug-Resistant Acinetobacter Baumannii. Sci Rep (2016) 6:22332. doi: 10.1038/srep22332
12. Garcia-Quintanilla M, Pulido MR, Pachon J, McConnell MJ. Immunization With Lipopolysaccharide-Deficient Whole Cells Provides Protective Immunity in an Experimental Mouse Model of Acinetobacter Baumannii Infection. PloS One (2014) 9(12):e114410. doi: 10.1371/journal.pone.0114410
13. Ainsworth S, Ketter PM, Yu JJ, Grimm RC, May HC, Cap AP, et al. Vaccination With a Live Attenuated Acinetobacter Baumannii Deficient in Thioredoxin Provides Protection Against Systemic Acinetobacter Infection. Vaccine (2017) 35(26):3387–94. doi: 10.1016/j.vaccine.2017.05.017
14. McConnell MJ, Dominguez-Herrera J, Smani Y, Lopez-Rojas R, Docobo-Perez F, Pachon J. Vaccination With Outer Membrane Complexes Elicits Rapid Protective Immunity to Multidrug-Resistant Acinetobacter Baumannii. Infect Immun (2011) 79(1):518–26. doi: 10.1128/IAI.00741-10
15. Pulido MR, Garcia-Quintanilla M, Pachon J, McConnell MJ. Immunization With Lipopolysaccharide-Free Outer Membrane Complexes Protects Against Acinetobacter Baumannii Infection. Vaccine (2018) 36(29):4153–6. doi: 10.1016/j.vaccine.2018.05.113
16. Huang W, Yao Y, Long Q, Yang X, Sun W, Liu C, et al. Immunization Against Multidrug-Resistant Acinetobacter Baumannii Effectively Protects Mice in Both Pneumonia and Sepsis Models. PloS One (2014) 9(6):e100727. doi: 10.1371/journal.pone.0100727
17. Luo G, Lin L, Ibrahim AS, Baquir B, Pantapalangkoor P, Bonomo RA, et al. Active and Passive Immunization Protects Against Lethal, Extreme Drug Resistant-Acinetobacter Baumannii Infection. PloS One (2012) 7(1):e29446. doi: 10.1371/journal.pone.0029446
18. Wang-Lin SX, Olson R, Beanan JM, MacDonald U, Balthasar JP, Russo TA. The Capsular Polysaccharide of Acinetobacter Baumannii Is an Obstacle for Therapeutic Passive Immunization Strategies. Infect Immun (2017) 85(12):591–17. doi: 10.1128/IAI.00591-17
19. Jahangiri A, Owlia P, Rasooli I, Salimian J, Derakhshanifar E, Naghipour Erami A, et al. Specific Egg Yolk Antibodies (IgY) Confer Protection Against Acinetobacter Baumannii in a Murine Pneumonia Model. J Appl Microbiol (2019) 126(2):624–32. doi: 10.1111/jam.14135
20. Huang W, Yao Y, Wang S, Xia Y, Yang X, Long Q, et al. Immunization With a 22-kDa Outer Membrane Protein Elicits Protective Immunity to Multidrug-Resistant Acinetobacter Baumannii. Sci Rep (2016) 6:20724. doi: 10.1038/srep20724
21. Huang W, Wang S, Yao Y, Xia Y, Yang X, Long Q, et al. Ompw is a Potential Target for Eliciting Protective Immunity Against Acinetobacter Baumannii Infections. Vaccine (2015) 33(36):4479–85. doi: 10.1016/j.vaccine.2015.07.031
22. Chiang MH, Sung WC, Lien SP, Chen YZ, Lo AF, Huang JH, et al. Identification of Novel Vaccine Candidates Against Acinetobacter Baumannii Using Reverse Vaccinology. Hum Vaccin Immunother (2015) 11(4):1065–73. doi: 10.1080/21645515.2015.1010910
23. Guo SJ, Ren S, Xie YE. Evaluation of the Protective Efficacy of a Fused OmpK/Omp22 Protein Vaccine Candidate Against Acinetobacter Baumannii Infection in Mice. BioMed Environ Sci (2018) 31(2):155–8. doi: 10.3967/bes2018.019
24. Yang AQ, Yang HY, Guo SJ, Xie YE. MF59 Adjuvant Enhances the Immunogenicity and Protective Immunity of the OmpK/Omp22 Fusion Protein From Acineterbacter Baumannii Through Intratracheal Inoculation in Mice. Scand J Immunol (2019) 90(1):e12769. doi: 10.1111/sji.12769
25. Bentancor LV, Routray A, Bozkurt-Guzel C, Camacho-Peiro A, Pier GB, Maira-Litran T. Evaluation of the Trimeric Autotransporter Ata as a Vaccine Candidate Against Acinetobacter Baumannii Infections. Infect Immun (2012) 80(10):3381–8. doi: 10.1128/IAI.06096-11
26. Singh R, Garg N, Shukla G, Capalash N, Sharma P. Immunoprotective Efficacy of Acinetobacter Baumannii Outer Membrane Protein, FilF, Predicted in Silico as a Potential Vaccine Candidate. Front Microbiol (2016) 7:158. doi: 10.3389/fmicb.2016.00158
27. Garg N, Singh R, Shukla G, Capalash N, Sharma P. Immunoprotective Potential of in Silico Predicted Acinetobacter Baumannii Outer Membrane Nuclease, NucAb. Int J Med Microbiol (2016) 306(1):1–9. doi: 10.1016/j.ijmm.2015.10.005
28. Singh R, Capalash N, Sharma P. Immunoprotective Potential of Bama, the Outer Membrane Protein Assembly Factor, Against MDR Acinetobacter Baumannii. Sci Rep (2017) 7(1):12411. doi: 10.1038/s41598-017-12789-3
29. Fattahian Y, Rasooli I, Mousavi Gargari SL, Rahbar MR, Darvish Alipour Astaneh S, Amani J. Protection Against Acinetobacter Baumannii Infection via its Functional Deprivation of Biofilm Associated Protein (Bap). Microb Pathog (2011) 51(6):402–6. doi: 10.1016/j.micpath.2011.09.004
30. Esmaeilkhani H, Rasooli I, Hashemi M, Nazarian S, Sefid F. Immunogenicity of Cork and Loop Domains of Recombinant Baumannii Acinetobactin Utilization Protein in Murine Model. Avicenna J Med Biotechnol (2019) 11(2):180–6.
31. Ramezanalizadeh F, Owlia P, Rasooli I. Type I Pili, CsuA/B and FimA Induce a Protective Immune Response Against Acinetobacter Baumannii. Vaccine (2020) 38(34):5436–46. doi: 10.1016/j.vaccine.2020.06.052
32. Qamsari MM, Rasooli I, Chaudhuri S, Astaneh SDA, Schryvers AB. Hybrid Antigens Expressing Surface Loops of Znud From Acinetobacter Baumannii Is Capable of Inducing Protection Against Infection. Front Immunol (2020) 11:158. doi: 10.3389/fimmu.2020.00158
33. Mahmoudi Z, Rasooli I, Jahangiri A, Darvish Alipour Astaneh S. Prevention of Nosocomial Acinetobacter Baumannii Infections With a Conserved Immunogenic Fimbrial Protein. APMIS (2020) 128(7):476–83. doi: 10.1111/apm.13061
34. Nielsen TB, Pantapalangkoor P, Luna BM, Bruhn KW, Yan J, Dekitani K, et al. Monoclonal Antibody Protects Against Acinetobacter Baumannii Infection by Enhancing Bacterial Clearance and Evading Sepsis. J Infect Dis (2017) 216(4):489–501. doi: 10.1093/infdis/jix315
35. Russo TA, Beanan JM, Olson R, MacDonald U, Cox AD, St Michael F, et al. The K1 Capsular Polysaccharide From Acinetobacter Baumannii Is a Potential Therapeutic Target via Passive Immunization. Infect Immun (2013) 81(3):915–22. doi: 10.1128/IAI.01184-12
36. Singh JK, Adams FG, Brown MH. Diversity and Function of Capsular Polysaccharide in Acinetobacter Baumannii. Front Microbiol (2018) 9:3301. doi: 10.3389/fmicb.2018.03301
37. Loraine J, Heinz E, Soontarach R, Blackwell GA, Stabler RA, Voravuthikunchai SP, et al. Genomic and Phenotypic Analyses of Acinetobacter Baumannii Isolates From Three Tertiary Care Hospitals in Thailand. Front Microbiol (2020) 11:548. doi: 10.3389/fmicb.2020.00548
38. Lindahl G. Subdominance in Antibody Responses: Implications for Vaccine Development. Microbiol Mol Biol Rev (2020) 85(1):78–80. doi: 10.1128/MMBR.00078-20
39. Mangas EL, Rubio A, Alvarez-Marin R, Labrador-Herrera G, Pachon J, Pachon-Ibanez ME, et al. Pangenome of Acinetobacter Baumannii Uncovers Two Groups of Genomes, One of Them With Genes Involved in CRISPR/Cas Defence Systems Associated With the Absence of Plasmids and Exclusive Genes for Biofilm Formation. Microb Genom (2019) 5(11):309. doi: 10.1099/mgen.0.000309
40. Bentancor LV, Camacho-Peiro A, Bozkurt-Guzel C, Pier GB, Maira-Litran T. Identification of Ata, a Multifunctional Trimeric Autotransporter of Acinetobacter Baumannii. J Bacteriol (2012) 194(15):3950–60. doi: 10.1128/JB.06769-11
41. Wyres KL, Cahill SM, Holt KE, Hall RM, Kenyon JJ. Identification of Acinetobacter Baumannii Loci for Capsular Polysaccharide (KL) and Lipooligosaccharide Outer Core (OCL) Synthesis in Genome Assemblies Using Curated Reference Databases Compatible With Kaptive. Microb Genom (2020) 6(3):339. doi: 10.1099/mgen.0.000339
42. Hyams C, Camberlein E, Cohen JM, Bax K, Brown JS. The Streptococcus Pneumoniae Capsule Inhibits Complement Activity and Neutrophil Phagocytosis by Multiple Mechanisms. Infect Immun (2010) 78(2):704–15. doi: 10.1128/IAI.00881-09
43. Wilson R, Cohen JM, Reglinski M, Jose RJ, Chan WY, Marshall H, et al. Naturally Acquired Human Immunity to Pneumococcus Is Dependent on Antibody to Protein Antigens. PloS Pathog (2017) 13(1):e1006137. doi: 10.1371/journal.ppat.1006137
44. Zurawski DV, McLendon MK. Monoclonal Antibodies as an Antibacterial Approach Against Bacterial Pathogens. Antibiotics (Basel) (2020) 9(4):155. doi: 10.3390/antibiotics9040155
45. Yang FL, Lou TC, Kuo SC, Wu WL, Chern J, Lee YT, et al. A Medically Relevant Capsular Polysaccharide in Acinetobacter Baumannii Is a Potential Vaccine Candidate. Vaccine (2017) 35(10):1440–7. doi: 10.1016/j.vaccine.2017.01.060
46. Schultz MB, Pham Thanh D, Tran Do Hoan N, Wick RR, Ingle DJ, Hawkey J, et al. Repeated Local Emergence of Carbapenem-Resistant Acinetobacter Baumannii in a Single Hospital Ward. Microb Genom (2016) 2(3):e000050. doi: 10.1099/mgen.0.000050
47. Delany I, Rappuoli R, Seib KL. Vaccines, Reverse Vaccinology, and Bacterial Pathogenesis. Cold Spring Harb Perspect Med (2013) 3(5):a012476. doi: 10.1101/cshperspect.a012476
48. Krammer F. Strategies to Induce Broadly Protective Antibody Responses to Viral Glycoproteins. Expert Rev Vaccines (2017) 16(5):503–13. doi: 10.1080/14760584.2017.1299576
49. Zhou H, Huang Y, Yuan S, Li Y, Wu S, Xu J, et al. Sequential Immunization With Consensus Influenza Hemagglutinins Raises Cross-Reactive Neutralizing Antibodies Against Various Heterologous HA Strains. Vaccine (2017) 35(2):305–12. doi: 10.1016/j.vaccine.2016.11.051
50. Moe GR, Zuno-Mitchell P, Hammond SN, Granoff DM. Sequential Immunization With Vesicles Prepared From Heterologous Neisseria Meningitidis Strains Elicits Broadly Protective Serum Antibodies to Group B Strains. Infect Immun (2002) 70(11):6021–31. doi: 10.1128/IAI.70.11.6021-6031.2002
51. Hyams C, Trzcinski K, Camberlein E, Weinberger DM, Chimalapati S, Noursadeghi M, et al. Streptococcus Pneumoniae Capsular Serotype Invasiveness Correlates With the Degree of Factor H Binding and Opsonization With C3b/iC3b. Infect Immun (2013) 81(1):354–63. doi: 10.1128/IAI.00862-12
52. van Faassen H, KuoLee R, Harris G, Zhao X, Conlan JW, Chen W. Neutrophils Play an Important Role in Host Resistance to Respiratory Infection With Acinetobacter Baumannii in Mice. Infect Immun (2007) 75(12):5597–608. doi: 10.1128/IAI.00762-07
53. Breslow JM, Meissler JJ Jr., Hartzell RR, Spence PB, Truant A, Gaughan J, et al. Innate Immune Responses to Systemic Acinetobacter Baumannii Infection in Mice: Neutrophils, But Not Interleukin-17, Mediate Host Resistance. Infect Immun (2011) 79(8):3317–27. doi: 10.1128/IAI.00069-11
54. McConnell MJ, Actis L, Pachon J. Acinetobacter Baumannii: Human Infections, Factors Contributing to Pathogenesis and Animal Models. FEMS Microbiol Rev (2013) 37(2):130–55. doi: 10.1111/j.1574-6976.2012.00344.x
55. Wong D, Nielsen TB, Bonomo RA, Pantapalangkoor P, Luna B, Spellberg B. Clinical and Pathophysiological Overview of Acinetobacter Infections: A Century of Challenges. Clin Microbiol Rev (2017) 30(1):409–47. doi: 10.1128/CMR.00058-16
Keywords: A. baumannii, antibodies, growth inhibition, neutrophils, heterologous protection
Citation: Kamuyu G, Suen Cheng Y, Willcocks S, Kewcharoenwong C, Kiratisin P, Taylor PW, Wren BW, Lertmemongkolchai G, Stabler RA and Brown J (2021) Sequential Vaccination With Heterologous Acinetobacter baumannii Strains Induces Broadly Reactive Antibody Responses. Front. Immunol. 12:705533. doi: 10.3389/fimmu.2021.705533
Received: 05 May 2021; Accepted: 08 July 2021;
Published: 30 July 2021.
Edited by:
Mario Alberto Flores-Valdez, CONACYT Centro de Investigación y Asistencia en Tecnología y Diseño del Estado de Jalisco (CIATEJ), MexicoReviewed by:
Jonathan Cohen, Evelina London Children’s Hospital, United KingdomIraj Rasooli, Shahed University, Iran
Copyright © 2021 Kamuyu, Suen Cheng, Willcocks, Kewcharoenwong, Kiratisin, Taylor, Wren, Lertmemongkolchai, Stabler and Brown. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jeremy Brown, amVyZW15LmJyb3duQHVjbC5hYy51aw==
 Gathoni Kamuyu
Gathoni Kamuyu Yat Suen Cheng
Yat Suen Cheng Sam Willcocks2
Sam Willcocks2 Pattarachai Kiratisin
Pattarachai Kiratisin Peter W. Taylor
Peter W. Taylor Brendan W. Wren
Brendan W. Wren Ganjana Lertmemongkolchai
Ganjana Lertmemongkolchai Richard A. Stabler
Richard A. Stabler Jeremy Brown
Jeremy Brown