- 1Department of Ophthalmology, Baylor College of Medicine, Houston, TX, United States
- 2Center for Metagenomics and Microbiome Research, Department of Molecular Virology and Microbiology, Baylor College of Medicine, Houston, TX, United States
- 3Grupo de Inmunodeficiencias Primarias, Facultad de Medicina, Universidad de Antioquia UdeA, Medellín, Colombia
Sjögren syndrome (SS) is an autoimmune condition that targets the salivary and lacrimal glands, with cardinal clinical signs of dry eye (keratoconjunctivitis sicca, KCS) and dry mouth. The conjunctiva of SS patients is often infiltrated by immune cells that participate in the induction and maintenance of local inflammation. The purpose of this study was to investigate immune-related molecular pathways activated in the conjunctiva of SS patients. Female SS patients (n=7) and controls (n=19) completed a series of oral, ocular surface exams. Symptom severity scores were evaluated using validated questionnaires (OSDI and SANDE). All patients fulfilled the ACR/EULAR criteria for SS and the criteria for KCS. Fluorescein and lissamine green dye staining evaluated tear-break-up time (TBUT), corneal and conjunctival disease, respectively. Impression cytology of the temporal bulbar conjunctiva was performed to collect cells lysed and subjected to gene expression analysis using the NanoString Immunology Panel. 53/594 differentially expressed genes (DEGs) were observed between SS and healthy controls; 49 DEGs were upregulated, and 4 were downregulated (TRAF5, TGFBI, KLRAP1, and CMKLRI). The top 10 DEGs in descending order were BST2, IFITM1, LAMP3, CXCL1, IL19, CFB, LY96, MX1, IL4R, CDKN1A. Twenty pathways had a global significance score greater or equal to 2. Spearman correlations showed that 29/49 upregulated DEGs correlated with either TBUT (inverse) or OSDI or conjunctival staining score (positive correlations). Venn diagrams identified that 26/29 DEGs correlated with TBUT, 5/26 DEGs correlated with OSDI, and 16/26 correlated with conjunctival staining scores. Five upregulated DEGs (CFB, CFI, IL1R1, IL2RG, IL4R) were uniquely negatively correlated with TBUT. These data indicate that the conjunctiva of SS patients exhibits a phenotype of immune activation, although some genes could be inhibitory. Some of the DEGs and pathways overlap with previous DEGs in salivary gland biopsies, but new DEGs were identified, and some of these correlated with symptoms and signs of dry eye. Our results indicate that gene analysis of conjunctiva imprints is a powerful tool to understand the pathogenesis of SS and develop new therapeutic targets.
Introduction
Patients with Sjögren syndrome (SS) develop an ocular surface disease called keratoconjunctivitis sicca (KCS). Pathological changes in the conjunctiva in KCS include altered epithelial differentiation with increased expression of cornified envelope precursors (1) that are found in the epidermis (involucrin, SPRR-2G) (2, 3) and dysfunction and loss of the goblet cells that secrete gel-forming mucin that is essential for maintaining tear film stability (4). Evidence from clinical and mouse model studies indicates innate, and adaptive immune mediators and cells contribute to these pathological changes based on clinical correlation with disease severity and improvement following topical treatment with the immunomodulatory agent cyclosporine A (5–8). Current methods to diagnose SS KCS include the use of dyes that stain conjunctival epithelial cells lacking a mucin coating or impression cytology to measure conjunctival goblet cell density, but they do not measure disease-relevant biomarkers. Additionally, the mechanism(s) by which this epithelial pathology develops is not entirely understood. An increased number of activated antigen-presenting cells (APCs) and increased interferon-γ expression have been found in the conjunctival epithelium in KCS (1, 6, 8–10). Interferon IFN-γ disrupts cholinergic mediated secretion and causes unfolded protein response (UPR) and apoptosis of conjunctival goblet cells (11, 12). Additional inflammatory mediators have been detected in the conjunctiva (1, 13, 14).
The purpose of this study was to compare the expression of immune-related molecular factors in the conjunctiva of normal subjects and SS KCS using the NanoString® Immunology panel.
Methods
The study was conducted following the Declaration of Helsinki, and the Baylor College of Medicine Institutional Review Board approved the protocol and informed consent form before study initiation. Written informed consent was obtained from all participants after explaining the purpose and possible consequences of the study. Female control subjects and patients with SS were enrolled in the study from January 2019 to January 2021. SS patients were recruited from the multispecialty SS clinic at Baylor College of Medicine (BCM) and had a complete ocular, oral, and rheumatologic evaluation, including a panel of serum autoantibodies, and met proposed the American College of Rheumatology/European League Against Rheumatism diagnostic criteria for SS (15).
Symptom assessment in dry eye (SANDE) and Ocular Surface Disease Index (OSDI) symptom questionnaires, fluorescein tear break-up time (TBUT), Schirmer I test, cornea fluorescein and conjunctival lissamine green dye staining, and tear meniscus height (TMH) measurement using optical coherence tomography (OCT) were performed as previously described (16, 17). The ocular surface clinical parameters were all measured by the same observer (S.C.P.). Dry Eye Workshop (DEWS) criteria were used to grade clinical severity (18).
Healthy control subjects had no eye irritation, a TBUT ≥ 7 s, Schirmer 1 ≥ 10 mm, corneal fluorescein score ≤2, conjunctival lissamine score ≤ 2, and no meibomian gland disease. Subjects were excluded if they had prior laser-assisted in situ keratomileusis or corneal transplantation surgery, cataract surgery in the past year, punctal occlusion with plugs or cautery, a history of contact lens wear, use of topical medications other than preservative-free artificial tears, or chronic use of systemic medications known to reduce tear production. They were instructed not to instill any eye drops on the day of the evaluation.
All SS KCS patients had a severity score ≥of 3, and healthy controls had a severity score = 0.
Conjunctival Impression Cytology and RNA Extraction
Cells were obtained by impression cytology of the temporal bulbar conjunctiva using the EyePrim™ device (Opia Tech, Paris, France) that applies a porous membrane with uniform pressure to the conjunctival surface. After collection, the membrane was placed in 0.5 mL RNA lysis buffer (Qiagen, Valencia, CA, USA) containing 1% 2-mercaptoethanol and immediately stored at -80°C. Total RNA was isolated using an RNeasy Mini Kit (Qiagen) following the manufacturer’s protocol. The RNA was eluted in 15 μL RNase-free water- The RNA concentration was measured by its absorption at 260 nm using a spectrophotometer (NanoDrop 2000; Thermo Scientific, Wilmington, DE, USA) and Agilent Bioanalyzer.
NanoString® and Data Analysis Using ROSALIND® NanoSstring Gene Expression Methods
Five hundred ninety-four transcripts were quantified with the NanoString® nCounter multiplexed target platform using the Human Immunology V2 panel (www.nanostring.com). nCounts of mRNA transcripts were normalized using the geometric means of 15 housekeeping genes (ABCF1, ALAS1, EEF1G, G6PD, GAPDH, GUSB, HPRT1, OAZ1, POLR1B, POLR2A, PPIA, RPL19, SDHA, TBP, TUBB). Data were analyzed by ROSALIND® (https://rosalind.onramp.bio/), with a HyperScale architecture developed by ROSALIND, Inc. (San Diego, CA). Read Distribution percentages, violin plots, identity heatmaps, and sample MDS plots were generated as part of the QC step. Normalization, fold changes, and p-values were calculated using criteria provided by NanoString®. ROSALIND® follows the nCounter® Advanced Analysis protocol of dividing counts within a lane using the same lane’s geometric mean of the normalizer probes. Housekeeping probes to be used for normalization are selected based on the geNorm algorithm as implemented in the NormqPCR R library (19). The abundance of various cell populations is calculated on ROSALIND using the NanoString® Cell Type Profiling Module. ROSALIND performs filtering of Cell Type Profiling results to include scores with a p-value lower than or equal to 0.05. Fold changes and P-values are calculated using the fast method described in the nCounter® Advanced Analysis 2.0 User Manual. The Gene Set Analysis (GSA) module from NanoString® is incorporated into ROSALIND to summarize the global significance score and the directed global significance score. GSA summarizes the change in regulation within each defined gene set relative to the baseline, as described in the nCounter® Advanced Analysis 2.0 User Manual. P-value adjustment is performed using the Benjamini-Hochberg method of estimating false discovery rates (FDR). Clustering genes for the final heatmap of differentially expressed genes was done using the PAM (Partitioning Around Medoids) method using the FPC R library (20) that considers the direction and type of all signals pathway the position, role, and type of every gene. Hypergeometric distribution was used to analyze the enrichment of pathways, gene ontology, domain structure, and other ontologies. The topGO R library (20) was used to determine local similarities and dependencies between GO terms to perform Elim pruning correction. Several database sources were referenced for enrichment analysis, including Interpro (21), NCBI5 (22), MSigDB (23, 24), REACTOME (25), WikiPathways (26). Enrichment was calculated relative to a set of background genes relevant for the experiment. Data analyzed in ROSALIND was downloaded, and Volcano plots of differential expression data were plotted using the −log10 (P-value) and log2 fold change using GraphPad Prism (Version 9, GraphPad Software, San Diego, CA). Heatmaps were also constructed using GraphPad Prism. Venn diagrams were made using Venny 2.1 (27). Over-represented pathway investigation was performed using www.innatedb.com (28).
Statistical Analysis
Comparisons of OSDI, SANDE, TMH, TBUT, corneal and conjunctival scores between HC and SS subjects were performed using non-parametric Mann Whitney U test using GraphPad Prism version 9.0 (GraphPad Sofware). Non-parametric Spearman correlations using GraphPad Prism investigated the correlations of OSDI, TBUT, corneal and conjunctival score with up or down-regulated DEGs.
Results
Patient Demographics
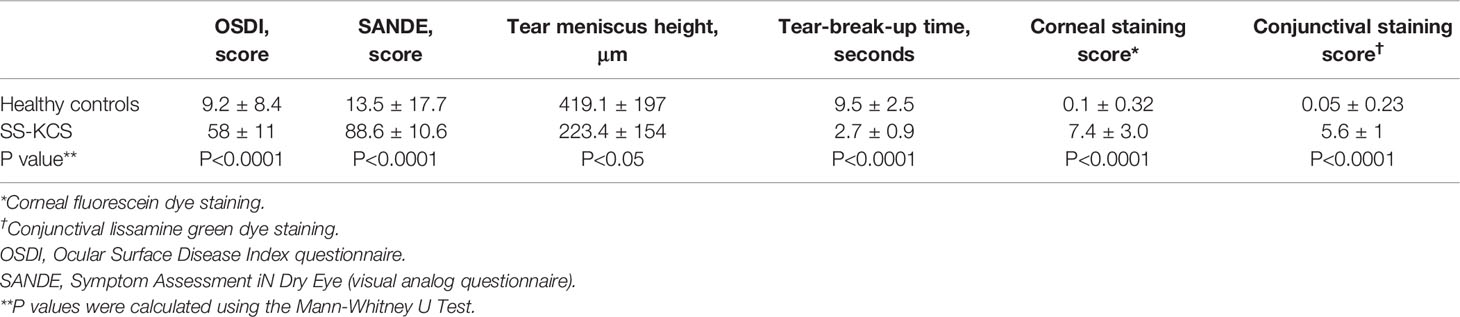
Nineteen control subjects and seven age-and-sex-matched SS KCS patients were enrolled from January 2019 to January 2021 (Table 1). The control subjects had minimal eye irritation symptoms and no clinical signs of KCS, while SS KCS patients had significant symptoms and clinical signs. Statistical comparison of OSDI and SANDE scores, tear meniscus height (TMS), tear-break-up time (TBUT), corneal and conjunctival staining scores between controls and SS KCS are shown in Table 2.
Gene Expression Analysis in the Conjunctiva of SS KCS Identifies Upregulated Targets
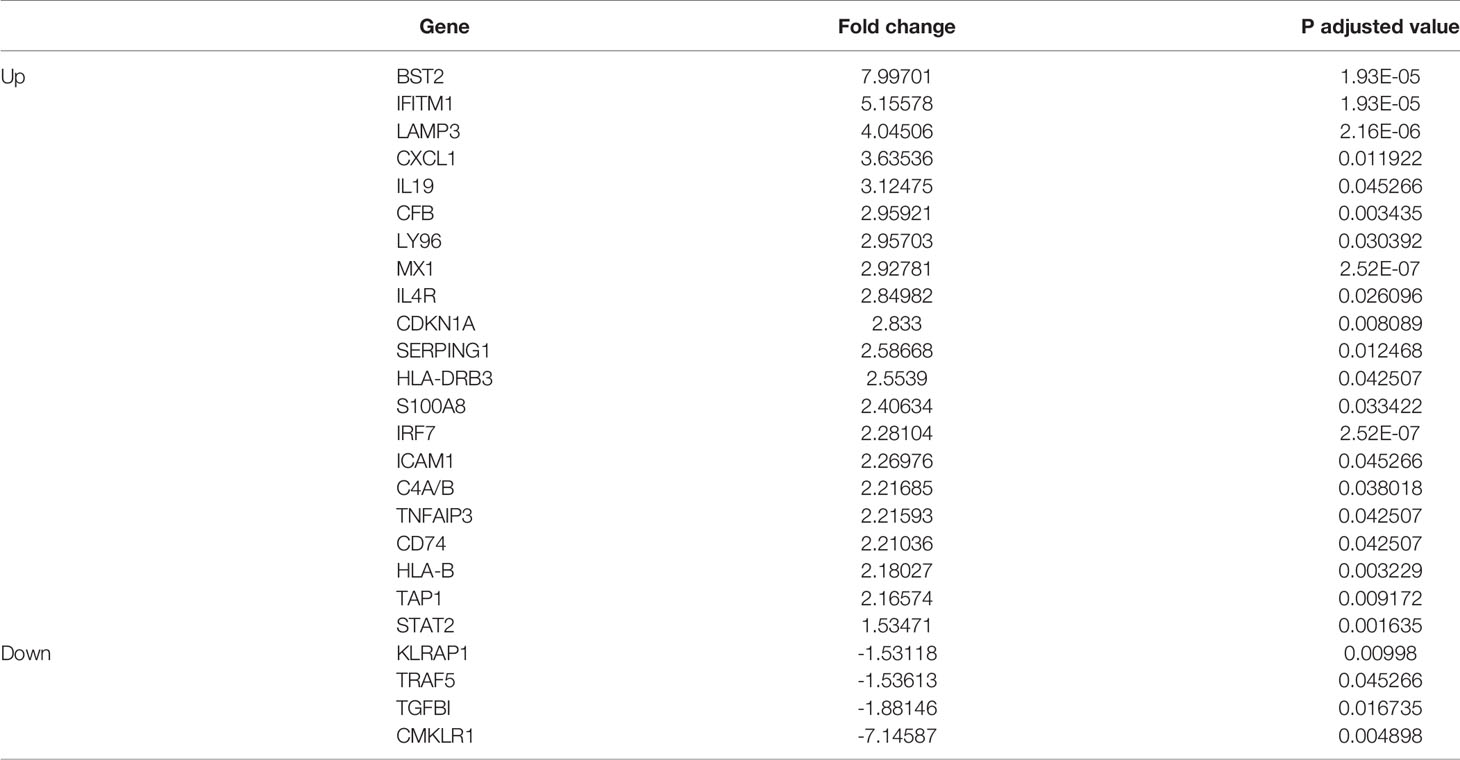
The conjunctiva of SS KCS shows epithelial metaplasia, goblet cell loss, T cell infiltration, and increased expression of HLA-DR and IFNG mRNA (6, 10, 29–32). We and others have shown that impression cytology of the conjunctiva collects a mix of epithelial and immune cells (8, 14, 33). To gain insights into the molecular mechanisms involved locally, impression cytology of the conjunctiva was used to collect conjunctival cells, which were then lysed and processed for RNA analysis using a NanoString® nCounter panel enriched for immune genes (Immunology V2). This panel allows for the simultaneous evaluation of 594 genes. Gene expression analysis was quantified in the conjunctiva of healthy and SS KCS patients using nCounter and ROSALIND software, as described in methods. Out of the 594 genes, eight genes were not detected; 53 were significantly modulated in SS KCS patients: 4 genes were downregulated (TRAF5, TGFBI, KLRAP1, and CMKLRI), and 49 genes were upregulated using a stringent 1.5-fold increase or decrease threshold (volcano plot in Figure 1A). Significance was plotted against fold change (log2 values) using a -log10 of the adjusted p-value for each dot. The top 20 upregulated differentially expressed genes (DEGs) in descending order were BST2, IFITM1, LAMP3, CXCL1, IL19, CFB, LY96, MX1, IL4R, CDKN1A, SERPING1, HLA-DRB3, S100A8, IRF7, ICAM1, C4A/B, TNFAIP3, CD74, HLA-B, and TAP1 (Table 3). A complete list of DEGs can be found in Supplementary Table 1.
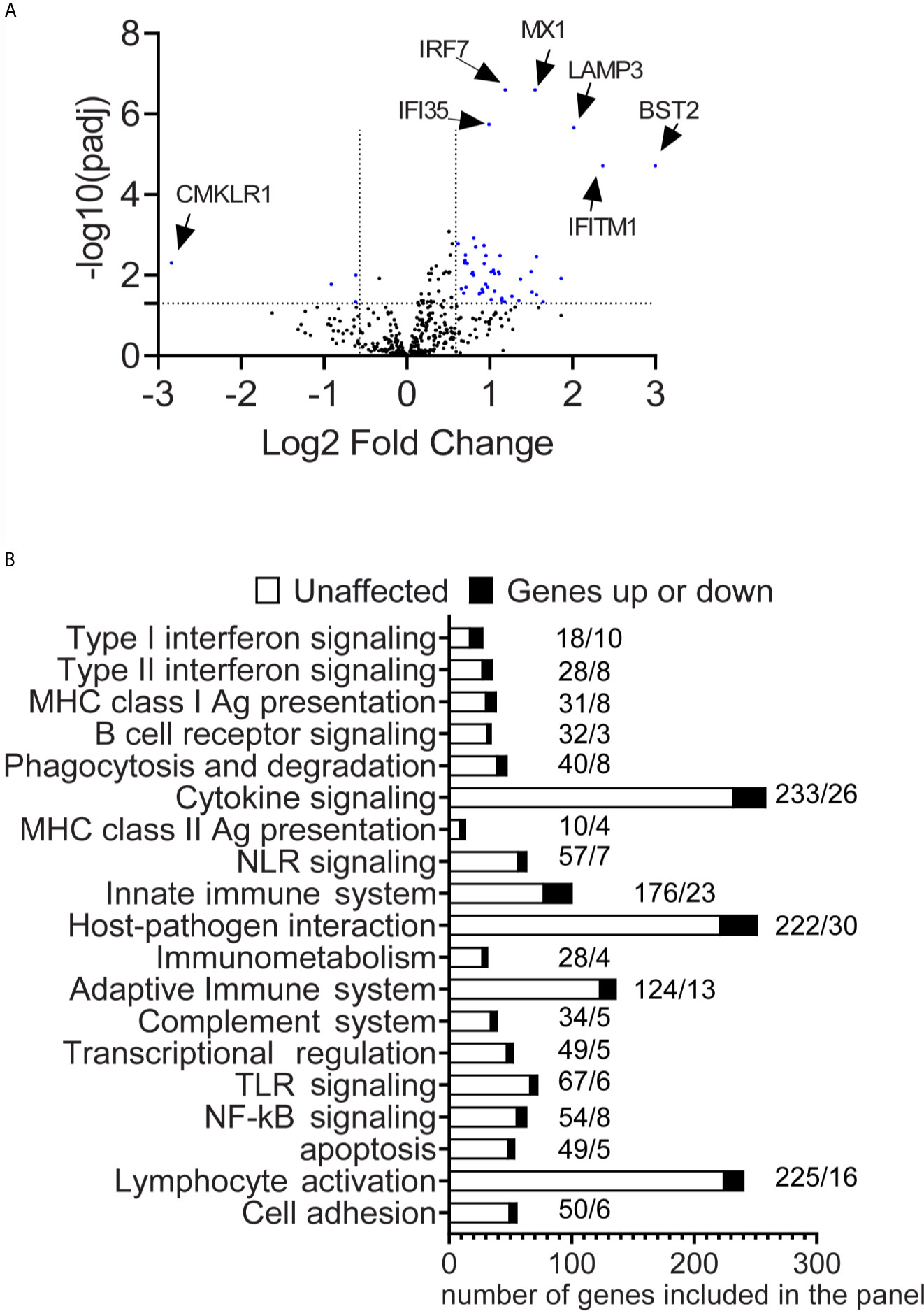
Figure 1 Differential expression of immune genes in the conjunctiva of SS patients. (A) A volcano plot shows −log10(p-value) and Log 2-fold change in gene expression in SS-KCS patients compared to control subjects (fold increase >1.5 or fold decrease <1.5 and p adjusted value of 0.05). P-value thresholds were adjusted using the Benjamini–Hochberg method of estimating false discovery rates. The horizontal line indicates p=0.05, and the two vertical lines indicate greater or lower than 1.5-fold. (B) Pathway descriptors are included in the panel. Dark bars indicate modulated genes (up or down), and white bars show unaffected genes. SS-KCS, Sjögren Syndrome keratoconjunctivitis sicca. Numbers above the bars indicate the number of unaffected genes (left) followed by the affected genes (right).
Differential Pathways Are Activated in the Conjunctiva of SS KCS Patients
Previous studies have identified a significant role for inflammation in the conjunctiva of SS KCS patients (6, 13, 14, 34). To gain insight into the pathways involved, we analyzed which DEGs have been annotated or identified in immune pathways using the ROSALIND software and nCounter software, using the gene set analysis. This algorithm calculates a T-statistic for each gene against each covariate in the model. A gene set’s global significance score for a covariate measures the cumulative evidence for the differential expression of specific genes in a pathway. Out of the 32 immune pathways included in the panel, the top 20 pathways are shown in Figure 1B (global significance range from 2.0 to 4.24). For each pathway, modulated genes and unaffected genes are shown. The pathways (Figures 2–4) and their respective global significance scores are Type I Interferonsignaling (4.24), Type II Interferon signaling (2.91), MHC Class I antigen presentation (2.84), B cell receptor signaling (2.44),Phagocytosis and degradation (2.44), Cytokine signaling (2.40), MHC Class II antigen presentation (2.37), NLR signaling (2.36), Innate immune system (2.32), Host-pathogen interaction (2.31), Immunometabolism (2.28), Adaptive immune system (2.28), Complement system (2.25), Transcriptional regulation (2.24), TLR signaling (2.23), NFkB signaling (2.23), Apoptosis (2.15), Lymphocyte activation (2). As expected, not all pathways in the panel were identified in our results.
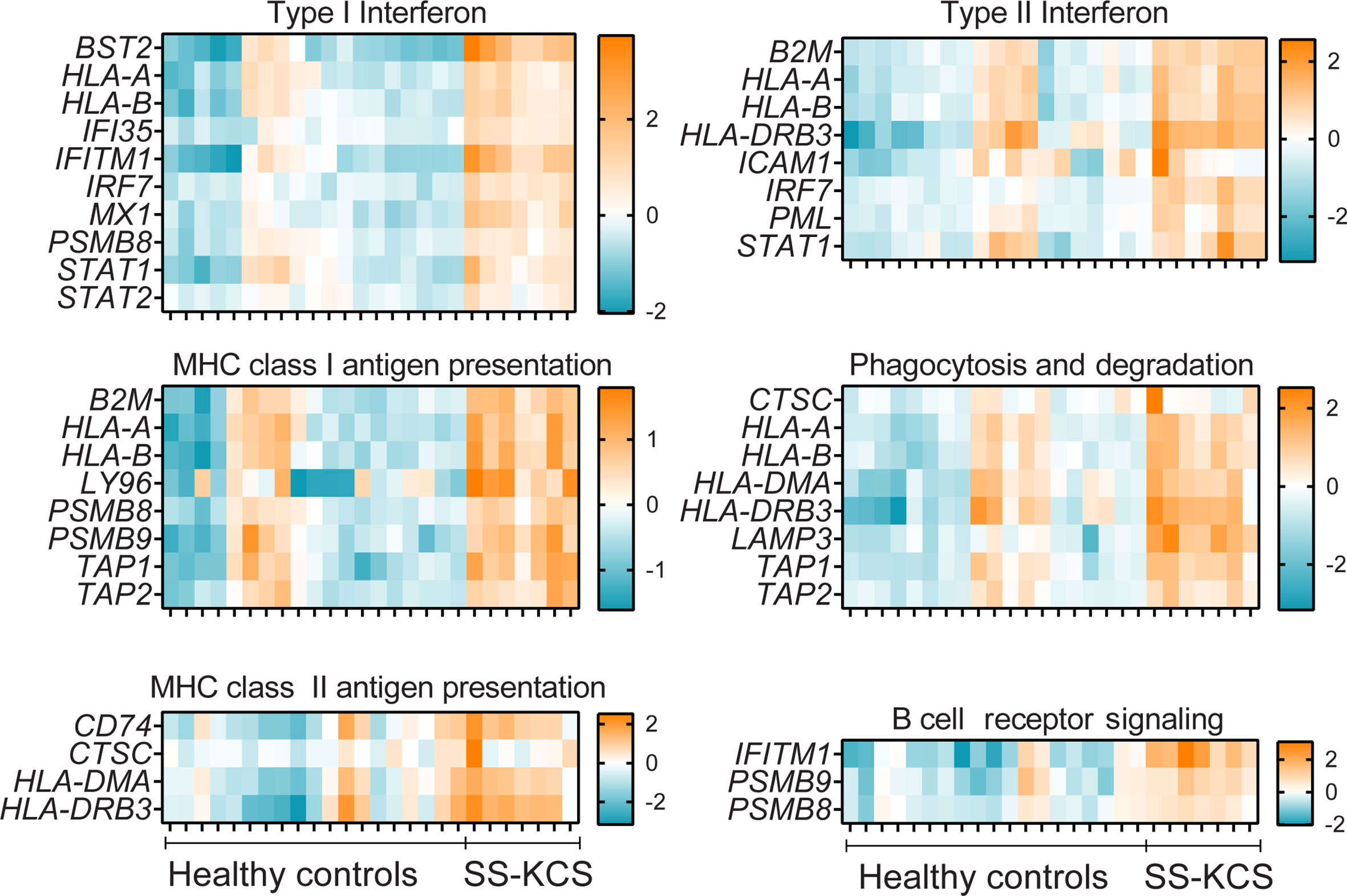
Figure 2 A heatmap of expression of immune pathways that were upregulated in SS KCS conjunctiva. Heatmaps show Type I and II Interferon, MHC class I and II antigen presentation, phagocytosis and degradation, and B cell receptor signaling pathways.
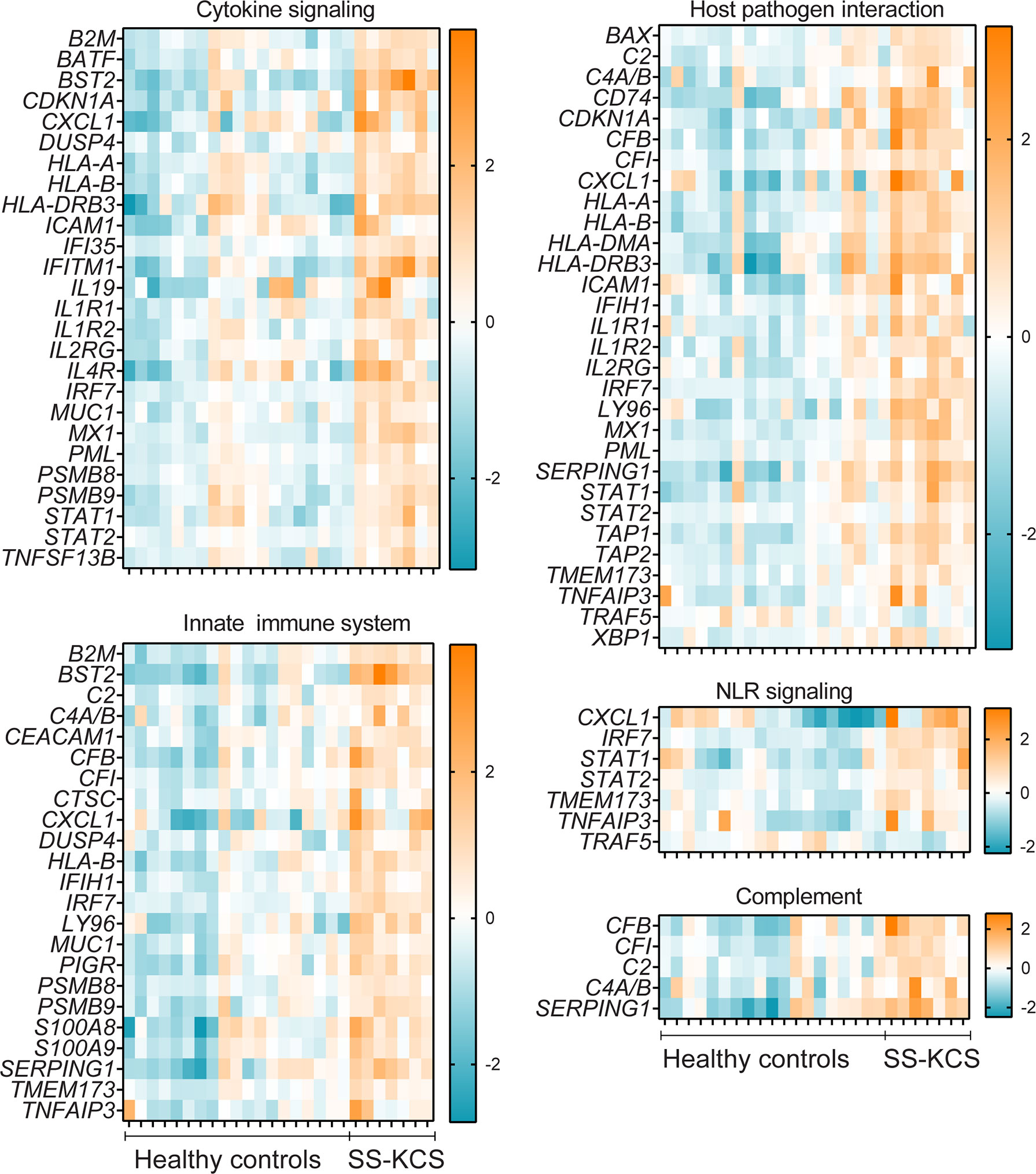
Figure 3 Heatmaps of expression of immune pathways with significant genes upregulated or downregulated gene (TRAF5) in SS KCS conjunctiva. Heatmaps show Cytokine signaling, Host-pathogen interaction, Innate immune system, Nod-like-receptor (NLR) signaling, and Complement pathways.
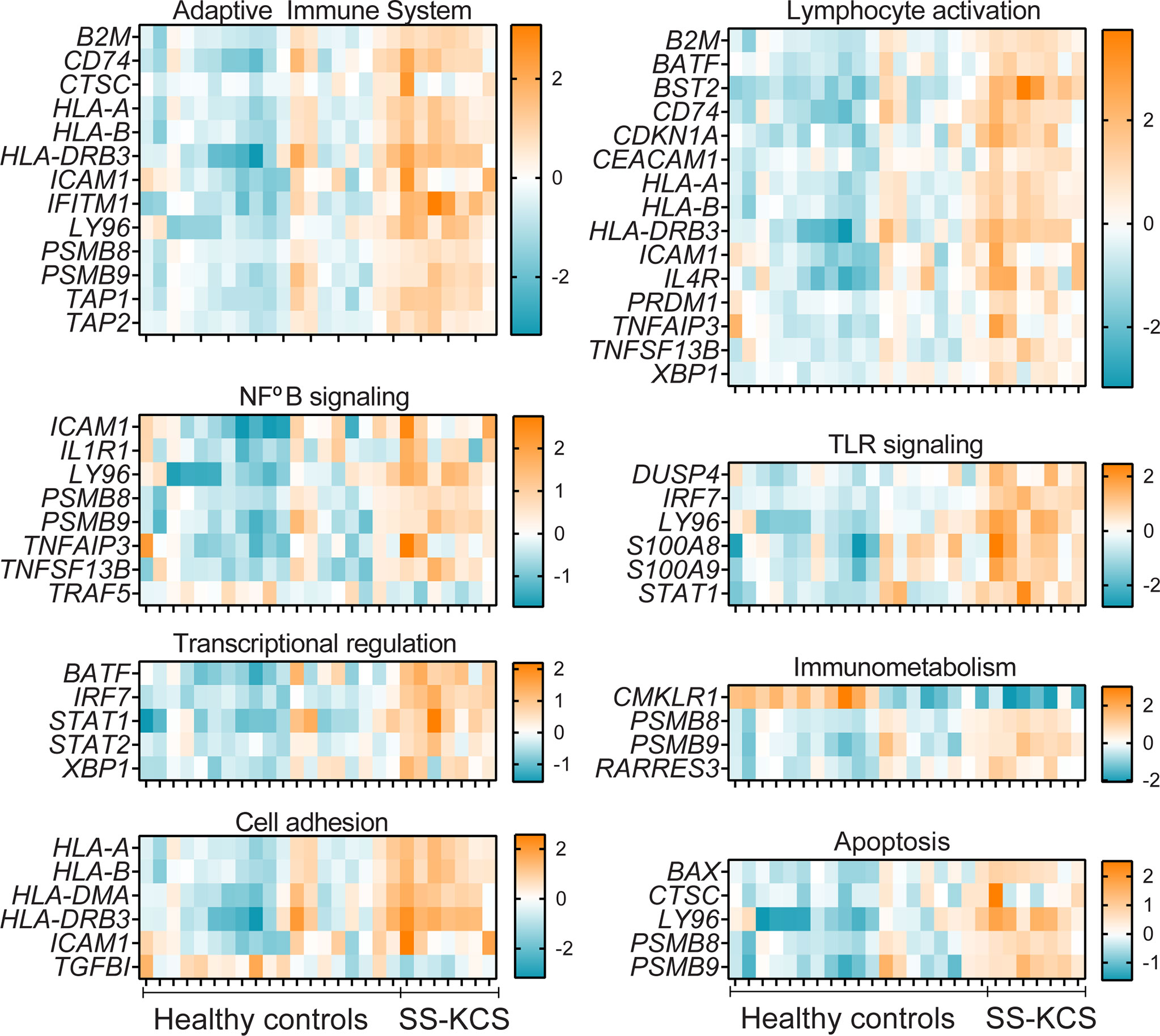
Figure 4 Heatmaps of Adaptive immune system, Lymphocyte activation, NFkB, TLR signaling, Transcriptional regulation, Immunometabolism, Cell adhesion, and apoptosis. The downregulated genes TRAF5, CMKLR1, and TGFBI, are found in the NFkB signaling, Immunometabolism, and Cell adhesion pathways.
Some DEGs overlapped in more than one pathway. For example, the proteasome subunits beta type-8 (PSMB8) and type-9 (PSMB9) were annotated in multiple pathways (type I interferon, MHC Class I antigen presentation, B cell receptor signaling, cytokine signaling, innate immune system, adaptive immune system, NFkB, immunometabolism, and apoptosis). Both PSMB8 and PSMB9 are integral parts of the immunoproteasome and, together with TAP1 and TAP2, are involved in the MHC class I presentation. TAP1 and TAP2 are transporters (-1,-2, respectively) and ATP binding cassette subfamily B members. Similar to PSMB8 and PSMB9, TAP1 and TAP2 were also annotated in multiple pathways (MHC class I presentation, phagocytosis, host-pathogen interaction, adaptive immune response, Figures 2–4). However, other DEGs were uniquely annotated. An example is LAMP3 (lysosome-associated membrane glycoprotein 3 or DC-LAMP), which was uniquely annotated in the phagocytosis pathway.
It has been demonstrated that SS patients have a Type I and II Interferon signature in their serum (35–38). Accordingly, we observed many DEGs involved in Type I and II interferon responses in the conjunctiva, such as BST2, IFITM1, IRF7, and MX1 (Figure 2). BST2 (bone marrow stromal antigen 2) was the most upregulated DEG in our results. Elevated BST2 levels are found in labial gland SS biopsies compared to controls and these levels correlated with rheumatoid factor and B2M serum levels (39). IFITM1 (interferon-induced transmembrane protein 1), IRF7 (interferon regulatory factor 7), and MX1 (interferon-induced GTP-binding protein Mx1) are also interferon-responsive genes (40) that were upregulated with greater significance in our results. IFITM1 was the second most upregulated DEGs in our results, and it is involved in Type I interferon and B cell receptor signaling, which also showed upregulation of PSMB8 and PSMB9.
The complement system participates in the lysis of infectious organisms by facilitating the binding of antibodies and phagocytic cells to microorganism membranes. It also promotes inflammation and immune clearance. There were five upregulated DEGs in our results, as shown in Figure 3 (CFB, CFI, C2, C4A/B, and SERPING1). CFB, a component of the alternative pathway of complement, is also increased in cerebrospinal fluid of patients with SS and fatigue (41). Both C2, C4A, and C4B proteins are known to participate in the generation of the classical complement pathway. The complement pathway is also part of the innate immune system, which showed many DEGs, such as CEACAM-1, CXCL1, S100A8, S100A9, PIGR, and MUC1. CEACAM-1 (carcinoembryonic antigen-related cell adhesion molecule 1) is a co-inhibitory molecule in mucosal immunology (42). CXCL1 is an antimicrobial protein that is a chemoattractant for neutrophils, which have been implicated in dry eye (43, 44). PIGR is a polymeric immunoglobulin receptor (also known as secretory component) that mediates polymeric IgA and IgM transport across mucosal epithelial cells. It is highly expressed in the lacrimal gland and tears (45) and, once secreted, can act as an antimicrobial agent on its own (46).
The involvement of the major inflammatory pathway NFkB in SS is well established in the literature (47, 48). Figure 3 shows the DEGs involved in this pathway: ICAM-1, IL1R1, Ly96, TNFAIP3 (encoding tumor necrosis factor, alpha-induced protein 3 or A20 protein), TNFSF13B, and downregulation of TRAF5. TNFSF13B encodes BAFF, B cell activation factor, a protein involved in B cell survival. BAFF SNPs and mutations have been shown in SS patients (49, 50). TRAF5, TNF receptor-associated factor 5, is a scavenger protein involved in TNF and NFkB activation and mediates IL-17-ACT1 interactions. TRAF5 SNPs showed an association with rheumatoid arthritis compared to control subjects (51). Genome-wide association studies have shown that TNFA1P3 is upregulated in Chinese SS patients (52). TNFAIP3, HLA-DMA, HLA-DR genes (HLA-DRB5 and HLA-DRB1), CDKN1B (cyclin-dependent kinase inhibitor 1B), IFITM1, NKFB1 (nuclear factor of kappa light polypeptide gene enhancer in B-cells 1), and IRF7 also elevated in labial salivary biopsies (53). Other small molecules and drugs that target the NFkB pathway in SS are undergoing clinical trials [reviewed in (54)].
Five transcription factors were upregulated and are shown in Figure 4: BATF, IRF7, STAT1, STAT2, XBP1. BATF (basic leucine zipper transcription factor, ATF-like) and is involved in Th17 differentiation. BATF-/- mice have decreased Th17 and ameliorated experimental autoimmune encephalitis (55), a model where Th17 cells are pathogenic. IL-17 has been implicated in dry eye and models of SS (56–61). IRF7 is a transcription factor that mediates IFN-α signaling. IRF7 has been found elevated in the SS patients, where it is thought to mediate the IFN signature (35–38, 62).
The immunometabolism pathway had two DEGS; CMKLR1 and RARRES3 (-7.1- and 1.6-fold change, respectively, Figure 4). CMKLR1 encodes chemerin chemokine-like receptor 1, also known as resolvin E1 receptor, a critical molecule in inflammation resolution (63). A CMKLR1 agonist antibody has shown promise in pre-clinical models of intestinal mucosal inflammation (64). RARRES3, retinoic acid receptor responder protein 3, is an intracellular retinoid receptor responder that belongs to RNA pattern recognition receptors. RARRES3 has been shown to interact with the immunoproteasome and upregulate IRF1 in breast cancer cells, and to stimulate the proliferation of keratinocytes (65).
TGBI and ICAM-1 were differentially modulated in our dataset and are annotated in the cell adhesion pathway (down and upregulated DEGs, respectively, Figure 4). Mutations of TGFBI (transforming growth factor beta-induced) have been implicated in cornea dystrophies (66). ICAM-1, intercellular adhesion molecule-1, is a critical molecule in the adaptive immune system that facilitates cell adhesion and promotes cell adhesion. ICAM-1 is elevated in SS serum and mouse models of dry eye disease (67–70). Lifitegrast, which blocks the immunological synapse of lymphocyte function-associated antigen/ICAM-1, is an FDA-approved eye drop for dry eye disease (71–73).
Pathway overrepresentation analysis using the InnateDB tool (28) showed that the “Antigen processing and presentation” pathway was over-represented in our results (10% of genes, P<0.00001, p-adjusted value), using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Genes involved in this pathway were B2M, CD74, HLA-A, HLA-B, HLA-DMA, TAP1, and TAP2. These same genes were annotated in Type I and II interferon responses, MHC class I and II antigen presentation, phagocytosis, and degradation in the NanoString® pathways (Figure 2). B2M (beta-2 microglobulin), involved in MHC Class I antigen presentation to CD8 T cells, has shown to be a hub for protein interaction in SS (74). HLA-A and HLA-B are major histocompatibility complexes that form a heterodimer with B2M.
These results demonstrate the pleiotropic abilities of specific genes, which participate in multiple pathways.
Correlation of Upregulated DEGs With Symptoms Severity Score and Signs
We performed Spearman correlation analysis between the normalized counts vs. OSDI, or TBUT, or corneal staining score, or conjunctival score to investigate which DEGs correlated with symptoms and signs of dry eye. No correlations between gene normalized counts and corneal fluorescein staining scores were observed in any of the genes in our results.
From the 54 DEGs, 29 DEGs correlated with either OSDI, TBUT, or conjunctival staining score (Table 4), and some DEGs correlated with more than one clinical sign. Only upregulated DEGs showed any significant correlation. Two representatives Spearman correlation graphs are shown in Figure 5A: 1) CD74, a critical step in MHC II pathway (75), inversely correlated with TBUT and positively with conjunctival staining score, 2) LAMP3, a lysosomal protein involved in autophagy (76), which correlated with OSDI, TBUT, and conjunctival staining score. Increased levels of CD74 have been previously reported in minor salivary gland SS biopsies (62, 77).
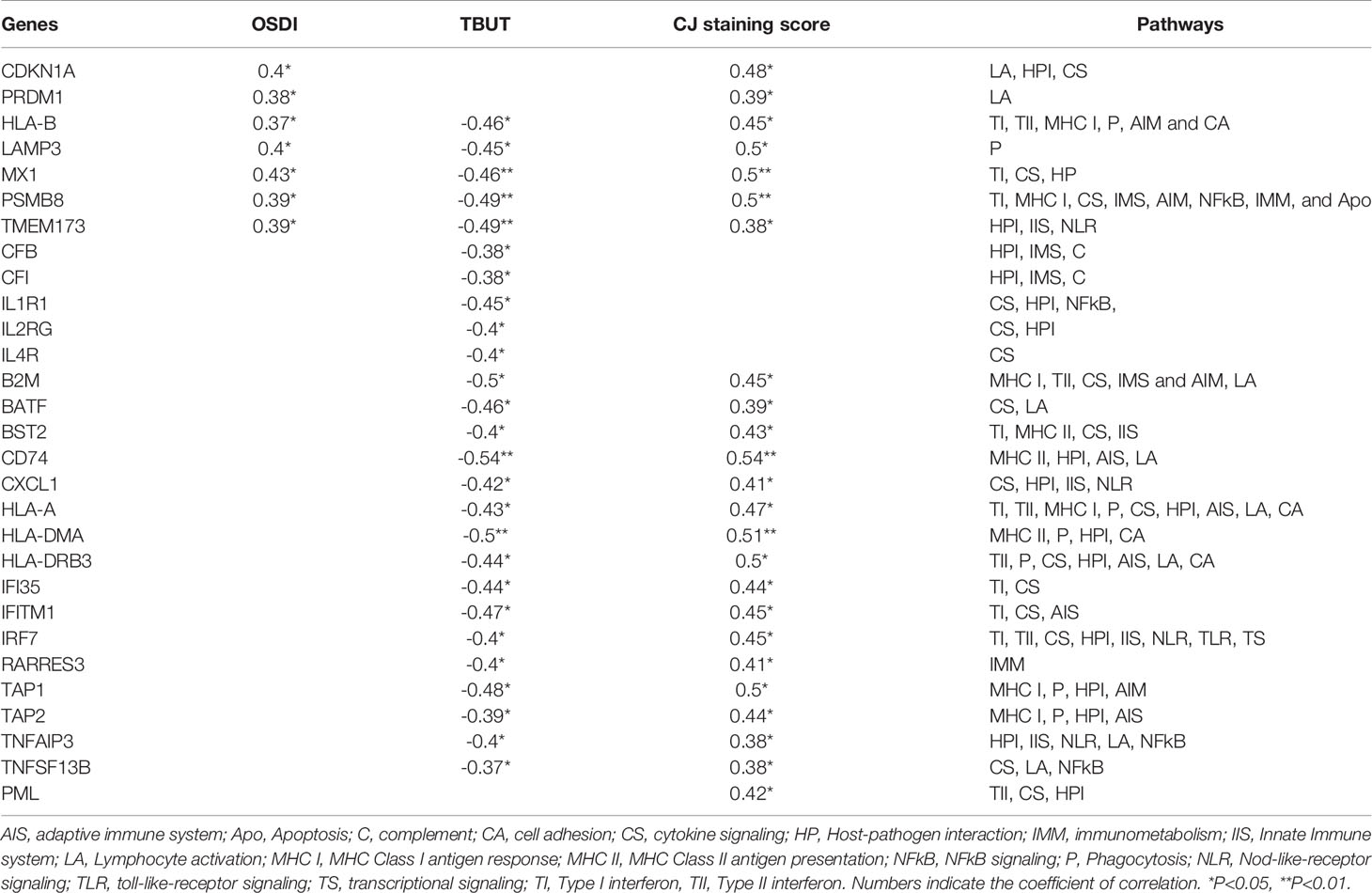
Table 4 Spearman correlations with clinical symptoms (OSDI), Tear-break-up time (TBUT), and conjunctival (CJ) staining score were calculated using GraphPad Prism software using clinical data from all subjects (19 healthy controls and 7 SS KCS subjects). Out of 54 DEGs, only 29 of the upregulated DEGs showed a correlation.
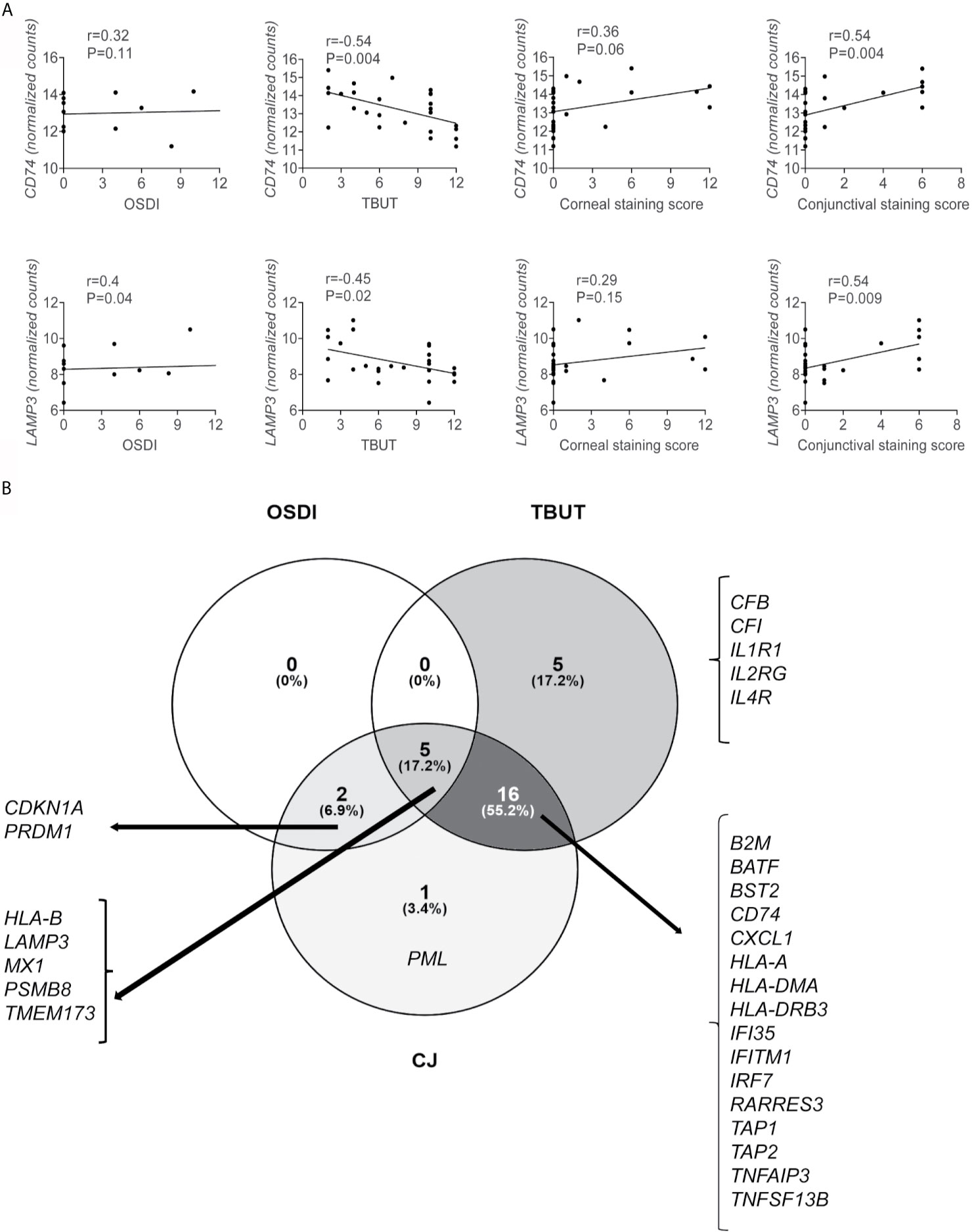
Figure 5 A subset of upregulated genes correlates with ocular symptoms and signs. Spearman correlations of clinical symptoms (OSDI), Tear-break-up time (TBUT), and conjunctival (CJ) staining score with up or downregulated DEGs were calculated using GraphPad Prism software using clinical data from all subjects (19 healthy controls and 7 SS KCS subjects). (A) Representative graph showing Spearman correlations of CD74 and LAMP3 vs. OSDI, TBUT, corneal staining score, and conjunctival staining score. Additional genes are shown in Table 3. (B) Venn diagram showing common and unique correlation of genes and OSDI, TBUT, and conjunctival staining score. R = coefficient of correlation. Please note that GraphPad Prism does not plot all dots if they overlap.
Next, we used the data in Table 4 to make Venn diagrams to identify the unique and shared correlations (Figure 5B). We identified that 26/29 DEGs showed an inverse correlation with TBUT; 5 of these 26 DEGs showed a positive correlation with OSDI, and 16/26 showed a positive correlation with conjunctival staining score. Five upregulated DEGs (CFB, CFI, IL1R1, IL2RG, IL4R) were uniquely inversely correlated with TBUT, while PML (promyelocytic leukemia protein) was uniquely correlated with conjunctival staining score (Table 4). PML is upregulated by STAT1 and participates in TNF-α and IFN-α induction of angiogenesis (78). Detection of PML in liver biopsies has been used as a diagnostic tool for autoimmune liver diseases, including primary biliary cirrhosis (79, 80). In addition to LAMP3, only an additional 6 DEGs correlated with OSDI (CDKN1A, PRDM1, HLA-B, MX1, PSMB8, and TMEM173). The disconnection between symptoms and signs in SS KCS is well described in the literature.
Discussion
The pathogenesis of SS has been a subject of intense investigation. The cornea and the conjunctiva develop pathological changes in SS KCS (81). Because it is not possible to obtain cells from the cornea noninvasively, we performed multiplex gene analysis of cells obtained from the conjunctiva using impression cytology using an Immunology panel. Impression cytology of conjunctiva has been used to investigate conjunctival goblet cell density, epithelial metaplasia, and obtain cells for flow cytometry (10, 82–87). It has also been used to investigate gene expression between different forms of dry eye disease and comparisons between healthy controls and SS KCS, as in our studies (6, 13, 14).
A careful investigation of pathways in our immunology panel confirmed some findings in literature but also unraveled new pathways and DEGs that had not been previously associated with SS. Pathways that have been previously associated with SS include Type I and Type II Interferon, MHC Class I and II presentation, Innate immune system, Phagocytosis, NFkB, B cell receptor to name a few. Increased expression of the interferon-inducible genes is consistent with our findings of increased IFN-γ expression in the conjunctiva of patients with SS KCS (1, 6) and that interferon-γ produced by NK and T cells increases chemokine expression and promotes goblet cell loss in conjunctival of mouse SS models (11, 88–92). Interestingly, the B cell receptor signaling pathway was upregulated in our results involving the conjunctiva. The involvement of B cells in SS is well established, as SS patients have B cell infiltration in salivary and lacrimal glands in humans and rodents. SS patients have an increased risk for non-Hodgkin lymphomas (93–96). The role of B cell receptor signaling in the conjunctiva of SS KCS patients has not been thoroughly investigated.
There were several upregulated genes in the host-pathogen, NLR signaling, and TLR signaling pathways. While no ocular surface pathogen has been identified in the pathogenesis of SS, there is a link between intestinal dysbiosis (with increased proteobacteria genera) and the severity of SS KCS (97, 98). Additionally, antibiotic-induced dysbiosis in mice increased the ocular surface inflammatory response to topically applied lipopolysaccharide (99) and led to a worse dry eye phenotype after desiccating stress (97). Upregulated genes in these pathways can heighten the TLR signaling (e.g., LY96) and innate and adaptive immune responses to microbial products that can be released into the circulation with intestinal dysbiosis. Several of the upregulated DEGs (IL-4R, IL-19, and DUSP) may have compensatory immunosuppressive functions. Th2 cytokines, such as IL-4 and IL-13, are usually implicated in allergic conjunctival inflammation (100), and IL-13 signaling through its heterodimeric receptor, which includes the IL-4R, stimulates goblet cell proliferation and mucin production in the conjunctiva (101, 102). Similarly, IL-19 is an anti-inflammatory cytokine that belongs to the IL-10 family and participates in the modulation of macrophage from M1 to M2, and DUSP4 negatively regulates phosphorylation of several MAPKs (103–105).
One pathway that was discovered in our panel was immunometabolism, with two unique DEGs (CMKLR1 and RARRES3). CMLKR1 also participates in the resolution of inflammation. Topically applied resolvin E1 analogs improved corneal staining, goblet cell density, and improved tear secretion while decreasing CD11b+ and CD4+ infiltration to the cornea in a murine model of dry eye disease (106, 107). A protective role for resolvin D has been shown in the salivary gland of SS animal models (108). Interestingly, RARRES1, another member of the same family, is elevated in SS labial salivary gland biopsy microarray studies (53). This is the first time that RARRES3 is associated with the conjunctiva of SS KCS patients.
Our results showing increased XBP-1 and STAT1 levels agree with the literature and with our previous publication that showed increased XBP-1 and GRP78 in the SS KCS conjunctiva compared to healthy controls (11). Increased levels of STAT1 have been previously reported in humans and animal models, also in the conjunctiva (13, 14, 62, 109). XBP-1 (X-box binding protein-1) participates in ER stress and UPR, decreasing salivary gland protein secretion (110). In the conjunctiva, XBP-1 participates in the IFN-γ-induced UPR by decreasing Muc5ac protein secretion from cultured conjunctival goblet cells (11).
Correlation analysis of the DEGs with clinical signs identified that 53% of DEGs correlated with at least one clinical sign. IFITM1, the second most upregulated DEG in our results, correlated with TBUT and conjunctival staining score. Elevated IFITM1 levels have been reported in the conjunctiva of dry eye patients (111). PSMB8 correlated with OSDI, TBUT, and conjunctival staining, while TAP1 and TAP2 correlated with TBUT and conjunctival staining. Epigenetic changes in PSMB8 and TAP1 have been reported in human labial gland biopsies in SS patients from the SICCA repository (112). Increased mRNA and protein expression in PSMB8 and PSMB9 in epithelial and immune cells of salivary glands have been reported (77, 113). LAMP3 was one of the only five DEGs correlated with three ocular parameters (OSDI, TBUT, and conjunctival staining score). LAMP3 is a marker of mature DCs in humans (114), and we and others have shown that the conjunctiva of SS KCS patients is rich in antigen-presenting cells (8, 33, 115, 116). LAMP3+ patients have increased levels of autoantibodies in their serum, and in-vitro, overexpression of LAMP3 induces apoptosis of minor salivary gland epithelial cells (117). Adenovirus transfection with LAMP3 in mice induces an SS-like phenotype accompanied by decreased salivary function (118). CDKN1A, cyclin-dependent kinase inhibitor 1, p21, correlated with OSDI and conjunctival staining score. Elevated senescence markers CDKN1A (119) and CDKN1B have been found in minor salivary gland SS biopsies (62). Genetic deletion of p21 in lupus-prone mice promoted apoptosis of long-lived T and B cells, decreasing autoimmunity (120). Our results are the first to show elevated CDKN1A, PSMB8, and PSMB9 in the conjunctiva of SS patients.
Our studies agree with Kessal and colleagues, who reported increased C2 and CFB (complement factor B) in the conjunctiva of SS KCS patients (13, 14). The complement system is activated at the ocular surface at low levels, where it is thought to participate in immunosurveillance (121). However, functional studies of the complement system in SS are primarily concentrated in the serum, where decreased complement levels have been found (122). Variants in C4 genes (which segregate with MHC II genes) increase the odds ratio of SS (123). A systematic review in the literature showed that a deficiency of C1-inhibitor (encoded by SERPING1) was present in autoimmune diseases (124). Our study adds CFI (complement factor I), C4/B, and SERPING1 to the list of upregulated genes in the complement cascade in the conjunctiva of SS KCS patients. The role of the complement cascade in the conjunctiva warrants further investigation.
Antigen presentation and processing, including MHC class I and class II pathways, were overrepresented in our results. Elevated HLA-A, HLA-B, HLA-DMA levels, together with other HLA-DR genes (HLA-DRB5 and HLA-DRB1), have been found in labial salivary biopsies in SS patients (53, 77). Genome-wide studies have shown that DEGs TAP1, TAP2, PSMB9, HLA-DR, HLA-DMA have been associated with SS in more than one study (52, 125, 126). LAMP3 was the third most upregulated DEG in our results, in agreement with previous studies that also reported elevated LAMP3 levels in SS (62, 117). MX1 has also been found elevated in other cohorts of SS patients, including conjunctiva (13, 14), labial salivary biopsies (53, 62, 127, 128), and serum (129). When comparing our DEGs with previous studies that performed comprehensive gene analysis of the conjunctiva, our studies identified four overlapping DEGs (MX1, C2, CFB, STAT1, STAT2) (13, 14). Compared to studies that performed gene analysis in labial gland biopsies from SS patients, our results identified some overlapping DEGs (62, 74, 77, 113, 127, 128). Unique DEGs not previously associated with SS were HLA-DRB3, CFI, CEACAM1, CXCL1, DUSP4, IL19, IL1R1, MUC1, SERPING1, S100A8, S100A9, PIGR, XBP1, although some proteins encoded by these DEGs have been associated with dry eye [XBP1, HL-A-DR (11, 33, 115)]. The majority of these DEGs participate in Innate immune response. Some of these proteins are secreted by epithelial cells and demonstrate the importance of the epithelial barrier in autoimmunity (130). Elevated levels of PIGR have been found in the SS saliva (131). MUC1, mucin 1, has been shown to regulate inflammation and TLR signaling after infections in mucosal sites (132). SNPs in the MUC1 gene have been identified in SS and dry eye subjects (133–135). Interestingly, 5/50 upregulated genes in the conjunctiva of a mouse model of SS were also included in our upregulated DEGs [MX1, ICAM1, S100A9, IRF7, BATF, (136)].
The strengths of this study include robust gene expression data via NanoString and comprehensive measures of signs and symptoms of dry eye disease to allow correlation of gene expression levels with different objective measures of the disease. The limitations of our study include the relatively limited set of genes evaluated by this NanoString® platform and the restriction to immune-related genes as compared to the more open approach of RNA sequencing, though the use of the NanoString platform allowed for a direct focus on immune-related genes, which was the purpose of the study.
Further studies are necessary to validate these DEGs identified in our study, which identified shared and unique DEGs and pathways in the conjunctiva of SS KCS patients. These findings highlight that some therapies targeting immune mediators might be efficacious for SS KCS. The ocular surface also has unique activated pathways that warrant further investigation if these DEGs can be used as valid biomarkers modulated by therapies such as cyclosporine A and short-term steroids. Investigation of immune pathways in the conjunctiva might yield novel therapeutic targets for SS KCS.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by Baylor College of Medicine Institutional Review Board. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
CP, CT-V, LS, SP, ZY, and RB were involved in the conception and design of the study. CP, CT-V, LS, SP, and ZY were involved in data acquisition. CP, CT-V, LS, SCP, ZY, and RB were involved in data analysis and interpretation. CP drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by NIH EY026893 (CP); NIH EY002520 (Core Grant for Vision Research Department of Ophthalmology); BCM Genomic & RNA Profiling Core GARP Core [P30 Digestive Disease Center Support Grant (NIDDKDK56338) and P30 Cancer Center Support Grant (NCICA125123), NIH S10 grant (1S10OD02346901)]. Further research support was provided by Research to Prevent Blindness Stein Award (RB), Research to Prevent Blindness (unrestricted grant to the Dept. Of Ophthalmology), The Hamill Foundation, and The Sid Richardson Foundation. Claudia M. Trujillo-Vargas received supplemental salary support from Facultad de Medicina, Universidad de Antioquia, UdeA, Medellin, Colombia.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work could not have been done without the voluntary participation of healthy subjects and SS patients. We acknowledge support from the Genomic and RNA Profiling Core at Baylor College of Medicine.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.702755/full#supplementary-material
References
1. Nakamura T, Nishida K, Dota A, Matsuki M, Yamanishi K, Kinoshita S. Elevated Expression of Transglutaminase 1 and Keratinization-Related Proteins in Conjunctiva in Severe Ocular Surface Disease. Invest Ophthalmol Vis Sci (2001) 42:549–56.
2. Kawasaki S, Kawamoto S, Yokoi N, Connon C, Minesaki Y, Kinoshita S, et al. Up-Regulated Gene Expression in the Conjunctival Epithelium of Patients With Sjogren’s Syndrome. Exp Eye Res (2003) 77:17–26. doi: 10.1016/S0014-4835(03)00087-3
3. Hirai N, Kawasaki S, Tanioka H, Connon CJ, Yamasaki K, Yokoi N, et al. Pathological Keratinisation in the Conjunctival Epithelium of Sjögren’s Syndrome. Exp Eye Res (2006) 82:371–8. doi: 10.1016/j.exer.2005.07.009
4. Pflugfelder SC, Stern ME. Biological Functions of Tear Film. Exp Eye Res (2020) 197:108115. doi: 10.1016/j.exer.2020.108115
5. Pflugfelder SC, de Paiva CS. The Pathophysiology of Dry Eye Disease: What We Know and Future Directions for Research. Ophthalmology (2017) 124:S4–s13. doi: 10.1016/j.ophtha.2017.07.010
6. Pflugfelder SC, De Paiva CS, Moore QL, Volpe EA, Li DQ, Gumus K, et al. Aqueous Tear Deficiency Increases Conjunctival Interferon-γ (IFN-γ) Expression and Goblet Cell Loss. Invest Ophthalmol Vis Sci (2015) 56:7545–50. doi: 10.1167/iovs.15-17627
7. de Paiva CS, Pflugfelder SC, Ng SM, Akpek EK. Topical Cyclosporine A Therapy for Dry Eye Syndrome. Cochrane Database Syst Rev (2019) 9:Cd010051. doi: 10.1002/14651858.CD010051.pub2
8. Pflugfelder SC, Bian F, Gumus K, Farley W, Stern ME, De Paiva CS. Severity of Sjogren’s Syndrome Keratoconjunctivitis Sicca Increases With Increased Percentage of Conjunctival Antigen-Presenting Cells. Int J Mol Sci (2018) 19. doi: 10.3390/ijms19092760
9. Ogawa Y, Shimizu E, Tsubota K. Interferons and Dry Eye in Sjogren’s Syndrome. Int J Mol Sci (2018) 19(11):3548. doi: 10.3390/ijms19113548
10. Tsubota K, Fukagawa K, Fujihara T, Shimmura S, Saito I, Saito K, et al. Regulation of Human Leukocyte Antigen Expression in Human Conjunctival Epithelium. Invest Ophthalmol Vis Sci (1999) 40:28–34.
11. Coursey TG, Henriksson JT, Barbosa FL, de Paiva CS, Pflugfelder SC. Interferon-Gamma-Induced Unfolded Protein Response in Conjunctival Goblet Cells as a Cause of Mucin Deficiency in Sjogren Syndrome. Am J Pathol (2016) 186:1547–58. doi: 10.1016/j.ajpath.2016.02.004
12. Garcia-Posadas L, Hodges RR, Li D, Shatos MA, Storr-Paulsen T, Diebold Y, et al. Interaction of IFN-Gamma With Cholinergic Agonists to Modulate Rat and Human Goblet Cell Function. Mucosal Immunol (2016) 9:206–17. doi: 10.1038/mi.2015.53
13. Kessal K, Liang H, Rabut G, Daull P, Garrigue JS, Docquier M, et al. Conjunctival Inflammatory Gene Expression Profiling in Dry Eye Disease: Correlations With HLA-DRA and HLA-Drb1. Front Immunol (2018) 9:2271. doi: 10.3389/fimmu.2018.02271
14. Liang H, Kessal K, Rabut G, Daull P, Garrigue JS, Melik Parsadaniantz S, et al. Correlation of Clinical Symptoms and Signs With Conjunctival Gene Expression in Primary Sjögren Syndrome Dry Eye Patients. Ocul Surf (2019) 17:516–25. doi: 10.1016/j.jtos.2019.03.005
15. Shiboski CH, Shiboski SC, Seror R, Criswell LA, Labetoulle M, Lietman TM, et al. 2016 American College of Rheumatology/European League Against Rheumatism Classification Criteria for Primary Sjogren’s Syndrome: A Consensus and Data-Driven Methodology Involving Three International Patient Cohorts. Ann Rheumatol Dis (2017) 76:9–16. doi: 10.1136/annrheumdis-2016-210571
16. Rao K, Farley WJ, Pflugfelder SC. Association Between High Tear Epidermal Growth Factor Levels and Corneal Subepithelial Fibrosis in Dry Eye Conditions. Invest Ophthalmol Vis Sci (2010) 51:844–9. doi: 10.1167/iovs.09-3875
17. Tung CI, Perin AF, Gumus K, Pflugfelder SC. Tear Meniscus Dimensions in Tear Dysfunction and Their Correlation With Clinical Parameters. Am J Ophthalmol (2014) 157:301–10.e301. doi: 10.1016/j.ajo.2013.09.024
18. Pflugfelder SC, Geerling G, Kinoshita S, Wilson C. Management and Therapy of Dry Eye Disease: Report of the Management and Therapy Subcommittee of the International Dry Eye WorkShop (2007). Ocul Surf (2007) 5:163–78. doi: 10.1016/S1542-0124(12)70085-X
19. Perkins JR, Dawes JM, McMahon SB, Bennett DL, Orengo C, Kohl M. ReadqPCR and NormqPCR: R Packages for the Reading, Quality Checking and Normalisation of RT-qPCR Quantification Cycle (Cq) Data. BMC Genomics (2012) 13:296. doi: 10.1186/1471-2164-13-296
21. Mitchell AL, Attwood TK, Babbitt PC, Blum M, Bork P, Bridge A, et al. InterPro in 2019: Improving Coverage, Classification and Access to Protein Sequence Annotations. Nucleic Acids Res (2019) 47:D351–d360. doi: 10.1093/nar/gky1100
22. Geer LY, Marchler-Bauer A, Geer RC, Han L, He J, He S, et al. The NCBI BioSystems Database. Nucleic Acids Res (2010) 38:D492–496. doi: 10.1093/nar/gkp858
23. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc Natl Acad Sci U S A (2005) 102:15545–50. doi: 10.1073/pnas.0506580102
24. Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular Signatures Database (MSigDB) 3.0. Bioinformatics (2011) 27:1739–40. doi: 10.1093/bioinformatics/btr260
25. Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res (2018) 46:D649–d655. doi: 10.1093/nar/gkx1132
26. Slenter DN, Kutmon M, Hanspers K, Riutta A, Windsor J, Nunes N, et al. WikiPathways: A Multifaceted Pathway Database Bridging Metabolomics to Other Omics Research. Nucleic Acids Res (2018) 46:D661–d667. doi: 10.1093/nar/gkx1064
27. Oliveros JC. Venny. An Interactive Tool for Comparing Lists With Venn’s Diagrams(2007). Available at: https://bioinfogp.cnb.csic.es/tools/venny/index.html.
28. Breuer K, Foroushani AK, Laird MR, Chen C, Sribnaia A, Lo R, et al. InnateDB: Systems Biology of Innate Immunity and Beyond–Recent Updates and Continuing Curation. Nucleic Acids Res (2013) 41:D1228–1233. doi: 10.1093/nar/gks1147
29. Raphael M, Bellefqih S, Piette JC, Le HP, Debre P, Chomette G. Conjunctival Biopsy in Sjogren’s Syndrome: Correlations Between Histological and Immunohistochemical Features. Histopathology (1988) 13:191–202. doi: 10.1111/j.1365-2559.1988.tb02024.x
30. Pflugfelder SC, Huang AJW, Schuchovski PT, Pereira IC, Tseng SCG. Conjunctival Cytological Features of Primary Sjogren Syndrome. Ophthalmology (1990) 97:985–91. doi: 10.1016/S0161-6420(90)32478-8
31. Aguilar AJ, Fonseca L, Croxatto JO. Sjogren’s Syndrome: A Comparative Study of Impression Cytology of the Conjunctiva and Buccal Mucosa, and Salivary Gland Biopsy. Cornea (1991) 10:203–6. doi: 10.1097/00003226-199105000-00003
32. Stern ME, Gao J, Schwalb TA, Ngo M, Tieu DD, Chan CC, et al. Conjunctival T-Cell Subpopulations in Sjogren’s and non-Sjogren’s Patients With Dry Eye. Invest Ophthalmol Vis Sci (2002) 43:2609–14.
33. Epstein SP, Gadaria-Rathod N, Wei Y, Maguire MG, Asbell PA. HLA-DR Expression as a Biomarker of Inflammation for Multicenter Clinical Trials of Ocular Surface Disease. Exp Eye Res (2013) 111:95–104. doi: 10.1016/j.exer.2013.03.018
34. Nicolle P, Liang H, Reboussin E, Rabut G, Warcoin E, Brignole-Baudouin F, et al. Proinflammatory Markers, Chemokines, and Enkephalin in Patients Suffering From Dry Eye Disease. Int J Mol Sci (2018) 19(4):1221. doi: 10.3390/ijms19041221
35. Peck AB, Nguyen CQ. Transcriptome Analysis of the Interferon-Signature Defining the Autoimmune Process of Sjogren’s Syndrome. Scand J Immunol (2012) 76:237–45. doi: 10.1111/j.1365-3083.2012.02749.x
36. Hall JC, Casciola-Rosen L, Berger AE, Kapsogeorgou EK, Cheadle C, Tzioufas AG, et al. Precise Probes of Type II Interferon Activity Define the Origin of Interferon Signatures in Target Tissues in Rheumatic Diseases. Proc Natl Acad Sci U S A (2012) 109:17609–14. doi: 10.1073/pnas.1209724109
37. Mavragani CP, Crow MK. Activation of the Type I Interferon Pathway in Primary Sjogren’s Syndrome. J Autoimmun (2010) 35:225–31. doi: 10.1016/j.jaut.2010.06.012
38. Emamian ES, Leon JM, Lessard CJ, Grandits M, Baechler EC, Gaffney PM, et al. Peripheral Blood Gene Expression Profiling in Sjögren’s Syndrome. Genes Immun (2009) 10:285–96. doi: 10.1038/gene.2009.20
39. Chen C, Shi H, Wang B, Cao N, Yu C, Zheng L. Aberrant Expression of the Innate Restriction Factor Bone Marrow Stromal Antigen-2 in Primary Sjögren’s Syndrome. J Craniomaxillofac Surg (2018) 46:1899–904. doi: 10.1016/j.jcms.2018.08.008
40. Gestermann N, Mekinian A, Comets E, Loiseau P, Puechal X, Hachulla E, et al. STAT4 is a Confirmed Genetic Risk Factor for Sjögren’s Syndrome and Could be Involved in Type 1 Interferon Pathway Signaling. Genes Immun (2010) 11:432–8. doi: 10.1038/gene.2010.29
41. Larssen E, Brede C, Hjelle A, Tjensvoll AB, Norheim KB, Bårdsen K, et al. Fatigue in Primary Sjögren’s Syndrome: A Proteomic Pilot Study of Cerebrospinal Fluid. SAGE Open Med (2019) 7:2050312119850390. doi: 10.1177/2050312119850390
42. Nagaishi T, Iijima H, Nakajima A, Chen D, Blumberg RS. Role of CEACAM1 as a Regulator of T Cells. Ann N Y Acad Sci (2006) 1072:155–75. doi: 10.1196/annals.1326.004
43. Gao Y, Min K, Zhang Y, Su J, Greenwood M, Gronert K. Female-Specific Downregulation of Tissue Polymorphonuclear Neutrophils Drives Impaired Regulatory T Cell and Amplified Effector T Cell Responses in Autoimmune Dry Eye Disease. J Immunol (2015) 195:3086–99. doi: 10.4049/jimmunol.1500610
44. Tibrewal S, Ivanir Y, Sarkar J, Nayeb-Hashemi N, Bouchard CS, Kim E, et al. Hyperosmolar Stress Induces Neutrophil Extracellular Trap Formation: Implications for Dry Eye Disease. Invest Ophthalmol Visual Sci (2014) 55:7961–9. doi: 10.1167/iovs.14-15332
45. Knop E, Knop N, Claus P. Local Production of Secretory IgA in the Eye-Associated Lymphoid Tissue (EALT) of the Normal Human Ocular Surface. Invest Ophthalmol Vis Sci (2008) 49:2322–9. doi: 10.1167/iovs.07-0691
46. Perrier C, Sprenger N, Corthésy B. Glycans on Secretory Component Participate in Innate Protection Against Mucosal Pathogens. J Biol Chem (2006) 281:14280–7. doi: 10.1074/jbc.M512958200
47. Vakrakou AG, Polyzos A, Kapsogeorgou EK, Thanos D, Manoussakis MN. Impaired Anti-Inflammatory Activity of Pparγ in the Salivary Epithelia of Sjögren’s Syndrome Patients Imposed by Intrinsic NF-κb Activation. J Autoimmun (2018) 86:62–74. doi: 10.1016/j.jaut.2017.09.007
48. Okuma A, Hoshino K, Ohba T, Fukushi S, Aiba S, Akira S, et al. Enhanced Apoptosis by Disruption of the STAT3-IkappaB-Zeta Signaling Pathway in Epithelial Cells Induces Sjogren’s Syndrome-Like Autoimmune Disease. Immunity (2013) 38:450–60. doi: 10.1016/j.immuni.2012.11.016
49. Nossent JC, Lester S, Zahra D, Mackay CR, Rischmueller M. Polymorphism in the 5’ Regulatory Region of the B-Lymphocyte Activating Factor Gene is Associated With the Ro/La Autoantibody Response and Serum BAFF Levels in Primary Sjogren’s Syndrome. Rheumatol (Oxford) (2008) 47:1311–6. doi: 10.1093/rheumatology/ken246
50. Papageorgiou A, Mavragani CP, Nezos A, Zintzaras E, Quartuccio L, De Vita S, et al. A BAFF Receptor His159Tyr Mutation in Sjögren’s Syndrome-Related Lymphoproliferation. Arthritis Rheumatol (Hoboken NJ) (2015) 67:2732–41. doi: 10.1002/art.39231
51. Potter C, Eyre S, Cope A, Worthington J, Barton A. Investigation of Association Between the TRAF Family Genes and RA Susceptibility. Ann Rheumatol Dis (2007) 66:1322–6. doi: 10.1136/ard.2006.065706
52. Li Y, Zhang K, Chen H, Sun F, Xu J, Wu Z, et al. A Genome-Wide Association Study in Han Chinese Identifies a Susceptibility Locus for Primary Sjögren’s Syndrome at 7q11.23. Nat Genet (2013) 45:1361–5. doi: 10.1038/ng.2779
53. Pérez P, Anaya JM, Aguilera S, Urzúa U, Munroe D, Molina C, et al. Gene Expression and Chromosomal Location for Susceptibility to Sjögren’s Syndrome. J Autoimmun (2009) 33:99–108. doi: 10.1016/j.jaut.2009.05.001
54. Sisto M, Ribatti D, Lisi S. Understanding the Complexity of Sjogren’s Syndrome: Remarkable Progress in Elucidating NF-kappaB Mechanisms. J Clin Med (2020) 9(9):2821. doi: 10.3390/jcm9092821
55. Schraml BU, Hildner K, Ise W, Lee WL, Smith WA, Solomon B, et al. The AP-1 Transcription Factor Batf Controls T(H)17 Differentiation. Nature (2009) 460:405–9. doi: 10.1038/nature08114
56. Zhang X, Volpe EA, Gandhi NB, Schaumburg CS, Siemasko KF, Pangelinan SB, et al. NK Cells Promote Th-17 Mediated Corneal Barrier Disruption in Dry Eye. PLoS One (2012) 7:e36822. doi: 10.1371/journal.pone.0036822
57. de Paiva CS, Chotikavanich S, Pangelinan SB, Pitcher JD III, Fang B, Zheng X, et al. IL-17 Disrupts Corneal Barrier Following Desiccating Stress. Mucosal Immunol (2009) 2:243–53. doi: 10.1038/mi.2009.5
58. Dohlman TH, Chauhan SK, Kodati S, Hua J, Chen Y, Omoto M, et al. The CCR6/CCL20 Axis Mediates Th17 Cell Migration to the Ocular Surface in Dry Eye Disease. Invest Ophthalmol Vis Sci (2013) 54:4081–91. doi: 10.1167/iovs.12-11216
59. Chauhan SK, El AJ, Ecoiffier T, Goyal S, Zhang Q, Saban DR, et al. Autoimmunity in Dry Eye is Due to Resistance of Th17 to Treg Suppression. J Immunol (2009) 182:1247–52. doi: 10.4049/jimmunol.182.3.1247
60. Nguyen CQ, Yin H, Lee BH, Carcamo WC, Chiorini JA, Peck AB. Pathogenic Effect of Interleukin-17A in Induction of Sjogren’s Syndrome-Like Disease Using Adenovirus-Mediated Gene Transfer. Arthritis Res Ther (2010) 12:R220. doi: 10.1186/ar3207
61. Nguyen CQ, Yin H, Lee BH, Chiorini JA, Peck AB. IL17: Potential Therapeutic Target in Sjogren’s Syndrome Using Adenovirus-Mediated Gene Transfer. Lab Invest (2011) 91:54–62. doi: 10.1038/labinvest.2010.164
62. Oyelakin A, Horeth E, Song EC, Min S, Che M, Marzullo B, et al. Transcriptomic and Network Analysis of Minor Salivary Glands of Patients With Primary Sjögren’s Syndrome. Front Immunol (2020) 11:606268. doi: 10.3389/fimmu.2020.606268
63. Pirault J, Bäck M. Lipoxin and Resolvin Receptors Transducing the Resolution of Inflammation in Cardiovascular Disease. Front Pharmacol (2018) 9:1273. doi: 10.3389/fphar.2018.01273
64. Trilleaud C, Gauttier V, Biteau K, Girault I, Belarif L, Mary C, et al. Agonist Anti-ChemR23 mAb Reduces Tissue Neutrophil Accumulation and Triggers Chronic Inflammation Resolution. Sci Adv (2021) 7:eabd1453. doi: 10.1126/sciadv.abd1453
65. Anderson AM, Kalimutho M, Harten S, Nanayakkara DM, Khanna KK, Ragan MA. The Metastasis Suppressor RARRES3 as an Endogenous Inhibitor of the Immunoproteasome Expression in Breast Cancer Cells. Sci Rep (2017) 7:39873. doi: 10.1038/srep39873
66. Han KE, Choi ,S-i, Kim T-i, Maeng Y-S, Stulting RD, Ji YW, et al. Pathogenesis and Treatments of TGFBI Corneal Dystrophies. Prog Retin Eye Res (2016) 50:67–88. doi: 10.1016/j.preteyeres.2015.11.002
67. Zinkevičienė A, Dumalakienė I, Mieliauskaitė D, Vilienė R, Narkevičiūtė I, Girkontaitė I. sICAM-1 as Potential Additional Parameter in the Discrimination of the Sjögren Syndrome and non-Autoimmune Sicca Syndrome: A Pilot Study. Clin Rheumatol (2019) 38:2803–9. doi: 10.1007/s10067-019-04621-2
68. Gao J, Morgan G, Tieu D, Schwalb TA, Luo JY, Wheeler LA, et al. ICAM-1 Expression Predisposes Ocular Tissues to Immune-Based Inflammation in Dry Eye Patients and Sjogrens Syndrome-Like MRL/lpr Mice. Exp Eye Res (2004) 78:823–35. doi: 10.1016/j.exer.2003.10.024
69. Hayashi Y, Haneji N, Yanagi K, Higashiyama H, Yagita H, Hamano H. Prevention of Adoptive Transfer of Murine Sjogren’s Syndrome Into Severe Combined Immunodeficient (SCID) Mice by Antibodies Against Intercellular Adhesion Molecule-1 (ICAM-1) and Lymphocyte Function-Associated Antigen-1 (LFA-1). Clin Exp Immunol (1995) 102:360–7. doi: 10.1111/j.1365-2249.1995.tb03790.x
70. Hsueh PY, Ju Y, Vega A, Edman MC, MacKay JA, Hamm-Alvarez SF. A Multivalent ICAM-1 Binding Nanoparticle Which Inhibits ICAM-1 and LFA-1 Interaction Represents a New Tool for the Investigation of Autoimmune-Mediated Dry Eye. Int J Mol Sci (2020) 21(8):2758. doi: 10.3390/ijms21082758
71. Semba CP, Gadek TR. Development of Lifitegrast: A Novel T-Cell Inhibitor for the Treatment of Dry Eye Disease. Clin Ophthalmol (2016) 10:1083–94. doi: 10.2147/OPTH.S110557
72. Pflugfelder SC, Stern M, Zhang S, Shojaei A. LFA-1/ICAM-1 Interaction as a Therapeutic Target in Dry Eye Disease. J Ocul Pharmacol Ther (2017) 33:5–12. doi: 10.1089/jop.2016.0105
73. Guimaraes de Souza R, Yu Z, Stern ME, Pflugfelder SC, de Paiva CS. Suppression of Th1-Mediated Keratoconjunctivitis Sicca by Lifitegrast. J Ocul Pharmacol Ther (2018) 34:543–9. doi: 10.1089/jop.2018.0047
74. Zhang L, Xu P, Wang X, Zhang Z, Zhao W, Li Z, et al. Identification of Differentially Expressed Genes in Primary Sjögren’s Syndrome. J Cell Biochem (2019) 120:17368–77. doi: 10.1002/jcb.29001
75. Geneve L, Chemali M, Desjardins M, Labrecque N, Thibodeau J. Human Invariant Chain Isoform P35 Restores Thymic Selection and Antigen Presentation in CD74-Deficient Mice. Immunol Cell Biol (2012) 90:896–902. doi: 10.1038/icb.2012.27
76. Cuervo AM. Autophagy and Aging: Keeping That Old Broom Working. Trends Genet (2008) 24:604–12. doi: 10.1016/j.tig.2008.10.002
77. Hjelmervik TO, Petersen K, Jonassen I, Jonsson R, Bolstad AI. Gene Expression Profiling of Minor Salivary Glands Clearly Distinguishes Primary Sjögren’s Syndrome Patients From Healthy Control Subjects. Arthritis Rheumatol (2005) 52:1534–44. doi: 10.1002/art.21006
78. Cheng X, Liu Y, Chu H, Kao H-Y. Promyelocytic Leukemia Protein (PML) Regulates Endothelial Cell Network Formation and Migration in Response to Tumor Necrosis Factor α (Tnfα) and Interferon α (Ifnα). J Biol Chem (2012) 287:23356–67. doi: 10.1074/jbc.M112.340505
79. Papamichalis PA, Zachou K, Papamichali RA, Ioannou M, Gatselis NK, Dalekos GN, et al. Promyelocytic Leukemia Antigen Expression: A Histological Marker for Primary Biliary Cholangitis Diagnosis? J Transl Int Med (2021) 9:43–51. doi: 10.2478/jtim-2021-0008
80. Andre C, Guillemin MC, Zhu J, Koken MH, Quignon F, Herve L, et al. The PML and PML/RARalpha Domains: From Autoimmunity to Molecular Oncology and From Retinoic Acid to Arsenic. Exp Cell Res (1996) 229:253–60. doi: 10.1006/excr.1996.0368
81. Kunert KS, Tisdale AS, Stern ME, Smith JA, Gipson IK. Analysis of Topical Cyclosporine Treatment of Patients With Dry Eye Syndrome: Effect on Conjunctival Lymphocytes. Arch Ophthalmol (2000) 118:1489–96. doi: 10.1001/archopht.118.11.1489
82. Pisella PJ, Brignole F, Debbasch C, Lozato PA, Creuzot-Garcher C, Bara J, et al. Flow Cytometric Analysis of Conjunctival Epithelium in Ocular Rosacea and Keratoconjunctivitis Sicca. Ophthalmology (2000) 107:1841–9. doi: 10.1016/S0161-6420(00)00347-X
83. Brignole F, Pisella PJ, Goldschild M, De Saint JM, Goguel A, Baudouin C. Flow Cytometric Analysis of Inflammatory Markers in Conjunctival Epithelial Cells of Patients With Dry Eyes. Invest Ophthalmol Vis Sci (2000) 41:1356–63.
84. Tsubota K, Fujihara T, Saito K, Takeuchi T. Conjunctival Epithelium Expression of HLA-DR in Dry Eye Patients. Ophthalmologica (1999) 213:16–9. doi: 10.1159/000027387
85. Reinoso R, Calonge M, Castellanos E, Martino M, Fernandez I, Stern ME, et al. Differential Cell Proliferation, Apoptosis, and Immune Response in Healthy and Evaporative-Type Dry Eye Conjunctival Epithelia. Invest Ophthalmol Vis Sci (2011) 52:4819–28. doi: 10.1167/iovs.10-6073
86. Tseng SCG. Staging of Conjunctival Squamous Metaplasia by Impression Cytology. Ophthalmology (1985) 92:728–33. doi: 10.1016/S0161-6420(85)33967-2
87. Wittpenn JR, Tseng SCG, Sommer A. Detection of Early Xerophthalmia by Impression Cytology. Arch Ophthalmol (1986) 104:237–9. doi: 10.1001/archopht.1986.01050140091027
88. de Paiva CS, Villarreal AL, Corrales RM, Rahman HT, Chang VY, Farley WJ, et al. Dry Eye-Induced Conjunctival Epithelial Squamous Metaplasia Is Modulated by Interferon-{Gamma}. Invest Ophthalmol Vis Sci (2007) 48:2553–60. doi: 10.1167/iovs.07-0069
89. Zhang X, de Paiva CS, Su Z, Volpe EA, Li DQ, Pflugfelder SC. Topical Interferon-Gamma Neutralization Prevents Conjunctival Goblet Cell Loss in Experimental Murine Dry Eye. Exp Eye Res (2014) 118:117–24. doi: 10.1016/j.exer.2013.11.011
90. Zhang X, Chen W, de Paiva CS, Corrales RM, Volpe EA, McClellan AJ, et al. Interferon-Gamma Exacerbates Dry Eye-Induced Apoptosis in Conjunctiva Through Dual Apoptotic Pathways. Invest Ophthalmol Vis Sci (2011) 52:6279–85. doi: 10.1167/iovs.10-7081
91. Chen Y, Chauhan SK, Saban DR, Sadrai Z, Okanobo A, Dana R. Interferon-{Gamma}-Secreting NK Cells Promote Induction of Dry Eye Disease. J Leuk Biol (2011) 89:965–72. doi: 10.1189/jlb.1110611
92. Volpe EA, Henriksson JT, Wang C, Barbosa FL, Zaheer M, Zhang X, et al. Interferon-Gamma Deficiency Protects Against Aging-Related Goblet Cell Loss. Oncotarget (2016) 7:64605–6461. doi: 10.18632/oncotarget.11872
93. Meng W, Li Y, Xue E, Satoh M, Peck AB, Cohen PL, et al. B-Cell Tolerance Defects in the B6.Aec1/2 Mouse Model of Sjogren’s Syndrome. J Clin Immunol (2012) 32:551–64. doi: 10.1007/s10875-012-9663-6
94. Tzioufas AG, Tatouli IP, Moutsopoulos HM. Autoantibodies in Sjogren’s Syndrome: Clinical Presentation and Regulatory Mechanisms. Presse Med (2012) 41:e451–460. doi: 10.1016/j.lpm.2012.05.022
95. Ogawa A, Ogawa Y, Mukai S, Shimizu E, Kuwana M, Kawakami Y, et al. Cluster of Differentiation 30 Expression in Lacrimal Gland and Conjunctival Tissues in Patients With Sjögren’s Syndrome: Case Series. Med (Baltimore) (2019) 98:e16390. doi: 10.1097/MD.0000000000016390
96. Pelegrino FS, Volpe EA, Gandhi NB, Li DQ, Pflugfelder SC, de Paiva CS. Deletion of Interferon-Gamma Delays Onset and Severity of Dacryoadenitis in CD25KO Mice. Arthritis Res Ther (2012) 14:R234. doi: 10.1186/ar4077
97. de Paiva CS, Jones DB, Stern ME, Bian F, Moore QL, Corbiere S, et al. Altered Mucosal Microbiome Diversity and Disease Severity in Sjogren Syndrome. Sci Rep (2016) 6:23561–71. doi: 10.1038/srep23561
98. Moon J, Choi SH, Yoon CH, Kim MK. Gut Dysbiosis is Prevailing in Sjogren’s Syndrome and is Related to Dry Eye Severity. PloS One (2020) 15:e0229029. doi: 10.1371/journal.pone.0229029
99. Wang C, Schaefer L, Bian F, Yu Z, Pflugfelder SC, Britton RA, et al. Dysbiosis Modulates Ocular Surface Inflammatory Response to Liposaccharide. Invest Ophthalmol Vis Sci (2019) 60:4224–33. doi: 10.1167/iovs.19-27939
100. Chen X, Deng R, Chi W, Hua X, Lu F, Bian F, et al. IL-27 Signaling Deficiency Develops Th17-Enhanced Th2-Dominant Inflammation in Murine Allergic Conjunctivitis Model. Allergy (2018) 74(5):910–21. doi: 10.1111/all.13691
101. Tukler Henriksson J, Coursey TG, Corry DB, De Paiva CS, Pflugfelder SC. IL-13 Stimulates Proliferation and Expression of Mucin and Immunomodulatory Genes in Cultured Conjunctival Goblet Cells. Invest Ophthalmol Vis Sci (2015) 56:4186–97. doi: 10.1167/iovs.14-15496
102. de Paiva CS, Raince JK, McClellan AJ, Shanmugam KP, Pangelinan SB, Volpe EA, et al. AndHomeostatic Control of Conjunctival Mucosal Goblet Cells by NKT-Derived IL-13. Mucosal Immunol (2011) 4:397–408. doi: 10.1038/mi.2010.82
103. Leigh T, Scalia RG, Autieri MV. Resolution of Inflammation in Immune and Nonimmune Cells by Interleukin-19. Am J Physiol Cell Physiol (2020) 319:C457–c464. doi: 10.1152/ajpcell.00247.2020
104. Ikezumi Y, Kondoh T, Matsumoto Y, Kumagai N, Kaneko M, Hasegawa H, et al. Steroid Treatment Promotes an M2 Anti-Inflammatory Macrophage Phenotype in Childhood Lupus Nephritis. Pediatr Nephrol (2021) 36:349–59. doi: 10.1007/s00467-020-04734-w
105. Chuang HC, Tan TH. MAP4K Family Kinases and DUSP Family Phosphatases in T-Cell Signaling and Systemic Lupus Erythematosus. Cells (2019) 8(11):1433. doi: 10.3390/cells8111433
106. de Paiva CS, Schwartz E, Gjorstrup P, Pflugfelder SC. Resolvin E1 (RX-10001) Reduces Corneal Epithelial Barrier Disruption and Protects Against Goblet Cell Loss in a Murine Model of Dry Eye. Cornea (2012) 31:1299–303. doi: 10.1097/ICO.0b013e31823f789e
107. Li N, He J, Schwartz CE, Gjorstrup P, Bazan HE. Resolvin E1 Improves Tear Production and Decreases Inflammation in a Dry Eye Mouse Model. J Ocul Pharmacol Ther (2010) 26:431–9. doi: 10.1089/jop.2010.0019
108. Parashar K, Schulte F, Hardt M, Baker OJ. Sex-Mediated Elevation of the Specialized Pro-Resolving Lipid Mediator Levels in a Sjögren’s Syndrome Mouse Model. FASEB J (2020) 34:7733–44. doi: 10.1096/fj.201902196R
109. Horvath S, Nazmul-Hossain ANM, Pollard RPE, Kroese FGM, Vissink A, Kallenberg CGM, et al. Systems Analysis of Primary Sjögren’s Syndrome Pathogenesis in Salivary Glands Identifies Shared Pathways in Human and a Mouse Model. Arthritis Res Ther (2012) 14:R238–8. doi: 10.1186/ar4081
110. Katsiougiannis S, Tenta R, Skopouli ,FN. Endoplasmic Reticulum Stress Causes Autophagy and Apoptosis Leading to Cellular Redistribution of the Autoantigens Ro/Sjögren’s Syndrome-Related Antigen A (SSA) and La/SSB in Salivary Gland Epithelial Cells. Clin Exp Immunol (2015) 181:244–52. doi: 10.1111/cei.12638
111. Mantelli F, Schaffer L, Dana R, Head SR, Argüeso P. Glycogene Expression in Conjunctiva of Patients With Dry Eye: Downregulation of Notch Signaling. Invest Ophthalmol Vis Sci (2009) 50:2666–72. doi: 10.1167/iovs.08-2734
112. Cole MB, Quach H, Quach D, Baker A, Taylor KE, Barcellos LF, et al. Epigenetic Signatures of Salivary Gland Inflammation in Sjögren’s Syndrome. Arthritis Rheumatol (Hoboken NJ) (2016) 68:2936–44. doi: 10.1002/art.39792
113. Egerer T, Martinez-Gamboa L, Dankof A, Stuhlmüller B, Dörner T, Krenn V, et al. Tissue-Specific Up-Regulation of the Proteasome Subunit Beta5i (LMP7) in Sjögren’s Syndrome. Arthritis Rheumatol (2006) 54:1501–8. doi: 10.1002/art.21782
114. Salaun B, de Saint-Vis B, Clair-Moninot V, Pin JJ, Barthélemy-Dubois C, Kissenpfennig A, et al. Cloning and Characterization of the Mouse Homologue of the Human Dendritic Cell Maturation Marker CD208/DC-LAMP. Eur J Immunol (2003) 33:2619–29. doi: 10.1002/eji.200324175
115. Baudouin C, Brignole F, Pisella PJ, De Jean MS, Goguel A. Flow Cytometric Analysis of the Inflammatory Marker HLA DR in Dry Eye Syndrome: Results From 12 Months of Randomized Treatment With Topical Cyclosporin a. Adv Exp Med Biol (2002) 506:761–9. doi: 10.1007/978-1-4615-0717-8_107
116. Brignole F, Pisella PJ, De Saint JM, Goldschild M, Goguel A, Baudouin C. Flow Cytometric Analysis of Inflammatory Markers in KCS: 6-Month Treatment With Topical Cyclosporin a. Invest Ophthalmol Vis Sci (2001) 42:90–5.
117. Tanaka T, Warner BM, Odani T, Ji Y, Mo Y-Q, Nakamura H, et al. LAMP3 Induces Apoptosis and Autoantigen Release in Sjögren’s Syndrome Patients. Sci Rep (2020) 10:15169. doi: 10.1038/s41598-020-71669-5
118. Nakamura H, Tanaka T, Pranzatelli T, Ji Y, Yin H, Perez P, et al. Lysosome-Associated Membrane Protein 3 Misexpression in Salivary Glands Induces a Sjögren’s Syndrome-Like Phenotype in Mice. Ann Rheumatol Dis (2021). annrheumdis-2020-219649. doi: 10.1136/annrheumdis-2020-219649
119. Marthandan S, Baumgart M, Priebe S, Groth M, Schaer J, Kaether C, et al. Conserved Senescence Associated Genes and Pathways in Primary Human Fibroblasts Detected by RNA-Seq. PloS One (2016) 11:e0154531. doi: 10.1371/journal.pone.0154531
120. Lawson BR, Baccala R, Song J, Croft M, Kono DH, Theofilopoulos AN. Deficiency of the Cyclin Kinase Inhibitor P21(WAF-1/CIP-1) Promotes Apoptosis of Activated/Memory T Cells and Inhibits Spontaneous Systemic Autoimmunity. J Exp Med (2004) 199:547–57. doi: 10.1084/jem.20031685
121. Bora NS, Jha P, Bora PS. The Role of Complement in Ocular Pathology. Semin Immunopathol (2008) 30:85–95. doi: 10.1007/s00281-008-0110-y
122. Ramos-Casals M, Brito-Zerón P, Yagüe J, Akasbi M, Bautista R, Ruano M, et al. Hypocomplementaemia as an Immunological Marker of Morbidity and Mortality in Patients With Primary Sjogren’s Syndrome. Rheumatol (Oxford) (2005) 44:89–94. doi: 10.1093/rheumatology/keh407
123. Kamitaki N, Sekar A, Handsaker RE, de Rivera H, Tooley K, Morris DL, et al. Complement Genes Contribute Sex-Biased Vulnerability in Diverse Disorders. Nature (2020) 582:577–81. doi: 10.1038/s41586-020-2277-x
124. Levy D, Craig T, Keith PK, Krishnarajah G, Beckerman R, Prusty S. Co-Occurrence Between C1 Esterase Inhibitor Deficiency and Autoimmune Disease: A Systematic Literature Review. Allergy Asthma Clin Immunol (2020) 16:41. doi: 10.1186/s13223-020-00437-x
125. Kumagai S, Kanagawa S, Morinobu A, Takada M, Nakamura K, Sugai S, et al. Association of a New Allele of the TAP2 Gene, TAP2*Bky2 (Val577), With Susceptibility to Sjögren’s Syndrome. Arthritis Rheumatol (1997) 40:1685–92. doi: 10.1002/art.1780400919
126. Taylor KE, Wong Q, Levine DM, McHugh C, Laurie C, Doheny K, et al. Genome-Wide Association Analysis Reveals Genetic Heterogeneity of Sjögren’s Syndrome According to Ancestry. Arthritis Rheumatol (Hoboken NJ) (2017) 69:1294–305. doi: 10.1002/art.40040
127. Min HK, Moon SJ, Park KS, Kim KJ. Integrated Systems Analysis of Salivary Gland Transcriptomics Reveals Key Molecular Networks in Sjögren’s Syndrome. Arthritis Res Ther (2019) 21:294. doi: 10.1186/s13075-019-2082-9
128. Liu Z, Li F, Pan A, Xue H, Jiang S, Zhu C, et al. Elevated CCL19/CCR7 Expression During the Disease Process of Primary Sjögren’s Syndrome. Front Immunol (2019) 10:795. doi: 10.3389/fimmu.2019.00795
129. Aydemir YG, Kocakusak A. The Evaluation of the Myxovirus Resistance 1 Protein in Serum and Saliva to Monitor Disease Activation in Primary Sjögren’s Syndrome. Clinics (Sao Paulo) (2019) 74:e631–1. doi: 10.6061/clinics/2019/e631
130. Akdis CA. Does the Epithelial Barrier Hypothesis Explain the Increase in Allergy, Autoimmunity and Other Chronic Conditions? Nat Rev Immunol (2021). doi: 10.1038/s41577-021-00538-7
131. Hu S, Wang J, Meijer J, Ieong S, Xie Y, Yu T, et al. Salivary Proteomic and Genomic Biomarkers for Primary Sjögren’s Syndrome. Arthritis Rheumatol (2007) 56:3588–600. doi: 10.1002/art.22954
132. Dhar P, McAuley J. The Role of the Cell Surface Mucin MUC1 as a Barrier to Infection and Regulator of Inflammation. Front Cell Infect Microbiol (2019) 9:117. doi: 10.3389/fcimb.2019.00117
133. Imbert Y, Foulks GN, Brennan MD, Jumblatt MM, John G, Shah HA, et al. MUC1 and Estrogen Receptor Alpha Gene Polymorphisms in Dry Eye Patients. Exp Eye Res (2009) 88:334–8. doi: 10.1016/j.exer.2008.05.019
134. Imbert Y, Darling DS, Jumblatt MM, Foulks GN, Couzin EG, Steele PS, et al. MUC1 Splice Variants in Human Ocular Surface Tissues: Possible Differences Between Dry Eye Patients and Normal Controls. Exp Eye Res (2006) 83:493–501. doi: 10.1016/j.exer.2006.01.031
135. Shah HA, Imbert Y, Foulks GN, Brennan MD, Jumblatt MM, John G, et al. MUC1 Gene Polymorphisms in Patients With Dry Eye Disease. Invest Ophthalmol Vis Sci (2008) 49:5306–6. doi: 10.1016/j.exer.2008.05.019
Keywords: Sjögren syndrome, dry eye, gene expression, conjunctiva, immune pathways
Citation: de Paiva CS, Trujillo-Vargas CM, Schaefer L, Yu Z, Britton RA and Pflugfelder SC (2021) Differentially Expressed Gene Pathways in the Conjunctiva of Sjögren Syndrome Keratoconjunctivitis Sicca. Front. Immunol. 12:702755. doi: 10.3389/fimmu.2021.702755
Received: 29 April 2021; Accepted: 01 July 2021;
Published: 19 July 2021.
Edited by:
Alan Baer, Johns Hopkins University, United StatesReviewed by:
Aleksandra Antovic, Karolinska Institutet, SwedenScott M. Lieberman, The University of Iowa, United States
Copyright © 2021 de Paiva, Trujillo-Vargas, Schaefer, Yu, Britton and Pflugfelder. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cintia S. de Paiva, Y2ludGlhZHBAYmNtLmVkdQ==
 Cintia S. de Paiva
Cintia S. de Paiva Claudia M. Trujillo-Vargas
Claudia M. Trujillo-Vargas Laura Schaefer2
Laura Schaefer2 Robert A. Britton
Robert A. Britton Stephen C. Pflugfelder
Stephen C. Pflugfelder