
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 20 August 2021
Sec. Cancer Immunity and Immunotherapy
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.704942
This article is part of the Research Topic The Intricate Innate Immune-Cancer Cell Relationship in the Context of Tumor Angiogenesis, Immunity and Microbiota: the Angiogenic Switch in the Tumor Microenvironment as a Key Target for Immunotherapies View all 12 articles
 Michela Roberto1,2
Michela Roberto1,2 Catia Carconi3
Catia Carconi3 Micaela Cerreti3
Micaela Cerreti3 Francesca Matilde Schipilliti1
Francesca Matilde Schipilliti1 Andrea Botticelli1,2*
Andrea Botticelli1,2* Federica Mazzuca1
Federica Mazzuca1 Paolo Marchetti1,2
Paolo Marchetti1,2The human microbiota and its functional interaction with the human body were recently returned to the spotlight of the scientific community. In light of the extensive implementation of newer and increasingly precise genome sequencing technologies, bioinformatics, and culturomic, we now have an extraordinary ability to study the microorganisms that live within the human body. Most of the recent studies only focused on the interaction between the intestinal microbiota and one other factor. Considering the complexity of gut microbiota and its role in the pathogenesis of numerous cancers, our aim was to investigate how microbiota is affected by intestinal microenvironment and how microenvironment alterations may influence the response to immune checkpoint inhibitors (ICIs). In this context, we show how diet is emerging as a fundamental determinant of microbiota’s community structure and function. Particularly, we describe the role of certain dietary factors, as well as the use of probiotics, prebiotics, postbiotics, and antibiotics in modifying the human microbiota. The modulation of gut microbiota may be a secret weapon to potentiate the efficacy of immunotherapies. In addition, this review sheds new light on the possibility of administering fecal microbiota transplantation to modulate the gut microbiota in cancer treatment. These concepts and how these findings can be translated into the therapeutic response to cancer immunotherapies will be presented.
Over the past few decades, significant progress has been achieved in cancer treatment, with immunotherapy becoming a research hotspot in recent years (1). The last years have seen unprecedented clinical responses and rapid drug development, accumulating reports of advanced cancer patients defying the odds and achieving complete remissions with immunotherapy treatments (2).
Immunotherapy is a powerful strategy to treat cancer by harnessing the body’s immune system to generate or augment an immune response against it (3). This is accomplished by either training resident immune cells to recognize and eliminate cells bearing tumor specific antigens, providing external stimuli to enhance immune mediated tumor cell lysis or abrogating signals directed by tumor cells to dampen immune responsiveness (4). Both cellular and molecular components of the tumor microenvironment can affect the efficacy of immunotherapy (5).
The tumor microenvironment has been recognized as a key factor in tumor development and progression (6). Many of its components influence cancer cell malignant behavior, within its three-dimensional structure (1, 2). Non-malignant cells include immune cells, cells of the vasculature and lymphatic system, cancer-associated fibroblasts, pericytes, and adipocytes (7). The communication between cell types is driven by an extremely complex network of cytokines, chemokines, growth factors, other inflammatory mediators, and matrix remodeling enzymes (8).
The intestinal microbiota is the collection of all microorganisms (eukaryotes, bacteria, virus) living in human gastrointestinal tract. Microbiome may be very different between individuals, and it is constantly influenced by age, nutrition, antibiotic use, smoking, alcohol. There is a continuous interaction and interplay between microbiome and the immune system, and the microbiota seems to play a role in the pathogenesis of various inflammatory diseases such as NASH, inflammatory bowel disease and obesity (9).
The human microbiome has recently been described as a component of various tumor microenvironments, due to its ability to impair tumor cell metabolism by maintaining a healthy mucosal barrier, to induce inflammation, and to produce genotoxins and different bacterial metabolites (10). It has been estimated that the total number of bacteria in the 70 kg average human male is 3.8·1013 and that 10% of metabolites found in mammalian blood are derived from the gut microbiota (11, 12). Indeed, humans and their microbiome are considered to form a composite organism, a so-called holobiont, that defines humans together with their connected microbial network, instead of merely autonomous eukaryotic organisms (13, 14). Furthermore, a clear interplay between the local microbiome, the intestinal epithelium, and resident immune cells has recently begun to emerge, where all participants actively foster gastrointestinal homeostasis. In this system, bacterially derived metabolites serve as important signals that continuously contribute to the proper function of the epithelial barrier and immune cells (14).
Over the last decade, researchers have found a consistent connection between a dysfunctional gut microbiota (dysbiosis) and various cancers, such as cancers of the urinary tract, cervix, skin, airways, colon, breast, and lymphomas (10, 15). Considering that the primary characteristics of microbiota dysbiosis are alterations of bacterial species and the increase of pathogenic bacteria (16), studying the microbial communities in the tumor microenvironment may shed light on the role of host-bacteria interactions in cancer.
The relation between cancer and microbiota is also influenced by other factors. Out of the multiple host-endogenous and host-exogenous factors involved in the modulation of the composition of gut microbiota, such as diseases, drugs, and smoke (17), diet emerges as a pivotal determinant of its community structure and function (18). Considering that the populations of dominant species within the human colonic microbiota can potentially be modified by dietary intake to influence health (19), the responses of the gut microbiota to various factors are considered to be a valuable tool to exploit in order to develop new strategies to promote human health.
Therefore, it is important to identify gut resident bacteria. Metagenomics and culturomics are the tools used to study human microbiota, to understand and detect gut microbes, to identify their specific role in the microenvironment and correlate all data with clinical specifical situations (20, 21).
Considering the increasing interest in the microbiota composition of oncological patients, the aim of this review is to analyze the role of microbiota in cancer promotion, its effects on the immune system and its emerging role as a response modulator to immunotherapy-based cancer treatments. In this perspective, this review focuses on understanding how the diet and the use of probiotics, prebiotics, postbiotics and antibiotics might modify the composition of the gut microbiota and, consequently, the therapeutic response to cancer immunotherapies (Figure 1).
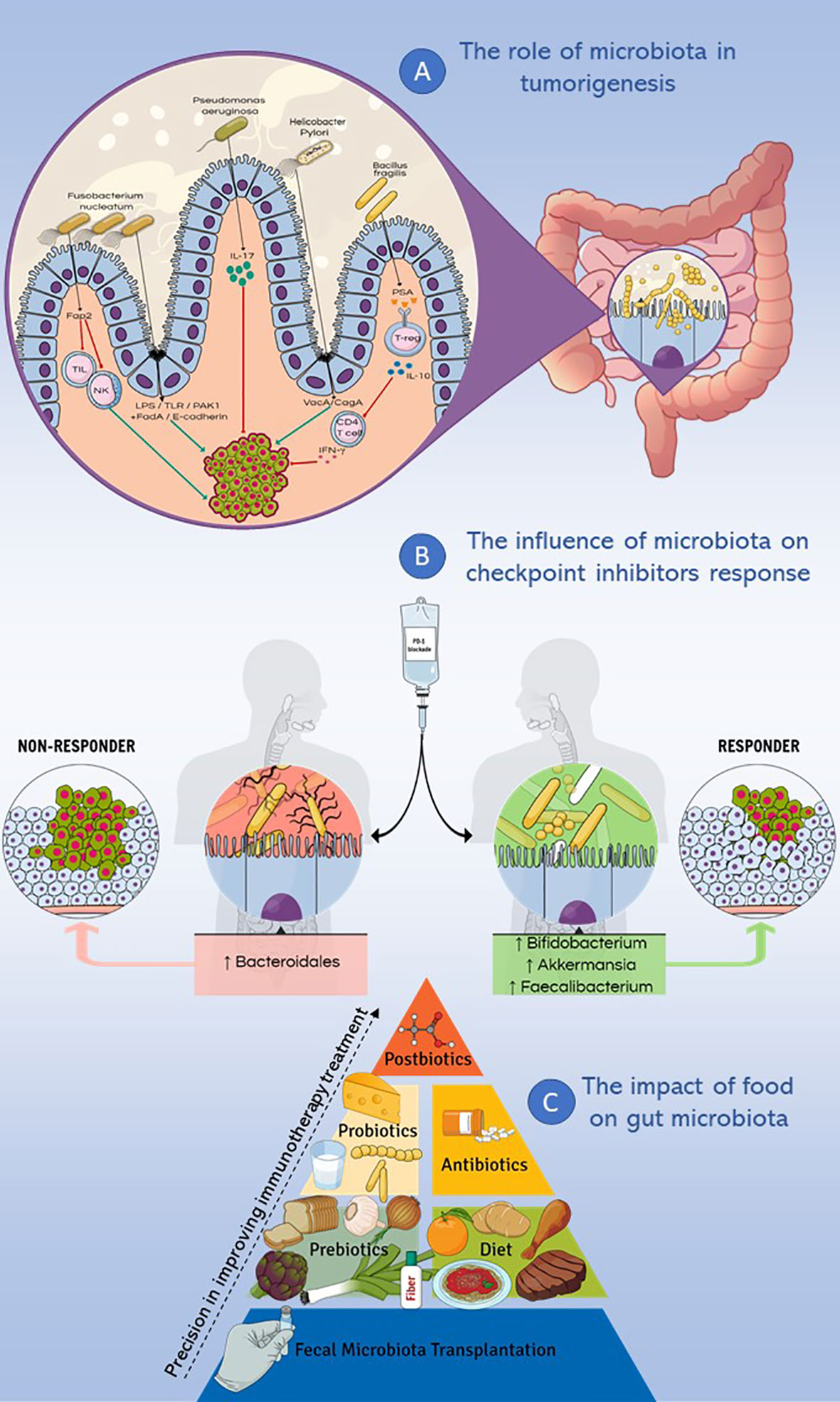
Figure 1 Microbiota and immunotherapy resistance. This figure summarizes the main topics discussed in the review. (A) Different genera such as Fusobacterium nucleatum, Pseudomonas aeruginosa, Helicobacter Pylori and Bacillus fragilis were studied for their implication in cancer pathogenesis, causing inflammatory and/or immune response, DNA damage and modulating cell proliferation. (B) Microbiota influences the response to checkpoint inhibitors therapy: the enrichment of fecal microbiota with Akkermansia muciniphila, Faecalibacterium spp and Bifidobacterium spp correlates with a positive response to PD-1 immune-checkpoint blockade, while a higher abundance of Bacteroidales correlates with a deficient response to the same treatment. (C) Different dietary nutrients modify the response to immunotherapy, ranging from fecal microbiota transplantation to the use of postbiotics, with increasingly precise effects on the treatment response.
Given the variability of gut microbiota between individuals due to external influences such as diet (22), host genetic background and other environmental factors, many studies employed both tumor and normal tissue samples taken from the same individual, in order to provide a more accurate view of the tumor-associated shifts in the microbiome (22, 23). The general conclusion is that tumor microenvironments harbor microbiomes distinct from those of normal tissue microenvironments. Various analyses consistently showed variation in the bacterial phyla abundance when comparing the matched normal and tumor tissues, demonstrating that there is indeed a cancer-associated signature in the tumor microbiome (24–26).
Gut microbiota can be divided into 3 clusters according to the effects of the microbes on the human body: beneficial, neutral, and pathogenic (27). The first group comprehends Bifidobacterium and Lactobacillus, which can protect the intestinal tract, produce beneficial metabolites, and detoxify the human gut. Neutral microbes, such as Enterococcus, have dual characteristics, being beneficial to human health in normal growth conditions and being able to cause different degrees of diseases when exceeding a certain standard growth or transferred to other parts of the body (28). Pathogenic microbes, such as Salmonella and Helicobacter pylori, secrete toxins and thus might cause disease (29).
The gut microbiota has differential effects on tumorigenesis, in fact bacteria may be tumour suppressive for cancer, especially at distal sites by releasing metabolites and immune modulators such as histone deacetylase (HDACi), hypoxia induced factor (HIF), interkeukin-10 (IL-10) that enrich gut barrier function and have an antioxidant effect (30). Moreover, it is important to consider the role of TME and the gut mucosal barrier: the increased permeability of gut mucosal barrier is correlated with inflammation and development of cancer. Literature data describes a link between integrity of gut mucosal barrier and differential faecal bacteria (31).
Lacking bacterial diversity in the intestine is the key feature for many intestinal and extraintestinal disorders. Considering the evident differences in the nutrient composition of the tumor microenvironment and the metabolic activity of microbiota, there is an unquestionable metabolic interaction between the tumor and its own microbiota (32). It is suggested that tumorigenesis is promoted by a combination of intestinal microbiota alterations (e.g., increased abundance of Escherichia coli and Fusobacterium nucleatum), rather than a difference in the abundance of a specific strain (33).
New evidence points to the association between the gut microbiota and the development and progression of gastrointestinal cancers such as colorectal cancer and hepatocellular carcinoma (34), as well as cancers of the respiratory system, where microbiota’s dysbiosis in heavy smokers, together with the epithelial integrity loss, could initiate inflammation in lung cancer (35). Moreover, the relationship between human microbiota and other types of cancers, such as breast cancer, is starting to emerge (36).
As an example of the role of microbiota in cancerogenesis, here it is described the hypothesis that emerged to explain the contribution of bacteria to colorectal cancer (CRC) carcinogenesis. On one hand, the presence of a dysbiotic microbial community with pro-carcinogenic features can remodel the microbiome towards pro-inflammatory responses and epithelial cell transformation, thus leading to cancer. On the other hand, the “driver-passenger” theory states that the so-called “bacteria drivers” could initiate CRC by inducing epithelial DNA damage leading to tumors with indigenous ability to promote the proliferation of “passenger bacteria”, by means of a growth advantage in the tumoral microenvironment (37, 38). These bacteria hardly colonize a healthy colon and cannot breach the intact colon wall, but they can easily invade a broken colon wall in the context of adenoma or carcinoma (37, 39). A highly diverse gut microbiota might be a key feature of a healthy gut, a balance between driver and passenger bacteria might create a species-rich ecosystem which is able to deal with environmental stresses that promote CRC (40).
Different studies aimed to identify potential “driver” bacteria. Bradyrhizobium japonicum was found to be increased in lung cancer patients with early-stage tumors (stages I and II) when compared to patients with advanced-stage tumors (III and IV) (41). Moreover, in patients with breast cancer, the analysis of 16S rRNA showed a higher relative abundance of Bacillus spp. compared with healthy samples, and Methanobacteriaceae was richer in malignant disease compared to benign disease (42, 43). The abundances of driver and passenger bacteria may serve as a primary indicator of cancer initiation risk and development.
The human gut microbiota is dominated by 3 primary phyla: Firmicutes (30%-50%), Bacteroidetes (20%-40%) and Actinobacteria (1% - 10%). Some strict anaerobes, as well as Bacteroides, Eubacterium, Bifidobacterium, Fusobacterium, Peptostreptococcus and Atopobium (44), constitute a major portion of the gut microbiota, while facultative anaerobes, such as Lactobacilli, Enterococci, Streptococci and Enterobacteriaceae, represent a minor proportion (45).
During their phylogenetic evolution, bacteria progressively acquired virulence factors that conferred pathogenicity. In this regard, bacteria developed the ability to penetrate the gut mucosal barrier, as well as the ability to adhere to and invade intestinal epithelial cells, using flagella, pili, and adhesins (46–48). These virulence factors are considered to be one of the elements that determine disease-promoting and pro-carcinogenic effects of pathogens (49).
Intestinal bacteria contribute to carcinogenesis in different ways, causing inflammatory and/or immune response, DNA damage and modulating cell proliferation. Different genera were studied to prove their implication in cancer pathogenesis, especially in CRC. A recent study showed how colorectal cancer samples were dominated by Firmicutes, Bacteroidetes, and Proteobacteria (22). Tumors showed an enrichment of Proteobacteria and a depletion of Firmicutes and Bacteroidetes, underlining the evident and significant changes in these phyla between the normal and cancer states. There was also an increase in the phylum Fusobacteria in the tumor-associated microbiome (22). The important findings were that two of the genera that have been found to be enriched in the tumor microbiome, Providencia and Fusobacteria, are already known to be pathogenic. Moreover, Fusobacteria has been implicated in CRC by many other studies (50, 51). The presence of species belonging to the genera Providencia and Fusobacterium in the tumor microenvironment may suggest that they could have a role in oncogenesis or tumor progression, or that the tumor’s niche favors them.
Several studies suggest that Fusobacteria is likely a cancer driver and its carcinogenic mechanism has been unveiled (52, 53). The discovery of Providencia in the tumor microbiome is interesting as it produces an immunogenic lipopolysaccharide that participates in epithelial barrier dysfunction and endothelial apoptosis (54). These factors generally lead to gastroenteritis, but its association with the tumor environment may suggest that it should be studied as a cancer-promoting pathogen. Interestingly, Fusobacteria and Providencia share many important phenotypic characteristics such as the ability to damage colorectal tissue and to encode several virulence genes that are responsible for lipopolysaccharide biosynthesis, which are also significantly increased in the tumor microenvironment (22).
In the same way, certain CRC-associated Escherichia coli strains acquired virulence factors, such as the afa and eae adhesins, which conferred the ability to adhere to and invade the intestinal epithelium (55, 56). E. Coli is indeed a common gut commensal bacterium, but it has been shown to be able to colonize the colonic mucosa; it increases mucosal permeability through the activation of Wnt mitogenic signaling, it damages the DNA and interferes with the DNA repair process, hence inducing CRC development (57).
Other common pathogenic bacteria have been studied for their association with carcinogenesis. A study showed that CRC patients and precancerous lesions had a higher expression level of Salmonella flagella antibodies than healthy controls, with diet differences being one of the mediating factors, suggesting a potential link between Salmonella and CRC (58). Furthermore, Salmonella can secrete the effector protein AvrA to promote acetylation and ubiquitination of target proteins. AvrA inhibits β-catenin degradation, maintains β-catenin stability, and promotes intestinal epithelial cell proliferation, thereby facilitating tumorigenesis, increasing tumor diversity, and driving tumor progression (59).
It has recently been shown that gut microbiota influences the host immune response to different cancer therapies, such as radiotherapy, chemotherapy, stem cell transplant and immunotherapy, by upsetting drug metabolism, the anti-tumor effects and the toxicity of the medications currently used (60).
ICIs immunotherapy is based on using natural and artificial components in order to promote or induce the natural immune system to neutralize cancer cells (61, 62). Since the introduction of ICIs, there has been a change in the treatment of advanced cancer by introducing immunotherapy as a recognized first and second-line therapies. ICIs are monoclonal antibodies which target inhibitory receptors on the surface of T cells. Checkpoint blockade therapies release the inhibitory mechanism that control T-cell mediated immunity. The immune checkpoints are inhibitory pathways of immune cell that are important to regulate immune response and maintaining self-tolerance.
Once T cells are activated, they strengthen the immune system and boost an immune-mediated eradication of cancer cells (63). Immune checkpoints expressed on cytotoxic and regulatory T cells include programmed cell death protein-1 (PD-1 or CD279) and cytotoxic T lymphocyte associated antigen 4 (CTLA-4 or CD152) (64, 65) that interact with ligands cluster differential 80 (CD80), cluster differential 86 (CD86) and programmed death ligand-1 (PDL-1) on antigen presenting cells (APCs). ICIs prevent receptors and ligands from binding to each other, interrupting signals. In line with these considerations, the host immune system provides a powerful therapeutic target, thanks to its ability to precisely focus on tumor cells (66).
Despite the abovementioned advantages of immunotherapy, patients respond to ICIs heterogeneously and with a short-term efficacy (67). The reason why some tumors lack response is still unclear, although it probably depends on antigenicity and adjuvanticity defects, which are key factors in shaping the immunogenicity of tumor cells (68). Despite the fact that several biomarkers (PD-L1 expression, tumor-infiltrating lymphocytes, mutational burden, immune gene signatures and microsatellite instability) have been proposed, their sensibility and sensitivity are limited (69). Given that tumors with a high number of somatic mutations are more responsive to immunotherapies than the ones with a lower rate, the level of somatic mutations seems to be a crucial factor (70).
Preliminary data indicate that enteric microbiota may affect the efficiency of immunotherapy (71). It is well known that gut microbiota can modulate the peripheral immune system and that its diversity plays a crucial role in the maturation, development and function of both the innate and the adaptive immune systems (66, 72). Given the crosstalk between gut microbiota and immunity and considering that T cell infiltration of solid tumors, such as metastatic melanoma, is associated with favorable outcomes (73), microbiota could be considered as an important modulator of response to immunotherapy.
Along these lines, remarkable studies have demonstrated how the gut microbiota and its composition play a major role in the response to immunotherapy with ICIs, targeting the PD-1 and the CTLA-4 (74, 75).
With regards to the influence of gut microbiota on therapies targeting the PD-1/PD-L1 axis, Sivan et al. have provided important insights from murine models in 2015 (74). Indeed, they have demonstrated how genetically similar mice with different microbiota composition exhibited significant immune-mediated differences in melanoma growth rate. The intratumoral CD8+ T cell accumulation was found to be significantly lower in mice with a more aggressive tumor growth and a remarkable reduction in the difference of antitumor immunity was shown after cohousing, suggesting an environmental influence. Moreover, fecal suspensions derived from mice with less aggressive tumor growth were able to delay tumor growth and to enhance the induction and infiltration of tumor-specific CD8+ T cells in the other group of mice, thus supporting a microbe-derived effect. Microbiota composition could also influence the response to immunotherapy with antibodies targeting PD-L1. These abovementioned data support the idea that microbiota might be a source of intersubjective heterogeneity regarding spontaneous antitumor immunity and therapeutic effects of antibodies targeting the PD-1/PD-L1 axis.
A related research revealed how the antitumor effects of CTLA-4 blockade depend on distinct Bacteroides species, with a lack of response to CTLA-4 blockade in antibiotic-treated or germ-free mice (75). The analysis of microbiota composition showed Bifidobacterium being positively associated with antitumor T cell responses. Furthermore, Bifidobacterium-treated mice showed better tumor surveillance compared to their non-Bifidobacterium treated counterparts, together with a high increase of tumor-specific T cells in the periphery and a significant increase of antigen-specific CD8+ T cells within the tumor (74).
On the other hand, the treatment itself may affect microbiota composition. Indeed, in patients with metastatic melanoma, Ipilimumab can alter the abundance of gut Bacteroides spp. with an immunogenic power, especially B. thetaiotaomicron and B. fragilis, which, in turn, can affect its therapeutic effect. Feces rich in B. fragilis (except B. distasonis or B. uniformis) were negatively associated with tumor dimension after the therapy. Hence, the efficacy of CTLA-4 blockade is influenced by the microbiota composition (75). The gut microbiome and antibiotic therapies appear to impact the response to adoptive cell therapies in murine models (76, 77) and preliminary studies on haematological and solid tumor case series seem to align with this data (78).
Recent studies on humans have reported an unexpected role of specific members of the gut microbiota as predictors of response to immunotherapy in a distinctive series of epithelial tumors (NSCLC, renal cell carcinoma, and urothelial carcinoma) and melanoma patients (79–81). Routy et al. recently demonstrated how patients with epithelial tumors that responded to PD-1 blockade had differential composition of gut bacteria, being enriched in Akkermansia and Alistipes. Moreover, by performing a fecal microbial transplantation in mice it was demonstrated how there were enhanced responses related to the responders’ fecal material. In addition, the efficacy of anti-PD-1 in GF mice receiving non-responders’ transplantation could be restored by the administration of Akkermansia muciniphila alone or in combination with Enterococcus hirae (79). Regarding metastatic melanoma, a study by Gopalakrishan et al. revealed that responders to anti-PD-1 therapy not only had a significantly higher diversity of bacteria in their gut microbiota, but also had a higher relative abundance of Clostridiales, Ruminococcaceae, and Faecalibacterium spp. On the other hand, non-responders had significantly lower diversity of gut bacteria and a higher abundance of Bacteroidales. The composition of microbiota was related to the expression of cytotoxic T cell markers and the mechanism of antigen processing and presentation, which was increased in the first group of patients (80). In addition, another study has shown how the transplantation of stool to germ-free mice could improve the efficacy of anti-PD-L1 immunotherapy in mice that received responder-stool by increasing the density of CD8+ T-cells and reducing FoxP3+ CD4+ Tregs in the tumor microenvironment.
Given the recent findings of the microbiota being a significant modulator of response to ICIs, important insights are provided into the possibility of intervening on the composition of the intestinal microbiota to affect the ability to modulate antitumor immune responses. The crosstalk between microbiota and the immune system may allow a microbiota-based selection of patients that might benefit from a specific immunotherapy treatment, boosting their anticancer response. The prospect of being able to manipulate gut microbiota in order to modify the response to checkpoint inhibitors, serves as a continuous stimulus future research.
Besides regulating the response to checkpoint blockade therapies, gut microbiota can also take part in resistance to this kind of treatment, crowding out its therapeutic benefits. Xiaochang Xue et al. indicated that commensal bacteria act in a direct way on our immune cells, down-regulating the intestinal miR-10a expression. As they have shown, E. coli and flagellated A4 commensal bacteria manage to recognize and engage TLR1/2, TLR4, TLR5, TLR9 and NOD2 on dendritic cells (DCs), resulting in a down-regulation of miR-10a via the MyD88-dependent pathway (82). Considering that miR-10a inhibits DC production of IL-12/IL-23p40, miR-10a itself acts as a negative regulator of both innate and adaptive immune responses to microbiota (82). It is known that IL-12/IL-23p40 gene has a key role in the stimulation of Th1 cell-mediated immune responses and cytotoxic activity of CD8+ T and natural killer cells (83). Thus, their absence threatens the effectiveness of the anticancer immune response.
Furthermore, both Gram-positive and Gram-negative bacteria are able to produce extracellular vesicles (EVs), which carry carbohydrates, signaling molecules, metabolites, proteins, DNA, RNA, in order to create a cell-to-cell communication through the transport of their content (84). Bacterial EVs contain short RNAs (85) (sRNAs) and miRNA-sized sRNAs (msRNAs) (86), which have regulatory functions as well as miRNA in eukaryotic cells. Different studies (87, 88) confirm that the exchange of information between bacterial EVs and host cells through the modulation of the gene expression, might be involved in inducing resistance to chemotherapy and immunotherapy. On the other hand, even human intestinal epithelial cells release miRNAs encapsulated in EVs, which, as it has been demonstrated by S. Liu et al., may promote the growth F. nucleatus and E. coli, in order to maintain a physiological balance of our intestinal microbiota (89).
In conclusion, it is clear that there is a mutual influence between bacteria and human host cells, thus, it is conceivable that further studies could provide additional findings to better understand EV-mediated inter-cell communication and, perhaps, a new opportunity to reduce the resistance to cancer therapies by using specific probiotics, antibiotics or focusing on the composition of microbiome to personalize therapies.
The contribution of diet to the modulation of microbiota and its crucial role in orchestrating the host–microbiota crosstalk is evident since the beginning of a human life when there is a microbiota-dependent relationship between milk oligosaccharides and growth promotion (90). This crosstalk between diet and microbiota continues and becomes more complex with the increased bacterial richness associated with the introduction of solid foods (91), and keeps affecting our lives until the end, with a decreased richness in the microbiota of frail ageing populations living in long-stay care, probably due to reduced food diversity (92).
A study demonstrated how the gut microbiome can respond to dietary interventions in humans in a rapid, diet-specific manner and how a diet composed entirely of animal products is able to trigger enrichment in bile-tolerant bacteria (Alistipes, Bilophila and Bacteroides) and depletion in Firmicutes that metabolize plant polysaccharides (Roseburia, Eubacterium rectale and Ruminococcus bromii) (93). Some more metagenomic and metabolomic analyses confirmed this trade-off between protein fermentation and degradation in protein-rich, animal-based diets, as opposed to carbohydrate fermentation and amino acid biosynthesis in plant-based diets (94). For example, the elimination of animal fats in the human diet was associated with a decrease in harmful Bacteroidales bacteria (95).
One of the dietary components that has shown to have a significant impact on the microbiota’s composition is fiber. Indeed, taking into consideration the different diet styles, it was shown how administering to mice a typical Western-style diet, that contains a relatively lower amount of fiber, could reduce the amount of Bifidobacterium and the gut microbiota diversity, leading to increased penetrability, and a reduced production rate of the inner mucus layer (96). Another study in healthy human volunteers (97), showed how the reduction in the amount of fiber intake led to a statistically significant reduction in the abundance of Faecalibacterium prausnitzii and Roseburia spp, which were positively correlated with the proportion of butyrate during both baseline normal diets. Moreover, a chronic lack of dietary fiber intake could lead to a reduced diversity in the gut microbiota (98). Preliminary data suggest that diet fiber intake could even impact the likelihood of response to anti-PD-1 treatment (99), providing interesting insights into the possible role of diet in the response to cancer therapies.
Many other dietary nutrients were studied for their roles in the modulation of gut microbiota, for example major groups of polyphenols assayed in both in vitro and preclinical studies have shown their ability to modulate the gut microbiota to a beneficial pool characterized by the abundance of Bifidobacterium, Lactobacillus, Akkermansia, and Faecalibacterium sp (100). Resveratrol is a naturally occurring polyphenol produced by some dietary botanicals, including red grapes (101), as a self-defence agent. Together with its cardio-protective and neuro-protective properties, it also serves as an antitumoral agent (102) which has shown the ability to induce antioxidant enzymes that attenuate oxidative stress (103).
Given the importance of these bacteria and their implications in cancer therapy, it is possible that diet could improve the patients’ outcomes through the modulation of their microbiome. Furthermore, considering that diet interacts with the human ‘holobiont’ in a person-specific way, being able to obtain multiple parameters from the host and its resident microbiota could assist in devising precision dietary interventions (104). This would provide a safe and simple opportunity for assessing the implication of microbiota and downstream immune manipulation in cancer patient populations.
Ongoing trials are currently exploring the impact that diet could have on the gut microbiota of oncologic patients. A randomized clinical trial that started in 2013 (NCT02079662) is currently studying how an integrative oncological program, that aims to make changes in the patients’ lifestyles and behaviors, including dietary recommendations and meal delivery, could influence long-term treatment results in patients with stage III breast cancer initiating radiotherapy. Interestingly, longitudinal gut and oral microbiome samples, along with a battery of questionnaires, are listed as secondary outcomes in order to better gauge how the microbiome might change in relation to behavioral patterns in cancer patients. A second trial (105) was designed to investigate fiber supplementation in patients with a previous history of colorectal cancer, through supplementation of beans into the normal diet for 8 weeks, to measure shifts in bacterial populations after a diet alteration. Even though both studies are not finalized yet, they will provide valuable information on how lifestyle factors can modulate the gut microbiome and its interaction with diet. A better understanding of the impact that diet has on microbiota will likely be key to the future of clinical and public health approaches to cancer.
Despite the impact of dietary nutrients seems relatively simple and fast to design, it may be hard to monitor the patient’s compliance in dietary description intake; the effect of food on the microbiota might be modest and heavily host related. An alternative method that could provide much more control towards microbial manipulation could be the administration of probiotics.
Probiotics are living microorganisms that, when balanced in terms of quantity, grant beneficial effects to the host (106). It is well-established that probiotics act in different ways to prevent the colonization of pathogens, such as Clostridium difficile and Staphylococcus aureus, and, consequently, dysbiosis (107). Indeed, probiotics antagonize pathogen colonization by competing for nutrients (108), sticking to the epithelial cell surfaces or to the mucus (109) and creating clusters with pathogens themselves (110). They also have a role in producing metabolites, such as lactic acid, acetic acid and bacteriocins, which are able to lower luminal pH (111) and unleash a direct antimicrobial activity (112), in order to inhibit pathogen growth.
There has been an increasing interest towards probiotics potential role in improving antitumor immunity, considering their ability to repress colonic inflammation and to stimulate immunosurveillance (113).
Bifidobacterium and Lactobacillus are two of the most active probiotics, which have been identified as regulators of gut homeostasis (114, 115). Moreover, other probiotics improve gut barrier function, by restoring epithelial integrity (116). An innovative approach could consist of administering probiotics before, during, or after potentially “microbiota-disrupting” or “microbiota-modulated” treatments. There have been several clinical trials administering probiotics in CRC patients. One that was completed in 2017 (117), aimed to unveil the change in fecal and tumor microbiota from the baseline, after using probiotics containing strains of L. acidophilus and B. lactis. The results showed an increased abundance of butyrate-producing bacteria (above all Faecalibacterium and other Clostridiales) within the tumor, and its associated non-tumor colonic mucosa and stool. This is a demonstration that probiotic therapy can change colonic mucosa. Some other ongoing trials are assessing the impact of probiotic therapy on different types of cancer, including the change on CD8+ T cell infiltrate in patients with stage I-III breast cancer (NCT03358511), and thus, providing a perspective for a future better understanding of their influence on microbiome.
Nevertheless, even though probiotics are deemed safe and well-tolerated by healthy subjects, in patients with damaged intestinal barrier or compromised immunity, such as cancer patients, their physiological protection may fail (118), resulting in bacteremia, fungemia, endocarditis, liver abscess and pneumonia (119). In fact, many of the ongoing trials mentioned before, have focused on safety endpoints. There is definitely wide variability regarding the stability and composition of the available probiotic therapies’ formulations (120), and despite caution should be taken towards their use in cancer patients, the use of probiotics is not absolutely forbidden (113).
Prebiotics, introduced by Gibson and Roberfroid in 1995, are non-viable food components, which can stimulate the growth and the activity of specific gut bacteria, improving the host’s health (121).
Probiotics produce some kinds of prebiotics, such as short-chain fatty acids (SCFAs) (122). SCFAs are indeed produced by several bacteria in the gut that ferment fibers. Many SCFAs, such as acetate, butyrate and propionate, are important in maintaining intestinal homeostasis (123). Because of their ubiquitous presence, they are being studied for their potential as universal metabolic regulators of the immune system. Among them, it has been noticed that butyrate has a relevant role in CRC patients, inducing the apoptosis of cancer cells and inhibiting inflammation as well as oxidative stress (124). Though, it needs to be considered that every host has a different genetic background, which may interfere with butyrate beneficial effects (125).
Furthermore, prebiotic oligosaccharides with a low grade of polymerization may induce CD4+ T cells to produce IFN-γ and IL-10 (126). Besides, two different studies in which mice with a transplantable liver tumor have received inulin or oligofructose together with subtherapeutic doses of six chemotherapeutics, pointed out boosted chemotherapeutic effects and observed an increased lifespan (127, 128).
Despite the positive effects mentioned above, Singh et al. have also reported a harmful microbial fermentation as a result of prebiotic supplementation (129). Firstly, they tried to examine whether inulin has a mitigating effect towards metabolic syndrome in Toll-like receptor 5 (TLR5) knockout mice. Unfortunately, even though a long-term inulin enriched diet alleviates metabolic dysfunctions, concurrently, it promotes cholestasis and necroinflammation, and therefore it can induce hepatocellular carcinoma (HCC). However, a constant supplementation of inulin in drinking water revealed to trigger hepatic inflammation and fibrosis, but it did not promote tumor development. Additionally, similar effects have been induced by other soluble fiber, such as pectin and fructo-oligosaccharide, in contrast with some non-fermentable and insoluble fiber, such as cellulose, for instance. Interestingly, Clostridia species are highly present in mice which develop an HCC and a depletion in butyrate-producing bacteria has been reported to reduce the incidence of the hepatocellular carcinoma in TLR5 knockout mice (129).
In conclusion, the above submissions suggest that prebiotic fermentation and butyrate production have a partial contribution in the hepatocellular carcinoma development, although not being the decisive driver (113).
In addition to probiotics and prebiotics, an interesting role in the modulation of gut homeostasis and patients’ outcome is played by postbiotics, which are soluble products and metabolites derived from microorganisms (130). Instead of relying on bacteria supported by prebiotics or introduced through probiotics, postbiotics represent the microbial product itself, thus surpassing the bacteria (131). Despite the advantage of not being dependent on the cultivation of specific microbiota compositions, further characterization of postbiotic mechanism of action is still required.
In fact, it has been noted that S. thermophilus (132) and E. coli (133) generate supernatants, which protect rat gut from 5-FU-induced mucositis. In addition, p40, a soluble protein produced by Lactobacillus rhamnosus GG, avoids cytokine-induced epithelial apoptosis, prevents gut barrier dissolution (134, 135) and raises immunoglobulin A secretion (136). Moreover, an example of a molecule that can induce an immune phenotype in the absence of the microorganism is polysaccharide A (PSA) derived from Bacterioides fragilis. A study reported how this prominent human commensal can direct the conversion of CD4+ T cells into Foxp3+ Treg cells with the immunomodulatory molecule being polysaccharide A. Interestingly, polysaccharide A administration alone was sufficient to induce expansion of Tregs and to increase the production of anti-inflammatory IL-10 in mice via TLR2 activation. Furthermore, PSA was not only able to prevent, but also cure experimental colitis in animals (137). Despite microbial products are considered to be adjuvants stimulating the immune response, this study provides an insight into their ability to promote immune suppression as well.
Moreover, as mentioned before, SCFAs are gut microbiota-derived bacterial fermentation products that are being studied for their effect on the immune system. A study demonstrated how short-chain fatty acids regulate the size and function of the colonic Treg pool and protect against colitis in a Ffar2-dependent manner in mice (138). Another study showed that butyrate, produced by commensal microorganisms during starch fermentation, facilitated extrathymic generation of Treg cells and de novo Treg-cell generation in the periphery was potentiated by propionate (139).
In oncologic patients, postbiotics induce antitumor effects (140). In support of this possibility, a study published by Konishi et al. in 2016, showed that Lactobacillus casei ATCC334 supernatant contained a powerful tumor-suppressive molecule, identified as ferrichrome. Ferrichrome treatment could induce apoptosis through the activation of c-jun N-terminal kinase (JNK). Interestingly, despite the tumor-suppressive effect of ferrichrome on colon cancer cells was found to be greater than or equal to that of conventional CRC drugs, this postbiotic showed less of an effect on healthy intestinal cells (140).
Overall, these data demonstrate that exogenous bacterial metabolites mediate the communication between the commensal microbiota and the immune system and can be utilized to influence immune activity in order to maintain homeostasis and promote health.
The putative mechanisms of actions of probiotics, prebiotics and postbiotics are shown in Figure 2.
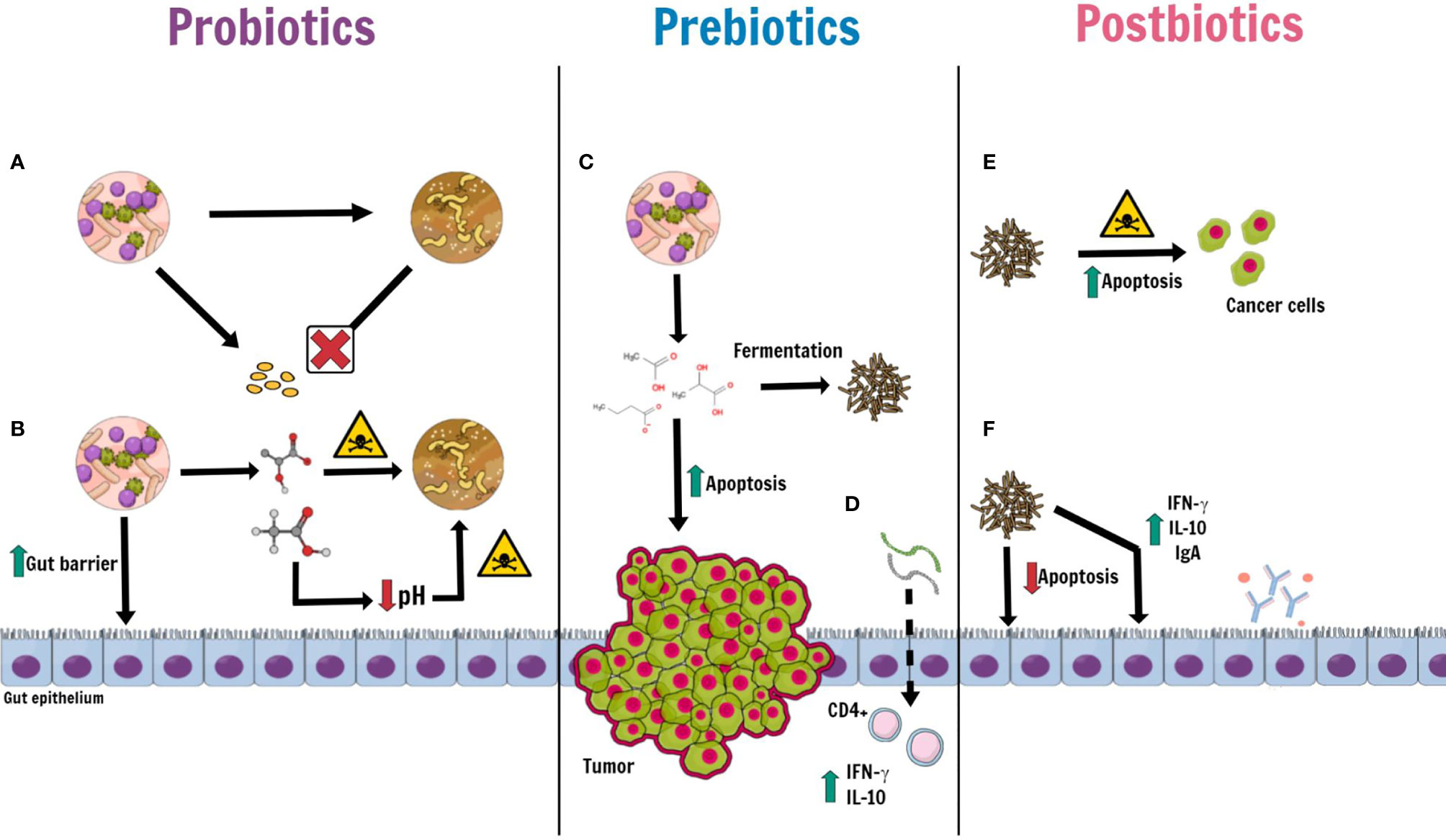
Figure 2 Putative mechanisms of actions of probiotics, prebiotics and postbiotics. Some kinds of probiotic, such as Bifidubacterium, Lactobacillus, Faecalibacterium and Clostridiales, may take an active role in maintaining gut homeostasis by: (A) preventing the proliferation and colonization of pathogens by competing for nutrients and microenvironment; (B) releasing antimicrobial peptides (such as lactic acid, acetic acid and bacteriocins) with a direct bactericidal effect and, indirectly, by lowering luminal pH. Moreover, probiotics induce an increase of mucin production, promote epithelial restoration and can enhance the expression of tight junctions. Prebiotics (inulin, oligofructose, soybean and oat fiber, pectin and non-digestible carbohydrates), derived from probiotics, (C) produce postbiotics through a fermentation process. Among prebiotics, Butyrate not only has an anti-inflammatory and antioxidative effect, but also an apoptotic effect against cancer cells, in CRC patients. (D) Oligosaccharides with a low grade of polymerization, directly absorbed by gut epithelium, stimulate T-cell CD4+ to release IFN-γ and IL-10. Postbiotics, prebiotics-derived products, on the one hand, (E) play a cytotoxic role against cancer cells, which increase their apoptosis; on the others (F) Lactobacillus rhamnosus GG and Bacteriodes fragilis, for instance, provide the wellness of the intestinal epithelium by inhibiting apoptosis of normal epithelial cells and raising the level of Ig A, IFN-γ and IL-10.
Even though probiotics and prebiotics bring numerous modifications to the human gut microbiota, unluckily, all their benefits are transient (141–144). Evidence sustains that intestinal microbiota alterations, provided by antibiotics injection, result in an enduring loss of the original human microbiota diversity (145). Considering that patients’ response to immunotherapy partly depends on the varied composition of their microbiota, a loss in terms of abundance and types of microorganism species could affect therapeutic outcome.
A retrospective study investigated the negative association between the administration of antibiotics and ICIs. Patients that were recently given antibiotic therapy (ATB) had shorter Progression Free Survival (PFS) and Overall Survival (OS) when compared to those who did not receive ATB (146). Furthermore, the combination of ATBs and proton pump inhibitors has also been associated with gut dysbiosis, decreased bacterial richness, and the promotion of T-cell tolerance (147). It seems that antibiotic treatment might reduce the efficacy of ICIs by modifying the patient’s microbiota (80).
Ipilimumab is a wholly human monoclonal antibody against CTLA-4 that was approved in 2011 for the treatment of unresectable and metastatic melanoma, as well as adjuvant treatment for melanoma (148). It was found that patients on treatment with Ipilimumab developed antibodies against some elements of gut microbiota (149). On the other hand, a combination of broad-spectrum antibiotics, such as Ampicillin, Colistin and Streptomycin could compromise the antitumoral effects of CTLA–4–specific antibodies, suggesting that gut microbiota is crucial to set up the best anticancer treatment outcome through CTLA-4 blockade (75). Indeed, it has been shown that the administration of antibiotics interferes with the clinical benefit of anti-CTLA-4 therapy in mouse models and also PD-1-based immunotherapy both in mice and in humans (75, 79, 150). In a study involving a group of 74 patients with a stage IV melanoma, 10 of them received ATB 30 days prior to the administration of ICI, while the rest of the group has been treated with a single-agent ICI, among Pembrolizumab, Nivolumab and Ipilimumab, as first-line therapy. Patients of the ATB group had a PFS and an OS meaningfully shorter than those in the non-ATB group (151).
Another study examined the impact of broad-spectrum antibiotic treatments administered 1 month before the initiation ICI to 3 months thereafter, in patients with metastatic non-small cell lung cancer. Interestingly, a shorter duration of ATB did not impact patient prognosis when compared with a longer course, bringing light on the potential importance of the duration of antibiotic treatments (152). The abovementioned data suggest that the duration of broad-spectrum antibiotic treatments with respect to the initiation of ICI-based immunotherapy is important.
In conclusion, it needs to be considered that patients that need antibiotic therapies may have an enfeebled immune system and are therefore more likely to be subjected to bacterial infections and to be refractory to anticancer immunotherapy. Consequently, in order to reduce the negative impact of ATB on ICI treatments, it will be important to define the specific antibiotics that are more likely to negatively impact on the clinical outcome. Thus, using prebiotics and probiotics during ATB might be solicited to reduce the negative impact on microbiome composition induced by antibiotic therapy.
Fecal microbiota transplantation (FMT) represents the most direct way to affect microbiota, using complete normal human flora as a therapeutic probiotic mixture of living organisms. This type of bacteriotherapy has a longstanding history in animal health and is used against chronic infections of the bowel, including those infected by Clostridium difficile resistant to conventional therapies as well as other patient populations (153). Nonetheless, fecal microbiota transplantation is also one of the most used ways to prove that microbiota is able to upset the outcome of immunotherapy (74, 75, 80, 154–156).
Several studies aimed to show the impact of fecal microbiota transplantation in mice. Germ-free or antibiotic-treated mice that had received a fecal microbiota transplantation from patients who had a response to immune-checkpoint blockade, were enriched in CD45+ and CD8+ T cells, indeed correlating with a positive response to PD-1 immune-checkpoint blockade (80, 157) (Figure 3). On the other hand, fecal microbiota transplantation with feces from non-responders led to resistance to ICIs, with tumors having a high density of immunosuppressive CD4+ Treg cells (157).
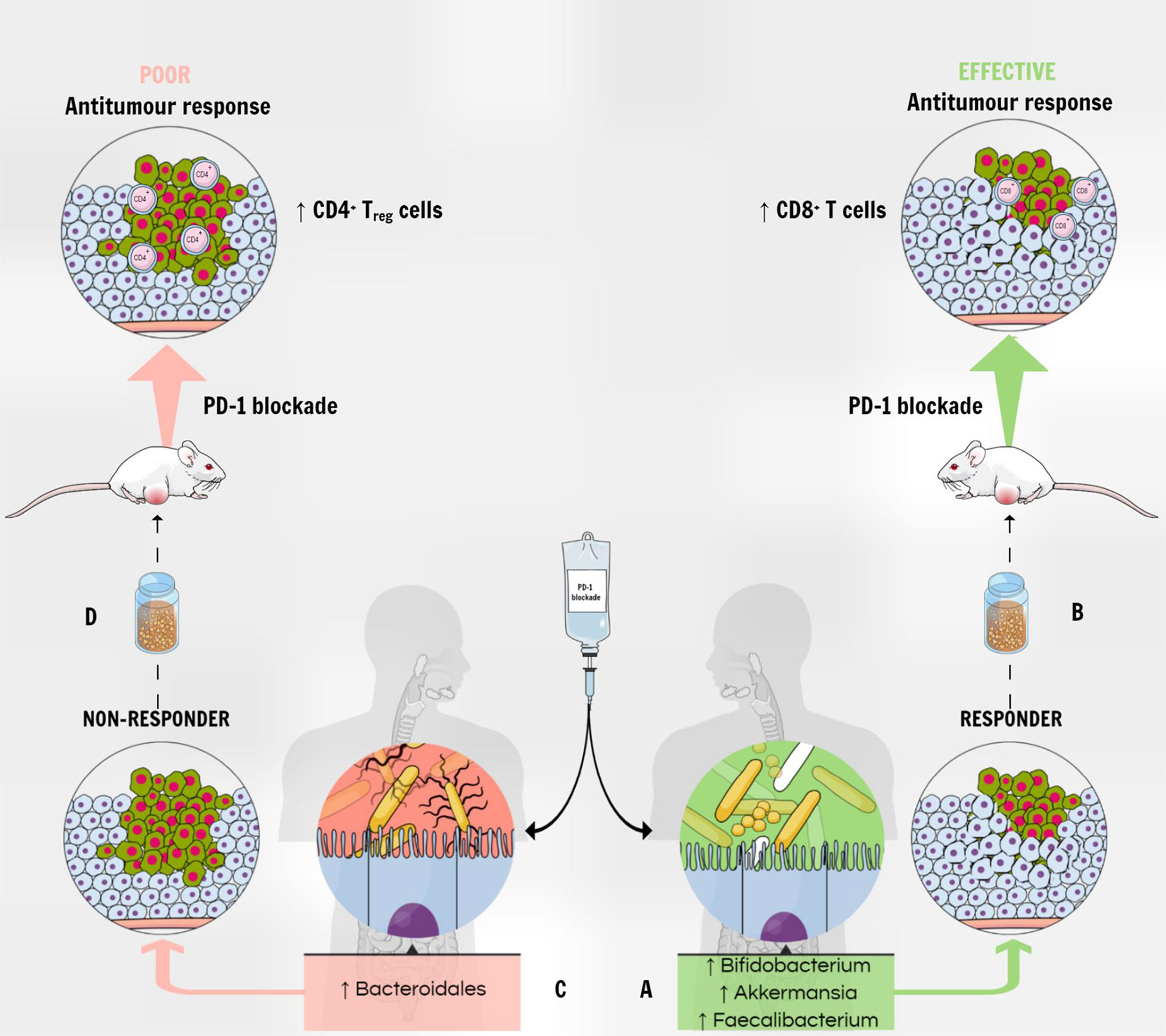
Figure 3 The gut microbiota modulates the response to PD-1 blockade therapy. (A) The enrichment of fecal microbiota with Akkermansia muciniphila, Faecalibacterium spp and Bifidobacterium spp correlates with a positive response to PD-1 immune-checkpoint blockade in patients with various types of tumors. (B) A fecal microbiota transplantation from responders into tumor-bearing mice correlates with increased antitumor CD8+ T cells in the tumor and improved response to anti–PD-1 therapy. (C) On the other hand, the higher abundance of Bacteroidales correlates with a deficient response to PD-1 blockade therapy in humans. (D) Mice receiving FMT from non-responders show poor anti-tumor response to anti–PD-1 therapy, and tumors show a higher density of immunosuppressive CD4+ Treg cells.
Moreover, mice transplanted with feces from responders developed a higher response to anti-PD-L1 therapy (80, 154). It is noteworthy that, when fecal microbiota is enriched with A. muciniphila, as well as with Faecalibacterium spp and Bifidobacterium spp (80, 157), it correlates with a positive response to PD-1 immune-checkpoint blockade in patients with various types of tumors. Thus, Bifidobacterium in the gut is positively related to anti-tumor activity, especially by stimulating CD8+ T cells and DCs (60). In line with these observations, the use of antibiotics is related to lower clinical efficiency of immune-checkpoint blockade in different kinds of tumor tested in mice and patients (157).
Furthermore, clinical FMT trials are being considered in patients with both hematologic malignancies and solid tumors. The single-arm study “ODYSSEE” (158), explored the use of autologous fecal microbiota transplantation in acute myeloid leukemia patients treated with intensive chemotherapy and antibiotics. The aim was to restore the balance of their intestinal microbiome and thereby eradicate treatment-induced multidrug resistant bacteria, infection-related complications, as well as sequelae to the gastrointestinal tract. Moreover, in a Phase 1 clinical trial, FMT from patients that responded to immunotherapy is being administered to refractory patients with metastatic melanoma and unresectable stage III melanoma who failed at least one line of PD-1 blockade (159).
Recently, Baruch et al. reported the first-in-human clinical trials to test whether fecal microbiota transplantation can affect the response to anti–PD-1 immunotherapy in melanoma patients. In their phase 1 clinical trial, they investigated the safety and feasibility of FMT and the combination of FMT and reinduction of anti–PD-1 immunotherapy in 10 patients with anti–PD-1–refractory metastatic melanoma. They observed clinical responses in three patients, with FMT being associated with favorable changes in immune cell infiltrates and gene expression profiles in both the gut lamina propria and the tumor microenvironment (160). The design of new additional trials is currently underway, in order to test the hypothesis that the modulation of the gut microbiota can improve the response to treatment with ICIs (80).
These interesting preliminary findings offer compelling evidence for the ability of FMT to affect immunotherapy response in cancer patients, supporting the concept of overcoming resistance to immunotherapy by modulating the gut microbiota.
The microbiome era has begun, and we have obtained substantial results on the influence of microbiota on cancer progression and treatment, including ICIs. The crosstalk between the host immune system and microbiota may allow a microbiota-based selection of patients that might benefit from a specific immunotherapy treatment, boosting their anticancer response. However, more studies on the topic are needed in order to better elucidate the microbial communities that colonize the tumor microenvironment, as well as the approaches to modulate the composition of gut microbiota.
Many dietary nutrients were studied for modulating gut microbiota, with fiber having shown a significant impact on the maintenance of microbiota diversity and the response to anti-PD-1 treatment. Since patients’ compliance might be hard to monitor and the effect of food on microbiota might be modest and heavily host related. An alternative method that could provide control towards gut homeostasis could be the use of prebiotic, postbiotic, probiotic and the administration of specific therapeutic schemes, for example with antibiotics. However, broader research is needed to determine the impact of these environmental factors on cancer therapy.
Satisfactory results offer compelling evidence on the ability of FMT to affect immunotherapy response in cancer patients. Further clinical trials with the use of FMT in cancer patients during ICIs are needed to better identify a strategy to overcome resistance to immunotherapy and improve patients’ outcomes.
Exploring the individual microbial profile and having a clear understanding of its interactions with various environmental factors could be a useful step to better modulate the gut microbiota. The prospect of being able to manipulate gut microbiota in order to modify the response to checkpoint inhibitors and set up personalized strategies serves as a continuous stimulus future research.
The authors contributed equally to this review. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Marin-Acevedo JA, Soyano AE, Dholaria B, Knutson KL, Lou Y. Cancer Immunotherapy Beyond Immune Checkpoint Inhibitors. J Hematol Oncol (2018) 11(1):8. doi: 10.1186/s13045-017-0552-6
2. Kelly PN. The Cancer Immunotherapy Revolution. Sci (N Y NY) (2018) 359(6382):1344–5. doi: 10.1126/science.359.6382.1344
3. Khalil DN, Smith EL, Brentjens RJ, Wolchok JD. The Future of Cancer Treatment: Immunomodulation, CARs and Combination Immunotherapy. Nature Reviews. Clin Oncol (2016) 13(5):273–90. doi: 10.1038/nrclinonc.2016.25
4. Frankel T, Lanfranca MP, Zou W. The Role of Tumor Microenvironment in Cancer Immunotheraphy. Advances in Experimental Medicine and Biology. Springer Nature: Cham (2017) 1036: 51–64. doi: 10.1007/978-3-319-67577-0_4
5. Jiao S, Subudhi SK, Aparicio A, Ge Z, Guan B, Miura Y, et al. Differences in Tumor Microenvironment Dictate T Helper Lineage Polarization and Response to Immune Checkpoint Therapy. Cell (2019) 179(5):1177–1190.e13. doi: 10.1016/j.cell.2019.10.029
6. Chen F, Zhuang X, Lin L, Yu P, Wang Y, Shi Y, et al. New Horizons in Tumor Microenvironment Biology: Challenges and Opportunities. BMC Med (2015) 13:45. doi: 10.1186/s12916-015-0278-7
7. Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell (2011) 144(5):646–74. doi: 10.1016/j.cell.2011.02.013
8. Balkwill FR, Capasso M, Hagemann T. The Tumor Microenvironment at a Glance. J Cell Sci (2012) 125(Pt 23):5591–6. doi: 10.1242/jcs.116392
9. Jain T, Sharma P, Are AC, Vickers SM, Dudeja V. New Insights Into the Cancer-Microbiome-Immune Axis: Decrypting a Decade of Discoveries. Front Immunol (2021) 12:622064. doi: 10.3389/fimmu.2021.622064
10. Kovács T, Mikó E, Ujlaki G, Sári Z, Bai P. The Microbiome as a Component of the Tumor Microenvironment. In: Birbrair A, editor. Tumor Microenvironment. Advances in Experimental Medicine and Biology, vol. 1225 . Cham: Springer (2020). doi: 10.1007/978-3-030-35727-6_10.
11. Sender R, Fuchs S, Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PloS Biol (2016) 14(8):e1002533. doi: 10.1371/journal.pbio.1002533
12. Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, et al. Metabolomics Analysis Reveals Large Effects of Gut Microflora on Mammalian Blood Metabolites. Proc Natl Acad Sci United States America (2009) 106(10):3698–703. doi: 10.1073/pnas.0812874106
13. Bordenstein SR, Theis KR. Host Biology in Light of the Microbiome: Ten Principles of Holobionts and Hologenomes. PloS Biol (2015) 13:e1002226. doi: 10.1371/journal.pbio.1002226
14. Postler TS, Ghosh S. Understanding the Holobiont: How Microbial Metabolites Affect Human Health and Shape the Immune System. Cell Metab (2017) 26:110–30. doi: 10.1016/j.cmet.2017.05.008
15. Nakatsu G, Li X, Zhou H, Sheng J, Wong SH, Wu WKK, et al. Gut Mucosal Microbiome Across Stages of Colorectal Carcinogenesis. Nat Commun (2015) 6:8727. doi: 10.1038/ncomms9727
16. Holmes E, Li JV, Athanasiou T, Ashrafian H, Nicholson JK. Understanding the Role of Gut Microbiome-Host Metabolic Signal Disruption in Health and Disease. Trends Microbiol (2011) 19(7):349–59. doi: 10.1016/j.tim.2011.05.006
17. Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, et al. Population-Based Metagenomics Analysis Reveals Markers for Gut Microbiome Composition and Diversity. Sci (N Y NY) (2016) 352(6285):565–9. doi: 10.1126/science.aad3369
18. Zmora N, Suez J, Elinav E. You are What You Eat: Diet, Health and the Gut Microbiota. Nat Rev Gastroenterol Hepatol (2019) 16:35–56. doi: 10.1038/s41575-018-0061-2
19. Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X, et al. Dominant and Diet-Responsive Groups of Bacteria Within the Human Colonic Microbiota. ISME J (2011) 5(2):220–30. doi: 10.1038/ismej.2010.118
20. Chang Y, Hou F, Pan Z, Huang Z, Han N. Optimization of Culturomics Strategy in Human Fecal Samples. Front Microbiol (2019) 10:2891. doi: 10.3389/fmicb.2019.02891
21. Lagier JC, Khelaifia S, Alou MT, Ndongo S, Dione N. Culture of Previously Uncultured Members of the Human Gut Microbiota by Culturomics. Nat Microbiol (2016) 1:16203. doi: 10.1038/nmicrobiol.2016.203
22. Burns MB, Lynch J, Starr TK, Knights D, Blekhman R. Virulence Genes are a Signature of the Microbiome in the Colorectal Tumor Microenvironment. Genome Med (2015) 7(1):55. doi: 10.1186/s13073-015-0177-8
23. Mira-Pascual L, Cabrera-Rubio R, Ocon S, Costales P, Parra A, Suarez A, et al. Microbial Mucosal Colonic Shifts Associated With the Development of Colorectal Cancer Reveal the Presence of Different Bacterial and Archaeal Biomarkers. J Gastroenterol (2015) 50(2):167–79. doi: 10.1007/s00535-014-0963-x
24. Bonnet M, Buc E, Sauvanet P, Darcha C, Dubois D, Pereira B, et al. Colonization of the Human Gut by E. Coli and Colorectal Cancer Risk. Clin Cancer Res: Off J Am Assoc Cancer Res (2014) 20(4):859–67. doi: 10.1158/1078-0432.CCR-13-1343
25. Kostic AD, Gevers D, Pedamallu CS, Michaud MM, Duke F, Earl AM, et al. Genomic Analysis Identifies Association of Fusobacterium With Colorectal Carcinoma. Genome Res (2012) 22(2):292–8. doi: 10.1101/gr.126573.111
26. Wu N, Yang X, Zhang R, Li J, Xiao C, Hu Y, et al. Dysbiosis Signature of Fecal Microbiota in Colorectal Cancer Patients. Microbial Ecol (2013) 66(2):462–70. doi: 10.1007/s00248-013-0245-9
27. Kich DM, Vincenzi A, Majolo F, Volken de Souza CF, Goettert MI. Probiotic: Effectiveness Nutrition in Cancer Treatment and Prevention. Nutricion Hospitalaria (2016) 33(6):1430–7. doi: 10.20960/nh.806
28. The Human Microbiome Project Consortium, Methé B, Nelson K, et al. A Framework for Human Microbiome Research. Nature (2012) 486:215–21. doi: 10.1038/nature11209
29. Gagnière J, Raisch J, Veziant J, Barnich N, Bonnet R, Buc E, et al. Gut Microbiota Imbalance and Colorectal Cancer. World J Gastroenterol (2016) 22(2):501–18. doi: 10.3748/wjg.v22.i2.501
30. Bhatt AP, Redinbo MR, Bultman SJ. The Role of the Microbiome in Cancer Development and Therapy. CA: Cancer J Clin (2017) 67(4):326–44. doi: 10.3322/caac.21398
31. Liu X, Cheng Y, Shao L, Ling Z. Alterations of the Predominant Fecal Microbiota and Disruption of the Gut Mucosal Barrier in Patients With Early-Stage Colorectal Cancer. BioMed Res Int (2020) 2020:2948282. doi: 10.1155/2020/2948282
32. Yuan C, Subramanian S. microRNA-Mediated Tumor-Microbiota Metabolic Interactions in Colorectal Cancer. DNA Cell Biol (2019) 38(4):281–5. doi: 10.1089/dna.2018.4579
33. Tilg H, Adolph TE, Gerner RR, Moschen AR. The Intestinal Microbiota in Colorectal Cancer. Cancer Cell (2018) 33:954–64. doi: 10.1016/j.ccell.2018.03.004
34. Jia W, Xie G, Jia W. Bile Acid-Microbiota Crosstalk in Gastrointestinal Inflammation and Carcinogenesis. Nat Rev Gastroenterol Hepatol (2018) 15(2):111–28. doi: 10.1038/nrgastro.2017.119
35. Carbone C, Piro G, Di Noia V, D’Argento E, Vita E, Ferrara M, et al. Lung and Gut Microbiota as Potential Hidden Driver of Immunotherapy Efficacy in Lung Cancer. Mediators Inflammation 2019 (2019) 2019:7652014. doi: 10.1155/2019/7652014
36. Fernández MF, Reina-Pérez I, Astorga JM, Rodríguez-Carrillo A, Plaza-Díaz J, Fontana L. Breast Cancer and Its Relationship With the Microbiota. Int J Environ Res Public Health (2018) 15(8):1747. doi: 10.3390/ijerph15081747
37. Sears CL, Garrett WS. Microbes, Microbiota, and Colon Cancer. Cell Host Microbe (2014) 15(3):317–28. doi: 10.1016/j.chom.2014.02.007
38. Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A Bacterial Driver-Passenger Model for Colorectal Cancer: Beyond the Usual Suspects. Nature Reviews. Microbiology (2012) 10(8):575–82. doi: 10.1038/nrmicro2819
39. Proctor LM. The Human Microbiome Project in 2011 and Beyond. Cell Host Microbe (2011) 10(4):287–91. doi: 10.1016/j.chom.2011.10.001
40. Wang Y, Zhang C, Hou S, Wu X, Liu J, Wan X. Analyses of Potential Driver and Passenger Bacteria in Human Colorectal Cancer. Cancer Manage Res (2020) 12:11553–61. doi: 10.2147/CMAR.S275316
41. Jin J, Gan T, Liu H, Wang Z, Yuan J, Deng T, et al. Diminishing Microbiome Richness and Distinction in the Lower Respiratory Tract of Lung Cancer Patients: A Multiple Comparative Study Design With Independent Validation. Lung Cancer (Amsterdam Netherlands) (2019) 136:129–35. doi: 10.1016/j.lungcan.2019.08.022
42. Eslami-S Z, Majidzadeh-A K, Halvaei S, Babapirali F, Esmaeili R. Microbiome and Breast Cancer: New Role for an Ancient Population. Front Oncol (2020) 10:120. doi: 10.3389/fonc.2020.00120
43. Meng S, Chen B, Yang J, Wang J, Zhu D, Meng Q, et al. Study of Microbiomes in Aseptically Collected Samples of Human Breast Tissue Using Needle Biopsy and the Potential Role of In Situ Tissue Microbiomes for Promoting Malignancy. Front Oncol (2018) 8:318. doi: 10.3389/fonc.2018.00318
44. Tlaskalová-Hogenová H, Štěpánková R, Hudcovic T, Tučková L, Cukrowska B, Lodinová-Žádnı́ková R, et al. Commensal Bacteria (Normal Microflora), Mucosal Immunity and Chronic Inflammatory and Autoimmune Diseases. Immunol Lett (2004) 93(2-3):97–108. doi: 10.1016/j.imlet.2004.02.005
45. Gagnière J, Raisch J, Veziant J, Barnich N, Bonnet R, Buc E, et al. Gut Microbiota Imbalance and Colorectal Cancer. World J Gastroenterol (2016) 22(2):501–18. doi: 10.3748/wjg.v22.i2.501
46. Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser AL, Barnich N, et al. High Prevalence of Adherent-Invasive Escherichia Coli Associated With Ileal Mucosa in Crohn’s Disease. Gastroenterology (2004) 127(2):412–21. doi: 10.1053/j.gastro.2004.04.061
47. Escobar-Páramo P, Grenet K, Le Menac'h A, Rode L, Salgado E, Amorin C, et al. Large-Scale Population Structure of Human Commensal Escherichia Coli Isolates. Appl Environ Microbiol (2004) 70(9):5698–700. doi: 10.1128/AEM.70.9.5698-5700.2004
48. Le Gall T, Clermont O, Gouriou S, Picard B, Nassif X, Denamur E, et al. Extraintestinal Virulence is a Coincidental by-Product of Commensalism in B2 Phylogenetic Group Escherichia Coli Strains. Mol Biol Evol (2007) 24(11):2373–84. doi: 10.1093/molbev/msm172
49. Schwabe RF, Jobin C. The Microbiome and Cancer. Nat Rev Cancer (2013) 13(11):800–12. doi: 10.1038/nrc3610
50. Luzzaro F, Mezzatesta M, Mugnaioli C, Perilli M, Stefani S, Amicosante G, et al. Trends in Production of Extended-Spectrum Beta-Lactamases Among Enterobacteria of Medical Interest: Report of the Second Italian Nationwide Survey. J Clin Microbiol (2006) 44(5):1659–64. doi: 10.1128/JCM.44.5.1659-1664.2006
51. Marchesi JR, Dutilh BE, Hall N, Peters WH, Roelofs R, Boleij A, et al. Towards the Human Colorectal Cancer Microbiome. PloS One (2011) 6(5):e20447. doi: 10.1371/journal.pone.0020447
52. Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, et al. Fusobacterium Nucleatum Infection is Prevalent in Human Colorectal Carcinoma. Genome Res (2012) 22(2):299–306. doi: 10.1101/gr.126516.111
53. Asakura H, Momose Y, Ryu C-H, Kasuga F, Yamamoto S, Kumagai S, et al. Providencia Alcalifaciens Causes Barrier Dysfunction and Apoptosis in Tissue Cell Culture: Potent Role of Lipopolysaccharides on Diarrheagenicity. Food Addit Amp Contam Part A Chem Anal Control Expo Risk Assess (2013) 30:1459–66. doi: 10.1080/19440049.2013.790086
54. Chen T, Li Q, Zhang X, Long R, Wu Y, Wu J, et al. TOX Expression Decreases With Progression of Colorectal Cancers and is Associated With CD4 T-Cell Density and Fusobacterium Nucleatum Infection. Hum Pathol (2018) 79:93–101. doi: 10.1016/j.humpath.2018.05.008
55. Maddocks OD, Short AJ, Donnenberg MS, Bader S, Harrison DJ. Attaching and Effacing Escherichia Coli Downregulate DNA Mismatch Repair Protein In Vitro and are Associated With Colorectal Adenocarcinomas in Humans. PloS One (2009) 4(5):e5517. doi: 10.1371/journal.pone.0005517
56. Prorok-Hamon M, Friswell MK, Alswied A, Roberts CL, Song F, Flanagan PK, et al. Colonic Mucosa-Associated Diffusely Adherent Afac+ Escherichia Coli Expressing lpfA and Pks are Increased in Inflammatory Bowel Disease and Colon Cancer. Gut (2014) 63(5):761–70. doi: 10.1136/gutjnl-2013-304739
57. Raskov H, Burcharth J, Pommergaard HC. Linking Gut Microbiota to Colorectal Cancer. J Cancer (2017) 8(17):3378–95. doi: 10.7150/jca.20497
58. Kato I, Boleij A, Kortman GA, Roelofs R, Djuric Z, Severson RK, et al. Partial Associations of Dietary Iron, Smoking and Intestinal Bacteria With Colorectal Cancer Risk. Nutr Cancer (2013) 65(2):169–77. doi: 10.1080/01635581.2013.748922
59. Liu X, Lu R, Wu S, Sun J. Salmonella Regulation of Intestinal Stem Cells Through the Wnt/β-Catenin Pathway. FEBS Lett (2010) 584(5):911–6. doi: 10.1016/j.febslet.2010.01.024
60. Singh RP, Bashir H, Kumar R. Emerging Role of Microbiota in Immunomodulation and Cancer Immunotherapy. Semin Cancer Biol (2020) 70:37–52. doi: 10.1016/j.semcancer.2020.06.008
61. Klener P.JR, Otáhal P, Lateckova L, Klener P. Immunotherapy Approaches in Cancer Treatment. Curr Pharm Biotechnol (2015) 16(9):771–81. doi: 10.2174/1389201016666150619114554
62. Alatrash G, Jakher H, Stafford PD, Mittendorf EA. Cancer Immunotherapies, Their Safety and Toxicity. Expert Opin Drug Saf (2013) 12(5):631–45. doi: 10.1517/14740338.2013.795944
63. Matzinger P, Kamala T. Tissue-Based Class Control: The Other Side of Tolerance. Nat Rev Immunol (2011) 11(3):221–30. doi: 10.1038/nri2940
64. Leach DR, Krummel MF, Allison JP. Enhancement of Antitumor Immunity by CTLA-4 Blockade. Sci (N Y NY) (1996) 271(5256):1734–6. doi: 10.1126/science.271.5256.1734
65. Wing K, Onishi Y, Prieto-Martin P, Yamaguchi T, Miyara M, Fehervari Z, et al. CTLA-4 Control Over Foxp3+ Regulatory T Cell Function. Sci (N Y NY) (2008) 322(5899):271–5. doi: 10.1126/science.1160062
66. Schluter J, Peled JU, Taylor BP, Markey KA, Smith M, Taur Y, et al. The Gut Microbiota is Associated With Immune Cell Dynamics in Humans. Nature (2020) 588(7837):303–7. doi: 10.1038/s41586-020-2971-8
67. Naidoo J, Page DB, Li BT, Connell LC, Schindler K, Lacouture ME, et al. Toxicities of the Anti-PD-1 and Anti-PD-L1 Immune Checkpoint Antibodies. Ann Oncol: Off J Eur Soc Med Oncol (2015) 26(12):2375–91. doi: 10.1093/annonc/mdv383
68. Galluzzi L, Buqué A, Kepp O, Zitvogel L, Kroemer G. Immunogenic Cell Death in Cancer and Infectious Disease. Nat Rev Immunol (2017) 17:97–111. doi: 10.1038/nri.2016.107
69. Gibney GT, Weiner LM, Atkins MB. Predictive Biomarkers for Checkpoint Inhibitor-Based Immunotherapy. Lancet Oncol (2016) 17:e542–51. doi: 10.1016/S1470-2045(16)30406-5
70. Australian Pancreatic Cancer Genome Initiative, ICGC Breast Cancer Consortium, ICGC MMML-Seq Consortium, ICGC PedBrain, Alexandrov LB, Nik-Zainal S, et al. Signatures of Mutational Processes in Human Cancer. Nature (2013) 500:415–21. doi: 10.1038/nature12477
71. Fessler J, Matson V, Gajewski TF. Exploring the Emerging Role of the Microbiome in Cancer Immunotherapy. J Immunother Cancer (2019) 7(1):108. doi: 10.1186/s40425-019-0574-4
72. Macpherson AJ, Harris NL. Interactions Between Commensal Intestinal Bacteria and the Immune System. Nature Reviews. Immunology (2004) 4(6):478–85. doi: 10.1038/nri1373
73. Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, et al. PD-1 Blockade Induces Responses by Inhibiting Adaptive Immune Resistance. Nature (2014) 515(7528):568–71. doi: 10.1038/nature13954
74. Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal Bifidobacterium Promotes Antitumor Immunity and Facilitates Anti–PD-L1 Efficacy. Science (2015) 350:1084–9. doi: 10.1126/science.aac4255
75. Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer Immunotherapy by CTLA-4 Blockade Relies on the Gut Microbiota. Sci (N Y NY) (2015) 350(6264):1079–84. doi: 10.1126/science.aad1329
76. Kuczma MP, Ding Z-C, Li T, Habtetsion T, Chen T, Hao Z, et al. The Impact of Antibiotic Usage on the Efficacy of Chemoimmunotherapy Is Contingent on the Source of Tumor-Reactive T Cells. Oncotarget (2017) 8:111931–42. doi: 10.18632/oncotarget.22953
77. Uribe-Herranz M, Bittinger K, Rafail S, Guedan S, Pierini S, Tanes C, et al. Gut Microbiota Modulates Adoptive Cell Therapy. Via CD8α Dendritic Cells IL-12 JCI Insight (2018) 3(4). doi: 10.1172/jci.insight.94952
78. Smith M, Littmann ER, Slingerland JB, Clurman A, Slingerland AE. Intestinal Microbiota Composition Prior to CAR T Cell Infusion Correlates With Efficacy and Toxicity. Blood (2018) 132:3492. doi: 10.1182/blood-2018-99-118628
79. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, et al. Gut Microbiome Influences Efficacy of PD-1–Based Immunotherapy Against Epithelial Tumors. Science (2018) 359:91–7. doi: 10.1126/science.aan3706
80. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut Microbiome Modulates Response to Anti–PD-1 Immunotherapy in Melanoma Patients. Science (2017) 359:97–103. doi: 10.1126/science.aan4236
81. Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre M-L, et al. The Commensal Microbiome is Associated With Anti–PD-1 Efficacy in Metastatic Melanoma Patients. Science (2018) 359:104–8. doi: 10.1126/science.aao3290
82. Xue X, Feng T, Yao S, Wolf KJ, Liu CG, Liu X, et al. Microbiota Downregulates Dendritic Cell Expression of miR-10a, Which Targets IL-12/IL-23p40. J Immunol (Baltimore Md: 1950) (2011) 187(11):5879–86. doi: 10.4049/jimmunol.1100535
83. Anfossi S, Calin GA. Gut Microbiota: A New Player in Regulating Immune- and Chemo-Therapy Efficacy. Cancer Drug Resist (Alhambra Calif) (2020) 3:356–70. doi: 10.20517/cdr.2020.04
84. van Niel G, D’Angelo G, Raposo G. Shedding Light on the Cell Biology of Extracellular Vesicles. Nature Reviews. Mol Cell Biol (2018) 19(4):213–28. doi: 10.1038/nrm.2017.125
85. Gottesman S. Micros for Microbes: non-Coding Regulatory RNAs in Bacteria. Trends Genet: TIG (2005) 21(7):399–404. doi: 10.1016/j.tig.2005.05.008
86. Kang SM, Choi JW, Lee Y, Hong SH, Lee HJ. Identification of microRNA-Size, Small RNAs in Escherichia Coli. Curr Microbiol (2013) 67(5):609–13. doi: 10.1007/s00284-013-0411-9
87. Koeppen K, Hampton TH, Jarek M, Scharfe M, Gerber SA, Mielcarz DW, et al. A Novel Mechanism of Host-Pathogen Interaction Through sRNA in Bacterial Outer Membrane Vesicles. PloS Pathog (2016) 12(6):e1005672. doi: 10.1371/journal.ppat.1005672
88. Choi JW, Kim SC, Hong SH, Lee HJ. Secretable Small RNAs via Outer Membrane Vesicles in Periodontal Pathogens. J Dental Res (2017) 96(4):458–66. doi: 10.1177/0022034516685071
89. Liu S, da Cunha AP, Rezende RM, Cialic R, Wei Z, Bry L, et al. The Host Shapes the Gut Microbiota via Fecal MicroRNA. Cell Host Microbe (2016) 19(1):32–43. doi: 10.1016/j.chom.2015.12.005
90. Charbonneau MR, O’Donnell D, Blanton LV, Totten SM, Davis JC, Barratt MJ, et al. Sialylated Milk Oligosaccharides Promote Microbiota-Dependent Growth in Models of Infant Undernutrition. Cell (2016) 164(5):859–71. doi: 10.1016/j.cell.2016.01.024
91. Laursen MF, Bahl MI, Michaelsen KF, Licht TR. First Foods and Gut Microbes. Front Microbiol (2017) 8:356. doi: 10.3389/fmicb.2017.00356
92. Claesson M, Jeffery I, Conde S, Power SE, O'Connor EM, Cusack S, et al. Gut Microbiota Composition Correlates With Diet and Health in the Elderly. Nature (2012) 488:178–84. doi: 10.1038/nature11319
93. David L, Maurice C, Carmody R, Gootenberg DB, Button JE, Wolfe BE, et al. Diet Rapidly and Reproducibly Alters the Human Gut Microbiome. Nature (2014) 505:559–63. doi: 10.1038/nature12820
94. Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, Fontana L, et al. Diet Drives Convergence in Gut Microbiome Functions Across Mammalian Phylogeny and Within Humans. Sci (N Y NY) (2011) 332(6032):970–4. doi: 10.1126/science.1198719
95. Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The Human Microbiome Project. Nature (2007) 449(7164):804–10. doi: 10.1038/nature06244
96. Schroeder BO, Birchenough GM, Ståhlman M, Arike L, Johansson ME, Hansson GC, et al. Bifidobacteria or Fiber Protects Against Diet-Induced Microbiota-Mediated Colonic Mucus Deterioration. Cell Host Microbe (2018) 23:27–40. doi: 10.1016/j.chom.2017.11.004
97. Benus RF, van der Werf TS, Welling GW, Judd PA, Taylor MA, Harmsen HJ, et al. Association Between Faecalibacterium Prausnitzii and Dietary Fibre in Colonic Fermentation in Healthy Human Subjects. Br J Nutr (2010) 104(5):693–700. doi: 10.1017/S0007114510001030
98. Prajapati B, Rajput P, Kumar Jena P, Seshadri S. Investigation of Chitosan for Prevention of Diabetic Progression Through Gut Microbiota Alteration in Sugar Rich Diet Induced Diabetic Rats. Curr Pharm Biotechnol (2016) 17:173–84. doi: 10.2174/1389201017666151029110505
99. Spencer CN, Gopalakrishnan V, McQuade J, Andrews MC, Helmink B, Khan MAW, et al. (2019). The Gut Microbiome (GM) and Immunotherapy Response are Influenced by Host Lifestyle Factors, in: Proceedings: AACR Annual Meeting 2019, Atlanta, GA, USA: American Association for Cancer Research 29 March–3 April doi: 10.1158/1538-7445.AM2019-2838.
100. Yang Q, Liang Q, Balakrishnan B, Belobrajdic DP, Feng Q-J, Zhang W. Role of Dietary Nutrients in the Modulation of Gut Microbiota: A Narrative Review. Nutrients (2020) 12:381. doi: 10.3390/nu12020381
101. Singh CK, Liu X, Ahmad N. Resveratrol, in its Natural Combination in Whole Grape, for Health Promotion and Disease Management. Ann New Y Acad Sci (2015) 1348(1):150–60. doi: 10.1111/nyas.12798
102. Gatouillat G, Balasse E, Joseph-Pietras D, Morjani H, Madoulet C. Resveratrol Induces Cell-Cycle Disruption and Apoptosis in Chemoresistant B16 Melanoma. J Cell Biochem (2010) 110(4):893–902. doi: 10.1002/jcb.22601
103. Di Renzo L, Marsella LT, Carraro A, Valente R, Gualtieri P, Gratteri S, et al. Changes in LDL Oxidative Status and Oxidative and Inflammatory Gene Expression After Red Wine Intake in Healthy People: A Randomized Trial. Mediators Inflamm (2015) 2015:317–48. doi: 10.1155/2015/317348
104. Zmora N, Suez J, Elinav E. You are What You Eat: Diet, Health and the Gut Microbiota. Nat Rev Gastroenterol Hepatol (2019) 16:35–56. doi: 10.1038/s41575-018-0061-2
105. Zhang X, Browman G, Siu W, Basen-Engquist KM, Hanash SM, Hoffman KL, et al. The BE GONE Trial Study Protocol: A Randomized Crossover Dietary Intervention of Dry Beans Targeting the Gut Microbiome of Overweight and Obese Patients With a History of Colorectal Polyps or Cancer. BMC Cancer (2019) 19(1):1233. doi: 10.1186/s12885-019-6400-z
106. Geier MS, Butler RN, Howarth GS. Probiotics, Prebiotics and Synbiotics: A Role in Chemoprevention for Colorectal Cancer? Cancer Biol Ther (2006) 5(10):1265–9. doi: 10.4161/cbt.5.10.3296
107. Mills JP, Rao K, Young VB. Probiotics for Prevention of Clostridium Difficile Infection. Curr Opin Gastroenterol (2018) 34(1):3–10. doi: 10.1097/MOG.0000000000000410
108. Kamada N, Kim YG, Sham HP, Vallance BA, Puente JL, Martens EC, et al. Regulated Virulence Controls the Ability of a Pathogen to Compete With the Gut Microbiota. Sci (N Y NY) (2012) 336(6086):1325–9. doi: 10.1126/science.1222195
109. Tuomola EM, Ouwehand AC, Salminen SJ. The Effect of Probiotic Bacteria on the Adhesion of Pathogens to Human Intestinal Mucus. FEMS Immunol Med Microbiol (1999) 26(2):137–42. doi: 10.1111/j.1574-695X.1999.tb01381.x
110. Campana R, van Hemert S, Baffone W. Strain-Specific Probiotic Properties of Lactic Acid Bacteria and Their Interference With Human Intestinal Pathogens Invasion. Gut Pathog (2017) Mar 69:12. doi: 10.1186/s13099-017-0162-4
111. Fayol-Messaoudi D, Berger CN, Coconnier-Polter MH, Liévin-Le Moal V, Servin AL. pH-, Lactic Acid-, and Non-Lactic Acid-Dependent Activities of Probiotic Lactobacilli Against Salmonella Enterica Serovar Typhimurium. Appl Environ Microbiol (2005) 71(10):6008–13. doi: 10.1128/AEM.71.10.6008-6013.2005
112. Gillor O, Etzion A, Riley MA. The Dual Role of Bacteriocins as Anti- and Probiotics. Appl Microbiol Biotechnol (2008) 81(4):591–606. doi: 10.1007/s00253-008-1726-5
113. Fong W, Li Q, Yu J. Gut Microbiota Modulation: A Novel Strategy for Prevention and Treatment of Colorectal Cancer. Oncogene (2020) 39(26):4925–43. doi: 10.1038/s41388-020-1341-1
114. van Baarlen P, Wells JM, Kleerebezem M. Regulation of Intestinal Homeostasis and Immunity With Probiotic Lactobacilli. Trends Immunol (2013) 34(5):208–15. doi: 10.1016/j.it.2013.01.005
115. Ruiz L, Delgado S, Ruas-Madiedo P, Sánchez B, Margolles A. Bifidobacteria and Their Molecular Communication With the Immune System. Front Microbiol (2017) 8:2345. doi: 10.3389/fmicb.2017.02345
116. Kumar M, Kissoon-Singh V, Coria AL, Moreau F, Chadee K. Probiotic Mixture VSL3 Reduces Colonic Inflammation and Improves Intestinal Barrier Function in Muc2 Mucin-Deficient Mice. Am J Physiol Gastrointestinal Liver Physiol (2017) 312(1):G34–45. doi: 10.1152/ajpgi.00298.2016
117. Hibberd AA, Lyra A, Ouwehand AC, Rolny P, Lindegren H, Cedgård L, et al. Intestinal Microbiota is Altered in Patients With Colon Cancer and Modified by Probiotic Intervention. BMJ Open Gastroenterol (2017) 4(1):e000145. doi: 10.1136/bmjgast-2017-000145
118. Cannon JP, Lee TA, Bolanos JT, Danziger LH. Pathogenic Relevance of Lactobacillus: A Retrospective Review of Over 200 Cases. Eur J Clin Microbiol Infect Dis: Off Publ Eur Soc Clin Microbiol (2005) 24(1):31–40. doi: 10.1007/s10096-004-1253-y
119. Doron S, Snydman DR. Risk and Safety of Probiotics. Clin Infect Dis: Off Publ Infect Dis Soc America (2015) 60 Suppl 2(Suppl 2):S129–34. doi: 10.1093/cid/civ085
120. Huys G, Botteldoorn N, Delvigne F, De Vuyst L, Heyndrickx M, Pot B, et al. Microbial Characterization of Probiotics–Advisory Report of the Working Group “8651 Probiotics” of the Belgian Superior Health Council (SHC). Mol Nutr Food Res (2013) 57(8):1479–504. doi: 10.1002/mnfr.201300065
121. Gibson GR, Roberfroid MB. Dietary Modulation of the Human Colonic Microbiota: Introducing the Concept of Prebiotics. J Nutr (1995) 125(6):1401–12. doi: 10.1093/jn/125.6.1401
122. Wong JM, de Souza R, Kendall CW, Emam A, Jenkins DJ. Colonic Health: Fermentation and Short Chain Fatty Acids. J Clin Gastroenterol (2006) 40(3):235–43. doi: 10.1097/00004836-200603000-00015
123. Parada Venegas D, de la Fuente MK, Landskron G, González MJ, Quera R, Dijkstra G, et al. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front Immunol (2019) 10:277. doi: 10.3389/fimmu.2019.00277
124. Canani RB, Costanzo MD, Leone L, Pedata M, Meli R, Calignano A. Potential Beneficial Effects of Butyrate in Intestinal and Extraintestinal Diseases. World J Gastroenterol (2011) 17(12):1519–28. doi: 10.3748/wjg.v17.i12.1519
125. Bultman SJ, Jobin C. Microbial-Derived Butyrate: An Oncometabolite or Tumor-Suppressive Metabolite? Cell Host Microbe (2014) 16(2):143–5. doi: 10.1016/j.chom.2014.07.011
126. Ito H, Takemura N, Sonoyama K, Kawagishi H, Topping DL, Conlon MA, et al. Degree of Polymerization of Inulin-Type Fructans Differentially Affects Number of Lactic Acid Bacteria, Intestinal Immune Functions, and Immunoglobulin A Secretion in the Rat Cecum. J Agric Food Chem (2011) 59(10):5771–8. doi: 10.1021/jf200859z
127. Taper HS, Roberfroid MB. Nontoxic Potentiation of Cancer Chemotherapy by Dietary Oligofructose or Inulin. Nutr Cancer (2000) 38(1):1–5. doi: 10.1207/S15327914NC381_1
128. Taper HS, Roberfroid MB. Possible Adjuvant Cancer Therapy by Two Prebiotics–Inulin or Oligofructose. In Vivo (Athens Greece) (2005) 19(1):201–4. https://iv.iiarjournals.org/content/19/1/201.long
129. Singh V, Yeoh BS, Chassaing B, Xiao X, Saha P, Aguilera Olvera R, et al. Dysregulated Microbial Fermentation of Soluble Fiber Induces Cholestatic Liver Cancer. Cell (2018) 175(3):679–694.e22. doi: 10.1016/j.cell.2018.09.004
130. Panebianco C, Andriulli A, Pazienza V. Pharmacomicrobiomics: Exploiting the Drug-Microbiota Interactions in Anticancer Therapies. Microbiome (2018) 6(1):92. doi: 10.1186/s40168-018-0483-7
131. Aguilar-Toalá JE, Garcia-Varela R, Garcia HS, Mata-Haro V, González-Córdova AF, Vallejo-Cordoba B, et al. Postbiotics: An Evolving Term Within the Functional Foods Field. Trends Food Sci Technol (2018) 75:105–14. doi: 10.1016/j.tifs.2018.03.009
132. Whitford EJ, Cummins AG, Butler RN, Prisciandaro LD, Fauser JK, Yazbeck R, et al. Effects of Streptococcus Thermophilus TH-4 on Intestinal Mucositis Induced by the Chemotherapeutic Agent, 5-Fluorouracil (5-Fu). Cancer Biol Ther (2009) 8(6):505–11. doi: 10.4161/cbt.8.6.7594
133. Prisciandaro LD, Geier MS, Butler RN, Cummins AG, Howarth GS. Probiotic Factors Partially Improve Parameters of 5-Fluorouracil-Induced Intestinal Mucositis in Rats. Cancer Biol Ther (2011) 11(7):671–7. doi: 10.4161/cbt.11.7.14896
134. Wang L, Cao H, Liu L, Wang B, Walker WA, Acra SA, et al. Activation of Epidermal Growth Factor Receptor Mediates Mucin Production Stimulated by P40, a Lactobacillus Rhamnosus GG-Derived Protein. J Biol Chem (2014) 289(29):20234–44. doi: 10.1074/jbc.M114.553800
135. Yan F, Polk DB. Characterization of a Probiotic-Derived Soluble Protein Which Reveals a Mechanism of Preventive and Treatment Effects of Probiotics on Intestinal Inflammatory Diseases. Gut Microbes (2012) 3(1):25–8. doi: 10.4161/gmic.19245
136. Wang Y, Liu L, Moore DJ, Shen X, Peek RM, Acra SA, et al. An LGG-Derived Protein Promotes IgA Production Through Upregulation of APRIL Expression in Intestinal Epithelial Cells. Mucosal Immunol (2017) 10(2):373–84. doi: 10.1038/mi.2016.57
137. Round JL, Mazmanian SK. Inducible Foxp3+ Regulatory T-Cell Development by a Commensal Bacterium of the Intestinal Microbiota. Proc Natl Acad Sci United States America (2010) 107(27):12204–9. doi: 10.1073/pnas.0909122107
138. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly- Y, et al. The Microbial Metabolites, Short-Chain Fatty Acids, Regulate Colonic Treg Cell Homeostasis. Sci (N Y NY) (2013) 341(6145):569–73. doi: 10.1126/science.1241165
139. Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites Produced by Commensal Bacteria Promote Peripheral Regulatory T-Cell Generation. Nature (2013) 504(7480):451–5. doi: 10.1038/nature12726
140. Konishi H, Fujiya M, Tanaka H, Ueno N, Moriichi K, Sasajima J, et al. Probiotic-Derived Ferrichrome Inhibits Colon Cancer Progression via JNK-Mediated Apoptosis. Nat Commun (2016) 7:12365. doi: 10.1038/ncomms12365
141. Manichanh C, Reeder J, Gibert P, Varela E, Llopis M, Antolin M, et al. Reshaping the Gut Microbiome With Bacterial Transplantation and Antibiotic Intake. Genome Res (2010) 20(10):1411–9. doi: 10.1101/gr.107987.110
142. Jernberg C, Löfmark S, Edlund C, Jansson JK. Long-Term Ecological Impacts of Antibiotic Administration on the Human Intestinal Microbiota. ISME J (2007) 1(1):56–66. doi: 10.1038/ismej.2007.3
143. Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB. Reproducible Community Dynamics of the Gastrointestinal Microbiota Following Antibiotic Perturbation. Infection Immun (2009) 77(6):2367–75. doi: 10.1128/IAI.01520-08
144. Dethlefsen L, Huse S, Sogin ML, Relman DA. The Pervasive Effects of an Antibiotic on the Human Gut Microbiota, as Revealed by Deep 16S rRNA Sequencing. PloS Biol (2008) 6(11):e280. doi: 10.1371/journal.pbio.0060280
145. Löfmark S, Jernberg C, Jansson JK, Edlund C. Clindamycin-Induced Enrichment and Long-Term Persistence of Resistant Bacteroides Spp. And Resistance Genes. J Antimicrobial Chemother (2006) 58(6):1160–7. doi: 10.1093/jac/dkl420
146. Derosa L, Hellmann MD, Spaziano M, Halpenny D, Fidelle M, Rizvi H, et al. Negative Association of Antibiotics on Clinical Activity of Immune Checkpoint Inhibitors in Patients With Advanced Renal Cell and non-Small-Cell Lung Cancer. Ann Oncol: Off J Eur Soc Med Oncol (2018) 29(6):1437–44. doi: 10.1093/annonc/mdy103
147. Chalabi M, Cardona A, Nagarkar DR, Dhawahir Scala A, Gandara DR, Rittmeyer A, et al. Efficacy of Chemotherapy and Atezolizumab in Patients With non-Small-Cell Lung Cancer Receiving Antibiotics and Proton Pump Inhibitors: Pooled Post Hoc Analyses of the OAK and POPLAR Trials. Ann Oncol (2020) 31:525–31. doi: 10.1016/j.annonc.2020.01.006
148. Peggs KS, Quezada SA, Korman AJ, Allison JP. Principles and Use of Anti-CTLA4 Antibody in Human Cancer Immunotherapy. Curr Opin Immunol (2006) 18(2):206–13. doi: 10.1016/j.coi.2006.01.011
149. Berman D, Parker SM, Siegel J, Chasalow SD, Weber J, Galbraith, et al. Blockade of Cytotoxic T-Lymphocyte Antigen-4 by Ipilimumab Results in Dysregulation of Gastrointestinal Immunity in Patients With Advanced Melanoma. Cancer Immun (2010) 10:11. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2999944/
150. Derosa L, Routy B, Mezquita L, Naltet C, Enot D, Fidelle M, et al. Antibiotics Prescription to Decrease Progression-Free Survival (PFS) and Overall Survival (OS) in Patients With Advanced Cancers Treated With PD1/PDL1 Immune Checkpoint Inhibitors. J Clin Oncol (2017) 35(15):3015–5. doi: 10.1200/JCO.2017.35.15_suppl.3015
151. Elkrief A, El Raichani L, Richard C, Messaoudene M, Belkaid W, Malo J, et al. Antibiotics are Associated With Decreased Progression-Free Survival of Advanced Melanoma Patients Treated With Immune Checkpoint Inhibitors. Oncoimmunology (2019) 8(4):e1568812. doi: 10.1080/2162402X.2019.1568812
152. Galli MPG, Poggi M, Fucà G, Imbimbo M, Proto C, Signorelli D, et al. Effects of Antibiotic Use During Immunotherapy in Metastatic non Small Cell Lung Cancer. Ann Oncol (2018) 29:493–547. doi: 10.1093/annonc/mdy292.088
153. Borody TJ, Warren EF, Leis SM, Surace R, Ashman O, Siarakas S. Bacteriotherapy Using Fecal Flora: Toying With Human Motions. J Clin Gastroenterol (2004) 38(6):475–83. doi: 10.1097/01.mcg.0000128988.13808.dc
154. Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA. The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell (2018) 33(4):570–80. doi: 10.1016/j.ccell.2018.03.015
155. Elkrief A, Derosa L, Zitvogel L, Kroemer G, Routy B. The Intimate Relationship Between Gut Microbiota and Cancer Immunotherapy. Gut Microbes (2019) 10(3):424–8. doi: 10.1080/19490976.2018.1527167
156. Routy B, Gopalakrishnan V, Daillère R, Zitvogel L, Wargo JA, Kroemer G, et al. The Gut Microbiota Influences Anticancer Immunosurveillance and General Health. Nat Rev Clin Oncol (2018) 15:382–96. doi: 10.1038/s41571-018-0006-2
157. Chaput N, Lepage P, Coutzac C, Soularue E, Le Roux K, Monot C, et al. Baseline Gut Microbiota Predicts Clinical Response and Colitis in Metastatic Melanoma Patients Treated With Ipilimumab. Ann Oncol: Off J Eur Soc Med Oncol (2017) 28(6):1368–79. doi: 10.1093/annonc/mdx108
158. Mohty M, Malard F, Vekhoff A, Lapusan S, Isnard F, D'Incan E, et al. The Odyssee Study: Prevention of Dysbiosis Complications With Autologous Fecal Microbiota Transfer (FMT) in Acute Myeloid Leukemia (AML) Patients Undergoing Intensive Treatment: Results of a Prospective Multicenter Trial. Blood (2018) 132:1444–4. doi: 10.1182/blood-2018-99-112825
159. Baruch EN, Youngster I, Ortenberg R, Ben-Betzalel G, et al. Abstract CT042: Fecal Microbiota Transplantation (FMT) and Re-Induction of Anti-PD-1 Therapy in Refractory Metastatic Melanoma Patients - Preliminary Results From a Phase I Clinical Trial (NCT03353402). (2019), CT042–2. doi: 10.1158/1538-7445.SABCS18-CT042
Keywords: microbiota, immunotherapy, immune checkpoint inhibitors (ICIs), fecal microbiota transplantation (FMT), diet, nutrients
Citation: Roberto M, Carconi C, Cerreti M, Schipilliti FM, Botticelli A, Mazzuca F and Marchetti P (2021) The Challenge of ICIs Resistance in Solid Tumours: Could Microbiota and Its Diversity Be Our Secret Weapon? Front. Immunol. 12:704942. doi: 10.3389/fimmu.2021.704942
Received: 04 May 2021; Accepted: 03 August 2021;
Published: 20 August 2021.
Edited by:
Lorenzo Mortara, University of Insubria, ItalyReviewed by:
Steven F. Gameiro, McMaster University, CanadaCopyright © 2021 Roberto, Carconi, Cerreti, Schipilliti, Botticelli, Mazzuca and Marchetti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrea Botticelli, YW5kcmVhLmJvdHRpY2VsbGlAdW5pcm9tYTEuaXQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.