
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 30 June 2021
Sec. Mucosal Immunity
Volume 12 - 2021 | https://doi.org/10.3389/fimmu.2021.702378
This article is part of the Research TopicImmunoregulation at Mucosal SurfacesView all 36 articles
The field of microbiome research has developed rapidly over the past decades and has become a topic of major interest to basic, preclinical, and clinical research, the pharmaceutical industry as well as the general public. The microbiome is a complex and diverse ecosystem and defined as the collection of all host-associated microorganisms and their genes. It is acquired through vertical transmission and environmental exposure and includes microbes of all kingdoms: bacteria, archaea, prokaryotic and eukaryotic viruses, fungi, protozoa, and the meiofauna. These microorganisms co-evolved with their respective hosts over millions of years, thereby establishing a mutually beneficial, symbiotic relationship on all epithelial barriers. Thus, the microbiome plays a pivotal role in virtually every aspect of mammalian physiology, particularly in the development, homeostasis, and function of the immune system. Consequently, the combination of the host genome and the microbial genome, together referred to as the metagenome, largely drives the mammalian phenotype. So far, the majority of studies have unilaterally focused on the gastrointestinal bacterial microbiota. However, recent work illustrating the impact of viruses, fungi, and protozoa on host immunity urges us towards a holistic view of the mammalian microbiome and the appreciation for its non-bacterial kingdoms. In addition, the importance of microbiota on epithelial barriers other than the gut as well as their systemic effects via microbially-derived biologically active compounds is increasingly recognized. Here, we want to provide a brief but comprehensive overview of the most important findings and the current knowledge on how microbes of all kingdoms and microbial niches shape local and systemic immunity in health and disease.
Multicellular organisms are not only composed of their individual cells, but also of the microorganisms that inhabit ecological niches such as the gastrointestinal tract, the skin, the respiratory tract, and the genitourinary system. These microbes do not just passively colonize their hosts, they rather established a symbiotic relationship on all epithelial barrier sites during millions of years of co-evolution. The host provides its resident microbes with a habitat as well as nutrients, and in return, they help in digesting food, providing vitamins, and protecting their host from diseases (1–4).
Microbiota is defined as the ecological community of all host-associated microbes within a particular niche, whereas the collection of those microorganisms and their genomes is known as the microbiome (5). The host genome is the building plan for mammalian organisms, thereby creating well-defined ecological niches for microbes. Nonetheless, the mammalian phenotype itself within a given environment is rather driven by the combination of the host genome and the microbiome together referred to as the metagenome. Therefore, the microbiome plays a pivotal role in virtually every aspect of mammalian physiology, particularly in the development, maturation, homeostasis, and ultimately the function of the immune system (6–11). This mutually beneficial coexistence is also acknowledged in the holobiont theory or the metaorganism concept, emphasizing the intimate relationship between the host and its inhabiting microorganisms (12–14).
Over the last decades, microbiome research has primarily focused on the gastrointestinal bacterial community and its effects on the host health and disease. This line of research provided insights into mechanisms of the bidirectional crosstalk between the host and its bacterial subtenants. However, many disease states are paralleled with changes in the bacterial microbiome and vice versa, leaving the question of whether the microbiome is responsible for the disorder or whether the disease influences microbial composition, placing researchers into a challenging chicken and egg situation. In this context, it is pivotal to acknowledge that the microbiome is a complex and diverse ecosystem comprising microorganisms of all kingdoms, namely bacteria, archaea, eukaryotes like protozoa and fungi, and even multicellular eukaryotes such as helminths (15). In addition, eukaryotic viruses, phages, and endogenous retroviral elements are also crucial members of the mammalian microbiome (16). These microbial communities are present not only in the gut, but at all epithelial barrier sites, and the overall complexity is likely potentiated by trans-kingdom interactions between these commensals (17). As explained above, this complex ecosystem together with the host genome shapes the metaorganism and its physiology through multifactorial and nonlinear interactions. Thus, experimental models of translational research that are aiming to identify causal relationships and to unravel underlying mechanisms must be capable of navigating this complexity. Therefore, focusing on the gut bacteria, merely one component of the entire microbiome, makes it exceedingly difficult to fully decipher complex microbiota-related physiological mechanisms. Instead, a full description of the microbiome at distinct epithelial barrier sites and a more comprehensive view of mammalian organisms as holobionts might be a key to successfully conduct mechanistic studies, thereby opening up a promising window of opportunity to solve the chicken and egg question.
Ever since, most studies investigating the microbial influence on the host’s physiology focused on bacteria of the gastrointestinal tract, particularly highly abundant bacteria. The primary reason for this bacteriocentric approach was a major technological restriction: pioneer studies on mammalian microbiota almost exclusively relied on culture-based approaches that were more commonly available for bacterial organisms, specifically gastrointestinal microbes. Moreover, the gut incorporates high microbial biomass and is thus much easier to study than niches with low microbial biomass like the skin, the respiratory tract, or the genitourinary system.
Technological advances, especially the common use of sequencing methods and a continuous reduction in the associated costs alongside novel computational bioinformatics tools, have enabled the scientific community to explore the bacterial microbiome as well as other microbial kingdoms in more detail, unveiling previously unappreciated microorganisms (18, 19). However, the focus on bacterial organisms of the gut remained vastly unchanged. Thus, the commonly used methods were optimized for detecting bacterial DNA, which causes problems when focusing on viruses, archaea, or eukaryotes like fungi. When using 16S rRNA gene profiling, microorganisms other than bacteria are vastly neglected as the primers fail to cover multi-kingdom diversity. Nowadays, the probably least biased approach to study highly diverse microbial communities is shotgun metagenomic sequencing. However, the predominance of bacteria leads to an unfavorable bacteria-to-archaea or bacteria-to-fungi ratio, and thus a high sequencing depth is needed to detect comparably rare fungal or archaeal signatures (20, 21). The differences in the cellular structures between bacteria, archaea, or eukaryotes are another technological hurdle in analyzing microbial DNA. For example, commercial DNA extraction kits contain lysozyme that cuts bacterial peptidoglycan but not archaeal pseudopeptidoglycan (21). Likewise, the efficiency of isolating fungal DNA significantly differs between several isolation methods for microbial DNA (22). Another challenge comes when analyzing viral communities, as the genome of many viruses is not composed of DNA but RNA, so that the study design requires the inclusion of RNA in its analyses (23). Thus, the research community urgently needs tailored protocols to enrich, extract and study non-bacterial kingdoms such as the archaeome, the virome, the mycobiome, and other eukaryotes like protozoa and the meiofauna.
Another crucial component to ensure future success in microbiome research will be the availability of high-quality databases. Although current databases for bacteria are already refined and accurate, they certainly need further optimization. This is substantially different for other microbial kingdoms, as reliable and well-annotated databases are urgently needed for analyses at the species level, but often not available (21, 24). Many sequences included in databases for fungal communities are annotated as uncultured or are incorrect at the species level (25). This is further complicated by the fact that sexual and asexual forms of the very same fungal species are frequently classified as different taxa (26, 27). In virome studies, only around half of the sequences can be aligned to reference databases, indicating an enormous amount of viral “dark matter” that needs further exploration (23, 28). Hence, the scientific community imperatively needs further optimization of databases, particularly as regards non-bacterial kingdoms. This could be achieved by studies shedding more light on the “dark matter” still obscuring vast parts of the mammalian microbiome, thereby empowering mechanistic studies on host-microbe interactions.
The gastrointestinal tract certainly is the most intensively studied microbial niche and considering its impact on local and systemic immunity, the most remarkable epithelial barrier site of mammalian organisms. Further, the bacterial microbiome of the gastrointestinal tract is the so far best characterized microbial community that mainly comprises anaerobic bacteria of the phyla Bacteroidetes and Firmicutes (29). It plays an essential role in the development, maturation, aging, and homeostasis of the host’s immune system and thus the orchestration and functionality of host immune responses in steady state and during various inflammatory events (Figure 1) (30, 31).
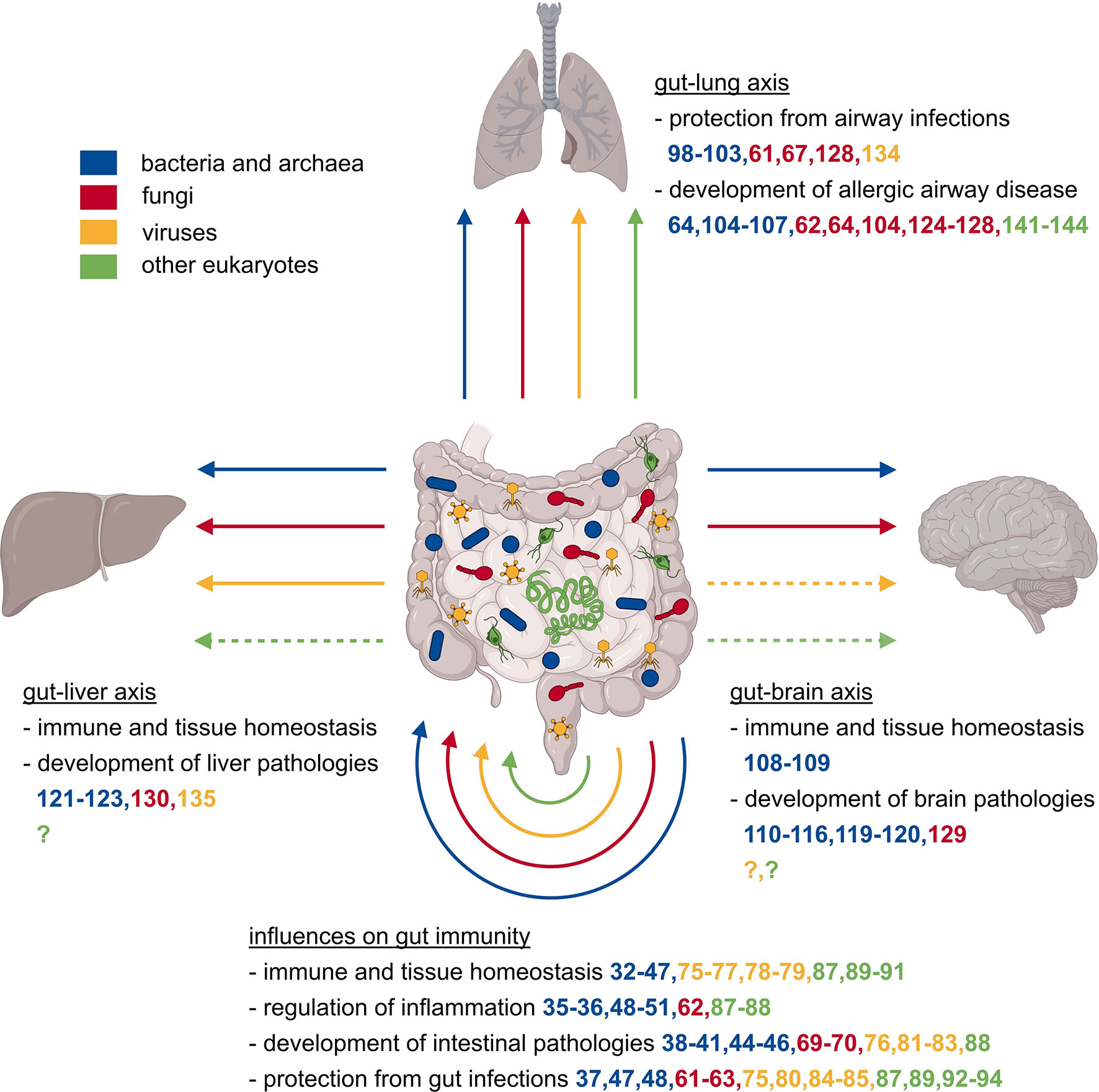
Figure 1 Local and systemic effects of the gut microbiome on the immune system. The mammalian gut is populated by a plethora of microbes comprising representatives of all kingdoms, specifically bacteria, archaea, eukaryotes like fungi, protozoa, and helminths, as well as eukaryotic viruses and bacterial phages. The collectivity of all gut-associated microorganisms affects the development, maturation, homeostasis, and consequently functionality of the immune system, which in turn has local and systemic consequences. Changes in the microbial composition might affect the protection against or acceleration of inflammatory diseases, regulation of intestinal pathologies, and the protection against intestinal infections. Microbiota influence not only the gut tissue itself, but also other organs like the liver, lung, and brain via the so-called gut-liver, gut-lung, or gut-brain axes. These distant organs are also affected positively or negatively in their homeostasis, defense against infections, or development of organ-specific pathologies.
Several studies showed that certain gut bacteria strongly shape local immunity at the corresponding barrier site. An example are segmented filamentous bacteria (SFB), gram-positive bacteria that grow close to the intestinal epithelium and that induced the formation of T helper 17 (TH17) cells, which are crucial for tissue homeostasis at barrier sites (32). However, not only commensal SFB but also other bacteria like murine enteropathogenic Citrobacter rodentium, a human pathogenic Escherichia coli strain, or the human symbiont Bifidobacterium adolescentis, were able to induce an accumulation of TH17 cells (33, 34). Adhesion to the intestinal epithelial cells seems to be a key factor for gut TH17 cell differentiation, as this is a shared feature of the above-mentioned bacteria, and adhesive-defect mutants did not induce TH17 cell development (33, 34). However, further studies suggested that TH17 cells induced by different bacteria display divergent inflammatory phenotypes (35). SFB are not only important in TH17 responses but also seem to induce T follicular helper cell development in the Peyer’s patches, which leads to the aggravation of inflammation in an autoimmune arthritis model (36). Not only bacteria themselves, but also their products such as metabolites affect immune development and functionality. For instance, microbiota-derived short-chain fatty acids (SCFA) are needed for antigen-activated CD8+ T cells to develop into long-living memory cells (37). SCFA produced by Clostridia of the Cluster IV, XIVa, and XVIII promote the accumulation of interleukin (IL)-10 producing regulatory T (Treg) cells in the colon and attenuate pathology in a colitis model (38–40). A bacterial polysaccharide of Bacteroides fragilis also induced IL-10 producing Treg cells, which protect against experimental colitis induced by Helicobacter hepaticus (41). Bile acids, further microbial-derived metabolites, play a protective role in the intestine by interacting with the farnesoid X receptor (FXR) (42, 43). Additionally, recent studies suggested bacterially produced secondary bile acids to regulate the differentiation of colonic Treg cells, which have a positive impact on dextran sulfate sodium (DSS)-induced colitis (44–46). Not only the gastrointestinal T cell compartment but also B cell development in the intestinal mucosa is affected by commensal colonization (47). Other microbial metabolites, like aryl hydrocarbon receptor (AhR) ligands, produced by tryptophan-metabolizing microbes, induce the production of IL-22, thereby providing resistance to Candida colonization and gut inflammation (48). Additionally, various microbial molecules can influence macrophage polarization towards either a pro-inflammatory or an anti-inflammatory state, depending on the stimulating metabolites and the context (49–51). Gut-resident bacteria and their metabolites thus have a substantial effect on gastrointestinal health and disease.
Over 50 years ago, the first methanogenic archaeal microorganism was isolated from human feces, ever since we know that the gastrointestinal tract is also inhabited by representatives of the archaeal domain (52). Further methanogenic and to a lesser extent also non-methanogenic archaeal species were detected in the human gut, supporting initial findings that methanogenic archaea are commensal inhabitants of the human gastrointestinal tract (53). Some methanoarchaeal species showed immunogenic properties (54, 55), however, the interaction between archaea and the immune system, as well as the involvement of archaea in human physiology remains blurry (21). A recent study suggested trans-kingdom interactions between bacteria of the Christensenellaceae family and archaea of the Methanobrevibacter family to be associated with a lean body mass index in humans (56). Interestingly, no human pathogenic archaeal species are known today, and there is only little to no data available on how archaea might affect the physiology of their respective hosts (21). Consequently, a substantial amount of research is required to further unravel the relationship between the host and its archaeal inhabitants.
The gut mycobiome is a diverse community mainly comprising Saccharomyces, especially Candida species, as well as yeasts of the family Dipodcaceae, and occasionally also Malassezia and Cladosporium (57–59). Comparable to the gut bacterial microbiome, gastrointestinal fungi were also shown to strongly shape local immunity at the corresponding barrier site (60). Candida albicans colonization in the gut induced IL-17 and IL-22 production of TH17 cells that protect from systemic infection with Candida albicans and Staphylococcus aureus (61, 62). However, Candida albicans gut colonization also promoted susceptibility to TH17 cell-mediated airway inflammation and correlated with systemic levels of TH17 cell inflammation (62). In humans, the gut mycobiome induced antifungal antibody production that protects against systemic fungal infections in a caspase recruitment domain-containing protein 9 (CARD9) dependent way (63). Commensal fungi seem to have certain anti-inflammatory properties as a disturbance of the fungal gut community aggravated colitis in a mouse model (64). Similarly, the interaction of gut fungi with C-type lectin receptors on macrophages seems to protect from DSS-induced colitis and colon cancer (65, 66). Fungal colonization after antibiotic treatment can even recapitulate the beneficial effect of bacterial gut colonization on DSS-induced colitis (67). However, these anti-inflammatory capabilities appear to be context-sensitive, since fungal colonization seemed to be detrimental in colitis in specific-pathogen-free (SPF) mice (68). Patients suffering from Crohn’s disease frequently show elevated anti-Saccharomyces cerevisiae antibodies (ASCA), leading to the assumption that commensal fungi might play a role in human gastrointestinal diseases (69). Importantly, ASCA can even be detected before disease onset and are highly predictive for receiving a diagnosis of Crohn’s disease within the next five years (70). Thus, the gut mycobiome also appears to be an essential component in shaping local immunity in steady state as well as during inflammation.
The gastrointestinal virome also plays a pivotal role in shaping local gut immunity. A substantial proportion of viruses detected in fecal samples are bacteriophages that are likely to influence the composition and functional properties of the gastrointestinal bacterial community (71). Phages might do so by predating susceptible bacterial strains, which confers an advantage to the growth of others (72). Another mechanism is horizontal gene transfer that changes genetic diversity and thereby influences virulence, antibiotic resistance, and metabolic determinants of the bacterial community (73). Thus, phages capable of affecting the composition of gastrointestinal bacterial communities exert an indirect impact on the function of the immune system (72, 74). This is further illustrated by a study that suggested bacteriophage diversity and colonization level to be a key factor in the success of fecal microbiota transplantation (FMT) in the therapy of recurrent Clostridium difficile infection (75). Besides their indirect effects, phages can also directly exacerbate colitis by inducing host immunity via toll-like receptor (TLR)-9 stimulated interferon (IFN)-γ production (76). Bacteria-infecting prophages initiate viral gut colonization during the first months of life; eukaryotic viruses follow a few months later, depending on breastfeeding (77). The presence of enteric viruses in the gut is imperative for tissue homeostasis and the prevention of overt inflammation. Type I IFN stimulated by activation of the viral pattern recognition receptors (PRRs) retinoic acid-inducible gene I (RIG-I) and stimulator of interferon genes (STING) is protective against intestinal barrier damage and prevented graft-versus-host disease (GvHD) in mice (78). RIG-I activated by enteric viruses also stimulates IL-15 secretion, which is important for maintaining tissue-regenerative intraepithelial lymphocytes (79). Another virus-sensing PRR, TLR7, enhances resistance to vancomycin-resistant Enterococcus infection of antibiotic-treated mice via secretion of anti-microbial peptides stimulated by IL-22 producing innate lymphoid cells (ILC) (80). Recognition of gut viruses by TLR3 and TLR7 also protects against DSS-induced colitis via a type-I IFN mediated mechanism (81). IFN-λ induced by enteric viruses was suggested as an additional protective factor against DSS-induced colitis by preventing reactive oxygen species (ROS) production and neutrophil degranulation (82). Infection with murine norovirus could even compensate for the absence of gut bacteria regarding their immune-promoting function and the protective capacities against Citrobacter rodentium induced pathologies (83). Another study found murine astrovirus, a commensal gastrointestinal virus in mice, to protect against murine norovirus and rotavirus in immunodeficient mice by increasing IFN-λ levels (84). This effect is mediated by stimulation of type I IFN that leads to the recruitment of C-C chemokine receptor type 2 (CCR2) dependent monocytes and the production of IL-22 by type 3 ILC (85). Hence, besides gut bacteria and fungi, the gut virome is also a crucial factor for immunity at the barrier site, not only by direct interaction but also indirectly by affecting other microbial kingdoms. This is a relevant and increasingly recognized phenomenon known as trans-kingdom interactions (17).
The impact of eukaryotic multicellular organisms like protozoa and helminths on local and systemic immunity is still largely unrecognized (86). Some organisms such as Entamoeba histolytica and Ascaris lumbricoides are obligatory pathogens, while others, for example, Blastocystis, are associated with disease but also found in healthy people (86). The commensal murine protist Tritrichomonas musculis induced an accumulation of T helper 1 (TH1) and TH17 cells in an inflammasome and IL-18 dependent mechanism (87). This induction of adaptive immunity enhanced anti-bacterial defense but also increased intestinal inflammation (87). Similarly, Tritrichomonas muris was found to induce a TH1 response in the cecum leading to accelerated gastrointestinal inflammation in a colitis mouse model (88). Different studies found the SCFA succinate, produced by Tritrichomonas muris, to activate tuft cells in the intestinal epithelium. Tuft cells subsequently induced the secretion of IL-13, IL-4, and IL-5 by type 2 ILC in an IL-25 dependent manner (89–91). This cytokine milieu promoted the expansion of tuft and goblet cells, a common mechanism for clearing helminth infection and protecting from subsequent colonization by other parasites (89). Interestingly, tuft cell expansion promoted norovirus infection as the norovirus receptor is specifically expressed on tuft cells (92). Moreover, helminth infection can manipulate the immune system leading to an impaired antiviral immunity or even viral reactivation (93, 94). Non-fungal eukaryotes are regularly found in the gut and seem to have beneficial as well as detrimental effects on local immunity by deteriorating intestinal inflammation but also preventing secondary helminth infection.
Taken together, a rapidly growing body of literature points towards a pivotal role of the microbial community including representatives of all microbial kingdoms in the homeostasis of the gut. Particularly, the development, maturation, aging, and thus the functionality of host immune responses at the barrier site in steady state as well as during various inflammatory events are largely orchestrated by the gut microbiome.
The gastrointestinal bacterial microbiome does not only shape immunity at its corresponding barrier site but also exerts powerful effects on systemic immune responses (Figure 1). Among the most affected organs are the lung, the brain, and the liver whose physiology is influenced via the so-called gut-lung (95), gut-brain (96), or gut-liver axis (97). The gut-lung axis has a significant influence on the susceptibility to respiratory infections and allergic airway diseases. For example, germ-free mice are more susceptible to lethal Klebsiella pneumoniae infection, an effect that can be reversed by transient TLR activation through administration of several TLR agonists usually produced by indigenous microbiota (98). A comparable effect of a reduced microbial load was observed in the context of Influenza A virus (IAV) infection. Mice pre-treated with antibiotics showed an aggravation of IAV infection due to a diminished T cell response because of poorly activated dendritic cells (DC) and a higher activation threshold of innate immunity (99, 100). Additionally, TLR5 activation by bacterial flagellin in the gut is essential for the development of an efficient antibody response to IAV vaccination (101). Another study found the metabolite desaminotyrosine produced by Clostridium orbiscindens to be a protective factor against IAV infection by modulating type I IFN signaling (102). A further mechanism that might prevent overt immunopathology in the lung following IAV infection was SCFA-mediated alteration of bone marrow hematopoiesis leading to increased numbers of anti-inflammatory macrophages in the lungs (103). Gastrointestinal bacteria not only play a pivotal role during respiratory infections, but they also appear to be crucial in allergic airway disorders. Gut dysbiosis induced by antibiotic or antifungal treatment can aggravate allergen-induced airway inflammation (64, 104). Otherwise, infection with Helicobacter pylori conferred protection towards experimentally induced lung inflammation, a Treg cell-mediated effect (105, 106). A comparable protection from allergic airway disease was found in mice supplemented with Lactobacillus johnsonii, a bacterium that is enriched in the gastrointestinal tract of mice that were previously exposed to house dust mites (106). However, the protective effect of the bacterial microbiome might be reversed in the context of chronic pulmonary diseases. A study using a mouse model of cystic fibrosis showed amelioration of airway hyperresponsiveness after lowering the enteric bacterial burden by antibiotic treatment with Streptomycin (107).
Besides the lung, the gut bacterial microbiome is very well known to affect the development, maturation, normal aging, homeostasis, and function of the brain, which is accomplished through communication along the gut-brain axis (96). Important mediators for this are neuroactive metabolites produced by gut-resident microbiota. Good examples are the influence of gastrointestinal bacteria on microglial development and homeostasis, an important phenomenon likely mediated through microbially-produced, neuro-modulatory SCFA (108). This study also highlights that the complex and diverse gut community, rather than single gut-resident bacteria, is fundamental for proper microglial development and function (108). Not only microglial development but also axogenesis is affected by gut bacteria and their metabolites. Axogenesis was markedly reduced in offspring of germ-free or antibiotics-treated damns, leading to sensation impairment in the offspring (109). This effect could be reversed by colonizing the pregnant damns with spore-forming bacteria or treatment with selected metabolites (109). The microbiota-mediated influence on brain homeostasis also affects the progression of several brain pathologies including psychiatric disorders like autism spectrum disorder (ASD), as well as neurological diseases such as Alzheimer’s disease (AD), Parkinson’s diseases (PD), multiple sclerosis (MS), and stroke. To mention some examples, microbiota depletion attenuated brain inflammation and pathologies in mice with experimental autoimmune encephalitis (EAE), a gold-standard model of human MS as well as in an AD mouse model (110–112). Susceptibility to developing EAE symptoms could be transferred from MS patients to transgenic mice spontaneously developing EAE, which showed an increased incidence of disease symptoms after FMT from MS diseased donors (113). Moreover, in a mouse model of ASD, oral treatment with Bacteroides fragilis ameliorated ASD-related behavioral abnormalities, an effect that is most likely mediated by microbiota-dependent metabolites (114). Another study using a PD mouse model has underlined the microbial influence on neurological disorders by identifying gut microbiota to enhance α-synuclein-mediated motor dysfunction, an effect probably also mediated by microbially-produced SCFA (115). Further, the gut bacterial microbiota affects meningeal IL-17 producing gamma delta T (γδT) cells, which worsened the outcome of experimentally induced strokes (116). Similar to the intestine, another part of the digestive tract, the oral cavity, has a niche-specific bacterial microbiome (117). Interestingly, the amount and diversity of oral colonization seem to decline during weaning, coinciding with upregulation of saliva production and salivary antimicrobial components (118). Moreover, in several studies, periodontitis and tooth loss were associated with the development of dementia and Alzheimer’s disease (119, 120).
There also is an interplay between the gut bacteria and the liver. So, common liver diseases like alcoholic liver disease (ALD), non-alcoholic fatty liver disease (NAFLD), liver cirrhosis, or hepatocellular carcinoma (HCC) are associated with changes in the bacterial gut microbiome (97). Changes in the microbiome can also change liver cancer by bile acids transformation that affect CXCR6+ natural killer T (NKT) cells in the liver (121). A therapeutic option using the microbiome-related effect on liver immunity is FMT, which showed promising results in an ALD mouse model and was recently also explored in a first clinical trial in humans (122, 123). Even though, science has already gained substantial knowledge on how gut bacterial microbiota influence immunity in distant organs, there is still extensive research required to fully decipher these complex interactions.
Like the gut bacterial microbiome, the gut mycobiome can also shape systemic immune responses, particularly in the lung, brain, and liver. Regarding the gut-lung axis, fungal dysbiosis caused by antibiotics or antifungal treatment can exacerbate experimentally induced allergic airway disease (64, 124). Allergic airway inflammation was significantly aggravated through sensing of fungal dysbiosis by gut resident mononuclear phagocytes (MNPs) and a subsequent increase in pulmonary T helper 2 (TH2) cells and eosinophils (125). The beneficial effect of fungi on allergic airway disease seems to be a tightly balanced equilibrium, as fungal overgrowth following antibiotic treatment leads to a promotion of allergic airway inflammation (126). One of the proposed mechanisms was the elevation of prostaglandin E2 (PGE2) plasma levels by the overgrowth of Candida species and the promotion of macrophage polarization towards an alternatively activated M2 phenotype (104). Additionally, the expansion of a single fungal species in the gut, Wallemia mellicola, could aggravate allergic airway disease (127). In humans, Candida albicans, a common gut fungal species, seems to be the major direct inducer of anti-fungal TH17 cells in peripheral blood (61). These TH17 cells are cross-reactive to inhaled Aspergillus fumigatus and are activated and expanded in patients with airway inflammation (61). Likewise, intestinal colonization with Candida albicans in mice promoted susceptibility to airway inflammation (62). Candida albicans strains adapted to the mouse gastrointestinal tract conferred enhanced protection against systemic infection with several fungi and bacteria, but this effect required IL-6 and was also observed in lymphocyte-deficient mice (128). Moreover, fungal colonization following antibiotic treatment recapitulates the beneficial effect of bacterial gut colonization in lethal IAV infection (67).
Like gut bacteria, fungi can also affect brain and liver homeostasis. An example of this is the improvement of symptoms of EAE, after oral supplementation of mice with Candida kefyr (129). Regarding influences on the liver, gut-resident fungi promote the development of ALD, an effect that is mediated by increased translocation of fungal β-glucan to the systemic circulation, which induced IL-1β-mediated liver inflammation through binding to C-type lectin domain family 7 member A (CLEC7A) on Kupffer cells (130).
Furthermore, the gut mycobiome has a fundamental influence on the maturation of the immune system itself. The gastrointestinal fungal species Candida tropicalis was found to play a substantial role in the early life maturation of secondary lymphoid organs (131). Recent studies transferring mice into a natural environment supported this by showing an increased fungal diversity and an elevated fungal load, especially in Aspergillus species. These alterations of the fungal community were accompanied by an increase in peripheral granulocytes and activated T cells, illustrating an enhanced immune maturation (132, 133). Not only gut bacteria, but also gut fungi have systemic effects on immunity. Even though there is only limited data on fungal metabolites, their involvement in this communication seems likely.
The gastrointestinal virome also shapes immunity at gut-distal sites such as the lung and the liver. For example, an infection with murine norovirus protects against lung infection with Pseudomonas aeruginosa and alleviates lung inflammation (134). A liver-related example is that a decreased diversity of the intestinal virome is associated with an increase in the severity of NAFLD in humans (135). Further research is required to fully understand how intestinal viral communities shape peripheral immune reactions at various sites.
In addition to gut-resident viruses, there is increasing evidence that systemic chronic viral infections are not only pathogenic but can also have advantages to the host (136). Latent herpesvirus infection mediates resistance to bacterial infection with Listeria monocytogenes or Yersinia pestis and results in increased resistance to tumor grafts (137–139). Type I IFN production induced by chronic murine cytomegalovirus (MCMV) infection stimulated epithelial proliferation and intestinal wound repair (140). Latent infection with chronic viruses has thus been considered as an integral part of the microbiome that has a substantial influence on the host’s immune system.
Apart from local effects on the gastrointestinal immune system, multicellular eukaryotes were also found to influence immunity in the lung. In mouse models, infection with Heligmosomoides polygyrus leads to a decreased development of allergic airway inflammation, probably through a mechanism that involves Treg cells and is independent of IL-10 (141). Interestingly, Heligmosomoides polygyrus derived egg-shell products were sufficient to prevent experimental allergic airway inflammation, probably by directly inhibiting IL-33 release (142, 143). In humans, reports are still inconsistent regarding a correlation between gastrointestinal helminth infection and the occurrence of allergic airway disease (144). More research on protist and helminth members of the gut microbiome is required for a better description of their influence on immunity.
Taken together, the gut bacterial microbiome exerts many important effects on various gut-distant organs such as the lung, the brain, and the liver. However, current data also clearly indicate a critical involvement of nearly all microbial kingdoms in the functionality of the host’s immune system, and thus the orchestration of systemic host immune responses in health and disease. Importantly, biologically active microbial compounds appear to be essential in mediating the communication between the host and its microbiota along the corresponding axis.
The skin is the outermost barrier of our body and is in constant contact with multiple environmental influences. To maintain this barrier, our skin works together with its residing microbiota (Figure 2) (145, 146). Various species are adapted to the specific properties of the respective site and thus inhabit different cutaneous microenvironments (147). For example, sebaceous sites are dominated by lipophilic Propionibacterium species, while Staphylococcus and Corynebacterium species colonize the moist areas (148). The initial microbial skin colonization depends on the delivery mode. Vaginally delivered babies acquire the mother’s vaginal microbiome, while cesarean section leads to the acquisition of skin-associated microbiota (149, 150). A major shift in microbial skin communities occurs during puberty: several taxa disappear and the microbiota becomes dominated by lipophilic species (151). During cutaneous immune homeostasis, skin commensal bacteria maintain the host-microbial mutualism by protective and regulatory responses. Commensal bacteria affect the immune system in the absence of inflammation and independent of changes in the gut microbiome (152, 153). Cutaneous colonization with the skin commensal Staphylococcus epidermidis leads to a non-inflammatory accumulation of IL-17A and IFN-γ expressing CD8+ T cells in the skin (152). Similarly, T cells in germ-free mice produced significantly lower cytokine levels, a phenotype that could be reversed by colonization with Staphylococcus epidermidis (153). Tolerance to commensal microbes is established during the postnatal period when developing hair follicles are colonized by microbes that induce commensal-specific Treg cells (154, 155). The complement system, which is part of this regulatory mechanism by maintaining microbial diversity, is also regulated by commensal microbes (156). The commensal Staphylococcus epidermidis seems to be an essential microbe capable of regulating immunity at this barrier site. In an in vitro study using human monocyte-derived DC, Staphylococcus epidermidis products stimulate DC to produce more IL-10 and lower the proliferation effect on CD4+ T cells (157). Additionally, Treg cells treated with Staphylococcus products have a higher immune-suppressive potential on T cells (157). Interestingly, this homeostatic immunity to microbiota is mediated by non-classical MHC class I molecules (158). Metabolites of commensal bacteria also have a suppressive effect on the development of skin inflammation. Treatment of atopic dermatitis with lysates of Vitreoscilla filiformis, a gram-negative bacterium present in thermal spa water, leads to pronounced amelioration of atopic dermatitis symptoms in a clinical trial and an experimental model (159, 160). Studies in mice suggested that this is an effect of IL-10 secretion from DC and accumulation of Treg cells, which have a suppressive effect on T cell expansion (160). The bacterial skin microbiome also affects the process of acute wound healing, specifically when the integrity of the skin barrier is breached. During inflammation caused by skin wounding, lipoteichoic acid of Staphylococcus epidermidis can mitigate skin inflammation in a TLR dependent manner (161). Wound closure is also accelerated by commensal-specific T cells that express tissue repair and immunoregulatory signatures (158). The influence of commensal microbes on local immunity confers resistance to potentially harmful pathogens. In vivo, skin colonization with Staphylococcus epidermidis led to CD103+ DC dependent formation of IL-17 producing CD8+ T cells leading to the production of alarmins by keratinocytes inhibiting Candida albicans outgrowth (152). Restoration of IL-17 production after Staphylococcus epidermidis colonization also helped in the control of Leishmania major infection but then again lead to an increased inflammatory response (153). In summary, the skin bacterial microbiome is a fundamental player in securing skin homeostasis, preventing inflammatory skin diseases, supporting wound healing, and protecting against pathogenic infections.
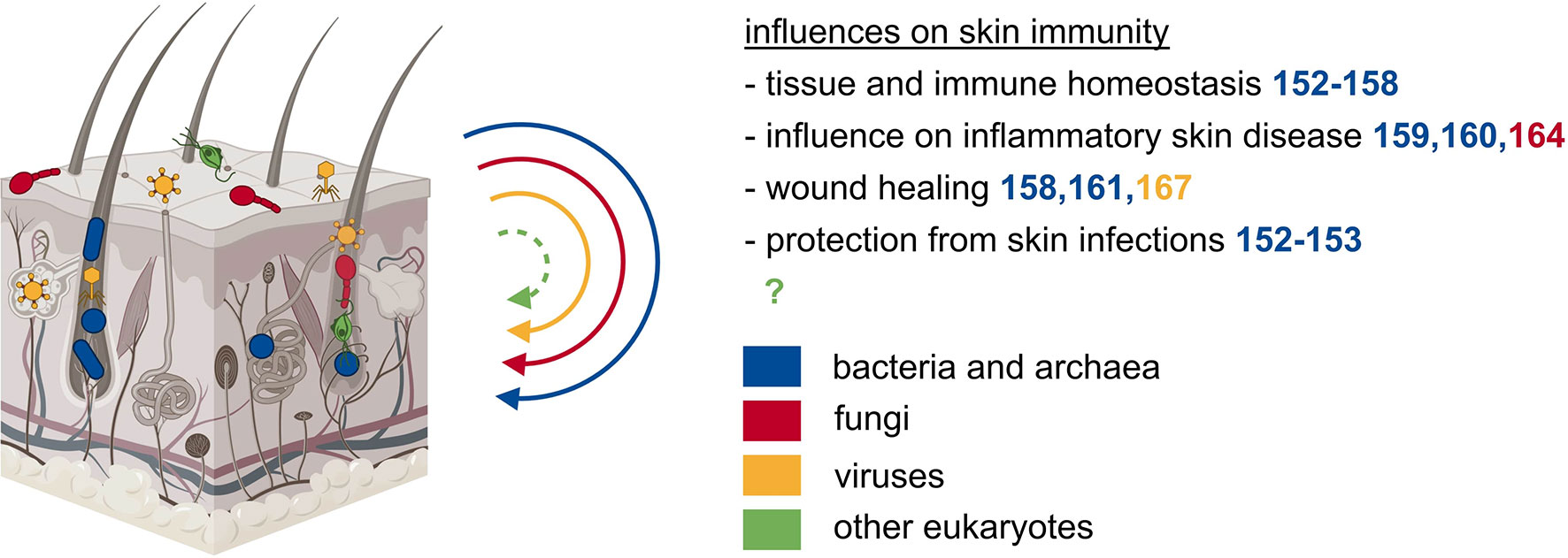
Figure 2 The microbiota of the skin exerts local effects on immunity. Besides the gut, also other epithelial barrier sites such as the skin are populated by diverse microbes of all kingdoms. The microbiota of the skin influences tissue and immune homeostasis, inflammatory skin diseases, wound healing, and protects from skin infections.
Besides multiple bacterial species, archaea of the phyla Thaumarchaeota and Euryarchaeota were detected on human skin samples (53, 162). Their abundance is correlated to dry skin occurrence, but mechanistic details of a potential causal relationship are missing (163).
Lipophilic fungi of the genus Malassezia are the dominant fungal microorganisms on the skin of most adults (148, 151). A recent publication suggested that immunity to commensal fungi might play a role in the aggravation of skin inflammation in psoriasis (164). Thus, recent literature shows an important role of fungi in skin immunity, thereby prompting us to more closely investigate the role of the skin mycobiome.
Eukaryotic viruses and phages are detected on human skin and are the most unstable part of the skin microbiome (165, 166). Bacteriophages were found to negatively interfere with chronic wound healing in Pseudomonas aeruginosa infected wounds, likely mediated by endocytosis and TLR3 (167).
In conclusion, even though microorganisms other than bacteria are highly abundant on the skin and their involvement in skin immunity has been shown in some promising studies, their local and especially systemic influence on the host’s immune system is still poorly explored. However, unraveling these interactions appears to be a promising field for future studies.
The initial dogma of the lung being devoid of microorganisms has recently shifted by advances in sequencing techniques, leading to improved microbial detection. These findings also indicate that lung immunity is influenced by the lung microbiome in several diseases (Figure 3). The respiratory bacterial microbiome differs markedly between the upper and lower respiratory tract and is probably influenced by the oral microbiome through frequent micro-aspirations and by airborne microorganisms present in the inhaled air (168). Enrichment of the bacterial lung microbiota with oral taxa is associated with an increased number of lymphocytes, especially TH17 cells, elevated cytokine levels, and a diminished TLR4 response by alveolar macrophages, thus influencing the basal inflammatory status in the lung (169). Staphylococcus aureus in the lung leads to TLR2-dependent recruitment of monocytes that differentiate into alveolar macrophages and exert a protective effect against IAV pathologies (170). The presence of lung microbiota in early life is essential for the immune maturation and attenuation of airway inflammation that is mediated by the interaction of Treg cells with programmed cell death 1 ligand 1 (PD-L1) expressing DC (171). A similar effect could be observed in children growing up on farms in Central Europe, that were protected from asthma and atopy supposedly due to an increased environmental microbial exposure (172). In line with this, maternal exposure to Acitenobacter lwoffii F78, a bacterium frequently found in cowsheds and farm dust samples leads to the protection of the offspring against experimental asthma (173). Thus, lung bacterial communities may mediate a regulatory function in the development of allergic airway diseases.
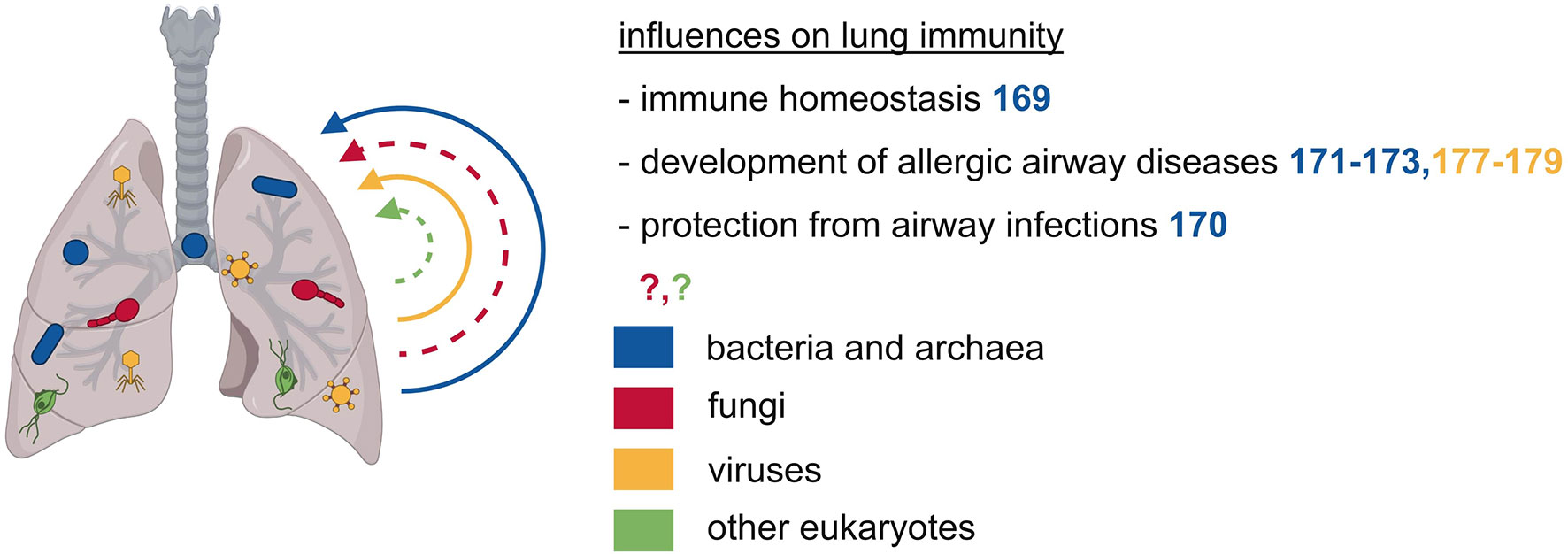
Figure 3 The microbiota of the respiratory tract exerts local effects on immunity. The respiratory tract is also inhabited by various microbes of different kingdoms. These microbes can prevent allergic airway diseases such as asthma and they can protect from airway infections.
The lung is also colonized by archaeal microbes, mainly by woesearchaeal species (53). However, there is little to nothing known about the function and effects of archaea in the respiratory tract.
There is only a low fungal burden in the lungs, consisting primarily of the division of Ascomycota and Basidiomycota with Candida species being the predominant organisms (174). So far, there is only sparse knowledge about fungal influences on the host’s immunity in the respiratory tract.
The virome in the respiratory tract is composed of intrinsic components with a high abundance of Anelloviruses and phages, and other respiratory viruses that are normally considered pathogens (175, 176). Early life infection with IAV protects against airway hyperreactivity, a protective effect primarily mediated through an expansion of NKT cells (177). On the contrary, early infection with respiratory syncytial virus increases susceptibility to allergic airway disease by impairing Treg cell function (178). Another study in mice found that infection with Sendai virus is associated with subsequent airway hyperreactivity that depends on IL-13 dependent activation of NKT cells and lung macrophages (179). Overall, there is only little knowledge regarding the impact of the lung-resident virome on local or systemic immunity.
In summary, the currently available data on lung microbiota are still limited, particularly as regards to microorganisms other than bacteria. This is likely due to the low microbial biomass present in the lung, thereby leading to profound technical challenges in reliably assessing the lung microbiota. However, studying the lung microbiome certainly is an interesting future topic, since the lung is the second largest barrier site and is affected by various diseases where the microbiome is known to be a crucial factor (e.g. infectious diseases, cancer, allergies, inflammatory and autoimmune diseases).
Similar to other microbial niches, the genitourinary tract is also colonized by microorganisms of different kingdoms that have direct or indirect effects on immunity (Figure 4). Compared to other microbial niches, the vaginal bacterial microbiome is a relatively basic community, characterized by comparably low diversity and is dominated by Lactobacillus species, which thrive in this anaerobic environment (24, 180). Some asymptomatic women carry a more diverse vaginal community containing bacteria of the genera Gardnerella and Prevotella (24, 180). However, Lactobacilli play the most pivotal role in maintaining the homeostasis of the vaginal tract through the production and secretion of anti-microbial compounds such as H2O2, bacteriocin, and lactic acid. These compounds serve as a first, effective line of defense by establishing a low vaginal pH and by creating an overall hostile environment for invading bacteria and other microorganisms (181). Another direct effect of lactic acid is its anti-inflammatory impact on vaginal epithelial cells, shown by a reduced production of pro-inflammatory cytokines and increased production of the IL-1 receptor antagonist (IL-1RA) (182). Therefore, a Lactobacilli-dominated vaginal microbiome protects against urogenital infections such as urinary tract infections and sexually transmitted pathogens like Chlamydia, human immunodeficiency virus (HIV), and herpes simplex virus 2 (HSV-2) (183–185). The increased occurrence of HIV infection in women with a high-diversity vaginal bacterial microbiome might be explained by an increased number of activated CD4+ T cells, also including CCR5+ CD4+ T cells, the HIV target cells (185). The increase in activated T cells might be induced by elevated cytokine and chemokine levels in the genitourinary tract of women with high-diverse vaginal bacterial communities (185, 186). The increased abundance of pro-inflammatory cytokines in vaginal fluid might also be a risk factor of preterm birth that was observed to be associated with a decreased vaginal colonization with Lactobacillus species (187). In summary, a vaginal bacterial microbiome rich in Lactobacillus species is important for the protection against urogenital infections as well as a beneficially balanced cytokine and chemokine profile in the genitourinary tract.
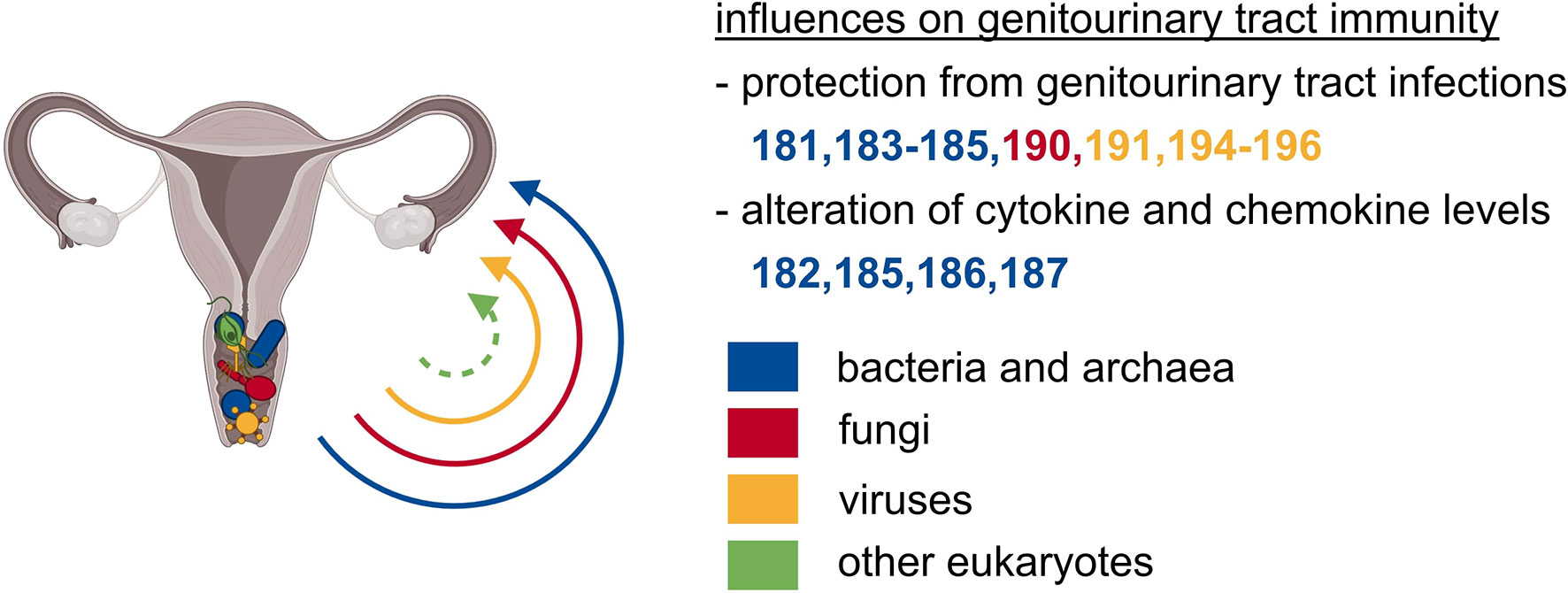
Figure 4 The microbiota of the genitourinary tract exerts local effects on immunity. Particularly the vagina is colonized with abundant microbes of unrelated kingdoms. The vaginal microbial communities safeguard from local, ascending, and subsequently systemic infections, and are crucial for beneficially balanced cytokine as well as chemokine profile.
The archaeal species Methanobrevibacter smithii was detected in vaginal samples and was associated with bacterial vaginosis (188, 189). Consequently, the presence of Methanobrevibacter smithii was proposed as a biomarker for the diagnosis of bacterial vaginosis (189). Apart from this, there is only little information available on associations between residing archaea and genitourinary tract diseases.
Apart from other commensal communities, fungi are a common constituent of the female vaginal microbiome in healthy women (24). The vaginal mycobiome is dominated by the division of Ascomycota, mainly the genus Candida, which was present in two-thirds of asymptomatic Estonian women (24, 190). Although candidiasis is one of the most frequent genital diseases, Candida species frequently colonize the vagina of healthy women, and only little is known about the influence of vaginal fungal communities on the host’s physiology (191).
Most vaginal DNA viruses that are identified today are double-stranded DNA bacteriophages, with eukaryotic viruses constituting only 4% of the total reads (192). Eukaryotic DNA viruses in the vagina are dominated by papillomavirus species, but also herpesviruses, polyomaviruses, and anelloviruses can be detected (192–194). Additionally, several groups have identified functional and nonfunctional prophages in the genomes of vaginal bacterial species, suggesting bacteriophages to play a role in shaping the vaginal bacterial microbiome, thereby influencing vaginal health (195–197). Another study also found links between eukaryotic viral and bacterial community composition and the occurrence of bacterial vaginosis (192). In conclusion, there are many potential trans-kingdom interactions between viral and bacterial communities in the vagina and thus indirect and potentially also direct influences of the vaginal virome on health and disease of the genitourinary system.
Endogenous retroviruses are a group of transposable elements that are stably integrated into the genome of their host. They were originally acquired by infection of the host’s germline cells with retroviruses and are vertically transmitted because of their permanent integration (198). The most prominent representatives of human endogenous retroviruses are syncytin-1 and syncytin-2, which are homologous to the surface proteins encoding env genes of human endogenous retroviruses (199). They have an essential role in the host’s physiology of the placenta by mediating cell fusion between cytotrophoblasts and placental syncytiotrophoblasts (200). Thus, these relicts of ancient viral infections play a fundamental role in the exchange of nutrients, gases, and hormones between the mother and the fetus. Additionally, they prevent fetal rejection by controlling the maternal immunosuppressive state (201, 202). Hence, these relicts of ancient viral infections were critically involved in mammalian evolution, a prime example of the intimate relationship between microorganisms and their hosts.
Taken together, the bacterial microbiome undoubtfully plays an essential role for the genitourinary tract. However, other microbial kingdoms and their important impact on health and diseases of the genitourinary tract are increasingly evident, appreciated, and also studied.
The past decade was characterized by substantial technological progress, allowing researchers to study the mammalian microbiome and its impact on the host in health and disease in a much more precise and mechanistic fashion. As a consequence, it has been established that the microbiota plays a pivotal role in virtually every aspect of mammalian physiology, particularly in the development, maturation, homeostasis, orchestration, and ultimately the function of the immune system (7, 8). By now, it can be considered as textbook knowledge that mammals are metaorganisms and that the combination of the host genome and the overall microbiome including all kingdoms at all epithelial barrier sites largely drives their phenotypes (15). This can be considered the most important conceptual advance in the field of microbiome research during the last decades.
The comparison of conventional SPF mice to germ-free, antibiotic-treated, and gnotobiotic mouse models was pivotal in illuminating the impact of the mammalian microbiome on host physiology. These proof-of-principal studies were essential in illustrating the therapeutic potential lying within microbiome research such as the remarkably diverse set of biologically active compounds produced by the microbiota. As illustrated throughout this review, these compounds can not only influence their corresponding barrier site but also establish axes of communication, thereby exerting crucial systemic effects. Translational microbiome research should aim to identify these compounds and to understand their biological function in health and disease. Among others, this may be a pathbreaking strategy to discover novel microbiota-based drugs (114, 203).
Aside from these important studies, a substantial body of literature emphasizes that the majority of rodent-based data could not be translated into clinical practice (204–211). Recent paradigm-shifting work illustrated that lab mice are too far removed from natural environmental conditions to reliably mirror the physiology of free-living mammals like humans (212–215). This circumstance distorts how the immune system of ultra-clean lab mice develops and functions, leading to false assumptions of how the human immune system works as reviewed elsewhere (216, 217). To address these shortcomings several approaches have been suggested: Cohousing of lab mice with pet store mice (212), sequential infections of lab mice (213), rewilding of lab mice in semi-natural habitats (214), engraftment of wild mouse gut microbiota into lab mice (215) and the transfers of lab mouse embryos into wild mouse surrogate mothers, the so-called “wildling” model (218). Indeed, compared to conventional lab mice, the resulting animals were protected in models of infectious diseases and cancer and displayed an increased translational research value (212, 215). Particularly, wildlings phenocopied the human outcome and could have prevented catastrophically failed clinical trials, where conventional rodent and non-human primate models had failed to predict the human response to harmful drug treatments (218–220). Thus, utilizing these microbially diverse models in translational microbiome research may help to discover novel disease treatment options that cannot be found in conventional mouse models and increase the safety and success rate of bench-to-bedside efforts.
As mentioned above, only a few findings in microbiome research could be directly translated into the clinic so far and many of them originate from human research. For example, there are ideas to use bacterial products in inflammatory skin disease, and promising studies have already proposed a beneficial outcome (159). Moreover, the injection of beta-glucans from fungi cell walls alongside therapeutic antibodies or chemotherapy in cancer treatment shows promising results (221, 222). Another well-known example is allogenic FMT, a potent treatment of antibiotic-refractory Clostridium difficile infection (223, 224). Recent studies also showed a beneficial combination of FMT with immunotherapy to overcome the initial resistance to immunotherapy in melanoma patients (225, 226). Allogeneic hematopoietic stem cell transplantation (allo-HSCT) is accompanied by the usage of broad-spectrum antibiotics leading to a low diversity of the gut microbiome which can be successfully treated by autologous FMT (227). This might be a promising treatment option as low-diversity microbiota correlate with increased mortality in allo-HSCT patients (228). Eran Elinav and colleagues published an encouraging exploratory study utilizing vaginal microbiome transplantation to efficiently treat recurrent bacterial vaginosis (229). Even though important steps have already been made, the journey of microbiome research and the successful transfer of microbiota-related therapies into the clinic has only begun.
Thus, besides further technological advances, the key to tap into the full therapeutic potential of translational microbiome research may be: (I) a stronger appreciation of mammals as metaorganisms; (II) the more pronounced investigation of non-bacterial members of the microbiome as well as trans-kingdom interactions at all epithelial barrier sites; (III) a focus on how the bidirectional crosstalk between the host and its microbiota works from a mechanistic standpoint of view, particularly (IV) how microbial biologically active compounds affect the health and disease of the host and (V) take advantage of newly developed translational microbiome research mouse models that more closely resemble the human metaorganism.
This approach will open up a promising window of opportunity to discover novel treatments for a wide range of human diseases of global relevance including transplant rejection, GvHD, cancer, infectious diseases, allergies, autoimmune and inflammatory diseases, psychiatric and neurological disorders as well as cardiovascular diseases.
SR wrote the manuscript. SR and SPR designed the figures. SPR supervised and edited the manuscript. All authors contributed to the article and approved the submitted version.
SPR was supported by the Deutsche Forschungsgemeinschaft DFG (German Research Foundation): Emmy Noether-Programm RO 6247/1-1, SFB 1160/2 IMPATH and IMM-PACT-Programme for Clinician Scientists, Department of Medicine II, Medical Center – University of Freiburg and Faculty of Medicine, University of Freiburg, 413517907.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All Figures were created with BioRender.com.
AD, Alzheimer’s disease; ALD, Alcoholic liver disease; allo-HSCT, Allogeneic hematopoietic stem cell transplantation; ASCA, Anti-saccharomyces cerevisiae antibodies; AhR, aryl hydrocarbon receptor; ASD, autism spectrum disorder; CARD, caspase recruitment domain-containing protein; CCR, C-C chemokine receptor; CLEC7A, C-type lectin domain family 7 member A; DC, dendritic cell; DSS, Dextran sulfate sodium; γδT cell, gamma delta T cell; EAE, experimental autoimmune encephalitis; FMT, fecal microbiota transplantation; FXR, farnesoid X receptor; GvHD, Graft-versus-host disease; HCC, hepatocellular carcinoma; HIV, human immunodeficiency virus; HSV-2, herpes simplex virus 2; IAV, influenza type A virus; IL-1RA, IL-1 receptor antagonist; ILC, innate lymphoid cell; IFN, interferon; IL, interleukin; MNP, mononuclear phagocyte; MS, multiple sclerosis; MCMV, murine cytomegalovirus; NAFLD, non-alcoholic fatty liver disease; NKT cell, natural killer T cell; PD, Parkinson’s disease; PD-L1, programmed cell death 1 ligand 1; PRR, pattern recognition receptor; PGE2, prostaglandin E2; RIG-I, retinoic acid-inducible gene I; ROS, reactive oxygen species; Treg cell, regulatory T cell; SFB, segmented filamentous bacteria; SCFA, short-chain fatty acids; SPF, Specific-pathogen-free; STING, Stimulator of interferon genes; TH1 cell, T helper 1 cell; TH2 cell, T helper 2 cell; TH17 cell, T helper 17 cell; TLR, toll-like receptor.
1. Sommer F, Bäckhed ,F. The Gut Microbiota - Masters of Host Development and Physiology. Nat Rev Microbiol (2013) 11:227–38. doi: 10.1038/nrmicro2974
2. Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds Within Worlds: Evolution of the Vertebrate Gut Microbiota. Nat Rev Microbiol (2008) 6:776–88. doi: 10.1038/nrmicro1978
3. Dethlefsen L, McFall-Ngai M, Relman DA. An Ecological and Evolutionary Perspective on Human-Microbe Mutualism and Disease. Nature (2007) 449:811–8. doi: 10.1038/nature06245
4. O’Hara AM, Shanahan F. The Gut Flora as a Forgotten Organ. EMBO Rep (2006) 7:688–93. doi: 10.1038/sj.embor.7400731
5. Berg G, Rybakova D, Fischer D, Cernava T, Vergès M-CC, Charles T, et al. Microbiome Definition Re-Visited: Old Concepts and New Challenges. Microbiome (2020) 8:103. doi: 10.1186/s40168-020-00875-0
6. Zheng D, Liwinski T, Elinav E. Interaction Between Microbiota and Immunity in Health and Disease. Cell Res (2020) 30:492–506. doi: 10.1038/s41422-020-0332-7
7. Hooper LV, Littman DR, Macpherson AJ. Interactions Between the Microbiota and the Immune System. Science (2012) 336:1268–73. doi: 10.1126/science.1223490
8. Belkaid Y, Hand TW. Role of the Microbiota in Immunity and Inflammation. Cell (2014) 157:121–41. doi: 10.1016/j.cell.2014.03.011
9. Cerf-Bensussan N, Gaboriau-Routhiau V. The Immune System and the Gut Microbiota: Friends or Foes? Nat Rev Immunol (2010) 10:735–44. doi: 10.1038/nri2850
10. Thaiss CA, Zmora N, Levy M, Elinav E. The Microbiome and Innate Immunity. Nature (2016) 535:65–74. doi: 10.1038/nature18847
11. Honda K, Littman DR. The Microbiota in Adaptive Immune Homeostasis and Disease. Nature (2016) 535:75–84. doi: 10.1038/nature18848
12. Zilber-Rosenberg I, Rosenberg E. Role of Microorganisms in the Evolution of Animals and Plants: The Hologenome Theory of Evolution. FEMS Microbiol Rev (2008) 32:723–35. doi: 10.1111/j.1574-6976.2008.00123.x
13. Bordenstein SR, Theis KR. Host Biology in Light of the Microbiome: Ten Principles of Holobionts and Hologenomes. PloS Biol (2015) 13:e1002226. doi: 10.1371/journal.pbio.1002226
14. Theis KR, Dheilly NM, Klassen JL, Brucker RM, Baines JF, Bosch TC, et al. Getting the Hologenome Concept Right: An Eco-Evolutionary Framework for Hosts and Their Microbiomes. mSystems (2016) 1:e00028-16. doi: 10.1128/mSystems.00028-16
15. Norman JM, Handley SA, Virgin HW. Kingdom-Agnostic Metagenomics and the Importance of Complete Characterization of Enteric Microbial Communities. Gastroenterology (2014) 146:1459–69. doi: 10.1053/j.gastro.2014.02.001
16. Virgin HW. The Virome in Mammalian Physiology and Disease. Cell (2014) 157:142–50. doi: 10.1016/j.cell.2014.02.032
17. Pfeiffer JK, Virgin HW. Viral Immunity. Transkingdom Control of Viral Infection and Immunity in the Mammalian Intestine. Science (2016) 351:aad5872. doi: 10.1126/science.aad5872
18. Song E-J, Lee E-S, Nam Y-D. Progress of Analytical Tools and Techniques for Human Gut Microbiome Research. J Microbiol (2018) 56:693–705. doi: 10.1007/s12275-018-8238-5
19. Arnold JW, Roach J, Azcarate-Peril MA. Emerging Technologies for Gut Microbiome Research. Trends Microbiol (2016) 24:887–901. doi: 10.1016/j.tim.2016.06.008
20. Limon JJ, Skalski JH, Underhill DM. Commensal Fungi in Health and Disease. Cell Host Microbe (2017) 22:156–65. doi: 10.1016/j.chom.2017.07.002
21. Borrel G, Brugère J-F, Gribaldo S, Schmitz RA, Moissl-Eichinger C. The Host-Associated Archaeome. Nat Rev Microbiol (2020) 18:622–36. doi: 10.1038/s41579-020-0407-y
22. Vesty A, Biswas K, Taylor MW, Gear K, Douglas RG. Evaluating the Impact of DNA Extraction Method on the Representation of Human Oral Bacterial and Fungal Communities. PloS One (2017) 12:e0169877. doi: 10.1371/journal.pone.0169877
23. Wang D. 5 Challenges in Understanding the Role of the Virome in Health and Disease. PloS Pathog (2020) 16:e1008318. doi: 10.1371/journal.ppat.1008318
24. Drell T, Lillsaar T, Tummeleht L, Simm J, Aaspõllu A, Väin E, et al. Characterization of the Vaginal Micro- and Mycobiome in Asymptomatic Reproductive-Age Estonian Women. PloS One (2013) 8:e54379. doi: 10.1371/journal.pone.0054379
25. Nilsson RH, Ryberg M, Kristiansson E, Abarenkov K, Larsson K-H, Kõljalg U. Taxonomic Reliability of DNA Sequences in Public Sequence Databases: A Fungal Perspective. PloS One (2006) 1:e59. doi: 10.1371/journal.pone.0000059
26. Underhill DM, Iliev ID. The Mycobiota: Interactions Between Commensal Fungi and the Host Immune System. Nat Rev Immunol (2014) 14:405–16. doi: 10.1038/nri3684
27. Hawksworth DL. Separate Name for Fungus’s Sexual Stage may Cause Confusion. Nature (2009) 458:29. doi: 10.1038/458029c
28. Tisza MJ, Pastrana DV, Welch NL, Stewart B, Peretti A, Starrett GJ, et al. Discovery of Several Thousand Highly Diverse Circular DNA Viruses. eLIFE (2020) 9:e51971. doi: 10.7554/eLife.51971
29. The Human Microbiome Project Consortium. Structure, Function and Diversity of the Healthy Human Microbiome. Nature (2012) 486:207–14. doi: 10.1038/nature11234
30. Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How Colonization by Microbiota in Early Life Shapes the Immune System. Science (2016) 352:539–44. doi: 10.1126/science.aad9378
31. Kamada N, Núñez G. Regulation of the Immune System by the Resident Intestinal Bacteria. Gastroenterology (2014) 146:1477–88. doi: 10.1053/j.gastro.2014.01.060
32. Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of Intestinal Th17 Cells by Segmented Filamentous Bacteria. Cell (2009) 139:485–98. doi: 10.1016/j.cell.2009.09.033
33. Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, et al. Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell (2015) 163:367–80. doi: 10.1016/j.cell.2015.08.058
34. Tan TG, Sefik E, Geva-Zatorsky N, Kua L, Naskar D, Teng F, et al. Identifying Species of Symbiont Bacteria From the Human Gut That, Alone, can Induce Intestinal Th17 Cells in Mice. Proc Natl Acad Sci USA (2016) 113:E8141–50. doi: 10.1073/pnas.1617460113
35. Omenetti S, Bussi C, Metidji A, Iseppon A, Lee S, Tolaini M, et al. The Intestine Harbors Functionally Distinct Homeostatic Tissue-Resident and Inflammatory Th17 Cells. Immunity (2019) 51:77–89.e6. doi: 10.1016/j.immuni.2019.05.004
36. Teng F, Klinger CN, Felix KM, Bradley CP, Wu E, Tran NL, et al. Gut Microbiota Drive Autoimmune Arthritis by Promoting Differentiation and Migration of Peyer’s Patch T Follicular Helper Cells. Immunity (2016) 44:875–88. doi: 10.1016/j.immuni.2016.03.013
37. Bachem A, Makhlouf C, Binger KJ, Souza DP de, Tull D, Hochheiser K, et al. Microbiota-Derived Short-Chain Fatty Acids Promote the Memory Potential of Antigen-Activated CD8+ T Cells. Immunity (2019) 51:285–97. doi: 10.1016/j.immuni.2019.06.002
38. Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg Induction by a Rationally Selected Mixture of Clostridia Strains From the Human Microbiota. Nature (2013) 500:232–6. doi: 10.1038/nature12331
39. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, et al. The Microbial Metabolites, Short-Chain Fatty Acids, Regulate Colonic Treg Cell Homeostasis. Science (2013) 341:569–73. doi: 10.1126/science.1241165
40. Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites Produced by Commensal Bacteria Promote Peripheral Regulatory T-Cell Generation. Nature (2013) 504:451–5. doi: 10.1038/nature12726
41. Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An Immunomodulatory Molecule of Symbiotic Bacteria Directs Maturation of the Host Immune System. Cell (2005) 122:107–18. doi: 10.1016/j.cell.2005.05.007
42. Inagaki T, Moschetta A, Lee Y-K, Peng L, Zhao G, Downes M, et al. Regulation of Antibacterial Defense in the Small Intestine by the Nuclear Bile Acid Receptor. Proc Natl Acad Sci USA (2006) 103:3920–5. doi: 10.1073/pnas.0509592103
43. Vavassori P, Mencarelli A, Renga B, Distrutti E, Fiorucci S. The Bile Acid Receptor FXR Is a Modulator of Intestinal Innate Immunity. J Immunol (2009) 183:6251–61. doi: 10.4049/jimmunol.0803978
44. Song X, Sun X, Oh SF, Wu M, Zhang Y, Zheng W, et al. Microbial Bile Acid Metabolites Modulate Gut Rorγ+ Regulatory T Cell Homeostasis. Nature (2020) 577:410–5. doi: 10.1038/s41586-019-1865-0
45. Hang S, Paik D, Yao L, Kim E, Trinath J, Lu J, et al. Bile Acid Metabolites Control TH17 and Treg Cell Differentiation. Nature (2019) 576:143–8. doi: 10.1038/s41586-019-1785-z
46. Campbell C, McKenney PT, Konstantinovsky D, Isaeva OI, Schizas M, Verter J, et al. Bacterial Metabolism of Bile Acids Promotes Generation of Peripheral Regulatory T Cells. Nature (2020) 581:475–9. doi: 10.1038/s41586-020-2193-0
47. Wesemann DR, Portuguese AJ, Meyers RM, Gallagher MP, Cluff-Jones K, Magee JM, et al. Microbial Colonization Influences Early B-Lineage Development in the Gut Lamina Propria. Nature (2013) 501:112–5. doi: 10.1038/nature12496
48. Zelante T, Iannitti RG, Cunha C, Luca A, Giovannini G, Pieraccini G, et al. Tryptophan Catabolites From Microbiota Engage Aryl Hydrocarbon Receptor and Balance Mucosal Reactivity via interleukin-22. Immunity (2013) 39:372–85. doi: 10.1016/j.immuni.2013.08.003
49. Danne C, Ryzhakov G, Martínez-López M, Ilott NE, Franchini F, Cuskin F, et al. A Large Polysaccharide Produced by Helicobacter Hepaticus Induces an Anti-Inflammatory Gene Signature in Macrophages. Cell Host Microbe (2017) 22:733–45.e5. doi: 10.1016/j.chom.2017.11.002
50. Schulthess J, Pandey S, Capitani M, Rue-Albrecht KC, Arnold I, Franchini F, et al. The Short Chain Fatty Acid Butyrate Imprints an Antimicrobial Program in Macrophages. Immunity (2019) 50:432–45.e7. doi: 10.1016/j.immuni.2018.12.018
51. Wu K, Yuan Y, Yu H, Dai X, Wang S, Sun Z, et al. The Gut Microbial Metabolite Trimethylamine N-Oxide Aggravates GVHD by Inducing M1 Macrophage Polarization in Mice. Blood (2020) 136:501–15. doi: 10.1182/blood.2019003990
52. Nottingham PM, Hungate RE. Isolation of Methanogenic Bacteria From Feces of Man. J Bacteriol (1968) 96:2178–9. doi: 10.1128/jb.96.6.2178-2179.1968
53. Koskinen K, Pausan MR, Perras AK, Beck M, Bang C, Mora M, et al. First Insights Into the Diverse Human Archaeome: Specific Detection of Archaea in the Gastrointestinal Tract, Lung, and Nose and on Skin. mBio (2019) 8:e00824-17. doi: 10.1128/mBio.00824-17
54. Bang C, Weidenbach K, Gutsmann T, Heine H, Schmitz RA. The Intestinal Archaea Methanosphaera Stadtmanae and Methanobrevibacter Smithii Activate Human Dendritic Cells. PloS One (2014) 9:e99411. doi: 10.1371/journal.pone.0099411
55. Blais Lecours P, Duchaine C, Taillefer M, Tremblay C, Veillette M, Cormier Y, et al. Immunogenic Properties of Archaeal Species Found in Bioaerosols. PloS One (2011) 6:e23326. doi: 10.1371/journal.pone.0023326
56. Ruaud A, Esquivel-Elizondo S, La Cuesta-Zuluaga J, Waters JL, Angenent LT, Youngblut ND, et al. Syntrophy via Interspecies H2 Transfer Between Christensenella and Methanobrevibacter Underlies Their Global Cooccurrence in the Human Gut. mBio (2020) 11:e03235-19. doi: 10.1128/mBio.03235-19
57. Hoffmann C, Dollive S, Grunberg S, Chen J, Li H, Wu GD, et al. Archaea and Fungi of the Human Gut Microbiome: Correlations With Diet and Bacterial Residents. PloS One (2013) 8:e66019. doi: 10.1371/journal.pone.0066019
58. Hallen-Adams HE, Kachman SD, Kim J, Legge RM, Martínez I. Fungi Inhabiting the Healthy Human Gastrointestinal Tract: A Diverse and Dynamic Community. Fungal Ecol (2015) 15:9–17. doi: 10.1016/j.funeco.2015.01.006
59. Suhr MJ, Banjara N, Hallen-Adams HE. Sequence-Based Methods for Detecting and Evaluating the Human Gut Mycobiome. Lett Appl Microbiol (2016) 62:209–15. doi: 10.1111/lam.12539
60. Li XV, Leonardi I, Iliev ID. Gut Mycobiota in Immunity and Inflammatory Disease. Immunity (2019) 50:1365–79. doi: 10.1016/j.immuni.2019.05.023
61. Bacher P, Hohnstein T, Beerbaum E, Röcker M, Blango MG, Kaufmann S, et al. Human Anti-Fungal Th17 Immunity and Pathology Rely on Cross-Reactivity Against Candida Albicans. Cell (2019) 176:1340–55.e15. doi: 10.1016/j.cell.2019.01.041
62. Shao T-Y, Ang WX, Jiang TT, Huang FS, Andersen H, Kinder JM, et al. Commensal Candida Albicans Positively Calibrates Systemic Th17 Immunological Responses. Cell Host Microbe (2019) 25:404–17.e6. doi: 10.1016/j.chom.2019.02.004
63. Doron I, Leonardi I, Li XV, Fiers WD, Semon A, Bialt-DeCelie M, et al. Human Gut Mycobiota Tune Immunity Via CARD9-Dependent Induction of Anti-Fungal IgG Antibodies. Cell (2021) 184:1017–31.e14. doi: 10.1016/j.cell.2021.01.016
64. Wheeler ML, Limon JJ, Bar AS, Leal CA, Gargus M, Tang J, et al. Immunological Consequences of Intestinal Fungal Dysbiosis. Cell Host Microbe (2016) 19:865–73. doi: 10.1016/j.chom.2016.05.003
65. Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, et al. Interactions Between Commensal Fungi and the C-Type Lectin Receptor Dectin-1 Influence Colitis. Science (2012) 336:1314–7. doi: 10.1126/science.1221789
66. Wang T, Pan D, Zhou Z, You Y, Jiang C, Zhao X, et al. Dectin-3 Deficiency Promotes Colitis Development Due to Impaired Antifungal Innate Immune Responses in the Gut. PloS Pathog (2016) 12:e1005662. doi: 10.1371/journal.ppat.1005662
67. Jiang TT, Shao T-Y, Ang WX, Kinder JM, Turner LH, Pham G, et al. Commensal Fungi Recapitulate the Protective Benefits of Intestinal Bacteria. Cell Host Microbe (2017) 22:809–16.e4. doi: 10.1016/j.chom.2017.10.013
68. Chiaro TR, Soto R, Zac Stephens W, Kubinak JL, Petersen C, Gogokhia L, et al. A Member of the Gut Mycobiota Modulates Host Purine Metabolism Exacerbating Colitis in Mice. Sci Transl Med (2017) 9:eaaf9044. doi: 10.1126/scitranslmed.aaf9044
69. Standaert-Vitse A, Jouault T, Vandewalle P, Mille C, Seddik M, Sendid B, et al. Candida Albicans Is an Immunogen for Anti-Saccharomyces Cerevisiae Antibody Markers of Crohn’s Disease. Gastroenterology (2006) 130:1764–75. doi: 10.1053/j.gastro.2006.02.009
70. Torres J, Petralia F, Sato T, Wang P, Telesco SE, Choung RS, et al. Serum Biomarkers Identify Patients Who Will Develop Inflammatory Bowel Diseases Up to 5 Years Before Diagnosis. Gastroenterology (2020) 159:96–104. doi: 10.1053/j.gastro.2020.03.007
71. Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, et al. Viruses in the Faecal Microbiota of Monozygotic Twins and Their Mothers. Nature (2010) 466:334–8. doi: 10.1038/nature09199
72. Duerkop BA, Clements CV, Rollins D, Rodrigues JL, Hooper LV. A Composite Bacteriophage Alters Colonization by an Intestinal Commensal Bacterium. Proc Natl Acad Sci USA (2012) 109:17621–6. doi: 10.1073/pnas.1206136109
73. Canchaya C, Fournous G, Chibani-Chennoufi S, Dillmann M-L, Brüssow H. Phage as Agents of Lateral Gene Transfer. Curr Opin Microbiol (2003) 6:417–24. doi: 10.1016/S1369-5274(03)00086-9
74. Hsu BB, Gibson TE, Yeliseyev V, Liu Q, Lyon L, Bry L, et al. Dynamic Modulation of the Gut Microbiota and Metabolome by Bacteriophages in a Mouse Model. Cell Host Microbe (2019) 25:803–14.e5. doi: 10.1016/j.chom.2019.05.001
75. Zuo T, Wong SH, Lam K, Lui R, Cheung K, Tang W, et al. Bacteriophage Transfer During Faecal Microbiota Transplantation in Clostridium Difficile Infection Is Associated With Treatment Outcome. Gut (2018) 67:634–43. doi: 10.1136/gutjnl-2017-313952
76. Gogokhia L, Buhrke K, Bell R, Hoffman B, Brown DG, Hanke-Gogokhia C, et al. Expansion of Bacteriophages Is Linked to Aggravated Intestinal Inflammation and Colitis. Cell Host Microbe (2019) 25:285–99.e8. doi: 10.1016/j.chom.2019.01.008
77. Liang G, Zhao C, Zhang H, Mattei L, Sherrill-Mix S, Bittinger K, et al. The Stepwise Assembly of the Neonatal Virome Is Modulated by Breastfeeding. Nature (2020) 581:470–4. doi: 10.1038/s41586-020-2192-1
78. Fischer JC, Bscheider M, Eisenkolb G, Lin C-C, Wintges A, Otten V, et al. RIG-I/MAVS and STING Signaling Promote Gut Integrity During Irradiation- and Immune-Mediated Tissue Injury. Sci Transl Med (2017) 9:eaag2513. doi: 10.1126/scitranslmed.aag2513
79. Liu L, Gong T, Tao W, Lin B, Li C, Zheng X, et al. Commensal Viruses Maintain Intestinal Intraepithelial Lymphocytes via noncanonical RIG-I signaling. Nat Immunol (2019) 20:1681–91. doi: 10.1038/s41590-019-0513-z
80. Abt MC, Buffie CG, Sušac B, Becattini S, Carter RA, Leiner I, et al. TLR-7 Activation Enhances IL-22-Mediated Colonization Resistance Against Vancomycin-Resistant Enterococcus. Sci Transl Med (2016) 8:327ra25. doi: 10.1126/scitranslmed.aad6663
81. Yang J-Y, Kim M-S, Kim E, Cheon JH, Lee Y-S, Kim Y, et al. Enteric Viruses Ameliorate Gut Inflammation Via Toll-Like Receptor 3 and Toll-Like Receptor 7-Mediated Interferon-β Production. Immunity (2016) 44:889–900. doi: 10.1016/j.immuni.2016.03.009
82. Broggi A, Tan Y, Granucci F, Zanoni I. IFN-λ Suppresses Intestinal Inflammation by Non-Translational Regulation of Neutrophil Function. Nat Immunol (2017) 18:1084–93. doi: 10.1038/ni.3821
83. Kernbauer E, Ding Y, Cadwell K. An Enteric Virus can Replace the Beneficial Function of Commensal Bacteria. Nature (2014) 516:94–8. doi: 10.1038/nature13960
84. Ingle H, Lee S, Ai T, Orvedahl A, Rodgers R, Zhao G, et al. Viral Complementation of Immunodeficiency Confers Protection Against Enteric Pathogens. Via Interferon-λ Nat Microbiol (2019) 4:1120–8. doi: 10.1038/s41564-019-0416-7
85. Neil JA, Matsuzawa-Ishimoto Y, Kernbauer-Hölzl E, Schuster SL, Sota S, Venzon M, et al. IFN-I and IL-22 Mediate Protective Effects of Intestinal Viral Infection. Nat Microbiol (2019) 4:1737–49. doi: 10.1038/s41564-019-0470-1
86. Lukeš J, Stensvold CR, Jirků-Pomajbíková K, Wegener Parfrey L. Are Human Intestinal Eukaryotes Beneficial or Commensals? PloS Pathog (2015) 11:e1005039. doi: 10.1371/journal.ppat.1005039
87. Chudnovskiy A, Mortha A, Kana V, Kennard A, Ramirez JD, Rahman A, et al. Host-Protozoan Interactions Protect From Mucosal Infections Through Activation of the Inflammasome. Cell (2016) 167:444–456.e14. doi: 10.1016/j.cell.2016.08.076
88. Escalante NK, Lemire P, Cruz Tleugabulova M, Prescott D, Mortha A, Streutker CJ, et al. The Common Mouse Protozoa Tritrichomonas Muris Alters Mucosal T Cell Homeostasis and Colitis Susceptibility. J Exp Med (2016) 213:2841–50. doi: 10.1084/jem.20161776
89. Schneider C, O’Leary CE, Moltke Jv, Liang H-E, Ang QY, Turnbaugh PJ, et al. A Metabolite-Triggered Tuft Cell-ILC2 Circuit Drives Small Intestinal Remodeling. Cell (2018) 174:271–84.e14. doi: 10.1016/j.cell.2018.05.014
90. Gerbe F, Sidot E, Smyth DJ, Ohmoto M, Matsumoto I, Dardalhon V, et al. Intestinal Epithelial Tuft Cells Initiate Type 2 Mucosal Immunity to Helminth Parasites. Nature (2016) 529:226–30. doi: 10.1038/nature16527
91. Howitt MR, Lavoie S, Michaud M, Blum AM, Tran SV, Weinstock JV, et al. Tuft Cells, Taste-Chemosensory Cells, Orchestrate Parasite Type 2 Immunity in the Gut. Science (2016) 351:1329–33. doi: 10.1126/science.aaf1648
92. Wilen CB, Lee S, Hsieh LL, Orchard RC, Desai C, Hykes BL, et al. Tropism for Tuft Cells Determines Immune Promotion of Norovirus Pathogenesis. Science (2018) 360:204–8. doi: 10.1126/science.aar3799
93. Osborne LC, Monticelli LA, Nice TJ, Sutherland TE, Siracusa MC, Hepworth MR, et al. Virus-Helminth Coinfection Reveals a Microbiota-Independent Mechanism of Immunomodulation. Science (2014) 345:578–82. doi: 10.1126/science.1256942
94. Reese TA, Wakeman BS, Choi HS, Hufford MM, Huang SC, Zhang X, et al. Helminth Infection Reactivates Latent γ-Herpesvirus Via Cytokine Competition at a Viral Promoter. Science (2014) 345:573–7. doi: 10.1126/science.1254517
95. Dang AT, Marsland BJ. Microbes, Metabolites, and the Gut-Lung Axis. Mucosal Immunol (2019) 12:843–50. doi: 10.1038/s41385-019-0160-6
96. Morais LH, Schreiber HL, Mazmanian SK. The Gut Microbiota-Brain Axis in Behaviour and Brain Disorders. Nat Rev Microbiol (2020) 19:241–55. doi: 10.1038/s41579-020-00460-0
97. Tripathi A, Debelius J, Brenner DA, Karin M, Loomba R, Schnabl B, et al. The Gut-Liver Axis and the Intersection With the Microbiome. Nat Rev Gastroenterol Hepatol (2018) 15:397–411. doi: 10.1038/s41575-018-0011-z
98. Fagundes CT, Amaral FA, Vieira AT, Soares AC, Pinho V, Nicoli JR, et al. Transient TLR Activation Restores Inflammatory Response and Ability to Control Pulmonary Bacterial Infection in Germfree Mice. J Immunol (2012) 188:1411–20. doi: 10.4049/jimmunol.1101682
99. Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, et al. Microbiota Regulates Immune Defense Against Respiratory Tract Influenza A Virus Infection. Proc Natl Acad Sci USA (2011) 108:5354–9. doi: 10.1073/pnas.1019378108
100. Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF, et al. Commensal Bacteria Calibrate the Activation Threshold of Innate Antiviral Immunity. Immunity (2012) 37:158–70. doi: 10.1016/j.immuni.2012.04.011
101. Oh JZ, Ravindran R, Chassaing B, Carvalho FA, Maddur MS, Bower M, et al. TLR5-Mediated Sensing of Gut Microbiota Is Necessary for Antibody Responses to Seasonal Influenza Vaccination. Immunity (2014) 41:478–92. doi: 10.1016/j.immuni.2014.08.009
102. Steed AL, Christophi GP, Kaiko GE, Sun L, Goodwin VM, Jain U, et al. The Microbial Metabolite Desaminotyrosine Protects From Influenza Through Type I Interferon. Science (2017) 357:498–502. doi: 10.1126/science.aam5336
103. Trompette A, Gollwitzer ES, Pattaroni C, Lopez-Mejia IC, Riva E, Pernot J, et al. Dietary Fiber Confers Protection Against Flu by Shaping Ly6c- Patrolling Monocyte Hematopoiesis and CD8+ T Cell Metabolism. Immunity (2018) 48:992–1005.e8. doi: 10.1016/j.immuni.2018.04.022
104. Kim Y-G, Udayanga KG, Totsuka N, Weinberg JB, Núñez G, Shibuya A. Gut Dysbiosis Promotes M2 Macrophage Polarization and Allergic Airway Inflammation Via Fungi-Induced PGE₂. Cell Host Microbe (2014) 15:95–102. doi: 10.1016/j.chom.2013.12.010
105. Arnold IC, Dehzad N, Reuter S, Martin H, Becher B, Taube C, et al. Helicobacter Pylori Infection Prevents Allergic Asthma in Mouse Models Through the Induction of Regulatory T Cells. J Clin Invest (2011) 121:3088–93. doi: 10.1172/JCI45041
106. Fujimura KE, Demoor T, Rauch M, Faruqi AA, Jang S, Johnson CC, et al. House Dust Exposure Mediates Gut Microbiome Lactobacillus Enrichment and Airway Immune Defense Against Allergens and Virus Infection. Proc Natl Acad Sci USA (2014) 111:805–10. doi: 10.1073/pnas.1310750111
107. Bazett M, Bergeron M-E, Haston CK. Streptomycin Treatment Alters the Intestinal Microbiome, Pulmonary T Cell Profile and Airway Hyperresponsiveness in a Cystic Fibrosis Mouse Model. Sci Rep (2016) 6:19189. doi: 10.1038/srep19189
108. Erny D, Hrabě de Angelis AL, Jaitin D, Wieghofer P, Staszewski O, David E, et al. Host Microbiota Constantly Control Maturation and Function of Microglia in the CNS. Nat Neurosci (2015) 18:965–77. doi: 10.1038/nn.4030
109. Vuong HE, Pronovost GN, Williams DW, Coley EJ, Siegler EL, Qiu A, et al. The Maternal Microbiome Modulates Fetal Neurodevelopment in Mice. Nature (2020) 586:281–6. doi: 10.1038/s41586-020-2745-3
110. Berer K, Mues M, Koutrolos M, Rasbi ZA, Boziki M, Johner C, et al. Commensal Microbiota and Myelin Autoantigen Cooperate to Trigger Autoimmune Demyelination. Nature (2011) 479:538–41. doi: 10.1038/nature10554
111. Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-Cell Responses to Gut Microbiota Promote Experimental Autoimmune Encephalomyelitis. Proc Natl Acad Sci USA (2011) 108(Suppl 1):4615–22. doi: 10.1073/pnas.1000082107
112. Mezö C, Dokalis N, Mossad O, Staszewski O, Neuber J, Yilmaz B, et al. Different Effects of Constitutive and Induced Microbiota Modulation on Microglia in a Mouse Model of Alzheimer’s Disease. Acta Neuropathol Commun (2020) 8:119. doi: 10.1186/s40478-020-00988-5
113. Berer K, Gerdes LA, Cekanaviciute E, Jia X, Xiao L, Xia Z, et al. Gut Microbiota From Multiple Sclerosis Patients Enables Spontaneous Autoimmune Encephalomyelitis in Mice. Proc Natl Acad Sci USA (2017) 114:10719–24. doi: 10.1073/pnas.1711233114
114. Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, et al. Microbiota Modulate Behavioral and Physiological Abnormalities Associated With Neurodevelopmental Disorders. Cell (2013) 155:1451–63. doi: 10.1016/j.cell.2013.11.024
115. Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, et al. Gut Microbiota Regulate Motor Deficits and Neuroinflammation in a Model of Parkinson’s Disease. Cell (2016) 167:1469–1480.e12. doi: 10.1016/j.cell.2016.11.018
116. Benakis C, Brea D, Caballero S, Faraco G, Moore J, Murphy M, et al. Commensal Microbiota Affects Ischemic Stroke Outcome by Regulating Intestinal γδ T Cells. Nat Med (2016) 22:516–23. doi: 10.1038/nm.4068
117. Bik EM, Long CD, Armitage GC, Loomer P, Emerson J, Mongodin EF, et al. Bacterial Diversity in the Oral Cavity of 10 Healthy Individuals. ISME J (2010) 4:962–74. doi: 10.1038/ismej.2010.30
118. Koren N, Zubeidat K, Saba Y, Horev Y, Barel O, Wilharm A, et al. Maturation of the Neonatal Oral Mucosa Involves Unique Epithelium-Microbiota Interactions. Cell Host Microbe (2021) 29:197–209. doi: 10.1016/j.chom.2020.12.006
119. Stein PS, Desrosiers M, Donegan SJ, Yepes JF, Kryscio RJ. Tooth Loss, Dementia and Neuropathology in the Nun Study. J Am Dent Assoc (2007) 138:1314–22. doi: 10.14219/jada.archive.2007.0046
120. Ide M, Harris M, Stevens A, Sussams R, Hopkins V, Culliford D, et al. Periodontitis and Cognitive Decline in Alzheimer’s Disease. PloS One (2016) 11:e0151081. doi: 10.1371/journal.pone.0151081
121. Ma C, Han M, Heinrich B, Fu Q, Zhang Q, Sandhu M, et al. Gut Microbiome-Mediated Bile Acid Metabolism Regulates Liver Cancer Via NKT Cells. Science (2018) 360:806–15. doi: 10.1126/science.aan5931
122. Ferrere G, Wrzosek L, Cailleux F, Turpin W, Puchois V, Spatz M, et al. Fecal Microbiota Manipulation Prevents Dysbiosis and Alcohol-Induced Liver Injury in Mice. J Hepatol (2017) 66:806–15. doi: 10.1016/j.jhep.2016.11.008
123. Philips CA, Pande A, Shasthry SM, Jamwal KD, Khillan V, Chandel SS, et al. Healthy Donor Fecal Microbiota Transplantation in Steroid-Ineligible Severe Alcoholic Hepatitis: A Pilot Study. Clin Gastroenterol Hepatol (2017) 15:600–2. doi: 10.1016/j.cgh.2016.10.029
124. Noverr MC, Falkowski NR, McDonald RA, McKenzie AN, Huffnagle GB. Development of Allergic Airway Disease in Mice Following Antibiotic Therapy and Fungal Microbiota Increase: Role of Host Genetics, Antigen, and Interleukin-13. Infect Immun (2005) 73:30–8. doi: 10.1128/IAI.73.1.30-38.2005
125. Li X, Leonardi I, Semon A, Doron I, Gao IH, Putzel GG, et al. Response to Fungal Dysbiosis by Gut-Resident CX3CR1+ Mononuclear Phagocytes Aggravates Allergic Airway Disease. Cell Host Microbe (2018) 24:847–856.e4. doi: 10.1016/j.chom.2018.11.003
126. Noverr MC, Noggle RM, Toews GB, Huffnagle GB. Role of Antibiotics and Fungal Microbiota in Driving Pulmonary Allergic Responses. Infect Immun (2004) 72:4996–5003. doi: 10.1128/IAI.72.9.4996-5003.2004
127. Skalski JH, Limon JJ, Sharma P, Gargus MD, Nguyen C, Tang J, et al. Expansion of Commensal Fungus Wallemia Mellicola in the Gastrointestinal Mycobiota Enhances the Severity of Allergic Airway Disease in Mice. PloS Pathog (2018) 14:e1007260. doi: 10.1371/journal.ppat.1007260
128. Tso GH, Reales-Calderon JA, Tan AS, Sem X, Le GT, Tan TG, et al. Experimental Evolution of a Fungal Pathogen Into a Gut Symbiont. Science (2018) 362:589–95. doi: 10.1126/science.aat0537
129. Takata K, Tomita T, Okuno T, Kinoshita M, Koda T, Honorat JA, et al. Dietary Yeasts Reduce Inflammation in Central Nerve System. via Microflora Ann Clin Transl Neurol (2015) 2:56–66. doi: 10.1002/acn3.153
130. Yang A-M, Inamine T, Hochrath K, Chen P, Wang L, Llorente C, et al. Intestinal Fungi Contribute to Development of Alcoholic Liver Disease. J Clin Invest (2017) 127:2829–41. doi: 10.1172/JCI90562
131. Zhang Z, Li J, Zheng W, Zhao G, Zhang H, Wang X, et al. Peripheral Lymphoid Volume Expansion and Maintenance Are Controlled by Gut Microbiota via RALDH+ Dendritic Cells. Immunity (2016) 44:330–42. doi: 10.1016/j.immuni.2016.01.004
132. Yeung F, Chen Y-H, Lin J-D, Leung JM, McCauley C, Devlin JC, et al. Altered Immunity of Laboratory Mice in the Natural Environment Is Associated With Fungal Colonization. Cell Host Microbe (2020) 27:809–22. doi: 10.1016/j.chom.2020.02.015
133. Lin J-D, Devlin JC, Yeung F, McCauley C, Leung JM, Chen Y-H, et al. Rewilding Nod2 and Atg16l1 Mutant Mice Uncovers Genetic and Environmental Contributions to Microbial Responses and Immune Cell Composition. Cell Host Microbe (2020) 27:830–840.e4. doi: 10.1016/j.chom.2020.03.001
134. Thépaut M, Grandjean T, Hober D, Lobert P-E, Bortolotti P, Faure K, et al. Protective Role of Murine Norovirus Against Pseudomonas Aeruginosa Acute Pneumonia. Vet Res (2015) 46:91. doi: 10.1186/s13567-015-0239-3
135. Lang S, Demir M, Martin A, Jiang L, Zhang X, Duan Y, et al. Intestinal Virome Signature Associated With Severity of Nonalcoholic Fatty Liver Disease. Gastroenterology (2020) 159:1839–52. doi: 10.1053/j.gastro.2020.07.005
136. Virgin HW, Wherry EJ, Ahmed R. Redefining Chronic Viral Infection. Cell (2009) 138:30–50. doi: 10.1016/j.cell.2009.06.036
137. Barton ES, White DW, Cathelyn JS, Brett-McClellan KA, Engle M, Diamond MS, et al. Herpesvirus Latency Confers Symbiotic Protection From Bacterial Infection. Nature (2007) 447:326–9. doi: 10.1038/nature05762
138. Yager EJ, Szaba FM, Kummer LW, Lanzer KG, Burkum CE, Smiley ST, et al. Gamma-Herpesvirus-Induced Protection Against Bacterial Infection is Transient. Viral Immunol (2009) 22:67–72. doi: 10.1089/vim.2008.0086
139. White DW, Keppel CR, Schneider SE, Reese TA, Coder J, Payton JE, et al. Latent Herpesvirus Infection Arms NK Cells. Blood (2010) 115:4377–83. doi: 10.1182/blood-2009-09-245464
140. Sun L, Miyoshi H, Origanti S, Nice TJ, Barger AC, Manieri NA, et al. Type I Interferons Link Viral Infection to Enhanced Epithelial Turnover and Repair. Cell Host Microbe (2015) 17:85–97. doi: 10.1016/j.chom.2014.11.004
141. Wilson MS, Taylor MD, Balic A, Finney CA, Lamb JR, Maizels RM. Suppression of Allergic Airway Inflammation by Helminth-Induced Regulatory T Cells. J Exp Med (2005) 202:1199–212. doi: 10.1084/jem.20042572
142. McSorley HJ, Blair NF, Smith KA, McKenzie AN, Maizels RM. Blockade of IL-33 Release and Suppression of Type 2 Innate Lymphoid Cell Responses by Helminth Secreted Products in Airway Allergy. Mucosal Immunol (2014) 7:1068–78. doi: 10.1038/mi.2013.123
143. McSorley HJ, O’Gorman MT, Blair N, Sutherland TE, Filbey KJ, Maizels RM. Suppression of Type 2 Immunity and Allergic Airway Inflammation by Secreted Products of the Helminth Heligmosomoides Polygyrus. Eur J Immunol (2012) 42:2667–82. doi: 10.1002/eji.201142161
144. Wammes LJ, Mpairwe H, Elliott AM, Yazdanbakhsh M. Helminth Therapy or Elimination: Epidemiological, Immunological, and Clinical Considerations. Lancet Infect Dis (2014) 14:1150–62. doi: 10.1016/S1473-3099(14)70771-6
145. Belkaid Y, Tamoutounour S. The Influence of Skin Microorganisms on Cutaneous Immunity. Nat Rev Immunol (2016) 16:353–66. doi: 10.1038/nri.2016.48
146. Belkaid Y, Segre JA. Dialogue Between Skin Microbiota and Immunity. Science (2014) 346:954–9. doi: 10.1126/science.1260144
147. Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, et al. Topographical and Temporal Diversity of the Human Skin Microbiome. Science (2009) 324:1190–2. doi: 10.1126/science.1171700
148. Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, et al. Topographic Diversity of Fungal and Bacterial Communities in Human Skin. Nature (2013) 498:367–70. doi: 10.1038/nature12171
149. Biasucci G, Rubini M, Riboni S, Morelli L, Bessi E, Retetangos C. Mode of Delivery Affects the Bacterial Community in the Newborn Gut. Early Hum Dev (2010) 86(Suppl 1):13–5. doi: 10.1016/j.earlhumdev.2010.01.004
150. Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery Mode Shapes the Acquisition and Structure of the Initial Microbiota Across Multiple Body Habitats in Newborns. Proc Natl Acad Sci USA (2010) 107:11971–5. doi: 10.1073/pnas.1002601107
151. Oh J, Byrd AL, Deming C, Conlan S, NISC Comparative Sequencing Program, Kong HH, et al. Biogeography and Individuality Shape Function in the Human Skin Metagenome. Nature (2014) 514:59–64. doi: 10.1038/nature13786
152. Naik S, Bouladoux N, Linehan JL, Han S-J, Harrison OJ, Wilhelm C, et al. Commensal-Dendritic-Cell Interaction Specifies a Unique Protective Skin Immune Signature. Nature (2015) 520:104–8. doi: 10.1038/nature14052
153. Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, et al. Compartmentalized Control of Skin Immunity by Resident Commensals. Science (2012) 337:1115–9. doi: 10.1126/science.1225152
154. Scharschmidt TC, Vasquez KS, Pauli ML, Leitner EG, Chu K, Truong H-A, et al. Commensal Microbes and Hair Follicle Morphogenesis Coordinately Drive Treg Migration Into Neonatal Skin. Cell Host Microbe (2017) 21:467–77. doi: 10.1016/j.chom.2017.03.001
155. Scharschmidt TC, Vasquez KS, Truong H-A, Gearty SV, Pauli ML, Nosbaum A, et al. A Wave of Regulatory T Cells Into Neonatal Skin Mediates Tolerance to Commensal Microbes. Immunity (2015) 43:1011–21. doi: 10.1016/j.immuni.2015.10.016
156. Chehoud C, Rafail S, Tyldsley AS, Seykora JT, Lambris JD, Grice EA. Complement Modulates the Cutaneous Microbiome and Inflammatory Milieu. Proc Natl Acad Sci USA (2013) 110:15061–6. doi: 10.1073/pnas.1307855110
157. Laborel-Préneron E, Bianchi P, Boralevi F, Lehours P, Fraysse F, Morice-Picard F, et al. Effects of the Staphylococcus Aureus and Staphylococcus Epidermidis Secretomes Isolated From the Skin Microbiota of Atopic Children on CD4+ T Cell Activation. PloS One (2015) 10:e0141067. doi: 10.1371/journal.pone.0141067
158. Linehan JL, Harrison OJ, Han S-J, Byrd AL, Vujkovic-Cvijin I, Villarino AV, et al. Non-Classical Immunity Controls Microbiota Impact on Skin Immunity and Tissue Repair. Cell (2018) 172:784–96.e18. doi: 10.1016/j.cell.2017.12.033
159. Gueniche A, Knaudt B, Schuck E, Volz T, Bastien P, Martin R, et al. Effects of Nonpathogenic Gram-Negative Bacterium Vitreoscilla Filiformis Lysate on Atopic Dermatitis: A Prospective, Randomized, Double-Blind, Placebo-Controlled Clinical Study. Br J Dermatol (2008) 159:1357–63. doi: 10.1111/j.1365-2133.2008.08836.x
160. Volz T, Skabytska Y, Guenova E, Chen K-M, Frick J-S, Kirschning CJ, et al. Nonpathogenic Bacteria Alleviating Atopic Dermatitis Inflammation Induce IL-10-Producing Dendritic Cells and Regulatory Tr1 Cells. J Invest Dermatol (2014) 134:96–104. doi: 10.1038/jid.2013.291
161. Lai Y, Di Nardo A, Nakatsuji T, Leichtle A, Yang Y, Cogen AL, et al. Commensal Bacteria Regulate Toll-Like Receptor 3-Dependent Inflammation After Skin Injury. Nat Med (2009) 15:1377–82. doi: 10.1038/nm.2062
162. Probst AJ, Auerbach AK, Moissl-Eichinger C. Archaea on Human Skin. PloS One (2013) 8:e65388. doi: 10.1371/journal.pone.0065388
163. Moissl-Eichinger C, Probst AJ, Birarda G, Auerbach A, Koskinen K, Wolf P, et al. Human Age and Skin Physiology Shape Diversity and Abundance of Archaea on Skin. Sci Rep (2017) 7:4039. doi: 10.1038/s41598-017-04197-4
164. Hurabielle C, Link VM, Bouladoux N, Han S-J, Merrill ED, Lightfoot YL, et al. Immunity to Commensal Skin Fungi Promotes Psoriasiform Skin Inflammation. Proc Natl Acad Sci USA (2020) 117:16465–74. doi: 10.1073/pnas.2003022117
165. Hannigan GD, Meisel JS, Tyldsley AS, Zheng Q, Hodkinson BP, SanMiguel AJ, et al. The Human Skin Double-Stranded DNA Virome: Topographical and Temporal Diversity, Genetic Enrichment, and Dynamic Associations With the Host Microbiome. mBio (2015) 6:e01578–15. doi: 10.1128/mBio.01578-15
166. Oh J, Byrd AL, Park M, Kong HH, Segre JA. Temporal Stability of the Human Skin Microbiome. Cell (2016) 165:854–66. doi: 10.1016/j.cell.2016.04.008
167. Sweere JM, van Belleghem JD, Ishak H, Bach MS, Popescu M, Sunkari V, et al. Bacteriophage Trigger Antiviral Immunity and Prevent Clearance of Bacterial Infection. Science (2019) 363:eaat9691. doi: 10.1126/science.aat9691
168. Dickson RP, Erb-Downward JR, Freeman CM, McCloskey L, Falkowski NR, Huffnagle GB, et al. Bacterial Topography of the Healthy Human Lower Respiratory Tract. mBio (2017) 8:e02287-16. doi: 10.1128/mBio.02287-16
169. Segal LN, Clemente JC, Tsay J-CJ, Koralov SB, Keller BC, Wu BG, et al. Enrichment of the Lung Microbiome With Oral Taxa is Associated With Lung Inflammation of a Th17 Phenotype. Nat Microbiol (2016) 1:16031. doi: 10.1038/nmicrobiol.2016.31
170. Wang J, Li F, Sun R, Gao X, Wei H, Li L-J, et al. Bacterial Colonization Dampens Influenza-Mediated Acute Lung Injury via Induction of M2 Alveolar Macrophages. Nat Commun (2013) 4:2106. doi: 10.1038/ncomms3106
171. Gollwitzer ES, Saglani S, Trompette A, Yadava K, Sherburn R, McCoy KD, et al. Lung Microbiota Promotes Tolerance to Allergens in Neonates Via PD-L1. Nat Med (2014) 20:642–7. doi: 10.1038/nm.3568
172. Ege MJ, Mayer M, Normand A-C, Genuneit J, Cookson WO, Braun-Fahrländer C, et al. Exposure to Environmental Microorganisms and Childhood Asthma. N Engl J Med (2011) 364:701–9. doi: 10.1056/NEJMoa1007302
173. Conrad ML, Ferstl R, Teich R, Brand S, Blümer N, Yildirim AO, et al. Maternal TLR Signaling Is Required for Prenatal Asthma Protection by the Nonpathogenic Microbe Acinetobacter Lwoffii F78. J Exp Med (2009) 206:2869–77. doi: 10.1084/jem.20090845
174. Rubio-Portillo E, Orts D, Llorca E, Fernández C, Antón J, Ferrer C, et al. The Domestic Environment and the Lung Mycobiome. Microorganisms (2020) 8:1717. doi: 10.3390/microorganisms8111717
175. Willner D, Furlan M, Haynes M, Schmieder R, Angly FE, Silva J, et al. Metagenomic Analysis of Respiratory Tract DNA Viral Communities in Cystic Fibrosis and Non-Cystic Fibrosis Individuals. PloS One (2009) 4:e7370. doi: 10.1371/journal.pone.0007370
176. Young JC, Chehoud C, Bittinger K, Bailey A, Diamond JM, Cantu E, et al. Viral Metagenomics Reveal Blooms of Anelloviruses in the Respiratory Tract of Lung Transplant Recipients. Am J Transplant (2015) 15:200–9. doi: 10.1111/ajt.13031
177. Chang Y-J, Kim HY, Albacker LA, Lee HH, Baumgarth N, Akira S, et al. Influenza Infection in Suckling Mice Expands an NKT Cell Subset That Protects Against Airway Hyperreactivity. J Clin Invest (2011) 121:57–69. doi: 10.1172/JCI44845
178. Krishnamoorthy N, Khare A, Oriss TB, Raundhal M, Morse C, Yarlagadda M, et al. Early Infection With Respiratory Syncytial Virus Impairs Regulatory T Cell Function and Increases Susceptibility to Allergic Asthma. Nat Med (2012) 18:1525–30. doi: 10.1038/nm.2896
179. Kim EY, Battaile JT, Patel AC, You Y, Agapov E, Grayson MH, et al. Persistent Activation of an Innate Immune Response Translates Respiratory Viral Infection Into Chronic Lung Disease. Nat Med (2008) 14:633–40. doi: 10.1038/nm1770
180. Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, et al. Vaginal Microbiome of Reproductive-Age Women. Proc Natl Acad Sci USA (2011) 108(Suppl 1):4680–7. doi: 10.1073/pnas.1002611107
181. Aroutcheva A, Gariti D, Simon M, Shott S, Faro J, Simoes JA, et al. Defense Factors of Vaginal Lactobacilli. Am J Obstet Gynecol (2001) 185:375–9. doi: 10.1067/mob.2001.115867
182. Hearps AC, Tyssen D, Srbinovski D, Bayigga L, Diaz DJ, Aldunate M, et al. Vaginal Lactic Acid Elicits an Anti-Inflammatory Response From Human Cervicovaginal Epithelial Cells and Inhibits Production of Pro-Inflammatory Mediators Associated With HIV Acquisition. Mucosal Immunol (2017) 10:1480–90. doi: 10.1038/mi.2017.27
183. Borgdorff H, Tsivtsivadze E, Verhelst R, Marzorati M, Jurriaans S, Ndayisaba GF, et al. Lactobacillus-Dominated Cervicovaginal Microbiota Associated With Reduced HIV/STI Prevalence and Genital HIV Viral Load in African Women. ISME J (2014) 8:1781–93. doi: 10.1038/ismej.2014.26
184. Kirjavainen PV, Pautler S, Baroja ML, Anukam K, Crowley K, Carter K, et al. Abnormal Immunological Profile and Vaginal Microbiota in Women Prone to Urinary Tract Infections. Clin Vaccine Immunol (2009) 16:29–36. doi: 10.1128/CVI.00323-08
185. Gosmann C, Anahtar MN, Handley SA, Farcasanu M, Abu-Ali G, Bowman BA, et al. Lactobacillus-Deficient Cervicovaginal Bacterial Communities Are Associated With Increased HIV Acquisition in Young South African Women. Immunity (2017) 46:29–37. doi: 10.1016/j.immuni.2016.12.013
186. Anahtar MN, Byrne EH, Doherty KE, Bowman BA, Yamamoto HS, Soumillon M, et al. Cervicovaginal Bacteria Are a Major Modulator of Host Inflammatory Responses in the Female Genital Tract. Immunity (2015) 42:965–76. doi: 10.1016/j.immuni.2015.04.019
187. Fettweis JM, Serrano MG, Brooks JP, Edwards DJ, Girerd PH, Parikh HI, et al. The Vaginal Microbiome and Preterm Birth. Nat Med (2019) 25:1012–21. doi: 10.1038/s41591-019-0450-2
188. Belay N, Mukhopadhyay B, Conway de Macario E, Galask R, Daniels L. Methanogenic Bacteria in Human Vaginal Samples. J Clin Microbiol (1990) 28:1666–8. doi: 10.1128/JCM.28.7.1666-1668.1990
189. Grine G, Drouet H, Fenollar F, Bretelle F, Raoult D, Drancourt M. Detection of Methanobrevibacter Smithii in Vaginal Samples Collected From Women Diagnosed With Bacterial Vaginosis. Eur J Clin Microbiol Infect Dis (2019) 38:1643–9. doi: 10.1007/s10096-019-03592-1
190. Guo R, Zheng N, Lu H, Yin H, Yao J, Chen Y. Increased Diversity of Fungal Flora in the Vagina of Patients With Recurrent Vaginal Candidiasis and Allergic Rhinitis. Microb Ecol (2012) 64:918–27. doi: 10.1007/s00248-012-0084-0
191. Bauters TG, Dhont MA, Temmerman MI, Nelis HJ. Prevalence of Vulvovaginal Candidiasis and Susceptibility to Fluconazole in Women. Am J Obstet Gynecol (2002) 187:569–74. doi: 10.1067/mob.2002.125897
192. Jakobsen RR, Haahr T, Humaidan P, Jensen JS, Kot WP, Castro-Mejia JL, et al. Characterization of the Vaginal DNA Virome in Health and Dysbiosis. Viruses (2020) 12:1143. doi: 10.3390/v12101143
193. Wylie KM, Mihindukulasuriya KA, Zhou Y, Sodergren E, Storch GA, Weinstock GM. Metagenomic Analysis of Double-Stranded DNA Viruses in Healthy Adults. BMC Biol (2014) 12. doi: 10.1186/s12915-014-0071-7
194. Wylie KM, Wylie TN, Cahill AG, Macones GA, Tuuli MG, Stout MJ. The Vaginal Eukaryotic DNA Virome and Preterm Birth. Am J Obstet Gynecol (2018) 219:189.e1–189.e12. doi: 10.1016/j.ajog.2018.04.048
195. Damelin LH, Paximadis M, Mavri-Damelin D, Birkhead M, Lewis DA, Tiemessen CT. Identification of Predominant Culturable Vaginal Lactobacillus Species and Associated Bacteriophages From Women With and Without Vaginal Discharge Syndrome in South Africa. J Med Microbiol (2011) 60:180–3. doi: 10.1099/jmm.0.024463-0
196. Kiliç AO, Pavlova SI, Alpay S, Kiliç SS, Tao L. Comparative Study of Vaginal Lactobacillus Phages Isolated From Women in the United States and Turkey: Prevalence, Morphology, Host Range, and DNA Homology. Clin Diagn Lab Immunol (2001) 8:31–9. doi: 10.1128/CDLI.8.1.31-39.2001
197. Martín R, Soberón N, Escobedo S, Suárez JE. Bacteriophage Induction Versus Vaginal Homeostasis: Role of H(2)O(2) in the Selection of Lactobacillus Defective Prophages. Int Microbiol (2009) 12:131–6. doi: 10.2436/20.1501.01.90
198. Grandi N, Tramontano E. Human Endogenous Retroviruses Are Ancient Acquired Elements Still Shaping Innate Immune Responses. Front Immunol (2018) 9:2039. doi: 10.3389/fimmu.2018.02039
199. Perron H, Lazarini F, Ruprecht K, Péchoux-Longin C, Seilhean D, Sazdovitch V, et al. Human Endogenous Retrovirus (HERV)-W ENV and GAG Proteins: Physiological Expression in Human Brain and Pathophysiological Modulation in Multiple Sclerosis Lesions. J Neurovirol (2005) 11:23–33. doi: 10.1080/13550280590901741
200. Mi S, Lee X, Li X, Veldman GM, Finnerty H, Racie L, et al. Syncytin is a Captive Retroviral Envelope Protein Involved in Human Placental Morphogenesis. Nature (2000) 403:785–9. doi: 10.1038/35001608
201. Tolosa JM, Schjenken JE, Clifton VL, Vargas A, Barbeau B, Lowry P, et al. The Endogenous Retroviral Envelope Protein Syncytin-1 Inhibits LPS/PHA-Stimulated Cytokine Responses in Human Blood and is Sorted Into Placental Exosomes. Placenta (2012) 33:933–41. doi: 10.1016/j.placenta.2012.08.004
202. Cianciolo GJ, Copeland TD, Oroszlan S, Snyderman R. Inhibition of Lymphocyte Proliferation by a Synthetic Peptide Homologous to Retroviral Envelope Proteins. Science (1985) 230:453–5. doi: 10.1126/science.2996136
203. Brown JM, Hazen SL. Targeting of Microbe-Derived Metabolites to Improve Human Health: The Next Frontier for Drug Discovery. J Biol Chem (2017) 292:8560–8. doi: 10.1074/jbc.R116.765388
204. Herrath MGv, Nepom GT. Lost in Translation: Barriers to Implementing Clinical Immunotherapeutics for Autoimmunity. J Exp Med (2005) 202:1159–62. doi: 10.1084/jem.20051224
205. Hay M, Thomas DW, Craighead JL, Economides C, Rosenthal J. Clinical Development Success Rates for Investigational Drugs. Nat Biotechnol (2014) 32:40–51. doi: 10.1038/nbt.2786
206. Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, et al. Genomic Responses in Mouse Models Poorly Mimic Human Inflammatory Diseases. Proc Natl Acad Sci USA (2013) 110:3507–12. doi: 10.1073/pnas.1222878110
207. Mak IW, Evaniew N, Ghert M. Lost in Translation: Animal Models and Clinical Trials in Cancer Treatment. Am J Transl Res (2014) 6:114–8.
208. Mestas J, Hughes CC. Of Mice and Not Men: Differences Between Mouse and Human Immunology. J Immunol (2004) 172:2731–8. doi: 10.4049/jimmunol.172.5.2731
209. Payne KJ, Crooks GM. Immune-Cell Lineage Commitment: Translation From Mice to Humans. Immunity (2007) 26:674–7. doi: 10.1016/j.immuni.2007.05.011
210. Shay T, Jojic V, Zuk O, Rothamel K, Puyraimond-Zemmour D, Feng T, et al. Conservation and Divergence in the Transcriptional Programs of the Human and Mouse Immune Systems. Proc Natl Acad Sci USA (2013) 110:2946–51. doi: 10.1073/pnas.1222738110
211. Wong CH, Siah KW, Lo AW. Estimation of Clinical Trial Success Rates and Related Parameters. Biostatistics (2019) 20:273–86. doi: 10.1093/biostatistics/kxx069
212. Beura LK, Hamilton SE, Bi K, Schenkel JM, Odumade OA, Casey KA, et al. Normalizing the Environment Recapitulates Adult Human Immune Traits in Laboratory Mice. Nature (2016) 532:512–6. doi: 10.1038/nature17655
213. Reese TA, Bi K, Kambal A, Filali-Mouhim A, Beura LK, Bürger MC, et al. Sequential Infection With Common Pathogens Promotes Human-Like Immune Gene Expression and Altered Vaccine Response. Cell Host Microbe (2016) 19:713–9. doi: 10.1016/j.chom.2016.04.003
214. Leung JM, Budischak SA, Chung The H, Hansen C, Bowcutt R, Neill R, et al. Rapid Environmental Effects on Gut Nematode Susceptibility in Rewilded Mice. PloS Biol (2018) 16:e2004108. doi: 10.1371/journal.pbio.2004108
215. Rosshart SP, Vassallo BG, Angeletti D, Hutchinson DS, Morgan AP, Takeda K, et al. Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance. Cell (2017) 171:1015–28.e13. doi: 10.1016/j.cell.2017.09.016
216. Graham AL. Naturalizing Mouse Models for Immunology. Nat Immunol (2021) 22:111–7. doi: 10.1038/s41590-020-00857-2
217. Masopust D, Sivula CP, Jameson SC. Of Mice, Dirty Mice, and Men: Using Mice To Understand Human Immunology. J Immunol (2017) 199:383–8. doi: 10.4049/jimmunol.1700453
218. Rosshart SP, Herz J, Vassallo BG, Hunter A, Wall MK, Badger JH, et al. Laboratory Mice Born to Wild Mice Have Natural Microbiota and Model Human Immune Responses. Science (2019) 365:eaaw4361. doi: 10.1126/science.aaw4361
219. Suntharalingam G, Perry MR, Ward S, Brett SJ, Castello-Cortes A, Brunner MD, et al. Cytokine Storm in a Phase 1 Trial of the Anti-CD28 Monoclonal Antibody TGN1412. N Engl J Med (2006) 355:1018–28. doi: 10.1056/NEJMoa063842
220. Fisher CJ, Agosti JM, Opal SM, Lowry SF, Balk RA, Sadoff JC, et al. Treatment of Septic Shock With the Tumor Necrosis Factor Receptor:Fc Fusion Protein. The Soluble TNF Receptor Sepsis Study Group. N Engl J Med (1996) 334:1697–702. doi: 10.1056/NEJM199606273342603
221. Thomas M, Sadjadian P, Kollmeier J, Lowe J, Mattson P, Trout JR, et al. A Randomized, Open-Label, Multicenter, Phase II Study Evaluating the Efficacy and Safety of BTH1677 (1,3-1,6 Beta Glucan; Imprime PGG) in Combination With Cetuximab and Chemotherapy in Patients With Advanced Non-Small Cell Lung Cancer. Invest New Drugs (2017) 35:345–58. doi: 10.1007/s10637-017-0450-3
222. Engel-Riedel W, Lowe J, Mattson P, Richard Trout J, Huhn RD, Gargano M, et al. A Randomized, Controlled Trial Evaluating the Efficacy and Safety of BTH1677 in Combination With Bevacizumab, Carboplatin, and Paclitaxel in First-Line Treatment of Advanced Non-Small Cell Lung Cancer. J Immunother Cancer (2018) 6:16. doi: 10.1186/s40425-018-0324-z
223. Hui W, Li T, Liu W, Zhou C, Gao F. Fecal Microbiota Transplantation for Treatment of Recurrent C. Difficile Infection: An Updated Randomized Controlled Trial Meta-Analysis. PloS One (2019) 14:e0210016. doi: 10.1371/journal.pone.0210016
224. Quraishi MN, Widlak M, Bhala N, Moore D, Price M, Sharma N, et al. Systematic Review With Meta-Analysis: The Efficacy of Faecal Microbiota Transplantation for the Treatment of Recurrent and Refractory Clostridium Difficile Infection. Aliment Pharmacol Ther (2017) 46:479–93. doi: 10.1111/apt.14201
225. Davar D, Dzutsev AK, McCulloch JA, Rodrigues RR, Chauvin J-M, Morrison RM, et al. Fecal Microbiota Transplant Overcomes Resistance to Anti-PD-1 Therapy in Melanoma Patients. Science (2021) 371:595–602. doi: 10.1126/science.abf3363
226. Baruch EN, Youngster I, Ben-Betzalel G, Ortenberg R, Lahat A, Katz L, et al. Fecal Microbiota Transplant Promotes Response in Immunotherapy-Refractory Melanoma Patients. Science (2021) 371:602–9. doi: 10.1126/science.abb5920
227. Taur Y, Coyte K, Schluter J, Robilotti E, Figueroa C, Gjonbalaj M, et al. Reconstitution of the Gut Microbiota of Antibiotic-Treated Patients by Autologous Fecal Microbiota Transplant. Sci Transl Med (2018) 10:eaap9489. doi: 10.1126/scitranslmed.aap9489
228. Peled JU, Gomes AL, Devlin SM, Littmann ER, Taur Y, Sung AD, et al. Microbiota as Predictor of Mortality in Allogeneic Hematopoietic-Cell Transplantation. N Engl J Med (2020) 382:822–34. doi: 10.1056/NEJMoa1900623
Keywords: bacterial microbiome, mycobiome, virome, archaeome, gut microbiota, lung microbiota, skin microbiota, vaginal microbiota
Citation: Runge S and Rosshart SP (2021) The Mammalian Metaorganism: A Holistic View on How Microbes of All Kingdoms and Niches Shape Local and Systemic Immunity. Front. Immunol. 12:702378. doi: 10.3389/fimmu.2021.702378
Received: 29 April 2021; Accepted: 17 June 2021;
Published: 30 June 2021.
Edited by:
Stefan Jordan, Charité – Universitätsmedizin Berlin, GermanyReviewed by:
Francesca Ronchi, University of Bern, SwitzerlandCopyright © 2021 Runge and Rosshart. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Solveig Runge, c29sdmVpZy5ydW5nZUB1bmlrbGluaWstZnJlaWJ1cmcuZGU=; Stephan Patrick Rosshart, c3RlcGhhbi5yb3NzaGFydEB1bmlrbGluaWstZnJlaWJ1cmcuZGU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.